Abstract
Carcinoma of the prostate is the most commonly diagnosed cancer in men. The current pharmacological treatment of choice for progressive androgen-dependent prostate cancer is the nonsteroidal antiandrogen, bicalutamide, either as monotherapy or with adjuvant castration or luteinizing hormone-releasing hormone superagonists to block the synthesis of endogenous testosterone. To date, no nonsteroidal or antagonist-bound androgen receptor (AR) structure is available. We solved the x-ray crystal structure of the mutant W741L AR ligand-binding domain bound to R-bicalutamide at 1.8-Å resolution. This mutation confers agonist activity to bicalutamide and is likely involved in bicalutamide withdrawal syndrome. The three-dimensional structure demonstrates that the B ring of R-bicalutamide in the W741L mutant is accommodated at the location of the indole ring of Trp-741 in the WT AR bound to dihydrotestosterone. Knowledge of the binding mechanism for R-bicalutamide will provide molecular rationale for the development of new antiandrogens and selective AR modulators.
Keywords: androgen receptor, crystallography
The androgen receptor (AR) is a ligand-inducible hormone receptor of the nuclear receptor superfamily that plays a role in development and regulation of male secondary characteristics, spermatogenesis, bone and muscle mass, and androgenic tissue. Agents that block the actions (i.e., antiandrogens) of endogenous androgens (e.g., testosterone) are highly effective and routinely used for the treatment of prostate cancer. The first nonsteroidal antiandrogen, flutamide (Eulexin) was approved for prostate cancer in 1989 (1) and the structurally related compounds, bicalutamide (Casodex) and nilutamide (Nilandron) (2), were later launched in 1995 and 1996, respectively. Nonsteroidal ligands are more favorable for clinical applications because of the lack of crossreactivity with other steroid receptors (e.g., progesterone receptor) and improved oral bioavailability. Of this structural class of antiandrogens, bicalutamide is the most potent (3) and best tolerated (4-6). Emerging data from clinical studies further reinforce the idea that bicalutamide is a drug of choice for progressive androgen-dependent prostate cancers often in conjunction with leuprolide or other luteinizing hormone-releasing hormone superagonists as a component of total androgen blockade (7-12). Although antiandrogen treatment and androgen blockade usually exhibit favorable responses, prostate cancers often become refractory, as evidenced by increasing prostate-specific antigen levels (i.e., progression) and/or regression upon cessation of antiandrogen therapy (i.e., antiandrogen withdrawal syndrome) (13). AR gene mutations in the ligand-binding domain (LBD) that alter ligand specificity and/or functional activity exist and are thought to contribute to the ability of some antiandrogens to exhibit androgenic activity. The LNCaP prostate cancer cell line expresses AR with a T877A point mutation that causes proliferation in the presence of the antiandrogens hydroxyflutamide and cyproterone acetate. As expected, this mutation has also been discovered in patients with antiandrogen withdrawal syndrome (14) being treated with these compounds. Recently, Hara et al. (15) reported two LBD mutations, W741L and W741C, that occurred upon androgen deprivation and bicalutamide treatment of LNCaP cells. Agonist activity of bicalutamide in these mutants was further confirmed through transactivation studies in cotransfected COS-7 cells (15).
Structure-activity relationships have been extensively studied for bicalutamide analogs (16), yet no structural evidence for receptor interactions exist. Bicalutamide is available as a racemic mixture. However, the R isomer has an ≈30-fold higher binding affinity to the AR than the S isomer (17) and was therefore used in this study. Mutation of Trp-741 to leucine to invoke an agonist conformation significantly facilitated purification and crystallization of the AR nonsteroidal ligand complex. Thus, we herein describe the x-ray crystal structure of R-bicalutamide bound to the AR mutant W741L LBD and its implications for antiandrogen withdrawal syndrome and rational drug design.
