Abstract
The ceroid lipofuscinosis neuronal 1 (CLN1) disease, formerly called infantile neuronal ceroid lipofuscinosis, is a fatal hereditary neurodegenerative lysosomal storage disorder. This disease is caused by loss-of-function mutations in the CLN1 gene, encoding palmitoyl-protein thioesterase-1 (PPT1). PPT1 catalyzes depalmitoylation of S-palmitoylated proteins for degradation and clearance by lysosomal hydrolases. Numerous proteins, especially in the brain, require dynamic S-palmitoylation (palmitoylation-depalmitoylation cycles) for endosomal trafficking to their destination. While 23 palmitoyl-acyl transferases in the mammalian genome catalyze S-palmitoylation, depalmitoylation is catalyzed by thioesterases such as PPT1. Despite these discoveries, the pathogenic mechanism of CLN1 disease has remained elusive. Here, we report that in the brain of Cln1−/− mice, which mimic CLN1 disease, the mechanistic target of rapamycin complex-1 (mTORC1) kinase is hyperactivated. The activation of mTORC1 by nutrients requires its anchorage to lysosomal limiting membrane by Rag GTPases and Ragulator complex. These proteins form the lysosomal nutrient sensing scaffold to which mTORC1 must attach to activate. We found that in Cln1−/− mice, two constituent proteins of the Ragulator complex (vacuolar (H+)-ATPase and Lamtor1) require dynamic S-palmitoylation for endosomal trafficking to the lysosomal limiting membrane. Intriguingly, Ppt1 deficiency in Cln1−/− mice misrouted these proteins to the plasma membrane disrupting the lysosomal nutrient sensing scaffold. Despite this defect, mTORC1 was hyperactivated via the IGF1/PI3K/Akt-signaling pathway, which suppressed autophagy contributing to neuropathology. Importantly, pharmacological inhibition of PI3K/Akt suppressed mTORC1 activation, restored autophagy, and ameliorated neurodegeneration in Cln1−/− mice. Our findings reveal a previously unrecognized role of Cln1/Ppt1 in regulating mTORC1 activation and suggest that IGF1/PI3K/Akt may be a targetable pathway for CLN1 disease.
Keywords: lysosomal storage disease, neurodegeneration, CLN1 disease, S-palmitoylation, IGF1 signaling, mTORC1 activation
Dysregulated signaling pathway of the mechanistic (or mammalian) target of rapamycin complex 1 (mTORC1) kinase has been linked to the development of human diseases, including cancer, diabetes, and neurodegeneration (1). In a group of >60 inherited metabolic diseases called lysosomal storage disorders (LSDs) (2), neurodegeneration is a devastating manifestation (3, 4). The lysosome (5) is the dynamic regulator of function of many proteins (6), especially in the brain.
Impaired lysosomal function underlies pathogenesis in the majority of the LSDs (2). Neuronal ceroid lipofuscinoses (NCLs) (7, 8, 9, 10, 11) commonly known as Batten disease (12), constitute a group of the most common neurodegenerative LSDs mostly affecting children. Mutations in at least 14 different genes ceroid lipofuscinosis neuronal(CLN1–CLN14) (13) underlie various forms of NCLs. The CLN1 disease, formerly called infantile neuronal ceroid lipofuscinosis (INCL) (14), is a devastating neurodegenerative LSD, caused by inactivating mutations in the CLN1 gene (15). Despite this discovery, the mechanism of pathogenesis underlying CLN1 disease has remained elusive for more than 2 decades. Children with CLN1 disease are phenotypically normal at birth, but by 6 to 18 months of age they manifest psychomotor retardation. Around 2 years of age, these children are completely blind due to retinal degeneration and develop frequent seizures. MRI of the brain shows marked cortical atrophy. At around 4 years of age, an isoelectric EEG attests to a vegetative state. They remain in this condition for several more years before eventual death (16, 17). These grim facts underscore an urgent need for understanding the mechanism of pathogenesis of CLN1 disease, which may facilitate the development of curative therapies.
The CLN1 gene encodes palmitoyl-protein thioesterase-1 (PPT1) (18) which catalyzes the removal of palmitate from S-palmitoylated proteins (constituents of ceroid lipofuscin) preventing their accumulation in lysosomes. Although PPT1 was originally reported to be localized in the lysosome (19, 20), recently, extra lysosomal presence of catalytically active PPT1 has been clearly demonstrated (21). Numerous proteins in the brain undergo S-palmitoylation (also called S-acylation), a posttranslational modification in which a saturated fatty acid (generally palmitate) is attached to specific cysteine residues in polypeptides via thioester linkage (22, 23). S-palmitoylation confers hydrophobicity to proteins, increasing their membrane-affinity and promoting protein–protein interactions (22, 24, 25). Dynamic S-palmitoylation (palmitoylation-depalmitoylation) (26) facilitates endosomal trafficking of many proteins for their trafficking to the appropriate cellular destinations (24). While S-palmitoylation is catalyzed by 23 palmitoyl acyltransferases (called ZDHHC-PATs or simply ZDHHCs) (22, 24), depalmitoylation is mediated by thioesterases (25). Recently, in a detailed unbiased proteomics study >100 novel proteins, suggested to be PPT1 substrates, have been identified (27). Although the physiological functions of these proteins in the context of CLN1 disease remain largely unclear, the loss-of function mutations of the Cln1 gene may disrupt the functions of at least some of those genes and activate various pathways of mTORC1 activation contributing to pathogenesis.
The lysosome has been suggested to play critical roles in multiple cellular functions, including signaling in response to environmental cues (28, 29). Emerging evidence has remarkably transformed our understanding of the lysosome from a terminal degradative organelle to a critical signaling hub for fundamental metabolic processes (30). Sensing essential nutrients is an important function of the lysosome to coordinate cellular metabolism and growth (31). Signals from nutrients such as glucose, amino acids, fatty acids, and cholesterol are integrated by the lysosome, turning the cellular events from anabolic to catabolic processes like autophagy (3, 32, 33). Whereas materials from extracellular sources are transported to the lysosome via endocytosis, those originating from intracellular sources are delivered by autophagy (32). Notably, activation of the mTORC1, situated at the crossroads of nutrient signaling (34), suppresses autophagy (35). The loss of autophagy in the central nervous system has been reported to cause neurodegeneration (36). There are three types of autophagy: macroautophagy, microautophagy, and chaperone-mediated autophagy in all of which the lysosome plays a pivotal role in degrading the cargo contained in the autophagosomes (37). Indeed, dysregulation of autophagy has been implicated not only in the pathogenesis of common neurodegenerative diseases like Alzheimer’s and Parkinson’s (38, 39) but also in most LSDs in which neurodegeneration is a devastating manifestation (3, 4).
In the present study, we used tissues from the cerebral cortex of the brain of Cln1−/− mice (40), a reliable animal model of CLN1 disease (41). Cortical tissues were used because in a previous clinical study (www.clinicaltrials.gov; NCT00028262), MRI of the brain of children affected with CLN1 disease showed a rapid degeneration of the cerebral cortex (16). Since CLN1/PPT1 is ubiquitously expressed in all tissues and cells, we also used cultured cells from INCL patients to determine whether the loss of Cln1/Ppt1 causes aberrant activation of the mTORC1 kinase and suppresses autophagy contributing to pathogenesis of CLN1 disease.
Results
Loss-of-function mutations in the Cln1 gene in mice hyperactivates mTORC1 signaling
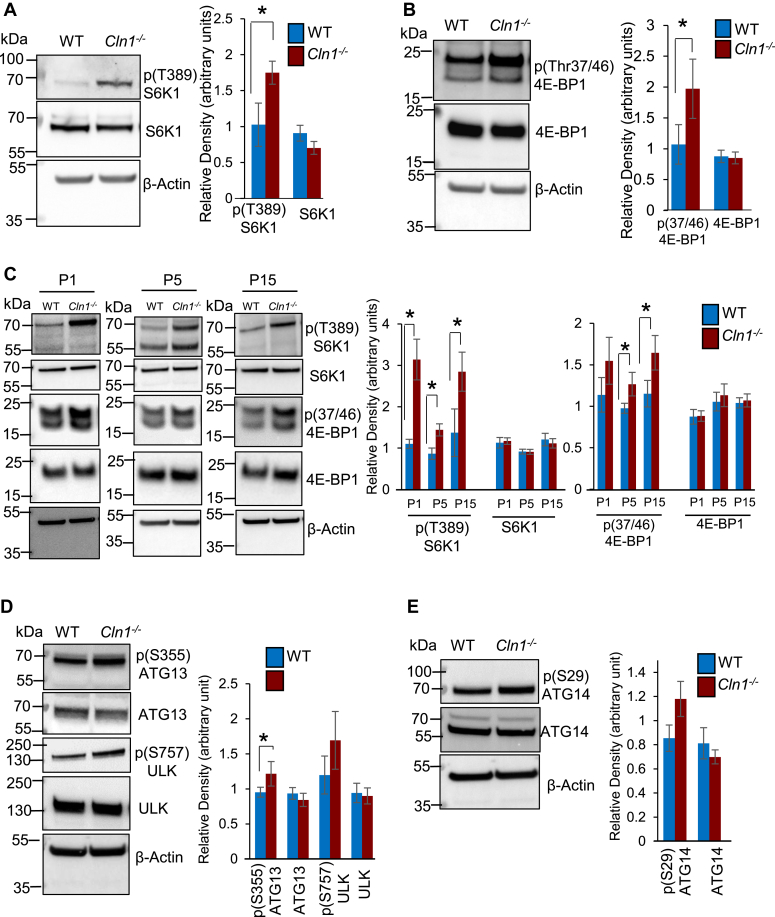
The mTOR (42, 43, 44), a serine/threonine kinase, is a master regulator of cellular growth and metabolism (28). The activation of mTORC1 signaling has been reported to suppress autophagy (34, 35, 45), which is a conserved pathway for the degradation of aged proteins and organelles (32, 37, 38). Autophagy plays an essential role for neuronal survival (46). Moreover, suppression of autophagy by activated mTORC1 underlies neuropsychiatric and neurodegenerative disorders (47, 48, 49). Thus, we first sought to determine whether the level of activated mTORC1 in the brain of Cln1−/− mice is higher than that in their WT littermates. Accordingly, we measured the levels of phosphorylated 70/S6 kinase-1 (pS6K1) and eukaryotic translation initiation factor 4E-binding protein-1 (p4E-BP1), which are the canonical substrates of mTORC1 kinase (28, 34). The results of Western blot analyses of total lysates of cortical tissues from Cln1−/− mice revealed that the levels of both pS6K1 and p4E-BP1 were significantly higher than those in their WT littermates (Fig. 1, A and B). Since the Cln1/Ppt1 gene is expressed in all cells and tissues in mice and in humans, we also determined the levels of pS6K1 and p4E-BP1 in cultured lymphoblasts from patients with CLN1 disease and those from age- and sex-matched normal subjects. The results showed that the levels of pS6K1 and p4E-BP1 in cultured lymphoblasts from patients with CLN1 disease were substantially higher than those in their normal counterparts (Fig. S1A). Taken together, these results demonstrated that the activation of mTORC1 signaling is significantly higher in the brain of Cln1−/− mice, as well as in cultured lymphoblasts from patients with CLN1 disease.
Figure 1.
Aberrant mTORC1 activation and autophagy in WT and Cln1−/−mouse brain.A, Western blot analyses and densitometric quantitation of phosphorylated-S6K1 (pS6K1) and B) phosphorylated- 4E-BP1 (p4E-BP1), the two canonical substrates of mTORC1, in the cortical homogenate from WT and Cln1−/−mice (n = 4). C, Western blots and densitometric analyses of pS6K1 and p4E-BP1 in the cortical lysates from postnatal WT and Cln1−/− mice (n = 4). Also see Figs. S1A and S2C. D, levels of phospho- and nonphospho-ATG13 and ULK in the brain of WT and Cln1−/− mice (n = 4). E, levels of phospho- and nonphospho-ATG14 in cortical tissue-lysates from WT and Cln1−/− mice (n = 4). Also see Fig. S1, D and E.Two-sample permutation t test with complete enumeration was used to calculate the p-values. Data presented as the mean ± SD and the p values (∗p < 0.05) represent WT versus Cln1−/− mice. CLN, ceroid lipofuscinosis neuronal; mTORC, mechanistic/mammalian target of rapamycin complex; pS6K1, phosphorylated 70/S6 kinase-1; p4E-BP1, eukaryotic translation initiation factor 4E-binding protein-1.
Activation of mTORC1 signaling in developing and adult brain of WT and Cln1−/− mice
During their lifetime, all organisms encounter some environmental stress (50). To cope with these stresses, most organisms have evolved survival mechanisms. In all placental mammals, a stressful event occurs during parturition when the transplacental nutrient supply is abruptly disrupted for a brief period. To cope with this brief but critical period of starvation, the neonates have evolved autophagy (51, 52) for the supply of nutrients. Several studies have demonstrated that mTORC1 senses and responds to cellular nutrient status via Rag family of GTPases (53, 54, 55, 56). The importance of lysosomal nutrient sensing in mammalian physiology has been elegantly demonstrated using a knockin mouse model that expressed a constitutively active form of Rag A (Rag AGTP) (52). The results of this study showed that although the Rag AGTP/GTP mice were normal during in utero development, these pups succumb to death on postnatal day 1 (P1).
Since the activation of mTORC1 signaling suppresses autophagy and dysregulation of autophagy underlies neurodegeneration in most LSDs (3, 4), we sought to determine at what stage during early development of WT and Cln1−/− mice mTORC1 signaling was initiated. Accordingly, we measured the levels of pS6K1 and p4E-BP1 in cortical tissues from WT and Cln1−/− fetuses on gestational days 17 (G17) and 18 (G18). We chose these two dates because previously we found that the expression of Cln1/Ppt1 gene is detectable from as early as the 17th-18th day of gestation in mice (57). Our results showed that there were no significant differences in the level of pS6K1 and p4E-BP1 in the brain of G17 and G18 fetuses from WT and Cln1−/− mice (Fig. S1B). Remarkably, compared with WT pups the levels of these markers of mTORC1 activation in the brain of Cln1−/− pups were consistently higher from P1, which persisted throughout adulthood (Figs. 1C and S1C). These results suggested that although during prenatal development, there were no significant differences in mTORC1 signaling between WT and Cln1−/− pups, it was substantially increased in those of P1-Cln1−/− pups and persisted throughout adulthood.
