Abstract
As current chemo- and photodynamic cancer therapies are associated with severe side effects due to a lack of specificity and to systemic toxicity, innovative solutions in terms of targeting and controlled functionality are in high demand. Here, we present the development of a polymersome nanocarrier equipped with targeting molecules and loaded with photosensitizers for efficient uptake and light-activated cell killing. Polymersomes were self-assembled in the presence of photosensitizers from a mixture of nonfunctionalized and functionalized PDMS-b-PMOXA diblock copolymers, the latter designed for coupling with targeting ligands. By encapsulation inside the polymersomes, the photosensitizer Rose Bengal was protected, and its uptake into cells was mediated by the nanocarrier. Inhibitor of fibroblast activation protein α (FAPi), a ligand for FAP, was attached to the polymersomes’ surface and improved their uptake in MCF-7 breast cancer cells expressing relatively high levels of FAP on their surface. Once internalized by MCF-7, irradiation of Rose Bengal-loaded FAPi-polymersomes generated reactive oxygen species at levels high enough to induce cell death. By combining photosensitizer encapsulation and specific targeting, polymersomes represent ideal candidates as therapeutic nanocarriers in cancer treatment.
Introduction
Chemotherapy, besides many benefits,1 still displays numerous side effects, including systemic toxicity and the inhibition of fast-growing cells and tissues such as hair follicles, gastrointestinal tract cells, and bone marrow.2 One key factor in causing side effects is the unspecific internalization by noncancerous cells and tissues.3 In this respect, the encapsulation or entrapment of therapeutic compounds inside nanocarriers is essential to avoid their intrinsic toxicity for cells and tissues other than the tumor site. Because cancer cells often become nonresponsive to chemotherapy, photodynamic therapy (PDT) has emerged as an alternative and/or complementary treatment modality for various malignant neoplasia and tumors.4,5 In PDT, photosensitizers (PSs) excited by light of an appropriate wavelength in the presence of molecular oxygen produce reactive oxygen species (ROS) to induce a tumor cell’s death.6 As PSs themselves are often cytotoxic, by encapsulating them inside nanocarriers, the side effects associated with PDT are diminished, and at the same time, the PS is protected from premature degradation.7,8 Various nanoassemblies, including liposomes,9,10 nanoparticles,11 micelles,12 and polymersomes,13 have been used as delivery systems that shield and release PS. A significant improvement was achieved by encapsulation of PS inside oxygen permeable polymersomes, where irradiation led to enhanced ROS production without releasing the PS.7 The light-activated ROS production by per se nontoxic polymersomes was sufficient to induce cell death under PDT conditions. However, the unspecific uptake of such nanocarriers decreases their efficacy at the tumor site.
Targeting represents a key approach to improve the localization of the nanocarriers at the desired site, resulting in an increase of the PDT efficacy.14−17 For example, core–shell polymeric nanoparticles with PS-conjugated hyaluronic acid shell produced for targeted PDT showed excellent singlet oxygen elevation in breast cancer cells and significant in vivo antitumor activity.14 Nevertheless, to unlock the full potential of PDT there is a need for targeting nanoplatforms that promote the desired, specific cellular uptake. While targeting of polymersomes after functionalization with antibodies,18 peptides,19 small molecules,20 aptamers,21 and carbohydrates22 has been applied in different cancer models and inflammatory tissues,23 there is still a large variety of PSs, cells, and conditions to be explored to increase the efficacy of PDT. Of particular interest for targeting is the fibroblast activation protein alpha (FAP), a type II transmembrane serine protease expressed almost exclusively in pathological conditions such as cancer. While the mechanisms underlying the pathological functions of FAP are not fully established, its role in tumor growth and progression through remodeling of the extracellular matrix was evidenced.24 Besides cancer-associated fibroblasts (CAFs),25 many cancer types express FAP at different levels,26 and across most cancer types, elevated FAP is associated with worse clinical outcomes.27−29 Therefore, targeting FAP represents a key aspect in improving the uptake of PSs. While it has been reported that inhibitors of FAP can improve nanoparticle targeting to CAFs,30 their potential to increase the efficacy of PS-loaded nanocarriers and thus offer efficient PDT solutions has not yet been explored.
Here, we present the generation of PS-loaded polymersomes equipped with specific targeting molecules that can differentially recognize and internalize into tumor cells with high FAP expression levels. When irradiated, cells harboring a large number of PS-loaded polymersomes increase the level of ROS production to levels that cause cell death. As the targeting molecule, we selected FAPi—a (4-quinolinoyl)glycyl-2-cyanopyrrolidine-based inhibitor of FAP, which was reported for the specific inhibition of FAP at nanomolar concentrations and demonstrated good biocompatibility in a human lung 3D tissue model.31 For the conjugation of FAPi, the covalent strain-promoted azide to alkyne [3 + 2] cycloaddition (SPAAC) was implemented, using a derivative of FAPi functionalized with a 4-dibenzocyclooctyne (DIBO) unit (DIBO-PEG3-FAPi). Polymersomes were self-assembled from a mixture of −OH and azide-terminated poly(dimethylsiloxane)-block-poly(2-methyl-2-oxazoline) (PDMS-b-PMOXA) diblock copolymers, the azide groups promoting the SPAAC reaction with DIBO-PEG3-FAPi. To show the efficacy of our approach, we used as a model Rose Bengal (4,5,6,7-tetrachloro-2′,4′,5′,7′-tetraiodofluorescein, RB), an anionic water-soluble xanthene PS. While RB increases cellular ROS upon activation with light in the visible range,32 it is known to have very limited cellular uptake because of its anionic character and low lipid solubility blocking cell membrane penetration.33 In addition, the therapeutic efficiency of RB is still limited, despite being encapsulated in liposomes34 and polymersomes that allowed only unspecific internalization.7,35
We generated polymersomes encapsulating a fluorescent dye (Atto633) and RB and subsequently decorated them with DIBO-PEG3-FAPi. FAPi-functionalized polymersomes loaded with Atto633 (FAPi-Pol-Atto633) served to optimize the encapsulation efficiency of the payload and then evaluate the internalization of the system in cells. The architecture of FAPi-functionalized polymersomes was characterized by a combination of light scattering, nanoparticle tracking analysis (NTA), and transmission electron microscopy (TEM), while absorption spectroscopy served to evaluate the cargo encapsulation and polymersome stability. The uptake of FAPi-Pol-Atto633 in cancer cell lines expressing different levels of FAP (MCF-7, A549, HepG2, and HeLa) was observed by confocal laser scanning microscopy (CLSM), which in addition served to analyze the role of FAPi in the escape of polymersomes from the endosomes. Finally, the capability of RB-loaded FAPi-polymersomes (FAPi-Pol-RB) to produce ROS under irradiation with white light was evaluated by an ROS detection assay and in terms of cell viability. We compared two cancer cell lines with different FAP expression levels and determined the ROS levels generated by FAPi-Pol-RB in order to establish a correlation between targeting and cytotoxicity associated with the functionality of the FAPi-Pol-RB. The correlation between superior targeting and cytotoxicity suggests that targeted polymersomes loaded with a photosensitizer represent an ideal candidate to improve the efficiency of PDT.
Experimental Section
Materials
Ethanol (≥99.9%, #1117272500), acetone (≥99.9%, #650501), trypsin–EDTA solution (#T3924), bovine serum albumin (#A2153), Rose Bengal (#330000), and Atto633 (#18620) were purchased from Merck. Fetal calf serum (FCS, #2-01F10-I), penicillin–streptomycin solution (Pen-Strep, #4-01F00-H), RPMI-1640 with stable glutamine (#1–41F50-I), and Dulbecco’s phosphate-buffered saline (PBS) without Ca2+/Mg2+ (#3-05F29-I) were purchased from BioConcept. Dulbecco’s Modified Eagle Medium (DMEM) high glucose (1×) with GlutaMAX (#61965-026) was purchased from Gibco. The ROS detection assay kit was purchased from Abcam (#ab287839). Calcein was purchased from ThermoFisher (#C481). All other chemicals were purchased from Merck at the highest available grade and used as received, unless stated otherwise. Hydroxyl-functionalized PDMS25-b-PMOXA10-OH (Mn = 2850 g mol–1) was synthesized and purified as previously described.36 Terminally azide-functionalized PDMS-b-PMOXA was synthesized following the method described by Meyer et al.371H NMR (Figure S1) revealed the polymer block length to be PDMS27-b-PMOXA7-PEG3-N3 (Mn = 3000 g mol–1).
Cell Culture
MCF-7, HepG2, A549, and HeLa cancer cell lines were subcultured in DMEM supplemented with 10% FCS and Pen-Strep (10000 units/mL of penicillin and 10000 μg/mL of streptomycin). A549 cells were cultured in RPMI-1640 supplemented with 10% FCS and Pen-Strep (10000 units/mL of penicillin and 10000 μg/mL of streptomycin). Cells were cultured at 37 °C in a humidified atmosphere with 5% CO2. For passaging, cells were rinsed with PBS, detached from the culture vessel surface by trypsinization and diluted at appropriated concentrations.
Formation and Surface Modification of Polymersomes
For self-assembly of polymersomes, a previously established thin film rehydration method was followed with minor modifications.37 Briefly, a volume of 25 μL of ethanol containing 2 mg of PDMS25-b-PMOXA10-OH and 0.5 mg of PDMS27-b-PMOXA7-PEG3-N3 was transferred to a round-bottom flask. The dry thin polymer film was obtained by evaporating ethanol under rotation. Polymersome self-assembly was induced by adding 0.5 mL of Atto633 in PBS (100 μM) or RB in PBS (1 mg/mL). The solution was stirred overnight at room temperature (RT) and then extruded 15 times through a 100 nm pore size polycarbonate membrane. Nonencapsulated Atto633/RB was removed by size exclusion chromatography (SEC) on a Sepharose 4B column, and purified polymersomes were stored at 4 °C until further use.
