Summary
Tauopathies are a group of neurodegenerative diseases defined by abnormal aggregates of tau, a microtubule-associated protein encoded by MAPT. MAPT expression is near absent in neural progenitor cells (NPCs) and increases during differentiation. This temporally dynamic expression pattern suggests that MAPT expression could be controlled by transcription factors and cis-regulatory elements specific to differentiated cell types. Given the relevance of MAPT expression to neurodegeneration pathogenesis, identification of such elements is relevant to understanding disease risk and pathogenesis. Here, we performed chromatin conformation assays (HiC & Capture-C), single-nucleus multiomics (RNA-seq+ATAC-seq), bulk ATAC-seq, and ChIP-seq for H3K27ac and CTCF in NPCs and differentiated neurons to nominate candidate cis-regulatory elements (cCREs). We assayed these cCREs using luciferase assays and CRISPR interference (CRISPRi) experiments to measure their effects on MAPT expression. Finally, we integrated cCRE annotations into an analysis of genetic variation in neurodegeneration-affected individuals and control subjects. We identified both proximal and distal regulatory elements for MAPT and confirmed the regulatory function for several regions, including three regions centromeric to MAPT beyond the H1/H2 haplotype inversion breakpoint. We also found that rare and predicted damaging genetic variation in nominated CREs was nominally depleted in dementia-affected individuals relative to control subjects, consistent with the hypothesis that variants that disrupt MAPT enhancer activity, and thereby reduced MAPT expression, may be protective against neurodegenerative disease. Overall, this study provides compelling evidence for pursuing detailed knowledge of CREs for genes of interest to permit better understanding of disease risk.
Keywords: Alzheimer disease, MAPT, gene regulation, neuron, enhancer
This study uses functional genomics approaches to assess candidate cis-regulatory elements (cCREs) for MAPT in neurons. The study suggests that impactful rare non-coding variants in MAPT cCREs (hypothesized to result in lower tau expression) may be protective against neurodegeneration, emphasizing the potential importance of rare non-coding variants in disease risk.
Introduction
Aggregation of the microtubule-associated protein tau (encoded by MAPT) is a defining pathological feature of neurodegenerative tauopathies like Alzheimer disease (AD) and tau-positive frontotemporal dementia (FTD) including progressive supranuclear palsy (PSP). Tau is highly abundant in the brain and functions to stabilize microtubules, which are critical for axonal growth and guidance.1,2,3 However, the physiological role of tau extends well beyond its role in microtubule stabilization, with evidence supporting roles for tau in insulin signaling, synaptic plasticity, excitatory postsynaptic transmission, and the generation and maintenance of brain rhythms (reviewed by Chang et al.4). There are six isoforms of tau expressed in the brain and different isoforms have either three (3R) or four (4R) microtubule-binding domains that are regulated by phosphorylation at sites within or adjacent to the binding domains. In pathological conditions, tau undergoes hyperphosphorylation at these sites, permitting it to become unbound from microtubules and promoting tau oligomerization.1,5 Hyperphosphorylation of tau leads to the mislocalization of tau from axons and into the soma and dendrites.6,7 Somatodendritic mislocalization of tau occurs early in disease pathogenesis before neurodegeneration and mediates synaptic dysfunction leading to neuronal loss.6,7,8,9 Tau hyperphosphorylation-induced oligomers subsequently form large insoluble fibrils that are the major components of neurofibrillary tangles (NFTs). These intracellular NFTs can be found in the hippocampus during normal aging, but abnormal loads elsewhere in the cortex are pathological hallmarks of tauopathies.1,10,11 In humans, regions with higher expression of MAPT exhibit more robust accumulation of pathological tau marked by tau PET in disease.12 In animal models, reducing endogenous tau has been successful in preventing early mortality, cognitive deficits, and excitotoxicity even in the presence of amyloid-beta.13,14,15,16,17,18 These studies in animal models have provided the basis for clinical trials that are currently underway aimed to reduce tau via antisense oligonucleotides.19
While studies examining altered expression levels of MAPT in AD have produced mixed results,20,21,22,23 rare copy number events point to the relevance of MAPT gene expression in pathogenesis. For example, MAPT duplication resulting in 1.6- to 1.9-fold higher levels of MAPT mRNA leads to a tauopathy with AD-like presentation.24 More generally beyond MAPT, efforts to better understand genetic contributors to disease have resulted in large-scale genome-wide association studies (GWASs) which have identified ∼83 common variants that are associated with late-onset AD (LOAD).25,26,27,28,29 The majority of GWAS-nominated variants are located in non-coding regions of the genome, pointing to the importance of understanding the roles these regions have in contributing to disease.30,31 The leading hypothesis is that these regions are cis-regulatory elements (CREs), or enhancers, contributing to gene expression.32,33 Large efforts, like ENCODE, have been made to identify CREs; however, experiments/data enabling CRE prediction in brain cell types are sparse, resulting in false negatives when attempting to identify enhancers for cell types specific to the brain. Chromatin looping, which can physically connect loci that are tens or hundreds of thousands of kilobases apart, also makes assigning non-coding GWAS single-nucleotide variants (SNVs) to genes difficult. SNVs are typically assigned to the nearest gene in the locus, but because of chromatin looping, enhancers may actually be in closer three-dimensional proximity with promoters of genes farther away in linear distance.34,35,36,37 A further complication is linkage disequilibrium (LD), in which multiple nearby alleles tend to be inherited together in haplotype blocks, which makes causal variant identification difficult. In fact, MAPT falls within one of the largest LD blocks known in the human genome, spanning 1.8 Mb.38 The locus can be divided into two haplotypes, defined by a 900 kb inversion event encompassing several genes, including MAPT. The “H1” haplotype is the most common haplotype (and is also the haplotype of the reference genome hg38) and its presence relative to the “H2” haplotype has been associated with PSP, corticobasal degeneration (CBD), and Parkinson disease (PD).39,40,41,42,43,44 The haplotype-tagging SNVs have also been associated with AD, but the causal gene driving these disease associations is still an ongoing area of research with evidence for both MAPT and KANSL1.25,45,46,47 For risk of Parkinson disease, additional genes such as LRRC37A/2 may account for this signal.48 Still, for AD, with the H2 haplotype, which carries the inversion relative to H1, evidence points to a possible reduction in MAPT expression along with a lower risk for AD.49
Here, we sought to nominate all possible putative cis-regulatory elements of MAPT using orthogonal genomic approaches. We integrated single-nucleus multiomics (snRNA-seq + snATAC-seq) from both cultured neurons and previously published dorsolateral prefrontal cortex (DLPFC) tissue23 to correlate chromatin accessibility with MAPT expression. We performed chromatin conformation assays (HiC and Capture-C) to determine the 3D interactions with the MAPT promoter in NPCs and matched differentiated neurons, as well as pure populations of human glutamatergic and GABAergic neurons. Finally, we manually inspected regions previously nominated by other studies,50,51 and to nominate more proximal regions with (1) characteristic histone modifications (H3K27ac & H3K4me1), (2) high conservation, (3) transcription factor (TF) binding motifs, or (4) ENCODE cCRE annotation. We assessed these nominated regions for sufficiency to induce transcriptional activity through reporter assays and for necessity to MAPT expression using CRISPR interference (CRISPRi) experiments.
Material and methods
Cell lines
XCL4 Neural Progenitor Cells (NPCs) were obtained from StemCell Technologies. BC1 NPCs were obtained from MTI GlobalStem. KOLF2.1J NPCs were generated from KOLF2.1J iPSCs, obtained from Jackson Laboratory, using the StemCell Technologies STEMdiff SMADi Neural Induction kit (StemCell Technologies 08581). All NPCs were maintained in NPC Media: 2:1 DMEM (high glucose, L-glutamine, 100 mg/L sodium pyruvate): Ham’s F-12 Nutrient Mix, and supplemented with 50× serum-free B-27 Supplement (ThermoFisher Scientific). hFGF (40 μg/mL), hEGF (20 μg/mL), and heparin (5 μg/mL) were added daily to NPC media. iCell GlutaNeurons and iCell GABA Neurons were obtained from FujiFilm and maintained according to the manufacturer protocols for 14 days 293FT cells were obtained from ThermoFisher Scientific (R70007) and maintained in DMEM (high glucose, L-glutamine, 100 mg/L sodium pyruvate) supplemented with 10% FBS, 1% Glutamax, 1% non-essential amino acids (NEAA), and 500 mg/mL Geneticin (G418 sulfate) (ThermoFisher Scientific). KOLF2.1J and KOLF2.1J-hNGN2 iPSCs were obtained from the Jackson Laboratory and maintained in mTeSR Plus medium (StemCell Technologies). All cells were cultured at 37°C with 5% CO2. KOLF2.1J NPCs were generated following the StemCell Technologies SMADi neural induction protocol.
Neuron differentiation
NPCs were differentiated into a mixed culture of neurons and astrocytes according to the Bardy et al. protocol.52 Cells were maintained in the neuronal differentiation medium for 14 or 21 days iCell GlutaNeurons and GABAneurons were differentiated according to the manufacturer recommendations (FujiFilm Cellular Dynamics). Cells were maintained in their respective BrainPhys complete media for 14 days. KOLF2.1J-hNGN2 iPSCs were dissociated using Accutase and plated as single cells at 50,000 cells/cm2 in induction medium: DMEM/F12 medium with HEPES (ThermoFisher), 100× N2 supplement (StemCell Technologies), 100× non-essential amino acids (NEAA, ThermoFisher), and 100× Glutamax (ThermoFisher). For plating cells, induction medium was supplemented with 10 μM ROCK inhibitor (Y-27632) and 2 mg/mL doxycycline. Induction medium supplemented with doxycycline was renewed daily for two days. On day 3, neurites were present and cells were renewed with cortical neuron culture media: BrainPhys neuronal medium (StemCell Technologies), 50× SM1 supplement (StemCell Technologies), 10 μg/mL BDNF (StemCell Technologies), 10 μg/mL NT-3 (StemCell Technologies), and 1 mg/mL mouse Laminin (Gibco). On day 3, cortical neuron culture media was supplemented with 2 mg/mL doxycycline. One-half media changes were performed every other day for a total of 14 days.
Single nucleus multiomics
Nuclei from cultured neurons were isolated as detailed in 10x Genomics demonstrated protocol CG000375 Rev A with modifications. Cells were washed with cold 1× DPBS and then 500 μL of NP-40 lysis buffer was added. Cells were gently scraped off the dish while on ice, transferred to a 1.5 mL tube, and incubated 5 min on ice. Cells were centrifuged at 500 × g for 5 min at 4°C and 500 μL 1× PBS +1% BSA + DAPI + 0.5 U/μL Protector RNAse Inhibitor (Roche) was added. Cells were spun again, gently resuspended 5× in 0.1× lysis buffer, and incubated 2 min on ice. After incubation, 1 mL of wash buffer was added and cells were immediately spun at 500 × g for 5 min at 4°C. Nuclei were resuspended in <100 μL of Diluted Nuclei Buffer and counted using Countess FL II aiming for ∼3,000–5,000 nuclei/μL. Transposition, barcoding, and library preparation were performed according to the 10× Genomics Chromium Next GEM Single Cell Multiome protocol CG000338 Rev E. Protocols for single nucleus multiomics in DLPFC tissue were performed as previously described.23
Joint snRNA-seq and snATAC-seq workflow
Count matrices were obtained for each time point using cellranger-arc count (v.2.0.1) then aggregated using cellranger-arc aggr. Low-quality cells were filtered on gene expression (nFeatures > 200, nFeatures < 10,000, and mitochondrial percent < 5) and chromatin accessibility (nucleosome signal < 2 and TSS enrichment > 2) metrics. Peaks that were present in fewer than 10 cells were removed from the ATAC matrix. RNA counts were normalized with SCTransform53 with mitochondrial percent per cell regressed out. Principal component analysis (PCA) was performed on RNA, and UMAP was run on the first 30 principal components (PCs). The optimal number of PCs was determined to be 30 PCs using an elbow plot. The ATAC counts were normalized with term-frequency inverse-document-frequency (TFIDF). Dimension reduction was performed with singular value decomposition (SVD) of the normalized ATAC matrix. The ATAC UMAP was created using the 2nd through the 50th LSI components. The weighted nearest neighbor (WNN) graph was determined with Seurat’s54,55 FindMultiModalNeighbors to represent a weighted combination of both modalities. The first 30 dimensions of the RNA PCA and the 2nd through the 50th dimensions from the ATAC LSI were used to create the graph. The WNN UMAP was created using the wknn (k = 20). Clusters were identified from the wknn graph, and markers for each cluster were determined using a Wilcoxon rank-sum test. Clusters with marker genes that were enriched for ribosomal genes were filtered from the data (Tables S1–S3). After filtering low-quality clusters, all normalization and dimension reduction were repeated as previously described.
Differential expression from snRNA-seq
Differentially expressed genes were determined for one cluster versus all other clusters using a Wilcoxon Rank-Sum test. Only genes that were expressed in at least 10% of cells in a cluster were tested. Genes with a Bonferroni adjusted p value < 0.01 were considered to be significant. Cluster markers were found for initial clusters to define and filter low-quality clusters. Cluster DEGs were re-called with the same method after filtering.
Gene set enrichment
The R package enrichR56,57,58 was used for gene set enrichment analyses. Sets of positive DEGs for each cluster were used as input to look for enrichment in GO Biological Process 2021, GO Molecular Function 2021, GO Cellular Component 2021, and KEGG 2021 databases. Terms with an adjusted p value less than 0.05 were considered to be enriched.
Feature linkage analysis
Feature linkages were called using cellranger-arc. The maximum interaction distance was restricted to 1 Mb, and feature linkages with an absolute correlation score < 0.2 were removed and not used for downstream analysis. For feature linkage calculation, ATAC and GEX counts were normalized independently using depth-adaptive negative binomial normalization. To account for sparsity in the data, the normalized counts were smoothed by taking the weighted sum of the 30 closest neighbors from the KNN graph. The cell weights are determined by using a Guassian kernel transformation of the euclidean distance. Feature linkage scores were calculated by taking the Pearson correlation between the smoothed counts, while the significance of the correlation was determined using the Hotspot algorithm.59 For cultured multiomics, the ATAC peaks were called jointly across all time points. Cells from all time points were used to call links. For links called from AD and control tissue, ATAC peaks were called for each cell type and the union of these peaks was used to call links. Cells from all cell types were used in the feature linkage calculation.
