Abstract
GrimAge acceleration has previously predicted age-related morbidities and mortality. In the current study, we sought to examine how GrimAge is associated with genetic predisposition for systemic inflammation and whether psychosocial factors moderate this association. Military veterans from the National Health and Resilience in Veterans study, which surveyed a nationally representative sample of European American male veterans, provided saliva samples for genotyping (N = 1135). We derived polygenic risk scores (PRS) from the UK Biobank as markers of genetic predisposition to inflammation. Results revealed that PRS for three inflammatory PRS markers—HDL (lower), apolipoprotein B (lower), and gamma-glutamyl transferase (higher)—were associated with accelerated GrimAge. Additionally, these PRS interacted with a range of potentially modifiable psychosocial variables, such as exercise and gratitude, previously identified as associated with accelerated GrimAge. Using gene enrichment, we identified anti-inflammatory and antihistamine drugs that perturbate pathways of genes highly represented in the inflammatory PRS, laying the groundwork for future work to evaluate the potential of these drugs in mitigating epigenetic aging.
Subject terms: DNA methylation, Predictive markers
Introduction
A host of psychological and physical health comorbidities accompany advancing age. To characterize biological changes involved in age-related morbidities, investigators have identified markers of ‘biological aging’ (i.e., the gradual deterioration of integrity of cells and organs with increasing age) that better capture the aging process than chronological age [1]. For example, DNA methylation (DNAm) epigenetic clock algorithms [2–4] predict age-related clinical phenotypes [5]. The ‘GrimAge’ algorithm is an epigenetic clock designed to examine DNAm changes associated with morbidity and mortality [6]. GrimAge predicts time-to-death independent of chronological age [7] and is associated with age-related health outcomes such as walking speed, polypharmacy, and frailty [4].
The National Health and Resilience in Veterans Study (NHRVS) reported a range of psychological, health, and lifestyle factors associated with ‘accelerated’ GrimAge: GrimAge that exceeded chronological age by 5 or more years [8]. Specifically, accelerated GrimAge was independently associated with lower physical exercise, history of lifetime substance use disorder, greater number of lifetime traumas, lower gratitude, reduced sleep quality, lower openness to experience, and unmarried/partnered status. Interestingly, many of these predictors of accelerated GrimAge are also risk factors for immune system dysregulation, particularly low-level inflammation that is common with advancing age (‘inflammaging’) [9–11].
Inflammation may be associated with accelerated GrimAge indirectly via risk for medical conditions, in addition to possible direct associations. For example, inflammation is associated with mortality [12], cardiovascular illness [13], impaired sleep quality, and psychiatric disorders including depression [14], posttraumatic stress disorder, and substance use disorders [9, 15–22]. Inflammation may also be directly associated with DNAm changes such as GrimAge acceleration. Accelerated epigenetic aging has been associated with increased C-reactive protein levels (CRP), interleukin-6 (IL-6), and tumor necrosis factor-alpha (TNF-alpha) [23–25]. These associations have also been observed in GrimAge in some studies [6, 26, 27], though others have shown null findings [26]. Inconsistent findings may be explained in part by prior work restricting analyses to few variables of inflammatory markers instead of considering a wider range of markers. In addition, circulatory levels of inflammatory markers are limited by the impact of reverse causation and poor reliability. Levels impacted by a range of factors that include variations in circadian rhythms, genetics, and laboratory practices, among others [28]. For example, one study found that salivary levels of immune markers were only weakly correlated across 18-month follow-ups (r = 0.18) with one additional assessment only increasing reliability to r = 0.27 [29]. Conversely, a recent paper has shown that a DNAm based measure outperforms circulating levels of molecular and cellular CRP as a marker of chronic inflammation [28], making gene-based measures of inflammatory markers a useful alternative to elucidate inconsistent findings of the association between accelerated GrimAge and inflammatory markers.
Research using gene-based measures can inform mechanisms of accelerated epigenetic aging. Because individuals’ genotypes do not change over the course of their lifetimes, polygenic risk scores (PRS) can be used to estimate the relationship between inflammatory biomarkers and accelerated GrimAge. This approach allows one to examine whether genetic susceptibility for inflammation present at birth may be associated with accelerated GrimAge, thereby circumventing the issue of reverse causation inherent in circulatory biomarkers. Toward this end, we sought to examine the relationship between inflammatory-biomarker PRS and GrimAge in a population-based cohort of male European-American U.S. military veterans who participated in the National Health and Resilience in Veterans Study. To investigate the relationship between inflammation and accelerated GrimAge comprehensively, we analyzed PRS related to a wider range of blood biomarkers than often examined [30–54]. Additionally, to understand how modifiable factors can alter the association of inflammatory biomarkers with biological aging, we tested the interaction of psychosocial variables with inflammatory-biomarker PRS with respect to accelerated GrimAge derived from saliva samples. Accelerated GrimAge was defined a priori as 5 or more years of accelerated epigenetic relative to chronological age to capture a clinically meaningful magnitude of difference (i.e., mean 5–10 year greater acceleration of epigenetic relative to chronological aging) and allows us to compare findings with prior studies that have employed this operationalization of accelerated GrimAge [55–58].
We hypothesized that PRS of inflammatory biomarkers would be associated with accelerated GrimAge based on previous studies examining how each of these markers is related to immune function and inflammation [30–54] and studies finding a relationship between inflammatory markers and accelerated biological aging [23–25] (see Table 1). Second, we predicted that this relationship would be moderated by psychosocial factors. Specifically, we expected that positive psychosocial factors such as gratitude, good sleep quality, openness to experience, and married/partnered status would reduce the strength of the association of inflammatory PRS markers with accelerated GrimAge. This hypothesis is based on findings that positive psychosocial factors such as social support are associated with reduced inflammation and moderate risk for inflammation [59–62]. In contrast, we expected that negative psychosocial factors such as substance use disorder and cumulative trauma burden would be associated with increased strength of association between inflammatory PRS markers and accelerated GrimAge [9, 15–22].
Table 1.
Details of biomarkers included in UKB blood analysis and our hypothesized direction of association with GrimAge for each biomarker.
| UKB Field ID | Biomarker | N | Heritability (SAIGE) | Hypothesized association with GrimAge |
|---|---|---|---|---|
| 30600 | Albumin | 367,192 | 0.064 | Negative |
| 30610 | Alkaline phosphatase | 400,988 | 0.131 | Positive |
| 30630 | Apolipoprotein A | 364,987 | 0.122 | Positive |
| 30640 | Apolipoprotein B | 399,003 | 0.079 | Positive |
| 30710 | C-reactive protein | 400,094 | 0.106 | Positive |
| 30720 | Cystatin C | 400,940 | 0.142 | Positive |
| 30730 | Gamma glutamyl transferase | 400,751 | 0.114 | Positive |
| 30760 | HDL cholesterol | 367,021 | 0.143 | Negative |
| 30770 | IGF-1 | 398,797 | 0.110 | Positive |
| 30790 | Lipoprotein A | 318,922 | 0.082 | Positive |
| 30810 | Phosphate | 366,484 | 0.061 | Positive |
| 30820 | Rheumatoid factor | 35,667 | <0.001 | Positive |
Materials and methods
Study population
Participants were 1135 U.S. military Veterans who participated in the National Health and Resilience in Veterans Study (NHRVS), a contemporary, nationally representative study of health and functioning in this population [63]. This sample was obtained from KnowledgePanel, a nationally representative survey research panel representing approximately 98% of U.S. households. Post-stratification weights were applied to inferential analyses to permit generalizability of results to the U.S. Veteran population; these weights were computed based on the demographic distribution of U.S. Veterans from concurrent U.S. Census Bureau Current Population Survey data. The analyzed sample was a sub-cohort of the overall sample (N = 3157) that provided saliva samples for epigenetic analyses. The Human Subjects Subcommittee of the VA Connecticut Healthcare System approved the study. All participants provided informed consent and provided saliva samples.
NHRVS participants completed a comprehensive survey including the assessment of demographics and military history, medical and psychiatric status, and psychosocial functioning [63]. A detailed description of the assessment measures is available in Supplementary Table 1.
DNA extraction, genotyping, imputation, and methylation assessment
Oragene kits (DNA Genotek, Ottawa, Ontario, Canada) were used to extract genomic DNA from the saliva samples. Samples were genotyped on the Illumina PsychChip microarray (San Diego, CA, USA) at the Gelernter Lab in West Haven, CT. NHRVS participants with mismatched sex or genotype call rate <95% were excluded from the analysis. Also, we investigated only unrelated participants. For each pair of related individuals (pihat > 0.2), the individual with the lowest call rate was removed from the analysis. Next, SNPs with genotype call rate <95% or minor allele frequency (MAF) < 0.01 were removed before imputation. Ancestry assignment was defined via principal component analysis combining NHRVS sample with 1,000 Genomes Project reference populations [64]. Because 85% of NHRVS sample is of European descent (EUR), we could not investigate other ancestry groups due to their limited sample size. Imputation was performed using the genotype imputation software Minimac4 [65] and the Haplotype Reference Consortium [66] as reference panel through the Michigan Imputation Server (available at https://imputationserver.sph.umich.edu/) [67]. After imputation, we retained a total of 7,974,742 variants with MAF > 1% and INFO score >0.6.
