Abstract
Lipid accumulation in macrophages (Mφs) is a hallmark of atherosclerosis, yet how lipid accumulation affects inflammatory responses through rewiring of Mφ metabolism is poorly understood. We modeled lipid accumulation in cultured wild type mouse thioglycolate-elicited peritoneal Mφs and bone marrow-derived Mφs with conditional (Lys2-Cre) or complete genetic deficiency of Vhl, Hif1a, Nos2 and Nfe2l2. Transfection studies employed RAW264.7 cells. Mφs were cultured for 24 hours with oxidized LDL (oxLDL) or cholesterol and then were stimulated with LPS. Transcriptomics revealed that oxLDL accumulation in Mφs down-regulated inflammatory, hypoxia and cholesterol metabolism pathways, while antioxidant pathway, fatty acid oxidation and ABC family proteins were up-regulated. Metabolomics and extracellular metabolic flux assays showed that oxLDL accumulation suppressed LPS-induced glycolysis. Intracellular lipid accumulation in Mφs impaired LPS-induced inflammation by reducing both HIF-1α stability and transactivation capacity; thus, the phenotype was not rescued in Vhl−/− Mφs. Intracellular lipid accumulation in Mφs also enhanced LPS-induced Nrf2-mediated antioxidative defense that destabilizes HIF-1α, and Nrf2-deficient Mφs resisted the inhibitory effects of lipid accumulation on glycolysis and inflammatory gene expression. Furthermore, oxLDL shifted NADPH consumption from HIF-1α- to Nrf2-regulated apoenzymes. Thus, we postulate that repurposing NADPH consumption from HIF-1α to Nrf2 transcriptional pathways is critical in modulating inflammatory responses in Mφs with accumulated intracellular lipid. The relevance of our in vitro models was established by comparative transcriptomic analyses, which revealed that Mφs cultured with oxLDL and stimulated with LPS shared similar inflammatory and metabolic profiles with foamy Mφs derived from the atherosclerotic mouse and human aorta.
Keywords: Atherosclerosis, Oxidized LDL, Macrophage foam cells, Immunometabolism, HIF-1α, Nrf2, NADPH
Introduction
The intracellular accumulation of neutral lipid in macrophages (Mφs) is a hallmark of many inflammatory diseases, including atherosclerosis, multiple sclerosis, and cancer (1). Therefore, significant effort has been devoted in the past to determine the possible links between Mφ lipid accumulation and the onset of inflammation. In atherosclerosis, hypercholesterolemia is a key risk factor and lipid accumulation in resident aortic intimal Mφs is one of the first steps of atherogenesis (2). Given their importance, it was postulated that inflammation in atherosclerosis is a direct consequence of intrinsic lipid accumulation in intimal Mφs (3, 4). However, there are also reports that lipid accumulation reduced Mφ inflammatory responses, partially due to desmosterol-induced Liver X receptors activation (5), oxidized low-density lipoprotein (oxLDL)-induced reduction of p65/RelA binding to chromatin (6), and a defective pentose phosphate pathway (7). In vivo studies have also examined the link between intracellular lipid accumulation and inflammation. Our group has shown evidence of lipid accumulation in mouse intimal Mφs within days of hypercholesteremic diet initiation (2, 8), yet increased monocyte recruitment was not detected until 2 weeks (9). The delayed onset of inflammation suggests that lipid accumulation in Mφs alone is insufficient to drive inflammation. This notion was recently supported by other in vivo studies that showed the transcriptomic profile of “foamy” Mφs derived from atherosclerotic mouse and human aortas are less inflammatory than the “non-foamy” Mφs (10, 11).
While the emerging in vivo evidence has implicated that lipid accumulation in Mφs is intrinsically not inflammatory, mechanistic evidence to support this notion is still lacking. In recent years, advances in the immunometabolism field have shown how activated immune cells rewire their metabolism to induce a fine-tuned inflammatory response. For instance, lipopolysaccharide (LPS) stimulation of Mφs through Toll-like receptor 4 (TLR4) reconfigures the Krebs cycle leading to the accumulation of succinate and itaconate, which is vital for activating hypoxia-inducible factor 1-alpha (HIF-1α)-dependent glycolytic reprogramming and nuclear factor erythroid 2-related factor 2 (Nrf2)-dependent antioxidative defense respectively (12, 13).
Here, we show how the rewiring of metabolic circuits in Mφs following oxLDL or cholesterol accumulation underlies their impaired inflammatory response to LPS. Specifically, we found that the oxidative stress induced by oxLDL accumulation activated a primary Nrf2-dependent response in Mφs, which was then enhanced by subsequent LPS stimulation. This secondary response increased the expression of NADPH-requiring detoxification enzymes, which repurposed NADPH consumption for detoxification of reactive radicals, rather than fueling their synthesis. Together, this suppressed LPS-induced HIF-1α-dependent activation of glycolysis and inflammation in a dual manner. Finally, comparative transcriptomic analysis between our in vitro oxLDL-loaded LPS-activated peritoneal Mφs (PMφs) and foamy Mφs derived from atherosclerotic mouse and human aortas confirmed similar inflammatory and metabolic profiles.
Materials and Methods
Mouse strains.
8 – 12 weeks-old mice were used. C57BL/6J (Strain #000664), B6.129S4(C)-Vhltm1Jae/J (Vhlfl/fl) (Strain #012933), B6.129X1-Nfe2l2tm1Ywk/J (Nfe2l2−/−) (Strain #017009), B6.129P2-Lyz2tm1(cre)Ifo/J (Strain #004781), B6.129P2-Nos2tm1Lau/J (Nos2−/−) (Strain#002609) and B6.129-Hif1atm3Rsjo/J (Hif1afl/fl) (Strain #007561) mice were purchased from The Jackson Laboratory. Nfe2l2−/− and Nos2−/− mice were generated by first crossing with wild type (WT) C57BL/6J mice, followed by heterozygote intercrossing. Lys2-Cre:Vhlfl/fl and Lys2-Cre:Hif1afl/fl mice were generated by backcrossing a single Lys2-Cre transgene into Vhlfl/fl and Hif1afl/fl mice. Breeding for experiments consisted of crosses between Cre-positive and Cre-negative Vhlfl/fl or Hif1afl/fl mice. All mice were maintained in a pathogen-free, temperature-regulated environment with a 12-hour light and dark cycle and were fed a normal chow diet (NCD, 16 kcal% fat). All studies were performed under the approval of Animal User Protocols by the Animal Care Committee at the University Health Network according to the guidelines of the Canadian Council on Animal Care. Littermates were used for experiments (see below).
Thioglycolate-elicited peritoneal Mφ (PMφ) isolation.
For each experiment, 1 or 2 litters of male and female WT C57BL/6J mice 8 to 12 weeks of age (up to 10 mice in total) were injected intraperitoneally with 1 mL of 4% aged thioglycolate (ThermoFisher Cat#211716) and PMφs were harvested after 4 days by lavage with cold PBS containing 2% FBS. Cells were pooled, counted, divided into 4 experimental groups and cultured (37°C, 5% CO2) in DMEM supplemented with 10% FBS, 2 mM L-glutamine, 10,000 U/mL penicillin/streptomycin). Adherent PMφs were used in experiments after 18 h.
Bone marrow derived Mφ (BMDMφ) generation.
BMDMφs were obtained from mice with a genetic deficiency (see above). Sex-matched littermates of up to 5 mice per experiment (+/+ and −/− or Cre+ and Cre-) were used. Mice were euthanized in a CO2 chamber and bone marrow cells were isolated from leg bones. Cells were cultured (37°C, 5% CO2) in RPMI supplemented with 10% FBS, 2 mM L-glutamine, 10,000 U/ml penicillin/streptomycin, and 40 ng/mL of M-CSF (PeproTech; Cat#AF-315–02) for 7 days. Cells were counted and replated for experiments. Male and female mice were used in different experiments. The results were comparable and therefore the data were combined.
Stable transfection of RAW264.7 Mφs.
RAW264.7 Mφs (2 × 106, ATCC, Cat# ATCC TIB-71) were electroporated (Amaxa® Cell Line Nucleofector® Kit V, LONZA; Cat#VCA-1003) with 1 – 5 µg of control plasmid (pcDNA3), HA-HIF1alpha-pcDNA3 (Addgene#18949), HA-HIF1alpha P402A/P564A-pcDNA3 (Addgene#18955), pcDNA3 mHIF-1α MYC-P402A/P577A/N813A (Addgene#44028), Mitochondrial mTurq2-Apollo-NADP+, Cytoplasmic mVenus-Apollo-NADP+) (14). Transfected cells were seeded in 6-well plates and cultured in recovery medium (DMEM, 20% FBS) for 3 h, then DMEM, 10% FBS for 48 h. G418 (400 µg/mL, replaced every 3 days, Gibco) was used for selection of stable-transfected cells After one week of selection, cells were used for experiments or cultured in DMEM,10% FBS, G418 (400 µg/mL).
Lipid loading, LPS stimulation and inhibitor studies.
PMφs, BMDMφs or RAW264.7 Mφs were cultured for 24 h with human medium oxidized low-density lipoprotein (100 μg/mL, Kalen Biomedical Cat#770202) or cholesterol (50 μg/mL, Sigma Cat#C3045), followed by ultrapure LPS stimulation (10 ng/mL, InvivoGen, Cat#tlrl-3pelps) for up to 8 h. Ethanol (0.5%) was used as a carrier control for cholesterol. For inhibitor experiments, 2-DG (25mM) (Sigma, D8375), Echinomycin (5 nM) (Cayman Chemical, Cat#11049), G6PDi-1 (25 µM) (Cayman Chemical, Cat#31484), MitoTEMPO Hydrate (25 µM) (Cayman Chemical, Cat#16621), DPI chloride (10 µM) (Sigma, Cat#) was added 1 h prior to LPS stimulation.
Immunoblotting.
PMφs and BMDMφs (2 × 106) cultured in 12-well plates, were incubated with or without oxLDL or cholesterol and stimulated with LPS for indicated times. Cells were lysed in ice-cold RIPA buffer (1% NP40, 0.1% SDS, 0.5% deoxycholate in PBS, supplemented with 1 mM PMSF, 1X cOmplete™, EDTA-free Mini Protease Inhibitor Cocktail (Sigma Cat#11873580001) and 1X PhosSTOP™ (Sigma Cat#4906845001)) for 15 minutes. Protein concentrations in lysates were determined by Protein Assay Dye Reagent (BioRad #5000006), diluted in 2× Laemmli sample buffer (BioRad Cat#161–0737) with fresh β-mercaptoethanol (BioRad #1610710), and heated at 95°C for 5 minutes. Samples (20 μg of protein per lane) were resolved on 8%−15% SDS-PAGE gels and transferred to polyvinylidene difluoride membranes (Sigma #IPVH00010) using a wet transfer system. Membranes were blocked with 5% skim milk non-fat powder or 3% BSA (Bioshop #ALB003) in Tris-buffered saline-Tween (TBST) for 1 h at room temperature. Membranes were incubated with primary antibodies overnight: anti-HIF-1α (Cell Signaling Technology (CST)#36169), anti-HIF-2α (Novus Biologicals (NB), NB100–122), anti-HIF-3α (ThermoFisher, PA5–36531), anti-HIF-1β (CST#5537), anti-Actin (Sigma, A2066), anti-Hexokinase II (CST#2867), anti-Lamin A/C (CST#2032), anti-Gapdh (CST#5174), anti-Egln2 (Bethyl Laboratories, Cat#A300–326A), anti-Egln2 (NB100–138), anti-Egln3 (NB100–303), anti-Txnrd1 (Abcam, Ab124954), anti-GSR (ThermoFisher, PA5–29945), anti-Nrf2 (CST#12721), anti-iNOS (Transduction Laboratories, #N32030), anti-Keap1 (ThermoFisher, #PA5–99434), followed by washing and incubation with HRP-conjugated anti-rabbit IgG (CST#7074) (22°C, 1 h). Blots were developed using Immobilon Forte Western HRP substrate (Sigma, WBLUF0100), imaged with Microchemi 4.2 (BioRad) and analyzed with ImageJ.
Cytoplasmic and nuclear subcellular fractionation.
PMφs and BMDMφs (6 × 106) were first seeded in 6-well plates, loaded with oxLDL or cholesterol, followed by LPS stimulation as described above. Cells were lysed and scraped with extraction buffer (10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, 1 mM NaF, 0.1% NP40, 1× cOmplete™, EDTA-free mini protease inhibitor cocktail). Lysates were centrifuged (1200 × g, 4°C, 10 minutes) to pellet nuclei and separate cytoplasmic fractions. To purify the nuclear fraction, the pellet was resuspended with extraction buffer and layered on top of a 30% sucrose solution, then centrifuged (3000 × g, 4°C, 20 minutes). The supernatant was discarded and nuclei in the pellet were washed once with extraction buffer, and then lysed with a detergent-rich buffer (10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, 1 mM NaF, 0.1% NP40, 1% deoxycholate, 0.1% SDS, 1× cOmplete™, EDTA-free mini protease inhibitor cocktail).
Cycloheximide chase assays.
PMφs (2 × 106) or BMDMφs were (2 × 106) were cultured in 12-well plates, loaded with oxLDL or cholesterol and stimulated for 6h or 8h with LPS (as above). Cycloheximide (CHX, 3 µg/mL, Sigma, Cat#239765) was added to wells in a time-dependent manner. For certain experiments, MG132 (10 µM, Peptide Institute, Cat#3175-v) or Bafilomycin A1 (40 nM, CST#54645) was added 1 h prior to CHX treatment.
Extracellular acidification rate (ECAR) measurement.
PMφs (2 × 105) and BMDMφs (3 × 105) were cultured in XF24 well plates (Agilent Technologies, Cat#102342-100), then loaded with oxLDL or cholesterol for 24 h. For glycolysis stress tests, cells were stimulated for 6 h with LPS, washed three times with Seahorse XF DMEM medium (Agilent Technologies, 103334–100) supplemented with 1 mM glutamine and 2 mM pyruvate, and incubated (37°C, 0% CO2) for 30 minutes prior to the test. During the test, glucose (Agilent Technologies Cat#103577–100), oligomycin A (Cayman Chemical Cat#11342) and 2-deoxyglucose (Sigma Cat#D8375) were added by the XFe24 Seahorse Analyzer (final concentrations were 25 mM, 2 μM and 25 mM, respectively). For real-time ECAR measurements, cells were washed three times with Seahorse XF DMEM medium supplemented with 25 mM glucose, 1 mM glutamine and 2 mM pyruvate, and incubated (37°C, 0% CO2) for 30 minutes prior to the addition of LPS (10 ng/mL final concentration) or PBS (equal volume) by the XFe24 Seahorse Analyzer.
Glucose uptake and reactive oxygen species measurement.
PMφs (3 × 106) and BMDMs (3 × 105) were first seeded in 35 mm petri dish, with 14 mm microwell (MatTek, P35G-1.5–14-C) or 8-well chamber slides (ThermoFisher Cat#154453) respectively, then cultured with oxLDL or cholesterol overnight. Cells were stimulated with LPS for 6 h the next day, washed three times with pre-warmed HBSS (Wisent, Cat#311–513-CL). For glucose uptake assay, cells were cultured for 1 h at 37°C, 5% CO2in DMEM without glucose (ThermoFisher #11966025) supplemented with 10% FBS, 2-NBDG (Cayman Chemicals #11046) (100 μg/mL) and 32.4 μM of Hoechst nuclear staining reagent (ThermoFisher #H3570). For reactive oxygen species assay, cells were cultured for 1 h at 37°C, 5% CO2 in HBSS, supplemented with CellRox Orange (10 μM, ThermoFisher #C10443) or MitoSox (5 μM, ThermoFisher #M36008). Nuclei were stained with Hoechst. Cells were then washed three times with pre-warmed HBSS. Cells that were stained with CellRox Orange were imaged live, while cells that were stained with MitoSox Red were fixed with 4% PFA for 1 h (4°C) prior to imaging. Cells were imaged with Olympus FluoView 1000 Laser Scanning Confocal Microscope (Olympus America) or A1R Confocal microscope with resonant scanner (Nikon). Mean fluorescence intensity measurements represented the ratio of total fluorescence intensity for each field to the number of nuclei in that field.
