Abstract
Flavonoids and isoflavonoids are major plant secondary metabolites that mediate diverse biological functions and exert significant ecological impacts. These compounds play important roles in many essential physiological processes. In addition, flavonoids and isoflavonoids have direct but complex effects on human health, ranging from reducing cholesterol levels and preventing certain cancers to improving women's health. In this study, we cloned and functionally characterized five soybean (Glycine max) chalcone isomerases (CHIs), key enzymes in the phenylpropanoid pathway that produces flavonoids and isoflavonoids. Gene expression and kinetics analysis suggest that the soybean type I CHI, which uses naringenin chalcone as substrate, is coordinately regulated with other flavonoid-specific genes, while the type II CHIs, which use a variety of chalcone substrates, are coordinately regulated with an isoflavonoid-specific gene and specifically activated by nodulation signals. Furthermore, we found that some of the newly identified soybean CHIs do not require the 4′-hydroxy moiety on the substrate for high enzyme activity. We then engineered yeast (Saccharomyces cerevisiae) to produce flavonoid and isoflavonoid compounds. When one of the type II CHIs was coexpressed with an isoflavone synthase, the enzyme catalyzing the first committed step of isoflavonoid biosynthesis, various chalcone substrates added to the culture media were converted to an assortment of isoflavanones and isoflavones. We also reconstructed the flavonoid pathway by coexpressing CHI with either flavanone 3β-hydroxylase or flavone synthase II. The in vivo reconstruction of the flavonoid and isoflavonoid pathways in yeast provides a unique platform to study enzyme interactions and metabolic flux.
Flavonoids and isoflavonoids are major plant secondary metabolites that mediate diverse biological functions and exert significant ecological impacts. Approximately 20% of the carbon fixed by photosynthesis is believed to be channeled into the phenylpropanoid pathway, generating the majority of the phenolic compounds found in nature, including flavonoids and isoflavonoids (Weisshaar and Jenkins, 1998). These compounds play important roles in many essential physiological processes. For example, lignins are major cell wall components, flavonols are UV protectants, and anthocyanins are the primary attractants for pollinators in many species (Dixon and Steele, 1999). In addition, flavonoids and isoflavonoids have direct but complex effects on human health. For example, resveratrol found in red wine has been shown to double the life span of yeast (Saccharomyces cerevisiae; Howitz et al., 2003), the flavonols and tannins found in green tea can drastically reduce the likelihood of stroke and heart disease, and the isoflavones produced by soybean (Glycine max) can prevent many hormone-dependent cancers and improve women's health (Beecher, 2003). Despite being one of the most studied plant secondary-metabolic pathways, surprising new functions and characteristics of these compounds are still being discovered.
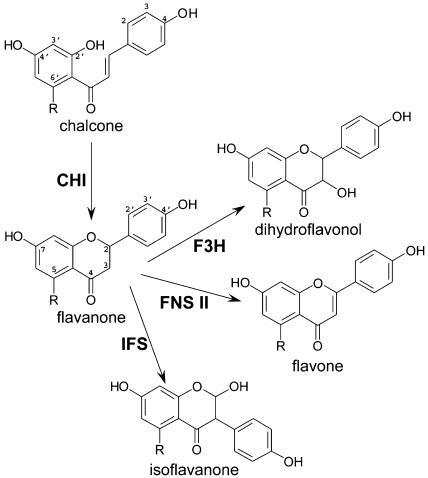
The general phenylpropanoid pathway is conserved in all plant species. In this core pathway, the amino acid Phe is converted into p-coumaroyl-CoA in three enzymatic steps. In most species, chalcone synthase (CHS; EC 2.3.1.74) then condenses p-coumaroyl-CoA with 3 molecules of malonyl-CoA to form 4, 2′, 4′, 6′-tetrahydroxychalcone (naringenin chalcone). As shown in Figure 1, naringenin chalcone can be further metabolized to (2S)-5, 7, 4′-trihydroxyflavanone (naringenin) by chalcone isomerase (CHI; EC 5.5.1.6) to form the primary C15 flavonoid skeleton. Recently, Shimada et al. (2003) reported that two types of CHIs with distinctive phylogenic lineages coexist in the legume Lotus japonicus. The type I CHIs are ubiquitous in the plant kingdom and convert naringenin chalcone to naringenin. In contrast, the type II CHIs appear to be legume specific and possess additional catalytic activity, which allows them to convert 4, 2′, 4′-trihydroxychalcone (isoliquiritigenin) into (2S)-7, 4′-dihydroxyflavanone (liquiritigenin). A type II CHI isolated from alfalfa (Medicago sativa) has been extensively studied structurally and mechanistically (Jez et al., 2000, 2002; Hur et al., 2004). These structure-function analyses suggest that the formation of a hydrogen bond network between the active site of CHI and its substrates is crucial for the enzyme's catalytic activity (Jez et al., 2002; Hur et al., 2004). Naringenin and liquiritigenin are the precursors of many flavonoid and isoflavonoid compounds. One reaction using naringenin as a substrate is the addition of a hydroxyl group at the C3 position to form dihydrokaempferol, catalyzed by flavanone 3β-hydroxylase (F3H; EC 1.14.11.9), a 2-oxoglutartate-dependent dioxygenase (Deboo et al., 1995; Fig. 1). This modification is essential for the production of anthocyanins and condensed tannins. For flavone biosynthesis, flavone synthases (FNS) convert naringenin into the flavone apigenin. Two types of FNS have been reported. However, FNSII, a P450 monooxygenase, appears to exist in the majority of plant species (Martens and Forkmann, 1999; Fig. 1). Naringenin is a substrate for a number of other enzymes. For example, in maize (Zea mays), in addition to F3H and FNSII, flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase both modify naringenin with additional B-ring hydroxylations, producing the precursors of maysins. Dihydroflavonol 4-reductase (EC1.1.1.219) can also utilize naringenin to initiate the tissue-specific production of phlobaphenes (Grotewold et al., 1994). In species that synthesize isoflavones, the enzyme isoflavone synthase (IFS), another Cyt P450 monooxygenase, acts as the key metabolic entry point for the formation of all isoflavonoid compounds (Akashi et al., 1999; Steele et al., 1999; Jung et al., 2000). IFS mediates the intramolecular aryl migration of both liquiritigenin and naringenin to form 2-hydroxyisoflavanones (Fig. 1). The isoflavanones are unstable under ambient conditions and can convert to isoflavones spontaneously (Sawada et al., 2002) or with the assistance of a putative isoflavanone dehydratase (Hakamatsuka et al., 1998).
Figure 1.
Partial flavonoid and isoflavonoid pathways. The numbering schemes of carbons for chalcone and flavanone are marked. The −R group is either H or OH. When R=H (OH), the common names are isoliquiritigenin (naringenin-chalcone), liquiritigenin (naringenin), DAI (genistein), dihydroxyflavone (apigenin), and kaempherol (quercetin) for chalcone, flavanone, isoflavone, flavone, and dihydroflavonol, respectively.
During plant-microbe interactions, many of the genes encoding the pathway enzymes are coordinately regulated at the transcriptional level, and a number of the transcription factors that bind to the conserved cis-elements of flavonoid pathway genes have been identified (Ni et al., 1996; Weisshaar and Jenkins, 1998). In addition, several enzymes of the phenylpropanoid pathway were shown to form protein complexes that efficiently direct metabolic flux toward the production of a particular compound (Dixon and Steele, 1999; Ovadi and Srere, 2000; Achnine et al., 2004). For example, Arabidopsis (Arabidopsis thaliana) CHS, CHI, and dihydroflavonol 4-reductase may interact during flavonoid biosynthesis (Winkel-Shirley, 1999). Understanding the partitioning of metabolites between the flavonoid and isoflavonoid pathways is essential to uncover and utilize the mechanisms of pathway regulation. For instance, to engineer the production of economically important isoflavones in nonlegume plants, the regulation of naringenin accumulation is critical, because heterologously expressed IFS must directly compete with endogenous enzymes for this substrate (Yu et al., 2000; Liu et al., 2002; Yu and McGonigle, 2005).
