Abstract
Pharmacogenetics is a key component of precision medicine. Genetic variation in drug metabolism enzymes can lead to variable exposure to drugs and metabolites, potentially leading to inefficacy and drug toxicity. Although the evidence for pharmacogenetic associations in children is not as extensive as for adults, there are several drugs across diverse therapeutic areas with robust pediatric data indicating important, and relatively common, drug–gene interactions. Guidelines to assist gene-based dose optimization are available for codeine, thiopurine drugs, selective serotonin reuptake inhibitors, atomoxetine, tacrolimus, and voriconazole. For each of these drugs, there is an opportunity to clinically implement precision medicine approaches with children for whom genetic test results are known or are obtained at the time of prescribing. For many more drugs that are commonly used in pediatric patients, additional investigation is needed to determine the genetic factors influencing appropriate dose.
Keywords: pharmacogenomics, pediatrics, infectious disease, psychiatry, oncology, immunology
1. INTRODUCTION
Precision medicine has been described as the “right drug for the right patient at the right time” (1, p. 11) and has received over $4 billion in global financial support (2). One important component of precision medicine is pharmacogenetics. Pharmacogenetic research aims to explain the heritable portion of individual variability in medication response. Clinical implementation incorporates information from inherited genetic variants into prescribing decisions for drug selection or dosing strategies. Although the bulk of pharmacogenetic research has been performed in adults, many medications with known drug–gene interactions are also used in children, and recommendations to alter dosage, or change the drug choice altogether, may apply to children as well as adults.
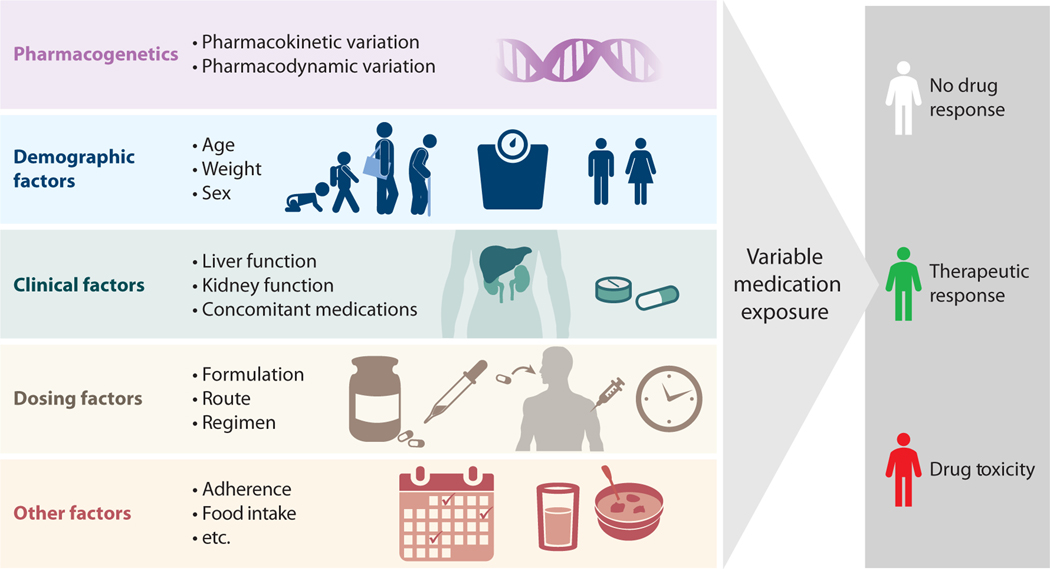
Pharmacogenetics is one of myriad factors that contribute to interindividual variability in drug exposure and response (Figure 1), and there are pediatric-specific nuances to these factors. For example, some hepatic metabolic enzymes have expression patterns that change throughout development, particularly in infancy (e.g., CYP3A enzymes) (3); thus, genetic variation in these enzymes will not explain individual differences in drug exposure until the enzymes are expressed. Weight, body surface area, and lean body mass can also influence variability in exposure and change significantly over the course of infancy, childhood, and adolescence. Drug exposure is potentially affected by kidney and liver dysfunction; developmental trajectories (e.g., pubertal status), chronic disease, and acute processes such as inflammation can impact organ function in children. Concomitant medications have long been recognized as influencing drug exposure, as they can inhibit or enhance metabolic capacity. Some foods can also influence medication exposure through changes in drug absorption, altered drug metabolism, or differences in drug response. Other external factors that can influence medication exposure are adherence, formulation, route of administration, dosing regimen, and time of day, all of which can be influenced by the age and developmental stage of the pediatric patient. This review focuses on the impact of genetic variation on dosing for children, as this is an important, nonmodifiable risk factor for drug inefficacy or toxicity.
Figure 1.
Illustration of factors contributing to individual differences in drug exposure. In addition to pharmacogenetics, demographic factors (age, weight, and sex), clinical factors (organ function and other drugs), dosing factors (formulation, route, and timing), and issues such as adherence and food intake are important determinants of drug exposure. Interindividual differences in drug exposure can lead to differences in drug response, including inefficacy or toxicity.
There have been several recent advances that have increased the interest in and implementation of gene-guided prescribing for children. These advances include the decreased cost of genotyping, increased knowledge of how genetic variants influence medication exposure such as that curated by the Pharmacogenomics Knowledge Base (PharmGKB) (4), establishment of clinical guidelines such as those from the Clinical Pharmacogenetics Implementation Consortium (CPIC) (5) and the Dutch Pharmacogenetics Working Group (6), and availability of commercial and direct-to-consumer genetic testing. CPIC guidelines specifically address pediatrics and include resources for implementation, although not all of their guidelines contain pediatric-specific recommendations. Here we review several drugs with potential for the clinical implementation of gene-guided dosing in children, including codeine, thiopurines, selective serotonin reuptake inhibitors (SSRIs), atomoxetine, tacrolimus, and voriconazole. We also identify common themes among the evidence and clinical implementations in pediatrics and strategies for advancing gene-based dosing for children.
2. BASIC PRINCIPLES OF PHARMACOGENETIC MECHANISMS
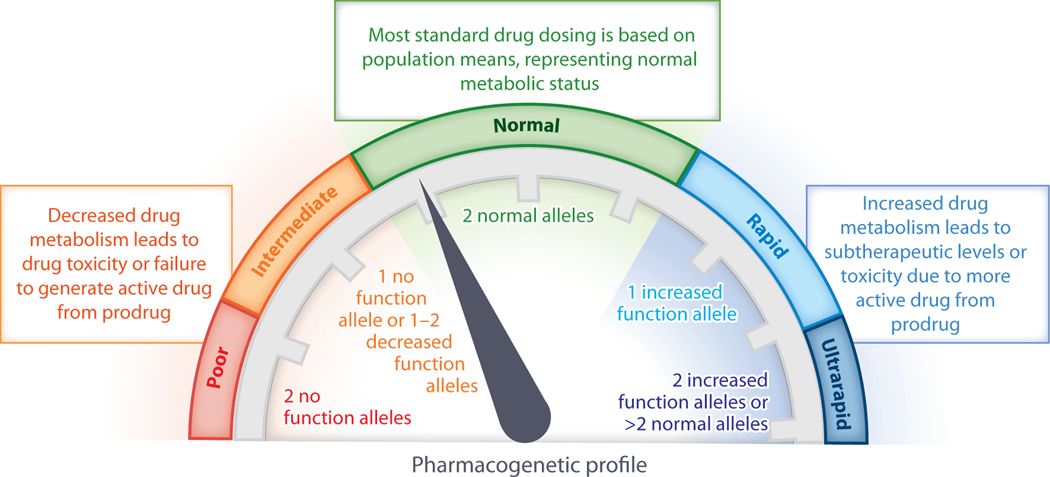
Genetic variants influence the outcomes of drug therapy through multiple mechanisms. Most well-established pharmacogenetic associations involve genes encoding drug metabolism enzymes or drug transporters. These variants have the potential to impact pharmacokinetics. For example, genetic variants that decrease enzyme function can lead to slower conversion of the active drug to inactive metabolites, contributing to high active drug concentrations and the potential for toxicity (Figure 2). Genetic variants that increase enzyme function lead to faster inactivation and may be associated with inefficacy. Examples of well-established drug–gene interactions of this type include thiopurine drugs with TPMT and NUDT15; escitalopram, sertraline, and voriconazole with cytochrome P450 (CYP)2C19; fluvoxamine and atomoxetine with CYP2D6; and tacrolimus with CYP3A5. For prodrugs that are converted from inactive compounds to active forms by drug metabolism enzymes, the relationships between enzyme function and active drug levels are reversed. Decreased enzyme function prevents formation of the active form, leading to inefficacy, while increased enzyme function generates high levels of the active form and the potential for toxicity. An example of this type of drug–gene interaction is codeine with CYP2D6. For pharmacokinetic drug–gene interactions, dose optimization or alternative medications can be recommended for individuals with atypical function. The proportion of individuals with atypical function is highly variable across genes and across populations. For example, fewer than 1% of individuals with European or African ancestry lack NUDT15 enzyme function, whereas the no function allele is seen in nearly 10% of East Asians (7). In contrast, 80–85% of individuals with European ancestry are poor metabolizers for CYP3A5, while most individuals with African ancestry have two functional alleles (8). Most individuals have at least one actionable pharmacogenetic variant; an assessment of nearly 10,000 individuals genotyped for variants for five drug-related genes revealed actionable variants in 91% of all individuals and in 96% of African American individuals (9). Throughout this review, we use the consensus terms for enzyme functional status (poor, intermediate, normal, rapid, and ultrarapid metabolizers) whenever possible (10).
Figure 2.
The impact of genetic variants on metabolizer status. Due to genetic differences, the functional activity of a drug metabolism enzyme may be absent, decreased, normal, or increased. Depending on the combination of alleles present, each individual can be classified regarding their functional status for each drug metabolic enzyme as a poor, intermediate, normal, rapid, or ultrarapid metabolizer.
Some genetic variants affect pharmacodynamics. For example, individuals may have specific genetic variants that affect the affinity of a drug for the target receptor or an off-target protein binding site. For these drug–gene interactions, clinical implementation often includes recommendations to use alternative medications in individuals who harbor variants associated with hypersensitivity or inefficacy (Table 1).
