Abstract
Purpose
To evaluate the expression of sry-box transcription factor 9 (SOX9) in orbital fibroblasts (OFs) of thyroid eye disease (TED) and to find its potential role and underlying mechanism in orbital fibrosis.
Methods
OFs were cultured from orbital connective tissues obtained from patients with TED (n = 10) and healthy controls (n = 6). SOX9 was depleted by small interfering RNA or overexpressed through lentivirus transduction in OFs. Fibroblast contractile activity was measured by collagen gel contraction assay and proliferation was examined by EdU assay. Transcriptomic changes were assessed by RNA sequencing.
Results
The mRNA and protein levels of SOX9 were significantly higher in OFs cultured from patients with TED than those from healthy controls. Extracellular matrix-related genes were down-regulated by SOX9 knockdown and up-regulated by SOX9 overexpression in TED-OFs. SOX9 knockdown significantly decrease the contraction and the antiapoptotic ability of OFs, whereas the overexpression of SOX9 increased the ability of transformation, migration, and proliferation of OFs. SOX9 knockdown suppressed the expression of phosphorylated ERK1/2, whereas its overexpression showed the opposite effect. Epidermal growth factor receptor (EGFR) is one of the notably down-regulated genes screened out by RNA sequencing. Chromatin immunoprecipitation-qPCR demonstrated SOX9 binding to the EGFR promoter.
Conclusions
A high expression of SOX9 was found in TED-OFs. SOX9 can activate OFs via MAPK/ERK1/2 signaling pathway, which in turn promotes proliferation and differentiation of OFs. EGFR was a downstream target gene of SOX9. SOX9/EGFR can be considered as therapeutic targets for the treatment of orbital fibrosis in TED.
Keywords: sry-box transcription factor 9, epidermal growth factor receptor, thyroid eye disease, orbital fibroblast, fibrosis
Thyroid eye disease (TED) is a common autoimmune disease involving the orbital tissue with the potential risk of cosmetic disfigurement and vision loss.1 Research on TED pathogenesis has largely focused on the orbital fibroblasts (OFs) as these are one of the major target cells in the pathological progression.1–3 Activated by immune cells and the secretion of inflammatory cytokines, fibroblasts were differentiated into adipocytes and myofibroblasts, synthesizing a large amount of extracellular matrix (ECM), thereby causing tissue remodeling and fibrosis in TED.4,5 The pathology of orbital fibrosis would contribute to several hallmark clinical features, including eyelid retraction, restricted eye movement, and strabismus,6 resulting in deterioration of quality of life.7,8 At present, the clinical treatment options are targeted primarily to the inflammatory process; the therapeutic effect of treatments for orbital fibrosis is not as effective as expected, and the pathogenesis of orbital fibrosis is not clear.
Sry-box transcription factor 9 (SOX9), a member of the sry-related high mobility group box-containing proteins family, is crucial for multiple organ development including the heart, lung, pancreas, and retina.9 Extensive experimental evidence indicated that the overexpression of SOX9 promoted fibrosis in the lung, kidney, and cardioid, and experimental reduction of SOX9 suppressed fibrosis.10–13 Biologically TGF-β accumulation is the crucial initiating step of myofibroblast transdifferentiation.14 SOX9 was up-regulated by TGF-β and induced fibrosis by participating in the TGF-β signaling pathway.15 SOX9 positively contributes to fibrosis by stimulating the expression of a range of genes encoding pathological ECM components.13
The activation of mitogen-activated protein kinase (MAPK) signalings was indispensable for the disassembly of adherens junctions and cell motility during TGF-β–induced epithelial–mesenchymal transition.16,17 Several studies demonstrated that there appeared to be a link between the functional role of SOX9 and MAPK pathways.18,19 Depletion of SOX9 attenuated protein kinase B (Akt) and MAPK activity. E-cadherin/SOX9 axis regulated cell growth and self-renewal of cancer stem cells partially through Akt and MAPK signaling in multiple myeloma cells.20 In addition, the activity of the SOX9-dependent collagen type II alpha 1 enhancer was increased upon the coexpression of a constitutively active MAPK kinase 1 mutant both in primary chondrocytes and C3H10T1/2 cells.21 Therefore, we hypothesized that SOX9 might promote fibrosis via the MAPK pathways.
Currently, no prior studies have yet been performed to evaluate the role of SOX9 in the pathogenesis of orbital fibrosis. This study aims to detect SOX9 levels and explore its profibrotic effect and the underlying mechanisms in the fibrotic pathogenesis of TED in primary cultured OFs.
Methods
Subjects
Orbital adipose tissue samples were collected from 10 patients with TED who underwent orbital decompression. Normal control (NC) tissues were collected from 6 healthy controls who underwent blepharoplasty. The baseline characteristics of TED patients and healthy controls are given in Table. No significant differences were observed between the two groups with regard to demographic characteristics (Supplementary Table S1). Exclusion criteria were listed as follows: use of glucocorticoids, immunosuppressive agents, or radiotherapy within the past 3 months; a history of ocular surgery and trauma; and other systemic diseases such as rheumatoid arthritis or systemic lupus erythematosus.
Table.
Clinical Characteristics of the Patient Samples Used in this Study
| Age | Sex (Male/Female) | TED History (Months) | Clinical Activity Score | TED Severity Assessment | Previous TED Treatment | Surgery Performed | |
|---|---|---|---|---|---|---|---|
| Patients | |||||||
| 1 | 46 | Male | 12 | 3/7 | VI | GCs | Decompression |
| 2 | 56 | Male | 10 | 1/7 | VI | GCs | Decompression |
| 3 | 26 | Male | 24 | 4/7 | III | GCs | Decompression |
| 4 | 26 | Female | 72 | 0/7 | III | GCs | Decompression |
| 5 | 36 | Female | 12 | 1/7 | III | GCs | Decompression |
| 6 | 54 | Female | 60 | 1/7 | III | GCs | Decompression |
| 7 | 52 | Male | 12 | 1/7 | IV | GCs | Decompression |
| 8 | 28 | Female | 156 | 0/7 | III | GCs | Decompression |
| 9 | 39 | Male | 12 | 2/7 | VI | GCs | Decompression |
| 10 | 54 | Female | 10 | 3/7 | IV | GCs | Decompression |
| Controls | |||||||
| 1 | 51 | Male | / | / | / | / | Blepharoplasty |
| 2 | 58 | Male | / | / | / | / | Blepharoplasty |
| 3 | 34 | Female | / | / | / | / | Blepharoplasty |
| 4 | 52 | Male | / | / | / | / | Blepharoplasty |
| 5 | 27 | Female | / | / | / | / | Blepharoplasty |
| 6 | 33 | Female | / | / | / | / | Blepharoplasty |
The ethical permission was granted by the Ethics Committee of Zhongshan Ophthalmic Center, Sun-Yat Sen University. Written informed consent was signed by all tissue donors, and the tenets of the Declaration of Helsinki were followed throughout.
