Summary
The calcium/calmodulin-dependent protein kinase type 2 (CAMK2) family consists of four different isozymes, encoded by four different genes—CAMK2A, CAMK2B, CAMK2G, and CAMK2D—of which the first three have been associated recently with neurodevelopmental disorders. CAMK2D is one of the major CAMK2 proteins expressed in the heart and has been associated with cardiac anomalies. Although this CAMK2 isoform is also known to be one of the major CAMK2 subtypes expressed during early brain development, it has never been linked with neurodevelopmental disorders until now. Here we show that CAMK2D plays an important role in neurodevelopment not only in mice but also in humans. We identified eight individuals harboring heterozygous variants in CAMK2D who display symptoms of intellectual disability, delayed speech, behavioral problems, and dilated cardiomyopathy. The majority of the variants tested lead to a gain of function (GoF), which appears to cause both neurological problems and dilated cardiomyopathy. In contrast, loss-of-function (LoF) variants appear to induce only neurological symptoms. Together, we describe a cohort of individuals with neurodevelopmental disorders and cardiac anomalies, harboring pathogenic variants in CAMK2D, confirming an important role for the CAMK2D isozyme in both heart and brain function.
Keywords: calcium/calmodulin-dependent protein kinase 2 delta, CAMK2D, intellectual disability, cardiomyopathy, neurodevelopment
CAMK2 disorder is a relatively new disorder where three of the four CAMK2s (CAMK2A, B, and G) are shown to cause neurodevelopmental disorders. In this paper we describe a cohort of eight individuals with neurodevelopmental disorders and cardiac abnormalities, having CAMK2D variants, proving all CAMK2s are important for normal development.
Introduction
The family of calcium/calmodulin-dependent protein kinase II (CAMK2) mediates calcium-dependent signaling and consists of four members: CAMK2A, CAMK2B, CAMK2G, and CAMK2D. All four proteins are highly homologous and share an evolutionarily conserved structure consisting of a kinase domain, regulatory segment, linker region, and association domain. The kinase domain is the catalytic component and binds ATP; the regulatory domain enforces autoinhibition of kinase activity in the basal state, binds to calcium/calmodulin (Ca2+/CaM), and harbors the Thr286/287 autophosphorylation site, which generates autonomous activity; the linker region (or variable domain) is highly susceptible to alternative splicing and shows the most variability between the different CAMK2 members; the association domain (or hub domain) is where the same or different CAMK2 molecules oligomerize to form the functional holoenzyme that generally consists of 12–14 subunits.1,2,3,4,5
CAMK2’s central role in synaptic plasticity, learning, and memory was established in various animal models, with a prime focus on CAMK2A,6,7,8,9 followed by CAMK2B.10,11,12,13 Although these two isozymes have unique functions in the brain, as shown by the phenotypes of the different knockout mouse models, they can also partially compensate for each other’s loss. Simultaneous deletion of Camk2a and Camk2b results in early postnatal lethality in mice, as well as lethality in the adult stage upon adult-induced deletion.14 Additionally, simultaneous deletion of Camk2a and Camk2b in the adult completely abolished long-term potentiation (LTP) at the CA3-CA1 synapse in the hippocampus.14 CAMK2G and CAMK2D are also expressed in the brain, albeit at much lower levels compared to CAMK2A and CAMK2B, and cannot compensate for absence of both CAMK2A and CAMK2B in survival or plasticity despite their high homology. Interestingly, a few recently published studies do point toward an important role for CAMK2G and CAMK2D in learning and memory in addition to their better defined non-neuronal functions.15,16,17 In mice, Camk2g has been shown to participate in excitation transcription coupling in excitatory neurons and in hippocampal synaptic plasticity in inhibitory neurons.15,16,18 Moreover, Camk2g has been shown to be involved in normal development of neurons and innate behaviors that have been associated with critical windows during development.19,20 Camk2d, on the other hand, is suggested to play a role in long-term memory consolidation, since in mice its RNA levels are upregulated up to seven days after executing a memory task, showing a positive correlation with the mouse’s memory performance.17 Correspondingly, knockdown of Camk2d after training impairs memory performance specifically when tested multiple days later.17
The importance of CAMK2 in normal human brain functioning has been highlighted by the identification of individuals with neurodevelopmental disorders carrying pathogenic variants in one of the CAMK2 genes. The first identified variant, discovered by exome sequencing, was located in CAMK2G (intellectual developmental disorder, autosomal dominant 59 [MIM: 618522]),19,21 which is somewhat surprising considering that CAMK2G is much less abundant in the brain, compared to CAMK2A or CAMK2B, but proving a function for this kinase in normal human brain function. Next, cohorts of individuals with neurodevelopmental disorders were identified with causative variants in CAMK2A and CAMK2B (intellectual developmental disorder, autosomal dominant 53 [MIM: 617798] and 54 [MIM: 617798], respectively).22,23,24,25,26,27,28 For CAMK2D, a role in human brain function has not been established thus far.
CAMK2D is expressed in multiple organs, including the brain, but has the highest expression levels in the heart.29,30 CAMK2D has multiple isoforms due to alternative splicing, which can result in different cellular localization.31 Deep sequencing of human samples of the hippocampus consistently detected six CAMK2D splice variants, of which CAMK2D (6v1,14b,18) and (6v1,14a,18) were most prevalent32 (GenBank: XP_047272177.1 and NM_001321571.2, respectively).
The expression pattern of Camk2d in rodent brain is widespread, showing elevated levels in granule cells of the cerebellum, a laminar distribution in layers II/III, IV, and deep VI of the cortex, and a global distribution in the cell layers of the hippocampus.29,33,34 During rodent brain development, Camk2d expression can be detected around embryonic day 10 and increases rapidly postnatally.30,33 It is then expressed in multiple cell types, such as excitatory neurons, interneurons, and astrocytes.35,36,37 During human brain development, stable RNA levels are detectable throughout gestation, starting in the first trimester.19 This expression profile hints at a possible neurodevelopmental role for CAMK2D.
In this manuscript, we sought to determine whether CAMK2D indeed plays a role in neurodevelopment. Our data provide evidence for the importance of CAMK2D in neurodevelopment in vitro and in vivo. Additionally, we present pathogenic variants in the CAMK2D gene identified in individuals with neurodevelopmental disorders, confirming its clinical relevance for cognitive function.
Subjects and methods
Genetic identification of the variants
The eight affected individuals participating in the study come from seven unrelated families. They were clinically assessed by centers internationally. All participants presented with unexplained congenital cardiomyopathy and/or developmental delay or intellectual disability. As relevant routine genetic testing, including copy number variation (CNV) analysis by high-resolution array-based comparative genomic hybridization (aCGH), had provided no diagnosis, exome sequencing or genome sequencing was performed in a diagnostic or research setting, which highlighted rare CAMK2D candidate variants. Segregation analysis was performed by Sanger or exome/genome sequencing. Contact between participating centers and teams was aided by the web-based tool GeneMatcher.38 Written informed consent was obtained from each affected individual or her/his guardian and available family members for use of medical history, genetic testing report, and photograph (if applicable), in accordance with the respective human ethics committees of each participating institution. Authorization to publish photographs was obtained from the parents of individuals 1, 2, 4, and 5.
Cloning for the stability assay and the in vivo and in vitro neuronal experiments
CAMK2DWT sequence (GenBank: NM_001321571.2) was obtained from the human brain cDNA library and immediately tagged with restriction sites AscI and PacI by PCR, which made it possible to ligate the CAMK2DWT sequence into the TOPO vector and our dual promoter expression vector.19 For the primers used, see Table S1. For overexpression, single nucleotide variants were introduced in the CAMK2DWT sequence in TOPO, using site-directed PCR mutagenesis. For the primers used for mutagenesis, see Table S2. Once successfully mutated, the sequence was ligated into the multiple cloning site (MCS) of the expression vector. The exact same expression vector, lacking an insert in the MCS, was used as a negative control throughout the experiments and referred to as “empty vector control.” For knockdown experiments, three different shRNAs that specifically target the coding sequence of mouse Camk2d, were obtained from the MISSION shRNA library for mouse genomes of Sigma Life Sciences and The RNAi Consortium (TRC): (1) 5′-GCAACTGATTGAAGCTATCAA-3′; (2) 5′-GCATAGACTGTATCAGCAGAT-3′; (3) 5′-CCTGAAGCATTGGGCAACTTA-3′. The control shRNA plasmid is the MISSION non-target shRNA control vector: 5′-CAACAAGATGAAGAGCACCAA-3′.
HEK293T transfections
Plasmids encoding the different CAMK2D variants were transfected in HEK293T cells. These cells were not authenticated. HEK293T cells were cultured in DMEM/10% Fetal Bovine Serum (FBS)/1% penicillin/streptomycin. Transfection was performed at 60%–70% confluency. Each well (6-well plate) was transfected with 3 μg DNA construct. The following constructs were transfected using polyethyleneimine (PEI) in a 1:3 (DNA:PEI) ratio: Empty vector control; CAMK2DWT; CAMK2DS79N; CAMK2DP139L; CAMK2DG210R; CAMK2DQ274P; CAMK2DR275H; CAMK2DL291F. To assess the efficiency of the Camk2d shRNAs, 2 μg shRNA construct was co-transfected with 1 μg mouse Camk2d construct (6v1, 14a, 18; for nomenclature see Sloutsky and Stratton39). To test its specificity, co-transfections of the shRNAs were also executed with mouse Camk2a (14, 18) and Camk2b (13, 14b, 16, 17, 18) plasmids. To avoid toxicity of the PEI, the medium was refreshed 4–6 h post-transfection. Transfected cells were used to prepare protein lysates for western blot.
Sample preparation and immunoblotting
The cultures were incubated for 48–72 h post-transfection, until full confluency. At this point the cells were harvested in ice-cold PBS, by scraping them off the bottom of the wells. After spinning down, the cell pellet was enriched with a mixture of Laemmli buffer (4% SDS, 0.1M Tris [pH 6.8])/protease inhibitor (Cat. nr. P3840, Sigma)/phosphatase inhibitor cocktails 2 and 3 (resp. Cat. nr. P5726, Sigma; Cat. nr. P0044, Sigma). After the samples were briefly sonicated, the concentrations of the protein lysates were measured, using the BCA assay kit (Cat. nr. 23225, Thermo Fisher Scientific). Samples were diluted to a concentration of 1 μg/μL and prepared for loading with DTT and XT Sample buffer (Cat. nr. 1610791, Biorad). 15 μg protein/lane was loaded in precast Bis-Tris 4%–12% gels (Cat. nr. 3450124, Biorad). After running, the proteins were transferred to the nitrocellulose membrane (Cat. nr. 1704159, Biorad) using the TurboBlot (Cat. nr.1704150, Biorad). Membranes were blocked with 5% milk in TBS-T and incubated with primary antibodies: rabbit-anti-phThr286/7, 1:2,000 dilution (Cat. nr. ab124880, Abcam); mouse-anti-CAMK2D, 1:500 dilution (Cat. nr. sc-100362, Santa Cruz); rabbit-anti-RFP, 1:2,000 diluted (Cat. nr. 600-401-379, Rockland), mouse-anti-CAMK2A, 1:20,000 diluted (Cat. nr. NB100-1983, Novus Biologicals); mouse-anti-CAMK2B, 1:10,000 diluted (Cat. nr. 13-9800, Invitrogen). For visualization with the Odyssey scanning device, secondary antibodies with fluorescent tags were used, 1:15,000 diluted: goat-anti-rabbit (Cat. nr. 926-68021, Westburg); goat-anti-mouse (Cat. nr. 926-32210, Westburg).
Plasmid construction for the kinase assay
Constructs were built utilizing exon gene blocks (IDT) and pre-existing pSMT3 vectors containing N-terminal 6xHis followed by a SUMO tag and full-length CAMK2D variants via Gibson Assembly. Mutants were cloned via site-directed mutagenesis and confirmed via Sanger sequencing (GeneWiz).