Chromatin conformation assays
HiC was performed in iCell GlutaNeurons, and Capture-C was performed in NPCs, Day 14 BrainPhys differentiated neurons, iCell GlutaNeurons and GABANeurons. All experiments were performed in triplicate with independent cell grow-ups of approximately 15 million cells for biological replication. For HiC, we followed manufacturer recommendations from the Arima HiC kit User Guide for Mammalian cell lines (A510008, V.Oct.2019) using the Arima HiC Library Prep with Swift Biosciences Accel-NGS 2S plus DNA library kit (A510008, V. Nov2018). Libraries were sequenced using an Illumina Nova Seq S4 with XP kit for 2.5 billion reads total, which is ∼833 million reads per replicate for HiC.
Capture-C was performed in both iCell GlutaNeurons and GABANeurons following the HiC protocols, with the addition of the Agilent Technology SureSelect XT HS/SureSelect XT Low input target enrichment with pre-capture pooling protocol. 102 SureSelect DNA probes spanning a total of 2,073 bp (chr17:45,892,780–45,893,184 and chr17:45,893,527-45,895,196; hg38) were ordered from AgilentTechnologies. The region between the two probe sets could not be synthesized because of low sequence complexity. Libraries were sequenced using an Illumina Nova Seq S4 with XP kit with ∼417 million reads per replicate.
NPC and Day 14 BrainPhys differentiated neuron Capture-C was performed using both the BC1 and XCL4 NPC lines with matched differentiated neurons. Capture-C was performed following the NG Capture-C Protocol (v.2.4).60 For this experiment, enrichment for MAPT promoter contacts was performed by double capture using two biotinylated oligonucleotides targeting chr17:45,892,836–45,892,923 and chr17:45,895,092–45,895,179 (both hg38). Libraries were sequenced using an Illumina NovaSeq S4 with XP kit yielding ∼208 million reads per replicate.
HiC and Capture-C analysis
HiC data were analyzed using the Juicer61 pipeline (v.1.6) with Juicer Tools (v.1.22.01). Capture-C data were analyzed using the Juicer Tools pipeline (v.1.21). Libraries from each replicate were first run individually. Restriction site positions were generated with generate_site_positions.py with Arima specified as the enzyme. Reads were aligned to hg38 for HiC, but only to chr17 of hg38 for Capture-C. For both HiC and Capture-C, all three replicates were merged to create a combined hic file with mega.sh. Loops were called on the Knight-Ruiz normalized combined hic files using HiCCUPs at a resolution of 5 kb. Window width and peak width were set to 20 and 10, respectively. Loops with a false discovery rate (FDR) < 0.2 were determined to be significant. Resulting loops were then filtered to a maximum interaction distance of 1 Mb.
Differential Capture-C analysis
Knight-Ruiz normalized counts were pulled from the .hic file for each replicate for Gluta and GABA Capture-C at a resolution of 5 kb and merged by contact positions. Counts that were not connected to the MAPT promoter were removed from the count matrix. Differential regions were tested using DESeq262 for 5 kb bins. Neuron and NPC Capture-C were processed using capC-MAP63 with the target set at the MAPT Promoter (chr17:45,892,837–45,895,177) and the restriction enzyme set as DpnII. Contact counts were taken from capC-MAP output at a step size of 500 bp and window size of 1 kb. Differential regions were tested using DESeq2 with cell line as a covariate. For both analyses, significant regions were defined as those with an adjusted p value < 0.01. DESeq2 results were formatted as a bigwig for plotting using the directional p value: −log10(adjusted p value) ∗ sign(log2(FC)).
ATAC-seq protocol
ATAC-seq was performed in the KOLF2.1J-hNGN2 cell line after 14 days of neuronal differentiation with three biological replicates per cell line. This protocol is similar to those previously published by Buenrostro et al.64,65 Briefly, we harvested 100,000 cells by mechanical dissociation and washed with 50 μL cold 1× PBS and centrifuged at 300 × g for 5 min. The cell pellet was then resuspended in 50 μL cold lysis buffer (10 mM Tris-HCl [pH 7.5], 10 mM NaCl, 3 mM MgCl2, 0.1% NP-40, 0.1% Tween 20, 0.01% Digitonin, and nuclease-free H2O) and incubated on ice for 3 min. After incubation, 1 mL of wash buffer (10 mM Tris-HCl [pH 7.5], 10 mM NaCl, 3 mM MgCl2, 0.1% Tween 20, and nuclease-free H2O) was added, samples were inverted gently to mix, and centrifuged at 500 × g for 10 min at 4°C. The supernatant was discarded and the remaining nuclei were resuspended in 50 μL Transposition Mix (2× TD Buffer, TDE1, and nuclease-free H2O) and incubated at 37°C on shaker at 1,000 rpm for 30 min. Samples were immediately purified using the Qiagen MinElute Reaction Cleanup kit and eluted in a final volume of 11 μL. Recovered DNA was then used to generate sequencing libraries using primers from the Nextera XT Index Kit (15055293) and Q5 Hot Start Master Mix and amplified (30 s at 98°C [10 s at 98°C, 30 s at 63°C, and 72°C for 1 min] × 10 cycles). Libraries were quantified with Qubit dsDNA HS Assay kit and visualized with BioAnalyzer High Sensitivity DNA Analysis kit (Agilent 5067-4626) and 2100 BioAnalyzer Instrument (Agilent). Libraries were sequenced using Illumina NovaSeq flow cell with 50-bp paired-end runs. Reads were processed using the standard ENCODE ATAC-seq pipeline (v.1.7.0, https://github.com/ENCODE-DCC/atac-seq-pipeline).
ChIP-seq protocol
ChIP-seq for H3K27Ac and CTCF were performed using chromatin from neurons differentiated from KOF2.1J NPCs (day 14) with biological replicates. Protocols for ChIP-seq are consistent with techniques previously described by our lab and available from the ENCODE Consortium (https://www.encodeproject.org/documents/73c95206-fc02-41ea-93e0-a929a6939aaf/).66,67,68 Antibodies targeting H3K27Ac (ActiveMotif, Cat: 39133) or CTCF (ActiveMotif, Cat: 61311) were used. Libraries were prepared by blunting and ligating ChIP DNA fragments to sequencing adapters for amplification with barcoded primers (30 s at 98°C [10 s at 98°C, 30 s at 65°C, 30 s at 72°C] × 15 cycles; 5 min at 72°C). Libraries were quantified with Qubit dsDNA HS Assay kit and visualized with Standard Sensitivity NGS Fragment Analysis Kit (Advanced Analytical DNF-473) and Fragment Analyzer 5200 (Agilent). Libraries were sequenced using Illumina NovaSeq flow cell with 100 bp single-end runs.
ChIP-seq analysis
Prior to analysis, reads were processed to remove optical duplicates with clumpify (BBMap v.38.20; https://sourceforge.net/projects/bbmap/) [dedupe = t, optical = t, dupedist = 2500] and remove adapter reads with Cutadapt (v.1.16) [-a AGATCGGAAGAGC -m 40].58 Input reads were capped at 40 million using Seqtk (v.1.2; https://github.com/lh3/seqtk). Individual experiments were constructed following ENCODE guidelines (https://www.encodeproject.org/about/experiment-guidelines/) and analyzed with the chip-seq-pipeline2 processing pipeline (https://github.com/ENCODE-DCC/chip-seq-pipeline2). Final peaks were called using pseudoreps and the IDR naive overlapping method with a threshold of 0.05.
Plasmids
The pNL1.1.CMV [Nluc/CMV] and pGL4.23 [luc2/minP] vectors were obtained from Promega. Luciferase elements were generated by selecting 467 bp of the nominated region and both the forward and reverse complement sequences were ordered as gBlocks from Integrated DNA Technologies (IDT). Elements were cloned into the pGL4.23 [luc2/minP] vector digested with EcoRV by Gibson Assembly. Element insertion was confirmed by Sanger sequencing (MCLAB). Each element was individually prepped 3 times for a total of 6 individual plasmid preparations per nominated region. The pMD2.G, psPAX2, FUGW-H1-GFP-neomycin, and pLV hU6-sgRNA hUbC-dCas9-KRAB-T2A-Puro plasmids were obtained from Addgene (#12259, #12260, #37632, and #71236, respectively). sgRNA sequences were designed using https://benchling.com and ordered as premixed primer pools from IDT (Table S4). The sgRNA sequences were then inserted into the pLV hU6-sgRNA hUbC-dCas9-KRAB-T2A-Puro plasmid by digesting with Esp3I and subsequent ligation.69,70
Lentivirus production
293FT cells were plated at 70,000 cells/cm2 in poly-L-ornithine coated 6-well culture plates. The next day, the media was renewed with OptiMEM Reduced Serum Media supplemented with 300 mg D-glucose. Cells were transfected with 1 μg pLV hU6-sgRNA hUbC-dCas9-KRAB-T2A-Puro plasmid with inserted sgRNA sequence using Lipofectamine LTX with Plus Reagent, following manufacturer recommendations. 48 h after transfection, supernatant was harvested and filtered through a 0.45 μm syringe filter into a 15 mL conical tube on ice.
CRISPRi experiments
NPCs were plated at 57,000 cells/cm2 in reduced growth factor Matrigel (Corning #354230)-coated 12-well culture plates. The next day, culture media was renewed with NPC media supplemented with protamine sulfate (10 mg/mL). To transduce NPCs, 500 μL of cold, filtered lentivirus was added to the NPC media and plates were centrifuged at 2,000 rpm for 1 h. Twenty-four hours after transduction, cells were renewed with fresh NPC media with growth factors and 0.5 μg/mL puromycin for selection of successfully transduced cells. Cells were selected for 24–48 h, until the control well (transduced with FUGW-H1-GFP-neomycin) had minimal remaining living cells. NPCs were then swapped to neuronal differentiation media following the Bardy et al. protocol.52 Neurons were harvested for RNA isolation at day 14 of differentiation, following the Norgen Total RNA Purification kit (Norgen 37500) with the RNase-free DNase I kit (Norgen 25720). sgRNAs were designed upstream of the MAPT TSS in the promoter region (∼chr17:45,884,000; hg38) as a positive control. sgRNAs targeting the AAVS1 safe harbor locus were used as non-targeting controls.
RT-qPCR
MAPT expression was also determined by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). cDNA was generated from 100 ng RNA using SuperScript IV Master Mix (Thermo 11756050). Two Taqman probes were used to measure MAPT relative abundance (Thermo Hs00213491_m1 and Hs00902194_m1). Taqman probes to AP1G1 (Thermo Hs00964419_m1) and GAPDH (Thermo Hs99999905_m1) were used as housekeeping controls (AP1G1 was determined in pilot RNA-seq experiments to be a well-expressed, low-variability transcript between both NPCs and neurons). For each sample, ΔΔCt values were calculated using the average of the median housekeeper Ct values. Samples with average housekeeper Ct values ±1 standard deviation from the average of the other samples in each run were removed.
3′mRNA-seq library generation
RNA isolation was performed using the Norgen Total RNA isolation and RNase-Free DNase I kits (Norgen 17250 and 25720, respectively) and quantified using the Qubit RNA HS Assay Kit (Thermo Q32855). Libraries were prepared using the QuantSeq 3′ mRNA-Seq Library Prep Kit FWD for Illumina and UMI Second Strand Synthesis Module for QuantSeq FWD (Illumina, Read 1) from Lexogen (015.96 and 081.96, respectively). Libraries were quantified using the Qubit DNA HS Assay Kit (Thermo Q32854) and visualized with the BioAnalyzer High Sensitivity DNA Analysis kit (Agilent 5067-4626) and 2100 BioAnalyzer Instrument (Agilent). Libraries were sequenced by HudsonAlpha Discovery using Illumina NovaSeq S1 100 cycle or S4 200 cycle flow cells.
RNA-seq analysis
UMIs were first extracted from the reads with UMI-tools extract with the extraction method set as regex. Reads were trimmed with bbduk then aligned to hg38 with STAR71 using the Lexogen recommended parameters for QuantSeq. Bams were deduplicated by UMI and mapping coordinate using UMI-tools72 dedup. Counts were generated with htseq-count73 using the intersection-nonempty method. The count matrix was normalized to counts per million (CPM). After normalization, each sample was scored based on the level of differentiation and astrocyte presence using Seurat’s AddModuleScore.54 Module scores were calculated using a list of markers for differentiation (CACNA1C, ENO2, MAP2) and astrocytes (AQP4, SLC1A3) (Figure S1). Differentially expressed genes (DEGs) were determined for each target region versus non-targeting controls for all genes in the MAPT locus (±1 Mb from TSS). Differential expression was assessed with a linear model with differentiation score, astrocyte score, and batch as covariates. Genes with a p value < 0.05 were determined to be significant.
Western blot
Following CRISPRi experiments, human iPSC-derived neurons were washed with PBS and harvested in 200 μL RIPA Buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 1.0% Triton X-100, 0.5% sodium deoxycholate, and 0.1% SDS) supplemented with Roche Complete Protease inhibitors (Roche # 60159800), 1 mM EDTA, and 1 mM DTT. Lysates were mixed for 30 min at 4°C in the presence of 25 U Benzonase nuclease (Sigma #E1014-25KU) to degrade genomic DNA and RNA. Following a 15-min centrifugation at 21,300 × g at 4°C, clarified lysate protein concentrations were quantified in triplicate using Bio Rad Protein Assay Dye Reagent (Bio Rad # 5000006) and BSA protein standards (Bio Rad # 5000207) by measuring absorbance at 595 nm on a Synergy H4 Hybrid Reader (BioTek). Biological quadruplicates of CRISPRi samples were analyzed by western analysis. For each western blot, 2 μg of lysate was loaded per lane on 4%–20% SDS-PAGE gels (Bio Rad # 4568096). Additionally, each gel was loaded with a 2-fold titration series of the AAVS1 safe harbor control lysate (0.125 μg–4 μg) for use in western quantification. Gels were transferred to nitrocellulose (Bio Rad # 1620115), blocked with 5% BSA in PBS +0.05% Tween 20, and probed with either the MED1 loading control antibody (Bethyl A300-793A used at 1:1,000 dilution on 100–300 kDa region of blot) or the Tau antibody (DAKO/Agilent # A0024 used at 1:5,000 dilution on the 37–100 kDa region of the blot). After washing with PBS + 0.2% Tween 20, the blots were probed with LI-COR IR Dye 800CW Secondary antibody-dye conjugate (LI-COR # 926–32213 used at 1:10,000 dilution) following the LI-COR suggested protocol. Western blots were imaged on an ODYSSEY CLx (LI-COR) and quantified using Image Studio (LI-COR) and Prism 10 (GraphPad) software packages.