Materials and Methods
Cloning, Expression, and Purification. An AR-LBD (663-919) was obtained by PCR amplification from a full-length AR expression vector with primers containing flanking restriction sites and inserted into the pGEX6P-1 plasmid vector (Amersham Pharmacia). The W741L mutation was created in the pGEX6P1-LBD plasmid through the Stratagene QuikChange mutagenesis kit according to the manufacturer's instructions. AR LBD expression and purification were performed similar to refs. 18 and 19. The AR LBD was expressed as a GST fusion protein in Escherichia coli BL21 DE3 at 15°C for 16 h by induction with 30 μM IPTG. Cells were lysed in a buffer containing 150 mM NaCl, 50 mM Tris (pH 8.0), 5 mM EDTA, 10% glycerol, 1 mg/ml lysozyme, 10 units/ml DNase I, 10 mM MgCl2, 10 mM DTT, 0.5% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, 100 μM R-bicalutamide, and 100 μM PMSF by three cycles of freeze-thaw. The supernatant from ultracentrifugation was incubated for 1 h at 4°C with glutathione Sepharose (Amersham Pharmacia) and washed with 150 mM NaCl/50 mM Tris, pH 8.0/5 mM EDTA/10% glycerol/10 μM R-bicalutamide/0.1% n-octyl-β-glucoside/1 mM DTT. The GST-LBD fusion protein was cleaved in a buffer containing 150 mM NaCl, 50 mM Tris (pH 7.0), 10% glycerol, 10 μM R-bicalutamide, 0.1% n-octyl-β-glucoside, 1 mM DTT, and 5 units of PreScission protease (Amersham Pharmacia) per mg of protein at 4°C overnight, releasing the AR LBD from the glutathione Sepharose. The supernatant was then diluted 3-fold in 10 mM Hepes, pH 7.2/10% glycerol/10 μM R-bicalutamide/0.1% n-octyl-β-glucoside/1 mM DTT and loaded onto an HP SP cation-exchange column (Amersham Pharmacia). Protein was eluted with a gradient of 50-500 mM NaCl in the same dilution buffer. The buffer was exchanged in a Millipore 10-kDa cutoff concentrator to 150 mM Li2SO4/50 mM Hepes, pH 7.2/10% glycerol/100 μM R-bicalutamide/0.1% n-octyl-β-glucoside/10 mM DTT, and protein was concentrated to 8 mg/ml.
Receptor Binding Affinity Determination. Recombinant GST-LBD was expressed as described above but in the absence of ligand. Aliquots of lysate were used to determine the equilibrium dissociation (Kd) of 3H-mibolerone (PerkinElmer) by Scatchard plot analysis. The equilibrium dissociation (Ki) values for dihydrotestosterone (DHT) and R-bicalutamide were determined for both the WT and W741L mutant by competitive binding with 3H-mibolerone according to methods in ref. 20.
Crystallization, Data Collection, and Structure Determination. Crystals formed in 1-2 days by using the sitting drop vapor diffusion method in 0.1 M Hepes, pH 7.5/0.75 M Li2SO4 and were transferred to a solution consisting of 0.1 M Hepes, pH 7.5/0.75 M Li2SO4/25% ethylene glycol before flash freezing in liquid nitrogen. Diffraction data were collected to 1.8-Å resolution by using a RU300 rotating anode generator (Rigaku, Tokyo) and an R-axis IV++ image plate (Rigaku) and processed with crystal clear (Molecular Structure, The Woodlands, TX). The DHT-LBD (PDB ID code 1I37) was used as a starting structure for refinement by using Crystallography & NMR System (CNS) (21). After an initial round of refinement, electron density maps allowed for accurate fitting of R-bicalutamide. Model building and water molecules were added by using the program o (22) and further rounds of refinement were performed by using rigid body, torsion angle simulated annealing, and individual temperature factor modules of CNS to an R factor of 0.223 and free R factor of 0.256 (Table 1). Surface area buried was calculated with a probe radius of 1.4 Å by using CNS. Figures were prepared with molscript (23) and raster3d (24).
Table 1. Crystallographic data and refinement statistics.
| Measurement | Value |
|---|---|
| Spacegroup | P212121 |
| Unit cell dimensions, Å | a = 56.5, b = 66.5, c = 72.2 |
| Resolution range, Å | 24.5−1.8 (1.9−1.8) |
| No. of unique reflections | 24,747 |
| Average redundancy | 3.79 (1.89) |
| Completeness, % | 95.7 (72.7) |
| Rmerge | 0.041 (0.245) |
| l/σ | 17.9 (3.3) |
| R factor | 0.223 (0.335) |
| Rfree | 0.256 (0.372) |
| rms deviation bonds, Å | 0.058 |
| rms deviation angles, ° | 1.11 |
| mean B value, Å2 | 31.2 |
Values for data in the last resolution shell shown in parentheses.
