Abstract
Objective
Clear cell renal cell carcinoma (ccRCC) is the most common subtype among renal cell carcinomas and has the worst prognosis, originating from renal tubular epithelial cells. Toll-like receptor 4 (TLR4) plays a crucial role in ccRCC proliferation, infiltration, and metastasis. The aim of this study was to construct a prognostic scoring model for ccRCC based on TLR4 expression heterogeneity and to explore its association with immune infiltration, thereby providing insights for the treatment and prognostic evaluation of ccRCC.
Methods
Using R software, a differential analysis was conducted on normal samples and ccRCC samples, and in conjunction with the KEGG database, a correlation analysis for the clear cell renal cell carcinoma pathway (hsa05211) was carried out. We observed the expression heterogeneity of TLR4 in the TCGA-KIRC cohort and identified its related differential genes (TRGs). Based on the expression levels of TRGs, consensus clustering was employed to identify TLR4-related subtypes, and further clustering heatmaps, principal component, and single-sample gene set enrichment analyses were conducted. Overlapping differential genes (ODEGs) between subtypes were analysed, and combined with survival data, univariate Cox regression, LASSO, and multivariate Cox regression were used to establish a prognostic risk model for ccRCC. This model was subsequently evaluated through ROC analysis, risk factor correlation analysis, independent prognostic factor analysis, and intergroup differential analysis. The ssGSEA model was employed to explore immune heterogeneity in ccRCC, and the performance of the model in predicting patient prognosis was evaluated using box plots and the oncoPredict software package.
Results
In the TCGA-KIRC cohort, TLR4 expression was notably elevated in ccRCC samples compared to normal samples, correlating with improved survival in the high-expression group. The study identified distinct TLR4-related differential genes and categorized ccRCC into three subtypes with varied survival outcomes. A risk prognosis model based on overlapping differential genes was established, showing significant associations with immune cell infiltration and key immune checkpoints (PD-1, PD-L1, CTLA4). Additionally, drug sensitivity differences were observed between risk groups.
Conclusion
In the TCGA-KIRC cohort, the expression of TLR4 in ccRCC samples exhibited significant heterogeneity. Through clustering analysis, we identified that the primary immune cells across subtypes are myeloid-derived suppressor cells, central memory CD4 T cells, and regulatory T cells. Furthermore, we successfully constructed a prognostic risk model for ccRCC composed of 17 genes. This model provides valuable references for the prognosis prediction and treatment of ccRCC patients.
Keywords: TLR4, Renal clear cell carcinoma, Consensus clustering, Prognostic model
1. Introduction
Renal cell carcinoma (RCC) is a malignant tumour originating from renal tubular epithelial cells; among them, clear cell renal cell carcinoma (ccRCC) is the most prevalent subtype, accounting for 80%–90% of all RCC cases [1]. Over the past few decades, the incidence of this tumour has been on the rise [2]. Although the rate of early diagnosis for ccRCC has improved, approximately one-third of patients are already metastatic at the time of diagnosis [3]. In recent years, scholars have delved deeply into the molecular mechanisms and therapeutic strategies of ccRCC. Trac et al. (2022) investigated certain key gene mutations in ccRCC and their associated therapeutic strategies [4], while Che et al. (2022) analysed another set of gene mutations and gene expression changes in ccRCC [5]. While molecular targeted therapies have shown efficacy in other cancers [6], developing targeted treatments for ccRCC remains challenging due to the lack of ideal targets. Moreover, numerous studies have indicated that the postoperative metastasis rate for ccRCC patients can be significant [7]. Some studies have begun to explore immune genes associated with ccRCC, their roles in the tumour immune microenvironment, and their potential value in prognosis prediction [8,9].
TLR4 is a member of the Toll-like receptor family (TLRs) [10]. Recent research has identified a close relationship between TLR4 expression in various tumour cells and tumour development and metastasis [11]. This relationship is particularly significant in prevalent cancers such as lung and breast cancer [12]. In clear cell renal cell carcinoma (ccRCC), TLR4 mRNA expression is elevated and correlates with the differential expression of other genes in ccRCC samples. Higher TLR4 expression is linked to a decreased overall survival rate for ccRCC patients [13]. Furthermore, the specific roles of Toll-like receptors (TLRs) in kidney clear cell carcinoma (KIRC) warrant additional investigation. Studies have shown that when compared to adjacent normal tissues, TLR expression levels in KIRC differ. Notably, TLR3 and TLR4 expression is significantly increased during the early stages of KIRC [14]. Within the renal cancer tumour microenvironment, TLR4 interacts with endogenous ligands such as VEGF and HMGB1, which are released during matrix degradation and play pivotal roles in tumour progression [15].
The Kyoto Encyclopedia of Genes and Genomes (KEGG) database is a bioinformatics database used for studying the genomics, metabolism, and signalling pathways of biological systems. The has05211 pathway describes the molecular mechanisms and pathways of renal cell carcinoma development and progression [16]. This pathway involves various biological processes, including cell proliferation, apoptosis, and angiogenesis, and is related to cell cycle regulatory factors such as Von Hippel–Lindau (VHL) and mesenchymal-epithelial transition factor (MET). Previous studies have explored the role of VHL in clear cell renal cell carcinoma, especially its positive feedback loop with the PDGFRβ signalling pathway [17], as well as the relationship between lncRNA APCDD1L-AS1 and the loss of VHL protein expression [18]. Although these studies have provided in-depth insights into this field, the specific role of thashsa05211 pathway in renal cell carcinoma remains unclear.
Consensus clustering is an integrated clustering algorithm widely used in processing high-dimensional data, such as gene expression data and proteomics data. Past research has employed consensus clustering to analyse gene expression data of kidney cancer. For instance, studies have explored gene expression related to ferroptosis in clear cell renal cell carcinoma [19], while in renal papillary cell carcinoma, gene expression associated with flame cell death has been researched [20]. Moreover, another study described the role of serine metabolism in papillary renal cell carcinoma and reported its relationship with tumour progression and immune suppression [21]. These studies suggest that consensus clustering can effectively identify different subgroups in datasets. These subgroups may exhibit distinct phenotypic features in different samples or datasets and could hold significant biological implications.
This study integrated the differential expression of Toll-like receptor 4 (TLR4) in ccRCC with the renal cell carcinoma pathway dataset in the KEGG database. Through consensus clustering methods, we constructed a scoring model that revealed potential relationships with immune infiltration, offering new insights into the biological mechanisms of ccRCC. Furthermore, this research provides a robust tool for further studies on ccRCC, with the aim of delivering more accurate and targeted references for prognosis assessment and treatment strategy selection in clinical practice, thereby enhancing patient treatment outcomes and quality of life.
2. Material and methods
2.1. Dataset source and preprocessing
The UCSC Xena database (http://xena.ucsc.edu/), established by bioinformatics researchers from the University of California, Santa Cruz, is a genome-related database that has collected approximately 200 public datasets, including TCGA, ICGC, TARGET, GTEx, CCLE, and others [22]. In this study, we selected the RNA-seq data (HTSeq counts) of clear cell renal cell carcinoma (KIRC) from the TCGA database as the training set, which consisted of 72 normal samples and 531 ccRCC samples. Concurrently, we obtained corresponding clinical pathological data, including age, sex, life status, survival time, and copy number variation (CNV) data, among others. The test set in this study was derived from the ArrayExpress database (https://www.ebi.ac.uk/arrayexpress/) and comprises the clear cell renal cell carcinoma dataset (ID: E-MTAB-1980) with a total of 101 cases.
To facilitate downstream data analysis, we normalized the raw counts of the RNA sequencing data using the transcripts per million (TPM) and the log2-based transformation (log2TPM) method. To align the scales of the training set and test set data, we utilized the scale function in R software to standardize the RNA transcription data. Specifically, the test set data were standardized prior to univariate Cox regression analysis and standardized again before analysing the test set data.
2.2. Differential and prognostic analysis of TLR4 in the KIRC cohort
Utilizing the ggstatsplot package in R, we observed the differential expression of TLR4 in the TCGA-KIRC cohort between normal samples and ccRCC samples and employed the Cairo package along with the ggbetweenstats function to present the intergroup differential expression boxplot [23]. In ccRCC samples, based on TLR4 expression levels, we demarcated high and low groups using the median value (4.52) as the threshold, used the survival package for statistical analysis of intergroup survival differences, and the survminer package for constructing Kaplan‒Meier survival curves [24].
2.2.1. Correlation analysis of TLR4 with the clear cell renal cell carcinoma pathway
We used the limma, ggsci, and org.Hs.eg.db packages to analyse the correlation between TLR4 expression levels in ccRCC and the renal cell carcinoma pathway (KEGG ID: hsa05211). We utilized the ggplot2 and patchwork packages to plot a single gene GSEA (Gene Set Enrichment Analysis) graph to demonstrate the correlation degree between TLR4 and the hsa05211 pathway. Using the DESeq2 package, we conducted a differential analysis of the global gene expression levels between the high and low TLR4 expression groups, setting |logFC| > 0.3 and p value < 0.05 as limiting conditions to obtain differentially expressed genes [25], named TLR4-Deg. The tinyarray package was utilized to construct a volcano plot, illustrating the up- and downregulated TLR4-Deg. We collected genes currently included in the KEGG renal cell carcinoma pathway (hsa05211), named RCC genes. The intersection of TLR4-Deg and RCC genes was obtained and named TRGs (Toll-Like Receptor 4-related Renal Cell Carcinoma differential genes).
