ABSTRACT
The acyl-homoserine lactones (AHLs)-mediated LuxI/LuxR quorum sensing (QS) system orchestrates diverse bacterial behaviors in response to changes in population density. The role of the BjaI/BjaR1 QS system in Bradyrhizobium diazoefficiens USDA 110, which shares homology with LuxI/LuxR, remains elusive during symbiotic interaction with soybean. Here this genetic system in wild-type (WT) bacteria residing inside nodules exhibited significantly reduced activity compared to free-living cells, potentially attributed to soybean-mediated suppression. The deletion mutant strain ΔbjaR1 showed significantly enhanced nodulation induction and nitrogen fixation ability. Nevertheless, its ultimate symbiotic outcome (plant dry weight) in soybeans was compromised. Furthermore, comparative analysis of the transcriptome, proteome, and promoter activity revealed that the inactivation of BjaR1 systematically activated and inhibited genomic modules associated with nodulation and nitrogen metabolism. The former appeared to be linked to a significant decrease in the expression of NodD2, a key cell-density-dependent repressor of nodulation genes, while the latter conferred bacterial growth and nitrogen fixation insensitivity to environmental nitrogen. In addition, BjaR1 exerted a positive influence on the transcription of multiple genes involved in a so-called central intermediate metabolism within the nodule. In conclusion, our findings highlight the crucial role of the BjaI/BjaR1 QS circuit in positively regulating bacterial nitrogen metabolism and emphasize the significance of the soybean-mediated suppression of this genetic system for promoting efficient symbiotic nitrogen fixation by B. diazoefficiens.
IMPORTANCE
The present study demonstrates, for the first time, that the BjaI/BjaR1 QS system of Bradyrhizobium diazoefficiens has a significant impact on its nodulation and nitrogen fixation capability in soybean by positively regulating NodD2 expression and bacterial nitrogen metabolism. Moreover, it provides novel insights into the importance of suppressing the activity of this QS circuit by the soybean host plant in establishing an efficient mutual relationship between the two symbiotic partners. This research expands our understanding of legumes’ role in modulating symbiotic nitrogen fixation through rhizobial QS-mediated metabolic functioning, thereby deepening our comprehension of symbiotic coevolution theory. In addition, these findings may hold great promise for developing quorum quenching technology in agriculture.
KEYWORDS: Bradyrhizobium diazoefficiens, LuxI/LuxR, quorum sensing, BjaR1, symbiosis, nitrogen fixation, metabolic modeling, soybean
INTRODUCTION
Quorum sensing (QS) is a coordinated gene regulation system that governs the biological behavior of bacterial communities, rapidly responds to changes in the surrounding microenvironment, and mediates bacteria’s interaction with their eukaryotic hosts (1). Bacteria secrete autoinducers (AIs), which bind to specific receptors and activate the transcription of target genes. The most extensively studied AIs are acyl-homoserine lactone (AHL or HSL) molecules containing conserved lactone rings linked to structurally diverse side chain groups. AHLs are synthesized by S-adenosylmethionine (SAM) and an acylated acyl carrier protein (acyl-ACP) from the LuxI family and can be recognized by signal receptors from the LuxR family (2, 3). AHLs-based canonical luxI/luxR circuits are widely distributed among gram-negative species and hold a central position in bacterial hierarchical regulatory networks. However, an increasing number of diverse families of QS signals with varying chemical structures, such as cyclic furanone compound, have been identified in numerous bacterial species, facilitating inter- and intra-species communication (4).
Rhizobia is a group of bacteria that can either exist freely in the soil or form a symbiotic association with legumes. The host plant secretes isoflavones, which stimulate the expression of nodule genes (nod, noe, or nol) in rhizobia to produce Nod factors (NFs). Subsequently, NFs trigger a complex series of genetic and physiological reactions in the host plant, ultimately leading to the development of specialized root organs known as nodules (5–7). Within these nodules, rhizobial bacteria differentiate into bacteroids and convert ambient N2 into NH3 for assimilation by the host plant. Bradyrhizobium diazoefficiens USDA 110 (formerly B. japonicum USDA 110) represents an extensively studied type strain of the novel species Bradyrhizobium diazoefficiens sp. nov., which serves as a crucial endosymbiont for soybean (8). In this strain, cell density negatively affects the induction of nod genes, and a unique cell density factor (CDF), bradyoxetin, mediates this process (9). Initially, it was believed that bradyoxetin’s structure resembled that of the siderophore mugineic acid, and its synthesis is regulated by Fe3+ (9); however, it was recently concluded that bradyoxetin must be chloramphenicol after reinterpreting the stated nuclear magnetic resonance spectroscopy data (10). At high cell densities, bradyoxetin progressively activates the synthesis of itself and its response regulators, NwsB, NolA, and NodD2. NodD2 eventually represses the common nodYABC gene expression and nodulation in the soybean plant (11, 12).
A set of luxI/luxR-type genes, designated blr1063/blr1062, was discovered as early as during B. diazoefficiens USDA 110 genome sequencing (13). Subsequent studies revealed that AHL-like autoinducer synthesis was prevalent in B. diazoefficiens and B. elkanii strains (14). Later, this pair of genes was named bjaI/bjaR1. The LuxI-type BjaI enzyme catalyzes the synthesis of isovaleryl-HSL (IV-HSL), which then binds to the LuxR-type BjaR1 regulator at extremely low concentrations (pM) (15). However, the symbiotic roles of this bacterial genetic system remain elusive. Specifically, it is unclear whether cross-talk exists between bradyoxetin and the isovaleryl-HSL-mediated QS signaling pathway, potentially impacting soybean nodulation. Increasing evidence has revealed that the HSLs-mediated QS system is crucial in maintaining bacterial metabolic homeostasis, extending beyond its involvement in transcriptional regulation (16). In the nonsymbiotic rice pathogen Burkholderia glumae, deficiency of luxI/luxR QS disrupts nutrient acquisition, leading to downregulation of glucose uptake, substrate-level oxidative phosphorylation, and de novo nucleotide biosynthesis (17). The ability of rhizobial bacteroids to fix nitrogen is tightly regulated by oxygen availability, metabolite transfer, and variations in the metabolism of both bacteria and their plant hosts (18, 19). Recent findings have demonstrated that nitrogen transport and assimilation in ex-planta B. diazoefficiens USDA110 bacteroids are governed by oxygen levels, L-malate concentration, and bacteroid density (20). Interestingly, the rhizosphere bacterial quorum sensing system has been found to significantly influence soil nitrogen mineralization (21), suggesting its potential role in modulating nitrogen metabolism within naturally occurring bacterial populations. Consequently, it is worth considering that bjaI/bjaR1 QS deficiency may affect bacteroid metabolic function and subsequent nitrogen fixation.
This study aimed to elucidate the symbiotic role of the BjaR1 regulator. We have demonstrated that the BjaR1-mediated BjaI/BjaR1 QS system exhibits activity at the symbiotic phase of bacteria (inside the soybean nodule) and, as a global activator, BjaR1 exerts positive influences on nodulation and nitrogen fixation by regulating nodD2 expression and bacterial nitrogen metabolic functioning, particularly nitrogen metabolism. Furthermore, we propose a hypothetical model suggesting that host-mediated suppression of this bacterial genetic system is crucial for efficient symbiotic nitrogen fixation, thereby playing a significant role in the mutualistic evolution between Bradyrhizobium diazoefficiens and soybean.
RESULTS
Soybean host plants suppressed the activity of the wild allelic form of the bjaI/bjaR1 circuit
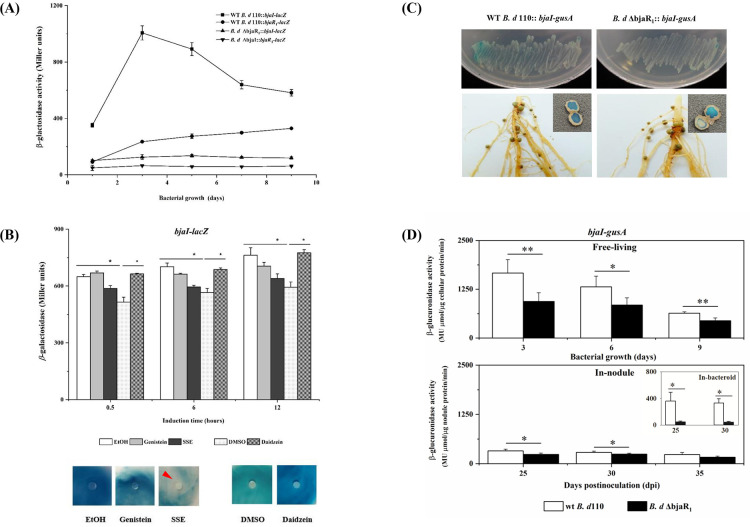
To investigate the functional redundancy of the BjaI/BjaR1 system and its similarity to the canonical LuxI/LuxR QS system, which is characterized by a positive regulatory feedback loop, we generated B. diazoefficiens mutant strains designated as B. diazoefficiens ΔbjaI and B. diazoefficiens ΔbjaR1 through targeted deletion of the bjaI and bjaR1 genes, respectively. Subsequently, the promoters of bjaI and bjaR1 gene in wild-type (WT) B. diazoefficiens USDA110, as well as the mutant strains lacking counterpart gene, were fused with reporter genes (lacZ or gusA) at the transcriptional level. The bjaI- and bjaR1-lacZ activity in the WT bacteria, as depicted in Fig. 1A, exhibited a growth-dependent increase, with the former reaching its peak early in the logarithmic phase (day 3) and the latter progressively rising throughout the entire growth period. The activity of bjaI-lacZ in the ΔbjaR1 mutant and bjaR1-lacZ in the ΔbjaI mutant was significantly reduced to a very low level. This suggests that deleting either the bjaI or bjaR1 gene could cause functional inactivation of nearly the entire QS system, without any observed functional redundancy on the chromosome. However, leaky expression of the bjaI gene was detected, as evidenced by the persistent GUS staining in the ΔbjaR1 mutant harboring a bjaI-gusA transcriptional fusion, both under free-living conditions and within soybean nodules at 40 days post-inoculation (dpi) (Fig. 1C). These findings corroborate an earlier study (13, 15) and suggest that the bjaI/bjaR1 QS circuit is functionally non-redundant in the B. diazoefficiens USDA110 genome, rendering it amenable to elucidating its symbiotic role.
Fig 1.
(A) The promoter activity of the bjaI and bjaR1 genes in B. diazoefficiens strains grown in yeast-extract mannitol (YEM) liquid medium. The wt B. d 110 :: bjaI-lacZ or wt B. d 110 :: bjaR1-lacZ denotes strains harboring a transcriptional lacZ fusion with the promoter region of either the bjaI or bjaR1 gene in the wild-type (WT) B. diazoefficiens USDA110, while B. d ΔbjaR1::bjaI-lacZ and B. d ΔbjaI::bjaR1-lacZ are derived from corresponding mutant backgrounds. (B) The bjaI-lacZ activity in WT B. diazoefficiens USDA110 grown in liquid YEM medium (upper panel) and GUS stain (bjaI-gusA) through agar plate-sensitivity assay (lower panel). Pure isoflavonoids (i.e., genistein or daidzein) at a final concentration of 5.0 µM, as well as soybean seed extract (SSE) (20 µL/mL culture), were applied. DMSO, dimethyl sulphoxide, the solvent for daidzein. The substrates for the lacZ activity assay and GUS-staining were 13.0 mM O-nitrophenyl-D-galactopyranoside (ONPG) and 38.0 mM 5-bromo-4-chloro-3-indolyl-glucronide (X-Gluc), respectively. For agar plate-sensitivity testing, the reporter bacteria suspension and X-Gluc were spread on the plate surface, while the chemical compounds were dropped on the filter paper. The red arrow indicates the inhibition zone. (C) The GUS stain in B. diazoefficiens strains grown in the YEM plate (upper panel) and nodules (lower panel) formed at 40 days post-inoculation (dpi). (D) The quantified GUS activity in bacteria growing in YEM liquid media and soybean nodule (i.e., utilizing nodule extract and bacteroid for determination). 4-methylumbelliferyl-D-glucuronide (MUG_GUS) was used as a substrate. The data represent the means ± standard errors (SEs) of six samples from two independent experiments, with statistically significant differences indicated by asterisks (* or **) (P-value ≤ 0.05 or 0.01).
