Abstract
In breast cancers, aberrant activation of the RAS/MAPK pathway is strongly associated with mesenchymal features and stemness traits, suggesting an interplay between this mitogenic signaling pathway and epithelial-to-mesenchymal plasticity (EMP). By using inducible models of human mammary epithelial cells, we demonstrate herein that the oncogenic activation of RAS promotes ZEB1-dependent EMP, which is necessary for malignant transformation. Notably, EMP is triggered by the secretion of pro-inflammatory cytokines from neighboring RAS-activated senescent cells, with a prominent role for IL-6 and IL-1α. Our data contrast with the common view of cellular senescence as a tumor-suppressive mechanism and EMP as a process promoting late stages of tumor progression in response to signals from the tumor microenvironment. We highlighted here a pro-tumorigenic cooperation of RAS-activated mammary epithelial cells, which leverages on oncogene-induced senescence and EMP to trigger cellular reprogramming and malignant transformation.
Transformation of human mammary epithelial cells is set off by paracrine signals from vicinal RAS-induced senescent cells.
INTRODUCTION
Epithelial-to-mesenchymal transition (EMT) is a fundamental embryonic process enabling the reprogramming of epithelial cells toward a mesenchymal phenotype (1, 2). During EMT, cells lose epithelial features including cell-cell junctions and apical-basal polarity and acquire mesenchymal phenotypic traits and increased cell motility (3). The activation of this transdifferentiation program is orchestrated by a network of EMT-inducing transcription factors (EMT-TFs) including members of the Zinc Finger E-Box Binding Homeobox (ZEB), SNAIL, and TWIST families that can repress the expression of epithelial genes and induce the expression of mesenchymal genes (4–9). EMT-TFs have been shown to be aberrantly reactivated in a series of pathogenic conditions, including tissue fibrosis and cancer (1, 10). In the context of tumor development, reactivation of EMT-TFs triggers an EMT-like process designated as epithelial-to-mesenchymal plasticity (EMP) (3). EMP was recently defined as a set of multiple and dynamic steps between epithelial and mesenchymal phenotypes, leading to carcinoma cells with hybrid epithelial-to-mesenchymal characteristics in which individual cells co-express markers of both epithelial and mesenchymal lineages (3, 11–14). Over the course of this dynamic transdifferentiation process, malignant cells acquire motility capabilities and stemness characteristics, promoting tumor invasion, metastases formation, and therapy resistance (10, 15, 16). It is generally accepted that the aberrant reactivation of EMT-TFs is mainly triggered by signals from the tumor microenvironment (TME), which include growth factors and pro-inflammatory cytokines (17, 18). Notably, apart from their role over the course of tumor progression, several in vitro and in vivo studies highlighted the reactivation of EMT-TFs in early steps of malignant transformation (19–22). In early tumorigenesis, because the microenvironment has not yet been shaped by cancer cells, these observations suggest that cell-autonomous mechanisms may be involved in EMP induction.
Although the precise role of EMP in the early stages of tumorigenesis remains to be determined, EMT-TFs have been shown in vitro to cooperate with mitogenic oncoproteins, including RAS, for malignant transformation by attenuating key oncosuppressive barriers (22–25). Furthermore, in mouse models, ectopic expression of EMT-TFs in mammary epithelial cells cooperates with mutant Kras to promote tumorigenesis (22, 26). Further supporting a link between the RAS/mitogen-activated protein kinase (MAPK) signaling pathway and EMP in vivo, molecular analysis of human tumors demonstrated common activation of the mitogenic pathway in the “claudin-low” (CL) molecular subtype of breast cancers, a subtype defined by its mesenchymal and stem cell signatures (27, 28). Together, these observations led us to test the hypothesis of a cell-autonomous interaction between oncogenic activation of RAS and EMP during early steps of breast tumorigenesis. Here, using human mammary epithelial (HME) cells with inducible RAS activation, we uncover an unexpected interplay between oncogene-induced senescence (OIS) and EMP during malignant transformation.
RESULTS
RAS activation in differentiated mammary cells induces a phenotypic switch associated with EMT-TF expression
To confirm the association between RAS/MAPK activation and mesenchymal features in breast cancers, single-sample gene set enrichment analysis (ssGSEA) scores were generated for the two pathways (RAS/MAPK and EMT, hereon designated as EMP) using Molecular Signatures Database (MSigDB) C2-curated gene sets (Kyoto Encyclopedia of Genes and Genomes, Gene Ontology, Reactome, etc.), and EMP scores were generated using transcriptomic EMP (EMT) signature defined by Tan et al. (29) on three breast cancer cohorts [The Cancer Genome Atlas Network (TCGA; www.cancer.gov/tcga), Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) (30), and Cancer Cell Line Encyclopaedia (CCLE) (31)]. As expected from recent reports, signatures of RAS/MAPK signaling as well as EMP were overrepresented in the CL breast cancer subtype compared to other breast cancer subtypes (fig. S1, A to F) (27). Notably, the RAS/MAPK pathway was highly correlated with EMP in the CL cell lines, excluding any stroma contamination bias (fig. S1, G to I). Nevertheless, although these data confirm a link between activated RAS/MAPK signaling and EMP features in breast cancers, they do not demonstrate that EMP reactivation is a consequence of oncogenic RAS activation.
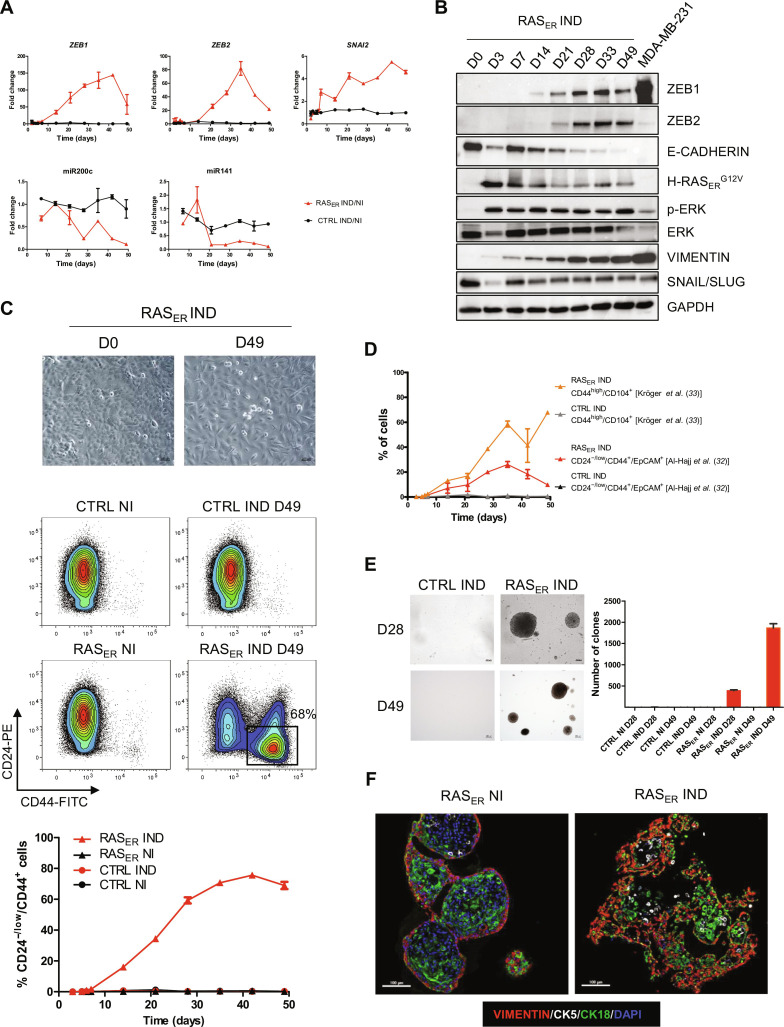
To test whether the correlation between RAS activation and EMP signatures in human breast cancers may imply causation, we carried out kinetics analysis in a RAS-inducible cellular model, HME-RASER, generated by introducing a 4-hydroxytamoxifen (4-OHT)–inducible form of HRASG12V into HME cells (HMECs). After 7 days of RAS activation in HME-RASER, an increase in expression of EMT-TFs ZEB1 and ZEB2 as well as a concomitant decrease in the expression of ZEB negative regulators, the miR-200 mRNA family members, were revealed (Fig. 1, A and B, and fig. S2A). Moreover, a clear change in cellular morphology was noted at day 49 (D49) following RAS induction, with HME-RASER cells losing their cobblestone-like epithelial morphology and gaining a spindle-like morphology suggestive of mesenchymal features (Fig. 1C). Engagement of HME-RASER cells to undergo EMP following RAS activation was confirmed by decreased E-cadherin expression and increased vimentin expression (Fig. 1B). It was previously demonstrated that cell commitment to EMP was associated with the acquisition of features of cancer stem cells (CSCs) (15, 16, 32, 33). Therefore, the expression of CD24, CD44, Epithelial Cell Adhesion Molecule (EpCAM), and CD104 was evaluated by flow cytometry analysis in HME-RASER cells ensuing RAS activation. The emergence of the three main phenotypes associated with breast CSC, namely, CD24−/low/CD44+ (Fig. 1C) and CD24−/low/CD44+/EpCAM+ and CD44+/CD104+ (Fig. 1D and fig. S2B), was observed as early as D7 following RAS activation, suggesting the onset of a reprogramming process. These results were confirmed using two other independent HME-RASER models (fig. S3). The emergence of these CSC phenotypes was accompanied by an increased capacity to grow in soft agar, a gold standard assay for cellular transformation (Fig. 1E). We next examined whether RAS activation affected the structural and morphological organization in three-dimensional (3D) culture. Here, HME-RASER cells were grown in 3D organoid culture conditions for 10 days, without 4-OHT activation, to obtain a typical organoid structure before RAS activation. A disruption in the structural organization associated with the acquisition of EMP features was observed in 4-OHT–induced HME-RASER organoids compared to non-induced organoids (Fig. 1F). Collectively, these data suggest that RAS activation in HMECs induces EMT-TF expression and promotes cell plasticity associated with the acquisition of stemness markers.
Fig. 1. RAS activation in differentiated mammary cells induces a phenotypic switch in 2D associated with the expression of EMT-TFs and a disruption of organoid formation in 3D.
(A) mRNA relative expression fold change across time for EMT-TFs ZEB1, ZEB2, and SNAI2; miR200c; and miR141 in HME-RASER (RASER) or HME-CTRL (CTRL) cells induced with 4-OHT (IND) or not (NI) (ratio IND/NI). Median ± range (n = 2 independent experiments in duplicate). (B) Immunoblot showing the expression of HRASERG12V, pERK1/2, ERK, ZEB1, ZEB2, SNAIL/SLUG, E-CADHERIN, and VIMENTIN in HME-RASER (RASER) cells at the indicated days following 4-OHT treatment. MDA-MB 231 cell line was used as a positive control for ZEB1, ZEB2, and SNAIL/SLUG expression. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) level was used as a loading control. (C) Top: Representative bright field images of HME-RASER cells at D0 and D49 after RAS activation (scale bars, 200 μm). Bottom: Representative fluorescence-activated cell sorting (FACS) analysis of CD24 and CD44 markers in HME-RASER (RASER) and HME-CTRL (CTRL) cells at D0 and D49 of induction with 4-OHT (IND) or not (NI). Kinetics analysis across time of the percentage population of CD24−/low/CD44+ in HME-RASER cells and HME-CTRL cells after 4-OHT treatment or not. Median ± range (n = 4 independent experiments). FITC, fluorescein isothiocyanate; PE, phycoerythrin. (D) Kinetics FACS analysis of cancer stem cell (CSC) markers CD24−/low/CD44+/EpCAM+ [as described by (32)] and CD44+/CD104+ [as described by (33)] in HME-RASER cells and HME-CTRL cells after 4-OHT treatment. Median ± range (n = 2 independent experiments). (E) Transformation potential analysis by soft agar colony formation assay. Representative images of phase-contrast colonies (scale bars, 200 μm). The number of colonies (defined by >20 cells) is indicated. Median ± SD (n = 3 independent experiments). (F) Representative images of multi-immunofluorescence staining for the indicated markers in cross sections of organoids generated from HME-RASER cells (RASER) that were induced with 4-OHT (IND) or not (NI) for 21 days (scale bars, 100 μm) (n = 3 independent experiments). DAPI, 4′,6-diamidino-2-phenylindole.