Results
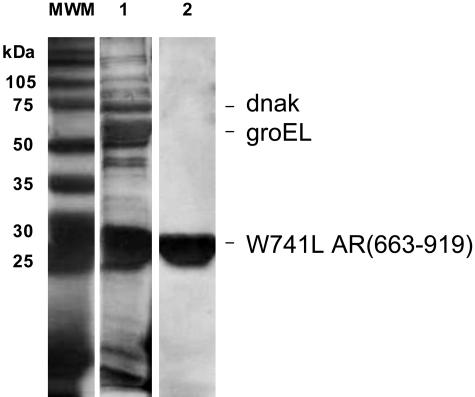
Protein Purification. Purification of the WT AR LBD complexed with R-bicalutamide has been unsuccessful in our laboratory. Previous reports of AR LBD preparations for crystallography use low induction temperatures and low concentrations of IPTG for protein expression followed by affinity chromatography and cation-exchange chromatography (18, 19, 25, 26). Low soluble yields of the GST-LBD in the presence of R-bicalutamide were obtained even after taking such measures to avoid inclusion body formation. Our failure was further documented by the lack of LBD bound R-bicalutamide retention on cation-exchange columns. This result appears to be caused by tight association with the bacterial chaperonin, groEL, which is easily separated from AR LBD preparations with agonist ligands (25). To circumvent these difficulties, we purified the W741L AR LBD in association with R-bicalutamide. The SDS/PAGE gel in Fig. 1 demonstrates the presence of a large amount of AR LBD and groEL and a smaller amount of dnak, similar to previously reported AR LBD purifications (25) after affinity chromatography and cleavage of the W741L-R-bicalutamide LBD from the GST tag. These chaperone contamination proteins were successfully removed by cation-exchange chromatography.
Fig. 1.
Purification of W741L AR LBD with R-bicalutamide shown by silver stained 12% SDS/PAGE Molecular Weight Marker (MWM). After PreScission Protease cleavage from the glutathione Sepharose, LBD, groEL, and dnak were observed in the supernatant (lane 1). Only LBD was present in the cation-exchange eluent (lane 2).
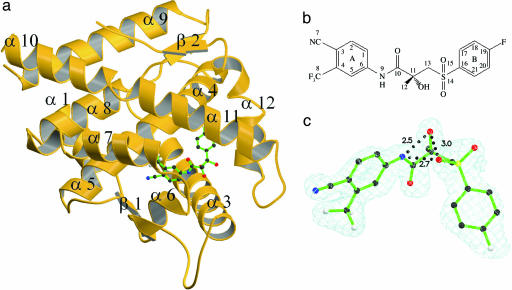
Overall Structure. The W741L-R-bicalutamide complex demonstrates the same overall fold as the WT-DHT complex (Fig. 2a) (26). This finding is not surprising because this mutation imparts agonist activity to R-bicalutamide. The W741L-R-bicalutamide complex superimposed to a root mean squared deviation of 0.33 Å for all main chain atoms to the WT-DHT structure (26). As expected, R-bicalutamide buries a greater surface area of 893 Å2 upon binding to the AR LBD as compared with 626 Å2 for the smaller DHT molecule. Like the WT complex solved by Sack et al. (26), we did not see clear electron density for the N-terminal (663-671) and C-terminal (918 and 919) residues or the loop between helices 9 and 10 (residues 844-851). We thus excluded these regions from the model.
Fig. 2.
Structure and binding conformation of R-bicalutamide. (a) W741L AR LBD-R-bicalutamide complex. (b) Structure and numbering scheme of R-bicalutamide (c) R-bicalutamide within the Fo-Fc simulated annealing omit map with R-bicalutamide omitted. Intramolecular hydrogen bonds are denoted by distances in Å. Nitrogen, blue; oxygen, red; carbon, black; sulfur, yellow; fluorine, white.
