Abstract
Global gene expression patterns of Bacillus subtilis in response to subinhibitory concentrations of protein synthesis inhibitors (chloramphenicol, erythromycin, and gentamicin) were studied by DNA microarray analysis. B. subtilis cultures were treated with subinhibitory concentrations of protein synthesis inhibitors for 5, 15, 30, and 60 min, and transcriptional patterns were measured throughout the time course. Three major classes of genes were affected by the protein synthesis inhibitors: genes encoding transport/binding proteins, genes involved in protein synthesis, and genes involved in the metabolism of carbohydrates and related molecules. Similar expression patterns for a few classes of genes were observed due to treatment with chloramphenicol (0.4× MIC) or erythromycin (0.5× MIC), whereas expression patterns of gentamicin-treated cells were distinct. Expression of genes involved in metabolism of amino acids was altered by treatment with chloramphenicol and erythromycin but not by treatment with gentamicin. Heat shock genes were induced by gentamicin but repressed by chloramphenicol. Other genes induced by the protein synthesis inhibitors included the yheIH operon encoding ABC transporter-like proteins, with similarity to multidrug efflux proteins, and the ysbAB operon encoding homologs of LrgAB that function to inhibit cell wall cleavage (murein hydrolase activity) and convey penicillin tolerance in Staphylococcus aureus.
Over the past decade, bacterial resistance to antibiotics has risen dramatically and “superbugs” resistant to most or all available agents have appeared in hospitals. Thus, there is an urgent need to discover and develop novel classes of antibiotics (11). The availability of the complete genome sequence of many pathogenic microbes provides information on potential drug targets. Approaches such as bioinformatics or whole-genome transcriptional analysis are now used to search for drug targets (reviewed in references 34 and 37). Genome-wide expression profiling generated by DNA microarrays can be used to observe differential gene expression in response to environmental stimuli or genetic alterations. Studies of transcriptional profiles of bacteria treated with an inhibitor can provide valuable information useful for both pathway characterization and for determination of the mechanism of the inhibitor (14, 42). In addition, differential expression of essential genes or previously uncharacterized genes caused by an inhibitor may identify the corresponding gene products as potential drug targets (22). Recently, several studies have been performed to study transcriptional or translational profiles of microorganisms subjected to one class of antibiotics (2, 8, 14, 23, 28, 40), or several classes of antibiotics (3, 6, 12, 16-18, 33), to generate databases that can be used to distinguish modes of actions of antimicrobial agents.
In the present study, we used DNA microarray analysis to determine the global gene expression pattern of Bacillus subtilis in response to subinhibitory concentrations of protein synthesis inhibitors: chloramphenicol, erythromycin, and gentamicin. Chloramphenicol is known to block peptidyl transferase activity by hindering the binding of tRNA to the A site (24, 32). Erythromycin, a macrolide, is believed to block the tunnel that channels the nascent peptides away from the peptidyl transferase center, thereby preventing movement and release of the nascent peptide (32). Gentamicin is an aminoglycoside that binds to a conserved sequence of rRNA that is near the site of codon-anticodon recognition in the aminoacyl-tRNA site (A site) of the 30S ribosomal subunit. This interaction in turn interferes with proofreading steps that ensure translational fidelity (9, 44). Both chloramphenicol and erythromycin target the 50S ribosomal subunit and inhibit translation elongation, whereas gentamicin targets the 30S ribosomal subunit and affects translational accuracy. We would like to generate transcriptional profiles of cells treated with protein synthesis inhibitors or other classes of antibiotics to create a library of signature responses for the determination of modes of action of new antimicrobial compounds. Microbial transcriptional or translational profiles after treatment with antibiotics or drugs have been determined recently, and statistical or mathematical tools can be applied to predict modes of actions of different compounds (3, 6, 17). Genes highly induced or repressed by protein synthesis inhibitors can potentially be used as signature genes to identify new drugs that inhibit translation. B. subtilis is an ideal microorganism for microarray analysis because its genome has been sequenced (20). Gene expression responses to higher concentrations of inhibitors have been shown to cause a broader effect on cellular processes, thereby giving much more complex response patterns (14, 34). We applied subinhibitory concentrations (concentrations that cause little growth inhibition) of protein synthesis inhibitors with the goal of observing antibiotic-specific primary expression profiles and that growth-inhibition-related secondary responses would be underrepresented. Multiple concentrations of inhibitors were applied so that dose-specific effects could be examined. Furthermore, a time course of 5 to 60 min was applied so that the kinetics of the transcription response over time could be studied. Several common major functional classes of genes were affected by the protein synthesis inhibitors used. These classes include transport/binding proteins and lipoproteins, metabolism of carbohydrates and related molecules, and protein synthesis.
MATERIALS AND METHODS
Bacterial strain and media.
B. subtilis 1A757, a prototroph of B. subtilis 168, was obtained from Bacillus Genetic Stock Center (Columbus, Ohio). Mueller-Hinton medium, purchased from Benton Dickson (Sparks, MD), was used to grow B. subtilis cultures.
Bacterial growth conditions.
Standard broth microdilution assays were performed to determine the MICs of B. subtilis against chloramphenicol, erythromycin, or gentamicin (27). Overnight cultures of B. subtilis 1A757 were diluted 1:100 in Mueller-Hinton medium and incubated at 37°C with shaking at 200 rpm. When cultures reached an optical density at 600 nm (OD600) of 0.2, chloramphenicol at 0.05× (0.2 μg/ml), 0.25× (1 μg/ml), or 0.4× (1.6 μg/ml) MIC; erythromycin at 0.1× (0.0125 μg/ml), 0.25× (0.03125 μg/ml), or 0.5× (0.0625 μg/ml) MIC; or gentamicin at 0.1× (0.0125 μg/ml), 0.25× (0.03125 μg/ml), or 0.4× (0.05 μg/ml) MIC was added to the cultures. Untreated cultures were used as controls. Samples were collected at 5, 15, 30, and 60 min after treatment for subsequent RNA isolation. The samples were processed immediately by centrifugation, and pellets were stored at −80°C. Growth and viability were monitored for at least 2.5 h posttreatment by measuring both the OD600 and CFU at several time points.
RNA isolation, probe preparation, and hybridization for DNA microarrays.
RNA samples were extracted independently from two experiments, and isolation was performed with the FastRNA Blue Kit (Qbiogene, Carlsbad, CA) according to the manufacturer's instructions. Fluorescent probes were prepared by reverse transcription (RT) of 25 μg of total RNA to incorporate aminoallyl-dUTP into first strand cDNA. The amino-cDNA was then labeled by direct coupling to either Cy3 (cDNA from untreated sample) or Cy5 (cDNA from antibiotic-treated sample) monofunctional reactive dyes (Amersham Biosciences, Piscataway, NJ). DNA microarrays consisting of PCR-amplified B. subtilis 168 open reading frames (ORFs) or ORF-specific oligonucleotides (60-mers) at a 10 μM concentration in 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate; Compugen, Jamesburg, NJ) were also used as described previously (4, 5). All labeled cDNA probes were hybridized to oligonucleotide slides with the exception of one chloramphenicol-treated experiment that was hybridized to slides spotted with PCR products. Hybridizations were performed as previously described (4, 5). The corresponding cDNA samples for treatment and control were mixed and hybridized to the microarray slides in replicates of a total of six genomes. The slide images were scanned and edited by using an Axon 4000B scanner (Axon Instruments, Union City, CA).
DNA microarray data analysis and clustering analysis.
The fluorescent signal ratios (Cy5/Cy3) were subjected to Lowess normalization with background correction in the GeneSpring software (Silicon Genetics, Redwood City, CA). The normalized data was then analyzed by a statistical technique, significance analysis for microarrays (SAM), to identify significantly up- or downregulated genes with the exclusion of invariant genes (39). SAM assigns a score to each gene on the basis of its change in expression relative to the standard deviation of repeated measurements for that gene. A “q value” assigned to each gene corresponds to the lowest false discovery rate at which the gene is called significant. The “one class response” was used, and the number of falsely significant genes was set to be less than one in SAM. The differentially expressed genes identified by SAM were further filtered to identify genes whose ratios of expression in treated versus untreated control were more than 1.5 or less than 0.67, indicating at least a 1.5-fold change of expression. Clustering analysis was performed by using best k-means provided in GeneSpring (reviewed in reference 36). We used the “best k-means” script in GeneSpring to determine the optimal number of the clusters for each analysis. The data used for clustering analysis was from treatment with each of the three concentrations of antibiotics over the time series (5, 15, 30, and 60 min). Genes used for clustering had to meet the following requirements: (i) genes had to be determined by SAM to be significantly variant; (ii) gene expression levels has to be above or below the cutoff range (1.5-fold); and (iii) the amount of up- or downregulation had to be consistent in two independent experiments and at least one condition. Expression ratios used for clustering analysis were average fluorescence intensity ratios from two independent experiments, each consisting of data from six microarray replicates (a total of 12 datum points per gene).
