Abstract
The problem of whether recombinant mtDNAs are created in mammalian cells has been controversial for many years. We show convincing evidence for the very rare creation of recombinant mtDNA haplotypes by isolating human somatic hybrid cells and by generating mice carrying two different mtDNA haplotypes. To avoid misinterpretation of PCR-jumping products as recombinants, we used purified mtDNAs for cloning and sequencing. The results showed that only three of 318 clones of mtDNA purified from mouse tissues corresponded to recombinant mtDNA haplotypes, whereas no recombinants were found in human somatic hybrid cells. Such an extremely low frequency of mtDNA recombination does not require any revision of important concepts on human evolution that are based on its absence. Considering the high concentration of reactive oxygen species around the mtDNA and its frequent strand breakage, recombinant clones would correspond to gene conversion products created by repair of nucleotide mismatches.
Keywords: mammalian mtDNA, gene conversion, somatic hybrids, mito-mice, PCR jumping
Extensive recombination between mtDNAs from both parental cells has been proved in yeast (1) and plant cells (2, 3). Moreover, the presence of recombinant mtDNA haplotypes was reported in mussel species (4). In mammalian species, however, the issue of mtDNA recombination has been controversial (5). Some reports provided indirect evidence for their creation (6, 7). One study also suggested the presence of mtDNA recombination by using linkage disequilibrium analysis of human mtDNA haplotypes (8), but subsequent studies did not support this idea (9-12).
This issue has to be resolved because mtDNA recombination could be important in phylogenetic studies. If mtDNA recombinants are created extensively in mammals, this case could require reassessment of the conventional “mitochondrial eve” hypothesis (13), which is based on the assumption that no extensive mtDNA recombination occurs in mammalian species.
Our previous studies showed extensive exchanges of mtDNAs between mitochondria in human hybrid cells (14, 15) and in mouse tissues (16, 17), suggesting exclusion of physical barriers against recombination between heteroplasmic mtDNAs derived from different individuals or even from different species. Therefore, this issue could be resolved by precise sequence analysis of the heteroplasmic mtDNAs. Recently, sequence analysis of PCR products of heteroplasmic human mtDNAs suggested the presence of recombination between maternal and leaked paternal mtDNAs in skeletal muscles of a patient with mitochondrial disease (18). However, studies using PCR for identification of recombinant mtDNA haplotypes could not completely exclude the possibility that apparent recombinants corresponded to PCR-jumping products.
To obtain convincing evidence for creation of recombinant mtDNA haplotypes in mammalian cells, we used two different procedures for mtDNA cloning. One was cloning of PCR products of mtDNAs, and the other was cloning of mtDNA purified by EtBr-CsCl centrifugation. Because strictly maternal inheritance of mtDNA inhibits the coexistence of mtDNAs from different individuals within the same cells in a natural population (19, 20), we isolated somatic hybrid cells within which two human mtDNAs possessing many different mutation sites coexisted. For further examination of mtDNA recombination in germ cells, we generated mice carrying different types of mouse mtDNAs by microinjection of mitochondria into mouse zygotes.
Materials and Methods
Cells and Cell Culture. All of the cell lines and hybrid cells used in this study were grown in normal RPMI medium 1640 with 0.1 mg/ml pyruvate/50 μg/ml uridine/10% FBS.
Analysis of mtDNA Genotypes in Human Hybrid Cells. Total DNA extracted from 2 × 105 human cultured cells was used for analysis of their mtDNA genotypes. A4269G mtDNA was identified by the PCR method by using a specific primer set [nucleotide position (np) 4,116-4,135 and np 4,299-4,270], and the A3243G mtDNA was identified by the PCR method by using a specific primer set (np 3,153-3,174 and np 3,551-3,528) as described in ref. 15.
As the result of a gain of an ApaI site by an A3243G substitution, ApaI digestion of PCR products from A3243G mtDNA produces a 309-bp fragment, whereas A4269G and wild-type mtDNAs without the mutation produce a 399-bp fragment. As the result of the loss of an SspI site by an A4269G substitution, SspI digestion of PCR products from A4269G mtDNA produces a 184-bp fragment, whereas PCR of A3243G and wild-type mtDNAs without the mutation produce a 153-bp fragment. Signal calculation was carried out by using an nih image program (http://rsb.info.nih.gov/nihimage).
Generation of Heteroplasmic Mice. Mice with heteroplasmic mtDNAs were generated as described in ref. 16 with slight modifications. Briefly, mitochondrial fractions were microinjected into pronuclear stage embryos by using a Piezo micromanipulator (PrimeTech, Tsuchiura, Japan).