Hyperactivation of mTORC1 suppresses autophagy in Cln1−/− mouse brain
Lysosomes degrade endocytosed material from both extracellular sources as well as dysfunctional intracellular macromolecules and organelles accumulated in autophagosomes (32, 37). For this reason, the LSDs have been called the disorders of dysregulated autophagy (2, 3, 4). Indeed, dysfunctional autophagy underlies virtually all neurodegenerative LSDs (3, 4). One of the roles mTORC1 activation is to maintain nutrient homeostasis through lysosomal biogenesis (6). The activation of mTORC1 signaling also inhibits the recycling of damaged organelles and aged macromolecules via autophagy (37). Emerging evidence indicates that mTORC1 and autophagy are closely integrated processes. Moreover, dysregulated mTORC1 signaling may lead to a range of human ailments like cancer, neurodevelopmental, and neurodegenerative diseases (28, 47, 58). The activation of mTORC1 negatively regulates autophagy by phosphorylating several autophagy-related proteins (59). Phosphorylation of some of these proteins like ULK and ATG-13 by mTORC1 results in inhibition of the ULK complex, thereby, preventing the initiation of autophagy. Further, the VPS34 complex plays a critical role in the induction of autophagy and vesicle trafficking (59). The mTORC1 also regulates autophagy by phosphorylating ATG14L in the VPS34 complex to inhibit it. Therefore, we sought to determine whether the activation of mTORC1 signaling in Cln1−/− mice inhibited autophagy by phosphorylating the autophagy-related proteins. Our results showed that compared with WT controls, the phosphorylation of ATG13 of the ULK complex was significantly higher in the brain of Cln1−/− mice (Fig. 1D). The level of phospho-ATG14 of the VPS34 complex, however, was not significantly higher in Cln1−/− mice compared with that of the WT controls (Fig. 1E). We also measured the levels of autophagy markers, LC3-II and p62/SQSTM1, to evaluate the status of autophagy in the brain of Cln1−/− mice. The levels of both LC3-II and p62/SQSTM1 were significantly higher in Cln1−/− mice than those in their WT littermates (Fig. S1, D and E). Taken together, these results suggested that in the brain of Cln1−/− mice aberrant hyperactivation of mTORC1 signaling dysregulated autophagy most likely contributing to neuropathology in this CLN1 disease model.
Genetic loss of Cln1/Ppt1 disrupts lysosomal nutrient sensing scaffold in Cln1−/− mice
Emerging evidence indicates that sensing of essential nutrients is an important function of the lysosome (54, 60). The lysosome integrates nutrient and metabolic cues to control cellular functions via mTORC1 signaling pathway (54). Signals from nutrients regulate the activation of mTORC1 through the Rag GTPases. The Rag GTPases are obligate heterodimers of Rag A or Rag B complexed with Rag C or Rag D. The Ragulator complex (composed of Lamtor 1–5), vacuolar (H+)-ATPase (vATPase), and SLC38A9 on the lysosomal limiting membrane, which constitute the lysosomal nutrient sensing scaffold (34). The mTORC1 must attach to the lysosomal nutrient sensing scaffold on lysosomal membrane (Fig. 2A) to manifest its kinase activity (54, 55, 61). The acidic pH of the lysosomal lumen facilitates the degradation and clearance of cargo delivered to it from intracellular and extracellular sources (62). The acidification of the lysosomal lumen is achieved primarily by the action of vATPase (63), a large multisubunit protein localized to the lysosomal membrane. The vATPase is also an important component of the lysosomal nutrient sensing scaffold (64, 65) as it catalyzes the hydrolysis of ATP, which is essential for the vATPase–Ragulator interaction by nutrients such as amino acids. The lysosomal nutrient sensing scaffold also facilitates the translocation of mTORC1 from the cytosol to the lysosomal membrane where it undergoes activation (30, 34).
Figure 2.
Levels of Rag/Ragulator complex proteins in lysosomes from WT and Cln1−/−mouse brain.A, schematic explaining how the RagGTPase/Ragulator complex facilitates mTORC1 activation. The Rag/Ragulator complex senses the presence of lysosomal amino acids and activates the Rag-GTPase tethered to lysosomal membrane by Ragulator, followed by docking of mTORC1 on lysosomal surface for activation. B, Western blot analysis and densitometric quantitation showing the total level of RAG-GTPase (RagA and RagC) in the cortical section of WT and Cln1−/− mice (n = 4). C, Western blot analysis of total level of RagC and RagD in the cortical section of WT and Cln1−/− mice (n = 4). D, levels of Rag GTPase (i.e., RagA, RagB, RagC, and RagD) in purified lysosomal fractions from the cortical homogenate of WT and Cln1−/− mice (n = 4). Also see Fig. S2A. E, Western blot analysis of Ragulator complex (Lamtor1, Lamtor2, Lamtor3, Lamtor4, and Lamtor5) in the cortical homogenate of WT and Cln1−/− mice (n = 4). F, the levels of Ragulator complex proteins (Lamtor1–Lamtor5) in lysosomal fractions from cortical tissues of WT and Cln1−/− mice (n = 4). Also see Fig. S2B. G, Western blot analysis of SLC38A9 in the cortical homogenates of WT and Cln1−/− mice (n = 4). H, Western blot analysis of lysosomal level of SLC38A9 in the cortical homogenates of WT and Cln1−/−mice (n = 4). Two-sample permutation t test with complete enumeration was used to calculate the p values. Data presented as the mean ± SD and p values (∗p < 0.05) represent WT versus Cln1−/− mice, respectively. CLN, ceroid lipofuscinosis neuronal;mTORC, mechanistic/mammalian target of rapamycin complex.
Previously, we reported that a critical subunit of vATPase, V0a1, required dynamic S-palmitoylation (palmitoylation-depalmitoylation) for endocytic trafficking to the lysosomal membrane (66). Intriguingly, we also found that in Ppt1-deficient Cln1−/− mice, V0a1 was misrouted to the plasma membrane instead of its normal location on lysosomal membrane, which dysregulated lysosomal acidification (66). Therefore, we sought to determine whether other constituent proteins of the lysosomal nutrient sensing scaffold such as the Rag-GTPases and the Ragulator complex also require dynamic S-palmitoylation and are misrouted to the plasma membrane in Cln1−/− mice. Accordingly, we first determined the mRNA and protein levels of the Rag-GTPases and the Ragulator complex proteins in cortical tissues from WT and Cln1−/− mice. We found that the mRNA levels of Rag A and Rag B in Cln1−/− mouse brain were significantly higher than those in their WT littermates (Fig. S2A). However, the protein levels of all the Rag-GTPases in total cortical lysates from WT and Cln1−/− mice were virtually identical (Fig. 2, B and C). Remarkably, the purified lysosomal fractions from cortical tissues of Cln1−/− mice contained significantly lower levels of Rag-GTPases (Fig. 2D). Most notably, while the mRNA and protein levels of some of the Ragulator complex proteins were substantially higher in total cortical lysates from Cln1−/− mice (Fig. S2B and E), those in the purified lysosomal fractions were significantly lower (Fig. 2F). Intriguingly, SLC38A9, a component of the lysosomal nutrient sensing scaffold (67, 68), also failed to localize on lysosomal membrane of Cln1−/− mice (Fig. 2, G and H). These results suggested that the lysosomal nutrient sensing scaffold in Cln1−/− mice may be disrupted. This may be because one of the constituent proteins requiring S-palmitoylation (i.e., vATPase) is misrouted to the plasma membrane (66). In addition, another S-palmitoylated constituent protein (i.e., Lamtor 1) (69, 70) may be misrouted and a third protein (i.e., SLC38A9), may also require dynamic S-palmitoylation for endosomal trafficking and Ppt1 deficiency may disrupt its localization on lysosomal membrane.
Lamtor 1-S-palmitoylation promotes lysosomal localization misrouted in Cln1−/− mice
Numerous proteins in the brain undergo S-palmitoylation (24, 25, 27), which enhances membrane affinity, promotes protein–protein interactions, and facilitates protein trafficking (24), especially in the brain. The p18/Lamtor1 plays critical roles in organizing the Ragulator–Rag GTPase complex (71), essential for the assembly of the lysosomal nutrient sensing scaffold and for nutrient-dependent activation of mTORC1 signaling (34, 61). We reasoned that Lamtor1, like V0a1, may also require S-palmitoylation for trafficking to the lysosomal membrane and Ppt1 deficiency in Cln1−/− mice may misroute it, thereby, disrupting the lysosomal nutrient sensing scaffold.
Previously, in a low-throughput proteomic study, Lamtor1 was predicted to be S-palmitoylated (70). This prediction was confirmed by acyl biotin exchange assay (72), although the specific S-palmitoylated cysteine residue(s) in Lamtor1 was not identified. To identify the specific cysteine residue(s) that may be S-palmitoylated, we first analyzed the peptide sequence of Lamtor1 using CSS-Palm (73), a computer program, which predicts the specific cysteine residues in polypeptides that are likely to be S-palmitoylated. The results of this analysis identified Cys3 and Cys4 in Lamtor1 from both humans and mice as potential S-palmitoylation sites (Fig. 3A). Notably, these Cys residues in Lamtor1 are also evolutionarily conserved (Fig. 3B). To confirm whether Cys3 and Cys4 are indeed the S-palmitoylated residues, we transfected HEK293T cells with complementary deoxy ribonucleic acid constructs of Myc-tagged WT- and mutant-Lamtor1 (Cys3Ala, Cys4Ala, or Cys3Ala + Cys4Ala). We then analyzed the proteins for S-palmitoylation using acyl Rac assay (74). We used Cys to Ala mutations because Ala does not undergo S-palmitoylation. We found that Cys3Ala or Cys4Ala mutants alone only partially reduced the level of S-palmitoylation. However, Cys3Ala and Cys4Ala mutations in the same construct completely abolished S-palmitoylation of Lamtor 1 (Fig. 3C). These results confirmed that both Cys3 and Cys4 are the S-palmitoylation sites in Lamtor 1. Notably, the level of S-palmitoylated Lamtor1 in the brain of Cln1−/− mice was significantly higher than that in their WT littermates (Fig. 3D). The reason behind this result is unclear. We also analyzed the peptide sequence of SLC38A9 using CSS-Palm and performed acyl Rac assay using native SLC38A9 protein from mouse brain lysate. The results showed that SLC38A9 does not undergo S-palmitoylation despite the prediction by CSS-Palm analysis (Fig. S3, A and B).
Figure 3.
Lamtor 1 requires dynamic S-palmitoylation for lysosomal localization and is misrouted in Cln1−/−mice.A, potential S-palmitoylation sites in Lamtor 1 as predicted by CSS-palm-4 analysis. B, amino acid sequence similarity among various species across phyla showing Cys3 and Cys4 in Lamtor 1 are evolutionarily conserved. C, acyl-RAC assay and Western blot analysis, respectively, of total lysates of HEK-293T cells transfected with Myc-Lamtor1, Myc-Lamtor1-mutant (Cys 3 Ala), and Myc-Lamtor1-mutant (Cys 4 Ala) constructs to confirm that Cys 3 and Cys 4 in Lamtor 1 are the residues that undergo S-palmitoylation. D, acyl-RAC assay and Western blot analysis to detect the endogenous level of S-palmitoylated Lamtor1in the brain cortical homogenate from WT and Cln1−/− mice (n = 4). E, confocal imaging showing colocalization of Lamtor1 with lysosomal marker, Lamp2, in Cln1−/− mouse neurons (n = 22) and WT mouse neurons (n = 18). Data were pooled from four independent experiments. F, Western blot analysis and densitometric quantitation showing the level of Lamtor1 in plasma membrane fractions from the cortical homogenate of WT and Cln1−/− mice (n = 4). Two-sample permutation t test with complete enumeration was used to calculate the p values. Data presented as the mean ± SD and p values (∗p < 0.05 and ∗∗p < 0.01) represent WT versus Cln1−/− mice, respectively. CLN, ceroid lipofuscinosis neuronal.
Next, we sought to confirm the lysosomal localization of Lamtor1 using confocal imaging of cultured cortical neurons from WT and Cln1−/− mice. We found that in neurons from Cln1−/− mice, there was a substantially reduced colocalization of Lamtor1 with the lysosomal membrane marker, Lamp2 (Fig. 3E). We also found that the level of Lamtor1 in enriched plasma membrane fractions of brain tissues from Cln1−/− mice was significantly higher than that of their WT littermates (Fig. 3F). Taken together, these results raised the possibility that like V0a1 subunit of vATPase (66), Lamtor1 in neurons from Cln1−/− mice may also be misrouted to the plasma membrane. Therefore, at least two of the most important constituent proteins in the lysosomal nutrient sensing scaffold (i.e., vATPase and Lamtor1), which require S-palmitoylation for trafficking to the lysosomal surface, were misrouted to the plasma membrane in Cln1−/− mice. Cumulatively, these findings suggested that most likely the organization of the lysosomal nutrient sensing scaffold is disrupted in Cln1−/− mice but the pathway underlying aberrant activation of mTORC1 signaling remained unclear. Thus, we sought to identify alternative pathways that may mediate aberrant activation of mTORC1 signaling in the brain of this mouse model of CLN1 disease.
Growth factor–mediated pathway underlies mTORC1 activation in Cln1−/− mice
The lysosomal degradative function and the activation of mTORC1 signaling are highly coordinated processes. Whereas signals from nutrients within the cell facilitate the translocation of mTORC1 from the cytoplasm to the lysosomal membrane (34), those from growth factors can mediate mTORC1 activation via PI3K/Akt pathway (75, 76). Moreover, genetic studies in mice and humans have also revealed the important physiological functions of the PI3K/Akt network in virtually all organ systems (77). Dysfunction of this network underlies diverse pathological conditions, including developmental abnormalities, cardiovascular diseases, insulin-resistance in type 2 diabetes, inflammatory, and neurological disorders (28, 47). One potential pathway that may mediate aberrant activation of mTORC1 signaling in Cln1−/− mouse brain may involve tuberous sclerosis complex (TSC) genes. Indeed, the knockdown of TSC2 gene in fetal tubers and in mouse neuroprogenitor cells has been reported to cause aberrant activation of mTORC1 signaling (78). The TSC is an autosomal dominant neurodevelopmental disorder, caused by mutations in either the TSC1 or TSC2 gene (79). It has also been reported that mTOR signaling plays a pivotal regulatory role during the development of cerebral cortex in mammals including humans. Consistent with this finding, rapid degeneration of neurons in the cerebral cortex of patients with CLN1 disease (16) has been observed. Moreover, the loss of function mutations of TSC1, TSC2, PTEN, and STRADα genes mediate mTORC1 activation, which causes epilepsy and neurobehavioral problems (80, 81). Furthermore, suppression of mTORC1 activation has been reported to ameliorate seizure activity in the brain (82), which is a common manifestation in CLN1 disease.
