Abstract
We investigate how matrix stiffness regulates chromatin reorganization and cell reprogramming and find that matrix stiffness acts as a biphasic regulator of epigenetic state and fibroblast-to-neuron conversion efficiency, maximized at an intermediate stiffness of 20 kPa. ATAC sequencing analysis shows the same trend of chromatin accessibility to neuronal genes at these stiffness levels. Concurrently, we observe peak levels of histone acetylation and histone acetyltransferase (HAT) activity in the nucleus on 20 kPa matrices, and inhibiting HAT activity abolishes matrix stiffness effects. G-actin and cofilin, the cotransporters shuttling HAT into the nucleus, rises with decreasing matrix stiffness; however, reduced importin-9 on soft matrices limits nuclear transport. These two factors result in a biphasic regulation of HAT transport into nucleus, which is directly demonstrated on matrices with dynamically tunable stiffness. Our findings unravel a mechanism of the mechano-epigenetic regulation that is valuable for cell engineering in disease modeling and regenerative medicine applications.
Matrix stiffness biphasically modulates histone acetylation and chromatin accessibility to enhance cell reprogramming.
INTRODUCTION
Biophysical factors such as the mechanical property and microtopography of cell adhesive substrates have been shown to regulate a variety of cellular functions such as migration, proliferation, and differentiation, which, in turn, can modulate wound healing, tissue remodeling, and tumor growth (1–10). There is also accumulative evidence that biophysical cues can be engineered to improve cell reprogramming efficiency (11–13), as exemplified by the effects of micro/nanotopography and nuclear deformation on the conversion of fibroblasts into induced neuronal (iN) cells (14–16). However, how matrix stiffness regulates the epigenetic state during cell reprogramming is not well understood. A better understanding of the epigenetic changes that occur during cell reprogramming in response to matrix stiffness would enable the engineering of biomaterials that can promote cell reprogramming, with the derived cells offering great potential for disease modeling, drug discovery, and tissue engineering applications (17–19).
Cell reprogramming involves extensive epigenetic changes to activate lineage-specific genes in heterochromatin and reorganize the chromatin structure (20), which is essential to change the cell memory and identity. Although the roles of biological and chemical factors in epigenetic changes during cell reprogramming have the widely studied, the effects of biophysical cues on cell reprogramming and the mechanotransduction to the nucleus are less well understood (21, 22). While recent studies have reported the effects of stiff and soft matrix on epigenetic changes in various cell types (23–25), the mechanotransduction mechanism from cell surface to nucleus is not clear. The actin cytoskeleton plays an important role in sensing and transducing extracellular biophysical signals to modulate intracellular signaling and cell functions (2, 26). Actin filaments can transmit mechanical signals to the nucleus to modulate gene expression and chromatin organization (22, 27, 28). On the other hand, actin polymerization/depolymerization can regulate G-actin–mediated transport of transcriptional factors such as myocardin-related transcription factor (MRTF) into the nucleus (29–32), and actin force generation has been reported to modulate nuclear pore size and Yes-associated protein (YAP) translocation (33). However, whether the transport of epigenetic enzymes that modulate histone acetylation, histone methylation, and DNA methylation is regulated by mechanisms similar to these transcriptional factors remains to be determined. Moreover, most studies of stiffness effects assume that stiffness regulation of cell functions are monotonic, leading to the simplification of experiments to compare two conditions (stiff versus soft) as representatives, which awaits further investigations.
In this study, we investigated the effect of matrix stiffness on the epigenetic state during iN reprograming by using polyacrylamide (PAAm) hydrogels, which are composed of a crosslinked acrylamide network where the stiffness of the hydrogel can be tuned by adjusting the amount of bis-acrylamide crosslinker and the acrylamide monomers during fabrication (34). In addition, PAAm hydrogels offer several advantages such as these hydrogels are easy to fabricate, biologically inert, allow for precise control and tuning of the stiffness during fabrication, offer a range of stiffness that covers most soft tissues, and have been extensively used by many researchers to study the effect of matrix stiffness on cell functions (34, 35). We reveal a role of matrix stiffness as a biphasic (instead of monotonic) regulator of epigenetic state and cell reprogramming through actin-mediated histone acetyltransferase (HAT) translocation into the nucleus, with the highest iN reprogramming at an intermediate stiffness, which is distinguishable from the soft surface known to facilitate neural differentiation (3).
RESULTS
Matrix stiffness enhances iN reprogramming efficiency in a biphasic manner
To investigate the role of matrix stiffness on iN reprogramming, adult mouse fibroblasts were transduced with doxycycline-inducible lentiviruses containing the three reprogramming factors Ascl1, Brn2, and Myt1l (BAM) and then seeded onto fibronectin-coated PAAm gels of various stiffness (40, 20, and 1 kPa) that mimic the physiological stiffness of the microenvironment where fibroblasts (36, 37) and neuronal cells (38, 39) typically reside in vivo (Fig. 1A). In addition, this stiffness range has been reported to regulate various cell functions and can be used to engineer cell behavior and reprogramming ex vivo. Glass coverslips coated with the same extracellular matrix (ECM) protein were used as a rigid surface control. We first confirmed that elastic modulus of the PAAm gels was in the desired range and found no major difference in the structural features or pore size of the gels upon examination with scanning electron microscopy (figs. S1 and S2). These substrates were nontoxic as there were no detrimental effects on cell viability when fibroblasts were cultured on the various surfaces (fig. S3). As shown in figs. S4 to S7, both the cell spreading area and nuclear volume of fibroblasts decreased with the stiffness of the PAAm gel, which appeared to correlate with a decrease in focal adhesions, e.g., paxillin-positive puncta (fig. S7). After fibroblasts were seeded on PAAm gels for 48 hours, the number of cells in the S phase of the cell cycle also decreased with matrix stiffness (figs. S8 and S9). These results suggest that matrix stiffness significantly affects fibroblast spreading and proliferation.
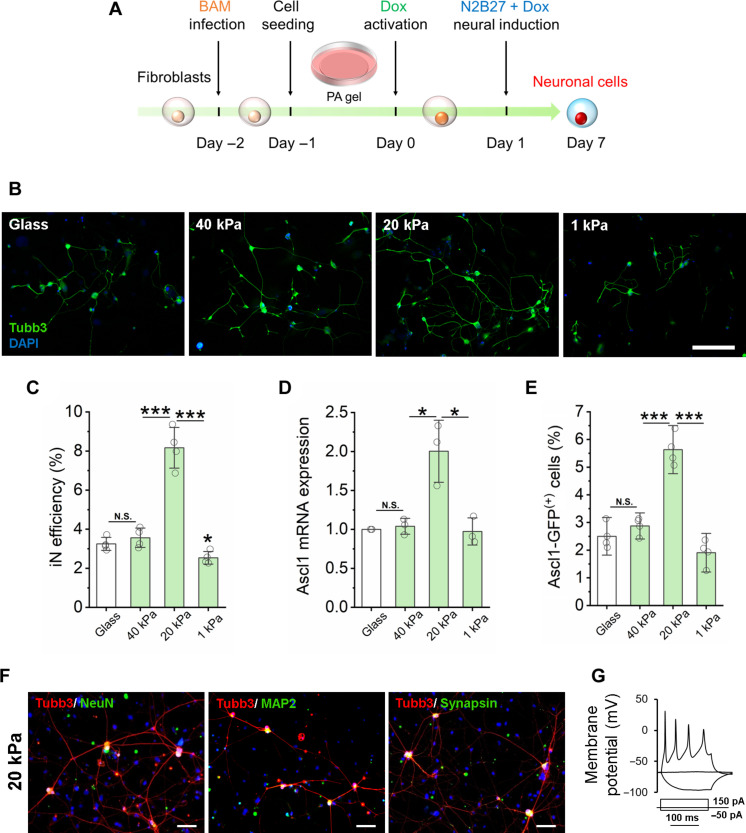
Fig. 1. Substrate stiffness-induced biphasic enhancement of iN reprogramming efficiency.
(A) Experimental timeline for substrate stiffness-induced iN reprogramming. (B) Immunofluorescent mages of Tubb3+ iN cells derived on glass or PAAm gels of varying stiffness on day 7. Scale bar, 200 μm. DAPI, 4′,6-diamidino-2-phenylindole. (C) Reprogramming efficiency of BAM-transduced fibroblasts that were cultured on glass and PAAm gels of varying stiffness (n = 4 independently prepared gel surfaces). On day 7, the cells were fixed and stained for Tubb3, followed by immunofluorescence microscopy to quantify Tubb3+ iN cells. (D) Relative Ascl1 mRNA expression in BAM-transduced fibroblasts seeded on glass and PAAm gels of varying stiffness at day 1 following Dox activation (n = 3). (E) Fibroblasts transduced with BAM and an Ascl1 promoter–GFP construct were seeded on glass and PAAm gels of varying stiffness for 24 hours and activated by Dox for another 24 hours. The cells were fixed and observed by immunofluorescence microscopy, showing that 20-kPa PAAm gel induced more Ascl1 promoter–GFP+ cells at day 1, as quantified in the bar graph (n = 3). (F) Representative images of Tubb3+ cells expressing mature neuronal markers, NeuN, MAP2, and synapsin at 21 days after cells were cultured on 20-kPa gels. Scale bars, 100 μm. (G) Representative trace showing spontaneous changes in membrane potential in response to current injection from iNs derived on 20-kPa gels. In (C) to (E), statistical significance was determined by a one-way analysis of variance (ANOVA) and Tukey’s multiple comparison test (N.S.: not significant, *P ≤ 0.05 and ***P ≤ 0.001). In (C) to (E), bar graphs show means ± SD.
To determine the effect of matrix stiffness on the direct conversion of fibroblasts into neurons, BAM-transduced fibroblasts were cultured on glass and PAAm gels of varying stiffness. After 7 days, cultures were fixed and immunostained for neuronal marker, neuron-specific class III β-tubulin (Tubb3) to identify iN cells and determine the reprogramming efficiency by counting Tubb3+ cells with a neuronal morphology (Fig. 1, A and B). Matrices of intermediate stiffness at 20 kPa significantly enhanced the reprogramming efficiency, while stiffer (40 kPa, glass) or softer (1 kPa) surfaces had modest effects (Fig. 1C). In addition, this biphasic enhancement of iN conversion at the intermediate stiffness is independent of ECM protein components (Fig. 1C and fig. S10). Intermediate stiffness (~20 kPa) significantly enhanced the mRNA level of pioneer factor Ascl1 compared to glass, 40- and 1-kPa gels (Fig. 1D). To determine whether matrix stiffness could modulate the activation of endogenous Ascl1, fibroblasts were transduced with an Ascl1 promoter–driven green fluorescent protein (GFP) construct and reprogrammed for 2 days. We found a significant increase in the number of Ascl1 promoter–GFP+ cells in the intermediate stiffness group compared to glass and other stiffness groups (Fig. 1E). Further characterization of the derived cells by immunostaining and electrophysiology (patch clamp) analysis revealed that mature iN cells were obtained on PAAm gels with various stiffness (Fig. 1, F and G, and figs. S11 and S12). Higher density of iN cells on 20 kPa surfaces also resulted in higher frequency of spontaneous excitatory postsynaptic currents (sEPSCs) (fig. S10C). Together, these results suggest that an intermediate matrix stiffness promotes endogenous Ascl1 expression that may enhance iN reprogramming.
