ABSTRACT
Halogenated aromatic compounds are used in a variety of industrial applications but can be harmful to humans and animals when released into the environment. Microorganisms that degrade halogenated aromatic compounds anaerobically have been isolated but the evolutionary path that they may have taken to acquire this ability is not well understood. A strain of the purple nonsulfur bacterium, Rhodopseudomonas palustris, RCB100, can use 3-chlorobenzoate (3-CBA) as a carbon source whereas a closely related strain, CGA009, cannot. To reconstruct the evolutionary events that enabled RCB100 to degrade 3-CBA, we isolated an evolved strain derived from CGA009 capable of growing on 3-CBA. Comparative whole-genome sequencing of the evolved strain and RCB100 revealed both strains contained large deletions encompassing badM, a transcriptional repressor of genes for anaerobic benzoate degradation. It was previously shown that in strain RCB100, a single nucleotide change in an alicyclic acid coenzyme A ligase gene, named aliA, gives rise to a variant AliA enzyme that has high activity with 3-CBA. When the RCB100 aliA allele and a deletion in badM were introduced into R. palustris CGA009, the resulting strain grew on 3-CBA at a similar rate as RCB100. This work provides an example of pathway evolution in which regulatory constraints were overcome to enable the selection of a variant of a promiscuous enzyme with enhanced substrate specificity.
IMPORTANCE
Biodegradation of man-made compounds often involves the activity of promiscuous enzymes whose native substrate is structurally similar to the man-made compound. Based on the enzymes involved, it is possible to predict what microorganisms are likely involved in biodegradation of anthropogenic compounds. However, there are examples of organisms that contain the required enzyme(s) and yet cannot metabolize these compounds. We found that even when the purple nonsulfur bacterium, Rhodopseudomonas palustris, encodes all the enzymes required for degradation of a halogenated aromatic compound, it is unable to metabolize that compound. Using adaptive evolution, we found that a regulatory mutation and a variant of promiscuous enzyme with increased substrate specificity were required. This work provides insight into how an environmental isolate evolved to use a halogenated aromatic compound.
KEYWORDS: Rhodopseudomonas palustris, 3-chlorobenzoate, 4-fluorobenzoate, anaerobic aromatic compound degradation
INTRODUCTION
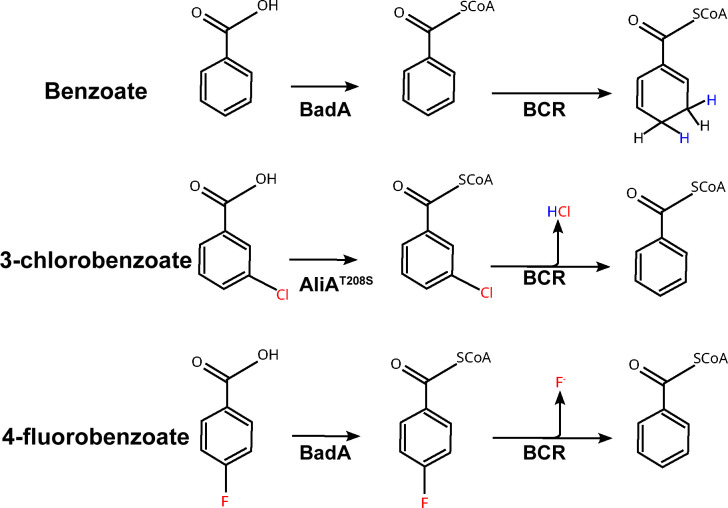
Halogenated aromatic compounds are widely used in a variety of applications but also have harmful effects on human and animal health (1, 2). Their widespread use has led to their accumulation in the environment, and much research has been done to understand the role microorganisms play in degrading these compounds (2). One such example is the purple nonsulfur bacterium, Rhodopseudomonas palustris RCB100. R. palustris RCB100 has been shown to metabolize 3-chlorobenzoate (3-CBA) during photoheterotrophic growth using the benzoyl-CoA degradation pathway (3). In this pathway, aromatic compounds are first activated by a coenzyme A (CoA) ligase to benzoyl-CoA and then reduced by the enzyme benzoyl-CoA reductase (Fig. 1) (4). In R. palustris RCB100, the CoA ligase responsible for activating 3-CBA shows a higher specificity for chlorobenzoates, and this specificity is due to a single nucleotide change in aliA, which encodes an alicyclic acid CoA ligase (5). This change results in a variant of AliA with a serine instead of a threonine found in other R. palustris strains at amino acid 208 (T208S). AliAT208S has 10-fold more activity with 3-CBA than AliA from R. palustris CGA009 (5). After activation by a CoA ligase, 3-chlorobenzoyl-CoA is dearomatized by benzoyl-CoA reductase leading to spontaneous elimination of the halogen and rearomatization to form benzoyl-CoA (6, 7). In addition to 3-chlorobenzoyl-CoA, benzoyl-CoA reductase can dehalogenate other halobenzoyl-CoA substrates like 4-fluorobenzoyl-CoA (Fig. 1) (6–8).
Fig 1.
Proposed pathways for benzoate, 3-CBA, and 4-fluoroacetate degradation in R. palustris. Although not shown, the 1,5-dienoyl-CoA formed by benzoyl-CoA reductase (BCR) is further metabolized to acetyl-CoA. Both 3-CBA and 4-flourobenzoate generate benzoyl-CoA after dehalogenation, which is then metabolized as shown in the top reaction.
Most strains of R. palustris, including the well-known strain CGA009, cannot use 3-CBA as a primary carbon source (9, 10). However, it has been observed that many of these strains can acquire the ability to metabolize 3-CBA within a relatively short period of time (11). This indicates that only one or a few genetic mutations may be necessary to enable 3-CBA metabolism. Presumably, any R. palustris strain containing the benzoyl-CoA degradation pathway would be capable of using 3-CBA if the aliA allele from RCB100 is introduced. However, introduction of the aliAT208S allele in R. palustris CGA009 is not sufficient to confer the ability to use 3-CBA as a sole carbon source (5) (Fig. S1). This suggests the presence of another unidentified factor that is required for 3-CBA metabolism in R. palustris RCB100.
