Abstract
Previous reports demonstrate that metformin, an anti-diabetic drug, can decrease the risk of cancer and inhibit cancer cell growth. However, its mechanism in cancer cells is still unknown. Metformin significantly blocks cell cycle and inhibits cell proliferation and colony formation of leukemic cells. However, the apoptotic response to metformin varies. Furthermore, daily treatment with metformin induces apoptosis and reduces tumor growth in vivo. While metformin induces early and transient activation of AMPK, inhibition of AMPKα1/2 does not abrogate anti-proliferative or pro-apoptotic effects of metformin. Metformin decreases electron transport chain complex I activity, oxygen consumption and mitochondrial ATP synthesis, while stimulating glycolysis for ATP and lactate production, pentose phosphate pathway for purine biosynthesis, fatty acid metabolism, as well as anaplerotic and mitochondrial gene expression. Importantly, leukemic cells with high basal AKT phosphorylation, glucose consumption or glycolysis exhibit a markedly reduced induction of the Pasteur effect in response to metformin and are resistant to metformin-induced apoptosis. Accordingly, glucose starvation or treatment with deoxyglucose or an AKT inhibitor induces sensitivity to metformin. Overall, metformin elicits reprogramming of intermediary metabolism leading to inhibition of cell proliferation in all leukemic cells and apoptosis only in leukemic cells responding to metformin with AKT phosphorylation and a strong Pasteur effect.
Keywords: metabolism, mitochondria, apoptosis, therapeutics, metformin
INTRODUCTION
An expanding body of evidence indicates that numerous metabolic pathways are altered in cancer cells.1,2 The Warburg effect, or aerobic glycolysis, is a common characteristic of cancer cells. However, metabolic reprogramming of transformed cells extends far beyond glucose metabolism.3,4 For example, numerous perturbations in the Krebs cycle, fatty acid and lipid metabolism and NADPH/ROS pathways have been identified in cancer cells, possibly as adaptations to their microenvironment, oncogenic stress or other factors. Understanding metabolic differences between normal and tumor cells offers promising possibilities for therapeutic strategies.
Acute myeloid leukemia (AML) is the most common adult acute leukemia and is characterized by clonal expansion of immature myeloblasts, initiating from rare leukemic stem or progenitor cells. Previous works have suggested that energetic metabolism has a role in cellular differentiation and chemoresistance in AML in vitro.5-7 Leukemic cells, like most cancer cells, may be addicted to glucose for generation of ATP and key biosynthetic intermediates, but recent data show that these cells also rely on fatty acid metabolism to grow and evade apoptosis.8 Furthermore, several groups9-12 found that 20–25% of AML patients harbor mutations in isocitrate dehydrogenase (IDH1 or IDH2), inducing a neomorphic enzymatic activity and production of oncometabolite, 2-hydroxyglutarate.13,14 These findings have changed our vision about the importance of cell metabolism in leukemia.
Metformin is a biguanide molecule used for treatment of diabetes mellitus. Several epidemiological studies have shown that although patients with type 2 diabetes have a higher risk for cancer, treatment with metformin is linked to a reduction in the risk of developing various types of cancers.15-17 Both in vitro and in vivo, metformin inhibits growth of pancreatic, colon, prostate, ovarian and breast cancer cells.18,19 Additionally, studies have shown that patients with breast cancer treated with a combination of chemotherapy and metformin have higher pathological response rates compared with those treated with chemotherapy alone.18,19 To date, two major effects of metformin have been described: inhibition of mitochondrial electron transport chain complex I (ETCI) and LKB1-dependent and independent activation of AMPK, a key energy sensor in cells (Supplementary Figure 1). Metformin can also induce a decrease in protein synthesis through a direct and AMPK-dependent activation of tuberous sclerosis complex 1/2 (TSC 1/2), which inhibits mammalian target of rapamycin.20-22 By targeting complex I, metformin mediates changes in AMP/ATP ratios, calcium levels, mitochondrial matrix pH and transmembrane potential, magnesium ions, inorganic phosphate, cyclophilin D and adenine nucleotides concentrations that commonly correlate with increased oxidative stress.23-25 These effects lead to a local and whole-body metabolic shift away from energy-consuming biochemical pathways to an energy-producing status, which profoundly reprograms cell metabolism. In brief, metformin activates catabolism and mitochondrial biogenesis while inhibiting protein synthesis and anabolic pathways. Unlike other indirect AMPK agonists (for example, AICAR), metformin inhibits ETCI, thereby preventing fatty acid oxidation and restricting glucose oxidation by mitochondria. The therapeutic potential of metformin might stem from this dual mechanism of action, which increases catabolism and limits substrate availability and oxidation. Importantly, both the effect of and sensitivity to metformin vary from cell to cell, providing a rationale for a therapeutic index for use of metformin in cancer therapy.
Herein, we investigate the metabolic, signaling and cytotoxic responses of diverse human leukemic cells to metformin. Metformin induces an early metabolic shift (for example, decreased oxygen consumption and mitochondrial ATP production while increasing glucose uptake and glycolytic ATP synthesis) and transient AMPK phosphorylation in all leukemic cells. Metformin inhibits cell proliferation in an AMPK-independent manner. The inhibition of mitochondrial respiratory capacities triggers a delayed and profound adaptation in the carbon flux balance and central metabolism in all leukemic cells, while inducing apoptosis in only some leukemic cells. Sensitivity to metformin-induced cell death is mediated by the metabolic capability to elicit the Pasteur effect, as well as by the basal level of AKT phosphorylation and glucose consumption. Collectively, these data demonstrate that metformin is a promising therapeutic drug and a useful tool for determining the contribution of each major metabolic pathway to leukemic cell survival.
MATERIALS AND METHODS
Cell proliferation and apoptosis
Cell proliferation was calculated by measuring the cell density using a Nexcelom cellometer with trypan blue dye. The cytotoxic response was determined by measuring apoptosis by flow cytometry after staining with AnnexinV-APC (Invitrogen, Saint Aubin, France) and 7AAD (BD Biosciences-Pharmingen, Le Pont de Claix, France). Samples were washed with cold HBSS (Invitrogen) and resuspended in 100 ul 1 × annexin-V-binding buffer (BD Biosciences). Cells were stained, on ice and in the dark, with Annexin V-APC for 15 min followed by 7AAD for another 15 min. We added 200 ul of 1 × Annexin-V-Binding buffer. Samples were analyzed using a FACSCalibur flow cytometer and BD CELLQuest-Pro software (BD Biosciences-Pharmingen).
Tumor xenografts
Xenograft tumors were generated by injecting 2 × 106 MOLM14 cells (in 100 ml of PBS) subcutaneously on both flanks of NU/NU Nude mice (Charles River, Wilmington, MA, USA) (n = 5, mice (10 tumors) per group). Mice were given daily intraperitoneal injections with 200 μl of 300 mg/kg/day metformin or vehicle (PBS). Tumor dimensions were measured with a caliper on days indicated and volume calculated using the formula: v = π/6 × A × B × B, where A is the larger diameter and B is the smaller diameter. At the end of the experiment, tumors were dissected, weighed, photographed and fixed with alcohol formalin acetic acid fixative for 48 h. Tumors were embedded in paraffin and sectioned for immunohistochemistry.
Immunohistochemistry
Apoptosis in xenografts was determined by immunohistochemistry for Active Caspase-3 (R&D Systems, Lille, France). For quantification, positively stained cells in six consecutive fields at × 20 magnification were counted from the edge towards the center of each section. Photographs for quantification were taken with a Leica DM4000B microscope (Leica, Saint Jorioz, France).
