Abstract
The mapping of impaired brain glucose metabolism is often used to understand brain diseases. However, current methods for imaging metabolism are limited by low sensitivities, the use of intravenous administration of radiotracers or specialized hardware. Deuterium (2H)-magnetic resonance spectroscopic imaging (2H-MRSI) uses orally administered, non-radioactive substrates providing a safe method to quantify the metabolism of 2H-labelled glucose and its downstream metabolism including neurotransmitter synthesis, yet still requires specialized hardware, and only measures a limited number of deuterated compounds. Here, we show an indirect dynamic proton (1H)-MRSI method at 7T, in participants, that provides higher sensitivity than direct 2H-MRSI with improved spatiotemporal resolution and higher chemical specificity to differentiate glutamate, glutamine, gamma-aminobutyric acid, and glucose deuterated at specific molecular positions while allowing simultaneous mapping of both labelled and unlabelled metabolites without the need for specialized hardware. We also show significant glutamate, glutamine, and glucose decreases, with an 18% faster glutamate reduction in the grey matter than white matter after ingestion of deuterated glucose. Thus, 1H-MRSI can detect downstream glucose metabolism non-invasively using clinically available hardware.
Non-invasive, affordable, and reliable mapping of glucose metabolism in the human brain is critically needed for both clinical and neuroscientific studies. While current approaches, such as fluorodeoxyglucose ([18F]FDG) positron emission tomography (PET), glucose chemical exchange saturation transfer,1 carbon-13 (13C) magnetic resonance spectroscopy (13C-MRS),2 and direct deuterium magnetic resonance spectroscopic imaging (2H-MRSI)3 of glucose metabolism, are technically challenging and require special hardware or expensive synthesis of intravenous radioactive tracers, a recent animal study has shown the potential of the quantitative exchanged-label turnover (QELT) technique for the indirect detection of deuterated compounds via proton magnetic resonance spectroscopy (1H-MRS) to quantify glucose metabolism in the rat brain.4 Orally administered, deuterium-labeled glucose is readily taken up by brain cells, and the deuterons are incorporated into downstream glucose metabolites.5 Since deuterons (2H) substitute protons (1H) in the molecule, they do not contribute to the proton (1H) spectrum. Hence, the increase in deuterium-labeled metabolites is reflected by the decrease in metabolite signals in 1H-MRS.
About ninety-five percent of the glucose in the healthy brain enters the tricarboxylic acid (TCA) cycle for glutamate cycling. The rest converts anaerobically to lactate (Lac).6 Glutamate (Glu) is then involved in the glutamate/glutamine cycle, ammonia detoxification, and GABA synthesis to supply excitatory and inhibitory neurotransmitters. The balance between aerobic and anaerobic glycolysis is critical for maintaining brain energy homeostasis, and the shift from aerobic glucose (Glc) metabolism to anaerobic pathways (i.e., Warburg effect) characterizes pathological conditions seen in tumors and ischemia.7 Anaerobic metabolism also plays a crucial role in mitochondrial dysfunction, an early condition in the pathophysiology of severe neurological diseases, such as Alzheimer’s disease8 and neuropsychiatric disorders such as major depression and schizophrenia.9
The gold standard for the clinical examination of metabolism, i.e., positron emission tomography (PET), with the glucose analog 18F-fluorodeoxyglucose ([18F]FDG) indeed provides quantitative measurements of the cerebral metabolic rate of Glc (CMRGlc) with excellent molecular sensitivity and test-retest variability.10,11 However, [18F]FDG cannot assess downstream metabolism, which is potentially relevant for diagnosis and treatment evaluation.12,13 13C-MRS is a research tool, which can quantify these compounds of the brain’s metabolic cycles;13,14 but suffers from poor spatial localization. Hyperpolarized carbon-13 (13C)-MRSI is another emerging molecular MRI method for rapid, pathway-specific investigation of dynamic metabolic and physiologic processes that are currently evaluated in clinical studies, but not yet available in clinical routine.15 The power of 13C-MRSI lies in its ability to investigate the entire metabolic pathway, including downstream metabolites, which is not possible via [18F]FDG PET. However, currently, the widely used substance, i.e., 13C1-labeled pyruvate, mainly measures the conversion rate of pyruvate to lactate and the entry into the TCA cycle with the formation of carbon dioxide, while it cannot provide information about glutamate/glutamine cycling and GABA synthesis. Both PET and hyperpolarized 13C-MRSI require additional costly hardware and invasive intravenous substrate administration, with PET also requiring harmful radioactive and unstable tracers. However, parallel measurements on PET/MR hybrid systems with ([18F]FDG using functional PET and DMI will provide the unique opportunity to evaluate different aspects of metabolism.16,17
Recently, direct 2H-MRS detection (deuterium metabolic imaging, DMI) provided alternative measures to address the aerobic/anaerobic imbalance, but the technique requires special hardware as well and estimates only a limited number of deuterated compounds, which diminishes its clinical applicability. In contrast, the proposed QELT-MRS promises to overcome these limitations by utilizing harmless and stable tracers and allowing quantification of both cerebral metabolic rates of Glc (CMRGlc) as well as the turnover of downstream intracellular Glc metabolism.4
Thus, since the deuterium method enables quantification of both the oxidative and anaerobic glucose utilization and assesses neurotransmitter synthesis, we aimed to demonstrate the capabilities of QELT-MRSI after peroral administration of 6,6’−2H2-Glc (2H-Glc) in the healthy human brain. Compared to direct 2H-MRS detection, the indirect technique allows quantification of extended metabolic profiles while reliably reflecting downstream metabolism, as shown in a single animal study.4 Motivated by the desire to establish an easy-to-apply approach for metabolic mapping in humans, we employed state-of-the-art methods of single-voxel 1H-MRS18 and echo-less 3D 1H-MRSI,19 which have previously demonstrated high sensitivity for the detection of small functional responses to physiological stimulation.20,21 Our current objective was to trace and map the decaying metabolite signals after non-invasive oral administration of deuterated Glc, utilizing widely available hardware. The experiments were conducted in two sessions with 2H-Glc oral administration to determine between-session repeatability and in another session after non-deuterated Glc (normal dextrose, 1H-Glc) ingestion to mitigate the possible metabolic effects of the Glc load, particularly those of hyperglycemia and insulin release.22 We also acquired one session with direct 2H-MRSI and a dedicated coil to directly compare metabolic effects of 2H-Glc to those obtained indirectly with QELT.
Results
SV-MRS: time-course analysis
While the signal amplitudes at the deuterated C4 position dropped for Glu (Glu4), Gln (Gln4), and Glc6 by 14.9% ± 2.8% (p = 0.0003), 14.4% ± 3.7% (p=0.003) and 21.8% ± 10.3% (p=0.009) respectively, following 2H-Glc ingestion (session 1) in accord with previous animal data,4 Glu4, Gln4 and Glc6 were stable during the control experiment using the administration of 1H-Glc (session 3, Table 1, Fig.1). The robust signal change of Glu4/Gln4 is visually discernible on individual subject spectra (Fig.2a). 2H-MRS spectra in the bottom row (Fig. 2b) additionally show an apparent 2H-Glc peak, whereas the spectral resolution of the Glu/Gln peak is compromised compared to 1H-MRS difference spectra. Asp, Glu2+3, Gln2+3, GABA2, GABA3+4, and Lac, as well as all other metabolites, did not change significantly during both sessions (Suppl. Fig.1). While the Glc1–5 values represent the non-deuterated part of the Glc molecule and the Glc1–5+Tau average time-course resembles the average glycemia time-course (Suppl. Fig. 2) as expected,23 Glc6 underwent deuteration and yielded significant concentration differences after 2H-Glc ingestion (Fig 1, Table1).
Table 1 |.
Metabolite concentration estimates, stability, and temporal changes with and without deuterated glucose all healthy volunteers (N = 5). Data are calculated from concentrations quantified in μmol/g for single-voxel 1H-MRS and referenced to total creatine (tCr) for 3D-1H-MRSI. The regional means from the gray and white matter voxels were used to calculate between-subject averages in metabolite concentrations. The average concentrations and their respective coefficients of variations were measured immediately after 1H-Glc administration (the first time-point). The within-session differences were calculated by subtracting the concentrations of the first and last data point, and their statistical significance was assessed via a standard, two-tailed, paired t-test (separately for 2H-Glc and 1H-Glc scans). The asterisks indicate statistical significance after correction for multiple comparisons with the false discovery rate method, which limited the likelihood of false positives to 7% (single-voxel data) and 3% (MRSI data).