Real-time RT-PCR.
Real-time RT-PCR was performed on an ABI Prism 7700 Sequence Detector (Applied Biosystems, Foster City, CA) with the SYBR Green detection method (Applied Biosystems, Foster City, CA). Primers used for RT-PCR were as follows: rplF, TCCTGAAGAAGGCATCGAAATC and CGCGGATGTTAGCAGCAATAG; rbsB, CGGCTCAGGATTCCATAACATC and GCAGCAGGTTTTCCATGACAGT; groES, GGTGATCGCGTTGTCATTGA and TGCCACGATTTTGCCTTCTT; clpP, TGGCGATCTATGATACCATGCA and AGCGCATAGCGTTTGCCTT; cspB, TTCTCTGCTATTCAAGGCGAAG and AACGTTAGCAGCTTGTGGTCC; cspD, AGTTGAAGGCGGAGACGATGT and GGTCCACGATTACCTTCGACAA; yecA, CTTGTTTTTGGCAGCGTCTTG and GCGATGCAATTTCTCCATTCTC; and yvaN, CCAAACCTGCCACCATCAA and CCTGGCTTCATGCACTTCTTC.
The RT-PCRs contained serial dilutions of RNA templates, 500 nM concentrations of each pair of primers, 1× SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA), and 0.24 U of Moloney murine leukemia virus reverse transcriptase (Invitrogen, Carlsbad, CA)/μl.Reverse transcription was performed at 50°C for 30 min, followed by inactivation of reverse transcriptase and activation of AmpliTaq DNA polymerase at 95°C for 15 min. For each dilution triplicate reactions were prepared. Forty thermocycles were performed as follows: 94°C for 15 s, 55°C for 30 s, and 72°C for 30 s. The RNA samples were also subjected to RT-PCR amplification with the yecA or yvaN primers. The amount of PCR product at each cycle was recorded by measuring fluorescence generated by binding of the SYBR dye to double-stranded DNA. The amplification plot generated with untreated (time zero) samples was used as the reference curve to standardize amplification results generated from the untreated and antibiotic-treated samples. The results were then normalized to the data generated from the yecA or yvaN amplification for chloramphenicol and erythromycin or gentamicin-treated samples, respectively.
RESULTS
Growth of B. subtilis in the presence of subinhibitory concentrations of chloramphenicol, erythromycin, and gentamicin.
The MICs of B. subtilis against chloramphenicol, erythromycin, and gentamicin were determined to be 4, 0.125, and 0.125 μg/ml, respectively. B. subtilis cultures were treated with 0.05×, 0.25×, or 0.4× MIC of chloramphenicol; 0.1×, 0.25×, or 0.5× MIC of erythromycin; or 0.1×, 0.25×, or 0.4× MIC of gentamicin. The concentrations were selected based on their subinhibitory phenotypes after antibiotic treatment measured by OD600 and CFU (data not shown). When chloramphenicol was applied to B. subtilis cultures, all treatments caused growth inhibitory effect after 60 min. When erythromycin was applied, 0.1× and 0.25× MIC treatments caused inhibitory effect after 60 min, whereas the 0.5× MIC-treated cultures showed growth inhibition as early as 15 min. Finally, when gentamicin was applied, 0.4× MIC treatment led to inhibition as early as 15 min, and 0.1× MIC- and 0.25× MIC-treated cultures displayed growth inhibition after 60 min. In summary, most conditions studied did not cause strong growth inhibition of B. subtilis.
Microarray experimental design and data analysis.
B. subtilis cultures were treated with subinhibitory concentrations of chloramphenicol, erythromycin, or gentamicin. Untreated cultures were used as control. Total RNA was extracted from each culture 5, 15, 30, and 60 min after treatment. Subinhibitory concentrations of inhibitors were used to avoid secondary responses caused by high concentrations of antibiotics. Multiple concentrations of antibiotics were tested to study dose-dependent effects. Furthermore, a time course of 5 to 60 min was applied so that the kinetics of the transcription response to time could be determined. RNA samples were used to prepare cDNA for subsequent Cy dye labeling and hybridization to microarrays. The control (no treatment) samples were labeled with Cy3, and treated samples were labeled with Cy5. The corresponding samples for each time point and treatment were mixed and hybridized to multiple microarray slides equivalent to at least six genomes.
The fluorescence intensity values, calculated by dividing the Cy5 signal by the Cy3 signal, were quantified, normalized, and assessed by SAM software to identify genes whose expression was significantly altered by treatment with antibiotics. The false discovery rate at which the gene is called significant was less than 0.5% for genes accepted to be significant with a few exceptions. The genes determined by SAM to be significant were then filtered to identify genes whose Cy5/Cy3 ratios were more than 1.5 or less than 0.67 (indicating more than a 1.5-fold change of expression).
Overview of transcriptional profiles of chloramphenicol-, erythromycin-, or gentamicin-treated B. subtilis cells.
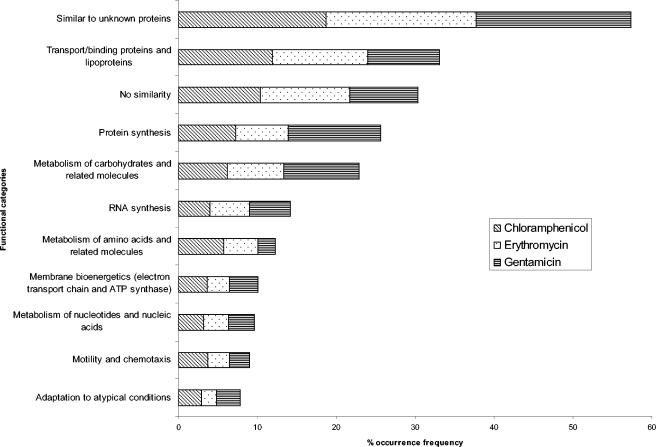
Treatment of B. subtilis with chloramphenicol, erythromycin, or gentamicin revealed a total of 856, 1,233, or 462 genes, respectively, up- or downregulated by at least 1.5-fold in two independent experiments and in at least one condition. Approximately 20% of the differentially expressed genes encode proteins with similarity to those with unknown functions in other organisms, whereas ca. 10% encode proteins with no homologs. Analysis of genes with known functions revealed that all three antibiotics primarily affected the transcription of genes from the following three categories: transport/binding protein genes, genes involved in protein synthesis, and genes involved in metabolism of carbohydrates and related molecules (Fig. 1). In addition, for all three antibiotics the frequencies of occurrence of genes in other functional categories whose expression was affected were similar. We also found subtle differences in abundance of genes in the major categories for chloramphenicol and erythromycin versus gentamicin. For example, there was a larger percentage of genes involved in transport/binding proteins and metabolism of amino acids for chloramphenicol- or erythromycin-treated cultures than for gentamicin-treated cultures. In contrast, larger percentages of genes involved in protein synthesis and metabolism of carbohydrates were affected by gentamicin than by chloramphenicol or erythromycin.
FIG. 1.
Distribution of functions of genes whose expression levels were affected by treatment with antibiotics. The top 11 functional categories affected by the protein synthesis inhibitors are illustrated. The category of each gene was assigned by SubtiList (http://genolist.pasteur.fr/SubtiList/), a website created by B. subtilis Genome Sequencing Project. The percent occurrence frequency is the percentage of the total number of genes in each functional category affected by each protein synthesis inhibitor.
Since chloramphenicol, erythromycin, and gentamicin primarily affected transcription of genes from the same three major functional categories, we studied expression patterns of genes from each of these functional groups. Average expression levels of category genes grouped by k-means clustering at each concentration were studied. Expression ratios of representative genes in each major functional category affected by the highest concentration of antibiotics are listed in Table 1. The representative genes shown in Table 1 were among those considered significant by SAM at a given time and dose in both experiments and were consistently regulated (either up- or downregulated) in both experiments. Expression patterns were similar for a few gene classes when cells were treated with the highest concentration tested for chloramphenicol (0.4× MIC) or erythromycin (0.5× MIC). When treated with the 0.4× or 0.5× MIC of chloramphenicol or erythromycin, respectively, many genes encoding transport/binding proteins were upregulated at 15 min. In addition, genes involved in protein synthesis showed a similar expression profile peaking at 15 min, followed by a gradual decrease after 30 min. Genes involved in the metabolism of carbohydrates were upregulated at 30 min by treatment with chloramphenicol and downregulated at 60 min by treatment with erythromycin. Gene expression patterns observed in gentamicin-treated cultures differed from those due to chloramphenicol or erythromycin treatment. When treated with gentamicin, many genes encoding transport/binding proteins or genes involved in metabolism of carbohydrates were downregulated at 15 min, whereas some protein synthesis genes were upregulated at 5 and 30 min and downregulated at 15 and 60 min. The similarity of expression patterns affected by highest tested concentrations of chloramphenicol and erythromycin may reflect the similar modes of action of the two antibiotics, since both antibiotics target the 50S ribosomal subunit.
