Abstract
Mycoplasma pneumoniae infections represent a major primary cause of human respiratory diseases, exacerbate other respiratory disorders, and are associated with extrapulmonary pathologies. Cytadherence is a critical step in mycoplasma colonization, aided by a network of mycoplasma adhesins and cytadherence accessory proteins which mediate binding to host cell receptors. Furthermore, the respiratory mucosa is enriched with extracellular matrix components, including surfactant proteins, fibronectin, and mucin, which provide additional in vivo targets for mycoplasma parasitism. In this study we describe interactions between M. pneumoniae and human surfactant protein-A (hSP-A). Initially, we found that viable M. pneumoniae cells bound to immobilized hSP-A in a dose- and calcium (Ca2+)-dependent manner. Mild trypsin treatment of intact mycoplasmas reduced binding markedly (80 to 90%) implicating a surface-associated mycoplasma protein(s). Using hSP-A-coupled Sepharose affinity chromatography and polyacrylamide gel electrophoresis, we identified a 65-kDa hSP-A binding protein of M. pneumoniae. The presence of Ca2+ enhanced binding of the 65-kDa protein to hSP-A, which was reduced by the divalent cation-chelating agent, EDTA. The 65-kDa hSP-A binding protein of M. pneumoniae was identified by sequence analysis as a novel protein (MPN372) possessing a putative S1-like subunit of pertussis toxin at the amino terminus (amino acids 1 to 226), with the remaining amino acids (227 to 591) exhibiting no homology with other subunits of pertussis toxin, other known toxins, or any reported proteins. Recombinant MPN372 (MPN372) bound to hSP-A in a dose-dependent manner, which was markedly reduced by preincubation with mouse recombinant MPN372 antisera. Also, adherence of viable M. pneumoniae cells to hSP-A was inhibited by recombinant MPN372 antisera, demonstrating that MPN372, a previously designated hypothetical protein, is surface exposed and mediates mycoplasma attachment to hSP-A.
The importance of Mycoplasma pneumoniae as a causative agent of acute and chronic human respiratory diseases and extrapulmonary pathologies has been well documented by epidemiological studies in many settings (8). M. pneumoniae is the etiologic agent of primary atypical pneumonia and is also responsible for a spectrum of other respiratory tract infections, including tracheobronchitis, bronchiolitis, pharyngitis, and croup in children and young adults (8). This bacterial pathogen accounts for 20 to 30% of all community-acquired pneumonia and has been implicated in asthma and chronic obstructive pulmonary disease (26). Furthermore, M. pneumoniae infections can lead to gastrointestinal, hematologic, neurologic, dermatologic, musculoskeletal, joint, and cardiovascular pathologies, in part manifested as arthritis, pericarditis, and central nervous system disorders (7). Therefore, the pathogenic potential of M. pneumoniae is substantial, and it is becoming clear that M. pneumoniae-associated diseases are greatly underreported because of the wide diversity of clinical manifestations linked to M. pneumoniae infections, difficulties in cultivating these pathogens directly from clinical specimens, and inadequate diagnostic tools.
M. pneumoniae is considered among the smallest self-replicating prokaryotic pathogens, with a genome size of 816 kb. It utilizes a unique terminal tip organelle composed of a network of specialized proteins which mediate adherence to sialylated and sulfated receptors on target cells (4, 27). Many of the tip-associated, adherence-related mycoplasma proteins appear to be homologues of cytoskeletal proteins of eukaryotes, consistent with their function to mobilize and cluster proteins to the mycoplasma tip structure and promote colonization of host tissues. Recently, we described a novel fibronectin-mediated pathway of M. pneumoniae-host cell interaction (13), suggesting that pathogenic mycoplasmas possess multiple and distinct mechanisms of parasitism.
Surfactant protein A (SP-A) is the major protein associated with pulmonary surfactant. SP-A is a multimeric protein composed of 28- to 36-kDa peptides with structural and functional similarities to other members of the collectin family, including pulmonary SP-D, serum mannose binding protein, conglutinin, and CL-43 (10). This category of molecules contains an NH2-terminal collagenous domain and a COOH-terminal carbohydrate recognition domain capable of binding ligands via Ca2+-dependent mechanisms (10, 11, 37). SP-A is expressed in alveolar type II cells, Clara cells, and submucosal glands of the respiratory airways and has also been detected in epithelial cells lining extrapulmonary sites (2, 25).
In vitro studies have demonstrated that SP-A binds to specific strains of respiratory bacterial pathogens, including Staphylococcus aureus (29), Streptococcus pneumoniae (29), group B streptococci (28), Haemophilus influenzae (33), Pseudomonas aeruginosa (24), Klebsiella pneumoniae (23), Mycobacterium bovis bacillus Calmette-Guérin (34), and Mycobacterium tuberculosis (16, 17, 31). Since SP-A stimulates phagocytosis by binding to alveolar macrophages, it is considered an important contributor to innate lung immunity. However, SP-A also promotes attachment and entry of M. tuberculosis into alveolar macrophages, thereby assisting mycobacteria in their infectious process (16, 17, 31). Similarly, SP-A can enhance antiviral activity or viral infectivity through increased binding and internalization of virus particles (9, 35). Thus, SP-A can serve a double-edged function, by either assisting in pathogen elimination or facilitating infection, depending on the biological properties of the infectious agent and the targeted host cell.
