Abstract
The pfcrt allelic type and adjacent microsatellite marker type were determined for 82 Plasmodium falciparum isolates from the Philippines. Mutant pfcrt allelic types P1a and P2a/P2b were dominant in different locations. Microsatellite analysis revealed that P2a/P2b evolved independently in the Philippines, while P1a shared common ancestry with Papua New Guinea chloroquine-resistant parasites.
Mutations in the pfcrt gene have been associated with chloroquine resistance in Plasmodium falciparum (4, 6, 8, 12). Fifteen amino acid mutations at positions 72, 74, 75, 76, 97, 144, 148, 160, 194, 220, 271, 326, 333, 356, and 371 have been identified in pfcrt of chloroquine-resistant (CQR) parasites from various regions (3, 7-9, 15). In general, the CQR isolates from Southeast Asia and Africa have pfcrt alleles with seven to nine mutated codons that are linked to a pattern of C/I/E/T/H(L)/A(F)/L(I)/L/I(T)/S/E/S/T(S)/T/I (boldface italics indicate the mutated codons), from positions 72 to 371, while the CQR parasites from South America and Papua New Guinea (PNG) possess pfcrt alleles with four or five mutated codons forming a pattern of S(C)/M/N/T/H/A/L/L/I/S/Q/D/T/L/R (3, 4, 8, 9, 15). Our previous study identified novel pfcrt allelic types in parasites from Morong, the Philippines, with four or five mutated amino acids linked into a pattern of S(C)/M/N/T/H/T/L/Y/I/A/Q/D/T/I/R. In vitro chloroquine susceptibility testing indicated that these parasites with the novel pfcrt allelic types were CQR (3). Among the mutated codons, A144T and L160Y were exclusively identified in the Philippine isolates, suggesting that CQR pfcrt alleles evolved independently in the Philippines (3). The novel Philippine pfcrt allelic types were named P2a and P2b according to the nomenclature used by Wellems and Plowe (3, 14).
In this study, we analyzed pfcrt allelic types in P. falciparum isolates from several locations within the Philippines to ascertain the geographical distribution of these novel and other pfcrt allelic types. The ancestral origin of the observed CQR pfcrt allelic types was investigated by microsatellite marker analysis (15).
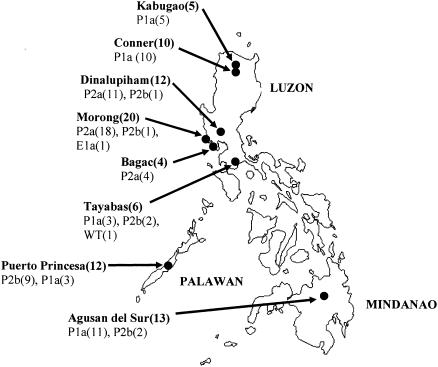
Eighty-two P. falciparum isolates were collected from the following locations in the Philippines between 1989 and 2002 (Fig. 1): Morong (n = 20) (3), Bagac (n = 4), and Dinalupiham (n = 12) in Bataan Province, Central Luzon; Puerto Princesa, Palawan (n = 12), and Tayabas (n = 6) in Quezon Province, South Luzon; Conner (n = 10) and Kabugao (n = 5) in Kalinga-Apayao Province, Northern Luzon; and Agusan del Sur, Mindanao (n = 13). Genomic DNA was extracted using a Wizard Plus Minipreps DNA Purification System (Promega), and DNA fragments covering the 12 known mutations in pfcrt were amplified by PCR (3). PCR products were purified with a NucleoSpin extraction kit (Macherey-Nagel) and sequenced. Microsatellite markers flanking the pfcrt gene, B5M77(−20 kb), 2E10 (−5 kb), PE12A (+6 kb), 2H4 (+22 kb), and PE14F (+106 kb), were amplified and analyzed as previously described (15).
FIG. 1.
Locations in the Philippines where parasite isolates were examined for pfcrt allelic types and microsatellite marker types. Numbers in parentheses following the name of a district/municipality indicate the total number of parasite samples. pfcrt allelic types and numbers detected at each location are indicated below the name of each district/municipality.