TABLE 1.
Expression ratios of genes involved in transport/binding, metabolism of carbohydrates, protein synthesis, metabolism of amino acids, purine/pyrimidine synthesis, and heat shock proteins in response to the highest tested concentrations of chloramphenicol, erythromycin, or gentamicin
| Function, antibiotic, and genea | Product or function | Expression ratiosb at:
|
|||
|---|---|---|---|---|---|
| 5 min | 15 min | 30 min | 60 min | ||
| Transport/binding | |||||
| Cm | |||||
| pbuG | Hypoxanthine/guanine permease | 1.7 | 3.2 | 1.0 | 1.5 |
| pbuX | Xanthine permease | 1.4 | 7.2 | 1.0 | 1.2 |
| pyrP | Uracil permease | 1.8 | 7.6 | 1.9 | 3.0 |
| yhcA | Similar to multidrug resistance protein | 1.1 | 2.4 | 1.8 | 2.1 |
| yxjA | Similar to pyrimidine nucleoside transport | 1.1 | 3.1 | 1.5 | 2.1 |
| Em | |||||
| pbuG | Hypoxanthine/guanine permease | 1.1 | 5.7 | 1.1 | 0.1 |
| pbuX | Xanthine permease | 1.0 | 3.4 | 1.7 | 0.1 |
| opuAB | Glycine betaine ABC transporter (permease) | 1.3 | 3.7 | 1.3 | 0.4 |
| ycgO | Similar to proline permease | 0.8 | 2.4 | 1.6 | 0.6 |
| yxjA | Similar to pyrimidine nucleoside transport | 1.6 | 10.0 | 2.1 | 2.4 |
| Gm | |||||
| dppB | Dipeptide ABC transporter (permease) | 0.6 | 0.2 | 1.2 | 1.8 |
| malP | Phosphotransferase system maltose-specific enzyme IICB component | 0.8 | 0.5 | 0.8 | 1.4 |
| nagP | Phosphotransferase system N-acetylglucosamine-specific enzyme IICB component | 0.8 | 0.4 | 1.1 | 1.7 |
| ptsI | Phosphotransferase system enzyme I | 1.5 | 0.6 | 1.2 | 1.0 |
| yclN | Similar to ferrichrome ABC transporter (permease) | 0.8 | 0.5 | 0.6 | 0.8 |
| Metabolism of carbohydrates | |||||
| Cm | |||||
| mleA | Malolactic enzyme | 1.3 | 6.1 | 28.4 | 0.5 |
| mtlD | Mannitol-1-phosphate dehydrogenase | 0.8 | 1.3 | 1.8 | 2.5 |
| nagB | N-Acetylglucosamine-6-phosphate isomerase | 1.0 | 1.8 | 4.7 | 5.3 |
| pckA | Phosphoenolpyruvate carboxykinase | 1.0 | 1.3 | 1.5 | 1.3 |
| yvfV | Similar to glycolate oxidase | 1.3 | 3.4 | 4.7 | 0.4 |
| Em | |||||
| citB | Aconitate hydratase (aconitase) | 2.7 | 1.0 | 1.2 | 0.4 |
| iolB | Myoinositol catabolism | 1.0 | 0.8 | 1.1 | 0.3 |
| iolD | Myoinositol catabolism | 1.0 | 0.9 | 0.9 | 0.4 |
| iolE | Myoinositol catabolism | 1.1 | 0.6 | 1.4 | 0.3 |
| lacA | β-Galactosidase | 1.0 | 0.9 | 1.1 | 0.4 |
| Gm | |||||
| citB | Aconitate hydratase (aconitase) | 1.3 | 0.6 | 0.7 | 1.4 |
| icd | Isocitrate dehydrogenase | 1.3 | 0.5 | 0.7 | 1.2 |
| mdh | Malate dehydrogenase | 1.5 | 0.5 | 0.6 | 1.2 |
| pckA | Phosphoenolpyruvate carboxykinase | 1.5 | 0.7 | 0.6 | 0.9 |
| pfkA | 6-Phosphofructokinase | 1.9 | 0.4 | 0.7 | 2.9 |
| Protein synthesis | |||||
| Cm | |||||
| infC | Initiation factor IF-3 | 1.2 | 1.9 | 2.1 | 1.5 |
| rplA | Ribosomal protein L1 (BL1) | 1.3 | 3.3 | 1.7 | 1.6 |
| rplK | Ribosomal protein L11 (BL11) | 1.6 | 3.4 | 1.5 | 2.1 |
| rplT | Ribosomal protein L20 | 1.1 | 3.4 | 1.6 | 1.9 |
| rplU | Ribosomal protein L21 (BL20) | 1.5 | 2.0 | 1.3 | 1.5 |
| Em | |||||
| rplC | Ribosomal protein L3 (BL3) | 1.4 | 3.1 | 1.6 | 0.3 |
| rplU | Ribosomal protein L21 (BL20) | 1.3 | 4.3 | 1.7 | 0.5 |
| rpsD | Ribosomal protein S4 (BS4) | 1.3 | 5.9 | 1.2 | 0.5 |
| rpsF | Ribosomal protein S6 (BS9) | 1.9 | 9.4 | 3.5 | 2.9 |
| rpsL | Ribosomal protein S12 (BS12) | 1.5 | 2.8 | 1.3 | 0.2 |
| Gm | |||||
| rplB | Ribosomal protein L2 (BL2) | 1.7 | 0.6 | 1.5 | 0.7 |
| rplJ | Ribosomal protein L10 (BL5) | 2.2 | 0.5 | 2.4 | 0.4 |
| rplQ | Ribosomal protein L17 (BL15) | 1.3 | 0.4 | 1.5 | 0.6 |
| rplR | Ribosomal protein L18 | 1.8 | 0.5 | 1.6 | 0.7 |
| rpsK | Ribosomal protein S11 (BS11) | 1.7 | 0.4 | 1.6 | 0.7 |
| Metabolism of amino acids | |||||
| Cm | |||||
| glnA | Required for transduction of the nitrogen regulation signal to GlnR and TnrA | 0.9 | 0.9 | 0.7 | 0.4 |
| glyA | Glycine requirement; indirect activation of KinB | 0.9 | 1.0 | 0.6 | 0.5 |
| goxB | Oxidation of sarcosine (N-methylglycine), N-ethylglycine, and glycine; lower activities on d-alanine, d-valine, and d-proline | 1.1 | 0.9 | 0.7 | 0.2 |
| pepT | Putative; Zn2+-dependent metalloenzyme; N-terminal region stabilizes Zn2+-binding | 0.9 | 0.4 | 0.6 | 0.4 |
| rocA | Sigma-L-dependent; positively regulated by RocR and AhrC (ARG recognition site located immediately upstream of the transcriptional start point) | 1.0 | 1.7 | 2.7 | 0.6 |
| Em | |||||
| glyA | Serine hydroxymethyltransferase | 1.1 | 1.0 | 1.4 | 0.4 |
| hisD | Histidinol dehydrogenase | 1.0 | 1.4 | 1.6 | 0.5 |
| rocA | Pyrroline-5 carboxylate dehydrogenase | 1.5 | 0.9 | 1.1 | 0.2 |
| rocD | Ornithine aminotransferase | 1.0 | 1.2 | 1.3 | 0.3 |
| rocG | Glutamate dehydrogenase | 0.9 | 1.0 | 1.4 | 0.3 |
| Purine and pyrimidine biosynthesis | |||||
| Cm | |||||
| purD | Phosphoribosylglycinamide synthetase | 1.1 | 2.2 | 1.7 | 1.4 |
| pyrB | Aspartate carbamoyltransferase | 1.1 | 1.8 | 1.2 | 1.1 |
| pyrF | Orotidine 5′-phosphate decarboxylase | 1.0 | 1.8 | 1.3 | 1.8 |
| pyrG | CTP synthetase | 1.8 | 7.6 | 1.9 | 3.0 |
| pyrP | Uracil permease | 0.8 | 1.7 | 0.9 | 0.8 |
| Em | |||||
| purC | Phosphoribosylaminoimidazole succinocarboxamide synthetase | 0.9 | 1.2 | 1.2 | 0.5 |
| purE | Phosphoribosylaminoimidazole carboxylase I | 1.0 | 2.1 | 1.2 | 0.3 |
| purN | Phosphoribosylglycinamide formyltransferase | 1.2 | 1.6 | 1.8 | 0.3 |
| pyrAB | Carbamoyl-phosphate synthetase (catalytic subunit) | 1.1 | 1.8 | 1.7 | 0.5 |
| pyrD | Dihydroorotate dehydrogenase (catalytic subunit) | 1.0 | 1.6 | 1.4 | 0.3 |
| Heat shock proteins | |||||
| Cm | |||||
| clpQ | Two-component ATP-dependent protease (N-terminal serine protease) | 0.8 | 0.9 | 0.6 | 1.0 |
| clpX | ATP-dependent Clp protease ATP-binding subunit (class III heat shock protein) | 1.2 | 0.6 | 0.6 | 0.8 |
| dnaK | Class I heat shock protein (molecular chaperone) | 1.2 | 0.7 | 1.0 | 0.7 |
| groES | Class I heat shock protein (chaperonin) | 1.1 | 0.2 | 0.3 | 0.3 |
| nadE | NH3-dependent NAD+ synthetase; strongly induced in response to heat, ethanol, and salt stress or after starvation for glucose | 1.0 | 1.0 | 0.7 | 0.7 |
| Gm | |||||
| clpP | ATP-dependent Clp protease proteolytic subunit (class III heat shock protein) | ||||
| 0.1× MIC | 1.4 | 0.6 | 0.7 | 2.1 | |
| 0.25× MIC | 1.3 | 0.8 | 1.1 | 3.7 | |
| 0.4× MIC | 1.4 | 1.4 | 1.2 | 4.5 | |
| clpX | ATP-dependent Clp protease ATP-binding subunit (class III heat shock protein) | 1.4 | 0.6 | 0.9 | 1.5 |
| ykrL | Similar to heat shock protein | 0.8 | 1.3 | 1.1 | 1.5 |
Cm, chloramphenicol; Em, erythromycin; Gm, gentamicin. Concentrations of 0.4×, 0.5×, and 0.4× MIC, respectively, were used unless indicated otherwise.
