Abstract
Efficient human T-cell leukemia virus type 1 (HTLV-1) replication and viral gene expression are dependent upon the virally encoded oncoprotein Tax. To activate HTLV-1 transcription, Tax interacts with the cellular DNA binding protein cyclic AMP-responsive element binding protein (CREB) and recruits the coactivator CREB binding protein (CBP), forming a nucleoprotein complex on the three viral cyclic AMP-responsive elements (CREs) in the HTLV-1 promoter. Short stretches of dG-dC-rich (GC-rich) DNA, immediately flanking each of the viral CREs, are essential for Tax recruitment of CBP in vitro and Tax transactivation in vivo. Although the importance of the viral CRE-flanking sequences is well established, several studies have failed to identify an interaction between Tax and the DNA. The mechanistic role of the viral CRE-flanking sequences has therefore remained enigmatic. In this study, we used high resolution methidiumpropyl-EDTA iron(II) footprinting to show that Tax extended the CREB footprint into the GC-rich DNA flanking sequences of the viral CRE. The Tax-CREB footprint was enhanced but not extended by the KIX domain of CBP, suggesting that the coactivator increased the stability of the nucleoprotein complex. Conversely, the footprint pattern of CREB on a cellular CRE lacking GC-rich flanking sequences did not change in the presence of Tax or Tax plus KIX. The minor-groove DNA binding drug chromomycin A3 bound to the GC-rich flanking sequences and inhibited the association of Tax and the Tax-CBP complex without affecting CREB binding. Tax specifically cross-linked to the viral CRE in the 5′-flanking sequence, and this cross-link was blocked by chromomycin A3. Together, these data support a model where Tax interacts directly with both CREB and the minor-groove viral CRE-flanking sequences to form a high-affinity binding site for the recruitment of CBP to the HTLV-1 promoter.
The virally encoded Tax protein is implicated in the various clinical manifestations associated with infection by human T-cell leukemia virus type 1 (HTLV-1), including an aggressive and fatal T-cell malignancy (for a review, see reference 18). The mechanism of lymphocyte transformation by Tax is not known, although it appears to be linked to the pleiotropic transcriptional deregulation properties associated with the Tax oncoprotein. Following T-cell infection, the retrovirus appears to establish and maintain a state of latency, with very low levels of viral gene expression (31, 34). The transition from latency to high-level viral RNA synthesis is initiated by an ill-defined signal which triggers Tax expression. Tax protein is transported into the nucleus, where it interacts with the three conserved 21-bp repeat elements located in the transcriptional control region of the virus, strongly activating viral gene expression (11, 19, 32, 48, 50, 51).
The interaction between Tax and the 21-bp repeats is facilitated by cellular transcription factors, which provide the primary recognition of these Tax-responsive elements (3, 9, 23, 43, 44). Centered within each 21-bp repeat is an off-consensus cyclic AMP (cAMP) response element, termed a viral CRE, which serves as the recognition element for members of the activating transcription factor/CRE binding protein (ATF/CREB) family of cellular transcription factors. The CRE binding activity of these proteins is derived from their basic amino acid DNA binding domain and adjacent leucine zipper dimerization domain (bZIP domain). While several members of the ATF/CREB family bind to the CREs in the viral promoter, CREB appears to play the most prominent role in mediating Tax transactivation (1, 2, 4, 9, 12, 17, 24, 36, 56, 59–63).
Although the precise molecular events which lead to Tax transactivation mediated through CREB are not well understood, several studies have established that Tax enters into a stable ternary complex with CREB and the viral CRE (12, 26, 46, 54, 61, 63). In the absence of Tax, CREB forms an unstable complex with the viral CRE; however, in the presence of Tax, the dissociation rate of CREB is decreased (12). Studies with a variety of bZIP family members suggest that Tax interacts primarily, although not exclusively, with the bZIP region of CREB (1, 7, 12, 17, 49, 56, 60). This interaction may stabilize the α-helical structure of the parallel bZIP dimers, resulting in both enhanced DNA binding and dimerization of CREB (7, 49, 56). Together, these observations led to the suggestion that transactivation may occur as a consequence of Tax stabilization of CREB bound to the off-consensus viral CREs (12), leading to increased viral mRNA synthesis through the activation properties associated with CREB.
This model for Tax transactivation was incomplete, because it did not address the role of CREB phosphorylation, which is necessary for CREB transcriptional activity (25). Recently, we and others (24, 36) have shown Tax directly interacts with the cellular coactivator CREB binding protein (CBP), recruiting it directly to CREB and the viral promoter and bypassing the requirement for protein kinase A phosphorylation of CREB. Once tethered to the promoter by Tax, CBP appears to facilitate transcriptional activation through chromatin remodeling and recruitment of the general transcription machinery (6, 35, 45, 58). A small region of CBP (amino acids [aa] 455 to 719), called the KIX domain, is sufficient for binding to both Tax and the protein kinase-phosphorylated form of CREB (24, 25, 36, 47). Tax recruits the KIX domain of CBP directly to complexes containing the viral CRE and CREB (24, 36), resulting in the formation of a large nucleoprotein complex.
Anchoring of CBP to the HTLV-1 promoter is believed to promote the strong transcriptional activation observed in the presence of Tax; however the precise molecular events involved in Tax recruitment of CBP are not well understood. It is known that full-length CREB, a truncated form of CREB containing principally the bZIP domain, and the related bZIP protein ATF-1 are all competent for Tax recruitment of CBP to a viral CRE (24, 38). Additionally, efficient CBP recruitment by Tax requires a short run of highly conserved dG-dC (GC) sequences immediately adjacent to the CRE core in each viral CRE (24, 36). These GC-rich flanking nucleotides are also critical for the entry of Tax into protein-DNA complexes containing CREB (12, 46, 63). Both the Tax-dependent decrease in the dissociation rate of CREB from the CRE (12) and Tax transactivation in vivo (12, 20, 32, 42, 46) require the GC flanking sequences. In contrast, the CRE from the human chorionic gonadotropin gene, which carries primarily dA-dT (AT) sequences immediately flanking the CRE core, does not confer responsiveness to any of the above-mentioned properties associated with Tax. We will refer to this sequence as the cellular CRE.
The role of the GC base pairs in mediating the activities of Tax is not known, because several studies have been unsuccessful in identifying a direct interaction between Tax and the viral CRE DNA (3, 8, 43, 46). In this study, we further examined the role of the GC base pairs in mediating Tax function. Using high-resolution methidiumpropyl-EDTA iron(II) (MPE:Fe) footprinting (28, 29), we make the unique observation that Tax extends the protection of CREB from the viral CRE core into the GC-rich flanking sequences. Without affecting the dissociation rate of the nucleoprotein complexes, the KIX domain of CBP enhances but does not further extend the Tax footprint observed with CREB. We also show that the GC-specific minor-groove binding drug chromomycin A3, when bound to the viral CRE flanks, inhibits Tax recruitment of CBP to the CREB-viral CRE complex. Using a cross-linking strategy that probes for both major- and minor-groove interactions, we demonstrate that Tax cross-links to the 5′-flanking sequence of the viral CRE and that this cross-link is inhibited by chromomycin A3. Together, these data support a model where Tax directly interacts with both the bZIP domain of CREB and the GC sequences flanking the viral CRE. This Tax-containing complex then serves as a high-affinity binding site for CBP, anchoring the coactivator to the HTLV-1 promoter.
MATERIALS AND METHODS
Protein expression and purification.
The GST-KIX bacterial expression plasmid was expressed in Escherichia coli XL1Blue and purified to >90% homogeneity as previously described (24). The purified protein was stored at −70°C in 1× TM buffer (50 mM Tris-HCl, 100 mM KCl, 12.5 mM MgCl2, 1 mM dithiothreitol [DTT], 1 mM EDTA, 0.1% [vol/vol] Nonidet P-40, 20% [vol/vol] glycerol). Bacterial expression plasmids for human CREB (17), CREB Δ57-132, and CREB binding region (bZIP aa 254 to 327) (30) were transformed into E. coli BL21(pLysS) (53), and the proteins were expressed and purified as previously described (24, 30). Each protein was purified to >90% homogeneity and was stored at −70°C in TM buffer. Sequence-specific binding was confirmed by a competition electrophoretic mobility shift assay (EMSA) (references 5 and 17 and data not shown). The HTLV-1 Tax protein was expressed in E. coli XL1Blue from the pTaxH6 expression plasmid (62) and purified to >90% homogeneity by nickel chelate chromatography as previously described (24). Purified Tax was dialyzed against TM buffer and stored at −70°C.
In vitro transcription.
The HTLV-1 promoter DNA template fragment was isolated and purified after HindIII-PvuII digestion of pU3RCATdl10-1 plasmid containing the −306 promoter derivative (11). The viral and cellular CRE promoter templates were derived from the pminCAT 21-bp repeat and pminCAT-CRE (12) and were isolated and purified after PvuII digestion. Preinitiation complexes were formed on the purified DNA templates by the addition of the indicated amounts of CREB and/or Tax and 31.5 μg of CEM nuclear extract (15) in a final reaction volume of 25 μl. The reaction mixtures were incubated for 60 min at 30°C. RNA synthesis was initiated by the addition of 250 μM each ATP, CTP, and UTP and 12 μM GTP plus 0.07 μM [α-32P]GTP (3,000 Ci/mmol), and the mixture was incubated for an additional 30 min at 30°C. A constant amount of a purified, labeled 622-bp DNA fragment (isolated from an HpaII digest of pBR322) was added to each reaction mixture as a recovery standard. The RNA was isolated, purified, and analyzed by urea-polyacrylamide gel electrophoresis (PAGE). Gels were dried and visualized by PhosphorImager analysis. Radiolabeled fragments isolated from HpaII-digested pBR322 were used to estimate the size of the runoff transcripts.