2.2.2. Copy number variation and survival prognosis analysis of TRGs
Copy number variation (CNV) refers to deletions or amplifications of DNA fragments with a length not less than 1 kbp compared to the reference genome, representing an important type of structural variation [26]. We filtered the CNV values of TRGs from the clinical information of TCGA-KIRC and utilized R software to create a mutation dumbbell plot illustrating the CNV situation of TRGs in KIRC. For the differential expression of TRGs between the normal sample group and the clear cell renal cell carcinoma sample group, we continued to showcase the results via boxplots. Regarding the impact of TRGs on the survival prognosis of ccRCC, we employed the survminer package in R to construct Kaplan‒Meier survival curves, separately displaying the influence of the expression levels of each TRG on the prognosis of ccRCC.
2.3. Consensus clustering analysis
We applied consensus clustering using the K-means method to identify different TLR4-related subtypes in ccRCC samples of the TCGA-KIRC cohort associated with TRG expression. The number of samples and their stability in each subtype were determined by the consensus clustering algorithm using the ConsensuClusterPlus package. We conducted 1000 iterations to ensure the stability of the classification. Survival analysis was performed on each clustering subtype to ascertain which subtype was associated with poor survival rates [27].
2.4. Data visualization and differential analysis
We implemented a suite of complex analysis tools in this study to reveal the multidimensional characteristics of TLR4-related subtypes in ccRCC. With the assistance of the stringr and pheatmap packages, we combined the TRG expression data with each subtype and clinical information (such as age, sex, tumour TMN staging, and survival outcomes) and presented them in a multipanel heatmap format, providing convenience for observing potential expression trends.
Furthermore, we implemented principal component analysis (PCA) to reduce the dimensionality of these high-dimensional data, thereby revealing the similarities and differences in gene expression profiles. Additionally, drawing on the research of Pornpimol Charoentong et al., we employed the single-sample gene set enrichment analysis (ssGSEA) algorithm to analyse the enrichment level of 28 immune cells in ccRCC, further elucidating the differences in immune cell enrichment among different subtypes.
Finally, we utilized the limma package to analyse gene expression levels in all ccRCC samples, identifying genes with differential expression between each pair of subtypes. Subsequently, the VennDiagram package was used to filter overlapping differentially expressed genes (ODEGs). We set the screening parameter standards (|log2FC| > 0.3, adjusted p value < 0.05), thereby pinpointing the ODEGs related to TLR4-associated ccRCC subtypes.
2.5. Development and validation of ODEG-based risk scoring model
Initially, we used the TCGA-KIRC cohort as the training set. To keep in line with the ArrayExpress test dataset, we normalized both the TCGA training set and the ArrayExpress test set using the scale function before performing univariate Cox regression. Then, based on ODEGs and survival information of the TCGA training set samples, we used the survival package for univariate Cox regression analysis to screen statistically significant ODEGs. To prevent overfitting, we conducted Lasso regression on the model to further narrow the gene scope for model construction [28]. We then incorporated the selected ODEGs into multivariate Cox regression analysis using the stepwise regression method (My.stepwise package) [29]. Through these steps, we determined the optimal ccRCC prognosis model. It consists of the expression level of each TLR4-related ODEG and the corresponding regression coefficient (coef) value, forming the prognostic risk score calculation formula:
| RiskScore = ∑ (Coef * Exp) |
where Coef refers to the regression coefficient related to TLR4-associated ODEGs, Exp refers to the expression level, and the sum is taken over all relevant ODEGs.
According to this calculation formula, we computed the risk score (RiskScore) for each sample in the training cohort. Then, using the median risk score as the cut-off, we divided the patients in the training cohort into low-risk and high-risk subgroups.
In the test set, after normalization using the scale function, we applied the aforementioned formula to analyse the E-MTAB-1980 cohort. Crucially, the cut-off value for dividing the validation cohort into high-risk and low-risk subgroups was set to the median risk score of the training set; this ensures that the criteria for distinguishing between high-risk and low-risk subgroups remain consistent across multiple cohort analyses.
2.6. Deep evaluation and risk analysis
In this study, we thoroughly evaluated the predictive accuracy of the model and delved into the relationship between risk factors and prognosis. Using the timeROC, survival, and survminer packages, we conducted ROC analysis, calculated the AUC for 1-year, 3-year, and 5-year survival times, and compared the model's risk score with other clinical information to assess its difference in predicting the prognosis of AML patients.
Through the risk_plot. R program, we created a risk factor association diagram, more intuitively understanding the differences in population proportions, survival status, and model gene expression distribution among different risk subgroups. Combined with information such as age, sex, and TNM stage, we used the survival package for univariate and multivariate Cox analyses to verify the possibility of the riskScore as an independent prognostic factor.
Finally, we studied the riskScore differences between different categories. We plotted box plots and scatter plots to observe the riskScore differences between different subtypes and plotted trend charts and stacked histograms to test whether the riskScore between different survival outcomes had significant differences. In conjunction with TMB from TCGA-KIRC, we further elucidated their mutual relationships and conducted joint analysis of prognostic outcomes.
2.7. Immune heterogeneity and treatment risk prediction
Through the utilization of the ssGSEA method and lollipop charts, we performed a statistical analysis on the infiltration level of 28 immune cells in both the high- and low-risk groups. Employing the estimate and CIBERSORT software packages, we calculated the immune scores and matrix scores of ccRCC samples, estimated the content of immune cells in the tissue, and further investigated their relationship with risk scores. The analysis results unveiled the correlation between the genes used for model construction, immune checkpoint genes, and the degree of immune cell infiltration.
Next, we evaluated the predictive capacity of the riskScore in the treatment of ccRCC. We computed the degree of correlation between the riskScore and the expression levels of immune cells and immune checkpoint genes, and through box plot analysis, we examined their significant differences between the high- and low-risk groups. Leveraging the oncoPredict package, we assessed the sensitivity of ccRCC samples to various drugs and scrutinized the significant differences in these sensitivities between the high- and low-risk groups.
2.8. Statistical analysis
All data analysis and graphical representations in this study were facilitated using the R programming language (version 4.1.0). Intergroup comparisons of continuous data were carried out utilizing either the t-test or the Mann‒Whitney U test. When comparisons involved three or more groups, the Kruskal‒Wallis test was employed. Survival curves (overall survival, OS) were scrutinized using the Kaplan‒Meier approach, accompanied by a log-rank test to discern group disparities. Through the use of R statistical software, the ggforest and survival packages, alongside the survminer method, forest plots were generated to portray models. Risk scores and prognosis were assessed through both univariate and multivariate Cox proportional regression methods. A p value of less than 0.05 was indicative of a statistically significant research outcome. In graphical representations, “****” signified p < 0.0001, “***” denoted p < 0.001, “**” represented p < 0.01, “*” stood for p < 0.05, while “ns” designated a lack of statistical significance.
3. Result
3.1. Heterogeneity of TLR4 in the TCGA-KIRC cohort
Within the TCGA-KIRC cohort, we scrutinized the expression matrix of TLR4. Our findings revealed that the average TLR4 expression level in normal tissue samples was 16.36, whereas in ccRCC samples, it was 24.37 (Fig. 1A). These data suggest a marked elevation in TLR4 expression in ccRCC compared to normal samples, a difference that is statistically significant (p < 0.05).
Fig. 1.
Heterogeneity of TLR4 in TCGA-KIRC. A: Inter-group comparison between normal and ccRCC TLR4 expression, with light green representing the normal group, and brown representing the ccRCC group. B: Kaplan-Meier survival analysis diagram comparing high and low TLR4 expression levels in ccRCC. Blue represents the low TLR4 expression group, while yellow signifies the high TLR4 expression group. C: Single-gene GSEA chart, with the peak on the left indicating that the number of positively correlated genes in the differentially expressed genes between TLR4 high and low expression groups exceeds the number of negatively correlated genes. D: Volcano plot, with red representing 3557 upregulated genes, and blue representing 1119 downregulated genes. E: Venn diagram. Blue denotes high and low TLR4 differentially expressed genes (TLR4_Deg), and red denotes genes recorded in renal cell carcinoma (hsa05211; RCC_gene). The overlapping nine genes are therapeutic response genes (TRGs).
Subsequent KM prognosis analysis highlighted that the survival prognosis for the group with high TLR4 expression was notably superior to that of the low TLR4 expression group (Fig. 1B). This observation stands in contrast to our initial hypotheses. To better understand this phenomenon, we employed the single-gene GSEA method to assess the correlation between TLR4 expression levels and the renal cell carcinoma pathway (hsa05211) in the KEGG database. The analysis underscored a significant positive linkage between TLR4 and the hsa05211 pathway (Fig. 1C).
