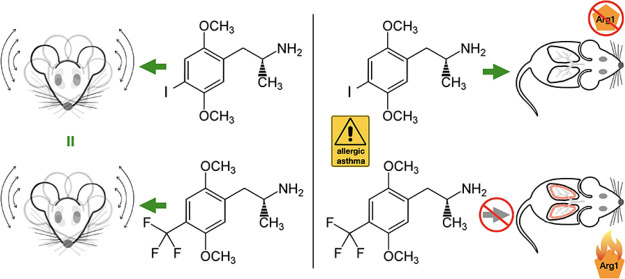
Abstract
Functional selectivity in the context of serotonin 2A (5-HT2A) receptor agonists is often described as differences psychedelic compounds have in the activation of Gq vs β-arrestin signaling in the brain and how that may relate to inducing psychoactive and hallucinatory properties with respect to each other. However, the presence of 5-HT2A receptors throughout the body in several cell types, including endothelial, endocrine, and immune-related tissues, suggests that functional selectivity may exist in the periphery as well. Here, we examine functional selectivity between two 5-HT2A receptor agonists of the phenylalkylamine class: (R)-2,5-dimethoxy-4-iodoamphetamine [(R)-DOI] and (R)-2,5-dimethoxy-4-trifluoromethylamphetamine [(R)-DOTFM]. Despite comparable in vitro activity at the 5-HT2A receptor as well as similar behavioral potency, (R)-DOTFM does not exhibit an ability to prevent inflammation or elevated airway hyperresponsiveness (AHR) in an acute murine ovalbumin-induced asthma model as does (R)-DOI. Furthermore, there are distinct differences between protein expression and inflammatory-related gene expression in pulmonary tissues between the two compounds. Using (R)-DOI and (R)-DOTFM as tools, we further elucidated the anti-inflammatory mechanisms underlying the powerful anti-inflammatory effects of certain psychedelics and identified key mechanistic components of the anti-inflammatory effects of psychedelics, including suppression of arginase 1 expression.
Keywords: psychedelic, functional selectivity, asthma, 5-HT2A, inflammation, anti-inflammatory
Serotonin 5-HT2A receptor agonists have recently emerged as an exciting new target for therapeutic development in the treatment of neuropsychiatric disorders like depression and substance use disorder.2 5-HT2A receptors are the primary site of action for the behavioral effects associated with psychedelic drugs,3 and because of their powerful behavioral effects, the vast majority of research concerning psychedelics has focused on their central nervous system (CNS) modulation.4 However, the wide prevalence of 5-HT2A receptors in peripheral tissues underscores the need for further research into their function and therapeutic potential in non-CNS tissues.5 Because of its high selectivity for the 5-HT2 receptor family,6 the agonist (R)-2,5-dimethoxy-4-iodoamphetamine [(R)-DOI] has received the most widespread utilization in preclinical studies of 5-HT2A receptor pharmacology. We previously discovered that (R)-DOI has potent anti-inflammatory activity in vitro(7) and across several rodent inflammatory systems8−11 through 5-HT2A receptor activation. We have examined the anti-inflammatory effects of (R)-DOI the most within the ovalbumin (OVA)-sensitization model of allergic asthma, a system with ease of use9 and high degree of reproducibility.12, 13 When administered intranasally at sub-behavioral doses prior to allergen challenge, (R)-DOI reduces both airway hyperresponsiveness (AHR) as well as resistance, prevents mucus overproduction that accompanies repeated allergen challenge, and prevents the increase in the mRNA expression of a specific subset of genes representing OVA-induced proinflammatory biomarkers (including Il5, Il13, Mcp-1, and Gm-csf) but not others (e.g., Il4).9 Further, intranasally administered (R)-DOI reverses collagen deposition and airway remodeling in addition to reducing inflammation in established asthma (i.e., a chronic model).11 Interestingly, we previously found that the anti-inflammatory efficacy of psychedelics in the acute OVA allergic asthma model does not correlate with either behavioral or canonical signaling potency or efficacy at 5-HT2A receptors.12 The effects of certain psychedelics to prevent inflammation and allergic asthma phenotypes are different from those of serotonin, which are known to be proinflammatory and increase AHR through activation of 5-HT2A receptors in models of allergic asthma.14
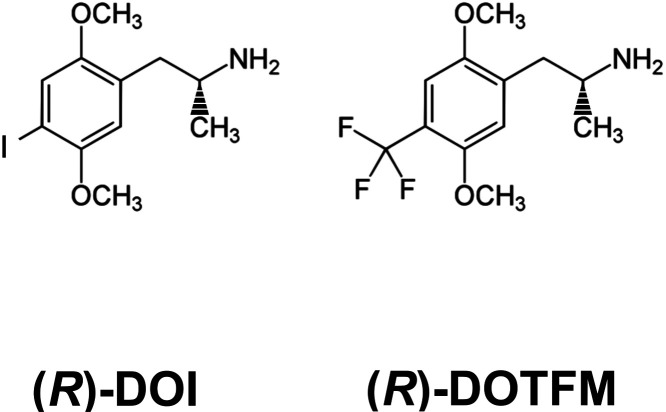
Here, we sought to further elucidate mechanisms underlying the anti-inflammatory effects of 5-HT2A receptor activation by certain psychedelics by capitalizing on the functionally selective properties of two specific ligands. The first is (R)-DOI, which we described above. The second is (R)-2,5-dimethoxy-4-trifluoromethylamphetamine [(R)-DOTFM]. (R)-DOTFM is very similar in structure to (R)-DOI, only differing at the 4’ position of their phenylalkylamine core15 (Figure 1). Furthermore, both are potent and full agonists at 5-HT2A receptors with virtually identical pharmacological properties and potent behavioral effects in rodents.15 However, whereas (R)-DOI is a potent anti-inflammatory, (R)-DOTFM has no measurable anti-inflammatory activity even at relatively high doses in our asthma model.12 In this work, we leverage these tools to examine physiological and molecular outcomes to elucidate mechanisms relevant for anti-inflammatory efficacy downstream of 5-HT2A receptor activation with the underlying hypothesis that effectors and pathways activated or inhibited specifically by (R)-DOI, but not by (R)-DOTFM, in OVA-treated animals represent those critical for anti-inflammatory effects. We demonstrate that (R)-DOTFM fails to prevent elevated AHR in our acute murine asthma model, fails to prevent the expression of OVA-induced proinflammatory cytokines, and induces a different proteomic response in the lung from that of (R)-DOI in our mouse OVA allergic asthma model. Our results suggest that the suppression of expression of certain proteins, such as arginase 1 (Arg1), may contribute to reduced airway pathophysiology and the anti-inflammatory effects of (R)-DOI. Further, our work demonstrates that functional selectivity exists between psychedelic compounds in vivo with respect to physiological outcomes, a finding that should be taken into account when designing novel 5-HT2A agonists for clinical applications.
Figure 1.
Structures of compounds. (R)-2,5-Dimethoxy-4-iodoamphetamine [(R)-DOI]. (R)-2,5-Dimethoxy-4-trifluoromethylamphetamine [(R)-DOTFM].
Results and Discussion
Functional Assessment of (R)-DOI and (R)-DOTFM on 5-HT2-Mediated Signaling Pathways
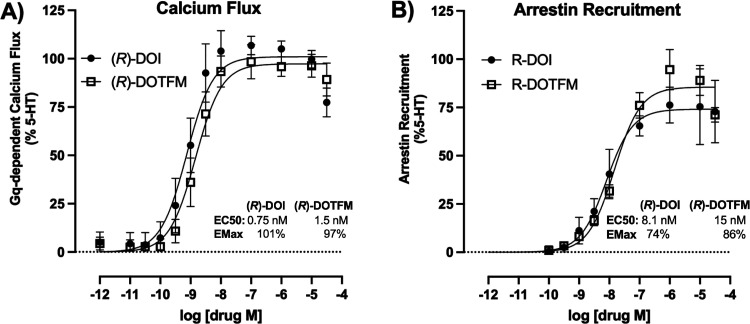
For Gq-mediated calcium flux downstream of the activation of human 5-HT2A receptors,16 we found that both (R)-DOI and (R)-DOTFM have virtually identical properties, both being full and potent agonists (Figure 2A). Further, β-arrestin recruitment to 5-HT2A receptors following the administration of (R)-DOI and (R)-DOTFM was not significantly different between the two compounds (Figure 2B). That (R)-DOTFM has no measurable anti-inflammatory activity while still being a potent and full agonist at these signaling pathways further underscores our earlier observation that these canonical pathways do not underlie the anti-inflammatory effects of psychedelics.12
Figure 2.
Concentration–response curves for signaling pathways in HEK cell lines stably expressing human 5-HT2A receptors. (A) Cells were administered varying concentrations of (R)-DOI or (R)-DOTFM, and fluorescence was monitored for 5 min. Both drugs are nearly identical at this Gq-mediated signaling pathway. (B) Cells were administered varying concentrations of (R)-DOI or (R)-DOTFM, and luminescence was monitored for 120 min. Experiments were performed in parallel and represent three or more independent experiments; error bars indicate ± SEM.
Head-Twitch Response
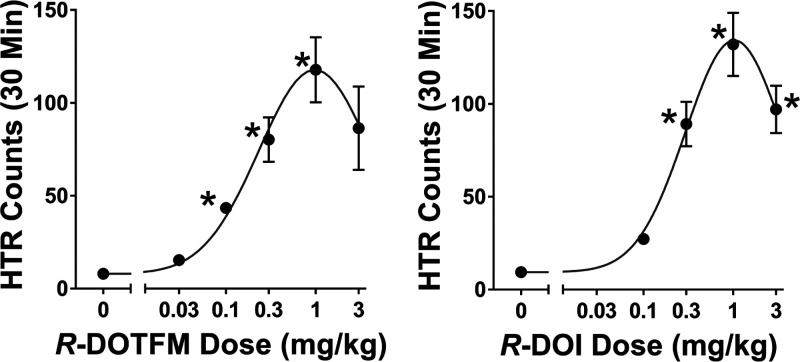
(R)-DOTFM is a potent activator of the HTR, with an ED50 of 0.60 μmol/kg (Figure 3). This is nearly identical to the 0.66 μmol/kg ED50 of (R)-DOI in the HTR assay as previously determined using the same protocols and number of animals as described by Halberstadt et al.1
Figure 3.
Head-twitch analysis of (R)-DOTFM. The effects of (R)-DOTFM were examined in the head-twitch response (HTR) assay. (R)-DOTFM produced a strong HTR with a potent ED50 of 0.60 μmol/kg. These values are equivalent to (R)-DOI, as previously determined.16 Values are mean HTR counts ± SEM.