Values shown are the relative transcript levels of treated samples over untreated samples, with values more than one indicating induction and values less than one indicating repression in the antibiotic-treated samples. Bold numbers represent the average expression ratios from two experiments considered significant by SAM and were consistently up- or downregulated in both experiments. The “q values” range from 0.0003 to 0.01, 0.0002 to 0.0017, and 0.0004 to 0.0025 under the conditions shown for chloramphenicol, erythromycin, and gentamicin, respectively.
Transcriptional response to different concentrations of antibiotics.
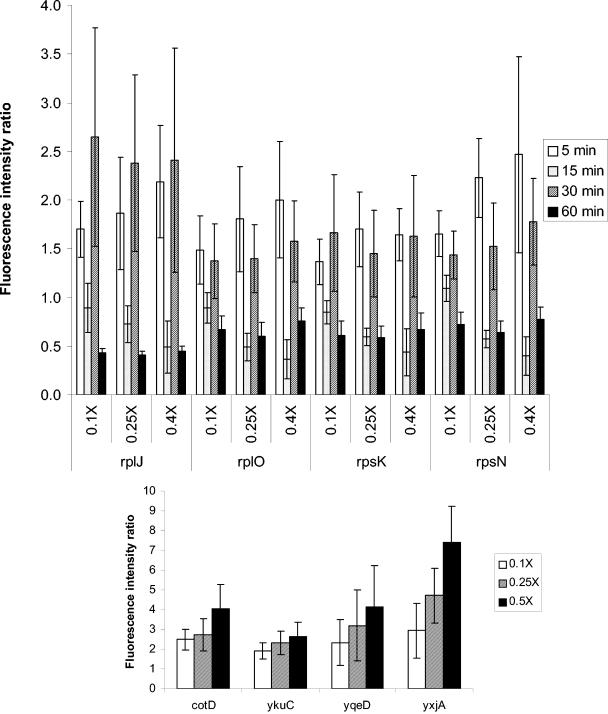
Erythromycin and gentamicin treatment usually generated expression profiles that were similar regardless of dose. However, expression patterns due to chloramphenicol treatment usually showed a dose-dependent difference (data not shown). Occasionally, more than one trend of expression patterns was observed, such as expression of genes encoding transport proteins or genes involved in protein synthesis upon treatment with chloramphenicol, and expression patterns of protein synthesis genes due to treatment with erythromycin. Expression patterns of four genes involved in protein synthesis due to treatment with three different concentrations of gentamicin are shown in Fig. 2, top panel. Similar expression patterns (upregulated at 5 and 30 min and downregulated at 15 and 60 min) were observed regardless of concentrations in both experiments, a finding consistent with the patterns shown in Table 1. Moreover, dose-dependent effects were observed for rplJ, rplO, and rpsN especially at the 5- and 15-min time points. Upon treatment with erythromycin, many genes consistently upregulated at 15 min in all three treated concentrations were identified. Representative genes (cotD, ykuC, yqeD, and yxjA) that displayed dose dependence at 15 min are depicted in Fig. 2, bottom panel. Similarly, they were repressed or invariant by 60 min, except for yxjA, which remained upregulated throughout treatment (data not shown).
FIG. 2.
Average gene expression ratios upon treatment with gentamicin or erythromycin. (Top panel) Average expression ratios of rplJ, rplO, rpsK, and rpsN genes upon treatment with 0.1×, 0.25×, or 0.4× MIC of gentamicin for 5, 15, 30, or 60 min. (Bottom panel) Dose-dependent expression ratios of cotD (spore coat protein), ykuC (similar to macrolide-efflux protein), yqeD (unknown), and yxjA (similar to pyrimidine nucleoside transport) genes upon treatment with 0.1×, 0.25×, or 0.5× MIC of erythromycin for 15 min. Values of more than one indicate induction, and values of less than one indicate repression. The error bars depict the standard deviation from 12 datum points in two independent experiments.
Validation of gene expression by real-time RT-PCR analysis.
The real-time RT-PCR analysis was chosen as an independent method to validate microarray results since it measures product accumulation during the linear phase of the PCR and is an accurate and reproducible approach to gene quantification (30, 31). We quantified transcripts of genes involved in protein synthesis (rplF, ribosomal protein L6), transport proteins (rbsB, ribose ABC transporters), heat shock (groES, class I heat shock protein; clpP, class III heat shock protein), and cold shock (cspB and cspD, cold shock proteins) due to treatment with chloramphenicol, erythromycin, or gentamicin in at least one condition. Relative quantities of each gene's transcription level in untreated and treated samples were determined and normalized by using the internal control genes, which were invariant on treatment with chloramphenicol and erythromycin (yecA, encoding a unknown protein that is similar to amino acid permease) or gentamicin (yvaN gene, encoding a unknown protein that is similar to immunity repressor protein). Finally, the ratios from treated/untreated samples were determined. The results are shown in Table 2. All genes were validated except for groES under one condition. Consistent with previous observations, differences in the magnitude of expression ratios between microarray analysis and real-time RT-PCR were observed (30).
TABLE 2.
Validation of microarray-based expression profiles by real-time RT-PCR
| Gene | Product/function | Antibiotica | Condition (MIC, time [min]) | Relative expression levelb at:
|
Validationc | |
|---|---|---|---|---|---|---|
| Microarray | Real-time PCR | |||||
| rplF | Ribosomal protein L6 | Cm | 0.25×, 15 | 3.8 | 7.3 | Yes |
| Em | 0.25×, 15 | 1.5 | 3.9 | Yes | ||
| Em | 0.25×, 60 | 0.6 | 0.7 | Yes | ||
| Gm | 0.25×, 5 | 2.2 | 4.3 | Yes | ||
| Gm | 0.25×, 60 | 0.5 | 0.3 | Yes | ||
| rbsB | Ribose ABC transporters; ribose-binding protein | Cm | 0.25×, 15 | 3.8 | 4.4 | Yes |
| Em | 0.25×, 15 | 0.5 | 0.3 | Yes | ||
| Em | 0.25×, 60 | 1.7 | 1.6 | Yes | ||
| Gm | 0.25×, 15 | 0.4 | 0.4 | Yes | ||
| groES | Class I heat shock protein | Cm | 0.25×, 15 | 0.3 | 0.3 | Yes |
| Cm | 0.25×, 60 | 0.3 | 0.4 | Yes | ||
| Em | 0.25×, 15 | 0.5 | 0.3 | Yes | ||
| Em | 0.25×, 60 | 1.6 | 0.1 | No | ||
| Gm | 0.25×, 60 | 3.9 | 9.8 | Yes | ||
| clpP | Class III heat shock protein | Cm | 0.25×, 15 | 0.6 | 0.6 | Yes |
| Gm | 0.1×, 60 | 2.0 | 2.3 | Yes | ||
| Gm | 0.25×, 60 | 3.8 | 4.0 | Yes | ||
| Gm | 0.4×, 60 | 4.3 | 4.7 | Yes | ||
| cspB | Cold shock protein | Cm | 0.4×, 60 | 3.4 | 6.5 | Yes |
| cspD | Cold shock protein | Em | 0.25×, 15 | 2.9 | 4.4 | Yes |
Cm, chloramphenicol; Em, erythromycin; Gm, gentamicin.