For DNAm assessment, DNA collected from whole blood was treated with bisulfite reagents (EZ-96 DNA methylation kit - Zymo Research, Orange, CA, USA) according to standard protocols. After bisulfite treatment, DNAm was assessed using the Illumina Infinium Human MethylationEPIC BeadChip v1.0 (Illumina, San Diego, CA, USA) that evaluates methylation in over 850,000 sites. DNAm assay performed at Yale Center for Genome Analysis in Orange, CT. β values were generated using GenomeStudio software (Illumina), considering the total methylated signal (M) and the unmethylated signal (U), where β = M/(M + U). See Supplementary Text for information about quality control and normalization.
GrimAge
We evaluated epigenetic aging by utilizing the DNA methylation aging calculator (https://dnamage.genetics.ucla.edu/new) and following the tutorial guidelines. This tool allowed us to estimate predicted aging and accelerated aging for the GrimAge clock, which is based on DNAm surrogate biomarkers associated with morbidity and mortality, as well as the quantification of smoking pack-years.
Data analysis
Calculation of PRS
Blood biomarkers were selected from UK Biobank (UKB) [68] by reviewing available biomarkers and identifying those previously linked to inflammatory processes, including [30–54] alkaline phosphatase, albumin, apolipoprotein A, apolipoprotein B, cystatin C, C reactive protein (CRP), gamma-glutamyl transferase, high-density lipoprotein cholesterol (HDL), insulin-like growth factor 1 (IGF-1), lipoprotein A, phosphate, rheumatoid factor, and sex hormone binding globulin (SHBG) (see Table 1 and Supplementary Fig. 1). The biomarkers were selected from biochemistry markers measured in blood samples collected from UKB participants. Details related to UKB biomarker measurements are available at https://biobank.ndph.ox.ac.uk/showcase/showcase/docs/serum_biochemistry.pdf. We used data generated from genome-wide association studies (GWAS) of biomarkers conducted in the Pan-UKB analysis (Table 1). A description of the quality control of the genetic data and the genetic association analysis is available at https://github.com/Nealelab/UK_Biobank_GWAS. Briefly, the genome-wide association analyses were conducted using regression models available in Hail (available at https://github.Com/hail-is/hail) and including the top-20 within-ancestry principal components (PC), sex, age, age2, sex × age, and sex × age2 as covariates. Using PRSice2 [69], Pan-UKB genome-wide association statistics were used to calculate PRS in the NHRVS cohort considering multiple p-value threshold (Pt) for the variant inclusion (i.e., clumping-thresholding method) [70]. Thresholds considered were Pt 5e08, 1e07, 1e06, 1e05, 0.0001, 0.001, 0.05, 0.1, 0.2, 0.3, 0.4, 0.5, and 1.
Selection of PRS variables
The PRS generated were tested by examining correlations with accelerated GrimAge (i.e., ≥5 years greater GrimAge than chronological age), controlling for the top five within-ancestry principal components. Considering the strongest result for each blood biomarker (Supplementary Table 2), we observed four PRS correlations surviving false discovery rate multiple testing correction (FDR q < 0.05): Three PRS were identified: Apolipoprotein B (Pt 5e08: p < 0.001, alpha = −0.12); HDL (Pt 0.1: p < 0.001, alpha = −0.11); Gamma-glutamyl transferase: Pt 0.05 correlation, p < 0.001, alpha = 0.12). All analyses were conducted with post-stratification weights applied. Forced enter regression was used for binary logistic regression analyses. To facilitate comparability across PRS, inflammatory PRS were standardized by z-scoring. This allowed us one to estimate the change in the odds of having 5+ years accelerated GrimAge associated with each SD unit increase in the inflammatory PRS investigated.
Selection of behavioral variables
Based on the results of a previous findings in NHRVS cohort [8], we included days of physical exercise, history of lifetime substance use disorder (including alcohol use disorder and drug use disorder), cumulative trauma burden, sleep quality, openness to experience, gratitude, and marital status in a logistic regression model in addition to the main effects of inflammatory markers PRS and interactions between inflammatory markers and these psychosocial variables. Additional covariates included income, sum of medical conditions, combat status, and top five within-ancestry genetic principal components. A list of psychosocial measures used can be found in Supplementary Table 1. We used a p ≤ 0.023 FDR-corrected level for the logistic regression analysis.
Gene-set enrichment analysis
Gene variants that were included in the inflammatory PRS associated with accelerated GrimAge (i.e., Apolipoprotein B Pt 5e08; HDL Pt 01; Gamma-glutamyl transferase Pt 0.05) were used as input for a gene-based analysis with the Versatile Gene-based Association Study 2 tool [71]. We used the “best SNP test” which uses a set of markers instead of genome-wide data while accounting for linkage disequilibrium. Based on the results of the gene-based associations, we performed an enrichment analysis based on gene ontology, biological pathways, and molecular functions [71]. Considering a Bonferroni-significant threshold, we identified gene-set overrepresented for genetic associations within the inflammatory PRS linked to accelerated biological aging estimated with epigenetic information.
Drug repurposing analysis
In an exploratory analysis, we used a novel methodology—drug repurposing analyses—to identify possible drugs that target perturbated pathways in inflammatory and immune processes. The Gene2drug tool was used for drug repurposing [72]. This tool implements Pathway-Set Enrichment Analysis, which identifies up- or down-regulated gene pathways by drugs. Gene2drug converts Connectivity Map [73] gene expression to pathway expression profiles, then ranks p values based on the Kolmogorov-Smirnov test. Finally, an enrichment score (EScore) is assigned to each drug which represents up- or down-regulation and its magnitude. Semantically reduced gene ontology (GO) terms (Bonferroni p < 0.05) were used as input for Gene2drug. A Bonferroni-corrected P-value was calculated for each drug considering the number of drugs tested.
Results
Prevalence and correlates of accelerated GrimAge
As reported elsewhere [74], a total of 178 Veterans (weighted 18.3%, 95% confidence interval = 15.4–21.6%) in the sample had accelerated GrimAge. The mean age of the full sample was 63 years (SD = 13.0). Most Veterans were married or cohabiting with a partner (76%), retired (52.9%), and had some college or higher education (67.9%). Approximately one-third were combat Veterans. Table 2 reports descriptive statistics and differences between NHRVS participants with vs. without accelerated GrimAge.
Table 2.
Sociodemographic, military, and trauma characteristics of the sample by accelerated GrimAge status.