DNA probes.
Footprinting and EMSA fragments were prepared by cloning complementary double-stranded oligonucleotides into the BamHI site in the polylinker of pUC19. The nucleotide sequences of the viral and cellular CREs are as follows (the viral CRE is from the HTLV-1 promoter-proximal 21-bp repeat, and the cellular CRE is from the human chorionic gonadotropin gene): Viral CRE AGGCGTTGACGACAACCCC Cellular CRE GATCCATGACGTCAATTGA
The CRE and CRE-like octanucleotide core sequences are shown in boldface type. The ∼75-bp EcoRI and HindIII fragments from these plasmids were isolated, and their concentrations were determined by measurement of UV absorbance at 260 nm. The purified fragments were 5′-end labeled on one strand with 32P for use in both the EMSA and footprinting reactions. Nucleotide sequences in the footprinting reactions were assigned by Maxam-Gilbert chemical sequencing of the labeled DNA fragments (40).
MPE:Fe footprinting analysis.
Footprinting experiments were carried out in a 20-μl reaction volume (pH 7.9) in 0.5× TM buffer. The indicated proteins or DNA binding drugs were incubated with 10 fmol of the DNA probe and 1 mg of the alternating copolymer poly(dA-dT) per ml for 20 min at room temperature. MPE:Fe digestion (28, 29) of the protein-DNA or drug-DNA complexes was initiated by the addition of 1 μl of 0.25 M DTT and 100 ng of MPE complexed with Fe [final concentrations, 6.0 μM MPE and 2.5 μM Fe(NH4)2(SO4)2]. Digestions were carried out for 8 min and terminated with Bathophenanthroline added to a final concentration of 10 mM. DNA fragments were purified, analyzed by urea-PAGE, and visualized by PhosphorImager analysis. Phoretix Photometrics 1D was used to determine the relative radioactive signal for each nucleotide band. To normalize each sample, an individual band outside of the protected region was used as a reference. The normalized radioactive signal for each nucleotide in the DNA ladder was given a value of 1.0, and the nucleotide signals from the protein-containing lanes were assigned values relative to the corresponding DNA ladder bands (DNA ladder band signal divided by protein-DNA complex band signal). Values above 1.0 were considered evidence of protection, and values below 1.0 were considered evidence of hypersensitivity. The results of three independent experiments were quantitated and averaged.
EMSA.
EMSAs were performed by incubation of the indicated amount of purified proteins and DNA binding drugs (Sigma) under the precise buffer conditions used in the footprinting reactions described above. The appropriate 32P-end-labeled DNA (2 fmol) and 250 ng of poly(dA-dT) per ml were used as probes in a 20-μl reaction mixture. The drugs were incubated with the DNA for 20 min at room temperature before protein addition. The appropriate proteins were added, the reaction tube was incubated on ice for 30 min, and its contents were analyzed on 5% nondenaturing polyacrylamide gels (acrylamide/N,N′-methylenebisacrylamide, 49:1 [wt/wt]) in buffer containing 0.04 M Tris · HCl, 0.306 M glycine (pH 8.5), and 0.1% (vol/vol) Nonidet P-40. The gels were visualized with a PhosphorImager. For the kinetic binding studies, the viral CRE was incubated with appropriate concentrations of CREB bZIP in the presence or absence of Tax and/or KIX to produce less than 50% of the probe bound in complexes. The binding-reaction mixtures were challenged with a 1,000-fold molar excess of unlabeled cellular CRE binding site and loaded onto a running gel at appropriate times following challenge. The protein-DNA complexes were resolved by EMSA, and the percent bound was determined by Imagequant and graphed with Cricket Graph.
Protein-DNA cross-linking.
Complementary oligonucleotides containing the sequence 5′-AGGCGTTGACGACAACCCCGATC-3′ (top strand) were obtained from Macromolecular Resource Facilities (Fort Collins, Colo.). Eight distinct top-strand oligonucleotides, each containing a single phosphorothioate substitution at the indicated scissile linkage, were synthesized (see Fig. 5A). The oligonucleotides were suspended in 10 mM Tris (pH 8.0)–1 mM EDTA (TE), and their concentrations were determined by measuring the absorbance at 260 nm. Subdued lighting was used in all subsequent steps. Attachment of an azidophenacyl group to the phosphorothioate was performed for 3 h in a 130-μl reaction mixture (58% methanol in water) containing 38.5 μM phosphorothioate-substituted top-strand oligonucleotide, 38.5 mM potassium phosphate buffer (pH 7.0), and 1.15 mM p-azidophenacyl bromide (Sigma) (37, 41). Following incubation, the derivatized oligonucleotides were precipitated, resuspended in 50 μl of TE, and 5′-32P-end labeled with T4 polynucleotide kinase. The resulting end-labeled, derivatized top-strand oligonucleotides were annealed with an equimolar amount of unmodified, complementary bottom strand.
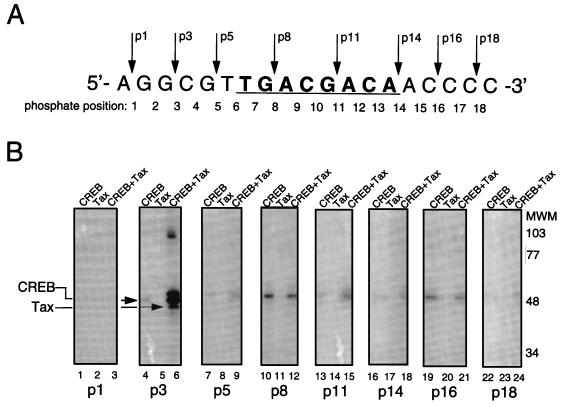
FIG. 5.
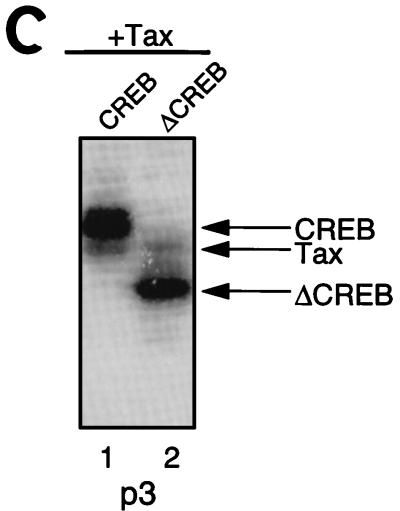
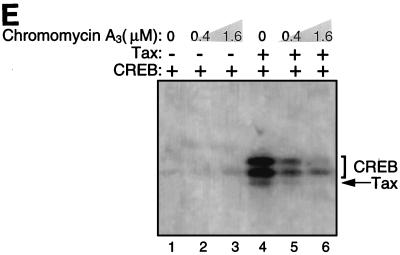
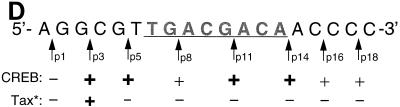
Tax cross-links to the 5′-flanking region of the viral CRE. (A) Schematic representation of the individual phosphate positions derivatized within the top strand of the viral CRE. The numbers below the sequence refer to the phosphate positions. The position of each cross-linking arm is indicated by arrows above the sequence. For example, p1 represents the oligomer which contains a single azidophenacyl cross-linking moiety attached to the phosphorothioate positioned at the first phosphate in the DNA backbone. (B) SDS-PAGE analysis of protein-DNA cross-linking. Cross-linking reaction mixtures contained 10 ng of CREB and/or 250 ng of Tax in the presence of 25 to 40 fmol of viral CRE. Each binding-reaction mixture contained the photoreactive cross-linking arm in the position indicated below the lane numbers. The positions of the proteins cross-linked to the derivatized viral CRE are indicated, and molecular weight markers (MWM) are shown on the right. (C) Identification of the cross-linked polypeptides with truncated CREB. Cross-linking reaction mixtures contained 10 ng of CREB or 2 ng of CREB Δ57–132 and 250 ng of Tax in the presence of 30 fmol of the viral CRE. The binding-reaction mixtures contained the photoreactive cross-linking arm at position 3. The positions of the proteins cross-linked to the derivatized viral CRE are indicated on the right. (D) Summary of the cross-linking studies. A schematic representation of all the phosphate positions individually examined in this study is shown at the top of the figure. The presence (+) or absence (−) of a CREB or Tax cross-link is indicated below each phosphate position. Boldface type indicates specific positions where Tax enhanced the CREB cross-link. The asterisk indicates that all Tax cross-links were observed only in the presence of CREB. (E) Chromomycin A3 inhibits cross-linking of Tax to the viral CRE. Reaction mixtures contained 30 fmol of p3 probe (preincubated with the indicated amount of chromomycin A3) and 10 ng of CREB (lanes 1 to 3) and/or 250 ng of Tax (lanes 4 to 6). The complexes were analyzed by SDS-PAGE.
Cross-linking binding-reaction mixtures contained 25 to 40 fmol of derivatized, singly end-labeled, annealed oligonucleotide, 10 ng of CREB or 2 ng of CREB Δ57-132 and/or 250 ng of TaxH6 (each protein purified as described above, omitting the addition of DTT in the final dialysis step), 0.5× TM buffer (containing 0.0125% Tween 20), and 40 ng of poly(dA-dT). The binding-reaction mixtures were incubated for 20 min at room temperature under subdued lighting. Following incubation, photoaffinity cross-linking was performed by irradiating samples for 5.5 min at room temperature with a model UVGL-58 Mineralight hand-held UV lamp (366 nm) positioned 7 cm above the sample. Sodium dodecyl sulfate (SDS) sample dyes were added, and the reaction mixtures were analyzed by SDS-PAGE (12% polyacrylamide). After electrophoresis, the gels were dried and protein-DNA cross-links were visualized by PhosporImager analysis. Reactions involving chromomycin A3 were performed as described above, except that twice the indicated amount of chromomycin A3 was preincubated with the p3 oligonucleotide for 20 min before protein addition. The volume of the protein mixture brought the final chromomycin A3 concentration to that indicated in Fig. 5E.