Table 1.
Selected drug–gene interactions with clinical guidelines relevant to pediatric populations
| Drug | FDA-approved pediatric indications (age) | Additional clinical uses | Gene(s) | Gene-based dose adjustments |
|---|---|---|---|---|
| Codeine | Pain (ages 12+ as part of combination drug) | Nonea | CYP2D6 | Use alternate therapy for CYP2D6 UM and PM |
| Azathioprine | None | Renal transplant Inflammatory bowel disease Autoimmune disease Other malignancies |
TPMT NUDT15 | Drastically reduce dose for TPMT PM Reduce dose for TPMT IM Reduce dose for NUDT15 IM and PM |
| Mercaptopurine | Acute lymphocytic leukemia (all ages) | |||
| Thioguanine | Acute nonlymphocytic leukemia (all ages) | |||
| Escitalopram | Major depression (ages 12+) | Anxiety Autism and pervasive developmental disorders |
CYP2C19 | Use alternate therapy for CYP2C19 UM 50% dose reduction for CYP2C19 PM |
| Sertraline | Obsessive-compulsive disorder (ages 6+) | |||
| Fluvoxamine | Obsessive-compulsive disorder (ages 8+) | Anxiety Major depression |
CYP2D6 | 25% dose reduction for CYP2D6 PM |
| Atomoxetine | Attention deficit hyperactivity disorder (ages 6+) | None | CYP2D6 | Aggressive dose titration (increase) for CYP2D6 NM, UM |
| Tacrolimus | Liver transplant (all ages) | Heart and kidney transplant Nephrotic syndrome |
CYP3A5 | Increase dose 1.5–2 times for CYP3A5 expressers |
| Voriconazole | Invasive fungal disease (ages 12+) | Antifungal prophylaxis | CYP2C19 | Use alternate therapy for CYP2C19 UM Decrease dose for CYP2C19 PM |
Children under 18 years of age should not be given prescription cough and cold medicines containing codeine.
Abbreviations: FDA, Food and Drug Administration; IM, intermediate metabolizer; NM, normal metabolizer; PM, poor metabolizer; UM, ultrarapid metabolizer.
3. EXAMPLES OF CLINICALLY IMPLEMENTED GENE-GUIDED DOSING OPTIMIZATION IN CHILDREN
3.1. Codeine
Codeine is an opioid commonly used for the relief of mild to moderate pain. Single-ingredient codeine is US Food and Drug Administration (FDA) approved only for use in adults (11, 12), whereas codeine with acetaminophen was previously approved by the FDA in patients over three years of age (13). Codeine is O-demethylated to morphine by CYP2D6. Morphine binds to μ-opioid receptors to provide analgesia. Codeine has been used as an antitussive agent, although early studies indicating efficacy across common causes of cough (14, 15) have been contradicted by more recent placebo-controlled studies (16–18).
Codeine metabolism is complex, and the majority of parent drug is converted to inactive metabolites by CYP3A4 and UGT2B7. An estimated 5–10% of the drug is biotransformed to morphine by CYP2D6. The morphine compound is further metabolized by UGT enzymes to the active compound morphine-6-glucuronide and inactive morphine-3-glucuronide. Morphine and morphine-6-glucuronide have 200-fold higher affinity for the μ-opioid receptor than does codeine (19, 20).
Since codeine is a prodrug dependent on CYP2D6 for activation, CYP2D6 poor metabolizers are unable to convert codeine to morphine and thus have no therapeutic effect. Depending on the population, the frequency of poor metabolizers varies from ≤1% (East Asian, South Central Asian, and Oceanian populations) to ~5% (European and Ashkenazi Jewish populations) (21). Conversely, CYP2D6 ultrarapid metabolizers generate excess morphine and are at risk for toxicity, including respiratory depression and death. The frequency of ultrarapid metabolizers is ~1% in East Asians; 3–5% in Africans, Europeans, South Central Asians, and Americans; and >10% in Middle Eastern, Ashkenazi Jewish, and Oceanian populations (21). Although much of the high-quality evidence for the drug–gene interaction comes from adults (22, 23), there is ample evidence from pediatric studies. There are several case reports of infant mortality and respiratory failure after exposure to morphine via breastmilk, following biotransformation of codeine in infants whose mothers were CYP2D6 ultrarapid metabolizers, and of additional children with codeine toxicity, some after adeno/tonsillectomy (24–29). Furthermore, higher morphine levels and risk for adverse events are observed in children with ultrarapid CYP2D6 function (30–35).
The FDA added a black box warning to the codeine label in February of 2013, stating that respiratory depression and death have occurred in children who received codeine following adeno/tonsillectomy and that CYP2D6 inhibitors may impact drug response (36). In April of 2017, a contraindication (the FDA’s strongest warning) was added, stating that codeine should not be used for pain or cough in children younger than 12 years old. The new guidance also includes a warning against codeine use in adolescents 12–18 years of age who are obese, have obstructive sleep apnea, or have severe lung disease and a strengthened warning against codeine use while breastfeeding.
Guidelines for codeine use based on CYP2D6 metabolizer status have been published (22, 23), and multiple centers have implemented CYP2D6 testing to guide codeine prescribing CYP2D6 genotyping for children with sickle cell disease has been successfully implemented to ensure that patients with ultrarapid or poor metabolizer phenotypes are not prescribed codeine (38). Some advocate for the continued use of codeine for analgesia in children when CYP2D6 functional status can be assessed (39). With the removal of codeine from the formulary for many children’s hospitals (40), the use of codeine in children is on the decline (41, 42), but it may be replaced by other opioid drugs that are also metabolized by CYP2D6.
3.2. Thiopurine Drugs
The thiopurine drugs azathioprine, mercaptopurine, and thioguanine are key components of therapy for pediatric patients with acute lymphoblastic leukemia, inflammatory bowel disease, and autoimmune disorders. The principal cytotoxic effect of thiopurine drugs is the result of the production of 6-thioguanine nucleotides, thioguanine mono- and di-phosphates, which are converted to thioguanine triphosphates. Thioguanine triphosphates are incorporated into RNA, while thio-deoxyguanosine triphosphates are incorporated into DNA, resulting in cytotoxicity (43).
The metabolism of thiopurine drugs is highly complex, involving multiple competing enzymatic steps of the salvage purine pathway (43). Azathioprine, mercaptopurine, and thioguanine are all inactive prodrugs that require intracellular activation by multiple enzymes. The first extracellular step of azathioprine activation involves conversion to mercaptopurine via metabolism by GSTM1, GSTA1, and GSTA2 (44). Once mercaptopurine or thioguanine are transported into the hepatocyte, there are multiple metabolic pathways. One important pathway involves thiopurine methyltransferase (TPMT), which methylates mercaptopurine and thioguanine compounds (45). Another important component of thiopurine metabolism involves the NUDT15 enzyme, which converts the active 6-thioguanine nucleotide into inactive metabolites (7).
The complex and competing nature of the thiopurine metabolic pathway can make achieving the clinical balance between efficacy and toxicity challenging. To date, preemptive testing of two genes, TPMT and NUDT15, has been clinically implemented to achieve that balance. There is an inverse relationship between TPMT activity, a heritable trait, and the level of the cytotoxic 6-thioguanine nucleotide metabolites (46). There are three well-characterized variants in TPMT that result in unstable TPMT protein and enhanced protein degradation. These three variants account for 90% of TPMT low-activity phenotypes (47–49) and are present in 5% of white populations, 3% of Asian populations, and 6% of black populations (47).
Patients who are TPMT poor metabolizers have a significantly increased risk of hematopoietic toxicity and cytopenia when compared to patients with normal TPMT activity, with dose reductions due to mercaptopurine toxicity required in 100% of poor metabolizers versus 35% of intermediate metabolizers and 7% of normal metabolizers (46). Thus, the recommendation for TPMT poor metabolizers requiring this drug for treatment of malignancy is to drastically reduce the thiopurine dose (e.g., a tenfold dose reduction) and to consider alternative therapy for nonmalignant conditions (47). TPMT intermediate metabolizers also have a significantly increased risk of hematopoietic toxicity and cytopenia when compared to patients with normal TPMT activity (46). However, 40–70% of TPMT intermediate metabolizers tolerate full doses of thiopurine drugs (47), demonstrating the complex nature of thiopurine metabolism. Therefore, the dosing recommendation for TPMT intermediate metabolizers is a more moderate dose reduction (e.g., 50–80% of the typical dose, depending on dose and indication) (47). All active Children’s Oncology Group protocols for acute lymphocytic leukemia currently recommend testing for TPMT variants at diagnosis and adjusting initial doses of thiopurine drugs accordingly.
Clinical decision support for TPMT-based thiopurine dosing is available via the PharmGKB website (https://www.pharmgkb.org/) for 34 different TPMT star alleles and their combinations, based on CPIC guidelines. Some institutions have also incorporated electronic health record–based decision support into their practice, which automatically advises providers ordering thiopurine drugs for patients with TPMT variants that their patient may require a dose adjustment based on their genotype or phenotype and provides links to current guidelines, such as CPIC (47).
NUDT15, which inactivates 6-thioguanine nucleotides, was initially recognized as a clinically important enzyme in the thiopurine pathway via genome-wide association study of patients treated with thiopurine drugs for acute lymphocytic leukemia and inflammatory bowel disease (7, 50). The NUDT15 variant rs116855232 was associated with thiopurine-related hematopoietic toxicity in several studies of mercaptopurine, and further studies identified a similar toxicity profile for azathioprine and thioguanine. Additional variants have been identified, with varying degrees of effect on NUDT15 activity (47). The frequency of poor metabolizer phenotypes ranges from 1% to 10%, with higher frequencies reported among Asian and Hispanic populations (51, 52).
NUDT15 poor metabolizers who require thiopurines for treatment of malignancy are recommended to start at a significantly decreased dose, and those being treated for nonmalignant conditions are recommended to use an alternate therapy. For NUDT15 intermediate metabolizers and those with variants with uncertain functional activity, reduced dosing is recommended, with careful dose adjustment after 2–4 weeks based on toxicity and response (47). CPIC guidelines also include guidance for patients for whom both TPMT and NUDT15 genotypes are known. All recommended dose reductions are based on a standard mercaptopurine starting dose of 75 mg/m2/day; lower starting doses may not require a dose reduction, particularly in intermediate metabolizers (47).