Values shown are the relative transcript levels of treated sample over untreated sample. The same RNA samples were used for both microarray and RT-PCR experiments. Values more than one indicate induction and values less than one indicate repression in the antibiotic-treated samples.
“Yes” indicates validation of microarray data of up- or downregulation.
Transcription responses specific to erythromycin, chloramphenicol, and/or gentamicin.
We examined additional functional category genes that showed consistent regulation patterns with treatment by chloramphenicol, erythromycin, or gentamicin. Expression of genes involved in metabolism of amino acids was consistently affected by erythromycin and chloramphenicol, sometimes in a dose-dependent manner when treated with erythromycin. Genes involved in the metabolism of amino acids, including glnA, glyA, goxB, pepT, and rocA, were downregulated at 60 min at higher concentrations of chloramphenicol, whereas glyA, hisD, rocA, rocD, and rocG were downregulated at 60 min by treatment with 0.5× erythromycin (Table 1).
Expression of genes involved in purine and pyrimidine synthesis was altered upon treatment with either chloramphenicol or erythromycin. Genes involved in purine and pyrimidine synthesis were upregulated at 15 min upon treatment with chloramphenicol, whereas expression was downregulated at 60 min with erythromycin treatment at all concentrations. Expression levels of representative genes involved in purine and pyrimidine biosynthesis are listed in Table 1. We found that many genes involved in purine/pyrimidine biosynthesis were regulated similarly to protein synthesis genes when treated with 0.4× MIC of chloramphenicol (Table 1). The coregulation of genes involved in purine/pyrimidine biosynthesis and those in protein synthesis was also observed in B. subtilis cultures treated with 0.5× MIC of cephalothin (M. Maranta and D. S. Yaver, unpublished data).
Heat shock genes were upregulated by gentamicin, especially at later time points, in all three concentrations. Notably, a dose-dependent regulatory effect was observed for the expression of clpP at 60 min (Tables 1 and 2). Furthermore, an upregulation in the expression of heat shock genes was also observed due to treatment with streptomycin (another aminoglycoside, data not shown). In contrast, expression of heat shock genes was repressed by treatment with chloramphenicol (Tables 1 and 2), with maximum repression observed after 15 min due to treatment with 0.25× or 0.4× MIC. Heat shock genes appeared to be sparingly affected by treatment with erythromycin (data not shown). Finally, the expression of ykuC, which is similar to macrolide efflux protein, was induced by erythromycin at 15 min in a dose-dependent manner (Fig. 2, bottom panel).
Genes most highly induced with chloramphenicol, erythromycin, or gentamicin treatment.
We analyzed genes that were highly induced due to antibiotic treatment at any concentration or time point. Genes highly induced by erythromycin included dctP, dppB, rpsF, ycnB, yonS, ysbA, ysbB, ytiP, and yvsH. These genes were usually upregulated at 15 or 60 min, with dctP most highly induced. Genes highly induced by chloramphenicol included gapB, mcpB, rbsB, yheH, yheI, yrzI, ysbA, and ysbB. Expression of these genes was upregulated after 15 min with treatment at various concentrations. Interestingly, yheI and yheH form an operon and encode ABC transporter-like proteins with similarity to multidrug efflux proteins (29). Genes highly induced by gentamicin include clpP, ysbA, ysbB, and yxiE. These genes were highly expressed at 60 min posttreatment. Expression of ysbA and ysbB were consistently highly induced by all three antibiotics tested and are located in an operon. They are currently annotated in the SubtiList website (http://genolist.pasteur.fr/SubtiList/) as unknown proteins or similar to proteins of unknown functions. We performed a homology search by using the blastp program and determined that ysbAB gene products are homologous to the lrgAB gene products in Staphylococcus aureus (15). The amino acid sequences of ysbA- and ysbB-encoding proteins are 44 and 57% identical to the LrgA and LrgB proteins, respectively, from S. aureus. The LrgAB proteins inhibit extracellular murein hydrolase activity (the enzymes that cleave structural components of the bacterial cell wall) as well as convey penicillin tolerance in S. aureus (15).
Genes with similar expression patterns with yheH or ysbAB were further analyzed by best k-means clustering analysis. The genes that clustered with yheH or ysbAB, and their relative expression levels are listed in Table 3. The yheH gene was highly induced after 15 min with chloramphenicol treatment at all concentrations tested. The ysbA and ysbB gene were induced after 30 min of treatment with any concentration of chloramphenicol or erythromycin. Finally, gentamicin treatment resulted in an induction of ysbA and ysbB at 60 min. The yheI gene was expressed similarly with yheH, which is in the same operon, at all chloramphenicol concentrations tested. The dctP gene, encoding a C4-dicarboxylate transport protein, displayed an expression profile similar to yheH with 0.4× MIC chloramphenicol treatment. Interestingly, the dctP gene also exhibited expression patterns similar to those for ysbA and ysbB when treated with higher concentrations (0.25× and 0.4×/0.5×) of chloramphenicol, erythromycin, or gentamicin. The ysbA and ysbB genes were also regulated similarly to yheH upon treatment with 0.4× MIC of chloramphenicol. In all erythromycin and gentamicin experiments, ysbA and ysbB had expression profiles similar to those of yolF and yxiE, respectively. The function of yolF is unknown, whereas yxiE is known to be induced by phosphate starvation (1). Homology searches with yolF and yxiE failed to reveal any similarity to known proteins (data not shown).
TABLE 3.