To determine what this factor may be, R. palustris CGA009 was experimentally evolved to grow photoheterotrophically using 3-CBA as a sole source of carbon. Whole-genome sequencing of the evolved strain and R. palustris RCB100 led to the discovery of a large deletion that included badM, a transcriptional repressor of genes involved in anaerobic benzoate degradation. Deletion of badM is sufficient to allow R. palustris CGA009 to degrade 3-CBA, and introduction of the aliAT208S allele from R. palustris RCB100 into R. palustris CGA009 ΔbadM results in a strain with a faster growth rate on 3-CBA. We present a model in which RCB100 initially acquired a deletion that led to constitutive expression of CoA ligases and benzoyl-CoA reductase and then accumulated other mutations that enhanced its ability to metabolize 3-CBA.
RESULTS
Strains capable of degrading 3-CBA contain large deletions near the bad operon
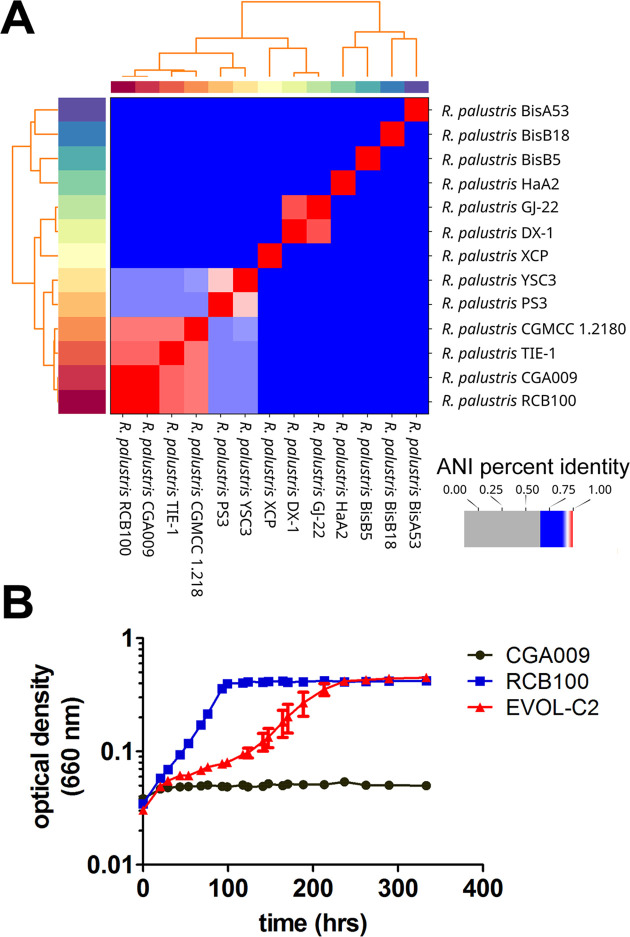
To understand what other factors may be required to enable R. palustris to use 3-CBA, a strain closely related to R. palustris RCB100 that is unable to metabolize 3-CBA was identified for use in adaptive evolution. The average nucleotide identity (ANI) of 13 R. palustris strains was analyzed with BLASTn. As shown in Fig. 2A, R. palustris CGA009 is the most closely related to R. palustris RCB100 with an ANI score of 0.99, corresponding to 99% identity between the two genomes. R. palustris CGA009 is unable to grow with 3-CBA as a carbon source, and we confirmed that introduction of aliAT208S into R. palustris CGA009 is not sufficient to enable R. palustris CGA009 to grow with 3-CBA as its sole carbon source (Fig. S1) (5).
Fig 2.
Isolation of a strain of R. palustris CGA009 capable of growing with 3-CBA as its sole carbon source. (A) As shown by ANI, the genomes of the closely related R. palustris RCB100 and R. palustris CGA009 are >95% identical. (B) Growth of R. palustris CGA009, R. palustris RCB100, and a strain of R. palustris CGA009 adapted to grow with 3-CBA (EVOL-C2) in minimal medium with 1 mM 3-CBA. Data are from three independent cultures, and error bars represent standard deviation.
To select an adapted strain that can grow with 3-CBA as the sole carbon source, CGA009 was grown in a medium with a mixture of benzoate and 3-CBA for several months in the light. After three successive transfers of enrichment cultures into a liquid medium with an increasing ratio of 3-CBA to benzoate, an evolved strain (EVOL-C2) derived from R. palustris CGA009 was isolated that was able to grow with 3-CBA as the sole carbon source. Unlike R. palustris CGA009, EVOL-C2 can use 3-CBA as a sole source of carbon, although it grew much more slowly than R. palustris RCB100 (Fig. 2B; Table 1).
TABLE 1.
Growth rates of R. palustris strains on aromatic compounds used in this study
| Doubling time (h)a | ||||
|---|---|---|---|---|
| Genotype | Benz | 3-CBA | 4-FlB | 4-HBA |
| RCB100 | 10 (0.3) | 35 (1.7) | 33 (0.9) | NGb |
| CGA009 | 10 (0.2) | NG | NG | 14 (0.4) |
| CGA009 EVOL-C2 | 9 (1.3) | 89 (5.1) | NDc | 35 (0.6) |
| CGA009 ΔbadM | 11 (0.6) | 178 (6.8) | 43 (2.6) | ND |
| CGA009 ΔbadM aliAT208S | 12 (2.2) | 39 (3.1) | 46 (0.9) | ND |
All doubling times are the averages from three independent experiments and are shown in hours with standard deviation in parentheses. Benz, 5.7 mM benzoate; 3-CBA, 3 mM 3-chlorobenzoate; 4-FlB, 5 mM 4-fluorobenzoate; and 4-HBA, 6 mM 4-hydroxybenzoate.
NG, no growth observed.
ND, not determined.