We undertook this study to establish a system for the in vivo functional analysis of metabolic enzymes. Initially, we cloned and characterized the soybean CHIs responsible for naringenin and liquiritigenin production. We found that the type II CHIs were coordinately regulated developmentally and during plant-microbe interactions with other isoflavonoid biosynthetic enzymes, whereas the type I CHI was coordinately regulated with other flavonoid pathway enzymes. When we expressed the CHIs in yeast and added chalcones to the culture media, the chalcones were taken up by the yeast and rapidly converted to flavanones. Moreover, when the CHIs were coexpressed with IFS in yeast, chalcone substrates were converted to flavanone and then to isoflavones as a result of the coordinate activity of both enzymes. Coexpression of CHI with F3H or FNSII resulted in the production of dihydroflavonols or flavones, respectively, from chalcones. Novel compounds were also produced through the addition of a number of chalcones not common to most plants. This in vivo assay system thus provides a unique platform for the functional analysis of interactions between metabolic enzymes, particularly for the membrane-bound Cyt P450s.
RESULTS
Identification of CHI Expressed Sequence Tags in Soybean
Seven different CHI tentative unique contigs were identified in soybean using the Plant Genome Database (Dong et al., 2004). These contigs were identified based on similarity to the alfalfa CHI DNA sequence (MsaCHI1, M91079; McKhann and Hirsch, 1994). Six of these (CHI1A, CHI1B1, CHI1B2, CHI2, CHI3, and CHI4A) appeared to contain intact coding regions, including complete open reading frames (ORFs) and partial 5′- and 3′-untranslated regions (UTRs). The CHI1B1 and CHI1B2 contigs were very similar. While their 5′-UTRs and ORFs were not noticeably different, their 3′-UTRs were highly divergent. The CHI4A and CHI4B contigs were also very similar. While the CHI4B contig was missing the 3′ end of its predicted coding region, the 5′-UTRs of CHI4A and CHI4B were 92.9% identical, and their ORFs were 96.8% identical at the nucleic acid level.
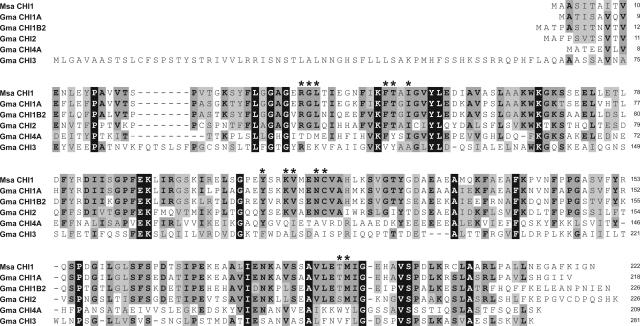
The deduced amino acid sequences of CHI1A, CHI1B2, CHI2, CHI3, and CHI4A were aligned to MsaCHI1 using the ClustalW algorithm (Fig. 2). Of the contigs identified, CHI1A was most homologous to MsaCHI1 with 79.3% identity. MsaCHI1 was 62.8% identical to CHI1B2, 46.9% identical to CHI2, 27.4% identical to CHI3, and 19.3% identical to CHI4. Twenty residues were absolutely conserved between MsaCHI1 and the soybean CHIs (Fig. 2). Surprisingly, none of these residues were active site residues. Residues lining the active site of the MsaCHI1 crystal structure (Jez et al., 2000) are identical in CHI1A, CHI1B2, and CHI2, with the exception of T190. Ser was substituted at this position in CHI2. Only a few of these residues are conserved in CHI3 (R36) and CHI4A (F46, I50, and Y106). Given their low homology to MsaCHI1 and lack of conservation of active site residues, CHI3 and CHI4A are not likely to possess CHI activity.
Figure 2.
Amino acid sequence alignment of the alfalfa and soybean CHIs. Alfalfa MsaCHI1 and five soybean (GmaCHIs) are indicated. Light gray boxes surround similar residues. Bold residues surrounded by light gray boxes are conserved in greater than 50% of the sequences pictured. Bold white residues enclosed by dark gray boxes are absolutely conserved. Residues that line the active site are indicated by bold asterisks.
CHI3 is predicted to be between 55 and 72 residues longer at the N terminus than the other CHIs identified in this study (Fig. 2). Four Web-based programs, ChloroP, iPSORT, Predotar, and TargetP, were used to ascertain whether the elongated N terminus of CHI3 encodes a signal peptide that might control subcellular targeting. The nuclear-encoded, plastid-targeted soybean ribulose 1,5-bisphosphate carboxylase small subunit was included for comparison. All four programs predicted both CHI3 and ribulose 1,5-bisphosphate carboxylase small subunit to be targeted to the chloroplast (see Supplemental Table I).
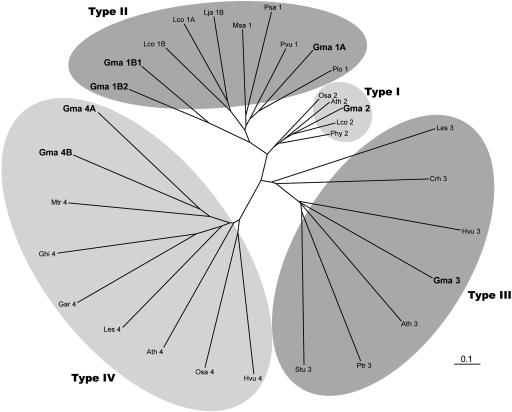
Comparison of the putative soybean CHI with previously characterized CHIs and CHI orthologs from other species shows that these sequences fell into four different subfamilies (Fig. 3). CHIs in the first subfamily (CHI1) were found only in legumes and were at least 70% similar to MsaCHI1. Some members of this subfamily have previously been demonstrated to metabolize both naringenin chalcone, which is found in all plants, and isoliquiritigenin, which is found primarily in legumes (Dewick, 1993; Yu et al., 2003). Members of the second subfamily (CHI2) are at least 60% similar to each other and occur in many distantly related plant species. All members of this subfamily that have been biochemically assayed metabolize only naringenin chalcone (Kimura et al., 2001). Based on sequence homology, the soybean CHI1 subfamily belongs to type II CHIs, and the CHI2 subfamily belongs to the type I CHIs.
Figure 3.
Phylogenetic comparison of deduced amino acid sequences of select CHI-like genes from higher plants and green algae. CHIs were grouped into families based on sequence homology and substrate specificity. The CHI sequences and corresponding GenBank accession numbers, TIGR Gene Index Numbers (TC), or PlantGDB EST Contig ID numbers (XXtuc) represented in this figure are as follows: Arabidopsis Ath2 (M86358), Ath3 (AY084729), Ath4 (AY063786); Chlamydomonas reinhardtii Chr3 (TC10476); soybean Gma1A (AY595413), Gma1B1 (AY595414), Gma1B2 (AY595419), Gma2 (AY595415), Gma3 (AY595416), Gma4 (AY595417); Gossypium arboreum Gar4 (GAtuc03-04-25.4968); Gossypium hirsutum Ghi4 (GHtuc03-04-25.551); Hordeum vulgare Hvu3 (HVtuc02-11-10.14749), Hvu4 (HVtuc02-11-10.15863); Lycopersicon esculentum Les3 (AY348871), Les4 (LEtuc02-10-21.8002); Lotus corniculatus Lco1A (AB073787), Lco1B (AB054801), Lco2 (AB054802); L. japonicus Lja1 (AJ548840); M. sativa Msa1 (M91079); M. truncatula Mtr1 (MTtuc03-04-26.11086); Oryza sativa Ora2 (AF474922), Ora4 (OStuc03-04-25.4740); Petunia hybrida Phy2 (AF233637); Phaseolus vulgaris Pvu1 (X16470); Pisum sativum Psa1 (U03433); Populus tremuloides Ptr3 (BU814723); Pueria lobata Plo1 (D63577); Solanum tuberosum Stu3 (STtuc02-10-23.12425). Soybean CHI sequences are bolded. Shaded ovals signify subfamily groupings.
