Abstract
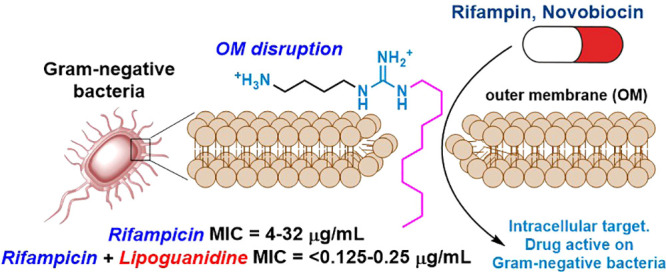
A new class of amphiphilic molecules, the lipoguanidines, designed as hybrids of guanidine and fatty acid compounds, has been synthesized and developed. The new molecules present both a guanidine polar head and a lipophilic tail that allow them to disrupt bacterial membranes and to sensitize Gram-negative bacteria to the action of the narrow-spectrum antibiotics rifampicin and novobiocin. The lipoguanidine 5g sensitizes Klebsiella pneumonia, Acinetobacter baumannii, Pseudomonas aeruginosa, and Escherichia coli to rifampicin, thereby reducing the antibiotic minimum inhibitory concentrations (MIC) up to 256-fold. Similarly, 5g is able to potentiate novobiocin up to 64-fold, thereby showing a broad spectrum of antibiotic potentiating activity. Toxicity and mechanism studies revealed the potential of 5g to work synergistically with rifampicin through the disruption of bacterial membranes without affecting eukaryotic cells.
Keywords: Lipoguanidine, Guanidine, Antibacterials, Antibiotic, Synergistic Activity
Antibiotics have long been a powerful tool in modern medicine, but their effectiveness is increasingly being undermined by the emergence of antibiotic-resistant bacteria.1 The overuse and misuse of antibiotics have led to the alarming situation where pathogens are becoming resistant to many commonly used antibiotic regimes.2 The scarcity of new antibiotic development is a significant contributing factor to the rise of antibiotic resistance,3,4 and thus, the development of new treatments for bacterial infections is considered critical by the World Health Organization (WHO). While some new antibiotic classes targeting Gram-positive bacteria have been introduced to the market within the last 20 years,5−8 only a few antibiotics that target Gram-negative bacteria are currently in the clinical pipeline.9−13 Such failure is largely attributed to the inability of many drugs to cross both the Gram-negative outer membrane (OM) and the inner membrane (IM) and to accumulate within these bacteria. Indeed, even though the molecular targets of many Gram-positive active antibiotics are also present in Gram-negative bacteria, the Gram-negative OM acts as a further barrier preventing the entry of such drugs to the cell.14,15 OM disrupting or perturbing agents, such as peptides,16 the chelating agent ethylenediaminetetraacetic acid (EDTA),17 the antiprotozoal drug pentamidine,18 the antifungal agent amphotericin B,19 or the cationic antibiotics,20−22 have shown their potential in sensitizing Gram-negative bacteria to the action of Gram-positive active antibiotics. The development of drugs able to disrupt bacterial membranes and to sensitize Gram-negative bacteria to antibiotics offers the possibility of turning narrow-spectrum antibiotics, like rifampicin or novobiocin, into broad-spectrum antibacterials.
Herein, we report the design and discovery of a new class of membrane-disrupting molecules, which we named lipoguanidines, presenting a general amphiphilic structure, A (Figure 1). The new molecules have been designed as hybrids of guanidines and fatty acids and they present both a guanidine polar head and a lipophilic tail.
Figure 1.
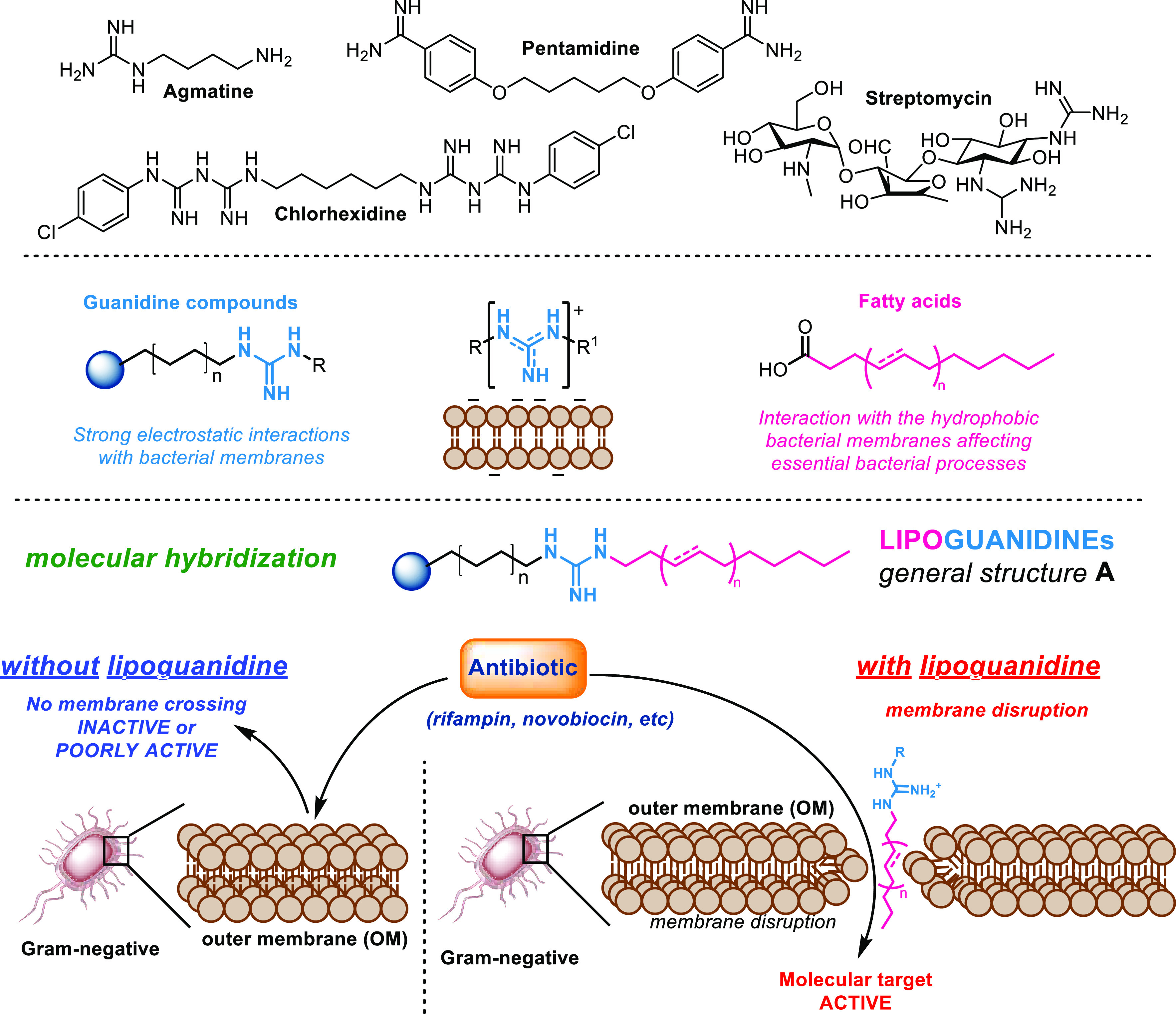
Guanidine antimicrobials and lipoguanidine design.
