ABSTRACT
We have optimized a clustered regularly interspaced short palindromic repeat (CRISPR) interference system to facilitate gene knockdown in the Gram-positive bacterial pathogen Staphylococcus aureus. Our approach used a CRISPRi system derived from Streptococcus pyogenes, which involves the co-expression of the dcas9 gene encoding a catalytically inactive Cas9 protein and a customizable single guide RNA (sgRNA). In our system, dcas9 is expressed from a single copy in the chromosome of methicillin-resistant S. aureus strains COL or JE2, under the control of a tightly regulated promoter, inducible by anhydrotetracycline. The sgRNAs are expressed from a replicative plasmid under the control of a constitutively active promoter. This system enables efficient, inducible, knockdown of both essential and non-essential genes. Using this approach, we constructed the Lisbon CRISPRi Mutant Library comprising 261 strains, in the JE2 background, containing sgRNAs targeting 200 essential genes/operons. This library facilitates the study of the function of essential S. aureus genes and is complementary to the Nebraska Transposon Mutant Library, which consists of nearly 2,000 strains, each carrying a transposon insertion within a non-essential gene. The availability of these two libraries will facilitate the study of S. aureus pathogenesis and biology.
IMPORTANCE
Staphylococcus aureus is an important clinical pathogen that causes a high number of antibiotic-resistant infections. The study of S. aureus biology, and particularly of the function of essential proteins, is of particular importance to develop new approaches to combat this pathogen. We have optimized a clustered regularly interspaced short palindromic repeat interference (CRISPRi) system that allows efficient targeting of essential S. aureus genes. Furthermore, we have used that system to construct a library comprising 261 strains, which allows the depletion of essential proteins encoded by 200 genes/operons. This library, which we have named Lisbon CRISPRi Mutant Library, should facilitate the study of S. aureus pathogenesis and biology.
KEYWORDS: Staphylococcus aureus, CRISPRi, essential genes
INTRODUCTION
Staphylococcus aureus is a Gram-positive bacterium that frequently colonizes the skin and nares of both humans and animals. It is also an opportunistic pathogen in community and hospital settings, causing a range of clinical conditions such as skin and soft tissue infections, bacteremia, or endocarditis (1). The emergence of multi-drug resistant strains, particularly methicillin-resistant S. aureus (MRSA), has complicated the treatment of S. aureus infections, and MRSA strains are currently the second most common cause of global deaths associated with bacterial antimicrobial resistance (2). The discovery of novel antibiotics with unique modes of action against S. aureus is crucial for combating multidrug-resistant strains. One approach to address this challenge is the study of essential genes that encode core proteins required for bacterial survival, as these genes may encode potential targets for new antimicrobial drugs.
Despite their importance, the function of many essential genes in S. aureus remains poorly understood. Gene inactivation or repression are common approaches to elucidate the molecular function of genes in bacteria. Genetic tools to disrupt S. aureus genes or impair their transcription have become increasingly available (3). However, classical approaches such as gene deletion or transposon mutagenesis cannot be used to study essential genes, as they play indispensable roles in bacterial survival. Several alternative strategies have been developed including the exchange of the endogenous promoter of a gene by an exogenous inducible promoter via allelic exchange, or the use of antisense RNA technology (4, 5). However, the former approach is labor-intensive and unsuitable for large-scale studies, while the latter has limitations due to variable efficacy.
In recent years, new tools based on CRISPR (clustered regularly interspaced short palindromic repeats) interference systems have emerged as powerful tools to control gene expression. The most widely used CRISPRi system is derived from Streptococcus pyogenes and involves the co-expression of a gene encoding a catalytically inactive or dead (d)Cas9Spy protein, which lacks endonuclease activity, and a customizable single guide RNA (sgRNA) (6, 7). The dCas9-sgRNA complex binds to the DNA target that is complementary to the sgRNA. Instead of introducing double-strand breaks, this complex causes a steric block that halts transcription by RNA polymerase, leading to repression of the target gene or operon. Besides the complementary nucleotide sequence, the only other prerequisite for the complex to bind DNA is a three nucleotide long (NGG) recognition motif downstream of the complementary region of the targeted DNA strand, known as the protospacer-adjacent motif (PAM).
CRISPRi systems based on S. pyogenes Cas9 (a type II CRISPR) have been established in several model organisms, including Escherichia coli (8), Bacillus subtilis (9), Streptococcus pneumoniae (10), or S. aureus (11–14). Type II CRISPR systems have been found in other bacteria besides S. pyogenes, including in S. aureus. Similar to CRISPR-Cas9Spy, these systems can be reprogrammed to serve as molecular tools for genome editing in eukaryotic cells (15, 16). The Cas9 from S. aureus (Cas9Sa) has raised some interest due to its smaller size compared to S. pyogenes Cas9Spy (1,053 versus 1,368 amino acids), hence allowing the use of adeno-associated viruses with restrictive cargo sizes as vehicles for delivering Cas9 to animal cells (16). Additionally, Cas9Sa requires a longer PAM sequence (NNGRRT) (17), which increases the specificity of DNA targeting and, therefore, reduces the potential for off-target effects, a concern when manipulating large genomes.
Previous CRISPRi systems for the genetic manipulation of S. aureus use plasmids to encode the sgRNA and the dCas9, enabling the system to be easily moved into various strains (11–14). However, the use of multicopy plasmids for dCas9 expression, combined with the fact that most inducible promoters used in S. aureus cannot be fully repressed, often leads to systems in which dCas9 production cannot be completely shut down. This becomes particularly problematic when targeting essential genes, as basal levels of dCas9 can result in impaired growth or cell death. Here, we report the construction of two variations of a gene knockdown system in S. aureus, based on the CRISPRi system originally established in E. coli by Qi and colleagues (8). The first system consists of two shuttle vectors, one encoding the dCas9 of either S. pyogenes or S. aureus and the other encoding the corresponding sgRNA with the gene-specific target sequence. The second system allows for efficient regulation of dCas9Spy production, as the gene encoding this protein was integrated into the genome under the control of a tightly controlled inducible promoter. This system was subsequently used to generate a knockdown library of 261 genes from 200 reported essential genes/operons in S. aureus, which we named the Lisbon CRISPRi Mutant Library (LCML). The LCML serves as a complementary resource to the widely used Nebraska Transposon Mutant Library, which includes mutants in virtually all non-essential genes in S. aureus (18). The use of both libraries will facilitate comprehensive functional studies of staphylococcal genes/operons.
RESULTS
Construction of two-plasmid systems for CRISPR interference in Staphylococcus aureus
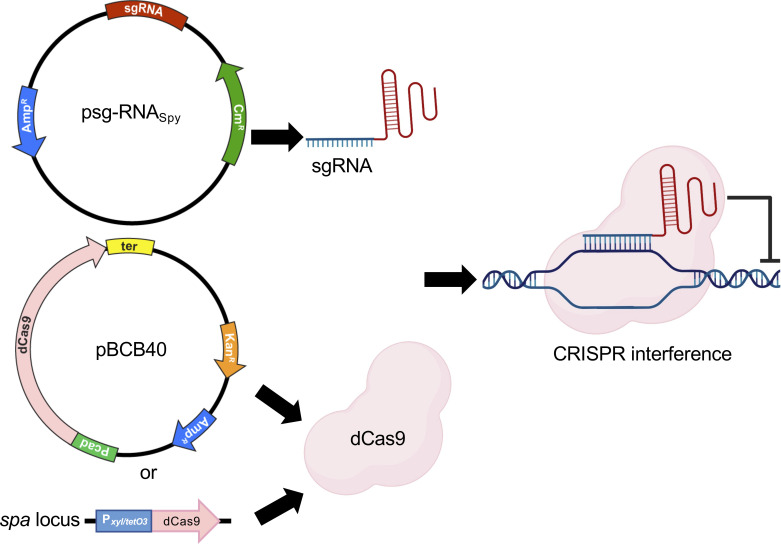
To establish a CRISPRi system in S. aureus, we chose two E. coli-S. aureus shuttle vectors to enable initial propagation and genetic manipulation in E. coli, followed by introduction into S. aureus by electroporation. S. aureus is not naturally competent, and transformation efficiency is very low compared to other model organisms (19, 20). The genes encoding a catalytically dead Cas9 from either S. pyogenes (dCas9Spy) or S. aureus (dCas9Sa), fused or not with superfast green fluorescent protein (sGFP), were cloned downstream of the cadmium-inducible promoter of the pCNX vector (21), which harbors a kanamycin resistance cassette for selection in S. aureus, generating the plasmids pBCB40 (dcas9Spy), pBCB41 (dcas9Spy-sgfp), and pBCB42 (dcas9Sa-sgfp). The tailor-made single guide RNAs from S. pyogenes (sgRNASpy) or S. aureus (sgRNASa) were cloned downstream of a constitutively active promoter in the pGC2 vector (22), which contains a chloramphenicol resistance cassette for selection in S. aureus (Fig. 1). Both sgRNASpy and sgRNASa consist of two regions: the 5′ end contains the variable region for targeted DNA binding (first 20 nucleotides of sgRNASpy or first 22 nucleotides of sgRNASa), while the downstream region contains a constant region for dCas9 binding (different for dCas9Spy and dCas9Sa) and transcription termination. Targeting of specific genes of interest can be accomplished by replacing the 20 (or 22) nucleotides of the variable region, using inverse PCR, for the specific sequence of interest. In the presence of dCas9, the sgRNA forms a complex with the inactive nuclease and guides it to the complementary DNA, where the complex acts as a physical blockage for RNA polymerase, impairing transcription (8).
Fig 1.
CRISPRi systems for S. aureus. The sgRNAs for S. pyogenes Cas9 are constitutively expressed from the psg-RNASpy plasmid. dCas9 can either be expressed from a second plasmid, under the control of a cadmium-inducible promoter (pBCB40), or from the spa locus in the S. aureus chromosome. Transcription inhibition occurs when a complex of dCas9 and sgRNA binds specific target genes, thus blocking the RNA polymerase.
