Abstract
Studies were undertaken to examine hepatocyte CD14 expression during endotoxemia. Our results show that lipopolysaccharide (LPS) treatment in vivo caused a marked upregulation in CD14 mRNA and protein levels in rat hepatocytes. Detectable increases in mRNA were seen as early as 1.5 h after LPS treatment; these increases peaked at 20-fold by 3 h and returned to baseline levels by 24 h. In situ hybridization localized the CD14 mRNA expression to hepatocytes both in vitro and in vivo. Increases in hepatic CD14 protein levels were detectable by 3 h and peaked at 12 h. Hepatocytes from LPS-treated animals expressed greater amounts of cell-associated CD14 protein, and more of the soluble CD14 was released by hepatocytes from LPS-treated rats in vitro. The increases in hepatocyte CD14 expression during endotoxemia occurred in parallel to increases of CD14 levels in plasma. To provide molecular identification of the hepatocyte CD14, we cloned the rat liver CD14 cDNA. The longest clone consists of a 1,591-bp insert containing a 1,116-bp open reading frame. The deduced amino acid sequence is 372 amino acids long, has 81.8 and 62.8% homology to the amino acid sequences of mouse and human CD14, respectively, and is identical to the rat macrophage CD14. The expressed CD14 protein from this clone was functional, as indicated by NF-κB activation in response to LPS and fluorescein isothiocyanate-LPS binding in CHO cells stably transfected with rat CD14. A nuclear run-on assay showed that CD14 transcription rates were significantly increased in hepatocytes from LPS-treated animals, indicating that the upregulation in CD14 mRNA levels observed in rat hepatocytes after LPS treatment is dependent, in part, on increased transcription. In vitro and in vivo experiments indicated that interleukin-1β and/or tumor necrosis factor α participate in the upregulation of CD14 mRNA levels in hepatocytes. Our data indicate that hepatocytes express CD14 and that hepatocyte CD14 mRNA and protein levels increase rapidly during endotoxemia. Our observations also support the idea that soluble CD14 is an acute-phase protein and that hepatocytes could be a source for soluble CD14 production.
Sepsis due to gram-negative bacterial infection remains a major cause of mortality. Gram-negative bacteria release endotoxins (lipopolysaccharide [LPS]) which elicit an acute inflammatory reaction (reviewed in reference 7). LPS exerts its profound effect on the host by activating LPS-sensitive cells such as monocytes and endothelial cells to release various cytokines, lipid mediators, and free radicals (reviewed in reference 52). During the past decade, great progress has been made in identifying LPS recognition and signaling molecules. Although several LPS-binding and putative signaling molecules have been discovered, including CD11b/CD18 (66), acetylated-low-density lipoprotein receptor (23), and an 80-kDa receptor (29), CD14 is thought to be the most important LPS recognition molecule that is responsible for the activation of cells by pathophysiological levels of LPS. CD14 was first identified as a monocyte differentiation marker expressed on the surface of macrophages, neutrophils, and other myeloid-linkage cells (60). Membrane-bound CD14 (mCD14), which attaches to the cell surface by a glycosyl-phosphatidylinositol anchor (24), initiates the activation of macrophages for cytokine synthesis by LPS (10). A soluble form of CD14 (sCD14) is present in plasma at concentrations of 3 to 5 μg/ml (5, 22, 26). Soluble CD14 is required for LPS-induced responses by endothelial cells (27), epithelial cells (46), and smooth-muscle cells (39). The interaction of LPS with mCD14 or sCD14 is greatly facilitated by another plasma protein, LPS-binding protein (53), which is thought to be derived primarily from hepatocytes (56). Experimental and clinical studies have indicated that the plasma levels of sCD14 can increase by up to 75% during infection or trauma (35–37). Although shedding from leukocytes has been proposed as the major source of systemic sCD14 levels, it is likely that other sources exist. Furthermore, plasma proteins which change in circulation by more than 25% during infection fit the definition of acute-phase reactants (65), suggesting that sCD14 behaves like other acute-phase proteins.
Hepatocytes are the major source of most acute-phase proteins (65). If in fact sCD14 is an acute-phase protein, then hepatocytes might be expected to express CD14, which is upregulated during endotoxemia. Furthermore, hepatocytes isolated from endotoxemic animals exhibit markedly enhanced responses to LPS (45, 61), raising the possibility that these cells could express CD14. To determine whether hepatocytes express CD14, experiments were undertaken to measure steady-state CD14 mRNA and protein levels in hepatocytes from normal and endotoxemic animals. We show here that rat hepatocytes express CD14 mRNA and protein and that these levels are markedly upregulated during endotoxemia. Furthermore, the upregulation is regulated by the cytokines interleukin-1β (IL-1β) and tumor necrosis factor alpha (TNF-α), at least in part through transcriptional mechanisms. Our data provide evidence that hepatocytes exhibit the regulated expression of CD14.
MATERIALS AND METHODS
Reagents.
LPS (Escherichia coli O111:B4) and fluorescein isothiocyanate (FITC)-LPS were purchased from Sigma Chemical Company (St. Louis, Mo.); William’s medium E was purchased from Gibco (Grand Island, N.Y.); recombinant human IL-6 was purchased from Genzyme (Boston, Mass.); recombinant murine IL-1β and TNF-α were obtained from the National Cancer Institute (Craig Reynolds); fetal bovine serum was purchased from HyClone Laboratories (Logan, Utah). All tissue culture plates and flasks were purchased from Corning (Corning, N.Y.); [α-32P]dCTP and [α-32P]UTP were purchased from NEN Life Science (Boston, Mass.); the ribonucleotide triphosphates (ATP, GTP, and UTP) and poly(A) were purchased from Boehringer Mannheim (Indianapolis, Ind.). The antibodies against NF-κB P65, P50 subunits, and NF-κB consensus oligonucleotide were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). A hamster anti-mouse CD14 monoclonal antibody, G5A10, was generously provided by Regine Landmann (Department of Research and Internal Medicine, University Hospital, Basel, Switzerland).
Animals.
Male Sprague-Dawley rats, which were pathogen-free and weighed approximately 200 g each, were purchased from Harlan Sprague-Dawley (Indianapolis, Ind.). The rats were exposed each day to 12 h of light and darkness. Rodent chow and water were provided ad libitum. Experimental protocols were approved by the Institutional Animal Care and Use Committee of the University of Pittsburgh.
Hepatocyte isolation.
Hepatocytes were isolated from normal and LPS-injected rats by an in situ collagenase (type VI; Sigma) perfusion technique, modified as described previously (54, 64). Hepatocytes were separated from the nonparenchymal cells by two cycles of differential centrifugation (50 × g for 2 min) and further purified over a 30% Percoll gradient. Hepatocyte purity exceeded 98% as assessed by light microscopy, and viability was typically greater than 95% as determined by trypan blue exclusion assay.
Cell culture and treatment.
Hepatocytes (5 × 106) were plated onto 10-cm, gelatin-coated, plastic tissue culture dishes. The initial culture medium was William’s medium E containing 10% calf serum (CS), 15 mM HEPES, 10−6 M insulin, 2 mM l-glutamine, and 100 U of penicillin and streptomycin per ml. Hepatocytes were allowed to attach to plates overnight and then were washed three times with serum-free medium prior to cytokine treatment. Cytokine treatments were performed in serum-free medium. All cytokine concentrations were 500 U/ml. The LPS concentration was 0.1 μg/ml. Chinese Hamster ovary cells (CHO; American Type Culture Collection) were cultured in Ham F-12 medium containing 10% fetal bovine serum.
Total RNA isolation and Northern blot analysis.
Total RNA was extracted by RNAzol as described previously (61). For Northern blot analysis, total RNA (20 μg per lane) was resolved by electrophoresis in a 1% agarose gel containing 2.2 M formaldehyde prior to being transferred to the GeneScreen membrane (Dupont, NEN Research Products, Boston, Mass.). The mRNA was cross-linked to the membrane with UV Stratalinker (Stratagene, San Diego, Calif.) and then hybridized with 32P-labeled probes. The probe for CD14 was a 1,043-bp NotI fragment of mouse CD14 cDNA (15). The probe was labeled by random prime labeling (Boehringer Mannheim). Hybridization was carried out for 16 to 18 h in buffer containing 50% deionized formamide, 0.25 M sodium phosphate (pH 7.2), 0.25 M sodium chloride, 1 mM EDTA, 7% sodium dodecyl sulfate (SDS), and 100 μg of denatured salmon sperm DNA per ml. Blots were washed sequentially in 2× SSC (1× SSC is 0.015 M NaCl plus 0.015 M sodium citrate)–0.1% SDS, 25 mM NaHPO4–1 mM EDTA–0.1% SDS, and 25 mM NaHPO4–1 mM EDTA–1% SDS buffers. Blots were washed for 15 min twice in each buffer at 53°C prior to exposure to X-ray film for autoradiography. Blots were stripped before hybridizing with another probe.
Quantitation of mRNA levels.
mRNA levels on blots were quantitated with PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) scanning. The relative mRNA levels were normalized to 18S RNA. The mRNA levels in treatment groups were expressed as the fold increase over the controls.
Western blot analysis of CD14 protein in hepatocytes, whole liver extracts, culture supernatants, and plasma.