In light of these results, we delved deeper by analysing the differentially expressed genes between the high and low TLR4 expression groups and juxtaposed them with the genes of the hsa05211 pathway. Initially, we pinpointed 4676 differentially expressed genes (Fig. 1D). Of the 69 genes present in the hsa05211 pathway, 9 target genes (TRGs) were identified as being associated with TLR4 expression, namely, HGF, TGFB2, ELOB, PIK3CA, GAB1, BAD, AKT3, PIK3R3, and KRAS (Fig. 1E).
To gain a deeper understanding of TRG characteristics in ccRCC, we performed copy number variation analysis, differential analysis between normal control groups, and survival prognosis analysis. The copy number variation analysis demonstrated that all 9 TRGs displayed varying degrees of variation. Specifically, PIK3CA exhibited the most pronounced variation, near 5%, while the variations in GAB1, AKT3, TGFB2, and PIK3R3 were between 1 and 2% (Fig. 2A). The differential analysis revealed that HGF, TGFB2, ELOB, GAB1, BAD, and PIK3R3 had marked expression differences between ccRCC and normal samples (Fig. 2B). Furthermore, survival analysis underscored that the expression levels of all TRGs significantly influence the prognosis of ccRCC. Of particular note, elevated expression of ELOB and HGF correlated with improved survival outcomes, suggesting that they might act as protective genes in ccRCC (Fig. 2C–K).
Fig. 2.
Heterogeneity Analysis of TRGs. A: Dumbbell plot, where red dots represent copy number increase (GAIN) and blue dots represent copy number reduction (LOSS). The y-axis represents the proportion (%) of samples with gene alterations. B: Boxplot for intergroup differential analysis. Red represents ccRCC samples, and blue represents normal tissue samples. The x-axis indicates the names of TRGs genes, while the y-axis represents the expression levels of these genes.C–K: Kaplan-Meier survival plots for individual TRGs. Blue represents low expression group for a given TRG, and red indicates the high expression group for that TRG.
3.2. Consensus clustering reveals distinct ccRCC subtypes: results and assessment
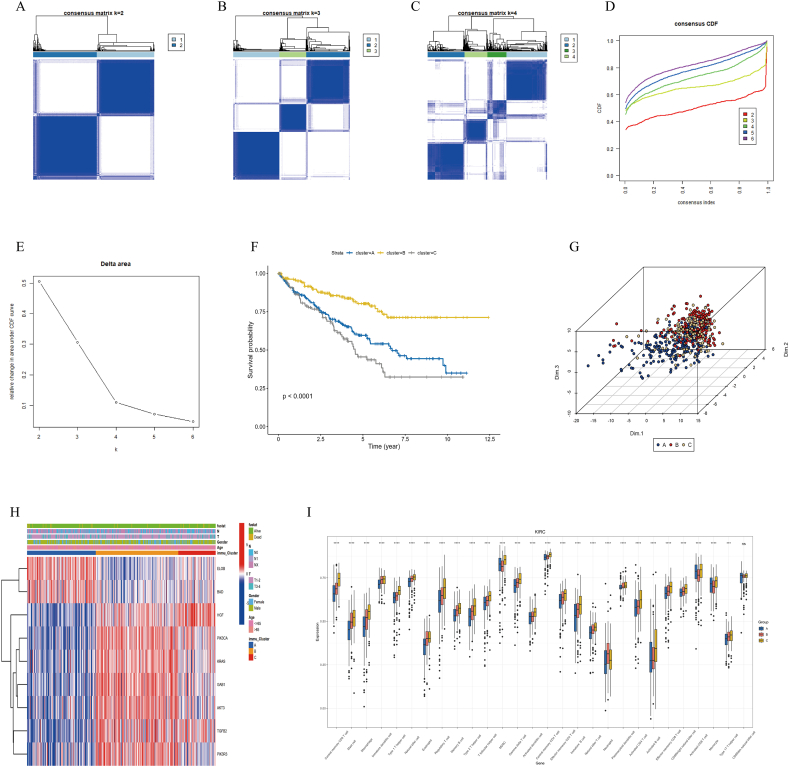
Based on the aforementioned results, we observed diverse expression patterns of TRGs in ccRCC, which implied that TLR4-associated genes exhibit multiple biological characteristics and functional states in tumour cells. Consensus clustering allows for the categorization of gene expression profiles, grouping samples based on similarities, and thereby revealing potential subtypes. Thus, based on the gene expression profile of TRGs, we employed consensus clustering to segregate 531 ccRCC samples into three stable subtypes: A (211 samples), B (197 samples), and C (123 samples) (Fig. 3A–E). These subtypes demonstrated pronounced intragroup similarities and marked intergroup differences.
Fig. 3.
Consensus Clustering Analysis and Evaluation. A–C: Consensus matrix plots when clustering into 2, 3, and 4 subclasses, respectively. Deeper shades of blue indicate higher similarity among base clusters, lighter shades of blue or colorless squares indicate lower similarity. The size of each square represents the weight of the cluster in the final classification. D: Consensus Cumulative Distribution Function (CDF) plot. The x-axis represents the similarity threshold of clustering results, and the y-axis represents the frequency of sample assignments to different clusters. A flatter curve in the plot suggests closer frequencies of sample assignments to clusters, indicating better clustering performance. E: Delta area plot in consensus clustering. When the Delta area value tends to stabilize, and the line in the plot becomes flat, the optimal number of clusters is considered to be reached. F: Kaplan-Meier survival plot. Different colors represent different cluster subtypes, where red signifies subtype A, yellow represents subtype B, and gray denotes subtype C. G: In the Principal Component Analysis (PCA) plot, each point represents the position of a sample in the principal component space, which in turn represents the concentration of gene expression for that sample. H: Multidimensional heatmap: red represents high gene expression, while blue indicates low gene expression. I: Single-sample Gene Set Enrichment Analysis (ssGSEA) plot. The x-axis represents the names of infiltrating immune cells in the tissue, while the y-axis represents the relative abundance of immune cell infiltration. “ns” denotes no statistical significance.
To evaluate the biological similarities within these classifications and the immune phenotype differences between them, we utilized Kaplan‒Meier survival analysis, PCA, multiple clustering heatmaps, and the ssGSEA algorithm. The Kaplan‒Meier study showed that subtype B has a significant advantage in terms of survival rate, while the prognosis for subtype C is relatively poorer (p < 0.001) (see Fig. 3F). The PCA plot visually depicts the heterogeneity of different subtypes at the whole-gene expression level (Fig. 3G).
Multiple clustering analysis revealed that the expression levels of ELOB and BAD in subtype C were notably higher than those in the other groups (Fig. 3H). The results from the ssGSEA study indicate that, except for mature cytotoxic cells (CD56dim natural killer cells), the infiltration levels of other immune cells have significant differences among the three subtypes (Fig. 3I). These data solidly support our consensus clustering, achieving its intended objective, showcasing pronounced biological similarities within groups and immune phenotype differences between them.
3.3. Construction and validation of a ccRCC prognostic risk model based on ODEGs
Addressing the distinct phenotypic cluster subtypes of ccRCC, we conducted a series of analyses to reveal their biological mechanisms and identify potential therapeutic targets and predictive model features. Through pairwise differential analysis, we obtained the intersection of three sets of differential genes, identifying 516 overlapping differential genes (ODEGs) (Fig. 4A). Through univariate Cox regression analysis, 225 ODEGs were confirmed to be related to survival outcomes. Using LASSO regression, we further narrowed the scope of the model genes to 34 (Fig. 4B and C). Ultimately, through multivariate Cox regression analysis and stepwise regression, based on the principles of model universality and the optimal C-index, we chose 17 genes to construct the best predictive model (Fig. 4D). Simultaneously, we obtained the risk score formula for the model:
Fig. 4.
Construction and Validation of the Prognostic Model. A: Venn diagram comparing the overlap of three groups of differentially expressed genes obtained from pairwise differential analysis. B: LASSO regression coefficient shrinkage path plot: shows how gene coefficients are reduced with the increase in regularization strength. C: Cross-validation error plot for LASSO regression: depicts the model's prediction error changing with the change in regularization strength. The prediction error is minimal when λ = λmin. D: Forest plot for the prognostic risk model: The x-axis represents the HR value. Each gene displays a 95% confidence interval. Genes are generally considered tumor suppressors when HR < 1, and oncogenes when HR > 1. E: Kaplan-Meier survival plot for the training set: Blue represents the low-risk group, yellow represents the high-risk group, and the shadow indicates the 95% confidence interval. F: Kaplan-Meier survival plot for the test set: The color scheme is the same as in Fig. 5E. Because the cutoff value is based on the training set, the number of samples in the high and low-risk groups differs.
riskScore = expPTPRB × (−0.319) + expLRP6 × 0.636 + expACVR2A × (−0.264) + expUSP2 × (−0.201) + expSLC38A5 × 0.176 + expNBEA × (−0.223) + expEMP1 × 0.394 + expZNF677 × (−0.274) + expMPZL2 × (−0.311) + expEVC × (−0.262) + expANKS1A × 0.380 + expTMEM72 × 0.253 + expMDK × 0.165 + expSCD5 × (−0.272) + expLIMCH1 × (−0.304) + expFAM13B × (−0.068) + expRUNX1 × 0.215.