As B. diazoefficiens USDA 110 is an endosymbiont, we investigated the potential impact of the soybean host on BjaI/BjaR1 activity by examining the effects of pure soybean-derived isoflavonoids (genistein and daidzein) and soybean seed extract (SSE) on bjaI-lacZ activity in the free-living WT cells (Fig. 1B). It became evident that daidzein, but not genistein, was capable of inducing the expression of the bjaI gene. According to a previous report (22), legume-released flavonoids can boost rhizobial total autoinducer output and the expression of the genes responsible for AHL synthesis. Interestingly, SSE significantly suppressed the activity of bjaI-lacZ without exerting any effect on bacterial growth. This was further confirmed using an additional agar plate-sensitivity approach, which involved the bjaI-gusA fusion (Fig. 1B). Actually, the inhibitory effect of SSE was also observed in our early investigation on the genome-wide expression profile of the WT strain, using macroarray analysis (Fig. S1). We also quantified the activity of bjaI-gusA in WT and ΔbjaR1 mutant strains grown freely and as bacteroids inside soybean nodules (Fig. 1D). The bjaI-gusA activity in WT bacteria was significantly lower in both root nodule extract and purified bacteroid than in free-living WT cells. In addition, it showed significantly higher activity in WT nodules formed at 30 and 35 dpi compared to ΔbjaR1 mutant nodules. These findings strongly suggest that the wild-type allelic BjaI/BjaR1 system exhibits activity during the symbiotic lifestyle of Bradyrhizobium diazoefficiens, albeit much lower than in free-living cells, most likely due to the suppression by the soybean host plant. This unique feature is important for understanding its role in symbiosis, which will be explored in subsequent experiments.
The symbiotic properties of the ΔbjaR1 mutant were significantly altered
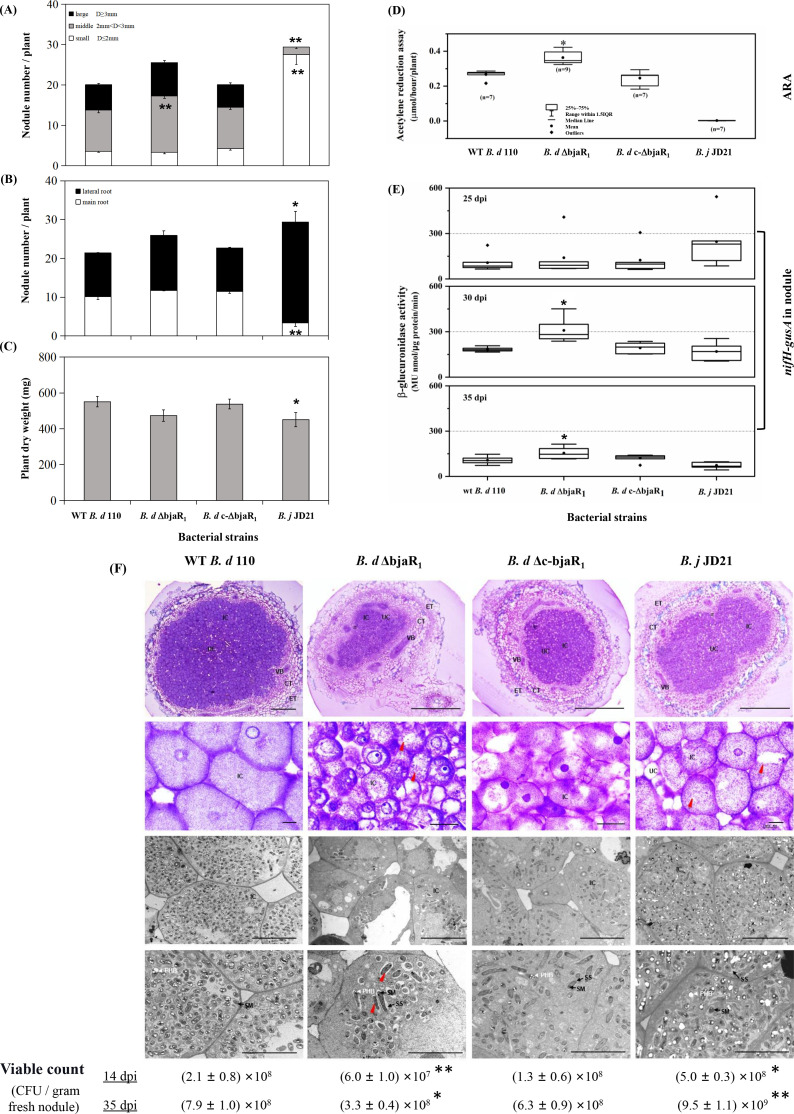
We assessed the effect of BjaR1 on bacterial symbiotic performance by conducting soybean plant tests under low-N conditions (Fig. 2A; Fig. S2). The reference strain used was B. j JD21 (12), a derivative of B. diazoefficiens USDA 110 harboring a Ω insertional mutation in the nodD2 gene. The ΔbjaR1 mutant induced a significantly higher number of nodules on soybean plants compared to both the WT and genetic complementation strain, B. d c-ΔbjaR1, with an increase of approximately 21.3% and 20.9%, respectively (Fig. 2A). The ΔbjaR1 mutant nodules typically ranged in size from 2 to 3 mm (diameter) and were predominantly localized on the basal region of the primary root or throughout the lateral root system. Conversely, the WT strain induced nodules within a similar size range but primarily clustered at the junction between roots and shoots. However, the dry weight of soybean plants infected by the ΔbjaR1 mutant strain was reduced by 14.2% compared to the WT strain (Fig. 2C). Despite producing 37.4% more nodules than the WT strain, the B. j JD 21 strain only formed small nodules (no larger than 2 mm) on lateral roots and resulted in an 18.1% decrease in dry weight of soybean plants. The results indicated that the ΔbjaR1 mutant exhibited a similar impact on nodule induction potential (i.e., the total number of nodules) and soybean dry weight as the nodD2 mutant, but otherwise closely resembled the WT parent strain.
Fig 2.
( A and B) Nodulation properties and (C) dry weight of the soybean [Glycine max (L.) Merr. cv. Williams 82] infected with B. diazoefficiens strains at 40 dpi. “D” in Fig 2A means the diameter of the nodule. Two independent experiments were conducted, with 10 replicates used for each treatment. (D) Acetylene reduction activity (ARA) and (E) nifH-gusA activity of in-nodule B. diazoefficiens at different dpis. The GUS activity was quantified using the method described above. Seven to nine plants in two independent experiments were used. (F) Light (top two panels) and electron (bottom two panels) micrographs of soybean root nodules induced by B. diazoefficiens at 14 dpi. ET, epidermal tissue. CT, cortical tissue. IT, infection thread. IC, infected cell; UC, uninfected cell. VB, vascular bundle. SM, symbiosome membrane. SS, symbiosome space. PHB, poly-β-hydroxybutyrate. Red and green arrows indicate the elongated and collapsing bacteroids, respectively. Ten fresh nodules per soybean plant were crushed and plated on an HM medium to determine colony-forming units (CFU). The data are presented as means ± SEs. Asterisks (*) and (**) indicate significant differences from the WT strain at P-values of ≤0.05 and ≤0.01, respectively.
The acetylene-reducing activity (ARA) was employed to assess the nitrogen-fixing capacity of these strains (Fig. 2D). The ARA of the ΔbjaR1 mutant at 40 dpi was 46% higher than that of the WT strain. However, overexpression of this QS circuit using a broad-host-range expression plasmid pTE3 (23) in the WT strain led to a significant decrease in nitrogenase activity and soybean dry weight (Fig. S3). Furthermore, we analyzed the promoter activity of the nifH gene, which encodes a nitrogenase reductase, by utilizing reporter strains carrying a chromosomally integrated nifH-gusA transcriptional fusion (Fig. 2E). The ΔbjaR1 mutant-induced nodules displayed significantly higher nifH-gusA activity at 30 and 35 dpi compared to those induced by other bacterial strains, consistent with the ARA results. Interestingly, the B. j JD 21 strain displayed a negligible ARA value at 40 dpi but showed certain nifH-gusA activity from 25 dpi onwards. The nodD2 mutation significantly enhances nodule induction capacity but the resulting nodules display abnormal nitrogen-fixing function (Fig. S2). Compared to WT bacteria, soybean plants inoculated with ΔbjaR1 mutant show an 8.2% increase in total nitrogen content, whereas a significant decrease was observed in plants inoculated with nodD2 mutant (Table S2).
By light and electron microscopy, we determined the effect of the ΔbjaR1 mutant on the morphological and structural characteristics of nodules and endosymbiotic tissues, including bacteroids and symbiosomes (Fig. 2F). Observation under light microscopy revealed that soybean nodules induced by the WT strain at 14 dpi contained uniform giant infected cells and a higher ratio of infected to uninfected cells compared to those produced by the ΔbjaR1 mutant. In ΔbjaR1 mutant nodules, the central infection area was smaller, the surrounding plant tissue (peripheral tissue and vascular bundles) was thicker, and the infected cells were heterogeneous. The transmission electron microscopy (TEM) analysis revealed a significantly reduced bacteroid density in plant cells infected with the ΔbjaR1 mutant strain compared to other strains, as indicated by the scarcity of bacteroids and a notably lower viable count within the nodule. The ΔbjaR1 bacteroids had very little polyhydroxybutyrate (PHB), in contrast to the WT and B. j JD21 bacteroids which accumulated significant amounts of PHB. Symbiosomes (SBs) containing one to two bacteroids were typically observed in plant cells infected by the wild-type strain, while SBs with only one bacteroid were characteristic of those infected by the ΔbjaR1 mutant. The presence of multiple (more than three) bacteroids in several SBs produced by B. j JD21 mutants indicates rapid differentiation of bacteroids. In contrast to the mainly globular shape of WT bacteroids, some ΔbjaR1 bacteroids exhibited an elongated shape and a few even displayed a collapsing phenotype, indicating that the absence of the bjaR1 gene resulted in slowed or halted division of bacteroids and SB formation. Furthermore, while the WT bacteroid had a relatively smooth surface and small symbiosome space (SS), the ΔbjaR1 bacteroid had a rough surface and larger SS, suggesting a likely defect in the bacterial capsule. The results demonstrated that, despite significantly higher levels of nodule induction and nitrogen fixation ability compared to the wild-type parent strain, the ΔbjaR1 mutant strain profoundly altered the morphology of bacteroid and symbiosome, ultimately compromising the symbiotic outcome on soybean such as plant dry weight.
