ABSTRACT
β-Lactamases can accumulate stepwise mutations that increase their resistance profiles to the latest β-lactam agents. CMY-185 is a CMY-2-like β-lactamase and was identified in an Escherichia coli clinical strain isolated from a patient who underwent treatment with ceftazidime-avibactam. CMY-185, possessing four amino acid substitutions of A114E, Q120K, V211S, and N346Y relative to CMY-2, confers high-level ceftazidime-avibactam resistance, and accumulation of the substitutions incrementally enhances the level of resistance to this agent. However, the functional role of each substitution and their interplay in enabling ceftazidime-avibactam resistance remains unknown. Through biochemical and structural analysis, we present the molecular basis for the enhanced ceftazidime hydrolysis and impaired avibactam inhibition conferred by CMY-185. The substituted Y346 residue is a major driver of the functional evolution as it rejects primary avibactam binding due to the steric hindrance and augments oxyimino-cephalosporin hydrolysis through a drastic structural change, rotating the side chain of Y346 and then disrupting the H-10 helix structure. The other substituted residues E114 and K120 incrementally contribute to rejection of avibactam inhibition, while S211 stimulates the turnover rate of the oxyimino-cephalosporin hydrolysis. These findings indicate that the N346Y substitution is capable of simultaneously expanding the spectrum of activity against some of the latest β-lactam agents with altered bulky side chains and rejecting the binding of β-lactamase inhibitors. However, substitution of additional residues may be required for CMY enzymes to achieve enhanced affinity or turnover rate of the β-lactam agents leading to clinically relevant levels of resistance.
IMPORTANCE
Ceftazidime-avibactam has a broad spectrum of activity against multidrug-resistant Gram-negative bacteria including carbapenem-resistant Enterobacterales including strains with or without production of serine carbapenemases. After its launch, emergence of ceftazidime-avibactam-resistant strains that produce mutated β-lactamases capable of efficiently hydrolyzing ceftazidime or impairing avibactam inhibition are increasingly reported. Furthermore, cross-resistance towards cefiderocol, the latest cephalosporin in clinical use, has been observed in some instances. Here, we clearly demonstrate the functional role of the substituted residues in CMY-185, a four amino-acid variant of CMY-2 identified in a patient treated with ceftazidime-avibactam, for high-level resistance to this agent and low-level resistance to cefiderocol. These findings provide structural insights into how β-lactamases may incrementally alter their structures to escape multiple advanced β-lactam agents.
KEYWORDS: ceftazidime, avibactam, cefiderocol, beta-lactamase, CMY-2 variant
INTRODUCTION
Multidrug resistance (MDR) in Gram-negative bacteria is a growing global public threat, of which resistance to β-lactam agents due to the production of β-lactamases is a major component. Avibactam is a potent inhibitor of the classes A, C, and some D β-lactamases, and is combined with the third-generation cephalosporin ceftazidime for the treatment of MDR Gram-negative bacterial infection. Ceftazidime-avibactam has robust in vitro activity against Klebsiella pneumoniae carbapenemase (KPC)-producing Enterobacterales, and favorable clinical outcomes have been reported for the patients infected with carbapenem-resistant pathogens and treated with this agent (1). Ceftazidime-avibactam is also active against non-carbapenemase-producing carbapenem-resistant Enterobacterales that produce class C β-lactamases, also known as AmpC β-lactamases, or extended-spectrum β-lactamases (ESBLs) (2). However, after the launch of ceftazidime-avibactam, emergence of ceftazidime-avibactam-resistant strains that produce mutant β-lactamases capable of efficiently hydrolyzing ceftazidime or resisting avibactam inhibition are increasingly reported (3–6).
CMY-2 is the most common plasmid-mediated AmpC β-lactamase and is inhibited by avibactam (7). CMY-185, a four amino acid variant of CMY-2, was identified in an Escherichia coli clinical strain isolated from a patient who underwent treatment with ceftazidime-avibactam. blaCMY-185 confers high-level ceftazidime-avibactam resistance with an MIC of 32 mg/L (8). CMY-185 possesses four amino acid substitutions of A114E, Q120K, V211S, and N346Y in comparison to the CMY-2 sequence. We previously reported the antibiotic susceptibility profiles of E. coli laboratory strain TOP10 producing the CMY-2 variants for every combination of the four amino acid substitutions A114E, Q120K, V211S, and N346Y observed in CMY-185 (8). These results suggested that the N346Y substitution in CMY-185 appeared to be a major driver of ceftazidime-avibactam resistance, but that additional substitutions were required for high-level resistance (8). The residues at position 346 among AmpC β-lactamases are either asparagine or isoleucine (9, 10). The N346 residue of AmpC β-lactamases directly interacts with the C4 carboxyl group on the dihydrothiazine ring of cephalosporin and the sulfate group of avibactam (11–15). We hypothesized that the N346Y substitution of CMY-185 disrupts these interactions, sterically clashes with the substrates, and reduces the binding affinity of both ceftazidime and avibactam. As a result, a strain producing the enzyme containing the N346Y substitution would be susceptible to ceftazidime due to a functional tradeoff between resistance to ceftazidime and avibactam. However, previous biochemical studies of AmpC β-lactamases possessing the N346Y substitution revealed that these enzymes retained the hydrolytic efficiency against cephalosporins including ceftazidime but had impaired avibactam inhibition (9). These findings prompted us to investigate the molecular mechanism underlying ceftazidime-avibactam resistance conferred by CMY-185 and reveal the functional role of each observed substitution, namely A114E, Q120K, V211S, and N346Y. Here, we report the biochemical and structural characterization of CMY-185 as well as the CMY-2 variants harboring each substitution.
RESULTS
Kinetic parameters of the CMY-2 variants
We tested the enzymes, CMY-2, two single mutants V211S (CMY-42) and N346Y, three double mutants A114E_N346Y, Q120K_N346Y, and V211S_N346Y, and the quadruplet mutant CMY-185 to elucidate the impact on the kinetic properties resulting from the four amino acid substitutions present in CMY-185 compared with CMY-2 (8). The steady-state kinetic parameters kcat and Km (or Ki app) for nitrocefin, cephalothin, ceftazidime, and cefiderocol and the inhibition parameters Ki app, k2/K, k−2, and koff for avibactam are summarized in Table 1. During the hydrolysis of nitrocefin, we observed the reduction of the initial velocity at concentrations above 50 µM particularly for the CMY-2 variants harboring the N346Y substitution. As this could have represented substrate inhibition, we then determined the kinetics parameters for nitrocefin using the Haldane equation (16). For ceftazidime or cefiderocol, no apparent hydrolysis was observed by CMY-2, N346Y, A114E_N346Y, and Q120K_N346Y (the latter only for cefiderocol) at the substrate concentration of 100 µM with 100–200 nM of the enzyme; thus, we determined each Ki app through the hydrolysis of reporter substrate cephalothin according to a previous report (17). In addition, during the hydrolysis of cefiderocol by CMY-185, the initial velocities at a concentration range of 3.7–59.4 µM were similar. These results indicated that the value of Km is very low and that it is difficult to determine the initial velocity at lower concentrations of cefiderocol than what we measured due to the undetectable absorbance changes caused by the hydrolysis of cefiderocol. Cefiderocol is a poorly hydrolyzed substrate for CMY-185 compared to cephalothin, thus we determined Ki app for cefiderocol of CMY-185 through the hydrolysis of reporter substrate cephalothin, and used it, which is assumed to be comparable to Km, to calculate the other kinetic parameters (18, 19).
TABLE 1.