Primary Cell Culture
Primary OFs were cultured following standard protocols as previously reported.22 Tissue samples were cut into 1- to 2-mm3 pieces and transferred into 10-cm plates, and OFs migrated out from these pieces. Cells were serially passaged with trypsinization after reaching 80% to 90% confluence. Passages 3 to 8 OFs with good viability were used during the whole experiment. OFs were treated with different experimental conditions, details were specified in the corresponding main text or respective figure legends. All experiments were repeated at least three times with different cell passage numbers, with at least three different individuals (OFs obtained from three or more different donors).
Immunofluorescence
OFs (1 × 104) were seeded in 20-mm confocal dishes, and adhered to the bottom after 12 hours. The OFs were rinsed with PBS and fixed with 4% paraformaldehyde for 20 minutes, then blocked and permeabilized in a mixture containing 3% BSA, 0.3% Triton X-100, and 5% normal donkey serum in PBS for 1 hour at room temperature. Afterward, the OFs were subjected to overnight incubation with the primary antibody, followed by incubation with the secondary antibody labeled with either Alexa Fluor 488 or Alexa Fluor 594 (Thermo Fisher Scientific, Waltham, MA, USA) for 2 hours at room temperature. After being counterstained with 4′,6-diamidino-2-phenylindole (DAPI), the OFs were visualized and photographed using a confocal microscope (Carl Zeiss 880, Oberkochen, Germany).
RNA Extraction and Quantitative Real-Time PCR
Total RNA from cultured OFs was isolated using an RNA Quick Purification kit (RN001, Esscience, Shanghai, China) and then reverse transcribed into cDNAs using the Evo M‐MLV RT Kit (Accurate Biology, Hunan, China). The quantitative real-time PCR (qRT-PCR) was performed on a Roche LightCycler 480 (Roche, Basel, Switzerland) using SYBR Premix Pro Taq HS (Accurate Biology, Hunan, China). To determine relative expression levels, the 2−ΔΔCT method was used with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) used as the endogenous control. Primer sequences used for qRT-PCR are shown in Supplementary Table S2.
RNA Sequencing and Bioinformatic Analysis
Total RNA was extracted using TRIzol reagent (RNAiso Plus, Takara, Japan). After this step, a NanoDrop spectrophotometer (Thermo Fisher Scientific) was used to assess the concentration, quality, and integrity of RNA. A total amount of 3 µg RNA per sample was used as input material for the library preparations. The DNA libraries were sequenced on NovaSeq 6000 platform (Illumina) Shanghai Personal Biotechnology Cp. Ltd. Hisat2 (v2.0.5) was used for aligning filtered reads to the reference genome sequence. HTSeq (version 0.9.1) was used for the analysis of Read Count values, providing the original expression levels of the genes. Subsequently, the expression levels were standardized using FPKM. Follow-up analyses were carried out using R software (The R Foundation for Statistical Computing, Vienna, Austria).
Western Blotting
The cellular lysates were obtained using radioimmunoprecipitation assay buffer supplemented with protease and phosphatase inhibitors (all from EpiZyme, Shanghai, China). Protein concentration was measured using the bicinchoninic acid protein assay kit (Beyotime, Shanghai, China). Equal amounts of total protein were separated on sodium dodecyl sulfate–polyacrylamide gels (8% or 12%) and electrotransferred onto polyvinylidene fluoride membranes (Millipore, Darmstadt, Germany). To block any nonspecific binding, the membranes were incubated in QuickBlock Blocking Buffer (Genscript, Nanjing, China) for 15 minutes and subsequently incubated overnight with primary antibodies against α-smooth muscle actin (α-SMA), collagen type I alpha 1 (COL1α1), connective tissue growth factor (CTGF), epidermal growth factor receptor (EGFR), extracellular signal-regulated kinase (ERK)1/2, p-ERK1/2, c-Jun N-terminal kinase (JNK), p-JNK, P38, p-P38 (all from CST, Beverly, MA, USA) and SOX9 (Abcam, Cambridge, MA, USA). GAPDH was used as endogenous control. After washing the membranes three times with tris buffered saline-TWEEN, they were incubated with horseradish peroxidase-conjugated anti-mouse or anti-rabbit secondary antibodies for 2 hours. Finally, the membrane was visualized by incubation with a chemiluminescence Western blot kit (Millipore) and captured using a chemiluminescence imager (Tanon Science & Technology Co., Ltd., Shanghai, China). Immunofluorescence images were analyzed using ImageJ software (NIH, Bethesda, MD, USA).
Wound Healing Assay
Wound healing assay was detected using the ibidi Culture-Insert 2 well (ibidi GmbH, Munich, Germany). OFs were cultured on Culture-Insert 2 well at a density of 2 × 104 cells/well. After 12 hours, the insert was removed and the medium was replaced with high-glucose Dulbecco's modified Eagle's medium (DMEM) with 1% fetal bovine serum. Micrographs were taken at 0 hour, 24 hours, and 48 hours to visualize and analyze the wound closure process.
Collagen Contraction Assay
The OFs in the logarithmic growth phase were treated with 0.25% trypsin and resuspended with serum-free DMEM medium to the final cell density of 2.5 × 105 cells/mL. A type-I collagen solution (Gibco, Carlsbad, CA, USA) mixture containing 250 µL of type I collagen solution, 32.5 µL of 10× DMEM, and 6.25 µL of 1N NaOH was prepared. The cell suspension (300 µL) was mixed with this collagen solution and then transferred into 24-well plates. The plates were incubated in a 37°C, 5% CO2 humidified incubator for 40 minutes to allow gelation. Subsequently, 500 µL of serum-free DMEM medium was added to each well and a pipette tip was used to release the collagen gel from the well. Photographs were taken at the time point (0 hour, 24 hours, and 48 hours) and analyzed using ImageJ software.
Apoptosis Assay
Apoptotic cells were visualized using a commercial TUNEL assay kit (Elabscience, Wuhan, China). Briefly, cells were fixed with 4% paraformaldehyde for 20 minutes and washed with PBS. Afterward, cells were treated with 0.2 % Triton X-100 for 10 minutes. Each sample was incubated in 100 µL TdT equilibration buffer for 30 minutes at 37°C. The newly configured TUNEL working solution was used for staining, and the incubation was carried out for 60 minutes at 37°C stored under light protection. Finally, nuclei were stained with DAPI. The number of apoptotic cells was indicated as the ratio of TUNEL-positive cells to the total nuclear number in each field.
Proliferation Assay
Proliferation experiments were analyzed using the 5-ethynyl-2′-deoxyuridine (EdU) Kit (C0078S, Beyotime). Cells were seeded into 48-well plates for 24 hours before the experiments and then incubated with EdU reagent (1:1000 dilution) for 2 hours. Cells were fixed with 4% formaldehyde for 15 minutes and permeabilized with 0.3% Triton X-100 for 15 minutes. After that, fluorescent dye and Hoechst were used to stain cells.
