Abstract
Plants have evolved diverse self-incompatibility (SI) systems for outcrossing. Since Darwin’s time, considerable progress has been made toward elucidating this unrivaled reproductive innovation. Recent advances in interdisciplinary studies and applications of biotechnology have given rise to major breakthroughs in understanding the molecular pathways that lead to SI, particularly the strikingly different SI mechanisms that operate in Solanaceae, Papaveraceae, Brassicaceae, and Primulaceae. These best-understood SI systems, together with discoveries in other “nonmodel” SI taxa such as Poaceae, suggest a complex evolutionary trajectory of SI, with multiple independent origins and frequent and irreversible losses. Extensive exploration of self-/nonself-discrimination signaling cascades has revealed a comprehensive catalog of male and female identity genes and modifier factors that control SI. These findings also enable the characterization, validation, and manipulation of SI-related factors for crop improvement, helping to address the challenges associated with development of inbred lines. Here, we review current knowledge about the evolution of SI systems, summarize key achievements in the molecular basis of pollen‒pistil interactions, discuss potential prospects for breeding of SI crops, and raise several unresolved questions that require further investigation.
Key words: self-incompatibility, S-RNase, evolution, genome editing, crop improvement
Self-incompatibility (SI) is an intraspecific reproductive barrier that promotes outbreeding. Molecular interactions between male and female S determinants trigger multiple signaling cascades and cellular events that lead to rejection of self-pollen and prevention of self-fertilization. Characterization of identity genes and modifier factors that control the SI response is crucial to understanding the complex evolutionary trajectories and breakdown of SI and provides guidance for SI crop breeding using biotechnology.
Introduction
Self-incompatibility (SI) is one of the most important reproductive innovations for prevention of self-fertilization. It enables a plant to recognize and reject self-pollen or pollen from genetically identical individuals, thereby promoting outcrossing (de Nettancourt, 2001). Particularly for many hermaphroditic species, SI represents the most widespread strategy for avoiding inbreeding depression and reduced genetic variability (Silva and Goring, 2001; Porcher and Lande, 2005). Approximately 40% of angiosperm lineages, representing at least 100 plant families, have developed SI systems (Igic et al., 2008). The diverse forms of SI systems and their scattered phylogenetic distribution suggest that SI has evolved de novo many times over the course of angiosperm evolution (Allen and Hiscock, 2008). The higher diversification rate observed in species with functional SI systems than in those with self-compatibility (SC) suggests strong species-level selection favoring obligate outcrossing (Goldberg et al., 2010). SI systems also frequently experience losses that are not subsequently regained (Barrett, 2013). Therefore, the ancestral conditions, the evolutionary dynamics of loss and gain, and the polymorphic nature of SI systems have been of long-standing interest.
In classic genetic studies, SI was classified into two homomorphic types, gametophytic self-incompatibility (GSI) and sporophytic self-incompatibility (SSI), as well as heteromorphic SI (HSI) characterized by heterostyly (de Nettancourt, 2001; Barrett and Shore, 2008). Since the advent of molecular biology, significant progress has been made in elucidating the genetic control of GSI in Solanaceae and Papaveraceae (Fujii et al., 2016), SSI in Brassicaceae (Goring et al., 2023), HSI in Primula (Huu et al., 2022), and other SI systems in which the S-components have not been well characterized (e.g., Poaceae) (Wang et al., 2022b). In well-characterized SI systems, SI is generally controlled by a single highly polymorphic locus (or two loci in Poaceae), referred to as the S-locus. The S-locus harbors tightly linked male and female identity genes (S-determinants) that engage in specific molecular interactions, enabling self-/nonself-discrimination during pollination (McCubbin and Kao, 2000). The functioning of these SI systems has provided insights into the shared features and unique characteristics of the signaling pathways and cellular processes that regulate the pollen‒pistil interaction, both within and across plant families. In the case of SI systems in Brassicaceae and Papaveraceae, a recurring observed pattern is the activation of destructive pathways designed to obstruct essential processes necessary for compatible pollen acceptance (Goring et al., 2023). In species with the S-RNase system, pollen tube growth is inhibited by cytotoxic ribonuclease activity, and distinct mechanisms exist to trigger this response (Kubo et al., 2015; Akagi et al., 2016; Li et al., 2018). These findings can also be extended to understand the evolutionary trajectories and breakdown of SI and guide future work on breeding of SI crops.
In this review, we present recent advances in SI systems and highlight key findings related to: (1) evolution and transition of SI systems, with a specific focus on S-RNase-based GSI and Brassicaceae SSI; (2) regulatory mechanisms that underlie the pollen‒pistil interaction, including SI systems in both eudicots and monocots; and (3) practical applications of molecular insights into SI, coupled with integration of contemporary biological techniques in SI crop breeding. We thus provide a clearer understanding of the evolutionary context of SI, the intricate molecular mechanisms at play, and the practical implications of recent advances.
Origins, evolution, and breakdown of SI systems
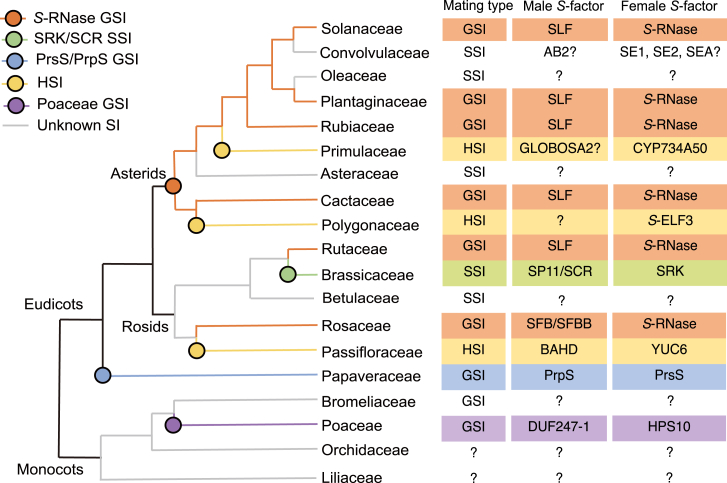
It has been suggested that SI systems originated from pathogen defense mechanisms because of the biochemical and molecular similarities observed between reactions to self-rejection and responses to pathogen invasion (Allen and Hiscock, 2008). Indeed, male S-determinants of Papaveraceae and Brassicaceae (PrsS and S-locus cysteine-rich protein [SCR/SP11]) both belong to the cysteine-rich peptide family, many of whose members function as antimicrobial peptides actively involved in pathogen defense (Silverstein et al., 2007). The female component S-locus receptor kinase (SRK) of Brassicaceae SSI is related to a large family of receptor-like kinases (RLKs), which also have crucial roles in defense responses (Shiu et al., 2004). S-RNases are class III members of Ribonuclease (RNase) T2, a family with various functions in plants, including defense against microorganisms (Maclntosh, 2011). The parallels between SI and pathogen defense, in terms of recognition mechanisms and cell‒cell signaling, present an interesting topic for future investigation. In addition, diverse SI mechanisms, highly polymorphic genes, and their scattered phylogenetic distribution have posed challenges to understanding the evolution of SI systems (Allen and Hiscock, 2008) (Figure 1). The maintenance of SI and the transition from one SI type to another have also been long-standing questions (Barrett, 2013). Over the past few decades, the use of molecular approaches, especially phylogenetics and genome-wide characterization of S-determinants, has considerably enhanced our understanding of these outstanding issues with regard to the evolution of two SI systems: S-RNase-based GSI and Brassicaceae SSI.
Figure 1.
Phylogenetic distribution of known SI systems in eudicots and monocots based on representative SI families.
Colored branches and circles indicate plausible evolutionary routes of different SI systems, with male and female SI components indicated on the right. The family tree construction is based on APG IV (Angiosperm Phylogeny Group, 2016).
Evolution of S-RNases and SLFs and their ancestral combination
Studies have suggested that S-RNase-based GSI is ancestral to 75% of all eudicots, with an independent monophyletic origin (Igic and Kohn, 2001; Ramanauskas and Igic, 2017). Recent identification of S-RNase in Citrus (Rutaceae) revealed that the S-RNase system existed prior to the divergence of asterids and eurosids I and II, thus providing additional evidence for a single origin of S-RNase in core eudicots (Liang et al., 2020). The increasing number of available plant genomes provides an opportunity to resolve the evolutionary relationships of the RNase T2 gene family and the origin of S-RNase SI on a broad scale. A study based on phylogenetics, syntenic analyses, and identification of whole gene duplication (WGD) events revealed that lineage-specific WGD resulted in expansion of the RNase T2 family (Lv et al. 2022). Although WGD or whole-genome triplication (WGT) has led to duplication of S-loci, S-RNase-based SI systems are still maintained in many lineages via deletion or inactivation of duplicate S-loci (Zhao et al., 2022). Loss of S-RNase GSI is caused mainly by inactivation, deletion, or duplication of S-loci rather than defects in S-RNase alone, possibly due to additional functions of S-RNase beyond its role in SI (Zhao et al., 2022). Phylogenetic studies have classified T2-RNases into three clades (class I, class II, class III); all functional and predicted S-RNases are members of the class III T2-RNase clade and are found exclusively in eudicots (Ramanauskas and Igic, 2017; Lv et al., 2022). Although there is evidence that an ancestral state of tightly linked pollen and pistil components may have existed prior to the eudicots, a genome study of pineapple (Ananas comosus) revealed that an RNase T2 family gene was closely linked to several F-box-associated genes (Chen et al., 2019a). This may suggest an ancestral state for the S-RNase/S-locus F-box protein (SLF) combination in monocots, although further genetic linkage studies are needed (Zhao et al., 2022). Like S-RNases, the male component genes SLF/SFB/SFBB are also clustered into a separate clade within eudicots, supporting a single origin in eudicots (Zhao et al., 2022). The S-haplotype-specific F-box protein (SFB) was found to have an independent evolutionary origin from SLF and S-locus F-box brother (SFBB), and some male components seem to have originated much more recently than their cognate S-RNases (Aguiar et al., 2015; Ashkani and Rees, 2016). Nevertheless, the core functions of SLF/SFB/SFBB in S-RNase detoxification may be conserved across different taxa. Functional analysis has demonstrated that introduction of SLF/SFB/SFBB/S-like SLF genes from species in Solanum (Solanaceae), Antirrhinum (Plantaginaceae), Prunus and Malus (Rosaceae), and Aquilegia (Ranunculaceae) can detoxify S-RNases in Petunia (Solanaceae) with 85% detoxification probability (Zhao et al., 2022). A possible explanation for this general inhibitory role is that SLFs act as E3 ligases to form an SCF complex for recognition and degradation of S-RNase (Entani et al., 2014). This auxiliary function for S-RNase degradation may be conserved in its homologs (SFB/SFBB/S-like SLF). Further research is needed to explore the evolutionary relationship between S-RNase and SLFs and the functional divergence among SLF/SFB/SFBB.
The ongoing release of plant whole genomes has enabled examination and comparison of S-components in a broad range of SI and SC species, providing a better understanding of the relationship between genome characteristics and S-RNase evolution. Identification of S-RNase and SLF in the Petunia genome and their chromosomal location suggested that recombination of SLF and S-RNase is suppressed as a mechanism to maintain their co-adaptation (Bombarely et al., 2016). In a study of the wolfberry (Lycium barbarum, Solanaceae) genome, an S-RNase and ten SLFs were found on chromosome 2 (Cao et al., 2021). Hybridization hotspot detection revealed that chromosome 2 exhibits a higher recombination frequency and the highest number of heterozygous sites per hotspot compared with other chromosomes, suggesting that SI components have contributed to elevated heterozygosity (Cao et al., 2021). The genome of selfing snapdragon (Antirrhinum majus, Plantaginaceae) contains a large number of SLFs in a pseudo (ψ) S-locus, likely resulting from gene duplication and negative selection related to the S-locus (Li et al., 2019). The divergence of S-RNase in Antirrhinum occurred approximately 62–120 million years ago (Mya), whereas that of SLFs was estimated at 4 Mya; these divergence times are much earlier and later, respectively, than the Plantaginaceae-specific WGD (46–49 Mya) (Li et al., 2019). In Jaltomata (Solanaceae), an SC genus with an ancestral loss of SI, genome-wide characterization revealed contractions in gene families related to pollen–pistil interactions and much lower heterozygosity compared with SI species (Wu et al., 2019). These genome studies have provided valuable insights into the evolution of the S-locus and the potential mechanisms underlying the origin, diversification, and deletion of S-genes in S-RNase-based GSI.
Multiple origins of the S-locus and the coevolution of SCR and SRK in Brassicaceae
Ancient and lineage-specific WGD events and polyploidization have given rise to multiple copies of S-loci in Brassicaceae (Wang et al., 2011; Haudry et al., 2013), including well-defined S-loci in Brassica (Br S-locus) (Takayama and Isogai, 2003), Arabidopsis (Al S-locus) (Kusaba et al., 2001), and Leavenworthia (La S-locus) (Chantha et al., 2013). These S-loci, together with the dynamics of S-genes, provide an opportunity to reconstruct the ancestral state and evolutionary path of the SSI system in Brassicaceae. Phylogenetic analysis based on syntenic genes extracted from 18 sequenced Brassicaceae species revealed different origins of S-loci in Brassicaceae descendant lineages I and II (Cui et al., 2020). Comparisons of the genomic locations of S-loci in Brassicaceae suggested that the Al S-locus is the most ancestral among the currently studied species in this family (Chantha et al., 2013; Vekemans et al., 2014; Cui et al., 2020). The genomic location of the Al S-locus is shared by all species in Brassicaceae lineage I, except Leavenworthia. The La S-locus was identified as being secondarily derived, characterized by the coexistence of SRK/SCR and the independent recruitment of additional pollen‒pistil recognition genes, Lal2 and SCRL, which together constitute a new functional SI system (Chantha et al., 2017). The genomic locations of the ψS-loci in three SC taxa of lineage II, Sisymbrium, Schrenkiella, and Eutrema, are shared with that of Arabidopsis (Chantha et al., 2013; Vekemans et al., 2014). However, the Br S-locus was suggested to have emerged recently after the Brassica-specific WGT because of the absence of syntenic relationships with S-loci in non-Brassicaceae species (Lysak et al., 2005; Cui et al., 2020). A possible explanation for the newly formed Br S-locus has been proposed (Cui et al., 2020). In this scenario, the ancestral Al S-locus was triplicated following the WGT in the ancestor of Brassica. Two of the triplicated Al S-loci were pseudogenized after the WGT; only one copy retained the SI function, and it was translocated to a new genomic region and retained as the Br S-locus. Hence, the presence of three distinct S-loci suggests a complex evolutionary trajectory in Brassicaceae.
As in S-RNase-based GSI, ancestral pistil and pollen components were also found to predate the formation of tightly linked SI specificities in Brassicaceae SSI (Zhao et al., 2022). It is plausible that strong selective pressure favoring outbreeding (Charlesworth, 1988) may have driven the tight linkage of genes that were initially engaged in receptor and signaling pathways related to defense responses. These genes may have undergone neofunctionalization, thus acquiring new roles in SI specificity. Indeed, SRKs are suggested to have originated from two ancient haplotypes that evolved from a pair of tandemly duplicated genes through neo-/subfunctionalization in the common ancestor of Brassicaceae (Xing et al., 2013). Phylogenetic analysis revealed that functional SRKs form three distinct clusters within Brassicaceae (Brassica class I, Brassica class II, Arabidopsis/Capsella) (Xing et al., 2013). The subdivision of extant Brassica SRKs is thought to have originated from the ancestor of both Arabidopsis and Brassica and to have undergone diversification after divergence of the two genera (Edh et al., 2009). Another interesting observation emerging from phylogenetic studies is that SCR exhibits a high congruency of genealogies with SRK (Sato et al., 2002; Edh et al., 2009). Their highly similar topology and specific S-haplotype interaction suggest that SCR and SRK are in strong linkage disequilibrium and have coevolved to stabilize self-combinations (Guo et al., 2011; Murase et al., 2020). In this coevolved SI system, intriguing questions persist about the generation of new S-alleles at the S-locus. An asymmetrical functional divergence model based on resurrection of ancestral SRK has been proposed to explain the diversification of SCR/SRK from their putative ancestor (Chantreau et al., 2019). According to this model, the two allelic variants involved in receptor‒ligand combinations have experienced asymmetric divergence. One variant has retained the same recognition specificity as their putative ancestor, whereas the other variant has functionally diverged, resulting in a novel specificity that was previously absent. This observation provides evidence for the hypothesis that disruption of the SRK–SCR interaction can be followed by a compensatory mutation that restores the interaction in a modified state, leading to new SI specificities (Uyenoyama et al., 2001; Gervais et al., 2011). Therefore, ancestral recognition specificity can be maintained over a long time span, and novel specificity occurs asymmetrically (Durand et al., 2020). Analysis of additional S-haplotypes and resurrection of the ancestral SCR are expected to provide a better understanding of the allelic diversification process.
Collectively, these studies have laid a foundation for understanding the evolutionary scenarios leading to the emergence and diversification of the S-locus and SI-specificity genes in Brassicaceae. In future studies, uncovering the components and downstream signaling events shared between SI and pathogen-defense recognition systems, particularly the role of defensin-like SCR/SRK, will be crucial. Such investigations may have profound implications for further understanding the origin of SI systems and the SI recognition repertoire.
SI to SC transitions in S-RNase-based GSI and Brassicaceae SSI
Although SI systems show independent origins in multiple lineages, loss of SI has been suggested to be a prevalent phenomenon (Barrett, 2013). SI loss is irreversible, and the same system is unlikely to be regained, as observed for families with different SI forms (Igic et al., 2008; Ferrer and Good, 2012). It has been reported that S-RNase-based GSI has been lost frequently, at least 60 times in Solanaceae alone (Igic et al., 2008). Likewise, genera within Brassicaceae also include many secondarily evolved SC species, predominantly SI species with SC strains or cultivars. Investigating the causes and consequences of these SI to SC transitions is crucial for understanding this important mating-system shift in plants.
Although the mechanisms underlying the SI to SC transition may vary among taxa, several key paths leading to SI breakdown can be gleaned from comparative and functional studies. The first path occurs through accumulated mutations in male and female determinants. Loss-of-function mutations and collapse of variation at the S-locus can confer SC, and these mutations may be fixed when SC is more advantageous in terms of male mating advantage, Fisher’s transmission advantage, and reproductive assurance (reviewed in Shimizu and Tsuchimatsu, 2015). In S-RNase-based GSI, breakdown of SI can be caused by defects in both pistil and pollen function. Phylogenetics, structural examination, and expression analysis of the tomato clade (Solanum section Lycopersicon) have attributed the SI to SC transition to deletion and dysfunction of S-RNase, which is followed by parallel mutations in pollen S-factors (Broz et al., 2017, 2021; Markova et al., 2017). SC in Citrus is caused by a truncated S-RNase resulting from a single-nucleotide deletion at position 443; this mutation initially occurred in mandarin (Citrus reticulata) and subsequently became fixed in populations through mating or introgression (Liang et al., 2020). Male SC-conferring mutations are found in Petunia and Prunus, in which mutations in the pollen S-component are closely related to SC (Tsukamoto et al., 2003a; Li et al., 2020). In Brassicaceae, the best-studied plant for SI to SC transition is A. thaliana, which is a predominantly selfing species, although its closest relatives Arabidopsis lyrata (with a minority as SC) and Arabidopsis halleri are mainly obligate outcrossers enforced by SI (Koch et al., 2000; Mable et al., 2005). Studies have provided evidence for three independent origins of SC in A. thaliana, with a recent transition to selfing: (1) three highly divergent, nonfunctional S-haplogroups have been identified and share high sequence similarity with those in A. lyrata and A. halleri (Bechsgaard et al., 2006; Shimizu et al., 2008); (2) these haplogroups exhibit various states of decay, with different gene-disruptive mutations of both SRK and SCR (Sherman-Broyles et al., 2007; Tsuchimatsu et al., 2017); (3) A. thaliana is estimated to have transitioned to SC at 1.5–1.0 Mya (Shimizu and Tsuchimatsu, 2015; Durvasula et al., 2017). This transition occurred much more recently than the divergence between A. thaliana and the A. lyrata/A. halleri clade (7–13 Mya) (Hohmann et al., 2015). Many studies have investigated the evolution and fixation of SC in A. thaliana by identifying prerequisites for re-establishment of SI in this species (reviewed in Shimizu and Tsuchimatsu, 2015; Ahmad et al., 2022). These studies have revealed that loss of SI in A. thaliana has been controlled predominantly by loss-of-function mutations or partial deletions on the male side (Tsuchimatsu et al., 2010; Suwabe et al., 2020). Pistil factors, including dominant SRK suppressors such as inverted repeats, are considered to have facilitated the rapid fixation of SC in A. thaliana (Fujii et al., 2020). However, phenotypic variation in SI among A. thaliana accessions and the presence of modifier genes still make it challenging to identify the primary mutation responsible for SI loss (Shimizu and Tsuchimatsu, 2015).