Kinetic and inhibition parameters of the CMY-2 variantsc
| CMY-2 variants | ||||||||
|---|---|---|---|---|---|---|---|---|
| Substrate | Kinetics parameters | Without mutation (CMY-2) | V211S (CMY-42) | N346Y | A114E_N346Y | Q120K_N346Y | V211S_N346Y | A114E_Q120K_ V211S_N346Y (CMY-185) |
| Nitrocefin | ||||||||
| kcat (s−1) | 2.4 × 102 ± 0.5 × 102 | 3.2 × 102 ± 1.02 × 102 | 61 ± 5 | 57 ± 10 | 1.2 × 102 ± 0.9 × 102 | 85 ± 18 | 17 ± 9 | |
| Km (µM) | 17 ± 5 | 29 ± 12 | 25 ± 3 | 16 ± 4 | 50 ± 40 | 35 ± 10 | 39 ± 24 | |
| Ki (µM) | 1.6 × 102 ± 1.0 × 102 | 1.3 × 102 ± 0.7 × 102 | 87 ± 25 | 68 ± 37 | 14 ± 12 | 1.0 × 102 ± 0.5 × 102 | 18 ± 13 | |
| kcat/Km (µM−1 s−1) | 14 ± 5 | 11 ± 5 | 2.5 ± 0.4 | 3.6 ± 1.2 | 2.4 ± 2.5 | 2.4 ± 0.9 | 0.4 ± 0.4 | |
| Cephalothin | ||||||||
| kcat (s−1) | 54 ± 0.8 | 83 ± 7 | 18 ± 7 | 3.3 ± 0.1 | 42 ± 5 | 9.6 ± 0.2 | 14 ± 1 | |
| Km (µM) | 4.1 ± 0.3 | 7.5 ± 1.1 | 3.9 ± 0.1 | 1.4 ± 0.1 | 18 ± 3 | 4.4 ± 0.3 | 19 ± 1 | |
| kcat/Km (µM−1 s−1) | 13 ± 1 | 11 ± 2 | 4.7 ± 1.8 | 2.3 ± 0.2 | 2.4 ± 0.5 | 2.2 ± 0.2 | 0.8 ± 0.1 | |
| Ceftazidime | ||||||||
| kcat (s−1) | N.D. | 1.5 ± 0.2 | N.D. | N.D. | 7.7 × 10−2 ± 0.3 × 10−2 | 0.3 ± 0.0 | 0.1 ± 0.0 | |
| Km (µM) | N.M. | 74 ± 10 | N.M. | N.M. | 5.3 ± 0.5 | 37 ± 2 | 2.2 ± 0.3 | |
| Ki app (µM) | 0.7 ± 0.1 | N.M. | 0.7 ± 0.2 | 0.3 ± 0.0 | N.M. | N.M. | N.M. | |
| kcat/Km (µM−1 s−1) | N.C. | 2.0 × 10−2 ± 0.3 × 10−2 | N.C. | N.C. | 1.5 × 10−2 ± 0.2 × 10−2 | 9.0 × 10−3 ± 0.7 × 10−3 | 5.3 × 10−2 ± 0.7 × 10−2 | |
| Cefiderocol | ||||||||
| kcat (s−1) | N.D. | 3.4 × 10−2 ± 0.2 × 10−2 | N.D. | N.D. | N.D. | 4.9 × 10−2 ± 0.3 × 10−2 | 6.9 × 10−3 ± 0.4 ×10−3 a | |
| Km (µM) | N.M. | 68 ± 11 | N.M. | N.M. | N.M. | 6.9 ± 1.5 | N.M. | |
| Ki app (µM) | 8.0 ± 1.3 | N.M. | 0.7 ± 0.0 | 0.4 ± 0.0 | 0.4 ± 0.0 | N.M. | 0.2 ± 0.0 | |
| kcat/Km (µM−1 s−1) | N.C. | 4.9 × 10−4 ± 0.9 × 10−4 | N.C. | N.C. | N.C. | 7.1 × 10−3 ± 1.6 × 10−3 | 3.0 × 10−2 ± 0.2 × 10−2 b | |
| Avibactam | ||||||||
| Ki app (µM) | 6.3 × 10−2 ± 1.2 × 10−2 | 3.4 × 10−2 ± 0.6 × 10−2 | 41 ± 6 | 80 ± 9 | 5.9 × 102 ± 0.2 × 102 | 14 ± 4 | 1.3 × 103 ± 0.1 × 103 | |
| k2/K (M−1 s−1) | 2.8 × 104 ± 0.1 × 104 | 2.3 × 104 ± 0.1 × 104 | 59 ± 6 | 41 ± 0.4 | 32 ± 0.4 | 66 ± 6 | 7.4 ± 0.3 | |
| k−2 (s−1) | 1.2 × 10−4 ± 0.2 × 10−4 | 1.5 × 10−4 ± 0.2 × 10−4 | 1 × 10−5 ± 16 × 10−5 | 1.1 × 10−4 ± 0.0 × 10−4 | 2.0 × 10−3 ± 0.1 × 10−3 | 4.4 × 10−4 ± 2.1 × 10−4 | 7.0 × 10−4 ± 0.5 × 10−4 | |
| koff (s−1) | 7.3 × 10−5 ± 0.0 × 10−5 | 6.0 × 10−5 ± 0.0 × 10−5 | 2.6 × 10−6 ± 0.0 × 10−6 | 9.0 × 10−7 ± 0.0 × 10−7 | 4.0 × 10−6 ± 0.0 ×10−6 | 2.4 × 10−6 ± 0.0 × 10−6 | 4.8 × 10−7 ± 0.0 × 10−7 | |
Due to the small Km value of CMY-185 for cefiderocol, the kcat value was determined with the measured velocities at the steady state (cefiderocol concentration range: 3.7–59.4 µM) and the Ki app value assumed to Km.
The kcat/Ki app value is shown.
The values are the mean ± standard error of three independent measurements. N.M. = not measured; N.D. = not detected; N.C. = not calculated.
V211S of CMY-2 variants augments the hydrolysis of ceftazidime and cefiderocol
The kinetics parameters for nitrocefin and cephalothin showed that hydrolytic efficiency of CMY-2 was at least threefold higher than that of the CMY-2 variants except for those with V211S, and the hydrolytic efficiency tended to decline as the mutations accumulated. The kcat and Km values for V211S were higher than those for CMY-2, resulting in comparable hydrolytic efficiency of nitrocefin and cephalothin between the two enzymes. For ceftazidime, the kcat and Km values of V211S and V211S_N346Y were higher than those of Q120K_N346Y and CMY-185. The Ki app values of CMY-2, N346Y, and A114E_N346Y were 0.3–0.7 µM and are an order magnitude lower than those for which the Km values could be determined. For AmpC β-lactamase, deacylation is considered to be a rate-limiting step for the third-generation cephalosporins (20, 21). Thus, the Ki app values of CMY-2, N346Y, and A114E_N346Y may be lower than actual Km value. For cefiderocol, we could determine the kinetic parameters kcat and Km on the CMY-2 variants harboring the V211S substitution. The kcat and Km values for V211S were (3.4 ± 0.2) × 10−2 s−1 and 68 ± 11 µM, respectively. The Km value of V211S_N346Y was 10-fold lower than that of V211S, while the kcat values were comparable. The Km (Ki app) value of CMY-185 was further lower, while the kcat value was also reduced. Comparing cefiderocol with ceftazidime, the Ki app value of CMY-2 was 10-fold higher, while the values of Ki app of N346Y and A114E_N346Y were comparable. These results indicated that the V211S substitution conferred superior turnover rates for most cephalosporins. The roles of the other substitutions A114E, Q120K, and N346Y in the hydrolysis of the cephalosporins are complex. In CMY-185, these substitutions appeared to enhance the hydrolysis of oxyimino-cephalosporins ceftazidime and cefiderocol due to the maintenance of the binding affinity. However, against the classical cephalosporins nitrocefin and cephalothin, it exhibited lower turnover rate or binding affinity, resulting in inferior hydrolysis compared to CMY-2.
The CMY-2 variants harboring N346Y and the Q120K_N346Y combination impair avibactam inhibition
The inhibitory constants Ki app of avibactam for CMY-2 and V211S were 62 ± 12 and 34 ± 6 nM, respectively. Compared to these enzymes, N346Y showed a 1,000-fold higher Ki app value, and Q120K_N346Y and CMY-185 showed 10,000- and 20,000-fold higher values, respectively. These results indicated that the N346Y substitution abolished avibactam inhibition, and the additional Q120K substitution contributed to enhanced resistance to avibactam inhibition. Avibactam is a covalent reversible inhibitor, and it binds to the enzyme, carbamoylates the active serine residue (S64 in CMY-185), then departs the enzyme by decarbamoylation through hydrolysis or intramolecular ring closure of avibactam, resulting in the release of regenerated avibactam (22). To reveal the mechanism underlying the impairment of avibactam inhibition, we determined the second-order carbamoylation rate constant k2/K and the decarbamoylation rate constants k−2 and koff for each enzyme (23). These results showed substantial reduction (~1,000-fold) in k2/K values for the CMY-2 variants harboring the N346Y substitution in comparison to those for CMY-2 or V211S, while the k−2 values were comparable with each other and the koff values were reduced as the substitutions were accumulated. The effect of the additional Q120K substitution was more complex in that the k2/K, k−2, and koff values were comparable with those of the CMY-2 variants harboring the N346Y substitution. However, the ratios of k−2 to k2/K of Q120K_N346Y and CMY-185 were slightly higher (~10-fold) than those for the CMY-2 variants harboring the N346Y substitution suggesting that, compared with the others, Q120K_N346Y and CMY-185 may release the avibactam molecule faster, before they are carbamoylated by avibactam.