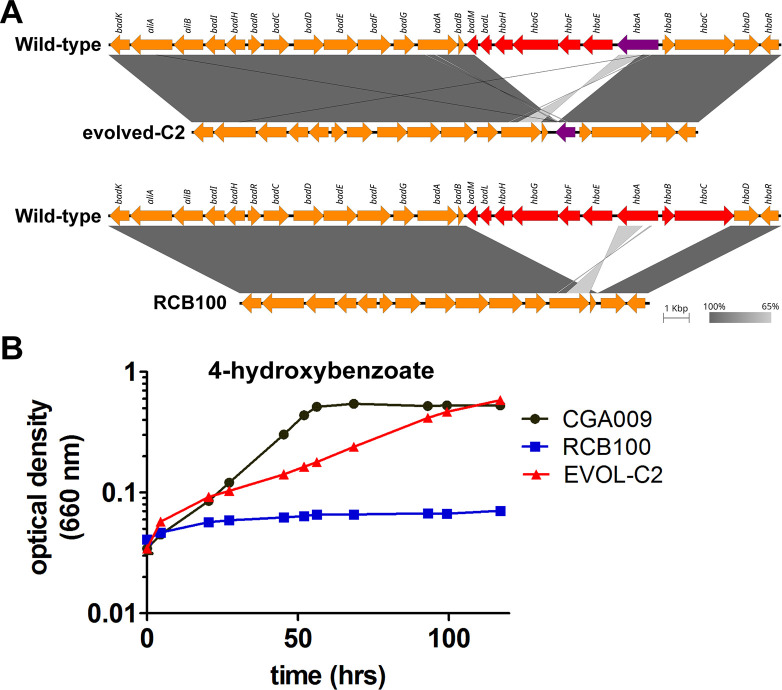
To determine the genetic basis of the evolved phenotype, the genome of EVOL-C2 was sequenced and compared to R. palustris CGA009. The sequenced genome of R. palustris RCB100 was also compared to R. palustris CGA009 (12). Unexpectedly, we found that both EVOL-C2 and R. palustris RCB100 contained large deletions near the bad operon, which encodes genes required for benzoyl-CoA degradation (Fig. 3A; Table S1). In EVOL-C2, the deleted region (6,521 bp) included partial deletion of badM and hbaA, and complete deletion of badL, hbaH, hbaG, hbaF, and hbaE. R. palustris RCB100 also contained a similar but significantly larger (10,387 bp) deletion that encompasses badM through hbaC (Fig. 3A; Table S1). This deletion was the only genetic change that was shared between EVOL-C2 and R. palustris RCB100 (Table S1).
Fig 3.
R. palustris RCB100 and R. palustris EVOL-C2 have large deletions near the bad operon. (A) Comparison of strains CGA009 and EVOL-C2 (top) and comparison of CGA009 with RCB100 (bottom). Arrows indicate genes and their direction with the gene names listed on top of the arrows. The deleted genes are shown in red whereas partially deleted genes are shown in purple. The shaded gray gradient represents the degree of homology between the compared regions. The plots were generated with Easyfig using the BLASTn algorithm. (B) Growth of R. palustris CGA009, R. palustris RCB100, and R. palustris CGA009 EVOL-C2 in minimal medium with 6 mM 4-HBA. Data are from three independent cultures, and error bars represent standard deviation.
The hba genes are associated with degradation of 4-hydroxybenzoate (4-HBA). Genes hbaHGFE encode a predicted ABC transport system for 4-HBA (4). Like benzoate, 4-HBA is activated by a 4-HBA-CoA ligase encoded by hbaA, and hbaBC encodes components of 4-HBA-CoA reductase that reductively removes the hydroxyl group from 4-HBA-CoA to yield benzoyl-CoA. Deletion of these genes would be expected to impact the ability of EVOL-C2 and R. palustris RCB100 to grow with 4-HBA as the sole carbon source. As shown in Fig. 3B and Table 1, EVOL-C2 grew more slowly on 4-HBA compared to its parent strain, R. palustris CGA009, and R. palustris RCB100 was unable to grow with 4-HBA as a sole carbon source, confirming the presence of deletions that disrupt 4-HBA metabolism.
Deletion of the transcriptional repressor badM in R. palustris CGA009 is sufficient to enable degradation of 3-CBA
Two genes that play a role in the regulation of the bad operon, badL and badM, were also deleted in EVOL-C2 and R. palustris RCB100 (Fig. 3A). The bad operon encodes the benzoyl-CoA ligase, BadA; a ferredoxin, BadB; and the subunits for benzoyl-CoA reductase, BadDEFG. BadM is a transcriptional repressor of the bad operon, and BadM bound to acetamidobenzoates leads to derepression of the bad operon (13–15). In a badM mutant, the bad operon is constitutively expressed (14). We hypothesized that constitutive expression of the bad operon in EVOL-C2 and R. palustris RCB100 may be sufficient to enable these strains to use 3-CBA as a sole carbon source, and the ability of R. palustris CGA009 ΔbadM to grow photoheterotrophically with 3-CBA as a carbon source was tested. As shown in Fig. 4A and B; Table 1, wild-type R. palustris CGA009 and R. palustris CGA009 ΔbadM grow at similar rates when benzoate is provided as a carbon source, but only R. palustris CGA009 ΔbadM can grow when 3-CBA is provided as a carbon source. This indicates that constitutive expression of the bad operon is sufficient for R. palustris to grow on 3-CBA. However, R. palustris CGA009 ΔbadM still grew slower than RCB100 or EVOL-C2 (Table 1).
Fig 4.
