SUMMARY
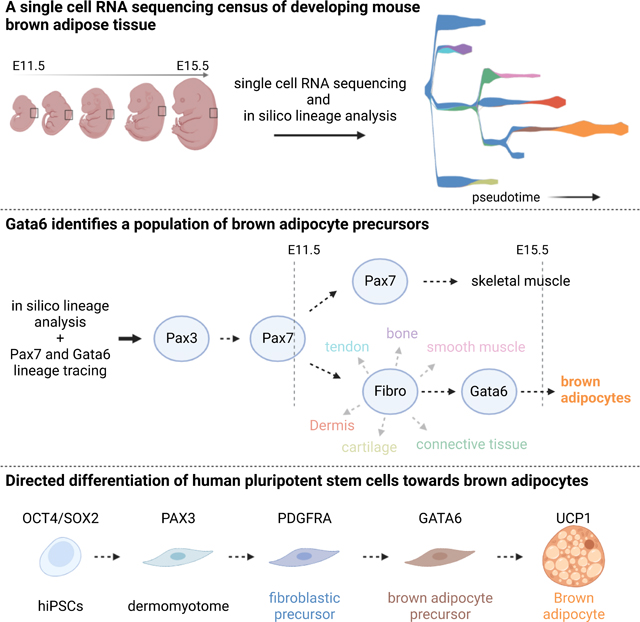
Brown adipocytes (BAs) represent a specialized cell type able to uncouple nutrient catabolism from ATP generation to dissipate energy as heat. In humans, the brown fat tissue is composed of discrete depots found throughout the neck and trunk region. BAs originate from a precursor common to skeletal muscle but their developmental trajectory remains poorly understood. Here, we used single cell RNA sequencing to characterize the development of interscapular brown fat in mouse. Our analysis identified a transient stage of BAs differentiation characterized by expression of the transcription factor GATA6. We show that recapitulating the sequence of signaling cues identified in mouse can lead to efficient differentiation of BAs in vitro from human pluripotent stem cells. These precursors can in turn be efficiently converted into functional BAs which can respond to signals mimicking adrenergic stimuli by increasing their metabolism, resulting in heat production.
ETOC BLURB
Brown adipose tissue controls thermogenesis and regulates metabolism. Rao et al. use single cell RNA sequencing to analyze its development in mouse embryos and recapitulate the developmental cues experienced by brown adipocytes in vitro to engineer a protocol to differentiate human pluripotent stem cells to functional brown adipocytes.
Graphical Abstract
INTRODUCTION
In humans, brown adipose tissue (BAT) is present most abundantly in the interscapular region at birth and in the supraclavicular region in adulthood 1–5. Brown adipocytes (BAs) derive from the paraxial mesoderm, an embryonic tissue that flanks the neural tube. In mammals, the paraxial mesoderm first forms through gastrulation in the primitive streak and generates the presomitic mesoderm (PSM) in the posterior region of the embryo. As it matures, the PSM forms epithelial somites which become organized in two parallel arrays of segments providing the blueprint for the future metameric organization of vertebrae and other trunk derivatives. Soon after their formation, the ventral part of somites forms the mesenchymal sclerotome which yields the axial skeleton. The dorsal part of the somite remains epithelial, forming the dermomyotome. This compartment is composed of cells expressing the transcription factor Pax3, and it generates the dermis of the back and skeletal muscles of the body.
Lineage tracing experiments have shown that murine interscapular brown fat arises from Pax3, Meox1 and Myf5-expressing dermomyotomal cells in the embryo 6–9. During development, Pax3-positive dermomyotome cells give rise to multipotential progenitor cells expressing Engrailed1 and Pax7. These multipotent cells develop into dermal cells, skeletal muscle and BAs 10,11. Lineage tracing of Pax7 multipotent progenitors in mouse shows that between E (embryonic day) 9.5 and E11.5 these cells can give rise to skeletal muscle, dermis and some BAs 9,11. The contribution of Pax7 progenitors becomes restricted to the skeletal muscle lineage at E11.5–12.5. Overall, the hierarchy of somitic precursors giving rise to this tissue is still poorly characterized 12.
Our knowledge of human BAT development is even more limited. Current in vitro models to study human BAT are mostly based on BAs differentiated in vitro from primary cell lines and stromal vascular cells 13. Several protocols have also been established to differentiate human pluripotent stem cells (hPSCs) into BAs using either transgenic overexpression of transcription factors 14 or treatment with growth factors and small molecules 15. Other reported methods rely on serum-based spontaneous differentiation or treatment of hPSCs with a cytokine cocktail 16–18. Recently, protocols allowing the generation of BAs from hPSCs by recapitulating developmental cues have also been reported 19,20. However, to date, a well characterized roadmap of brown fat lineage development is still missing. Therefore, benchmarking these protocols against the normal trajectory of BAs differentiation in vivo has not been possible.
In this study, we used single cell RNA sequencing (scRNAseq) to profile mouse somite-derived dorsal tissues (including brown fat, dermis and skeletal muscle) to investigate brown fat development. We identify a transient stage in BA development characterized by the selective expression of the transcription factor Gata6. We previously developed an efficient protocol recapitulating the development of somitic mesoderm and skeletal muscle progenitors from human and mouse PSCs in vitro 21–23. Building on this protocol and using developmental cues identified in our scRNAseq data, we now report an efficient strategy to generate GATA6-positive BA precursors and functional mature BAs in vitro from human induced pluripotent stem cells (hiPSCs).
RESULTS
Single-cell transcriptomic analysis of the dorsal trunk of mouse embryos captures the development of somitic lineages
In mouse, interscapular BAT develops in between the dermis, trapezius muscle, pectoralis muscle, and deep dorsal muscle bundles. We isolated dorsal tissues at the forelimb level at E11.5, E12.5, E13.5, E14.5, and E15.5 (Figure S1A) to perform single cell RNA sequencing (scRNAseq). For stages E11.5-E14.5, the isolated dorsal tissues include epidermis, dermis, mesenchyme, interscapular brown fat and skeletal muscle. For the E15.5 time point, to enrich for BAT precursors, we removed the epidermis and underlying dermis before dissociation. Using the inDrops workflow 24, we generated single-cell transcriptomes from at least 4000 cells per stage, resulting in a final dataset of 28244 cells after quality control filtering. Cells from the different time points largely segregate based on transcriptional similarity rather than with embryonic age (figure S1B-D). We identified 17 clusters representing the somitic and non-somitic cell types that reside in the dorsal trunk region of the murine embryo (Figure S1C). Among somite derivatives, we observed clusters of cells that originate from the dermomyotome, including BAs (Cidea, Ucp1), skeletal muscle (Myog, Ttn, Actn2, Myh3) and dermal fibroblasts (Twist2, Dpt, Crabp1) (Figure S1E-F). In addition, we could annotate clusters of smooth muscle cells (Cnn1, Acta2), muscle connective tissue (Ngfr, Osr1, Osr2), cartilage (Cnmd, Col2a1), endothelial cells (Cdh5, Kdr) and meninges (Foxc1, Cldn11, Aldh1a2) (Figure S1E-F). Among non-somitic cells, we found macrophages (Csf1r, C1qb), mast cells (Srgn, Kit), neutrophils (Ngp, Lccn2), Schwann cells (Sox10, Mpz), and epidermal cells (Krt5, Krt14) (Figure S1E-F). In conclusion, our single-cell transcriptomics analysis captured major dermomyotome derivatives including cells of the BA lineage present in the mouse dorsal trunk during embryonic development.
Characterization of the developmental trajectory of BA precursors
Most of the somite-derived cells, except endothelial and skeletal muscle cells, lie closely together on the UMAP embedding suggesting that they have related transcriptional signatures (Figure S1B-C). This large group of cells forms a single continuous ensemble including clusters corresponding to cartilage, bone, smooth muscle, connective tissue, dermis and BAs. All these cell types arise from fibroblastic precursors thus potentially explaining the similarity of their transcriptomes. We extracted the clusters representing this group of cells and reanalyzed this new dataset in detail. Most of the cells belong to a large central cluster already present at E11.5, which we name Fibroblastic Progenitors (FP) (Figure 1A-B). Cells of this cluster express fibroblasts markers including Pdgfra or Hic1 (Figure 1C). Differentially expressed genes of the FP cluster do not contain characteristic lineage markers (Table S1). Moreover, the FP cluster connects to all the different fates, suggesting that it corresponds to a multipotent progenitor population. The FP cluster is linked to a cluster of prospective BAs, which we termed BA Precursors (BAPre) (Figure 1A). These cells express BA precursor markers such as Ebf2, Cebpa and Pparg (Figure 1C, Figure S2A) 7. The BAPre cluster is connected to a cluster of BAs characterized by expression of Cidea or Ucp1, Figure 1C, Figure S2A). The BAPre cluster is first detected at day 13.5 and its size progressively increases until E15.5, while the BA cluster is first detected at E15.5 (Figure 1D). Pseudotime analysis supports the existence of a developmental trajectory for the BA lineage which starts from FP cells followed by BAPre and finally BAs (Figure 1F).
Figure 1 -. Embryonic development of mouse interscapular BAT.
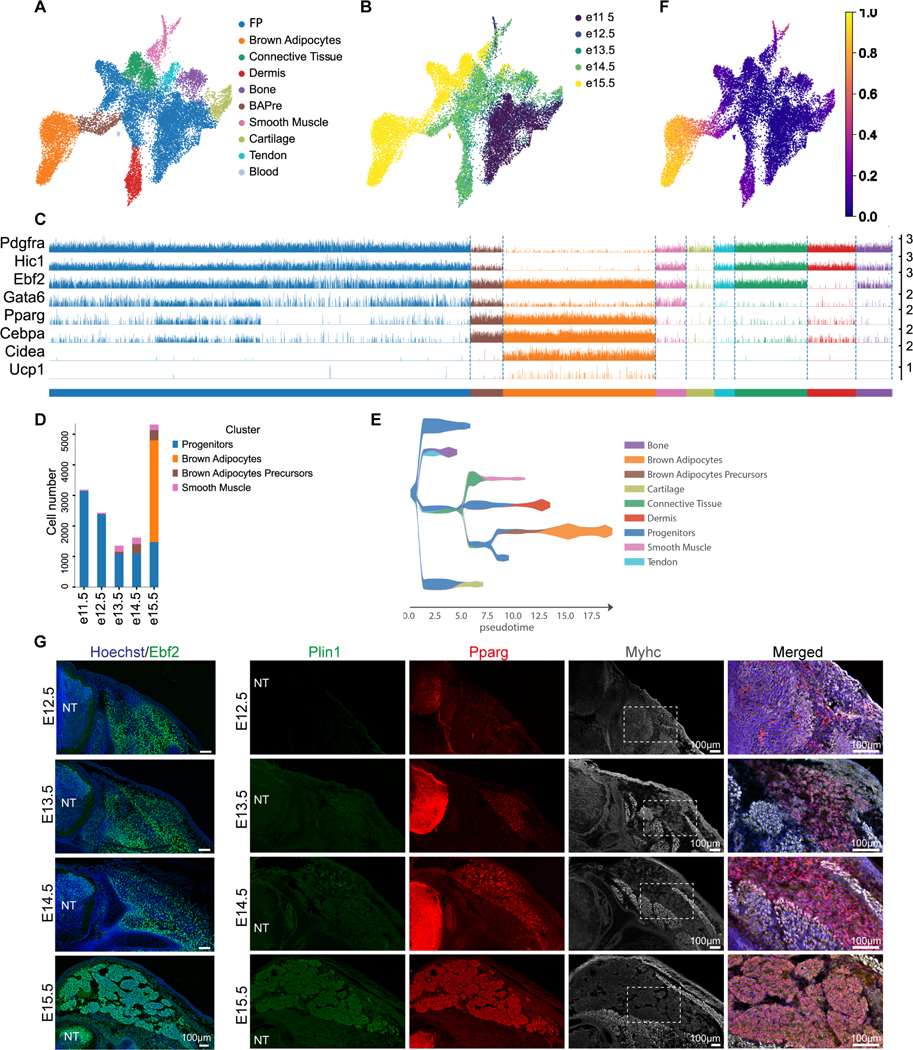
A- B UMAP embedding showing cell clusters (A) and embryonic days (B) from dorsal region of mouse embryos. (BApre=BA precursors FP= Fibroblasts precursors).
C – Tracksplot showing the expression of markers of BA differentiation in clusters shown in A. Bars show expression level in individual cells.
D – Barplot showing cell numbers in indicated clusters at 5 timepoints.
E –Putative developmental trajectories inferred by the STREAM algorithm.
F – UMAP embedding showing diffusion pseudotime.
G – Immunofluorescence analysis with antibodies against Ebf2, Plin1, Pparg and MyHC of the developing interscapular BAT in mouse. Transverse sections of mouse embryos at the forelimb level. NT = Neural tube, n≥3, scale bar=100μm.
We performed an analysis with the STREAM pipeline 25, which confirmed these putative lineage relationships among clusters (Figure 1E). These conclusions are further supported by an analysis using the Waddington OT pipeline which allows to characterize the ancestor/descendant sequence based on cell-to-cell transitions probabilities and gene expression similarities 26 (Figure S2B-C). These analyses support the notion that cartilage, bone, tendon, connective tissue, dermis and BAs all arise from the FP cluster and that the BA cluster derives from the BAPre cluster (Figure 1E-F). Immunofluorescence analysis of E12.5 to E15.5 embryos confirmed the expression of Ebf2 in the presumptive interscapular brown fat region but also in skeletal muscle and precartilage (Figure 1G) 7. Developing adipocyte precursors expressing Pparg probably corresponding to BAPre start to appear at E13.5 and develop in between the dermis and Myosin Heavy Chain (MyHC)-expressing muscle fibers (Figure 1G). Perilipin1 (Plin1) staining shows that a small number of preadipocytes start to accumulate lipid vesicles at E14.5. An increasing number of lipid-containing immature adipocytes appears by E15.5 (Figure 1G).
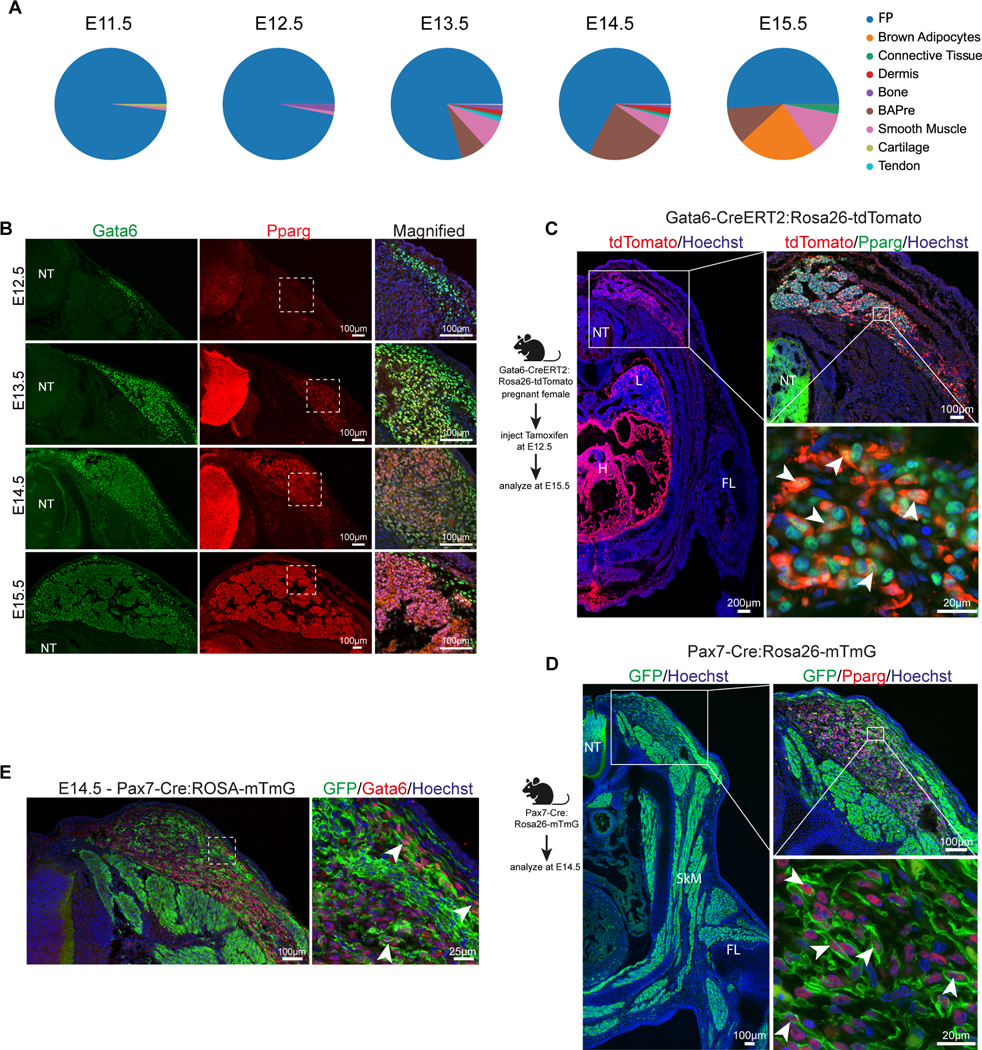
Angueira et al. (2021) recently identified progenitors that give rise to BAs in mouse periaortic (pVAT) brown adipose cells analyzed by scRNAseq at E18 and perinatal stages 27. We reanalyzed their E18 dataset and identified 13 clusters as described in the original study (Figure S2D). We identified a trajectory similar to that observed for the somitic lineages starting with a cluster of Fibroblastic Progenitors (Pdgfra) followed by BAPre (Pparg) and by BAs (Cidea) (Figure S2E-F). Pi16, Cd34 and Ly6a, which were identified as BA progenitor markers in the Angueira report are also differentially expressed FP cluster in our study (Figure S2G). Using a machine learning classifier trained on the Angueira dataset, we confirmed the similarity between progenitors and BAs fates identified in the two studies (Figure S2H). Immunofluorescence analysis at E15.5 confirmed the expression of the adipocyte precursors markers Dpp4, Cd34 and Ly6a at the periphery of the interscapular depot (Figure S2I-J). Their location suggested that they might correspond to precursor cells involved in the generation of mature brown fat. Together, these data suggest that the BAs of the interscapular region derive from a multipotent population of fibroblastic progenitors expressing markers such as Pdgfra which gives rise to an immature population of lineage restricted precursors (BAPre) appearing around E13.5. Around E15.5, this population starts giving rise to BAs identified by specific markers such as Cidea.