Many antimicrobial agents, including the cholic acid derivatives,20 contain a guanidine functional group.23−29 Guanidines can be protonated and positively charged at physiological pH in a similar way to the antibacterial quaternary ammonium compounds (QAC).30,31 Thus, a positively charged guanidine compound can facilitate the disruption of bacterial cell membranes through electrostatic interactions with the membrane anionic lipids.32 Since mammalian cells possess mainly neutral and uncharged lipids, guanidine compounds can selectively disrupt bacteria over mammalian cell membranes. Similarly, fatty acids exhibit antibacterial activity33−35 by interacting with hydrophobic bacterial membranes and impacting essential bacterial processes. The merging of hydrophilic guanidines with lipophilic fatty acids led to the design of novel lipoguanidine compounds that are able to sensitize Gram-negative bacteria to narrow-spectrum antibiotics.
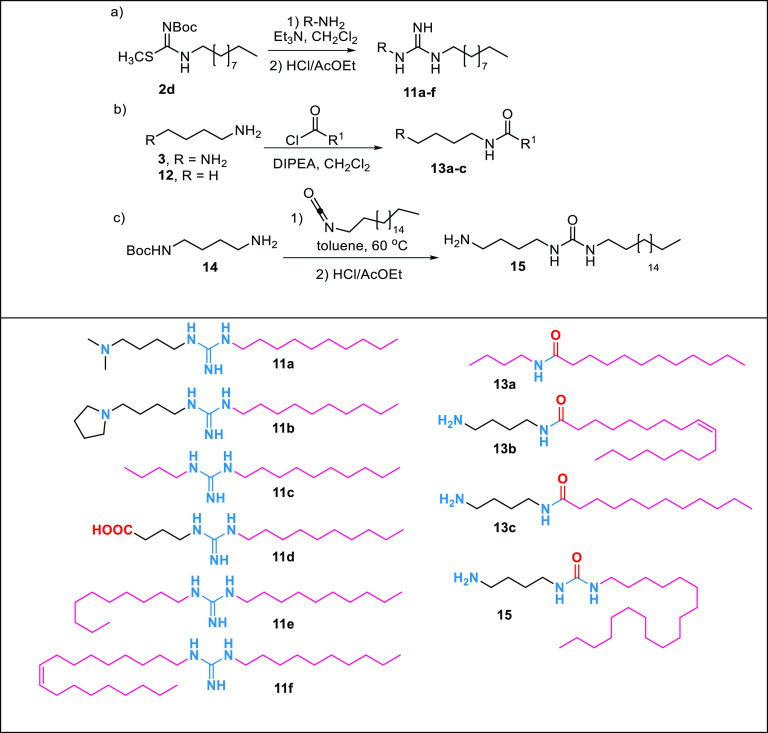
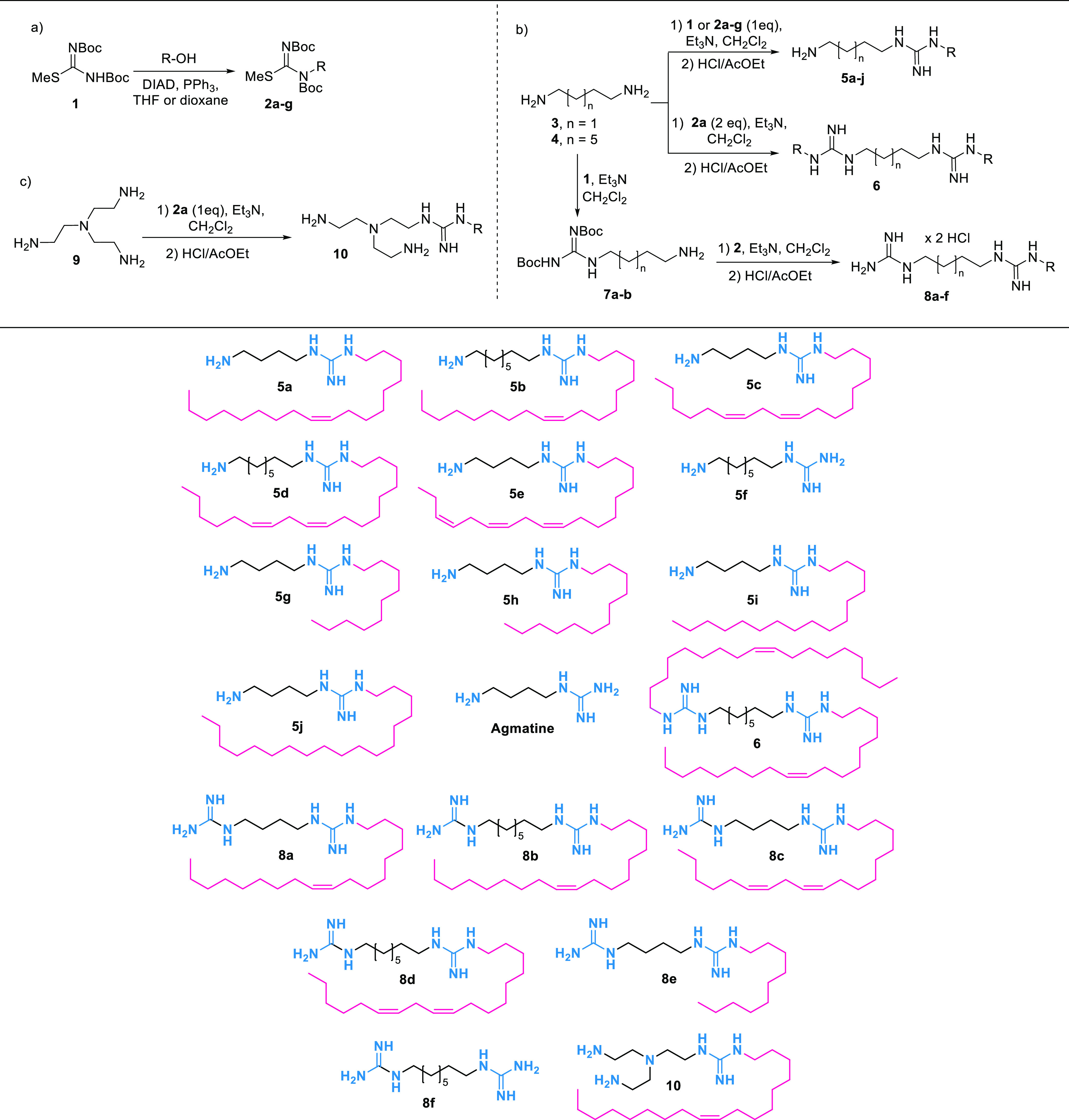
A series of lipoguanidine derivatives 5a–j was designed and synthesized according to Table 1. Agmatine, a natural guanidine compound bearing two protonable moieties, an amine, and a guanidine, was chosen as the template for the synthesis of lipoguanidine derivatives.26 Previous literature36−39 showed that agmatine and guanidines derivatives possess good antibacterial properties and that the guanidine group is essential for their activity. The 1,3-bis(tert-butoxycarbonyl)-2-methyl-2-thiopseudourea 1 was first reacted with different saturated and unsaturated fatty alcohols affording the alkyl-thiourea derivatives 2a–g. Thioureas 2a–g were then reacted with the diamines 3 and 4 to give, after acid-mediated Boc deprotection, the desired lipoguanidines 5a–j. The derivative 6 bearing a second alkylated guanidine moiety was prepared by reacting 4 with 2 equiv of 2a. A series of diguanidine compounds 8a–f equipped with a single lipophilic chain was then synthesized from the Boc-amino-guanidines 7a and 7b through guanylation with 2a–g followed by Boc deprotection. The derivatives 5k and 8f bearing no lipophilic chains were also prepared to evaluate the importance of the fatty chains for the lipoguanidine membrane disrupting activity. Finally, a branched triamine-lipoguanidine compound 10 was synthesized from the tetra-amine 9 with the aim to evaluate the role that multiple protonable amino groups might play on the lipoguanidines activity.