Lysosomal pH measurement with lysotracker.
PMφs were seeded at 2 × 105 cells/well in a 96-well black microplate (Sigma Aldrich, P8741), then cultured with oxLDL or cholesterol overnight. The next day after LPS treatment, cells were washed three times with pre-warmed HBSS. Cells were incubated with HBSS, cultured for 1 h (37°C, 5% CO2) with LysoTracker Red (50 nM, ThermoFisher #L7528), washed three times with pre-warmed HBSS, and the fluorescent absorbance values of wells were acquired in triplicates with a Cytation 5 Imaging Multi-Mode Reader (BioTek) (excitation and emission values with bandwidth were set as 571/9 and 590/9).
Quantification of total NADPH, NADP+ and glutathione.
Total NADPH assay kit (Abcam, #ab186031) and total NADP+ and NADPH assay kits (Abcam #ab186033) were used. To measure glutathione, glutathione assay kit (Cayman Chemicals #703002) was used. In brief, cells were seeded in 24-well (total NADP+ and NADPH, glutathione) or 6-well (NADPH) plates, then loaded with oxLDL for 24 h with subsequent 6 h LPS stimulation the next day. Cells were then lysed with respective lysis buffers provided in the kits and the abundance of metabolites was determined according to the manufacturers’ protocols.
Thioredoxin reductase and glutathione-disulfide reductase activity.
To measure thioredoxin reductase activity, a thioredoxin reductase colorimetric assay kit was used (Cayman Chemicals #10007892). In brief, cells (3.2 × 107) were plated in a 10 cm dish and cultured with oxLDL for 24 h, followed by LPS stimulation up to 6 h. Cells were lysed with TrxR assay buffer (1x) and the activity of thioredoxin reductase was measured according to manufacturer’s protocol. Specifically, the absorbance in duplicate samples was read at 405–414 nm every 2 minutes for a total of 10 minutes. The slope and the activity of thioredoxin reductase were calculated. To measure glutathione-disulfide reductase activity, NADPH and GSH were measured post 2-deoxyglucose (2DG) and diamide addition in a time-dependent manner. Specifically, DMEM cultured media in ± oxLDL loaded cells post LPS stimulation was replaced with 25 mM of 2DG and 1 mM of diamide (Sigma #D3648) in HBSS every 5 minutes for a total of 25 minutes. 2DG blocks de novo synthesis of NADPH while diamide oxidizes GSH to GSSG. Cells were lysed and NADPH and GSH were measured as described above.
RNA isolation and real-time (RT) PCR.
Total RNA was isolated with E.Z.N.A.® Total RNA Kit I (Omega Cat#R6834–01) and reverse transcription (RT) reactions were performed with High-Capacity cDNA Reverse Transcription Kit (ThermoFisher Cat#4368814) according to manufacturer’s protocol. RT quantitative-PCR (qPCR) was then performed using Roche LightCycler 480 with Luna® Universal qPCR Master Mix (New England Biolabs, Cat#M3003E). Quantification of mRNA was performed by using primers that span over two adjacent exons, quantified using the comparative standard curve method and normalized to hypoxanthine phosphoribosyltransferase (HPRT) as the housekeeping gene. Primer sequences used for qPCR are listed below:
Il1a ACGGCTGAGTTTCAGTGAGACC CACTCTGGTAGGTGTAAGGTGC
Il1b AGTTGACGGACCCCAAAAGA TGCTGCTGCGAGATTTGAAG
Il6 CTCCCAACAGACCTGTCTATACCA TGCCATTGCACAACTCTTTTCT
Il12b AAGTGGGCATGTGTTCC TCTTCCTTAATGTCTTCCACTT
Il15 GTAGGTCTCCCTAAAACAGAGGC TCCAGGAGAAAGCAGTTCATTGC
Il18 ACAGGCCTGACATCTTCTGC CCTTGAAGTTGACGCAAGAGT
Ccl3 CCCAGCCAGGTGTCATTT AGTTCCAGGTCAGTGATGTATTC
Ccl5 CCTGCTGCTTTGCCTACCTCTC ACACACTTGGCGGTTCCTTCGA
Ccl9 TCCAGAGCAGTCTGAAGGCACA CCGTGAGTTATAGGACAGGCAG
Ccl22 GTGGAAGACAGTATCTGCTGCC AGGCTTGCGGCAGGATTTTGAG
Tnfa GTAGCCCACGTCGTAGCAAAC GCACCACTAGTTGGTTGTCTTTGA
Nos2 AAACCCCTTGTGCTGTTCTC GGGATTCTGGAACATTCTGTGC
Slc2a1 GCTTCTCCAACTGGACCTCAAAC ACGAGGAGCACCGTGAAGATGA
Hk1 GAAAGGAGACCAACAGCAGAGC TTCGTTCCTCCGAGATCCAAGG
Hk2 CCCTGTGAAGATGTTGCCCACT CCTTCGCTTGCCATTACGCACG
Pfkfb3 TCATCGAGTCGGTCTGTGACGA CATGGCTTCTGCTGAGTTGCAG
Pfkp AAGAGGAAACCAAGCAGTGCGC TTCCTCGGAGTTTCACGGCTTC
Pgk1 GATGCTTTCCGAGCCTCACTGT ACCAGCCTTCTGTGGCAGATTC
Mct4 TCACGGGTTTCTCCTACGC GCCAAAGCGGTTCACACAC
Ldha ACGCAGACAAGGAGCAGTGGAA ATGCTCTCAGCCAAGTCTGCCA
Gclc ACACCTGGATGATGCCAACGAG CCTCCATTGGTCGGAACTCTAC
Gclm TCCTGCTGTGTGATGCCACCAG GCTTCCTGGAAACTTGCCTCAG
Gss CCAGGAAGTTGCTGTGGTGTAC GCTGTATGGCAATGTCTGGACAC
Gsr GTTTACCGCTCCACACATCCTG GCTGAAAGAAGCCATCACTGGTG
Hmox1 CACTCTGGAGATGACACCTGAG GTGTTCCTCTGTCAGCATCACC
Nqo1 GCCGAACACAAGAAGCTGGAAG GGCAAATCCTGCTACGAGCACT
Txnrd1 AGTCACATCGGCTCGCTGAACT GATGAGGAACCGCTCTGCTGAA
Srxn1 TACCAATCGCCGTGCTCATCCG CCTTTGATCCAGAGGACGTCGA
Fasn CACAGTGCTCAAAGGACATGCC CACCAGGTGTAGTGCCTTCCTC
Hmgcr GCTCGTCTACAGAAACTCCACG GCTTCAGCAGTGCTTTCTCCGT
Sqle TGTTGCGGATGGACTCTTCTCC GTTGACCAGAACAAGCTCCGCA
Cyp11a1 TGCTCAACCTGCCTCCAGACTT ACTGGCTGAAGTCTCGCTTCTG
Nox2 TGGCGATCTCAGCAAAAGGTGG GTACTGTCCCACCTCCATCTTG
Hif1a TCTCGGCGAAGCAAAGAGTC AGCCATCTAGGGCTTTCAGATAA
Hprt CAAGCTTGCTGGTGAAAAGGA TGAAGTACTCATTATAGTCAAGGGCATATC
Vhl GTTTGTGCCATCCCTCAATGTCG ACCTGACGATGTCCAGTCTCCT
Metabolite sample processing.
Extracellular (culture supernatants) and intracellular (detergent cell extracts) samples were centrifuged for 7 min at 14,000 × g, and supernatants were transferred to new tubes. Samples were then dried under nitrogen using a TurboVap. Samples were reconstituted in 75 μl of LC-MS grade water per 1 mL of original volume. Reconstitution volumes for cell extracts were calculated to reflect that 26.7 ng of RNA were reconstituted in 15 μL of LC-grade water. RNA content was measured using Quant-iT™ RiboGreen™ RNA Assay Kit. Cell pellets were reconstituted in 250 μL Elisa buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl,1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 1 mM sodium orthovanadate). Total RNA content of cell pellets was measured using Quant-iT™ RiboGreen™ RNA Assay Kit. Resuspended cell pellets were diluted 20× in 10mM TE (pH=7.5) and combined with the dye and incubated for 5 min at room temperature. Fluorescence was read using a Victor Wallac plate reader. Reconstituted samples were combined with an equal volume of 99.9% 13C15N labeled Saccharomyces cerevisiae standard extract to serve as an internal standard. Samples were centrifuged 7 minutes at 14,000 × g at 4°C and transferred to LC-MS polypropylene conical vials. Vials were stored at −80°C until analysis using LC-MS.
Liquid chromatography.
All metabolites except 9 dicarboxylic acids were analyzed essentially as described in Wan et al. (15). Briefly, tributylamine ion paired liquid chromatography was carried out using an Agilent 1290 UPLC system with a 0.25 mL/min flow rate, a 2 μL sample loop and an Extend C18 RRHD 1.8 mm, 2.1 × 150 mm column (Agilent). Nine carboxylic acids (malic acid, succinic acid, itaconic acid, citric acid, alpha-ketoglutaric acid, 2-hydroxyglutaric acid, isocitric acid, fumaric acid, and lactic acid) were analyzed using an acidic chromatographic method. Acidic chromatography was carried out on an Agilent 1290 UPLC system with a 0.4 mL flow rate using a 2.1 mm x150 mm Waters HSS-T3 C18 column with 1.8 μm packing thermostatted at 45°C. All listed metabolites eluted isocratically in 7 min using 0.5% Acetonitrile and 99.5% 20mM ammonium formate pH 2.9. Then the column was regenerated for 10 min using 99.5% Acetonitrile and 0.5% ammonium formate buffer (1 mL/min flow rate).
Mass spectrometry.
All data in this study was acquired using an Agilent 6550 QToF instrument using an Agilent Jet Stream electrospray ionization source (AJS ESI) in negative mode. Gas temperature in the ion source was 150°C with a flow rate of 14 L/min. Nebulizer pressure was 45 psig, sheath temperature was 325°C with a gas flow rate of 12L/min. Voltage for both capillary and nozzle were 2000V. The funnel DC voltage was −30V, funnel voltage drops were −100 and −50V in the high- and low-pressure funnels, respectively. The RF voltages were 110 and 60V in the high- and low-pressure funnels, respectively. Mass spectra were acquired between 50 and 1100 m/z with a rate of 2 spectra per second. Mass lock mixture was used (15).
Bulk RNA-seq library preparation.
Total RNA was extracted from 2 × 106 PMφs using RNeasy mini kit (Qiagen, #74104). To eliminate genomic DNA, 20 U of Roche RNase-free DNase I (Roche, Cat#4716728001) was added in the eluant (100 µl), followed by 20 min of incubation at 37°C. Samples were then passed through RNeasy columns. Total RNA was quantified by NanoDrop Spectrophotometer ND-1000 and 750 ng was used to enrich mRNA using NEBNext poly(A) mRNA magnetic isolation module (New England Biolabs Cat#E7490). Library construction followed immediately using NEBNext Ultra II directional RNA library prep kit (New England Biolabs Cat#E7760) and NEBNext multiple oligos for Illumina (New England Biolabs # E7335, E7500, E7710, E7730). Libraries were PCR-amplified for 10 cycles, quantified by MiSeq Nano V2, and sequenced on an Illumina NovaSeq S1 flow cell with 150 bp paired-end run to obtain about 30 million reads per sample. The complete RNA-seq data set has been deposited to the GEO repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE239696).
Bulk RNA-seq data acquisition and processing.
The adaptor sequences were removed using Trimmomatic (v0.32; recommended parameters). The reads were aligned to the mouse genome reference assembly mm10 (GRCm38) using STAR (v2.5.1b; default parameters). The quality metrics were done using MultiQC (v1.3). To count exonic reads we used featureCounts (http://subread.sourceforge.net/,v1.5.0) and annotated them for mouse genes using GENCODE vM4. The foamy and non-foamy intimal macrophage dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE116239) was downloaded from the GEO database. Intimal foamy and non-foamy macrophages were prepared from apolipoprotein E knockout mice fed a high fat diet for 28 weeks (10).
Bulk RNA-seq differential expression and pathway analysis.
The DEseq2 R/Bioconductor package was used to perform hierarchical cluster analysis, PCA, and to determine differential expression between -oxLDL +LPS and +oxLDL +LPS PMφs and foamy and non-foamy aortic Mφs (16). Differential expression was assessed using the Wald test with Benjamini-Hochberg correction; genes with an adjusted P value of <0.1 were considered significantly differentially expressed. EnhancedVolcano (Version 1.11.3) was used to create volcano plots (17). Genes were pre-ranked using the DEseq2 output (-log10(pvalue)*sign(logFC)) for enrichment analysis. Pathway enrichment and leading edge analysis was performed using Gene Set Enrichment Analysis software (GSEA version 4.1.0) from the Broad Institute (software.broadinstitute.org/GSEA) (18, 19). Related pathways were grouped and labeled by theme using Cytoscape (Version 3.9.0) and AutoAnnotate (Version 1.3.5) and visualized using Enrichment Map (Version 3.3.3) (20, 21).
Single-cell RNA-seq data processing.
We used Seurat (Version 4.0.4) (22, 23) to analyze an integrated single-cell RNA-seq data set containing mononuclear phagocytes from human carotid endarectomy specimens and coronary artery atherosclerotic lesions (11). The Seurat object of the integrated data was provided by Drs Alma Zernecke-Madsen and Clement Cochain and was generated by integrating data from 2 prior studies (24, 25). Average expression levels of NRF2-dependent genes were determined using AddModuleScore and projected to the UMAP using FeaturePlot. Differentially expressed genes between the Foamy_Mac and MHCII_Mac clusters were determined using FindMarkers and volcano plots were generated using EnhancedVolcano (17).
HIF-1α-ODD firefly luciferase reporter assay.
Using Amaxa® Cell Line Nucleofector® Kit V, RAW264.7 Mφs (2 × 106) were co-transfected with 1 µg of HIF-1α-ODD-luciferase plasmid (Addgene #18965) and 0.1 µg of CMV–Renilla luciferase plasmid. Transfected cells were recovered in DMEM with 20% FBS for 3 h, and media was replaced with DMEM with 10% FBS overnight. Cells were cultured with oxLDL or cholesterol the next day for 24 h and stimulated with LPS the subsequent day for 6 h. Cells were lysed and processed with Promega Dual-Luciferase™ Reporter (DLR™) Assay Systems (Promega Cat#E1910), and both the Firefly and Renilla luciferase were quantified by using a GloMax 20/20 luminometer (Promega). CMV-Renilla luciferase was used as internal normalization for transfection efficiency.
Cellular imaging of NADP+ sensors.
All live imaging was done in HBSS (with calcium, magnesium, no phenol red) at 37°C with 5% CO2. Images were collected using a custom-built widefield RAMM microscope (ASI) equipped with excitation LEDs (405, 505, and 590nm), excitation polarizing filter (Edmund Optics, Barrington, NJ), and a 40×/0.75 NA air objective lens (Olympus, Richmond Hill, Canada). Fluorescence was passed through a Cerulean (ET470/24M) or Venus (ET535/30M) emission filter on a filter wheel, and subsequently split using an Optosplit II (Cairn, Faversham, UK) to simultaneously collect parallel and perpendicular emission light on separate regions of an IRIS 15 CMOS camera (Teledyne Photometrics, Tucson, AZ).
Image analysis of NADP+ sensors.
Parallel (I‖) and perpendicular (I⊥) fluorescence intensity images were analyzed with a custom ImageJ plugin. These are available at https://github.com/RocheleauLab/Optosplit-Anisotropy-Analysis-scripts. The images were background corrected using a rolling ball filter. Pixel-by-pixel anisotropy (r) was calculated using the background corrected intensities: r = (I‖ – GI⊥)/(I ‖ + 2G⊥) (26). The G factor for the two-photon microscope was measured by exciting samples with both vertically (V) polarized and horizontally (H) polarized light and collecting the polarized emission (27, 28). The G-factor for the widefield microscope was calculated using fluorescein solutions, simplifying the standard anisotropy equation to G = I‖ / I⊥. For images collected using the 1.42 NA objective lens, we used additional correction factors (Ka, Kb, and Kc) to account for the blurring of parallel and perpendicular intensities (26). Cells were manually selected after discarding cells of dim or saturated intensity from analyses, both of which can skew anisotropy results. Regions of interest (ROI) were selected using the parallel intensity images to avoid selection bias.