Lentiviral Construct and Overexpression
SOX9 overexpression lentivirus and control lentivirus were purchased from Shanghai Jikai Gene Co., Ltd. Lentiviral transduction of OFs was performed at a multiplicity of infection of 100. Selection of stably transduced OFs was carried out using puromycin
RNA Interference
For gene knockdown, OFs were transfected with small interfering RNA (siRNA) purchased from RiboBio (Guangzhou, China). Transfection of siRNAs was conducted using lipo3000 (Thermo Fisher Scientific) as per the manufacturer's instructions.
Chromatin Immunoprecipitation (ChIP) Coupled With qPCR Assay
ChIP assays were performed using a ChIP Assay Kit (KT101-02, gzscbio, Guangzhou, China) following the manufacturer's protocol. Cells were cross-linked by adding 1% formaldehyde at room temperature. Then, cells were lysed and digested by micrococcal nuclease. Immunoprecipitation was carried out using either an anti-SOX9 antibody or control IgG. After washing the immunoprecipitates, DNA/antibody complexes were eluted twice with IP elution buffer. Finally, the eluted DNA was purified and analyzed by qPCR. Primers listed in Supplementary Table S2 are used in ChIP-qPCR assay.
Statistical Analyses
Statistical analysis and figure drawing were performed using GraphPad Prism version 9.5.0 (GraphPad Software, Inc., La Jolla, CA, USA). Data are presented as mean ± SD. The Student t test was used to analyze normally distributed continuous data between two groups, and the Mann–Whitney test was used for non-normally distributed continuous data. When comparing multiple groups, one-way ANOVA or the Kruskal–Wallis test was selected based on whether the data were normally distributed. A P value of less than 0.05 was regarded as statistically significant.
Results
Cell Identification
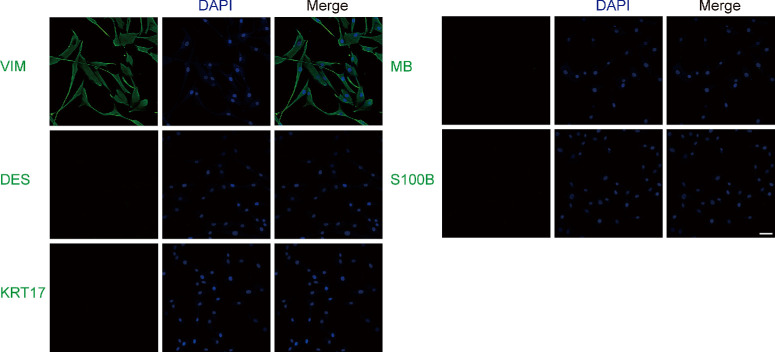
Primary cells from healthy controls and TED patients’ orbital connective tissues were cultured for the subsequent experiments. Cells expressing vimentin (VIM) indicated mesoderm differentiation, such as fibroblasts. The positive expression of VIM was observed in all cells, suggesting that they were fibroblasts. In addition, the cells stained negatively for desmin (DES), keratin (KRT17), myoglobin, and S100B, excluding the smooth muscle cells and cardiomyocytes, skin cells, muscle and striated muscle, nerve cells and skin melanocytes (Fig. 1). Therefore, these primary cells were identified as fibroblasts as they were positively marked with VIM and negatively marked with DES, KRT17, myoglobin, and S100B.
Figure 1.
Immunofluorescence staining showed that primary cells were stained positive for VIM, whereas negative for DES, keratin 17 (KRT17), myoglobin (MB), and S100B. Scale bar, 50 µm.
SOX9 Is Highly Expressed in OFs Cultured From Patients With TED
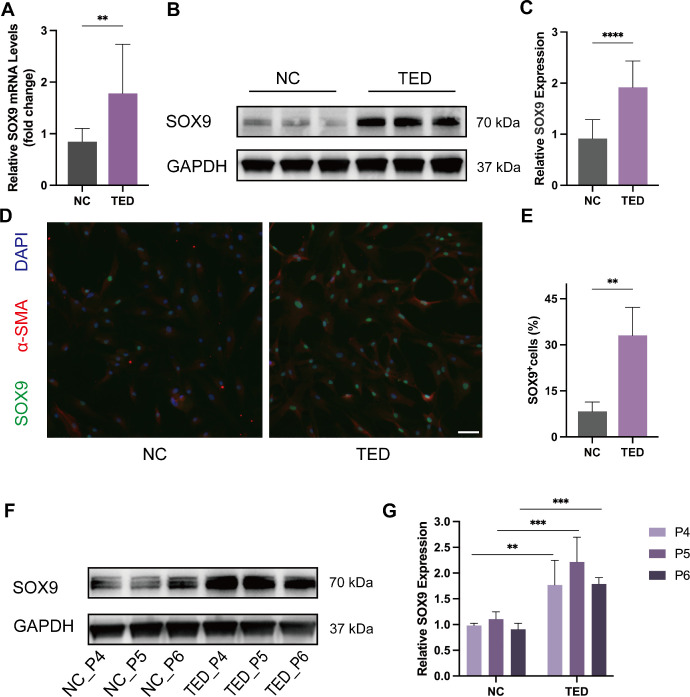
RNA was extracted from OFs derived from TED patients (n = 10) and healthy controls (n = 6), and SOX9 transcript levels were compared between the two groups using qRT-PCR. The qRT-PCR results showed that the expression of SOX9 mRNA was greater in the TED group compared with the control group (Fig. 2A). SOX9 protein expression was analyzed and quantified by Western blotting, and GAPDH served as the endogenous control. SOX9 protein expression was significantly higher in OFs from patients with TED (n = 10) than those from controls (n = 6) (Figs. 2B, 2C). Similarly, immunofluorescence staining revealed more SOX9-positive OFs in the TED group (n = 3) than in the control group (n = 3) (Figs. 2D, 2E). These results suggested that SOX9 is up-regulated in TED-OFs. To explore the effect of passages on the expression of SOX9, we analyzed the expression of SOX9 in the P4, P5, and P6 passages in OFs of the two groups (n = 3). The expression of SOX9 remained relatively constant through passages in OFs cultured from healthy controls and patients with TED. It is worth mentioning that OFs obtained from TED patients expressed a higher amount of SOX9 compared with those obtained from controls even as the passage number increased (Figs. 2F, 2G).
Figure 2.