The inactivation of BjaR1 resulted in a significant decrease in the expression of nodD2
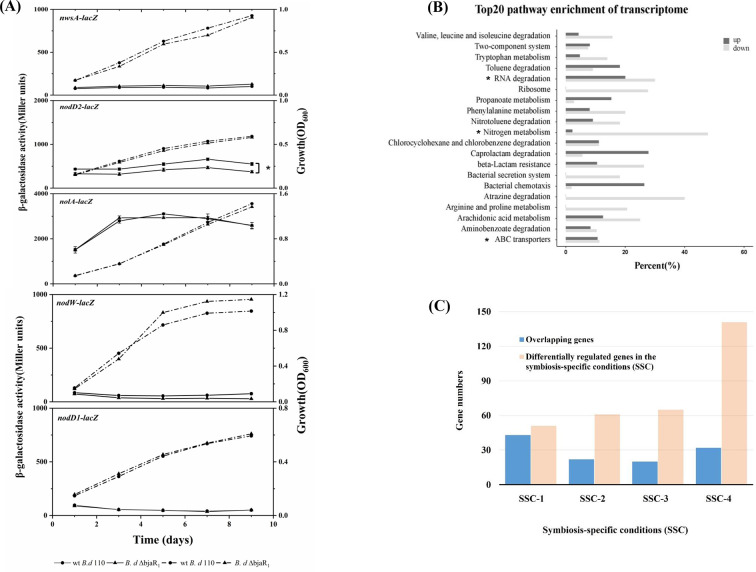
To investigate potential cross-talk between BjaR1 and regulators in the bradyoxetin-mediated QS pathway, we assessed the activity of nwsA-lacZ, nolA-lacZ, nodD2-lacZ, nodD1-lacZ, and nodW-lacZ in both WT and ΔbjaR1 mutant strains (Fig. 3A). Among these fusions, only nolA-lacZ and nodD2-lacZ exhibited robust activity during bacterial growth. However, the former maintained consistent activity in both strains, whereas the latter demonstrated a significantly reduced activity within ΔbjaR1 mutant cells. To assess the effect on nodYABC gene induction, we introduced the broad-host-range plasmid pZB32 containing the nodY-lacZ translational fusion into both strains. Upon the presence of genistein, a significant increase in the induction of nodY was observed in ΔbjaR1 cells compared to WT cells at a low cell density (Fig. S4). The nodW- and nodD1-lacZ activities of both strains remained consistently low throughout the bacterial growth curve. The nwsA-lacZ activity in the ΔbjaR1 cells exhibited a slight increase, whereas the nodW-lacZ activity showed a slight decrease compared to the WT cells. The NwsAB and NodVW systems are both two-component regulatory systems that functionally complement each other to induce nodulation genes (24, 25). The results suggest that while BjaR1 inactivation led to a significant decrease in nodD2 expression, it did not appear to affect the bradyxoetin-signaling transduction pattern in response to cell population density.
Fig 3.
Promoter activity of previously proposed cell-density-dependent regulator genes in free-living B. diazoefficiens strains. The solid and dashed lines represent the β-galactosidase activity and the optical density at 600 nm (OD600) measured during bacterial growth. These genes have been described elsewhere (11). (B) The percentage of differentially regulated genes in each metabolic pathway of B. diazoefficiens USDA110 and the top 20 most highly enriched KEGG pathways are listed here. (C) The number of overlapping genes significantly regulated in the free-living ΔbjaR1 mutant with those implicated in various symbiosis-specific conditions (SSCs). SSC-1, microoxic free-living ΔfixK2 mutants and associated bacteroid (26); SSC-2, ropN1+2 double mutant bacteroids (27); SSC-3, NifA, and RpoN1 regulon (28); SSC-4; B. diazoefficiens USDA110 bacteroid (27). The list of overlapping genes is available in Table S3. Statistically significant differences (P-value ≦ 0.05) are denoted by asterisks (*).
Transcriptome analysis of ΔbjaR1 mutant cells in free-living conditions
In the presence of genistein, we compared the transcriptomes between ΔbjaR1 mutant and wild-type cells under free-living conditions using RNA sequencing analysis. The expression change referred to in RNAseq analysis is represented by the ratio of the gene’s FPKM (fragments per kilobase of transcript sequence per millions base pairs) in the ΔbjaR1 mutant compared to that in WT cells. We identified 123 and 408 genes that were significantly upregulated and downregulated in the ΔbjaR1 mutant, respectively, using a critical ratio (2.5 or 0.4) with a significance level of P ≤ 0.05 (Table S3). The higher number of downregulated genes suggests that akin to the majority of LuxR family members, BjaR1 acts as a global transcriptional activator. The transcriptional profiles of the genomic module associated with nodulation and nitrogen metabolism are presented in Table 1. The bjaI transcript level exhibited a significant decrease, consistent with the observed decline in bjaI-lacZ activity shown in Fig. 1A. The nodulation gene cluster in rhizobia operates as a symbiotic module that primarily responds to legume-released isoflavonoid compounds. Among the 23 putatively annotated genes within this module, a collective activation of 16 structural genes, particularly the common nodYABC genes, was observed to varying extents. The expression of nodulation genes is under the coordinated regulation of three successive regulatory genes, nodD1, nolA, and nodD2. However, only the transcript of nodD2 displayed a significant decrease.
TABLE 1.
Expression change of genes involved in nodulation and nitrogen metabolism module in the free-living ΔbjaR1 mutant compared with wild-type cells
| Gene IDa | Gene namesb | Descriptionc | Ratiod | Function or pathway involved |
|---|---|---|---|---|
| BjaI/BjaR1 quorum sensing | ||||
| blr1063 | bjaI | Putative autoinducer synthase | 0.19* | IV- HSL synthesis |
| Nodulation genes set (or module) | ||||
| bll1475 | nodQ | NodQ bifunctional enzyme | 2.11 | |
| bll1630 | nolK | GDP-fucose synthetase | 1.86 | |
| bll1631 | noeL | GDP-mannose 4,6-dehydratase | 2.25 | |
| blr1632 | nodM | Putative glucosamine synthase | 1.28 | |
| bll1714 | nodW | Two component regulator | 0.94 | |
| bll1715 | nodV | Two component regulator | 0.79 | |
| bll2019 | nolA | Transcriptional regulatory protein | 0.65 | |
| bll2021 | nodD2 | Transcriptional regulatory protein | 0.28* | |
| bll2023 | nodD1 | LysR family transcriptional regulator | 1.44 | |
| blr2024 | nodY | 4.39* | ||
| blr2025 | nodA | Acyl transferase | 3.13* | |
| blr2026 | nodB | Chitooligosaccharide deacetylase | 2.30 | |
| blr2027 | nodC | Chitin synthase | 2.65* | |
| blr2028 | nodS | N-methyl transferase | 1.65 | |
| blr2029 | nodU | 6-O-carbamoyl transferase | 2.37 | |
| blr2030 | nodI | Transporter of Nod Factor | 2.33 | |
| blr2031 | nodJ | Lipooligosaccharide transport system permease protein | 1.65 | |
| blr2033 | nolN | Nol N protein | 1.87 | |
| blr2034 | nolO | Carbamoyltransferase | 2.46 | |
| blr2035 | nodZ | NodZ protein | 1.84 | |
| blr2062 | noeI | 2-O-methyltransferase | 1.98 | |
| blr4773 | nwsA | Two component regulator | 1.48 | |
| blr4774 | nwsB | Two component regulator | 1.51 | |
| Nitrogen metabolism module | ||||
| blr0613 | amtB | Ammonium transpoter | 0.10* | Uptake of NH4+ - N |
| blr1327 | Nitronate monooxygenase | 3.10* | Transformation of nitronate to nitrile | |
| blr2803 ~ blr2805 | nrtA/B/C | ABC transporter nitrate-binding protein | 0.28 ~ 0.40* | Nitrate assimilation |
| blr2808 | Putative nitrite reductase | 0.41 | Transformation of NO - - N to ammonia 2 | |
| blr2809 | nasA | Nitrate reductase large subunit | 0.31* | Assimilatory nitrate reduction |
| blr3214 | norC | Nitric oxide reductase subunit C | 0.24* | Denitrification |
| blr3215 | norB | Nitric oxide reductase subunit B | 0.37* | Denitrification |
| blr3397 | nit | Nitrilase | 0.15* | Transformation of nitrile - N to ammonia |
| blr4169 | glnII | Glutamine synthetase II | 0.28* | Glutamine synthesis |
| bll4571 | nirA | Putative ferredoxin—nitrite reductase | 0.35* | Assimilatory nitrate reduction |
| bll4798 | Putative glutaminase | 0.33* | Glutamine synthesis | |
| blr4949 | glnA | Glutamine synthetase I | 2.04 | Glutamine synthesis |
| bll5731 | cyns | Probable cyanate hydratase | 0.26* | Reaction of cyanate with bicarbonate to produce ammonia and carbon dioxide |
| bll5732 ~ blr5734 | nrtA/B/C | ABC transporter nitrate-binding protein | 0.18 ~ 0.26* | Nitrate assimilation |
| blr6402 | Nitrilase | 0.19* | Transformation of nitrile - N to ammonia | |
| bsr7036 | napE | Periplasmic nitrate reductase | 0.04* | |
| blr7037 | napD | Periplasmic nitrate reductase | 0.03* | |
| blr7038 | napA | Periplasmic nitrate reductase large | 0.02* | Dissimitory nitrate reduction to nitrite |
| blr7039 | napB | Periplasmic nitrate reductase small | 0.03* | Dissimitory nitrate reduction to nitrite |
| blr7040 | napC | Cytochrome C-type protein | 0.02* | |
| blr7089 | nirK | Respiratory nitrite reductase | 0.02* | Nitrite reductase (NO-forming) |
| blr0314 | nosR | Nitrous oxide reductase expression | 0.24* | Nitrous oxide reductase regulator |
| blr0606 | glnK | Nitrogen regulatory protein P-II 2 | 3.39* | Nitrogen regulation factor |
| blr0612 | glnK | Nitrogen regulatory protein P-II 2 | 0.12* | Nitrogen regulation factor |
| blr0723 | rpoN 2 | RNA polymerase sigma-54 subunit | 0.47 | Nitrogen regulation sigma factor |
| blr1883 | rpoN 1 | RNA polymerase sigma-54 subunit | 0.04* | Nitrogen regulation sigma factor |
| blr4486 | nifR | Nitrogen regulation protein | 0.31* | |
| blr4487 | ntrB | Two-component sensor histidine kinase | 0.31* | Global nitrogen regulatory system |
| blr4488 | ntrC | Two-component sensor histidine kinase | 0.40* | |
| blr4948 | glnB | Nitrogen regulatory protein P-II 1 | 0.38* | Nitrogen regulation factor |
| blr7084 | nnrR | Transcriptional regulatory protein | 0.03* | Nitrogen oxide reductase regulator |
Nomenclature according to Kaneko et al (2002).
Gene name according to the EMBL-EBI database.
Gene description according to GeneBank.
The transcript ratio of genes in the ΔbjaR1 mutant compared with those in the wild-type cells freely grown in the liquid yeast-extract mannitol (YEM) medium. Before RNA isolation, genistein was added to the culture with a final concentration of 5.0 μM for 12 h. “*” indicating the significantly up- or down-regulated genes with the transcription ratio values ≥ 2.5 or ≤ 0.4, respectively.