For surface modification of polymersomes with FAPi, 10 μL of DIBO-PEG3-FAPi (MW = 989.39 g mol–1)31 in dimethylformamide (DMF) was added to 500 μL of polymersome suspension to a final concentration of 8 μM under constant stirring. DIBO-PEG3-FAPi was added very slowly under intensive stirring to the polymersome suspension. The SPAAC reaction was performed overnight at RT. Unbound FAPi derivative was removed by SEC (Superdex 200 10/300 GL column), and samples were stored at 4 °C in PBS. For the quantification of FAPi ligand functionalization, a SEC fraction corresponding to nonconjugated ligand was collected and its absorbance at λ = 270 nm was measured using UV-transparent 96-well plates (#655801, Greiner Bio-One) in a SpectraMax iD3 plate reader. The concentration of unbound FAPi was calculated based on a standard curve established with DIBO-PEG3-FAPi concentrations ranging from 0.25 to 5 μM. The density of the surface conjugated FAPi was deduced by the subtraction of unbound FAPi from the initial concentration of DIBO-PEG3-FAPi used for the coupling.
Dynamic Light Scattering
The hydrodynamic mean diameter (z-average) of the Atto633-loaded polymersomes was measured by a Zeta Sizer Nano ZSP (Malvern Instruments Ltd., UK) using a laser wavelength of 633 nm and a detection angle of 173° at 25 °C. Each sample was diluted 1:500–1:1000 in PBS to avoid multiple scattering. The dynamic light scattering (DLS) measurements were performed in triplicate.
The Rh of the RB-loaded polymersomes (Pol-RB) and FAPi-Pol-RB was measured with a light scattering spectrometer (LS Instruments, Switzerland), equipped with a He–Ne (21 mW) laser (λ = 632.8 nm) at scattering angles from 20° to 150° at 25 °C. The samples were diluted 1:100–1:200 in PBS in order to reduce multiple scattering.
Static Light Scattering
Static light scattering (SLS) measurements were performed using a light scattering spectrometer (LS Instruments, Switzerland), equipped with a He–Ne (21 mW) laser (λ = 632.8 nm) at scattering angles from 20° to 150° at 25 °C. The samples were diluted 100–200 times in PBS in order to reduce multiple scattering. The radius of gyration (Rg) was obtained from the SLS data using Guinier plots, while the hydrodynamic radius (Rh) was obtained from DLS measured using the same instrument at scattering angles from 20° to 150°.
Zeta Potential
The zeta potential of polymersomes suspended in PBS was measured using a Zeta Sizer Nano ZSP (Malvern Instruments Ltd., UK). Zeta potential data represent the mean of three consecutive measurements. All measurements were performed at 25 °C.
Nanoparticle Tracking Analysis
Nanoparticle tracking analysis (NTA) was performed using a NanoSight NS 300 instrument (NanoSight Ltd., Amesbury, UK). Polymersome dispersions were diluted 4000-fold in PBS and injected into the flow cell at 100 μL min–1 using a 1 mL syringe. For each sample, three videos of 60 s were captured at RT. The concentration of polymersomes in solution was calculated by the NTA software (ver. 3.4, NanoSight) based on polymersome movement.
Transmission Electron Microscopy
For transmission electron microscopy (TEM), polymersome aliquots (5 μL) were adsorbed on 400-mesh square copper grids. Grids were negatively stained with 2% uranyl acetate. Transmission electron micrographs were recorded using a CM100 (Philips) instrument at an acceleration voltage of 80 kV.
Quantification of RB Concentration
The concentration of RB in Pol-RB and FAPi-Pol-RB samples was determined by absorption spectroscopy using a SpectraMax iD3 well plate reader. We used a standard curve established from a serial dilution of RB in PBS (0–1 mg/mL). Triton X-100 was added to RB dilutions and RB polymersome samples to a final concentration of 1%. Solubilized samples were incubated at 95 °C for 15 min and transferred to an ultrasonic bath for 5 min, and the absorbance intensity was measured at λ = 563 nm.
Purity and Stability Study of RB-Loaded Samples
To ensure no free RB was present in the sample after SEC purification, purified polymersomes were filtered through centrifugal regenerated cellulose filters with 3000 Da MWCO (Microcon YM-3, Merck). The centrifugation of 300 μL of sample (35 min, 10000g) resulted in filtering out approximately half of the initial volume. A 100 μL volume of the concentrated polymersome suspension and the filtrate were pipetted into a 96-well plate, and the absorbance at the spectrum of λ = 450–650 nm was recorded. The absence of absorbance intensity in the measured range indicated that no free RB was present in the sample (detection limit of 0.5 μg/mL). To investigate polymersome stability over time, samples stored at 4 °C were tested for the presence of free RB after four months.
Protein Extraction
MCF-7, HepG2, Hela, and A549 cells were cultured in T-25 flask to approximately 80% confluency. Cells were rinsed 3 times with ice-cold PBS and collected in 0.5 mL of RIPA buffer (Merck Millipore) containing EDTA free Halt protease inhibitor cocktail (no. 87785, ThermoFisher) using a rubber cell scrapper. Cell lysates were transferred to 1.5 mL Eppendorf tubes and extracted for 30 min at 4 °C with constant agitation. Extracts were centrifuged (16000g, 20 min at 4 °C) to pellet cellular debris. Supernatants were removed, and their protein content was determined using a Pierce bicinchoninic acid (BCA) protein assay kit (no. 23225, ThermoFisher) according to the manual provided by the manufacturer. Total cellular protein extracts were aliquoted and stored at −80 °C until further use.
Western Blotting
40 μg of total cellular protein extract was transferred to 1.5 mL Eppendorf tubes. The proteins were precipitated by adding four volumes of ice-cold acetone. The solution was then incubated for one h at −20 °C. Subsequently, samples were centrifuged (16000g, 20 min at 4 °C), the supernatants discarded, and the pellets dried to remove residual supernatant. Pellets were resuspended in Laemmli sodium dodecyl sulfate (SDS) sample buffer (no. J61337, Alfa Aesar) and denatured at 95 °C for 5 min. Equivalents of 40 μg of protein of each lysate were loaded onto any kD mini-PROTEAN TGX precast protein gels (#4569035, Bio-Rad). A PageRuler Plus prestained protein ladder, 10 to 250 kDa (no. 26619, ThermoFisher), was used as molecular mass standard. Samples were resolved by SDS–electrophoresis and electrotransferred onto Trans-Blot Turbo mini prewetted PVDF membranes (#1704156, Bio-Rad). The membrane was blocked using 5% milk in PBS with 0.1% Tween-20 (PBS-T) for 2 h at RT with gentle agitation. Then, the membrane was cut horizontally at an approximate 55 kDa. The upper part of the membrane was incubated with rabbit polyclonal IgG anti-FAP (1:500 in PBS-T, 5% milk; #PA5-99313, ThermoFisher) and the lower part with rabbit polyclonal IgG anti-GAPDH (1:5000 in PBS-T, 5% milk; #VPA00187, Bio-Rad). Both membranes were incubated overnight at 4 °C with gentle agitation, washed 4 times with PBS-T, and incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (1:10000 in PBS-T, 5% milk; #AC2114, Azure) for 2 h at RT. Membranes were washed 4 times with PBS-T, incubated for 30 s in Radiance Q-HRP chemiluminescence substrate (#AC2101, Azure), and imaged using a ChemiDoc MP imaging system. The images were acquired using Image Lab software from Bio-Rad and processed using Image Studio Lite software (ver. 5.2, LI-COR).
Cellular Uptake
Cells were plated on eight-well glass-bottom chamber slides (Ibidi, Germany) at a density of 2.5 × 104 cells/well (MCF-7), 3 × 104 cells/well (A549), 5 × 105 cells/well (HepG2), and 2.6 × 104 cells/well (HeLa). After 24 h incubation at 37 °C, culture medium was replaced with 200 μL of fresh medium containing Atto633-loaded polymersomes (Pol-Atto633) or FAPi-Pol-Atto633 at equal concentrations (1.74 × 107 polymersomes/mL, determined by NTA), and cells were incubated for another 24 h. Then, cells were carefully washed 3 times with PBS to remove excess polymersomes, fixed using 4% paraformaldehyde (PFA) in PBS, and cell nuclei were stained using Hoechst 33342 (#H3570, ThermoFisher). CLSM was performed on an CLSM 880, AxioObserver (Carl Zeiss), equipped with a water immersion objective C-Apochromat 40×/1.2 W Korr FCS M27. Samples were excited with a 405 nm diode laser collecting the emitted fluorescence in the range of 415–470 nm and with a 633 nm HeNe laser collecting the emitted fluorescence in the range of 643–758 nm. Samples were scanned unidirectionally, at 2048 × 2048 pixels, with a bit depth of 16 bit. Images were processed using ZEN Blue software (ver. 3.2, Carl Zeiss Microscopy GmbH). To quantify the uptake efficiency, 106 μm × 106 μm images of cells after 24 h uptake were analyzed. Arithmetic mean fluorescence (AMF) values of Atto633 channel were divided by the number of cells within the image. The AMF value obtained from untreated cells of each cell line was first subtracted from all samples from the same cell line. Error bars represent standard deviation of the mean of at least four images. The AMF values were obtained using ZEN Blue software. The statistical differences were calculated by two-way ANOVA comparison followed by Tukey’s HSD (honestly significant difference).
Endosomal Escape Assay
The endosomal escape properties of the FAPi-functionalized polymersomes were evaluated with a calcein assay. MCF-7 cells were plated on 8-well chamber glass slide (#C8-1.5H-N, Cellvis) at 2.5 × 104 cells per well and cultured for 24 h. Cells were placed on ice and incubated for 10 min. Then, the medium was replaced with 200 μL of ice-cold fresh medium containing 250 μM calcein and Pol-Atto633 and FAPi-Pol-Atto633 at equal concentrations (1.74 × 107 polymersomes/mL). Cells were incubated on ice for 30 min, transferred to the CO2 incubator, and incubated for 2 h at 37 °C. Then, cells were washed 3 times with PBS, 200 μL of phenol free DMEM was added to each well, and cells were imaged by CLSM. Samples were excited with a 488 nm argon laser, and emitted fluorescence was collected in the range of 498–543 nm. Samples were scanned unidirectionally, with a resolution of 1024 × 1024 pixels and bit depth of 16 bit. Images were processed using ZEN Blue software (ver. 3.2, Carl Zeiss Microscopy GmbH).