Cultured hepatocytes on plates were washed twice with phosphate-buffered saline (PBS), scraped, and pelleted by centrifugation. Cell pellets were resuspended in 50 μl of lysis buffer containing 20 mM HEPES (pH 7.9), 25% glycerol, 0.42 M NaCl, 15 mM MgCl2, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), and 0.5 mM dithiothreitol (DTT). After three freeze-thaw cycles, cell lysates were centrifuged at 12,000 × g for 30 min, and the supernatant was saved. The liver tissue was homogenized with a Dounce homogenizer before the freeze-thaw lysis, as described above for cultured hepatocytes. Blood samples were retrieved by cardiac puncture, and plasma was obtained following centrifugation at 1,000 × g for 5 min. Supernatants of cultured hepatocytes were concentrated by Centricon-30 (Amicon, Beverly, Mass.), according to the manufacturer’s instructions, prior to Western blot analysis.
For Western blot analysis, samples (50 μg per lane) were separated on an SDS–10% polyacrylamide gel and transferred to nitrocellulose membrane. The membrane was sequentially blocked in PBS-Tween (0.1%) containing 5% milk and then incubated with 5 μg of G5A10 (a hamster anti-mouse CD14 monoclonal antibody) per ml, washed, and further incubated with a goat anti-Armenian hamster immunoglobulin G horseradish peroxidase-conjugated secondary antibody (1:5,000; Jackson ImmunoResearch Laboratories, West Grove, Pa.). Blocking and antibody incubations each lasted 1 h at room temperature. After several washes, the membrane was developed with an enhanced luminol reagent (DuPont, NEN) and exposed to Kodak X-Omat film.
In situ hybridization for cultured hepatocytes.
Hepatocytes grown on glass coverslips were fixed in 2% paraformaldehyde in PBS for 10 min, permeated in buffer containing 2% paraformaldehyde–0.01% Triton X-100 for 10 min, washed in PBS, and then postfixed in PBS containing 4% paraformaldehyde. Prior to hybridization, the slides were treated with proteinase K (10 μg/ml) for 10 min, acetylated with 0.25% acetic anhydride, washed in 2× SSC, and dehydrated through graded alcohols. Radiolabeled riboprobe for CD14 was prepared as described previously (68) with cloned rat hepatocyte CD14 cDNA as a template as described below. Hybridization with sense and antisense probes, slide washings, and emulsion autoradiography were performed as described previously (49).
In situ hybridization for sectioned liver.
In situ hybridization on sectioned liver samples was performed with digoxigenin-labeled rather than radiolabeled probe. Linearized rat CD14 cDNA template was incubated in the presence of digoxigenin-labeled UTP and unlabeled CTP, ATP, and GTP, along with the relevant RNA polymerase for 2 h at 37°C. The labeled RNA was precipitated in ethanol, dried, and resuspended in diethyl pyrocarbonate-treated water. Whole livers were perfusion-fixed in 2% paraformaldehyde and cryoprotected by immersion in 30% sucrose overnight. The whole tissue was then frozen in liquid nitrogen-cooled isopentane. Frozen sections were cut (5 μm), fixed in 2% paraformaldehyde in PBS, washed twice in PBS, digested with proteinase K (10 mg/ml, 5 min), washed in PBS containing 1% glycine, and acetylated. After dehydration through graded alcohols, the sections were hybridized overnight at 42°C in digoxigenin-labeled, CD14-specific riboprobe (the controls were sense probe or no probe). Sections were then washed twice in 50% formamide–2× SSC for 15 min at 50°C and digested with RNase A for 30 min at 37°C. After further sequential washes in 50% formamide–2× SSC and then 2× SSC, sections were washed in Genius buffer I and Genius buffer II (Boehringer Mannheim) and incubated with alkaline phosphate-conjugated antibodies against digoxigenin for 1 h. Sections were then washed in Genius buffer III, developed in nitroblue tetrazolium (Sigma) overnight, washed in PBS, dehydrated, and mounted in Permount.
Preparation of rat hepatocyte nuclei.
Nuclei were isolated by the method described by Boggaram and Mendelson (4). Freshly isolated rat hepatocytes were homogenized with a Dounce homogenizer in a buffer containing 0.25 M sucrose, 10 mM HEPES (pH 8.0), 10 mM MgCl2, 0.1% Triton X-100, and 2 mM DTT. After five washes at 600 × g and 4°C, the nuclei were purified by centrifugation on a 1.3 M sucrose cushion in the homogenization buffer at 10,000 × g for 10 min. Finally, nuclei were resuspended in 50 mM HEPES (pH 8.0), 40% glycerol, 5 mM MgCl2, 0.1 mM EDTA, and 2 mM DTT (108 nuclei/ml) and then snap frozen in liquid nitrogen. The nuclei were stored at −80°C until use.
Nuclear run-on assay.
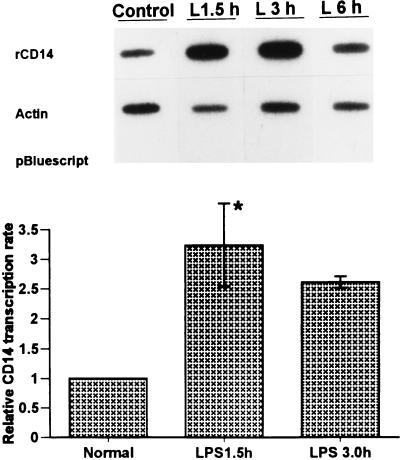
In order to determine the CD14 transcription rate, in vitro labeling of nascent nuclear RNA was performed as described previously (4, 8), with some modifications. Briefly, nuclei (2.1 × 107) were incubated at 30°C for 30 min in 300 μl of reaction mixture containing 5 mM Tris-HCl (pH 8.0); 2.5 mM MgCl2; 150 mM KCl; 0.25 mM (each) GTP, ATP, and CTP; 25 μl of [α-32P]UTP (250 μCi, specific activity of 3,000 Ci/mmol; NEN Life Science); and 1.0 μl of RNasin (40 U/μl; Promega). Radiolabeled nuclear RNA was extracted from the reaction mixture by Trizol (Gibco-BRL) according to the manufacturer’s instructions. The labeled nuclear RNA was purified on a G-50 Sephadex (Pharmacia) column.
Linearized plasmid containing rat CD14 cDNA was applied to GeneScreenPlus membrane (NEN Life Science) via a slot-blot apparatus (Schleicher & Schuell Minifold II; Keene, N.H.). Linearized vector pBluescript and plasmid-containing actin cDNA were also included as internal controls. Plasmids were denatured with 0.3 M NaOH at 60°C for 1 h before loading (5 μg per slot). After UV cross-linking, the blot was prehybridized at 43°C in buffer containing 50% deionized formamide, 4× SSC, 2× Denhardt’s solution, 50 mM PIPES (pH 7.0), 2.0 mM EDTA, 0.5% SDS, 200 μg of yeast tRNA per ml, 200 μg of salmon sperm DNA per ml, and 100 μg of poly(A) per ml for at least 2 h. Hybridization was carried out at 43°C for 48 h in the same buffer used for the prehybridization, with the equivalent counts per minute of 32P-labeled probe per milliliter added to each membrane. The membranes were washed in 2× SSC–0.5% SDS twice at 53°C for 15 min and then briefly rinsed in 2× SSC; this was followed by a 0.5-h digestion period with 10 μg of RNase A and 10 U of RNase T1 per ml. Finally, the blots were washed twice at 53°C for 15 min with 0.1× SSC–0.1% SDS buffer. The blots were exposed to X-ray film for autoradiography, and the signal intensity was determined with a PhosphorImager (Molecular Dynamics). CD14 transcription rates were normalized to that of β-actin.
In vivo experiments with IL-1ra and rsTNF-RI.
In order to test the roles of IL-1 and TNF in LPS-induced CD14 upregulation in vivo, rats were injected with a vehicle (saline), LPS (10 mg/kg, intravenously [i.v.]), or LPS plus either IL-1 receptor antagonist protein (IL-1ra; 100 mg/kg, subcutaneously [s.c.], at time 0 and then every 6 h), a polyethylene glycol-linked dimer of type I soluble TNF-α receptor (rsTNF-RI, 1.5 mg/kg, i.v., at time 0), or a combination of both. IL-1ra at 100 mg/kg and rsTNF-RI at 1.5 mg/kg are maximally efficacious doses for promoting survival at doses of LPS around 12.5 mg/kg (51) and were kindly provided by James L. Vannice (Synergen, Inc., Boulder, Colo.). The livers were collected at 6 h after treatment and snap frozen in liquid nitrogen for subsequent RNA isolation and Northern blot analysis.
Hepatocyte CD14 cDNA cloning.
An acute-phase rat liver cDNA library constructed in the Lambda Zap II/CIAP cloning vector was purchased from Stratagene. One million plaques were blotted onto Nytran membrane (Schleicher & Schuell) and hybridized with a 1,043-bp NotI fragment of mouse CD14 cDNA (15). The cDNA probe was labeled by random prime labeling (Boehringer Mannheim). Hybridization and blot washing were carried out under the conditions described above for total RNA isolation. Positive plaques were isolated and purified by two subsequent screenings. Rat CD14 cDNA inserts contained within the vector pBluescript were rescued by coinfection with helper phage. Plasmid DNA was prepared by using Qiagen Maxiprep columns (Chatsworth, Calif.). Sequencing was performed with an ABI automated DNA sequencer (core facility at the University of Pittsburgh School of Medicine) with the appropriate primers.