Members of the CHI3 and CHI4 subfamilies also occur in a broad cross section of plant species (Fig. 3). These appear to be distantly related orthologs, containing only a subset of motifs that are conserved in the first two CHI subfamilies. None of these orthologs have been characterized biochemically; most were assembled from the Plant Genome Database based upon homology to MsaCHI1. Interestingly, all members of the CHI3 subfamily contain putative chloroplast targeting peptides.
Expression Patterns of Flavonoid and Isoflavonoid Pathway Genes in Soybean
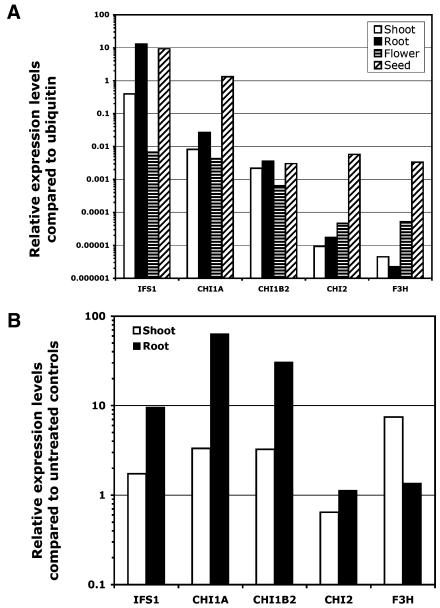
To understand the physiological role of each CHI subfamily in flavonoid and isoflavonoid biosynthesis, the transcripts of CHI1A, CHI1B2, CHI2, CHI3, and CHI4A were analyzed in soybean and compared to those of IFS (AF195789) and F3H genes (see below). To study the transcriptional regulation of these genes, total RNA was extracted from the roots, shoots, flowers, and seeds of soybean and quantitative reverse transcription (RT)-PCR was performed (Fig. 4). Transcript levels were normalized against those of soybean ubiquitin (D28123) in each tissue. The IFS gene showed root-specific expression with approximately 32-fold higher transcript levels in roots than in shoots. IFS1 was much less expressed in flowers, with an expression level less than 0.1% of that in the root (Fig. 4A). IFS expression levels in seeds were comparable to those in the roots. These data agree with our earlier observations (Subramanian et al., 2004). Similarly, CHI1A and CHI1B2 both showed higher expression in the roots and seeds than in the flowers. CHI1A transcript accumulation was 3-fold and 100-fold higher in the roots and seeds than in the shoots. CHI1A and CHI1B2 were least expressed in the flowers at levels approximately 10-fold less than in the roots (Fig. 4A). IFS and type II CHIs are required to synthesize the isoflavone daidzein (DAI), which is the precursor of the phytoalexin glyceollins in soybean. It has been reported that isoflavones, especially DAI, accumulate in roots but not in shoots or flowers (Graham, 1991). The expression patterns of IFS and the type II CHIs appear to support these earlier findings. The expression of IFS and type II CHIs in the seeds is consistent with the accumulation of isoflavonoid compounds in the seed coats and embryos.
Figure 4.
Quantitative RT-PCR analysis of the expression of soybean CHI gene family members compared with IFS and F3H in different tissues and in response to treatment with B. japonicum. A, The transcript levels of root, shoot, floral tissues, and immature embryos. B, The transcript levels after B. japonicum treatment compared to untreated controls. The relative transcript levels of IFS1, CHI1A, CHI1B2, CHI2, and F3H in the roots were 1,299.60%, 2.66%, 0.36%, 0.0018%, and 0.0002% compared to that of ubiquitin.
The type I CHI, CHI2, showed high expression levels in the flowers. CHI2 exhibited 5-fold higher expression in flowers compared to shoots or roots (Fig. 4A). F3H showed higher expression level in flowers as well, with more than 11-fold higher expression than in the shoots (Fig. 4A). Both these genes showed highest expression in the seeds. Since F3H is an essential component of the anthocyanin pathway, the high expression of the gene in flowers is not surprising. The coordinate expression of CHI2 and F3H suggested that the type I CHI in soybean is indeed involved in flavonoid biosynthesis. However, F3H and CHI2 exhibited different expression patterns in the roots. CHI2 showed about 2-fold stronger expression in the roots than in the shoots, whereas F3H expression in the roots was barely detectable, suggesting that CHI2 may serve additional roles in the roots. Neither CHI3 nor CHI4A were expressed at significant levels in any plant tissues, suggesting these 2 CHIs might not play important roles in the phenylpropanoid pathway in soybean under normal conditions (data not shown).
To further delineate the function of different soybean CHIs, their expression in response to symbiotic bacteria (Bradyrhizobium japonicum) was measured by quantitative RT-PCR. The steady-state transcript levels of type II CHI1A and CHI1B2 increased significantly in roots 8 h after the inoculation (90- and 60-fold, respectively). Similar induction was observed for the IFS gene as well, with IFS transcript levels increasing 10-fold in the roots (Fig. 4B). A slight increase was observed for all three genes in the Bradyrhizobium-treated shoot tissue, which may reflect a general defense response during plant-microbe interactions. After Bradyrhizobium treatment, CHI1A and CHI1B2 expression levels in the shoots increased by 3-fold, and IFS1 expression increased by 2-fold. As a consequence, the ratios of shoot to root transcript levels were altered; without Bradyrhizobium treatment, the CHI1A had about 8-fold higher expression in the roots than in the shoots, and CHI1B2 is up to 7-fold higher. After the treatment, the ratio became 140-fold and 61-fold higher, respectively. In contrast, Bradyrhizobium did not induce the type I CHI2 in either roots or shoots, although F3H expression increased significantly in the shoots (Fig. 4B). The results suggest that during the early nodulation response, the type II CHI was highly induced while the type I was not. The IFS expression in response to Bradyrhizobium was similar to our earlier observations (Subramanian et al., 2004).
Catalytic Properties of Soybean CHIs
The catalytic properties of representatives from each soybean CHI subfamily were assessed. The full-length coding sequences of CHI1A, CHI1B2, CHI2, and CHI4A were cloned into a bacterial expression vector and fused in-frame to a His8 epitope tag. CHI3 was truncated at its N terminus to remove its putative chloroplast targeting sequence, yielding CHI3Δ2-66. The His-tagged proteins were purified by affinity chromatography, and His-tags were subsequently removed by digestion with thrombin. All CHIs were expressed at high levels and could be isolated as soluble proteins in milligram quantities. The purified enzymes were then assayed for their ability to metabolize 10 different chalcones (Fig. 5). Neither CHI3Δ2-66 nor CHI4A metabolized any of the chalcones in vitro irrespective of enzyme or substrate concentration (data not shown). CHI2 was able to metabolize only naringenin chalcone (Ch10), whereas CHI1A and CHI1B2 were able to metabolize all but 3 chalcones: Ch2, Ch6, and Ch7 (Table I). The inability of CHI1A and CHI1B2 to metabolize these 3 substrates is most likely due to steric interference by the hydroxyl groups at C2 and C3 position of these chalcones.