Polymorphisms in pfcrt in P. falciparum collected from the Philippines are shown in Table 1. Novel pfcrt allelic types P2a and P2b with mutations at A144T and L160Y were identified in 18/20 and 1/20 Morong isolates (Central Luzon) (3), respectively. P2a was also the major pfcrt allelic type in Bagac (4/4) and Dinalupiham (11/12) in Central Luzon, while P2b was the major type in Puerto Princesa, Palawan (9/12). In comparison, P2b was only identified in a small proportion of the isolates from Tayabas (2/6) in Southern Tagalog and from Agusan del Sur, Mindanao (2/13). Neither P2a nor P2b was detected in isolates from Conner and Kabugao, Northern Luzon, where P1a, the PNG allelic type, dominated. The chloroquine-sensitive wild-type pfcrt was detected in one isolate from Tayabas. Overall, among the 82 Philippine isolates analyzed, 48 (58.5%) possessed the P2a/P2b alleles, 32 (39%) had the P1a allele, and 1 (1.2%) each had the E1a allele and the wild type.
TABLE 1.
pfcrt allelic types and polymorphisms in microsatellite markers flanking pfcrt in P. falciparum samples collected from different districts/municipalities in the Philippines compared to those from other areas
| Origin (total no. of isolates) and sample | No. of isolates | % | MSb marker size (bp)
|
PFCRT (allelic type)a | MS marker size (bp)
|
|||
|---|---|---|---|---|---|---|---|---|
| B5M77 (−20 kb) | 2F10 (−5 kb) | PE12A (+6 kb) | 2H4 (+22 kb) | PE14F (+106 kb) | ||||
| Philippines | ||||||||
| Morong (20) | ||||||||
| PH1 group | 18 | 90 | 149 | 182 | CMNTHTYAQDLR (P2a) | 314 | 228 | 136 |
| PH2 | 1 | 5 | 149 | 182 | SMNTHTYAQDIR (P2b) | 314 | 228 | 136 |
| PH4 | 1 | 5 | 149 | 170 | CIETHALSESTI (E1a) | 314 | 204 | 145 |
| Bagac (4), B1 group | 4 | 100 | 149 | 182 | CMNTHTYAQDLR (P2a) | 314 | 228 | 136 |
| Dinalupiham (12) | ||||||||
| D1 group | 10 | 83 | 149 | 182 | CMNTHTYAQDLR (P2a) | 314 | 228 | 136 |
| D16 | 1 | 8 | 149 | 182 | CMNTHTYAQDLR (P2a) | 314 | 220 | 142 |
| D3 | 1 | 8 | 149 | 182 | SMNTHTYAQDIR (P2b) | 314 | 228 | 136 |
| Palawan (12) | ||||||||
| PAL2 group | 5 | 42 | 149 | 182 | SMNTHTYAQDIR (P2b) | 314 | 228 | 136 |
| PAL12 group | 2 | 17 | 149 | 182 | SMNTHTYAQDIR (P2b) | 314 | 228 | 151 |
| PAL7 | 1 | 8 | 149 | 182 | SMNTHTYAQDIR (P2b) | 314 | 228 | 148 |
| PAL6 | 1 | 8 | 149 | 182 | SMNTHTYAQDIR (P2b) | 314 | 220 | 136 |
| PAL5 group | 2 | 17 | 147 | 194 | SMNTHALSQDLR (P1a) | 328 | 228 | 136 |
| PAL1 | 1 | 8 | 147 | 194 | SMNTHALSQDLR (P1a) | 328 | 228 | 148 |
| Tayabas (6) | ||||||||
| T1 group | 2 | 33 | 149 | 174 | SMNTHALSQDLR (P1a) | 328 | 218 | 148 |
| T4 | 1 | 17 | 149 | 174 | SMNTHALSQDLR (P1a) | 328 | 184 | 148 |
| T2 group | 2 | 33 | 149 | 182 | SMNTHTYAQDIR (P2b) | 314 | 228 | 136 |
| T3 | 1 | 17 | 149 | 176 | CMNKHALAQNIR (CQS) | 328 | 182 | 142 |
| Mindanao (13) | ||||||||
| L104 group | 4 | 31 | 149 | 174 | SMNTHALSQDLR (P1a) | 328 | 184 | 148 |
| L088 group | 2 | 15 | 149 | 174 | SMNTHALSQDLR (P1a) | 328 | 184 | 151 |