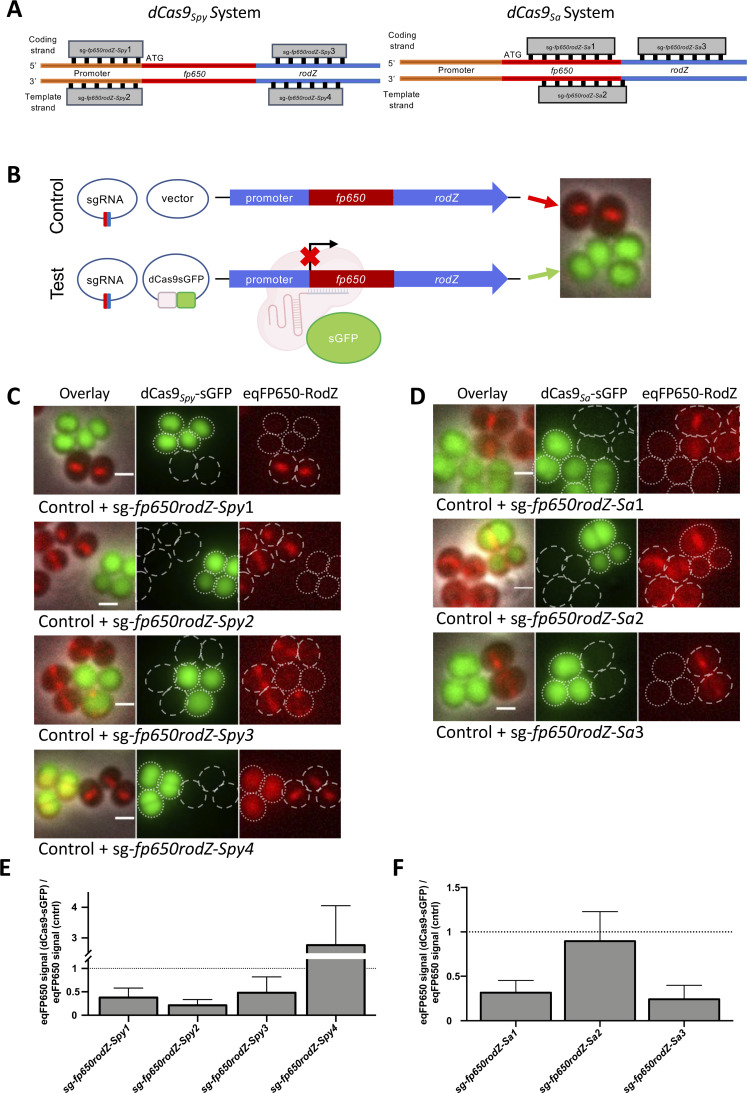
A key requirement for an efficient CRISPRi system is the ability to shut down dCas9 production so that the system can be turned ON/OFF. Western blot analysis of dCas9Spy levels in strain BCBMS02 (expressing cadmium-inducible dcas9Spy-sgfp) showed the presence of dCas9Spy-sGFP even in samples grown in the absence of inducer, indicating that the cadmium-inducible promoter is leaky (Fig. S1A). Despite this limitation, we proceeded to evaluate the efficiency of the system. For that, we constructed the reporter strains BCBMS05 (expressing dcas9Spy-sgfp) and BCBLS01 (expressing dcas9Sa-sgfp), in the background of a derivative of strain NCTC8325-4, where the non-essential gene rodZ, which encodes the septal protein RodZ, was fused to the gene encoding red fluorescent protein eqFP650. Both strains allow easy evaluation of eqFP650-RodZ (red fluorescence) depletion and dCas9-sGFP (green fluorescence) production by fluorescence microscopy. Deletion of rodZ or dCas9 production did not affect the growth of parental strain NCTC8325-4 (Fig. S2). We then designed four different dCas9Spy sgRNAs, targeting the coding or the template strand of fp650-rodZ, as well as its upstream region (Fig. 2A). We also designed three dCas9Sa sgRNAs, two targeting the coding strand and one targeting the template strand of fp650-rodZ (Fig. 2A). To analyze the efficiency of the two systems, we used samples with pairs of strains, both expressing the same sgRNA targeting fp650-rodZ plus either dcas9-sgfp expressed from a pCNX-based plasmid (test) or the empty pCNX vector (control). The two strains were grown in the presence of the inducer cadmium, and then mixed and placed on the same microscopy slide for direct comparison (Fig. 2B). With the dCas9Spy system, we observed that sgRNAs targeting the promoter sequence on either strand (sg-fp650rodZ-Spy1 and sg-fp650rodZ-Spy2) or the gene sequence on the coding strand (sg-fp650rodZ-Spy3) were efficient in depleting eqFP650-RodZ, leading to a strong reduction in red fluorescence intensity in the cells producing dCas9Spy-sGFP (dotted cell outlines, Fig. 2C) when compared to the corresponding control strains lacking dCas9Spy-sGFP (dashed cell outlines, Fig. 2C). On the contrary, the sgRNA that targeted the template strand of fp650-rodZ gene (sg-fp650rodZ-Spy4) was not effective, as cells expressing green fluorescence from dCas9Spy-sGFP also show red fluorescence from eqFP650-RodZ (Fig. 2C and E), in agreement with previous data for E. coli describing that targeting the template strand is not efficient (8). Notably, the eqFP650 signal in this strain was delocalized from the septum to the cytosol and it even increased 2.5-fold (Fig. 2E) compared to control cells (expressing sg-fp650rodZ-Spy4 but lacking dCas9Spy-sGFP). The sgRNA of this strain targets rodZ, downstream of fp650, so it is possible that transcription and translation of fp650 can still occur, producing a soluble, truncated eqFP650.
Fig 2.
Plasmid-based CRISPRi systems using either dCas9Spy or dCas9Sa inhibit gene expression in S. aureus. (A) Schematic overview of sgRNA target sites in fp650-rodZ used to test the dCas9Spy (left) or dCas9Sa (right) CRISPRi systems. (B) Experimental setup to test CRISPRi systems. Genetically modified NCTC8324-5 cells expressing an eqFP650 fusion to the non-essential septal protein RodZ were transformed with two plasmids encoding sgRNA against fp650-rodZ and either an empty vector control (top, control cells) or the same vector encoding dCas9-sGFP (bottom, test cells). Strains were grown separately in the presence of 0.1 µM CdCl2 and subsequently mixed to perform the microscope experiments. Control cells lack dCas9 and, therefore, do not impair the production of eqFP650-RodZ, which can be seen as a septal band (red). Test cells express a cytoplasmic sGFP fusion of dCas9 (green), which is targeted by sgRNA to specific regions of fp650-rodZ, blocking its expression. (C and D) Fluorescence microscopy images of mixtures of control cells (dashed cell outlines) and test cells (encoding dCas9-sGFP, dotted cell outlines) expressing sgRNAs shown in panel B, using either the dCas9Spy (C) or dCas9Sa (D) systems. From left to right, overlay image, green channel image showing dCas9-sGFP signal, and red channel image showing eqFP650-RodZ signal. Scale bars, 1 µm. (E and F) Bar charts showing the ratio of the median eqFP650 signal of cells expressing dCas9-sGFP versus the median eqFP650 signal of the corresponding control cells, both expressing the same sgRNA. Error bars represent the SEM calculated from three independent experiments.
We noticed that depletion of eqFP650-RodZ, resulting in loss of red fluorescence signal, occurred even in the absence of dcas9 induction (Fig. S1B). This indicates that the leaky dcas9 expression results in the production of low levels of dCas9 protein, as seen by western blot (Fig. S1A), which are sufficient to efficiently drive the CRISPRi system in the absence of inducer, i.e., that the system cannot be efficiently turned off.
Next, we tested the alternative version of the CRISPRi system that uses an inactive variant of a Cas9Sa native to S. aureus. Similarly to the dCas9Spy system, this system was also effective when the sgRNASa targeted the coding strand (sg-fp650rodZ-Sa1 and sg-fp650rodZ-Sa3) within the fp650-rodZ gene, as seen by the decreased red fluorescence of these strains (dotted cell outlines, Fig. 2D) compared to the corresponding control strains lacking dCas9Sa (dashed cell outlines, Fig. 2D). No inhibition of fp650-rodZ expression was observed when the sgRNA targeted the template strand (sg-fp650rodZ-Sa2). We achieved similar knockdown efficiencies with both the dCas9Spy and the dCas9Sa system, in both cases leading to a decrease in the relative fluorescence signal of eqFP650-RodZ (Fig. 2E and F). Therefore, we decided to proceed with S. pyogenes dCas9 as it is less limited in target selection due to the shorter PAM sequence, a relevant criterion for work in the low GC content bacterium S. aureus.
CRISPRi system with chromosome-encoded dCas9Spy enables the repression of essential genes’ transcription in S. aureus
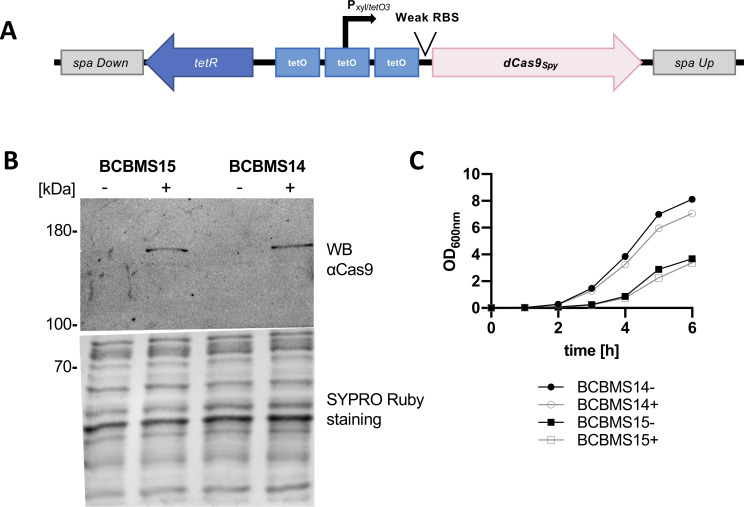
To construct an efficient knockdown system, which can be used to target essential genes in S. aureus, we re-designed the CRISPRi system described above to minimize the production of dCas9 in the absence of an inducer. For that, we integrated dcas9Spy into the spa locus of the staphylococcal genome to reduce the gene copy number to one (notice that inactivation of spa results in strains that do not produce Protein A). This chromosomal copy of dcas9Spy was placed under the control of a tetracycline-inducible xyl/tetO promoter (23). Furthermore, we used a weak ribosome binding site (RBS) (24) separated from the start codon by only three nucleotides to decrease the translation efficiency (Fig. 3A; Fig. S3). This Pxyl/tetO3-dcas9Spy construct was integrated into the spa locus of two MRSA strains, JE2 (generating strain BCBMS14) and COL (generating strain BCBMS15). In both backgrounds, dCas9 was detected by western blot analysis in cells grown in the presence of the inducer anhydrotetracycline (aTc), but not in its absence, indicating that dcas9Spy expression was now under tight control (Fig. 3B). Cells expressing dcas9Spy from the chromosome in the presence of aTc were only mildly impaired in growth (Fig. 3C).