| Posterior cingulum | Whole gray matter | Whole white matter | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Metabolite | conc. | CV | Δconc. 2H-Glc | Δconc. 1H-Glc | conc. | CV | Δconc. 2H-Glc | Δconc. 1H-Glc | conc. | CV | Δconc. 2H-Glc | Δconc. 1H-Glc | |||||||
| (mean±SD) | (mean±SD) | (mean±SD) | p-value | (mean±SD) | p-value | (mean±SD) | (mean±SD) | (mean±SD) | p-value | (mean±SD) | p-value | (mean±SD) | (mean±SD) | (mean±SD) | p-value | (mean±SD) | p-value | ||
| (/tCr) | (%) | (%) | (%) | (/tCr) | (%) | (%) | (%) | (/tCr) | (%) | (%) | (%) | ||||||||
| 1.29±0.23 | 15.0±6.2 | 19.4±27.6 | 0.2002 | 17.1±18.0 | 0.11 | 0.44±0.02 | 6.2±8.0 | −5.9±6.4 | 0.107 | −3.4±1.8 | 0.01 | 0.47±0.03 | 3.9±2.4 | 1.6±1.5 | 0.0848 | −2.3±4.7 | 0.36 | ||
| – | – | – | – | – | – | 0.23±0.02 | 2.9±1.7 | −3.0±4.4 | 0.215 | −5.1±7.4 | 0.20 | 0.23±0.01 | 2.8±1.1 | −2.2±4.8 | 0.3675 | −3.9±7.1 | 0.26 | ||
| 1.15±0.25 | 9.8±1.9 | 11.9±21.4 | 0.1909 | −3.6±15.6 | 0.60 | – | – | – | – | – | – | – | – | – | – | – | – | ||
| 2.38±0.43 | 14.1±7.5 | −2.9±16.8 | 0.5735 | −6.1±19.7 | 0.44 | – | – | – | – | – | – | – | – | – | – | – | – | ||
| 2.32±0.28 | 12±4.3 | −1.3±11.8 | 0.8136 | 9.2±17.4 | 0.22 | – | – | – | – | – | – | – | – | – | – | – | – | ||
| 1.06±0.23 | 23.9±9.6 | 1.0±24.4 | 0.8801 | 16.8±34.4 | 0.22 | – | – | – | – | – | – | – | – | – | – | – | – | ||
| 3.37±0.21 | 8.2±4.3 | 21.8±10.3 | 0.0086* | 4.5±10.8 | 0.38 | x | x | x | x | x | x | x | x | x | x | x | x | ||
| – | – | – | – | – | – | 0.30±0.03 | 6.3±4.1 | −4.8±7.9 | 0.288 | −3.1±8.5 | 0.46 | 0.30±0.02 | 5.5±3.1 | 0.3±6.2 | 0.8439 | −9.4±8.8 | 0.07 | ||
| – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | ||
| 2.60±0.31 | 4.5±1.6 | 14.4±3.7 | 0.0029* | 2.4±4.4 | 0.25 | – | – | – | – | – | – | – | – | – | – | – | – | ||
| – | – | – | – | – | – | 0.72±0.04 | 2.3±2.6 | −5.1±2.6 | 0.012 | −4.2±6.7 | 0.23 | 0.71±0.02 | 2.7±2.7 | −5.1±2.3 | 0.0068 | −3.9±6.8 | 0.28 | ||
| 7.67±0.22 | 1.4±0.1 | 14.9±2.8 | 0.0003* | −0.8±2.2 | 0.49 | 0.54±0.04 | 1.7±1.8 | 13.4±3.5 | 0.001* | −0.7±1.3 | 0.35 | 0.50±0.03 | 1.6±1.5 | 14.0±2.9 | 0.0007* | −2.4±3.8 | 0.24 | ||
| 13.26±0.57 | 1.1±0.1 | −3.4±3.0 | 0.0651 | −0.5±0.7 | 0.19 | – | – | – | – | – | – | – | – | – | – | – | – | ||
| 0.76±0.09 | 4.3±1.0 | 1.9±3.8 | 0.3373 | −2.4±4.0 | 0.30 | – | – | – | – | – | – | – | – | – | – | – | – | ||
| 0.74±0.11 | 8.0±4.4 | 4.8±7.7 | 0.2804 | 0.3±10.0 | 0.85 | – | – | – | – | – | – | – | – | – | – | – | – | ||
| 5.60±0.27 | 1.0±0.2 | −0.2±1.6 | 0.7935 | −0.8±1.1 | 0.20 | 0.82±0.05 | 2.3±0.9 | −0.1±2.7 | 0.931 | −0.6±2.7 | 0.65 | 0.84±0.07 | 2.2±0.6 | −0.4±2.6 | 0.7662 | −0.7±3.8 | 0.74 | ||
| 0.61±0.27 | 23.0±23.2 | −11.1±24.7 | 0.3110 | −18.6±54.1 | 0.95 | – | – | – | – | – | – | – | – | – | – | – | – | ||
| 9.72±0.38 | 1.1±0.7 | −1.6±2.3 | 0.1861 | −0.2±0.7 | 0.55 | – | – | – | – | – | – | – | – | – | – | – | – | ||
| 0.70±0.15 | 8.2±3.2 | 8.8±7.4 | 0.0527 | −2.7±5.8 | 0.34 | – | – | – | – | – | – | – | – | – | – | – | – | ||
| 0.28±0.04 | 12.0±3.5 | −9.1±8.1 | 0.0676 | −0.6±19.0 | 0.86 | – | – | – | – | – | – | – | – | – | – | – | – | ||
| 2.55±0.24 | 3.6±1.2 | 5.2±7.8 | 0.2085 | 0.4±6.8 | 0.82 | – | – | – | – | – | – | – | – | – | – | – | – | ||
| 0.10±0.05 | 12.7±6.3 | 2.4±14.1 | 0.6303 | 1.4±18.6 | 0.54 | – | – | – | – | – | – | – | – | – | – | – | – | ||
| 1.26±0.15 | 3.9±0.3 | −3.5±5.6 | 0.2474 | 2.1±4.0 | 0.26 | 0.35±0.02 | 4.2±1.2 | −5.9±5.6 | 0.077 | −1.2±6.2 | 0.69 | 0.38±0.03 | 3.6±1.5 | −3.4±9.7 | 0.4758 | −3.0±7.2 | 0.39 | ||
| 1.04±0.06 | 1.8±0.6 | −1.0±4.5 | 0.6782 | −1.2±3.9 | 0.53 | 0.33±0.02 | 0.9±0.8 | −2.3±1.8 | 0.050 | −0.5±1.9 | 0.60 | 0.35±0.23 | 1.0±0.6 | −1.7±1.8 | 0.1051 | −0.9±2.1 | 0.43 | ||
| 6.28±0.12 | 1.3±0.5 | −1.3±2.4 | 0.2870 | −1.8±1.5 | 0.05 | 1.00±0.00 | – | – | – | – | – | 1.00±0.00 | – | – | – | – | – | ||
| tNAA | 10.42±0.49 | 1.2±0.7 | −1.0±2.3 | 0.4015 | −0.3±0.5 | 0.19 | 1.47±0.08 | 1.9±2.5 | 1.1±3.9 | 0.575 | −1.2±2.2 | 0.30 | 1.59±0.06 | 1.7±2.2 | 1.4±3.2 | 0.4114 | −0.3±2.1 | 0.78 | |
Fig.1 |. Quantification of SV-MR spectra obtained in posterior cingulum with single-voxel proton-MRS.
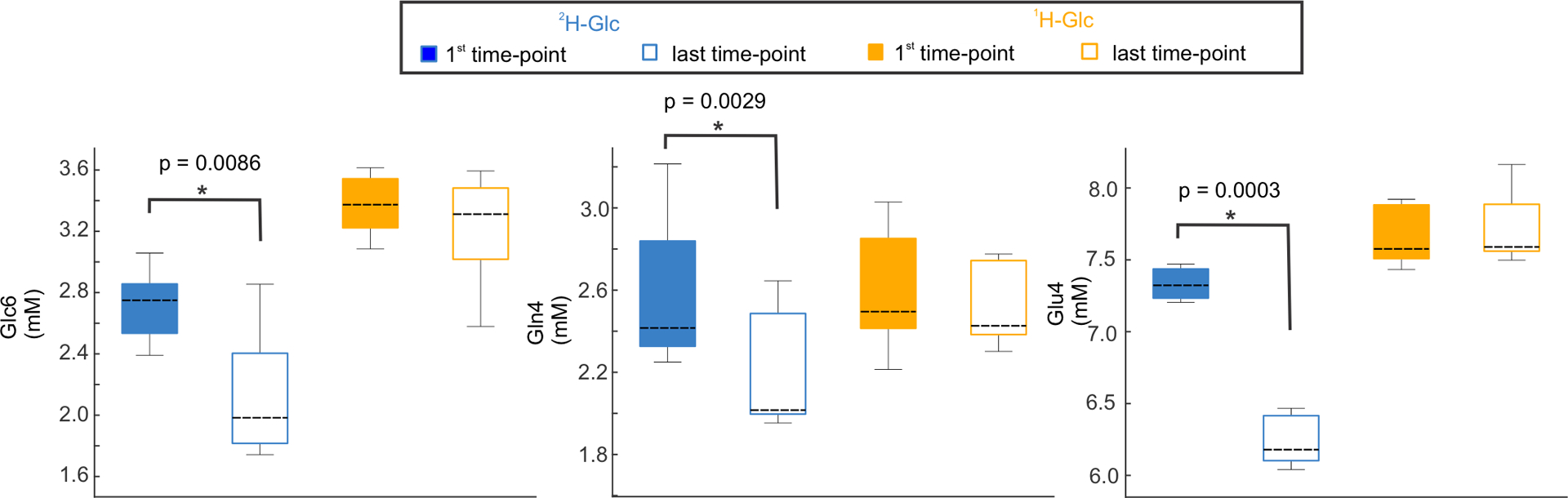
Bar diagrams demonstrate concentration comparisons quantified from the first and last time-point spectra following 2H-Glc and 1H-Glc administration for 5 healthy volunteers and reflect continuously decaying metabolite signals through the acquisition period. Peaks that originated from the specific carbon position undergoing deuteration, namely, the 6th carbon position for Glc (Glc6) the 4th carbon position for Glu (Glu4) and Gln (Gln4) and the 2nd carbon position for GABA (GABA2), were separated. Concentrations were compared with a standard, two-tailed, paired t-test between the first and last time-point within the 2H-Glc and 1H-Glc sessions. The metabolites that appeared significant (p < 0.05) are displayed here. The whiskers represent the minimal and maximal values, the boxes first and third quartile and the dashed lines are medians. P-values were not corrected for multiple comparisons. The full analyzed neurochemical profile is shown at Suppl. Fig.1.
Fig. 2 |. Example of MR spectra obtained from one voxel in the posterior cingulum with single voxel 1H-MRS (panel A) and with 2H-MRSI (panel B) in one participant at 7 Tesla.
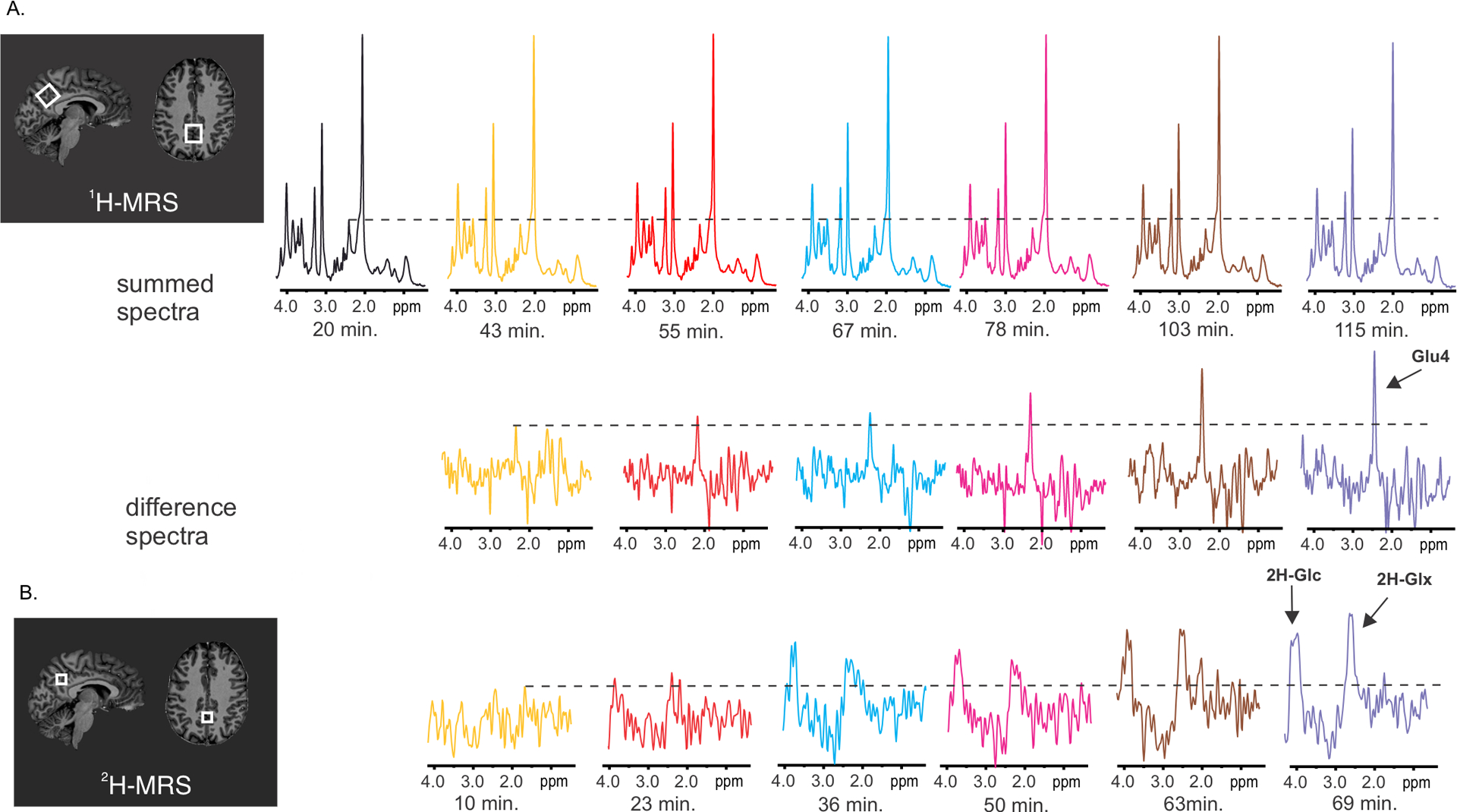
The displayed spectra were obtained after peroral administration of 2H-Glc. A. The summed spectra show decreasing signal at the resonance frequency of -C41H2- in the glutamate molecule (2.34 ppm) due to enrichment of the glutamate pool with 2H (either -C42H2H- or -C42H1H). The robust change at the 4th carbon position is documented by the increasing signal amplitude at 2.34 ppm in the difference spectra calculated by subtraction of the respective spectrum from the last session minus the first session. B. 2H-MR spectra reflect an increase in the deuterated fraction of Glc and Glx (Glu+Gln). The insets with anatomical images show location of voxels in the posterior cingulate gyrus. The voxel dimensions were 22×20×20 mm for single-voxel proton MRS, whereas voxel selected from 2H-MRSI data had volume of (12.5 mm)3.