Table 1. First Series of Lipoguanidine Compounds.
The antibacterial activity of compounds 5a–j, 6, 8a–f, and 10 was first evaluated against a panel of drug-susceptible and drug-resistant Gram-positive and Gram-negative bacterial strains (Table 2). All lipoguanidines showed good activity against Gram-positive bacteria. A significant structure–activity correlation was observed for lipoguanidines 5a–j, which exhibited good activity against wild-type and drug-resistant Staphylococcus aureus and Enterococci strains (Enterococci faecalis and Enterococci faecium) with minimum inhibitory concentrations (MICs) ranging between 2 and 8 μg/mL. Compounds 5g (containing a shorter side chain) and 5f (no lipophilic chain) were found to be inactive against Gram-positive bacteria, which suggests a correlation between the lipophilicity of the chain and the antibacterial activity. Compound 6, which contains two guanidine groups and two lipophilic chains, was found to be poorly active, while derivatives 8a–d and 10 showed a biological profile similar to lipoguanidines 5a–j. Such data indicate that the replacement of the amine group in 5a–j with a second guanidine moiety does not affect their antibacterial profile, whereas the introduction of a second lipophilic chain in 6 is not tolerated. Compound 8f containing no lipophilic chain proved to be inactive against all strains of bacteria, thereby confirming the key role of the fatty chain. As a general trend, the compounds bearing a guanidine moiety, a saturated or unsaturated lipophilic chain, and a second protonable group (guanidine or amine) showed the best activities against Gram-positive bacteria. However, most lipoguanidines proved to be moderately or poorly active against Gram-negative bacteria. Compounds 5a and 5b containing an oleyl chain showed activity against Acinetobacter baumannii, Klebsiella pneumoniae, and Escherichia coli strains with MIC = 8–16 μg/mL. A similar antibacterial profile was also observed for compounds 5c and 5d, which incorporate a linoleyl chain.
Table 2. Evaluation of the Antibacterial Activity of Lipoguanidines.
| compd | MIC (μg/mL) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gram-positive |
Gram-negative |
|||||||||||||
|
S. aureus |
E. faecalis |
E. faecium |
K. pneumoniae |
A. baumannii |
P. aeruginosa |
E. coli | ||||||||
| MSSA ATCC 9144 | MRSA NCTC 13616 | MRSA USA 300 | MRSA SA 1199B | VSE NCTC 775 | VRE NCTC 12201 | VRE NCTC 12204 | NCTC 13368 | M6 | AYE | ATCC 17978 | PAO1 | NCTC 13437 | NCTC 12923 | |
| 5a | 4 | 4 | 2 | 4 | 4 | 4 | 4 | 128 | 8 | 16 | 8 | 128 | >128 | 8 |
| 5b | 4 | 4 | 2 | 4 | 4–8 | 2 | 4 | 32–128 | >128 | 32 | 8–16 | >128 | >128 | 16 |
| 5c | 8 | 16 | 8 | 16 | 32 | 16 | 16 | 16 | 8 | 8 | 8 | 16 | >64 | 4 |
| 5d | 2–4 | 4 | 2–4 | 4 | 8 | 4 | 4 | 32 | 16 | 16 | 8 | >128 | >128 | 8 |
| 5e | 8 | 8 | 4 | 8 | 8 | 8 | 8 | >128 | 128 | 128 | 128 | >128 | >128 | 32 |
| 5f | 128 | >128 | 128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 128 |
| 5g | 16 | 32 | 32 | 32 | 128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | 128 |
| 5h | 2 | 4 | 4 | 4 | 8 | 16 | 16 | >128 | >128 | >128 | 128 | >128 | 64 | 16 |
| 5i | 2 | 2–4 | 2 | 4 | 2 | 2 | 2 | >128 | 8–64 | 32→128 | 4–8 | >128 | >128 | 32–64 |
| 5j | 8 | 4 | 2 | 4 | 2 | 2 | 2 | >128 | >128 | 128 | 16–32 | >128 | >128 | >128 |
| 6 | 64 | >64 | 64 | 32–64 | 64 | >64 | 32 | >128 | >128 | >128 | 128 | >128 | >128 | 128 |
| 8a | 4 | 2 | 2 | 2 | 2 | 2 | 2 | 128 | 16 | 32 | 8 | 128 | >128 | 8 |
| 8b | 4–8 | 4–8 | 2–4 | 4–8 | 4–8 | 4 | 4 | 128 | 64 | 16–64 | 16 | >128 | >128 | 32 |
| 8c | 4 | 4 | 4 | 4 | 8 | 8 | 8 | >64 | 32 | >64 | 32 | 64 | 64 | 8 |
| 8d | 2–4 | 4 | 2 | 2–4 | 4 | 4 | 2 | 32–64 | 16–32 | 16–64 | 4–8 | >128 | >128 | 8 |
| 8e | NDa | NDa | NDa | NDa | NDa | NDa | NDa | >128 | >128 | >128 | >128 | 64 | >128 | 64 |
| 8f | 128 | >128 | 128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 | >128 |
| 10 | 8 | 4 | 4 | 4 | 4 | 4 | 4 | >128 | >128 | 128 | 64 | 64 | 32–64 | 8–16 |
| agmatine | NDa | NDa | NDa | NDa | NDa | NDa | NDa | >128 | >128 | >128 | >128 | >128 | >128 | >128 |
Not determined.
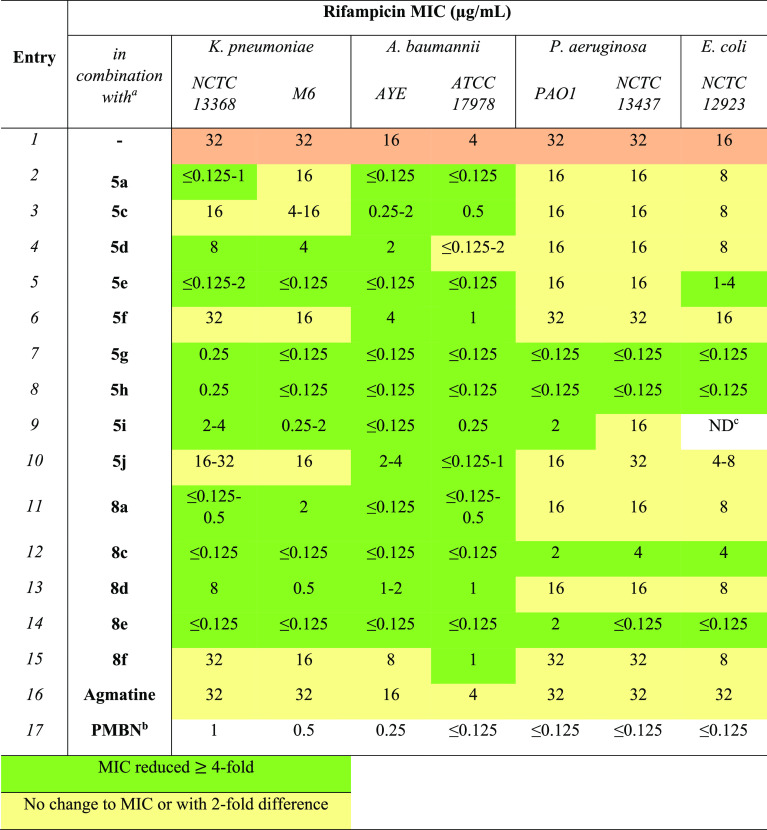
In contrast, compounds 5g and 5h bearing a saturated lipophilic chain were inactive against the Gram-negative strains, though the exception of 5i and 5j showed moderate activity against A. baumannii ATCC 17978 (MIC = 4–8 and 16–32 μg/mL, respectively). The lipoguanidines 8a–e bearing two guanidine groups showed a biological profile similar to that of their corresponding amino-lipoguanidines 5a–j. As observed in Gram-positive bacteria, both the aminoguanidines 5f and 8f bearing no lipophilic side chain, as well as agmatine, were completely inactive against all the Gram-negative strains, which confirmed the key role of the alkyl chain in improving the antibacterial profile of the lipoguanidines.