RESULTS
Efficient Tax transactivation is dependent upon the viral CRE in vitro.
The viral CREs are composed of an off-consensus core CRE octanucleotide, which serves as a recognition element for CREB (and other ATF/CREB proteins), and a short run of conserved G · C base pairs immediately upstream and downstream of the core (see Materials and Methods). The GC-rich flanking sequences are critical for mediating Tax transactivation in vivo (12, 20, 32, 42, 46), as well as for several biochemical functions of Tax in vitro. These include nucleoprotein complex formation between Tax, CREB, and the viral CRE (12, 46, 63); Tax stabilization of CREB binding (7, 12, 59, 60); and Tax recruitment of CBP (24, 36). A cellular CRE, which does not contain the GC-rich flanking sequences, is negative for these biochemical functions of Tax.
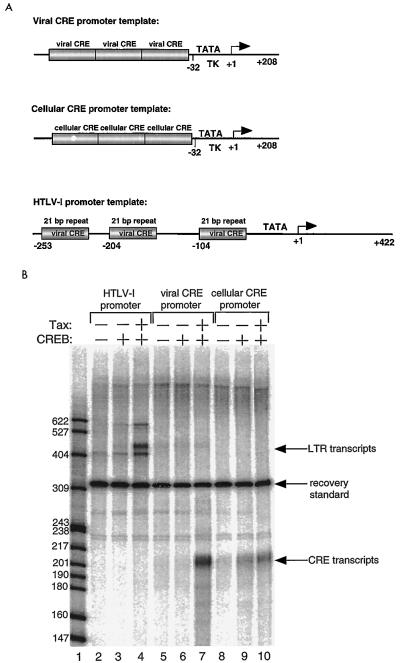
To begin dissecting the function of the GC sequences in the viral CRE, we first tested whether these elements mediate CREB-dependent Tax transactivation in vitro. We performed runoff transcription assays with heterologous promoters containing three copies of either the viral CRE or the cellular CRE (see Materials and Methods) cloned immediately upstream of the herpesvirus thymidine kinase minimal promoter (12). A schematic representation of each promoter template is shown in Fig. 1A. The activity of the chimeric promoters was compared directly with that of the natural viral promoter, which contains the entire HTLV-1 transcriptional control region, including the three Tax-responsive viral CREs (Fig. 1A). Promoter template fragments were incubated with purified recombinant Tax and/or CREB and nuclear extracts prepared from the HTLV-1-negative human T-lymphocyte cell line CEM. The radiolabeled runoff RNA transcripts were analyzed by denaturing PAGE. We used primer extension to confirm that the major runoff transcripts correctly initiated at +1 (with a minor HTLV-1 transcript initiating at −30), and we used α-amanitin sensitivity to show that the transcripts were products of RNA polymerase II (data not shown; see references 17 and 39). The addition of small amounts of CREB to the preincubation reaction mixtures did not significantly increase the amount of transcription from the three promoters (Fig. 1B, lanes 3, 6, and 9). However, the addition of Tax to the reaction mixtures containing CREB increased the level of RNA synthesis in a viral CRE-dependent manner, indicating that these flanking sequences are critical for Tax transactivation in vitro (Fig. 1B, compare lanes 3 and 6 and lanes 4 and 7). Under identical reaction conditions, Tax was unable to stimulate transcription from the promoter carrying the cellular CRE (lanes 8 and 9), which does not have GC-rich DNA adjacent to the CRE core. We did not observe significant Tax transactivation in the absence of exogenously added CREB (data not shown). These data indicate that in vitro, as in vivo, the GC sequences adjacent to the viral CRE core are critical for Tax transactivation.
FIG. 1.
Tax transactivation in vitro is dependent upon CREB and the viral CRE. (A) Schematic representation of the linearized promoter templates used in the in vitro transcription reactions. The HTLV-1 promoter carries the full transcriptional control region of the virus, including the three 21-bp repeats (viral CREs). The heterologous promoters each contained three tandem copies of either the viral CRE (from the promoter-proximal 21-bp repeat) or the cellular CRE (from the human chorionic gonadotropin gene), cloned immediately upstream of the herpesvirus thymidine kinase (TK) minimal promoter (at position −32). (B) In vitro transcription assay showing that Tax activation of transcription is dependent upon the viral CREs. Transcription reaction mixtures contained the HTLV-1 template fragment (50 ng) (lanes 2 to 4), the viral CRE promoter template fragment (100 ng) (lanes 5 to 7), or the cellular CRE promoter template fragment (25 ng) (lanes 8 to 10) in the presence of 32 μg of nuclear extract from the HTLV-1-negative human T-lymphocyte cell line CEM. A 50-ng amount of purified, recombinant CREB and/or 125 ng of purified recombinant Tax was added to the indicated reaction mixtures. A labeled DNA fragment was added to each reaction prior to RNA isolation to measure the recovery of the labeled RNA transcript. The positions of the recovery standard, the RNA transcripts, and the size of the radiolabeled DNA markers are indicated.
GC sequences in the viral CRE are protected from MPE:Fe digestion in a Tax-dependent manner.
The importance of the GC-rich flanking sequences for Tax transactivation strongly suggests that these DNA elements may stabilize the association of Tax with the viral promoter, possibly through sequence-specific contacts with Tax. Previous DNase I footprinting and methylation interference studies have been unable to show an interaction between Tax and the viral CRE (3, 43, 46). These negative results, however, do not rule out the possibility that direct contacts between Tax and the DNA exist but have been refractory to identification. To further examine whether Tax contacts the viral CRE DNA, we used MPE:Fe footprinting (28, 29), because this technique reveals sugar-backbone contacts and thus differs in cleavage chemistry from the footprinting methods used previously. The 75-bp singly end-labeled fragment containing the viral CRE DNA (see Materials and Methods) was incubated with purified recombinant CREB and/or Tax and treated with MPE, and the cleavage products were separated by denaturing PAGE. Chemical sequencing reactions were run adjacent to the cleavage reactions to allow nucleotide sequence assignment (40). As expected, the addition of CREB to the binding-reaction mixtures containing the viral CRE probe (labeled on the noncoding strand) produced an MPE:Fe footprint centered directly over the CRE octanucleotide (Fig. 2A, lane 2). Surprisingly, the addition of increasing amounts of Tax to the binding-reaction mixtures containing CREB extended the protection from the viral CRE core into the flanking sequences (lanes 3 to 5). Tax expanded the CREB footprint both 5′ and 3′ of the CRE core. We observed a similar expansion of the CREB footprint on the coding strand of the viral CRE (Fig. 2D). The alteration of the CREB footprint was Tax dependent, since the addition of either glutathione S-transferase (GST)–Tax, a fusion protein defective for ternary-complex formation (2), or 10 μg of bovine serum albumin did not produce a change in the CREB footprint (data not shown). Tax did not produce a footprint in the absence of CREB, indicating that CREB is required for the Tax-dependent change in the footprint pattern (Fig. 2A, lane 7).
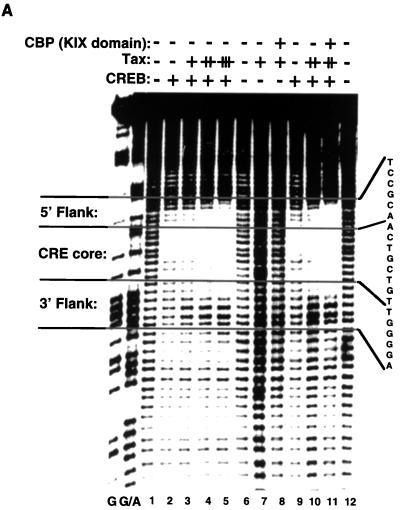
FIG. 2.
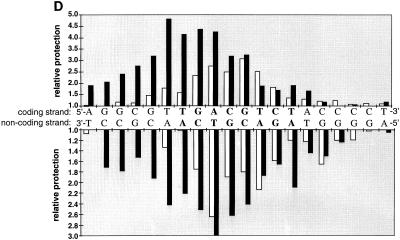
Tax protein alters the MPE:Fe footprint pattern of CREB on a viral CRE but not on a cellular CRE. (A) Tax expands and enhances the MPE:Fe footprint of CREB on the noncoding strand of the viral CRE. The binding-reaction mixtures contained 10 fmol of the viral CRE probe, singly end labeled on the noncoding strand, and 25 ng of purified, recombinant CREB (lanes 2 to 5 and 9 to 11). The reaction mixtures also contained 50 ng (lane 3), 100 ng (lane 4), or 200 ng (lanes 5, 7, 8, 10, and 11) of purified recombinant Tax and 200 ng of purified GST-KIX (lanes 8 and 11), as indicated. Maxam-Gilbert sequencing reactions for guanine and guanine/adenine were run adjacent to the MPE:Fe cleavage products to assign specific nucleotides (40). (B) The KIX domain of CBP is required for Tax expansion of the CREB bZIP footprint. Binding-reaction mixtures contained 10 fmol of the viral CRE probe, singly end labeled on the coding strand, and 5 ng of purified CREB bZIP domain (amino acids 254 to 327) (lanes 2 to 4 and 7 to 9). The reaction mixtures also contained 100 ng (lanes 3 and 9) or 200 ng (lane 4) of Tax and 1 μg of GST-KIX (lanes 8 and 9), as indicated. Maxam-Gilbert sequencing markers were run adjacent to the cleavage products to assign specific nucleotides. (C) Tax does not alter the footprint of CREB or CREB bZIP on the cellular CRE. The binding-reaction mixtures contained 10 fmol of the cellular CRE, singly end labeled on the coding strand, and 10 ng of CREB (lanes 2 to 6 and 14) or 2 ng of CREB bZIP (lanes 8 to 12 and 15). The reaction mixtures also contained 200 ng (lanes 3 and 9) or 400 ng (lanes 4 to 6, 10 to 12, 16 and 17) of Tax and 1 μg (lanes 5 and 11) or 2 μg (lanes 6, 12, 14 to 16, and 18) of GST-KIX as indicated. Maxam-Gilbert sequencing reactions were run adjacent to the MPE:Fe cleavage products to assign specific nucleotides. (D) Schematic representation of the relative protection produced by CREB or by CREB and Tax on the viral CRE. To account for Tax enhancement of CREB binding, experiments with high concentrations of CREB were directly compared to experiments with lower concentrations of CREB in the presence of Tax that gave similar degrees of protection in the core CREB binding site. The relative protection from MPE:Fe cleavage in the presence of CREB (open bars) or CREB and Tax (solid bars) was quantitated and averaged from three separate experiments. Bar heights are proportional to the degree of protection from the cleavage agent, with DNA-alone values equal to 1.0 and higher values corresponding to increased protection from MPE:Fe cleavage.