Intranasal Administration of (R)-DOI and (R)-DOTFM Differentially Impacts Anti-AHR Activity Associated with 5-HT2A Receptor Activation
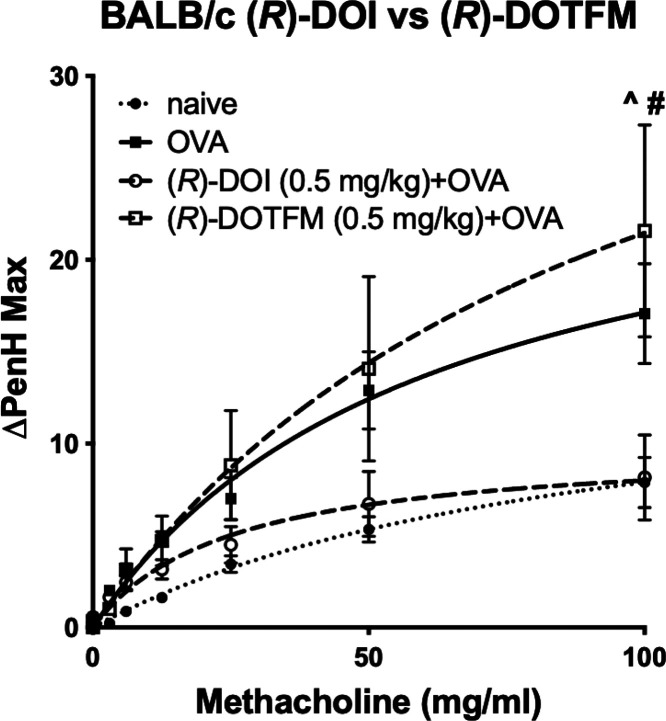
In the acute allergic OVA asthma model, we determined the enhanced pause (PenH) in wild-type BALB/c mice (Figure 4) for (R)-DOI and (R)-DOTFM. In agreement with our previous results, intranasally administered (R)-DOI (0.5 mg/kg) fully prevented ovalbumin-induced elevations in AHR following methacholine challenge. Conversely, intranasally administered (R)-DOTFM (0.5 mg/kg) was unable to prevent ovalbumin-induced elevations in AHR (Figure 4). Further, pretreatment with (R)-DOTFM elicited a trend for increased hyperresponsivity greater than OVA-alone.
Figure 4.
(R)-DOTFM fails to prevent OVA-mediated elevations in AHR compared to (R)-DOI in BALB/c mice. Results from whole body plethysmography on awake, freely moving mice exposed to 0.5 mg/kg (R)-DOI or 0.5 mg/kg DOTFM (inhaled). n = 7–8 mice/treatment group; ^p < 0.001 (R)-DOI + OVA vs naive, #p < 0.001 (R)-DOTFM + OVA vs naive; error bars represent ± SEM; two-way ANOVA with Bonferroni post hoc test.
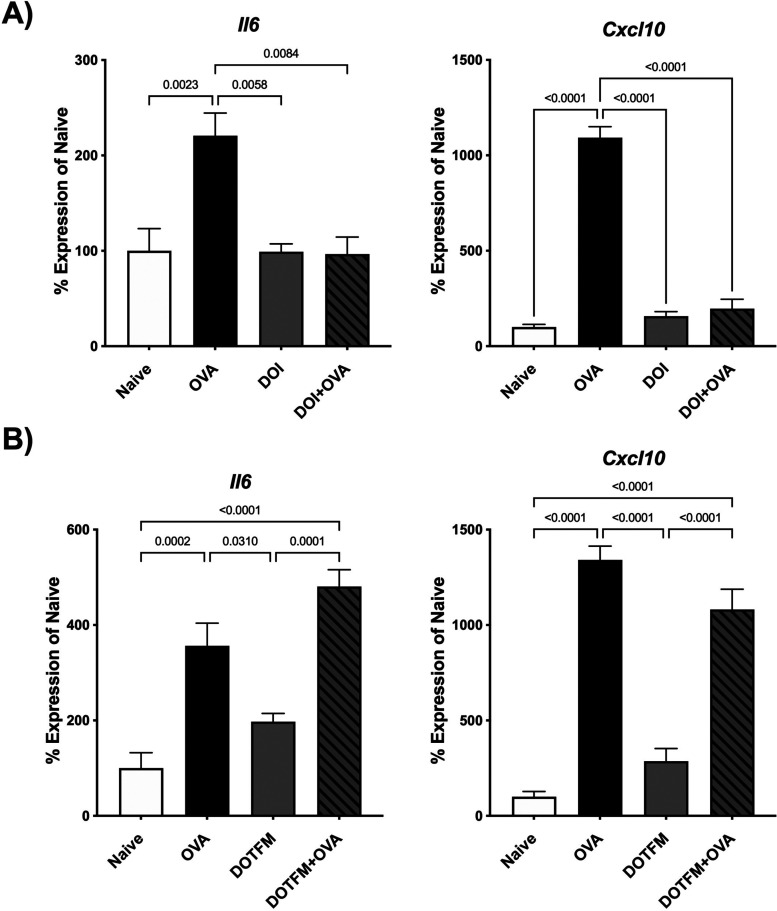
Effects of Intranasally Administered (R)-DOI and (R)-DOTFM on Proinflammatory Biomarker Gene Expression
In the standard acute OVA sensitization model of allergic asthma, pulmonary inflammation is at its peak 48 h after the third and final OVA exposure.13 In this experiment, similar to our previous experiment, appropriate groups of animals were pretreated with 0.5 mg/kg intranasal (R)-DOI or (R)-DOTFM for 30 min prior to allergen exposure. For proinflammatory biomarker analyses, two genes were selected: interleukin 6 (Il6) and C-X-C motif chemokine ligand 10 (Cxcl10) (Figure 5). IL-6 has well-defined roles in both the immune response and asthma;17 concordantly, CXCL10 expression is altered in several inflammatory models following 5-HT2 activation.10,11 OVA increased mRNA levels of both inflammatory markers (Figure 5). Exposure to (R)-DOI or (R)-DOTFM alone did not significantly alter the expression of Il6 and Cxcl10 (Figure 5). Pretreatment with (R)-DOI completely attenuated elevations of OVA-induced Il6 and Cxcl10 (Figure 5A), whereas (R)-DOTFM pretreatment produced no significant reduction (Figure 5B).
Figure 5.
Nebulized nose-only administration of (R)-DOTFM and (R)-DOI (0.5 mg/kg) differentially regulates inflammatory gene expression prior to and upon OVA exposure. Quantitative RT-PCR measurement of mRNA expression levels for interleukin 6 (Il6) and C-X-C motif chemokine ligand 10 (Cxcl10) following drug treatment 30 min prior to OVA exposure (Drug + OVA) ((A) (R)-DOI and (B) (R)-DOTFM, respectively) is shown. OVA alone increased the expression of both inflammatory markers, whereas nasal treatment with either drug produced no significant difference from naive controls. Pretreatment with (R)-DOI prior to allergen exposure completely blunted the expression of both inflammatory markers. (R)-DOTFM was unable to reduce inflammatory marker expression for either inflammatory marker. n = 6–7 mice/treatment group; error bars represent SEM; p values for all comparisons < 0.05 are shown; one-way ANOVA with Tukey post hoc test for multiple comparisons.
Quantitative Proteomics Analysis of OVA-Challenged Whole-Lung Homogenate from Mice Treated with Intranasal (R)-DOI or (R)-DOTFM
To gain insight into the potential mechanism of action, these two drugs were leveraged as tools in quantitative proteomic experiments. Our hypothesis was that proteins with shared regulation likely represent overall effects of 5-HT2 receptor activation, whereas those significantly influenced by (R)-DOI but not (R)-DOTFM represent those potentially involved in the mechanism of action for anti-inflammatory effects. Tandem mass tag-liquid chromatography–mass spectrometry (TMT-LC–MS/MS) analyses were performed for proteomic profiling of whole-lung homogenate from all treatment groups: naive mice (n = 5); OVA mice (n = 5); (R)-DOI alone mice (n = 5); (R)-DOTFM alone mice (n = 5); (R)-DOI + OVA (n = 6); and (R)-DOTFM + OVA (n = 6). About 5000 total proteins were quantifiably detected, with about another 800 proteins identified but not quantifiable.