RAS-induced cellular plasticity and transforming ability are ZEB1-dependent
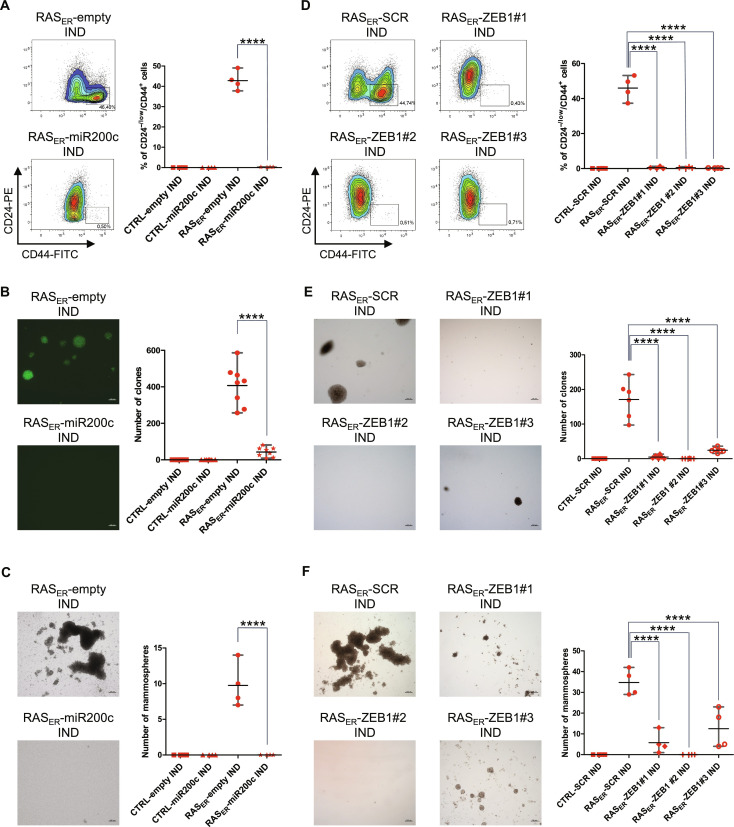
To determine whether RAS activation could trigger EMP in the absence of EMT-TFs, a vector allowing the constitutive expression of miR-200c, a major negative regulator of ZEB1 and ZEB2, was introduced into HME-RASER cells (fig. S4, A to C) (2, 34). As expected, an increase of both ZEB1 and ZEB2 gene expression and protein levels was dampened following RAS activation in this model (fig. S4, D and E). miR-200c expression further prevented the emergence of CD24−/low/CD44+ cells and the acquisition of properties associated with transformation and stemness (Fig. 2, A to C). Induced HME-RASER–miR-200c cells were neither able to form colonies in soft agar assays nor mammospheres in low-adherent culture conditions (Fig. 2, B and C). Hence, inhibiting the expression of ZEB1 and ZEB2 by overexpressing miR-200c prevented EMP induction following RAS activation.
Fig. 2. RAS-induced EMP and transformation capabilities are ZEB1-dependent.
(A and C) HME-RASER-miR200c cells (RASER-miR200c) or HME-RASER-empty cells (RASER-empty) after 28 days of induction with 4-OHT (IND). (D and F) HME-RASER-CRISPR ZEB1 clones (RASER-ZEB1#1, RASER-ZEB1#2, and RASER-ZEB1#3) or HME-RASER-CRISPR scramble (RASER-SCR) cells after 28 days of 4-OHT treatment (IND). (A) and (D) Flow cytometry analysis and quantification of CD24−/low/CD44+ cells. Data are presented as median ± range of four independent experiments (n = 4). (B and E) Transformation potential analysis by soft agar colony formation assay. The number of colonies (as defined by >20 cells) is indicated. Data are presented as median ± range of (B) four independent experiments in duplicate (n = 8), or (E) three independent experiments in duplicate (n = 6). (B) Images of GFP-positive colonies or (E) phase-contrast colonies (scale bars, 200 μm). (C) and (F) Quantification and phase-contrast images of mammospheres (scale bars, 200 μm). Data are presented as median ± range of four independent experiments (n = 4). P values are calculated by one-way ANOVA, Tukey multiple comparisons test (****P ≤ 0.0001).
Because ZEB1 is a known master regulator of EMP in mammary epithelial cells (35), a CRISPR-Cas9 knockdown approach was used to determine its role in the context of RAS activation. Three independent ZEB1-inactivated clones of HME-RASER cells were generated (HME-RASER-CRISPR ZEB1#1, HME-RASER-CRISPR ZEB1#2, and HME-RASER-CRISPR ZEB1#3) (fig. S4, F and G). ZEB1 inactivation alone inhibited the emergence of the CD24−/low/CD44+ population as well as the acquisition of transformation and stemness properties (Fig. 2, D to F). ZEB2 and SNAI2 induction was decreased in ZEB1-inactivated clones, suggesting that their up-regulation was at least partially ZEB1-dependent (fig. S4H). Together, these data demonstrate that, following RAS activation in differentiated HMECs, EMP induction relies on the ZEB1 TF.
Dual fate following RAS activation: Senescence or EMP
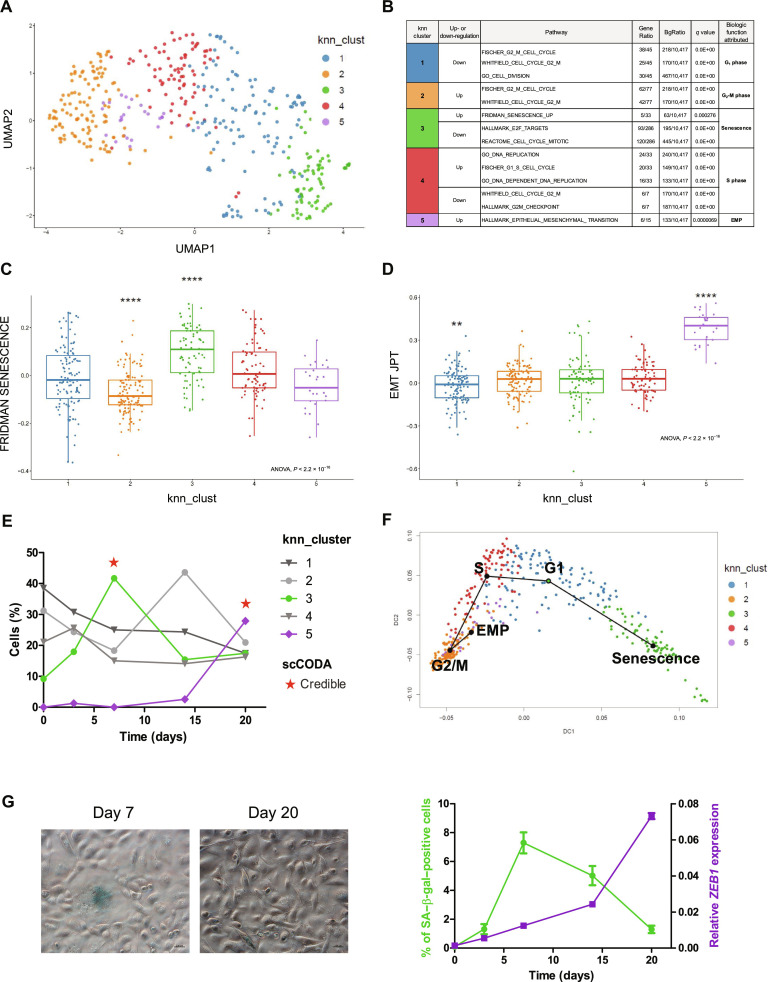
Because only a subpopulation of induced HME-RASER cells displays EMP features after RAS activation, we performed single-cell RNA sequencing (scRNA-seq) analysis at different time points (D0, D3, D7, D14, and D20) with the aim of deciphering the fate of individual cells. Expression profiles were obtained for 12,357 genes in 445 cells (117 cells at D0, 83 cells at D3, 74 cells at D7, 82 cells at D14, and 89 cells at D20). To analyze transcriptional variability over the complete scRNA-seq dataset, dimensions were reduced using principal components analysis (PCA) and uniform manifold approximation and projection (UMAP) (Fig. 3A) (36). A dimension reduction analysis and graph-based k-means clustering identified five clusters (numbered 1 to 5) based on differential gene expression (Fig. 3A and fig. S5A). For each cluster, specific marker genes were extracted to search for enriched pathways using MSigDB (tables S1 to S5). This allowed us to functionally annotate each cluster (Fig. 3B), resulting in clusters 1, 2, and 4 related to G1, G2-M, and S phases, respectively, and clusters 3 and 5 related to senescence and EMP, respectively (Fig. 3, C and D). Cluster 3, which represents less than 10% of cells at D0, constitutes more than 40% of cells at D7, while cluster 5 emerges at D14 and is the most represented at D20. Tests to assess compositional differences between cell types established them as credible for D7 and D20 [single-cell differential composition analysis (SCODA)] (Fig. 3E). We next interrogated a possible link between these two emerging clusters through evolutionary trajectory analysis. Given the uncertain origin of a potential trajectory, cluster 1 (related to G1 phase) was used as a starting point because it was the most represented at D0 (40%) (Fig. 3E). Trajectory analysis showed two different end points: cluster 3/senescence or cluster 5/EMP (Fig. 3F and fig. S5B), suggesting that, following RAS activation, cells would either undergo senescence or acquire a mesenchymal phenotype. Trajectory analysis using cluster 2 or 4 as a starting point gave identical results with regards to the evolutionary outcome: senescence or EMP (fig. S5C). Consistently, RNA velocity analysis corroborated the bidirectional nature discovered in the trajectory analysis, suggesting that cluster 3/senescence originated from cluster 1/G1, while cluster 5/EMP emerged from cluster 2/G2-M (fig. S5D). To mitigate the effects of cell cycle heterogeneity in scRNA-seq data, we regressed the cell cycle effect in our analysis, leading us to identify three clusters based on differential gene expression: cluster 1 related to proliferated epithelial cells, cluster 2 related to senescence, and cluster 3 related to EMP (fig. S6, A to F, and tables S6 to S8). Like for the first analysis, cluster 2/senescence was scarcely represented at D0 (5%), while being the major cell population at D7 (40%), whereas cluster 3/EMP emerged at D14 and was the most represented at D20 (fig. S6E). Thus, cell cycle regression analysis resulted in the same evolutionary trajectory supporting two distinct paths that cells take following RAS activation: senescence or EMP (fig. S6F). Independent of the analysis performed, the rise of the senescence cluster preceded that of the EMP one (D7 versus D20, respectively; Fig. 3, E and G, and fig. S6E), suggesting a causal relationship.
Fig. 3. Identification of distinct senescence and EMP clusters in induced HME-RASER.
(A) Unsupervised uniform manifold approximation and projection (UMAP) of the transcriptome of all cells at all time points (D0, D3, D7, D14, and D20). Cells are colored by their attributed cluster. (B) Main altered pathways by marker genes for each cluster. Gene ratio is presented as k/n, where k is the size of the overlap of our input with the specific gene set and n is the size of the overlap of our input with all members of the collection of gene sets. q Value refers to false discovery rate. (C and D) Scores per cell for two transcriptomic pathways: (C) FRIDMAN SENESCENCE (ssGSEA score) and (D) EMT JPT [EMT cell line score from (29)]. Cells are grouped by clusters, each box representing the median and interquartile ranges. Individual Wilcoxon tests, P value is represented by stars (**P ≤ 0.01 and ****P ≤ 0.0001). (E) Proportion of cells by cluster at each time point. Emerging clusters across time (cluster 3 in green and cluster 5 in purple) and statistical compositional single cells analysis by scCODA (* indicates credible change). (F) Trajectory curves projected onto a diffusion map for dimension reduction. Cells are colored by k-nearest neighbor (knn) clusters. Starting point of the trajectory analysis is cluster 1 (green circle). (G) Left: Representative images of senescence-associated β-galactosidase (SA–β-gal) activity in induced HME-RASER cells at D7 and D20 (scale bars, 200 μm). Right: Percentage of SA–β-gal–positive cells (green) and ZEB1 mRNA expression (purple) across time after RAS activation in induced HME-RASER cells. Median ± range (n = 3 independent experiments).
ZEB1-dependent EMP is driven by OIS
On the basis of scRNA-seq data, we sought to gain further insights into a potential cross-talk between OIS and EMP. We therefore analyzed the emergence of senescent cells, determined by β-galactosidase (SA–β-gal)–positive staining with respect to ZEB1 expression in a time-dependent manner. As shown in Fig. 3 (E and G), RAS-induced senescent cells appeared from D3 onward, with a peak at D7, whereas ZEB1 expression increased from D14 onward and was directly associated with the emergence of the CD24−/low/CD44+ population (Fig. 1, A to C).
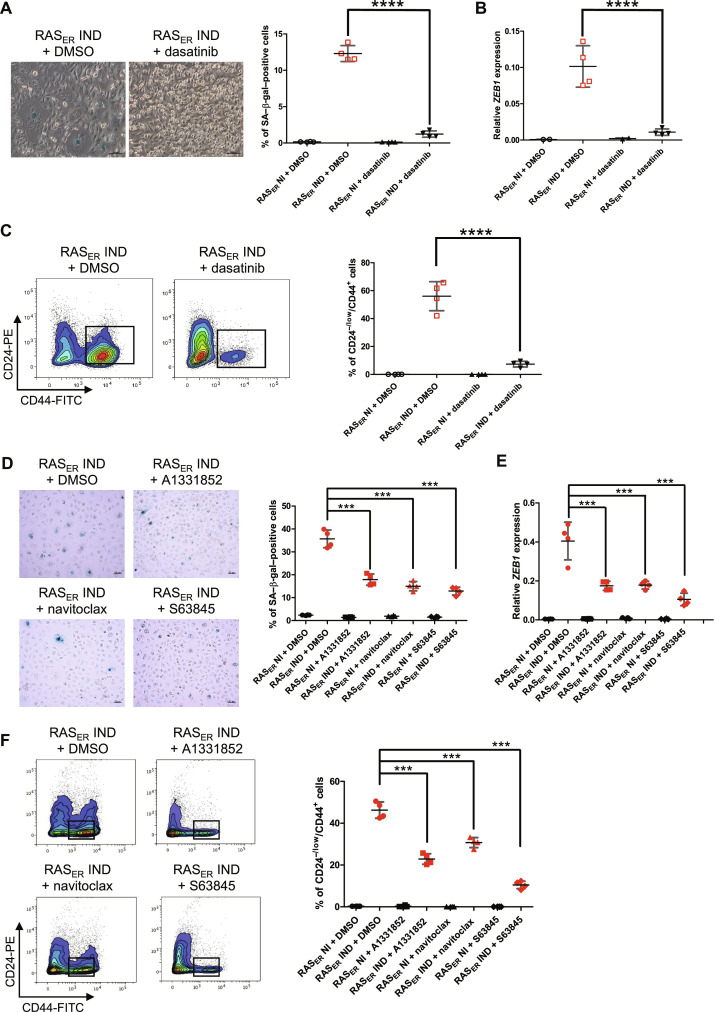
The initial emergence of OIS followed by the delayed occurrence of EMP led us to hypothesize that senescence may contribute to the induction of cellular plasticity. As a first approach to test this hypothesis, HME-RASER cells were treated with well-known senolytic drugs: dasatinib, an inhibitor of the Bcr-Abl protein kinase; A1331852 and navitoclax, both inhibitors of BCL-2, BCL-XL; and the MCL-1 inhibitor S63845 (37–41). Treatments with these different drugs significantly reduced the number of senescent cells at D7 of RAS-activation, along with a decrease in SA–β-gal–positive cells (Fig. 4, A and D). Notably, this senescence blockage was also associated with a decrease at D21 in both ZEB1 expression (Fig. 4, B and E) and the percentage of CD24−/low/CD44+ cells (Fig. 4, C and F), thus supporting ZEB1-dependent EMP to be reliant on the onset of senescence.