The term ‘exp’ in the formula denotes the scaled expression level of the gene.
Based on the established model, a risk score (riskScore) was calculated for the ccRCC samples. When the riskScore reached the median (−0.08), the samples were divided into high-risk and low-risk groups. The results of the Kaplan‒Meier survival analysis showed that the survival time of the high-risk group was significantly shorter than that of the low-risk group (p < 0.001, Fig. 4E). To further validate the applicability of the model, the E-MTAB-1980 dataset from the Arrayexpress database was selected, which includes 101 ccRCC samples. After normalizing these samples, their riskScores were calculated using the same model and grouped based on the median riskScore value from the TCGA-KIRC dataset. The results of the Kaplan‒Meier survival analysis were consistent with the TCGA-KIRC training set, showing that the clinical outcomes for the high-risk group were significantly worse (p = 0.021, Fig. 4F).
3.4. Performance evaluation of the prognostic risk model
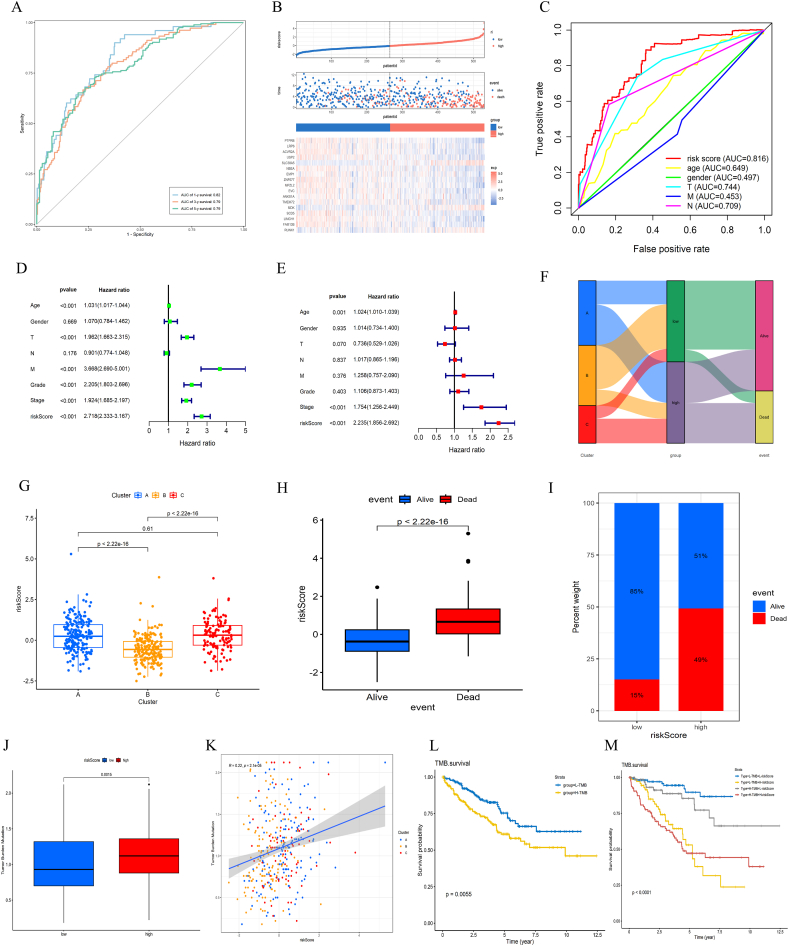
After an in-depth evaluation and validation of the risk model, the predictive performance of the riskScore for survival outcomes in the TCGA-KIRC cohort was studied using ROC curve analysis. The analysis results indicated that the AUC values for 1 year, 3 years, and 5 years were 0.82, 0.79, and 0.79, respectively, fully demonstrating the pivotal role of the riskScore in prognostic prediction (Fig. 5A). Further analysis revealed that with the increase in riskScore, the mortality rate of ccRCC patients notably increased, and the survival period correspondingly decreased (Fig. 5B). Compared to other clinical information, the AUC value of the riskScore was 0.816, indicating that its prediction performance is significantly superior to that of other metrics (Fig. 5C). The results of the Cox regression analysis showed that the riskScore is an independent indicator for prognosis, both in univariate and multivariate analyses (Fig. 5D and E). The Sankey diagram analysis results indicated that subtype B has more low-risk samples, while subtype C mainly comprises high-risk samples, which aligns with the KM curve results of the subtypes (Fig. 5F).
Fig. 5.
Series of Plots Evaluating the Performance of the Prognostic Risk Model. A: 1, 3, and 5-year ROC curves for riskScore. B: Risk factor correlation plot. The top diagram shows each ccRCC sample ordered from smallest to largest by riskScore. The middle diagram plots the prognostic outcomes of samples with riskScore on the x-axis and survival time on the y-axis. Red indicates death, blue indicates survival/loss to follow-up, which corresponds to the order of samples in the top diagram. The bottom diagram is a heatmap matrix of model genes in the samples, where red represents high expression, and blue indicates low expression, which also corresponds to the top and middle diagrams. C: Multi-ROC curve plot that combines age, sex, and TNM stage. D: Univariate prognostic analysis plot. E: Multivariate prognostic analysis plot. F: Sankey plot, from left to right: subtype classification, high and low-risk groups, and outcome classification. G: Box plot of riskScore among subtypes, with the y-axis representing riskScore and the x-axis representing subtype names. H: Box plot of prognostic grouping, with the y-axis representing riskScore and the x-axis representing survival outcomes. I: Stacked histogram for riskScore grouping. The y-axis represents percentage values, and the x-axis represents high and low-riskScore groups, showing the proportion of dead and living samples within each group. J: Box plot of TMB versus riskScore, showing a comparison of TMB values between high and low-riskScore groups. K: Trend plot among TMB, riskScore, and subtypes. The x-axis represents the specific values of riskScore, the y-axis represents TMB values, different colors represent samples from different subtypes, straight lines represent linear regression, and shadows represent the 95% confidence interval. L: KM survival plot for TMB. M: Joint KM survival analysis plot for TMB and riskScore.
The scatter box plot indicates that the median riskScore of subtype B is notably lower than that of the other subtypes (Fig. 5G). The analysis results revealed that the riskScore of deceased patients was significantly higher than that of surviving patients (Fig. 5H), and the proportion in the high-risk group was greater than that in the low-risk group (Fig. 5I). The TMB value in the high-risk group was significantly higher than that in the low-risk group and was positively correlated with the riskScore (Fig. 5J and K). TMB had a significant impact on survival prognosis (Fig. 5L), and when combined with different TMB and riskScore groupings, the survival prognosis still held statistical significance (Fig. 5M). Overall, the model constructed in this study exhibits good stability and broad applicability.
3.5. Correlation study between the prognostic risk model and tumour-infiltrating immune cells
The type and quantity of immune infiltrating cells in tumours have been proven to serve as crucial indicators for predicting patient prognosis [30]. To more accurately predict patient outcomes, prognostic risk models often consider the type, quantity, and distribution of immune infiltrating cells [31,32]. In this study, leveraging ssGSVA, the immune matrix score, and the correlation analysis between model construction genes and immune cells and checkpoints, we explored the relationship between the prognostic model and immune infiltrating cells in ccRCC tissues. ssGSEA results showed that the infiltration levels of 23 types of immune cells showed significant differences between the high- and low-risk groups. The primary infiltrating cells included myeloid-derived suppressor cells (MDSCs), central memory CD4 T cells, activated CD4 T cells, and activated CD8 T cells. Except for mature cytotoxic cells (CD56dim natural killer cells) and immature dendritic cells, the infiltration levels of the other 21 immune cell types in the high-risk group were noticeably higher than those in the low-risk group (Fig. 6A). In terms of the immune score, the median value in the high-risk group was significantly higher than that in the low-risk group. However, in the matrix score, the difference between the two groups was not statistically significant (Fig. 6B).
Fig. 6.
Study on the Correlation between the Prognostic Risk Model and Immune Infiltrating Cells. A: ssGSVA analysis plot between high and low-risk groups. The x-axis represents immune cells infiltrating ccRCC tissues, and the y-axis represents the relative abundance of infiltration. B: The left diagram is for the matrix score, and the right diagram is for the immune score. The x-axis represents the high and low-risk groups, and the y-axis represents the relative scores. C: Association plot of the model construction genes and immune infiltrating cells. The right-side y-axis represents the names of the genes used in model construction, and the x-axis represents the names of immune infiltrating cells. D: Correlation analysis plot of model construction genes and immune checkpoint genes: the x-axis represents the names of immune checkpoint genes. In both C and D, red indicates positive correlation, and blue indicates negative correlation.