The N346Y substitution induces a drastic structural change of the R2 loop in CMY-185 when ceftazidime is bound
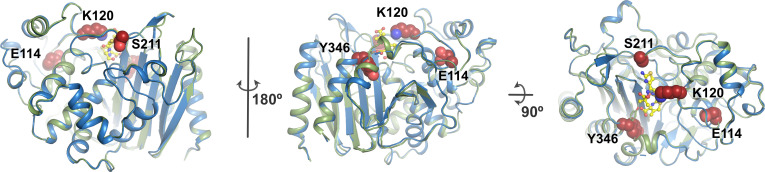
To obtain further structural insights on CMY-185, we determined the crystal structures of the CMY-185 free form and its complex with ceftazidime at the resolution of 1.35 and 2.40 Å, respectively. We prepared the crystals of the CMY-185-ceftazidime complex by soaking a crystal of the CMY-185 free-form in mother liquor containing 100 mM ceftazidime for 4 h. Data collection and refinement statistics are summarized in Table 2. To date, crystal structures of CMY-2 (PDB ID 1ZC2) and CMY-136 (PDB ID 6G9T) (24), a CMY-2 variant harboring the Y221H substitution, have been deposited in PDB database among the CMY-2-like β-lactamases. Structural comparison with these enzymes showed that the CMY-185 free-form structure is quite similar to the structure of CMY-2 and CMY-136 with the root-mean-square deviation (RMSD) values of 0.45 and 0.52 Å, respectively. The substituted residues K120, S211, and Y346 in CMY-185 are located adjacent to the substrate binding site (Fig. 1). The residues S211 and Y346 form hydrogen bonds with residues E61 and S318, respectively, while the side chain of K120 is exposed to the bulk solvent. The substituted residue E114 is located at the inner molecule, and it forms hydrogen bonds with the main chain nitrogen atoms of G116 and L117. The effect of the A114E substitution to the overall structure appears to be minimal, resulting in the loop structure around the P118 residue pushed up ~1 Å away.
TABLE 2.
Data collection and structure refinement statistics
| Data set | Free form | Ceftazidime complex |
|---|---|---|
| Data collection | ||
| Source | Photon Factory BL-17A | Photon Factory BL-17A |
| Wavelength (Å) | 0.9800 | 0.9800 |
| Space group | P212121 | P212121 |
| Unit-cell parameters | ||
| Length (Å) | a = 79.7, b = 89.5, c = 104.3, | a = 81.1, b = 89.3, c = 104.8, |
| Resolution range (Å) | 45.1–1.35 (1.43–1.35) | 45.2–2.40 (2.55–2.40) |
| No. of observed reflections | 2,207,927 (333,504) | 410,355 (57,630) |
| No. of unique reflections | 163,856 (26,083) | 30,373 (4,823) |
| Multiplicity | 13.5 (12.8) | 13.5 (11.9) |
| Completeness (%) | 99.8 (99.0) | 99.9 (99.4) |
| Rmerge (%)a | 8.1 (88.8) | 13.6 (87.8) |
| 〈I/σ(I)〉 | 17.42 (2.72) | 15.96 (2.96) |
| Refinement | ||
| Resolution (Å) | 39.1–1.35 (1.37–1.35) | 45.2–2.40 (2.48–2.40) |
| Reflection used | 163,761 (5,165) | 30,319 (2,700) |
| Rwork (%)b | 13.9 (22.0) | 18.6 (24.2) |
| Rfree (%)c | 17.7 (25.1) | 23.0 (30.6) |
| No. of non-hydrogen atoms | 7,055 | 5,740 |
| Protein | 5,965 | 5,498 |
| Ligands | 10 | 62 |
| Solvent | 1,080 | 180 |
| RMSD from ideality | ||
| Bond length (Å) | 0.008 | 0.005 |
| Bond angle (°) | 0.940 | 0.742 |
| Average B-factor | 20.4 | 43.3 |
| Protein | 18.1 | 43.4 |
| Ligands | 28.2 | 44.0 |
| Solvent | 33.0 | 41.3 |
| Ramachandran plot | ||
| Favored region (%) | 98.46 | 98.43 |
| Allowed region (%) | 1.54 | 1.57 |
| Outlier region (%) | 0.00 | 0.00 |
| Clashscore | 2.46 | 3.19 |
| PDB ID | 8JB7 | 8JB8 |
Rmerge = 100 × ∑hkl ∑i | Ii(hkl) − 〈I(hkl)〉 | / Σhkl Σi Ii(hkl), where 〈I(hkl)〉 is the mean value of I(hkl).
Rwork = 100 × ∑hkl| |Fo | – |Fc | | / ∑hkl |Fo| where Fo and Fc the observed and calculated structure factors, respectively.
Rfree is calculated as for Rwork, but for the test set comprising 5% reflections not used in refinement.
Fig 1.
Overall structure of CMY-185. Crystal structures of CMY-185 are shown as cartoon representations colored green for the free form and blue for the complex with ceftazidime. The substituted residues E114, K120, S211, and Y346 are shown as CPK representations colored red. The ceftazidime molecule is shown as a ball-and-stick representation colored yellow.
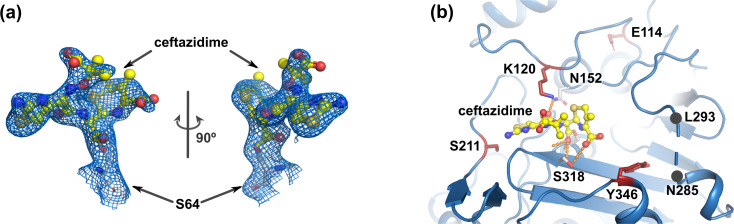
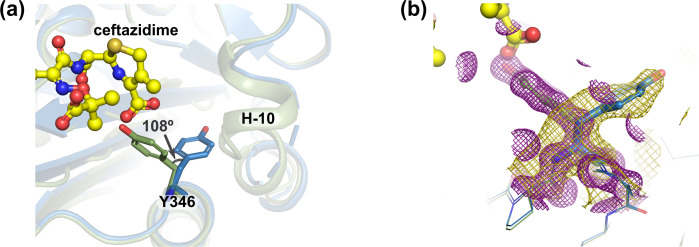
Crystal structure of the CMY-185-ceftazidime complex showed an acyl-enzyme intermediate where the S64 catalytic residue is covalently bound to the C8 atom of ceftazidime, and its β-lactam structure is cleaved (Fig. 3a). In contrast to the free form, crystal structure of the CMY-185-ceftazidime complex revealed a drastic structural change of the R2 loop in that the H-10 helix was disrupted and disordered at residues N285–L293 (Fig. 2). In comparing the crystal structures of the CMY-185 free form and its complex, the C4 carboxyl group on the dihydrothiazine ring of ceftazidime occupied the position of the side chain of Y346 observed in the free form. The side chain of Y346 in the complex in turn was rotated 108° toward the H-10 helix and positioned to clash with the S287 residue, and this appeared to cause the disruption of the H-10 helix structure (Fig. 3b). We did not observe overall structural changes except for the R2 loop with the RMSD value of 0.68 Å, and the positions of the substituted residues E114, K120, and S211 were almost identical. Structure comparisons of the CMY-185-ceftazidime complex with the other class C β-lactamase AmpCE. coli (PDB ID 1IEL) (11) and AmpCEnt385 (PDB ID 6LC9) (15) complex with ceftazidime showed that the binding position of the ceftazidime molecule was identical, and the hydrogen bond between Q120 and the amide group at the R1 side chain of ceftazidime disappeared by the substitutions, while S318 formed a hydrogen bond with the C4 carboxyl group of ceftazidime instead of N346 (Fig. 2).
Fig 2.