Figure 5.
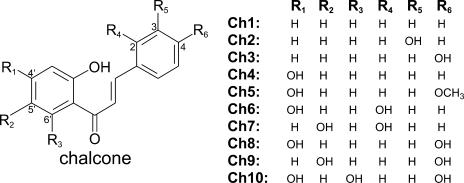
Schematic diagram of chalcones used in this study. Ch1, 2′-Hydroxychalcone; Ch2, 3,2′-dihydroxychalcone; Ch3, 4,2′-dihydroxychalcone; Ch4, 2′,4′-dihydroxychalcone; Ch5, 2′,4′-dihydroxy-4-methoxychalcone; Ch6, 2,2′,4′-trihydroxychalcone; Ch7, 2,2′,5′-trihydroxychalcone; Ch8, 4,2′,4′-trihydroxychalcone; Ch9, 4,2′, 5′-trihydroxychalcone; and Ch10, 4,2′,4′,6′-tetrahydroxychalcone.
Table I.
Comparison of kinetic properties of soybean CHIs with a variety of chalcone substrates
| Chalcone
|
CHI1A
|
CHI1B2
|
CHI2
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Km | kcat | Efficiency | Km | kcat | Efficiency | Km | kcat | Efficiency | |
| μm | s−1 | m−1 s−1 | μm | s−1 | m−1 s−1 | μm | s−1 | m−1 s−1 | |
| Ch1 | 21 | 59.9 | 2.83E + 06 | 10 | 37.4 | 3.70E + 06 | ND | ND | ND |
| Ch3 | 20 | 57.4 | 2.93E + 06 | 13 | 22.6 | 1.77E + 06 | ND | ND | ND |
| Ch4 | 150 | 42.0 | 2.80E + 05 | 42 | 86.3 | 2.05E + 06 | ND | ND | ND |
| Ch5 | 49 | 33.5 | 6.86E + 05 | 21 | 96.8 | 4.50E + 06 | ND | ND | ND |
| Ch8 | 30 | 51.3 | 1.72E + 06 | 20 | 84.5 | 4.12E + 06 | ND | ND | ND |
| Ch9 | 29 | 62.5 | 2.13E + 06 | 15 | 32.5 | 2.17E + 06 | ND | ND | ND |
| Ch10 | 15 | 152.2 | 9.88E + 06 | 7 | 219.1 | 3.04E + 07 | 2 | 478.8 | 1.92E + 08 |
The steady-state kinetics data obtained for CHI1A, CHI1B2, and CHI2 is presented in Table I. Based on the comparison of kcat/Km values, the CHIs exhibited a marked preference for naringenin chalcone over the other chalcones. This is likely due to the functional equivalence of the 2′- and 6′-hydroxyl groups in the isomerization of the chalcone to the flavanone. Of these 3 enzymes, CHI2 most efficiently catalyzed the conversion of naringenin chalcone to naringenin, with a catalytic efficiency more than 6 times greater than CHI1B2 and almost 20 times greater than that of CHI1A (Table I). As mentioned above, CHI2 does not metabolize any of the remaining chalcones in vitro. CHI1A and CHI1B2 metabolize 6 of the remaining chalcones, exhibiting different catalytic efficiencies for each. CHI1A most efficiently metabolizes Ch3, followed by Ch1, Ch9, Ch8, Ch5, and Ch4, whereas CHI1B2 most efficiently metabolizes Ch5 and Ch8, followed by Ch1, Ch9, Ch4, and Ch3. Besides naringenin chalcone, isoliquiritigenin (Ch8) is the other naturally occurring CHI substrate in legumes. As such, the expectation would be that Ch8 would be isomerized more efficiently than the other chalcones. This is the case with CHI1B2, but not with CHI1A. CHI1B2 metabolizes Ch8 and Ch5 with almost equal efficiency. The only difference between these 2 chalcones is that Ch8 contains a hydroxyl group at C4, whereas Ch5 contains a methoxyl group at this position. Compared to earlier reports, CHI1B2 and MsaCHI have similar substrate preferences. Surprisingly, CHI1A preferred 4′-deoxychalcones (Ch3, Ch1, and Ch9) over 4′-hydroxychalcones (Ch8, Ch4, and Ch5). The 4′-hydroxyl residue has been postulated to hydrogen bond with Asn-113 and Thr-190 of MsaCHI to stabilize the substrate during catalysis. Although these 2 amino acids are conserved in all soybean type II CHIs, the hydroxyl group at C4′ is not required to ensure high enzymatic activity.
In Vivo Activity of CHI Expressed in Yeast
As the first step in building the flavonoid/isoflavonoid pathway in yeast, we tested whether chalcones could be metabolized to flavanones in vivo using soybean CHIs. The full-length ORFs of CHI1A, CHI1B2, CHI2, and CHI4 and the truncated CHI3Δ2-66 were placed under the control of a Gal-inducible promoter in the pESC-TRP yeast expression vector. All constructs, including the empty vector, were transformed into yeast. Transformed yeast cultures were induced, fed with substrates, incubated 12 to 16 h, and then extracted with ethyl acetate. Aliquots of the organic extracts were analyzed by reverse-phase HPLC, or they were derivatized and analyzed by gas chromatography-mass spectrometry (GC-MS). When available, metabolites were compared to authentic standards.
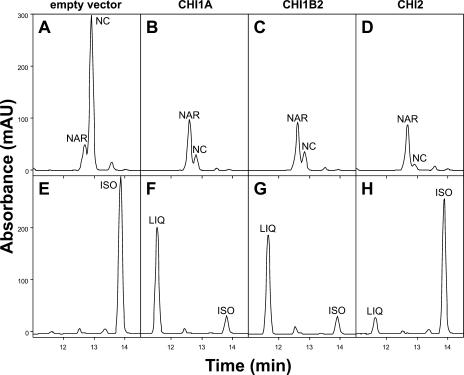
Yeast transformed by CHI1A and CHI1B2 metabolized naringenin chalcone to naringenin (Fig. 6, B and C) and isoliquiritigenin to liquiritigenin (Fig. 6, F and G). CHI2 was able to metabolize naringenin chalcone to naringenin more efficiently than CHI1A or CHI1B2 (Fig. 6D), but it converted only a small amount of isoliquiritigenin to liquiritigenin in 12-h assays (Fig. 6H). Even the metabolism of this small amount of chalcone was unexpected, since the purified CHI2 was unable to metabolize isoliquiritigenin in vitro (see above kinetics data). The control yeast carrying the empty vector did not show any liquiritigenin accumulation, suggesting the automatic conversion of isoliquiritigenin did not contribute to liquiritigenin accumulation (Fig. 6E). This low level of conversion may have been the noncatalytic active-site conversion by the CHI (Bednar and Hadcock, 1988). As observed in vitro, CHI3 and CHI4A did not metabolize either chalcone (data not shown). However, a small amount of naringenin chalcone spontaneously isomerized to the flavanone in all yeast lines (Fig. 6A).
Figure 6.
HPLC profiles from yeast transformed with the empty vector (A and E), CHI1A (B and F), CHI1B2 (C and G), and CHI2 (D and H). Yeast cultures were fed naringenin chalcone (NC; sections A–D) or isoliquiritigenin (ISO; sections E–H). Some naringenin (NAR) was formed by spontaneous cyclization in yeast transformed by the empty vector (A). CHI1A cyclized both chalcones to their corresponding flavanones (B and F), as did CHI1B2 (C and G). CHI2 cyclized naringenin chalcone (D), and to a lesser degree isoliquiritigenin (H, compare to section A). Top sections (A–D) display A344, and bottom sections (E–H) display A312.