Detection of Intracellular Reactive Oxygen Species
ROS detection was performed using a ROS detection assay kit (Abcam, #ab287839) according to the manufacturer’s instructions. Briefly, MCF-7 cells were plated on eight-well glass-bottom chamber slides (Ibidi, Germany) at 2.5 × 104 cells/well and cultured for 24 h. Culture medium was replaced with fresh medium containing Pol-RB, FAPi-Pol-RB, or free RB. The RB concentration was determined as described above and adjusted so all the samples contained the same RB concentration (1 μg/mL, 1.05 μM). For the control without PS, Pol-Atto633 and FAPi-Pol-Atto633 were applied at vesicle concentrations corresponding to those of Pol-RB and FAPi-Pol-RB, respectively. Cells were incubated for another 24 h to allow for uptake and washed with 100 μL of the ROS assay buffer. As positive control, untreated cells were incubated for 1 h in ROS assay buffer containing tert-butyl hydroperoxide (inducer, provided by the manufacturer). Then, all cells were incubated with a ROS-label solution containing 2′,7′-dichlorodihydrofluorescein diacetate (100 μL per well) at 37 °C for 45 min. Cells were washed with 100 μL of ROS assay buffer and irradiated for 30 min on ice with an LED lamp (HepatoChem HCK1012-01-005 EvoluChem LED, 18 W, 6200K) placed at a 10 cm distance. After irradiation, cells were imaged by CLSM as described above.
MTS Cell Viability Assay
Cytotoxicity was evaluated using a tetrazolium compound-based Cell Titer 96 AQueous One Solution cell proliferation assay (#G3580, Promega) according to the manufacturer’s instructions. Briefly, exponentially growing MCF-7 cells were plated on 96-well plates at 2.5 × 103 cells per well in 100 μL of culture medium. Cells were incubated for 24 h, treated with polymersome samples, and incubated for another 24 h. Then cells were washed with PBS to remove excess polymersomes, and phenol-free DMEM was added (100 μL/well). Plates were placed on ice 10 cm from the LED lamp and irradiated for 30 min. For the dark control, cells were treated under the same conditions but kept on ice for 30 min without irradiation. Subsequently, cells were cultured for 24 h, and then 20 μL of 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) reagent was added to each well, and plates were returned to the incubator for 2 h at 37 °C. The absorbance was measured at λ = 490 nm using a SpectraMax iD3 plate reader. The background signal of wells containing only medium and MTS reagent (without cells) was subtracted from each sample-treated well, and obtained values were normalized to untreated control cells. Viability measurements were performed in triplicate and repeated at least twice.
Results and Discussion
Formation of FAPi-Decorated Polymersomes
To generate polymersomes with surface-exposed functional groups for conjugation to FAPi, we used a mixture of the hydroxyl-terminated amphiphilic diblock copolymer PDMS25-b-PMOXA10-OH and terminally azide-functionalized PDMS27-b-PMOXA7-PEG3-N3 at a mass ratio of 4:1. For the polymersome formation we used the film rehydration method as a straightforward approach allowing for minimal organic solvent contamination. Because both of the used copolymers comprise a similar number of monomers and thus have a similar chain length, the azide functional groups are expected to be accessible for conjugation to FAPi, especially considering the additional poly(ethylene glycol) (PEG) linker in the azide-terminated PDMS-b-PMOXA polymer.38 To avoid locally high DMF concentrations that might affect polymersome formation, DIBO-PEG3-FAPi was added to the polymersome suspension very slowly with vigorous stirring. We generated two types of assemblies by film rehydration: one encapsulating Atto633 fluorescent dye, Pol-Atto633 (2.0 × 1012 ± 3.5 × 1010 vesicles/mL) (Figure 1b and Table S1), and the other encapsulating RB, Pol-RB (9.1 × 1011 ± 2.0 × 1010 vesicles/mL) (Figure 2a and Table S1). The resulting supramolecular assemblies were extruded 15 times through 100 nm size polycarbonate membrane to reduce size dispersity and purified by gel filtration chromatography to remove nonencapsulated dye or RB. Then, the assemblies were characterized by a combination of DLS, SLS, zeta potential measurements, NTA, and TEM.
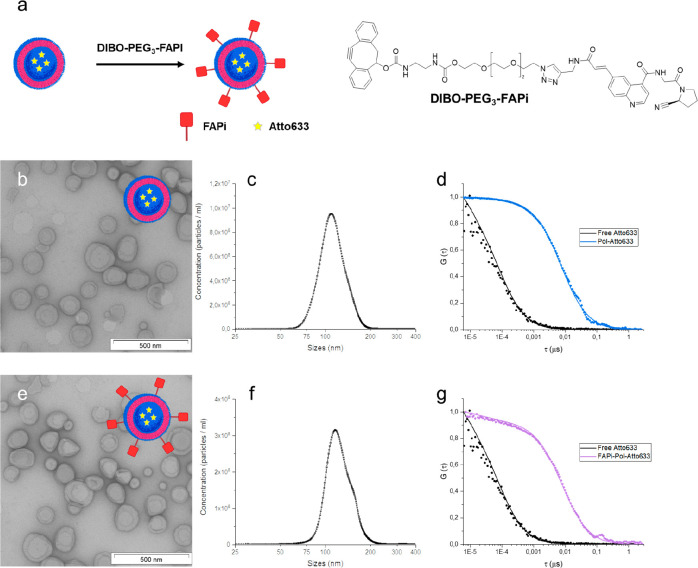
Figure 1.
Characterization of FAP-targeting polymersomes. (a) Schematic representation of polymersome surface functionalization with DIBO-PEG3-FAPi by SPAAC reaction and chemical structure of the Pol-Atto633-FAPi ligand (FAPi).31 (b) TEM images of Atto633-loaded polymersomes without the targeting ligand (Pol-Atto633). (c) Size distribution of Pol-Atto633 measured by NTA. (d) Normalized FCS autocorrelation curves of free Atto633 (black) and Pol-Atto633 (blue); dots represent experimental autocorrelation data, and solid lines are fitted curves. (e) TEM images of FAPi-Pol-Atto633. (f) Size distribution of FAPi-Pol-Atto633 measured by NTA. (g) Normalized FCS autocorrelation curves of free Atto633 (black) and FAPi-Pol-Atto633 (purple). Scale bars: 500 nm.
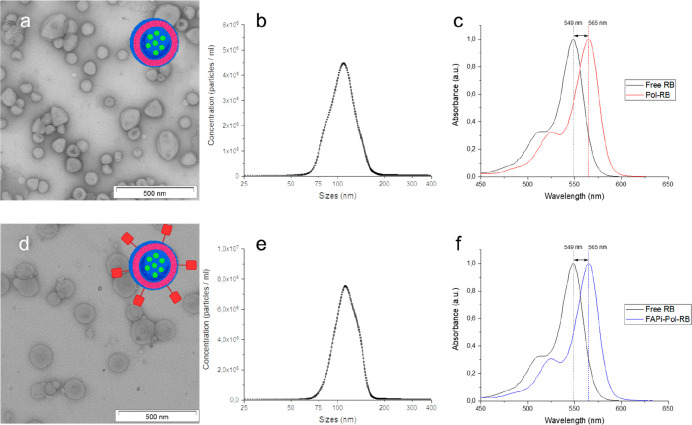
Figure 2.
Physicochemical characterization of RB-loaded polymersomes. (a, d) Schematic representation and TEM micrographs of RB-loaded polymersomes without the targeting ligand (Pol-RB) (a) and functionalized with FAPi (FAPi-Pol-RB) (d). Scale bars: 500 nm. (b, e) Size distribution of Pol-RB (b) and FAPi-Pol-RB (e) measured by NTA. (c, f) Absorption spectra of free RB (black), Pol-RB (red), and FAPi-Pol-RB (blue).