The successes of M. pneumoniae as a pulmonary pathogen indicate that these mycoplasmas are capable of parasitizing the airways by circumventing or exploiting host defense mechanisms. In the present study we initiated characterization of the interaction between M. pneumoniae and human SP-A (hSP-A) in order to clarify the dynamics of mycoplasma-associated human respiratory infections and the role of innate host defense mechanisms. Here, we describe a novel mycoplasma hSP-A binding protein which may play a significant role in disease pathogenesis.
MATERIALS AND METHODS
Bacterial culture conditions and radiolabeling.
Wild-type Mycoplasma pneumoniae reference strain B9 and clinical strain S1, the latter isolated from an outbreak of M. pneumoniae respiratory infections in San Antonio (unpublished data), were grown in SP-4 medium at 37°C for 72 to 96 h in 150-cm2 tissue culture flasks as described earlier (14). Mycoplasma monolayers in the mid- to late logarithmic growth phase were washed two times with 10 ml Tris-buffered saline (TBS, pH 7.4), one time with Dulbecco's modified Eagle's medium (DMEM) without l-methionine, and resuspended in 10 ml DMEM without L-methionine, which was supplemented with 10% heat-inactivated fetal bovine serum and 100 μCi [35S]methionine. After 4 h incubation at 37°C supernatants were removed, and biosynthetically radiolabeled mycoplasma monolayers were washed twice with 25 ml TBS. Mycoplasma cells were scraped off the flask into a volume of 10 ml sterile TBS and collected by centrifugation at 9,500 × g. Cell pellets were resuspended in TBS and dispersed through 25 gauge needles to reduce mycoplasma clumps before use in binding assays (see below). Other radiolabeled mycoplasma pellets were resuspended in complete lysis buffer (CLB: 150 mM NaCl, 10 mM Tris, 20 μM EGTA, 0.5 M Triton X-114, 1 mM CaCl2 and protease inhibitors) and sheared through 25 gauge needles to achieve clear lysis. These cell lysates were passed through control and hSP-A coupled Sepharose columns and analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) analysis.
Purification of SP-A.
SP-A was purified from the lavage fluid of patients with alveolar proteinosis as previously described (38). In brief, the lipid-rich pellet was extracted sequentially with butanol, octylglucoside, and low-salt buffer. SP-A preparations were then treated with polymyxin-agarose to reduce endotoxin contamination (39) and dialyzed against 5 mM Tris and centrifuged at 100,000 × g for 30 min before storage in 5 mM Tris, pH 7.4. All SP-A preparations were analyzed for the presence of endotoxin by the Limulus amebocyte lysate assay (Bio-Whittaker, Walkersville, MD), and endotoxin levels were routinely less than 0.1 pg/μg SP-A.
Binding of M. pneumoniae cells to immobilized hSP-A.
Individual wells of microtiter plates (Immunoplate I; Nunc) were coated overnight with 100 μl of 0.5 to 50 μg/ml solution of hSP-A in TBS. After air drying, wells were washed twice with TBS containing 0.05% (vol/vol) Tween 20 (TBST). Unoccupied sites were blocked with 200 μl of 1 mg/ml of bovine serum albumin (BSA) in TBST for 1 h at 37°C. Wells coated with BSA alone served as negative controls. Viable, [35S]methionine-labeled M. pneumoniae cells [100 μl in TBS with 1 mM CaCl2 (TBSC); 107 cells] were added to each well, and microtiter plates were incubated at 37°C for 2 h and washed four times with TBST to remove nonadherent mycoplasmas. Microtiter wells were detached and dissolved in scintillation fluid for radioactive determinations. Mannose competition studies were performed by coating microtiter wells with 100 μl of hSP-A (15 μg/ml) plus mannose (0 to 100 mM in washing buffer) at 37°C. After air drying, wells were incubated 2 h with blocking buffer and then extensively rinsed with TBS. Radiolabeled M. pneumoniae were added as described above, incubated overnight and radioactivity determined. Also, radiolabeled mycoplasmas plus mannose were added directly to hSP-A-coated wells to assess binding.
Purification of mycoplasma hSP-A binding proteins.
Glass columns (20 by 1.2 cm) were packed with either 3 ml uncoupled Sepharose CL-4B or Sepharose CL-4B coupled to hSP-A according to the manufacturer's instructions, except the coupling buffer was 10 mM sodium bicarbonate, pH 8.3 (30). Columns were equilibrated with 50 ml CLB prior to addition of radiolabeled mycoplasma cell lysates, which were collected and reapplied to each column three to four times. Individual columns were extensively washed with CLB to remove unbound proteins. M. pneumoniae hSP-A-binding proteins were eluted by NaCl gradient (0.2 to 3 M NaCl) containing 10 mM EDTA (30). Eluates were collected as 1-ml fractions, and 20 μl of each fraction was assayed for radioactivity. To determine the role of divalent cations in M. pneumoniae protein interactions with hSP-A, CaCl2 (1 mM) was replaced in CLB by 1 mM MgCl2 or 1 mM zinc acetate. For experiments employing EDTA, CLB was prepared with the addition of 10 mM EDTA.
SDS-PAGE and autoradiography.