While preemptive testing of both TPMT and NUDT15 prior to thiopurine drug exposure can mitigate some of the toxicity associated with these drugs, thiopurine metabolism is complex and involves multiple additional enzymes of potential significance (e.g., XO, ITPA, MTHFR, IMPDH1, and IMPDH2) (53–55). Therefore, it is crucial that providers initiating thiopurine therapy continue to monitor patients for toxicity rather than interpreting normal metabolizer status for TPMT or NUDT15 as a guarantee against encountering significant toxicity.
3.3. Antidepressant Medications: Selective Serotonin Reuptake Inhibitors
SSRIs are commonly prescribed for the treatment of mood and anxiety disorders. In children and adolescents, these medications are often trialed after nonpharmacological approaches (e.g., cognitive behavioral therapies) have been ineffective. Studies of adolescent depression suggest that the combination of an antidepressant and cognitive behavioral therapy is better than either intervention alone (56, 57). While there are many SSRIs approved for adults, only four currently have FDA indications for depression, anxiety, or obsessive-compulsive disorder in patients under 18 years of age: escitalopram, sertraline, fluvoxamine, and fluoxetine (although others are used off-label in young patients).
The metabolism of SSRIs is primarily mediated through hepatic enzymes, including CYP2D6 and CYP2C19. While each SSRI mentioned above is approved for use in adults and younger patient populations, it is important to recognize that there may be some differences in pharmacokinetic parameters across age ranges that impact dosing strategies. For example, escitalopram and sertraline have steady-state concentrations or exposures that are approximately 15–30% lower in children and adolescents than in adults due to faster clearance in the younger patients and clinically relevant shorter half-lives (~10 h or ~30% shorter) (58). On the other hand, fluoxetine and fluvoxamine may have two- to threefold higher steady-state plasma concentrations and exposure in younger patients (59, 60). Presently, the relationships between drug metabolism genotypes and clinical or pharmacokinetic phenotypes are largely informed by data from adults.
Among SSRIs, perhaps the best-described pharmacogenetic relationships are between CYP2C19 metabolizer status and escitalopram. A meta-analysis of nearly 1,000 individuals (patients and healthy controls) from pharmacokinetic studies identified 95% increases in overall exposure in poor metabolizers (2–15% of individuals, depending on the population) as compared to normal metabolizers. Rapid and ultrarapid metabolizers (∗1/ ∗17 or ∗17/ ∗17, 2–30% of individuals) had decreased exposure of 14–36% as compared to normal metabolizers (61). Additionally, in a retrospective study of more than 2,000 patients (including some adolescents), escitalopram discontinuation was more common in the extremes than normal metabolizers (62), a finding that was replicated in a study that included only children and adolescents (63). CPIC guidelines recommend alternative therapy (a drug metabolized by another enzyme) for individuals who are CYP2C19 ultrarapid metabolizers and consideration of an alternative therapy or a 50% dose reduction of the starting dose for CYP2C19 poor metabolizers (64).
Despite two major CYP pathways (CYP2C19 and CYP2D6) and a number of secondary pathways for sertraline metabolism, variants in the CYP2C19 gene appear to have the greatest impact on the pharmacokinetic parameters of this drug (64). CYP2C19 poor metabolizers have a slower rate of formation of the active metabolite (desmethylsertraline), a longer half-life (~36 h, or 50% longer than normal metabolizers), and an increased (~55%) overall exposure to the parent drug as compared to normal metabolizers (65, 66). Not surprisingly, and likely due to the contributions of multiple enzymatic pathways to sertraline’s metabolism, heterogeneity exists across pharmacogenetic studies, with other investigations not finding a significant impact of CYP2C19 variants on response to sertraline (67, 68). CPIC guidelines suggest the selection of an alternative therapy in CYP2C19 ultrarapid metabolizers and either the selection of an alternative therapy or a 50% reduction in the starting dose for poor metabolizers (64).
Fluvoxamine metabolism is mediated primarily through CYP1A2 and CYP2D6. There is moderate evidence that CYP2D6 poor metabolizer status leads to higher peak plasma concentrations (~50% higher), greater overall exposure (200%), or longer half-life (~60% longer) (60, 64). CPIC guidelines suggest a 25–50% reduction of the recommended starting dose or selection of an alternative therapy in patients who are known CYP2D6 poor metabolizers.
Arguably the most clinically relevant (for younger patients) and controversial SSRI, with respect to pharmacogenetic influence from CYP2D6, is fluoxetine. Fluoxetine is commonly recommended as the first-line SSRI for consideration in patients under 18 years of age who are in need of a pharmacological treatment for depression (69). While adult studies identify some small increases in concentrations of fluoxetine and the most active metabolite (S-norfluoxetine), comparisons of the total active components (parent + metabolites) between poor and nonpoor metabolizers were largely equivocal (59, 64). However, this is one example in which the relationship between genotypes and clinical and pharmacokinetic phenotypes has been directly examined in subjects under 18 years of age (70). In younger patients, CYP2D6 poor metabolizers had increased ratios of fluoxetine to S-norfluoxetine (the active metabolite), even after adjusting for patient weight and drug dose. However, the total active component (fluoxetine + S-norfluoxetine) did not differ across genotype groups, nor did clinical response measures that were examined at 8- and 12-week time points in this study. Based on these data, there are currently no genotype-informed dosing guidelines for fluoxetine.
As previously noted, much of the pharmacogenetic data for SSRIs have been derived in adult populations. At this time, there are few systematic implementation efforts to incorporate genotype-guided dosing of SSRIs into routine clinical care (71). Most clinical pharmacogenetic testing for children (and adults) being treated with an SSRI is currently done using commercial laboratories that examine a constellation of pharmacokinetic and pharmacodynamic genes. While drug dosing recommendations from these tests may be similar to those presented above, the added influences of pharmacodynamic genes (i.e., serotonin receptors and transporters) are also important and in some cases difficult to separate from drug metabolism pharmacogenetic results. The genes/alleles tested, interpretation of metabolizer phenotypes, and treatment recommendations also vary widely between commercial testing companies (72, 73). While the American Psychiatric Association Task Force for Biomarkers and Novel Treatments recognizes the potential influence of drug metabolism pharmacogenetics in relation to adverse effects, they have concluded that data determining how and when to obtain pharmacogenetic testing are lacking (74). Therefore, at this time, pharmacogenetic tests for SSRIs are not considered standard of care and, if ordered, are done so predominantly by prescribers who determine that genotype guidance may be informative for a specific patient.
3.4. Atomoxetine
Atomoxetine is a selective norepinephrine reuptake inhibitor (75) approved for the use of attention-deficit hyperactivity disorder (ADHD). Initially introduced in 2002, atomoxetine was the first nonstimulant option approved for the treatment of ADHD and is typically prescribed to children when stimulants are contraindicated or not tolerated. Atomoxetine is thought to exert its therapeutic effect primarily through increased extracellular concentrations of norepinephrine in the prefrontal cortex (76).
Atomoxetine is an active compound predominantly metabolized by CYP2D6 to the active metabolite 4-hydroxyatomoxetine; however, this metabolite is present at low concentrations and is rapidly glucuronidated to the inactive 4-hydroxyatomoxetine-O-glucuronide metabolite (77). To a lesser extent, CYP2C19 also contributes to atomoxetine metabolism, forming N-desmethylatomoxetine, which is further broken down by CYP2D6 to N-desmethyl-4-hydroxyatomoxetine (4).
CYP2D6 variation wields considerable impact on atomoxetine exposure. A single-dose pharmacokinetic study of 0.5 mg/kg of atomoxetine in 23 children with ADHD showed significantly higher atomoxetine exposures in poor metabolizers as compared to intermediate and normal metabolizers (78). CYP2D6 poor metabolizers experienced 11.4-fold higher dose-corrected exposures as compared to normal metabolizers, while there was a 30-fold range in exposures across all participants. Poor metabolizers have also been shown to require lower doses of atomoxetine, be more likely to have a therapeutic response to atomoxetine, and have lower discontinuation rates as compared to nonpoor metabolizers (79, 80).
Atomoxetine is one of the few CYP2D6 substrates with genotype-guided dosing recommendations for children in the FDA label. Standard dose recommendations in children and adolescents up to 70 kg are to initiate atomoxetine therapy at 0.5 mg/kg/day and titrate up to 1.2 mg/kg/day after a minimum of three days, while dose increases in known CYP2D6 poor metabolizers are recommended only after four weeks if a lack of response is observed. Although FDA labeling provides dose recommendations for poor metabolizers, it is important to recognize that, in clinical studies, CYP2D6 poor metabolizers were more likely to respond to treatment as compared to CYP2D6 normal or ultrarapid metabolizers. Thus, individuals who are normal or ultrarapid metabolizers should be closely monitored for a lack of efficacy. A recently published CPIC guideline for CYP2D6 and atomoxetine recommends initiating dosing at 0.5 mg/kg/day and increasing to 1.2 mg/kg/day after three days in normal and ultrarapid metabolizers, while also utilizing plasma concentrations to attain peak concentrations approaching 400 ng/mL if there is no clinical response or adverse events after two weeks. In poor or intermediate metabolizers, the recommendation is to initiate dosing at 0.5 mg/kg/day and wait two weeks before utilizing peak plasma concentrations to guide dose adjustments in the absence of clinical response and adverse events (81). The Royal Dutch Pharmacists Association Pharmacogenetics Working Group also provides therapeutic dose recommendations for atomoxetine (6). Specifically, they state that poor metabolizers should be closely monitored for adverse drug events and that dose increases are likely unnecessary in this subset. They also note that, while there are insufficient data to allow for dose adjustments in ultrarapid metabolizers, these individuals should be closely monitored for reduced efficacy or prescribed an alternative ADHD medication.