| Total Sample | <5 years Accelerated GrimAge | ≥5 years Accelerated GrimAge | Bivariate analyses | ||
|---|---|---|---|---|---|
| Weighted M (SD) or unweighted n (weighted %) N = 1135 | Weighted M (SD) or unweighted n (weighted %) N = 957 | Weighted M (SD) or unweighted n (weighted %) N = 178 | X2 or t statistic | p-value | |
| Sociodemographic Characteristics | |||||
| Age | 63.1 (14.0) | 63.1 (14.2) | 63.3 (13.0) | −0.22 | 0.83 |
| Some college or higher educationa | 981 (67.9%) | 847 (71%) | 134 (54.4%) | 19.15 | <0.001 |
| Married/cohabiting with partnerb | 893 (76.0%) | 765 (77.2%) | 128 (70.2%) | 4.14 | 0.047 |
| Currently employed | 418 (36.1%) | 356 (36.9%) | 62 (32.6%) | 1.32 | 0.28 |
| Combat veterana | 404 (32.4%) | 334 (31.0%) | 70 (38.6%) | 4.01 | 0.048 |
| Trauma and Clinical Characteristics | |||||
| Wave 1 lifetime PCL-4 scorea | 27.3 (11.5) | 26.9 (11.1) | 29.0 (12.8) | −2.29 | 0.01 |
| Number of lifetime potentially traumatic eventsb | 3.3 (2.7) | 3.2 (2.7) | 3.9 (2.9) | −3.53 | <0.001 |
| Lifetime substance use disorderb | 562 (49.6%) | 445 (46.1%) | 117 (65.1%) | 22.14 | <0.001 |
| Lifetime MDD or PTSD | 198 (18.6%) | 155 (18.0%) | 43 (21.7%) | 1.46 | 0.26 |
| Perceived stress | 0 (1.00) | −0.03 (0.99) | 0.13 (1.0) | −1.96 | 0.05 |
| Hostility | 0 (1.00) | −0.02 (0.97) | 0.11 (1.1) | −1.73 | 0.08 |
| Health/lifestyle Characteristics | |||||
| Body mass indexa | 28.7 (5.2) | 28.5 (4.8) | 29.5 (6.3) | −2.41 | 0.02 |
| Physical exercise (days per week)b | 2.6 (2.4) | 2.7 (2.3) | 1.9 (2.3) | 4.25 | <0.001 |
| Number of medical conditionsa | 2.85 (2.01) | 2.77 (1.94) | 3.15 (2.26) | −2.31 | 0.01 |
| Good sleep qualitya | 0 (1.00) | 0.05 (0.99) | −0.20 (1.03) | 3.07 | 0.001 |
| Inflammatory markers | |||||
| Apolipoprotein B (Pt 5e08)b | 0 (1.00) | 0.05 (0.99) | −0.24 (0.98) | 3.69 | <0.001 |
| Gamma-glutamyl transferase (Pt 0.05)b | 0 (1.00) | −0.09 (1.00) | 0.24 (1.11) | −3.73 | <0.001 |
| High density lipoprotein (Pt 0.1)a | 0 (1.00) | 0.06 (1.00) | −0.20 (1.11) | 2.96 | 0.003 |
| Psychosocial Characteristics | |||||
| Big 5 personality variables | |||||
| Openness to Experienceb | 0 (1.00) | 0.05(1.0) | −0.22(0.9) | 03.35 | <0.001 |
| Extraversion | 0 (1.00) | 0.03 (1.0) | −0.01 (0.9) | 0.13 | 0.90 |
| Emotional Stability | 0 (1.00) | 0.03 (1.0) | −0.12 (1.1) | 1.78 | 0.08 |
| Conscientiousness | 0 (1.00) | 0.01 (1.0) | −0.07 (1.0) | 1.01 | 0.32 |
| Agreeableness | 0 (1.00) | 0.00 (1.0) | −0.02 (0.9) | 0.32 | 0.75 |
| Dispositional Gratitudea | 0 (1.00) | 0.05 (1.0) | −0.21 (1.1) | 3.25 | 0.001 |
| Expectations regarding aging | 0 (1.00) | 0.01 (0.98) | −0.05 (1.09) | 0.82 | 0.41 |
| Dispositional Optimism | 0 (1.00) | 0.01 (1.00) | −0.05 (0.99) | 0.83 | 0.20 |
| Curiosityb | 0 (1.00) | 0.05 (0.98) | −0.21 (1.07) | 3.21 | <0.001 |
| Resilience | 0 (1.00) | 0.02 (0.98) | −0.10 (1.08) | 1.50 | 0.13 |
| Social network size | 8.8 (9.7) | 8.7 (9.5) | 9.3 (10.3) | −0.82 | 0.41 |
| Secure attachment style | 837 (71.4%) | 716 (72.3%) | 121 (67.2%) | 2.08 | 0.15 |
| Perceived social support | 0 (1.00) | 0.005 (0.999) | −0.02 (1.01) | 0.35 | 0.73 |
| GrimAge variables | |||||
| ≥ 5 years accelerated GrimAge | 178 (18.3%) | 0 (0%) | 178 (100%) | ||
| Years GrimAge acceleration | 0.42 (4.90) | −1.36 (3.30) | 8.40 (2.18) | ||
The following variables were standardized by z-scoring: Perceived stress; Hostility; Openness to Experience; Extraversion; inflammatory markers.
Emotional Stability; Conscientiousness; Agreeableness; Dispositional gratitude; Expectations regarding aging; Resilience; Perceived social support; sleep quality.
X2 df = 1; t statistic df = 1028.
Inflammatory variable thresholds were selected by examining correlations between all thresholds for each PRS and 5-plus years of GrimAge, controlling for the top five principal components. We selected those PRS that survived false discovery rate (FDR).
asignificant at the 0.05 level, determined by a two-sided test.
bsignificant at the <0.001 level, determined by a two-sided test.
Multivariate Logistic regression
Table 3 reports main and interaction effects of biomarker PRS and psychosocial variables on accelerated GrimAge estimated using a multivariate logistic regression model.
Table 3.
Main and interaction effects of PRS and psychosocial variables on accelerated GrimAge.
| Wald Χ2 (df = 1) | p-value | Odds Ratio | Upper 95% CI | Lower 95% CI | |
|---|---|---|---|---|---|
| Main effects | |||||
| Apolipoprotein B PRS | 5.10 | 0.024* | 0.48 | 0.25 | 0.91 |
| High density lipoprotein PRS | 0.42 | 0.52 | 1.23 | 0.66 | 2.32 |
| Gamma-glutamyl transferase PRS | 7.67 | 0.006* | 2.37 | 1.29 | 4.37 |
| Openness to experience | 6.67 | 0.010* | 0.73 | 0.57 | 0.93 |
| Gratitude | 0.01 | 0.91 | 0.99 | 0.78 | 1.24 |
| Days of exercise per week | 6.11 | 0.013* | 0.88 | 0.80 | 0.97 |
| Marital/cohabitation status | 1.23 | 0.27 | 0.73 | 0.42 | 1.27 |
| Lifetime substance use disorder | 8.57 | 0.003* | 2.09 | 1.28 | 3.43 |
| Cumulative trauma burden | 1.01 | 0.32 | 1.05 | 0.96 | 1.14 |
| Interaction effects | |||||
| Cumulative trauma burden x gamma-glutamyl transferase PRS | 0.24 | 0.62 | 0.98 | 0.91 | 1.06 |
| Cumulative trauma burden x high density lipoprotein PRS | 4.77 | 0.029* | 0.91 | 0.84 | 0.99 |
| Cumulative trauma burden x apolipoprotein B PRS | 1.69 | 0.19 | 1.06 | 0.97 | 1.15 |
| Marital/cohabitation status x gamma-glutamyl transferase PRS | 6.30 | 0.012* | 0.52 | 0.31 | 0.87 |
| Marital/cohabitation status x high density lipoprotein PRS | 0.90 | 0.34 | 0.79 | 0.48 | 1.29 |
| Marital/cohabitation status x apolipoprotein B PRS | 0.41 | 0.52 | 1.19 | 0.70 | 2.00 |
| Sleep quality x gamma-glutamyl transferase PRS | 0.47 | 0.49 | 0.92 | 0.73 | 1.17 |
| Sleep quality x high density lipoprotein PRS | 11.61 | 0.001* | 1.57 | 1.21 | 2.04 |
| Sleep quality x apolipoprotein B PRS | 1.06 | 0.30 | 0.87 | 0.66 | 1.14 |
| Days of exercise per week x gamma-glutamyl transferase PRS | 0.11 | 0.74 | 0.98 | 0.90 | 1.08 |
| Days of exercise per week x high density lipoprotein PRS | 0.05 | 0.83 | 1.01 | 0.91 | 1.12 |
| Days of exercise per week x apolipoprotein B PRS | 9.37 | 0.002* | 1.17 | 1.06 | 1.29 |
| Openness to experience x gamma-glutamyl transferase PRS | 1.40 | 0.24 | 0.86 | 0.67 | 1.11 |
| Openness to experience x high density lipoprotein PRS | 0.41 | 0.52 | 1.09 | 0.84 | 1.41 |
| Openness to experience x apolipoprotein B PRS | 0.02 | 0.90 | 1.02 | 0.79 | 1.32 |
| Lifetime substance use disorder x gamma-glutamyl transferase PRS | 0.09 | 0.77 | 0.94 | 0.60 | 1.46 |
| Lifetime substance use disorder x high density lipoprotein PRS | 0.04 | 0.84 | 1.05 | 0.66 | 1.67 |
| Lifetime substance use disorder x apolipoprotein B PRS | 8.14 | 0.004* | 0.50 | 0.32 | 0.81 |
| Gratitude x gamma-glutamyl transferase PRS | 0.00 | 0.99 | 1.00 | 0.79 | 1.27 |
| Gratitude x high density lipoprotein PRS | 7.00 | 0.008* | 0.74 | 0.58 | 0.92 |
| Gratitude x apolipoprotein B PRS | 1.14 | 0.29 | 1.14 | 0.90 | 1.45 |
Nagelkerke R2 = 0.13 for the full model. Covariates included income, sum of medical conditions, combat status and top five principal components. Inflammatory variables were standardized by z-scoring.
*significant at the p ≤ 0.023 FDR-corrected level.
Main effects of polygenic risk and psychosocial factors
Each standard deviation increase in apolipoprotein B PRS was associated with a 52% decrease in odds of 5+ years accelerated GrimAge (95% CI = 25 to 91%). Conversely, higher gamma-glutamyl transferase PRS was associated with increased odds of higher GrimAge, with each SD increase in this PRS increasing the odds of accelerated GrimAge by 137% (95% CI = 29 to 337%).