The second path is via polyploidy. For the S-RNase-based system, when polyploidy generates diploid pollen tube (PT) cells with duplicated S-loci, S-RNase can be detoxified, and SI is directly broken down (Dickinson et al., 2007; Robertson et al., 2011; Sutherland et al., 2018). This automatic transition of SI is frequently observed in Solanaceae species, which usually undergo a one-step transition from diploid to polyploid and thus from SI to SC, resulting in SC polyploid lineages (Robertson et al., 2011). In Brassicaceae, the transition to SC in polyploid species can be caused by dominance of nonfunctional alleles over S-alleles (Novikova et al., 2017) or by a single mutation in a dominant haplotype (Okamoto et al., 2007; Shimizu et al., 2011). In Arabidopsis suecica, the S-allele inherited from SI Arabidopsis arenosa (father) has been inactivated by loss-of-function S-alleles from A. thaliana (mother), making it an irrevocable selfer (Novikova et al., 2017). Insertion of transposons in the SP11 promoter region led to loss of SI in Brassica napus, and this insertion plausibly occurred after polyploidization, as it has not been found in the diploid progenitors (Gao et al., 2016; Bayer et al., 2021). In allotetraploid Arabidopsis kamchatica, degradation of male specificity genes has played a role in the transition to SC, and the presence of multiple S-haplogroups indicates recurrent evolution of SC, likely associated with multiple polyploidization events (Tsuchimatsu et al., 2012). Some polyploid lines also exhibit the SI phenotype (e.g., B. napus N110 and N343) in the absence of SP11 expression, suggesting that novel mechanisms may be involved in controlling self-recognition in these lines (Okamoto et al., 2020).
Other mechanisms, such as modifiers and epigenetic changes, can also influence the expression of S-locus genes and result in loss of SI. Multiple modifiers have been identified as being associated with loss of S-function in S-RNase-based GSI. A modifier locus (MDF) was found to be unlinked to the S-locus and to specifically suppress expression of S-RNase, resulting in loss of SI in some plants of Petunia axillaris (Tsukamoto et al., 2003b). The reduced SI activity of Citrus maxima is caused by a CgHB40 gene that inhibits S-RNase expression by regulating its promoter region (Hu et al., 2021). SC mutants of Prunus species are more likely to be caused by pollen-part modifier genes, with many modifiers having been reported to confer pollen-part SC (Ono et al., 2018). In addition to these cryptic modifiers, DNA methylation is thought to participate in modifying the expression and function of SI genes. In SC almond (Prunus dulcis), inactivation of S-RNase is associated with DNA methylation at its 5′-flanking region, which appears to be a crucial site for alteration of S-RNase expression (Fernández i Martí et al., 2014). Further molecular evidence is needed to better understand the role of DNA methylation in the SI to SC transition.
Interestingly, a study in SC tobacco (Nicotiana attenuata) revealed the intriguing phenomenon of polyandrous mate selection, which seems to be a by-product of the SI to SC transition (Guo et al., 2019). The SI machinery of S-RNase-like/SLF-like genes has been repurposed to serve as a mate-selection tool, enabling the plant to select certain pollen genotypes with desired traits (Guo et al., 2019). Moreover, intraspecific unilateral incompatibility (UI) in the tomato clade is thought to be linked to multiple independent SI to SC transitions, as discussed in a study by Bedinger et al. (2011), suggesting potential connections and complexities between different patterns of mate preference. Collectively, the prevalence of dominant SC mutations suggests that the evolutionary transition from SI to SC is irreversible. Ongoing endeavors to reconstruct the ancestral states of SI and SC and induction of gain- or loss-of-function mutations in S-genes hold great promise for investigating the origin, evolution, and fate of SI lineages.
Molecular basis of self-pollen rejection in different SI systems
S-RNase-based GSI: Poisoning incompatible pollen tubes by various pathways
The GSI system is the most common SI mechanism in flowering plants (Franklin-Tong and Franklin, 2003). In GSI, S-specificity is determined by the pollen’s own haploid genome, and pollen rejection occurs when the pollen S-haplotype matches one of the two pistil haplotypes. To date, the S-RNase-based GSI system has been described in six families: Solanaceae (Sijacic et al., 2004), Plantaginaceae (Li et al., 2019), Rubiaceae (Asquini et al., 2011), Cactaceae (Ramanauskas and Igic, 2021), Rutaceae (Liang et al., 2020), and Rosaceae (Vieira et al. 2021). The SI response is mediated by a pistil S-determinant (a glycoprotein S-RNase) that traffics within PTs and inhibits PT growth through its cytotoxic ribonuclease activity (McClure et al., 1990). Extensive investigations into Solanaceae- and Rosaceae-type SI have revealed the distinct molecular mechanisms and cellular events of S-RNase-mediated pollen rejection within and between these two plant groups.
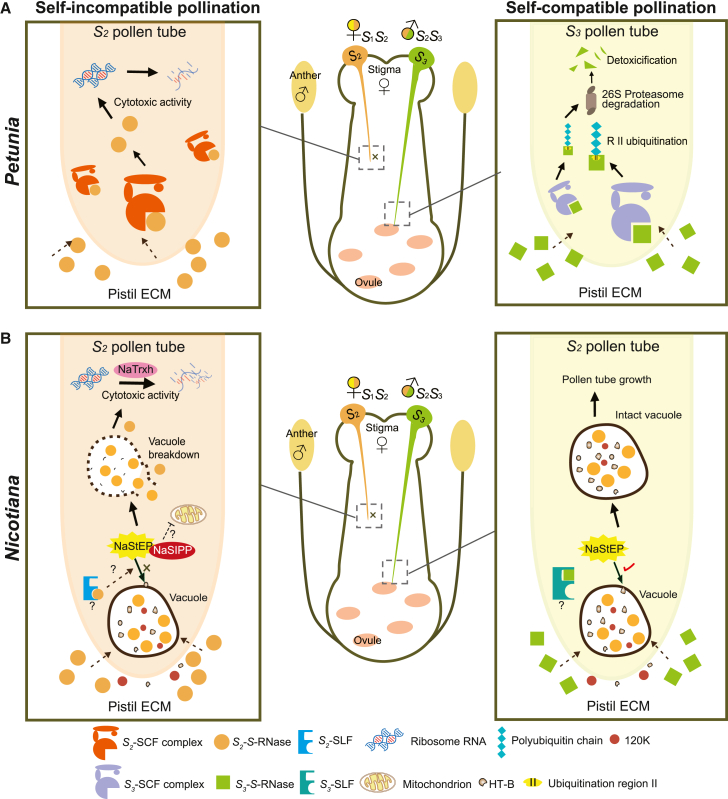
In Solanaceae, S-RNases are specifically recognized by SLFs, which serve as pollen S-determinants (Kubo et al., 2010). Studies of Petunia and Nicotiana have led to the proposal of two models (collaborative degradation and compartmentalization, respectively) for the S-RNase–SLF interaction. In the collaborative degradation model (Figure 2A), SLF is proposed to form an SCF (Skp1/Cullin1/F-box) complex, acting as an E3 ubiquitin ligase that can polyubiquitinate all nonself S-RNase proteins, with PhS3-RNase R II identified as a major region for ubiquitination (Zhao et al., 2021). The polyubiquitinated S-RNases are further degraded in a proteasome-dependent manner by the 26S proteasome, enabling the growth of compatible PTs (Entani et al., 2014). This process is considered a collaborative nonself-recognition process, in which a nonself S-RNase can be detoxified by multiple SLFs (Kubo et al., 2010, 2015). Conversely, in incompatible PTs, the specific interaction between self S-RNase and SLF fails to form a functional SCF complex, and self S-RNase thus evading degradation and acting as a cytotoxin to inhibit PT growth (Liu et al., 2014). The compartmentalization model suggests that nonself S-RNases are spatially sequestered in an intact vacuole (Goldraij et al., 2006) (Figure 2B). The vacuole is degraded in incompatible PTs, releasing self S-RNases into the cytoplasm and thus triggering cytotoxic activities and RNA degradation (Goldraij et al., 2006). Breakdown of vacuoles containing S-RNases relies on the specific interaction between SLF and S-RNase, as well as the involvement of modifier genes such as HT-B, 120K, NaStEP, NaSIPP, and NaTrxh (McClure et al., 1999; Nathan Hancock et al., 2005; Jiménez-Durán et al., 2013; García-Valencia et al., 2017; Torres-Rodríguez et al., 2020). HT-B is a key molecule in the S-RNase-containing vacuole, as it is degraded in incompatible PTs but stabilized in compatible PTs (Goldraij et al., 2006). The stability of HT-B relies on the presence of NaStEP, which protects HT-B from degradation by its proteinase inhibitor activity (Jiménez-Durán et al., 2013). NaStEP also interacts with NaSIPP, a mitochondrial phosphate carrier, to form a complex with a mitochondrial voltage-dependent channel, potentially leading to mitochondrial destabilization and triggering cell death (García-Valencia et al., 2017; Cruz-Zamora et al., 2020). Ultimately, the compartmentalization of self S-RNase is disrupted by a yet-unclear specific interaction among these modifier genes. Upon their release into the cytosol, the ribonuclease activity of free S-RNases is enhanced (sevenfold) by NaTrxh through the reduction of specific cysteine disulphide bonds (Torres-Rodríguez et al., 2020). However, the direct connections among these modifier genes and their biochemical roles have yet to be explored. In addition, it remains unknown whether the collaborative model and compartmentalization model are exclusively applicable to Petunia and Nicotiana, respectively.
Figure 2.
Two S-RNase-based SI models.
(A) The collaborative degradation model in Petunia. In compatible pollen tubes, SLF forms an SCF (Skp1/Cullin1/F-box) complex that polyubiquitinates all nonself S-RNase proteins, with PhS3-RNase R II as the main ubiquitination region. The polyubiquitinated S-RNases are further degraded by the 26S proteasome, enabling the growth of compatible pollen tubes. Conversely, in incompatible pollen tubes, the interaction between self S-RNase and SLF fails to produce a functional SCF complex; self S-RNase thus evading degradation and acting as a cytotoxin to inhibit pollen tube growth.
(B) The compartmentalization model in Nicotiana. Nonself S-RNases, along with HT-B and 120K, become sequestered within an intact vacuole upon uptake by the compatible pollen tube, preventing the cytotoxic activities of S-RNase. HT-B acts as a key molecule in the S-RNase-containing vacuole, as it is degraded in incompatible pollen tubes. The stability of HT-B relies on the presence of NaStEP, which protects HT-B from degradation by its proteinase inhibitor activity. NaStEP also interacts with NaSIPP, potentially leading to mitochondrial destabilization and eventual cell death. The recognition event between SLF and self S-RNase triggers specific interactions among these modifier genes, resulting in breakdown of the vacuole and disruption of self S-RNase compartmentalization. Once free S-RNases are in the cytosol, their ribonuclease activity is further enhanced by NaTrxh.
In contrast to the two models proposed for Solanaceae-type SI, novel mechanisms have been reported to explain the cytotoxicity of S-RNase in rosaceous species. In Maloideae, several signaling factors were found to be associated with S-RNase-induced PT growth arrest. S-RNase can inhibit the specific activity of a soluble inorganic pyrophosphatase (MdPPa), which disrupts tRNA aminoacylation and triggers programmed cell death (PCD) in incompatible PTs (Li et al., 2018). S-RNase also mediates actin cytoskeleton depolymerization by increasing pollen phosphatidic acid levels, repressing ABF–LRX signaling, and disrupting the interaction between MdMVG and F-actin (Chen et al., 2018; Yang et al., 2018; Wu et al., 2023). Two phytohormone signaling pathways, the jasmonic acid (JA)–MdMYC2–MdD1 pathway and the BZR1-mediated brassinosteroid (BR) pathway, have been shown to be directly and indirectly induced, respectively, by S-RNase to inhibit PT growth (Gu et al., 2019; Wang et al., 2023). Other cellular processes, such as alterations in cytoplasmic Ca2+ concentration and reactive oxygen species (ROS) levels, are also likely to be involved in the SI response (Jiang et al., 2014). Intriguingly, a self-recognition mechanism has been reported to induce the SI response in Prunus (Akagi et al., 2016). It has been suggested that its pollen S-determinant SFB interacts specifically with self-S-RNases to protect them from a general inhibitor such as pollen-specific SLF/SFB proteins that are conserved and expressed in all S-haplotypes, resulting in self-cytotoxicity (Matsumoto and Tao, 2019). This scenario is supported by the fact that loss-of-function SFB mutations can cause SC directly, whereas polyploidy does not directly confer SC in Prunus, in contrast to Solanaceae and Maloideae (reviewed in Tao et al., 2010). In addition, phylogenetic analyses indicated that S-specific genes of Malus and Prunus belong to distinct gene lineages, implying independent recruitment of SI genes and different recognition mechanisms in Prunus (Aguiar et al., 2015). Prunus-specific SI may be due to subfunctionalization of the newly generated pollen S-determinants, which differentiates them from the ancestral pollen. Further research is required to determine whether there are commonalities between Prunus and Maloideae in the cellular events induced by S-RNase.
In summary, Solanaceae and Rosaceae exhibit distinct pollen-rejection mechanisms, even though they share S-RNase as the female S-determinant. Despite extensive research, the intricacy and diversity of SI systems imply that novel mechanisms underlying S-RNase-mediated SI responses may still remain to be discovered. It will be essential to investigate components associated with S-RNase cytotoxicity in other plant groups (e.g., Rubiaceae, Cactaceae, Rutaceae) under S-RNase GSI control. In addition, the existence of S-RNase GSI in other taxa remains to be explored, as S-RNase orthologs have not yet been found outside the eudicot lineage (Niu et al., 2017; Ramanauskas and Igic, 2017; Lv et al., 2022). These studies will enhance our understanding of how S-RNase functions as a key player in PT growth arrest and will provide valuable information on the coevolutionary dynamics between S-RNases and their interaction partners.
Papaveraceae GSI: Ca2+ signaling cascade triggers programmed cell death in self-pollen
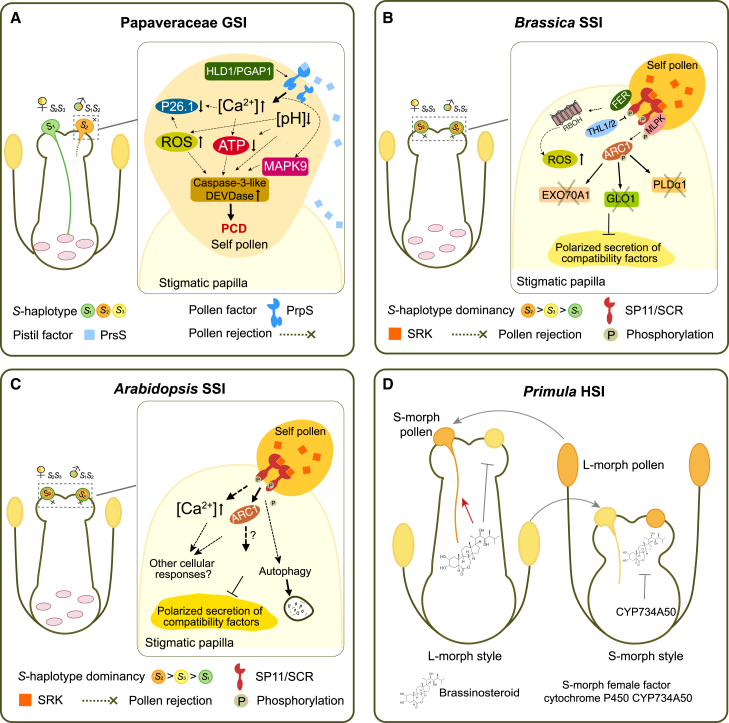
The SI mechanism identified in Papaver rhoeas (common poppy) is a Ca2+-based GSI system (Figure 3A). The pistil S-determinant (PrsS) is a small (∼15 kDa), unique protein secreted by stigmatic papilla cells and is considered to be a signaling ligand (Bosch and Franklin-Tong, 2008). The pollen S-determinant was suggested to be a transmembrane receptor and was designated PrpS (Wheeler et al., 2009, 2010). A transgenic experiment showed that coexpression of cognate PrpS and PrsS (pistil S determinant) in A. thaliana (self-compatible) conferred a “Papaver-like” SI response in plants, despite Pa. rhoeas and A. thaliana being distantly related species (140 million years) (Lin et al., 2015). In addition, ectopic expression of PrpS–PrsS in Arabidopsis roots led to root growth arrest and triggered an SI-like PCD response (Lin et al., 2020). Given the common and universal presence of downstream components of “receptor‒ligand” interactions in plants, it has been suggested that the two SI-identity genes of Papaver are sufficient to produce SI (Wang et al., 2018). In addition to known mechanisms led by PrpS and PrsS, novel Papaver SI factors have recently been identified and validated in transgenic SI Arabidopsis lines (Lin et al., 2022). Remodeling glycosylphosphatidylinositol-anchored proteins (GPI-APs) and their cleavage/release by GPI-inositol deacylase HLD1/AtPGAP1 was found to be crucial for self-pollen rejection, suggesting a role for GPI-APs as coreceptors in regulation of the PrpS–PrsS interaction (Lin et al., 2022; Goring et al., 2023).
Figure 3.
Mechanisms of different self-incompatibility systems.
(A) Papaveraceae GSI. The interaction between PrpS and PrsS activates Ca2+-dependent signaling events, including an increase in cytosolic free Ca2+ and ROS, a reduction in ATP depletion and acidification of the cytosol, the inhibition of soluble inorganic pyrophosphatase P26.1, and the initiation of two key regulators of SI-PCD, MAPK9 and caspase-3-like/DEVDase, thereby resulting in programmed cell death of self-pollen.
(B)Brassica SSI. Phosphorylated SRK/MLPK activates ARC1, which targets three compatibility factors, EXO70A1, GLO1, and PLDα1, for degradation. A second signaling pathway is mediated by the FER–Rac/Rop–Rboh module, which triggers a rapid increase in NADPH oxidase–produced ROS, leading to self-pollen rejection.