When the other chalcones listed in Figure 5 were added to yeast cultures, CHI1A and CHI1B2 converted all chalcones to their corresponding flavanones, including Ch2, Ch6, and Ch7, which we did not observe in vitro activity (see Supplemental Fig. 1 for CHI1B2 results). The products were confirmed by HPLC based on retention time and UV spectra, and by GC-MS analysis as reported in supplemental materials (Supplemental Table III, except for Ch1 derived compounds, which cannot be derivatized by trimethylsilyl [TMS]). For CHI2, only isoliquiritigenin and naringenin chalcone were converted as shown above (Fig. 6). The isomerization of the unexpected chalcones may reflect a low CHI activity toward these compounds that cannot be detected in the 5-min in vitro assays.
In Vivo Activities of IFS, FNSII, and F3H in Yeast
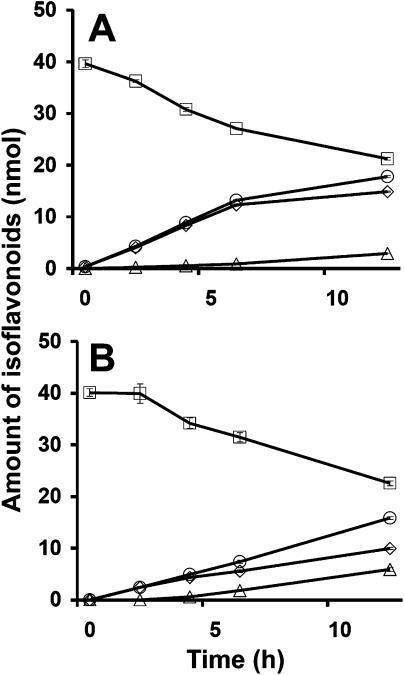
We further tested the individual in vivo activity of IFS, FNSII, and F3H in yeast. All three enzymes use naringenin as a substrate. For isoflavonoid synthesis, the soybean IFS was cloned into pYeDP60 expression vector, under the control of a Gal-inducible promoter and transformed into the WAT11 yeast strain. The WAT11 strain contains an Arabidopsis Cyt P450: NADPH reductase (Pompon et al., 1996; Urban et al., 1997), which provides the reducing equivalents essential for the activity of plant Cyt P450s such as IFS and FNSII. In the in vivo assay where the substrates naringenin and liquiritigenin were added to the culture medium, the conversion of flavanones to 2-hydroxyisoflavanones and eventually to isoflavones was observed. In a time course assay, the production of 2-hydroxyisoflavanones and isoflavones were monitored over 12 h. The accumulation of 2-hydroxyisoflavanones increased with the concomitant decrease of chalcone substrates for the first 6 h (Fig. 7, A and B). The accumulation of isoflavones, however, occurred much later. This accumulation of isoflavones is correlated with decreased accumulation rates of 2-hydroxyisoflavanones (Fig. 7, A and B). These results suggest that IFS catalyzed the conversion of flavanone to 2-hydroxyisoflavanone in yeast, and the slower conversion of 2-hydroxyisoflavanone to isoflavone appears to be noncatalytic.
Figure 7.
Time course of isoflavones and 2-hydroxyisoflavanones accumulation in yeast expressing IFS using naringenin (A) or liquiritigenin (B) as substrate. Each data point represents the average of three independent assays. Error bars represent sd. The 2-hydroxyisoflavanones are represented by diamonds, isoflavones by triangles, combined IFS products by squares, and chalcones by squares.
We sought to demonstrate 2-hydroxyisoflavanones converts to isoflavones spontaneously in yeast. Microsomes were extracted from IFS-containing yeast. The in vitro IFS reaction produced 2-hydroxyisoflavanones and isoflavones. The addition of the specific Cyt P450 inhibitor ancymidol prevented further production of 2-hydroxyisoflavanone, but the dehydration to isoflavone from existing 2-hydroxyisoflavanone continues (Table II). When microsomes or soluble proteins extracted from wild-type yeast were added to the 2-hydroxyisoflavanone, the rate of conversion was not altered compared to solvent controls, supporting the nonenzymatic mechanism of this conversion (Table II).
Table II.
In vitro dehydration of 4HI to genistein
4HI was synthesized in vitro by incubating microsomes isolated from yeast expressing IFS. Products were extracted and added separately to solvent controls, wild-type yeast protein extracts, wild-type yeast microsome extracts; IFS-containing microsomes plus ancymidol inhibitor; or IFS-containing microsomes. The dehydration is measured by HPLC analysis of final products from the time of adding 4HI (Time 0), to 30 min, or 90 min post incubation. Each data point represents the mean of three independent reactions and their sds.
| Incubation | Naringenin | 4HI | Genistein | |
|---|---|---|---|---|
| min | nmol | nmol | nmol | |
| Solvent | 0 | 31.5 ± 4.1 | 3.8 ± 0.7 | 0.7 ± 0.0 |
| 30 | 28.3 ± 1.8 | 3.0 ± 0.3 | 1.1 ± 0.1 | |
| 90 | 30.9 ± 1.3 | 3.0 ± 0.4 | 1.5 ± 0.2 | |
| Wild-type soluble | 30 | 30.0 ± 1.1 | 3.1 ± 0.3 | 1.3 ± 0.1 |
| 90 | 30.2 ± 1.0 | 2.8 ± 0.2 | 1.5 ± 0.1 | |
| Wild-type microsome | 30 | 29.5 ± 1.5 | 3.1 ± 0.2 | 1.2 ± 0.6 |
| 90 | 27.3 ± 3.3 | 2.5 ± 0.5 | 1.4 ± 0.3 | |
| IFS + ancymidol | 30 | 31.5 ± 1.1 | 3.6 ± 0.6 | 1.2 ± 0.1 |
| 90 | 32.7 ± 0.4 | 3.0 ± 0.2 | 1.5 ± 0.1 | |
| IFS microsome | 30 | 23.6 ± 1.0 | 8.2 ± 0.3 | 2.8 ± 0.1 |
| 90 | 19.0 ± 1.2 | 10.5 ± 0.2 | 4.3 ± 0.2 |
For in vivo flavone synthesis, a Gerbera hybrida FNSII gene previously cloned into a yeast expression vector under a Gal-inducible promoter (Martens and Forkmann, 1999) was transformed into WAT11 cells. As observed by HPLC, FNSII metabolizes externally added naringenin and liquiritigenin to the flavones apigenin and 7,4′-dihydroxyflavone, respectively (data not shown; see below).
In addition, 1 complete soybean F3H contig, including ORF and 5′- and 3′-UTRs, was identified from the Plant Genome Database, based on its 83.0% similarity to the alfalfa F3H (X78994; Charrier et al., 1995). To confirm its activity, this soybean F3H was expressed as a FLAG-tagged (N-terminus) protein using a coupled in vitro transcription and translation system. Purified F3H metabolized naringenin to dihydrokaempferol. When this F3H was expressed in yeast, the in vivo assay demonstrated the expected enzyme activity, where naringenin and liquiritigenin were metabolized to dihydrokaempferol and 3, 7, 4′-trihydroxyflavonol, respectively (data not shown; see below).
