Abstract
Nucleolar segregation is observed under some physiological conditions of transcriptional arrest. This process can be mimicked by transcriptional arrest after actinomycin D treatment leading to the segregation of nucleolar components and the formation of unique structures termed nucleolar caps surrounding a central body. These nucleolar caps have been proposed to arise from the segregation of nucleolar components. We show that contrary to prevailing notion, a group of nucleoplasmic proteins, mostly RNA binding proteins, relocalized from the nucleoplasm to a specific nucleolar cap during transcriptional inhibition. For instance, an exclusively nucleoplasmic protein, the splicing factor PSF, localized to nucleolar caps under these conditions. This structure also contained pre-rRNA transcripts, but other caps contained either nucleolar proteins, PML, or Cajal body proteins and in addition nucleolar or Cajal body RNAs. In contrast to the capping of the nucleoplasmic components, nucleolar granular component proteins dispersed into the nucleoplasm, although at least two (p14/ARF and MRP RNA) were retained in the central body. The nucleolar caps are dynamic structures as determined using photobleaching and require energy for their formation. These findings demonstrate that the process of nucleolar segregation and capping involves energy-dependent repositioning of nuclear proteins and RNAs and emphasize the dynamic characteristics of nuclear domain formation in response to cellular stress.
INTRODUCTION
The nucleus is a dynamic organelle consisting of interacting chromosomal and protein compartments. One of the major pathways of nuclear translocation is the movement of preribosomal particles from the nucleolus into the cytoplasm for the assembly of functional ribosomes. The main nucleolar functions involve RNA polymerase (pol) I transcription, posttranscriptional maturation of pre-rRNA transcripts and their subsequent assembly with ribosomal proteins into preribosomal particles. Other functions have been attributed to the nucleolus (for reviews, see Carmo-Fonseca et al., 2000; Olson, 2004b) and include the processing of RNA pol III transcripts, RNA editing, sequestration of cell cycle components in yeast, and Mdm2 protein in mammalian cells. The localization of telomere proteins and telomerase RNA in nucleoli suggests a role for the nucleolus in aging.
Nucleolar components are found in all cells and tissues although the size, shape, and number of nucleoli may change depending on the species, cell type, and functional state. Transmission electron microscopy (TEM) has revealed three major structures within nucleoli: fibrillar centers (FC), dense fibrillar components (DFC), and the granular component (GC; for reviews, see Busch and Smetana, 1970; Goessens, 1984; Shaw and Jordan, 1995; Scheer and Hock, 1999). rDNA transcription units are found in the FC and consist of tandem repeats of these genes. rRNAs are harbored within the DFC and are processed there. It is therefore thought that rRNA transcription occurs at the interface between the FC and the DFC. Later stages of rRNA processing take place in the GC. Thus, the processing of rRNA is spatially arranged in accordance to the ultrastructure of these compartments.
Great variability is found between nucleoli of cells observed at different stages of cellular metabolic activity. In quiescent cells or cells subjected to transcriptional arrest a phenotype of nucleolar segregation is observed, in which the fibrillar and granular zones disengage to form separate but juxtaposed structures (Smetana and Busch, 1974; Vera et al., 1993; Malatesta et al., 2000). In some cases, for example in developing Xenopus oocytes (Van Gansen and Schram, 1972), these structures resemble cap-like formations situated on the outer part of the segregated nucleolus. Although the processes of nucleolar segregation and nucleolar capping are physiological occurrences assumed to reflect the inhibition of RNA synthesis, they have not been pursued and have only been structurally characterized, mostly by TEM, using agents that induce transcriptional inhibition (for reviews, see Bernhard and Granboulan, 1968; Busch and Smetana, 1970; Simard et al., 1974; Smetana and Busch, 1974). Based on differences in phase contrast light microscopy, the formation of two types of “nucleolar caps” was observed during transcriptional arrest by inhibitors such as actinomycin D (ActD; Journey and Goldstein, 1961; Reynolds et al., 1963, 1964). Multiple “dark nucleolar caps” (DNCs) had a concave base and appeared to be pressed onto the surface of the nucleolar body, thus forming an interface between the two. The less frequent “light nucleolar caps” (LNCs) had a convex appearance without a clear margin between them and the nucleolar body, therefore seeming closely attached or protruding slightly into the nucleolar body. Time-lapse microscopy showed that this cap originated from the center of the nucleolus. Independently, Schoefl observed similar structures: RNP granules embedded in a protein matrix and a fibrillar RNP component (Schoefl, 1964). Another study called the granular structures the P2 fraction, forming on the surface of the nucleolar body termed P1 and separate from other smaller caps he termed the “fibrillar substance” (Recher et al., 1971).
These studies have led to the general assumption that nucleolar caps consist of nucleolar proteins originating from the disintegrating nucleolus. However, the static view of the nucleolus in 1960s and 70s has since been replaced by our knowledge that the nucleolus is a dynamic structure that has the ability to disassemble and reassemble (for review see Hernandez-Verdun, 2004). We have previously shown how a nucleoplasmic protein, normally excluded from the nucleolus, is highly enriched in the nucleolar region (Shav-Tal et al., 2001b). Another study, has shown by use of proteomics that a number of proteins are enriched in the nucleolar fraction during transcriptional arrest induced by ActD (Andersen et al., 2002). The purpose of the present study was to determine which nucleoplasmic proteins can be directed to the nucleolar caps and whether they belong to a specific family of proteins. The analysis of more than 70 endogenous nuclear proteins allows us to assemble a comprehensive picture of nuclear compartments during transcriptional arrest. By characterizing the different types of nucleolar caps and identifying their protein and RNA composition, we show that nucleolar segregation is a concerted process involving specific spatial reshuffling of nucleoplasmic and nucleolar components in response to transcriptional arrest. We find that nucleolar caps are dynamic structures constantly exchanging with the nucleoplasm and that their formation is an energy-dependent process.
MATERIALS AND METHODS
Cell Culture
For indirect immunofluorescence and in situ hybridization, HeLa cells were grown on glass coverslips in DMEM supplemented with 10% fetal calf serum (FCS). Human U2OS osteosarcoma cells were cultured in low-glucose DMEM (Invitrogen, Carlsbad, CA) with 10% FCS and for live cell experiments were maintained in phenol-red free Leibovitz's L15 medium at 37°C using a temperature-controlled Delta T4 culture dish system with a heated lid and an objective heater (Bioptechs, Butler, PA). For transcriptional inhibition of RNA polymerase I and II 5 μg/ml ActD (Sigma, St. Louis, MO) were added to cells for 2 h (Ochs, 1998). ActD at 0.01 μg/ml for 3–6 h was used for inhibition of RNA pol I only and 5,6-dicholoro-β-d-ribofuranosylbenzimidazole (DRB) at 25 μg/ml for 2 h or α-amanitin at 30 μg/ml for 6 h was used inhibition of pol II only. 2,3-butanedione monoxime (BDM, 20 mM) was used for myosin I inhibition. For full metabolic depletion, 6 mM 2-deoxyglucose (Sigma) and 10 mM Na-azide were added to cell cultures in either Hanks' balanced salt solution (HBSS) or DMEM. Glucose, 25 mM, or 2 mM pyruvate were added to HBSS as indicated.
Protein Extraction and Western Blotting
HeLa cells were lysed on ice in 50 mM Tris-HCl, pH 8, containing 1% Nonidet P-40 (NP-40), 150 mM NaCl, 5 mM EDTA, 1 mM phenyl methylsulfonyl fluoride, 3 μg/ml aprotinin, 20 μg/ml leupeptin, 10 mM iodoactetate, 1 mM sodium orthovanadate, 10 mM sodium fluoride. Protein extracts were boiled in SDS-sample buffer (5% glycerol, 2% SDS, 62.5 mM Tris-HCl, pH 6.8, 2% 2-mercaptoethanol, 0.01% bromophenol blue) and analyzed by SDS-PAGE. Immunoblot analysis of cell lysates (40 μg/lane) was performed using the anti-PSF B92 monoclonal antibody (mAb). Specific binding was detected with goat anti-mouse horseradish peroxidase (HRP)-coupled antibody (Sigma) and enhanced chemiluminescence (ECL) reagents.
Primary Antibodies to Nuclear Proteins
The primary antibodies to nuclear proteins are shown in Tables 1 and 2.
Table 1.
Antibodies to components of nucleolar caps
| Antigen in nucleolar caps | Antibody | Clone | Antibody provided by |
|---|---|---|---|
| ALL-1 | Rab | P4 | E. Canaani, Weizmann Institute, Israel |
| cdk2 | Rab | D. Ginsberg, Weizmann Institute, Israel | |
| Coilin | Rab | A. Lamond, University of Dundee, UK | |
| CstF-64 | MAb | Y. Takagaki, University of Virginia School of Medicine | |
| EWS | Rab | 677 | O. Delattre, Curie Institute, France |
| Fibrillarin | MAb | I. Raska, Academy of Sciences of the Czech Republic, Czech Republic | |
| gar1 | Rab | W. Filipowicz, Friedrich Miescher Institute for Biomedical Research, Switzerland | |
| hnRNP F and H | Rab | D. Black, University of California | |
| hnRNP K | Rab | K. Bomsztyk, University of Washington | |
| MPP10 | Guinea pig | S. Baserga, Yale University School of Medicine | |
| Nopp140 | Rab | RH10 | T. Meier, Albert Einstein College of Medicine |
| p110 | Rab | C. Adamson, University of Arizona | |
| p220NPAT | Rab | J. Harper, Baylor College of Medicine | |
| p54nrb/NonO | Rab | P. Tucker, University of Texas at Austin | |
| p68 | MAb | F. Fuller-Pace, University of Dundee, UK | |
| PML | MAb | 5E10 | R. Van Driel, University of Amsterdam, The Netherlands |
| PML | MAb | A. Ben-Ze'ev, Weizmann Institute, Israel | |
| PML | MAb | PGM3 | NeoMarkers, Fremont, CA |
| PSF | MAb | B92 | Lee et al. (1996) |
| PSF | Rab | 1121 | Shav-Tal et al. (2000) |
| Sm | MAb | Y12 | A. Lamond, University of Dundee, UK |
| Sp100 | Rat | rSp26 | H. Will, Heinrich-Pette Institute for Experimental Virology and Immunology at the University of Hamburg, Germany |
| TAFII70 | MAb | Transduction Labs, Lexington, KY | |
| TBP | Rab | Santa Cruz Biotechnology, Santa Cruz, CA | |
| TLS | Rab | D. Ron, NYU School of Medicine | |
| TLS | MAb | 4H11 | R. Goodman, Vollum Institute, Portland |
| U1-70K | MAb | H111 | R. Lührmann and E. Makarov, Max-Planck Institute for Biophysical Chemistry, Germany |
| U1-70K | MAb | A. Rosen, John Hopkins School of Medicine | |
| UBF | Human | E. Chan, Scripps Research Institute |
Table 2.