The supramolecular assemblies containing Atto633 revealed an average hydrodynamic radius of Rh = 68 ± 5 nm measured by DLS and 55 ± 3 nm measured by NTA (Figures S2 and 1c, Tables S2 and S1). The difference in Rh values obtained by NTA and DLS (Tables S1 and S2) is inherent to the respective methods.39 The spherical deflated morphology of the assemblies containing Atto633 revealed by TEM (Figure 1b) is consistent with the formation of polymersomes.40 No aggregation of polymersomes was observed by LS, TEM, and NTA. Pol-Atto633 exhibited a slightly negative zeta potential (−4.0 ± 0.5 mV), in good agreement with the values obtained for polymersomes assembled using mixtures of PDMS-b-PMOXA copolymers (Table S2).41 We calculated an encapsulation efficiency of Atto633 of 47% ± 3%, assessed by UV spectroscopy, which is in agreement with values obtained for the encapsulation of small molecular weight molecules inside polymersomes during the self-assembly process.42,43
We then coupled the fibroblast activation protein inhibitor (FAPi; Figure 1a) to the azide-functional groups exposed at the surface of polymersomes by click chemistry. We used a DIBO-functionalized, poly(ethylene glycol)-linked FAPi molecule (DIBO-PEG3-FAPi)31 where the strained cyclooctyne group allowed the conjugation of FAPi to the azide-terminated copolymer by SPAAC reaction.44,45 In addition, due to the length of the PEG3 linker, the FAPi will protrude from the surface of the polymersome and thereby support targeting. An increase in the Rh value upon attachment of FAPi was determined by DLS (Figure S2 and Table S2), in agreement with NTA analysis of FAPi-Pol-Atto633, which indicated a size of 58 ± 5 nm after FAPi attachment (Figure 1f). This slight increase in size compared to that of Pol-Atto633 (Figure 1c) gave evidence for the successful ligand attachment (Figure 1f and Table S1). We used TEM to visualize FAPi-Pol-Atto633 and compared them with Pol-Atto633. The targeting moiety neither changed the polymersome morphology nor induced aggregation of the polymersomes, which are important aspects for further application (Figure 1b,e). Zeta potential values of polymersomes decreased slightly after their functionalization with DIBO-PEG3-FAPi (−6.2 ± 0.5 mV) (Table S2). A lowered zeta potential value upon attachment of this ligand has been reported for bismuth ferrite harmonic nanoparticles31 and is another indication of the presence of the FAPi on the polymersome surface. The absorbance intensity of DIBO-PEG3-FAPi in UV, with its peak at 270 nm, allowed us to estimate the functionalization efficiency (Figure S3). After FAPi functionalization, the SEC fraction corresponding to nonconjugated ligand was collected. Based on a linear calibration curve of absorbance (λ = 270 nm) vs concentration of DIBO-PEG3-FAPi (Figure S3b) and the polymersome concentration obtained by NTA, we calculated 696 ± 11 FAPi molecules to be conjugated per polymersome, corresponding to a surface density of 1.6/100 nm2 FAPi/polymersomes. We then analyzed the encapsulation of Atto633 inside polymersomes with and without FAPi by fluorescence correlation spectroscopy (FCS). Compared with the diffusion time of free Atto633 in solution (τD = 82 ± 18 μs), both Atto633-loaded polymersomes and Atto633-loaded FAPi-polymersomes were characterized by significantly higher diffusion times (τD = 7.5 ± 1.4 ms and τD = 7.7 ± 1.2 ms, respectively). The comparable diffusion times indicated that the attachment of FAPi did not lead to aggregation (Figure 1d,g), in agreement with the TEM data (Figure 1b,e). In addition, FAPi attachment did not rupture the membrane of dye-loaded polymersomes as a minimal fraction of free dye (<0.1%) was obtained from the fit of the autocorrelation curve. The very small fraction of free dye in solutions of Atto633-loaded polymersomes (<1%) and FAPi-Pol-Atto633 (<0.1%) indicates that the purification efficiently removed nonencapsulated dye from Atto633-loaded polymersomes.
Formation of FAPi-Decorated Polymersomes Loaded with RB
To obtain the RB-loaded polymersomes, we performed film rehydration of the mixture of nonfunctionalized and functionalized copolymers in the presence of RB under similar conditions as for Atto633 encapsulation (Figure 2).
By using DLS we measured Rh = 60 ± 2 nm, and the radius of gyration (Rg) was obtained by SLS (Rg = 62 ± 3 nm). The value of the shape factor Rh/Rg of 1.03 indicates a vesicle-like morphology typical for polymersomes (Table S3 and Figure S4). The encapsulation of the PS, as a small molecular weight molecule, did not affect the self-assembly of polymersomes.46,47 Similarly, after ligand attachment, the slight increase of Rh (to 67 ± 4 nm) indicates persistent vesicle-like morphology (Table S3 and Figure S5). These data corroborate that 2% DMF final concentration is well tolerated by the polymersomes, and their membrane integrity is maintained under the conditions used for FAPi coupling. NTA measurements of Pol-RB indicated a size of 54 ± 4 nm (Figure 2b and Table S1), similar to that of Atto633-loaded polymersomes (Figure 1c), while after FAPi modification, the increase Rh to 58 ± 7 nm reflects the successful attachment of the targeting ligands on the polymersome surface (Figure 2e and Table S1). TEM revealed that the spherical morphology of polymersomes was not altered by RB encapsulation and that FAPi attachment did not affect then architecture of Pol-RB, exactly as was the case for encapsulation of Atto633 (Figure 2a,d).
Encapsulation of RB was evaluated by absorbance spectroscopy. The RB absorbance spectrum was reported to shift toward longer wavelengths upon encapsulation in DMPC liposomes and chitosan nanoparicles.34,48 The absorption spectra for RB-loaded polymersome, both without and with FAPi, displayed a maximum absorbance at λmax = 565 nm, which represented a red-shift of 16 nm compared to that of the free PS (λmax = 549 nm) (Figure 2c,f). This red-shift indicated the encapsulation of RB inside polymersomes during their formation process. In addition, we measured the absorbance spectra of the FAPi-Pol-RB after filtering the solution through centrifugal regenerated cellulose filter of 3000 Da MWCO. The lack of absorbance intensity in the range 450–650 nm demonstrated that there was no free RB present after the purification procedure. We evaluated the encapsulation efficiency of RB inside polymersomes by measuring the absorbance intensity associated with RB after addition of a nonionic detergent (Triton X-100) at increased temperature for 15 min, which was expected to induce polymersome rupture and concomitant release of RB. Although λmax of RB was red-shifted after encapsulation inside polymersomes, both free RB and RB-loaded polymersomes kept at 95 °C in the presence of 1% Triton X-100 display the same absorbance spectra (λmax) (Figure S6), indicating the release of RB upon the rupture of the polymersome membrane. The RB concentrations have been calculated based on a standard curve of free RB dilutions (incubated with detergent at 95 °C) using the absorbance intensity values at λ = 563 nm (Figure S6). The encapsulation efficiency of RB into polymersomes was calculated to be 31 ± 2%. However, we cannot exclude the presence of a very small fraction of RB at the external interface of polymersomes with the environment.
Stability of RB-Loaded Polymersomes
A key factor for further bioapplication was to determine the stability over time of Pol-RB and FAPi-Pol-RB. We measured their absorbance spectra after 4 months of storage at 4 °C and compared them to that of free RB (Figure S7). The absence of dye release indicated high stability of polymersomes and lack of undesired cargo release over extended time periods independent of the presence of the targeting ligand.
Cellular Uptake of FAPi-Pol-Atto633
We studied the uptake of polymersomes with FAPi targeting ligand exposed at their surface in relation to FAP expression in MCF-7, A549, HepG2, and HeLa cancer cell lines (Figure 3). Comparison of their FAP protein expression levels by Western blot (Figures 3a and S8) revealed different levels of FAP protein in these cell lines, with MCF-7 expressing the highest amounts of FAP. To investigate the targeting effect of FAPi exposed at the surface of polymersomes on cellular uptake, we treated the different cell lines with Pol-Atto633 without and with FAPi attached.
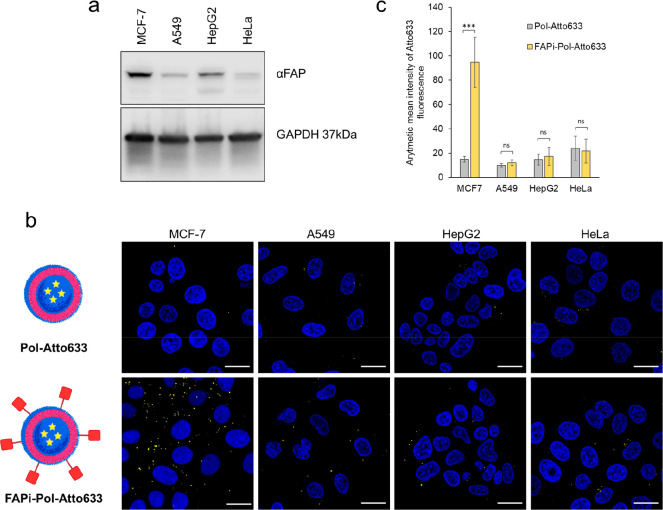
Figure 3.
Cellular uptake of polymersomes with and without targeting ligand in different cancer cell lines. (a) Western blot analysis of FAP protein expression in MCF-7, A549, HepG2, and HeLa cell extracts (40 μg of total protein loaded per lane). GAPDH was used as loading control. (b) CLSM images of polymersome-treated MCF-7, HeLa, HepG2, and A549 cells. Top: nonfunctionalized polymersomes; bottom: FAPi-functionalized polymersomes. Merged fluorescence images of polymersomes loaded with Atto633 (yellow) and Hoechst 33342 stained nuclei (blue) are shown. Scale bars: 20 μm. (c) Quantification of Pol-Atto633 and FAPi-pol-Atto633 uptake in different cell lines after 24 h incubation. Graph shows arithmetic mean fluorescence (AMF) values of the Atto633 channel of each image divided by the number of cells within the image. Error bars represent standard deviation of the mean of at least four images. The statistical differences were calculated by two-way ANOVA comparison followed by Tukey’s HSD (honestly significant difference); ns: p > 0.05; ∗∗∗: p ≤ 0.001.
The use of Atto633-loaded polymersomes allowed us to study their uptake by CLSM (Figure 3b). CLSM images of cells incubated for 24 h with Pol-Atto633 and FAPi-Pol-Atto633, respectively, indicate different internalization efficiencies depending on the carcinoma cell line. A weak fluorescent signal was detected in all four cell lines after incubation with polymersomes without the targeting ligand, indicating a minor unspecific uptake.49 While no apparent difference between the uptake of FAPi-Pol-Atto633 and Pol-Atto633 was observed for HeLa cells, measuring the mean fluorescence intensity revealed a slightly enhanced uptake for FAPi-Pol-Atto633 compared with Pol-Atto633 in HepG2 and A549 cells (Figure 3c). The marginal targeting effect observed in these three cell lines can be explained by their relatively low levels of FAP expression (Figure 3a). On the contrary, in MCF-7 cells that express large amounts of FAP, a significantly enhanced uptake was obtained for FAPi-conjugated polymersomes compared to polymersomes without FAPi (Figure 3b,c). This data indicate that an increased uptake is mediated by the FAPi targeting ligand and thus dependent on the FAP levels of the corresponding cell line. To ensure that FAPi-Pol-Atto633 were internalized by the cells rather than merely interacting with FAP on the cell surface, we captured a z-stack of optical sections through MCF-7 cells that were treated with the polymersomes for 24 h (Figure 4). The detection of FAPi-Pol-Atto633 fluorescence in several sections along the z-axis supports the intracellular localization of polymersomes. Additionally, the 3D reconstruction of z-stacks indicates that FAPi-Pol-Atto633 was located throughout the cytoplasm (Figure 4). Interaction of the surface-exposed FAPi with FAP mediated efficient polymersome binding to the cell membrane and eventually led to cellular uptake by energy-dependent pathways.50 As the size of polymersomes plays important role in their cellular uptake, the polydispersity resulting from film rehydration could possibly influence the results of subsequent experiments.49 However, FAPi-Pol-Atto633 was prepared by functionalization of Pol-Atto633, such that both systems were obtained from the same polymersomes and should display a similar size distribution. Therefore, the polydispersity only marginally contributes to differences in the rate of the uptake.