Fig. 4. Senescent cells drive ZEB1-dependent EMP.
(A and D) Representative phase-contrast images and quantification of SA–β-gal activity in HME-RASER (RASER) cells analyzed at D7 of induction with 4-OHT (IND) or not (NI) and treatment with senolytic drugs or the equivalent concentration of DMSO starting from D0. Median ± range (n = 4 independent experiments) [scale bars, (A) 100 μM and (D) 50 μm]. (B and E) ZEB1 mRNA expression in HME-RASER (RASER) cells at D21 of induction with 4-OHT (IND) or not (NI) and treatment with senolytic drugs or the equivalent concentration of DMSO starting from D0. Median ± range (n = 4 independent experiments). (C and F) Representative flow cytometry analysis on CD24 and CD44 markers. Quantification of CD24−/low/CD44+ population in HME-RASER (RASER) cells at D21 of induction with 4-OHT (IND) or not (NI) and treatment with senolytic drugs or the equivalent concentration of DMSO starting from D0. Median ± range (n = 4 independent experiments). P values are calculated by one-way ANOVA, Tukey multiple comparisons test (***P ≤ 0.001 and ****P ≤ 0.0001).
ZEB1-dependent EMP is driven by factors secreted by senescent cells
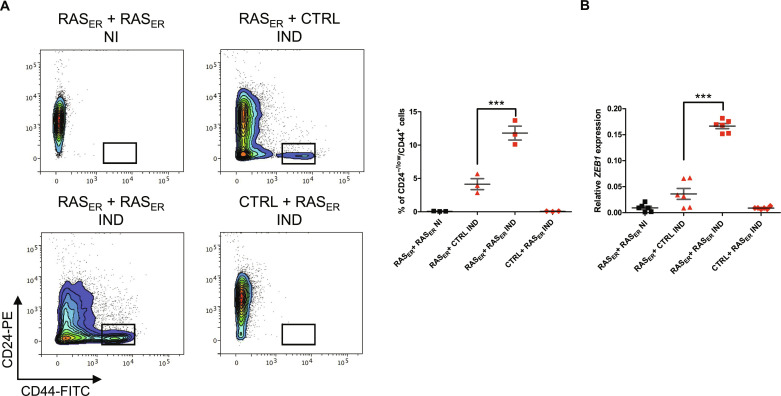
Upon entering senescence, cells produce a set of pro-inflammatory cytokines and chemokines, extracellular matrix proteins, growth factors, and metalloproteinases composing the senescence-associated secretory phenotype (SASP), which, through secretory signaling, affects the dedifferentiation of somatic cells (42). We therefore interrogated whether a secretory signaling process emanating from RAS-activated senescent cells may be responsible for ZEB1-mediated plasticity in neighboring RAS-activated cells. To test this hypothesis, HME-RASER or HME-CTRL cells previously treated for 7 days with 4-OHT were co-cultured with either HME-RASER or HME-CTRL cells seeded in a transwell insert. At D18, corresponding to 11 days of RAS activation for cells in the insert, we noted a threefold increase in the percentage of CD24−/low/CD44+ cells in the HME-RASER population that was co-cultured with pre-induced HME-RASER as compared to HME-RASER cells co-cultured with pre-induced HME-CTRL cells (Fig. 5A). This increase was associated with an upregulation of ZEB1 expression (Fig. 5B).
Fig. 5. Factors secreted by senescent cells favor the emergence of a cell population with EMP features.
(A) Representative flow cytometry analysis of CD24 and CD44 markers. Quantification of CD24−/low/CD44+ population in HME-RASER (RASER) or HME-CTRL (CTRL) cells after 11 days of transwell co-culture with HME-RASER (RASER) or HME-CTRL (CTRL) cells pre-induced for 7 days with 4-OHT (IND) or not (NI). Median ± range (n = 3 independent experiments). (B) Quantification of ZEB1 mRNA expression in HME-RASER (RASER) or HME-CTRL (CTRL) cells after 11 days of transwell co-culture with HME-RASER (RASER) or HME-CTRL (CTRL) cells pre-induced for 7 days with 4-OHT (IND) or not (NI). Median ± range (n = 6, 3 independent experiments in duplicate). P values are calculated by one-way ANOVA, Tukey multiple comparisons test (***P < 0.001).
To further investigate the involvement of secretory properties of RAS-activated senescent cells in EMP induction, we explored their impact on normal HMECs. For that purpose, we generated a cellular HME model (HME_d2GFP_200) that expressed a destabilized green fluorescent protein (GFP) (d2GFP_200) including a 3′ untranslated region containing five miR-200 target sequences (fig. S7A). This setup aimed to highlight cells undergoing EMP through an increased GFP signal underlying lower levels of expression of miR-200 family members (43). To determine whether senescent cells promote EMP in neighboring cells, HME-RASER cells tagged with dsRED (HME-RASER-dsRED cells), were treated with tamoxifen to initiate RAS-induced senescence and co-cultured with HME_d2GFP_200 cells (fig. S7B). An enrichment of GFPhigh cells associated with an emergence of a CD24−/low/CD44+ phenotype was observed in HME_d2GFP_200 cells (fig. S7, C and D), revealing that ZEB1 reactivation can be driven by secreted factors from senescent cells. Moreover, co-cultured HME_d2GFP_200 cells showed a significant increase in ZEB1 expression and were able to form colonies in soft agar assay, suggesting that they had acquired cellular properties associated with transformation (fig. S7, E and F). Last, we performed a transwell co-culture assay using HME_d2GFP_200 cells and RAS-induced HME-RASER-dsRED cells. The characterization of cells following co-culture showed a significant increase of GFPhigh cells enriched in a CD24−/low/CD44+ phenotype (fig. S7, G and H), thus evoking a paracrine signaling process.
IL-6 and IL-1α secreted by senescent cells following RAS activation promote ZEB1-induced EMP
Considering the well-known cooperation between activated RAS and transforming growth factor–β (TGF-β) ligands to promote cellular plasticity, the secretion of TGF-β1, TGF-β2, and TGF-β3 was assessed (44, 45). Unexpectedly, the secretion rate of these factors remained unchanged following RAS activation, suggesting the involvement of other cytokines capable of inducing EMP (fig. S8A).
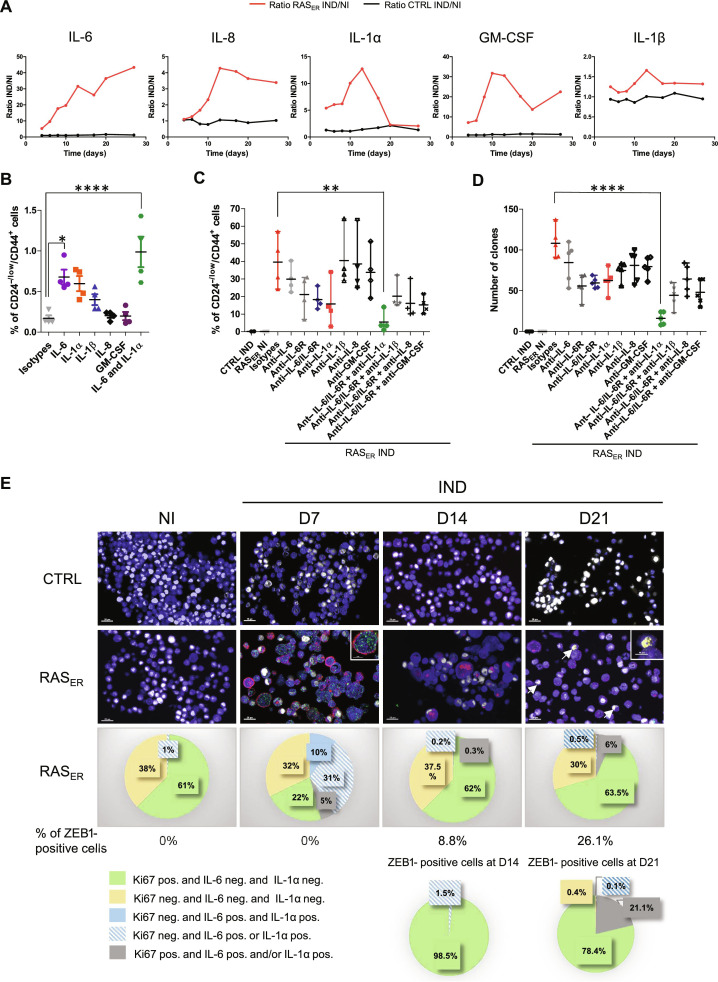
We next aimed to narrow down the specific factors produced by senescent cells leading to this effect, focusing specifically on SASP constituents. Multiplex technology was used to quantify the concentrations of SASP factors in RAS-induced HME-RASER cell culture supernatants. Quantifications revealed high levels of cytokines IL-6 (interleukin-6), IL-8, IL-1α, IL-1β, and granulocyte-macrophage colony-stimulating factor (GM-CSF) (Fig. 6A). Experiments based on the direct addition of these cytokines to HMECs at the highest concentration found in cell culture supernatants showed the ability of IL-6, IL-1α, and IL-1β to promote the acquisition of a plasticity phenotype (Fig. 6B). The combination of IL-6 and IL-1α had a cumulative effect on the emergence of CD24−/low/CD44+ cells. Next, to evaluate the role of these cytokines in RAS-induced plasticity, we used a depletive approach with cytokine-neutralizing antibodies or IL-6R antibody (tocilizumab). Double inhibition of IL-6/IL-6R or the inhibition of IL-1α alone decreased the CD24−/low/CD44+ population and ZEB1 protein levels as well as the transforming abilities of RAS-induced HME-RASER cells (Fig. 6, C and D, and fig. S8B). Notably, combination treatments with different cytokines showed that the concomitant inhibition of IL-6/IL-6R and IL-1α induced an almost 10-fold decrease in cellular plasticity and transformation capacities, suggesting that these two cytokines are major determinants of ZEB1-mediated plasticity and transformation (Fig. 6, C and D).
Fig. 6. IL-6 and IL-1α are major cytokines responsible for ZEB1-dependent EMP.
(A) Quantification of cytokine levels in HME-RASER supernatants following 4-OHT treatment compared to controls at D4, D6, D8, D10, D13, D17, D20, and D27. Data are presented as fold change of secreted cytokine concentrations in HME-RASER or HME-CTRL cells induced by 4-OHT (IND) or not (NI). (B) Quantification by flow cytometry analysis of CD24−/low/CD44+ population of HMECs after treatment by the indicated cytokine(s) or isotype controls analyzed at D40. Median ± range (n = 4 independent experiments). (C) Quantification by flow cytometry analysis of CD24−/low/CD44+ population of HME-RASER (RASER) cells induced by 4-OHT (IND) or not (NI) and treated by neutralizing antibodies or isotype controls analyzed at D28. Median ± range (n = 4 independent experiments). (D) Transformation potential analysis by soft agar colony formation assay in HME-RASER (RASER) cells induced by 4-OHT (IND) or not (NI) and treated by neutralizing antibodies or isotype controls analyzed at D28. Median ± range (n = 5 independent experiments). (E) Top: multi-fluorescence staining kinetics. HME-RASER (RASER) and HME-CTRL (CTRL) cells were induced by 4-OHT (IND) or not (NI) and stained with anti-Ki67 (white), anti–IL-6 (green), anti–IL-1α (red), and anti-ZEB1 (yellow) at D7, D14, and D21. Representative pictures are shown (scale bars, 20 and 10 μm in enlarged pictures). Bottom: Graphical representation of cellular phenotype distribution according to Ki67, IL-6, IL-1α, or ZEB1 staining. P values are calculated by one-way ANOVA, Tukey multiple comparisons test (**P < 0.01 and ****P < 0.0001).
To confirm that IL-1α and IL-6 secretion predominantly originates from senescent cells, the levels of these cytokines were assessed after senolytic treatment. Concomitant with senescence, secretion of IL-1α and IL-6 in the supernatant of induced HME-RASER cells was significantly reduced after senolytic treatment (fig. S8C).
