Abstract
Sulfated N-glycans play a crucial role in the interaction between influenza A virus (IAV) and its host. These glycans have been found to enhance viral replication, highlighting their significance in IAV propagation. This study investigated the expression of acidic N-glycans, specifically sulfated and phosphorylated glycans, in the egg whites of 72 avian species belonging to the Order Anseriformes (waterfowls). We used the glycoblotting-based sulphoglycomics approach to elucidate the diversity of acidic N-glycans and infer their potential role in protecting embryos from infections. Family-specific variations in sulfated and phosphorylated N-glycan profiles were identified in waterfowl egg whites. Different waterfowl species exhibited distinct expressions of sulfated trans-Gal(+) and trans-Gal(−) N-glycan structures. Additionally, species-specific expression of phosphorylated N-glycans was observed. Furthermore, it was found that waterfowl species with high avian influenza virus (AIV) prevalence displayed a higher abundance of phosphorylated hybrid and high-mannose N-glycans on their egg whites. These findings shed light on the importance of phosphorylated and sulfated N-glycans in understanding the role of acidic glycans in IAV propagation.
Keywords: glycoblotting, ovo-sulphoglycomics, on-bead esterification, MTT, virus prevalence
Influenza A virus (IAV) membrane proteins, hemagglutinin (HA) and neuraminidase (NA), play crucial roles in virus infectivity, transmissibility, pathogenicity, and host specificity.1 Combinations of different HA and NA subtypes give rise to some of the deadliest IAVs that have caused major pandemics.2 IAVs can evolve and evade neutralization by antibodies and vaccinations through antigenic evolution.3 In host specificity, human-adapted IAVs HA exhibit binding specificity toward sialyl-LacNAc, characterized by an α2,6 linkage between Sia and Gal, while avian IAVs bind to sialyl-LacNAc with α2,3 linkages.4,5
IAVs are spreading to a wide range of host species, with avian species belonging to the Order Anseriformes (waterfowls) serving as its natural reservoir.2 Waterfowl are resistant to avian influenza virus (AIV) and show no clinical symptoms despite harboring almost all subtypes of IAV.6,7 Avian species primarily experience IAV infections in their respiratory and intestinal tracts. In chickens, the respiratory tract is the initial site of infection, facilitating viral transmission through aerosol droplets. On the other hand, ducks transmit the virus through the oral–fecal route, as IAV replication occurs in their intestine, colon, and cloaca, typically observed in birds infected with low pathogenic avian influenza virus (LPAIV). In contrast, highly pathogenic avian influenza virus (HPAIV) causes systemic infections, affecting various tissues such as the heart, brain, spleen, liver, and oviduct.6−8
HPAIV has been observed and isolated in the oviducts of certain avian species. Additionally, LPIAV infection of avian oviduct explants has been demonstrated in chickens, turkeys, and ducks, with susceptibility observed in all oviduct sections, particularly the magnum cells.9 This susceptibility explains the detection of HPIAV in yolk and egg albumen, suggesting a potential role of the reproductive tract in the pathobiology of IAV in avian species. Furthermore, egg-borne influenza viruses not only impact wild and domesticated birds but also pose serious implications for viral dissemination to humans.10,11
Avian IAVs exhibit species-specific differences in the receptor binding specificities of HA. While Sia-α2,3Gal is considered the minimum essential glycan structure for binding, the fine details of HA specificity vary depending on the original host species. These binding specificities of avian IAVs are determined through synthetic glycan library evaluations using microarray or histochemical analyses.6,12,13 Therefore, it is crucial to determine the glycan structures expressed in host cells and tissues, as the receptor binding specificities of HA play a crucial role in determining tissue and species tropism of IAV. Information about the glycan structures on these tissues is valuable for identifying critical structures that IAVs would bind to, highlighting the significance of host cell glycans as natural barriers for transmission between different species.8
Recent studies have shed light on the importance of sulfated-glycan structures on IAV hosts. Notably, the inoculation of IAV into MDCK cells overexpressing sulfotransferase resulted in a 90-fold increase in viral replication. Fuc- and sulfated Sialyl-LacNAc moieties were found to be expressed in chicken embryos, suggesting their involvement in the efficient propagation of human H1N1 in chicken embryos.14,15 These findings underscore the importance of thoroughly evaluating different sulfated-glycan structures on IAV hosts, considering that the receptor binding affinity of each hemagglutinin subtype varies and greatly depends not only on α2,3/6-linked sialosides but also on the underlying glycan structures.13,14,16
Egg white samples offer valuable insights into glycan diversity and their roles in protecting embryos from infections. The structural diversity of egg white glycans is species-specific, resulting from environmental pressures, such as pathogenic invasions. Additionally, since embryos are unable to produce antibodies, immunoglobulins from the hen are transferred into the egg, providing a snapshot of the maternal immune system’s specific defense against pathogens.17 Hence, the diversity of glycans in egg whites can be inferred from the evolutionary history of antipathogenic offense and defense.18 In this study, we conducted a large-scale analysis of avian egg whites from the Order Anseriformes (Waterfowls) using the glycoblotting-based sulphoglycomics approach we previously described. Our findings unveiled a wide range of sulfated and phosphorylated N-glycan species that exhibit distinct expression patterns in waterfowl egg whites. Importantly, we inferred the potential significance of these glycan variations concerning influenza infection prevalence in waterfowl.
Results and Discussion
Sulphoglycomics Revealed Diverse Acidic N-Glycans in Waterfowl Egg Whites
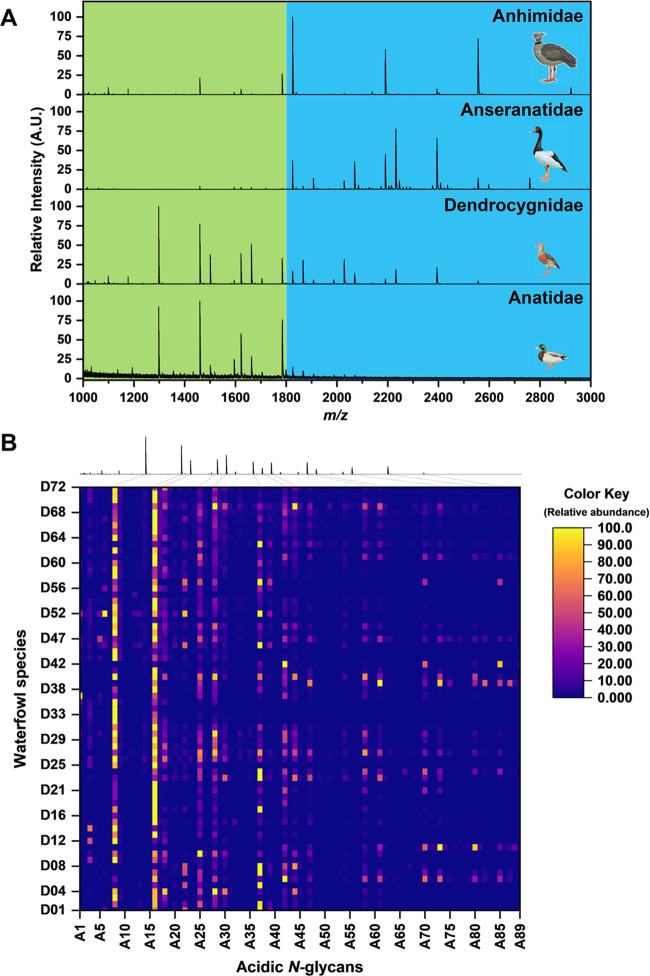
We analyzed egg whites from 72 avian species that belong to the order Anseriformes (waterfowls). The species were classified into different families based on Sibley’s DNA–DNA hybridization, wherein 64 of the species belong to Anatidae, 6 from Dendrocygnidae, 1 from Anhimidae, and 1 from Anseranatidae (Table S1). Acidic N-glycans (i.e., sulfated and phosphorylated) from egg whites were analyzed using the glycoblotting-based sulphoglycomics workflow we described previously.19 Briefly, egg white N-glycans were released using PNGase F, followed by glycan enrichment, methyl esterification, and benzyloxyamine (BOA)-labeling by glycoblotting. BOA-labeled N-glycans were then fractionated by using an amine-functionalized weak anion exchange (WAX) microcolumn. Neutral and monomethylated sialyl N-glycans were eluted first with 1% AcOH in 50% MeCN (neutral fraction), while sulfated and phosphorylated N-glycans were eluted next with 1% NH4OH in 5% MeCN (pH 10.5) (acidic fraction). MALDI-TOF MS analysis of the acidic fractions from the waterfowl egg whites (72 species) detected a total of 89 monoisotopic peaks in negative-ion mode [M – H]− identified as acidic N-glycan peaks (Table S2). However, 89 does not correspond to the number of N-glycan structures due to the presence of structural isomers for every m/z value observed. Figure 1A shows the representative acidic N-glycan profiles of each waterfowl family. Major acidic N-glycans expressed by Anhimidae (Chauna torquata) and Anseranatidae (Anseranas semipalmata) have monoisotopic masses above 1800 m/z. While acidic N-glycans with low m/z values (below 1800 m/z) are observed for Anatidae (Anas platyrhynchos) and Dendrocygnidae (Dendrocygna viduata). The relative abundance of each monoisotopic peak (m/z values) identified as N-glycans expressed in the egg whites of the 72 waterfowl species described in this work are shown as Heat Map in Figure 1B. It can be noticed that two acidic N-glycans (A8 and A16) are highly expressed across all 72 waterfowl species. A8 and A16 have monoisotopic masses of 1297 m/z and 1459 m/z, respectively. A8 (1297 m/z) was inferred to have a glycan composition of HexNAc1Su1 + Hex3GlcNAc2 (complex N-glycan), while A16 (1459 m/z) had Hex1HexNAc1Su1 + Hex3GlcNAc2 (hybrid/complex N-glycan). On the other hand, A37 (1783 m/z) was also noted to be abundant in some species. A37 glycan composition was inferred to be Hex3HexNAc1Su1 + Man3GlcNAc2, multiple hexoses with a single HexNAc residue on its antennae indicate that probably it has a monosulfated hybrid N-glycan structure.