CAMK2D expression and purification
A previously established bacterial expression system40 for CAMK2D by co-expression with Lambda Phosphatase was utilized. We expressed CAMK2D in Escherichia coli by co-expressing with λ phosphatase (from Kuriyan Lab) in Rosetta (DE3)pLysS competent cells (Novagen). λ phosphatase was expressed via a pCDFDuet1 vector and N-terminal tagged clones 6xHis AviTag CAMK2D was expressed in a pET287 vector. Cells were grown to an OD (595 nm) of 0.6 and induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG; GoldBio). Cells were grown for 16 h at 18°C and resuspended in buffer A (25 mM Tris-HCl [pH 8.5], 150 mM KCl, 50 mM imidazole, 10% glycerol; Sigma) with 25 mM magnesium chloride, containing a cocktail of protease inhibitors and deoxyribonuclease (DNase) (0.2 mM 4 benzenesulfonyl fluoride hydrochloride [AEBSF], 5.0 μM leupeptin, pepstatin [1 μg/mL], aprotinin [1 μg/mL], trypsin inhibitor [0.1 mg/mL], 0.5 mM benzamidine, DNase [1 μg/mL]) (Sigma). Resuspended cells were flash frozen until used.
Cells were lysed via a French press, and clarified cell lysate was aspirated from cell debris following centrifugation (18,000 rpm; 4°C; 60 min). All subsequent purification steps were performed with an ÄKTA pure chromatography system (Cytiva Life Sciences) at 4°C. Clarified cell lysate was loaded onto two 5-mL HisTrap FF NiNTA Sepharose columns (Cytiva Life Sciences) and eluted with a combination of 50% buffer A and 50% buffer B (25 mM Tris-HCl [pH 8.5], 150 mM KCl, 1 M imidazole, 10% glycerol) for a final concentration of 0.5 M imidazole. The eluate was desalted of residual imidazole with a HiPrep 26/10 Desalting column, and His SUMO tags were cleaved with Ulp1 protease overnight at 4°C in buffer C (25 mM Tris-HCl [pH 8.5], 150 mM KCl, 2 mM Tris(2-carboxyethyl)phosphine [TCEP] [GoldBio], 50 mM imidazole, 10% glycerol). Cleaved tags were removed by a subtractive NiNTA step. Subsequently, an anion exchange step was performed with two 5-mL HiTrap Q-FF (Cytiva Life Sciences) and protein was eluted with a KCl gradient. Eluted proteins were visualized via SDS-PAGE and select fractions were concentrated and further purified in gel filtration buffer (25 mM Tris-HCl [pH 8.0], 150 mM KCl, 1 mM TCEP, 10% glycerol) using a Superose 6 Increase 10/300 size exclusion column (Cytiva Life Sciences). Fractions were visualized by SDS-PAGE, and pure fractions were pooled, concentrated, aliquoted, flash-frozen in liquid nitrogen, and stored at −80°C until used. Protein concentration was calculated using absorbance (280 nm).
Calmodulin purification
Calmodulin (Gallus gallus) was recombinantly expressed from a pET-15b vector (a gift from A. Nairn, Yale School of Medicine) in BL21(DE3) cells (Millipore). Cells were grown to an OD (595 nm) of 0.6 and induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG; GoldBio). Cells were grown for 16 h at 18°C and resuspended in cell lysis buffer (40 mM Tris-HCl [pH 8.0], 100 mM KCl, 10 mM EDTA). Resuspended cells were flash frozen until used. Cells were lysed via a French press, and clarified cell lysate was aspirated from cell debris following centrifugation (18,000 rpm; 4°C; 60 min). All subsequent purification steps were performed with an ÄKTA pure chromatography system (Cytiva Life Sciences) at 4°C. Clarified cell lysate was loaded onto two 5-mL HiTrap Phenyl FF (Low Sub) columns (Cytiva Life Sciences). Column flow through was collected, and CaCl2 was added to the flow through (final concentration 20 mM CaCl2). The flow through was applied to a different pair of 5-mL HiTrap Phenyl FF (Low Sub) columns (Cytiva Life Sciences) with buffer A (50 mM Tris-HCl [pH 7.5], 1 mM CaCl2). The column was washed sequentially with buffer A, buffer B (50 mM Tris-HCl [pH 7.5], 500 mM NaCl, 1 mM CaCl2), and buffer A. Calmodulin was eluted with buffer C (50 mM Tris-HCl [pH 7.5], 2 mM EDTA). Purity was assessed via SDS-PAGE, and clean eluate fractions were pooled and concentrated. To quantify calmodulin concentration, we used circular dichroism on a Jasco J-1500 spectrophotometer to make a measurement in triplicate for our purified sample scanning a wavelength spectrum between 250 and 215 nm to measure the characteristic wavelength of 222 nm as previously described.41 We calculated the calmodulin concentration as follows:
where the circular dichroism at 222 nm (CD222nm) is expressed in mdeg, Θ is the molar ellipticity, and l is the path length in cm.
Coupled kinase assay
Kinase activity was monitored under previously described conditions40,42 with a Synergy H1 microplate reader (BioTek). Each well of the plate contained master mix composed of the following (all concentrations listed as final, working concentrations): 5 mM Tris (pH 7.5; Thermo Fisher Scientific), 150 mM KCl (Sigma), 10× Tris/MgCl2 buffer (50 mM/10 mM, respectively) (Thermo Fisher Scientific), 0.2 mM CaCl2 (Sigma), 1 mM phosphoenolpyruvate (Alfa Aesar), nicotinamide adenine dinucleotide (0.15 mg/mL; Sigma), pyruvate kinase (10.0 U/ml; Sigma), lactate dehydrogenase (30 U/ml; Millipore Sigma), 2.0 mM adenosine triphosphate (ATP) (Sigma), and 0.3 mM syntide (LifeTein). The final pH in each well of the reaction was ∼7.5–8, and the final enzyme concentration was 13.3 nM. The reagents were added to the well in the following order: Tris buffer, calmodulin (ranging from 0.4 nM to 2 μm final concentration), and master mix. The addition of CAMK2D was used to initiate the kinase reaction, after which absorbance (340 nm) was measured at 15-s intervals for 10 min. The change in absorbance over time was fitted with a straight line (y = mx + c) to obtain a slope (m) proportional to the kinetic rate of the reaction. For each time series, slopes were fitted to a sliding window of five points (1 min 15 s), and the maximum observed slope was used to represent the kinetic rate of that reaction. Kinetic rates across a series of calmodulin concentrations were fitted with the following equation:
to obtain EC50 (defined as the calmodulin concentration needed to reach the half-maximal reaction velocity) and cooperativity values (Hill coefficients, nH). Ninety-five percent confidence intervals for fit parameters (EC50 and nH) were determined using the following bootstrap procedure. Ten thousand replicate calmodulin concentration series were generated by randomly selecting one observed kinetic rate at each measured calmodulin concentration from the set of replicates for that variant. Each generated concentration series was fit with the equation described earlier. Parameter values at the 2.5th and 97.5th quantiles of the 10,000 fits were taken as the boundaries of the 95% confidence interval.
Mice
For the in utero electroporation, FvB/NHanHsd females (ordered at 6–8 weeks old from Envigo) were crossed with C57Bl6/J (ordered at 6–8 weeks old from Charles River). All mice were group-housed in IVC cages (Sealsage 1145T, Tecniplast) with bedding material (Lignocel BK 8/15 from Rettenmayer) on a 12/12 h light/dark cycle in 21°C (±1°C), humidity at 40%–70% and with food pellets (801727CRM(P) from Special Dietary Service) and water available ad libitum. All animal experiments were approved by the Local Animal Experimentation Ethical Committee, in accordance with Institutional Animal Care and Use Committee guidelines (IRN2019-0030).
In utero electroporation
The in utero electroporation was done as previously described.19 Briefly, adult pregnant female mice (FvB/NHanHsd) underwent surgery at E14.5 of gestation. After exposing the uterus, embryonic pups were injected intraventricularly with a mixture of DNA construct (1.5–3.0 μg/μL) and FastGreen (0.05%), using the Picospritzer III, after which a small electrical current was applied (five electrical square pulses of 45V; duration 50 ms/pulse; duration pulse interval 150 ms; driven by a pulse generator ECM 830, BTX Harvard Apparatus), orientating the tweezer-type pedestal with its positive pool on top of the developing somatosensory cortex. The following plasmids were electroporated: Empty vector control; CAMK2DWT; CAMK2DS79N; CAMK2DP139L; CAMK2DG210R; CAMK2DQ274P; CAMK2DR275H; CAMK2DL291F. For knockdown experiments, co-transfection of different Camk2d shRNAs or the control shRNA with an RFP or GFP plasmid (Addgene) were electroporated. After the procedure, the mice were returned to their home cage and went naturally into labor five days post-surgery.
Perfusion and immunohistochemistry
Pups were sacrificed at the date of birth or one day after birth (P0 or P1; postnatal day 0 or 1, respectively), through cardiac perfusion with saline solution, followed by 4% PFA. After perfusion, the brains were isolated and post-fixed in 4% PFA for 1 h at room temperature. The brains were then stored overnight in 30% sucrose in 0.1M PB. Perfused brains were embedded in 14% gelatin/30% sucrose - and free-floating sections were made using a cryo-microtome (40–50 μm thick). Sections were washed in 0.1M PB and counterstained with 4′,6-diamidino-2-phenylindole solution (DAPI, 1:10,000, Invitrogen) before being mounted on glass and covered with Mowiol (Sigma). Only a selection of sections was used for counterstaining. Stained and covered brain slices were used for confocal imaging.
Confocal microscopy
All images were acquired using a LSM700 confocal microscope (Zeiss). Images for the migration analysis were taken from two to three non-consecutive sections from at least three successfully targeted animals per condition (10× objective, 0.5 zoom, 1,024 × 1,024). Images from Zeiss were converted to .tif files and cropped in such a way that each cropped image resulted in a rectangle image reaching from the IZ (intermediate zone) to the CP (cortical plate). The cropped images were vertically divided over 10 equally sized bins, having bin 1 as first bin covering the outer layer of the CP and bin 10 as the most inner area of the cropped image (IZ). The number of transfected neurons in each bin was calculated using ImageJ and used for analysis.
Statistical analysis
Protein expression in HEK293T
Image Studio Lite (v.5.2.5) was used to quantify the amount of produced protein and autophosphorylation after transfections of the CAMK2D constructs in HEK293T cells. The bands were fenced in order to measure the fluorescent signal and background in the drawn square (2 pixels, top and bottom). All data were corrected for background signal. Signal of the phThr287 phosphorylation band was normalized for the signal of the CAMK2D band. To determine the CAMK2D protein level, the CAMK2D band was normalized over the RFP band. All data were then normalized to the autophosphorylation or protein levels of CAMK2DWT overexpression, which was set at 1. All data were assumed to be normally distributed with equal variances. Two-tailed unpaired t tests were performed between CAMK2DWT and each CAMK2D variant (dual comparison). Each condition was at least transfected in triplicate, in two separate batches of cells, for a minimum sample size of n = 6. To test the specificity of the shRNAs against Camk2d, statistical analysis was performed with one-way ANOVA followed by Dunnett’s posthoc test for multiple comparisons, comparing the conditions to the control shRNA.
Neuronal migration
All data were assumed to be normally distributed. Statistical analysis was performed on data of the first four bins of each cropped image, corresponding to the cortical plate (CP). CP is here defined as the most proximal 40% of the dorsoventral distance between the pia and ventricle (first 4 of 10 equally spaced bins). In these neuronal migration experiments, a one-way ANOVA followed by Dunnett’s posthoc test for multiple comparisons was performed. In the knockdown experiments, all conditions were compared to the scramble shRNA (control). In the overexpression experiments, all conditions were compared to empty vector control and to CAMK2DWT overexpression. Cumulative graphs were made as visual indication of the general migration pattern but were not used for statistical analysis. For all conditions, except shRNA3, images of at least 3 separate pups (2–4 images per pup) were used for the migration analysis. For shRNA 3, images taken of 2 separate pups (3 images per pup) were used for the analysis. Statistical tests and results have been described in Tables 1, 2, 3, 4, and 5. PRISM software (Graphpad 8.0) was used for the statistical tests, p values <0.05 were considered significant.