Intermediate stiffness increases chromatin accessibility and histone acetylation for iN conversion
We postulated that matrix stiffness modulated the epigenetic state and, thus, the reprogramming process. To directly determine whether matrix stiffness altered chromatin accessibility, we performed assay of transposase accessible chromatin sequencing (ATAC-seq) in fibroblast in the absence of BAM reprogramming factors and found that cells on matrices with different stiffness (1 kPa, 20 kPa, and glass) had distinguishable chromatin accessibility regions (Fig. 2A and fig. S13). Principal components analysis indicated that the independent samples of fibroblasts cultured on the same stiffness were similar as they were clustered together (fig. S13). Moreover, the genomic distribution of ATAC peaks across the conditions tested were similar, with most of the peaks corresponding to promoter, intron, and distal intergenic regions (fig. S13). Soft surfaces (20 and 1 kPa) increased the percentage of peaks annotated to promoter regions compared to stiff (glass) substrates (fig. S13). Volcano plots showed that an intermediate stiffness of 20 kPa increased chromatin accessibility at numerous genomic loci when compared to 1 kPa and glass, respectively (Fig. 2A). In addition, although the intermediate matrix stiffness of 20 kPa could decrease the accessibility of certain regions when compared to glass, there appeared to be less reduction in chromatin accessibility compared to soft surfaces (Fig. 2A). To determine how these accessibility differences might be relevant to iN reprogramming, we integrated our ATAC-seq results with previously published Ascl1 chromatin immunoprecipitation sequencing (ChIP-seq) data (40) and found that soft surfaces (both 20 and 1 kPa) increased the accessibility of genomic regions of Ascl1-target genes (Fig. 2B); in comparison to 1 kPa and stiff surfaces, the intermediate stiffness of 20 kPa had the highest accessibility at the promoter regions of these neuronal genes (Fig. 2B and fig. S14).
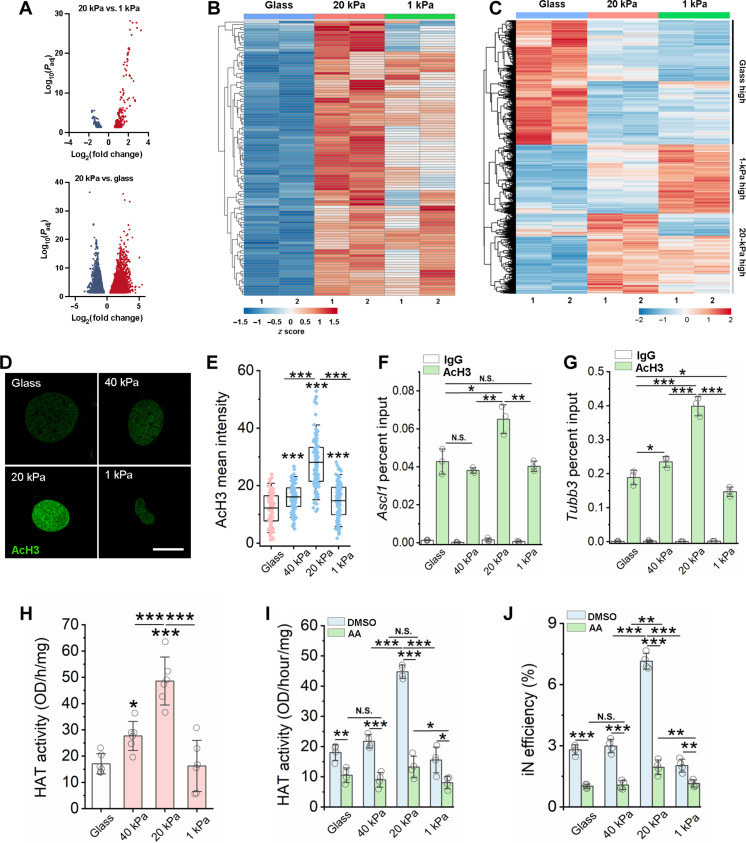
Fig. 2. Matrix stiffness modulates chromatin accessibility, gene expression, HAT activity, and histone acetylation.
(A) Volcano plot showing differential accessible regions. Red and blue dots indicate regions with increased or decreased chromatin accessibility, respectively. (B) Heatmap representation of differentially accessible regions that overlap with Ascl1 ChIP-seq peaks (GSE43916: SRX323557). Each row represents a differential region; each column is one biological replicate of the indicated condition. (C) Heatmap representation of differentially expressed genes at day 3. (D) Immunofluorescent images of AcH3 in non-transduced fibroblasts at day 2. Scale bar, 10 μm. (E) Quantification of AcH3 intensity based on experiments in (A) (n ≥ 116). (F and G) ChIP-qPCR analysis shows the percent input of AcH3 at the promoter regions of Ascl1 (F) and Tubb3 (G) in BAM-transduced fibroblasts at day 3 (n = 3). (H) Quantification of HAT activity at day 2 (n = 6). (I) Quantification of HAT activity in fibroblasts cultured on various substrates for 1 day followed by treatment with vehicle control [dimethyl sulfoxide (DMSO)] or a HAT inhibitor [anacardic acid (AA)] for 24 hours (n = 4). (J) Reprogramming efficiency of BAM-transduced fibroblasts cultured on matrices of varying stiffness and pre-treated with anacardic acid for 24 hours before adding Dox (n = 4). In (E) to (J), statistical significance was determined by a one-way ANOVA and Tukey’s multiple comparison test (*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001). In (E), box plots show the ends at the quartiles, the mean as a horizontal line in the box, and the whiskers represent the SD. In (F) to (J), bar graphs show means ± SD.
To determine whether matrix stiffness altered gene expression in the absence of BAM reprogramming factors, we performed bulk RNA sequencing and found that cells on matrices of varying stiffness (glass, 20 kPa, and 1 kPa) had discernible gene expression patterns (Fig. 2C and fig. S15). We found that different subsets of genes had highest expression levels on surfaces with different stiffness (Fig. 2C). Gene Ontology (GO) analysis revealed that an intermediate stiffness of 20 kPa could promote genes related to synapse organization and axonogenesis, which was not evident for glass or 1-kPa samples (fig. S16). On the other hand, a stiff surface (glass) enhances cell cycle genes and actin-related genes. It is worth noting that many of these changes of gene expression are related to the regulation of genes in euchromatin and may not be attributed to epigenetic changes. It is possible that both the gene expression changes and epigenetic changes may contribute to the reprogramming process when BAM are introduced.
Previous studies have reported that epigenetic modifications, such as histone methylation, histone acetylation, and DNA methylation, regulate chromatin accessibility and play an important role in cell reprogramming (41, 42). To determine whether matrix stiffness may modulate iN reprogramming through global histone modifications, we performed immunostaining of euchromatin marks histone H3 acetylation (AcH3), histone H4 acetylation (AcH4), acetylated histone H3 on lysine-27 (H3K27ac), and tri-methylated histone H3 on lysine-4 (H3K4me3), and heterochromatin marks tri-methylated histone H3 on lysine-9 (H3K9me3), tri-methylated histone H3 on lysine-27 (H3K27me3), and tri-methylated histone H4 on lysine-20 (H4K20me3) in nontransduced fibroblasts cultured on gels of various stiffness. As shown in Fig. 2 (D and E) and fig. S17, an intermediate stiffness of 20 kPa induced higher AcH3 and AcH4 levels compared to stiffer and softer surfaces. On the other hand, no change was apparent in heterochromatin and other euchromatin marks examined (fig. S18). These results suggest that an intermediate stiffness may induce a more open chromatin structure via AcH3 to facilitate cell reprogramming.
To determine whether there is an increase of AcH3 at the promoter of neuronal genes, we performed ChIP–quantitative polymerase chain reaction (qPCR) at day 3 and found an increase in AcH3 at the promoter regions of Ascl1 and Tubb3 in cells on 20-kPa surface, suggesting that an intermediate matrix stiffness can promote neuronal gene expression by modulating site-specific epigenetic changes (Fig. 2, F and G).
HAT mediates matrix stiffness-modulated AcH3
HATs are chromatin-modifying enzymes that regulate the acetylation of histone proteins, resulting in a loosely packed chromatin structure that allows for the binding of transcription factors (43). To elucidate how intermediate matrix stiffness promotes a more open chromatin state, we analyzed the activity of HAT in fibroblasts cultured on PAAm of varying stiffness for 2 days. Quantification of HAT activity revealed that gels of intermediate stiffness increased HAT activity compared to stiffer and soft surfaces (Fig. 2H). On the other hand, the activity of histone deacetylase (HDAC) in fibroblasts increased monotonically with the decrease of matrix stiffness (fig. S19). In addition, fibroblasts cultured on glass had significantly higher DNA methylation levels than cells cultured on PAAm gels with varying stiffness (fig. S20). We then used chemical inhibitors to test the relative contributions of these epigenetic regulators to iN conversion. Inhibition of HAT activity by anacardic acid inhibited the biphasic enhancement of HAT activity and the reprogramming efficiency (Fig. 2, I and J). However, inhibition of HDAC activity by valproic acid (VPA) (16) only slightly increased HAT activity and iN conversion efficiency of fibroblasts cultured on stiff gels (40 kPa) and glass but had no effect on cells cultured on soft and intermediate stiffness gels (figs. S21 and S22). Furthermore, inhibition of H3K9 and DNA methyltransferase activity using Bix01294 and RG108, respectively, had no notable effect on the biphasic enhancement in cell reprogramming (figs. S23 and S24). These results suggest that HATs play a major role in matrix stiffness-enhanced iN conversion.
Actin polymerization mediates the effect of matrix stiffness on HAT translocation into nucleus
There is accumulating evidence that biophysical signals can be transmitted through the actin cytoskeleton to the nucleus to elicit changes in gene expression (27, 44–48); in addition, actin polymerization/depolymerization can regulate G-actin–mediated transport of transcriptional factors into the nucleus (29–32). To determine whether actin plays a role in intermediate stiffness-mediated HAT activity, HAT1, a specific type of HAT, was costained with F-actin and β-actin, respectively. As shown in Fig. 3A and fig. S22, cells cultured on soft surfaces had less polymerized actin (F-actin). HAT1 and β-actin had a pronounced nuclear colocalization when cells were cultured on 20-kPa gels (Fig. 3A and fig. S25), suggesting that substrate stiffness modulates the nuclear translocation of actin and HAT1. Western blotting analysis of G-actin and F-actin content showed that F-actin decreased with matrix stiffness, whereas the opposite trend was observed for G-actin (Fig. 3, B and C), which could make more actin monomer available for nuclear translocation on soft surfaces.
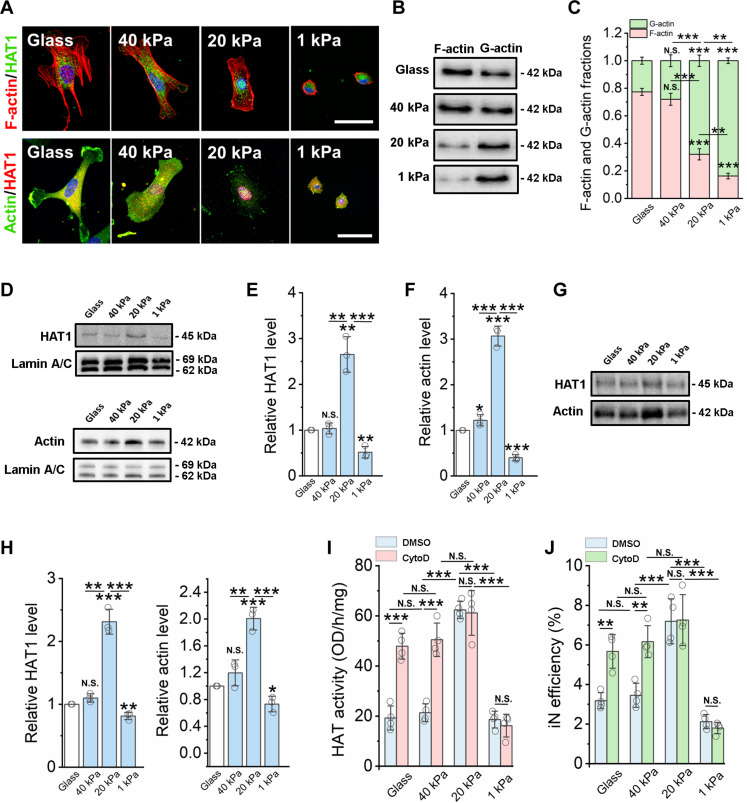
Fig. 3. Matrix stiffness modulates the nuclear translocation of HAT via actin assembly.