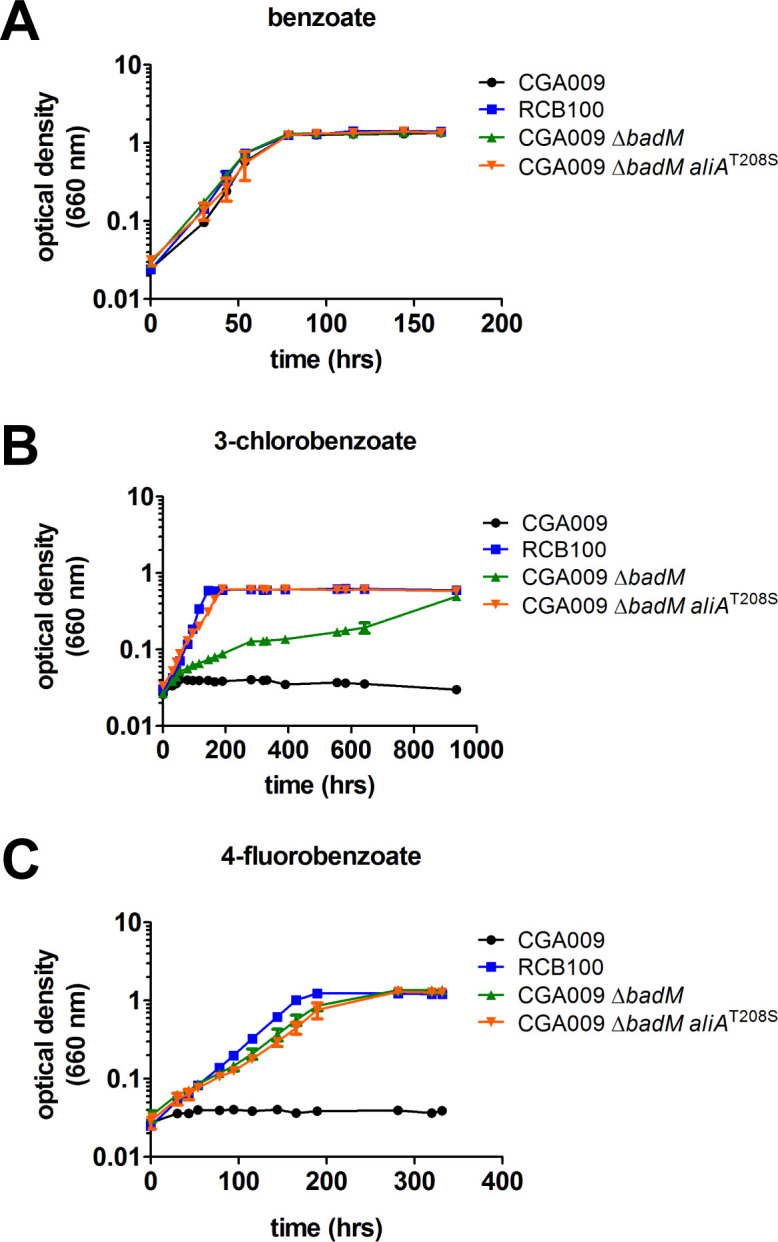
Deletion of badM enables growth of R. palustris on 3-CBA and 4-fluorobenzoate, and aliAT208S enhances growth of a badM mutant on 3-CBA. Growth of R. palustris strains CGA009, RCB100, CGA009 with a deletion in badM (CGA009 ΔbadM), and CGA009 ΔbadM encoding the allele from RCB100 (CGA009 ΔbadM aliAT208S) in (A) minimal medium supplemented with 5.7 mM benzoate; (B) minimal medium supplemented with 3 mM 3-CBA; and (C) minimal medium supplemented with 5 mM 4-fluorobenzoate. Data are from three independent cultures, and error bars represent standard deviation.
EVOL-C2 also contained a single mutation in the transcription regulator badR, which resulted in a protein variant with arginine 80 substituted for a leucine. BadR represses expression of the cyclohexanecarboxylate operon, which includes aliA (badHI-aliAB-badK) (16). Since a variant of AliA plays a role in 3-CBA metabolism in RCB100, we wanted to determine if inactivation of badR plays a role in 3-CBA metabolism. To test this, R. palustris CGA009 ΔbadM, R. palustris CGA009 ΔbadR, and R. palustris CGA009 ΔbadM badR were grown photoheterotrophically with 3-CBA as a carbon source. We found that R. palustris CGA009 ΔbadM and R. palustris CGA009 ΔbadM badR grew at similar rates on 3-CBA (Fig. S2; Table S2). However, R. palustris CGA009 ΔbadR was unable to grow on 3-CBA as a sole source of carbon (Fig. S2). This indicates that deletion of badR even when in combination with a deletion in badM does not enhance degradation of 3-CBA by R. palustris CGA009.
To determine if deletion of badM was sufficient to enable growth on other halogenated aromatic compounds, growth rates on 4-fluorobenzoate were also determined. Unlike 3-CBA, which requires a CoA ligase that has more activity with 3-CBA, 4-fluorobenzoate can be activated by benzoyl-CoA ligase at similar rates to benzoate, forming 4-fluorobenzoyl-CoA. 4-fluorobenzoyl-CoA is then reductively dehalogenated by benzoyl-CoA reductase to form benzoyl-CoA (Fig. 1) (8). We hypothesized that R. palustris CGA009 ΔbadM and R. palustris RCB100 would be able to grow with 4-fluorobenzoate as a carbon source since these strains constitutively express benzoyl-CoA ligase and benzoyl-CoA reductase. As shown in Fig. 4C, R. palustris CGA009 is unable to use 4-fluorobenzoate as growth substrate, but R. palustris CGA009 ΔbadM and RCB100 were able to grow. This indicates that deletion of badM enables CGA009 and RCB100 to metabolize another halogenated aromatic compound.
Introduction of aliAT208S in R. palustris CGA009 ΔbadM improves growth with 3-CBA but not 4-fluorobenzoate
Although R. palustris CGA009 ΔbadM can grow when 3-CBA is provided, it grows slowly compared to RCB100 (Fig. 4B; Table 1). This is likely because, in addition to constitutive expression of benzoyl-CoA reductase, RCB100 also expresses a CoA ligase (AliAT208S) that has 10 times more activity with 3-CBA than AliA from CGA009 (5). To test this, the aliAT208S allele was introduced into R. palustris CGA009 ΔbadM. As shown in Fig. 4A and Table 1, there is no difference in growth between these strains when they are provided with benzoate as a carbon source. However, when provided with 3-CBA as a carbon source, R. palustris CGA009 ΔbadM aliAT208S cells grew significantly faster than R. palustris CGA009 ΔbadM (Fig. 4B; Table 1). Both R. palustris CGA009 ΔbadM and R. palustris CGA009 ΔbadM aliAT208S grew at similar rates on 4-fluorobenzoate, confirming that AliAT208S is only required for 3-CBA degradation (Fig. 4C; Table 1). This indicates that deletion of badM and aliAT208S is sufficient to reproduce the ability of RCB100 to degrade 3-CBA in another R. palustris strain.