SOX9 is highly expressed in OFs cultured from patients with TED compared with those from NCs. (A) The expression of SOX9 mRNA in OFs obtained from the TED (n = 10) and NC (n = 6) was quantified by qRT-PCR. (B) Western blot analyses were conducted to compare the protein expression of SOX9 in OFs obtained from the TED (n = 10) and NC (n = 6). (C) Quantification and statistical analysis of relative protein expression. (D) Representative immunofluorescence pictures of OFs stained with SOX9 (green), α-SMA (red), and DAPI (blue). Scale bar, 100 µm (TED, n = 3; NC, n = 3). (E) Statistical analysis of the percentage of SOX9-positive OFs per visual field. (F) The expression of SOX9 in the P4, P5, and P6 passages in OFs cultured from the TED (n = 3) and NC (n = 3). (G) Quantification and statistical analysis of relative protein expression. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
SOX9 Is a Positive Regulator of OFs Migration, Survival, Transformation, and Proliferation
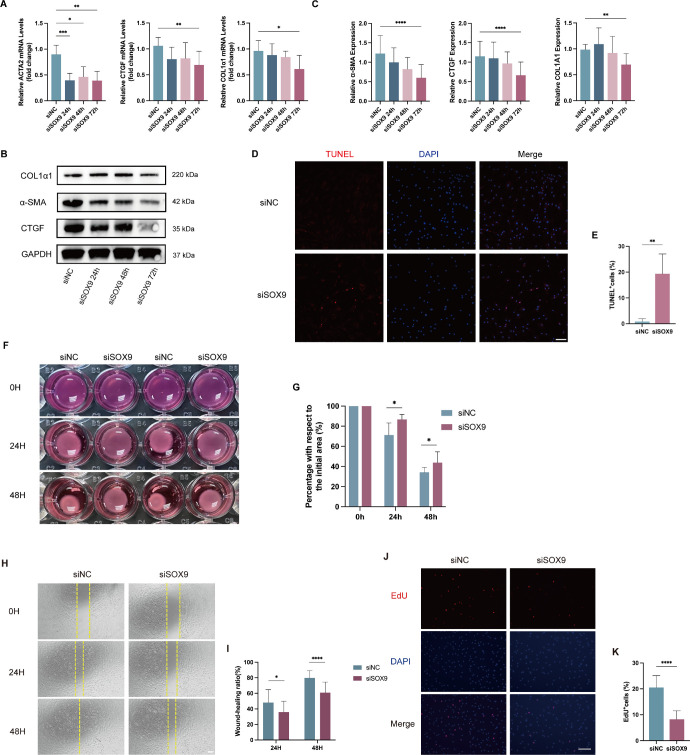
To evaluate the effect of SOX9 on fibrogenesis, we knocked down SOX9 expression by siRNA. OFs cultured from patients with TED were transfected with NC-siRNA or SOX9-siRNA for 24 hours, 48 hours, and 72 hours, then replaced the growth medium and cultured for an additional 24 hours with 10 ng/mL TGF-β1 (n = 6). The knockdown efficiency reached 40% at 24 hours, 70% at 48 hours, and 79% at 72 hours in OFs transfected with SOX9-siRNA compared with those transfected with NC-siRNA (Supplementary Fig. S1). ECM-related gene expression including COL1α1, ACTA2, and CTGF gradually decreased as the knockdown efficiency increased (Figs. 3A–C). TUNEL assay, collagen gel contraction assay, scratch migration assay, and EdU assay were carried out in OFs at 72 hours after transfection (n = 6). The results of TUNEL staining demonstrated that in contrast with the control group, more TUNEL-positive OFs were observed in SOX9 knockdown group (Figs. 3D, 3E). In the collagen gel contraction assay, SOX9 knockdown led to a significant reduction in fibroblast contractile ability, comparable to the control siRNA knockdown (Figs. 3F, 3G). The scratch migration assay showed that migrative capacity was impaired in SOX9 knockdown group (Figs. 3H, 3I). Furthermore, the EdU assay revealed that SOX9 knockdown markedly decreased the percentage of EdU‐positive cells (Figs. 3J, 3K).
Figure 3.
SOX9 knockdown attenuated the expression of ECM-related genes, the antiapoptotic effect, and the ability of migration, proliferation and collagen contraction in OFs cultured from patients with TED. OFs were transfected with SOX9-siRNA or control-siRNA for 24 hours, 48 hours, and 72 hours, then replaced the growth medium and cultured for an additional 24 hours with 10 ng/mL TGF-β1 (n = 6). (A) qRT-PCR results of the relative mRNA expression levels of ECM-related genes (COL1α1, ACTA2, and CTGF) after SOX9 knockdown. (B) Western blot results showed the relative protein expression of ECM-related proteins (COL1α1, α-SMA, and CTGF) after SOX9 knockdown. (C) Quantification and statistical analysis of relative protein expression. (D) TUNEL assay was carried out in OFs at 72 hours after transfection. Scale bar, 100 µm. (E) Statistical analysis of the percentage of TUNEL-positive OFs per visual field. (F) The representative images of the ability of collagen contraction of OFs at 0 hour, 24 hours, 48 hours after 72 hours after transfection. (G) Statistical analysis of the percentage contracted gel area relative to the initial area at different time points. (H) Representative scratch assay images of OFs at 0 hours, 24 hours, and 48 hours after 72 hours after transfection. Scale bar, 200 µm. (I) Statistical analysis of cell migration at different time points in the scratch wound healing assays. (J) EdU incorporation assay on OFs transfected with SOX9-siRNA or control-siRNA for 72 hours. Scale bar, 100 µm. (K) Statistical analysis of the percentage of EdU-positive OFs per visual field. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
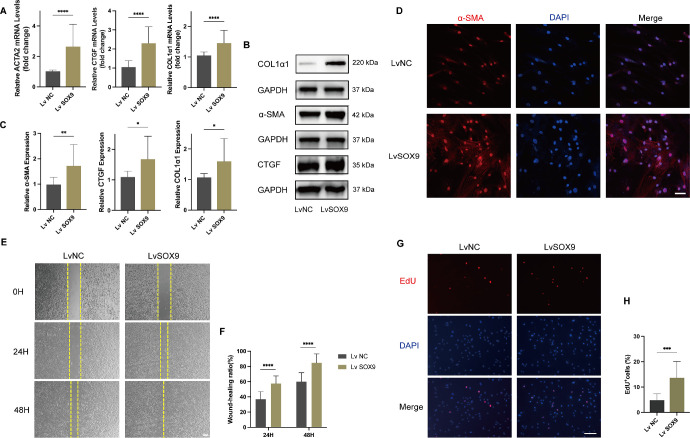
We then constructed SOX9 overexpressing system using a lentivirus vector. OFs cultured from patients with TED (n = 3) were infected with SOX9 overexpression lentivirus or control lentivirus (multiplicity of infection = 100), and qRT‐PCR and Western blot assay validated the overexpression efficiency (Supplementary Fig. S2). After successful SOX9 overexpression was confirmed, OFs were exposed to TGF-β1 (10 ng/mL) for 24 hours. As expected, the expression of COL1α1, α-SMA, and CTGF was significantly elevated in OFs after the overexpression of SOX9 (Figs. 4A–C). Immunofluorescence staining revealed that the expression of myofibroblast marker α-SMA was more intense in fibroblasts upon overexpression of SOX9 (Fig. 4D). The wound healing assay showed that cell mobility was significantly increased in SOX9 overexpression OFs, compared with the control overexpression OFs (Figs. 4E, 4F). Consistently, the EdU assay revealed that SOX9 overexpression increased EdU incorporated cell proportion (Figs. 4G, 4H).
Figure 4.