Analyses of Cytochrome c Oxidase (COX) Activity. COX activity was measured by examining the rate of cyanide-sensitive oxidation of reduced cytochrome c (21) with modifications. Biochemical analysis was based on the procedure described in ref. 22. COX electron micrographs were carried out as described in ref. 23 with slight modifications.
Analysis of Mitochondrial Translation Products. Mitochondrial translation products were labeled with [35S]methionine as described in ref. 14 with slight modifications. Proteins in the mitochondrial fraction were separated by 0.85% SDS/12% PAGE.
Analysis of mtDNA Genotypes in Mito-Mice and NZB/B6 Mice. For quantification of Mus musculus domesticus ΔmtDNA and M. spretus wild-type mtDNA, BglII fragments of total DNA extracted from brain and skeletal muscle were transferred to a nylon membrane and hybridized with alkaline phosphate-labeled mouse probe (np 1,895-2,762). Probe labeling and signal detection were carried out as described in the protocols of the AlkPhos Direct (Amersham Pharmacia Biotech). For quantification of B6 and NZB mtDNAs, PCR and restriction enzyme digestion were carried out as described in ref. 24. Signal calculation was carried out by using an nih image program.
PCR Amplification of mtDNA in Human Hybrid Cells for Cloning and Sequencing. Total DNA extracted from 2 × 105 human hybrid cells was used for amplification of mtDNA fragments, including pathogenic mutation sites and the D loop region by using a primer set of human mtDNA (np 15,994-16,024 and np 4,300-4,271). The 4,878-bp PCR products were ligated with pUC118 (Takara, Tokyo), and then introduced into DH5α (Takara).
Purification of mtDNA. Mitochondria and crude mtDNA were prepared from cultured human hybrid cells and mouse tissues as described in ref. 25, with a modification of additionally treating mitochondria with DNaseI to eliminate nuclear contaminants. The mtDNAs were purified by EtBr-CsCl density-gradient centrifugation at 36,000 rpm for 40 h (SW55Ti rotor, Beckman Coulter).
Nucleotide Sequence Analysis. Sequence templates were prepared with a TempliPhi DNA Sequencing Template Amplification Kit (Amersham Pharmacia Biosciences) following the manufacturer's protocol. Sequence reactions were performed by Dye Termination Methods (Takara PCR Thermal Cycler GP). Samples were then sequenced on MegaBACE1000 (Amersham Pharmacia Biosciences). To obtain whole-genome sequences of 11.6-kbp ΔmtDNA and 16.3-kbp NZB mtDNA clones, we used 70 and 96 sequence primers, respectively.
Amplification of Recombinant Molecules by Using PCR. Primers specific for M. spretus (np 4,141-4,166, CAATTATAAACAATTAGGAACATGGG) and M. m. domesticus (np 4,311-4,282, CATGTAAGAAGAATAAGTCCTATGTGCAGT) were used for selective amplification of recombinant molecules in mtDNA samples prepared from skeletal muscles, liver, kidney, and brain of an F3 mito-mouse. The cycle times were 20 sec for denaturation at 95°C, and 30 sec for annealing and extension at 62°C for 30 cycles.
Supporting Information. For more information on human and mouse mtDNA, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Results
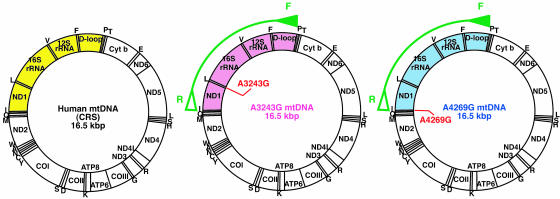
Isolation of Human Hybrid Cells Carrying Heteroplasmic mtDNAs Within the Same Mitochondrion. First, we examined mtDNAs in human hybrid cells, which we previously isolated by fusion of two types of respiration-deficient parental cells caused by different pathogenic mutations, A3243G in the tRNALeu(UUR) gene and A4269G in the tRNAIle gene of mtDNA, derived from patients with MELAS and cardiomyopathy, respectively (15). Accumulation of A3243G mtDNA and A4269G mtDNA in parental cells inhibited mitochondrial translation and respiratory function because of abnormal tRNALeu(UUR) and tRNAIle, respectively (see Fig. 5, which is published as supporting information on the PNAS web site). To allow sufficient time for recombination of parental mtDNAs in the hybrid cells, we cultivated these hybrid cells for 9 months after fusion.
The hybrid cells we examined possessed 36% A3243G mtDNA and 64% A4269G mtDNA (Fig. 5A), and their coexistence restored respiratory enzyme activity to the normal level (Fig. 5 B and C), suggesting the presence of mitochondrial complementation by extensive exchange of mtDNAs between the two parental mitochondria in the hybrid cells. This complementation indicated the coexistence of both parental A3243G and A4269G mtDNAs within the same mitochondrion. Therefore, physical barriers for recombination between mtDNAs derived from different patients were excluded in our hybrid cells. Genotype analysis of subclones isolated from the hybrid cells showed that heteroplasmy of both parental mtDNAs was maintained in all of the subclones during 9 months cultivation after the fusion (data not shown).