DISCUSSION
Using adaptive laboratory evolution, we reconstructed the evolutionary events that enabled a natural isolate to metabolize 3-CBA. We found that the environmental isolate R. palustris RCB100 and a laboratory strain evolved to metabolize 3-CBA, R. palustris EVOL-C2, contain deletions that encompass badLM and hba genes (Fig. 3). The genes in this region (badLM-hbaAEFGH) are encoded on the lagging strand (Fig. 3), which are sites of head-on, replication-transcription collisions. Such head-on collisions have been shown to promote the generation of duplications and deletions in bacteria, and this may explain why large deletions were found at this locus in both R. palustris EVOL-C2 and RCB100 (17). However, we found that deletion of a single gene in this region, badM, is sufficient to enable R. palustris to use 3-CBA as a carbon source, indicating that derepression of the bad genes is required for R. palustris to degrade 3-CBA. This is consistent with the observation that R. palustris strains can co-metabolize 3-CBA and benzoate despite being unable to metabolize 3-CBA as the sole source of carbon (3, 9, 10).
The fact that R. palustris CGA009 ΔbadM can grow on 3-CBA implies that native CoA ligases have sufficient activity with 3-CBA to enable 3-CBA metabolism. This is surprising because the ability to degrade 3-CBA is thought to be restricted to bacterial strains encoding a CoA ligase with increased affinity for 3-CBA (6, 18). R. palustris CGA009 encodes three CoA ligases involved in anaerobic aromatic compound degradation: benzoyl-CoA ligase, BadA; 4-hydroxybenzoyl-CoA ligase, HbaA; and cyclohexanecarboxylate-CoA ligase, AliA. HbaA is likely not involved since hbaA is deleted in R. palustris EVOL-C2 and R. palustris RCB100. However, both badA and aliA are intact in R. palustris EVOL-C2, and BadA and AliA have been shown to have some activity against 3-CBA in vitro (5, 19). BadA and AliA are also expressed in a badM mutant grown with 3-CBA as a carbon source. Deletion of badM leads to constitutive expression of BadA, and BadA activity is even higher in a badM mutant compared to induced wild-type cells (14, 20). AliA activity has also been observed in a badA mutant grown with benzoate, indicating that both AliA and BadA are present in cells carrying out benzoyl-CoA degradation (21). This suggests that there is enough promiscuous activity by BadA and AliA in a badM mutant to enable growth on 3-CBA, and a CoA ligase specific to 3-CBA is not a requirement per se.
Together, these data point toward a model wherein R. palustris RCB100 first acquired a deletion that inactivated badM, leading to constitutive expression of BadA, AliA, and benzoyl-CoA reductase. While the promiscuous activity of the CoA ligases is adequate for enabling 3-CBA degradation, CoA activation still represents a potential rate-limiting step, creating selective pressure to acquire a mutation that enhances CoA ligase activity with 3-CBA. This aligns with the presence of AliAT208S, a CoA ligase that has higher activity with 3-CBA, in R. palustris RCB100. Introduction of the aliA allele from R. palustris RCB100 into R. palustris CGA009 ΔbadM significantly increased growth on 3-CBA, resulting in a growth rate similar to R. palustris RCB100. This establishes these two mutations as sufficient to reconstruct the genetic requirements in R. palustris RCB100 for 3-CBA degradation.
Our findings also indicate that R. palustris is unable to sense halogenated aromatic compounds since deletion of badM is required for R. palustris to grow on these compounds. Repression of BadM is relieved when it is bound to acetamidobenzoates, which are produced by the acetylation of aminobenzoates by BadL (13). When grown with 3-CBA or 4-fluorobenzoate, accumulation of acetaminobenzoates must not occur in R. palustris, and BadM repression is not relieved. It is difficult to speculate why BadM repression is not relieved in the presence of halogenated compounds as it is still unclear why acetaminobenzoates accumulate when cells are grown with benzoate. The growth medium for R. palustris contains 4-aminobenzoate (11 mM) for folate biosynthesis, and 4-aminobenzoate is a substrate for BadL (13, 22). Why derepression of BadM only occurs in the presence of aromatic compounds like benzoate but not in the presence of halogenated aromatic compounds in a medium containing 4-aminobenzoate is unclear.
This is the first evolutionary reconstruction of an environmental isolate capable of using halogenated aromatic compounds. Adaptive laboratory evolution has provided numerous examples of how regulatory mutations or variants of a promiscuous enzyme can enable degradation of man-made compounds (23–31). Here, we show that both bottlenecks, gene regulation and substrate specificity of a promiscuous enzyme, were overcome to enable efficient degradation of 3-CBA.
MATERIAL AND METHODS
Bacterial strains and culture conditions
All bacterial strains used in the current study are listed in Table 2. R. palustris strains were grown under anoxic conditions in 10 mL of minimal mineral photosynthetic medium (PM) in sealed Hungate tubes (22, 32). Carbon substrates from anoxic, sterile stock solutions were added after autoclaving. Carbon sources were added to the following final concentrations where indicated: 10 mM succinate, 20 mM acetate, 5.7 mM benzoate, 1 to 3 mM 3-chlorobenzoate, 5 mM 4-fluorobenzoate, and 6 mM 4-HBA. All liquid cultures were grown at 30°C and provided light (30 µmol photons/m2/s) from a 60 W halogen light bulb (General Electric). Cultures were initially grown in PM with succinate and then diluted in PM with the stated carbon source for growth analysis. Optical density was determined at 660 nm using a Genesys 50 UV-visible spectrophotometer. Escherichia coli strains were grown in lysogeny broth medium at 37°C. When necessary, R. palustris and E. coli cultures were supplemented with gentamicin (Gm) at concentrations of 100 and 20 µg/mL, respectively.