Nucleofection for luciferase assays
iCell GlutaNeurons and KOLF2.1J-hNGN2 differentiated neurons were plated at 200,000 cells/cm2 in 24-well matrigel or matrigel and 0.01% poly-L-ornithine pre-coated plates, respectively. Cells were differentiated according to their respective protocols for 14 days. One hour before nucleofection, media was renewed to fresh media plus 10 μM Rock Inhibitor (Y27632). Neurons were nucleofected with 10 μg total DNA plasmid DNA using the AD1 4D-Nucleofector Y kit (Lonza V4YP-1A24) with the ED158 (iCell GlutaNeurons) and EH166 (KOLF2.1J-hNGN2 neurons) pulse settings. Per nucleofection, 9 μg of the pGL4.23 luciferase reporter vector containing the test element and 1 μg pNL1.1.CMV (Nluc/CMV) were used. A transfection reaction of 9 μg pGL4.23 (luc2/minP) and 1 μg pNL1.1.CMV (Nluc/CMV) was used as a baseline control. Both vectors were also transfected as background controls (1 μg) with pmaxGFP (9 μg, Lonza). Twenty-four hours after nucleofection, cells were renewed with fresh media without Rock inhibitor. Cell lysates were harvested by freezing at −80°C 48 h after transfection.
Luciferase assays
Luciferase assays were performed using the Nano-Glo Dual-Luciferase Reporter Assay System (Promega Cat#N1630) following the manufacturer’s protocol. Cell lysis was performed on the 24-well plate and aliquoted across 4 wells of a white 96-well plate for 4 technical replicates per biological replicate. Six biological replicates were performed per nominated region. For iCell GlutaNeuron samples, the ratio of firefly to nano luminescence was first determined. For KOLF2.1J-hNGN2 neurons, only firefly luminescence was used for analysis. Firefly luminescence was normalized across the average plate luminescence and then normalized to the average control luminescence. For each biological replicate, the median fold luminescence value was determined for the four technical replicates. Four biological replicates were compared to the pGL4.23 (luc2/minP)/pNL1.1.CMV (Nluc/CMV) control using an ordinary one-way ANOVA with Fisher’s LSD.
Genetic burden analysis
Sequencing data were obtained from the 10th release of the Alzheimer Disease Sequencing Project. Data for chromosome 17 were filtered to the region of interest (hg38 chromosome 17 from 44.8 to 47.0 megabases, which is inclusive of MAPT, nearby genes, and all nominated regulatory regions) and annotated with dbSNP v.154 (https://www.ncbi.nlm.nih.gov/snp/),74 TOPMed Bravo freeze 8 (https://bravo.sph.umich.edu/freeze8/hg38/), CADD v.1.6,75 and the ENSEMBL GRCh38.99 gene model (https://ftp.ensembl.org/pub/release-99/gtf/homo_sapiens/Homo_sapiens.GRCh38.99.gtf.gz). Burden analysis was conducted on the three largest self-reported race/ethnicity groups as reported in the results to avoid confounding results from population stratification issues. In addition, we analyzed the Accelerating Medicine Partnerships – Parkinson Disease AMP-PD dataset. For all data, we used GenoTools (https://github.com/dvitale199/GenoTools)76 to calculate genetic ancestry superpopulations for inclusion as covariates as well as the first 10 PCs which were also included as covariates along with sex, age, and H1/H2 haplotype status (both ADSP and AMP-PD) and also sample tissue, sequencer type, and sequencing center for ADSP (not available for AMP-PD). PASS filter variants meeting criteria described in the results were aggregated, and individuals were counted as qualifying if they harbored 1 or more qualifying variants based on the filter conditions in the results (thus, if an individual harbored multiple qualifying variants, they were only counted once, though most individuals harbored only one qualifying variant). Differential analysis was conducted using SKAT (sequence kernel association test) comparing the number of affected individuals and control subjects with and without a qualifying variant.
Results
Identification of candidate CREs by single nucleus multiomics
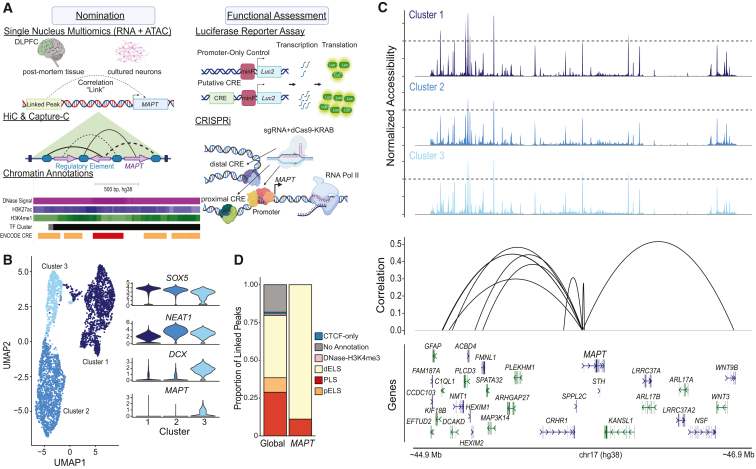
To identify regions that may regulate MAPT expression, we used orthogonal approaches to nominate candidate genomic regions interacting with the MAPT promoter (Figure 1A). First, we performed single-nucleus multiomics (snRNA-seq and snATAC-seq) using the 10x Genomics Multiome technology on nuclei isolated from neural progenitor cells (NPCs) and neurons differentiated from these NPCs for 14 and 21 days.52 This differentiation protocol produces a mixture of inhibitory and excitatory neurons and immature glial cells. Using this multiomics assay allows for direct mapping of gene expression and chromatin accessibility within the same nuclei without the need to computationally infer cell type identities prior to cross-modality integration. We removed low-quality nuclei and doublets (material and methods), and we retained a total of 3,881 nuclei with an average of 1,293 per time point (range of 856–1,601). We detected a median of 3,559 genes and 55,996 ATAC fragments per cell. We performed normalization and dimensionality reduction for snRNA-seq and snATAC-seq data using Seurat (v.4) and Signac (v.5), respectively. We used weighted-nearest neighbor (WNN) analysis to determine a joint representation of expression and accessibility and identified 3 distinct clusters (Figures 1B and S2A–S2D). Cluster 1 represents a mostly (98.1%) NPC cluster and is defined by expression of SOX5, which regulates embryonic development and cell fate determination.77 Cluster 2 is a mixture of neurons differentiated for 14 (43.8%) and 21 (56.2%) days and is defined by expression of NEAT1, which in iPSC-derived neurons has been shown to directly regulate neuronal excitability.78 Cluster 3 is also a mixture of time points of differentiated neurons (19.3% day 14 and 80.7% day 21) and is defined by expression of DCX, which encodes doublecortin that is expressed in migrating neurons through the central and peripheral nervous system during embryonic and postnatal development, representing an immature neuron population.79 Cluster 3 also is the only cluster that expresses MAPT. This clustering was expected given the resulting mixed culture produced from this differentiation protocol. Differentially expressed genes (DEGs) were identified for each cluster by comparing to the other two remaining clusters (Table S3). A total of 792 DEGs were identified, including MAPT (log2(Fold Change) = 0.75; adjusted p = 5.39 × 10−124).
Figure 1.
Nomination of candidate CREs by snRNA- + snATAC-seq in iPSC-derived neuronal cultures
(A) Experimental design.
(B) Left: UMAP visualization of the weighted nearest neighbor (WNN) of single nuclei colored by cluster assignment. Right: violin plot of expression of key genes by cluster assignment.
(C) Linkage plot for all links to MAPT. Top panels: coverage plot of normalized accessibility (range 0–8,900) separated by cluster assignment (light blue, NPC; blue, intermediate differentiation; dark blue, immature neurons). Bottom panel: significant peak-gene links. Arc height represents the strength of correlation shown as absolute correlation values.
(D) Comparison of proportion of linked peaks by ENCODE regulatory element annotation of global and MAPT links (dELS, distal enhancer-like sequence; PLS, promoter-like sequence; pELS, proximal enhancer-like sequence).
Using the cellranger-arc (v.2.0) analysis pipeline, we measured correlations, or “links,” between gene expression and chromatin accessibility to nominate CREs active in differentiated neurons. A “linked peak” is an ATAC-seq peak whose accessibility across all nuclei is significantly correlated with the expression of a “linked gene” (Figure 1A, top panel). We restricted this correlation analysis to consider only peaks within 1 megabase (Mb) of each transcription start site (TSS), given previous studies that the vast majority of distal regulatory elements are less than 1 Mb from their target genes (see material and methods).80,81,82,83,84,85,86,87,88 A total of 54,879 links were found, including 9 MAPT-specific links (Figure 1C; Tables 1, S5, and S6). We furthermore overlapped the linked peaks with ENCODE-curated CREs.89 The linked peaks were enriched for promoter-like sequences (odds ratio [OR] = 6.11; p < 2.2 × 10−16) and proximal enhancer-like sequences (OR = 1.59; p < 2.2 × 10−16), with the majority (8 of 9, 88.9%) of MAPT-linked peaks overlapping distal enhancer-like sequences (Figure 1D). ATAC peaks can be linked to one or more genes; we identified a median of 1 link per peak globally (range 1–31) and 2 links per peak for MAPT linked peaks (range 1–3; Figure S2E). Additionally, the mean peak distance from gene TSS was ∼440 kb globally, but for MAPT-linked peaks, the peaks tended to be farther from the TSS, with an average distance of ∼670 kb (Figure S2F).
Table 1.
Total number of MAPT nominated cis-regulatory elements by nomination method
| Nomination Method | Total # | # Overlapping |
|---|---|---|
| snMultiomics (DLPFC tissue) | 58 | 9 |
| snMultiomics (cultured neurons) | 9 | 2 |
| GABANeuron Capture-C | 6 | 4 |
| GlutNeuron Capture-C | 15 | 6 |
| GlutaNeuron HiC | 2 | 1 |
| Other | 22 | 0 |
| Total # of unique tested regions | 97 | |
For each nomination method, the total number of nominated regions and number of regions that were also nominated by another method are shown. The total number of unique regions tested across methods is shown at the bottom.
In addition to the single-nucleus multiomics data generated from cultured neurons for this study, we also identified MAPT links from our previously published data.23 Using nuclei from DLPFC of AD 7 and 8 control donors, we identified cell type- and disease-specific CREs and their target genes. Focusing on the MAPT locus, we expanded the original restriction of 500 kb from each gene’s TSS in our previous study to 1 Mb to match the current analysis. We identified 58 MAPT links; 28 of these links were called in both AD and control subjects (common), while there were 26 AD-specific and 4 control-specific links (Figure S3A; Tables S7 and S8). We overlapped the MAPT-linked peaks with ENCODE CREs and found that they were significantly enriched for both proximal (OR = 4.50; p = 1.58 × 10−13) and distal (OR = 1.98; p = 0.011) enhancer-like sequences and promoter-like sequences (OR = 4.51; p = 1.58 × 10−13; Figure S3B).89 MAPT-linked peaks had a median of 7.5 (range 1–15) links, while globally the median links per peak was only 3 (range 1–31; Figure S3C). MAPT links also tended to be farther away from the TSS in this dataset with an average peak distance of ∼500 kb compared to ∼440 kb globally (Figure S3D).
We found that many of the DLPFC-linked peaks were negatively correlated with MAPT expression. As with the published analysis method, links were called across all cells in the dataset. These negative correlations are likely driven in part by high accessibility of cCREs in cell types where MAPT is not expressed and the low variability of MAPT expression in neurons (example shown in Figure S4A). We also hypothesized that the number of negative correlations could be due to the large inversion of the MAPT locus in the H2 haplotype compared to H1 haplotype individuals. Therefore, we recalled links separately for H1/H1 (n = 8), H1/H2 (n = 5), and H2/H2 (n = 2) individuals (Figure S4B). While we observed fewer links called due to smaller sample sizes, we did not observe any differences in the direction of correlation at linked peaks, indicating that haplotype status was not affecting this trend. Since the majority of the negatively correlated links are very distal to the MAPT promoter (≥300 kb) and could be considered false positives, we performed permutation testing to evaluate the association between link distance and false positive rate (FPR).23 We found that on average links further from the TSS occur in fewer permutations (Kruskal-Wallis, p < 2.2 × 10−16); however, many of the distal links occur in 100% of the permutation tests, indicating a high likelihood of these links being true positives (Figure S4C). Therefore, we included all correlated regions in the validation set to comprehensively evaluate all potential links but excluded these negative correlation links from the genetic variant burden analysis (following) under the conservative assumption that these links may have artifactual properties.
Candidate CRE identification by structural analysis and chromatin marks
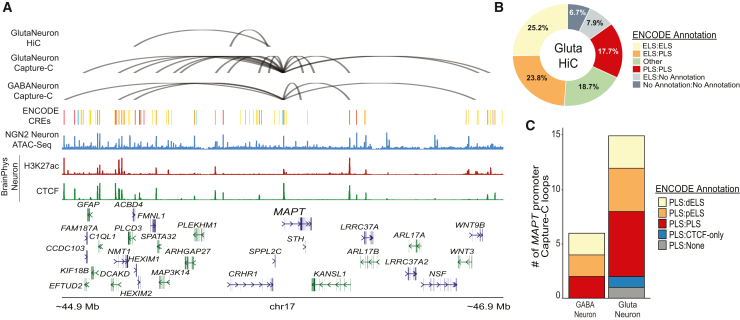
Enhancers can exert their function by being in physical contact with the target gene’s promoter, despite large intervening sequencing distances, via chromatin looping.34,35,36,88 We sought to identify looping events involving the MAPT promoter using HiC and Capture-C assays. We performed HiC in iCell GlutaNeurons, which produce a ≥90% pure population of human glutamatergic (excitatory) neurons. Using the Juicer Tools pipeline (v.1.21) and HiCCUPs, we identified 15,918 loops genome-wide with 2 of these loops contacting the MAPT promoter region (chr17:45,830,001–45,835,000 [hg38]; Figure 2A; Tables 1 and S9). When overlapping loop ends with ENCODE annotations,89 about half are interactions with enhancer-like sequences (enhancer:enhancer 25.2% and enhancer:promoter 23.8%) and 17.7% are promoter:promoter interactions (Figure 2B). Both loops interacting with the MAPT promoter are regions annotated as enhancer-like sequences.
Figure 2.
Nomination of candidate CREs using HiC and Capture-C in pure excitatory and inhibitory neuron cultures
(A) Loop plot of HiCCUPs nominated loops to the MAPT promoter. Top panel: nominated loops by dataset. Top: HiC loops to the MAPT promoter from iCell GlutaNeuron cultures. Middle: Capture-C loops to the MAPT promoter from iCell GlutaNeuron cultures. Bottom: Capture-C loops to the MAPT promoter from iCell GABANeuron cultures. Bottom panel, top: track of ENCODE-annotated candidate CREs. Middle: ATAC-seq peaks from KOLF2.1J-hNGN2 differentiated neurons. Bottom: H3K27ac and CTCF ChIP-seq peaks from day 14 BrainPhys differentiated neurons.
(B) Proportion of HiC global loop calls by ENCODE annotation.