The polyamino-lipoguanidine 10 also showed no activity against most of the Gram-negative strains apart from E. coli (MIC = 8–16 μg/mL). The derivatives 5a, 5d, 5g, 5h, 5j, 8a, and 8d were also screened against Gram-negative bacteria in combination with the membrane permeabilizing agent polymyxin B nonapeptide (PMBN) to determine if the lower activity observed against Gram-negative bacteria was ascribable to their inability to enter the cells by crossing their OM (Table S1). Gram-negative bacteria were treated with a range of concentrations of lipoguanidine compounds in the presence of PMBN at a fixed concentration of 30 μg/mL, which was able to permeabilize the OM without causing more than 20% growth inhibition for each strain tested. The observed MICs with and without PMBN were identical, and no improvement in activity was observed, which suggested that the lipoguanidines had already achieved the maximum OM penetration and bacterial inhibition.
Sensitization of Gram-Negative Bacteria to Rifampicin by Lipoguanidines
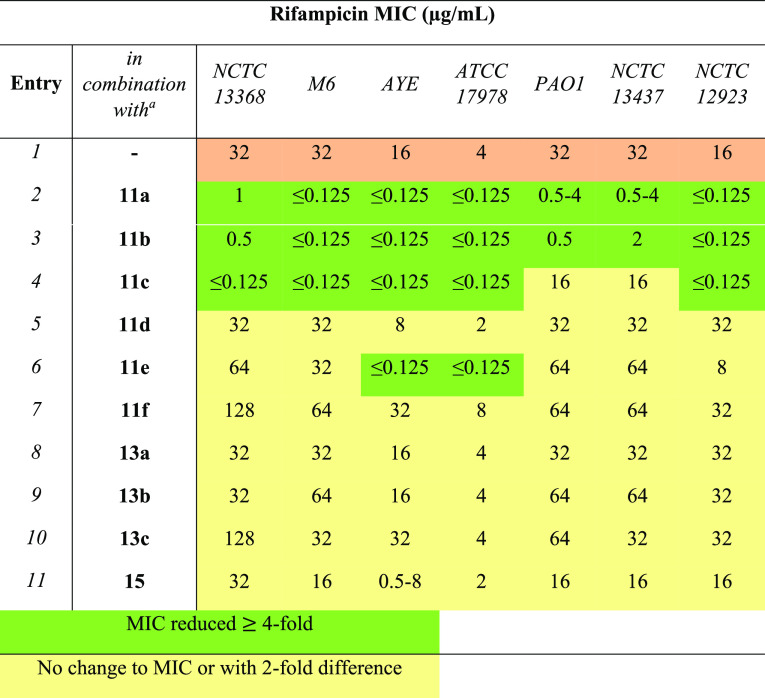
The ability of lipoguanidine compounds to sensitize Gram-negative bacteria to the action of the narrow-spectrum antibiotic rifampicin through membrane disruption was then evaluated (Table 3). Compounds 5a–j and 8a–f were tested against a panel of Gram-negative bacteria in combination with rifampicin. Rifampicin is poorly active against Gram-negative bacteria (entry 1, Table 3) because of its poor ability to cross the OM and, as a consequence, to reach its molecular target, as confirmed by the addition of the membrane permeabilizer PMBN (entry 17, Table 3).
Table 3. Synergistic Activity of Rifampicin in Combination with Lipoguanidines.
Lipoguanidine was added at a concentration of ≤0.25 × MIC. The exact concentration of lipoguanidines 5 and 8 used with each different bacterial strain for the synergistic study with rifampicin is reported in the Supporting Information Table S2.
PMBN (30 μg/mL) was added.
Not determined.
Gram-negative bacteria were treated with a range of concentrations of rifampicin in the presence of the lipoguanidine compounds 5a–j and 8a–f at a concentration of ≤0.25 × MIC. A ≥4-fold decrease in rifampicin MIC in the presence of a lipoguanidine derivative indicates that the bacteria are sensitized to the antibiotic. Lipoguanidines 5a, 5d–j, and 8a–e exhibited strong rifampicin potentiating activity against some or all K. pneumoniae and A. baumannii strains with a >4-fold reduction in rifampicin MIC. The oleyl lipoguanidine 5a demonstrated excellent rifampicin potentiating activity with rifampicin MIC reduced to ≤0.125 μg/mL against both drug-sensitive (ATCC 17978) and drug-resistant (AYE) A. baumannii strains and K. pneumoniae NCTC 13368 (entry 2, Table 3). A similar profile was observed for the linoleyl derivatives 5c and 5d (entries 3–4, Table 3), while the linolenyl 5e reduced the rifampin MIC to ≤0.125 μg/mL in both K. pneumoniae strains (entry 5, Table 3). None of the lipoguanidines 5a–e bearing an unsaturated lipophilic chain were able to improve the rifampicin activity against Pseudomonas aeruginosa, and only 5e showed a sensitizing action on E. coli. Remarkably, lipoguanidines 5g and 5h bearing, respectively, a decyl- and a dodecyl-saturated lipophilic chain exhibited an excellent sensitizing effect on all Gram-negative strains, including the P. aeruginosa strains, with rifampicin MIC ≤ 0.125 μg/mL (entries 6 and 7, Table 3). Interestingly, even though 5g and 5h did not have any activity against any Gram-negative bacteria, they showed a remarkable sensitizing effect on all Gram-negative strains, including P. aeruginosa and E. coli. Such data are remarkable, especially when compared with those of the lipoguanidines 5a, 5d, and 5e, which showed good Gram-negative inhibitory activity but no sensitizing activity on P. aeruginosa and E. coli species, thereby suggesting a correlation between the antibacterial activity/sensitizing effect and the unsaturated/saturated lipophilic chains. Interestingly, an increase in the length of the saturated lipophilic chain in 5i and 5j led to a decrease of the sensitizing action of lipoguanidines, especially against P. aeruginosa (entries 8 and 9, Table 3).
Derivatives 8 bearing two guanidine groups also showed, in many cases, a strong sensitizing effect against Gram-negative bacteria. The lipoguanidine 8e bearing a decyl chain like 5g showed the best sensitizing activity with a remarkable improvement of rifampicin MIC also against P. aeruginosa (entry 14, Table 3). As observed for compounds 5, the derivatives 8a–d bearing an unsaturated lipophilic chain showed good-to-moderate activity against Gram-negative bacteria and sensitizing activity against K. pneumoniae and A. baumannii species but were unable to improve the rifampicin MIC against P. aeruginosa and E. coli. Conversely, compound 8e bearing a fully saturated lipophilic chain like 5g and 5h showed no activity against Gram-negative bacteria but a strong sensitizing effect on P. aeruginosa and E. coli, thereby confirming the correlation between the sensitizing activity and the lipophilic chain degree of saturation. Notably, guanidines 5f and 8f did not show any rifampicin MIC reduction except for some mild improvement on A. baumannii, which clearly indicates the essential role of a lipophilic chain in the sensitization of Gram-negative bacteria (entries 10 and 15, Table 3).