Antibodies to proteins that are not components of nucleolar caps
| Antigen not in nucleolar caps | Species | Clone | Antibody provided by |
|---|---|---|---|
| 4.1R | Rab | 10b | I. Correas, Universidad Autonoma de Madrid, Spain |
| BAF155 | Goat | Santa Cruz | |
| CBP | Rab | Santa Cruz | |
| c-myc | MAb | NeoMarkers | |
| CrkRS | Rab | J. Pines, Wellcome/CRC Institute, UK | |
| FBP11 and FBP21 | Rab | M. Bedford, University of Texas, MD Andersen Cancer Center | |
| fos | MAb | NeoMarkers | |
| Gemin 2 | MAb | 2E17 | G. Dreyfuss, University of Pennsylvania School of Medicine |
| HDAC3 | MAb | Transduction Labs | |
| histone H3 | Rab | Upstate Biotechnology, Lake Placid, NY | |
| acetyl-K9 | |||
| dimethyl-K9 | |||
| dimethyl-K4 | |||
| HMGN1 and 2 | Rab, Goat | R. Hock, University of Würzburg, Germany | |
| hnRNP A1 | MAb | 4B10 | G. Dreyfuss, University of Pennsylvania School of Medicine |
| hnRNP A2/B1 | Goat | E. Canaani, Weizmann Institute, Israel | |
| hnRNP C1/C2 | MAb | 4F4 | G. Dreyfuss, University of Pennsylvania School of Medicine |
| hnRNP R & Q | Rab | M. Sendtner and W. Rossoll, University of Würzburg, Germany | |
| hPrp5 | Rab | C. Query, Albert Einstein College of Medicine | |
| HSF-1 | Rab | StressGen, Victoria, British Columbia, Canada | |
| Importin α | MAb | BD Biosciences, San Diego, CA | |
| JunB | Rab | Santa Cruz | |
| KSRP | MAb | Ab5 | D. Black, University of California |
| lamin A | MAb | LA-2H10 | V. Parnaik, Center for Cellular and Molecular Biology, India |
| lamin B1 | Rab | E. Canaani, Weizmann Institute, Israel | |
| Lsm10 | Rab | D. Schümperli, University of Bern, Germany | |
| MAQ1 | Rab | O. Bensaude, ENS-CNRS Paris, France | |
| Mdm2 | MAb | V. Rotter, Weizmann Institute, Israel | |
| NF-IL6 | Rab | Santa Cruz | |
| NSAP1 | Rab | B4 & 3E | C. Astell, University of British Columbia, Canada |
| nuclear myosin 1β | Rab | P. de Lanerolle, University of Illinois at Chicago | |
| Nucleolin | MAb | 7G2 | S. Piñol-Roma, Mount Sinai School of Medicine |
| Nucleophosmin | Rab | M. Olson, University of Mississippi Medical Center | |
| NuMA | Rab | D. Compton, Dartmouth Medical School | |
| Nup153 | MAb | QE5 | R. Bastos, Hospital Clinic Universitari, Spain |
| p/CAF | Rab | Santa Cruz | |
| p14(ARF) | MAb | 14PO2 | K. Wiman and M. Lindström, Karolinska Hospital, Sweden |
| Rab | 271, FL132 | ||
| p62 of TFIIH | MAb | J. Egly and B. Sandrock, Louis Pasteur University, France | |
| PABP-2 | Rab | E. Wahle and U. Kühn, Martin-Luther-Universität Halle-Wittenberg, Germany | |
| PACT | Rab | V. Rotter, Weizmann Institute, Israel | |
| PTB | MAb | D. Helfman, Cold Spring Harbor Laboratory | |
| RNA pol II | MAb | 8WG16 | R. Burgess, University of Wisconsin-Madison |
| RNA pol II | MAb | H5, H14 | Y. Shav-Tal, Albert Einstein College of Medicine |
| RNA pol III RPC155 and RPC53 | Rab | N. Hernandez, Cold Spring Harbor Laboratory | |
| RNA pol III RPC62 | Rab | R. Roeder, Rockefeller University | |
| SAF-A | Rab | F. Fackelmayer, Heinrich-Pette Institute for Experimental Virology and Immunology at the University of Hamburg, Germany | |
| SC-35 | MAb | E. Canaani, Weizmann Institute, Israel | |
| Sin3A | Rab, MAb | Santa Cruz | |
| SMN | MAb | 2B1 | G. Dreyfuss, University of Pennsylvania School of Medicine |
| TFIIFβ RAP74 and RAP30 | Rab | S. Kitajima, Tokyo Medical and Dental University, Japan | |
| U2AF65 | MAb | MC3 | M. Carmo-Fonseca, University of Lisbon, Portugal |
| U5 snRNP 116K | Rab | R. Lührmann, Max-Planck Institute for Biophysical Chemistry, Germany | |
| YT521-B | Rab | S. Stamm, Friedrich-Alexander-University Erlangen, Germany |
Secondary Antibodies
Fluorophore-labeled secondary antibody purchased from Jackson ImmunoResearch (West Grove, PA) were as follows: for double labelings with primary mouse and rabbit antibodies: fluorescein (FITC)-conjugated donkey anti-rabbit F(ab′)2, Cy3-conjugated goat anti-mouse IgG; for triple labeling: Cy5-conjugated goat anti-rabbit F(ab′)2; other antibodies used for verification of stainings: R-phycoerythrin (PE)-conjugated donkey anti-rabbit F(ab′)2, FITC-conjugated donkey anti-mouse IgG, Alexa 488 donkey anti-goat IgG (Molecular Probes, Eugene, OR). For other double labelings: FITC-conjugated mouse anti-rat IgG, Texas Red–conjugated goat anti-human IgG and Cy3-conjugated anti-mouse IgM (Jackson ImmunoResearch), FITC-conjugated anti-guinea pig IgG (B. Geiger, Weizmann Institute), FITC-conjugated anti-mouse IgM (Serotec, Oxford, United Kingdom).
Immunofluorescence and In Situ Hybridization
HeLa cells were fixed for 2 min in 4% paraformaldehyde with 0.5% Triton X-100 and for an additional 20 min in 4% paraformaldehyde. After washing and blocking in 5% bovine serum albumin (BSA), cells were stained with the indicated antibodies for 45 min, washed twice and then incubated with the appropriate secondary antibodies for 45 min, and counterstained with Hoechst for DNA labeling. Before all confocal double-labeling experiments each antibody was first checked on its own by immunofluorescent microscopy. Immunofluorescence was viewed and analyzed using a Bio-Rad confocal microscope (Bio-Rad, Richmond, CA) or a Zeiss Axioplan microscope equipped with SPOT-II (Diagnostic Instruments, Sterling Heights, MI) cooled CCD camera.
For in situ hybridization the following probes were synthesized, labeled with either Cy3 or Cy5, and hybridized as previously described (Chartrand et al., 2000): U3: CGCTACCTCTCTTCCTCGTGGTTTTCGGTG-CTCTACACGTTCAGAGAAAC; CTCCCTCTCACTCCCCAATACGGAGAGAAGAACGATCATCAATGGCTGAC (two probes); U2: AGTGGACGGAGCAAGCTCCTATTCCATCTCCCTGCTCCAAAAATCCATTT; U6: CGTGTCATCCTTGCGCAGGGGCCATGCTAATCTTCTCTGTATCGTTCCAA; U93: CATGCCTCAGCTTCCTCTGGAGTATGCTTTTTCTGCAAGTCTGGTGTGCT; ACCAGAAACTATCAGAGGAAAATTGCACATGGAACAGCTGGCTCTCGAGC; MRP: GGAACAGAGTCCTCAGTGTGTAGCCTAGGATACAGGCCTTCAGCACGAAC; RNAseP: ATTGAACTCACTTCGCTGGCCGTGAGTCTGTTCCAAGCTCCGGCAAAGGA; 5′ ETS: CTCTCAGATCGCTAGAGAAGGCTTTTCTCACCGAGGGTGGGTCACACTCC; 7SK: CTTGAGAGCTTGTTTGGAGGTTCTAGCAGGGGAGCGCAGCTACTCGTATA; CCACATGCAGCGCCTCATTTGGATGTGTCTGGAGTCTTGGAAGCTTGACT; and E3: CTGAGCAGGGGGAACGACAACACAGCACTGAGCAGCCATATTGTAGTAAA; CAAGCGTCCCTGGCTGCTACAGGTAGACAGCAGACAGGTATAGTTAAGAA
For poly(A) mRNA detection an oligo(dT) probe was used. After hybridization, immunofluorescence was performed from the blocking step as above.
For chromosomal DNA FISH, U2OS cells were seeded at low density in two-well chamber slides (Nunc, Napierville, IL) and grown over night. One well was treated with ActD for 2 h. Cells were fixed in ice cold methanol for 10 min. Chromosome painting probes were generated by degenerate oligonucleotide primed PCR amplification (DOP-PCR) of flow-sorted chromosomes (University of Cambridge, United Kingdom) with direct incorporation of Spectrum Orange-dUTP (Vysis, Abbot Laboratories, Abbott Park, IL). Detailed protocols for probes generation and FISH are available at (http://www.riedlab.nci.nih.gov). Slides were denatured in formaldehyde/SSC for 1.5 min and hybridized overnight at 37°C in a moist chamber.
Images were acquired with an Olympus BX61 epifluorescence microscope (Olympus America, Melville, NY) and a Roper Scientific CoolSNAP HQ camera (Roper Scientific, Tucson, AZ).
Transfection
For studies in fixed cells, HeLa cells were transfected with GFP-PSF constructs (Dye and Patton, 2001) or GFP-p54nrb (Peng et al., 2002) or GFP-PML (A. Ben-Ze'ev, Weizmann Institute; Shtutman et al., 2002) or YFP-ASF/SF2 (Bubulya et al., 2004) using either the calcium phosphate transfection method or Fugene. Cells were fixed and processed for immunofluorescence after overnight transfection. For live cell imaging, U2OS cells were electroporated with the following constructs: GFP-fibrillarin, GFP-Nopp140 (T. Meier, AECOM; Dundr et al., 2004), GFP-p14(ARF) (G. Peters, Cancer Research UK, London; Llanos et al., 2001). For GFP-TLS, the TLS sequence was amplified by PCR from pBS-TLS and subcloned into the SacI-BamHI sites of pEGFP-C3 (Clontech, Palo Alto, CA). Stable GFP-PSF U2OS cells were generated by electroporation and selected under G418.
Fluorescent Recovery after Photobleaching
Transfected U2OS cells, with or without ActD treatment, were imaged at 37°C with a Leica TCS SP2 AOBS laser scanning confocal microscope equipped with a 63×, 1.4 NA objective (Leica Microsystems, Exton, PA) and scanned using a 488-nm laser for the detection of GFP at low laser powers (<1.5%) to avoid bleaching and cytotoxicity. For photobleaching, one scan at 100% of the laser was required using the Leica fluorescence recovery after photobleaching (FRAP) module. Specific scanning times and intervals between images were used for each GFP-tagged protein depending on the measured recovery times. Measurements of intensity were performed using the Leica software. For each time point, the background taken from a ROI outside of the cell was subtracted from all other measurements. T(t) and I(t) were measured for each time point as the average intensity of the nucleus and the average intensity in the bleached ROI, respectively. One image was collected before bleaching and these initial conditions are referred to as Ti = nuclear intensity and Ii = intensity in ROI before bleaching. Ic(t) is the corrected intensity of the bleached ROI at time t (Phair and Misteli, 2000):
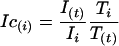 |
Transmission Electron Microscopy
HeLa cells were fixed with 4% paraformaldehyde, 0.1% glutaraldehyde in 0.1 M sodium cacodylate buffer, dehydrated through a graded series of ethanol using a progressively lowering of temperature technique, embedded in Lowicryl HM20 resin, and polymerized with UV light at -20°C. Ultrathin sections were cut on a Reichert Ultracut UCT and viewed on a JEOL 1200EX transmission electron microscope at 80 kV. For immunogold labeling, thin sections mounted on grids were incubated with blocking solution with the addition of 5% BSA and 1% cold water fish gelatin, followed by primary antibody and secondary antibody conjugated to 10 nm gold (Aurion, Wageningen, The Netherlands).