Statistical analysis.
All the statistical details of experiments can be found in the figure legends. In brief, all figures show pooled data from independent experiments. All experiments were repeated at least three times. All results are expressed as the mean ± SEM. The number of biological replicates is listed as the n value and each replicate was performed using a different Mφ preparation. Statistical analyses were performed using Prism software, unless otherwise specified in the figure legends.
Results
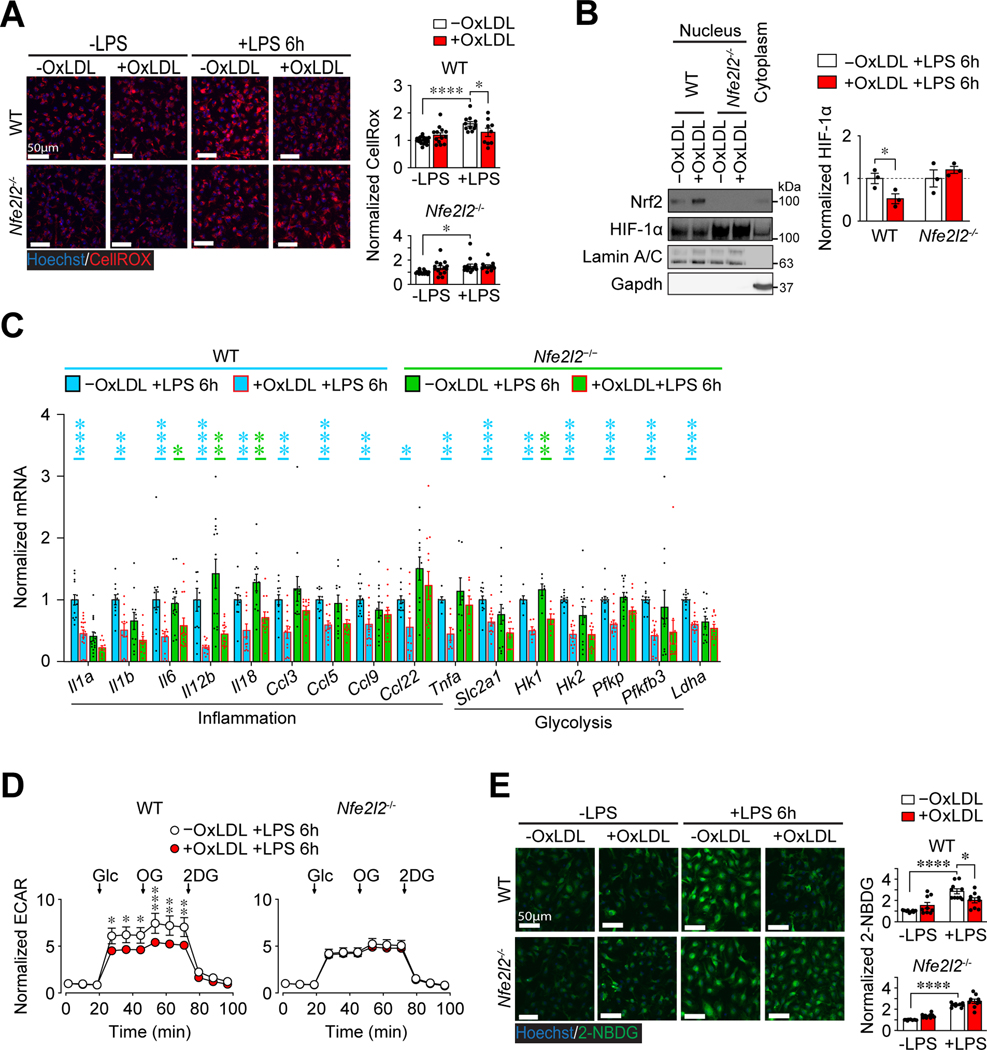
The accumulation of oxLDL in Mφs impairs LPS-induced glycolysis and inflammation.
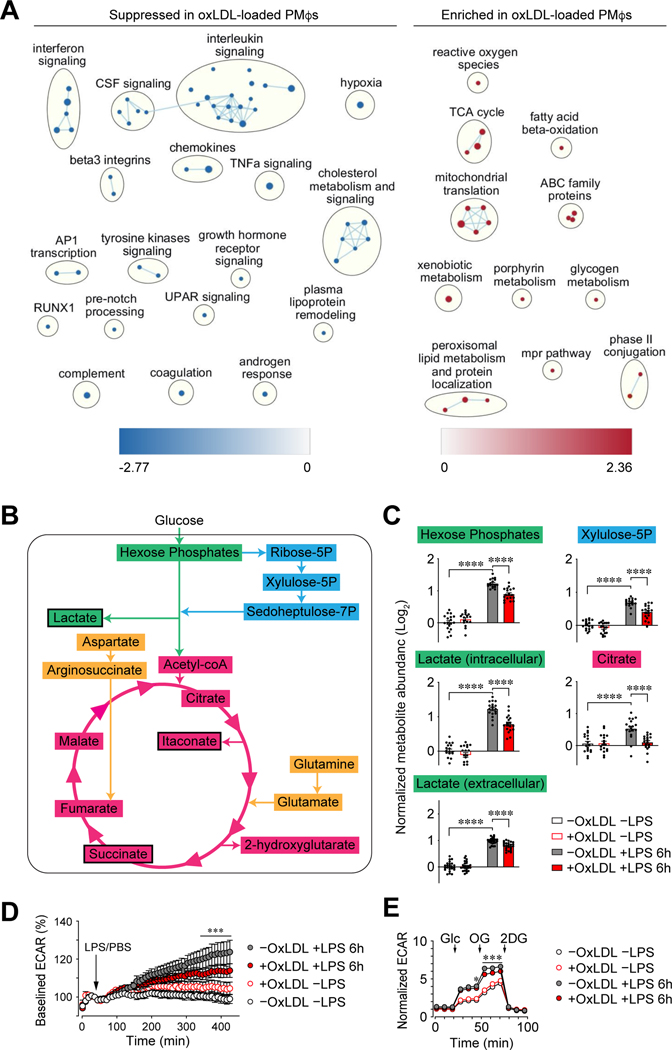
Consistent with our previous study (6), loading of PMφs with oxLDL or cholesterol for 24 h failed to induce the expression of proinflammatory genes and impaired a subsequent inflammatory response to LPS (Supplemental Figures 1A and 1B). To extend these observations, we performed RNA-seq and confirmed that oxLDL loading suppressed LPS-stimulated inflammatory pathways in PMφs, such as interleukin, interferon, TNFα signaling and hypoxia (Figure 1A, Supplemental Figures 1C and 1D). Since hypoxia-related pathways are regulated by HIF-1α, this opens the possibility that the suppression of HIF-1α-dependent processes, such as glycolysis, underlies the suppressed inflammatory response in Mφs with accumulated lipid.
Figure 1: The accumulation of oxLDL in Mφs impairs LPS-induced inflammation and glycolysis.
(A) Network analysis of gene sets in LPS-stimulated PMφs suppressed or enriched by oxLDL loading. Only gene sets with an FDR of <0.01 are included. Nodes represent individual gene sets and lines illustrate their overlap based on gene similarity. Related pathways are grouped in clusters. (B-C) Metabolomics of PMφs ±oxLDL loading (24h) and ±LPS stimulation (6 h) (n=18). OxLDL accumulation lowered LPS-induced intracellular abundance of shown metabolites involved in glycolysis (green), pentose phosphate pathway (cyan), Krebs cycle (magenta) and anaplerotic reactions (amber). Black outline indicates reduced extracellular abundance. (D-E) Seahorse analysis of PMφs (treatments are indicated). (D) Baselined ECAR values over time (normalized to the last pre-injection value, n=4). The addition of LPS or PBS is indicated. (E) Glycolysis stress tests showing ECAR (normalized to baseline, assigned a value of 1) in cells ±oxLDL and ±LPS (n=13). The addition of glucose (Glc), oligomycin (OG) and 2-deoxyglucose (2DG) is indicated. Values are normalized to -oxLDL -LPS. The mean ± SEM is plotted in all graphs. Statistical significance was determined by two-way ANOVA with Bonferroni correction (*P<0.05, **P<0.01, ***P<0.001).
LPS-induced glycolysis is vital for the expression of proinflammatory genes in Mφs (12, 29, 30). The blockade of glycolysis with 2DG in PMφs suppressed the induction of proinflammatory genes by LPS (Supplemental Figure 1E). The inhibition of proinflammatory gene induction by 2DG was similar to the effects of oxLDL or cholesterol loading, which opens the possibility that intracellular lipid accumulation in Mφs impairs LPS-induced inflammation by suppressing glycolysis. To evaluate this, we performed metabolomics and found that oxLDL accumulation in PMφs lowered LPS-induced metabolites involved in glycolysis, pentose phosphate pathway, Krebs cycle and anaplerotic reactions (Figure 1B and C, Supplemental Figure 2). Assessment of the extracellular acidification rate (ECAR) with the Seahorse Analyzer is a surrogate measure of glycolysis. We found that oxLDL accumulation in PMφs reduced LPS-induced ECAR in a time-dependent manner (Figure 1D). A glycolysis stress test confirmed that the accumulation in PMφs of oxLDL (Figure 1E) or cholesterol (Supplemental Figure 3A) impaired LPS-induced glycolysis.
Lipid accumulation in Mφs impairs LPS-induced HIF-1α-dependent glycolysis.
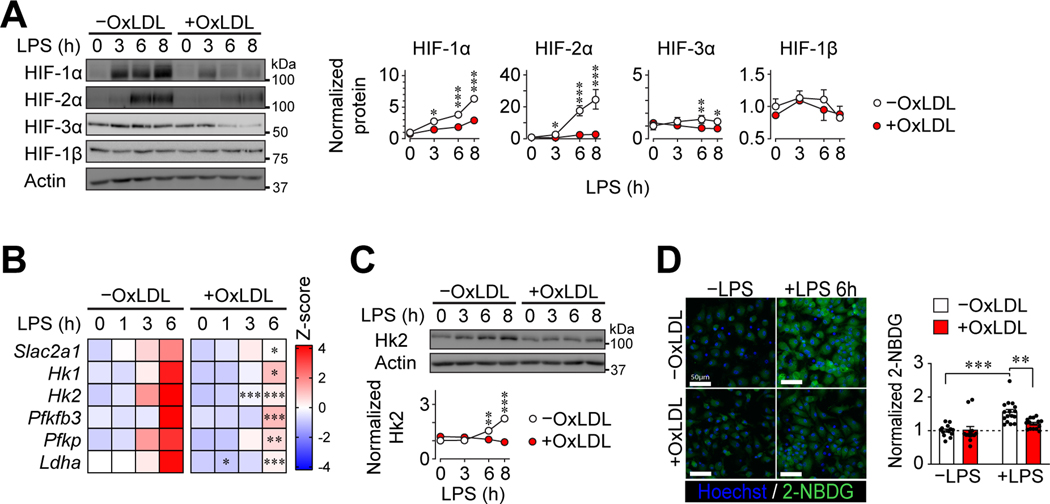
Next, we endeavored to elucidate the mechanism underlying glycolytic impairment. HIF-1-mediated transcription was shown to be vital for inflammation and glycolytic reprogramming in LPS-activated Mφs (12, 31, 32). Consistent with these studies, deletion of Hif1a (Supplemental Figure 3B) or blocking HIF-1α binding to DNA with echinomycin (Supplemental Figure 3C) lowered LPS-induced inflammatory and glycolytic gene mRNA expression, such as hexokinase 2 (Hk2) (Supplemental Figure 3D), the rate-limiting enzyme that phosphorylates glucose and mediates its cytosolic entrapment. Under unstimulated conditions, HIF-1 α subunits have a short half-life (33), which is a key mechanism for reducing its abundance and hence its transcriptional function. Therefore, we interrogated whether the accumulation or oxLDL (Figure 2A) or cholesterol (Supplemental Figure 3E) in PMφs modulates HIF-1α protein levels in response to LPS. The elevation of HIF-1α protein over 8 h following LPS stimulation was suppressed in PMφs with accumulated lipid. Similarly, HIF-2α and HIF-3α protein levels were lower in LPS-stimulated PMφs with accumulated lipid. However, the abundance of HIF-1β, a HIF family member whose protein expression levels are not altered by LPS, remained unchanged in PMφs with accumulated lipid. The abundance of LPS-induced glycolytic gene mRNAs was suppressed in PMφs with accumulated oxLDL (Figure 2B) or cholesterol (Supplemental Figure 3F), consistent with reduced HIF-1 α subunits. This correlated with a reduction in Hk2 protein (Figure 2C and Supplemental Figure 3G) and glucose uptake, as measured by 2-Deoxy-2-(7-nitro-2,1,3-benzoxadiazol-4-yl)amino)-D-glucose (2-NBDG), a fluorescent glucose analog (Figure 2D and Supplemental Figure 3H). Collectively, our data suggest that the accumulation of lipid in PMφs impairs LPS-induced HIF-1α-dependent glycolysis and inflammation.
Figure 2: OxLDL accumulation in Mφs impairs LPS-induced HIF-1α-dependent glycolysis.
(A) Effect of oxLDL accumulation and LPS stimulation on the expression of HIF family members in PMφs. Representative immunoblots and quantification of HIF-1α (n=6), HIF-2α (n=4), HIF-3α (n=5), HIF-1β (n=4) during a LPS time course. For each time point, values were normalized to the corresponding actin value and the 0 h LPS time point of PMφs without oxLDL (assigned a value of 1). (B) Effect of oxLDL accumulation and LPS stimulation on glycolysis gene mRNA expression in PMφs. mRNA abundance was analyzed by qPCR. For each gene, the data were normalized to values of PMφs without oxLDL and LPS stimulation (z-scores on heat map, n=5–11). (C) Representative immunoblots and quantification of Hk2 in LPS-stimulated PMφs (n=5, normalization as in A). (D) Effect of oxLDL loading and LPS stimulation on glucose accumulation in PMφs. Representative images and quantification of 2-NBDG uptake (green) by LPS-stimulated PMφs (n=5). Values are normalized to -oxLDL -LPS (assigned a value of 1, dashed line). The mean ± SEM is plotted in all graphs. Statistical significance was determined by two-way ANOVA with Bonferroni correction (*P<0.05, **P<0.01, ***P<0.001).
Intracellular lipid accumulation in Mφs impairs LPS-induced inflammation by reducing both HIF-1α stability and transactivation capacity.
