Abstract
Background:
DNA methylation-based measures of biological aging have been associated with air pollution and may link pollutant exposures to aging-related health outcomes. However, evidence is inconsistent and there is little information for Black women.
Objective:
We examined associations of ambient particulate matter <2.5 μm and <10 μm in diameter (PM2.5 and PM10) and nitrogen dioxide (NO2) with DNA methylation, including epigenetic aging and individual CpG sites, and evaluated whether associations differ between Black and non-Hispanic White (NHW) women.
Methods:
Validated models were used to estimate annual average outdoor residential exposure to PM2.5, PM10, and NO2 in a sample of self-identified Black (n=633) and NHW (n=3493) women residing in the contiguous US. We used sampling-weighted generalized linear regression to examine the effects of pollutants on six epigenetic aging measures (primary: DunedinPACE, GrimAgeAccel, and PhenoAgeAccel; secondary: Horvath intrinsic epigenetic age acceleration [EAA], Hannum extrinsic EAA, and skin & blood EAA) and epigenome-wide associations for individual CpG sites. Wald tests of nested models with and without interaction terms were used to examine effect measure modification by race/ethnicity.
Results:
Black participants had higher median air pollution exposure than NHW participants. GrimAgeAccel was associated with both PM10 and NO2 among Black participants, (Q4 versus Q1, PM10: β=1.09, 95% CI: 0.16–2.03; NO2: β=1.01, 95% CI 0.08–1.94) but not NHW participants (p-for-heterogeneity: PM10=0.10, NO2=0.20). In Black participants, we also observed a monotonic exposure–response relationship between NO2 and DunedinPACE (Q4 versus Q1, NO2: β=0.029, 95% CI: 0.004–0.055; p-for-trend=0.03), which was not observed in NHW participants (p-for-heterogeneity=0.09). In the EWAS, pollutants were significantly associated with differential methylation at 19 CpG sites in Black women and one in NHW women.
Conclusions:
In a US-wide cohort study, our findings suggest that air pollution is associated with DNA methylation alterations consistent with higher epigenetic aging among Black, but not NHW, women.
Keywords: Air pollution, DNA methylation, Particulate matter, Epigenetic age, Epigenome-wide association study
1. Introduction
Air pollution exposure is linked to many adverse age-related health outcomes. An estimated 4.2 million premature deaths globally were caused by ambient outdoor air pollution in 2019, with the majority of these deaths attributable to aging-related conditions, such as heart disease, stroke, and cancer (WHO, 2022). The link between air pollution and age-related diseases may be due, in part, to air pollution effects on biological processes that underlie aging (Peters et al., 2021). Biological age measures efficiently integrate molecular processes that underlie aging and may therefore be useful tools in understanding the relationship between air pollution and age-related disease risk.
Biological age can be estimated in a variety of ways, including telomere length and epigenetic markers such as histone modification and DNA methylation (Ferrucci et al., 2020). Several DNA methylation-based measures have been developed that use site-specific patterns of DNA methylation to predict chronological age (e.g., Hannum and Horvath), mortality risk (e.g., PhenoAge and GrimAge), or aging rates (e.g., DunedinPACE) (Belsky et al., 2022; Hannum et al., 2013; Horvath, 2013; Levine et al., 2018; Lu et al., 2019).
Several studies have suggested that ambient air pollution is associated with DNA methylation-based measures of biological age (Nwanaji-Enwerem et al., 2016, 2017; Ward-Caviness et al., 2016; White et al., 2019). However, the evidence is inconsistent and existing literature has mainly focused on the Hannum, Horvath, and PhenoAge clocks. To our knowledge no prior studies have yet examined the more recently developed DunedinPACE measure, which, along with GrimAge and PhenoAge, may be a more robust marker of age-related disease risk than the Horvath and Hannum clocks based on the outcomes they were designed to predict.
Prior studies have primarily been conducted among non-Hispanic White (NHW) populations, leaving knowledge gaps concerning other racial and ethnic groups. Racial and ethnic identity are social factors that, driven by historical and ongoing racism, are associated with differences in exposure levels, proximity, and chemical composition of ambient air pollution (Lane et al., 2022; Tessum et al., 2021) as well as susceptibility to the harmful effects of air pollution (Josey et al., 2023; Younan et al., 2021). Similar differences have been documented for chronic disease risk, biological age, and methylation patterns (Barcelona de Mendoza et al., 2018; Caraballo et al., 2022; Geronimus et al., 2010). Thus, associations between pollutants and epigenetic aging may vary by race and ethnicity and warrant exploration.
White et al. (2019) examined cross-sectional associations of air pollutants with age acceleration measured by the Hannum, Horvath, and PhenoAge clocks and conducted an epigenome-wide association study (EWAS) among a sample of NHW participants in the Sister Study. Here we extend this work to include Black participants (Hispanic or non-Hispanic) and to examine cross-sectional associations of PM2.5, PM10, and NO2 with two measures of epigenetic aging acceleration (PhenoAgeAccel and GrimAgeAccel) and a measure of aging rate (DunedinPACE). As a secondary analysis, we assess associations with three additional measures of epigenetic aging acceleration (Horvath intrinsic epigenetic age acceleration [IEAA], Hannum extrinsic epigenetic age acceleration [EEAA], and skin & blood clock epigenetic age acceleration [SBEAA]). For convenience, we refer to all six as epigenetic aging measures. Further, we explore if these associations differ among Black and NHW participants and evaluate pollutant associations with individual CpG sites by performing an EWAS.
2. Methods
2.1. Study sample
The Sister Study is an ongoing, prospective cohort of self-identified women from across the United States (US) that was established to study risk factors for breast cancer. Between 2003 and 2009, 50,884 females aged 35–74 who had at least one biological sister with a diagnosis of breast cancer and no history of breast cancer themselves were enrolled (Sandler et al., 2017). At enrollment, participants provided information through computer-assisted telephone interviews, including demographics, lifestyle factors, medical history, and residential history. Participants self-classified their race and ethnicity based on Census categories required by the Office of Management and Budget. While race and ethnicity are distinct concepts, given their combined use in subgroup classification for the present study, we henceforth refer to the subgroups as “race/ethnicity” subgroups. A trained examiner visited participants’ homes to collect anthropometric information and fasting blood samples (Sandler et al., 2017). All participants provided written informed consent and the study was approved by the National Institute of Environmental Health Institutional Review Board, and is currently overseen by the centralized NIH Institutional Review Board. The data used is from data release version 10.1.
Subsets of Black, Hispanic or non-Hispanic, and NHW participants with available blood samples (>99 % of cohort) were selected for assays at two times for separate case-cohort studies designed to assess the association between DNA methylation and incident breast cancer, as previously described (Kresovich et al., 2023). Briefly, 2875 NHW participants were selected for the case-cohort in 2014. In 2018, a set of 2166 participants were selected, specifically oversampling for Black participants (n=738), and including 541 participants that were also sampled for the first case-cohort. After accounting for overlap and excluding participants whose methylation data failed quality control procedures, 4482 participants with DNA methylation data remained.
Among this sample, we excluded participants who lived at their residence for less than one year prior to baseline (n=268), were missing all exposure data (n=28), or were missing covariate information (n=60). This resulted in 4126 participants who were eligible for at least one analysis (Supplemental Fig. S1).