Figure 1.
(A) Representative MALDI-TOF MS acidic N-glycan profiles (in negative-ion mode) of the four waterfowl families (anhimidae, anseranatidae, dendrocygnidae, and anatidae) belonged to the order Anseriformes. (B) Heat map representation of the area normalized acidic N-glycan profiles of the 72 waterfowl species depicting the relative abundance of each m/z value (total 89) identified as N-glycan peaks. MALDI-TOF MS profile of D. viduata is shown on top of the heat map.
MALDI-TOF/TOF MS Analysis of Acidic N-Glycans
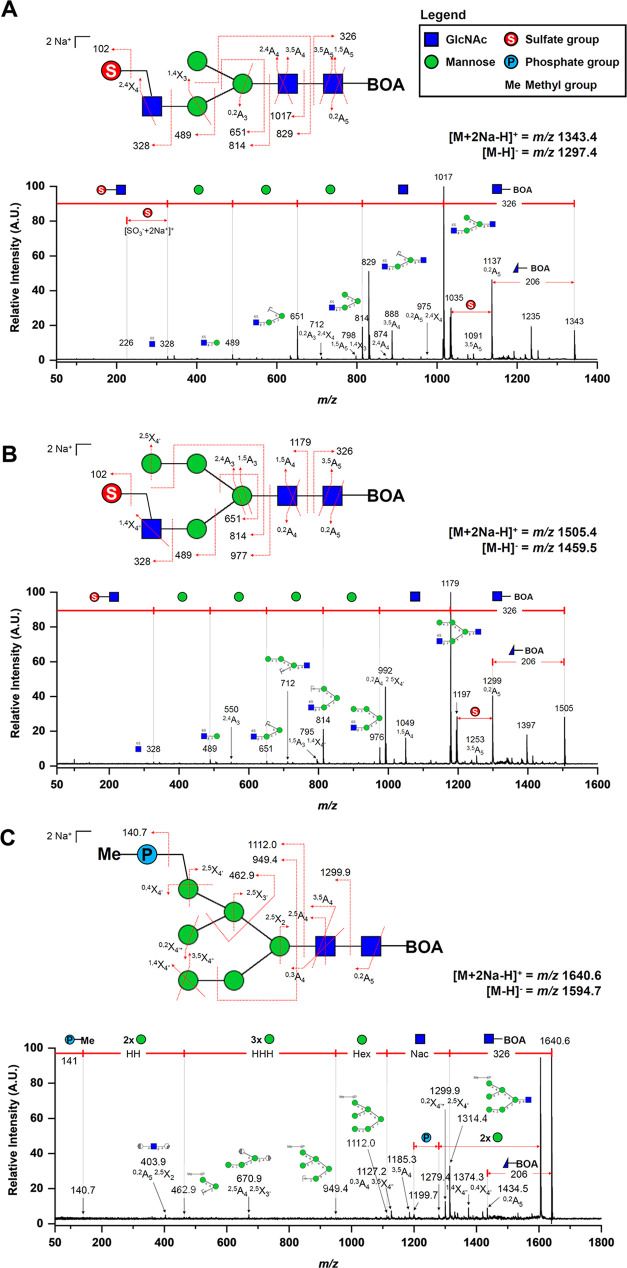
MALDI-TOF/TOF MS analysis of the acidic N-glycan species was performed in positive-ion reflectron mode on their corresponding single-charged disodiated molecular ions [M + 2Na – H]+. The common glycosidic bond cleavages (B and Y ions) and cross-ring cleavages (A and X ions) observed in the TOF/TOF spectra of acidic N-glycans enable an informative sequence assignment of their glycan structures. Diagnostic fragment ions and neutral losses implicating BOA-labeled sulfated N-glycan structures were observed. A neutral loss of 326 m/z was found across all TOF/TOF MS spectra indicating the loss of the reducing end GlcNAc terminus labeled with BOA. Followed by a subsequent neutral loss of another GlcNAc residue (m/z 203) at the N-glycan core structure. Also, a cross-ring fragment (0,2A) of the GlcNAc residue at the reducing end terminus was observed with a concurrent 206 m/z neutral loss. Fragment ions bearing the sulfate moiety were also detected in the TOF/TOF spectra. The fragment ion peak at m/z 328 corresponds to the loss of a monosulfated GlcNAc residue [SO3 + GlcNAc + 2Na]+ at the nonreducing end of the N-glycan structure. Fragment ions at m/z 489, 651, and 814 correspond to [SO3 + GlcNAc + Hex + 2Na]+, [SO3 + GlcNAc + Hex2 + 2Na]+, and [SO3 + GlcNAc + Hex3 + 2Na]+, respectively, indicating the sequential cleavages of the mannose residues of the N-glycan core. Furthermore, a neutral loss of sodium sulfite (m/z 102) occurs readily20 and observed cross-ring fragments (1,4X and 2,4X) of the GlcNAc antennae may suggest that the sulfate group is attached at C6 position (Figure 2A,B).
Figure 2.
MALDI-TOF/TOF MS profiles of selected BOA-labeled acidic N-glycans from waterfowl’s egg whites. TOF/TOF analysis was performed in positive-ion mode on the [M + 2Na – H]+ ion adducts of each N-glycans. (A) TOF/TOF MS profile of a complex N-glycan type—A8 (1297.4 m/z, [M – H]−; 1343.4 m/z, [M + 2Na – H]+). (B) TOF/TOF MS profile of a hybrid N-glycan type—A16 (1459.5 m/z, [M – H]−; 1505.4 m/z, [M + 2Na – H]+), wherein both A8 and A16 were identified to have monosulfation on its terminal GlcNAc. (C) TOF/TOF MS profile of a phosphorylated high-mannose-type N-glycan – A22 (1594.7 m/z, [M – H]−; 1640.6 m/z, [M + 2Na – H]+). Linkage-specific MS/MS structural analysis of each glycan species was not attempted.