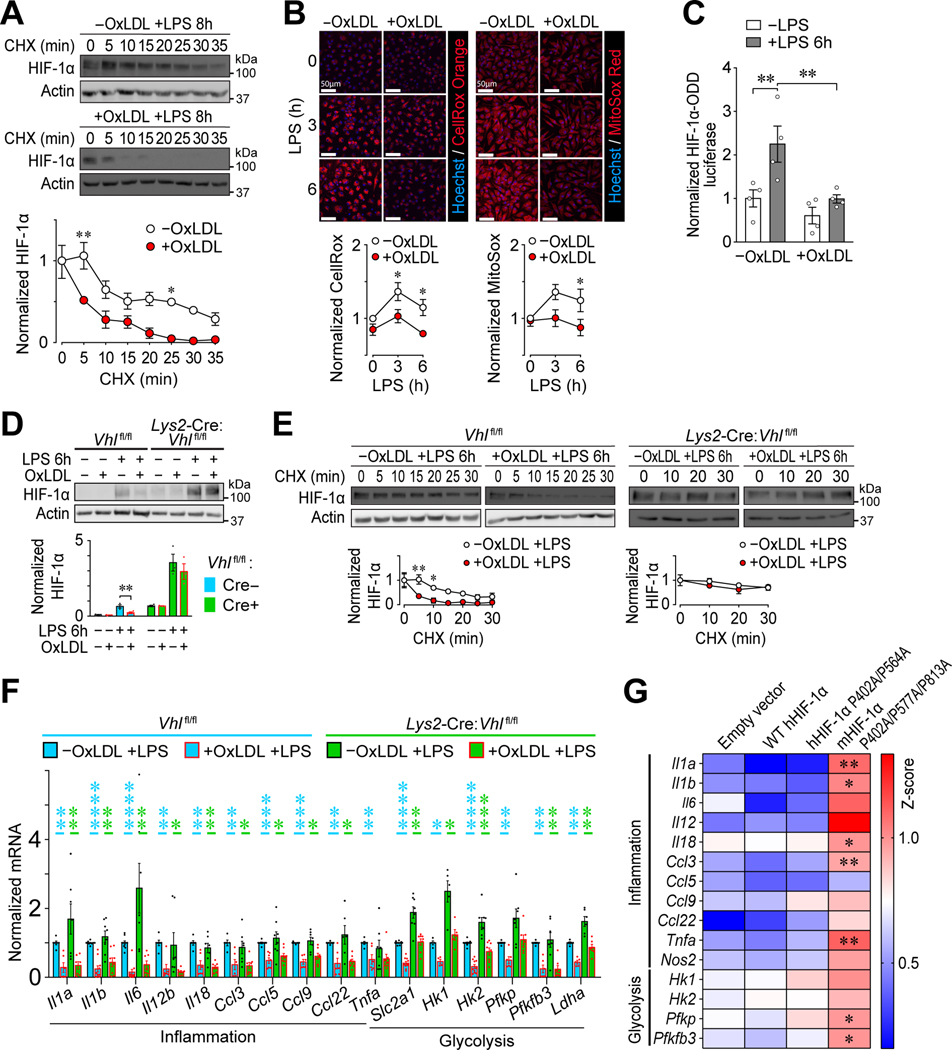
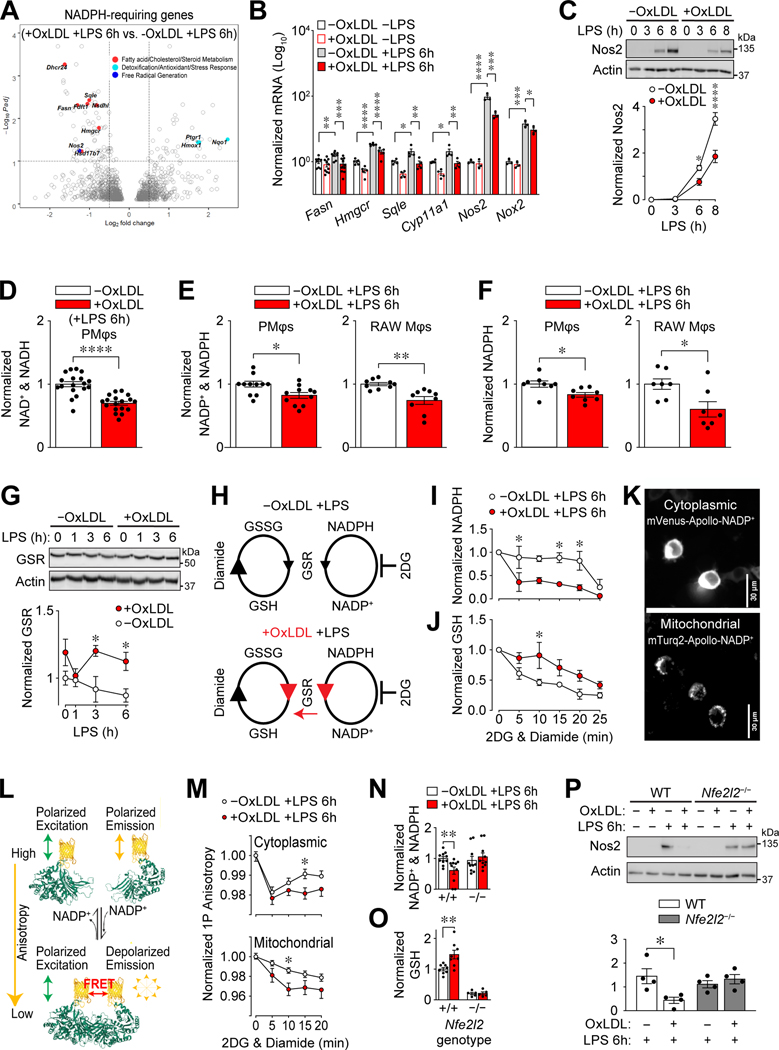
All α subunits of the HIF family harbor an oxygen-degradation-domain (ODD) (33–35); thus, their reduced protein expression (Figure 2A and Supplemental Figure 3E) implies increased degradation. We used a cycloheximide (CHX) chase assay to determine if lipid accumulation in PMφs reduces the stabilization of HIF-1α after LPS stimulation. We found that loading with oxLDL or cholesterol reduced the stability of total cellular (Figure 3A) and nuclear (Supplemental Figure 4A) HIF-1α protein. HIF-1α can be degraded by proteasomes (33) or lysosomes (36). Blocking proteasome function with MG132 maintained nuclear HIF-1α levels in cholesterol-loaded PMφs, whereas inhibition of lysosome function with Bafilomycin 1A did not (Supplemental Figure 4B). This suggests that proteasomes, but not lysosomes, enhanced HIF-1α degradation in PMφs with accumulated lipid. Since proteasomal degradation of HIF-1α is regulated by prolyl hydroxylases (PHDs) (37, 38), which can be inhibited by reactive oxygen species (ROS) (39), we examined if lipid accumulation in PMφs alters ROS production and the activity of PHDs. We first confirmed that HIF-1α protein abundance is dependent on ROS. Blocking NADPH oxidases with diphenyleneiodonium (DPI) or scavenging of mitochondria-derived ROS by mitoTEMPO lowered LPS-induced HIF-1α protein abundance (Supplemental Figures 4C and 4D), consistent with previous reports (13, 40). We then found that loading with oxLDL (Figure 3B) lowered total and mitochondrial ROS in LPS-stimulated PMφs. PHD activity was studied using RAW264.7 Mφs transfected with a firefly luciferase construct fused to the ODD of HIF-1α. We found that loading with oxLDL (Figure 3C) decreased LPS-induced luciferase activity. Collectively, our data show that intracellular lipid accumulation in Mφs impairs LPS-induced stabilization of HIF-1α by reducing ROS and increasing the activity of PHDs.
Figure 3: OxLDL accumulation in Mφs impairs LPS-induced inflammation by reducing HIF-1α stability and transactivation capacity.
(A) Effect of oxLDL accumulation on HIF-1α stability in LPS-stimulated (8 h) PMφs. Representative immunoblots showing a time course after CHX treatment and quantification. Values are normalized to corresponding actin and the pre-CHX time point (assigned a value of 1, n=3). (B) Effect of oxLDL loading on ROS in LPS-stimulated PMφs. Representative images and quantification of total ROS (CellRox Orange, Orange, n=5) and mitochondrial ROS (MitoSox, red, n=5). Data are normalized to the 0 h LPS time point of PMφs without oxLDL. (C) Effect of LPS stimulation (6 h) on PHD activity in RAW264.7 cells transfected with luciferase reporters. HIF-1α-ODD firefly luciferase activity is normalized to Renilla luciferase activity to account for transfection efficiency and to values found in control cells (n=4). (D) Assessment of HIF-1α abundance in unstimulated and LPS-stimulated (6 h) VHL-deficient (Lys2-Cre:Vhlfl/fl) and WT control (Vhlfl/fl) BMDMφs. Representative immunoblots and quantification are shown. Values are normalized to actin (n=3; unpaired Student’s t-test). (E) Assessment of HIF-1α stability in LPS-stimulated (6 h) VHL-deficient (Lys2-Cre:Vhlfl/fl) and WT control (Vhlfl/fl) BMDMφs. Representative immunoblots and quantification are shown. Values are normalized to actin and the first time point of CHX treatment (n=3–4). (F) Suppression of inflammatory and glycolysis gene mRNA expression by oxLDL accumulation in LPS-stimulated (6 h) Vhlfl/fl vs. Lys2-Cre:Vhlfl/fl BMDMφs analyzed by qPCR. Data were normalized to Vhlfl/fl cells without oxLDL (assigned a value of 1). Statistical comparisons using the Mann-Whitney U Test were within each genotype (p values are blue for Vhlfl/fl and green for Lys2-Cre:Vhlfl/fl, n=3–10 for each group). (G) Effect of HIF-1α mutations on proinflammatory and glycolysis gene mRNA expression in LPS-stimulated (6 h) stable transfected RAW264.7 cell lines. The effect of cholesterol accumulation was assessed for each cell line and all cell lines were compared to the empty vector line. Heat map z scores of qPCR data are plotted (n=3–9 per group, one-way ANOVA with Bonferroni correction). A higher score represents reduced inhibition of mRNA expression by cholesterol. In all graphs, the mean ± SEM is plotted. Unless indicated otherwise, statistical significance was determined by two-way ANOVA with Bonferroni correction (*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001).
Von Hippel–Lindau tumor suppressor (VHL) is an adaptor subunit of a ubiquitination complex that targets HIF-1α for proteasomal degradation (37, 38). To determine the causal link between intracellular lipid accumulation in Mφs and reduced HIF-1α, we characterized VHL-deficient bone marrow-derived Mφs (BMDMφs) from Lys2-Cre:Vhlfl/fl mice. Steady-state Vhl mRNA was dramatically lower in Lys2-Cre:Vhlfl/fl BMDMφs (Supplemental Figure 4E) and HIF-1α protein expression was elevated especially after LPS stimulation (Figure 3). The abundance of HIF-1α post-LPS was reduced in Cre-negative Vhlfl/fl BMDMφs with accumulated oxLDL (Figure 3D), similar to WT PMφs (Figure 2A and Supplemental Figure 3E). On the other hand, in Lys2-Cre:Vhlfl/fl BMDMφs with accumulated oxLDL, the LPS-induced increase in HIF-1α was comparable to cells without oxLDL (Figure 3D). The rate of HIF-1α degradation was increased by oxLDL accumulation in LPS-stimulated Cre-negative Vhlfl/fl BMDMφs, but was comparable in Lys2-Cre:Vhlfl/fl BMDMφs with or without lipid uptake (Figure 3E). Collectively the experiments with Vhl deficient BMDMφs confirm that lipid accumulation in Mφs reduces LPS-induced HIF-1α protein primarily by enhancing its degradation in a VHL-dependent manner. Thus, in spite of comparable HIF-1α protein abundance, we were surprised that LPS-induced proinflammatory and glycolysis gene expression remained suppressed in Lys2-Cre:Vhlfl/fl BMDMφs with accumulated oxLDL (Figure 3F). This suggests that in addition to an effect on HIF-1α stability, oxLDL accumulation impairs HIF-1α function by a different mechanism.
Under hypoxic conditions, the function of HIF-1α as a transcription factor depends on both protein stability and transactivation capacity. The latter is negatively regulated by factor inhibiting HIF-1 (FIH-1), which hydroxylates HIF-1α on asparagine 813 and blocks the recruitment of transcription co-activators (41). FIH-1 as well as PHDs are iron (II)-dependent dioxygenases (42). We therefore investigated if the activity of FIH-1 is increased by lipid accumulation, analogous to the activity of PHDs. We assessed this by generating 3 stable lines of RAW264.7 Mφs transfected with plasmids that encode different mutated forms of HIF-1α: (1) HA-hHIF-1α WT control, (2) HA-hHIF-1α (P402A/ P564A) and (3) Myc-mHIF-1α (P402A/P577A/N813A). Each line was cultured with cholesterol, and the gene expression profile of proinflammatory and glycolysis genes was determined after LPS stimulation. RAW264.7 cells transfected with HIF-1α with three mutations that impair the modifications catalyzed by both PHDs and FIH-1 (Line 3) were most resistant to the inhibitory effects of cholesterol accumulation (Figure 3G). Collectively, our data suggest that lipid loading of Mφs impairs LPS-induced inflammatory gene expression by reducing both HIF-1α stability and transactivation capacity.
Intracellular lipid accumulation in Mφs enhances LPS-induced Nrf2-mediated antioxidative defense, which destabilizes HIF-1α.
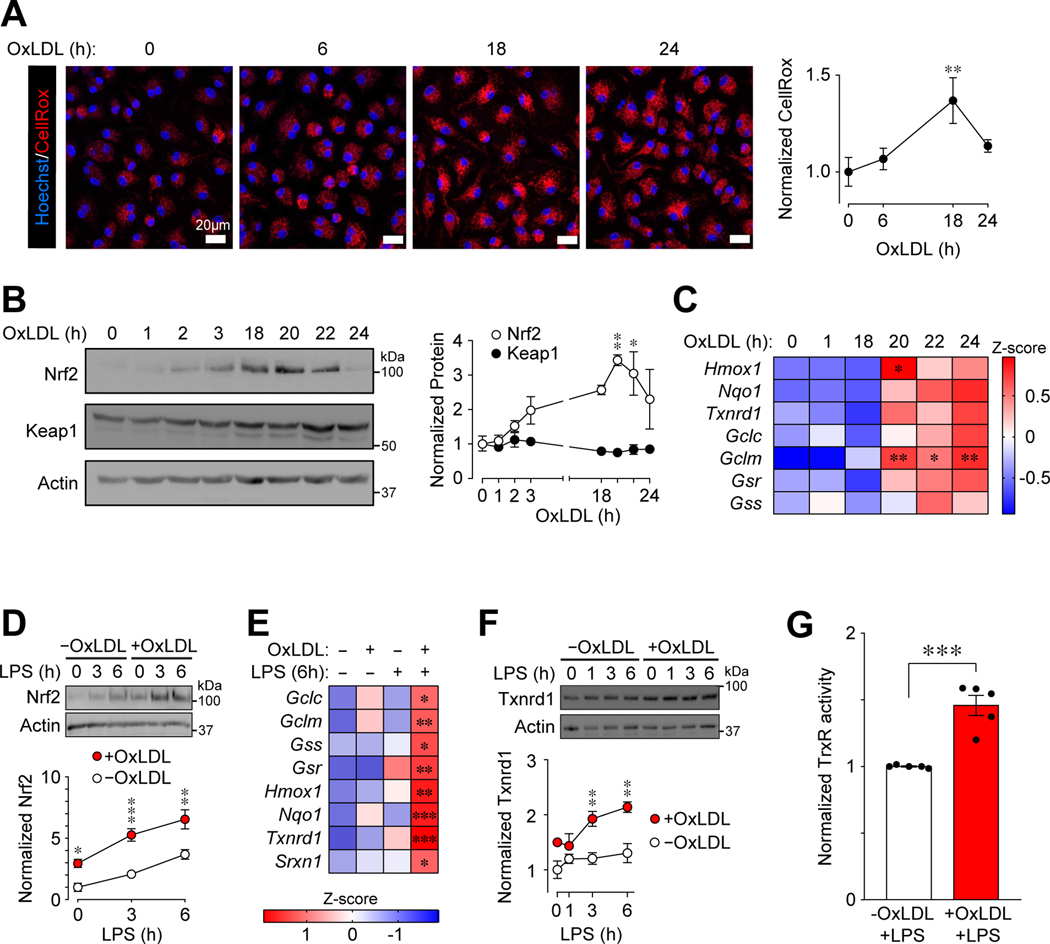
We next focused on determining why LPS-induced ROS was reduced in PMφs with accumulated oxLDL (Figure 3B). Our previous study (6) showed that oxLDL accumulation in Mφs can induce Nrf2-dependent transcription of detoxification enzymes, consistent with published reports (43–45); thus, the reduction of LPS-induced ROS may be due to an enhanced Nrf2-regulated antioxidant response. The main regulator of Nrf2 is Keap1, an electrophilic sensor with many cysteine residues (46). The cysteine residues of Keap1 can readily react with free radicals, including ROS, which alters the conformation of Keap1 and inhibits its Nrf2 degradative function (47). In the absence of LPS stimulation, culture of PMφs with oxLDL (Figure 4A) increased ROS in a time-dependent manner. The abundance of Nrf2 also increased (Figure 4B), as did the mRNA expression of Nrf2 target genes (Figure 4C), whereas Keap1 remained unchanged. The induction of Nrf2 (peak at 20 h) and its target genes (20–24 h) may account for the decline of ROS at 24 h.
Figure 4: OxLDL accumulation in Mφs induces oxidative stress that activates Nrf2-mediated antioxidative defense.