ZEB1-induced EMP relies on a paracrine effect mediated by senescent cells
To further characterize the process involved in ZEB1-mediated plasticity following RAS activation, we used multi-fluorescent labeling kinetics to visualize the expression of IL-6, IL-1α, ZEB1, and Ki67, a well-known proliferation marker, in HME-RASER cells (Fig. 6E). We observed that, at D7 of RAS activation, 10% of RAS-induced HME-RASER cells showed a negative Ki67 profile and were double-positive for IL-6 and IL-1α, consistent with a senescent state. Moreover, 31% of cells presented a negative Ki67 profile and were positive for either IL-6 or IL-1α, potentially indicating a pre-senescent state. None of these cells were positive for ZEB1. Fourteen days after RAS induction, 9% of cells were ZEB1-positive and less than 2% of them were both negative for Ki67 and positive for IL-6 and/or IL-1α (Fig. 6E). At D21, 26% of cells were ZEB1-positive with the majority being Ki67-positive. Overall, these data suggest that cells entering OIS do not express ZEB1, consistent with the hypothesis of the involvement of a paracrine process driving EMP in neighboring cells. Next, we co-cultured HME-d2GFP_200 cells with tamoxifen-induced HME-RASER-dsRED cells and treated them with IL-6/IL-6R and IL-1α neutralizing antibodies. EMP induction in GFPhigh cells was strongly impaired following treatment (fig. S8, D and E). Together, these results reveal that ZEB1 activation after RAS induction is driven by a paracrine process involving the secretion of IL-6 and IL-1α cytokines from senescent cells.
DISCUSSION
In cancer, EMP is generally considered to be a process induced during the progression of the disease in response to microenvironmental signals such as growth factor signaling and immune response pathways (9), which promotes key steps of the invasion-metastasis cascade and resistance to treatment (7). Our work reveals another aspect of EMP, both in terms of its mechanisms of reactivation and its role in tumorigenesis. First, we show that the activation of the RAS/MAPK signaling pathway is sufficient to induce the expression of EMT-TFs and to promote EMP in HMECs, the in vitro approach precluding the influence of other cell types. Second, inhibition of EMP prevents RAS-induced transformation of HMECs, suggesting a determinant role for this transdifferentiation process in early phases of tumorigenesis following the oncogenic hit. Consistent with this hypothesis, we have previously shown in a model of breast tumorigenesis that EMT-TFs of the TWIST and ZEB families cooperate with mitogenic oncoproteins for malignant transformation by alleviating OIS (22, 24). Therefore, our observations reinforce the notion that the induction of EMP may constitute an early event for cellular adaptation to certain oncogenic insults, thereby fostering malignant transformation. Further strengthening this notion, several in vivo studies demonstrated the cooperation between RAS activation and the expression of EMT-TFs in tumor initiation. As examples, in a mouse model of epithelial cancer driven by the activation of mutant Kras, depletion of Zeb1 significantly reduces neoplastic lesions (21, 46), and ectopic expression of Twist1 in differentiated mammary epithelial cells cooperates with mutant Kras for the development of CL tumors (22, 26). In addition, Kras activation in murine luminal mammary epithelial cells leads to early EMP engagement and promotes the development of CL tumors (26).
Our study further highlights a mechanistic link between OIS and EMP during malignant transformation. Strengthening our observations, recent studies have used a pan-cancer scRNA-seq approach to show recurrent signatures associated with EMP and senescence at tumor initiation and over the course of malignant transformation (47–49). Cellular senescence is a stress response that is generally considered to irreversibly arrest the cell cycle following a variety of intrinsic and extrinsic stressors such as shortening of chromosomal termini, DNA damage, oxidative stress, oncogenic insults, or inactivation of tumor suppressors (50). Notably, OIS or senescence induced by loss of a tumor suppressor was observed in vivo in human and murine preneoplastic lesions (51–53). Senescence is traditionally described as an innate anticancer mechanism because cells harboring oncogenic mutations are prevented from proliferating and transforming (54). Moreover, in addition to decreasing replicative capacity, activation of senescence in different contexts and tissues leads to increased expression of inflammatory cytokines that elicit immune-mediated tumor clearance (55, 56). However, recent studies challenge this conventional view, showing a paradoxical role for senescence emanating from its special secretory profile that can counterintuitively promote cancer stemness and aggressiveness (57, 58). In our study, single-cell analysis revealed OIS in a subset of RAS-activated cells and the subsequent induction of an EMP process in a separate subset. Senolytic treatments showed that senescent cells comprising the first subset play a causal role in the acquisition of plasticity by vicinal cells. This duality in cellular outcomes recalls previous studies showing that activation of the four TFs—Oct4, Sox2, Klf4, and c-Myc (OSKM)—in mouse tissue triggers opposite cellular fates, namely, senescence and reprogramming, fates that coexist in vivo within separate subpopulations of cells (59, 60). Consistent with a senescence response, overexpression of OSKM in human primary fibroblasts triggers replication stress and DNA damage characterized by the up-regulation of p53, p16INK4a, and p21CIP1, as well as impaired proliferation and formation of senescence-associated heterochromatin foci (61). With notable conceptual similarity, we have thus demonstrated that entry into senescence of a subset of mammary epithelial cells facing RAS-dependent oncogenic insult promotes the transdifferentiation of neighboring cells.
Cellular senescence creates a tissue context that favors OSKM-driven reprogramming in surrounding cells through the paracrine action of the SASP, with IL-6 being a key mediator (59, 60). A similar interplay also occurs in tissue aging and injury, where there is an accumulation of senescent cells that secrete pro-inflammatory cytokines, triggering OSKM-driven dedifferentiation and reprogramming in neighboring cells. Quantification of a series of SASP constituents and functional assays enabled us to determine the factors inherent to senescent mammary epithelial cells that are responsible for triggering a similar reprogramming process in the cellular vicinity. Namely, we identified proinflammatory cytokines IL-6 and IL-1α as major mediators of senescence-induced EMP. Of note, IL-6 has previously been shown to trigger EMP via Janus kinase–signal transducer and activator of transcription 3 or nuclear factor κB signaling, leading to the activation of EMT-TFs (62–69). Accordingly, IL-6 overexpression and/or hyperactivation has been reported in several human cancers (70–73). Relatively fewer studies have directly examined the role of IL-1α in cellular reprogramming and in cancer development and progression. Nevertheless, it is noteworthy that depletion of IL-1α in a murine pancreatic cancer model resulted in impaired immune cell infiltration and decreased Kras-induced dysplastic and neoplastic lesions (74). Moreover, a recent work examining the relationship between HER2 expression in breast cancer, inflammation, and expansion of cells exhibiting stemness properties highlighted an essential role for IL-1α (75). In light of these observations, further investigations will now be necessary to define the steps downstream of IL-6 and IL-1α secretion that lead to the induction of ZEB1 expression.
The present data suggest that OIS of only a subset of epithelial cells is sufficient to trigger the bypass of this same oncosuppressive barrier in neighboring cells via the action of released SASP factors, thereby favoring their malignant transformation. From a deterministic point of view, we are confronted here with a paradox: The attempt of cells to undergo senescence following an oncogenic hit may be interpreted as a mechanism of self-sacrifice to protect an organism as a whole, yet this desperate act may be the critical element that promotes the transformation of neighboring cells facing the same stress. This notion supports the hypothesis that OIS has contrasting cell-autonomous and non–cell-autonomous effects in early tumorigenesis, providing previously unknown clues to the ongoing debate on its tumor suppressor and pro-tumorigenic effects.
Last, this study suggests a reassessment of the microenvironmental components affecting malignant transformation. Oncogene-induced senescent cells constitute an adverse microenvironment inducing modifications to the identity of neighboring cells and the acquisition of EMT-related plasticity fostering tumor initiation. In addition to fibroblasts or immune cells, oncogene-induced senescent cells could thus also be considered as crucial players affecting tumor development and progression. This concept prompts further investigations to assess whether early EMP commitment of transformed cells would be imprinted as a memory of cell identity to dictate tumor evolution, including the formation of metastases and resistance to treatment.
MATERIALS AND METHODS
Cell culture
Primary HMECs (Lonza) were immortalized by human Telomerase Reverse Transcriptase (hTERT), and named HME thereafter (22). HME-derived cells (HME-RASER, HME-CTRL, HME-RASER–miR-200c, HME-RASER–empty, HME-RASER-CRISPR ZEB1, HME-RASER-CRISPR SCR, and HME_d2GFP_200) were cultured in Dulbecco’s modified Eagle’s medium (DMEM)/Ham′s F12 medium with 1% GlutaMAX (Thermo Fisher Scientific) supplemented with 10% heat-inactivated fetal calf serum (Eurobio Scientific), penicillin (100 IUml), streptomycin (100 μg/ml; Thermo Fisher Scientific), human epidermal growth factor (EGF; 10 ng/ml) (PeproTech), hydrocortisone (0.5 mg/ml; Sigma-Aldrich), and insulin (10 mg/ml; NovoRapid, Novo Nordisk), at 37°C in 5% CO2/95% air. Breast cancer cell lines were obtained from the American Tissue Culture Collection. MDA-MB-231, Hs578T, and human embryonic kidney (HEK) 293T cells were maintained in DMEM with 1% GlutaMAX (Thermo Fisher Scientific) with 10% heat-inactivated fetal calf serum (Eurobio Scientific), penicillin (100 IU/ml), and streptomycin (100 μg/ml; Thermo Fisher Scientific). All cell lines were regularly tested negative using a MycoAlert mycoplasma detection kit (Lonza).
Lentiviral and retroviral infections
Lentiviral particles were produced using non-confluent 2 × 106 HEK293T cells transfected with the GeneJuice Transfection Reagent (Merck Millipore) with 13.02 μg of total lentiviral expression vectors (5.1 μg of pCMVdeltaR8.91, 1.32 μg of phCMVG-VSVG, and 6.6 μg of plasmid of interest). The pCMVdeltaR8.91 and phCMVG-VSVG were gifts from D. Nègre (International Center for Infectiology Research, INSERM U1111–CNRS UMR5308–ENS de Lyon–UCB
Lyon1, EVIR Team, Lyon, France). Forty-eight hours after transfection, the supernatant was collected, filtered, and supplemented with polybrene (5 μg/ml; Merck Millipore) combined with cells for 6 hours. Cell selection was done 72 hours after infection through fluorescence-activated cell sorting (FACS) for GFP plasmids (pCDH-CMV-EF1-copGFP/MIR200c, pCDH-CMV-MCS-EF1-copGFP-empty, and FUGW-d2GFP-200), or dsRED plasmid (pLLRSV Red-empty), or by antibiotics (puromycin at 0.5 μg/ml or neomycin at 100 μg/ml; InvivoGen) depending on plasmid construct. To produce retrovirus particles, 2 × 106 Phoenix cells were transfected with the GeneJuice Transfection Reagent (Merck Millipore) and 10 μg of retroviral expression vectors (76). At 48 hours after transfection, the supernatant was collected, filtered, supplemented with polybrene (5 μg/ml; Merck Millipore) and combined with 106 targeted cells for 6 hours. Cells were selected 48 hours following infection with puromycin (0.5 μg/ml) and/or neomycin (100 μg/ml).
Plasmids
The lentiviral plasmids, pCDH-CMV-EF1-copGFP/MIR200c and pCDH-CMV-MCS-EF1-copGFP-empty, were gifts from T. Brabletz; the dsRED plasmid, pLLRSV Red-empty, was a gift from C. Ginestier; and the miR-200 sensor plasmid, FUGW-d2GFP-200, was purchased from Addgene (plasmid no. 79602) (43). Retroviral plasmids used were purchased from Addgene: pLNCX2 neo-RAS-ER (no. 67844) and pLNCX2 neo-empty (77, 78).
CRISPR-Cas9 knockdown
ZEB1-depletion model in HME (HME-RASER-CRISPR ZEB1) was generated using CRISPR-Cas9 gene editing technology. Scrambled single guide RNA/Cas9 All-in-One Lentivector (Applied Biological Materials, catalog no. K010) and ZEB1 sgRNA/Cas9 All-in-One Lentivector (Human) (target 1: 5′-CACCTGAAGAGGACCAG-3′) (Applied Biological Materials, catalog no. K2671006) lentiviral particles were used to infect HMECs. Scrambled sgRNA/Cas9 and ZEB1 sgRNA/Cas9 cells were selected with puromycin (InvivoGen) at 0.5 μg/ml 48 hours after infection. After cloning by limited dilution, single cells were grown for ~3 weeks and colonies were screened for knockouts by quantitative polymerase chain reaction (qPCR), genomic DNA sequencing, and Western blotting. Genomic DNA sequencing was performed using the sanger method with the following primers for amplification and sequencing 5′-TGAACTGAACGTCAGAGTGGT-3′ (forward) and 5′-TCACGTGCAGTGGCATTACT-3′ (reverse). Three different clones were lastly validated.