A greater number of days of exercise and openness to experience were associated with decreased odds of accelerated GrimAge (exercise OR = 0.88, 95% CI = 0.80–0.97; openness OR = 0.073, 95% CI = 0.57–0.93), whereas the presence of a lifetime substance use disorder was associated with increased odds (OR = 2.09, 95% CI = 1.28–3.43). Other psychosocial, lifestyle, and health variables had no statistically significant effects (see Table 3).
Moderating effects of psychosocial factors on the relationship between inflammation PRS and GrimAge
Psychosocial factors moderated the strength of the relationship between inflammation PRS’ and GrimAge. Overall, the correlations between the inflammation PRS’ and GrimAge were stronger in individuals with higher trauma, unmarried, have poor sleep, do less exercise, report low gratitude, or have a substance use disorder.
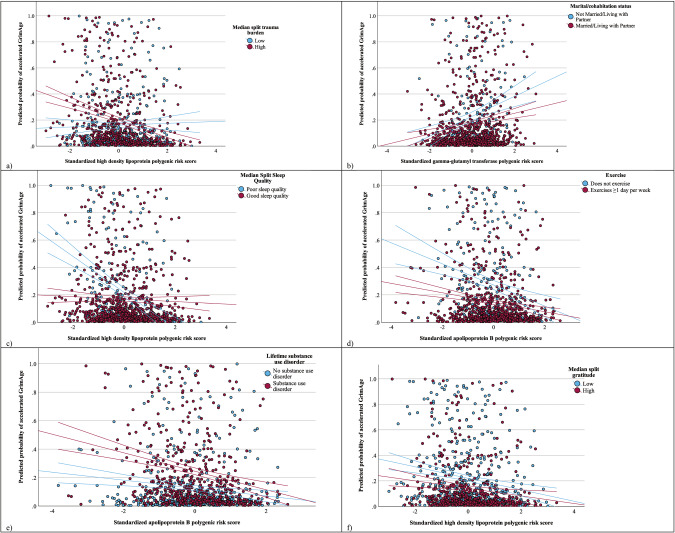
Firstly, we observed a trauma burden-by-HDL PRS interaction (OR = 0.91, 95% CI = 0.84–0.99). Specifically, the relationship between HDL PRS and GrimAge was stronger for individuals with higher cumulative trauma burden. Gamma-glutamyl transferase PRS by marital status interaction (OR = 0.52, 95% CI = 0.31–0.87) indicated that being unmarried strengthened the association between this PRS and accelerated GrimAge compared with Veterans who were married. Similarly, worse sleep quality strengthened the negative relationship between HDL PRS and odds of accelerated GrimAge (OR = 1.57, 95% CI = 1.21–2.04). We also found a statistically significant interaction between days of exercise and apolipoprotein B PRS (OR = 1.17, 95% CI = 1.06–1.29). This interaction reflected that the relationship between apolipoprotein B and accelerated GrimAge was strengthened by reporting fewer days of exercise (Fig. 1). The relationship between apolipoprotein B PRS and GrimAge was stronger in the context of a lifetime substance use disorder (OR = 0.50, 95% CI = 0.32–0.81). Lastly, lower levels of self-reported dispositional gratitude strengthened the negative correlation between HDL PRS and accelerated GrimAge (OR = 0.74, 95% CI = 0.58–0.92).
Fig. 1. Scatterplots for PRS x environment interactions.
Each illustrates the interactive effects between standardized polygenic risk score for (a) HDL PRS and trauma burden (median split); b gamma-glutamyl transferase PRS and marital status; c HDL PRS and sleep quality (median split); d apolipoprotein B PRS and 1 or more days of exercise; e apolipoprotein B PRS and lifetime substance abuse disorder; and f HDL PRS and median split self-reported gratitude score on predicted probability of accelerated GrimAge.
Post-hoc analyses on specific trauma types
To identify whether specific trauma types were driving any inflammatory PRS by lifetime trauma interactions, we entered the following individual trauma types from the Trauma History Screen–(see Supplementary Table 1)– into the model. Two statistically significant interactions emerged. Firstly, there was a statistically significant interaction between HDL and military trauma (Wald Χ2 (df = 1) = 4.06, p = 0.04, OR = 44% (95% CI = 12–68%). This relationship was such that the negative relationship between HDL PRS and accelerated GrimAge was stronger for Veterans who endorsed a military trauma than those who denied a military trauma. Similarly, there was an apolipoprotein B PRS interaction with sudden death of a close family member or friend trauma exposure. Only Veterans who endorsed this trauma showed a relationship between apolipoprotein B PRS and accelerated GrimAge, whereas Veterans who denied this trauma did not should this relationship (Wald Χ2 (df = 1) = 7.37, p = 0.007, OR = 87% (95% CI = 19 to 194%)).
Gene-set enrichment and drug repurposing
Considering Apolipoprotein B, Gamma-glutamyl transferase, and HDL PRS, 87, 6591, and 5557 genes were identified in the gene-based analysis, respectively (Supplementary Tables 3, 4, and 5, respectively). Using these genes as input for the pathway analysis, 516, 109, and 142 overrepresented pathways reached the Bonferroni-significant threshold accounting for the number of pathways tested (Supplementary Tables 6, 7, and 8, respectively). Due to the high number of Bonferroni-significant enrichments, we applied REVIGO [75] to remove redundant gene ontologies. After clumping the redundant terms, 267, 48, and 76 pathways were identified with respect to Apolipoprotein B, Gamma-glutamyl transferase, and HDL PRS, respectively (Supplementary Tables 9, 10, and 11). For Apolipoprotein B, the strongest Bonferroni-significant GO enrichments were related to reproduction (GO:0000003 p = 1.77 × 10−28), cell cycle processes (e.g., GO:0022402 p = 8.04 × 10−20, GO:0000278 p = 4.06 × 10−11), and lipid metabolism (e.g., GO:0008203 p = 1.77 × 10−28, GO:0006641 p = 8.04 × 10−20). For Gamma-glutamyl transferase, the strongest Bonferroni-significant significant enrichments included immune system process (GO:0002376 p < 5.86 × 10−308), regulation of different metabolic processes (e.g., GO:0006796 p < 5.86 × 10−308), and cellular processes (e.g., GO:0007049 p < 5.86 × 10−308). For HDL, the strongest Bonferroni-significant enrichments included reproduction (GO:0000003 p < 4.82 × 10−307), regulation of different metabolic processes (e.g., GO:0009892 p < 4.82 × 10−307), and protein regulation (e.g., GO:0006357 p < 4.82 × 10−307).
The semantically reduced GO terms were entered in the drug repurposing analysis. For Apolipoprotein B, four compounds survived Bonferroni multiple testing correction: molindone, an antipsychotic (p = 8.14 × 10−6, EScore = −0.986); phenoxybenzamine, an antihypertensive (p = 1.11 × 10−5, EScore = 0.743); metitepine, a psychotropic agent (p = 2.74 × 10−5, EScore = 0.979); and chloropyramine, an antihistamine (p = 3.29 × 10−5, EScore = 0.977). For Gamma-glutamyl transferase, acebutolol, a beta-blocker (p = 8.85 × 10 − 6, EScore = 0.608) remained significant after Bonferroni multiple testing correction. For HDL, Prestwick-1100 (p = 2.25 × 10−6, EScore = 0.992); oleandomycin, a macrolide antibiotic (p = 3.38 × 10−6, EScore = 0.991); quinostatin, a lipid lowering compound (p = 9.43 × 10−6, EScore = 0.394); flunixin, a nonsteroidal anti-inflammatory drug (p = 1.47 × 10−5, EScore = 0.983); doxylamine, an antihistamine (negative enrichment; p = 1.47 × 10−5, EScore = −0.385); metitepine, a psychotropic agent (p = 2.74 × 10−5, EScore = 0.979); phensuximide, an anticonvulsant (p = 2.74 × 10−5, EScore = 0.979); and alexidine, an antimicrobial drug (p = 3.55 × 10−5, EScore = 0.372).
Discussion
To our knowledge, the current study is the first to (i) use PRS to estimate the association between genetic predisposition for several selected inflammatory biomarkers on accelerated epigenetic aging and (ii) examine the role of psychosocial variables in moderating these relationships. We found that PRS for HDL (lower), gamma-glutamyl transferase (higher), and apolipoprotein B (lower), were associated with accelerated GrimAge. The former two inflammatory variables were each associated with the predicted direction of GrimAge (Table 1). However, contrary to our hypotheses, we observed an inverse association between apolipoprotein B PRS and accelerated GrimAge. Though this finding not align with our hypotheses, it is consistent with findings from a genetically informed study in UKB that showed that higher apolipoprotein B increases risk of heart disease and stroke, shortening lifespan [76]. Accordingly, the negative association observed in the NHRVS cohort may be due to survival bias, where individuals with high apolipoprotein B PRS and accelerated GrimAge may be underrepresented due to their increased mortality rate in this age group. Our findings build upon work examining circulatory inflammation markers, which are subject to unreliability and issues of reverse causation [28, 29]. Our findings contribute to this literature by showing that the association between inflammation and accelerated biological aging may, at least in part, be explained by genetic predisposition to inflammation.