Fig 3.
CRISPRi system with chromosome-encoded dCas9Spy allows tight regulation of dCas9Spy production. (A) Schematic representation of the genomic construct inserted into the spa locus of strains BCBMS14 (JE2 background) and BCBMS15 (COL background), which allows tight control of dCas9Spy production due to the presence of three TetR-binding sites (tetO) flanking and overlapping the xylose promoter and of an inefficient RBS. The sgRNA is expressed from plasmid psg-RNASpy (not shown). (B) Western blot, using an anti-Cas9 antibody, of protein extracts from induced (+) and non-induced (−) BCBMS15 and BCBMS14 cultures showing tight regulation of dCas9Spy expression. SYPRO Ruby staining was used as a loading control. (C) Growth curves of induced (+, 100 ng/mL aTc) and non-induced (−) BCBMS14 and BCBMS15.
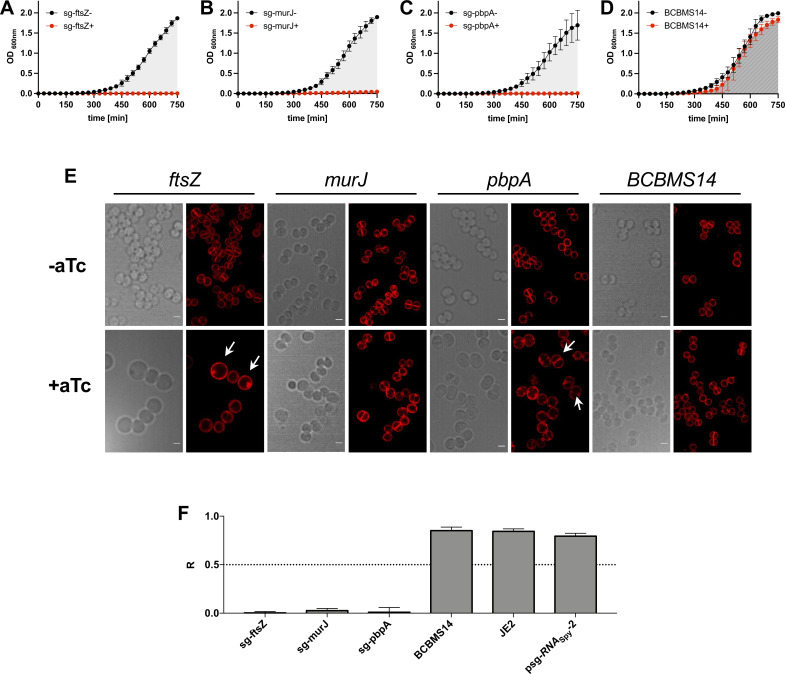
We then tested the efficiency of the improved CRISPRi system when targeting essential genes. For that, we designed sgRNAs targeting the essential cell division genes ftsZ, murJ, and pbpA, encoding the cell division cytoskeletal protein FtsZ (25, 26), the peptidoglycan precursor lipid II flippase MurJ (26, 27), and the septal penicillin-binding protein PBP1 (28, 29), respectively. The resulting CRISPRi strains were analyzed by growth assays and microscopy (see Fig. 4; Fig. S4 for strains in the background of JE2 and COL, respectively). Strains showed severe growth inhibition upon induction of dcas9Spy expression (but not in its absence), and the expected phenotype for the depletion of each gene was observed: while depletion of MurJ resulted in slightly larger cells, depletion of FtsZ led to greatly enlarged, spherical cells without detectable septa (30), and depletion of PBP1 led to enlarged, elongated cells (29). Importantly, these phenotypes were barely observed in the absence of an inducer, confirming that dcas9 is under tight regulation in this improved CRISPRi system.
Fig 4.
CRISPRi system with chromosome-encoded dCas9Spy is suitable to target essential genes in S. aureus. (A–D) Growth assays performed in 96-well plates at 30°C in tryptic soy broth (TSB), in the absence (−) or the presence (+) of aTc (200 ng/mL, inducer for dCas9 expression) of BCBMS14 strains expressing sgRNAs targeting the essential genes ftsZ (BCBMS16, A), murJ (BCBMS18, B), and pbpA (BCBMS17, C). The strain BCBMS14 (D) was used as a control. Cells were grown for 120 min (not shown) before being diluted 1:100 into fresh media with the same composition (growth curves shown). (E) Bright-field and fluorescence microscopy images of cells stained with Nile Red (membrane stain) are shown for each strain shown in panels A–D. Scale bars, 1 µm. Overnight cultures were diluted 1/1,000 in fresh TSB and grown at 37°C until OD600 0.6–0.8 for microscopy analysis. Notice that due to the absence of a second dilution to fully deplete essential proteins of interest, strains are able to initiate growth, allowing imaging of the cells during depletion of the protein of interest. Depletion of FtsZ results in greatly enlarged cells (white arrows), MurJ depletion leads to an increase in cell size, and PBP1 depletion leads to cells that are enlarged, elongated and show an invagination (white arrows). (F) Graphs show the ratio (R) of the area under the curve (AUC) of growth curves obtained in the presence (+) versus the absence (−) of aTc for strains shown in panels A–D, as well as for parental strain JE2 and for BCBMS14 containing psg-RNASpy lacking a specific sgRNA sequence. R can vary between 0 (complete growth inhibition in the presence of the inducer) and 1 (no growth inhibition). Error bars indicate the SEM calculated with data from three independent experiments.
Construction of a CRISPRi library of conditional mutants of essential S. aureus genes
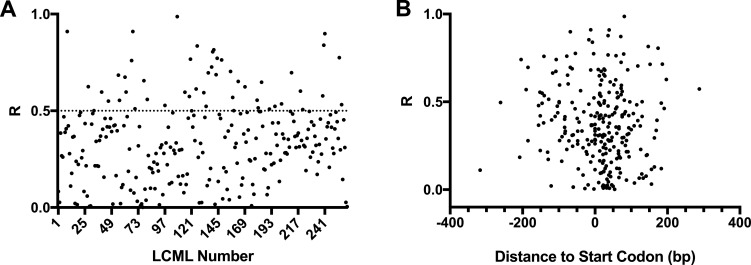
The Nebraska Transposon Mutant Library is an extremely useful resource for the study of S. aureus, as it contains transposon mutants in virtually all non-essential genes of MRSA strain JE2 (18). A complementary library containing depletion mutants for the remaining, essential, genes would be a valuable tool and, therefore, we decided to use the improved CRISPRi system for this purpose. Given that CRISPRi affects the expression of entire operons and not just of the target gene, we are unable to construct single mutants for each essential gene using this technology. We, therefore, targeted each essential operon, which allows users to perform initial screenings that may have to be followed up by detailed studies of individual genes in operons. We identified 200 predicted monocistronic or polycistronic operons (Table S1) containing genes originally described in the literature as essential using antisense RNA (31) or transposon library screenings (32, 33). The identity of genes belonging to each operon was based on the information available in aureowiki (34) (https://aureowiki.med.uni-greifswald.de/Main_Page). We designed sgRNAs targeting the first essential gene of each essential operon and sgRNAs targeting more than one gene in an operon were designed for 46 operons (genes in bold in Table S1) resulting in a total of 261 sgRNAs tested. sgRNAs were cloned into psg-RNASpy using inverse PCR with primer 5846-EcR and a primer containing the specific sgRNA sequence to be introduced (Table S2). The resulting library of plasmids encoding the sgRNAs was electroporated into BCBMS14, generating a library of 261 conditional mutant strains, targeting 200 operons, listed in Table S3, which we named the Lisbon CRISPRi Mutant Library. Growth of these strains in the presence or absence of the aTc inducer (200 ng/mL) was followed in a 96-well plate reader, for 750 min, at 30°C. The ratio (R) of the area under the curve (AUC) of the growth curve obtained in the presence versus the absence of an inducer was used to evaluate the efficiency of growth inhibition (Fig. S5). Of the 261 strains, 148 strains showed a clear growth reduction upon induction (R ≤ 0.5, an arbitrary threshold chosen to decide if further optimization was required). However, 113 of the strains had milder growth deficits (R > 0.5). For these, we designed a second sgRNA (sgRNA_B, B clones) targeting an alternative region of the gene. Of the 113 strains for which a second sgRNA was designed, 48 showed R lower than 0.5 (Fig. S5 shows R data for the strain with the sgRNA, A or B, with highest efficiency). For 65 strains for which R was higher than 0.5, growth analysis was repeated at 37°C, given that some mutants may show a stronger growth defect at 37°C than at 30°C. Figure S6 shows that inhibition of the expression of a further 20 genes led to impaired growth at 37°C. Overall, we achieved a success rate of ~83% as we observed strong growth inhibition (R ≤ 0.5) for 216 of the 261 strains constructed (Fig. 5A). The LCML clones showing the greatest growth impairment are listed in Table S4 with the sgRNA sequence, temperature used for growth inhibition, and R values listed.