Small interfering RNA transfections
Small interfering RNA transfection of AMPKα1/2 was performed using the NEON Transfection System (Sigma-Aldrich, Lyon, France). Small interfering RNA for AMPKα1 and α2 was purchased from Invitrogen and previously published.26 In summary, 1 million cells were transfected with siControl or siAMPKα1/2 at a final concentration of 700 nm and then resuspended in 2.0 ml of MEMα + 10% FBS. Cells were incubated for 24 h to obtain complete inhibition of AMPKα1/2. Cells were then resuspended in fresh MEMα + 10% FBS and treated with vehicle or 10 mm metformin for an additional 48 h.
Measurement of metabolic parameters
We measured ETCI activity, oxygen consumption, adenosine triphosphate (ATP), lactate production and flux levels of glucose, lactate and glutamine. An extended description of the material and methods is provided in Supplementary Information Materials and Methods.
Liquid chromatography coupled to electrospray-LTQ-Orbitrap mass spectrometry with data processing and multivariate statistical analysis
Intracellular metabolites were analyzed with high performance anion exchange chromatography coupled to a triple quadrupole QTrap 4000 mass spectrometer (Method 1) and with ultra-high performance liquid chromatography coupled to mass spectrometry (UHPLC/MS), performed using an LTQ-Orbitrap hybrid mass spectrometer (Method 2). Raw data were analyzed using the XCMS software Version 1.14.1 running under R Version 2.8.1,19 (http://www.bioconductor.org/packages/bioc/html/xcms.html). The data sets resulting from the XCMS process were then annotated using tools developed in-house. An extended description of the Material and Methods is provided in Supplementary Information Materials and Methods.
Statistical analysis
Unpaired t-test was used to calculate final P-values. Significance is represented by stars in which *P<0.05, **P<0.01 and ***P<0.005.
RESULTS
Metformin, independent of AMPK, inhibits cell growth in human leukemic cells
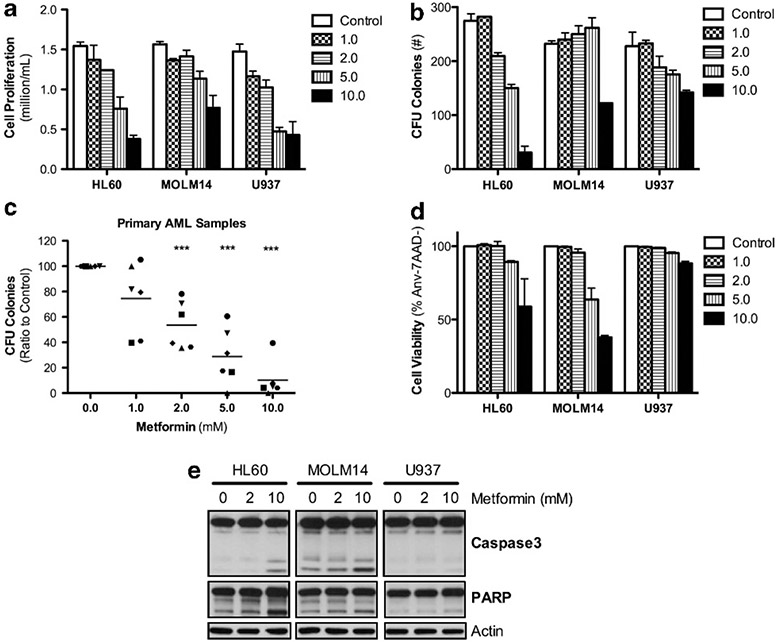
We first determined the in vitro effect of metformin on cell proliferation and apoptosis of three AML cell lines and six primary AML patient specimens (Supplementary Table 1 shows the characteristics of cell lines and patient samples). Metformin induced a strong inhibitory effect on cell proliferation in all leukemic cell lines (Figure 1a) and significantly reduced leukemia colony-forming units of three AML cell lines and six primary AML patient samples in a concentration-dependent manner (Figures 1b and c). Interestingly, metformin induced a cell cycle block in G0/G1 (U937) or S-G2/M (HL60 and MOLM14) phase in all three cell lines (Supplementary Figure S2A and B). However, we observed striking differences in metformin-induced cell death. HL60 and MOLM14 cells exhibited significant apoptosis-dependent cell death in response to metformin while U937 cells were resistant to this treatment (Figures 1d and e).
Figure 1.
Metformin inhibits cell proliferation in all human leukemic cell lines but has varying effects on apoptotic-dependent cell death. (a) HL60, MOLM14 and U937 cells after 48 h with 0.0, 1.0, 2.0, 5.0 or 10.0 mm metformin. Proliferation is calculated by ratio of final to initial cell count (with trypan blue exclusion) over the 48 h of incubation. (b) Average number of colony-forming units after 5-7 days incubation of HL60, MOLM14 and U937 cells in methylcellulose enriched media with 0.0, 1.0, 2.0, 5.0 or 10.0 mm metformin. (c) Average number of colony-forming units after 14 days incubation of six AML patient samples in methylcellulose enriched media with 0.0, 1.0, 2.0, 5.0 or 10.0 mm metformin, statistics by unpaired t-test with P-value as ***P < 0.005. (d, e) Viable cells as the percentage of Annexin-V negative and 7AAD negative cells carried out on the same cells listed for Figure 1a and the cleavage of capase-3 and PARP, respectively.
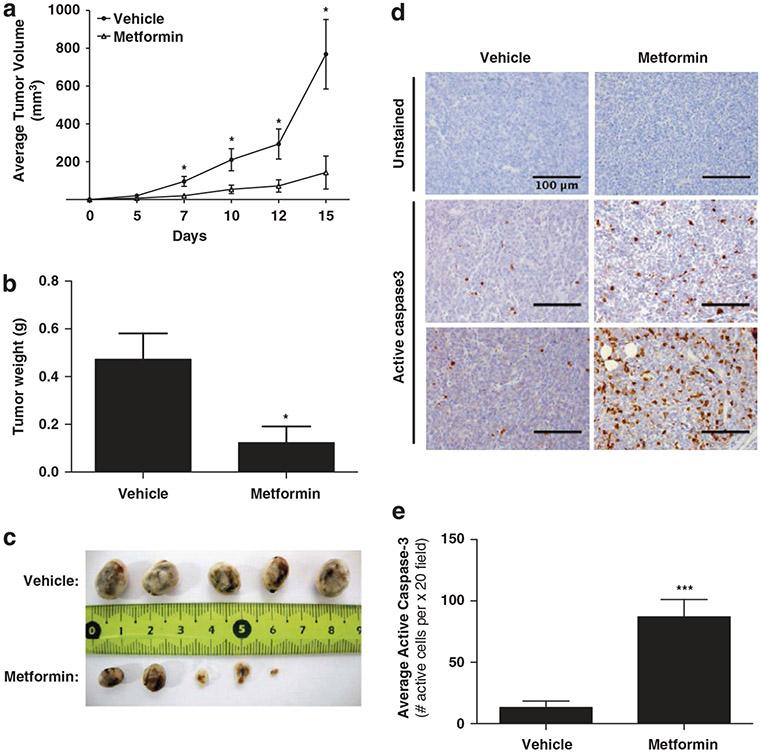
To determine if these results apply in vivo, we established a xenograft model for AML in female nude mice using MOLM14 cells and treated these mice with daily intraperitoneal injections of metformin (300 mg/kg/day). Metformin strongly reduced the tumor growth and weight in this model (Figures 2a and c). Additionally, analysis of treated tumors demonstrated that metformin induced apoptosis in vivo with an increase in active caspase-3 staining (Figures 2d and e). Thus, metformin is capable of inducing cell death in AML cells in vivo.
