Abstract
The nucleus is a mechanically stable compartment of the cell that contains the genome and performs many essential functions. Nuclear mechanical components chromatin and lamins maintain nuclear shape, compartmentalization, and function by resisting antagonistic actin contraction and confinement. Studies have yet to compare chromatin and lamins perturbations side-by-side as well as modulated actin contraction while holding confinement constant. To accomplish this, we used nuclear localization signal green fluorescent protein to measure nuclear shape and rupture in live cells with chromatin and lamin perturbations. We then modulated actin contraction while maintaining actin confinement measured by nuclear height. Wild type, chromatin decompaction, and lamin B1 null present bleb-based nuclear deformations and ruptures dependent on actin contraction and independent of actin confinement. Actin contraction inhibition by Y27632 decreased nuclear blebbing and ruptures while activation by CN03 increased rupture frequency. Lamin A/C null results in overall abnormal shape also reliant on actin contraction, but similar blebs and ruptures as wild type. Increased DNA damage is caused by nuclear blebbing or abnormal shape which can be relieved by inhibition of actin contraction which rescues nuclear shape and decreases DNA damage levels in all perturbations. Thus, actin contraction drives nuclear blebbing, bleb-based ruptures, and abnormal shape independent of changes in actin confinement.
Studies have yet to determine the separate roles of actin contraction and actin confinement in controlling nuclear shape and ruptures.
We find that actin contraction determines nuclear blebbing, bleb-based ruptures, and abnormal nuclear shape independent of changes in actin confinement.
These findings reshape our understanding of how actin antagonizes nuclear shape.
INTRODUCTION
The nucleus is the organelle that protects and compartmentalizes the genome and its vital functions that dictate cellular behavior. The nucleus and its main mechanical components chromatin and lamins must properly resist cytoskeletal and/or external forces to maintain the shape and integrity of the nucleus as a compartment (Kalukula et al., 2022). Abnormal nuclear morphology is a major hallmark of human disease spanning aging to heart disease to cancer and many others (Butin-Israeli et al., 2012; Stephens et al., 2019a; Stephens, 2020). In perturbed nuclei, actin running over the top and along the sides of the nucleus antagonizes and causes abnormal nuclear morphology and ruptures (Khatau et al., 2009; Le Berre et al., 2012; Hatch and Hetzer, 2016). Loss of nucleus compartmentalization via nuclear rupture results in spilling of nuclear contents into the cytoplasm and vice versa (De Vos et al., 2011; Vargas et al., 2012). Nuclear ruptures have been shown to lead to dysfunction that underlies human disease including causing DNA damage/genomic instability (Denais et al., 2016; Irianto et al., 2016; Raab et al., 2016; Chen et al., 2018; Xia et al., 2018; Stephens et al., 2019b; Shah et al., 2021), altering transcription (De Vos et al., 2011; Helfand et al., 2012), and disruption of cell-cycle control (Pfeifer et al., 2018). However, it remains unclear what the separate roles of actin contraction versus actin confinement are in relation to nuclear shape and rupture, respectively, the hallmark and driver of human disease.
Actin controls nuclear shape (Khatau et al., 2009), but it remains unclear how to separate the effects of actin contraction from actin confinement for both nuclear shape and ruptures. The current dogma is that the actin cytoskeleton compresses the nucleus to cause nuclear blebbing supported by experiments depolymerizing actin which restores shape and compartmentalization. Placing the nucleus under artificial confinement then causes the return of nuclear blebbing and rupture in perturbed nuclei (Le Berre et al., 2012; Hatch and Hetzer, 2016). However, in these studies loss of actin disrupts both actin contraction and confinement while artificial confinement induces greater levels of confinement than caused by actin in WT nuclei. Furthermore, pivotal studies of confined migration rely on both confinement as well as actin contraction for migration (Denais et al., 2016) because they rely on hormone gradients that both induce directed migration and increase actin contraction (Schneider et al., 2009). A recent paper revealed that indeed modulation of actin contraction while under artificial confinement leads to drastic changes in nuclear blebbing and rupture potential (Mistriotis et al., 2019). Thus, actin depolymerization and confined migration respectively remove or activate both actin contraction and confinement.
The alternative hypothesis is that actin contraction and not confinement controls nuclear shape and ruptures. Separation of these two roles of actin could be accomplished using a two-dimensional cell culture system (no artificial confinement), modulation of actin contraction, and the ability to measure actin confinement. Modulation of actin contraction can be accomplished by Rho Activator II, which we denote as CN03 (De Silva et al., 2015), and Rho-associated kinase inhibitor Y27632 (Rees et al., 2001; Inoue-Mochita et al., 2015) which both ultimately alter phosphorylation of myosin light chain 2 (pMLC2). To properly assay the independent role of actin contraction in nuclear shape and ruptures, one would need to determine its role across wild-type (WT) cells and the most reported nuclear perturbations.
The major components of the nucleus responsible for resisting antagonistic actin forces to maintain nuclear shape and compartmentalization include chromatin compaction, lamin B1, and lamin A/C. Perturbations of chromatin structure results in a weaker nucleus, nuclear blebbing, and rupture via altering levels of histone modification state (Stephens et al., 2018, 2019b; Tamashunas et al., 2020; Schibler et al., 2023), H1 dynamics (Furusawa et al., 2015; Senigagliesi et al., 2019), and most recently HP1α (Strom et al., 2021) and Hi-C (Belaghzal et al., 2021). Lamin B1 loss was one of the first reported perturbations that resulted in nuclear blebbing and rupture (Lammerding et al., 2006; Vargas et al., 2012). The mechanism remains unclear because different publications have reported conflicting roles of lamin B1 to nuclear mechanics as none (Lammerding et al., 2006), stronger in the lamin regime (Shin et al., 2013; Stephens et al., 2017), and weaker (Vahabikashi et al., 2022). Finally, perturbation of lamin A/C via knockdown (shRNAi) or knockout (LMNA−/−) is reported to result in reduced nuclear rigidity (Lammerding et al., 2006; Pajerowski et al., 2007; Swift et al., 2013; Stephens et al., 2017; Hobson et al., 2020) and overall abnormal nuclear shape including lamin A mutants associated with Progeria (Goldman et al., 2004; Lammerding et al., 2006; Robijns et al., 2016; Soria-Valles et al., 2016; Stephens et al., 2018; Köhler et al., 2020). However, depending on perturbation of lamin A/C or whether under confined migration the nucleus can also present nuclear blebbing (Goldman et al., 2004; Coffinier et al., 2010; De Vos et al., 2011; Chen et al., 2018). Recent work has detailed these different perturbations’ effect on overall cell and nuclear mechanics (Vahabikashi et al., 2022). Interestingly, all of these perturbations disrupt chromatin histone modification state, but do not necessarily show the same phenotype. Thus, individual studies focusing on either chromatin decompaction, lamin B1 loss, and lamin A/C loss are known to result in loss of nuclear shape and rupture, but they have yet to be compared side-by-side.
Here we image mouse embryonic fibroblasts (MEF) with NLS-GFP to track nuclear shape and rupture across multiple nuclear perturbations while modulating actin contraction. Rupture of the nucleus can be assayed by tracking nuclear localization signal green fluorescent protein (NLS-GFP), which provides a diffusible fluorescence marker that accumulates in the nucleus and spills into the cytoplasm upon nuclear envelope rupture. Using NLS-GFP we compare the nuclear shape and rupture behaviors across WT and the nuclear perturbations of chromatin decompaction, lamin B1 null cells (LMNB1−/−), and lamin A/C null cells (LMNA−/−). We establish that actin contraction can be increased (CN03) and decreased (Y27632) without affecting actin confinement in almost all cases. With this approach we find significant changes in nuclear shape and ruptures are driven by actin contraction. However, this change only impacts WT, chromatin decompaction, and lamin B1 null bleb-based nuclear shape change and ruptures, but not abnormally shaped nonbleb-based lamin A/C null. We reveal that lamin B1 is a chromatin decompaction perturbation while lamin A/C has the capacity to display bleb-based nuclear ruptures, but only when additionally perturbed with VPA. Overall, we provide novel data that actin contraction is essential to nuclear blebbing and rupture while actin confinement is not.
RESULTS
A cross comparison of nuclear shape and compartmentalization characteristics in chromatin and lamin perturbations
Loss of chromatin compaction, lamin B1, or lamin A/C are all known to cause abnormal nuclear morphology and rupture, but no studies have done a cross comparison. To accomplish this, we used stable MEF cell lines expressing NLS-GFP to provide live cell tracking over 3 h at 2-min intervals. NLS-GFP concentrates in the nucleus to provide nuclear shape measurements and spills into the cytoplasm upon nuclear rupture. Specifically, we scored nuclei as blebbed with > 1 µm diameter protrusion from the main nuclear body and ruptures as a > 25% increase in the NLS-GFP intensity ratio cell/nucleus (Supplemental Figure 1; Supplemental Movies 1 and 2; see Materials and Methods). NLS-GFP can then also reaccumulate post nuclear rupture, which reseal on the order of 10 min, to provide continual measurements of nuclear shape and the ability to track multiple ruptures for one nucleus (Supplemental Figure 1; Supplemental Movie 3; [Halfmann et al., 2019; Young et al., 2020]). Using NLS-GFP we tracked nuclear shape, rupture, type of rupture, and frequency of ruptures for WT and perturbations of chromatin decompaction (VPA), loss of lamin B1 (LMNB1−/−), and loss of lamin A/C (LMNA−/−).
Video S1.
Bleb‐based nuclear rupture. Example video of a MEF cell expressing NLS‐GFP which is concentrated in a blebbed nucleus but spills into the cytoplasm upon rupture. Quantification shown in Supplemental Figure 1.
Video S2.
Non‐bleb‐based nuclear rupture. Example video of a MEF cell expressing NLS‐GFP which is concentrated in a normally ellipse shaped nucleus but spills into the cytoplasm upon rupture. Quantification shown in Supplemental Figure 1.
Video S3.
Single nucleus multiple bleb‐based nuclear ruptures. Example video of a MEF cell expressing NLS‐GFP which is concentrated in a blebbed nucleus but spills into the cytoplasm upon rupture multiple times in the same nucleus. Each rupture is followed by healing and re‐concentration of the NLS‐GFP in the cytoplasm in about 10 minutes. Quantification shown in Supplemental Figure 1.