RESULTS
PSF and GFP-PSF Colocalize within ActD-induced Nucleolar Caps
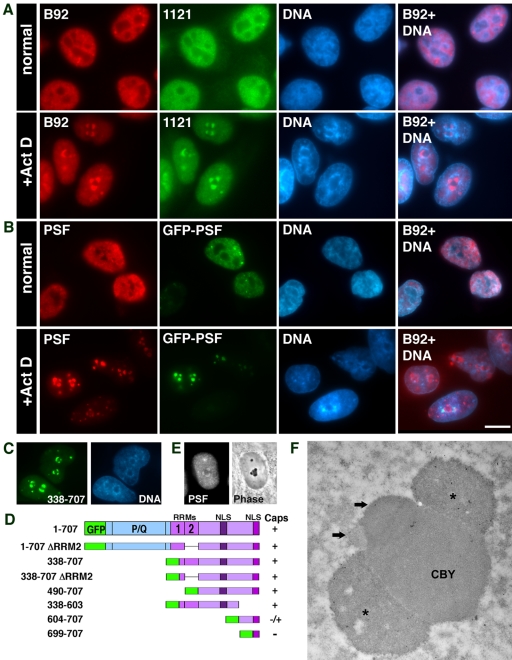
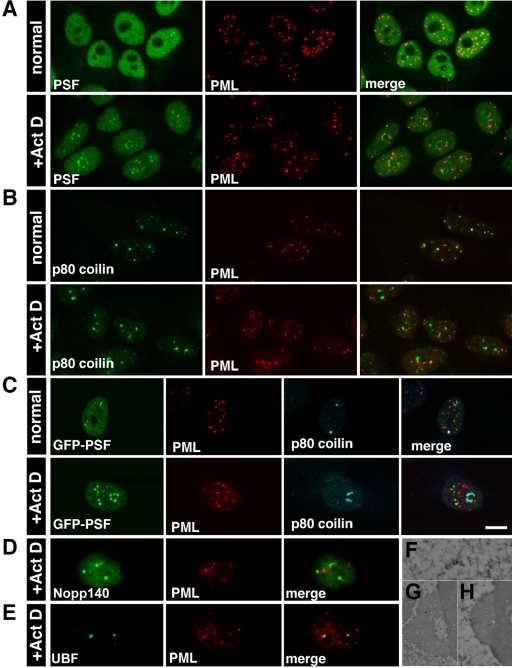
We have previously found that a nucleoplasmic protein, PSF (PTB-associated splicing factor; Shav-Tal and Zipori, 2002) formed peri-nucleolar structures in HL-60 cells treated with transcriptional inhibitors ActD and DRB (Shav-Tal et al., 2001b). This phenomenon occurred also in GFP-PSF transfected HeLa cells (Dye and Patton, 2001). This finding is of interest because nucleolar segregation is thought to involve separation of nucleolar components rather than addition of nucleoplasmic components. We were therefore interested in defining the conditions that caused PSF to relocalize to the nucleolus and the structures to which it was targeted. PSF in the nucleoplasm exhibited both diffuse and punctate staining that was excluded from the nucleolus (Figure 1A). In addition, several brighter foci were observed (Shav-Tal et al., 2000). During ActD treatment of HeLa cells, both monoclonal (B92) and polyclonal (1121) anti-PSF antibodies identified the PSF protein in 2–3 concave cap-like structures surrounding the nucleolus (Figure 1A). There was a consequent reduction in nucleoplasmic PSF and the bright foci. In addition, the nucleolar caps were surrounded by a dense chromatin ring. Concentration of PSF in nucleolar caps was observed during inhibition of RNA polymerases I and II using concentrations of ActD reported to result in fully segregated nucleoli (Ochs, 1998). Similarly, nucleolar relocalization of PSF was seen when the RNA pol II specific inhibitors DRB or α-amanitin were used, but not when only RNA pol I activity was inhibited by low concentrations of ActD (unpublished data). We therefore focused on conditions sufficient to inhibit both RNA pol I and II.
Figure 1.
PSF and GFP-PSF localize in the same nucleolar caps. (A) Immunofluorescence images of double staining for nucleoplasmic PSF with the B92 mAb and the 1121 polyclonal antibody in untreated and in ActD-treated cells. (B) GFP-PSF–transfected cells were labeled with the B92 antibody to PSF. Hoechst DNA counterstain is shown in blue. The forth column on the right is a computer-generated overlap of the PSF and DNA stain showing the dense chromatin ring surrounding the nucleolar caps. Bar, 10 μm. (C) GFP-PSF 338–707 forms nucleolar caps. (D) Other GFP-PSF constructs were tested for nucleolar cap formation: +, form caps; -, do not form caps. P/Q, proline- and glutamine-rich domain; RRM, RNA recognition motif; NLS, nuclear localization sequences. (E) PSF (left panel: immunofluorescence) is found in phase dark nucleolar caps (DNCs; right panel: phase contrast). (F) Immunogold labeling with anti-PSF showed localization in large concave caps (stars) situated on the central body (CBY). The smaller caps (arrows) represent the segregated DFC and FC.
GFP-PSF entered the same nucleolar caps identified by the anti-PSF antibodies (Figure 1B). This required the C-terminal portion of PSF (amino acids 338–707) containing two RNA binding domains (RRMs) and nuclear localization sequences (NLSs; Figure 1C). The N-terminal part of PSF did not enter the nucleus. GFP-PSF localized to nucleolar caps after deletion of either or both RRMs (amino acids 490–707), required for normal localization in nucleoplasmic foci (Dye and Patton, 2001). This targeting was reduced in constructs containing only the very C-terminal portion containing the last NLS (604–707 and 699–707; Figure 1D).
We proceeded to clarify which type of nucleolar cap contained PSF after ActD treatment. Using phase-contrast microscopy, PSF was detected in large and dark nucleolar caps (DNCs; Figure 1E). Immunogold labeling showed that PSF was enriched in the large nucleolar caps (Figure 1F, stars), previously described as having a concave base and appearing pressed onto the surface of the central body that was originally the GC (Reynolds et al., 1964). For the sake of clarity, we will refer to nucleolar caps using the original terminology (Reynolds et al., 1964). The above nucleolar caps will be referred to as concave caps or DNCs. This is in contrast to the smaller and convex shaped nucleolar caps termed “light nucleolar caps” (LNCs), closely attached to the central body (CBY) and occasionally protruding into it (Figure 1F, top arrow and see Figure 4D for phase). It is known that in the segregated nucleolus, the FC is removed from the DFC and forms nucleolar caps termed fibrillar caps that cannot be seen by light microscopy but that are in close association with LNCs (Figure 1F, bottom arrow and Figure 4D for phase). The remaining granular component will be called the central body (CBY; Figure 1F).
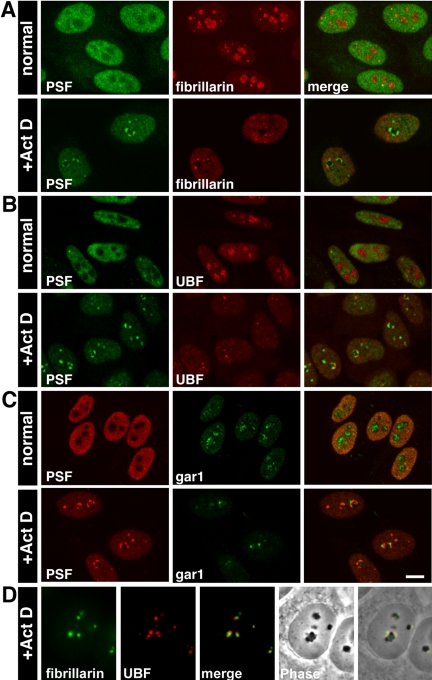
Figure 4.
DNCs, LNCs, and fibrillar caps are distinct compartments. Confocal images of (A) untreated: PSF is nucleoplasmic, whereas fibrillarin was in the nucleoli; treated: PSF and fibrillarin relocalized in the two different nucleolar caps. This was true also for (B) PSF and UBF, (C) PSF and gar1. Bar, 10 μm. (D) Fibrillarin is found in phase light nucleolar caps (LNCs), whereas UBF, representing the transcriptional machinery, is found in the adjacent fibrillar caps.
Composition of Dark Nucleolar Caps
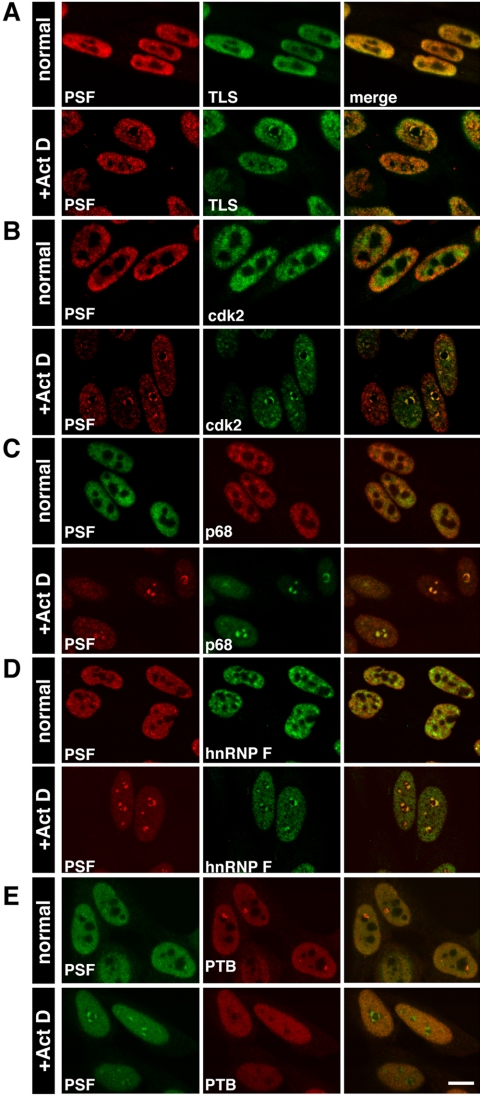
We identified components of either dark or light nucleolar caps using antibodies to more than 70 endogenous nucleoplasmic or nucleolar proteins. PSF was used as a reference for dark nucleolar caps. TLS/FUS is a nucleoplasmic protein that colocalized with PSF in dark nucleolar caps (DNCs; Figure 2A). It has previously been shown to enter nucleolar structures through its oncogenic N-terminus (Zinszner et al., 1997). This was also true for GFP-TLS and colocalization with PSF was also corroborated in cells cotransfected with GFP-PSF and HA-TLS (unpublished data). We also identified EWS, a homologue of TLS, and p54nrb/NonO (and GFP-p54nrb), a heterodimer of PSF, as components of DNCs (Table 3). The latter corroborates the localization of PSF to nucleolar caps, because p54nrb shares homology with the C-terminus of PSF.
Figure 2.
Nucleoplamic proteins that colocalize with PSF in concave DNCs. Confocal images of untreated and ActD-treated cells showing colocalization of (A) PSF and TLS, (B) PSF and cdk2, (C) PSF and p68, (D) PSF and hnRNP F. (E) PSF and PTB do not colocalize in concave nucleolar caps. The strong foci seen in PTB labeling of untreated cells are the perinucleolar compartments (PNC). Right-hand column in this and the following figures shows the merged output. Bar, 10 μm
Table 3.
Endogenous nuclear proteins and RNAs that we detected in nucleolar caps (using immunofluorescence and FISH)
| DNC | LNC | FC | Central body | Cajal body | PML body |
|---|---|---|---|---|---|
| PSF | Fibrillarin | UBF | p14(ARF) | p80 coilin | PML |
| p54nrb | Nopp140 | TBP | Sp100 | ||
| TLS/FUS | gar1 | ||||
| EWS | ALL-1 | ||||
| U1–70K | p110 | ||||
| cdk2 | |||||
| hnRNP K | |||||
| p68 helicase | |||||
| p220NPAT | |||||
| TAFII70 | |||||
| CstF-64 | |||||
| hnRNP F | |||||
| hnRNP H | |||||
| MPP10 | |||||
| Pre-rRNA | U14 snoRNA | MRP RNA | U93 scaRNA | ||
| E3 snoRNA | |||||
| U3 snoRNA | |||||
| U6 snRNA |
Other known cap-localizing proteins detected in concave nucleolar caps were the U1–70K component of U1 snRNP (Carmo-Fonseca et al., 1991), cdk2 (Liu et al., 2000; Figure 2B), and hnRNP K (Kamath et al., 2001; Table 3). Paraspeckle proteins PSP1, PSP2/CoAA, and p54nrb together with the helicases p68 and p72 are also present (Fox et al., 2002). Indeed, we confirmed that PSF, p68 (Figure 2C) and p54nrb colocalized to the same concave DNC and thus conclude that PSP1, PSP2, and p72 are also components of DNCs.
We identified previously unknown components of DNCs such as p220NPAT, which interacts with cdk2-cyclin E complex; the TFIID subunit TAFII70; CstF-64, which is involved in the polyadenylation process; the hnRNP F and H proteins (Figure 2D); and the SR splicing factor ASF/SF2 (Table 3). On the other hand, PTB (polypyrimidine tract-binding protein) known to be in complex with PSF under normal conditions (Patton et al., 1993; Meissner et al., 2000) remained dispersed in the nucleoplasm and did not relocalize with PSF to nucleolar caps (Figure 2E and Table 4). Interestingly, the peri-nucleolar compartment (PNC; Matera et al., 1995; Huang et al., 1997) normally containing PTB (Figure 2E) disappeared under these conditions. Sin3A, another nuclear factor directly associated with PSF (Mathur et al., 2001) was not found in DNCs either (Table 4).