(C) Proportion of Capture-C MAPT promoter loops by ENCODE annotation for both iCell GlutaNeurons and GABANeurons.
To more sensitively detect regions specifically interacting with the MAPT promoter, we used Agilent Technologies SureSelect DNA probes spanning a 2 kb region (chr17:45,892,780–45,893,184 and chr17:45,893527–45,895,196; hg38) to perform Capture-C in iCell GlutaNeurons and iCell GABANeurons (≥95% pure population of GABAergic (inhibitory) neurons) cultures. Loops were called on the Knight-Ruiz normalized combined .hic file using HiCCUPs at a resolution of 5 kb with the MAPT promoter region defined as chr17:45,890,001–45,895,000. Resulting loops were then filtered to a maximum interaction distance of 1 Mb, matching the chosen restriction space for single-nucleus multiomics nominations. We identified 15 and 6 MAPT promoter contact loops in GlutaNeuron and GABANeurons, respectively (Figure 2A; Tables 1, S10, and S11). About half of these loops are regions annotated as either proximal or distal enhancer-like sequences with the majority of the remaining loops classified as promoter-like sequences (Figure 2C).
We also nominated candidate CREs by manual inspection using the UCSC genome browser (http://genome.ucsc.edu) incorporating ENCODE chromatin annotations (DNase Hypersensitivity, H3K27ac, and H3K4me1),90 transcription factor clusters (ENCODE),90,91 conservation (Vertebrate Multiz Alignment & Conservation [100 Species]), and previously published data.50,51 Limiting the search area to the 1 Mb upstream or downstream of the MAPT promoter, we nominated an additional 22 regions not overlapping any of the experimental nomination methods (“Other” Method Tables 1 and S12). Of these nominated regions, most (56%) overlap distal enhancer-like sequences, but many are not annotated by ENCODE. However, since ENCODE annotations are defined by non-neuronal cell types, these nominated regions may be CREs specific to neuronal cell types (Figure S5A). To provide supporting evidence in neuronal cell types, we additionally overlapped all nominated regions with bulk and NeuN+ ChIP-seq of 103 TFs in postmortem DLPFC, occipital lobe (OL), and frontal pole (FP) tissue and NeuN+ ATAC-seq from postmortem entorhinal cortex (EC) tissue.92,93 Only 21 regions overlapped an ATAC-seq peak, but most (77.7%) overlapped a ChIP-seq peak for at least one TF (Figure S5B).
Differential analysis of CREs nominated by structural analysis
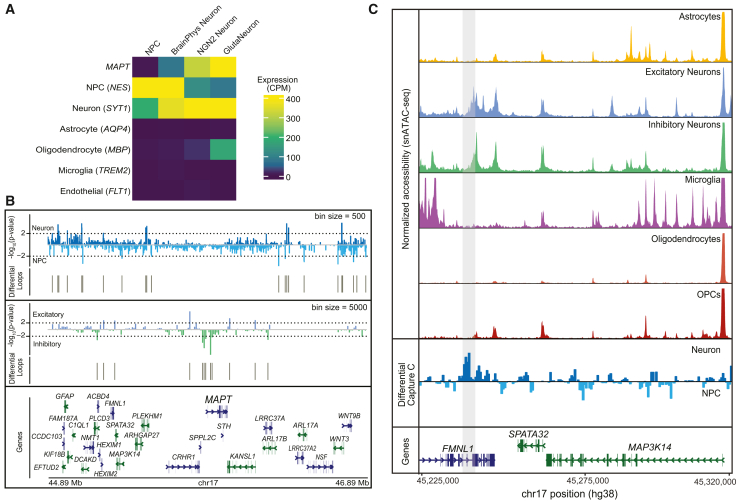
In the chosen culture system, MAPT is very lowly expressed at the NPC stage but is more highly expressed in the differentiated neuron state (Figure 3A). This expression switch makes this model very useful in examining MAPT regulation. To interrogate cCREs controlling this biological switch in MAPT expression, we performed differential analysis of Capture-C data generated in NPCs and differentiated neurons. For differential Capture-C, we chose bins of 500 bp, and we identified 49 regions differentially interacting with the MAPT promoter between NPC and neurons (Table S13). Of these interacting regions, 27 were specific to neurons (Figure 3B). One of these neuron-specific regions falls within a single-nucleus multiomics link located 933,730 bp upstream of MAPT in the 3′ UTR of C1QL1. We also performed differential analysis of the interacting regions identified in excitatory and inhibitory neurons and found that 12 regions differentially interacted with the MAPT promoter (Figure 3B; Table S14). Of these differential regions, nine overlapped the nominated MAPT regulatory regions with five being nominated from single nucleus links. For the remaining overlapping regions, three were previously identified in the excitatory neuron Capture-C and one was identified in inhibitory neuron Capture-C. One neuron-specific interacting region identified was located >650 kb upstream of the MAPT promoter in the FMNL1 gene body (Figure 3C). We evaluated chromatin accessibility within this locus using snATAC-seq data generated previously.23 We found that this region was specifically accessible in both excitatory and inhibitory neurons from adult DLPFC tissue, providing further evidence that this region likely functions as a neuron-specific regulatory element. In agreement with our findings, a previous study also found that this region harbored putative neuron-specific regulatory elements.51
Figure 3.
Identification of neuron-specific candidate regulatory regions
(A) Heatmap showing expression of MAPT and cell type marker genes in NPCs, day 14 BrainPhys differentiated neurons (BrainPhys Neurons), KOLF2.1J-hNGN2 Neurons (NGN2 Neuron), and iCell GlutaNeurons.
(B) Differential Capture-C directional p value track. Top panel: comparison of NPCs and day 14 BrainPhys neurons. Middle panel: comparison of iCell GlutaNeurons (excitatory) and iCell GABANeurons (inhibitory).
(C) Browser track view of chr17:45,225,000–45,320,000. Top panels: normalized chromatin accessibility (range 0–700) across cell types generated by snATAC-seq in DLPFC nuclei. Bottom panels: differential capture-C comparing BrainPhys differentiated neurons and NPCs with the neuron-specific region highlighted in gray.
Functional assessment of candidate CREs
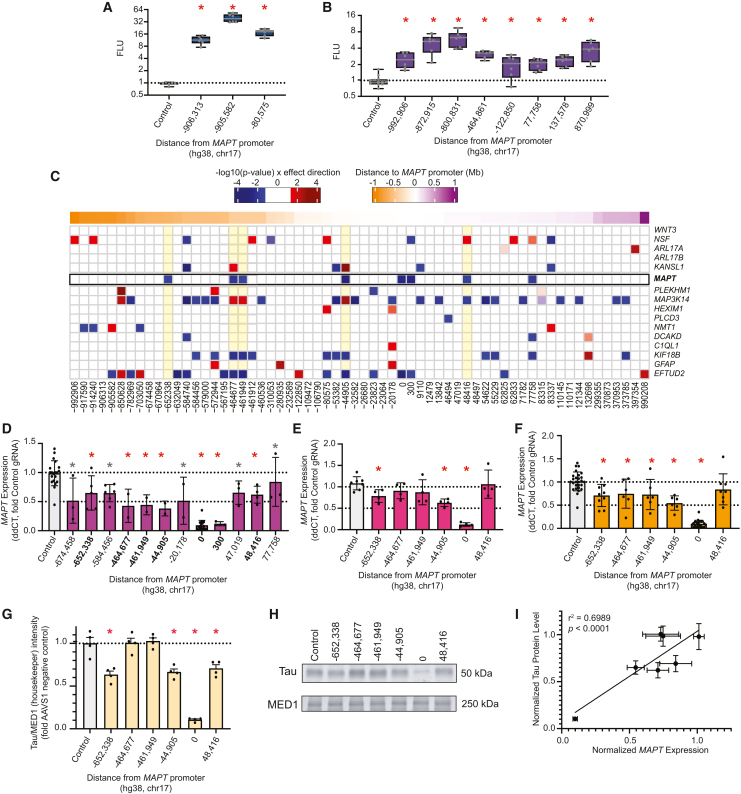
We sought to functionally assess the 97 nominated regions using human neurons differentiated from NPCs (BrainPhys), glutamatergic neurons generated from KOLF2.1J-hNGN2 iPSCs, and iCell GlutaNeurons. All of these models highly express the neuronal marker gene SYT194 and MAPT (Figure 3A). We first tested 39 of the nominated regions for enhancer-like activity by testing for sufficiency to induce transcriptional activity using a luciferase reporter assay (Table S15). We performed reporter assays in pure cultures of human iPSC-derived glutamatergic (excitatory) neurons (iCell GlutaNeurons and KOLF2.1J-hNGN2).95,96 We selected regions with high levels of conservation, high DNase hypersensitivity signal, histone modifications characteristic of active elements (H3K27ac and H3K4me1), and overlapping transcription factor motif clusters90 and ENCODE candidate CREs.89 We excluded regions annotated as promoter-like sequences because of the prior hypothesis that these regions would be active in a luciferase assay (as expected for a promoter sequence) and therefore would not provide useful information on the sufficiency of these sequences to induce transcription. Of the nominated regions, 21 were tested in iCell GlutaNeurons. During the course of this study, the KOLF2.1J-hNGN2 cell line was made publicly accessible and produced pure excitatory neurons.95,96 These neurons were more experimentally tractable than iCell GlutaNeurons, so the remaining 18 were tested in KOLF2.1J-hNGN2-derived neurons (Figures S6A and S6B). Eleven of the regions significantly increased activity of the luciferase reporter (Figures 4A and 4B).
Figure 4.
Functional validation of nominated CREs
(A) Boxplots showing statistically significant (∗p < 0.05, ANOVA with Fisher’s LSD) elements tested in luciferase assays in iCell GlutaNeurons.
(B) Boxplots showing statistically significant (∗p < 0.05, ANOVA with Fisher’s LSD) elements tested in luciferase assays in KOLF2.1J-hNGN2 neurons.
(C) Heatmap showing differential expression following CRISPRi experiments in day 14 XCL4 BrainPhys differentiated neurons of MAPT and all genes expressed (cpm > 30) within 1 Mb of MAPT TSS. X axis indicates the distance of the midpoint of the target region tested in CRISPRi experiments from the MAPT promoter (chr17:45,894,000).
(D) RT-qPCR of MAPT expression. Barplots show statistically significant regions tested. Red asterisk indicates regions significant by 3′ mRNA-seq and RT-qPCR. Gray asterisk indicates regions significant in either RT-qPCR or 3′ mRNA-seq. ∗p < 0.05, ANOVA with Fisher’s LSD.
(E) Replication of five key regions identified in (C) and (D) in KOLF2.1J BrainPhys differentiated neurons.
(F) Meta-analysis combining RT-qPCR of resulting neuronal MAPT expression data from both the XCL4 and KOLF2.1J BrainPhys neurons.
(G) Quantification of western blot (∗p < 0.05 by ANOVA with a Dunnett’s multiple comparisons post-hoc vs. AAVS1 safe harbor control).
(H) Western blot of Tau and MED1 (housekeeper) protein levels following CRISPRi perturbation.
(I) Correlation plot of MAPT RNA transcript abundance with tau protein level using simple linear regression analysis.
Since we tested a subset of the nominated regions for luciferase activity, we also assessed publicly accessible massively parallel reporter assay (MPRA) data to determine whether the nominated regions had enhancer-like activity. Seventeen of the nominated regions overlap significant elements from either the Cooper et al.50 MPRA or the van Arensbergen et al.97 SuRE-seq datasets. Given that these datasets are assessing cCREs across the genome and in non-neuronal cells, it is not unexpected that the overlap with our regions is only ∼18% as genome-wide assays may have lower sensitivity than targeted assays for MAPT. Therefore, we also overlapped nominations with HiChIP for H3K27ac data generated from multiple brain regions by Corces et al.51 Eighteen of the nominated regions overlapped a H3K27ac HiChIP peak (q < 0.01), including 5 regions overlapping either of the two MPRAs (Table S16). Additionally, we overlapped all nominated regions with significant (p < 5 × 10−8) SNVs from the largest AD GWASs from Bellenguez et al.25 and PD GWAS from Nalls et al.98 We found that 46 of the nominated regions overlapped a significant PD SNV and 30 regions overlapped a significant AD SNV. Importantly, the region corresponding to the element 77,758 bp away from the MAPT promoter that had significant luciferase activity in KOLF2.1J-hNGN2 neurons was also significant in both MPRAs analyzed and overlapped a H3K27ac HiChIP peak (Figure 4B). Together these pieces of evidence support that this region has enhancer-like activity and interacts with the MAPT promoter.
We used CRISPRi to determine the target gene of the nominated regulatory regions, as well as their necessity for that gene’s expression. CRISPRi employs a catalytically inactive Cas9 enzyme (dCas9) fused to a Krüppel-associated box (KRAB) domain that recruits transcriptional repressors, and it has been previously shown to be effective at reducing gene expression in neurons with minimal off-target effects.99 At least one guide RNA (gRNA) was tested per candidate region, and positive controls were designed to target a region encompassing the MAPT promoter and TSS. We also designed gRNAs to the AAVS1 safe harbor locus as negative controls where gRNAs should have no effect on gene expression (the AAVS1 safe harbor locus is on a different chromosome [19 vs. MAPT on 17] and has been extensively characterized to have limited cellular effects when edited).100,101,102 gRNAs were introduced into NPCs that were subsequently differentiated into neurons and harvested after two weeks. At this time point, differentiated neurons sufficiently express MAPT to confidently measure changes in its expression (Figure 3A). We isolated RNA and performed both quantitative reverse-transcription PCR (qRT-PCR) and 3′ mRNA-sequencing (RNA-seq) to assess gene expression changes for 62 of the nominated regions (Figures 4C, 4D, S6C, and S7A; Table S17). Some of the nominated regions could not be tested due to technical difficulties such as gRNA design, cloning failure, toxicity to cells following transduction, or not meeting quality control metrics for RNA-seq. We observed robust knockdown of MAPT expression when targeting the promoter and 300 bp downstream. We observed significant knockdown of MAPT after repressing 11 other regions, six of which were significant in either RNA-seq or RT-qPCR, but not both (Figures 4C, 4D [gray asterisks], S6C, and S7A). The remaining five regions were confirmed in both RNA-seq and RT-qPCR (Figures 4C and 4D). Overall, nomination by multiple methods did not correspond with a higher likelihood of validation (Fisher’s exact test p = 0.1014). However, permutation testing did prove to be a strong predictor of successful functional validation for single-nucleus multiomics links (F1 score = 0.857). When performing permutation testing of linked cCREs within 500 kb of the MAPT TSS, we found that the 4 dCas9-KRAB-validated regions meeting this criteria occur in ≥99% of permutation tests (Figure S7B).