Assessment of the Hemolytic Activity and Toxicity of Lipoguanidines 5 and 8
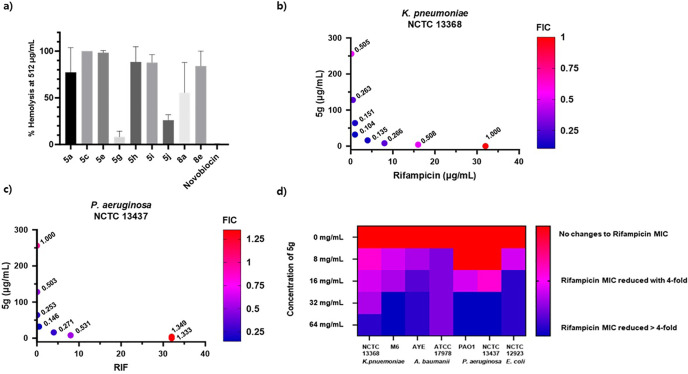
To evaluate the potential of lipoguanidines as therapeutic agents, the hemolytic activity of the most promising compounds was evaluated. All tested compounds demonstrated a noticeable hemolytic effect, with 5c being the most hemolytic (100.0%) and 5g the least (8.2%). Compounds 5a, 5c, 5e, and 8a bearing unsaturated lipophilic chains were found toxic to erythrocytes, which suggests a correlation between the degree of unsaturation in the lipophilic chain and hemolysis. Saturated derivatives 5h, 5i, and 8e, also exhibited high hemolytic activity, while 5g showed a better cytotoxic profile and a large difference between its antibiotic potentiation activity and hemolytic effect because it potentiates the activity of rifampicin against all strains of Gram-negative bacteria when used at 32 μg/mL and only causes 8.2% hemolysis at 512 μg/mL. Therefore, 5g emerged as the most promising lipoguanidine hit.
An in vivo toxicity assay using Galleria mellonella was then carried out to evaluate the toxicity of 5g in a nonanimal infection model.40,41 Ten wax moth larvae were injected with 5g at concentrations of 50 mg/kg, 20 mg/kg, and 10 mg/kg (Table S5, Supporting Information). By the end point of the assay at 120 h, 10 of 10 larvae were still alive at all doses. Such data are in line with the in vitro hemolysis assay data, which indicates that 5g is not toxic on eukaryotic cells and in Galleria models.
Assessment of the Antibiotic Synergism between Rifampicin and 5g by Checkerboard Assay
In the initial synergy screening, the sensitizing activity of lipoguanidines was assessed using a single concentration of lipoguanidine (generally corresponding to ≤0.25 × MIC). In the case of 5g, which showed no activity against most Gram-negative strains (>128 μg/mL) and moderate activity against A. baumannii ATCC 17978 (16–32 μg/mL), concentrations around 32 μg/mL were used across the panel. The different concentrations of 5g at which a sensitizing effect of Gram-negative bacteria to rifampicin can be observed were then evaluated (Figure 2d). When 5g was used at 32 μg/mL, a significant reduction in rifampicin MIC in all Gram-negative strains was observed. Although higher concentrations of 5g (64 μg/mL) showed an improved rifampicin potentiation activity in K. pneumoniae NCTC 13368, the concentration of 32 μg/mL was more efficient in producing significant changes in rifampicin MIC. At the lower concentration of 5g (8–16 μg/mL) a reduction in the rifampicin potentiation activity was still observed in K. pneumonia and P. aeruginosa strains. The optimal concentration of 5g for synergism activity was set at 32 μg/mL. Since 5g showed antimicrobial activity against A. baumannii ATCC 17978 at 32 μg/mL, it is not possible to unambiguously conclude that, for this strain, the concentration was potentiating.
Figure 2.
(a) % Hemolysis of lipoguanidines at 512 μg/mL; (b) synergism of 5g with rifampicin on K. pneumoniae NCTC 13368 and FIC value; (c) synergism of 5g with rifampicin on P. aeruginosa NCTC 13437 and FIC value; and (d) evaluation of the Gram-negative bacteria sensitizing activity of 5g at different concentrations.
A checkerboard assay was then carried out to assess the efficacy of the combination of rifampicin and 5g. The synergistic activity of the two agents was measured using the fractional inhibitory concentration (FIC) index.42 A FIC value of ≤0.5 indicates a strong synergistic effect, while values of 0.5 to <1 indicate weak synergism. The synergistic activity of the two compounds was tested against the drug-resistant strains K. pneumoniae NCTC 13368 and P. aeruginosa NCTC 13437. These strains were selected because previous research in antimicrobial evaluation studies has repeatedly suggested that they have less permeable membranes and, thus, were ideal worst-case models to evaluate the synergistic effect of membrane-disrupting agent 5g. A strong synergy between 5g and rifampicin was observed against both K. pneumoniae strain NCTC 13368 and P. aeruginosa strain NCTC 13437. The MIC values of both compounds were reduced when they were used in combination. FIC values of 0.10 for NCTC 13368 and 0.15 for NCTC 13437 indicate a strong synergistic effect between the two compounds (Figure 2b,c).
Mechanism of Action of 5g
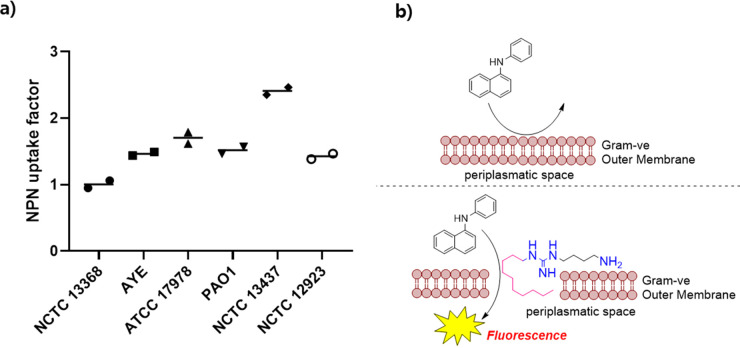
In order to confirm the mode of action of 5g, a 1-N-phenylnaphthylamine (NPN) uptake assay was carried out. NPN is a fluorescent probe that is unable to cross the Gram-negative bacteria OM because of its hydrophobic nature. When the outer membrane is damaged, the NPN gains access to the bacterial periplasmatic space, which results in a prominent fluorescent signal.
The NPN uptake factor for 5g at 128 μg/mL was calculated in relation to the quantity of NPN taken up by the cells when treated with 10 μg/mL of the outer membrane permeabilizer polymyxin B (PMB).43 An NPN uptake factor of 1 indicates that the bacteria took up the same amount of NPN as when they were treated with PMB, thereby suggesting that NPN gained access to the bacteria because of the membrane-permeabilizing activity of 5g. As shown in Figure 3, an NPN factor > 1 was observed against all Gram-negative strains, including K. pneumoniae NCTC 13368 and P. aeruginosa NCTC 13437, which are understood to have a more robust outer membrane. These data confirmed a membrane-disrupting activity for lipoguanidine 5g.
Figure 3.
Evaluation of the mechanism of action of 5g through the NPN assay. (A) OM permeabilization of Gram-negative bacteria by 5g, as measured through NPN uptake. (B) Cartoon describing the suspected mechanism of action of 5g on the OM of Gram-negative bacteria. For the time point, the NPN uptake was calculated using the following equations where Fobs is the observed fluorescence of the sample, F0 is the initial fluorescence of NPN with bacteria in the absence of any compound, and F100 is the fluorescence of NPN with bacteria upon the addition of 10 μg/mL of PMB. NPN uptake factor = (Fobs– F0)/(F100– F0).