Expression ratios of genes that were highly expressed or involved in transcription regulatory functions in response to various concentrations of chloramphenicol, erythromycin, or gentamicin
| Genea | Product or function | Antibioticb (MIC) | Relative expression levelsc at:
|
|||
|---|---|---|---|---|---|---|
| 5 min | 15 min | 30 min | 60 min | |||
| Highly expressed genes | ||||||
| dctP | C4-dicarboxylate transport protein | Cm (0.25×) | 0.4 | 1.1 | 0.7 | 2.1 |
| Cm (0.4×) | 0.4 | 1.4 | 3.4 | 3.5 | ||
| Em (0.25×) | 0.7 | 0.4 | 4.2 | 24.7 | ||
| Em (0.5×) | 0.8 | 0.9 | 2.8 | 20.4 | ||
| Gm (0.25×) | 0.5 | 0.2 | 1.2 | 3.9 | ||
| Gm (0.4×) | 0.6 | 0.4 | 1.1 | 4.3 | ||
| yheH | Similar to ABC transporter (ATP-binding protein) | Cm (0.05×) | 1.1 | 1.7 | 3.6 | 2.2 |
| Cm (0.25×) | 1.0 | 7.2 | 19.4 | 17.7 | ||
| Cm (0.4×) | 1.1 | 4.5 | 12.2 | 14.4 | ||
| yheI | Similar to ABC transporter (ATP-binding protein) | Cm (0.05×) | 1.3 | 1.5 | 3.5 | 2.1 |
| Cm (0.25×) | 1.2 | 8.4 | 16.7 | 21.3 | ||
| Cm (0.4×) | 1.1 | 7.0 | 19.1 | 20.7 | ||
| yolF | Unknown | Em (0.1× | 0.8 | 0.2 | 1.0 | 2.8 |
| Em (0.25×) | 0.9 | 0.3 | 1.5 | 5.4 | ||
| Em (0.5×) | 0.7 | 0.6 | 1.4 | 6.4 | ||
| ysbA | Antiholin-like membrane protein | Cm (0.05×) | 0.3 | 0.6 | 2.3 | 1.2 |
| Cm (0.25×) | 0.3 | 0.8 | 1.1 | 3.0 | ||
| Cm (0.4×) | 0.7 | 1.3 | 3.2 | 4.0 | ||
| Em (0.1×) | 1.1 | 0.2 | 2.0 | 3.1 | ||
| Em (0.25×) | 1.2 | 0.1 | 2.6 | 6.6 | ||
| Em (0.5×) | 1.3 | 0.2 | 1.8 | 7.8 | ||
| Gm (0.1×) | 0.8 | 0.2 | 1.0 | 2.7 | ||
| Gm (0.25×) | 0.9 | 0.3 | 1.1 | 6.4 | ||
| Gm (0.4×) | 0.9 | 0.2 | 1.0 | 5.0 | ||
| ysbB | Antiholin-like membrane protein | Cm (0.05×) | 0.3 | 0.8 | 6.0 | 1.1 |
| Cm (0.25×) | 0.3 | 0.8 | 1.2 | 4.2 | ||
| Cm (0.4×) | 0.8 | 1.1 | 7.2 | 8.6 | ||
| Em (0.1×) | 1.3 | 0.1 | 2.1 | 3.0 | ||
| Em (0.25×) | 1.3 | 0.1 | 2.9 | 8.2 | ||
| Em (0.5×) | 1.1 | 1.1 | 0.3 | 1.9 | ||
| Gm (0.1×) | 0.8 | 0.2 | 1.1 | 2.4 | ||
| Gm (0.25×) | 1.1 | 1.0 | 1.0 | 6.2 | ||
| Gm (0.4×) | 0.9 | 0.2 | 0.8 | 4.2 | ||
| yxiE | Induced by phosphate starvation independently of | Gm (0.1×) | 0.9 | 0.6 | 0.8 | 2.1 |
| sigma-B and PhoPR; similar to unknown proteins | Gm (0.25×) | 0.9 | 0.5 | 0.8 | 4.1 | |
| Gm (0.4×) | 1.2 | 0.5 | 0.7 | 3.9 | ||
| Transcriptional regulators | ||||||
| lmrA | Transcriptional repressor of the lincomycin operon | Cm (0.05×) | 1.3 | 1.0 | 1.2 | 1.1 |
| Cm (0.25×) | 0.8 | 1.0 | 1.2 | 0.9 | ||
| Cm (0.4×) | 1.3 | 1.1 | 1.0 | 1.0 | ||
| Em (0.1×) | 0.8 | 2.1 | 0.9 | 0.7 | ||
| Em (0.25×) | 1.0 | 2.6 | 1.1 | 1.0 | ||
| Em (0.5×) | 1.2 | 3.4 | 1.3 | 1.0 | ||
| Gm (0.1×) | 0.9 | 2.3 | 1.0 | 0.9 | ||
| Gm (0.25×) | 1.3 | 2.1 | 1.1 | 0.8 | ||
| Gm (0.4×) | 0.9 | 1.9 | 1.1 | 0.8 | ||
| lmrBd | Drug export protein | Cm (0.05×) | 0.9 | 0.9 | 0.8 | 1.3 |
| Cm (0.25×) | 0.8 | 0.7 | 0.6 | 0.9 | ||
| Cm (0.4×) | 0.9 | 0.6 | 0.6 | 1.1 | ||
| Em (0.1×) | 0.8 | 1.7 | 0.9 | 1.2 | ||
| Em (0.25×) | 1.1 | 2.6 | 1.0 | 1.1 | ||
| Em (0.5×) | 1.1 | 2.1 | 1.2 | 1.1 | ||
| Gm (0.1×) | 0.9 | 2.0 | 0.9 | 0.9 | ||
| Gm (0.25×) | 0.8 | 1.7 | 1.0 | 0.9 | ||
| Gm (0.4×) | 0.9 | 1.3 | 1.0 | 0.9 | ||
| pyrR | Transcriptional attenuator and uracil | Cm (0.05×) | 0.9 | 1.9 | 1.6 | 1.1 |
| phosphoribosyltransferase activity (minor) | Cm (0.25×) | 1.3 | 3.9 | 1.7 | 1.8 | |
| Cm (0.4×) | 1.4 | 5.7 | 1.7 | 2.3 | ||
| Em (0.1×) | 1.2 | 4.4 | 1.0 | 0.7 | ||
| Em (0.25×) | 1.3 | 4.8 | 1.0 | 0.4 | ||
| Em (0.5×) | 1.3 | 3.3 | 1.8 | 0.4 | ||
| Gm (0.1×) | 2.2 | 1.7 | 0.7 | 0.8 | ||
| Gm (0.25×) | 1.5 | 4.7 | 0.9 | 1.0 | ||
| Gm (0.4×) | 1.5 | 1.5 | 0.9 | 1.1 | ||
Representative genes in each category showed consistent and reproducible 1.5 fold regulation in at least one condition of each antibiotic treatment.
Cm, chloramphenicol; Em, erythromycin; Gm, gentamicin.
Values are relative transcript levels of treated samples over untreated samples, with values more than one indicating induction and values less than one indicating repression. Bold numbers represent the average expression ratios from two experiments that are considered significant by SAM and were consistently up- or downregulated in both experiments. The “q values” range from 0.0003 to 0.01, 0.0002 to 0.0017, and 0.0004 to 0.0025 under the conditions shown for chloramphenicol, erythromycin, and gentamicin, respectively.
The lmrB gene does not encode a transcriptional regulator but is in the same operon with lmrA, a transcriptional regulator.
Transcriptional regulators affected by protein synthesis inhibitors.
We investigated transcriptional regulators whose expression was altered by treatment with protein synthesis inhibitors. Expression levels of representative genes are listed in Table 3. Transcriptional regulators that were affected by all three protein synthesis inhibitors include PyrR. When pyrimidines are abundant, the PyrR protein binds to the conserved sequence in the pyr operon mRNA and disrupts the antiterminator, permitting terminator hairpin formation and promoting transcription termination (35, 38). Transcription of pyrR was induced at 15 min in all concentrations of erythromycin treatment. Similarly, pyrR gene expression peaked at 15 min and remained induced at 60 min after the addition of chloramphenicol. Gentamicin treatment resulted in increased expression of pyrR at 5 and 15 min at all concentrations. Genes in the pyrR regulon, the pyrRPBC(AA)(AB)KDFE operon, were induced at 15 min, but expression decreased after 15 min of treatment with all of the protein synthesis inhibitors tested (data not shown).
Expression of the lmrAB operon was induced at 15 min by at least twofold due to treatment with gentamicin or erythromycin at all concentrations. The lmrA gene encodes a negative regulator that autogenously represses the transcription of the operon, and the lmrB gene product is a drug efflux pump (19, 25, 43). The expression of the lmrAB operon was reduced after 15 min in erythromycin or gentamicin-treated cultures, probably due to repression by LmrA. Expression of lmrA and lmrB appeared to be unaffected by chloramphenicol treatment.
DISCUSSION
We used microarray analysis to study transcriptional profiles of B. subtilis cultures treated with subinhibitory concentrations of chloramphenicol, erythromycin, and gentamicin. In order to study the kinetics of the transcriptional response induced by protein synthesis inhibitors, a time course of 5 to 60 min and multiple concentrations of antibiotic were applied. We found that the largest number of genes affected by the three protein synthesis inhibitors included genes encoding transport/binding proteins, genes involved in metabolism of carbohydrates, and genes involved in protein synthesis. In addition, expression patterns were similar in two functional classes when cells were treated with the highest concentration of chloramphenicol and erythromycin. This similarity in expression patterns may reflect the fact that both antibiotics target the 50S ribosomal subunit.
Intriguingly, the same three functional classes of genes that were affected by the protein synthesis inhibitors were also affected by antibiotics inhibiting cell wall, RNA, or DNA synthesis (M. Maranta, C. Amolo, H. Ge, and D. S. Yaver, unpublished data). This presents the possibility that altered transcriptional expression of genes in these categories is a universal response to general stress caused by treatment with antibiotics. However, expression profiles were distinct among the antibiotics with different mechanisms of action, indicating that specific transcriptional responses resulted from different antibiotic treatments.