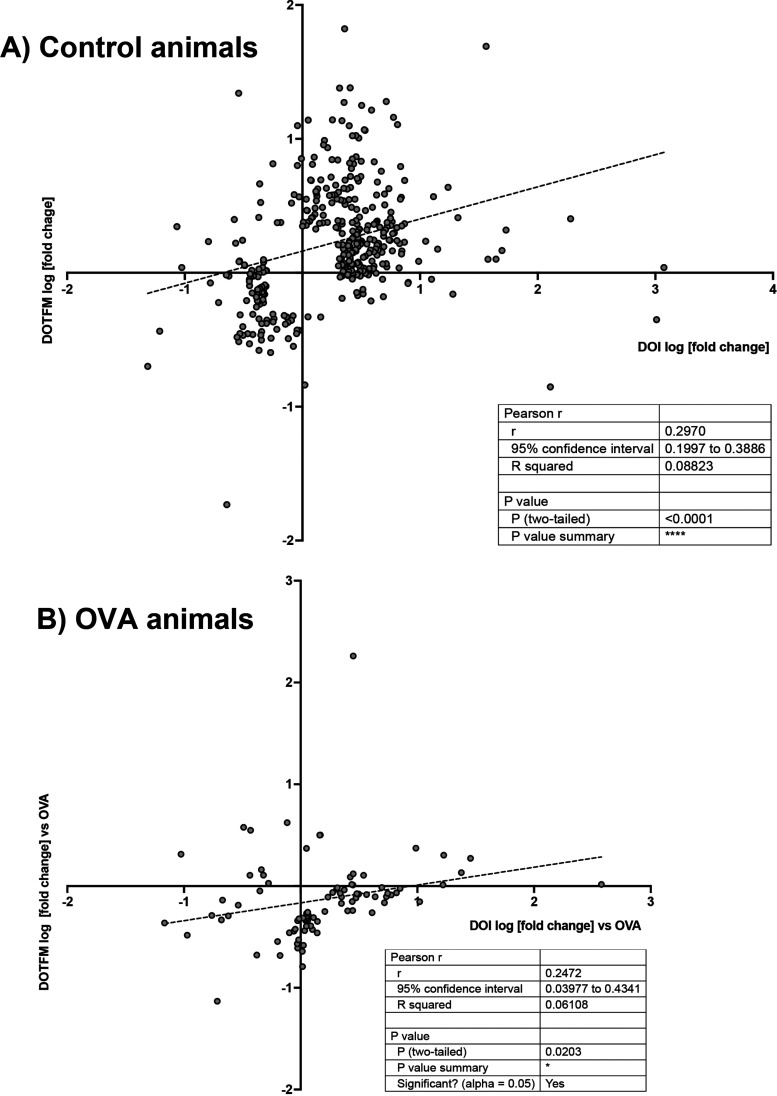
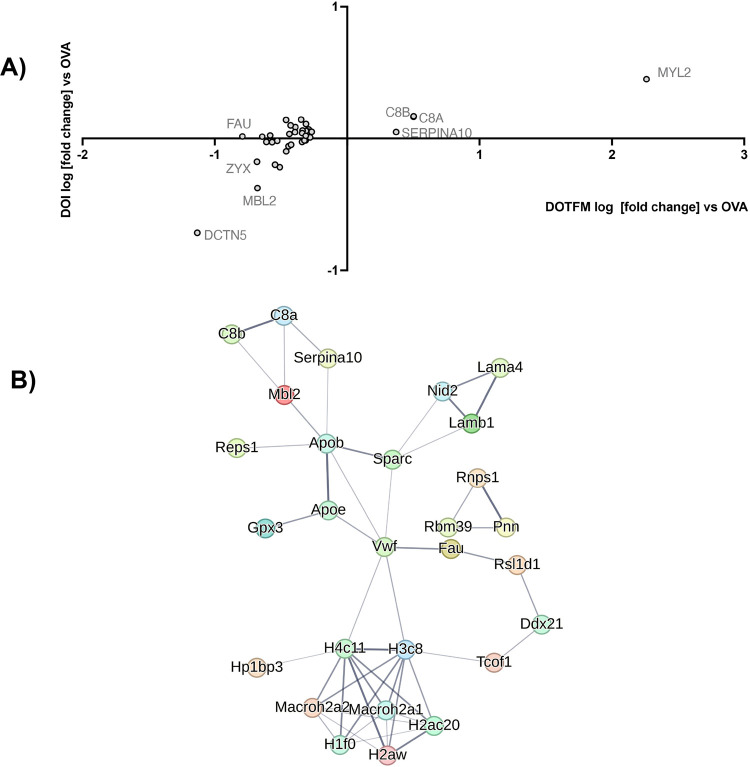
As expected, the treatment with an OVA alone induced several protein changes relevant to inflammation and asthma (Table S1). Both (R)-DOI and (R)-DOTFM had significant effects on protein expression in normal mice (non-OVA treated) (Tables S2 and S3). In comparing the effects of (R)-DOI to (R)-DOTFM in control mice for proteins differentially affected by >2-fold, p < 0.01, there was a weak correlation (r = 0.297, p < 0.0001) (Figure 6A). In OVA-treated animals, (R)-DOI and (R)-DOTFM had a similar weak correlation for differentially expressed proteins (r = 0.247, p < 0.0001) (Figure 6B). These results indicate that although (R)-DOI and (R)-DOTFM induce somewhat of a shared proteomic response in both control and OVA-treated animals, there is a considerable difference in the proteomic responses between the two drugs. Specifically, we identified several proteins that had a greater than 2-fold expression difference (p < 0.01) between (R)-DOI and (R)-DOTFM in OVA-treated animals (Table S4). These can be grouped into two sets: (1) proteins significantly altered by (R)-DOI but not (R)-DOTFM (Figure 7) and (2) proteins significantly altered by (R)-DOTFM but not (R)-DOI (Figure 8).
Figure 6.
Correlational analysis between the effects of (R)-DOI and (R)-DOTFM. There is a weak correlation between protein changes induced in control animals (>2-fold; p < 0.01) (A). There is a moderate correlation between protein changes in OVA-treated animals (B). The log [fold change] induced by (R)-DOI (DOI on plot) is plotted on the X axis, and the log [fold change] induced by (R)-DOTFM (DOTFM on plot) is plotted on the Y axis.
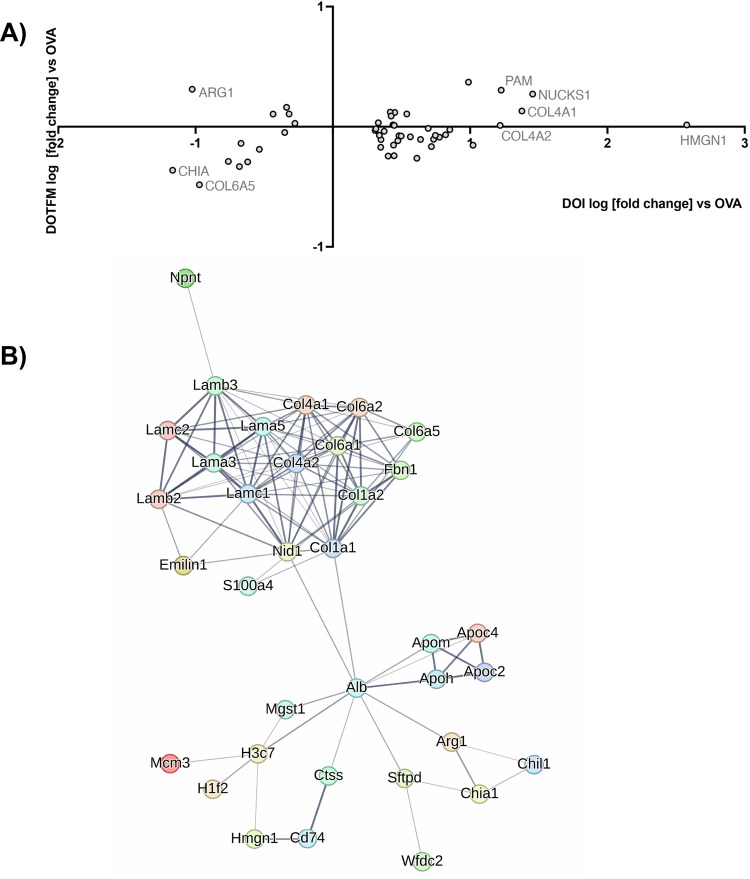
Figure 7.
Interaction networks: (R)-DOI. Proteins whose expression are significantly (p < 0.01) changed 2-fold or greater in the (R)-DOI + OVA treatment group compared to OVA alone but not significantly changed in the (R)-DOTFM + OVA group compared to OVA alone. (A) Log [fold change] plot, with those most differentially expressed labeled. (B) String db interaction map (default settings, disconnected nodes hidden, thickness of lines represents overall strength of connections across databases).
Figure 8.
Interaction networks: (R)-DOTFM. Proteins whose expression are significantly (p < 0.01) changed 2-fold or greater in the (R)-DOTFM + OVA treatment group compared to OVA alone but not significantly changed in the (R)-DOI + OVA group compared to OVA alone. (A) Log [fold change] plot, with those most differentially expressed labeled. (B) String db interaction map (default settings, disconnected nodes hidden, thickness of lines represents overall strength of connections across databases).
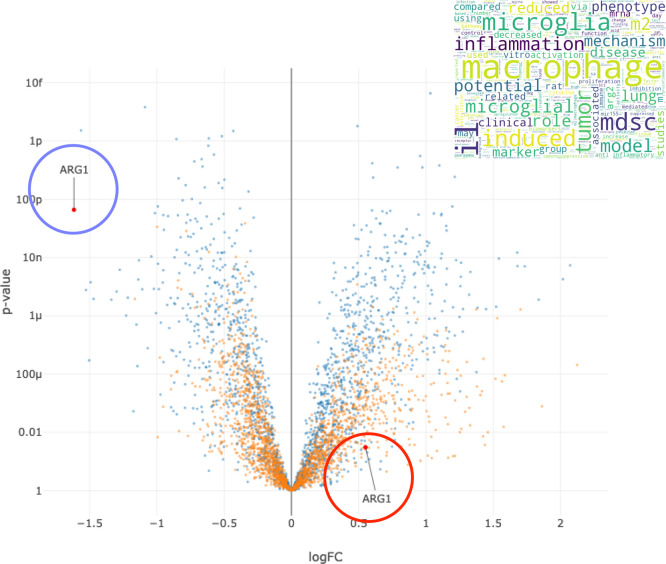
The protein that was most changed by (R)-DOI compared to (R)-DOTFM in the OVA-treated mice was the high mobility group nucleosome binding domain 1 (HMGN1), which increased by 378-fold in the (R)-DOI treatment vs no change in the (R)-DOTFM treatment (Figure 7A). Interestingly, several additional chromatin/epigenetic-related proteins were differentially altered between (R)-DOI and (R)-DOTFM. Another interesting protein preferentially increased by (R)-DOI compared to (R)-DOTFM in the OVA-treated animals is Nucks1, which is associated with chromatin accessibility,18 energy homeostasis,19 and DNA damage repair.20 Two of the most repressed proteins by (R)-DOI compared to (R)-DOTFM were arginase 1 (Arg1; 11-fold decrease vs no change) and chitinase, both of which have been tightly linked to the pathophysiology of allergen-induced asthma and proinflammatory processes.21,22Figure 9 shows a volcano plot of proteins in the (R)-DOI + OVA group compared to OVA alone and the (R)-DOTFM + OVA group compared to OVA alone, highlighting the marked decrease in expression produced by (R)-DOI treatment compared to (R)-DOTFM treatment. Cluster analysis using STRING highlighted a densely interconnected subset of proteins primarily consisting of an extracellular matrix and structural proteins including lamins and collagens (Figure 7B). A similar analysis of proteins only significantly affected by (R)-DOTFM revealed a loosely interconnected set of histone-related proteins (Figure 8B). Myosin light chain kinase 2 (Myl2) was the most increased by (R)-DOTFM, by 182-fold vs no change by (R)-DOI in the OVA-treated animals (Figure 8A).
Figure 9.
Arginase 1. Volcano plot of differential protein expression of the effects of each drug in OVA-treated animals (blue dots = (R)-DOI + OVA vs OVA alone; orange dots = (R)-DOTFM + OVA vs OVA alone). Arginase 1 (ARG1) is decreased >10-fold by (R)-DOI treatment in OVA-treated animals compared to OVA alone (blue circle), whereas (R)-DOTFM does not significantly change the expression of arginase 1, although there is a trend for a slight increase. X axis = log [fold change]; Y axis= log [p]. Inset = word cloud generated for arginase 1 based on associations in scientific databases.