Ceftazidime binding site. The ceftazidime molecule is shown as a ball-and-stick representation colored yellow. (a) The S64 residue is shown as a white stick representation. 2mFo-DFc map is shown as blue mesh contoured 1σ. (b) Hydrogen bonds are shown as orange dashed lines. Disordered residues are shown as blue dashed lines and the positions of N285 and L293 are marked as black dots.
Fig 3.
Structure comparison of the Y346 residue. (a) Structures of CMY-185 are colored green for the free form and blue for the complex with ceftazidime. (b) 2mFo-DFc map is shown as purple (free form) and gold (complex) mesh contoured 1.25σ. The maps are drawn within 3 Å radius of each Y346 residue.
The substituted residues E114, K120, and S211 form stable hydrogen bonds
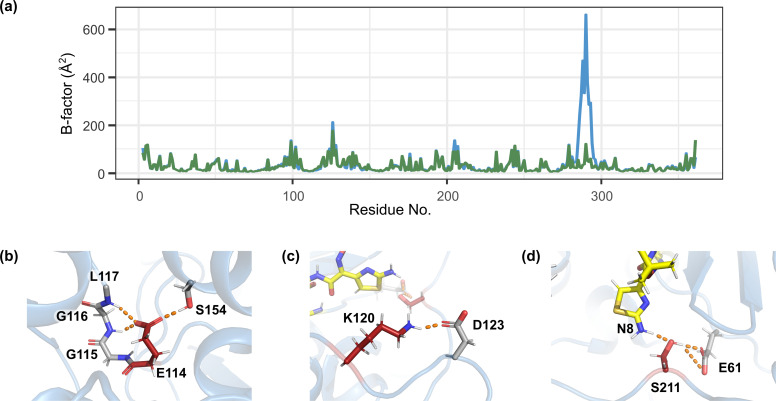
Crystal structures of CMY-185 revealed that the N346Y substitution induced the R2 loop structure change due to the complex formation with ceftazidime. To elucidate further characteristics and potential effect of the other substituted residues on CMY-185, we examined the molecular behaviors of the CMY-185-ceftazidime complex using molecular dynamics simulations. Five runs of 100 ns molecular dynamics simulations were performed for the free form and the acyl complex with ceftazidime each of CMY-2 and CMY-185 at 300 K. The RMSD analysis indicated that the systems for both CMY-2 and CMY-185 reached equilibrium within 1 ns of the simulation (Fig. S1) and a stable conformation over the simulations, with mean RMSD values of 1.12 Å for the free form and 1.09 Å for the complex (Fig. S2a and S2b). Compared to CMY-2, CMY-185 showed slightly greater fluctuation, with mean RMSD values of 1.55 Å for the free form and 1.58 Å for the complex (Fig. S2c and S2d). Both CMY-2 and CMY-185 exhibited similar overall fluctuations between the free form and the complex (Fig. S1). Fig. 4a shows the overall profiles of the temperature factors for each residue. Compared to the CMY-2-ceftazidime complex, the CMY-185-ceftazidime complex structure showed more vibrations in the residues I284–L293 on the R2 loop, whereas the overall vibrations outside the R2 loop were similar. The residues I284–L293 almost correspond to the disordered residues in the crystal structure of the CMY-185-ceftazidime complex, suggesting that, in the crystal, the fragile structure was entirely disrupted by the complex formation, without the influence of crystal packing. Molecular dynamics simulation of the CMY-185-ceftazidime complex showed that the substituted residues formed stable hydrogen bonds or salt bridges. The side chain of E114 interacted with the side chain of S154 in addition to the main chain nitrogen atoms of G116 and L117 (Fig. 4b; Fig. S2). The side chain of K120 formed a salt bridge with the side chain of D123 (Fig. 4b; Fig. S3), while the side chain of S211 maintained a hydrogen bond with the side chain of E61 (Fig. 4c; Fig. S4).
Fig 4.
Molecular dynamics simulation for the CMY-2 and CMY-185 complexes with ceftazidime. (a) B factors profiles of the CMY-2 (green) and the CMY-185 (blue) complexes with ceftazidime. (b–d) Close-up view of the hydrogen bond or salt bridge formation of E114 (b), K120 (c), and S211 (d). The hydrogen bonds or salt bridge were shown as orange dashed lines.
DISCUSSION
The N346 residue of typical AmpC β-lactamases directly interacts with cephalosporins and avibactam. Focusing on the disruption of the interaction, the N346Y substitution in the AmpC β-lactamases appears to confer functional tradeoff between resistance to cephalosporins and avibactam. However, in fact, this functional tradeoff is not always realized (8). Our structural studies presented here clearly demonstrate that the ceftazidime molecule is bound to CMY-185, and that this in turn leads to the drastic structural change of CMY-185 to accommodate the ceftazidime binding (Fig. 1 to 3). In addition, the kinetics studies indicated that, compared to CMY-2 or its V211S substitution mutant, the CMY-2 variants harboring the N346Y substitution had lower k2/K values (Table 1) and rejected the primary binding of avibactam like the other AmpC β-lactamases harboring the N346Y substitution (9). These results suggest that the sulfate group of avibactam is essential for its binding to the AmpC β-lactamases, while the cephalosporins are recognized through the overall interaction. This observation is supported by the number of hydrogen bonds between a typical AmpC β-lactamase and avibactam. The sulfate group of avibactam is recognized by three residues through ~5 hydrogen bonds, while carbamoyl group located at the opposite side of the sulfate group is recognized by the residues Q120 and N152 through two hydrogen bonds (12–15, 25). Moreover, in CMY-185, the Q120 residue is substituted to lysine, and the Ki app value of Q120K_N346Y is 10-fold higher compared to that of N346Y (Table 1). When the positions of nitrogen and oxygen atoms of the carbamoyl group of avibactam and the side chain of N152 were altered, the K120 residue in CMY-185 appeared to form a hydrogen bond with the oxygen atom of the carbamoyl group. However, the N152 residue also formed a hydrogen bond with the K67 residue conserved among the AmpC β-lactamases and could not rotate spontaneously. In addition, the molecular dynamics simulation indicated that the K120 residue formed a stable salt bridge with the D123 residue (Fig. 4c; Fig. S3), suggesting that the K120 residue was not primarily directed to interact with avibactam. These findings explain why the Q120K substitution combined with N346Y substitution enhances resistance to the avibactam inhibition.
The A114E substitution had a smaller effect on the enzyme reaction compared to the other substitutions. The kinetics studies of the CMY-2 variants harboring the A114E substitution showed that the values of Km or Ki app for the cephalosporins measured in this study were generally lower than the other enzymes. On the other hand, the Ki app value for avibactam was higher than those of N346Y and V211S_N346Y, indicating that the A114E substitution affected the avibactam binding to the enzyme. Structural studies indicated that the A114E substitution did not alter the overall structure, and it instead seemed to enhance the intramolecular interaction through the hydrogen bond formation compared to alanine. We could define the phenotype of the CMY-2 variants harboring the A114E substitution, but the functional significance of the A114E substitution remains unclear.