In the present investigation, we thoroughly examined the association between the infiltration degree of immune cells in ccRCC tissues and the expression levels of genes in our constructed model. We identified that PTPRB, EMP1, MPZL2, RUNX1, SLC38A5, and MDK exhibited a positive correlation with primary immune infiltrating cells. Conversely, USP2 and TMEM72 were negatively correlated with these primary immune cells (Fig. 6C). Furthermore, we observed that PD-1 was negatively correlated with the principal model genes, while PD-L1 and PD-L2 showed positive correlations (Fig. 6D). These analyses enriched our understanding of the mechanisms underlying tumour immune evasion and the potential avenues for immunotherapy, laying a theoretical foundation for personalized treatment.
3.6. The role of riskScore in immunotherapy and antitumour drug sensitivity
This study explored the correlation between model genes, immune-infiltrating cells, and immune checkpoint genes. Correlation analysis revealed that the infiltration of 22 types of immune cells had a statistically significant relationship with the riskScore. Among them, mast cells and immature dendritic cells were negatively correlated with the riskScore, while activated CD4 T cells showed the most significant positive correlation with the riskScore (Fig. 7A). Further analysis revealed that, compared to PD-L2, the correlation between PD-1, PD-L1, and CTLA4 and the riskScore was stronger (Fig. 7B). Intergroup difference analysis results showed that the expression of PDCD1 and CTLA4 in the high-risk group was significantly higher than that in the low-risk group, while the expression of CD274 was more pronounced in the low-risk group. The expression difference for PDCD1LG2 was not evident (Fig. 7C–F). Drug sensitivity analysis indicated that the high-risk group was more sensitive to cisplatin, gemcitabine, and topotecan but showed lower sensitivity to zoledronic acid (Fig. 7G–I). These findings suggest that the model holds potential applicability in tumour immunotherapy.
Fig. 7.
The role of riskScore in immunotherapy and anti-cancer drug sensitivity. A: Lollipop chart where the left y-axis represents immune cell types, and the right y-axis represents the p-value of correlation analysis, and the x-axis represents the correlation coefficient between the type of immune cell and the riskScore of the sample. B: Chord plot, where the size and depth of color of the red band respectively represent the strength of correlation between the modules. C–F: Box plots of risk model for immune checkpoint genes. Blue and red represent the low-risk and high-risk groups, respectively, and the y-axis represents the gene expression level. G–J: Box plots of drug sensitivity analysis risk model. Green and red represent the low-risk and high-risk groups, respectively, and the y-axis represents the drug sensitivity coefficient.
4. Discussion
While treatment for renal clear cell carcinoma has seen advancements, its complexity and heterogeneity continue to pose challenges in therapy and prognosis evaluation [33]. To better navigate treatment strategies and prognostic models, technologies such as artificial intelligence have begun to play a pivotal role [34]. Concurrently, consensus clustering has also played a key role in tumour gene research [35].
In renal clear cell carcinoma, the expression of TLR4 is closely associated with patient prognosis and survival rates [14]. Although inhibiting TLR4 can slow tumour growth, studies have found that high expression of TLR4 correlates with better survival outcomes [36]; this suggests that the role of TLR4 in renal clear cell carcinoma might be intricately tied to its expression levels. Moreover, TLR4 expression is intertwined with the biological pathways of renal cell carcinoma, underscoring its crucial role in this malignancy. Through integrated immune infiltration analysis and consensus clustering, we gleaned deeper insights into the mechanism of action of TLR4, offering theoretical support for the treatment and prognosis evaluation of renal clear cell carcinoma [3].
First, we performed a differential analysis on samples with varying TLR4 expression in the TCGA-KIRC cohort to identify differentially expressed genes. This approach aligns with recent research methodologies, wherein similar techniques have been employed to study other types of cancers [37]. Subsequently, we integrated genes from the renal cell carcinoma pathway in the KEGG database and identified TRGs through intersection analysis. This strategy has also been validated in recent literature [38]. Further analysis revealed that, whether in copy number variations, differential analysis between the normal and experimental groups, or prognosis outcome analysis, TRGs consistently yielded positive results. This evidence provided robust support for the subsequent consensus clustering, consistent with findings in the literature [39]. Based on TRGs, we categorized the ccRCC samples from the TCGA-KIRC cohort into three subtypes. These subtypes demonstrated their distinctiveness in survival analysis, principal component analysis, and multiple clustering heatmaps, aligning with recent research outcomes [40].
By employing these TRGs, we divided the ccRCC samples from the TCGA-KIRC cohort into three unique subtypes; their distinct characteristics were reflected in survival analysis, principal component analysis, and multiple cluster heatmaps.
Clearly, subtype C has an unfavourable prognosis. In this subtype, the expression of ELOB and BAD is significantly elevated, both of which are detrimental to the prognosis of ccRCC. The ELOB gene encodes Elongin B, a protein involved in multiple biological processes, including transcriptional regulation and protein degradation. Recent research, including a study by Jin et al. [41], has emphasized the role of ELOB in other malignancies, suggesting that it can regulate gene expression and subsequently promote tumour cell proliferation and metastasis. According to Samra Turajlic et al. [42], suppressing ELOB expression can curb the growth and invasion of renal clear cell carcinoma cells. In addition, the BAD gene encodes the Bcl-2-associated death promoter, a protein that facilitates cell apoptosis [43]. In the context of tumour progression, BAD has emerged as a significant player. Current studies have shown that the expression of BAD positively correlates with tumour staging. Specifically, the levels of BAD protein in high-grade tumours surpass those in low-grade tumours, further emphasizing its central role in tumour progression [44].
Based on the differences in the three subtypes, we successfully constructed a risk prediction model using univariate, LASSO, and multivariate Cox regression analyses. This model furnishes a risk score for each sample, quantifying the prognostic risk for ccRCC patients and offering guidance for both prognostic assessment and personalized treatment. In recent years, predicting the future development of tumours has been deemed critical for both patients and physicians by numerous scholars [45]. For instance, prognostic models rooted in immune and apoptotic signalling pathways have been shown to accurately forecast tumour survival rates [46]. Research by Shen et al. highlighted that immune genes serve as effective predictive markers for ccRCC prognosis [47]. Concurrently, researchers such as Bian Z and Yanhua Mou, utilizing death-related genes and ferroptosis-related genes, respectively, developed ccRCC prognosis models, underscoring their predictive efficacy [48,49]. Mirroring the aforementioned studies, for our study we also crafted a ccRCC prognosis model based on the differential expression of TLR4 and subsequently validated it using external datasets, which emphasizes its significance in the clinical appraisal of ccRCC patient prognosis.
The study identified marked differences in immune infiltration levels among the three subtypes through ssGSEA. Specifically, subtype B, which exhibited the most favourable prognosis, was predominantly characterized by immune-infiltrating cells such as myeloid-derived suppressor cells (MDSCs), central memory CD4 T cells, and regulatory T cells. Immune infiltration exerts a critical influence on the tumour microenvironment. For example, the activity of CD8 T cells is pivotal for immune regulation [50], and the activation of T cells is central to immune modulation [51]. Recent studies suggest that MDSCs can suppress CD8+ T cells and augment the count of tumour-specific regulatory T cells exhibiting robust immunosuppressive actions [52]. Conversely, regulatory T cells can restrain unchecked lymphocyte proliferation and the broader immune response [53]. Additionally, CD4+ memory T cells can stifle tumour cell growth by amplifying the proliferation of CD8+ memory T cells [54].
In this study, we selected 17 genes to create a prognostic model, with each gene playing diverse roles in the development of clear cell renal cell carcinoma (ccRCC) or other malignancies. Notably, the PTPRB gene, a crucial regulator of protein tyrosine kinase receptors, exhibits diminished expression in the vast majority of ccRCC cases [55]. Ren Y demonstrated that targeting LRP6 could inhibit ccRCC proliferation and metastasis, introducing a novel avenue for antitumour treatments [56]. Additionally, the AVCR 2A gene, which encodes the TGF-β receptor, is a focal point in contemporary tumour mutation research. Changes within the TGF-β superfamily are positively correlated with the expression of metastasis-associated genes and are associated with decreased tumour survival rates [57]. Furthermore, the USP2 gene has been employed to develop an immune-related ccRCC prognostic risk model aimed at predicting patient survival outcomes [58]. SLC38A5 is a Na+-coupled transporter that is upregulated in various cancers. It mediates the influx of glutamine, serine, glycine, and asparagine into cancer cells, playing a pivotal role in promoting cancer cell survival and growth [59]. Case reports have indicated that the fusion of the NBEA and EML4 genes can enhance the sensitivity of non-small cell lung cancer patients to alectinib [60]. EMP1, which expresses epithelial membrane protein 1 and is regulated by TAZ, can induce the expression of NADPH oxidase 4 (NOX4), which is crucial for haemochromatosis. Together, EMP1 and NOX4 can mediate the ferroptosis program in renal cell carcinoma, offering potential therapeutic value [61]. ZNF677 impedes the progression of renal cancer through the transcriptional suppression of N6-methyladenosine and CDKN3 [62]. In a study on breast cancer, Anks1a was identified as a Rac1 effector and was found to promote cancer cell migration by activating the human epidermal growth factor receptor 2 (HER2) switch [63], consistent with the findings of this research. TMEM72 belongs to the transmembrane protein family. Differential analysis between metastatic and nonmetastatic ccRCC revealed elevated expression of TMEM72, suggesting an unfavourable prognosis. MDK encodes Midkine, a heparin-binding growth factor that is aberrantly expressed at high levels in various human cancers; it fosters cancer cell growth, survival, metastasis, migration, and angiogenesis, establishing itself as a crucial cancer marker [64]. SCD5 has been instrumental in crafting a prognostic model for ccRCC using metabolic gene markers, showcasing its precision in predicting ccRCC outcomes [65]. In a cohort study led by Nicholas Rooney, decreased expression of RUNX1 was associated with reduced survival rates in ccRCC patients. Further experimental evidence confirmed that the absence of RUNX1 improves the survival rate in a genetic mouse model of kidney cancer [66]. These research findings align closely with the conclusions drawn in our study.