In addition, we add to extant work, providing the first known evidence of how inflammatory markers may interact with psychosocial factors in predicting accelerated epigenetic aging. We found evidence of psychosocial moderators of HDL, gamma-glutamyl transferase, and apolipoprotein B PRS. First, the association between increasing gamma-glutamyl transferase and accelerated GrimAge was stronger in Veterans who were unmarried/unpartnered relative to those who were married/partnered. Second, the presence of substance use disorder and reduced physical exercise strengthened the association of reduced apolipoprotein B PRS with accelerated GrimAge. Third, the relationship between lower HDL PRS and accelerated GrimAge was stronger in Veterans with poor sleep quality, lower gratitude, and higher cumulative trauma burden.
When specific trauma types were examined, we found interactions between military trauma by HDL PRS. U.S. Veterans have overall worse general health, greater numbers of health risk behaviors, and more chronic health conditions compared to civilians [77, 78]. The interaction between military trauma and HDL on accelerated epigenetic aging may be one factor that may account for these health disparities. Second, there was an interaction between sudden death of a close family member or friend by apolipoprotein PRS, suggesting a possible interplay between this biomarker and grief in relation to epigenetic aging. Grief has been previously reported to be associated with altered inflammatory processes [79].
The three PRS variables that emerged in main effects or interactions are associated with a variety of age-related morbidities and mortality, potentially providing insight into overall transdiagnostic biological aging vs. disease-specific biological aging. Gamma-glutamyl transferase is an enzyme needed for glutathione metabolism and has a role in antioxidant defense systems and inflammation. Gamma-glutamyl transferase has been associated with cardiovascular risk, diabetes, oxidative stress, excessive alcohol consumption, liver disease, and all-cause mortality [30–37]. Consistent with these findings, we showed that higher PRS for this variable was associated with greater odds of accelerated GrimAge. This PRS was enriched for genes involved in immune system processes, regulation of different metabolic processes, and cellular processes. HDL has a role in absorbing cholesterol and transporting it to the liver to be removed from the body, with high levels of HDL reducing risk of cardiovascular disease and stroke, and HDL having a role in female and male fertility [38–42]. In line with this role, we found that decreasing levels of HDL PRS were associated with greater odds of accelerated GrimAge. HDL PRS was enriched for genes involved in reproduction, regulation of different metabolic processes, and protein regulation.
Our HDL and gamma-glutamyl transferase findings highlight that certain psychosocial and behavior variables studied may moderate the relationship between genetic predisposition to inflammation and accelerated GrimAge. For instance, both traumatic stress and accelerated DNAm age have been related to increased inflammation in the context of diminished immune response [9, 15–19]. In this regard, one study found that many epigenetic clock CpG sites were located within glucocorticoid response elements, and that their methylation changed with dexamethasone treatment [80]. Thus, stress might influence epigenetic clocks via shared sensitivity to glucocorticoid modulation [80, 81], with implications for immune functioning. This is because the effects of hypothalamic-pituitary-adrenal axis functioning are implicated in immune function [82]. Consequently, immune system dysregulation and inflammation may also contribute to the relationship between trauma and accelerated epigenetic aging [9, 10]. Furthermore, sleep is thought to have a role in immune response, altering inflammatory and immune response profiles, and sleep disturbance has been linked with increased inflammation to the detriment of antiviral immune responses [20].
Using exploratory drug repurposing analyses, we identified drugs that target pathways perturbated in genes associated with inflammatory and immune processes. Several antihistamines, two anti-hypertensives (including a beta blocker and phenoxybenzamine), an anti-inflammatory drug, and drugs that target the immune response, such as antimicrobial and antibiotic drugs emerged for one or more of the PRS in these analyses. Interestingly, the antihistamine doxylamine emerged in the drug repurposing analysis for HDL, which aligns with our previous finding that genes implicated in PTSD PRS are perturbated by this drug [83]. Two drugs with antipsychotic properties—metitepine and molindone—were perturbated for apolipoprotein B and HDL PRS, respectively, which is consistent with other work showing that antipsychotic drugs (e.g., clozapine) are associated with up to 7-year epigenetic age deceleration in adult males [84] and in a cell culture model [85]. Taken together, these findings suggest future avenues of research into candidate drugs that may help mitigate epigenetic aging.
Our findings have implications for interventions via psychosocial factors in the interplay between inflammatory markers and accelerated GrimAge. This proposition is supported by the changes observed in accelerated epigenetic aging using lifestyle interventions [86–88]. Various psychosocial factors that emerged in interactions in the present study are potentially modifiable with interventions targeting these variables. For example, sleep and exercise are modifiable with interventions such as cognitive behavioral therapy for insomnia [89] and lifestyle changes. Consistent with this idea, in one randomized controlled trial, CBT for insomnia has been associated with reduced CRP and reduced expression of genes encoding proinflammatory markers [90].
Although our findings expand our knowledge regarding the interplay among inflammation, psychosocial factors, and biological aging, we must acknowledge several limitations. First, like most studies of accelerated epigenetic aging, our study is cross-sectional. We cannot accurately model how the interaction effects between inflammatory PRS, and psychosocial factors can modify accelerated GrimAge. This limits our ability to determine whether interventions targeting the variables that emerged in this study will change GrimAge. Longitudinal studies in large samples are needed to examine whether interventions targeting factors such as sleep, inflammatory markers, and gratitude are associated with a subsequent deceleration of GrimAge and whether these changes are linked with corresponding changes in risk for physical health morbidities and mortality.
Second, given that most U.S. Veterans are older European-Americans men, we restricted analyses to EUR male NHRVS participants due to the limited sample size of other ancestry groups in our cohort. Further, well-powered studies are needed to examine correlates of GrimAge in female and non-EA samples [91]. Additionally, the characteristics of the study population may have introduced survival bias when investigating population groups with high mortality risk in the age range targeted.
Third, we assessed inflammation biomarkers via PRS modeling to investigate their association with accelerated aging. Although UKB GWAS can generate highly predictive biomarker PRS [92], the lack of information about inflammatory biomarkers in the NHRVS cohort did not permit us to triangulate the effects on accelerated GrimAge to confirm possible causal effects. Relatedly, it should be noted that the GrimAge includes 7 DNAm-based surrogate markers of plasma proteins tied to mortality risk, including adrenomedullin levels (DNAm ADM), beta-2 microglobulin (DNAm B2M), cystatin C (DNAm Cystatin C), growth differentiation factor 15 (DNAm GDF-15), leptin (DNAm Leptin), plasminogen activation inhibitor 1 (DNAm PAI-1), issue inhibitor metalloproteinase 1 (DNAm TIMP-1). Except for Cystatin C (which did not survive multiple correction in the present study and thus is not included in the final set of findings), none of the surrogate markers included in the derivation of GrimAge are included in the present study. Nonetheless, it is possible that there is some circularity in predicting GrimAge from inflammatory PRS, though the correlations between these were generally small (r’s = 0.01 to 0.12, suggesting <1.5% overlapping variance). This is likely due to the fact that the PRS investigated reflect the individual predisposition to changes in inflammatory biomarkers while epigenetic associations with inflammatory biomarkers should mostly reflect the consequences of changes due to inflammatory processes. Last, it should be acknowledged that methylation patterns are cell-type-specific, necessitating future work to compare epigenetic clocks built from different tissues.
In conclusion, the current study is the first, to our knowledge, to examine how psychosocial and inflammatory variables may interact in relation to accelerated GrimAge. Results identified a range of potentially modifiable factors that interact with three inflammatory PRS markers—HDL, apolipoprotein B, and gamma-glutamyl transferase—to be associated with accelerated GrimAge. Specifically, our findings suggest that PRS for inflammation derived from the UK Biobank interact with cumulative trauma burden, marital/cohabitation status, sleep quality, physical exercise, substance use disorder, and gratitude to be associated with accelerated GrimAge. A notable strength of our study includes the consideration of a wide range of inflammatory markers in epigenetic aging, whereas previous work has mainly been restricted to CRP, IL-6, and TNF-alpha [6, 23–27]. Additionally, using PRS instruments, we circumvented possible biases due to environmental confounders and possible reverse causation. Further, using gene enrichment, we identified anti-inflammatory and antihistamine drugs, laying the groundwork for future work to test these drugs as potential forestallers of epigenetic age. Additional research is needed to evaluate the effects of these drugs on accelerated GrimAge using longitudinal designs; evaluate the efficacy of interventions designed to target the modifiable factors identified here, such as sleep and exercise; and replicate these findings in more race- and ethnic-, and sex-diverse samples.