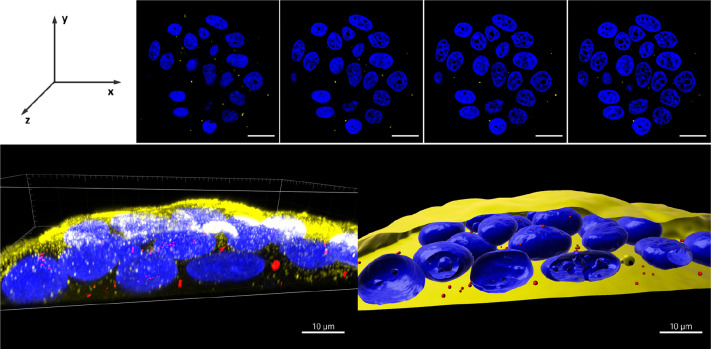
Figure 4.
3D reconstruction of intracellular location of Pol-FAPi in MCF-7 cells. (Top) z-stack images of cells captured with a 0.5 μm z-depth interval. Images present merged fluorescence of Hoechst 33342 stained cell nuclei (blue) and Atto633-loaded Pol-FAPi (yellow). Scale bars: 20 μm. (Bottom) 3D projection of confocal sections. Left: raw fluorescence of cell nuclei stained with Hoechst 33342 (blue), cell membrane stained with WGA-Alexa Fluor 488 (yellow), and FAPi-Pol-Atto633 (red). Right: fluorescence of polymersomes cell nuclei and cell membranes reconstructed and rendered using IMARIS.
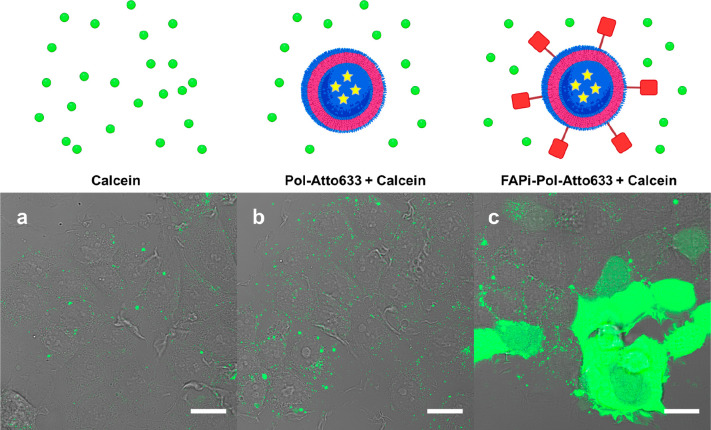
An important point in achieving high therapeutic efficiency is the ability of the nanocarriers to escape endosomes and to avoid possible degradation in the lysosomes.51,52 As the targeting of FAPi-polymersomes was significant in MCF-7 cells, we selected this cell line to follow their endosomal escape. The endosomal escape property of FAPi-Pol-Atto633 was examined by incubating cells with polymersomes together with membrane-impermeable fluorescent dye calcein. In cells with intact endosomes, calcein fluorescence is confined to the endosomal compartments. The release of calcein to the cytosol, accompanied by an equal distribution of fluorescence throughout the cell, is a marker for the rupture of endosomal membranes. When polymersomes taken up via endocytosis disrupt the endosomal membranes, their release to the cytoplasm is indicated by widespread calcein fluorescence (Figure 5).
Figure 5.
Endosomal escape assay. CLSM images of MCF-7 cells incubated with polymersome suspensions in the presence of calcein: (a) PBS, (b) Pol-Atto633 and (c) FAPi-Pol-Atto633. Images present transmission light channel merged with fluorescence light of calcein (green). Scale bars: 20 μm.
Following uptake, FAPi-Pol-Atto633 induced a pronounced cytoplasmic fluorescence compared to that of Pol-Atto633 and control MCF-7 cells incubated only with calcein in PBS. The high fluorescence intensity, when FAPi-Pol-Atto633 was internalized by MCF-7 cells, indicates an efficient release of FAPi-Pol-Atto633 from the endosomal compartment. The apparent absence of endosomal escape in cells treated with Pol-Atto633 at corresponding concentrations is related to a low cellular unspecific uptake of polymersomes without the FAPi targeting ligand.
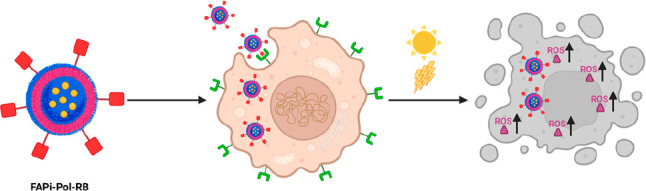
Functionality of RB-Loaded FAPi-Polymersomes
After demonstrating the role of FAPi in polymersome targeting, uptake, and endosomal release, we set out to test the ability of RB-loaded FAPi-polymersomes to produce singlet oxygen in situ upon irradiation with visible light. Except for hydrogen peroxide (H2O2), the membranes of PMOXA-b-PDMS-b-PMOXA polymersomes are permeable to oxygen species.53,54 Thus, irradiation of the PS will produce ROS in situ that will be released to the environment and gradually induce cell death.55 On the contrary, when RB-polymersomes (without or with FAPi) are not light-activated, the PS is not producing ROS. We studied ROS production in MCF-7 cells that had been prior incubated with RB-loaded polymersomes (FAPi-Pol-RB and Pol-RB) for 24 h. The cells were then washed with PBS to remove noninternalized polymersomes and a cell-permeant 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) probe was added. This probe converts to highly fluorescent 2′,7′-dichlorofluorescein (DCF) upon cleavage by intracellular esterases and oxidation by ROS. Subsequently, the cells were directly analyzed by CLSM and compared to cells that had been irradiated for 30 min with white light to activate RB (Figure 6).
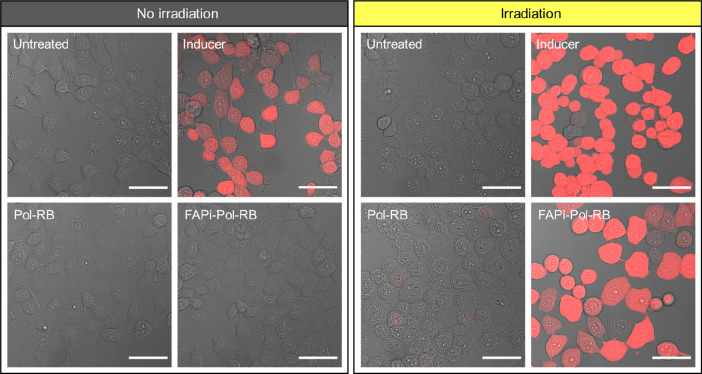
Figure 6.
Reactive oxygen species detection. CLSM images of MCF-7 cells after 24 h uptake of the polymersomes indicated were incubated for 45 min with H2DCFDA probe with (left panel) and without (right panel) subsequent irradiation. Images show transmitted light channel merged with fluorescence of the ROS detection agent (DCF, red). As a negative control, untreated cells were incubated only with the H2DCFDA probe (untreated). As a positive control, cells lacking polymersomes were exposed to tert-butyl hydroperoxide (inducer) which induces ROS production and subsequently incubated with H2DCFDA probe. Scale bars: 50 μm.
CLSM micrographs of MCF-7 cells with FAPi-Pol-RB showed strong DCF fluorescence after irradiation, indicating enhanced ROS production by the RB loaded inside of the polymersomes. In contrast, RB-loaded polymersomes without FAPi were not taken up to a degree sufficient for ROS production to be detected. No induction of ROS production was observed for untreated control cells (Figure 6) and for irradiated cells treated with polymersomes without PS (Figure S9). Comparison of CLSM micrographs of MCF-7 cells treated with free RB and with FAPi-Pol-RB (at comparable RB concentrations) indicated that higher ROS levels were produced upon irradiation when RB was encapsulated inside polymersomes (Figures 6 and S9). The significant increase in ROS production upon irradiation when RB is encapsulated inside FAPi-polymersomes is correlated to the increase of the PS uptake mediated by FAPi-Pol. Without irradiation, RB had no effect on the ROS levels in MCF-7 cells. Irradiated cells seem to display a more rounded morphology than nonirradiated cells which might result from the combined effect of irradiation and the incubation of cells in serum-free ROS assay buffer (Figure 6).
As enhanced ROS production is known to activate cell death signaling pathways,56 we tested whether the cellular uptake of RB-loaded polymersomes (without and with FAPi) followed by irradiation with white light affects the viability of cells expressing relatively high (MCF-7) and low (HeLa) amounts of FAP (Figure 7). We used an MTS assay to determine the viability of cells treated with RB-loaded polymersomes (without and with FAPi) and subsequent irradiation (Figure 7b) and compared it with the values obtained without irradiation (Figure 7a). Cells treated with equivalent amounts of free RB (2 μg/mL) without and after irradiation in similar conditions and cells incubated with PBS or polymersomes lacking RB served as controls.