A custom cDNA library derived from cytokine-treated cultured human hepatocytes (19) was constructed in Lambda ZapII vector and screened for the presence of human CD14 clones. The human hepatocyte CD14 cloning procedure was the same as that described for rat CD14.
FITC-LPS binding by rat CD14-transfected CHO cells.
The binding of fluorescein isothiocyanate (FITC)-LPS to rat CD14-transfected CHO cells was assessed by fluorescence-activated cell sorting (FACS; Becton Dickinson, Mountainview, Calif.). Cells grown as monolayers in culture flasks were detached by using PBS containing 2 mM EDTA and resuspended in 2% CS-PBS. Cells (2.5 × 105/well) were incubated in V-bottom 96-well plates with 2.5 μg of FITC-LPS per ml in 100 μl of 10% CS-PBS containing 0.1% sodium azide for 1 h at 4°C. Cells incubated with 10% CS-PBS alone were used as controls. After being washed the cells were analyzed by FACS.
Electrophoretic mobility shift assay.
Nuclear extracts were prepared as described previously (31). Cells were washed with PBS, resuspended in a five-pellet-volume of buffer A (10 mM HEPES, pH 7.9; 10 mM KCl; 1.5 mM MgCl2; 0.5 mM DTT; 0.2 mM PMSF; 0.5% Nonidet P-40), and then incubated on ice for 15 min before being disrupted with 10 strokes in a Dounce homogenizer. The nuclei were washed once with buffer B (same as buffer A, but without Nonidet P-40). Nuclear proteins were extracted by gently resuspending the nuclei in 150 μl of buffer C (20 mM HEPES, pH 7.9; 10 mM KCl; 1.5 mM MgCl2; 10% glycerol; 0.2 mM EDTA; 0.5 mM DTT; 0.2 mM PMSF) and 50 μl of buffer D (same as buffer C but with 400 mM KCl). Buffer D was added in a dropwise fashion. After the nuclei were gently shaken in buffer C plus buffer D for 1 h at 4°C, the supernatants were collected by centrifugation at 13,000 rpm for 30 min. Double-stranded NF-κB-specific oligonucleotide was end labeled with [γ-32P]ATP by using T4 polynucleotide kinase (USB, Cleveland, Ohio) and purified on a G-50 Sephadex spin column. Nuclear proteins (10 μg per lane) were incubated with ∼40,000 cpm of 32P-labeled oligonucleotide for 20 min at room temperature in a buffer containing 2 μg of poly(dI-dC), 4.2 mM HEPES (pH 7.4), 2.5% glycerol, 4.2 mM KCl, 1 mM MgCl2, 0.02 mM EDTA, 2% Ficoll, and 21 mM DTT (final volume, 30 μl). For the supershift assay, 2 μg of antibody was added to the reaction mixture, and the reaction was incubated for another 30 min at room temperature. DNA-protein complexes were resolved on a 5% nondenaturing polyacrylamide gel in 0.5× Tris-borate-EDTA buffer. The gel was then dried and subjected to autoradiography.
Statistical analysis.
Data are presented as the mean ± the standard error (SE). Experimental results were analyzed for their significance by analysis of variance with Statview statistic software (Abacus Concept, Inc., Berkeley, Calif.). Significance was established at a P value of <0.05.
RESULTS
Hepatocytes express CD14 mRNA that is upregulated during endotoxemia.
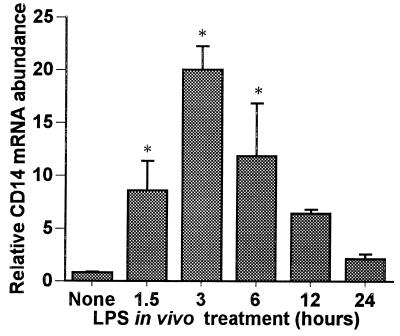
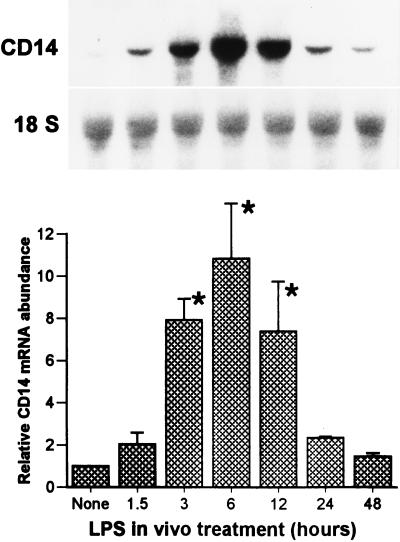
We postulated that hepatocytes could express CD14 which could be upregulated during endotoxemia. Rats were injected with LPS (10 mg/kg, given intraperitoneally [i.p.]), and total RNA was extracted from freshly isolated and purified hepatocytes at the time points indicated in Fig. 1. Northern blot analysis showed that hepatocytes isolated from controls had low but detectable levels of CD14 mRNA, which was visualized after prolonged exposure as a 1.6-kb transcript (data not shown). LPS treatment increased the steady-state CD14 mRNA levels in hepatocytes, inducing a ninefold elevation by as early as 1.5 h after LPS treatment. The levels increased with time, reaching a maximum induction (20-fold) by 3 h after treatment, and subsequently declined to near baseline levels by 24 h. We also examined the CD14 mRNA levels in RNA isolated from whole liver during endotoxemia (Fig. 2) and found that the pattern of CD14 mRNA induction by LPS was similar to that of the isolated hepatocytes, indicating that the upregulation of CD14 mRNA was not likely to be simply a consequence of the hepatocyte isolation procedure.
FIG. 1.
Time course of hepatocyte CD14 mRNA induction during endotoxemia. Sprague-Dawley rats were injected with LPS (10 mg/kg, i.p.). Hepatocytes were isolated at 1.5, 3, 6, 12, and 24 h from either LPS-treated or control rats. Total RNA was extracted, and Northern blot analysis was performed for CD14 mRNA. Membranes were rehybridized with probe for 18S rRNA. The levels of CD14 mRNA were normalized to 18S rRNA and are presented as the fold increase over the controls. Each column represents the mean ± the SE of triplicate samples at each time point (∗, P < 0.05 versus control).
FIG. 2.
Steady-state CD14 mRNA levels in liver tissue after LPS injection. Sprague-Dawley rats were injected with LPS (10 mg/kg, i.p.). Total RNA was extracted from the liver at 1.5, 3, 6, 12, and 24 h after LPS injection, and Northern blot analysis was performed for CD14 mRNA. Membranes were rehybridized with a probe for 18S rRNA. The bar graph shows the levels of CD14 mRNA normalized to 18S rRNA and presented as the fold increase over the controls. The Northern blot is from a representative experiment. Each column represents the mean ± the SE of three independent experiments (∗, P < 0.05 versus control).
In situ hybridization for CD14.
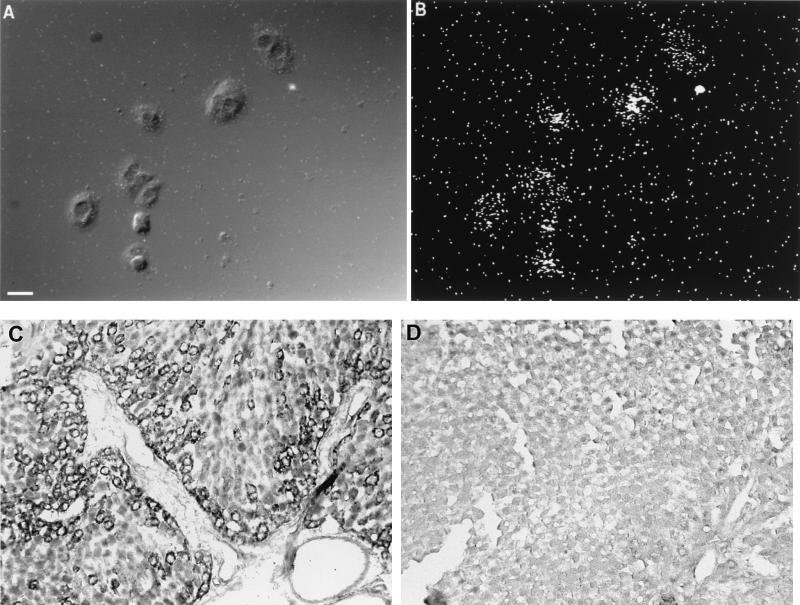
The liver contains a large number of macrophages that could provide a source of CD14 mRNA. To ensure that the CD14 mRNA was indeed expressed in hepatocytes, in situ hybridization was performed on isolated hepatocytes and whole-liver sections from LPS-treated rats. Consistent with the Northern blot results, CD14 mRNA was easily detected in cultured individual hepatocytes (Fig. 3A and B) and intact liver sections (Fig. 3C and D). Rat hepatocytes isolated from LPS-treated animals showed a strong cytoplasmic labeling (Fig. 3A). In situ hybridization performed on sectioned liver from LPS-treated rats (Fig. 3C) also showed a strong labeling in hepatocytes. In control livers, only a few labeled cells were seen (Fig. 3D). Notably, the labeling was most intense in cells close to the vasculature (either in the portal vein or in the hepatic triad).
FIG. 3.