Institutions that have currently implemented CYP2D6 pharmacogenetic testing into clinical care are well positioned to expand their utilization of this information for atomoxetine dosing. Clinical decision support tools can alert providers to individuals known to be CYP2D6 poor metabolizers and provide them with dose recommendations from both the FDA label and the CPIC guideline. Conversely, these tools may also alert clinicians to ultrarapid CYP2D6 metabolizers who may be at an increased risk of nonresponse to standard doses. Additionally, many commercially available pharmacogenetic testing companies include atomoxetine and CYP2D6 on their test panel and provide interpretation on its usage. To date, pharmacogenetic-guided dosing strategies for atomoxetine have predominantly focused on variants in the CYP2D6 gene that result in considerable differences in atomoxetine plasma concentrations; however, future studies assessing atomoxetine response and variation in the norepinephrine transporter gene (SLC6A2) may eventually provide additional insight into which individuals are most likely to respond favorably (82, 83).
3.5. Tacrolimus
Tacrolimus is the most widely used immunosuppressant after kidney transplant. This calcineurin inhibitor is also used after other solid organ transplantations to prevent and treat allograft rejection and to treat glomerulonephritis and graft-versus-host disease in recipients of blood and marrow transplants (84). The inhibition of calcineurin prevents T cell activation and interleukin-2 production, leading to immunosuppressant effects. Tacrolimus use is complicated by the narrow therapeutic window and high interindividual variability in drug disposition. Therapeutic drug monitoring (TDM) of tacrolimus concentrations is used to optimize exposure for each patient; however, under- and overexposure are still common and put patients at risk for graft rejection and toxicity.
The metabolism of tacrolimus is predominately through CYP3A5 (and some contribution of CYP3A4) in enterocytes and hepatocytes. Many metabolites are formed, with one minor metabolite, 31-O-demethyl-tacrolimus, having comparable immunosuppressive activity to tacrolimus. Other metabolites, including the most prevalent 13-O-demethyl-tacrolimus, have little pharmacological activity. Tacrolimus is effluxed by the P-glycoprotein transporter, which is expressed on epithelial cells, endothelial cells, and lymphocytes.
CYP3A5 variants explain 40–50% of the variability in blood concentrations of tacrolimus (85, 86). There are a multitude of studies of transplant patients demonstrating that carriers of the CYP3A5∗1 allele (CYP3A5 expressers) have significantly lower dose-adjusted trough concentrations of tacrolimus compared to noncarriers (8). A CPIC guideline exists for CYP3A5-based dosing of tacrolimus (8), recommending a one-and-a-half- to twofold increase in dose for CYP3A5 expressers for both pediatric and adult patients. One of two randomized clinical trials demonstrated that a higher proportion of patients in the CYP3A5 genotype–guided group achieved a therapeutic dose after three days of tacrolimus compared to standard bodyweight-based dosing in adults after kidney transplant (87), but the other did not (88). Of note, there were no differences between the genotype-guided versus bodyweight-based dosing in graft survival, acute rejection, delayed graft function, or tacrolimus-related toxicities in either trial. In the only pediatric trial to assess CYP3A5-guided tacrolimus dosing in solid organ transplant patients, the therapeutic concentration was reached sooner in the CYP3A5-guided group compared to the unguided group (3.4 days versus 4.7 days), and there were no differences in adverse events (89). CYP3A5-guided dosing is not intended to replace TDM but to be used in conjunction with TDM to achieve target concentrations more quickly and stably than TDM alone, as several studies have suggested (8). The addition of the CYP3A4 genotype may improve dose predictions, as one study found that tacrolimus dose requirement was better predicted by CYP3A4 and CYP3A5 alleles than either gene alone (90).
CYP3A5 genotyping is available clinically and has been implemented in some academic medical centers, including Vanderbilt’s PREDICT program (9), the Mayo Clinic’s Center for Individualized Medicine (91), and St. Jude Children’s Research Hospital’s PG4KDS program. Clinical decision support for tacrolimus dosing based on CYP3A5 is fairly straightforward in nonliver transplant patients (8), but in patients receiving liver transplants, the donor liver must be genotyped since it will influence the pharmacokinetics of tacrolimus posttransplant.
3.6. Voriconazole
Voriconazole is a broad-spectrum triazole antifungal agent. It is recommended as a first-line agent for the treatment and prophylaxis of invasive Aspergillus infections and an alternative therapy for Candida infections (92, 93). The mechanism of action is the inhibition of ergosterol synthesis via inhibition of lanosterol 14α-demethylase. Voriconazole has a narrow therapeutic window; subtherapeutic drug concentrations are associated with mortality from treatment failure, and supratherapeutic levels lead to toxicities, including neurotoxicity and hepatotoxicity (94–97). Due to significant interindividual variability in pharmacokinetics attributed to age, weight, liver function, concomitant medications, and genotype, TDM is recommended (96, 98).
Voriconazole is primarily inactivated by CYP2C19, with minor contributions by CYP3A enzymes and CYP2C9. Decreased CYP2C19 function is associated with higher concentrations of voriconazole and/or drug toxicity. Increased CYP2C19 function is associated with low voriconazole concentrations, subtherapeutic drug levels, and treatment failure. CPIC guidelines recommend that alternate agents be used for CYP2C19 rapid and ultrarapid metabolizers and that alternate agents or lower doses of voriconazole be used in CYP2C19 poor metabolizers (99).
Age is an important consideration for voriconazole dosing. The standard dosing to achieve therapeutic concentrations is higher for children than for adults. For example, for invasive Aspergillus infection treatment, adult maintenance requires a 4 mg/kg intravenous (IV) dose every 12 h, whereas children require an 8 mg/kg IV dose every 12 h (92). In children who are CYP2C19 ultrarapid metabolizers, even higher voriconazole doses are required (100, 101). Given the difficulty of achieving therapeutic concentrations in this group, alternate therapy is recommended when treatment is urgent (99). For children who are rapid metabolizers, guidelines suggest initiation of standard dosing with dose titration guided by TDM, in contrast to adult rapid metabolizers, where alternative agents are recommended (99). This pediatric-specific recommendation stems from a lack of data demonstrating a difference between CYP2C19 normal and rapid metabolizers in children. There is a case report of a 10-year-old CYP2C19 rapid metabolizer who required voriconazole dosing of 14 mg/kg twice daily to achieve therapeutic levels (102). Rapid metabolizers under 12 years of age were predicted to need higher doses of voriconazole than normal metabolizers (30 versus 20 mg/kg/day, respectively) (101). However, there is tremendous variability in voriconazole pharmacokinetics in children, making it difficult to accurately predict voriconazole dosing.
For CYP2C19 poor metabolizers, alternative therapy or lower doses (with careful TDM) are recommended. The limited data for this subgroup in children (101, 103, 104) support extrapolation from adults, as children without CYP2C19 function have high plasma concentrations, which may put them at risk for adverse effects.
Genotype-guided voriconazole dosing has been implemented by some medical centers. In pediatric patients, adjusting voriconazole dosing based on CYP2C19 metabolizer status in pediatric patients needing antifungal prophylaxis resulted in a significant reduction in the time required to achieve target drug concentrations (105). However, genotype is one of many factors influencing voriconazole pharmacokinetics. Ideally, individualized dose calculations will incorporate genotype and other factors to accurately forecast the dose required (98). Timely generation of these sophisticated models could be facilitated by consolidating data from many pediatric centers.
4. COMMON THEMES IN GENE-BASED DOSE OPTIMIZATION FOR CHILDREN
Review of the pediatric-specific evidence for pharmacogenetic associations that are well-established in adults reveals that, in most instances, few studies (and in only relatively small cohorts) of children have been published. These small studies, particularly if they fail to replicate the drug–gene interaction (perhaps due to inadequate power), may not be convincing to pediatric providers who are considering pharmacogenetic implementation in their clinical practice. Although each of the drug–gene interactions discussed herein has been clinically implemented for pediatric patients, most children who are treated with these drugs do not receive preprescription pharmacogenetic testing, as these tests are localized to a few early adopters.
The validation of pharmacogenetic associations in pediatric patients is, however, an important step prior to implementation. The complex physiology of growth and development can lead to pediatric-specific effects. As discussed above, voriconazole variability, after accounting for CYP2C19 variation, is more pronounced in children than adults, indicating additional genetic or nongenetic factors influencing exposure. SSRIs have age- and drug-specific pharmacokinetic profiles, which should ideally be incorporated into dosing recommendations. It has also been demonstrated that the effect size for the SLCO1B1 variant on simvastatin disposition in children is twofold that of adults (106), providing another example of the impact of age on drug–gene interactions.
The population health impact of gene-guided dose optimization for children depends on the frequency of drug exposures and genetic variants and the severity of the adverse outcomes. A study in one tertiary care children’s hospital demonstrated that exposures to sertraline and escitalopram were common (over 500 children exposed per year), atomoxetine and tacrolimus were less common (200–400 per year), and fluvoxamine and voriconazole were rare (30–40 per year) (107). Given the frequency of atypical CYP2D6 and CYP2C19 function, gene-guided therapy for drugs such as the SSRIs can improve therapeutic outcomes for many children. Conversely, while thiopurine drug exposures and the problematic genotypes are relatively uncommon in children, the potential adverse events (life-threatening cytopenias) are severe, emphasizing the value of pre-exposure genetic testing.
There are several drugs commonly used in children for which there is emerging evidence for pharmacogenetic associations. Methylphenidate, a mild central nervous system stimulant prescribed for ADHD (108), is predominantly metabolized through carboxylesterase 1 (CES1). One of the many variants in CES1, rs71647871, considerably reduces methylphenidate metabolism, increases methylphenidate exposure, and reduces the dose requirement for children (109–112). Risperidone is an atypical antipsychotic drug used in the management of schizophrenia, bipolar disorder, autism, and other mental/behavioral health diagnoses. Risperidone is metabolized by CYP2D6, and CYP2D6 metabolizer status is associated with adverse events, including weight gain (113–116). Proton pump inhibitors (PPIs) are one of the most commonly prescribed drugs in the United States and are metabolized predominately by CYP2C19 (117). CYP2C19 metabolizer status has been demonstrated to impact PPI exposure, drug efficacy, and adverse events in children (118–126).
For many drugs with well-established pharmacogenetic associations in adults, there are no data in children. In some cases, this is due to the infrequent use of the drug in pediatric patients (e.g., the antiplatelet drug clopidogrel), but other drugs with potential pharmacogenomic associations are commonly used in children (107). The anti-nausea drug ondansetron is metabolized by CYP2D6, and adult ultrarapid metabolizers are frequently nonresponders to therapy. Ondansetron is one of the most commonly used drugs in pediatrics, which may facilitate investigation of the impact of CYP2D6 function on ondansetron response in children.