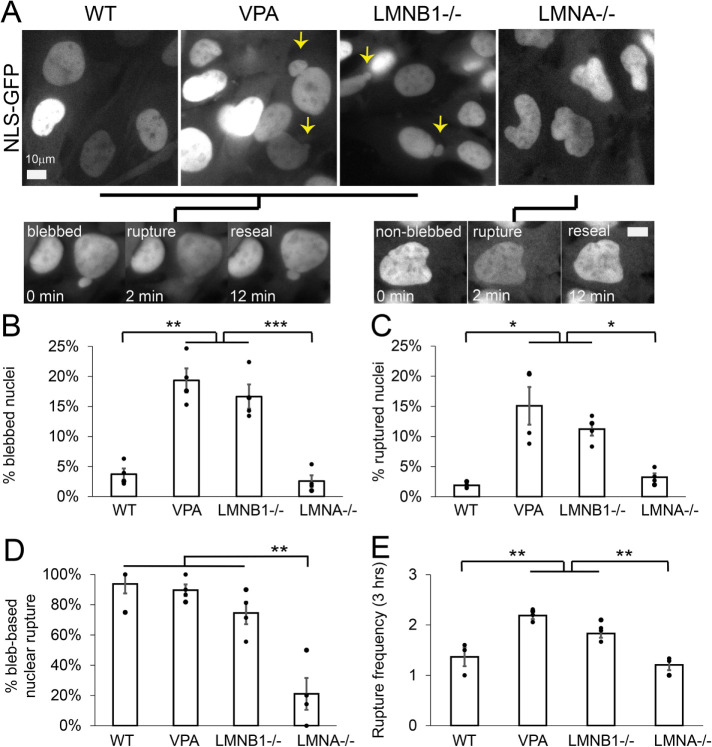
First, we tracked nuclear blebbing and rupture across all conditions (Figure 1, A–C). WT nuclei exhibit low levels of nuclear blebbing and rupture at 4 ± 1%. Chromatin decompaction, via histone deacetylase inhibitor valproic acid (VPA), and loss of lamin B1 (LMNB1−/−) both resulted in significantly increased nuclear blebbing (15–20%) and rupture (10–15%) compared with WT, consistent with previous reports (Lammerding et al., 2006; Vargas et al., 2012; Hatch and Hetzer, 2016; Chen et al., 2018; Stephens et al., 2019b; Berg et al., 2023). Interestingly, loss of lamin A/C (LMNA−/−) does not result in either an increase in nuclear blebbing or rupture compared with WT. Instead, LMNA−/− nuclei present as abnormally shaped nuclei that quantitatively measured decreased nuclear circularity (Supplemental Figure 2). This data is consistent with previous reports (Broers et al., 2004; Lammerding et al., 2006; Nmezi et al., 2019). Thus, loss of nuclear shape via nuclear blebbing correlates with increased nuclear rupture.
FIGURE 1:
Nuclear morphology and ruptures are bleb-based in WT, VPA, and LMNB1−/−, but LMNA−/− presents differently. (A) Example images of MEF WT, VPA, LMNB1−/−, and LMNA−/− via NLS-GFP and black lines connecting each to either an example of bleb-based or non-bleb-based nuclear rupture. Graphs of the percentage of nuclei that (B) display a nuclear bleb, (C) rupture at least once, (D) display bleb-based nuclear rupture, and (E) number of nuclear ruptures for a single nucleus that ruptures. All data was measured over a 3-h period of observation at 2-min imaging intervals. Each condition has four biological replicates shown as dots. n = WT, 102, 127, 139, and 158; VPA, 227, 146, 170, and 108; LMNB1–/–, 192, 216, 119, and 164; LMNA–/–, 111, 142, 149, and 101. Student’s t test p values reported as * < 0.05, ** < 0.01, *** < 0.001, no asterisk denotes no significance, p >0.05. Error bars represent standard error. Scale bar = 10 µm.
To determine whether nuclear blebbing could be the main mechanism for nuclear rupture, we tracked nuclear shape upon nuclear rupture. Bleb-based nuclear rupture accounted for > 75% of all nuclear ruptures in WT, VPA, and LMNB1−/− nuclei (Figure 1, A and D). Again, and interestingly, LMNA−/− cells displayed a different behavior where the majority of nuclear ruptures occurred in nonblebbed nuclei at ∼ 80% of the time, a complete reversal relative to the other conditions. Further analysis shows that nuclear circularity measurements are similar for nuclei that rupture and nuclei that do not rupture, suggesting ruptures do not only occur in abnormally shaped low circularity nuclei but evenly across the population of LMNA−/− nuclei (Supplemental Figure 3). Thus, nuclear blebbing is the main cause of nuclear rupture for both WT and most, but not all, nuclear perturbations.
Finally, to determine the frequency of nuclear ruptures from a single nucleus, we tracked the number of nuclear ruptures per nucleus that ruptures over a 3-h period (Figure 1E). The pattern remained consistent with WT having infrequent multiple nuclear ruptures per nucleus averaging 1.3 ± 0.2. Both VPA and LMNB1−/− nuclei significantly increased the frequency of nuclear ruptures per nucleus to 2.2 ± 0.1 and 1.9 ± 0.1, respectively. LMNA−/− nuclei displayed similar rupture frequency as WT, 1.2 ± 0.1. Thus, increased nuclear blebbing and bleb-based ruptures upon chromatin decompaction or loss of lamin B1 also results in increased frequency of ruptures in those nuclei.
Actin contraction modulation independent of actin confinement
Nuclear shape and compartmentalization are determined by the nucleus’ ability to resist antagonistic cytoskeletal and/or external forces working to compress, deform, and rupture the nucleus. Actin contraction is generated by activated myosin motors sliding actin filaments (Murrell et al., 2015) and can be measured by pMLC2 (Figure 2A). Actin confinement is due to actin cables running over the top of the nucleus to compress it, which can be measured by nuclear height (Figure 2B). Both actin contraction and confinement can antagonize nuclear shape and stability, but their relative roles remain unknown.
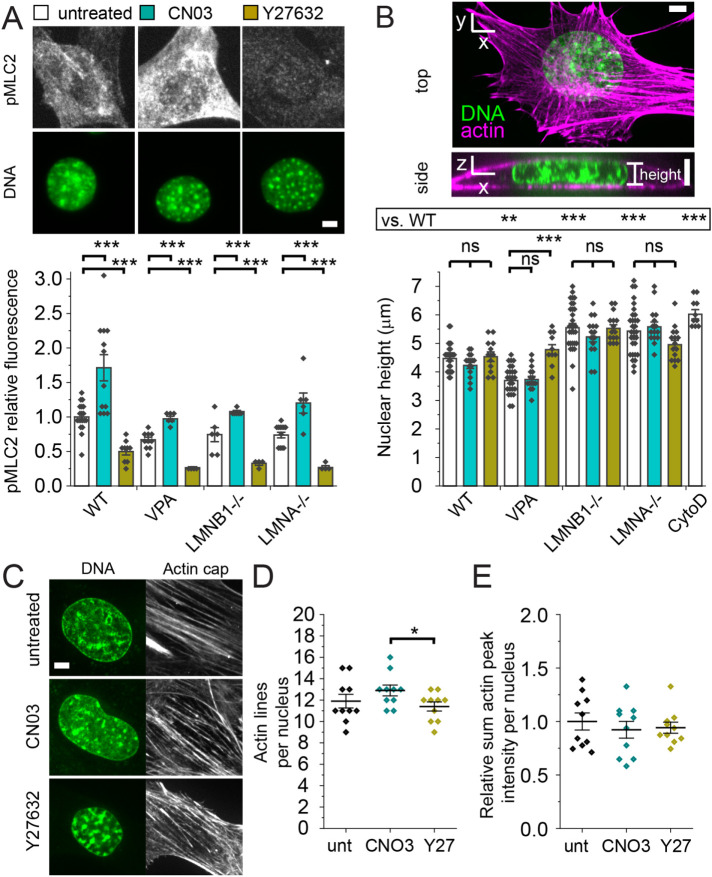
FIGURE 2:
Actin contraction can be modulated while keeping actin confinement constant. (A) Example images of cells labeled for pMLC2 (white) via immunofluorescence and nuclei stained with Hoechst (DNA, green). Bar graph of relative fluorescence of pMLC2 for WT, VPA, LMNB1−/−, and LMNA−/− without modulation (white bar), with Rho activator II CN03 (turquoise bar), or with ROCK inhibitor Y27632 (gold bar). Biological replicates (diamonds; unmodulated n ≥ 6, CNO3 n ≥ 6, and Y27632 n ≥ 4), each consists of ≥ 20 cells. (B) Representative images of DNA (green) and actin (purple) showing a vertical slice for height and a horizontal maximum projection. Graph of nuclear height measured by confocal imaging of DNA stained by Hoechst for WT, VPA, LMNB1−/−, and LMNA−/− without modulation (white bar), with Rho activator II CN03 (turquoise bar), or with ROCK inhibitor Y27632 (gold bar). Unmodulated n = 25–30 nuclei; CNO3 n = 15 nuclei, Y27632 n = 10–15 nuclei. WT treated with cytochalasin D (CytoD, actin depolymerization, n = 10) serves as a known control for increase in height. (C–E) Example confocal images of nucleus (DNA via Hoechst, green) and actin running over the top of the nucleus labeled via SPY555-Actin Probe (white). Line scans of actin on top of the nucleus provide (D) number of actin lines and (E) relative sum fluorescence intensity for MEF WT cells untreated (unt), CN03, and Y27632 (Y27), n = 10 nuclei each. Student’s t test p values reported as * < 0.05, ** < 0.01, *** < 0.001, or ns denotes no significance, p > 0.05. Error bars represent standard error. Scale bar = 5 µm.
To determine whether actin contraction and/or actin confinement are partially responsible for changes in nuclear shape and rupture upon nuclear perturbations, we tracked both in WT, VPA, LMNB1−/−, and LMNA−/−. First, we measured actin contraction levels by immunofluorescence of pMLC2. All nuclear perturbations measured a similar level of pMLC2, suggesting changes in nuclear shape via chromatin and lamin perturbations do not arise from changes in actin contraction (Figure 2A). Next, we measured nuclear height to determine the level of actin confinement from actin that runs over the top of the nucleus compressing it (Figure 2B). A control for loss of actin confinement is actin depolymerization by cytochalasin D, where nuclear height increases significantly (CytoD; Figure 2B). Nuclear height decreased upon chromatin decompaction via VPA, as expected because the nucleus is softer (Krause et al., 2013; Shimamoto et al., 2017; Stephens et al., 2017; Hobson et al., 2020). However, LMNB1−/− and LMNA−/− nuclei, both reported to be softer as well (Vahabikashi et al., 2022), did not decrease in nuclear height but instead increased in nuclear height relative to WT (Figure 2B). This data suggests that actin confinement is not a driver of nuclear shape change in loss of either lamin B1 or lamin A/C. More specifically, the data suggests that actin confinement is not a driver of nuclear blebbing because nuclear height of VPA-treated and LMNB1−/− nuclei differs by >1 µm or 25% but results in the same drastic increase to 15–20% nuclear blebbing. Measurements of nuclear height to assess actin confinement in normal versus blebbed LMNB1−/− nuclei reveal no change, further supporting the idea that actin confinement changes are not essential to nuclear blebbing (Supplemental Figure 4A). Overall, nuclear perturbations to chromatin or lamins do not impact actin contraction or confinement in a consistent manner.