TABLE 2.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Genotype, description, or primer sequence (5’ to 3’) | Reference, origin, or description |
|---|---|---|
| R. palustris strains | ||
| CGA009 | Wild-type strain; spontaneous CmR derivative of CGA001 | (32) |
| RCB100 | Isolated on 3-CBA near Cascadilla Creek, Ithaca, NY | (3) |
| EVOL-C2 | CGA009 adapted to grow on 3-CBA; genotype in Table S1 | This study |
| CGA131 | CGA009 ΔbadM; CGA009 containing an in-frame deletion of badM (rpa0663) | (14) |
| CGA609a | CGA009 ΔbadR; CGA009 containing an in-frame deletion of badR (rpa0655) | (16) |
| CGA139 | CGA009 ΔbadM badR; CGA009 containing an in-frame deletion of badR and badM | Gift from Caroline Harwood |
| aliAT208S | CGA009 encoding the aliA allele from RCB100 | This study |
| ΔbadM aliAT208S | CGA131 encoding the aliA allele from RCB100 | This study |
| E. coli strains | ||
| S17-1 | thi pro hdsR hdsM+ recA; chromosomal insertion of RP4-2 (Tc::Mu Km::Tn7) | (3) |
| Plasmids | ||
| pJQ200SK | GmR, sacB; mobilizable suicide vector | (4) |
| pJQ-aliAT208S | GmR; used for allelic exchange of the aliA allele from R. palustris RCB100 | This study |
| Primers | ||
| pJQ200SK-aliAF | GCAGCAGAACGGCGTCGAACTCCAT-CTCCAGCTTTTGTTCCCTTTAGTGAGGG | This study |
| pJQ200SK-badKR | GATGTGCTCAACGCGCTCAACGATG-GCTCTAGAACTAGTGGATCCCCCGG | This study |
| aliAF | TCACTAAAGGGAACAAAAGCTGGAG-ATGGAGTTCGACGCCGTTCTGC | This study |
| badKR | CCGGGGGATCCACTAGTTCTAGAGC-CATCGTTGAGCGCGTTGAGCACATC | This study |
Experimental evolution of R. palustris CGA009 and isolation of evolved R. palustris EVOL-C2
A single colony of R. palustris CGA009 was picked up with a sterilized toothpick and resuspended in 200 µL of PM medium in a sterile Eppendorf tube before the suspended cells were introduced into sealed Hungate tubes containing 10 mL of PM supplemented with acetate. To adapt R. palustris CGA009 to grow on 3-CBA, cells were inoculated into four tubes containing PM supplemented with 5.7 mM benzoate and 1 mM 3-CBA as described in reference (11). Cells were grown for 3 weeks under light, before they were transferred to PM medium supplemented with 2.85 mM benzoate and 1.5 mM 3-CBA. Cells were grown in the light for 5 weeks and then transferred into PM medium supplemented with 1.5 mM benzoate and 2.5 mM 3-CBA. Cells were grown in the light for 5 weeks before they were transferred to PM medium with 0.5 mM benzoate and 3.5 mM 3-CBA. Cells were grown in the light for 5 weeks and transferred into PM supplemented with 3.5 mM 3-CBA. After 9 weeks, a single replicate grew, and 100 µL of cells were spread on plates containing PM with 5.7 mM benzoate. After 3 weeks of incubation, 13 random colonies were selected and grown in Hungate tubes with PM supplemented with 1 mM 3-CBA. EVOL-C2 was one of the 13 colonies that consistently grew in PM supplemented with 1 mM 3-CBA. After repeated confirmation of growth on liquid PM supplemented with 1 mM 3-CBA, 100 µL of EVOL-C2 was transferred to PM liquid medium supplemented with 20 mM acetate and 0.1% yeast extract. Once fully grown, a glycerol stock was prepared and stored at −80°C, and genomic DNA was extracted for sequencing as described below.
Genomic DNA extraction and whole genome sequence analyses
Genomic DNA was extracted as described previously (33). Briefly, R. palustris EVOL-C2 was grown in 10 mL PM medium with 20 mL acetate. Freshly grown R. palustris cells were harvested with centrifugation at 12,000 rpm for 10 minutes, and genomic DNA was extracted from cell pellets as described (6, 8). Both Illumina and Nanopore libraries were prepared at Microbial Genome Sequencing Center (MiGS; https://www.migscenter.com/), using the workflows reported previously (6, 9), before sequencing on NextSeq 550 and MinION v.9.4.1 flow cell, respectively. Base calling was performed using guppy v.4.2.2 with default parameters + effbaf8. The quality control and adapter trimming for Illumina reads were performed with bcl2fastq v.2.20.0.445 (10), whereas porechop v.0.2.3_seqan2.1.1 (11) was used for Oxford Nanopore Technology (ONT) sequencing reads. The Illumina data set comprised 5,063,564 reads pairs and 1,632,037,238 bases whereas Nanopore sequencing produced 116,959 reads and 303,624,697 bases.
A hybrid genome assembly was generated from Illumina and ONT reads with Unicycler v.0.4.8 (12), and QUAST v.5.0.2 (13) was used to obtain genome assembly statistics. Genome assembly resulted in two circular contigs of 5,452,700 bp and 8,427 bp that represented chromosome and plasmid, respectively. The GC (%) nucleotide content of the assembled genome was 65.03, whereas the length of the genome was 5,461,127 bp. The assembly annotation was performed with NCBI Prokaryotic Genome Annotation Pipeline. To predict mutations in strains EVOL-C2 and RCB100 in comparison to strain CGA009, we used breseq v.0.36.0 (34).
We used PYANI v.0.2.10 [(35); https://github.com/widdowquinn/pyani] to calculate average nucleotide identity among 13 selected R. palustris genomes based on BLASTN+ (ANIb) method. Easyfig v.2.2.2 (36) was used to generate genome comparison visualization plots.
Genetic manipulation of R. palustris
A construct containing the aliA locus from R. palustris RCB100 was constructed for allelic exchange using the primers shown in Table 2. Genomic DNA purified from R. palustris RCB100 was used as a template for PCR amplification using Phusion High-Fidelity DNA polymerase (New England Biolabs) with primers aliAF and badKR (Table 2). Using homologous recombination as described in reference (37), the resulting product was incorporated into linearized pJQ200SK generated from PCR amplification with primers pJQ200SK-aliAF and pJQ200SK-badKR (Table 2). The resulting plasmid was mobilized into R. palustris CGA009 by conjugation with E. coli S17-1, and double-crossover events for allelic exchange were accomplished using a previously described selection and screening strategy (38). Replacement of aliA in R. palustris CGA009 with the aliA locus from R. palustris RCB100 was verified using PCR followed by Sanger sequencing (Azenta Life Sciences, Burlington, MA).