Examination of mtDNA Recombination in Human Somatic Hybrid Cells. Because many different mutation sites in parental mtDNAs were required to demonstrate the presence of their recombinants, we used 4,878-bp PCR products ranging from np 15,994 to 4,300 (Fig. 1). The products included both the pathogenic mutation sites and the D loop region, in which many polymorphic mutations were expected to exist (Fig. 1; Fig. 6, which is published as supporting information on the PNAS web site). We carried out sequence analyses of the 4,878-bp PCR products amplified from parental cells carrying A3243G and A4269G mtDNAs and compared their sequences with Cambridge reference sequences (CRS; ref. 26). The results showed that seven sites were specific to A3243G mtDNA and 11 sites to A4269G mtDNA (Fig. 6A).
Fig. 1.
Analysis of cloned PCR fragments from hybrid cells carrying parental A3243G and A4269G mtDNAs. Gene maps of human mtDNAs. Closed and open green arrowheads on the maps represent the forward primer (F) and reverse primer (R), respectively, used for amplification of 4,878-bp products (arcs). The cloned regions of 4,878-bp PCR products include the D loop and the pathogenic mutation sites of both parental A3243G and A4269G mtDNAs and are shown in pink and blue regions, respectively, on the gene maps. The corresponding region of the CRS (26) is shown in yellow.
Then, we carried out cloning and sequence analysis of the 4,878-bp PCR products of mtDNAs in the hybrid cells. Of the 100 clones we sequenced, 64 clones (clones 16-79) possessed mutation sites specific to parental A3243G mtDNA and to A4269G mtDNA (Fig. 6B), suggesting that they corresponded to recombinant mtDNA haplotypes.
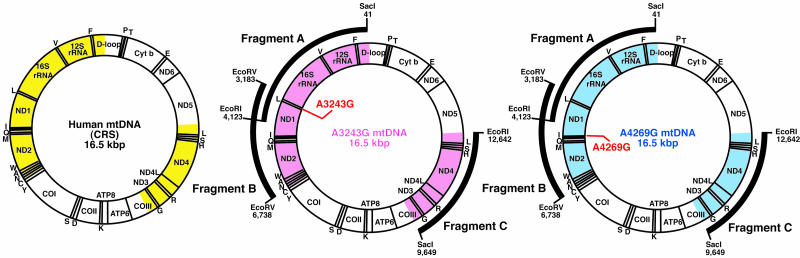
For confirmation of such extensive creation of mtDNA recombinants in the hybrid cells (Fig. 6B), we carried out cloning of purified mtDNA without using PCR amplification. The mtDNA of the hybrid cells was purified by EtBr-CsCl centrifugation, and the purified mtDNA was digested with restriction enzymes (Fig. 2). After agarose gel electrophoresis, three fragments, fragments A, B, and C, consisting of 4,083-, 3,556-, and 2,994-bp fragments, respectively, were extracted from the gels for cloning.
Fig. 2.
Analysis of purified mtDNA from hybrid cells carrying parental A3243G and A4269G mtDNAs. Gene maps of human mtDNAs indicating sequenced regions. Cloning and sequencing were carried out by using fragments A (4,083 bp), B (3,556 bp), and C (2,994 bp) purified from the hybrid cells carrying parental A3243G and A4269G mtDNAs (arcs). Sequenced regions corresponding to CRS, parental A3243G, and A4269G mtDNAs are shown in yellow, pink, and blue, respectively.
Fragment A included 11 CRS sites that were specific to either A4269G or A3243G mtDNA (see Fig. 7, which is published as supporting information on the PNAS web site). Of 85 clones in fragment A, 30 clones (clones 1-30) and 55 clones (clones 31-85) possessed sequences identical to those of parental A3243G and A4269G mtDNAs, respectively, although single somatic mutations were observed in 9 clones (clones 25-33) (Fig. 7). Fragment B included 10 CRS sites that were specific to either A4269G or A3243G mtDNA. Of 101 clones in fragment B, 44 clones (clones 1-44) and 57 clones (clones 45-101) possessed sequences identical to those of parental A3243G and A4269G mtDNA, respectively, whereas single somatic mutations were observed in 16 clones (clones 41-56) (Fig. 7). Fragment C included 7 CRS sites that were specific to either parental A4269G or A3243G. Of 85 clones in fragment C, 35 clones (clones 1-35) and 50 clones (clones 36-85) possessed sequences identical to those of parental A3243G and A4269G mtDNAs, respectively (Fig. 7). These observations suggested that there were no recombinant mtDNAs in any of the clones we examined. We sequenced >85 clones, which covered 60% of the whole mitochondrial genome (Figs. 2 and 7). Thus, even if recombinant mtDNA haplotypes were present in the hybrid cells, their frequency would be <2%. These observations suggested that most apparent recombinants observed in PCR products (Fig. 6B) corresponded to jumping products rather than to real recombinants.