2.2. Air pollution exposure estimates
Annual average outdoor air pollution concentrations for PM2.5 and NO2 were estimated at participants’ baseline residence using validated spatiotemporal models, while concentrations for PM10 were estimated using validated universal kriging models, as previously described (Kirwa et al., 2021; Sampson et al., 2013). Briefly, the models leveraged data from over 1000 regulatory monitors, such as Environmental Protection Agency Air Quality System monitoring data and interagency monitoring of protected visual environments network data. The spatiotemporal models additionally included data from over 900 research monitors, including residential monitoring campaigns and gradient monitoring near roadways for NO2. All models incorporated spatial smoothing and geographic covariates. We used concentrations from calendar-year 2006 for PM2.5 and NO2, as this was the middle of the Sister Study enrollment period. Data for PM10 was only available for 2000 and 2010, therefore, estimates for 2000 were used to capture PM10 exposure prior to enrollment and blood draw.
2.3. DNA methylation and epigenetic aging outcomes
DNA methylation was measured using the Illumina Infinium HumanMethylation450 BeadChip (450K array) and the Infinium MethylationEPIC V1 BeadChip (EPIC array) for participants assayed in 2014 and 2018, respectively. For participants whose methylation data was assayed on both platforms, we used data from the EPIC array. DNA methylation processing and quality control (QC) procedures have been described previously (Kresovich et al., 2023). Briefly, the ENmix R package was used for preprocessing and quality checks (Xu et al., 2016), and CpG sites and participants with low quality data were removed. Methylation was measured as a β value using the formula β=M/(U+M+100) where U and M were the individual’s proportion of unmethylated and methylated sites, respectively, at a given locus. β values were logit-transformed to M-values for the EWAS.
DunedinPACE (‘Pace of Aging Calculated from the Epigenome’) is proposed to estimate the pace, or rate, at which a person is aging (Belsky et al., 2022). DunedinPACE values are always positive, where a value of 1.0 signifies the expected amount of aging per chronological year of life; values greater than one represent faster rates of aging whereas values less than one represent slower rates of aging. DunedinPACE was calculated separately for the 450K and EPIC array samples using the methylAge function as part of the ENmix R package on Bioconductor (https://www.bioconductor.org/packages/release/bioc/html/ENmix.html).
Age acceleration as estimated by the GrimAge (GrimAgeAccel) and PhenoAge (PhenoAgeAccel) epigenetic clocks represents the interval between DNA methylation-predicted biological age and chronological age. GrimAgeAccel and PhenoAgeAccel were calculated as the residuals from regressing DNA methylation-predicted biological age onto chronological age, where positive values indicate epigenetic age is greater than chronological age and negative values indicate epigenetic age is less than chronological age. PhenoAgeAccel and GrimAgeAccel were estimated separately for the 450K and EPIC array samples using an online calculator without fast imputation (https://dnamage.genetics.ucla.edu/home). For both race/ethnicity groups, 9 out of 514 CpGs for PhenoAgeAccel and 4 out of 173 CpGs for DunedinPACE were missing; GrimAge component CpGs are not publicly available.
Three additional age acceleration measures were evaluated in secondary analyses as they offer different advantages than the primary aging measures: Horvath IEAA, which is proposed to be independent of blood cell composition; Hannum EEAA, which is proposed to incorporate aging of the immune system; and SBEAA, which is reported to have greater accuracy in blood samples compared to the original Horvath clock (Horvath et al., 2018; Smith et al., 2019). These measures are calculated similarly to GrimAgeAccel and PhenoAgeAccel and were estimated using the same online calculator.
2.4. Statistical analysis
Cross-sectional associations between residential air pollution concentrations and the epigenetic aging measures were estimated using sampling-weighted multivariable linear regression models for each air pollutant (PM2.5, PM10, NO2) and epigenetic aging outcome (DunedinPACE, GrimAgeAccel, PhenoAgeAccel). Air pollution concentrations were modeled in two ways: linearly scaled by an interquartile range (IQR) increase and as quartiles, defined in the overall sample and treated categorically, to assess possible exposure–response relationships or thresholds of effects. Confounders were identified using a directed acyclic graph and included DNA methylation array (450K, EPIC), chronological age, self-reported race/ethnicity (Black, NHW), highest attained education level (high school/GED or less, some college/associate or technical degree, college degree or more), body mass index at home exam (BMI, continuous), physical activity (hours per week, continuous), smoking status (never, past, current), total cigarette smoking pack-years (continuous), region (Midwest, Northeast, South, West), and a neighborhood-level measure of socioeconomic disadvantage, the area deprivation index (ADI, percentile, continuous) at the 2000 census block group level (Kind et al., 2014).
Among the 4126 participants eligible for at least one analysis, those who had acceleration measures more than 4 standard deviations away from the sample mean (n=3 for DunedinPACE, n=6 for GrimAgeAccel, n=3 for PhenoAgeAccel) or were missing one of the pollutant estimates (n=33 for PM2.5 and NO2) were excluded for the relevant models. We conducted complete case analysis due to the low percentage of missing data (<3.0 %).
P-for-heterogeneity values to assess effect measure modification by race/ethnicity were calculated using Wald tests of nested models with and without interaction terms between race/ethnicity and air pollutant exposures. P-for-trend values were determined using Wald tests on ordinal categories for exposure quartiles.
We conducted a sensitivity analysis where all pollutants were included in the same model to account for possible mutual confounding. Secondary analyses with the additional aging measures (IEAA, EEAA, SBEAA) were performed using the same approach as for the primary aging outcomes. Sampling-weighted Pearson correlation coefficients were calculated for all aging measures and chronological age by race/ethnicity to assess possible performance differences in the epigenetic age measures by race/ethnicity.
For the EWAS, we assessed associations between IQR increases in pollutants and individual CpG sites using sampling-weighted generalized linear regression, separately for each race/ethnicity group, adjusting for Houseman cell type proportions (B-cell, CD4+ T-cell, CD8+ T-cell, granulocyte, monocyte; natural killer was left out) (Houseman et al., 2012) in addition to the covariates used for the epigenetic aging analyses. For the NHW subgroup, probes from the 450K (469,412 post QC) and EPIC array (847,518 post QC) were analyzed separately and overlapping probes (431,879) were meta-analyzed using a random effects model as part of the metafor R package. EWAS results were additionally checked for differentially methylated regions (DMRs) using the ipDMR method from the ENmix R package (Xu et al., 2020a). We corrected for multiple testing using a false discovery rate (FDR) of q<0.05. The top 100 probes for each pollutant in each racial/ethnic group (i.e., six separate analyses) were analyzed for pathway enrichment using the Ingenuity Pathway Analysis (IPA) database, as previously described (Xu et al., 2020b).
In all analyses, participants were weighted by their inverse probability of selection into the case-cohort samples. Sampling weights were based on race/ethnicity as well as future breast cancer status and characteristics (i.e., estrogen receptor status), so that the study sample could represent the full sample of Black and NHW participants in the Sister Study (O’Brien et al., 2022). The weighted models for epigenetic aging outcomes were estimated using PROC SURVEYREG in SAS 9.4. Analyses were conducted using SAS 9.4 (Cary, NC) and R (4.1.0).
3. Results
3.1. Participant characteristics
Sample characteristics are displayed in Table 1. The median chronological age at baseline was 54.8 and 56.9 years among the Black and NHW participants, respectively, and more than half of all participants had a college degree or more. A majority of the Black participants resided in the South (59%), while the NHW participants resided more evenly across the four US Census regions (30% Midwest, 18% Northeast, 30% South, and 22% West). Black participants were more likely to be never smokers than NHW participants (64% versus 53%) and reside in a neighborhood with greater deprivation (median ADI 44.0 versus 27.0).
Table 1.
