Abstract
Phage predation of Vibrio cholerae has recently been reported to be a factor that influences seasonal epidemics of cholera in Bangladesh. To understand more about this phenomenon, we studied the dynamics of the V. cholerae-phage interaction during a recent epidemic in Dhaka. Because the outbreak strain causing this epidemic was resistant to multiple antibiotics, including streptomycin, we used a selective medium containing streptomycin to monitor accurately the abundance of this strain in the environment. The changing prevalence in the environment of the epidemic V. cholerae O1 strain and a particular lytic cholera phage (JSF4) to which it was sensitive was measured every 48-72 h for 17 weeks. We also monitored the incidence of phage excretion in stools of 387 cholera patients during the epidemic. The peak of the epidemic was preceded by high V. cholerae prevalence in the environment and was followed by high JSF4 phage levels as the epidemic ended. The buildup to the phage peak in the environment coincided with increasing excretion of the same phage in the stools of cholera patients. These results suggest that patients toward the end of the epidemic ingested both JSF4 phage and the outbreak V. cholerae strain. Host-mediated phage amplification during the cholera epidemic likely contributed to increased environmental phage abundance, decreased load of environmental V. cholerae and, hence, the collapse of the epidemic. Thus, in vivo phage amplification in patients and subsequent phage predation in the environment may explain the self-limiting nature of seasonal cholera epidemics in Bangladesh.
Keywords: Vibrio cholerae, vibriophage
Toxigenic strains of Vibrio cholerae belonging to the O1 and O139 serogroups cause cholera, a devastating diarrhea disease that occurs frequently as epidemics in many developing countries (1). Epidemics of cholera occur regularly in the Ganges Delta region of Bangladesh and India. In Bangladesh, outbreaks usually occur twice during a year, with the highest number of cases just after the monsoon and a somewhat smaller number of cholera cases during the spring. The occurrence of epidemics are known to coincide with increased prevalence of the causative V. cholerae strain in the aquatic environment (2). A variety of physical and biological parameters are likely to influence the survival and abundance of V. cholerae as a species in the environment, but these factors do not exclusively modulate the prevalence of toxigenic V. cholerae O1 and O139 strains. In contrast, bacterial viruses (phage or bacteriophage) in the environment have recently been found to inversely correlate with the abundance of toxigenic V. cholerae in water samples and the incidence rates of cholera (3). These data strongly suggest that phage predation in the environment likely influences the temporal dynamics of cholera epidemics. Phages also play a role in the emergence of pathogenic clones and may also be involved in territorialism between different strains of V. cholerae (3-5). For example, cholera toxin genes are transferred to nontoxigenic strains through a lysogenic filamentous phage (6), and the emergence and dominance of V. cholerae O139 in Bangladesh and India during 1992-1993 may have involved phages both as a means of horizontal gene transfer as well as a bacteriocidal selective mechanism (3-5, 7). Recently, it has also been recognized that some bacteriophages carry powerful diversity-generating retroelements (DGR) that are used to alter host range of the phages (8, 9). Thus, possible DGR-driven changes in the host range of vibriophages may lead to the emergence of new phage types that could impact the epidemiology of cholera or the emergence of new serogroups of V. cholerae (3).
Because cholera is a waterbourne disease, real-time environmental monitoring for the prevalence of V. cholerae strains with epidemic potential and lytic vibriophages is important to understand their precise role in cholera dynamics. However, quantitative estimation of pathogenic V. cholerae in the environment has been limited by the lack of a suitable technique to measure a low level of pathogenic strains in the presence of a large population of mostly nonpathogenic strains. Therefore, previous studies (10, 11) relied mostly on a qualitative assessment of environmental V. cholerae levels.
In recent times, epidemics of cholera in Bangladesh are caused by toxigenic V. cholerae O1 or O139 strains that are resistant to streptomycin, trimethoprim, and sulfamethoxazole. This resistance profile is encoded by the chromosomal SXT element carried by these strains (12). A recent cholera epidemic in Bangladesh that began in August 2004 was caused by such an antibiotic-resistant strain of V. cholerae O1. We exploited this resistance phenotype to significantly enhance the estimation of environmental prevalence of the epidemic V. cholerae strain by selection on streptomycin plates after enrichment in bile peptone medium. We also used this assay system to monitor the changing prevalence of the outbreak strain in relation to that of a lytic vibriophage during the course of the epidemic. At the same time, we monitored the excretion of the phage in stools of cholera patients. This study provides evidence for the role of phage predation in controlling the dynamics of epidemic cholera and has significant implications in devising cholera control measures by possible phage-mediated interventions.