| L016 | 1 | 8 | 149 | 174 | SMNTHALSQDLR (P1a) | 328 | 184 | 142 |
| M013 group | 2 | 15 | 149 | 174 | SMNTHALSQDLR (P1a) | 328 | 206 | 148 |
| M007 | 1 | 8 | 149 | 174 | SMNTHALSQDLR (P1a) | 328 | 186 | 148 |
| X039 | 1 | 8 | 149 | 174 | SMNTHALSQDLR (P1a) | 328 | 204 | 148 |
| X098 | 1 | 8 | 149 | 182 | SMNTHTYAQDIR (P2b) | 314 | 202 | 148 |
| X097 | 1 | 8 | 149 | 182 | SMNTHTYAQDIR (P2b) | 314 | 202 | 151 |
| Connor (10) | ||||||||
| C3 group | 7 | 70 | 149 | 174 | SMNTHALSQDLR (P1a) | 328 | 184 | 148 |
| C1 group | 3 | 30 | 149 | 174 | SMNTHALSQDLR (P1a) | 328 | 228 | 136 |
| Kabugao (5), Kab1 group | 5 | 100 | 149 | 174 | SMNTHALSQDLR (P1a) | 328 | 184 | 148 |
| Other areas | ||||||||
| Brazil (1), 7G8 | 1 | 151 | 190 | SMNTHALSQDLR (W1a) | 314 | 194 | 142 | |
| Thailand (5) | ||||||||
| Dd2 group | 2 | 149 | 170 | CIETHALSESTI (E1a) | 314 | 204 | 145 | |
| K1 | 1 | 149 | 170 | CIETHALSESII (E1b) | 314 | 204 | 145 | |
| TM93-C1088 | 1 | 149 | 170 | CIETLALSESTI (E1c) | 314 | 204 | 145 | |
| C2B | 1 | 149 | 170 | CIETHALSESTI (E1a) | 314 | 184 | 148 | |
| Solomon (4) | ||||||||
| N18 group | 2 | 149 | 174 | SMNTHALSQDLR (P1a) | 328 | 184 | 142 | |
| N70 group | 2 | 147 | 174 | SMNTHALSQDLR (P1a) | 328 | 184 | 142 | |
| PNG (6) | ||||||||
| AN001 group | 4 | 149 | 174 | SMNTHALSQDLR (P1a) | 328 | 184 | 142 | |
| AN018 group | 2 | 149 | 174 | SMNTHALSQDLR (P1a) | 328 | 192 | 139 | |
Genotyping using msp1, msp2, and glurp (10) was performed to examine the heterogeneity of the isolates on loci not linked with pfcrt. Various banding patterns were observed among isolates with identical novel P2a and P2b pfcrt allelic types, indicating the isolates were diverse rather than clonal (data not shown). This is highlighted in 18 Morong isolates, where nine distinct banding patterns identified for msp1, msp2, and glurp corresponded to the P2a pfcrt allele.
The dissemination of mutant pfcrt allelic types in the Philippines is shown in Fig. 1. P2a was the major type in isolates from Central Luzon, while P2b predominated in those from Palawan. P1a was found to be dominant in isolates from Northern Luzon and Mindanao. Although geographically the provinces of Bataan and Quezon are situated in Luzon, they are well separated by mountain ranges. This may provide an explanation for the dominance of P2a and P1a, respectively, in these provinces. Palawan, though separated by sea, shares a closely related dominant pfcrt allelic type with Central Luzon. The novel mutations in P2a and P2b alleles (A144T and L160Y) were not found in a wild-type pfcrt gene, indicating that these mutations in the CQR isolates were not inherited from chloroquine-susceptible parasites in the Philippines. In addition, none of the Philippine isolates with A144T and L160Y mutations (n = 48) carried the A220S mutation very commonly seen in CQR parasites elsewhere. The mutually exclusive presence of A144T/L160Y with A220S suggests that A144T and L160Y may play a similar role as A220S in CQR.