(C)Arabidopsis SSI. The SRK/SCR interaction triggers a rapid increase in cytosolic free Ca2+ and the activation of autophagy, eventually resulting in inhibition of compatibility factors for pollen acceptance.
(D)Primula HSI. SI occurs in short (S)-style morphs with stigmas positioned above the anther or long (L)-style morphs with stigmas situated below the anther. In the S-morph, the cytochrome P450 CYP734A50 inhibits cell elongation by repressing brassinosteroid.
Papaver SI involves a complex signal transduction network that first inhibits PT tip growth and then triggers PCD of pollen (Thomas and Franklin-Tong, 2004). The interaction of PrpS and PrsS immediately increases levels of cytosolic free Ca2+ (Franklin-Tong et al., 2002) and ROS (Wilkins et al., 2011; Haque et al., 2020). Papaver SI also triggers a reduction in ATP depletion (Wang et al., 2022a) and acidification of the cytosol (Wang et al., 2018). These SI-induced components lead to inhibition of soluble inorganic pyrophosphatase P26.1 (de Graaf et al., 2006) and depolymerization of actin cytoskeletons (Snowman et al., 2002), which rapidly inhibit PT growth. They also create an intracellular environment that is critical for initiation of cell death in incompatible pollen. Two key regulators of SI-PCD, mitogen-activated protein kinase 9 (MAPK9) and caspase-3-like/DEVDase, are activated in SI-induced pollen (Chai et al., 2017). These downstream PCD-related signals are in place to kill pollen cells, thus preventing self-fertilization. As PCD is a common and important process in plant reproduction, how this self-pollen PCD evolved with an SI-induced function remains an intriguing question. Systematic comparative analysis could be performed to identify shared PCD executers in reproductive PCD.
In summary, Papaver SI involves Ca2+-dependent signaling cascades followed by several intracellular dynamics that contribute to PT growth arrest and self-pollen PCD.
The success of transgenic Arabidopsis SI lines carrying the Papaver SI system (Lin et al., 2015) and the availability of the Pa. rhoeas genome (Yang et al., 2021) should provide more opportunities for investigating new mechanisms and genetic components of cell‒cell signaling during Papaver SI.
Brassicaceae SSI: Degradation of compatibility factors for pollen acceptance
In Brassicaceae SSI, pollen S-specificity is determined by the genotype of the diploid donor tissues (sporophyte), and self-pollen or genetically similar pollen is rejected when there is an S-haplotype match between the pistil and the anther. The SI response occurs as a rapid cessation of pollen hydration/germination and initial stages of PT growth (Takayama and Isogai, 2005). Similar to SI in Papaver, self-pollen rejection in Brassicaceae is also mediated by ligand–receptor interactions in which two tightly linked and highly polymorphic proteins, SRK and SCR/SP11, are the female and male determinants, respectively (Takayama and Isogai, 2003). Differences between multiple contact regions and some conserved residues in SRK and SCR/SP11 are key for guiding self-/nonself-discrimination (Murase et al., 2020). Following incompatible pollination, the SRK–SCR/SP11 interaction triggers a cascade of intracellular molecular events that disrupt secretion and homeostasis in the stigmatic papillae (reviewed in Goring et al., 2023). These events result in proteasomal degradation of stigmatic compatibility factors required for pollen hydration and/or PT growth, leading to pollen rejection (Abhinandan et al., 2022). Considerable progress has been made in elucidating the downstream molecular components of the SRK–SCR/SP11 interaction in Brassica and Arabidopsis. Studies have also suggested mechanistic and evolutionary differences in SI signaling within Brassicaceae due to certain components that seem to be genus specific in Arabidopsis and Brassica (Zhang et al., 2019; Fujii et al., 2020; Yamamoto et al., 2022).
In Brassica, THIOREDOXIN H-LIKE proteins (THL1 and THL2) (Cabrillac et al., 2001), M-locus protein kinase (MLPK) (Murase et al., 2004), and the E3 ubiquitin ligase ARC1 (Armadillo repeat-containing 1) (Stone et al., 1999) are three direct interactors of SRK. THL1/2 proteins are negative regulators of SRK prior to the SRK–SCR/SP11 interaction (Cabrillac et al., 2001). MLPK directly interacts with and is phosphorylated by SRK to activate SRK-dependent responses (Kakita et al., 2007). Phosphorylated SRK/MLPK activates ARC1, which targets stigmatic compatibility factors for proteasomal degradation (Stone et al., 2003). Knockout of ARC1 in B. napus can lead to complete loss of SI, demonstrating the indispensable role of ARC1 in mediating the SI pathway (Abhinandan et al., 2023). Three compatibility factors were found to be targeted by ARC1: exocyst complex subunit (EXO70A1) (Samuel et al., 2009), glyoxalase 1 (GLO1) (Sankaranarayanan et al., 2015), and phospholipase D alpha 1 (PLDα1) (Scandola and Samuel, 2019). Disruption of these factors, which are necessary for compatible pollen hydration and germination, results in self-pollen rejection. Recent studies on Brassica rapa have uncovered another signaling pathway led by FERONIA (FER) in the Brassica SI response (Zhang et al., 2021; Huang et al., 2023). The signaling pathway mediated by the FER–Rac/Rop GTPase-respiratory burst oxidase homologs (FER–Rac/Rop–Rboh) module triggers a rapid increase in NADPH oxidase–produced ROS, leading to self-pollen rejection (Zhang et al., 2021). Reducing ROS levels by scavengers or by suppressing expression of the FER signaling module can effectively break down SI (Zhang et al., 2021). However, it is unclear how FER–ROS signaling is initiated by SRK and whether it acts in parallel with MLPK- and ARC1-mediated pathways for ubiquitination and degradation of compatibility factors. Investigating their interactions during the SI response could provide more insights into the intracellular events involved in pollen rejection.
Compatibility factors for pollen hydration and/or germination in the stigmatic papillae appear to be shared targets for degradation in Arabidopsis SI (Goring et al., 2023). The Arabidopsis SI response is characterized by a rapid increase in cytosolic free Ca2+ (Iwano et al., 2015) and the activation of autophagy (Safavian and Goring, 2013). Disruption of autophagy in autophagy-deficient mutants leads to only partial breakdown of SI, suggesting that additional signaling pathways function together with autophagy for pollen rejection (Macgregor et al., 2022). Studies of SI in A. lyrata and transgenic A. thaliana SI lines have not yet determined a clear role or necessity for Brassica SI components (e.g., ARC1, Exo70A1) in Arabidopsis SI (Kitashiba et al., 2011; Indriolo et al., 2014). The exact mechanisms of autophagy-induced pollen rejection are also unclear. There is still much to be learned about the downstream signaling that operates in the Arabidopsis SI response and how it may differ from that of Brassica. Components of the Brassicaceae SI pathway identified in Arabidopsis and Brassica are shown in Figure 3B and 3C, respectively.
In addition to the signaling events that take place in the stigma, molecular factors have been identified on the pollen side that contribute to modulation of the SI response (Tarutani et al., 2010; Finnegan et al., 2011; Gao et al., 2016). The occurrence of dominant and recessive S-haplotypes is an intriguing feature of the SSI system (reviewed in Fujii and Takayama, 2018). Studies on Brassica and Arabidopsis have revealed that this dominance hierarchy is determined by small RNAs (sRNAs) linked to the S-locus (Tarutani et al., 2010; Durand et al., 2014). This role of sRNAs in controlling the dominance hierarchy of male SI determinants appears to be conserved in Brassicaceae (Yasuda et al., 2021). In Brassica, SP11 methylation inducer (Smi1) was found to induce DNA methylation of the recessive class II SP11/SCR promoter to regulate monoallelic gene silencing during dominant–recessive interactions (Shiba et al., 2006; Tarutani et al., 2010). Smi2 and its polymorphic targets govern the linear dominance hierarchy of the class II SP11/SCR alleles (Yasuda et al., 2016). Dominant–recessive relationships identified in SI Arabidopsis species were found to be more complex than those observed in Brassica, including the existence of multiple dominance classes and their nonlinear dominant–recessive interactions (Durand et al., 2014; Jany et al., 2019). In A. halleri, sRNA-mediated transcriptional silencing of recessive SCR alleles involves base-pairing requirements, with a given sRNA required to target a specific SCR sequence (Burghgraeve et al., 2020). Two polymorphic sRNAs, AlSmi1 and AlSmi2, were suggested to control the dominance hierarchy of A. lyrata, and their predicted targets were as follows: class IV > class III > class II > class I (Yasuda et al., 2021). Further analysis of interactions between sRNAs and their target sites in other SI species should provide more information on dominance–recessiveness mechanisms in Brassicaceae (Fujii and Takayama, 2018; Duan et al., 2023).
In summary, self-pollen rejection in Brassicaceae is governed by the SRK–SCR/SP11 interaction, which triggers multiple signaling events in the stigmatic papillae to prevent self-pollen from receiving compatible factors that would normally be needed for acceptance and growth. To fully comprehend the SSI system, it is necessary to gain a deeper understanding of the molecular basis of SI responses at both the stigma and pollen sides.
Diverse SSI systems and mechanisms in other families
Although the molecular mechanism of SSI has been most extensively characterized in Brassicaceae, SI under sporophytic control has also been reported for species in Asteraceae (Ferrer and Good-Avila, 2007), Convolvulaceae (Kowyama, 2000), Oleaceae (Saumitou-Laprade et al., 2017a), and Betulaceae (Hill et al., 2021). Genetic studies have suggested the presence of distinct mechanisms within and between these families, which are less likely to adopt the SRK–SCR/SP11 system that operates in Brassicaceae (Rahman et al., 2007a, 2007b; Gonthier et al., 2013; Saumitou-Laprade et al., 2017b; Hu et al., 2021).
Approximately 63% of Asteraceae species are estimated to possess SI (Ferrer and Good-Avila, 2007). Genetic mapping of chicory (Cichorium intybus) and Silphium integrifolium has identified quantitative trait loci (QTLs) (a 0.8-cM region on linkage group [LG] 2 and a 0–27.7-cM region on LG 6, respectively) that are likely to contain the S-locus (Gonthier et al., 2013; Price et al., 2022). Recent genome mapping of chicory has identified MDIS1 INTERACTING RECEPTOR-LIKE KINASE 2 (MIK2) as a candidate gene associated with the S-locus (Palumbo et al., 2023). A transcriptome-based study revealed membrane-associated protein (MAP), nodulin/mtn3 (Nod), and sunflower-21 (SF21) as three candidates involved in pollen‒pistil interactions of Senecio squalidus (Allen et al., 2011). In addition, SRK-like genes identified in S. squalidus, chicory, and Si. integrifolium were not linked to the S-locus, suggesting that they are unlikely to function as S-determinants (Tabah et al., 2004; Gonthier et al., 2013; Price et al., 2022). However, functional identification of SRK and SCR/SP11 homologs (CmSRK1 and CmPCP1) in Chrysanthemum morifolium suggested that they may be S-determinants in this species (Wang et al., 2021). Hybridization of transgenic Arabidopsis using CmSRK1 as the female parent and CmPCP1 as the male parent resulted in extremely low seed set (11.64%–19.62%) compared with those resulting from either self-pollination or cross-pollination of two independent transgenic lines (∼90%) (Wang et al., 2021). These findings suggest that there are multiple SSI systems in Asteraceae, with a high diversity of specific genes involved in pollen‒pistil interactions.
A linkage map of DNA markers showed that the S-locus is located in the S-haplotype-specific divergent region (SDR) of Ipomoea (Convolvulaceae) (Kowyama et al., 2008). Three novel stigma-specific genes, SE1, SE2, and SEA, and an anther-specific gene, AB2, were identified in the SDR of Ipomoea (Rahman et al., 2007a, 2007b). These tissue-specific expressed genes are considered S-determinant candidates for SI in Ipomoea. Analysis of genotypes and S-allelic interactions in Ipomoea trifida populations and their crossed F1 progeny revealed a linear dominant–recessive hierarchy with multiple levels of codominance among the S-alleles (Kowyama, 2000). However, the molecular bases of the Ipomoea SSI system remain to be characterized. The availability of three chromosome-level Ipomoea genomes (I. trifida, I. triloba, and I. batatas) (Wu et al., 2018) should provide more information for determining the precise role of SI candidates and further delineating the molecular mechanisms that regulate SI in Ipomoea.
SSI has also been reported in several members of the Oleaceae family, where it is characterized by the presence of only two S-alleles, the diallelic SSI (DSI) system (Vernet et al., 2016; Saumitou-Laprade et al. 2018; Besnard et al. 2020; De Cauwer et al., 2021). The diallelic SI locus of olive (Olea europaea) is located at 5.4 cM on LG 18, and two genes related to flower development (STYLISH1 and FAR1) were identified within the DSI region (Mariotti et al., 2020). Examining the interactions between six S-alleles in olive varieties revealed dominance relationships of R6 > R2, R2 > R3, R3 = R1 = R5, and R5 > R4 (Breton and Bervillé, 2012), and the dominant alleles take precedence over others in determining mate compatibility during pollination (Breton et al., 2014). Cellular events reported to be involved in the SI response of Oleaceae include PCD (Serrano et al., 2012). A study has shown that olive SI is mediated by a peroxynitrite-dependent PCD signaling pathway that fine-tunes ROS and nitric oxide production to trigger cell death (Serrano et al., 2012). Although DSI is shared by all SI species studied to date in Oleaceae, the SI activities in other genera remain unknown. Comparative analyses of the diallelic interaction, SI response, and evolutionary conservation of DSI in other genera should enable a deeper understanding of the DSI system in Oleaceae.
The genus Corylus (Betulaceae) includes several important nut-producing species that exhibit SSI systems (Ma et al., 2013). In European hazelnut (Corylus avellana), the S-locus was identified within a 193.5-kb region of LG 5, at which 33 alleles have been identified (Mehlenbacher, 2014; Hill et al., 2021). The S-alleles in Corylus exhibit a linear codominance relationship, and the dominance hierarchy includes eight levels (Mehlenbacher, 2014). Several genes encoding RLKs have been identified at the S-locus (e.g., MIK2 and Putative Interactor of XopAC 7 [PIX7]) (Hill et al., 2021; Hou et al., 2022a). Expression analyses and yeast two-hybrid assays confirmed that Corylus does not employ the SRK-mediated signaling pathway of Brassica SSI (Hill et al., 2021; Hou et al., 2022a, 2022b). Recent phylogenetic analysis provided additional evidence supporting the genus-specific evolution of SSI in Corylus, which is not shared by other members of Betulaceae (Brainard et al., 2023).
Overall, families that possess the SSI system exhibit diverse SI mechanisms. It is likely that their SSI mechanisms and those of Brassicaceae have undergone convergent evolution, and these families are thus less likely to use the same SRK–SCR/SP11 system that operates in Brassicaceae. Investigating the S-determinants and downstream signaling networks in these families will provide new insights into the evolutionary origins of SSI systems.
Heteromorphic self-incompatibility characterized by heterostyly
Heterostyly refers to the presence of two or more floral morphs (e.g., distyly or tristyly) that exhibit spatial separation of pollen and stigma, usually in the form of a short-style (thrum) morph with the stigma positioned above the anther and a long-style (pin) morph with the stigma situated below the anther (Barrett and Shore, 2008). This reciprocal floral trait, in which the stigmatic surface and the anther are spatially separated (kerkogamy), promotes efficient cross-pollination and is often associated with a form of SI known as heteromorphic SI (HSI) (Barrett, 2000). The most distinguishing feature of HSI compared with homomorphic SI is the tight association between floral morphology and female compatibility type (McCubbin, 2008). HSI is thought to have originated independently multiple times, with a scattered phylogenetic distribution in 28 families (Barrett, 2002). The recent availability of molecular techniques has led to fundamental advances in understanding the molecular mechanisms of HSI, including characterization of S-locus and female SI types in Primula (Primulaceae) (Li et al., 2011, 2015; Huu et al., 2022), identification of male and female determinants at the S-locus and generation of SC mutants in Turnera (Passifloraceae) (Chafe et al., 2015; Matzke et al., 2021; Henning et al., 2022), identification of floral morph–related genes in Linum grandiflorum (Linaceae) (Ushijima et al., 2012, 2015), and analysis of S-locus genes in the buckwheat Fagopyrum esculentum (Yasui et al., 2012; Ueno et al., 2016). Over 91% of species in the genus Primula have dimorphic HSI to promote legitimate pollination between thrum (S) and pin (L) morphs, making it the most studied model for HSI (McCubbin, 2008). The dimorphs of Primula are determined by dominant and recessive genotypes at the single S-locus, which is a supergene containing at least two tightly linked genes that control different traits (Kappel et al., 2017). In short-styled Primula flowers, the female SI type is determined by the expression of the cytochrome P450 CYP734A50 in the style, which inhibits cell elongation by inactivating brassinosteroids (BRs) (Huu et al., 2016, 2022) (Figure 3D). The GLOBOSA2 (GLO2) gene at the S-locus may control the position and elevation of the anther (Huu et al., 2020), but the gene that determines male incompatibility types remains unclear. A recent study showed that virus-induced gene silencing of CYP734A50 in S-morph plants altered both style length and female compatibility, resulting in homostyled flowers (Huu et al., 2022). This study also found that BRs play a role in determining female SI by functioning both indirectly in pollen germination and directly in PT growth, with S- and L-morph pollen responding differently to BR treatment. This system of female SI and style-length determination by a BR-inactivating enzyme has also been reported in Turnera, in which the BR-inactivating activity of BAHD, an S-locus gene of the BAHD acyltransferase family, pleiotropically controls style length and female mating specificity (Shore et al., 2019; Matzke et al., 2020). BRs play a role in mediating these traits by differentially regulating gene expression in the style (Matzke et al., 2021). Recent work suggests that the male mating type and pollen size of Turnera are controlled by the YUCCA family member YUC6 at the S-locus (Henning et al., 2022). YUC6 is thought to have undergone neofunctionalization in Turnera and acts as a male determination factor by affecting auxin production in the anther. In contrast to the system mediated by BR inactivation in Primula and Turnera, the style length of short-styled Fagopyrum is controlled by Early flowering 3 at the S-locus (S-ELF3) (Yasui et al., 2012), suggesting that HSI may be controlled by different systems in different heterostyled species. Use of molecular techniques to identify and characterize additional genes involved in female specificity and heterostyly in model systems such as Primula and Turnera will likely lead to more exciting discoveries in this field.
SI in monocots, using the grass family as an example
As another important angiosperm lineage, monocots also include many taxa that exhibit SI with vastly diverse SI systems based on different types of incompatible reactions (Sage et al., 2000). SI types in monocots span GSI in the grass family (Poaceae) (de Nettancourt, 2001), heteromorphic SSI in Pontederiaceae (Bianchi et al., 2000), late-acting prezygotic SI, also known as ovarian SI, in Velloziaceae, Iridaceae, Amaryllidaceae, and Xanthorrhoeaceae (Gibbs, 2014), indeterminate GSI in pineapple (Ananas comosus) (Chen et al., 2019a), and uncharacterized but non-S-RNase-type SI in Orchidaceae (Niu et al., 2017). Despite the widespread occurrence of SI in monocots, little is known about the molecular mechanisms and related physiological and biochemical processes involved in SI within this group. Here, we review research progress on monocots, using the grass family as an example.