The cellular coactivator CBP binds to Tax in the presence of CREB and the viral CRE and, through this interaction, appears to play a prominent role in mediating Tax transactivation (24, 36). We were interested in testing whether CBP might alter the pattern of MPE:Fe protection observed in the presence of Tax and CREB on the viral CRE probe. The addition of purified, recombinant KIX domain of CBP (GST-KIX; aa 451 to 720 [24]) to binding-reaction mixtures containing the viral CRE, CREB, and Tax did not further extend the pattern of MPE:Fe protection observed with just CREB and Tax (Fig. 2A, compare lane 11 with lanes 4, 5, and 10). KIX did, however, enhance the protection observed, suggesting that KIX may stabilize the Tax-CREB-DNA complex without altering specific protein interactions with the DNA. KIX produced no change in the MPE:Fe cleavage pattern in the absence of CREB and produced no change in the cleavage pattern of CREB in the absence of Tax (lane 8, and data not shown).
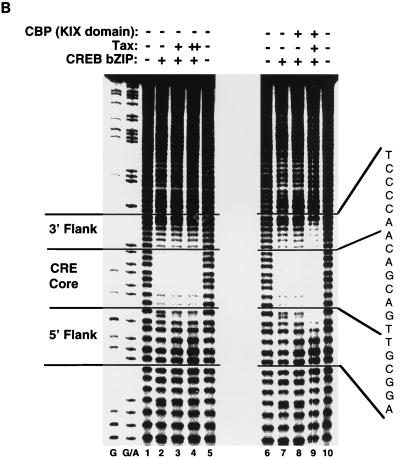
The Tax-dependent change in MPE:Fe protection on the viral CRE strongly suggests a Tax-DNA interaction; however, we cannot rule out the possibility that Tax alters the conformation of CREB, promoting novel contacts between CREB and the DNA flanking the octanucleotide CRE. To address this possibility, we used a truncated form of CREB, containing only the N-terminal 73-aa DNA binding and dimerization domain (bZIP domain) (CREB bZIP; aa 254 to 327 [30]). Several reports have demonstrated that Tax interacts with the bZIP domain of CREB (7, 12, 17, 49, 56, 59). While the structure of the bZIP CREB-DNA complex is not known, the crystal structure of the 56-aa bZIP domain of the related protein GCN4, bound to its cognate recognition element, has been determined (16). The GCN4 bZIP dimer forms a pair of parallel α-helices that contact the major groove of the DNA and extend into the leucine zipper dimerization interface. The GCN4 α-helices form a smooth, continuous dimeric structure that is oriented perpendicular to the axis of the DNA. The structure of the DNA-bound CREB bZIP is probably comparable to that of the DNA-bound GCN4 bZIP, since the proteins are similar in their amino acid sequence and the sequence of their recognition elements. Based on these observations, we hypothesized that the CREB bZIP domain, bound perpendicular to the axis of the viral CRE, would be unlikely to make additional DNA contacts in the presence of Tax. Figure 2B shows that CREB bZIP protected the viral CRE core indistinguishably from the protection observed with full-length CREB (lane 2; compare with Fig. 2D). However, titration of Tax into binding-reaction mixtures containing CREB bZIP produced no change in the MPE:Fe footprint pattern of the viral CRE (Fig. 2B, lanes 3 and 4). While this result was unexpected, it is known that the interaction between Tax and truncated CREB is considerably weaker than the interaction between Tax and full-length CREB, since EMSA ternary complexes are not observed with Tax and CREB bZIP (12). Since we have previously shown that the addition of KIX to EMSAs containing Tax, CREB bZIP, and the viral CRE produces a stable quaternary complex (24), we hypothesized that CBP might stabilize the Tax-CREB bZIP-viral CRE interaction. Figure 2B shows that the addition of the KIX domain to the binding-reaction mixtures containing CREB bZIP and Tax resulted in an extension of the CREB bZIP protection pattern that was nearly indistinguishable from that observed with Tax and full-length CREB. The same result was obtained with the viral CRE noncoding strand (data not shown). The amino terminus of CREB therefore does not appear to be responsible for the additional protein-DNA contacts observed in the footprint. These data provide further support for the hypothesis that CBP stabilizes the Tax-CREB bZIP-DNA complex.
To test whether the observed Tax-dependent changes in the MPE:Fe footprint pattern of CREB required the GC-rich flanking sequences of the viral CRE, we performed parallel experiments with the 75-bp singly end-labeled cellular CRE probe. Figure 2C shows that the addition of CREB and CREB bZIP to the cellular CRE probe each produced the expected protection of the CRE core (lanes 2 and 8). However, the addition of Tax, KIX, or Tax plus KIX did not extend the CREB protection or alter the footprint pattern in any detectable manner (lanes 3 to 6 and 9 to 12). Identical results were obtained with the opposite strand of the cellular CRE DNA labeled (data not shown). These data suggest that the specific GC-rich flanking sequences of the viral CRE are required for the altered footprint pattern produced by Tax.
A summary schematic showing quantitation of the MPE:Fe protection pattern observed in the presence of Tax on the third viral CRE is shown in Fig. 2D. This analysis revealed that both the 5′ and 3′ viral CRE flanking sequences were protected in the presence of Tax. We were concerned, however, that Tax enhancement of CREB DNA binding activity may account for expansion of the CREB footprint into the flanking sequences. To eliminate this possible effect, we compared footprints obtained by using high concentrations of CREB alone with footprints where a similar degree of core CRE protection was observed with less CREB in the presence of Tax. As shown in Fig. 2D, the expansion of the CREB footprint by Tax is observed primarily 5′ of the viral CRE core. Similar Tax-dependent expansions of the CREB MPE:Fe footprint were also observed on the first and second viral CREs in the context of the full HTLV-1 promoter (data not shown).
The KIX domain of CBP does not alter the dissociation kinetics of CREB bZIP in the presence or absence of Tax.
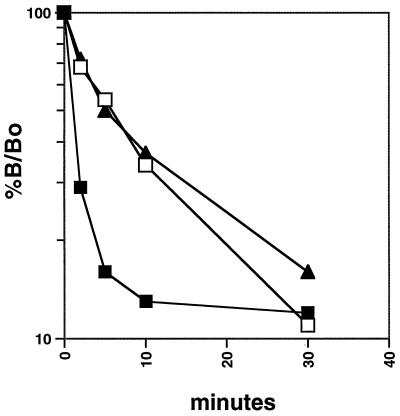
The observation that the KIX domain of CBP enhanced but did not extend the protection pattern observed with Tax, CREB bZIP, and the viral CRE DNA suggests that CBP increases the binding affinity of Tax for the CREB bZIP-DNA complex. To investigate the basis for this stabilization, we analyzed the dissociation kinetics of CREB bZIP binding to the viral CRE probe in the presence of Tax and/or KIX. The labeled viral CRE probe was incubated with the appropriate proteins, and the binding-reaction mixtures were then challenged with a large excess of unlabeled cellular CRE and loaded onto a nondenaturing polyacrylamide gel. The kinetics of CREB bZIP dissociation were determined by quantitative analysis of the EMSA. A plot of CREB bZIP dissociation from the DNA is shown in Fig. 3. In the absence of Tax, the dissociation of CREB bZIP from the viral CRE was rapid (t1/2 = 1 min). The addition of Tax to the binding-reaction mixture increased the stability of the CREB bZIP-DNA complex (t1/2 = 5.5 min). We have previously observed a similar yet more dramatic stabilization of full-length CREB by Tax (12). Surprisingly, the addition of KIX to binding-reaction mixtures containing CREB bZIP and DNA (data not shown) or CREB bZIP, Tax, and DNA (Fig. 3) had no effect on the dissociation rate of the complexes. These data indicate that, as with full-length CREB (24), KIX has no detectable effect on the dissociation kinetics of CREB bZIP from the viral CRE. Furthermore, they strongly suggest that the observed stabilization by KIX must derive from an increase in the on-rate. We have examined the kinetics of CREB bZIP association in the presence of Tax and KIX and have found that the binding reactions have reached equilibrium by the earliest time point (10 s), thus making kinetic analysis difficult (data not shown).
FIG. 3.
Kinetics of CREB bZIP dissociation in the presence of Tax and KIX. The 73-aa bZIP domain of CREB was incubated with the viral CRE probe in the absence or presence of Tax or of Tax plus KIX. Binding-reaction mixtures were then challenged with a 1,000-fold molar excess of unlabeled cellular CRE binding site, and the kinetics of dissociation were analyzed by EMSA and quantitated by ImageQuant. The concentration of protein-DNA complexes remaining bound, relative to the concentration bound at time zero (no added competitor) (B/B0), was plotted as a function of the time after challenge. The dissociation of CREB bZIP from the viral CRE probe (solid squares) in the presence of Tax (open squares) or in the presence of Tax plus KIX (solid triangles) is shown.