Localization of CD14 mRNA in rat primary hepatocytes and sectioned liver. Hepatocytes grown on coverslips or liver tissue sections were subjected to in situ hybridization with antisense and sense riboprobes for CD14. (A) A differential interference contrast image of individual rat primary hepatocytes (the size bar indicates 20 μm). (B) Dark-field view of the in situ signal (35S). Primary hepatocytes isolated from LPS-treated rats (10 mg/kg, i.p., 24-h treatment) show strong cytoplasmic labeling. In the lower panels, in situ hybridizations performed on sectioned livers from LPS-treated (C) and normal (D) rats are shown. The signal detected with the digoxigenin-labeled riboprobe and colorimetric technique shows labeling in individual hepatocytes around blood vessels, in this case a portal triad. (Positive cells on the original slides were blue and, as shown here, are black.) The number of positive cells decreases with distance from the vasculature. In the control liver (panel D), very few or no positively labeled cells are seen. The sense probe showed little or no labeling (data not shown).
CD14 protein expression correlates with upregulated hepatic CD14 mRNA expression.
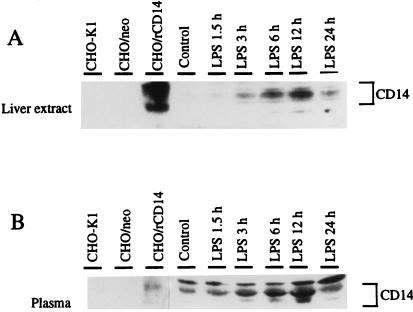
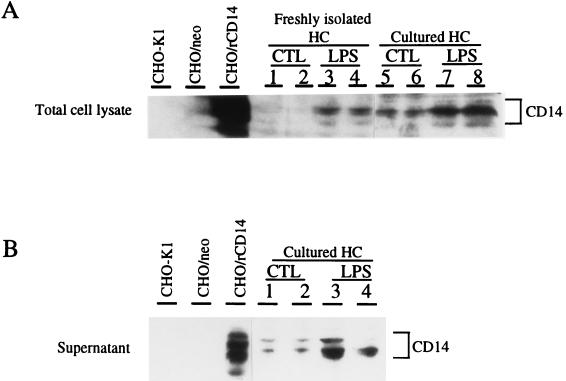
To determine if the upregulation of CD14 expression could also be appreciated in protein levels, Western blot analysis was performed on both whole liver and plasma samples from control or LPS-treated animals. Whole-cell extracts of parental CHO cells and neotransfected CHO cells served as negative controls, and rat CD14-transfected CHO cells served as a positive control. As indicated in Fig. 4, whole-cell extracts from rat CD14-transfected CHO cells showed a strong cross-reactivity to anti-mouse CD14 monoclonal antibody, but parental CHO or neotransfected CHO cells did not. Both liver homogenates (Fig. 4A) and plasma (Fig. 4B) displayed CD14-like bands with molecular masses of approximately 50 kDa that aligned with the positive control. The appearance of multiple isoforms of CD14 is consistent with the fact that CD14 is a highly glycosylated protein (19). In whole-liver extracts, increases of CD14 protein expression were seen 3 h after LPS treatment, peaked at 12 h, and declined thereafter (Fig. 4A). A similar transient increase in sCD14 in plasma was observed (Fig. 4B). To establish that hepatocytes express and release more CD14 after LPS treatment, Western blot analysis was performed on isolated hepatocytes. Lysate from freshly isolated hepatocytes from LPS-treated rats contained more CD14 than control cells at 12 h after LPS treatment (Fig. 5A, lanes 1 and 2 versus lanes 3 and 4) when placed in culture. The cells from LPS-treated rats continued to exhibit higher CD14 levels (Fig. 5A, lanes 5 and 6 versus lanes 7 and 8) and release more CD14 into the culture supernatant over time than did the control hepatocytes (Fig. 5B, lanes 1 and 2 versus lanes 3 and 4).
FIG. 4.
Western blot analysis of CD14 protein levels in whole-liver extracts (A) and plasma (B) during endotoxemia. Whole livers and plasma samples from LPS-treated rats at 1.5, 3, 6, 12, and 24 h (10 mg/kg, i.p.) or from control rats were prepared. Whole-cell extracts of parental CHO cells and neotransfected CHO cells served as negative controls, and rat CD14-transfected CHO cells were used as a positive control. Proteins (50 μg of whole-liver homogenate or plasma, 1 μg of recombinant proteins per lane) were separated on an SDS–10% polyacrylamide gel and transferred to nitrocellulose membrane. For the detection of CD14 protein, a hamster anti-mouse CD14 monoclonal antibody (G5A10) was incubated at 5 μg/ml with the membranes for 1 h. Immune complexes were detected by enhanced luminol reagent (Dupont, NEN) as described by the manufacturer. The blots are representative of at least three independent experiments. Abbreviations: CHO/rCD14, stably transfected rat CD14 CHO cells; CHO/neo, stably neotransfected CHO cells.
FIG. 5.
(A) CD14 protein levels in freshly isolated and cultured hepatocytes. Western blot procedures and abbreviations are as described for Fig. 4. Each group contains duplicate samples from individual animals. Lanes 1 and 2, freshly isolated control hepatocytes; lanes 3 and 4, freshly isolated hepatocytes from LPS-treated rats (10 mg/kg, i.p., 12-h treatment); lanes 5 and 6, control hepatocytes plated for 24 h; lanes 7 and 8, cultured hepatocytes from LPS-treated rats (10 mg/kg, i.p., 12-h treatment) plated for 24 h. (B) CD14 protein levels in hepatocyte culture supernatant. Each group contains duplicate samples from individual animals. Lanes 1 and 2, control hepatocytes plated for 24 h; lanes 3 and 4, cultured hepatocytes from LPS-treated rats (10 mg/kg, i.p., 12-h treatment) plated for 24 h. The data are representative of three independent experiments.
Cloning and sequence analysis of rat hepatocyte CD14 cDNA.
The human and rodent CD14 cDNA nucleotide sequences have been reported by Ferrero and coworkers (14, 15) and others (42). These clones were isolated from libraries established from monocytes or macrophages. A partial rat CD14 cDNA clone isolated from an astrocyte library has been reported (17), and a full-length CD14 cDNA has been cloned from rat macrophages (57). In order to determine the molecular identity of the rat hepatocyte CD14, an acute-phase rat liver cDNA library was screened at high stringency with a 1,043-bp mouse CD14 cDNA NotI fragment. Screenings from a total of 106 plaques yielded multiple overlapping cDNA clones. The longest clone contained a 1,591-bp insert. This fragment contains a 65-bp 5′-untranslated region (5′-UTR), a 1,116-bp entire portion of the protein-coding region, and a 410-bp 3′-UTR. Our rat hepatocyte CD14 sequence from nucleotides 1 to 1508 matched exactly with the published rat macrophage CD14 cDNA sequence from nucleotides 40 to 1547 (57); therefore, the sequence is not shown here. The translation initiation site was assigned to the first ATG triplet at nucleotides 66 to 68 since the initiation codon is flanked by a sequence (CGACCATGC) which has only one nucleotide mismatch to the consensus sequence of functional initiation codon [C(C)A/GCCATGG] defined by Kozak (34). The TAA termination codon is at position 1181. This open reading frame could therefore encode a 372-amino-acid primary translation protein. One copy of the sequence AUUUA, a common mRNA destabilizing sequence found in many inflammation-related genes (6), is located within the 3′-UTR. Two putative polyadenylation signals (AAUAAA) were found at positions 1393 to 1398 and positions 1491 to 1496, respectively, the latter being followed 7 bp further downstream by the polyadenylate tail.
The deduced amino acid sequence contains five putative N-glycosylation sites conforming to the canonical Asn-X-Ser sequence and a hydrophobic region preceded by a potential cleavage site (data not shown). The rat CD14 cDNA encodes a polypeptide that is similar to that of mouse CD14 (GenBank access number M34510), showing 81.8% amino acid sequence identity. Alignment of the amino acid sequences of rat and human CD14 (GenBank access number X06882) reveals 62.8% homology. We also screened a cDNA library derived from cytokine-stimulated, purified human hepatocytes (19). Two of eleven positive clones were sequenced. Sequence alignment (data not shown) showed near identity (96%) to the published human monocyte CD14 (14). Taken together, these data strongly suggest that hepatocytes express a CD14 that is identical to macrophages.
Functional analysis of the cloned rat CD14 cDNA.
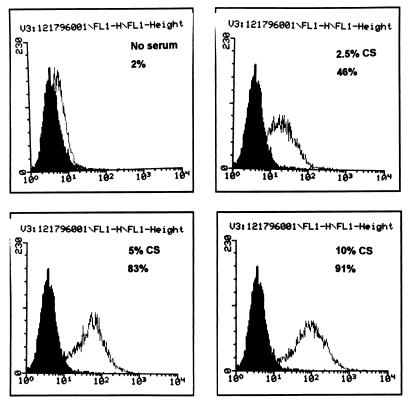
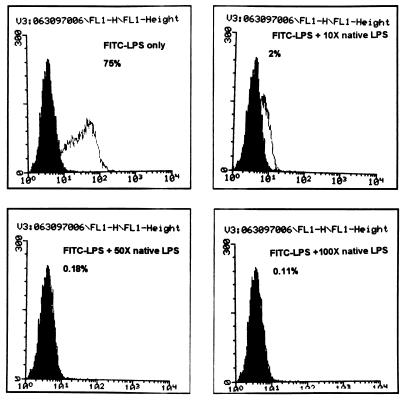
Functional analysis of expressed rat CD14 protein was examined. The 1,591-bp full-length rat CD14 cDNA was inserted downstream of the cytomegalovirus promoter in a pcDNA3 vector (Invitrogen, San Diego, Calif.) and used to transfect CHO cells that lack CD14 expression by Northern blot analysis (data not shown). Stably transfected cell lines were established by G418 selection and limiting dilution. One clone (CHO/rCD14) was selected, and Northern blot analysis showed abundant CD14 mRNA expression (data not shown). The CHO/rCD14 cells also showed strong binding by G5A10, a hamster anti-mouse CD14 monoclonal antibody, by flow cytometry analysis (35a). FITC-LPS binding assay exhibited a marked enhancement of LPS binding to the CD14-transfected cells, and serum was a limiting factor for FITC-LPS binding (Fig. 6). The binding could be inhibited by an excess of nonlabeled LPS (Fig. 7). These observations are consistent with the LPS binding characteristics of membrane-bound CD14 (32, 59).