Figure 2.
Metformin inhibits tumor growth of AML cells in vivo. (a) Mean tumor size (mm3) and (b) tumor weight (g) was assessed in nude mice xenografted with MOLM14 cells and treated with either vehicle or 300 mg/kg/day metformin (n = 10, tumors in each group). (c) Representative photographs of xenografted tumors at day 17. (d) Tumor sections of mice injected with either vehicle or metformin were stained for active caspase-3. Representative photos of three metformin and three vehicle tumors are shown. (e) Quantification of the average number of active caspase-3 cells in 6 consecutive fields at x 20 magnification in tumors treated with either vehicle or metformin. Statistics were performed with an unpaired t-test with P-values represented as *P<0.05 and **P<0.005.
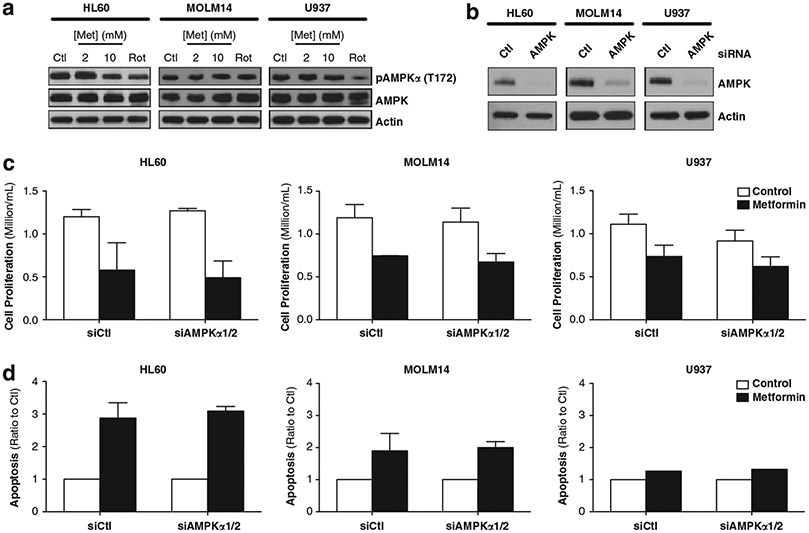
We next studied the biochemical effects of metformin on AML cells to understand the mechanism of sensitivity. As Green et al. (21) showed that metformin activates AMPK in various AML cell lines, we analyzed expression of AMPK and phospho-T172-AMPK in HL60, MOLM14 and U937 cells following metformin treatment. We noted no changes in phospho-T172-AMPK after 24 h metformin in all leukemic cells (Figure 3a). However, metformin transiently increased phosphorylation of AMPK in all AML cell lines as early as 1 h after incubation with 1 and 10 mm metformin with a maximum activation at 1 and 6 h for U937 and MOLM14 cells, respectively (Supplementary Figure S3). To determine whether the anti-proliferative and apoptotic effects of metformin are secondary to this transient activation of AMPK, we blocked this pathway using small interfering RNA of the two catalytic subunits AMPKα1 and α2. While we strongly diminished expression of AMPK in all three AML cell lines (Figure 3b), we did not prevent either inhibition of cell proliferation or induction of apoptosis in response to metformin (Figures 3c and d). Thus, AMPK activation by metformin is not necessary for metformin-induced cell death in AML cells.
Figure 3.
AMPKα1/2 does not effect the proliferation inhibition or apoptosis induced by metformin. (a) Western blots in HL60, MOLM14 and U937 cells after 24 h with untreated, 2 mm metformin, 10 mm metformin or 200 nm rotenone (Rot). Western blots were analyzed using primary antibodies for pAMPKα (T172) and AMPKα, all normalized to β-actin. (b) Western blots in HL60, MOLM14 and U937 cells 24 h after control small interfering RNA or AMPKα1/2 small interfering RNA. Following 24 h inhibition, these cells were treated for 48 h with control or 10 mm metformin and then analyzed for (c) cell proliferation, calculated by ratio of final to initial cell count (with trypan blue exclusion) and (d) apoptosis, measured as 7AAD-positive and AnnexinV-positive cells and presented as ratio to control.
Metformin inhibits ETCI in all leukemic cells but resistance to metformin-induced cell death correlates with a weak induction of the Pasteur effect
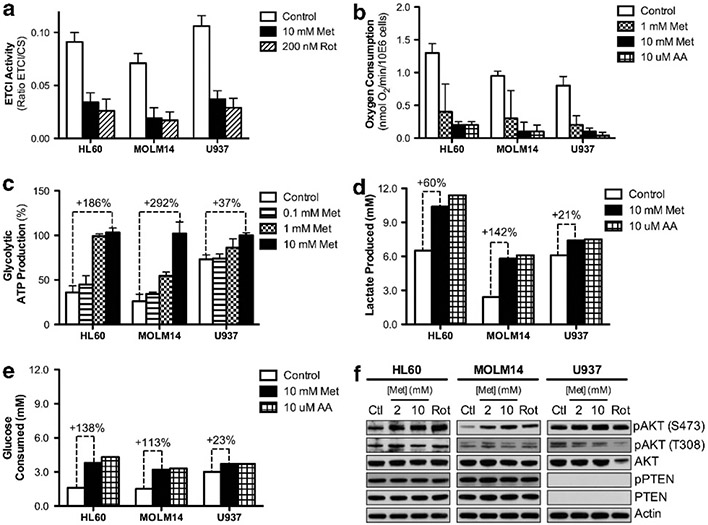
As metformin is known to inhibit ETCI, we examined the correlation between cell survival and respiratory characteristics of AML cell lines grown under the same conditions (MEMα containing 5.6 mm glucose, 2.5 mm glutamine + glutamate and 1 mm pyruvate). First, we assessed ETCI activity following metformin and observed a significant decrease at a similar level to rotenone, a specific ETCI inhibitor (Figure 4a). Accordingly, determination of oxygen consumption showed that metformin elicited a concentration-dependent decrease in the respiratory rate in HL60, MOLM14 and U937 cells (Figure 4b). Metformin inhibited oxygen consumption by 90% in leukemic cells after 24 h, an effect comparable to that observed with specific ETCIII inhibitor, antimycin A. Furthermore, all measured rates of oxygen consumption (oligomycin-sensitive, oligomycin-insensitive, uncoupler-stimulated (maximal) respiration and spare respiratory capacity of cells) decreased upon treatment with either 10 mm metformin or 10 μm antimycin A (Supplementary Figure S4A). Hence, our data is consistent with previously published in vitro studies showing a similar concentration-dependent inhibition of oxygen consumption over a range of 1 to 15 mm metformin after 24 h in several cell types.23,27
Figure 4.
Inhibition of oxygen consumption by metformin induces a stronger Pasteur Effect in HL60 and MOLM14 cells due to lower basal glucose consumption for ATP production. (a) ETCI activity measured as a ratio of ETCI activity to citrate synthase activity in HL60, MOLM14 and U937 cells after 24 h with control, 10 mm metformin or 200 nm rotenone (Rot). (b) The basal oxygen consumption rate, measured as nmol/min/million cells, in HL60, MOLM14 and U937 cells after 24 h with control, 1.0 mm metformin, 10.0 mm metformin or 10 mm antimycin A (AA). (c) Glycolytic ATP production measured as a percentage of total ATP production in HL60, MOLM14 and U937 cells after 24 h with 0.0, 0.1, 1.0 or 10.0 mm metformin. Comparison of (d) lactate produced (mm) and (e) glucose consumed (mm) in HL60, MOLM14 and U937 cells after 24 h with control, 10 mm metformin and 10 mm antimycin A. (f) Western blots in HL60, MOLM14 and U937 cells after 24 h with control, 2 mm metformin, 10 mm metformin or 200 nm rotenone. Western blots were analyzed using primary antibodies for pAKT (S473), pAKT (T308), AKT, pPTEN, and PTEN, all normalized to β-actin.