A complex interplay between TSC2 and PI3K/Akt have also been reported to cause hyperactivation of mTORC1 signaling (83). Accordingly, we investigated the growth factor–mediated pathway of mTORC1 signaling via PI3K/Akt, which phosphorylates TSC2 activating the small GTPase, Ras homolog enriched in brain (Rheb) (Fig. 4A). Notably, phosphorylation of TSC2 by PI3K/Akt promotes the activation of Rheb to the lysosomal membrane, thereby, activating mTORC1 signaling (84). We reasoned that aberrant activation of mTORC1 signaling in Cln1−/− mouse brain may be mediated via TSC1–TSC2/Rheb pathway despite the disruption of the lysosomal nutrient sensing scaffold. To test this hypothesis, we determined the levels of TSC complex consisting of TSC1, TSC2, and TBCD17 in the brain of WT and Cln1−/−mice. The results showed that while there was no difference in the basal level of the constituent proteins of the TSC complex between WT and Cln1−/−mice (Fig. S4A), the levels of phosphorylated TSC2 (Ser939) were significantly higher in Cln1−/− mouse brain (Fig. 4B).
Figure 4.
Growth factor–mediated pathway of mTORC1 activation in Cln1−/−mouse brain.A, schematic representation of AKT/TSC pathway activating mTORC1 signaling. AKT inhibits the TSC complex by phosphorylation of TSC2. This promotes the activation of Rheb GTPase by conversion of Rheb-GDP to Rheb-GTP essential for mTORC1 activation on lysosomal membrane. B, determination of the phosphorylation status of TSC2 in total cortical lysates from WT and Cln1−/− mice (n = 4). Also see Fig. S4A. C, immunoprecipitation and Western blot analysis showing the level of GTP-Rheb in the cortical homogenate of WT and Cln1−/− mice (n = 4). D, Western blot analysis of total cortical lysates from WT and Cln1−/− mice to determine the level of phosphorylated PRAS-40 (n = 4). E, Western blot analysis of cortical homogenate showing the levels of phosphorylated AKT in WT and Cln1−/− mice (n = 4). Also see Fig. S4, B and C. F, level of IGF1 by ELISA in cortical lysates from WT and Cln1−/− mice (n = 4). Also see Fig. S4, D–F. G, Western blot analysis of cortical homogenate from WT and Cln1−/− mice (n = 4) showing the phosphorylation status of P13K kinase. Also see Fig. S4G. Two-sample permutation test with complete enumeration was used to calculate the p values. Data presented as the mean ± SD. The p values (∗p < 0.05 and ∗∗p < 0.01) represent WT versus Cln1−/− mice, respectively. CLN, ceroid lipofuscinosis neuronal; IGF1, insulin-like growth factor-1; mTORC, mechanistic/mammalian target of rapamycin complex; PRAS40, proline-rich Akt-substrate-40; Rheb, Ras homolog enriched in brain; TSC, tuberous sclerosis complex.
Since the activation of Rheb requires the conversion of GDP-Rheb (inactive) to GTP-Rheb (active), we measured the level of active Rheb by pull-down assay using antibody to GTP-Rheb. Our results showed that although there was no difference in the total level of Rheb between WT and Cln1−/−mice, there was a significantly higher level of active Rheb in Cln1−/− mouse brain (Fig. 4C). We also evaluated the phosphorylation status of a 40-kDa proline-rich Akt-substrate-40 (PRAS40), which releases its inhibitory effects on Raptor promoting the activation of mTORC1 signaling (85). The results showed that in the brain of Cln1−/− mice, the levels of pPRAS40 were significantly higher than those in their WT littermates (Fig. 4D). Since phosphorylated Akt (pAkt) catalyzes the phosphorylation of TSC2 and PRAS40, we measured the levels of pAkt in the brain of WT and Cln1−/− mice. We found that compared with WT mice, the pAkt levels in Cln1−/− littermates were significantly higher (Fig. 4E). Our results also showed that in Cln1−/− mice, there was a markedly higher level of pGSK-3β (Fig. S4, B and C), which is a substrate of Akt. Taken together, these results suggested that PI3K/Akt is at least one of the pathways that mediate aberrant activation of mTORC1 signaling in this mouse model of CLN1 disease.
Insulin-like growth factor-1 mediates aberrant mTORC1 signaling in Cln1−/− mice
The lysosomal degradative function is highly integrated with cues from intracellular nutrients, which promote translocation of mTORC1 from the cytoplasm to the lysosomal membrane. Signals generated from growth factors are relayed by phosphorylation of PI3K/Akt-mediating mTORC1 activation (86). Both insulin and insulin-like growth factor-1 (IGF1) can mediate the activation of PI3K/Akt (Fig. S4D). To delineate whether mTORC1 signaling is mediated via growth factor pathway, we first measured the levels of both insulin and IGF1 in cortical tissues from WT and Cln1−/− mice. We found that while there was no significant difference in the levels of insulin among these animals (Fig. S4E), the level of IGF1 in the brain of Cln1−/− mice was significantly higher than that in their WT littermates (Fig. 4F). Interestingly, even though the IGF1-receptor levels were unaltered in Cln1−/− mouse brain (Fig. S4F), the phosphorylated PI3K level was appreciably higher (Fig. 4G). The PI3K catalyzes the conversion of phosphatidylinositol 4, 5-bisphosphate to phosphatidylinositol 3, 4, 5-trisphosphate (PIP3). While PIP3 is required for the activation of AKT by PDK1, PTEN catalyzes the conversion of PIP3 back to phosphatidylinositol 4, 5-bisphosphate (Fig. S4D). Notably, the levels of phosphorylated PTEN were significantly reduced in Cln1−/− mouse brain (Fig. S4G). Cumulatively, these results suggested that in the brain of Cln1−/− mice, the aberrant activation of mTORC1 signaling, at least in part, is mediated by IGF1 via PI3K/Akt pathway.
Earlier studies have indicated that mTORC1 can be activated by spatial association with focal adhesions near the plasma membrane. Coupling of mTORC1 with growth factor signaling and amino acid input into the cell may be sufficient to activate mTORC1, even when it is not on the lysosomes (87). It is therefore possible that mTORC1 in Cln1−/− cells are activated near the plasma membrane in response to IGF1. We therefore determined the plasma membrane level Rheb, Raptor, and mTOR to determine if it was the site of mTORC1 activation in Cln1−/− mouse brain. The results however indicated no difference in the plasma membrane level of Rheb, Raptor, or mTOR between WT and Cln1−/− mouse (Fig. S5A). Earlier studies on mTOR localization using live and fixed cell imaging have shown that translocation of mTORC1 to lysosome is a transient event and occurs within minutes of amino acid addition. However, the chain of events following mTORC1 translocation is delayed suggesting a dynamic process of mTORC1 translocation for its activation (88). Therefore, it may not be possible to capture a dynamic event from isolated plasma membrane fractions from mouse brain.
Pharmacological inhibitors of PI3K/Akt suppresses mTORC1 activation
Thus far, our results have shown that Ppt1 deficiency in Cln1−/− mice, via growth factor (IGF1)-mediated pathway, activated aberrant mTORC1 signaling. Next, we sought to determine whether nontoxic, pharmacological inhibitors of Akt may suppress aberrant mTORC1 activation and rescue autophagy in PPT1-deficient cells. Since Cln1/Ppt1 gene is universally expressed in all cells and tissues, we used cultured lymphoblasts from patients with CLN1 disease and those from age- and sex-matched healthy subjects to test the effects of these inhibitors (e.g., A674563, MK2206, Afuresertib, or Uprosertib). We quantified the levels of pS6K1 and p4E-BP1 before and after treatment of the cells with AKT inhibitors. The results showed that treatment of cultured lymphoblasts from CLN1 disease patients with recombinant PPT1 (positive control) significantly suppressed pS6K1 and p4E-BP1 levels (Fig. S5B). Notably, treatment of these cells with pharmacological inhibitors of Akt also significantly suppressed pS6K1 and p4E-BP1 levels compared with those in dimethyl sulphoxide (DMSO)-treated controls (Figs. 5A and S5, B–D). Since AKT phosphorylates GSK3β we also measured the level pGSK3β to determine the extent of AKT inhibition. The results showed that treatment of cultured CLN1 disease lymphoblasts with Akt inhibitors also significantly reduced the level of pGSK3β (Figs. 5A and S5C), indicating the suppression of Akt activity. Furthermore, since the Akt inhibitors, Afuresertib and Uprosertib, are nontoxic and orally administrable (89), we treated Cln1−/− mice with these drugs by gavage feeding. The results showed that after only 14 days of treatment, the levels of mTORC1 signaling in the brain of these mice were significantly lower than those in their untreated counterparts (Fig. 5B). As expected, the levels of pS6K1 and p4E-BP1 in the brain of vehicle-treated mice were unaltered (Fig. S5E). We did not observe any adverse effects during this short period of treatment. It should be noted, however, that the Akt inhibitor, Afuresertib, is currently undergoing phase-2 clinical trial (NCT04374630) for the treatment of ovarian cancer.
Figure 5.
AKT inhibitors suppress mTORC1 activation in CLN1 patient lymphoblasts and Cln1−/−mice.A, detection and quantitation of pS6K1, p4E-BP1, and pGSK3β in lymphoblasts from normal individuals (NL) (1) and patient with CLN1 disease lymphoblast without (2) or treated with AKT inhibitors, Afuresertib [10 μM (3) and 30 μM (4)] and Uprosertib [10 μM (5) and 30 μM (6)] (n = 3). B, the levels of pS6K1, p4E-BP1, and pGSK3β in the brain cortical homogenate of untreated or AKT inhibitor, Afuresertib (Afu)-treated WT and Cln1−/− mice (n = 5). AKT inhibitors, Afuresertib (Afu 50 mg/kg body weight and Uprosertib (Upr 50 mg/kg body weight) for 2 weeks. C, the levels of S6K1, p4E-BP1, and pGSK3β in untreated or Afuresertib (Afu)-treated (50 mg/kg body weight for 5 months) WT and Cln1−/− mice (n = 4). Two-sample permutation t test with complete enumeration was used to calculate the p values (∗p < 0.05 and ∗∗p < 0.01). Data presented as the mean ± SD. CLN, ceroid lipofuscinosis neuronal; mTORC1, mechanistic target of rapamycin complex-1; pS6K1, phosphorylated 70/S6 kinase-1; p4E-BP1, eukaryotic translation initiation factor 4E-binding protein-1.
To further evaluate the beneficial and/or adverse effects of these drugs, we conducted a long-term (5 months) treatment of Cln1−/− mice with Afuresertib starting at 1 month of age. Importantly, the results showed that long-term treatment of Cln1−/− mice with Afuresertib significantly reduced the levels of pS6K1, p4E-BP1, and pGSK3β in these mice compared with those in the untreated controls (Fig. 5C). Notably, we did not observe any apparent adverse effects. Taken together, these results suggested that treatment of cells from CLN1 disease patients in vitro as well as Cln1−/− mice with nontoxic pharmacological inhibitors of Akt such as Afuresertib suppressed aberrant activation of mTORC1 signaling with no apparent adverse effects.
Pharmacological inhibitors of PI3K/Akt rescues autophagy
Since the activation of mTORC1 signaling suppresses autophagy (35) and loss of autophagy in the brain causes neurodegeneration (3, 4), we sought to determine whether pharmacological inhibitors of Akt may rescue autophagy in PPT1-deficient cells and in Cln1−/− mice. Accordingly, we determined the level of LC3-II and p62/SQSTM1 to evaluate the status of autophagy in CLN1 disease lymphoblasts with/without treatment with Akt inhibitors. The results showed that the levels of both autophagy markers, LC3-II and p62/SQSTM1 in treated cultured lymphoblasts from CLN1 disease patients were reduced in a dose-dependent manner and comparable to levels in lymphoblasts from healthy adults (Fig. S6A). Since phosphorylation of ULK and ATG13 is directly regulated by mTORC1, we measured the levels of pULK and pATG13 levels in cultured lymphoblasts from healthy adults and CLN1 disease patients with/without Akt inhibitors. The results showed a significant reduction in the levels of phosphorylated, ULK and ATG13 in treated cultured lymphoblasts from CLN1 disease patients in a dose-dependent manner (Fig. S6A).
Next, we determined the level of LC3-II and p62/SQSTM1 to evaluate the status of autophagy in untreated and Afuresertib-treated Cln1−/−mice. The results showed that the levels of LC3-II and p62/SQSTM1 in treated Cln1−/−mice were substantially reduced compared with those in the untreated controls (Fig. S6, B and C). We also determined the phosphorylation status of the autophagy-related proteins ULK, ATG13, and ATG14 in cortical tissues from untreated and Afuresertib-treated mice. The results showed a significant reduction in the levels of phosphorylated, ATG13 and ATG14 in the brain of Cln1−/− mice treated with Akt inhibitor compared with those of their untreated Cln1−/− littermates (Fig. S6D). Taken together, these results showed that pharmacological suppression of mTORC1 activation in Cln1−/− mice rescued autophagy.
Afuresertib-treatment ameliorates neuroinflammation in Cln1−/− mice
Despite the notion that the brain is an immune privileged site, it is not uncommon to encounter immune responses in this organ (90). The microglia and astrocytes are the primary constituents of the innate immune system in the central nervous system (91). Activation of these innate immune cells is a major contributor of neuroinflammation, which contributes to neurodegeneration. Recently, we reported that activated astrocytes stimulating microglia proliferation contributed to neuroinflammation mediating neuronal death in Cln1−/− mice (92). Therefore, we sought to determine if suppression of mTORC1 by Afuresertib arrested the progression of neuroinflammatory changes characteristically found in these animals (41, 93). Accordingly, we evaluated the levels of the astrocyte marker, glial fibrillary acidic protein, and the microglial markers, CD68 and IBA1, respectively, by Western blot analysis. Our results showed a significantly higher levels of these proteins in the brain of untreated Cln1−/− mice than those in the WT mice (Figu. 6, A–C). Remarkably, treatment of the Cln1−/− mice with Afuresertib resulted in a substantially reduced levels of these neuroinflammatory markers (Fig. 6, A–C). Confocal imaging of brain sections from Afuresertib-treated animals further confirmed a significant reduction in glial fibrillary acidic protein–, CD68-, and IBA1-positive cells compared with those in untreated Cln1−/− mice (Figs. 6, D and E, and S6E).