Gata6 marks early BAT precursors
To identify specific markers for BA precursors, we examined the genes differentially expressed in the BAPre cluster. We noticed that the transcription factor Gata6 whose expression in BA development has not been characterized, is found in the top 60 differentially expressed genes. In the BAPre cluster differentially expressed gene list, Gata6 ranks before Ebf2 which has been described a selective marker for brown adipogenic precursor cells 7. Gata6 is restricted to the BA and smooth muscle lineages (Figure 1C). Extraction of the Gata6-expressing cells from the mouse dataset shows that these cells mostly belong to these two lineages with the majority of them belonging to the FP cluster (Figure 2A). Gata6-expressing cells are first detected in the FP, then in the BApre and in the BA and smooth muscle clusters. Gata6 expression progressively decreased during the differentiation of BAs (Figure 1C). These observations were confirmed at the protein level by immunofluorescence analysis which indicated that Gata6 is detected at E12.5 in the presumptive brown fat area under the dermis (Figure 2B). Differentiating Pparg-positive immature adipocytes as well as Pparg-negative adipocyte precursors expressed Gata6 at E14.5 (Figure 2B). At E15.5, most Pparg-positive cells of the forming interscapular BAT had downregulated the Gata6 protein. Strong expression of Gata6 was still observed in Pparg-negative cells at the periphery of BAT, possibly corresponding to BA precursors (Figure 2B). Downregulation of both Gata6 mRNA and protein in BAs at E15.5 suggests that Gata6 is transiently expressed during BA differentiation and is downregulated in more mature BAs. Similar expression of Gata6 during BA differentiation was observed in the Angueira dataset (Figure S2F).
Figure 2 -. Gata6 is expressed by developing BA precursors.
A – Proportion of Gata6-positive cells in the clusters.
B –Immunohistochemical analysis of transverse sections of mouse embryos at the forelimb level with Pparg and Gata6 antibodies. n≥4.
C – Lineage tracing experiments using Gata6-CreERT2:Rosa26-tdTomato mice. Transverse section at the forelimb level of E15.5 embryos stained with anti-RFP antibody to detect tdTomato-positive cells (left panel). Magnified image of the transverse section from the interscapular region showing BAT stained with Pparg and RFP (tdTomato) antibody (upper right panel). Higher magnification image of BAs in the interscapular region showing double positive cells for Pparg and tdTomato. Arrowheads mark double positive cells (lower right panel). n≥3.
D – Lineage tracing strategy to label Pax7 progeny during mouse development. Transverse section at the forelimb level of E14.5 embryos stained with anti-GFP antibody (left panel). Transverse section of E14.5 Pax7-Cre:Rosa26-mTmG embryo showing interscapular region stained with antibodies against Pparg and GFP (upper right panel). Representative high magnification image showing double positive cells stained for nuclear Pparg and membranous GFP. Arrowheads mark double positive cells (lower right panel). n≥3.
E – Transverse section of E14.5 Pax7-Cre:Rosa26-mTmG embryo showing interscapular region stained with anti-GFP antibody and anti-Gata6. Inset on right panel shows magnified view. Arrowheads: double positive cells. n≥3.
H: heart, L: lung, NT: Neural tube, SkM: skeletal muscle.
To demonstrate that Gata6 is expressed by precursors giving rise to BAs, we used a Gata6-CreERT2:Rosa26-tdTomato mouse line to label Gata6-expressing cells during embryonic development 28. We induced recombination using tamoxifen injection at E12.5 and analyzed the contribution of labelled cells to BAT at E15.5 (Figure 2C). Immunofluorescence analysis for tdTomato, which labels the descendants of Gata6-expressing cells confirmed that at E12.5, they give rise to heart and lung bud cells as expected (Figure 2C). Among somite-derived cells, tdTomato positive cells were mostly found in the interscapular BAT region and not in other somite derivatives such as skeletal muscle, cartilage or endothelial cells (Figure S3A). No tdTomato-positive cells were detected in skeletal muscle, and we did not observe any overlap with the CD31+ population of endothelial cells in BAT (Figure 2C, Figure S3A). tdTomato-positive cells in the interscapular BAT also stained positive for Pparg at E15.5 confirming that these cells are adipocytes (Figure 2C). We also found rare tdTomato-Myh11 double positive cells next to blood vessels in the interscapular BAT (Figure S3B). This is consistent with previously reported expression of Gata6 by vascular smooth muscle cells 29. The number of such potential smooth muscle cells labelled was lower than expected from the scRNAseq analysis. These cells could correspond to the recently identified BA precursors deriving from the vascular smooth muscle lineage 30. Also, cells expressing both tdTomato and Dpp4 were observed in the periphery of the forming brown fat mass, supporting the expression of Gata6 by brown fat precursors (Figure S3C). In conclusion, our lineage tracing analysis argues that Gata6-expressing cells present at E12.5 in the mouse dorsal trunk mostly belong to the BA lineage.
To further investigate the origin of these Gata6 precursors, we used a Pax7-Cre:Rosa26-mTmG mouse line to analyze the contribution of Pax7 descendants to Gata6-positive brown fat precursors at E14.5 31. As expected, dorsal, ventral and limb skeletal muscle as well as Pparg-expressing prospective BAs precursors were identified as Pax7 descendants (Figure 2D). Membrane GFP also labeled Gata6-positive cells in the brown fat region indicating that these cells derive from an earlier Pax7-expressing population (Figure 2E). Taken together, our experiments indicate that Pax7-positive somitic progenitors give rise to a Gata6-positive precursor population that differentiates into BAs.
To further examine whether Gata6 expression is conserved in human brown adipogenesis, we performed immunofluorescence analysis on 98- and 135-day-old (estimated post-conceptual age) human fetal tissue from interscapular and scapular regions. We detected expression of EBF2 and GATA6 in cells adjacent to skeletal muscle tissue (detected by MYHC antibody) under the dorsal dermis and dispersed between deeper dorsal muscle bundles at 98 days (Figure 3A). As in mouse, EBF2 was expressed in a broader territory in the dermis and deeper muscle areas. We also observed a few lipid-filled UCP1- and PPARG-positive adipocytes in the deeper muscle area (Figure 3B-C). In these regions, we detected PPARG and GATA6 double-positive cells, supporting the existence of GATA6-positive BA progenitors in humans (Figure 3B-C). More mature BAs identified by the presence of lipid droplets, expression of PLIN1 and high expression of PPARG did not express GATA6 suggesting that GATA6 expression is also restricted to an early stage of BA development in humans.
Figure 3 -. Expression of GATA6 in developing human BAT.
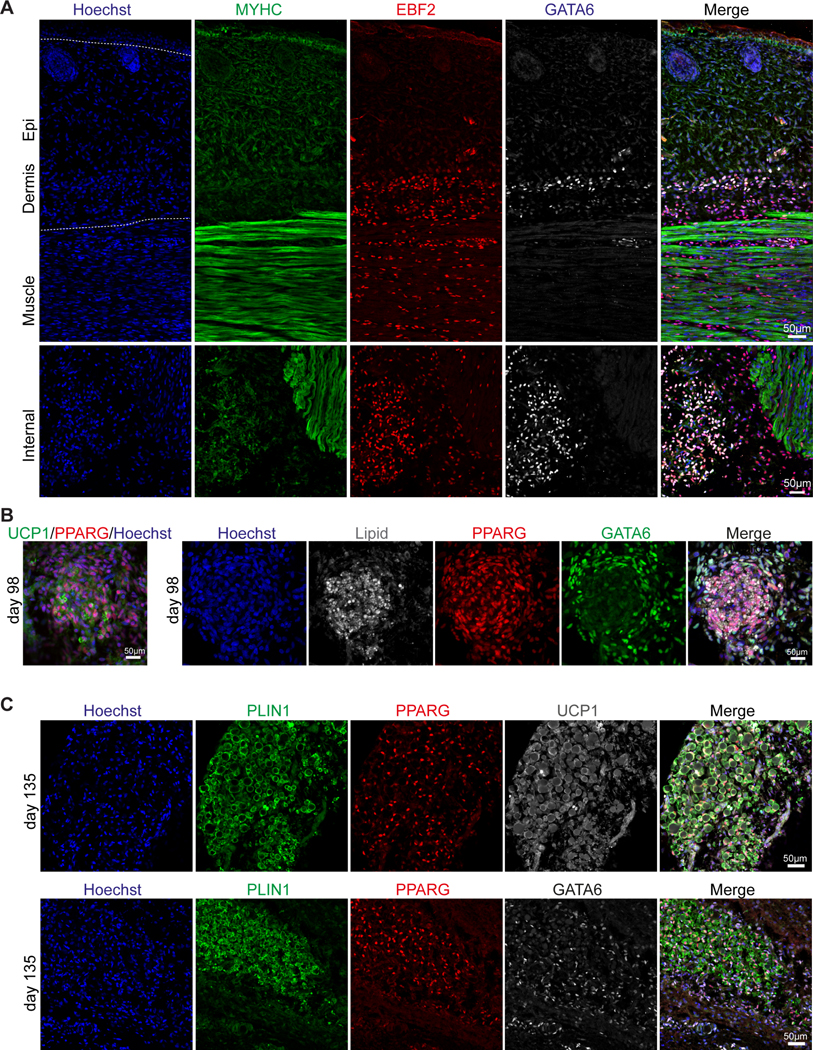
A – Immunofluorescence analysis with anti-MYHC, EBF2 and GATA6 antibodies of human fetal tissues (upper panel – skin, lower panel - dorsal muscles) isolated from scapular region of a 98-day old fetus. n≥3 sections.
B – Immunofluorescence analysis of human fetal BAT isolated from the scapular region of a 98-day old fetus stained with anti-UCP1 and PPARG antibodies (left panel). Consecutive section stained with antibodies against GATA6 and lipid droplets containing adipocytes (right panel), n≥3 sections.
C – Immunofluorescence analysis of human fetal BAT isolated from scapular region of a 135-day old fetus. Expression of PLIN1, PPARG, UCP1 (upper panel), and GATA6 (lower panel) detected with specific antibodies, n≥3 sections.
Recapitulation of human BA development in vitro from hiPSCs
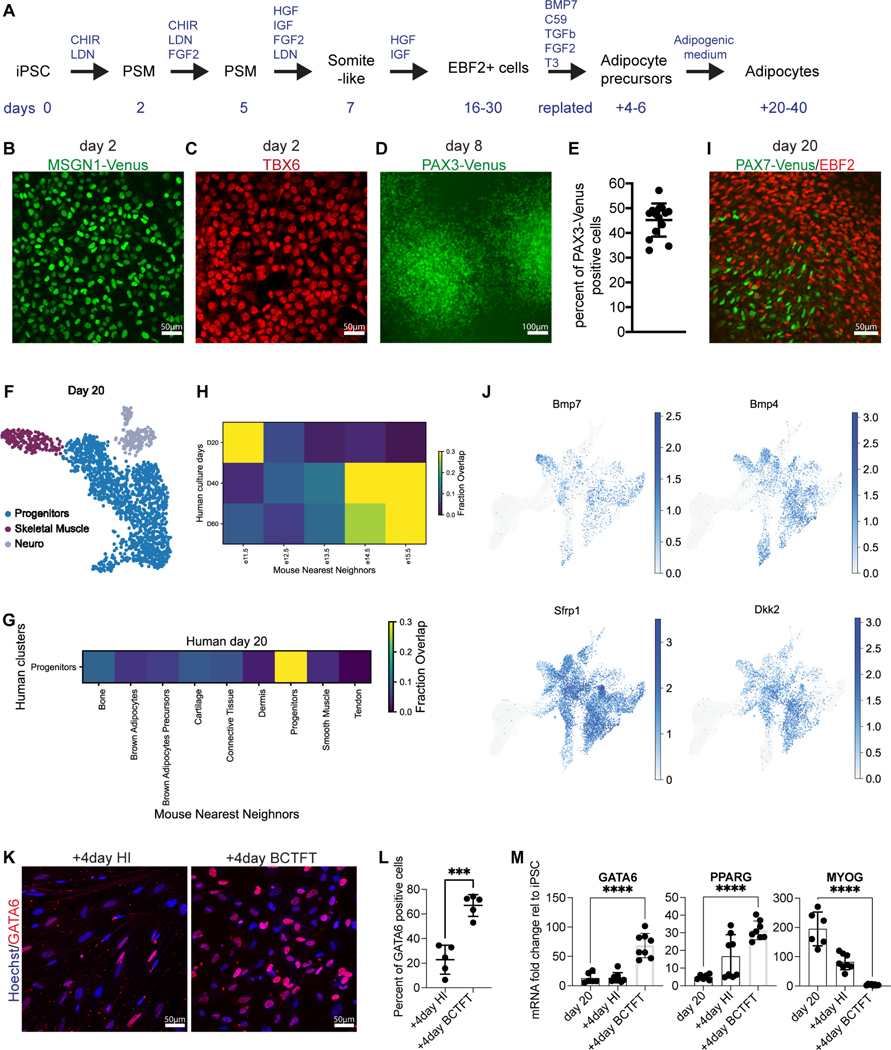
We next used these data to inform the design of a differentiation protocol aiming to recapitulate this process from hiPSCs. We previously developed efficient protocols to differentiate human paraxial mesoderm and skeletal muscle from hiPSCs in vitro 21–23. As BAs originate mostly from the paraxial mesoderm and share a common precursor with skeletal muscles in the dermomyotome 6, we first aimed to recapitulate the development of Pax-3 positive dermomyotomal cells in vitro. To achieve this, we treated hiPSCs with the Wnt agonist CHIR and the BMP inhibitor LDN (Figure 4A). At day 2–3, most of the cells expressed the PSM markers MSGN1 (95±0.4%) and TBX6 (Figure 4B-C, Figure S4A). To further differentiate the PSM cells into dermomyotomal PAX3-positive cells, we treated cultures with IGF, FGF, HGF, and LDN 21,22. To monitor cell differentiation towards the dermomyotome fate, we used CRISPR-Cas9 to knock-in a Venus fluorescent reporter into the PAX3 locus (figure S4B). On day 8, 45±6.7 % of the cells started to express Venus (Figure 4D-E, Figure S4C). We further differentiated these cultures in medium containing HGF and IGF as this treatment promotes the differentiation of somitic PAX7 precursor cells 21,22. PAX7 appeared around day 14 and at day 20 (d20) around 20–30 % of the mononucleated population expressed PAX7 (Figure 4I) 32. To investigate the cell composition of such cultures, we analyzed 1671 cells from d20 cultures by scRNAseq. We observed a major cluster containing cells expressing fibroblastic markers such as PDGFRA, HIC1 and EBF2 (Figure 4F, Figure S5D). This cluster also contained a subpopulation of PAX7-positive cells connected to a smaller cluster of differentiating myogenic cells (MYOD1, MYH3) (Figure S4D). A small cluster of SOX2+ neural cells was also observed at this stage. Using a machine-learning classifier trained on the mouse dataset, we observed that the identity of this human progenitor cluster is related to the mouse FP cluster (Figure 4G). Using the classifier trained on mouse dataset, we found that these d20 iPSC-BA cultures were closely related to mouse E11.5 cells (Figure 4H) 11. BAs and myogenic precursors have undergone lineage commitment by the time Ebf2 protein expression is detected in Myf5Cre+ lineage cells (Wang et al, 2014). Most of the cells of the FP cluster expressing EBF2 are negative for PAX7 at the mRNA and protein levels and MYF5 is known to be largely co-expressed with PAX7 in our cultures around d20 32. This suggests that the adipogenic and myogenic lineages are already separated or in the process of segregating in our 20-day cultures.