Changes in the concentration of GABA and GABA2 were not statistically significant, possibly due to higher variance in GABA and GABA2 typical for non-edited 1H-MRS methodology. However, GABA2 changes in difference spectra were quantified with mean within-session Cramer-Rao-Lower-Bounds (CRLB) <20%, which is generally acceptable for analysis.
To corroborate the LCModel analysis of single-subject data, high-SNR spectra were calculated by summing the spectra from all subjects (N = 5). The summed spectra and their difference displayed in Fig. 3 clearly demonstrated the effect of deuterium enrichment (Fig. 3A–D). While the quantification of single-subject time-courses did not reveal a change in GABA2, the quantification of difference spectra yielded acceptable Cramér-Rao lower bounds, i.e., the estimates of quantification error, of 16% for GABA2. While the SNR of 1H-MRS difference spectrum (Fig.3C) does not differ much from the 2H-MRS spectrum, the 1H-MRS difference spectrum shows substantially higher spectral resolution than the 2H-MRS spectrum (Fig 3E).
Fig.3 |. 1H-MRS difference spectra and their quantification.
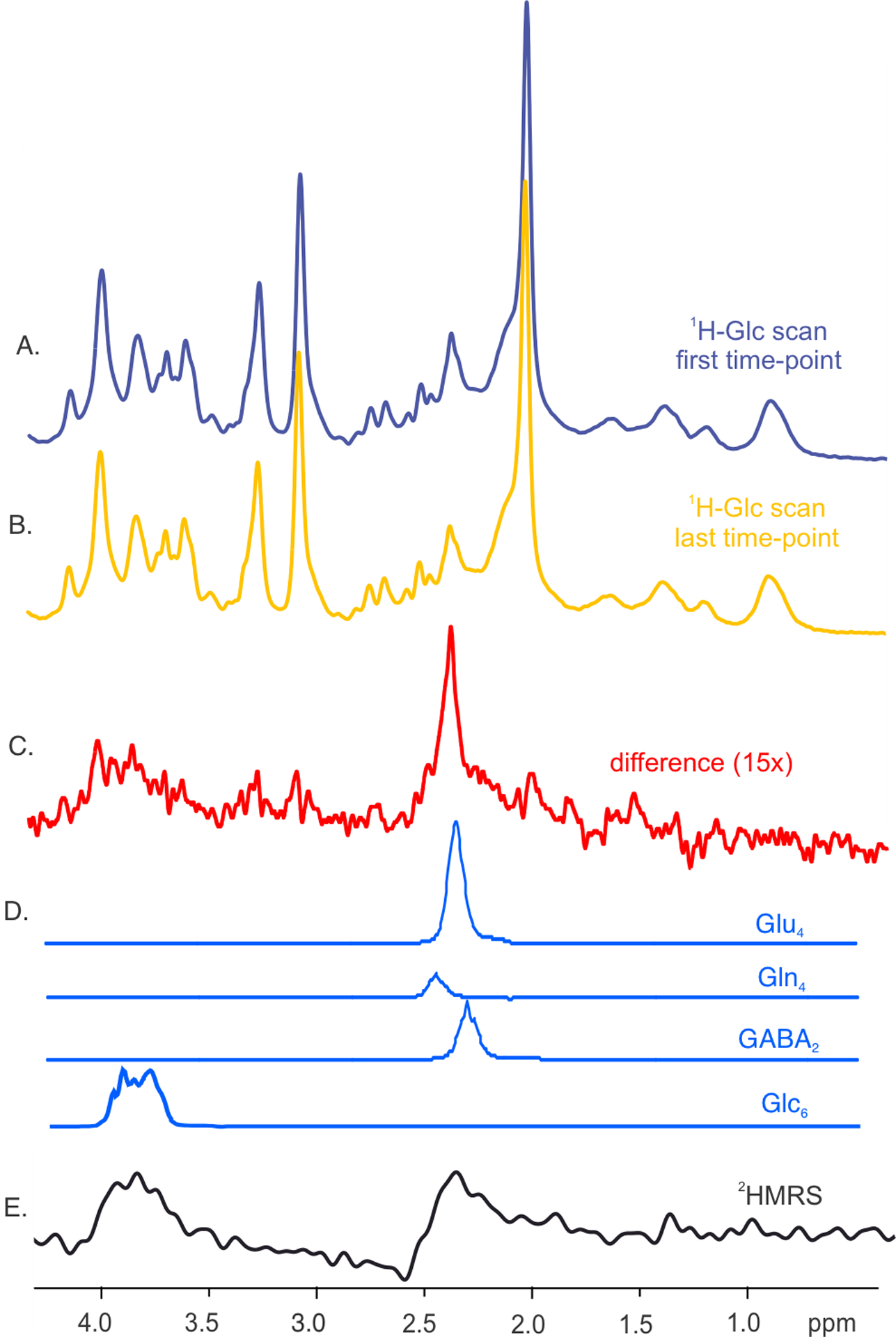
Summed spectra from all subjects (N=5) represent the first and last time-points after 2H-Glc ingestion. The spectra (A and B) were linewidth-matched with exponential line-broadening and subtracted. The resulting difference spectrum in panel C represents the effect of metabolite 2H enrichment. The metabolite components (D) were obtained via LCModel analysis using a basis set containing simulated spectra of neurochemicals that were undergoing deuteration (i.e., Glu, Gln, and GABA and Glc). The proton signals that originated from different carbon groups were separated. The summed 2H-MRS spectrum (N=5) from the last acquired time-point resembles the 1H-MR spectrum except for the spectral resolution (E).
The exponential and linear fits of Glu4, Gln4, and Glc6 data clearly demonstrated decaying signals in the 2H-Glc session (session #1, Fig. 4, left column). The slopes were reproduced in session #4 with direct measurement of deuterated compounds i.e., Glx4 (Glu4+Gln4) and Glc6 (Fig.4, right column). While the slopes of directly measured 2H-Glx4 (0.03±0.006) had a between-subject variance of 19% similar to indirectly measured linearly fitted slopes of Glu4 (−0.01± 0.003, CV=18%), Glc6 had a higher between-subject variance in the slopes measured with both direct (0.009±0.004, CV=46%) and indirect (−0.007±0.002, CV=34%) methods. Fig.4. demonstrates similar concentration differences (1st minus last time-point) of ~2 mM for Glx (Glu and Gln) and 0.6 mM for Glc with both methods (QELT and DMI). The changes in Glu4 and Gln4 were in quantitative agreement also with the previous 13C study conducted with orally administered labeled Glc, in similar amounts used in the current study, i.e., the fractional isotopic enrichments were ~1.5mM for Glu, ~0.5 mM for Gln in the brain.24
Fig. 4 |. Fitting of time courses obtained by quantification of 1H (left column) and 2H single-voxel MR spectra (right column).
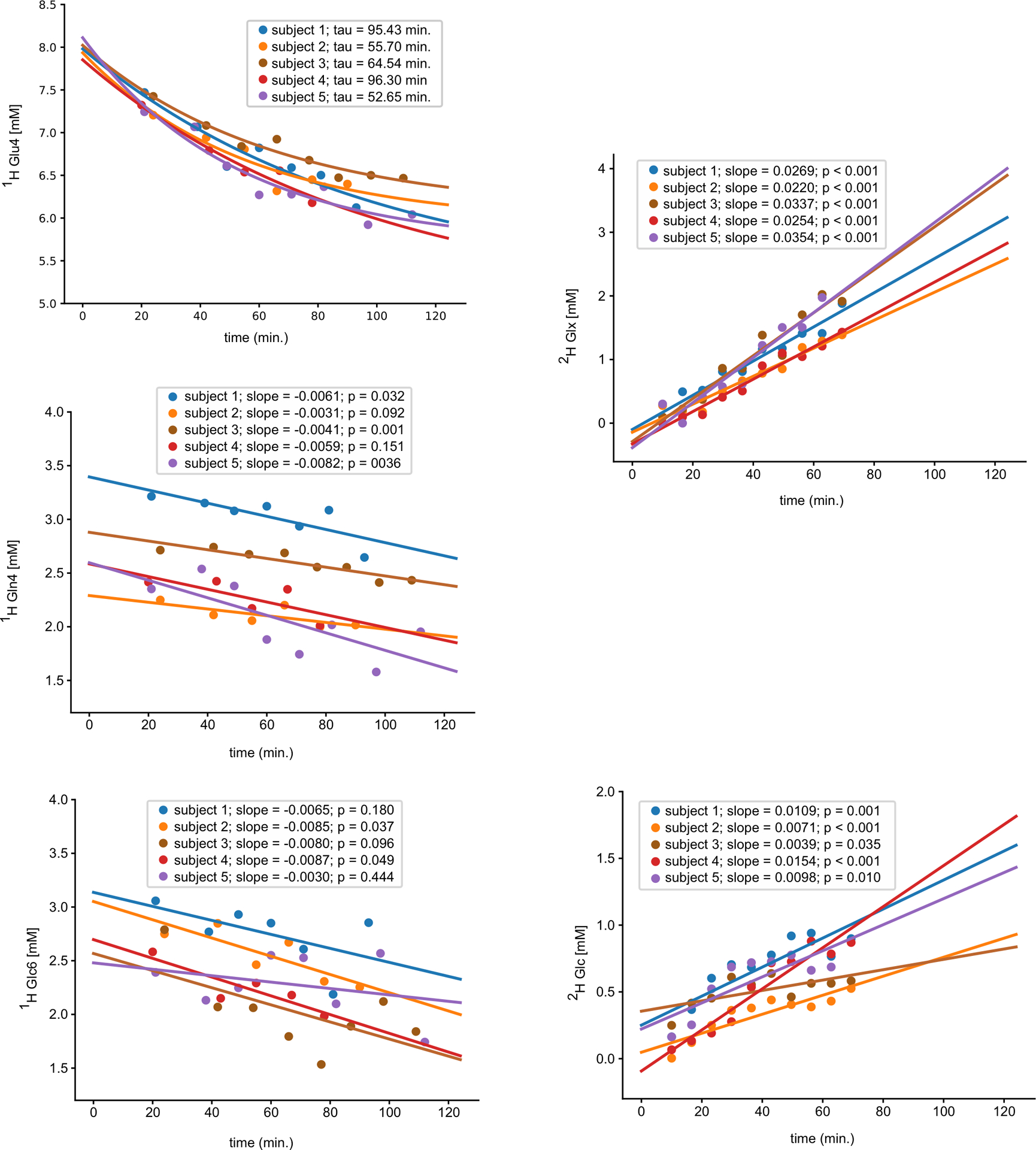
The spectra were acquired in a single PCC voxel and concentrations were obtained by LCmodel. Decay in the concentrations of glutamate (Glu4) and glutamine (Gln4), glucose (Glc6), Glx (Glu+Gln) were fitted using the exponential function Y=Y0(−t/tau)+c (Glu4), and linear regression Y = a + bX (Gln4,Glc6, 2H-Glc, and 2H-Glx). The 1H-MRS time-courses and their fits following 2H-Glc ingestion are in line with those obtained by quantification of 2H-MR spectra. Two-tailed test was used to calculate significance of the slopes (p-values).
While, the slopes were obtained per subject from Glu4 with a CV of 18% (session #1, linear fits), the mean between-session CVs in the slopes were 11% ± 9% (session #1 vs. session #2, linear fitting, Suppl. Fig 3). The decaying slopes of Gln4 and Glc6 were obtained with the between-subject variance of 36% and 34%, whereas the between-session variance was 61% ± 37% (Gln4) and 30% ± 13% (Glc6). The linear fitting of the paired 1H-Glc data showed stable Glu4, Gln4, and Glc6 concentrations during the control condition (session #3) scanned after 1H-Glc ingestion (Suppl. Fig. 3).