Figure 6.
Treatment with AKT inhibitor suppresses glial cell levels in Cln1−/−mice.A, level of astrocyte marker, GFAP, in WT, untreated (Un), and Afuresertib (Afu) (50 mg/kg body weight for 5 months) Cln1−/− mice (n = 4). B, detection of microglia marker, CD68, in WT, untreated (Un)- or Afuresertib (Afu) (50 mg/kg body weight)-treated Cln1−/− mice (n = 4). Mice were treated for 5 months. C, Western blot analysis of microglia marker, IBA1, in WT, untreated (Un)- or Afuresertib (Afu)- treated (for 5 months) Cln1−/− mice (n = 4). Also see Fig. S6E. D, fluorescence imaging of GFAP in WT, untreated (Un)- and Afuresertib (Afu)-treated (for 5 months) Cln1−/− mouse cortical sections of untreated (Un) or treated with Afuresertib (Afu) for 5 months, (n = 12). The scale bar represents 50 μm. E, fluorescence imaging of CD68 in WT & Cln1−/− mouse brain cortical sections of untreated (Un) or treated with Afuresertib (Afu) for 5 months, (n = 12). The scale bar represents 50 μm. Two-sample permutation t test with complete enumeration was used to calculate the p values (∗p < 0.05 and ∗∗∗p < 0.001). Data are presented as the mean ± SD. CLN, ceroid lipofuscinosis neuronal.
Next, we sought to evaluate whether Afuresertib treatment reduced brain atrophy by measuring the cortical thickness and neuron density in Cln1−/− mice. Accordingly, we evaluated the cortical thickness of WT, untreated, and Afuresertib-treated Cln1−/− mice using H&E staining. The results showed that compared with WT mice, there was significant reduction in cortical thickness in untreated Cln1−/− mice (Fig. S7A). Remarkably, treatment of the Cln1−/− mice with Afuresertib showed a modest but significant preservation of cortical thickness compared with that in untreated mice (Fig. S7A). We also analyzed the level of neurons by Nissl staining and detection of neuronal marker, NeuN, in cortical tissues of these animals by Western blot analysis. We found that treatment with Afuresertib significantly prevented the decline in the level Nissl-positive cells in the brain of Cln1−/− mice (Fig. 7A). Moreover, the level of NeuN was also significantly higher in Cln1−/− mice treated with Afuresertib than untreated Cln1−/− mice (Fig. 7B). Furthermore, immunohistochemical analysis of brain sections for layer-specific neuronal markers, using Cux1 (for cortical layers II–IV) and Ctip2 (for cortical layers IV–VI) showed that there was a substantial reduction in the number of both Cux1- and Ctip2-positive neurons in the untreated Cln1−/− mouse brain. Importantly, compared with untreated controls, Afuresertib-treatment provided significant protection of Cux1- and Ctip2-positive neurons (Fig. S7B). Taken together, these findings demonstrated that the pharmacological inhibitors of Akt are effective in suppressing aberrant activation of mTORC1 signaling, which suppressed neuroinflammation, thereby, protecting the neurons from degeneration in this mouse model of CLN1 disease.
Figure 7.
Suppression of brain atrophy and improved motor function in Cln1−/−mice treated with AKT inhibitor.A, representative images of Nissl-stained neurons in the cerebral cortex of WT and Cln1−/− mice (n = 4). Mice were either untreated (Un) or treated with Afuresertib (Afu) for 5 months. Two-sample permutation t test with complete enumeration was used to calculate the p values (∗p < 0.05). Data are presented as the mean ± SD. B, detection of NeuN-positive cells in the cortical section of WT and Cln1−/− mouse (n = 4), which were either untreated (Un) or treated with Afuresertib (Afu) (50 mg/kg body weight for 5 months). Two-sample permutation t test with complete enumeration was used to calculate the p-values (∗p < 0.05). Data are presented as the mean ± SD. Also see Fig. S7. C, Rotarod test of untreated (Un) or Afuresertib (Afu)-treated WT and Cln1−/− mice (n = 7) for 5 months, and 7 months (n = 7 in each group). Two-sample permutation t test with complete enumeration was used to calculate the p values (∗∗p < 0.01). Data presented are the mean ± SD. D, schematic showing mTORC1 activation in WT (left panel) and Cln1−/− (right panel) mouse brain. mTORC1 integrates signals from growth factors, nutrition, energy, and stress-regulating cell growth and proliferation through phosphorylation of substrates that activate various anabolic processes like protein, lipid, and nucleotide synthesis and inhibit catabolic processes like autophagy. The lysosomal nutrition sensing scaffold (LNSS) (consisting of V-ATPase and Rag–Ragulator complex), plays critical roles in amino acid sensing and mTORC1 activation. The growth factor pathway consisting of the PI3K and AKT also regulates mTORC1 activation through the TSC complex (TSC1, TSC2, TBCD17) and RheB. Upon activation, AKT phosphorylates TSC2 and inhibits it promoting the conversion of Rheb-GDP (inactive) to Rheb-GTP (active), which can then activate mTORC1 on the lysosomal surface. Despite misvocalization of the components of the LNSS complex in Cln1−/− cells the mTORC1 is activated via the growth factor–mediated pathway. Constitutive activation of mTORC1 in Cln1−/− cells contributes to the suppression of autophagy and increased cell proliferation contributing to neurodegeneration in CLN1 disease. CLN, ceroid lipofuscinosis neuronal; AKT, protein kinase B; TSC, tuberous sclerosis complex; IGFR, insulin-like growth factor receptor; IGF1, insulin-like growth factor-1; mTORC1, mechanistic target of rapamycin complex-1; Rheb, Ras homolog enriched in brain.
Afuresertib treatment of Cln1−/− mice show improvement in motor function
One of the early clinical manifestations of CLN1 disease is psychomotor retardation (8, 15). We sought to determine whether treatment of Cln1−/− mice with Akt inhibitors improve motor function. Accordingly, we performed Rotarod test (94) of untreated and Afuresertib-treated Cln1−/− mice at 6 months and 8 months of age. The results showed that in Rotarod test, both 6- and 8-month-old Cln1−/− mice treated with Afuresertib performed significantly better compared with those of the untreated controls (Fig. 7C). These results suggested that treatment of Cln1−/− mice with a nontoxic, orally administrable inhibitor of Akt, Afuresertib, not only suppressed aberrant activation of mTORC1 signaling but also sustained the motor function in these animals.
Discussion
In this study, we sought to understand the mechanism underlying pathogenesis of CLN1 disease, a devastating neurodegenerative LSD, which mostly affect children and young adults (9, 14, 16, 17). We uncovered that some of the proteins that are constituents of the lysosomal nutrient sensing scaffold (e.g., vATPase and Lamtor1), require dynamic S-palmitoylation for their localization on lysosomal membrane where mTORC1 must attach for its activation (34). We also found that the loss of Cln1/Ppt1 in Cln1−/− mice caused misrouting of vATPase and Lamtor1, which most likely disrupted the lysosomal nutrient sensing scaffold impairing lysosomal nutrient sensing. Interestingly, a component of the lysosomal nutrient sensing scaffold (e.g., SLC38A9), which does not undergo S-palmitoylation, also failed to localize on lysosomal surface, although the mechanism of this defect remained unclear. It has been reported that disruption of any component of the Ragulator complex suppresses the activation of mTORC1 by mistargeting the Rags as well as mTORC1 from the lysosomal surface (95, 96). Notably, the disruption of the Rag–Ragulator complex constituting the lysosomal nutrient sensing scaffold has been reported to suppress mTORC1 activation (65). We reasoned that in Cln1−/− mice failure of these Ragulator proteins to localize on lysosomal surface disrupted the lysosomal nutrient sensing scaffold. However, we found that despite this defect there was hyperactivation of mTORC1 signaling in the brain of Cln1−/− mice from P1, which persisted throughout adulthood.
How might loss-of-function mutations in the Cln1 gene lead to mTORC1 hyperactivation and contribute to pathogenesis of CLN1 disease? In a recent unbiased proteomic study, using synaptosomes from WT and Cln1−/− mice >100 proteins have been identified as PPT1 substrates (27). Although the physiological functions of these proteins remain largely unknown, some of them may require dynamic S-palmitoylation for their localization in various cellular compartments. Thus, it is conceivable that Cln1 mutations may impair dynamic S-palmitoylation of some of these proteins rendering them to promote various pathways of mTORC1 activation suppressing autophagy (3, 4) contributing to neurodegeneration in CLN1 disease. If our reasoning is correct, one would expect multiple pathways of mTORC1 activation, which suppresses autophagy contributing to pathogenesis of this disease.
A major downstream effect of mTORC1 activation is the suppression of autophagy (4, 35). Moreover, loss of autophagy in the brain of mice has been reported to cause neurodegeneration (36). Moreover, in common neurodegenerative diseases like Alzheimer’s (37, 38, 39) and in most of the LSDs (2, 97) dysregulated autophagy leads to neurodegeneration (3, 4). Furthermore, impaired autolysosome acidification is found to be dysregulated in several models of Alzheimer’s disease (38) as well as in Cln1−/− mice (66). Recently, by isolating autophagic vesicles from mouse brain it has been demonstrated that nucleoid-associated proteins are enriched in these vesicles (96). Moreover, the efficient autophagic turnover of nucleoids prevents the accumulation of mitochondrial DNA in the neurons, which mitigates the activation of proinflammatory pathways, which contributes to neuroinflammation leading to neurodegeneration (98).
The activation of mTORC1 signaling by nutrients is initiated following its attachment to the lysosomal nutrient sensing scaffold on lysosomal surface (34). The lysosomal nutrient sensing scaffold on lysosomal membrane is composed of the vATPase, the Ragulator (a pentameric protein complex consisting of Lamtor 1–5), the Rag GTPases and SLC38A9. The attachment of mTORC1 on the lysosomal nutrient sensing scaffold is essential for its nutrient-mediated activation (34, 52, 55, 60). The V0a1 subunit of vATPase requires dynamic S-palmitoylation for trafficking to the lysosomal surface and in PPT1-deficient Cln1−/− mice, this critical subunit of vATPase is misrouted to the plasma membrane instead of its normal location on lysosomal membrane (66). While the specific substrates of PPT1 are yet to be identified and characterized, it is possible that this enzyme may have a role in promoting dynamic S-palmitoylation of some of the proteins that are constituents of the lysosomal nutrient sensing scaffold. Consequently, PPT1 deficiency may misroute these proteins to unintended targets. Our results confirmed that Cys3 and Cys4 in both human and mouse Lamtor 1 are S-palmitoylated. Furthermore, our study revealed that PPT1 deficiency in Cln1−/− mice also misrouted Lamtor 1 to the plasma membrane. Since lysosomal localization of Lamtor1 as well as vATPase is critical for the organization of the Ragulator complex, misrouting of Lamtor 1, and vATPase to the plasma membrane most likely disrupted the lysosomal nutrient sensing scaffold. Notably, the Rag GTPases tethered to the lysosomal membrane by a Rag GTPase/Ragulator scaffold brings mTORC1 to the lysosomal surface where its signaling pathway is activated. Although the disruption of lysosomal nutrient sensing scaffold has been reported to inhibit mTORC1 activation (65), our results showed that despite this defect, mTORC1 was aberrantly activated in the brain of Cln1−/− mice. We reasoned that even though the lysosomal nutrient sensing scaffold may be disrupted, a cell may utilize alternative pathways for the activation of mTORC1 signaling in Cln1−/− mouse brain.
The convergence of a complex molecular machinery on the lysosomal surface ensures that signals generated from growth factors can activate mTORC1 and this pathway is tightly regulated. A small GTPase, Rheb, allosterically binds to lysosomal membrane and activates mTORC1 in a highly coordinated manner (84). Moreover, the growth factors like insulin and IGF1 have been reported to mediate the activation of mTORC1 signaling via TSC1, TSC2, and Rheb GTPase (28, 83, 86). Our results are consistent with this mechanism as they show that the level of IGF1 in Cln1−/− mouse brain is significantly than that in the brain of their WT littermates. Moreover, we found that IGF1-mediated activation of mTORC1 signaling via phosphorylation of PI3K/Akt causing Rheb to localize on the lysosomal membrane. Therefore, even though the lysosomal nutrient sensing scaffold may be disrupted in Cln1−/− mice, IGF1 via PI3K/Akt provided an alternative pathway to mediate the aberrant activation of mTORC1 signaling.
How might aberrant activation of mTORC1 signaling in the brain leads to neuropathology in this mouse model of CLN1 disease? One of the consistent clinical findings in CLN1 disease is the development of seizures (7, 14). Recently, it has been reported that hyperactive mTORC1 plays a critical role in the development of epilepsy, ameliorated by fasting (82). Indeed, it has been shown that hyperactive mTORC1 signaling causes epilepsy and inhibition of mTORC1 reduces seizures in this model of epilepsy (82, 99). Persistent mTORC1 activation has been reported to mediate both neurodegenerative and neurodevelopmental disorders (49). The activation of mTORC1 in dividing cells as well as in postmitotic cells like the neurons have been reported to dysregulate autophagy (35). Indeed, suppression of autophagy is found in virtually all LSDs in which neurodegeneration is a devastating manifestation (3, 4, 39). It is noteworthy that the molecular details of mTORC1 activation and its regulation have been investigated almost exclusively using in vitro experimental models (29, 46, 100). While the in vitro cell culture models provide important information, the results may not always be extrapolated in vivo. Thus, studies using animal models provide an additional measure of confirmation of the results obtained from the in vitro experiments.