Materials and Methods
Clinical Surveillance for Cholera. The International Centre for Diarrhoeal Disease Research, Bangladesh (ICDDRB) maintains a 2% surveillance system at its Dhaka hospital in which data from every 50th patient presenting to the hospital is collected, including clinical information and biological specimens. Data from this surveillance system was used to extrapolate the overall numbers of patients with cholera, and specimens from these patients were used to obtain clinical V. cholerae and phage strains as required for the studies described.
Environmental Sampling Sites. Eleven different sites along three major rivers, one lake, and marshy lands in and around Dhaka were selected for sampling (Fig. 1). Cholera patients from these areas normally seek treatment in the Dhaka hospital of ICDDRB. Water samples for the study were collected from each of these sites 2-3 times in a week at 48- to 72-h intervals during the duration of the epidemic. All water samples were collected in sterile containers and transported to the laboratory for initial processing within 2 h of collection.
Fig. 1.
Map of Dhaka showing the environmental sampling sites (•) and the location of the ICDDRB cholera hospital at the center of the city.
Detection and Analysis of Phage. A panel of recipient V. cholerae strains, including isolates from the recent cholera epidemic, were used to estimate environmental phage concentration by plaque assay as described in ref. 3. Briefly, logarithmic-phase cells (500 μl) of each host bacterial strain in nutrient broth (Difco, Detroit) were mixed with 3.5-ml aliquots of soft agar (nutrient broth containing 0.8% Bactoagar, Difco), and the mixtures were overlayed on nutrient agar plates. Aliquots of water (10-50 μl), which were prefiltered through 0.22-μm pore-size filters (Millipore) to make bacteria-free were inoculated on the plates, and the plates were incubated for 16 h at 37°C. Aliquots of water samples mixed with serial dilutions of a control phage strain (10 to 104 particles per ml) were used as positive controls in all phage assays. For detection of phage in cholera stools, aliquots of watery stools were filtered by using the 0.22-μm filter, and dilutions of this filtrate were inoculated on plates. A sample was scored positive for phages when a plaque was observed on the bacterial lawn. Plaques were counted to estimate the concentration of phage particles in the sample. Phages from representative plaques were further purified and stocked for subsequent analysis. Nucleic acids were prepared from representative phage isolates by using methods in ref. 3 and were analyzed for restriction endonuclease cleavage patterns of the phage genomes by using standard procedures in ref. 13.
Culture of Environmental Water Samples for V. cholerae. Water samples were analyzed for the presence of V. cholerae by modifications of methods described in ref. 11. Briefly, an aliquot (50 ml) of each water sample was added to 25 ml of 3× concentrated bile peptone medium (BP) (1% peptone/0.5% tauracholic acid/1% NaCl, pH 9.0) and incubated for 6 h. Dilutions of this BP culture were plated on tauracholate tellarite gelatin agar (TTGA) plates (14) containing streptomycin (70 μg/ml) as well as on TTGA plates devoid of the antibiotic. Suspected Vibrio colonies were picked and subjected to biochemical and serological tests to identify V. cholerae belonging to the O1 or O139 serogroups. All isolates were saved for further genetic and phenotypic studies, including analysis of drug resistance, presence of the SXT element, virulence gene content, and ribotype analysis.
Probes and PCR Assays. Representative V. cholerae O1 strains derived from each water sample were analyzed for different virulence associated genes and were compared by ribotyping for clonal relationship. Presence of virulence genes was determined by using specific PCR assays for the tcpA, tcpI, and acfB genes of the TCP pathogenicity island (11) and the ctxA and zot genes of the CTX prophage (6, 11). The SXT probe was a NotI fragment of pSXT1 (12), and the rRNA gene probe consisted of a BamHI fragment of the Escherichia coli rRNA clone pKK3535 (15, 16). Probes were labeled by using a random primers DNA labeling kit (Invitrogen) and [α-32P]-deoxycytidine triphosphate (3,000 Ci/mmol, 1 Ci = 37 GBq) (Amersham Pharmacia Biosciences). Colony blots or Southern blots were prepared and hybridized with the labeled probes by following standard methods in ref. 13.
Data Analysis. Frequency of isolation of V. cholerae O1 from different water samples by selecting on streptomycin plates and on plates without streptomycin were summarized into tables for group comparisons. Statistical analyses were done by using epi info 6.0 (USD, Stone Mountain, GA). The significance of difference in proportions was evaluated by the χ2 test.