Figure 4 -. Development of human BA precursors in vitro.
A – Differentiation protocol. + indicates additional days after replating.
B – Venus signal in MSGN1-Venus hiPSC line on day 2 of differentiation, n=4.
C – Immunofluorescence analysis for transcription factor TBX6 on day 2 of differentiation, n=4.
D – Venus signal in PAX3-Venus hiPSC line on day 8 of differentiation, n=7.
E – FACS analysis quantification of PAX3-Venus positive cells on day 8 of differentiation. Mean ± SD, n=14.
F – UMAP embedding showing cell clusters in the human in-vitro cultured cells at d20.
G –Results of analysis with a k-NN classifier trained on of mouse embryonic clusters used to predict identity of the human in-vitro cultured progenitors.
H –Results of analysis with a k-NN classifier trained on clusters of E11.5, E12.5, E13.5, E14.5, E15.5 mouse embryos used to predict identity of the human in-vitro cultured timepoints at d20, 40 and 60.
I – Immunofluorescence analysis using antibodies against EBF2 and PAX7 on d20 of differentiation, n=8.
J – UMAP embedding showing expression of a curated list of signaling genes.
K – Immunofluorescence analysis using an antibody against GATA6 in d40 cultures in HI medium or adipogenic BCTFT medium for 4 days (+4 represents 4 days after replating). n=5.
L – Quantification of GATA6-positive cells in panel G. Mean ± SD, n=5, t-test, p≤0.001.
M – RT-qPCR analysis for GATA6, PPARG and MYOG on d20 of differentiation and 4 days after replating in HI medium or adipogenic BCTFT medium (+4 represents 4 days after replating). Mean ± SD, n=6–8, t-test, p≤0.0001.
We next sought to identify potential signaling pathways involved in brown adipogenesis from the mouse dataset. Mouse knock-out experiments have identified BMP activation and Wnt inhibition as important signals required for BA development 10,33,34. However, these studies have not precisely defined the time window when these cues act on BA precursors. We observed expression of the BMP ligands (Bmp7, Bmp4) and of the Wnt signaling antagonists (Sfrp1, Sfrp2, Sfrp4, Dkk2) mostly at the FP stage in the mouse dataset (Figure 4J, Figure S5). Expression of these secreted cues was downregulated in more mature cells of the BA lineage. These data suggest that BMP activation and Wnt inhibition are prominently required at the FP stage. Using KEGG pathway analysis, we identified several pathway-specific genes differentially expressed in the FP cluster (Figure S5A). These include genes involved in TGFbeta signaling (Tgfbr2, Gdf10), Thyroid hormone signaling (Creb3l1, Gnas, Gpx7, Creb5, Hsp90b1) and FGF/PI3K-AKT pathway (Akt1, Igf1, Fgfr1) (Figure S5A-C). Thus, to recapitulate the sequence of signaling cues to which BA precursors are exposed during their differentiation in vivo, we dissociated the primary cultures around d20 of differentiation to eliminate the myofibers and replated the cells in a medium containing BMP7, the Wnt antagonist C-59, TGFb1, FGF2 and Triiodothyronine (BCTFT medium) for 4–6 days (Figure 4A). This led to a strong increase in number of GATA6-expressing cells compared to cells cultured in medium containing HGF and IGF (Figure 4K-L). The cells also showed an upregulation of GATA6 and PPARG and a downregulation of MYOG mRNA, suggesting that the BCTFT medium can promote the adipogenic fate while repressing the myogenic fate (Figure 4M).
To follow the differentiation of hiPSCs into BAs, we generated a UCP1-mCherry fluorescent hiPSC reporter line using CRISPR-Cas9. In this knock-in line, an mCherry construct was introduced immediately 5’ to the start codon of UCP1 from which it was separated by a P2A peptide. Thus, the reporter is produced from the same transcript as UCP1 and targeted to the nucleus using an H2B tag (Figure 5A). Using this reporter line, we further differentiated the GATA6-positive progenitors derived in BCTFT medium. We treated the progenitor cultures with an adipogenic cocktail containing 3-Isobutyl-1-methylxanthine, Ascorbic acid, Triiodothyronine, TGFb inhibitor SB431542, Dexamethasone, EGF, Hydrocortisone and Rosiglitazone for another 20–40 days (Figure 4A) 16. In cultures examined after 40 days in differentiation, large (50–100 microns) UCP1-positive lipid-filled adipocytes started to appear, and the adipocyte number steadily increased over the following 20 days. On day 60 (d60) of differentiation, the cultures contained 46±12% UCP1-mCherry positive cells (Figure 5B-C). Using immunohistochemistry, we showed that 95% of the mCherry-positive cells expressed UCP1 (Figure 5D). PLIN1 and BODIPY staining confirmed that hiPSC-derived BAs (iPSC-BAs) contain multilocular lipid droplets (Figure 5E-F). We further confirmed expression of BA markers such as UCP1, PPARG, CIDEA and PLIN1 after 60 days of differentiation using RT-qPCR in cultures (Figure 5G).
Figure 5 -. Differentiation of human mature BA in vitro.
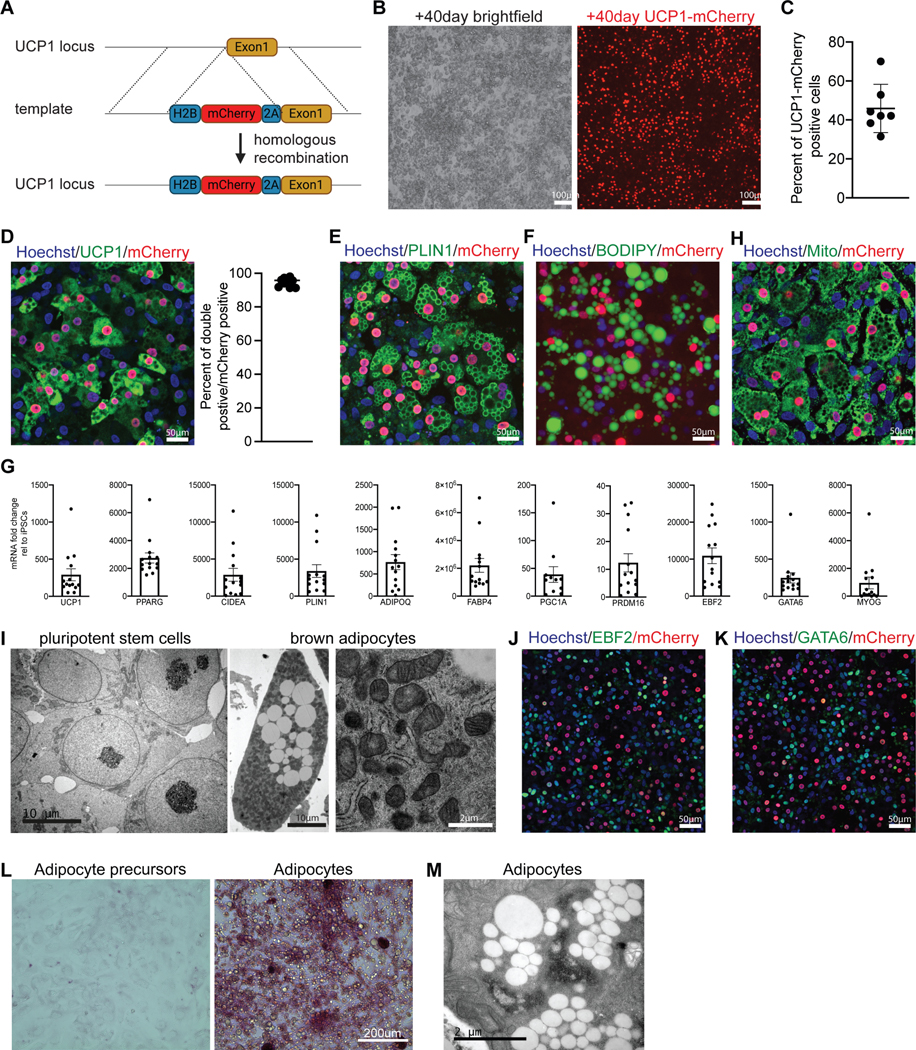
A – Targeting strategy used to generate UCP1-mCherry hiPSC line.
B – Brightfield and mCherry signal in iPSC-BA cultures on d60, n≥20.
C – Quantification of mCherry-positive cells in iPSC-BA cultures on d60. Mean ± SD, n=7.
D– Immunofluorescence staining with UCP1 and mCherry antibodies in the UCP1-mCherry knock-in iPSC-BA cultures on d60. Fraction of UCP1 and mCherry double positive cells from all mCherry-positive cells. n=10.
E– Immunofluorescence staining for PLIN1 antibody in the UCP1-mCherry knock-in iPSC-BA cultures on d60. n=6.
F – Neutral lipid staining using 0.5mM BODIPY in iPSC-BA cultures on d60, n=3.
G – RT-qPCR analysis of iPSC-BA cultures on d60. Mean ± SD, n=11–14.
H – Immunofluorescence staining for mitochondria on iPSC-BA cultures on d60, n=5.
I – Representative transmission electron micrographs of hiPSCs (left) and iPSC-BA (middle and right), n=3.
J–K – Immunofluorescence staining with antibodies against EBF2 (O), GATA6 (P) in UCP1mCherry iPSC-BA cultures on d60, n=4.
L – Periodic Acid staining on BA precursors (d20) and BAs (d60), n=3.
M – Transmission electron microscopy micrographs demonstrating glycogen accumulation in iPSC-BA, n=3.
See also Figure S5.
iPSC-BAs contain abundant mitochondria (a characteristic of BAs) as revealed by immunofluorescence for mitochondrially encoded cytochrome c oxidase II (MT-CO2) and electron microscopy analysis at 60 days of differentiation (Figure 5H-I). These cultures contained a fraction of UCP1-negative cells, which expressed EBF2 and/or GATA6 cells suggesting these cells might correspond to BA precursor cells (Figure 5J-K). Periodic acid staining and electron microscopy showed that at this stage, many cells accumulate glycogen, a characteristic of differentiating BAs (Figure 5L-M) 35. Thus, our protocol recapitulates a developmental trajectory similar to that observed for BAT differentiation in mouse embryos, leading to efficient differentiation of hiPSCs into UCP1-expressing BAs in serum free culture conditions.
scRNAseq analysis of the human brown adipogenic lineage differentiated in vitro
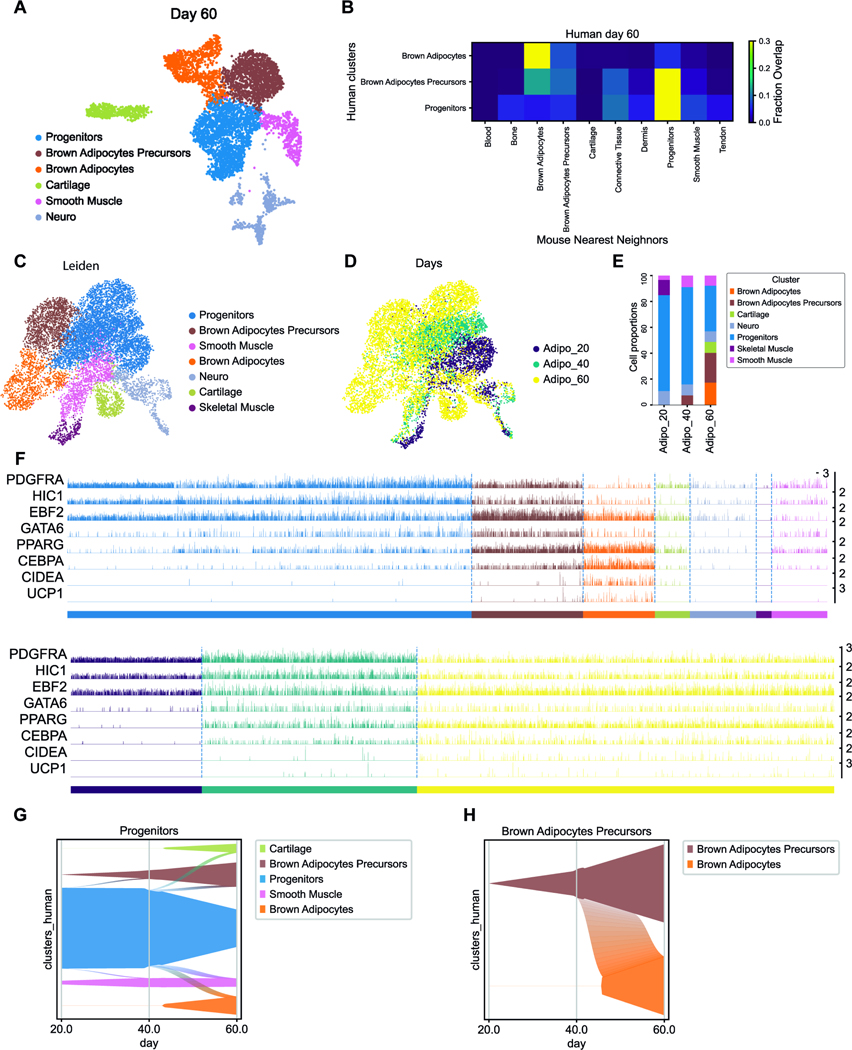
To further characterize the differentiation of hiPSCs into BAs, we performed scRNAseq of the cultures at d40 and d60 of differentiation. We analyzed 2744 and 5322 cells from d40 and d60 samples, respectively. At d40, most of the cells belong to a large cluster which expresses fibroblastic markers such as PDGFRA and HIC1 (figure S6A-B). Using the machine learning classifier trained on the mouse dataset, we confirmed that this cluster, which we term progenitors, is most similar to the mouse FP cluster (Figure S6C). We also detected a cluster of cells expressing BAPre markers (EBF2, CEBPA, PPARG) most closely related to BA precursors and BAs clusters according to the classifier. These two clusters express GATA6. We also observed a small cluster of skeletal muscle cells (MYOD1) and one of neural cells (SOX2). At this stage PAX7 expression was undetectable. At d60, we still observed a large fibroblastic cluster expressing markers such as PDGFRA and HIC1 (Figure 6A-B). Comparison to the mouse dataset with the classifier shows that it is closest to the FP cluster (Figure 6C). This cluster is connected to a cluster which expresses GATA6, EBF2 and PPARG resembling the mouse BAPre cluster. While the classifier analysis shows that it is more similar to the FP cluster, it also shows similarity with the mouse BA precursors and BA clusters suggesting that it corresponds to immature cells of the BA lineage. We also identified a cluster of BAs (CEBPA, CIDEA, UCP1) most similar to the mouse BA cluster (Figure 6B). Small separate clusters of cartilage (SOX9), smooth muscle (CNN1, ACTA2) and neural cells (SOX2) were also observed (Figure S6D-E). We next merged the three time points (d20, d40 and d60) to generate a UMAP projection and performed a Leiden clustering analysis on this combined dataset (Figure 6C-E). The cell types identified in the clustering analysis of individual time points mostly merged into single clusters defined by cell identity rather than age (compare Figure 6C and Figure S6F). The largest cluster corresponds to the Progenitors cluster which is connected to clusters of more mature fates. This suggests that the Progenitor cluster generated in vitro represents multipotent precursors similar to the mouse FP cluster. This cluster is already present at d20 and maintained during later stages (Figure 6E-F), like the FP cluster of the mouse dataset. Waddington OT analysis of the developmental trajectories of cells differentiating in vitro suggests that the progenitors cluster contributes to BA precursors and smooth muscle clusters observed at d40 and d60 as well as to the cartilage identified at d60 (Figure 6G). It also shows that the d60 BA cluster likely derives from the BA precursor cluster (Figure 6H).
Figure 6 -. ScRNAseq analysis of human BA generated in vitro.
A – UMAP embedding showing cell clusters in human in-vitro differentiated cells at d60.
B –Result of analysis with a k-NN classifier trained on mouse embryo clusters used to predict the identity of human in-vitro cultured clusters.
C – UMAP embedding showing the identity of cell clusters in the merged human in-vitro dataset of cultured cells at d20, 40 and 60.
D – UMAP embedding showing timepoints in the merged human in-vitro dataset of cultured cells at d20, 40 and 60.
E – Proportions of cell clusters per timepoints.
F –Tracksplot showing the expression level of markers genes of the BA lineage in individual cells in the clusters shown in C (top) and at the different ages as shown in D (bottom).
G – H Graphs showing how probability mass flows from the progenitor cluster (G) or from the BAPre cluster (H) to other clusters as a function of time based on Waddington-OT transition matrix.
See also Figure S6.