(A) Immunofluorescent images of fibroblasts cultured on glass and PAAm gels of various stiffness for 2 days and stained for F-actin (phalloidin, red), HAT1, and β-actin (green). Scale bars, 50 μm. (B) Western blotting analysis shows F-actin and G-actin levels in fibroblasts cultured on glass and PAAm gels for 2 days. (C) Quantification of F-actin and G-actin fractions from Western blots (n = 3). (D) Western blotting analysis shows HAT1 and β-actin levels from nuclear fractions of fibroblasts cultured on glass and PAAm gels of varying stiffness for 2 days, where lamin A/C serves as a housekeeping protein. (E) Quantification of HAT1 level from Western blots (n = 3). (F) Quantification of actin level from Western blots (n = 3). (G) Co-immunoprecipitation of HAT1 and actin from nuclear fractions of fibroblasts on glass or varying matrix stiffness. (H) Quantification of HAT1 and actin levels from co-immunoprecipitation Western blot (n = 3). (I) Quantification of HAT activity in fibroblasts cultured on various substrates for 1 day, followed by treatment with vehicle control (DMSO) or an actin polymerization inhibitor [cytochalasin D (CytoD); 0.5 μM] for 24 hours (n = 4). (J) Reprogramming efficiency of BAM-transduced fibroblasts cultured on various substrates and pretreated with cytochalasin D (0.5 μM) for 24 hours before adding Dox (n = 4). In (C), (E), (F) and (H) to (J), statistical significance was determined by a one-way ANOVA and Tukey’s multiple comparison test (*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001). In (C), (E), (F) and (H) to (J), bar graphs show means ± SD.
To further determine whether matrix stiffness regulated the translocation of actin and HAT1, Western blotting analysis was used to detect actin and HAT1 levels in nuclear fractions and whole-cell lysates. As shown in Fig. 3 (D to F), β-actin and HAT1 were significantly higher in nuclear fractions of cells cultured on 20-kPa surfaces compared to other substrates but did not show significant changes at the whole-cell level in fibroblasts cultured on glass and gels of varying stiffness (fig. S26). This finding could be extended beyond HAT1 as an intermediate stiffness of 20 kPa also promoted the translocation of p300/CBP-associated factor (PCAF), another HAT, into the nucleus, compared to soft and stiff surfaces (fig. S27). Immunoprecipitation of nuclear actin demonstrated that, on the surface with an intermediate stiffness of 20 kPa, there was the highest HAT1 and actin association within the nucleus (Fig. 3, G and H), coinciding with the nuclear localization of actin and HAT1. Furthermore, the inhibition of actin polymerization using cytochalasin D significantly increased HAT activity (Fig. 3I) and reprogramming efficiency on stiff gels (40 kPa) and glass but had no effect on cells cultured on softer gels (20- and 1-kPa gels) (Fig. 3J). These results suggest that actin depolymerization may mediate matrix stiffness-induced HAT1 activity and iN reprogramming, but it is not clear why the translocation of HAT1 and actin was limited on 1-kPa surface.
Cofilin serves as a cotransporter of actin/HAT translocation into nucleus
Cofilin, an actin-binding and depolymerization protein, has a nuclear localization signal (NLS) domain and is required to shuttle actin into the nucleus through importin-9 (49, 50). Recent studies suggest that nucleocytoplasmic transport plays a role in mechanotransduction, whereby mechanoresponsive transcription factors transduce mechanical signals to the nucleus via their transport through nuclear pore complexes (51, 52). Moreover, cofilin-1 has been recently identified as a mechanosensitive regulator of transcription (53), suggesting that biophysical cues can potentially modulate cofilin and actin transport to control cell fate and function. Therefore, cofilin expression and localization was determined by immunostaining and Western blot analysis. As shown in Fig. 4 (A and B) and figs. S28 and S29, consistent with actin and HAT1, cofilin also had a pronounced nuclear localization and was significantly higher in nuclear fractions of cells on gels of intermediate stiffness (20 kPa) compared with the other groups. In addition, upon performing actin immunoprecipitation of nuclear fractions followed by cofilin analysis, we found that cells on an intermediate matrix stiffness had the highest cofilin-actin association within the nucleus (Fig. 4, C and D), which coincides with nuclear actin-HAT1 association and cofilin nuclear localization.
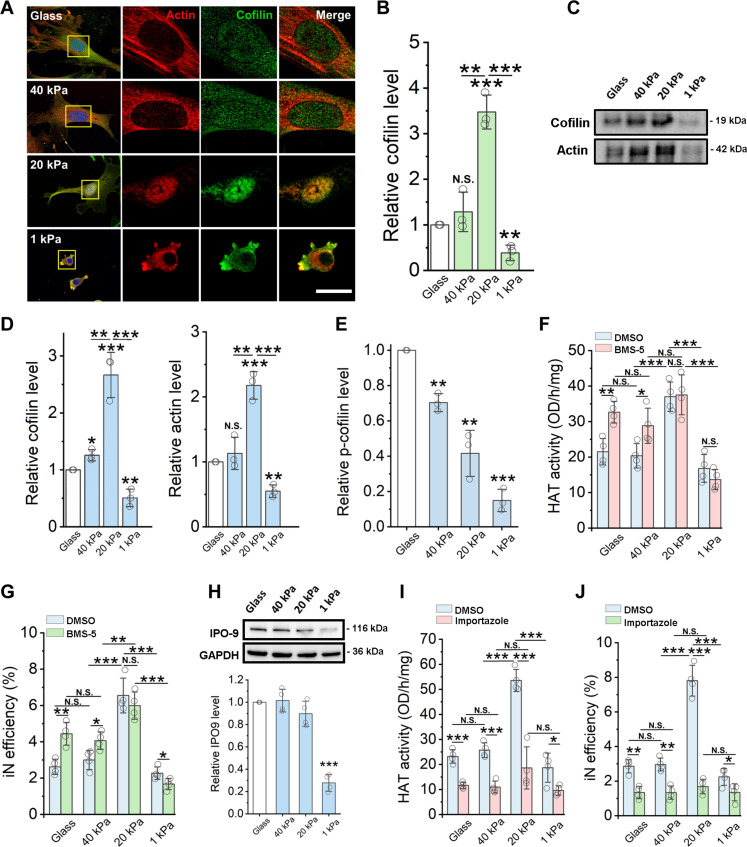
Fig. 4. Matrix stiffness regulates cofilin and actin nuclear translocation to modulate HAT activity.
Fibroblasts were cultured on glass or PAAm gels of various stiffness. (A) Immunofluorescent images of actin (red) and cofilin-1 (green) at day 2. Scale bar, 5 μm. (B) Cofilin level in nuclear fractions of fibroblasts at day 2, normalized by lamin A/C level (n = 3). (C) Co-immunoprecipitation of cofilin and actin from nuclear fractions of fibroblasts. (D) Quantification of cofilin and actin levels from co-immunoprecipitation Western blots (n = 3). (E) Phospho-cofilin (Ser3) level at day 2, normalized to cofilin level (n = 3). (F) HAT activity in fibroblasts cultured for 1 day followed by treatment with vehicle control (DMSO) or a LIMK inhibitor (BMS-5) for 24 hours (n = 4). (G) Reprogramming efficiency of BAM-transduced fibroblasts treated with BMS-5 for 24 hours before adding Dox (n = 4). (H) Western blotting analysis of importin 9 (IPO-9) levels in whole-cell lysates of fibroblasts, normalized by glyceraldehyde-3-phosphate dehydrogenase (GAPDH) level (n = 4). (I) HAT activity in fibroblasts cultured on various substrates for 1 day, followed by treatment with DMSO or a nuclear import inhibitor (importazole) for 48 hours (n = 4). (J) Reprogramming efficiency of BAM-transduced fibroblasts treated with importazole before adding Dox and during the reprogramming process (n = 4). In (B) and (D) to (J), bar graphs show means ± SD. Statistical significance was determined by a one-way ANOVA and Tukey’s multiple comparison test (*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001).
Because cofilin phosphorylation at Ser3 can inhibit the activity of cofilin during actin depolymerization (50, 54), we further analyzed phospho-cofilin (Ser3) level in fibroblasts cultured on different matrix stiffness. Western blotting analysis showed that the level of p-cofilin decreased with matrix stiffness (Fig. 4E and fig. S30), consistent with our finding that the F-actin decreases with matrix stiffness (Fig. 3, B and C). To determine whether cofilin phosphorylation plays a role in regulating HAT activity and, therefore, iN reprogramming, we treated the cells with BMS-5, a LIM kinase (LIMK)–1/2 inhibitor that inhibits the phosphorylation of cofilin by LIMK (55, 56). As shown in Fig. 4 (F and G), pretreatment with BMS-5 significantly increased HAT activity and iN reprogramming efficiency of fibroblasts on glass and 40-kPa gels. However, there was no apparent difference for fibroblasts cultured on 20- or 1-kPa gels (Fig. 4, F and G) as cofilin phosphorylation was already low in the absence of BMS-5 (fig. S30). As free cofilin and G-actin increased with the disassembly of actin filaments on soft surfaces (Fig. 3, B and C), presumably the transport of HAT1 and cofilin/actin should increase with decreasing matrix stiffness, yet the nuclear translocation of cofilin on 1 kPa was low (Fig. 4, A to C). Therefore, it is likely that other cellular changes on soft matrices might limit HAT activity and the reprogramming efficiency.
To directly test the role of actin polymerization, we used cytochalasin D at various concentrations to induce different extents of F-actin depolymerization that did not notably affect cell viability (fig. S31). As shown in fig. S32, cofilin and HAT1 translocated into the nucleus in a dose-dependent manner when the cytochalasin D concentration increased from 0.25 to 0.75 μM, thereby increasing HAT activity and iN reprogramming. However, treating the cells with higher doses of cytochalasin D (i.e., from 1.5 to 3 μM) suppressed cofilin and HAT1 translocation, HAT activity, and iN reprogramming (fig. S32), consistent with the low level of HAT1 translocation into nucleus on 1-kPa surfaces.
To look for other factors accounting for the low translocation level of HAT and actin on soft surface of 1 kPa, we investigated whether nuclear import was modulated by matrix stiffness. The analysis of nuclear pore protein expression revealed that importin-9 levels significantly decreased on soft substrates (1 kPa) when compared to stiff and intermediate surfaces (Fig. 4H), and this appears to be specific to importin-9 as we observed no apparent difference in nuclear pore complex protein levels across the various matrix stiffness examined (figs. S33 and S34). To further determine whether cofilin translocation via importin-9 plays a role in nuclear actin transport of HAT1 and, thus, iN reprogramming, importazole was used to block the importin-9 nuclear receptor, which is responsible for the nuclear translocation of actin-cofilin complexes (49, 57). As expected, inhibiting the importin-9–mediated nuclear transport impaired the biphasic enhancement in HAT activity and iN reprogramming efficiency of fibroblasts cultured on intermediate stiffness gels and significantly decreased both HAT activity and iN reprogramming efficiency across all the surfaces tested (Fig. 4, I and J). These results demonstrate a potential mechanism of how the softest surface (1 kPa) limits nuclear translocation of cofilin-actin-HAT complexes (Fig. 4, C and D).