Because enrichment culture was necessary to recover viable V. cholerae O1 cells from water samples, the absolute concentration of V. cholerae O1 in water samples could not be determined during the study. Accordingly, we defined a quantitative parameter “index of V. cholerae concentration,” whose value reflects the dynamic changes in V. cholerae O1 concentration based on quantitative yield of viable cells after a standard enrichment procedure. This index was calculated as follows. For each week of the study period, the mean of the observed V. cholerae O1 concentration in different water samples after standard 6-h enrichment culture was determined. We also determined the median value by analyzing the range of weekly mean values over the entire study period of 17 weeks. Each weekly mean value was then divided by the study period median value to determine a weekly index of V. cholerae O1 concentration. Thus, the index represents the extent of increase or decrease in the mean concentration of V. cholerae O1 during any given week of the study period.
Results
Environmental Prevalence of Epidemic V. cholerae Is High When Measured by a Selective Enrichment Technique. Environmental water samples collected from 11 different sites around Dhaka (Fig. 1) were analyzed for the presence of V. cholerae by using two different approaches. After enrichment in bile peptone medium for 6 h, aliquots of the culture were inoculated on conventional TTGA plates (14) and on a set of TTGA plates that contained streptomycin (70 μg/ml). Selection on streptomycin plates allowed isolation of V. cholerae O1 from 141 of 339 samples tested, whereas only 60 samples were positive for V. cholerae O1 when selected on TTGA plates without streptomcycin (Table 1). All V. cholerae O1 strains that were initially isolated on nonantibiotic plates were also found to be resistant to streptomycin, whereas non-O1 strains isolated from the same plates were sensitive to the antibiotic. In addition, all V. cholerae O1 strains isolated by both methods were also resistant to trimethoprim, and sulfamethoxazole and were positive for the SXT element (12). The difference between the proportion of water samples found to be positive for V. cholerae O1 by the two assay methods was highly significant (P < 0.001). Whereas only 17.6% of samples analyzed yielded V. cholerae O1 by conventional enrichment culture, as high as 41.5% of the same water samples were positive for the organism when selected on streptomycin plates. These positive water samples included 81 samples that were found to be negative by conventional culture (Table 1).
Table 1. Detection of V. cholerae O1 in environmental water samples by selection on TTGA plates containing streptomycin after enrichment in bile peptone medium as compared with culture on TTGA plates devoid of the antibiotic.
| No. of water samples (%) positive for V. cholerae O1
|
||||
|---|---|---|---|---|
| Month of sampling in 2004 | No. of water samples analyzed* | Selected on TTGA plates without antibiotic* | Selected on TTGA plates containing streptomycin | P value† |
| August | 33 | 2 (6.0) | 12 (36.3) | 0.006 |
| September | 98 | 22 (22.4) | 48 (48.9) | 0.0002 |
| October | 109 | 26 (23.8) | 45 (41.2) | 0.009 |
| November | 66 | 7 (10.6) | 26 (39.3) | 0.0002 |
| December | 33 | 3 (9.0) | 10 (30.3) | 0.063 |
| Total | 339 | 60 (17.6) | 141 (41.5) | <0.00001 |
All V. cholerae O1 strains isolated on TTGA plates devoid of antibiotics were also found to be resistant to streptomycin.
Difference between the proportion of water samples found positive for V. cholerae O1 by the two assay methods was statistically significant at P <0.001. Statistical analyses were done by using epi info 6.0 (USD). The significance of difference in proportions was evaluated by the χ2 test.
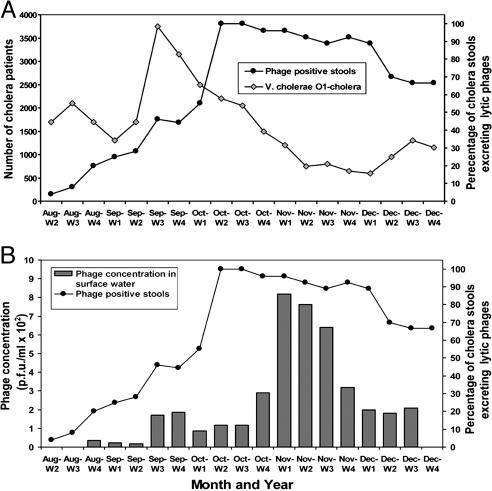
Prevalence of V. cholerae and Vibriophages Changes Rapidly During a Cholera Epidemic. Previous studies involving environmental monitoring of V. cholerae strains with epidemic potential relied mostly on a qualitative assessment, due mainly to inadequacies in techniques to quantify V. cholerae in environmental samples (10, 11). Because V. cholerae O1 is estimated after enrichment, the observed counts, although expected to be proportional to the actual concentration, are by no means the real concentration in water samples. However, in this study, we used an index of epidemic V. cholerae O1 concentration rather than the real concentration to be able to record possible changes in the environmental prevalence of the strain during the epidemic. The index of V. cholerae O1 concentration in different waters varied between 0 and 42.8, and the weekly mean index of all samples taken together varied between 0.16 and 34.38 during the sampling period. The weekly mean V. cholerae index gradually increased as the epidemic progressed and reached a maximum of 215-fold more than the minimum index during the second week of September 2004, i.e., immediately before the peak of the epidemic (Fig. 2).