Overall, concentrations of 14 metabolites were quantified with average within-session CVs below 5% (MM, Cr, GPC, myo-Ins, PCr, PE, NAA, Tau, Glu2+3, Glu4, Gln4, tCho, tCr, tNAA), and another eight (Asp, Glc, GSH, scyllo-Ins, PCh, NAAG, GABA2, Glc+Tau) with CVs below 20% as assessed from concentrations obtained from the 1H-Glc scan (Table 1).
3D-MRSI: voxel-wise time-course analysis
Analysis of time-courses on a per-voxel basis revealed a drop in Glu signal at the C4 position (Glu4) by 13.4%±3.5% in the gray matter (GM) (p = 0.001) and 14.0%±2.9% in the white matter (WM) (p = 0.0007, Table 1). For all other reliably quantified metabolites, no significant changes were found during the 2H-Glc scan (session #1, Table 1). As expected, all metabolites were stable during the 1H-Glc session #3 (tCho, tNAA, GABA, Gln, Glu4, Glu2+3, myo-Ins). The regional difference in the Glu4 drop between GM and WM during the 2H-Glc session was reflected by the spectra shown in Fig. 5A. The signal attenuation around 2.34 ppm (i.e., Glu4 resonance) and no other major signal changes in other LCModel-quantified spectral regions (1.9–4.2 ppm) corroborated the robust detection of ΔGlu4 with single-subject spectral analysis during the 2H-Glc experiment. The regional averages of the 2H-MRS spectra measured in the same subject (the last time-point, session #4) are shown in the bottom raw (Fig 5B). The 1H-MR spectra from a single MRSI voxel acquired in the PCC region are shown in Suppl. Fig. 4 to demonstrate the spectral quality of single-subject data.
Fig. 5 |. Effect of Deu-Glc on the spectra obtained from the gray and white matter with 3D multi-voxel 1H-MRSI and 2H-MRSI data.
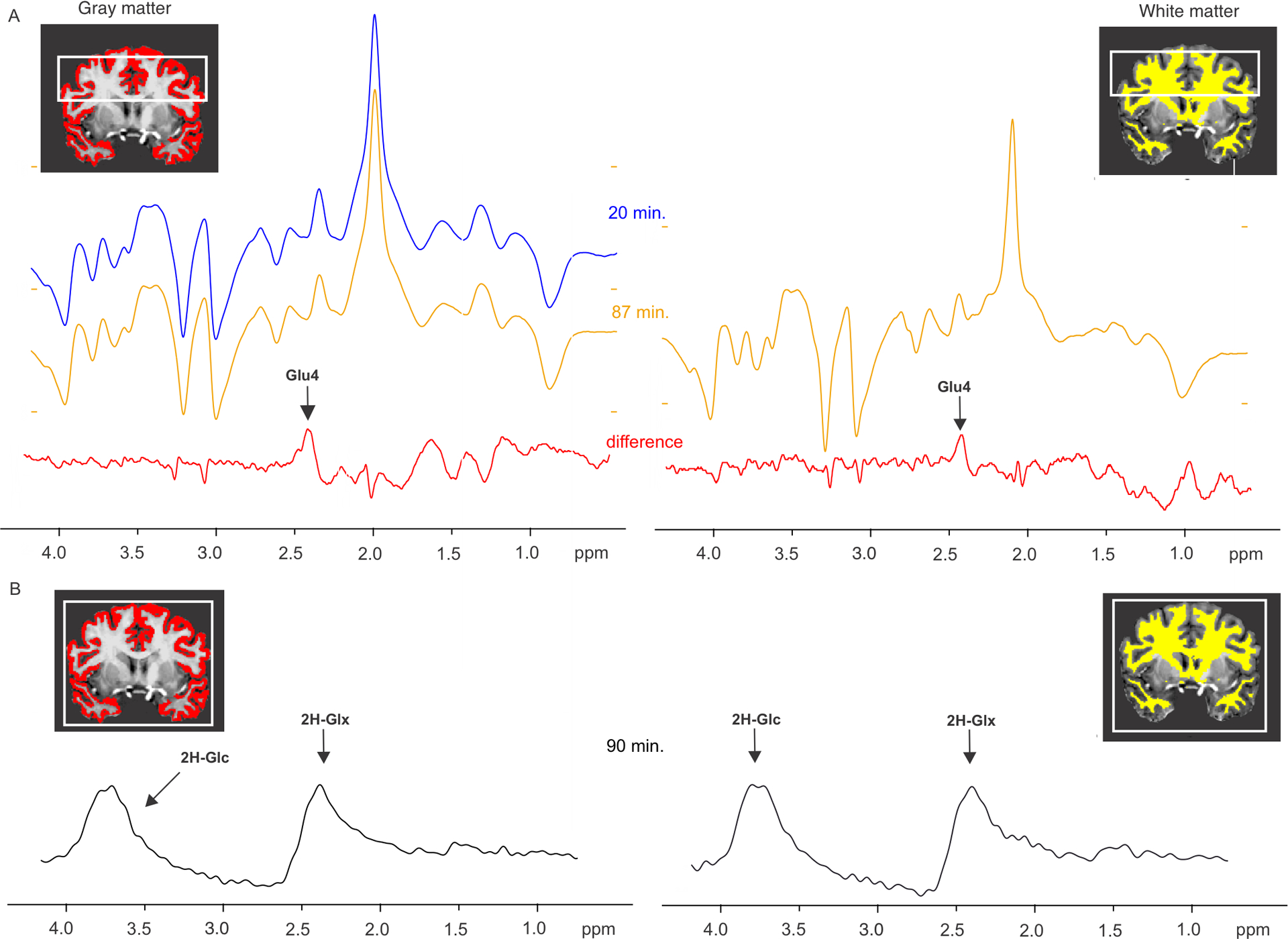
Difference spectra (red) in the panel A were calculated by subtraction of spectra obtained from the first (purple) and last (green) time-point after deuterium ingestion in one healthy volunteer. The spectra were selected using a quality control mask and segmented gray or white matter masks within the 40 mm-thick volume-of-interest (180×180×40 mm) (white box). The signal loss at 2.34 ppm reflects the exchange of protons and deuterons at the 4th carbon position in the glutamate molecule and is displayed as a positive peak in the difference spectra. The signal decay is convincingly found in both gray and white matter. Lack of unwanted signals in the quantified range 1.9–4.2 ppm verifies good spectral quality and stability during the acquisition. The 2H MRS spectra in panel B were acquired in the last time point in the whole brain (the white box, 200×200×175 mm) and averaged from the voxels with most gray and white matter content. While the 1H-MRSI data were acquired with spatial resolution of ~(5 mm)3, the nominal voxel dimension for 2H-MRSI were ~(12.5 mm)3.
As the magnitude of Glu4 signal attenuation is influenced by the variable initiation and duration of the MRS data acquisition among scans, each single-subject temporal MRSI dataset was fitted with an exponential function to characterize the speed of signal decay—the time constant (tau)—which is well characterized in each scan session. The averaged fits are shown in Fig.6A along with averaged fits of the 2H-MRSI data (Fig. 6B). The exponential fit of Glu4 concentrations yielded tau values (the rate constants) of 44±22 minutes and 52±23 minutes in GM and WM, respectively, in the 2H-Glc sessions (session #1). While the between-subject CVs for the concentration differences in Glu4 were 26% (GM) and 21% (WM), the respective CVs calculated from the rate constants (tau) were 50% (GM) and 45% (WM). The decay was 18% faster in GM than in WM, on average. On the other hand, the slopes of linearly fitted 2H-MRSI yielded minimal difference between the GM and WM, likely due to the substantial partial volume effect given by the nominal spatial resolution of ~(12.5 mm)3 vs, ~(5 mm)3 for 1H-MRSI. The between-session CVs in the slopes between session #1 and session #2 were 18.3%±12.4% and 12.4%±7.5% in GM and WM, respectively (Suppl. Fig. 5).
Fig. 6 |. Fitting of time courses from averaged regional MRSI maps after 2H-Glc ingestion:
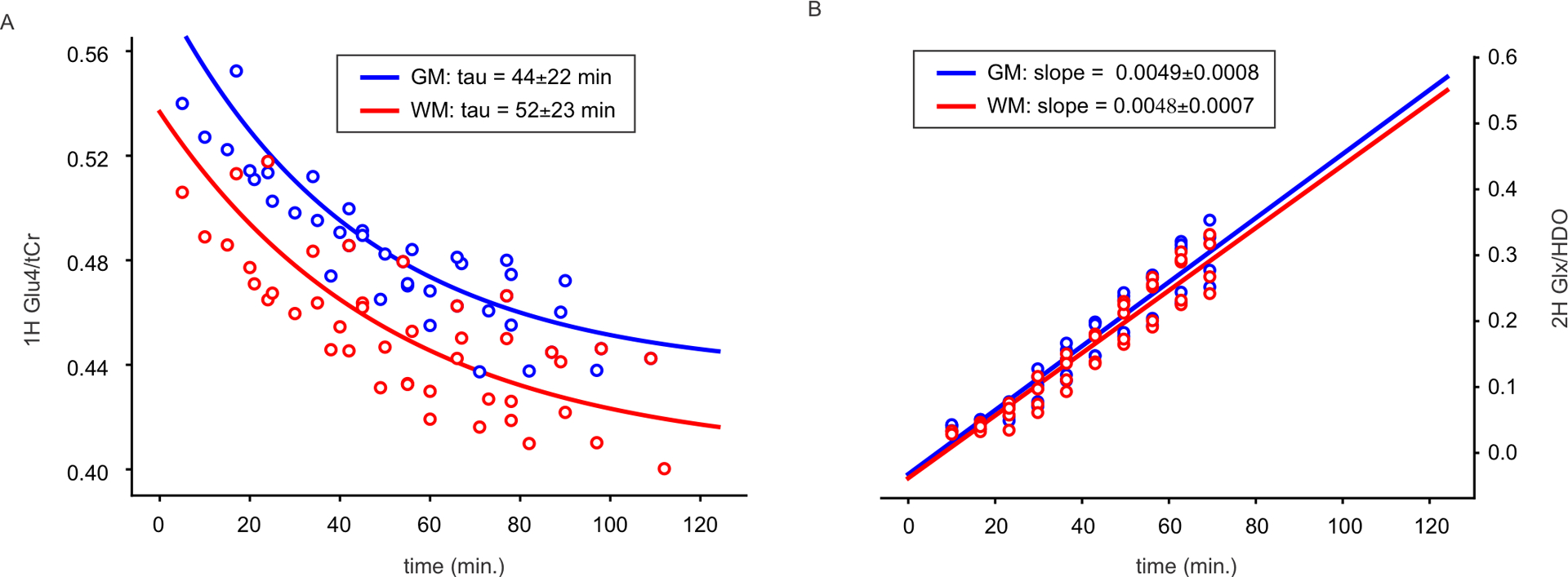
While the exponential fits of 1H-MRS data (panel A) showed 18% faster decay (smaller constant of the decay – tau, M=M0(−t/tau)+c) in the gray (GM, blue) than in the white matter (WM, red), the linear fits of 2H-MRS data (panel B) yielded only minimal differences between GM and WM.
The linear regression of the Glu4/tCr time-courses measured during the 1H-Glc session #3 yielded non-significant slopes of 0.00002±0.00006 and 0.0001±0.0002 (p>0.07), thus indicating no effect of glucose load on the Glu4 and good stability of the measurements (Suppl. Fig. 5).
In GM and WM, 372±169 and 737±280 voxels, respectively, fulfilled the quality assessment criteria and were, thus, used to calculate regional means for each time-point and their CVs per session (Table 1). LCModel quantified 10 metabolites referenced to tCr with within-session CVs below 7%, i.e., Asp, myo-Ins, Tau, Glu2+3, Glu4, Gln, GABA, tCho, and tNAA as assessed on the data measured in the 1H-Glc sessions. Namely, Glu4 was consistently quantified with CVs below 2% in both GM and WM.