Supplementary information
Combined supplementary text, table, and figure
Author contributions
Formal analysis: AJFT, DK, RP, and SN; Investigation: RHP; Resources: RHP; Writing – original draft: AJFT, DK, and RHP; Visualization: AJFT, DK, and RHP; Writing – review and editing: RHP, JK, JG, JLMO, SN, CA, RP, and BCM; Supervision: RHP; Funding acquisition: RHP; Conceptualization: JK, AJFT, and RHP; Methodology: RP and JG.
Funding
RP would like to acknowledge the following funding sources: Horizon 2020 Marie Sklodowska-Curie Individual Fellowship (101028810), the National Institutes of Drug Abuse (R33 DA047527), the National Institute of Mental Health (RF1 MH132337).
Competing interests
This manuscript contains original research, has not been published elsewhere and has not been submitted simultaneously for publication elsewhere. We have the following disclosures to report: JG is named as an inventor on PCT patent application #15/878,640 entitled: “Genotype-guided dosing of opioid agonists,” filed January 24, 2018. JG and RP are paid for their editorial work on the journal Complex Psychiatry. RP received a research grant from Alkermes. JHK has served as a scientific consultant to the following companies (the Individual Consultant Agreements listed are less than $5000 per year): AMGEN; AstraZeneca Pharmaceuticals; Bigen, Idec, MA; Biomedisyn Corporation; Forum Pharmaceuticals; Janssen Research & Development; Otsuka America Pharmaceutical, Inc.; Sunovion Pharmaceuticals, Inc.; Takeda Industries; Taisho Pharmaceutical Co., Ltd. He is on the Scientific Advisory Board for the following companies: Biohaven Pharmaceuticals; Blackthorn Therapeutics,Inc.; Lohocla Research Corporation; Luc Therapeutics, Inc.; Pfizer Pharmaceuticals; TRImaran Pharma. He holds stock in Biohaven Pharmaceuticals Medical Sciences and stock options in Blackthorn Therapeutics, Inc. and Luc Therapeutics, Inc. He is the editor of Biological Psychiatry (Income greater than $10,000). CGA has served as a consultant and/or on advisory boards for Aptinyx, Genentech, Janssen, Psilocybin Labs, Lundbeck, Guidepoint, and FSV7, and as editor of Chronic Stress for Sage Publications, Inc. He also filed a patent for using mTORC1 inhibitors to augment the effects of antidepressants (Aug 20, 2018). AJFT, BCM, DK, RHP, SN, and JLMO reported no biomedical financial interest.
Footnotes
The original online version of this article was revised: the author name Sheila Nagamatsu was incorrectly given as ‘Sheila Nagamastu’.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
1/5/2024
A Correction to this paper has been published: 10.1038/s41386-023-01786-y
Supplementary information
The online version contains supplementary material available at 10.1038/s41386-023-01747-5.
References
- 1.Kennedy BK, Berger SL, Brunet A, Campisi J, Cuervo AM, Epel ES, et al. Aging: a common driver of chronic diseases and a target for novel interventions. Cell. 2014;159:709. doi: 10.1016/j.cell.2014.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–67. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:1–20. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCrory C, Fiorito G, Hernandez B, Polidoro S, O’Halloran AM, Hever A, et al. GrimAge outperforms other epigenetic clocks in the prediction of age-related clinical phenotypes and all-cause mortality. J Gerontol A Biol Sci Med Sci. 2021;76:741–9. doi: 10.1093/gerona/glaa286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oblak L, van der Zaag J, Higgins-Chen AT, Levine ME, Boks MP. A systematic review of biological, social and environmental factors associated with epigenetic clock acceleration. Ageing Res Rev. 2021;69:101348. doi: 10.1016/j.arr.2021.101348. [DOI] [PubMed] [Google Scholar]
- 6.Lu AT, Quach A, Wilson JG, Reiner AP, Aviv A, Raj K, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY) 2019;11:303. doi: 10.18632/aging.101684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, Ploner A, Wang Y, Magnusson PK, Reynolds C, Finkel D, et al. Longitudinal trajectories, correlations and mortality associations of nine biological ages across 20-years follow-up. Elife. 2020;9:e51507. doi: 10.7554/eLife.51507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamman AJF, Nagamatsu S, Krystal JH, Gelernter J, Montalvo-Ortiz JL, Pietrzak RH. Psychosocial factors associated with accelerated GrimAge in male U.S. military veterans. Am J Geriatr Psychiatry. 2023;31:97–109. doi: 10.1016/j.jagp.2022.09.002. [DOI] [PubMed] [Google Scholar]
- 9.Wolf EJ, Logue MW, Morrison FG, Wilcox ES, Stone A, Schichman SA, et al. Posttraumatic psychopathology and the pace of the epigenetic clock: a longitudinal investigation. Psychol Med. 2019;49:791–800. doi: 10.1017/S0033291718001411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Declerck K, Berghe WV. Back to the future: epigenetic clock plasticity towards healthy aging. Mech Ageing Dev. 2018;174:18–29. doi: 10.1016/j.mad.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 11.Baylis D, Bartlett DB, Patel HP, Roberts HC. Understanding how we age: insights into inflammaging. Longev Healthspan. 2013;2:1–8. doi: 10.1186/2046-2395-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dugué P-A, Hodge AM, Ulvik A, Ueland PM, Midttun Ø, Rinaldi S, et al. Association of markers of inflammation, the kynurenine pathway and B vitamins with age and mortality, and a signature of inflammaging. J Gerontol A Biol Sci Med Sci. 2022;77:826–36. doi: 10.1093/gerona/glab163. [DOI] [PubMed] [Google Scholar]
- 13.Ziv-Baran T, Shenhar-Tsarfaty S, Etz-Hadar I, Goldiner I, Gottreich A, Alcalay Y, et al. The ability of the wide range CRP assay to classify individuals with low grade inflammation into cardiovascular risk groups. Clin Chim Acta. 2017;471:185–90. doi: 10.1016/j.cca.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 14.Miller AH. Beyond depression: the expanding role of inflammation in psychiatric disorders. World Psychiatry. 2020;19:108. doi: 10.1002/wps.20723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sommershof A, Aichinger H, Engler H, Adenauer H, Catani C, Boneberg E-M, et al. Substantial reduction of naive and regulatory T cells following traumatic stress. Brain Behav Immun. 2009;23:1117–24. doi: 10.1016/j.bbi.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 16.Miller MW, Lin AP, Wolf EJ, Miller DR. Oxidative stress, inflammation, and neuroprogression in chronic PTSD. Harv Rev Psychiatry. 2018;26:57. doi: 10.1097/HRP.0000000000000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quach A, Levine ME, Tanaka T, Lu AT, Chen BH, Ferrucci L, et al. Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging (Albany NY) 2017;9:419. doi: 10.18632/aging.101168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolf EJ, Maniates H, Nugent N, Maihofer AX, Armstrong D, Ratanatharathorn A, et al. Traumatic stress and accelerated DNA methylation age: a meta-analysis. Psychoneuroendocrinology. 2018;92:123–34. doi: 10.1016/j.psyneuen.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horsburgh S, Robson-Ansley P, Adams R, Smith C. Exercise and inflammation-related epigenetic modifications: focus on DNA methylation. Exerc Immunol Rev. 2015;21:26–41. [PubMed] [Google Scholar]
- 20.Irwin MR. Sleep and inflammation: partners in sickness and in health. Nat Rev Immunol. 2019;19:702–15. doi: 10.1038/s41577-019-0190-z. [DOI] [PubMed] [Google Scholar]