Fig. 2.
Dynamics of epidemic V. cholerae O1 and vibriophage concentration in surface water during a cholera epidemic in Bangladesh. Weekly mean V. cholerae O1 levels (A) and lytic vibriophage levels (B) in the aquatic environment were taken during the course of a cholera epidemic in Bangladesh. Environmental sampling was conducted from the third week of August to the third week of December 2004. The number of cholera patients are extrapolated from 2% surveillance samples.
V. cholerae O1 strain causing this epidemic was found to be killed by an environmental phage, previously designated as JSF4 (3). The concentration of this phage in the environment was also found to fluctuate during the study period. Phage concentration in different water samples varied between an undetectable level (<50 phage particles per ml) and 4.5 × 103 pfu/ml during the study, and the weekly mean phage concentration of all samples taken together ranged from an undetectable level to 8.18 × 102 pfu/ml. The concentration of the phage increased rapidly during the third week of October and the first week of November 2004, just after the peak of the cholera epidemic. During the same period, epidemic V. cholerae O1 concentration tended to vary inversely with phage concentration, as has been previously reported for other study periods (3). The mean V. cholerae O1 level in the environment and the number of cholera cases rapidly decreased after the phage peak (Fig. 2). In the following weeks, the phage prevalence also decreased considerably, suggesting that the duration of the phage peak may depend on the stability of the phage particles in environmental waters and, thus, may be influenced by particular physicochemical parameters of the water.
The Lytic Phage Is Excreted in Cholera Stools. During the study period, we examined available cholera stools for the presence of lytic cholera phages. Between 25 and 30 randomly obtained cholera stools (each from a different patient and later confirmed by culture) were examined each week during the first 12 weeks of the study. However, toward the end of the epidemic (the second week of November to the fourth week of December 2004) when cholera cases decreased, 9-20 stool samples were available for analysis each week. We found that as the epidemic progressed, an increasingly higher proportion of patients were excreting phage particles in their stools (Fig. 3), and molecular analysis of representative isolates indicated that this phage was JSF4 (see below). The phage positive stools contained between 4.0 × 102 and 1.1 × 108 phage particles per ml of watery stools. The proportion of stool samples excreting phage was minimal (<5%) at the beginning of the epidemic and gradually increased to reach a peak when 100% of the stools tested were positive for the phage. The timing of the phage peak in stools overlapped with that of the observed environmental phage peak (Fig. 3). Interestingly, despite the presence of high titers of lytic phage particles in the stools, the stools were also culture positive for V. cholerae O1. The V. cholerae strain isolated from the stools were completely susceptible to lysis by the phage from the same stools when examined by a plaque assay. Analysis of representative V. cholerae isolates from cholera stools by using DNA probes derived from the phage genome did not show the presence of lysogenic phage genome in the V. cholerae strain. This finding ruled out the possibility that the phage in stools might have originated from induction of lysogenic cells in vivo. These data suggested that the observed excretion of phage in cholera stools was possibly due to ingestion of both V. cholerae and cholera phage JSF4 as a mixture by susceptible individuals. The excretion of high counts of both phage and V. cholerae in stools may be explained by assuming that one intestinal compartment favors rapid growth of V. cholerae cells, whereas another compartment preferentially favor phage amplification. The former compartment may be under the protective mucus layer near the intestinal brush border, and the latter may be within the intestinal lumen.
Fig. 3.
Vibriophage excretion by cholera patients and its correlation with environmental prevalence of the same phage. Excretion of lytic vibriophages in cholera stools (A) and weekly mean vibriophage levels in the aquatic environment (B) were taken during the course of a cholera epidemic.
V. cholerae and Phages Excreted in Stools May Contribute to Their Environmental Prevalence. We used molecular epidemiological analysis to compare both V. cholerae and phage isolates from clinical and environmental sources. All V. cholerae O1 strains isolated during the epidemic from environmental and clinical sources carried the known virulence gene clusters, including the TCP pathogenicity island (17) and the CTX prophage (6). In addition, all strains from the current epidemic carried the SXT element and showed an identical antibiotic resistance pattern. All strains were analyzed for BglI restriction patterns of their rRNA genes to understand their clonal relationship. We found that V. cholerae strains isolated from different water samples during the epidemic belong to a single ribotype that was identical to that of the clinical strains isolated from cholera stools (Fig. 5, which is published as supporting information on the PNAS web site). These observations epidemiologically linked the strains isolated from the environment and the patients and supported the assumption that V. cholerae strains excreted in cholera stools had contributed to the environmental prevalence of the pathogen. This finding is also consistent with the suggestion that human infections causes enrichment of the strain and helps to increase its concentration in the aquatic environment through fecal contamination of surface water (1, 11).