Fig 5.
Efficiency of growth inhibition by CRISPRi system with chromosome-encoded dCas9Spy (A). The ratio (R) of the AUC of growth rates obtained in the presence versus the absence of aTc (inducer for dCas9 expression) was calculated for LCML strains expressing 261 different sgRNAs. Graph shows that R is lower than 0.5 for ~80% of the strains. (B) R was plotted against the distance between the sgRNA target position and the start codon of the targeted gene, showing that there is no correlation between this distance and the efficiency of growth inhibition by the CRISPRi system.
To determine if there were target regions for sgRNA that resulted in higher efficiency of the CRISPRi system, we plotted R for each sgRNA against their target position relative to the start codon of the corresponding gene (Fig. 5B) and tested those targeting the coding strand for statistically relevant correlation using a Spearman correlation test. For that, we split the data into two sets: the first set included the 87 sgRNAs that bind upstream of the start codon, and the second set included the 174 sgRNAs targeting in the gene. sgRNAs targeting the template strand were excluded from the analyses due to the small sample size (n = 8). We found no statistically significant correlation between the efficiency of growth inhibition and localization of sgRNAs targeted sequence relative to the start codon, for either the first set (r: −0.06365; P-value: 0.5581) or the second set (r: 0.09877; P-value: 0.1947).
DISCUSSION
Construction of S. aureus conditional knockouts or gene deletion mutants using traditional approaches that require recombination events is labor-intensive and time-consuming. In contrast, the use of a CRISPRi system allows the generation of conditional mutants through the simple exchange of the target sequence of the sgRNA, so that it specifically inactivates any target gene/operon. In this study, we developed different CRISPRi systems for S. aureus, using dCas9 proteins originating from both S. pyogenes and S. aureus. In our first approach, sgRNAs and dcas9 from either S. pyogenes or S. aureus were expressed from two replicative vectors, allowing rapid introduction into different S. aureus strains and mutants. Staphylococcal Cas9Sa is significantly smaller than Cas9 from S. pyogenes (1,053 versus 1,368 amino acid residues) and it requires a longer PAM sequence. The smaller size of the nuclease is important when used in eukaryotes because of the limited packaging size of adenovirus systems and lower transfection efficiency of very large plasmids. Additionally, a longer PAM sequence should decrease off-target effects, which is a problem when manipulating eukaryotic cells due to their large genome size. Both advantages are less important in bacteria. Nevertheless, we hypothesized that the expression of the smaller, native nuclease could be advantageous in S. aureus. However, the two CRISPRi systems, with either S. pyogenes or S. aureus dCas9, worked similarly. Given that S. pyogenes dCas9 is less limited in target selection due to the shorter PAM sequence, we decided to proceed with dCas9Spy for further optimization of the system. Optimization was required because low levels of dCas9 were produced even in the absence of an inducer when using these two-plasmid systems. To reduce gene transcription of dcas9 in the absence of an inducer to a minimum we (i) integrated dcas9 into the genome of S. aureus MRSA strains JE2 and COL to reduce its copy number to one, (ii) exchanged the leaky cadmium-inducible promoter with a tighter xyl/tetO promoter, and (iii) decreased the translation efficiency by switching to an inefficient ribosome binding site. The resulting strains, BCBMS14 (JE2 background) and BCBMS15 (COL background), produced dCas9 under tight control, as it was not detected by western blot in the absence of aTc inducer, but reached sufficient levels to silence essential genes upon addition of the inducer. Targeting of essential genes ftsZ, pbpA, or murJ with this improved CRISPRi system led to a severe decrease in growth, showing the efficiency of the system.
Given the efficacy of the improved CRISPRi system to silence essential genes, we constructed a library of CRISPRi conditional mutants in the strain BCBMS14, each containing a specific sgRNA targeting 1 of 261 genes, corresponding to 200 operons, which harbor at least one essential gene. Overall, we were able to construct strains to silence genes in 169 of these essential operons. It is possible that some of the tested genes for which expression inhibition did not result in severe growth defects (R > 0.5) are not actually essential for bacterial survival under the growth conditions tested, which would justify the lack of efficiency of some sgRNAs. For example, it was reported that recU (R = 0.595) can be depleted in S. aureus, although mutants display chromosome segregation and DNA damage repair defects (35). Similarly, a ltaS mutant (R = 0.597) is viable in S. aureus but displays temperature sensitivity (36). Viable (although often exhibiting growth defects) mutants of other genes that displayed R higher than 0.5 when gene expression was inhibited with CRISPRi can also be found in the literature, namely, parE (37), dnaK (38), secDF (39), ackA (40), murA (41), ureA (42), xpt (43), mraZ (44), and gdpP (45).
Large variation in sgRNA efficiency has been reported in several published studies (8, 46–48). In E. coli, the distance between the transcription start site and the sgRNA target was reported by Qi and colleagues to affect knockdown efficiency (8) but this effect was not observed in genome-wide screens (49, 50). We selected PAM sequences close to the start codon, so all the designed sgRNAs target regions between −400 and +400 nucleotides from the start codon. Given the large number of sgRNAs tested, we evaluated if there were specific locations that corresponded to more efficient sgRNAs. However, as shown in Fig. 5B, we could not find a correlation between the distance of the sgRNA target to the start codon of the corresponding gene and sgRNA efficiency and therefore could not establish guiding principles for sgRNA design.
The constructed Lisbon CRISPRi Mutant Library of conditional mutants in essential genes/operons of S. aureus is complementary to the widely used Nebraska Transposon Mutant library, which includes mutants in virtually all non-essential genes in S. aureus (18). After the submission of this work, a genome-wide CRISPR interference library was reported by Liu and colleagues, which should also constitute a useful tool for S. aureus studies (51). The combined use of these libraries will allow the evaluation of the function of any gene/operon of this important pathogen.
MATERIALS AND METHODS
Bacterial strains and growth conditions
Plasmids and strains used in this study are described in Tables 1 and 2. S. aureus cells were grown at 37°C with aeration in liquid tryptic soy broth (TSB; Difco) or on solid tryptic soy agar (VWR) medium, supplemented, when necessary, with antibiotics (chloramphenicol [Cm], Sigma-Aldrich, 10 µg/mL in liquid media, 20 µg/mL in solid medium; kanamycin [Kan], Apollo Scientific, 50 µg/mL; and neomycin [Neo], Sigma-Aldrich, 50 µg/mL) and inducer (cadmium chloride, Sigma-Aldrich, 0.1 µM or anhydrotetracycline [aTc], Sigma-Aldrich, 100–200 ng/mL). E. coli cells were grown at 37°C with aeration in either Luria-Bertani broth (VWR) or Luria Agar (VWR), supplemented with ampicillin (Apollo Scientific, 100 µg/mL) when required. Strain growth in liquid media was followed by measurement of optical density at 600 nm (OD600).
TABLE 1.
Plasmids used and constructed in this study
| Name | Description | Reference |
|---|---|---|
| pTRC99a-P7 | Vector containing sgfp-p7; AmpR | (52) |
| pFP60-F | pTnT with fp650 in forward orientation | (53) |
| pgRNA_bacteria | Vector containing sgRNASpy | (8, 54) |
| pdCas9_bacteria | Vector containing dcas9Spy | (8, 54) |
| pMAD | E. coli/S. aureus shuttle vector with a thermosensitive origin of replication for Gram-positive bacteria; AmpR; EryR; lacZ | (55) |
| pGC2 | Replicative E. coli/S. aureus shuttle vector; AmpR; CmR | (22) |
| pBCB13 | pMAD with up- and downstream regions of spa and Pspac-lacI region from pDH88 | (56) |
| pCNX | Replicative E. coli/S. aureus shuttle vector containing the cadmium-inducible Pcad promoter; AmpR; KanR/NeoR | (21) |
| pCNX_sGFP | pCNX derivative containing sgfp from plasmid pTRC99a-P7 | This study |
| pBCB40 | pCNX derivative containing S. pyogenes dcas9; AmpR; KanR/NeoR | This study |
| pBCB41 | pCNX derivative containing S. pyogenes dcas9-sgfp; AmpR; KanR/NeoR | This study |
| pBCB42 | pCNX derivative containing S. aureus dcas9-sgfp; AmpR; KanR/NeoR | This study |
| pBCB43 | pBCB13 derivative lacking Pspac and lacI, containing tetR, PxyltetO, and a downstream located tetO site, AmpR; EryR; lacZ | This study |
| pBCB44 | pBCB43 derivative containing dcas9Spy under Pxyl/tetO3 control; AmpR; EryR; lacZ | This study |
| psg-RNASpy | pGC2 derivative containing the sgRNA cloned from pgRNA_bacteria plasmid; CmR | This study |
| psg-RNASpy-2 | psg-RNASpy derivative lacking the 20-target specific base pairing nucleotides; CmR | This study |
| psg-fp650rodZ-Spy1 | psg-RNASpy derivative with sgRNA altered to sg-fp650rodZ-Spy1; AmpR; CmR | This study |
| psg-fp650rodZ-Spy2 | psg-RNASpy derivative with sgRNA altered to sg-fp650rodZ-Spy2; AmpR; CmR | This study |
| psg-fp650rodZ-Spy3 | psg-RNASpy derivative with sgRNA altered to sg-fp650rodZ-Spy3; AmpR; CmR | This study |
| psg-fp650rodZ-Spy4 | psg-RNASpy derivative with sgRNA altered to sg-fp650rodZ-Spy4; AmpR; CmR | This study |
| psg-fp650rodZ-Sa1 | pGC2 derivative containing the sg-fp650rodZ-Sa1 under the control of a constitutive active promoter; AmpR; CmR | This study |
| psg-fp650rodZ-Sa2 | pGC2 derivative containing the sg-fp650rodZ-Sa2 under the control of a constitutive active promoter; AmpR; CmR | This study |
| psg-fp650rodZ-Sa3 | pGC2 derivative containing the sg-fp650rodZ-Sa3 under the control of a constitutive active promoter; AmpR; CmR | This study |
| psg-ftsZ | psg-RNASpy derivative with sgRNA altered to sg-ftsZ; AmpR; CmR | This study |
| psg-pbpA | psg-RNASpy derivative with sgRNA altered to sg-pbpA; AmpR; CmR | This study |
| psg-murJ | psg-RNASpy derivative with sgRNA altered to sg-murJ; AmpR; CmR | This study |
| pMAD_ΔrodZ | pMAD containing rodZ upstream and downstream regions; AmpR EryR | (57) |
| pMAD_fp650rodZ | pMAD containing rodZ upstream region-eqfp650–rodZ-rodZ downstream region; AmpR EryR | This study |
TABLE 2.