Kinetics, co-culture, and transwell assay
For all kinetics, 27,000 cells were seeded at D0 in a six-well plate with 2 ml of medium under all conditions. At D4, medium was changed, and, at D7, cells were trypsinized (0.05% trypsin and 0.53 mM EDTA; Thermo Fisher Scientific); this cycle was repeated until D49. For transwell kinetics, 21,000 cells (HME-RASER or HME-CTRL) were first plated with or without 4-OHT; 7 days later, 7000 cells (HME-RASER or HME-CTRL) were plated in the insert of the plate. At D18, corresponding to 11 days of RAS activation for the cells on the insert, FACS analyses were performed. For co-culture, HME_d2GFP_200 cells were mixed 20:80 with HME-RASER-dsRED cells. For 1-week kinetics in a six-well plate, around 5000 HME_d2GFP_200 cells were mixed with 22,000 HME-RASER-dsRED cells per well. Cells were trypsinized at D7 and D14. For transwell kinetics, 7000 HME_d2GFP_200 cells were plated in the insert for 27,000 HME-RASER-dsRED cells per well at D0. Cells were trypsinized at D7 and D14.
Mammospheres
Cells were suspended in DMEM-F12 medium and counted. A total of 10,000 cells were seeded into an ultralow-attachment 24-well plate (Corning). In necessary cases, 4-OHT was added to each well every 4 days (without removing the old medium), and 100 μl of fresh medium was added every 3 to 4 days. Mammospheres were collected after 7 to 10 days by centrifugation (800g), washed two times in phosphate-buffered saline (PBS), trypsinized (100 μl, 30 min), and dissociated mechanically through pipetting. Cells were assessed microscopically for single-cellularity. Next, cells were resuspended with 900 μl of fresh medium and sieved through a 40-μm sieve. Three rounds of mammosphere formation of 7 to 10 days were conducted. The number of spheres (nonadherent spherical clusters of cells with basal membrane) for each well was evaluated under the microscope after round three. Quantification of formed mammospheres was performed microscopically by manual counting. Images of mammospheres were acquired using a ZEISS Axio Vert.A1 inverted microscope.
Organoids
Cell suspension was resuspended in Reduced Growth Factor Basement Membrane Matrix (BME; Bio-Techne). A 50-μl drop of this BME suspension containing 1000 cells was placed in the center of a well of a prewarmed ultralow-attachment 96-well plate (Corning), allowed to harden at 37°C for 20 min. Upon complete gelation, 200 μl of organoid medium supplemented with different factors was added to each well (79). The plate was incubated in a humidified 37°C/5% CO2 incubator. Culture medium was changed every 2 days. After 10 days, 4-OHT was added to the medium every 3 days in induced RAS condition. After 21 days of 4-OHT treatment or nontreatment, organoids were fixed in buffered formalin for multiplexed immunofluorescence staining.
Multiplexed immunofluorescence
For histological examination, tissue samples (organoids or cell pellets) were fixed in 10% buffered formalin and embedded in paraffin using a cytoblock kit (Microm Microtech France, E/7401150). Four-micrometer-thick sections of formalin-fixed, paraffin-embedded tissue were prepared according to conventional procedures.
Multiplexed immunofluorescence was performed on a Bond RX automated immunostainer (Leica Biosystems) using Opal detection kits (AKOYA Biosciences). Sequential immunofluorescence stainings were performed using the following: organoid panel: Opal 520 (anti-CK5; Leica, no. NCL-L-CK5), Opal 690 (anti-CK18; Agilent, no. M7010), and Opal 780 (anti-VIMENTIN; Agilent, no. M0725); cell panel: Opal 520 (anti–IL-1α; LSBio, no. LS-C33821), Opal 620 (anti–IL-6; ABclonal, no. A0286), Opal 570 (anti-ZEB1; Bethyl, no. IHC-00419), and Opal 690 (anti-Ki67 Mib1; Agilent, no. M7240).
Samples were counterstained with 4′,6-diamidino-2-phenylindole (Sigma-Aldrich, D8417). Fluorescent slides were mounted using ProLong Gold Antifade Reagent (Invitrogen, ref. no. P36930).
Sections were scanned using the Vectra POLARIS device (AKOYA Biosciences). An autofluorescence treatment was carried out on images using the Inform software (PerkinElmer). For analysis of cells, an evaluation of the staining was carried out using the HALO Image Analysis Software (Indica Labs). Cells were considered Ki67-positive if the fluorescence intensity value was >30, IL-6–positive if the fluorescence intensity value was >25, IL-1α–positive if the fluorescence intensity value was >75, and ZEB1-positive if the fluorescence intensity value was >32.
Soft agar colony formation assay
A 2× DMEM/F12 powder including l-glutamine (Sigma-Aldrich, no. D0547) was resuspended with sterile water and completed with sodium bicarbonate (1.2 g/liter; Sigma-Aldrich, no. S5761). This prewarmed medium was supplemented with 20% heat-inactivated fetal calf serum, penicillin (200 IU/ml), streptomycin (200 μg/ml), hydrocortisone (1 mg/ml), and EGF (20 ng/ml) and mixed 1:1 with melted SeaPlaque Agarose (Lonza) solution to obtain a final 0.75% low-melting agarose base that was then overlaid with a suspension of cells in 0.45% low-melting agarose (10,000 cells/well). Six-well plates were incubated for 4 weeks at 37°C with 5% CO2/95% air, and colonies defined as a group of >20 cells were counted under the microscope. In experiments using GFP-tagged cells, only GFP-positive cells were counted. Experiments were performed in triplicate.
SA–β-gal staining
Cells were rinsed with PBS and fixed for 4 min in 2% formaldehyde/0.2% glutaraldehyde, then washed twice with PBS, and incubated at 37°C for 18 hours in a buffer containing 40 mM sodium phosphate (pH 6.0), 5 mM K4Fe(CN)6, 5 mM K3Fe(CN)6, 150 mM NaCl, 2 mM MgCl2, and X-Gal (1 mg/ml). All reagents were purchased at Sigma-Aldrich except for X-Gal powder (VWR).
Multiplex cytokine assay
We assessed 31 cytokines in each sample with the Panel (human) kit [Meso Scale Discovery (MSD)], which measures IL-1α, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12p40, IL-12p70, IL-13, IL-15, IL-16, IL-17A, GM-CSF, tumor necrosis factor–α (TNF-α), TNF-β, interferon-γ, TGF-β1, TGF-β2, TGF-β3, Interferon γ-induced Protein 10 kDa (IP-10), Monocyte Chemoattractant Protein-1 (MCP-1), Monocyte Chemoattractant Protein-4 (MCP-4), Macrophage Inflammatory Protein-1 Alpha (MIP-1α), Macrophage Inflammatory Protein-1 Beta (MIP-1β), Macrophage-Derived Chemokine (MDC), C-C Motif Chemokine Ligand 17 (CCL17), eotaxin 1, eotaxin 3, and vascular EGF. MSD plates were analyzed on the MS2400 imager (MSD). Calibrator dilutions and samples were prepared according to the manufacturer’s recommendations.
Determination of the secreted level of IL-6 and IL-1α in cells after senolytic treatment
We assessed concentrations of secreted IL-6 and IL-1α in each supernatant by performing enzyme-linked immunosorbent assay (ELISA) kits (Quantikine ELISA, R&D Systems, Bio-Techne, no. D6050 (IL-6) and no. DLA50 (IL-1α). Calibrator dilutions and samples were prepared according to the manufacturer’s recommendations. The concentration of the secreted cytokine was normalized according to the number of cells in each condition.
Reagent, antibodies, and cytokines
To induce RAS activation in HME-derived cells, (Z)–4-OHT (Sigma-Aldrich, no. H7904) was added in the medium at 500 nM every 3 to 4 days. Cells were treated with dasatinib (1.5 μM), A1331852 (1 μM), navitoclax (2 μM), S63845 (2 μM), or dimethyl sulfoxide (DMSO) every 3 days.
All antibodies except Tocilizumab-Roactemera were purchased at R&D Systems (Bio-Techne): IL-1α /IL-1F1 antibody (MAB200), IL-1β /IL-1F2 antibody (MAB201), IL-6 antibody (MAB2061), IL-8/CXCL8 antibody (MAB208), GM-CSF antibody (MAB215), and normal human immunoglobulin G (IgG) control (1-001-A). Tocilizumab (anti-human IL-6R, humanized antibody IgG1; Tocilizumab-Roactemera, 20 mg/ml; Roche) was stored at 4°C and used at a final concentration of 50 μg/ml. Other antibodies were reconstituted in aliquots stored at −80°C and, once used, were stored at 4°C for no more than 2 weeks. Antibodies were used at the following final concentrations guided by MSD/ELISA highest concentrations measured: anti–IL-6 (0.6 μg/ml), anti–IL-8 (1 μg/ml), anti–IL-1α (1 μg/ml), anti–IL-1β (0.3 μg/ml), anti–GM-CSF (4 μg/ml), and normal human IgG control (50 μg/ml). Medium was changed every 3 to 4 days, and antibodies were added at the same time.
Cytokines were purchased at R&D Systems (Bio-Techne): recombinant human IL-1α/IL-1F1 protein (200-LA), recombinant human IL-1β/IL-1F2 protein (201-LB), recombinant human IL-6 protein (206-IL), recombinant human IL-8/CXCL8 protein (208-IL), and recombinant human GM-CSF protein (215-GM). These cytokines were reconstituted in sterile PBS with 0.1% bovine serum albumin (Sigma-Aldrich) and stored at −80°C. Once thawed, cytokines were kept at 4°C up to 2 weeks. Cytokines were used at the following final concentrations: IL-6 (2 ng/ml), IL-8 (10 ng/ml), IL-1α (1.5 ng/ml), IL-1β (50 pg/ml), GM-CSF (1 ng/ml), and normal human IgG control (10 ng/ml). Cytokines were added every 2 days, and medium was changed every 3 to 4 days.
RNA extraction, reverse transcription, and PCR
Adherent cells were washed in PBS (Eurobio Scientific); TRIzol reagent (Merck Millipore) was then added. After sorting, cells were directly centrifuged and washed before TRIzol was added. Total RNA was isolated using the TRIzol reagent according to the manufacturer’s instructions. Total RNA concentration and purity were determined from absorbance at 260 and 280 nm using a ND-1000 NanoDrop spectrophotometer. Reverse transcription (RT) was performed with 300 ng to 1 μg of RNA using a cDNA RT kit (Maxima cDNA synthesis kit, Thermo Fisher Scientific). qPCR analysis was performed with 3 μl of a 1:10 dilution of the resulting cDNA. DNA amplification was monitored by real-time PCR with a CFX96 instrument (Bio-Rad) and analyzed with Bio-Rad CFX manager software. The relative quantification of gene expression was performed using the comparative threshold cycles (CT) method, with normalization of the target gene to Hypoxanthine Phosphoribosyltransferase 1 (HPRT1) housekeeping gene.
The list of primer sequences used for Q-PCR analysis is as follows: human ZEB1, 5′-AGGGCACACCAGAAGCCAG-3′ (forward) and 3′-GAGGTAAAGCGTTTATAGCCTCTATCA-5′ (reverse); human ZEB2, 5′-AAGCCAGGGACAGATCAGC-3′ (forward) and 3′-GCCACACTCTGTGCATTTGA-5′ (reverse); human SNAI1, 5′-GCTGCAGGACTCTAATCCAGA-3′ (forward) and 3′-ATCTCCGGAGGTGGGATG-5′ (reverse); human SNAI2, 5′-TGGTTGCTTCAAGGACACAT-3′ (forward) and 3′-GTTGCAGTGAGGGCAAGAA-5′ (reverse); human TWIST1, 5′-GGCTCAGCTACGCCTTCTC-3′ (forward) and 3′-CCTTCTCTGGAAACAATGACATCT-5′ (reverse); human TWIST2, 5′-CATGTCCGCCTCCCACTA-3′ (forward) and 3′-GCATCATTCAGAATCTCCTCCT-5′ (reverse); and human HPRT1, 5′-TGACCTTGATTTATTTTGCATACC3′ (forward) and 3′-CGAGCAAGACGTTCAGTC-5′ (reverse).
TaqMan qRT-PCR assay was used for detection of mature microRNAs. Reagents, primers, and specific probes for miR-200c and miR-141 were obtained from Applied Biosystems. RT reactions and real-time qPCR were performed according to the manufacturer’s protocols from 40 ng of RNA per sample. RNU48 was used as a loading control. Relative expression was determined by the ΔΔCt method, normalized to the expression of HPRT1 housekeeping gene for ZEB1, ZEB2, and SNAI2 mRNA, and to RNU-48 for miR200c and miR141.
Immunoblot analysis
Cells were washed in PBS with CaCl2 and lysed in a radioimmunoprecipitation assay buffer [100 mM NaCl, 1% NP-40, 0.1% SDS, and 50 mM tris (pH 8)] supplemented with protease inhibitor cocktail (tablets, Roche), phenylmethylsulfonyl fluoride (Merck Millipore), and a phosphatase inhibitor cocktail (Merck Millipore). Cells were scratched at 4°C, and lysed cells were sonicated. Protein concentration was dosed with the Bio-Rad Protein Assay Dye Reagent (Bio-Rad).
Primary antibodies used include anti-ZEB1 (Merck Millipore, no. HPA027524), anti-ZEB2 (Merck Millipore, no. HPA003456), anti-SNAIL+SLUG (Abcam, no. ab85936), anti–phospho-p44/42 MAPK [extracellular signal–regulated kinase 1/2 (Erk1/2)] (Thr202/Tyr204) (Cell Signaling Technology, no. 4370), anti-p44/42 MAPK (Erk1/2) (Cell Signaling Technology, no. 9102), anti-RAS (G12V mutant specific) (Cell Signaling Technology, no. 14412), anti–glyceraldehyde-3-phosphate dehydrogenase (Merck Millipore, no. ABS16), and anti-tubulin (Sigma-Aldrich, no. T7816).