Comparative analysis of the DNA derived from phage particles isolated from the environment and stools based on restriction patterns and DNA hybridization identified the same phage from both these sources (Fig. 6, which is published as supporting information on the PNAS web site). This phage was previously designated as JSF4 and specifically kills V. cholerae O1 strains (3). Indeed, in a previous study, this phage was most consistently found in water samples that contained no detectable V. cholerae O1 (3). These results thus suggest that phage amplification in patients may account for upsurges in phage abundance in the environment and subsequent killing of environmental V. cholerae.
Discussion
The hypothesis that cholera epidemics are triggered by environmental factors has been embraced by many environmental microbiologists, climatologists, and ecologists during the last decade (18-20). The fact that cholera epidemics seem to occur in several different foci simultaneously (21, 22) is often cited as evidence for an environmental component to its epidemiology. However, it is seldom appreciated that most of the time, an epidemic is caused by a single bacterial clone, and only a few epidemics have involved more than one clone in the same geographic location (23, 24). All V. cholerae O1 isolates from the epidemic analyzed here also belonged to a single clone, suggesting that the outbreak probably began from a point source and then spread rapidly. Contrary to common assumptions, it therefore seems possible that most cholera epidemics begin from a point source. However, in rural areas of a developing country such as Bangladesh, where health care infrastructure is inadequate, the first few cases are likely to remain unnoticed, and cholera would spread rapidly to other locations before an outbreak is reported. In this study, streptomycin enrichment culture made it possible for us to monitor the temporal and population dynamics of a single clone of V. cholerae O1 causing a large epidemic in Dhaka that followed a severe flood.
The cholera incidence curve for this epidemic was typical in shape (Fig. 2) and showed that the number of patients increased steadily until a peak was reached and was then followed by a period of rapid decrease in the number of cases. Thus, this epidemic, like most cholera epidemics, was self-limiting despite the fact that epidemics of this sort reoccur at regular intervals in an endemic area like Bangladesh. Although climatic factors have been implicated in cholera epidemics (25), once an epidemic is initiated, these factors do not adequately explain the rapid collapse of the epidemic. For example, in a temperate region, the climatic factors such as temperature or rainfall may not substantially change during the course of an epidemic. Development of adequate immunity by the population as a cause for the epidemic to end is also ruled out because in an endemic area such as Bangladesh, epidemics are known to reoccur regularly.
The present study found that as the epidemic under study progressed, more and more cholera patients excreted both V. cholerae and the lytic phage JSF4. Both V. cholerae and JSF4 phage also increased in concentration in the environment as the epidemic progressed and ultimately ended (Figs. 2 and 3). The collapse of the environmental V. cholerae population coincided with the plateau of environmental phage. The data rather strongly suggest that the phage's killing action on the epidemic strain was probably responsible for the collapse of the environmental V. cholerae population and termination of the epidemic. Our data further suggest that amplification of first V. cholerae, and then the phage in the human host, led to the observed increase in the environmental prevalence of both V. cholerae and the phage in their respective temporal waves (Figs. 2 and 3). This assumption was further supported by the observation that the phage excreted in cholera stools and those isolated from the environment during the epidemic were the same phage (Fig. 6). We propose that coexcretion of JSF4 phage and V. cholerae was probably a result of victims ingesting both phage and V. cholerae at the time of infection. A corollary to this scenario is that a considerably higher dose of V. cholerae would likely be needed to establish an acute infection in the individual who had also ingested a lytic phage. This assumption is simply because the coingested phage should kill some fraction of the infectious innoculum and could in theory abort the infection. This concept could be easily validated by testing whether coingested phage have a protective effect in appropriate volunteer models for cholera.