Generation of Mito-Mice Carrying Heteroplasmic mtDNAs Within the Same Mitochondrion. In these experiments, we used human hybrid cells isolated by the fusion of somatic cells and found no mtDNA recombination. Therefore, it is still possible that mtDNA recombination is an event that frequently occurs in tissues or in female germ-line cells but not in cultivated somatic cells. For examination of recombination in living mice, we have to generate mice with heteroplasmic mtDNAs carrying different sequences. However, most laboratory mouse strains are derived from a single female M. m. domesticus, and their mtDNA sequences are almost identical (27, 28). Another problem was how to obtain clear evidence for the coexistence of both parental mtDNAs within the same mitochondrion, because recombinant haplotypes could not be created unless the parental mtDNAs coexist within the same mitochondrion.
To solve these problems, we used zygotes of congenic B6mtspr strain mice carrying wild-type mtDNA of M. spretus, which is a different species from M. m. domesticus. Then, we generated mito-mice by introducing respiration-deficient mitochondria carrying pathogenic ΔmtDNA of M. m. domesticus into zygotes of B6mtspr mice carrying wild-type mtDNA, but not wild-type mtDNA of M. spretus. Accumulated ΔmtDNA progressively reduced COX activity in the mitochondria (16, 17) because of a large-scale (4,696-bp) deletion mutation (Fig. 3A). Because the exo- and endogenous mtDNAs were derived from different mouse species, they differed at many nucleotide positions, which were useful for identification of their recombinants.
Fig. 3.
Sequence analysis of ΔmtDNA purified from skeletal muscles of an F3 mito-mouse. (A) Gene maps of wild-type mtDNA of M. spretus (green circle) and ΔmtDNA of M. m. domesticus (smaller red circle). The open arc in the ΔmtDNA map corresponds to the deleted region expanded from tRNALys to ND5 genes (np 7,759-12,454). No MluI site is present in wild-type mtDNA of M. spretus, whereas ΔmtDNA of M. m. domesticus possesses one MluI site. (B) Sequences of the cloned ΔmtDNA purified from skeletal muscles of an F3 mito-mouse possessing 30.7% ΔmtDNA. Green and red lanes correspond to mtDNA sequences of M. spretus and M. m. domesticus (38), respectively. Of 210 clones of ΔmtDNA from skeletal muscles, two clones (clones 17 and 18) possess two regions carrying sequences specific to M. spretus mtDNA in np 3,350-3,386 (ND1 gene) and np 4,093-4,213 (ND2 gene), and one clone (clone 19) possesses a region carrying sequences specific to M. spretus mtDNA in np 6,627-6,638 (CO1 gene). Of the remaining 207 clones, 191 possess completely identical sequences to those of M. m. domesticus mtDNA, whereas 16 clones (clones 1-16) possess somatic mutations. (C) Gene maps of clones 17 and 18 (Left) and clone 19 (Right). Green and red regions correspond to sequences of M. spretus and M. m. domesticus mtDNAs, respectively.
After three generations, we obtained an F3 mito-mouse (6 months old) carrying 30.0% ΔmtDNA, but not wild-type mtDNA, of M. m. domesticus and 70.0% wild-type mtDNA of M. spretus in its tail. Southern blot analysis of BglII digests showed that mtDNA samples purified from mito-mouse skeletal muscles and brain contained 30.7% and 34.2% ΔmtDNA of M. m. domesticus, respectively. COX electronmicrographs of skeletal muscles with 30.7% ΔmtDNA identified COX activity at the individual mitochondrial level and showed that all 5,160 mitochondria in sectioned areas of 10 muscle fibers possessed COX activity. If exogenous COX-negative mitochondria with ΔmtDNA and endogenous COX-positive mitochondria with wild-type mtDNA do not fuse with each other, 30.7% of the mitochondria should be COX negative, but results in the F3 mito-mouse was not the case. The presence of COX activity in all of the mitochondria suggested mixing of exogenous ΔmtDNA of M. m. domesticus and endogenous wild-type mtDNA of M. spretus and/or their gene products within the same mitochondrion in mito-mice tissues.