Mycoplasma protein fractions eluted from specific columns were individually desalted in P10 columns (Amersham) against TBS and concentrated using Amicon concentrators to 1/30th of the original volume. Each sample was resolved in 12% SDS-PAGE and stained with Coomassie brilliant blue or transferred to nitrocellulose and exposed to Kodak XRP-40 X-ray film (Kodak, Rochester, NY) for 2 to 8 days.
Amino acid sequencing.
The dominant M. pneumoniae 65-kDa protein, which was purified by hSP-A column chromatography and separated by 12% SDS-PAGE, was excised and destained, and matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis was performed at the Baylor College of Medicine Protein Core Laboratory. Amino acid identity matches were evaluated using the National Center for Biotechnology Information's sequence similarity search tool designed to support analysis of nucleotide and protein databases at http://www.ncbi.nlm.nih.gov/BLAST/. All M. pneumoniae sequence data used in this study were downloaded from the database at http://www.zmbh.uni-heidelberg.de/M_pneumoniae/genome/Results.html and analyzed using http://www.bork.embl-heidelberg.de/Annot/MP/.
Binding of hSP-A mycoplasma binding protein (MPN372) to immobilized hSP-A.
The ability of recombinant MPN372 (rMPN372; details of rMPN372 construction and expression and its other properties are the focus of a separate publication) to bind to immobilized hSP-A was assayed by enzyme-linked immunosorbent assay (ELISA). Wells of microtiter plates were coated with 2 μg hSP-A in TBS, pH 7.5, for 18 h at 4°C and washed three times with TBST. Remaining protein binding sites were blocked with 3% (wt/vol) BSA in TBS for 2 h at room temperature, and wells were subsequently washed three times with TBST. Purified rMPN372 and M. pneumoniae recombinant EF-Tu [rEF-Tu; served as a negative control (13)] were diluted in TBSC containing 0.1% (wt/vol) BSA and added to individual wells for 1 h at room temperature. After wells were thoroughly washed with TBST, bound protein was incubated with 1:3,000 dilution of anti-His tag monoclonal antibody (Clonetech) in TBST containing 0.1% (wt/vol) BSA for one hour at room temperature, followed by three washes with TBST. Then, 1:2,000 dilution of goat anti-mouse alkaline phosphatase-conjugated polyclonal Abs (Zymed) in TBST containing 0.1% (wt/vol) BSA and p-nitrophenyl phosphate (Sigma) in 1 M diethanolamine, 0.5 mM MgCl2, pH 9.0, was added at room temperature for 15 to 30 min. Plates were read at 405 nm using an ELISA plate reader (Dynatech).
For antibody-mediated blocking studies, viable radiolabeled mycoplasmas or rMPN372 were preincubated with pooled, heat inactivated prebleed or anti-rMPN372 mouse sera (final dilutions of 1:10, 1:100, and 1:1000) for one hour, prior to their addition to hSP-A (2 μg/well) on microtiter plates for 2 h at 37°C. Microtiter wells were washed four times with TBST to remove nonadherent mycoplasmas or rMPN372 and dissolved in scintillation fluid for radioactive determinations or prepared for spectrophotometric assessment using goat anti-mouse antibody reagent.
Generation of mouse anti-MPN372 polyclonal antibodies.
Six-week-old BALB/c female mice (n = 10) were bled and screened by immunoblot using rMPN372 to determine preexisting antibodies. No immune reactivity was observed, and mice were immunized with 95 μg (3 mice) or 47.5 μg (4 mice) of rMPN372 mixed with complete Freund's adjuvant intraperitoneally (I.P.). Three weeks postimmunization animals were boosted with 50 μg rMPN372 mixed with incomplete Freund's adjuvant I.P. Three weeks later animals were screened by immunoblot using rMPN372 or whole M. pneumoniae cell lysates. All mice exhibited strong immune reactivity to rMPN372 and MPN372 in whole cell mycoplasma lysates. Mice were again immunized with 50 μg rMPN372 mixed with Freund's incomplete adjuvant I.P. Sera were collected 2 weeks later, rescreened by immunoblot and ELISA, and pooled.
Statistical analysis.
All data are expressed as the mean ± standard error of the mean, and statistical analysis was based upon one-tailed Student's t test for unpaired samples.
RESULTS
hSP-A as substrate for mycoplasma cell binding.
Our initial experiments were designed to assess whether viable M. pneumoniae cells interact specifically with hSP-A. Mycoplasmas bound hSP-A in a concentration-dependent manner, which was saturated at ≈25 μg/ml hSP-A. Although a modest increase in binding of mycoplasmas to SP-A occurred in the presence of Tris, the addition of 1 mM Ca2+ markedly improved binding while the presence of 10 mM EDTA reduced binding by ≈70% (Fig. 1). Minimal mycoplasma binding to BSA was observed under these conditions and this value, which represented less than 2.5% of Ca2+-dependent binding, was subtracted from all data points. Furthermore, since hSP-A is a glycoprotein with high-mannose-containing complex oligosaccharide moieties and since mannose-dependent interactions are known to occur between SP-A and specific bacteria (19), we added mannose to the binding assay. Mannose had no effect at all concentrations tested (1 to 100 mM).
FIG. 1.