Finally, single-subject MRSI time-courses obtained with high time resolution were fitted with linear regression per voxel (Fig. 7A, B). While the time course slopes (p<0.05, r<−0.8) were steeper in the GM than WM by 19% for 1H-MRSI and yielded a clear distinction between GM and WM on the slope map (Fig. 7A). The smaller WM/GM difference of 6 % for 2H-MRSI data can be ascribed to a greater partial volume effect than in 1H-MRSI obtained with substantially higher spatial resolution.
Fig. 7 |. Voxel-wise fitting of Glu4 and Glx4 time-courses obtained with high time resolution.
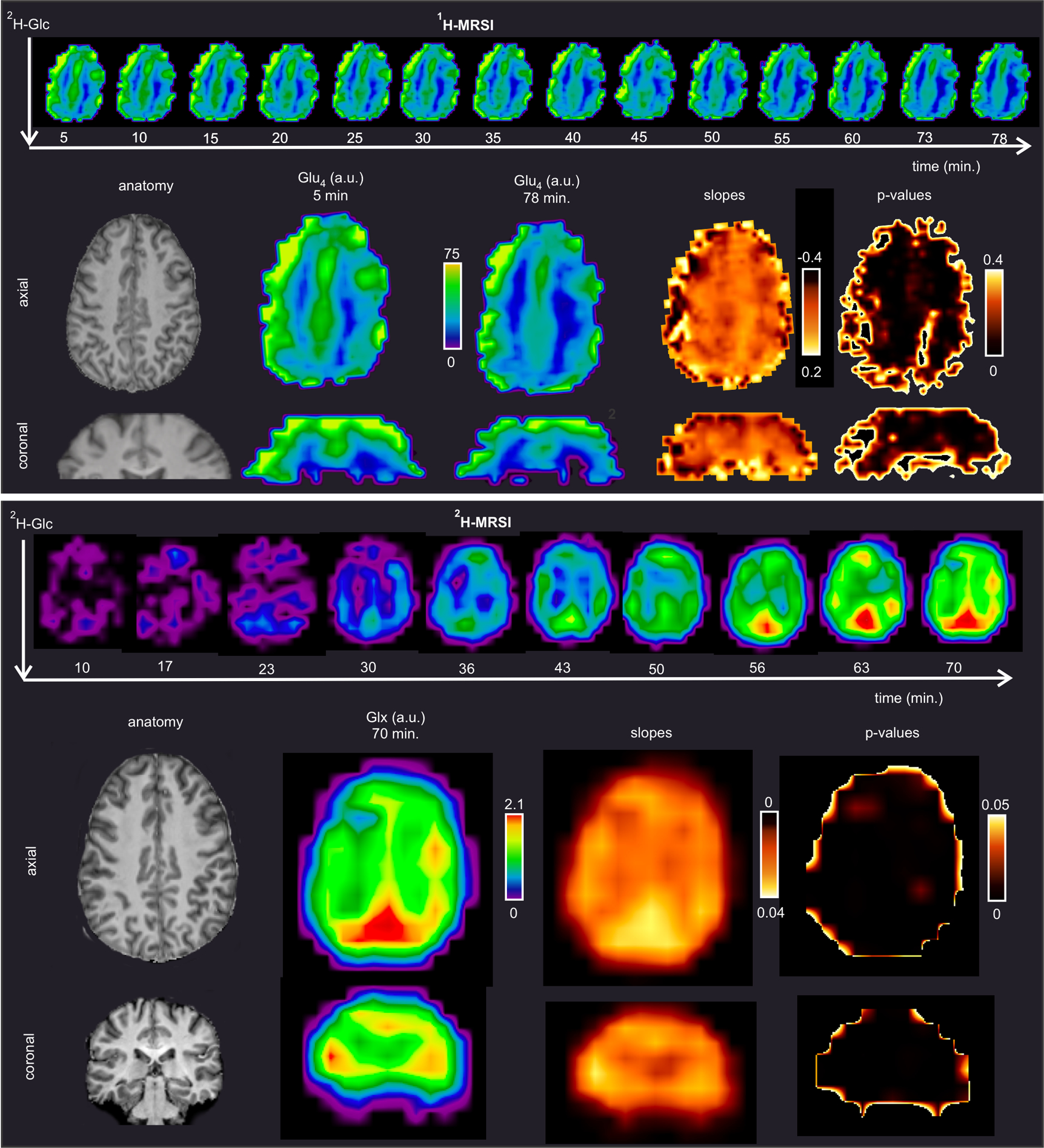
MRSI data were acquired in one participant with a time-resolution of five (1H-MRSI, panel A.) and 6 minutes (2H-MRSI, panel B.) after peroral administration of 2H-Glc. Maps demonstrate the signal decay of protons on the 4th carbon position in the glutamate molecule (Glu4, 1H-MRSI) and in Glx4 (2H-MRSI). Voxel-wise linear regression was applied to Glu4 and Glx4 time-courses. Respective slopes tend to be higher in the gray matter, which suggests a higher glutamate turnover in the gray than in the white matter. The p-values refer to the significance of the slopes. Spectral postprocessing of 1H-MRSI included channel-wise L2-regularization for suppression of unwanted lipid signals.
Discussion
The current work reveals the potential of deuterium labeling to measure the turnover of metabolites involved in oxidative glucose metabolism in the human brain. Impaired glucose homeostasis and mitochondrial dysfunction are key components in the pathophysiology of neurodegenerative diseases, such as Alzheimer’s and other forms of dementia, and also in metabolic disorders, including obesity and insulin resistance,25 moving the imaging of brain glucose metabolism to center stage.26 The development of non-invasive methods that enable objective, dynamic, and longitudinal metabolic tracking in health, disease, and aging are urgently needed to track drug treatment effects and investigate emerging therapies.
Here, we demonstrated dynamic downstream Glu mapping after 2H-Glc administration in humans using a multi-voxel 1H-MRSI sequence, which was corroborated by the well-established, single-voxel, functional MRS methodology,20,27 as well as direct measurements of deuterated compounds with 2H-MRSI (DMI). Our work benefited from the combination of ultra-high-field (7T) and a high natural abundance of protons in the human body. The 1H-based QELT-MRS(I) approach utilized commercially available radiofrequency coils and optimized 1H-MRS approaches to overcome the major drawbacks of direct techniques tuned to the deuterium frequency (2H-MRS) that require dedicated coils and pulse sequences.28 Compared to 2H-MRS, the indirect QELT approach also enabled the detection of GABA (despite a non-significant GABA2 signal drop) and the separation of Glu from Gln, which is not feasible using 2H-MRS even at ultra-high MR fields (16.4T) due to the limited spectral resolution of 2H-spectra.29 The orally administered deuterated Glc is safe, stable, and affordable compared to radioactive PET tracers with short half-lives, which require technically challenging onsite preparation and expensive cyclotrons. We have also shown the ability of QELT-MRS to overcome challenges of 13C-MRS, which was previously the only method for quantitative assessment of TCA cycle kinetics and Glu and Gln cycling. The modified basis sets with separated Glu and Gln components on the C4 position enabled the extraction of quantitative information about Glu and Gln turnover in a manner similar to that of the technically challenging 13C-MRS. Although GABA changes have been tracked in individual subject data in a preclinical study at 9.4T,4 but not in our data, our results depicted signal changes in GABA2 using difference spectra calculated from group-averaged data with acceptable quantification errors (CRLB of 16%). The analysis of group difference spectra convincingly corroborated Gln4, Glu4, and Glc6 changes captured in individual data. The fitting of difference spectra constructed by subtracting spectra pooled from multiple subjects indeed benefits from a high signal-to-noise ratio and elimination of signals from static metabolites, including the molecular background. The same analysis approach was previously used to unravel metabolite consequences of chromatic and achromatic stimulation and27 thus analysis of pooled spectral differences can be analogically utilized to compare cohorts of controls and patients. We expect a further boost of quantification of J-coupled relatively low-abundant metabolites such as GABA and Gln by optimization of the sequences for higher fields (i.e., above 7T), possibly enabling robust GABA detection even at low-field MR scanners.30
The deuteration of molecules (i.e., replacing 1H with 2H nuclei) is a simple chemical procedure that allows the labeling of a broad range of substances with minimal influence on their in vivo kinetics within metabolite cycles.31 Other deuterated tracers, such as deuterated choline, could be indirectly detected with 1H-MRS in brain tumors.32 Other potential candidates are deuterated-ketone bodies, such as beta-hydroxybutyrate (BHB)33,34 which serve as alternative brain fuels, especially during fasting, with critical implications in AD,35 some forms of epilepsy36, and brain tumors. Acetate is preferentially utilized by astrocytes and thus could be used to probe energy metabolism in astrocytes.37 While we did not detect any lactate changes due to minimal activity of anaerobic glycolysis in the healthy brain, 1H-MRS is highly sensitive to lactate.20 Indirect QELT-MRS(I) can be thus used to measure the Warburg effect, a shift toward anaerobic glycolysis due to mitochondrial failure in ischemia and brain tumors, as shown in animal glioma models.4 Increased lactate is also a hallmark of aging-related mitochondrial dysfunction,38 and it is conceivable that lactate production accompanies deteriorated Glu production via the TCA cycle in neurodegeneration.39 Thus, the characterization of the main Glc pathways through Lac and Glu dynamics extends the clinical utility of this method.
Our study demonstrated several methodical advancements compared to the initial preclinical animal QELT at 9.4T work that are critical for future applications in humans. We achieved substantial signal gain due to the use of an echo-less 1H-MRSI sequence with concentric ring trajectory readout as well as high spatial and temporal resolution 3D imaging at 7T.19,40 Preliminary 7T QELT-MRS(I) results in humans41 we obtained utilizing a standard vendor-provided single-voxel PRESS MRS sequence which suffers from poor spatial selection and TE-/J-evolution related signal losses at 7T,42 along with a slow phase-encoded CSI sequence with a 4×4 cm box within a single slice subdivided into 4×4 voxels of 1cm3 volume in the center of the brain in ~4min per time frame. In contrast, the semi-LASER single-voxel sequence utilized in the current study provides accurate quantification of extended neurochemical profile even at 7T43 and our multi-voxel 1H-MRSI sequence achieved 8-times higher spatial resolution (i.e., 0.12 cm3) in 3 min per time frame and almost 10,000-times more voxels over a 3D-brain volume using a fast spatial-spectral encoded echo-less FID-MRSI alternative, which is recommended for 7T.43 FID-MRSI offered several-fold increased signal amplitudes for our target J-coupled metabolites compared to the TE of 28ms. Also, our spatial resolution was 67-times higher than in previous direct 2H-MRSI studies at 4T (0.12 mL vs. 8 mL)28 and 16-times higher compared to our 2H-MRSI scans at 7T (0.12 mL vs. 2 mL). Finally, our whole brain 2H-MRSI was obtained with a spatial resolution similar to a recent human study at 9.4T (3.4mL vs. 3.0 mL).3 The differences in obtained spatial and temporal resolution can be explained by the different sensitivities of 2H- and 1H-detection, which are estimated and experimentally validated to be at least ~5 folder higher SNR for 1H4, when considering the ~6.5-fold higher gyromagnetic ratio, half the nuclear spin, and ten-times slower T1 relaxation, neglecting differences in coil hardware (e.g., ~3-folder higher for array coils). However, a realistic comparison must also consider the fact that 2H-MRS detects only the signal of interest (=2H-labeled compounds), whereas 1H-MRS detects also signals from >80% non-labeled substances (which are not of primary interest).