Intense research for more than several decades have shown that the PI3K signaling regulates virtually all aspects of normal cellular function and dysregulation of one or more proteins in the PI3K-signaling pathway invariably leads to human pathology (101, 102). Our results show that despite disruption of the lysosomal nutrient sensing scaffold, aberrant activation of mTORC1 signaling in Cln1−/− mouse brain occurs via growth factor (IGF1)-mediated pathway. Since this pathway required phosphorylation of Akt, we sought to determine whether pharmacological inhibitors of Akt-phosphorylation may suppress the aberrant activation of mTORC1 signaling. Accordingly, we treated both Cln1−/− mice and cultured lymphoblasts from CLN1 disease patients with A674563, MK2206, Afuresertib, or Uprosertib and evaluated the activation of mTORC1 signaling by quantitating pS6K1 and p4E-BP1 levels. The results showed that Akt inhibitors were effective in significantly reducing the levels of phosphorylated canonical substrates of mTORC1 kinase. These results suggested that inhibition of growth factor–mediated pathway suppresses mTORC1 activation. To further confirm the effects of these inhibitors in vivo, we treated 1-month-old Cln1−/− mice for 2 weeks with the orally administrable AKT inhibitors (Afuresertib and Uprosertib) and measured pS6K1 and p4E-BP1 levels. We found that the levels of pS6K1 and p4E-BP1 were substantially downregulated suggesting the effectiveness of these inhibitors in vivo. Studies with various Akt-inhibitors have been reported to reduce glial activation (90, 103). Further, we found that long-term treatment with Afuresertib not only suppressed aberrant mTORC1 signaling but also reduced glial activation, microglial proliferation, and improved motor function in the treated Cln1−/− mice.
Numerous proteins in the brain require dynamic S-palmitoylation (palmitoylation-depalmitoylation) for endosomal trafficking and function. While S-palmitoylation is catalyzed by ZDHHCs (22, 24), depalmitoylation is mediated by thioesterases (25). The results of our present investigation support a model in which the activation of mTORC1 signaling in WT mice is regulated primarily by lysosomal nutrient sensing via the Rag–Ragulator pathway (Fig. 7D, left panel). For example, when the nutritional content in the cell is low, as happens during parturition, activation of autophagy generates energy required for survival of the fetus. However, in PPT1-deficient Cln1−/− mice misrouting of several constituent proteins of the lysosomal nutrient sensing scaffold to the plasma membrane most likely disrupts the lysosomal nutrient sensing scaffold disabling the nutrition-sensing pathway of mTORC1 activation. In the PPT1-deficient Cln1−/− mice, impaired nutrient-mediated mTORC1 signaling promotes its activation via growth factor (i.e., IGF1)-mediated pathway, which requires phosphorylation of Akt (Fig. 7D, right panel). Our results have shown that in Cln1−/− mice, mTORC1 remains persistently activated from P1, which continues through adulthood. Activated mTORC1 in Cln1−/− mouse brain suppresses autophagy, contributing to neurodegeneration. Interestingly, activation of mTORC1 signaling in both hippocampal neurons and non-neuronal cells has been reported to require dynamic S-palmitoylation of Lamtor1 as well as mTOR (70). We confirmed that Lamtor1 is S-palmitoylated but have not determined whether mTOR is also required S-palmitoylation for localization on lysosomal surface. Recently, it has been shown that S-palmitoylation plays a regulatory role on Akt (103). It remains unclear, however, whether Akt undergoes dynamic S-palmitoylation for its role in IGF-mediated mTORC1 activation and how this pathway may be affected by PPT1 deficiency in Cln1−/− mice. The phosphorylation of PI3K/Akt, localized upstream of the IGF1-mediated mTORC1 activation, when inhibited by a pharmacological inhibitor of this pathway suppressed mTORC1 signaling, which rescued autophagy. Moreover, this treatment also inhibited neuroinflammation, which protected neurons and preserved motor function in Cln1−/− mice. Our results reveal a previously unrecognized role of CLN1/PPT1 in regulating mTORC1 activation and how PPT1 deficiency may contribute to a pathway leading to neurodegeneration in CLN1 disease.
Experimental procedures
Animals and treatments
All animal experiments were performed according to an animal study protocol (ASC#10-012) approved by the Animal Care and Use Committee of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, Cln1−/− mice were generated by targeted disruption of the last exon in the Ppt1 gene in embryonic stem cells (129 strain) as previously reported (40). These mice were subsequently backcrossed for ten generations with WT C57BL/6 mice to obtain congenic C57 background and a breeding pair was kindly provided to us by Dr M.S. Sands (Washington University School of Medicine) to start our Ppt1−/−mouse colony at the National Institute of Child Health and Human Development. Both male and female mice were used for the study. Equal numbers of male and female mice were used in each experiment and the “n” number represents the total number of mice (male and female) used in each experiment. Mice were housed and maintained in a pathogen-free facility under a 12-h light and 12-h dark cycle. Mice were provided with food and water ad libitum. In this study, we used WT mice (C57BL/6J; Stock No. 000664; Jackson Laboratory) and C57 Cln1−/− mice. Six-month-old mice were used for most of the experiments unless otherwise stated. For short term treatment, three and half months old Cln1−/− mice were gavage fed for 2 weeks with Afuresertib or Uprosertib (50 mg/Kg body weight) prepared in 5% DMSO, 40% PEG in saline. For long-term treatment, the Cln1−/− mice were treated with Afuresertib (50 mg/Kg body weight) starting at 1 month of age (three times/week). The animals were sacrificed at 6 months of age for evaluating the biochemical and histological parameters.
Lymphoblast culture and treatment
Normal (C9955, C9982) and immortalized INCL lymphoblast [Patient-1, C11796 Nonsense C451T (R151X) Nonsense C451T (R151X), Patient-2, C10949 Nonsense C490T (R161X) Nonsense C490T (R161X)] were obtained from the laboratory of late Dr Krystina E. Wisniewski and maintained with RPMI containing 15% fetal bovine serum and Pen-Strep. All cell lines were used for initial experiments, but most of the experiments were conducted using C9955 and C11796. The lymphoblasts were treated with recombinant PPT1 (5ug/ml) or Akt inhibitors (A674563, MK2206, Afuresertib, and Uprosertib) or DMSO control for 48 h. The cells were washed twice with RPMI and the media was replaced with RPMI containing 15% fatty acid–free bovine serum albumin and the inhibitors 2 h before harvesting the cells to deplete the media of any PPT1 activity.
Primary neuronal culture
Primary neurons from cortical tissues were isolated using Neuronal Dissociation kit (130-094-802, Miltenyl Biotec) and Neuron isolation kit, Mouse (130-115-389, Miltenyl Biotec) from P2 pups using the standard manufacturer’s protocol with minor modifications. Briefly, brain from P2 pups was isolated and dissociated using Neuronal Dissociation kit and passed through 70 μm strainer to generate single-cell suspension. The cells were first incubated with a cocktail of biotin labeled antibodies for non-neuronal cells and then incubated with microbeads coated with biotin antibody. The cells were then passed through an LS column on a magnetic rack so that the labeled non-neuronal cells are retained in the column. The neurons, which are unlabeled with biotin, were collected in the flow through. These neurons were then cultured in chamber slides using neurobasal media containing B27 supplement and glutamine for at least 7 days before fixing with methanol.
Phosphoprotein analysis
For preparation of phosphoproteins, care was taken to inhibit the protease and phosphatase activities. The protease and phosphatase inhibitors were present throughout sample preparation. The samples were further processed by boiling with SDS sample buffer before storing them in (100 μl) aliquots. Briefly, the mouse brain cortex was dissected and homogenized with homogenizing media (0.32 M Sucrose, 1 mM EDTA, 10 mM Hepes) containing 1× Halt protease and phosphatase inhibitors (Thermo Fisher Scientific, Cat #-78440). The homogenate was then centrifuged at 1000g to pellet the nuclear fraction. The supernatant containing the post nuclear fraction was immediately diluted with 4× LDS sample Buffer (Thermo Fisher Scientific, Cat # NP0007) and heated at 95 °C for 10 min in a heating block and stored in aliquots (100 μl) at −80 °C to avoid multiple freezing and thawing. An aliquot of the post nuclear fraction was saved for protein estimation. For cell lysate preparation, the cells were harvested and washed with PBS (three times) and the cell pellet was collected by centrifugation. The cells were then sonicated in radioimmunoprecipitation assay buffer containing 1× Halt protease and phosphatase inhibitors and 50 U benzonaze/ml. The cell lysates were then processed with 4× LDS sample buffer.
Isolation of plasma membrane fractions
For isolation of the plasma membrane fractions from cortical tissues, a plasma membrane isolation kit (Abcam, Cat# ab65400) was used following the manufacturer’s protocol. Briefly, freshly isolated cortical tissues were homogenized in four volumes of 1× homogenizing buffer and centrifuged at 10,000g for 30 min in 1.5 ml tubes. The supernatants were discarded, and the pellets containing total membrane fractions were resuspended in 200 μl of upper phase solution. Two hundred microliters of lower phase solution was then added to each tube (tube A), mixed thoroughly, and kept in ice for 5 min. A fresh tube with 200 μl of upper and lower phase solution was similarly prepared for reference (tube B). The tubes were then centrifuged at 1000g for 5 min at 4 °C. The upper phase was carefully transferred from the sample tubes to new tubes (tube C). The extraction process was repeated by adding 100 μl of upper phase buffer to the sample tube (tube A) for maximizing the yield. The upper phase collected from two rounds of extraction was combined and mixed with 100 μl of lower phase solution and centrifuged at 1000g (10 min). The upper phase was carefully collected, diluted with five volumes of water, and kept in ice for 5 min. The samples were then centrifuged at 20,000g for 10 min at 4 °C, and the pellets enriched in plasma membrane were collected. The pellets were then resuspended in homogenising buffer and processed with 4× LDS sample buffer.
Lysosome purification
Lysosomes were purified from cortical tissues using Optiprep density gradient media provided in the lysosome isolation kit (LYSISO1; Sigma-Aldrich) following the supplier’s protocol with minor modifications as follows: freshly isolated cortical tissues were homogenized in four volumes of 1 × extraction buffer containing protease inhibitor and centrifuged for 1000g for 10 min. The supernatant was then further centrifuged at 20,000g for 20 min at 4 °C. The resultant pellet containing the Crude lysosomal fraction was resuspended in 1X extraction buffer layered over a multistep Optiprep gradient consisting of 27%, 22.5%, 19%, 16%, 12%, and 8% Optiprep. The gradient was centrifuged for 4 h at 150,000g using a swing-out rotor. The various layers, F1 (top layer) to F5 (bottom layer), formed at the junction of each gradient were collected after centrifugation. Fraction 2 and fraction 3 were found to be lysosome-enriched fractions and were combined and used for all assays including the immunoblotting experiments.
Plasmid constructs and transfection
Myc-tagged Lamtor1 plasmid (EX-Mm19894-M09) was purchased from GeneCopoeia. Lamtor1 mutant constructs were generated by Bioinnovatise using the primers listed in the Table 1. HEK293T cells were transfected with the plasmids using lipofectamine 3000 (Cat# L3000015, Invitrogen) as per the manufacturer’s protocol.
Table 1.