Binding of viable M. pneumoniae cells to hSP-A. [35S]methionine-labeled M. pneumoniae cells were incubated with increasing concentrations of hSP-A with or without 1 mM CaCl2 for 2 h at 37°C. BSA served as the control for nonspecific binding, and this value represented less than 2.5% of Ca2+-dependent binding. Values represent the means and standard deviations of triplicate wells from which background counts were subtracted. In parallel experiments the effect of EDTA (10 mM) on mycoplasma binding was compared. Also, radiolabeled mycoplasma cells were pretreated with trypsin (10 μg/ml, 10 min) to determine the extent of protease-sensitive binding. P ≤ 0.02 versus CaCl2.
To further determine whether surface proteins of M. pneumoniae mediated hSP-A binding, 35S-labeled viable mycoplasmas were treated with trypsin. Mild trypsin digestion markedly reduced binding to hSP-A by 80 to 90%, suggesting that an M. pneumoniae hSP-A binding protein(s) was surface exposed and protease sensitive (Fig. 1).
Characteristics of hSP-A binding proteins of M. pneumoniae.
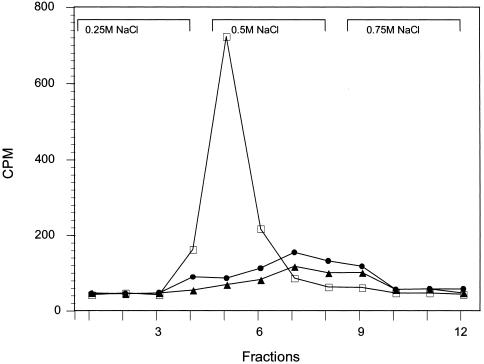
Since the interaction of M. pneumoniae with hSP-A was mediated by trypsin-sensitive and Ca2+-dependent mechanisms, we examined the binding characteristics of mycoplasma proteins to hSP-A in the presence of 1 mM Ca2+. Representative elution profiles of M. pneumoniae radiolabeled total protein lysates appear in Fig. 2. One predominant mycoplasma protein peak eluted in fractions 4 to 6 in the presence of 0.5 M NaCl plus 10 mM EDTA. However, replacement of Ca2+ with the divalent cations Zn2+ and Mg2+ did not enhance hSP-A binding of total mycoplasma protein lysate (Fig. 2).
FIG. 2.
Binding of M. pneumoniae total protein lysates to hSP-A. Radiolabeled mycoplasma total protein lysates, which were prepared in the presence of Ca2+ (open squares), Zn2+ (solid triangles), or Mg2+ (solid circles) were applied sequentially to control and hSP-A-coupled Sepharose columns. The NaCl gradient elution patterns of M. pneumoniae proteins which bound to hSP-A columns were generated by measuring radioactivity in 20-μl samples from 1-ml fractions.
Purification of M. pneumoniae hSP-A binding proteins.
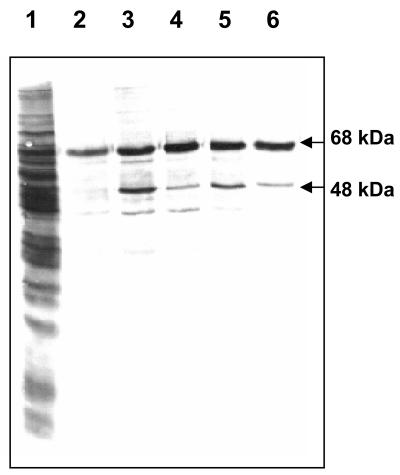
To remove nonspecific mycoplasma binding proteins (i.e., proteins that bound to uncoupled Sepharose in the presence of Ca2+) and to increase the specificity of M. pneumoniae proteins that selectively bound hSP-A, we repeatedly passed the [35S]-methionine-labeled M. pneumoniae total cell lysate through uncoupled Sepharose in the presence of Ca2+ and collected elutes, which were subsequently passed through the hSP-A-coupled Sepharose column. Autoradiogram analysis of mycoplasma proteins eluted with 0.5 M NaCl and 10 mM EDTA revealed a prominent 65-kDa protein, along with a weak 48-kDa protein (Fig. 3).
FIG. 3.
Purification of hSP-A binding proteins of M. pneumoniae. Individual Ca2+-containing fractions (Fig. 2) were desalted, concentrated, and resolved using 12% SDS-PAGE. Lane 1, whole-cell lysate; lanes 2-6, individual fractions eluted with 0.5 M NaCl-10 mM EDTA.
MALDI-TOF and BLAST analyses of the hSP-A column-purified 65-kDa protein revealed 100% identity to a 591-amino-acid (68-kDa) hypothetical protein, MPN372 (A19_orf591) of M. pneumoniae. The protein size implicated by microsequencing (68 kDa) correlated directly with the observed hSP-A binding protein profile determined by SDS-PAGE (65 kDa; Fig. 3). The amino-terminal region of MPN372 exhibited amino acid identity (27% of amino acids 1 to 226) with the Bordetella pertussis pertussis toxin S1 subunit (Fig. 4). However, the remaining amino acid residues of MPN372 (amino acids 227 to 591) shared no homologies with other subunits of pertussis toxin, any known toxins, or reported protein sequences (http://www.ncbi.nlm.nih.gov/BLAST/).
FIG. 4.
Predicted homologies between hSP-A binding protein (MPN372) of M. pneumoniae and Bordetella pertussis pertussis toxin S1 subunit. A) Schematic presentation of conserved domain of B. pertussis pertussis toxin S1 subunit in hSPA-mycoplasma binding protein MPN372. B) Amino acid sequence homologies between B. pertussis pertussis toxin S1 subunit (BORPES1) and MPN372. Identical amino acid residues and similar amino acids are marked with asterisks (*) and plus signs (+), respectively.