Secondary antibodies used were horseradish peroxidase–conjugated goat anti-mouse (Santa Cruz Biotechnology, no. sc-2005) or mouse anti-rabbit polyclonal antibodies (Santa Cruz Biotechnology, no. sc-2357). Western blots were revealed using a Western blot luminol reagent kit (Bio-Rad). Luminal signal was detected by ChemiDoc (Bio-Rad). Post-analysis was done with ImageLab software 6.0.1 (Bio-Rad).
Flow cytometry analysis and sorting
For CD44/CD24/CD104 or EpCAM staining, cells were stained with primary antibodies from Miltenyi Biotec: anti–CD44-labeled fluorescein-5-isothiocyanate (clone REA690, dilution 1:400), anti–CD24-labeled phycoerythrin (clone 32D12, dilution 1:50), anti-CD104 (Integrin β4)–allophycocyanin (clone: REA236, dilution 1:400), or anti-CD326 (EpCAM)–allophycocyanin (clone REA764, dilution 1:400). For analyzing GFP- or dsRED-tagged cells, anti–CD44–Brilliant Violet 421 (clone BJ18, dilution 1:50; Ozyme) and anti CD24-allophycocyanin (clone ML5, dilution 1:100; Ozyme) were used.
Fresh cells were washed once in PBS, stained protected from light at 4°C for at least 1 hour, and then washed again with PBS three times. A FACSCanto II cytometer (Becton Dickinson) or BD LSR Fortessa (Becton Dickinson) was used for data acquisition alongside Diva software. The cytometer was calibrated daily using Cytometer, Setup, and Tracking (CST) beads (Becton Dickinson) according to the manufacturer’s instructions. Sample acquisition was made at a medium flow rate and set to record the highest number and identical number of events possible for each condition. Post-analysis was done using FlowLogic software (Miltenyi Biotec). Antibody validation information is available on the manufacturers’ websites. Cell sorting was performed using Bio-Rad S3 (Bio-Rad) for GFP- and dsRED-tagged cells after cell infection. For all other sorting experiments, FACSAria III (BD Biosciences) was used.
Statistical analysis
For all biological experiments, significance testing was performed using Prism Software version 7.0 (GraphPad). P values were calculated by one-way analysis of variance (ANOVA) and Tukey multiple comparisons test or Student’s t test as specified in the figure legends. Kinetics analysis data were plotted as median ± range and showing all points. P values of ≤ 0.05 were considered significant. All statistical tests are two-tailed.
Bioinformatics analysis on breast cancer dataset
Samples and statistics
Breast tumor samples used in this study are from METABRIC (30) and TCGA Research Network (www.cancer.gov/tcga). Cohorts and breast cancer cell lines are from CCLE database (31). All analyses, statistical tests, and figures were realized using either the R software (version 3.6.1) or GraphPad Prism 8.0 (GraphPad Software Inc., San Diego, USA). All statistical tests are two-tailed.
Expression data processing
METABRIC microarray expression data from discovery and validation sets were extracted from the EMBL-EBI archive (EGA; www.ebi.ac.uk/ega/; accession number: EGAS00000000083) (normalized expression data files) (30). Normalized expression data per probe of the discovery set and the validation set were combined after independent normalization of each set with a median z-score calculation for each probe. The expression levels of different probes associated with the same Entrez Gene ID were averaged for each sample to obtain a single expression value by gene.
TCGA Breast Cancer (BRCA) RNA-seq expression data were extracted as fragments per kilobase million (FPKM) values from the Genomic Data Commons (GDC) data portal (https://portal.gdc.cancer.gov/). FPKM data by gene were converted to transcript per million (TPM) as follows: for each gene g ∈ G and each sample s ∈ S
RNA-seq expression data from the CCLE breast cell lines were extracted as reads per kilobase million (RPKM) values from the CCLE data portal (https://portals.broadinstitute.org/ccle). RPKM data by gene were converted to TPM as follows: for each gene g ∈ G and each sample s ∈ S
Molecular breast cancer subtype assignment
For both tumors and cell lines, attribution of breast cancer molecular subtypes was performed using the R package genefu (80). Basal-like, luminal A, luminal B, Her2, and normal-like subtype assignments were computed from five different algorithms (PAM50, AIMS, SCMGENE, SSP2006, and SCMOD2) (81–85). An assignment was considered final if defined by at least three different algorithms. In case of divergence between classifiers, PAM50-subtype attribution was used. For METABRIC and TCGA breast tumors, classification as a CL subtype was defined by nearest centroid method. For this, the Euclidean distance between each tumor sample and the previously described CL and non-CL tumor centroids was determined using the 1667 genes defined by Prat et al. (28) as “significantly differentially expressed” between CL tumors and all other molecular subtypes.
For CCLE breast cancer cell lines, CL status assignment was performed using the nine–cell line predictor via the R package genefu (80).
Pathway analysis
ssGSEA scores were computed through gsva R package (86) using gene signatures from the MSigDB (msigdbr R package) (87). Pan-cancer transcriptomic EMT signature defined by Tan et al. (29) was used to compute EMT scores for each sample.
Single-cell transcriptome analysis
Sample preparation
Full-length scRNA-seq libraries were prepared using the Smart-seq2 protocol (88) with minor modifications. Briefly, freshly harvested single cells were sorted into 96-well plates containing the lysis buffer [0.2% Triton X-100 and RNase inhibitor (1 U/μl); Thermo Fisher Scientific). RT was performed using SuperScript II (Thermo Fisher Scientific) in the presence of 1 μM oligo-dT30VN (IDT), 1 μM template-switching oligonucleotides (QIAGEN), and 1 M betaine. cDNA was amplified using the KAPA Hifi Hotstart ReadyMix (Roche) and IS PCR primer [Integrated DNA Technologies (IDT)], with 23 cycles of amplification. Following purification with Agencourt Ampure XP beads (Beckmann Coulter), product size distribution and quantity were assessed on a Bioanalyzer using the High Sensitivity DNA Kit (Agilent Technologies). A total of 140 pg of the amplified cDNA was fragmented using Nextera XT (Illumina) and amplified with Nextera XT indexes (Illumina). Products of each well of the 96-well plate were pooled and purified twice with Agencourt Ampure XP beads (Beckmann Coulter). Final libraries were quantified and checked for fragment size distribution using the Bioanalyzer High Sensitivity DNA Kit (Agilent Technologies). Pooled sequencing of Nextera libraries was carried out using a HiSeq4000 (Illumina) to an average sequencing depth of 0.5 million reads per cell. Sequencing was carried out as paired-end reads of 75–base pair length (PE75) with library indexes corresponding to cell barcodes.
Data processing
For the Smart-seq2 single-cell samples, quality check of the sequenced reads was performed with the FastQC v0.11.8 software to ensure the quality standards. Next, reads were aligned on the human genome (hg38) using STAR v2.7.0f (default parameters) (89). Raw counts for each gene were also computed by STAR, using the gencode V29 annotations.
Counts data for each cell of the same plate were grouped together.
Single-cell analysis
Quality control for the data was performed using the R packages scater (90) and scran (91). To eliminate low-quality data, we excluded cells with gene counts less than 3000 or greater than 7000. Genes expressed in less than 5% of cells were also removed. No further filtering was necessary. Subsequently, all cell data were consolidated into a unified matrix and normalized using the logNormCounts method. Using the 500 most variable genes, we conducted PCA and UMAP dimension reduction. Data clustering was carried out using the buildKNNGraph method with a parameter of K = 15. Signature scores for individual cells were obtained through the ssGSEA method from the GSVA package (86), except for the Jean-Paul Thierry EMT signature, for which we used the published cell line method (29).
The identification of gene markers for each data cluster was accomplished using the Seurat (92) package’s FindMarkers method, using the Model-based Analysis of Single-cell Transcriptomics (MAST) method. Top marker genes from each cluster were selected on the basis of criteria of a P value less than 0.001 and a fold change greater than 1 or less than −1. Enriched pathways within these genes were investigated using the clusterProfiler package (93) and the Hallmark, C2, and C5 genesets from MSigSB (94).
We generated a diffusion map using the R package Destiny (95), using the 50 principal component axes as input and setting a k parameter of 15. For trajectory analysis on the established data clusters that underwent PCA dimension reduction, the slingshot method (96) was used. Significance of cluster cell composition at different time points was assessed using scCODA (97). Velocity analyses were carried out by applying velocyto (98) to the BAM files, followed by analysis using the scVelo Python package (99).
Single-cell analysis with partial cell-cycle regression
Data from all cells were pooled into a single matrix and normalized using Scater logNormCounts method. The data were then scaled, and the cell cycle effect was reduced by regressing with the difference between the S and G2-M scores of each cell. Using the 300 most variable genes, we calculated the PCA and UMAP dimension reductions. We then clustered the data with Seurat’s FindClusters method, using the first 15 PCA axes, a k parameter of 15, and Leiden’s algorithm with a resolution of 0.25. Marker genes were called for each cluster using the FindMarkers method and the parameters test.used = MAST and min.pct = 0.1, logfc.threshold = 0.1. Genes were then selected for pathway analysis with a minimum log2 fold change of 0.6 and an adjusted P value ≤ 0.001. Overrepresentation analysis of pathways in marker genes was performed with the enricher function of the clusterProfiler R package, using gene sets from the Hallmark, C2, and C5 MSigDB database.
Acknowledgments
We thank A. P. Senaratne for critical reading of the manuscript. We are grateful to the Cancer Research Centre of Lyon - Centre Léon Bérard core facilities, the Cell Imaging Platform, and the Flow Cytometry Core Facility. We acknowledge the help of N. Gadot and C. Leneve (Research Pathology Platform East) for multiplex immunofluorescence stainings and H. Heyn, S. R. Gil, and G. Rodriguez-Esteban (CNAG-CRG, Barcelona Institute of Science and Technology, Spain) for single-cell analyses.
Funding: This work was supported by funding from Fondation pour la Recherche Médicale (FRM) and LabEx DEVweCAN (HDB), the Ligue Nationale Contre le Cancer (EL2016.LNCC/AIP and EL2019.LNCC/AIP), and SIRIC LYriCAN (INCa-DGOS-Inserm_12563). This work was additionally supported by funding from Inserm within the framework of the International Associated Laboratory between the Cancer Research Centre of Lyon, France, and the Victorian Comprehensive Cancer Centre of Melbourne, Australia (LIA/LEA 2016 and ASC17019CSA), as well as from Le Cancer du sein. Parlons-en! association.
Author contributions: A.P. and A.-P.M. conceived the study, supervised the work, and analyzed the data; H.D.B. designed and performed most experiments and analyzed the data; L.T. and R.M.P. performed the bioinformatics analyses; F.F., C.L., R.B., F.A., B.G., and A.P.M. contributed to experiments setup, methodology optimization, validation, and resources; M.O. and R.R. contributed to discussions about the project and exchanged ideas about the manuscript review and editing; A.P. and A.P.M. wrote the manuscript with inputs from H.D.B., M.O., and R.R. All authors discussed the results and commented on the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The raw FASTQ files and the processed single-cell sequencing data of this study can be obtained from Gene Expression Omnibus with the accession number GSE201536, accessible with the token evuzkskkttyhzwd. METABRIC data are available in the European Molecular Biology Laboratory–European Bioinformatics Institute (EMBL-EBI) archive (accession number EGAS00000000083) and from supplementary information of (30) and (100). TCGA BRCA RNA-seq expression data were extracted as FPKM values from the GDC data portal (https://portal.gdc.cancer.gov/). RNA-seq expression data from the CCLE breast cell lines were extracted as RPKM values from the CCLE data portal (https://portals.broadinstitute.org/ccle). Raw data used in Figs. 1 to 6, figs. S1 to S9, and raw data of Western blots and qPCRs can be obtained from Mendeley Data Sets (https://data.mendeley.com/datasets/gcy3g4g42j/2).