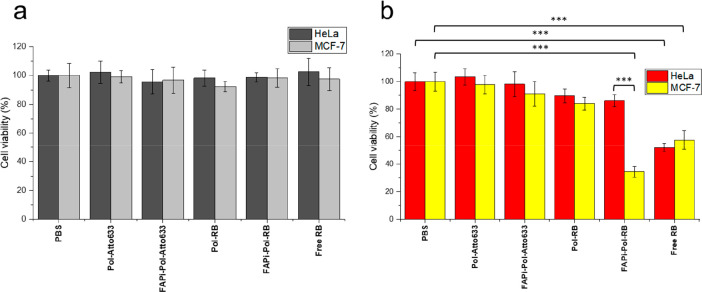
Figure 7.
MTS cell viability assay. MCF-7 and HeLa cells were treated with polymersomes with and without RB/FAPi targeting ligand or with free RB for 24 h and then washed 3 times with PBS and either kept in the dark (a) or irradiated on ice with white light for 30 min (b). All RB-containing samples were adjusted to have equal RB concentration (2 μg/mL, measured by absorbance at λ = 563 nm), and the polymersomes without RB were adjusted to the vesicle concentration of the corresponding RB-loaded polymersome samples (measured by NTA). The statistical differences were calculated by two-way ANOVA comparison followed by Tukey’s HSD (honestly significant difference); ns: p > 0.05; ∗∗∗: p ≤ 0.001. All statistically significant differences are shown.
In the absence of irradiation (Figure 7a), MCF-7 and HeLa cells treated with RB-loaded polymersomes (without and with FAPi) or with free RB were equally viable to untreated cells at the RB concentrations used (2 μg/mL). Also, the uptake of polymersomes without RB had no effect on viability. This is a key aspect because it indicates that the RB-loaded and FAPi-polymersomes are not cytotoxic without irradiation. Upon irradiation (Figure 7b), cells treated with free RB showed a significant decrease of viability in both cell lines, confirming that at the concentrations used, the PS was generating enough singlet oxygen under illumination to induce cell death. Light-activated cytotoxicity induced by free RB is similar to cytotoxicity studies conducted on Chinese hamster ovary cells57 and human fibroblasts58 using similar RB concentrations. Likewise, irradiation of MCF-7 cells incubated for 24 h with RB-loaded FAPi-polymersomes resulted in significantly reduced cell viability, while the cytotoxicity of Pol-RB was rather low under the same conditions. The strong light-activated cell killing caused by FAPi-Pol-RB is due to the FAP targeting, which led to high amounts of encapsulated RB inside cells. Consistent with this notion, HeLa cells treated with FAPi-Pol-RB showed only a small decrease in viability compared to untreated HeLa cells. This can be attributed to the lower FAP expression, resulting in less efficient targeting of FAPi-Pol-RB and subsequently, lower uptake. The FAP level dependent difference in uptake of FAPi-Pol-RB between HeLa and MCF-7 cells was also reflected by a significantly higher level of light-activated killing of the latter. Notably, in MCF-7 cells, the cytotoxicity of free RB is higher than that of FAPi-Pol-RB. Therefore, the encapsulation of the PS inside FAPi-polymersomes plays a dual role: it reduces unspecific toxicity and specifically locates it to desired sites for a better control of its therapeutic function. As the maximum of absorption of RB loaded into FAPi-polymersomes is at λ = 565 nm, the light penetration depth seems to be a limiting factor for their use in the treatment of deep-seated tumors. The penetration depth of light at this wavelength is around 4 mm, sufficient to penetrate the epidermis and to reach the dermis, making our system suitable for testing on skin cancer models.59 Furthermore, the encapsulation of various water-soluble molecules can be readily achieved by the polymersome formation method used. Therefore, RB can be easily replaced by PSs absorbing in the near-infrared region which allows for the treatment of deeply located cancer sites.60
Conclusion
Here we present a polymersome platform that unites targeting properties with the cytoplasmic delivery of light-activated cancer photosensitizers. We used PDMS-b-PMOXA-based polymersomes exposing azide groups on their surface to couple an inhibitor of FAP via straightforward click chemistry. The attachment of FAPi affected neither the self-assembly process of polymersome formation nor the encapsulation of the payload. Cancer cell lines expressing different levels of FAP showed that the FAPi-mediated uptake of polymersomes is enhanced according to the amount of FAP exposed on the surface of the cells. In addition, FAPi-functionalized polymersomes efficiently escaped the endosomal compartment. The suitability of FAPi-polymersomes to transport the payload to the desired site of action was supported by loading the polymersomes with a photosensitizer. Delivery of encapsulated RB to the cytoplasm resulted in enhanced ROS levels upon light irradiation. Moreover, under the effect of light, the photosensitizer-loaded polymersomes showed remarkable cytotoxicity, which was dependent on targeting FAP and uptake efficiency. The activation of the PS inside polymersomes without release to the environment is essential for future therapeutic applications as it minimizes potential side effects of the drug. In addition, inside polymersomes, RB’s photodynamic action was protected from inhibition, e.g., by l-Trp,61 and from inactivation when traveling through the endocytic pathway.
Taken together, PS-loaded polymersomes targeting FAP by the surface-attached FAPi offer several properties sought after in PDT including specific on-site and on-demand cell killing. Besides, FAPi-polymersomes loaded with a fluorescent dye suggest that this platform could also be used as diagnostic probe or further developed by co-loading a contrast agent with PS for theranostic applications. As FAP is highly upregulated in a wide variety of cancers, and its overexpression promotes tumor development and metastasis, we believe that our FAP targeting polymersome-based delivery system has the potential to become an excellent tool in cancer treatment.
Acknowledgments
The authors acknowledge the financial support of NCCR Molecular Systems Engineering, University of Basel, and Ecole Polytechnique Fédérale de Lausanne. The authors thank Dr Voichita Mihali for TEM imaging, Lukas Heuberger for statistical analysis of the experimental data, and Dr Daniel Messmer for NMR measurements and data interpretation. The authors kindly thank Dr Riccardo Wehr for providing the polymers and Dr Raphaël De Matos and Adrian Gheata for the synthesis and purification of DIBO-PEG3-FAPi. We also thank the Imaging Core facility (IMCF, Biozentrum, University of Basel) for assistance provided with the image processing. Schematics in this article were created with BioRender.com.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biomac.3c00943.
1H NMR of PDMS27-PMOXA7-PEG3-N3, size distribution of polymersomes measured by DLS and NTA, SLS measurements, polymersome concentrations determined by NTA, UV absorbance spectra of DIBO-PEG3-FAPi, RB concentration measurements, RB-loaded polymersome stability study, CLSM images presenting ROS detection in MCF-7 cells (PDF)
Author Contributions
∥ M.S. and D.H. contributed equally to this work. Conceptualization: C.G.P.; methodology: M.S., C.G.P.; investigation: M.S., D.H., S.T., M.K.; resources: C.G.P., S.G.-L; writing–original draft: M.S., D.H., C.S., S.T., C.G.P.; writing–review and editing: C.S., C.G.P., M.S., D.H., S.T., S.G.-L.; visualization: M.S., S.T., C.S.; supervision, project administration and funding acquisition: C.G.P.
The authors declare no competing financial interest.
Special Issue
Published as part of Biomacromoleculesvirtual special issue “Functional Compartmentalized Polymeric Systems - In Honor of Wolfgang Meier”.