Strains used in this study
| Name | Description | Reference |
|---|---|---|
| NCTC8325-4 | MSSA strain NCTC8325-4 | (58) |
| COL | Hospital-acquired homogeneous MRSA | (59) |
| JE2 | Derivative of community-acquired MRSA | (18) |
| BCBMS01 | JE2 with pBCB40; expressing S. pyogenes dcas9Spy; Kan/NeoR | This study |
| BCBMS02 | JE2 with pBCB41; expressing S. pyogenes dcas9Spy-sgfp; Kan/NeoR | This study |
| NCTC ΔrodZ | rodZ deletion mutant in NCTC8325-4 | This study |
| BCBHV200 | NCTC8325-4 ΔrodZ:fp650rodZ; expressing rodZ translationally fused to fluorescent protein eqFP650 |
This study |
| BCBMS04 | BCBHV200 with pCNX; serves as control; Kan/NeoR | This study |
| BCBMS05 | BCBHV200 with pBCB41, expressing S. pyogenes dcas9Spy-sgfp; Kan/NeoR | This study |
| BCBMS06 | BCBMS04 with psg-fp650rodZ-Spy1; CmR, Kan/NeoR | This study |
| BCBMS07 | BCBMS04 with psg-fp650rodZ-Spy2; CmR, Kan/NeoR | This study |
| BCBMS08 | BCBMS04 with psg-fp650rodZ-Spy3; CmR, Kan/NeoR | This study |
| BCBMS09 | BCBMS04 with psg-fp650rodZ-Spy4; CmR, Kan/NeoR | This study |
| BCBMS10 | BCBMS05 with psg-fp650rodZ-Spy1; CmR, Kan/NeoR | This study |
| BCBMS11 | BCBMS05 with psg-fp650rodZ-Spy2; CmR, Kan/NeoR | This study |
| BCBMS12 | BCBMS05 with psg-fp650rodZ-Spy3; CmR, Kan/NeoR | This study |
| BCBMS13 | BCBMS05 with psg-fp650rodZ-Spy4; CmR, Kan/NeoR | This study |
| BCBLS01 | BCBHV200 with pBCB42; expressing S. aureus dcas9Sa-sgfp; Kan/NeoR | This study |
| BCBLS02 | BCBMS04 with psg-fp650rodZ-Sa1; CmR, Kan/NeoR | This study |
| BCBLS03 | BCBMS04 with psg-fp650rodZ-Sa2; CmR, Kan/NeoR | This study |
| BCBLS04 | BCBMS04 with psg-fp650rodZ-Sa3; CmR, Kan/NeoR | This study |
| BCBLS05 | BCBLS01 with psg-fp650rodZ-Sa1; CmR, Kan/NeoR | This study |
| BCBLS06 | BCBLS01 with psg-fp650rodZ-Sa2; CmR, Kan/NeoR | This study |
| BCBLS07 | BCBLS01 with psg-fp650rodZ-Sa3; CmR, Kan/NeoR | This study |
| BCBMS14 | JE2 Δspa: Pxyl/tetO3-dcas9Spy; expressing dcas9Spy under the control of an aTc-inducible promoter Pxyl/tetO3 from the spa locus |
This study |
| BCBMS14 psg-RNASpy-2 | BCBMS14 with psg-RNASpy-2; CmR | This study |
| BCBMS15 | COL Δspa: Pxyl/tetO3-dcas9Spy; expressing dcas9Spy under the control of an aTc-inducible promoter Pxyl/tetO3 from spa locus |
This study |
| BCBMS16 | BCBMS14 with psg-ftsZ; CmR (same as LCML 261) | This study |
| BCBMS17 | BCBMS14 with psg-pbpA; CmR (same as LCML 76) | This study |
| BCBMS18 | BCBMS14 with psg-murJ; CmR (same as LCML 260) | This study |
| BCBMS20 | BCBMS15 with psg-ftsZ; CmR | This study |
| BCBMS21 | BCBMS15 with psg-pbpA; CmR | This study |
| BCBMS22 | BCBMS15 with psg-murJ; CmR | This study |
| BCBMS25 | BCBHV200 with pBCB40; Kan/NeoR | This study |
| BCBMS26 | BCBHV200 with pBCB40 and psg-fp650rodZ-Spy2; CmR, Kan/NeoR | This study |
DNA purification and manipulation
Plasmid DNA was extracted from E. coli cells using the Wizard SV Plus Miniprep Kit (Promega) or the QIAprep Spin Miniprep Kit (Qiagen), following the manufacturer’s instructions. DNA was digested with FastDigest restriction enzymes (ThermoFisher) by incubating 0.5–1 µg DNA at 37°C for 1 hour, with 1× FastDigest buffer, 1 µL of the selected endonuclease, and 1 µL of Fast Alkaline Phosphatase (ThermoFisher), when necessary. DNA fragments were purified using the Wizard SV Plus Cleanup Kit (Promega), according to the manufacturer’s instructions. DNA ligations were performed by standard molecular biology techniques using T4 DNA Ligase (ThermoFisher). PCR reactions were performed using either Phusion DNA Polymerase for cloning purposes or DreamTaq Polymerase for colony screenings (both ThermoFisher), following the manufacturer’s instructions. For inverse PCR, one primer was 5′ phosphorylated using a T4 Polynucleotide Kinase (ThermoFisher) using recommended reaction conditions. After PCR amplification, 1 µL DpnI (NEB) was added to the PCR mix, which was incubated for 1 hour at 37°C followed by purification. Five microliters of the purified PCR product was used for ligation using T4 DNA ligase followed by transformation.
Plasmid and strain construction
Plasmids and primers used in this study are described in Table 1; Table S5, and primers used to generate the different sgRNAs for the LCML are described in Table S2.
To construct the pBCB40 plasmid, dcas9Spy was amplified from plasmid pdCas9_bacteria (8, 54) using primers 5450/5451. The PCR product and the vector pCNX (21) were digested with PstI and SalI, and the fragment containing dcas9Spy was ligated downstream of the cadmium-inducible promoter from pCNX. To construct pBCB41, primers 3714/5450 were used to amplify dcas9Spy with a linker insertion, and sgfp was amplified from the plasmid pTRC99a-P7 (52) using primers 3715/5452. The two fragments were joined by overlap PCR using primers 5450/5452, digested with PstI and SalI, and ligated into pCNX, digested with the same enzymes.
To construct the pBCB42, we first constructed the pCNX_sGFP plasmid by amplifying the sgfp gene sequence from pTRC99a-P7 plasmid using the primer pair 5602/5603 and cloning it into pCNX using EcoRI and BamHI restriction sites. The dcas9Sa gene sequence was kindly provided by F. Ann Ran (16). The sequence, comprising a ribosome binding site and the dcas9Sa gene plus restriction sites SalI and BamHI, was synthesized (NZYTech), digested with the respective restriction enzymes, and cloned into the pCNX_sGFP plasmid, resulting in plasmid pBCB42.
To construct plasmid pBCB43, a fragment of the antisense secY expression cassette in pIMAY (60) was amplified using primers 7015/7016. The PCR product, comprising tetR, Pxyl-tetO, and a downstream located tetO site, was digested with EcoRI/NheI and ligated into similarly digested pBCB13 (56) lacking lacI and Pspac. The resulting plasmid, pBCB43, was SmaI/EagI digested to clone dcas9Spy, amplified from pBCB40 with the primer pair 7268/7272, downstream of tetO of pBCB43, via Gibson assembly (61), generating plasmid pBCB44, verified by sequencing.
Plasmid psg-RNASpy was constructed by cloning the sgRNA sequence including the minimal constitutive promoter and transcription terminator from plasmid pgRNA_bacteria (8, 54) into pGC2 (22). For that, the multiple cloning site of pGC2 was altered via inverse PCR with primer pair 5922/5923 (5′ phosphorylated) and for a second time with primer pair 5745/5746 (5′ phosphorylated) to obtain a BglBrick (62) composition with restriction sites for EcoRI, BglII, BamHI, and XhoI. The vector pgRNA_bacteria was digested with EcoRI and XhoI, and the fragment was ligated into the new multiple cloning site of altered pGC2 resulting in the plasmid psg-RNASpy, which was verified by sequencing. To construct control plasmid psg-RNASpy-2, encoding a truncated sgRNA that lacks the target-specific base-pairing region but contains the dCas9-binding hairpin and S. pyogenes terminator sequence, the region was removed from plasmid psg-RNASpy via inverse PCR using primers 5846 and 5845. The plasmid sequence was verified.
Plasmid pBCB44 was electroporated into RN4220 and transduced into JE2 or COL. The replacement of the spa gene for Pxyl/tetO3-dcas9 was completed after a two-step homologous recombination event and the resulting strains BCBMS14 and BCBMS15 were verified by PCR and sequencing.