While actin contraction stays constant in nuclear perturbations, we hypothesized that it may have an important role in antagonizing nuclear shape and compartmentalization. To modulate actin contraction, we used drugs previously established to affect the Rho pathway via activator CN03 (De Silva et al., 2015) and Rho-associated kinase inhibitor Y27632 (Rees et al., 2001; Inoue-Mochita et al., 2015). Immunofluorescence measurements of pMLC2 confirm across WT and all nuclear perturbations in our study that activator CN03 significantly increases actin contraction while inhibitor Y27632 drastically decreases it (Figure 2A), in agreement with other reports (Hernandez et al., 2020). Thus, we established methodology to modulate actin contraction to study its effect on nuclear shape and ruptures.
Alterations in actin contraction could also affect actin confinement. To determine whether actin confinement changes upon modulation of action contraction via activator CN03 and inhibitor Y27632, we measured nuclear height in all conditions. In WT and all nuclear perturbations, except VPA Y27632, nuclear height did not change significantly upon increased or decreased actin contraction (p > 0.05; Figure 2B). Further imaging of actin running over the top of the nucleus revealed no change in number and fluorescence intensity of actin lines upon actin contraction modulation (p > 0.05; Figure 2, C–E). Qualitative imaging of cellular actin structure also appears unchanged upon modulation of actin contraction (Supplemental Figure 5). Thus, actin contraction in most cases can be modulated independently of actin confinement, actin levels on top of the nucleus, and macro actin structure. This data supports an approach that allows us to determine the role of actin contraction in antagonizing nuclear shape and stability.
Actin contraction is an essential determinant of bleb-based nuclear shape deformations and ruptures
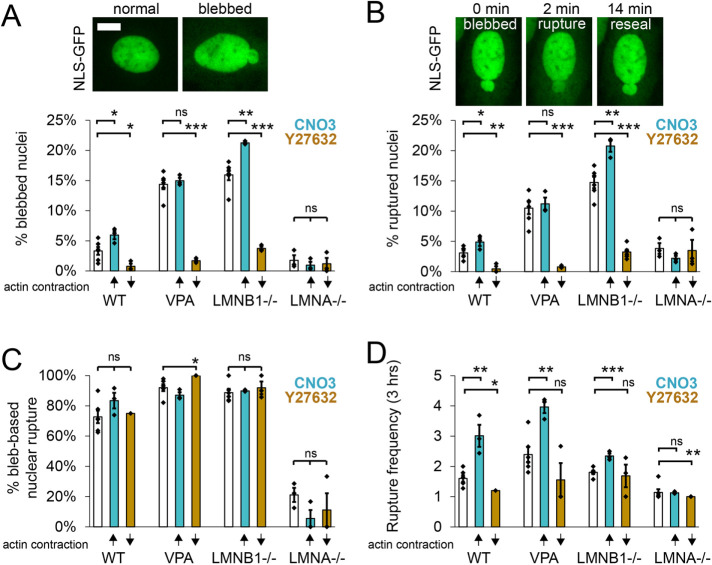
To determine the effect of actin contraction in nuclear blebbing and rupture, we first increased actin contraction via CN03 across all conditions and imaged NLS-GFP for 3 h at 2-min intervals. Both WT and lamin B1 null treated with CN03 to increase actin contraction resulted in both increased percentage of nuclei that blebbed and ruptured (teal; Figure 3, A and B), in agreement with the use of calyculin A (Nmezi et al., 2019). However, chromatin decompaction via VPA showed no change in either blebbing or rupture upon activation of actin contraction via CN03. Tracking nuclear shape in which nuclei ruptured reveals no change in the base behavior of each condition (bleb-based vs. nonbleb-based rupture; Figure 3C). Finally, CN03 increased the frequency of ruptures for a single nucleus for all bleb-based nuclear rupture conditions WT, VPA, and LMNB1−/− (Figure 3D). This is interesting because while CN03 did not increase the number of nuclei that blebbed or ruptured for VPA, it does nearly double the number of ruptures for a single nucleus that does rupture over a 3-h period. Again, non-bleb-based LMNA−/− showed no change upon CN03 treatment, suggesting it was insensitive. Overall, increased actin contraction via CN03 can increase nuclear blebbing, rupture, and frequency of ruptures for perturbations presenting a bleb-based nuclear rupture phenotype.
FIGURE 3:
Actin contraction controls nuclear blebbing and bleb-based ruptures. Example images of (A) normal and blebbed nuclei as well as (B) bleb-based nuclear rupture via NLS-GFP time lapse imaging. (A–D) Graphs showing the percentage of (A) blebbed nuclei, (B) nuclear ruptures, and (C) bleb-based ruptures along with (D) the number of ruptures (frequency) of a single nucleus that ruptures over 3 h imaged at 2-min intervals for WT, VPA, LMNB1−/−, and LMNA−/− without modulation (white bar), with increased actin contraction (turquoise bar, CN03), or with decreased actin contraction (gold bar, Y27632). Six biological replicates for unmodulated and three biological replicates for increased or decreased actin contraction, experiments represented by black dots, n = 75–400 cells each experiment. Student’s t test p values reported as * < 0.05, ** < 0.01, *** < 0.001, or ns denotes no significance, p > 0.05. Error bars represent standard error. Scale bar = 10 µm.
Next, we determined the effects of actin contraction inhibition on nuclear blebbing and ruptures in WT and nuclear perturbations. Decreased actin contraction via Y27632 revealed a drastic and consistent response decreasing the percentage of nuclei that presented blebs and ruptures for bleb-based conditions WT, VPA, and LMNB1−/− (gold, Y27632; Figure 3, A and B). Even WT’s low levels of nuclear blebbing and ruptures decreased significantly from ∼3% to < 0.5% upon actin contraction inhibition, suggesting it is essential for these behaviors. Both VPA and LMNB1−/− perturbations showed a similar nearly essential need for actin contraction to form blebs and cause ruptures as these dropped from 10–20% to < 1–3%. This data agrees with published data that actin contraction inhibition decreased nuclear blebbing and rupture in melanoma cell culture model (Jung-Garcia et al., 2023). However, LMNA−/− cells’ low percentages of nuclear blebs and ruptures remained unchanged upon decreased actin contraction, continuing the trend of insensitivity to changes in actin contraction. However, nuclear shape measured by circularity in LMNA−/− was significantly improved upon actin contraction inhibition (Supplemental Figure 2), showing that LMNA−/− is not completely insensitive. The type of rupture, bleb-based or non, remained similar as well as number of ruptures per nucleus that ruptures (Figure 3, C and D), with minor exceptions. Thus, bleb-based nuclear rupture conditions WT, chromatin decompaction, and lamin B1 null require actin contraction for nuclear blebbing and rupture.
In summary, we find that actin contraction is essential for the bleb-based nuclear rupture phenotype. Furthermore, increased actin contraction can drive more cells to bleb and rupture while driving the number of ruptures for a single nucleus higher as well. Oppositely, actin contraction has little effect on the percentage of nuclei that bleb and rupture in the non-blebbed rupture phenotype of LMNA−/−. Taken together this data supports that, independent of actin confinement, actin contraction is a major determinant of bleb-based nuclear shape and rupture.
Blebbed or abnormal nuclei present higher levels of DNA damage
Loss of nuclear shape and compartmentalization causes nuclear dysfunction. One of the most common measures of nuclear dysfunction upon abnormal nuclear deformation and rupture is DNA damage. Thus, we tracked DNA damage via γH2AX foci relative to nuclear shape in WT, VPA, LMNB1−/−, and LMNA−/−. To determine the number of DNA damage foci in a nucleus, we experimentally measured the Gaussian full width half maximum of a diffraction limited focus and empirically determined intensity threshold (see Materials and Methods). Relative to normally shaped nuclei, blebbed nuclei in WT, VPA, and LMNB1−/− displayed greater than a twofold increase in DNA damage on average (Figure 4, A and B), in agreement with previous work (Stephens et al., 2019b). LMNA−/− abnormally shaped nuclei, determined by circularity <0.9, also displayed a significant increase in DNA damage relative to normally shaped nuclei. Thus, across WT and different nuclear perturbations deformed nuclei present drastically increased levels of DNA damage.
FIGURE 4:
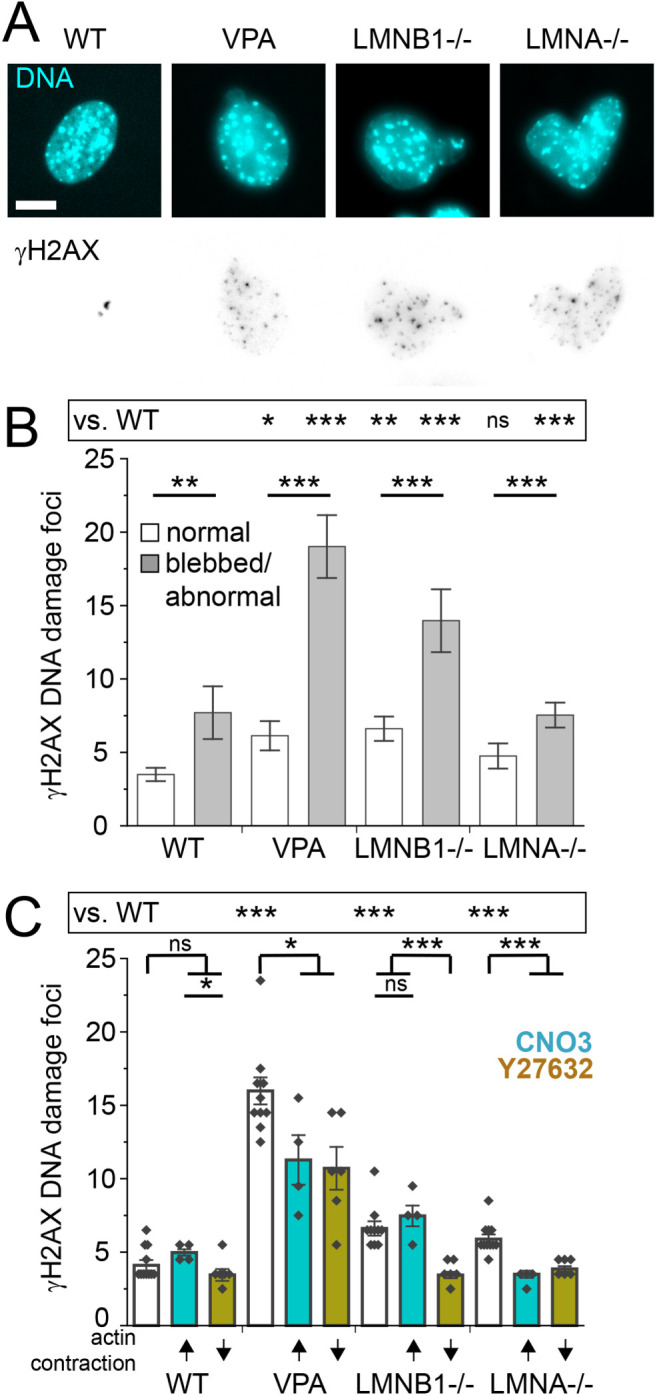
Increased DNA damage is associated with nuclear shape and can be rescued by actin contraction inhibition. (A) Representative images of nucleus shape via labeling DNA (Hoechst, Cyan) and DNA damage foci (γH2AX, inverted gray scale). (B) Graph of number of γH2AX DNA damage foci for normally shaped nuclei versus blebbed/abnormal (WT normal n = 115, blebbed n = 17; VPA normal n = 119, blebbed n = 55; LMNB1−/− normal n = 103, blebbed n = 32, LMNA–/– normal n = 73, abnormal circularity < 0.9 n = 214 [see Supplemental Table 1]). (C) Graph of number of γH2AX DNA damage foci for WT, VPA, LMNB1−/−, and LMNA−/− without modulation (white bar), with increased actin contraction (turquoise bar, CN03), or with decreased actin contraction (gold bar, Y27632). Multiple biological replicates of unmodulated (n = 10), increased (n = 4) and decreased (n = 6) actin contraction. Experiments represented by black dots, where n > 40 cells per experiment. Student’s t test p values reported as * < 0.05, ** < 0.01, *** < 0.001, or ns denotes no significance, p > 0.05. Error bars represent standard error. Scale bar = 10 µm.