Baseline study population characteristics stratified by race/ethnicity in the Sister Study 2003–2009 (n=4126).
| Black (n=633) |
NHW (n=3493) |
|
|---|---|---|
| Age (years), median (IQR) | 54.8 (48.6, 59.6) | 56.9 (50.6, 63.6) |
| Education level, n (%) | ||
| High school/GED or less | 63 (10%) | 537 (15%) |
| Some college/associate or technical degree | 215 (34%) | 1135 (33%) |
| College degree or more | 355 (56%) | 1821 (52%) |
| Region, n (%) | ||
| Midwest | 141 (22%) | 1040 (30%) |
| Northeast | 61 (10%) | 639 (18%) |
| South | 374 (59%) | 1046 (30%) |
| West | 57 (9%) | 768 (22%) |
| BMI (kg/m 2 ), median (IQR) | 30.2 (26.4, 35.4) | 26.4 (23.3, 30.5) |
| Smoking status, n (%) | ||
| Never | 405 (64%) | 1844 (53%) |
| Past | 176 (28%) | 1393 (40%) |
| Current | 52 (8%) | 256 (7%) |
| Total pack years among current and past smokers, median (IQR) | 8.6 (2.9, 18.1) | 10.5 (3.1, 23.0) |
| Physical activity (hours per week), median (IQR) | 11.0 (6.9, 16.0) | 12.5 (7.7, 19.0) |
| ADI of primary residence (percentile), median (IQR) | 44.0 (24.0, 69.0) | 27.0 (12.0, 48.0) |
| Air pollutants, median (IQR) | ||
| PM2.5 2006, μg/m3 | 12.0 (10.9, 13.2) | 10.4 (8.5, 11.7) |
| PM10 2000, μg/m3 | 23.4 (20.7, 26.6 | 21.4 (18.4, 24.2 |
| NO2 2006, ppb | 10.8 (8.0, 14.7) | 8.0 (5.5, 11.2) |
| Epigenetic aging measures, median (IQR) | ||
| DunedinPACE | 1.10 (1.05, 1.17) | 1.05 (1.00, 1.11) |
| GrimAgeAccel | 0.16 (−1.86, 2.89) | −0.81 (−2.51, 1.48) |
| PhenoAgeAccel | −0.08 (−4.91, 4.45) | −0.39 (−4.17, 3.69) |
Abbreviations: ADI, area deprivation index; BMI, body mass index; IQR, interquartile range; NHW, non-Hispanic White; NO2, nitrogen dioxide; PM10, particulate matter<10 μm in diameter; PM2.5, particulate matter<2.5 μm in diameter; Ppb, parts per billion.
3.2. Distribution of exposure and outcome by race/ethnicity
Black participants were exposed to higher median air pollutant concentrations than NHW participants for all three pollutant categories, with the greatest difference in median NO2 level (Black: 10.8 ppb; NHW: 8.0 ppb; Table 1). Black participants also exhibited modestly higher median epigenetic aging by all three primary measures (median DunedinPACE: 1.10 versus 1.05, GrimAgeAccel: 0.16 versus −0.81, PhenoAgeAccel: −0.08 versus −0.39).
3.3. Air pollutants and epigenetic age measures
Overall, there was little evidence of associations between PM2.5 and the primary epigenetic aging measures for either Black or NHW participants (Fig. 1A and Supplemental Table S1). In Black participants, some quartiles exhibited higher aging values compared to quartile 1, but there was no evidence of monotonic trends. Similarly, in the NHW subgroup there were modest inverse associations between some quartiles of PM2.5, but no monotonic trends.
Fig. 1.
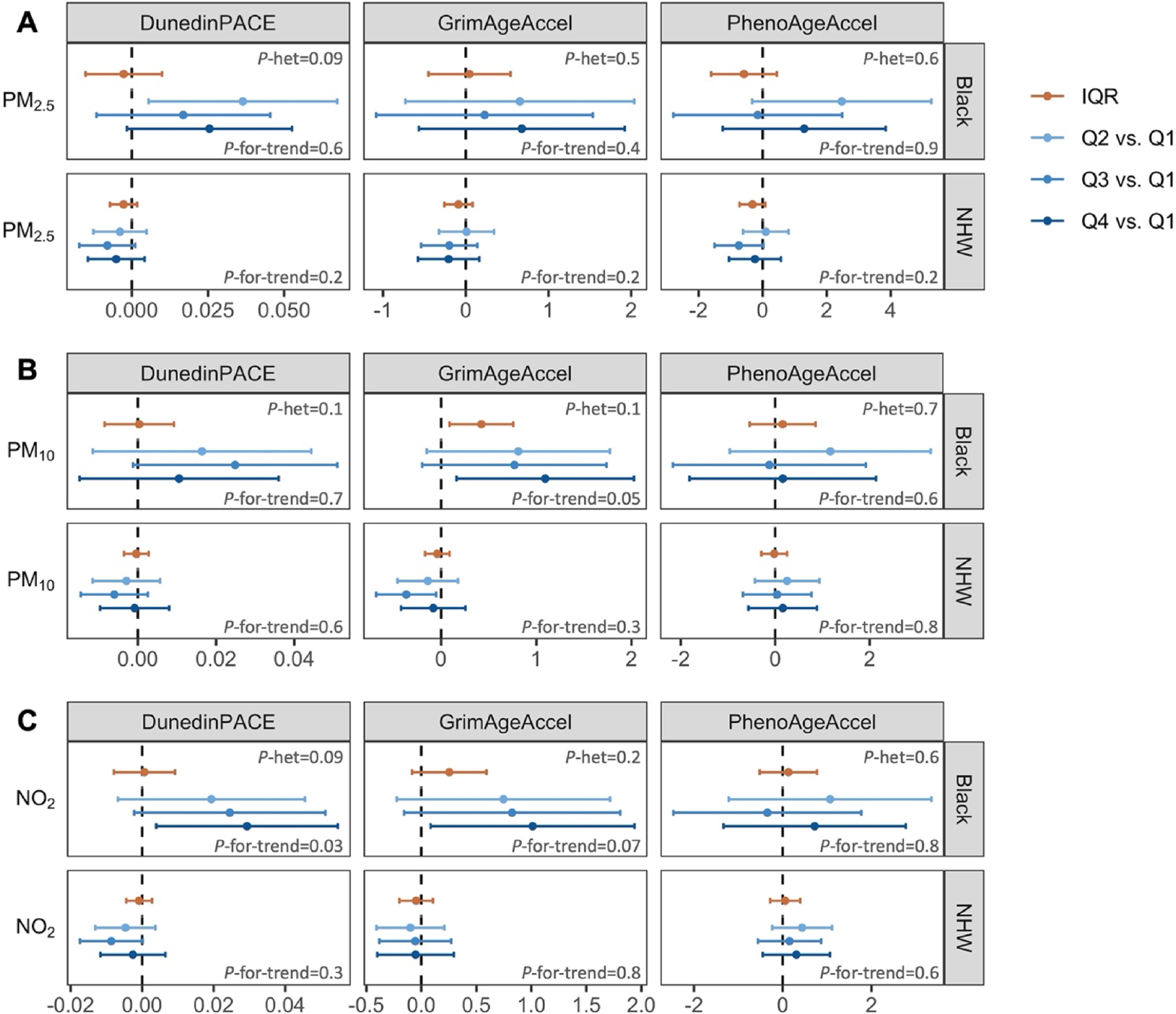
Effect estimates and 95% CIs for the associations between (A) PM2.5, (B) PM10, and (C) NO2 and epigenetic aging measures, by race/ethnicity. Notes: [a] β estimates are the result of linear regression for the association between either an IQR increase in air pollutants or quartiles 2–4 (compared to quartile 1) of air pollutants and epigenetic aging adjusting for methylation array, chronological age, race/ethnicity, education, BMI, physical activity, smoking status, cigarette smoking pack-years, region, and ADI at the census block group level. [b] P-for-trend values are from models where quartiles are treated ordinally. [c] P-het values displayed are from Wald tests of nested models with and without interaction terms for quartiles and race/ethnicity. P-het values for the IQR models are from an interaction term between pollutants and race/ethnicity: Panel A. P-het DunedinPACE=1.0, GrimAgeAccel=0.6, PhenoAgeAccel=0.6. Panel B. P-het DunedinPACE=0.9, GrimAgeAccel=0.01, PhenoAgeAccel=0.6. Panel C. P-het DunedinPACE=0.8, GrimAgeAccel=0.1, PhenoAgeAccel=0.8. Abbreviations: ADI, area deprivation index; BMI, body mass index; CI, confidence interval; IQR, interquartile range; NHW, non-Hispanic White; NO2, nitrogen dioxide; P-het, p-for-heterogeneity; PM10, particulate matter<10 μm in diameter; PM2.5, particulate matter<2.5 μm in diameter; Q1–4, quartile 1–4.