The dynamic change of matrix stiffness modulates HAT translocation
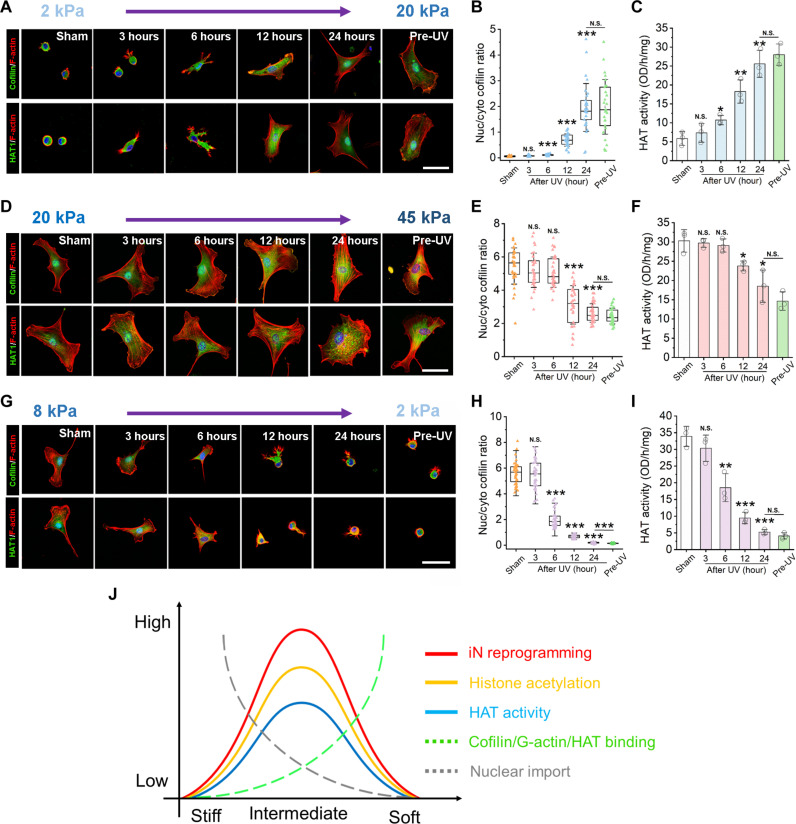
Moreover, to directly determine how matrix stiffening can regulate cofilin and HAT1 nuclear translocation, we sought out a tunable hydrogel system that would allow for matrix stiffening in response to a stimulus, such as light. Because a commercially available PAAm gel system that can be dynamically stiffened does not currently exist, tunable methacrylated hyaluronic (MeHA) hydrogels were prepared to dynamically increase the surface stiffness from 2 to 20 kPa and from 20 to 45 kPa upon exposure to ultraviolet (UV) light (figs. S35 and S36). When matrix stiffness was increased from 2 to 20 kPa, cofilin and HAT1 translocated from the cytoplasm to the nucleus, and HAT activity significantly increased after 6 hours of matrix stiffening (Fig. 5, A to C). On the other hand, when matrix stiffness increased from 20 to 45 kPa, cofilin and HAT1 relocated from the nuclei to the cytosol, and HAT activity significantly decreased 12 hours after surface stiffness was increased to 45 kPa (Fig. 5, D to F). Similarly, when matrix stiffness was softened from 8 to 2 kPa using tunable ortho-nitrobenzyl (o-NB) PAAm gels that have an o-NB-bis-acrylate crosslinker that cleaves and leads to gel softening after exposure to light (58), cofilin was translocated from the nucleus to the cytoplasm and HAT activity significantly decreased 6 hours after inducing matrix softening (Fig. 5, G to I). These results directly demonstrate that an intermediate level of substrate stiffness (~20 kPa) is optimal to promote HAT1 translocation into the nucleus to increase HAT activity.
Fig. 5. Actin assembly, HAT translocation, and HAT activity are modulated by dynamically tunable gels.
(A) Representative immunofluorescent images of F-actin, cofilin, and HAT1 in fibroblasts at the indicated time points after UV radiation to stiffen the gels (from 2 to 20 kPa). Scale bar, 50 μm. (B) Nuclear to cytoplasmic ratio of cofilin based on images in (A) (n = 30 cells). (C) HAT activity in fibroblasts cultured on dynamically stiffening gels (2 to 20 kPa) at the indicated time points after UV radiation (n = 3). (D) Images of F-actin, cofilin, and HAT1 in fibroblasts at the indicated time points after UV radiation to stiffen the gels (20 to 45 kPa). Scale bar, 50 μm. (E) Nuclear to cytoplasmic ratio of cofilin based on images in (D) (n = 30 cells). (F) HAT activity in fibroblasts cultured on dynamically stiffening gels (20 to 45 kPa) after UV radiation (n = 3). (G) Images of F-actin with cofilin or F-actin with HAT-1 in fibroblasts at the indicated time points after UV radiation to soften the gels (from 8 to 2 kPa). Scale bar, 50 μm. (H) The nuclear to cytoplasmic ratio of cofilin based on images in (G) (n = 30 cells). (I) HAT activity in fibroblasts cultured on dynamically softening gels (8 to 2 kPa) after UV radiation (n = 3). (J) Summary of hypothesis for matrix stiffness-mediated iN reprogramming. In (B), (C), (E), (F), and (H) to (I), statistical significance was determined by a one-way ANOVA and Tukey’s multiple comparison test (*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001).
DISCUSSION
Our work demonstrates a biphasic dependence of cell reprogramming on matrix stiffness through the regulation of the epigenetic state and suggests that matrix stiffness can regulate actin assembly and nuclear transport, which, in turn, modulates HAT activity and histone acetylation. These changes in the epigenetic state, especially on surfaces of intermediate stiffness (~20 kPa), promote a more open chromatin structure that facilitates neuronal gene induction and, thus, an enhancement in iN reprogramming. On the basis of the effects of matrix stiffness on actin polymerization and nuclear transport of cofilin/actin/HAT, we propose a two-factor hypothesis (Fig. 5J) to explain the biphasic regulation of epigenetic state and cell reprogramming by matrix stiffness. On the one hand, cells on a stiff matrix (40 kPa or higher) have a limited amount of cofilin and G-actin to mediate HAT translocation into the nucleus, although nuclear importer such as importin-9 is available, resulting in low levels of histone acetylation and iN reprogramming. On the other hand, cells on soft matrices (1 kPa) have low levels of actin polymerization and higher levels of cofilin and G-actin, which would facilitate HAT translocation to nucleus; however, the down-regulation of nuclear transporters such as importin-9, especially on 1-kPa surface, may limit the nuclear transport of cofilin/actin/HAT, resulting in a low iN reprogramming efficiency. Therefore, when cells are cultured on an intermediate matrix stiffness, the formation of cofilin-actin-HAT complexes and the nuclear transport via importin-9 are balanced at an optimal level, allowing for the most efficient shuttling of actin and HAT1 into the nucleus, thereby increasing AcH3 that is conducive to iN reprogramming.
In addition, there may be other mechanisms accounting for the stiffness effects on cell reprogramming. For instance, higher cell proliferation rate is correlated with higher reprogramming efficiency (59), and the decrease of cell proliferation on soft surfaces would lower the reprogramming efficiency, although this does not explain the low reprogramming efficiency on stiff surfaces where cell proliferation is high. The decrease in cell spreading on soft surfaces may decrease actin assembly and increase G-actin availability but decrease the contraction force in the cells, which may affect the mechanotransduction process. Nuclear size may affect the molecular transport through nuclear membrane and inside the nucleus. There are also previous reports that nuclear shape regulates the epigenetic state and induced pluripotent stem cell (iPSC) reprogramming (13) and that an intermediate nuclear shape enhances collagen I synthesis (60).
Consistently, the biphasic regulation of HAT activity can be observed through the disruption of the actin cytoskeleton and the mechanical tuning of extracellular matrix stiffness. Notably, the effect of cytochalasin D treatment on HAT activity is both dose dependent and biphasic; HAT activity increases from 0 to 0.75 μM cytochalasin D but decreases at higher doses (fig. S32). It is likely that higher doses of cytochalasin D (>0.75 μM) also inhibit the nuclear import of cofilin/actin/HAT via importin-9, similar to a 1-kPa surface. In addition, the disruption of actin cytoskeleton may decrease the size of nuclear pores(33) to suppress nuclear transport. Inhibiting actomyosin contraction using blebbistatin or decreasing focal adhesions using a focal adhesion kinase inhibitor also demonstrates a biphasic effect on iN reprogramming (61). This further confirms that HAT activity, and thus the epigenetic state, is regulated by cell adhesion–mediated actin polymerization. Furthermore, matrices with tunable stiffness, whether increased or decreased, directly influences HAT translocation and activity (Fig. 5). Note that disrupting the actin cytoskeleton, such as with blebbistatin treatment, may not induce the exact same epigenetic changes as altering matrix stiffness. Our previous studies have shown that blebbistatin induces global changes not only in AcH3 but also in H3K4me3, H3K9me3, and DNA methylation, leading to a more open chromatin state (61).
Our findings on the nuclear transport of cofilin/actin/HAT provide a novel mechanism that directly links actin cytoskeleton organization to epigenetic changes. This discovery may have broad implications for understanding the mechanical regulation of the epigenetic state by various intracellular and extracellular factors. Our findings contribute to the existing knowledge that actin dynamics (i.e., polymerization/depolymerization) can mediate the nuclear transport of transcription factors, such as MRTF (29, 30, 32) and, more recently, β-catenin (62). The nuclear transport of β-catenin in response to dynamic strain was shown to be dependent on cofilin and importin-9 (62), which is consistent with our results. Several previous studies have shown that matrix stiffness can modulate chromatin accessibility, DNA methylation, and heterochromatin in various cell types (23–25, 63). Our findings are in agreement with other studies that have shown that an intermediate stiffness (i.e., 13 kPa) can promote histone acetylation (23), while soft substrates increase HDAC expression (64) and decrease DNA methylation (24). However, these studies generally make comparisons between two stiffness (i.e., soft versus stiff), which assume a monotonic regulation of epigenetic state by matrix stiffness, and thus, further investigations are necessary to examine the potential biphasic regulation. In addition, the range of stiffness studied and cell types used could all lead to potential differences in epigenetic findings. By examining a broad range of matrix stiffness during iN reprogramming, we show that matrix stiffness regulates direct reprogramming through a biphasic mechanism. This finding may not be limited to only direct reprogramming as we found that an intermediate matrix stiffness of 20 kPa could promote iPSC reprogramming (fig. S37). However, one should note that cell reprogramming is a complex and multifaceted process that most likely involves various interconnected mechanisms, and future studies are necessary to fully elucidate all aspects of these mechanisms.
In this study, PAAm gels were used to investigate how matrix stiffness regulates the epigenetic state during iN reprogramming. While PAAm gels have several advantages, their use is limited to two-dimensional (2D) culture as acrylamide is toxic before polymerization (34). Thus, embedding cells to study the effects of substrate stiffness in 3D culture or for therapeutic purposes is not feasible using this hydrogel system. Alternative biomaterials would need to be considered, particularly when potentially translating these findings for regenerative medicine applications. Although PAAm and MeHA gels may differ in their chemical composition, we found that dynamic stiffening of MeHA gels could modulate the translocation of actin-cofilin-HAT complexes (Fig. 5, A and B), suggesting that the observed responses were due to changes in matrix stiffness and not limited to PAAm gels. As o-NB PAAm gels have a similar composition to PAAm gels and only differ in that they have a photodegradable crosslinker(58), this provided us an appropriate and comparable tool to study the effects of matrix softening, which also confirmed that our findings were attributed to changes in matrix stiffness. Furthermore, as cells experience a viscoelastic microenvironment in vivo, there is growing interest to engineer materials with viscoelastic properties (65, 66). While our study provides insights into the effects of elastic hydrogels on direct reprograming, future directions could include to investigate the influence of viscoelasticity on cell reprogramming and whether our findings would be consistent using viscoelastic gels.
Moreover, our findings indicate that soft surfaces of both 20 and 1 kPa increased the accessibility of neuronal genes, with the highest accessibility around Ascl1 target genes on the surface of 20 kPa. This is distinguishable from stem cell differentiation where soft surfaces (less than 20 kPa) appear to promote neural differentiation more efficiently (3, 67). First, cell reprogramming and differentiation are distinct processes with different rate-limiting factors. Second, the chromatin structure, the accessibility of neuronal genes, and signaling pathways in fibroblasts and neural stem cells are different, resulting in cell type–specific mechanical regulation of epigenetic state and signaling.
Our findings indicate that intermediate matrix stiffness of 20 kPa can increase iN reprogramming efficiency. While our findings contribute to our understanding of the mechano-epigenetic mechanisms during cell reprogramming, the identification of biophysical cues that can promote cell reprogramming has important implications for regenerative medicine applications. In addition to direct reprogramming being a faster method to obtain desired cell types, increasing the yield of target cells is highly beneficial for disease modeling and drug discovery, particularly for high-throughput drug screening platforms. Moreover, the translation of our findings to in vivo reprogramming paradigms could be potentially valuable for tissue engineering applications. Together, our findings shed light on how matrix stiffness biophysically regulates the epigenetic state and cell reprogramming, providing insights for engineering biomaterials for cell engineering.
MATERIALS AND METHODS
All experiments were performed in accordance with relevant guidelines and ethical regulations approved by the University of California, Los Angeles (UCLA) Institutional Biosafety Committee (BUA-2016-222). All animal experiments, including breeding, maintenance, and euthanasia, were performed in accordance with relevant guidelines and ethical regulations approved by the UCLA Institutional Animal Care and Use Committee (protocol nos. ARC-2016-036 and ARC-2016-101).