Phosphorylated N-glycan structures were also observed on egg whites as previously reported.19Figure 2C shows the MALDI-TOF/TOF MS profile of a monomethylated phosphorylated high mannose (Man6). Similar to the sulfated N-glycans TOF/TOF profiles, a neutral loss of m/z 326 and subsequent neutral loss of the second GlcNAc residue on the N-glycan core were also observed. The cross-ring fragment ion 0,2A was also present. This suggests that these neutral losses (m/z 326, 203, and 206) are characteristics of a BOA-labeled N-glycan TOF/TOF MS fragmentation.18,21 On the other hand, diagnostic fragmentation ions bearing the phosphate group were detected. The molecular ion peak of the monomethylated phosphate group [PO3Me + 2Na]+ was observed at m/z 140.7, while [PO3Me + Hex2 + 2Na]+, [PO3Me + Hex5 + 2Na]+, and [PO3Me + Hex6 + 2Na]+ were also observed at m/z 462.9, 949.4, and 1112.0, respectively. Cross-ring fragment ions 0,4X4′ and 2,5X4′ of the mannose residue at the α1–6 antennae may suggest that the phosphate moiety is attached to the C6 position.
It should be noted that there are a few unassigned fragment ions on each MALDI-TOF/TOF MS spectra. Thus, care must be taken when interpreting these TOF/TOF data due to the possibility that the parent ion is a composite of several isomeric forms of each glycan structure, which are commonly found in biological samples. Lastly, an MS/MS linkage-specific analysis of the glycan structures was not attempted due to the low signal intensities of cross-ring fragments.
Differential Expression Profiles of Acidic N-Glycans in Waterfowls Egg Whites
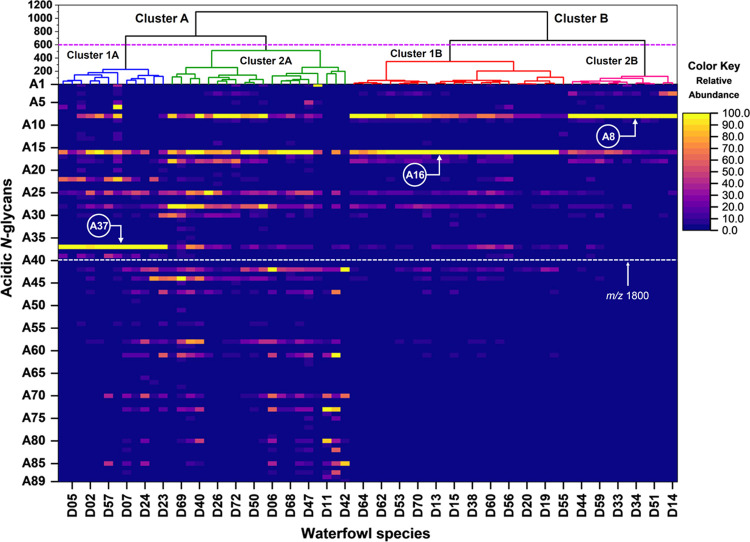
Multivariate analysis was employed to explore and understand the relationships among the Anseriformes species’ different acidic N-glycan expressions of egg whites. Agglomerative hierarchical clustering analysis (HCA) employing the Ward linkage method and Euclidean distance metric was used to group the egg whites from 72 waterfowl species based on the relative abundance of the 89 monoisotopic peaks (acidic N-glycan peaks) (see Table S2). The dendrogram (Figure 3) shows that the 72 waterfowl species are grouped into two major clusters (A and B) based on their acidic N-glycan expressions. Waterfowl species belonging to Cluster A expressed acidic N-glycans having monoisotopic masses above 1800 m/z, whereas Cluster B expressed little to none. Waterfowl species in Cluster B were grouped based on the expressed N-glycans having m/z values below 1800 m/z. These two major clusters were further subgrouped into 4 clusters (Clusters 1A, 2A, 1B, and 2B). Clusters 1A and 2A split from the branch of Cluster A, while Clusters 1B and 2B split from Cluster B. The branch differentiation between Clusters 1A and 2A is due to the expression of the monosulfated hybrid N-glycan (A37) having a monoisotopic mass of 1783 m/z. Clusters 1B and 2B were grouped according to the relative abundances of 1297 m/z (A8) and 1459 m/z (A16) expressions. Interestingly, Anseriformes species belonging to Cluster A is a mixture of the four (4) waterfowl families, while Cluster B was exclusively Anatidae family species.
Figure 3.
Multivariate hierarchical clustering analysis (HCA) of the 72 egg white samples from Anseriformes species. Waterfowl species were clustered according to the differential expression of acidic N-glycan (i.e., sulfated and phosphorylated) expressed on egg whites. Relative abundance of the 89 monoisotopic masses identified as acidic N-glycans on egg white samples are shown as a heat map.
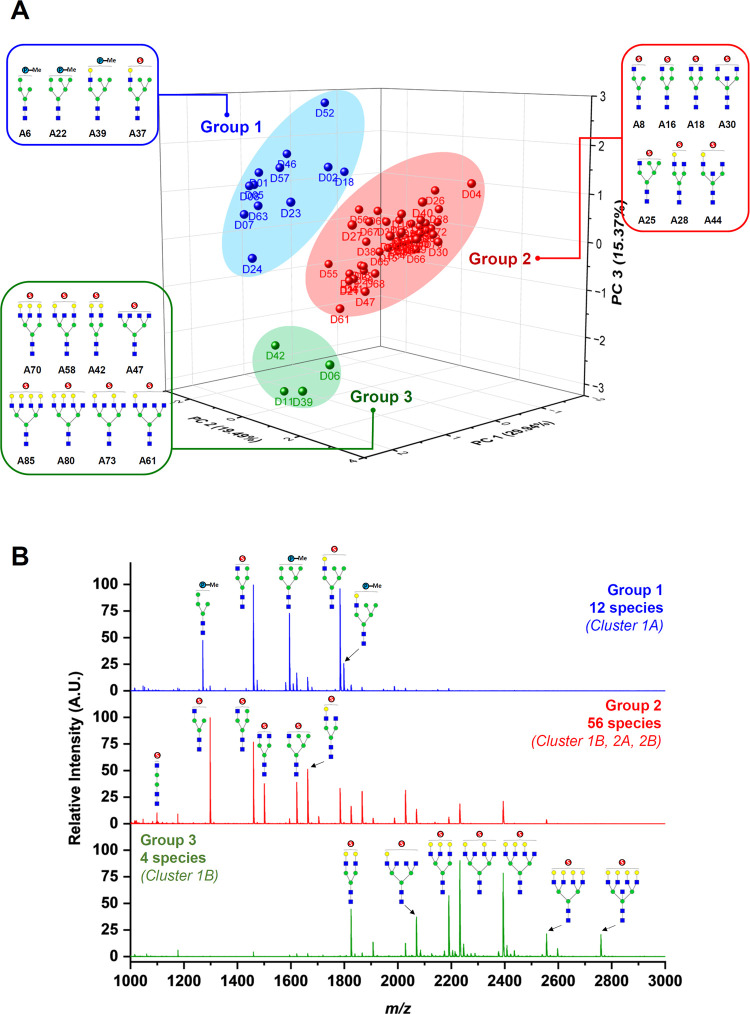
Principal component analysis (PCA) was performed to visualize the variance of the acidic N-glycans expressed on waterfowl egg whites. The PCA is an unsupervised model that finds natural variation in the data without overfitting on new vectors (principal components, PCs). The observations (waterfowl species) are displayed in the score plots, while the variables (acidic N-glycans) are shown as loading plots. The analysis resulted in a PCA model explaining 90.52% of the variance within the data set using 8 principal components. Figure 4A displays the score plot using PC1 (29.9%), PC2 (19.5%), and PC3 (15.4%), which accounts for 64.8% of the total variance between the differential expressions of acidic N-glycans in egg whites from the 72 waterfowl species. The 72 waterfowl species were classified into three major groups, wherein 12 species clustered in Group 1 (Blue), while Group 2 (Red) had 56 species and Group 3 (Green) only had 4 species. Interestingly, waterfowl species belonging to Group 1 are the same species that belonged to Cluster 1A from hierarchical clustering analysis. Group 2 is a mix of species that belong to Clusters 2A, 1B, and 2B. While Group 3 species belonged to a specific branch of Cluster 2A. The group classification of every waterfowl species was due to some specific acidic N-glycans expressed on their respective egg whites. Figure 4B displays a representative MALDI-TOF MS spectrum of Groups 1, 2, and 3, which clearly shows the acidic N-glycans that cause the variation of each group. Phosphorylated high-mannose and hybrid sulfated N-glycans were observed in Group 1. Complex and hybrid sulfated N-glycans without terminal galactose were the abundant glycan structures present in Group 2. Group 3 expressed multiantennary complex N-glycans having terminal galactose on their antennae. It was also noted that some waterfowl species belonging to Group 2 expressed multiantennary complex N-glycans similar to Group 3 but of lower abundance. The list of acidic N-glycan species that gave the variance between each group on PCA analysis is listed in Table S3. Furthermore, each N-glycan structure in Table S3 was classified as trans-Gal(±) based on Hirose et al.’s18 classification of N-glycans expressed in avian egg whites. Accordingly, trans-Gal(+) is an N-glycan structure with terminal galactose, which is abundantly expressed in large waterfowls as hyperbranched structures, while trans-Gal(−) are N-glycans with little to no Gal-modified structures on their reducing terminus, commonly expressed in small waterfowl egg whites.