- 21.Namba MD, Leyrer-Jackson JM, Nagy EK, Olive MF, Neisewander JL. Neuroimmune mechanisms as novel treatment targets for substance use disorders and associated comorbidities. Front Neurosci. 2021;15:427. doi: 10.3389/fnins.2021.650785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costello EJ, Copeland WE, Shanahan L, Worthman CM, Angold A. C-reactive protein and substance use disorders in adolescence and early adulthood: a prospective analysis. Drug Alcohol Depend. 2013;133:712–7. doi: 10.1016/j.drugalcdep.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY) 2018;10:573. doi: 10.18632/aging.101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irvin MR, Aslibekyan S, Do A, Zhi D, Hidalgo B, Claas SA, et al. Metabolic and inflammatory biomarkers are associated with epigenetic aging acceleration estimates in the GOLDN study. Clin Epigenetics. 2018;10:9. doi: 10.1186/s13148-018-0481-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevenson AJ, McCartney DL, Harris SE, Taylor AM, Redmond P, Starr JM, et al. Trajectories of inflammatory biomarkers over the eighth decade and their associations with immune cell profiles and epigenetic ageing. Clin Epigenetics. 2018;10:10. doi: 10.1186/s13148-018-0585-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cribb L, Hodge AM, Yu C, Li SX, English DR, Makalic E, et al. Inflammation and epigenetic aging are largely independent markers of biological aging and mortality. J Gerontol A Biol Sci Med Sci. 2022;77:2378–86. doi: 10.1093/gerona/glac147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hawn SE, Zhao X, Sullivan DR, Logue M, Fein-Schaffer D, Milberg W, et al. For whom the bell tolls: psychopathological and neurobiological correlates of a DNA methylation index of time-to-death. Transl Psychiatry. 2022;12:406. doi: 10.1038/s41398-022-02164-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verschoor CP, Vlasschaert C, Rauh MJ, Paré G. A DNA methylation based measure outperforms circulating CRP as a marker of chronic inflammation and partly reflects the monocytic response to long‐term inflammatory exposure: A Canadian longitudinal study of aging analysis. Aging Cell. 2023;22:e13863. doi: 10.1111/acel.13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shields GS, Slavich GM, Perlman G, Klein DN, Kotov R. The short-term reliability and long-term stability of salivary immune markers. Brain Behav Immun. 2019;81:650–4. doi: 10.1016/j.bbi.2019.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mason NL, Szabo A, Kuypers KP, Mallaroni PA, de la Torre R, Reckweg JT, et al. Psilocybin induces acute and persisting alterations in immune status in healthy volunteers: An experimental, placebo-controlled study. Brain Behav Immun. 2023;114:299–310. [DOI] [PubMed]
- 31.Mellner C, Dahlen M, Simonsson O. Association between lifetime classic psychedelic use and sick leave in a population-based sample. Int J Environ Res Public Health. 2022;19:11353. doi: 10.3390/ijerph191811353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee D-H, Ha M-H, Kim J-H, Christiani D, Gross M, Steffes M, et al. Gamma-glutamyltransferase and diabetes—a 4 year follow-up study. Diabetologia. 2003;46:359–64. doi: 10.1007/s00125-003-1036-5. [DOI] [PubMed] [Google Scholar]
- 33.Jiang S, Jiang D, Tao Y. Role of gamma-glutamyltransferase in cardiovascular diseases. Exp Clin Cardiol. 2013;18:53. [PMC free article] [PubMed] [Google Scholar]
- 34.Lee D-H, Blomhoff R, Jacobs DR. Review is serum gamma glutamyltransferase a marker of oxidative stress? Free Radic Res. 2004;38:535–9. doi: 10.1080/10715760410001694026. [DOI] [PubMed] [Google Scholar]
- 35.Silva IS, Ferraz MLC, Perez RM, Lanzoni VP, Figueiredo VM, Silva AE. Role of γ‐glutamyl transferase activity in patients with chronic hepatitis C virus infection. J Gastroenterol Hepatol. 2004;19:314–8. doi: 10.1111/j.1440-1746.2003.03256.x. [DOI] [PubMed] [Google Scholar]
- 36.Ndrepepa G, Colleran R, Kastrati A. Gamma-glutamyl transferase and the risk of atherosclerosis and coronary heart disease. Clin Chim Acta. 2018;476:130–8. doi: 10.1016/j.cca.2017.11.026. [DOI] [PubMed] [Google Scholar]
- 37.Kazemi-Shirazi L, Endler G, Winkler S, Schickbauer T, Wagner O, Marsik C. Gamma glutamyltransferase and long-term survival: is it just the liver? Clin Chem. 2007;53:940–6. doi: 10.1373/clinchem.2006.081620. [DOI] [PubMed] [Google Scholar]
- 38.Ibrahim HA, Zhu Y, Wu C, Lu C, Ezekwe MO, Liao SF, et al. Selenium-enriched probiotics improves murine male fertility compromised by high fat diet. Biol Trace Elem Res. 2012;147:251–60. doi: 10.1007/s12011-011-9308-2. [DOI] [PubMed] [Google Scholar]
- 39.Sedes L, Thirouard L, Maqdasy S, Garcia M, Caira F, Lobaccaro J-MA, et al. Cholesterol: a gatekeeper of male fertility? Front Endocrinol (Lausanne) 2018;9:369. doi: 10.3389/fendo.2018.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miettinen HE, Rayburn H, Krieger M. Abnormal lipoprotein metabolism and reversible female infertility in HDL receptor (SR-BI)–deficient mice. J Clin Investig. 2001;108:1717–22. doi: 10.1172/JCI13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bacchetti T, Morresi C, Vignini A, Tiano L, Orlando P, Montik N, et al. HDL functionality in follicular fluid in normal-weight and obese women undergoing assisted reproductive treatment. J Assist Reprod Genet. 2019;36:1657–64. doi: 10.1007/s10815-019-01523-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rader DJ, Hovingh GK. HDL and cardiovascular disease. Lancet. 2014;384:618–25. doi: 10.1016/S0140-6736(14)61217-4. [DOI] [PubMed] [Google Scholar]
- 43.Bilski J, Mazur-Bialy A, Wojcik D, Zahradnik-Bilska J, Brzozowski B, Magierowski M, et al. The role of intestinal alkaline phosphatase in inflammatory disorders of gastrointestinal tract. Mediators Inflamm. 2017;2017:9074601. doi: 10.1155/2017/9074601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lallès J-P. Recent advances in intestinal alkaline phosphatase, inflammation, and nutrition. Nutr Rev. 2019;77:710–24. doi: 10.1093/nutrit/nuz015. [DOI] [PubMed] [Google Scholar]
- 45.Sheinenzon A, Shehadeh M, Michelis R, Shaoul E, Ronen O. Serum albumin levels and inflammation. Int J Biol Macromol. 2021;184:857–62. doi: 10.1016/j.ijbiomac.2021.06.140. [DOI] [PubMed] [Google Scholar]
- 46.Don BR, Kaysen G. Poor nutritional status and inflammation: serum albumin: relationship to inflammation and nutrition. In Seminars in dialysis. Oxford, UK: Blackwell Science Inc.; 2004. Vol. 17, pp. 432–37. [DOI] [PubMed]
- 47.Burger D, Dayer J-M. High-density lipoprotein-associated apolipoprotein AI: the missing link between infection and chronic inflammation? Autoimmun Rev. 2002;1:111–7. doi: 10.1016/S1568-9972(01)00018-0. [DOI] [PubMed] [Google Scholar]
- 48.Georgila K, Vyrla D, Drakos E. Apolipoprotein AI (ApoA-I), immunity, inflammation and cancer. Cancers (Basel) 2019;11:1097. doi: 10.3390/cancers11081097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh D, Whooley MA, Ix JH, Ali S, Shlipak MG. Association of cystatin C and estimated GFR with inflammatory biomarkers: the Heart and Soul Study. Nephrol Dial Transplant. 2007;22:1087–92. doi: 10.1093/ndt/gfl744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamada J, Tomiyama H, Yambe M, Koji Y, Motobe K, Shiina K, et al. Elevated serum levels of alanine aminotransferase and gamma glutamyltransferase are markers of inflammation and oxidative stress independent of the metabolic syndrome. Atherosclerosis. 2006;189:198–205. doi: 10.1016/j.atherosclerosis.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 51.Pelosi L, Giacinti C, Nardis C, Borsellino G, Rizzuto E, Nicoletti C, et al. Local expression of IGF‐1 accelerates muscle regeneration by rapidly modulating inflammatory cytokines and chemokines. FASEB J. 2007;21:1393–402. doi: 10.1096/fj.06-7690com. [DOI] [PubMed] [Google Scholar]
- 52.Lang F, Leibrock C, Pandyra AA, Stournaras C, Wagner CA, Föller M. Phosphate homeostasis, inflammation and the regulation of FGF-23. Kidney Blood Press Res. 2018;43:1742–8. doi: 10.1159/000495393. [DOI] [PubMed] [Google Scholar]
- 53.Sokolove J, Johnson DS, Lahey LJ, Wagner CA, Cheng D, Thiele GM, et al. Rheumatoid factor as a potentiator of anti–citrullinated protein antibody–mediated inflammation in rheumatoid arthritis. Arthritis Rheumatol. 2014;66:813–21. doi: 10.1002/art.38307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao C-H, Li H-Y, Yu H-J, Chiang H-S, Lin M-S, Hua C-H, et al. Low serum sex hormone-binding globulin: marker of inflammation? Clin Chim Acta. 2012;413:803–7. doi: 10.1016/j.cca.2012.01.021. [DOI] [PubMed] [Google Scholar]
- 55.Marioni RE, Shah S, McRae AF, Chen BH, Colicino E, Harris SE, et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015;16:1–12. doi: 10.1186/s13059-015-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Christiansen L, Lenart A, Tan Q, Vaupel JW, Aviv A, McGue M, et al. DNA methylation age is associated with mortality in a longitudinal Danish twin study. Aging Cell. 2016;15:149–54. doi: 10.1111/acel.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perna L, Zhang Y, Mons U, Holleczek B, Saum K-U, Brenner H. Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clin Epigenetics. 2016;8:1–7. doi: 10.1186/s13148-016-0228-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tamman AJ, Nagamatsu S, Krystal JH, Gelernter J, Montalvo-Ortiz JL, Pietrzak RH. Psychosocial factors associated with accelerated GrimAge in male US military veterans. Am J Geriatr Psychiatry. 2023;31:97–109. doi: 10.1016/j.jagp.2022.09.002. [DOI] [PubMed] [Google Scholar]