However, MPZL2 is regarded as a poor prognostic indicator in paediatric acute myeloid leukaemia [67], but in this study, it acted as a protective marker, which might be related to the complex regulatory environment of tumours. In studies of renal clear cell carcinoma associated with smoking, LIMCH1 was identified as a significantly expressed gene, but its mechanism of action is still unclear [68], differing from the current research, which might be related to the reference used. The EVC gene is associated with recessive genetic diseases, often leading to skeletal dysplasia [69], but it has not been linked to tumours. FAM13B is a type of circular DNA reported in diseases associated with neonatal death, but no abnormalities have been found in the field of oncology [70]. These findings suggest that these genes play different roles in the onset and development of tumours and might play a crucial role in the prognosis of KIRC, but further verification is needed.
We conducted a comprehensive analysis of renal cell carcinoma gene alterations, expression discrepancies, and TLR4-related immune infiltration within the TCGA-KIRC cohort. By discerning TLR4-associated subtypes, we underscored the heterogeneity inherent in renal clear cell carcinoma. Leveraging these subtype distinctions, we formulated a scoring model encompassing 17 genes. This model exhibited robust and consistent predictive efficacy across both training and validation datasets, offering a formidable tool for forecasting the prognosis of ccRCC and guiding immunotherapy decisions. Recent studies have posited that TLR family expression in renal cell carcinoma correlates with prognosis [71], and the degree of immune infiltration within the tumour microenvironment is another predictive indicator [72]. Such insights resonate with our conclusions, reinforcing the potential clinical significance and utility of our proposed model.
While it is true that this study was primarily focused on the expression and function of TLR4 and its associated genes, we must recognize that the onset and progression of renal clear cell carcinoma is a complex process involving multiple factors. Therefore, future research should not only consider mechanisms related to TLR4 but also incorporate more potential molecules and biological factors; this will aid in our achievement of a more comprehensive and in-depth understanding of the pathogenesis of renal clear cell carcinoma.
5. Conclusions
This study opens new perspectives for the prognosis assessment and treatment of renal clear cell carcinoma (ccRCC). By delving into the role of TLR4 in ccRCC, we identified tumor-related genes (TRGs) associated with renal cell carcinoma. Based on these 17 genes, we constructed a risk prediction model that displayed exceptional stability and predictive capability in both training and validation sets. This provides an innovative assessment method for predicting the prognosis of ccRCC and lays a solid foundation for future clinical studies. Furthermore, this discovery offers practical value for clinical practice. Through this, understanding TLR4 and its related genes enables doctors to more accurately assess the prognosis of ccRCC patients, providing them with personalized treatment strategies to enhance survival rates and quality of life. The study also highlights some genes that have not yet been thoroughly investigated, offering new directions for future basic and clinical research, giving ccRCC patients a broader range of treatment options and optimized therapeutic outcomes.
Funding
This research was financially supported by the Science Technology Bureau of Shaoxing (Grant/Award Number: 2023A14035) and the Zhejiang Province Traditional Chinese Medicine Science and Technology Plan Project (Grant/Award Number: 2024ZF168).
Ethics statement
The data used in this study are sourced from public databases, and hence, specific ethical review requirements are not applicable.
Data availability statement
Our dataset is primarily composed of two main components: Raw Data: This data is sourced from a genomic-related database curated by bioinformatics researchers at the University of California, Santa Cruz (UCSC). For more details, you can visit the official UCSC website at http://xena.ucsc.edu/. The specific data we utilized can be accessed directly from this link: https://xenabrowser.net/datapages/?dataset=TCGA-KIRC.htseq_fpkm.tsv&host=https%3A%2F%2Fgdc.xenahubs.net&removeHub=https%3A%2F%2Fxena.treehouse.gi.ucsc.edu%3A443. Analysis Code: We have archived our analysis code on Nutstore cloud storage. It can be accessed and downloaded directly from the provided link: https://www.jianguoyun.com/p/DdnXlgYQxoCACxiThosFIAA,TCGA-KIRC.
CRediT authorship contribution statement
Qingbo Zhou: Writing – review & editing, Writing – original draft, Methodology, Conceptualization. Qiang Sun: Formal analysis, Data curation. Qi Shen: Writing – original draft. Xinsheng Li: Investigation. Jijiang Qian: Writing – review & editing, Writing – original draft, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Qingbo Zhou reports financial support was provided by the Science Technology Bureau of Shaoxing (NO. 2023A14035). Qingbo Zhou reports financial support was provided by Zhejiang Province Traditional Chinese Medicine Science and Technology Plan Project (NO. 2024ZF168). If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors extend their sincere gratitude to the patients who participated in the study and provided samples. We deeply thank Professor Qiang He, my esteemed Master's thesis advisor, for his continuous guidance, invaluable insights, and unwavering support throughout this research journey. We acknowledge the invaluable databases TCGA and ArrayExpress for their support in facilitating this research. Special thanks to Dr. Jianming Zeng from the University of Macau, and all the members of his bioinformatics team, Biotrainee, for generously sharing their experience and codes. Your contribution has significantly enriched this work.
References
- 1.Wei H., Miao J., Cui J., Zheng W., Chen X., Zhang Q., Liu F., Mao Z., Qiu S., Zhang D. The prognosis and clinicopathological features of different distant metastases patterns in renal cell carcinoma: analysis based on the SEER database. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-97365-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu H., Xu W.H., Ren F., Wang J., Wang H.K., Cao D.L., Shi G.H., Qu Y.Y., Zhang H.L., Ye D.W. Prognostic value of epithelial-mesenchymal transition markers in clear cell renal cell carcinoma. Aging (Albany NY) 2020;12:866–883. doi: 10.18632/aging.102660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suh J.Y., Maeng S.J., Kim M., Kang S.J., Choi Y.W., Chang I.H. Current trends in liquid biopsy technology for early diagnosis of metastatic renal cell carcinoma. Korean J. Urol. Oncol. 2022;20:223–234. doi: 10.22465/kjuo.2022.20.4.223. [DOI] [Google Scholar]
- 4.Trac N., Oh H.S., Jones L.I., Caliliw R., Ohtake S., Shuch B., Chung E.J. CD70-Targeted micelles enhance HIF2α siRNA delivery and inhibit oncogenic functions in patient-derived clear cell renal carcinoma cells. Molecules. 2022;27 doi: 10.3390/molecules27238457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Che X., Li J., Xu Y., Wang Q., Wu G. Analysis of genomes and transcriptomes of clear cell renal cell carcinomas identifies mutations and gene expression changes in the TGF-beta pathway. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.953322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naoum G.E., Morkos M., Kim B., Arafat W. Novel targeted therapies and immunotherapy for advanced thyroid cancers. Mol. Cancer. 2018;17:51. doi: 10.1186/s12943-018-0786-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vitruk I., Voylenko O., Stakhovsky O., Kononenko O., Pikul M., Semko S., Hrechko B., Koshel D., Stakhovsky E. Advantages of organ-sparing treatment approaches in metastatic kidney cancer. J. Cancer Res. Clin. Oncol. 2023;149:3131–3137. doi: 10.1007/s00432-022-04216-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Şenbabaoğlu Y., Gejman R.S., Winer A.G., Liu M., Van Allen E.M., de Velasco G., Miao D., Ostrovnaya I., Drill E., Luna A., et al. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol. 2016;17:231. doi: 10.1186/s13059-016-1092-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Büttner F.A., Winter S., Stühler V., Rausch S., Hennenlotter J., Füssel S., Zastrow S., Meinhardt M., Toma M., Jerónimo C., et al. A novel molecular signature identifies mixed subtypes in renal cell carcinoma with poor prognosis and independent response to immunotherapy. Genome Med. 2022;14:105. doi: 10.1186/s13073-022-01105-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y., Liang X., Bao X., Xiao W., Chen G. Toll-like receptor 4 (TLR4) inhibitors: current research and prospective. Eur. J. Med. Chem. 2022;235 doi: 10.1016/j.ejmech.2022.114291. [DOI] [PubMed] [Google Scholar]