For the five regions confirmed by both RNA-seq and RT-qPCR, we repeated the CRISPRi experiments in quadruplicate in a second NPC line, generated from KOLF2.1J iPSCs, and we measured MAPT expression by RT-qPCR and western blot (Figures 4E–4I). Targeting the region located in the first intron of tau (region 48,416) resulted in no difference of MAPT expression. We observed a trend of tau reduction when targeting the other four regions, with significant reduction in expression when targeting regions −652,338 (∼30% reduction, p = 0.02) and −44,905 (∼45% reduction, p = 0.001, Figure 4E). We combined these data with the XCL4 BrainPhys RT-qPCR results to gain power in detecting MAPT reduction (Figure 4F). In this analysis, only the region 48,416 bp upstream did not reach statistical significance for MAPT reduction. To determine whether the changes in RNA transcript abundance corresponded to a reduction in tau protein levels, we also performed western blot analysis (Figures 4G, 4H, and S7C). We observed a significant reduction in tau protein levels when targeting the promoter (∼90% reduction, p < 0.0001). There was little difference in protein levels when targeting regions −464,677 and −461,949 but we observed significant reduction of tau protein when targeting regions −652,338 (∼37% reduction, p < 0.0001), −44,905 (∼33% reduction, p = 0.0002), and 48,416 (∼30% reduction, p = 0.0009). Overall, tau protein levels significantly correlated with MAPT RNA transcript abundance (R2 = 0.6989, p < 0.0001; Figure 4I).
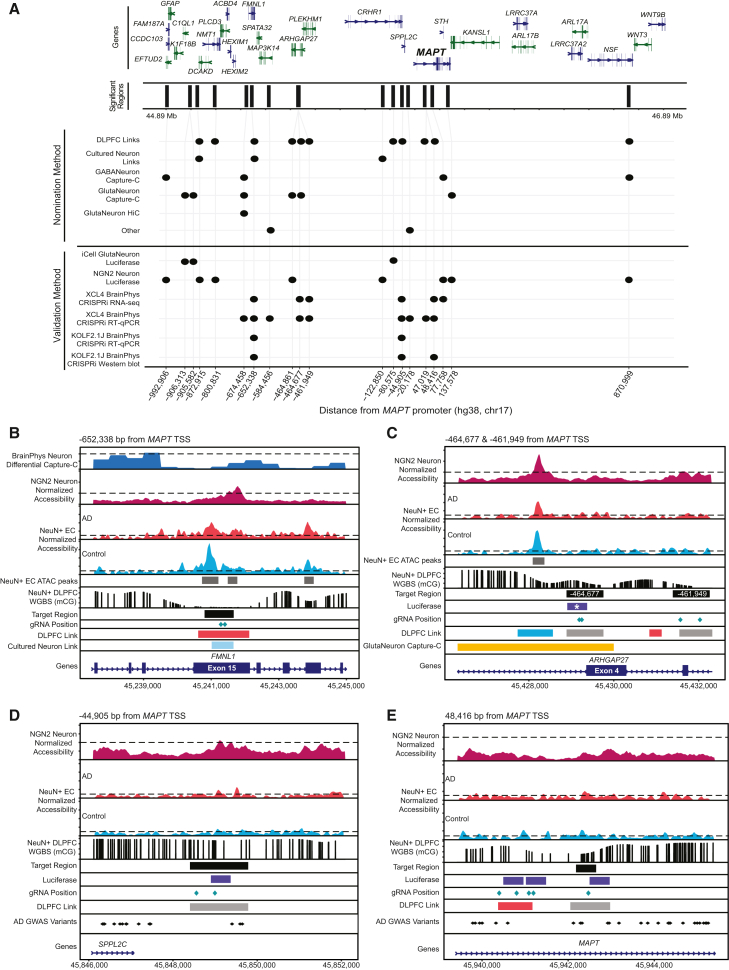
We validated regulatory regions spanning from >650 kb upstream to ∼50 kb downstream of the MAPT promoter, with three of these high-confidence regions being beyond the centromeric H1/H2 inversion breakpoint (Figure 5A). The gRNA target region located 652,338 bp upstream of the MAPT promoter resulted in significant knockdown of MAPT expression in both XCL4 (33%; p = 0.0358) and KOLF2.1J (30%; p = 0.0238) BrainPhys neurons (Figures 4D, 4E, and 5B). Importantly, MAPT was the only differentially expressed gene identified when targeting this region for repression (Figure 4C). This region was nominated in both cultured neuron and DLPFC single-nucleus multiomics datasets; it lies within exon 15 of FMNL1, although FMNL1 is not expressed in our cell lines (average cpm < 1). Two regions within the ARHGAP27 locus each resulted in more than 50% knockdown of MAPT expression in XCL4 BrainPhys neurons (−464,677, p = 0.0195; −461,949, p = 0.0233) and about a 20% reduction in KOLF2.1J BrainPhys neurons (−464,677, p = 0.1688; −461,949, p = 0.1020; Figures 4C–4F and 5C). Targeting both of these regions also significantly reduced expression of EFTUD2 (log2FC = −0.414; p = 0.004, log2FC = −0.515; p = 0.003) and KIF18B (log2FC = −1.367; p = 0.006, log2FC = −1.411; p = 0.006), while MAP3K14 (log2FC = 0.766; p = 0.011, log2FC = 1.114; p = 0.015) expression was significantly increased (Figure 4C; Table S17). ARHGAP27 is very lowly expressed in BrainPhys neurons (average cpm ∼2), so we could not evaluate any effect on its expression. These regions were both nominated by DLPFC single-nucleus multiomics, and the linked peaks identified were also linked to MAP3K14. The region 464,677 bp upstream from the MAPT TSS was additionally nominated by Capture-C and showed cell type-specific contact with the MAPT promoter in excitatory neurons. This region also had enhancer-like activity in the KOLF2.1J-hNGN2 neuron luciferase dataset (Figure 4B [region −464,861]). Together these data indicate that this region is an enhancer regulating MAPT. The closest validated region (region −44,905) lies within the first intron of MAPT-AS1 and was nominated by DLPFC single-cell multiomics with links to 11 genes including MAPT, MAPT-AS1, and KANSL1 (Figure 5D). Targeting this region resulted in significant reduction of MAPT expression in both XCL4 (60%; p = 0.0115) and KOLF2.1J BrainPhys neurons (45%; p = 0.0011), significant reduction in EFTUD2 (log2FC = −0.479; p = 0.0002) and KIF18B (log2FC = −0.932; p = 0.008), and significant increase in MAP3K14 (log2FC = 0.934; p = 0.0025) and KANSL1 (log2FC = 1.135; p = 4.74 × 10−5) expression (Figures 4C–4I). The region 48,416 downstream of the MAPT promoter was the only downstream element to validate in both RNA-seq and RT-qPCR (Figure 5E). This region lies within the first intron of MAPT, a region with established importance due to the presence of the H1c tagging variant rs242557.103 This region was nominated by single-nucleus multiomics and is linked to 10 genes, including MAPT and NSF. Targeting this region resulted in ∼37% (p = 0.0373) reduction of MAPT expression, significant reduction in MAP3K14 (log2FC = −0.82; p = 0.0169), and significant increase in NSF (log2FC = 0.232; p = 0.0351) expression (Figures 4C and 4D). While this region was not validated by RT-qPCR in KOLF2.1J BrainPhys neurons (p = 0.8727), targeting it did result in significant reduction of tau protein levels (∼30%; p = 0.009). This region was previously established as interacting with MAPT based on its overlap with a H3K27Ac HiChIP peak.51
Figure 5.
Overview of regions with sufficient evidence of CRE activity
(A) Browser view of significant nominated regions by functional assay. Top: gene model of genes within the locus (hg38, chr17: 44.89–46.89 Mb). Middle: regions with significant activity in functional assays denoted by their distance from the MAPT TSS. Bottom: upset plot showing functional assays in which cCREs had significant activity.
(B–E) Key CREs regulating MAPT expression. Zoom in on regions validated by qPCR and CRISPRi in XCL4 BrainPhys neurons (B) −652,338, (C) −464,677 and −461,949, (D) −44,905, and (E) 48,416. Top panels: differential Capture-C in BrainPhys Neurons (range 0–2), normalized accessibility from KOLF2.1J-hNGN2 differentiated Neurons (range 0–6), and normalized accessibility from NeuN+ entorhinal cortex (EC) postmortem tissue from individuals with AD (red) and healthy control subjects (blue, range 0–20). Called peaks are shown as gray bars. Bottom panels: NeuN+ DLPFC methylated CpGs from whole-genome bisulfite sequencing (WGBS, range 0–1). Tracks of target region, luciferase region (∗p < 0.05, ANOVA with Fisher’s LSD), gRNA position, significant AD GWAS variants, and nominated regions (DLPFC Links: blue, control-specific link; gray, common link; red, AD-specific link).
MAPT cCREs are depleted of rare, deleterious non-coding variants in dementia
We next asked whether the 97 cCREs we identified exhibited differential burden of rare non-coding genetic variation between neurodegeneration-affected individuals and control subjects. We first evaluated individuals in the Alzheimer Disease Sequencing Project case/control dataset. The data available to us at the time of analysis based on consent group and filtering by the consortium-determined integrated phenotypes—which accounts for relatedness, only one individual per family for family groups, sample technical problems and duplicates, etc. (https://github.com/NIAGADS/ADSPIntegratedPhenotypes)—yielded 10,668 affected individuals and 13,637 control subjects across all self-reported ancestries. We chose to constrain our analysis to the three largest self-reported race/ethnicity groups (self-reported Hispanic, non-Hispanic white, and non-Hispanic black) because of large case/control imbalance in self-reported Asian (247 affected individuals and 2,508 control subjects) and small sample size of all other combined self-reported ancestries (501 affected individuals and 186 control subjects), yielding a final set of 9,920 affected individuals and 10,943 control subjects available for analysis. We also analyzed the Accelerating Medicine Partnerships – Parkinson Disease (AMP-PD) (6,142 affected individuals and 4,066 control subjects) cohort filtered to self-reported non-Hispanic white individuals only (5,946 affected individuals and 3,939 control subjects) because they made up the vast majority of the cohort. Filtering conditions are described in Tables 2 and S18 for all candidate regulatory regions. We note that the associations reported in Table 2 meet only nominal significance, were selected from two segmentation approaches and six filtering conditions, and are not adjusted for multiple comparisons. Furthermore, they do not meet classic genome-wide speculative (1 × 10−5) or significant (5 × 10−8) thresholds. Therefore, these signals should be considered to be speculative or exploratory. Because the regions nominated are often much larger than expected for a regulatory element (e.g., 5 kb for chromatin capture-nominated regions), we applied two sets of filtering conditions to nominated elements. First, we manually curated elements by inspecting each region for regulatory marks nominated from single-nucleus multiomics, chromatin conformation, or “other” nomination methods (e.g., cCREs proximal to MAPT with strong regulatory marks but lacking multiomic and chromatin capture linkages) as described previously for design of luciferase elements and guide RNAs (Tables S19 and S20). Second, to assess how results would translate without manual curation (thus allowing broader deployment of these approaches to other genes and/or for genome-wide approaches in future studies), we applied an automated approach where we considered only multiomics and chromatin conformation links and filtered them by ENCODE cCREs v.3 (Table S21). In each case, we excluded multiomics linkages with a negative correlation with MAPT expression levels under the conservative assumption that these may be artifacts. We identified several filter conditions with a nominally significant depletion of rare and damaging variation in cCREs for MAPT in ADSP and in the subset of AMP-PD affected individuals aggregated as “other” (90% dementia with Lewy bodies), but not in Parkinson. Furthermore, we were able to detect a significant effect only on the H1/H1 background. Qualifying variants for nominally significant conditions are listed in Tables S22–S24). This effect direction is expected, as we would hypothesize that rare and damaging genetic variation in these regulatory elements would be associated with a reduction of their function, which in turn would lead to lower expression of MAPT and therefore be protective against neurodegenerative disease. Importantly, it is clear that experimental identification of regulatory regions implicated for MAPT adds information, as simply assessing the burden of genetic variation across the entire region, or filtering by ENCODE v.3 cCREs alone (Table S25) using the same allele frequency and CADD filtering conditions does not reveal a depletion of burden of qualifying genetic variants in affected individuals. Additionally, we conducted an analysis of non-coding variants in the ADSP dataset that had a TOPMed allele frequency less than 1 in 10,000 and a CADD score >20 and found that they disrupted a motif 57% of the time (64/111 variants). While the aggregate evidence for the genetic variation in these elements suggests a likely role in impacting MAPT expression levels, we note that this is not explicitly tested. It is possible that these variants could have effects on other nearby genes, especially those that have already been implicated in neurodegeneration disease risk,48 or could lack effect entirely when considered individually through an eQTL analysis.
Table 2.
Burden analysis of rare non-coding genetic variants from individuals with neurodegenerative diseases and control subjects in MAPT candidate cis-regulatory elements (cCREs) and associated controls regions
| Condition | SKAT p | OR (95% CI) | Affected w/ | Affected w/o | Control w/ | Control w/o |
|---|---|---|---|---|---|---|
| Alzheimer Disease Sequencing Project (ADSP) | ||||||
| AF 1 in 10k, CADD 20: manual curation | ||||||
| All nominated | 0.026∗ | 0.70 (0.49–0.99) | 56 | 9,864 | 88 | 10,855 |
| Overall region | 0.534 | 1.07 (0.93–1.24) | 394 | 9,526 | 407 | 10,536 |
| Nom. H1/H1 | 0.017∗ | 0.61 (0.39–0.94) | 34 | 6,375 | 61 | 6,975 |
| Nom. H1/H2 | 0.481 | 0.84 (0.44–1.60) | 19 | 3,082 | 25 | 3,422 |
| Nom. H2/H2 | 0.681 | 1.69 (0.19–20.3) | 3 | 407 | 2 | 458 |
| AF 1 in 100k, CADD 10: automated selection | ||||||
| All nominated | 0.007∗ | 0.79 (0.65–0.96) | 193 | 9,727 | 267 | 10,676 |
| ENCODE cCREs | 0.108 | 0.95 (0.86–1.05) | 846 | 9,074 | 977 | 9,966 |
| Nom. H1/H1 | 0.014∗ | 0.74 (0.58–0.95) | 120 | 6,289 | 176 | 6,860 |
| Nom. H1/H2 | 0.261 | 0.92 (0.65–1.29) | 68 | 3,033 | 82 | 3,365 |
| Nom. H2/H2 | 0.734 | 0.62 (0.16–2.08) | 5 | 405 | 9 | 451 |
| AMP-PD | ||||||
| AF 1 in 10k, CADD 10: manual curation | ||||||
| All nominated | 0.530 | 0.89 (0.74–1.06) | 303 | 5,643 | 225 | 3,714 |
| Parkinson | 0.167 | 0.94 (0.76–1.16) | 166 | 2,912 | 225 | 3,714 |
| Other (90% DLB) | 0.035∗ | 0.83 (0.66–1.03) | 137 | 2,731 | 225 | 3,714 |
Related to Table S18. Rare (allele frequency less than listed threshold in the TOPMed Bravo population database) and predicted damaging (CADD score exceeding listed threshold) variants are depleted in affected individuals vs. control subjects across implicated regulatory regions for MAPT in ADSP (Alzheimer Disease Sequencing Project), but not across the region as a whole without enrichment for cCREs nor the region as a whole solely filtered by Encyclopedia of DNA elements (ENCODE) cCREs v.3. Manual curation indicates manually set bounds applied to multiomics, chromatin conformation, and other nominated elements based on chromatin marks and ENCODE cCRE predictions. Automated selection indicates filtering of multiomic and chromatin conformation nominations only via ENCODE v.3 predicted cCREs. When stratified by H1/H2 haplotype status (Nom. HX/HX indicates all nominated elements stratified by the listed H1/H2 status), the effect is detectable only in haplotype H1/H1 homozygous individuals. In the Accelerating Medicine Partnerships – Parkinson Disease (AMP-PD) dataset, there is no detectable overall effect, but there is a nominal effect in the non-Parkinson’s (90% dementia with Lewy bodies (DLB)) sub-group. p value is indicative of a SKAT (sequence kernel association test) adjusted for sex, age, sample tissue, sequencer type, sequencing center, genetic ancestry superpopulation, 1st 10 common variation PCs, and H1/H2 haplotype status (with exception of H1/H2 stratified analyses). CADD, scaled Combined Annotation Dependent Depletion score; AF, allele frequency; OR (95% CI), odds ratio (95% confidence interval).