Overexpression of SOX9 promoted the transformation, migration, and proliferation of OFs. OFs obtained from patients with TED were transfected with SOX9 overexpression lentivirus or negative lentivirus and then exposed to TGF-β1 (10 ng/mL) for 24 hours (n = 6). (A) qRT-PCR was performed to detect the expression of ECM-related genes (COL1α1, ACTA2, and CTGF) transcripts in OFs after SOX9 was overexpressed. (B) The expression of ECM-related proteins (COL1α1, α-SMA, and CTGF) was detected by western blot assays. (C) Quantification and statistical analysis of relative protein expression. (D) Representative confocal images of OFs stained with α-SMA (red) and DAPI (blue) after SOX9 overexpression. Scale bar, 50 µm. (E) Representative scratch assay images at 0 hour, 24 hours, and 48 hours. Scale bar, 200 µm. (F) Statistical analysis of cell migration at different time points in the scratch wound healing assays. (G) Evaluation of OFs proliferation by EdU incorporation assay. Scale bar, 100 µm. (H) Statistical analysis of the percentage of EdU-positive OFs per visual field. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
SOX9 Regulates the MAPK/ERK1/2 Pathway During Fibrosis
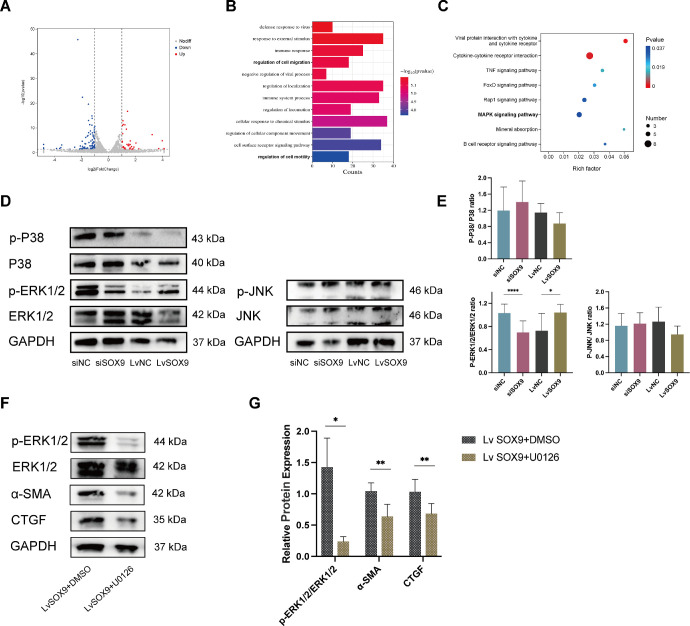
To further demonstrate the mechanism of SOX9 in orbital fibrosis, we performed transcriptome sequencing on OFs transfected with SOX9-siRNA in comparison with those transfected with NC-siRNA for 72 hours (n = 3). Differentially expressed genes (DEGs) were visually represented using volcano plots (Fig. 5A). The top 10 up-regulated and down-regulated genes are listed in Supplementary Table S3. Gene Ontology enrichment analysis revealed that these DEGs were predominantly enriched in biological processes such as regulation of cell migration and regulation of cell motility (Fig. 5B). The MAPK signaling pathways were identified as one of the top-enriched Kyoto Encyclopedia of Genes and Genomes pathways (Fig. 5C). To verify the involvement of the MAPK signaling pathways, total and phosphorylated protein of three MAPK subfamilies (p-ERK1/2, ERK1/2, p-P38, P38, p-JNK, and JNK) were explored in the SOX9 knockdown and SOX9 overexpressing systems after 1 hours of TGF-β1 (10 ng/mL) stimulation (n = 3). SOX9 knockdown abrogated ERK1/2 phosphorylation, while overexpression of SOX9 exhibited the opposite effect. However, overexpression or knockdown of SOX9 did not affect the phosphorylation of P38 and JNK (Figs. 5D, 5E). To further support the involvement of the ERK 1/2 pathway, we used U0126 (CST, Beverly, MA, USA), a highly selective inhibitor of MAPK kinase 1/2 acting upstream ERK1/2. OFs (n = 3) were treated with 10 ng/mL TGF-β1 for 90 minutes with pretreatment with 20 µM U0126 or vehicle (0.1% DMSO) for 1 hour. The inhibitor of ERK1/2, U0126, significantly decreased the expression of ECM-related genes caused by SOX9 overexpression (Figs. 5F, 5G). These data further confirmed that SOX9 promoted orbital fibrosis by regulating the MAPK/ERK1/2 signaling pathway.
Figure 5.
SOX9-mediated the phosphorylation of ERK1/2 but not P38 and JNK. (A) Volcano plot showing the DEGs with the knockdown of SOX9 in OFs cultured from patients with TED by RNA sequencing (n = 3). (B) Gene Ontology (GO) term enrichment analysis of DEGs. (C) Signaling pathways in Kyoto Encyclopedia of Genes and Genomes (KEGG) database enrichment analysis. (D) OFs cultured from TED patients were transfected with siRNA or lentivirus prior to stimulation with 10 ng/mL TGF-β1 for 1 hour (n = 3). Protein levels of p-P38, P38, p-ERK1/2, ERK1/2, p-JNK, JNK, and GAPDH were determined by Western blot. (E) Quantification and statistical analysis of relative protein expression. (F) SOX9-overexpressing OFs were treated with 10 ng/mL TGF-β1 for 90 minutes with pretreatment with 20 µM U0126 or vehicle (0.1% DMSO) for 1 hour (n = 3). The expression of ECM-related proteins (α-SMA and CTGF) was detected by western blot assays. (G) Quantification and statistical analysis of relative protein expression. *P < 0.05, **P < 0.01, ****P < 0.0001.
EGFR Was a Downstream Target Gene of SOX9 in Regulating Fibrosis
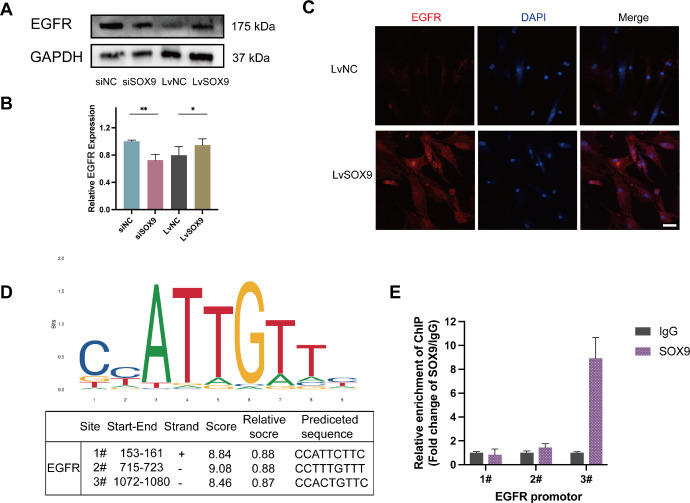
Key downstream target genes were identified by taking the intersection of DEGs with predicted target genes generated by CHEA, JASPAR, and MotifMap datasets (https://maayanlab.cloud/Harmonizome/). The intersection results are shown in Supplementary Figure S3 in the Supplementary file. EGFR is one of the notably down-regulated genes in our RNA sequencing results (Supplementary Table S3). After transfection with siRNA or lentivirus, the OFs were stimulated with 10 ng/mL TGF-β1 for 24 hours (n = 3). Immunoblotting data illustrated that overexpression of SOX9 led to an increase in EGFR expression, whereas knockdown of SOX9 impeded this induction, which suggested that the expression of EGFR was regulated by SOX9 in fibroblasts (Figs. 6A, 6B). The EGFR immunofluorescence intensity was significantly higher in the SOX9 overexpression group compared with the control group (Fig. 6C). The binding sites of SOX9 in the potential promoter region of EGFR were predicted by JASPAR website (http://jaspar.Genereg.net/) (Fig. 6D). The ChIP-qPCR results demonstrated that EGFR promoter region #3 (−1072 to −1080 before the TSS) was the most abundant among regions enriched by the anti-SOX9 antibody (Fig. 6E).