We next directly compared the d60 iPSC-BA cultures to the mouse scRNaseq dataset. The list of expressed mouse genes was converted into their human orthologs, and the two datasets were merged and analyzed together using UMAP projection followed by Leiden clustering. Human and mouse cells identified as Fibroblastic Progenitors, BAPre and BAs were found in the same clusters in the merged dataset indicating the similarity of the human cells differentiated in vitro to the mouse BA lineage differentiating in vivo (Figure S7A-C). Clusters of other lineages such a cartilage or smooth muscle also contained both mouse and human cells. Side by side comparison of the expression of specific markers of BA differentiation in the same clusters in mouse and human cells shows very similar expression patterns (Figure S7D). Finally, we used the classifier trained on the mouse dataset to compare each time point analyzed in vitro to each of the different mouse embryonic ages analyzed. This suggests that the d40 cells are closer to mouse E14.5–15.5 and d60 to E15.5 (Figure 4G).
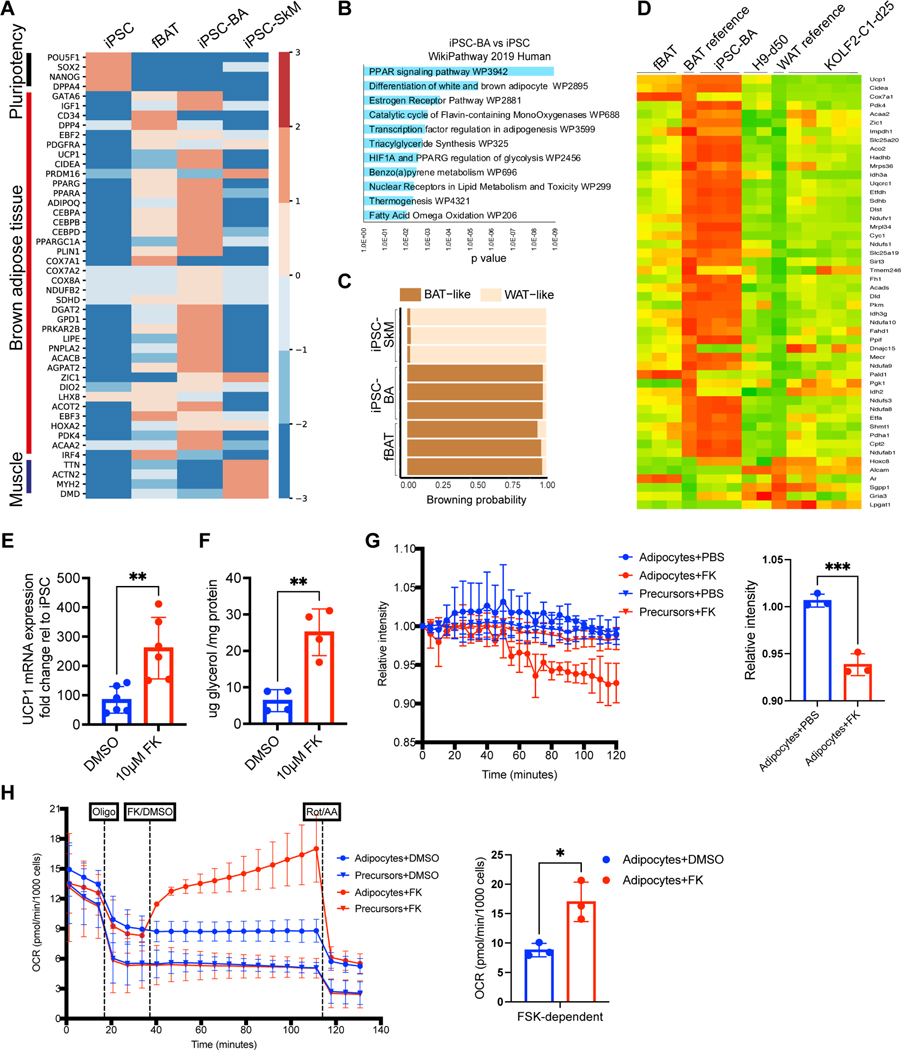
To molecularly characterize the iPSC-BAs which are mostly lost during dissociation in the scRNAseq analysis, we performed bulk mRNA sequencing of undifferentiated hiPSCs and cultures at d60. In parallel, as a reference for differentiated adipocytes, we performed bulk RNAseq of human fetal BAT (fBAT) isolated from 115, 122 and 125-day post-conceptual age fetuses from the interscapular and scapular regions. As an outgroup, we used bulk RNAseq data from iPSC-derived skeletal muscle cultures (iPSC-SkM) 36. BA-specific genes such as UCP1, CIDEA, PARGC1A and DIO2 were specifically expressed in iPSC-BAs and fBAT (Figure 7B). GATA6 were also detected suggesting the presence of adipocyte precursors in differentiating cultures (Figure 7A). The top 200 differentially expressed genes in iPSC-BAs corresponded to human WikiPathways related to ‘PPARG signaling pathway’, ‘Estrogen receptor pathway’ and ‘Thermogenesis’ (Figure 7B). When analyzed using the ProFat database 37, the browning probability of iPSC-BAs was comparable to fBAT (Figure 7C). Recently, two studies have described the directed differentiation of BAs from hPSCs 19,20. Comparison of our iPSC-BAs with the terminally differentiated adipocytes from the two studies – 50-day old adipocytes from Zhang et al. (H9-d50) and 25-day old adipocytes from Carobbio et al. (KOLF2-C1-d25) showed that cells differentiated according to our protocol exhibit a higher degree of similarity to the fBAT and BAs gene expression profile (Figure 7D, Figure S7E).
Figure 7 -. Human BA generated in vitro are functional.
A – Heatmap showing differentially expressed genes Scale bar represents log normalized read counts.
B – WikiPathway 2019 Human analysis of top 200 differentially expressed genes (FDR<0.05, log2|FC|>6) in iPSC-BAs compared to iPSCs.
C – Estimation of browning probability of iPSC-BA, iPSC-SkM and fBAT using PROFAT database. Numbers represent replicates.
D – Heatmap comparing transcriptomic profiles of iPSC-BA, fBAT, hPSC-BAs in Zhang et al. 2020 (H9-d50, 2 replicates) and iPSC-BAs in Carobbio et al. 2021 (KOLF2-C1-d25, 5 replicates).
E – RT-qPCR analysis for UCP1 of iPSC-BAs treated with 10μM forskolin for 4 hours. Relative gene expression is shown as relative change to undifferentiated iPSC. Mean± SD, n= 5, t-test, p≤0.01.
F – Measurement of glycerol release in iPSC-BAs treated with 10μM forskolin for 4 hours. Glycerol amount normalized using total protein amount. Mean± SD, n= 4, t-test, p≤0.01.
G – Fluorescent intensity of thermosensitive ERthermAC dye in d20 (precursors) and d60 (BAs) cultures in response to 10μM forskolin. Each data point represents Mean± SD, n= 3, t-test, p=0.0009.
H – Oxygen consumption rate in d20 (precursors) and d60 (BAs) cultures in response to 1.5μM oligomycin, 10μM forskolin and 1μM Rotenone and Antimycin (Rot/AA). Oxygen consumption rate was normalized by number of cells per well. Each data point represents Mean± SD, n= 3, t-test, p=0.015.
iPSCs: undifferentiated hiPSCs, fBAT: human fetal BAT from 115, 122 and 125-day post-conceptual age, iPSC-BA: 60-day old iPSC-derived BAs, iPSC-SkM: iPSC derived skeletal muscle.
See also Figure S7.
In conclusion, this analysis suggests that a BA lineage developmental trajectory highly similar to that observed for mouse interscapular fat in vivo can be recapitulated in vitro from human iPS cells. This results in the production of immature BAs approximately equivalent to those of the E15.5 mouse embryo.
Functionality of iPSC-BAs
Generally, BAs are characterized by their potential to generate heat in response to a beta-adrenergic stimulus and downstream activation of the cAMP pathway 38. This activation can be mimicked by treatment with Forskolin which activates cAMP signaling 19,39,40. d60 cultures treated with forskolin for 4 hours show increased UCP1 mRNA levels compared to vehicle-treated cells (Figure 7E). To evaluate the lipolytic activity of the cells, we measured the amount of glycerol released in the medium after forskolin treatment. In comparison to vehicle control treated cells, forskolin-treated cells showed an increased glycerol release (Figure 7F). To measure the ability of iPSC-BAs to generate heat, we used ERThermAC, a thermosensitive vital fluorescent dye 39. As temperature increases, the cells display lower ERthermAC fluorescence intensity. We preincubated iPSC-BAs with ERThermAC, treated with PBS or forskolin and measured the fluorescence intensity of the dye over time. We observed a reduction in dye intensity in forskolin-treated cells compared to vehicle-treated cells (Figure 7G). In contrast, no change in intensity was observed for d20 BA precursors in either condition. Overall, our data demonstrate that iPSC-BAs induce UCP1 expression, undergo lipolysis and generate heat in response to forskolin activation as normal human BAs.
The process of thermogenesis is driven by the flow of protons through UCP1 in the mitochondrial inner membrane. To examine whether forskolin-induced thermogenesis in iPSC-BAs is coupled to proton leak in mitochondria, we measured oxygen consumption rate (OCR) with a Seahorse metabolic flux analyzer in d60 cultures (Figure 7H). Upon treatment with Oligomycin, an ATP synthase inhibitor, the OCR in cells decreased. When iPSC-BAs were treated with forskolin together with Oligomycin, the OCR increased suggesting that proton leak is increased, leading to higher oxygen consumption (Figure 7H). In contrast, the d20 precursors showed lower basal respiration level and did not show increased OCR upon forskolin treatment. As expected, inhibiting the electron transport chain using Rotenone (mitochondrial complex I inhibitor) or Antimycin (mitochondrial complex III inhibitor), resulted in a drop of OCR (Figure 7H). These analyses support the functionality of human iPSC-BAs differentiated in vitro.
DISCUSSION
Studying brown fat lineage development has proven complicated due to the difficulty of identifying intermediate stages in differentiation. In this report, we first characterized the development of interscapular mouse brown fat using scRNAseq. We analyzed the development of paraxial mesoderm-derived cell populations in the dorsal trunk at the forelimb level of mouse embryos which contains the interscapular brown fat. This analysis identified a previously unrecognized BA differentiation stage characterized by expression of the transcription factor Gata6. Using bioinformatics tools and lineage tracing analysis in mouse, we show that these Gata6+ cells derive from Pax7-precursor cells and generate Pparg-positive preadipocytes which differentiate into Ucp1-positive BAs (Figure 1E, 2B). This scRNAseq analysis also allowed the identification of specific signaling pathways associated to the Gata6-positive BAs’ progenitor state. We used these signaling cues to modify a protocol previously established to differentiate human paraxial mesoderm in vitro 22. This allowed highly efficient generation of functional human BAs in vitro using defined serum-free culture conditions. ScRNAseq analysis of the human cells differentiating in vitro show that they recapitulate a developmental sequence very similar to that observed in mouse in vivo. This sequence results in the production of GATA6-positive BA precursors and then to PPARG-positive preadipocytes and finally to UCP1-positive adipocytes which exhibit functional properties characteristic of BAs.
Descriptions of brown fat development were mostly done in mouse by lineage tracing, using early Cre drivers such as En1, Pax3, Pax7, myf5 or Pdgfra followed by late analyses identifying brown fat based on morphology or after in vitro culture and qPCR for Ucp1 6–11,41. These studies showed that brown fat cells mostly derive from Pax3 and Pax7/Myf5-positive progenitors which subsequently down-regulate these two genes while expressing Ebf2 and Pdgfra. These analyses identified the origin of BAs in early mouse embryos, but provided little information on the developmental trajectory followed by their precursors. Mouse brown fat differentiation has been examined using histochemistry for Pparg, perilipin or Pdgfra 7 and in situ hybridization with genes such as Cebpa which is selectively expressed by the interscapular depot 35. These analyses demonstrated that formation of the lipid droplets in brown fat precursors is initiated within glycogen clusters starting around E14.5. This precedes expression of Ucp1 which occurs around E16.5 35.
In this report, we perform a direct analysis of the transcriptome of the developing interscapular BA depot in mouse. Based on this data, we describe the early stages of BA differentiation trajectory. Our analyses using the STREAM or Waddington OT pipelines 25,26 suggest that BA precursors share a common Pdgfra+ precursor with other somite-derived mesenchymal lineages such as dermis, muscle connective tissue, cartilage precursors and smooth muscle (Figure 1E, Figure S2B-C). This is in line with lineage tracing studies showing that BAs derive from Pdgfra+ cells (Wang et al, 2014). In contrast, the skeletal muscle cluster is clearly segregated from the mesenchymal cluster in our dataset. This is consistent with the early loss of adipogenic potential of the dermomyotomal Pax7-expressing cells which occurs at E11.5 in mouse 11. Our lineage tracing experiments confirm that most of the mouse interscapular brown fat derives from Pax7-expressing precursors 8,11 which can give rise to the Gata6-positive BA precursor population. These early Pax7 precursors could correspond to the En1-positive cells of the central dermomyotome previously identified in mouse 10. We reanalyzed a scRNAseq dataset of developing mouse periaortic BAT and observed a population of Gata6-positive BA precursors closely related to the one identified in the interscapular BAT 27. Interestingly, periaortic BAT receives little contribution from Pax3+ cells and has no contribution from Myf5+ cells 8. This therefore suggests that different embryonic populations can contribute to the Gata6-positive BA precursors.
We identified Gata6 (GATA binding protein 6) as a gene enriched in the BA cluster in mouse. Gata6 is a zinc finger transcription factor which plays an essential role in the development of extraembryonic tissues 42. Its null mutation in mouse leads to lethality shortly after implantation. Gata6 has also been implicated in the development of several organs including heart, lung, gut, pancreas, skeleton and skin but its expression during brown fat development has so far not been reported 28,29,42–45. Gata6 was however identified in silico as a transcription factor potentially regulating the expression of brown fat specific genes such as Zic1 and TCA cycle enzymes such as citrate synthase (Cs) in the ProFat database 37. Also, a GATA recognition motif was identified in ATACseq data of differentiating human adipocytes in vitro 19. In cardiomyocytes, GATA-6 was shown to associate with Ppara to regulate the expression of the glucose transporter Glut4 46. Together, these data suggest that Gata6 could play a role in the regulation of energy metabolism in differentiating BAs.
We show that Bmp4/7 and Bmpr1a/2 are enriched in the Gata6-positive progenitors. This is consistent with the well-established role of Bmps in the promotion of brown adipogenesis 33,47. We also observed transient expression of the Wnt inhibitors Sfrp1,2 and 4 and Dkk2 in the Gata6-positive precursors, consistent with the well-known anti-adipogenic role of Wnt signaling 34,48. These signals are downregulated as cell differentiate further in the BA lineage. The thyroid hormone and the FGF/IGF1/PI3K pathways which play a role in the control of thermogenesis and BAT development and physiology were also activated in the Gata6 precursors 49,50. Thus, our data suggest that signaling cues known to be important for brown adipogenesis such as Wnt inhibition and BMP activation act at the Gata6-positive precursor stage.
We previously established an efficient protocol to differentiate hiPSCs to skeletal muscle fate 21,22. These cells first differentiate to the presomitic (MSGN1) stage 23, then to the dermomyotome (PAX3) stage and after two-weeks in vitro, they start to differentiate to PAX7-positive cells. After ~20 days in vitro, up to 25 % of the culture mononucleated fraction express PAX7 32. Our classifier analyses comparing the mouse embryo dataset to the hiPSCs differentiated in vitro suggest that at d20 of the myogenic differentiation protocol, cells are at stage equivalent to the E11.5 mouse embryo (Figure 4H) when Pax7-precursors lose their potential to contribute to the adipogenic lineage11. Moreover, a majority of d20 cells appear closely related to the fibroblastic precursors identified in mouse (Figure 4G). We therefore decided to treat d20 cultures with the signals acting on the fibroblastic population identified in the mouse embryo. Treating replated d20 cultures for 4–6 days with BMP7, the Wnt antagonist C-59, TGFb1, FGF2 and Triiodothyronine (BCTFT medium) leads to a significant increase in Gata6 expression while Myog is decreasing. When, these cultures are subsequently exposed to a classical adipogenic medium, they differentiate very efficiently to the BA lineage after d40–60. Counting the UCP1-positive cells using fluorescence microscopy in d60 cultures yields an estimate of ~40–50% positive cells. In contrast, we only detect ~1% of cells expressing the UCP1 transcript in the d60 scRNAseq dataset. This is because single cell dissociation required for scRNAseq disrupts the mature lipid-filled BAs 51,52. Nevertheless, our scRNAseq analysis suggests that in d60 cultures, ~40% of the sequenced cells belong to the BAPre and BA clusters while another 40% of the cells are in the Progenitors cluster. Together, this suggests that the vast majority (possibly up to 80–90%) of the d60 cultures corresponds to cells at various stages of the differentiation of the BA lineage. We also performed bulk RNAseq of d60 differentiated cultures in order to include the population of mature BAs which is absent in the scRNAseq data. Using the Profat database 37, we observed that these cells exhibit a browning probability similar to fBAT (Figure 7B, E). Finally, these human BAs generated in vitro are able to respond to a signal mimicking adrenergic stimulation by increasing Ucp1 expression and glycerol release as well as by increasing temperature and oxygen consumption as their in vivo counterparts 12. This showed that these cells are closely related to fetal and adult human BAs. The comparison of these iPSC-BA cultures with recently generated datasets of mature BAs will require to generate single nuclei analyses 51,53.