The two-locus (S-Z) GSI system in Poaceae is the most studied SI system in monocots (Langridge and Baumann, 2008). The GSI system is controlled by two multiple-allelic loci, S and Z (Li et al., 1997). Each locus contains two male and one female determinant, and SI occurs when the pollen S- and Z- genotypes are identical to the corresponding pistil genotypes (Rohner et al., 2023). The downstream reaction upon pollen–stigma interaction in grass appears to involve Ca2+ signal transduction, protein phosphorylation, and proteolysis pathways (Yang et al., 2008; Klaas et al., 2011; Chen et al., 2019b). Comparative genomic analysis suggested a duplicated origin of the grass SI system owing to the similar gene composition and structure of the S-locus and Z-locus, and the high synteny between these two loci in the Poaceae tribe supported the hypothesis that grasses share the same SI determinants (Rohner et al., 2023). Fine-mapping and comparative studies identified two DUF247 family genes (SDUF247-I and SDUF247-II) and an sS gene as the male and female determinants, respectively, at the S-locus (Manzanares et al., 2016). Likewise, pollen and stigma components are determined by two DUF247 family genes (ZDUF247-I and ZDUF247-II) and the sZ gene at the Z-locus (Rohner et al., 2023). Two linked glycerol kinase-like genes (LpGK1 and LpGK2) were reported as Z-candidates, but further validation experiments remain to be performed (Studer and Torben, 2014). The HPS10 expression pattern and the results of CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9) knockout confirmed that HPS10 (which encodes a small extracellular pistil peptide) is the sS gene of the S-locus in African wild rice (Oryza longistaminata) (Lian et al., 2021; Wang et al., 2022b). Loss of function of either the female or male component at the S-locus can cause breakdown of SI, which functions in a cognate manner to reject self-pollen. This specific linkage between HPS-10 and DUF247 is found exclusively in Poaceae, suggesting the independent origin of the S-Z GSI system (Wang et al., 2022b). However, the sZ gene at the Z-locus and the downstream components of pollen‒pistil interactions remain to be identified. With the availability of high-quality Poaceae genome assemblies and recent advances in gene engineering techniques (Zhang et al., 2015, 2020; Melonek et al., 2021), a comprehensive comparative genomics approach can now be used to resolve these questions in grass SI and beyond.
Breeding strategies for SI crops
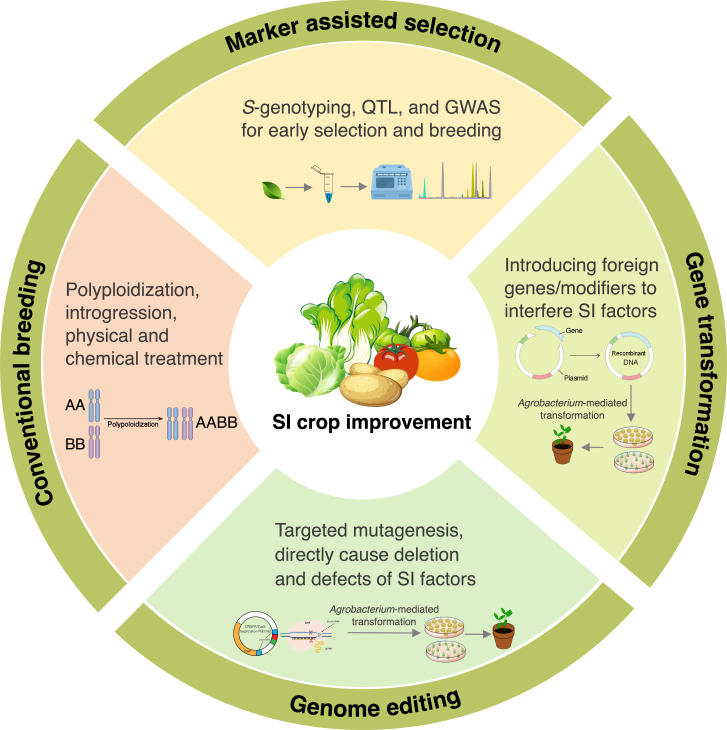
Obligate outcrossing is favored in SI species because of its evolutionary advantage in preventing inbreeding depression and maintaining a high level of diversity (Barrett, 2002). However, SI may impose constraints on crop production and horticultural practices owing to the limited availability of mating partners and pollination vectors, which inevitably results in low fertilization efficiency and disparities in fruit quality. This is a significant barrier, particularly when the commercial product is seeds, and intense management and higher costs are required to achieve good yields (reviewed in Ahmad et al., 2022). Hence, developing SC lines that bypass outcrossing is an important objective for some SI crops to promote efficient breeding and achieve and maintain elite genotypes. With a large number of molecular techniques now available, our current understanding of SI is increasingly being applied in a crop-breeding context to address three main objectives: overcoming SI for crop improvement, marker-assisted selection and breeding, and interspecific UI.
Crop improvement
Conventionally, most SC cultivars are derived from spontaneous mutations or physiological induction/chemical treatment by breeders and growers and are therefore strongly dependent on artificial evaluation and empirical selection (de Nettancourt, 2001). Since the first attempt to break down SI in B. rapa using Agrobacterium-mediated transformation (Shiba et al., 1995), there has been growing interest in the production of SC crops through biotechnology (Figure 4 and Table 1). Modern genetic engineering approaches such as transgenic breeding, breeding by genome editing, and gene silencing have enabled SI to SC transitions (and vice versa) through intentional manipulation of target SI factors. Among these techniques, genome editing enables unprecedented advances in the introduction of new traits into SI species and the generation of SC lines, revolutionizing crop improvement through precise DNA manipulation. CRISPR/Cas9, an advanced genome-editing technology, has been used in important food crops, fruit crops, and forage crops to perform precise genetic modifications, leading to both gain-of-function outcomes through incorporation of desired mutations and loss-of-function effects by creation of knockouts (Nadakuduti et al., 2018; Chen et al., 2020; Zhou et al., 2020).
Figure 4.
Four breeding strategies commonly used for SI crop improvement.
Table 1.
Examples of conferring self-compatibility through biotechnology.
| Species/cultivars | Methods | Target gene | Purpose of the study | Reference |
|---|---|---|---|---|
| Brassica rapa | Agrobacterium tumefaciens–mediated transformation | S-locus glycoproteins (SLGs) | Break down self-incompatibility (SI) in B. rapa | Shiba et al. (2000) |
| Brassica napus | A. tumefaciens strain COR338–mediated transformation | SLG, S-locus receptor kinase (SRK) | Generate SI in B. napus | Cui et al. (2000) |
| Brassica napus | A. tumefaciens–mediated transformation | SRK | Examine the role of SRK in inducing SI | Silva et al. (2001) |
| Arabidopsis thaliana | A. tumefaciens–mediated transformation | Glyoxalase I (GLO1) | Break down artificial SI in A. thaliana | Kenney et al. (2020) |
| Solanum chacoense lines 314, 582, G4, V22 | RNA interference (RNAi) | HT | Gene silencing of HT to impart SC in S. chacoense | O’Brien et al. (2002) |
| Petunia hybrida | A. tumefaciens–mediated transformation | S-RNase, SLF | Break down pollen SI function in P. hybrida | Qiao et al. (2004) |
| Solanum tuberosum RH89-039-16 (RH) | A. tumefaciens strain GV3101–mediated transformation | S-locus inhibitor (Sli) | Develop inbred potato lines | Ma et al. (2021) |
| Solanum tuberosum cultivars | A. tumefaciens strain AGL0 and AGL1–mediated transformation, CRISPR/Cas9 | S-locus inhibitor (Sli) | Develop inbred potato lines | Eggers et al. (2021) |
In SI crops such as potato (Solanum tuberosum), crop quality can be improved by introducing genes from wild species with resistance traits, and the development of inbred lines can further accelerate fixation of desirable allelic combinations (Enciso-Rodriguez et al., 2019). Although biotechnology has greatly promoted the production of clonally propagated potato, sexual reproduction via seeds still offers advantages for new variety selection and pathogen avoidance (Kardile et al., 2022). In this context, CRISPR/Cas9 has been effectively used to knock out S-RNase genes for direct breakdown of SI and development of SC diploid potato (Ye et al., 2018; Enciso-Rodriguez et al., 2019). Notably, the SI trait can be transmitted to T1 progeny, suggesting the possibility of transgenerational deletion (Enciso-Rodriguez et al., 2019). In a recent study, double knockouts of HT-B and S-RNase led to enhanced SC phenotypes in two potato lines (124_001 and 124_137), which can serve as additional genetic resources for future diploid breeding (Lee et al., 2023). Development of inbred diploid lines via genome editing appears to offer tremendous opportunities for crop improvement, providing an alternative to clonally propagated tetraploid potatoes (Jansky et al., 2016).
Genome editing has also been used for trait improvement in Brassica, a genus with many agriculturally important SI crops. CRISPR/Cas9 knockout of BnS6-Smi2, the suppressor of BnSCR, generated a novel SI phenotype in rapeseed (B. napus) and thus represents a promising method for creating more SI lines for breeding (Dou et al., 2021). In addition to manipulation of S-determinants, CRISPR/Cas9-based knockout of the gene encoding PP2A 55-kDa B regulatory subunit (PR55/B) conferred SC in Chinese cabbage (B. rapa ssp. pekinensis), highlighting opportunities for development of SC lines by targeting non-S loci and SI-related genes (Shin et al., 2022). CRISPR/Cas9 can also be used to induce multiple simultaneous mutations for trait improvement (Ma et al., 2019). The obligate outcrossing and prolonged vernalization time of cabbage (Brassica oleracea var. capitata) have complicated breeding efforts to create new germplasm (Ma et al., 2019). To achieve trait improvement in cabbage (B. oleracea var. capitata), an endogenous tRNA processing-based CRISPR/Cas9 system was used to induce mutagenesis in the S-receptor kinase gene BoSRK and the male-sterility-associated gene BoMS1 (Ma et al., 2019). This multiple editing strategy that produces SC and male-sterile lines can overcome breeding barriers in cabbage, enabling the stacking of desirable traits in a single operation.
Other species in which CRISPR/Cas9 has been used for trait improvement include cultivated alfalfa (Medicago sativa), one of the world’s most important forage crops that exhibits recalcitrant characteristics such as SI and tetrasomic inheritance (Chen et al., 2020), apple (Malus domestica) (Osakabe et al., 2018), and Prunus species (Prudencio et al., 2022), suggesting that precise gene modifications are increasingly feasible for SI crops. Although routine protocols remain to be developed for more SI crops, these studies highlight the great potential of genetic engineering techniques such as CRISPR/Cas9 to confer desirable traits for crop improvement without lengthy procedures.
Marker-assisted selection and breeding
For woody perennials such as fruit trees, long juvenile periods and large plant sizes have greatly hindered progress in cultivar selection and improvement of elite breeding lines (Dirlewanger et al., 2004). In many rosaceous species that exhibit obligate outcrossing, the presence of suitable pollinizers (compatible pollen donors) or SC cultivars is an important determinant of fruit set, and it may take years for these traits to be evaluated in the field (Schneider et al., 2005; Dirlewanger et al., 2009). Traditional selection based on controlled pollination and observation of PT growth is laborious and time consuming, with unexpected variables due to varying environmental conditions. As information on the S-specificity of each SI system has become available, molecular genetics approaches (e.g., genotyping, linkage maps, QTLs) have been progressively used in marker-assisted selection (MAS) and breeding to precisely target the genotypes associated with SI traits.
Allele-specific polymerase chain reaction is routinely used to identify S-genotypes and new S-alleles in important fruit crops, including apple (M. domestica) (Sheick et al., 2018), apricot (Prunus armeniaca) (Orlando Marchesano et al., 2022), cherry (Prunus avium) (Kivistik et al., 2022), almond (Prunus dulcis) (Gouta et al., 2021), plum (diploid Pyrus salicina, hexaploid Pyrus domestica and Pyrus insititia) (Abdallah et al., 2019), and pear (Pyrus communis) (Claessen et al., 2022). These S-genotypes have been particularly useful in commercial orchard management, making it possible to rapidly determine compatible combinations among cultivars and shorten the generation cycle needed to characterize a genotype (Dirlewanger et al., 2004). Built upon DNA-based markers, advances in next-generation sequencing technologies have enabled the use of single-nucleotide polymorphism (SNP) markers (Mammadov et al., 2012) in genetic mapping and diversity screening for the breeding of SI crops. Examples of this can be seen in cacao (Theobroma cacao) and wheatgrass (Thinopyrum intermedium), in which genome-wide studies have identified SNPs associated with SI or SC factors and fine-mapped the regions containing the candidate genes, thus facilitating early selection and development of inbred lines (da Silva et al., 2016; Crain et al., 2020; Lopes et al., 2022).
QTL mapping is another important tool for MAS. Although QTL characterization is more commonly used for quantitative traits with intricate inheritance patterns such as fruit quality and bloom time (Dirlewanger et al., 2009), mapping QTLs associated with SI or SC can aid in the selection of lines with target traits. For instance, Xiao et al. (2019) performed recurrent backcrossing for rapid introgression using two SC-QTL-specific markers, BoID0709 and BoID0992, thereby overcoming crossing incompatibility in SI cabbage. In a related approach, QTLs linked to self-fertility were mapped in SI alfalfa to reduce the occurrence of self-seeded cultivars (Robins and Brummer, 2010). In B. rapa, five QTLs associated with SI were identified by constructing a linkage map for MAS of stable SI parental lines (Hatakeyama et al., 2010). In sunflower (Helianthus annuus), two QTLs associated with self-pollination were detected in elite crosses of wild populations to understand the genetic control of SC in inbred lines and accelerate gene introgression via MAS (Gandhi et al., 2005).
The successful application of molecular markers to SI crops underscores their potential to expedite the development of inbred lines with desired traits. By harnessing the genetic information encoded in these markers, breeders can make informed decisions, prioritize valuable traits, and navigate complex inheritance patterns with greater efficiency.
Interspecific unilateral incompatibility
In addition to SI, the best-known intraspecific reproduction barrier, plants also use a variety of interspecific reproductive barriers (IRBs) to maintain species identity, hindering interspecific hybridization by breeders (Baek et al., 2016). Because these two reproductive barriers are mechanistically related, knowledge gained from well-studied SI systems has provided many insights into mechanisms of hybrid incompatibility (Bedinger et al., 2017). One of the IRBs is interspecific UI, a unidirectional interspecific pollen rejection that typically occurs when the female parent is SI and the male parent is SC (the SI × SC rule) and is thought to have a close connection to SI in some lineages (Chalivendra et al., 2013).
Extensive work has been undertaken to unravel the mechanisms underlying UI in two prominent crop genera, Brassica and Solanum. In B. rapa, UI is triggered by recognition between genes encoding the pistil receptor SUI1 (STIGMATIC UNILATERAL INCOMPATIBILITY 1) and the pollen ligand PUI1 (POLLEN UNILATERAL INCOMPATIBILITY 1) that display genetic control similar to that of the SI genes SRK and SP11, respectively (Takada et al., 2017). In a very recent Brassicaceae study, SRK was shown to mediate rejection of UI pollen by recruiting FER and rapidly increasing FER-mediated ROS levels in stigmas in a process that resembled SRK signaling in the SI response but occurred via different intracellular pathways (Huang et al., 2023). A study in A. thaliana showed that STIGMATIC PRIVACY 1 (SPRI1), which encodes a stigma-specific plasma membrane protein, is a stigmatic UI factor that rejects pollen from distantly related Brassicaceae species (Fujii et al., 2019). Although there is no evidence for shared mechanisms between SI and UI, SI components such as MLPK have been found to be involved in UI signaling pathways in Brassica (Takada et al., 2013), indicating that UI occurs in a species-specific manner in Brassicaceae. Nevertheless, the parallel between UI and SI is more evident in Solanum species, as several SI factors, including S-RNase, CUL1, and HT, also function in the UI system (Hancock et al., 2003). Studies on tomato cultivars and related Solanum species showed that genes encoding SLF proteins and Cullin1 protein function as pollen factors in UI and that pollen recognition involves mechanisms related to SI (Li and Chetelat, 2014, 2015). Tovar-Méndez et al. (2014) performed a gain-of-function experiment with S-RNase and HT genes in cultivated tomato and found that they were sufficient to confer UI between red- and green-fruited species. These results point to the possibility that some types of UI are genetically associated with SI, but a solid conclusion on this issue must await the characterization and validation of more genes underlying UI. Existing knowledge of SI systems undoubtedly presents an avenue to reveal the hidden interplay between these two reproductive barriers, ultimately enhancing our understanding of interspecific hybridization possibilities.
Concluding remarks and prospects
SI has long captivated scientists because of its pivotal role in evolutionary processes that lead to speciation and diversification, as well as the important consequences of mating-system shifts. Since Darwin’s time, our understanding of diverse SI systems and their underlying mechanisms has been greatly enhanced by advances in molecular and evolutionary studies. Undoubtedly, genomic approaches stand as the emerging forefront in the study of SI and its integration into breeding practices. These techniques offer unprecedented opportunities to unravel the intricate signaling pathways triggered by SI responses, illuminating the trajectory of SI evolution and the transitions between SI and SC. The extensive repository of molecular and genetic insights has also paved the way for precise engineering of SI crops, building a solid foundation for crop enhancement and molecular-assisted selection.
Although numerous SI mechanisms have been investigated, tremendous variation in the genetic control of SI poses ongoing challenges. This is particularly evident in the necessity to decipher SI systems and S-determinants in monocots and other angiosperm lineages. Although molecular techniques have been extensively applied to well-characterized SI systems such as S-RNase-based GSI and Brassica SSI, outstanding questions persist, especially concerning their origin, evolution, and gain and loss, as well as the identification of novel regulatory factors. Addressing differences in downstream signaling components among taxa that use the same SI systems (e.g., the SCR–SRK system in Brassica and Arabidopsis or the S-RNase-based system in Petunia, Nicotiana, and Prunus) presents another critical challenge. There is also compelling evidence for parallels in the mechanisms that govern self-/nonself-discrimination and pathogen defense, as well as shared components of SI and IRBs. A crucial direction for future research is to identify the potential intersections and explore the potential functional redundancy of these pathways. Furthermore, with genome editing playing a central role in crop engineering, precise DNA manipulation to confer desired traits and disrupt SI holds great promise for tackling various breeding challenges in the foreseeable future. Development of routine protocols for genome-editing techniques such as CRISPR/Cas9 will be a pivotal step toward realizing these goals. The combination of molecular tools and cutting-edge techniques promises to propel future research forward, ushering in a new era in our understanding of SI.
Funding
This work was supported by the Forestry Peak Discipline Construction Project of Fujian Agriculture and Forestry University (Grant number 72202200205).
Author contributions
D.Z. wrote the manuscript and drew the figures. Y.-Y.L., X.Z., C.Z., and D.-K.L. collected the references. S.L., W.Y., and Z.-J.L. supervised and revised the manuscript.
Acknowledgments
The authors declare that they have no competing interests.
Published: September 16, 2023
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Contributor Information
Siren Lan, Email: lkzx@fafu.edu.cn.
Weilun Yin, Email: yinwl@bjfu.edu.cn.
Zhong-Jian Liu, Email: zjliu@fafu.edu.cn.