The nearly twenty percent difference in the glutamate turnover in GM and WM is lower compared to the current gold standard ([18F]FDG-PET studies, which measured the differences in the CMRGlc (oxidative) at 0.18 and 0.24 μmol/g/min,44,45 i.e., ~33% higher oxidative Glc consumption ascribed to the higher inhibitory and excitatory demands of the gray matter.46 A 13C study estimated the rate of TCA cycle (VTCA) at 0.88 ± 0.12 in the gray and 0.28 ± 0.13 in the white matter, i.e, 68% difference between GM and WM47 and the outcomes were corroborated by other studies reviewed herein.14 The Glu4 decays in our study do not directly reflect VTCA due to the compartmental nature of Glu4 metabolism. Labeled Glu4 is released to the synaptic cleft and taken up by glia (astrocytes), where Glu is converted to Gln and returned to neurons.48 Thus, the Glu4 decaying constants reflect both the VTCA and Glu/Gln cycle. The enrichment of the glial glutamate pool with neuronal Glu might explain lower differences in Glu4 turnover between GM and WM. While we observed differences between Glu4 slopes in GM and WM, the between-subject differences (last minus first point) were similar in the PCC (ΔGlu4=15%±3, 80% GM/WM fraction, SV-MRS) and GM (ΔGlu4=13%±4, MRSI) and WM (ΔGlu4=14%±3, MRSI). This indicates that similar relative differences between the first point and steady-state are achieved at different speeds. The within-session differences are in agreement with a previous 13C study, which reported peak fractional enrichment of Glu4 of 16%±2% 130 min. after oral administration of 13C-Glc.49 Thus, the fractional enrichment in our data is very similar considering the shorter acquisition window of (98±7 min on average) with 0.8 g/kg of 2H-Glc vs. lower dose of 13C-Glc (0.65 g/kg) and yields also similar between-subject variance. The robust detection of within-session differences in Glu4 was enabled by excellent CVs of 2% for both 1H-MRS and 1H-MRSI, which were well below the detected differences. While the between-subject variance in the concentration differences between the first point and the last point was 19% (PCC - SV-MRS), 26% (GM - MRSI), and 21% (WM – MRSI), the between-subject variance in Glu rates appeared similar for MRS (~ 29%) and higher for MRSI (50% for GM and 45% for WM). Thus, MRS fitting yielded a similar variance as the gold standard ([18F]FDG-PET measures of CMRGlc, reported in the range of 19%−29% (mean – median)) between subjects.16,50 This can be mainly ascribed to high physiological variation in resting brain metabolism. The same applies to the between-session variance, which was 14%±8% for ([18F]FDG-PET and 11%±9% for Glu4 in single-voxel 1H-MRS data.51 The current work is a preliminary study, and further technical improvements are expected to yield higher reproducibility, and time/spatial resolution by integrating techniques for instability correction,52 advanced B0 shimming, or (k,t)-undersampling.53 Despite these methodological limitations, we clearly proved that observed metabolite changes are related to deuterium enrichment and the measured signal time courses are suitable for quantitative modeling of metabolite kinetics. The repeated session after 1H-Glc ingestion demonstrated that metabolite changes were not related to instability in the spectral quality or the metabolic effects of hyperglycemia.
Increasing concentration of 2H-Glc in the brain, reproducibly measured with both QELT and DMI, is likely caused by continuous substrate supply during mild hyperglycemia, which resembles the glucose excursion of an oral glucose tolerance test. Our 2H-Glc and Glc6 brain time-courses differ from the time-courses measured previously with DMI, due to different routes of administration of the 2H-Glc i.e. oral vs. intravenous bolus.29 The negligible blood Glc enrichment with deuterium does not contribute to the 2H-Glc delivery and transport to the brain and therefore brain 2H-Glc levels reflect only the conversion of the 2H-Glc to the downstream metabolites. Thus, directly-measured brain 2H-Glc decreased shortly after initiation of the scans in the previous intravenous experiment, whereas indirectly and directly-measured 2H-Glc grows throughout our scans. The same effects of both intravenous and oral administration of 1-13C-Glc on isotopic brain Glc levels were reported.49 Despite MR methods are not able to distinguish compartmental origin of Glc signals, dominant contribution comes from the extracellular compartment, since there is ~3 order lower concentration intracellularly due to rapid Glc metabolism within brain cells.54 Even though the calculation of metabolic fluxes is rather complex due to the non-steady-state of blood 2H-Glc levels this can be solved in future studies once the blood 2H-Glc enrichment is known. Another study utilized 13C-Glu and 13C-Gln time-courses measured after oral administration of 13C-Glc to calculate rates of TCA and Gln synthesis (VTCA and Vgln).24 Thus, future QELT studies will benefit from blood sampling to measure blood glucose deuterium enrichment as a function of time and quantitative compartmental modeling approaches.55
The practicality of non-invasive peroral tracer administration in our study, which is more comfortable for patients, highlights the clinical feasibility of our approach.24 Despite the delay in the blood enrichment with the labeled substrate, our between-subject variance in Glu4 decay constants of 29% corroborates previous work in which oral administration of 13C-Glc provided similar results compared to an intravenous clamp, albeit with lower precision24 estimating VTCA with between-subject CVs (n=4) of 62% and 23% for oral and intravenous administration, respectively. The somatostatin-free protocol could contribute to a higher variance of the modeling outcomes compared to 2H-Glc works with somatostatin infusion (frequently used in pancreatic clamps)24 that blocks endogenous insulin/glucagon release. The use of clamp protocols with continuous tracer application in future studies could thus further improve reproducibility and/or shorten the acquisition window due to longer periods near the “steady-state” 2H-Glc blood enrichment and might along with higher fields above 7T further improve detection of Gln4 and GABA2.
While the measurements of deuterated Glc and Glx with DMI in an animal experiment allowed to calculate CMRGlc and VTCA,29 QELT also concomitantly quantifies Glc and separates Glu and Gln. Thus, QELT can estimate Glu/Gln cycling in combination with measurements of Glc turnover, which is challenging with 13C methods due to overlapping signals of Glc with residual water signal and high chemical shift between Glc and Glu/Gln.29 Recent 13C study demonstrated substantial improvement of reproducibility of Gln time-courses individual-subject level by prolonged acquisitions of 3h that minimizes scatter of delayed Gln4 increase24, while others suggested “snapshot” acquisition during 30–60 and 120–150 minutes with a one hour break.56
While 7T MR scanners have been recently approved for clinical use, they are still not as widely available as lower field 3T scanners.18,57 Advancements of 3T SV-M that detected subtle 3% Glu and ~28% lactate responses to visual stimulation and 1H-MRSI,58 which allowed whole-brain Glu mapping within 4 minutes57 promise implementation of the indirect QELT-MRS(I) at 3T despite the lower spectral and reduced spatial resolution.
Non-invasive quantification and imaging of metabolite changes relevant to the Glc metabolism and neurotransmitter synthesis (Glu/Gln, GABA) are crucial for the understanding of many brain disorders. Here, we have demonstrated the capabilities of affordable QELT-MRS(I) approach for metabolic measures in humans that utilizes widely available 1H-MR scanner equipment and time-resolved whole-brain mapping that allows to indirectly detect deuterated compounds also in a clinical setting. The negligible risk associated with deuterium administration compared to radioactive ([18F]FDG predetermines the methodology, especially for studies with a multi-session longitudinal design to track treatment effects or disease progression over time. In contrast to direct 2H-MRS, QELT-MRS allows the quantification of an extended neurochemical profile, including non-deuterated compounds. Thus, the current methodology provides a critical step forward for future metabolic projects in the resting or activated human brain in disease, health, and aging, which will offer relevant metabolic information using widely available hardware in a single MR session.
Methods
Study design
Five healthy, right-handed volunteers (30±4 y.o., 4 males and 1 female) were scanned on a 7T whole-body MR scanner (Siemens Healthcare, Erlangen, Germany) utilizing a commercially available 32-channel receive-array coil (Nova Medical, Wilmington, MA, USA) for 1H-MRS and a dual-tuned (2H/1H) quadrature transmit-receive birdcage coil specifically developed for the project (Stark Contrast MRI Coils Research, Germany) for 2H-MRSI acquisitions. All participants were lean (BMI = 22.6±1.4 kg/m2) without a history of diabetes or other metabolic and severe diseases. Blood glucose measurements were performed with a standard strip glucometer in triplicates after capillary blood sampling from the leg of the volunteers. The study was approved by the Ethical Commission at the Medical University of Vienna. All participants signed informed consent. Each participant underwent four MR scans in total:
The 1st session (“test” session) utilized an interleaved single-voxel 1H-MRS/1H-MRSI (QELT) protocol performed after oral administration of 2H-Glc.
The 2nd session (“retest” session) performed identically to the 1st session to assess test-retest repeatability.
The 3rd session (“control” session) performed the same interleaved single-voxel 1H-MRS/1H-MRSI (QELT) protocol, but after oral intake of non-deuterated D-Glc (normal dextrose, 1H-Glc) as a control condition.
The 4th session (“reproducibility” session) acquired direct 2H-MRSI dynamically using the dual-tuned head coil following oral administration of 2H-Glc to directly compare the results with those obtained in the 1st session (i.e., QELT).
One subject was additionally scanned after 2H-Glc administration with a 1H-MRSI-only protocol to obtain the time-course with high temporal resolution. All sessions were conducted in the morning after an overnight fast. Both compounds (2H-Glc and 1H-Glc) were dissolved in ~300 mL of water and ingested in equal amounts (0.8 g/kg body weight) immediately before the scan was initiated. MRSI and MRS data were interleaved with navigator images obtained prior to each 1H-MRSI/1H-MRS or 2H-MRSI block to assure the stable position of the localized volume. The navigator images were registered to the atlas space with Autoalign methodology implemented on the MR scanner.59 The coordinates of the MRS volume of interests (MRS-VOI) were later used in the second MR session for consistent MRS-VOI placement. The first MRS/MRSI block was acquired after calibrating MRS radio-frequency pulses and B0-shimming within 30 minutes after 2H-Glc/1H-Glc administration. The following MRS/MRSI blocks were obtained after the acquisition of the T1-weighted MP2RAGE image with an isotropic resolution of 1.1×1.1×1.1 mm3; TR, 3900 ms; TE, 2.8 ms; flip angle, 4°/5°; and acquisition time 3:52 min. Our goal was to cover a time window of 120 minutes following the ingestion of the tracer, assuming slower dynamics of deuterium enrichment in the human brain compared to that of rats.4
MRI and 1H/2H MRS acquisitions
Standard 2nd-order B0-shimming was performed via an imaging-based approach (two iterations) and FASTMAP60 for MRSI and MRS acquisition, respectively. Multi-voxel (MRSI) and single-voxel (SV-MRS) acquisitions were interleaved except for one study session, where only the MRSI data were acquired with a high time resolution of five minutes after 2H-Glc ingestion.
1H-MRSI data were obtained via an FID-MRSI sequence19 with an ultra-short acquisition delay of 1.3 ms, a short TR of 320 ms, and ellipsoidal 3D k-space encoding using concentric ring trajectories (CRT), variable temporal interleaves, 36×36×26 matrix, 5×5×4.8 mm3 voxel size, 2:58 min per block, 558 complex points, 34° (Ernst) excitation flip angle, 600 μs pulse duration, and 7 kHz pulse bandwidth. The excited 40 mm-thick slab was centered around the posterior cingulate region.
2H-MRSI data were acquired using a phase-encoded non-selective FID-MRSI sequence with an acquisition delay of 1.5 ms after a 500μs block excitation pulse adjusted to a 86° (Ernst) flip angle and a TR of 290ms. An ellipsoidal k-space with a matrix size of 16×16×14 was acquired with two weighted averages (and retrospective Hamming filtering), with 12.5×12.5×12.5 mm3 voxel size, a vector size of 256, and a spectral bandwidth of 500Hz in 6:37 min per block.
Single-voxel MRS data were obtained from the posterior cingulate (PCC) region using a semi-LASER sequence (TR 7 s, TE 28 ms, 3:43 min per block, 2048 complex points, 90° asymmetric sinc pulse with a duration of 2.5 ms, FOCI pulse bandwidth, and a duration of 4.2 ms).61 A 22×20×20 mm (AP×LR×SI) voxel was placed mid-sagittally, based on anatomical landmarks. The voxel was rotated in the sagittal plane by 30° such that it was aligned with the posterior border of the splenium. To mitigate possible effects of patient motion and chemical shift displacement, the voxel was backed away anteriorly from the splenium and caudally from the occipital-parietal fissure by 2 mm.62 All spectra were collected with water suppression63 and outer volume suppression (number of excitations, NEX = 32) along with unsuppressed water spectra utilized to remove residual eddy currents (NEX = 2) and as a reference from which to derive metabolite concentration estimates (NEX = 2).