Reagent resources and list of primers
| Reagent or Resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| 4E-BP antibody sampler kit | Cell Signaling | Cat# 9955T; RRID: AB_823409 |
| Rabbit monoclonal anti-Akt (11E7) (Pan) | Cell Signaling | Cat#4685S; RRID: AB_2225340 |
| Rabbit monoclonal anti-phospho-Akt (S473) (D9E) | Cell Signaling | Cat#4060; RRID: AB_2315049 |
| Rabbit monoclonal anti-phospho-Akt (Thr308) (D25E6) | Cell Signaling | Cat#13038; RRID: AB_2629477 |
| Rabbit polyclonal anti-Atg13 | Proteintech | Cat#18258-1-AP; RRID: AB_2130658 |
| Rabbit monoclonal anti-phospho-Atg13 (Ser355) (E4D3T) | Cell Signaling | Cat#46329; RRID: AB_ |
| Rabbit polyclonal anti-Atg14/Barkor (N-treminal) | Proteintech | Cat#19491-1-AP; RRID: AB_10642701 |
| Rabbit monoclonal anti-phospho-Atg14 (Ser29) (D4B8M) | Cell Signaling | Cat#92340; RRID: AB_2800182 |
| Mouse monoclonal anti-CD68 (3F7D3) | Abcam | Cat# ab201973; RRID: AB_ |
| Rat monoclonal anti-Ctip2 | Abcam | Cat# ab18465; RRID: AB_2064130 |
| Rabbit polyclonal anti-CUX1 | Proteintech | Cat#11733-1-AP; RRID: AB_2086995 |
| Mouse monoclonal anti-GFAP (GA5) | Cell signaling | Cat#3670; RRID: AB_561049 |
| Mouse monoclonal anti-GSK-3ß (3D10) | Cell signaling | Cat#9832; RRID: AB_ |
| Rabbit monoclonal anti-GSK-3ß (D5C5Z) | Cell signaling | Cat#12456; RRID: AB_2636978 |
| Rabbit monoclonal anti-phospho-GSK-3ß (Ser9) (D85E12) | Cell signaling | Cat#5558; RRID: AB_ |
| Rabbit polyclonal anti-IBA1 | Invitrogen | Cat# PA5-27436; RRID: AB_ |
| Rabbit monoclonal IGF-1 receptor ß (D23H3) | Cell signaling | Cat#9750; RRID: |
| Mouse monoclonal anti-LAMP2 | Abcam | Cat# ab25631; RRID: AB_470709 |
| Rabbit monoclonal anti-LAMTOR1/C11 orf59 (D11H6) | Cell signaling | Cat#8975; RRID: AB_10860252 |
| Rabbit monoclonal anti-LAMTOR4/C7 orf59 (D4P6O) | Cell signaling | Cat#13140; RRID: AB_2798129 |
| Rabbit monoclonal Anti-LAMTOR5/HBXIP (D4V4S) | Cell signaling | Cat#14633; RRID: AB_2798547 |
| Rabbit monoclonal anti-LC3A/B (D3U4C) | Cell signaling | Cat#12741; RRID: AB_2617131 |
| Mouse monoclonal anti-Myc-Tag (9B11) | Cell signaling | Cat#2276; RRID: AB_331783 |
| Rabbit polyclonal anti-Na,K-ATPase | Cell signaling | Cat#3010; RRID: AB_2060983 |
| Mouse monoclonal anti-NeuN (E4M5P) | Cell signaling | Cat#94403; RRID: AB_2904530 |
| Rabbit monoclonal anti-PI3 Kinase p110α (C73F8) | Cell signaling | Cat#4249; RRID: AB_2165248 |
| Rabbit polyclonal anti-SQSTM1/p62 | Abcam | Cat#ab91526; RRID: AB_2050336 |
| Phospho-TSC2 antibody sampler kit | Cell signaling | Cat#8350; RRID: AB_10949106 |
| Rabbit polyclonal anti-phospho-PI3 Kinase p85 (Tyr458) p55 (Tyr199) | Cell signaling | Cat#4228; RRID: AB_659940 |
| Rabbit monoclonal anti-PRAS40 (D23C7) | Cell signaling | Cat#2691; RRID: AB_2225033 |
| Rabbit monoclonal anti-phospho-PRAS40 (Thr246) (C77D7) | Cell signaling | Cat#2997; RRID: AB_2258110 |
| Rabbit monoclonal anti-PTEN (D4.3) | Cell signaling | Cat#9188; RRID: AB_2253290 |
| Rabbit monoclonal anti-phospho-PTEN (Ser380/Thr382/383) (44A7) | Cell signaling | Cat#9549; RRID: AB_659891 |
| Rag and Lamtor antibody sampler kit | Cell signaling | Cat#8665; RRID: AB_ |
| Rabbit monoclonal anti-Rheb (E1G1R) | Cell signaling | Cat#13879; RRID: AB_2721022 |
| Rabbit monoclonal anti-p70 S6 kinase (49D7) | Cell signaling | Cat#2708; RRID: AB_390722 |
| Rabbit polyclonal anti-phospho- p70 S6 kinase (Thr389) | Cell signaling | Cat#9205; RRID: AB_330944 |
| Mouse monoclonal anti-β-Actin (8H10D10) | Cell signaling | Cat#3700; RRID: AB_2242334 |
| Rabbit monoclonal anti-TBC1D7 (D8K1Y) | Cell signaling | Cat#14949; RRID: AB_2749838 |
| Rabbit monoclonal anti-Hamartin/TSC1 (D43E2) | Cell signaling | Cat#6935; RRID: AB_10860420 |
| Rabbit monoclonal anti-ULK1 (D8H5) | Cell signaling | Cat#8054; RRID: AB_11178688 |
| Rabbit monoclonal anti-phospho-UKL1 (Ser757) (D7O6U) | Cell signaling | Cat#14202; RRID: AB_2665508 |
| Chemicals | ||
| B27 supplement | Invitrogen | Cat# 17504044 |
| GlutaMAX | Invitrogen | Cat#35050 |
| N-[6-(biotinamido) hexyl]-3′-(2′-pyridyldithio) propionamide-biotin | Thermo Fisher Scientific | Cat#21341 |
| PBS | Thermo Fisher Scientific | Cat#10010023 |
| Benzonase | Sigma-Aldrich | Cat# E1014 |
| β-Mercaptoethanol | Sigma-Aldrich | Cat #M6250 |
| Na2CO3 | Sigma-Aldrich | Cat#222321 |
| NaHCO3 | Sigma-Aldrich | Cat#1063290 |
| Hepes | Quality Biologicals | Cat#118–089–721 |
| EDTA | Corning | Cat#46–034-CI |
| Paraformaldehyde | Electron Microscopy Sciences | Cat#15710 |
| PEG-400 | Med Chem Express | Cat# HY-Y0873A |
| Afuresertib | Med Chem Express | Cat# HY-15727 |
| Uprosertib | Med Chem Express | Cat# HY-15965 |
| A-674563 | Selleckchem | Cat#S2670 |
| MK2206 | Selleckchem | Cat#S1078 |
| Recombinant Human PPT1 | Creative Biomart | Cat#PPT1-367H |
| RPMI | Gibco | Cat#11875-093 |
| Fetal bovine serum | Hyclone | Cat#SH30071.03 |
| Pen-Strep | Gibco | Cat#15140-122 |
| Fatty acid–free BSA | ChemCruz | Cat#SC-2323A |
| Halt protease inhibitor | Thermo Fisher Scientific | Cat#1861279 |
| Halt protease-phosphatase inhibitor | Thermo Fisher Scientific | Cat#-78440 |
| LDS sample buffer | Thermo Fisher Scientific | Cat# NP0007 |
| RIPA buffer | Thermo Fisher Scientific | Cat#89901 |
| N-Ethylmaleimide | Sigma-Aldrich | Cat#E3876 |
| Hydroxylamine hydrochloride | Sigma-Aldrich | Cat#159417 |
| Thiopropyl Sepharose | GE-Amersham | Cat#17-0420-01 |
| SuperSignal West Pico PLUS chemiluminescent substrate | Pierce, Thermo Fisher Scientific | Cat#34578 |
| SuperSignal West Femto maximum sensitivity substrate | Pierce, Thermo Fisher Scientific | Cat#34096 |
| Methanol | Sigma-Aldrich | Cat#34860 |
| Triton X-100 | Sigma-Aldrich | Cat#T8787 |
| TrueBlack® lipofuscin autofluorescence quencher | Biotium | Cat # 23007 |
| DAPI-fluoromount G | Thermo Fisher Scientific | Cat #010020 |
| Cresyl violet stain solution | Abcam | Cat#ab246817 |
| Sucrose | Sigma-Aldrich | Cat#S0389 |
| Critical commercial kits | ||
| Neuronal dissociation kit | Miltenyl Biotec | Cat#130-094-802 |
| Neuron isolation kit | Miltenyl Biotec | Cat#130-115-389 |
| Plasma membrane isolation kit | Abcam | Cat# ab65400 |
| Lysosome isolation kit | Sigma=-Aldrich | Cat#LYSISO1 |
| RNA easy Mini kit | QIAGEN | |
| Superscript III First–strand synthesis kit | Invitrogen | Cat#18080051 |
| RNeasy Mini Kit | Qiagen | Cat#74104 |
| IGF1 ELISA kits | Thermo Fisher Scientific | Cat# EMIGF1 |
| RheB activation assay kit | New East Biosciences | Cat# 81201 |
| Insulin ELISA kits | Thermo Fisher Scientific | Cat# EMINS |
| H&E kit | Abcam | Cat#ab245880 |
| Primers used for Lamtor1 mutagenesis | ||
| Lamtor1-C3A-5 CTTGAAGGAATTCGGTACCATGGGGGCCTGCTATAGCAGCGAAAACGAGGACTC |
||
| Lamtor1-C3A-3 GAGTCCTCGTTTTCGCTGCTATAGCAGGCCCCCATGGTACCGAATTCCTTCAAG |
||
| Lamtor1-C4A-5 GAAGGAATTCGGTACCATGGGGTGCGCCTATAGCAGCGAAAACGAGGACTCGG Lamtor1-C4A-3 CCGAGTCCTCGTTTTCGCTGCTATAGGCGCACCCCATGGTACCGAATTCCTTC |
||
| C3AC4A-5 GGCTTGAAGGAATTCGGTACCATGGGGGCCGCCTATAGCAGCGAAAACGAGGACTCGGAC C3AC4A-3 GTCCGAGTCCTCGTTTTCGCTGCTATAGGCGGCCCCCATGGTACCGAATTCCTTCAAGCC |
||
| Primers used for qRT-PCR | ||
| Lamtor1 F-CATTGATGTGTCTGCCGCAGAC R-CTTCCAATGGGTCAGACTGCTG |
||
| Lamtor2 F-ACCAAGCGTTTAATGAAGACAGTC R-TGAGCATTCCGAAGCCTACGGT |
||
| Lamtor3 F-AGACCTGGCTTCCTATCCACGT R-AACTCACCACCAGAGGCAAACG |
||
| Lamtor4 F-AGTGCCATCTCGGAGTTGGTCA R-ACCTCGGTTCTGCCTCTTCACT |
||
| Lamtor5 F-GTCCTATGCACAGATTCACAAGG R-ACATACCACAGGGATGTCGGTG |
| Primers used for qRT-PCR | Source | Identifier |
|---|---|---|
| Rag A F-TGCAAAGAGCAGCGAGATGT R-CATGCTCTGGAAAGAAGCGG |
||
| Rag B F-ATCGCGTCCGCTAGAATGTT R-TGGGCATCTCGCTGTTCTTT |
||
| Rag C F-GCAGTCATCAGACCAGTGCT R-CAGCAGAGAAACTCCACCGT |
||
| Rag D F-CCTTTGCAGTCACCTTGTGC R-GTGTTTTAGACCCCTCGGCA |
||
| GAPDH F-TGGCCTCCAAGGAGTAAGAA R-TGTGAGGGAGATGCTCAGTG |
||
| Plasmid | ||
| Myc-tagged Lamtor1 | GeneCopoeia | Cat#EX-Mm19894-M09 |
| Software and algorithm | ||
| ABI Prism software version 1.01 | Applied Biosystems | N/A |
| Image quant IQTL software | GE Healthcare Lifesciences | N/A |
| ZenDesk | Zeiss | N/A |
BSA, bovine serum albumin; DAPI, 4',6-diamidino--2-phenylindole; IGF1, insulin-like growth factor-1; PPT1, palmitoyl-protein thioesterase-1; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; RIPA, radioimmunoprecipitation assay.
RNA isolation and real-time RT-PCR
Total RNA was isolated from cortical tissues of WT and Cln1−/−mice using RNA easy Mini kit (QIAGEN). This was followed by complementary deoxy ribonucleic acid synthesis using Superscript III First-Strand Synthesis kit (Invitrogen) according to manufacturer’s instructions. The mRNAs were quantified and compared by real time RT-PCR with SYBR Green PCR mix using ABI Prism 7000 (https://resource.thermofisher.com/pages2013/WE111944/) Sequence detection system and analyzed by using ABI Prism software version 1.01 (Applied Biosystems). The Ct values were calculated using GAPDH as control. The primers used for quatitative reverse transcription-polymerase chain reaction are provided in the Table 1.
Acyl-RAC assay
Acyl Rac assay (74) was used to determine the S-palmitoylation of Lamtor1 with minor modifications. Briefly, 1 mg of protein (from cell lysate or tissue lysate) was diluted to 1 ml in blocking buffer (100 mM Hepes, 1.0 mM EDTA, 2.5% SDS, 50 mM N-ethylmaleimide, pH 7.5) and incubated at 50 °C (60 min) with mixing every 15 min. Proteins were extracted using three volumes of ice-cold acetone at −20 °C for 20 min. This was followed by centrifugation at 5000g for 10 min, and the pellet was extensively washed with 70% acetone (four times). The pellet was then resuspended in 250 μl of binding buffer (100 mM Hepes, 1.0 mM EDTA, 1% SDS, pH 7.5). Approximately 50 μl of resuspended protein was saved as the total input. The remaining protein sample was divided in two aliquots (100 μl each). One aliquot was treated with 100 μl of 2 M NH2OH (freshly prepared, pH 7.5), while the other aliquot was treated with 100 μl PBS (for control). Thiopropyl Sepharose (GE-Amersham) was activated by letting it swell in ultrapure water for 15 to 20 min and washed (2–3 times) with ultrapure water. Approximately 40 μl of prewashed and activated thiopropyl Sepharose was added to each of these aliquots, and the binding reactions were carried out on a rotator at room temperature for around 4 h. The unbound fraction was collected carefully to a separate tube as a nonpalmitoylated protein fraction. Resins were washed at least five times with binding buffer and then boiled (95 °C for 5 min) with 60 μl of 4 × loading buffer (NuPage 4 × LD). The bound and unbound fractions were then resolved by SDS-PAGE.
Western blot analysis
Protein samples (10–20 μg) were resolved by electrophoresis using 4 to 12% or 12% SDS-polyacrylamide gels under reducing and denaturing conditions, followed by electro-transfer to nitrocellulose or PVDF membranes. The membranes were blocked with 2% bovine serum albumin or 5% nonfat dry milk (Bio-Rad) and then subjected to immunoblot analysis using standard methods. The primary antibodies used for the immunoblots are listed in the Table 1. The blots were then probed with horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology) followed by detection using SuperSignal west femto or pico solution (Pierce, Thermo Scientific) according to the manufacturer's instructions. Image quant 4000 mini (GE Healthcare Lifesciences) was used to capture the chemiluminescent signals and the immunoblots were quantified using the image quant IQTL software (https://www.cytivalifesciences.com/en/us/shop/protein-analysis/molecular-imaging-for-proteins/imaging-software/imagequant-tl-10-2-analysis-software-p-28619). All experiments were repeated at least three to five times to confirm the reproducibility.
RheB activation assay
For RheB activation assay we used RheB activation assay kit (Cat # 81201, New East Biosciences) which uses confirmation-specific anti-RheB-GTP mouse mAb to measure RheB-GTP levels. Briefly, freshly prepared protein lysates (1 ml containing 1 mg/ml protein) from mouse cortical tissue were incubated with anti-RheB-GTP mouse mAb (2 ul) and 20 ul of protein A/G Agarose bead slurry for 1 h at 4 °C. Next, the protein A/G agarose beads were washed three times with 1X assay buffer and centrifugation at 5000g for 1 min. Finally, the precipitated RheB-GTP is detected through immunoblot analysis using anti-RheB antibody.
Quantitation of IGF1 and insulin levels
IGF1 and insulin in cortical lysates from mouse brain were measured using ELISA kits (Cat No # EMIGF1, Thermo Fisher Scientific) and (Cat No # EMINS, Thermo Fisher Scientific), respectively, using manufacturers protocol.
Immunocytochemistry and confocal imaging of cultured neurons
Primary Neurons isolated from P2 pups were cultured on chamber slides for 7 days in Neurobasal media containing B27 supplement. The cells were then washed two to three times with PBS, incubated at 37 °C and fixed using 4% paraformaldehyde or 100% methanol (in −20 °C for 15 min). Paraformaldehyde-fixed cells were permeabilized with 0.1% Triton X-100 in PBS. After blocking with 10% normal goat serum, the cells were incubated with primary antibodies (Table 1) overnight at 4 °C. The cells were then washed with PBS and incubated with Alexa Fluor–conjugated secondary antibodies (Life Technologies, Thermo Fisher Scientific) for 1 h at room temperature. Cells were mounted using 4',6-diamidino--2-phenylindole-fluoromount G (Thermo Fisher Scientific, 010020), and fluorescence was visualized with the Zeiss LSM 710 Inverted Meta confocal microscope (Carl Zeiss). The images were processed with the LSM image software (Carl Zeiss). Overlap colocalization coefficient was calculated using Zen Desk software from Zeiss (104). We have used the weighted colocalization coefficient for analysis.