References
- Abdallah D., Baraket G., Perez V., Ben Mustapha S., Salhi-Hannachi A., Hormaza J.I. Analysis of self-incompatibility and genetic diversity in diploid and hexaploid plum genotypes. Front. Plant Sci. 2019;10:896. doi: 10.3389/fpls.2019.00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abhinandan K., Hickerson N.M., Lan X., Samuel M.A. Disabling of ARC1 through CRISPR–Cas9 leads to a complete breakdown of self-incompatibility responses in Brassica napus. Plant Commun. 2023;4 doi: 10.1016/j.xplc.2022.100504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abhinandan K., Sankaranarayanan S., Macgregor S., Goring D.R., Samuel M.A. Cell–cell signaling during the Brassicaceae self-incompatibility response. Trends Plant Sci. 2022;27:472–487. doi: 10.1016/j.tplants.2021.10.011. [DOI] [PubMed] [Google Scholar]
- Aguiar B., Vieira J., Cunha A.E., Fonseca N.A., Iezzoni A., van Nocker S., Vieira C.P. Convergent evolution at the gametophytic self-incompatibility system in Malus and Prunus. PLoS One. 2015;10 doi: 10.1371/journal.pone.0126138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad M.H., Rao M.J., Hu J., Xu Q., Liu C., Cao Z., Larkin R.M., Deng X., Bosch M., Chai L. Systems and breakdown of self-incompatibility. CRC Crit. Rev. Plant Sci. 2022;41:209–239. [Google Scholar]
- Akagi T., Henry I.M., Morimoto T., Tao R. Insights into the Prunus-specific S-RNase-based self-incompatibility system from a genome-wide analysis of the evolutionary radiation of S locus-related F-box genes. Plant Cell Physiol. 2016;57:1281–1294. doi: 10.1093/pcp/pcw077. [DOI] [PubMed] [Google Scholar]
- Allen A.M., Hiscock S.J. Self-Incompatibility in Flowering Plants. Springer Berlin Heidelberg; Heidelberg: 2008. Evolution and phylogeny of self-incompatibility systems in angiosperms; pp. 73–101. [Google Scholar]
- Allen A.M., Thorogood C.J., Hegarty M.J., Lexer C., Hiscock S.J. Pollen–pistil interactions and self-incompatibility in the Asteraceae: new insights from studies of Senecio squalidus (Oxford ragwort) Ann. Bot. 2011;108:687–698. doi: 10.1093/aob/mcr147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Angiosperm Phylogeny Group An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016;181:1–20. [Google Scholar]
- Ashkani J., Rees D.J.G. A comprehensive study of molecular evolution at the self-incompatibility locus of Rosaceae. J. Mol. Evol. 2016;82:128–145. doi: 10.1007/s00239-015-9726-4. [DOI] [PubMed] [Google Scholar]
- Asquini E., Gerdol M., Gasperini D., Igic B., Graziosi G., Pallavicini A. S-RNase-like sequences in styles of coffea (Rubiaceae). Evidence for S-RNase based gametophytic self-incompatibility? Trop. Plant Biol. 2011;4:237–249. [Google Scholar]
- Baek Y.S., Royer S.M., Broz A.K., Covey P.A., López-Casado G., Nuñez R., Kear P.J., Bonierbale M., Orillo M., van der Knaap E., et al. Interspecific reproductive barriers between sympatric populations of wild tomato species (Solanum section Lycopersicon) Am. J. Bot. 2016;103:1964–1978. doi: 10.3732/ajb.1600356. [DOI] [PubMed] [Google Scholar]
- Barrett S. The evolution and function of stylar polymorphisms in flowering plants. Ann. Bot. 2000;85:253–265. [Google Scholar]
- Barrett S.C.H. The evolution of plant sexual diversity. Nat. Rev. Genet. 2002;3:274–284. doi: 10.1038/nrg776. [DOI] [PubMed] [Google Scholar]
- Barrett S.C.H. The evolution of plant reproductive systems: how often are transitions irreversible? Proc. Biol. Sci. 2013;280 doi: 10.1098/rspb.2013.0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett S.C.H., Shore J.S. Self-Incompatibility in Flowering Plants. Springer Berlin Heidelberg; Heidelberg: 2008. New insights on heterostyly: comparative biology, ecology and genetics; pp. 3–32. [Google Scholar]
- Bayer P.E., Scheben A., Golicz A.A., Yuan Y., Faure S., Lee H., Chawla H.S., Anderson R., Bancroft I., Raman H., et al. Modelling of gene loss propensity in the pangenomes of three Brassica species suggests different mechanisms between polyploids and diploids. Plant Biotechnol. J. 2021;19:2488–2500. doi: 10.1111/pbi.13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechsgaard J.S., Castric V., Charlesworth D., Vekemans X., Schierup M.H. The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 Myr. Mol. Biol. Evol. 2006;23:1741–1750. doi: 10.1093/molbev/msl042. [DOI] [PubMed] [Google Scholar]
- Bedinger P.A., Broz A.K., Tovar-Mendez A., McClure B. Pollen-pistil interactions and their role in mate selection. Plant Physiol. 2017;173:79–90. doi: 10.1104/pp.16.01286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedinger P.A., Chetelat R.T., McClure B., Moyle L.C., Rose J.K.C., Stack S.M., van der Knaap E., Baek Y.S., Lopez-Casado G., Covey P.A., et al. Interspecific reproductive barriers in the tomato clade: opportunities to decipher mechanisms of reproductive isolation. Sex. Plant Reprod. 2011;24:171–187. doi: 10.1007/s00497-010-0155-7. [DOI] [PubMed] [Google Scholar]
- Besnard G., Cheptou P.O., Debbaoui M., Lafont P., Hugueny B., Dupin J., Baali-Cherif D. Paternity tests support a diallelic self-incompatibility system in a wild olive (Olea europaeasubsp.laperrinei, Oleaceae) Ecol. Evol. 2020;10:1876–1888. doi: 10.1002/ece3.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi M., Vesprini J., Barrett S.C.H. Trimorphic incompatibility in Eichhornia azurea (Pontederiaceae) Sex. Plant Reprod. 2000;12:203–208. [Google Scholar]
- Bombarely A., Moser M., Amrad A., Bao M., Bapaume L., Barry C.S., Bliek M., Boersma M.R., Borghi L., Bruggmann R., et al. Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nat. Plants. 2016;2 doi: 10.1038/nplants.2016.74. [DOI] [PubMed] [Google Scholar]
- Bosch M., Franklin-Tong V.E. Self-incompatibility in Papaver: signalling to trigger PCD in incompatible pollen. J. Exp. Bot. 2008;59:481–490. doi: 10.1093/jxb/erm195. [DOI] [PubMed] [Google Scholar]
- Brainard S.H., Sanders D.M., Bruna T., Shu S., Dawson J.C. The first two chromosome-scale genome assemblies of American hazelnut enable comparative genomic analysis of the genus Corylus. bioRxiv. 2023 doi: 10.1101/2023.04.27.537858. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton C.M., Bervillé A. New hypothesis elucidates self-incompatibility in the olive tree regarding S-alleles dominance relationships as in the sporophytic model. C. R. Biol. 2012;335:563–572. doi: 10.1016/j.crvi.2012.07.006. [DOI] [PubMed] [Google Scholar]
- Breton C.M., Farinelli D., Shafiq S., Heslop-Harrison J.S., Sedgley M., Bervillé A.J. The self-incompatibility mating system of the olive (Olea europaea L.) functions with dominance between S-alleles. Tree Genet. Genomes. 2014;10:1055–1067. [Google Scholar]
- Broz A.K., Miller C.M., Baek Y.S., Tovar-Méndez A., Acosta-Quezada P.G., Riofrío-Cuenca T.E., Rusch D.B., Bedinger P.A. S-RNase alleles associated with self-compatibility in the tomato clade: structure, origins, and expression plasticity. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.780793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz A.K., Randle A.M., Sianta S.A., Tovar-Méndez A., McClure B., Bedinger P.A. Mating system transitions in Solanum habrochaites impact interactions between populations and species. New Phytol. 2017;213:440–454. doi: 10.1111/nph.14130. [DOI] [PubMed] [Google Scholar]
- Burghgraeve N., Simon S., Barral S., Fobis-Loisy I., Holl A.C., Ponitzki C., Schmitt E., Vekemans X., Castric V. Base-pairing requirements for small RNA-mediated gene silencing of recessive self-incompatibility alleles in Arabidopsis halleri. Genetics. 2020;215:653–664. doi: 10.1534/genetics.120.303351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrillac D., Cock J.M., Dumas C., Gaude T. The S-locus receptor kinase is inhibited by thioredoxins and activated by pollen coat proteins. Nature. 2001;410:220–223. doi: 10.1038/35065626. [DOI] [PubMed] [Google Scholar]
- Cao Y.L., Li Y.L., Fan Y.F., Li Z., Yoshida K., Wang J.Y., Ma X.K., Wang N., Mitsuda N., Kotake T., et al. Wolfberry genomes and the evolution of Lycium (Solanaceae) Commun. Biol. 2021;4:671. doi: 10.1038/s42003-021-02152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chafe P.D.J., Lee T., Shore J.S. Development of a genetic transformation system for distylous Turnera joelii (Passifloraceae) and characterization of a self-compatible mutant. Plant Cell Tissue Organ Cult. 2015;120:507–517. [Google Scholar]
- Chai L., Tudor R.L., Poulter N.S., Wilkins K.A., Eaves D.J., Franklin F.C.H., Franklin-Tong V.E. MAP kinase PrMPK9-1 contributes to the self-incompatibility response. Plant Physiol. 2017;174:1226–1237. doi: 10.1104/pp.17.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalivendra S.C., Lopez-Casado G., Kumar A., Kassenbrock A.R., Royer S., Tovar-Mèndez A., Covey P.A., Dempsey L.A., Randle A.M., Stack S.M., et al. Developmental onset of reproductive barriers and associated proteome changes in stigma/styles of Solanum pennellii. J. Exp. Bot. 2013;64:265–279. doi: 10.1093/jxb/ers324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantha S.C., Herman A.C., Castric V., Vekemans X., Marande W., Schoen D.J. The unusual S locus of Leavenworthia is composed of two sets of paralogous loci. New Phytol. 2017;216:1247–1255. doi: 10.1111/nph.14764. [DOI] [PubMed] [Google Scholar]
- Chantha S.-C., Herman A.C., Platts A.E., Vekemans X., Schoen D.J. Secondary evolution of a self-incompatibility locus in the Brassicaceae genus Leavenworthia. PLoS Biol. 2013;11 doi: 10.1371/journal.pbio.1001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantreau M., Poux C., Lensink M.F., Brysbaert G., Vekemans X., Castric V. Asymmetrical diversification of the receptor-ligand interaction controlling self-incompatibility in Arabidopsis. Elife. 2019;8 doi: 10.7554/eLife.50253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D. Evolution of homomorphic sporophytic self-incompatibility. Heredity. 1988;60:445–453. [Google Scholar]
- Chen H., Zeng Y., Yang Y., Huang L., Tang B., Zhang H., Hao F., Liu W., Li Y., Liu Y., et al. Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa. Nat. Commun. 2020;11:2494. doi: 10.1038/s41467-020-16338-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wang P., de Graaf B.H.J., Zhang H., Jiao H., Tang C., Zhang S., Wu J. Phosphatidic acid counteracts S-RNase signaling in pollen by stabilizing the actin cytoskeleton. Plant Cell. 2018;30:1023–1039. doi: 10.1105/tpc.18.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.Y., VanBuren R., Paris M., Zhou H., Zhang X., Wai C.M., Yan H., Chen S., Alonge M., Ramakrishnan S., et al. The bracteatus pineapple genome and domestication of clonally propagated crops. Nat. Genet. 2019;51:1549–1558. doi: 10.1038/s41588-019-0506-8. [DOI] [PubMed] [Google Scholar]
- Chen S., Jia J., Cheng L., Zhao P., Qi D., Yang W., Liu H., Dong X., Li X., Liu G. Transcriptomic analysis reveals a comprehensive calcium- and phytohormone-dominated signaling response in Leymus chinensis self-incompatibility. Int. J. Mol. Sci. 2019;20:2356. doi: 10.3390/ijms20092356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen H., Van de Poel B., Keulemans W., De Storme N. A semi in vivo pollination technique to assess the level of gametophytic self-incompatibility and pollen tube growth in pear (Pyrus communis L.) Plant Reprod. 2022;35:127–140. doi: 10.1007/s00497-021-00435-y. [DOI] [PubMed] [Google Scholar]
- Crain J., Larson S., Dorn K., Hagedorn T., DeHaan L., Poland J. Sequenced-based paternity analysis to improve breeding and identify self-incompatibility loci in intermediate wheatgrass (Thinopyrum intermedium) Theor. Appl. Genet. 2020;133:3217–3233. doi: 10.1007/s00122-020-03666-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Zamora Y., Nájera-Torres E., Noriega-Navarro R., Torres-Rodríguez M.D., Bernal-Gracida L.A., García-Valdés J., Juárez-Díaz J.A., Cruz-García F. NaStEP, an essential protein for self-incompatibility in Nicotiana, performs a dual activity as a proteinase inhibitor and as a voltage-dependent channel blocker. Plant Physiol. Biochem. 2020;151:352–361. doi: 10.1016/j.plaphy.2020.03.052. [DOI] [PubMed] [Google Scholar]
- Cui Y., Bi Y.M., Brugière N., Arnoldo M., Rothstein S.J. The S locus glycoprotein and the S receptor kinase are sufficient for self-pollen rejection in Brassica. Proc. Natl. Acad. Sci. USA. 2000;97:3713–3717. doi: 10.1073/pnas.050480297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Zhuang M., Wu J., Liu J., Zhang Y., Zhang L., Huang Y., Cai X., Liang J., Zhang K., et al. Segmental translocation contributed to the origin of the Brassica S-locus. Hortic. Plant J. 2020;6:167–178. [Google Scholar]
- da Silva M.R., Clément D., Gramacho K.P., Monteiro W.R., Argout X., Lanaud C., Lopes U. Genome-wide association mapping of sexual incompatibility genes in cacao (Theobroma cacao L.) Tree Genet. Genomes. 2016;12:62. [Google Scholar]
- De Cauwer I., Vernet P., Billiard S., Godé C., Bourceaux A., Ponitzki C., Saumitou-Laprade P. Widespread coexistence of self-compatible and self-incompatible phenotypes in a diallelic self-incompatibility system in Ligustrum vulgare (Oleaceae) Heredity. 2021;127:384–392. doi: 10.1038/s41437-021-00463-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf B.H.J., Rudd J.J., Wheeler M.J., Perry R.M., Bell E.M., Osman K., Franklin F.C.H., Franklin-Tong V.E. Self-incompatibility in Papaver targets soluble inorganic pyrophosphatases in pollen. Nature. 2006;444:490–493. doi: 10.1038/nature05311. [DOI] [PubMed] [Google Scholar]
- de Nettancourt D. Springer; Berlin Heidelberg, New York: 2001. Incompatibility and Incongruity in Wild and Cultivated Plants. [Google Scholar]
- Dickinson T.A., Lo E., Talent N. Polyploidy, reproductive biology, and Rosaceae: understanding evolution and making classifications. Plant Systemat. Evol. 2007;266:59–78. [Google Scholar]
- Dirlewanger E., Claverie J., Iezzoni A.F., Wünsch A. In: Genetics and Genomics of Rosaceae. Folta K.M., Gardiner S.E., editors. Springer New York; New York: 2009. Sweet and Sour Cherries: Linkage Maps, QTL Detection and Marker Assisted Selection; pp. 291–313. [Google Scholar]
- Dirlewanger E., Graziano E., Joobeur T., Garriga-Calderé F., Cosson P., Howad W., Arús P. Comparative mapping and marker-assisted selection in Rosaceae fruit crops. Proc. Natl. Acad. Sci. USA. 2004;101:9891–9896. doi: 10.1073/pnas.0307937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou S., Zhang T., Tu J., Shen J., Yi B., Wen J., Fu T., Dai C., Ma C. Generation of novel self-incompatible Brassica napus by CRISPR/Cas9. Plant Biotechnol. J. 2021;19:875–877. doi: 10.1111/pbi.13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan T., Zhang Z., Genete M., Poux C., Sicard A., Lascoux M., Castric V., Vekemans X. Dominance between self-incompatibility alleles determines the mating system of Capsella allopolyploids. bioRxiv. 2023:04. doi: 10.1101/2023.04.17.537155. 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand E., Chantreau M., Le Veve A., Stetsenko R., Dubin M., Genete M., Llaurens V., Poux C., Roux C., Billiard S., et al. Evolution of self-incompatibility in the Brassicaceae: lessons from a textbook example of natural selection. Evol. Appl. 2020;13:1279–1297. doi: 10.1111/eva.12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand E., Méheust R., Soucaze M., Goubet P.M., Gallina S., Poux C., Fobis-Loisy I., Guillon E., Gaude T., Sarazin A., et al. Dominance hierarchy arising from the evolution of a complex small RNA regulatory network. Science. 2014;346:1200–1205. doi: 10.1126/science.1259442. [DOI] [PubMed] [Google Scholar]
- Durvasula A., Fulgione A., Gutaker R.M., Alacakaptan S.I., Flood P.J., Neto C., Tsuchimatsu T., Burbano H.A., Picó F.X., Alonso-Blanco C., et al. African genomes illuminate the early history and transition to selfing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2017;114:5213–5218. doi: 10.1073/pnas.1616736114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edh K., Widén B., Ceplitis A. The evolution and diversification of S-locus haplotypes in the Brassicaceae family. Genetics. 2009;181:977–984. doi: 10.1534/genetics.108.090837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers E.-J., van der Burgt A., van Heusden S.A.W., de Vries M.E., Visser R.G.F., Bachem C.W.B., Lindhout P. Neofunctionalisation of the Sli gene leads to self-compatibility and facilitates precision breeding in potato. Nat. Commun. 2021;12:4141. doi: 10.1038/s41467-021-24267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enciso-Rodriguez F., Manrique-Carpintero N.C., Nadakuduti S.S., Buell C.R., Zarka D., Douches D. Overcoming self-incompatibility in diploid potato using CRISPR-Cas9. Front. Plant Sci. 2019;10:376. doi: 10.3389/fpls.2019.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entani T., Kubo K.i., Isogai S., Fukao Y., Shirakawa M., Isogai A., Takayama S. Ubiquitin-proteasome-mediated degradation of S-RNase in a solanaceous cross-compatibility reaction. Plant J. 2014;78:1014–1021. doi: 10.1111/tpj.12528. [DOI] [PubMed] [Google Scholar]
- Fernández i Martí A., Gradziel T.M., Socias i Company R. Methylation of the Sf locus in almond is associated with S-RNase loss of function. Plant Mol. Biol. 2014;86:681–689. doi: 10.1007/s11103-014-0258-x. [DOI] [PubMed] [Google Scholar]
- Ferrer M.M., Good S.V. Self-sterility in flowering plants: preventing self-fertilization increases family diversification rates. Ann. Bot. 2012;110:535–553. doi: 10.1093/aob/mcs124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer M.M., Good-Avila S.V. Macrophylogenetic analyses of the gain and loss of self-incompatibility in the Asteraceae. New Phytol. 2007;173:401–414. doi: 10.1111/j.1469-8137.2006.01905.x. [DOI] [PubMed] [Google Scholar]