Table 4.
Endogenous nuclear proteins and RNAs that did not compartmentalize in nucleolar caps
| RNA processing and spliceosome | Transcription related factors | Nuclear structure and chromatin | Nucleolar processes | Other nuclear factors |
|---|---|---|---|---|
| SC-35 | RNA pol II | Lamin A | Nucleolin | CrkRS (kinase) |
| PTB | RNA pol III | Lamin B1 | Nucleophosmin | Nup153 (pore) |
| hnRNP A1 | Sin3A | 4.1R | PABP-2 (polyA) | |
| hnRNP A2/B1 | p/CAF | HDAC3 | PACT | |
| hnRNP C1/C2 | CBP | SAF-A | YT521-B | |
| hnRNP R and Q | JunB | BAF155 | NSAP1 | |
| Lsm 10 | C/EBPβ | NuMA | HSF1 (heatshock) | |
| U2AF65 | Mdm2 | HMGN1 and 2 | Imp-α (import) | |
| U5 snRNP116K | TFIIFα, TFIIFβ | |||
| SMN | TFIIH | |||
| Gemin2 | Nuclear myosin I | |||
| FBP11, FBP21 | MAQ1 | |||
| KSRP | c-myc, fos | |||
| hPrp5 | ||||
| U2 snRNA | 7SK RNA | RNaseP RNA | ||
| mRNAs | ||||
| β-actin mRNA |
Because many of the identified DNC proteins are normally associated with mRNAs, we were interested to determine whether other components of the transcriptional machinery can be found in DNCs. A recent study has identified proteins of the transcription and splicing machineries in functional complexes (termed X1 and X2) formed at the mRNA 5′ splice site (Kameoka et al., 2004). Some of the RNA-binding proteins identified were PSF, p54nrb, TLS, and U1–70K, all components of DNCs. We therefore tested the presence of other X1/X2 proteins in the segregated nucleolus. X1/X2 complexes contain the active forms of RNA pol II together with transcription, splicing, and elongation factors. However, we did not detect the spliceosome-associated proteins hPrp5, U5–166K, FBP11, and FBP21, or the hypo- or hyperphosphorylated forms of RNA pol II CTD, or the basal transcription factors TFIIF and TFIIH, or the elongation factor P-TEFb in DNCs (Table 4). These data indicate that even though an influx of a unique group of nucleoplasmic proteins into the region of the segregating nucleolus is observed, and many of them are RNA binding proteins, this relocation is not a general trait of all the RNA transcribing and processing machinery. Moreover, even though nucleolar caps are commonly regarded as part of the nucleolus, we could not detect nucleolar proteins in the large and concave (DNC) caps.
Composition of LNCs and Fibrillar Caps
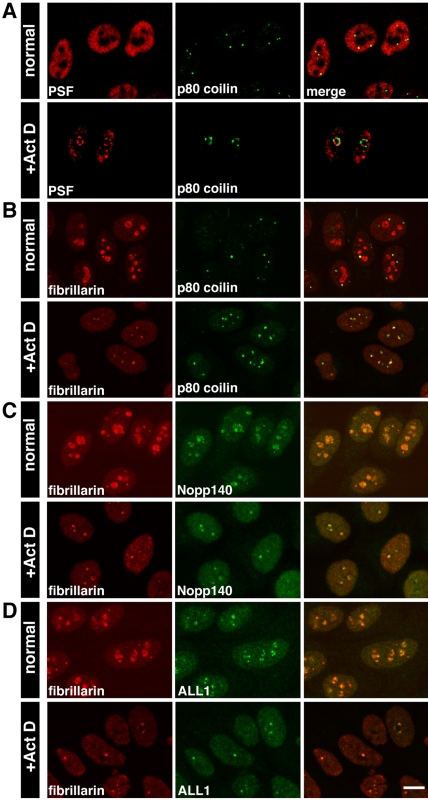
p80 coilin, a nucleoplasmic protein, is normally found in Cajal bodies, which redistribute to the nucleolar periphery during inhibition of transcription (Raska et al., 1990). It was shown to form nonconcave caps distinct from U1–70K caps (Carmo-Fonseca et al., 1992). We found that PSF and p80 coilin each formed two distinct nucleolar caps, observed tandemly around the nucleolar body (Figure 3A). p80 coilin positive areas around the nucleolus were a peripheral subset (Raska et al., 1990) distinct from nucleolar caps formed by fibrillarin, a DFC marker (Ochs et al., 1985). The incomplete overlap between fibrillarin and p80 coilin can be seen in Figure 3B. Fibrillarin containing nucleolar caps are termed “light nucleolar caps” (LNCs; see Figure 4D for phase).
Figure 3.
Nuclear proteins that colocalize in LNCs. Confocal images of untreated and ActD-treated cells showing: (A) Untreated: PSF nucleoplasmic staining and p80 coilin labeling of Cajal bodies. Treated: PSF and p80 coilin were found in two distinct nucleolar caps. (B) Untreated: fibrillarin was found mainly in nucleoli but also in Cajal bodies. Treated: partial overlap between p80 coilin caps and fibrillarin caps. (C) Fibrillarin and Nopp140 were found in the same nucleolar caps. (D) Fibrillarin and ALL-1 also colocalized in these caps. Bar, 10 μm.
Other nucleolar proteins that compartmentalize with fibrillarin in LNCs upon transcriptional inhibition are Nopp140 (Chen et al., 1999; Figure 3C), gar1 (Pellizzoni et al., 2001), and MSP58 (Ren et al., 1998; Table 3). Interestingly, we found that the Drosophila trithorax homologue, ALL-1, normally found in a nucleoplasmic complex consisting of RNA-binding proteins (Yano et al., 1997; Nakamura et al., 2002) also was found in LNCs (Figure 3D). Another nucleolar protein, p110, which is associated with U3 snoRNP, was not found in fibrillarin containing caps at low ActD concentrations (Adamson et al., 2001), but did colocalize with high levels of ActD (Table 3).
Proteins related to the RNA pol I transcriptional machinery have been shown to colocalize in small nucleolar caps in regions juxtaposed to fibrillarin (LNCs). These caps have been termed on occasion fibrillar caps as they contain FC proteins. These proteins include: RNA pol I, UBF, TBP, TAFI63, and TAFI110 (Reimer et al., 1987; Zatsepina et al., 1993; Jordan et al., 1996) and topoisomerase I (Christensen et al., 2004; Table 3). It has been shown that some nucleolar proteins can be found in both types of cap regions that contain DFC or FC proteins, such as Nopp140 (Chen et al., 1999) and MPP10. The latter was seen to partially colocalize with fibrillarin caps during incubation with low levels of ActD (Westendorf et al., 1998); however, it remained partially in DNCs using high levels of ActD (Table 3). This is the only nucleolar protein we identified in DNCs.
The presence of three distinct nucleolar caps was then confirmed by costaining with antibodies to PSF, fibrillarin, UBF and gar1 (Figure 4, A–D). Fibrillarin and UBF segregate to distinct but adjacent nucleolar caps; fibrillarin is in phase-dense LNCs, whereas caps containing UBF were not observed (Figure 4D). To summarize, concave DNCs were larger and wider and contained incoming nucleoplasmic proteins. The smaller and rounder caps found in between DNCs contained two regions with either nucleolar DFC or FC proteins, whereas p80 coilin formed a separate cap. In addition, although DNCs were always found on the periphery of the nucleolar body, fibrillar caps were occasionally situated on top of concave caps (Figure 4C).
p14(ARF) Remains in the Central Body after ActD Treatment
As it became apparent that the formation of nucleolar caps involved nucleoplasmic proteins, we were interested in identifying proteins in the central body. As known, nucleolin (C23), which is found both in the DFC and GC, and nucleophosmin (B23) which is a GC protein, dispersed in the nucleoplasm after ActD treatment (Figure 5A, Table 4). Staining of PSF, nucleolin and p80 coilin showed nucleolar caps situated on the periphery of the central body (Figure 5A, inset). RNA helicase RH-II/Gu normally found in the GC has been described to disperse in a similar manner (Valdez et al., 1998).
Figure 5.
Nuclear proteins that do not localize in nucleolar caps during transcriptional inhibition. Confocal images of (A) GFP-PSF–transfected cells were labeled with antibodies to nucleolin and p80 coilin. Untreated: GFP-PSF was nucleoplasmic, nucleolin was in nucleoli and p80 coilin was in Cajal bodies. No colocalization was seen in the merged image. Treated: GFP-PSF concave nucleolar caps were situated on the central body and p80 coilin fibrillar caps were found inbetween the concave caps. A segregated nucleolus is seen in the enlarged merged image. (B) Untreated: PSF was nucleoplasmic, whereas p14(ARF) was in the GC. Treated: p14(ARF) was both in the central body and in the nucleoplasm. Concave PSF caps formed on this body. A segregated nucleolus is seen in the enlarged merged image. (C) Untreated: PSF foci dispersed throughout the nucleoplasm in comparison to the large SC-35 speckles. Treated: PSF and SC-35 localized in distinct compartments. Bar, 10 μm.
p14(ARF) was the only GC nucleolar protein that remained in the central body during ActD treatment, although some of it was dispersed in the nucleoplasm (Figure 5B, Table 3). GFP-p14(ARF) behaved in the same manner (unpublished data). ARF is involved in the attenuation of the Mdm2-mediated ubiquitination and degradation of p53. During cellular stress ARF binds to Mdm2, resulting in an upregulation of p53. It has been suggested that this interaction occurs in the nucleolus (Weber et al., 1999; Llanos et al., 2001). However, we found by antibody staining, that Mdm2 did not show any accumulation in the central body of the segregated nucleolus (Table 4).
A large number of nucleoplasmic proteins related to different nuclear functions were then tested and most were found to remain in the nucleoplasm without compartmentalization during transcriptional inhibition (Table 4). Nuclear speckles, which normally contain proteins like SC-35, 4.1R, lamin A, hPrp5, FBP11, and FBP21, became rounded and sometimes enlarged, but were distinct and spatially distant from nucleolar caps (Figure 5C). The relative concentration of SC-35 in speckles is only twice the intensity of the nucleoplasmic pool (Fay et al., 1997). Measurements of the average intensities of PSF staining in the nucleolplasm and nucleolar caps using confocal microscope line scans showed that in ActD-treated cells PSF was 3–5 times more concentrated in concave caps than in the nucleoplasm, whereas SC-35 in speckles was 2–3 times more concentrated than the corresponding nucleoplasmic signal (unpublished data).
Unique Nucleolar Cap for PML Body Components
Because PML bodies are normally associated with regions of active transcription (Wang et al., 2004), we tested whether proteins commonly found in PML bodies would move to nucleolar caps. Indeed, a portion of the PML protein moved to the nucleolar periphery in distinct nucleolar caps either adjacent to PSF caps or on top of them (Figure 6A). This was also seen with GFP-PML (unpublished data). Similarly, a portion of Sp100, another component of PML bodies, was also found in these caps (Table 3). Because p80 coilin from Cajal bodies formed similar nucleolar caps, we checked whether these proteins share the same nucleolar cap. However, this was not the case. PML protein formed a novel nucleolar cap distinct from p80 coilin (Figure 6, B and C), Nopp140 (LNCs; Figure 6D), and UBF (fibrillar caps; Figure 6E). Using an antibody that detects PML bodies in EM preparations (Figure 6F), we could detect PML both in small structures adjacent to large nucleolar caps (Figure 6G) or on top of DNCs (Figure 6H).
Figure 6.
PML protein is found in distinct nucleolar caps. Confocal images of (A) Untreated: PSF was nucleoplasmic, whereas PML was found in PML bodies. Treated: PSF and PML were found in two different nucleolar caps. (B) Untreated: p80 coilin was in Cajal bodies and PML was in PML bodies. Occasionally, there was close association between the two bodies. Treated: p80 coilin and PML were in two separate caps. (C) GFP-PSF–transfected cells were labeled with antibodies to PML and p80 coilin. The three different cap structures were seen in the treated cells. Bar, 10 μm. (D) PML caps were distinct from Nopp140 LNCs and (E) UBF fibrillar caps. (F) Immunogold labeling with anti-PML detected PML bodies in the nucleoplasm of untreated cells and (G) PML in small cap structures adjacent to DNCs or (H) on top of DNCs.