Examination of mtDNA Recombination in Tissues from a Living Mito-Mouse. To avoid PCR jumping artifacts, we did not use PCR products but used mtDNA purified from mito-mouse tissues by EtBr-CsCl centrifugation for cloning and sequencing. To exclude the possibility of recombination between two plasmids carrying ΔmtDNA and M. spretus mtDNA after cloning in E. coli cytoplasm, we carried out exclusive cloning of ΔmtDNA by digestion of purified mouse mtDNA with the restriction enzyme MluI, so that M. spretus mtDNA could not be cloned because of the absence of the MluI site (Fig. 3A).
After agarose gel electrophoresis, an 11.6-kbp fragment corresponding to whole linear ΔmtDNA was extracted from the gels for cloning. Then, we sequenced 210 and 108 clones of whole ΔmtDNA purified from skeletal muscles (Fig. 3B) and brain (see Fig. 8, which is published as supporting information on the PNAS web site), respectively. The results showed that two clones (clones 17 and 18) of muscle ΔmtDNA contained two regions, and one clone (clone 19) of muscle ΔmtDNA contained one region, in which the sequences were identical to those of M. spretus mtDNA (Fig. 3 B and C). The remaining 315 ΔmtDNA clones did not contain any sequences that were specific to M. spretus mtDNA (Figs. 3B and 8). These observations suggested that the frequency of recombinant mtDNA haplotypes in mito-mice tissues was <1%.
Then, to obtain evidence for the presence of the recombinant molecules corresponding to clones 17 and 18 in the mtDNA samples before cloning (see Fig. 9, which is published as supporting information on the PNAS web site), we carried out their selective amplification by using a mismatch primer set containing sequences specific to M. spretus and M. m. domesticus, respectively (Fig. 9A). In 30 cycles of PCR, an expected 171-bp fragment was amplified from the mito-mouse skeletal muscle mtDNA and not from either parental mtDNA or from their mixture (Fig. 9B). However, 50 cycles of PCR produced the 171-bp fragment from the mixture (Fig. 9B), indicating that all of the 171-bp fragments corresponded to PCR jumping products.
This possibility was excluded by cloning and sequencing the 171-bp fragments amplified from the mito-mouse mtDNA and from the mixture of parental mtDNAs, respectively. The results showed that all clones from the mito-mouse mtDNA had an identical sequence to that of the recombinant clones 17 and 18, whereas the clones from the mixture of mtDNAs each had different sequences from that of recombinant clones 17 and 18 (Fig. 9C). Therefore, the former clones corresponded to real recombinants, whereas the latter clones corresponded to PCR jumping artifacts. These observations provided unambiguous evidence for the rare creation of recombinant molecules identical to clones 17 and 18 in skeletal muscles of the mito-mouse (Fig. 3C).
The same 171-bp fragment was amplified from liver and kidney mtDNA samples but not from a brain mtDNA sample (Fig. 9B). The cloned 171-bp fragments from liver and kidney mtDNA had an identical sequence to that of the recombinant clones 17 and 18, suggesting creation of recombinants in female germ cells or during early embryogenesis, followed by their propagation to various tissues.
Generation of NZB/B6 Mice for Examination of mtDNA Recombinants. However, this low frequency for creation of mtDNA recombinants in mito-mice tissues (Fig. 3B) could be due to a large sequence divergence (7.1%) between heteroplasmic mtDNAs from different mouse species or to their different sizes. For resolving this issue, we generated NZB/B6 mice carrying heteroplasmic mtDNAs with a small sequence divergence (0.6%) from NZB and B6 strain mice, which belong to the same mouse species, M. m. domesticus. NZB strain mice possess mtDNA with a slightly different sequence from mtDNAs of other inbred strain mice (24, 28) and, thus, their recombinants could be identified by sequence analysis.
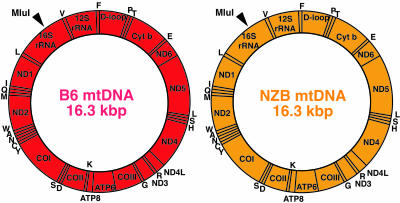
First, we introduced mitochondria carrying NZB mtDNA into zygotes of B6 mice (Fig. 4) and obtained an F3 NZB/B6 mouse (6 months old) carrying 24.0% NZB mtDNA and 76.0% B6 mtDNA in its skeletal muscles. Purified mtDNA from the skeletal muscles by EtBr-CsCl centrifugation was digested with MluI and used for cloning in E. coli. We selected colonies with NZB mtDNA by the difference of the RsaI digestion patterns of PCR products, because selective sequencing of colonies with NZB mtDNA would be more effective to find recombinants due to the smaller proportion of NZB mtDNA (24.0% NZB and 76.0% B6 mtDNA) in the NZB/B6 mouse.
Fig. 4.