Bound Conformation of R-Bicalutamide. In concordance with previously reported solution NMR data with free ligand (27), the distance between the amide nitrogen and chiral hydroxyl group (2.5 Å), as well as the chiral hydroxyl group and the oxygen sulfonyl denoted O14 (3.0 Å) in R-bicalutamide (Fig. 2b) are suitable for intramolecular hydrogen bonding in the x-ray structure. Additionally, the bent conformation adopted by R-bicalutamide within the binding pocket (Fig. 2c) positions the amide nitrogen 2.7 Å away from the O14 sulfonyl oxygen, allowing another potential intramolecular hydrogen bond. Because the chiral hydroxyl group seems to act as a hydrogen bond donor upon receptor binding and the sulfonyl of R-bicalutamide is bound in a more hydrophobic receptor environment, the hydrogen bond between the sulfonyl group and the amide nitrogen is more likely.
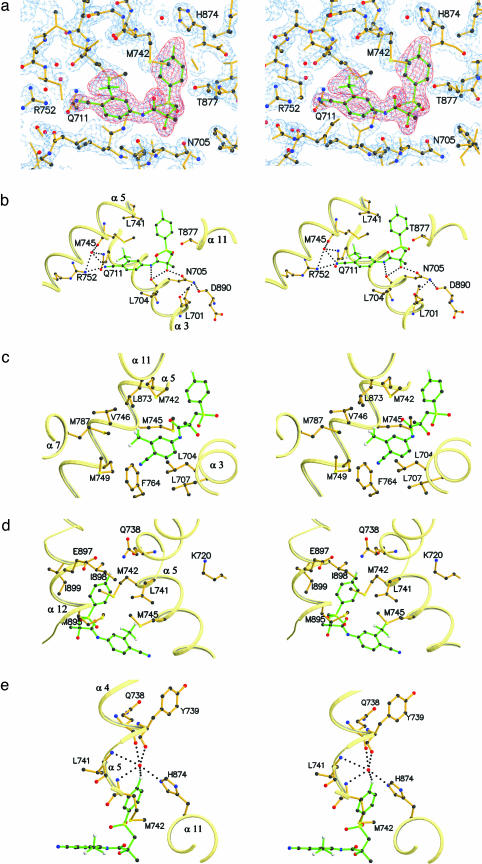
Binding of R-Bicalutamide to the W741L AR. Hydrogen bonds are present between R-bicalutamide and the AR binding pocket in two different regions. The cyano group on the A ring of R-bicalutamide is located at a distance of 3.0 Å from Arg-752 Nη2, indicative of hydrogen bonding. Conversely, the Gln-711 Nε2 is located 3.7 Å from the cyano group and may be slightly out of hydrogen bond range. Similar to other AR structures and in the progesterone receptor crystal structures (18, 26), the well conserved water molecule is present at a distance of 3.0 Å from the cyano group, 3.0 Å from Arg-752 Nη2, 2.8 Å from Gln-711 Nε2, and 2.7 Å from Met-745 O suggesting the possible hydrogen bonds depicted in Fig. 3b. The amide nitrogen and the chiral hydroxyl group of R-bicalutamide are also seen in range for hydrogen bonding to the AR. The Leu-704 backbone oxygen is situated 3.2 Å from the amide nitrogen and 3.2 Å from the chiral hydroxyl group of R-bicalutamide, whereas Asn-705 Oδ1 was observed 2.5 Å away from the chiral hydroxyl group (Fig. 3b). To position Asn-705 Nδ2 for hydrogen bond formation with the backbone oxygen atoms of Leu-701 (3.2 Å) and Asp-890 (2.8 Å), the Asn-705 side chain was rotated 180° about the χ2 dihedral from the starting model (PDB ID code 1I37). Contrary to steroidal bound AR structures, the Oγ of Thr-877 clearly does not hydrogen bond with R-bicalutamide, with its closest ligand contact being 3.7 and 3.4 Å from the carbon atoms denoted C12 and C13, respectively.
Fig. 3.