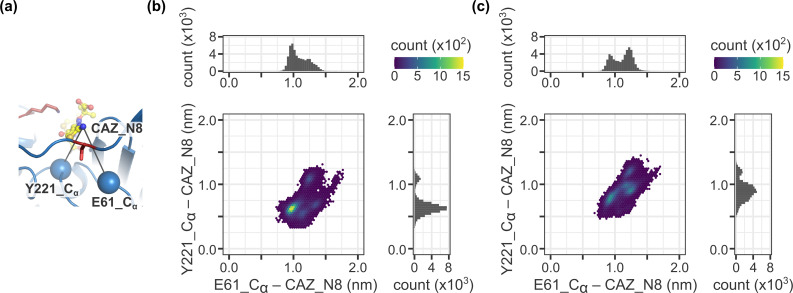
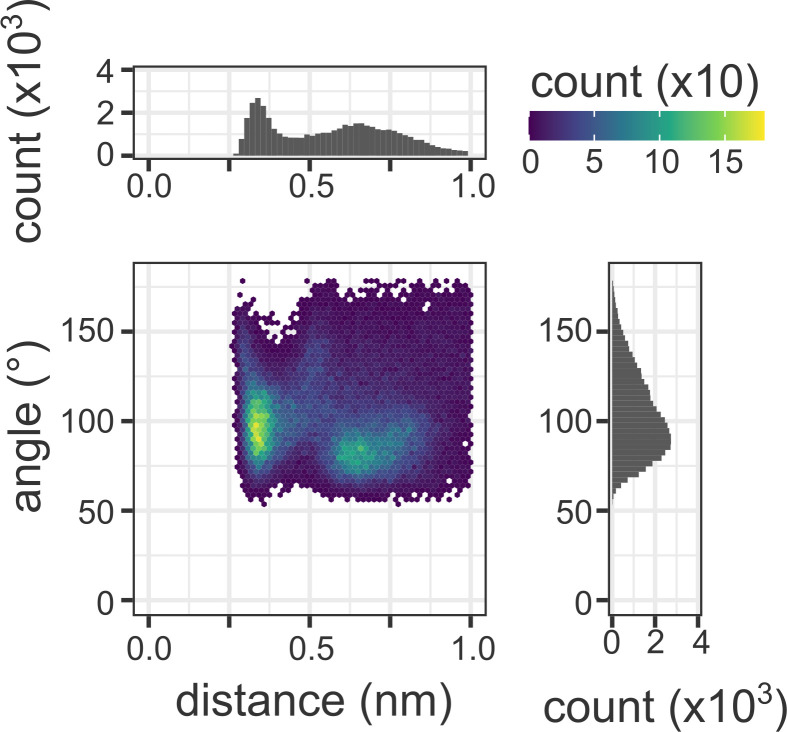
CMY-30 and CMY-42 are single mutants of CMY-2 harboring the substitutions of V211 to glycine and serine, respectively (17, 26). CMY-30 and CMY-42 are reported to hydrolyze oxyimino-cephalosporins such as ceftazidime more efficiently than CMY-2 with higher turnover rates. According to the studies of the molecular dynamics simulation for CMY-30 and CMY-42, the V211 substitution increases the fluctuation of the structure spanning the Ω loop to the Q120-loop, resulting in better accommodation of the substrate cephalosporin for the deacylation reaction and leading to efficient release of the product (27, 28). This increase in fluctuation is caused by the removal of restriction on the Y221 residue, which is usually restrained by V211 in CMY-2. In addition, CMY-107, which is a single mutant of CMY-2 harboring the Y199C substitution, possesses similar hydrolytic efficiency to that of CMY-30 or CMY-42, and it is also reported to display incremental fluctuation of the structure from the Ω loop to the Q120-loop due to the motile side chain of the unrestrained Y221 residue (29). We could not observe an increase in the fluctuation of the structure around the Y221 residue during the molecular dynamics simulations for the CMY-185-ceftazidime complex (Fig. 4a). In the CMY-185-ceftazidime complex, the K120 residue formed a stable salt bridge with the D123 residue. Both residues K120 and D123 are located at the Q120 loop, and the Q120 loop structure is rigidified due to this interaction, suggesting a cause for the restriction of fluctuation. In fact, the kcat value of CMY-185 was lower than that of V211S (CMY-42) or V211S_N346Y (Table 1). However, fluctuation of the structure of the aminothiazole ring included in the R1 side chain of ceftazidime notably increased in comparison to that of the CMY-2-ceftazidime complex. Fig. 5 shows the distance distribution between the aminothiazole ring of ceftazidime and each enzyme. A single peak was observed in the CMY-2-ceftazidime complex and positioned around (0.95, 0.62) nm for the distances between the N8 atom of ceftazidime and the Cα atoms for E61 or Y221, respectively (Fig. 5b). On the other hand, the 2D histogram of the CMY-185-ceftazidime complex showed the broad distribution of the distances, and at least four peaks were observed with similar levels around (0.92, 0.78), (1.21, 0.90), (1.22, 1.19), and (1.25, 0.90) nm for the distances between the N8 atom of ceftazidime and the Cα atoms of E61 or Y221, respectively (Fig. 5c). These findings indicate that the ceftazidime molecule in the CMY-185 complex is more flexible than that in the CMY-2 complex. In addition, the Oγ atom of S211 formed a stable hydrogen bond with the side chain of E61 and was sandwiched between the N8 atom of ceftazidime and the side chain of E62 (Fig. 4d). Fifteen percent of the snapshots during the molecular dynamics simulation showed that the distances between the Oγ atom of S211 and the N8 atom of ceftazidime were within 3.5 Å (Fig. 6). Hydrogen bonds were observed in 51% of these distances within 3.5 Å (8% of overall distances). These results suggest that the interaction between S211 and N8 atom of ceftazidime through a weak hydrogen bond might contribute to stimulate the fluctuation of ceftazidime to pull the R1 side chain of ceftazidime toward the Ω loop.
Fig 5.
Fluctuations of the ceftazidime molecule bound to CMY-2 and CMY-185. (a) The Cα atoms for E61 and Y221 are shown as blue spheres. (b, c) 2D histogram for the distance distribution between the N8 atom of ceftazidime (CAZ_N8) and the Cα atoms of E61 (E61_Cα) or Y221 (Y221_Cα). Each point represents a snapshot from the molecular dynamics simulations, colored according to its counts shown in the color bar on the top right corner. (b) CMY-2. (c) CMY-185.
Fig 6.
Interaction between S211 and the N8 atom of ceftazidime. 2D histogram for the distribution of the distance and angle between the Oγ atom of S211 and the N8 atom of ceftazidime. “Distance” axis indicates the distance between the Oγ atom of S211 and the N8 atom of ceftazidime. “Angle” axis indicates the angle between the atoms of Oγ and HG of S211 and the N8 atom of ceftazidime. Each point represents a snapshot from the molecular dynamics simulations, colored according to its counts shown in the color bar on the top right corner.
The structural change of the R2 loop in CMY-185 leads to further expansion of the substrate binding site for cephalosporins, and this expansion is a major cause for the reduced susceptibility to cefiderocol of the strain. A similar expansion of the substrate binding site is observed in AmpCEnt385, which contains the deletion of residues A294 and P295 and also confers reduced susceptibility to cefiderocol (6, 15). In addition, CMY-172 and CMY-178 contain the deletion of three residues K290, V291, and A292 on the R2 loop, resulting in the higher MICs of 16 and 64 mg/L for ceftazidime-avibactam, respectively, and 1 mg/L for cefiderocol (30). Moreover, our results indicated that the CMY-2-like β-lactamases strongly recognize cefiderocol compared to the other β-lactamases. To our knowledge to date, the lowest Km value against cefiderocol is 49.8 µM for AmpCEnt385 (15) and the Km values for other β-lactamases are over 190 µM (19, 31). Compared to these values, CMY-2 had a notably lower Ki app value of 8.0 µM, and N346Y and A114E_N346Y showed even lower Ki app values of 0.7 and 0.4 µM, respectively. In addition, the Ki app values for N346Y and A114E_N346Y against cefiderocol were comparable to those against ceftazidime. These findings suggest that the CMY-2-like enzymes strongly recognize the structure around the R1 side chain and the cephalosporin cephem core, which is shared between ceftazidime and cefiderocol, and this may serve as the driving force for the rotation of Y346. We observed that the CMY-2 variants harboring the N346Y substitution exhibit enhanced substrate inhibition during the nitrocefin hydrolysis. The Y346 residue is required to rotate for recognition of cephalosporins; therefore, this rotation may be a rate-limiting step of the enzyme reaction for the substrates exhibiting high turnover rate such as nitrocefin. Before the substrate is accommodated, there would be a pre-binding state, where the CMY-2 variants harboring the N346Y substitution only recognize the structure around the R1 side chain of cephalosporin, resulting in the substrate inhibition. Thus, we propose the following enzyme reaction scheme: the CMY-2 variant harboring the N346Y substitution initially recognizes the structure around R1 side chain of cephalosporin, then the steric hindrance between the cephalosporin and the enzyme induces the rotation of Y346 to accommodate the substrate, and finally the activated S64 residue nucleophilically attacks the C8 atom of the cephalosporin to cleave the β-lactam ring. This hypothesis also appears to be consistent with the rejection of avibactam binding to the CMY-2 variants harboring N346Y substitution. The oxyimino group is the major R1 side chain of oxyimino-cephalosporins (32); thus, the diazabicyclooctane derivatives with substitution of 2-carbamoyl group with the oxyimino group may be potential inhibitors of mutated β-lactamases conferring resistance to valuable antibiotics like ceftazidime-avibactam or cefiderocol.