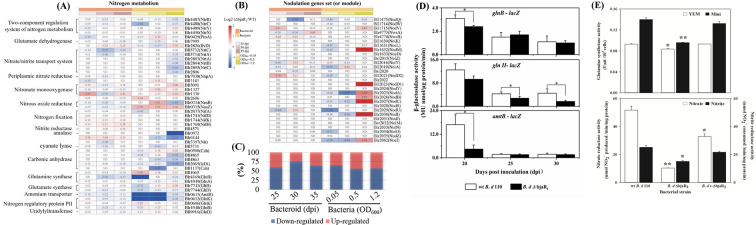
Based on the metabolic pathway enrichment analysis from KEGG (Kyoto Encyclopedia of Genes and Genomes), we observed repression of nitrogen metabolism processes at the entire pathway level (pathway ID: bja00910) (Fig. 3B). As this metabolic pathway does not include many N-regulatory genes, such as the ammonium transporter amtB gene and genes with regulatory function, we compiled expression profiles of nearly all 58 putative N-regulatory genes in the genome (Table S4). Out of these genes, a total of 47 were downregulated, with the majority (61%, or 30 genes) showing statistically significant decreases in expression levels (P value ≦ 0.05) ( Table 1). The products of these genes participate in diverse branches of the nitrogen metabolism pathway. The gene clusters blr2803-blr2805 and bll5732-bll5734 (nrtABC) encode the essential components responsible for facilitating extracellular nitrate uptake. The genes blr4169 (glnII) and blr4949 (glnA) encode isoforms of glutamine synthetase, namely GSII and GSI, respectively. Like the ΔbjaR1 mutant, the mutant strain lacking either of these genes exhibited significantly higher levels of nodulation induction and N2 fixation capability than the WT parent strain (29, 30). The operon consisting of the genes blr0612 (glnK) and blr0613 (amtB) enables bacteria to take up environmental NH4+. The AmtB system in rhizobia is typically activated under nitrogen-limited conditions, while in Rhizobium etli, the downregulation of this homologous operon is crucial for proper bacteroid function (31, 32). In addition, the transcription of genes blr4886-blr4887-blr4888 (nifR-ntrB-ntrC) was significantly reduced. The NtrB/NtrC two-component regulatory system is highly conserved and widely distributed in the bacterial domain, activating promoters for various N-regulatory genes globally. Surprisingly, a significant decrease in transcription was observed for numerous denitrification-related genes, including metabolic enzyme-coding genes such as blr3214-blr3215 (norCB), bsr7036-blr7039 (napEAB), and blr7089 (nirK), along with a critical regulatory gene blr7048 (nnrR). In B. diazoefficiens USDA 110, the transcription of denitrification genes is regulated by a FixLJ-FixK2-NnrR cascade (33, 34). In addition to two directly low-oxygen-responsive regulator genes, bll2759-bll2760 (fixJL), several other genes in the vicinity, specifically blr2757 to blr2769 (fixK2, fixNOQP, and fixGHIS), as well as a distantly located gene, blr6061 (fixK1), exhibited noticeable downregulation (Table S3). These findings indicate that the deletion of the bjaR1 gene resulted in the systematic activation and suppression, at the transcriptional level, of genomic modules associated with nodulation and nitrogen metabolism, including anaerobic denitrification.
Compared with the numerous microarray platform-based transcription studies on B. diazoefficiens, USDA 110 conducted under various symbiosis-specific conditions (SSC) can offer insights into the function of BjaR1 (Fig. 3C; Table S3). The results show varying degrees of overlap between the genes regulated by ΔbjaR1 mutant and those involved in these SSCs. In both microoxic free-living ΔfixK2 mutants and associated bacteroids, Mesa et al. (26) identified 51 promoter regions (or genes) that were significantly downregulated. Interestingly, approximately 80% (42 genes) of these genes were also markedly repressed in oxic free-living ΔbjaR1 cells. Two nitrogen-responsive sigma δ54-factor genes, blr1883 (rpoN1) and blr0723 (rpoN2), particularly the former, exhibited downregulation (Table 1). RpoN1 is not only regulated by FixK2 but also essential for RNA polymerase to initiate transcription at numerous NifA-dependent -24/–12 type promoters of genes that are directly or indirectly involved in N2-fixation (e.g., nif and fix genes as well as groESL3) (26, 35). In a previous study (27), 61 genes displayed reduced expression in nodules infected by a ropN1+2 double mutant. The present study shared 22 genes, including the hupFHDCLS gene cluster for hydrogen uptake, glnK-amtB, blr4169 (glnII), bll5733-bll5734 (nrtA-nrtB), a dicarboxylate transporter gene, blr6145 (dctA), and blr1719 (modB) (Table S3). Among the 65 genes regulated by the NifA and RpoN1 regulon (28), a total of 20 genes exhibited overlap with those identified in our study. Moreover, out of the 141 highly induced genes in WT B. diazoefficiens USDA110 bacteroids (27), we identified 33 genes overlapping with differentially expressed genes in ΔbjaR1 cells. However, most of these genes (27 genes), including bll3998 encoding succinate hemialdehyde dehydrogenase and chaperone genes grolE3/S3 (bll2059/bll2060) as well as nodD2, exhibited downregulation. It is reasonable to infer that BjaR1, as a global activator with activity in WT nodules (Fig. 1C), exerts positive regulation on the transcription of these genes. FixK2, NifA, and sigma σ54-factor RopN1 are well-established key positive regulators in a so-called central intermediate metabolism process that operates specifically in response to oxygen-limiting conditions in bacteroids. Therefore, BjaR1 represents a novel factor that exerts a positive influence on bacteroid metabolism.
Proteomic analysis of ΔbjaR1 mutant cells under both free-living and symbiotic conditions
The translational impact of BjaR1 inactivation was investigated through a proteomics analysis using mass spectrometry (MS). The MS data were collected from bacteroids formed at different time points (25, 30, and 35 dpi) and free-living cells cultured at varying population densities (OD600 = 0.05, 0.5, and 1.2). The proteomic profiles of the aforementioned modules are depicted in Fig. 4. The expression of NifHDK proteins in the nitrogenase complex was significantly upregulated in the ΔbjaR1 bacteroid, consistent with its significantly higher level of ARA and nifH expression shown in Fig. 2D and E. However, regardless of the ΔbjaR1 mutant’s lifestyle, more N-regulatory proteins were downregulated compared to the WT strain. More than 56% of detected proteins were observed to be repressed, and this proportion was as high as 75% in the ΔbjaR1 bacteroids formed at 30 dpi (Fig. 4C). Some remarkably repressed proteins, such as NtrBC, GlnII, and AmtB/GlnK, are particularly interesting due to their vital roles in NH4+ assimilation. Since nodulation proteins are primarily produced in the rhizobial bacteria during the early stages of the symbiosis, most of them were not detected in the bacteroids (Fig. 4B). In contrast to the gradual increase in activity observed in housekeeping nitrogen metabolism with increasing cell population density, the genistein-induced nodulation module exhibited transient inducibility at each cell density, resulting in irregular proteomic profiles (Fig. 4B; Fig. S5). Most nodulation proteins were detected in the low-density ΔbjaR1 cells (OD600 = 0.05) but showed general downregulation. However, in cells with an OD600 of 1.2, a collective activation of many nodulation proteins was observed, particularly NodABC, accompanied by systematic repression of nitrogen metabolism (Fig. S5). The expression of both genomic modules exhibited contrasting trends (activation and repression) in both the high-density cells used for proteome analysis and cells for RNA-seq experiments, suggesting that the intracellular nitrogen metabolic status may influence the induction of nodulation genes. The results here offer additional elucidation for previous research observations that the enhancement of NH4+ assimilation in rhizobial bacteria hinders the flavonoid-mediated induction of nod genes, thereby restricting nodulation in legume host plants (36, 37).
Fig 4.
The proteomic profiles of nitrogen metabolism (A) and nodulation (B) genomic module in B. diazoefficiens bacteroids formed at different days post-inoculation (dpi) and bacterial culture grown at various optical densities at 600 nm (OD600). The value within each box represents the log2 (fold change, FC) of protein abundance in ΔbjaR1 mutant compared to that in WT cells. Protein abundance is indicated as an average mass spectrometry signal for proteins from three samples determined by label-free quantification (LFQ) intensities obtained from DIA-NN output. ND indicates not detected. (C) The proportion of downregulated or upregulated proteins among all detected proteins in the nitrogen metabolism module in Fig. 4A. (D) Promoter activities of N-responsive genes in soybean root nodules formed at various days post-inoculation (dpi). The genes glnB (blr4948), glnII (blr4169), and amtB (blr0613) encode the N-regulatory protein PII, glutamine synthetase, and an ammonium transporter, respectively. Nodules from each soybean plant were collected and crushed for lacZ fusion activity assays using 4-methylumbelliferyl-β-D-galactopyranoside (MUG_LACZ) as a fluorogenic substrate. Data represent means ± SEs of six plants across two independent experiments. (E) Glutamine synthetase activity in B. diazoefficiens cells grown in YEM and minimal (Mini) liquid media (38). One unit of enzyme activity is defined as a change of 0.01 in the OD540 value per minute for every 104 cells. The data presented are the means ± SEs of six samples from two independent experiments. Nitrate/nitrite reductase activity was determined in anaerobically grown B. diazoefficiens cells after 4 days in a YEM liquid medium containing 10 mM KNO3. Asterisks (*or**) denote statistically significant differences compared to the WT strain, with a P-value ≤ 0.05 or 0.01 significance level.
To validate these findings, we utilized transcriptional lacZ fusion to assess the promoter activity of three representative N-regulatory genes, blr4948 (glnB), blr4169 (glnII), and blr0613 (amtB). Regardless of the type of nutrient medium, these fusions in ΔbjaR1 mutant cells exhibited lower activity than WT cells (Fig. S6). The addition of various N-containing compounds did not alter this trend except for amtB-lacZ (Fig. S7). Their activities in ΔbjaR1 mutant nodules were also generally lower than those in WT nodules (Fig. 4D). Subsequently, we evaluated the enzymatic activity of glutamine synthetase (GS) involved in NH4+ assimilation, as well as the activity of nitrate and nitrite reductases during denitrification process. Their activity in ΔbjaR1 mutant cells was generally lower than in WT cells (Fig. 4E). Taken together, the absence of the bjaR1 gene results in intrinsic repression of nitrogen metabolic functioning in bacteria, both in their free-living and symbiotic states, at the translational level.
The distinct proteomic profiles of the two genomic modules in free-living cells with an OD600 of 1.2 suggest a strong correlation between the whole-cell proteome and transcriptome. Therefore, we focused on the proteome at this specific cell density point and compared both datasets. The analysis revealed that 131 proteins were significantly upregulated, while 207 proteins were downregulated, with a threshold ratio value of 1.50 or 0.67 (P value ≤ 0.05) (Table S5). The entire nitrogen metabolism pathway was also notably repressed at the translational level (Fig. S5). Interestingly, the activation of QS metabolism (ID: bja02024) occurs at the level of the entire pathway, indicating that this QS system may primarily function at the post-transcriptional level. We have identified a total of 50 genes, including multiple ABC transporter genes, nodA, fixK2, and nnrR, which exhibit significant alterations in both transcriptional and translational expression (Table S6). The observed inadequate PHB accumulation in the ΔbjaR1 bacteroid (Fig. 2F) may be attributed to the substantial reduction in the expression of bll6073 (phbC) and its protein product, polyhydroxybutyric acid (PHB) polymerase. Simultaneous activation of two genes encoding acyl-CoA synthase (bll0994) and acyl-CoA dehydrogenase (blr7270) suggests a likely rapid change in fatty acid metabolism. The product of the latter gene participates in leucine degradation (pathway ID: bja00280) as an intermediate and a substrate for both leucine metabolism and the BjaI enzyme.
Physiological and symbiotic response of the ΔbjaR1 mutant to environmental nitrogen
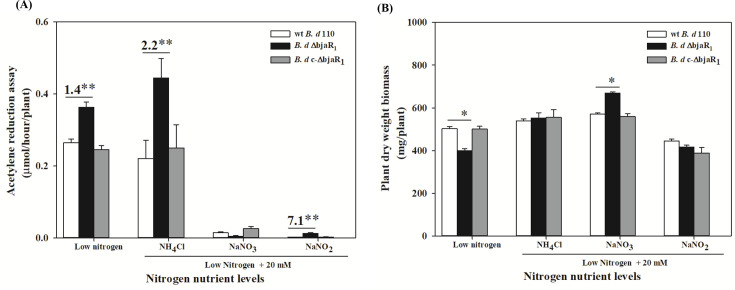
We investigated the physiological and symbiotic responses of the ΔbjaR1 mutant to environmental nitrogen sources. The ΔbjaR1 mutant grew faster in the eutrophic medium, with a doubling time of 24.7 h during the initial 4-day period, which was shorter than that (26.3 hours) of the WT strain. However, under nutrient-restricted conditions, its growth was slower compared to the WT strain, with doubling times of 34.4 and 32.8 h, respectively (Fig. S8). This result indicates that BjaR1 confers bacterial metabolic adaptation to environmental nutrition status. Subsequently, we measured the bacterial growth in a defined media (39) containing various nitrogenous compounds (ammonium chloride, sodium nitrate, sodium nitrite, and glutamine) (Fig. S9). All N-containing compounds, except for sodium nitrite (5.0 mM), enhanced the growth of all bacterial strains within the concentration range of 0.1–5.0 mM, particularly in the case of the ΔbjaR1 mutant strain. Notably, even when NH4+, NO3-, and glutamine concentrations were increased to 20 mM, the ΔbjaR1 mutant still displayed a slight growth advantage. The ΔbjaR1 mutant, with suppressed intrinsic nitrogen metabolism, shows improved efficiency and preference in utilizing external nitrogen sources or is less sensitive toward the inhibitory effects produced by these nitrogenous compounds.