Synthesis, Biological Evaluation, and Structure–Activity Relationships (SARs) of Lipoguanidine Analogues
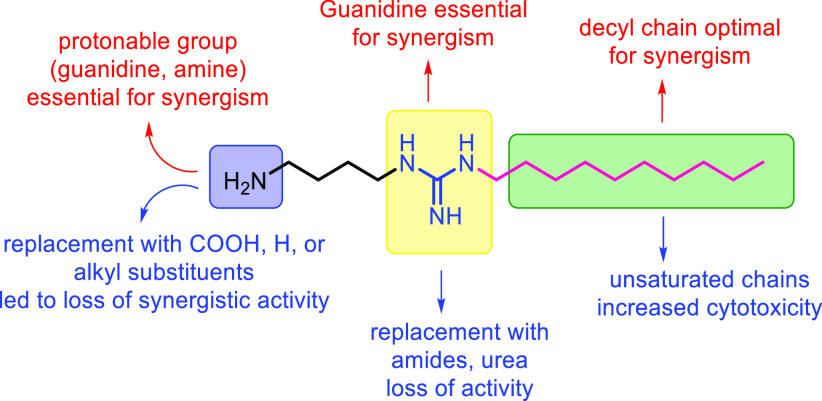
With the aim to further elucidate the SAR of the new lipoguanidine compounds and to identify key features requisite for synergistic activity, a series of derivatives was designed and synthesized (Table 4). The derivatives 11a and 11b bearing a tertiary amine group were synthesized through reaction of the decyl-thiourea 2d with 4-dimethylaminobutylamine and pyrrolidine-butylamine. Derivative 11d, which was designed as a zwitterionic lipoguanidine incorporating a carboxylate moiety, was obtained through the reaction of 2g with 4-aminobutanoic acid. Lipoguanidines 11c, 11e, and 11f lacking an amine group were synthesized by reacting 2g with butyl-, decyl-, and oleylamine. Lastly, butylamine and diaminobutane were reacted with dodecyl-chloride and oleyl-chloride, respectively, to afford the amides 13a–c, while the N-Boc-diamine 14 was reacted with octadecyl-isocyanate followed by Boc deprotection to obtain the urea 15.
Table 4. Synthesis of Lipoguanidine Analogues 11, 13, and 15.
All derivatives 11, 13, and 15 were first assessed for their activity against Gram-negative bacteria (Table S3). Only the dialkyl-lipoguanidines 11c and 11e showed moderate activity against strains of A. baumannii, K. pneumoniae, and E. coli with MICs of 16–32 μg/mL.
Derivatives 11, 13, and 15 were then tested at a concentration of ≤0.25 × MIC in combination with rifampicin to assess their ability to sensitize Gram-negative bacteria to the antibiotic (Table 5). Lipoguanidines 11a and 11b were able to sensitize all Gram-negative bacteria to rifampicin with a decrease of the rifampicin MIC > 4 fold. However, compared with the lipoguanidine 5g bearing a primary amine as a second protonable group, 11a and 11b showed a lower sensitizing effect against P. aeruginosa strains PAO1 and NCTC 13437 (entries 2 and 3, Table 5). Guanidine 11c showed excellent activity with rifampicin against K. pneumoniae, A. baumannii, and E. coli but no sensitizing effect on P. aeruginosa (entry 4, Table 5).
Table 5. Synergistic Activity of Rifampicin in Combination with Lipoguanidines and SARs.
Lipoguanidine/guanidine/amide/urea was added at a concentration of ≤0.25 × MIC. The exact concentration of lipoguanidine/guanidine/amide/urea used with each different bacterial strain for the synergistic study with rifampicin is reported in the Supp. Information in Table S4.
Guanidine 11e showed sensitizing activity only on A. baumannii, while no activity was observed on K. pneumoniae and E. coli. (entry 6, Table 5). Such data confirm that the presence of a second hydrophilic/protonable moiety in the lipoguanidine compounds is crucial in improving the sensitization of bacteria to rifampicin. Finally, the derivatives 11f and 11d, as well as the amides 13a–c and urea 15, showed no reduction of the rifampicin MIC, thereby confirming the crucial role of the guanidine group for the activity.
Sensitization of Gram-Negative Bacteria to Novobiocin by Lipoguanidines
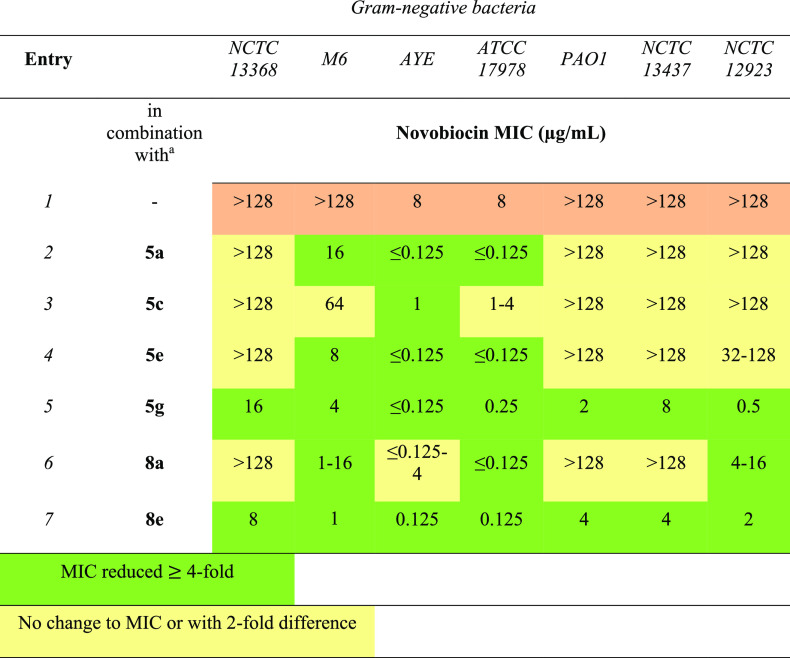
Lipoguanidines were finally assessed in combination with novobiocin against Gram-negative bacteria (Table 6). Compounds 5a, 5e, and 8a effectively sensitized A. baumannii strains and K. pneumoniae M6 to novobiocin (entries 1, 3, and 6, Table 6). Remarkably, the less cytotoxic lipoguanidine 5g and compound 8e broadened novobiocin’s activity across various Gram-negative species (entries 5 and 7, Table 6). The lower potentiating effect of 5g on P. aeruginosa with novobiocin than with rifampicin may be attributed to underlying mutations in these strains (i.e., DNA gyrase or efflux pump mutations) that may impact the novobiocin activity.
Table 6. Sensitization of Gram-Negative Bacteria to Novobiocin by Lipoguanidines33.
Lipoguanidine was added at concentration of ≤0.25 × MIC. The exact concentration of lipoguanidines 5 and 8 used with each different bacterial strain for the synergistic study with rifampicin is reported in the Supporting Information Table S2.
The potentiating effect of lipoguanidine 5g in combination with the antibiotics ampicillin, ceftazidime, doxycycline, and tobramycin was also investigated (Table S5). Only a potentiating effect of 5g on doxycycline on Klebsiella, Pseudomonas, and E. coli species was observed.