Supplementary Material
References
- Arruebo M.; Vilaboa N.; Sáez-Gutierrez B.; Lambea J.; Tres A.; Valladares M.; González-Fernández Á. Assessment of the Evolution of Cancer Treatment Therapies. Cancers 2011, 3 (3), 3279–3330. 10.3390/cancers3033279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z.; Li M.; Dey R.; Chen Y. Nanomaterials for Cancer Therapy: Current Progress and Perspectives. J. Hematol. Oncol. J. Hematol Oncol 2021, 14 (1), 85. 10.1186/s13045-021-01096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. T.; Tan Y. J.; Oon C. E. Molecular Targeted Therapy: Treating Cancer with Specificity. Eur. J. Pharmacol. 2018, 834, 188–196. 10.1016/j.ejphar.2018.07.034. [DOI] [PubMed] [Google Scholar]
- Kruijt B.; Van Der Snoek E. M.; Sterenborg H. J. C. M.; Amelink A.; Robinson D. J. A Dedicated Applicator for Light Delivery and Monitoring of PDT of Intra-Anal Intraepithelial Neoplasia. Photodiagnosis Photodyn. Ther. 2010, 7 (1), 3–9. 10.1016/j.pdpdt.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Hu T.; Wang Z.; Shen W.; Liang R.; Yan D.; Wei M. Recent Advances in Innovative Strategies for Enhanced Cancer Photodynamic Therapy. Theranostics 2021, 11 (7), 3278–3300. 10.7150/thno.54227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perillo B.; Di Donato M.; Pezone A.; Di Zazzo E.; Giovannelli P.; Galasso G.; Castoria G.; Migliaccio A. ROS in Cancer Therapy: The Bright Side of the Moon. Exp. Mol. Med. 2020, 52 (2), 192–203. 10.1038/s12276-020-0384-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P.; Spulber M.; Dinu I. A.; Palivan C. G. Cellular Trojan Horse Based Polymer Nanoreactors with Light-Sensitive Activity. J. Phys. Chem. B 2014, 118 (31), 9361–9370. 10.1021/jp505152u. [DOI] [PubMed] [Google Scholar]
- Hong E. J.; Choi D. G.; Shim M. S. Targeted and Effective Photodynamic Therapy for Cancer Using Functionalized Nanomaterials. Acta Pharm. Sin. B 2016, 6 (4), 297–307. 10.1016/j.apsb.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Wang L.; Cao H.; Li Q.; Li Y.; Han M.; Wang H.; Li J. Photodynamic Therapy with Liposomes Encapsulating Photosensitizers with Aggregation-Induced Emission. Nano Lett. 2019, 19 (3), 1821–1826. 10.1021/acs.nanolett.8b04875. [DOI] [PubMed] [Google Scholar]
- Alimu G.; Yan T.; Zhu L.; Du Z.; Ma R.; Fan H.; Chen S.; Alifu N.; Zhang X. Liposomes Loaded with Dual Clinical Photosensitizers for Enhanced Photodynamic Therapy of Cervical Cancer. RSC Adv. 2023, 13 (6), 3459–3467. 10.1039/D2RA03055A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous A.; Abrahamse H. Photodynamic Therapy with an AlPcS4Cl Gold Nanoparticle Conjugate Decreases Lung Cancer’s Metastatic Potential. Coatings 2022, 12 (2), 199. 10.3390/coatings12020199. [DOI] [Google Scholar]
- Zhen S.; Yi X.; Zhao Z.; Lou X.; Xia F.; Tang B. Z. Drug Delivery Micelles with Efficient Near-Infrared Photosensitizer for Combined Image-Guided Photodynamic Therapy and Chemotherapy of Drug-Resistant Cancer. Biomaterials 2019, 218, 119330 10.1016/j.biomaterials.2019.119330. [DOI] [PubMed] [Google Scholar]
- Cao S.; Xia Y.; Shao J.; Guo B.; Dong Y.; Pijpers I. A. B.; Zhong Z.; Meng F.; Abdelmohsen L. K. E. A.; Williams D. S.; van Hest J. C. M. Biodegradable Polymersomes with Structure Inherent Fluorescence and Targeting Capacity for Enhanced Photo-Dynamic Therapy. Angew. Chem. 2021, 133 (32), 17770–17778. 10.1002/ange.202105103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J.; Park W.; Park S.; Na K. Photosensitizer-Conjugated Hyaluronic Acid-Shielded Polydopamine Nanoparticles for Targeted Photomediated Tumor Therapy. ACS Appl. Mater. Interfaces 2016, 8 (12), 7739–7747. 10.1021/acsami.6b01664. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Chen S.; Sun J.; Zhu S.; Chen C.; Xie W.; Zheng J.; Zhu Y.; Xiao L.; Hao L.; Wang Z.; Chang S. Folate-Targeted and Oxygen/Indocyanine Green-Loaded Lipid Nanoparticles for Dual-Mode Imaging and Photo-Sonodynamic/Photothermal Therapy of Ovarian Cancer in Vitro and in Vivo. Mol. Pharmaceutics 2019, 16 (10), 4104–4120. 10.1021/acs.molpharmaceut.9b00339. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Zhang L.; Zhao G.; Zhang Y.; Zhan F.; Chen Z.; He T.; Cao Y.; Hao L.; Wang Z.; Quan Z.; Ou Y. Homologous Targeting Nanoparticles for Enhanced PDT against Osteosarcoma HOS Cells and the Related Molecular Mechanisms. J. Nanobiotechnology 2022, 20 (1), 83. 10.1186/s12951-021-01201-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X.; Zebibula A.; Dong X.; Li G.; Zhang G.; Zhang D.; Qian J.; He S. Targeted and Imaging-Guided in Vivo Photodynamic Therapy for Tumors Using Dual-Function, Aggregation-Induced Emission Nanoparticles. Nano Res. 2018, 11 (5), 2756–2770. 10.1007/s12274-017-1906-7. [DOI] [Google Scholar]
- Pourtau L.; Oliveira H.; Thevenot J.; Wan Y.; Brisson A. R.; Sandre O.; Miraux S.; Thiaudiere E.; Lecommandoux S. Antibody-Functionalized Magnetic Polymersomes: In Vivo Targeting and Imaging of Bone Metastases Using High Resolution MRI. Adv. Healthc. Mater. 2013, 2 (11), 1420–1424. 10.1002/adhm.201300061. [DOI] [PubMed] [Google Scholar]
- Wei Y.; Gu X.; Sun Y.; Meng F.; Storm G.; Zhong Z. Transferrin-Binding Peptide Functionalized Polymersomes Mediate Targeted Doxorubicin Delivery to Colorectal Cancer in Vivo. J. Controlled Release 2020, 319, 407–415. 10.1016/j.jconrel.2020.01.012. [DOI] [PubMed] [Google Scholar]
- Xie S.; Gong Y. C.; Xiong X. Y.; Li Z. L.; Luo Y. Y.; Li Y. P. Targeted Folate-Conjugated Pluronic P85/Poly(Lactide- Co -Glycolide) Polymersome for the Oral Delivery of Insulin. Nanomed. 2018, 13 (19), 2527–2544. 10.2217/nnm-2017-0372. [DOI] [PubMed] [Google Scholar]
- Alibolandi M.; Ramezani M.; Abnous K.; Hadizadeh F. AS1411 Aptamer-Decorated Biodegradable Polyethylene Glycol–Poly(Lactic-Co-Glycolic Acid) Nanopolymersomes for the Targeted Delivery of Gemcitabine to Non–Small Cell Lung Cancer In Vitro. J. Pharm. Sci. 2016, 105 (5), 1741–1750. 10.1016/j.xphs.2016.02.021. [DOI] [PubMed] [Google Scholar]
- Zhu D.; Hu C.; Fan F.; Qin Y.; Huang C.; Zhang Z.; Lu L.; Wang H.; Sun H.; Leng X.; Wang C.; Kong D.; Zhang L. Co-Delivery of Antigen and Dual Agonists by Programmed Mannose-Targeted Cationic Lipid-Hybrid Polymersomes for Enhanced Vaccination. Biomaterials 2019, 206, 25–40. 10.1016/j.biomaterials.2019.03.012. [DOI] [PubMed] [Google Scholar]
- Anajafi T.; Mallik S. Polymersome-Based Drug-Delivery Strategies for Cancer Therapeutics. Ther. Delivery 2015, 6 (4), 521–534. 10.4155/tde.14.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedo A. Targeting Fibroblast Activation Protein in Cancer Ndash Prospects and Caveats. Front. Biosci. 2018, 23 (10), 1933–1968. 10.2741/4682. [DOI] [PubMed] [Google Scholar]
- Yanagawa N.; Sugai M.; Shikanai S.; Sugimoto R.; Osakabe M.; Uesugi N.; Saito H.; Maemondo M.; Sugai T. High Expression of Fibroblast-activating Protein Is a Prognostic Marker in Non-small Cell Lung Carcinoma. Thorac. Cancer 2022, 13 (16), 2377–2384. 10.1111/1759-7714.14579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y.; Zheng J.; Ma L.; Liu X.; Xu S.; Wang S.; Pei J.; Cheng K.; Yuan S.; Yu J. [18F]AlF-NOTA-FAPI-04: FAP-Targeting Specificity, Biodistribution, and PET/CT Imaging of Various Cancers. Eur. J. Nucl. Med. Mol. Imaging 2022, 49 (8), 2761–2773. 10.1007/s00259-022-05758-0. [DOI] [PubMed] [Google Scholar]
- Fitzgerald A. A.; Weiner L. M. The Role of Fibroblast Activation Protein in Health and Malignancy. Cancer Metastasis Rev. 2020, 39 (3), 783–803. 10.1007/s10555-020-09909-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olumi A.; Grossfeld G.; Hayward S.; Carroll P.; Cunha G.; Hein P.; Tlsty T. Carcinoma-Associated Fibroblasts Stimulate Tumor Progression of Initiated Human Epithelium. Breast Cancer Res. 2000, 2 (S1), S.19. 10.1186/bcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi R. S.; Kanugula S. S.; Sudhir S.; Pereira M. P.; Jain S.; Aghi M. K. The Role of Cancer-Associated Fibroblasts in Tumor Progression. Cancers 2021, 13 (6), 1399. 10.3390/cancers13061399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Q.; Liu S.; Cui J.; Zhao R.; Han Q.; Hou P.; Li Y.; Lv J.; Zhang X.; Luo Q.; Wang X. Mesoporous Polydopamine Loaded Pirfenidone Target to Fibroblast Activation Protein for Pulmonary Fibrosis Therapy. Front. Bioeng. Biotechnol. 2022, 10, 920766 10.3389/fbioe.2022.920766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Matos R.; Vuilleumier J.; Mas C.; Constant S.; Staedler D.; Gerber-Lemaire S. Inhibitor-Conjugated Harmonic Nanoparticles Targeting Fibroblast Activation Protein. RSC Adv. 2019, 9 (54), 31659–31669. 10.1039/C9RA05299B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastav A. K.; Mujtaba S. F.; Dwivedi A.; Amar S. K.; Goyal S.; Verma A.; Kushwaha H. N.; Chaturvedi R. K.; Ray R. S. Photosensitized Rose Bengal-Induced Phototoxicity on Human Melanoma Cell Line under Natural Sunlight Exposure. J. Photochem. Photobiol., B 2016, 156, 87–99. 10.1016/j.jphotobiol.2015.12.001. [DOI] [PubMed] [Google Scholar]