An inoculation experiment was conducted to assess the effect of excessively supplied N on the symbiotic efficacy of the ΔbjaR1 mutants. The soybean plants were cultivated in nutrient solutions containing nitrogenous compounds mentioned above, excluding glutamine, at a concentration of 20 mM. A low-N solution (40) was used as the control. The bacterial nitrogenase activity (indicated as ARA value) was inhibited to varying degrees by all forms of nitrogenous compounds, with the exception observed in the case of the ΔbjaR1-soybean combination treated with 20 mM NH4+-N (Fig. 5A). Under low-N conditions, the ARA of the ΔbjaR1 mutant was 1.4 times higher than that of the WT strain. This value increased to 2.2 times when exposed to 20 mM NH4+ -N, surpassing even its ARA under low-N conditions. The presence of 20 mM NO3− - and NO2− -N significantly inhibited the nitrogenase activity of all bacterial strains. However, the inoculation of ΔbjaR1 mutant appeared to alleviate the inhibitory effect of nitrite on the nitrogenase activity. These results demonstrate that the N2-fixing process in ΔbjaR1 bacteroid became less sensitive to high levels of NH4+- and NO2−-N. Under low-N conditions, the dry weight of ΔbjaR1-inoculated soybean inoculated was significantly lower than that of WT-infected soybean, but this difference disappeared under high NH4+ and NO3−-N cultivation conditions. Interestingly, the growth of soybean plants inoculated with the ΔbjaR1 strain displayed resistance to high levels of NO3− -N, resulting in significantly higher biomass than those infected with the WT strain, despite experiencing pronounced nitrogenase activity inhibition. The supplementary file (Table S7) summarizes additional symbiotic properties such as nodule number, dry mass, nodule activity, and root volume.
Fig 5.
(A) Acetylene reduction activity (ARA) and (B) dry weight of soybean infected with B. diazoefficiens strains at 30 dpi under low- and high-N cultivation conditions. The composition of the low-N nutrient solution was previously described (40). Data represent mean ± SEs of nine plants in three independent experiments. Asterisks (* or **) denote statistically significant differences based on t-tests, with P ≦ 0.05 or 0.01 significance levels, respectively.
DISCUSSION
Many other rhizobia possess multiple sets of luxI/luxR QS systems, leading to cross-talk and functional redundancy that often complicate the interpretation of symbiotic phenotypes in relevant mutants (16). The BjaI/BjaR1 QS circuit is not only functionally redundant in B. diazoefficiens but also active inside soybean nodules, which helps elucidate its role in symbiosis. Modulation of QS systems in various symbiotic and pathogenic bacteria by host-released substances has been reported (41–43). The present study demonstrates for the first time that the bjaI/bjaR1 activity is inhibited by SSE, and its activity in the bacteroid is significantly lower than in free-living bacteria (Fig. 1B and D). The latter finding is surprising, as the density of B. diazoefficiens USDA 110 bacteroids was reported to be quite high inside soybean nodules (44), which could achieve a “quorum” to trigger robust bjaI/bjaR1 activity. The specific inhibitors in SSE remain undefined, and their presence in root nodule plant tissues is yet to be ascertained. If they are indeed present, it is unclear whether they are transported to bacteroids through symbiosome membranes, ultimately impacting the QS system’s functionality within nodules. However, both overexpression and deficiency of bjaI/bjaR1 QS significantly altered the nitrogenase activity and ultimately compromised symbiotic outcomes in soybean plants. This strongly indicates that this rhizobial genetic system strictly regulates bacteroid nitrogen fixation, soybean-mediated repression being crucial for the process. It was revealed that the luminescence of various AHL-reporter strains was significantly inhibited on the surface of pea seedlings (45). The inhibition of BjaI/BjaR1 activity may occur during the initial stages of symbiosis, potentially causing the metabolic shift from a free-living to a symbiotic state within the B. diazoefficiens community. Elucidating the regulatory mechanisms of soybean plants on the QS circuit will enhance our comprehension of bacterial metabolic control of nitrogen fixation and advance quorum-quenching technologies for agricultural applications. This encompasses the identification of inhibitory substances, determination of their genetic backgrounds, and investigation of environmental factors influencing their production.
Like the nodD2 mutant, the ΔbjaR1 mutant exhibited enhanced nodule induction ability (e.g., increased nodule numbers) on soybean plants (Fig. 2A). As nodD2 is situated within the nodulation gene cluster, direct mutation of this gene resulted in more pronounced effects on bacterial nodule induction ability compared to indirect mutations of bjaR1, as shown by the disparity in nodule numbers formed by both mutants (Fig. 2A). The literature (11) and our results (Fig. 3A) overwhelmingly confirm that NodD2 activity positively correlates with bacterial growth, which is intricately linked to cellular metabolic activity. Therefore, NodD2 may affect the isoflavonoid-mediated nodulation gene expression by sensing bacterial metabolic state. Regarding the significant role of Nod Factors synthesis in initiating nodulation and the necessity of housekeeping nitrogen metabolism for bacterial proliferation during the early stage of symbiosis, NodD2 may serve as a regulatory mechanism to meticulously coordinate these two distinct processes. The former is a symbiotic module obtained through genetic lateral transfer during evolution, while the latter constitutes a highly conserved genomic module. It provides an explanation for the previous report that the N-dependent regulatory circuit modulates the expression of nod genes as a function of the N status of the cells (36). As BjaI/BjaR1 QS deficiency leads to excessive nodulation, soybean suppression of the activity of this QS system may have a feedback effect on the nodulation process, promoting a symbiotic relationship. We performed the electrophoretic mobility shift assays (EMSAs) to investigate the direct activation of NodD2 by BjaR1. However, no binding signal was detected between the E. coli-expressed BjaR1 protein and the upstream DNA regions of this regulator. LuxR-type regulators typically activate target genes in conjunction with corresponding LuxI-produced HSL signals (2–4). As isovaleryl-HSL signaling compounds produced by BjaI are unavailable, further genetic analyses will be necessary to address this issue. The direct regulation of NodD2 activation by BjaR1 seems unlikely, as evidenced by the distinct symbiotic phenotypes observed in the two mutant strains. Although similar effects were observed on nodule number and soybean dry weight, significant differences were noted regarding nodule size, bacteroid morphology, and particularly nitrogen-fixing ability (Fig. 2).
Genetic analysis of a massive number of single N-regulatory genes across various rhizobial species has suggested that the downregulation of NH4+ assimilation in their bacteroids is essential for efficient symbiotic nitrogen fixation (refer to comprehensive review papers 29). Our omics analysis demonstrated that BjaR1 inactivation globally repressed the intrinsic nitrogen metabolism network, particularly key components involved in NH4+ assimilation like NtrBC and AmtB/GS, at both transcriptional and translational levels. However, the complexity of the regulatory network for nitrogen metabolism renders our omics data insufficient for deciphering the intricate interplay between genes accurately. The underlying mechanism for the significantly enhanced nitrogen-fixing capacity of ΔbjaR1 bacteroid remains unclear. The inhibition of nitrogen metabolism due to bjaI/bjaR1 deficiency enhances nitrogen fixation, a highly energy-intensive process, by redirecting energy consumption allocation. Interestingly, compared to soybeans inoculated with the WT strain, the total carbon content of ΔbjaR1-infected plants remained unchanged, but a significant increase in total nitrogen content was observed (Table S2). However, the excessive nitrogen fixation by rhizobial bacteroids is widely acknowledged to impose a great metabolic burden on the legume host plant. In addition, the attenuation of denitrification may confer benefits to N2 fixation, as NO, one of its byproducts, has been demonstrated to exert a potent inhibitory effect on nitrogenase activity (46). It is advantageous for the soybean host plant to effectively inhibit the activity of B. diazoefficiens BjaI/BjaR1 to achieve coordinated metabolic regulation of nitrogen fixation. From this point of view, it is of co-evolutionary significance concerning the evidence that the soybean host plant modulates B. diazoefficiens community behaviors via the suppression of the BjaI/BjaR1 QS system.
Surprisingly, there was significant overlap between the genes regulated by FixK2, NifA, and sigma δ54-factor RopN1—which primarily function in microoxic conditions—and a multitude of differentially expressed genes in the aerobic free-living ΔbjaR1 cells. The number of overlapping genes is likely to be underestimated due to differences in analysis platforms and critical values for detecting differentially expressed genes. We observed that the transcript levels of fixK2 (FPKM values) in wild-type cells reached high magnitudes, indicating potential oxygen limitation during growth, thereby triggering microoxic respiration and denitrification mediated by FixK2. Bacterial capsules may function similarly to extracellular polysaccharide (EPS)-based biofilms, serving as a barrier to oxygen diffusion. The luxI/luxR QS system is known to regulate EPS synthesis in many gram-negative bacteria. Our findings demonstrate that the ΔbjaR1 mutant not only failed to form a biofilm when grown freely (data not shown) but also exhibited an abnormal bacteroid surface and symbiosome space (Fig. 2F). Therefore, it is reasonable to hypothesize that the deformation of mutant bacterial capsules leads to aberrant intracellular oxygen tension, consequently diminishing fixK2 transcription. It is widely acknowledged that Azotobacter vinelandii can effectively regulate the thickness of its bacterial capsule in response to nitrogen limitation, thereby maintaining optimal intracellular oxygen levels for N2 fixation. Mesa et al. (34) reported that certain FixK2-regulated genes in ΔfixK2 mutant bacteroids differed from those observed in the same mutant grown freely under microoxic conditions, suggesting integration of an unknown signal other than oxygen limitation at the level of fixK2. BjaR1 is a likely candidate for this novel factor, although its expression may be relatively low in the bacteroid or primarily function at the translational level (Fig. S5). In Anabaena sp. PCC7120, a range of AHLs with varying acyl side chain lengths strongly represses N2-fixation at the translational level (47). Our findings indicate that the bjaR1 mutation leads to a reduction in the bacteroid density (Fig. 2F). Recent studies have unveiled that bacteroid density plays a crucial role in determining oxygen availability within individual bacteroids during ex planta assays (20). Therefore, it is highly likely that the BjaI/BjaR1 QS system is closely linked with an oxygen-sensing mechanism or microxic respiration metabolism within nodule in response to B. diazoefficiens bacteroid density.