ACKNOWLEDGMENTS
We thank Caroline Harwood and Yasuhiro Oda for providing mutant strains of R. palustris used in this study. We also thank Nathan Lewis, Nicholas Haas, and Jack Reddan for valuable discussions during the course of this study.
This work was supported through the Biocatalysis Initiative grant awarded to K.R.F. by the BioTechnology Institute, University of Minnesota.
Contributor Information
Kathryn R. Fixen, Email: kfixen@umn.edu.
Ning-Yi Zhou, Shanghai Jiao Tong University, Shanghai, China.
DATA AVAILABILITY
The R. palustris RCB100 genome sequence is available at NCBI under the accession number GCF_016584445.1. The R. palustris CGA009 genome sequence is available under the accession number GCF_000195775.2, and the R. palustris CGA009 EVOL-C2 genome sequence is available under the accession number GCA_031600295.1. Raw sequencing data for EVOL-C2 is available at NCBI under the accession numbers SRR17562209 (Oxford Nanopore) and SRR17562208 (Illumina). The accession number for the EVOL-C2 sequencing BioProject is PRJNA796295, and the Biosample accession number is SAMN24839554.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aem.02104-23.
Supplemental material.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Olaniran AO, Igbinosa EO. 2011. Chlorophenols and other related derivatives of environmental concern: properties, distribution and microbial degradation processes. Chemosphere 83:1297–1306. doi: 10.1016/j.chemosphere.2011.04.009 [DOI] [PubMed] [Google Scholar]
- 2. Pimviriyakul P, Wongnate T, Tinikul R, Chaiyen P. 2020. Microbial degradation of halogenated aromatics. Microb Biotechnol 13:67–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Egland PG, Gibson J, Harwood CS. 2001. Reductive, coenzyme A-mediated pathway for 3-chlorobenzoate degradation in the phototrophic bacterium Rhodopseudomonas palustris. Appl Environ Microbiol 67:1396–1399. doi: 10.1128/AEM.67.3.1396-1399.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harwood CS. 2009. Degradation of aromatic compounds in purple nonsulfur bacteria, p. 577–594. In The Purple Phototrophic Bacteria. Springer International Publishing. [Google Scholar]
- 5. Samanta SK, Harwood CS. 2005. Use of the Rhodopseudomonas palustris genome sequence to identify a single amino acid that contributes to the activity of a coenzyme A ligase with chlorinated substrates. Mol Microbiol 55:1151–1159. doi: 10.1111/j.1365-2958.2004.04452.x [DOI] [PubMed] [Google Scholar]
- 6. Kuntze K, Kiefer P, Baumann S, Seifert J, von Bergen M, Vorholt JA, Boll M. 2011. Enzymes involved in the anaerobic degradation of meta-substituted halobenzoates. Mol Microbiol 82:758–769. doi: 10.1111/j.1365-2958.2011.07856.x [DOI] [PubMed] [Google Scholar]
- 7. Tiedt O, Fuchs J, Eisenreich W, Boll M. 2018. A catalytically versatile benzoyl-CoA reductase, key enzyme in the degradation of methyl- and halobenzoates in denitrifying bacteria. J Biol Chem 293:10264–10274. doi: 10.1074/jbc.RA118.003329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tiedt Oliver, Mergelsberg M, Boll K, Müller M, Adrian L, Jehmlich N, von Bergen M, Boll M. 2016. ATP-dependent C–F bond cleavage allows the complete degradation of 4-fluoroaromatics without oxygen. mBio 7:e00990-16. doi: 10.1128/mBio.00990-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Oda Y, Meijer WG, Gibson JL, Gottschal JC, Forney LJ. 2004. Analysis of diversity among 3-Chlorobenzoate-degrading strains of Rhodopseudomonas palustris. Microb Ecol 47:68–79. doi: 10.1007/s00248-003-1028-5 [DOI] [PubMed] [Google Scholar]
- 10. Kamal VS, Wyndham RC. 1990. Anaerobic phototrophic metabolism of 3-chlorobenzoate by Rhodopseudomonas palustris WS17. Appl Environ Microbiol 56:3871–3873. doi: 10.1128/aem.56.12.3871-3873.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oda Y, Vries YP, Forney LJ, Gottschal JC. 2001. Acquisition of the ability for Rhodopseudomonas palustris to degrade chlorinated Benzoic acids as the sole carbon source. FEMS Microbiol Ecol 38:133–139. doi: 10.1111/j.1574-6941.2001.tb00891.x [DOI] [Google Scholar]
- 12. Haq IU, Fixen KR, Maresca JA. 2021. Complete genome sequence of Rhodopseudomonas palustris RCB100, an anoxygenic phototroph that degrades 3-chlorobenzoate. Microbiol Resour Announc 10. doi: 10.1128/MRA.00043-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. VanDrisse CM, Escalante-Semerena JC. 2018. Small-molecule acetylation controls the degradation of benzoate and photosynthesis in Rhodopseudomonas palustris. mBio 9:e01895-18. doi: 10.1128/mBio.01895-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peres CM, Harwood CS. 2006. BadM is a transcriptional Repressor and one of three regulators that control benzoyl coenzyme A reductase gene expression in Rhodopseudomonas palustris. J Bacteriol 188:8662–8665. doi: 10.1128/JB.01312-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hirakawa H, Schaefer AL, Greenberg EP, Harwood CS. 2012. Anaerobic p-coumarate degradation by Rhodopseudomonas palustris and identification of CouR, a MarR repressor protein that binds p-coumaroyl coenzyme A. J Bacteriol 194:1960–1967. doi: 10.1128/JB.06817-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hirakawa H, Hirakawa Y, Greenberg EP, Harwood CS. 2015. BadR and BadM proteins transcriptionally regulate two operons needed for anaerobic benzoate degradation by Rhodopseudomonas palustris. Appl Environ Microbiol 81:4253–4262. doi: 10.1128/AEM.00377-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sankar TS, Wastuwidyaningtyas BD, Dong Y, Lewis SA, Wang JD. 2016. The nature of mutations induced by replication–transcription collisions. Nature 535:178–181. doi: 10.1038/nature18316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Louie TS, Pavlik EJ, Häggblom MM. 2021. Genome analysis of Thauera chlorobenzoica strain 3CB-1T, a halobenzoate-degrading bacterium isolated from aquatic sediment. Arch Microbiol 203:5095–5104. doi: 10.1007/s00203-021-02550-w [DOI] [PubMed] [Google Scholar]