- 11.Ahmed A., Wang J.H., Redmond H.P. Silencing of TLR4 increases tumor progression and lung metastasis in a murine model of breast cancer. Ann. Surg. Oncol. 2013;20(Suppl 3):S389–S396. doi: 10.1245/s10434-012-2595-9. [DOI] [PubMed] [Google Scholar]
- 12.Li G., Sun Y., Huang Y., Lian J., Wu S., Luo D., Gong H. Fusobacterium nucleatum-derived small extracellular vesicles facilitate tumor growth and metastasis via TLR4 in breast cancer. BMC Cancer. 2023;23:473. doi: 10.1186/s12885-023-10844-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z., Wang S., Lu Y., Xia D., Liu Y. TLR4 predicts patient prognosis and immunotherapy efficacy in clear cell renal cell carcinoma. J. Cancer. 2023;14:2181–2197. doi: 10.7150/jca.84502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou X., Guo B., Ling Q., Mo Z. Toll-like receptors serve as biomarkers for early diagnosis and prognosis assessment of kidney renal clear cell carcinoma by influencing the immune microenvironment: comprehensive bioinformatics analysis combined with experimental validation. Front. Mol. Biosci. 2022;9 doi: 10.3389/fmolb.2022.832238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kashani B., Zandi Z., Pourbagheri-Sigaroodi A., Bashash D., Ghaffari S.H. The role of toll-like receptor 4 (TLR4) in cancer progression: a possible therapeutic target? J. Cell Physiol. 2021;236:4121–4137. doi: 10.1002/jcp.30166. [DOI] [PubMed] [Google Scholar]
- 16.Linehan W.M., Rubin J.S., Bottaro D.P. VHL loss of function and its impact on oncogenic signaling networks in clear cell renal cell carcinoma. Int. J. Biochem. Cell Biol. 2009;41:753–756. doi: 10.1016/j.biocel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J., Luo L., Zhao C., Li X., Wang Z., Zeng Z., Yang X., Zheng X., Jie H., Kang L., et al. A positive feedback loop between inactive VHL-triggered histone lactylation and PDGFRβ signaling drives clear cell renal cell carcinoma progression. Int. J. Biol. Sci. 2022;18:3470–3483. doi: 10.7150/ijbs.73398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang W., Zhou J., Zhang Z., Zhang K., Xu Y., Li L., Cai L., Gong Y., Gong K. Downregulation of lncRNA APCDD1L-AS1 due to DNA hypermethylation and loss of VHL protein expression promotes the progression of clear cell renal cell carcinoma. Int. J. Biol. Sci. 2022;18:2583–2596. doi: 10.7150/ijbs.71519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun Z., Li T., Xiao C., Zou S., Zhang M., Zhang Q., Wang Z., Zhan H., Wang H. Prediction of overall survival based upon a new ferroptosis-related gene signature in patients with clear cell renal cell carcinoma. World J. Surg. Oncol. 2022;20:120. doi: 10.1186/s12957-022-02555-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu J., Chen Y., Gao L., Ge C., Xie X., Lei P., Zhang Y., Liang P. A novel pyroptosis-related gene signature for predicting prognosis in kidney renal papillary cell carcinoma. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.851384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong W., Wang Z., Chen N., Mei Y., Li Y., Yue Y. SHMT2 regulates serine metabolism to promote the progression and immunosuppression of papillary renal cell carcinoma. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.914332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldman M.J., Craft B., Hastie M., Repečka K., McDade F., Kamath A., Banerjee A., Luo Y., Rogers D., Brooks A.N., et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020;38:675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai L., Liu H., Minna J.D., DeBerardinis R.J., Xiao G., Xie Y. Assessing consistency across functional screening datasets in cancer cells. Bioinformatics (Oxford, England) 2021;37:4540–4547. doi: 10.1093/bioinformatics/btab423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brentnall A.R., Cuzick J., Buist D.S.M., Bowles E.J.A. Long-term accuracy of breast cancer risk assessment combining classic risk factors and breast density. JAMA Oncol. 2018;4 doi: 10.1001/jamaoncol.2018.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lye Z.N., Purugganan M.D. Copy number variation in domestication. Trends Plant Sci. 2019;24:352–365. doi: 10.1016/j.tplants.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Coleman S., Kirk P.D.W., Wallace C. Consensus clustering for Bayesian mixture models. BMC Bioinformat. 2022;23:290. doi: 10.1186/s12859-022-04830-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedman J., Hastie T., Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 29.Genell A., Nemes S., Steineck G., Dickman P.W. Model selection in medical research: a simulation study comparing Bayesian model averaging and stepwise regression. BMC Med. Res. Methodol. 2010;10:108. doi: 10.1186/1471-2288-10-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galon J., Pagès F., Marincola F.M., Thurin M., Trinchieri G., Fox B.A., Gajewski T.F., Ascierto P.A. The immune score as a new possible approach for the classification of cancer. J. Transl. Med. 2012;10:1. doi: 10.1186/1479-5876-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giraldo N.A., Becht E., Pagès F., Skliris G., Verkarre V., Vano Y., Mejean A., Saint-Aubert N., Lacroix L., Natario I., et al. Orchestration and prognostic significance of immune checkpoints in the microenvironment of primary and metastatic renal cell cancer. Clin. Cancer Res. 2015;21:3031–3040. doi: 10.1158/1078-0432.Ccr-14-2926. [DOI] [PubMed] [Google Scholar]
- 32.Mahmoud S.M., Paish E.C., Powe D.G., Macmillan R.D., Grainge M.J., Lee A.H., Ellis I.O., Green A.R. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J. Clin. Oncol. 2011;29:1949–1955. doi: 10.1200/jco.2010.30.5037. [DOI] [PubMed] [Google Scholar]
- 33.Peng Q., Shen Y., Fu K., Dai Z., Jin L., Yang D., Zhu J. Artificial intelligence prediction model for overall survival of clear cell renal cell carcinoma based on a 21-gene molecular prognostic score system. Aging (Albany NY) 2021;13:7361–7381. doi: 10.18632/aging.202594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herrgott G.A., Snyder J.M., She R., Malta T.M., Sabedot T.S., Lee I.Y., Pawloski J., Podolsky-Gondim G.G., Asmaro K.P., Zhang J., et al. Detection of diagnostic and prognostic methylation-based signatures in liquid biopsy specimens from patients with meningiomas. Nat. Commun. 2023;14:5669. doi: 10.1038/s41467-023-41434-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giulietti M., Cecati M., Sabanovic B., Scirè A., Cimadamore A., Santoni M., Montironi R., Piva F. The role of artificial intelligence in the diagnosis and prognosis of renal cell tumors. Diagnostics (Basel) 2021;11 doi: 10.3390/diagnostics11020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pastille E., Faßnacht T., Adamczyk A., Ngo Thi Phuong N., Buer J., Westendorf A.M. Inhibition of TLR4 signaling impedes tumor growth in colitis-associated colon cancer. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.669747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He M., Hu F. TF-RBP-AS triplet analysis reveals the mechanisms of aberrant alternative splicing events in kidney cancer: implications for their possible clinical use as prognostic and therapeutic biomarkers. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22168789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X., Wei X., Wang Y., Wang S., Ji C., Yao L., Song N. Pyroptosis regulators and tumor microenvironment infiltration characterization in clear cell renal cell carcinoma. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.774279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang S., Zhao Z., Wang Y., El Akkawi M.M., Tan Z., Liu D., Chen G., Liu H. DHRS7 is an immune-related prognostic biomarker of KIRC and pan-cancer. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.1015844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Long Z., Sun C., Tang M., Wang Y., Ma J., Yu J., Wei J., Ma J., Wang B., Xie Q., Wen J. Single-cell multiomics analysis reveals regulatory programs in clear cell renal cell carcinoma. Cell Discov. 2022;8:68. doi: 10.1038/s41421-022-00415-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bian Z., Fan R., Xie L. A novel cuproptosis-related prognostic gene signature and validation of differential expression in clear cell renal cell carcinoma. Genes (Basel) 2022;13 doi: 10.3390/genes13050851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mou Y., Wu J., Zhang Y., Abdihamid O., Duan C., Li B. Low expression of ferritinophagy-related NCOA4 gene in relation to unfavorable outcome and defective immune cells infiltration in clear cell renal carcinoma. BMC Cancer. 2021;21:18. doi: 10.1186/s12885-020-07726-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y., Misumi I., Gu A.D., Curtis T.A., Su L., Whitmire J.K., Wan Y.Y. GATA-3 controls the maintenance and proliferation of T cells downstream of TCR and cytokine signaling. Nat. Immunol. 2013;14:714–722. doi: 10.1038/ni.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Y., Meng Z., Zhang L., Liu F. CD2 is a novel immune-related prognostic biomarker of invasive breast carcinoma that modulates the tumor microenvironment. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.664845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goldberg H. The suggested importance of PBRM1 mutation in predicting postoperative recurrence of localized clear cell renal cell carcinoma. Ann. Surg. Oncol. 2021;28:1889–1891. doi: 10.1245/s10434-021-09661-0. [DOI] [PubMed] [Google Scholar]