Five microsatellite markers flanking the pfcrt gene, B5M77, 2E10, PE12A, 2H4, and PE14F (15), were analyzed for all the Philippine isolates included in this study, as well as several standard laboratory lines and isolates from Southeast Asia, Oceania, and South America. As shown in Table 1, all isolates with the P2a pfcrt allele, except for one from Dinalupiham (D16), gave the identical size for all five microsatellite markers, and the banding pattern was unique compared to that in parasites from other regions. While some isolates with the P2b pfcrt allele had microsatellite markers identical to those in P2a, others showed variations in some microsatellite markers. The unique microsatellite marker patterns of P2a/P2b pfcrt allelic types demonstrate that P2a/P2b alleles evolved independently in the Philippines. The similarity between the microsatellite patterns of P2a and P2b and fact that P2b carried an extra mutation (C72S) suggest that the P2b allelic type was derived from P2a. We hypothesize that CQR parasites (P2a) originated in Central Luzon and spread south to Palawan and Mindanao with the addition of an extra mutation in pfcrt (P2b). This is consistent with field observation of chloroquine resistance in Philippines: first documented in Luzon from the early 1970s to the 1980s (2, 5, 11, 13) and subsequently in Palawan (1).
In contrast, the microsatellite marker patterns of the Philippine isolates with P1a pfcrt type closely resembled that of the PNG and the Solomon Islands isolates, suggesting that P1a pfcrt alleles in parasites from the Philippines and Oceania evolved from a common origin and may have spread from neighboring countries. The only Philippine isolate with the E1a pfcrt type (PH4) was identical to Dd2 (Thailand) in microsatellite marker pattern (Table 1). Significantly, this was obtained in close proximity to the site of the main transit camp for Indochinese refugees in the Philippines.
This study confirms that at least two founder events of chloroquine resistance have occurred in the Pacific region, one in PNG and one in the Philippines, and demonstrates that under chloroquine selection pressure, P. falciparum parasites with various genetic backgrounds have developed chloroquine resistance independently by mutating different positions in the pfcrt gene.
Acknowledgments
This research was funded in part by the U.S. Army Medical Research and Materiel Command, Ft. Detrick, MD, and an NIH grant (2RO1 AI047500) and by the National Health and Medical Research Council, Australia, and assisted by AusAID through the Research Institute for Tropical Medicine, Philippines.
The opinions expressed herein are those of the authors and do not necessarily reflect those of the Defence Health Service or any extant policy of the Department of Defence, Australia or United States.
We thank Rouel Go and colleagues from the Malaria Study Group, the Research Institute for Tropical Medicine, Manila, Philippines, for collecting malaria-infected samples in the Philippines.