Table 1.
Overview of statistical analysis on the knockdown experiments in HEK293T cells
| p value | F (df) | shRNA 1 | shRNA 2 | shRNA 3 | |
|---|---|---|---|---|---|
| Camk2d knockdown | <0.0001∗ | (3,12) 436.1 | <0.0001∗ | <0.0001∗ | <0.0001∗ |
| Camk2a knockdown | 0.0179∗ | (3,11) 5.18 | 0.1397 | 0.0086∗ | 0.7684 |
| Camk2b knockdown | 0.0617 | (3,11) 3.29 |
One-way ANOVA, Dunnet’s multiple comparison test compared to control scramble shRNA. Asterisks (∗) indicate a statistically significant difference.
Table 2.
Overview of statistical analysis on the in utero electroporation experiments: Sum percentage of targeted cells in bins 1–4
| p value | F (df) | |
|---|---|---|
| Camk2d knockdown (shRNA) | <0.0001∗ | (3,44) 22.69 |
| CAMK2D overexpression | <0.0001∗ | (7,107) 51.07 |
One-way ANOVA. Asterisks (∗) indicate a statistically significant difference.
Table 3.
Post-hoc analysis of the in utero electroporation experiments: Sum percentage of targeted cells in bins 1–4
| Camk2d knockdown (shRNA) | p value |
|---|---|
| Scramble shRNA (ctrl) versus shRNA 1 | <0.0001∗ |
| Scramble shRNA (ctrl) versus shRNA 2 | <0.0001∗ |
| Scramble shRNA (ctrl) versus shRNA 3 | <0.0001∗ |
Dunnett’s multiple comparison test. Asterisks (∗) indicate a statistically significant difference.
Table 4.
Clinical features of the individuals with variants in CAMK2D
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | Total | |
|---|---|---|---|---|---|---|---|---|---|
| Variant nomenclature chr4 (GRCh37)a | |||||||||
| Genome | g.114530307C>A | g.114530347C>T | g.114458598G>A | g.114438787C>T | g.114435068T>G | g.114435065C>T | g.114435065C>T | g.114435016C>G | 7 variants 1 SS, 6 MS |
| Nucleotides | c.275+1G>T | c.236G>A | c.416C>T | c.628G>A | c.821A>C | c.824G>A | c.824G>A | c.873G>C | |
| Amino acids | p.? | p.Ser79Asn | p.Pro139Leu | p.Gly210Arg | p.Gln274Pro | p.Arg275His | p.Arg275His | p.Leu291Phe | |
| Variant annotation | |||||||||
| MobiDetailsb | 92725 | 93616 | 93619 | 272319 | 93654 | 93656 | 93656 | 93659 | |
| CADD (v.1.6) | 34 | 25.90 | 33 | 26.30 | 24.90 | 24.60 | 24.60 | 22.50 | |
| Metadome (tolerance score) | N/A | neutral | highly intolerant | unknown | intolerant | intolerant | intolerant | highly intolerant | |
| MPA | 10 (high splice) | 6 (moderate missense) | 7 (moderate missense) | 7 (moderate missense) | 5 (low missense) | 5 (low missense) | 5 (low missense) | 5 (low missense) | |
| Revel | N/A | uncertain | damaging | damaging | uncertain | uncertain | uncertain | uncertain | |
| ClinPred | N/A | damaging | damaging | damaging | damaging | damaging | damaging | damaging | |
| Mistic | N/A | damaging | damaging | damaging | damaging | damaging | damaging | damaging | |
| gnomAD v.2.1.1 | absent | absent | absent | absent | absent | absent | absent | absent | |
| ClinVar | SCV002103284 | SCV002103285 | SCV002103286 | SCV003799177 | SCV002103287 | SCV002103288 | SCV002103288 | SCV002103289 | |
| Mode of inheritance | de novo | de novo | de novo | de novo | de novo | dominant | dominant | de novo | |
| Method of mutation detection | ES | ES | ES | ES | ES | ES | ES | GS | |
| Gender | female | female | male | male | female | male | female | male | 4 F, 3 M |
| Age at last investigation | 11 years | 2 years | 4 years | 6 years | 20 years | 12 years | 17 years | 5 weeks | 5 weeks to 19 years |
| Height at age last investigation (SD) | +1.69 | −1.14 | −2.86 | −1.37 | −3.28 | −0.94 | −2.1 | −1.14 | −3.28 to −0.94 |
| Developmental delay or ID | +; mild | +; moderate to severe | +; severe | +; severe | +; profound | +; mild | +; severe | ND | 7/7 |
| Delay in walking | + | + | + | + | + | + | + | ND | 7/7 |
| Speech delay | + | +; non-verbal | +; non-verbal | +; vocalizations only | +; non-verbal | +; mixed receptive-expressive language disorder | +; non-verbal | ND | 7/7 |
| Dysmorphic facial features | + | + | + | + | + | + | + | – | 7/8 |
| Skeletal anomalies | +; hands, feet | +; spine | +; thorax | +; hands, feet, thorax, palate | +; feet, thorax | +; hands | +; hands, feet, palate | – | 7/8 |
| Dilated cardiomyopathy | – | +; cardiac transplantation | +; planned cardiac transplantation | – | +; severe, cardiac transplantation impossible; VSD repaired, PDA | +; cardiac transplantation | +; severe; AtSD and PAPVR repaired | +; severe | 6/8 |
| Abnormal muscle tone | + | + | + | + | + | – | – | – | 5/8 |
| Seizures | + | – | – | – | + | – | – | – | 2/8 |
| Behavioral anomalies | +; ASD | +; ASD | + | + | +; ASD | +; ADHD | +; ASD | ND | 7/7 |
| Digestive problems | – | +; GJ tube feeding | +; NJ feeding | +; chronic constipation | +; tube feeding, EoE, dysphagia, megacolon | ND | ND | – | 4/6 |
| Visual anomalies | + | + | ND | + | + | ND | + | ND | 5/5 |
| Urogenital/kidney anomalies | – | – | + | – | + | ND | ND | – | 2/6 |
| Anomalies in brain imaging | +; enlarged ventricles | +; enlarged ventricles | +; hemorrhagic infarcts | +; enlarged ventricles | +; decreased brain volume | ND | ND | ND | 5/5 |
| EEG anomalies | + | – | ND | + | ND | ND | ND | ND | 2/3 |
N/A, not applicable; ND, not determined; SD, standard deviation; ES, exome sequencing; GS, genome sequencing; VSD, ventricular; AtSD, atrial septal defect septal defect; PDA, patent ductus arteriosus; ASD, autism spectrum disorder; ADHD, attention deficit/hyperactivity disorder; GJ tube feeding, gastrostomy-jejunostomy tube feeding; NJ feeding, naso-jejunal feeding; EoE, eosinophilic esophagitis; PAPVR, partial anomalous pulmonary venous return.
Nomenclature HGVS v.2.0 according to mRNA reference sequence GenBank: NC_000004.11 (NM_001321579.2) for the splice site variant in individual 1 and GenBank: NM_001321579.2 for missense variants in individuals 2–9. Nucleotide numbering uses +1 as the A of the ATG translation initiation codon in the reference sequence, with the initiation codon as codon 1.
For online access, insert the accession number in place of <XXX> at https://mobidetails.iurc.montp.inserm.fr/MD/api/variant/<XXX>/browser/
Table 5.
Overview of statistical analysis on the western blot experiments for CAMK2D protein levels and Thr287 autophosphorylation
|
CAMK2D expression |
Thr287 phosphorylation |
|||||
|---|---|---|---|---|---|---|
| p value | df | t value | p value | df | t value | |
| WT versus p.Ser79Asn | 0.0831 | 50 | 1.768 | 0.0368∗ | 51 | 2.145 |
| WT versus p.Pro139Leu | 0.7043 | 47 | 0.3818 | 0.0002∗ | 50 | 4.064 |
| WT versus p.Gly210Arg | <0.0001∗ | 44 | 25.08 | <0.0001∗ | 48 | 7.877 |
| WT versus p.Gln274Pro | 0.0004∗ | 47 | 3.813 | 0.0007∗ | 57 | 3.566 |
| WT versus p.Arg275His | 0.0032∗ | 47 | 3.112 | 0.0175∗ | 59 | 2.446 |
| WT versus p.Leu291Phe | <0.0001∗ | 39 | 6.455 | <0.0001∗ | 43 | 8.059 |
Two-tailed unpaired t test was performed. Asterisks (∗) indicate a statistically significant difference.
Results
Knockdown of Camk2d during brain development disrupts neuronal migration
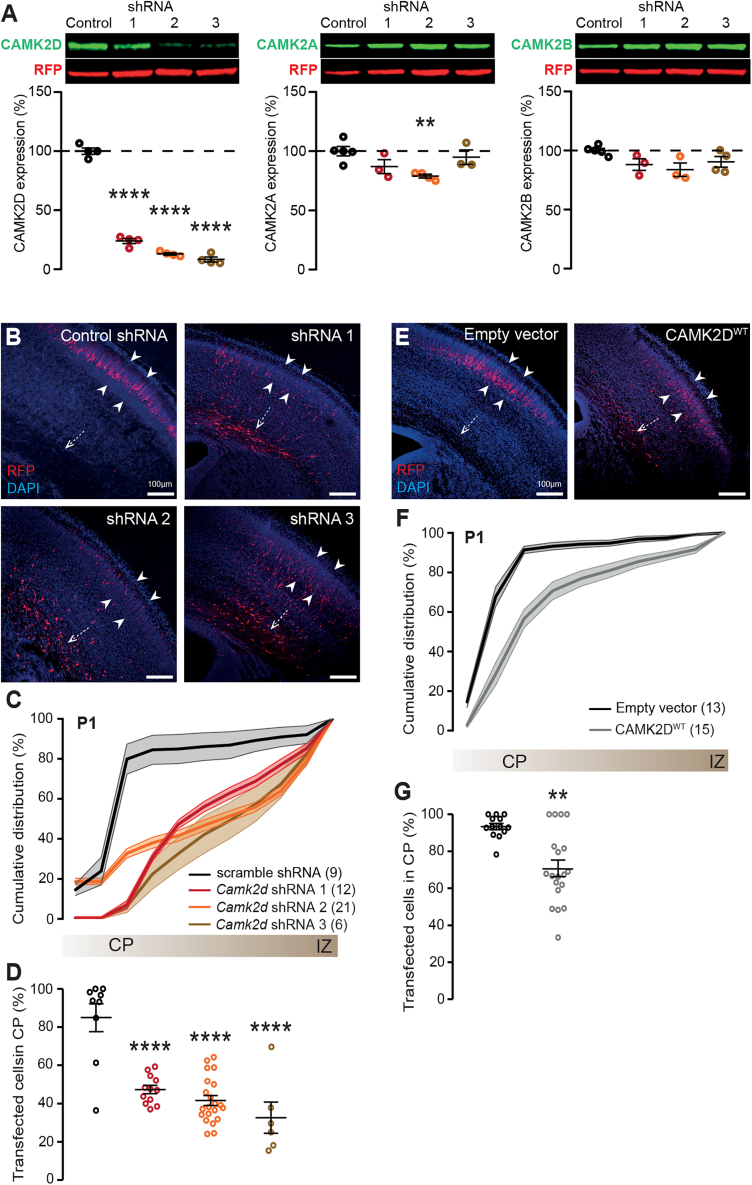
Expression of CAMK2D in the brain starts during early prenatal development, suggesting an important role for CAMK2D in neurodevelopment. To investigate this, we used shRNAs to silence endogenous Camk2d in mice. Specificity of the three shRNAs used was tested by co-transfecting mouse Camk2d, Camk2a, or Camk2b together with the individual shRNAs in HEK293T cells, a cell line that has very little or no endogenous production of CAMK2. A non-specific shRNA was taken along as control. All Camk2d shRNAs reduced CAMK2D protein levels significantly compared to control, of which shRNA 3 proved most efficient with 91.75% protein expression downgraded (Figure 1A, statistical analysis in Table 1). CAMK2A levels were slightly but significantly reduced upon co-transfection with shRNA 2, but not with shRNA 1 or 3, whereas CAMK2B levels were unaltered in all conditions (Figure 1A).