Validation of Arg1 Expression
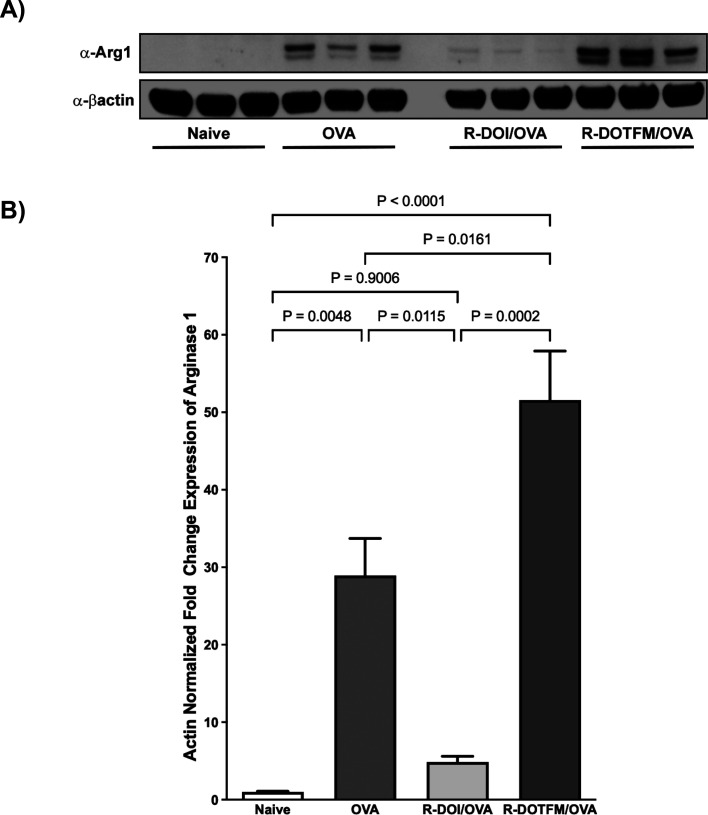
In allergic asthma, Arg1 expression levels correlate with AHR, airway inflammation, and airway remodeling.23,24 To validate quantitative proteomic results for Arg1 expression, we performed Western blot analysis on lung homogenates. As expected, Arg1 expression was significantly and robustly increased in the OVA-treated animals compared to the control group (Figure 10A,B; Figure S1). Pretreatment with (R)-DOI blocked OVA-induced Arg1 expression. In contrast, pretreatment with (R)-DOTFM significantly increased Arg1 expression above that of OVA-alone.
Figure 10.
Western blot analysis of Arg1 expression. Lung homogenates were analyzed by Western blot to validate proteomic results for Arg1. (A) Representative samples from treatment groups showing Arg1 expression and β-actin used for normalization. (B) Quantification of results showing that OVA treatment robustly increased Arg1 expression; pretreatment with (R)-DOI reduced Arg1 expression in OVA-treated animals to baseline levels. (R)-DOTFM, however, significantly increased Arg1 levels above that of even OVA-alone (error bars = SEM; p values shown; one-way ANOVA with Tukey post hoc analysis).
GSEA
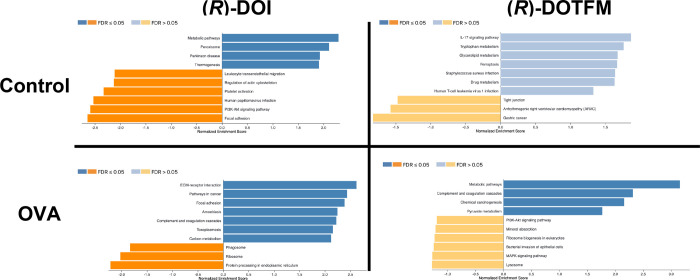
Applying GSEA to protein data sets, several sets of proteins were found to be significantly increased and decreased by (R)-DOI in the control animals (Figure 11). Those pathways with proteins increased by (R)-DOI include “metabolic pathways” (Table S5) and “peroxisome”, with significantly decreased sets including “focal adhesion” (Table S6) and “PI3K-Akt signaling pathway”. There were no sets significantly changed by (R)-DOTFM in the control animals. In the OVA-treated animals, (R)-DOI had a very different set of proteins significantly altered that included the increased sets of “ECM-receptor interaction” (Table S7) and “focal adhesion”, and the decreased sets of “protein processing in endoplasmic reticulum” (Table S8) and “phagosome”. In the OVA-treated animals, (R)-DOTFM only significantly increased protein sets, including “metabolic pathways” (Table S9), and none were significantly decreased. These results together underscore that these two drugs are eliciting quite different responses despite their similar pharmacology.
Figure 11.
Gene Set Enrichment Analysis. GSEA found several sets significantly altered by drug treatment. (R)-DOI had more significant effects (FDR < 0.05) than (R)-DOTFM and significantly altered sets by both increases and decreases in expression. (R)-DOTFM elicited no significant changes in set expression at an FDR > 0.05 control animals (all FDRs > 0.05), and only sets significantly increased (FDR < 0.05) were identified in the OVA-treated animals given (R)-DOTFM.
We have previously demonstrated in a rodent acute asthma model that 5-HT2A receptor activation mediates the anti-inflammatory effects of psychedelics and that 5-HT2A agonist behavioral effects do not correlate with anti-inflammatory efficacy.12 In this study, we examined two structurally similar phenylalkylamine psychedelics, (R)-DOI and (R)-DOTFM, that are virtually identical with respect to physiochemical properties, 5-HT2A receptor affinity and pharmacology, as well as behavioral effects.15 We directly compared the two drugs in the canonical 5-HT2A-coupled signaling pathway of Gq-mediated calcium flux and recruitment of β-arrestin2 and found no significant differences in either potency or efficacy (Figure 2). We also found that (R)-DOI and (R)-DOTFM have virtually identical activity in the HTR assay (Figure 3), a 5-HT2A receptor-mediated behavior in mice. However, despite having similar effects on in vitro and in vivo measures of 5-HT2A receptor activation, (R)-DOI is a potent anti-inflammatory compound, whereas (R)-DOTFM is devoid of anti-inflammatory activity (Figures 4 and 5). These two drugs therefore represent ideal tools to tease apart aspects of the mechanism of action for the anti-inflammatory effects of psychedelics and inform future drug discovery. Further, this work represents the first in-depth analysis of the functional selectivity of 5-HT2 receptor agonists outside the CNS.
In agreement with previous results, nasally administered (R)-DOI completely prevented the development of airway hyperresponsiveness and suppressed the expression of proinflammatory biomarker mRNA in lung tissues in animals sensitized and exposed to OVA in our allergic asthma model. At the same concentration, nasally administered (R)-DOTFM did not significantly attenuate either OVA-induced airways hyperresponsiveness or mRNA expression of proinflammatory biomarkers. The dose of drug that we used, 0.5 mg/kg, is nearly 2 orders of magnitude fold greater than the EC50 anti-inflammatory dose of (R)-DOI in this model (0.006 mg/kg).12 Because of the virtually identical nature of the receptor pharmacology between (R)-DOI and (R)-DOTFM, we do not believe that this lack of anti-inflammatory activity for (R)-DOTFM is due to not using enough drug.
We hypothesize that functional selectivity underlies the differences in anti-inflammatory effects with (R)-DOTFM stabilizing the 5-HT2A receptor in a slightly different conformation than (R)-DOI, which ultimately retains coupling to canonical signaling effectors but lacks coupling to anti-inflammatory relevant effectors. Further, we hypothesize that (R)-DOI potently recruits a yet to be identified signal transduction effector that leads to its anti-inflammatory effects that serotonin and some agonists like (R)-DOTFM do not recruit. We employed quantitative proteomic analysis of lung tissues from animals treated with these drugs to reveal key mechanistic components of the anti-inflammatory effects of (R)-DOI. These would likely be proteins whose expression was significantly altered more by (R)-DOI than by (R)-DOTFM in OVA-treated animals. In the OVA-alone-treated animals compared to control, as anticipated, the top differentially expressed proteins are all relevant to inflammation and/or asthma (e.g., Clca1,25 Chia,22,26 Bpifb1,27 Epx,28 and Arg123,24,29). Somewhat surprisingly, given their conserved structures and pharmacology, there were numerous and substantial differences between the proteomic responses to these drugs in both control and the OVA-treated animals. In control animals, GSEA found that (R)-DOI treatment led to the most significant decrease in the focal adhesion set (Table S6). Among the decreased proteins in this set are those associated with Rho kinase (ROCK; Rho-associated coiled-coil forming kinases) signaling and subsequent downstream targets such as myosin light chain and actin, suggesting that (R)-DOI may abrogate some of the effects of an inflammatory agent on tissues through limiting cytoskeletal or cellular structure remodeling that exposure to an inflammatory agent may induce. Indeed, pharmacological inhibition or genetic deletion of ROCK reduces airway stiffness and abolishes airway hyperresponsiveness. (R)-DOTFM did not affect these proteins.
To narrow candidates to those potentially involved in the anti-inflammatory effects of (R)-DOI, we filtered differentially expressed proteins for those where there was a 2-fold or greater difference in the expression change induced by either drug compared to the OVA-only treated animals (p < 0.01) and whose difference in expression between that induced by (R)-DOI and (R)-DOTFM was also 2-fold or greater. We further grouped these proteins into those that (R)-DOI, but not (R)-DOTFM, altered levels and those that (R)-DOTFM, but not (R)-DOI, altered levels in the OVA-treated animals. One of the most interesting proteins identified is arginase 1 (Arg1). This enzyme is upregulated by OVA treatment and significantly downregulated by (R)-DOI treatment (>10-fold), but not by (R)-DOTFM, where it remains upregulated. In agreement with the previously established role of elevated Arg1 in the pathophysiology of asthma, Arg1 levels are significantly elevated in the group of Arg1 treated above levels induced by OVA alone. Arg1 catalyzes the production of l-ornithine and urea from l-arginine, leading to cellular proliferation and collagen production.30 In the context of asthma, its upregulation results in inflammation, increased formation of peroxinitrites, and airways remodeling that are associated with AHR and airway obstruction.23 In the absence of Arg1 activity, l-arginine is instead converted to NO via NOS, which inhibits inflammation and contributes to the relaxation of the bronchial smooth muscle.24 As such, inhibitors of Arg1 have been explored for asthma therapeutics.31 In animal models of ischemic heart disease, high Arg1 expression is associated with a macrophage phenotype shift from the “classical activated” phenotype (M1) to the “alternatively activated” phenotype (M2).32 Misbalance of the M1/M2 polarization equilibrium or misregulated macrophage polarization switching can exacerbate chronic inflammatory pathologies.33 Accordingly, Arg1-expressing M2 macrophages produce the precursors necessary for collagen synthesis and cell proliferation.33,34 As 5-HT2A receptor activation impacts both elements of macrophage polarization9,35 (unpublished data) and collagen deposition,11 a proteomics approach has allowed for the identification of Arg1 and its suppression as a potential mechanism underlying (R)-DOI’s antiasthma effects.