To construct plasmids expressing different sgRNAs in the S. pyogenes CRISPRi systems, psg-RNASpy was used as a template and the 20 nt target-specific sequence present in psg-RNASpy was replaced by the 20 nt sequence of interest via inverse PCR as described (54). To generate the initial test sg-RNASpy by inverse PCR, reverse primer 5846 and forward primers 5887, 5849, 5890, and 5889 (Table S5) were used for the construction of psg-fp650rodZ-Spy1-4. Similarly, reverse primer 5846 and forward primers 6424 (sg-ftsZ), 6426 (sg-pbpA), and 6423 (sg-murJ) were used to construct plasmids psg-ftsZ, psg-pbpA, and psg-murJ encoding sgRNAs against selected essential genes. These three plasmids were introduced into strain BCBMS14, generating strains BCBMS16–18 and into strain BCBMS15 generating strains BCBMS20–22. For the LCML, gene-specific base-pairing regions were selected by searching for PAM sequences in/upstream of essential genes/operons, and the 20 nt upstream of the first PAM sequence in the coding strand of a gene were chosen as target DNA in most cases. To generate sg-RNASpy for the library construction, we always used 5′ phosphorylated reverse primer 5846 together with a gene-specific forward primer (see Table S2) containing the base-pairing region as an overhang in combination with the constant part of the sgRNA gttttAGAGCTAGAAATAGCAAGTTAAAATAAGGC.
To construct psg-fp650rodZ-Sa1, the pbpB promoter sequence (63) was amplified from NCTC8325-4 genomic DNA using primer pair 5639/5838. Primer 5838 contains the sgRNASa (64), a region targeting the eqFP650-RodZ-specific gene sequence and a transcription terminator. The resulting PCR product was digested with SmaI and EcoRI and cloned into pGC2. Plasmid psg-fp650rodZ-Sa2 was constructed by amplifying the pbpB promoter from NCTC8325-4 genomic DNA using primer pair 5639/5839 and the sgRNASa from psg-fp650rodZ-Sa1 using the primer pair 5840/5837. The fragments were joined by overlap PCR with primers 5639/5837 and cloned into pCG2 using SmaI and EcoRI enzymes identical to psg-fp650rodZ-Sa1. For the construction of psg-fp650rodZ-Sa3, the pbpB promoter was exchanged for the synthetic minimal promoter used in psg-RNASpy plasmids. Primer pair 6013/5837 was used to amplify the sgRNASa with primer 6013 containing the minimal promoter and target-specific nucleotide sequence. The resulting PCR product was digested with SmaI and EcoRI and cloned into pGC2. All constructed sgRNAs were confirmed by sequencing before further utilization.
All plasmids except those encoding the sgRNA for the library were initially transformed into E. coli DC10B (60) with ampicillin (100 µg/mL) selection. Constructs were verified by PCR and sequencing. Plasmids were electroporated into the S. aureus strain RN4220 as previously described (20), followed by phage lysate preparation and transduction into other S. aureus strains (65). Following this protocol, the plasmids pCNX, pBCB40, pBCB41, pBCB42, psg-fp650rodZ-Spy1-4, and psg-fp650rodZ-Sa1-3 were electroporated into RN4220 with subsequent preparations of phage lysates. Plasmids pBCB40 and pBCB41 were transduced into JE2, generating strains BCBMS01 and BCBMS02, respectively. Strains BCBMS04, BCBMS05, and BCBLS01 were created by introducing pCNX, pBCB41, or pBCB42, respectively, into the strain BCBHV200 (see below). Plasmids psg-fp650rodZ-Spy1-4 were transduced either into BCBMS05, creating the strains BCBMS10-13, or into BCBMS04, creating control strains BCBMS06-09. Plasmids psg-fp650rodZ-Sa1-3 were transduced either into BCBLS01, creating the strains BCBLS05-07, or into BCBMS04, creating control strains BCBLS02-04. pBCB40 and psg-fp650rodZ-Spy2 were also transduced into BCBHV200, creating strain BCBMS26.
The plasmids for the sgRNA library based on psg-RNASpy were initially transformed into E. coli strain IM08B (66). IM08B has a modified DNA methylation pattern compatible with that of S. aureus JE2 increasing the efficiency of transformation into this strain (66). Plasmids propagated in IM08B could therefore be directly electroporated into competent BCBMS14 cells, bypassing the time-consuming steps of electroporation into RN4220 and subsequent phage transduction, frequently used to transfer plasmids into S. aureus. Preparation of S. aureus competent cells and electroporation protocol were performed as described by Johnston and colleagues (67).
To construct a S. aureus strain encoding an N-terminal eqFP650-RodZ fusion as the only rodZ copy in the cell, three DNA fragments were amplified. Fragment 1, containing a 1,037-bp upstream region of rodZ, and fragment 3, containing 1,039 bp encompassing the rodZ gene and its downstream region, were amplified from NCTC8325-4 genome using the primers pairs 3627/3628 and 3382/3630, respectively. Fragment 2, encompassing the fp650 gene without its stop codon and encoding a five amino acid linker (SCGAS) at the 3′ end, was amplified from plasmid pFP650-F (53) using primers 3629/3381. The three fragments were then joined by overlap PCR using primers 3627/3630, and the resulting fragment was digested with SalI and NcoI and cloned into pMAD (55). The resulting plasmid was named pMAD_fp650rodZ and was verified by sequencing. The pMAD_fp650rodZ plasmid was electroporated into RN4220 at 30°C and subsequently transduced to NCTC8325-4 (selection at 30°C with erythromycin in both steps). The replacement of the rodZ gene for fp650-rodZ was completed after a two-step homologous recombination event and was confirmed by PCR. The strain was named BCBHV200. Similarly, pMAD_ΔrodZ (57) was used to delete the rodZ gene from the genome of NCTC8325-4, resulting in strain NCTC ΔrodZ.
Fluorescence microscopy NCTC fp650-rodZ conditional mutant
Overnight cultures of strains expressing dcas9-sgfp (from S. pyogenes or S. aureus) with corresponding sgRNA targeting fp650-rodZ (BCBMS10-13; BCBLS05-07) and control strains (BCBMS06-09; BCBLS02-04, lacking dcas9) were back-diluted 1:500 in 10 mL of TSB, with 10 µg/mL of Cm, 50 µg/mL of both Neo and Kan, and 0.1 µM CdCl2 and incubated at 37°C, with aeration, to an OD600 of 0.8. A 1 mL aliquot of culture was pelleted and re-suspended in 30 µL of phosphate-buffered saline (PBS). A 5 µL sample of each dCas9-producing culture expressing a specific sgRNA was mixed with an equal volume of the corresponding control culture lacking dCas9 but expressing the same sgRNA (see experimental setup in Fig. 2B). One microliter of the mixed culture was placed on a thin layer of 1.2% agarose in PBS and imaged using an Axio Observer.Z1 microscope equipped with a Photometrics CoolSNAP HQ2 camera (Roper Scientific), using phase contrast objective Plan Apo 100×/1.4 oil Ph3. Cells were imaged using the ZEN software (Zeiss). The median fluorescence of sGFP and eqFP650 in each cell was determined using eHooke (68).
Microscopy analysis of conditional mutants in essential genes
Overnight cultures of derivatives of BCBMS14 (JE2 background) expressing sgRNAs against ftsZ (BCBMS16), pbpA (BCBMS17), or murJ (BCBMS18) were each back-diluted 1:1,000 into fresh media containing 10 µg/mL Cm with (induced) or without (non-induced) 100 ng/mL aTc and grown at 37°C to exponential phase (OD600 0.6–0.8). A 1 mL aliquot of each culture was incubated with 2.5 µg/mL Nile Red (membrane stain, Invitrogen) for 5 min at 37°C with shaking. The culture was then pelleted and resuspended in 30 µL PBS. One microliter of cell culture was placed on a thin layer of 1.2% agarose in PBS and imaged via structured illumination microscopy (SIM) using an Elyra PS.1 microscope (Zeiss) with a Plan-Apochromat 63×/1.4 oil DIC M27 objective. SIM images were acquired using five grid rotations, with 34 µm grating period for the 561 nm laser (100 mW), and captured using a Pco.edge 5.5 camera. Images were reconstructed using ZEN software (black edition, 2012, version 8.1.0.484).
Western blot
S. aureus strains were grown overnight, diluted 1:500 in fresh medium and incubated at 37°C with aeration. When necessary, cultures were supplemented with required antibiotics and aTc or CdCl2. At an OD600 of approximately 0.6, cells were harvested and broken with glass beads in a SpeedMill (Analytik Jena). Samples were centrifuged for 10 min at 16,000 × g and resuspended in 300 µL of PBS containing protease inhibitors (cOmplete, Roche). A 16 µL sample was added to 5 µL of 5× SDS sample buffer (300 mM Tris-HCl, pH 6.8, 50% glycerol, 10% SDS, 0.01% bromophenol blue, 10% beta-mercaptoethanol), boiled for 5 min, and samples were separated by SDS-PAGE (12% gel) at 120 V. Proteins were then transferred to Hybond-P PVDF membrane (GE Healthcare) using a BioRad Transblot Turbo transfer system, according to the manufacturer’s instructions. After transfer, the membrane was cut at the 100 kDa marker band. The upper part was used for protein detection using a specific monoclonal antibody against Cas9 (1:1,000 dilution, #MAC133, Sigma-Aldrich) and Alexa555 secondary antibody for fluorescence detection (iBright, ThermoFisher), while the lower part of the membrane was stained with Sypro Ruby Protein Blot Stain (Thermo Fisher), a total protein stain for western blot normalization.
Growth curves
For the analysis of growth of the COL CRISPRi conditional mutants BCBMS20-22, cells were grown overnight at 37°C in TSB medium supplemented with 10 µg/mL Cm and diluted 1:500 into 100 mL Erlenmeyer flasks containing fresh TSB medium with 10 µg/mL Cm, with or without 100 ng/ mL aTc, which induces the expression of dcas9. After 4 hours, cultures were diluted a second time 1:100 in a fresh medium, and growth was followed by measuring the OD600 every hour. All cultures were incubated at 37°C with agitation, and the OD600 was recorded.
The growth of NCTC8325-4 ΔrodZ and BCBMS25 strains (with or without 0.1 mM CdCl2) was analyzed in a 96-well plate reader (Biotek Synergy Neo2). Overnight cultures were diluted 1:1,000 in fresh media, 200 µL of each culture was added to wells in a 96-well plate, and growth was followed at 37°C with agitation for 750 min. The OD600 was measured every 30 min.