The stringent response is a process that enhances survival during starvation stress and coincides with the rapid accumulation of guanosine 3′-5′-bispyrophosphate (ppGpp). The hallmark of the stringent response is the negative regulation of components of the translational apparatus including rRNAs, tRNAs, ribosomal proteins, and translation factors (10, 13). Several studies have shown that while transcriptional and translational inhibitors cause decreased synthesis of the stringent factor ppGpp in Escherichia coli, some aminoglycosides such as neomycin, streptomycin, or spectinomycin had little effect on the level of ppGpp (21, 26). A similar effect was observed in Haemophilus influenzae when transcriptional and proteomic analyses were performed in the presence of transcriptional and translational inhibitors (12). The authors of that study found that ribosomal protein synthesis rates were increased by treatment with most protein synthesis inhibitors. However, aminoglycoside such as streptomycin had little effect. These researchers further demonstrated that the transcriptional and translational responses to translational inhibitors were coordinately mediated by the synthesis of ppGpp. Proteomic studies in B. subtilis also showed that inhibitors of translation elongation (such as tetracycline, chloramphenicol, and erythromycin) induced the rate of synthesis of the stringently controlled ribosomal proteins and elongation factors, whereas aminoglycosides (such as gentamicin, kanamycin, and streptomycin), which interfere with ribosomal translation accuracy, did not (3). Consistently, we found that expression of ribosomal protein and elongation factor genes was induced by chloramphenicol and erythromycin, whereas genes coding for elongation factors were not affected by gentamicin. However, our results showed that gentamicin can trigger transcriptional induction of genes encoding ribosomal proteins even 5 min posttreatment (Table 1). A real-time RT-PCR experiment validated that one of the ribosomal protein genes, rplF, was indeed induced at the early time point (Table 2). Many ribosomal protein genes showed dynamic gene regulation after treatment with gentamicin (i.e., up at 5 min, down at 15 min, up at 30 min, and then down at 60 min) (Tables 1 and 2). This discrepancy could be due to our use of subinhibitory concentrations of antibiotic (versus the higher dosages used in the other studies) or to other differences in experimental conditions.
Other functional categories affected by both chloramphenicol and erythromycin included genes involved in metabolism of amino acids (Table 1). The similar expression profiles between genes encoding ribosomal proteins and genes involved in purine/pyrimidine biosynthesis due to treatment with chloramphenicol could be due to coregulation (i.e., repression of the stringent response). Genes involved in purine/pyrimidine biosynthesis were shown to be repressed under conditions that provoke stringent response, but regulation was relA independent (13).
Previous studies in E. coli showed that the H group antibiotics (such as aminoglycosides) induce heat shock response, whereas the C group antibiotics (such as erythromycin and chloramphenicol) induce cold shock and repress heat shock genes (7, 41). A recent study also confirmed that the heat shock response was triggered by treatment with kanamycin (33). A proteomic study in B. subtilis confirmed that aminoglycosides induced expression of heat shock proteins, but the authors did not observe induction of cold shock proteins by chloramphenicol or erythromycin (3). In our study, a few cold shock genes were induced by treatment with chloramphenicol or erythromycin in some of the experiments, but induction was not consistent for most cold shock genes (Table 2 and data not shown). A whole-genome transcriptional analysis of a gram-positive bacterium Streptococcus pneumoniae also showed that, although streptomycin induced heat shock genes, erythromycin and chloramphenicol did not alter heat shock gene expression (28). Our data are consistent with the Bacillus and Streptococcus studies in that gentamicin, as well as streptomycin, induced heat shock genes. The delayed induction (at 60 min) of the heat shock genes on treatment with gentamicin is consistent with studies by VanBogelen and Neidhardt in E. coli (41). These researchers found that, whereas temperature shifts resulted in an immediate induction of heat shock proteins, the addition of antibiotics such as aminoglycosides resulted in a much delayed heat shock response. In contrast to the studies described above, we show here that the expression of heat shock genes was repressed by chloramphenicol. These microarray results were validated by real-time RT-PCR analysis (Table 2). Again, the discrepancy could be due to experimental design and conditions. We could be observing a more primary effect, since altered expression of heat shock genes by treatment with chloramphenicol was observed after 15 min when the growth inhibitory effect was still minimal.
The effects of sublethal concentrations of translation inhibitors (chloramphenicol, erythromycin, tetracycline, and puromycin) on global transcription patterns of S. pneumoniae R6 was previously studied (28). These researchers found that genes from the major biological categories that were affected by translation inhibitors included genes involved in translation, transport and carrier proteins, and in amino acid biosynthesis. As mentioned above, we found similar classes of genes being regulated in chloramphenicol- and erythromycin-treated B. subtilis cultures.
We examined genes that were highly induced due to treatment with chloramphenicol, erythromycin, or gentamicin. Of particular interest were the yheIH and ysbAB operons. The expression of yheIH was highly induced by chloramphenicol after 15 min, whereas the expression of ysbAB was highly induced by all three antibiotics tested. The yheI and yheH genes were previously shown to encode ABC transporter-like proteins which were classified in subfamily 6 of B. subtilis ATP-binding proteins (29). Proteins in subfamily 6 are similar to multidrug resistance proteins of eukaryotes and prokaryotes. The YheI/YheH proteins might constitute four heterodimer ABC transporters. The ysbAB gene products are homologous to the lrgAB gene products in S. aureus. The LrgAB proteins confer negative control on extracellular murein hydrolase activity (the enzymes that cleave structural components of the bacterial cell wall), as well as decreased sensitivity to penicillin-induced killing in S. aureus (15). The lrgAB genes form an operon and encode for antiholin-like membrane proteins that were hypothesized to inhibit the formation of murein hydrolase transport channels (holin) in the bacterial membrane. Since ysbAB homologs function to inhibit cell wall cleavage (the murein hydrolase activity) and convey penicillin tolerance in S. aureus, it is slightly surprising to observe that ysbAB were induced by protein synthesis inhibitors. Our observation suggests that ysbA and ysbB may be involved in tolerance to protein synthesis inhibitors as well. Furthermore, the ysbAB genes were coregulated with yheH under 0.4× MIC chloramphenicol treatment, implying that they could share a similar function with the putative multidrug resistance protein. Sensitivity toward penicillin and protein synthesis inhibitors in B. subtilis strains overexpressing ysbAB is currently being studied.
We also investigated transcriptional regulators that were affected by chloramphenicol, erythromycin, and gentamicin. The pyrR gene, the transcription attenuation factor of the pyrR operon involved in pyrimidine biosynthesis, was induced after 15 min by all three protein synthesis inhibitors. As expected, the increased expression of the PyrR transcription attenuator leads to repression of the genes in the pyrR regulon after 15 min of antibiotic treatment. Expression of the lmrA gene, encoding a negative regulator of transcription of the lmrAB operon, was readily induced at 15 min by treatment with erythromycin or gentamicin. The lmrB gene, encoding a multidrug-resistant efflux protein, of the lmrAB operon was also induced at 15 min by erythromycin or gentamicin (25). Expression of the lmrAB operon was decreased after 15 min, possibly due to repression by the negative transcription regulator LmrA. Induction of lmrB may indicate a self-defense mechanism induced by treatment with erythromycin and gentamicin.
In summary, we determined the expression profiles of chloramphenicol-, erythromycin-, and gentamicin-treated B. subtilis cultures and showed that transcription of several major functional classes of genes was affected. Genes that were specifically affected by treatment with protein synthesis inhibitors could potentially be used as signature genes, along with signature genes from cultures treated with other antibiotics for determination of modes of action of new drugs.
Acknowledgments
We thank Randy Berka and Alan Sloma for critical reading of the manuscript.