Figure 1.
Reduced as well as increased CAMK2D protein level during prenatal neurodevelopment results in a migration delay
(A) Knockdown of Camk2d with shRNAs proves to be efficient and mostly specific. Top: representative western blots of protein lysates from HEK293T cells, which were transfected with either control (scramble) or Camk2d specific shRNA’s. Blots were probed with an antibody against the specific CAMK2 (green), and RFP (red). Bottom: Quantification of CAMK2D (left), CAMK2A (middle), or CAMK2B (right) protein levels normalized against RFP.
(B) Representative images of coronal brain slices from P0/P1 pups that were transfected with scramble or Camk2d-specific shRNAs at E14.5, using in utero electroporation. White dashed arrow indicates the subventricular zone (SVZ); white arrowheads indicate the cortical plate (CP). DAPI is in blue, RFP in red.
(C) Cumulative graph indicating the migration pattern from the SVZ to the CP in presence of scramble or Camk2d-specific shRNAs.
(D) Quantification of tdTomato-positive cells that have successfully migrated to the CP, revealing that knockdown of Camk2d leads to a delay in migration.
(E) Representative images of coronal brain slices from P0/P1 pups that were transfected with a control empty vector or CAMK2DWT at E14.5, using in utero electroporation.
(F) Cumulative graph indicating the migration pattern from the SVZ to the CP upon overexpression of the empty vector or CAMK2DWT.
(G) Quantification of tdTomato-positive cells that have successfully migrated to the CP, showing that overexpression of CAMK2DWT leads to a delay in migration as well. Number in parentheses represents the number of images used for the quantification; dots represent data points and error bars indicate SEM; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001.
To assess the developmental role for CAMK2D, an in utero electroporation (IUE) was performed at E14.5 with the different Camk2d shRNAs and the control shRNA. In the control shRNA condition, 84.93% of all transfected cells migrated to the cortical plate (CP) at postnatal day 1 (P1). Remarkably, knockdown of Camk2d resulted in less than 50% of all transfected neurons migrating to the CP (Figures 1B–1D, statistical analysis in Tables 2 and 3). Additionally, we found that CAMK2D protein levels need to be tightly regulated, as overexpression of this isozyme using IUE also resulted in a migration deficit. Where in the empty vector (EV) control condition an average of 93.32% of all transfected neurons had successfully migrated to the CP, only an average of 70.77% of the neurons reached the CP when CAMK2DWT was overexpressed (Figures 1E–1G, statistical analysis in Tables 2, and 5). These results indicate that CAMK2D plays a role in neurodevelopment.
Affected individuals harboring CAMK2D variants present with a developmental condition characterized by developmental delay/intellectual disability and cardiomyopathy
The first individual identified, individual #5 in Table 6, is a young adult woman with a congenital multiple malformations syndrome. She presented with congenital heart disease (dilated cardiomyopathy with heart failure, large ventricular septal defect requiring surgical repair), renal failure due to prolonged cardiopulmonary bypass, neurological and neurodevelopmental impairment (profound intellectual disability, almost total lack of spoken language, cerebral palsy with mixed hypotonia and hypertonia, abnormally high pain tolerance), intestinal abnormalities (Chilaiditi syndrome complicated by a megacolon, associated with dysphagia and necessitating tube feeding), compromised immune system (eosinophilic esophagitis due to atopic predisposition combined with severe allergies, asthma, eczema, rosacea), skeletal abnormalities (scoliosis, kyphosis), and strabismus. Trio-based exome sequencing highlighted a nonsynonymous de novo variant in CAMK2D (GenBank: NM_001321571.2): c.821A>C (p.Gln274Pro). Via the data sharing platform GeneMatcher38 and direct requests in variant databases from participating centers, five additional distinct rare missense CAMK2D variants and one splice CAMK2D variant could be identified in seven individuals with neurodevelopmental disorders. Five variants were de novo, whereas the sixth one was present in two siblings and likely inherited from the deceased father who had similar symptoms.
Table 6.
Post-hoc analysis of the in utero electroporation experiments: Sum percentage of targeted cells in bins 1–4
| p value | |
|---|---|
| Empty vector (ctrl) versus CAMK2D overexpression | |
| Ctrl versus WT | 0.0021∗ |
| Ctrl versus p.Ser79Asn | 0.9842 |
| Ctrl versus p.Pro139Leu | <0.0001∗ |
| Ctrl versus p.Gly210Arg | <0.0001∗ |
| Ctrl versus p.Gln274Pro | <0.0001∗ |
| Ctrl versus p.Arg275His | <0.0001∗ |
| Ctrl versus p.Leu291Phe | <0.0001∗ |
| CAMK2DWT(WT) overexpression versus CAMK2Dmutantoverexpression | |
| WT versus p.Ser79Asn | 0.0024∗ |
| WT versus p.Pro139Leu | <0.0001∗ |
| WT versus p.Gly210Arg | <0.0001∗ |
| WT versus p.Gln274Pro | 0.5225 |
| WT versus p.Arg275His | 0.1682 |
| WT versus p.Leu291Phe | <0.0001∗ |
Dunnett’s multiple comparison test. Asterisks (∗) indicate a statistically significant difference.
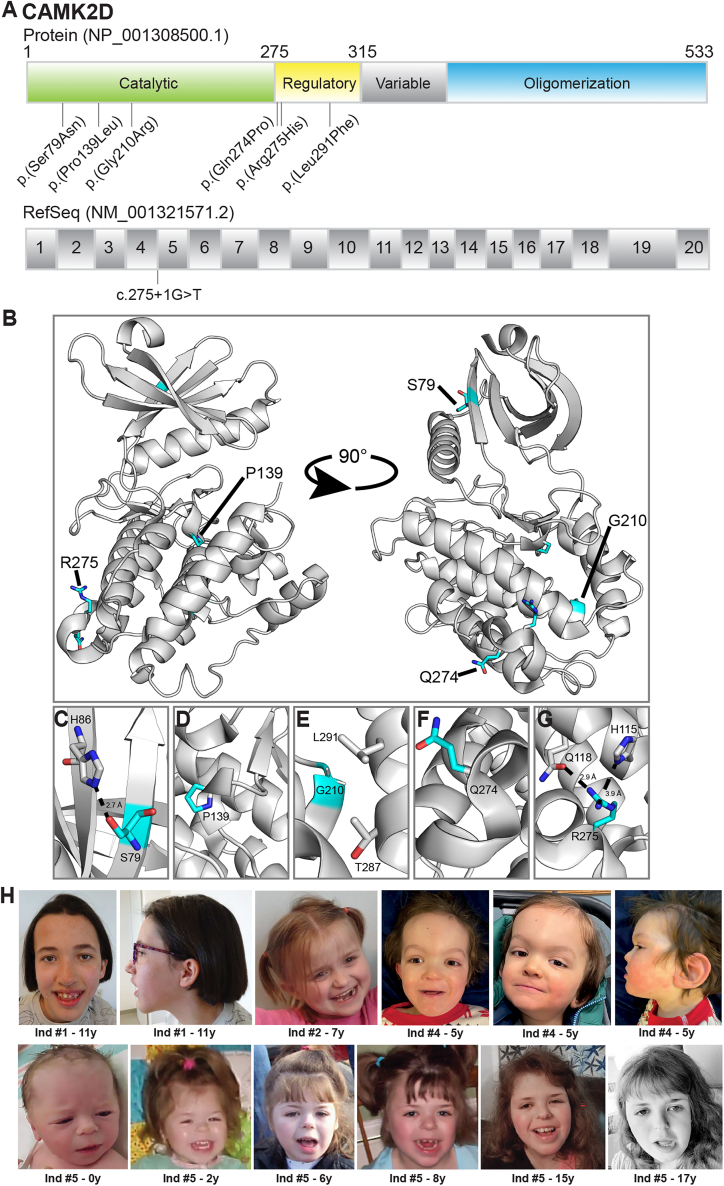
Six of the seven variants are missense substitutions located in either the catalytic domain (p.Ser79Asn, p.Pro139Leu, and p.Gly210Arg) or the regulatory domain (p.Gln274Pro, p.Arg275His, and p.Leu291Phe), whereas the seventh variant is located at a splice site, potentially leading to frameshift variant (p.Val74Glufs∗11) and thus to a premature truncation of the protein, or nonsense-mediated RNA decay, as a consequence of skipping of exon 4 (c.275+1G>T) (Figure 2A). Predicted conformational changes on tertiary protein structure of the missense variants are shown in Figures 2B–2G. All affected amino acid residues were absent in public exome and genome databases (gnomAD, >246,000 chromosomes; NHLBI Exome Variant Server, >13,000 alleles; Bravo, 125,568 alleles), highly conserved across species from mammals down to Caenorhabditis elegans and Saccharomyces cerevisiae, and predicted as pathogenic by bioinformatics programs compiled by MobiDetails43 and including CADD, REVEL, SIFT, and PolyPhen 2 (Tables 6 and S3).
Figure 2.
CAMK2D missense variants identified in our cohort
(A) Schematic overview of the CAMK2D protein domain organization (top) and the corresponding messenger CAMK2D RNA structure (bottom), showing the location of the different missense variants as well as the location of the splice site variant. Four of the variants are located in the catalytic domain (c.275+1G>T, p.Ser79Asn, p.Pro139Leu, and p.Gly210Arg) and three are located in the regulatory domain (p.Gln274Pro, p.Arg275His, and p.Leu291Phe).
(B) Crystal structure of inhibited CAMK2D and regulatory segment (PDB: 2VN944), with the positions of the human variants highlighted as cyan sticks.
(C) The hydroxyl side chain of Ser79 forms an electrostatic interaction with the sidechain of His86. Asn at this position would be too bulky to accommodate this interaction.
(D) Pro139 facilitates a short helical turn in the C-lobe. Substitution to Leu would disrupt this.
(E) Gly210 is located at the base of a helix directly facing the regulatory segment. Arg at this position would be too bulky and likely dislodge the regulatory segment.
(F) Gln274 is located within a helical turn. The positioning of a Pro in this helix would disrupt folding of this region.
(G) Arg275 is located within the same turn and is proximal enough to Gln118 for hydrogen bonding (2.9 Å) and His115 (3.9 Å) for like-charged pair interaction45 between the two nitrogen atoms. Thus, substitution to a His would disrupt these interactions and the position of the alpha helix.
(H) Pictures of four individuals carrying a variant in CAMK2D. Note the coarse features and down-slanting palpebral fissures visible especially in individuals #2, #4, and #5, who also have a short nose and thin lips. Individual #1 has a long face with a high forehead, and she also has low-set ears, like individual #4.