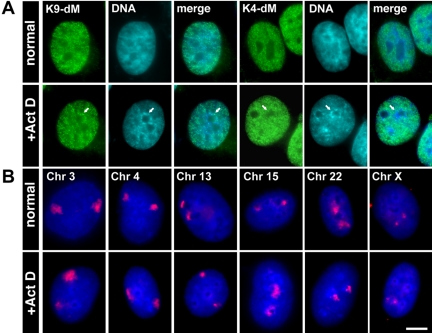
Nucleolar Caps Are Surrounded by Heterochromatin
In ActD-treated cells, the segregated nucleolar regions became surrounded by dense chromatin (Figure 1, A and B). Antibodies specific for dimethylation of lysine 9 on histone H3 (Figure 7A), a characteristic of heterochromatin, stained the dense chromatin regions. These domains remained unstained with antibodies that detect either dimethylation of lysine 4 (Figure 7A) or acetylation of lysine 9 on histone H3, characteristic of euchromatin. Using DNA FISH against human chromosomes, we determined that chromosomes containing nucleolar organizer regions (NORs; chr 13, 14, 15, 21, 22) retained their nucleolar proximity during transcriptional inhibition (Figure 7B), except for chromosome 13 that was not always positioned in contact with nucleoli. Other chromosomes that normally showed peripheral positioning in the nucleus did not tend to associate with nucleolar regions during transcriptional arrest (Figure 7B).
Figure 7.
(A) The chromatin surrounding the segregated nucleolus showed characteristics of heterochromatin via staining with an antibody against dimethylated lysine 9 of histone H3, but not with the euochromatin marker: dimethyled lysine 4 of histone H3. Bar, 5 μm. (B) Chromosome labeling by DNA FISH showed that peripheral chromosomes (3, 4) retained this positioning during transcriptional arrest, whereas NOR-containing chromosomes (15, 22) remained associated with the nucleolar region. In many cells, NOR containing chromosome 13 was not associated with nucleolar region after transcriptional arrest. Chromosome X was not associated with the nucleolus either. Bar, 10 μm
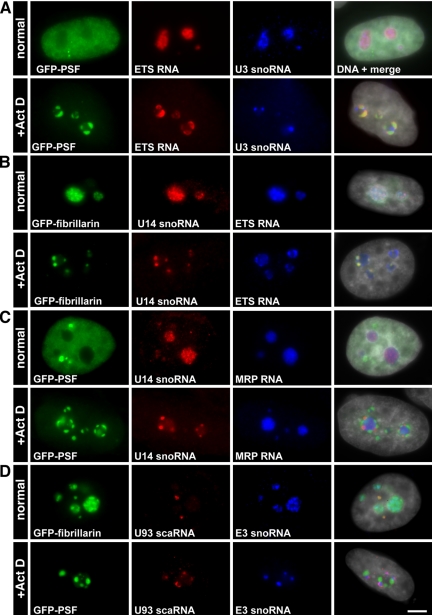
Nucleolar RNAs Segregate to Different Subtypes of Nucleolar Caps
Our finding that the DNCs contain nucleoplasmic RNA binding proteins led us to examine the distribution of several functional nuclear RNAs during the process of nucleolar segregation. The ribosomal genes transcribe a large precursor RNA that is cleaved at the A0 site, which removes the 5′ external transcribed sequence (5′ETS). This step occurs in the nucleolus and the 5′ETS is then degraded by the exosome. Using a probe to the 5′ETS, which specifically recognizes pre-rRNAs, we found that the precursor rRNAs segregated to the DNCs enriched in nucleoplasmic proteins and colocalized with PSF (Figure 8A). U1 snRNA also localized to these nucleolar caps (Carmo-Fonseca et al., 1991), whereas U2 snRNA remained in speckles as described (Carmo-Fonseca et al., 1992) and 7SK RNA was enriched in nuclear speckles, after ActD treatment (Table 4). Nucleoplasmic mRNAs identified using an oligo(dT) probe or a probe to the abundant β-actin transcript were not detected in nucleolar caps (Table 4).
Figure 8.
Nucleolar RNAs are found in the different nucleolar caps. (A and B) U3 and U14 snoRNAs were found to localize in fibrillarin containing caps, whereas 5′ETS rRNA was observed with GFP-PSF in concave caps. (C) MRP RNA remained in the central body. (D) U93 scaRNA was typically found in Cajal bodies as was fibrillarin, but not E3 snoRNA. After ActD treatment, U93 scaRNA was juxtaposed to E3 snoRNA in nucleolar caps and distinct from concave caps. Bar, 5 μm.
Small nucleolar RNAs (snoRNAs), required for the process of ribosome assembly, colocalized with fibrillarin in LNCs: U14 snoRNA (Figure 8B), U3 snoRNA (Carmo-Fonseca et al., 1991; Puvion-Dutilleul et al., 1997; Leary et al., 2004), and E3 snoRNA (Figure 8D; Table 3). The nucleolus plays a role also in the formation of RNA pol III transcripts. Three of these transcripts were tested, and we found that MRP RNA remained in the central body (Figure 8C), U6 snRNA colocalized with fibrillarin (Table 3; Carmo-Fonseca et al., 1991), and RNase P RNA did not enter nucleolar caps (Table 4).
Cajal body–specific RNAs (scaRNAs) have been found in Cajal bodies (Darzacq et al., 2002) and so we were interested to see whether they would follow a similar fate as described above for Cajal body proteins. Indeed, U93 scaRNA, typically found in Cajal bodies (Kiss et al., 2002), as seen by colocalization with GFP-fibrillarin in Cajal bodies but not with GFP-fibrillarin and E3 snoRNA in the nucleolus (Figure 8D, top), was found to localize on top of E3 snoRNA containing caps (that also contain fibrillarin; Figure 8D, bottom) and not with GFP-PSF–containing caps (Figure 8D, bottom). A similar distribution was described also for p80 coilin (Figure 3B).
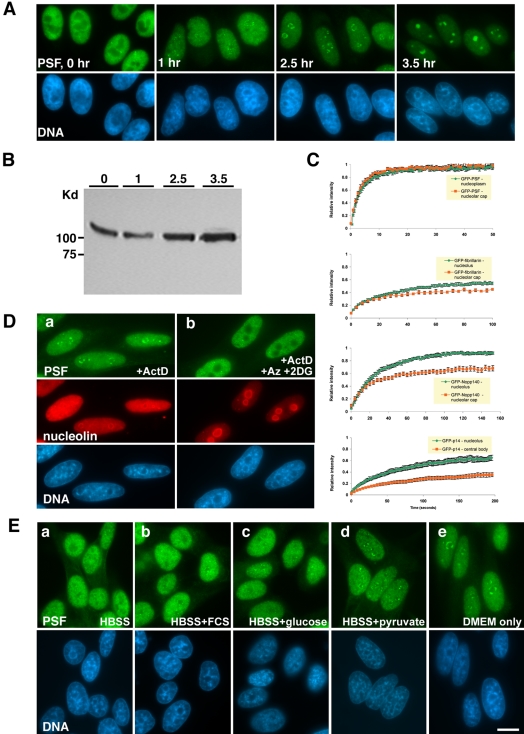
Nucleolar Caps Are Dynamic Structures
Nuclei observed at different time points after addition of ActD indicated that the process of nucleolar capping was dynamic. One hour posttreatment with ActD, PSF staining showed delicate bead-like structures around the nucleolus (Figure 9A). These increased in volume and became cap-like after 2–2.5 h. In parallel, condensation of chromatin was observed. After longer exposure to the drug these compartments took upon a more rounded structure. We have previously shown that GFP-PSF in apoptotic cells, induced by prolonged exposure to ActD, formed large, rounded structures (Shav-Tal et al., 2001a). This correlated with hyperphosphorylation of PSF in apoptotic and mitotic cells. However, no change in the mobility of the PSF band was observed during nucleolar capping upon short exposure to ActD (Figure 9B). This meant that the hyper-phosphorylation of PSF was a later step in the molecular pathway induced by ActD.
Figure 9.
Formation of nucleolar caps is a dynamic energy-requiring process. Immunofluorescence microscopy of (A) The formation of PSF concave nucleolar caps at different time points after addition of ActD. Hoechst DNA counterstain is seen in blue. (B) Western blot analysis of PSF from protein extracts of cells at time points as in A. (C) Recovery curves after photobleaching of GFP-PSF, GFP-fibrillarin, GFP-Nopp140, and GFP-p14(ARF) in the nucleoplasm or nucleolus before ActD treatment (green) and in nucleolar caps after ActD treatment (orange). (D) PSF and nucleolin labeling of cells treated with: (a) ActD only, (b) ActD plus Na-azide and 2-deoxyglucose. (E) Formation of PSF concave caps. ActD treatment was performed under different conditions: (a) HBSS medium, (b) HBSS medium plus 10% FCS, (c) HBSS medium with glucose, (d) HBSS medium with pyruvate, (e) DMEM without FCS. Bar, 10 μm.
The dynamic characteristics of nucleolar caps were then examined using FRAP. Several GFP-tagged proteins, which were found to localize to segregated nucleoli as their endogenous counterparts, were photobleached, either in the nucleoplasm/nucleolus of untreated cells or in segregated nucleoli of ActD-treated cells, and the recovery of the fluorescent signal was monitored over time (Figure 9C). GFP-PSF demonstrated a half-time of recovery of ∼2.7 s, in contrast to the nucleolar proteins GFP-fibrillarin (t1/2 ∼15 s; Phair and Misteli, 2000), GFP-Nopp140 (t1/2 ∼ 17 s), and GFP-p14(ARF) (t1/2 ∼ 43 s), which also exhibited an immobile nondynamic nucleolar fraction (45, 10, and 35%, respectively; Figure 9C, green curves). Surprisingly, under ActD conditions, the rate of exchange of GFP-PSF in concave DNCs was as rapid as the nucleoplasmic pool of GFP-PSF under unperturbed conditions (t1/2 ∼ 2.4 s; Figure 9C, orange curves). The recovery of GFP-fibrillarin (t1/2 ∼ 10 s) and GFP-Nopp140 (t1/2 ∼ 14 s) in LNCs was similar, yet an increased immobile fraction was seen for both proteins (55 and 30%, respectively). Notably, GFP-p14(ARF) in the central nucleolar body of ActD-treated cells exhibited an extremely slow turnover (t1/2 ∼ 40 s) and the fixed fraction was greatly enhanced (from 50 to 80%). This analysis shows that nucleolar caps contain two distinct protein pools, a portion in constant exchange with the nucleoplasm and a fixed immobile fraction. This demonstrates that the dynamics of nuclear domains are at least partially retained also under conditions of transcriptional inhibition.
The Formation of Nucleolar Caps Is an Energy-dependent Process
It has been shown that nuclear dynamics is sensitive to metabolic stress (Muratani et al., 2002; Platani et al., 2002; Shav-Tal et al., 2004). We therefore investigated nucleolar cap formation under low cellular energy levels. When cells were incubated at 4°C and concurrently treated with ActD, no formation of nucleolar caps was observed (unpublished data). Inhibition of metabolism by addition of Na-azide and 2-deoxyglucose to ActD-treated cells showed that nuclear proteins remained in their normal distributions (compare Figure 9D, a and b). The same phenomenon was observed with other components of nucleolar caps (unpublished data). This indicated that the redistribution of the proteins required energy. In contrast, chromatin condensation seen after ActD treatment was still observed (see DNA panels in Figure 9D). It has been shown that the nucleus contains a unique myosin 1β form (Pestic-Dragovich et al., 2000). Because this motor protein might be involved in the translocation of proteins in the nucleus, cells were treated with 2,3-butanedione monoxime (BDM), an inhibitor of actin-dependent myosins. No change in cap formation was seen (unpublished data).
To identify the energy source required for cap formation, treatment with ActD was performed in minimal medium (HBSS) without metabolic supplements. In this case, most nuclei did not contain nucleolar caps (Figure 9Ea), indicating that reduction in metabolic levels due to lack in energy sources did not allow the formation of caps. The addition of either protein in the form of FCS (Figure 9Eb) or amino acids (unpublished data) or glucose (Figure 9Ec) did not change this condition. However, when pyruvate was added to the minimal medium, all cells formed nucleolar caps. These however, were not fully formed as those seen after 2.5 h of treatment (Figure 9Ed). Finally, when ActD treatment was performed in a carbohydrate-rich medium but without FCS, nucleolar caps formed as usual (Figure 9Ee). In all the above cases, condensation of chromatin was seen. We therefore conclude that energy produced by the phosphorylative oxidation pathway is a prerequisite in the process of reshuffling of nuclear proteins during transcriptional stress.