We next asked which metabolic alterations are involved in metformin-induced cell death. As observed in proliferating cells undergoing inhibition of mitochondrial OXPHOS, the decrease in mitochondrial ATP production was compensated by an increase in ATP production through glycolysis, the so-called Pasteur effect. Therefore, we first measured glycolytic ATP following metformin-induced inhibition of oxygen consumption and found that glycolytic ATP production is increased to a larger extent in HL60 (+ 186%) and MOLM14 (+ 292%) compared with U937 (+ 37%) after 24 h treatment with 10 mm metformin (Figure 4c). As other surrogates of the Pasteur effect, we also measured lactate production and release to the extracellular medium (Figure 4d). HL60 and MOLM14 cells showed a large (+ 60% and + 142%, respectively) increase in lactate production when treated with either 10 mm metformin or 10 μm antimycin A, while U937 cells only exhibited a limited (+ 21%) increase. This demonstrated that sensitive HL60 and MOLM14 cells show a more significant induction of the Pasteur effect in response to metformin than less sensitive U937 cells. To understand why U937 cells did not undergo apoptosis and did not exhibit a pronounced Pasteur effect, we analyzed different oxidizable substrates in the culture medium (glutamine, glucose) after treating HL60, MOLM14 and U937 cells with metformin or antimycin A. A significant increase was observed in glutamine consumption upon treatment in all cell types (Supplementary Figure S4B). Consistent with the increase in glycolytic ATP and lactate production, glucose consumption was markedly increased in metformin- and antimycin-treated HL60 and MOLM14 cells (Figure 4e). Interestingly, both metformin and antimycin A only mildly increased glucose utilization in U937 cells (+ 23% versus + 138% and + 113% in HL60 and MOLM14, respectively; Figure 4e). These results again demonstrate that metformin-sensitive HL60 and MOLM14 cells have a pronounced induction of the Pasteur effect following metformin treatment.
Interestingly, basal glucose consumption was two-fold higher in untreated U937 cells compared to MOLM14 cells, which correlated with higher basal lactate production in untreated U937 cells (Supplementary Figure S5A). This difference in basal glucose metabolism between MOLM14 and U937 cells was also evident in their respective glycolytic and oxidative phosphorylation contributions to ATP synthesis (26%/74% for MOLM14 versus 73%/27% for U937 of ATP produced by glycolysis/OXPHOS; Supplementary Figure S5B). These results demonstrated that the weak Pasteur effect observed in U937 cells in response to metformin is correlated to a decreased dependency on mitochondrial energetic metabolism in the basal state.
To ascertain whether mitochondrial changes and inhibition of ETC complex I account for the observed Pasteur effect and cell survival of metformin-treated AML cells, we examined ATP production at different time points (1–3–6 h compared with 24 h metformin). Our data indicated that the global ATP content of metformin-treated MOLM14 and U937 cells did not change during the experiment (Supplementary Figure S6A). In addition, we found that metformin-induced glycolytic ATP production occurred as early as 1 h with a maximum at 3 h of metformin treatment in MOLM14 and U937 (Supplementary Figure S6B). However, the induction of the Pasteur effect was much stronger in MOLM14 than U937 cells as above-mentioned. Of note, the increase in lactate production in the culture medium in response to metformin was significantly delayed when compared with the increase of glycolytic ATP production and occurred only after 24 h treatment, and markedly in MOLM14 cells (Supplementary Figure S6C compared with Supplementary Figure S6B). The discrepancy between the kinetics of these two readouts of the Pasteur effect is likely due to the time required to induce expression of plasma membrane monocarboxylic acid transporters implicated in lactate release.
Because basal glucose uptake and glycolytic rate seem to be critical parameters of the Pasteur effect induction, we asked how this balance is regulated. Thus, we analyzed the activation of the PI3K/AKT pathway, a key metabolic regulator of aerobic glycolysis in cancer cells. Interestingly, while metformin did not affect the expression of PTEN, metformin treatment increased and sustained phosphorylation of AKT at Ser473 (and not Thr308) only in metformin-sensitive HL60 and MOLM14 cells (Figure 4f). In contrast, we detected strong basal AKT phosphorylation in untreated, PTEN-null U937 cells and did not see further increase upon metformin treatment (Figure 4f). This is consistent with the notion that U937 cells have a strong glycolytic basal status, as illustrated by higher basal levels of lactate secretion and glucose uptake, lower levels of oxygen consumption, and lower dependence on mitochondrial ATP. In summary, these data suggest that U937 cells are resistant to metformin-induced cell death because they constitutively exhibit a strong glycolytic phenotype in the basal state and thus derive a significantly smaller fraction of ATP from mitochondrial OXPHOS, thereby requiring a smaller compensatory response to metformin-driven inhibition of ETCI.
Metformin-induced mitochondrial changes lead to profound metabolic adaptations in leukemic cells
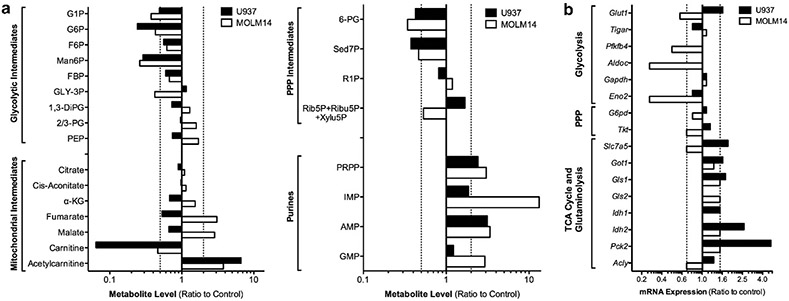
To better characterize the metabolic adaptations of AML cells in response to metformin we analyzed the metabolomic signature of metformin using quantitative LC-MS/MS mass spectrometry (Figure 5a and supplementary Figure S7A), as well as protein and gene expression analysis of key metabolic enzymes (Figure 5b and Supplementary Figure 7B). Genome-scale metabolic network of Homo sapiens28 was downloaded from BiGG database29 to perform integrated analysis of the data using the Gene to Protein to Reaction association for each leukemic cell line treated with 10 mm metformin for 24 h. The results showed a significant decrease in intermediates of the upper segment of glycolysis (G1P, G6P, F6P, FBP, Man6P) and the oxidative segment of the pentose phosphate pathway (6-PG, Sed7P) and a strong accumulation of purines (PRPP, IMP, AMP) in both MOLM14 and U937 cells after 24 h metformin (Figure 5a). We also found a strong increase in acetylcarnitine and decrease in carnitine, suggesting that metformin promotes high mitochondrial fatty acid shuttling (Figure 5a). Interestingly, we observed a significant difference between these two cell types, decreased in U937 and increased in MOLM14, in TCA metabolites (α-KG, fumarate, malate) and glycolytic intermediates (Gly-3P, 2/3PG, PEP) after metformin. Finally, comparing U937 to MOLM14 cells, we found a significant reduction in basal metabolites in the lower segment of glycolysis (Gly3P, 1,3-DiPG, 2/3-PG, PEP), pentose phosphate pathway (6-PG, Rib5P + Ribu5P + Xylu5P) and amino acid biosynthesis (methionine, glutamine), but an increase of purines (orotate, IMP, GMP, total NAD + /NADH pool), glutathione, carnitine and acetylcarnitine (Supplementary Figure 8), suggesting that control of flux through key metabolic pathways is significantly different between MOLM14 and U937.