R-bicalutamide interactions with the W741L binding pocket in multiple orientations. (a) Side chains fit into the 2Fo-Fc electron density maps contoured at the 2σ level (blue) with R-bicalutamide shown in the Fo-Fc simulated annealing omit map contoured at the 4σ level (red). (b) Possible hydrogen bonds depicted between R-bicalutamide and the W741L AR LBD in a similar orientation as Fig. 1a. (c) View from the bottom relative to Fig. 1a to emphasize the van der Waals interactions with the A ring of R-bicalutamide. (d) View from the back relative to Fig. 1a portrays the B ring binding pocket formed by residues of helices 5 and 12 in relation to AF-2 charge clamp residues K720 and E897. (e) View from the top relative to Fig. 1a demonstrates the location of the para position of the R-bicalutamide B ring in a hydrophilic environment.
As expected from the hydrophobic character of the AR binding pocket observed with steroids, van der Waals forces comprise the majority of interactions with R-bicalutamide. The trifluoromethyl group on the metaposition of the A ring is situated in a hydrophobic environment surrounded by Met-742, Val-746, Met-787, and Leu-873 as seen in Fig. 3c. Other contacts with the A ring of R-bicalutamide include Leu-704, Leu-707, Met-745, and Phe-764. The carbonyl oxygen of the amide moiety on R-bicalutamide lacks H bond partners, the closest atom being the Sδ of Met-742. In addition, Met-895 comes into close contact (3.3 Å) with the sulfonyl oxygen denoted O15 of R-bicalutamide. Met-895 also participates in the formation of a hydrophobic pocket enclosing the B ring of the ligand along with other helix 12 residues, Ile-898 and Ile-899, and helix 5 residues Leu-741 and Met-742 (Fig. 3d). The fluorine at the para position of the B ring however is bound in a more hydrophilic environment, located 2.9 Å from a water molecule. The backbone oxygens of Gln-738 (2.8 Å) and Tyr-739 (2.7 Å), backbone nitrogens of Leu-741 (3.3 Å) and Met-742 (2.9 Å), and the His-874 Nε2 (2.8 Å) are all situated in suitable hydrogen bonding distance with this same water molecule (Fig. 3e).
Binding Affinities. The binding affinities of R-bicalutamide, DHT, and the synthetic steroidal androgen, mibolerone, were determined to further investigate changes in ligand binding induced by the W741L AR mutation. A 3- to 4-fold loss in affinity to the W741L mutant AR LBD as compared with the WT AR LBD was observed with 3H-mibolerone (Table 2). Surprisingly, the binding affinity for DHT to the W741L mutant showed a decrease of nearly 40-fold relative to the WT. This unexpected result suggests that DHT and mibolerone interact differently with the Trp-741 side chain. Alternatively, R-bicalutamide demonstrated a 2-fold increase in binding affinity to the W741L mutant. Therefore, this mutation only slightly alters the binding affinity of R-bicalutamide to the AR, although it drastically changes the activity (15).
Table 2. Binding affinities.
| WT | W741L | |
|---|---|---|
| 3H-Mibolerone, Kd | 2 ± 1 | 7 ± 1 |
| DHT Ki | 2 ± 1 | 81 ± 7 |
| R-bicalutamide Ki | 145 ± 30 | 76 ± 26 |
Discussion
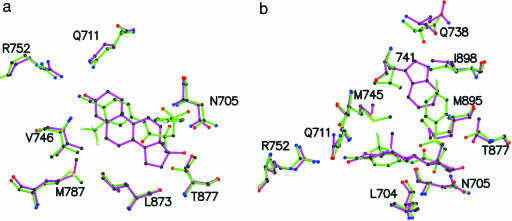
Comparison with WT-DHT Complex. In general, residues are positioned similarly in the W741L bound R-bicalutamide complex as compared with the WT-DHT LBD (26). The cyano group of R-bicalutamide mimics the 3-keto functionality of DHT (Fig. 4a). The A ring of R-bicalutamide binds in a region overlapping both the A and B rings of DHT, whereas the trifluoromethyl group bulges out slightly into an area not occupied by the steroid. The C6, C7, and C8 atoms on the B ring and C15 and C16 atoms on the D ring of DHT bind in a location not occupied by atoms of R-bicalutamide. This finding suggests that more bulk in these regions may enhance the binding affinity of nonsteroidal AR ligands. Hydrogen bonding differs for the chiral hydroxyl group of R-bicalutamide and the 17β-OH of DHT. For R-bicalutamide, Leu-704 and Asn-705 are in close proximity to the hydroxyl functionality, whereas the 17β-OH group of DHT hydrogen bonds to Asn-705 and Thr-877. Further, in the W741L-R-bicalutamide structure, the Thr-877 side chain is rotated 180° situating the Oγ in close proximity to the R-bicalutamide C12 and C13 atoms. Formation of a hydrogen bond to Thr-877 may significantly improve nonsteroidal binding affinities.