Dose-dependent binding of recombinant hSP-A binding protein (rMPN372).
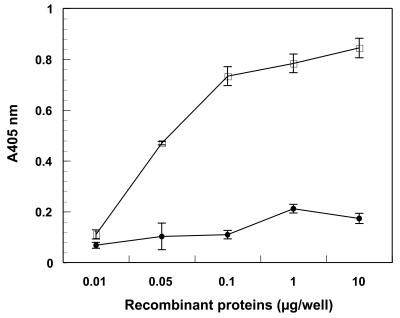
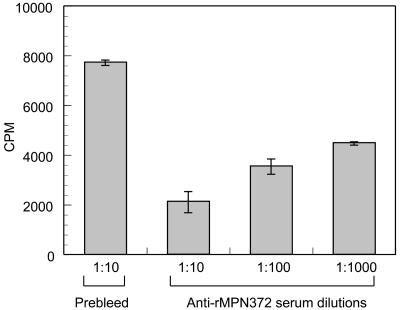
Binding of rMPN372 to hSP-A was assessed by ELISA and compared to that of rEF-TuMp, a recombinant M. pneumoniae protein that adheres specifically to fibronectin (13). rMPN372 bound to immobilized hSP-A in a dose-dependent, saturable manner, in contrast to rEF-Tu (Fig. 5). Preincubation of [35S]methionine-labeled viable M. pneumoniae cells with anti-rMPN mouse sera (1:1,000, 1:100, and 1:10) blocked mycoplasma binding to hSP-A by 45%, 55%, and 72%, respectively (Fig. 6). Preincubation of rMPN372 with the same dilutions of anti-rMPN serum reduced binding of rMPN372 to hSP-A by 21%, 31%, and 45%, respectively. No binding inhibition of rMPN372 or mycoplasmas to hSP-A was observed with prebleed sera at all dilutions.
FIG. 5.
Binding of recombinant MPN372 to immobilized hSP-A. Microtiter wells were coated with 2 μg hSP-A, and increasing concentrations of rMPN372 (open squares) or rEF-Tu (solid circles; negative control) were added to individual wells for 1 h at room temperature. Bound protein was detected with anti-His tag monoclonal antibody and goat anti-mouse alkaline phosphatase-conjugated polyclonal antibodies, followed by development with p-nitrophenyl phosphate substrate. Values represent the means of triplicate wells from three separate experiments. P ≤ 0.01 versus prebleed.
FIG. 6.
Inhibition of viable M. pneumoniae binding to immobilized hSP-A by antiserum to rMPN372. Microtiter wells were coated with 2 μg of hSP-A. [35S]methionine-labeled M. pneumoniae cells were preincubated with serial dilutions of heat-inactivated anti-rMPN372 and prebleed sera (1:10, 1:100, and 1:1,000) for 1 h at room temperature. Bound rMPN372 was detected with anti-His tag monoclonal antibody and goat anti-mouse alkaline phosphatase-conjugated polyclonal antibodies (described in Fig. 5 legend and Materials and Methods), followed by development with p-nitrophenyl phosphate. Values represent the means and standard deviations of triplicate wells from three separate experiments. P ≤ 0.01 versus prebleed.
DISCUSSION
Human SP-A is the major protein component of pulmonary surfactant. It is secreted by several respiratory cell types and reduces surface tension at alveolar air-liquid interfaces (36). Furthermore, interactions between surfactant protein A and microbial pathogens are considered part of the innate host lung defense mechanism (10, 19). It has been proposed that hSP-A functions as an antibody-independent opsonin for rapid recognition and elimination of a wide range of pathogens, including viruses, bacteria, and fungi (19). However, hSP-A may be deleterious to the host by facilitating entry of specific infectious agents into critical host target cells, as described for respiratory syncytial virus (20), cytomegalovirus (35), M. tuberculosis (16), and Pneumocystis carinii (40) More recently, hSP-A has been detected in nonalveolar sites, such as the eustachian tube, middle ear, and paranasal sinuses as well as in other organs, including the gastrointestinal tract, prostate gland, thymus, spleen, mesothelium, synovium, and lacrimal and salivary glands (25). Interestingly, mycoplasmas are widely distributed throughout the human body, both as commensals and as primary and opportunistic pathogens, and hSP-A distribution may contribute to successful mycoplasma colonization.
Cytadherence plays an essential role in the survival and propagation of M. pneumoniae. Earlier, we had suggested that multiple pathways of mycoplasma parasitism exist, including colonization and invasion through use of the unique specialized mycoplasma tip organelle (3, 5, 7) and distinct fibronectin-mediated mechanisms (13, 18). In order to uncover additional host targets, we monitored the interaction between M. pneumoniae and hSP-A. We observed the Ca2+-dependent binding of viable mycoplasmas to hSP-A (Fig. 1), which was markedly reduced by EDTA. Calcium could not be replaced by other divalent cations (Fig. 1). Additionally, pretreatment of viable mycoplasmas with low levels of trypsin decreased binding by 80 to 90% (Fig. 1), implicating surface-exposed, membrane-localized mycoplasma proteins as mediators of the mycoplasma-hSP-A interaction. Also, mannose did not inhibit mycoplasma binding to hSP-A, indicating that neither the carbohydrate recognition domain nor mannose-related moieties were directly involved in the interaction.