Figure 4.
(A) Principal component analysis of the acidic N-glycans expressed on egg whites from 72 waterfowl species. The model resulted in 8 principal components (PCs) explaining 90.52% of the variance within the data set. The 72 waterfowl species were also classified into 3 groups based on their acidic N-glycan expressions. (B) Representative MALDI-TOF MS spectra of each group revealed specific acidic N-glycan species that cause the variation between groups.
Phosphorylated N-Glycans Suggest AIV Prevalence in Waterfowl
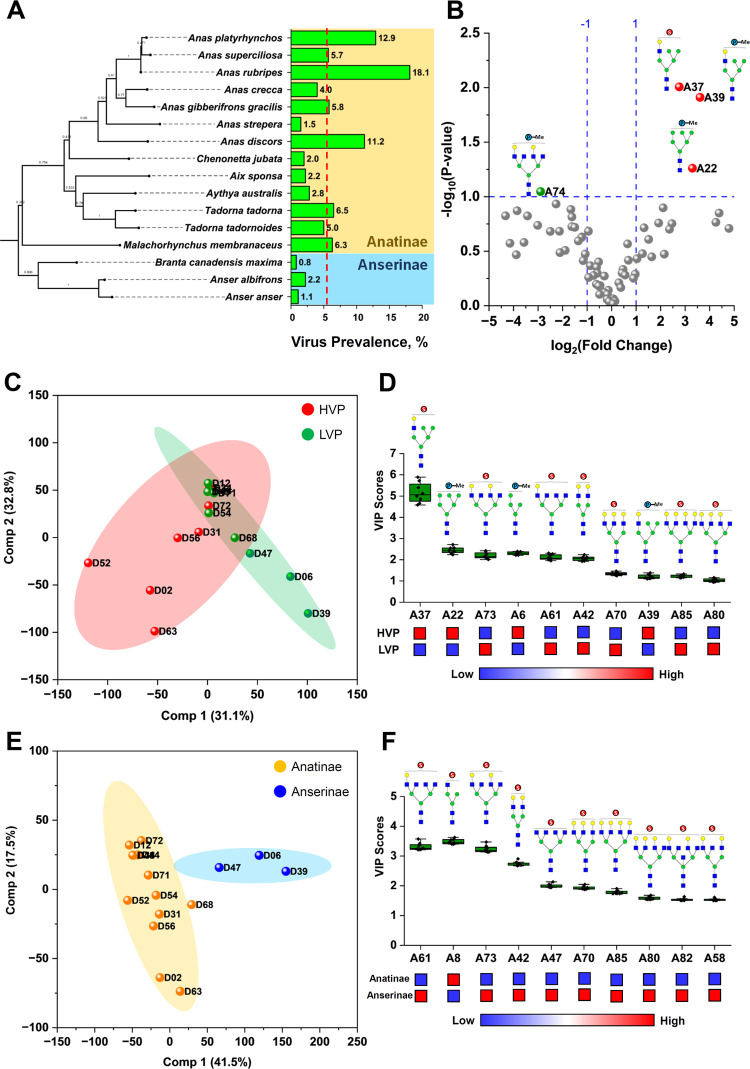
Previous studies revealed that the host phylogeny is a crucial driver in the influenza A virus (IAV) host range, indicating that phylogeny may be an essential factor in host–virus co-evolution, which may explain the variability in the host response to infection.22 Furthermore, species variation in IAV prevalence is often associated with host susceptibility and ecology.23,24 Since glycans are traits that vary between organisms as a direct response to physiological and ecological conditions, their structures are physical records due to genetic and environmental influences.25 Here, we scrutinized the acidic N-glycan expressions of egg whites from waterfowl species with respect to the species’ virus prevalence and lineage. The virus prevalence (VP) data used in this study was taken from the work of Wille et al.22 and Olsen et al.26 Virus prevalence data of Anseriformes species with >50 samples were used. A total of 16 Anatidae species belonging to Anatinae (13) and Anserinae (3) subfamilies were included. The phylogenetic tree of these 16 waterfowl species was then reconstructed based on the mitochondrial Cytochrome b (cty b) gene.27−29 The gene sequences were obtained from NCBI GenBank (Table S4) and then analyzed using Maximum Likelihood method and Tamura–Nei model (TN93 G) in MEGA11.30,31 The tree was rooted using Struthio camelus (ostrich) as an out-group. Waterfowl species were then classified based on their VPs as low viral prevalence (LVP) and high viral prevalence (HVP). Classification was made by taking the average values of the 16 VPs. Based on the average VP (5.50%), species with VPs above 5.50% were classified as HVP, while those below it were classified as LVP (Table S5). It can be clearly inferred from Figure 5A that Anatinae species generally have a higher viral prevalence compared to Anserinae species. Furthermore, Anas rubripes (D34, 18.1%), A. platyrhynchos (D2, 12.9%), and Anas discors (D52, 11.2%) are the top 3 species with HVP and belong to the genus Anas.
Figure 5.
(A) Phylogenetic tree of 16 waterfowl species reconstructed based on the mitochondrial Cytochrome b (cty b) gene using the maximum likelihood method and Tamura–Nei model (TN93 G) in MEGA11. Influenza virus prevalence (VP) of each species is shown as bar graph.22,26 (B) Differential expression of acidic N-glycans between waterfowl species with low viral prevalence (LVP) and high viral prevalence (HVP) is shown as a volcano plot. Partial least-squares discriminant analysis (PLS-DA) of the 16 waterfowl species based on viral prevalence (C, D) and based on their lineage classification (E, F). Variable importance to projection (VIP) scores show important glycoforms that strongly influenced the PLS-DA score plot.
Acidic N-glycan expressions of egg whites from the 16 waterfowl species were further analyzed according to their VP classifications (LVP and HVP). Fold change (FC) analysis of the glycan expression between HVP and LVP groups shown as a volcano plot (Figure 5B) revealed three acidic N-glycans that were highly expressed in egg whites of waterfowl species with high virus prevalence. These differentially expressed N-glycans are 1594 m/z (A22, phosphorylated high-mannose), 1783 m/z (A37, sulfated hybrid N-glycan), and 1797 m/z (A39, phosphorylated hybrid N-glycan). Furthermore, partial least-squares-discriminant analysis (PLS-DA) was used to assess the significant difference of N-glycans expressed between species belonging to HVP and LVP groups. Figure 5C shows the PLS-DA score plot using PC1 (31.1%) and PC2 (32.8%), which explains 63.9% of the total variance between the groups. Variable importance to projection (VIP) scores was also determined to identify the acidic N-glycans that strongly influence the separation of the two groups in the PLS-DA score plot. Acidic N-glycans with VIP scores >1 are shown in Figure 5D. The expression of these acidic N-glycans in waterfowl egg whites provided the discriminating information between HVP and LVP groups. Similar to FC analysis, VIP scores identified m/z 1594, 1783, and 1797 as important discriminating variables for species belonging to HVP. In addition, another phosphorylated high-mannose N-glycan (A6, 1270 m/z) was also identified as an important variable for HVP, while VIP scores identified bi-, tri-, and tetra-antennary sulfated trans-Gal(+) N-glycans as important variables for waterfowl species with low virus prevalence. Interestingly, these sulfated multiantennary trans-Gal(+) N-glycans are identified as characteristic N-glycan structures expressed by waterfowl species belonging to Anserinae subfamily (Figure 5E,F) with a relatively low percentage of viral prevalence (Figure 5A).