Fibroblast isolation, culture, and reprogramming
Fibroblasts were isolated from ear tissues of adult C57BL/6 mice (1 month old) and expanded in fibroblast medium containing Dulbecco’s modified Eagle medium (DMEM) (Gibco, 11965), 10% fetal bovine serum (FBS; Gibco, 26140079), and 1% penicillin/streptomycin (Gibco, 15140122). For all experiments, passage-2 cells were used and synchronized upon reaching 80% confluency using DMEM with 1% FBS for 24 hours before transduction with viruses containing BAM constructs. Twenty-four hours later, cells were seeded onto PAAm gels coated with fibronectin (0.1 mg/ml; Thermo Fisher Scientific, 33016015) at a density of 3000 cells/cm2. The following day (day 0), the medium was replaced to fibroblast medium containing doxycycline (2 ng/ml; Sigma-Aldrich) to initiate the expression of the transgenes and thus reprogramming. Twenty-four hours later (day 1), cells were cultured in N2B27 medium containing DMEM/F12 (Gibco, 11320033), N-2 supplement (Gibco, 17502048), B-27 supplement (Gibco, 17504044), 1% penicillin/streptomycin, and doxycycline (2 ng/ml), and half medium changes were performed every 2 days. On day 7, cells were fixed and stained for Tubb3 to determine the reprogramming efficiency. iN cells were identified on the basis of positive Tubb3 staining and a neuronal morphology. The reprogramming efficiency was determined as the percentage of iN cells on day 7 relative to the number of the cells initially seeded. For long-term studies where maturation and functionality of the iN cells were examined, cells were kept in culture for 3 or 5 weeks, respectively.
To determine the role of histone acetylation-modifying enzymes in stiffness-mediated iN reprogramming, BAM-transduced fibroblasts were treated with HAT inhibitor anacardic acid (1 μM; Cayman chemical, 13144) or HDAC inhibitor VPA (500 μM; Cayman chemical, 13033), respectively, for 24 hours before seeding the cells on substrates. To investigate the role of actin polymerization in stiffness-mediated iN reprogramming, BAM-transduced fibroblasts were treated with actin polymerization inhibitor cytochalasin D (0.5 μM; Cayman chemical, 11330) for 24 hours before seeding the cells on substrates. To determine whether the translocation of the actin-cofilin complex regulates stiffness-mediated iN reprogramming, cells were treated with importin-9 inhibitor importazole (10 μM; Cayman chemical, 21491) for 24 hours before seeding the cells on gels. In addition, fibroblasts were treated with LIMK-1/2 inhibitor BMS-5 (4 μM; Cayman chemical, 21072) for 24 hours before seeding the cells on the gels to further elucidate the role of phospho-cofilin in stiffness-mediated iN reprogramming. Parallel treatments with dimethyl sulfoxide (DMSO) served as controls.
Lentiviral production and transduction
Doxycycline-inducible lentiviral vectors for Tet-O-FUW-Brn2, Tet-O-FUW-Ascl1, Tet-O-FUW-Myt1l, and FUW-rtTA plasmids were used to transduce fibroblasts for ectopic expression of Brn2, Ascl1, Myt1L, and rtTA. The Ascl1–enhanced GFP promoter lentiviral vector (Genecopoeia, MPRM39894-LvPF02) was used to identify the activation of the Ascl1 promoter. Lentivirus was produced by using established calcium phosphate transfection methods, and Lenti-X Concentrator (Clontech, 631232) was used to concentrate viral particles according to the manufacturer’s protocol. Stable virus was aliquoted and stored at −80°C. Fibroblasts were plated and synchronized for 24 hours before viral transduction in the presence of polybrene (8 μg/ml; Sigma-Aldrich, H9268). Cells were incubated with the virus for 24 hours before performing matrix stiffness-mediated experiments.
PAAm gel fabrication
PAAm gels were used in most experiments except the living cell studies using dynamically tunable gels. PAAm gels were fabricated according to established protocols, and gels of different stiffness were obtained by varying the ratio of acrylamide to bisacrylamide as previously documented (6, 68). Briefly, glass coverslips (12 mm) were sonicated in 70% ethanol for 10 min and then allowed to air dry. The coverslips were oxygen plasma–treated for 5 min, followed by incubation with a methacryloxypropyl-trimethoxysilane (Gelest) solution for 5 min. Coverslips were washed three times with methanol and incubated at 110°C for 30 min. PAAm gel solution with the desired concentration of acrylamide and bis-acrylamide was allowed to polymerize for 20 to 30 min to form 100-μm-thick gels. Sulfo-SANPAH (1 mg/ml in Hepes buffer; Thermo Fisher Scientific, 22589) was used to crosslink fibronectin (0.1 mg/ml) to the gel surface. The conjugation of matrix proteins such as fibronectin onto the surface of PAAm gels have been well established, and previous studies have shown that fibronectin is incorporated at the hydrogel surface, and the amount of fibronectin does not change as hydrogel stiffness varies (69). On the other hand, surface conjugation does not significantly affect the stiffness PAAm substrates (70). Stiff substrates (glass slides) were coated with the same density of fibronectin. All substrates were sterilized with 70% ethanol for 15 min, followed by three phosphate-buffered saline (PBS) washes. Cells were then seeded onto substrates at a density of 3000 cells/cm2.
Methacrylated hyaluronic acid hydrogel fabrication
Methacrylated hyaluronic acid (MeHA) gels were prepared to study the effect of matrix stiffening on HAT/actin translocation into nucleus in living cells. MeHA gels were prepared using the PhotoHA-IRG Kit (Advanced BioMatrix, #5220) according to the manufacturer’s instructions and as previously described (71). Briefly, glass coverslips were prepared as aforementioned for PAAm hydrogels. MeHA powder was dissolved in PBS (1% solution) and mixed on shaker for 1 hour at 4°C. Irgacure (photoinitiator) was dissolved in 100% methanol and then diluted to 0.01% (w/v) in the MeHA solution. Eighteen microliters of the hydrogel solution was sandwiched between methacryloxypropyl-trimethoxysilane–treated glass coverslip and Gel slick (Lonza, #50640)–activated glass slide and photopolymerized using a 365-nm Black Ray Bench Lamp light source (7.8 mW/cm2, UVP, LLC). Tunable gels were initially UV-radiated for 30 or 60 s to yield ~2- and ~ 20-kPa gels. To further stiffen the hydrogels, gels were radiated from 60 to 120 s. For protein conjugation, 100 μl of an 1-ethyl-carbodiimide hydrochloride (EDC)/N-hydroxysuccinimide (NHS) solution (76 mg of EDC + 115 mg of NHS + 2 ml of PBS buffer) was added to the surface of the gel and incubated for 30 min. Gels were washed in PBS for 5 min, and then 100 μl of fibronectin (0.1 mg/ml) was added to the gel surface and incubated overnight at 37°C. All substrates were sterilized with 70% ethanol for 15 min, followed by three PBS washes. Cells were then seeded onto substrates at a density of 3000 cells/cm2. In one set of experiments, cells cultured on 2-kPa gels with no UV radiation and 20-kPa gels (pre-exposed to UV radiation, i.e., Pre-UV) were used as controls. In a second set of experiments, cells cultured on 20-kPa gels with no UV radiation and 45-kPa gels (pre-exposed to UV radiation, i.e., Pre-UV) were used as controls.
Photodegradable o-NB PAAm hydrogel fabrication
Photodegradable PAAm gels were used to study the effects of gel softening (i.e., stiffness decrease) on HAT/actin translocation into the nucleus in living cells. Photodegradable o-NB PAAm hydrogels were prepared as previously described by replacing bisacrylamide with a photocleavable o-NB–bis-acrylate crosslinker (58). Briefly, glass coverslips were prepared as aforementioned for PAAm hydrogels and PAAm gel solution with the desired concentration of acrylamide, bis-acrylamide, o-NB–bis-acrylate, and 6-acrylamidohexylaminohexanoic acid (N6) was allowed to polymerize for 20 min to form 100-μm-thick gels. Gels were then washed on a shaker plate in a PBS/isopropanol solution for 5 days with two changes of the wash solution per day. Before conducting protein conjugation, gels were washed with PBS twice, with the second wash spanning overnight. The next day, gels were submersed in a 2-(N-morpholino)ethanesulfonic acid (MES) buffer solution (0.1 M, pH = 6) for at least 2 hours. The MES buffer was aspirated off, and 100 μl of 3-(3-dimethylaminopropyl)–EDS and NHS solution (76 mg of EDC + 115 mg of NHS + 2 ml of MES buffer) was added to the surface of the gel and incubated for 30 min. The EDC/NHS solution was then aspirated off, and the gels were washed in PBS for 5 min. After PBS was aspirated off, 100 μl of fibronectin (0.1 mg/l) was added to the gel surface. Gels were placed into the cell culture incubator overnight. The following day, gels were sterilized in 70% ethanol and rehydrated in sterile PBS. Cells were cultured on the gels the same day after sterilization at a density of 3000 cells/cm2. For gel degradation studies, a 365-nm Black Ray Bench Lamp light source [115 V 60 Hz, 0.68 A (UVP, LLC)] with an output intensity of 7.8 mW/cm2 (integrated between 300 and 500 nm) as measured by a spectroradiometer (International Light Technologies, ILT950) was used. All samples receiving irradiation were exposed for 40 min in four increments of 10 min. Between degradations, the samples were placed in the incubator for 10 min to minimize changes in culture temperature and pH. For these experiments, cells cultured on 8-kPa gels with no UV radiation and 2-kPa gels (pre-exposed to UV radiation, i.e., Pre-UV) served as controls.
Immunofluorescence staining and microscopy
Samples collected for immunofluorescence staining at the indicated time points were washed once with PBS and fixed in 4% paraformaldehyde for 15 min. Samples were washed three times with PBS for 5 min each and permeabilized using 0.5% Triton X-100 for 10 min. After three subsequent PBS washes, samples were blocked with 5% normal donkey serum (NDS; Jackson Immunoresearch, 017000121) in PBS for 1 hour. Samples were incubated with primary antibodies (refer to table S1) in antibody dilution buffer (1% NDS + 0.1% Triton X-100 in PBS) for either 1 hour or overnight at 4°C followed by three PBS washes and a 1-hour incubation with Alexa Fluor 488– and/or Alexa Fluor 546–conjugated secondary antibodies (Molecular Probes). F-actin was labeled by incubating with Alexa Fluor 546 Phalloidin (Invitrogen, A22283) for 1 hour, and nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) in PBS for 10 min. Epifluorescence images were collected using a Zeiss Axio Observer Z1 inverted fluorescence microscope and analyzed using ImageJ. Confocal images were collected using a Leica SP8-STED/FLIM/FCS Confocal and analyzed using ImageJ.
Cell area and nuclear volume measurements were derived from images of phalloidin and DAPI-labeled cells, respectively, which were analyzed using ImageJ. Average AcH3 and AcH4 intensities per nuclei were quantified using an ImageJ macro. Gaussian blur, thresholding, watershed, and analyze particle functions were applied to the DAPI channel to create individual selections for each nucleus. This mask was applied to the corresponding stain image to measure the average fluorescence intensity within each nucleus. In addition, ImageJ was used to analyze the fluorescence intensity within the nucleus and out of the nuclear area to quantify the protein translocation.
Quantitative reverse transcription PCR
Fibroblasts cultured on glass or PAAm gels of varying stiffness for 1 day were lysed using TRIzol (Invitrogen, 15596026), and then RNA was isolated using the PureLink RNA Mini Kit (Thermo Fisher Scientific, #121830020) as we have previously reported [15]. After RNA extraction, Thermo Scientific Maxima First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, K1641) was used for first-strand cDNA synthesis. Quantitative reverse transcription PCR (qRT-PCR) was performed using a CFX96 Real-Time PCR Detection System to detect the gene expression levels of Ascl1 (forward primer: GAAGCAGGATGGCAGCAGAT; reverse primer: TTTTCTGCCTCCCCATTTGA), where 18S (forward primer: GCCGCTAGAGGTGAAATTCTTG; reverse primer: CATTCTTGGCAAATGCTTTCG) served as a housekeeping gene and was used for normalization.