Analysis of NZB mtDNA purified from skeletal muscles of an F3 NZB/B6 mouse. Gene maps of mtDNAs of B6 strain mice (red circle) and NZB strain mice (orange circle). One MluI site, indicated by an arrowhead, is present in both B6 and NZB mtDNAs and was used for cloning.
On sequencing of 114 NZB mtDNA clones, no clones contained sequences identical to those of B6 mtDNA (see Fig. 10, which is published as supporting information on the PNAS web site). These observations suggested that the frequency of mtDNA recombination, even if it occurred, should be <1% in mouse tissues. On the other hand, 18 clones possessed new somatic mutations (Fig. 10), but no clones shared the same mutations, suggesting the absence of clonal expansion of NZB mtDNA haplotypes carrying specific somatic mutations in NZB/B6 mouse.
Discussion
This study provided convincing evidence for the presence of an extremely low level of recombinant mtDNA haplotypes in mouse tissues. Then, what is the biological significance of creating mtDNA recombinants? One explanation is complementation of mitochondrial dysfunction caused by pathogenic mutations in mtDNA. In this case, the restoration of respiratory function in the hybrid cells (Fig. 5C) could be attained by production of recombinant haplotypes with neither A3243G nor A4269G mutations, because the accumulation of >5% recombinant haplotypes without either mutation may restore respiratory function in the hybrid cells (29). However, this study unambiguously showed that no such forms were present in 101 clones of fragment B (Figs. 2 and 7), although the hybrid cells showed restoration from respiration deficiency (Fig. 5C). These observations suggest that restoration in human hybrid cells was entirely due to the exchange of mtDNAs or their products between mitochondria, not to recombination between mtDNAs. Thus, an extremely low level of mtDNA recombination, even if it occurred, would not complement respiration defects caused by various mutations in mtDNAs.
Another explanation for creating mtDNA recombinants is to generate sequence divergence in the mammalian mtDNA population. Creation of recombinant mtDNA haplotypes was reported in species that show biparental mtDNA inheritance (1) or leakage of paternal mtDNA (4). In these cases, recombination occurred between maternal and paternal mtDNAs, and its biological meaning should be to provide extensive variations of mtDNA haplotypes in the species. In mammalian species, however, rapid and complete elimination of paternal mtDNA from zygotes (19, 20) prevents the coexistence of mtDNAs from both parents within individuals, resulting in the homoplasmic nature of the maternal mtDNA population in each individual. Thus, recombination of homoplasmic mtDNAs, even if it occurred, to create homoplasmic mtDNAs does not seem to have any biological significance.
A low incidence of mtDNA recombination would also not be effective to shed the burden of Mueller's ratchet, which irreversibly accumulates deleterious mutations in such an asexual genetic system as mammalian mtDNA. In fact, replication and transmission of mtDNAs were shown to be under relaxed control (30), and Takahata and Slatkin (31) showed by using a mathematical model that random segregation of mtDNA was sufficient to reset Muller's ratchet even in the absence of mtDNA recombination.
Therefore, the frequency of recombination in mammalian mtDNA is not sufficient for complementing respiration defects caused by deleterious mutations in mtDNA (Fig. 5C), for the generation of sequence variations in the mammalian mtDNA population, and for resetting Mueller's ratchet.
On the other hand, we speculate that the mtDNA recombinants simply reflect products resulting from the mtDNA repair reaction upon strand breakage. Because of the high concentration of reactive oxygen species in mitochondria, there should be frequent creation of single-strand breakage in mtDNA, resulting in some strand invasion and displacement between homologous or heterologous mtDNA molecules in heteroplasmic cells. In the case of strand invasion between heterologous mtDNA molecules, nucleotide mismatches in the heteroduplex should be subsequently repaired by mismatch repair activity in mitochondria (32). If invading strands are repaired, there should be no sequence changes, but in the case of repair of recipient strands, the repair process should create sequences identical to those of the invading mtDNA molecules. In fact, the mtDNA recombination we observed was restricted to very small regions (Fig. 3C). Thus, the observed recombinant mtDNA haplotypes might simply reflect the gene conversion products in the repair of damaged mtDNA molecules.