In conclusion, we have designed and developed a novel class of structurally simple amphiphilic lipoguanidine compounds through the hybridization of aminoalkylguanidine derivatives and fatty acids. While lipoguanidines are not highly effective against Gram-negative bacteria, they exhibit synergy in combination with narrow-spectrum antibiotics, such as rifampicin and novobiocin. Lipoguanidine 5g significantly lowered the MIC of rifampicin and novobiocin against both wild-type and drug-resistant Gram-negative bacteria (up to ≤0.125 μg/mL) through the permeabilization of the bacterial membranes, as confirmed through an NPN assay. Finally, 5g demonstrated low toxicity to eukaryotic cells and living organisms, as indicated by hemolysis and in vivo toxicity assays.
Acknowledgments
For G.F.S.F. and D.C. this project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No. 101027065. D.S. and D.C. acknowledge the South African National Research Foundation-SARChI for financial support to D.S. D.C. acknowledges BBSRC LIDo (BB/M009513/1) for Ph.D. Studentships to S.A. A.J.M. and D.C. acknowledge NC3Rs (NC/T001240/1) for financial support. D.C. and J.W. acknowledge China Scholarship Council programme (CSC, scholarship No. 202008060037) for financial support to J.W. The Royal Society is gratefully acknowledged for financial support (IEC\R2\202028). Work at UKHSA was supported by Grant-in-Aid project, Open Innovation in AMR; 113361. Any opinions expressed in the paper are those of the authors and not necessarily UKHSA or the Department for Health and Social Care.
Glossary
Abbreviations
- OM
outer membrane
- IM
inner membrane
- MIC
minimum inhibitory concentration
- QAC
quaternary ammonium compounds
- PMBN
polymyxin B nonapeptide
- FIC
fractional inhibitory concentration
- NPN
1-N-phenylnaphthylamine
- SARs
structure–activity relationships
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.3c00460.
Experimental procedures, characterization of chemical compounds, microbiological procedures and data, analytical data, HPLC traces, and NMR spectra (PDF)
Author Contributions
The manuscript was written through contributions of all authors.
The authors declare no competing financial interest.
Supplementary Material
References
- Chellat M. F.; Raguž L.; Riedl R. Targeting antibiotic resistance. Angew. Chem., Int. Ed. 2016, 55, 6600–6626. 10.1002/anie.201506818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden R.; Kelly R.; Davies S. Combatting antimicrobial resistance globally. Nat. Microbiol. 2016, 1, 16187. 10.1038/nmicrobiol.2016.187. [DOI] [PubMed] [Google Scholar]
- Beyer P.; Paulin S. The antibacterial research and development pipeline needs urgent solutions. ACS Infect. Dis. 2020, 6, 1289–1291. 10.1021/acsinfecdis.0c00044. [DOI] [Google Scholar]
- Theuretzbacher U.; Outterson K.; Engel A.; Karlén A. The global preclinical antibacterial pipeline. Nat. Rev. Microbiol. 2020, 18, 275–285. 10.1038/s41579-019-0288-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Ruiz A.; Seaton R. A.; Hamed K. Daptomycin: an evidence-based review of its role in the treatment of Gram-positive infections. Infect. Drug Resist. 2016, 9, 47–58. 10.2147/IDR.S99046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemian S. M. R.; Farhadi T.; Ganjparvar M. Linezolid: a review of its properties, function, and use in critical care. Drug Des. Devel. Ther. 2018, 12, 1759–1767. 10.2147/DDDT.S164515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paukner S.; Riedl R. Pleuromutilins: potent drugs for resistant bugs-mode of action and resistance. Cold Spring Harb. Perspect. Med. 2017, 7, a027110 10.1101/cshperspect.a027110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goethe O.; Heuer A.; Ma X.; Wang Z.; Herzon S. B. Antibacterial properties and clinical potential of pleuromutilins. Nat. Prod. Rep. 2019, 36, 220–247. 10.1039/C8NP00042E. [DOI] [PubMed] [Google Scholar]
- Shi Z.; Zhang J.; Tian L.; Xin L.; Liang C.; Ren X.; Li M. A comprehensive overview of the antibiotics approved in the last two decades: retrospects and prospects. Molecules 2023, 28, 1762. 10.3390/molecules28041762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler M. S.; Henderson I. R.; Capon R. J.; Blaskovich M. A. T. Antibiotics in the clinical pipeline as of December 2022. J. Antibiot. 2023, 76, 431–473. 10.1038/s41429-023-00629-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuf E.; Bax H. I.; Verkaik N. J.; van Westreenen M. An update on eight “new” antibiotics against multidrug-resistant gram-negative bacteria. J. Clin. Med. 2021, 10, 1068. 10.3390/jcm10051068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaiskos I.; Lagou S.; Pontikis L.; Rapti V.; Poulakou G. The “old” and the “new” antibiotics for MDR Gram-negative pathogens: for whom, when, and how. Front. Public Health 2019, 7, 151. 10.3389/fpubh.2019.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad N. K.; Seiple I. B.; Cirz R. T.; Rosenberg O. S. Leaks in the pipeline: a failure analysis of Gram-negative antibiotic development from 2010 to 2020. Antimicrob. Agents Chemother. 2022, 66, e0005422 10.1128/aac.00054-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breijyeh Z.; Jubeh B.; Karaman R. Resistance of Gram-negative bacteria to current antibacterial agents and approaches to resolve it. Molecules 2020, 25, 1340. 10.3390/molecules25061340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcour A. H. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta Proteins Proteom. 2009, 1794, 808–816. 10.1016/j.bbapap.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L.; Chi J.; Yan Y.; Luo R.; Feng X.; Zheng Y.; Xian D.; Li X.; Quan G.; Liu D.; Wu C.; Lu C.; Pan X. Membrane-disruptive peptides/peptidomimetics-based therapeutics: promising systems to combat bacteria and cancer in the drug-resistant era. Acta Pharm. Sin. B 2021, 11, 2609–2644. 10.1016/j.apsb.2021.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan S.; Percival S. L. EDTA: an antimicrobial and antibiofilm agent for use in wound care. Adv. Wound Care (New Rochelle) 2015, 4, 415–421. 10.1089/wound.2014.0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes J. M.; MacNair C. R.; Ilyas B.; French S.; Côté J. P.; Bouwman C.; Farha M. A.; Sieron A. O.; Whitfield C.; Coombes B. K.; Brown E. D. Pentamidine sensitizes Gram-negative pathogens to antibiotics and overcomes acquired colistin resistance. Nat. Microbiol. 2017, 2, 17028. 10.1038/nmicrobiol.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis D. Amphotericin B: spectrum and resistance. J. Antimicrob. Chemother. 2002, 49, 7–10. 10.1093/jac/49.suppl_1.7. [DOI] [PubMed] [Google Scholar]
- Li C.; Peters A. S.; Meredith E. L.; Allman G. W.; Savage P. B. Design and synthesis of potent sensitizers of Gram-negative bacteria based on a cholic acid scaffolding. J. Am. Chem. Soc. 1998, 120, 2961–2962. 10.1021/ja973881r. [DOI] [Google Scholar]