Figure 6.
EGFR is a downstream target of SOX9. After transduction with siRNA or lentivirus, the OFs cultured from TED were stimulated with 10 ng/mL TGF-β1 for 24 hours (n = 3). (A, B) Representative images of Western blotting results and the quantitative analysis of EGFR in OFs treated with siRNA or lentivirus. (C) Immunofluorescence showing the EGFR expression with SOX9 overexpression in OFs scale bar, 50 µm. (D) JASPAR websites predicted the possible binding sequences of SOX9 to the EGFR promoter region. (E) Enrichment of SOX9 protein on the EGFR promoter was determined by ChIP-qPCR assay. *P < 0.05, **P < 0.01.
Discussion
In the current study, SOX9 was aberrantly up-regulated in the OFs cultured from patients with TED. We revealed that SOX9 is a positive regulator of migration, survival, differentiation, and proliferation in TED-OFs by SOX9 knockdown and SOX9-overexpressing systems. SOX9 might exert its profibrotic effects partially by activating the MAPK/ERK1/2 pathway, and partially by activation of the downstream target gene EGFR.
SOX9 was found to be overexpressed in tissues or cells in a number of human fibrotic diseases.12,15,23 In line with those studies, we also observed higher SOX9 expression in TED-OFs compared with those from controls, even as the passage number increased. It has been remarked on several occasions that TED-OFs exhibit different characteristics compared with OFs from healthy controls. In addition to highly expressing some cell surface molecules such as CD40 and Thy-1,24,25 TED-OFs exhibited stronger proliferative activity when stimulated with certain cytokines and growth factors.26 We suggest that TED-OFs show hyper-responsiveness to certain cytokines and growth factors might be a key mechanism in the pathogenesis of TED. Furthermore, high expression of these proteins, such as SOX9, CD40, and Thy-1, may be responsible for the hyper-responsiveness. Crucial points of fibrosis involved the aberrant conversion of fibroblasts into α-SMA–expressing myofibroblasts and increased collagen deposition.27,28 We investigated the in vitro effect of SOX9 in TGF-β1–induced myofibroblast transdifferentiation. SOX9 knockdown caused down-regulation of TGF-β1 induced α-SMA, COL1α1, and CTGF expression, whereas overexpression resulted in an increase, suggesting SOX9 promoted myofibroblast transdifferentiation and ECM remodeling. Activated fibroblasts are hyper-proliferative, hyper-secretory, migrative,29 and resistant to apoptosis.30 Our experiments showed that SOX9 deficiency attenuated the contraction, survival, proliferation, and migration capacity of OFs, whereas SOX9 overexpression increased the differentiation, migration, and proliferation of OFs. The present results demonstrated that SOX9 is an important factor in the promotion of OFs activation and deposition of ECM during the orbital fibrotic process. Additionally, our study adds to evidence that SOX9 can drive fibrosis in multiple organs.
To probe the underlying mechanisms by which SOX9 enforces fibrosis, we conducted an RNA-sequencing analysis of SOX9 knockdown in TED-OFs and identified gene regulatory networks and biological processes. Functional enrichment analysis revealed that DEGs were mainly enriched in the regulation of migration and cell motility and immune system process. Kyoto Encyclopedia of Genes and Genomes pathways identified modulation of MAPK pathways by SOX9 in the process of fibrosis. TGF-β–induced myofibroblast differentiation can be activated by Smad signaling or non-Smad signaling pathways, such as MAPK pathways (including ERK1/2 and JNK ) and phosphatidylinositol 3-kinase–Akt pathway.31 MAPK pathways regulate diverse cellular activities in multiple cell types, including proliferation, differentiation, and apoptosis.32 Numerous experiments demonstrated that antifibrotic drugs exerted their effect through MAPK signaling pathways.33–35 Simvastatin inhibits myofibroblast differentiation induced by TGF-β through suppression of the Ras homolog family member A/Rho-associated protein kinase/ERK and p38 MAPK signaling pathways, what's more, inhibitors of p38 and ERK reduced the α-SMA expression mediated by TGF-β.36 Intercellular cell adhesion molecule-1 induced by CD40L, which is expressed in several types and associated with various inflammatory and autoimmune diseases,37 was blocked by the ERK1/2, p38, and JNK antagonists in fibroblast of TED.38 Gajjala et al.12 clarified that SOX9 was up-regulated via MAPK/phosphatidylinositol 3-kinase–dependent signaling in idiopathic pulmonary fibrosis; furthermore, they found that transcription factor Wilms’ tumor 1 may directly bind to SOX9 promoter enhancing the fibroblast activation. We observed that the overexpression of SOX9 markedly promoted the phosphorylation of ERK1/2, whereas its knockdown yielded the opposite effect. However, SOX9 expression did not affect the phosphorylation of P38 and JNK. Using specific inhibitors of the ERK1/2 pathway attenuated the expression of profibrotic proteins induced by SOX9 overexpression. The ERK1/2 pathway is predominantly implicated in the regulation of growth and proliferation, whereas the P38 and JNK pathways are activated by different stress-inducing stimuli and play vital roles in inflammation, apoptosis, and stress responses.39–41 Most likely, SOX9 activates fibroblast and promotes its growth and proliferation through the MAPK/ERK1/2 signaling pathway in fibrosis induced by TGF-β1. We demonstrated that ERK1/2, but not p38 and JNK, was implicated in the fibrotic process mediated by SOX9.