To determine whether actin contraction-based changes significantly impact DNA damage levels, we measured the number of γH2AX foci upon activation and inhibition of actin contraction. DNA damage measured by γH2AX foci did not significantly increase upon CN03 activation of actin contraction, suggesting that the slight yet significant increase in nuclear blebbing, rupture, and rupture frequency from CN03 did not have a significant impact on DNA damage (Figure 4, C and B). Treatment of all conditions with actin contraction inhibitor Y27632 significantly decreased DNA damage levels (gold, Figure 4C). These results might be expected given that Y27632 drastically decreases nuclear blebbing and abnormal nuclear morphology across all conditions (Figure 3; Supplemental Figure 2), recapitulating decreased DNA damage in normally shaped nuclei (Figure 4, A and B). Overall, this data shows actin contraction driven antagonism of nuclear shape results in nuclear dysfunction measured by increased DNA damage, independent of changes in nuclear confinement.
LMNB1–/– loss of heterochromatin is sufficient to generate nuclear blebs and ruptures
We hypothesized that the similarities between VPA and LMNB1−/− might be due to the shared perturbation of changes in histone modification state. While VPA treatment does not cause lamin B1 loss (Stephens et al., 2018), it has been reported by many that LMNB1−/− nuclei display chromatin decompaction via loss of facultative heterochromatin (Camps et al., 2014; Stephens et al., 2018; Vahabikashi et al., 2022). To determine whether LMNB1−/− nuclei bleb-based phenotype could be due to facultative heterochromatin loss, we recapitulated this facultative heterochromatin loss via treatment with EZH2 methyltransferase inhibitor GSK126 (McCabe et al., 2012). Immunofluorescence measurements confirm a significant decrease of facultative heterochromatin marker H3K27me3 in LMNB1−/− nuclei relative to WT (53 ± 3% loss, Figure 5A). Treatment with GSK126 revealed a significant decrease in H3K27me3 on the order of LMNB1–/– (39 ± 2% loss, Figure 5A). Thus, GSK126 treatment roughly recapitulates LMNB1−/− loss of facultative heterochromatin.
FIGURE 5:
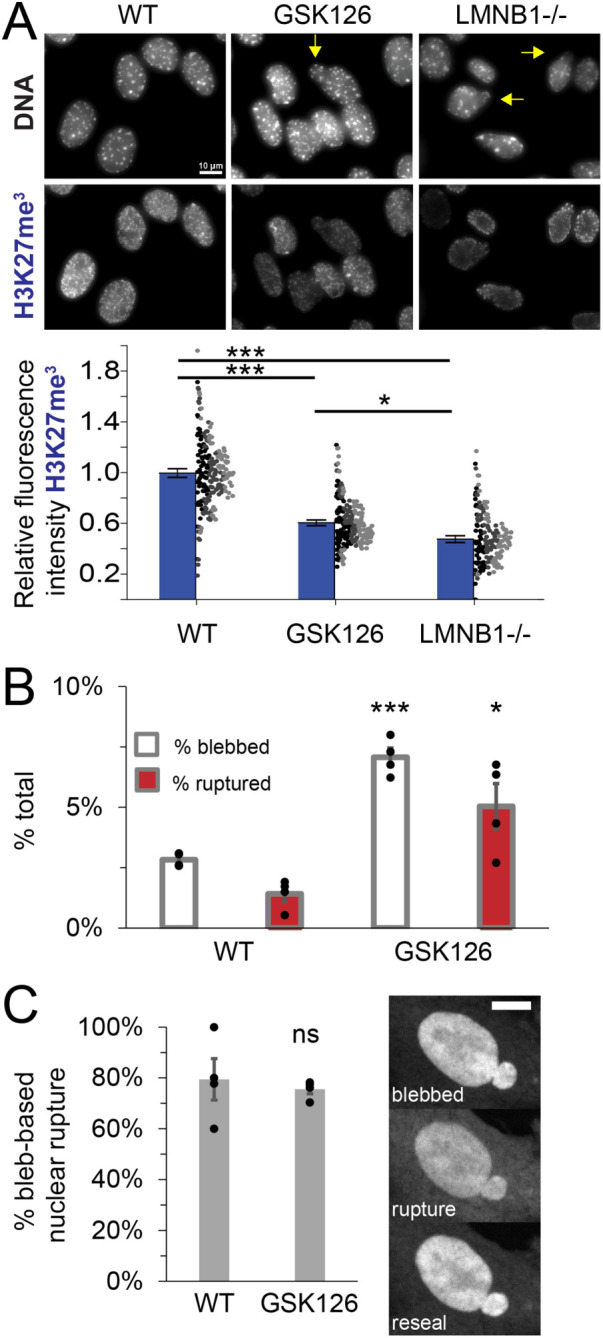
Recapitulating LMNB1−/− loss of facultative heterochromatin alone is sufficient to phenocopy increased nuclear blebbing and rupture. (A) Representative images of nuclei stained for DNA via Hoechst and facultative heterochromatin via H3K27me3 (blue). Yellow arrow denotes blebbed nucleus. Graph of relative fluorescence intensity of H3K27me3 for WT, EZH2 inhibitor (GSK126), and lamin B1 null (LMNB1−/−). Dots are individual measures n > 50 nuclei per experiment from three biological triplicates, denoted by color (black, dark gray, light gray), were averaged (blue bar). (B) Graph of percentage total of cells displaying nuclear blebs (white) or ruptures (red) for WT and GSK 126. (C) Graph of percentage bleb-based nuclear ruptures for WT and GSK 126. Four biological replicates displayed as dots with > 250 cells each. Blebbing and ruptures in GSK126 are dependent on actin contraction as Y27632 decreases blebbing to 1% and ruptures to 0% (GSK126 + Y27632 n = 226 cells, p > 0.01, Supplemental Table 1), data not graphed. Student’s t test p values reported as * < 0.05, ** < 0.01, *** < 0.001, no asterisk or ns denotes no significance, p > 0.05. Error bars represent standard error. Scale bar = 10 µm.
To determine whether loss of facultative heterochromatin is sufficient to induce the bleb-based phenotype of LMNB1−/−, we measured nuclear shape and ruptures in GSK126-treated nuclei. NLS-GFP nuclear shape and rupture tracking in GSK126-treated cells reveals a significant increase in nuclear blebbing and nuclear ruptures compared with WT (Figure 5B). Nuclear ruptures were also found to be bleb-based in the majority of cases similar to WT (>80%, Figure 5C). Actin contraction was essential for nuclear bleb formation in GSK126-treated cells, as treatment with inhibitor Y27632 decreased nuclear blebbing and rupture to 1% or less (Supplemental Table 1). Overall, this data suggests that most of LMNB1−/− nuclear phenotype could be due to loss of facultative heterochromatin. Thus, our data supports that VPA and LMNB1−/− show similar nuclear behaviors because both perturbations are based in histone modification changes that decompact chromatin.
LMNA−/− nuclei are capable of bleb-based deformation and ruptures
We hypothesized that LMNA−/− nuclei do not show bleb-based behaviors because loss of lamin A/C disrupts nuclear to actin cytoskeleton connections (Broers et al., 2004; Vahabikashi et al., 2022) which is essential to transmit tension (Arsenovic et al., 2016) and cause blebbing and rupture (Hatch and Hetzer, 2016). To determine whether LMNA−/− nuclei have the capacity to form nuclear blebs and ruptures, we treated non-bleb-based LMNA−/− with VPA, a condition that causes bleb-based nuclear shape change and rupture. Upon VPA treatment of LMNA−/− MEF cells nuclear blebbing, ruptures, percentage bleb-based ruptures, and rupture frequency all significantly increased relative to LMNA−/− untreated (Figure 6). Interestingly, this increase in bleb-based behaviors was not reliant on changes in actin confinement, as nuclear height did not decrease in LMNA−/− with VPA treatment (Supplemental Figure 4B), further suggesting changes in actin confinement are unimportant to nuclear blebbing. Thus, LMNA−/− nuclei have the capacity to display increased nuclear blebbing and ruptures and suggests a more complex reason for why this perturbation behaves differently.