REFERENCES
- 1.Baird, J. K., M. E. Caneta, S. Masbar, D. G. Bustos, J. A. Abrenica, A. V. Layawen, J. M. Calulut, B. Leksana, and F. S. Wignall. 1996. Survey of resistance to chloroquine of falciparum and vivax malaria in Palawan, The Philippines. Trans. R. Soc. Trop. Med. Hyg. 90:413-414. [DOI] [PubMed] [Google Scholar]
- 2.Buck, R. L., A. K. Alcantara, C. V. Uylangco, and J. H. Cross. 1983. Malaria at San Lazaro Hospital, Manila, Philippines, 1979-1981. Am. J. Trop. Med. Hyg. 32:212-216. [DOI] [PubMed] [Google Scholar]
- 3.Chen, N., D. E. Kyle, C. Pasay, E. V. Fowler, J. Baker, J. M. Peters, and Q. Cheng. 2003. pfcrt allelic types with two novel amino acid mutations in chloroquine-resistant Plasmodium falciparum isolates from the Philippines. Antimicrob. Agents Chemother. 47:3500-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, N., B. Russell, J. Staley, B. Kotecka, P. Nasveld, and Q. Cheng. 2001. Sequence polymorphisms in pfcrt are strongly associated with chloroquine resistance in Plasmodium falciparum. J. Infect. Dis. 183:1543-1545. [DOI] [PubMed] [Google Scholar]
- 5.Clyde, D. F., G. T. Shute, V. C. McCarthy, and R. P. Sangalang. 1971. Characterization of a drug resistant strain of Plasmodium falciparum from the Philippines. J. Trop. Med. Hyg. 74:101-105. [PubMed] [Google Scholar]
- 6.Djimde, A., O. K. Doumbo, J. F. Cortese, K. Kayentao, S. Doumbo, Y. Diourte, A. Dicko, X. Z. Su, T. Nomura, D. A. Fidock, T. E. Wellems, C. V. Plowe, and D. Coulibaly. 2001. A molecular marker for chloroquine-resistant falciparum malaria. N. Engl. J. Med. 344:257-263. [DOI] [PubMed] [Google Scholar]
- 7.Durrand, V., A. Berry, R. Sem, P. Glaziou, J. Beaudou, and T. Fandeur. 2004. Variations in the sequence and expression of the Plasmodium falciparum chloroquine resistance transporter (Pfcrt) and their relationship to chloroquine resistance in vitro. Mol. Biochem. Parasitol. 136:273-285. [DOI] [PubMed] [Google Scholar]
- 8.Fidock, D. A., T. Nomura, A. K. Talley, R. A. Cooper, S. M. Dzekunov, M. T. Ferdig, L. M. Ursos, A. B. Sidhu, B. Naude, K. W. Deitsch, X. Z. Su, J. C. Wootton, P. D. Roepe, and T. E. Wellems. 2000. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol. Cell 6:861-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehlotra, R. K., H. Fujioka, P. D. Roepe, O. Janneh, L. M. Ursos, V. Jacobs-Lorena, D. T. McNamara, M. J. Bockarie, J. W. Kazura, D. E. Kyle, D. A. Fidock, and P. A. Zimmerman. 2001. Evolution of a unique Plasmodium falciparum chloroquine-resistance phenotype in association with pfcrt polymorphism in Papua New Guinea and South America. Proc. Natl. Acad. Sci. USA 98:12689-12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ranford-Cartwright, L. C., J. Taylor, T. Umasunthar, L. H. Taylor, H. A. Babiker, B. Lell, J. R. Schmidt-Ott, L. G. Lehman, D. Walliker, and P. G. Kremsner. 1997. Molecular analysis of recrudescent parasites in a Plasmodium falciparum drug efficacy trial in Gabon. Trans. R. Soc. Trop. Med. Hyg. 91:719-724. [DOI] [PubMed] [Google Scholar]
- 11.Shute, G. T., A. P. Ray, and R. Sangalang. 1972. Preliminary studies on a Philippine strain of Plasmodium falciparum resistant to amodiaquine. J. Trop. Med. Hyg. 75:125-132. [PubMed] [Google Scholar]
- 12.Sidhu, A. B., D. Verdier-Pinard, and D. A. Fidock. 2002. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science 298:210-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valera, C. V., and G. T. Shute. 1976. Preliminary studies on the response of Plasmodium falciparum to chloroquine in the Philippines, with the in vitro technique. Bull. W. H. O. 53:391-398. [PMC free article] [PubMed] [Google Scholar]
- 14.Wellems, T. E., and C. V. Plowe. 2001. Chloroquine-resistant malaria. J. Infect. Dis. 184:770-776. [DOI] [PubMed] [Google Scholar]
- 15.Wootton, J. C., X. Feng, M. T. Ferdig, R. A. Cooper, J. Mu, D. I. Baruch, A. J. Magill, and X. Z. Su. 2002. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature 418:320-323. [DOI] [PubMed] [Google Scholar]