Comparison of the BAs generated with our protocol with those generated by two other recently published directed differentiation protocols 19,20 suggest that the transcriptional signature of the adipocytes generated in our conditions is closer to endogenous fetal BAs. The choice of agonists and antagonists and their timing of administration described in these papers is not informed by the signaling pattern during BA differentiation as in our protocol. Zhang et al. (2020) used BMP7 and FGF2 in combination with an adipogenic cocktail on day 8 of differentiation to induce BA specification. They also inhibit TGFbeta (our study suggests that activation of Tgfbeta signaling promotes adipogenesis at the progenitor stage) and they do not manipulate Wnt signaling. This study claims a differentiation efficiency of 63% of UCP1+ cells based on immunostaining and FACS analysis. However, FACS analysis of lipid containing BAs is challenging as lipid containing cells burst open during the dissociation process 54. In the Carobbio et al. 2021 report, the authors do not inhibit Wnt and do not activate BMP signaling. In this report, the efficiency of adipocyte generation is measured using UCP1 western blotting. However, surprisingly UCP1 is already detected at d12 when the human cultures are still extremely immature.
In humans, BAT activity shows an inverse correlation with body mass index and percentage of total body fat 55,56. Moreover, the graft of ectopic BAT leads to improvement of glucose metabolism in diabetic or obese mice 57,58. Our work could therefore help the development of cell therapy strategies involving the graft of human BAs generated in vitro for the treatment of obesity, metabolic syndrome and diabetes.
LIMITATIONS OF STUDY
Our scRNAseq analysis of the iPSC-BA cultures detects few UCP1-positive cells. This is because single cell dissociation required for scRNAseq disrupts the mature lipid-filled BAs [54, 55], which are large buoyant cells that cannot be encapsulated using the microfluidics platform used for the Indrops pipeline and are thus absent from the scRNA dataset. This limitation can be overcome by performing single nuclei RNA sequencing. We estimated the number of UCP1+ cells by quantifying the UCP1-mCherry-positive cells. We propose that Gata6 labels the BA progenitors of the interscapular BAT, however we did not investigate other BAT depots to test if similar lineage dynamics apply.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Olivier Pourquié (pourquie@genetics.med.harvard.edu).
Data and Code Availability
Single-cell RNA-seq data have been deposited at NCBI GEO under accession number GSE185518. Accession number is also listed in the key resources table.
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Cd34 | Thermo Fisher Scientific | 14-0341-82 |
| Dpp4 | R&D | AF954-SP |
| EBF-2 | R&D | AF7006 |
| Gata6 | RnD | AF1700-SP |
| Gata6 | Cell Signaling Technology | 5851T |
| GFP | SantaCruz | SC-101536 |
| GFP | Abcam | AB13970 |
| Keratin 14 | Biolegend | 906001 |
| mCherry | OriGene | AB0040-200 |
| mCherry | Neuromics | CH22115 |
| MTC02 | Abcam | AB3298 |
| Myh11 | Alfa Aesar | J64817 |
| MyHCfast | Sigma-Aldrich | M4276 |
| Perilipin 1 | Progen | GP29 |
| Perilipin-1 | Abcam | AB61682 |
| PPARG | Thermo Fisher Scientific | MA5-14889 |
| RFP | Rockland | 600-901-379 |
| RFP | Rockland | 600-401-379 |
| Sca-1/Ly6 | R&D | AF1226-SP |
| TBX6 | Abcam | AB38883 |
| UCP1 | R&D | MAB6158 |
| Donkey Anti-Rat IgG H&L (Alexa Fluor® 647) preadsorbed | Abcam | A-11015 |
| Donkey anti-Goat IgG (H+L) Secondary Antibody, Alexa Fluor® 488 conjugate | Thermo Fisher Scientific | A21206 |
| Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | A-21207 |
| Donkey anti-Sheep IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 | Thermo Fisher Scientific | A-31573 |
| Donkey anti-Sheep IgG (H+L) Secondary Antibody, Alexa Fluor® 488 conjugate | Life Technologies | A-31572 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | 703-545-155 |
| Donkey anti-Rabbit IgG (H+L) Secondary Antibody, Alexa Fluor® 594 | Thermo Fisher Scientific | 703-165-155 |
| Donkey anti-Rabbit IgG (H+L) Secondary Antibody, Alexa Fluor® 647 | Thermo Fisher Scientific | A-31571 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 | Thermo Fisher Scientific | 706-545-148 |
| Alexa Fluor® 488 AffiniPure Donkey Anti-Chicken IgY (IgG) (H+L) | Jackson ImmunoResearch Laboratories | 703-545-155 |
| Cy™3 AffiniPure Donkey Anti-Chicken IgY (IgG) (H+L) | Jackson ImmunoResearch Laboratories | 703-165-155 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | A-31571 |
| Alexa Fluor® 488 AffiniPure Donkey Anti-Guinea Pig IgG (H+L) | Jackson ImmunoResearch Laboratories | 706-545-148 |
| Bacterial and virus strains | ||
| Promega JM109 Competent E.Coli Cells | Thermo Fisher Scientific | PR-L1001 |
| Biological samples | ||
| Human fetal tissues | University of Washington Birth Defects Research Laboratory (BDRL) under a protocol approved by the University of Washington Institutional Review Board | 98 days (H28540), 115 days (H28572), 122 days (H28560), 125 days (H28626) |
| Chemicals, peptides, and recombinant proteins | ||
| Y-27632 dihydrochloride | R&D Systems | 1254/10 |
| CHIR 99021 | R&D Systems | 4423 |
| LDN-193189 | Stemgent | 04-0074 |
| FGF-2 | PeproTech | 450-33 |
| HGF | PeproTech | 315-23 |
| IGF-1 | PeproTech | 250-19 |
| TGFb1 | PeproTech | 100-21-10 |
| BMP7 | Thermo Fisher Scientific | PHC9544 |
| 3,3′,5-Triiodo-L-thyronine sodium salt | Sigma-Aldrich | T6397 |
| Porcn Inhibitor II C59 | Millipore Sigma | 500496 |
| SB431542 | Selleck Chemicals | S1067 |
| Dexamethasone | Sigma-Aldrich | D4902 |
| Hydrocortisone | Sigma-Aldrich | H0888 |
| Rosiglitazone | Sigma-Aldrich | R2408 |
| EGF | PeproTech | AF-100-15 |
| 3-Isobutyl-1-methylxanthine | Sigma-Aldrich | I7018 |
| ERthermAC | Sigma | SCT057 |
| Oligomycin | Tocris | 4110 |
| Forskolin | Sigma-Aldrich | F6886 |
| Antimycin A | Sigma-Aldrich | A8674 |
| Rotenone | Sigma-Aldrich | R8875 |
| 4% Paraformaldehyde | Electron Microscopy Sciences | 15710 |
| Critical commercial assays | ||
| NucleoSpin® RNA kit | Macherey and Nagel | 740955 |
| iScript™ cDNA Synthesis Kit | Bio-Rad | 1708891 |
| iTaq™ Universal SYBR® Green Supermix | Bio-Rad | 172-5124 |
| Deposited data | ||
| Single cell RNA seq data | This paper | GEO (GSE185623) |
| Bulk RNA seq data | This paper | GEO (GSE185518) |
| Experimental models: Cell lines | ||
| NCRM-1 (NIH CRM control iPSC line (male)) human iPSC | RUCDR Infinite Biologics | ND50028 |
| NCRM-1-UCP1-H2B-mCherry - 2 clones | This paper | N.A. |
| NCRM-1-PAX3-NLS-Venus - 2 clones | This paper | N.A. |
| Experimental models: Organisms/strains | ||
| Mouse : Pax7-iCre/+;Rosa26-mTmG/+ | This study; Gabrielle Kardon Lab | N.A. |
| Mouse : Gata6-EGFPCreERT2;Rosa26-fl/STOP/fl-tdTomato | This study; Fiona Watt Lab | N.A. |
| Mouse : wildtype CD1 IGS mice | Charles River | 022 |
| Oligonucleotides | ||
| RT-qPCR primers | This study | Table S2 |
| Recombinant DNA | ||
| PSpCas9 (BB)-2A-GFP(PX458) | Addgene | 48138 |
| pUC19 vector backbone | New England Biolabs | E5510S |
| pUC19-5’HA-H2B-mCherry-P2A-3’HA | This study | N.A. |
| Software and algorithms | ||
| ImageJ | Image J | https://imagej.nih.gov/ij/ |
| Prism 9 | GraphPad | https://www.graphpad.com |
| Single cell analysis code | This study | https://doi.org/10.5281/zenodo.8071624 |
| kNN-classifier | Diaz-Cuadros et al., 23 | N.A. |
| Stream | Chen et al., 25 | N.A. |
| Waddington-OT kernel | Schiebinger et al., 26 | N.A. |
| ProFAT webtool | Cheng et al., 37 | N.A. |
| FastQC (v0.11.9) and used STAR RNA-seq aligner (v2.7.3a) | Dobin et al., 61 | N.A. |
| featureCounts (v2.0.1) | Liao et al., 62 | N.A. |
| DESeq2 package (v 1.22.2) | Love et al., 63 | N.A. |
| EnrichR | Kuleshov et al., 64 | N.A. |
| Scrublet | Wolock et al., 65 | N.A. |
| Scanpy (v.1.6.0) | Wolf et al., 70 | N.A. |
| Cellrank | Lange et al., 71 | N.A. |
Microscopy data reported in this paper will be shared by the lead contact upon request. Raw and processed Bulk RNA-seq data have been deposited at NCBI GEO under accession number GSE185623 and are publicly available as of the date of publication. Accession number is also listed in the key resources table.
All original code has been deposited at https://github.com/PourquieLab/Djeffal_Rao_2023.git and is publicly available. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Materials Availability
Plasmids and cell lines generated in this study are available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Human induced pluripotent stem cells
NCRM1 (NIH CRM control iPSC line (male)) human iPSC and other cell lines were routinely cultured on Matrigel (Corning, 354263) coated culture plates (Corning, 353046) in mTeSR (Stemcell Technologies, 85850) at 37C and normaxia.. Upon confluency, cultures were dissociated into single cells using Accutase (Corning, 25–058-C). Cells were seeded on Matrigel coated culture plates in mTeSR supplemented with 10μM Y-27632 dihydrochloride (R&D Systems, 1254/10) at a density of 56,000/cm2 for routine maintenance of iPSC lines. On the following day, medium was replaced with mTeSR only and medium was changed every day. Cultures became confluent every fourth day and were passaged as described above. For freezing, cultures were dissociated into single cells with Accutase and frozen in NutriFreez D10 Cryopreservation Medium (Biological Industries, 01–0020-50).
Mouse lines
Mice were bred and maintained under standard conditions. Pax7-iCre/+;Rosa26-mTmG/+ embryos were generated by crossing Rosa26-mTmG/mTmG 59 males with Pax7-iCre/+ females in timed matings and embryos were collected at E14.5. Gata6-EGFPCreERT2 mice were generated by knocking an EGFPCreERT2 cassette into the endogenous Gata6 locus as reported previously 28. They were crossed with Rosa26-fl/STOP/fl-tdTomato mice 60. For lineage tracing experiments, pregnant females were injected intraperitoneally with 50 lg/g of tamoxifen (Sigma) at E12.5 and embryos were collected at E15.5. For experiments other than lineage tracing, wildtype CD1 IGS mice (Charles River) were used.
Human fetal tissue
Human fetal tissues were obtained by the University of Washington Birth Defects Research Laboratory (BDRL) under a protocol approved by the University of Washington Institutional Review Board. BAT tissues were isolated at 98 days (H28540), 115 days (H28572), 122 days (H28560), 125 days (H28626) and 135 days estimated post-conceptual age from the interscapular and scapular region. Informed consent was obtained for use of human tissues in research.
METHOD DETAILS
Brown adipocyte differentiation
iPSCs were differentiated into presomitic mesoderm as described previously (Chal et al., 2016). Briefly, confluent maintenance cultures were dissociated into single cells using Accutase (Corning, 25–058-C) and cells were seeded at a density of 30,000–33,000/cm2 on Matrigel (Corning, 354263) coated culture plates (Corning, 353046) in mTeSR (Stemcell Technologies, 85850) supplemented with 10μM Y-27632 dihydrochloride (R&D Systems, 1254/10). Next day, the cells formed small compact colonies. On day 0 of differentiation, to induce presomitic mesoderm, cells were treated with CL medium [DMEM/F12 GlutaMAX (Thermo Fisher Scientific, 10565042) + 1% Insulin-Transferrin-Selenium (Gibco, 41400045) + 3μM CHIR 99021 (R&D Systems, 4423) + 0.5μM LDN-193189 (Stemgent, 04–0074)]. Cells were treated with CL medium for 3 days and medium was refreshed every day. On day 3 of differentiation, cells were changed to CLF medium [CL medium + 20ng/ml FGF-2 (PeproTech, 450–33)]. Cells were treated with CLF medium for 3 days and medium was changed every day.
To further differentiate presomitic mesoderm cells to dermomyotomal fate, cells were changed to HIFL medium [DMEM high glucose (Thermo Fisher Scientific, 11965–118) + Penicillin/Streptomycin (Life Technologies, 15140122) + 15% KnockOut™ Serum Replacement (Life Technologies, 10828–028) + NEAA (Thermo Fisher Scientific, 11140–050) + 0.01mM 2-Mercaptoethanol (Life Technologies, 21985–023) + 10ng/ml HGF (PeproTech, 315–23) + 2ng/ml IGF-1 (PeproTech, 250–19) + 20ng/ml FGF-2 (PeproTech, 450–33) + 0.5μM LDN-193189 (Stemgent, 04–0074)]. Cells were treated with HIFL for two days and medium was changed every day. To let the PAX3 expressing dermomyotomal cells into myogenic and non-myogenic lineages, cells were changed to HI medium (HIFL medium without FGF-2 and LDN-193189) and medium was changed every day.
On day 16–30 of differentiation, cells were replated to differentiate cells into BA lineage. Cultures were dissociated into single cells with 2.5mg/ml Collagenase, (Type IV, Thermo Fisher Scientific, 17104019) and 0.05% Trypsine EDTA (Thermo Fisher Scientific, 25200–056) in PBS (Gibco, 14190) and filtered through a 30 μm (CellTrics, 04–0042-2316) filter and seeded on Matrigel coated plates at a density of 60,000–100,000/cm2 in BCTFT medium [DMEM high glucose (Thermo Fisher Scientific, 11965–118) + Penicillin/Streptomycin (Life Technologies, 15140122) + 5% KnockOut™ Serum Replacement (Life Technologies, 10828–028) + 1% Insulin-Transferrin-Selenium (Gibco, 41400045) + 10 ng/ml FGF-2 (PeproTech, 450–33) + 10ng/ml BMP7 (Thermo Fisher Scientific, PHC9544) + 20nM Porcn Inhibitor II, C59 ( Millipore Sigma, 500496) + 10ng/ml TGFb1 (PeproTech, 100–21-10) + 2nM 3,3′,5-Triiodo-L-thyronine sodium salt (Sigma-Aldrich, T6397)]. Cells were treated with BCTFT for 4–6 days and medium was refreshed every day. Adipocyte precursors were either frozen to be used later or differentiated into adipocytes. For freezing, cultures were dissociated into single cells using 0.05% Trypsine EDTA (Thermo Fisher Scientific, 25200–056) in PBS (Gibco, 14190) and cells were frozen in NutriFreez D10 Cryopreservation Medium (Biological Industries, 01–0020-50).
To differentiate adipocyte precursors into adipocytes, cells were cultured in adipogenic medium [DMEM high glucose (Thermo Fisher Scientific, 11965–118) + Penicillin/Streptomycin (Life Technologies, 15140122) + 5% KnockOut™ Serum Replacement (Life Technologies, 10828–028) + 1% Insulin-Transferrin-Selenium (Gibco, 41400045) + 500μM IBMX (3-Isobutyl-1-methylxanthine, Sigma-Aldrich, I7018) + 25.5μg/ml L-Ascorbic acid (Sigma-Aldrich, A4544) + 2nM T3 (3,3′,5-Triiodo-Lthyronine sodium salt, Sigma-Aldrich, T6397) + 5μM TGFb inhibitor SB431542 (Selleck Chemicals, S1067) + 1μM Dexamethasone (Sigma-Aldrich, D4902) + 10ng/ml EGF (Peprotech, AF-100–15) + 4μg/ml Hydrocortisone (Sigma-Aldrich, H0888) + 1μM Rosiglitazone (Sigma-Aldrich, R2408)] for 30–40 days. Medium was changed every third day.