Immunohistochemistry of brain sections
Frozen whole-brain sections (WT, untreated, and Afuresertib-treated Cln1−/− mice) were mounted with optimal cutting temperature compound and the specimens were kept at −80 ° C for at least 12 h before sectioning. Mounted samples (10 μm thick) were sectioned using a cryostat. For immunohistochemical analyses, tissue sections were washed with PBS and then all the sections were kept in methanol at −20 °C for 20 min. The tissue sections were then washed with 1 × PBS three times. The paraffin embedded sections (10 μm thick) were deparaffinized and hydrated. Then antigen retrieval was performed by using 5 mM Hepes buffer with 1 mM EDTA and 0.05% Triton X-100 at pH 8. They were then incubated at 80 °C for 10 min and blocked with 10% normal goat serum for 1 h. The sections were then incubated with TrueBlack Lipofuscin Autofluorescence Quencher (Biotium. Cat # 23007) for 30 s and washed three times with 1 × PBS to quench any background autofluorescence. The slides were then incubated with primary antibodies, overnight at 4 °C. Then slides were washed three times with 1 × PBS and incubated with Alexa Fluor–conjugated secondary antibodies (AF 488 and AF555, diluted 1:200) (Invitrogen) for 2 h at room temperature in the dark. Then the slides were washed with 1 × PBS (three times), and the sections were mounted with 4',6-diamidino--2-phenylindole-fluoromount-G mounting medium (Southern Biotech. Cat # 0100-20). Fluoromount G (Thermo Fisher Scientific, 010020) and fluorescence was visualized with the Zeiss LSM 710 Inverted Meta confocal microscope (Carl Zeiss). The images were processed with the LSM image software (https://www.zeiss.com/microscopy/en/products/software/zeiss-zen-lite.html, Carl Zeiss). Overlap colocalization coefficient was calculated using Zen Desk software (https://www.zeiss.com/microscopy/en/products/software/zeiss-zen-desk.html) from Zeiss (104).
H&E staining of brain sections
Whole-brain sagittal sections (5 μm thick) from 6-month-old WT, untreated, and Afuresertib-treated Cln1−/− mice were deparaffinized, hydrated and stained with H&E kit (ab245880, Abcam) using manufacturer’s protocol. At least four comparable sections from each brain were used for measurement of cortical thickness and a similar area (secondary visual cortex mediolateral) for all the sections was used.
Cresyl violet staining for Nissl bodies
Five micron thick whole-brain sagittal sections from 6-month-old WT, untreated, and Afuresertib-treated Cln1−/− mice were deparaffinized, hydrated and stained with Cresyl Violet Stain Solution (ab246817, Abcam) using manufacturer’s protocol. Nissl-positive cells were counted from four comparable sections from each group from similar area of the cortex for all the sections.
Evaluation of motor function by rotarod endurance test
Motor coordination of the WT, untreated, and Afuresertib-treated Cln1−/− mice was assessed using Rotarod) (UGO Basile) (91). For testing the endurance on rotarod, we used 6- and 8-month-old WT (n = 7), untreated (n = 7) and Afuresertib-treated Cln1−/− (n = 7) mice. Animals were habituated in the training room and trained (at Rotarod speed 8 rpm) twice for 1 min each for 4 days. Animals were given 1 min rest between two trials. On the 5th and the final day, the animals were designated as group A (WT), group B (untreated Cln1−/−) and group C (Afuresertib-treated Cln1−/−) The tests were performed in the same training room, by a single operator who was unaware of the identity of the groups. The animals were allowed to stay on the rotating rod for up to 300 s. All the training sessions and the final rotarod test were conducted at the same time in the morning between 9 AM and 11 AM.
Mycoplasma testing
All cell lines were tested for Mycoplasma using MycoFluor Mycoplasma Detection Kit (M-7006, Molecular Probes) and found to be free of contamination.
Statistical analysis
The data are represented as the mean ± SD and the “n” numbers denote the number of biological replicates for each experiment unless otherwise stated. For confocal imaging shown in Figure 3E "n" is the total number of images of cells used for analysis was acquired from four biological replicates. For Figure 6, D and E, Extended data Figs. S6D and S7B "n" numbers are the total number of fields used for analysis, which were acquired from four biological replicates. We used two-sample permutation t test with complete enumeration to calculate the p values to examine the differences between two independent groups. For n 3 we used t test to calculate the p value. p values <0.05 were considered statistically significant.
Data availability
Most of the raw data are provided with the manuscript. Any additional data will be shared upon request addressed to Dr Maria B Bagh (e-mail: baghmb@mail.nih.gov)/Dr Abhilash P Appu (e-mail: abhilash.appu@nih.gov).
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Supporting information
Supplemental information can be found online.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Dr Janice Y. Chou for critical review of the manuscript and helpful suggestions. The Cln1−/− mice, which established our mouse colony, were a generous gift from Dr Sandra L. Hofmann, Southwestern Medical Center, Dallas, Texas. We also thank Dr Vincent Schram, Microscopy and Imaging Core, for the expert assistance in confocal microscopy. We are also thankful to the Animal care facility, NICHD for their continuous support which made the animal experiments possible. This research was supported in full by the intramural program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), National Institutes of Health (NIH).
Author contributions
M. B. B. and A. B. M. conceptualization; M. B. B. and A. B. M. methodology; M. B. B., A. P. A., T. S., A. M., N. P., and S. S. investigation; M. B. B., A. P. A., T. S., A. M., N. P., M. R., A. M. S., S. S., A. L., and A. B. M. formal analysis; A. B. M. funding acquisition; M. B. B., A. P. A., T. S., A. M., N. P., M. R., A. M. S., S. S., A. L., and A. B. M. writing-original draft; M. B. B., A. P. A., T. S., A. M., N. P., M. R., A. M. S., S. S., A. L., and A. B. M. writing-review and editing.
Funding and additional information
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Biographies

Abhilash P. Appu completed his postdoctoral training at the NICHD, NIH, where he is promoted to a Research Fellow position. His research focuses on understanding the molecular mechanism(s) of pathogenesis of a group of the most common childhood neurodegenerative lysosomal storage disorders (LSDs), commonly known as neuronal ceroid lipofuscinoses (NCLs) or Batten disease. His current research focuses on understanding the mechanism of neurodegeneration in CLN1 disease. His areas of expertise include endosomal protein-trafficking, cholesterol homeostasis and mTOR activation.

Maria B. Bagh completed her postdoctoral fellowship at the NICHD, NIH, following which she was promoted to Staff Scientist. Her research was focused on understanding the pathogenic mechanism underlying a fatal childhood neurodegenerative lysosomal storage disorder (LSD), called CLN1 disease. She was interested in understanding the mechanism of neurodegeneration in this disease. Her areas of expertise include endo-lysosomal sorting, post-translational lipid modification (S-palmitoylation) and protein-protein interactions. Presently, Maria is a regulatory scientist at the Food and Drug Administration.
Reviewed by members of the JBC Editorial Board. Edited by Elizabeth J. Coulson
Supporting information
References
- 1.Laplante M., Sabatini D.M. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Platt F.M. Emptying the stores: lysosomal diseases and therapeutic strategies. Nat. Rev. Drug Discov. 2018;17:133–150. doi: 10.1038/nrd.2017.214. [DOI] [PubMed] [Google Scholar]
- 3.Fraldi A., Klein A.D., Medina D.L., Settembre C. Brain disorders due to lysosomal dysfunction. Annu. Rev. Neurosci. 2016;39:277–295. doi: 10.1146/annurev-neuro-070815-014031. [DOI] [PubMed] [Google Scholar]
- 4.Seranova E., Connolly K.J., Zatyka M., Rosenstock T.R., Barrett T., Tuxworth R.I., et al. Dysregulation of autophagy as a common mechanism in lysosomal storage diseases. Essays Biochem. 2017;61:733–749. doi: 10.1042/EBC20170055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Duve C. The lysosome turns fifty. Nat. Cell Biol. 2005;7:847–849. doi: 10.1038/ncb0905-847. [DOI] [PubMed] [Google Scholar]
- 6.Ballabio A., Bonifacino J.S. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Biol. 2020;21:101–118. doi: 10.1038/s41580-019-0185-4. [DOI] [PubMed] [Google Scholar]
- 7.Anderson G.W., Goebel H.H., Simonati A. Human pathology in NCL. Biochim. Biophys. Acta. 2013;1832:1807–1826. doi: 10.1016/j.bbadis.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 8.Haltia M., Goebel H.H. The neuronal ceroid lipofuscinoses: a historical introduction. Biochim. Biophys. Acta. 2013;1832:1795–1800. doi: 10.1016/j.bbadis.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Mole S.E., Anderson G., Band H.A., Berkovic S.F., Cooper J.D., Kleine Holthaus S.M., et al. Clinical challenges and future therapeutic approaches for neuronal ceroid lipofuscinosis. Lancet Neurol. 2019;18:107–116. doi: 10.1016/S1474-4422(18)30368-5. [DOI] [PubMed] [Google Scholar]
- 10.Mukherjee A.B., Appu A.P., Sadhukhan T., Casey S., Mondal A., Zhang Z., et al. Emerging new roles of the lysosome and neuronal ceroid lipofuscinoses. Mol. Neurodegener. 2019;14:1–23. doi: 10.1186/s13024-018-0300-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Specchio N., Ferretti A., Trivisano M., Pietrafusa N., Pepi C., Calabrese C., et al. Neuronal ceroid lipofuscinosis: potential for targeted therapy. Drugs. 2021;81:101–123. doi: 10.1007/s40265-020-01440-7. [DOI] [PubMed] [Google Scholar]
- 12.Rider J.A., Rider D.L. Batten disease: past, present, and future. Am. J. Med. Genet. Suppl. 1988;5:21–26. doi: 10.1002/ajmg.1320310606. [DOI] [PubMed] [Google Scholar]
- 13.Kousi M., Lehesjoki A.E., Mole S.E. Update of the mutation spectrum and clinical correlations of over 360 mutations in eight genes that underlie the neuronal ceroid lipofuscinoses. Hum. Mutat. 2012;33:42–63. doi: 10.1002/humu.21624. [DOI] [PubMed] [Google Scholar]
- 14.Santavuori P., Haltia M., Rapola J. Infantile type of so-called neuronal ceroid-lipofuscinosis. Dev. Med. Child Neurol. 1974;16:644–653. doi: 10.1111/j.1469-8749.1974.tb04183.x. [DOI] [PubMed] [Google Scholar]
- 15.Vesa J., Hellsten E., Verkruyse L.A., Camp L.A., Rapola J., Santavuori P., et al. Mutations in the palmitoyl protein thioesterase gene causing infantile neuronal ceroid lipofuscinosis. Nature. 1995;376:584–587. doi: 10.1038/376584a0. [DOI] [PubMed] [Google Scholar]
- 16.Levin S.W., Baker E.H., Zein W.M., Zhang Z., Quezado Z.M., Miao N., et al. Oral cysteamine bitartrate and N-acetylcysteine for patients with infantile neuronal ceroid lipofuscinosis: a pilot study. Lancet Neurol. 2014;13:777–787. doi: 10.1016/S1474-4422(14)70142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mole S.E. Development of new treatments for Batten disease. Lancet Neurol. 2014;13:749–751. doi: 10.1016/S1474-4422(14)70151-6. [DOI] [PubMed] [Google Scholar]
- 18.Camp L.A., Hofmann S.L. Purification and properties of a palmitoyl-protein thioesterase that cleaves palmitate from H-Ras. J. Biol. Chem. 1993;268:22566–22574. [PubMed] [Google Scholar]
- 19.Hellsten E., Vesa J., Olkkonen V.M., Jalanko A., Peltonen L. Human palmitoyl protein thioesterase: evidence for lysosomal targeting of the enzyme and disturbed cellular routing in infantile neuronal ceroid lipofuscinosis. EMBO J. 1996;15:5240–5245. [PMC free article] [PubMed] [Google Scholar]
- 20.Verkruyse L.A., Hofmann S.L. Lysosomal targeting of palmitoyl-protein thioesterase. J. Biol. Chem. 1996;271:15831–15836. doi: 10.1074/jbc.271.26.15831. [DOI] [PubMed] [Google Scholar]
- 21.Sapir T., Segal M., Grigoryan G., Hansson K.M., James P., Segal M., et al. The interactome of palmitoyl-protein thioesterase 1 (PPT1) affects neuronal morphology and function. Front. Cell Neurosci. 2019;13:92. doi: 10.3389/fncel.2019.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linder M.E., Deschenes R.J. Palmitoylation: policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt M.F. Fatty acylation of proteins. Biochim. Biophys. Acta. 1989;988:411–426. doi: 10.1016/0304-4157(89)90013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukata Y., Fukata M. Protein palmitoylation in neuronal development and synaptic plasticity. Nat. Rev. Neurosci. 2010;11:161–175. doi: 10.1038/nrn2788. [DOI] [PubMed] [Google Scholar]
- 25.Zhang M.M., Hang H.C. Protein S-palmitoylation in cellular differentiation. Biochem. Soc. Trans. 2017;45:275–285. doi: 10.1042/BST20160236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salaun C., Greaves J., Chamberlain L.H. The intracellular dynamic of protein palmitoylation. J. Cell Biol. 2010;191:1229–1238. doi: 10.1083/jcb.201008160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorenberg E.L., Massaro Tieze S., Yücel B., Zhao H.R., Chou V., Wirak G.S., et al. Identification of substrates of palmitoyl protein thioesterase 1 highlights roles of depalmitoylation in disulfide bond formation and synaptic function. PLoS Biol. 2022;20 doi: 10.1371/journal.pbio.3001590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saxton R.A., Sabatini D.M. mTOR signaling in growth, metabolism, and disease. Cell. 2017 doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Settembre C., Fraldi A., Medina D.L., Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 2013;14:283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawrence R.E., Zoncu R. The lysosome as a cellular centre for signalling, metabolism and quality control. Nat. Cell Biol. 2019;21:133–142. doi: 10.1038/s41556-018-0244-7. [DOI] [PubMed] [Google Scholar]