Figure 5.
Metformin induces a metabolic adaptation in all leukemic cells, but insensitive cells have additional adaptive mechanisms to sustain cell survival. (a) Quantified intracellular concentrations of metabolites and (b) gene expression, calculated as ratio of 10 mm metformin to control, for MOLM14 and U937 cells after 24 h. For metabolite abbreviations, glucose-1-phosphate (G1P); glucose-6-phosphate (G6P); fructose-6-phosphate (F6P); mannose-6-phosphate (Man6P); fructose-1,6-bisphosphate (FBP); glyceraldehyde-3-phosphate (GLY-3P); 1,3-phosphodiglycerate (1,3-DiPG); 2/3-phosphoglycerate (2/3-PG); phosphoenolpyruvate (PEP); alpha-ketoglutarate (α-KG), 6-phosphogluconate (6-PG); sedoheptulose-7-phosphate (Sed7P); ribose-1-phosphate (R1P), ribose-5-phosphate (Rib5P); ribulose-5-phosphate (Ribu5P); xylulose-5-phosphate (Xylu5P); phosphoribosyl pryphosphate (PRPP); inosine monophosphate (IMP); adenosine monophosphate (AMP); guanine monophosphate (GMP). For gene expression abbreviations, glucose transporter 1 (Glut1); TP53 (Tumor Protein 53)-induced glycolysis and apoptosis regulator (Tigar); 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 (Pfkfb4); aldolase c (Aldoc); glyceraldehyde-3-phosphate dehydrogenase (Gapdh); enolase 2 (Eno2); glucose-6-phosphate dehydrogenase (G6pd); transketolase (Tkt), solute carrier family 7 (amino acid transporter light chain, L system); member 5 (Slc7a5), glutamate oxaloacetate transaminase 1 (Got1); glutaminase 1 (Gls1); glutaminase 2 (Gls2); isocitrate dehydrogenase 1 (Idh1); isocitrate dehydrogenase 2 (Idh2); phosphoenolpyruvate carboxykinase 2 (Pck2); ATP citrate lyase (Acly).
Metformin globally increased gene expression of enzymes involved in anaplerotic reactions (Pck2, Idh2, Got1 and Gls1) in MOLM14 and U937 cells (Figure 5b) and downregulated Pfkfb4, AldoC, Eno2, Glut1, Slc7A5, Tkt and Acly only in metformin-treated MOLM14 cells (Figure 5b). Interestingly, we also observed that metformin decreased phosphorylation of key rate-limiting enzyme of mitochondrial pyruvate oxidation, pyruvate dehydrogenase only in metformin-sensitive leukemic cells (Supplementary Figure S7B). However, we observed no significant changes for HK-II, GAPDH, PK-M2, LDHA, IDH1, IDH2, CPT1 or ACC (Supplementary Figure S7B). Of note, the calculated free energy changes for three key reactions (GPI, PFK, ENO) in the glycolytic pathway were similar between treated and untreated MOLM14 and U937 cells (Supplementary Figure S9). However, the PRPP biosynthetic equilibrium was significantly displaced by metformin in MOLM14 and the free energy change of the PFK reaction was significantly more negative in U937 under basal growth conditions, suggesting FBP formation by PFK is strongly favored in U937 cells.
Inhibition of glycolysis or AKT induces cell death upon metformin treatment of resistant U937 cells
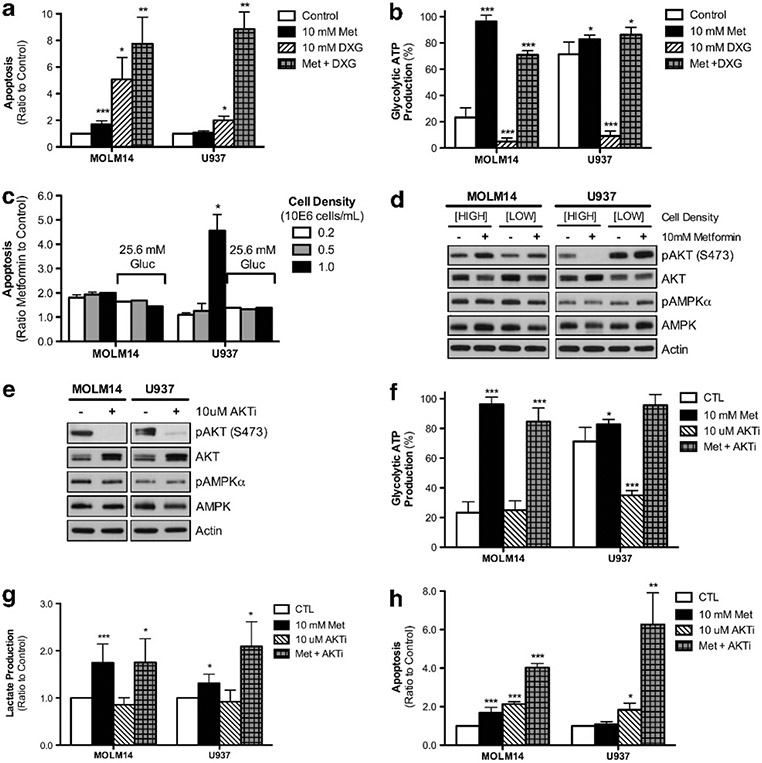
Finally, we tested whether the strong basal AKT activation and glycolysis protects U937 cells from metformin-induced apoptosis. First, we inhibited glycolytic metabolism with deoxyglucose (DXG) to determine if this would preferentially sensitize U937 cells to metformin-induced apoptosis following the shift from non-glucose oxidizing substrates (glutamine, fatty acids) to glucose oxidation through mitochondria. We observed marked sensitivity to DXG alone in MOLM14 without any synergistic effect with metformin. In U937 cells, by contrast, DXG had no effect on apoptosis alone even though DXG significantly decreased global and glycolytic ATP production. However, in combination with metformin, DXG induced marked apoptosis in U937 cells and significantly induced the Pasteur effect (that is, glycolytic ATP production) (Figures 6a and b; Supplementary Figure S10A). These results suggest that U937 cells depend heavily on glycolysis after metformin treatment. We confirmed these results by starving U937 of glucose by culturing the cells at high cell density in media with 5.6 mm glucose. In these conditions, U937 cells are highly sensitive to metformin, which can be abrogated by growing high cell density cells in the presence of 25.6 mm glucose (Figure 6c). Culturing U937 at high cell density also significantly decreased AKT phosphorylation, suggesting the key role of glucose uptake and the AKT pathway in metformin sensitivity (Figure 6d).
Figure 6.