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Legends for tables S1 to S8
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S8
REFERENCES AND NOTES
- 1.Thiery J. P., Acloque H., Huang R. Y. J., Nieto M. A., Epithelial-mesenchymal transitions in development and disease. Cell 139, 871–890 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Puisieux A., Brabletz T., Caramel J., Oncogenic roles of EMT-inducing transcription factors. Nat. Cell Biol. 16, 488–494 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Yang J., Antin P., Berx G., Blanpain D., Brabletz T., Bronner M., Campbell K., Cano A., Casanova J., Christofori G., Dedhar S., Derynck R., Ford H. L., Fuxe J., Garcamp A., Goodall G. J., Hadjantonakis A.-K., Huang R. Y. J., Kalcheim C., Kalluri R., Kang Y., Khew-Goodall Y., Levine H., Liu J., Longmore G. D., Mani S. A., Massaguamp J., Mayor R., McClay D., Mostov K. E., Newgreen D. F., Angela Nieto M., Puisieux A., Runyan R., Savagner P., Stanger B., Stemmler M. P., Takahashi Y., Takeichi M., Theveneau E., Paul Thiery J., Thompson E. W., Weinberg R. A., Williams E. D., Xing J., Zhou B. P., Sheng G., EMT International Association (TEMTIA) , Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 21, 341–352 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nieto M. A., Huang R. Y.-J., Jackson R. A., Thiery J. P., Emt: 2016. Cell 166, 21–45 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Batlle E., Sancho E., Francí C., Domínguez D., Monfar M., Baulida J., De Herreros A. G., The transcription factor Snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2, 84–89 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Cano A., Pérez-Moreno M. A., Rodrigo I., Locascio A., Blanco M. J., Del Barrio M. G., Portillo F., Nieto M. A., The transcription factor Snail controls epithelial–mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2, 76–83 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Yang J., Mani S. A., Donaher J. L., Ramaswamy S., Itzykson R. A., Come C., Savagner P., Gitelman I., Richardson A., Weinberg R. A., Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117, 927–939 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Comijn J., Berx G., Vermassen P., Verschueren K., Van Grunsven L., Bruyneel E., Mareel M., Huylebroeck D., Van Roy F., The two-handed E box binding zinc finger protein SIP1 downregulates E-Cadherin and induces invasion. Mol. Cell 7, 1267–1278 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Dongre A., Weinberg R. A., New insights into the mechanisms of epithelial–Mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 20, 69–84 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Brabletz S., Schuhwerk H., Brabletz T., Stemmler M. P., Dynamic EMT: A multi-tool for tumor progression. EMBO J. 40, e108647 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jolly M. K., Tripathi S. C., Jia D., Mooney S. M., Celiktas M., Hanash S. M., Mani S. A., Pienta K. J., Ben-Jacob E., Levine H., Stability of the hybrid epithelial/mesenchymal phenotype. Oncotarget 7, 27067–27084 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer-Schaller N., Cardner M., Diepenbruck M., Saxena M., Tiede S., Lüönd F., Ivanek R., Beerenwinkel N., Christofori G., A hierarchical regulatory landscape during the multiple stages of EMT. Dev. Cell 48, 539–553.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Pastushenko I., Brisebarre A., Sifrim A., Fioramonti M., Revenco T., Boumahdi S., Van Keymeulen A., Brown D., Moers V., Lemaire S., De Clercq S., Minguijón E., Balsat C., Sokolow Y., Dubois C., De Cock F., Scozzaro S., Sopena F., Lanas A., D’haene N., Salmon I., Marine J.-C., Voet T., Sotiropoulou P. A., Blanpain C., Identification of the tumour transition states occurring during EMT. Nature 556, 463–468 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Zhang J., Tian X.-J., Zhang H., Teng Y., Li R., Bai F., Elankumaran S., Xing J., TGF-β-induced epithelial-to-mesenchymal transition proceeds through stepwise activation of multiple feedback loops. Sci. Signal. 7, ra91 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Mani S. A., Guo W., Liao M.-J., Eaton E. N., Ayyanan A., Zhou A. Y., Brooks M., Reinhard F., Zhang C. C., Shipitsin M., Campbell L. L., Polyak K., Brisken C., Yang J., Weinberg R. A., The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morel A.-P., Lièvre M., Thomas C., Hinkal G., Ansieau S., Puisieux A., Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLOS ONE 3, e2888 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L., Cao L., Wang H., Liu B., Zhang Q., Meng Z., Wu X., Zhou Q., Xu K., Wang L., Cao L., Wang H., Liu B., Zhang Q., Meng Z., Wu X., Zhou Q., Xu K., Cancer-associated fibroblasts enhance metastatic potential of lung cancer cells through IL-6/STAT3 signaling pathway. Oncotarget 8, 76116–76128 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aggarwal V., Montoya C. A., Donnenberg V. S., Sant S., Interplay between tumor microenvironment and partial EMT as the driver of tumor progression. iScience 24, 102113 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caramel J., Papadogeorgakis E., Hill L., Browne G. J., Richard G., Wierinckx A., Saldanha G., Osborne J., Hutchinson P., Tse G., Lachuer J., Puisieux A., Pringle J. H., Ansieau S., Tulchinsky E., A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell 24, 466–480 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Rhim A. D., Mirek E. T., Aiello N. M., Maitra A., Bailey J. M., McAllister F., Reichert M., Beatty G. L., Rustgi A. K., Vonderheide R. H., Leach S. D., Stanger B. Z., EMT and dissemination precede pancreatic tumor formation. Cell 148, 349–361 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y., Lu X., Huang L., Wang W., Jiang G., Dean K. C., Clem B., Telang S., Jenson A. B., Cuatrecasas M., Chesney J., Darling D. S., Postigo A., Dean D. C., Different thresholds of ZEB1 are required for Ras-mediated tumour initiation and metastasis. Nat. Commun. 5, 5660 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Morel A.-P., Hinkal G. W., Thomas C., Fauvet F., Courtois-Cox S., Wierinckx A., Devouassoux-Shisheboran M., Treilleux I., Tissier A., Gras B., Pourchet J., Puisieux I., Browne G. J., Spicer D. B., Lachuer J., Ansieau S., Puisieux A., EMT inducers catalyze malignant transformation of mammary epithelial cells and drive tumorigenesis towards claudin-low tumors in transgenic mice. PLOS Genet. 8, e1002723 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valsesia-Wittmann S., Magdeleine M., Dupasquier S., Garin E., Jallas A.-C., Combaret V., Krause A., Leissner P., Puisieux A., Oncogenic cooperation between H-Twist and N-Myc overrides failsafe programs in cancer cells. Cancer Cell 6, 625–630 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Ansieau S., Bastid J., Doreau A., Morel A.-P., Bouchet B. P., Thomas C., Fauvet F., Puisieux I., Doglioni C., Piccinin S., Maestro R., Voeltzel T., Selmi A., Valsesia-Wittmann S., Fromentel C. C., Puisieux A., Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell 14, 79–89 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Zhong X., Zheng L., Shen J., Zhang D., Xiong M., Zhang Y., He X., Tanyi J. L., Yang F., Montone K. T., Chen X., Xu C., Xiang A. P., Huang Q., Xu X., Zhang L., Suppression of microRNA 200 family expression by oncogenic KRAS activation promotes cell survival and epithelial-mesenchymal transition in KRAS-driven cancer. Mol. Cell. Biol. 36, 2742–2754 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rädler P. D., Wehde B. L., Triplett A. A., Shrestha H., Shepherd J. H., Pfefferle A. D., Rui H., Cardiff R. D., Perou C. M., Wagner K.-U., Highly metastatic claudin-low mammary cancers can originate from luminal epithelial cells. Nat. Commun. 12, 3742 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pommier R. M., Sanlaville A., Tonon L., Kielbassa J., Thomas E., Ferrari A., Sertier A. S., Hollande F., Martinez P., Tissier A., Morel A.-P., Ouzounova M., Puisieux A., Comprehensive characterization of claudin-low breast tumors reflects the impact of the cell-of-origin on cancer evolution. Nat. Commun. 11, 3431 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prat A., Parker J. S., Karginova O., Fan C., Livasy C., Herschkowitz J. I., He X., Perou C. M., Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 12, R68 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan T. Z., Miow Q. H., Miki Y., Noda T., Mori S., Huang R. Y.-J., Thiery J. P., Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol. Med. 6, 1279–1293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Curtis C., Shah S. P., Chin S.-F., Turashvili G., Rueda O. M., Dunning M. J., Speed D., Lynch A. G., Samarajiwa S., Yuan Y., Gräf S., Ha G., Haffari G., Bashashati A., Russell R., McKinney S., METABRIC Group, Langerød A., Green A., Provenzano E., Wishart G., Pinder S., Watson P., Markowetz F., Murphy L., Ellis I., Purushotham A., Børresen-Dale A.-L., Brenton J. D., Tavaré S., Caldas C., Aparicio S., Aparicio S., The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A. A., Kim S., Wilson C. J., Lehár J., Kryukov G. V., Sonkin D., Reddy A., Liu M., Murray L., Berger M. F., Monahan J. E., Morais P., Meltzer J., Korejwa A., Jané-Valbuena J., Mapa F. A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I. H., Cheng J., Yu G. K., Yu J., Aspesi P., de Silva M., Jagtap K., Jones M. D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R. C., Liefeld T., MacConaill L., Winckler W., Reich M., Li N., Mesirov J. P., Gabriel S. B., Getz G., Ardlie K., Chan V., Myer V. E., Weber B. L., Porter J., Warmuth M., Finan P., Harris J. L., Meyerson M., Golub T. R., Morrissey M. P., Sellers W. R., Schlegel R., Garraway L. A., The Cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al-Hajj M., Wicha M. S., Benito-Hernandez A., Morrison S. J., Clarke M. F., Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 100, 3983–3988 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kröger C., Afeyan A., Mraz J., Eaton E. N., Reinhardt F., Khodor Y. L., Thiru P., Bierie B., Ye X., Burge C. B., Weinberg R. A., Acquisition of a hybrid E/M state is essential for tumorigenicity of basal breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 116, 7353–7362 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burk U., Schubert J., Wellner U., Schmalhofer O., Vincan E., Spaderna S., Brabletz T., A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 9, 582–589 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caramel J., Ligier M., Puisieux A., Pleiotropic roles for ZEB1 in cancer. Cancer Res. 78, 30–35 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Becht E., McInnes L., Healy J., Dutertre C.-A., Kwok I. W. H., Ng L. G., Ginhoux F., Newell E. W., Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Zhu Y., Tchkonia T., Pirtskhalava T., Gower A. C., Ding H., Giorgadze N., Palmer A. K., Ikeno Y., Hubbard G. B., Lenburg M., O’Hara S. P., LaRusso N. F., Miller J. D., Roos C. M., Verzosa G. C., LeBrasseur N. K., Wren J. D., Farr J. N., Khosla S., Stout M. B., McGowan S. J., Fuhrmann-Stroissnigg H., Gurkar A. U., Zhao J., Colangelo D., Dorronsoro A., Ling Y. Y., Barghouthy A. S., Navarro D. C., Sano T., Robbins P. D., Niedernhofer L. J., Kirkland J. L., The Achilles’ heel of senescent cells: From transcriptome to senolytic drugs. Aging Cell 14, 644–658 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirkland J. L., Tchkonia T., Senolytic drugs: From discovery to translation. J. Intern. Med. 288, 518–536 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Y., Tchkonia T., Fuhrmann-Stroissnigg H., Dai H. M., Ling Y. Y., Stout M. B., Pirtskhalava T., Giorgadze N., Johnson K. O., Giles C. B., Wren J. D., Niedernhofer L. J., Robbins P. D., Kirkland J. L., Identification of a novel senolytic agent, navitoclax, targeting the Bcl-2 family of anti-apoptotic factors. Aging Cell 15, 428–435 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu Y., Doornebal E. J., Pirtskhalava T., Giorgadze N., Wentworth M., Fuhrmann-Stroissnigg H., Niedernhofer L. J., Robbins P. D., Tchkonia T., Kirkland J. L., New agents that target senescent cells: The flavone, fisetin, and the BCL-XL inhibitors, A1331852 and A1155463. Aging 9, 955–963 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Troiani M., Colucci M., D’Ambrosio M., Guccini I., Pasquini E., Varesi A., Valdata A., Mosole S., Revandkar A., Attanasio G., Rinaldi A., Rinaldi A., Bolis M., Cippà P., Alimonti A., Single-cell transcriptomics identifies Mcl-1 as a target for senolytic therapy in cancer. Nat. Commun. 13, 2177 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mosteiro L., Pantoja C., Alcazar N., Marión R. M., Chondronasiou D., Rovira M., Fernandez-Marcos P. J., Muñoz-Martin M., Blanco-Aparicio C., Pastor J., Gómez-López G., De Martino A., Blasco M. A., Abad M., Serrano M., Tissue damage and senescence provide critical signals for cellular reprogramming in vivo. Science 354, aaf4445 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Toneff M. J., Sreekumar A., Tinnirello A., Den Hollander P., Habib S., Li S., Ellis M. J., Xin L., Mani S. A., Rosen J. M., The Z-cad dual fluorescent sensor detects dynamic changes between the epithelial and mesenchymal cellular states. BMC Biol. 14, 47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Janda E., Lehmann K., Killisch I., Jechlinger M., Herzig M., Downward J., Beug H., Grünert S., Ras and TGFβ cooperatively regulate epithelial cell plasticity and metastasis. J. Cell Biol. 156, 299–314 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oft M., Peli J., Rudaz C., Schwarz H., Beug H., Reichmann E., TGF-β1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev. 10, 2462–2477 (1996). [DOI] [PubMed] [Google Scholar]
- 46.Krebs A. M., Mitschke J., Losada M. L., Schmalhofer O., Boerries M., Busch H., Boettcher M., Mougiakakos D., Reichardt W., Bronsert P., Brunton V. G., Pilarsky C., Winkler T. H., Brabletz S., Stemmler M. P., Brabletz T., The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat. Cell Biol. 19, 518–529 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Kinker G. S., Greenwald A. C., Tal R., Orlova Z., Cuoco M. S., McFarland J. M., Warren A., Rodman C., Roth J. A., Bender S. A., Kumar B., Rocco J. W., Fernandes P. A. C. M., Mader C. C., Keren-Shaul H., Plotnikov A., Barr H., Tsherniak A., Rozenblatt-Rosen O., Krizhanovsky V., Puram S. V., Regev A., Tirosh I., Pan-cancer single-cell RNA-seq identifies recurring programs of cellular heterogeneity. Nat. Genet. 52, 1208–1218 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.A. Gavish, M. Tyler, D. Simkin, D. Kovarsky, L. N. Gonzalez Castro, D. Halder, R. Chanoch-Myers, J. Laffy, M. Mints, A. R. Greenwald, A. Wider, R. Tal, A. Spitzer, T. Hara, A. Tirosh, S. V Puram, M. L. Suva, I. Tirosh, The transcriptional hallmarks of intra-tumor heterogeneity across a thousand tumors. bioRxiv 10.1101/2021.12.19.473368 [Preprint]. 2021.