- Finnegan E.J., Liang D., Wang M.-B. Self-incompatibility: Smi silences through a novel sRNA pathway. Trends Plant Sci. 2011;16:238–241. doi: 10.1016/j.tplants.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Franklin-Tong N.V.E., Franklin F.C.H. Gametophytic self-incompatibility inhibits pollen tube growth using different mechanisms. Trends Plant Sci. 2003;8:598–605. doi: 10.1016/j.tplants.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Franklin-Tong V.E., Holdaway-Clarke T.L., Straatman K.R., Kunkel J.G., Hepler P.K. Involvement of extracellular calcium influx in the self-incompatibility response of Papaver rhoeas. Plant J. 2002;29:333–345. doi: 10.1046/j.1365-313x.2002.01219.x. [DOI] [PubMed] [Google Scholar]
- Fujii S., Takayama S. Multilayered dominance hierarchy in plant self-incompatibility. Plant Reprod. 2018;31:15–19. doi: 10.1007/s00497-017-0319-9. [DOI] [PubMed] [Google Scholar]
- Fujii S., Kubo K.I., Takayama S. Non-self- and self-recognition models in plant self-incompatibility. Nat. Plants. 2016;2 doi: 10.1038/nplants.2016.130. [DOI] [PubMed] [Google Scholar]
- Fujii S., Shimosato-Asano H., Kakita M., Kitanishi T., Iwano M., Takayama S. Parallel evolution of dominant pistil-side self-incompatibility suppressors in Arabidopsis. Nat. Commun. 2020;11:1404. doi: 10.1038/s41467-020-15212-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S., Tsuchimatsu T., Kimura Y., Ishida S., Tangpranomkorn S., Shimosato-Asano H., Iwano M., Furukawa S., Itoyama W., Wada Y., et al. A stigmatic gene confers interspecies incompatibility in the Brassicaceae. Nat. Plants. 2019;5:731–741. doi: 10.1038/s41477-019-0444-6. [DOI] [PubMed] [Google Scholar]
- Gandhi S.D., Heesacker A.F., Freeman C.A., Argyris J., Bradford K., Knapp S.J. The self-incompatibility locus (S) and quantitative trait loci for self-pollination and seed dormancy in sunflower. Theor. Appl. Genet. 2005;111:619–629. doi: 10.1007/s00122-005-1934-7. [DOI] [PubMed] [Google Scholar]
- Gao C., Zhou G., Ma C., Zhai W., Zhang T., Liu Z., Yang Y., Wu M., Yue Y., Duan Z., et al. Helitron-like transposons contributed to the mating system transition from out-crossing to self-fertilizing in polyploid Brassica napus L. Sci. Rep. 2016;6 doi: 10.1038/srep33785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Valencia L.E., Bravo-Alberto C.E., Wu H.M., Rodríguez-Sotres R., Cheung A.Y., Cruz-García F. SIPP, a novel mitochondrial phosphate carrier, mediates in self-incompatibility. Plant Physiol. 2017;175:1105–1120. doi: 10.1104/pp.16.01884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervais C.E., Castric V., Ressayre A., Billiard S. Origin and diversification dynamics of self-incompatibility haplotypes. Genetics. 2011;188:625–636. doi: 10.1534/genetics.111.127399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs P.E. Late-acting self-incompatibility–the pariah breeding system in flowering plants. New Phytol. 2014;203:717–734. doi: 10.1111/nph.12874. [DOI] [PubMed] [Google Scholar]
- Goldberg E.E., Kohn J.R., Lande R., Robertson K.A., Smith S.A., Igić B. Species selection maintains self-incompatibility. Science. 2010;330:493–495. doi: 10.1126/science.1194513. [DOI] [PubMed] [Google Scholar]
- Goldraij A., Kondo K., Lee C.B., Hancock C.N., Sivaguru M., Vazquez-Santana S., Kim S., Phillips T.E., Cruz-Garcia F., McClure B. Compartmentalization of S-RNase and HT-B degradation in self-incompatible Nicotiana. Nature. 2006;439:805–810. doi: 10.1038/nature04491. [DOI] [PubMed] [Google Scholar]
- Gonthier L., Blassiau C., Mörchen M., Cadalen T., Poiret M., Hendriks T., Quillet M.C. High-density genetic maps for loci involved in nuclear male sterility (NMS1) and sporophytic self-incompatibility (S-locus) in chicory (Cichorium intybus L., Asteraceae) Theor. Appl. Genet. 2013;126:2103–2121. doi: 10.1007/s00122-013-2122-9. [DOI] [PubMed] [Google Scholar]
- Goring D.R., Bosch M., Franklin-Tong V.E. Contrasting self-recognition rejection systems for self-incompatibility in Brassica and Papaver. Curr. Biol. 2023;33:R530–R542. doi: 10.1016/j.cub.2023.03.037. [DOI] [PubMed] [Google Scholar]
- Gouta H., Ksia E., Laaribi I., Bennici S., La Malfa S., Gentile A., Distefano G. Efficiency of S-genotyping for diversity screening and self-incompatible group identification of almond cultivars within the Mediterranean basin. J. Hortic. Sci. Biotechnol. 2021;96:338–343. [Google Scholar]
- Gu Z., Li W., Doughty J., Meng D., Yang Q., Yuan H., Li Y., Chen Q., Yu J., Liu C.s., Li T. A gamma-thionin protein from apple, MdD1, is required for defence against S-RNase-induced inhibition of pollen tube prior to self/non-self recognition. Plant Biotechnol. J. 2019;17:2184–2198. doi: 10.1111/pbi.13131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Halitschke R., Wielsch N., Gase K., Baldwin I.T. Mate selection in self-compatible wild tobacco results from coordinated variation in homologous self-incompatibility genes. Curr. Biol. 2019;29:2020–2030.e5. doi: 10.1016/j.cub.2019.05.042. [DOI] [PubMed] [Google Scholar]
- Guo Y.L., Zhao X., Lanz C., Weigel D. Evolution of the S-locus region in Arabidopsis relatives. Plant Physiol. 2011;157:937–946. doi: 10.1104/pp.111.174912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock C.N., Kondo K., Beecher B., McClure B. The S-locus and unilateral incompatibility. Phil. Trans. Roy. Soc. Lond. B. 2003;358:1133–1140. doi: 10.1098/rstb.2003.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque T., Eaves D.J., Lin Z., Zampronio C.G., Cooper H.J., Bosch M., Smirnoff N., Franklin-Tong V.E. Self-Incompatibility triggers irreversible oxidative modification of proteins in incompatible pollen. Plant Physiol. 2020;183:1391–1404. doi: 10.1104/pp.20.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama K., Horisaki A., Niikura S., Narusaka Y., Abe H., Yoshiaki H., Ishida M., Fukuoka H., Matsumoto S. Mapping of quantitative trait loci for high level of self-incompatibility in Brassica rapa. Genome. 2010;53:257–265. doi: 10.1139/g10-001. [DOI] [PubMed] [Google Scholar]
- Haudry A., Platts A.E., Vello E., Hoen D.R., Leclercq M., Williamson R.J., Forczek E., Joly-Lopez Z., Steffen J.G., Hazzouri K.M., et al. An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat. Genet. 2013;45:891–898. doi: 10.1038/ng.2684. [DOI] [PubMed] [Google Scholar]
- Henning P.M., Shore J.S., McCubbin A.G. The S-gene YUC6 pleiotropically determines male mating type and pollen size in heterostylous Turnera (Passifloraceae): A novel neofunctionalization of the YUCCA gene family. Plants. 2022;11:2640. doi: 10.3390/plants11192640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill R.J., Baldassi C., Snelling J.W., Vining K.J., Mehlenbacher S.A. Fine mapping of the locus controlling self-incompatibility in European hazelnut. Tree Genet. Genomes. 2021;17:6–14. [Google Scholar]
- Hohmann N., Wolf E.M., Lysak M.A., Koch M.A. A time-calibrated road map of Brassicaceae species radiation and evolutionary history. Plant Cell. 2015;27:2770–2784. doi: 10.1105/tpc.15.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S., Zhao T., Yang Z., Yang D., Li Q., Liang L., Wang G., Ma Q. Molecular cloning and yeast two-hybrid provide new evidence for unique sporophytic self-incompatibility system of Corylus. Plant Biol. 2022;24:104–116. doi: 10.1111/plb.13347. [DOI] [PubMed] [Google Scholar]
- Hou S., Zhao T., Yang Z., Liang L., Ma W., Wang G., Ma Q. Stigmatic Transcriptome analysis of self-incompatible and compatible pollination in Corylus heterophylla Fisch. × Corylus avellana L. Front. Plant Sci. 2022;13 doi: 10.3389/fpls.2022.800768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Xu Q., Liu C., Liu B., Deng C., Chen C., Wei Z., Ahmad M.H., Peng K., Wen H., et al. Downregulated expression of S2-RNase attenuates self-incompatibility in “Guiyou No. 1” pummelo. Hortic. Res. 2021;8:199. doi: 10.1038/s41438-021-00634-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Yang L., Yang L., Wu X., Cui X., Zhang L., Hui J., Zhao Y., Yang H., Liu S., et al. Stigma receptors control intraspecies and interspecies barriers in Brassicaceae. Nature. 2023;614:303–308. doi: 10.1038/s41586-022-05640-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huu C.N., Kappel C., Keller B., Sicard A., Takebayashi Y., Breuninger H., Nowak M.D., Bäurle I., Himmelbach A., Burkart M., et al. Presence versus absence of CYP734A50 underlies the style-length dimorphism in primroses. Elife. 2016;5 doi: 10.7554/eLife.17956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huu C.N., Keller B., Conti E., Kappel C., Lenhard M. Supergene evolution via stepwise duplications and neofunctionalization of a floral-organ identity gene. Proc. Natl. Acad. Sci. USA. 2020;117:23148–23157. doi: 10.1073/pnas.2006296117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huu C.N., Plaschil S., Himmelbach A., Kappel C., Lenhard M. Female self-incompatibility type in heterostylous Primula is determined by the brassinosteroid-inactivating cytochrome P450 CYP734A50. Curr. Biol. 2022;32:671–676.e5. doi: 10.1016/j.cub.2021.11.046. [DOI] [PubMed] [Google Scholar]
- Igic B., Kohn J.R. Evolutionary relationships among self-incompatibility RNases. Proc. Natl. Acad. Sci. USA. 2001;98:13167–13171. doi: 10.1073/pnas.231386798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igic B., Lande R., Kohn J. Loss of self-incompatibility and its evolutionary consequences. IJPS. 2008;169:93–104. [Google Scholar]
- Indriolo E., Safavian D., Goring D.R. The ARC1 E3 ligase promotes two different self-pollen avoidance traits in Arabidopsis. Plant Cell. 2014;26:1525–1543. doi: 10.1105/tpc.114.122879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwano M., Ito K., Fujii S., Kakita M., Asano-Shimosato H., Igarashi M., Kaothien-Nakayama P., Entani T., Kanatani A., Takehisa M., et al. Calcium signalling mediates self-incompatibility response in the Brassicaceae. Nat. Plants. 2015;1 doi: 10.1038/nplants.2015.128. [DOI] [PubMed] [Google Scholar]
- Jansky S.H., Charkowski A.O., Douches D.S., Gusmini G., Richael C., Bethke P.C., Spooner D.M., Novy R.G., De Jong H., De Jong W.S., et al. Reinventing potato as a diploid inbred line–based crop. Crop Sci. 2016;56:1412–1422. [Google Scholar]
- Jany E., Nelles H., Goring D.R. The molecular and cellular regulation of Brassicaceae self-incompatibility and self-pollen rejection. Int. Rev. Cell Mol. Biol. 2019;343:1–35. doi: 10.1016/bs.ircmb.2018.05.011. [DOI] [PubMed] [Google Scholar]
- Jiang X., Gao Y., Zhou H., Chen J., Wu J., Zhang S. Apoplastic calmodulin promotes self-incompatibility pollen tube growth by enhancing calcium influx and reactive oxygen species concentration in Pyrus pyrifolia. Plant Cell Rep. 2014;33:255–263. doi: 10.1007/s00299-013-1526-y. [DOI] [PubMed] [Google Scholar]
- Jiménez-Durán K., McClure B., García-Campusano F., Rodríguez-Sotres R., Cisneros J., Busot G., Cruz-García F. NaStEP: A proteinase inhibitor essential to self-incompatibility and a positive regulator of HT-B stability in Nicotiana alata pollen tubes. Plant Physiol. 2013;161:97–107. doi: 10.1104/pp.112.198440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakita M., Shimosato H., Murase K., Isogai A., Takayama S. Direct interaction between S-locus receptor kinase and M-locus protein kinase involved in Brassica self-incompatibility signaling. Plant Biol. 2007;24:185–190. [Google Scholar]
- Kappel C., Huu C.N., Lenhard M. A short story gets longer: recent insights into the molecular basis of heterostyly. J. Exp. Bot. 2017;68:5719–5730. doi: 10.1093/jxb/erx387. [DOI] [PubMed] [Google Scholar]
- Kardile H.B., Yilma S., Sathuvalli V. Molecular approaches to overcome self-incompatibility in diploid potatoes. Plants. 2022;11:1328. doi: 10.3390/plants11101328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney P., Sankaranarayanan S., Balogh M., Indriolo E. Expression of Brassica napus GLO1 is sufficient to breakdown artificial self-incompatibility in Arabidopsis thaliana. Plant Reprod. 2020;33:159–171. doi: 10.1007/s00497-020-00392-y. [DOI] [PubMed] [Google Scholar]
- Kitashiba H., Liu P., Nishio T., Nasrallah J.B., Nasrallah M.E. Functional test of Brassica self-incompatibility modifiers in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2011;108:18173–18178. doi: 10.1073/pnas.1115283108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivistik A., Jakobson L., Kahu K., Laanemets K. Wild and rare self-incompatibility allele S17 found in 24 sweet cherry (Prunus avium L.) cultivars. Plant Mol. Biol. Rep. 2022;40:376–388. [Google Scholar]
- Klaas M., Yang B., Bosch M., Thorogood D., Manzanares C., Armstead I.P., Franklin F.C.H., Barth S. Progress towards elucidating the mechanisms of self-incompatibility in the grasses: further insights from studies in Lolium. Ann. Bot. 2011;108:677–685. doi: 10.1093/aob/mcr186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M.A., Haubold B., Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae) Mol. Biol. Evol. 2000;17:1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248. [DOI] [PubMed] [Google Scholar]
- Kowyama Y. Sporophytic Self-incompatibility in Ipomoea trifida, a Close Relative of Sweet Potato. Ann. Bot. 2000;85:191–196. [Google Scholar]
- Kowyama Y., Tsuchiya T., Kakeda K. Self-Incompatibility in Flowering Plants. Springer Berlin Heidelberg; Heidelberg: 2008. Molecular genetics of sporophytic self-Incompatibility in Ipomoea, a member of the Convolvulaceae; pp. 259–274. [Google Scholar]
- Kubo K.i., Entani T., Takara A., Wang N., Fields A.M., Hua Z., Toyoda M., Kawashima S.i., Ando T., Isogai A., et al. Collaborative non-self recognition system in S-RNase–based self-incompatibility. Science. 2010;330:796–799. doi: 10.1126/science.1195243. [DOI] [PubMed] [Google Scholar]
- Kubo K.I., Paape T., Hatakeyama M., Entani T., Takara A., Kajihara K., Tsukahara M., Shimizu-Inatsugi R., Shimizu K.K., Takayama S. Gene duplication and genetic exchange drive the evolution of S-RNase-based self-incompatibility in Petunia. Nat. Plants. 2015;1 doi: 10.1038/nplants.2014.5. [DOI] [PubMed] [Google Scholar]
- Kusaba M., Dwyer K., Hendershot J., Vrebalov J., Nasrallah J.B., Nasrallah M.E. Self-incompatibility in the genus Arabidopsis: characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. Plant Cell. 2001;13:627–643. [PMC free article] [PubMed] [Google Scholar]
- Langridge P., Baumann U. Self-Incompatibility in Flowering Plants. Springer Berlin Heidelberg; Heidelberg: 2008. Self-Incompatibility in the Grasses; pp. 275–287. [Google Scholar]
- Lee S., Enciso-Rodriguez F.E., Behling W., Jayakody T., Panicucci K., Zarka D., Nadakuduti S.S., Buell C.R., Manrique-Carpintero N.C., Douches D.S. HT-B and S-RNase CRISPR-Cas9 double knockouts show enhanced self-fertility in diploid Solanum tuberosum. Front. Plant Sci. 2023;14 doi: 10.3389/fpls.2023.1151347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Webster M.A., Smith M.C., Gilmartin P.M. Floral heteromorphy in Primula vulgaris: progress towards isolation and characterization of the S locus. Ann. Bot. 2011;108:715–726. doi: 10.1093/aob/mcr181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Webster M.A., Wright J., Cocker J.M., Smith M.C., Badakshi F., Heslop-Harrison P., Gilmartin P.M. Integration of genetic and physical maps of the Primula vulgaris S locus and localization by chromosome in situ hybridization. New Phytol. 2015;208:137–148. doi: 10.1111/nph.13373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Zhang D., Gao Q., Luo Y., Zhang H., Ma B., Chen C., Whibley A., Zhang Y., Cao Y., et al. Genome structure and evolution of Antirrhinum majus L. Nat. Plants. 2019;5:174–183. doi: 10.1038/s41477-018-0349-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Chetelat R.T. The role of a pollen-expressed cullin1 protein in gametophytic self-incompatibility in Solanum. Genetics. 2014;196:439–442. doi: 10.1534/genetics.113.158279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Chetelat R.T. Unilateral incompatibility gene ui1.1 encodes an S-locus F-box protein expressed in pollen of Solanum species. Proc. Natl. Acad. Sci. USA. 2015;112:4417–4422. doi: 10.1073/pnas.1423301112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Lu Y., Han R., Yue Q., Song X., Wang F., Wu R., Hou F., Yang L., Xu L., et al. Apple S-RNase triggers inhibition of tRNA aminoacylation by interacting with a soluble inorganic pyrophosphatase in growing self-pollen tubes in vitro. New Phytol. 2018;47:579–589. doi: 10.1111/nph.15028. [DOI] [PubMed] [Google Scholar]
- Li X., Paech N., Nield J., Hayman D., Langridge P. Self-incompatibility in the grasses: evolutionary relationship of the S gene from Phalaris coerulescens to homologous sequences in other grasses. Plant Mol. Biol. 1997;34:223–232. doi: 10.1023/a:1005802327900. [DOI] [PubMed] [Google Scholar]
- Li Y., Duan X., Wu C., Yu J., Liu C., Wang J., Zhang X., Yan G., Jiang F., Li T., et al. Ubiquitination of S4 -RNase by S-LOCUS F-BOX LIKE2 contributes to self-compatibility of sweet cherry ‘Lapins’. Plant Physiol. 2020;184:1702–1716. doi: 10.1104/pp.20.01171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X., Zhang S., Huang G., Huang L., Zhang J., Hu F. Confirmation of a gametophytic self-incompatibility in Oryza longistaminata. Front. Plant Sci. 2021;12 doi: 10.3389/fpls.2021.576340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M., Cao Z., Zhu A., Liu Y., Tao M., Yang H., Xu Q., Wang S., Liu J., Li Y., et al. Evolution of self-compatibility by a mutant Sm-RNase in citrus. Nat. Plants. 2020;6:131–142. doi: 10.1038/s41477-020-0597-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Eaves D.J., Sanchez-Moran E., Franklin F.C.H., Franklin-Tong V.E. The Papaver rhoeas S determinants confer self-incompatibility to Arabidopsis thaliana in planta. Science. 2015;350:684–687. doi: 10.1126/science.aad2983. [DOI] [PubMed] [Google Scholar]