Identification of M. pneumoniae hSP-A binding proteins was accomplished by sequential column chromatography using uncoupled Sepharose, followed by hSP-A-coupled Sepharose chromatography. This protocol resulted in removal of almost all nonspecific binding (Fig. 2), leading to the enrichment of a dominant 65-kDa protein and a much less intense 48-kDa protein (Fig. 3). None of the previously identified mycoplasma tip-localized adhesins or cytadherence accessory proteins or mycoplasma fibronectin binding proteins possessed similar molecular masses (6, 8, 13, 27). Microsequencing identified the purified protein as MPN372, which was reported as a 65-kDa hypothetical protein (22) later annotated as a putative pertussis toxin-like molecule (15). Based on BLAST analysis of MPN372, we observed the conserved amino-terminal 226-amino-acid domain, which exhibits 27% identity and 41% similarity to the S1 subunit of B. pertussis pertussis toxin. No further homologies were detected with any other reported protein sequences (http://www.ncbi.nlm.nih.gov/BLAST/). MPN372 did not share homology with other Mycoplasma spp. proteins, except for one orthologue protein in Mycoplasma penetrans, MYPE9110 (≈26.7% identity) (32).
M. pneumoniae is a facultative intracellular pathogen with a very limited genome (5, 7, 12), and its only known host is the human. Therefore, its abilities to infect and establish residence in privileged sites, circumvent host defense mechanisms, and facilitate transmission are essential to its survival. We have described several distinct and novel pathways of mycoplasma parasitism (1, 3, 5, 7, 13, 18) and, based upon the current study, further suggest that binding of M. pneumoniae to hSP-A may serve as an additional and nonredundant mechanism to successfully cross host permeability barriers and colonize diverse target sites.
The precise biological roles and benefits of the interaction between viable M. pneumoniae, its unique hSP-A binding protein (MPN372) and hSP-A remain unclear. This specific binding event may indeed be a mechanism by which the host eliminates or reduces the number of pathogenic mycoplasmas. For example, in the case of Mycoplasma pulmonis, a murine pulmonary pathogen, the addition of SP-A to infected mouse alveolar macrophages increases nitric oxide production with concomitant reduction in mycoplasma numbers, but not complete elimination (21). Also, the binding of MPN372 to hSP-A may neutralize or destroy a potential toxin-like, virulence determinant.
Alternatively, M. pneumoniae may utilize the hSP-A binding mechanism to introduce and enhance uptake of both mycoplasma cells and MPN372 into an array of host cells and tissues that possess hSP-A receptors, like alveolar macrophages, alveolar type II cells and other respiratory and extrapulmonary targets. By so doing, mycoplasmas or the toxin-like MPN372 protein itself may establish privileged residence, which could lead to dissemination of the primary infection, or the establishment of chronic disease and sequelae, all of which are consistent with published reports and case histories linked to M. pneumoniae infections.
In several of the scenarios presented above, benefits to both host and pathogen may be realized, orchestrating a double-edged cross talk between host and pathogen, where mechanisms of host defense, host circumvention, or both merge. The final outcome may be determined by variables, such as mycoplasma infectious load, tropism, survival and spread via alveolar macrophages and other cell types, host immune and nutritional status, concomitant infections, etc. We have reported that M. pneumoniae can invade, persist, and replicate in human cells (12). Clearly, the physiological importance of the interaction between viable M. pneumoniae cells and hSP-A via MPN372 and the possible role of MPN372 in virulence need to be clarified. Furthermore, the relationships between specific domains of MPN372 and their binding to hSP-A are of primary interest. Since no bona fide pathogenic determinants other than cytadherence-related proteins have been identified in M. pneumoniae, the highly specific binding of MPN372 to hSP-A and its pertussis-like domain may offer new insights into the processes of mycoplasma pathogenesis and the development of new diagnostic, prognostic and therapeutic strategies to reduce or eliminate disease progression.
Acknowledgments
This work was supported by grants AI 45737 and HL 68072 from the National Institutes of Health.
We thank Marianna Cagle for generating mouse polyclonal antibodies.