Although both proteomic and transcriptomic analysis revealed the unique expression characteristics of two modules, certain regulatory proteins such as RopN1 and NodD2, as well as other proteins, were not consistently detected. In addition, the abundance of some proteins did not align with the transcript levels due to limitations in the resolution of MS techniques or challenges caused by low protein content and rapid degradation. Here, we tend to discuss the issue of impaired soybean growth. One plausible explanation is that the significantly enhanced N2 fixation may overpower the reducing power of photosynthesis required for plant growth, or that soybeans cannot efficiently and systematically utilize excess fixed nitrogen. Interestingly, soybean roots inoculated with the ΔbjaR1 strain exhibited a significant increase in volume under low-N conditions compared to those infected with the WT strain (Table S8), indicating potentially localized utilization of excess fixed nitrogen by roots. The transport and distribution of nitrogen between symbiotic partners may be hindered, as indicated by the profoundly altered symbiosome membrane and space (Fig. 2F). This article highlights the significant omics features of two genomic modules in the ΔbjaR1 mutant and logically connects them to the greatly enhanced nodulation induction and nitrogen fixation performance, drawing on existing literature. We propose a model for the symbiotic role of the BjaI/BjaR1 QS system, as illustrated in Fig. 6, and suggest that soybean-mediated suppression of this QS system is crucial for effective symbiotic interaction with B. diazoefficiens. However, it does not rule out the effect of BjaR1 on other metabolic pathways and genetic systems within the intricate network of the bacterium. In this paper, we have identified some genes that are subject to both transcriptional and translational regulation by BjaR1 (Table S5). They are valuable for elucidating the regulatory mechanism of BjaR1. This study provides novel insights into the metabolic regulation of nitrogen fixation in B. diazoefficiens mediated by the BjaI/BjaR1 QS system. Moreover, it underscores the need for soybean host plants to suppress this genetic system to establish a proficient symbiotic relationship.
Fig 6.
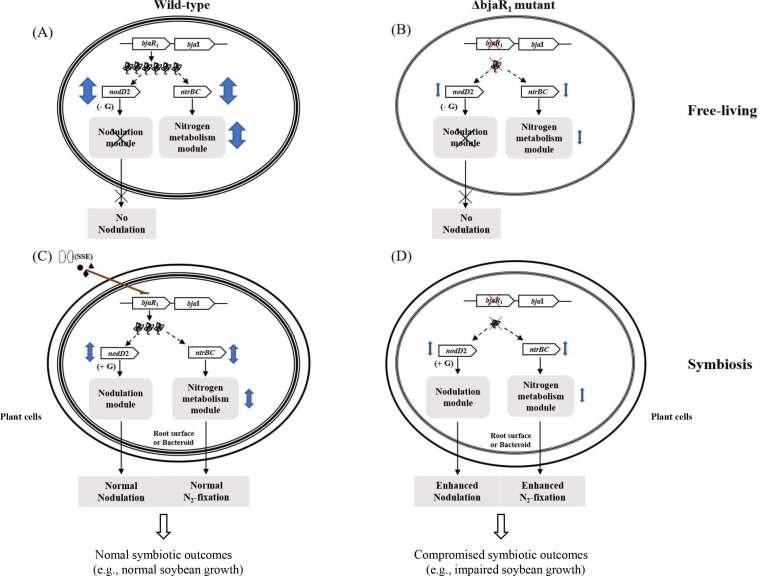
A proposed hypothetical symbiotic role of BjaR1 in B. diazoefficiens at the free-living and symbiotic state. (A) In the wild-type strain, BjaR1 positively influences nodD2 expression, and bacterial nitrogen metabolism likely via the NtrBC two-component regulatory system. (B) Inactivation of BjaR1 notably suppresses these physiological processes. Bacteria may employ this QS regulator to coordinate their metabolic function, enhancing adaptability to the free-living microenvironment. (D) Loss of BjaR1 function during symbiosis leads to excessive nodulation and nitrogen fixation, triggered by the alleviation of NodD2’s inhibitory influence on isoflavonoid (i.e., genistein)-mediated nodulation gene induction, and the suppression of nitrogen metabolism (or central intermediate metabolism) in bacteroids. However, the symbiotic outcomes in soybeans are compromised, as evidenced by the remarkably reduced plant dry weight. (C) The soybean-mediated suppression of BjaI/BjaR1 activity in the WT strain is crucial for efficient symbiotic nitrogen fixation, providing tight control over these two fundamental processes. The double arrow symbolizes the extent of BjaR1’s positive influence on NodD2 and NtrBC activities, both of which are known to potentially regulate genomic modules associated with bacterial nodulation and nitrogen metabolism. The diagram does not illustrate the regulatory positive feedback loop between BjaR1 and BjaI. G, genistein.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions
The bacterial strains and plasmids used in this study are listed in Table S1 of the supplementary material. B. diazoefficiens strains were cultured at 28°C on either yeast-extract mannitol (YEM) or two types of minimal medium (38, 39). Parental mating and conjugate selection were performed using HM salts (48) and arabinose-gluconate (AG) medium (49), respectively. Escherichia coli strains were grown with Luria-Bertani (LB) medium at 37°C, while appropriate antibiotics were added to the media as previously reported (50). Isoflavonoids such as genistein and daidzein were purchased from Solarbio Life Sciences Co., Ltd., Beijing, China, whereas soybean seed extract was prepared with 50% alcohol according to previous descriptions (50).
Construction of mutants and reporter strains
The primers used for genetic manipulation are listed in Table S1. The PCR-driven overlap extension technique (51) was employed to amplify a DNA fragment containing the deleted bjaR1 (or bjaI) gene, using B. diazoefficiens USDA110 chromosomal DNA as a template and the corresponding primers. Subsequently, the amplified DNA fragments were cloned into the EcoRI-XbaI site of the pK18mobSacB plasmid (52), and the resulting recombinant plasmid was conjugated into B. diazoefficiens USDA110 with assistance from the pRK2013 plasmid. After counter-selection with 10% sucrose, the deletion in ΔbjaR1 and ΔbjaI mutant strains was confirmed by DNA sequencing. To generate a genetically complementary strain of B. diazoefficiens c-ΔbjaR1, a DNA fragment containing the bjaR1 gene and its native promoter region was amplified using WT bacterial DNA as a template and CΔbjaR1_for/_ rev as primers; this fragment was then cloned into the PmeI site of broad-host-range plasmid pKS800 (53) and transferred into ΔbjaR1 mutant strain. All reporter strains were constructed following similar experimental procedures: amplification of a DNA fragment containing part of the 5′-end of the target gene and its promoter region, cloning it into SmaI site upstream of lacZ (or gusA) gene on R6K-replicon plasmids pVIK112 (54) or pVIK166, mobilization of recombinant plasmids into both B. diazoefficiens USDA110 and corresponding mutant strains, respectively, selection of correct chromosomally integrated fusions by colony PCR followed by sequencing verification. Plasmid pVIK166 was generated by replacing the gusA gene with the gfp gene from the pVIK165 (54) plasmid in this study; briefly, the coding sequence for gusA gene in the pCAM20 (55) plasmid was PCR-amplified using gusA_for/_rev primers and ligated with larger fragment cut from SmaI-digested pVIK165.
Beta-galactosidase and beta-glucuronidase activity assay
The activity of β-galactosidase was assessed using O-nitrophenyl-β-D-galactopyranoside (ONPG) and 4-methylumbelliferyl-β-D-galactopyranoside (MUG_LACZ), while the activity of β-glucuronidase was measured with 4-methylumbellliferyl-β-D-glucuronide (MUG_GUS) as a substrate. The enzymatic reaction product, methylumbelliferone (MU), generated from MUG_LACZ and MUG_GUS, can be quantified through fluorescence detection at an excitation wavelength of 365 nm and an emission wavelength of 455 nm. The fluorometric assay is a more sensitive and efficient method for quantifying gene transcription in cases of weak promoter activity (56). The bacteria were cultured in 20 mL of liquid YEM medium at a temperature of 28℃, followed by the collection of 250 µL of culture to measure β-galactosidase activity using ONPG as a substrate according to the method described by Miller (57). The fluorescence determination was performed as previously described (58). The nodules on one soybean root or cells in 1.0 mL of bacterial culture were lysed in 1.0 mL extraction buffer, rapidly frozen with liquid nitrogen, and subsequently pulverized using a tissue grinder (IANGEN Biotech, Ltd., Beijing, China) at 11,000 rpm for 30 seconds on ice. After centrifugation at 12,000 rpm for 5 min, the supernatant was mixed with 100 µL extraction buffer containing 1.0 mM fluoric substrate and incubated at 37 ℃ for 15 min. The fluorescence measurement was performed using a Florescence Microplate Reader, Varioscan Flash (Thermo Fisher Scientific, Shanghai, China), with a reaction solution of 40 µL. The standard curve for different concentrations of standard MU was generated based on the determined fluorescence readings. Total proteins in the extract were quantified by the dye-binding method of Bradford using a kit supplied by Vazyme Biotech Co., Ltd., Nanjing, China.
RNA extraction, library construction, sequencing, and bioinformatic analysis
The B. diazoefficiens bacteria were pre-cultured in 20 mL of YEM liquid medium until reaching the logarithmic growth phase, followed by transfer to 200 mL of fresh YEM medium for further growth until the bacterial cells reached an OD600 of 0.4–0.5. Then, the cultures were exposed to genistein at a final concentration of 5.0 µM for 12 h. To prevent RNA degradation, a cold 5% phenol/ethanol (vol/vol) solution of 50 mL was immediately added before cell collection, and total RNA was isolated using a Trizol kit from Invitrogen. The input material for cDNA library construction consisted of 3 µg of RNA per sample, which was confirmed for purity, concentration, and integrity before being subjected to NEBNext UltraTM directional RNA Library Prep Kit for Illumina (NEB, USA). Subsequently, the library quality was evaluated using the Agilent Bioanalyzer 2100 system. The construction of cDNA libraries was performed using three independent experimental RNA samples. Following the manufacturer’s instructions, the index-coded samples were clustered using the TruSeq PE Cluster Kit v3-cBot-HS (Illumina) on a cBot Cluster Generation System. Subsequently, sequencing of the libraries was performed on an Illumina Hiseq 2500 platform (Novogene Biotech Co., Ltd. Beijing, China). Illumina reads in FASTQ format were trimmed to ensure high sequence quality with a Phred score of 20. The reference genome and gene model annotation files for B. diazoefficiens USDA110 were retrieved from the NCBI database. Reads were aligned to the B. diazoefficiens USDA110 genome using Bowtie2-2.2.3 (59). Gene expression levels were quantified using HTSeq v0.6.1 to count the number of reads mapped to each gene, and FPKM values (Fragments Per Kilobase of transcript sequence per Millions base pairs) were calculated based on gene length and read counts (60). Differential expression analysis was conducted using the DESeq R package (1.18.0). P-values were adjusted using Benjamini and Hochberg’s approach for controlling the false discovery rate; genes with an adjusted P-value of ≦0.05 found by DE Seq were considered differentially expressed. KEGG enrichment analysis was performed using KOBAS software (61) to test the statistical enrichment of differentially expressed genes.
Total protein extraction, digestion, and mass spectrometry analysis
For the proteomic analysis under free-living conditions, B. diazoefficiens bacteria were individually cultured in 50 mL of liquid YEM medium to log phase (OD600 = 0.5). Subsequently, they were diluted 10-fold and divided into triplicates (50 mL each). One portion of the bacterial cultures was then induced with genistein at a final concentration of 5.0 μ M for 12 h. This procedure was applied to the remaining two cultures after their OD600 reached 0.5 and 1.2, respectively. The soybean inoculation procedures are described below. Bacteroids were isolated from fresh nodules (4–5 g) using the Percoll gradient centrifugation method, as previously reported (20), resulting in an extract solution with a density of 2–3 mg/mL of total cellular protein. The proteins from 0.2 mL bacterial culture or bacteroid suspension were extracted, digested, and analyzed by mass spectrometry following the established protocols (62). To acquire label-free quantification (LFQ) intensities, we employed the DIA-NN (63) computational proteomics platform to process the mass spectrometry (MS) raw data against the protein sequence of B. diazoefficiens USDA110 available in the UniProt database. A protein was considered to have undergone differential changes if its coefficient of variance (CV) in abundance, calculated from triplicate samples, was less than 0.5 and if the t-test comparing its ratio in ΔbjaR1 mutant to WT cells yielded a P-value of no more than 0.05.