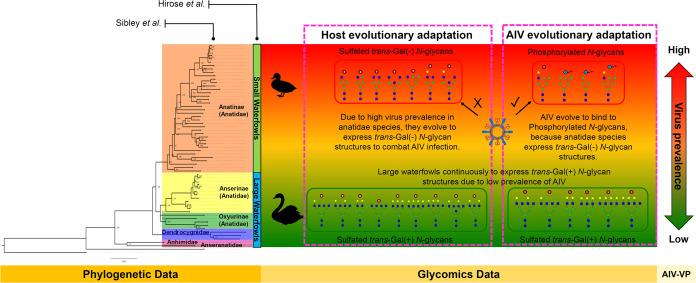
The differential expression of phosphorylated high-mannose and hybrid N-glycan structures suggests that these glycans may serve as potential determinants of the prevalence of influenza A virus in waterfowl species. This observation provides profound insight into the crucial role played by acidic N-glycans (i.e., sulfated and phosphorylated) as receptors for IAV tropism and infection. In Figure 6, when integrated with phylogenetic and virus prevalence information, glycomics data offer additional knowledge of the intricate dynamics of IAV in waterfowl populations.
Figure 6.
Glycomics data complement phylogenetic and virus prevalence data and together provide important insights into the dynamics of IAV in waterfowl species. The phylogenetic tree was reconstructed based on the mitochondrial Cytochrome b (Cty b) gene of 60 Anseriformes species obtained from NCBI GenBank.
Notably, the expression of acidic N-glycans in waterfowl egg whites implies possible aspects of host and virus co-evolution. Waterfowl species with a high virus prevalence probably have evolved to express sulfated trans-Gal(−) N-glycan structures, serving as an adaptive mechanism that helps small waterfowls evade IAV infection. The lack of terminal galactose expression in small waterfowl species may hinder the efficient binding of IAV to sulfated trans-Gal(−) N-glycans, wherein previous research has demonstrated the affinity of influenza virus for sulfated LacNAc glycan epitopes.6,32,6,32 Also, the limited expression of galactosyltransferase (GalT1, GalT2) and the absence of enzymatic activity of GalT3 in the magnum region of avian oviducts result in incomplete galactosylation of egg white proteins, aligning with findings from phylogenomic and morphological studies.18,33 This may underscore the evolutionary necessity of IAV for alternative binding interactions. Consequently, IAV may have adapted to selectively bind to phosphorylated N-glycans, as these glycan structures are predominantly expressed in Anatidae species with high virus prevalence. Furthermore, a similar co-evolutionary pattern has been observed in human IAV, as demonstrated by Byrd-Leotis et al., where IAV exhibits binding affinity to phosphorylated high-mannose N-glycans in human lungs.34,35 This possible co-evolutionary relationship emphasizes the intricate molecular interactions governing viral infection.
In contrast, large waterfowls continuously express sulfated trans-Gal(+) N-glycans, a pattern observed that is attributed to the low virus prevalence of IAV in these species. Notably, the sustained expression of these glycan structures in large waterfowls suggests the presence of a robust defense mechanism. Consequently, these species may mount a more vigorous immune defense compared to smaller waterfowls,7,17,36 given that IAV effectively binds to sulfated trans-Gal(+) N-glycans structures.6,14,32 This effective binding potentially obviates the need for further viral adaptation in large waterfowl populations.
However, care must be taken when interpreting these results because the virus prevalence data used in the study was limited to 16 Anatidae species. Furthermore, the glycomics data was limited to waterfowl species with available VP data. We believe that these findings are significant in understanding the dynamics of IAV infection in waterfowl, but they may not completely reflect the 174 waterfowl species (53 Genera) belonging to the Order Anseriformes. Furthermore, the implications of this work extend beyond avian populations, offering valuable insights into influenza transmission and infection dynamics, potentially affecting humans as well. Especially that overexpression of high-mannose N-glycans in humans was identified as a marker for IAV pathogenesis and infection severity outcome.37 This study may improve surveillance and control strategies by understanding the intricate relationships between viral infection and glycan structures, mitigating the risk of zoonotic transmission from birds to humans.
Finally, the study of acidic N-glycan structures in waterfowl species unravels the important role of acidic N-glycans as determinants of IAV prevalence. Phylogenetic and prevalence data complemented by glycomics data strengthens our grasp of the co-evolutionary adaptations between the virus and its avian hosts, illuminating the molecular intricacies governing IAV dynamics in waterfowl populations.
Conclusion
The glycoblotting-based sulphoglycomics approach revealed a diverse array of sulfated and phosphorylated N-glycans in waterfowl egg whites, providing meaningful insights into influenza A virus (IAV) dynamics. Distinct variations in acidic N-glycan expressions were observed among the four families (Anhimidae, Anseranatidae, Dendrocygnidae, and Anatinae) within the order Anseriformes. Waterfowl species were differentiated based on their expressions of sulfated trans-Gal(+) and trans-Gal(−) N-glycan structures, as well as phosphorylated N-glycans. Moreover, phosphorylated hybrid and high-mannose N-glycans were found to be highly expressed in the egg whites of waterfowl species with a high prevalence of the virus. These findings underscore the importance of phosphorylated N-glycans, in addition to sulfated N-glycans, in comprehending the dynamics of the IAV in waterfowl species. Understanding these glycan structures provides valuable insights into the factors influencing the transmission and evolution of IAV within avian populations.
Materials and Methods
Egg Whites
Egg whites from various species of Order Anseriformes (4 families, 27 genera, 66 species) were collected by Laskowski Jr.,38−40 and were maintained at −20 °C. The scientific names of the birds were adapted from Sibley and Monroe. The DNA–DNA hybridization of Sibley et al. was used as our primary reference because their classification of birds worldwide is complementary to the phylogenetic analysis.41,42
Materials
Peptide N-glycosidase F (PNGase F) was acquired from New England BioLabs (Ipswich, MA), proteinase K was from Roche (Germany), trypsin was from Sigma-Aldrich Corp. (St. Loius, MO), and the bacterial alkaline phosphatase (BAP) was from Nippon Gene, Ltd. (Tokyo, Japan). Ammonium carbamate, benzyloxyamine hydrochloride (BOA), 3-methyl-1-p-tolyltriazene (MTT), disialyloctasaccharide (SGP-10), hexa-N-acetylchitohexaose, 2,5-dihydroxybenzoic acid (DHB), sodium bicarbonate (NaHCO3), 3,4-diaminobenzophenone (DABP), and trifluoroacetic acid (TFA) were obtained from Tokyo Chemical Industry Co. (Tokyo, Japan). BlotGlycoH bead was acquired from Sumitomo Bakelite, Co., Ltd. (Tokyo, Japan).
N-Glycan Release18,21
Lyophilized egg whites (approximately 1 mg) were dissolved in 50 μL of 200 mM NH4HCO3, followed by the addition of 4 μL of denaturation buffer (5% SDS, 0.4 M DTT). The mixture was denatured for 10 min at 100 °C. After denaturation, 10 μL of 123 mM iodoacetamide was added to the mix and incubated in the dark at room temperature for 1 h. Tryptic digestion was achieved by adding 10 μL of 40 U/μL sequence-grade Trypsin (Sigma-Aldrich) in 1 mM HCl; the mixture was then incubated overnight at 37 °C, followed by heat inactivation of the enzyme at 90 °C for 10 min. The tryptic digest was allowed to cool at room temperature, then 8 μL of reaction buffer (0.5 M Na3PO4, pH 7.5), 8 μL of 10% NP-40, and 2 μL of 5 U/μL PNGase F (New England BioLabs) were added, and the mixture was incubated overnight at 37 °C. The mixture was further digested with 10 μL of 0.5 U/μL Proteinase K (Roche, Germany) at 37 °C for 3 h, followed by heat inactivation of the enzyme at 90 °C for 10 min. The sample was dried in a SpeedVac and stored at −20 °C until use.