- 59.Nelson BW, Wright DB, Allen NB, Laurent HK. Maternal stress and social support prospectively predict infant inflammation. Brain Behav Immun. 2020;86:14–21. doi: 10.1016/j.bbi.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 60.Runsten S, Korkeila K, Koskenvuo M, Rautava P, Vainio O, Korkeila J. Can social support alleviate inflammation associated with childhood adversities? Nord J Psychiatry. 2014;68:137–44. doi: 10.3109/08039488.2013.786133. [DOI] [PubMed] [Google Scholar]
- 61.Tomfohr LM, Edwards KM, Madsen JW, Mills PJ. Social support moderates the relationship between sleep and inflammation in a population at high risk for developing cardiovascular disease. Psychophysiology. 2015;52:1689–97. doi: 10.1111/psyp.12549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang YC, Schorpp K, Harris KM. Social support, social strain and inflammation: evidence from a national longitudinal study of US adults. Soc Sci Med. 2014;107:124–35. doi: 10.1016/j.socscimed.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fogle BM, Tsai J, Mota N, Harpaz-Rotem I, Krystal JH, Southwick SM, et al. The national health and resilience in veterans study: a narrative review and future directions. Front Psychiatry. 2020;11:538218. doi: 10.3389/fpsyt.2020.538218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Consortium GP. A global reference for human genetic variation. Nature. 2015;526:68. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fuchsberger C, Abecasis GR, Hinds DA. minimac2: faster genotype imputation. Bioinformatics. 2014;31:782–4. doi: 10.1093/bioinformatics/btu704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279–83. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Das S, Forer L, Schonherr S, Sidore C, Locke AE, Kwong A, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–7. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–9. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choi SW, O’Reilly PF. PRSice-2: polygenic risk score software for biobank-scale data. Gigascience. 2019;8:giz082. doi: 10.1093/gigascience/giz082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen W, Zeng Y, Suo C, Yang H, Chen Y, Hou C, et al. Genetic predispositions to psychiatric disorders and the risk of COVID-19. BMC Med. 2022;20:1–9. doi: 10.1186/s12916-022-02520-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mishra A, Macgregor S. VEGAS2: software for more flexible gene-based testing. Twin Res Hum Genet. 2015;18:86–91. doi: 10.1017/thg.2014.79. [DOI] [PubMed] [Google Scholar]
- 72.Napolitano F, Carrella D, Mandriani B, Pisonero-Vaquero S, Sirci F, Medina DL, et al. gene2drug: a computational tool for pathway-based rational drug repositioning. Bioinformatics. 2018;34:1498–505. doi: 10.1093/bioinformatics/btx800. [DOI] [PubMed] [Google Scholar]
- 73.Subramanian A, Narayan R, Corsello S, Peck D, Natoli T, Lu X, et al. A next generation connectivity map: L1000 Platform and the first 1,000,000 profiles. Cell. 2017;171:1437–1452.e17. [DOI] [PMC free article] [PubMed]
- 74.Tamman AJ, Nagamatsu S, Krystal JH, Gelernter J, Montalvo-Ortiz JL, Pietrzak RH. Psychosocial factors associated with accelerated GrimAge in male US military veterans. Am J Geriatric Psychiatry. 2022;31:97–109. [DOI] [PubMed]
- 75.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Richardson TG, Wang Q, Sanderson E, Mahajan A, McCarthy MI, Frayling TM, et al. Effects of apolipoprotein B on lifespan and risks of major diseases including type 2 diabetes: a mendelian randomisation analysis using outcomes in first-degree relatives. Lancet Healthy Longev. 2021;2:e317–e26. doi: 10.1016/S2666-7568(21)00086-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lehavot K, Hoerster KD, Nelson KM, Jakupcak M, Simpson TL. Health indicators for military, veteran, and civilian women. Am J Prev Med. 2012;42:473–80. doi: 10.1016/j.amepre.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 78.Hoerster KD, Lehavot K, Simpson T, McFall M, Reiber G, Nelson KM. Health and health behavior differences: US Military, veteran, and civilian men. Am J Prev Med. 2012;43:483–9. doi: 10.1016/j.amepre.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 79.Fagundes CP, Brown RL, Chen MA, Murdock KW, Saucedo L, LeRoy A, et al. Grief, depressive symptoms, and inflammation in the spousally bereaved. Psychoneuroendocrinology. 2019;100:190–7. doi: 10.1016/j.psyneuen.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zannas AS, Arloth J, Carrillo-Roa T, Iurato S, Röh S, Ressler KJ, et al. Correction to: Lifetime stress accelerates epigenetic aging in an urban, African American cohort: relevance of glucocorticoid signaling. Genome Biol. 2018;19:1–1. doi: 10.1186/s13059-018-1441-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Davis E, Humphreys K, McEwen L, Sacchet M, Camacho M, MacIsaac J, et al. Accelerated DNA methylation age in adolescent girls: associations with elevated diurnal cortisol and reduced hippocampal volume. Transl Psychiatry. 2017;7:e1223–e23. doi: 10.1038/tp.2017.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Silverman MN, Sternberg EM. Glucocorticoid regulation of inflammation and its functional correlates: from HPA axis to glucocorticoid receptor dysfunction. Ann N Y Acad Sci. 2012;1261:55–63. doi: 10.1111/j.1749-6632.2012.06633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tamman AJ, Wendt FR, Pathak GA, Krystal JH, Montalvo-Ortiz JL, Southwick SM, et al. Attachment style moderates polygenic risk for posttraumatic stress in United States military veterans: results from the national health and resilience in veterans study. Biol Psychiatry. 2021;89:878–87. doi: 10.1016/j.biopsych.2020.09.018. [DOI] [PubMed] [Google Scholar]
- 84.Higgins-Chen AT, Boks MP, Vinkers CH, Kahn RS, Levine ME. Schizophrenia and epigenetic aging biomarkers: increased mortality, reduced cancer risk, and unique clozapine effects. Biol Psychiatry. 2020;88:224–35. doi: 10.1016/j.biopsych.2020.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Du J, Nakachi Y, Fujii A, Fujii S, Bundo M, Iwamoto K. Antipsychotics function as epigenetic age regulators in human neuroblastoma cells. Schizophrenia. 2022;8:69. doi: 10.1038/s41537-022-00277-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fitzgerald KN, Hodges R, Hanes D, Stack E, Cheishvili D, Szyf M, et al. Potential reversal of epigenetic age using a diet and lifestyle intervention: a pilot randomized clinical trial. Aging (Albany NY) 2021;13:9419. doi: 10.18632/aging.202913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fiorito G, Caini S, Palli D, Bendinelli B, Saieva C, Ermini I, et al. DNA methylation‐based biomarkers of aging were slowed down in a two‐year diet and physical activity intervention trial: the DAMA study. Aging Cell. 2021;20:e13439. doi: 10.1111/acel.13439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fahy GM, Brooke RT, Watson JP, Good Z, Vasanawala SS, Maecker H, et al. Reversal of epigenetic aging and immunosenescent trends in humans. Aging Cell. 2019;18:e13028. doi: 10.1111/acel.13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cunningham JE, Shapiro CM. Cognitive behavioural therapy for insomnia (CBT-I) to treat depression: a systematic review. J Psychosom Res. 2018;106:1–12. doi: 10.1016/j.jpsychores.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 90.Irwin MR, Olmstead R, Breen EC, Witarama T, Carrillo C, Sadeghi N, et al. Cognitive behavioral therapy and tai chi reverse cellular and genomic markers of inflammation in late-life insomnia: a randomized controlled trial. Biol Psychiatry. 2015;78:721–29. doi: 10.1016/j.biopsych.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Simons RL, Lei M-K, Klopack E, Beach SR, Gibbons FX, Philibert RA. The effects of social adversity, discrimination, and health risk behaviors on the accelerated aging of African Americans: further support for the weathering hypothesis. Soc Sci Med. 2021;282:113169. doi: 10.1016/j.socscimed.2020.113169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sinnott-Armstrong N, Tanigawa Y, Amar D, Mars N, Benner C, Aguirre M, et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat Genet. 2021;53:185–94. doi: 10.1038/s41588-020-00757-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Combined supplementary text, table, and figure