Conclusion
Evolution of antimicrobial resistance caused by antimicrobial selection pressure is a significant threat and requires close monitoring. The CMY-185 enzyme is a CMY-2 variant possessing four amino acid substitutions A114E, Q120K, V211S, and N346Y, and was identified in an E. coli strain isolated from a patient who was treated with the ceftazidime-avibactam combination. CMY-185 is a unique enzyme in which the level of resistance to ceftazidime-avibactam is incrementally enhanced with the accumulation of amino acid substitutions. Here, we revealed the functional role of each substituted residue in CMY-185 using biochemical and structural analyses. The N346Y substitution confers resistance to avibactam inhibition and also induces a drastic structural change of CMY-185 with the expansion of the substrate binding site. The Q120K substitution enhances resistance to the avibactam inhibition, while the V211S substitution increases the turnover rate of the cephalosporin hydrolysis. The A114E substitution combined with the other substituted residue appears to adjust the binding affinity of the substrate. These findings demonstrate that CMY-185 has achieved an optimal balance between the turnover rate and the binding affinity for hydrolyzing ceftazidime while maintaining high-level resistance to avibactam inhibition. CMY-185 has acquired these enzymatic characteristics and is able to confer high-level resistance to ceftazidime-avibactam. The findings in this study provide new insights into how β-lactamases evolve in bacteria by accumulating beneficial substitutions to survive the selective pressure of the latest β-lactam agents.
MATERIALS AND METHODS
Cloning, expression, and purification of the CMY-2 variants
In this study, all CMY-2 variant genes encoding the mature enzyme without a signal peptide were cloned into the protein expression plasmid vector pET-30b between the restriction enzyme sites of NdeI and EcoRI. The expression and purification of all CMY-2 variants were performed in the same way as follows. Lysogenic broth plus kanamycin (30 µg/mL) was inoculated with E. coli BL21 (DE3) harboring each pET-30b-CMY-2 variant recombinant plasmid and grown at 30°C to an OD600 of 0.5–0.7. A final concentration of 0.1 mM IPTG was added, and the culture was incubated for an additional 3 h. The cells were harvested by centrifugation at 3,500 × g for 10 min at 15°C, and the pellet was resuspended in 20 mM Tris-HCl pH 7.0 buffer, sonicated, and centrifuged at 8,000 × g for 30 min at 4°C. The supernatant was transferred to the new centrifuge tube and further centrifuged at 48,000 × g for 90 min at 4°C. The crude extract was loaded onto a HiTrap SP HP (Cytiva, Tokyo, Japan) previously equilibrated with 20 mM Tris-HCl pH 7.0 buffer. The CMY-2 variant was then eluted with a NaCl linear gradient of 0–0.5 M. The peak fractions containing the CMY-2 variant were pooled and then dialyzed overnight with 20 mM Tris-HCl pH 7.0 buffer. Further purification was performed with a HiTrap Blue HP column (Cytiva, Tokyo, Japan) chromatography, and the CMY-2 variant was eluted with a linear gradient of 0–2 M NaCl in 20 mM Tris-HCl pH 7.0 buffer. The active fractions were confirmed with BD BBL Cefinase Paper Disc (Becton Dickinson, Sunnyvale, CA, USA), pooled and concentrated using a Vivaspin turbo 4 centrifugal concentrator (MWCO 10,000, Sartorius, Gottingen, Germany). The buffer was exchanged to 20 mM Tris-HCl pH 7.0 by several rounds of dilution and concentration. The purified active CMY-2 variant solution was finally concentrated to 10–20 mg/mL and stored in 20 µL aliquots at −80°C until used in crystallization and kinetics experiments.
Kinetic measurements
The steady-state kinetics were carried out on a UV-Visible spectrometer (UV-1280, Shimadzu, Japan) in PBS pH 7.2 at 25°C with a constant amount of enzyme and varying concentrations of the substrates. We tested nitrocefin, cephalothin, ceftazidime, and cefiderocol as the substrates and avibactam as the inhibitor. The steady-state kinetics parameters Km and kcat were determined with the Michaelis-Menten equation. For poorly hydrolyzed substrates, the Km value was assumed to be the competitive inhibition constant (Ki app) value according to previous studies (17, 19). Inhibition parameter values of Ki app, k2/K, k−2, and koff for avibactam were determined using a method as described previously (33). Cephalothin was used as the reporter substrate for competitive inhibition experiments, and its concentration was fixed at 100 µM. The enzyme concentration was 1–10 nM. Inverse steady-state initial velocities were plotted against the concentration of the competitive inhibitor agents, such as ceftazidime, cefiderocol, and avibactam. The data were analyzed by linear regression analysis of the Dixon plot, and Ki app (observed) was determined by dividing the y-intercept value by the slope of the line. It was then corrected to account for the affinity of cephalothin for each enzyme using the Equation (1):
| (1) |
where [S] represents the concentration of the reporter substrate cephalothin, and Km (cephalothin) is the Km value of cephalothin for each enzyme.
The koff value was determined using a jump dilution assay. The enzyme and avibactam were mixed and preincubated at 25°C for 5 min. The concentration of avibactam depended on the enzyme and concentrations 10-fold higher from each Ki app (observed) value were adopted, which were determined based on the corrected competitive inhibition constants Ki app. Preincubated enzyme-avibactam mixture was then diluted 2,000-fold in the 100 µM cephalothin solution, and the hydrolysis of cephalothin was measured. Three independent measurements were performed for each enzyme and substrate or inhibitor combination. Curve fittings using linear or non-linear regression were performed with R 4.1.3 (R Core Team, 2022).
Crystallization of CMY-185
Prior to the crystallization, the purified CMY-185 recombinant enzyme was diluted to the final concentration of 15 mg/mL in 20 mM Tris-HCl pH 7.0. Crystals of CMY-185 were obtained using hanging-drop vapor diffusion method with the crystallization condition in which the CMY-185 solution was mixed with an equal volume of a reservoir solution containing 24% PEG 20,000, 0.1 M Tris-HCl pH 8.5, and 0.2 M lithium sulfate at 20°C. Crystals of the CMY-185-ceftazidime complex were obtained by soaking the CMY-185 crystals in a solution containing 100 mM ceftazidime, 26% PEG 20,000, 0.1 M Tris-HCl pH 7.0, and 0.2 M lithium sulfate at 4°C for 4 h.
Data collection, structure determination, and refinement
The CMY-185 crystals were transferred into a cryoprotectant solution composed of reservoir solution containing 15% glycerol, and then flash-frozen in liquid nitrogen. Synchrotron experiments were performed at Photon Factory BL-17A (High Energy Accelerator Research Organization, Tsukuba, Japan). Diffraction data sets were collected at −173°C using an EIGER X16M detector, and were processed and scaled using XDS (34). The initial phase of the CMY-185 structure was determined by the molecular replacement method using Molrep (35) from the CCP4 program suite (36), with the coordinate (PDB ID: 6G9T) serving as the search model (24). Manual model rebuilding was performed with COOT (37). Structure refinements were performed with phenix.refine from the PHENIX package (38). The CMY-185 free-form structure was refined with the anisotropic atomic displacement parameters for the heavy atoms of protein. The CMY-185-ceftazidime complex structure was refined with the atomic displacement parameters using the translation, liberation, and screw (TLS) method, and the TLS groups were determined by using phenix.find_tls_groups. The stereochemical quality of the final structures were evaluated by MolProbity (39). All molecular graphics were prepared using PyMOL (Schrödinger, L. & DeLano, W., 2020. PyMOL, Available at: http://www.pymol.org/pymol.).