Most clinically implemented pharmacogenetic drug-gene pairs involve drug metabolism genes. The sentinel observations of genetic variation influencing drug response were in the drug metabolism enzymes and set the precedent for the importance of these enzymes (127–129). The ability to analyze drug concentrations as an outcome in pharmacokinetic studies has facilitated further discoveries. Given the complexity of drug responses, it is likely that genetic variation in drug targets and the downstream pathways make significant contributions to variability in therapeutic outcomes. Modern genomic techniques and the definition of pharmacodynamic outcomes have the potential to fuel additional discoveries of important drug–gene interactions in adults and children.
5. STRATEGIES FOR ADVANCING GENE-BASED DOSE OPTIMIZATION FOR CHILDREN
The generation of high-quality, validated, generalizable evidence for pediatric pharmacogenetics is necessary for pediatric patients to reap the benefits of precision medicine. One strategy for gathering this evidence is to use real-world data generated during the routine care of pediatric patients to validate known pharmacogenetic findings and discover novel associations. Real-world data have the advantage of representing the target population for therapy, thus including the appropriate ages, demographics, and disease states. Real-world data also may overcome some practical barriers to pediatric research studies, using strategies such as analysis of remnant blood specimens to avoid additional blood draws. Careful attention must be paid to data quality to ensure that findings are robust (130).
Establishing evidence for the benefits of gene-based dosing for children is perhaps the most effective way to facilitate the implementation of this approach. Demonstration of the costs and benefits (e.g., decreased length of stay for oncology patients who undergo gene-guided dosing of chemotherapy) will increase enthusiasm among health-care institutions and payers. Demonstration of improved outcomes (e.g., reduction in organ rejection for transplant patients with gene-based tacrolimus dosing) will convince health-care providers, patients, and parents of the utility of pharmacogenetic testing. Pragmatic trials of gene-based dosing may be an efficient way to generate this evidence (131, 132). However, given that only about 1–10% of patients (depending on the relevant gene) harbor an actionable pharmacogenetic variant, the sample size needed to evaluate these outcomes is large. Collaboration across pediatric centers through national networks will facilitate the timely accumulation of data.
The implementation of pharmacogenetic testing and clinical decision support is not a straightforward task (71, 133–138). Resources for implementation are being developed by CPIC (139) and IGNITE. The implementation barrier may be difficult to overcome in many settings with limited laboratory resources and information technology support. The generation of low-cost, easy-to-interpret pharmacogenetic testing technology and interoperable clinical decision support for pharmacogenetic test results generated from a variety of sources are required for widespread adoption of gene-based dose optimization outside of major academic children’s hospitals.
ACKNOWLEDGMENTS
S.L.V.D. is funded by Burroughs Wellcome Innovation in Regulatory Science award 1015006 and Doris Duke Clinical Scientist Development award 2017075.
Glossary
- Pharmacogenetics
the effects of genetic variation on an individual’s response to a drug
- CPIC
international team facilitating genotype-guided therapy by creating, curating, and posting peer-reviewed, evidence-based, updatable, and detailed gene/drug clinical practice guidelines
- Pharmacokinetics
movement of drugs/metabolites within the body, including absorption, distribution, metabolism, and excretion; “What the body does to the drug”
- Cytochrome P450
family of enzymes that metabolize drug compounds; star allele nomenclature used to denote variations within the gene, with ∗1 typically indicating fully functional protein product
- Pharmacodynamics
effects of a drug through the mechanism of action, e.g., binding to a drug receptor leading to downstream signaling cascades; “What the drug does to the body”
- Therapeutic drug monitoring
clinical practice of measuring a specific drug’s concentration in a patient’s blood to achieve a target concentration through dose titration
Footnotes
DISCLOSURE STATEMENT
The authors are members of CPIC and the Pharmacogenomic Research Network. The authors have been funded by a Burroughs Wellcome Innovation in Regulatory Science award, a Doris Duke Clinical Scientist Development award, and the NIH/NHGRI Implementing Genomics in Practice (IGNITE) Network.
LITERATURE CITED
- 1.Abrahams E. 2008. Right drug—right patient—right time: personalized medicine coalition. Clin. Transl. Sci 1(1):11–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stark Z, Dolman L, Manolio TA, Ozenberger B, Hill SL, et al. 2018. Integrating genomics into health-care: a global responsibility. Am. J. Hum. Genet 104(1):13–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hines RN. 2013. Developmental expression of drug metabolizing enzymes: impact on disposition in neonates and young children. Int. J. Pharm 452(1–2):3–7 [DOI] [PubMed] [Google Scholar]
- 4.Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, et al. 2012. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther 92(4):414–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Relling MV, Klein TE. 2011. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin. Pharmacol. Ther 89(3):464–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swen JJ, Nijenhuis M, de Boer A, Grandia L, Maitland-van der Zee AH, et al. 2011. Pharmacogenetics: from bench to byte—an update of guidelines. Clin. Pharmacol. Ther 89(5):662–73 [DOI] [PubMed] [Google Scholar]
- 7.Yang JJ, Landier W, Yang W, Liu C, Hageman L, et al. 2015. Inherited NUDT15 variant is a genetic determinant of mercaptopurine intolerance in children with acute lymphoblastic leukemia. J. Clin. Oncol 33(11):1235–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birdwell KA, Decker B, Barbarino JM, Peterson JF, Stein CM, et al. 2015. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for CYP3A5 genotype and tacrolimus dosing. Clin. Pharmacol. Ther 98(1):19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Driest SL, Shi Y, Bowton E, Schildcrout J, Peterson J, et al. 2014. Clinically actionable genotypes among 10,000 patients with preemptive pharmacogenomic testing. Clin. Pharmacol. Ther 95(4):423–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caudle KE, Dunnenberger HM, Freimuth RR, Peterson JF, Burlison JD, et al. 2017. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet. Med 19(2):215–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DailyMed. 2019. Codeine sulfate—codeine sulfate tablet. Fact Sheet, DailyMed, Natl. Lib. Med., Bethesda, MD. https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=010905f9-3bcb-4b50-9fe8-a3ad0010f14c [Google Scholar]
- 12.DailyMed. 2019. Oxycodone hydrochloride—oxycodone hydrochloride solution. Fact Sheet, DailyMed, Natl. Lib. Med., Bethesda, MD. https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=c193a56a-bb8f-476d-b1ec-789fd2aaef7e [Google Scholar]
- 13.DailyMed. 2019. Acetaminophen and codeine phosphate—acetaminophen and codeine phosphate liquid. Fact Sheet, DailyMed, Natl. Lib. Med., Bethesda, MD. https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=7bca1d24-a317-44c4-8db2-68030eb961ed [Google Scholar]
- 14.Sevelius H, Colmore JP. 1966. Objective assessment of antitussive agents in patients with chronic cough. J. New Drugs 6(4):216–23 [PubMed] [Google Scholar]
- 15.Sevelius H, McCoy JF, Colmore JP. 1971. Dose response to codeine in patients with chronic cough. Clin. Pharmacol. Ther 12(3):449–55 [DOI] [PubMed] [Google Scholar]
- 16.Freestone C, Eccles R. 1997. Assessment of the antitussive efficacy of codeine in cough associated with common cold. J. Pharm. Pharmacol 49(10):1045–49 [DOI] [PubMed] [Google Scholar]
- 17.Hutchings HA, Eccles R. 1994. The opioid agonist codeine and antagonist naltrexone do not affect voluntary suppression of capsaicin induced cough in healthy subjects. Eur. Respir. J 7(4):715–19 [DOI] [PubMed] [Google Scholar]
- 18.Smith J, Owen E, Earis J, Woodcock A. 2006. Effect of codeine on objective measurement of cough in chronic obstructive pulmonary disease. J. Allergy Clin. Immunol 117(4):831–35 [DOI] [PubMed] [Google Scholar]
- 19.Mignat C, Wille U, Ziegler A. 1995. Affinity profiles of morphine, codeine, dihydrocodeine and their glucuronides at opioid receptor subtypes. Life Sci. 56(10):793–99 [DOI] [PubMed] [Google Scholar]
- 20.Volpe DA, McMahon Tobin GA, Mellon RD, Katki AG, Parker RJ, et al. 2011. Uniform assessment and ranking of opioid μ receptor binding constants for selected opioid drugs. Regul. Toxicol. Pharmacol 59(3):385–90 [DOI] [PubMed] [Google Scholar]
- 21.Gaedigk A, Sangkuhl K, Whirl-Carrillo M, Klein T, Leeder JS. 2017. Prediction of CYP2D6 phenotype from genotype across world populations. Genet. Med 19(1):69–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crews KR, Gaedigk A, Dunnenberger HM, Leeder JS, Klein TE, et al. 2014. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin. Pharmacol. Ther 95(4):376–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crews KR, Gaedigk A, Dunnenberger HM, Klein TE, Shen DD, et al. 2012. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for codeine therapy in the context of cytochrome P450 2D6 (CYP2D6) genotype. Clin. Pharmacol. Ther 91(2):321–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koren G, Cairns J, Chitayat D, Gaedigk A, Leeder SJ. 2006. Pharmacogenetics of morphine poisoning in a breastfed neonate of a codeine-prescribed mother. Lancet 368(9536):704. [DOI] [PubMed] [Google Scholar]