DISCUSSION
We have found that the process of nucleolar segregation caused by the transcriptional arrest of RNA polymerases I and II was accompanied by the sorting and rearranging of nuclear proteins and RNAs into defined nuclear subdomains. The course of events was a dynamic and specific process, encompassing many major players that define intranuclear structure. Several main themes of nuclear rearrangement were observed: 1) the segregation of the three defining regions of the nucleolus (FC, DFC, GC) into three distinct but juxtaposed domains that retain many of their original protein and RNA components and that are termed nucleolar caps and central body, 2) this segregation is accompanied by the release of several GC proteins into the nucleoplasm, 3) the disassembly of nuclear bodies such as Cajal bodies, SMN bodies, and PML bodies and the relocation of their protein and RNA components to discrete nucleolar caps, 4) the formation of a large heterochromatin domain surrounding the segregated nucleolus, 5) the influx of a significant number of nucleoplasmic proteins, many of which are RNA binding proteins, into large nucleolar caps, whereas 6) most nucleoplasmic proteins and nuclear speckle proteins retained their localization.
The nucleolus is sensitive to the transcriptional profile of the cell, and the status of transcriptional activity is reflected in nucleolar structure. Nucleolar segregation and capping is a normal cellular process occurring under physiological circumstances that involve transcriptional shut down (Smetana and Busch, 1974) and can be mimicked by drug-induced transcriptional arrest (Bernhard and Granboulan, 1968; Zinszner et al., 1997; Dousset et al., 2000; Andersen et al., 2002; Fox et al., 2002; Ospina and Matera, 2002). In the developing Xenopus oocyte dramatic changes in nucleolar structure were observed, which included a stage of nucleolar segregation and cap formation reminiscent of ActD treatment (Van Gansen and Schram, 1972). During ovulation of Xenopus eggs, nucleoli disappear and transcription is shut off, later to return in the embryo. In the procedure of nuclear cloning, nuclei from somatic cells are injected into interphase eggs and the somatic nucleoli are then found to segregate and finally disassemble (Gonda et al., 2003). This process is triggered by two germ cell proteins FRGY2a and FRGY2b and is independent of rRNA transcription. Other natural instances of segregation and capping occur in oocytes (Mirre et al., 1980; Crozet et al., 1981), spermatocytes (Stahl et al., 1991), developing embryos (Hyttel et al., 2000), hepatocytes (Reddy and Svoboda, 1972), keratinocytes (Karasek et al., 1972), mycoplasma infection (Jezequel et al., 1967), and certain diseases (Karasek et al., 1970; Smetana et al., 1972). Nucleolar capping of p80 coilin naturally occurs in normal mouse tissues (Tucker et al., 2001) and in neuronal cells (Raska et al., 1990; Carmo-Fonseca et al., 1993; Janevski et al., 1997).
The study of electron micrographs of transcriptionally arrested cells in the 1960s lead to the simple assumption that the material found in nucleolar caps originated from the segregation of granular and fibrillar components of the nucleolus. Yet, one study noted that the granular material of P2 (concave) caps observed by TEM was not found in untreated nucleoli (Recher et al., 1971). In other studies, nucleoplasmic splicing factors were found associated with segregated nucleoli of hibernating rodents (Malatesta et al., 2000) and SR proteins were transiently associated with the nucleolus after mitosis (Bubulya et al., 2004). Our study shows that DNCs consisted mainly of nucleoplasmic proteins, whereas LNCs and fibrillar caps evolved from nucleolar proteins. A few RNA-binding nucleoplasmic proteins that were found in nucleolar caps have been detected in the nucleolus by proteomic analysis: PSF, p54nrb, hnRNP K, hnRNP H, and SF2/ASF (Andersen et al., 2002; Scherl et al., 2002). However, these proteins are normally found in the nucleoplasm and are excluded from the nucleolus as observed either by immunofluorescent stainings or GFP tagging. Interestingly, a kinetic proteomic analysis has shown PSF to be one of the highly enriched proteins in the nucleolus during ActD treatment (Andersen et al., 2005). In the case of PSF, GFP-PSF constructs showed that the C-terminal half of the protein is important for the localization in caps. This property was independent of the two RRMs in this region. From analyzing constructs of the C-terminus we conclude that the middle part of the C-terminal half of PSF is required for this localization. The C-terminus is homologous to p54nrb, the heterodimer of PSF, which also translocated to DNCs. Alignment of amino acid sequences of DNC proteins did not reveal any “cap localization signals,” although most of these proteins contain RNA-binding domains such as RRMs or RGG boxes, shown to be the most highly abundant motifs in proteins identified in the nucleolar proteome (Leung et al., 2003). Yet, these motifs are probably not sufficient for nucleolar cap targeting because other proteins tested, which also contain RRM or RGG boxes, were not localized to nucleolar caps (Table 4). The unexpected finding that nucleolar pre-rRNA was preferentially detected in the DNCs that harbored mainly nucleoplasmic proteins still implies that the localization of nucleoplasmic RNA-binding proteins to DNCs was a consequence of the RNA-binding properties of these proteins. FRAP experiments revealed that these structures were not static depots of proteins but were highly dynamic structures in constant exchange with the nucleoplasm, complementary to FRAP experiments performed in transcriptionally active cells (Janicki and Spector, 2003).
Our data strongly suggest that nucleolar segregation is part of a general concerted process of nuclear rearrangement taking place during transcriptional shut down and comprises both preexisting protein-RNA interactions and newly established interactions. For example, we have detected a trend in the association of nuclear body proteins with the segregated nucleolus. Components of SMN bodies have been shown to transiently pass through nucleolar caps (Pellizzoni et al., 2001). p80 coilin from Cajal bodies was found on the peripheral part of nucleolar caps that contain fibrillarin, which is also, in part, a Cajal body component. Nucleolar and Cajal body RNAs were also found to closely segregate during ActD treatment. A Cajal body scaRNA was found, as described above for p80 coilin, to be peripherally situated on fibrillarin. Small nucleolar RNAs were found in fibrillarin containing LNCs. We also detected a novel nucleolar cap to which a portion of PML and Sp100 proteins localized that were distinct from DNCs, LNCs, or fibrillar caps. PML caps were usually the smallest of the above and formed on top or in between the other caps. Interestingly, a different pathway of cellular stress involving the inhibition of proteosome action caused PML and Sp100 to relocalize inside the nucleolus (Mattsson et al., 2001). PML translocated to the nucleolar periphery also in response to DNA damage and colocalized there with Mdm2, but in an ARF-independent manner (Bernardi et al., 2004). Another study has shown that a variety of stress responses activating the p53 pathway affect nucleolar integrity (Rubbi and Milner, 2003). The nucleolus therefore probably plays an important role in sensing these stresses (Olson, 2004a) and can act as a docking site for many proteins released from disrupted structures and complexes.
The central body, on which nucleolar caps are formed, is an intriguing structure. It was assumed to originate from the GC region of the nucleolus. MRP RNA was detected in the central body, whereas RNase P RNA was not. Although different in sequence, these RNAs are structurally similar, whereas MRP is involved in cleavage and maturation of the precursor rRNA and RNase P acts in the processing of pre-tRNA (van Eenennaam et al., 2000). However, under these conditions MRP RNA and rRNAs were segregated in distinct compartments. As for the protein composition of the central body, we could only detect p14(ARF) in the central body, whereas proteins found in the GC such as nucleophosmin and nucleolin dispersed throughout the nucleoplasm. FRAP experiments showed that p14(ARF) had the slowest recovery rates in comparison to the other nucleolar proteins tested, fibrillarin and Nopp140. These rates all differed from the nuclear mobility of PSF in DNCs, which was comparable to its mobility in the nucleoplasm of untreated cells and did not show a fixed fraction. Moreover, most of p14(ARF) protein was not dynamic as observed by the increase in the immobile fraction from 35 to 65%. This might indicate that p14(ARF) has a function in retaining the structural integrity of nucleolus.
Our study shows that the process of nucleolar segregation and capping is an active process that requires active metabolism of the cell. However, it does not require active protein synthesis (Goldblatt et al., 1970). The addition of metabolic inhibitors to cells being treated with ActD hindered the formation of nucleolar caps. The energy source required for nucleolar cap formation was not in the form of amino acids, proteins, or growth factors but was carbohydrate based. We found that the addition of pyruvate (citric acid cycle) but not glucose (glycolysis) to minimal medium could lead to the formation of caps, although this was probably not the only requisite. Because pyruvate is the end product of glycolysis, it is possible that ActD is also inhibiting a certain step in this process, as previously suggested (Laszlo et al., 1966). Pyruvate is also the input molecule for the citric acid cycle and oxidative phosphorylation, the main producers of cellular energy in the form of ATP and GTP. Azide is a metabolic poison that blocks oxidative phosphorylation in the mitochondria, the organelle in which pyruvate is metabolized. It was described for the TLS protein (DNC) that its entrance into nucleolar caps is an active process requiring the integrity of the Ran/TC4-RCC1 nuclear transport cycle (Zinszner et al., 1997) and that the drug-induced translocation of nucleophosmin from nucleoli to the nucleoplasm requires ATP (Wu et al., 1995; Finch and Chan, 1996). These findings indicate that the relocalization and nucleolar capping of nuclear proteins is not just a byproduct of transcriptional arrest but actually an active mechanism driving the reshaping of nuclear compartments.
Although the physiological significance of nucleolar segregation and capping is unclear, a correlation between reduction in RNA transcription and the formation of these structures can be drawn, especially during differentiation and development. In the normal situation, much of the nuclear activity is devoted to transcription. In active cells, transcriptional and posttranscriptional components are in equilibrium; being recruited to DNA or to nascent mRNA transcripts, respectively. Under ActD-induced transcriptional arrest, the RNA polymerase II complexes are blocked during the elongation process, thus titrating one fraction of the transcription machinery to the DNA. Because mRNA production has ceased, there exists an excess pool of free posttranscriptional factors. In conjunction, in the nucleolus, separation of the RNA polymerase I transcription machinery from the rRNA occurs. The observed clustering of nucleoplasmic RNA-binding proteins, in part of the segregated nucleolus that contains pre-rRNA, could be the result of the high abundance of these proteins and the existence of newly exposed RNA partners. A recent study showed that during telophase, as RNA pol I activity in nucleolar NORs resumed, nucleoplasmic SR-splicing factors become transiently associated with the NORs (Bubulya et al., 2004). The interactions we observe have specificity, as only a subset of nucleoplasmic RNA-binding proteins relocalize to these regions and may reflect some of the protein complexes present in transcriptionally active cells. Taken together, these data argue for an additional pathway that RNA-binding proteins can take when RNA pol II transcription is arrested. It has been suggested that nuclear organization is driven by the state of gene expression in the cell (Singer and Green, 1997) and our data support this notion. Because nucleolar segregation can occur under physiological states and can be followed by nucleolar reassembly, it stands to reason that the nucleus evolved a mechanism that uniquely redistributes specific nuclear components during transcriptional arrest, while simultaneously up keeping certain basic interactions. Such a mechanism would provide the flexibility required for responding to metabolic cues and would maintain a certain degree of structure necessary for the efficient reassembly once the transcriptional status of the cell changes.
Acknowledgments
We are immensely grateful to the researchers listed in Materials and Methods who generously shared their antibodies and constructs with us. We thank Tom Meier for critical comments on this manuscript. We thank Leslie Cummings, Juan Jimenez, and Frank Macaluso of the Albert Einstein Imaging Facility for assistance in the electron microscopy work and Jenetta Smith for the DNA FISH. D.Z. is an incumbent of the Joe and Celia Weinstein professorial chair at the Weizmann Institute of Science. R.H.S. is supported by National Institutes of Health grants EB2060 and DOE63056.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-11-0992) on March 9, 2005.
Abbreviations used: ActD, actinomycin D.