(A) The accumulation of oxLDL increases ROS in PMφs in a time-dependent manner. Representative images and quantification showing CellRox Orange staining (red) at different time points after addition of oxLDL to cultured PMφs. Data are normalized to the 0 h time point (assigned a value of 1, n=3). (B) Nrf2 (n=3) and Keap1 (n=4) protein in PMφs at different time points after addition of oxLDL. Representative immunoblots and quantification. Data are normalized and compared to the 0 h time point (assigned a value of 1). (C) OxLDL induces Nrf2-regulated gene expression in PMφs in a time-dependent manner. A heatmap shows mRNA measured by qPCR. All time points were compared to 0 h (n=6). (D) Effect of oxLDL accumulation over 24 h on Nrf2 protein in LPS-stimulated PMφs. Representative immunoblots and quantification. Values are normalized to actin and the 0 h LPS time point in cells without oxLDL (assigned a value of 1, n=4). (E) Effect of oxLDL accumulation (24 h) and LPS stimulation (6 h) on Nrf2-dependent gene expression (qPCR analysis of mRNA plotted as a heatmap). Comparisons are to PMφs without oxLDL and LPS stimulation (n=4–11). (F, G) Effect of oxLDL accumulation on Txnrd1 protein expression (F) and thioredoxin reductase (TrxR) activity (G) in LPS-stimulated PMφs. (F) Representative immunoblots and quantification normalized to actin and the 0 h LPS time point without oxLDL (assigned a value of 1, n=3). (G) Txnrd1 activity assays normalized to the group without oxLDL (assigned a value of 1, n=4, unpaired Student’s t-test). The mean ± SEM is plotted in all graphs. Unless indicated otherwise, statistical significance was determined by one-way (A-C) or two-way ANOVA (D-F), with Bonferroni correction (*P<0.05, **P<0.01, ***P<0.001).
We found that oxLDL accumulation in PMφs increased not just basal but also LPS-induced Nrf2 protein levels (Figure 4D). This correlated with increased mRNA expression of Nrf2-dependent genes (Figure 4E), as well as protein expression (Figure 4F) and activity (Figure 4G) of Trxnrd1, a Nrf2-dependent detoxification enzyme. These data support the concept that oxLDL accumulation in PMφs enhances the Nrf2-dependent antioxidant defense in LPS-stimulated Mφs.
We then tested if Nrf2-mediated reduction of ROS impairs LPS-induced HIF-1α protein expression in lipid-loaded Mφs. Using BMDMφs from Nrf2-deficient (Nfe2l2−/−) mice, we first showed that in the absence of Nrf2, oxLDL accumulation did not reduce ROS in response to LPS in contrast BMDMφs from WT littermates (Figure 5A). Since ROS up-regulates HIF-1α abundance through inhibition of PHD function, we assessed nuclear HIF-1α in LPS-stimulated BMDMφs and found comparable HIF-1α in Nrf2-deficient Mφs with and without oxLDL accumulation (Figure 5B). As expected, HIF-1α was reduced in oxLDL-loaded BMDMφs from WT littermates (Figure 5B). This finding was supported by restored LPS-induced mRNA expression of several inflammatory and glycolysis genes in oxLDL-loaded Nfe2l2−/− BMDMφs (Figure 5C). LPS-induced glycolytic response (Figure 5D) and glucose uptake (Figure 5E) were also restored in Nrf2-deficient BMDMφs with accumulated oxLDL.
Figure 5: Nrf2 deficiency reverses the suppression of LPS-induced oxidative stress, HIF-1α-mediated glycolysis and inflammation in Mφs with accumulated oxLDL.
(A) Effect of oxLDL accumulation and LPS stimulation (6 h) on ROS in BMDMφs with Nrf2 deficiency. Representative images and quantification of total ROS (CellRox, red; n=3) in cells derived from Nfe2l2+/+ (WT) and Nfe2l2−/− littermates. Data are normalized to BMDMφs without oxLDL and LPS. (B) Effect of oxLDL accumulation on HIF-1α abundance in nuclear extracts obtained from LPS-stimulated (6 h) WT and Nfe2l2−/− BMDMφs. A representative immunoblot and quantification. For each genotype, values are normalized to the nuclear protein Lamin A/C and values from cells without oxLDL (assigned a value of 1, dashed line, n=3). (C) Expression of inflammatory and glycolysis gene mRNA in LPS-stimulated (6 h) Nfe2l2+/+ (WT) vs. Nfe2l2−/− BMDMφs ±oxLDL. Data analyzed by qPCR are normalized to WT cells without oxLDL (assigned a value of 1). Statistical comparisons using the Mann-Whitney U Test were within each genotype (p values are blue for WT and green for Nfe2l2−/−, n=6–17 for each group). (D) Glycolysis stress tests showing normalized ECAR values over time in LPS-stimulated (6 h) WT and Nfe2l2−/− BMDMφs ±oxLDL (n=17). The addition of Glc, OG and 2DG is indicated. (E) 2-NBDG uptake (green) by WT and Nfe2l2−/− BMDMφs. Representative images and quantification. For each genotype, values are normalized to cells without oxLDL and LPS (assigned a value of 1, n=3 for all groups). In all graphs, the mean ± SEM is shown. Unless indicated otherwise, statistical significance was determined by two-way ANOVA with Bonferroni correction (*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001).
Altered NADPH metabolism in Mφs with accumulated lipid fuels Nrf2-dependent antioxidative defense.
NADPH is a vital co-factor for Mφs as it is required for the enzymatic functions related to inflammatory pathways, such as free radical and de novo lipid synthesis, as well as anti-inflammatory pathways, such as antioxidant and free radical detoxification. Given the vital role that NADPH plays in mediating redox homeostasis, we reasoned that NADPH metabolism is altered in Mφs with accumulated oxLDL. We evaluated our RNA-seq data of LPS-stimulated PMφs ±oxLDL (Supplemental Figure 1A) and found differential expression among genes encoding NADPH-requiring enzymes (Figure 6A). Specifically, oxLDL loading reduced the expression of inflammatory NADPH-requiring enzymes (Fasn, Hmgcr, Nos2). In contrast, the expression of anti-inflammatory NADPH-requiring enzymes (Nqo1, Hmox1 and Ptgr1) regulated by Nrf2 was increased. This differential expression pattern was confirmed and extended by qPCR and immunoblotting assays (Figure 6B, 6C, 4E, 4F).
Figure 6: Altered NADPH metabolism in Mφs with accumulated oxLDL is Nrf2-dependent.
(A) OxLDL accumulation induces differential expression of NADPH-requiring enzymes in LPS-stimulated (6 h) PMφs. A volcano plot shows differentially expressed genes (identified using DESeq2, n=3). NADPH-requiring enzymes are indicated as filled circles, each color corresponding to a different pathway (indicated). (B) Effect of oxLDL accumulation in PMφ and LPS stimulation (6 h) on mRNA expression of NADPH-requiring enzymes. For each gene, values were determined by qPCR and normalized to PMφs without oxLDL and LPS (assigned a value of 100, n=3–8). (C) Effect of oxLDL accumulation on LPS-induced Nos2 protein in PMφs. Representative immunoblots showing a time course after LPS stimulation and quantification (Nos2 values are normalized to actin, n=3). (D-F) Effect of oxLDL accumulation on metabolites in LPS-stimulated (6 h) Mφs. (D) LC-MS analysis of total NAD+ and NADH in PMφs (n=18), (E) assay of NADP+ and NADPH in PMφs (n=11) and RAW264.7 cells (n=9), and (F) assay of total NADPH in PMφs (n=8) and RAW264.7 cells (n=7). Values were normalized to the corresponding -oxLDL group (assigned a value of 1). (G) Effect of oxLDL accumulation on Glutathione-disulfide reductase (GSR) protein in LPS-stimulated RAW264.7 cells. A representative immunoblot shows a time course after LPS stimulation. GSR values were quantified and normalized to actin and the -oxLDL, 0 h LPS group (assigned a value of 1, n=4). (H-J) Assessment of GSR activity by measuring the rate of NADPH consumption. (H) Schematic illustrating conversion of glutathione disulfide (GSSG) to glutathione (GSH) by GSR and its cofactor NADPH. Diamide drives this reaction by converting GSH to GSSG, and 2DG blocks NADPH replenishment. Anticipated effects of oxLDL accumulation are shown in red. (I, J) Effect of oxLDL accumulation on the rate of NADPH consumption (I) and GSH production (J) in LPS-stimulated (6 h) RAW264.7 cells. Data are normalized to the 0 min time point i.e., just prior to diamide and 2DG addition (assigned a value of 1, n=3 for -oxLDL and +oxLDL groups). (K) Representative images of stable-transfected RAW264.7 cells expressing cytoplasmic mVenus-Apollo-NADP+ (top) and mitochondrial mTurq2-Apollo-NADP+ (bottom) sensors. (L) Schematic of the Apollo-NADP+ sensor, comprised of enzymatically inactivated human G6PD tagged with fluorescent protein, which responds to free NADP+ through allosteric dimerization. (M) 1P anisotropy quantification using stable-transfected RAW264.7 cells. Anisotropy values are normalized to the 0 min time point (assigned a value of 1, n=3). (N, O) Effect of oxLDL accumulation on the total NADP+ and NADPH (N) and GSH (O) in LPS-stimulated (6h) Nfe2l2+/+ (WT) and Nfe2l2−/− BMDMφs. Data are normalized to WT cells without oxLDL (assigned a value of 1). Comparisons within each genotype using the Mann-Whitney U test for NADP+ and NADPH (n=6–7) and unpaired Student’s t-test for GSH (n=5–8). (P) OxLDL accumulation suppresses LPS-inducible Nos2 protein in WT, but not Nfe2l2−/−, BMDMφs. Representative immunoblots and quantification (Nos2 values are normalized to actin; n=4, comparisons within each genotype using an unpaired Student’s t-test). Nos2 was not detected in cells without LPS stimulation (6h) and this condition was omitted from the graphs. The mean ± SEM is plotted in all graphs. Unless indicated otherwise, statistical significance was determined by a two-way ANOVA with Bonferroni correction (*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001).
Next, we determined if NADPH abundance was altered by intracellular oxLDL accumulation. The steady-state abundance of NAD+ and NADH (Figure 6D), NADP+ and NADPH (Figure 6E), and NADPH (Figure 6F) was reduced after LPS stimulation in PMφs and RAW264.7 cells with oxLDL accumulation. The increased expression of NADPH-requiring antioxidative enzymes may account for NADPH consumption. Indeed, LPS stimulation of RAW264.7 cells increased glutathione-disulfide reductase (GSR) protein over time in oxLDL-loaded cells (Figure 6G). We devised an assay to determine if increased GSR, a Nrf2-targeted enzyme that reduces glutathione disulfide (GSSG) to glutathione (GSH), accelerates the consumption of NADPH. We added 2DG to block the reduction of NADP+ to NADPH and diamide to oxidize GSH to GSSG (48), then measured GSH and NADPH over time (Figure 6H). In this assay, NADPH was depleted more rapidly in RAW264.7 cells with accumulated oxLDL (Figure 6I), while GSH declined more slowly (Figure 6J), suggesting a direct relationship between GSR expression and NADPH consumption. Since GSR is expressed in the cytoplasm and the mitochondria (49), we next determined if oxLDL loading differentially regulates NADPH in these compartments. We generated stably transfected RAW264.7 cell lines that expressed Apollo-NADP+ sensors in the cytoplasm and the mitochondria (Figure 6K). The quantification of NADP+ is possible by using Apollo-NADP+ sensors imaged with steady-state fluorescence anisotropy (Figure 6L) (14, 50). LPS-stimulated cells were treated with 2DG and diamide and anisotropy in both compartments was significantly lower in cells with accumulated oxLDL (Figure 6M). This is indicative of higher NADP+ and thus depletion of NADPH.
To determine if an enhanced Nrf2-response in Mφs with accumulated intracellular oxLDL underlies a shift in NADPH metabolism from inflammatory to antioxidative purposes, we characterized NADPH metabolism in Nrf2-deficient BMDMφs. We did not observe a reduction of NADP+ and NADPH upon LPS stimulation of cells with accumulated oxLDL, in contrast to WT BMDMφs (Figure 6N). GSH was higher in LPS-stimulated WT vs. Nfe2l2−/− BMDMφs and increased in WT, but not Nfe2l2−/− cells with accumulated oxLDL (Figure 6O). Analysis of mRNA expression of NADPH-requiring inflammatory enzymes revealed comparable expression in LPS-stimulated Nfe2l2−/− BMDMφs with or without oxLDL accumulation (Supplemental Figure 4F). As above (Figure 6B), expression was lower in LPS-stimulated WT BMDMφs cultured with oxLDL. The expression of Nos 2 protein was similar in Nfe2l2−/− BMDMφs with or without oxLDL accumulation (Figure 6P). Collectively, we show a metabolic adaptation in Mφs with accumulated intracellular oxLDL, where LPS stimulation shifts NADPH consumption from inflammatory to antioxidative purposes in a Nrf2-dependent manner.
NADPH metabolism regulates a positive feedback loop between HIF-1α and Nos2.
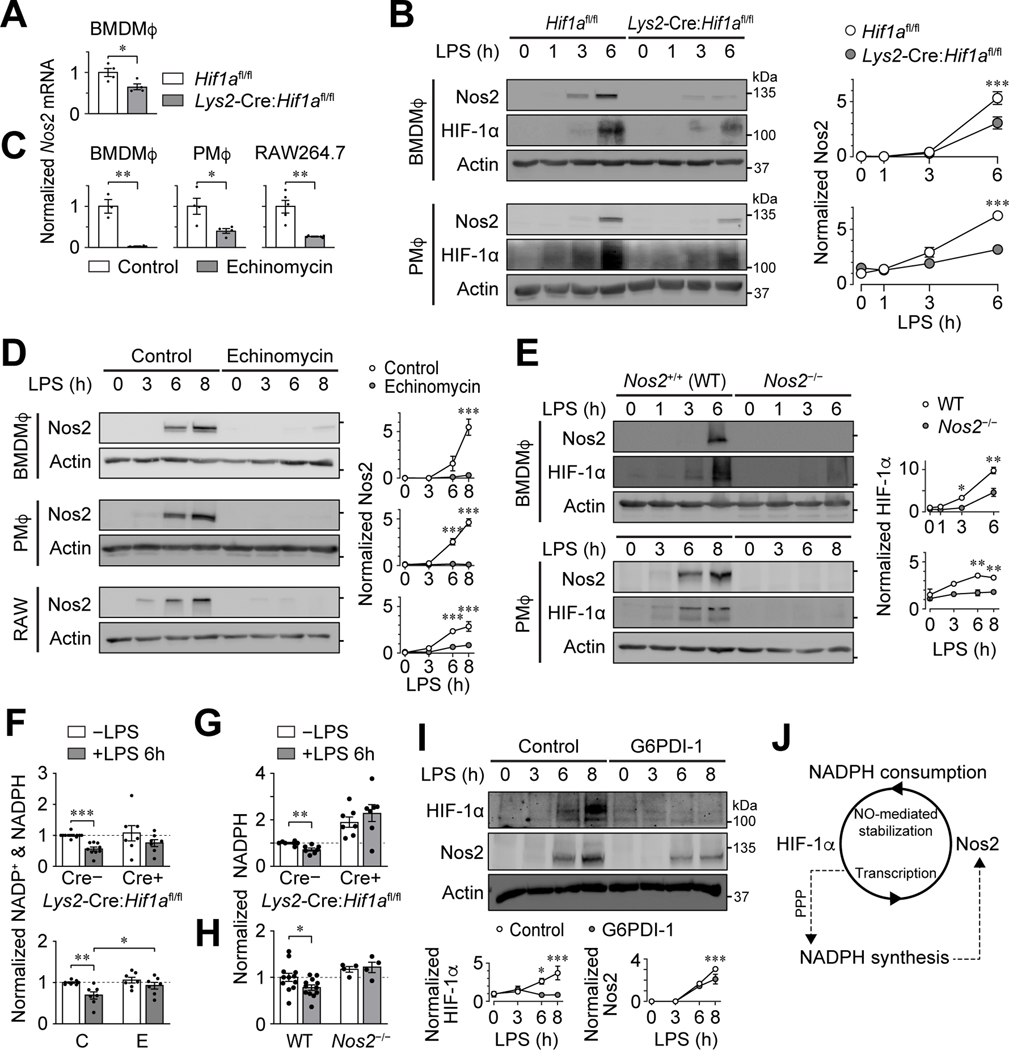
The transcription of inflammatory NADPH-requiring enzymes, such as Nos2, is regulated by HIF-1α (51). In turn, Nos2 utilizes NADPH to produce reactive radicals that stabilize HIF-1α by inhibiting PHDs (52, 53). These studies suggest a HIF-1α – Nos2 positive feedback loop. We established that the expression of Nos2 is dependent on HIF-1α by showing that LPS-induced Nos2 mRNA and protein were reduced in Hif1a-deficient PMφs and BMDMφs (Figure 7A and B) or WT PMφs, BMDMφs and RAW264.7 cells treated with echinomycin (Figure 7C and D). We also established that the abundance of HIF-1α is dependent on Nos2 by showing that LPS-induced HIF-1α protein accumulation is impaired in Nos2-deficient PMφs and BMDMφs (Figure 7E).