To perform functional assays and immunofluorescence analysis, 30–40 day old replated cultures were dissociated with 2.5mg/ml Collagenase, (Type IV, Thermo Fisher Scientific, 17104019) and 0.05% Trypsine EDTA (Thermo Fisher Scientific, 25200–056) in 1X Phosphate Buffered Saline (Gibco, 14190) and cells were seeded onto suitable plate format required for the assay.
Human fetal tissue processing
Human fetal tissues were obtained by the University of Washington Birth Defects Research Laboratory (BDRL) under a protocol approved by the University of Washington Institutional Review Board. BAT tissues were isolated at 98 days (H28540), 115 days (H28572), 122 days (H28560), 125 days (H28626) and 135 days estimated post-conceptual age from the interscapular and scapular region. Since adipose depot are relatively small at these stages and are in close contact with adjacent tissues, some surrounding tissue (including connective tissue, skeletal muscle) was also present in the dissected tissue. Tissues were rinsed in 1X Hanks’ Balanced Salt Solution (HBSS, Thermo Fisher Scientific, 14185052) before processing. For immunofluorescence analysis, tissues were fixed with 4% Paraformaldehyde (Electron Microscopy Sciences, 15710) overnight at 4°C, washed 3 times in 1X Phosphate Buffered Saline (PBS, Sigma-Aldrich, P5493) and stored at 4°C in PBS supplemented with 0.1% Sodium Azide (Sigma-Aldrich, 71290) until further processing. For bulk mRNA sequencing, tissues were snap frozen in liquid nitrogen and stored at −80°C until further processing. 28,59,60
Generation of UCP1-mCherry and PAX3-Venus reporter line
UCP1 gene was targeted using the CRISPR-Cas9 system-based genome editing to generate a reporter NCRM1 iPSC line which already had been modified at PAX7 locus to introduce Venus. To target the UCP1 locus, a single guide RNA (sense = CACCGGGTTTGCTGCCCGGCGGAC, antisense = AAACGTCCGCCGGGCAGCAAACCC ) targeting the 5’ region of the gene was designed using the MIT Crispr Design Tool (www.crispr.mit.edu). The guide RNA was cloned into PSpCas9 (BB)-2A-GFP(PX458) (Addgene, 48138) following protocol from Ran et al., 2013. The final vector was sequence to ensure no mutations were generated during cloning. To generate a targeting vector for homology dependent repair, we cloned 5’ and 3’ 1kb long homology sequence (HA) flanking a (nuclear localization sequence) NLS region from H2B gene sequence, fluorescent protein mCherry sequence and self-cleaving P2A peptide sequence (5’HA-H2B-mCherry-P2A-3’HA) in a pUC19 vector backbone using Gibson Assembly (New England Biolabs (NEB), E5510S). To mutate the PAM site for the guide RNA, the assembled targeting vector was mutated using site-directed mutagenesis using In-Fusion HD Cloning Plus (Takara, 638909). NCRM1 iPSCs were transfected with both Guide RNA vector (PSpCas9 (BB)-2A-GFP) and targeting vector (pUC19–5’HA-H2B-mCherry-P2A-3’HA) using Lipofectamine™ Stem Transfection Reagent (Thermo Fisher Scientific, STEM00001). 24-hours after transfection, cells were sorted using flow cytometry (S3 cell sorter, Biorad) for GFP positive cells and plated at low density for clonal expansion in Matrigel (Corning, #354263) coated culture plates (Corning, 353046) in mTeSR (Stemcell Technologies, 85850) supplemented with 10μM Y-27632 dihydrochloride (R&D Systems, 1254/10) and Penicillin/Streptomycin (Life Technologies, 15140122). After appearance of small colonies, the colonies were sub-cultured and genotyped using PCR for targeted homozygous insertion of the H2B-mCherry-P2A in the UCP1 locus before the transcription start site. Positive clones were sequenced and clones with no undesired mutation were further validated using immunofluorescence and RT-qPCR. Two positive clones were differentiated into adipocytes and expression of UCP1 in mitochondria and mCherry in the nucleus was confirmed. To generate the human PAX3-Venus iPSC line using the CRISPR/Cas9 technology we followed a similar strategy. We generated a Venus knock-in allele by inserting the Venus sequence in front of the coding sequence of exon 1. Guide RNA (sense = 5’-CCGGCCAGCGTGGTCATCCT GGG-3’, antisense = 5’-TGCCCCCAGGATGACCACGC TGG-3’) targeting the 5’ region of the gene was designed using the MIT Crispr Design Tool (www.crispr.mit.edu). The targeting plasmid (pBSKS-2A-3xNLS) was designed to contain NLS sequence, 1.5 kb of the 5’ genomic region of the PAX3 gene, 1 kb of the 3’ sequence and 2A sequence. Targeting vector, together with the Cas9 plasmid was electroporated into cells by nucleofection and clones were sub-cultured and genotyped using PCR for targeted homozygous insertion of the NLS-Venus-2A in the PAX3 locus.
RNA extraction, Reverse transcription and real time quantitative PCR
Samples were harvested and RNA was extracted using NucleoSpin® RNA kit (Macherey and Nagel, 740955) following manufacturer’s instructions. DNA digestion was performed on column and RNA quality and concentration was measured using Nanodrop. For reverse transcription, 1ug RNA was in a 20ul reaction volume to generate cDNA using iScript™ cDNA Synthesis Kit (Bio-Rad, 1708891). Typically, cDNA was diluted 1:10 with nuclease free water. For real time quantitative PCR, 3.5ul of cDNA, 1.5ul of 10μM forward and reverse primer mix (300μM of forward and reverse primers) and 5ul of iTaq™ Universal SYBR® Green Supermix (Bio-Rad, 172–5124) was used for 10ul reaction volume. For each sample and gene primer set, 3 technical replicates were performed. PCR primers were designed using primer 3, typically spanning splice junctions wherever possible. Primers were validated for amplification efficiency and specificity using a standard curve and melting curves, respectively. PCRs were run on a Bio-Rad CFX384 thermocycler with the following cycling program: initial denaturation step (95°C for 1 minute), 40 cycles of amplification and SYBR green signal detection (denaturation at 95°C for 5 seconds, annealing/extension and plate read at 60°C for 40 seconds), followed by final rounds of gradient annealing from 65°C to 95°C to generate dissociation curves. Relative gene expression was calculated using ΔΔCt method and RPL37A (ribosomal protein L37a) was used as a housekeeping gene. A list of qPCR primers is provided in Table S2.
Immunocytochemistry
Cultured cells:
Cells were cultured on Matrigel coated plastic culture plates (Corning, #353046), glass bottom 24-well plates (Cellvis, P24–1.5H-N), μ-Dish 35 mm (Ibidi, 81156) or μ-Plate 24 Well Black (Ibidi, 82406). Cultures were washed with 1X Phosphate Buffered Saline (PBS, Sigma-Aldrich, P5493) and fixed with 4% Paraformaldehyde (Electron Microscopy Sciences, 15710) for 20 minutes at room temperature. Cultures were washed with PBS and stored in PBS supplemented with 0.2% Sodium Azide (Sigma-Aldrich, 71290) until used for immunostaining.
Mouse tissues:
Mouse embryos were dissected out at different stages in Hanks’ Balanced Salt Solution (HBSS) (Gibco, 14185–052). After several washes in HBBS, embryos were fixed in 4% Paraformaldehyde (Electron Microscopy Sciences, 15710) overnight at 4°C. Embryos were then washed in PBS and transferred to 15% sucrose solution (Sigma-Aldrich, 84097) in PBS overnight at 4°C followed by incubation in 30% sucrose solution for overnight at 4°C. Embryos were then embedded in Tissue-Tek O.C.T. Compound (VWR, 25608–930) and stored at −80°C until sectioned. 10–20μm thick sections were cut and stored at −20°C until used for immunostaining.
Fixed cells or sections were permeabilized using 1% Triton-X (Millipore Sigma, T8787) for 10 minutes at room temperature. Samples were washed in PBS and were incubated in blocking solution [PBS supplemented with 3% donkey serum (Jackson ImmunoResearch, 017–000-121) and 0.1% Triton-X] for 1 hour at room temperature. Samples were then incubated with primary antibody diluted in blocking solution at 4°C overnight. Next day, samples were washed 3 times with PBS for 5 minutes. Samples were then incubated with secondary antibody and Hoechst 33342 (ThermoFisher Scientific, H3570) diluted in blocking solution for 1 hour at room temperature followed by 3 washes with PBS for 5 minutes each. Cultured cell samples were stored in PBS until imaged. Mouse tissue slides were mounted using Fluoromount aqueous mounting medium (Sigma-Aldrich, F4680) and were stored at 4°C until analyzed. Samples were imaged using EVOS FL imaging system (Thermo Fisher Scientific) or LSM780 confocal microscope (Zeiss). A list of primary and secondary antibodies is provided in the Key Resource Table.
Lipid staining
iPSC-BA cultures were washed with 1X Phosphate Buffered Saline (PBS, Sigma-Aldrich, P5493) and fixed with 4% Paraformaldehyde (Electron Microscopy Sciences, 15710) for 20 minutes at room temperature. Cell were then incubated with 0.5mM BODIPY™ (Invitrogen™, D3922) in PBS for 20 minutes at room temperature. Cells were washed 3 times with PBS and visualized with EVOS FL imaging system (Thermo Fisher Scientific).
Seahorse assay
Agilent Seahorse XFe96 Analyzer was used to measure oxygen consumption in live iPSC-BAs and precursor cells following manufacturer’s instructions. 30,000 iPSC-BAs and precursor cells were cultured on Matrigel (Corning, 354263) coated Seahorse XF96 cell culture microplates (Agilent Technologies, 101085–004). Assay medium was prepared using Seahorse XF DMEM (Agilent Technologies, 103575–100) supplemented with 1 mM pyruvate (Agilent Technologies, 103578–100), 2 mM glutamine (Agilent Technologies, 103579–100), and 10 mM glucose (Agilent Technologies, 103577–100). Basal respiration was measured 3 times in the assay medium. Cells were then treated with 1.5μM Oligomycin (Tocris, 4110) and 3 measurements were taken. Cells were then treated with 10μM Forskolin (Sigma-Aldrich, F6886) or DMSO (Sigma-Aldrich, D2650) and oxygen consumption rate (OCR) was measured for 70 minutes (12 measurements). To block the electron transport chain, cells were then treated with 1μM Rotenone (Sigma-Aldrich, R8875) and Antimycin A (Sigma-Aldrich, A8674) for 3 measurements. Cells were then washed with 1X Phosphate Buffered Saline (PBS, Sigma-Aldrich, P5493) and fixed with 4% Paraformaldehyde (Electron Microscopy Sciences, 15710) for 10 minutes at room temperature. Seahorse XF96 cell culture microplates were imaged for DAPI and mCherry using GE INCELL Analyzer 2200 Widefield High-Content Imager. Cells were automatically segmented using custom code in Fiji and number of DAPI and mCherry positive cells were quantified. OCR data from each well was normalized with DAPI positive cell number in each well. Four replicates were performed. Each experiment consisted of 40 technical replicates for each cell type.
Thermogenesis assay
iPSC-BAs and iPSC precursors were cultured on Matrigel (Corning, 354263) coated 96-well Black Clear-Bottom Plates (Corning, 3603) and were incubated in DMEM/H with 250 nM ERthermAC (Sigma, SCT057) for 30 min at 37°C. After washing with PBS, fresh 90μl DMEM/H without phenol red was added prior to imaging. Fluorescence in stained cells were detected with a GloMax Discover Multimode Detection System (Promega) using 520 nm excitation and emission at 590 nm. Temperature inside the machine was equilibrated at 25°C. After measuring 3 points of basal fluorescence, 10μl forskolin (final concentration 10μM, F6886, Sigma) was added to initiate thermogenesis.10 μl PBS was added for vehicle control group. Fluorescence was recorded every 5 min over 120 min. Results are interpreted as relative intensity (intensity was normalized to basal measurements).
Electron microscopy
iPSC-BAs were rinsed with 0.1M Phosphate buffer pH 7.5 and fixed in 2.5% glutaraldehyde (Sigma, G7651) in 0.1M phosphate buffer at 4°C, for 2 days. Samples were washed in 0.1M phosphate buffer and post-fixed in 1% osmic acid in 0.1M phosphate buffer for 1 hour at room temperature. Dehydration was performed by serial incubation in 50%, 70%, 80%, 95% and 100% ethanol before incubation in propylene oxide for 30 minutes. Samples were impregnated by incubation in propylene oxide/araldite (1:1 v/v) for 60–90 min, in propylene oxide/araldite (1:2 v/v) for 1 hour and in 100% araldite overnight at 4°C. The samples were finally incubated in Araldite which was allowed to polymerize for 24 hours at 56°C. Ultrathin sections (85nm) were cut with a ultracut Leica EM UC. Sections were contrasted 8 min with 2% uranyl acetate and 8 min with lead citrate (Reynolds). All pictures were taken with the Zeiss EM 900 microscope and a GATAN camera (Orius SC 1000).
Glycerol release assay
iPSC-BAs were cultured in a (Corning, 354263) coated 96-well culture plate (Genesee Scientific, 25–109). Cells were serum starved for 16 hours and cultured in serum-free DMEM/F12 GlutaMAX (Thermo Fisher Scientific, 10565042) containing 0.5% BSA. Next day, cells were washed with KRB-HEPES buffer [118.5 mM NaCl, 4.75 mM KCl, 1.92 mM CaCl2, 1.19 mM KH2PO4, 1.19 mM MgSO4, 25 mM NaHCO3, 6 mM glucose and 10 mM HEPES, pH 7.4] containing 4% fatty-acid-free BSA (Bioworld, 22070017–1). Cells were treated with DMSO or 10μM Forskolin in KRB-HEPES buffer supplemented with 4% fatty-acid-free BSA for at 37°C, 5% CO2 for 4 hours. Cell culture medium was collected for glycerol measurement using the free glycerol reagent (Sigma-Aldrich, F6428) following manufacturer’s instructions. A standard curve was generated using Glycerol Standard Solution (Sigma-Aldrich, G7793). Test samples, standards and water control were incubated with the Free Glycerol Reagent for 5 minutes at 37°C and absorbance was recorded at A540 using a spectrophotometer. To normalize with total protein content, Bradford assay was performed using DC Protein Assay (Biorad, 500–0116) following manufacturers protocol.
Periodic Acid-Schiff staining
To stain glycogen in differentiating iPSC-BAs, cells were treated with Periodic Acid-Schiff stain (Sigma, 395B). iPSC-BAs were fixed with 4% paraformaldehyde (Electron Microscopy Sciences, 15710) at room temperature for 15 minutes. Fixed cells were washed several times with distilled water and immersed in Periodic Acid Solution for 5 minutes at room temperature. Cells were then washed in distilled water and incubated with Schiff’s Reagent for 15 minutes at room temperature. Cells were washed in running tap water for 5 minutes. Stained cells were visualized in brightfield using EVOS FL imaging system (Thermo Fisher Scientific).
Bulk mRNA sequencing
UCP1-mCherry knock-in line was differentiated into BAs as described above. Cells were harvested on day +40 of differentiation and RNA was extracted using NucleoSpin® RNA kit (Macherey and Nagel, 740955) following manufacturer’s instructions. DNA digestion was performed on column and RNA quality and concentration was measured using Nanodrop. Human fetal tissues were harvested as described above and RNA was extracted using NucleoSpin® RNA kit following manufacturer’s instructions.
RNA library preparations and sequencing reactions were conducted at GENEWIZ, LLC. (South Plainfield, NJ, USA). RNA samples received were quantified using Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA) and RNA integrity was checked using Agilent TapeStation 4200 (Agilent Technologies, Palo Alto, CA, USA). RNA sequencing libraries were prepared using the NEBNext Ultra II RNA Library Prep Kit for Illumina using manufacturer’s instructions (NEB, Ipswich, MA, USA). Briefly, mRNAs were initially enriched with Oligod(T) beads. Enriched mRNAs were fragmented for 15 minutes at 94°C. First strand and second strand cDNA were subsequently synthesized. cDNA fragments were end repaired and adenylated at 3’ends, and universal adapters were ligated to cDNA fragments, followed by index addition and library enrichment by PCR with limited cycles. The sequencing library was validated on the Agilent TapeStation (Agilent Technologies, Palo Alto, CA, USA), and quantified by using Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA) as well as by quantitative PCR (KAPA Biosystems, Wilmington, MA, USA).