- 46.Zhang X., Zhu Y., Zhou Y., Fei B. Interleukin 37 (IL-37) reduces high glucose-induced inflammation, oxidative stress, and apoptosis of podocytes by inhibiting the STAT3-cyclophilin A (CypA) signaling pathway. Med. Sci. Monit. 2020;26 doi: 10.12659/msm.922979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wan B., Liu B., Huang Y., Yu G., Lv C. Prognostic value of immune-related genes in clear cell renal cell carcinoma. Aging (Albany NY) 2019;11:11474–11489. doi: 10.18632/aging.102548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao G.J., Wu Z., Ge L., Yang F., Hong K., Zhang S., Ma L. Ferroptosis-related gene-based prognostic model and immune infiltration in clear cell renal cell carcinoma. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.650416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mou Y., Zhang Y., Wu J., Hu B., Zhang C., Duan C., Li B. The landscape of iron metabolism-related and methylated genes in the prognosis prediction of clear cell renal cell carcinoma. Front. Oncol. 2020;10:788. doi: 10.3389/fonc.2020.00788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lees J.R. CD8+ T cells: the past and future of immune regulation. Cell Immunol. 2020;357 doi: 10.1016/j.cellimm.2020.104212. [DOI] [PubMed] [Google Scholar]
- 51.Hwang J.R., Byeon Y., Kim D., Park S.G. Recent insights of T cell receptor-mediated signaling pathways for T cell activation and development. Exp. Mol. Med. 2020;52:750–761. doi: 10.1038/s12276-020-0435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagaraj S., Schrum A.G., Cho H.I., Celis E., Gabrilovich D.I. Mechanism of T cell tolerance induced by myeloid-derived suppressor cells. J. Immunol. 2010;184:3106–3116. doi: 10.4049/jimmunol.0902661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woo E.Y., Yeh H., Chu C.S., Schlienger K., Carroll R.G., Riley J.L., Kaiser L.R., June C.H. Cutting edge: regulatory T cells from lung cancer patients directly inhibit autologous T cell proliferation. J. Immunol. 2002;168:4272–4276. doi: 10.4049/jimmunol.168.9.4272. [DOI] [PubMed] [Google Scholar]
- 54.Lai Y.-P., Jeng C.-J., Chen S.-C. The roles of CD4+ T cells in tumor immunity. ISRN Immunol. 2011;2011 doi: 10.5402/2011/497397. [DOI] [Google Scholar]
- 55.Casagrande S., Ruf M., Rechsteiner M., Morra L., Brun-Schmid S., von Teichman A., Krek W., Schraml P., Moch H. The protein tyrosine phosphatase receptor type J is regulated by the pVHL-HIF axis in clear cell renal cell carcinoma. J. Pathol. 2013;229:525–534. doi: 10.1002/path.4107. [DOI] [PubMed] [Google Scholar]
- 56.Ren Y., Zhang L., Zhang W., Gao Y. MiR-30a suppresses clear cell renal cell carcinoma proliferation and metastasis by targeting LRP6. Hum. Cell. 2021;34:598–606. doi: 10.1007/s13577-020-00472-1. [DOI] [PubMed] [Google Scholar]
- 57.Korkut A., Zaidi S., Kanchi R.S., Rao S., Gough N.R., Schultz A., Li X., Lorenzi P.L., Berger A.C., Robertson G., et al. A pan-cancer analysis reveals high-frequency genetic alterations in mediators of signaling by the TGF-β superfamily. Cell Syst. 2018;7 doi: 10.1016/j.cels.2018.08.010. 422-437.e427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He J., Zhong Y., Sun Y., Xie C., Yu T. Construction of an immune-related prognostic model by exploring the tumor microenvironment of clear cell renal cell carcinoma. Anal. Biochem. 2022;643 doi: 10.1016/j.ab.2022.114567. [DOI] [PubMed] [Google Scholar]
- 59.Sniegowski T., Korac K., Bhutia Y.D., Ganapathy V. SLC6A14 and SLC38A5 drive the glutaminolysis and serine-glycine-one-carbon pathways in cancer. Pharmaceuticals (Basel) 2021;14 doi: 10.3390/ph14030216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liang Q., Xu H., Liu Y., Zhang W., Sun C., Hu M., Zhu Y., Tan S., Xu X., Wang S., Liu L. Coexistence of a novel NBEA-ALK, EML4-ALK double-fusion in a lung adenocarcinoma patient and response to alectinib: a case report. Lung Cancer. 2021;162:86–89. doi: 10.1016/j.lungcan.2021.10.015. [DOI] [PubMed] [Google Scholar]
- 61.Yang W.H., Ding C.C., Sun T., Rupprecht G., Lin C.C., Hsu D., Chi J.T. The hippo pathway effector TAZ regulates ferroptosis in renal cell carcinoma. Cell Rep. 2019;28 doi: 10.1016/j.celrep.2019.07.107. 2501-2508.e2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li A., Cao C., Gan Y., Wang X., Wu T., Zhang Q., Liu Y., Yao L., Zhang Q. ZNF677 suppresses renal cell carcinoma progression through N6-methyladenosine and transcriptional repression of CDKN3. Clin. Transl. Med. 2022;12 doi: 10.1002/ctm2.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zholudeva A.O., Lomakina M.E., Orlova E.A., Wang Y., Fokin A.I., Polesskaya A., Gautreau A.M., Alexandrova A.Y. The role of the adapter protein Anks1a in the regulation of breast cancer cell motility. Biochemistry (Mosc) 2022;87:1651–1661. doi: 10.1134/s0006297922120203. [DOI] [PubMed] [Google Scholar]
- 64.Filippou P.S., Karagiannis G.S., Constantinidou A. Midkine (MDK) growth factor: a key player in cancer progression and a promising therapeutic target. Oncogene. 2020;39:2040–2054. doi: 10.1038/s41388-019-1124-8. [DOI] [PubMed] [Google Scholar]
- 65.Wu Y., Wei X., Feng H., Hu B., Liu B., Luan Y., Ruan Y., Liu X., Liu Z., Wang S., et al. An eleven metabolic gene signature-based prognostic model for clear cell renal cell carcinoma. Aging (Albany NY) 2020;12:23165–23186. doi: 10.18632/aging.104088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rooney N., Mason S.M., McDonald L., Däbritz J.H.M., Campbell K.J., Hedley A., Howard S., Athineos D., Nixon C., Clark W., et al. RUNX1 is a driver of renal cell carcinoma correlating with clinical outcome. Cancer Res. 2020;80:2325–2339. doi: 10.1158/0008-5472.Can-19-3870. [DOI] [PubMed] [Google Scholar]
- 67.Nakase K., Tanaka I., Tawara I., Shiku H. CD22 expression in acute myeloid leukemia: close correlation with interleukin-2 receptor α-chain (CD25) expression and poor prognosis. Leuk Lymphoma. 2022;63:2251–2253. doi: 10.1080/10428194.2022.2062346. [DOI] [PubMed] [Google Scholar]
- 68.Eshibona N., Giwa A., Rossouw S.C., Gamieldien J., Christoffels A., Bendou H. Upregulation of FHL1, SPNS3, and MPZL2 predicts poor prognosis in pediatric acute myeloid leukemia patients with FLT3-ITD mutation. Leuk Lymphoma. 2022;63:1897–1906. doi: 10.1080/10428194.2022.2045594. [DOI] [PubMed] [Google Scholar]
- 69.Lamuedra A., Gratal P., Calatrava L., Ruiz-Perez V.L., Palencia-Campos A., Portal-Núñez S., Mediero A., Herrero-Beaumont G., Largo R. Blocking chondrocyte hypertrophy in conditional Evc knockout mice does not modify cartilage damage in osteoarthritis. Faseb J. 2022;36 doi: 10.1096/fj.202101791RR. [DOI] [PubMed] [Google Scholar]
- 70.Ran Y., Yin N., Huang D., Zhao Y., Yang J., Zhang H., Qi H. Identification and characterization of circular RNA as a novel regulator and biomarker in preterm birth. Front. Bioeng. Biotechnol. 2020;8 doi: 10.3389/fbioe.2020.566984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liao G., Lv J., Ji A., Meng S., Chen C. TLR3 serves as a prognostic biomarker and associates with immune infiltration in the renal clear cell carcinoma microenvironment. J. Oncol. 2021;2021 doi: 10.1155/2021/3336770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li H., Hu J., Yu A., Othmane B., Guo T., Liu J., Cheng C., Chen J., Zu X. RNA modification of N6-methyladenosine predicts immune phenotypes and therapeutic opportunities in kidney renal clear cell carcinoma. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.642159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Our dataset is primarily composed of two main components: Raw Data: This data is sourced from a genomic-related database curated by bioinformatics researchers at the University of California, Santa Cruz (UCSC). For more details, you can visit the official UCSC website at http://xena.ucsc.edu/. The specific data we utilized can be accessed directly from this link: https://xenabrowser.net/datapages/?dataset=TCGA-KIRC.htseq_fpkm.tsv&host=https%3A%2F%2Fgdc.xenahubs.net&removeHub=https%3A%2F%2Fxena.treehouse.gi.ucsc.edu%3A443. Analysis Code: We have archived our analysis code on Nutstore cloud storage. It can be accessed and downloaded directly from the provided link: https://www.jianguoyun.com/p/DdnXlgYQxoCACxiThosFIAA,TCGA-KIRC.