GSEA revealed that ECM-related proteins are significantly increased by (R)-DOI relative to that of (R)-DOTFM in OVA-treated animals. In asthma, dysregulation of airway and basement membrane remodeling are critical processes in the progression and chronicity of asthma.36,37 As such, fibrillar collagens (i.e., collagens I and III) and network-forming collagens (type IV) all contribute to airway structure and are misregulated in asthma. Our work here shows that unlike (R)-DOTFM, (R)-DOI both positively and negatively regulates collagen associated proteins associated with the preservation of homeostatic basement membrane conditions (e.g., Serpinh1, Col14a1, Col15a1, Col6a1, Col6a2, and Col6a6). Importantly, collagen fibers undergo cross-linking processes, which play critical roles in the stiffness of the lung structure.37 A recent study utilized vitamin B2 and ultraviolet A radiation to cross-link collagen in bovine lungs, ultimately stiffening their structure.38 Interestingly, even without an inflammatory stimulus, cross-linked lungs constricted more quickly and to a greater degree than control samples. Our work suggests that the maintenance of airway structure integrity via (R)-DOI treatment is likely a mechanism of negating cytokine-mediated alterations to membrane structure, which prophylactically may provide a noninflamed microenvironment that prevents increased misregulation. Similar mechanisms may underlie our previous finding that (R)-DOI can reverse and reduce collagen deposition and airway structural remodeling in a chronic asthma model.11
Our results represent the first in-depth study of the functional selectivity of 5-HT2 receptor agonists in peripheral tissues. Manipulating elements of functional selectivity to improve therapeutic properties of compounds is a strategy currently being employed in drug discovery with cardiovascular medications.39 Results from our previous work investigating the SAR of psychedelics identified 2,5-dimethoxyphenethylamine (2C-H) as the pharmacophore for psychedelic anti-inflammatory activity,12 which ultimately can be integrated with mechanistic findings here toward drug discovery and development efforts to develop next-generation anti-inflammatories based on 5-HT2A receptor agonism for clinical use. Leveraging the functional selectivity between (R)-DOI and (R)-DOTFM, we have identified proteins and molecular processes likely contributing to the anti-inflammatory effects of certain psychedelics like (R)-DOI. These include reductions in the levels of proteins associated with inflammation, such as Arg1; increases in levels of proteins involved in maintaining physical cellular integrity; and changes in the levels of proteins involved in epigenetic mechanisms and chromatin structure.
Methods
Drugs and Reagents
(R)-DOI hydrochloride was synthesized by Onyx (London, UK). (R)-DOTFM hydrochloride was synthesized and kindly provided by Dr. Jason Wallach (University of the Sciences, Philadelphia, PA). All drugs for asthma studies were dissolved in 0.9% sterile saline solution (Baxter Healthcare Corp., Deerfield, IL) immediately prior to use at a concentration of 1.0 mg/mL.
Animals for Use in the Allergic Asthma Assay
For all inflammatory experiments, pathogen-free wild-type BALB/c mice were obtained from Charles River Laboratories (Wilmington, MA). Animals were maintained in a pathogen-free environment within the Division of Animal Care vivarium at the Louisiana State University Health Sciences Center (New Orleans, LA) in ventilated cages (Allentown Caging Equipment Co., Inc., Allentown, NJ) with ad libitum access to food and water on a 12 h light/dark cycle (lights on at 7:00 a.m. and off at 7:00 p.m.). All animal care and experimental procedures were approved by the Institutional Animal Care and Use Committee at the Louisiana State University Health Sciences Center.
OVA-Induced Acute Allergic Airway Inflammation and Drug Exposure
Forty-eight 6–8 week old male mice weighing between 20 and 25 g were sensitized and challenged with chicken OVA grade V (Sigma-Aldrich, St. Louis, MO) as previously described.9 Briefly, on days 0 and 7, mice were sensitized by intraperitoneal injection (IP) of 0.1 mL solution containing 20 μg OVA emulsified in 2 mL Imject Alum [Al(OH)3/Mg(OH)2; Pierce, Rockford, IL]. On days 14–16, 30 min prior to saline/OVA challenge, each animal was placed in a nylon-coated stainless-steel wire restraint with a nose cone for administration of nebulized drug (SoftRestraints; SciReq, Montreal, QC, Canada), with efforts taken to minimize stress and the amount of time in the restraint. Animals were treated with 0.5 mg/kg of (R)-DOI, (R)-DOTFM, or sterile saline control (nose-only inhalation) using an ultrasonic nebulizer (Aeroneb Pro; Aerogen, Galway, Ireland). Mice were placed in groups of no more than 20 animals into a 7 L (21 × 22 × 15 cm) clear plexiglass induction chamber (Vet Equip, Inc., Pleasonton, CA) and challenged for 20 min with either sterile saline (Baxter Healthcare Corp.) or a 1% (wt/vol) OVA solution in sterile saline. Aerosol was generated using a VixOne reusable nebulizer (Westmed, Inc., Tucson, AZ) driven by a Pari Proneb Ultra compressor (Pari Proneb, Midlothian, WA).
Noninvasive Whole-Body Plethysmography and Methacholine Challenge
Forty-eight hours following the final OVA challenge, airway function was measured in a single-chamber WBP system (Buxco Electronics, Troy, NY, and EMKA Technologies, Falls Church, VA) in unrestrained, conscious mice. The enhanced pause (PenH) was used as a proxy for AHR, as previously described.40 Animals were allowed to acclimate by first being placed in a single chamber for 10 min; baseline PenH was subsequently measured for 3 min prior to the methacholine (MeCh; Sigma-Aldrich) challenge. Mice were exposed to the bronchoconstrictor MeCh for 1 min at increasing concentrations (0, 3.125, 6.25, 12.5, 50, and 100 mg/mL in isotonic saline), and PenH was recorded for 3 min. Animals displaying signs of distress were immediately removed from the protocol. Maximal PenH obtained from the IOX2 software (SciReq) was analyzed by using the Datanalyst software (Datanalyst v2.6.1.14; EMKA Technologies, Paris, France). Maximal PenH values were averaged for each dose and plotted as percent change from vehicle control, as described.9,11
mRNA Expression Analysis of Inflammatory Biomarkers
All animals were humanely euthanized using a small mammalian anesthetic device (Handlebar Anesthesia Service, Austin, TX) to first induce deep anesthesia at 4% inhaled isoflurane (Piramal Enterprises Ltd., Telangana, India) with a continuous oxygen flow rate of 0.5–1 L/min followed by cardiac desanguination. Lungs were harvested 48 h after the final OVA exposure and immediately immersed in isopentane (2-methylbutane; Sigma-Aldrich), chilled on dry ice, and stored at −80 °C until processing. Total RNA was extracted from the left lobe of each animal with the TRIzol reagent (Life Technologies; Carlsbad, CA) per the manufacturer’s instruction. RNA pellets were resuspended in nuclease-free H2O and quantified by a spectrophotometer (NanoDrop ND-1000; NanoDrop Technologies, Inc., Wilmington, DE). RNA was quantified at A260 and A280, with a minimum 260/280 absorbance ratio of 1.6 used for all assays to maintain purity. Suitable RNA was processed into first-strand cDNA using the ImProm-II cDNA synthesis kit (Promega; Madison, WI) following the manufacturer’s instruction. Input cDNA for each reaction was 500 ng of total RNA. Inflammatory gene expression was examined by probe-based qRT-PCR; primers were synthesized by Integrated DNA Technologies (Coralville, IA) and designed to be compatible with the Universal ProbeLibrary system using the Universal ProbeLibrary Assay Design Center (Roche Diagnostics; Indianapolis, IN). Primer sequences used in this study are as follows: Il6 forward 5′-ctggggatgtctgtagctcat-3′ and reverse 5′-ggcaactggatggaagtctc-3′; Cxcl10 forward 5′-gctgccgtcattttctgc-3′ and reverse 5′-tctcactggcccgtcatc-3′. Probes used were from the Universal ProbeLibrary (Roche Diagnostics, Indianapolis, IN) and are listed with the following numbers: U50 and U18 for Il6 and Cxcl10. Gene expression quantification was performed on a Roche LightCycler 480 Instrument II LC instrument (Roche Diagnostics). Gene expression levels were calculated using the comparative threshold cycle method and normalized to internal Gapdh expression, as determined using the Mouse GAPD Gene Assay (cat. no. 05046211001; Roche Diagnostics) in multiplex format. GAPDH was selected as an internal control due to minimal impacts on gene41 and protein42 expression following 5-HT2 receptor activation as found by others and our previous studies using this gene as a reference for pulmonary analysis of gene expression in the OVA model as its expression remains stable between treatment groups11,.12
Calcium Flux Assay
Calcium flux downstream of 5-HT2A receptor activation was determined using human embryonic kidney cells (HEK293) stably expressing the human 5-HT2A receptor (HEKH2A; Bmax: 1600 fmol/mg protein). The generation of this cell line has been previously reported.16 Cells were maintained in DMEM (Gibco, cat. no. 10569-010) supplemented with 10% fetal bovine serum (Gibco, cat. no. 16000-044; lot no. 2344386RP), 100 U/mL penicillin, 100 mg/mL streptomycin, and 100 μg/mL Zeocin. Cells were seeded in 200 μL DMEM supplemented with 1% dialyzed fetal bovine serum (Gibco, cat. no. A33820-01; lot no. 2244935P) onto black 96-well poly-d-lysine coated plates with clear bottoms (30,000 cells/well) and maintained overnight in a humidified atmosphere at 37 °C and 5% CO2. The following day, the medium was aspirated and replaced with HBSS supplemented with 30 mM HEPES, loaded with 5 mM Fluo-2 AM HA (ION Biosciences, LLC) and 2.5 mM water-soluble probenecid (ThermoFisher Scientific), incubated for 1 h at 37 °C, washed with HBSS–HEPES, and maintained in 100 μL of HBSS–HEPES supplemented with 2.5 mM water-soluble probenecid. The plates of dye-loaded cells were placed into a FlexStation 3 microplate reader (Molecular Devices, LLC) set at 37 °C to monitor fluorescence (excitation, 485 nm; emission, 525 nm; cutoff, 515 nm). Plates were read for 30 s (2 s interval) to establish baseline fluorescence, then administered 50 μL of test compounds diluted for a final concentration range of 1 pM to 30 μM, and read for an additional 300 s. After obtaining a calcium flux trace for each sample, the mean baseline fluorescence (F) was subtracted from the peak fluorescence (ΔF) in each well, and the product was normalized by F(ΔF/F). The data were analyzed using three-parameter nonlinear regression curve-fitting routines in GraphPad Prism 9.2 (GraphPad Software, Inc.) to generate EC50 values. EMAX values were determined by normalization to the maximum 5-HT response (100%) and minimum 5-HT response (at 10–12 M concentration; 0%) on the same plate. Reported values are the average of each drug concentration tested in duplicate over three independent experiments.