Chromatin immunoprecipitation–qPCR
ChIP-qPCR analysis was performed as described previously (61). Briefly, 3 days post-Dox addition, 10 million BAM-transduced fibroblasts cultured on glass or PAAm gels of varying stiffness were fixed by using 1% formaldehyde in PBS (Thermo Fisher Scientific, BP531) for 10 min. Glycine was added at a final 1× concentration for 5 min at room temperature, followed by two washes with cold PBS. Cells were scraped and collected in PBS containing protease inhibitors. Samples were centrifuged at 2000g at 4°C for 5 min, the supernatant was removed, and samples were stored at −80°C before further processing. The cells were resuspended and lysed in cell lysis buffer and resuspended in nuclei lysis buffer before sonication using a Branson SFX250 Sonifier at 40% amplitude, 0.7-s on, and 1.3-s off, for a total of 8 min. Samples were spun down at maximum speed in a 4°C centrifuge, and the supernatant was collected. Fifty microliters was removed from each sample and stored at 4°C as a downstream internal control.
Normal rabbit IgG (Millipore, CS200581) or anti-rabbit histone 3 acetylation (Millipore, 06-599) were added to samples and incubated in a rotator overnight at 25 rpm in a 4°C refrigerator. Pierce Protein A/G Magnetic Agarose Beads (Thermo Fisher Scientific, #78610) were washed with ChIP dilution buffer using a magnetic separation rack and added to each sample and incubated in a rotator for 2 hours. Following washing steps, the beads were resuspended in 50 μl of freshly prepared ChIP elution buffer (1% SDS and 0.1 M NaHCO3) and placed in a 65°C bath for 10 min. The supernatant was collected, and this elution step was performed once more and the corresponding eluates were combined.
qRT-PCR was performed on input and ChIP DNA samples using a CFX qPCR machine (Bio-Rad) and the following primers: Ascl1 (forward primer: AACCCCATATGGCTGCAGAG; reverse primer: GGGAGAGCGTTTGCACACTA) and Tubb3 (forward primer: AGAGGTCTCAAGAAGGGTTTCGC; reverse primer: AGAGGGTCTTTCTTTCTCTCAAGTG). ChIP-qPCR data were analyzed by normalizing the DNA concentration to percent input using the relative standard curve method.
ATAC sequencing
A total of 1,000,000 fibroblasts were cultured on matrices of varying stiffness for 3 days and stored at −80°C before sample processing. ATAC-seq was performed as described previously (61, 72). Briefly, frozen cells were thawed and washed once with PBS and then resuspended in 500 μl of cold PBS. The cell number was assessed by Cellometer Auto 2000 (Nexcelom Bioscience, Massachusetts, USA), and 100,000 cells were then added to ATAC lysis buffer and centrifuged at 500g in a prechilled centrifuge for 5 min. Supernatant was removed, and the nuclei were resuspended in 50 μl of tagmentation reaction mix by pipetting up and down. The reactions were incubated at 37°C for 30 min in a thermomixer with shaking at 1000 rpm and then cleaned up using the MiniElute Reaction Clean Up Kit (QIAGEN). Tagmented DNA was amplified with barcoded primers. Library quality and quantity were assessed with Qubit 2.0 DNA HS Assay (Thermo Fisher Scientific), Tapestation High Sensitivity D1000 Assay (Agilent Technologies), and QuantStudio 5 System (Applied Biosystems). Equimolar pooling of libraries was performed on the basis of quality control values and sequenced on an Illumina NovaSeq (Illumina, California, USA) with a read length configuration of 150 paired-end (PE) for [100]M PE reads (50 M in each direction) per sample.
For mapping and peak analysis, FASTQ files were trimmed with Trim Galore and cutadapt (73). Pair-ended reads were then aligned to the mouse reference genome (mm10) with Bowtie2 (74). Mitochondrial reads and PCR duplicates were removed using SAMtools (75) and Picard (http://broadinstitute.github.io/picard/), respectively. Peaks were called over input using MACS3 (76), and only peaks outside the ENCODE blacklist region were kept. All peaks from the samples were combined and merged using bedtools, and featureCount (77) was used to count the mapped reads for each sample. Peaks that were up- or down-regulated under different conditions were defined by using DESeq2 (78) with Padj = 0.05 as the threshold. Peaks located at cis-regulatory elements related to genes of interest (±5-kb region) were visualized by using Integrative Genomics Viewer (79) to demonstrate differentially up- or down-regulated peaks at genomic regions of Ascl1-target sites (40, 80). To identify differentially accessible regions around Ascl1-targeting genes, we used an Ascl1 ChIP-seq dataset that was previously published (40) (GSE43916, Ascl1 MEF). Genome coordinates of peaks were converted from mm9 to mm10 reference genome by using UCSC liftOver tools. Peaks identified in both Ascl1 ChIP-seq and ATAC-seq datasets were selected for heatmap visualization.
RNA sequencing and analysis
A total of 500,000 fibroblasts were cultured on matrices of varying stiffness for 3 days and stored at −80°C before sample processing. Upon isolating RNA from the samples, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. After fragmentation, the first strand cDNA was synthesized using random hexamer primers, followed by the second strand cDNA synthesis. The library was ready after end repair, A-tailing, adapter ligation, size selection, amplification, and purification. The library was checked with Qubit and real-time PCR for quantification and bioanalyzer for size distribution detection. Quantified libraries were pooled and sequenced on the NovaSeq PE150 (Illumina). Raw reads were trimmed with Trim Galore and cutadapt (73) based on quality. Pair-ended reads were then aligned to mouse reference genome (mm10) with STAR v2.7.10b (81). Transcriptome alignments were quantified using featureCount (77) using the gene annotation file from GENCODE. Differentially expressed genes (DEGs) were defined using DESeq2 (78) with Padj = 0.05 as the threshold. GO enrichment analysis was performed using the enrichGO function in clusterProfiler package (82). Generally, ggplot2 and gplots packages were used to generate data graphs. For heatmap of DEGs, values were normalized using size factor estimated from gene count matrix by DESeq2 and then z-scored by row.
HAT and HDAC activity assays
Nuclear protein extractions were isolated from 105 fibroblasts cultured on glass, PAAm, MeHA, or o-NB PAAm gels of varying stiffness for the indicated time points by using the nuclear extraction kit (EpiGentek, OP-0002, USA), according the manufacturer’s instructions. HAT and HDAC activity were measured by using the HAT activity/inhibition assay kit (EpiGentek, P-4003-48, USA) and HDAC activity/inhibition assay kit (EpiGentek, P-4034-96, USA), respectively. Following the manufacturer’s instructions, 5 μg of nuclear extract was added into the assay wells and incubated at 37°C for 60 min. After adding the color developer solution, the absorbance was measured using a plate reader (Infinite 200Pro, 30050303) at 450 nm. For inhibitor experiments, fibroblasts were cultured on glass and PA gels of varying stiffness for 1 day, followed by treatment with vehicle control (DMSO), anacardic acid (1 μM; Cayman chemical, 13144), VPA (500 μM; Cayman chemical, 13033), cytochalasin D (0.5 μM or indicated concentrations; Cayman chemical, 11330), importazole (10 μM; Cayman chemical, 21491), or BMS-5 (4 μM; Cayman chemical, 21072) for 24 hours before isolating nuclear protein extracts and measuring HAT or HDAC activity as aforementioned.
Western blotting analysis
Fibroblasts cultured on glass or PAAm gels of varying stiffness for 2 days were lysed using radioimmunoprecipitation assay buffer with protease inhibitors. The protein concentration of each sample was measured using a NanoDrop Spectrophotometer (Life technology, USA). Equal amounts of total protein (50 μg) from each sample were separated in a 10% SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred to a polyvinylidene difluoride membrane at 120 V for 2 hours at room temperature. The blot was blocked with 3% nonfat dry milk suspended in TBS-T (Thermo Fisher Scientific, J77500.K2) for 1 hour at room temperature. Membranes were incubated with primary antibodies overnight at 4°C (table S1). The resulting blots were incubated with secondary antibodies (Anti-mouse IgG HRP-linked antibody, Cell Signaling Technology, 7076S and Anti-mouse IgG HRP-linked antibody, Cell Signaling Technology, 7074S) at 1:3000 dilution. Bands were scanned using a densitometer (Bio-Rad) and quantified using the Quantity One 4.6.3 software (Bio-Rad). Original blots can be found in fig. S38.
Nuclear and cytosol protein extraction
Nuclear and cytosol protein extractions were isolated from 105 fibroblasts cultured on glass or PAAm gels of varying stiffness for 48 hours using the nuclear extraction kit (EpiGentek, OP-0002, USA), according the manufacturer’s instructions. Briefly, after removal of the growth medium, cells were washed twice with PBS. One milliliter of fresh PBS was added and cells were scraped into a 15-ml conical tube, followed by centrifugation at 1000 rpm for 5 min. Cytoplasmic extracts were isolated by incubating samples with pre-extraction buffer containing DTT and protease inhibitors for 10 min at 4°C and centrifuged at 12,000 rpm for 10 min. Upon removing the cytoplasmic extract (supernatant), samples were incubated with extraction buffer containing dithiothreitol and protease inhibitors for 15 min at 4°C and centrifuged at 14,000 rpm for 10 min to obtain nuclear protein extracts.
Immunoprecipitation
For immunoprecipitation studies, the immunoprecipitation kit (Abcam, ab206996, USA) was used to isolate the protein complex. After cells were cultured on gels for 48 hours, nuclear and cytosol protein were isolated using the nuclear extraction kit. Then, 20 μl of actin primary antibody (Santa Cruz Biotechnology, USA, sc-1616 and sc-47778) was mixed with the cell extract, and the volume was made up to 500 μl with lysis buffer containing the protease inhibitor cocktail, followed by an overnight incubation at 4°C on a rotary mixer. The next day, 40 μl of protein A/G sepharose beads slurry was added to each sample and incubated for 1 hour at 4°C. Beads were collected by low-speed centrifugation at 4°C and washed three times with the wash buffer. Upon removing the wash buffer, 40 μl of 2X SDS-PAGE loading buffer was added to the beads and samples were boiled for five min to elute the complex. Samples were then subjected to SDS-PAGE and Western blotting to visualize the results.
F-actin and G-actin isolation
To study the effects of matrix stiffness on F-actin and G-actin ratio, F-actin and G-actin content were isolated from nuclear and cytoplasmic extracts using the G-actin/F-actin In Vivo Assay Biochem Kit according to manufacturer’s instructions (Cytoskeleton, USA). Briefly, 5 × 106 cells were cultured on gels for 48 hours, and nuclear and cytoplasmic extracts were isolated by using the nuclear extraction kit. Proteins were then transferred to a prewarmed (37°C) ultracentrifuge (Beckman, Fullerton, CA) and spun at 150,000g for 1 hour at 37°C to separate G-actin (supernatant) and F-actin (pellet) fractions. After collecting the G-actin content, the pellets were resuspended in ice-cold deionized water with 10 μM cytochalasin D. The pellets were dissolved by triturating with a pipette and left on ice for 1 hour, vortexing every 10 min to dissociate F-actin. Resuspended solutions were centrifuged at 2300g for 5 min at 4°C. The second supernatants were collected as F-actin samples. All samples were diluted with appropriate loading buffer and boiled for 5 min. The samples were then subjected to SDS-PAGE and Western blotting to analyze actin content.