- 25.Ciszkowski C, Madadi P, Phillips MS, Lauwers AE, Koren G. 2009. Codeine, ultrarapid-metabolism genotype, and postoperative death. N. Engl. J. Med 361(8):827–28 [DOI] [PubMed] [Google Scholar]
- 26.Ferreirós N, Dresen S, Hermanns-Clausen M, Auwaerter V, Thierauf A, et al. 2009. Fatal and severe codeine intoxication in 3-year-old twins—interpretation of drug and metabolite concentrations. Int. J. Legal Med 123(5):387–94 [DOI] [PubMed] [Google Scholar]
- 27.Kelly LE, Rieder M, van den Anker J, Malkin B, Ross C, et al. 2012. More codeine fatalities after tonsillectomy in North American children. Pediatrics 129(5):e1343–47 [DOI] [PubMed] [Google Scholar]
- 28.Friedrichsdorf SJ, Nugent AP, Strobl AQ. 2013. Codeine-associated pediatric deaths despite using recommended dosing guidelines: three case reports. J. Opioid Manag 9(2):151–55 [DOI] [PubMed] [Google Scholar]
- 29.Voronov P, Przybylo HJ, Jagannathan N. 2007. Apnea in a child after oral codeine: a genetic variant—an ultra-rapid metabolizer. Pediatr. Anaesth 17(7):684–87 [DOI] [PubMed] [Google Scholar]
- 30.Madadi P, Ross CJD, Hayden MR, Carleton BC, Gaedigk A, et al. 2009. Pharmacogenetics of neonatal opioid toxicity following maternal use of codeine during breastfeeding: a case-control study. Clin. Pharmacol. Ther 85(1):31–35 [DOI] [PubMed] [Google Scholar]
- 31.Prows CA, Zhang X, Huth MM, Zhang K, Saldaña SN, et al. 2014. Codeine-related adverse drug reactions in children following tonsillectomy: a prospective study. Laryngoscope 124(5):1242–50 [DOI] [PubMed] [Google Scholar]
- 32.Khetani JD, Madadi P, Sommer DD, Reddy D, Sistonen J, et al. 2012. Apnea and oxygen desaturations in children treated with opioids after adenotonsillectomy for obstructive sleep apnea syndrome: a prospective pilot study. Pediatr. Drugs 14(6):411–15 [DOI] [PubMed] [Google Scholar]
- 33.Yue QY, Svensson JO, Alm C, Sjöqvist F, Säwe J. 1989. Codeine O-demethylation co-segregates with polymorphic debrisoquine hydroxylation. Br. J. Clin. Pharmacol 28(6):639–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams DG, Patel A, Howard RF. 2002. Pharmacogenetics of codeine metabolism in an urban population of children and its implications for analgesic reliability. Br. J. Anaesth 89(6):839–45 [DOI] [PubMed] [Google Scholar]
- 35.Sistonen J, Madadi P, Ross CJ, Yazdanpanah M, Lee JW, et al. 2012. Prediction of codeine toxicity in infants and their mothers using a novel combination of maternal genetic markers. Clin. Pharmacol. Ther 91(4):692–99 [DOI] [PubMed] [Google Scholar]
- 36.Food Drug Admin. 2013. Safety review update of codeine use in children; new Boxed Warning and Contraindication on use after tonsillectomy and/or adenoidectomy. Drug Saf. Commun., US Food Drug Admin., Silver Spring, MD. https://www.fda.gov/media/85072/download [Google Scholar]
- 37.Cavallari LH, Van Driest SL, Prows CA, Bishop JR, Limdi NA, et al. 2019. Multi-site investigation of strategies for the clinical implementation of CYP2D6 genotyping to guide drug prescribing. Genet. Med 21:2255–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gammal RS, Crews KR, Haidar CE, Hoffman JM, Baker DK, et al. 2016. Pharmacogenetics for safe codeine use in sickle cell disease. Pediatrics 138(1):e20153479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gammal RS, Caudle KE, Quinn CT, Wang WC, Gaedigk A, et al. 2019. The case for pharmacogenetics-guided prescribing of codeine in children. Clin. Pharmacol. Ther 105(6):1300–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jerome J, Solodiuk JC, Sethna N, McHale J, Berde C. 2014. A single institution’s effort to translate codeine knowledge into specific clinical practice. J. Pain Symptom Manag 48(1):119–26 [DOI] [PubMed] [Google Scholar]
- 41.Livingstone MJ, Groenewald CB, Rabbitts JA, Palermo TM. 2017. Codeine use among children in the United States: a nationally representative study from 1996 to 2013. Pediatr. Anaesth 27(1):19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Cleve WC. 2017. Pediatric posttonsillectomy analgesia before and after the black box warning against codeine use. JAMA Otolaryngol. 143(10):1052–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Beaumais TA, Jacqz-Aigrain E. 2012. Intracellular disposition of methotrexate in acute lymphoblastic leukemia in children. Curr. Drug Metab 13(6):822–34 [DOI] [PubMed] [Google Scholar]
- 44.Zaza G, Cheok M, Krynetskaia N, Thorn C, Stocco G, et al. 2010. Thiopurine pathway. Pharmacogenet. Genom 20(9):573–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weinshilboum R. 2001. Thiopurine pharmacogenetics: clinical and molecular studies of thiopurine methyltransferase. Drug Metab. Dispos 29(4):601–5 [PubMed] [Google Scholar]
- 46.Relling MV, Hancock ML, Rivera GK, Sandlund JT, Ribeiro RC, et al. 1999. Mercaptopurine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus. J. Natl. Cancer Inst 91(23):2001–8 [DOI] [PubMed] [Google Scholar]
- 47.Relling MV, Schwab M, Whirl-Carrillo M, Suarez-Kurtz G, Pui C-H, et al. 2018. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for thiopurine dosing based on TPMT and NUDT15 genotypes: 2018 update. Clin. Pharmacol. Ther 105(5):1095–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Relling MV, Gardner EE, Sandborn WJ, Schmiegelow K, Pui C-H, et al. 2013. Clinical pharmacogenetics implementation consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing: 2013 update. Clin. Pharmacol. Ther 93(4):324–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weinshilboum RM, Sladek SL. 1980. Mercaptopurine pharmacogenetics: monogenic inheritance of erythrocyte thiopurine methyltransferase activity. Am. J. Hum. Genet 32(5):651–62 [PMC free article] [PubMed] [Google Scholar]
- 50.Walker GJ, Harrison JW, Heap GA, Voskuil MD, Andersen V, et al. 2019. Association of genetic variants in NUDT15 with thiopurine-induced myelosuppression in patients with inflammatory bowel disease. JAMA 321(8):773–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanaka Y, Kato M, Hasegawa D, Urayama KY, Nakadate H, et al. 2015. Susceptibility to 6-MP toxicity conferred by a NUDT15 variant in Japanese children with acute lymphoblastic leukaemia. Br. J. Haematol 171(1):109–15 [DOI] [PubMed] [Google Scholar]
- 52.Moriyama T, Nishii R, Perez-Andreu V, Yang W, Klussmann FA, et al. 2016. NUDT15 polymorphisms alter thiopurine metabolism and hematopoietic toxicity. Nat. Genet 48(4):367–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matimba A, Li F, Livshits A, Cartwright CS, Scully S, et al. 2014. Thiopurine pharmacogenomics: association of SNPs with clinical response and functional validation of candidate genes. Pharmacogenomics 15(4):433–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stocco G, Franca R, Verzegnassi F, Londero M, Rabusin M, Decorti G. 2012. Multilocus genotypes of relevance for drug metabolizing enzymes and therapy with thiopurines in patients with acute lymphoblastic leukemia. Front. Genet 3:309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hareedy MS, El Desoky ES, Woillard J-B, Thabet RH, Ali AM, et al. 2015. Genetic variants in 6-mercaptopurine pathway as potential factors of hematological toxicity in acute lymphoblastic leukemia patients. Pharmacogenomics 16(10):1119–34 [DOI] [PubMed] [Google Scholar]
- 56.March J, Silva S, Petrycki S, Curry J, Wells K, et al. 2004. Fluoxetine, cognitive-behavioral therapy, and their combination for adolescents with depression: Treatment for Adolescents with Depression Study (TADS) randomized controlled trial. JAMA 292(7):807–20 [DOI] [PubMed] [Google Scholar]
- 57.Brent D, Emslie G, Clarke G, Wagner KD, Asarnow JR, et al. 2008. Switching to another SSRI or to venlafaxine with or without cognitive behavioral therapy for adolescents with SSRI-resistant depression: the TORDIA randomized controlled trial. JAMA 299(8):901–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Findling RL, McNamara NK, Stansbrey RJ, Feeny NC, Young CM, et al. 2006. The relevance of pharmacokinetic studies in designing efficacy trials in juvenile major depression. J. Child Adolesc. Psychopharmacol 16(1–2):131–45 [DOI] [PubMed] [Google Scholar]
- 59.DailyMed. 2019. Fluoxetine—fluoxetine capsules capsule. Fact Sheet, DailyMed, Natl. Lib. Med., Bethesda, MD. https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=59de2889-c3d3-4ebf-8826-13f30a3fa439 [Google Scholar]
- 60.DailyMed. 2019. Fluvoxamine maleate—fluvoxamine maleate tablet, film coated. Fact Sheet, DailyMed, Natl. Lib. Med., Bethesda, MD. https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=85b61f83-ab47-470d-8673-02fea29247be [Google Scholar]
- 61.Chang M, Tybring G, Dahl M-L, Lindh JD. 2014. Impact of cytochrome P450 2C19 polymorphisms on citalopram/escitalopram exposure: a systematic review and meta-analysis. Clin. Pharmacokinet 53(9):801–11 [DOI] [PubMed] [Google Scholar]
- 62.Jukić MM, Haslemo T, Molden E, Ingelman-Sundberg M. 2018. Impact of CYP2C19 genotype on escitalopram exposure and therapeutic failure: a retrospective study based on 2,087 patients. Am. J. Psychiatry 175(5):463–70 [DOI] [PubMed] [Google Scholar]