- Fadel M.; Kassab K. Evaluation of the Photostability and Photodynamic Efficacy of Rose Bengal Loaded in Multivesicular Liposomes. Trop. J. Pharm. Res. 2011, 10 (3), 289–297. 10.4314/tjpr.v10i3.5. [DOI] [Google Scholar]
- Chang C.-C.; Yang Y.-T.; Yang J.-C.; Wu H.-D.; Tsai T. Absorption and Emission Spectral Shifts of Rose Bengal Associated with DMPC Liposomes. Dyes Pigments 2008, 79 (2), 170–175. 10.1016/j.dyepig.2008.02.003. [DOI] [Google Scholar]
- Sztandera K.; Gorzkiewicz M.; Wang X.; Boye S.; Appelhans D.; Klajnert-Maculewicz B. PH-Stable Polymersome as Nanocarrier for Post-Loaded Rose Bengal in Photodynamic Therapy. Colloids Surf. B Biointerfaces 2022, 217, 112662 10.1016/j.colsurfb.2022.112662. [DOI] [PubMed] [Google Scholar]
- Meyer C. E.; Craciun I.; Schoenenberger C.-A.; Wehr R.; Palivan C. G. Catalytic Polymersomes to Produce Strong and Long-Lasting Bioluminescence. Nanoscale 2021, 13 (1), 66–70. 10.1039/D0NR07178A. [DOI] [PubMed] [Google Scholar]
- Meyer C. E.; Liu J.; Craciun I.; Wu D.; Wang H.; Xie M.; Fussenegger M.; Palivan C. G. Segregated Nanocompartments Containing Therapeutic Enzymes and Imaging Compounds within DNA-Zipped Polymersome Clusters for Advanced Nanotheranostic Platform. Small 2020, 16 (27), 1906492. 10.1002/smll.201906492. [DOI] [PubMed] [Google Scholar]
- LoPresti C.; Massignani M.; Fernyhough C.; Blanazs A.; Ryan A. J.; Madsen J.; Warren N. J.; Armes S. P.; Lewis A. L.; Chirasatitsin S.; Engler A. J.; Battaglia G. Controlling Polymersome Surface Topology at the Nanoscale by Membrane Confined Polymer/Polymer Phase Separation. ACS Nano 2011, 5 (3), 1775–1784. 10.1021/nn102455z. [DOI] [PubMed] [Google Scholar]
- Kim A.; Ng W. B.; Bernt W.; Cho N.-J. Validation of Size Estimation of Nanoparticle Tracking Analysis on Polydisperse Macromolecule Assembly. Sci. Rep. 2019, 9 (1), 2639. 10.1038/s41598-019-38915-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli S.; Nussbaumer M. G.; Balasubramanian V.; Chami M.; Bruns N.; Palivan C.; Meier W. Biocompatible Functionalization of Polymersome Surfaces: A New Approach to Surface Immobilization and Cell Targeting Using Polymersomes. J. Am. Chem. Soc. 2011, 133 (12), 4476–4483. 10.1021/ja110275f. [DOI] [PubMed] [Google Scholar]
- Mihali V.; Skowicki M.; Messmer D.; Palivan C. G. Clusters of Polymersomes and Janus Nanoparticles Hierarchically Self-Organized and Controlled by DNA Hybridization. Nano Today 2023, 48, 101741 10.1016/j.nantod.2022.101741. [DOI] [Google Scholar]
- Guinart A.; Korpidou M.; Doellerer D.; Pacella G.; Stuart M. C. A.; Dinu I. A.; Portale G.; Palivan C.; Feringa B. L. Synthetic Molecular Motor Activates Drug Delivery from Polymersomes. Proc. Natl. Acad. Sci. U. S. A. 2023, 120 (27), e2301279120 10.1073/pnas.2301279120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oz U. C.; Kucukturkmen B.; Ozkose U. U.; Gulyuz S.; Bolat Z. B.; Telci D.; Sahin F.; Yilmaz O.; Bozkir A. Design of Colloidally Stable and Non-Toxic Petox-Based Polymersomes for Cargo Molecule Encapsulation. ChemNanoMat 2019, 5 (6), 766–775. 10.1002/cnma.201800606. [DOI] [Google Scholar]
- Ning X.; Temming R. P.; Dommerholt J.; Guo J.; Ania D. B.; Debets M. F.; Wolfert M. A.; Boons G.-J.; van Delft F. L. Protein Modification by Strain-Promoted Alkyne-Nitrone Cycloaddition. Angew. Chem., Int. Ed. 2010, 49 (17), 3065–3068. 10.1002/anie.201000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R.; Jansen D. J.; Datta A. Intramolecular Azide-Alkyne [3 + 2] Cycloaddition: Versatile Route to New Heterocyclic Structural Scaffolds. Org. Biomol. Chem. 2009, 7 (9), 1921. 10.1039/b818962e. [DOI] [PubMed] [Google Scholar]
- Stauch O.; Schubert R.; Savin G.; Burchard W. Structure of Artificial Cytoskeleton Containing Liposomes in Aqueous Solution Studied by Static and Dynamic Light Scattering. Biomacromolecules 2002, 3 (3), 565–578. 10.1021/bm0200074. [DOI] [PubMed] [Google Scholar]
- Egelhaaf S. U.; Schurtenberger P. Shape Transformations in the Lecithin-Bile Salt System: From Cylinders to Vesicles. J. Phys. Chem. 1994, 98 (34), 8560–8573. 10.1021/j100085a041. [DOI] [Google Scholar]
- Bekmukhametova A.; Antony A.; Halliday C.; Chen S.; Ho C.; Uddin M. M. N.; Longo L.; Pedrinazzi C.; George L.; Wuhrer R.; Myers S.; Mawad D.; Houang J.; Lauto A. Rose Bengal–Encapsulated Chitosan Nanoparticles for the Photodynamic Treatment of Trichophyton Species. Photochem. Photobiol. 2023, php.13839. 10.1111/php.13839. [DOI] [PubMed] [Google Scholar]
- Roobol S. J.; Hartjes T. A.; Slotman J. A.; De Kruijff R. M.; Torrelo G.; Abraham T. E.; Bruchertseifer F.; Morgenstern A.; Kanaar R.; Van Gent D. C.; Houtsmuller A. B.; Denkova A. G.; Van Royen M. E.; Essers J. Uptake and Subcellular Distribution of Radiolabeled Polymersomes for Radiotherapy. Nanotheranostics 2020, 4 (1), 14–25. 10.7150/ntno.37080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesniak A.; Salvati A.; Santos-Martinez M. J.; Radomski M. W.; Dawson K. A.; Åberg C. Nanoparticle Adhesion to the Cell Membrane and Its Effect on Nanoparticle Uptake Efficiency. J. Am. Chem. Soc. 2013, 135 (4), 1438–1444. 10.1021/ja309812z. [DOI] [PubMed] [Google Scholar]
- Wu X. A.; Choi C. H. J.; Zhang C.; Hao L.; Mirkin C. A. Intracellular Fate of Spherical Nucleic Acid Nanoparticle Conjugates. J. Am. Chem. Soc. 2014, 136 (21), 7726–7733. 10.1021/ja503010a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleron J.; Querbes W.; Zeigerer A.; Borodovsky A.; Marsico G.; Schubert U.; Manygoats K.; Seifert S.; Andree C.; Stöter M.; Epstein-Barash H.; Zhang L.; Koteliansky V.; Fitzgerald K.; Fava E.; Bickle M.; Kalaidzidis Y.; Akinc A.; Maier M.; Zerial M. Image-Based Analysis of Lipid Nanoparticle–Mediated SiRNA Delivery, Intracellular Trafficking and Endosomal Escape. Nat. Biotechnol. 2013, 31 (7), 638–646. 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- Baumann P.; Balasubramanian V.; Onaca-Fischer O.; Sienkiewicz A.; Palivan C. G. Light-Responsive Polymer Nanoreactors: A Source of Reactive Oxygen Species on Demand. Nanoscale 2013, 5 (1), 217–224. 10.1039/C2NR32380J. [DOI] [PubMed] [Google Scholar]
- Axthelm F.; Casse O.; Koppenol W. H.; Nauser T.; Meier W.; Palivan C. G. Antioxidant Nanoreactor Based on Superoxide Dismutase Encapsulated in Superoxide-Permeable Vesicles. J. Phys. Chem. B 2008, 112 (28), 8211–8217. 10.1021/jp803032w. [DOI] [PubMed] [Google Scholar]
- Fujii J.; Homma T.; Osaki T. Superoxide Radicals in the Execution of Cell Death. Antioxidants 2022, 11 (3), 501. 10.3390/antiox11030501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redza-Dutordoir M.; Averill-Bates D. A. Activation of Apoptosis Signalling Pathways by Reactive Oxygen Species. Biochim. Biophys. Acta BBA - Mol. Cell Res. 2016, 1863 (12), 2977–2992. 10.1016/j.bbamcr.2016.09.012. [DOI] [PubMed] [Google Scholar]
- Hunter-Ellul L.; Wang S.; Wickliffe J.; Wilkerson M. Is the Photosuturing Agent, Rose Bengal, a Mutagen?. J. Cosmet. Dermatol. Sci. Appl. 2014, 04 (01), 24–30. 10.4236/jcdsa.2014.41004. [DOI] [Google Scholar]
- Goulart R. D. C.; Bolean M.; Paulino T. D. P.; Thedei G.; Souza S. L. S.; Tedesco A. C.; Ciancaglini P. Photodynamic Therapy in Planktonic and Biofilm Cultures of Aggregatibacter Actinomycetemcomitans. Photomed. Laser Surg. 2010, 28 (S1), S-53–S-60. 10.1089/pho.2009.2591. [DOI] [PubMed] [Google Scholar]
- Francisco M. D.; Chen W.-F.; Pan C.-T.; Lin M.-C.; Wen Z.-H.; Liao C.-F.; Shiue Y.-L. Competitive Real-Time Near Infrared (NIR) Vein Finder Imaging Device to Improve Peripheral Subcutaneous Vein Selection in Venipuncture for Clinical Laboratory Testing. Micromachines 2021, 12 (4), 373. 10.3390/mi12040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinna Ayya Swamy P.; Sivaraman G.; Priyanka R. N.; Raja S. O.; Ponnuvel K.; Shanmugpriya J.; Gulyani A. Near Infrared (NIR) Absorbing Dyes as Promising Photosensitizer for Photo Dynamic Therapy. Coord. Chem. Rev. 2020, 411, 213233 10.1016/j.ccr.2020.213233. [DOI] [Google Scholar]
- Sabbahi S.; Ben Ayed L.; Jemli M. Staphylococcus Aureus Photodynamic Inactivation Mechanisms by Rose Bengal: Use of Antioxidants and Spectroscopic Study. Appl. Water Sci. 2018, 8 (2), 56. 10.1007/s13201-018-0693-y. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.