Figure 7: NADPH metabolism regulates a positive feedback loop between HIF-1α and Nos2.
(A, B) Effect of Hif1a-deficiency on Mφ Nos2 expression. Nos2 mRNA (A) was measured by qPCR in LPS-stimulated (6 h) Hif1afl/fl and Lys2-Cre:Hif1afl/fl BMDMφs. Values are normalized to Hif1afl/fl (assigned a value of 1, n=4, unpaired two-tailed student’s t-test). Representative immunoblots and corresponding quantification showing a LPS time course in Hif1afl/fl and Lys2-Cre:Hif1afl/fl BMDMφs and PMφs (B). The HIF-1α blots show a reduction of expression in Lys2-Cre:Hif1afl/fl Mφs, indicative of a significant but incomplete deletion of Hif1a. Nos2 protein is normalized to actin (n=3). (C, D) Effect of echinomycin treatment on Mφ Nos2 expression. Nos2 mRNA 6 h after LPS stimulation (C). qPCR data are normalized to the control group (assigned a value of 1, n=5 for PMφs, n=3 for BMDMφs, n=4 for RAW264.7 cells, unpaired two-tailed student’s t-test). Representative immunoblots and corresponding quantification showing a LPS time course (D). Nos2 protein is normalized to actin (n=3 for all groups). (E) Effect of Nos2-deficiency on Mφ HIF-1α protein expression. Representative immunoblots and corresponding quantification showing a LPS time course in Nos2+/+ (WT) and Nos2−/− BMDMφs and PMφs. The Nos2 blots show undetectable expression in Nos2−/− Mφs, consistent with a gene knockout. HIF-1α protein is normalized to actin (n=3). (F) Effect of Hif1a deficiency or function blockade by echinomycin on NADP+ and NADPH in unstimulated and LPS-stimulated (6 h) PMφs. NADP+ and NADPH is normalized to Hif1afl/fl -LPS or control -LPS groups, respectively (assigned a value of 1, dashed lines, n=7–9 for Hif1afl/fl and Lys2-Cre:Hif1afl/fl, n=7 for control (C) and echinomycin (E), unpaired two-tailed student’s t-test). (G, H) Effect of Hif1a or Nos2 deficiency on NADPH in unstimulated and LPS-stimulated (6 h) PMφs. NADPH is normalized to Hif1afl/fl -LPS (G) or WT -LPS (H) (assigned a value of 1, dashed line, n=7 for Hif1afl/fl vs. Lys2-Cre:Hif1afl/fl, n=4–12 for WT vs. Nos2−/−, unpaired two-tailed student’s t-test). (I) Effect of blocking of de novo NADPH synthesis in PMφs with G6PDi-1 on LPS-induced Nos2 and HIF-1α protein. A representative immunoblot showing a LPS time course and quantification of Nos2 and HIF-1α protein. Data are normalized to actin and the 0 h LPS time point for Nos2 (assigned a value of 1, n=4 for Nos2 and HIF-1α). (J) Schematic illustrating how NADPH mediates the positive feedback loop between HIF-1α-regulated transcription of Nos2 and Nos2-dependent HIF-1α stabilization. The mean ± SEM is plotted in all graphs. Unless indicated otherwise, statistical significance was determined by a two-way ANOVA with Bonferroni correction (*P<0.05, **P<0.01, ***P<0.001).
Next, we determined if the HIF-1α – Nos2 positive feedback loop regulates the abundance of NADPH. The abundance of combined NADP+ and NADPH was reduced 6 h post-LPS in WT PMφs but remained unchanged in Hif1a-deficient cells or cells treated with echinomycin (Figure 7F). Similarly, NADPH was reduced 6 h post-LPS in WT PMφs, but was unchanged in Hif1a-deficient and Nos2-deficient PMφs (Figure 7G and H). Together, these data suggest that NADPH consumption in LPS-stimulated PMφs is dependent on HIF-1α-regulated transcription of Nos2. Finally, we assessed whether NADPH is critical for regulating the HIF-1α – Nos2 positive feedback loop. Blockade of de novo NADPH synthesis with G6PDi-1, an inhibitor of glucose-6-phosphate dehydrogenase, abrogated LPS-induced expression of HIF-1α and Nos2 protein (Figure 7I). Collectively, our data support NADPH being a cofactor that regulates the positive feedback between HIF-1α-dependent transcription of Nos2, and Nos2-dependent stabilization of HIF-1α (Figure 7J).
PMφs cultured with oxLDL and stimulated with LPS share similar inflammatory and metabolic profiles with foamy Mφs derived from the atherosclerotic mouse and human aorta.
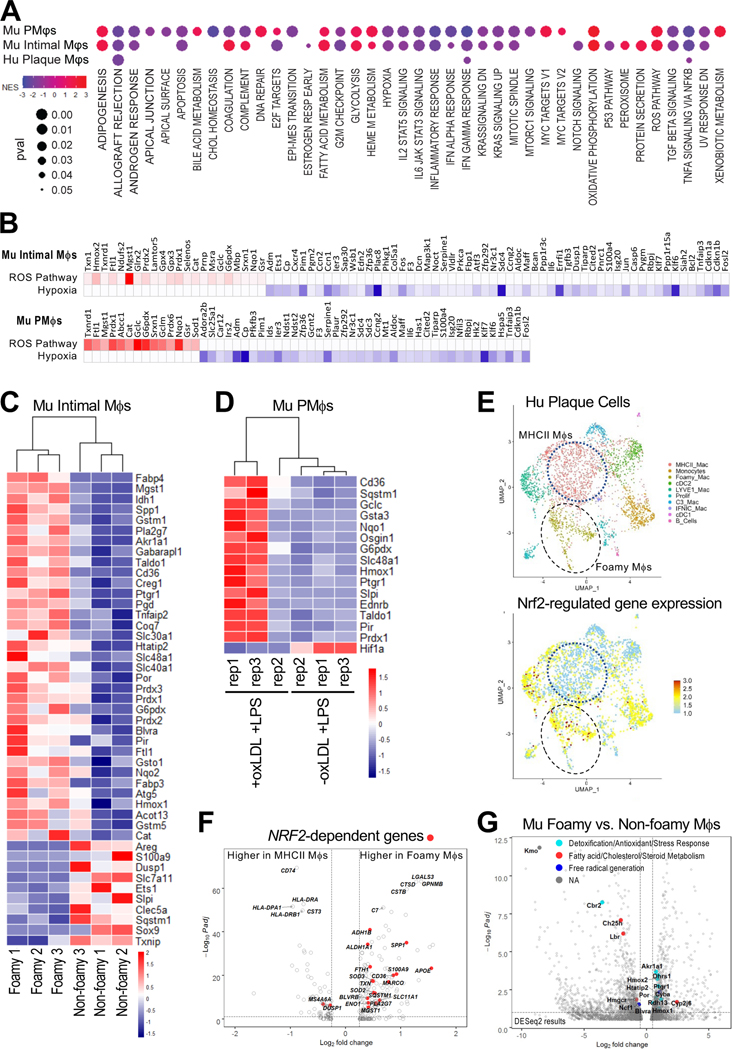
Recent in vivo transcriptomic studies have shown that foamy Mφs derived from atherosclerotic lesions are less inflammatory than non-foamy Mφs (10, 11), yet the mechanism responsible for this phenotype remains unclear. We performed comparative transcriptomic analysis of LPS-stimulated PMφs with and without intracellular oxLDL accumulation, and foamy versus non-foamy Mφs derived from atherosclerotic mouse and human aortas (10, 11). Hallmark pathway analysis revealed very similar patterns in both +oxLDL+LPS PMφs and foamy mouse Mφs (Figure 8A). Importantly, we observed downregulation of multiple inflammatory pathways, and the hypoxia pathway and upregulation of the ROS pathway. Some of the inflammatory pathways were downregulated in foamy human Mφs (Figure 8A). Leading edge analysis identified genes that contribute most to enrichment of these pathways. Many genes that were increased in the ROS pathway are Nrf2-targets related to antioxidative defense (Nqo1, Txnrd1), while genes that were reduced in the hypoxia pathway are HIF-1α-targets related to glycolysis (Aldoc) and hypoxia (Adm) (Figure 8B).
Figure 8: Transcriptomic profiling reveals similar alterations of inflammatory and metabolic pathways in cultured oxLDL-loaded LPS-stimulated PMφs and foamy Mφs from atherosclerotic mouse and human aortas.
(A) Dot plots comparing Hallmark pathway analysis of datasets from cultured murine (Mu) PMφs (+oxLDL +LPS vs. -oxLDL +LPS) and Mφs isolated from atherosclerotic mouse aorta (foamy vs. non-foamy) and Mφs isolated from human (Hu) plaques (foamy vs. non-foamy MHCII+). Only pathways with FDR <0.15 and p <0.05 are shown. Dots are colored by enrichment score and significance is proportional to the dot size. (B) Leading edge analysis of the Hallmark reactive oxygen species (ROS) and hypoxia pathways in mouse intimal Mφs and mouse PMφs. Genes that contributed most to the pathway enrichment are identified and expression values are colored to represent enrichment from low (blue) to high (red). (C, D) Heatmaps of Nrf2-regulated genes in mouse intimal foamy and non-foamy Mφs (C) and mouse PMφs (D). All represented genes had a log2FC >0.5 and Padj <0.1. (E) UMAP clustering analysis of human mononuclear phagocytes from atherosclerotic lesions and a feature plot of Nrf2-regulated gene expression projected onto the UMAP space. (F) Volcano plot of genes enriched in the human foamy and MHCII+ Mφ clusters. Differentially expressed Nrf2-regulated genes are indicated in red. The top10 differentially expressed genes (DEGs) are also labelled. G) Volcano plot of genes enriched in the mouse foamy versus non-foamy Mφ clusters identified with DESeq2. NADPH-requiring enzymes are identified among the significant DEGs.
A subset of Mφs in mouse atherosclerotic lesions expresses Nrf2-regulated markers (44). However, this study did not determine if these markers were expressed by foamy Mφs and whether Nrf2 regulates their impaired inflammatory responses. We assessed the expression of genes with a functional antioxidant response element (i.e., ARE Nrf2 binding site) in foamy mouse Mφs and +oxLDL+LPS PMφs (Figure 8C, 8D). In line with our leading edge analysis, enhanced expression of Nrf2-targets linked to antioxidative defense was observed in both populations. Similar findings were also found in foamy human Mφs in that Nrf2-dependent genes were more abundant in the Foamy Mac cluster than the inflammatory MHCII Mac cluster (Figure 8E, 8F). Finally, we assessed the expression of NADPH-requiring enzymes in foamy Mφs from the mouse aorta, and found the inflammatory enzymes (Ch25h, Hmgcr, Ncf1) were reduced, while the anti-inflammatory enzymes (Hmox1, Blvra) were increased (Figure 8G).
Discussion
Hypercholesterolemia is a key risk factor in atherosclerosis, yet its link with inflammation in the artery wall is unclear. The concept that loading of Mφs with oxLDL is intrinsically inflammatory has been pervasive in the atherosclerosis field; however, recent in vivo studies showing minimal inflammatory gene expression in foamy Mφs (10) and in vitro reports, that lipid accumulation in Mφs actually down-regulates the response to inflammatory stimuli (5, 6), are changing the paradigm. The main mechanisms underlying the suppression of inflammation in Mφs with accumulated oxLDL had remained unknown. Our current study elucidates the key targets. Analysis of RNA-seq data from LPS-stimulated PMφs revealed that oxLDL accumulation down-regulated inflammatory, hypoxia and cholesterol metabolism pathways, while ROS and some metabolic pathways were up-regulated (Figure 1). Metabolomics and Seahorse assays revealed that LPS-induced glycolysis was suppressed by oxLDL accumulation (Figure 1), as was glucose uptake, HIF-1α stabilization and expression of glycolysis genes (Figs 2, 3). Our new insights are important as previous studies established that LPS-induced glycolysis and HIF-1-mediated transcription are vital for the expression of pro-inflammatory genes in Mφs (12, 29–32).
The quality of the oxLDL preparation may be a confounding factor in the role of oxLDL accumulation on inflammation (54). We therefore used high quality medium-oxidized LDL from a reliable commercial source that specializes in lipoprotein purification and each batch was tested in house. We also cultured Mφs with 50 μg/mL of cholesterol to promote passive uptake of lipid and found that this approach yielded similar results to scavenger receptor-mediated uptake of oxLDL (cholesterol data are shown in the Supplemental Figures).
Baardman et al (7) reported previously that thioglycolate-elicited PMφs isolated from hyperlipidemic LDL receptor knockout mice displayed diminished pentose phosphate pathway metabolites and suggested a link to reduced Nrf2 response. We also found reduced levels of pentose phosphate pathway metabolites in LPS-stimulated PMφs with accumulated oxLDL (Figure 1 and Supplemental Figure 2); however, we found that an enhanced Nrf2 response was critical for suppressing LPS-induced glycolysis and inflammation. The differences between our studies may be due to different experimental conditions and timing of assays.
Our metabolomics studies showed reduced LPS-induced citrate and succinate by oxLDL accumulation. The reduction of these metabolites as well as ROS (Figure 3B) suggest that these metabolites may contribute to the suppression of HIF-1α function in LPS-stimulated oxLDL-loaded PMφs (13, 39). It seemed paradoxical that the induction of LPS-induced itaconate and fumarate was reduced in PMφs with accumulated oxLDL because an anti-inflammatory role has been established for itaconate through inhibition of succinate dehydrogenase and stimulation of Nrf2 (55, 56). Fumarate may also activate Nrf2 (57). Recently, citraconate, mesaconate and itaconate, naturally occurring isomers that differs only by the location of a double bond, were shown to profoundly modulate cell metabolism and act as strong Nrf2 agonists with anti-oxidative and anti-inflammatory functions (58). Citraconate is the strongest electrophile and Nrf2 agonist, reduces itaconate through competitive inhibition of cis-aconitate decarboxylase and prevents the accumulation of itaconate and mesaconate in activated Mφs. Unfortunately, citrocanate and mesaconate were not part of our metabolomics panel; therefore, it remains to be determined if the levels of these metabolites are higher in LPS-stimulated PMφs with accumulated oxLDL.
Although vital for inflammatory responses in Mφs, the regulation of HIF-1α-mediated transcription following TLR4 signaling by LPS is not fully defined. ROS and succinate can stabilize HIF-1α by inhibiting the activity of PHDs (13, 39); however, the incomplete rescue of glycolysis and inflammatory gene expression in our lipid loaded Vhl-deficient Mφs (Figure 3F) implies the existence of additional means to activate HIF-1α function. The repressive effects of cholesterol loading were overridden only when an engineered HIF-1α resistant to the inhibitory effects of both PHDs and FIHs was expressed (Figure 3G). This finding reveals that HIF-1α function following LPS stimulation relies both on its stability and transactivation capacity, analogous to HIF-1α function during hypoxia (41, 59). Although our study has implicated the role that FIH plays in LPS-stimulated Mφs, how FIH is inhibited by local inflammatory factors remains unclear. Interestingly, metabolites and oxidative stress could differentially inhibit PHDs and FIH activities. For instance, while citrate and oxaloacetate can inhibit both PHDs and FIHs, succinate and fumarate can only inhibit PHDs but not FIHs (60). In contrast, peroxide inhibits FIHs more than PHDs (59).