References
- Adamson, C., Niu, S., Bahl, J. J., and Morkin, E. (2001). Cloning and characterization of P110, a novel small nucleolar U3 ribonucleoprotein, expressed in early development. Exp. Cell Res. 263, 55-64. [DOI] [PubMed] [Google Scholar]
- Andersen, J. S., Lam, Y. W., Leung, A. K., Ong, S. E., Lyon, C. E., Lamond, A. I., and Mann, M. (2005). Nucleolar proteome dynamics. Nature 433, 77-83. [DOI] [PubMed] [Google Scholar]
- Andersen, J. S., Lyon, C. E., Fox, A. H., Leung, A. K., Lam, Y. W., Steen, H., Mann, M., and Lamond, A. I. (2002). Directed proteomic analysis of the human nucleolus. Curr. Biol. 12, 1-11. [DOI] [PubMed] [Google Scholar]
- Bernardi, R., Scaglioni, P. P., Bergmann, S., Horn, H. F., Vousden, K. H., and Pandolfi, P. P. (2004). PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat. Cell Biol. 6, 665-672. [DOI] [PubMed] [Google Scholar]
- Bernhard, W., and Granboulan, N. (1968). The nucleolus in vertebrate cells. In: The Nucleus, ed. A. J. Dalton and F. Haguenau, New York: Academic Press.
- Bubulya, P. A., Prasanth, K. V., Deerinck, T. J., Gerlich, D., Beaudouin, J., Ellisman, M. H., Ellenberg, J., and Spector, D. L. (2004). Hypophosphorylated SR splicing factors transiently localize around active nucleolar organizing regions in telophase daughter nuclei. J. Cell Biol. 167, 51-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch, H., and Smetana, K. (eds.) (1970). The Nucleolus, New York: Academic Press.
- Carmo-Fonseca, M., Ferreira, J., and Lamond, A. I. (1993). Assembly of snRNP-containing coiled bodies is regulated in interphase and mitosis— evidence that the coiled body is a kinetic nuclear structure. J. Cell Biol. 120, 841-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca, M., Mendes-Soares, L., and Campos, I. (2000). To be or not to be in the nucleolus. Nat. Cell Biol. 2, E107-E112. [DOI] [PubMed] [Google Scholar]
- Carmo-Fonseca, M., Pepperkok, R., Carvalho, M. T., and Lamond, A. I. (1992). Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J. Cell Biol. 117, 1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca, M., Pepperkok, R., Sproat, B. S., Ansorge, W., Swanson, M. S., and Lamond, A. I. (1991). In vivo detection of snRNP-rich organelles in the nuclei of mammalian cells. EMBO J. 10, 1863-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrand, P., Bertrand, E., Singer, R. H., and Long, R. M. (2000). Sensitive and high-resolution detection of RNA in situ. Methods Enzymol. 318, 493-506. [DOI] [PubMed] [Google Scholar]
- Chen, H. K., Pai, C. Y., Huang, J. Y., and Yeh, N. H. (1999). Human Nopp140, which interacts with RNA polymerase I: implications for rRNA gene transcription and nucleolar structural organization. Mol. Cell. Biol. 19, 8536-8546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, M. O., Krokowski, R. M., Barthelmes, H. U., Hock, R., Boege, F., and Mielke, C. (2004). Distinct effects of topoisomerase I and RNA polymerase I inhibitors suggest a dual mechanism of nucleolar/nucleoplasmic partitioning of topoisomerase I. J. Biol. Chem. 279, 21873-21882. [DOI] [PubMed] [Google Scholar]
- Crozet, N., Motlik, J., and Szollosi, D. (1981). Nucleolar fine structure and RNA synthesis in porcine oocytes during the early stages of antrum formation. Biol. Cell 41, 35-42. [Google Scholar]
- Darzacq, X., Jady, B. E., Verheggen, C., Kiss, A. M., Bertrand, E., and Kiss, T. (2002). Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 21, 2746-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dousset, T., Wang, C., Verheggen, C., Chen, D., Hernandez-Verdun, D., and Huang, S. (2000). Initiation of nucleolar assembly is independent of RNA polymerase I transcription. Mol. Biol. Cell 11, 2705-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr, M., Hebert, M. D., Karpova, T. S., Stanek, D., Xu, H., Shpargel, K. B., Meier, U. T., Neugebauer, K. M., Matera, A. G., and Misteli, T. (2004). In vivo kinetics of Cajal body components. J. Cell Biol. 164, 831-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye, B. T., and Patton, J. G. (2001). An RNA recognition motif (RRM) is required for the localization of PTB-associated splicing factor (PSF) to subnuclear speckles. Exp. Cell Res. 263, 131-144. [DOI] [PubMed] [Google Scholar]
- Fay, F. S., Taneja, K. L., Shenoy, S., Lifshitz, L., and Singer, R. H. (1997). Quantitative digital analysis of diffuse and concentrated nuclear distributions of nascent transcripts, SC35 and poly(A). Exp. Cell Res. 231, 27-37. [DOI] [PubMed] [Google Scholar]
- Finch, R. A., and Chan, P. K. (1996). ATP depletion affects NPM translocation and exportation of rRNA from nuclei. Biochem. Biophys. Res. Commun. 222, 553-558. [DOI] [PubMed] [Google Scholar]
- Fox, A. H., Lam, Y. W., Leung, A. K., Lyon, C. E., Andersen, J., Mann, M., and Lamond, A. I. (2002). Paraspeckles: a novel nuclear domain. Curr. Biol. 12, 13-25. [DOI] [PubMed] [Google Scholar]
- Goessens, G. (1984). Nucleolar structure. Int. Rev. Cytol. 87, 107-158. [DOI] [PubMed] [Google Scholar]
- Goldblatt, P. J., Verbin, R. S., and Sullivan, R. J. (1970). Induction of nucleolar segregation by actinomycin D following inhibition of protein synthesis with cycloheximide. Exp. Cell Res. 63, 117-123. [DOI] [PubMed] [Google Scholar]
- Gonda, K., Fowler, J., Katoku-Kikyo, N., Haroldson, J., Wudel, J., and Kikyo, N. (2003). Reversible disassembly of somatic nucleoli by the germ cell proteins FRGY2a and FRGY2b. Nat. Cell Biol. 5, 205-210. [DOI] [PubMed] [Google Scholar]
- Hernandez-Verdun, D. (2004). Behavior of the nucleolus during mitosis. In: The Nucleolus, ed. M. O. J. Olson, New York: Kluwer/Plenum, 41-57.
- Huang, S., Deerinck, T. J., Ellisman, M. H., and Spector, D. L. (1997). The dynamic organization of the perinucleolar compartment in the cell nucleus. J. Cell Biol. 137, 965-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyttel, P., Laurincik, J., Rosenkranz, C., Rath, D., Niemann, H., Ochs, R. L., and Schellander, K. (2000). Nucleolar proteins and ultrastructure in preimplantation porcine embryos developed in vivo. Biol. Reprod. 63, 1848-1856. [DOI] [PubMed] [Google Scholar]
- Janevski, J., Park, P. C., and De Boni, U. (1997). Changes in morphology and spatial position of coiled bodies during NGF-induced neuronal differentiation of PC12 cells. J. Histochem. Cytochem. 45, 1523-1531. [DOI] [PubMed] [Google Scholar]
- Janicki, S. M., and Spector, D. L. (2003). Nuclear choreography: interpretations from living cells. Curr. Opin. Cell Biol. 15, 149-157. [DOI] [PubMed] [Google Scholar]
- Jezequel, A. M., Shreeve, M. M., and Steiner, J. W. (1967). Segregation of nucleolar components in mycoplasma-infected cells. Lab. Invest. 16, 287-304. [PubMed] [Google Scholar]
- Jordan, P., Mannervik, M., Tora, L., and Carmo-Fonseca, M. (1996). In vivo evidence that TATA-binding protein/SL1 colocalizes with UBF and RNA polymerase I when rRNA synthesis is either active or inactive. J. Cell Biol. 133, 225-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journey, L. J., and Goldstein, M. N. (1961). Electron microscope studies on HeLa cell lines sensitive and resistant to actinomycin D. Cancer Res. 21, 929-932. [PubMed] [Google Scholar]
- Kamath, R. V., Leary, D. J., and Huang, S. (2001). Nucleocytoplasmic shuttling of polypyrimidine tract-binding protein is uncoupled from RNA export. Mol. Biol. Cell 12, 3808-3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameoka, S., Duque, P., and Konarska, M. M. (2004). p54(nrb) associates with the 5′ splice site within large transcription/splicing complexes. EMBO J. 23, 1782-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasek, J., Smetana, K., Hrdlicka, A., Dubinin, I., Hornak, O., and Ohelert, W. (1972). Nuclear and nucleolar ultrastructure during the late stages of normal human keratinocyte maturation. Br. J. Dermatol. 86, 601-607. [DOI] [PubMed] [Google Scholar]
- Karasek, J., Smetana, K., Oehlert, W., and Konrad, B. (1970). The ultrastructure of Bowen's disease: nuclear and nucleolar lesions. Cancer Res. 30, 2791-2795. [PubMed] [Google Scholar]
- Kiss, A. M., Jady, B. E., Darzacq, X., Verheggen, C., Bertrand, E., and Kiss, T. (2002). A Cajal body-specific pseudouridylation guide RNA is composed of two box H/ACA snoRNA-like domains. Nucleic Acids Res. 30, 4643-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laszlo, J., Miller, D. S., McCarty, K. S., and Hochstein, P. (1966). Actinomycin D: inhibition of respiration and glycolysis. Science 151, 1007-1010. [DOI] [PubMed] [Google Scholar]
- Leary, D. J., Terns, M. P., and Huang, S. (2004). Components of U3 snoRNA-containing complexes shuttle between nuclei and the cytoplasm and differentially localize in nucleoli: implications for assembly and function. Mol. Biol. Cell 15, 281-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B. C., Shav-Tal, Y., Peled, A., Gothelf, Y., Jiang, W., Toledo, J., Ploemacher, R. E., Haran-Ghera, N., and Zipori, D. (1996). A hematopoietic organ-specific 49-kD nuclear antigen: predominance in immature normal and tumor granulocytes and detection in hematopoietic precursor cells. Blood 87, 2283-2291. [PubMed] [Google Scholar]
- Leung, A. K., Andersen, J. S., Mann, M., and Lamond, A. I. (2003). Bioinformatic analysis of the nucleolus. Biochem. J. 376, 553-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Hebert, M. D., Ye, Y., Templeton, D. J., Kung, H., and Matera, A. G. (2000). Cell cycle-dependent localization of the CDK2-cyclin E complex in Cajal (coiled) bodies. J. Cell Sci. 113(Pt 9), 1543-1552. [DOI] [PubMed] [Google Scholar]
- Llanos, S., Clark, P. A., Rowe, J., and Peters, G. (2001). Stabilization of p53 by p14ARF without relocation of MDM2 to the nucleolus. Nat. Cell Biol. 3, 445-452. [DOI] [PubMed] [Google Scholar]
- Malatesta, M., Gazzanelli, G., Battistelli, S., Martin, T. E., Amalric, F., and Fakan, S. (2000). Nucleoli undergo structural and molecular modifications during hibernation. Chromosoma 109, 506-513. [DOI] [PubMed] [Google Scholar]
- Matera, A. G., Frey, M. R., Margelot, K., and Wolin, S. L. (1995). A perinucleolar compartment contains several RNA polymerase III transcripts as well as the polypyrimidine tract-binding protein, hnRNP I. J. Cell Biol. 129, 1181-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur, M., Tucker, P. W., and Samuels, H. H. (2001). PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol. Cell. Biol. 21, 2298-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson, K., Pokrovskaja, K., Kiss, C., Klein, G., and Szekely, L. (2001). Proteins associated with the promyelocytic leukemia gene product (PML)-containing nuclear body move to the nucleolus upon inhibition of proteasome-dependent protein degradation. Proc. Natl. Acad. Sci. USA 98, 1012-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner, M., Dechat, T., Gerner, C., Grimm, R., Foisner, R., and Sauermann, G. (2000). Differential nuclear localization and nuclear matrix association of the splicing factors PSF and PTB. J. Cell. Biochem. 76, 559-566. [PubMed] [Google Scholar]
- Mirre, C., Hartung, M., and Stahl, A. (1980). Association of ribosomal genes in the fibrillar center of the nucleolus: a factor influencing translocation and nondisjunction in the human meiotic oocyte. Proc. Natl. Acad. Sci. USA 77, 6017-6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratani, M., Gerlich, D., Janicki, S. M., Gebhard, M., Eils, R., and Spector, D. L. (2002). Metabolic-energy-dependent movement of PML bodies within the mammalian cell nucleus. Nat. Cell Biol. 4, 106-110. [DOI] [PubMed] [Google Scholar]
- Nakamura, T., Mori, T., Tada, S., Krajewski, W., Rozovskaia, T., Wassell, R., Dubois, G., Mazo, A., Croce, C. M., and Canaani, E. (2002). ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol. Cell 10, 1119-1128. [DOI] [PubMed] [Google Scholar]
- Ochs, R. L. (1998). Methods used to study structure and function of the nucleolus. Methods Cell Biol. 53, 303-321. [DOI] [PubMed] [Google Scholar]
- Ochs, R. L., Lischwe, M. A., Spohn, W. H., and Busch, H. (1985). Fibrillarin: a new protein of the nucleolus identified by autoimmune sera. Biol. Cell 54, 123-133. [DOI] [PubMed] [Google Scholar]
- Olson, M. O. (2004a). Sensing cellular stress: another new function for the nucleolus? Sci. STKE 2004, pe10. [DOI] [PubMed] [Google Scholar]
- Olson, M. O. J. (2004b). Nontraditional roles of the nucleolus. In: The Nucleolus, ed. M. O. J. Olson, New York: Kluwer/Plenum, 329-342.