FIGURE 6:
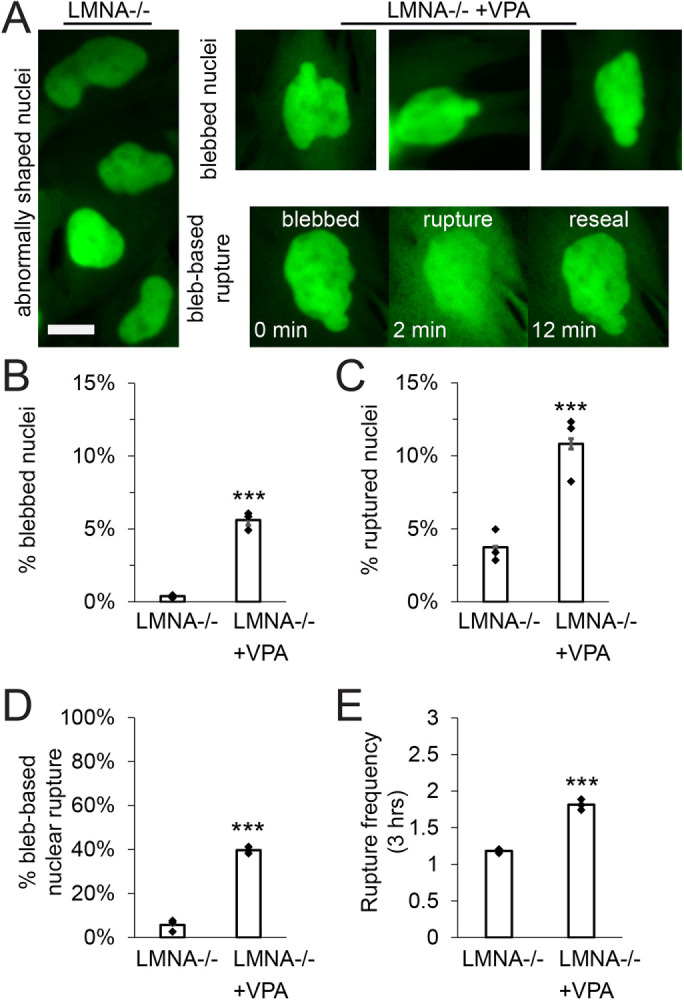
LMNA−/− nuclei have the capacity to form blebs demonstrated by treatment with VPA. (A) Representative images of LMNA−/− abnormally shaped nuclei (left) and LMNA−/− nuclei treated with VPA that display nuclear blebs and bleb-based rupture (right). Graphs of (B) percentage of blebbed nuclei, (C) percentage of ruptured nuclei, (D) percentage of total ruptures that were bleb-based, and (E) nuclear rupture frequency for LMNA−/− and LMNA−/− + VPA imaged for 3 h at 2-min intervals via NLS-GFP. Three biological replicates n ≥ 100 cells each for LMNA−/− without or with VPA. Student’s t test p values report significance *** < 0.001. Error bars represent standard error. Scale bar = 10 µm.
DISCUSSION
Maintenance of nuclear shape and compartmentalization is determined by a balance between nuclear components resisting deformations induced by the cytoskeleton and external forces. Here we provide novel evidence that actin contraction independent of actin confinement controls nuclear blebbing and ruptures in both WT and many prominent nuclear perturbations seen in human disease. Through modulation of actin contraction, we show in almost all cases that actin confinement, measured by nuclear height, is unchanged. Inhibition of actin contraction reveals it is essential for nuclear blebbing and ruptures, while activation of actin contraction drastically increased nuclear rupture frequency of single nuclei. Interestingly, actin contraction has less effect on LMNA−/− non-bleb-based behavior but is partially responsible for abnormal nuclear shape in this perturbation. We go on to show that the similarities between VPA and LMNB1−/− are both due to underlying histone modification changes that lead to decompact chromatin. On the other hand, our data show that LMNA−/− nuclei have the capacity to present nuclear blebbing and bleb-based ruptures, revealing that this perturbation will require future studies to further understand its different and complex phenotype. Overall, our work reveals that actin contraction is a major determinant of nuclear blebbing and bleb-based ruptures.
Actin contraction not confinement controls nuclear shape and ruptures
The previous dogma was that actin fibers running over the top of the nucleus control nuclear shape and compartmentalization via confinement/compression. Pivotal studies of nuclear shape determination reported that actin was a major antagonistic factor to nuclear shape. Loss of nuclear shape overall in progeria cells or nuclear blebs in lamin B1–null cells could be alleviated by removing actin via actin depolymerization drugs such as latrunculin A/B and cytochalasin D (Le Berre et al., 2012; Hatch and Hetzer, 2016). In these actin depolymerization treatments, nuclear height was reported to increase because actin was no longer compressing the nucleus, which provided data supporting that actin confinement might control nuclear shape. Abnormal deformations could then be reestablished whether actin depolymerizers were removed and actin reformed on top of the nucleus causing actin confinement. To prove that actin confinement was responsible, cells with depolymerized actin were compressed via glass plate on top of the cell to restore artificial confinement which resulted in abnormal shape, nuclear blebbing, and nuclear ruptures. These experiments have two weaknesses. First, use of drugs that cause actin depolymerization disrupts both actin contraction as well as confinement. Second, artificial confinement studies overcompress the nucleus significantly more than compared with the nucleus before actin depolymerization. Overcompression of the nucleus causes material failure, which agrees with experiments where nuclei migrate between narrow channels resulting in deformation and rupture (Denais et al., 2016; Raab et al., 2016), but it does not support that confinement is the major determinant. To truly test the relative roles of actin contraction versus confinement, a different approach emerged.
Our work and others show that actin confinement is not the major mechanism of abnormal nuclear shape and ruptures. Increased nuclear blebbing and ruptures in artificially confined cells might actually be dependent on actin contraction (see Discussion below [Mistriotis et al., 2019]). Similarly, in unconfined cells our findings show that actin confinement measured by nuclear height matters very little to nuclear shape, blebbing, and ruptures in nuclear perturbations where it can vary by >1 µm or 25% but have the same outcome (Figure 2B). In contradiction with the current view, actin confinement is less (increased nuclear height) in both LMNB1−/−, which displays increased nuclear blebbing and ruptures, and LMNA−/−, which displays decreased nuclear circularity. Furthermore, in the one perturbation where actin confinement increases (VPA), this change is dependent on actin contraction. Thus, actin contraction might control actin confinement, in agreement with artificial confinement studies (Mistriotis et al., 2019). Furthermore, nuclear height remained similar in normally shaped nuclei versus blebbed nuclei (Supplemental Figure 4A). Finally, increased nuclear blebbing and ruptures in LMNA−/− and VPA dual perturbation did not change actin confinement (nuclear height) relative to LMNA−/− (Supplemental Figure 4B). Thus, our data and others clearly show how actin confinement is not the main antagonistic factor working to deform and rupture the nucleus in chromatin or lamin perturbations.
Actin contraction is the major antagonistic factor of nuclear shape and compartmentalization. Actin contraction inhibition via Y27632 resulted in a near total loss of nuclear blebs and ruptures in all bleb-based conditions (Figure 3). Specifically, already low levels of WT blebbing and ruptures significantly decreased to a fraction of a percent, strongly suggesting actin contraction is the main antagonist. Our data agrees nicely with work in an artificial confinement system where actin contraction inhibition suppresses nuclear blebbing back to unconfined levels (Mistriotis et al., 2019) and nuclear dysfunction from cell migration through pores (Cho et al., 2019; Xia et al., 2019). Recent work shows that transcription is also a major contributor to nuclear blebbing and ruptures in VPA and LMNB1−/− (Berg et al., 2023). However, transcription had no effect on WT nuclear blebbing percentage, further supporting actin contraction as the main determinant. Mitotic failures can also cause abnormal nuclear shape (Chiu et al., 2023), but have not been shown to cause nuclear blebbing. Finally, increased contraction via CN03 showed increased nuclear rupture frequency with the capability to increase nuclear blebbing and ruptures in most bleb-based conditions. This again agrees with studies showing increased actin contraction in cells under confinement increases nuclear blebbing (Mistriotis et al., 2019) and that actin contraction causes nuclear strain (Alam et al., 2015). Overall, the data strongly support that actin contraction is the main antagonistic force deforming and rupturing the nucleus.
Nuclear blebbing is the base phenotype for MEF cells
While many papers have studied the effects of chromatin- or lamin-based perturbations on nuclear shape and ruptures, most have only focused on one perturbation. Excitingly, many current gene screens have provided some measure of nuclear characteristics across many chromatin and lamins protein knockdowns and/or drug treatments (Tamashunas et al., 2020; Schibler et al., 2023). Other studies have worked to extensively characterize the loss of different lamins (Chen et al., 2018; Vahabikashi et al., 2022). Here we provide one of the first detailed studies of both a direct chromatin perturbation and lamin perturbations. Interestingly, we find that nuclear bleb formation and bleb-based nuclear ruptures are the dominant form of nuclear shape and compartmentalization loss, at least for MEF cells. These findings are supported by many other studies of individual perturbations but specifically provided for the first time the ability to compare across both chromatin and lamin perturbations.
Chromatin decompaction is now well-known to lead to abnormal nuclear morphologies and ruptures (Stephens et al., 2019a). Chromatin decompaction by histone modification alterations or other proteins provides a perturbation of chromatin without altering lamin levels (Furusawa et al., 2015; Stephens et al., 2018; Strom et al., 2021). Oppositely, almost all lamin perturbations, but specifically LMNB1−/− and LMNA−/−, have a secondary effect of altered chromatin, usually through change to histone modifications (Stephens et al., 2018, 2019b; Vahabikashi et al., 2022). Our data here, provide insight that chromatin decompaction simply exacerbates the WT phenotype of nuclear blebbing and majority bleb-based ruptures, which both can be almost fully suppressed by an actin contractility inhibitor Y27632 (Figures 1 and 3).
Loss of lamin B1 was one of the first and most studied nuclear blebbing perturbations (Lammerding et al., 2006; Shimi et al., 2008; Vargas et al., 2012). More recently, many have shown that loss of lamin B1 results in loss of facultative heterochromatin (Camps et al., 2014; Stephens et al., 2018, 2019b; Chang et al., 2022; Vahabikashi et al., 2022), which we recapitulated (Figure 5A). Our data reveal that simply decreasing levels of facultative heterochromatin via methyltransferase inhibitor GSK126 is sufficient to increase nuclear blebbing and rupture (Figure 5, B and C). This new data agrees with our previous data showing that increased heterochromatin levels via histone demethylase inhibition by methylstat treatment (Stephens et al., 2018) and mechanotransduction (Stephens et al., 2019b) rescues nuclear shape in LMNB1−/− nuclei. However, loss of facultative heterochromatin alone does not match the high levels of nuclear blebbing and rupture in LMNB1−/−, suggesting loss of lamin B1 causes an additive effect. To ultimately determine whether lamin B1 is a chromatin perturbation, we would need to conduct micromanipulation force measurements to assay the separate short-extension chromatin-based regime versus long-extension lamin-based regime (Stephens et al., 2017; Currey et al., 2022). Lamin B1’s role in nuclear mechanics, morphology, and compartmentalization remains to be settled, but our data clearly provide evidence that changes to histone modification state are a major contributor to its phenotype.