Editor: A. D. O'Brien
REFERENCES
- 1.Alvarez, R. A., M. W. Blaylock, and J. B. Baseman. 2003. Surface localized glyceraldehyde-3-phosphate dehydrogenase of Mycoplasma genitalium binds mucin. Mol.. Microbiol. 48:1417-1425. [DOI] [PubMed] [Google Scholar]
- 2.Balis, J. U., J. F. Paterson, J. E. Paciga, E. M. Haller, and S. A. Shelley. 1985. Distribution and subcellular localization of surfactant-associated glycoproteins in human lung. Lab. Investig. 52:657-669. [PubMed] [Google Scholar]
- 3.Baseman, J. B. 1993. The cytadhesins of Mycoplasma pneumoniae and Mycoplasma genitalium. Subcell. Biochem. 20:243-259. [DOI] [PubMed] [Google Scholar]
- 4.Baseman, J. B., R. M. Cole, D. C. Krause, and D. K. Leith. 1982. Molecular basis for cytadsorption of Mycoplasma pneumoniae. J. Bacteriol. 151:1514-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baseman, J. B., M. Lange, N. L. Criscimagna, J. A. Giron, and C. A. Thomas. 1995. Interplay between mycoplasmas and host target cells. Microb. Pathog. 19:105-116. [DOI] [PubMed] [Google Scholar]
- 6.Baseman, J. B., J. Morrison-Plummer, D. Drouillard, B. Puleo-Scheppke, V. V. Tryon, and S. C. Holt. 1987. Identification of a 32-kilodalton protein of Mycoplasma pneumoniae associated with hemadsorption. Isr. J. Med. Sci. 23:474-479. [PubMed] [Google Scholar]
- 7.Baseman, J. B., S. P. Reddy, and S. F. Dallo. 1996. Interplay between mycoplasma surface proteins, airway cells, and the protean manifestations of mycoplasma-mediated human infections. Am. J. Respir. Crit. Care Med. 154:S137-144. [DOI] [PubMed] [Google Scholar]
- 8.Baseman, J. B., and J. G. Tully. 1997. Mycoplasmas: sophisticated, reemerging, and burdened by their notoriety. Emerg. Infect. Dis. 3:21-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benne, C. A., B. Benaissa-Trouw, J. A. van Strijp, C. A. Kraaijeveld, and J. F. van Iwaarden. 1997. Surfactant protein A, but not surfactant protein D, is an opsonin for influenza A virus phagocytosis by rat alveolar macrophages. Eur. J. Immunol. 27:886-890. [DOI] [PubMed] [Google Scholar]
- 10.Crouch, E., and J. R. Wright. 2001. Surfactant proteins A and D and pulmonary host defense. Annu. Rev. Physiol. 63:521-554. [DOI] [PubMed] [Google Scholar]
- 11.Crouch, E. C. 1998. Collectins and pulmonary host defense. Am. J. Respir. Cell Mol. Biol. 19:177-201. [DOI] [PubMed] [Google Scholar]
- 12.Dallo, S. F., and J. B. Baseman. 2000. Intracellular DNA replication and long-term survival of pathogenic mycoplasmas. Microb. Pathog. 29:301-309. [DOI] [PubMed] [Google Scholar]
- 13.Dallo, S. F., T. R. Kannan, M. W. Blaylock, and J. B. Baseman. 2002. Elongation factor Tu and E1 beta subunit of pyruvate dehydrogenase complex act as fibronectin binding proteins in Mycoplasma pneumoniae. Mol. Microbiol. 46:1041-1051. [DOI] [PubMed] [Google Scholar]
- 14.Dallo, S. F., A. L. Lazzell, A. Chavoya, S. P. Reddy, and J. B. Baseman. 1996. Biofunctional domains of the Mycoplasma pneumoniae P30 adhesin. Infect. Immun. 64:2595-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dandekar, T., M. Huynen, J. T. Regula, B. Ueberle, C. U. Zimmermann, M. A. Andrade, T. Doerks, L. Sanchez-Pulido, B. Snel, M. Suyama, Y. P. Yuan, R. Herrmann, and P. Bork. 2000. Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function and reading frames. Nucleic Acids Res. 28:3278-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Downing, J. F., R. Pasula, J. R. Wright, H. L. Twigg, 3rd, and W. J. Martin, 2nd. 1995. Surfactant protein a promotes attachment of Mycobacterium tuberculosis to alveolar macrophages during infection with human immunodeficiency virus. Proc. Natl. Acad. Sci. USA 92:4848-4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaynor, C. D., F. X. McCormack, D. R. Voelker, S. E. McGowan, and L. S. Schlesinger. 1995. Pulmonary surfactant protein A mediates enhanced phagocytosis of Mycobacterium tuberculosis by a direct interaction with human macrophages. J. Immunol. 155:5343-5351. [PubMed] [Google Scholar]
- 18.Giron, J. A., M. Lange, and J. B. Baseman. 1996. Adherence, fibronectin binding, and induction of cytoskeleton reorganization in cultured human cells by Mycoplasma penetrans. Infect. Immun. 64:197-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haagsman, H. P. 1998. Interactions of surfactant protein A with pathogens. Biochim. Biophys. Acta. 1408:264-277. [DOI] [PubMed] [Google Scholar]
- 20.Hickling, T. P., R. Malhotra, H. Bright, W. McDowell, E. D. Blair, and R. B. Sim. 2000. Lung surfactant protein A provides a route of entry for respiratory syncytial virus into host cells. Viral Immunol. 13:125-135. [DOI] [PubMed] [Google Scholar]