Molecular dynamics simulations
Molecular dynamics simulations were run for the free forms and the ceftazidime complexes with CMY-2 and CMY-185 using GROMACS (40) and the ff14SB forcefield (41). Prior to the simulation, the loop and the side chain structures disordered in the crystal structure of the CMY-185-ceftazidime complex were modeled using MODELLER (42). The model structure of the CMY-2-ceftazidime complex was generated by superimposing the structures of the CMY-185-ceftazidime complex on CMY-2 (PDB ID 1ZC2). The protonation state of each residue was predicted with PROPKA 3 (43). During the deacylation reaction in the AmpC β-lactamases, Y150 is assumed to be deprotonated and acts as a general base in activating hydrolyzing water molecule (44), therefore the Y150 residue instead of the K67 residue in the model structure of each complex was deprotonated. The parameters for the residues S64 acylated with ceftazidime and deprotonated Y150 were generated with the GAFF forcefield (45) and AM1-BCC partial charges (46) using Antechamber (47) from the AmberTools 20 software (48). After adding hydrogen atoms, the modeled structure of each CMY-2 and CMY-185 was placed in a cubic box with a boundary of 10 Å under a periodic boundary condition. The system was solvated with TIP3P water, and then neutralized by adding sodium and chloride ions using LEaP (49). Amber topology files were converted to GROMACS topology files using ACPYPE (50). The energy minimization of the system was performed with the steepest descent algorithm, and then the system was heated gradually from 0 to 300 K in NVT ensemble for 200 ps with harmonic positional restraints on the heavy atoms of protein and ligand (force constant, 10 kcal/mol/Å2). The system was relaxed at 300 K in NPT ensemble for 100 ps with the positional restraints maintained, and the force constants of the positional restraints were gradually reduced to 0 kcal/mol/Å2 for further 700 ps. The equilibrated system was then subjected to molecular dynamics simulation for 100 ns. All bonds were constrained with the LINCS algorithm (51). Cutoffs of 1.0 nm were used for the neighbor list, Coulomb interactions and Van der Waals interactions. Long-range electrostatic interactions were measured using the particle-mesh Ewald algorithm (52). The time step of the molecular dynamics simulation was set to 2 fs, and the snapshots were stored every 10 ps. The simulations were performed with five runs for each system and all data were subjected to analysis.
ACKNOWLEDGMENTS
Synchrotron experiments were performed with the approval of the Photon Factory Program Advisory Committee (Proposal nos. 2017G554 and 2019G592). The authors are grateful to the beamline staff for their support of our synchrotron experiments, and to Shionogi Inc. for the provision of cefiderocol.
This work was supported by grant R21AI151362 from the National Institute of Allergies and Infectious Diseases (NIAID), JSPS KAKENHI Grant number 23K06267, and a grant from the Takeda Science Foundation. W.C.S. is supported by the Texas Medical Center Training Program in Antimicrobial Resistance (TPAMR) T32 Fellowship Program (T32AI141349) from the NIAID. S.A.S. is supported by grant P01AI152999 and Y.D. by grant R01AI104895, both from the NIAID.
Contributor Information
Akito Kawai, Email: kawai-a@fujita-hu.ac.jp.
Yohei Doi, Email: yod4@pitt.edu.
Alejandro J. Vila, IBR (CONICET) University of Rosario, Rosario, Santa Fe, Argentina
DATA AVAILABILITY
The atomic coordinates of the CMY-185 free form and the CMY-185-ceftazidime complex have been submitted to the Protein Data Bank under PDB accession numbers 8JB7 and 8JB8, respectively.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.02874-23.
Fig. S1 to S4.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Soriano A, Carmeli Y, Omrani AS, Moore LSP, Tawadrous M, Irani P. 2021. Ceftazidime-avibactam for the treatment of serious gram-negative infections with limited treatment options: a systematic literature review. Infect Dis Ther 10:1989–2034. doi: 10.1007/s40121-021-00507-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Castanheira M, Doyle TB, Deshpande LM, Mendes RE, Sader HS. 2021. Activity of ceftazidime/avibactam, meropenem/vaborbactam and imipenem/relebactam against carbapenemase-negative carbapenem-resistant enterobacterales isolates from US hospitals. Int J Antimicrob Agents 58:106439. doi: 10.1016/j.ijantimicag.2021.106439 [DOI] [PubMed] [Google Scholar]
- 3. Aitken SL, Tarrand JJ, Deshpande LM, Tverdek FP, Jones AL, Shelburne SA, Prince RA, Bhatti MM, Rolston KVI, Jones RN, Castanheira M, Chemaly RF. 2016. High rates of nonsusceptibility to ceftazidime-avibactam and identification of New Delhi metallo-beta-lactamase production in Enterobacteriaceae bloodstream infections at a major cancer center. Clin Infect Dis 63:954–958. doi: 10.1093/cid/ciw398 [DOI] [PubMed] [Google Scholar]
- 4. Shields RK, Chen L, Cheng S, Chavda KD, Press EG, Snyder A, Pandey R, Doi Y, Kreiswirth BN, Nguyen MH, Clancy CJ. 2017. Emergence of ceftazidime-avibactam resistance due to plasmid-borne bla(KPC-3) mutations during treatment of carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob Agents Chemother 61:e02097-16. doi: 10.1128/AAC.02097-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Papp-Wallace KM, Mack AR, Taracila MA, Bonomo RA. 2020. Resistance to novel beta-lactam-beta-lactamase inhibitor combinations: the "price of progress” Infect Dis Clin North Am 34:773–819. doi: 10.1016/j.idc.2020.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shields RK, Iovleva A, Kline EG, Kawai A, McElheny CL, Doi Y. 2020. Clinical evolution of AmpC-mediated ceftazidime-avibactam and cefiderocol resistance in Enterobacter cloacae complex following exposure to cefepime. Clin Infect Dis 71:2713–2716. doi: 10.1093/cid/ciaa355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jacoby GA. 2009. AmpC Β-Lactamases. Clin Microbiol Rev 22:161–182. doi: 10.1128/CMR.00036-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shropshire WC, Endres BT, Borjan J, Aitken SL, Bachman WC, McElheny CL, Wu C-T, Egge SL, Khan A, Miller WR, Bhatti MM, Saharasbhojane P, Kawai A, Shields RK, Shelburne SA, Doi Y. 2023. High-level ceftazidime/avibactam resistance in Escherichia coli conferred by the novel plasmid-mediated β-lactamase CMY-185 variant. J Antimicrob Chemother 78:2442–2450. doi: 10.1093/jac/dkad249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Compain F, Debray A, Adjadj P, Dorchêne D, Arthur M. 2020. Ceftazidime-avibactam resistance mediated by the N346Y substitution in various AmpC β-Lactamases. Antimicrob Agents Chemother 64:1–9. doi: 10.1128/AAC.02311-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mack AR, Barnes MD, Taracila MA. 2020. A standard numbering scheme for class C beta-lactamases. Antimicrob Agents Chemother 64. doi: 10.1016/j.diagmicrobio.2020.115242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Powers RA, Caselli E, Focia PJ, Prati F, Shoichet BK. 2001. Structures of ceftazidime and its transition-state analogue in complex with AmpC beta-lactamase: implications for resistance mutations and inhibitor design. Biochemistry 40:9207–9214. doi: 10.1021/bi0109358 [DOI] [PubMed] [Google Scholar]
- 12. Lahiri SD, Mangani S, Durand-Reville T, Benvenuti M, De Luca F, Sanyal G, Docquier J-D. 2013. Structural insight into potent broad-spectrum inhibition with reversible recyclization mechanism: avibactam in complex with CTX-M-15 and Pseudomonas aeruginosa AmpC β-lactamases. Antimicrob Agents Chemother 57:2496–2505. doi: 10.1128/AAC.02247-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lahiri SD, Johnstone MR, Ross PL, McLaughlin RE, Olivier NB, Alm RA. 2014. Avibactam and class C β-Lactamases: mechanism of inhibition, conservation of the binding pocket, and implications for resistance. Antimicrob Agents Chemother 58:5704–5713. doi: 10.1128/AAC.03057-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pozzi C, Di Pisa F, De Luca F, Benvenuti M, Docquier JD, Mangani S. 2018. Atomic-resolution structure of a class C β-lactamase and its complex with avibactam. ChemMedChem 13:1437–1446. doi: 10.1002/cmdc.201800213 [DOI] [PubMed] [Google Scholar]
- 15. Kawai A, McElheny CL, Iovleva A, Kline EG, Sluis-Cremer N, Shields RK, Doi Y. 2020. Structural basis of reduced susceptibility to ceftazidime-avibactam and cefiderocol in Enterobacter cloacae due to AmpC R2 loop deletion. Antimicrob Agents Chemother 64:e00198-20. doi: 10.1128/AAC.00198-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haldane J. 1930. Enzymes Longmans. Green and Co, UK. [Google Scholar]