Partial Reconstitution of Flavonoid and Isoflavonoid Pathways in Yeast
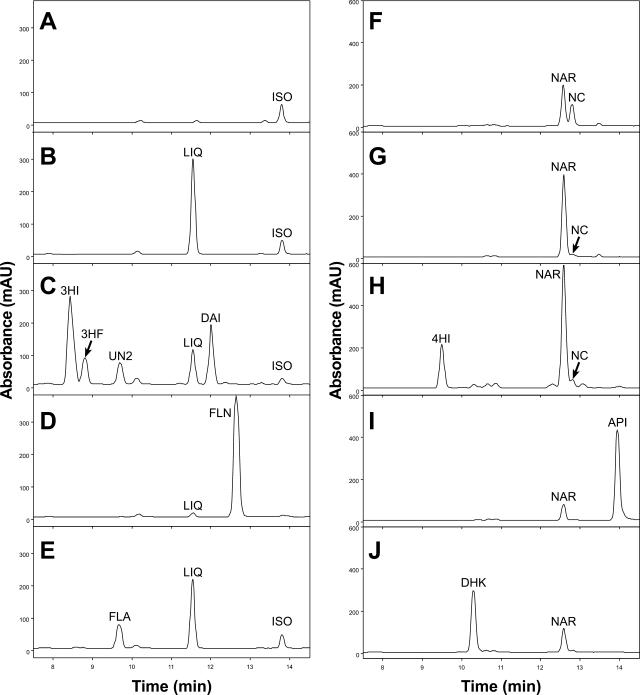
Having demonstrated the abilities of CHIs and other key flavonoid and isoflavonoid biosynthetic enzymes to metabolize chalcones and flavanones in vitro and in vivo, we tried to reconstitute the initiation points of the isoflavonoid and flavonoid pathways in vivo. When the chalcone substrates were added to the culture media of yeast cotransformed with IFS and CHI1A, the chalcones were first converted to flavanones by CHI and subsequently converted to 2-hydroxyisoflavanones and isoflavones by IFS. For example, when isoliquiritigenin was added, the 2, 7, 4′-trihydroxyisoflavanone product was readily detectable after 12 h (Fig. 8C). The 3, 7, 4′-trihydroxyflavanone side product was also detected (Sawada et al., 2002), in addition to the expected isoflavone DAI. Similarly, when naringenin chalcone was fed to the same yeast, the 2, 5, 7, 4′-tetrahydroxyisoflavanone (4HI) was detected (Fig. 8H). The isoflavone genistein (not shown in Fig. 8H) can be detected at 261 nm chromatogram. These products were confirmed by GC-MS (Supplemental Table III).
Figure 8.
HPLC profiles from yeast expressing the empty vector (A and F), CHI1A (B and G), coexpressing CHI1A and IFS (C and H), CHI1A and FNSII (D and I), or CHI1A and F3H (E and J). Yeast cultures were fed isoliquiritigenin (ISO; sections A–E) or naringenin chalcone (NC; sections F–J). Some naringenin (NAR) was formed by spontaneous cyclization in the yeast transformed by the empty vector (F). CHI1A cyclized both chalcones to the corresponding flavanone (B and G). IFS metabolized liquiritigenin (LIQ) to 2,7,4′-trihydroxyisoflavanone (3HI), 3,7,4′-trihydroxyflavanone (3HF), an unknown compounds (UN2), and DAI (C); and naringenin to 4HI and genistein (H). FNSII metabolized liquiritigenin to 7,4′-dihydroxyflavone (FLN; D); and naringenin to apigenin (API; I). F3H metabolized liquiritigenin to 3,7,4′-trihydroxyflavonol (FLA; E); and naringenin to dihydrokaempferol (DHK; J). All metabolite traces are displayed at 312 nm, because all metabolites except genistein are visible at this wavelength.
The Gerbera FNSII or soybean F3H gene was independently cotransformed with CHI1A into yeast. In these yeasts, flavanones produced by CHI were converted to flavones by FNSII or to dihydroflavonols by F3H. For example, when yeast cotransformed with CHI1A and FNSII was fed isoliquiritigenin and naringenin chalcone, they produced 7, 4′-dihydroxyflavone (Fig. 8D) and apigenin (Fig. 8I), respectively. Similarly, when the same substrates were fed to the yeast cotransformed with CHI1A and F3H, 3, 7, 4′-trihydroxyflavonol (Fig. 8E) and dihydrokaempferol (Fig. 8J) were detected by HPLC and confirmed by GC-MS (Supplemental Table III).
DISCUSSION
Classification and Regulation of CHI
CHI catalyzes the stereospecific isomerization of chalcones into corresponding (2S)-flavanones. The in vitro enzyme kinetic assays indicate that CHI operates at the upper limit of the turnover rate for all enzymes, i.e. approaching the diffusion limit (Jez et al., 2000). Some chalcones (e.g. naringenin chalcone) in aqueous solution can be spontaneously isomerized into (2RS)-flavanones with a fairly high turnover rate. CHI ensures that the reaction produces only (2S)-flavanones, which are the biological substrates for downstream enzymes. The only functional CHI gene in Arabidopsis (tt5) is essential for the biosynthesis of anthocyanin and other flavonoid compounds (Shirley et al., 1992).
CHIs have been divided into two classes based on substrate specificity, and their distribution is highly family specific. Type I CHIs are found in both legumes and nonlegumes and isomerize only naringenin chalcone to naringenin. Type II CHIs are found exclusively in leguminous plants and have activities toward both naringenin chalcone and isoliquiritigenin, yielding naringenin and liquiritigenin, respectively. Genes encoding both types of CHIs have been cloned from various plant species (Kimura et al., 2001; Shimada et al., 2003), and the deduced amino acid sequences within the same type of CHI share high levels of homology (>70%), while the identity between type I and II CHIs is only about 50%. Shimada et al. (2003) recently reported that L. japonicus contains both type I and type II CHIs. We confirm in this report that soybean also contains both types of CHIs. In fact, we now speculate that all legume plants contain both types of CHIs. When Medicago truncatula expressed sequence tag (EST) databases at MtDB (http://www.medicago.org/MtDB2/Queries/SimilarityDB2.html) and The Institute for Genomic Research (TIGR; http://www.tigr.org/tigr-scripts/tgi/T_index.cgi?species=soybean) were queried with the keyword “chalcone isomerase,” a total of 15 singletons and contigs were collected. When these ESTs were aligned with soybean CHI cDNA sequences, all 4 subfamilies were conserved, including the subfamily of 1A and 1B (see Supplemental Fig. 4).
Type I and II CHIs have different expression patterns. In L. japonicus, type I and II CHIs are differentially regulated after fungal elicitor treatment (Shimada et al., 2003). Our soybean data indicates that the type II CHIs and IFS are coordinately regulated. They both exhibit root-specific expression and can be highly induced by nodulation signals. In contrast, the type I CHI is coordinately regulated with the flavonoid pathway gene F3H. These genes are expressed primarily in floral tissues and do not respond to Bradyrhizobium inoculation. This is consistent with earlier reports that isoflavonoids DAI and genistein are the main inducers of nodulation-related genes in B. japonicum, the major symbiont of soybean (Smit et al., 1992). Significantly, legume isoflavonoids and flavonoids share one important intermediate, naringenin. The coordinated differential regulation of type I and type II CHIs may imply that instead of sharing a common pool of intermediates and competing only with enzyme kinetics, the flux of substrates toward each pathway may be controlled by separate biosynthetic machinery involving multiple enzyme complexes.
Interestingly, not only type I and II CHIs seem to be conserved in M. truncatula; the homologs of soybean CHI3 and CHI4 were also discovered, suggesting physiological roles for these nonenzymatic CHIs may exist (Supplemental Fig. 4). Recent evidence suggests that some CHIs may have functions other than those of catalysts. Mutant maize CHIs with only 3% to 5% of wild-type CHI activity are able to complement the Arabidopsis tt5 mutant (Dong et al., 2001). Either a small amount of CHI activity is sufficient to drive flux, or the enzymatic activity of CHI is actually not its sole function (Gensheimer and Mushegian, 2004). However, additional evidence is required to confirm these noncatalytic functions, such as serving as a structural scaffold for the enzymes involved in the various branches of the pathway (Irani and Grotewold, 2003) or functioning as flavonoid chaperons and/or transporters.