Several studies have demonstrated that SOX9 promotes fibrosis by regulating downstream genes.11,42 Osteopontin is a vital component of the ECM with profibrotic and proinflammatory effects.43–45 SOX9 combines with a conserved upstream region of the osteopontin gene and modulates its expression, which in turn contributes to fibrosis progression.42 Interestingly, we found that EGFR was dramatically down-regulated after SOX9 knockdown by high-throughput sequencing analysis. By using the ChIP-qPCR assay, we further confirmed that SOX9 can directly bind to the EGFR promoter. EGFR belongs to the family of the ErbB tyrosine kinase receptors, which is a critical regulator of cellular growth and proliferation.46,47 Activation of the EGFR can phosphorylate multiple effector proteins, triggering multiple downstream signal pathways, including the MAPK/ERK1/2 pathway and the PIK3 (phosphatidylinositol 3-kinase)/Akt pathway, in turn a series of tissue regeneration and wound healing processes.48,49 Growing evidence indicates the pivotal role of EGFR in the pathophysiology of hyperproliferative diseases, such as cardiac fibrosis and lung fibrosis.50,51 Suppression of EGFR signaling promotes liver progenitor cells mediated liver regeneration through the MAPK kinase–ERK–SOX9 cascade in vitro.52 A pan-EGFR inhibitor (afatinib) down-regulated forkhead box protein A2 transcript, which in turn decreased the expression of SOX9.53 Previous experiments indicated that SOX9 is regulated positively by EGFR through activating specific mediators. Combined with previous research results, it is speculated that SOX9 and EGFR positively co-regulate with each other, and activate fibroblasts gradually induce the development of orbital fibrosis through SOX9-EGFR circuitry. SOX9 might exert profibrotic effects either through activating MAPK/ERK/2 pathway or by binding to the downstream target gene EGFR.
Taken together, we confirmed by SOX9 knockdown and overexpression experiments that SOX9 promoted OFs activation via MAPK/ERK1/2 pathway or activating the downstream target gene EGFR, which enhanced fibrosis progression. Our data provide insight into the roles of SOX9 in the regulation of orbital fibrosis and may serve as a potential therapeutic target or as a useful starting point to identify downstream targets for the discovery of future drug candidates.
Supplementary Material
Acknowledgments
Supported by the Guangdong Provincial Natural Science Foundation (2021A1515010467), Guangdong Basic and Applied Basic Research Foundation (2019A1515110012), and the National Natural Science Foundation of China (82201200).
Disclosure: M. Zhou, None; B. Lin, None; P. Wu, None; Y. Ke, None; S. Huang, None; F. Zhang, None; X. Hei, None; Z. Mao, None; X. Li, None; P. Wan, None; T. Chen, None; H. Yang, None; D. Huang, None
References
- 1. Taylor PN, Zhang L, Lee RWJ, et al.. New insights into the pathogenesis and nonsurgical management of Graves orbitopathy. Nat Rev Endocrinol. 2020; 16: 104–116. [DOI] [PubMed] [Google Scholar]
- 2. Bahn RS. Graves’ ophthalmopathy. N Engl J Med. 2010; 362: 726–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dik WA, Virakul S, van Steensel L.. Current perspectives on the role of orbital fibroblasts in the pathogenesis of Graves’ ophthalmopathy. Exp Eye Res. 2016; 142: 83–91. [DOI] [PubMed] [Google Scholar]
- 4. Smith TJ, Hegedüs L, Douglas RS.. Role of insulin-like growth factor-1 (IGF-1) pathway in the pathogenesis of Graves’ orbitopathy. Best Pract Res Clin Endocrinol Metab. 2012; 26: 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Neag EJ, Smith TJ.. 2021 update on thyroid-associated ophthalmopathy. J Endocrinol Invest. 2022; 45: 235–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ludgate M. Fibrosis in dysthyroid eye disease. Eye (Lond). 2020; 34: 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cockerham KP, Padnick-Silver L, Stuertz N, Francis-Sedlak M, Holt RJ.. Quality of life in patients with chronic thyroid eye disease in the United States. Ophthalmol Ther. 2021; 10: 975–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ponto KA, Hommel G, Pitz S, Elflein H, Pfeiffer N, Kahaly GJ.. Quality of life in a German graves orbitopathy population. Am J Ophthalmol. 2011; 152: 483–490.e481. [DOI] [PubMed] [Google Scholar]
- 9. Ming Z, Vining B, Bagheri-Fam S, Harley V.. SOX9 in organogenesis: shared and unique transcriptional functions. Cell Mol Life Sci. 2022; 79: 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lacraz GPA, Junker JP, Gladka MM, et al.. Tomo-Seq identifies SOX9 as a key regulator of cardiac fibrosis during ischemic injury. Circulation. 2017; 136: 1396–1409. [DOI] [PubMed] [Google Scholar]
- 11. Raza S, Jokl E, Pritchett J, et al.. SOX9 is required for kidney fibrosis and activates NAV3 to drive renal myofibroblast function. Sci Signal. 2021; 14: eabb4282. [DOI] [PubMed] [Google Scholar]
- 12. Gajjala PR, Kasam RK, Soundararajan D, et al.. Dysregulated overexpression of Sox9 induces fibroblast activation in pulmonary fibrosis. JCI Insight. 2021; 6:e152503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Scharf GM, Kilian K, Cordero J, et al.. Inactivation of Sox9 in fibroblasts reduces cardiac fibrosis and inflammation. JCI Insight. 2019; 5: e126721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shu DY, Lovicu FJ.. Myofibroblast transdifferentiation: the dark force in ocular wound healing and fibrosis. Prog Retin Eye Res. 2017; 60: 44–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang H, Chen Y, Zhao S, Wang X, Lu K, Xiao H.. Effect of Sox9 on TGF-β1-mediated atrial fibrosis. Acta Biochim Biophys Sin (Shanghai). 2021; 53: 1450–1458. [DOI] [PubMed] [Google Scholar]
- 16. Zavadil J, Bitzer M, Liang D, et al.. Genetic programs of epithelial cell plasticity directed by transforming growth factor-beta. Proc Natl Acad Sci USA. 2001; 98: 6686–6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xie L, Law BK, Chytil AM, Brown KA, Aakre ME, Moses HL.. Activation of the Erk pathway is required for TGF-beta1-induced EMT in vitro. Neoplasia. 2004; 6: 603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Song YW, Zhang T, Wang WB.. Gluococorticoid could influence extracellular matrix synthesis through Sox9 via p38 MAPK pathway. Rheumatol Int. 2012; 32: 3669–3673. [DOI] [PubMed] [Google Scholar]
- 19. Hoffman LM, Weston AD, Underhill TM.. Molecular mechanisms regulating chondroblast differentiation. J Bone Joint Surg Am. 2003; 85-A(Suppl 2): 124–132. [DOI] [PubMed] [Google Scholar]