The pro-apoptotic effect of metformin can be increased or induced in AML cells by specific metabolic manipulation. After a 4 h preincubation with or without 10 mm DXG, U937 and MOLM14 cells were then incubated for 24 h with control, 10 mm metformin, 10 mm DXG, or the combination of metformin and DXG and then analyzed for (a) apoptosis, measured as 7AAD-positive and AnnexinV-positive cells and presented as ratio to control and (b) glycolytic ATP production, presented as a percentage of total ATP production. (c) Apoptosis, measured as 7AAD-positive and AnnexinV-positive cells and presented as ratio of 10 mm metformin-treated to untreated, in MOLM14 and U937 cells at starting cell densities of 0.2, 0.5 or 1.0 million cells/mL either in MEMα (5.6 mm) + 10% FBS or MEMα (25.6 mm) + 10% FBS. (d) Western blots in MOLM14 and U937 cells after 24 h with control or 10 mm metformin with either high or low starting cell densities. Western blots were analyzed using primary antibodies for pAKT (S473), AKT, pAMPKα (T172) and AMPKα, all normalized to β-actin. (e) Western blot data in MOLM14 and U937 cells after 4 h pretreatment with untreated or 10 μm AKTi to confirm AKT inhibition. Western blots were analyzed using primary antibodies for pAKT (S473), AKT, pAMPKα (T172) and AMPKα, all normalized to β-actin. Following the 4 h preincubation with either control or 10 μm AKTi, U937 and MOLM14 cells were then incubated for 24 h with control, 10 mm metformin, 10 μm AKTi, or the combination of metformin and AKTi and then analyzed for (f) glycolytic ATP production, presented as a percentage of total ATP production, (g) lactate production, presented as a ratio to control, and (h) apoptosis, measured as 7AAD-positive and AnnexinV-positive cells and presented as ratio to control. Statistics were performed with an unpaired t-test with P-values represented as ***P<0.005, **P<0.01 and *P<0.05.
To test the hypothesis that high AKT activation can drive glycolysis and consequently metformin insensitivity in U937 cells, we studied the effects of AKT inhibition on cell metabolism and survival. We observed that 4 h-pretreatment with an AKT inhibitor completely reduced AKT phosphorylation in both leukemic cells (Figure 6e) and induced a strong Pasteur effect (increased glycolytic ATP and lactate production Figures 6f and g) and apoptosis in response to metformin that was markedly higher in U937 (Figure 5h). Overall, these results support our hypothesis that high pAKT leads to high basal glycolysis and insensitivity to metformin-induced cell death in U937 cells.
DISCUSSION
While metformin inhibited cell cycle progression, cell proliferation and leukemia colony-forming activities in all leukemic cells, metformin induced apoptosis only in certain cell types. Furthermore, daily treatment of xenograft mice with metformin induced apoptosis and decreased tumor growth in vivo. To understand the pro-apoptotic effect of metformin, we analyzed mitochondrial function and signaling as well as global metabolism in three different (sensitive, HL60 and MOLM14 versus insensitive, U937) leukemic cell lines. We demonstrated that metformin, similar to rotenone or antimycin A, inhibited both ETCI and oxygen consumption and shifted energetic ATP production from oxidative to glycolytic. Of note, we observed significant and transient AMPK phosphorylation at the Thr172 site in all leukemic cell lines in response to metformin, with a time course similar to inhibition of oxygen consumption and mitochondrial ATP production, but without any changes in global ATP content, as previously described in cells with defective AMPK signaling or effectors pathways.30,31 However, the anti-proliferative and pro-apoptotic effects of metformin are not mediated by activation of AMPK, which is also consistent with recent studies.20,26,32,33
Next, we interrogated the role of metabolism on metformin-induced cell death. In all leukemic cells, metformin induced a common metabolic adaptation to its early inhibitory effects on mitochondrial energetics with increased glucose uptake and consumption, as well as redirection of carbon flux towards lactate, the pentose phosphate pathway and purine biosynthesis. Furthermore, metformin increased mitochondrial and anaplerotic reactions (for example, fatty acid shuttling and oxidation, glutamine consumption and glutaminolysis) in all leukemic cells. However, between the two cell lines, metformin had the opposite effect on TCA cycle intermediates (α-ketoglutarate, fumarate, malate) and pyruvate dehydrogenase phosphorylation, confirming the importance of mitochondria on both basal energetic status and metformin sensitivity. Accordingly, metformin-insensitive U937 cells exhibited lower basal OCR and are less oxidative and more glycolytic under basal conditions, thereby requiring a smaller metabolic response (for example, Pasteur effect) following ETC1 inhibition by metformin.
The basal metabolome also showed significantly reduced carbon metabolites in the lower segment of glycolysis (Gly-3P to PEP) of U937 compared with MOLM14. This is consistent with our thermodynamic analysis showing FBP formation by PFK is strongly favored in U937 and rapidly consumed in the lower segment of glycolysis as intermediates are withdrawn for biosynthetic reactions. In addition to being converted to lactate, glycolytic intermediates in U937 might be diverted to increase flux through the PPP, to operate the glycerol-3-phosphate shuttle (dihydroxyacetone phosphate) and/or to support glycerol metabolism for biosynthetic purposes, including amino acids and triglycerides. Increased flow through the PPP increases production of purines and NADPH to maintain high GSH levels, which provides a high antioxidant capacity and links energetic reprogramming and redox balance in cancer cells.34,35 Rerouting glucose flux from glycolysis to PPP does not limit substrate availability in U937 because they sustain survival by alternative, non-glycolytic pathways and substrates (for example, glutamine or fatty acids) as supported by the marked basal accumulation of carnitine and acetylcarnitine. Accordingly, and as recently discovered in other leukemic cells,36 we observed significant phosphorylation of mitochondrial pyruvate dehydrogenase in both U937 and MOLM14, which suggests mitochondrial acetyl-CoA is derived from glutaminolysis (MOLM14) or glutaminolysis and FA oxidation (U937) rather than pyruvate oxidation. Additionally, accumulation of fumarate and malate in MOLM14 following metformin also suggests these cells might preferentially use glutamine-derived reductive carboxylation of α-KG to produce acetyl-CoA for four carbon intermediates for the TCA cycle, fatty acids, lipids, sterols and lactic acid, as very recently observed in cancer cells upon mitochondrial mutation, acute pharmacological ETC inhibition or hypoxia.37-39 This is also consistent with high sensitivity to metformin of cMyc-driven glutamine addicted cells (such as HL60).40
Finally, and of prime importance, we showed that metformin-insensitive U937 cells have high basal pAKT that has a pivotal role in metformin resistance, especially during early stages of treatment. Increased pAKT might also occur in sensitive cells following metformin treatment as an adaptive mechanism to early AMPK- and mitochondrial-induced metabolic responses. As a key regulator of glucose metabolism, AKT increases translocation of glucose transporters, GLUT1 and GLUT4, to the plasma membrane to increase glucose uptake and aerobic glycolysis.41-43 AKT also favors hexokinase-II activity and its association with the outer mitochondrial membrane, thereby increasing ATP affinity and providing direct access to mitochondrial ATP.44 Additionally, hexokinase-II binds specifically to the mitochondrial voltage-dependent anion channel, preventing opening of the permeabilization membrane pore, Cytochrome c release and subsequent apoptosis.45-48 In U937 cells, constitutive activation of the PI3K/AKT pathway by PTEN deletion leads to AKT-dependent stimulation of glucose uptake and metabolism. This confers resistance to the apoptotic and Pasteur effects of metformin in U937 cells, but renders them susceptible to death following glucose withdrawal or glycolytic inhibitors.49,50 Supporting this conclusion, metformin-mediated apoptosis was induced in U937 and augmented in MOLM14 by metabolic manipulations with AKT and glycolytic inhibitors and glucose starvation. These results show that the basal AKT status of leukemic cells is crucial in determining the ability to elicit a Pasteur effect and to regulate apoptosis in response to metformin.