In all higher plants, isomerization of naringenin chalcone into naringenin by CHI occurs rapidly. However, the conversion of isoliquiritigenin to liquiritigenin, has relatively slower kinetics, because of differences in hydrogen bonding between the active site of CHI and the substrate (Jez et al., 2002). Earlier reports using substrate Ch3, Ch4, Ch8, and Ch10 suggested that the catalytic activity of alfalfa type II CHI requires Asn-113 and Thr-190, which stabilize the substrate by forming hydrogen bonds with the hydroxyl residue at C4′ of chalcone (Jez et al., 2000). Surprisingly, our data indicate that although these 2 amino acids are conserved in soybean type II CHIs, the hydroxyl group at C4′ is not required to ensure high enzymatic activity. Based on kinetic data, CHI1A prefer 4′-deoxychalcones over 4′-hydroxychalcones, suggesting a novel enzyme-substrate interaction may be involved at the C4′ for some CHIs. Amino acid residues outside of the active site may contribute to substrate binding and recognition. These nonactive site interactions may also explain the exclusion of any 2- or 3-hydroxychalcones as substrates in our in vitro assays.
Potential Applications of in Vivo Reconstruction of the Flavonoid/Isoflavonoid Pathway
Functional reconstruction of the pathways in vivo has been attempted in several heterologous systems with different pathways. For example, Phe ammonia lyase and C4H were functionally coexpressed in yeast recently and worked in concert to produce p-coumarate from endogenous Trp pools (Ro and Douglas, 2004). Additionally, when Phe ammonia lyase, 4CL, and CHS were introduced into Escherichia coli as an artificial gene cluster, (2RS)-naringenin was detected in these engineered bacteria (Hwang et al., 2003). Here, we demonstrate that flavonoid and isoflavonoid biosynthetic pathways can be introduced into yeast as well. The amphipathic nature of chalcones and flavanones allows us to feed substrates directly into the media. After multiple enzymes were cointroduced, the transformed yeasts were able to carry out sequential enzymatic reactions to synthesize the final products. Therefore, it is now possible to engineer de novo biosynthesis of flavonoid/isoflavonoid compounds in yeast.
This in vivo system opens numerous possibilities for interesting biochemical studies. First, protein-protein interactions between pathway enzymes can be easily studied in yeast. Currently, we have coexpressed CHI, IFS, and FNSII/F3H to reconstitute the branching of phenylpropanoid pathway in yeast. This in vivo system allows us to easily measure and characterize the products of each enzyme. Thus, the flux of each pathway can be quantified, without interference from other flavonoid/isoflavonoid pathway enzymes. Second, the ease of generating and screening large numbers of yeast lines provides an opportunity to conduct mutagenesis of membrane-bound enzymes for structure-function analyses, or protein engineering. Third, since the consumption of flavonoid and isoflavonoid compounds is often associated with health benefits, introducing the biosynthetic capacity of these compounds into yeast may have direct economic impacts. It would allow for the production of specific flavonoid/isoflavonoid compounds in large scale. Additionally, producing novel flavonoids during fermentation may have many industrial applications, including the biosynthesis of novel pharmaceuticals.
To demonstrate this point, yeast cotransformed with type II CHI1B2 and IFS were fed with each of the 10 chalcones mentioned above. CHI1B2 was found to convert many uncommon chalcones into their corresponding flavanones, thus providing uncommon substrates for IFS. In addition to Ch8 (isoliquiritigenin) and Ch10 (naringenin chalcone), the yeast produced isoflavonoids from all remaining chalcones (see Supplemental Fig. 1). This is the first time that IFS was shown to metabolize substrates other than isoliquiritigenin or naringenin chalcone. These results were subsequently confirmed by in vitro microsome assays (data not shown).
Interestingly, the GC-MS data indicated that both 2-hydroxyisoflavanone and isoflavone products could be detected when Ch2, Ch6, Ch7, and Ch9 were the substrates. For the remaining Ch3, Ch4, and Ch5 reactions, only 2-hydroxyisoflavanones were detected (Supplemental Table III). It suggests that the dehydration of 2-hydroxyisoflavanones requires at least one hydroxyl group on both A and B rings of isoflavanones. Furthermore, a hydroxyl group at the C4′ or C5′ of chalcones appears to be important for IFS activity. Without any hydroxyl groups at these positions, Ch1 and Ch2 could not be efficiently converted to the corresponding isoflavonoids. Since the mechanism of IFS reaction is not clear, further studies are needed to illuminate the possible mechanism of this conversion in yeast.
This yeast system also provided an opportunity to study the substrate specificity of FNSII and F3H. When the 10 chalcones were fed to the yeast cotransformed with CHI1B2 and FNSII, Ch1, Ch3, Ch4, Ch5, Ch8, and Ch10 were converted to their respective flavone compounds (see Supplemental Fig. 2). In the yeast cotransformed with CHI1B2 and F3H, only Ch3, Ch8, Ch9, and Ch10 were metabolized by the F3H (see Supplemental Fig. 3). In these assays, FNSII had a broader substrate spectrum than F3H.
In conclusion, we have identified an entire family of CHIs in soybean. The kinetics and expression profiles suggest that the type I CHI is associated with flavonoid biosynthesis, while the type II CHIs are closely associated with isoflavonoid biosynthesis. For some CHIs, the 4′-hydoxyl residues of chalcones may not be essential for high enzyme activities. Functional reconstruction of flavonoid and isoflavonoid biosynthesis from a shared chalcone precursor proved successful with various enzyme combinations. The broad substrate specificity of soybean type II CHIs resulted in the production of several flavonoid/isoflavonoid compounds in yeast. This in vivo system provides multiple opportunities for research and industries.
MATERIALS AND METHODS
EST Sequence Analysis and Cloning of cDNAs
CHI and F3H cDNA sequences were identified from the Plant Genome Database (http://www.plantgdb.org/; Dong et al., 2004) based on homology to the alfalfa (Medicago sativa) CHI (GenBank accession no. M91079) and F3H (X78994; Charrier et al., 1995). Sequence alignments were performed using the AlignX module of Vector NTI (Invitrogen, Carlsbad, CA). Non-soybean (Glycine max) CHIs were identified by searching TIGR gene indices and the Plant Genome Database using the TBLASTN algorithm. E values were set at 1e−40. Representative family members were grouped into radial tree phylogenies, generated using TreeView (Page, 1996). Identification of the putative signal peptide and prediction of chloroplast localization of CHI3 was performed using ChloroP 1.1 (Emanuelsson et al., 1999), iPSORT (Bannai et al., 2002), Predotar (http://www.inra.fr/predotar), and TargetP 1.0 (Emanuelsson et al., 2000).
CHI1A and CHI4A were cloned previously (O. Yu and J.T. Odell, unpublished data). ESTs containing CHI1B2 (BQ786323), CHI2 (AW733840), CHI3 (BM885445), and F3H (BI894148) were obtained from the Washington University School of Medicine in St. Louis. All of these clones were sequenced to verify that they contained complete ORFs as well as 5′- and 3′-UTRs. Appropriate restriction sites for subsequent cloning steps were introduced immediately upstream of the ATG start codon and downstream of the stop codon via PCR, with the exception of CHI3. The 66 amino acid N-terminal extension was removed from CHI3 by PCR to create CHI3Δ2-66 prior to expression studies. Amplicons were then cloned directly using the Zero Blunt TOPO PCR Cloning kit (Invitrogen) and verified by DNA sequencing before subcloning into bacterial or yeast (Saccharomyces cerevisiae) expression vectors. The CHIs and F3H sequences have been submitted to GenBank and assigned the following accession numbers: CHI1A (AY595413), CHI1B1 (AY595414), CHI1B2 (AY595419), CHI2 (AY595415), CHI3 (AY595416), CHI4A (AY595417), CHI4B (AY595418), and F3H (AY595420).