Among Black participants, an IQR increase in PM10 was associated with higher GrimAgeAccel of 0.42 years (95% CI: 0.09, 0.76) whereas no effect was observed among NHW participants (β=−0.04 [95% CI: −0.17, 0.09]; p-for-heterogeneity=0.01) (Fig. 1B and Supplemental Table S1). Quartile analysis among Black women found the increase was most evident among the highest quartile of exposure which, relative to quartile 1 was associated with 1.09 years (95% CI: 0.16, 2.03) of acceleration with evidence of an exposure–response relationship (p-for-trend 0.05). In contrast, among NHW participants, we found negligible or inconsistent associations of higher quartiles of exposure with GrimAgeAccel (p-for-heterogeneity=0.1) and no evidence of trend (p-for-trend=0.3).
Higher levels of NO2 were associated with GrimAgeAccel among Black participants (quartile 4 versus quartile 1: β=1.01 [95% CI: 0.08, 1.94]; p-for-trend=0.07), whereas no association was observed in NHW participants (β=−0.05 [95% CI: −0.40, 0.30]; p-for-trend=0.8; p-for-heterogeneity=0.2) (Fig. 1C and Supplemental Table S1). Similarly, among Black participants the highest quartile of exposure was associated with higher DunedinPACE (β=0.029 95% CI: 0.004, 0.055; p-for-trend=0.03). In contrast, in NHW participants there was no evidence of an exposure trend (p-for-trend=0.3; p-for-heterogeneity=0.09).
In a sensitivity analysis where all pollutants were included in the same model, the findings were similar except the association between quartile 4 versus quartile 1 of NO2 and GrimAgeAccel among Black participants was attenuated towards the null (Supplemental Table S2). No associations were identified between pollutants and IEAA, EEAA, or SBEAA measures (Supplemental Table S3). The measures overall demonstrated high magnitudes of correlation with chronological age for both race/ethnicity groups, except for DunedinPACE which, unlike the other measures, was not designed to be a measure of chronological or biological age (Supplemental Fig. S2).
3.4. EWAS of air pollutants
The EWAS of Black women identified 19 CpG sites associated (FDR q<0.05) with pollutant exposures (Table 2). An IQR increase in NO2 was associated with lower methylation at one CpG site (cg24269657) on chromosome 13, mapped near the F7 gene; and an IQR increase in PM2.5 was associated with lower methylation at 15 CpG sites and higher methylation at 3 CpG sites. Of these 18 CpG sites, the site with the smallest q-value was cg07635198, whose closest gene is PRPF40A on chromosome 2. Given possible inflation in the EWAS of PM2.5 among Black participants (λ=1.41; Supplemental Fig. S3), we performed a sensitivity analysis using surrogate variable analysis methods to check if the EWAS was affected by unobserved confounders (Leek and Storey, 2007). The surrogate variable analysis (adjusting for 68 surrogate variables) produced a similar QQ-plot (λ=1.25), and the results were correlated with the original analysis (adjusting for 13 covariates), especially for the set of statistically significant probes (Supplemental Fig. S6). The EWAS among NHW women identified no sites among the separately analyzed 450K and EPIC probes and a single site from the meta-analyzed probes, cg24537688, on chromosome 2 for which methylation was inversely associated with NO2 exposure (Table 2). None of the 20 statistically significant CpG sites are included in calculation of PhenoAge or DunedinPACE; GrimAge component CpGs are not publicly available.
Table 2.
| Associated pollutant | Probe | Mapped gene | Chr | Relation to CpG island | Black |
NHW |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Effect estimate | 95 % CI | P-value | Effect estimate | 95 % CI | P-value | |||||
| NO2 | cg24537688 | – | 2 | −0.006 | (−0.039, 0.027) | 0.725 | −0.049 | (−0.066, −0.031) | 8.47E-08 | |
| cg24269657 | F7 | 13 | North shore | −0.118 | (−0.159, −0.077) | 2.90E-08 | 0.012 | (−0.015, 0.039) | 0.373 | |
| PM2.5 | cg07635198 | PRPF40A | 2 | −0.115 | (−0.154, −0.076) | 1.11E-08 | 0.014c | (−0.017, 0.045) | 0.370 | |
| cg08097847 | ZCCHC24 | 10 | North shore | 0.207 | (0.136, 0.278) | 1.76E-08 | −0.061c | (−0.124, 0.001) | 0.055 | |
| cg01593570 | MARCH10 | 17 | −0.133 | (−0.182, −0.084) | 1.18E-07 | −0.009c | (−0.046, 0.029) | 0.643 | ||
| cg13538431 | RP11–1252I4.2;RP11–423H2.1;RP11–423H2.3 | 5 | −0.164 | (−0.225, −0.103) | 2.00E-07 | 0.027c | (−0.020, 0.074) | 0.261 | ||
| cg23717809 | RP11–510 M2.1 | 16 | South shore | −0.114 | (−0.157, −0.071) | 2.44E-07 | −0.030c | (−0.070, 0.010) | 0.143 | |
| cg06738602 | PTGER2 | 14 | North shore | 0.185 | (0.115, 0.256) | 3.05E-07 | −0.008 | (−0.053, 0.036) | 0.711 | |
| cg00212245 | – | 8 | −0.103 | (−0.142, −0.063) | 3.73E-07 | −0.011c | (−0.042, 0.019) | 0.465 | ||
| cg01906102 | SSX1; SSX5 | X | −0.121 | (−0.168, −0.074) | 4.89E-07 | 0.004 | (−0.022, 0.030) | 0.761 | ||
| cg04902542 | TBC1D16 | 17 | North shelf | −0.077 | (−0.106, −0.047) | 5.00E-07 | 0.007 | (−0.009, 0.024) | 0.361 | |
| cg05411829 | – | 16 | −0.088 | (−0.122, −0.054) | 5.08E-07 | −0.004 | (−0.022, 0.013) | 0.615 | ||
| cg08052751 | SETBP1 | 18 | −0.128 | (−0.177, −0.078) | 5.92E-07 | −0.002c | (−0.039, 0.034) | 0.907 | ||
| cg11514293 | GALC | 14 | −0.093 | (−0.129, −0.056) | 8.01E-07 | 0.005c | (−0.026, 0.035) | 0.762 | ||
| cg10011083 | VOPP1 | 7 | Island | 0.173 | (0.104, 0.241) | 9.71E-07 | −4.87E-04 | (−0.026, 0.025) | 0.971 | |
| cg16722016 | – | 12 | −0.125 | (−0.175, −0.075) | 1.02E-06 | −1.58E-05c | (−0.038, 0.038) | 0.999 | ||
| cg18580650 | – | 5 | North shelf | −0.083 | (−0.116, −0.050) | 1.08E-06 | −0.014 | (−0.030, 0.002) | 0.085 | |
| cg11130461 | ZBTB20 | 3 | Island | −0.161 | (−0.225, −0.097) | 1.10E-06 | −0.003 | (−0.062, 0.056) | 0.931 | |
| cg24544356 | FSTL5 | 4 | −0.087 | (−0.122, −0.053) | 1.15E-06 | −0.010c | (−0.041, 0.021) | 0.528 | ||
| cg17608585 | RP11–1055B8.3 | 17 | North shore | −0.114 | (−0.160, −0.069) | 1.23E-06 | 0.016c | (−0.018, 0.050) | 0.356 | |
Survey-weighted generalized linear regression models were performed separately by race/ethnicity subgroup. Models adjusted for Houseman cell type proportions, chronological age, education, BMI, physical activity, smoking status, cigarette pack-years, region, and ADI at the census block group level.