- 17. Hentschke M, Kotsakis SD, Wolters M, Heisig P, Miriagou V, Aepfelbacher M. 2011. CMY-42, a novel plasmid-mediated CMY-2 variant AmpC beta-lactamase. Microb Drug Resist 17:165–169. doi: 10.1089/mdr.2010.0137 [DOI] [PubMed] [Google Scholar]
- 18. Ito-Horiyama T, Ishii Y, Ito A, Sato T, Nakamura R, Fukuhara N, Tsuji M, Yamano Y, Yamaguchi K, Tateda K. 2016. Stability of novel siderophore cephalosporin S-649266 against clinically relevant carbapenemases. Antimicrob Agents Chemother 60:4384–4386. doi: 10.1128/AAC.03098-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ito A, Nishikawa T, Ota M, Ito-Horiyama T, Ishibashi N, Sato T, Tsuji M, Yamano Y. 2018. Stability and low induction propensity of cefiderocol against chromosomal AmpC beta-lactamases of Pseudomonas aeruginosa and Enterobacter cloacae. J Antimicrob Chemother 73:3049–3052. doi: 10.1093/jac/dky317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mazzella LJ, Pratt RF. 1989. Effect of the 3'-leaving group on turnover of cephem antibiotics by a class C beta-lactamase. Biochem J 259:255–260. doi: 10.1042/bj2590255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nukaga M, Kumar S, Nukaga K, Pratt RF, Knox JR. 2004. Hydrolysis of third-generation cephalosporins by class C beta-lactamases. structures of a transition state analog of cefotoxamine in wild-type and extended spectrum enzymes. J Biol Chem 279:9344–9352. doi: 10.1074/jbc.M312356200 [DOI] [PubMed] [Google Scholar]
- 22. Ehmann DE, Jahić H, Ross PL, Gu R-F, Hu J, Kern G, Walkup GK, Fisher SL. 2012. Avibactam is a covalent, reversible, non-beta-lactam beta-lactamase inhibitor. Proc Natl Acad Sci USA 109:11663–11668. doi: 10.1073/pnas.1205073109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lang PA, Leissing TM, Page MGP, Schofield CJ, Brem J. 2021. Structural investigations of the inhibition of Escherichia coli AmpC beta-lactamase by Diazabicyclooctanes. Antimicrob Agents Chemother 65:e02073-20. doi: 10.1128/AAC.02073-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zavala A, Retailleau P, Elisée E, Iorga BI, Naas T. 2019. Genetic, biochemical, and structural characterization of CMY-136 β-lactamase, a peculiar CMY-2 variant. ACS Infect Dis 5:528–538. doi: 10.1021/acsinfecdis.8b00240 [DOI] [PubMed] [Google Scholar]
- 25. Nukaga M, Papp-Wallace KM, Hoshino T, Lefurgy ST, Bethel CR, Barnes MD, Zeiser ET, Johnson JK, Bonomo RA. 2018. Probing the mechanism of inactivation of the FOX-4 cephamycinase by avibactam. Antimicrob Agents Chemother 62:e02371-17. doi: 10.1128/AAC.02371-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kotsakis SD, Papagiannitsis CC, Tzelepi E, Tzouvelekis LS, Miriagou V. 2009. Extended-spectrum properties of CMY-30, a Val211Gly mutant of CMY-2 cephalosporinase. Antimicrob Agents Chemother 53:3520–3523. doi: 10.1128/AAC.00219-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kotsakis SD, Tzouvelekis LS, Petinaki E, Tzelepi E, Miriagou V. 2011. Effects of the Val211Gly substitution on molecular dynamics of the CMY-2 cephalosporinase: implications on hydrolysis of expanded-spectrum cephalosporins. Proteins 79:3180–3192. doi: 10.1002/prot.23150 [DOI] [PubMed] [Google Scholar]
- 28. Kotsakis SD, Caselli E, Tzouvelekis LS, Petinaki E, Prati F, Miriagou V. 2013. Interactions of oximino-substituted boronic acids and beta-lactams with the CMY-2-derived extended-spectrum cephalosporinases CMY-30 and CMY-42. Antimicrob Agents Chemother 57:968–976. doi: 10.1128/AAC.01620-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kotsakis SD, Miriagou V, Vetouli EE, Bozavoutoglou E, Lebessi E, Tzelepi E, Tzouvelekis LS. 2015. Increased hydrolysis of oximino-beta-lactams by CMY-107, a Tyr199Cys mutant form of CMY-2 produced by Escherichia coli. Antimicrob Agents Chemother 59:7894–7898. doi: 10.1128/AAC.01793-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhou J, Wang W, Liang M, Yu Q, Cai S, Lei T, Jiang Y, Du X, Zhou Z, Yu Y, Zhou X. 2023. A novel CMY variant confers transferable high-level resistance to ceftazidime-avibactam in multidrug-resistant Escherichia coli. Microbiol Spectr 11:e0334922. doi: 10.1128/spectrum.03349-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ito A, Nishikawa T, Yamano Y, Matsumoto S, Yoshizawa H, Sato T, Nakamura R, Tsuji M. 2016. Transporter systems for antibacterial activity against Pseudomonas aeruginosa. Antimicrob Agents Chemother 60:7396–7401. doi: 10.1128/AAC.01405-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chaudhry SB, Veve MP, Wagner JL. 2019. Cephalosporins: a focus on side chains and beta-lactam cross-reactivity. Pharmacy (Basel) 7:103. doi: 10.3390/pharmacy7030103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Papp-Wallace KM, Winkler ML, Gatta JA, Taracila MA, Chilakala S, Xu Y, Johnson JK, Bonomo RA. 2014. Reclaiming the efficacy of β-lactam-β-lactamase inhibitor combinations: avibactam restores the susceptibility of CMY-2-producing Escherichia coli to ceftazidime. Antimicrob Agents Chemother 58:4290–4297. doi: 10.1128/AAC.02625-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66. doi: 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vagin A, Teplyakov A. 2010. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr 66:22–25. doi: 10.1107/S0907444909042589 [DOI] [PubMed] [Google Scholar]
- 36. Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, et al. 2019. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in phenix. Acta Crystallogr D Struct Biol 75:861–877. doi: 10.1107/S2059798319011471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. Molprobity: All-atom structure validation for macromolecular crystallography . Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. 2015. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19–25. doi: 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- 41. Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C. 2015. ff14Sb: improving the accuracy of protein side chain and backbone parameters from ff99Sb. J Chem Theory Comput 11:3696–3713. doi: 10.1021/acs.jctc.5b00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Webb B, Sali A. 2016. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics 54:5. doi: 10.1002/cpbi.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Søndergaard CR, Olsson MHM, Rostkowski M, Jensen JH. 2011. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J Chem Theory Comput 7:2284–2295. doi: 10.1021/ct200133y [DOI] [PubMed] [Google Scholar]
- 44. Tripathi R, Nair NN. 2016. Deacylation mechanism and kinetics of acyl-enzyme complex of class C β-lactamase and cephalothin. J Phys Chem B 120:2681–2690. doi: 10.1021/acs.jpcb.5b11623 [DOI] [PubMed] [Google Scholar]
- 45. Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. 2004. Development and testing of a general amber force field. J Comput Chem 25:1157–1174. doi: 10.1002/jcc.20035 [DOI] [PubMed] [Google Scholar]
- 46. Jakalian A, Jack DB, Bayly CI. 2002. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. parameterization and validation. J Comput Chem 23:1623–1641. doi: 10.1002/jcc.10128 [DOI] [PubMed] [Google Scholar]
- 47. Wang J, Wang W, Kollman PA, Case DA. 2006. Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graph Model 25:247–260. doi: 10.1016/j.jmgm.2005.12.005 [DOI] [PubMed] [Google Scholar]
- 48. Case DA, Aktulga HM, Belfon K. 2020. Amber 2020. University of California, San Francisco [Google Scholar]
- 49. Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ. 2005. The amber biomolecular simulation programs. J Comput Chem 26:1668–1688. doi: 10.1002/jcc.20290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sousa da Silva AW, Vranken WF. 2012. ACPYPE - AnteChamber PYthon Parser interfacE. BMC Res Notes 5:367. doi: 10.1186/1756-0500-5-367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hess B. 2008. P-LINCS: a parallel linear constraint solver for molecular simulation. J Chem Theory Comput 4:116–122. doi: 10.1021/ct700200b [DOI] [PubMed] [Google Scholar]
- 52. Salomon-Ferrer R, Götz AW, Poole D, Le Grand S, Walker RC. 2013. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. explicit solvent particle mesh ewald. J Chem Theory Comput 9:3878–3888. doi: 10.1021/ct400314y [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4.
Data Availability Statement
The atomic coordinates of the CMY-185 free form and the CMY-185-ceftazidime complex have been submitted to the Protein Data Bank under PDB accession numbers 8JB7 and 8JB8, respectively.