- 20. Samart P, Rojanasakul Y, Issaragrisil S, Luanpitpong S.. A novel E-cadherin/SOX9 axis regulates cancer stem cells in multiple myeloma by activating Akt and MAPK pathways. Exp Hematol Oncol. 2022; 11: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Murakami S, Kan M, McKeehan WL, de Crombrugghe B.. Up-regulation of the chondrogenic Sox9 gene by fibroblast growth factors is mediated by the mitogen-activated protein kinase pathway. Proc Natl Acad Sci USA. 2000; 97: 1113–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu PS, Lin BY, Huang SY, et al.. IL-11 is elevated and drives the profibrotic phenotype transition of orbital fibroblasts in thyroid-associated ophthalmopathy. Front Endocrinol. 2022; 13: 846106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Athwal VS, Pritchett J, Martin K, et al.. SOX9 regulated matrix proteins are increased in patients serum and correlate with severity of liver fibrosis. Sci Rep. 2018; 8: 17905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Khoo TK, Coenen MJ, Schiefer AR, Kumar S, Bahn RS.. Evidence for enhanced Thy-1 (CD90) expression in orbital fibroblasts of patients with Graves’ ophthalmopathy. Thyroid. 2008; 18: 1291–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hwang CJ, Afifiyan N, Sand D, et al.. Orbital fibroblasts from patients with thyroid-associated ophthalmopathy overexpress CD40: CD154 hyperinduces IL-6, IL-8, and MCP-1. Invest Ophthalmol Vis Sci. 2009; 50: 2262–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heufelder AE, Bahn RS.. Modulation of Graves’ orbital fibroblast proliferation by cytokines and glucocorticoid receptor agonists. Invest Ophthalmol Vis Sci. 1994; 35: 120–127. [PubMed] [Google Scholar]
- 27. Eyden B. The myofibroblast: phenotypic characterization as a prerequisite to understanding its functions in translational medicine. J Cell Mol Med. 2008; 12: 22–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Darby I, Skalli O, Gabbiani G.. Alpha-smooth muscle actin is transiently expressed by myofibroblasts during experimental wound healing. Lab Invest. 1990; 63: 21–29. [PubMed] [Google Scholar]
- 29. Lehmann GM, Feldon SE, Smith TJ, Phipps RP.. Immune mechanisms in thyroid eye disease. Thyroid. 2008; 18: 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sgalla G, Iovene B, Calvello M, Ori M, Varone F, Richeldi L.. Idiopathic pulmonary fibrosis: pathogenesis and management. Respir Res. 2018; 19: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang YE. Non-Smad signaling pathways of the TGF-β family. Cold Spring Harb Perspect Biol. 2017; 9: a022129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Seger R, Krebs EG.. The MAPK signaling cascade. Faseb J. 1995; 9: 726–735. [PubMed] [Google Scholar]
- 33. Zeng J, Huang H, Zhang Y, et al.. Dapagliflozin alleviates renal fibrosis in a mouse model of adenine-induced renal injury by inhibiting TGF-β1/MAPK mediated mitochondrial damage. Front Pharmacol. 2023; 14: 1095487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ji P, Shi Q, Liu Y, et al.. Ginsenoside Rg1 treatment alleviates renal fibrosis by inhibiting the NOX4-MAPK pathway in T2DM mice. Ren Fail. 2023; 45: 2197075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang D, Deng B, Cheng L, et al.. A novel and low-toxic peptide DR3penA alleviates pulmonary fibrosis by regulating the MAPK/miR-23b-5p/AQP5 signaling axis. Acta Pharm Sin B. 2023; 13: 722–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wei YH, Liao SL, Wang SH, Wang CC, Yang CH.. Simvastatin and ROCK inhibitor Y-27632 inhibit myofibroblast differentiation of Graves’ ophthalmopathy-derived orbital fibroblasts via RhoA-mediated ERK and p38 signaling pathways. Front Endocrinol (Lausanne). 2020; 11: 607968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Roebuck KA, Finnegan A.. Regulation of intercellular adhesion molecule-1 (CD54) gene expression. J Leukoc Biol. 1999; 66: 876–888. [DOI] [PubMed] [Google Scholar]
- 38. Zhao LQ, Wei RL, Cheng JW, Cai JP, Li Y.. The expression of intercellular adhesion molecule-1 induced by CD40-CD40L ligand signaling in orbital fibroblasts in patients with Graves’ ophthalmopathy. Invest Ophthalmol Vis Sci. 2010; 51: 4652–4660. [DOI] [PubMed] [Google Scholar]
- 39. Johnson GL, Lapadat R.. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002; 298: 1911–1912. [DOI] [PubMed] [Google Scholar]
- 40. Phan T, Zhang XH, Rosen S, Melstrom LG.. P38 kinase in gastrointestinal cancers. Cancer Gene Ther. 2023; 30: 1181–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Evens L, Beliën H, Deluyker D, et al.. The Impact of Advanced Glycation End-Products (AGEs) on Proliferation and Apoptosis of Primary Stem Cells: a systematic review. Stem Cells Int. 2020; 2020: 8886612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pritchett J, Harvey E, Athwal V, et al.. Osteopontin is a novel downstream target of SOX9 with diagnostic implications for progression of liver fibrosis in humans. Hepatology. 2012; 56: 1108–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moorman HR, Poschel D, Klement JD, Lu C, Redd PS, Liu K.. Osteopontin: a key regulator of tumor progression and immunomodulation. Cancers (Basel). 2020; 12: 3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sabo-Attwood T, Ramos-Nino ME, Eugenia-Ariza M, et al.. Osteopontin modulates inflammation, mucin production, and gene expression signatures after inhalation of asbestos in a murine model of fibrosis. Am J Pathol. 2011; 178: 1975–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Merszei J, Wu J, Torres L, et al.. Osteopontin overproduction is associated with progression of glomerular fibrosis in a rat model of anti-glomerular basement membrane glomerulonephritis. Am J Nephrol. 2010; 32: 262–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Harskamp LR, Gansevoort RT, van Goor H, Meijer E.. The epidermal growth factor receptor pathway in chronic kidney diseases. Nat Rev Nephrol. 2016; 12: 496–506. [DOI] [PubMed] [Google Scholar]
- 47. Herbst RS. Review of epidermal growth factor receptor biology. Int J Radiat Oncol Biol Phys. 2004; 59: 21–26. [DOI] [PubMed] [Google Scholar]
- 48. Schramm F, Schaefer L, Wygrecka M.. EGFR signaling in lung fibrosis. Cells. 2022; 11: 986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jorissen RN, Walker F, Pouliot N, Garrett TP, Ward CW, Burgess AW.. Epidermal growth factor receptor: mechanisms of activation and signalling. Exp Cell Res. 2003; 284: 31–53. [DOI] [PubMed] [Google Scholar]
- 50. Martin R, Gutierrez B, Cordova C, et al.. Secreted phospholipase A(2)-IIA modulates transdifferentiation of cardiac fibroblast through EGFR transactivation: an inflammation-fibrosis link. Cells. 2020; 9: 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hardie WD, Davidson C, Ikegami M, et al.. EGF receptor tyrosine kinase inhibitors diminish transforming growth factor-alpha-induced pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2008; 294: L1217–L1225. [DOI] [PubMed] [Google Scholar]
- 52. So J, Kim M, Lee SH, et al.. Attenuating the epidermal growth factor receptor-extracellular signal-regulated kinase-sex-determining region Y-Box 9 axis promotes liver progenitor cell-mediated liver regeneration in Zebrafish. Hepatology. 2021; 73: 1494–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kaushik G, Seshacharyulu P, Rauth S, et al.. Selective inhibition of stemness through EGFR/FOXA2/SOX9 axis reduces pancreatic cancer metastasis. Oncogene. 2021; 40: 848–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.