CpG sites that were significant for at least one of the race/ethnicity subgroups are presented for both subgroups, for comparison.
Effect estimates, CIs, and p-values for probes that were not meta-analyzed among NHW participants are from the EWAS of EPIC probes only (n=1311). Abbreviations: ADI, area deprivation index; BMI, body mass index; Chr, chromosome; CI, confidence interval; CpG, Cytosine-phosphate-Guanine; FDR, false discovery rate; NHW, non-Hispanic White; NO2, nitrogen dioxide; PM2.5, particulate matter<2.5 μm in diameter.
Among Black women, we found 272 DMRs for PM2.5, 8 DMRs for PM10, and 15 DMRs for NO2 (Supplemental Table S4). Among NHW women we identified only 2 DMRs, both of which were associated with PM2.5; none were observed for NO2 or PM10. One DMR (Chr8: 2238275–2238306) was associated with both PM10 and NO2 among Black women. Four CpG sites identified in the EWAS of PM2.5 among Black participants were also present in significant DMRs identified in association with PM2.5 (cg08097847, cg13538431, cg06738602, cg10011083) and one was present in a DMR associated with NO2 among Black participants (cg23717809).
Using the EWAS results for Black women, IPA pathway analysis of the top 100 CpGs associated with each exposure suggested enrichment of 17 pathways for NO2, 1 for PM2.5, and none for PM10 (Supplemental Table S5). Using the EWAS results for NHW women, IPA pathway analysis of the top 100 CpGs associated with each exposure suggested enrichment of 5 pathways for NO2, 3 for PM2.5, and 17 for PM10 (Supplemental Table S5). Five of the pathways identified were found in more than one of the exposure and race/ethnicity-specific analyses. Among the 38 distinct pathways identified in the six IPA analyses, the most common categories included intracellular and second messenger signaling (n=6) and cardiovascular signaling (n=5).
4. Discussion
This study presents novel air pollution associations with DNA methylation and recently developed measures of epigenetic aging among Black women. We found evidence of heterogeneity by race/ethnicity in air pollution exposure associations with epigenetic aging measures. Among Black participants, we found positive associations between PM10 and GrimAgeAccel as well as NO2 and GrimAgeAccel and DunedinPACE. In contrast, in NHW women we observed little evidence of association between any pollutant and epigenetic aging outcomes. In EWAS analysis, there was little evidence of associations among NHW women, but a number of individual CpGs and DMRs were associated with the three different exposures among Black women.
Previous studies of PM2.5, PM10, and NO2 and epigenetic aging have produced mixed findings. Prior research conducted in the Sister Study among NHW women, who were also included in the present study, found no overall association of PM10 or PM2.5 with acceleration based on the Hannum, Horvath, or PhenoAge clocks and an inverse association between NO2 and acceleration using the Hannum clock (White et al., 2019). A study of Black and White participants of the Health and Retirement Study (n=2960) yielded no associations between NO2 or PM2.5 with GrimAgeAccel and DunedinPACE’s precedessor, DunedinPoAm, in the overall sample but found a statistically significant interaction between race and PM2.5 with DunedinPoAm suggesting greater vulnerability to PM2.5 among Black participants (Yannatos et al., 2023). While less directly comparable to the present study, results from the Cooperative Health Research in the Region of Augsburg (KORA, n=1777) and the Normative Aging Study (NAS, n=589), a cohort of elderly males, both reported positive associations between PM2.5 and accelerated age on the Horvath clock (Nwanaji-Enwerem et al., 2016; Ward-Caviness et al., 2016). In the KORA study, a subgroup analysis among females (n=855) found that NOx, but not PM10, were associated with accelerated age using the Horvath clock.
The different patterns we observed between pollutants and the epigenetic aging measures among Black and NHW participants could relate to a variety of race-related environmental and social factors that increase Black individuals’ susceptibility to harmful exposures, such as lower health care quality or heightened psychosocial stress (Bailey et al., 2017). For example, several studies have found stronger associations between air pollution and adverse health outcomes among those experiencing greater psychosocial stress (Ailshire et al., 2017; Astell-Burt et al., 2013; Padula et al., 2020). We also observed higher median air pollution concentrations in Black than NHW women, which could account for variation observed with quartile 4 of exposures by race/ethnicity. Such differences in exposure levels aligns with previous research that observed disparities in residential pollution levels by neighborhood racial composition (Jbaily et al., 2022; Lane et al., 2022).
For PM10, the different associations with epigenetic aging measures by race/ethnicity may also reflect differential exposure sources and thus chemical composition of PM10. Relative to NHW participants in our sample, Black participants were more likely to reside in an urban setting and live in the South, and there are documented differences in air pollution profiles by geographic region and urbanicity. For example, Kundu and Stone (2014) showed variations between PM2.5 sources and components between urban and rural areas in Iowa (Kundu and Stone, 2014), while White et al. (2019) found variation in the association between PM2.5 and epigenetic aging by geographic region and by PM2.5 component profiles in the Sister Study (White et al., 2019), which could apply similarly to PM10. Another publication in NAS similarly found different associations with age acceleration by specific PM components (Wang et al., 2020). While of interest, we did not have sufficient sample size, particularly among Black participants, to explore possible heterogeneity by region or PM component clusters in the present study.
Among the Black subgroup, we observed a signal for faster aging based on GrimAgeAccel associated with PM10, but negligible evidence for PM2.5, though PM2.5 is included in PM10 and considered a greater threat due to its ability to penetrate farther into the lungs (Ferrari et al., 2019). This may indicate that the harm of PM10 is driven by PMcoarse (PM2.5–10). Studies in Boston and Los Angeles revealed differences of the elemental composition and sources of PM2.5 and PMcoarse in these urban areas (Masri et al., 2015; Oroumiyeh et al., 2022), while another study in Colorado found that PMcoarse comprises approximately 50% of PM10 in urban residential and rural areas and up to 70% of PM10 in urban roadside areas (Clements et al., 2016). These findings support the plausibility that PM10 could have different effects than PM2.5. These studies also support that PM10 may be a proxy for traffic-related pollution, which would align with the attenuated association of NO2 and GrimAgeAccel in the sensitivity analysis after including all pollutants in the same model.
Inconsistent findings across the aging measures may reflect differences in how the measures were designed. DunedinPACE estimates the pace of aging and was developed using indicators of aging among a single-year birth cohort from Dunedin, New Zealand (Belsky et al., 2022). PhenoAge was designed to predict phenotypic age based on clinical markers of aging-related outcomes, including physical functioning and mortality (Levine et al., 2018). GrimAge estimates age based on predicted healthy lifespan and incorporates methylation-based estimators of circulating protein concentrations and cigarette smoking pack-years (Lu et al., 2019). Low to moderate correlations across the different epigenetic aging measures have been shown previously, suggesting they capture distinct features of aging (Belsky et al., 2022; Kresovich et al., 2022). The inclusion of CpG sites specifically related to smoking may make GrimAge more sensitive to air pollution exposure than the other measures given shared constituents and pathologic pathways between air pollution and cigarette smoking (Forman and Finch, 2018).