REFERENCES
- 1.Antelmann, H., C. Scharf, and M. Hecker. 2000. Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis. J. Bacteriol. 182:4478-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bandow, J. E., H. Brotz, and M. Hecker. 2002. Bacillus subtilis tolerance of moderate concentrations of rifampin involves the σB-dependent general and multiple stress response. J. Bacteriol. 184:459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bandow, J. E., H. Brotz, L. I. Leichert, H. Labischinski, and M. Hecker. 2003. Proteomic approach to understanding antibiotic action. Antimicrob. Agents Chemother. 47:948-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berka, R. M., X. Cui, and C. Yanofsky. 2003. Genomewide transcriptional changes associated with genetic alterations and nutritional supplementation affecting tryptophan metabolism in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 100:5682-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berka, R. M., J. Hahn, M. Albano, I. Draskovic, M. Persuh, X. Cui, A. Sloma, W. Widner, and D. Dubnau. 2002. Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol. Microbiol. 43:1331-1345. [DOI] [PubMed] [Google Scholar]
- 6.Betts, J. C., A. McLaren, M. G. Lennon, F. M. Kelly, P. T. Lukey, S. J. Blakemore, and K. Duncan. 2003. Signature gene expression profiles discriminate between isoniazid-, thiolactomycin-, and triclosan-treated Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 47:2903-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bianchi, A. A., and F. Baneyx. 1999. Stress responses as a tool to detect and characterize the mode of action of antibacterial agents. Appl. Environ. Microbiol. 65:5023-5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao, M., T. Wang, R. Ye, and J. D. Helmann. 2002. Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis sigma(W) and sigma(M) regulons. Mol. Microbiol. 45:1267-1276. [DOI] [PubMed] [Google Scholar]
- 9.Carter, A. P., W. M. Clemons, D. E. Brodersen, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407:340-348. [DOI] [PubMed] [Google Scholar]
- 10.Chatterji, D., and A. K. Ojha. 2001. Revisiting the stringent response, ppGpp and starvation signaling. Curr. Opin. Microbiol. 4:160-165. [DOI] [PubMed] [Google Scholar]
- 11.Espinal, M. A., A. Laszlo, L. Simonsen, F. Boulahbal, S. J. Kim, A. Reniero, S. Hoffner, H. L. Rieder, N. Binkin, C. Dye, R. Williams, M. C. Raviglione, et al. 2001. Global trends in resistance to antituberculosis drugs. N. Engl. J. Med. 344:1294-1303. [DOI] [PubMed] [Google Scholar]
- 12.Evers, S., K. Di Padova, M. Meyer, H. Langen, M. Fountoulakis, W. Keck, and C. P. Gray. 2001. Mechanism-related changes in the gene transcription and protein synthesis patterns of Haemophilus influenzae after treatment with transcriptional and translational inhibitors. Proteomics 1:522-544. [DOI] [PubMed] [Google Scholar]
- 13.Eymann, C., G. Homuth, C. Scharf, and M. Hecker. 2002. Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. J. Bacteriol. 184:2500-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gmuender, H., K. Kuratli, K. Di Padova, C. P. Gray, W. Keck, and S. Evers. 2001. Gene expression changes triggered by exposure of Haemophilus influenzae to novobiocin or ciprofloxacin: combined transcription and translation analysis. Genome Res. 11:28-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Groicher, K. H., B. A. Firek, D. F. Fujimoto, and K. W. Bayles. 2000. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 182:1794-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutter, B., C. Fischer, A. Jacobi, C. Schaab, and H. Loferer. 2004. Panel of Bacillus subtilis reporter strains indicative of various modes of action. Antimicrob. Agents Chemother. 48:2588-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutter, B., C. Schaab, S. Albrecht, M. Borgmann, N. A. Brunner, C. Freiberg, K. Ziegelbauer, C. O. Rock, I. Ivanov, and H. Loferer. 2004. Prediction of mechanisms of action of antibacterial compounds by gene expression profiling. Antimicrob. Agents Chemother. 48:2838-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaldalu, N., R. Mei, and K. Lewis. 2004. Killing by ampicillin and ofloxacin induces overlapping changes in Escherichia coli transcription profile. Antimicrob. Agents Chemother. 48:890-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumano, M., M. Fujita, K. Nakamura, M. Murata, R. Ohki, and K. Yamane. 2003. Lincomycin resistance mutations in two regions immediately downstream of the −10 region of lmr promoter cause overexpression of a putative multidrug efflux pump in Bacillus subtilis mutants. Antimicrob. Agents Chemother. 47:432-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 21.Lund, E., and N. O. Kjeldgaard. 1972. Metabolism of guanosine tetraphosphate in Escherichia coli. Eur. J. Biochem. 28:316-326. [DOI] [PubMed] [Google Scholar]
- 22.Marton, M. J., J. L. DeRisi, H. A. Bennett, V. R. Iyer, M. R. Meyer, C. J. Roberts, R. Stoughton, J. Burchard, D. Slade, H. Dai, D. E. Bassett, Jr., L. H. Hartwell, P. O. Brown, and S. H. Friend. 1998. Drug target validation and identification of secondary drug target effects using DNA microarrays. Nat. Med. 4:1293-1301. [DOI] [PubMed] [Google Scholar]
- 23.Mascher, T., N. G. Margulis, T. Wang, R. W. Ye, and J. D. Helmann. 2003. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol. 50:1591-1604. [DOI] [PubMed] [Google Scholar]
- 24.Moazed, D., and H. F. Noller. 1987. Chloramphenicol, erythromycin, carbomycin and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S rRNA. Biochimie 69:879-884. [DOI] [PubMed] [Google Scholar]
- 25.Murata, M., S. Ohno, M. Kumano, K. Yamane, and R. Ohki. 2003. Multidrug resistant phenotype of Bacillus subtilis spontaneous mutants isolated in the presence of puromycin and lincomycin. Can. J. Microbiol. 49:71-77. [DOI] [PubMed] [Google Scholar]
- 26.Muto, A., A. Kimura, and S. Osawa. 1975. Effects of some antibiotics on the stringent control of RNA synthesis in Escherichia coli. Mol. Gen. Genet. 139:321-327. [DOI] [PubMed] [Google Scholar]
- 27.NCCLS. 2000. Methods for dilution antimicrobial susceptibility test for bacterial that grow aerobically, 5th ed. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 28.Ng, W. L., K. M. Kazmierczak, G. T. Robertson, R. Gilmour, and M. E. Winkler. 2003. Transcriptional regulation and signature patterns revealed by microarray analyses of Streptococcus pneumoniae R6 challenged with sublethal concentrations of translation inhibitors. J. Bacteriol. 185:359-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quentin, Y., G. Fichant, and F. Denizot. 1999. Inventory, assembly, and analysis of Bacillus subtilis ABC transport systems. J. Mol. Biol. 287:467-484. [DOI] [PubMed] [Google Scholar]
- 30.Rajeevan, M. S., D. G. Ranamukhaarachchi, S. D. Vernon, and E. R. Unger. 2001. Use of real-time quantitative PCR to validate the results of cDNA array and differential display PCR technologies. Methods 25:443-451. [DOI] [PubMed] [Google Scholar]
- 31.Rajeevan, M. S., S. D. Vernon, N. Taysavang, and E. R. Unger. 2001. Validation of array-based gene expression profiles by real-time (kinetic) RT-PCR. J. Mol. Diagn. 3:26-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlunzen, F., R. Zarivach, J. Harms, A. Bashan, A. Tocilj, R. Albrecht, A. Yonath, and F. Franceschi. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814-821. [DOI] [PubMed] [Google Scholar]
- 33.Shaw, K. J., N. Miller, X. Liu, D. Lerner, J. Wan, A. Bittner, and B. J. Morrow. 2003. Comparison of the changes in global gene expression of Escherichia coli induced by four bactericidal agents. J. Mol. Microbiol. Biotechnol. 5:105-122. [DOI] [PubMed] [Google Scholar]
- 34.Shaw, K. J., and B. J. Morrow. 2003. Transcriptional profiling and drug discovery. Curr. Opin. Pharmacol. 3:508-512. [DOI] [PubMed] [Google Scholar]
- 35.Switzer, R. L., R. J. Turner, and Y. Lu. 1999. Regulation of the Bacillus subtilis pyrimidine biosynthetic operon by transcriptional attenuation: control of gene expression by an mRNA-binding protein. Prog. Nucleic Acids Res. Mol. Biol. 62:329-367. [DOI] [PubMed] [Google Scholar]
- 36.Tamames, J., D. Clark, J. Herrero, J. Dopazo, C. Blaschke, J. M. Fernandez, J. C. Oliveros, and A. Valencia. 2002. Bioinformatics methods for the analysis of expression arrays: data clustering and information extraction. J. Biotechnol. 98:269-283. [DOI] [PubMed] [Google Scholar]
- 37.Tang, C. M., and E. R. Moxon. 2001. The impact of microbial genomics on antimicrobial drug development. Annu. Rev. Genomics Hum. Genet. 2:259-269. [DOI] [PubMed] [Google Scholar]
- 38.Turner, R. J., Y. Lu, and R. L. Switzer. 1994. Regulation of the Bacillus subtilis pyrimidine biosynthetic (pyr) gene cluster by an autogenous transcriptional attenuation mechanism. J. Bacteriol. 176:3708-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Utaida, S., P. M. Dunman, D. Macapagal, E. Murphy, S. J. Projan, V. K. Singh, R. K. Jayaswal, and B. J. Wilkinson. 2003. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 149:2719-2732. [DOI] [PubMed] [Google Scholar]
- 41.VanBogelen, R. A., and F. C. Neidhardt. 1990. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc. Natl. Acad. Sci. USA 87:5589-5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson, M., J. DeRisi, H. H. Kristensen, P. Imboden, S. Rane, P. O. Brown, and G. K. Schoolnik. 1999. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc. Natl. Acad. Sci. USA 96:12833-12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshida, K., Y. H. Ohki, M. Murata, M. Kinehara, H. Matsuoka, T. Satomura, R. Ohki, M. Kumano, K. Yamane, and Y. Fujita. 2004. Bacillus subtilis LmrA is a repressor of the lmrAB and yxaGH operons: identification of its binding site and functional analysis of lmrB and yxaGH. J. Bacteriol. 186:5640-5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshizawa, S., D. Fourmy, and J. D. Puglisi. 1998. Structural origins of gentamicin antibiotic action. EMBO J. 17:6437-6448. [DOI] [PMC free article] [PubMed] [Google Scholar]