Our data suggest further refinements to a model for cholera epidemiology (1, 11). In the newest reiteration (Fig. 4), an epidemic begins when human victims further amplify a V. cholerae strain that initially caused an index case. Other cholera victims eventually amplify phages and the epidemic clone, leading to increases in phage concentration in the environment where predation subsequently decreases the V. cholerae concentration. Cholera cases diminish because (i) in the face of phage predation, the environment fails to support a heavy load of viable V. cholerae, and (ii) the infectious dose increases dramatically because of the deleterious effect of coingesting phage with the innoculum. Thus, the equilibrium shifts rapidly in favor of the phage and leads to the collapse of the epidemic. However, subsequently the concentration of phages diminishes because the phages are not indefinitely stable under particular physicochemical conditions in the water, or they may be washed away by floods or rain waters. Consequently, V. cholerae strains with epidemic potential can grow again fostered by other environmental factors. An early index case leads to the start of the next epidemic cycle dominated by a single clone of V. cholerae. If this model is correct, it seems apparent that cholera phages like JSF4 might be used as biological control agents to interrupt epidemics before they run their natural course. This approach might be most fruitful if applied to epidemics occurring in “cholera naive” geographical locales where V. cholerae O1 phages might be low in prevalence or even absent from the environment. Such a situation may have existed during the earliest stage of the Latin American O1 epidemic that began with the introduction of the seventh pandemic El Tor O1 V. cholerae clone into Lima, Peru, in 1991 (1).
Fig. 4.
Schematic model for factors influencing seasonal epidemics of cholera. Events occurring in the environment are shown in circles, whereas those occurring in human cholera victims are shown in rectangles. 1, Seasonal conditions (e.g., floods or physical-chemical factors causing phage instability) lead to the reduction of viable lytic phages in the aquatic environment. 2, Other seasonal factors (e.g., elevated temperature or biological-cofactors) lead to a bloom of nonpathogenic and pathogenic Vibrio species in local water sources. 3, A favorable aquatic environment facilitates infection by a pathogenic V. cholerae strain (black vibrios), causing an asymptomatic or undetected symptomatic infection in an index case. 4, Amplification of the pathogenic clone in the index case seeds the environment. 5, An epidemic begins, triggered by the index case leading to host amplification of the pathogenic clone and massive environmental contamination. 6, Phages produced by environmental Vibrio species (e.g., through induction of lysogens or lytic growth) begin to amplify on the pathogenic clone in the environment. 7, Cholera victims increasingly ingest both the pathogenic clone and the lytic phage from environmental sources leading to amplification of the phage in vivo. 8, Environmental contamination with phage shed by cholera victims and further amplification of phage on the pathogenic clone within the environment lead to collapse of the epidemic. Factors contributing to the collapse of the epidemic include the decreasing concentration of the pathogenic clone in the environment coincident with a upsurge in environmental phage concentration that together contribute to a dramatic reduction in the rate of transmission of cholera in the interepidemic period.
Critically in this study, we exploited the drug resistance phenotype of the epidemic V. cholerae strain to be able to monitor its presence and fluctuation in concentration in the aquatic environment. The V. cholerae O1 strain was present in the environment throughout the sampling period, although the concentration varied considerably (Fig. 2). The usual standard enrichment culture technique seems to have a lower sensitivity for detection of this epidemic V. cholerae strain, and, thus, a large number of samples analyzed by standard methods were scored as negative despite the presence of viable V. cholerae in these samples. Although selection on antibiotics cannot be generalized for all V. cholerae O1 strains, we used the antibiotic resistance property of the prevailing strain as an opportunity to understand the dynamics of epidemic V. cholerae O1 in the environment. Our selection method confirmed the presence of the pathogenic clone in the water coincident with the epidemic under study.
Previously, volunteer challenge studies with V. cholerae suggested that generally a high dose of V. cholerae is required to cause clinical disease (26, 27). Although cholera is known to spread through contaminated environmental water, the presence of toxigenic V. cholerae usually detected in surface water is far less than that required to induce infection under experimental conditions. Interestingly, this study suggests that in an endemic area, the concentration of a toxigenic V. cholerae O1 in the environment could be quite high during an epidemic and its distribution more widespread than previously appreciated. However, the relative infectivity for humans of these environmental cells is unknown but likely to be high given the coincidence of their appearance with the epidemic.
It has been suggested that under unfavorable environmental conditions, V. cholerae can enter into a viable but “nonculturable” state (18). However, because the nonculturable state by definition is a condition when cells are incapable of forming a colony on commonly used media, it has been impossible to isolate the strains responsible for generating such cells in the environment. Thus, reported demonstrations of the existence of viable but nonculturable (VBNC) V. cholerae cells in the environment relied on fluorescent antibody techniques (19, 20). Some reports suggest that V. cholerae can be converted to a VBNC state under defined laboratory conditions, and in one such case, nonculturable cells were putatively converted to viable cells by passage through the guts of human subjects (28). In contrast, experiments performed by Bogosian et al. (29) suggested that VBNC cells they observed were simply dead and that when “resuscitation” was observed, it was merely due to the growth of rare viable cells present in the preparation. Therefore, in our experiments, we did not attempt to differentiate between resuscitation of VBNC cells in aquatic samples and the recovery of rare viable cells in these samples. Nonetheless, our streptomycin enrichment technique may provide a potent tool to study the physiological state of viable V. cholerae in aquatic environments and perhaps VBNC cells that resist quantification and identification by direct culture.