The exact biochemical mechanisms linking air pollution to DNA methylation and specific DNA methylation patterns associated with pollutants remains uncertain. Nonetheless, the biological connection between air pollution and DNA methylation is supported by several EWAS and global methylation studies that have found that medium and long-term exposures to PM and NO2, among other pollutants, are associated with methylation changes (Chi et al., 2022; Poursafa et al., 2022; Wu et al., 2021). Randomized crossover trials and animal studies of short-term exposure to pollutants further support a causal relationship between exposure to pollutants and methylation alterations (Chen et al., 2016; Ding et al., 2016; Du et al., 2022).
To our knowledge, none of the 20 CpG sites identified in our EWAS have been reported in previous research on PM2.5, PM10, or NO2 and methylation. However, several significant sites from the PM2.5 analysis among the Black subgroup map near genes (ZCCHC24, MARCH10, TBC1D16, SETBP1, GALC, VOPP1) that have been found to have other CpG sites differentially methylated in association with PM2.5 and NO2 in studies of the KORA, NAS, and LifeLines cohorts (de F.C.Lichtenfels et al., 2018; Panni et al., 2016; Wang et al., 2022). Several of the identified CpG sites have been previously associated with aging: an EWAS of epigenetic changes associated with age from birth through adolescence reported both of the CpG sites associated with NO2 in the Black and NHW subgroups (cg24537688, cg24269657) and 6 of the 18 CpG sites associated with PM2.5 in the Black subgroup (cg06738602, cg04902542, cg05411829, cg10011083, cg18580650, cg11130461) (Mulder et al., 2021). Additionally, one CpG site associated with PM2.5 in the Black subgroup (cg06738602) is included in the Horvath clock (Horvath, 2013). Many of the pathways identified in IPA analysis align with previously reported systems implicated in human response to environmental pollutants, such as cellular immune response and intercellular communication (Peters et al., 2021). Several pathways also relate to aging-related outcomes, such as cardiovascular disease and cancer (Jaul and Barron, 2017), and may point to the molecular underpinnings linking air pollution to these diseases. However, findings from the EWAS, DMR, and IPA analyses for PM2.5 among Black participants should be interpreted with caution considering inflation detected in this analysis.
As mentioned, one limitation of the present study is that we did not have sufficient sample size to assess possible heterogeneity by factors that may clarify the differences we observed between race/ethnicity groups, including region or PM component profiles. Another limitation is the cross-sectional nature of the analyses, which prevents us from exploring trends of aging-related DNA methylation patterns over time. Additionally, though the Sister Study is a US-wide sample, generalizability of findings may be limited by the high level of educational attainment and the requirement that all participants have a biological sister with breast cancer. Finally, all three primary epigenetic aging measures were developed in predominantly White cohorts. However, PhenoAge, GrimAge, and DunedinPACE’s predecessor, DunedinPoAm have been used and demonstrated validity in samples of Black participants (Belsky et al., 2022; Levine et al., 2018; Lu et al., 2019).
On the other hand, ours is one of the largest studies to date of air pollution and methylation-based age and is among the first to include Black women, who are often underrepresented in epigenetics research (Breeze et al., 2022). Additionally, the pollutant exposure estimates are highly specific to each participant’s baseline residence and based on sophisticated spatiotemporal and universal kriging models. We also use multiple measures of methylation-based biological aging, including the more recently developed GrimAge clock and DunedinPACE metric that may capture effects of aging better than previous estimates based on measures of chronological age. Finally, comprehensive data collection at Sister Study baseline allowed for a high degree of confounder control.
5. Conclusions
Using recently developed DNA methylation-based measures of aging, this study presents evidence of novel associations between ambient air pollution and DNA methylation. We observed that higher PM10 and NO2 levels were associated with higher epigenetic aging among Black women, but not among NHW women. We also identified 19 differentially methylated CpG sites associated with pollutants among Black participants and one site among NHW participants. These findings underscore the importance of ongoing environmental justice efforts to reduce harmful exposures in Black communities.
Supplementary Material
Funding sources
This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (grant number Z01-ES044005, Z1A-ES103332).
Abbreviations:
- 450K
Illumina Infunium HumanMethylation450 BeadChip
- ADI
area deprivation index
- BMI
body mass index
- Chr
chromosome
- CI
confidence interval
- CpG
Cytosine-phosphate-Guanine
- DMR
differentially methylated region
- DNA
deoxyribonucleic acid
- EEAA
extrinsic epigenetic age acceleration
- EPIC
Illumina Infinium MethylationEPIC V1 BeadChip
- EWAS
epigenome-wide association study
- FDR
false discovery rate
- IEAA
intrinsic epigenetic age acceleration
- IPA
Ingenuity Pathway Analysis
- IQR
interquartile range
- KORA
Cooperative Health Research in the Region Augsburg study
- NAS
Normative Aging Study
- NHW
non-Hispanic White
- NO2
nitrogen dioxide
- P-het
p-for-heterogeneity
- PM10
particulate matter <10 μm in diameter
- PM2.5
particulate matter <2.5 μm in diameter
- PMcoarse
particulate matter >2.5 and <10 μm in diameter
- Ppb
parts per billion
- Q1–4
quartile 1–4
- QC
quality control
- SBEAA
skin & blood epigenetic age acceleration
- US
United States
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envint.2023.108270.
Data availability
All data necessary to reproduce the current analysis are publicly available following procedures described on the Sister Study website (https://sisterstudy.niehs.nih.gov/English/data-requests.htm).
References
- Ailshire J, Karraker A, Clarke P, 2017. Neighborhood social stressors, fine particulate matter air pollution, and cognitive function among older U.S. adults. Soc. Sci. Med 172, 56–63. 10.1016/j.socscimed.2016.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astell-Burt T, Maynard MJ, Lenguerrand E, Whitrow MJ, Molaodi OR, Harding S, 2013. Effect of air pollution and racism on ethnic differences in respiratory health among adolescents living in an urban environment. Health Place 23, 171–178. 10.1016/j.healthplace.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey ZD, Krieger N, Agénor M, Graves J, Linos N, Bassett MT, 2017. Structural racism and health inequities in the USA: evidence and interventions. Lancet 389 (10077), 1453–1463. 10.1016/S0140-6736(17)30569-X. [DOI] [PubMed] [Google Scholar]
- Barcelona de Mendoza V, Huang Y, Crusto CA, Sun YV, Taylor JY, 2018. Perceived racial discrimination and DNA methylation among african american women in the InterGEN study. Biol. Res. Nurs 20 (2), 145–152. 10.1177/1099800417748759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky DW, Caspi A, Corcoran DL, Sugden K, Poulton R, Arseneault L, Baccarelli A, Chamarti K, Gao X, Hannon E, Harrington HL, Houts R, Kothari M, Kwon D, Mill J, Schwartz J, Vokonas P, Wang C, Williams BS, Moffitt TE, 2022. DunedinPACE, a DNA methylation biomarker of the pace of aging. Elife 11. 10.7554/eLife.73420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze CE, Beck S, Berndt SI, Franceschini N, 2022. The missing diversity in human epigenomic studies. Nat. Genet 54 (6), 737–739. 10.1038/s41588-022-01081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caraballo C, Herrin J, Mahajan S, Massey D, Lu Y, Ndumele CD, Drye EE, Krumholz HM, 2022. Temporal Trends in Racial and Ethnic Disparities in Multimorbidity Prevalence in the United States, 1999–2018. Am. J. Med 135 (9), 1083–1092.e1014. 10.1016/j.amjmed.2022.04.010. [DOI] [PubMed] [Google Scholar]