- 31.Liu G.Y., Sabatini D.M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 2020;21:183–203. doi: 10.1038/s41580-019-0199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cuervo A.M. Autophagy: in sickness and in health. Trends Cell Biol. 2004;14:70–77. doi: 10.1016/j.tcb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 33.Shin H.R., Zoncu R. The lysosome at the intersection of cellular growth and destruction. Dev. Cell. 2020;54:226–238. doi: 10.1016/j.devcel.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawrence R.E., Cho K.F., Rappold R., Thrun A., Tofaute M., Kim D.J., et al. A nutrient-induced affinity switch controls mTORC1 activation by its Rag GTPase-Ragulator lysosomal scaffold. Nat. Cell Biol. 2018;20:1052–1063. doi: 10.1038/s41556-018-0148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martina J.A., Chen Y., Gucek M., Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 37.Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee J.H., Yang D.S., Goulbourne C.N., Im E., Stavrides P., Pensalfini A., et al. Faulty autolysosome acidification in Alzheimer's disease mouse models induces autophagic build-up of Aβ in neurons, yielding senile plaques. Nat. Neurosci. 2022;25:688–701. doi: 10.1038/s41593-022-01084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nixon R.A. The role of autophagy in neurodegenerative disease. Nat. Med. 2013;19:983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- 40.Gupta P., Soyombo A.A., Atashband A., Wisniewski K.E., Shelton J.M., Richardson J.A., et al. Disruption of PPT1 or PPT2 causes neuronal ceroid lipofuscinosis in knockout mice. Proc. Natl. Acad. Sci. U. S. A. 2001;98:13566–13571. doi: 10.1073/pnas.251485198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bible E., Gupta P., Hofmann S.L., Cooper J.D. Regional and cellular neuropathology in the palmitoyl protein thioesterase-1 null mutant mouse model of infantile neuronal ceroid lipofuscinosis. Neurobiol. Dis. 2004;16:346–359. doi: 10.1016/j.nbd.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 42.Brown E.J., Albers M.W., Shin T.B., Ichikawa K., Keith C.T., Lane W.S., et al. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 43.Sabatini D.M., Erdjument-Bromage H., Lui M., Tempst P., Snyder S.H. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 44.Sabers C.J., Martin M.M., Brunn G.J., Williams J.M., Dumont F.J., Wiederrecht G., et al. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J. Biol. Chem. 1995;270:815–822. doi: 10.1074/jbc.270.2.815. [DOI] [PubMed] [Google Scholar]
- 45.Dibble C.C., Manning B.D. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat. Cell Biol. 2013;15:555–564. doi: 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deleyto-Seldas N., Efeyan A. The mTOR-autophagy Axis and the control of metabolism. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.655731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lipton J.O., Sahin M. The neurology of mTOR. Neuron. 2014;84:275–291. doi: 10.1016/j.neuron.2014.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Menzies F.M., Fleming A., Caricasole A., Bento C.F., Andrews S.P., Ashkenazi A., et al. Autophagy and neurodegeneration: pathogenic mechanisms and therapeutic opportunities. Neuron. 2017;93:1015–1034. doi: 10.1016/j.neuron.2017.01.022. [DOI] [PubMed] [Google Scholar]
- 49.Fleming A., Bourdenx M., Fujimaki M., Karabiyik C., Krause G.J., Lopez A., et al. The different autophagy degradation pathways and neurodegeneration. Neuron. 2022;110:935–966. doi: 10.1016/j.neuron.2022.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoffmann A.A., Willi Y. Detecting genetic responses to environmental change. Nat. Rev. Genet. 2008;9:421–432. doi: 10.1038/nrg2339. [DOI] [PubMed] [Google Scholar]
- 51.Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 52.Efeyan A., Zoncu R., Chang S., Gumper I., Snitkin H., Wolfson R.L., et al. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature. 2013;493:679–683. doi: 10.1038/nature11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bar-Peled L., Chantranupong L., Cherniack A.D., Chen W.W., Ottina K.A., Grabiner B.C., et al. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Efeyan A., Comb W.C., Sabatini D.M. Nutrient-sensing mechanisms and pathways. Nature. 2015;517:302–310. doi: 10.1038/nature14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim E., Goraksha-Hicks P., Li L., Neufeld T.P., Guan K.L. Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sancak Y., Peterson T.R., Shaul Y.D., Lindquist R.A., Thoreen C.C., Bar-Peled L., et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Z., Mandal A.K., Wang N., Keck C.L., Zimonjic D.B., Popescu N.C., et al. Palmitoyl-protein thioesterase gene expression in the developing mouse brain and retina: implications for early loss of vision in infantile neuronal ceroid lipofuscinosis. Gene. 1999;231:203–211. doi: 10.1016/s0378-1119(99)00050-5. [DOI] [PubMed] [Google Scholar]
- 58.Parenti G., Medina D.L., Ballabio A. The rapidly evolving view of lysosomal storage diseases. EMBO Mol. Med. 2021;13 doi: 10.15252/emmm.202012836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu L., Chen Y., Tooze S.A. Autophagy pathway: cellular and molecular mechanisms. Autophagy. 2018;14:207–215. doi: 10.1080/15548627.2017.1378838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perera R.M., Zoncu R. The lysosome as a regulatory hub. Annu. Rev. Cell Dev. Biol. 2016;32:223–253. doi: 10.1146/annurev-cellbio-111315-125125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim Y.C., Guan K.L. mTOR: a pharmacologic target for autophagy regulation. J. Clin. Invest. 2015;125:25–32. doi: 10.1172/JCI73939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mindell J.A. Lysosomal acidification mechanisms. Annu. Rev. Physiol. 2012;74:69–86. doi: 10.1146/annurev-physiol-012110-142317. [DOI] [PubMed] [Google Scholar]
- 63.Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 64.Abu-Remaileh M., Wyant G.A., Kim C., Laqtom N.N., Abbasi M.S., Chan H., et al. Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science. 2017;358:807–813. doi: 10.1126/science.aan6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schweitzer L.D., Comb W.C., Bar-Peled L., Sabatini D.M. Disruption of the rag-ragulator complex by c17orf59 inhibits mTORC1. Cell Rep. 2015;12:1445–1455. doi: 10.1016/j.celrep.2015.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bagh M.B., Peng S., Chandra G., Zhang Z., Singh S.P., Pattabiraman N., et al. Misrouting of v-ATPase subunit V0a1 dysregulates lysosomal acidification in a neurodegenerative lysosomal storage disease model. Nat. Commun. 2017;8 doi: 10.1038/ncomms14612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rebsamen M., Pochini L., Stasyk T., de Araújo M.E., Galluccio M., Kandasamy R.K., et al. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature. 2015;519:477–481. doi: 10.1038/nature14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang S., Tsun Z.Y., Wolfson R.L., Shen K., Wyant G.A., Plovanich M.E., et al. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 2015;347:188–194. doi: 10.1126/science.1257132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martin B.R., Cravatt B.F. Large-scale profiling of protein palmitoylation in mammalian cells. Nat. Methods. 2009;6:135–138. doi: 10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanders S.S., De Simone F.I., Thomas G.M. mTORC1 signaling is palmitoylation-dependent in hippocampal neurons and non-neuronal cells and involves dynamic palmitoylation of LAMTOR1 and mTOR. Front. Cell Neurosci. 2019;13:115. doi: 10.3389/fncel.2019.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yonehara R., Nada S., Nakai T., Nakai M., Kitamura A., Ogawa A., et al. Structural basis for the assembly of the Ragulator-Rag GTPase complex. Nat. Commun. 2017;8:1625. doi: 10.1038/s41467-017-01762-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brigidi G.S., Bamji S.X. Detection of protein palmitoylation in cultured hippocampal neurons by immunoprecipitation and acyl-biotin exchange (ABE) J. Vis. Exp. 2013;72 doi: 10.3791/50031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ren J., Wen L., Gao X., Jin C., Xue Y., Yao X. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng. Des. Sel. 2008;21:639–644. doi: 10.1093/protein/gzn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Forrester M.T., Hess D.T., Thompson J.W., Hultman R., Moseley M.A., Stamler J.S., et al. Site-specific analysis of protein S-acylation by resin-assisted capture. J. Lipid Res. 2011;52:393–398. doi: 10.1194/jlr.D011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dibble C.C., Cantley L.C. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 2015;25:545–555. doi: 10.1016/j.tcb.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Menon S., Dibble C.C., Talbott G., Hoxhaj G., Valvezan A.J., Takahashi H., et al. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell. 2014;156:771–785. doi: 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Manning B.D., Toker A. AKT/PKB signaling: navigating the network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Orlova K.A., Crino P.B. The tuberous sclerosis complex. Ann. N. Y Acad. Sci. 2010;1184:87–105. doi: 10.1111/j.1749-6632.2009.05117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Crino P.B. mTOR: a pathogenic signaling pathway in developmental brain malformations. Trends Mol. Med. 2011;17:734–742. doi: 10.1016/j.molmed.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 80.Bassetti D., Luhmann H.J., Kirischuk S. Effects of mutations in TSC genes on neurodevelopment and synaptic transmission. Int. J. Mol. Sci. 2021;22:7273. doi: 10.3390/ijms22147273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zordan P., Cominelli M., Cascino F., Tratta E., Poliani P.L., Galli R. Tuberous sclerosis complex-associated CNS abnormalities depend on hyperactivation of mTORC1 and Akt. J. Clin. Invest. 2018;128:1688–1706. doi: 10.1172/JCI96342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yuskaitis C.J., Modasia J.B., Schrötter S., Rossitto L.A., Groff K.J., Morici C., et al. DEPDC5-dependent mTORC1 signaling mechanisms are critical for the anti-seizure effects of acute fasting. Cell Rep. 2022;40 doi: 10.1016/j.celrep.2022.111278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang J., Manning B.D. A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem. Soc. Trans. 2009;37:217–222. doi: 10.1042/BST0370217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang H., Jiang X., Li B., Yang H.J., Miller M., Yang A., et al. Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature. 2017;552:368–373. doi: 10.1038/nature25023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang L., Harris T.E., Lawrence J.C., Jr. Regulation of proline-rich Akt substrate of 40 kDa (PRAS40) function by mammalian target of rapamycin complex 1 (mTORC1)-mediated phosphorylation. J. Biol. Chem. 2008;283:15619–15627. doi: 10.1074/jbc.M800723200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Alessi D.R., Andjelkovic M., Caudwell B., Cron P., Morrice N., Cohen P., et al. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 87.Rabanal-Ruiz Y., Byron A., Wirth A., Madsen R., Sedlackova L., Hewitt G., et al. mTORC1 activity is supported by spatial association with focal adhesions. J. Cell Biol. 2021;220 doi: 10.1083/jcb.202004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Manifava M., Smith M., Rotondo S., Walker S., Niewczas I., Zoncu R., et al. Dynamics of mTORC1 activation in response to amino acids. Elife. 2016;5 doi: 10.7554/eLife.19960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Uko N.E., Güner O.F., Matesic D.F., Bowen J.P. Akt pathway inhibitors. Curr. Top Med. Chem. 2020;20:883–900. doi: 10.2174/1568026620666200224101808. [DOI] [PubMed] [Google Scholar]
- 90.Labzin L.I., Heneka M.T., Latz E. Innate immunity and neurodegeneration. Annu. Rev. Med. 2018;69:437–449. doi: 10.1146/annurev-med-050715-104343. [DOI] [PubMed] [Google Scholar]
- 91.Ransohoff R.M., Brown M.A. Innate immunity in the central nervous system. J. Clin. Invest. 2012;122:1164–1171. doi: 10.1172/JCI58644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sadhukhan T., Bagh M.B., Appu A.P., Mondal A., Zhang W., Liu A., et al. In a mouse model of INCL reduced S-palmitoylation of cytosolic thioesterase APT1 contributes to microglia proliferation and neuroinflammation. J. Inherit. Metab. Dis. 2021;44:1051–1069. doi: 10.1002/jimd.12379. [DOI] [PubMed] [Google Scholar]
- 93.Shyng C., Sands M.S. Astrocytosis in infantile neuronal ceroid lipofuscinosis: friend or foe? Biochem. Soc. Trans. 2014;42:1282–1285. doi: 10.1042/BST20140188. [DOI] [PubMed] [Google Scholar]
- 94.Hamm R.J., Pike B.R., O'Dell D.M., Lyeth B.G., Jenkins L.W. The rotarod test: an evaluation of its effectiveness in assessing motor deficits following traumatic brain injury. J. Neurotrauma. 1994;11:187–196. doi: 10.1089/neu.1994.11.187. [DOI] [PubMed] [Google Scholar]
- 95.Bar-Peled L., Schweitzer L.D., Zoncu R., Sabatini D.M. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sancak Y., Bar-Peled L., Zoncu R., Markhard A.L., Nada S., Sabatini D.M. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Parenti G., Andria G., Ballabio A. Lysosomal storage diseases: from pathophysiology to therapy. Annu. Rev. Med. 2015;66:471–486. doi: 10.1146/annurev-med-122313-085916. [DOI] [PubMed] [Google Scholar]
- 98.Goldsmith J.,A., Ordureau J.W., Harper E., Holzbaur L.F. Brain-derived autophagosome profiling reveals the engulfment of nucleoid-enriched mitochondrial fragments by basal autophagy in neurons. Neuron. 2022;110:967–976.e8. doi: 10.1016/j.neuron.2021.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Switon K., Kotulska K., Janusz-Kaminska A., Zmorzynska J., Jaworski J. Molecular neurobiology of mTOR. Neuroscience. 2016;341:112–153. doi: 10.1016/j.neuroscience.2016.11.017. [DOI] [PubMed] [Google Scholar]
- 100.Zoncu R., Bar-Peled L., Efeyan A., Wang S., Sancak Y., Sabatini D.M. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Toker A. Phosphoinositide 3-kinase- a historical perspective. Subcell. Biochem. 2012;58:95–110. doi: 10.1007/978-94-007-3012-0_4. [DOI] [PubMed] [Google Scholar]
- 102.Cianciulli A., Porro C., Calvello R., Trotta T., Lofrumento D.D., Panaro M.A. Microglia mediated neuroinflammation: focus on PI3K modulation. Biomolecules. 2020;10:137. doi: 10.3390/biom10010137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Blaustein M., Piegari E., Martínez Calejman C., Vila A., Amante A., Manese M.V., et al. Akt is S-palmitoylated: a new layer of regulation for Akt. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.626404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bolte S., Cordelières F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Most of the raw data are provided with the manuscript. Any additional data will be shared upon request addressed to Dr Maria B Bagh (e-mail: baghmb@mail.nih.gov)/Dr Abhilash P Appu (e-mail: abhilash.appu@nih.gov).