The GC-specific minor-groove DNA binding drug chromomycin A3 inhibits the putative Tax interaction with the viral CRE.
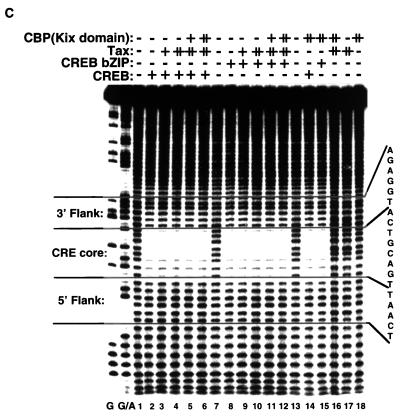
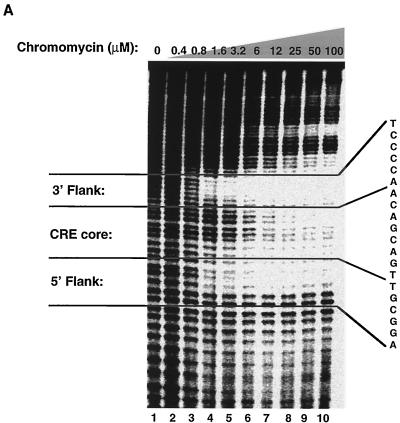
To further characterize the interaction between Tax and the DNA sequences in the viral CRE, we used a panel of specific minor-groove binding drugs to determine if they might block access of Tax to the GC nucleotides in the viral CRE. These nonintercalating DNA binding drugs are aureolic acid derivatives that have antibiotic and antitumor activity (for a review, see reference 64) and have previously been used to inhibit transcription factor interactions with DNA (10, 13, 14, 22, 57). The minor-groove binding drug chromomycin A3 was selected because it preferentially binds dG-dC base pairs with ∼1 μM binding affinity (57). To establish whether chromomycin A3 bound specifically to the viral CRE flanking sequences, we used MPE:Fe footprinting to identify the nucleotide sequences protected by the drug. Figure 4A shows that the lower concentrations of chromomycin A3 (0.8 to 1.6 μM) produced MPE:Fe protection specifically in the GC-rich flanking sequences adjacent to the viral CRE core. This data suggests that chromomycin A3 may be a useful reagent to test whether Tax requires access to the GC-rich viral sequences to recruit CBP.
FIG. 4.
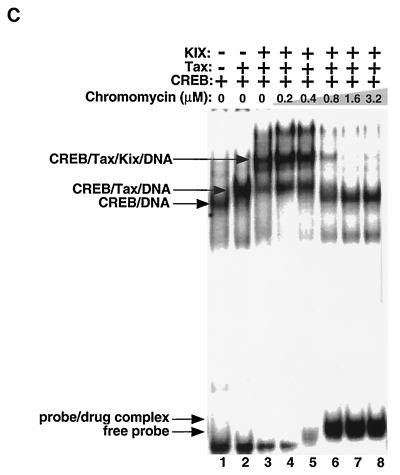
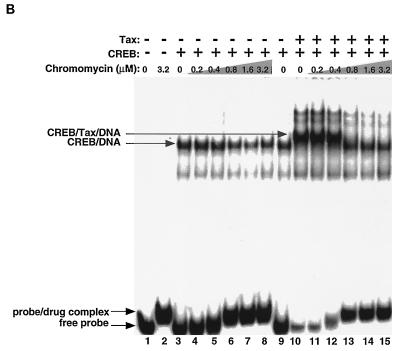
Chromomycin A3 binds to the GC-rich flanking sequences and inhibits the association of Tax and KIX with CREB and the viral CRE. (A) MPE:Fe footprinting shows that chromomycin A3 binds preferentially to the viral CRE flanking sequences. Increasing concentrations of chromomycin A3 (lanes 2 to 10) were incubated with 10 fmol of the viral CRE probe singly end labeled on the coding strand. Nucleotide positions were assigned based on previous footprinting experiments with Maxam-Gilbert sequencing markers. (B) Chromomycin A3 inhibits association of Tax with CREB and the viral CRE. Increasing concentrations of chromomycin A3 were incubated with the viral CRE probe for 20 min before the addition of 5 ng of CREB (lanes 3 to 15) and 50 ng of Tax (lanes 10 to 15), as indicated. The binding-reaction products were analyzed by EMSA. The positions of the free probe and the protein-DNA and drug-DNA complexes are indicated. (C) Chromomycin A3 inhibits the association of Tax and KIX with the viral CRE. Increasing concentrations of chromomycin A3 were incubated with the DNA for 20 min before the addition of 5 ng of CREB (lanes 1 to 8), 50 ng of Tax (lanes 2 to 8), and/or 70 ng of GST-KIX (lanes 3 to 8), as indicated. The binding-reaction products were analyzed by EMSA. The positions of the free probe and the protein-DNA and drug-DNA complexes are indicated.
Several previous studies have used EMSA to show the presence of Tax complexed with CREB and the viral CRE (12, 46, 63). In the EMSA, Tax forms a slower-migrating ternary complex in the presence of full-length CREB and the viral CRE (Fig. 4B, compare lanes 9 and 10). The ternary complex is dependent upon the G · C base pairs flanking the viral CRE octanucleotide, since it does not form with the cellular CRE. We used EMSA to determine whether the binding of chromomycin A3 to the GC sequences in the viral CRE inhibits Tax association with the CREB-viral CRE complex. Increasing concentrations of chromomycin A3 were titrated into binding-reaction mixtures containing CREB and the viral CRE probe, in the presence or absence of Tax. Figure 4B shows that at 0.8 μM chromomycin A3, the drug bound to the viral CRE DNA, since we observed a modest “supershift” of the free probe (compare lanes 5 and 6 and lanes 12 and 13). There was also complete loss of the ternary complex containing Tax at this concentration of chromomycin A3, with conversion to the binary complex containing CREB and the viral CRE probe (lanes 10 to 15). The loss of Tax from the nucleoprotein complex occurred at precisely the same concentration of chromomycin A3 which produced MPE:Fe protection of the GC nucleotides in the viral CRE (Fig. 4A). Additionally, at this concentration, we observed that Tax enhancement of CREB binding was lost, since the fraction of CREB bound to DNA returned to that observed in the absence of Tax (Fig. 4B, compare lanes 6 to 8 with lanes 13 to 15; quantitation not shown). Under identical EMSA reaction conditions, chromomycin A3 did not affect the binding of CREB to the viral CRE probe (Fig. 4B, lanes 3 to 8), consistent with the interaction of the DNA binding domain of CREB with the major groove of the CRE core (16). As expected, chromomycin A3 had no effect on EMSA binding-reaction mixtures containing Tax, CREB, and the cellular CRE probe, since Tax does not form a ternary complex with CREB bound to a CRE lacking the GC-rich flanking sequences (data not shown).
We also tested whether chromomycin A3 would inhibit the association of the KIX domain of CBP with the Tax-CREB-viral CRE complex. Figure 4C shows that, as expected, the addition of Tax promoted ternary-complex formation with CREB on the viral CRE (lane 2). The addition of KIX to the binding-reaction mixture produced a much more slowly migrating quaternary complex, containing CREB, Tax, KIX, and the viral CRE (lane 3). We and others have previously shown that this quaternary complex is composed of these proteins (24, 36). The addition of chromomycin A3 to the binding-reaction mixtures converted this quaternary complex directly to a CREB-viral CRE binary complex at precisely the concentration of drug which bound the viral CRE probe (Fig. 4C, lanes 4 to 8). Chromomycin A3 also inhibited Tax recruitment of KIX to the CREB bZIP domain-viral CRE complex at exactly the same concentrations that inhibited Tax-DNA interactions in the presence of full-length CREB (data not shown). Significantly higher concentrations of chromomycin A3 were needed to disrupt Tax-DNA interactions if the proteins were preincubated with the DNA prior to drug addition. This data suggests that the mechanism of chromomycin A3 inhibition of Tax is direct competition for access to the viral CRE-flanking sequences and not disruption of protein-protein interactions (data not shown). We have obtained identical results with the dG-dC minor-groove binding drug olivomycin (data not shown), and as a negative control, we used the dA-dT-specific minor-groove binding drugs netropsin and distamycin A. These two drugs bound preferentially to the viral CRE core and did not specifically disrupt ternary or quaternary complexes formed in the presence of Tax (data not shown). Together, these data strongly support the conclusion that CBP is directly recruited to the viral CRE through interaction with Tax and that disruption of the Tax-DNA interaction by GC-specific minor-groove binding drugs abolishes recruitment of CBP.
Tax covalently cross-links to the 5′ viral CRE-flanking sequence.