Fig. 4.
Overlay of the R-bicalutamide-W741L complex (green) and DHT-WT complex (magenta). (a) Overview of the steroidal plane. Notice the similar positioning of the cyano group of R-bicalutamide to the 3-keto group of DHT and the differences in the location of bulk from these ligands. (b) Side view of the steroidal plane. The R-bicalutamide B ring in the W741L AR binds in the region occupied by the Trp-741 indole ring in the WT AR bound to DHT. Also notice differences in the locations of Met-895, Met-745, and Thr-877.
The majority of the R-bicalutamide molecule binds in a similar plane as DHT. However, at the chiral hydroxyl group, R-bicalutamide bends into a region not occupied by DHT and makes direct contacts with residues of helix 12 (Fig. 4b). In the WT receptor, Trp-741 is seen in this location likely providing favorable van der Waals contacts with DHT. Conversely, loss of bulk in the W741L allows accommodation of the larger R-bicalutamide molecule while maintaining the same fold seen with other agonist bound AR LBD structures. The Leu-741 side chain is displaced toward the Leu-712 side chain but does not affect its location relative to the WT-DHT complex (26). Additionally, Met-895 is considerably displaced by bulk from the sulfonyl linkage group of R-bicalutamide as compared with the WT-DHT structure. The Met-745 side chain is positioned differently in the two complexes, seemingly because of the 19-methyl group on DHT. Previously reported AR LBD crystal structures complexed with R1881 (28), a synthetic steroidal androgen that lacks the 19-methyl group, demonstrate a similar positioning for the Met-745 as with the W741L-R-bicalutamide structure. More interestingly, the location of Met-745 in R1881 AR complexes appears to result in the change of the Trp-741 indole ring to a more distant location from the steroidal binding pocket. This observation could explain the reason for the greater loss in affinity of DHT to the W741L mutant as compared with mibolerone, which also lacks the 19-methyl group.
Insight for R-Bicalutamide Antagonism in WT AR. Although the antagonist AR LBD structure has not been reported, the W741L-R-bicalutamide complex provides insight into the mechanism of bicalutamide antagonism. R-bicalutamide binding induces few changes to residue positions in the W741L AR LBD relative to the WT AR LBD associated with steroidal androgens. Close contacts with Thr-877 may induce its rotation; however, mutations that alleviate bulk in this region do not cause resistance to bicalutamide as with hydroxyflutamide (15). The Met-895 side chain is seemingly displaced by the sulfonyl group oxygen denoted O15 of R-bicalutamide and is accommodated near Leu-741. When R-bicalutamide is bound to the WT receptor, the presence of the Trp-741 indole ring would further increase the bulk in this region. The sulfonyl-linked phenyl ring portion of R-bicalutamide therefore is unlikely accommodated in the agonist conformation and may promote partial unfolding of the AR. This finding offers an explanation for bicalutamide antagonism and stabilized association with heat shock proteins (29). Further studies are needed to uncover the precise antagonist mechanism of bicalutamide.