The main clinical features of individuals with CAMK2D variants are summarized in Table 6. Additional details are provided in Table S3. With the exception of individual #8 who was too young at the time of consultation to have a cognitive assessment, all other individuals had mild to profound developmental delay or intellectual disability (7/7) characterized by speech (7/7, including 4 non-verbal individuals) and motor delay (7/7, including one individual unable to walk and 3 requiring assistance to walk), as well as behavioral problems (7/7, including 4 individuals with autism spectrum disorder and 1 with attention deficit hyperactivity disorder). Brain magnetic resonance imaging performed in 5/8 individuals revealed variable structural brain abnormalities, particularly a dilatation of lateral ventricles. Hypotonia was noted in 5/8 and seizures in 2/8 of the individuals. Strikingly, all but one of the individuals identified with a missense variant had dilated cardiomyopathy (6/7), in contrast to the only individual identified with a gene disruptive variant (#1), who had no cardiac symptoms or anomalies seen by echocardiography. Cardiomyopathy was severe in all the patients concerned, and cardiac transplantation was performed or planned in most cases after failure of symptomatic treatments (including angiotensin-converting enzyme inhibitors). Almost all individuals showed dysmorphic facial features (7/8; Figure 2H). The facial analysis by GestaltMatcher46,47 and GestaltMatcher Database48 showed that although the facial phenotype of CAMK2D was not similar to CAMK2A, CAMK2B, and CAMK2G on the cohort level analysis, the four individuals of CAMK2D showed a moderate degree of facial similarity (see GestaltMatcher analysis and Figures S1 and S2). Most individuals exhibited variable skeletal malformations (7/8), involving spine, hands, feet, or palate. Additional recurrent issues were severe gastrointestinal problems—especially dysphagia—often requiring tube feeding (4/6) and correlating with low muscle tone, short stature (3/8; from −2.1 to −3.3 SD), and visual anomalies (5/5, including 2 individuals with astigmatism and individuals with either cortical vision impairment, myopia, or strabismus).
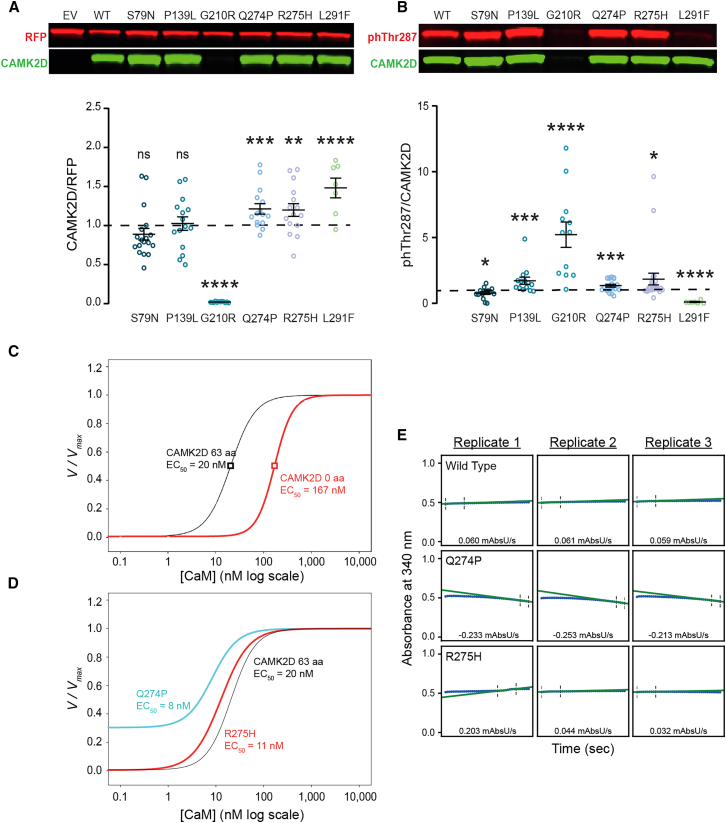
CAMK2D variants affect CAMK2D protein levels and kinase activity
To investigate how the variants of the eight individuals affected protein levels and/or kinase activity of CAMK2D, we expressed them in HEK293T cells and performed western blot analysis on the lysates. All data were normalized to CAMK2DWT expression (dotted horizontal line in Figures 3A and 3B). A significant increase in CAMK2D protein levels in HEK293T cells was observed for CAMK2DQ274P, CAMK2DR275H, and CAMK2DL291F, whereas CAMK2DG210R showed a significant decrease, i.e., very little expression. CAMK2DS79N and CAMK2DP139L showed similar protein levels to CAMK2DWT (Figure 3A). Statistical analysis can be found in Table 4.
Figure 3.
Assessment of protein levels and kinase activity
(A) Top: representative western blot of protein lysates from HEK293T cells that were transfected with either EV (empty control vector), CAMK2DWT, CAMK2DS79N, CAMK2DP139L, CAMK2DG210R, CAMK2DQ274P, CAMK2DR275H, or CAMK2DL291F, probed with an antibody against CAMK2D (green) and RFP (red). Bottom: Quantification of CAMK2D expression normalized against RFP, showing the effects on protein of the variants. Black dashed line represents CAMK2DWT.
(B) Top: representative western blot of protein lysates from HEK293T cells which were transfected with either EV (empty control vector), CAMK2DWT, CAMK2DS79N, CAMK2DP139L, CAMK2DG210R, CAMK2DQ274P, CAMK2DR275H, or CAMK2DL291F, probed with an antibody against CAMK2D (green), phThr287 (red). Bottom: Quantification of autophosphorylated CAMK2D (phThr287) normalized against CAMK2D protein level, showing the effects on autophosphorylation of the variants. All data were normalized against level of autophosphorylation in CAMK2DWT, represented by the black dashed line.
(C and D) On the y axis is the normalized rate of phosphorylation and on the x axis the respective calmodulin concentration. Data were normalized within the triplicate to the maximal rate. The hill coefficient (nH) and EC50 determined by the fits are listed below the corresponding CAMK2D variant for each plot. (C) Fits (see subjects and methods) of CAMK2D without linker (red) or with a 63-amino-acid linker region (black). (D) Fits of CAMK2DQ274P (cyan) and CAMK2DR275P (red) each with a 63-amino acid linker.
(E) Individual replicates of the coupled kinase assay in absence of CaM, to assess baseline kinase activity. A negative slope indicates increased kinase activity (blue line is the raw data; green line is the linear fit from which maximal slopes are calculated). Dots represent data points. Error bars indicate SEM; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Next, we assessed the enzymatic activity of the different CAMK2D constructs during expression in transfected cells by using autophosphorylation at Thr287 as a readout, i.e., the ratio of autophosphorylated vs. total expression of each protein. Most of the variants revealed increased levels of phThr287, varying from a small but significant increase for CAMK2DP139L, CAMK2DQ274P, and CAMK2DR275H to a robust phThr287 increase of the small amount of CAMK2DG210R that is expressed (Figure 3B). Overexpressing CAMK2DS79N resulted in a small but significant reduction of phThr287, whereas overexpression of CAMK2DL291F showed very little phThr287. For statistics, see Table 4.
To further investigate the kinase activity of the CAMK2DQ274P and CAMK2DR275H variants, where the increase in autophosphorylation appeared rather mild, in vitro kinase assays were performed. First, similar to CAMK2A data shown previously,32,40 a CAMK2D variant containing a 63-residue linker (6v1-14a-16-17-18, 63 aa) was more sensitive to Ca2+/CaM (lower EC50) compared to a CAMK2D variant containing no linker (Figure 3C). The CAMK2D-63 with an EC50 for Ca2+/CaM of 20 nM was used to assess the effect of the human variants on kinase activity.32 CAMK2DQ274P and CAMK2DR275H displayed slightly lower EC50 values of 8 nM and 11 nM, respectively (Figure 3D). These results suggest that the variants make it easier for CAMK2D to become enzymatically active, requiring lower levels of Ca2+/CaM. Strikingly, CAMK2DQ274P showed an elevated baseline, indicating there is kinase activity in the absence of Ca2+/CaM (Figure 3D). We compared the phosphorylation rates at zero Ca2+/CaM and found that CAMK2DQ274P consistently showed a negative slope (indicating kinase activity) whereas CAMK2DWT and CAMK2DR275H did not (Figure 3E). These data suggest that under conditions where calcium levels are low, p.Gln274Pro likely has elevated activity, which would be detrimental to the cell.
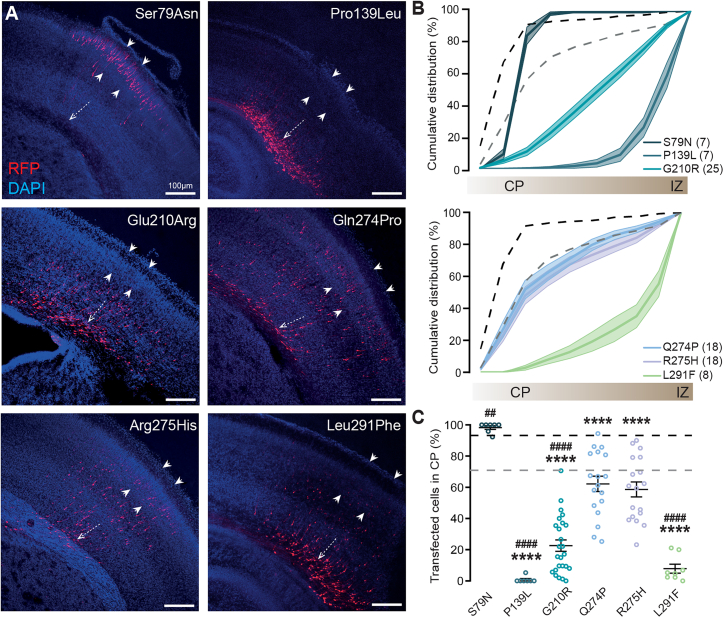
Overexpression of CAMK2D variants in utero affects neuronal migration
Having shown that neuronal migration is sensitive to altered protein levels of CAMK2D, the IUE assay was used as an additional functional assessment for pathogenicity of the variants. We compared the effect of overexpression of the different CAMK2D variants to overexpression of CAMK2DWT in the IUE assay. When overexpressed during development, CAMK2DP139L and CAMK2DG210R showed severe migration deficits compared to CAMK2DWT, with only 0.76% and 22.64% of the transfected neurons migrating to the outer cortical layers at P1 (Figure 4). Interestingly, CAMK2DS79N significantly improved neuronal migration compared to CAMK2DWT, where 98.31% of the neurons reached the CP, which is comparable to the empty vector control condition. CAMK2DQ274P and CAMK2DR275H showed similar behavior in the migration assay as CAMK2DWT, with an average of 62.21% and 58.67%, respectively, of transfected neurons migrated to their final destination (Figure 4). For statistics, see Tables 2 and 5. An overview of the functional assessments on the CAMK2D missense variants can be found in Table 7.
Figure 4.
Overexpression of CAMK2D variants during prenatal neurodevelopment causes a migration deficit
(A) Representative images of coronal brain slices from P0/P1 pups that were transfected with either CAMK2DS79N, CAMK2DP139L, CAMK2DG210R, CAMK2DQ274P, CAMK2DR275H, or CAMK2DL291F at E14.5, using in utero electroporation. White dashed arrow indicates the SVZ; white arrowheads indicate the CP. DAPI in blue, RFP in red. Scale bar indicates 100 μm.
(B) Cumulative graph indicating the migration pattern from the SVZ to the CP in presence of overexpressed CAMK2DS79N, CAMK2DP139L, CAMK2DG210R, CAMK2DQ274P, CAMK2DR275H, or CAMK2DL291F. Number in parentheses is the number of images used for the quantification. Black dashed line indicates EV (n = 12); gray dashed line indicates CAMK2DWT (n = 19) overexpression.
(C) Quantification of RFP-positive cells that have successfully migrated to the CP, showing the different effects of the variants on migration. # indicates the levels of significance compared to CAMK2DWT overexpression. ∗ indicates the levels of significance compared to the empty vector control (EV). Black dashed line represents EV, gray dashed line represents CAMK2DWT overexpression. Dots represent data points. Error bars indicate SEM; ∗∗/##p < 0.01; ∗∗∗∗/####p < 0.0001.
Table 7.