Arrestin Recruitment Assay
Recruitment of the cytosolic chaperone protein arrestin-3 (β-arrestin2) to 5-HT2A receptors was determined using an HEK cell line containing stable expressions of human 5-HT2A receptors (550 fmol/mg protein) and human β-arrestin2 tagged with components of the NanoLuc Binary Technology (NanoBiT) functional complementation system (Promega Corporation, Madison, WI). The cell line was a gift from Drs. Eline Pottie and Christophe Stove (University of Ghent, Belgium), and its generation has been described in the literature.43 Cells were seeded in 200 μL of DMEM supplemented with 1% (v/v) dialyzed fetal bovine serum (Gibco, cat. no. A33820-01; lot no. 2244935P) onto white 96-well poly-d-lysing coated plates with clear bottoms (40,000 cells/well) and maintained overnight in a humidified atmosphere at 37 °C and 5% CO2. The following day, the medium was aspirated, and wells were washed once with HBSS supplemented with 30 mM HEPES (pH 7.4); then, 100 μL HBSS–HEPES buffer and 25 μL of Nano-Glo Live Cell Reagent (diluted 1/20 in the manufacturer supplied LCS buffer) were added to each well. Test plates were then placed into a FlexStation 3 microplate reader (Molecular Devices, Sunnyvale, CA) set at 37 °C to monitor luminescence. Plates were read for 15 min (2 min interval) to allow for the stabilization of luminescent signal and establishing baseline luminescence. Test compounds were diluted in the HBSS–HEPES buffer and manually added to wells. Plates were then returned to FlexStation 3 to read luminescence for an additional 120 min (2 min interval). After obtaining raw luminescence kinetic traces, the AUC of the 2 h run was calculated, and the data were analyzed using a three-parameter nonlinear regression curve-fitting function in GraphPad Prism 9.3.1 (GraphPad Software, San Diego, CA), where the Hill coefficient is assumed to be unity (n = 1), to generate potency and maximal response values. MAX values were normalized to the maximum 5-HT response (100%) and minimum 5-HT response (0%) on the same plate. Three independent experiments were carried out, each performed in duplicate.
Head-Twitch Response
Male C57BL/6 J mice (Jackson Laboratory, Bar Harbor, ME) were housed in a vivarium at the University of California San Diego (UCSD), which is an AAALAC-approved animal facility that complies with Federal and State requirements for the care and treatment of laboratory animals. The mice (6–8 weeks old) were housed in a climate-controlled room with a reversed light cycle (lights on at 19:00 h and off at 07:00 h) up to four animals per cage. Food and water were provided ad libitum, except during behavioral testing, which occurred between 10:00 and 18:00 h. All experiments were conducted according to NIH guidelines and were approved by the UCSD animal care committee. Head movement was recorded using a head-mounted magnet and a magnetometer coil.44 Briefly, mice were anesthetized; a small incision was made in the scalp, and a neodymium magnet was attached to the dorsal surface of the cranium using dental cement. Following a 1–2 week recovery period, behavioral experiments were conducted in a well-lit room with at least 7 days between sessions to avoid any carryover effects. Mice (n = 5–7/group) were treated with vehicle or test compound and then placed in a glass cylinder surrounded by a magnetometer coil, and activity was recorded for 30 min. (R)-DOTFM was administered IP dissolved in saline using an injection volume of 5 mL/kg. Coil voltage was filtered (5 to 10 kHz low-pass), digitized (20 kHz sampling rate), and saved to a disk using a Powerlab/8SP with LabChart v 7.3.2 (ADInstruments, Colorado Springs, CO). To detect head twitches, events in the recordings were converted into scalograms using a wavelet transform, and then the images were classified using a multistage approach combining the deep convolutional neural network (CNN) ResNet-50 with a support vector machine (SVM) algorithm.45 HTR counts were analyzed using a one-way analysis of variance (ANOVA) (or a one-way Welch ANOVA in experiments where variances are not equal). Post hoc pairwise comparisons were performed using Dunnett’s test. Significance was demonstrated using an α-level of 0.05. Median effective doses (ED50 values) and 95% confidence intervals were calculated using nonlinear regression (Prism ver. 9.0.2, GraphPad Software Inc., San Diego, CA, USA).
Lung Homogenate Preparation and Quantitative Shotgun Proteomics
Proteomics analysis was performed as described by Yue and Guidry.46 Animals were humanely sacrificed with lungs removed en bloc 48 h post final OVA exposure, transferred to a lysis buffer (1% w/v SDS [Invitrogen] in 1× Tris-HCL pH 7.4 [Gibco, Bleiswijk]) in a gentleMACS C Tube (Miltenyi Biotec Gmbh, Bergisch Gladbach, Germany), and homogenized using the gentleMACS Octo Dissociator (Miltenyi Biotec, Germany) via the program “lung_02” (37 s, 165 rpr). Single lung samples were labeled using a TMT 16-plex Reagent set (Thermo Fisher Scientific, Waltham, MA) per the manufacturer’s protocol. Sixteen samples were analyzed on three fluidics chips and included naive (n = 5), OVA (n = 5), (R)-DOI (n = 5), (R)-DOTFM (n = 5), (R)-DOI+OVA (n = 6), and (R)-DOTFM+OVA (n = 6).
An equal amount of TMT-labeled sample was pooled in a single tube and SepPak purified (Waters Chromatography, Dublin, Ireland) under acidic reverse phase conditions. Following a drying period, an offline fractionation step was utilized to reduce the complexity of the sample. Samples were brought up in 260 μL of 10 mM ammonium hydroxide, pH 10, and subjected to basic pH reverse phase chromatography (Dionex U300, Thermo Fisher). UV was monitored at 215 nm for 100 μL injections at 0.1 mL/min under a 10 nM ammonium hydroxide, pH 10, to 100% acetonitrile (ACN, pH 10) gradient for 90 min. Fractions were collected in a 96-well microplate and recombined in checkerboard fashion to create “super fractions”.
“Super fractions” were run on a Dionex U3000 nano flow system coupled to a Thermo Fisher Orbitrap Fusion Tribrid Mass Spectrometer. Over the course of 65 min, fractions were first subjected to a 90 min chromatographic method employing a 2–25% ACN in 0.1% formic acid (FA) gradient followed by a 10 min 50% ACN/FA gradient, a 5 min 90% ACN/FA step, and a final 10 min re-equilibration into 2% ACN/FA. A PicoChip source (New Objective, Woburn, MA) carried out all chromatography in a “trap-and-load” format, with a C18 PepMap 100 (5 μm, 100 Å) trap column and a PicoChip REPROSIL-Pur C18-AQ (3 μm, 120 Å, 105 mm) separation column, at a flow rate of 3 μL/min, with electrospray achieved at 2.6 kV.
An MS3 approach employing TMT data acquisition was utilized for data acquisition. Survey scans were performed in an Orbitrap at a resolution of 120,000, with data-dependent MS2 scans performed in a linear ion trap using a 25% collision induced dissociation rate. Reporter ions were fragmented using high energy collision dissociation at 65% and detected in an Orbitrap at a resolution of 50,000. This was repeated for a total of three technical replicates. The Proteome Discoverer 2.2 performed the TMT data analysis. The three runs of each “super fraction” were merged and searched using SEQUEST.
For simple analysis, data were analyzed on the basis of mean log fold changes (logFCs) vs treatment parameter.
Bioinformatics of Protein Expression
Protein abundances were batch corrected at the fluidics chip level using Combat.47 Proteins with missing data were excluded, as Combat batch-correction requires complete data. LogFCs and p values between experimental conditions were generated using LIMMA.48 Volcano plots were then generated, and results for differential expression were calculated as shown in the supplementary tables.
Network Analysis
Interaction networks for specific subsets of proteins were determined using the online STRING Database resource (https://string-db.org)49 using default Web-UI settings. Gene Set Analysis (GSEA) was performed using the online interface WebGestalt50 using input lists of protein names converted to gene identifiers.
Western Blot
Homogenized mouse lung samples were suspended in a lysis buffer containing the mammalian protein extraction reagent (MPER), 0.1% sodium dodecyl sulfate (SDS), and 0.1 M Halt protease and phosphatase inhibitor cocktail. The protein content of mouse lung samples was determined by the bicinchoninic acid (BCA) assay. Normalized protein samples were separated on 4–12% continuous gradient Bolt-Bis Tris Plus polyacrylamide gels, transferred to nitrocellulose membranes, and blocked with 5% nonfat milk for 1 h. Blots were probed with rabbit α-Arg1 antibodies (CellSignal, 93668, 1:2000, 2.25% BSA, overnight at 4 °C), washed with Tris-buffered saline 0.1% Tween20 (TBS-T), probed with goat α-rabbit-HRP antibodies (Kindlebio, R1006, 1:1000, 1 h at RT), and then washed again prior to adding the KwikQuant Ultra HRP substrate solution (Kindlebio, R1002) for image acquisition. Following image acquisition, blots were stripped with the Restore PLUS Western Blot stripping buffer (Thermoscientific, 46430, 30 min) and reblocked in 5% nonfat milk for 30 min. Blots were reprobed with mouse α–β-actin (Sigma, clone AC-15, 1:25,000) and goat α-mouse-HRP (Kindlebio, R1005, 1:2000) antibodies for 30 min, washed with TBS-T, and then developed with a 1:10 dilution of the KwikQuant Ultra HRP substrate solution. Protein bands were visualized by chemiluminescence on an Azure 600 with 1 × 1 pixel binning and a wide dynamic range exposure. Densitometric analysis was conducted using ImageJ. Individual bands, or an area of representative background, were encompassed within a fixed defined area using the selection tool, and inverted mean gray values for each were determined (inverted mean gray value = 65,535 – measured mean gray value). Subsequently, the background inverted mean gray value was subtracted from all band measurements, and values between samples were normalized by ratioing the background adjusted Arg1 densitometric value relative to corresponding β-actin densitometric values. Normalized levels of Arg1 expression were expressed relative to the naive controls.
Statistics
PenH, gene, and protein statistical analysis was performed using GraphPad Prism v9 (GraphPad Software, La Jolla, CA) with post hoc analysis for multiple comparisons where appropriate.
Acknowledgments
Funding for the majority of this work was provided by a generous donation to the LSUHSC Foundation for C.D.N. Additional support was provided by 2A Biosciences and Eleusis Therapeutics to C.D.N. and T.P.F. We thank the LSUHSC proteomics core for performing the quantitative proteomic screen.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsptsci.3c00297.