Electrophysiology
Samples were treated with a standard bath solution containing 145 mM NaCl, 3 mM KCl, 10 mM Hepes, 3 mM CaCl2, 8 mM glucose, and 2 mM MgCl2 at pH 7.4. Whole-cell recording was made using a patch clamp amplifier (MultiClamp 700B, Axon Instruments) under infrared differential interference contrast optics. Microelectrodes were made from borosilicate glass capillaries, with a resistance of 4 to 5 megohm. For recording action potentials, cells were held at −70 mV in voltage-clamp mode. The intracellular solution for whole-cell recording of excitatory postsynaptic potentials and action potentials contained 140 mM potassium gluconate, 5 mM KCl, 10 mM Hepes, 0.2 mM EGTA, 2 mM MgCl2, 4 mM Mg–adenosine 5′-triphosphate (ATP), 0.3 mM Na2–guanosine 5′-triphosphate (GTP), and 10 mM Na2-phosphocreatine at pH 7.2. For recording sEPSCs, cells were pretreated with the extracellular bath solution containing 50 μM picrotoxin to exclude an inhibitory synaptic activity and held at −70 mV in voltage-clamp mode with the intracellular solution containing 30 mM CsMeSO4, 7 mM CsCl, 10 mM Hepes, 1 mM EGTA, 4 mM Mg-ATP, 0.3 mM Na2-GTP, and 10 mM Na2-phosphocreatine at pH 7.3. After recording basal sEPSC responses for 5 min, both 10 μM CNQX and 100 μM d,l-APV were applied to test a dependency of sEPSC responses on AMPA- and N-methyl-d-aspartate type of glutamate receptors. For measuring spontaneous inhibitory postsynaptic currents (sIPSCs), cells were pretreated with the bath solution containing 10 μM CNQX and 100 μM d,l-APV and held at −70 mV with the intracellular solution containing 137 mM CsCl, 10 mM Hepes, 1 mM EGTA, 4 mM Mg-ATP, 0.3 mM Na2-GTP, and 10 mM Na2-phosphocreatine at pH 7.3. Picrotoxin (50 μM) was then added to test a dependency of sIPSC responses on γ-aminobutyric acid receptors after acquiring basal sIPSC responses for 5 min. Series resistance (15 to 30 megohm) and input resistance (~200 megohm using potassium-based internal solution; 1 to 2 gigaohm using Cs-based internal solution) were monitored throughout the whole-cell recording or compared before and after sEPSC/IPSC recordings. Off-line analysis of spontaneous EPSC and IPSC were performed by using a threshold event detection function of Clampfit software (Molecular Devices).
DNA methylation assay
Fibroblasts plated on glass or PAAm gels of varying stiffness for 2 days were trypsinized, and DNA was extracted by using the Invitrogen PureLink Genomic DNA Mini Kit (Invitrogen, K1820-01). The 5-mC level was analyzed by using the MethylFlash Global DNA Methylation (5-mC) ELISA Easy Kit (Epigentek, P-1030) according to the manufacturer’s instructions. Briefly, 100 ng of sample DNA was bonded into the assay wells and incubated with a 5-mC detection complex solution for 60 min. Then, color developer solution was added into assay wells, and the absorbance at 450 nm was measured using a plate reader (Infinite 200Pro, 30050303).
Atomic force microscopy
To determine the elastic modulus of MeHA gels, mechanical measurements of hydrogels were performed using atomic force microscopy (AFM). Uncoated MeHA gels were fabricated by exposure to UV light as indicated and incubated in PBS for several hours before performing the AFM measurements. Mechanical measurements were performed on a JPK Nanowizard 4a AFM with a colloidal probe (sphere Ø = 3.5 μm) (CP-qp-CONT-SiO-B, NanoAndMore Corp., USA), a highly sensitive cantilever k = 0.1 N/m, and sample Poisson’s ratio of 0.5 at the UCLA Nano and Pico Characterization facility. During the measurement, hydrogels were incubated on a glass-bottom dish with prewarmed PBS and set on a temperature-controlled stage at 37°C. The force-distance curves were recorded, and the elastic modulus of hydrogels was calculated by NanoScope Analysis using the Hertz model.
Compression testing
Compression test was performed on a Chatillon TCD 225 series force measurement system. After measuring the thickness of the PAAm gels (8 mm in diameter, equilibrated in deionized water for 24 hours), the samples were compressed with a deformation rate of 0.85 mm min−1 and a sampling rate of 1 Hz. Stress and strain were calculated from the load/displacement data. Elastic modulus were then computed from the stress-strain curve to compare the mechanical properties of samples.
Scanning electron microscopy
After equilibrating PAAm gels in deionized water for 24 hours, gels were put in liquid nitrogen and freeze-dried. The freeze-dried gels were mounted on stubs for scanning electron microscopy (SEM) with the aid of conductive adhesive tapes and then sputter-coated with gold palladium (Ted Pella Inc.) at 40 mA for 60 s to achieve satisfactory conductivity with minimal damage to the specimen. The coated samples were observed in a Zeiss Supra 40 VP SEM, operated at 10 kV.
Cell viability
Cell viability was assayed using the PrestoBlue Cell Viability Reagent (Invitrogen, A13261) according to the manufacturer’s protocol. Cells were incubated with the PrestoBlue Reagent for 2 hours. Absorbance was measured by a plate reader (Infinite 200PRO) at excitation/emission = 560/590 nm. Results were normalized to control (i.e., DMSO) or glass samples.
Statistical analysis
All data are presented as means ± 1 SD, where sample size (n) ≥ 3. Box plots show the ends at the quartiles and the mean as a horizontal line in the box, and the whiskers represent the SD. For most of the experiments, the sample size (n) refers to the number of independently prepared hydrogels analyzed. When sample size indicates a certain number of cells were examined, these results were obtained from analyzing cells across three hydrogels. Comparisons among values for groups greater than two were performed using a one-way analysis of variance (ANOVA), followed by a Tukey’s post hoc test. For two group analysis, a two-tailed, unpaired Student’s t test was used to analyze differences. For all cases, P values less than 0.05 were considered statistically significant. Origin 2018 software was used for all statistical evaluations.
Acknowledgments
We would like to thank J. Yu, S. Duan, Y. Teng, and R. Fu for the advice and assistance with SEM sample preparation.
Funding: The authors were supported, in part, by a UCLA Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research Innovation Award; grants from the National Institute of Health (GM143485 and NS130677 to S.L.); a National Science Scholarship (PhD) from the Singapore Agency for Science, Technology and Research (to S.Y.W.); a fellowship from the National Science Foundation GRFP (to J.S.); a postdoctoral fellowship from the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the NIH under the Ruth L. Kirschstein National Research Service Award (T32AR059033, to J.S.); a National Institutes of Health (NIH) New Innovator Award (DP2) Grant (DP2CA250382-01 to T.L.D); a National Science Foundation (NSF) grant (DMS1763272 to T.L.D.); a grant from the Simons Foundation (594598 QN to T.L.D.); and a T32 training grant from the NIH (HL116270 to N.A.). The authors acknowledge the support by the NIH/NCI under award number P30CA016042 and the Genomics core at the Jonsson Comprehensive Cancer Center, and the Advanced Light Microscopy and Spectroscopy Laboratory at the California NanoSystems Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author contributions: Y.S., J.S., and S.L. designed the experiments. Y.S., J.S., S.Y.W., N.A., S.N., J.C., H.P., D.O.K., and C.E.A. performed the experiments. Y.S., J.S., S.Y.W., Y.W., T.H., J.C., H.P., and D.O.K. analyzed the data. Y.S., J.S., M.W., A.K., T.L.D., M.-m.P., and S.L. contributed to data interpretation and discussion. Y.S., J.S., and S.L. wrote the manuscript.
Competing interests: Y.S., J.S., B.C., Y.W., and S.L. have filed a patent application. M.W. is cofounder and owns stock at Neucyte Inc. and is a scientific advisor and was offered stock options at bit.bio Inc. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Sequencing datasets are available from the GEO repository (GSE250069).
Supplementary Materials
This PDF file includes:
Figs. S1 to S38
Table S1
REFERENCE AND NOTES
- 1.Chowdhury F., Na S., Li D., Poh Y.-C., Tanaka T. S., Wang F., Wang N., Material properties of the cell dictate stress-induced spreading and differentiation in embryonic stem cells. Nat. Mater. 9, 82–88 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DuFort C. C., Paszek M. J., Weaver V. M., Balancing forces: Architectural control of mechanotransduction. Nat. Rev. Mol. Cell Biol. 12, 308–319 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engler A. J., Sen S., Sweeney H. L., Discher D. E., Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Guilak F., Cohen D. M., Estes B. T., Gimble J. M., Liedtke W., Chen C. S., Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell 5, 17–26 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McBeath R., Pirone D. M., Nelson C. M., Bhadriraju K., Chen C. S., Cell shape, cytoskeletal tension, and RhoA Regulate Stem Cell Lineage Commitment. Dev. Cell 6, 483–495 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Park J. S., Chu J. S., Tsou A. D., Diop R., Tang Z., Wang A., Li S., The effect of matrix stiffness on the differentiation of mesenchymal stem cells in response to TGF-β. Biomaterials 32, 3921–3930 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das R. K., Gocheva V., Hammink R., Zouani O. F., Rowan A. E., Stress-stiffening-mediated stem-cell commitment switch in soft responsive hydrogels. Nat. Mater. 15, 318–325 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Watt F. M., Huck W. T. S., Role of the extracellular matrix in regulating stem cell fate. Nat. Rev. Mol. Cell Biol. 14, 467–473 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Wen J. H., Vincent L. G., Fuhrmann A., Choi Y. S., Hribar K. C., Taylor-Weiner H., Chen S., Engler A. J., Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat. Mater. 13, 979–987 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang C., Tibbitt M. W., Basta L., Anseth K. S., Mechanical memory and dosing influence stem cell fate. Nat. Mater. 13, 645–652 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowder S. W., Leonardo V., Whittaker T., Papathanasiou P., Stevens M. M., Material cues as potent regulators of epigenetics and stem cell function. Cell Stem Cell 18, 39–52 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong S. Y., Soto J., Li S., Biophysical regulation of cell reprogramming. Curr. Opin. Chem. Eng. 15, 95–101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Downing T. L., Soto J., Morez C., Houssin T., Fritz A., Yuan F., Chu J., Patel S., Schaffer D. V., Li S., Biophysical regulation of epigenetic state and cell reprogramming. Nat. Mater. 12, 1154–1162 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulangara K., Adler A. F., Wang H., Chellappan M., Hammett E., Yasuda R., Leong K. W., The effect of substrate topography on direct reprogramming of fibroblasts to induced neurons. Biomaterials 35, 5327–5336 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoo J., Noh M., Kim H., Jeon N. L., Kim B.-S., Kim J., Nanogrooved substrate promotes direct lineage reprogramming of fibroblasts to functional induced dopaminergic neurons. Biomaterials 45, 36–45 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Song Y., Soto J., Chen B., Hoffman T., Zhao W., Zhu N., Peng Q., Liu L., Ly C., Wong P. K., Wang Y., Rowat A. C., Kurdistani S. K., Li S., Transient nuclear deformation primes epigenetic state and promotes cell reprogramming. Nat. Mater. 21, 1191–1199 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grath A., Dai G., Direct cell reprogramming for tissue engineering and regenerative medicine. J. Biol. Eng. 13, 14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vasan L., Park E., David L. A., Fleming T., Schuurmans C., Direct neuronal reprogramming: Bridging the gap between basic science and clinical application. Front. Cell Dev. Biol. 9, 681087 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H., Yang Y., Liu J., Qian L., Direct cell reprogramming: Approaches, mechanisms and progress. Nat. Rev. Mol. Cell Biol. 22, 410–424 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith Z. D., Sindhu C., Meissner A., Molecular features of cellular reprogramming and development. Nat. Rev. Mol. Cell Biol. 17, 139–154 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Uhler C., Shivashankar G. V., Regulation of genome organization and gene expression by nuclear mechanotransduction. Nat. Rev. Mol. Cell Biol. 18, 717–727 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Song Y., Soto J., Chen B., Yang L., Li S., Cell engineering: Biophysical regulation of the nucleus. Biomaterials 234, 119743 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Walker C. J., Batan D., Bishop C. T., Ramirez D., Aguado B. A., Schroeder M. E., Crocini C., Schwisow J., Moulton K., Macdougall L., Weiss R. M., Allen M. A., Dowell R., Leinwand L. A., Anseth K. S., Extracellular matrix stiffness controls cardiac valve myofibroblast activation through epigenetic remodeling. Bioeng. Transl. Med. 7, e10394 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao X., Chen Y., Tan M., Zhao L., Zhai Y., Sun Y., Gong Y., Feng X., Du J., Fan Y., Extracellular matrix stiffness regulates DNA methylation by PKCα-dependent nuclear transport of DNMT3L. Adv. Healthc. Mater. 10, e2100821 (2021). [DOI] [PubMed] [Google Scholar]