Our data clearly suggest that environmental water samples contain more viable V. cholerae cells than are recovered by usual enrichment culture techniques. Streptomycin enrichment proves these cells are present in the water samples, viable, and thus potentially able to cause disease. It seems possible that in the environment, these pathogenic O1 V. cholerae cells exist in a metabolically less-active state and, hence, initially grow slower than normal cells when nutrients are added. This assumption is consistent with results reported by Baker and colleagues (30), who reported that laboratory-grown, nutrient-starved V. cholerae cells assumed an unusual coccoid shape and required incubation for 2 h in rich medium to reassume normal size and shape. Remarkably, such cells only began to divide after 5 h of incubation (30), a time frame remarkably close to the 6 h of enrichment culture we used before selection on streptomycin containing media. During conventional enrichment culture, environmental O1 V. cholerae might become undetectable because of overgrowth of more rapidly growing bacterial species (or even nonpathogenic environmental non-O1 V. cholerae strains) when cultured on plates devoid of streptomycin. Thus, the nonpathogenic environmental bacterial flora, including non-O1 V. cholerae, might interfere with direct cultivation of damaged, pathogenic O1 cells or even kill these O1 cells by production of broad-host range phages (5).
We hypothesize that the water samples characterized in this study probably contain starved but viable V. cholerae cells similar to those described by Baker et al. (30). For future clarity, the viable V. cholerae in these water samples should be differentiated from VBNC (18) because we demonstrated that the V. cholerae in our samples could in fact be cultured. Thus, these environmental V. cholerae cells are at least “conditionally viable” if one uses the appropriate selective procedure. Accordingly, we propose to call such cells “conditionally viable environmental cells” or CVEC. There is much to elucidate regarding V. cholerae CVEC including their structure, metabolic state, infectivity, phage sensitivity, and ultimately their relationship to the epidemiology of cholera.
Supplementary Material
Acknowledgments
This research was funded by National Institutes of Health Research Grant GM068851 under a subagreement between Harvard Medical School and ICDDRB and by the Swedish International Development Agency under an agreement with ICDDRB. ICDDRB is supported by countries and agencies that share its concern for the health problems of developing countries.
Author contributions: S.M.F. and J.J.M. designed research; S.M.F., M.J.I., Q.S.A., A.S.G.F., D.A.S., G.B.N., and J.J.M. performed research; S.M.F., A.S.G.F., D.A.S., G.B.N., and J.J.M. analyzed data; S.M.F. and J.J.M. wrote the paper; and A.S.G.F. and D.A.S. conducted surveillance for cholera.
Abbreviations: ICDDRB, International Centre for Diarrhoeal Disease Research, Bangladesh; TTGA, tauracholate tellarite gelatin agar; VBNC, viable but nonculturable.
References
- 1.Faruque, S. M., Albert, M. J. & Mekalanos, J. J. (1998) Microbiol. Mol. Biol. Rev. 62, 1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khan, M. U., Shahidullah, M., Haque, M. S. & Ahmed, W. U. (1984) Trop. Geogr. Med. 36, 335-340. [PubMed] [Google Scholar]
- 3.Faruque, S. M., Naser, I. B. Islam, M. J., Faruque, A. S. G., Ghosh, A. N., Nair, G. B., Sack, D. A. & Mekalanos, J. J. (2005) Proc. Natl. Acad. Sci. USA 102, 1702-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jouravleva, E. A., McDonald, G. A., Gaon, C. F., Boesman-Finkelstein, M. & Finkelstein, R. A. (1998) Microbiology 144, 315-324. [DOI] [PubMed] [Google Scholar]
- 5.Faruque, S. M. & Mekalanos, J. J. (2003) Trends Microbiol. 11, 505-510. [DOI] [PubMed] [Google Scholar]
- 6.Waldor, M. K. & Mekalanos, J. J. (1996) Science 272, 1910-1914. [DOI] [PubMed] [Google Scholar]