- 49.C. Landragin, M. Saichi, P. Prompsy, A. Durand, J. Mesple, A. Trouchet, M. Faraldo, H. Salmon, C. Vallot, Luminal progenitors undergo partial epithelial-to-mesenchymal transition at the onset of basal-like breast tumorigenesis. bioRxiv 10.1101/2022.06.08.494710 [Preprint]. 2022.
- 50.Campisi J., Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 75, 685–705 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collado M., Blasco M. A., Serrano M., Cellular senescence in cancer and aging. Cell 130, 223–233 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Gorgoulis V., Adams P. D., Alimonti A., Bennett D. C., Bischof O., Bishop C., Campisi J., Collado M., Evangelou K., Ferbeyre G., Gil J., Hara E., Krizhanovsky V., Jurk D., Maier A. B., Narita M., Niedernhofer L., Passos J. F., Robbins P. D., Schmitt C. A., Sedivy J., Vougas K., von Zglinicki T., Zhou D., Serrano M., Demaria M., Cellular senescence: Defining a path forward. Cell 179, 813–827 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Kuilman T., Peeper D. S., Senescence-messaging secretome: SMS-ing cellular stress. Nat. Rev. Cancer 9, 81–94 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Rodier F., Campisi J., Four faces of cellular senescence. J. Cell Biol. 192, 547–556 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xue W., Zender L., Miething C., Dickins R. A., Hernando E., Krizhanovsky V., Cordon-Cardo C., Lowe S. W., Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445, 656–660 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kang T.-W., Yevsa T., Woller N., Hoenicke L., Wuestefeld T., Dauch D., Hohmeyer A., Gereke M., Rudalska R., Potapova A., Iken M., Vucur M., Weiss S., Heikenwalder M., Khan S., Gil J., Bruder D., Manns M., Schirmacher P., Tacke F., Ott M., Luedde T., Longerich T., Kubicka S., Zender L., Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature 479, 547–551 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Milanovic M., Fan D. N. Y., Belenki D., Däbritz H. M., Zhao Z., Yu Y., Dörr J. R., Dimitrova L., Lenze D., Monteiro Barbosa I. A., Mendoza-Parra M. A., Kanashova T., Metzner M., Pardon K., Reimann M., Trumpp A., Dörken B., Zuber J., Gronemeyer H., Hummel M., Dittmar G., Lee S., Schmitt C. A., Senescence-associated reprogramming promotes cancer stemness. Nature 553, 96–100 (2018). [DOI] [PubMed] [Google Scholar]
- 58.De Blander H., Morel A. P., Senaratne A. P., Ouzounova M., Puisieux A., Cellular plasticity: A route to senescence exit and tumorigenesis. Cancer 13, 4561 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chiche A., Le Roux I., von Joest M., Sakai H., Aguín S. B., Cazin C., Salam R., Fiette L., Alegria O., Flamant P., Tajbakhsh S., Li H., Injury-induced senescence enables in vivo reprogramming in skeletal muscle. Cell Stem Cell 20, 407–414.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Mosteiro L., Pantoja C., de Martino A., Serrano M., Senescence promotes in vivo reprogramming through p16INK4a and IL-6. Aging Cell 17, e12711 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Banito A., Rashid S. T., Acosta J. C., De Li S., Pereira C. F., Geti I., Pinho S., Silva J. C., Azuara V., Walsh M., Vallier L., Gil J., Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 23, 2134–2139 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu Y., Deng J., Rychahou P. G., Qiu S., Evers B. M., Zhou B. P., Stabilization of snail by NF-kappaB is required for inflammation-induced cell migration and invasion. Cancer Cell 15, 416–428 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yadav A., Kumar B., Datta J., Teknos T. N., Kumar P., IL-6 promotes head and neck tumor metastasis by inducing epithelial-mesenchymal transition via the JAK-STAT3-SNAIL signaling pathway. Mol. Cancer Res. 9, 1658–1667 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burkholder B., Huang R.-Y., Burgess R., Luo S., Jones V. S., Zhang W., Lv Z.-Q., Gao C.-Y., Wang B.-L., Zhang Y.-M., Huang R.-P., Tumor-induced perturbations of cytokines and immune cell networks. Biochim. Biophys. Acta 1845, 182–201 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Chang Q., Daly L., Bromberg J., The IL-6 feed-forward loop: A driver of tumorigenesis. Semin. Immunol. 26, 48–53 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Sullivan N. J., Sasser A. K., Axel A. E., Vesuna F., Raman V., Ramirez N., Oberyszyn T. M., Hall B. M., Interleukin-6 induces an epithelial-mesenchymal transition phenotype in human breast cancer cells. Oncogene 28, 2940–2947 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Al-Ismaeel Q., Neal C. P., Al-Mahmoodi H., Almutairi Z., Al-Shamarti I., Straatman K., Jaunbocus N., Irvine A., Issa E., Moreman C., Dennison A. R., Emre Sayan A., McDearmid J., Greaves P., Tulchinsky E., Kriajevska M., ZEB1 and IL-6/11-STAT3 signalling cooperate to define invasive potential of pancreatic cancer cells via differential regulation of the expression of S100 proteins. Br. J. Cancer 121, 65–75 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carpenter R. L., Lo H.-W., STAT3 target genes relevant to human cancers. Cancers 6, 897–925 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pires B. R. B., Mencalha A. L., Ferreira G. M., De Souza W. F., Morgado-Díaz J. A., Maia A. M., Corrêa S., Abdelhay E. S. F. W., NF-kappaB is involved in the regulation of EMT genes in breast cancer cells. PLOS ONE 12, e0169622 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grivennikov S., Karin E., Terzic J., Mucida D., Yu G. Y., Vallabhapurapu S., Scheller J., Rose-John S., Cheroutre H., Eckmann L., Karin M., IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell 15, 103–113 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sansone P., Storci G., Tavolari S., Guarnieri T., Giovannini C., Taffurelli M., Ceccarelli C., Santini D., Paterini P., Marcu K. B., Chieco P., Bonafè M., IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J. Clin. Invest. 117, 3988–4002 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lederle W., Depner S., Schnur S., Obermueller E., Catone N., Just A., Fusenig N. E., Mueller M. M., IL-6 promotes malignant growth of skin SCCs by regulating a network of autocrine and paracrine cytokines. Int. J. Cancer 128, 2803–2814 (2011). [DOI] [PubMed] [Google Scholar]
- 73.Walter M., Liang S., Ghosh S., Hornsby P. J., Li R., Interleukin 6 secreted from adipose stromal cells promotes migration and invasion of breast cancer cells. Oncogene 28, 2745–2755 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lau L., Porciuncula A., Yu A., Iwakura Y., David G., Uncoupling the senescence-associated secretory phenotype from cell cycle exit via Interleukin-1 inactivation unveils its protumorigenic role. Mol. Cell. Biol. 39, e00586-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu S., Lee J. S., Jie C., Park M. H., Iwakura Y., Patel Y., Soni M., Reisman D., Chen H., HER2 overexpression triggers an IL1α proinflammatory circuit to drive tumorigenesis and promote chemotherapy resistance. Cancer Res. 78, 2040–2051 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morgenstern J. P., Land H., Advanced mammalian gene transfer: High titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18, 3587–3596 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Young A. R. J., Narita M., Ferreira M., Kirschner K., Sadaie M., Darot J. F. J., Tavaré S., Arakawa S., Shimizu S., Watt F. M., Narita M., Autophagy mediates the mitotic senescence transition. Genes Dev. 23, 798–803 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.García-Prat L., Martínez-Vicente M., Perdiguero E., Ortet L., Rodríguez-Ubreva J., Rebollo E., Ruiz-Bonilla V., Gutarra S., Ballestar E., Serrano A. L., Sandri M., Muñoz-Cánoves P., Autophagy maintains stemness by preventing senescence. Nature 529, 37–42 (2016). [DOI] [PubMed] [Google Scholar]
- 79.Sachs N., de Ligt J., Kopper O., Gogola E., Bounova G., Weeber F., Balgobind A. V., Wind K., Gracanin A., Begthel H., Korving J., van Boxtel R., Duarte A. A., Lelieveld D., van Hoeck A., Ernst R. F., Blokzijl F., Nijman I. J., Hoogstraat M., van de Ven M., Egan D. A., Zinzalla V., Moll J., Boj S. F., Voest E. E., Wessels L., van Diest P. J., Rottenberg S., Vries R. G. J., Cuppen E., Clevers H., A living biobank of breast cancer organoids captures disease heterogeneity. Cell 172, 373–386.e10 (2018). [DOI] [PubMed] [Google Scholar]
- 80.Gendoo D. M. A., Ratanasirigulchai N., Schröder M. S., Paré L., Parker J. S., Prat A., Haibe-Kains B., Genefu: An R/Bioconductor package for computation of gene expression-based signatures in breast cancer. Bioinformatics 32, 1097–1099 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Haibe-Kains B., Desmedt C., Loi S., Culhane A. C., Bontempi G., Quackenbush J., Sotiriou C., A three-gene model to robustly identify breast cancer molecular subtypes. J. Natl. Cancer Inst. 104, 311–325 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wirapati P., Sotiriou C., Kunkel S., Farmer P., Pradervand S., Haibe-Kains B., Desmedt C., Ignatiadis M., Sengstag T., Schütz F., Goldstein D. R., Piccart M., Delorenzi M., Meta-analysis of gene expression profiles in breast cancer: Toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 10, R65 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Parker J. S., Mullins M., Cheung M. C. U., Leung S., Voduc D., Vickery T., Davies S., Fauron C., He X., Hu Z., Quackenhush J. F., Stijleman I. J., Palazzo J., Matron J. S., Nobel A. B., Mardis E., Nielsen T. O., Ellis M. J., Perou C. M., Bernard P. S., Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 27, 1160–1167 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hu Z., Fan C., Oh D. S., Marron J. S., He X., Qaqish B. F., Livasy C., Carey L. A., Reynolds E., Dressler L., Nobel A., Parker J., Ewend M. G., Sawyer L. R., Wu J., Liu Y., Nanda R., Tretiakova M., Orrico A., Dreher D., Palazzo J. P., Perreard L., Nelson E., Mone M., Hansen H., Mullins M., Quackenbush J. F., Ellis M. J., Olopade O. I., Bernard P. S., Perou C. M., The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics 7, 96 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Paquet E. R., Hallett M. T., Absolute assignment of breast cancer intrinsic molecular subtype. J. Natl. Cancer Inst. 107, 357 (2015). [DOI] [PubMed] [Google Scholar]
- 86.Hänzelmann S., Castelo R., Guinney J., GSVA: Gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics 14, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Picelli S., Björklund Å. K., Faridani O. R., Sagasser S., Winberg G., Sandberg R., Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013). [DOI] [PubMed] [Google Scholar]
- 89.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mccarthy D. J., Campbell K. R., Lun A. T. L., Wills Q. F., Scater: Pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lun A. T. L., McCarthy D. J., Marioni J. C., A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 5, 2122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W. M. III, Hao Y., Stoeckius M., Smibert P., Satija R., Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yu G., Wang L.-G., Han Y., He Q.-Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liberzon A., Subramanian A., Pinchback R., Thorvaldsdóttir H., Tamayo P., Mesirov J. P., Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Angerer P., Haghverdi L., Büttner M., Theis F. J., Marr C., Buettner F., Destiny: Diffusion maps for large-scale single-cell data in R. Bioinformatics 32, 1241–1243 (2016). [DOI] [PubMed] [Google Scholar]
- 96.Street K., Risso D., Fletcher R. B., Das D., Ngai J., Yosef N., Purdom E., Dudoit S., Slingshot: Cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Büttner M., Ostner J., Müller C. L., Theis F. J., Schubert B., scCODA is a Bayesian model for compositional single-cell data analysis. Nat. Commun. 12, 6876 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.La Manno G., Soldatov R., Zeisel A., Braun E., Hochgerner H., Petukhov V., Lidschreiber K., Kastriti M. E., Lönnerberg P., Furlan A., Fan J., Borm L. E., Liu Z., van Bruggen D., Guo J., He X., Barker R., Sundström E., Castelo-Branco G., Cramer P., Adameyko I., Linnarsson S., Kharchenko P. V., RNA velocity of single cells. Nature 560, 494–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bergen V., Lange M., Peidli S., Wolf F. A., Theis F. J., Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 38, 1408–1414 (2020). [DOI] [PubMed] [Google Scholar]
- 100.Pereira B., Chin S.-F., Rueda O. M., Vollan H.-K. M., Provenzano E., Bardwell H. A., Pugh M., Jones L., Russell R., Sammut S.-J., Tsui D. W. Y., Liu B., Dawson S.-J., Abraham J., Northen H., Peden J. F., Mukherjee A., Turashvili G., Green A. R., McKinney S., Oloumi A., Shah S., Rosenfeld N., Murphy L., Bentley D. R., Ellis I. O., Purushotham A., Pinder S. E., Børresen-Dale A.-L., Earl H. M., Pharoah P. D., Ross M. T., Aparicio S., Caldas C., The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 7, 11479 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Legends for tables S1 to S8
Tables S1 to S8