- 25.Heo S. J., Thakur S., Chen X., Loebel C., Xia B., McBeath R., Burdick J. A., Shenoy V. B., Mauck R. L., Lakadamyali M., Aberrant chromatin reorganization in cells from diseased fibrous connective tissue in response to altered chemomechanical cues. Nat. Biomed. Eng. 7, 177–191 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vining K. H., Mooney D. J., Mechanical forces direct stem cell behaviour in development and regeneration. Nat. Rev. Mol. Cell Biol. 18, 728–742 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang N., Tytell J. D., Ingber D. E., Mechanotransduction at a distance: Mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 10, 75–82 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Kirby T. J., Lammerding J., Emerging views of the nucleus as a cellular mechanosensor. Nat. Cell Biol. 20, 373–381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikeda T., Hikichi T., Miura H., Shibata H., Mitsunaga K., Yamada Y., Woltjen K., Miyamoto K., Hiratani I., Yamada Y., Hotta A., Yamamoto T., Okita K., Masui S., Srf destabilizes cellular identity by suppressing cell-type-specific gene expression programs. Nat. Commun. 9, 1387 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alisafaei F., Jokhun D. S., Shivashankar G. V., Shenoy V. B., Regulation of nuclear architecture, mechanics, and nucleocytoplasmic shuttling of epigenetic factors by cell geometric constraints. Proc. Natl. Acad. Sci. U.S.A. 116, 13200–13209 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pereira D., Richert A., Medjkane S., Hénon S., Weitzman J. B., Cell geometry and the cytoskeleton impact the nucleo-cytoplasmic localisation of the SMYD3 methyltransferase. Sci. Rep. 10, 20598 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dupont S., Wickström S. A., Mechanical regulation of chromatin and transcription. Nat. Rev. Genet. 23, 624–643 (2022). [DOI] [PubMed] [Google Scholar]
- 33.Elosegui-Artola A., Andreu I., Beedle A. E. M., Lezamiz A., Uroz M., Kosmalska A. J., Oria R., Kechagia J. Z., Rico-Lastres P., Le Roux A.-L., Shanahan C. M., Trepat X., Navajas D., Garcia-Manyes S., Roca-Cusachs P., Force triggers YAP nuclear entry by regulating transport across nuclear pores. Cell 171, 1397–1410.e14 (2017). [DOI] [PubMed] [Google Scholar]
- 34.L. Vincent, A. J. Engler, in Comprehensive Biomaterials II (Elsevier, 2017), pp. 88–101. [Google Scholar]
- 35.Yi B., Xu Q., Liu W., An overview of substrate stiffness guided cellular response and its applications in tissue regeneration. Bioact. Mater. 15, 82–102 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petrie R. J., Gavara N., Chadwick R. S., Yamada K. M., Nonpolarized signaling reveals two distinct modes of 3D cell migration. J. Cell Biol. 197, 439–455 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Achterberg V. F., Buscemi L., Diekmann H., Smith-Clerc J., Schwengler H., Meister J. J., Wenck H., Gallinat S., Hinz B., The nano-scale mechanical properties of the extracellular matrix regulate dermal fibroblast function. J. Invest. Dermatol. 134, 1862–1872 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Antonovaite N., Beekmans S. V., Hol E. M., Wadman W. J., Iannuzzi D., Regional variations in stiffness in live mouse brain tissue determined by depth-controlled indentation mapping. Sci. Rep. 8, 12517 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Viji Babu P. K., Radmacher M., Mechanics of brain tissues studied by atomic force microscopy: A perspective. Front. Neurosci. 13, 453881 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wapinski O. L., Vierbuchen T., Qu K., Lee Q. Y., Chanda S., Fuentes D. R., Giresi P. G., Ng Y. H., Marro S., Neff N. F., Drechsel D., Martynoga B., Castro D. S., Webb A. E., Südhof T. C., Brunet A., Guillemot F., Chang H. Y., Wernig M., Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell 155, 621–635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cherry A. B. C., Daley G. Q., Reprogramming cellular identity for regenerative medicine. Cell 148, 1110–1122 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Theunissen T. W., Jaenisch R., Molecular control of induced pluripotency. Cell Stem Cell 14, 720–734 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee K. K., Workman J. L., Histone acetyltransferase complexes: One size doesn’t fit all. Nat. Rev. Mol. Cell Biol. 8, 284–295 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Chambliss A. B., Khatau S. B., Erdenberger N., Robinson D. K., Hodzic D., Longmore G. D., Wirtz D., The LINC-anchored actin cap connects the extracellular milieu to the nucleus for ultrafast mechanotransduction. Sci. Rep. 3, 1087 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dahl K. N., Ribeiro A. J. S., Lammerding J., Nuclear shape, mechanics, and mechanotransduction. Circ. Res. 102, 1307–1318 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khatau S. B., Hale C. M., Stewart-Hutchinson P. J., Patel M. S., Stewart C. L., Searson P. C., Hodzic D., Wirtz D., A perinuclear actin cap regulates nuclear shape. Proc. Natl. Acad. Sci. U.S.A. 106, 19017–19022 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simon D. N., Wilson K. L., The nucleoskeleton as a genome-associated dynamic “network of networks”. Nat. Rev. Mol. Cell Biol. 12, 695–708 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Tajik A., Zhang Y., Wei F., Sun J., Jia Q., Zhou W., Singh R., Khanna N., Belmont A. S., Wang N., Transcription upregulation via force-induced direct stretching of chromatin. Nat. Mater. 15, 1287–1296 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dopie J., Skarp K.-P., Kaisa Rajakylä E., Tanhuanpää K., Vartiainen M. K., Active maintenance of nuclear actin by importin 9 supports transcription. Proc. Natl. Acad. Sci. 109, E544–E552 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanellos G., Frame M. C., Cellular functions of the ADF/cofilin family at a glance. J. Cell Sci. 129, 3211–3218 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Hoffman L. M., Smith M. A., Jensen C. C., Yoshigi M., Blankman E., Ullman K. S., Beckerle M. C., Mechanical stress triggers nuclear remodeling and the formation of transmembrane actin nuclear lines with associated nuclear pore complexes. Mol. Biol. Cell 31, 1774–1787 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kassianidou E., Kalita J., Lim R. Y. H., The role of nucleocytoplasmic transport in mechanotransduction. Exp. Cell Res. 377, 86–93 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Domingues C., Geraldo A. M., Anjo S. I., Matos A., Almeida C., Caramelo I., Lopes-da-Silva J. A., Paiva A., Carvalho J., das Neves R. P., Manadas B., Grãos M., Cofilin-1 is a mechanosensitive regulator of transcription. Front. Cell Dev. Biol. 8, 678 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu J., Huang Y., Zhao J., Wu L., Qi Q., Liu Y., Li G., Li J., Liu H., Wu H., Cofilin: A promising protein implicated in cancer metastasis and apoptosis. Front. Cell Dev. Biol. 9, 599065 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park J.-B., Agnihotri S., Golbourn B., Bertrand K. C., Luck A., Sabha N., Smith C. A., Byron S., Zadeh G., Croul S., Berens M., Rutka J. T., Transcriptional profiling of GBM invasion genes identifies effective inhibitors of the LIM kinase-Cofilin pathway. Oncotarget 5, 9382–9395 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang W., Halasz E., Townes-Anderson E., Actin dynamics, regulated by RhoA-LIMK-cofilin signaling, mediates rod photoreceptor axonal retraction after retinal injury. Invest. Ophthalmol. Vis. Sci. 60, 2274–2285 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rajakylä E. K., Vartiainen M. K., Rho, nuclear actin, and actin-binding proteins in the regulation of transcription and gene expression. Small GTPases. 5, e27539 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Norris S. C. P., Soto J., Kasko A. M., Li S., Photodegradable polyacrylamide gels for dynamic control of cell functions. ACS Appl. Mater. Interfaces 13, 5929–5944 (2021). [DOI] [PubMed] [Google Scholar]
- 59.Guo S., Zi X., Schulz V. P., Cheng J., Zhong M., Koochaki S. H. J., Megyola C. M., Pan X., Heydari K., Weissman S. M., Gallagher P. G., Krause D. S., Fan R., Lu J., Nonstochastic reprogramming from a privileged somatic cell state. Cell 156, 649–662 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thomas C. H., Collier J. H., Sfeir C. S., Healy K. E., Engineering gene expression and protein synthesis by modulation of nuclear shape. Proc. Natl. Acad. Sci. U.S.A. 99, 1972–1977 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soto J., Song Y., Wu Y., Chen B., Park H., Akhtar N., Wang P., Hoffman T., Ly C., Sia J., Wong S., Kelkhoff D. O., Chu J., Poo M., Downing T. L., Rowat A. C., Li S., Reduction of intracellular tension and cell adhesion promotes open chromatin structure and enhances cell reprogramming. Adv. Sci.(Weinh) 10, e2300152 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sen B., Xie Z., Howard S., Styner M., van Wijnen A. J., Uzer G., Rubin J., Mechanically induced nuclear shuttling of β-catenin requires Co-transfer of actin. Stem Cells 40, 423–434 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Song Y., Soto J., Li S., Mechanical regulation of histone modifications and cell plasticity. Curr. Opin. Solid State Mater. Sci. 24, 100872 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Y., Tang C. B., Kilian K. A., Matrix mechanics influence fibroblast–myofibroblast transition by directing the localization of histone Deacetylase 4. Cell. Mol. Bioeng. 10, 405–415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chaudhuri O., Cooper-White J., Janmey P. A., Mooney D. J., Shenoy V. B., Effects of extracellular matrix viscoelasticity on cellular behaviour. Nature 584, 535–546 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cacopardo L., Guazzelli N., Ahluwalia A., Characterizing and engineering biomimetic materials for viscoelastic mechanotransduction studies. Tissue Eng. Part B Rev. 28, 912–925 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saha K., Keung A. J., Irwin E. F., Li Y., Little L., Schaffer D. V., Healy K. E., Substrate modulus directs neural stem cell behavior. Biophys. J. 95, 4426–4438 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tse J. R., Engler A. J., Preparation of hydrogel substrates with tunable mechanical properties. Curr. Protoc. Cell Biol. 47, 10.16 1–10.16.16 (2010). [DOI] [PubMed] [Google Scholar]
- 69.Sunyer R., Jin A. J., Nossal R., Sackett D. L., Fabrication of hydrogels with steep stiffness gradients for studying cell mechanical response. PLOS ONE 7, e46107 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee I. N., Dobre O., Richards D., Ballestrem C., Curran J. M., Hunt J. A., Richardson S. M., Swift J., Wong L. S., Photoresponsive hydrogels with photoswitchable mechanical properties allow time-resolved analysis of cellular responses to matrix stiffening. ACS Appl. Mater. Interfaces 10, 7765–7776 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ondeck M. G., Engler A. J., Mechanical characterization of a dynamic and tunable methacrylated hyaluronic acid hydrogel. J. Biomech. Eng. 138, 021003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Corces M. R., Trevino A. E., Hamilton E. G., Greenside P. G., Sinnott-Armstrong N. A., Vesuna S., Satpathy A. T., Rubin A. J., Montine K. S., Wu B., Kathiria A., Cho S. W., Mumbach M. R., Carter A. C., Kasowski M., Orloff L. A., Risca V. I., Kundaje A., Khavari P. A., Montine T. J., Greenleaf W. J., Chang H. Y., An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J. 17, 10–12 (2011). [Google Scholar]
- 74.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Danecek P., Bonfield J. K., Liddle J., Marshall J., Ohan V., Pollard M. O., Whitwham A., Keane T., McCarthy S. A., Davies R. M., Li H., Twelve years of SAMtools and BCFtools. Gigascience 10, giab008 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nussbaum C., Myers R. M., Brown M., Li W., Shirley X. S., Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, 1–9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 78.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., Getz G., Mesirov J. P., Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Treutlein B., Lee Q. Y., Camp J. G., Mall M., Koh W., Shariati S. A., Sim S. K., Neff N., Skotheim J., Wernig M., Quake S., Quake S., Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq. Nature 534, 391–395 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., Fu X., Liu S., Bo X., Yu G., clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation(Camb) 2, 100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S38
Table S1