Given that HIF-1α stability takes precedence over its transactivation capacity during HIF-1 activation, it is counterintuitive for FIH to be more active than PHDs unless it can regulate HIF-1 function independent of its hydroxylation activity. Indeed, overexpressing FIH could reduce GLUT-1 mRNA even in hypoxia, and FIH could block histone deacetylases recruitment and reduce gene expression (41, 61). Overall, these studies have shown how local inflammatory factors can differentially inhibit FIH and PHDs activities, which confer possible means to differentially regulate HIF-1α transcriptional output. Not surprisingly, this is analogous to how the differences in O2 binding affinities between PHDs and FIHs underlie the canonical graded HIF-1α response to a range of hypoxic conditions.
In addition to stabilization of HIF-1α, we also showed that LPS-induced HIF-2α expression is down-regulated by oxLDL or cholesterol accumulation to a similar extent as HIF-1α (Figure 2 and Suppl Figure 3E). Both HIFα isoforms are stabilized by the inhibition of the PHD/VHL proteolytic machinery and were shown to be critical for LPS-induced inflammation (62, 63). However, the divergence in the genes that they bind to and their distinct functions in Mφ polarization suggest that further studies are required to elucidate the impact of HIF-2α down-regulation by lipid accumulation in LPS-induced Mφs (64).
HIF-1α is expressed in different vascular cells and plays a critical role in atherogenesis by promoting increased glucose uptake and inflammation in the lesions (65). For instance, in LDL receptor knockout mice reconstituted with bone marrow cells deficient in myeloid HIF-1α, a robust reduction in advanced lesions was detected in the aortic arch and root after 16 weeks of hypercholesterolemic diet. While necrotic core area was diminished, Mφ content remained the same in myeloid HIF-1α-deficient atheromas. Conversely, reconstitution with myeloid VHL deficiency yielded constitutively expressed HIF-1α and significantly increased lesions (66). Deficiency of HIF-1α in smooth muscle cells or endothelial cells in apolipoprotein E knockout mice with atherosclerosis induced by aortic constriction and/or high fat or high cholesterol diets resulted in reduced atherosclerotic lesions and Mφ content (67, 68). In view of these studies and reduced inflammation and hypoxia gene expression in foamy Mφs from murine atheromas (Figure 8) as well as oxLDL-loaded LPS-stimulated PMφs (Figure 1 and 8), it is likely that oxidative stress and HIF-1α-mediated inflammation in vascular SMC and EC may have a prominent role in fueling atherogenesis, while Mφ foam cells dampen oxidative stress and glycolysis to enhance their survival in a milieu of chronic inflammation.
We found that destabilization of HIF-1α was accompanied by enhanced Nrf2 activity, which confers antioxidant response against the oxidative radicals. (Figs. 4, 5) (69). The accumulation of oxLDL in cultured Mφs progressively induced oxidative stress that stabilized Nrf2 and activated a primary Nrf2-antixodiative response without inducing an inflammatory response (Figure 4A-C). Bae et al (2009) showed that treatment of J774 cells with minimally modified LDL (mmLDL) or oxLDL induced ROS within 10 minutes and ROS production in response to mmLDL was dependent on Nox2 activation and TLR4 signaling (70). This study raises the possibility that during the early stages of lipid loading Mφs responses to pro-inflammatory stimuli may be accentuated. We, however, focused on later stages of lipid accumulation. We observed that ROS increased over time and peaked at 18 hours (Figure 4A). Mφs adapted to this by activating Nrf2 and expressing Nrf2-dependent antioxidant genes (Figure 4B, C), without an induction of pro-inflammatory gene expression. Upon LPS stimulation, the subsequent Nrf2 response was enhanced in cells with accumulated oxLDL (Figure 4D-G). We showed a link between enhanced Nrf2 and reduced HIF-1α by using Nrf2-deficient mice where ROS levels as well as glycolysis and inflammatory genes were rescued and HIF-1α levels were stabilized (Figure 5). Overall, the enhancement of Nrf2-dependent antioxidative defense in Mφs with accumulated lipid impaired LPS-induced ROS production, thereby destabilizing HIF-1α-dependent glycolytic and proinflammatory responses.
NADPH plays a multifunctional role in regulating Mφ inflammation. While it is consumed by apoenzymes for the synthesis of lipids and ROS, it is also used for detoxification and to maintain redox homeostasis. NADPH can support inflammatory and anti-inflammatory processes simultaneously, but how NADPH-dependent pathways coordinate to fine-tune an inflammatory response remains to be determined. Here, we show that NADPH usage is shared between HIF-1α and Nrf2-dependent responses. Upon LPS stimulation, NADPH is consumed by NOSs to generate reactive radicals and by detoxification enzymes to limit the abundance of radicals. Shifts in these opposing processes fine-tune reactive radical levels, the stabilization of HIF-1α, and optimize the inflammatory response. The accumulation of oxLDL in Mφs significantly upregulated LPS-induced expression of NADPH-requiring detoxification enzymes. This not only repurposed NADPH consumption for the detoxification of reactive radicals, but also reduced the availability of NADPH for reactive radical synthesis, ultimately deactivating inflammation. Overall, we argue that under conditions where NADPH is limited, inflammatory and anti-inflammatory processes compete for NADPH to the detriment of each other. This competition is important, not only because it reveals how NADPH-dependent pathways coordinate to regulate inflammation, but more importantly, other cofactors may also be consumed in a competitive manner by biologically opposing pathways. Indeed, an example of this is NADH consumption, which is vital for LPS-induced glycolysis and poly (ADP-ribose) polymerase (PARP)-mediated DNA repair response (71), where PARP hyperactivation can impair glycolysis (72, 73). Along with these reports, our study reveals an emerging biological principle, which proposes that the competition for cofactor consumption between inflammatory and anti-inflammatory pathways is a naturally evolved mechanism to regulate inflammation efficiently. Specifically, the extent of this competition underlies the magnitude of an inflammatory response. Future studies are warranted to identify other cofactors shared between biologically opposing pathways. Since the consumption rate of cofactors relies on the differential transcription of apoenzymes in response to external stimuli, this also shows how the sensing of extracellular cues by transcription factors regulates inflammation.
Comparative transcriptomic analysis of LPS-stimulated PMφs with accumulated oxLDL and foamy Mφs from mouse and human atherosclerotic aortas revealed similar metabolic and inflammatory profiles (Figure 8A), demonstrating the pathophysiological relevance of our in vitro findings. Comparison of bulk seq data sets is robust, whereas comparison of bulk and single cell transcriptomic data is limited by the relatively lower depth of the single cell data. Thus, although similar trends were identified in mouse and human data sets, only several inflammatory pathways were reduced in foamy human plaque Mφs. Downregulation of the hypoxia and multiple inflammatory pathways and upregulation of the reactive oxygen species (ROS) pathway in the mouse data sets (Figure 8B) is consistent with the involvement of HIF-1α and Nrf2, respectively. Subsequently we established the upregulation of Nrf2-dependent genes in Mφs with accumulated oxLDL and foamy Mφs from mouse and human atherosclerotic aortas (Fig 8C-G). Our observations are consistent with studies on atherosclerosis where myeloid Nrf2 deficiency in LDL receptor knockout mice resulted in increased atherosclerosis both in early and late stages, a finding that was consistent with studies interrogating the role of anti-oxidant genes by genetic approaches (74–76).
While hypercholesterolemia and foamy Mφs are key players in atherosclerosis, our study argues that intracellular lipid accumulation in Mφs is insufficient to drive inflammation and in fact dampens their inflammatory response to external stimuli because enhanced Nrf2-regulated transcription of antioxidative enzymes impairs a HIF-1α-regulated glycolytic response. This implicates the existence of other factors, such as oxidized phospholipids, or other vascular cells as potential drivers of inflammation in atherosclerotic lesions (77).
Supplementary Material
Key points:
OxLDL accumulation in Mφs impairs LPS-induced HIF-1α-dependent glycolysis.
OxLDL accumulation in Mφs enhances LPS-induced Nrf2-dependent antioxidative response.
Competition for NADPH between HIF-1 and Nrf2 processes fine-tunes inflammation.
Acknowledgements
We thank Dr. Alma Zernecke-Madsen and Dr. Clément Cochain for sharing their bioinformatics data prior to the acceptance of their publication. Dr. Michael Wilson and Mr. Azad Alizada assisted with the analysis of RNA-sequencing. Dr. Jason Fish provided the CMV–Renilla luciferase plasmid construct and access to a GloMax 20/20 luminometer. Dr. Tracy McGaha provided us the Lys2-Cre:Hif1afl/fl mice.
This study was supported by the Canadian Institutes of Health Research (CIHR) grant FDN-154299 (M.I.C.). Additional support was from the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences (Z01 ES102005) (M.B.F.), CIHR project grant (DOL 409157) (J.R.) and NSERC Research Tools and Instrumentation Grant (2018–00846) (J.R.). M.I.C. holds a Tier 1 Canada Research Chair. K.K.Y.T and C.A.S. were supported by fellowships from Canadian Institutes of Health Research. K.K.Y.T. and H.I. were supported by Ontario Graduate Scholarship. K.K.Y.T. was supported by fellowships from University of Toronto and The Peterborough K. M. Hunter Charitable Foundation.
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Guerrini V, and Gennaro ML. 2019. Foam cells: One size doesn’t fit all. Trends Immunol 40: 1163–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paulson KE, Zhu SN, Chen M, Nurmohamed S, Jongstra-Bilen J, and Cybulsky MI. 2010. Resident intimal dendritic cells accumulate lipid and contribute to the initiation of atherosclerosis. Circ Res 106: 383–390. [DOI] [PubMed] [Google Scholar]
- 3.Stewart CR, Stuart LM, Wilkinson K, van Gils JM, Deng J, Halle A, Rayner KJ, Boyer L, Zhong R, Frazier WA, Lacy-Hulbert A, El Khoury J, Golenbock DT, and Moore KJ. 2010. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nat Immunol 11: 155–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duewell P, Kono H, Rayner KJ, Sirois CM, Vladimer G, Bauernfeind FG, Abela GS, Franchi L, Nunez G, Schnurr M, Espevik T, Lien E, Fitzgerald KA, Rock KL, Moore KJ, Wright SD, Hornung V, and Latz E. 2010. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature 464: 1357–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spann NJ, Garmire LX, McDonald JG, Myers DS, Milne SB, Shibata N, Reichart D, Fox JN, Shaked I, Heudobler D, Raetz CR, Wang EW, Kelly SL, Sullards MC, Murphy RC, Merrill AH Jr., Brown HA, Dennis EA, Li AC, Ley K, Tsimikas S, Fahy E, Subramaniam S, Quehenberger O, Russell DW, and Glass CK. 2012. Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell 151: 138–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jongstra-Bilen J, Zhang CX, Wisnicki T, Li MK, White-Alfred S, Ilaalagan R, Ferri DM, Deonarain A, Wan MH, Hyduk SJ, Cummins CL, and Cybulsky MI. 2017. Oxidized low-density lipoprotein loading of macrophages downregulates TLR-induced proinflammatory responses in a gene-specific and temporal manner through transcriptional control. J Immunol 199: 2149–2157. [DOI] [PubMed] [Google Scholar]
- 7.Baardman J, Verberk SGS, Prange KHM, van Weeghel M, van der Velden S, Ryan DG, Wust RCI, Neele AE, Speijer D, Denis SW, Witte ME, Houtkooper RH, O’Neill L A, Knatko EV, Dinkova-Kostova AT, Lutgens E, de Winther MPJ, and Van den Bossche J. 2018. A Defective Pentose Phosphate Pathway Reduces Inflammatory Macrophage Responses during Hypercholesterolemia. Cell Rep 25: 2044–2052 e2045. [DOI] [PubMed] [Google Scholar]
- 8.Ikeda J, Scipione CA, Hyduk SJ, Althagafi MG, Atif J, Dick SA, Rajora M, Jang E, Emoto T, Murakami J, Ikeda N, Ibrahim HM, Polenz CK, Gao X, Tai K, Jongstra-Bilen J, Nakashima R, Epelman S, Robbins CS, Zheng G, Lee WL, MacParland SA, and Cybulsky MI. 2021. Radiation Impacts Early Atherosclerosis by Suppressing Intimal LDL Accumulation. Circ Res 128: 530–543. [DOI] [PubMed] [Google Scholar]
- 9.Zhu SN, Chen M, Jongstra-Bilen J, and Cybulsky MI. 2009. GM-CSF regulates intimal cell proliferation in nascent atherosclerotic lesions. J Exp Med 206: 2141–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim K, Shim D, Lee JS, Zaitsev K, Williams JW, Kim KW, Jang MY, Seok Jang H, Yun TJ, Lee SH, Yoon WK, Prat A, Seidah NG, Choi J, Lee SP, Yoon SH, Nam JW, Seong JK, Oh GT, Randolph GJ, Artyomov MN, Cheong C, and Choi JH. 2018. Transcriptome Analysis Reveals Nonfoamy Rather Than Foamy Plaque Macrophages Are Proinflammatory in Atherosclerotic Murine Models. Circ Res 123: 1127–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zernecke A, Erhard F, Weinberger T, Schulz C, Ley K, Saliba AE, and Cochain C. 2022. Integrated single-cell analysis based classification of vascular mononuclear phagocytes in mouse and human atherosclerosis. Cardiovasc Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, Zheng L, Gardet A, Tong Z, Jany SS, Corr SC, Haneklaus M, Caffrey BE, Pierce K, Walmsley S, Beasley FC, Cummins E, Nizet V, Whyte M, Taylor CT, Lin H, Masters SL, Gottlieb E, Kelly VP, Clish C, Auron PE, Xavier RJ, and O’Neill LA. 2013. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature 496: 238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mills EL, Kelly B, Logan A, Costa ASH, Varma M, Bryant CE, Tourlomousis P, Dabritz JHM, Gottlieb E, Latorre I, Corr SC, McManus G, Ryan D, Jacobs HT, Szibor M, Xavier RJ, Braun T, Frezza C, Murphy MP, and O’Neill LA. 2016. Succinate Dehydrogenase Supports Metabolic Repurposing of Mitochondria to Drive Inflammatory Macrophages. Cell 167: 457–470 e413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang HH, Bennett AM, Cameron WD, Floro E, Au A, McFaul CM, Yip CM, and Rocheleau JV. 2022. Targeting Apollo-NADP(+) to Image NADPH Generation in Pancreatic Beta-Cell Organelles. ACS Sens 7: 3308–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wan LC, Mao DY, Neculai D, Strecker J, Chiovitti D, Kurinov I, Poda G, Thevakumaran N, Yuan F, Szilard RK, Lissina E, Nislow C, Caudy AA, Durocher D, and Sicheri F. 2013. Reconstitution and characterization of eukaryotic N6-threonylcarbamoylation of tRNA using a minimal enzyme system. Nucleic Acids Res 41: 6332–6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Love MI, Huber W, and Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blighe K, Rana S, and Lewis M. 2018. EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling. R/Bioconductor package. [Google Scholar]
- 18.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, and Groop LC. 2003. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34: 267–273. [DOI] [PubMed] [Google Scholar]