FIG. 6.
Serum-dependent binding of FITC-LPS to CHO/rCD14 cells. Cells were stained as described in Materials and Methods and subjected to flow cytometry analysis. The serum concentrations contained in the binding buffer and the percentage of FITC-positive cells are shown. The open histogram represents the FITC-LPS staining. x axis, log fluorescence intensity; y axis, cell number. No FITC-LPS staining was found in CHO-K1 and CHO/neo cells (data not shown). The data are representative of at least three independent experiments.
FIG. 7.
Competition of FITC-LPS binding to CHO/rCD14 cells by excess of native-form LPS. The binding buffer is PBS with 10% CS and 0.1% sodium azide. The fold excess of native-form LPS added in the binding buffer and the respective percentage of FITC-positive cells are shown. The open histogram represents the FITC-LPS staining. x axis, log fluorescence intensity; y axis, cell number. The data are representative of three independent experiments.
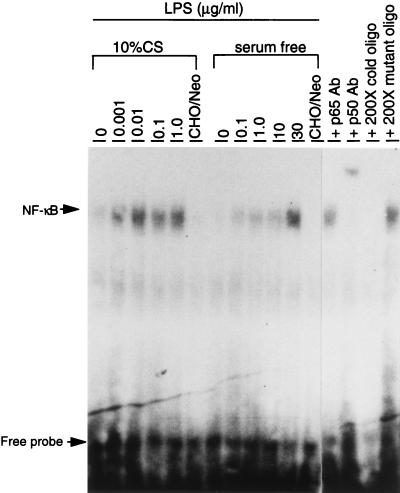
It has been previously shown that transfection of human or mouse CD14 cDNA into CHO cells renders the cells responsive to LPS for nuclear translocation of NF-κB (9, 21, 67). As shown in Fig. 8, the CHO/rCD14 cells responded to LPS at concentrations as low as 1 ng/ml in the presence of serum and 30 μg/ml in the absence of serum. Thus, rat CD14 mediates serum-dependent responses to LPS in a way similar to that reported for human and mouse CD14 (8, 21, 48). The response could be detected by as early as 30 min and peaked at 60 min (data not shown). The specificity of NF-κB-shifted bands was confirmed by cold and mutant oligonucleotide competitions. Supershift assay with specific NF-κB subunit antibody indicated that this DNA-protein binding complex contains at least p50 NF-κB subunits.
FIG. 8.
NF-κB activation by LPS in CHO/rCD14 cells in serum and serum-free conditions. The experiment was carried out as described in Materials and Methods. Cells were treated with different doses of LPS prior to nuclear protein extraction. The doses of LPS for treatment are indicated, except in CHO/neo cells (0.1 μg/ml). Abbreviations: CHO/neo, stably neotransfected CHO cells; p65 Ab, anti-NF-κB p65 subunit antibody; p50 Ab, anti-NF-κB p65 subunit antibody. The data are representative of two independent experiments.
The increase in hepatocyte CD14 mRNA levels by LPS in vivo is regulated at the transcriptional level.
To determine whether transcription plays a role in the large increase in CD14 expression in hepatocytes by LPS in vivo, nuclei were isolated from hepatocytes of control and LPS-treated rats for the nuclear run-on assay. The elongated nascent RNA was hybridized to a plasmid containing CD14 cDNA immobilized to membrane. Plasmid containing β-actin cDNA and the vector pBluescript were included as internal controls. The transcription rates of CD14 were normalized to that of β-actin. As indicated in Fig. 9, basal CD14 transcription was observed in nuclei from control hepatocytes. However, in the nuclei of hepatocytes isolated from LPS-treated rats, the transcription rates for CD14 were increased by 3.2- and 2.6-fold at 1.5 and 3.0 h, respectively (P < 0.05). After 6 h of LPS treatment the level of transcription for CD14 had decreased, and at 24 h it had returned almost to basal levels (data not shown). Thus, the upregulation of CD14 mRNA levels during endotoxemia involves increased transcription.
FIG. 9.
Nuclear run-on assay in hepatocyte nuclei from LPS-treated rats. Nuclei from control or LPS-injected rats were incubated with [α-32P]UTP to elongate the nascent RNA. The elongated nascent RNA was hybridized to plasmid containing CD14 cDNA immobilized to GeneScreen plus membrane (Dupont, NEN) by a slot-blot apparatus (5 μg/per slot). Equivalent counts per minute of 32P-labeled nuclear RNA probe per milliliter were added to each membrane. Plasmid containing β-actin cDNA and empty vector pBluescript were included as the internal controls. The CD14 transcription rates were quantitated with a PhosphorImager and normalized to that of β-actin. Each column represents the mean ± the SE of duplicate samples (∗, P < 0.05 versus control).
IL-1β and TNF-α upregulate CD14 expression in hepatocytes in vitro.
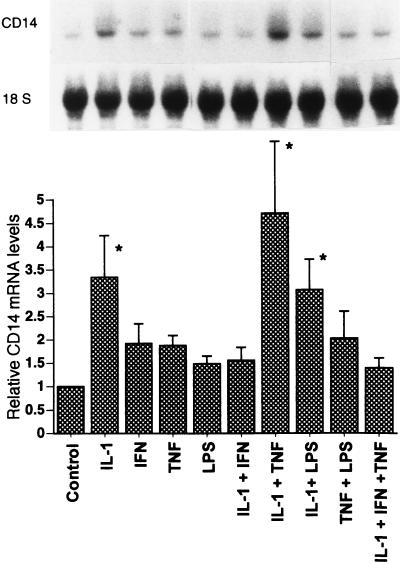
It is well known that LPS elicits the synthesis of cytokines such as IL-1, TNF, and IL-6, as well as other inflammatory mediators that are known to regulate the hepatic acute-phase response (33, 44). To determine whether CD14 expression is regulated by cytokines in vitro, cultured hepatocytes were exposed to various cytokines. The CD14 mRNA levels were examined by Northern blot analysis at 12 h (initial results showed maximal induction was at 12 h; data not shown). As shown in Fig. 10, cultured rat hepatocytes expressed basal levels of a 1.6-kb CD14 mRNA transcript. IL-1β caused a 3.3-fold increase in the CD14 mRNA levels compared to the control. IL-1β combined with TNF-α caused a greater increase in CD14 mRNA accumulation (4.7-fold increase) than did IL-1β alone, although TNF-α alone did not significantly increase the CD14 mRNA levels. When IFN-γ was combined with IL-1β or IL-1β–TNF-α, the CD14 mRNA levels were reduced. IL-6 has been reported to be a strong inducer of many acute-phase reactants (44); however, either alone or combined with other cytokines, IL-6 did not affect the accumulation of CD14 mRNA in cultured hepatocytes (data not shown), whereas IFN-γ or LPS alone had negligible effects in vitro.
FIG. 10.
Effects of cytokines on steady-state CD14 mRNA levels in cultured rat hepatocytes. Hepatocyte isolation and Northern blot analysis were carried out as described in Materials and Methods. Rat primary hepatocytes were incubated with single or combined cytokines (500 U/ml) for 12 h. The levels of CD14 mRNA were normalized to 18S rRNA and expressed as the fold increase over the control. Each column represents the mean ± the SE of three independent experiments (∗, P < 0.05 versus control).
IL-1β and TNF-α contribute to increased hepatic CD14 expression in vivo.
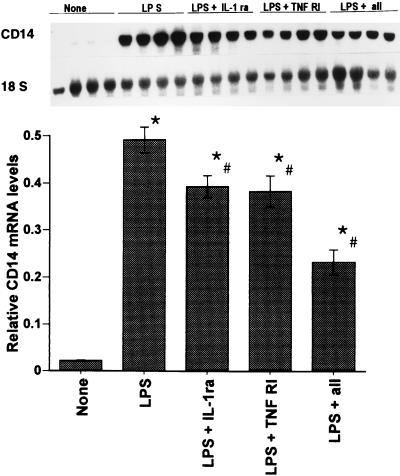
To establish the involvement of IL-1β and/or TNF-α in the upregulation of hepatic CD14 in vivo during endotoxemia, experiments were carried out with IL-1ra or rsTNF-RI to block IL-1β or TNF-α, respectively. The doses and methods of administration of IL-1ra and rsTNF-RI have proven to be maximally efficacious for preventing mortality in rats during endotoxemia (51). Northern blot analysis was performed to measure the hepatic CD14 mRNA levels in rats treated with LPS plus IL-1ra, rsTNF-RI, or both in vivo. As shown in Fig. 11, animals treated with LPS and IL-1ra or with LPS and rsTNF-RI exhibited a 20% decrease (P < 0.05) in hepatic CD14 mRNA levels compared to rats given LPS alone 6 h previously. LPS plus both IL-1ra and rsTNF-RI showed a further decrease (50%, P < 0.05) in hepatic CD14 mRNA levels, implicating both IL-1 and TNF in the upregulation of hepatic CD14 mRNA during endotoxemia.