Determination of glutamine synthetase (GS) activity and nitrate/nitrite reductase activities
Bacteria were cultured in 20 mL of liquid YEM medium until reaching the logarithmic phase, followed by treatment with genistein using a similar procedure as outlined above. Subsequently, 0.5 mL of bacterial culture was utilized to determine GS (EC 6.3.1.2) activity via an assay kit (BC0910) from Solarbio Life Science, Beijing, China. The assay kit operates using NH4+ and glutamate as substrates to synthesize glutamine in the presence of ATP and Mg2+ in the reaction catalyzed by GS. Glutamine is then transformed into glutamic-γ-hydroxamic acid under acidic conditions, forming a red complex with Fe3+ that has a maximum absorption peak value at 540 nm. Bacteria cultivated aerobically in YEM medium were collected by centrifugation (8,000 g for 10 min at 4 ℃), washed twice with YEM, resuspended in 20 mL of the same medium supplemented with 10 mM KNO3, and purged of oxygen by injecting 100% N2 using a syringe three times. The bacterial culture was then incubated under anaerobic conditions. To determine the activities of methyl viologen-dependent periplasmic nitrate reductase (Nap) and succinate-dependent nitrite reductase (Nir), 1.0 mL of bacterial culture was used following previously described methods (64, 65).
Plant test and microscopic examination
Soybean [Glycine max (L.) Merr. cv. Williams 82] seeds were disinfected by immersing them in 70% alcohol for 1 min, followed by a 30-second treatment with 10% sodium hypochlorite and thorough washing with sterilized water. Subsequently, three disinfected seeds were then sown in a 1,000-mL glass bottle containing vermiculite pre-soaked in a mixture of 300 mL distilled water and 100 mL of a low-N nutrient solution (40). This solution consisted of the following components (g/L): 0.34 CaSO4.2H2O, 0.17 K2HPO3, 0.25 MgSO4.7H2O, 0.075 KCl, and 0.002 Fe-citrate; while minor elements were added as 0.5 mL stock/L. The stock solution was prepared by dissolving the following compounds (g/100 mL): 0.203 MnSO4.4H2O, 0.0078 CuSO4.5H2O, 0.0022 ZnSO4.7H2O, 0.143 H3BO3, and 0.001 (NH4)Mo4O2.4H2O. Each seed was inoculated with 1.0 mL of WT or ΔbjaR1 cell suspension, which was equally adjusted in advance with sterilized water based on the OD600 value. The plants were cultivated under controlled greenhouse conditions (25/22℃ day/night temperature, 55% relative humidity, and a 16/8 h day/night photoperiod) and received regular watering. After 21 days post-inoculation (dpi), an additional 100 mL of low-N solution was applied. The same planting regime was followed for soybean cultivation under high-N conditions, except that growth solutions were prepared by adding 20 mM N-containing compounds (NH4Cl, NaNO3, or NaNO2) to the low-N solution. We investigated the number of nodules and the dry weight of both nodules and plants. To determine acetylene reduction activity (ARA), plant roots were cut and incubated with 10% (vol/vol) acetylene in a slender glass container (60 mL) at 25°C for 30 min. Ethylene was monitored using gas chromatography equipped with FID (FL9720) fitted with a CP-PoraBOND column CP7381 (Agilent Technologies, Inc.).
For the microscopic examination, soybean root nodules at 14 dpi were isolated and prefixed with formaldehyde-acetic acid-ethanol (FAA) solution twice for 30 min each under vacuum. Then, after being progressively dehydrated by ethanol with concentration gradient (30%, 50%, 70%, and 90%) for 30 min each, they were embedded with Technovit 7100 Embedding Kits (Technovit, Germany) and cut into thin sections (5–10 mm) by a Fully Motorized Rotary Microtome (Leica RM2265, Germany). The Technovit-embedded sections were stained with methylene blue (0.2%) and toluidine blue (1%) in water before observations with a light microscope (Nikon Ecllipse Ni-U, Japan). For the TEM analysis, the nodules were prefixed in 2.5% glutaraldehyde (Sigma-Aldrich, Beijing, China) diluted in 0.1 M phosphate buffer (pH 7.0) for 24 h. Then, they were rinsed in the same buffer and post-fixed in 1% osmium tetroxide (diluted in 0.2 M sodium cacodylate) for about 2–3 h, rinsed again, and progressively dehydrated with ethanol (50%, 70%, and 90% for 15 min), ethanol:acetone (1:1, 15 min) and pure acetone (20 min ×3) and propylene oxide. Finally, they were embedded in an Epon 812 resin following the manufacturer’s instructions for a Pelco Eponate 12TM assay kit (Ted Pella. Inc., CA, USA). Ultra-thin sections (50–60 nm) were prepared using an ultramicrotome (Leica EM UC7/FC7, Germany) and were stained with uranium acetate (3%) and lead citrate before observations with a transmission electron microscope (Hitachi HT7700, Japan).
ACKNOWLEDGMENTS
The research was supported by grants from the National Natural Science Foundation of China (Nos. 31670504, 32325036) and National Key Research and Development Program of China (2022YFF1003200).
Contributor Information
Min Wei, Email: weim@lzu.edu.cn.
Gladys Alexandre, University of Tennessee at Knoxville, Knoxville, Tennessee, USA.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aem.01374-23.
Inoculation experiment, promoter analysis, bacterial growth, etc.
Bacterial strains, Omics data, etc.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. González JE, Keshavan ND. 2006. Messing with bacterial quorum sensing. Microbiol Mol Biol Rev 70:859–875. doi: 10.1128/MMBR.00002-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fuqua C, Parsek MR, Greenberg EP. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet 35:439–468. doi: 10.1146/annurev.genet.35.102401.090913 [DOI] [PubMed] [Google Scholar]
- 3. Hense BA, Schuster M. 2015. Core principles of bacterial autoinducer systems. Microbiol Mol Biol Rev 79:153–169. doi: 10.1128/MMBR.00024-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Papenfort K, Bassler BL. 2016. Quorum sensing signal-response systems in gram-negative bacteria. Nat Rev Microbiol 14:576–588. doi: 10.1038/nrmicro.2016.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schultze M, Kondorosi A. 1998. Regulation of symbiotic root Nodule development. Annu Rev Genet 32:33–57. doi: 10.1146/annurev.genet.32.1.33 [DOI] [PubMed] [Google Scholar]
- 6. Poole P, Ramachandran V, Terpolilli J. 2018. Rhizobia: from saprophytes to endosymbionts. Nat Rev Microbiol 16:291–303. doi: 10.1038/nrmicro.2017.171 [DOI] [PubMed] [Google Scholar]
- 7. Ferguson BJ, Mens C, Hastwell AH, Zhang M, Su H, Jones CH, Chu X, Gresshoff PM. 2019. Legume nodulation: the host controls the party. Plant Cell Environ 42:41–51. doi: 10.1111/pce.13348 [DOI] [PubMed] [Google Scholar]
- 8. Delamuta JRM, Ribeiro RA, Ormeño-Orrillo E, Melo IS, Martínez-Romero E, Hungria M. 2013. Polyphasic evidence supporting the reclassification of Bradyrhizobium japonicum group Ia strains as Bradyrhizobium diazoefficiens sp. nov. Int J Syst Evol Microbiol 63:3342–3351. doi: 10.1099/ijs.0.049130-0 [DOI] [PubMed] [Google Scholar]
- 9. Loh J, Carlson RW, York WS, Stacey G.. 2002a. Bradyoxetin, a unique chemical signal involved in symbiotic gene regulation. Proc Natl Acad Sci USA 99: 14446-14451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yajima A, Katsuta R, Shimura M, Yoshihara A, Saito T, Ishigami K, Kai K. 2021. Disproof of the proposed structures of bradyoxetin, a putative Bradyrhizobium japonicum signaling molecular, and HMCP, a putative Ralstonia solanacearum quorum-sensing molecular. J Nat Prod 84:495–502. doi: 10.1021/acs.jnatprod.0c01369 [DOI] [PubMed] [Google Scholar]
- 11. Loh J, Stacey G. 2003. Nodulation gene regulation in Bradyrhizobium japonicum: a unique integration of global regulatory circuits. Appl Environ Microbiol 69:10–17. doi: 10.1128/AEM.69.1.10-17.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jitacksorn S, Sadowsky MJ. 2008. Nodulation gene regulation and quorum sensing control density-dependent suppression and restriction of nodulation in the Bradyrhizobium japonicum-soybean symbiosis. Appl Environ Microbiol 74:3749–3756. doi: 10.1128/AEM.02939-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kaneko T, Nakamura Y, Sato S, Minamisawa K, Uchiumi T, Sasamoto S, Watanabe A, Idesawa K, Iriguchi M, Kawashima K, Kohara M, Matsumoto M, Shimpo S, Tsuruoka H, Wada T, Yamada M, Tabata S. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res 9:225–256. doi: 10.1093/dnares/9.6.225 [DOI] [PubMed] [Google Scholar]
- 14. Pongsilp N, Triplett EW, Sadowsky MJ. 2005. Detection of homoserine lactone-like quorum sensing molecules in Bradyrhizobium strains. Curr Microbiol 51:250–254. doi: 10.1007/s00284-005-4550-5 [DOI] [PubMed] [Google Scholar]
- 15. Lindemann A, Pessi G, Schaefer AL, Mattmann ME, Christensen QH, Kessler A, Hennecke H, Blackwell HE, Greenberg EP, Harwood CS. 2011. Isovaleryl-homoserine lactone, an unusual branched-chain quorum-sensing signal from the soybean symbiont Bradyrhizobium japonicum. Proc Natl Acad Sci USA 108:16765–16770. doi: 10.1073/pnas.1114125108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Calatrava-Morales N, McIntosh M, Soto MJ. 2018. Regulation mediated by N-Acyl homoserine lactone quorum sensing signals in the rhizobium-legume symbiosis. Genes (Basel) 9:263. doi: 10.3390/genes9050263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. An JH, Goo E, Kim H, Seo Y-S, Hwang I. 2014. Bacterial quorum sensing and metabolic slowing in a cooperative population. Proc Natl Acad Sci USA 111:14912–14917. doi: 10.1073/pnas.1412431111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dixon R, Kahn D. 2004. Genetic regulation of biological nitrogen fixation. Nat Rev Microbiol 2:621–631. doi: 10.1038/nrmicro954 [DOI] [PubMed] [Google Scholar]
- 19. Udvardi M, Poole PS. 2013. Transport and metabolism in legume-rhizobia symbioses. Annu Rev Plant Biol 64:781–805. doi: 10.1146/annurev-arplant-050312-120235 [DOI] [PubMed] [Google Scholar]
- 20. Waters JK, Mawhinney TP, Emerich DW. 2020. Nitrogen assimilation and transport by ex planta nitrogen-fixing Bradyrhizobium diazoefficiens bacteroids is modulated by oxygen, bacteroid density and L-malate. Int J Mol Sci 21:7542. doi: 10.3390/ijms21207542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. DeAngelis KM, Lindow SE, and Firestone MK.. 2008. Bacterial quorum sensing and nitrogen cycling in rhizosphere soil. FEMS Microbiol Ecol 66: 197-207. [DOI] [PubMed] [Google Scholar]