- 20.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merico D, Isserlin R, Stueker O, Emili A, and Bader GD. 2010. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 5: e13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satija R, Farrell JA, Gennert D, Schier AF, and Regev A. 2015. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33: 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R. 2019. Comprehensive integration of single-cell data. Cell 177: 1888–1902 e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez DM, Rahman AH, Fernandez NF, Chudnovskiy A, Amir ED, Amadori L, Khan NS, Wong CK, Shamailova R, Hill CA, Wang Z, Remark R, Li JR, Pina C, Faries C, Awad AJ, Moss N, Bjorkegren JLM, Kim-Schulze S, Gnjatic S, Ma’ayan A, Mocco J, Faries P, Merad M, and Giannarelli C. 2019. Single-cell immune landscape of human atherosclerotic plaques. Nat Med 25: 1576–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wirka RC, Wagh D, Paik DT, Pjanic M, Nguyen T, Miller CL, Kundu R, Nagao M, Coller J, Koyano TK, Fong R, Woo YJ, Liu B, Montgomery SB, Wu JC, Zhu K, Chang R, Alamprese M, Tallquist MD, Kim JB, and Quertermous T. 2019. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat Med 25: 1280–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Axelrod D. 1979. Carbocyanine dye orientation in red cell membrane studied by microscopic fluorescence polarization. Biophys J 26: 557–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blackman SM, Cobb CE, Beth AH, and Piston DW. 1996. The orientation of eosin-5-maleimide on human erythrocyte band 3 measured by fluorescence polarization microscopy. Biophys J 71: 194–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rocheleau JV, Edidin M, and Piston DW. 2003. Intrasequence GFP in class I MHC molecules, a rigid probe for fluorescence anisotropy measurements of the membrane environment. Biophys J 84: 4078–4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langston PK, Nambu A, Jung J, Shibata M, Aksoylar HI, Lei J, Xu P, Doan MT, Jiang H, MacArthur MR, Gao X, Kong Y, Chouchani ET, Locasale JW, Snyder NW, and Horng T. 2019. Glycerol phosphate shuttle enzyme GPD2 regulates macrophage inflammatory responses. Nat Immunol 20: 1186–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lauterbach MA, Hanke JE, Serefidou M, Mangan MSJ, Kolbe CC, Hess T, Rothe M, Kaiser R, Hoss F, Gehlen J, Engels G, Kreutzenbeck M, Schmidt SV, Christ A, Imhof A, Hiller K, and Latz E. 2019. Toll-like Receptor Signaling Rewires Macrophage Metabolism and Promotes Histone Acetylation via ATP-Citrate Lyase. Immunity 51: 997–1011 e1017. [DOI] [PubMed] [Google Scholar]
- 31.Wang T, Liu H, Lian G, Zhang SY, Wang X, and Jiang C. 2017. HIF1alpha-Induced Glycolysis Metabolism Is Essential to the Activation of Inflammatory Macrophages. Mediators Inflamm 2017: 9029327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peyssonnaux C, Datta V, Cramer T, Doedens A, Theodorakis EA, Gallo RL, Hurtado-Ziola N, Nizet V, and Johnson RS. 2005. HIF-1alpha expression regulates the bactericidal capacity of phagocytes. J Clin Invest 115: 1806–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang LE, Gu J, Schau M, and Bunn HF. 1998. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci U S A 95: 7987–7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Rourke JF, Tian YM, Ratcliffe PJ, and Pugh CW. 1999. Oxygen-regulated and transactivating domains in endothelial PAS protein 1: comparison with hypoxia-inducible factor-1alpha. J Biol Chem 274: 2060–2071. [DOI] [PubMed] [Google Scholar]
- 35.Gu YZ, Moran SM, Hogenesch JB, Wartman L, and Bradfield CA. 1998. Molecular characterization and chromosomal localization of a third alpha-class hypoxia inducible factor subunit, HIF3alpha. Gene Expr 7: 205–213. [PMC free article] [PubMed] [Google Scholar]
- 36.Hubbi ME, Hu H, Kshitiz I Ahmed A. Levchenko, and G. Semenza L. 2013. Chaperone-mediated autophagy targets hypoxia-inducible factor-1α (HIF-1α) for lysosomal degradation. J Biol Chem 288: 10703–10714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, and Ratcliffe PJ. 2001. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292: 468–472. [DOI] [PubMed] [Google Scholar]
- 38.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, and Kaelin WG Jr. 2001. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292: 464–468. [DOI] [PubMed] [Google Scholar]
- 39.Pan Y, Mansfield KD, Bertozzi CC, Rudenko V, Chan DA, Giaccia AJ, and Simon MC. 2007. Multiple factors affecting cellular redox status and energy metabolism modulate hypoxia-inducible factor prolyl hydroxylase activity in vivo and in vitro. Mol Cell Biol 27: 912–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishi K, Oda T, Takabuchi S, Oda S, Fukuda K, Adachi T, Semenza GL, Shingu K, and Hirota K. 2008. LPS induces hypoxia-inducible factor 1 activation in macrophage-differentiated cells in a reactive oxygen species-dependent manner. Antioxid Redox Signal 10: 983–995. [DOI] [PubMed] [Google Scholar]
- 41.Mahon PC, Hirota K, and Semenza GL. 2001. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev 15: 2675–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, and Bruick RK. 2002. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev 16: 1466–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ishii T, Itoh K, Ruiz E, Leake DS, Unoki H, Yamamoto M, and Mann GE. 2004. Role of Nrf2 in the regulation of CD36 and stress protein expression in murine macrophages: activation by oxidatively modified LDL and 4-hydroxynonenal. Circ Res 94: 609–616. [DOI] [PubMed] [Google Scholar]
- 44.Kadl A, Meher AK, Sharma PR, Lee MY, Doran AC, Johnstone SR, Elliott MR, Gruber F, Han J, Chen W, Kensler T, Ravichandran KS, Isakson BE, Wamhoff BR, and Leitinger N. 2010. Identification of a novel macrophage phenotype that develops in response to atherogenic phospholipids via Nrf2. Circ Res 107: 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, and Nabeshima Y. 1997. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 236: 313–322. [DOI] [PubMed] [Google Scholar]
- 46.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, and Yamamoto M. 1999. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev 13: 76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baird L, and Yamamoto M. 2020. The Molecular Mechanisms Regulating the KEAP1-NRF2 Pathway. Mol Cell Biol 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kosower NS, Kosower EM, Wertheim B, and Correa WS. 1969. Diamide, a new reagent for the intracellular oxidation of glutathione to the disulfide. Biochemical and Biophysical Research Communications 37: 593–596. [DOI] [PubMed] [Google Scholar]
- 49.Mari M, de Gregorio E, de Dios C, Roca-Agujetas V, Cucarull B, Tutusaus A, Morales A, and Colell A. 2020. Mitochondrial Glutathione: Recent Insights and Role in Disease. Antioxidants (Basel) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cameron WD, Bui CV, Hutchinson A, Loppnau P, Gräslund S, and Rocheleau JV. 2016. Apollo-NADP+: a spectrally tunable family of genetically encoded sensors for NADP+. Nature Methods 13: 352–358. [DOI] [PubMed] [Google Scholar]
- 51.Melillo G, Musso T, Sica A, Taylor LS, Cox GW, and Varesio L. 1995. A hypoxia-responsive element mediates a novel pathway of activation of the inducible nitric oxide synthase promoter. J Exp Med 182: 1683–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bailey JD, Diotallevi M, Nicol T, McNeill E, Shaw A, Chuaiphichai S, Hale A, Starr A, Nandi M, Stylianou E, McShane H, Davis S, Fischer R, Kessler BM, McCullagh J, Channon KM, and Crabtree MJ. 2019. Nitric Oxide Modulates Metabolic Remodeling in Inflammatory Macrophages through TCA Cycle Regulation and Itaconate Accumulation. Cell Rep 28: 218–230.e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kasuno K, Takabuchi S, Fukuda K, Kizaka-Kondoh S, Yodoi J, Adachi T, Semenza GL, and Hirota K. 2004. Nitric oxide induces hypoxia-inducible factor 1 activation that is dependent on MAPK and phosphatidylinositol 3-kinase signaling. J Biol Chem 279: 2550–2558. [DOI] [PubMed] [Google Scholar]
- 54.Kannan Y, Sundaram K, Aluganti Narasimhulu C, Parthasarathy S, and Wewers MD. 2012. Oxidatively modified low density lipoprotein (LDL) inhibits TLR2 and TLR4 cytokine responses in human monocytes but not in macrophages. J Biol Chem 287: 23479–23488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mills EL, Ryan DG, Prag HA, Dikovskaya D, Menon D, Zaslona Z, Jedrychowski MP, Costa ASH, Higgins M, Hams E, Szpyt J, Runtsch MC, King MS, McGouran JF, Fischer R, Kessler BM, McGettrick AF, Hughes MM, Carroll RG, Booty LM, Knatko EV, Meakin PJ, Ashford MLJ, Modis LK, Brunori G, Sevin DC, Fallon PG, Caldwell ST, Kunji ERS, Chouchani ET, Frezza C, Dinkova-Kostova AT, Hartley RC, Murphy MP, and O’Neill LA. 2018. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 556: 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lampropoulou V, Sergushichev A, Bambouskova M, Nair S, Vincent EE, Loginicheva E, Cervantes-Barragan L, Ma X, Huang SC, Griss T, Weinheimer CJ, Khader S, Randolph GJ, Pearce EJ, Jones RG, Diwan A, Diamond MS, and Artyomov MN. 2016. Itaconate Links Inhibition of Succinate Dehydrogenase with Macrophage Metabolic Remodeling and Regulation of Inflammation. Cell Metab 24: 158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Linker RA, Lee DH, Ryan S, van Dam AM, Conrad R, Bista P, Zeng W, Hronowsky X, Buko A, Chollate S, Ellrichmann G, Bruck W, Dawson K, Goelz S, Wiese S, Scannevin RH, Lukashev M, and Gold R. 2011. Fumaric acid esters exert neuroprotective effects in neuroinflammation via activation of the Nrf2 antioxidant pathway. Brain 134: 678–692. [DOI] [PubMed] [Google Scholar]
- 58.Chen F, Elgaher WAM, Winterhoff M, Bussow K, Waqas FH, Graner E, Pires-Afonso Y, Casares Perez L, de la Vega L, Sahini N, Czichon L, Zobl W, Zillinger T, Shehata M, Pleschka S, Bahre H, Falk C, Michelucci A, Schuchardt S, Blankenfeldt W, Hirsch AKH, and Pessler F. 2022. Citraconate inhibits ACOD1 (IRG1) catalysis, reduces interferon responses and oxidative stress, and modulates inflammation and cell metabolism. Nat Metab 4: 534–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Masson N, Singleton RS, Sekirnik R, Trudgian DC, Ambrose LJ, Miranda MX, Tian YM, Kessler BM, Schofield CJ, and Ratcliffe PJ. 2012. The FIH hydroxylase is a cellular peroxide sensor that modulates HIF transcriptional activity. EMBO Rep 13: 251–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koivunen P, Hirsilä M, Remes AM, Hassinen IE, Kivirikko KI, and Myllyharju J. 2007. Inhibition of hypoxia-inducible factor (HIF) hydroxylases by citric acid cycle intermediates: possible links between cell metabolism and stabilization of HIF. J Biol Chem 282: 4524–4532. [DOI] [PubMed] [Google Scholar]
- 61.Wang E, Zhang C, Polavaram N, Liu F, Wu G, Schroeder MA, Lau JS, Mukhopadhyay D, Jiang SW, O’Neill BP, Datta K, and Li J. 2014. The role of factor inhibiting HIF (FIH-1) in inhibiting HIF-1 transcriptional activity in glioblastoma multiforme. PLoS One 9: e86102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Imtiyaz HZ, Williams EP, Hickey MM, Patel SA, Durham AC, Yuan LJ, Hammond R, Gimotty PA, Keith B, and Simon MC. 2010. Hypoxia-inducible factor 2alpha regulates macrophage function in mouse models of acute and tumor inflammation. J Clin Invest 120: 2699–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McGettrick AF, and O’Neill LAJ. 2020. The Role of HIF in Immunity and Inflammation. Cell Metab 32: 524–536. [DOI] [PubMed] [Google Scholar]
- 64.Smythies JA, Sun M, Masson N, Salama R, Simpson PD, Murray E, Neumann V, Cockman ME, Choudhry H, Ratcliffe PJ, and Mole DR. 2019. Inherent DNA-binding specificities of the HIF-1alpha and HIF-2alpha transcription factors in chromatin. EMBO Rep 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thomas C, Leleu D, and Masson D. 2022. Cholesterol and HIF-1alpha: Dangerous Liaisons in Atherosclerosis. Front Immunol 13: 868958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aarup A, Pedersen TX, Junker N, Christoffersen C, Bartels ED, Madsen M, Nielsen CH, and Nielsen LB. 2016. Hypoxia-Inducible Factor-1alpha Expression in Macrophages Promotes Development of Atherosclerosis. Arterioscler Thromb Vasc Biol 36: 1782–1790. [DOI] [PubMed] [Google Scholar]
- 67.Liu D, Lei L, Desir M, Huang Y, Cleman J, Jiang W, Fernandez-Hernando C, Di Lorenzo A, Sessa WC, and Giordano FJ. 2016. Smooth Muscle Hypoxia-Inducible Factor 1alpha Links Intravascular Pressure and Atherosclerosis--Brief Report. Arterioscler Thromb Vasc Biol 36: 442–445. [DOI] [PubMed] [Google Scholar]
- 68.Akhtar S, Hartmann P, Karshovska E, Rinderknecht FA, Subramanian P, Gremse F, Grommes J, Jacobs M, Kiessling F, Weber C, Steffens S, and Schober A. 2015. Endothelial Hypoxia-Inducible Factor-1alpha Promotes Atherosclerosis and Monocyte Recruitment by Upregulating MicroRNA-19a. Hypertension 66: 1220–1226. [DOI] [PubMed] [Google Scholar]
- 69.Kuhn AM, Tzieply N, Schmidt MV, von Knethen A, Namgaladze D, Yamamoto M, and Brune B. 2011. Antioxidant signaling via Nrf2 counteracts lipopolysaccharide-mediated inflammatory responses in foam cell macrophages. Free Radic Biol Med 50: 1382–1391. [DOI] [PubMed] [Google Scholar]
- 70.Bae YS, Lee JH, Choi SH, Kim S, Almazan F, Witztum JL, and Miller YI. 2009. Macrophages Generate Reactive Oxygen Species in Response to Minimally Oxidized Low-Density Lipoprotein. Circulation Research 104, 210–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cameron AM, Castoldi A, Sanin DE, Flachsmann LJ, Field CS, Puleston DJ, Kyle RL, Patterson AE, Hässler F, Buescher JM, Kelly B, Pearce EL, and Pearce EJ. 2019. Inflammatory macrophage dependence on NAD+ salvage is a consequence of reactive oxygen species–mediated DNA damage. Nature Immunology 20: 420–432. [DOI] [PubMed] [Google Scholar]
- 72.Murata MM, Kong X, Moncada E, Chen Y, Imamura H, Wang P, Berns MW, Yokomori K, and Digman MA. 2019. NAD+ consumption by PARP1 in response to DNA damage triggers metabolic shift critical for damaged cell survival. Mol Biol Cell 30: 2584–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Szántó M, and Bai P. 2020. The role of ADP-ribose metabolism in metabolic regulation, adipose tissue differentiation, and metabolism. Genes Dev 34: 321–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mimura J, and Itoh K. 2015. Role of Nrf2 in the pathogenesis of atherosclerosis. Free Radic Biol Med 88: 221–232. [DOI] [PubMed] [Google Scholar]
- 75.Ruotsalainen AK, Inkala M, Partanen ME, Lappalainen JP, Kansanen E, Makinen PI, Heinonen SE, Laitinen HM, Heikkila J, Vatanen T, Horkko S, Yamamoto M, Yla-Herttuala S, Jauhiainen M, and Levonen AL. 2013. The absence of macrophage Nrf2 promotes early atherogenesis. Cardiovasc Res 98: 107–115. [DOI] [PubMed] [Google Scholar]
- 76.Collins AR, Gupte AA, Ji R, Ramirez MR, Minze LJ, Liu JZ, Arredondo M, Ren Y, Deng T, Wang J, Lyon CJ, and Hsueh WA. 2012. Myeloid deletion of nuclear factor erythroid 2-related factor 2 increases atherosclerosis and liver injury. Arterioscler Thromb Vasc Biol 32: 2839–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Que X, Hung M-Y, Yeang C, Gonen A, Prohaska TA, Sun X, Diehl C, Määttä A, Gaddis DE, Bowden K, Pattison J, MacDonald JG, Ylä-Herttuala S, Mellon PL, Hedrick CC, Ley K, Miller YI, Glass CK, Peterson KL, Binder CJ, Tsimikas S, and Witztum JL. 2018. Oxidized phospholipids are proinflammatory and proatherogenic in hypercholesterolaemic mice. Nature 558: 301–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.