- Chen R, Meng X, Zhao A, Wang C, Yang C, Li H, Cai J, Zhao Z, Kan H, 2016. DNA hypomethylation and its mediation in the effects of fine particulate air pollution on cardiovascular biomarkers: a randomized crossover trial. Environ. Int 94, 614–619. 10.1016/j.envint.2016.06.026. [DOI] [PubMed] [Google Scholar]
- Chi GC, Liu Y, MacDonald JW, Reynolds LM, Enquobahrie DA, Fitzpatrick AL, Kerr KF, Budoff MJ, Lee SI, Siscovick D, Kaufman JD, 2022. Epigenome-wide analysis of long-term air pollution exposure and DNA methylation in monocytes: results from the multi-ethnic study of atherosclerosis. Epigenetics 17 (3), 297–313. 10.1080/15592294.2021.1900028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements N, Hannigan MP, Miller SL, Peel JL, Milford JB, 2016. Comparisons of urban and rural PM10 – 2.5 and PM2.5 mass concentrations and semi-volatile fractions in northeastern Colorado. Atmos. Chem. Phys 16 (11), 7469–7484. 10.5194/acp-16-7469-2016. [DOI] [Google Scholar]
- de F.C.Lichtenfels AJ, van der Plaat DA, de Jong K, van Diemen CC, Postma DS, Nedeljkovic I, van Duijn CM, Amin N, la Bastide-van Gemert S, de Vries M, Ward-Caviness CK, Wolf K, Waldenberger M, Peters A, Stolk RP, Brunekreef B, Boezen HM, Vonk JM, 2018. Long-term air pollution exposure, genome-wide DNA methylation and lung function in the lifelines cohort study. Environ. Health Perspect 126 (2), 027004 10.1289/ehp2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R, Jin Y, Liu X, Zhu Z, Zhang Y, Wang T, Xu Y, 2016. Characteristics of DNA methylation changes induced by traffic-related air pollution. Mutat. Res./gene. Toxicol. Environ. Mutagen 796, 46–53. 10.1016/j.mrgentox.2015.12.002. [DOI] [PubMed] [Google Scholar]
- Du X, Jiang Y, Li H, Zhang Q, Zhu X, Zhou L, Wang W, Zhang Y, Liu C, Niu Y, Chu C, Cai J, Chen R, Kan H, 2022. Traffic-related air pollution and genome-wide DNA methylation: a randomized, crossover trial. Sci. Total Environ 850, 157968 10.1016/j.scitotenv.2022.157968. [DOI] [PubMed] [Google Scholar]
- Ferrari L, Carugno M, Bollati V, 2019. Particulate matter exposure shapes DNA methylation through the lifespan. Clin. Epigenetics 11 (1), 129. 10.1186/s13148-019-0726-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrucci L, Gonzalez-Freire M, Fabbri E, Simonsick E, Tanaka T, Moore Z, Salimi S, Sierra F, de Cabo R, 2020. Measuring biological aging in humans: a quest. Aging Cell 19 (2), e13080. 10.1111/acel.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman HJ, Finch CE, 2018. A critical review of assays for hazardous components of air pollution. Free Radic. Biol. Med 117, 202–217. 10.1016/j.freeradbiomed.2018.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geronimus AT, Hicken MT, Pearson JA, Seashols SJ, Brown KL, Cruz TD, 2010. Do US Black women experience stress-related accelerated biological aging? Hum. Nat 21 (1), 19–38. 10.1007/s12110-010-9078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y, Deconde R, Chen M, Rajapakse I, Friend S, Ideker T, Zhang K, 2013. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 49 (2), 359–367. 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, 2013. DNA methylation age of human tissues and cell types. Genome Biol 14 (10), 3156. 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Oshima J, Martin GM, Lu AT, Quach A, Cohen H, Felton S, Matsuyama M, Lowe D, Kabacik S, Wilson JG, Reiner AP, Maierhofer A, Flunkert J, Aviv A, Hou L, Baccarelli AA, Li Y, Stewart JD, Raj K, 2018. Epigenetic clock for skin and blood cells applied to Hutchinson Gilford Progeria Syndrome and ex vivo studies. Aging (Albany NY) 10 (7), 1758–1775. 10.18632/aging.101508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT, 2012. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinf 13, 86. 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaul E, Barron J, 2017. Age-related diseases and clinical and public health implications for the 85 years old and over population. Front. Public Health 5, 335. 10.3389/fpubh.2017.00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jbaily A, Zhou X, Liu J, Lee T-H, Kamareddine L, Verguet S, Dominici F, 2022. Air pollution exposure disparities across US population and income groups. Nature 601 (7892), 228–233. 10.1038/s41586-021-04190-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josey KP, Delaney SW, Wu X, Nethery RC, DeSouza P, Braun D, Dominici F, 2023. Air pollution and mortality at the intersection of race and social class. N. Engl. J. Med 388 (15), 1396–1404. 10.1056/NEJMsa2300523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind AJ, Jencks S, Brock J, Yu M, Bartels C, Ehlenbach W, Greenberg C, Smith M, 2014. Neighborhood socioeconomic disadvantage and 30-day rehospitalization: a retrospective cohort study. Ann. Intern. Med 161 (11), 765–774. 10.7326/m13-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirwa K, Szpiro AA, Sheppard L, Sampson PD, Wang M, Keller JP, Young MT, Kim SY, Larson TV, Kaufman JD, 2021. Fine-scale air pollution models for epidemiologic research: insights from approaches developed in the multi-ethnic study of atherosclerosis and air pollution (MESA Air). Curr. Environ. Health Rep 8 (2), 113–126. 10.1007/s40572-021-00310-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresovich JK, Park YM, Keller JA, Sandler DP, Taylor JA, 2022. Healthy eating patterns and epigenetic measures of biological age. Am. J. Clin. Nutr 115 (1), 171–179. 10.1093/ajcn/nqab307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresovich JK, Sandler DP, Taylor JA, 2023. Methylation-based biological age and hypertension prevalence and incidence. Hypertension 80 (6), 1213–1222. 10.1161/hypertensionaha.122.20796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu S, Stone EA, 2014. Composition and sources of fine particulate matter across urban and rural sites in the Midwestern United States. Environ. Sci. Process Impacts 16 (6), 1360–1370. 10.1039/c3em00719g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane HM, Morello-Frosch R, Marshall JD, Apte JS, 2022. Historical redlining is associated with present-day air pollution disparities in U.S Cities. Environ. Sci. Technol. Lett 9 (4), 345–350. 10.1021/acs.estlett.1c01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Storey JD, 2007. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet 3 (9), 1724–1735. 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, Hou L, Baccarelli AA, Stewart JD, Li Y, Whitsel EA, Wilson JG, Reiner AP, Aviv A, Lohman K, Liu Y, Ferrucci L, Horvath S, 2018. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY) 10 (4), 573–591. 10.18632/aging.101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu AT, Quach A, Wilson JG, Reiner AP, Aviv A, Raj K, Hou L, Baccarelli AA, Li Y, Stewart JD, Whitsel EA, Assimes TL, Ferrucci L, Horvath S, 2019. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY) 11 (2), 303–327. 10.18632/aging.101684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S, Kang CM, Koutrakis P, 2015. Composition and sources of fine and coarse particles collected during 2002–2010 in Boston, MA. J. Air Waste Manag. Assoc 65 (3), 287–297. 