- Ospina, J. K., and Matera, A. G. (2002). Proteomics: the nucleolus weighs in. Curr. Biol. 12, R29-R31. [DOI] [PubMed] [Google Scholar]
- Patton, J. G., Porro, E. B., Galceran, J., Tempst, P., and Nadal-Ginard, B. (1993). Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 7, 393-406. [DOI] [PubMed] [Google Scholar]
- Pellizzoni, L., Baccon, J., Charroux, B., and Dreyfuss, G. (2001). The survival of motor neurons (SMN) protein interacts with the snoRNP proteins fibrillarin and GAR1. Curr. Biol. 11, 1079-1088. [DOI] [PubMed] [Google Scholar]
- Peng, R., Dye, B. T., Perez, I., Barnard, D. C., Thompson, A. B., and Patton, J. G. (2002). PSF and p54nrb bind a conserved stem in U5 snRNA. RNA 8, 1334-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestic-Dragovich, L., Stojiljkovic, L., Philimonenko, A. A., Nowak, G., Ke, Y., Settlage, R. E., Shabanowitz, J., Hunt, D. F., Hozak, P., and de Lanerolle, P. (2000). A myosin I isoform in the nucleus. Science 290, 337-341. [DOI] [PubMed] [Google Scholar]
- Phair, R. D., and Misteli, T. (2000). High mobility of proteins in the mammalian cell nucleus. Nature 404, 604-609. [DOI] [PubMed] [Google Scholar]
- Platani, M., Goldberg, I., Lamond, A. I., and Swedlow, J. R. (2002). Cajal body dynamics and association with chromatin are ATP-dependent. Nat. Cell Biol. 4, 502-508. [DOI] [PubMed] [Google Scholar]
- Puvion-Dutilleul, F., Puvion, E., and Bachellerie, J. P. (1997). Early stages of pre-rRNA formation within the nucleolar ultrastructure of mouse cells studied by in situ hybridization with a 5′ETS leader probe. Chromosoma 105, 496-505. [DOI] [PubMed] [Google Scholar]
- Raska, I., Ochs, R. L., Andrade, L. E., Chan, E. K., Burlingame, R., Peebles, C., Gruol, D., and Tan, E. M. (1990). Association between the nucleolus and the coiled body. J. Struct. Biol. 104, 120-127. [DOI] [PubMed] [Google Scholar]
- Recher, L., Briggs, L. G., and Parry, N. T. (1971). A reevaluation of nuclear and nucleolar changes induced in vitro by actinomycin D. Cancer Res. 31, 140-151. [PubMed] [Google Scholar]
- Reddy, J. K., and Svoboda, D. J. (1972). Natural segregation of nucleolar components of liver cells in newts. J. Ultrastruct. Res. 38, 608-613. [DOI] [PubMed] [Google Scholar]
- Reimer, G., Rose, K. M., Scheer, U., and Tan, E. M. (1987). Autoantibody to RNA polymerase I in scleroderma sera. J. Clin. Invest. 79, 65-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, Y., Busch, R. K., Perlaky, L., and Busch, H. (1998). The 58-kDa microspherule protein (MSP58), a nucleolar protein, interacts with nucleolar protein p120. Eur. J. Biochem. 253, 734-742. [DOI] [PubMed] [Google Scholar]
- Reynolds, R. C., Montgomery, P. O., and Hughes, B. (1964). Nucleolar “Caps” produced by Actinomycin D. Cancer Res. 24, 1269-1277. [PubMed] [Google Scholar]
- Reynolds, R. C., Montgomery, P. O., and Karney, D. H. (1963). Nucleolars “caps”—a morphologic entity produced by the carcinogen 4-nitroquinoline N-oxide. Cancer Res. 23, 535-538. [PubMed] [Google Scholar]
- Rubbi, C. P., and Milner, J. (2003). Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 22, 6068-6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer, U., and Hock, R. (1999). Structure and function of the nucleolus. Curr. Opin. Cell Biol. 11, 385-390. [DOI] [PubMed] [Google Scholar]
- Scherl, A., Coute, Y., Deon, C., Calle, A., Kindbeiter, K., Sanchez, J. C., Greco, A., Hochstrasser, D., and Diaz, J. J. (2002). Functional proteomic analysis of human nucleolus. Mol. Biol. Cell 13, 4100-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoefl, G. I. (1964). The effect of Actinomycin D on the fine structure of the nucleolus. J. Ultrastruct. Res. 10, 224-243. [DOI] [PubMed] [Google Scholar]
- Shav-Tal, Y., Cohen, M., Lapter, S., Dye, B., Patton, J. G., Vandekerckhove, J., and Zipori, D. (2001a). Nuclear relocalization of the pre-mRNA splicing factor PSF during apoptosis involves hyperphosphorylation, masking of antigenic epitopes, and changes in protein interactions. Mol. Biol. Cell 12, 2328-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal, Y., Darzacq, X., Shenoy, S. M., Fusco, D., Janicki, S. M., Spector, D. L., and Singer, R. H. (2004). Dynamics of single mRNPs in nuclei of living cells. Science 304, 1797-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal, Y., Lee, B., Bar-Haim, S., Vandekerckhove, J., and Zipori, D. (2000). Enhanced proteolysis of pre-mRNA splicing factors in myeloid cells. Exp. Hematol. 28, 1029-1038. [DOI] [PubMed] [Google Scholar]
- Shav-Tal, Y., Lee, B. C., Bar-Haim, S., Schori, H., and Zipori, D. (2001b). Reorganization of nuclear factors during myeloid differentiation. J. Cell. Biochem. 81, 379-392. [DOI] [PubMed] [Google Scholar]
- Shav-Tal, Y., and Zipori, D. (2002). PSF and p54(nrb)/NonO—multi-functional nuclear proteins. FEBS Lett. 531, 109-114. [DOI] [PubMed] [Google Scholar]
- Shaw, P. J., and Jordan, E. G. (1995). The nucleolus. Annu. Rev. Cell Dev. Biol. 11, 93-121. [DOI] [PubMed] [Google Scholar]
- Shtutman, M., Zhurinsky, J., Oren, M., Levina, E., and Ben-Ze'ev, A. (2002). PML is a target gene of beta-catenin and plakoglobin, and coactivates beta-catenin-mediated transcription. Cancer Res. 62, 5947-5954. [PubMed] [Google Scholar]
- Simard, R., Langelier, Y., Mandeville, R., Maestracci, N., and Royal, A. (1974). Inhibitors as tools in elucidating the structure and function of the nucleus. New York: Academic Press.
- Singer, R. H., and Green, M. R. (1997). Compartmentalization of eukaryotic gene expression: causes and effects. Cell 91, 291-294. [DOI] [PubMed] [Google Scholar]
- Smetana, K., and Busch, H. (1974). The nucleolus and nucleolar DNA. In: The Cell Nucleus, vol. 1, ed. H. Busch, New York: Academic Press, 73-147. [Google Scholar]
- Smetana, K., Gyorkey, F., Gyorkey, P., and Busch, H. (1972). Studies on nucleoli and cytoplasmic fibrillar bodies of human hepatocellular carcinomas. Cancer Res. 32, 925-932. [PubMed] [Google Scholar]
- Stahl, A., Wachtler, F., Hartung, M., Devictor, M., Schofer, C., Mosgoller, W., de Lanversin, A., Fouet, C., and Schwarzacher, H. G. (1991). Nucleoli, nucleolar chromosomes and ribosomal genes in the human spermatocyte. Chromosoma 101, 231-244. [DOI] [PubMed] [Google Scholar]
- Tucker, K. E., Berciano, M. T., Jacobs, E. Y., LePage, D. F., Shpargel, K. B., Rossire, J. J., Chan, E. K., Lafarga, M., Conlon, R. A., and Matera, A. G. (2001). Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J. Cell Biol. 154, 293-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez, B. C., Perlaky, L., Cai, Z. J., Henning, D., and Busch, H. (1998). Green fluorescent protein tag for studies of drug-induced translocation of nucleolar protein RH-II/Gu. Biotechniques 24, 1032-1036. [DOI] [PubMed] [Google Scholar]
- van Eenennaam, H., Jarrous, N., van Venrooij, W. J., and Pruijn, G. J. (2000). Architecture and function of the human endonucleases RNase P and RNase MRP. IUBMB Life 49, 265-272. [DOI] [PubMed] [Google Scholar]
- Van Gansen, P., and Schram, A. (1972). Evolution of the nucleoli during oogenesis in Xenopus laevis studied by electron microscopy. J. Cell Sci. 10, 339-367. [DOI] [PubMed] [Google Scholar]
- Vera, M. I., Norambuena, L., Alvarez, M., Figueroa, J., Molina, A., Leon, G., and Krauskopf, M. (1993). Reprogramming of nucleolar gene expression during the acclimatization of the carp. Cell Mol. Biol. Res. 39, 665-674. [PubMed] [Google Scholar]
- Wang, J., Shiels, C., Sasieni, P., Wu, P. J., Islam, S. A., Freemont, P. S., and Sheer, D. (2004). Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J. Cell Biol. 164, 515-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, J. D., Taylor, L. J., Roussel, M. F., Sherr, C. J., and Bar-Sagi, D. (1999). Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1, 20-26. [DOI] [PubMed] [Google Scholar]
- Westendorf, J. M., Konstantinov, K. N., Wormsley, S., Shu, M. D., Matsumoto-Taniura, N., Pirollet, F., Klier, F. G., Gerace, L., and Baserga, S. J. (1998). M phase phosphoprotein 10 is a human U3 small nucleolar ribonucleoprotein component. Mol. Biol. Cell 9, 437-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, M. H., Lam, C. Y., and Yung, B. Y. (1995). Translocation of nucleophosmin from nucleoli to nucleoplasm requires ATP. Biochem. J. 305(Pt 3), 987-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, T., Nakamura, T., Blechman, J., Sorio, C., Dang, C. V., Geiger, B., and Canaani, E. (1997). Nuclear punctate distribution of ALL-1 is conferred by distinct elements at the N terminus of the protein. Proc. Natl. Acad. Sci. USA 94, 7286-7291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatsepina, O. V., Voit, R., Grummt, I., Spring, H., Semenov, M. V., and Trendelenburg, M. F. (1993). The RNA polymerase I-specific transcription initiation factor UBF is associated with transcriptionally active and inactive ribosomal genes. Chromosoma 102, 599-611. [DOI] [PubMed] [Google Scholar]
- Zinszner, H., Immanuel, D., Yin, Y., Liang, F. X., and Ron, D. (1997). A topogenic role for the oncogenic N-terminus of TLS: nucleolar localization when transcription is inhibited. Oncogene 14, 451-461. [DOI] [PubMed] [Google Scholar]