Full scans of Western blot analysis for Arg1 quantification (PDF)
Tables derived from bioinformatics results involving differences in protein expression: (S1) differential protein expression between control and OVA treated groups; (S2) differential protein expression between control and (R)-DOI treated groups; (S3) differential protein expression between control and (R)-DOTFM treated groups; (S4) differential protein expression between OVA-only and (R)-DOI + OVA treated groups; (S5) differential protein expression between OVA-only and (R)-DOTFM + OVA treated groups; (S6) differential protein expression of greater than 2-fold between (R)-DOI and (R)-DOTFM in OVA treated mice compared to OVA-alone treated mice; (S7) GSEA of proteins significantly increased by (R)-DOI in mice not treated with OVA; (S8) GSEA of proteins significantly decreased by (R)-DOI in mice not treated with OVA; (S9) GSEA of proteins significantly increased by (R)-DOI in mice treated with OVA; (S10) GSEA of proteins significantly decreased by (R)-DOI in mice treated with OVA; and (S11) GSEA of proteins significantly increased by (R)-DOTFM in mice treated with OVA (XLSX)
Author Contributions
Thomas Flanagan designed and performed the in vivo experiments, analyzed data, and wrote the manuscript. Timothy Foster and Thomas Galbato performed and analyzed Arg1 expression by Western blot and contributed to the writing and editing of the manuscript. Pek Lum, Brent Louie, and Gavin Song performed the raw data analysis of the quantitative proteomics data sets. Adam Halberstadt performed the Head-Twitch Assay and its data analysis. Gerald Billac performed the calcium flux and arrestin recruitment experiments and data analysis. Charles Nichols designed experiments, performed proteomic data analysis, and edited the manuscript.
The authors declare the following competing financial interest(s): CDN, TPF and ALH had a sponsored research contract with Eleusis Therapeutics and CDN and TPF were members of its scientific advisory board when portions of this study were performed. CDN and TPF are founders of 2A Biosciences and receive sponsored research funding from 2A Biosciences.
Supplementary Material
References
- Halberstadt A. L.; Chatha M.; Klein A. K.; Wallach J.; Brandt S. D. Correlation between the potency of hallucinogens in the mouse head-twitch response assay and their behavioral and subjective effects in other species. Neuropharmacology 2020, 167, 107933 10.1016/j.neuropharm.2019.107933. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.
- Kyzar E. J.; Nichols C. D.; Gainetdinov R. R.; Nichols D. E.; Kalueff A. V. Psychedelic Drugs in Biomedicine. Trends in pharmacological sciences 2017, 38 (11), 992–1005. 10.1016/j.tips.2017.08.003. [DOI] [PubMed] [Google Scholar]; From NLM.
- Nichols D. E. Psychedelics. Pharmacol. Rev. 2016, 68 (2), 264–355. 10.1124/pr.115.011478. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.
- Nichols D. E.; Walter H. The History of Psychedelics in Psychiatry. Pharmacopsychiatry 2021, 54, 151. 10.1055/a-1310-3990. [DOI] [PubMed] [Google Scholar]; From NLM.
- Nichols D. E.; Johnson M. W.; Nichols C. D. Psychedelics as Medicines: An Emerging New Paradigm. Clinical pharmacology and therapeutics 2017, 101 (2), 209–219. 10.1002/cpt.557. [DOI] [PubMed] [Google Scholar]; From NLM.
- Canal C. E.; Morgan D. Head-twitch response in rodents induced by the hallucinogen 2,5-dimethoxy-4-iodoamphetamine: a comprehensive history, a re-evaluation of mechanisms, and its utility as a model. Drug Test Anal 2012, 4 (7–8), 556–576. 10.1002/dta.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.
- Yu B.; Becnel J.; Zerfaoui M.; Rohatgi R.; Boulares A. H.; Nichols C. D. Serotonin 5-hydroxytryptamine(2A) receptor activation suppresses tumor necrosis factor-alpha-induced inflammation with extraordinary potency. Journal of pharmacology and experimental therapeutics 2008, 327 (2), 316–323. 10.1124/jpet.108.143461. [DOI] [PubMed] [Google Scholar]; From NLM.
- Nau F. Jr.; Yu B.; Martin D.; Nichols C. D. Serotonin 5-HT2A receptor activation blocks TNF-alpha mediated inflammation in vivo. PloS one 2013, 8 (10), e75426 10.1371/journal.pone.0075426. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.
- Nau F. Jr.; Miller J.; Saravia J.; Ahlert T.; Yu B.; Happel K. I.; Cormier S. A.; Nichols C. D. Serotonin 5-HT(2) receptor activation prevents allergic asthma in a mouse model.. American journal of physiology. Lung cellular and molecular physiology 2015, 308 (2), L191–198. 10.1152/ajplung.00138.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.
- Flanagan T. W.; Sebastian M. N.; Battaglia D. M.; Foster T. P.; Maillet E. L.; Nichols C. D. Activation of 5-HT2 Receptors Reduces Inflammation in Vascular Tissue and Cholesterol Levels in High-Fat Diet-Fed Apolipoprotein E Knockout Mice. Sci. Rep. 2019, 9 (1), 13444. 10.1038/s41598-019-49987-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.
- Flanagan T. W.; Sebastian M. N.; Battaglia D. M.; Foster T. P.; Cormier S. A.; Nichols C. D. 5-HT2 receptor activation alleviates airway inflammation and structural remodeling in a chronic mouse asthma model. Life sciences 2019, 236, 116790 10.1016/j.lfs.2019.116790. [DOI] [PubMed] [Google Scholar]; From NLM.
- Flanagan T. W.; Billac G. B.; Landry A. N.; Sebastian M. N.; Cormier S. A.; Nichols C. D.. Structure–Activity Relationship Analysis of Psychedelics in a Rat Model of Asthma Reveals the Anti-Inflammatory Pharmacophore ACS Pharmacology & Translational Science 2020488. 10.1021/acsptsci.0c00063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan T. W.; Nichols C. D. Psychedelics and Anti-inflammatory Activity in Animal Models. Current topics in behavioral neurosciences 2022, 56, 229–245. 10.1007/7854_2022_367. [DOI] [PubMed] [Google Scholar]; From NLM.
- Mendez-Enriquez E.; Alvarado-Vazquez P. A.; Abma W.; Simonson O. E.; Rodin S.; Feyerabend T. B.; Rodewald H. R.; Malinovschi A.; Janson C.; Adner M.; et al. Mast cell-derived serotonin enhances methacholine-induced airway hyperresponsiveness in house dust mite-induced experimental asthma. Allergy 2021, 76 (7), 2057–2069. 10.1111/all.14748. [DOI] [PubMed] [Google Scholar]; From NLM.
- Nichols D. E.; Frescas S.; Marona-Lewicka D.; Huang X.; Roth B. L.; Gudelsky G. A.; Nash J. F. 1-(2,5-Dimethoxy-4-(trifluoromethyl)phenyl)-2-aminopropane: A Potent Serotonin 5-HT2A/2C Agonist. J. Med. Chem. 1994, 37 (25), 4346–4351. 10.1021/jm00051a011. [DOI] [PubMed] [Google Scholar]
- Braden M. R.; Parrish J. C.; Naylor J. C.; Nichols D. E. Molecular interaction of serotonin 5-HT2A receptor residues Phe339(6.51) and Phe340(6.52) with superpotent N-benzyl phenethylamine agonists. Molecular pharmacology 2006, 70 (6), 1956–1964. 10.1124/mol.106.028720. [DOI] [PubMed] [Google Scholar]; From NLM.
- Busse W. W.; Lemanske R. F. Jr. Asthma. New England journal of medicine 2001, 344 (5), 350–362. 10.1056/NEJM200102013440507. [DOI] [PubMed] [Google Scholar]; From NLM.; Naik S. R.; Wala S. M. Inflammation, allergy and asthma, complex immune origin diseases: mechanisms and therapeutic agents. Recent patents on inflammation & allergy drug discovery 2013, 7 (1), 62–95. 10.2174/187221313804004682. [DOI] [PubMed] [Google Scholar]; From NLM.; Hamid Q.; Tulic M. Immunobiology of asthma. Annual review of physiology 2009, 71, 489–507. 10.1146/annurev.physiol.010908.163200. [DOI] [PubMed] [Google Scholar]; From NLM.; Barnes P. J. Pathophysiology of allergic inflammation. Immunological reviews 2011, 242 (1), 31–50. 10.1111/j.1600-065X.2011.01020.x. [DOI] [PubMed] [Google Scholar]; From NLM.
- Østvold A. C.; Grundt K.; Wiese C. NUCKS1 is a highly modified, chromatin-associated protein involved in a diverse set of biological and pathophysiological processes. Biochemical journal 2022, 479 (11), 1205–1220. 10.1042/BCJ20220075. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.; Huang P.; Cai Y.; Zhao B.; Cui L. Roles of NUCKS1 in Diseases: Susceptibility, Potential Biomarker, and Regulatory Mechanisms. BioMed. research international 2018, 2018, 7969068. 10.1155/2018/7969068. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.
- Qiu B.; Shi X.; Wong E. T.; Lim J.; Bezzi M.; Low D.; Zhou Q.; Akıncılar S. C.; Lakshmanan M.; Swa H. L.; et al. NUCKS is a positive transcriptional regulator of insulin signaling. Cell Rep 2014, 7 (6), 1876–1886. 10.1016/j.celrep.2014.05.030. [DOI] [PubMed] [Google Scholar]; From NLM.
- Maranon D. G.; Sharma N.; Huang Y.; Selemenakis P.; Wang M.; Altina N.; Zhao W.; Wiese C.. NUCKS1 promotes RAD54 activity in homologous recombination DNA repair. J. Cell Biol. 2020, 219 ( (10), ). 10.1083/jcb.201911049. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.; Hume S.; Grou C. P.; Lascaux P.; D’Angiolella V.; Legrand A. J.; Ramadan K.; Dianov G. L. The NUCKS1-SKP2-p21/p27 axis controls S phase entry. Nat. Commun. 2021, 12 (1), 6959. 10.1038/s41467-021-27124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.
- Donnelly L. E.; Barnes P. J. Acidic mammalian Chitinase--a potential target for asthma therapy. Trends in pharmacological sciences 2004, 25 (10), 509–511. 10.1016/j.tips.2004.08.002. [DOI] [PubMed] [Google Scholar]; From NLM.; Mazur M.; Olczak J.; Olejniczak S.; Koralewski R.; Czestkowski W.; Jedrzejczak A.; Golab J.; Dzwonek K.; Dymek B.; Sklepkiewicz P. L.; et al. Targeting Acidic Mammalian Chitinase Is Effective in Animal Model of Asthma. J. Med. Chem. 2018, 61 (3), 695–710. 10.1021/acs.jmedchem.7b01051. [DOI] [PubMed] [Google Scholar]; From NLM.
- Zhu Z.; Zheng T.; Homer R. J.; Kim Y. K.; Chen N. Y.; Cohn L.; Hamid Q.; Elias J. A. Acidic mammalian Chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science (New York, N.Y.) 2004, 304 (5677), 1678–1682. 10.1126/science.1095336. [DOI] [PubMed] [Google Scholar]; From NLM.