The MPE:Fe footprinting and minor-groove binding studies above suggest that Tax may directly bind to GC-rich sequences in the minor groove of the viral CRE-flanking sequences. To more directly test this hypothesis, we performed protein-DNA cross-linking analysis with eight distinct photoreactive DNA derivatives with availability for both minor- and major-groove interactions (37, 41). The site-specific placement of a single 9.7-Å azidophenacyl cross-linking arm at various positions along the top strand of the viral CRE is shown in Fig. 5A. These derivatized, 5′-end-labeled oligonucleotides were annealed and incubated in individual binding-reaction mixtures containing CREB and/or Tax. The binding-reaction mixtures were exposed to 366-nm UV light, and the protein-oligonucleotide (full length) products were directly analyzed by SDS-PAGE. Figure 5B shows that CREB cross-linked at nearly all positions within the viral CRE, with the exception of the most 5′ position, p1 (lanes 4, 7, 10, 13, 16, 19, and 22). The most intense CREB cross-links were generally observed within and near the CRE core. This extensive CREB cross-linking may result from positioning of the 9.7-Å arm directed toward the CRE core. Interestingly, we observed a covalent Tax cross-link only at the p3 position in the 5′ flank, within the conserved AGGC sequence of the viral CRE (lane 6). As expected, the Tax-DNA cross-link was dependent upon the presence of CREB in the binding reaction (compare lane 5 to lane 6). While only a single Tax cross-link was observed, Tax slightly enhanced the cross-linking of CREB at several positions within the viral CRE, with very significant enhancement of CREB binding at p3 (lanes 6, 9, 15, and 18). This is consistent with previous observations showing that Tax enhances the equilibrium binding affinity of CREB to the viral CRE (7, 12, 49, 56, 61). Enhancement of both the Tax and CREB cross-links was observed in the presence of the KIX domain of CBP (data not shown). Since Tax and CREB are similar in molecular weight, their cross-linked products migrate in close proximity on SDS-PAGE. To establish the identity of the cross-linked proteins, we used a deletion mutant of CREB (CREB Δ57–133, which behaves as the wild type in interactions with Tax [unpublished data]) which migrated significantly faster in SDS-PAGE (Fig. 5C). This approach also enabled the identification of cross-linked Tax, since the position of the Tax-dependent band remained unaffected. Although the Tax used in this assay was purified to greater than 95% homogeneity, definite conformation of Tax in the cross-linking experiment could not be obtained by immunodepleting Tax from the binding-reaction mixture, since our anti-Tax antibody recognizes only denatured Tax (data not shown). The cross-linked species migrating slightly slower than CREB in the p3 reaction appears to be composed of CREB, since the migration of this species depends on the molecular weight of CREB (Fig. 5C). The high-molecular-weight species in p3 (Fig. 5B, lane 6) appears to be a cross-linked CREB dimer, since its molecular weight is also altered when CREB Δ57–133 is used in the reaction (data not shown). A summary of the cross-linking data is shown in Fig. 5D.
To identify whether the Tax-DNA cross-link was specific to the minor groove, we performed protein-DNA cross-linking reactions in the presence of the minor-groove binding drug chromomycin A3. Increasing amounts of chromomycin A3 were preincubated with the photoreactive p3 probe, and binding-reaction mixtures containing CREB and/or Tax were analyzed. Figure 5E shows that increasing concentrations of chromomycin A3 significantly reduced the Tax-DNA cross-link (lanes 4 to 6) while the CREB-DNA cross-link in the absence of Tax remained unchanged (lanes 1 to 3). Interestingly, Tax enhancement of the CREB cross-link was also eliminated in the presence of chromomycin A3, suggesting that this property of Tax is also abolished if Tax does not have access to the DNA. Together, these data suggest that Tax interacts specifically with GC sequences in the minor groove of the viral CRE.
DISCUSSION
The mechanistic role of the viral CRE-flanking sequences in mediating Tax transactivation has remained an unresolved question in HTLV-1 biology for many years. The GC-rich sequences flanking the viral CRE are critical for mediating Tax activation of HTLV-1 transcription in vivo (12, 20, 32, 42, 46). Several in vitro studies have corroborated the in vivo studies, including demonstration that the flanking sequences are required for Tax stabilization of CREB on the viral CRE (7, 12, 61). The flanking sequences are also required for efficient Tax recruitment of the pleiotropic coactivator CBP to the HTLV-1 promoter (24, 36), which appears to mediate Tax transactivation through chromatin remodeling and nucleation of the general transcription machinery (6, 35, 45, 58). The GC-rich sequences are conserved at all three 21-bp repeats in HTLV-1 and in the related virus HTLV-2, further supporting their functional significance (see reference 18 for a review). The most plausible interpretation of these observations is that Tax binds specifically to the GC-rich DNA, with the resulting Tax-DNA contacts contributing to the stability of the nucleoprotein complex. However, several studies have examined this possibility by using DNase I footprinting and methylation interference and have been unable to identify a direct interaction between Tax and the viral DNA (3, 43, 46).
In this study, we have used a variety of high-resolution approaches to dissect the role of the GC-rich flanking sequences in mediating the functions of Tax. We demonstrate that the viral CRE flanking sequences are required for Tax transactivation in a runoff transcription assay, confirming a functional role for the GC sequences in vitro. We also provide evidence that Tax may directly interact with DNA. With MPE:Fe as a DNA cleavage agent, we use a cutting agent that is 45 times smaller than DNase I, allowing a much higher resolution analysis of the Tax-CREB complex on the DNA than was previously obtained (3, 43, 46). Additionally, the MPE:Fe cleavage mechanism probes sugar-phosphate backbone interactions that were not detectable by methylation interference (46). We show that Tax enhances the CREB footprint on the viral CRE and extends the protection from MPE:Fe cleavage into the 5′ GC-rich flanking sequences, with additional protein-DNA contacts made by CREB and/or Tax within the 3′ flank. Tax extension of the CREB footprint is dependent upon the GC-rich flanking sequences, since Tax does not change the footprint pattern of CREB on a cellular CRE lacking these sequences. These data, coupled with the mechanism of MPE:Fe cleavage (29), suggest a putative Tax interaction with DNA that involves base recognition as well as phosphate backbone interactions within the 5′ region of the viral CRE.
A possible role for CBP in the putative Tax-DNA interaction was explored by carrying out footprinting reactions in the presence of the KIX domain of CBP. The KIX domain (aa 451 to 719) has previously been shown to interact with Tax (24, 36, 38). KIX enhanced the degree of protection of the Tax-CREB footprint, but it did not alter the nucleotides protected from MPE:Fe cleavage by CREB and Tax on the viral CRE. The footprint enhancement by KIX is probably due to an increase in the on-rate of complex formation, since KIX does not change the rate of dissociation of Tax and CREB from a viral CRE (24). Surprisingly, KIX was required for the extended footprint pattern observed with Tax and the bZIP domain of CREB. This KIX-induced expansion of the bZIP footprint was dependent upon Tax in the binding reaction. Furthermore, the pattern of protection was nearly indistinguishable from the footprint protection observed with full-length CREB and Tax. This is consistent with the observation that Tax forms a detectable EMSA complex with CREB bZIP only in the presence of KIX (24). These data support the idea that KIX strongly stabilizes the Tax-bZIP interaction without directly interacting with the DNA, thus enabling detection of the expanded footprint produced by Tax and the bZIP domain of CREB. Consistent with full-length CREB studies (24), KIX stabilization of the Tax-bZIP-viral CRE interaction must be due to an increase in the on-rate, since we did not observe a KIX-dependent change in the off-rate of bZIP from the DNA in the presence of Tax. Together, these data suggest that while KIX forms a highly stable complex with Tax and CREB on the viral CRE, it does so without directly contacting the DNA. Future studies with full-length CBP may reveal additional interactions with the Tax-CREB-viral promoter complex.
The Tax-dependent expansion of the CREB footprint into the GC-rich flanking sequences suggests that Tax directly binds the viral CRE DNA. However, footprinting data alone does not confirm that Tax is making the additional contacts. We provide further support for a Tax-DNA interaction by using the GC-specific minor-groove binding drug chromomycin A3. This drug, as well as the related drug olivomycin, inhibits the association of Tax and of the Tax-KIX complex with CREB (or bZIP) and the viral DNA. Concomitantly, chromomycin A3 negates Tax enhancement of CREB binding. Inhibition of Tax function is observed at the precise concentrations where chromomycin A3 preferentially protects the GC-rich flanking sequences from MPE:Fe cleavage. Since chromomycin A3 specifically binds the minor groove, Tax recognition of the GC DNA sequences is likely to occur through base-specific contacts in the minor groove. We cannot, however, rule out the possibility that chromomycin A3 blocks Tax interactions by distorting DNA structure (21), thus inhibiting Tax contacts with the GC-rich flanking sequences. This scenario, however, is unlikely. The degree of DNA distortion by chromomycin A3 at this concentration cannot be significant, since CREB binding to the adjacent CRE sequences is unaffected. In summary, these experiments show that when the minor-groove viral CRE-flanking sequences are physically blocked, the biochemical functions associated with Tax are abolished. Not surprisingly, these are the same viral CRE-flanking sequences that we show are protected from MPE cleavage in the presence of Tax. Together, these two distinct methods provide strong evidence that the GC-rich flanking sequences directly interact with Tax.
Further evidence to support the Tax-DNA interaction was obtained in protein-DNA photo-cross-linking studies. Probing simultaneously for major- and minor-groove interactions, we detected a specific CREB-dependent covalent cross-link of Tax to the 5′ flank of the viral CRE. This observation directly shows that Tax is present in the ternary complex. Tax cross-links within the AGGC sequence that is conserved at each of the three viral CREs in HTLV-1 and HTLV-2 promoters and maps to the region of the viral CRE where the Tax-dependent MPE:Fe footprint is observed. These data strongly suggest that Tax more intimately associates with the 5′-flanking sequence, although the absence of a 3′ cross-link does not preclude a Tax interaction within this region. Recent studies have provided evidence that Tax may exist as a homodimer both in vivo and in vitro (33, 52, 55). Our observation of an asymmetric association of Tax with the viral CRE is not consistent with a dimeric form of Tax in our binding reactions. Furthermore, stoichiometric quantitation of Tax in EMSA quaternary complexes suggests that a single molecule of Tax is present (24). Perhaps under our reaction conditions, the dimeric form of Tax is not favored, and therefore we observe cross-linking and significant MPE:Fe protection only within the 5′ flank.