- Lin Z., Xie F., Triviño M., Karimi M., Bosch M., Franklin-Tong V.E., Nowack M.K. Ectopic expression of a self-incompatibility module triggers growth arrest and cell death in vegetative cells. Plant Physiol. 2020;183:1765–1779. doi: 10.1104/pp.20.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Xie F., Triviño M., Zhao T., Coppens F., Sterck L., Bosch M., Franklin-Tong V.E., Nowack M.K. Self-incompatibility requires GPI anchor remodeling by the poppy PGAP1 ortholog HLD1. Curr. Biol. 2022;32:1909–1923.e5. doi: 10.1016/j.cub.2022.02.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Fan J., Li J., Song Y., Li Q., Zhang Y., Xue Y. SCFSLF-mediated cytosolic degradation of S-RNase is required for cross-pollen compatibility in S-RNase-based self-incompatibility in Petunia hybrida. Front. Genet. 2014;5:228. doi: 10.3389/fgene.2014.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes U.V., Pires J.L., Gramacho K.P., Grattapaglia D. Genome-wide SNP genotyping as a simple and practical tool to accelerate the development of inbred lines in outbred tree species: An example in cacao (Theobroma cacao L.) PLoS One. 2022;17 doi: 10.1371/journal.pone.0270437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv S., Qiao X., Zhang W., Li Q., Wang P., Zhang S., Wu J. The origin and evolution of RNase T2 Family and gametophytic self-incompatibility system in plants. Genome Biol. Evol. 2022;14 doi: 10.1093/gbe/evac093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak M.A., Koch M.A., Pecinka A., Schubert I. Chromosome triplication found across the tribe Brassiceae. Genome Res. 2005;15:516–525. doi: 10.1101/gr.3531105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Zhu C., Zheng M., Liu M., Zhang D., Liu B., Li Q., Si J., Ren X., Song H. CRISPR/Cas9-mediated multiple gene editing in Brassica oleracea var. capitata using the endogenous tRNA-processing system. Hortic. Res. 2019;6:20. doi: 10.1038/s41438-018-0107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Zhang C., Zhang B., Tang F., Li F., Liao Q., Tang D., Peng Z., Jia Y., Gao M., et al. A non S-locus F-box gene breaks self-incompatibility in diploid potatoes. Nat. Commun. 2021;12:4142. doi: 10.1038/s41467-021-24266-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q.H., Wang G.X., Liang W.J., Chen X., Liang L.S., Zhao T.T. Progress on pollen-stigma compatibility in Corylus (hazelnuts): a review. J. Forestry Res. 2013;24:397–402. [Google Scholar]
- Mable B.K., Robertson A.V., Dart S., Di Berardo C., Witham L. Breakdown of self-incompatibility in the perennial Arabidopsis Lyrata (Brassicaceae) and its genetic consequences. Evolution. 2005;59:1437–1448. [PubMed] [Google Scholar]
- Macgregor S.R., Lee H.K., Nelles H., Johnson D.C., Zhang T., Ma C., Goring D.R. Autophagy is required for self-incompatible pollen rejection in two transgenic Arabidopsis thaliana accessions. Plant Physiol. 2022;188:2073–2084. doi: 10.1093/plphys/kiac026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclntosh G.C. In: RNase T2 Family: Enzymatic Properties, Functional Diversity, and Evolution of Ancient Ribonucleases. Ribonucleases, Nicholson A.W., editor. Springer Berlin Heidelberg; Heidelberg: 2011. pp. 89–114. [Google Scholar]
- Mammadov J., Aggarwal R., Buyyarapu R., Kumpatla S. SNP Markers and Their Impact on Plant Breeding. Int. J. Pharmagenesis (IJPG) 2012;2012:1–11. doi: 10.1155/2012/728398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanares C., Barth S., Thorogood D., Byrne S.L., Yates S., Czaban A., Asp T., Yang B., Studer B. A gene encoding a duf247 domain protein cosegregates with the S self-incompatibility locus in Perennial Ryegrass. Mol. Biol. Evol. 2016;33:870–884. doi: 10.1093/molbev/msv335. [DOI] [PubMed] [Google Scholar]
- Mariotti R., Fornasiero A., Mousavi S., Cultrera N.G.M., Brizioli F., Pandolfi S., Passeri V., Rossi M., Magris G., Scalabrin S., et al. Genetic mapping of the incompatibility locus in olive and development of a linked sequence-tagged site marker. Front. Plant Sci. 2020;10:1760. doi: 10.3389/fpls.2019.01760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markova D.N., Petersen J.J., Yam S.E., Corral A., Valle M.J., Li W., Chetelat R.T. Evolutionary history of two pollen self-incompatibility factors reveals alternate routes to self-compatibility within Solanum. Am. J. Bot. 2017;104:1904–1919. doi: 10.3732/ajb.1700196. [DOI] [PubMed] [Google Scholar]
- Matsumoto D., Tao R. Recognition of S-RNases by an S locus F-box like protein and an S haplotype-specific F-box like protein in the Prunus-specific self-incompatibility system. Plant Mol. Biol. 2019;100:367–378. doi: 10.1007/s11103-019-00860-8. [DOI] [PubMed] [Google Scholar]
- Matzke C.M., Hamam H.J., Henning P.M., Dougherty K., Shore J.S., Neff M.M., McCubbin A.G. Pistil mating type and morphology are mediated by the brassinosteroid inactivating activity of the S-locus gene BAHD in heterostylous Turnera species. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms221910603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke C.M., Shore J.S., Neff M.M., McCubbin A.G. The Turnera style S-locus gene TsBAHD possesses brassinosteroid-inactivating activity when expressed in Arabidopsis thaliana. Plants. 2020;9:1566. doi: 10.3390/plants9111566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcclure B.A., Gray J.E., Anderson M.A., Clarke A.E. Self-incompatibility in Nicotiana alata involves degradation of pollen rRNA. Nature. 1990;347:757–760. [Google Scholar]
- McClure B., Mou B., Canevascini S., Bernatzky R. A small asparagine-rich protein required for S-allele-specific pollen rejection in Nicotiana. Proc. Natl. Acad. Sci. USA. 1999;96:13548–13553. doi: 10.1073/pnas.96.23.13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCubbin A. Self-Incompatibility in Flowering Plants. Springer Berlin Heidelberg; Heidelberg: 2008. Heteromorphic self-incompatibility in Primula: twenty-first century tools promise to unravel a classic nineteenth century model system; pp. 289–308. [Google Scholar]
- McCubbin A.G., Kao T. Molecular recognition and response in pollen and pistil interactions. Annu. Rev. Cell Dev. Biol. 2000;16:333–364. doi: 10.1146/annurev.cellbio.16.1.333. [DOI] [PubMed] [Google Scholar]
- Mehlenbacher S.A. Geographic Distribution of Incompatibility Alleles in Cultivars and Selections of European Hazelnut. J. Am. Soc. Hortic. Sci. 2014;139:191–212. [Google Scholar]
- Melonek J., Korzun V., Hackauf B. The Rye Genome. Springer; Cham: 2021. Genomics of self-incompatibility and male-fertility restoration in rye; pp. 181–212. [Google Scholar]
- Murase K., Moriwaki Y., Mori T., Liu X., Masaka C., Takada Y., Maesaki R., Mishima M., Fujii S., Hirano Y., et al. Mechanism of self/nonself-discrimination in Brassica self-incompatibility. Nat. Commun. 2020;11:4916. doi: 10.1038/s41467-020-18698-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K., Shiba H., Iwano M., Che F.-S., Watanabe M., Isogai A., Takayama S. A membrane-anchored protein kinase involved in Brassica self-incompatibility signaling. Science. 2004;303:1516–1519. doi: 10.1126/science.1093586. [DOI] [PubMed] [Google Scholar]
- Nadakuduti S.S., Buell C.R., Voytas D.F., Starker C.G., Douches D.S. Genome editing for crop improvement – applications in clonally propagated polyploids with a focus on potato (Solanum tuberosum L.) Front. Plant Sci. 2018;9:1607. doi: 10.3389/fpls.2018.01607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan Hancock C., Kent L., McClure B.A. The stylar 120 kDa glycoprotein is required for S-specific pollen rejection in Nicotiana. Plant J. 2005;43:716–723. doi: 10.1111/j.1365-313X.2005.02490.x. [DOI] [PubMed] [Google Scholar]
- Niu S.C., Huang J., Zhang Y.Q., Li P.-X., Zhang G.Q., Xu Q., Chen L.-J., Wang J.-Y., Luo Y.-B., Liu Z.-J. Lack of S-RNase-based gametophytic self-incompatibility in orchids suggests that this system evolved after the monocot-eudicot split. Front. Plant Sci. 2017;8:1106. doi: 10.3389/fpls.2017.01106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova P.Y., Tsuchimatsu T., Simon S., Nizhynska V., Voronin V., Burns R., Fedorenko O.M., Holm S., Säll T., Prat E., et al. Genome sequencing reveals the origin of the allotetraploid Arabidopsis suecica. Mol. Biol. Evol. 2017;34:957–968. doi: 10.1093/molbev/msw299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien M., Kapfer C., Major G., Laurin M., Bertrand C., Kondo K., Kowyama Y., Matton D.P. Molecular analysis of the stylar-expressed Solanum chacoense small asparagine-rich protein family related to the HT modifier of gametophytic self-incompatibility in Nicotiana. Plant J. 2002;32:985–996. doi: 10.1046/j.1365-313x.2002.01486.x. [DOI] [PubMed] [Google Scholar]
- Okamoto S., Odashima M., Fujimoto R., Sato Y., Kitashiba H., Nishio T. Self-compatibility in Brassica napus is caused by independent mutations in S-locus genes: S-locus mutations cause self-compatibility in B. napus. Plant J. 2007;50:391–400. doi: 10.1111/j.1365-313X.2007.03058.x. [DOI] [PubMed] [Google Scholar]
- Okamoto T., Okamoto M., Hikichi E., Ogawa M., Takada Y., Suzuki G., Takayama S., Watanabe M. Characterization of self-incompatible Brassica napus lines lacking SP11 expression. Genes Genet. Syst. 2020;95:111–118. doi: 10.1266/ggs.19-00050. [DOI] [PubMed] [Google Scholar]
- Ono K., Akagi T., Morimoto T., Wünsch A., Tao R. Genome re-sequencing of diverse sweet cherry (Prunus avium) individuals reveals a modifier gene mutation conferring pollen-part self-compatibility. Plant Cell Physiol. 2018;59:1265–1275. doi: 10.1093/pcp/pcy068. [DOI] [PubMed] [Google Scholar]
- Orlando Marchesano B.M., Chiozzotto R., Baccichet I., Bassi D., Cirilli M. Development of an hrma-based marker assisted selection (mas) approach for cost-effective genotyping of S and M loci controlling self-compatibility in apricot (Prunus armeniaca L.) Genes. 2022;13:548. doi: 10.3390/genes13030548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y., Liang Z., Ren C., Nishitani C., Osakabe K., Wada M., Komori S., Malnoy M., Velasco R., Poli M., et al. CRISPR–Cas9-mediated genome editing in apple and grapevine. Nat. Protoc. 2018;13:2844–2863. doi: 10.1038/s41596-018-0067-9. [DOI] [PubMed] [Google Scholar]
- Palumbo F., Draga S., Magon G., Gabelli G., Vannozzi A., Farinati S., Scariolo F., Lucchin M., Barcaccia G. MIK2 is a candidate gene of the S-locus for sporophytic self-incompatibility in chicory (Cichorium intybus, Asteraceae) Front. Plant Sci. 2023;14 doi: 10.3389/fpls.2023.1204538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcher E., Lande R. Loss of gametophytic self-incompatibility with evolution of inbreeding depression. Evolution. 2005;59:46–60. [PubMed] [Google Scholar]
- Price J.H., Raduski A.R., Brandvain Y., Van Tassel D.L., Smith K.P. Development of first linkage map for Silphium integrifolium (Asteraceae) enables identification of sporophytic self-incompatibility locus. Heredity. 2022;128:304–312. doi: 10.1038/s41437-022-00530-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudencio A.S., Devin S.R., Mahdavi S.M.E., Martínez-García P.J., Salazar J.A., Martínez-Gómez P. Spontaneous, artificial, and genome editing-mediated mutations in Prunus. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms232113273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H., Wang F., Zhao L., Zhou J., Lai Z., Zhang Y., Robbins T.P., Xue Y. The F-Box protein AhSLF-S 2 controls the pollen function of S-RNase-based self-incompatibility. Plant Cell. 2004;16:2307–2322. doi: 10.1105/tpc.104.024919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M.H., Tsuchiya T., Suwabe K., Kohori J., Tomita R.N., Kagaya Y., Kobayashi I., Kakeda K., Kowyama Y. Physical size of the S locus region defined by genetic recombination and genome sequencing in Ipomoea trifida. Sex. Plant Reprod. 2007;20:63–72. [Google Scholar]
- Rahman M.H., Uchiyama M., Kuno M., Hirashima N., Suwabe K., Tsuchiya T., Kagaya Y., Kobayashi I., Kakeda K., Kowyama Y. Expression of stigma- and anther-specific genes located in the S locus region of Ipomoea trifida. Sex. Plant Reprod. 2007;20:73–85. doi: 10.1007/s00497-007-0045-9. [DOI] [Google Scholar]
- Ramanauskas K., Igić B. The evolutionary history of plant T2/S-type ribonucleases. PeerJ. 2017;5 doi: 10.7717/peerj.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanauskas K., Igić B. RNase-based self-incompatibility in cacti. New Phytol. 2021;231:2039–2049. doi: 10.1111/nph.17541. [DOI] [PubMed] [Google Scholar]
- Robertson K., Goldberg E.E., Igić B. Comparative evidence for the correlated evolution of polyploidy and self-compatibility in Solanaceae. Evolution. 2011;65:139–155. doi: 10.1111/j.1558-5646.2010.01099.x. [DOI] [PubMed] [Google Scholar]
- Robins J.G., Brummer E.C. QTL underlying self-fertility in tetraploid alfalfa. Crop Sci. 2010;50:143–149. [Google Scholar]
- Rohner M., Manzanares C., Yates S., Thorogood D., Copetti D., Lübberstedt T., Asp T., Studer B., Purugganan M. Fine-mapping and comparative genomic analysis reveal the gene composition at the S and Z self-incompatibility loci in Grasses. Mol. Biol. Evol. 2023;40:msac259. doi: 10.1093/molbev/msac259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safavian D., Goring D.R. Secretory activity is rapidly induced in stigmatic papillae by compatible pollen, but inhibited for self-incompatible pollen in the Brassicaceae. PLoS One. 2013;8 doi: 10.1371/journal.pone.0084286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage T.L., Pontieri V., Christopher R. In: Monocots: Systematics and Evolution. Wilson K.L., Morrison D.A., editors. CSIRO; Melbourne: 2000. Incompatibility and mate recognition in monocotyledons; pp. 268–275. [Google Scholar]
- Samuel M.A., Chong Y.T., Haasen K.E., Aldea-Brydges M.G., Stone S.L., Goring D.R. Cellular pathways regulating responses to compatible and self-incompatible pollen in Brassica and Arabidopsis stigmas intersect at Exo70A1, a putative component of the exocyst complex. Plant Cell. 2009;21:2655–2671. doi: 10.1105/tpc.109.069740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaranarayanan S., Jamshed M., Samuel M.A. Degradation of glyoxalase I in Brassica napus stigma leads to self-incompatibility response. Nat. Plants. 2015;1 doi: 10.1038/nplants.2015.185. [DOI] [PubMed] [Google Scholar]
- Sato K., Nishio T., Kimura R., Kusaba M., Suzuki T., Hatakeyama K., Ockendon D.J., Satta Y. Coevolution of the S-locus genes SRK, SLG and SP11/SCR in Brassica oleracea and B. rapa. Genetics. 2002;162:931–940. doi: 10.1093/genetics/162.2.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saumitou-Laprade P., Vernet P., Dowkiw A., Bertrand S., Billiard S., Albert B., Gouyon P.-H., Dufay M. Polygamy or subdioecy? The impact of diallelic self-incompatibility on the sexual system in Fraxinus excelsior (Oleaceae) Proc. Biol. Sci. 2018;285 doi: 10.1098/rspb.2018.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saumitou-Laprade P., Vernet P., Vekemans X., Billiard S., Gallina S., Essalouh L., Mhaïs A., Moukhli A., El Bakkali A., Barcaccia G., et al. Elucidation of the genetic architecture of self-incompatibility in olive: Evolutionary consequences and perspectives for orchard management. Evol. Appl. 2017;10:867–880. doi: 10.1111/eva.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saumitou-Laprade P., Vernet P., Vekemans X., Castric V., Barcaccia G., Khadari B., Baldoni L. Controlling for genetic identity of varieties, pollen contamination and stigma receptivity is essential to characterize the self-incompatibility system of Olea europaea L. Evol. Appl. 2017;10:860–866. doi: 10.1111/eva.12498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scandola S., Samuel M.A. A flower-specific phospholipase D is a stigmatic compatibility factor targeted by the self-incompatibility response in Brassica napus. Curr. Biol. 2019;29:506–512.e4. doi: 10.1016/j.cub.2018.12.037. [DOI] [PubMed] [Google Scholar]
- Schneider D., Stern R.A., Goldway M. A comparison between semi- and fully compatible apple pollinators grown under suboptimal pollination conditions. Hortscience. 2005;40:1280–1282. [Google Scholar]
- Serrano I., Romero-Puertas M.C., Rodríguez-Serrano M., Sandalio L.M., Olmedilla A. Peroxynitrite mediates programmed cell death both in papillar cells and in self-incompatible pollen in the olive (Olea europaea L.) J. Exp. Bot. 2012;63:1479–1493. doi: 10.1093/jxb/err392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheick R., Serra S., De Franceschi P., Dondini L., Musacchi S. Characterization of a novel self-incompatibility allele in Malus and S-genotyping of select crabapple cultivars. Sci. Hortic. 2018;240:186–195. [Google Scholar]
- Sherman-Broyles S., Boggs N., Farkas A., Liu P., Vrebalov J., Nasrallah M.E., Nasrallah J.B. S locus genes and the evolution of self-fertility in Arabidopsis thaliana. Plant Cell. 2007;19:94–106. doi: 10.1105/tpc.106.048199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba H., Hinata K., Suzuki A., Isogai A. Breakdown of self-incompatibility in Brassica by the antisense RNA of the SLG gene. Proc. Jpn. Acad. Series B. 1995;71:81–83. [Google Scholar]
- Shiba H., Kakizaki T., Iwano M., Tarutani Y., Watanabe M., Isogai A., Takayama S. Dominance relationships between self-incompatibility alleles controlled by DNA methylation. Nat. Genet. 2006;38:297–299. doi: 10.1038/ng1734. [DOI] [PubMed] [Google Scholar]
- Shiba H., Kimura N., Takayama S., Hinata K., Suzuki A., Isogai A. Alteration of the self-incompatibility phenotype in Brassica by transformation of the antisense SLG gene. Biosci. Biotechnol. Biochem. 2000;64:1016–1024. doi: 10.1271/bbb.64.1016. [DOI] [PubMed] [Google Scholar]
- Shimizu K.K., Tsuchimatsu T. Evolution of selfing: recurrent patterns in molecular adaptation. Annu. Rev. Ecol. Evol. Syst. 2015;46:593–622. [Google Scholar]
- Shimizu K.K., Shimizu-Inatsugi R., Tsuchimatsu T., Purugganan M.D. Independent origins of self-compatibility in Arabidopsis thaliana: self-compatibility of Arabidopsis thaliana. Mol. Ecol. 2008;17:704–714. doi: 10.1111/j.1365-294X.2007.03605.x. [DOI] [PubMed] [Google Scholar]