Lamin A/C loss was the outlier in this group of nuclear perturbations because it presents with a nonbleb-based loss of nuclear shape and compartmentalization. This finding is somewhat confusing as it is well-reported that loss of lamin A/C already causes loss of heterochromatin, a condition shown to cause bleb-based behavior. Our finding of loss of overall shape in LMNA−/−, we report as decreased nuclear circularity, is consistent with many other studies (Lammerding et al., 2006; Robijns et al., 2016; Chen et al., 2018). Thus, while LMNA−/− presents a different phenotype from WT, VPA, and LMNB1−/−, this different overall loss of shape is consistently reported across other publications in both MEFs and other cell lines. Another study in MEFs reported that loss of all lamins resulted in no nuclear blebs but overall loss of nuclear shape and many nuclear ruptures (Chen et al., 2018). Interestingly, shape was restored after adding back lamin A and rupture was suppressed by adding back lamin B1. LMNA−/− cells plus VPA treatment results in increased nuclear blebbing and bleb-based ruptures (Figure 6), a phenotype reliant on actin contraction. LMNA−/− nuclei are reported to have disrupted nuclear-actin attachments, specifically due to lamin A/C’s role interacting with SUN1/2 (Broers et al., 2004; Chen et al., 2018; Vahabikashi et al., 2022), which could disrupt actin contraction–based nuclear bleb formation. Recently, the ability to separate lamin A and C has gained new tools and insights (Wong et al., 2021; Vahabikashi et al., 2022) which will be vital to future studies needed to better understand the roles of each lamin and chromatin histone modification state. Overall, loss of both lamin A and C provides a different phenotype that may provide the key to how the chromatin and lamins resist actin to maintain nuclear shape and stability.
Nuclear shape and ruptures determine increased DNA damage
Previous studies have worked to determine the role of both nuclear deformation and ruptures to increased DNA damage (reviewed in [Miroshnikova and Wickström, 2022]). One prominent idea is that DNA damage occurs due to nuclear ruptures because of loss of nuclear repair proteins from the nucleus (Xia et al., 2018) and allowing cytoplasmic DNA cutting enzyme TREX1 into the nucleus (Nader et al., 2021). Live cell imaging of a nucleus undergoing confined migration shows that the nuclear deformation before rupture induces DNA damage foci formation (Denais et al., 2016). However, these studies and many others require the nucleus to transit a confined pore/space resulting in plastic deformation of the nucleus and rupture – an event where drastic shape change, and rupture are interwind. Alternatively, deformation without rupture can also increase DNA damage levels (Shah et al., 2021). Thus, it is possible that nuclear deformations segregate DNA repair factors away from sites in need of repair (Irianto et al., 2017), without necessarily requiring rupture. Our data in WT, VPA, and LMNB1−/− cannot decouple the roles of nuclear shape and ruptures, which are intertwined, causing increased DNA damage (Figure 4). However, we provide novel data that actin contraction is necessary for the behaviors of nuclear blebbing, rupture, and increased DNA damage, independent of changes in actin confinement. Our data does provide one novel insight via LMNA−/− nuclei which display no increase in nuclear blebbing or rupture but do show drastically decreased circularity (Figure 1; Supplemental Figure 2). These abnormally shaped nuclei also present increased levels of DNA damage (Figure 4). We also present evidence that the abnormal shape is the cause of the increased DNA damage as inhibition of action contraction that rescues nuclear shape also rescues DNA damage levels. This is consistent with previously published work showing that actin contraction inhibition with blebbistatin rescued DNA damage as well as cell-cycle disruption (Cho et al., 2019; Xia et al., 2019). More work will be needed to continue to understand the relative roles of nuclear blebbing, rupture, and abnormal shape in increasing DNA damage in perturbed nuclei.
CONCLUSION
Nuclear rupture dynamics studies seeking to understand the role of actin provide insight into an important contributor to disease, loss of nuclear shape and compartmentalization, which then causes nuclear dysfunction. Past studies have focused on the repair of nuclear ruptures and ruptures in confined migration (Denais et al., 2016; Raab et al., 2016; Halfmann et al., 2019; Young et al., 2020; Sears and Roux, 2022). However, our data on the dynamics of nuclear blebbing and ruptures in nonmigrating and nonartificially confined cells provides insights that help clarify lessons learned from both approaches. One of those main lessons is that there is a clear need to update the model for how actin deforms and ruptures the nucleus. First, actin contraction should now be included as a major antagonist in both unconfined and confined cells. Second, chromatin’s contribution to nuclear shape and rupture must be included, as it is a major mechanical component (Pajerowski et al., 2007; Krause et al., 2013; Schreiner et al., 2015; Shimamoto et al., 2017; Stephens et al., 2017; Melters et al., 2019; Hobson et al., 2020; Nava et al., 2020; Strickfaden et al., 2020). Here we show that chromatin changes are both possibly responsible for some lamin perturbation phenotypes (e.g., LMNB1−/−) and an enhancement of the underlying WT of behavior. Past schematics and theoretical models of nuclear rupture only account for lamin (lamin A) behavior, which we now know presents differently than WT, chromatin perturbations, and loss of lamin B1. The ability to incorporate these ideas into both base and theoretical models will aid further investigations aimed at understanding the mechanisms that can disrupt nuclear stability. The relationship between antagonistic external/cytoskeleton forces and the resistive nuclear components to maintain nuclear shape and stability is essential to both basic cell biology and human diseases presenting abnormalities.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Cell culture and drug treatments
MEF WT, MEF LMNB1−/−, and MEF LMNA−/−, were cultured in DMEM (Corning) containing 10% fetal bovine serum (FBS, HyClone) and 1% penicillin/streptomycin (Corning). The cells were incubated at 37°C and 5% CO2, passaged every 2–3 d and kept for no more than 30 generations. MEFs were immortalized with SV40 large T antigen by retroviral transduction of the gene encoding the SV40 large T antigen as previously described (Shimi et al., 2011, 2015).
To treat with drugs, the cells were first plated in DMEM complete and incubated overnight. Cells were then treated with 4 mM VPA (1069-66-5, Sigma), 10 µM Y27632 (129830-38-2, Tocris), 10 nM of Rho Activator II CN03 (Cytoskeleton), or 2 µM of CytoD (22144-77-0, Tocris). Cells were imaged after 12–24 h of treatment with VPA and Y27, 3 h of treatment with CN03, and 1 h of treatment with CytoD. Cells were not serum starved before treatment with CN03.
Immunofluorescence
Cells were grown in 8-well cover glass chambers (Cellvis) and treated as above. After reaching 80% confluency, cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences) in phosphate-buffered saline (PBS; Corning) at room temperature for 15 min. The cells were then washed three times with PBS, 5 min per wash. After fixation, cells were permeabilized with 0.1% Triton X-100 (US Biological) with PBS for 15 min at room temperature. The cells were then washed with 0.06% Tween 20 (US Biological) in PBS for 5 min. Cells were washed two more times with PBS, 5 min per wash. Cells were blocked in 10% goat serum (Sigma-Aldrich) with PBS for 1 h at room temperature.
Primary, secondary, and conjugate antibodies were all diluted using the blocking solution (10% goat serum in PBS, Sigma). The primary antibodies used were γMLC2 rabbit Ab 1:100 (3672, Cell Signaling Technologies) and H3K27me3 1:100 (9733, Cell Signaling Technologies). Primary antibodies were added to the dish for 12 h at 4°C. The cells were then washed three times with PBS for 5 min. The secondary antibody used was Alexa Fluor 647 Anti-Rabbit IgG 1:1000 (4414, Cell Signaling Technologies). Secondary antibodies were added to the dish and left to sit at room temperature for 1 h. Afterward, the cells were washed with PBS three times. The conjugate antibody used was γH2AX-647 rabbit mAb 1:300 (9720, Cell Signaling Technologies), treated and washed as written above for the primaries.
Next, the cells were stained with a 1 µg/ml dilution of Hoechst 33342 (Life Technologies) in PBS for 5 min and then washed with PBS three times. The dish was then mounted using ProLong Gold antifade (Life Technologies) and allowed to cure for 12 h at room temperature.
Imaging
Images were acquired with Nikon Elements software on a Nikon Instruments Ti2-E microscope with Crest V3 Spinning Disk Confocal, Orca Fusion Gen III camera, Lumencor Aura III light engine, TMC CLeanBench air table, with 40× air objective (N.A 0.75, W.D. 0.66, MRH00401) or Plan Apochromat Lambda 100× Oil Immersion Objective Lens (N.A. 1.45, W.D. 0.13 mm, F.O.V. 25 mm, MRD71970). Live cell time lapse imaging was possible using Nikon Perfect Focus System and Okolab heat, humidity, and CO2 stage top incubator (H301). Images were captured via camera 16 bit for population images or 12 bit sensitive for time lapse live cell imaging with 40× air objective N.A 0.75 (Nikon MRH00401). Cells were imaged in either four-well cover glass dishes or 8-well cover glass chambers (Cellvis). For time lapse data, images were taken in 2-min intervals during 3 h with six fields of view for each condition single plane. Immunofluorescence images were acquired with the 40× air objective at 0.5 µm z-steps over 4.5 µm (nine steps) and maximum intensity compiled post acquisition. Nuclear height measurements were captured using the 100× oil objective using 0.2-µm z-steps over 15 µm.
pMLC2 analysis
Z-stacks were compiled into a maximum projection and background intensity was measured using a 30 × 30-pixel area containing no cells. A 30 × 30-pixel ROI was drawn around the cell to capture the average intensity of Cy5 fluorescence, then exported from the NIS-Elements software to Excel. Background intensity was subtracted to determine the average levels of pMLC2. Statistical significance was determined using the t test.
DNA damage foci analysis
Z-stacks were compiled into a maximum projection and average background fluorescence was subtracted using a 30 × 30-pixel area containing no cells. Individual nuclei were selected using the NIS-Elements threshold or hand drawn over Hoechst fluorescent images if auto selection was unable to separate between adjacent nuclei. Circularity measurements were taken from auto selected and hand drawn ROIs, then exported from the NIS-Elements software to Excel. We use the bright spots program in Nikon Elements that requires a size and contrast to determine what is a focus. The size of the diffraction limited focus selected was 0.5 µm based on full-width half-maximum experimental measurement of a gaussian from a 175-nm fluorescent bead and used across all conditions and biological replicates. Contrast was determined empirically in WT cells and kept constant between treatments in the same replicate. Using the bright spots program Nikon Elements output the number of DNA damage foci per nucleus which was marked by the selected ROI. These data was then exported to Excel for averages and statistical significance determined using the t test.