The sequencing libraries were clustered on a single lane of a flowcell. After clustering, the flowcell was loaded on the Illumina HiSeq instrument (4000 or equivalent) according to manufacturer’s instructions. The samples were sequenced using a 2×150bp Paired End (PE) configuration. Image analysis and base calling were conducted by the HiSeq Control Software (HCS). Raw sequence data (.bcl files) generated from Illumina HiSeq was converted into fastq files and de-multiplexed using Illumina’s bcl2fastq 2.17 software. One mismatch was allowed for index sequence identification.
For bulk RNA-seq data analysis, we performed quality control on the sequence data (FastQ files) using FastQC (v0.11.9) and used STAR RNA-seq aligner (v2.7.3a) 61 to map the sequenced reads to the human reference genome (GRCh38 release 101 from ENSEMBL). Mapped reads were quantified using featureCounts (v2.0.1) 62. Starting from the raw gene counts, normalization and differential expression analysis were done in the R environment (v3.6.0), using the DESeq2 package (v 1.22.2) 63. Genes were defined as differentially expressed when the false discovery rate was lower than 0.05. Gene Ontology (GO) enrichment analysis was performed on the differentially expressed genes using EnrichR 64, choosing the WikiPathway 2019 Human resource.
To quantify the thermogenic potential of each sample we relied on the ProFAT webtool 37. Starting from the raw count matrix, barplot and heatmap, representing the adipose browning capacity of each sample, were generated as standard output of ProFAT.
To produce the heat-map for genes of interest (linked to muscle, pluripotency and BAT) we used normalized read counts produced by DESeq2. To calculate up- or downregulation, we computed the log difference of the average of the biological replicates against the baseline value (average over all conditions) for each gene. All data generated in this study are available at GSE185518.
Single cell RNA sequencing
Preparation of single cell suspension
Single cell analysis of differentiating iPSCs and mouse embryos cells was performed using inDrops as previously reported (Klein et al.).
UCP1-mCherry iPSC line was differentiated into BAs as described above. Cultures were harvested on d20, d40 and d60.
The cultures were washed with PBS (Gibco, 14190) and dissociated into single cells using 2.5mg/ml Collagenase, (Type IV, Thermo Fisher Scientific, 17104019) and 0.05% Trypsine EDTA (Thermo Fisher Scientific, 25200–056) in PBS for 5–15 minutes at 37ºC. Dissociated cells were run through 70μm cell strainer (Celltreat, 229483) followed by 30μm cell strainer (CellTrics, 04–0042-2316), spun at 300g for 5 minutes and resuspended in DMEM (Thermo Fisher Scientific, 11965–118) + 5% fetal bovine serum (VWR, 89510–186). Cells were again spun at 300g for 5 minutes and resuspended in DMEM + 5% fetal bovine serum, this was repeated 2–3 times to remove debris. Final resuspension was made in PBS + 0.1% bovine serum albumin (Gibco, 15260–037). Cell density was adjusted to 200,000 cells/ml. For day 20, 2500 cells were collected from each sample. For samples at d40 and d60, at least 3000 cells were collected from each sample from two independent differentiations.
For analyzing mouse embryonic tissues, embryos were collected on embryonic day (E) 11.5, 12.5, 13.5, 14.5 and 15.5 from wildtype CD1 IGS mice (Charles River). For each stage, back tissue dorsal to rib cage at the level of forelimb was dissected out. Neural tube/spinal cord and dorsal root ganglion were removed wherever possible. For E15.5, epidermis and underlying dermis was removed before dissociating the tissue. Dissected tissues were washed several times in Hanks’ Balanced Salt Solution (HBSS, Thermo Fisher Scientific, 14170112). Tissues were transferred to microfuge tube with 200μl of 2.5mg/ml Collagenase, (Type IV, Thermo Fisher Scientific, 17104019) and 0.05% Trypsine EDTA (Thermo Fisher Scientific, 25200–056) in PBS (Gibco, 14190), chopped into small pieces using scissors and incubated at 37ºC for 15 minutes with intermittent shaking. Tissues were mechanically dissociated by triturating several times using wide-bore 1ml pipette. The resulting cell suspension was mixed with HBBS + 10% FBS, filtered through a 30μm cell strainer and spun at 300g for 5 minutes. To remove red blood cells, cell pellet was resuspended and incubated with RBC Lysis Solution (Qiagen, 158902) for 5 minutes at room temperature. Cells were washed with HBSS + 10% FBS for 2–3 times to remove debris. Final suspension was made in PBS + 0.1% BSA. Cell density was adjusted to 200,000/ml. For each stage, cells were collected from two littermate embryos. For E11.5–14.5 embryo, 3000 cells were encapsulated and sequenced from each embryo. For E15.5, 5000 cells were collected and sequenced from each embryo.
Encapsulation, sequencing and analysis
Single cells were encapsulated using inDrops technique as reported previously (Klein et al.). Cells were barcoded using v3 sequencing adapters and were sequenced on an Illumina NextSeq 500 using the NextSeq 75 High Output Kits using standard Illumina sequencing primers and 61 cycles for Read1, 14 cycles for Read2, 8 cycles each for IndexRead1 and IndexRead2. Sequence FASTQ files were processed according to indrops.py pipeline (available at github.com/indrops/indrops). Single cell transcriptomes were mapped to mouse (GRCm38/mm10) and human (GRCh38/hg19) reference transcriptomes. Samtools version 1.3.1, rsem version 1.3.0 and Bowtie version 1.2.2 was used with parameter –e 100.
A weighted histogram of transcript counts per cell barcode vs cell barcode abundance was used to identify transcripts originating from abundant cell barcodes. Only transcript counts originating from abundant cell barcodes were included in downstream analysis. Basic filtering parameters were used to exclude cells expressing <500 genes and genes expressed in less than 3 cells. The filtered counts were normalized by total number of counts for each biological sample. Top 1000 variable genes were identified according to Satija 2015. Cell doublets were identified using Scrublet and filtered out 65. For each cell, fraction of counts due to mitochondrial genes was determined and cells with high fraction were filtered out. The cell cycle was scored as in Tirosh et al. 2016 66. Each cell was given a cell cycle score based on the expression of G2/M and S phase markers. The cells not expressing the markers from G2/M and S phase were identified to be in G0/G1 stage. Source of variation between the libraries were regressed out using bkknn batch correction function 67. Single cell data were projected into a low dimensional space by principal component analysis (PCA). UMAP 68 was used to embed the neighborhood graph. Cell clusters were identified using Leiden graph-clustering method 69. Differentially expressed genes were identified by a Wilcoxon rank-sum test by comparing cells of each cluster with cells of all the other clusters.
Tracksplot were made using each gene are plotted as a filled line plot where the y are the genes processed expression values and x is each of the cells 70. Stream: Stream pipeline was used to layout the trajectories inside each dataset. Original code and documentation: 25 Neural cells were removed from the dataset analyzed with stream as their developmental origin differ from the rest of the clusters.
Trajectory inference analysis was done using Cellrank 71. Waddington-OT kernel 26was used to infer the temporal couplings of cells from the different samples collected independently at various timepoints. Transition matrices were calculated to show the amount of mass transported from a cell type to another from a start and an end point.
Single cell RNA sequencing data from mouse E18 periaortic (pVAT) BAT described in Angueira et al. 2021 was processed through the same processing pipeline as described above. Cell state predictions on single cell clusters from pVAT described in Angueira et al. 2021 were made using a kNN-classifier trained on our E13.5-E15.5 mouse single cell clusters 23.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed using GraphPad Prism (v8.1.1). Statistical parameters such as the value of n, mean, standard deviation (SD), p values, and the statistical tests used are reported in the figure legends. For hiPSC differentiation experiments, the “n” refers to the number of independent hiPSC differentiation experiments analyzed. In mouse embryo studies, “n” indicates the number of embryos from which samples were obtained. In human primary tissue studies, “n” indicates the number of randomly selected imaging regions from each specimen. All bar graphs and line graphs are displayed as mean ± SD. Paired or unpaired student’s t-tests were used for two-sample comparisons.
Supplementary Material
Table S1 – List of marker genes in Leiden clusters identified in E11.5-E15.5 mouse single cell sequencing data. cluster_n =cluster name, cluster_s=cluster z-score, cluster_l=cluster log fold change, cluster_p=cluster adjusted p value (Wilcoxon-Rank-Sum test, Benjamini-Hochberg correction method). Related to Figure S1, Figure 4 and Figure S5.
Table S2 – List of RT-qPCR primer used in this study.
HIGHLIGHTS.
A single cell RNA sequencing census of developing mouse brown adipose tissue
GATA6 identifies a population of brown adipocyte precursors
Directed differentiation of human pluripotent stem cells towards brown adipocytes
ACKNOWLEDGEMENTS
We thank the Pourquié lab members for helpful discussions. We also acknowledge Sam Wolock and Dan Wagner for their help with the scRNAseq analysis. We thank Anne Loyens for help with electron microscopy and Matteo Battilocchi for help with the Gata6 lineage tracing experiments in mouse. We thank the NeuroTechnology Studio at Brigham and Women’s Hospital for providing instrument access and consultation on data analysis.
INCLUSION AND DIVERSITY
We support inclusive, diverse and equitable conduct of research.
Footnotes
DECLARATION OF INTERESTS
J.R. and O.P. have filed a patent related to this work. The authors declare no other competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Merklin RJ (1974). Growth and distribution of human fetal brown fat. Anat Rec 178, 637–645. 10.1002/ar.1091780311. [DOI] [PubMed] [Google Scholar]
- 2.Heaton JM (1972). The distribution of brown adipose tissue in the human. J Anat 112, 35–39. [PMC free article] [PubMed] [Google Scholar]
- 3.Lidell ME (2019). Brown Adipose Tissue in Human Infants. Handb Exp Pharmacol 251, 107–123. 10.1007/164_2018_118. [DOI] [PubMed] [Google Scholar]
- 4.van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, Schrauwen P, and Teule GJ (2009). Cold-activated brown adipose tissue in healthy men. The New England journal of medicine 360, 1500–1508. 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 5.Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, Taittonen M, Laine J, Savisto NJ, Enerback S, and Nuutila P. (2009). Functional brown adipose tissue in healthy adults. The New England journal of medicine 360, 1518–1525. 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 6.Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. (2008). PRDM16 controls a brown fat/skeletal muscle switch. Nature 454, 961–967. 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang W, Kissig M, Rajakumari S, Huang L, Lim HW, Won KJ, and Seale P. (2014). Ebf2 is a selective marker of brown and beige adipogenic precursor cells. Proc Natl Acad Sci U S A 111, 14466–14471. 10.1073/pnas.1412685111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez-Gurmaches J, and Guertin DA (2014). Adipocytes arise from multiple lineages that are heterogeneously and dynamically distributed. Nature communications 5, 4099. 10.1038/ncomms5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sebo ZL, Jeffery E, Holtrup B, and Rodeheffer MS (2018). A mesodermal fate map for adipose tissue. Development 145. 10.1242/dev.166801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atit R, Sgaier SK, Mohamed OA, Taketo MM, Dufort D, Joyner AL, Niswander L, and Conlon RA (2006). Beta-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Dev Biol 296, 164–176. 10.1016/j.ydbio.2006.04.449. [DOI] [PubMed] [Google Scholar]
- 11.Lepper C, and Fan CM (2010). Inducible lineage tracing of Pax7-descendant cells reveals embryonic origin of adult satellite cells. Genesis 48, 424–436. 10.1002/dvg.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schulz TJ, and Tseng YH (2013). Brown adipose tissue: development, metabolism and beyond. Biochem J 453, 167–178. 10.1042/BJ20130457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samuelson I, and Vidal-Puig A. (2020). Studying Brown Adipose Tissue in a Human in vitro Context. Frontiers in endocrinology 11, 629. 10.3389/fendo.2020.00629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahfeldt T, Schinzel RT, Lee YK, Hendrickson D, Kaplan A, Lum DH, Camahort R, Xia F, Shay J, Rhee EP, et al. (2012). Programming human pluripotent stem cells into white and brown adipocytes. Nat Cell Biol 14, 209–219. 10.1038/ncb2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeda Y, Harada Y, Yoshikawa T, and Dai P. (2017). Direct conversion of human fibroblasts to brown adipocytes by small chemical compounds. Sci Rep 7, 4304. 10.1038/s41598-017-04665-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hafner AL, Contet J, Ravaud C, Yao X, Villageois P, Suknuntha K, Annab K, Peraldi P, Binetruy B, Slukvin II, et al. (2016). Brown-like adipose progenitors derived from human induced pluripotent stem cells: Identification of critical pathways governing their adipogenic capacity. Sci Rep 6, 32490. 10.1038/srep32490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guenantin AC, Briand N, Capel E, Dumont F, Morichon R, Provost C, Stillitano F, Jeziorowska D, Siffroi JP, Hajjar RJ, et al. (2017). Functional Human Beige Adipocytes From Induced Pluripotent Stem Cells. Diabetes 66, 1470–1478. 10.2337/db16-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishio M, Yoneshiro T, Nakahara M, Suzuki S, Saeki K, Hasegawa M, Kawai Y, Akutsu H, Umezawa A, Yasuda K, et al. (2012). Production of functional classical brown adipocytes from human pluripotent stem cells using specific hemopoietin cocktail without gene transfer. Cell Metab 16, 394–406. 10.1016/j.cmet.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, Avery J, Yin A, Singh AM, Cliff TS, Yin H, and Dalton S. (2020). Generation of Functional Brown Adipocytes from Human Pluripotent Stem Cells via Progression through a Paraxial Mesoderm State. Cell stem cell 27, 784–797 e711. 10.1016/j.stem.2020.07.013. [DOI] [PubMed] [Google Scholar]
- 20.Carobbio S, Guenantin AC, Bahri M, Rodriguez-Fdez S, Honig F, Kamzolas I, Samuelson I, Long K, Awad S, Lukovic D, et al. (2021). Unraveling the Developmental Roadmap toward Human Brown Adipose Tissue. Stem cell reports 16, 641–655. 10.1016/j.stemcr.2021.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chal J, Oginuma M, Al Tanoury Z, Gobert B, Sumara O, Hick A, Bousson F, Zidouni Y, Mursch C, Moncuquet P, et al. (2015). Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy. Nat Biotechnol 33, 962–969. 10.1038/nbt.3297. [DOI] [PubMed] [Google Scholar]
- 22.Chal J, Al Tanoury Z, Hestin M, Gobert B, Aivio S, Hick A, Cherrier T, Nesmith AP, Parker KK, and Pourquie O. (2016). Generation of human muscle fibers and satellite-like cells from human pluripotent stem cells in vitro. Nature protocols 11, 1833–1850. 10.1038/nprot.2016.110. [DOI] [PubMed] [Google Scholar]
- 23.Diaz-Cuadros M, Wagner DE, Budjan C, Hubaud A, Tarazona OA, Donelly S, Michaut A, Al Tanoury Z, Yoshioka-Kobayashi K, Niino Y, et al. (2020). In vitro characterization of the human segmentation clock. Nature 580, 113–118. 10.1038/s41586-019-1885-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, and Kirschner MW (2015). Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201. 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H, Albergante L, Hsu JY, Lareau CA, Lo Bosco G, Guan J, Zhou S, Gorban AN, Bauer DE, Aryee MJ, et al. (2019). Single-cell trajectories reconstruction, exploration and mapping of omics data with STREAM. Nature communications 10, 1903. 10.1038/s41467-019-09670-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schiebinger G, Shu J, Tabaka M, Cleary B, Subramanian V, Solomon A, Gould J, Liu S, Lin S, Berube P, et al. (2019). Optimal-Transport Analysis of Single-Cell Gene Expression Identifies Developmental Trajectories in Reprogramming. Cell 176, 928–943 e922. 10.1016/j.cell.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Angueira AR, Sakers AP, Holman CD, Cheng L, Arbocco MN, Shamsi F, Lynes MD, Shrestha R, Okada C, Batmanov K, et al. (2021). Defining the lineage of thermogenic perivascular adipose tissue. Nat Metab 3, 469–484. 10.1038/s42255-021-00380-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donati G, Rognoni E, Hiratsuka T, Liakath-Ali K, Hoste E, Kar G, Kayikci M, Russell R, Kretzschmar K, Mulder KW, et al. (2017). Wounding induces dedifferentiation of epidermal Gata6(+) cells and acquisition of stem cell properties. Nat Cell Biol 19, 603–613. 10.1038/ncb3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrisey EE, Ip HS, Lu MM, and Parmacek MS (1996). GATA-6: a zinc finger transcription factor that is expressed in multiple cell lineages derived from lateral mesoderm. Dev Biol 177, 309–322. 10.1006/dbio.1996.0165. [DOI] [PubMed] [Google Scholar]