Discussion
Many studies have been successful in reducing tau levels by targeting either tau modulators or knocking out tau itself.13,14,15,17,104 In mice, some studies of total tau ablation noted subtle motor deficits later in life; however, these deficits were not observed with partial tau reduction, which still provides therapeutic benefit.13,105 Targeting tau through regulatory mechanisms controlling its expression may be a tractable method for reducing, instead of ablating, MAPT expression with limited off-target effects. Further, in modern genetics, understanding how non-coding regions of the genome contribute to disease has been a major area of focus. Many methods have been developed to better identify candidate regulatory regions by assessing chromatin accessibility, modifications, and structure to determine interactions contributing to gene expression. However, each of these methods has limitations that lead to both false positives and negatives. By combining orthogonal genomics methods, we identified 97 MAPT candidate cis-regulatory elements and then sought to functionally interrogate these regions to determine which regulate MAPT. We tested nominated regions using CRISPRi to determine their necessity for MAPT expression, and using 3′ RNA-seq, we determined whether these regions were necessary for the expression of any other genes within the region. A subset of our nominated regions were selected for testing in a luciferase reporter assay to determine whether these regions were sufficient for enhancer-like activity (Figure 5A).
We identified two regions in the FMNL1 locus as putative regulators of MAPT expression. We provide evidence in three independent chromatin conformation datasets that the region corresponding to 674,458 bp upstream of MAPT interacts with the MAPT promoter in excitatory and inhibitory neurons. While this region was not confirmed by RNA-seq, we observed significant reduction of MAPT expression by RT-qPCR. Importantly, we identified a regulator of MAPT in a region 20 kb downstream (region −652,338; Figure 5B) within exon 15 of FMNL1. A study by Birnbaum et al.106 demonstrated that DNA sequences could act as a protein-coding sequence in one tissue but regulate the expression of a nearby gene in another tissue. FMNL1 is not expressed in any cell type in the DLPFC snRNA-seq data (data not shown) and is detected at very low rates in the brain (gtexportal.org/home/gene/FMNL1). Additionally, this region was specifically hypomethylated compared to the rest of the FMNL1 gene body, consistent with previous evidence of hypomethylated exons having a functional role in transcriptional regulation.107,108 Therefore, this region could have dual function where it acts as a regulatory element of MAPT in tissues where the gene harboring the sequence is not expressed. This region was linked to MAPT in nuclei from AD donors, and we found that this region is ∼1.5 kb downstream of a neuron-specific Capture-C interaction (chr17:45,239,001–45,239,500) with the MAPT promoter. We confirmed using snATAC-seq from DLPFC tissue that this region was specifically accessible in excitatory and inhibitory neurons. This region was previously reported by Corces et al. to have elevated interaction with the MAPT promoter by H3K27ac HiChIP specifically in the H1 haplotype, which is associated with an increased risk for AD.51 Corces et al. also provided single-cell ATAC-seq evidence that this region harbors neuron-specific regulatory elements. While we only examined the function of this region in H1 haplotype neurons, we provide strong evidence that this region harbors an important CRE for MAPT regulation. Specific testing in H2 haplotype cell lines would be required to validate this cell type- and haplotype-specific regulation.
Within the ARHGAP27 locus, we identified two regions as high-confidence CREs of MAPT, corresponding to regions 464,677 bp and 461,949 bp upstream of MAPT (Figure 5C). Both of these regions were identified as links to MAPT in both AD and control DLPFC tissues. The region 464,677 bp upstream of MAPT had enhancer-like activity in the luciferase reporter assay and showed significant knockdown of MAPT expression in CRISPRi experiments. Together these two lines of evidence prove that this region is an enhancer that regulates MAPT expression. Interestingly, both regions also showed a significant increase in MAP3K14 expression upon targeting with CRISPRi. We also observe this inverse differential expression change when targeting a region within the first intron of MAPT-AS1 (region −44,905; Figure 5D). This region was also nominated by single-nucleus multiomics and linked to several other genes, but not to MAP3K14. Additionally, in the RNA-seq analysis, we observe that 19 regions tested significantly reduce MAP3K14 expression. MAP3K14 is located more than 500 kb upstream of MAPT near FMNL1. Several of these regions also show a trend of decreased MAPT expression that would suggest that they may be involved in MAPT regulation but not required for its expression. These observations suggest that there may be significant interaction between the MAP3K14/FMNL1 region and MAPT, but this relationship requires further investigation.
Intronic CREs have been previously shown to regulate the expression of their host gene, and recently it was reported that intronic enhancers are enriched for tissue-specific activity.109 The proportion of intronic enhancer-like sequences was shown to be enriched in the most specialized cell types, like neurons, and they were shown to regulate genes involved with cell-type-specific functions. We confirmed two intronic CREs. The closest validated region (region −44,905) lies within the first intron of MAPT-AS1 and was nominated by DLPFC single-cell multiomics with links to 11 genes including MAPT, MAPT-AS1, and KANSL1. Targeting this region resulted in significant reduction of MAPT, EFTUD2, and KIF18B, but a significant increase in MAP3K14 and KANSL1 expression (Figure 5D). We also identified a CRE within the first intron of MAPT (region 48,416; Figure 5E). This region was linked to MAPT in both AD and control DLPFC tissues. Targeting this region with CRISPR dCas9-KRAB resulted in significant knockdown of both MAPT and MAP3K14 expression, but it resulted in a significant increase in NSF expression. This intronic cCRE is particularly important because the H1c haplotype-tagging SNP rs242557 is located within this region. In our luciferase data, we tested the reference allele and did not observe a significant increase in luminescence signal. However, the alternate allele has been previously shown to increase luciferase activity when present and is associated with an increased risk of AD.103 Taken together, these results suggest that this specific CRE may have differing contributions to MAPT expression dependent on the haplotype context.
We observed a nominal depletion of rare and predicted damaging non-coding genetic variation in AD-affected individuals as well as in “other” individuals from AMP-PD (90% dementia with Lewy bodies), but not Parkinson disease, in nominated CREs. While opposite to typical variant-burden studies where one may expect enrichment of rare deleterious variants, this effect direction would be expected under a model wherein damaging variation in these elements impairs enhancer function and thereby reduces expression of MAPT, which would likely be protective against AD. The observation of depletion of rare and predicted damaging genetic variation in dementia in both nominated and confirmed CREs is particularly important evidence that supports the value of detailed identification of CREs for dementia-associated genes. We do not observe an effect in Parkinson disease despite the strong GWAS signal for Parkinson disease implicating this region, which we speculate is likely due to other genes in this region more strongly contributing to the signal observed in PD.48 Interestingly, when stratifying based on H1/H2 haplotype status, we observed a detectable effect only on the H1/H1 background. This could be simply due to reduced numbers in the analysis, though the smaller (though non-significant) effect size in the H1/H2 condition implies that these variants may indeed have greater impact on the H1 background.
This study has several limitations. First, all functional assessment experiments were performed in H1 haplotype cell lines. Therefore, regulatory regions identified here may not translate to the H2 haplotype, as the inversion of the locus may result in the loss of interaction of these regions with the MAPT promoter.51 Further study for this important aspect is an active area of research being led by a large consortium, the Tau Center Without Walls (Tau CWOW; NIH: U54NS123746). Additionally, the cell lines used for functional assessment experiments, like most iPSC models, predominantly express 3R isoforms of tau, specifically 99% for KOLF2.1J-hNGN2 neurons. These cells express predominantly 0N (98%), 2% 1N, and undetectable 2N. Two recently developed iPSC models also express 4R isoforms of tau.110,111 However, since our focus is on transcription of MAPT that does not depend on isoform composition (all are expressed from the same promoter), we assert that this important question of the potential impact of isoform splicing/ratios beyond MAPT transcription is peripheral to the main question we seek to address here, which focuses on MAPT transcription. Another limitation is that we performed CRISPRi experiments in a mixed culture of inhibitory and excitatory neurons and astrocytes by transducing cells at the NPC stage. Genetic manipulation at the NPC stage could affect neuronal differentiation; however, transduction of differentiated neurons and subsequent antibiotic selection was not technically tractable. Therefore, we relied on gene expression data from RNA-seq to determine that perturbed NPCs were successfully differentiated. Additionally, given the variability in purity of this culture system, it is possible that there are false negatives for some neuron-specific regulatory regions. Future experiments examining these putative CREs in high-throughput CRISPR screens, like Perturb-seq,112 using pure neuronal cultures might reveal more neuron-specific regulatory regions. Additionally, due to the low-throughput nature of luciferase assays, we were not able to assess all regions for enhancer-like activity. Therefore, further investigation of these regions through methods like massively parallel reporter assays (MPRAs) executed in relevant cell types such as neurons are warranted to fully understand their function. While we focused on identifying CREs, specifically enhancers, there are several other non-coding regulatory mechanisms that are not assessed here. Furthermore, the focus on CRISPRi could lead to false negatives for regulatory regions that are not required for MAPT expression, but could still be involved if activated (which could be addressed with an alternative approach, CRISPRa), or that only have measurable effects on MAPT expression in the presence of specific alleles. A final limitation is that, due to the preference for an experimental design as a screen that is as comprehensive as possible, some experiments were limited in the number of experimental replicates.
In conclusion, this study is a comprehensive evaluation of cis-regulatory elements important for expression of MAPT. Identification of these CREs not only facilitates better understanding of how genetic variants around MAPT contribute to disease risk, but also provides important foundational knowledge of regulation of an important marker gene in the central nervous system. Future studies aimed at identifying TFs bound to these regulatory regions could point to new therapeutic targets, and because we have identified neuron-specific regulatory regions, drug screens targeting TFs bound to these regions could provide therapeutic targets with both target gene and cell-type specificity. This study also lays the groundwork for high-throughput approaches for fine mapping and/or combinatorial knockdown of CREs as well as for studies beyond the assessment conducted here on the effects of genetic variation in regulatory regions that are important for MAPT expression.
Data and code availability
The raw and processed data generated are available through NCBI GEO under series accession number GSE228121. All original code generated during this study is publicly available at github.com/aanderson54/MAPT_cre. Variant data were obtained from the Alzheimer Disease Sequencing Project (ADSP) (NIAGADS accession number: NG00067) and the Accelerating Medicine Partnerships – Parkinson Disease (AMP-PD) (amp-pd.org).
Acknowledgments
We thank Jacob M. Loupe for assisting A.G.A. with ChIP-seq analysis. We thank Yann Le Guen at Stanford University for helpful discussions regarding ADSP data. We thank Caroline Pantazis, Cornelis Blauwendraat, and Pilar Alvarez Jerez at the Center for Alzheimer’s and Related Dementias for data for investigating MAPT isoform expression levels in KOLF2.1J-derived neurons.
This study was supported by BrightFocus fellowship A2019129F and NIH grants K99AG068271/5R00AG068271 awarded to J.N.C. as well as donors to the HudsonAlpha Foundation Memory and Mobility Program and Leo Fund.
Author contributions
Conceptualization, J.N.C.; formal analysis, A.G.A., B.B.R., J.N.C., L.F.R., and M.M..; investigation, B.B.R., S.N.L., S.C.R., B.S.R., M.N.D., K.T.-L., M.T.K., M.M., I.R.-N., R.M.H., J.N.C., E.A.B., and P.I.H.; data curation, A.G.A., B.B.R., and J.W.T.; writing – original draft, B.B.R., A.G.A., and J.N.C.; writing – review and editing, L.F.R., J.N.C., and R.M.M.; supervision, S.J.C., R.M.M., and J.N.C.; funding, R.M.M. and J.N.C.
Declaration of interests
The authors declare no competing interests.
Published: January 16, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2023.12.015.
Contributor Information
Richard M. Myers, Email: rmyers@hudsonalpha.org.