- 7.Mooi, F. R. & Bik, E. M. (1997) Trends Microbiol. 5, 161-165. [DOI] [PubMed] [Google Scholar]
- 8.Doulatov, S., Hodes, A., Dai, L., Mandhana, N., Liu, M., Deora, R., Simons, R. W., Zimmerly, S. & Miller, J. F. (2004) Nature 431, 476-481. [DOI] [PubMed] [Google Scholar]
- 9.Liu, M., Deora, R., Doulatov, S. R., Gingery, M., Eiserling, F. A., Preston, A., Maskell, D. J., Simons, R. W., Cotter, P. A., Parkhill, J. & Miller, J. F. (2002) Science 295, 2091-2094. [DOI] [PubMed] [Google Scholar]
- 10.Madico, G., Checkley, W., Gilman, R. H., Bravo, N., Cabrera, L., Calderon, M. & Ceballos, A. (1996) J. Clin. Microbiol. 34, 2968-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faruque, S. M., Chowdhury, N., Kamruzzaman, M., Dziejman, M., Rahman, M. H., Sack, D. A., Nair, G. B. & Mekalanos, J. J. (2004) Proc. Natl. Acad. Sci. USA 101, 2123-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waldor, M. K., Tschape, H. & Mekalanos, J. J. (1996) J. Bacteriol. 178, 4157-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maniatis, T., Fritsch, E. F. & Sambrook, J. (1982) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY)
- 14.Monsur, K. A. (1961) Trans. R. Soc. Trop. Med. Hyg. 55, 440-442. [DOI] [PubMed] [Google Scholar]
- 15.Brosius, J., Ullrich, A., Raker, M. A., Gray, A., Dull, T.J., Gutell, R. R. & Noller, H. F. (1981) Plasmid 6, 112-118. [DOI] [PubMed] [Google Scholar]
- 16.Faruque, S. M., Roy, S. K., Alim, A. R. M. A., Siddique, A. K. & Albert, M. J. (1995) J. Clin. Microbiol. 33, 2833-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovach, M. E., Shaffer, M. D. & Peterson, K. M. (1996) Microbiology 142, 2165-2174. [DOI] [PubMed] [Google Scholar]
- 18.Colwell, R. R. & Huq, A. (1994) in Vibrio cholerae and cholera: Molecular to Global Perspectives, eds. Wachsmuth, I. K., Blake, P. A. & Olsvik, O. (Am. Soc. Microbiol., Washington, DC) pp. 117-133.
- 19.Huq, A., Colwell, R., Rahman, R., Ali, A., Chowdhury, M., Parveen, S., Sack, D. & Russek-Cohen, E. (1990) Appl. Environ. Microbiol. 56, 2370-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Binsztein, N., Costagliola, M., Pichel, M., Jurquiza, V., Ramirez, F., Akselman, R., Vacchino, M., Huq, A. & Colwell, R. (2004) Appl. Environ. Microbiol. 70, 7481-7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaper, J. B., Morris, J. G. & Levine, M. M. (1995) Clin. Microbiol. Rev. 8, 48-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glass, R. I., Becker, S., Huq, M. I., Stoll, S. B., Khan, M. U., Merson, M. H., Lee, J. V. & Black, R. E. (1982) Am. J. Epidemiol. 116, 959-970. [DOI] [PubMed] [Google Scholar]
- 23.Islam, M. S., Talukder, K. A., Khan, N. H., Mahmud, Z. H., Rahman, M. Z., Nair, G. B., Siddique, A. K., Yunus, M., Sack, D. A., Sack, R. B., Huq, A. & Colwell, R. R. (2004) Microbiol. Immunol. 48, 773-777. [DOI] [PubMed] [Google Scholar]
- 24.Siddique, A. K., Zaman, K. & Mazumder, Y. (1989) Lancet. 2, 396. [DOI] [PubMed] [Google Scholar]
- 25.Lobitz, B., Beck, L., Huq, A., Wood, B., Fuchs, G., Faruque, A. S. G. & Colwell, R. (2000) Proc. Natl. Acad. Sci. USA 97, 1438-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cash, R. A., Music, S. I., Libonati, J. P., Snyder, M. J., Wenzel, R. P. & Hornick, R. B. (1974) J. Infect. Dis. 129, 45-52. [DOI] [PubMed] [Google Scholar]
- 27.Levine, M. M., Kaper, J. B. Herrington, D., Losonsky, G., Morris, J. G., Clements, M. L., Black, R. E., Tell, B. & Hall, R. (1988) Infect. Immun. 56, 161-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colwell, R. R., Brayton, P., Huq, A., Tall, B., Harrington, P. & Levine, M. (1996). World J. Microbiol. Biotechnol. 12, 28-31. [DOI] [PubMed] [Google Scholar]
- 29.Bogosian, G., Morris, P. J. L. & O'Neil, J. P. (1998) Appl. Environ. Microbiol. 57, 875-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baker, E. M., Singleton, F. L. & Hood, M. A. (1983) Appl. Environ. Microbiol. 46, 930-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.