- Shimizu K.K., Kudoh H., Kobayashi M.J. Plant sexual reproduction during climate change: gene function in natura studied by ecological and evolutionary systems biology. Ann. Bot. 2011;108:777–787. doi: 10.1093/aob/mcr180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin N.-R., Shin Y.-H., Kim H.-S., Park Y.-D. Function analysis of the pr55/b gene related to self-incompatibility in Chinese cabbage Using CRISPR/Cas9. Int. J. Mol. Sci. 2022;23:5062. doi: 10.3390/ijms23095062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S.-H., Karlowski W.M., Pan R., Tzeng Y.-H., Mayer K.F.X., Li W.-H. Comparative analysis of the receptor-like kinase family in Arabidopsis and Rice. Plant Cell. 2004;16:1220–1234. doi: 10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore J.S., Hamam H.J., Chafe P.D.J., Labonne J.D.J., Henning P.M., McCubbin A.G. The long and short of the S-locus in Turnera (Passifloraceae) New Phytol. 2019;224:1316–1329. doi: 10.1111/nph.15970. [DOI] [PubMed] [Google Scholar]
- Sijacic P., Wang X., Skirpan A.L., Wang Y., Dowd P.E., McCubbin A.G., Huang S., Kao T.H. Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature. 2004;429:302–305. doi: 10.1038/nature02523. [DOI] [PubMed] [Google Scholar]
- Silva N.F., Goring D.R. Mechanisms of self-incompatibility in flowering plants. Cell. Mol. Life Sci. 2001;58:1988–2007. doi: 10.1007/PL00000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva N.F., Stone S.L., Christie L.N., Sulaman W., Nazarian K.A., Burnett L.A., Arnoldo M.A., Rothstein S.J., Goring D.R. Expression of the S receptor kinase in self-compatible Brassica napus cv. Westar leads to the allele-specific rejection of self-incompatible Brassica napus pollen. Mol. Genet. Genom. 2001;265:552–559. doi: 10.1007/s004380100446. [DOI] [PubMed] [Google Scholar]
- Silverstein K.A.T., Moskal W.A., Jr., Wu H.C., Underwood B.A., Graham M.A., Town C.D., VandenBosch K.A. Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 2007;51:262–280. doi: 10.1111/j.1365-313X.2007.03136.x. [DOI] [PubMed] [Google Scholar]
- Snowman B.N., Kovar D.R., Shevchenko G., Franklin-Tong V.E., Staiger C.J. Signal-mediated depolymerization of actin in pollen during the self-incompatibility response. Plant Cell. 2002;14:2613–2626. doi: 10.1105/tpc.002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Anderson E.M., Mullen R.T., Goring D.R. ARC1 Is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica Pollen. Plant Cell. 2003;15:885–898. doi: 10.1105/tpc.009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Arnoldo M., Goring D.R. A breakdown of Brassica self-incompatibility in ARC1 antisense transgenic plants. Science. 1999;286:1729–1731. doi: 10.1126/science.286.5445.1729. [DOI] [PubMed] [Google Scholar]
- Studer B., Torben A.S.P. 2014. Z Locus Self-Incompatibility Alleles in Poaceae. Patent Application; pp. 2–27. [Google Scholar]
- Sutherland B.L., Quarles B.M., Galloway L.F. Intercontinental dispersal and whole-genome duplication contribute to loss of self-incompatibility in a polyploid complex. Am. J. Bot. 2018;105:249–256. doi: 10.1002/ajb2.1027. [DOI] [PubMed] [Google Scholar]
- Suwabe K., Nagasaka K., Windari E.A., Hoshiai C., Ota T., Takada M., Kitazumi A., Masuko-Suzuki H., Kagaya Y., Yano K., et al. Double-locking mechanism of self-compatibility in Arabidopsis thaliana: the synergistic effect of transcriptional depression and disruption of coding region in the male specificity gene. Front. Plant Sci. 2020;11 doi: 10.3389/fpls.2020.576140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabah D.A., McInnis S.M., Hiscock S.J. Members of the S-receptor kinase multigene family in Senecio squalidus L. (Asteraceae), a species with sporophytic self-incompatibility. Sex. Plant Reprod. 2004;17:131–140. [Google Scholar]
- Takada Y., Murase K., Shimosato-Asano H., Sato T., Nakanishi H., Suwabe K., Shimizu K.K., Lim Y.P., Takayama S., Suzuki G., Watanabe M. Duplicated pollen–pistil recognition loci control intraspecific unilateral incompatibility in Brassica rapa. Nat. Plants. 2017;3 doi: 10.1038/nplants.2017.96. [DOI] [PubMed] [Google Scholar]
- Takada Y., Sato T., Suzuki G., Shiba H., Takayama S., Watanabe M. Involvement of MLPK pathway in intraspecies unilateral incompatibility regulated by a single locus with stigma and pollen factors. G3 (Bethesda). 2013;3:719–726. doi: 10.1534/g3.113.005892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S., Isogai A. Molecular mechanism of self-recognition in Brassica self-incompatibility. J. Exp. Bot. 2003;54:149–156. doi: 10.1093/jxb/erg007. [DOI] [PubMed] [Google Scholar]
- Takayama S., Isogai A. Self-incompatibility in plants. Annu. Rev. Plant Biol. 2005;56:467–489. doi: 10.1146/annurev.arplant.56.032604.144249. [DOI] [PubMed] [Google Scholar]
- Tao R., Emslie G.J., Mayes T.L., Nakonezny P.A., Kennard B.D. The S-RNase-based gametophytic self-incompatibility system in Prunus exhibits distinct genetic and molecular features. Sci. Hortic. 2010;20:423–430. [Google Scholar]
- Tarutani Y., Shiba H., Iwano M., Kakizaki T., Suzuki G., Watanabe M., Isogai A., Takayama S. Trans-acting small RNA determines dominance relationships in Brassica self-incompatibility. Nature. 2010;466:983–986. doi: 10.1038/nature09308. [DOI] [PubMed] [Google Scholar]
- Thomas S.G., Franklin-Tong V.E. Self-incompatibility triggers programmed cell death in Papaver pollen. Nature. 2004;429:305–309. doi: 10.1038/nature02540. [DOI] [PubMed] [Google Scholar]
- Torres-Rodríguez M.D., Cruz-Zamora Y., Juárez-Díaz J.A., Mooney B., McClure B.A., Cruz-García F. NaTrxh is an essential protein for pollen rejection in Nicotiana by increasing S-RNase activity. Plant J. 2020;103:1304–1317. doi: 10.1111/tpj.14802. [DOI] [PubMed] [Google Scholar]
- Tovar-Méndez A., Kumar A., Kondo K., Ashford A., Baek Y.S., Welch L., Bedinger P.A., McClure B.A. Restoring pistil-side self-incompatibility factors recapitulates an interspecific reproductive barrier between tomato species. Plant J. 2014;77:727–736. doi: 10.1111/tpj.12424. [DOI] [PubMed] [Google Scholar]
- Tsuchimatsu T., Goubet P.M., Gallina S., Holl A.C., Fobis-Loisy I., Bergès H., Marande W., Prat E., Meng D., Long Q., et al. Patterns of polymorphism at the self-incompatibility locus in 1,083 Arabidopsis thaliana genomes. Mol. Biol. Evol. 2017;34:1878–1889. doi: 10.1093/molbev/msx122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimatsu T., Kaiser P., Yew C.L., Bachelier J.B., Shimizu K.K. Recent loss of self-incompatibility by degradation of the male component in allotetraploid Arabidopsis kamchatica. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimatsu T., Suwabe K., Shimizu-Inatsugi R., Isokawa S., Pavlidis P., Städler T., Suzuki G., Takayama S., Watanabe M., Shimizu K.K. Evolution of self-compatibility in Arabidopsis by a mutation in the male specificity gene. Nature. 2010;464:1342–1346. doi: 10.1038/nature08927. [DOI] [PubMed] [Google Scholar]
- Tsukamoto T., Ando T., Kokubun H., Watanabe H., Sato T., Masada M., Marchesi E., Kao T.h. Breakdown of self-incompatibility in a natural population of Petunia axillaris caused by a modifier locus that suppresses the expression of an S-RNase gene. Sex. Plant Reprod. 2003;15:255–263. [Google Scholar]
- Tsukamoto T., Ando T., Takahashi K., Omori T., Watanabe H., Kokubun H., Marchesi E., Kao T.H. Breakdown of self-incompatibility in a natural population of Petunia axillaris caused by loss of pollen function. Plant Physiol. 2003;131:1903–1912. doi: 10.1104/pp.102.018069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno M., Yasui Y., Aii J., Matsui K., Sato S., Ota T. In: Molecular Breeding and Nutritional Aspects of Buckwheat. Rabanus-Wallace M.T., Stein N., editors. Springer; Cham: 2016. Genetic analyses of the heteromorphic self-incompatibility (S) locus in buckwheat; pp. 411–421. [Google Scholar]
- Ushijima K., Ikeda K., Nakano R., Matsubara M., Tsuda Y., Kubo Y. Genetic control of floral morph and petal pigmentation in Linum grandiflorum Desf., a heterostylous flax. Horticulture J. 2015;84:261–268. [Google Scholar]
- Ushijima K., Nakano R., Bando M., Shigezane Y., Ikeda K., Namba Y., Kume S., Kitabata T., Mori H., Kubo Y. Isolation of the floral morph-related genes in heterostylous flax (Linum grandiflorum): the genetic polymorphism and the transcriptional and post-transcriptional regulations of the S locus. Plant J. 2012;69:317–331. doi: 10.1111/j.1365-313X.2011.04792.x. [DOI] [PubMed] [Google Scholar]
- Uyenoyama M.K., Zhang Y., Newbigin E. On the origin of self-incompatibility haplotypes: transition through self-compatible intermediates. Genetics. 2001;157:1805–1817. doi: 10.1093/genetics/157.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vekemans X., Poux C., Goubet P.M., Castric V. The evolution of selfing from outcrossing ancestors in Brassicaceae: what have we learned from variation at the S-locus? J. Evol. Biol. 2014;27:1372–1385. doi: 10.1111/jeb.12372. [DOI] [PubMed] [Google Scholar]
- Vernet P., Lepercq P., Billiard S., Bourceaux A., Lepart J., Dommée B., Saumitou-Laprade P. Evidence for the long-term maintenance of a rare self-incompatibility system in Oleaceae. New Phytol. 2016;210:1408–1417. doi: 10.1111/nph.13872. [DOI] [PubMed] [Google Scholar]
- Vieira J., Pimenta J., Gomes A., Laia J., Rocha S., Heitzler P., Vieira C.P. The identification of the Rosa S-locus and implications on the evolution of the Rosaceae gametophytic self-incompatibility systems. Sci. Rep. 2021;11:3710–3712. doi: 10.1038/s41598-021-83243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Xu S., Wu Z., Zhong X., Fang W., Chen F., Teng N. Screening and functional analysis of potential S genes in Chrysanthemum morifolium. O. 2021;1:1–11. [Google Scholar]
- Wang L., Lin Z., Carli J., Gladala-Kostarz A., Davies J.M., Franklin-Tong V.E., Bosch M. ATP depletion plays a pivotal role in self-incompatibility, revealing a link between cellular energy status, cytosolic acidification and actin remodelling in pollen tubes. New Phytol. 2022;236:1691–1707. doi: 10.1111/nph.18350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Ding Q., Triviño M., Pan Y., Song Z., Qin Y., Yan X. Self-incompatibility in Papaver pollen: programmed cell death in an acidic environment. Oncol. Lett. 2018;16:2113–2118. doi: 10.1093/jxb/ery406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang H., Wang J., Sun R., Wu J., Liu S., Bai Y., Mun J.H., Bancroft I., Cheng F., et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011;43:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- Wang Y., Liu P., Cai Y., Li Y., Tang C., Zhu N., Wang P., Zhang S., Wu J. PbrBZR1 interacts with PbrARI2. 3 to mediate brassinosteroid-regulated pollen tube growth during self-incompatibility signaling in pear. Plant Physiol. 2023;192:2356–2373. doi: 10.1093/plphys/kiad208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhao H., Zhu S., Zhang H., Chen Y., Tao X., Tong W., Tian H., Guan Y., Huang H., et al. Control of gametophytic self-incompatibility in the African wild rice. Preprint. Research Square. 2022 doi: 10.21203/rs.3.rs-2121145/v1. [DOI] [Google Scholar]
- Wheeler M.J., de Graaf B.H.J., Hadjiosif N., Perry R.M., Poulter N.S., Osman K., Vatovec S., Harper A., Franklin F.C.H., Franklin-Tong V.E. Identification of the pollen self-incompatibility determinant in Papaver rhoeas. Nature. 2009;459:992–995. doi: 10.1038/nature08027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler M.J., Vatovec S., Franklin-Tong V.E. The pollen S-determinant in Papaver: comparisons with known plant receptors and protein ligand partners. J. Exp. Bot. 2010;61:2015–2025. doi: 10.1093/jxb/erp383. [DOI] [PubMed] [Google Scholar]
- Wilkins K.A., Bancroft J., Bosch M., Ings J., Smirnoff N., Franklin-Tong V.E. Reactive oxygen species and nitric oxide mediate actin reorganization and programmed cell death in the self-incompatibility response of Papaver. Plant Physiol. 2011;156:404–416. doi: 10.1104/pp.110.167510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Liu X., Zhang M.Y., Qi K.J., Jiang X.T., Yao J.L., Zhang S.L., Gu C. Self S-RNase inhibits ABF–LRX signaling to arrest pollen tube growth to achieve self-incompatibility in pear. Plant J. 2023;113:595–609. doi: 10.1111/tpj.16072. [DOI] [PubMed] [Google Scholar]
- Wu M., Kostyun J.L., Moyle L.C. Genome sequence of Jaltomata addresses rapid reproductive trait evolution and enhances comparative genomics in the hyper-diverse Solanaceae. Genome Biol. Evol. 2019;11:335–349. doi: 10.1093/gbe/evy274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Lau K.H., Cao Q., Hamilton J.P., Sun H., Zhou C., Eserman L., Gemenet D.C., Olukolu B.A., Wang H., et al. Genome sequences of two diploid wild relatives of cultivated sweet potato reveal targets for genetic improvement. Nat. Commun. 2018;9:4580. doi: 10.1038/s41467-018-06983-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z., Han F., Hu Y., Xue Y., Fang Z., Yang L., Zhang Y., Liu Y., Li Z., Wang Y., et al. Overcoming cabbage crossing incompatibility by the development and application of self-compatibility-QTL-specific markers and genome-wide background analysis. Front. Plant Sci. 2019;10:189. doi: 10.3389/fpls.2019.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S., Li M., Liu P. Evolution of S-domain receptor-like kinases in land plants and origination of S-locus receptor kinases in Brassicaceae. BMC Evol. Biol. 2013;13:69. doi: 10.1186/1471-2148-13-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Kitashiba H., Nishio T. Generation of Arabidopsis thaliana transformants showing the self-recognition activity of Brassica rapa. Plant J. 2022;111:496–507. doi: 10.1111/tpj.15811. [DOI] [PubMed] [Google Scholar]
- Yang B., Thorogood D., Armstead I., Barth S. How far are we from unravelling self-incompatibility in grasses? New Phytol. 2008;178:740–753. doi: 10.1111/j.1469-8137.2008.02421.x. [DOI] [PubMed] [Google Scholar]
- Yang Q., Meng D., Gu Z., Li W., Chen Q., Li Y., Yuan H., Yu J., Liu C., Li T. Apple S-RNase interacts with an actin-binding protein, MdMVG, to reduce pollen tube growth by inhibiting its actin-severing activity at the early stage of self-pollination induction. Plant J. 2018;95:41–56. doi: 10.1111/tpj.13929. [DOI] [PubMed] [Google Scholar]
- Yang X., Gao S., Guo L., Wang B., Jia Y., Zhou J., Che Y., Jia P., Lin J., Xu T., et al. Three chromosome-scale Papaver genomes reveal punctuated patchwork evolution of the morphinan and noscapine biosynthesis pathway. Nat. Commun. 2021;12:6030. doi: 10.1038/s41467-021-26330-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S., Kobayashi R., Ito T., Wada Y., Takayama S. Homology-based interactions between small RNAs and their targets control dominance hierarchy of male determinant alleles of self-incompatibility in Arabidopsis lyrata. Int. J. Mol. Sci. 2021;22:6990. doi: 10.3390/ijms22136990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S., Wada Y., Kakizaki T., Tarutani Y., Miura-Uno E., Murase K., Fujii S., Hioki T., Shimoda T., Takada Y., et al. A complex dominance hierarchy is controlled by polymorphism of small RNAs and their targets. Nat. Plants. 2016;3:16206–16215. doi: 10.1038/nplants.2016.206. [DOI] [PubMed] [Google Scholar]
- Yasui Y., Mori M., Aii J., Abe T., Matsumoto D., Sato S., Hayashi Y., Ohnishi O., Ota T. S-locus early flowering 3 is exclusively present in the genomes of short-styled buckwheat plants that exhibit heteromorphic self-incompatibility. PLoS One. 2012;7 doi: 10.1371/journal.pone.0031264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye M., Peng Z., Tang D., Yang Z., Li D., Xu Y., Zhang C., Huang S. Generation of self-compatible diploid potato by knockout of S-RNase. Nat. Plants. 2018;4:651–654. doi: 10.1038/s41477-018-0218-6. [DOI] [PubMed] [Google Scholar]
- Zhang L., Huang J., Su S., Wei X., Yang L., Zhao H., Yu J., Wang J., Hui J., Hao S., et al. FERONIA receptor kinase-regulated reactive oxygen species mediate self-incompatibility in Brassica rapa. Curr. Biol. 2021;31:3004–3016.e4. doi: 10.1016/j.cub.2021.04.060. [DOI] [PubMed] [Google Scholar]
- Zhang T., Zhou G., Goring D.R., Liang X., Macgregor S., Dai C., Wen J., Yi B., Shen J., Tu J., et al. Generation of transgenic self-incompatible Arabidopsis thaliana shows a genus-specific preference for self-incompatibility genes. Plants. 2019;8:570. doi: 10.3390/plants8120570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Ran Y., Nagy I., Lenk I., Qiu J.L., Asp T., Jensen C.S., Gao C. Targeted mutagenesis in ryegrass (Lolium spp.) using the CRISPR/Cas9 system. Plant Biotechnol. J. 2020;18:1854–1856. doi: 10.1111/pbi.13359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang S., Liu H., Fu B., Li L., Xie M., Song Y., Li X., Cai J., Wan W., et al. Genome and comparative transcriptomics of African wild rice Oryza longistaminata provide insights into molecular mechanism of rhizomatousness and self-incompatibility. Mol. Plant. 2015;8:1683–1686. doi: 10.1016/j.molp.2015.08.006. [DOI] [PubMed] [Google Scholar]
- Zhao H., Song Y., Li J., Zhang Y., Huang H., Li Q., Zhang Y., Xue Y. Primary restriction of S-RNase cytotoxicity by a stepwise ubiquitination and degradation pathway in Petunia hybrida. New Phytol. 2021;231:1249–1264. doi: 10.1111/nph.17438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Zhang Y., Zhang H., Song Y., Zhao F., Zhang Y., Zhu S., Zhang H., Zhou Z., Guo H., et al. Origin, loss, and regain of self-incompatibility in angiosperms. Plant Cell. 2022;34:579–596. doi: 10.1093/plcell/koab266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li D., Wang G., Wang F., Kunjal M., Joldersma D., Liu Z. Application and future perspective of CRISPR/Cas9 genome editing in fruit crops. J. Integr. Plant Biol. 2020;62:269–286. doi: 10.1111/jipb.12793. [DOI] [PMC free article] [PubMed] [Google Scholar]