CREB broadly cross-links throughout the viral CRE, with the strongest interactions generally observed within and near the CRE core. Although we expected to observe CREB cross-linking only in the core, careful examination of the crystal structure of the GCN4 bZIP bound to the major groove of the DNA shows additional phosphate backbone interactions outside of the DNA recognition element (16). The observed CREB interactions with the flanking sequences are therefore likely to occur through the phosphate backbone, since CREB interactions in the major groove would have previously been detected by methylation interference (46). CREB interactions in the minor groove would require that basic-domain amino acids contact specific base pairs in the minor-groove flanking sequences. This structural change would require dramatic and thermodynamically unfavorable changes in the structure of CREB. Furthermore, a panel of four distinct minor-groove binding drugs had little or no effect on CREB-DNA interactions. Together, these data support a model where CREB interactions in the flanking sequences occur through the phosphate backbone, solidifying the observation that Tax is primarily responsible for minor-groove contacts in the flanking sequence.
This model is further supported by evidence that in a cross-linking reaction, chromomycin A3 blocks the interaction of Tax with the 5′-flanking region, without eliminating the CREB cross-link at the same position. This observation is significant, since it demonstrates that the minor-groove contacts must be due to Tax and not to an altered structure of CREB. Interestingly, at this cross-link position (p3), we observed the most significant Tax enhancement of the CREB cross-link. Titration of chromomycin A3 into these reaction mixtures also eliminated Tax enhancement of CREB binding, suggesting that the effect of Tax on CREB binding activity is tightly coupled with stable Tax interaction with the flanking sequences. Therefore, it is likely that Tax stabilization of CREB binding is a consequence of the simultaneous association of Tax with the DNA and the parallel α-helices of CREB bZIP.
In summary, the experiments presented in this paper provide a strong body of evidence for a direct interaction between Tax and the minor groove of the viral CRE-flanking sequences. This previously uncharacterized interaction appears to be critical to Tax recruitment of CBP and the strong transactivation of the HTLV-1 genome. The identification of a DNA binding drug that blocks Tax function in vitro is significant, since Tax activity is crucial to the life cycle of the virus and the pathogenesis associated with HTLV-1 infection. We tested the effect of chromomycin A3 on Tax transactivation in an in vitro transcription assay and found that it abolished basal HTLV-1 transcription (data not shown). This result agrees with previous studies showing that chromomycin A3 and other naturally occurring DNA minor-groove binding drugs pleiotropically inhibit cellular gene transcription, consistent with their minimal DNA sequence recognition (reviewed in reference 64). Recently, high-affinity sequence-specific DNA minor-groove binding drugs have been synthesized and shown to successfully inhibit the expression of the 5S RNA gene in vitro and in vivo (27). The delineation of the base pairs putatively contacted by Tax, coupled with the strong evidence that Tax interacts with the minor groove, may provide the framework for development of rationally designed nontoxic drugs that specifically disrupt the interaction between Tax and the viral CREs.
ACKNOWLEDGMENTS
We thank Duane Ruffner and members of the Marv Paule laboratory, especially Cathy Radebaugh and Gary Geiss, for their generous advice.
This work was supported by Public Health Service grant CA-55035 from the National Cancer Institute.
REFERENCES
- 1.Adya N, Zhao L-J, Huang W, Boros I, Giam C-Z. Expansion of CREB’s DNA recognition specificity by Tax results from interaction with Ala-Ala-Arg at position 282-284 near the conserved DNA-binding domain of CREB. Proc Natl Acad Sci USA. 1994;91:5642–5646. doi: 10.1073/pnas.91.12.5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adya N, Giam C-Z. Distinct regions in human T-cell lymphotropic virus type I Tax mediate interactions with activator protein CREB and basal transcription factors. J Virol. 1995;69:1834–1841. doi: 10.1128/jvi.69.3.1834-1841.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altman R, Harrich D, Garcia J A, Gaynor R B. Human T-cell leukemia virus types I and II exhibit different DNase I protection patterns. J Virol. 1988;62:1339–1346. doi: 10.1128/jvi.62.4.1339-1346.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson M G, Dynan W S. Quantitative studies of the effect of HTLV-1 Tax protein on CREB protein-DNA binding. Nucleic Acids Res. 1994;22:3194–3201. doi: 10.1093/nar/22.15.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armstrong A P, Franklin A A, Uittenbogaard M N, Giebler H A, Nyborg J K. Pleiotropic effect of the human T-cell leukemia virus Tax protein on the DNA binding activity of eucaryotic transcription factors. Proc Natl Acad Sci USA. 1993;90:7303–7307. doi: 10.1073/pnas.90.15.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature (London) 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 7.Baranger A M, Palmer C R, Hamm M K, Giebler H A, Brauweiler A, Nyborg J K, Schepartz A. Mechanism of DNA binding enhancement by the HTLV-I transactivator Tax. Nature (London) 1995;376:606–608. doi: 10.1038/376606a0. [DOI] [PubMed] [Google Scholar]
- 8.Beimling P, Moelling K. Isolation and characterization of the Tax protein of HTLV-1. Oncogene. 1989;4:511–516. [PubMed] [Google Scholar]
- 9.Beimling P, Moelling K. Direct interaction of CREB protein with 21 base pair Tax-responsive elements of HTLV-1 LTR. Oncogene. 1992;7:257–262. [PubMed] [Google Scholar]
- 10.Bellorini M, Moncollin V, D’Incalci M, Mongelli N, Manovani R. Distamycin A and tallimustine inhibit TBP binding and basal in vitro transcription. Nucleic Acids Res. 1995;23:1657–1663. doi: 10.1093/nar/23.10.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brady J, Jeang K-T, Duvall J, Khoury G. Identification of p40x-responsive regulatory sequences within the human T-cell leukemia virus type I long terminal repeat. J Virol. 1987;61:2175–2181. doi: 10.1128/jvi.61.7.2175-2181.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brauweiler A, Garl P, Franklin A A, Giebler H A, Nyborg J K. A molecular mechanism for HTLV-I latency and Tax transactivation. J Biol Chem. 1995;270:12814–12822. doi: 10.1074/jbc.270.21.12814. [DOI] [PubMed] [Google Scholar]
- 13.Broggini M, D’Incalci M. Modulation of transcription factor-DNA interactions by anticancer drugs. Anti-Cancer Drug Design. 1994;9:373–387. [PubMed] [Google Scholar]
- 14.Chiang S-Y, Welch J, Rauscher III F J, Beerman T A. Effects of minor groove binding drugs on the interaction of TATA box binding protein and TFIIA with DNA. Biochemistry. 1994;33:7033–7040. doi: 10.1021/bi00189a003. [DOI] [PubMed] [Google Scholar]
- 15.Dynan W S. DNase I footprinting as an assay for mammalian gene regulatory proteins. Genet Eng. 1987;9:75–87. [Google Scholar]
- 16.Ellenberger T E, Brandel C J, Struhl K, Harrison S C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted α helices: crystal structure of the protein-DNA complex. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- 17.Franklin A A, Kubik M F, Uittenbogaard M N, Brauweiler A, Utaisincharoen P, Matthews M-A H, Dynan W S, Hoeffler J P, Nyborg J K. Transactivation by the human T-cell leukemia virus Tax protein is mediated through enhanced binding of activating transcription factor-2 (ATF-2) ATF-2 response and cAMP element-binding protein (CREB) J Biol Chem. 1993;268:21225–21231. [PubMed] [Google Scholar]
- 18.Franklin A A, Nyborg J K. Mechanisms of Tax regulation of human T-cell leukemia virus type I gene expression. J Biomed Sci. 1995;2:17–29. doi: 10.1007/BF02257921. [DOI] [PubMed] [Google Scholar]
- 19.Fujisawa J-I, Seiki M, Sato M, Yoshida M. A transcriptional enhancer sequence of HTLV-1 is responsible for transactivation mediated by p40x of HTLV-1. EMBO J. 1986;5:713–718. doi: 10.1002/j.1460-2075.1986.tb04272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujisawa J-I, Toita M, Yoshida M. A unique enhancer element for the trans activator (p40tax) of human T-cell leukemia virus type I that is distinct from cyclic AMP- and 12-O-tetradecanoylphorbol-13-acetate-responsive elements. J Virol. 1989;63:3234–3239. doi: 10.1128/jvi.63.8.3234-3239.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao X, Patel D J. Solution structure of the chromomycin-DNA complex. Biochemistry. 1989;28:751–762. doi: 10.1021/bi00428a051. [DOI] [PubMed] [Google Scholar]
- 22.Geiss G K, Radebaugh C A, Paule M P. The fundamental ribosomal transcription initiation factor, TIF-1B (SL1, factor D), binds to the rRNA core promoter primarily by minor groove contacts. J Biol Chem. 1997;272:29243–29254. doi: 10.1074/jbc.272.46.29243. [DOI] [PubMed] [Google Scholar]
- 23.Giam C-Z, Xu Y-L. HTLV-I Tax gene product activates transcription via pre-existing cellular factors and cAMP responsive element. J Biol Chem. 1989;264:15236–15241. [PubMed] [Google Scholar]
- 24.Giebler H A, Loring J E, van Orden K, Colgin M M, Garrus J E K, Escudero W, Brauweiler A, Nyborg J K. Anchoring of CREB binding protein to the human T-cell leukemia virus type 1 promoter: a molecular mechanism of Tax transactivation. Mol Cell Biol. 1997;17:5156–5164. doi: 10.1128/mcb.17.9.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez G A, Montminy M R. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 26.Goren I, Semmes O J, Jeang K-T, Moelling K. The amino terminus of Tax is required for interaction with the cyclic AMP response element binding protein. J Virol. 1995;69:5806–5811. doi: 10.1128/jvi.69.9.5806-5811.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gottesfeld J M, Neely L, Trauger J W, Baird E E, Dervan P B. Regulation of gene expression by small molecules. Nature (London) 1997;387:202–205. doi: 10.1038/387202a0. [DOI] [PubMed] [Google Scholar]