H3K27me 3 analysis
As described above, Z-stacks were compiled into a maximum projection and average background fluorescence was subtracted using a 30 × 30-pixel area containing no cells. ROIs were drawn around individual nuclei by hand or using the NIS-Elements threshold. Average intensity of the nucleus was used to determine relative levels of heterochromatin between conditions. Statistical significance was determined using the t test.
Live cell NLS-GFP imaging and analysis
Images were captured via camera 12 bit sensitive with 40× air objective N.A 0.75. Time lapse imaging parameters used were FITC Wide Field light modality at 4% power, 30 ms exposure time at 2-min intervals during 3 h with six adjacent fields of view for each condition. Images were saved within the NIS-Elements AR Analysis software. Images were observed to record total number of nuclei, number of blebs, and number of ruptures in each field of view. Number of ruptures recorded included each nuclear rupture observed, whether the rupture was bleb-based or nonbleb based, and how frequently each nucleus ruptured throughout the 3-h duration. Data collected was then compiled and averaged in Excel (Microsoft) to determine percent blebbing, percent rupture, percent bleb-based rupture, and rupture frequency for each condition. Statistical significance was determined by conducting t tests between the two conditions.
Live cell imaging of nuclear height and actin analysis
Cells were grown in dishes divided into four glass wells (Cellvis) and treated as above. After reaching 80% confluency, the cells were treated with 1 µg/ml dilution of Hoechst 33342 (Life Technologies) for 10 min before being imaged on a wide-field microscope. For nuclear height measurements, cells were treated with a 1:1000 dilution of SPY555-Actin Probe (CY-SC202, Cytoskeleton) in complete DMEM at 37°C for 3 h before imaging. Live cell images were taken on a Nikon Eclipse Ti2 wide-field microscope using a 100× oil objective. Image stacks with 0.2 µm steps were also taken using a spinning disk confocal microscope with a 100× oil objective. Exposure times for Hoechst (DAPI), NLS-GFP (FITC), and SPY555-Actin Probe (TRITC) were between 30 and 100 ms. To determine nuclear height in Z, intensity line scans were taken in Hoechst fluorescence, values were exported to excel and relative positions of the top and bottom of the nucleus were determined using the full-width half-max of the intensity graph in Excel. Two line scan measurements were taken per nucleus and the two values were averaged to determine the height. Statistical significance was determined using the t test.
Actin images were also analyzed for number of actin lines and relative fluorescence via line scans of actin on top of the nucleus. Each line scan was quantified for number of actin lines determined by number of fluorescence peaks > 1.5 signal to noise. The relative sum intensity was quantified by measuring the peak fluorescence intensity for each peak. Statistical significance was determined using the t test. Raw data can be found in Supplemental Table 1.
Supplementary Material
Acknowledgments
We would like to thank Edward J. Banigan for helpful and insightful discussions and Jeri Ruth Moskowitz for editing support. M.P., Y.B., A.G., A.L., M.L.C., K.C., A.P., A.D.S. are supported by the Pathway to Independence Award (R00GM123195) and Center for three-dimensional Structure and Physics of the Genome 4DN2 grant (1UM1HG011536). The authors declare not competing interests.
Abbreviations used:
- LMNA–/–
lamin A/C knockout
- LMNB1–/–
lamin B1 knockout
- MEF
mouse embryonic fibroblasts
- NLS-GFP
nuclear localization signal green fluorescent protein
- VPA
valproic acid
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E23-07-0292) on December 13, 2023.
REFERENCES
- Alam SG, Lovett D, Kim DI, Roux KJ, Dickinson RB, Lele TP (2015). The nucleus is an intracellular propagator of tensile forces in NIH 3T3 fibroblasts. J Cell Sci 128, 1901–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsenovic PT, Ramachandran I, Bathula K, Zhu R, Narang JD, Noll NA, Lemmon CA, Gundersen GG, Conway DE (2016). Nesprin-2G, a component of the nuclear LINC complex, is subject to myosin-dependent tension. Biophys J 110, 34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaghzal H, Borrman T, Stephens AD, Lafontaine DL, Venev SV, Weng Z, Marko JF, Dekker J (2021). Liquid chromatin Hi-C characterizes compartment-dependent chromatin interaction dynamics. Nat Genet 53, 367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg IK, Currey ML, Gupta S, Berrada Y, Nguyen BV, Pho M, Patteson AE, Schwarz JM, Banigan EJ, Stephens AD (2023). Transcription inhibition suppresses nuclear blebbing and rupture independent of nuclear rigidity. J Cell Sci 136, jcs261547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broers JLV, Peeters EAG, Kuijpers HJH, Endert J, Bouten CVC, Oomens CWJ, Baaijens FPT, Ramaekers FCS (2004). Decreased mechanical stiffness in LMNA–/– cells is caused by defective nucleo-cytoskeletal integrity: implications for the development of laminopathies. Hum Mol Genet 13, 2567–2580. [DOI] [PubMed] [Google Scholar]
- Butin-Israeli V, Adam SA, Goldman AE, Goldman RD (2012). Nuclear lamin functions and disease. Trends Genet 28, 464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps J, Wangsa D, Falke M, Brown M, Case CM, Erdos MR, Ried T (2014). Loss of lamin B1 results in prolongation of S phase and decondensation of chromosome territories. FASEB J 28, 3423–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Li M, Shao S, Li C, Ai S, Xue B, Hou Y, Zhang Y, Li R, Fan X, et al. (2022). Nuclear peripheral chromatin-lamin B1 interaction is required for global integrity of chromatin architecture and dynamics in human cells. Protein Cell 13, 258–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen NY, Kim P, Weston TA, Edillo L, Tu Y, Fong LG, Young SG (2018). Fibroblasts lacking nuclear lamins do not have nuclear blebs or protrusions but nevertheless have frequent nuclear membrane ruptures. Proc Natl Acad Sci USA 115, 10100–10105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu K, Berrada Y, Eskndir N, Song D, Fong C, Naughton S, Chen T, Moy S, Gyurmey S, James L, et al. (2023). CTCF is essential for proper mitotic spindle structure and anaphase segregation. Chromosoma, 10.1007/s00412-023-00810-w. [DOI] [PubMed] [Google Scholar]
- Cho S, Vashisth M, Abbas A, Majkut S, Vogel K, Xia Y, Ivanovska IL, Irianto J, Tewari M, Zhu K, et al. (2019). Mechanosensing by the lamina protects against nuclear rupture, DNA damage, and cell-cycle arrest. Dev Cell 49, 920–935.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffinier C, Jung H-J, Li Z, Nobumori C, Yun UJ, Farber EA, Davies BS, Weinstein MM, Yang SH, Lammerding J, et al. (2010). Direct synthesis of lamin A, bypassing prelamin a processing, causes misshapen nuclei in fibroblasts but no detectable pathology in mice. J Biol Chem 285, 20818–20826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currey ML, Kandula V, Biggs R, Marko JF, Stephens AD (2022). A versatile micromanipulation apparatus for biophysical assays of the cell nucleus. Cell Mol Bioeng 15, 303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva TM, Ketsawatsomkron P, Pelham C, Sigmund CD, Faraci FM (2015). Genetic interference with peroxisome proliferator-activated receptor γ in smooth muscle enhances myogenic tone in the cerebrovasculature via a Rho kinase-dependent mechanism. Hypertension 65, 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos WH, Houben F, Kamps M, Malhas A, Verheyen F, Cox J, Manders EMM, Verstraeten VLRM, van Steensel MAM, Marcelis CLM, et al. (2011). Repetitive disruptions of the nuclear envelope invoke temporary loss of cellular compartmentalization in laminopathies. Hum Mol Genet 20, 4175–4186. [DOI] [PubMed] [Google Scholar]
- Denais CM, Gilbert RM, Isermann P, McGregor AL, te Lindert M, Weigelin B, Davidson PM, Friedl P, Wolf K, Lammerding J (2016). Nuclear envelope rupture and repair during cancer cell migration. Science 352, 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa T, Rochman M, Taher L, Dimitriadis EK, Nagashima K, Anderson S, Bustin M (2015). Chromatin decompaction by the nucleosomal binding protein HMGN5 impairs nuclear sturdiness. Nat Commun 6, 6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman RD, Shumaker DK, Erdos MR, Eriksson M, Goldman AE, Gordon LB, Gruenbaum Y, Khuon S, Mendez M, Varga R, Collins FS (2004). Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci USA 101, 8963–8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann CT, Sears RM, Katiyar A, Busselman BW, Aman LK, Zhang Q, O’Bryan CS, Angelini TE, Lele TP, Roux KJ (2019). Repair of nuclear ruptures requires barrier-to-autointegration factor. J Cell Biol 218, 2136–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Hetzer MW (2016). Nuclear envelope rupture is induced by actin-based nucleus confinement. J Cell Biol 215, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfand BT, Wang Y, Pfleghaar K, Shimi T, Taimen P, Shumaker DK (2012). Chromosomal regions associated with prostate cancer risk localize to lamin B-deficient microdomains and exhibit reduced gene transcription. J Pathol 226, 735–745. [DOI] [PubMed] [Google Scholar]
- Hernandez PA, Jacobsen TD, Chahine NO (2020). Actomyosin contractility confers mechanoprotection against TNFα-induced disruption of the intervertebral disc. Sci Adv 6, eaba2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson CM, Kern M, O’Brien ET, 3rd, Stephens AD, Falvo MR, Superfine R (2020). Correlating nuclear morphology and external force with combined atomic force microscopy and light sheet imaging separates roles of chromatin and lamin A/C in nuclear mechanics. Mol Biol Cell 31, 1788–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue-Mochita M, Inoue T, Fujimoto T, Kameda T, Awai-Kasaoka N, Ohtsu N, Kimoto K, Tanihara H (2015). p38 MAP kinase inhibitor suppresses transforming growth factor-β2-induced type 1 collagen production in trabecular meshwork cells. PLoS One 10, e0120774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irianto J, Pfeifer CR, Bennett RR, Xia Y, Ivanovska IL, Liu AJ, Greenberg RA, Discher DE (2016). Nuclear constriction segregates mobile nuclear proteins away from chromatin. Mol Biol Cell 27, 4011–4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irianto J, Xia Y, Pfeifer CR, Athirasala A, Ji J, Alvey C, Tewari M, Bennett RR, Harding SM, Liu AJ, et al. (2017). DNA damage follows repair factor depletion and portends genome variation in cancer cells after pore migration. Curr Biol 27, 210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung-Garcia Y, Maiques O, Monger J, Rodriguez-Hernandez I, Fanshawe B, Domart M-C, Renshaw MJ, Marti RM, Matias-Guiu X, Collinson LM, et al. (2023). LAP1 supports nuclear adaptability during constrained melanoma cell migration and invasion. Nat Cell Biol 25, 108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalukula Y, Stephens AD, Lammerding J, Gabriele S (2022). Mechanics and functional consequences of nuclear deformations. Nat Rev Mol Cell Biol 23, 583–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, Searson PC, Hodzic D, Wirtz D (2009). A perinuclear actin cap regulates nuclear shape. Proc Natl Acad Sci USA 106, 19017–19022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler F, Bormann F, Raddatz G, Gutekunst J, Corless S, Musch T, Lonsdorf AS, Erhardt S, Lyko F, Rodríguez-Paredes M (2020). Epigenetic deregulation of lamina-associated domains in Hutchinson-Gilford progeria syndrome. Genome Med 12, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Te Riet J, Wolf K (2013). Probing the compressibility of tumor cell nuclei by combined atomic force-confocal microscopy. Phys Biol 10, 065002. [DOI] [PubMed] [Google Scholar]