FIG. 11.
In vivo experiments with IL-1β and TNF-α antagonists. Rats were injected with either saline (vehicle), LPS (10 mg/kg, i.v.), LPS plus IL-1ra (IL-1ra 100 mg/kg, s.c., at time 0 and then every 6 h), LPS plus rsTNF-RI (rsTNF-RI, 1.5 mg/kg, i.v., at time 0), or LPS plus IL-1ra plus sTNF-RI. Total RNA was extracted from the liver 6 h later and Northern blot analysis was performed for CD14 mRNA. The levels of CD14 mRNA were normalized to 18S rRNA and presented as the fold increase over the control. Each column represents the mean ± the SE of four animals in each group (∗, P < 0.05 versus the control; #, P < 0.05 versus the LPS group).
DISCUSSION
CD14 as a key LPS signaling molecule has been well documented in vitro in many cell systems (1, 38, 59, 63). In vivo experimental data have begun to accumulate, defining the critical role of CD14 in the host response to LPS. Transgenic mice that overexpress human CD14 on bone marrow-derived cells are highly sensitive to LPS (16), providing in vivo evidence that endogenous overexpression of CD14 can enhance LPS-induced activation. Conversely, CD14 knockout mice are at least 10 times less sensitive to LPS with regard to both lethality and TNF and IL-6 production (25). The 20-fold upregulation of hepatocyte CD14 mRNA and the significant increase in CD14 protein expression and release during endotoxemia suggest that hepatic CD14 expression is part of the systemic response to infection.
In the studies reported here, we examined rat hepatocyte CD14 expression and regulation both in vivo and in vitro. We also cloned a full-length rat hepatic CD14 cDNA. Our findings demonstrate the following. (i) Isolated rat hepatocytes express basal levels of CD14 mRNA, and that expression is markedly upregulated during endotoxemia. (ii) In situ hybridization with riboprobe for rat CD14 identified CD14 mRNA in cultured hepatocytes and parenchymal liver cells in whole-liver sections. (iii) Both whole liver and plasma display an LPS-dependent increase in CD14 protein levels that correlates with the increases in hepatocyte CD14 expression. (iv) Hepatocytes from LPS-treated animals contain more CD14 and release more sCD14 in culture than do control hepatocytes. (v) CD14 mRNA upregulation includes increased transcription. (vi) The cloned full-length rat hepatocyte CD14 cDNA has a sequence identical to that of the rat macrophage CD14 and encodes a functional cell surface LPS recognition molecule. (vii) Both in vitro and in vivo data indicate that IL-1β and/or TNF-α regulate the expression of CD14 mRNA in hepatocytes. CD14 and LBP are the two key LPS recognition and signaling molecules, and production of LBP by primary hepatocytes has been well characterized both by us (18, 56, 61) and by others (47). Our data indicate that hepatocytes also express CD14, raising the possibility that hepatocytes are a source of CD14 protein during endotoxemia.
Although liver is a potentially important source for elevated sCD14 in plasma during endotoxemia, there are limited reports concerning the expression of CD14 in the liver. Matsuura et al. (41) reported a time- and dose-dependent induction of CD14 mRNA in mouse liver after LPS administration. Fearns et al. (11) demonstrated that extramyeloid expression of CD14 took place in selected organs, including the liver, and that plasma CD14 levels are elevated during endotoxemia. We verified these previous findings by showing that hepatocyte CD14 mRNA and protein expression is increased during endotoxemia by using isolated purified hepatocytes. Resident Kupffer cells (3, 30, 41), as well as polymorphonuclear monocytes (28, 62) that accumulate in the liver during endotoxemia, represent potential sources of CD14 in whole liver. Our purified hepatocyte preparations contained less than 2% contaminating cells. Thus, it is unlikely that these small numbers of cells contribute a significant level of mRNA or protein. Furthermore, we used in situ hybridization to definitively localize CD14 mRNA to hepatocytes in vitro and in vivo (Fig. 3).
Soluble CD14 has been found to bind LPS with high affinity and mediates activation in many CD14-negative cells (20, 27, 39, 46). Indirect evidence for sCD14’s participation in sepsis comes from studies showing that plasma CD14 levels are significantly elevated during gram-negative sepsis (25, 37), gram-positive sepsis (5), trauma, or burns (35). Levels of sCD14 in plasma, which ranged from 3 to 5 μg/ml (5, 22), are increased during sepsis by 45 to 75% over those for normal controls and nonseptic patients. Thus, sCD14 fits the criteria for an acute-phase reactant. A significant feature of many acute-phase proteins is that their genes are usually regulated by cytokines and other mediators. The best-characterized cytokines and other mediators are IL-1, TNF, IL-6, leukemia inhibitory factor, and IL-11, with IL-1 and TNF typically inducing a similar spectrum of changes (2, 7). Both our in vitro and our in vivo data indicate that hepatocyte CD14 mRNA is upregulated effectively by IL-1β and/or TNF-α (Fig. 10 and 11), which is consistent with many other genes of acute-phase proteins. The in vivo results cannot prove that IL-1β and TNF-α directly act upon hepatocytes but only that these cytokines directly or indirectly participate in the upregulation of CD14. IL-1β and/or TNF-α did directly increase hepatocyte CD14 mRNA levels in vitro at the doses and time point (12 h) chosen. The changes in steady-state mRNA in vitro were not of the same magnitude as that seen with LPS treatment in vivo. Whether this was due to suboptimal conditions in vitro is unknown. The roles of IL-1 and TNF in the upregulation of CD14 mRNA in the liver and kidney have also been demonstrated by studies from Fearns and coworkers (12, 13) and Takakuwa et al. (58).
Although we do not provide direct evidence here that sCD14 in plasma originates from mCD14 on hepatocytes during endotoxemia, our data (Fig. 4) clearly demonstrate that there is a correlated expression of CD14 in the liver and plasma in a time-dependent manner, raising the possibility that the liver is an important source for elevated plasma CD14 levels during endotoxemia. Fearns and Loskutoff (12) observed that in a murine endotoxemic model increases in plasma CD14 levels occurred at times when the epithelial CD14 expression was maximal. Their data support the idea that hepatocytes and tubular epithelium may contribute to the pool of sCD14 in the plasma after LPS stimulation. Our data (Fig. 5A) indicate that hepatocytes from LPS-treated animals express higher amounts of mCD14 and, most importantly, release more sCD14 (Fig. 5B).
Up to 80% of injected LPS concentrates in the liver within 20 to 30 min (50), and within hours LPS can be found in the bile (40). Recent studies have shown that LPS can be found in hepatocytes just 5 min following an intraportal endotoxin injection (43). Although some of the LPS clearly interacts with the Kupffer cells and endothelial cells (41), there is also strong evidence that hepatocytes can directly respond to LPS (45, 61). Whether hepatocyte CD14 contributes to this interaction is unclear. However, it is interesting to note that the hepatocytes close to the portal triad showed greater CD14 mRNA expression by in situ hybridization. These cells would be exposed first to LPS entering the liver, where either mCD14 on the cells or sCD14 released by the cells would interact with the endotoxin, thus facilitating uptake or initiation with other cell types. Another possibility would be that the hepatocytes in the periportal regions have a higher LPS elimination rate than the central regions. Studies are underway to determine how hepatocyte CD14 is processed.
ACKNOWLEDGMENTS
This work was supported in part by NIH grant R01-GM-50441.
We wish to acknowledge the technical assistance of Debra Williams, Suhua Nie, and Angela Green and the secretarial assistance of Margaret Bullers, as well as the assistance of Rosemary Hoffman with the flow cytometry.
S.L. and L.S.K. contributed equally to this work.