- 19. Harwood CS, Gibson J. 1988. Anaerobic and aerobic metabolism of diverse aromatic compounds by the photosynthetic bacterium Rhodopseudomonas palustris. Appl Environ Microbiol 54:712–717. doi: 10.1128/aem.54.3.712-717.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Geissler JF, Harwood CS, Gibson J. 1988. Purification and properties of benzoate-coenzyme A ligase, a Rhodopseudomonas palustris enzyme involved in the anaerobic degradation of benzoate. J Bacteriol 170:1709–1714. doi: 10.1128/jb.170.4.1709-1714.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Egland PG, Gibson J, Harwood CS. 1995. Benzoate-coenzyme A ligase, encoded by badA, is one of three ligases able to catalyze benzoyl-coenzyme A formation during anaerobic growth of Rhodopseudomonas palustris on benzoate. J Bacteriol 177:6545–6551. doi: 10.1128/jb.177.22.6545-6551.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lewis NM, Sarne A, Fixen KR. 2023. Evolving a new electron transfer pathway for nitrogen fixation uncovers an electron bifurcating-like enzyme involved in anaerobic aromatic compound degradation. mBio 14:e0288122. doi: 10.1128/mbio.02881-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Deng S-K, Zhang W-M, Wang J-P, Gao Y-Z, Xu Y, Zhou N-Y. 2019. Single point Mutation in the transcriptional regulator PnpR renders Pseudomonas sp. strain WBC-3 capable of utilizing 2-chloro-4-nitrophenol. Inter Biod & Biod 143:104732. doi: 10.1016/j.ibiod.2019.104732 [DOI] [Google Scholar]
- 24. Ng LC, Poh CL, Shingler V. 1995. Aromatic effector activation of the ntrc-like transcriptional regulator PhhR limits the catabolic potential of the (methyl)phenol degradative pathway it controls. J Bacteriol 177:1485–1490. doi: 10.1128/jb.177.6.1485-1490.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wise AA, Kuske CR. 2000. Generation of novel bacterial regulatory proteins that detect priority pollutant phenols. Appl Environ Microbiol 66:163–169. doi: 10.1128/AEM.66.1.163-169.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Beggah S, Vogne C, Zenaro E, Van Der Meer JR. 2008. Mutant HbpR transcription activator isolation for 2-chlorobiphenyl via green fluorescent protein-based flow cytometry and cell sorting. Microb Biotechnol 1:68–78. doi: 10.1111/j.1751-7915.2007.00008.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ju K-S, Parales JV, Parales RE. 2009. Reconstructing the evolutionary history of nitrotoluene detection in the transcriptional regulator NtdR. Mol Microbiol 74:826–843. doi: 10.1111/j.1365-2958.2009.06904.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wackett LP. 2004. Evolution of enzymes for the metabolism of new chemical inputs into the environment. J Biol Chem 279:41259–41262. doi: 10.1074/jbc.R400014200 [DOI] [PubMed] [Google Scholar]
- 29. Aharoni A, Gaidukov L, Khersonsky O, McQ Gould S, Roodveldt C, Tawfik DS. 2005. “The “evolvability” of promiscuous protein functions”. Nat Genet 37:73–76. doi: 10.1038/ng1482 [DOI] [PubMed] [Google Scholar]
- 30. Mahan KM, Penrod JT, Ju K-S, Al Kass N, Tan WA, Truong R, Parales JV, Parales RE. 2015. Selection for growth on 3-nitrotoluene by 2-nitrotoluene-utilizing Acidovorax sp. strain JS42 identifies nitroarene dioxygenases with altered specificities. Appl Environ Microbiol 81:309–319. doi: 10.1128/AEM.02772-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Copley SD. 2009. Evolution of efficient pathways for degradation of anthropogenic chemicals. Nat Chem Biol 5:559–566. doi: 10.1038/nchembio.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim M-K, Harwood CS. 1991. Regulation of benzoate-CoA Ligase in Rhodopseudomonas palustris. FEMS Microbiol Lett 83:199–203. doi: 10.1111/j.1574-6968.1991.tb04440.x-i1 [DOI] [Google Scholar]
- 33. Oda Y, Larimer FW, Chain PSG, Malfatti S, Shin MV, Vergez LM, Hauser L, Land ML, Braatsch S, Beatty JT, Pelletier DA, Schaefer AL, Harwood CS. 2008. Multiple genome sequences reveal adaptations of a phototrophic bacterium to sediment microenvironments. Proc Natl Acad Sci U S A 105:18543–18548. doi: 10.1073/pnas.0809160105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Deatherage DE, Barrick JE. 2014. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using Breseq, p 165–188. In Sun L, Shou W (ed), Engineering and analyzing Multicellular systems. Springer, New York. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. 2016. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods 8:12–24. doi: 10.1039/C5AY02550H [DOI] [Google Scholar]
- 36. Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kostylev M, Otwell AE, Richardson RE, Suzuki Y. 2015. Cloning should be simple: Escherichia coli Dh5α-mediated assembly of multiple DNA fragments with short end homologies. PLoS ONE 10:e0137466. doi: 10.1371/journal.pone.0137466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rey FE, Harwood CS. 2010. FixK, a global regulator of microaerobic growth, controls photosynthesis in Rhodopseudomonas Palustris. Mol Microbiol 75:1007–1020. doi: 10.1111/j.1365-2958.2009.07037.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material.
Data Availability Statement
The R. palustris RCB100 genome sequence is available at NCBI under the accession number GCF_016584445.1. The R. palustris CGA009 genome sequence is available under the accession number GCF_000195775.2, and the R. palustris CGA009 EVOL-C2 genome sequence is available under the accession number GCA_031600295.1. Raw sequencing data for EVOL-C2 is available at NCBI under the accession numbers SRR17562209 (Oxford Nanopore) and SRR17562208 (Illumina). The accession number for the EVOL-C2 sequencing BioProject is PRJNA796295, and the Biosample accession number is SAMN24839554.