The analysis of the library of CRISPRi conditional mutants (derived from strain BCBMS14) was performed in a 96-well plate reader. Overnight cultures were diluted 1:1,000 in fresh media containing 10 µg/mL Cm. Samples (200 µL) of each culture, with or without the addition of 200 ng/mL aTc inducer, were added to wells in a 96-well plate. The plates were incubated at 30°C with shaking for 120 min, and the OD600 was measured every 30 min. After 2 hours, the samples were diluted 1:100 into fresh media (with 10 µg/mL Cm), with or without aTc, and growth was followed in all cultures for a further 750 min. For CRISPRi conditional mutants showing <50% growth reduction at 30°C, growth analysis was also performed at 37°C.
ACKNOWLEDGMENTS
We thank Vincent de Bakker for advice on statistical analysis. Figure 1 was created with Biorender.com.
This study was funded by the European Research Council through grant ERC-2017-CoG-771709 (to M.G.P.), by national funds through FCT–Fundação para a Ciência e a Tecnologia through MOSTMICRO-ITQB R&D Unit (UIDB/04612/2020 and UIDP/04612/2020 to ITQB) and LS4FUTURE Associated Laboratory (LA/P/0087/2020 to ITQB), FCT fellowships PD/BD/128408/2017 (to M.S.) and 2022.12215.BD (to D.A.), by the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No. 839596 (to S.S.) and by the European Molecular Biology Organization through award ALTF 673-2018 (to S.S.).
Contributor Information
Mariana G. Pinho, Email: mgpinho@itqb.unl.pt.
Steven J. Projan, MedImmune, Gaithersburg, Maryland, USA
DATA AVAILABILITY
Data and strains will be made available upon reasonable request to the corresponding author.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/mbio.02773-23.
Supplemental figures and tables.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Turner NA, Sharma-Kuinkel BK, Maskarinec SA, Eichenberger EM, Shah PP, Carugati M, Holland TL, Fowler VG. 2019. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol 17:203–218. doi: 10.1038/s41579-018-0147-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Murray CJL, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A, Han C, Bisignano C, Rao P, Wool E, et al. 2022. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet 399:629–655. doi: 10.1016/S0140-6736(21)02724-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Prax M, Lee CY, Bertram R. 2013. An update on the molecular genetics toolbox for staphylococci. Microbiology (Reading) 159:421–435. doi: 10.1099/mic.0.061705-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thomason MK, Storz G. 2010. Bacterial antisense RNAs: how many are there, and what are they doing? Annu Rev Genet 44:167–188. doi: 10.1146/annurev-genet-102209-163523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lucks JB, Qi L, Mutalik VK, Wang D, Arkin AP. 2011. Versatile RNA-sensing transcriptional regulators for engineering genetic networks. Proc Natl Acad Sci USA 108:8617–8622. doi: 10.1073/pnas.1015741108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. doi: 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. 2013. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res 41:7429–7437. doi: 10.1093/nar/gkt520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152:1173–1183. doi: 10.1016/j.cell.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Peters JM, Colavin A, Shi H, Czarny TL, Larson MH, Wong S, Hawkins JS, Lu CHS, Koo BM, Marta E, Shiver AL, Whitehead EH, Weissman JS, Brown ED, Qi LS, Huang KC, Gross CA. 2016. A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell 165:1493–1506. doi: 10.1016/j.cell.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu X, Gallay C, Kjos M, Domenech A, Slager J, van Kessel SP, Knoops K, Sorg RA, Zhang J-R, Veening J-W. 2017. High-throughput CRISPRi phenotyping identifies new essential genes in Streptococcus pneumoniae. Mol Syst Biol 13:931. doi: 10.15252/msb.20167449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dong X, Jin Y, Ming D, Li B, Dong H, Wang L, Wang T, Wang D. 2017. CRISPR/Dcas9-mediated inhibition of gene expression in Staphylococcus aureus. J Microbiol Methods 139:79–86. doi: 10.1016/j.mimet.2017.05.008 [DOI] [PubMed] [Google Scholar]
- 12. Zhao C, Shu X, Sun B, Schottel JL. 2017. Construction of a gene knockdown system based on catalytically inactive ("dead") Cas9 (Dcas9) in Staphylococcus aureus. Appl Environ Microbiol 83:e00291-17. doi: 10.1128/AEM.00291-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stamsås GA, Myrbråten IS, Straume D, Salehian Z, Veening J-W, Håvarstein LS, Kjos M. 2018. CozEa and CozEb play overlapping and essential roles in controlling cell division in Staphylococcus aureus. Mol Microbiol 109:615–632. doi: 10.1111/mmi.13999 [DOI] [PubMed] [Google Scholar]
- 14. Chen W, Zhang Y, Yeo WS, Bae T, Ji Q. 2017. Rapid and efficient genome editing in Staphylococcus aureus by using an engineered CRISPR/Cas9 system. J Am Chem Soc 139:3790–3795. doi: 10.1021/jacs.6b13317 [DOI] [PubMed] [Google Scholar]
- 15. Nishimasu H, Cong L, Yan WX, Ran FA, Zetsche B, Li Y, Kurabayashi A, Ishitani R, Zhang F, Nureki O. 2015. Crystal structure of Staphylococcus aureus Cas9. Cell 162:1113–1126. doi: 10.1016/j.cell.2015.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA, Zhang F. 2015. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520:186–191. doi: 10.1038/nature14299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xie H, Tang L, He X, Liu X, Zhou C, Liu J, Ge X, Li J, Liu C, Zhao J, Qu J, Song Z, Gu F. 2018. SaCas9 requires 5'-NNGRRT-3' PAM for sufficient cleavage and possesses higher cleavage activity than SpCas9 or FnCpf1 in human cells. Biotechnology Journal 13. doi: 10.1002/biot.201700561 [DOI] [PubMed] [Google Scholar]
- 18. Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW, Bush K. 2013. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4:e00537–12. doi: 10.1128/mBio.00537-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kraemer GR, Iandolo JJ. 1990. High-frequency transformation of Staphylococcus aureus by electroporation. Current Microbiology 21:373–376. doi: 10.1007/BF02199440 [DOI] [Google Scholar]
- 20. Veiga H, Pinho MG. 2009. Inactivation of the SauI type I restriction-modification system is not sufficient to generate Staphylococcus aureus strains capable of efficiently accepting foreign DNA. Appl Environ Microbiol 75:3034–3038. doi: 10.1128/AEM.01862-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Monteiro JM, Fernandes PB, Vaz F, Pereira AR, Tavares AC, Ferreira MT, Pereira PM, Veiga H, Kuru E, VanNieuwenhze MS, Brun YV, Filipe SR, Pinho MG. 2015. Cell shape dynamics during the staphylococcal cell cycle. Nat Commun 6:8055. doi: 10.1038/ncomms9055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu S, de Lencastre H, Sali A, Tomasz A. 1996. A phosphoglucomutase-like gene essential for the optimal expression of methicillin resistance in Staphylococcus aureus: molecular cloning and DNA sequencing. Microb Drug Resist 2:277–286. doi: 10.1089/mdr.1996.2.277 [DOI] [PubMed] [Google Scholar]
- 23. Corrigan RM, Foster TJ. 2009. An improved tetracycline-inducible expression vector for Staphylococcus aureus. Plasmid 61:126–129. doi: 10.1016/j.plasmid.2008.10.001 [DOI] [PubMed] [Google Scholar]
- 24. Vellanoweth RL, Rabinowitz JC. 1992. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol Microbiol 6:1105–1114. doi: 10.1111/j.1365-2958.1992.tb01548.x [DOI] [PubMed] [Google Scholar]
- 25. Bi EF, Lutkenhaus J. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161–164. doi: 10.1038/354161a0 [DOI] [PubMed] [Google Scholar]
- 26. Monteiro JM, Pereira AR, Reichmann NT, Saraiva BM, Fernandes PB, Veiga H, Tavares AC, Santos M, Ferreira MT, Macário V, VanNieuwenhze MS, Filipe SR, Pinho MG. 2018. Peptidoglycan synthesis drives an FtsZ-treadmilling-independent step of cytokinesis. Nature 554:528–532. doi: 10.1038/nature25506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ruiz N. 2008. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc Natl Acad Sci USA 105:15553–15557. doi: 10.1073/pnas.0808352105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pereira SFF, Henriques AO, Pinho MG, de Lencastre H, Tomasz A. 2007. Role of PBP1 in cell division of Staphylococcus aureus. J Bacteriol 189:3525–3531. doi: 10.1128/JB.00044-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reichmann NT, Tavares AC, Saraiva BM, Jousselin A, Reed P, Pereira AR, Monteiro JM, Sobral RG, VanNieuwenhze MS, Fernandes F, Pinho MG. 2019. SEDS-bPBP pairs direct lateral and septal peptidoglycan synthesis in Staphylococcus aureus. Nat Microbiol 4:1368–1377. doi: 10.1038/s41564-019-0437-2 [DOI] [PubMed] [Google Scholar]
- 30. Pinho MG, Errington J. 2003. Dispersed mode of Staphylococcus aureus cell wall synthesis in the absence of the division machinery. Mol Microbiol 50:871–881. doi: 10.1046/j.1365-2958.2003.03719.x [DOI] [PubMed] [Google Scholar]
- 31. Ji Y, Zhang B, Van SF, Warren P, Woodnutt G, Burnham MKR, Rosenberg M. 2001. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science 293:2266–2269. doi: 10.1126/science.1063566 [DOI] [PubMed] [Google Scholar]