REFERENCES
- 1.Arditi M, Zhou J, Torres M, Durden D L, Stins M, Kim K S. Lipopolysaccharide stimulates the tyrosine phosphorylation of mitogen-activated protein kinase p44, p42, and p41 in vascular endothelial cells in a soluble CD14-dependent manner. J Immunol. 1995;155:3994–4003. [PubMed] [Google Scholar]
- 2.Baumann H, Morella K K, Wong G H W. TNFα, IL-1β, and hepatocyte growth factor cooperate in stimulating specific acute-phase plasma protein gene in rat hepatoma cells. J Immunol. 1993;151:4248–4255. [PubMed] [Google Scholar]
- 3.Bellezzo J M, Britton R S, Bacon B R, Fox E S. LPS-mediated NF-kappa beta activation in rat Kupffer cells can be induced independently of CD14. Am J Physiol. 1996;270:G956–G961. doi: 10.1152/ajpgi.1996.270.6.G956. [DOI] [PubMed] [Google Scholar]
- 4.Boggaram V, Mendelson C R. Transcriptional regulation of the gene coding the major surfactant protein (SPA) in rabbit fetal lung. J Biol Chem. 1988;263:19060–19065. [PubMed] [Google Scholar]
- 5.Burgmann H, Winkler S, Locker G J, Presterl E, Laczika K, Staudinger T, Knapp S, Thalhammer F, Wenisch C, Zedwitz-Liebenstein K, Frass M, Graninger W. Increased serum concentration of soluble CD14 is a prognostic marker in gram-positive sepsis. Clin Immunol Immunopathol. 1996;80:307–310. doi: 10.1006/clin.1996.0128. [DOI] [PubMed] [Google Scholar]
- 6.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci USA. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cybulsky M I, Chan M K W, Movat H Z. Acute inflammation and microthrombosis induced by endotoxin, interleukin-1, and tumor necrosis factor and their implication in gram-negative infection. Lab Investig. 1988;58:365–376. [PubMed] [Google Scholar]
- 8.Davis L G, Kuehl W M, Battey J F. Nuclear run-on for determining rate for transcription. In: Davis L, Kuehl M, Battey J, editors. Basic methods in molecular biology. 2nd ed. Stamford, Conn: Appleton & Lange Press; 1994. pp. 384–395. [Google Scholar]
- 9.Delude R L, Fenton M J, Savedra R, Jr, Perera P-Y, Vogel S N, Thieringer R, Golenbock D T. CD14-mediated translocation of nuclear factor-κB induced by lipopolysaccharide does not require tyrosine kinase activity. J Biol Chem. 1994;269:22253–22260. [PubMed] [Google Scholar]
- 10.Dentener M A, Bazil V, Von Asmuth E J, Ceska M, Buurman W A. Involvement of CD14 in lipopolysaccharide-induced tumor necrosis factor-alpha, IL-6 and IL-8 release by human monocytes and alveolar macrophages. J Immunol. 1993;150:2885–2891. [PubMed] [Google Scholar]
- 11.Fearns C, Kravchenko V V, Ulevitch R J, Loskutoff D J. Murine CD14 gene expression in vivo: extramyeloid synthesis and regulation by lipopolysaccharide. J Exp Med. 1995;181:857–866. doi: 10.1084/jem.181.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fearns C, Loskutoff D J. Role of tumor necrosis factor alpha in induction of murine CD14 gene expression by lipopolysaccharide. Infect Immun. 1997;65:4822–4831. doi: 10.1128/iai.65.11.4822-4831.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fearns C, Ulevitch R J. Effect of recombinant interleukin-1beta on murine CD14 gene expression in vivo. Shock. 1998;9:157–163. doi: 10.1097/00024382-199803000-00001. [DOI] [PubMed] [Google Scholar]
- 14.Ferrero E, Goyert S M. Nucleotide sequence of the gene encoding the monocyte differentiation antigen, CD14. Nucleic Acids Res. 1988;16:4173. doi: 10.1093/nar/16.9.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrero E, Hsieh C-L, Francke U, Goyert S M. CD14 is a member of the family of leucine-rich proteins and is encoded by a gene syntenic with multiple receptor genes. J Immunol. 1990;145:331–336. [PubMed] [Google Scholar]
- 16.Ferrero E, Jiao D, Tsuberi B Z, Tesio L, Rong G-W, Hazior A, Goyert S M. Transgenic mice expressing human CD14 are hypersensitive to lipopolysaccharide. Proc Natl Acad Sci USA. 1993;90:2380–2384. doi: 10.1073/pnas.90.6.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galea E, Reis D J, Fox E S, Xu H, Feinstein D L. CD14 mediate endotoxin induction of nitric oxide synthase in cultured brain glial cells. J Neuroimmunol. 1996;64:19–28. doi: 10.1016/0165-5728(95)00143-3. [DOI] [PubMed] [Google Scholar]
- 18.Geller D A, Kispert P H, Su G L, Wang S C, Di Silvio M, Tweardy D J, Billiar T R, Simmons R L. Induction of hepatocyte lipopolysaccharide binding protein in models of sepsis and the acute-phase response. Arch Surg. 1993;128:22–27. doi: 10.1001/archsurg.1993.01420130026005. [DOI] [PubMed] [Google Scholar]
- 19.Geller D A, Lowenstein C J, Shapiro R A, Nussler A K, Di Silvio M, Wang S C, Nakayama D K, Simmons R L, Snyder S H, Billiar T R. Molecular cloning and expression of inducible nitric oxide from human hepatocytes. Proc Natl Acad Sci USA. 1993;90:3491–3495. doi: 10.1073/pnas.90.8.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldblum S E, Brann T W, Ding X, Tobias P S. Lipopolysaccharide (LPS)-binding protein and soluble CD14 function as accessory molecules for LPS-induced changes in endothelial barrier function in vitro. J Clin Investig. 1994;93:692–702. doi: 10.1172/JCI117022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golenbock D T, Liu Y, Millham F H, Freeman M W, Zoeller R A. Surface expression of human CD14 in Chinese hamster ovary fibroblasts imparts macrophage-like responsiveness to bacterial endotoxin. J Biol Chem. 1993;268:22055–22059. [PubMed] [Google Scholar]
- 22.Grumwald U, Krüger C, Schütt C. Endotoxin-neutralizing capacity of soluble CD14 is a highly conserved specific function. Circ Shock. 1993;39:220–225. [PubMed] [Google Scholar]
- 23.Hampton R Y, Golenbock D T, Penman M, Krieger M, Raetz C R. Recognition and plasma clearance of endotoxin by scavenger receptors. Nature. 1991;353:342–344. doi: 10.1038/352342a0. [DOI] [PubMed] [Google Scholar]
- 24.Haziot A, Chen S, Ferrero F, Low M G, Silber R, Goyert S M. The monocyte differentiation antigen, CD14, is anchored to the cell membrane by a phosphatidyl-inositol linkage. J Immunol. 1988;141:547–552. [PubMed] [Google Scholar]
- 25.Haziot A, Ferrero E, Kontgen F, Hijiya N, Yamamoto S, Silver S, Stewart C L, Goyert S M. Resistance to endotoxin shock and reduced dissemination of gram-negative bacteria in CD14-deficient mice. Immunity. 1996;4:407–414. doi: 10.1016/s1074-7613(00)80254-x. [DOI] [PubMed] [Google Scholar]
- 26.Haziot A, Rong G W, Lin X Y, Silver J, Goyert S M. Recombinant soluble CD14 prevents mortality in mice treated with endotoxin (lipopolysaccharide) J Immunol. 1995;154:6529–6532. [PubMed] [Google Scholar]
- 27.Haziot A, Rong G W, Silver J, Goyert S M. Recombinant soluble CD14 mediates the activation of endothelial cells by lipopolysaccharide. J Immunol. 1993;151:1500–1507. [PubMed] [Google Scholar]
- 28.Hirano S. Migratory responses of PMN after intraperitoneal and intratracheal administration of lipopolysaccharide. Am J Physiol. 1996;270:L836–L845. doi: 10.1152/ajplung.1996.270.5.L836. [DOI] [PubMed] [Google Scholar]
- 29.Hunt J S, Soares M J, Lei M-G, Smith R N, Wheaton D, Atherton R A, Morrison D C. Products of lipopolysaccharide modify DNA synthesis by rat trophoblast cells exhibiting the 80-kDa lipopolysaccharide-binding protein. J Immunol. 1989;143:1606–1613. [PubMed] [Google Scholar]
- 30.Jarvelainen H A, Oinonen T, Lindros K O. Alcohol-induced expression of the CD14 endotoxin receptor protein in rat Kupffer cells. Alcohol Clin Exp Res. 1997;21:1547–1551. [PubMed] [Google Scholar]
- 31.Kim Y M, de Vera M E, Watkins S C, Billiar T R. Nitric oxide protects cultured rat hepatocytes from TNFα-induced apoptosis by inducing heat shock protein 70 expression. J Biol Chem. 1996;272:1402–1411. doi: 10.1074/jbc.272.2.1402. [DOI] [PubMed] [Google Scholar]
- 32.Kirkland T N, Finley F, Leturcq D, Moriarty A, Lee J D, Ulevitch R J, Tobias P S. Analysis of lipopolysaccharide binding by CD14. J Biol Chem. 1993;268:24818–24823. [PubMed] [Google Scholar]
- 33.Koj A, Gauldie J, Baumann H. Biological perspectives of cytokines and hormone networks. In: Mackeiwicz A, Kushner I, Baumann H, editors. Acute phase proteins: molecular biology, biochemistry, clinical applications. Boca Raton, Fla: CRC Press, Inc.; 1993. p. 275. [Google Scholar]
- 34.Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984;12:857–871. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krüger C, Schütt C, Obertacke U, Joka T, Müller T E, Knöller J, Knöller K, König W, Schöndeld W. Serum CD14 levels in polytraumatized and severely burned patients. Clin Exp Immunol. 1991;85:297–301. doi: 10.1111/j.1365-2249.1991.tb05722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Landmann, R. Personal communication.