- 63.Aldrich SL, Poweleit EA, Prows CA, Martin LJ, Strawn JR, Ramsey LB. 2019. Influence of CYP2C19 metabolizer status on escitalopram/citalopram tolerability and response in youth with anxiety and depressive disorders. Front. Pharmacol 10:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hicks JK, Bishop JR, Sangkuhl K, Müller DJ, Ji Y, et al. 2015. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin. Pharmacol. Ther 98(2):127–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang JH, Liu ZQ, Wang W, Chen XP, Shu Y, et al. 2001. Pharmacokinetics of sertraline in relation to genetic polymorphism of CYP2C19. Clin. Pharmacol. Ther 70(1):42–47 [DOI] [PubMed] [Google Scholar]
- 66.Rudberg I, Hermann M, Refsum H, Molden E. 2008. Serum concentrations of sertraline and N-desmethyl sertraline in relation to CYP2C19 genotype in psychiatric patients. Eur. J. Clin. Pharmacol 64(12):1181–88 [DOI] [PubMed] [Google Scholar]
- 67.Brandl EJ, Tiwari AK, Zhou X, Deluce J, Kennedy JL, et al. 2014. Influence of CYP2D6 and CYP2C19 gene variants on antidepressant response in obsessive-compulsive disorder. Pharmacogenomics J. 14(2):176–81 [DOI] [PubMed] [Google Scholar]
- 68.Yuce-Artun N, Baskak B, Ozel-Kizil ET, Ozdemir H, Uckun Z, et al. 2016. Influence of CYP2B6 and CYP2C19 polymorphisms on sertraline metabolism in major depression patients. Int. J. Clin. Pharm 38(2):388–94 [DOI] [PubMed] [Google Scholar]
- 69.Vitiello B, Ordóñez AE. 2016. Pharmacological treatment of children and adolescents with depression. Expert Opin. Pharmacother 17(17):2273–79 [DOI] [PubMed] [Google Scholar]
- 70.Gassó P, Rodríguez N, Mas S, Pagerols M, Blázquez A, et al. 2014. Effect of CYP2D6, CYP2C9 and ABCB1 genotypes on fluoxetine plasma concentrations and clinical improvement in children and adolescent patients. Pharmacogenomics J. 14(5):457–62 [DOI] [PubMed] [Google Scholar]
- 71.Ramsey LB, Prows CA, Zhang K, Saldaña SN, Sorter MT, et al. 2019. Implementation of pharmacogenetics at Cincinnati Children’s Hospital Medical Center: lessons learned over 14 years of personalizing medicine. Clin. Pharmacol. Ther 105(1):49–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bousman CA, Dunlop BW. 2018. Genotype, phenotype, and medication recommendation agreement among commercial pharmacogenetic-based decision support tools. Pharmacogenomics J. 18(5):613–22 [DOI] [PubMed] [Google Scholar]
- 73.Bousman CA, Jaksa P, Pantelis C. 2017. Systematic evaluation of commercial pharmacogenetic testing in psychiatry: a focus on CYP2D6 and CYP2C19 allele coverage and results reporting. Pharmacogenetics Genom. 27(11):387–93 [DOI] [PubMed] [Google Scholar]
- 74.Zeier Z, Carpenter LL, Kalin NH, Rodriguez CI, McDonald WM, et al. 2018. Clinical implementation of pharmacogenetic decision support tools for antidepressant drug prescribing. Am. J. Psychiatry 175(9):873–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zerbe RL, Rowe H, Enas GG, Wong D, Farid N, Lemberger L. 1985. Clinical pharmacology of tomoxetine, a potential antidepressant. J. Pharmacol. Exp. Ther 232(1):139–43 [PubMed] [Google Scholar]
- 76.Bymaster FP, Katner JS, Nelson DL, Hemrick-Luecke SK, Threlkeld PG, et al. 2002. Atomoxetine increases extracellular levels of norepinephrine and dopamine in prefrontal cortex of rat: a potential mechanism for efficacy in attention deficit/hyperactivity disorder. Neuropsychopharmacology 27(5):699–711 [DOI] [PubMed] [Google Scholar]
- 77.Dinh JC, Pearce RE, Van Haandel L, Gaedigk A, Leeder JS. 2016. Characterization of atomoxetine biotransformation and implications for development of PBPK models for dose individualization in children. Drug Metab. Dispos. Biol. Fate Chem 44(7):1070–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brown JT, Abdel-Rahman SM, van Haandel L, Gaedigk A, Lin YS, Leeder JS. 2016. Single dose, CYP2D6 genotype-stratified pharmacokinetic study of atomoxetine in children with ADHD. Clin. Pharmacol. Ther 99(6):642–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Michelson D, Read HA, Ruff DD, Witcher J, Zhang S, McCracken J. 2007. CYP2D6 and clinical response to atomoxetine in children and adolescents with ADHD. J. Am. Acad. Child Adolesc. Psychiatry 46(2):242–51 [DOI] [PubMed] [Google Scholar]
- 80.Trzepacz PT, Williams DW, Feldman PD, Wrishko RE, Witcher JW, Buitelaar JK. 2008. CYP2D6 metabolizer status and atomoxetine dosing in children and adolescents with ADHD. Eur. Neuropsychopharmacol 18(2):79–86 [DOI] [PubMed] [Google Scholar]
- 81.Brown JT, Bishop JR, Sangkuhl K, Nurmi EL, Mueller DJ, et al. 2019. Clinical Pharmacogenetics Implementation Consortium Guideline for cytochrome P450 (CYP)2D6 genotype and atomoxetine therapy. Clin. Pharmacol. Ther 106:94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ramoz N, Boni C, Downing AM, Close SL, Peters SL, et al. 2009. A haplotype of the norepinephrine transporter (Net) gene Slc6a2 is associated with clinical response to atomoxetine in attention-deficit hyperactivity disorder (ADHD). Neuropsychopharmacology 34(9):2135–42 [DOI] [PubMed] [Google Scholar]
- 83.Brown JT, Bishop JR. 2015. Atomoxetine pharmacogenetics: associations with pharmacokinetics, treatment response and tolerability. Pharmacogenomics 16(13):1513–20 [DOI] [PubMed] [Google Scholar]
- 84.Barbarino JM, Staatz CE, Venkataramanan R, Klein TE, Altman RB. 2013. PharmGKB summary: cyclosporine and tacrolimus pathways. Pharmacogenet. Genom 23(10):563–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Birdwell KA, Grady B, Choi L, Xu H, Bian A, et al. 2012. The use of a DNA biobank linked to electronic medical records to characterize pharmacogenomic predictors of tacrolimus dose requirement in kidney transplant recipients. Pharmacogenet. Genom 22(1):32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Haufroid V, Mourad M, Van Kerckhove V, Wawrzyniak J, De Meyer M, et al. 2004. The effect of CYP3A5 and MDR1 (ABCB1) polymorphisms on cyclosporine and tacrolimus dose requirements and trough blood levels in stable renal transplant patients. Pharmacogenetics 14(3):147–54 [DOI] [PubMed] [Google Scholar]
- 87.Thervet E, Loriot MA, Barbier S, Buchler M, Ficheux M, et al. 2010. Optimization of initial tacrolimus dose using pharmacogenetic testing. Clin. Pharmacol. Ther 87(6):721–26 [DOI] [PubMed] [Google Scholar]
- 88.Pallet N, Etienne I, Buchler M, Bailly E, Hurault de Ligny B, et al. 2016. Long-term clinical impact of adaptation of initial tacrolimus dosing to CYP3A5 genotype. Am. J. Transplant 16(9):2670–75 [DOI] [PubMed] [Google Scholar]
- 89.Min S, Papaz T, Lafreniere-Roula M, Nalli N, Grasemann H, et al. 2018. A randomized clinical trial of age and genotype-guided tacrolimus dosing after pediatric solid organ transplantation. Pediatr. Transplant 22(7):1–9 [DOI] [PubMed] [Google Scholar]
- 90.Elens L, Bouamar R, Hesselink DA, Haufroid V, Van Der Heiden IP, et al. 2011. A new functional CYP3A4 intron 6 polymorphism significantly affects tacrolimus pharmacokinetics in kidney transplant recipients. Clin. Chem 57(11):1574–83 [DOI] [PubMed] [Google Scholar]
- 91.Caraballo PJ, Hodge LS, Bielinski SJ, Stewart AK, Farrugia G, et al. 2017. Multidisciplinary model to implement pharmacogenomics at the point of care. Genet. Med 19(4):421–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Patterson TF, Thompson GR, Denning DW, Fishman JA, Hadley S, et al. 2016. Practice guidelines for the diagnosis and management of aspergillosis: 2016 update by the Infectious Diseases Society of America. Clin. Infect. Dis 63(4):e1–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pappas PG, Kauffman CA, Andes DR, Clancy CJ, Marr KA, et al. 2016. Clinical practice guideline for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin. Infect. Dis 62(4):e1–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Denning DW, Ribaud P, Milpied N, Caillot D, Herbrecht R, et al. 2002. Efficacy and safety of voriconazole in the treatment of acute invasive aspergillosis. Clin. Infect. Dis 34(5):563–71 [DOI] [PubMed] [Google Scholar]
- 95.Hamada Y, Seto Y, Yago K, Kuroyama M. 2012. Investigation and threshold of optimum blood concentration of voriconazole: a descriptive statistical meta-analysis. J. Infect. Chemother 18(4):501–7 [DOI] [PubMed] [Google Scholar]
- 96.Pascual A, Calandra T, Bolay S, Buclin T, Bille J, Marchetti O. 2008. Voriconazole therapeutic drug monitoring in patients with invasive mycoses improves efficacy and safety outcomes. Clin. Infect. Dis 46(2):201–11 [DOI] [PubMed] [Google Scholar]
- 97.Tan K, Brayshaw N, Tomaszewski K, Troke P, Wood N. 2006. Investigation of the potential relationships between plasma voriconazole concentrations and visual adverse events or liver function test abnormalities. J. Clin. Pharmacol 46(2):235–43 [DOI] [PubMed] [Google Scholar]
- 98.Kadam RS, Van Den Anker JN. 2016. Pediatric clinical pharmacology of voriconazole: role of pharmacokinetic/pharmacodynamic modeling in pharmacotherapy. Clin. Pharmacokinet 55(9):1031–43 [DOI] [PubMed] [Google Scholar]
- 99.Moriyama B, Obeng AO, Barbarino J, Penzak SR, Henning SA, et al. 2017. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for CYP2C19 and voriconazole therapy. Clin. Pharmacol. Ther 102(1):45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cendejas-Bueno E, Borobia AM, Gomez-Lopez A, Escosa-García L, Río-García M, et al. 2016. Invasive aspergillosis in a paediatric allogeneic stem cell transplantation recipient owing to a susceptible Aspergillus fumigatus: treatment failure with high doses of voriconazole and influence of CYP2C19 polymorphisms. Int. J. Antimicrob. Agents 47(5):410–11 [DOI] [PubMed] [Google Scholar]