- 21.Hickman-Davis, J., J. Gibbs-Erwin, J. R. Lindsey, and S. Matalon. 1999. Surfactant protein A mediates mycoplasmacidal activity of alveolar macrophages by production of peroxynitrite. Proc. Natl. Acad. Sci. USA 96:4953-4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Himmelreich, R., H. Hilbert, H. Plagens, E. Pirkl, B. C. Li, and R. Herrmann. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24:4420-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabha, K., J. Schmegner, Y. Keisari, H. Parolis, J. Schlepper-Schaeffer, and I. Ofek. 1997. SP-A enhances phagocytosis of Klebsiella by interaction with capsular polysaccharides and alveolar macrophages. Am. J. Physiol. 272:L344-352. [DOI] [PubMed] [Google Scholar]
- 24.Khubchandani, K. R., R. E. Oberley, and J. M. Snyder. 2001. Effects of surfactant protein A and NaCl concentration on the uptake of Pseudomonas aeruginosa by THP-1 cells. Am. J. Respir. Cell Mol. Biol. 25:699-706. [DOI] [PubMed] [Google Scholar]
- 25.Khubchandani, K. R., and J. M. Snyder. 2001. Surfactant protein A (SP-A): the alveolus and beyond. FASEB J. 15:59-69. [DOI] [PubMed] [Google Scholar]
- 26.Kraft, M., G. H. Cassell, J. E. Henson, H. Watson, J. Williamson, B. P. Marmion, C. A. Gaydos, and R. J. Martin. 1998. Detection of Mycoplasma pneumoniae in the airways of adults with chronic asthma. Am. J. Respir. Crit.Care Med. 158:998-1001. [DOI] [PubMed] [Google Scholar]
- 27.Krause, D. C. 1996. Mycoplasma pneumoniae cytadherence: unravelling the tie that binds. Mol. Microbiol. 20:247-253. [DOI] [PubMed] [Google Scholar]
- 28.LeVine, A. M., K. E. Kurak, J. R. Wright, W. T. Watford, M. D. Bruno, G. F. Ross, J. A. Whitsett, and T. R. Korfhagen. 1999. Surfactant protein-A binds group B streptococcus enhancing phagocytosis and clearance from lungs of surfactant protein-A-deficient mice. Am. J. Respir. Cell Mol. Biol. 20:279-286. [DOI] [PubMed] [Google Scholar]
- 29.McNeely, T. B., and J. D. Coonrod. 1993. Comparison of the opsonic activity of human surfactant protein A for Staphylococcus aureus and Streptococcus pneumoniae with rabbit and human macrophages. J. Infect. Dis. 167:91-97. [DOI] [PubMed] [Google Scholar]
- 30.Michelis, D., M. Z. Kounnas, W. S. Argraves, E. D. Sanford, J. D. Borchelt, and J. R. Wright. 1994. Interaction of surfactant protein A with cellular myosin. Am. J. Respir. Cell Mol. Biol. 11:692-700. [DOI] [PubMed] [Google Scholar]
- 31.Pasula, R., J. F. Downing, J. R. Wright, D. L. Kachel, T. E. Davis, Jr., and W. J. Martin, 2nd. 1997. Surfactant protein A (SP-A) mediates attachment of Mycobacterium tuberculosis to murine alveolar macrophages. Am. J. Respir. Cell Mol. Biol. 17:209-217. [DOI] [PubMed] [Google Scholar]
- 32.Sasaki, Y., J. Ishikawa, A. Yamashita, K. Oshima, T. Kenri, K. Furuya, C. Yoshino, A. Horino, T. Shiba, T. Sasaki, and M. Hattori. 2002. The complete genomic sequence of Mycoplasma penetrans, an intracellular bacterial pathogen in humans. Nucleic Acids Res. 30:5293-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tino, M. J., and J. R. Wright. 1996. Surfactant protein A stimulates phagocytosis of specific pulmonary pathogens by alveolar macrophages. Am. J. Physiol. 270:L677-688. [DOI] [PubMed] [Google Scholar]
- 34.Weikert, L. F., K. Edwards, Z. C. Chroneos, C. Hager, L. Hoffman, and V. L. Shepherd. 1997. SP-A enhances uptake of bacillus Calmette-Guerin by macrophages through a specific SP-A receptor. Am. J. Physiol. 272:L989-995. [DOI] [PubMed] [Google Scholar]
- 35.Weyer, C., R. Sabat, H. Wissel, D. H. Kruger, P. A. Stevens, and S. Prosch. 2000. Surfactant protein A binding to cytomegalovirus proteins enhances virus entry into rat lung cells. Am. J. Respir. Cell Mol. Biol. 23:71-78. [DOI] [PubMed] [Google Scholar]
- 36.White, R. T., D. Damm, J. Miller, K. Spratt, J. Schilling, S. Hawgood, B. Benson, and B. Cordell. 1985. Isolation and characterization of the human pulmonary surfactant apoprotein gene. Nature 317:361-363. [DOI] [PubMed] [Google Scholar]
- 37.Wright, J. R., P. Borron, K. G. Brinker, and R. J. Folz. 2001. Surfactant Protein A: regulation of innate and adaptive immune responses in lung inflammation. Am. J. Respir. Cell Mol. Biol. 24:513-517. [DOI] [PubMed] [Google Scholar]
- 38.Wright, J. R., R. E. Wager, S. Hawgood, L. Dobbs, and J. A. Clements. 1987. Surfactant apoprotein MR = 26,000-36,000 enhances uptake of liposomes by type II cells. J. Biol. Chem. 262:2888-2894. [PubMed] [Google Scholar]
- 39.Wright, J. R., D. F. Zlogar, J. C. Taylor, T. M. Zlogar, and C. I. Restrepo. 1999. Effects of endotoxin on surfactant protein A and D stimulation of NO production by alveolar macrophages. Am. J. Physiol. 276:L650-658. [DOI] [PubMed] [Google Scholar]
- 40.Zimmerman, P. E., D. R. Voelker, F. X. McCormack, J. R. Paulsrud, and W. J. Martin, 2nd. 1992. 120-kD surface glycoprotein of Pneumocystis carinii is a ligand for surfactant protein A. J. Clin. Investig 89:143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]