- Li C.; Budge L. P.; Driscoll C. D.; Willardson B. M.; Allman G. W.; Savage P. B. Incremental conversion of outer-membrane permeabilizers into potent antibiotics for Gram-negative bacteria. J. Am. Chem. Soc. 1999, 121, 931–940. 10.1021/ja982938m. [DOI] [Google Scholar]
- Savage P. B.; Li C.; Taotafa U.; Ding B.; Guan Q. Antibacterial properties of cationic steroid antibiotics. FEMS Microbiol. Lett. 2002, 217, 1–7. 10.1111/j.1574-6968.2002.tb11448.x. [DOI] [PubMed] [Google Scholar]
- Sączewski F.; Balewski Ł. Biological activities of guanidine compounds, 2008 - 2012 update. Expert Opin. Ther. Pat. 2013, 23, 965–995. 10.1517/13543776.2013.788645. [DOI] [PubMed] [Google Scholar]
- Kim S. H.; Semenya D.; Castagnolo D. Antimicrobial drugs bearing guanidine moieties: a review. Eur. J. Med. Chem. 2021, 216, 113293 10.1016/j.ejmech.2021.113293. [DOI] [PubMed] [Google Scholar]
- Berlinck R. G. S.; Trindade-Silva A. E.; Santos M. F. C. The chemistry and biology of organic guanidine derivatives. Nat. Prod. Rep. 2012, 29, 1382–1406. 10.1039/c2np20071f. [DOI] [PubMed] [Google Scholar]
- Castagnolo D.; Schenone S.; Botta M. Guanylated diamines, triamines, and polyamines: chemistry and biological properties. Chem. Rev. 2011, 111, 5247–5300. 10.1021/cr100423x. [DOI] [PubMed] [Google Scholar]
- Saeed A.; Bosch A.; Bettiol M.; Nossa González D. L.; Erben M. F.; Lamberti Y. Novel guanidine compound against multidrug-resistant cystic fibrosis-associated bacterial species. Molecules 2018, 23, 1158. 10.3390/molecules23051158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H. T.; Venter H.; Veltman T.; Williams R.; O’Donovan L. A.; Russell C. C.; McCluskey A.; Page S. W.; Ogunniyi A. D.; Trott D. J. In vitro synergistic activity of NCL195 in combination with colistin against Gram-negative bacterial pathogens. Int. J. Antimicrob. Agents. 2021, 57, 106323 10.1016/j.ijantimicag.2021.106323. [DOI] [PubMed] [Google Scholar]
- Li J.; Zhang X.; Han N.; Wan P.; Zhao F.; Xu T.; Peng X.; Xiong W.; Zeng Z. Mechanism of action of isopropoxy benzene guanidine against multidrug-resistant pathogens. Microbiol Spectr. 2023, 11, e0346922 10.1128/spectrum.03469-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings M. C.; Minbiole K. P. C.; Wuest W. M. Quaternary ammonium compounds: an antimicrobial mainstay and platform for innovation to address bacterial resistance. ACS Infect. Dis. 2015, 1, 288–303. 10.1021/acsinfecdis.5b00047. [DOI] [PubMed] [Google Scholar]
- Leong J.; Yang C.; Tan J.; Tan B. Q.; Hor S.; Hedrick J. L.; Yang Y. Y. Combination of guanidinium and quaternary ammonium polymers with distinctive antimicrobial mechanisms achieving a synergistic antimicrobial effect. Biomater. Sci. 2020, 8, 6920–6929. 10.1039/D0BM00752H. [DOI] [PubMed] [Google Scholar]
- Andreev K.; Bianchi C.; Laursen J. S.; Citterio L.; Hein-Kristensen L.; Gram L.; Kuzmenko I.; Olsen C. A.; Gidalevitz D. Guanidino groups greatly enhance the action of antimicrobial peptidomimetics against bacterial cytoplasmic membranes. Biochim. Biophys. Acta 2014, 1838, 2492–2502. 10.1016/j.bbamem.2014.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broxton P.; Woodcock P. M.; Gilbert P. A study of the antibacterial activity of some polyhexamethylene biguanides towards Escherichia coli ATCC 8739. J. Appl. Bacteriol. 1983, 54, 345–353. 10.1111/j.1365-2672.1983.tb02627.x. [DOI] [PubMed] [Google Scholar]
- McGaw L. J.; Jäger A. K.; van Staden J. Antibacterial effects of fatty acids and related compounds from plants. S. Afr. J. Bot. 2002, 68, 417–423. 10.1016/S0254-6299(15)30367-7. [DOI] [Google Scholar]
- Desbois A. P.; Smith V. J. Antibacterial free fatty acids: activities, mechanisms of action and biotechnological potential. Appl. Microbiol. Biotechnol. 2010, 85, 1629–1642. 10.1007/s00253-009-2355-3. [DOI] [PubMed] [Google Scholar]
- Casillas-Vargas G.; Ocasio-Malavé C.; Medina S.; Morales-Guzmán C.; Del Valle R. G.; Carballeira N. M.; Sanabria-Ríos D. J. Antibacterial fatty acids: an update of possible mechanisms of action and implications in the development of the next-generation of antibacterial agents. Prog. Lipid Res. 2021, 82, 101093 10.1016/j.plipres.2021.101093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manetti F.; Castagnolo D.; Raffi F.; Zizzari A. T.; Rajamäki S.; D’Arezzo S.; Visca P.; Cona A.; Fracasso M. E.; Doria D.; Posteraro B.; Sanguinetti M.; Fadda G.; Botta M. Synthesis of new linear guanidines and macrocyclic amidinourea derivatives endowed with high antifungal activity against Candida spp. and Aspergillus spp. J. Med. Chem. 2009, 52, 7376–7379. 10.1021/jm900760k. [DOI] [PubMed] [Google Scholar]
- Maccari G.; Sanfilippo S.; De Luca F.; Deodato D.; Casian A.; Lang M. C. D.; Zamperini C.; Dreassi E.; Rossolini G. M.; Docquier J. D.; Botta M. Synthesis of linear and cyclic guazatine derivatives endowed with antibacterial activity. Bioorg. Med. Chem. Lett. 2014, 24, 5525–5529. 10.1016/j.bmcl.2014.09.081. [DOI] [PubMed] [Google Scholar]
- Pasero C.; D’Agostino I.; De Luca F.; Zamperini C.; Deodato D.; Truglio G. I.; Sannio F.; Del Prete R.; Ferraro T.; Visaggio D.; Mancini A.; Guglielmi M. B.; Visca P.; Docquier J. D.; Botta M. Alkyl-guanidine compounds as potent broad-spectrum antibacterial agents: chemical library extension and biological characterization. J. Med. Chem. 2018, 61, 9162–9176. 10.1021/acs.jmedchem.8b00619. [DOI] [PubMed] [Google Scholar]
- Allegra E.; Titball R. W.; Carter J.; Champion O. L. Galleria mellonella larvae allow the discrimination of toxic and non-toxic chemicals. Chemosphere 2018, 198, 469–472. 10.1016/j.chemosphere.2018.01.175. [DOI] [PubMed] [Google Scholar]
- Touitou M.; Manetti F.; Ribeiro C. M.; Pavan F. R.; Scalacci N.; Zrebna K.; Begum N.; Semenya D.; Gupta A.; Bhakta S.; McHugh T. D.; Senderowitz H.; Kyriazi M.; Castagnolo D. Improving the potency of N-aryl-2,5-dimethylpyrroles against multidrug-resistant and intracellular mycobacteria. ACS Med. Chem. Lett. 2020, 11, 638–644. 10.1021/acsmedchemlett.9b00515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botelho M. G. Fractional inhibitory concentration index of combinations of antibacterial agents against cariogenic organisms. J. Dent. 2000, 28, 565–570. 10.1016/S0300-5712(00)00039-7. [DOI] [PubMed] [Google Scholar]
- Saravanan R.; Mohanram H.; Joshi M.; Domadia P. N.; Torres J.; Ruedl C.; Bhattacharjya S. Structure, activity and interactions of the cysteine deleted analog of tachyplesin-1 with lipopolysaccharide micelle: Mechanistic insights into outer-membrane permeabilization and endotoxin neutralization. Biochim. Biophys. Acta 2012, 1818, 1613–24. 10.1016/j.bbamem.2012.03.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.