- 101.Hicks JK, Crews KR, Flynn P, Haidar CE, Daniels CC, et al. 2014. Voriconazole plasma concentrations in immunocompromised pediatric patients vary by CYP2C19 diplotypes. Pharmacogenomics 15(8):1065–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hicks JK, Gonzalez BE, Zembillas AS, Kusick K, Murthy S, et al. 2016. Invasive Aspergillus infection requiring lobectomy in a CYP2C19 rapid metabolizer with subtherapeutic voriconazole concentrations. Pharmacogenomics 17(7):663–67 [DOI] [PubMed] [Google Scholar]
- 103.Mori M, Kobayashi R, Kato K, Maeda N, Fukushima K, et al. 2015. Pharmacokinetics and safety of voriconazole intravenous-to-oral switch regimens in immunocompromised Japanese pediatric patients. Antimicrob. Agents Chemother 59(2):1004–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Narita A, Muramatsu H, Sakaguchi H, Doisaki S, Tanaka M, et al. 2013. Correlation of CYP2C19 phenotype with voriconazole plasma concentration in children. J. Pediatr. Hematol. Oncol 35(5):e219–23 [DOI] [PubMed] [Google Scholar]
- 105.Teusink A, Vinks A, Zhang K, Davies S, Fukuda T, et al. 2016. Genotype-directed dosing leads to optimized voriconazole levels in pediatric patients receiving hematopoietic stem cell transplantation. Biol. Blood Marrow Transplant 22(3):482–86 [DOI] [PubMed] [Google Scholar]
- 106.Wagner JB, Abdel-Rahman S, Van Haandel L, Gaedigk A, Gaedigk R, et al. 2018. Impact of SLCO1B1 genotype on pediatric simvastatin acid pharmacokinetics. J. Clin. Pharmacol 58(6):823–33 [DOI] [PubMed] [Google Scholar]
- 107.Aka I, Bernal CJ, Carroll R, Maxwell-Horn A, Oshikoya KA, Van Driest SL. 2017. Clinical pharmacogenetics of cytochrome P450-associated drugs in children. J. Pers. Med 7(4):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chai G, Governale L, McMahon AW, Trinidad JP, Staffa J, Murphy D. 2012. Trends of outpatient prescription drug utilization in US children, 2002–2010. Pediatrics 130(1):23–31 [DOI] [PubMed] [Google Scholar]
- 109.Marsh S, Xiao M, Yu J, Ahluwalia R, Minton M, et al. 2004. Pharmacogenomic assessment of carboxylesterases 1 and 2. Genomics 84(4):661–68 [DOI] [PubMed] [Google Scholar]
- 110.Zhu H-J, Patrick KS, Yuan H-J, Wang J-S, Donovan JL, et al. 2008. Two CES1 gene mutations lead to dysfunctional carboxylesterase 1 activity in man: clinical significance and molecular basis. Am. J. Hum. Genet 82(6):1241–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nemoda Z, Angyal N, Tarnok Z, Gadoros J, Sasvari-Szekely M. 2009. Carboxylesterase 1 gene polymorphism and methylphenidate response in ADHD. Neuropharmacology 57(7–8):731–33 [DOI] [PubMed] [Google Scholar]
- 112.Stage C, Jürgens G, Guski LS, Thomsen R, Bjerre D, et al. 2017. The impact of CES1 genotypes on the pharmacokinetics of methylphenidate in healthy Danish subjects. Br. J. Clin. Pharmacol 83(7):1506–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Youngster I, Zachor DA, Gabis LV, Bar-Chaim A, Benveniste-Levkovitz P, et al. 2014. CYP2D6 genotyping in paediatric patients with autism treated with risperidone: a preliminary cohort study. Dev. Med. Child Neurol 56(10):990–94 [DOI] [PubMed] [Google Scholar]
- 114.Nussbaum LA, Dumitrasçu V, Tudor A, Grãdinaru R, Andreescu N, Puiu M. 2014. Molecular study of weight gain related to atypical antipsychotics: clinical implications of the CYP2D6 genotype. Rom. J. Morphol. Embryol 55(3):877–84 [PubMed] [Google Scholar]
- 115.dos Santos A Jr., Henriques TB, de Mello MP, Ferreira Neto AP, Paes LA, et al. 2015. Hyperprolactinemia in children and adolescents with use of risperidone: clinical and molecular genetics aspects. J. Child Adolesc. Psychopharmacol 25(10):738–48 [DOI] [PubMed] [Google Scholar]
- 116.Oshikoya KA, Neely KM, Carroll RJ, Aka IT, Maxwell-Horn AC, et al. 2019. CYP2D6 genotype and adverse events to risperidone in children and adolescents. Pediatr. Res 85(5):602–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ward RM, Kearns GL. 2013. Proton pump inhibitors in pediatrics: mechanism of action, pharmacokinetics, pharmacogenetics, and pharmacodynamics. Pediatr. Drugs 15(2):119–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kearns GL, Blumer J, Schexnayder S, James LP, Adcock KG, et al. 2008. Single-dose pharmacokinetics of oral and intravenous pantoprazole in children and adolescents. J. Clin. Pharmacol 48(11):1356–65 [DOI] [PubMed] [Google Scholar]
- 119.Kearns GL, Leeder JS, Gaedigk A. 2010. Impact of the CYP2C19∗17 allele on the pharmacokinetics of omeprazole and pantoprazole in children: evidence for a differential effect. Drug Metab. Dispos 38(6):894–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gumus E, Karaca O, Babaoglu MO, Baysoy G, Balamtekin N, et al. 2012. Evaluation of lansoprazole as a probe for assessing cytochrome P450 2C19 activity and genotype-phenotype correlation in childhood. Eur. J. Clin. Pharmacol 68(5):629–36 [DOI] [PubMed] [Google Scholar]
- 121.Shakhnovich V, Smith PB, Guptill JT, James LP, Collier DN, et al. 2018. Obese children require lower doses of pantoprazole than nonobese peers to achieve equal systemic drug exposures. J. Pediatr 193:102–8.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lima JJ, Lang JE, Mougey EB, Blake KB, Gong Y, et al. 2013. Association of CYP2C19 polymorphisms and lansoprazole-associated respiratory adverse effects in children. J. Pediatr 163(3):686–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lang JE, Holbrook JT, Mougey EB, Wei CY, Wise RA, et al. 2015. Lansoprazole is associated with worsening asthma control in children with the CYP2C19 poor metabolizer phenotype. Ann. Am. Thorac. Soc 12(6):878–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Franciosi JP, Mougey EB, Williams A, Gomez-Suarez RA, Thomas C, et al. 2018. Association between CYP2C19∗17 alleles and pH probe testing outcomes in children with symptomatic gastroesophageal reflux. J. Clin. Pharmacol 58(1):89–96 [DOI] [PubMed] [Google Scholar]
- 125.Franciosi JP, Mougey EB, Williams A, Gomez Suarez RA, Thomas C, et al. 2018. Association between CYP2C19 extensive metabolizer phenotype and childhood anti-reflux surgery following failed proton pump inhibitor medication treatment. Eur. J. Pediatr 177(1):69–77 [DOI] [PubMed] [Google Scholar]
- 126.Molina-Infante J, Rodriguez-Sanchez J, Martinek J, van Rhijn BD, Krajciova J, et al. 2015. Long-term loss of response in proton pump inhibitor-responsive esophageal eosinophilia is uncommon and influenced by CYP2C19 genotype and rhinoconjunctivitis. Am. J. Gastroenterol 110(11):1567–75 [DOI] [PubMed] [Google Scholar]
- 127.Price Evans DA, Manley KA, McKusick VA. 1960. Genetic control of isoniazid metabolism in man. Br. Med. J 2(5197):485–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Evans FT, Gray PWS, Lehmann H, Silk E. 1952. Sensitivity to succinylcholine in relation to serumcholinesterase. Lancet 1(6721):1229–30 [DOI] [PubMed] [Google Scholar]
- 129.Mahgoub A, Idle JR, Dring LG, Lancaster R, Smith RL. 1977. Polymorphic hydroxylation of Debrisoquine in man. Lancet 2(8038):584–86 [DOI] [PubMed] [Google Scholar]
- 130.Van Driest SL, Choi L. 2019. Real world data for pediatric pharmacometrics: Can we upcycle clinical data for research use? Clin. Pharmacol. Ther 106(1):84–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lee CR, Sriramoju VB, Cervantes A, Howell LA, Varunok N, et al. 2018. Clinical outcomes and sustainability of using CYP2C19 genotype-guided antiplatelet therapy after percutaneous coronary intervention. Circ. Genom. Precis. Med 11(4):e002069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Williams AK, Klein MD, Martin J, Weck KE, Rossi JS, et al. 2019. CYP2C19 genotype-guided antiplatelet therapy and 30-day outcomes after percutaneous coronary intervention. Circ. Genom. Precis. Med 12(2):e002441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Empey PE, Stevenson JM, Tuteja S, Weitzel KW, Angiolillo DJ, et al. 2018. Multisite investigation of strategies for the implementation of CYP2C19 genotype-guided antiplatelet therapy. Clin. Pharmacol. Ther 104(4):664–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bell GC, Crews KR, Wilkinson MR, Haidar CE, Hicks JK, et al. 2013. Development and use of active clinical decision support for preemptive pharmacogenomics. J. Am. Med. Inform. Assoc 21(e1):e93–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bielinski SJ, Olson JE, Pathak J, Weinshilboum RM, Wang L, et al. 2014. Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time—using genomic data to individualize treatment protocol. Mayo Clin. Proc 89(1):25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Danahey K, Borden BA, Furner B, Yukman P, Hussain S, et al. 2017. Simplifying the use of pharmacogenomics in clinical practice: building the genomic prescribing system. J. Biomed. Inform 75:110–21 [DOI] [PubMed] [Google Scholar]
- 137.Leeder JS, Brown JT, Soden SE. 2014. Individualizing the use of medications in children: making Goldilocks happy. Clin. Pharmacol. Ther 96(3):304–6 [DOI] [PubMed] [Google Scholar]
- 138.Manzi SF, Fusaro VA, Chadwick L, Brownstein C, Clinton C, et al. 2017. Creating a scalable clinical pharmacogenomics service with automated interpretation and medical record result integration—experience from a pediatric tertiary care facility. J. Am. Med. Inform. Assoc 24(1):74–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Traynor K. 2019. CPIC secures $5 million for pharmacogenetics work. Am. J. Health. Syst. Pharm 76(4):197. [DOI] [PubMed] [Google Scholar]