Segmentation of MPRAGE scans
The T1w-MRI images were segmented in Freesurfer (v.5.3) to obtain masks of the brain gray matter (GM) and white matter (WM). The masks were resampled to the MRSI space and used to obtain tissue-specific averages of metabolite levels. In addition, probabilistic maps of the GM, WM, and cerebrospinal fluid (CSF) were derived by segmenting the T1w-MRI images using the SPM12 software package. The probabilistic tissue maps were thresholded with an in-house-written MATLAB script using the iterative method of threshold selection64 to determine the within-PCC-VOI fraction of GM, WM, and CSF.
Processing of MRSI
MRSI data were reconstructed offline with an in-house-developed software pipeline consisting of MATLAB (R2013a, MathWorks, Natick, MA, USA), BASH (v4.2.25, Free Software Foundation, Boston, MA, USA), and MINC (MINC tools, v2.0, McConnell Brain Imaging Center, Montreal, QC, Canada). Data processing included an iMUSICAL coil combination,65,66 water normalization,67 k-space reconstruction with in-plane convolution gridding, spatial Hamming filtering, channel-wise noise-decorrelation, and off-resonance correction.40 Due to the short TR, obtaining metabolite concentration estimates would require strong assumptions about T1 relaxation. Therefore, the metabolite concentrations were instead quantified in institutional units using LCModel (v6.3–1, LCModel Inc, ONT, CA) with a basis set that included 17 simulated brain metabolites and a measured macromolecular background (detailed below).68 Data processing of 2H-MRSI data was simpler, involving only spatial Hamming filtering prior Fourier transform.
Processing of SV-MRS
Single shots were corrected for small frequency and phase fluctuations, and residual eddy currents and 32 single shots were summed per block. The frequency and phase of the spectra obtained for each block were aligned to each other within the scanning session and quantified in LCModel. The metabolite concentrations were corrected for variable water content in the gray and white matter, as well as for the within-voxel CSF fraction.69
Finally, the spectra measured in session #1 representing the first time-points (FIRST) and the last time-points (LAST) were pooled together and summed, resulting in two sums (FIRST and LAST). The FIRST and LAST from the 2H-Glc session were subtracted, and a difference spectrum was calculated. The difference spectrum characterized metabolite changes following 2H-Glc ingestion. The difference spectra were quantified in LCModel utilizing the basis sets that contained only the metabolites enriched with deuterium (detailed below) to estimate quantification errors, i.e., relative CRLB.27
Quantification of MR spectra
Basis sets previously routinely used to quantify 1H-MRS/MRSI data were modified to reflect the fact that deuterium is incorporated at certain carbon positions in the molecules. Modifications to the basis sets were performed based on the previous animal experiments, theoretical predictions, and preliminary analysis of the current data. Glu, Gln, and GABA included the split of proton signals that originated from different carbon positions for C2, C3, and C4. Thus, the basis set included all molecular variants that occurred in the brain in a dynamically changing ratio and took into account the homonuclear (1H-1H) and heteronuclear (1H-2H) coupling constants. For instance, the basis set of glutamate included six components, i.e., those present in the molecule with all positions occupied with protons (C21H, C31H2, C41H2), in the molecule with one deuteron on the C4 (C21H, C31H2, C41H2H), and two deuterons on C4 (C21H, C31H2). Thus, we distinguished three variants of the C3 resonance affected by homo- and heteronuclear coupling with 1H and 2H at C4. While the couplings between protons and deuterons on the C3 and C4 (Glu and Gln) and C2 and C3 (GABA) were simulated, the couplings between C2 and C4 were minimal and were neglected. Yet, the number of components had to be reduced for MRS and MRSI data to preserve the stability of the fits. This was performed by neglecting the heteronuclear and homonuclear coupling effects of the deuteration, assuming that the dominant signal change occurred on the position where proton(s) is/are replaced by deuteron(s), i.e., C4 for Glu and Gln and C2 for GABA. Thus, we used only two components per metabolite (Glu2+3, Glu4; Gln2+3, Gln4; and GABA2, GABA3+4) to quantify SV-MRS data. The basis set was further simplified for MRSI, where we split only Glu2+3 and Glu4 resonances since the fitting with more components yielded a less stable quantification of neurochemical profiles. Finally, we used the components whose signals were expected to change due to progressive deuteration during the scans (i.e., for Glu and Gln: C41H2, C41H2H, and two variants of C31H2 present in the molecule with one and two deuterons; and for GABA: C21H2, C21H2H, and two variants C31H2) to quantify the difference spectra. As difference spectra do not contain the background of signals from the metabolites that were stable over the task, we could account for the subtle hetero- and homonuclear coupling effects. The 2H-labeled metabolite concentrations, acquired directly with 2H-MRSI, were quantified using a basis set containing only simulated 2H-Glx, 2H-Glc, 2H-Lac, and 2H-water resonances.
Quality control of MRS(I) data
Voxels with sufficient temporal stability and spectral quality were selected using masks. The masks included voxels with CVs below 12% for the three main metabolites that remained stable during the scan, i.e., tCr, tCho, and tNAA. The criteria also utilized parameters provided by LCModel (FWHM<0.1 ppm, SNR>5, zero-order phase <40°) in line with expert recommendations.70 These criteria were utilized to calculate regional GM and WM means in metabolite concentrations, as well as to select the spectra used for the calculation of high-SNR sums that represented either GM or WM. The metabolite concentrations quantified with all CRLBs were used for further analysis except those that could not be quantified with CRLBs of 999%. This criterion avoided bias due to an arbitrarily set CRLB threshold, cutting off lower concentrations with higher relative CRLB.71 The metabolites quantified with a CRLB of 999% in most of the time-points (MRS), or consistently in more than 10% of voxels (MRSI), were not analyzed.72
Statistical analysis
Differences in metabolite concentrations were calculated between the first and last time-point, and their significance was tested with the standard paired t-test separately within both sessions (2H-Glc, 1H-Glc). Time-dependent decay in the Glu4 concentration after 2H-Glc administration was fitted with an exponential function M=M0(−t/tau)+c per subject. The coefficient of variations in the time constants of the decay (tau) were calculated per session to express the between-subject variation of the metabolic rates between subjects in the region of interest (PCC, GM, WM). The concentrations of Gln4 (both sessions) and Glu4 (1H-Glc) were instead fitted with linear functions (Y=slopes*t+a).
Supplementary Material
Acknowledgements
We thank P. Bolan of the Center for Magnetic Resonance Research, University of Minnesota, and C. Rogers, University of Cambridge, for providing a tool to store and apply 7 T B0-shims for the 7 T MR scanner; V. Mlynarik for helpful discussions; and the study participants whose help is greatly appreciated. P.B. was supported by the European Union’s Horizon 2020 research and innovation programme under a Marie Skłodowska-Curie grant (agreement no. 846793), and by a NARSAD Young Investigator Grant from the Brain and Behavior Research Foundation (no. 27238). A.S. received funding from the European Union’s Horizon 2020 research and from an innovation programme under a Marie Skłodowska-Curie grant (agreement no. 794986). The authors acknowledge support from the Austrian Science Fund (FWF) (grants P 30701 and KLI 718 to W.B., I 6037 to B.S., KLI 782 to T.S., and KLI 646 to G.H.). W.B. acknowledges the support of the following NIH grant: R01EB031787. D.K.D. acknowledges support from the following National Institutes of Health grants: BTRC P41 EB027061 and P30 NS076408
Competing interests
R. Lanzenberger received travel grants and/or conference speaker honoraria within the last three years from Bruker BioSpin MR andHeel, and has served as a consultant for Ono Pharmaceutical. He received investigator-initiated research funding from Siemens Healthcare regarding clinical research using PET/MR. He is a shareholder of the start-up company BM Health GmbH since 2019.
The other authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Footnotes
Code Availability
The custom code for time course analysis using linear and exponential fits was performed using custom-made python code (version 3.10) available at https://github.com/MRSI-HFMR-Group Vienna/DeuteriumToProtonExchangeMRS.
Data Availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. Source data for Figs. 1, 4, 5 and for Supplementary Figs. 1,2,3 and 5 are provided with this paper. The raw data acquired for the study are too large to be publicly shared, yet they are available for research purposes from the corresponding authors on reasonable request. The data generated by postprocessing methods (i.e., metabolite maps, MR spectra, and outcomes of their quantification in LCModel) are available at https://doi.org/10.5281/zenodo.5705959. The shared data are in the minc, niifti, and MRSpa data formats. A priory information (“the basis sets”) needed for MRS/MRSI data quantification in LCModel is also available.