Glycan Enrichment Using Glycoblotting18,21,43
A 250 μL aliquot of a 10 mg/mL BlotGlycoH bead (Sumitomo Bakelite, Co.) suspension was dispensed into each well of the 96-well multiScreen Solvinert filter plate (Millipore, Billerica, MA). The filter plate was then attached to a vacuum manifold to remove water. The dried sample containing released N-glycans from egg whites was reconstituted with 40 μL of Milli-Q water. A 20 μL aliquot of the reconstituted sample was added into the wells with 10 μL of 100 μM disialyloctasaccharide, SGP-10 (Tokyo Chemical Industry Co., Ltd.) internal standard, and 180 μL of 2% AcOH in acetonitrile (MeCN). The 96-well filter plate was incubated at 80 °C for 45 min until dry. Each sample well was washed with 200 μL of 2 M guanidine-HCl in 16 mM NH4HCO3, water, and 1% triethylamine in methanol (MeOH) sequentially. Each solvent washing was performed twice and vacuumed after each washing step. Unreacted hydrazide functional groups on the beads were capped with an acetyl group by incubating each sample well with 100 μL of 10% acetic anhydride in MeOH for 30 min at room temperature. The capping solution was then removed by vacuum and sequentially washed twice with 200 μL of 10 mM HCl, MeOH, and dioxane. On-bead methyl esterification of the carboxyl groups of acidic glycans (e.g., sialic acid) was performed by adding 100 μL of 100 mM 3-methyl-1-p-tolyltriazene (MTT) in dioxane into the sample and incubated at 60 °C for 90 min until dry.44 The 96-well plate was washed twice with 200 μL of dioxane, water, MeOH, and water sequentially. The captured glycans on the BlotGlycoH beads were labeled with benzyloxyamine (BOA) via trans-iminization reaction. The labeling was performed by adding 20 μL of 50 mM BOA-HCl and 180 μL of 2% AcOH in MeCN at 80 °C for 45 min. BOA-labeled glycans were eluted with 150 μL of water twice. The sample was dried in a SpeedVac and stored at −20 °C until use.
Anionic-Glycan Separation Using WAX20,45
50 μL of 100 mg/mL 3-aminopropyl silica gel suspension (1 mmol/mg, Tokyo Chemical Industry Co. Ltd.) was packed into a 200 μL micropipette tip with a cotton plug. The packed weak anion exchange (WAX) microcolumn was conditioned and washed sequentially with 100 μL of water, MeCN, and 1% AcOH in 95% MeCN twice. After every conditioning and washing step, the column was centrifugated at 500 rpm for 2 min. BOA-labeled N-glycans were reconstituted with 20 μL of water. A 5 μL sample aliquot was dissolved in 150 μL of 1% AcOH in 95% MeCN and then loaded into the column. The sample was allowed to elute by gravity, and the collected eluate was reloaded back into the column; this step was repeated three times. The column was washed with 1% AcOH in 95% MeCN to remove unbound and hydrophobic contaminants. BOA-labeled neutral and monomethylated sialyl N-glycans were eluted with 1% AcOH in 50% MeCN (neutral N-glycan fraction), while BOA-labeled sulfated and phosphorylated N-glycans were eluted with 1% NH4OH in 5% MeCN (pH 10.5) (acidic N-glycan fraction). The eluates were then dried in a SpeedVac and stored at −20 °C until use.
Mass Spectrometric Analysis
MALDI-TOF MS analysis of BOA-labeled N-glycans was performed using Ultraflex III (Bruker, Bremen, Germany) operated on reflectron mode on both positive- and negative-ion acquisition mode. Neutral and monomethylated sialyl-glycans were analyzed in positive-ion mode using 10 mg/mL DHB/NaHCO3 (10:1) in 50% MeCN matrix.46,47 While sulfated glycans were analyzed in negative-ion mode using the DABP matrix (3,4-diaminobenzophenone, 10 mg/mL in 75% MeCN with 0.1% TFA).20,45,48,49 The MALDI-TOF and MALDI-TOF/TOF MS data were annotated using the Bruker FlexAnalysis 3.0 software package. Experimental m/z values were used to predict possible glycan composition using the Expasy GlycoMod Tool and Glyconnect Database of the Swiss Institute of Bioinformatics (https://web.expasy.org/glycomod/) and GlycoWorkbench.50,51
Statistical Analysis
Multivariate analysis was performed by using OriginPro statistical software (OriginLab Corp.). The N-glycan peaks detected on MALDI-TOF MS spectra were picked and annotated using FlexAnalysis software (Bruker Daltonics). The peak area of each N-glycan monoisotopic peak was normalized to the area of the most abundant N-glycan in the MS spectra, and the peak areas were then expressed as relative abundance. MS data sets used for the subsequent multivariate analyses contain the relative abundance of monoisotopic peaks (total of 89 m/z) identified as N-glycan peaks from each egg white of 72 waterfowl species. Hierarchical clustering analysis (HCA) was performed to investigate the similarity of the glycan expression profiles based on statistical distance. Principal component analysis (PCA) was also used to explore the glycan profiles based on their variation-covariance (information on each glycan); score plots are provided for visual inspection of the relationships of principal components. While MetaboAnalyst ver. 5.0 was used to perform fold change analysis (FC), partial least-squares-discriminant analysis (PLS-DA) and determined variable importance to projection (VIP) scores to identify N-glycan species that strongly influence the group separation in the PLS-DA score plot.
Acknowledgments
The authors sincerely acknowledge Dr. Y. C. Lee and the late Dr. M. Lascowsky, Jr., for providing the invaluable egg white collection. This research was supported by A-STEP (JPMJTM20JB and JPMJTM20A7) from the Japan Science and Technology Agency (JST) and a Grant-in-Aid for Scientific Research (B: 22H02191) from the Ministry of Education, Culture, Sports, Sciences, and Technology of Japan.
Glossary
Abbreviations
- BOA
benzyloxyamine
- DABP
3,4-diaminobenzophenone
- MTT
3-methyl-1-p-tolyltriazene
- LVP
low viral prevalence
- HVP
high viral prevalence
- VIP
variable importance to projection
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsinfecdis.3c00520.