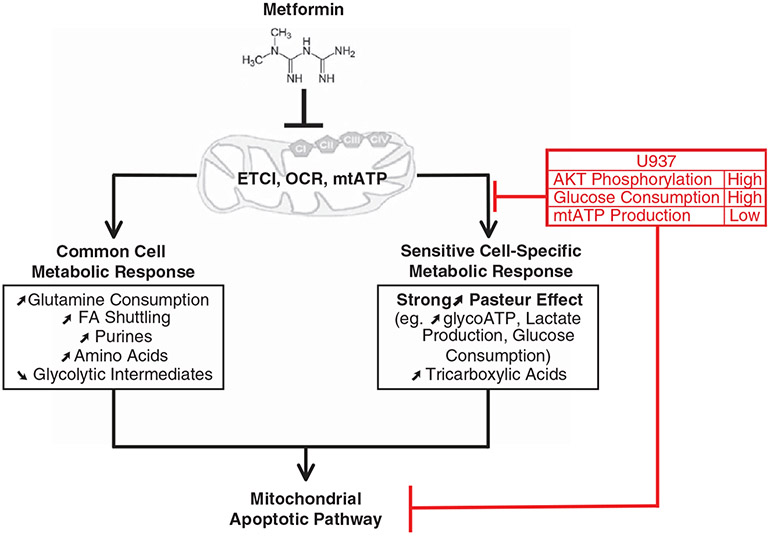
In conclusion, and represented in Supplementary Figure 1 and Figure 7, metformin induces an early (within 3 h) effect on mitochondrial energetics leading to long-term (6–24 h) metabolic adaptations (common- and sensitive-cell specific) followed by inhibition of cell cycle progression and cell proliferation and induction of the caspase-3 dependent mitochondrial apoptotic pathway (24–48 h) in leukemic cells. However, the late specific metabolic adaptation (strong induction of the Pasteur effect) and apoptosis do not occur in U937 cells, which already exhibit high basal glucose consumption, glycolysis and AKT activation and are thus metabolically poised to withstand inhibition of ETCI by metformin. Taken together, our study allows for a more general conclusion: metabolic fluxes of leukemic cells for energy production and generation of biosynthetic precursors are important determinants in metformin sensitivity. Our findings concerning the integration and regulation of energetic and metabolic cross-talks between mitochondria and cytosol, and other recent discoveries of cancer-related changes in metabolic pathways,38,39,51-53 demonstrate that metabolic and energetic flexibility are a common feature of tumor cell metabolism. A deeper understanding of this intrinsic capacity and its targeting are crucial steps to establish new therapeutic strategies in oncology.
Supplementary Material
Figure 7.
Working model of the multifacted mechanism of action of metformin in leukemic cells. In vitro metformin induces early energetic changes through the inhibition of ETCI, mitochondrial oxygen consumption (OCR) and mitochondrial ATP production (mtATP). The resulting metabolic response in all cells is an increase in glutamine consumption, fatty acid (FA) shuttling (acetylcarnitine), purines (PRPP, IMP, AMP) and amino acids (methionine), as well as a decrease in glycolytic intermediates. For cells that are sensitive to metformin-induced apoptosis, including HL60 and MOLM14, the specific metabolic response includes a strong increase in the Pasteur effect (for example, increased glycolytic ATP production, glucose consumption and lactate production), as well as an increase in tricarboxylic acids (fumarate and malate) which leads to activation of the mitochondrial apoptotic pathway. Cells that are insensitive to metformin-induced apoptosis, such as U937 cells, block this sensitive cell-specific metabolic response and apoptosis due to high AKT phosphorylation, high glucose consumption and low mitochondrial ATP production in the basal state.
ACKNOWLEDGEMENTS
We thank Clément Larrue, Zhu Wang, Frédéric Lagarrigue and Drs Marion David, Christian Touriol and Stéphane Rocchi for technical assistance, and Dr Laurent Le Cam, Professor Louis Casteilla and Hugues Chap for helpful discussion and reading of the manuscript. We thank the Platform of Experimental Histophathology of INERM/UPS for technical assistance. We thank Valérie Duplan-Eche, Delphine Lestrade and Fatima L’Faqihi-Olive for technical assistance at the flow cytometry core facility of INSERM UMR1043. SS is a recipient of the American Society of Hematology Trainee Research Award and Chateaubriand Fellowship from the French Embassy at Washington DC. YM was a recipient of Fondation de France post-doctoral fellowship. MC is supported by the Veterans Affairs Administration (1I01BX000918-01) and National Institute of Health (1R01CA149566-01A1). This work was supported by grants from CHOP Mitochondrial Interest Group (J-ES), Association Laurette Fugain (J-ES) and Fondation InNaBioSanté (CR, J-ES).
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell 2008; 13: 472–482. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- 3.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009; 324: 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov 2011; 10: 671–684. [DOI] [PubMed] [Google Scholar]
- 5.Ahmed N, Williams JF, Weidemann MJ. Glycolytic, glutaminolytic and pentose-phosphate pathways in promyelocytic HL60 and DMSO-differentiated HL60 cells. Biochem Mol Biol Int 1993; 29: 1055–1067. [PubMed] [Google Scholar]
- 6.Samudio I, Fiegl M, McQueen T, Clise-Dwyer K, Andreeff M. The warburg effect in leukemia-stroma cocultures is mediated by mitochondrial uncoupling associated with uncoupling protein 2 activation. Cancer Res 2008; 68: 5198–5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samudio I, Fiegl M, Andreeff M. Mitochondrial uncoupling and the Warburg effect: molecular basis for the reprogramming of cancer cell metabolism. Cancer Res 2009; 69: 2163–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samudio I, Harmancey R, Fiegl M, Kantarjian H, Konopleva M, Korchin B et al. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J Clin Invest 2010; 120: 142–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 2009; 361: 1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou WC, Hou HA, Chen CY, Tang JL, Yao M, Tsay W et al. Distinct clinical and biologic characteristics in adult acute myeloid leukemia bearing the isocitrate dehydrogenase 1 mutation. Blood 2010; 115: 2749–2754. [DOI] [PubMed] [Google Scholar]
- 11.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 2010; 17: 225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med 2010; 207: 339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science 2010; 330: 1340–1344. [DOI] [PubMed] [Google Scholar]
- 14.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 2010; 18: 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Libby G, Donnelly LA, Donnan PT, Alessi DR, Morris AD, Evans JM. New users of metformin are at low risk of incident cancer: a cohort study among people with type 2 diabetes. Diabetes Care 2009; 32: 1620–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Decensi A, Puntoni M, Goodwin P, Cazzaniga M, Gennari A, Bonanni B et al. Metformin and cancer risk in diabetic patients: a systematic review and meta-analysis. Cancer Prev Res (Phila) 2010; 3: 1451–1461. [DOI] [PubMed] [Google Scholar]
- 17.Currie CJ, Poole CD, Jenkins-Jones S, Gale EA, Johnson JA, Morgan CL. Mortality after incident cancer in people with and without type 2 diabetes: impact of metformin on survival. Diabetes Care 2012; 35: 299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirsch HA, Iliopoulos D, Tsichlis PN, Struhl K. Metformin selectively targets cancer stem cells, and acts together with chemotherapy to block tumor growth and prolong remission. Cancer Res 2009; 69: 7507–7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iliopoulos D, Hirsch HA, Struhl K. Metformin decreases the dose of chemotherapy for prolonging tumor remission in mouse xenografts involving multiple cancer cell types. Cancer Res 2011; 71: 3196–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalender A, Selvaraj A, Kim SY, Gulati P, Brule S, Viollet B et al. Metformin, independent of AMPK, inhibits mTORC1 in a rag GTPase-dependent manner. Cell Metab 2010; 11: 390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green AS, Chapuis N, Maciel TT, Willems L, Lambert M, Arnoult C et al. The LKB1/AMPK signaling pathway has tumor suppressor activity in acute myeloid leukemia through the repression of mTOR-dependent oncogenic mRNA translation. Blood 2010; 116: 4262–4273. [DOI] [PubMed] [Google Scholar]