J. Nicholas Cochran, Email: ncochran@hudsonalpha.org.
Supplemental information
References
- 1.Chong F.P., Ng K.Y., Koh R.Y., Chye S.M. Tau Proteins and Tauopathies in Alzheimer’s Disease. Cell. Mol. Neurobiol. 2018;38:965–980. doi: 10.1007/s10571-017-0574-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coupland K.G., Kim W.S., Halliday G.M., Hallupp M., Dobson-Stone C., Kwok J.B.J. Role of the long non-coding RNA MAPT-AS1 in regulation of microtubule associated protein tau (MAPT) expression in Parkinson’s disease. PLoS One. 2016;11:e0157924. doi: 10.1371/journal.pone.0157924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hefti M.M., Farrell K., Kim S., Bowles K.R., Fowkes M.E., Raj T., Crary J.F. High-resolution temporal and regional mapping of MAPT expression and splicing in human brain development. PLoS One. 2018;13 doi: 10.1371/journal.pone.0195771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang C.-W., Shao E., Mucke L. Tau: enabler of diverse brain disorders and target of rapidly evolving therapeutic strategies. Science. 2021;371:eabb8255. doi: 10.1126/science.abb8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cherry J.D., Esnault C.D., Baucom Z.H., Tripodis Y., Huber B.R., Alvarez V.E., Stein T.D., Dickson D.W., McKee A.C. Tau isoforms are differentially expressed across the hippocampus in chronic traumatic encephalopathy and Alzheimer’s disease. Acta Neuropathol. Commun. 2021;9:86. doi: 10.1186/s40478-021-01189-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoover B.R., Reed M.N., Su J., Penrod R.D., Kotilinek L.A., Grant M.K., Pitstick R., Carlson G.A., Lanier L.M., Yuan L.-L., et al. Tau mislocalization to dendritic spines mediates synaptic dysfunction independently of neurodegeneration. Neuron. 2010;68:1067–1081. doi: 10.1016/j.neuron.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cochran J.N., Hall A.M., Roberson E.D. The dendritic hypothesis for Alzheimer’s disease pathophysiology. Brain Res. Bull. 2014;103:18–28. doi: 10.1016/j.brainresbull.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller E.C., Teravskis P.J., Dummer B.W., Zhao X., Huganir R.L., Liao D. Tau phosphorylation and tau mislocalization mediate soluble Aβ oligomer-induced AMPA glutamate receptor signaling deficits. Eur. J. Neurosci. 2014;39:1214–1224. doi: 10.1111/ejn.12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ittner L.M., Ke Y.D., Delerue F., Bi M., Gladbach A., van Eersel J., Wölfing H., Chieng B.C., Christie M.J., Napier I.A., et al. Dendritic Function of Tau Mediates Amyloid-β Toxicity in Alzheimer’s Disease Mouse Models. Cell. 2010;142:387–397. doi: 10.1016/j.cell.2010.06.036. [DOI] [PubMed] [Google Scholar]
- 10.Braak H., Alafuzoff I., Arzberger T., Kretzschmar H., Del Tredici K. Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol. 2006;112:389–404. doi: 10.1007/s00401-006-0127-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braak H., Thal D.R., Ghebremedhin E., Tredici K.D. Stages of the Pathologic Process in Alzheimer Disease: Age Categories From 1 to 100 Years. J. Neuropathol. Exp. Neurol. 2011;70:960–969. doi: 10.1097/NEN.0b013e318232a379. [DOI] [PubMed] [Google Scholar]
- 12.Zheng L., Rubinski A., Denecke J., Luan Y., Smith R., Strandberg O., Stomrud E., Ossenkoppele R., Svaldi D.O., Higgins I.A., et al. Combined Connectomics, MAPT Gene Expression, and Amyloid Deposition to Explain Regional Tau Deposition in Alzheimer Disease. Ann. Neurol. 2023 doi: 10.1002/ana.26818. [DOI] [PubMed] [Google Scholar]
- 13.Roberson E.D., Scearce-Levie K., Palop J.J., Yan F., Cheng I.H., Wu T., Gerstein H., Yu G.-Q., Mucke L. Reducing endogenous tau ameliorates amyloid beta-induced deficits in an Alzheimer’s disease mouse model. Science. 2007;316:750–754. doi: 10.1126/science.1141736. [DOI] [PubMed] [Google Scholar]
- 14.Roberson E.D., Halabisky B., Yoo J.W., Yao J., Chin J., Yan F., Wu T., Hamto P., Devidze N., Yu G.-Q., et al. Amyloid-β/Fyn-induced synaptic, network, and cognitive impairments depend on tau levels in multiple mouse models of Alzheimer’s disease. J. Neurosci. 2011;31:700–711. doi: 10.1523/JNEUROSCI.4152-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeVos S.L., Miller R.L., Schoch K.M., Holmes B.B., Kebodeaux C.S., Wegener A.J., Chen G., Shen T., Tran H., Nichols B., et al. Tau reduction prevents neuronal loss and reverses pathological tau deposition and seeding in mice with tauopathy. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aag0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wegmann S., Maury E.A., Kirk M.J., Saqran L., Roe A., Devos S.L., Nicholls S., Fan Z., Takeda S., Cagsal-Getkin O., et al. Removing endogenous tau does not prevent tau propagation yet reduces its neurotoxicity. EMBO J. 2015;34:3028–3041. doi: 10.15252/embj.201592748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeVos S.L., Corjuc B.T., Commins C., Dujardin S., Bannon R.N., Corjuc D., Moore B.D., Bennett R.E., Jorfi M., Gonzales J.A., et al. Tau reduction in the presence of amyloid-β prevents tau pathology and neuronal death in vivo. Brain. 2018;141:2194–2212. doi: 10.1093/brain/awy117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang C.-W., Evans M.D., Yu X., Yu G.-Q., Mucke L. Tau reduction affects excitatory and inhibitory neurons differently, reduces excitation/inhibition ratios, and counteracts network hypersynchrony. Cell Rep. 2021;37 doi: 10.1016/j.celrep.2021.109855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mummery C.J., Börjesson-Hanson A., Blackburn D.J., Vijverberg E.G.B., De Deyn P.P., Ducharme S., Jonsson M., Schneider A., Rinne J.O., Ludolph A.C., et al. Tau-targeting antisense oligonucleotide MAPTRx in mild Alzheimer’s disease: a phase 1b, randomized, placebo-controlled trial. Nat. Med. 2023;29:1437–1447. doi: 10.1038/s41591-023-02326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huin V., Deramecourt V., Caparros-Lefebvre D., Maurage C.-A., Duyckaerts C., Kovari E., Pasquier F., Buée-Scherrer V., Labreuche J., Behal H., et al. The MAPT gene is differentially methylated in the progressive supranuclear palsy brain. Mov. Disord. 2016;31:1883–1890. doi: 10.1002/mds.26820. [DOI] [PubMed] [Google Scholar]
- 21.Fukasawa J.T., de Labio R.W., Rasmussen L.T., de Oliveira L.C., Chen E., Villares J., Tureck G., de Arruda C Smith M., Payao S.L.M. CDK5 and MAPT Gene Expression in Alzheimer’s Disease Brain Samples. Curr. Alzheimer Res. 2018;15:182–186. doi: 10.2174/1567205014666170713160407. [DOI] [PubMed] [Google Scholar]
- 22.Jiang J., Wang C., Qi R., Fu H., Ma Q. scREAD: A Single-Cell RNA-Seq Database for Alzheimer’s Disease. iScience. 2020;23:101769. doi: 10.1016/j.isci.2020.101769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson A.G., Rogers B.B., Loupe J.M., Rodriguez-Nunez I., Roberts S.C., White L.M., Brazell J.N., Bunney W.E., Bunney B.G., Watson S.J., et al. Single nucleus multiomics identifies ZEB1 and MAFB as candidate regulators of Alzheimer’s disease-specific cis-regulatory elements. Cell Genomics. 2023;3 doi: 10.1016/j.xgen.2023.100263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Guennec K., Quenez O., Nicolas G., Wallon D., Rousseau S., Richard A.-C., Alexander J., Paschou P., Charbonnier C., Bellenguez C., et al. 17q21.31 duplication causes prominent tau-related dementia with increased MAPT expression. Mol. Psychiatry. 2017;22:1119–1125. doi: 10.1038/mp.2016.226. [DOI] [PubMed] [Google Scholar]
- 25.Bellenguez C., Küçükali F., Jansen I.E., Kleineidam L., Moreno-Grau S., Amin N., Naj A.C., Campos-Martin R., Grenier-Boley B., Andrade V., et al. New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat. Genet. 2022;54:412–436. doi: 10.1038/s41588-022-01024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wightman D.P., Jansen I.E., Savage J.E., Shadrin A.A., Bahrami S., Holland D., Rongve A., Børte S., Winsvold B.S., Drange O.K., et al. A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer’s disease. Nat. Genet. 2021;53:1276–1282. doi: 10.1038/s41588-021-00921-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunkle B.W., Grenier-Boley B., Sims R., Bis J.C., Damotte V., Naj A.C., Boland A., Vronskaya M., van der Lee S.J., Amlie-Wolf A., et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 2019;51:414–430. doi: 10.1038/s41588-019-0358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Novikova G., Kapoor M., Tcw J., Abud E.M., Efthymiou A.G., Chen S.X., Cheng H., Fullard J.F., Bendl J., Liu Y., et al. Integration of Alzheimer’s disease genetics and myeloid genomics identifies disease risk regulatory elements and genes. Nat. Commun. 2021;12:1610. doi: 10.1038/s41467-021-21823-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrews S.J., Renton A.E., Fulton-Howard B., Podlesny-Drabiniok A., Marcora E., Goate A.M. The complex genetic architecture of Alzheimer’s disease: novel insights and future directions. EBioMedicine. 2023;90 doi: 10.1016/j.ebiom.2023.104511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe K., Stringer S., Frei O., Umićević Mirkov M., de Leeuw C., Polderman T.J.C., van der Sluis S., Andreassen O.A., Neale B.M., Posthuma D. A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet. 2019;51:1339–1348. doi: 10.1038/s41588-019-0481-0. [DOI] [PubMed] [Google Scholar]
- 31.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J., et al. Systematic Localization of Common Disease-Associated Variation in Regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trynka G., Sandor C., Han B., Xu H., Stranger B.E., Liu X.S., Raychaudhuri S. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet. 2013;45:124–130. doi: 10.1038/ng.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reed K.S.M., Davis E.S., Bond M.L., Cabrera A., Thulson E., Quiroga I.Y., Cassel S., Woolery K.T., Hilton I., Won H., et al. Temporal analysis suggests a reciprocal relationship between 3D chromatin structure and transcription. Cell Rep. 2022;41:111567. doi: 10.1016/j.celrep.2022.111567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carter D., Chakalova L., Osborne C.S., Dai Y.f., Fraser P. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 36.Chakraborty S., Kopitchinski N., Zuo Z., Eraso A., Awasthi P., Chari R., Mitra A., Tobias I.C., Moorthy S.D., Dale R.K., et al. Enhancer–promoter interactions can bypass CTCF-mediated boundaries and contribute to phenotypic robustness. Nat. Genet. 2023;55:280–290. doi: 10.1038/s41588-022-01295-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whalen S., Truty R.M., Pollard K.S. Enhancer–promoter interactions are encoded by complex genomic signatures on looping chromatin. Nat. Genet. 2016;48:488–496. doi: 10.1038/ng.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caffrey T.M., Wade-Martins R. Functional MAPT haplotypes: Bridging the gap between genotype and neuropathology. Neurobiol. Dis. 2007;27:1–10. doi: 10.1016/j.nbd.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dickson D.W., Rademakers R., Hutton M.L. Progressive Supranuclear Palsy: Pathology and Genetics. Brain Pathol. 2007;17:74–82. doi: 10.1111/j.1750-3639.2007.00054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kouri N., Ross O.A., Dombroski B., Younkin C.S., Serie D.J., Soto-Ortolaza A., Baker M., Finch N.C.A., Yoon H., Kim J., et al. Genome-wide association study of corticobasal degeneration identifies risk variants shared with progressive supranuclear palsy. Nat. Commun. 2015;6:7247. doi: 10.1038/ncomms8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vandrovcova J., Anaya F., Kay V., Lees A., Hardy J., de Silva R. Disentangling the role of the tau gene locus in sporadic tauopathies. Curr. Alzheimer Res. 2010;7:726–734. doi: 10.2174/156720510793611619. [DOI] [PubMed] [Google Scholar]
- 42.Strickland S.L., Reddy J.S., Allen M., N’songo A., Burgess J.D., Corda M.M., Ballard T., Wang X., Carrasquillo M.M., Biernacka J.M., et al. MAPT haplotype–stratified GWAS reveals differential association for AD risk variants. Alzheimers Dement. 2020;16:983–1002. doi: 10.1002/alz.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nalls M.A., Pankratz N., Lill C.M., Do C.B., Hernandez D.G., Saad M., DeStefano A.L., Kara E., Bras J., Sharma M., et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 2014;46:989–993. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tauber C.V., Schwarz S.C., Rösler T.W., Arzberger T., Gentleman S., Windl O., Krumbiegel M., Reis A., Ruf V.C., Herms J., Höglinger G.U. Different MAPT haplotypes influence expression of total MAPT in postmortem brain tissue. Acta Neuropathol. Commun. 2023;11:40. doi: 10.1186/s40478-023-01534-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jun G., Ibrahim-Verbaas C.A., Vronskaya M., Lambert J.-C., Chung J., Naj A.C., Kunkle B.W., Wang L.S., Bis J.C., Bellenguez C., et al. A novel Alzheimer disease locus located near the gene encoding tau protein. Mol. Psychiatry. 2016;21:108–117. doi: 10.1038/mp.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong X., Liao Z., Gritsch D., Hadzhiev Y., Bai Y., Locascio J.J., Guennewig B., Liu G., Blauwendraat C., Wang T., et al. Enhancers active in dopamine neurons are a primary link between genetic variation and neuropsychiatric disease. Nat. Neurosci. 2018;21:1482–1492. doi: 10.1038/s41593-018-0223-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soutar M.P.M., Melandri D., O’Callaghan B., Annuario E., Monaghan A.E., Welsh N.J., D’Sa K., Guelfi S., Zhang D., Pittman A., et al. Regulation of mitophagy by the NSL complex underlies genetic risk for Parkinson’s disease at 16q11.2 and MAPT H1 loci. Brain. 2022;145:4349–4367. doi: 10.1093/brain/awac325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bowles K.R., Pugh D.A., Liu Y., Patel T., Renton A.E., Bandres-Ciga S., Gan-Or Z., Heutink P., Siitonen A., Bertelsen S., et al. 17q21.31 sub-haplotypes underlying H1-associated risk for Parkinson’s disease are associated with LRRC37A/2 expression in astrocytes. Mol. Neurodegener. 2022;17:48. doi: 10.1186/s13024-022-00551-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allen M., Kachadoorian M., Quicksall Z., Zou F., Chai H.S., Younkin C., Crook J.E., Pankratz V.S., Carrasquillo M.M., Krishnan S., et al. Association of MAPT haplotypes with Alzheimer’s disease risk and MAPT brain gene expression levels. Alzheimer's Res. Ther. 2014;6:39. doi: 10.1186/alzrt268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cooper Y.A., Teyssier N., Dräger N.M., Guo Q., Davis J.E., Sattler S.M., Yang Z., Patel A., Wu S., Kosuri S., et al. Functional regulatory variants implicate distinct transcriptional networks in dementia. Science. 2022;377:eabi8654. doi: 10.1126/science.abi8654. [DOI] [PubMed] [Google Scholar]