List of egg whites from various species of Order Anseriformes used in this study (Table S1); list of 89 acidic N-glycans identified from the egg whites of 72 waterfowl species (Table S2); list of acidic N-glycans that gave the variation between groups in PCA (Table S3); GenBank accession numbers for various genes of the 72 Anseriformes species in this study (Table S4); waterfowl classification based on their virus prevalence (Table S5); and evolutionary history was inferred by using the maximum likelihood method and Tamura–Nei model (Figures S1 and S2) (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Sakai T.; Nishimura S. I.; Naito T.; Saito M. Influenza A Virus Hemagglutinin and Neuraminidase Act as Novel Motile Machinery. Sci. Rep. 2017, 7, 45043 10.1038/srep45043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horimoto T.; Kawaoka Y. Influenza: Lessons from Past Pandemics, Warnings from Current Incidents. Nat. Rev. Microbiol. 2005, 3, 591–600. 10.1038/nrmicro1208. [DOI] [PubMed] [Google Scholar]
- Broszeit F.; van Beek R. J.; Unione L.; Bestebroer T. M.; Chapla D.; Yang J. Y.; Moremen K. W.; Herfst S.; Fouchier R. A. M.; de Vries R. P.; Boons G. J. Glycan Remodeled Erythrocytes Facilitate Antigenic Characterization of Recent A/H3N2 Influenza Viruses. Nat. Commun. 2021, 12 (1), 5449 10.1038/s41467-021-25713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N.; Abe T.; Natsuka S. Structural Analysis of N-Glycans in Chicken Trachea and Lung Reveals Potential Receptors of Chicken Influenza Viruses. Sci. Rep. 2022, 12 (1), 2081 10.1038/s41598-022-05961-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C.; Pu J. Influence of Host Sialic Acid Receptors Structure on the Host Specificity of Influenza Viruses. Viruses 2022, 14, 2141 10.3390/v14102141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi D.; Hiono T.; Ichii O.; Nishihara S.; Takase-Yoden S.; Yamamoto K.; Kawashima H.; Isoda N.; Sakoda Y. Turkeys Possess Diverse Siaα2–3Gal Glycans That Facilitate Their Dual Susceptibility to Avian Influenza Viruses Isolated from Ducks and Chickens. Virus Res. 2022, 315, 198771. 10.1016/j.virusres.2022.198771. [DOI] [PubMed] [Google Scholar]
- Yang J.; Cui H.; Teng Q.; Ma W.; Li X.; Wang B.; Yan D.; Chen H.; Liu Q.; Li Z. Ducks Induce Rapid and Robust Antibody Responses than Chickens at Early Time after Intravenous Infection with H9N2 Avian Influenza Virus. Virol. J. 2019, 16 (1), 46 10.1186/s12985-019-1150-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreman S.; Bergervoet S. A.; Zwart R.; Stockhofe-Zurwieden N.; Beerens N. Tissue Tropism and Pathology of Highly Pathogenic Avian Influenza H5N6 Virus in Chickens and Pekin Ducks. Res. Vet. Sci. 2022, 146, 1–4. 10.1016/j.rvsc.2022.03.010. [DOI] [PubMed] [Google Scholar]
- Sid H.; Hartmann S.; Winter C.; Rautenschlein S. Interaction of Influenza A Viruses with Oviduct Explants of Different Avian Species. Front. Microbiol. 2017, 8, 1338. 10.3389/fmicb.2017.01338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai S. P. S.; Saif Y. M.; Lee C. W. Detection of Influenza A Viruses in Eggs Laid by Infected Turkeys. Avian Dis. 2010, 54, 830–833. 10.1637/9102-101209-Reg.1. [DOI] [PubMed] [Google Scholar]
- Uchida Y.; Takemae N.; Tanikawa T.; Kanehira K.; Saito T. Transmission of an H5N8-Subtype Highly Pathogenic Avian Influenza Virus from Infected Hens to Laid Eggs. Avian Dis. 2016, 60 (2), 450–453. 10.1637/11312-110315-Reg. [DOI] [PubMed] [Google Scholar]
- Hiono T.; Okamatsu M.; Nishihara S.; Takase-Yoden S.; Sakoda Y.; Kida H. A Chicken Influenza Virus Recognizes Fucosylated A2,3 Sialoglycan Receptors on the Epithelial Cells Lining Upper Respiratory Tracts of Chickens. Virology 2014, 456–457 (1), 131–138. 10.1016/j.virol.2014.03.004. [DOI] [PubMed] [Google Scholar]
- Gambaryan A.; Yamnikova S.; Lvov D.; Tuzikov A.; Chinarev A.; Pazynina G.; Webster R.; Matrosovich M.; Bovin N. Receptor Specificity of Influenza Viruses from Birds and Mammals: New Data on Involvement of the Inner Fragments of the Carbohydrate Chain. Virology 2005, 334 (2), 276–283. 10.1016/j.virol.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Ichimiya T.; Okamatsu M.; Kinoshita T.; Kobayashi D.; Ichii O.; Yamamoto N.; Sakoda Y.; Kida H.; Kawashima H.; Yamamoto K.; Takase-Yoden S.; Nishihara S. Sulfated Glycans Containing NeuAcα2–3Gal Facilitate the Propagation of Human H1N1 Influenza A Viruses in Eggs. Virology 2021, 562, 29–39. 10.1016/j.virol.2021.06.008. [DOI] [PubMed] [Google Scholar]
- Ichimiya T.; Nishihara S.; Takase-Yoden S.; Kida H.; Aoki-Kinoshita K. Frequent Glycan Structure Mining of Influenza Virus Data Revealed a Sulfated Glycan Motif That Increased Viral Infection. Bioinformatics 2014, 30 (5), 706–711. 10.1093/bioinformatics/btt573. [DOI] [PubMed] [Google Scholar]
- Broszeit F.; Tzarum N.; Zhu X.; Nemanichvili N.; Eggink D.; Leenders T.; Li Z.; Liu L.; Wolfert M. A.; Papanikolaou A.; Martínez-Romero C.; Gagarinov I. A.; Yu W.; García-Sastre A.; Wennekes T.; Okamatsu M.; Verheije M. H.; Wilson I. A.; Boons G. J.; de Vries R. P. N-Glycolylneuraminic Acid as a Receptor for Influenza A Viruses. Cell Rep. 2019, 27 (11), 3284–3294. 10.1016/j.celrep.2019.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hincke M. T.; Da Silva M.; Guyot N.; Gautron J.; McKee M. D.; Guabiraba-Brito R.; Réhault-Godbert S. Dynamics of Structural Barriers and Innate Immune Components during Incubation of the Avian Egg: Critical Interplay between Autonomous Embryonic Development and Maternal Anticipation. J. Innate Immun. 2019, 11, 111–124. 10.1159/000493719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose K.; Amano M.; Hashimoto R.; Lee Y. C.; Nishimura S. I. Insight into Glycan Diversity and Evolutionary Lineage Based on Comparative Avio-N-Glycomics and Sialic Acid Analysis of 88 Egg Whites of Galloanserae. Biochemistry 2011, 50 (21), 4757–4774. 10.1021/bi101940x. [DOI] [PubMed] [Google Scholar]
- Montalban B. M.; Hinou H. Glycoblotting Enables Seamless and Straightforward Workflow for MALDI-TOF/MS-Based Sulphoglycomics of N- and O-Glycans. Proteomics 2023, 23, e2300012 10.1002/pmic.202300012. [DOI] [PubMed] [Google Scholar]
- Yu S. Y.; Wu S. W.; Hsiao H. H.; Khoo K. H. Enabling Techniques and Strategic Workflow for Sulfoglycomics Based on Mass Spectrometry Mapping and Sequencing of Permethylated Sulfated Glycans. Glycobiology 2009, 19 (10), 1136–1149. 10.1093/glycob/cwp113. [DOI] [PubMed] [Google Scholar]
- Sanes J. T.; Hinou H.; Lee Y. C.; Nishimura S. I. Glycoblotting of Egg White Reveals Diverse N-Glycan Expression in Quail Species. J. Agric. Food Chem. 2019, 67 (1), 531–540. 10.1021/acs.jafc.8b04782. [DOI] [PubMed] [Google Scholar]
- Wille M.; Lisovski S.; Roshier D.; Ferenczi M.; Hoye B. J.; Leen T.; Warner S.; Fouchier R. A. M.; Hurt A. C.; Holmes E. C.; Klaassen M. Strong Host Phylogenetic and Ecological Effects on Host Competency for Avian Influenza in Australian Wild Birds. Proc. R. Soc. B: Biol. Sci. 2023, 290 (1991), 2237. 10.1098/rspb.2022.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk J. G.; Verhagen J. H.; Wille M.; Waldenström J. Host and Virus Ecology as Determinants of Influenza A Virus Transmission in Wild Birds. Curr. Opin. Virol. 2018, 28, 26–36. 10.1016/j.coviro.2017.10.006. [DOI] [PubMed] [Google Scholar]