- 32. Chaudhuri RR, Allen AG, Owen PJ, Shalom G, Stone K, Harrison M, Burgis TA, Lockyer M, Garcia-Lara J, Foster SJ, Pleasance SJ, Peters SE, Maskell DJ, Charles IG. 2009. Comprehensive identification of essential Staphylococcus aureus genes using transposon-mediated differential hybridisation (TMDH). BMC Genomics 10:291. doi: 10.1186/1471-2164-10-291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Coe KA, Lee W, Stone MC, Komazin-Meredith G, Meredith TC, Grad YH, Walker S. 2019. Multi-strain Tn-Seq reveals common daptomycin resistance determinants in Staphylococcus aureus. PLoS Pathog 15:e1007862. doi: 10.1371/journal.ppat.1007862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fuchs S, Mehlan H, Bernhardt J, Hennig A, Michalik S, Surmann K, Pané-Farré J, Giese A, Weiss S, Backert L, Herbig A, Nieselt K, Hecker M, Völker U, Mäder U. 2018. Aureowiki the repository of the Staphylococcus aureus research and annotation community. Int J Med Microbiol 308:558–568. doi: 10.1016/j.ijmm.2017.11.011 [DOI] [PubMed] [Google Scholar]
- 35. Pereira AR, Reed P, Veiga H, Pinho MG. 2013. The holliday junction resolvase RecU is required for chromosome segregation and DNA damage repair in Staphylococcus aureus. BMC Microbiol 13:18. doi: 10.1186/1471-2180-13-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oku Y, Kurokawa K, Matsuo M, Yamada S, Lee BL, Sekimizu K. 2009. Pleiotropic roles of polyglycerolphosphate synthase of lipoteichoic acid in growth of Staphylococcus aureus cells. J Bacteriol 191:141–151. doi: 10.1128/JB.01221-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang W, Chen J, Chen G, Du X, Cui P, Wu J, Zhao J, Wu N, Zhang W, Li M, Zhang Y. 2015. Transposon mutagenesis identifies novel genes associated with Staphylococcus aureus persister formation. Front. Microbiol 6:1437. doi: 10.3389/fmicb.2015.01437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Singh VK, Utaida S, Jackson LS, Jayaswal RK, Wilkinson BJ, Chamberlain NR. 2007. Role for dnaK locus in tolerance of multiple stresses in Staphylococcus aureus. Microbiology (Reading) 153:3162–3173. doi: 10.1099/mic.0.2007/009506-0 [DOI] [PubMed] [Google Scholar]
- 39. Quiblier C, Zinkernagel AS, Schuepbach RA, Berger-Bächi B, Senn MM. 2011. Contribution of SecDF to Staphylococcus aureus resistance and expression of virulence factors. BMC Microbiol 11:72. doi: 10.1186/1471-2180-11-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sadykov MR, Thomas VC, Marshall DD, Wenstrom CJ, Moormeier DE, Widhelm TJ, Nuxoll AS, Powers R, Bayles KW. 2013. Inactivation of the Pta-AckA pathway causes cell death in Staphylococcus aureus. J Bacteriol 195:3035–3044. doi: 10.1128/JB.00042-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Blake KL, O’Neill AJ, Mengin-Lecreulx D, Henderson PJF, Bostock JM, Dunsmore CJ, Simmons KJ, Fishwick CWG, Leeds JA, Chopra I. 2009. The nature of Staphylococcus aureus MurA and MurZ and approaches for detection of peptidoglycan biosynthesis inhibitors. Mol Microbiol 72:335–343. doi: 10.1111/j.1365-2958.2009.06648.x [DOI] [PubMed] [Google Scholar]
- 42. Zhou C, Bhinderwala F, Lehman MK, Thomas VC, Chaudhari SS, Yamada KJ, Foster KW, Powers R, Kielian T, Fey PD, Peschel A. 2019. Urease is an essential component of the acid response network of Staphylococcus aureus and is required for a persistent murine kidney infection. PLoS Pathog 15:e1007538. doi: 10.1371/journal.ppat.1007538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kofoed EM, Yan D, Katakam AK, Reichelt M, Lin B, Kim J, Park S, Date SV, Monk IR, Xu M, Austin CD, Maurer T, Tan M-W. 2016. De novo guanine biosynthesis but not the riboswitch-regulated purine salvage pathway is required for Staphylococcus aureus infection in vivo. J Bacteriol 198:2001–2015. doi: 10.1128/JB.00051-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang B, Duan J, Jin Y, Zhan Q, Xu Y, Zhao H, Wang X, Rao L, Guo Y, Yu F. 2021. Functional insights of MraZ on the pathogenicity of Staphylococcus aureus Infect Drug Resist 14:4539–4551. doi: 10.2147/IDR.S332777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Poon R, Basuino L, Satishkumar N, Chatterjee A, Mukkayyan N, Buggeln E, Huang L, Nair V, Argudín MA, Datta SK, Chambers HF, Chatterjee SS. 2022. Loss of GdpP function in Staphylococcus aureus leads to beta-lactam tolerance and enhanced evolution of beta-lactam resistance. Antimicrob Agents Chemother 66:e0143121. doi: 10.1128/AAC.01431-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guo J, Wang T, Guan C, Liu B, Luo C, Xie Z, Zhang C, Xing XH. 2018. Improved sgRNA design in bacteria via genome-wide activity profiling. Nucleic Acids Res 46:7052–7069. doi: 10.1093/nar/gky572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yao L, Cengic I, Anfelt J, Hudson EP. 2016. Multiple gene repression in cyanobacteria using CRISPRi. ACS Synth Biol 5:207–212. doi: 10.1021/acssynbio.5b00264 [DOI] [PubMed] [Google Scholar]
- 48. Spoto M, Guan C, Fleming E, Oh J. 2020. A universal, genomewide guidefinder for CRISPR/Cas9 targeting in microbial genomes. mSphere 5:e00086-20. doi: 10.1128/mSphere.00086-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cui L, Vigouroux A, Rousset F, Varet H, Khanna V, Bikard D. 2018. A CRISPRi screen in E. coli reveals sequence-specific toxicity of dCas9. Nat Commun 9:1912. doi: 10.1038/s41467-018-04209-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Spoto M, Riera Puma JP, Fleming E, Guan C, Ondouah Nzutchi Y, Kim D, Oh J. 2022. Large-scale CRISPRi and transcriptomics of Staphylococcus epidermidis identify genetic factors implicated in lifestyle versatility. mBio 13:e0263222. doi: 10.1128/mbio.02632-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu X, Vd B, Heggenhougen MV, Mårli MT, Frøynes AH, Salehian Z, Porcellato D, Angeles DM, Veening J-W, Kjos M. 2023. Genome-wide CRISPRi screens reveal the essentialome and determinants for susceptibility to dalbavancin in Staphylococcus aureus. bioRxiv. doi: 10.1101/2023.08.30.555613:2023.08.30.555613 [DOI] [Google Scholar]
- 52. Fisher AC, DeLisa MP. 2008. Laboratory evolution of fast-folding green fluorescent protein using secretory pathway quality control. PLoS ONE 3:e2351. doi: 10.1371/journal.pone.0002351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bose JL, Fey PD, Bayles KW. 2013. Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl Environ Microbiol 79:2218–2224. doi: 10.1128/AEM.00136-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS. 2013. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc 8:2180–2196. doi: 10.1038/nprot.2013.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Arnaud M, Chastanet A, Débarbouillé M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol 70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pereira PM, Veiga H, Jorge AM, Pinho MG. 2010. Fluorescent reporters for studies of cellular localization of proteins in Staphylococcus aureus. Appl Environ Microbiol 76:4346–4353. doi: 10.1128/AEM.00359-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Costa SF, Saraiva BM, Veiga H, Marques LB, Schäper S, Sporniak M, Vega DE, Jorge AM, Duarte AM, Brito AD, Tavares AC, Reed P, Pinho MG. 2023. The role of GpsB in cell morphogenesis of Staphylococcus aureus. bioRxiv. doi: 10.1101/2023.06.16.545294:2023.06.16.545294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Novick R. 1967. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33:155–166. doi: 10.1016/0042-6822(67)90105-5 [DOI] [PubMed] [Google Scholar]
- 59. Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M, Dodson RJ, Daugherty SC, Madupu R, Angiuoli SV, Durkin AS, Haft DH, Vamathevan J, Khouri H, Utterback T, Lee C, Dimitrov G, Jiang L, Qin H, Weidman J, Tran K, Kang K, Hance IR, Nelson KE, Fraser CM. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol 187:2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Monk IR, Shah IM, Xu M, Tan MW, Foster TJ. 2012. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio 3:e00277-11. doi: 10.1128/mBio.00277-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kKilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- 62. Anderson JC, Dueber JE, Leguia M, Wu GC, Goler JA, Arkin AP, Keasling JD. 2010. BglBricks: a flexible standard for biological part assembly. J Biol Eng 4:1. doi: 10.1186/1754-1611-4-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pinho MG, de Lencastre H, Tomasz A. 1998. Transcriptional analysis of the Staphylococcus aureus penicillin binding protein 2 gene. J Bacteriol 180:6077–6081. doi: 10.1128/JB.180.23.6077-6081.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales APW, Li Z, Peterson RT, Yeh J-RJ, Aryee MJ, Joung JK. 2015. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523:481–485. doi: 10.1038/nature14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Oshida T, Tomasz A. 1992. Isolation and characterization of a Tn551-Autolysis mutant of Staphylococcus aureus. J Bacteriol 174:4952–4959. doi: 10.1128/jb.174.15.4952-4959.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Monk IR, Tree JJ, Howden BP, Stinear TP, Foster TJ. 2015. Complete bypass of restriction systems for major Staphylococcus aureus lineages. mBio 6:e00308-15. doi: 10.1128/mBio.00308-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Johnston CD, Cotton SL, Rittling SR, Starr JR, Borisy GG, Dewhirst FE, Lemon KP. 2019. Systematic evasion of the restriction-modification barrier in bacteria. Proc Natl Acad Sci USA 116:11454–11459. doi: 10.1073/pnas.1820256116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Saraiva BM, Krippahl L, Filipe SR, Henriques R, Pinho MG. 2021. eHooke: a tool for automated image analysis of spherical bacteria based on cell cycle progression. Biol Imaging 1:e3. doi: 10.1017/S2633903X21000027 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figures and tables.
Data Availability Statement
Data and strains will be made available upon reasonable request to the corresponding author.