Proposed Mechanism for Selective AR Modulator Binding and Activity. Recently we discovered a class of compounds that resemble R-bicalutamide in structure (30, 31) but act as agonists for the AR in a tissue-selective manner (32, 33). The most crucial structural alteration to obtain agonist activity is the replacement of the sulfonyl linkage of bicalutamide with an ether or thio linkage. One explanation for this switch in activity is that the ether- and thio-linked compounds form an intramolecular hydrogen bond with the amide nitrogen similar to the sulfonyl hydrogen bond to the amide nitrogen seen in R-bicalutamide. This interaction would require a more bent ligand conformation than that of R-bicalutamide to position the linkage groups within hydrogen bonding range. In addition, the lack of the O15 sulfonyl oxygen as in the R-bicalutamide structure would further decrease the bulk in this region and may allow accommodation of the Met-895 side chain in a similar region as with the WT-DHT complex. Agonist activity of selective AR modulators could therefore be attributed to their lack of bulk in the region that is occupied by the R-bicalutamide sulfonyl group.
Here we have presented the clinically relevant structural data for nonsteroidal AR ligand binding and offered suggestions of structural modifications to increase binding affinity. Additionally, we demonstrated the means by which bicalutamide is able to acquire agonist activity to the AR and portrayed a structural explanation for bicalutamide withdrawal syndrome. Knowledge of R-bicalutamide binding to the W741L mutant offers insight to the antagonist mechanism of bicalutamide and agonist properties of recently reported selective AR modulators (30-33).
Acknowledgments
This work was supported by National Institutes of Health Grants R01 DK59800 and R01 DK065227.
Author contributions: C. E. Bohl, W.G., and J.T.D. designed research; C. E. Bohl performed research; D.D.M. and C. E. Bell contributed new reagents/analytic tools; C. E. Bohl and C. E. Bell analyzed data; and C. E. Bohl wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AR, androgen receptor; LBD, ligand-binding domain; DHT, dihydrotestosterone.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1Z95).
References
- 1.Anonymous (1989) Med. Lett. Drugs Ther. 31, 72 (Clinical trial).2501635 [Google Scholar]
- 2.Anonymous (1996) Am. J. Health Syst. Pharm. 53, 2784 (News). [Google Scholar]
- 3.Kolvenbag, G. J. & Furr, B. J. (1999) Urology 54, 194-197. [PubMed] [Google Scholar]
- 4.Kolvenbag, G. J. & Blackledge, G. R. (1996) Urology 47, 70-79, discussion 80-84. [DOI] [PubMed] [Google Scholar]
- 5.Schellhammer, P. F. & Davis, J. W. (2004) Clin. Prostate Cancer 2, 213-219. [DOI] [PubMed] [Google Scholar]
- 6.Fradet, Y. (2004) Expert Rev. Anticancer Ther. 4, 37-48. [DOI] [PubMed] [Google Scholar]
- 7.See, W. A., Wirth, M. P., McLeod, D. G., Iversen, P., Klimberg, I., Gleason, D., Chodak, G., Montie, J., Tyrrell, C., Wallace, D. M., et al. (2002) J. Urol. 168, 429-435. [PubMed] [Google Scholar]
- 8.Akaza, H., Yamaguchi, A., Matsuda, T., Igawa, M., Kumon, H., Soeda, A., Arai, Y., Usami, M., Naito, S., Kanetake, H. & Ohashi, Y. (2004) Jpn. J. Clin. Oncol. 34, 20-28. [DOI] [PubMed] [Google Scholar]
- 9.Smith, M. R., Goode, M., Zietman, A. L., McGovern, F. J., Lee, H. & Finkelstein, J. S. (2004) J. Clin. Oncol. 22, 2546-2553. [DOI] [PubMed] [Google Scholar]
- 10.Iversen, P. (2003) J. Urol. 170, S48-S52; discussion S52-S4. [DOI] [PubMed] [Google Scholar]