If extensive mtDNA recombination occurred in mammalian species, it could have important influences on phylogenetic studies. For example, it requires the reassessment of the conventional mitochondrial eve hypothesis (13), because this idea was proposed based on the assumption that no mtDNA recombination occurred in mammalian species because of clonal inheritance of maternal mtDNA. However, this study shows that the frequency of creation of recombinant haplotypes is very low in mammalian species (Figs. 3B, 7, 8, and 10) even under very favorable conditions, which are unlikely to occur in mammals given apparent mechanisms for exclusion of paternal mtDNA/mitochondria (19, 20). Moreover, although the transmission of paternal mtDNA was reported in a patient with mitochondrial disease (33) and in mouse interspecies F1 hybrids (20), subsequent studies showed no paternal mtDNA leakage in patients with mitochondrial diseases (34, 35) and no transmission of the leaked paternal mtDNA to following generations (20). Because paternal mtDNA leakage is rare and its transmission is even rarer in mammalian species, creation and transmission of recombinant mtDNA haplotypes through female germ lines to the following generation is an extremely rare event, even when paternal mtDNA leaks by some defect of the machinery required to ensure complete elimination of sperm mtDNA from zygotes. Therefore, these observations do not require any revision of the important hypothesis (13).
This study resolves another controversial issue with respect to mitochondrial genetic complementation (36, 37). Our previous reports provided evidence for genetic complementation between mitochondria based on the observation that expression of respiration defects induced by exogenous ΔmtDNA was prevented by endogenous wild-type mtDNA of the same mouse species in mito-mice (17). Although the prevention of respiration defects could be explained by assuming clonal expansion of wild-type mtDNA preexisting in exogenous mitochondria (36), this possibility was completely excluded by the findings that the exo- and endogenous mtDNA genotypes come into close enough proximity to convert their sequences (Fig. 3B) and that clonal expansion of specific mtDNA haplotypes did not occur in mouse tissues (Figs. 3B, 8, and 10). Therefore, mammalian mitochondria have the abilities not only to exchange their genetic contents but also to create gene conversion products (Fig. 3B). These observations support our idea that free mixing of mitochondrial genetic components throughout the mitochondrial network would protect mammalian mitochondria from direct expression of respiration defects caused by accumulated somatic mutations in mtDNAs or by reactive oxygen species-induced mtDNA strand breakage (15-17).
Supplementary Material
Acknowledgments
This work was supported in part by grants for a Research Fellowship from the Japan Society for Promotion of Science for Young Scientists (to A.S. and T.O.), a grant for the Hayashi project of Tsukuba Advanced Research Alliance, University of Tsukuba, and by Grants-in-Aid for Creative Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to J.-I.H.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: COX, cytochrome c oxidase; CRS, Cambridge reference sequences; np, nucleotide position.
References
- 1.Dujon, B., Slonimski, P. P. & Weill, L. (1974) Genetics 78, 415-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belliard, G., Vadel, F. & Pelletier, G. (1979) Nature 281, 401-403. [Google Scholar]
- 3.Boeshore, M. L., Lifshitz, I., Maureen, R., Hanson, M. & Izhar, S. (1983) Mol. Gen. Genet. 190, 459-467. [Google Scholar]
- 4.Ladoukakis, E. D. & Zouros, E. (2001) Mol. Biol. Evol. 18, 1168-1175. [DOI] [PubMed] [Google Scholar]
- 5.Rokas, A., Ladoukakis, E. & Zouros, E. (2003) Trends Ecol. Evol. 18, 411-417. [Google Scholar]
- 6.Hayashi, J.-I., Tagashira, Y. & Yoshida, M. C. (1985) Exp. Cell Res. 160, 387-395. [DOI] [PubMed] [Google Scholar]
- 7.Thyagarajan, B., Padua, R. A. & Campbell, C. (1996) J. Biol. Chem. 271, 27536-27543. [DOI] [PubMed] [Google Scholar]
- 8.Awadalla, P., Eyre-Walker, A. & Smith, J. M. (1999) Science 286, 2524-2525. [DOI] [PubMed] [Google Scholar]