Overview of functional assessments of CAMK2D missense variants in molecular assays
| Protein stability compared to WT | Phosphotransferase activity compared to WT | Neuronal migration compared to WT | Inferred neuropathological mechanism | |
|---|---|---|---|---|
| Empty vector | increased | |||
| p.Ser79Asn | unaffected | mildly decreased | increased | loss-of-function |
| p.Pro139Leu | unaffected | increased | severely impaired | gain-of-function |
| p.Gly210Arg | severely decreased | increased | impaired | gain-of-function |
| p.Gln274Pro | increased | increased | similar | gain-of-function |
| p.Arg275His | increased | increased | similar | gain-of-function |
| p.Leu291Phe | increased | severely decreased | impaired | dominant negative |
Discussion
The CAMK2 protein family consists of four different isozymes, of which CAMK2A, CAMK2B, and CAMK2G have been shown to play a role in the human brain, causing neurodevelopmental disorders.19,22,23,24,25,26,27 In this study, we provide evidence for a role for the CAMK2D protein in neurodevelopment. First, we show that knockdown of Camk2d during neurodevelopment severely disrupts the neuronal migration in vivo, and second, we identified eight individuals with neurodevelopmental disorders carrying variants in CAMK2D, all presenting with a neurodevelopmental phenotype with significant delay across all domains (cognitive, motor, speech-language) and dysmorphic facial features. Additionally, the majority of individuals presented with dilated cardiomyopathy, further emphasizing the role of CAMK2D in cardiac functioning. Similar to what has been found for CAMK2A and CAMK2B, the variants are found in the kinase domain and in the regulatory domain and cause both loss-of-function and gain-of-function effects of CAMK2D. Although the total cohort of individuals harboring variants in one of the CAMK2 genes is small (44 published in total) and the clinical spectrum is broad, some overlapping and some unique clinical manifestations can be identified. Two common clinical manifestations seen in all CAMK2 individuals are intellectual disability, ranging from mild to profound, and delay in speech development. Similar to individuals with variants in CAMK2D, those associated with CAMK2A and CAMK2B variants may display seizures, brain imaging abnormalities, and digestive disorders, although this is not constant in all individuals and is more often seen with CAMK2B than CAMK2A variants.22,23,24,26,27,49,50 Interestingly, the two individuals carrying the same variant in CAMK2G do not show seizures or abnormalities of the digestive system.19 Distinct facial features and hypotonia have also been described for CAMK2A, CAMK2B, and CAMK2G, but again they are neither systematic nor homogeneous. Cardiac abnormalities are unique for the individuals harboring CAMK2D variants, as they are not found in CAMK2A-, CAMK2B-, or CAMK2G-related conditions. Identification and deep clinical phenotyping of additional individuals harboring variants in CAMK2 genes will further clarify the differences between and specificities of the different CAMK2-related conditions.
Our functional analyses indicate that CAMK2DS79N results in a possible loss of function (LoF) of CAMK2D, with slightly reduced phosphotransferase activity in HEK293T cells and improved migration compared to CAMK2DWT overexpression; CAMK2DP139L, CAMK2DG210R, CAMK2DQ274P, CAMK2DR275H, and CAMK2DL291F cause a GoF, all showing increased protein expression and/or phosphotransferase activity, and overexpression of CAMK2DP139L, CAMK2DG210R, and CAMK2DL291F induced a severe migration deficit, whereas for CAMK2DQ274P and CAMK2DR275H this migration deficit was milder, but still significantly worse than CAMK2DWT overexpression. Furthermore, CAMK2DQ274P and CAMK2DR275H had a reduced threshold to become enzymatically active (i.e., requiring less Ca2+/CaM). The CAMK2DQ274P variant revealed increased autonomous baseline activity compared to CAMK2DWT, showing enzymatic activity even in the absence of Ca2+/CaM. Interestingly, CAMK2DG210R had increased phosphotransferase activity but was expressed poorly in HEK293T cells, suggesting protein instability. Overexpression of this variant led to a severe migration deficit, suggesting that this variant potentially disrupts the CAMK2 holoenzymes endogenously expressed in the neurons that were transfected. However, it might still be that in the individual carrying this mutation, the expression levels are so low that the net effect in this individual is a LoF instead of a GoF effect.
Our findings confirm previous results, where the residue Arg274 in CAMK2A, equivalent to Arg275 in CAMK2D, was shown to be important for autoinhibition.51 Interestingly, Lys291 in CAMK2A, equivalent to Lys292 in CAMK2D, right next to the p.Leu291Phe variant identified in our cohort, is also found to play a role in autoinhibition. Finally, Glu139 in CAMK2A, equivalent to Glu140 in CAMK2D, is shown to be important for substrate binding and is right next to the p.Pro139Leu variant identified in our cohort.51 Our results suggest that residues Leu291 and Pro139 in CAMK2D have similar roles as their neighboring residues, but the exact mechanism remains to be studied. Of note, the CAMK2D p.Pro139Leu variant identified here is the equivalent to the CAMK2B p.Pro139Leu variant, which is the most recurrent variant previously identified by us and others in individuals with neurodevelopmental disorders.26,27 Whereas neurologically the phenotypes partially overlap (severe intellectual disability, delayed speech, behavioral anomalies), affected individuals with pathogenic CAMK2B variants do not show cardiac anomalies, consistent with the finding that CAMK2B is not expressed in the heart.52,53 CAMK2D p.Gln274Pro was found to have increased baseline autonomous Ca2+/CaM-independent activity. The importance of this region for autonomous activity has been shown for O-GlcNAc-modified Ser280, a few amino acids downstream.54 It could be hypothesized that the addition of a proline at this position alters the conformation of the CAMK2D regulatory domain so that binding to the catalytic domain necessary for autoinhibition is weakened.
CAMK2D is the major CAMK2 isozyme present in the heart30,52 and plays an important role in cardiac function. CAMK2D activity and expression is increased upon human heart failure,55,56 as well as upon experimentally induced heart failure.53,57,58 Elevated CAMK2D activity and/or expression has been associated in many studies with cardiac malfunctioning. Overexpression of a nuclear isoform of Camk2d induces hypertrophy in vitro and in vivo,59,60 and overexpression of a cytosolic isoform of rat Camk2d or human CAMK2D induces dilated cardiomyopathy and higher mortality,58,61 which is alleviated by CAMK2 inhibition.62 For arrhythmias and myocardial apoptosis, CAMK2D activation or overexpression is detrimental as well, whereas inhibition or knockdown is protective.61,63,64 Interestingly, knockout of Camk2d in mice does not alter heart function under basal conditions53,65,66 but rather protects against adverse cardiac remodeling and increases survival.66 In a preclinical model, inducible deletion of Camk2d and Camk2g (also expressed in the heart) after heart failure slowed down the progression of cardiac dysfunction.67 Finally, CRISPR-Cas9 gene therapy targeting Camk2d protected against cardiac dysfunction.68 This is consistent with our finding, which shows that most CAMK2D variants associated with dilated cardiomyopathy have GoF effects, and provides a clinical validation for CAMKD hyperactivity and cardiomyopathy. The only exception is individual #2, harboring the CAMK2DS79N variant, who also has dilated cardiomyopathy. This variant shows a LoF effect in our functional assays. It may be that this variant has a dominant-negative effect, undetected by our functional assays, or that other factors are at play. The individuals who do not have dilated cardiomyopathy carry the variant CAMK2DG210R and the frameshift variant CAMK2DV74E∗. CAMK2DV74E∗ likely causes loss of protein expression, due to the premature stop, hence haploinsufficiency, which is consistent with the studies mentioned above that suggest that loss of CAMK2D is not detrimental for cardiac function. However, the lack of cardiomyopathy in individual #4 harboring the CAMK2DG210R variant might be more puzzling, as we find a GoF effect. We found that upon expression in HEK293T cells, the expression levels were very low. Hence the GoF effect might be overruled by the low expression levels of this protein, resulting in a significant LoF.
We describe here a role for CAMK2D in human brain functioning that we did not find in our literature review. Indeed, all individuals identified in this cohort show intellectual disability, delayed speech, and gross motor delay, which (because of the severity) cannot be explained by or as a consequence of their cardiac phenotype. Whether the delay in speech and walking are indeed of neurological origin or alternatively caused by affected muscle tone remains to be understood.
That this so-called non-neuronal CAMK2 protein plays a role in neurodevelopment is not completely unexpected. Whereas CAMK2A and CAMK2B are the most abundant CAMK2 transcripts in the adult brain, during early neurodevelopment, CAMK2D and CAMK2G are the most abundant, both in rodents and humans.19,30 Indeed, CAMK2G has been shown previously to play a role in neurodevelopment, with pathogenic variants causing developmental delay, severe intellectual disability, hypotonia, and facial dysmorphic features.19 Similar clinical phenotypes are found in our CAMK2D cohort, described here. It could be hypothesized that the clinical phenotypes in CAMK2D- and CAMK2G-affected individuals are caused by alteration of early prenatal neurodevelopment, due to changes in early CAMK2 signaling, but that at later stages, the role of CAMK2D and CAMK2G in normal brain functioning diminishes, as CAMK2A and CAMK2B take over. For Camk2a and Camk2b it has been shown that they play an important role in the adult brain, using mouse models to induce deletion or bring back expression in adulthood.14,69,70,71 Similar inducible studies have not been done yet for Camk2d and Camk2g. Hence, further research is required to gain insight into the specific roles of the CAMK2 isozymes during different stages of neurodevelopment and in the adult brain.
Taken together we report a cohort of individuals harboring variants in CAMK2D, presenting with neurodevelopmental disorders and dilated cardiomyopathy. Our findings further emphasize the important roles for CAMK2D in heart function and in normal brain development and function.
Appendix A
Details about the statistical tests that have been performed on results in the molecular assays can be found in Tables 1, 2, 3, 4, and 5.
Data and code availability
All variants identified are submitted to ClinVar. The functional datasets used for Figures 1, 3, and 4, supporting the current study have not been deposited in a public repository because sequences for cloning was obtained from public databases, and no public repository exist to upload raw western blot or other experimental data, but are available from the corresponding author on request.
Acknowledgments
We would like to thank the families of the affected individuals described in this paper for their help and their willingness to contribute to this study. This research was supported by the NWO-VIDI (016.Vidi.188.014 to G.M.v.W.). Individual 8 was tested as part of the Acute Care Genomics study (GHFM76747).
Author contributions
G.M.v.W., S.K., M.M.S., and D.C.M.V. designed the study. P.M.F.R., C.d.K., M.J.D., M.P.O., E.H., C.N.J., L.A., B.Y., S.P., M.M.S., and G.M.v.W. performed functional experiments and analysis. T.-C.H., P.M.K., and M.E. performed the facial analysis. A.B. and I.M.W. performed analysis and reporting of GeneDx cases, J.A.R. and F.X. handled the Baylor Genetics cases. S.K., J.B.H., M.T., C.B., C.E.P., K.N.W., T.D.R., O.C., J.C., E.C., C.-T.F., W.W., M.M., D.B., Z.S., E.S., C.P., F.M., E.B., C.P.B., S.B., and S.M. were involved in case identification and/or data collection and contributed to the clinical information of patients. P.M.F.R., C.d.K., M.M.S., S.K., H.S., and G.M.v.W. drafted the manuscript. All authors contributed to the final version of the manuscript.
Declaration of interests
The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing completed at Baylor Genetics Laboratories. A.B. and I.M.W. are employees of GeneDx, LLC; H.S. is a consultant for Vasa Therapeutics (Poland).
Published: January 24, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2023.12.016.
Contributor Information
Sébastien Küry, Email: sebastien.kury@chu-nantes.fr.
Geeske M. van Woerden, Email: g.vanwoerden@erasmusmc.nl.