- 11.Iversen, P., Johansson, J. E., Lodding, P., Lukkarinen, O., Lundmo, P., Klarskov, P., Tammela, T. L., Tasdemir, I., Morris, T. & Carroll, K. (2004) J. Urol. 172, 1871-1876. [DOI] [PubMed] [Google Scholar]
- 12.Wirth, M. P., See, W. A., McLeod, D. G., Iversen, P., Morris, T. & Carroll, K. (2004) J. Urol. 172, 1865-1870. [DOI] [PubMed] [Google Scholar]
- 13.Scher, H. I., Steineck, G. & Kelly, W. K. (1995) Urology 46, 142-148. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki, H., Akakura, K., Komiya, A., Aida, S., Akimoto, S. & Shimazaki, J. (1996) Prostate 29, 153-158. [DOI] [PubMed] [Google Scholar]
- 15.Hara, T., Miyazaki, J., Araki, H., Yamaoka, M., Kanzaki, N., Kusaka, M. & Miyamoto, M. (2003) Cancer Res. 63, 149-153. [PubMed] [Google Scholar]
- 16.Bohl, C. E., Chang, C., Mohler, M. L., Chen, J., Miller, D. D., Swaan, P. W. & Dalton, J. T. (2004) J. Med. Chem. 47, 3765-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukherjee, A., Kirkovsky, L., Yao, X. T., Yates, R. C., Miller, D. D. & Dalton, J. T. (1996) Xenobiotica 26, 117-122. [DOI] [PubMed] [Google Scholar]
- 18.Matias, P. M., Donner, P., Coelho, R., Thomaz, M., Peixoto, C., Macedo, S., Otto, N., Joschko, S., Scholz, P., Wegg, A., et al. (2000) J. Biol. Chem. 275, 26164-26171. [DOI] [PubMed] [Google Scholar]
- 19.Hur, E., Pfaff, S. J., Payne, E. S., Gron, H., Buehrer, B. M. & Fletterick, R. J. (2004) PLoS Biol. 2, E274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalton, J. T., Mukherjee, A., Zhu, Z., Kirkovsky, L. & Miller, D. D. (1998) Biochem. Biophys. Res. Commun. 244, 1-4. [DOI] [PubMed] [Google Scholar]
- 21.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 22.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 23.Kraulis, P. J. (1991) J. Appl. Crystallogr. 24, 946-950. [Google Scholar]
- 24.Merritt, E. A. & Murphy, M. E. (1994) Acta Crystallogr. D 50, 869-873. [DOI] [PubMed] [Google Scholar]
- 25.Matias, P. M., Carrondo, M. A., Coelho, R., Thomaz, M., Zhao, X. Y., Wegg, A., Crusius, K., Egner, U. & Donner, P. (2002) J. Med. Chem. 45, 1439-1446. [DOI] [PubMed] [Google Scholar]
- 26.Sack, J. S., Kish, K. F., Wang, C., Attar, R. M., Kiefer, S. E., An, Y., Wu, G. Y., Scheffler, J. E., Salvati, M. E., Krystek, S. R., Jr., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 4904-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tucker, H., Crook, J. W. & Chesterson, G. J. (1988) J. Med. Chem. 31, 954-959. [DOI] [PubMed] [Google Scholar]
- 28.He, B., Gampe, R. T., Jr., Kole, A. J., Hnat, A. T., Stanley, T. B., An, G., Stewart, E. L., Kalman, R. I., Minges, J. T. & Wilson, E. M. (2004) Mol. Cell 16, 425-438. [DOI] [PubMed] [Google Scholar]
- 29.Masiello, D., Cheng, S., Bubley, G. J., Lu, M. L. & Balk, S. P. (2002) J. Biol. Chem. 277, 26321-26326. [DOI] [PubMed] [Google Scholar]
- 30.Yin, D., He, Y., Perera, M. A., Hong, S. S., Marhefka, C., Stourman, N., Kirkovsky, L., Miller, D. D. & Dalton, J. T. (2003) Mol. Pharmacol. 63, 211-223. [DOI] [PubMed] [Google Scholar]
- 31.Marhefka, C. A., Gao, W., Chung, K., Kim, J., He, Y., Yin, D., Bohl, C., Dalton, J. T. & Miller, D. D. (2004) J. Med. Chem. 47, 993-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen, J., Hwang, D. J., Bohl, C. E., Miller, D. D. & Dalton, J. T. (2005) J. Pharmacol. Exp. Ther. 312, 546-553. [DOI] [PubMed] [Google Scholar]
- 33.Gao, W., Kearbey, J. D., Nair, V. A., Chung, K., Parlow, A. F., Miller, D. D. & Dalton, J. T. (2004) Endocrinology 145, 5420-5428. [DOI] [PMC free article] [PubMed] [Google Scholar]