References
- 1.Kim M et al. What do we know about dynamic glucose-enhanced (DGE) MRI and how close is it to the clinics? Horizon 2020 GLINT consortium report. Magn. Reson. Mater. Physics, Biol. Med 35, 87–104 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Graaf RA, Mason GF, Patel AB, Behar KL & Rothman DL In vivo 1H-[13 C]-NMR spectroscopy of cerebral metabolism. NMR Biomed. 16, 339–357 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Ruhm L et al. Deuterium metabolic imaging in the human brain at 9.4 Tesla with high spatial and temporal resolution. Neuroimage (2021) doi: 10.1016/j.neuroimage.2021.118639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rich LJ et al. 1H magnetic resonance spectroscopy of 2H-to-1H exchange quantifies the dynamics of cellular metabolism in vivo. Nat. Biomed. Eng 4, 335–342 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Zijl PCM & Brindle KM Spectroscopic measurements of metabolic fluxes. Nat. Biomed. Eng 4, 254–256 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Zhu X-H, Lu M & Chen W Quantitative imaging of brain energy metabolisms and neuroenergetics using in vivo X-nuclear 2H, 17O and 31P MRS at ultra-high field. J. Magn. Reson 292, 155–170 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koppenol WH, Bounds PL & Dang CV Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Cancer 11, 325–337 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Norat P et al. Mitochondrial dysfunction in neurological disorders: Exploring mitochondrial transplantation. NPJ Regen, Med (2020) doi: 10.1038/s41536-020-00107-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manji H et al. Impaired mitochondrial function in psychiatric disorders. Nat. Rev. Neurosci (2012) doi: 10.1038/nrn3229. [DOI] [PubMed] [Google Scholar]
- 10.Hahn A et al. Quantification of Task-Specific Glucose Metabolism with Constant Infusion of 18F-FDG. J. Nucl. Med 57, 1933–1940 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Rischka L et al. Reliability of task-specific neuronal activation assessed with functional PET, ASL and BOLD imaging. J. Cereb. Blood Flow Metab (2021) doi: 10.1177/0271678X211020589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hesketh RL et al. Magnetic Resonance Imaging Is More Sensitive Than PET for Detecting Treatment-Induced Cell Death–Dependent Changes in Glycolysis. Cancer Res. 79, 3557–3569 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rothman DL et al. Methods | 13C MRS Measurements of in Vivo Rates of the Glutamate/Glutamine and GABA/Glutamine Neurotransmitter Cycles. in Encyclopedia of Biological Chemistry III 688–700 (Elsevier, 2021). doi: 10.1016/B978-0-12-819460-7.00341-8. [DOI] [Google Scholar]
- 14.Shulman RG, Rothman DL, Behar KL & Hyder F Energetic basis of brain activity: Implications for neuroimaging. Trends Neurosci. 27, 489–495 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Wang ZJ et al. Hyperpolarized 13C MRI: State of the art and future directions. Radiology (2019) doi: 10.1148/radiol.2019182391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rischka L et al. Reduced task durations in functional PET imaging with [18F]FDG approaching that of functional MRI. Neuroimage 181, 323–330 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Stiernman LJ et al. Dissociations between glucose metabolism and blood oxygenation in the human default mode network revealed by simultaneous PET-fMRI. Proc. Natl. Acad. Sci. U. S. A (2021) doi: 10.1073/pnas.2021913118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terpstra M et al. Test-retest reproducibility of neurochemical profiles with short-echo, single-voxel MR spectroscopy at 3T and 7T. Magn. Reson. Med, 76, 1083–1091 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hingerl L et al. Clinical High-Resolution 3D-MR Spectroscopic Imaging of the Human Brain at 7 T. Invest. Radiol 55, 239–248 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Bednarik P et al. Neurochemical and BOLD responses during neuronal activation measured in the human visual cortex at 7 Tesla. J, Cereb, Blood Flow Metab 35, 601–610 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seuwen A, Schroeter A, Grandjean J, Schlegel F & Rudin M Functional spectroscopic imaging reveals specificity of glutamate response in mouse brain to peripheral sensory stimulation. Sci. Rep 9, 10563 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scherer T, Sakamoto K & Buettner C Brain insulin signalling in metabolic homeostasis and disease. Nature Reviews Endocrinology (2021) doi: 10.1038/s41574-021-00498-x. [DOI] [PubMed] [Google Scholar]
- 23.Gruetter R et al. Localized in vivo 13C NMR spectroscopy of the brain. NMR Biomed. 16, 313–338 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mason GF et al. A comparison of 13C NMR measurements of the rates of glutamine synthesis and the tricarboxylic acid cycle during oral and intravenous administration of [1–13C]glucose. Brain Res. Protoc 10, 181–190 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Iozzo P & Guzzardi MA Imaging of brain glucose uptake by PET in obesity and cognitive dysfunction: Life-course perspective. Endocr. Connect 8, R169–R183 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuehn BM In Alzheimer Research, Glucose Metabolism Moves to Center Stage. JAMA 323, 297 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Bednařík P et al. Neurochemical responses to chromatic and achromatic stimuli in the human visual cortex. J. Cereb. Blood Flow Metab 38, 347–359 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Feyter HM et al. Deuterium metabolic imaging (DMI) for MRI-based 3D mapping of metabolism in vivo. Sci. Adv 4, eaat7314 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu M, Zhu X-H, Zhang Y, Mateescu G & Chen W Quantitative assessment of brain glucose metabolic rates using in vivo deuterium magnetic resonance spectroscopy. J. Cereb. Blood Flow Metab 37, 3518–3530 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tiwari V, An Z, Wang Y & Choi C Distinction of the GABA 2.29 ppm resonance using triple refocusing at 3 T in vivo. Magn. Reson. Med 80, 1307–1319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Graaf RA, Thomas MA, Behar KL & De Feyter HM Characterization of Kinetic Isotope Effects and Label Loss in Deuterium-Based Isotopic Labeling Studies. ACS Chem. Neurosci 12, 234–243 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veltien A et al. Simultaneous Recording of the Uptake and Conversion of Glucose and Choline in Tumors by Deuterium Metabolic Imaging. Cancers (Basel). 13, 4034 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plecko B et al. Oral β-Hydroxybutyrate Supplementation in Two Patients with Hyperinsulinemic Hypoglycemia: Monitoring of β-Hydroxybutyrate Levels in Blood and Cerebrospinal Fluid, and in the Brain by In Vivo Magnetic Resonance Spectroscopy. Pediatr. Res (2002) doi: 10.1203/00006450-200208000-00025. [DOI] [PubMed] [Google Scholar]
- 34.Scafidi S, Jernberg J, Fiskum G & McKenna MC Metabolism of Exogenous [2,4–13C]β-Hydroxybutyrate following Traumatic Brain Injury in 21–22-Day-Old Rats: An Ex Vivo NMR Study. Metabolites 12, 710 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Craft S et al. The ketogenic diet as a potential prevention or therapeutic strategy for AD. Alzheimer’s Dement. (2020) doi: 10.1002/alz.038148. [DOI] [Google Scholar]
- 36.Wright JN, Saneto RP & Friedman SD Hydroxybutyrate detection with proton MR spectroscopy in children with drug-resistant epilepsy on the ketogenic diet. Am. J. Neuroradiol 39, 1336–1340 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lebon V et al. Astroglial contribution to brain energy metabolism in humans revealed by 13C nuclear magnetic resonance spectroscopy: Elucidation of the dominant pathway for neurotransmitter glutamate repletion and measurement of astrocytic oxidative metabolism. J. Neurosci (2002) doi: 10.1523/jneurosci.22-05-01523.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ross JM et al. High brain lactate is a hallmark of aging and caused by a shift in the lactate dehydrogenase A/B ratio. Proc. Natl. Acad. Sci. U. S. A (2010) doi: 10.1073/pnas.1008189107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liguori C et al. CSF lactate levels, τ proteins, cognitive decline: A dynamic relationship in Alzheimer’s disease. J. Neurol. Neurosurg. Psychiatry (2015) doi: 10.1136/jnnp-2014-308577. [DOI] [PubMed] [Google Scholar]
- 40.Hingerl L et al. Density-weighted concentric circle trajectories for high resolution brain magnetic resonance spectroscopic imaging at 7T. Magn. Reson. Med (2018) doi: 10.1002/mrm.26987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cember ATJ et al. Integrating 1H MRS and deuterium labeled glucose for mapping the dynamics of neural metabolism in humans. Neuroimage 251, 118977 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maudsley AA et al. Advanced magnetic resonance spectroscopic neuroimaging: Experts’ consensus recommendations. NMR Biomed. (2021) doi: 10.1002/nbm.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson M et al. Methodological consensus on clinical proton MRS of the brain: Review and recommendations. Magn. Reson. Med 82, 527–550 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hyder F, Fulbright RK, Shulman RG & Rothman DL Glutamatergic function in the resting awake human brain is supported by uniformly high oxidative energy. J. Cereb. Blood Flow Metab 33, 339–47 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hyder F & Rothman DL Quantitative fMRI and oxidative neuroenergetics. NeuroImage (2012) doi: 10.1016/j.neuroimage.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu Y, Herman P, Rothman DL, Agarwal D & Hyder F Evaluating the gray and white matter energy budgets of human brain function. J. Cereb. Blood Flow Metab (2018) doi: 10.1177/0271678X17708691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pan JW et al. Spectroscopic imaging of glutamate C4 turnover in human brain. Magn. Reson. Med (2000) doi:. [DOI] [PubMed] [Google Scholar]
- 48.de Graaf RA, Mason GF, Patel AB, Behar KL & Rothman DL In vivo 1H-[13C]-NMR spectroscopy of cerebral metabolism. NMR Biomed. 16, 339–357 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Moreno A, Blüml S, Hwang JH & Ross BD Alternative 1–13C glucose infusion protocols for clinical 13C MRS examinations of the brain. Magn. Reson. Med (2001) doi: 10.1002/mrm.1158. [DOI] [PubMed] [Google Scholar]
- 50.Sundar LKS et al. Towards quantitative [18F]FDG-PET/MRI of the brain: Automated MR-driven calculation of an image-derived input function for the non-invasive determination of cerebral glucose metabolic rates. J. Cereb. Blood Flow Metab 39, 1516–1530 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shiyam Sundar LK et al. Fully Integrated PET/MR Imaging for the Assessment of the Relationship Between Functional Connectivity and Glucose Metabolic Rate. Front. Neurosci 14, 252 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andronesi OC et al. Motion correction methods for MRS: experts’ consensus recommendations. NMR Biomed. 34, e4364 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dikaios N, Arridge S, Hamy V, Punwani S & Atkinson D Direct parametric reconstruction from undersampled (k, t)-space data in dynamic contrast enhanced MRI. Med. Image Anal (2014) doi: 10.1016/j.media.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 54.Knutsson L, Xu X, van Zijl PCM & Chan KWY Imaging of sugar‐based contrast agents using their hydroxyl proton exchange properties. NMR Biomed. 1–25 (2022) doi: 10.1002/nbm.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mason GF et al. Simultaneous determination of the rates of the TCA cycle, glucose utilization, α-ketoglutarate/glutamate exchange, and glutamine synthesis in human brain by NMR. J. Cereb. Blood Flow Metab (1995) doi: 10.1038/jcbfm.1995.2. [DOI] [PubMed] [Google Scholar]
- 56.Ross B, Lin A, Harris K, Bhattacharya P & Schweinsburg B Clinical experience with 13C MRS in vivo. NMR Biomed. 16, 358–369 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Moser P et al. Intra‐session and inter‐subject variability of 3D‐FID‐MRSI using single‐echo volumetric EPI navigators at 3T. Magn. Reson. Med 83, 1920–1929 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dinuzzo M et al. Perception is associated with the brain’s metabolic response to sensory stimulation. Elife (2022) doi: 10.7554/ELIFE.71016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dou W et al. Automatic voxel positioning for MRS at 7 T. Magn. Reson. Mater. Physics, Biol. Med (2015) doi: 10.1007/s10334-014-0469-9. [DOI] [Google Scholar]
- 60.Gruetter R & Tkac I Field mapping without reference scan using asymmetric echo-planar techniques. Magn. Reson. Med 43, 319–323 (2000). [DOI] [PubMed] [Google Scholar]
- 61.Oz G & Tkac I Short-Echo, Single-Shot, Full-Intensity Proton Magnetic Resonance Spectroscopy for Neurochemical Profiling at 4 T: Validation in the Cerebellum and Brainstem. Magn. Reson. Med 65, 901–910 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bednarik P et al. Effect of Ketamine on Human Neurochemistry in Posterior Cingulate Cortex: A Pilot Magnetic Resonance Spectroscopy Study at 3 Tesla. Front. Neurosci (2021) doi: 10.3389/fnins.2021.609485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tkac I, Starcuk Z, Choi IY & Gruetter R In vivo H-1 NMR spectroscopy of rat brain at 1 ms echo time. Magn. Reson. Med 41, 649–656 (1999). [DOI] [PubMed] [Google Scholar]
- 64.Ridler TW & Calvard S Picture Thresholding Using an Iterative Selection Method. Ieee Trans. Syst. Man Cybern 8, 630–632 (1978). [Google Scholar]
- 65.Strasser B et al. Coil combination of multichannel MRSI data at 7 T: MUSICAL. NMR Biomed. 26, 1796–1805 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moser P et al. Non-Cartesian GRAPPA and coil combination using interleaved calibration data – application to concentric-ring MRSI of the human brain at 7T. Magn. Reson. Med (2019) doi: 10.1002/mrm.27822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maudsley AA et al. Mapping of brain metabolite distributions by volumetric proton MR spectroscopic imaging (MRSI). Magn. Reson. Med (2009) doi: 10.1002/mrm.21875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Považan M et al. Mapping of brain macromolecules and their use for spectral processing of 1 H-MRSI data with an ultra-short acquisition delay at 7 T. Neuroimage 121, 126–135 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Gröhn H et al. Influence of Repetitive Transcranial Magnetic Stimulation on Human Neurochemistry and Functional Connectivity: A Pilot MRI/MRS Study at 7 T. Front. Neurosci 13, 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oz G et al. Clinical Proton MR Spectroscopy in Central Nervous System Disorders. Radiology 270, 658–679 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kreis R The trouble with quality filtering based on relative Cramer-Rao lower bounds. Magn. Reson. Med 75, 15–18 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Hangel G et al. Inter-subject stability and regional concentration estimates of 3D-FID-MRSI in the human brain at 7 T. NMR Biomed. (2021) doi: 10.1002/nbm.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The main data supporting the results in this study are available within the paper and its Supplementary Information. Source data for Figs. 1, 4, 5 and for Supplementary Figs. 1,2,3 and 5 are provided with this paper. The raw data acquired for the study are too large to be publicly shared, yet they are available for research purposes from the corresponding authors on reasonable request. The data generated by postprocessing methods (i.e., metabolite maps, MR spectra, and outcomes of their quantification in LCModel) are available at https://doi.org/10.5281/zenodo.5705959. The shared data are in the minc, niifti, and MRSpa data formats. A priory information (“the basis sets”) needed for MRS/MRSI data quantification in LCModel is also available.