- Garamszegi L. Z.; Møller A. P. Prevalence of Avian Influenza and Host Ecology. Proc. R. Soc. B: Biol. Sci. 2007, 274 (1621), 2003–2012. 10.1098/rspb.2007.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer S. A.; Gagneux P. Glycomics: Revealing the Dynamic Ecology and Evolution of Sugar Molecules. J. Proteomics 2016, 135, 90–100. 10.1016/j.jprot.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen B.; Munster V. J.; Wallensten A.; Waldenström J.; Osterhaus A. D. M. E.; Fouchier R. A. M. Global Patterns of Influenza A Virus in Wild Birds. Science 2006, 312 (5772), 384–388. 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- Gonzalez J.; Düttmann H.; Wink M. Phylogenetic Relationships Based on Two Mitochondrial Genes and Hybridization Patterns in Anatidae. J. Zool. 2009, 279 (3), 310–318. 10.1111/j.1469-7998.2009.00622.x. [DOI] [Google Scholar]
- Donne-Goussé C.; Laudet V.; Hanni C. A Molecular Phylogeny of Anseriformes Based on Mitochondrial DNA Analysis. Mol. Phylogenet. Evol. 2002, 23, 339–356. 10.1016/S1055-7903(02)00019-2. [DOI] [PubMed] [Google Scholar]
- Sun Z.; Pan T.; Hu C.; Sun L.; Ding H.; Wang H.; Zhang C.; Jin H.; Chang Q.; Kan X.; Zhang B. Rapid and Recent Diversification Patterns in Anseriformes Birds: Inferred from Molecular Phylogeny and Diversification Analyses. PLoS One 2017, 12 (9), e0184529 10.1371/journal.pone.0184529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K.; Nei M. Estimation of the Number of Nucleotide Substitutions in the Control Region of Mitochondrial DNA in Humans and Chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Tamura K.; Stecher G.; Kumar S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38 (7), 3022–3027. 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriwilaijaroen N.; Kondo S.; Yagi H.; Wilairat P.; Hiramatsu H.; Ito M.; Ito Y.; Kato K.; Suzuki Y. Analysis of N-Glycans in Embryonated Chicken Egg Chorioallantoic and Amniotic Cells Responsible for Binding and Adaptation of Human and Avian Influenza Viruses. Glycoconjugate J. 2009, 26 (4), 433–443. 10.1007/s10719-008-9193-x. [DOI] [PubMed] [Google Scholar]
- Mizutani A.; Tsunashima H.; Nishijima K.; Sasamoto T.; Yamada Y.; Kojima Y.; Motono M.; Kojima J.; Inayoshi Y.; Miyake K.; Park E. Y.; Iijima S. Genetic Modification of a Chicken Expression System for the Galactosylation of Therapeutic Proteins Produced in Egg White. Transgenic Res. 2012, 21 (1), 63–75. 10.1007/s11248-011-9511-0. [DOI] [PubMed] [Google Scholar]
- Byrd-Leotis L.; Jia N.; Dutta S.; Trost J. F.; Gao C.; Cummings S. F.; Braulke T.; Müller-Loennies S.; Heimburg-Molinaro J.; Steinhauer D. A.; Cummings R. D. Influenza Binds Phosphorylated Glycans from Human Lung. Sci. Adv. 2019, 5 (2), eaav2554 10.1126/sciadv.aav2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia N.; Byrd-Leotis L.; Matsumoto Y.; Gao C.; Wein A. N.; Lobby J. L.; Kohlmeier J. E.; Steinhauer D. A.; Cummings R. D. The Human Lung Glycome Reveals Novel Glycan Ligands for Influenza A Virus. Sci. Rep. 2020, 10 (1), 5320 10.1038/s41598-020-62074-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris K. M.; Mishra A.; Raut A. A.; Gaunt E. R.; Borowska D.; Kuo R. I.; Wang B.; Vijayakumar P.; Chingtham S.; Dutta R.; Baillie K.; Digard P.; Vervelde L.; Burt D. W.; Smith J. The Molecular Basis of Differential Host Responses to Avian Influenza Viruses in Avian Species with Differing Susceptibility. Front. Cell. Infect. Microbiol. 2023, 13, 1067993 10.3389/fcimb.2023.1067993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heindel D. W.; Koppolu S.; Zhang Y.; Kasper B.; Meche L.; Vaiana C. A.; Bissel S. J.; Carter C. E.; Kelvin A. A.; Elaish M.; Lopez-Orozco J.; Zhang B.; Zhou B.; Chou T. W.; Lashua L.; Hobman T. C.; Ross T. M.; Ghedin E.; Mahal L. K. Glycomic Analysis of Host Response Reveals High Mannose as a Key Mediator of Influenza Severity. Proc. Natl. Acad. Sci. U.S.A. 2020, 117 (43), 26926–26935. 10.1073/pnas.2008203117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski M. Jr.; Kato I.; Ardelt W.; Cook J.; Denton A.; Empie M. W.; Kohr W. J.; Park J.; Parks K.; Schatzley B. L.; Schoenberger O. L.; Tashiro M.; Vichot G.; Whatley H. E.; Wieczorek A.; Wieczorek M. Ovomucoid Third Domains from 100 Avian Species: Isolation, Sequences, and Hypervariability of Enzyme-Inhibitor Contact Residues. Biochemistry 1987, 26, 202–221. 10.1021/bi00375a028. [DOI] [PubMed] [Google Scholar]
- Laskowski M. Jr.; Apostol I.; Ardelt W.; Cook J.; Giletto A.; Kelly C. A.; Lu W.; Park S. J.; Qasim M. A.; Whatley H. E.; Wieczorek A.; Wynn R. Amino Acid Sequences of Ovomucoid Third Domain from 25 Additional Species of Birds. J. Protein Chem. 1990, 9 (6), 715. 10.1007/BF01024766. [DOI] [PubMed] [Google Scholar]
- Apostol I.; Giletto A.; Komiyama T.; Zhang W.; Laskowski M. Jr. Amino Acid Sequences of Ovomucoid Third Domains from 27 Additional Species of Birds. J. Protein Chem. 1993, 12 (4), 419. 10.1007/BF01025042. [DOI] [PubMed] [Google Scholar]
- Sibley C. G.; Monroe B. L.. Distribution and Taxonomy of Birds of the World; Yale University Press: New Haven, CT, 1990. [Google Scholar]
- Sibley C. G.; Monroe B. L.. A Supplement to Distribution and Taxonomy of Birds of the Worlds; Yale University Press: New Haven, CT, 1993. [Google Scholar]
- Gizaw S. T.; Ohashi T.; Tanaka M.; Hinou H.; Nishimura S. I. Glycoblotting Method Allows for Rapid and Efficient Glycome Profiling of Human Alzheimer’s Disease Brain, Serum and Cerebrospinal Fluid towards Potential Biomarker Discovery. Biochim. Biophys. Acta 2016, 1860 (8), 1716–1727. 10.1016/j.bbagen.2016.03.009. [DOI] [PubMed] [Google Scholar]
- Miura Y.; Shinohara Y.; Furukawa J. I.; Nagahori N.; Nishimura S. I. Rapid and Simple Solid-Phase Esterification of Sialic Acid Residues for Quantitative Glycomics by Mass Spectrometry. Chem. – Eur. J. 2007, 13 (17), 4797–4804. 10.1002/chem.200601872. [DOI] [PubMed] [Google Scholar]
- Yu S.-Y.; Snovida S.; Khoo K.-H. Permethylation and Microfractionation of Sulfated Glycans for MS Analysis. Bio-Protocol 2020, 10 (10), e3617 10.21769/BioProtoc.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinou H. Aniline Derivative/DHB/Alkali Metal Matrices for Reflectron Mode MALDI-TOF and TOF/TOF MS Analysis of Unmodified Sialylated Oligosaccharides and Glycopeptides. Int. J. Mass Spectrom. 2019, 443, 109–115. 10.1016/j.ijms.2019.06.006. [DOI] [Google Scholar]
- Barada E.; Hinou H. BOA/DHB/Na: An Efficient UV-MALDI Matrix for High-Sensitivity and Auto-Tagging Glycomics. Int. J. Mol. Sci. 2022, 23 (20), 12510. 10.3390/ijms232012510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. Y.; Huang H. H.; Yu S. Y.; Wu S. J.; Kannagi R.; Khoo K. H. Concerted Mass Spectrometry-Based Glycomic Approach for Precision Mapping of Sulfo Sialylated N-Glycans on Human Peripheral Blood Mononuclear Cells and Lymphocytes. Glycobiology 2018, 28 (1), 9–20. 10.1093/glycob/cwx091. [DOI] [PubMed] [Google Scholar]
- Kuo C. W.; Guu S. Y.; Khoo K. H. Distinctive and Complementary MS 2 Fragmentation Characteristics for Identification of Sulfated Sialylated N-Glycopeptides by NanoLC-MS/MS Workflow. J. Am. Soc. Mass Spectrom. 2018, 29 (6), 1166–1178. 10.1007/s13361-018-1919-9. [DOI] [PubMed] [Google Scholar]
- Ceroni A.; Maass K.; Geyer H.; Geyer R.; Dell A.; Haslam S. M. GlycoWorkbench: A Tool for the Computer-Assisted Annotation of Mass Spectra of Glycans. J. Proteome Res. 2008, 7 (4), 1650–1659. 10.1021/pr7008252. [DOI] [PubMed] [Google Scholar]
- Cooper C. A.; Gasteiger E.; Packer N. H. GlycoMod-A Software Tool for Determining Glycosylation Compositions from Mass Spectro-Metric Data. Proteomics 2001, 1, 340–349. . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.