- 28.Hertzberg R P, Dervan P B. Cleavage of double helical DNA by (methidiumpropyl-EDTA) iron (II) J Am Chem Soc. 1982;104:313–315. [Google Scholar]
- 29.Hertzberg R P, Dervan P B. Cleavage of DNA with methidiumpropyl-EDTA-iron(II): reaction conditions and product analyses. Biochemistry. 1984;23:3934–3945. doi: 10.1021/bi00312a022. [DOI] [PubMed] [Google Scholar]
- 30.Hoeffler J P, Lustbader J W, Chen C-Y. Identification of multiple nuclear factors that interact with cyclic adenosine 3′,5′-monophosphate response element-binding protein and activating transcription factor-2 by protein-protein interactions. Mol Endocrinol. 1991;5:256–266. doi: 10.1210/mend-5-2-256. [DOI] [PubMed] [Google Scholar]
- 31.Ishibashi K, Hanada S, Hashimoto S. Expression of HTLV-I in serum of HTLV-I-related subjects and the early detection of overt ATL in HTLV-I carriers. J Immunol. 1987;139:1509–1513. [PubMed] [Google Scholar]
- 32.Jeang K-T, Boros I, Brady J, Radonovich M, Khoury G. Characterization of cellular factors that interact with the human T-cell leukemia virus type I p40x-responsive 21-base-pair sequence. J Virol. 1988;62:4499–4509. doi: 10.1128/jvi.62.12.4499-4509.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin D-Y, Jeang K-T. HTLV-I Tax self-association in optimal trans-activation function. Nucleic Acids Res. 1997;25:379–387. doi: 10.1093/nar/25.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinoshita T, Shimoyama M, Tobinai K, Oto M, Ito S-I, Ikeda S, Tajima K, Shimotohno K, Sugimura T. Detection of mRNA for the Tax/Rex gene of HTLV-I in fresh peripheral blood mononuclear cells of ATL patients and viral carriers, using the PCR reaction. Proc Natl Acad Sci USA. 1989;86:5620–5624. doi: 10.1073/pnas.86.14.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwok R P S, Lundblad J R, Chrivia J C, Richards J P, Bächinger H P, Brennan R G, Roberts S G E, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature (London) 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 36.Kwok R P S, Laurance M E, Lundblad J R, Goldman P S, Shih H-M, Connor L M, Marriott S J, Goodman R H. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the coactivator CBP. Nature (London) 1996;380:642–646. doi: 10.1038/380642a0. [DOI] [PubMed] [Google Scholar]
- 37.Lagrange T, Kim T-K, Orphanides G, Ebright Y W, Ebright R H, Reinberg D. High-resolution mapping of nucleoprotein complexes by site-specific protein-DNA photocrosslinking: organization of the human TBP-TFIIA-TFIIB-DNA quaternary complex. Proc Natl Acad Sci USA. 1996;93:10620–10625. doi: 10.1073/pnas.93.20.10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laurance M E, Kwok R P S, Huang M S, Richards J P, Lundblad J R, Goodman R H. Differential activation of viral and cellular promoters by human T-cell lymphotropic virus-I Tax and cAMP-responsive element modulator isoforms. J Biol Chem. 1997;272:2646–2651. doi: 10.1074/jbc.272.5.2646. [DOI] [PubMed] [Google Scholar]
- 39.Lenzmeier B A, Nyborg J K. In vitro transcription of human T-cell leukemia virus type 1 is RNA polymerase II dependent. J Virol. 1997;71:2577–2580. doi: 10.1128/jvi.71.3.2577-2580.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 41.Mayer A N, Barany R. Photoaffinity crosslinking of TaqI restriction endonuclease using an aryl azide linked to the phosphate backbone. Gene. 1995;153:1–8. doi: 10.1016/0378-1119(94)00752-e. [DOI] [PubMed] [Google Scholar]
- 42.Numata N, Ohtani K, Niki M, Nakamura M, Sugamura K. Synergism between two distinct elements of the HTLV-I enhancer during activation by the trans-activator of HTLV-I. New Biol. 1991;3:896–906. [PubMed] [Google Scholar]
- 43.Nyborg J K, Dynan W S, Chen I S Y, Wachsman W. Binding of host-cell factors to DNA sequences in the long terminal repeat of human T-cell leukemia virus type 1: implications for viral gene expression. Proc Natl Acad Sci USA. 1988;85:1457–1461. doi: 10.1073/pnas.85.5.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nyborg J K, Dynan W S. Interaction of cellular proteins with the human T-cell leukemia virus type I transcriptional control region: purification of cellular proteins that bind the 21-base pair repeat elements. J Biol Chem. 1990;265:8230–8236. [PubMed] [Google Scholar]
- 45.Ogryzko V V, Schiltz R L, Russanove V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 46.Paca-Uccaralertkun S, Zhao L-J, Adya N, Cross J V, Cullen B R, Boros I, Giam C-Z. In vitro selection of DNA elements highly responsive to the human T-cell lymphotropic virus type I transcriptional activator, Tax. Mol Cell Biol. 1994;14:456–462. doi: 10.1128/mcb.14.1.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parker D, Ferreri K, Nakajima T, LaMorte V J, Evans R, Koerber S C, Hoeger C, Montminy M R. Phosphorylation of CREB at Ser-133 induces complex formation with CREB-binding protein via a direct mechanism. Mol Cell Biol. 1996;16:694–703. doi: 10.1128/mcb.16.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paskalis H, Felber B K, Pavlakis G N. cis-Acting sequences responsible for the transcriptional activation of human T-cell leukemia virus type I constitute a conditional enhancer. Proc Natl Acad Sci USA. 1986;83:6558–6562. doi: 10.1073/pnas.83.17.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perini G, Wagner S, Green M R. Recognition of bZIP proteins by the human T-cell leukemia virus transactivator Tax. Nature (London) 1995;376:602–605. doi: 10.1038/376602a0. [DOI] [PubMed] [Google Scholar]
- 50.Rosen C Z, Sodroski J G, Haseltine W A. Location of cis-acting regulatory sequences in the human T-cell leukemia virus type I long terminal repeat. Proc Natl Acad Sci USA. 1985;82:6502–6506. doi: 10.1073/pnas.82.19.6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimotohno K, Takano M, Teruuchi T, Miwa M. Requirement of multiple copies of a 21-nucleotide sequence in the U3 region of human T-cell leukemia virus type I and type II long terminal repeats for trans-acting activation of transcription. Proc Natl Acad Sci USA. 1986;83:8112–8116. doi: 10.1073/pnas.83.21.8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shnyreva M, Munder T. The oncoprotein Tax of the human T-cell leukemia virus type 1 activates transcription via interactions with cellular ATF-1/CREB factors in Saccharomyces cerevisiae. J Virol. 1996;70:7478–7484. doi: 10.1128/jvi.70.11.7478-7484.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Studier F W, Moffat B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 54.Suzuki T, Fujisawa J-I, Toita M, Yoshida M. The transactivator Tax of human T-cell leukemia virus type I (HTLV-I) interacts with cAMP-response element (CRE) binding and CRE modulator proteins that bind to the 21-base pair enhancer of HTLV-I. Proc Natl Acad Sci USA. 1993;90:610–614. doi: 10.1073/pnas.90.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tie R, Adya N, Greene W C, Giam C-Z. Interaction of the human T-lymphotropic virus type 1 Tax dimer with CREB and the viral 21-base-pair repeat. J Virol. 1996;70:8368–8374. doi: 10.1128/jvi.70.12.8368-8374.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wagner S, Green M R. HTLV-I Tax protein stimulation of DNA binding of bZIP proteins by enhancing dimerization. Science. 1993;262:395–399. doi: 10.1126/science.8211160. [DOI] [PubMed] [Google Scholar]
- 57.Welch J J, Rauscher III F J, Beerman T A. Targeting DNA-binding drugs to sequence-specific transcription factor-DNA complexes. J Biol Chem. 1994;269:31051–31058. [PubMed] [Google Scholar]
- 58.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 59.Yin M-J, Paulssen E J, Seeler J S, Gaynor R B. Chimeric proteins composed of Jun and CREB define domains required for interaction with the human T-cell leukemia virus type 1 Tax protein. J Virol. 1995;69:6209–6218. doi: 10.1128/jvi.69.10.6209-6218.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yin M-J, Paulssen E J, Seeler J S, Gaynor R B. Protein domains involved in both in vivo and in vitro interactions between human T-cell leukemia virus type I Tax and CREB. J Virol. 1995;69:3420–3432. doi: 10.1128/jvi.69.6.3420-3432.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yin M-J, Gaynor R B. Complex formation between CREB and Tax enhances the binding affinity of CREB for the human T-cell leukemia virus type 1 21-base-pair repeats. Mol Cell Biol. 1996;16:3156–3168. doi: 10.1128/mcb.16.6.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao L-J, Giam C-Z. Interaction of the human T-cell lymphotrophic virus type I (HTLV-I) transcriptional activator Tax with cellular factors that bind specifically to the 21-base-pair repeats in the HTLV-I enhancer. Proc Natl Acad Sci USA. 1991;88:11445–11449. doi: 10.1073/pnas.88.24.11445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao L-J, Giam C-Z. Human T-cell lymphotropic virus type I (HTLV-1) transcriptional activator, Tax, enhances CREB binding to HTLV-1 21-base-pair repeats by protein-protein interaction. Proc Natl Acad Sci USA. 1992;89:7070–7074. doi: 10.1073/pnas.89.15.7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zimmer C, Wahnert U. Nonintercalating DNA-binding ligands: specificity of the interaction and their use as tools in biophysical, biochemical, and biological investigations of the genetic material. Prog Biophys Mol Biol. 1986;47:31–112. doi: 10.1016/0079-6107(86)90005-2. [DOI] [PubMed] [Google Scholar]