- 22.Grimaldi C, Chiarini F, Tabellini G, Ricci F, Tazzari PL, Battistelli M et al. AMP-dependent kinase/mammalian target of rapamycin complex 1 signaling in T-cell acute lymphoblastic leukemia: therapeutic implications. Leukemia 2012; 26: 91–100. [DOI] [PubMed] [Google Scholar]
- 23.Owen MR, Doran E, Halestrap AP. Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem J 2000; 348: 607–614. [PMC free article] [PubMed] [Google Scholar]
- 24.Guigas B, Detaille D, Chauvin C, Batandier C, De Oliveira F, Fontaine E et al. Metformin inhibits mitochondrial permeability transition and cell death: a pharmacological in vitro study. Biochem J 2004; 382: 877–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaw RJ, Lamia KA, Vasquez D, Koo SH, Bardeesy N, Depinho RA et al. The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science 2005; 310: 1642–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomic T, Botton T, Cerezo M, Robert G, Luciano F, Puissant A et al. Metformin inhibits melanoma development through autophagy and apoptosis mechanisms. Cell Death Dis 2011; 2: e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El-Mir MY, Nogueira V, Fontaine E, Averet N, Rigoulet M, Leverve X. Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. J Biol Chem 2000; 275: 223–228. [DOI] [PubMed] [Google Scholar]
- 28.Duarte NC, Becker SA, Jamshidi N, Thiele I, Mo ML, Vo TD et al. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci USA 2007; 104: 1777–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schellenberger J, Park JO, Conrad TM, Palsson BO. BiGG: a Biochemical Genetic and Genomic knowledgebase of large scale metabolic reconstructions. BMC Bioinformatics 2010; 11: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buzzai M, Jones RG, Amaravadi RK, Lum JJ, DeBerardinis RJ, Zhao F et al. Systemic treatment with the antidiabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer Res 2007; 67: 6745–6752. [DOI] [PubMed] [Google Scholar]
- 31.Algire C, Amrein L, Bazile M, David S, Zakikhani M, Pollak M. Diet and tumor LKB1 expression interact to determine sensitivity to anti-neoplastic effects of metformin in vivo. Oncogene 2011; 30: 1174–1182. [DOI] [PubMed] [Google Scholar]
- 32.Ben Sahra I, Laurent K, Loubat A, Giorgetti-Peraldi S, Colosetti P, Auberger P et al. The antidiabetic drug metformin exerts an antitumoral effect in vitro and in vivo through a decrease of cyclin D1 level. Oncogene 2008; 27: 3576–3586. [DOI] [PubMed] [Google Scholar]
- 33.Ben Sahra I, Regazzetti C, Robert G, Laurent K, Le Marchand-Brustel Y, Auberger P et al. Metformin, independent of AMPK, induces mTOR inhibition and cell-cycle arrest through REDD1. Cancer Res 2011; 71: 4366–4372. [DOI] [PubMed] [Google Scholar]
- 34.Hamanaka RB, Chandel NS. Cell biology. Warburg effect and redox balance. Science 2011; 334: 1219–1220. [DOI] [PubMed] [Google Scholar]
- 35.Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 2011; 334: 1278–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal 2009; 2: ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci USA 2011; 108: 19611–19616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 2012; 481: 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 2012; 481: 385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Javeshghani S, Zakikhani M, Austin S, Bazile M, Blouin MJ, Topisirovic I et al. Carbon source and myc expression influence the antiproliferative actions of metformin. Cancer Res 2012; 72: 6257–6267. [DOI] [PubMed] [Google Scholar]
- 41.Barthel A, Okino ST, Liao J, Nakatani K, Li J, Whitlock JP Jr et al. Regulation of GLUT1 gene transcription by the serine/threonine kinase Akt1. J Biol Chem 1999; 274: 20281–20286. [DOI] [PubMed] [Google Scholar]
- 42.Kohn AD, Summers SA, Birnbaum MJ, Roth RA. Expression of a constitutively active Akt Ser/Thr kinase in 3T3-L1 adipocytes stimulates glucose uptake and glucose transporter 4 translocation. J Biol Chem 1996; 271: 31372–31378. [DOI] [PubMed] [Google Scholar]
- 43.Wieman HL, Wofford JA, Rathmell JC. Cytokine stimulation promotes glucose uptake via phosphatidylinositol-3 kinase/Akt regulation of Glut1 activity and trafficking. Mol Biol Cell 2007; 18: 1437–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mathupala SP, Ko YH, Pedersen PL. Hexokinase-2 bound to mitochondria: cancer’s stygian link to the "Warburg Effect" and a pivotal target for effective therapy. Semin Cancer Biol 2009; 19: 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Majewski N, Nogueira V, Bhaskar P, Coy PE, Skeen JE, Gottlob K et al. Hexokinase-mitochondria interaction mediated by Akt is required to inhibit apoptosis in the presence or absence of Bax and Bak. Mol Cell 2004; 16: 819–830. [DOI] [PubMed] [Google Scholar]
- 46.Arzoine L, Zilberberg N, Ben-Romano R, Shoshan-Barmatz V. Voltage-dependent anion channel 1-based peptides interact with hexokinase to prevent its anti-apoptotic activity. J Biol Chem 2009; 284: 3946–3955. [DOI] [PubMed] [Google Scholar]
- 47.McCubrey JA, Steelman LS, Abrams SL, Bertrand FE, Ludwig DE, Basecke J et al. Targeting survival cascades induced by activation of Ras/Raf/MEK/ERK, PI3K/PTEN/Akt/mTOR and Jak/STAT pathways for effective leukemia therapy. Leukemia 2008; 22: 708–722. [DOI] [PubMed] [Google Scholar]
- 48.Martelli AM, Evangelisti C, Chappell W, Abrams SL, Basecke J, Stivala F et al. Targeting the translational apparatus to improve leukemia therapy: roles of the PI3K/PTEN/Akt/mTOR pathway. Leukemia 2011; 25: 1064–1079. [DOI] [PubMed] [Google Scholar]
- 49.Buzzai M, Bauer DE, Jones RG, Deberardinis RJ, Hatzivassiliou G, Elstrom RL et al. The glucose dependence of Akt-transformed cells can be reversed by pharmacologic activation of fatty acid beta-oxidation. Oncogene 2005; 24: 4165–4173. [DOI] [PubMed] [Google Scholar]
- 50.Ben Sahra I, Laurent K, Giuliano S, Larbret F, Ponzio G, Gounon P et al. Targeting cancer cell metabolism: the combination of metformin and 2-deoxyglucose induces p53-dependent apoptosis in prostate cancer cells. Cancer Res 2010; 70: 2465–2475. [DOI] [PubMed] [Google Scholar]
- 51.Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 2011; 476: 346–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frezza C, Zheng L, Folger O, Rajagopalan KN, MacKenzie ED, Jerby L et al. Haem oxygenase is synthetically lethal with the tumour suppressor fumarate hydratase. Nature 2011; 477: 225–228. [DOI] [PubMed] [Google Scholar]
- 53.Zhang F, Du G. Dysregulated lipid metabolism in cancer. World J Biol Chem 2012; 3: 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.