10.1080/10962247.2014.982307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder RH, Neumann A, Cecil CAM, Walton E, Houtepen LC, Simpkin AJ, Rijlaarsdam J, Heijmans BT, Gaunt TR, Felix JF, Jaddoe VWV, Bakermans-Kranenburg MJ, Tiemeier H, Relton CL, van IMH, Suderman M, 2021. Epigenome-wide change and variation in DNA methylation in childhood: trajectories from birth to late adolescence. Hum. Mol. Genet 30 (1), 119–134. 10.1093/hmg/ddaa280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwanaji-Enwerem JC, Colicino E, Trevisi L, Kloog I, Just AC, Shen J, Brennan K, Dereix A, Hou L, Vokonas P, Schwartz J, Baccarelli AA, 2016. Long-term ambient particle exposures and blood DNA methylation age: findings from the VA normative aging study. Environ. Epigenet 2 (2) 10.1093/eep/dvw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwanaji-Enwerem JC, Dai L, Colicino E, Oulhote Y, Di Q, Kloog I, Just AC, Hou L, Vokonas P, Baccarelli AA, Weisskopf MG, Schwartz JD, 2017. Associations between long-term exposure to PM(2.5) component species and blood DNA methylation age in the elderly: The VA normative aging study. Environ. Int 102, 57–65. 10.1016/j.envint.2016.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien KM, Lawrence KG, Keil AP, 2022. The case for case-cohort: an applied epidemiologist’s guide to reframing case-cohort studies to improve usability and flexibility. Epidemiology 33 (3), 354–361. 10.1097/ede.0000000000001469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oroumiyeh F, Jerrett M, Del Rosario I, Lipsitt J, Liu J, Paulson SE, Ritz B, Schauer JJ, Shafer MM, Shen J, Weichenthal S, Banerjee S, Zhu Y, 2022. Elemental composition of fine and coarse particles across the greater Los Angeles area: Spatial variation and contributing sources. Environ. Pollut 292, 118356 10.1016/j.envpol.2021.118356. [DOI] [PubMed] [Google Scholar]
- Padula AM, Monk C, Brennan PA, Borders A, Barrett ES, McEvoy CT, Foss S, Desai P, Alshawabkeh A, Wurth R, Salafia C, Fichorova R, Varshavsky J, Kress A, Woodruff TJ, Morello-Frosch R, on behalf of program collaborators for Environmental influences on Child Health Outcomes, 2020. A review of maternal prenatal exposures to environmental chemicals and psychosocial stressors—implications for research on perinatal outcomes in the ECHO program. J. Perinatol 40 (1), 10–24. 10.1038/s41372-019-0510-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panni T, Mehta AJ, Schwartz JD, Baccarelli AA, Just AC, Wolf K, Wahl S, Cyrys J, Kunze S, Strauch K, Waldenberger M, Peters A, 2016. Genome-wide analysis of DNA methylation and fine particulate matter air pollution in three study populations: KORA F3, KORA F4, and the normative aging study. Environ. Health Perspect 124 (7), 983–990. 10.1289/ehp.1509966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, Nawrot TS, Baccarelli AA, 2021. Hallmarks of environmental insults. Cell 184 (6), 1455–1468. 10.1016/j.cell.2021.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poursafa P, Kamali Z, Fraszczyk E, Boezen HM, Vaez A, Snieder H, 2022. DNA methylation: a potential mediator between air pollution and metabolic syndrome. Clin. Epigenetics 14 (1), 82. 10.1186/s13148-022-01301-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson PD, Richards M, Szpiro AA, Bergen S, Sheppard L, Larson TV, Kaufman JD, 2013. A regionalized national universal kriging model using Partial Least Squares regression for estimating annual PM(2.5) concentrations in epidemiology. Atmos. Environ 75, 383–392. 10.1016/j.atmosenv.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler DP, Hodgson ME, Deming-Halverson SL, Juras PS, D’Aloisio AA, Suarez LM, Kleeberger CA, Shore DL, DeRoo LA, Taylor JA, Weinberg CR, 2017. The Sister Study Cohort: Baseline Methods and Participant Characteristics. Environ. Health Perspect 125 (12), 127003 10.1289/ehp1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JA, Raisky J, Ratliff SM, Liu J, Kardia SLR, Turner ST, Mosley TH, Zhao W, 2019. Intrinsic and extrinsic epigenetic age acceleration are associated with hypertensive target organ damage in older African Americans. BMC Med. Genom 12 (1), 141. 10.1186/s12920-019-0585-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessum CW, Paolella DA, Chambliss SE, Apte JS, Hill JD, Marshall JD, 2021. PM2.5 polluters disproportionately and systemically affect people of color in the United States. Sci. Adv 7 (18), eabf4491 10.1126/sciadv.abf4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Koutrakis P, Gao X, Baccarelli A, Schwartz J, 2020. Associations of annual ambient PM(2.5) components with DNAm PhenoAge acceleration in elderly men: the Normative Aging Study. Environ. Pollut 258, 113690 10.1016/j.envpol.2019.113690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Cardenas A, Hutchinson JN, Just A, Heiss J, Hou L, Zheng Y, Coull BA, Kosheleva A, Koutrakis P, Baccarelli AA, Schwartz JD, 2022. Short- and intermediate-term exposure to ambient fine particulate elements and leukocyte epigenome-wide DNA methylation in older men: the Normative Aging Study. Environ. Int 158, 106955 10.1016/j.envint.2021.106955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward-Caviness CK, Nwanaji-Enwerem JC, Wolf K, Wahl S, Colicino E, Trevisi L, Kloog I, Just AC, Vokonas P, Cyrys J, Gieger C, Schwartz J, Baccarelli AA, Schneider A, Peters A, 2016. Long-term exposure to air pollution is associated with biological aging. Oncotarget 7 (46), 74510–74525. 10.18632/oncotarget.12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AJ, Kresovich JK, Keller JP, Xu Z, Kaufman JD, Weinberg CR, Taylor JA, Sandler DP, 2019. Air pollution, particulate matter composition and methylation-based biologic age. Environ. Int 132, 105071 10.1016/j.envint.2019.105071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Ambient (Outdoor) Air Pollution, 2022. https://www.who.int/news-room/fact-sheets/detail/ambient-(outdoor)-air-quality-and-health (Retrieved May 26).
- Wu Y, Qie R, Cheng M, Zeng Y, Huang S, Guo C, Zhou Q, Li Q, Tian G, Han M, Zhang Y, Wu X, Li Y, Zhao Y, Yang X, Feng Y, Liu D, Qin P, Hu D, Zhang M, 2021. Air pollution and DNA methylation in adults: a systematic review and meta-analysis of observational studies. Environ. Pollut 284, 117152 10.1016/j.envpol.2021.117152. [DOI] [PubMed] [Google Scholar]
- Xu Z, Niu L, Li L, Taylor JA, 2016. ENmix: a novel background correction method for Illumina HumanMethylation450 BeadChip. Nucl. Acids Res 44 (3), e20–e. 10.1093/nar/gkv907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Sandler DP, Taylor JA, 2020b. Blood DNA methylation and breast cancer: a prospective case-cohort analysis in the sister study. J. Natl Cancer Inst 112 (1), 87–94. 10.1093/jnci/djz065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Xie C, Taylor JA, Niu L, 2020a. ipDMR: identification of differentially methylated regions with interval P-values. Bioinformatics 37 (5), 711–713. 10.1093/bioinformatics/btaa732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yannatos I, Stites S, Brown RT, McMillan CT, 2023. Contributions of neighborhood social environment and air pollution exposure to Black-White disparities in epigenetic aging. PLoS One 18 (7), e0287112. 10.1371/journal.pone.0287112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younan D, Wang X, Gruenewald T, Gatz M, Serre ML, Vizuete W, Braskie MN, Woods NF, Kahe K, Garcia L, Lurmann F, Manson JE, Chui HC, Wallace RB, Espeland MA, Chen J-C, 2021. Racial/ethnic disparities in Alzheimer’s disease risk: role of exposure to ambient fine particles. J. Gerontol.: Ser. A 77 (5), 977–985. 10.1093/gerona/glab231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data necessary to reproduce the current analysis are publicly available following procedures described on the Sister Study website (https://sisterstudy.niehs.nih.gov/English/data-requests.htm).