- 30.Shamsi F, Piper M, Ho LL, Huang TL, Gupta A, Streets A, Lynes MD, and Tseng YH (2021). Vascular smooth muscle-derived Trpv1(+) progenitors are a source of cold-induced thermogenic adipocytes. Nat Metab 3, 485–495. 10.1038/s42255-021-00373-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hutcheson DA, Zhao J, Merrell A, Haldar M, and Kardon G. (2009). Embryonic and fetal limb myogenic cells are derived from developmentally distinct progenitors and have different requirements for beta-catenin. Genes Dev 23, 997–1013. 10.1101/gad.1769009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al Tanoury Z, Rao J, Tassy O, Gobert B, Gapon S, Garnier JM, Wagner E, Hick A, Hall A, Gussoni E, and Pourquie O. (2020). Differentiation of the human PAX7-positive myogenic precursors/satellite cell lineage in vitro. Development. 10.1242/dev.187344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tseng YH, Kokkotou E, Schulz TJ, Huang TL, Winnay JN, Taniguchi CM, Tran TT, Suzuki R, Espinoza DO, Yamamoto Y, et al. (2008). New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature 454, 1000–1004. 10.1038/nature07221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Longo KA, Wright WS, Kang S, Gerin I, Chiang SH, Lucas PC, Opp MR, and MacDougald OA (2004). Wnt10b inhibits development of white and brown adipose tissues. J Biol Chem 279, 35503–35509. 10.1074/jbc.M402937200. [DOI] [PubMed] [Google Scholar]
- 35.Mayeuf-Louchart A, Lancel S, Sebti Y, Pourcet B, Loyens A, Delhaye S, Duhem C, Beauchamp J, Ferri L, Thorel Q, et al. (2019). Glycogen Dynamics Drives Lipid Droplet Biogenesis during Brown Adipocyte Differentiation. Cell reports 29, 1410–1418 e1416. 10.1016/j.celrep.2019.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Al Tanoury Z, Zimmermann J, Rao J, Siero D, McNamara HM, Cherrrier T, and al., E. (2021). Prednisolone rescues Duchenne Muscular Dystrophy phenotypes in human pluripotent stem cells-derived skeletal muscle in vitro PNAS in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng Y, Jiang L, Keipert S, Zhang S, Hauser A, Graf E, Strom T, Tschop M, Jastroch M, and Perocchi F. (2018). Prediction of Adipose Browning Capacity by Systematic Integration of Transcriptional Profiles. Cell reports 23, 3112–3125. 10.1016/j.celrep.2018.05.021. [DOI] [PubMed] [Google Scholar]
- 38.Wang W, and Seale P. (2016). Control of brown and beige fat development. Nat Rev Mol Cell Biol 17, 691–702. 10.1038/nrm.2016.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang CH, Lundh M, Fu A, Kriszt R, Huang TL, Lynes MD, Leiria LO, Shamsi F, Darcy J, Greenwood BP, et al. (2020). CRISPR-engineered human brown-like adipocytes prevent diet-induced obesity and ameliorate metabolic syndrome in mice. Science translational medicine 12. 10.1126/scitranslmed.aaz8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kriszt R, Arai S, Itoh H, Lee MH, Goralczyk AG, Ang XM, Cypess AM, White AP, Shamsi F, Xue R, et al. (2017). Optical visualisation of thermogenesis in stimulated single-cell brown adipocytes. Sci Rep 7, 1383. 10.1038/s41598-017-00291-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanchez-Gurmaches J, and Guertin DA (2014). Adipocyte lineages: tracing back the origins of fat. Biochim Biophys Acta 1842, 340–351. 10.1016/j.bbadis.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koutsourakis M, Langeveld A, Patient R, Beddington R, and Grosveld F. (1999). The transcription factor GATA6 is essential for early extraembryonic development. Development 126, 723–732. [PubMed] [Google Scholar]
- 43.Freyer L, Schroter C, Saiz N, Schrode N, Nowotschin S, Martinez-Arias A, and Hadjantonakis AK (2015). A loss-of-function and H2B-Venus transcriptional reporter allele for Gata6 in mice. BMC developmental biology 15, 38. 10.1186/s12861-015-0086-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kozhemyakina E, Ionescu A, and Lassar AB (2014). GATA6 is a crucial regulator of Shh in the limb bud. PLoS genetics 10, e1004072. 10.1371/journal.pgen.1004072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alexandrovich A, Qureishi A, Coudert AE, Zhang L, Grigoriadis AE, Shah AM, Brewer AC, and Pizzey JA (2008). A role for GATA-6 in vertebrate chondrogenesis. Dev Biol 314, 457–470. 10.1016/j.ydbio.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 46.Yao CX, Xiong CJ, Wang WP, Yang F, Zhang SF, Wang TQ, Wang SL, Yu HL, Wei ZR, and Zang MX (2012). Transcription factor GATA-6 recruits PPARalpha to cooperatively activate Glut4 gene expression. J Mol Biol 415, 143–158. 10.1016/j.jmb.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 47.Schulz TJ, Huang P, Huang TL, Xue R, McDougall LE, Townsend KL, Cypess AM, Mishina Y, Gussoni E, and Tseng YH (2013). Brown-fat paucity due to impaired BMP signalling induces compensatory browning of white fat. Nature 495, 379–383. 10.1038/nature11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bagchi DP, and MacDougald OA (2021). Wnt Signaling: From Mesenchymal Cell Fate to Lipogenesis and Other Mature Adipocyte Functions. Diabetes 70, 1419–1430. 10.2337/dbi20-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mullur R, Liu YY, and Brent GA (2014). Thyroid hormone regulation of metabolism. Physiol Rev 94, 355–382. 10.1152/physrev.00030.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohta H, and Itoh N. (2014). Roles of FGFs as Adipokines in Adipose Tissue Development, Remodeling, and Metabolism. Frontiers in endocrinology 5, 18. 10.3389/fendo.2014.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Loureiro Z, Solivan-Rivera J, and Corvera S. (2022). Adipocyte Heterogeneity Underlying Adipose Tissue Functions. Endocrinology 163. 10.1210/endocr/bqab138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emont MP, Jacobs C, Essene AL, Pant D, Tenen D, Colleluori G, Di Vincenzo A, Jorgensen AM, Dashti H, Stefek A, et al. (2022). A single-cell atlas of human and mouse white adipose tissue. Nature 603, 926–933. 10.1038/s41586-022-04518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun W, Dong H, Balaz M, Slyper M, Drokhlyansky E, Colleluori G, Giordano A, Kovanicova Z, Stefanicka P, Balazova L, et al. (2020). snRNA-seq reveals a subpopulation of adipocytes that regulates thermogenesis. Nature 587, 98–102. 10.1038/s41586-020-2856-x. [DOI] [PubMed] [Google Scholar]
- 54.Aad G, Abbott B, Abdallah J, Abdelalim AA, Abdesselam A, Abdinov O, Abi B, Abolins M, Abramowicz H, Abreu H, et al. Observation of a centrality-dependent dijet asymmetry in lead-lead collisions at sqrt[S(NN)] =2.76 TeV with the ATLAS detector at the LHC. Physical review letters 105, 252303. [DOI] [PubMed] [Google Scholar]
- 55.Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, et al. (2009). Identification and importance of brown adipose tissue in adult humans. The New England journal of medicine 360, 1509–1517. 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saito M, Okamatsu-Ogura Y, Matsushita M, Watanabe K, Yoneshiro T, Nio-Kobayashi J, Iwanaga T, Miyagawa M, Kameya T, Nakada K, et al. (2009). High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes 58, 1526–1531. 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stanford KI, Middelbeek RJ, Townsend KL, An D, Nygaard EB, Hitchcox KM, Markan KR, Nakano K, Hirshman MF, Tseng YH, and Goodyear LJ (2013). Brown adipose tissue regulates glucose homeostasis and insulin sensitivity. J Clin Invest 123, 215–223. 10.1172/JCI62308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gunawardana SC, and Piston DW (2012). Reversal of type 1 diabetes in mice by brown adipose tissue transplant. Diabetes 61, 674–682. 10.2337/db11-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muzumdar MD, Tasic B, Miyamichi K, Li L, and Luo L. (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605. 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 60.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13, 133–140. 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liao Y, Smyth GK, and Shi W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 63.Love MI, Huber W, and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44, W90–97. 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wolock SL, Lopez R, and Klein AM (2019). Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Syst 8, 281–291 e289. 10.1016/j.cels.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K, Fisher JM, Rodman C, Mount C, Filbin MG, et al. (2016). Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 539, 309–313. 10.1038/nature20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Polanski K, Young MD, Miao Z, Meyer KB, Teichmann SA, and Park JE (2020). BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics 36, 964–965. 10.1093/bioinformatics/btz625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McInnes L. (2018). UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv. 10.48550/arXiv.1802.03426. [DOI] [Google Scholar]
- 69.Blondel VD, Guillaume JL, Hendrickx JM, de Kerchove C, and Lambiotte R. (2008). Local leaders in random networks. Physical review 77, 036114. 10.1103/PhysRevE.77.036114. [DOI] [PubMed] [Google Scholar]
- 70.Wolf FA, Angerer P, and Theis FJ (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19, 15. 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lange M, Bergen V, Klein M, Setty M, Reuter B, Bakhti M, Lickert H, Ansari M, Schniering J, Schiller HB, et al. (2022). CellRank for directed single-cell fate mapping. Nature methods 19, 159–170. 10.1038/s41592-021-01346-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 – List of marker genes in Leiden clusters identified in E11.5-E15.5 mouse single cell sequencing data. cluster_n =cluster name, cluster_s=cluster z-score, cluster_l=cluster log fold change, cluster_p=cluster adjusted p value (Wilcoxon-Rank-Sum test, Benjamini-Hochberg correction method). Related to Figure S1, Figure 4 and Figure S5.
Table S2 – List of RT-qPCR primer used in this study.
Data Availability Statement
Single-cell RNA-seq data have been deposited at NCBI GEO under accession number GSE185518. Accession number is also listed in the key resources table.
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Cd34 | Thermo Fisher Scientific | 14-0341-82 |
| Dpp4 | R&D | AF954-SP |
| EBF-2 | R&D | AF7006 |
| Gata6 | RnD | AF1700-SP |
| Gata6 | Cell Signaling Technology | 5851T |
| GFP | SantaCruz | SC-101536 |
| GFP | Abcam | AB13970 |
| Keratin 14 | Biolegend | 906001 |
| mCherry | OriGene | AB0040-200 |
| mCherry | Neuromics | CH22115 |
| MTC02 | Abcam | AB3298 |
| Myh11 | Alfa Aesar | J64817 |
| MyHCfast | Sigma-Aldrich | M4276 |
| Perilipin 1 | Progen | GP29 |
| Perilipin-1 | Abcam | AB61682 |
| PPARG | Thermo Fisher Scientific | MA5-14889 |
| RFP | Rockland | 600-901-379 |
| RFP | Rockland | 600-401-379 |
| Sca-1/Ly6 | R&D | AF1226-SP |
| TBX6 | Abcam | AB38883 |
| UCP1 | R&D | MAB6158 |
| Donkey Anti-Rat IgG H&L (Alexa Fluor® 647) preadsorbed | Abcam | A-11015 |
| Donkey anti-Goat IgG (H+L) Secondary Antibody, Alexa Fluor® 488 conjugate | Thermo Fisher Scientific | A21206 |
| Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | A-21207 |
| Donkey anti-Sheep IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 | Thermo Fisher Scientific | A-31573 |
| Donkey anti-Sheep IgG (H+L) Secondary Antibody, Alexa Fluor® 488 conjugate | Life Technologies | A-31572 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | 703-545-155 |
| Donkey anti-Rabbit IgG (H+L) Secondary Antibody, Alexa Fluor® 594 | Thermo Fisher Scientific | 703-165-155 |
| Donkey anti-Rabbit IgG (H+L) Secondary Antibody, Alexa Fluor® 647 | Thermo Fisher Scientific | A-31571 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 | Thermo Fisher Scientific | 706-545-148 |
| Alexa Fluor® 488 AffiniPure Donkey Anti-Chicken IgY (IgG) (H+L) | Jackson ImmunoResearch Laboratories | 703-545-155 |
| Cy™3 AffiniPure Donkey Anti-Chicken IgY (IgG) (H+L) | Jackson ImmunoResearch Laboratories | 703-165-155 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | A-31571 |
| Alexa Fluor® 488 AffiniPure Donkey Anti-Guinea Pig IgG (H+L) | Jackson ImmunoResearch Laboratories | 706-545-148 |
| Bacterial and virus strains | ||
| Promega JM109 Competent E.Coli Cells | Thermo Fisher Scientific | PR-L1001 |
| Biological samples | ||
| Human fetal tissues | University of Washington Birth Defects Research Laboratory (BDRL) under a protocol approved by the University of Washington Institutional Review Board | 98 days (H28540), 115 days (H28572), 122 days (H28560), 125 days (H28626) |
| Chemicals, peptides, and recombinant proteins | ||
| Y-27632 dihydrochloride | R&D Systems | 1254/10 |
| CHIR 99021 | R&D Systems | 4423 |
| LDN-193189 | Stemgent | 04-0074 |
| FGF-2 | PeproTech | 450-33 |
| HGF | PeproTech | 315-23 |
| IGF-1 | PeproTech | 250-19 |
| TGFb1 | PeproTech | 100-21-10 |
| BMP7 | Thermo Fisher Scientific | PHC9544 |
| 3,3′,5-Triiodo-L-thyronine sodium salt | Sigma-Aldrich | T6397 |
| Porcn Inhibitor II C59 | Millipore Sigma | 500496 |
| SB431542 | Selleck Chemicals | S1067 |
| Dexamethasone | Sigma-Aldrich | D4902 |
| Hydrocortisone | Sigma-Aldrich | H0888 |
| Rosiglitazone | Sigma-Aldrich | R2408 |
| EGF | PeproTech | AF-100-15 |
| 3-Isobutyl-1-methylxanthine | Sigma-Aldrich | I7018 |
| ERthermAC | Sigma | SCT057 |
| Oligomycin | Tocris | 4110 |
| Forskolin | Sigma-Aldrich | F6886 |
| Antimycin A | Sigma-Aldrich | A8674 |
| Rotenone | Sigma-Aldrich | R8875 |
| 4% Paraformaldehyde | Electron Microscopy Sciences | 15710 |
| Critical commercial assays | ||
| NucleoSpin® RNA kit | Macherey and Nagel | 740955 |
| iScript™ cDNA Synthesis Kit | Bio-Rad | 1708891 |
| iTaq™ Universal SYBR® Green Supermix | Bio-Rad | 172-5124 |
| Deposited data | ||
| Single cell RNA seq data | This paper | GEO (GSE185623) |
| Bulk RNA seq data | This paper | GEO (GSE185518) |
| Experimental models: Cell lines | ||
| NCRM-1 (NIH CRM control iPSC line (male)) human iPSC | RUCDR Infinite Biologics | ND50028 |
| NCRM-1-UCP1-H2B-mCherry - 2 clones | This paper | N.A. |
| NCRM-1-PAX3-NLS-Venus - 2 clones | This paper | N.A. |
| Experimental models: Organisms/strains | ||
| Mouse : Pax7-iCre/+;Rosa26-mTmG/+ | This study; Gabrielle Kardon Lab | N.A. |
| Mouse : Gata6-EGFPCreERT2;Rosa26-fl/STOP/fl-tdTomato | This study; Fiona Watt Lab | N.A. |
| Mouse : wildtype CD1 IGS mice | Charles River | 022 |
| Oligonucleotides | ||
| RT-qPCR primers | This study | Table S2 |
| Recombinant DNA | ||
| PSpCas9 (BB)-2A-GFP(PX458) | Addgene | 48138 |
| pUC19 vector backbone | New England Biolabs | E5510S |
| pUC19-5’HA-H2B-mCherry-P2A-3’HA | This study | N.A. |
| Software and algorithms | ||
| ImageJ | Image J | https://imagej.nih.gov/ij/ |
| Prism 9 | GraphPad | https://www.graphpad.com |
| Single cell analysis code | This study | https://doi.org/10.5281/zenodo.8071624 |
| kNN-classifier | Diaz-Cuadros et al., 23 | N.A. |
| Stream | Chen et al., 25 | N.A. |
| Waddington-OT kernel | Schiebinger et al., 26 | N.A. |
| ProFAT webtool | Cheng et al., 37 | N.A. |
| FastQC (v0.11.9) and used STAR RNA-seq aligner (v2.7.3a) | Dobin et al., 61 | N.A. |
| featureCounts (v2.0.1) | Liao et al., 62 | N.A. |
| DESeq2 package (v 1.22.2) | Love et al., 63 | N.A. |
| EnrichR | Kuleshov et al., 64 | N.A. |
| Scrublet | Wolock et al., 65 | N.A. |
| Scanpy (v.1.6.0) | Wolf et al., 70 | N.A. |
| Cellrank | Lange et al., 71 | N.A. |
Microscopy data reported in this paper will be shared by the lead contact upon request. Raw and processed Bulk RNA-seq data have been deposited at NCBI GEO under accession number GSE185623 and are publicly available as of the date of publication. Accession number is also listed in the key resources table.
All original code has been deposited at https://github.com/PourquieLab/Djeffal_Rao_2023.git and is publicly available. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.