- 9.Kivisild, T. & Villems, R. (2004) Science 288, 1931a. [PubMed] [Google Scholar]
- 10.Jorde, L. B. & Bamshad, M. (2004) Science 288, 1931a. [PubMed] [Google Scholar]
- 11.Kumar, S., Hedrick, P., Dowling, T. & Stoneking, M. (2004) Science 288, 1931a. [PubMed] [Google Scholar]
- 12.Parsons, T. J. & Irwin, J. A. (2004) Science 288, 1931a. [Google Scholar]
- 13.Cann, R. L., Stoneking, M. & Wilson, A. C. (1987) Nature 325, 31-36. [DOI] [PubMed] [Google Scholar]
- 14.Takai, D., Inoue, K., Goto, Y.-i., Nonaka, I. & Hayashi, J.-I. (1997) J. Biol. Chem. 272, 6028-6033. [DOI] [PubMed] [Google Scholar]
- 15.Ono, T., Isobe, K., Nakada, K. & Hayashi, J.-I. (2001) Nat. Genet. 28, 272-275. [DOI] [PubMed] [Google Scholar]
- 16.Inoue, K., Nakada, K., Ogura, A., Isobe, K., Goto, Y., Nonaka, I. & Hayashi, J.-I. (2000) Nat. Genet. 26, 176-181. [DOI] [PubMed] [Google Scholar]
- 17.Nakada, K., Inoue, K., Ono, T., Isobe, K., Ogura, A., Goto, Y., Nonaka, I. & Hayashi, J.-I. (2001) Nat. Med. 7, 934-940. [DOI] [PubMed] [Google Scholar]
- 18.Kraytsberg, Y., Schwartz, M., Brown, T. A., Ebralidse, K., Kunz, W. S., Clayton, D. A., Vissing, J. & Khrapko, K. (2004) Science 304, 981. [DOI] [PubMed] [Google Scholar]
- 19.Kaneda, H., Hayashi, J.-I., Takahama, S., Taya, C., Lindahl, K. F. & Yonekawa, H. (1995) Proc. Natl. Acad. Sci. USA 92, 4542-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shitara, H., Hayashi, J.-I., Takahama, S., Kaneda, H. & Yonekawa, H. (1998) Genetics 148, 851-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seligman, A. M., Karnovsky, M. J., Wasserkrug, H. L. & Hanker, J. S. (1968) J. Cell. Biol. 38, 1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyabayashi, S., Narisawa, K., Iinuma, K., Tada, K., Sakai, K., Kobayashi, K., Kobayashi, Y. & Morinaga, S. (1984) Brain Dev. 6, 362-372. [DOI] [PubMed] [Google Scholar]
- 23.Nonaka, I., Koga, Y., Ohtake, E. & Yamamoto, M. (1998) J. Neurol. Sci. 92, 193-203. [DOI] [PubMed] [Google Scholar]
- 24.Jenuth, J. P., Peterson, A. C., Fu, K. & Shoubridge, E. A. (1996) Nat. Genet. 14, 146-151. [DOI] [PubMed] [Google Scholar]
- 25.Yonekawa, H., Gotoh, O., Motohashi, J., Hayashi, J.-I. & Tagashira, Y. (1978) Biochim. Biophys. Acta 251, 510-519. [DOI] [PubMed] [Google Scholar]
- 26.Andrews, R. M., Kubacka, I., Chinnery, P. F., Lightowlers, R. N., Turnbull, D. M. & Howell, N. (1999) Nat. Genet. 23, 147. [DOI] [PubMed] [Google Scholar]
- 27.Ferris, S. D., Sage, R. D. & Wilson, A. C. (1982) Nature 295, 163-165. [DOI] [PubMed] [Google Scholar]
- 28.Yonekawa, H., Moriwaki, K., Gotoh, O., Miyashita, N., Migita, S., Bonhomme, F., Hjorth, J. P., Petras, M. L. & Tagashira, Y. (1982) Differentiation (Berlin) 22, 222-226. [DOI] [PubMed] [Google Scholar]
- 29.Chomyn, A., Martinuzzi, A., Yoneda, M., Daga, A., Hurko, O., Johns, D., Lai, S. T., Nonaka, I., Angelini, C. & Attardi, G. (1992) Proc. Natl. Acad. Sci. USA 89, 4221-4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Birky, C. W., Jr. (1983) Science 222, 468-475. [DOI] [PubMed] [Google Scholar]
- 31.Takahata, N. & Slatkin, M. (1983) Genet. Res. 42, 257-265. [Google Scholar]
- 32.Mason, P. A., Matheson, E. C., Hall, A. G. & Lightowlers, R. N. (2003) Nucleic Acids Res. 31, 1052-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwartz, M. & Vissing, J. (2002) N. Engl. J. Med. 347, 576-580. [DOI] [PubMed] [Google Scholar]
- 34.Filosto, M., Mancuso, M., Vives-Bauza, C., Vila, M. R., Shanske, S., Hirano, M., Andreu, A. L. & DiMauro, S. (2003) Ann. Neurol. 54, 524-526. [DOI] [PubMed] [Google Scholar]
- 35.Taylor, R. W., McDonnell, M. T., Blakely, E. L., Chinnery, P. F., Taylor, G. A., Howell, N., Zeviani, M., Briem, E., Carrara, F. & Turnbull, D. M. (2003) Ann. Neurol. 54, 521-524. [DOI] [PubMed] [Google Scholar]
- 36.Attardi, G., Enriquez, J. A. & Cabezas-Herrera, J. (2002) Nat. Genet. 30, 360. [DOI] [PubMed] [Google Scholar]
- 37.Hayashi, J.-I., Nakada, K. & Ono, T. (2002) Nat. Genet. 30, 361. [DOI] [PubMed] [Google Scholar]
- 38.Bibb, M. J., Van Etten, R. A., Wright, C. T., Walberg, M. W. & Clayton, D. A. (1981) Cell 26, 167-180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.