- 51.Corces M.R., Shcherbina A., Kundu S., Gloudemans M.J., Frésard L., Granja J.M., Louie B.H., Eulalio T., Shams S., Bagdatli S.T., et al. Single-cell epigenomic analyses implicate candidate causal variants at inherited risk loci for Alzheimer’s and Parkinson’s diseases. Nat. Genet. 2020;52:1158–1168. doi: 10.1038/s41588-020-00721-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bardy C., Van Den Hurk M., Eames T., Marchand C., Hernandez R.V., Kellogg M., Gorris M., Galet B., Palomares V., Brown J., et al. Neuronal medium that supports basic synaptic functions and activity of human neurons in vitro. Proc. Natl. Acad. Sci. USA. 2015;112:E2725–E2734. doi: 10.1073/pnas.1504393112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hao Y., Stuart T., Kowalski M.H., Choudhary S., Hoffman P., Hartman A., Srivastava A., Molla G., Madad S., Fernandez-Granda C., Satija R. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 2023:1–12. doi: 10.1038/s41587-023-01767-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stuart T., Srivastava A., Madad S., Lareau C.A., Satija R. Single-cell chromatin state analysis with Signac. Nat. Methods. 2021;18:1333–1341. doi: 10.1038/s41592-021-01282-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xie Z., Bailey A., Kuleshov M.V., Clarke D.J.B., Evangelista J.E., Jenkins S.L., Lachmann A., Wojciechowicz M.L., Kropiwnicki E., Jagodnik K.M., et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021;1:e90. doi: 10.1002/cpz1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DeTomaso D., Yosef N. Hotspot identifies informative gene modules across modalities of single-cell genomics. Cell Syst. 2021;12:446–456.e9. doi: 10.1016/j.cels.2021.04.005. [DOI] [PubMed] [Google Scholar]
- 60.Downes D.J., Smith A.L., Karpinska M.A., Velychko T., Rue-Albrecht K., Sims D., Milne T.A., Davies J.O.J., Oudelaar A.M., Hughes J.R. Capture-C: a modular and flexible approach for high-resolution chromosome conformation capture. Nat. Protoc. 2022;17:445–475. doi: 10.1038/s41596-021-00651-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Durand N.C., Shamim M.S., Machol I., Rao S.S.P., Huntley M.H., Lander E.S., Aiden E.L. Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Syst. 2016;3:95–98. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buckle A., Gilbert N., Marenduzzo D., Brackley C.A. capC-MAP: software for analysis of Capture-C data. Bioinformatics. 2019;35:4773–4775. doi: 10.1093/bioinformatics/btz480. [DOI] [PubMed] [Google Scholar]
- 64.Buenrostro J.D., Wu B., Chang H.Y., Greenleaf W.J. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr. Protoc. Mol. Biol. 2015;109:21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Partridge E.C., Chhetri S.B., Prokop J.W., Ramaker R.C., Jansen C.S., Goh S.-T., Mackiewicz M., Newberry K.M., Brandsmeier L.A., Meadows S.K., et al. Occupancy maps of 208 chromatin-associated proteins in one human cell type. Nature. 2020;583:720–728. doi: 10.1038/s41586-020-2023-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reddy T.E., Pauli F., Sprouse R.O., Neff N.F., Newberry K.M., Garabedian M.J., Myers R.M. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19:2163–2171. doi: 10.1101/gr.097022.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Johnson D.S., Mortazavi A., Myers R.M., Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 69.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G., Zhang F. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith T., Heger A., Sudbery I. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 2017;27:491–499. doi: 10.1101/gr.209601.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinforma. Oxf. Engl. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sherry S.T., Ward M.-H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rentzsch P., Schubach M., Shendure J., Kircher M. CADD-Splice—improving genome-wide variant effect prediction using deep learning-derived splice scores. Genome Med. 2021;13:31. doi: 10.1186/s13073-021-00835-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bandres-Ciga S., Faghri F., Majounie E., Koretsky M.J., Kim J., Levine K.S., Leonard H., Makarious M.B., Iwaki H., Crea P.W., et al. NeuroBooster Array: A Genome-Wide Genotyping Platform to Study Neurological Disorders Across Diverse Populations. medRxiv. 2023 doi: 10.1101/2023.11.06.23298176. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.She Z.-Y., Yang W.-X. SOX family transcription factors involved in diverse cellular events during development. Eur. J. Cell Biol. 2015;94:547–563. doi: 10.1016/j.ejcb.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 78.Barry G., Briggs J.A., Hwang D.W., Nayler S.P., Fortuna P.R.J., Jonkhout N., Dachet F., Maag J.L.V., Mestdagh P., Singh E.M., et al. The long non-coding RNA NEAT1 is responsive to neuronal activity and is associated with hyperexcitability states. Sci. Rep. 2017;7 doi: 10.1038/srep40127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gleeson J.G., Lin P.T., Flanagan L.A., Walsh C.A. Doublecortin Is a Microtubule-Associated Protein and Is Expressed Widely by Migrating Neurons. Neuron. 1999;23:257–271. doi: 10.1016/s0896-6273(00)80778-3. [DOI] [PubMed] [Google Scholar]
- 80.Snetkova V., Skok J.A. Enhancer talk. Epigenomics. 2018;10:483–498. doi: 10.2217/epi-2017-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fulco C.P., Nasser J., Jones T.R., Munson G., Bergman D.T., Subramanian V., Grossman S.R., Anyoha R., Doughty B.R., Patwardhan T.A., et al. Activity-by-contact model of enhancer–promoter regulation from thousands of CRISPR perturbations. Nat. Genet. 2019;51:1664–1669. doi: 10.1038/s41588-019-0538-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lettice L.A., Heaney S.J.H., Purdie L.A., Li L., de Beer P., Oostra B.A., Goode D., Elgar G., Hill R.E., de Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 83.Bahr C., von Paleske L., Uslu V.V., Remeseiro S., Takayama N., Ng S.W., Murison A., Langenfeld K., Petretich M., Scognamiglio R., et al. A Myc enhancer cluster regulates normal and leukaemic haematopoietic stem cell hierarchies. Nature. 2018;553:515–520. doi: 10.1038/nature25193. [DOI] [PubMed] [Google Scholar]
- 84.Jin F., Li Y., Dixon J.R., Selvaraj S., Ye Z., Lee A.Y., Yen C.-A., Schmitt A.D., Espinoza C.A., Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ron G., Globerson Y., Moran D., Kaplan T. Promoter-enhancer interactions identified from Hi-C data using probabilistic models and hierarchical topological domains. Nat. Commun. 2017;8:2237. doi: 10.1038/s41467-017-02386-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Doyle B., Fudenberg G., Imakaev M., Mirny L.A. Chromatin Loops as Allosteric Modulators of Enhancer-Promoter Interactions. PLoS Comput. Biol. 2014;10 doi: 10.1371/journal.pcbi.1003867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Symmons O., Uslu V.V., Tsujimura T., Ruf S., Nassari S., Schwarzer W., Ettwiller L., Spitz F. Functional and topological characteristics of mammalian regulatory domains. Genome Res. 2014;24:390–400. doi: 10.1101/gr.163519.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.ENCODE Project Consortium. Moore J.E., Purcaro M.J., Pratt H.E., Epstein C.B., Shoresh N., Adrian J., Kawli T., Davis C.A., Dobin A., et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 2020;583:699–710. doi: 10.1038/s41586-020-2493-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Luo Y., Hitz B.C., Gabdank I., Hilton J.A., Kagda M.S., Lam B., Myers Z., Sud P., Jou J., Lin K., et al. New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res. 2020;48:D882–D889. doi: 10.1093/nar/gkz1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Loupe J.M., Anderson A.G., Rizzardi L.F., Rodriguez-Nunez I., Moyers B., Trausch-Lowther K., Jain R., Bunney W.E., Bunney B.G., Cartagena P., et al. Extensive profiling of transcription factors in postmortem brains defines genomic occupancy in disease-relevant cell types and links TF activities to neuropsychiatric disorders. bioRxiv. 2023 doi: 10.1101/2023.06.21.545934. Preprint at. [DOI] [Google Scholar]
- 93.Bendl J., Hauberg M.E., Girdhar K., Im E., Vicari J.M., Rahman S., Fernando M.B., Townsley K.G., Dong P., Misir R., et al. The three-dimensional landscape of cortical chromatin accessibility in Alzheimer’s disease. Nat. Neurosci. 2022;25:1366–1378. doi: 10.1038/s41593-022-01166-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fernández-Chacón R., Königstorfer A., Gerber S.H., García J., Matos M.F., Stevens C.F., Brose N., Rizo J., Rosenmund C., Südhof T.C. Synaptotagmin I functions as a calcium regulator of release probability. Nature. 2001;410:41–49. doi: 10.1038/35065004. [DOI] [PubMed] [Google Scholar]
- 95.Pantazis C.B., Yang A., Lara E., McDonough J.A., Blauwendraat C., Peng L., Oguro H., Kanaujiya J., Zou J., Sebesta D., et al. A reference human induced pluripotent stem cell line for large-scale collaborative studies. Cell Stem Cell. 2022;29:1685–1702.e22. doi: 10.1016/j.stem.2022.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schmid B., Holst B., Poulsen U., Jørring I., Clausen C., Rasmussen M., Mau-Holzmann U.A., Steeg R., Nuthall H., Ebneth A., Cabrera-Socorro A. Generation of two gene edited iPSC-lines carrying a DOX-inducible NGN2 expression cassette with and without GFP in the AAVS1 locus. Stem Cell Res. 2021;52 doi: 10.1016/j.scr.2021.102240. [DOI] [PubMed] [Google Scholar]
- 97.van Arensbergen J., Pagie L., FitzPatrick V.D., de Haas M., Baltissen M.P., Comoglio F., van der Weide R.H., Teunissen H., Võsa U., Franke L., et al. High-throughput identification of human SNPs affecting regulatory element activity. Nat. Genet. 2019;51:1160–1169. doi: 10.1038/s41588-019-0455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nalls M.A., Blauwendraat C., Vallerga C.L., Heilbron K., Bandres-Ciga S., Chang D., Tan M., Kia D.A., Noyce A.J., Xue A., et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. doi: 10.1016/S1474-4422(19)30320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zheng Y., Shen W., Zhang J., Yang B., Liu Y.-N., Qi H., Yu X., Lu S.-Y., Chen Y., Xu Y.-Z., et al. CRISPR interference-based specific and efficient gene inactivation in the brain. Nat. Neurosci. 2018;21:447–454. doi: 10.1038/s41593-018-0077-5. [DOI] [PubMed] [Google Scholar]
- 100.Hayashi H., Kubo Y., Izumida M., Matsuyama T. Efficient viral delivery of Cas9 into human safe harbor. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-78450-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Smith J.R., Maguire S., Davis L.A., Alexander M., Yang F., Chandran S., ffrench-Constant C., Pedersen R.A. Robust, Persistent Transgene Expression in Human Embryonic Stem Cells Is Achieved with AAVS1-Targeted Integration. Stem Cell. 2008;26:496–504. doi: 10.1634/stemcells.2007-0039. [DOI] [PubMed] [Google Scholar]
- 102.Hockemeyer D., Soldner F., Beard C., Gao Q., Mitalipova M., DeKelver R.C., Katibah G.E., Amora R., Boydston E.A., Zeitler B., et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Myers A.J., Pittman A.M., Zhao A.S., Rohrer K., Kaleem M., Marlowe L., Lees A., Leung D., McKeith I.G., Perry R.H., et al. The MAPT H1c risk haplotype is associated with increased expression of tau and especially of 4 repeat containing transcripts. Neurobiol. Dis. 2007;25:561–570. doi: 10.1016/j.nbd.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 104.Kim J., de Haro M., Al-Ramahi I., Garaicoechea L.L., Jeong H.-H., Sonn J.Y., Tadros B., Liu Z., Botas J., Zoghbi H.Y. Evolutionarily conserved regulators of tau identify targets for new therapies. Neuron. 2023;111:824–838.e7. doi: 10.1016/j.neuron.2022.12.012. [DOI] [PubMed] [Google Scholar]
- 105.Morris M., Hamto P., Adame A., Devidze N., Masliah E., Mucke L. Age-appropriate cognition and subtle dopamine-independent motor deficits in aged Tau knockout mice. Neurobiol. Aging. 2013;34:1523–1529. doi: 10.1016/j.neurobiolaging.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Birnbaum R.Y., Clowney E.J., Agamy O., Kim M.J., Zhao J., Yamanaka T., Pappalardo Z., Clarke S.L., Wenger A.M., Nguyen L., et al. Coding exons function as tissue-specific enhancers of nearby genes. Genome Res. 2012;22:1059–1068. doi: 10.1101/gr.133546.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Singer M., Kosti I., Pachter L., Mandel-Gutfreund Y. A diverse epigenetic landscape at human exons with implication for expression. Nucleic Acids Res. 2015;43:3498–3508. doi: 10.1093/nar/gkv153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rizzardi L.F., Hickey P.F., Rodriguez DiBlasi V., Tryggvadóttir R., Callahan C.M., Idrizi A., Hansen K.D., Feinberg A.P. Neuronal brain region-specific DNA methylation and chromatin accessibility are associated with neuropsychiatric trait heritability. Nat. Neurosci. 2019;22:307–316. doi: 10.1038/s41593-018-0297-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Borsari B., Villegas-Mirón P., Mirón M., Laayouni H., Segarra-Casas A., Bertranpetit J., Guigó R., Guigó G., Acosta S. Intronic enhancers regulate the expression of genes involved in tissue-specific functions and homeostasis. bioRxiv. 2020 doi: 10.1101/2020.08.21.260836. Preprint at. [DOI] [Google Scholar]
- 110.Bravo C.P., Giani A.M., Perez J.M., Zhao Z., Samelson A., Wong M.Y., Evangelisti A., Fan L., Pozner T., Mercedes M., et al. Human iPSC 4R tauopathy model uncovers modifiers of tau propagation. bioRxiv. 2023 doi: 10.1101/2023.06.19.544278. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bowles K.R., Pugh D.A., Pedicone C., Oja L., Weitzman S.A., Liu Y., Chen J.L., Disney M.D., Goate A.M. Development of MAPT S305 mutation models exhibiting elevated 4R tau expression, resulting in altered neuronal and astrocytic function. bioRxiv. 2023 doi: 10.1101/2023.06.02.543224. Preprint at. [DOI] [Google Scholar]
- 112.Dixit A., Parnas O., Li B., Chen J., Fulco C.P., Jerby-Arnon L., Marjanovic N.D., Dionne D., Burks T., Raychowdhury R., et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell. 2016;167:1853–1866.e17. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw and processed data generated are available through NCBI GEO under series accession number GSE228121. All original code generated during this study is publicly available at github.com/aanderson54/MAPT_cre. Variant data were obtained from the Alzheimer Disease Sequencing Project (ADSP) (NIAGADS accession number: NG00067) and the Accelerating Medicine Partnerships – Parkinson Disease (AMP-PD) (amp-pd.org).