Web resources
OMIM, https://www.omim.org/
Supplemental information
References
- 1.Bhattacharyya M., Stratton M.M., Going C.C., McSpadden E.D., Huang Y., Susa A.C., Elleman A., Cao Y.M., Pappireddi N., Burkhardt P., et al. Molecular mechanism of activation-triggered subunit exchange in Ca2+/calmodulin-dependent protein kinase II. Elife. 2016;5 doi: 10.7554/eLife.13405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lisman J., Yasuda R., Raghavachari S. Mechanisms of CaMKII action in long-term potentiation. Nat. Rev. Neurosci. 2012;13:169–182. doi: 10.1038/nrn3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hell J.W. CaMKII: Claiming center stage in postsynaptic function and organization. Neuron. 2014;81:249–265. doi: 10.1016/j.neuron.2013.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaertner T.R., Kolodziej S.J., Wang D., Kobayashi R., Koomen J.M., Stoops J.K., Waxham M.N. Comparative Analyses of the Three-dimensional Structures and Enzymatic Properties of α, β, γ, and δ Isoforms of Ca2+-Calmodulin-dependent Protein Kinase II. J. Biol. Chem. 2004;279:12484–12494. doi: 10.1074/jbc.M313597200. [DOI] [PubMed] [Google Scholar]
- 5.Hudmon A., Schulman H. Neuronal Ca2+/Calmodulin-Dependent Protein Kinase II: The Role of Structure and Autoregulation in Cellular Function. Annu. Rev. Biochem. 2002;71:473–510. doi: 10.1146/annurev.biochem.71.110601.135410. [DOI] [PubMed] [Google Scholar]
- 6.Silva A.J., Paylor R., Wehner J.M., Tonegawa S. Impaired spatial learning in α-calcium-calmodulin kinase II mutant mice. Science. 1992;257:206–211. doi: 10.1126/science.1321493. [DOI] [PubMed] [Google Scholar]
- 7.Giese K.P., Fedorov N.B., Filipkowski R.K., Silva A.J. Autophosphorylation at Thr286 of the α calcium-calmodulin kinase II in LTP and learning. Science. 1998;279:870–873. doi: 10.1126/science.279.5352.870. [DOI] [PubMed] [Google Scholar]
- 8.Elgersma Y., Fedorov N.B., Ikonen S., Choi E.S., Elgersma M., Carvalho O.M., Giese K.P., Silva A.J. Inhibitory autophosphorylation of CaMKII controls PSD association, plasticity, and learning. Neuron. 2002;36:493–505. doi: 10.1016/s0896-6273(02)01007-3. [DOI] [PubMed] [Google Scholar]
- 9.Silva A.J., Stevens C.F., Tonegawa S., Wang Y. Deficient hippocampal long-term potentiation in α-calcium-calmodulin kinase II mutant mice. Science. 1992;257:201–206. doi: 10.1126/science.1378648. [DOI] [PubMed] [Google Scholar]
- 10.Van Woerden G.M., Hoebeek F.E., Gao Z., Nagaraja R.Y., Hoogenraad C.C., Kushner S.A., Hansel C., De Zeeuw C.I., Elgersma Y. βCaMKII controls the direction of plasticity at parallel fiber-Purkinje cell synapses. Nat. Neurosci. 2009;12:823–825. doi: 10.1038/nn.2329. [DOI] [PubMed] [Google Scholar]
- 11.Borgesius N.Z., van Woerden G.M., Buitendijk G.H.S., Keijzer N., Jaarsma D., Hoogenraad C.C., Elgersma Y. βCaMKII plays a nonenzymatic role in hippocampal synaptic plasticity and learning by targeting αCaMKII to synapses. J. Neurosci. 2011;31:10141–10148. doi: 10.1523/JNEUROSCI.5105-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho M.H., Cao X., Wang D., Tsien J.Z. Dentate gyrus-specific manipulation of β-Ca2+/calmodulin-dependent kinase II disrupts memory consolidation. Proc. Natl. Acad. Sci. USA. 2007;104:16317–16322. doi: 10.1073/pnas.0703344104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bachstetter A.D., Webster S.J., Tu T., Goulding D.S., Haiech J., Watterson D.M., Van Eldik L.J. Generation and Behavior Characterization of CaMKIIβ Knockout Mice. PLoS One. 2014;9 doi: 10.1371/journal.pone.0105191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kool M.J., Proietti Onori M., Borgesius N.Z., Van De Bree J.E., Elgersma-Hooisma M., Nio E., Bezstarosti K., Buitendijk G.H.S., Aghadavoud Jolfaei M., Demmers J.A.A., et al. CAMK2-Dependent Signaling in Neurons Is Essential for Survival. J. Neurosci. 2019;39:5424–5439. doi: 10.1523/JNEUROSCI.1341-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen S.M., Suutari B., He X., Wang Y., Sanchez S., Tirko N.N., Mandelberg N.J., Mullins C., Zhou G., Wang S., et al. Calmodulin shuttling mediates cytonuclear signaling to trigger experience-dependent transcription and memory. Nat. Commun. 2018;9:2451. doi: 10.1038/s41467-018-04705-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He X., Li J., Zhou G., Yang J., McKenzie S., Li Y., Li W., Yu J., Wang Y., Qu J., et al. Gating of hippocampal rhythms and memory by synaptic plasticity in inhibitory interneurons. Neuron. 2021;109:1013–1028.e9. doi: 10.1016/j.neuron.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zalcman G., Federman N., Fiszbein A., de la Fuente V., Ameneiro L., Schor I., Romano A. Sustained CaMKII Delta Gene Expression Is Specifically Required for Long-Lasting Memories in Mice. Mol. Neurobiol. 2019;56:1437–1450. doi: 10.1007/s12035-018-1144-3. [DOI] [PubMed] [Google Scholar]
- 18.Ma H., Groth R.D., Cohen S.M., Emery J.F., Li B., Hoedt E., Zhang G., Neubert T.A., Tsien R.W. γCaMKII shuttles Ca2+/CaM to the nucleus to trigger CREB phosphorylation and gene expression. Cell. 2014;159:281–294. doi: 10.1016/j.cell.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Proietti Onori M., Koopal B., Everman D.B., Worthington J.D., Jones J.R., Ploeg M.A., Mientjes E., van Bon B.W., Kleefstra T., Schulman H., et al. The intellectual disability-associated CAMK2G p.Arg292Pro mutation acts as a pathogenic gain-of-function. Hum. Mutat. 2018;39:2008–2024. doi: 10.1002/humu.23647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rigter P.M.F., de Konink C., van Woerden G.M. Loss of CAMK2G affects intrinsic and motor behavior but has minimal impact on cognitive behavior. Front. Neurosci. 2023;16:1–14. doi: 10.3389/fnins.2022.1086994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Ligt J., Willemsen M.H., Van Bon B.W.M., Kleefstra T., Yntema H.G., Kroes T., Vulto-van Silfhout A.T., Koolen D.A., de Vries P., Gilissen C., et al. Diagnostic Exome Sequencing in Persons with Severe Intellectual Disability. N. Engl. J. Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 22.Akita T., Aoto K., Kato M., Shiina M., Mutoh H., Nakashima M., Kuki I., Okazaki S., Magara S., Shiihara T., et al. De novo variants in CAMK2A and CAMK2B cause neurodevelopmental disorders. Ann. Clin. Transl. Neurol. 2018;5:280–296. doi: 10.1002/acn3.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chia P.H., Zhong F.L., Niwa S., Bonnard C., Utami K.H., Zeng R., Lee H., Eskin A., Nelson S.F., Xie W.H., et al. A homozygous loss-of-function CAMK2A mutation causes growth delay, frequent seizures and severe intellectual disability. Elife. 2018;7:e32519. doi: 10.7554/eLife.32451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heiman P., Drewes S., Ghaloul-Gonzalez L. A familial case of CAMK2B mutation with variable expressivity. SAGE Open Med. Case Reports. 2021;9 doi: 10.1177/2050313X21990982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iossifov I., O’Roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D., Stessman H.A., Witherspoon K.T., Vives L., Patterson K.E., et al. The contribution of de novo coding mutations to autism spectrum disorder. Nat. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Küry S., van Woerden G.M., Besnard T., Proietti Onori M., Latypova X., Towne M.C., Cho M.T., Prescott T.E., Ploeg M.A., Sanders S., et al. De Novo Mutations in Protein Kinase Genes CAMK2A and CAMK2B Cause Intellectual Disability. Am. J. Hum. Genet. 2017;101:768–788. doi: 10.1016/j.ajhg.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rizzi S., Spagnoli C., Salerno G.G., Frattini D., Caraffi S.G., Trimarchi G., Moratti C., Pascarella R., Garavelli L., Fusco C. Severe intellectual disability, absence of language, epilepsy, microcephaly and progressive cerebellar atrophy related to the recurrent de novo variant p.(P139L) of the CAMK2B gene: A case report and brief review. Am. J. Med. Genet. 2020;182:2675–2679. doi: 10.1002/ajmg.a.61803. [DOI] [PubMed] [Google Scholar]
- 28.Proietti Onori M., van Woerden G.M. Role of calcium/calmodulin-dependent kinase 2 in neurodevelopmental disorders. Brain Res. Bull. 2021;171:209–220. doi: 10.1016/j.brainresbull.2021.03.014. [DOI] [PubMed] [Google Scholar]
- 29.Tobimatsu T., Fujisawa H. Tissue-specific expression of four types of rat calmodulin-dependent protein kinase II mRNAs. J. Biol. Chem. 1989;264:17907–17912. [PubMed] [Google Scholar]
- 30.Bayer K.U., Löhler J., Schulman H., Harbers K. Developmental expression of the CaM kinase II isoforms: Ubiquitous γ- and δ-CaM kinase II are the early isoforms and most abundant in the developing nervous system. Mol. Brain Res. 1999;70:147–154. doi: 10.1016/s0169-328x(99)00131-x. [DOI] [PubMed] [Google Scholar]
- 31.Srinivasan M., Edman C.F., Schulman H. Alternative Splicing Introduces a Nuclear Localization Signal That Targets Multifunctional CaM Kinase to the Nucleus. J. Cell Biol. 1994;126:839–852. doi: 10.1083/jcb.126.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sloutsky R., Dziedzic N., Dunn M.J., Bates R.M., Torres-Ocampo A.P., Boopathy S., Page B., Weeks J.G., Chao L.H., Stratton M.M. Heterogeneity in human hippocampal CaMKII transcripts reveals allosteric hub-dependent regulation. Sci. Signal. 2020;13 doi: 10.1126/scisignal.aaz0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakagami H., Kondo H. Differential expression of mRNAs encoding γ and δ subunits of Ca2+/calmodulin-dependent protein kinase type II (CaM kinase II) in the mature and postnatally developing rat brain. Mol. Brain Res. 1993;20:51–63. doi: 10.1016/0169-328x(93)90109-3. [DOI] [PubMed] [Google Scholar]
- 34.Murray K.D., Isackson P.J., Jones E.G. N-methyl-d-aspartate receptor dependent transcriptional regulation of two calcium/calmodulin-dependent protein kinase type II isoforms in rodent cerebral cortex. Neuroscience. 2003;122:407–420. doi: 10.1016/j.neuroscience.2003.07.015. [DOI] [PubMed] [Google Scholar]
- 35.Huntley M.A., Srinivasan K., Friedman B.A., Wang T.M., Yee A.X., Wang Y., Kaminker J.S., Sheng M., Hansen D.V., Hanson J.E. Genome-wide analysis of differential gene expression and splicing in excitatory neurons and interneuron subtypes. J. Neurosci. 2020;40:958–973. doi: 10.1523/JNEUROSCI.1615-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vallano M.L., Beaman-Hall C.M., Mathur A., Chen Q. Astrocytes Express Specific Variants of CaM KII δ and γ, But Not α and β, That Determine Their Cellular Localizations. Glia. 2000;30:154–164. doi: 10.1002/(sici)1098-1136(200004)30:2<154::aid-glia5>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 37.Takeuchi Y., Yamamoto H., Fukunaga K., Miyakawa T., Miyamoto E. Identification of the Isoforms of Ca2+/Calmodulin-Dependent Protein Kinase II in Rat Astrocytes and Their Subcellular Localization. J. Neurochem. 2000;74:2557–2567. doi: 10.1046/j.1471-4159.2000.0742557.x. [DOI] [PubMed] [Google Scholar]