- 36.Landmann R, Reber A M, Sansano S, Zimmerli W. Function of soluble CD14 in serum from patients with septic shock. J Infect Dis. 1996;173:661–668. doi: 10.1093/infdis/173.3.661. [DOI] [PubMed] [Google Scholar]
- 37.Landmann R, Zimmerli W, Sansano S, Link S, Hahn A, Glauser M P, Calandra T. Increased circulating soluble CD14 is associated with high mortality in gram-negative septic shock. J Infect Dis. 1995;171:639–644. doi: 10.1093/infdis/171.3.639. [DOI] [PubMed] [Google Scholar]
- 38.LeGrand C B, Thieringer R. CD14-dependent induction of protein tyrosine phosphorylation by lipopolysaccharide in murine B-lymphoma cells. Biochim Biophys Acta. 1994;1223:36–46. doi: 10.1016/0167-4889(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 39.Loppnow H, Stelter F, Schönbeck U, Schlüter C, Ernst M, Schütt C, Flad H D. Endotoxin activates human vascular smooth muscle cells despite lack of expression of CD14 mRNA or endogenous membrane CD14. Infect Immun. 1995;63:1020–1026. doi: 10.1128/iai.63.3.1020-1026.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maitra S K, Rachmilewitz D, Eberle D, Kaplowitz N. The hepatocellular uptake and biliary excretion of endotoxin in the rat. Hepatology. 1981;1:401–407. doi: 10.1002/hep.1840010506. [DOI] [PubMed] [Google Scholar]
- 41.Matsuura K, Ishida T, Setoguchi M, Higuchi Y, Akizuki S, Yamamoto S. Upregulation of mouse CD14 expression in Kupffer cells by lipopolysaccharide. J Exp Med. 1994;179:1671–1676. doi: 10.1084/jem.179.5.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsuura K, Setoguchi M, Nasu N, Higuchi Y, Yoshida S, Akizuki S, Yamamoto S. Nucleotide and amino acid sequences of the mouse CD14 gene. Nucleic Acids Res. 1989;17:2132. doi: 10.1093/nar/17.5.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mimura Y, Sakisaka S, Harada M, Sata M, Tanikawa K. Role of hepatocytes in direct clearance of lipopolysaccharide in rats. Gastroenterology. 1995;109:1969–1976. doi: 10.1016/0016-5085(95)90765-3. [DOI] [PubMed] [Google Scholar]
- 44.Morrone G, Giliberto G, Oliviero S, Arcone R, Dente L, Content J, Cortese R. Recombinant IL-6 regulates the transcriptional activation of a set of human acute phase genes. J Biol Chem. 1988;263:12554–12558. [PubMed] [Google Scholar]
- 45.Pittner R A, Spitzer J A. Endotoxin and TNF alpha directly stimulate nitric oxide formation in cultured rat hepatocytes from chronically endotoxemic rats. Biochem Biophys Res Commun. 1992;185:430–435. doi: 10.1016/s0006-291x(05)81003-4. [DOI] [PubMed] [Google Scholar]
- 46.Pugin J, Schurer-Maly C C, Leturcq D, Moriarty A, Ulevitch R J, Tobias P S. Lipopolysaccharide activation of human endothelial and epithelial cells is mediated by lipopolysaccharide-binding protein and soluble CD14. Proc Natl Acad Sci USA. 1993;90:2744–2748. doi: 10.1073/pnas.90.7.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramadori G, Meyer zum Buschenfelde K-H, Tobias P S, Mathison J C, Ulevitch R J. Biosynthesis of lipopolysaccharide-binding protein in rabbit hepatocytes. Pathobiology. 1990;58:89–94. doi: 10.1159/000163569. [DOI] [PubMed] [Google Scholar]
- 48.Read M A, Cordle S R, Veach R A, Carlisle C D, Hawiger J. Cell-free pool of CD14 mediates activation of transcription factor NF-κB by lipopolysaccharide in human endothelial cells. Proc Natl Acad Sci USA. 1993;90:9887–9891. doi: 10.1073/pnas.90.21.9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roger M. Detection of hybridized probe. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons, Inc.; 1987. pp. 14.4.1–14.4.3. [Google Scholar]
- 50.Ruiter D J, van der Meulen J, Brouwer A, Hummel M, Jr, Mauw B J, van der Ploeg J C M, Wisse E. Uptake by liver cells of endotoxin following its intravenous injection. Lab Investig. 1981;45:38–45. [PubMed] [Google Scholar]
- 51.Russell D A, Tucker K K, Chinookoswong N, Thompson R C, Kohno T. Combined inhibition of interleukin-1 and tumor necrosis factor in rodent endotoxemia: improved survival and organ function. J Infect Dis. 1995;171:1528–1538. doi: 10.1093/infdis/171.6.1528. [DOI] [PubMed] [Google Scholar]
- 52.Schletter J, Heine H, Ulmer A J, Rietschel E T. Molecular mechanisms of endotoxin activity. Arch Microbiol. 1995;164:383–389. doi: 10.1007/BF02529735. [DOI] [PubMed] [Google Scholar]
- 53.Schumann R R, Leong S R, Flaggs G W, Gray P W, Wright S D, Mathison J C, Tobias P S, Ulevitch R J. Structure and function of lipopolysaccharide binding protein. Science. 1990;249:1429–1431. doi: 10.1126/science.2402637. [DOI] [PubMed] [Google Scholar]
- 54.Seglan P. Preparation of isolated rat liver cells. In: Prescott D, editor. Methods in cell biology. New York, N.Y: Academic Press, Inc.; 1976. pp. 28–83. [DOI] [PubMed] [Google Scholar]
- 55.Stelter F, Pfister M, Bernheiden M, Jack R S, Bufler P, Engelmann H, Schutt C. The myeloid differentiation antigen CD14 is N- and O-glycosylated. Contribution of N-linked glycosylation to different soluble CD14 isoforms. Eur J Biochem. 1996;236:457–464. doi: 10.1111/j.1432-1033.1996.00457.x. [DOI] [PubMed] [Google Scholar]
- 56.Su G L, Freeswick P D, Geller D A, Wang Q, Shapiro R A, Wan Y-H, Billiar T R, Tweardy D J, Simmons R L, Wang S C. Molecular cloning, characterization, and tissue distribution of rat lipopolysaccharide binding protein. J Immunol. 1994;153:743–752. [PubMed] [Google Scholar]
- 57.Takai N, Kataoka M, Higuchi Y, Matsuura K, Yamamoto S. Primary structure of rat CD14 and characteristics of rat CD14, cytokine, and NO synthase mRNA expression in monocyte system cells in response to LPS. J Leukoc Biol. 1997;61:736–744. doi: 10.1002/jlb.61.6.736. [DOI] [PubMed] [Google Scholar]
- 58.Takakuwa T, Knopf H P, Sing A, Carsetti R, Galanos C, Freudenberg M A. Induction of CD14 expression in Lpsn, Lpsd and tumor necrosis factor receptor-deficient mice. Eur J Immunol. 1996;26:2686–2692. doi: 10.1002/eji.1830261121. [DOI] [PubMed] [Google Scholar]
- 59.Tobias P S, Soldau K, Kline L, Lee J-D, Kato K, Martin T P, Ulevitch R J. Cross-linking of lipopolysaccharide (LPS) to CD14 on THP-1 cells mediated by LPS-binding protein. J Immunol. 1993;150:3011–3021. [PubMed] [Google Scholar]
- 60.Ulevitch R J, Tobias P S. Receptor-dependent mechanisms of cell stimulation by bacterial endotoxin. Annu Rev Immunol. 1995;13:437–457. doi: 10.1146/annurev.iy.13.040195.002253. [DOI] [PubMed] [Google Scholar]
- 61.Wan Y-H, Freeswick P D, Khemlani L S, Kispert P H, Wang S C, Su G L, Billiar T R. Role of lipopolysaccharide (LPS), interleukin-1, interleukin-6, tumor necrosis factor, and dexamethasone in regulation of LPS-binding protein expression in normal hepatocytes and hepatocytes from LPS-treated rats. Infect Immun. 1995;63:2435–2442. doi: 10.1128/iai.63.7.2435-2442.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J H, Redmond H P, Watson R W, Bouchier-Hayes D. Role of lipopolysaccharide and tumor necrosis factor-alpha in induction of hepatocyte necrosis. Am J Physiol. 1995;269:G297–G304. doi: 10.1152/ajpgi.1995.269.2.G297. [DOI] [PubMed] [Google Scholar]
- 63.Watanabe A, Takeshita A, Kitano S, Hanazawa S. CD14-mediated signal pathway of Porphyromanas gingivalis lipopolysaccharide in human gingival fibroblasts. Infect Immun. 1996;64:4488–4494. doi: 10.1128/iai.64.11.4488-4494.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.West M A, Billiar T R, Curran R D, Hyland B J, Simmons R L. Evidence that rat Kupffer cells stimulate and inhibit hepatocytes protein synthesis in vitro by different mechanisms. Gastroenterology. 1989;96:1572–1582. doi: 10.1016/0016-5085(89)90529-5. [DOI] [PubMed] [Google Scholar]
- 65.Whicher J T, Westacott I. The acute phase response. In: Whicher J T, Evans S W, editors. Biochemistry of inflammation. Vol. 18. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1992. pp. 243–269. [Google Scholar]
- 66.Wright S D, Levin S M, Jong M T, Chad Z, Kabbash L G. CR3 (CD11b/CD18) expresses one binding site for arg-gly-asp-containing peptides and a second site for bacterial lipopolysaccharide. J Exp Med. 1989;169:175–183. doi: 10.1084/jem.169.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamamoto H, Hanada K, Nishijima M. Involvement of diacylglycerol production in activation of nuclear factor kappaB by a CD14-mediated lipopolysaccharide stimulus. Biochem J. 1997;325:223–228. doi: 10.1042/bj3250223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zeller R, Roger M. In situ hybridization to cellular RNA. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons, Inc.; 1987. pp. 14.3.1–14.3.14. [Google Scholar]