- 22. Pérez-Montaño F, Guasch-Vidal B, González-Barroso S, López-Baena FJ, Cubo T, Ollero FJ, Gil-Serrano AM, Rodríguez-Carvajal MÁ, Bellogín RA, Espuny MR. 2011. Nodulation-gene-inducing flavonoids increase overall production of autoinducers and expression of N-acyl homoserine lactone synthesis genes in rhizobia. Res Microbiol 162:715–723. doi: 10.1016/j.resmic.2011.05.002 [DOI] [PubMed] [Google Scholar]
- 23. Egelhoff TT, Long SR. 1985. Rhizobium meliloti nodulation genes: identification of nodDABC gene products, purification of nodA protein, and expression of nodA in Rhizobium meliloti. J Bacteriol 164:591–599. doi: 10.1128/jb.164.2.591-599.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grob P, Hennecke H, Göttfert M. 1994. Cross-talk between the two-component regulatory systems NodVW and NwsAB of Bradyrhizobium japonicum. FEMS Microbiol Lett 120:349–353. doi: 10.1111/j.1574-6968.1994.tb07057.x [DOI] [Google Scholar]
- 25. Loh J, Garcia M, Stacey G. 1997. NodV and NodW, a second flavonoid recognition system regulating nod gene expression in Bradyrhizobium japonicum. J Bacteriol 179:3013–3020. doi: 10.1128/jb.179.9.3013-3020.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mesa S, Ucurum Z, Hennecke H, Fischer H-M. 2005. Transcription activation in vitro by the Bradyrhizobium japonicum regulatory protein Fixk2. J Bacteriol 187:3329–3338. doi: 10.1128/JB.187.10.3329-3338.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pessi G, Ahrens CH, Rehrauer H, Lindemann A, Hauser F, Fischer H-M, Hennecke H. 2007. Genome-wide transcript analysis of Bradyrhizobium japonicum bacteroids in soybean root nodules. Mol Plant Microbe Interact 20:1353–1363. doi: 10.1094/MPMI-20-11-1353 [DOI] [PubMed] [Google Scholar]
- 28. Hauser F, Pessi G, Friberg M, Weber C, Rusca N, Lindemann A, Fischer HM, Hennecke H. 2007. Dissection of the Bradyrhizobium japonicum Nifa+Sigma54 regulon, and identification of a ferredoxin gene (fdxN) for symbiotic nitrogen fixatio. Mol Genet Genomics 278:255–271. doi: 10.1007/s00438-007-0246-9 [DOI] [PubMed] [Google Scholar]
- 29. Carlson TA, Martin GB, Chelm BK. 1987. Differential transcription of the two glutamine synthetase genes of Bradyrhizobium japonicum. J Bacteriol 169:5861–5866. doi: 10.1128/jb.169.12.5861-5866.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carlson TA, Guerinot ML, Chelm BK. 1985. Characterization of the gene encoding glutamine synthetase I (glnA) from Bradyrhizobium japonicum. J Bacteriol 162:698–703. doi: 10.1128/jb.162.2.698-703.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Taté R, Riccio A, Merrick M, Patriarca EJ. 1998. The Rhizobium etli amtB gene coding for an NH4+ transporter is down-regulated early during bacteroid differentiation. Mol Plant Microbe Interact 11:188–198. doi: 10.1094/MPMI.1998.11.3.188 [DOI] [PubMed] [Google Scholar]
- 32. Patriarca EJ, Tatè R, Iaccarino M. 2002. Key role of bacterial NH4+ metabolism in rhizobium-plant symbiosis. Microbiol Mol Biol Rev 66:203–222. doi: 10.1128/MMBR.66.2.203-222.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cabrera JJ, Jiménez-Leiva A, Tomás-Gallardo L, Parejo S, Casado S, Torres MJ, Bedmar EJ, Delgado MJ, Mesa S. 2021. Dissection of Fixk2 protein-DNA interaction unveils new insights into Bradyrhizobium diazoefficiens lifestyles control. Environ Microbiol 23:6194–6209. doi: 10.1111/1462-2920.15661 [DOI] [PubMed] [Google Scholar]
- 34. Mesa S, Hauser F, Friberg M, Malaguti E, Fischer H-M, Hennecke H. 2008. Comprehensive assessment of the regulons controlled by the FixLJ-FixK2-FixK1 cascade in Bradyrhizobium japonicum. J Bacteriol 190:6568–6579. doi: 10.1128/JB.00748-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fischer HM. 1994. Genetic regulation of nitrogen fixation in rhizobia. Microbiol Rev 58:352–386. doi: 10.1128/mr.58.3.352-386.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang SP, Stacey G. 1990. Ammonia regulation of nod genes in Bradyrhizobium japonicum. Mol Gen Genet 223:329–331. doi: 10.1007/BF00265071 [DOI] [PubMed] [Google Scholar]
- 37. Mendoza A, Leija A, Martínez-Romero E, Hernández G, Mora J. 1995. The enhancement of ammonium assimilation in Rhizobium etli prevents nodulation of Phaseolus vulgaris. Mol Plant Microbe Interact 8:584–592. doi: 10.1094/mpmi-8-0584 [DOI] [PubMed] [Google Scholar]
- 38. Bergersen FJ. 1961. The growth of rhizobium in synthetic media. Aust Jnl Of Bio Sci 14:349–360. doi: 10.1071/BI9610349 [DOI] [Google Scholar]
- 39. Guerinot ML, Meidl EJ, Plessner O. 1990. Citrate as a siderophore in Bradyrhizobium japonicum. J Bacteriol 172:3298–3303. doi: 10.1128/jb.172.6.3298-3303.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Norris DO, Date RA. 1976. CAB bulletin, p 134–147. In Shaw NH, Bryan WW (ed), Legume Bacteriology, tropical pasture principles and methods. Vol. 51. [Google Scholar]
- 41. Gao M, Teplitski M, Robinson JB, Bauer WD. 2003. Production of substances by Medicago truncatula that affect bacterial quorum sensing. Mol Plant Microbe Interact 16:827–834. doi: 10.1094/MPMI.2003.16.9.827 [DOI] [PubMed] [Google Scholar]
- 42. Keshavan ND, Chowdhary PK, Haines DC, González JE. 2005. L-Canavanine made by Medicago sativa interferes with quorum sensing in Sinorhizobium meliloti. J Bacteriol 187:8427–8436. doi: 10.1128/JB.187.24.8427-8436.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pietschke C, Treitz C, Forêt S, Schultze A, Künzel S, Tholey A, et al. 2017. Host modification of a bacterial quorum-sensing signal induces a phenotypic switch in bacterial symbionts. Proc Natl Acad Sci USA 114: E8488-e8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bergersen FJ. 1982. Root nodules of legumes: structure and function, p 166. In Bergersen FJ (ed), Research studies press. Letchworth, UK. [Google Scholar]
- 45. Teplitski M, Robinson JB, Bauer WD. 2000. Plants Secrete substances that mimic bacterial N-acyl homoserine lactone signal activities and affect population density-dependent behaviors in associated bacteria. Mol Plant Microbe Interact 13:637–648. doi: 10.1094/MPMI.2000.13.6.637 [DOI] [PubMed] [Google Scholar]
- 46. Berger A, Boscari A, Frendo P, Brouquisse R. 2019. Nitric oxide signaling, metabolism and toxicity in nitrogen-fixing Symbiosis. J Exp Bot 70:4505–4520. doi: 10.1093/jxb/erz159 [DOI] [PubMed] [Google Scholar]
- 47. Romero M, Muro-Pastor AM, Otero A. 2011. Quorum sensing N-acyl homoserine lactone signals affect nitrogen fixation in the cyanobacterium anabaena sp. PCC7120. FEMS Microbiol Lett 315:101–108. doi: 10.1111/j.1574-6968.2010.02175.x [DOI] [PubMed] [Google Scholar]
- 48. Cole MA, Elkan GH. 1973. Transmissible resistance to penicillin G, neomycin, and chloramphenicol in Rhizobium japonicum. Antimicrob Agents Chemother 4:248–253. doi: 10.1128/AAC.4.3.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sadowsky MJ, Tully RE, Cregan PB, Keyser HH. 1987. Genetic diversity in Bradyrhizobium japonicum serogroup 123 and its relation to genotype-specific nodulation of soybean. Appl Environ Microbiol 53:2624–2630. doi: 10.1128/aem.53.11.2624-2630.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wei M, Yokoyama T, Minamisawa K, Mitsui H, Itakura M, Kaneko T, Tabata S, Saeki K, Omori H, Tajima S, Uchiumi T, Abe M, Ohwada T. 2008. Soybean seed extracts preferentially express genomic loci of Bradyrhizobium japonicum in the initial interaction with soybean, Glycine max (L.) Merr. DNA Res 15:201–214. doi: 10.1093/dnares/dsn012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Heckman KL, Pease LR. 2007. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc 2:924–932. doi: 10.1038/nprot.2007.132 [DOI] [PubMed] [Google Scholar]
- 52. Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. 1994. Small Mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids PK18 and PK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. doi: 10.1016/0378-1119(94)90324-7 [DOI] [PubMed] [Google Scholar]
- 53. Hattori Y, Omori H, Hanyu M, Kaseda N, Mishima E, Kaneko T, Tabata S, Saeki K. 2002. Ordered Cosmid library of the Mesorhizobium loti MAFF303099 genome for systematic gene disruption and complementation analysis. Plant Cell Physiol 43:1542–1557. doi: 10.1093/pcp/pcf175 [DOI] [PubMed] [Google Scholar]
- 54. Kalogeraki VS, Winans SC. 1997. Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create Fusions to target genes of diverse bacteria. Gene 188:69–75. doi: 10.1016/s0378-1119(96)00778-0 [DOI] [PubMed] [Google Scholar]
- 55. Wilson KJ, Sessitsch A, Corbo JC, Giller KE, Akkermans AD, Jefferson RA. 1995. Beta-glucuronidase (GUS) transposons for ecological and genetic studies of rhizobia and other gram-negative bacteria. Microbiology 141:1691–1705. doi: 10.1099/13500872-141-7-1691 [DOI] [PubMed] [Google Scholar]
- 56. Ramsay JP. 2013. High-throughput β -galactosidase and β-glucuronidase assays using fluorogenic substrates. Bio-protocol 3:e827. doi: 10.21769/BioProtoc.827 [DOI] [Google Scholar]
- 57. Miller JH. 1972. Experiments in molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 58. Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and Isoform switching during cell differentiation. Nat Biotechnol 28:511–515. doi: 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bu D, Luo H, Huo P, Wang Z, Zhang S, He Z, Wu Y, Zhao L, Liu J, Guo J, Fang S, Cao W, Yi L, Zhao Y, Kong L. 2021. KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res 49(W1):317–325. doi: 10.1093/nar/gkab447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Han J-T, Li D-Y, Zhang M-Y, Yu X-Q, Jia X-X, Xu H, Yan X, Jia W-J, Niu S, Kempher ML, Tao X, He Y-X. 2021. EmhR is an indole-sensing transcriptional regulator responsible for the indole-induced antibiotic tolerance in Pseudomonas fluorescens. Environ Microbiol 23:2054–2069. doi: 10.1111/1462-2920.15354 [DOI] [PubMed] [Google Scholar]
- 63. Demichev V, Messner CB, Vernardis SI, Lilley KS, Ralser M. 2020. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat Methods 17:41–44. doi: 10.1038/s41592-019-0638-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Delgado MJ, Bonnard N, Tresierra-Ayala A, Bedmar EJ, Müller P. 2003. The Bradyrhizobium japonicum napEDABC genes encoding the periplasmic nitrate reductase are essential for nitrate respiration. Microbiology 149:3395–3403. doi: 10.1099/mic.0.26620-0 [DOI] [PubMed] [Google Scholar]
- 65. Bueno E, Bedmar EJ, Richardson DJ, Delgado MJ. 2008. Role of Bradyrhizobium japonicum cytochrome C550 in nitrite and nitrate respiration. FEMS Microbiol Lett 279:188–194. doi: 10.1111/j.1574-6968.2007.01034.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Inoculation experiment, promoter analysis, bacterial growth, etc.
Bacterial strains, Omics data, etc.