- North M. L.; Khanna N.; Marsden P. A.; Grasemann H.; Scott J. A. Functionally important role for arginase 1 in the airway hyperresponsiveness of asthma.. American journal of physiology. Lung cellular and molecular physiology 2009, 296 (6), L911–920. 10.1152/ajplung.00025.2009. [DOI] [PubMed] [Google Scholar]; From NLM.
- Cloots R. H.; Sankaranarayanan S.; de Theije C. C.; Poynter M. E.; Terwindt E.; van Dijk P.; Hakvoort T. B.; Lamers W. H.; Köhler S. E. Ablation of Arg1 in hematopoietic cells improves respiratory function of lung parenchyma, but not that of larger airways or inflammation in asthmatic mice. American Journal of Physiology-Lung Cellular and Molecular Physiology 2013, 305 (5), L364–L376. 10.1152/ajplung.00341.2012. [DOI] [PubMed] [Google Scholar]
- Brett T. J. CLCA1 and TMEM16A: the link towards a potential cure for airway diseases. Expert Rev. Respir Med. 2015, 9 (5), 503–506. 10.1586/17476348.2015.1081064. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.; Centeio R.; Ousingsawat J.; Schreiber R.; Kunzelmann K.. CLCA1 Regulates Airway Mucus Production and Ion Secretion Through TMEM16A. Int. J. Mol. Sci. 2021, 22 ( (10), ). DOI: 5133. 10.3390/ijms22105133. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.
- Zhu Y.; Yan X.; Zhai C.; Yang L.; Li M. Association between risk of asthma and gene polymorphisms in CHI3L1 and CHIA: a systematic meta-analysis. BMC Pulm Med. 2017, 17 (1), 193. 10.1186/s12890-017-0515-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.
- Tomazic P. V.; Birner-Gruenberger R.; Leitner A.; Obrist B.; Spoerk S.; Lang-Loidolt D. Nasal mucus proteomic changes reflect altered immune responses and epithelial permeability in patients with allergic rhinitis. J. Allergy Clin Immunol 2014, 133 (3), 741–750. 10.1016/j.jaci.2013.09.040. [DOI] [PubMed] [Google Scholar]; From NLM.; Vizuet-de-Rueda J. C.; Montero-Vargas J. M.; Galván-Morales M.; Porras-Gutiérrez-de-Velasco R.; Teran L. M.. Current Insights on the Impact of Proteomics in Respiratory Allergies. Int. J. Mol. Sci. 2022, 23 ( (10), ). DOI: 5703. 10.3390/ijms23105703. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.
- Klonoff-Cohen H.; Polavarapu M. Eosinophil protein X and childhood asthma: A systematic review and meta-analysis. Immun Inflamm Dis 2016, 4 (2), 114–134. 10.1002/iid3.104. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.
- Pesce J. T.; Ramalingam T. R.; Mentink-Kane M. M.; Wilson M. S.; El Kasmi K. C.; Smith A. M.; Thompson R. W.; Cheever A. W.; Murray P. J.; Wynn T. A. Arginase-1–expressing macrophages suppress Th2 cytokine–driven inflammation and fibrosis. PLoS pathogens 2009, 5 (4), e1000371 10.1371/journal.ppat.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bando J. K.; Nussbaum J. C.; Liang H. E.; Locksley R. M. Type 2 innate lymphoid cells constitutively express arginase-I in the naive and inflamed lung. Journal of leukocyte biology 2013, 94 (5), 877–884. 10.1189/jlb.0213084. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.; Monticelli L. A.; Buck M. D.; Flamar A.-L.; Saenz S. A.; Tait Wojno E. D.; Yudanin N. A.; Osborne L. C.; Hepworth M. R.; Tran S. V.; Rodewald H.-R. Arginase 1 is an innate lymphoid-cell-intrinsic metabolic checkpoint controlling type 2 inflammation. Nature immunology 2016, 17 (6), 656–665. 10.1038/ni.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell L.; Saville C. R.; Murray P. J.; Cruickshank S. M.; Hardman M. J. Local arginase 1 activity is required for cutaneous wound healing. Journal of investigative dermatology 2013, 133 (10), 2461–2470. 10.1038/jid.2013.164. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.
- Meurs H.; Zaagsma J.; Maarsingh H.; van Duin M. Recent Patents in Allergy/Immunology: Use of arginase inhibitors in the treatment of asthma and allergic rhinitis. Allergy 2019, 74 (6), 1206–1208. 10.1111/all.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.
- Troidl C.; Möllmann H.; Nef H.; Masseli F.; Voss S.; Szardien S.; Willmer M.; Rolf A.; Rixe J.; Troidl K.; et al. Classically and alternatively activated macrophages contribute to tissue remodelling after myocardial infarction. J. Cell Mol. Med. 2009, 13 (9b), 3485–3496. 10.1111/j.1582-4934.2009.00707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.
- de las Casas-Engel M.; Corbí A. L.; Camps J.. Serotonin modulation of macrophage polarization: inflammation and beyond. Oxidative stress and inflammation in non-communicable diseases-molecular mechanisms and perspectives in therapeutics; Springer Cham: 2014, 89-115 10.1007/978-3-319-07320-0_9. [DOI] [PubMed] [Google Scholar]
- Odegaard J. I.; Chawla A. Alternative macrophage activation and metabolism. Annu. Rev. Pathol 2011, 6, 275–297. 10.1146/annurev-pathol-011110-130138. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.
- Flanagan T. W.; Nichols C. D. Psychedelics as anti-inflammatory agents. Int. Rev. Psychiatry 2018, 1–13. 10.1080/09540261.2018.1481827. [DOI] [PubMed] [Google Scholar]; From NLM.
- Lambrecht B. N.; Hammad H. The airway epithelium in asthma. Nat. Med. 2012, 18 (5), 684–692. 10.1038/nm.2737. [DOI] [PubMed] [Google Scholar]; From NLM.
- Mereness J. A.; Mariani T. J. The critical role of collagen VI in lung development and chronic lung disease. Matrix Biol. Plus 2021, 10, 100058 10.1016/j.mbplus.2021.100058. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.
- Jamieson R. R.; Stasiak S. E.; Polio S. R.; Augspurg R. D.; McCormick C. A.; Ruberti J. W.; Parameswaran H. Stiffening of the extracellular matrix is a sufficient condition for airway hyperreactivity. J. Appl. Physiol (1985) 2021, 130 (6), 1635–1645. 10.1152/japplphysiol.00554.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.
- Aplin M.; Christensen G. L.; Hansen J. L. Pharmacologic perspectives of functional selectivity by the angiotensin II type 1 receptor. Trends Cardiovasc Med. 2008, 18 (8), 305–312. 10.1016/j.tcm.2009.01.003. [DOI] [PubMed] [Google Scholar]; From NLM.; Christensen G. L.; Aplin M.; Hansen J. L. Therapeutic potential of functional selectivity in the treatment of heart failure. Trends Cardiovasc Med. 2010, 20 (7), 221–227. 10.1016/j.tcm.2011.11.005. [DOI] [PubMed] [Google Scholar]; From NLM.
- HAMELMANN E.; SCHWARZE J.; TAKEDA K.; OSHIBA A.; LARSEN G. L.; IRVIN C. G.; GELFAND E. W. Noninvasive measurement of airway responsiveness in allergic mice using barometric plethysmography. Ame. J. Respiratory Crit. Care Med. 1997, 156 (3 Pt 1), 766–775. 10.1164/ajrccm.156.3.9606031. [DOI] [PubMed] [Google Scholar]; From NLM.
- Derangeon M.; Bozon V.; Defamie N.; Peineau N.; Bourmeyster N.; Sarrouilhe D.; Argibay J. A.; Hervé J. C. 5-HT4 and 5-HT2 receptors antagonistically influence gap junctional coupling between rat auricular myocytes. Journal of molecular and cellular cardiology 2010, 48 (1), 220–229. 10.1016/j.yjmcc.2009.07.005. [DOI] [PubMed] [Google Scholar]; From NLM.
- Xiang M.; Jiang Y.; Hu Z.; Yang Y.; Du X.; Botchway B. O.; Fang M. Serotonin receptors 2A and 1A modulate anxiety-like behavior in post-traumatic stress disordered mice. Am. J. Transl Res. 2019, 11 (4), 2288–2303. [PMC free article] [PubMed] [Google Scholar]; From NLM.
- Pottie E.; Cannaert A.; Stove C. P. In vitro structure–activity relationship determination of 30 psychedelic new psychoactive substances by means of β-arrestin 2 recruitment to the serotonin 2A receptor. Archives of toxicology 2020, 94 (10), 3449–3460. 10.1007/s00204-020-02836-w. [DOI] [PubMed] [Google Scholar]
- Halberstadt A. L.; Geyer M. A. Characterization of the head-twitch response induced by hallucinogens in mice: detection of the behavior based on the dynamics of head movement. Psychopharmacology 2013, 227 (4), 727–739. 10.1007/s00213-013-3006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.
- Halberstadt A. L. Automated detection of the head-twitch response using wavelet scalograms and a deep convolutional neural network. Sci. Rep. 2020, 10 (1), 8344. 10.1038/s41598-020-65264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.
- Yue X.; Guidry J. J.. Differential Protein Expression Profiles of Bronchoalveolar Lavage Fluid Following Lipopolysaccharide-Induced Direct and Indirect Lung Injury in Mice. Int. J. Mol. Sci. 2019, 20 ( (14), ). DOI: 3401. 10.3390/ijms20143401. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.
- Behdenna A.; Haziza J.; Azencott C.-A.; Nordor A. pyComBat, a Python tool for batch effects correction in high-throughput molecular data using empirical Bayes methods. BioRxiv 2020, 2020 (2003), 2017.995431. 10.1186/s12859-023-05578-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M. E.; Phipson B.; Wu D.; Hu Y.; Law C. W.; Shi W.; Smyth G. K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research 2015, 43 (7), e47–e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D.; Gable A. L.; Lyon D.; Junge A.; Wyder S.; Huerta-Cepas J.; Simonovic M.; Doncheva N. T.; Morris J. H.; Bork P.; et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic acids research 2019, 47 (D1), D607–d613. 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.; Szklarczyk D.; Gable A. L.; Nastou K. C.; Lyon D.; Kirsch R.; Pyysalo S.; Doncheva N. T.; Legeay M.; Fang T.; Bork P.; et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic acids research 2021, 49 (D1), D605–d612. 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.
- Liao Y.; Wang J.; Jaehnig E. J.; Shi Z.; Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic acids research 2019, 47 (W1), W199–w205. 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]; From NLM.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.