- Lammerding J, Fong LG, Ji JY, Reue K, Stewart CL, Young SG, Lee RT (2006). Lamins A and C but not lamin B1 regulate nuclear mechanics. J Biol Chem 281, 25768–25780. [DOI] [PubMed] [Google Scholar]
- Le Berre M, Aubertin J, Piel M (2012). Fine control of nuclear confinement identifies a threshold deformation leading to lamina rupture and induction of specific genes. Integr Biol (Camb) 4, 1406–1414. [DOI] [PubMed] [Google Scholar]
- McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A 3rd, Diaz E, et al. (2012). EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492, 108–112. [DOI] [PubMed] [Google Scholar]
- Melters DP, Pitman M, Rakshit T, Dimitriadis EK, Bui M, Papoian GA, Dalal Y (2019). Intrinsic elasticity of nucleosomes is encoded by histone variants and calibrated by their binding partners. Proc Natl Acad Sci USA 116, 24066–24074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroshnikova YA, Wickström SA (2022). Mechanical forces in nuclear organization. Cold Spring Harb Perspect Biol 14, a039685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistriotis P, Wisniewski EO, Bera K, Keys J, Li Y, Tuntithavornwat S, Law RA, Perez-Gonzalez NA, Erdogmus E, Zhang Y, et al. (2019). Confinement hinders motility by inducing RhoA-mediated nuclear influx, volume expansion, and blebbing. J Cell Biol 218, 4093–4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell M, Oakes PW, Lenz M, Gardel ML (2015). Forcing cells into shape: the mechanics of actomyosin contractility. Nat Rev Mol Cell Biol 16, 486–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nader GP de F, Agüera-Gonzalez S, Routet F, Gratia M, Maurin M, Cancila V, Cadart C, Palamidessi A, Ramos RN, San Roman M, et al. (2021). Compromised nuclear envelope integrity drives TREX1-dependent DNA damage and tumor cell invasion. Cell 184, 5230–5246.e22. [DOI] [PubMed] [Google Scholar]
- Nava MM, Miroshnikova YA, Biggs LC, Whitefield DB, Metge F, Boucas J, Vihinen H, Jokitalo E, Li X, García Arcos JM, et al. (2020). Heterochromatin-driven nuclear softening protects the genome against mechanical stress-induced damage. Cell 181, 800–817.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nmezi B, Xu J, Fu R, Armiger TJ, Rodriguez-Bey G, Powell JS, Ma H, Sullivan M, Tu Y, Chen NY, et al. (2019). Concentric organization of A- and B-type lamins predicts their distinct roles in the spatial organization and stability of the nuclear lamina. Proc Natl Acad Sci USA 116, 4307–4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE (2007). Physical plasticity of the nucleus in stem cell differentiation. Proc Natl Acad Sci USA 104, 15619–15624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer CR, Xia Y, Zhu K, Liu D, Irianto J, García VMM, Millán LMS, Niese B, Harding S, Deviri D, et al. (2018). Constricted migration increases DNA damage and independently represses cell cycle. Mol Biol Cell 29, 1948–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M, Gentili M, de Belly H, Thiam H-R, Vargas P, Jimenez AJ, Lautenschlaeger F, Voituriez R, Lennon-Duménil A-M, Manel N, Piel M (2016). ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 352, 359–362. [DOI] [PubMed] [Google Scholar]
- Rees RW, Ralph DJ, Royle M, Moncada S, Cellek S (2001). Y-27632, an inhibitor of Rho-kinase, antagonizes noradrenergic contractions in the rabbit and human penile corpus cavernosum. Br J Pharmacol 133, 455–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robijns J, Molenberghs F, Sieprath T, Corne TDJ, Verschuuren M, De Vos WH (2016). In silico synchronization reveals regulators of nuclear ruptures in lamin A/C deficient model cells. Sci Rep 6, 30325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler A, Jevtic P, Pegoraro G, Levy DL, Misteli T (2023). Identification of epigenetic modulators as determinants of nuclear size and shape. eLife 12, e80653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider IC, Hays CK, Waterman CM (2009). Epidermal growth factor–induced contraction regulates paxillin phosphorylation to temporally separate traction generation from DE-adhesion. Mol Biol Cell 20, 3155–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner SM, Koo PK, Zhao Y, Mochrie SGJ, King MC (2015). The tethering of chromatin to the nuclear envelope supports nuclear mechanics. Nat Commun 6, 7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears RM, Roux KJ (2022). Mechanisms of A-type Lamin targeting to nuclear ruptures are disrupted in LMNA- and BANF1-associated progerias. Cells 11, 865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senigagliesi B, Penzo C, Severino LU, Maraspini R, Petrosino S, Morales-Navarrete H, Pobega E, Ambrosetti E, Parisse P, Pegoraro S, et al. (2019). The High Mobility Group A1 (HMGA1) chromatin architectural factor modulates nuclear stiffness in breast cancer cells. Int J Mol Sci 20, 2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Hobson CM, Cheng S, Colville MJ, Paszek MJ, Superfine R, Lammerding J (2021). Nuclear deformation causes DNA damage by increasing replication stress. Curr Biol 31, 753–765.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto Y, Tamura S, Masumoto H, Maeshima K (2017). Nucleosome-nucleosome interactions via histone tails and linker DNA regulate nuclear rigidity. Mol Biol Cell 28, 1580–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, Butin-Israeli V, Adam SA, Hamanaka RB, Goldman AE, Lucas CA, Shumaker DK, Kosak ST, Chandel NS, Goldman RD (2011). The role of nuclear lamin B1 in cell proliferation and senescence. Genes Dev 25, 2579–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, Kittisopikul M, Tran J, Goldman AE, Adam SA, Zheng Y, Jaqaman K, Goldman RD (2015). Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol Biol Cell 26, 4075–4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, Pfleghaar K, Kojima S-I, Pack C-G, Solovei I, Goldman AE, Adam SA, Shumaker DK, Kinjo M, Cremer T, Goldman RD (2008). The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev 22, 3409–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J-W, Spinler KR, Swift J, Chasis JA, Mohandas N, Discher DE (2013). Lamins regulate cell trafficking and lineage maturation of adult human hematopoietic cells. Proc Natl Acad Sci USA 110, 18892–18897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria-Valles C, Carrero D, Gabau E, Velasco G, Quesada V, Bárcena C, Moens M, Fieggen K, Möhrcken S, Owens M, et al. (2016). Novel LMNA mutations cause an aggressive atypical neonatal progeria without progerin accumulation. J Med Genet 53, 776–785. [DOI] [PubMed] [Google Scholar]
- Stephens AD (2020). Chromatin rigidity provides mechanical and genome protection. Mutat Res 821, 111712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Banigan EJ, Adam SA, Goldman RD, Marko JF (2017). Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol Biol Cell 28, 1984–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Banigan EJ, Marko JF (2019a). Chromatin’s physical properties shape the nucleus and its functions. Curr Opin Cell Biol 58, 76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Liu PZ, Banigan EJ, Almassalha LM, Backman V, Adam SA, Goldman RD, Marko JF (2018). Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Mol Biol Cell 29, 220–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Liu PZ, Kandula V, Chen H, Almassalha LM, Herman C, Backman V, O’Halloran T, Adam SA, Goldman RD, et al. (2019b). Physicochemical mechanotransduction alters nuclear shape and mechanics via heterochromatin formation. Mol Biol Cell 30, 2320–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickfaden H, Tolsma TO, Sharma A, Underhill DA, Hansen JC, Hendzel MJ (2020). Condensed chromatin behaves like a solid on the mesoscale in vitro and in living cells. Cell 183, 1772–1784.e13. [DOI] [PubMed] [Google Scholar]
- Strom AR, Biggs RJ, Banigan EJ, Wang X, Chiu K, Herman C, Collado J, Yue F, Ritland Politz JC, Tait LJ, et al. (2021). HP1α is a chromatin crosslinker that controls nuclear and mitotic chromosome mechanics. eLife 10, e63972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PCDP, Pinter J, Pajerowski JD, Spinler KR, Shin J-W, Tewari M, et al. (2013). Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science 341, 1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamashunas AC, Tocco VJ, Matthews J, Zhang Q, Atanasova KR, Paschall L, Pathak S, Ratnayake R, Stephens AD, Luesch H, et al. (2020). High-throughput gene screen reveals modulators of nuclear shape. Mol Biol Cell 31, 1392–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahabikashi A, Sivagurunathan S, Nicdao FAS, Han YL, Park CY, Kittisopikul M, Wong X, Tran JR, Gundersen GG, Reddy KL, et al. (2022). Nuclear lamin isoforms differentially contribute to LINC complex-dependent nucleocytoskeletal coupling and whole-cell mechanics. Proc Natl Acad Sci USA 119, e2121816119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas JD, Hatch EM, Anderson DJ, Hetzer MW (2012). Transient nuclear envelope rupturing during interphase in human cancer cells. Nucleus 3, 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong X, Hoskins VE, Melendez-Perez AJ, Harr JC, Gordon M, Reddy KL (2021). Lamin C is required to establish genome organization after mitosis. Genome Biol 22, 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Ivanovska IL, Zhu K, Smith L, Irianto J, Pfeifer CR, Alvey CM, Ji J, Liu D, Cho S, et al. (2018). Nuclear rupture at sites of high curvature compromises retention of DNA repair factors. J Cell Biol 217, 3796–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Pfeifer CR, Zhu K, Irianto J, Liu D, Pannell K, Chen EJ, Dooling LJ, Tobin MP, Wang M, et al. (2019). Rescue of DNA damage after constricted migration reveals a mechano-regulated threshold for cell cycle. J Cell Biol 218, 2545–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AM, Gunn AL, Hatch EM (2020). BAF facilitates interphase nuclear membrane repair through recruitment of nuclear transmembrane proteins. Mol Biol Cell 31, 1551–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.