- 38.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sloutsky R., Stratton M.M. Functional implications of CaMKII alternative splicing. Eur. J. Neurosci. 2021;54:6780–6794. doi: 10.1111/ejn.14761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chao L.H., Pellicena P., Deindl S., Barclay L.A., Schulman H., Kuriyan J. Intersubunit capture of regulatory segments is a component of cooperative CaMKII activation. Nat. Struct. Mol. Biol. 2010;17:264–272. doi: 10.1038/nsmb.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harmat V., Böcskei Z., Náray-Szabó G., Bata I., Csutor A.S., Hermecz I., Arányi P., Szabó B., Liliom K., Vértessy B.G., Ovádi J. A new potent calmodulin antagonist with arylalkylamine structure: crystallographic, spectroscopic and functional studies. J. Mol. Biol. 2000;297:747–755. doi: 10.1006/jmbi.2000.3607. [DOI] [PubMed] [Google Scholar]
- 42.Barker S.C., Kassel D.B., Weigl D., Huang X., Luther M.A., Knight W.B. Characterization of pp60c-src Tyrosine Kinase Activities Using a Continuous Assay: Autoactivation of the Enzyme Is an Intermolecular Autophosphorylation Process. Biochemistry. 1995;34:14843–14851. doi: 10.1021/bi00045a027. [DOI] [PubMed] [Google Scholar]
- 43.Baux D., Van Goethem C., Ardouin O., Guignard T., Bergougnoux A., Koenig M., Roux A.F. MobiDetails: online DNA variants interpretation. Eur J Hum Genet. 2021;29:356–360. doi: 10.1038/s41431-020-00755-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rellos P., Pike A.C.W., Niesen F.H., Salah E., Lee W.H., von Delft F., Knapp S. Structure of the CaMKIIδ/Calmodulin Complex Reveals the Molecular Mechanism of CaMKII Kinase Activation. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heyda J., Vincent J.C., Tobias D.J., Dzubiella J., Jungwirth P. Ion specificity at the peptide bond: Molecular dynamics simulations of N-methylacetamide in aqueous salt solutions. J. Phys. Chem. B. 2010;114:1213–1220. doi: 10.1021/jp910953w. [DOI] [PubMed] [Google Scholar]
- 46.Hsieh T.-C., Bar-Haim A., Moosa S., Ehmke N., Gripp K.W., Pantel J.T., Danyel M., Mensah M.A., Horn D., Rosnev S., et al. GestaltMatcher facilitates rare disease matching using facial phenotype descriptors. Nat. Genet. 2022;54:349–357. doi: 10.1038/s41588-021-01010-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hustinx A., Hellmann F., Sümer Ö., Javanmardi B., André E., Krawitz P., Hsieh T.-C. Improving Deep Facial Phenotyping for Ultra-rare Disorder Verification Using Model Ensembles. Proc. - 2023 IEEE Winter Conf. Appl. Comput. Vision, WACV. 2023:5017. [Google Scholar]
- 48.Lesmann H., Lyon G.J., Caro P., Abdelrazek I.M., Moosa S., Pantel J.T., Ten Hagen M., Rosnev S., Kamphans T., Meiswinkel W., et al. GestaltMatcher Database - a FAIR Database for Medical Imaging Data of Rare Disorders. medRxiv. 2023 doi: 10.1101/2023.06.06.23290887. Preprint at. [DOI] [Google Scholar]
- 49.Fujii H., Kidokoro H., Kondo Y., Kawaguchi M., Horigane S.I., Natsume J., Takemoto-Kimura S., Bito H. Förster resonance energy transfer-based kinase mutation phenotyping reveals an aberrant facilitation of Ca2+/calmodulin-dependent CaMKIIα activity in de novo mutations related to intellectual disability. Front. Mol. Neurosci. 2022;15:970031. doi: 10.3389/fnmol.2022.970031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dwyer B.K., Veenma D.C.M., Chang K., Schulman H., Van Woerden G.M. Case Report : Developmental Delay and Acute Neuropsychiatric Episodes Associated With a de novo Mutation in the CAMK2B Gene (c.328G>A p.Glu110Lys) Front. Pharmacol. 2022;13:1–9. doi: 10.3389/fphar.2022.794008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang E., Schulman H. Structural examination of autoregulation of multifunctional calcium/calmodulin-dependent protein kinase II. J. Biol. Chem. 1999;274:26199–26208. doi: 10.1074/jbc.274.37.26199. [DOI] [PubMed] [Google Scholar]
- 52.Tombes R.M., Faison M.O., Turbeville J.M. Organization and evolution of multifunctional Ca2+/CaM- dependent protein kinase genes. Gene. 2003;322:17–31. doi: 10.1016/j.gene.2003.08.023. [DOI] [PubMed] [Google Scholar]
- 53.Kreusser M.M., Lehmann L.H., Keranov S., Hoting M.O., Oehl U., Kohlhaas M., Reil J.C., Neumann K., Schneider M.D., Hill J.A., et al. Cardiac CaM kinase II genes δ and γ contribute to adverse remodeling but redundantly inhibit calcineurin-induced myocardial hypertrophy. Circulation. 2014;130:1262–1273. doi: 10.1161/CIRCULATIONAHA.114.006185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Erickson J.R., Pereira L., Wang L., Han G., Ferguson A., Dao K., Copeland R.J., Despa F., Hart G.W., Ripplinger C.M., Bers D.M. Diabetic hyperglycaemia activates CaMKII and arrhythmias by O-linked glycosylation. Nature. 2013;502:372–376. doi: 10.1038/nature12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kirchhefer U., Schmitz W., Scholz H., Neumann J. Activity of cAMP-dependent protein kinase and Ca2+/calmodulin-dependent protein kinase in failing and nonfailing human hearts. Cardiovasc. Res. 1999;42:254–261. doi: 10.1016/s0008-6363(98)00296-x. [DOI] [PubMed] [Google Scholar]
- 56.Hoch B., Meyer R., Hetzer R., Krause E.G., Karczewski P. Identification and Expression of δ-Isoforms of the Multifunctional Ca2+/Calmodulin-Dependent Protein Kinase in Failing and Nonfailing Human Myocardium. Circ. Res. 1999;84:713–721. doi: 10.1161/01.res.84.6.713. [DOI] [PubMed] [Google Scholar]
- 57.Colomer J.M., Mao L., Rockman H.A., Means A.R. Pressure Overload Selectively Up-Regulates Ca2+/Calmodulin-Dependent Protein Kinase II in Vivo. Mol. Endocrinol. 2003;17:183–192. doi: 10.1210/me.2002-0350. [DOI] [PubMed] [Google Scholar]
- 58.Zhang T., Maier L.S., Dalton N.D., Miyamoto S., Ross J., Bers D.M., Brown J.H. The δC Isoform of CaMKII Is Activated in Cardiac Hypertrophy and Induces Dilated Cardiomyopathy and Heart Failure. Circ. Res. 2003;92:912–919. doi: 10.1161/01.RES.0000069686.31472.C5. [DOI] [PubMed] [Google Scholar]
- 59.Ramirez M.T., Zhao X.L., Schulman H., Brown J.H. The nuclear δ(B) isoform of Ca2+/calmodulin-dependent protein kinase II regulates atrial natriuretic factor gene expression in ventricular myocytes. J. Biol. Chem. 1997;272:31203–31208. doi: 10.1074/jbc.272.49.31203. [DOI] [PubMed] [Google Scholar]
- 60.Zhang T., Johnson E.N., Gu Y., Morissette M.R., Sah V.P., Gigena M.S., Belke D.D., Dillmann W.H., Rogers T.B., Schulman H., et al. The Cardiac-specific Nuclear δB Isoform of Ca2+/Calmodulin-dependent Protein Kinase II Induces Hypertrophy and Dilated Cardiomyopathy Associated with Increased Protein Phosphatase 2A Activity. J. Biol. Chem. 2002;277:1261–1267. doi: 10.1074/jbc.M108525200. [DOI] [PubMed] [Google Scholar]
- 61.Zhang M., Gao H., Liu D., Zhong X., Shi X., Yu P., Jin L., Liu Y., Tang Y., Song Y., et al. CaMKII-δ9 promotes cardiomyopathy through disrupting UBE2T-dependent DNA repair. Nat. Cell Biol. 2019;21:1152–1163. doi: 10.1038/s41556-019-0380-8. [DOI] [PubMed] [Google Scholar]
- 62.Zhang M., Zhang J., Zhang W., Hu Q., Jin L., Xie P., Zheng W., Shang H., Zhang Y. CaMKII-δ9 Induces Cardiomyocyte Death to Promote Cardiomyopathy and Heart Failure. Front. Cardiovasc. Med. 2022;8:2163. doi: 10.3389/fcvm.2021.820416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu Y., Temple J., Zhang R., Dzhura I., Zhang W., Trimble R., Roden D.M., Passier R., Olson E.N., Colbran R.J., et al. Calmodulin Kinase II and Arrhythmias in a Mouse Model of Cardiac Hypertrophy. Circulation. 2002;106:1288–1293. doi: 10.1161/01.cir.0000027583.73268.e7. [DOI] [PubMed] [Google Scholar]
- 64.Vila-Petroff M., Salas M.A., Said M., Valverde C.A., Sapia L., Portiansky E., Hajjar R.J., Kranias E.G., Mundiña-Weilenmann C., Mattiazzi A. CaMKII inhibition protects against necrosis and apoptosis in irreversible ischemia-reperfusion injury. Cardiovasc. Res. 2007;73:689–698. doi: 10.1016/j.cardiores.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 65.Backs J., Backs T., Neef S., Kreusser M.M., Lehmann L.H., Patrick D.M., Grueter C.E., Qi X., Richardson J.A., Hill J.A., et al. The δ isoform of CaM kinase II is required for pathological cardiac hypertrophy and remodeling after pressure overload. Proc. Natl. Acad. Sci. USA USA. 2009;106:2342–2347. doi: 10.1073/pnas.0813013106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ling H., Zhang T., Pereira L., Means C.K., Cheng H., Gu Y., Dalton N.D., Peterson K.L., Chen J., Bers D., Brown J.H. Requirement for Ca2+/calmodulin–dependent kinase II in the transition from pressure overload–induced cardiac hypertrophy to heart failure in mice. J. Clin. Invest. 2009;119:1230–1240. doi: 10.1172/JCI38022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kreusser M.M., Lehmann L.H., Wolf N., Keranov S., Jungmann A., Gröne H.J., Müller O.J., Katus H.A., Backs J. Inducible cardiomyocyte-specific deletion of CaM kinase II protects from pressure overload-induced heart failure. Basic Res. Cardiol. 2016;111:65–69. doi: 10.1007/s00395-016-0581-2. [DOI] [PubMed] [Google Scholar]
- 68.Lebek S., Chemello F., Caravia X.M., Tan W., Li H., Chen K., Xu L., Liu N., Bassel-Duby R., Olson E.N. Ablation of CaMKIIδ oxidation by CRISPR-Cas9 base editing as a therapy for cardiac disease. Science. 2023;379:179–185. doi: 10.1126/science.ade1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Achterberg K.G., Buitendijk G.H.S., Kool M.J., Goorden S.M.I., Post L., Slump D.E., Silva A.J., van Woerden G.M., Kushner S.A., Elgersma Y. Temporal and region-specific requirements of αCaMKII in spatial and contextual learning. J. Neurosci. 2014;34:11180–11187. doi: 10.1523/JNEUROSCI.0640-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kool M.J., Van De Bree J.E., Bodde H.E., Elgersma Y., Van Woerden G.M. The molecular, temporal and region-specific requirements of the beta isoform of Calcium/Calmodulin-dependent protein kinase type 2 (CAMK2B) in mouse locomotion. Sci. Rep. 2016;6:26989. doi: 10.1038/srep26989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rigter P.M.F., Wallaard I., Aghadavoud Jolfaei M., Kingma J., Post L., Elgersma M., Elgersma Y., van Woerden G.M. Adult Camk2a gene reinstatement restores the learning and plasticity deficits of Camk2a knockout mice. iScience. 2022;25 doi: 10.1016/j.isci.2022.105303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All variants identified are submitted to ClinVar. The functional datasets used for Figures 1, 3, and 4, supporting the current study have not been deposited in a public repository because sequences for cloning was obtained from public databases, and no public repository exist to upload raw western blot or other experimental data, but are available from the corresponding author on request.