Quantitative RT-PCR Analysis of Gene Expression
RNA isolation and quantitative RT-PCR analysis were performed as described in Subramanian et al. (2004). Briefly, total RNA was isolated from soybean tissue using Trizol (Invitrogen). RNA was subsequently treated with DNase I to remove contaminating genomic DNA. First strand cDNA was prepared using Moloney murine leukemia virus reverse transcriptase (New England Biolabs, Beverly, MA). Quantitative RT-PCR was performed with SYBR-Green using an iCycler thermocycler (Bio-Rad, Hercules, CA). Estimates of initial transcript concentrations were performed using the comparative threshold cycle method (Bovy et al., 2002). PCR primers are listed in Supplemental Table II.
Bacterial Expression and Protein Purification
CHI1A, CHI1B2, CHI2, CHI3Δ2-66, and CHI4A were inserted into the pHIS8 bacterial expression vector at the NcoI and EcoRI sites (Jez et al., 2002). His8-CHIs were expressed in Rosetta (DE3) cells (Novagen, Madison, WI). Bacterial cultures were grown in Luria-Bertani media containing 50 mg L−1 kanamycin overnight at 37°C. The next day, the cultures were diluted and then grown to an A600 nm = 0.8 to 1. At this point, cultures were induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside and incubated overnight at 28°C. After induction, cultures were sonicated and His8-tagged CHIs were purified using Ni2+-NTA columns (Qiagen, Valencia, CA) according to manufacturer's recommendations. His-tags were subsequently removed by digestion with thrombin. Thrombin and undigested protein were removed using a mixed Ni2+-NTA/benzamidine-sepharose column (Qiagen).
Enzyme Assays
Chalcones, flavonoids, and isoflavonoids were purchased from Indofine Chemical, Hillsborough, NJ. Naringenin chalcone was chemically synthesized from naringenin (Shimokoriyama, 1957). CHI assays were carried out as described previously (Bednar and Hadcock, 1988; Jez and Noel, 2002) with minor modifications. Spectrophotometric assays were performed at 20°C in 0.5 mL reaction buffer (50 mm Tris-Cl, pH 8, 1% dimethyl sulfoxide, and 0.1 mg mL−1 bovine serum albumin) using 0.1 to 1 μg of enzyme and 50 μm isoliquiritigenin or naringenin chalcone as substrate. Initial velocity measurements were executed using 0.5 to 75 μm chalcones. Assays using naringenin chalcone as substrate contained 9 ng CHI1A, 6 ng CHI1B2, or 2 ng of CHI2. Assays using all other chalcone substrates contained 27 ng CHI1A, 16 ng CHI1B2, or 100 ng of CHI2. Assays were performed using a Varian Cary 50 Bio spectrophotometer (Palo Alto, CA). Reactions were initiated by the addition of enzyme, and initial rates were calculated based upon decrease in absorbance over time as described previously (Jez et al., 2002). Steady-state kinetic parameters (kcat and Km) were calculated by fitting the untransformed initial velocity data to the Michaelis-Menton equation with Kaleidagraph (Synergy Software, Reading, PA).
The in vitro IFS activity assay was carried out as described previously using microsomal extracts (Jung et al., 2000). First, 500 μL of IFS-containing microsomes were incubated with naringenin for 15 min to synthesize 2-hydroxyisoflavanone. The product was then extracted by ethyl acetate, dried, and redissolved in dimethyl sulfoxide. For inhibition assays, aliquots of 2-hydroxyisoflavanone were added to reactions containing: (1) wild-type microsome extracts, (2) wild-type total protein extracts, (3) IFS-containing microsomes, or (4) IFS-containing microsomes plus 5 μm ancymidol. The products from the second round of reactions were extracted with ethyl acetate and analyzed by HPLC.
For in vitro F3H activity assays, the F3H ORF was fused to an N-terminal FLAG tag and expressed using the in vitro Director PCR System (Sigma-Aldrich, St. Louis). The fusion protein was transcribed and translated in vitro and purified according to the manufacturer's protocol. Enzyme activity assays were carried out as described previously (Charrier et al., 1995).
Yeast Expression and in Vivo Characterization
CHI1A, CHI1B2, CHI2, CHI3Δ2-66, and CHI4A were cloned into the pESC-TRP yeast expression vector (Stratagene, La Jolla, CA) at the EcoRI and BglII sites under the control of the Gal10 promoter. IFS and F3H were cloned into the BamHI and EcoRI sites of the pYeDP60 expression vector (kindly provided by Dr. Philippe Urban), under the control of the Gal10 promoter (Pompon et al., 1996). FNSII was previously cloned into the pYES2 expression vector and kindly provided by Dr. Stefan Martens (Martens and Forkmann, 1999). CHIs were expressed singly, or in combination with IFS, FNSII, or F3H, in the WAT11 strain of yeast, also provided by Dr. Urban (Urban et al., 1997). The above constructs, as well as pESC-TRP and pYeDP60 empty vector controls, were transformed into WAT11 cells made competent with the Frozen-EZ Yeast Transformation II kit (Zymo Research, Orange, CA). Yeast cultures were grown overnight at 30°C in selective liquid media containing Glc as the carbon source. The next day, cultures were collected by centrifugation and washed in sterile water. Cultures were then resuspended to an A600 nm = 1 in selective media containing Gal. Chalcone or flavonoid substrates were then added to a final concentration of 50 to 100 μm. Approximately 12 to 15 h after induction, aliquots of cultures were sonicated and extracted with ethyl acetate to collect phenolic compounds. Extracts were concentrated to dryness with an Eppendorf Vacufuge (Eppendorf Scientific, Westbury, NY) at 30°C or 45°C and subsequently redissolved in methanol. Samples were then analyzed by HPLC and GC-MS.
Metabolite Analysis
HPLC samples were analyzed on a System Gold HPLC equipped with a photodiode-array detector (Beckman Coulter, Fullerton, CA), using a Luna C18 (2), 5-μm, 4.6- × 150-mm column (Phenomenex, Torrance, CA). Samples were diluted into ethyl acetate or methanol and then separated using an 18-min linear gradient from 20% methanol/80% 10 mm ammonium acetate, pH 5.6, to 100% methanol at a flow rate of 1 mL min−1. Elution of metabolites was monitored by photodiode array. Retention time and UV spectra were compared to those of authentic standards when available.
GC-MS analysis was used to confirm the identities of the various chalcone, flavonoid, and isoflavonoid metabolites. Samples were first converted to TMS ether derivatives by adding N,O-bis(TMS)-trifluoroacetamide (Sigma-Aldrich) and incubating at 37°C for 1 h. Samples were then dried under nitrogen and dissolved in chloroform. GC-MS analysis was conducted using a ThermoFinnigan Trace 2000 GC (Thermoquest, San Jose, CA) equipped with an AS2000 autoinjector and a GCQ-plus Mass Spectrometer System. Samples were resolved using a 5-μm, 15-m × 0.25-mm i.d. DB-5ms column (J&W Scientific, Folsom, CA). The oven temperature was programmed from 200°C (3.5 min hold) to 300°C at a rate of 5°C min−1 with a column flow rate of 0.75 mL min−1 He. The ionization potential of the mass selective detector was 70 eV. TMS derivatives were identified by comparison of retention times and mass spectra with that of standards derivatized with N,O-bis(TMS)-trifluoroacetamide when available.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY595413, AY595414, AY595419, AY595415, AY595416, AY595417, AY595418, and AY595420.
Supplementary Material
Acknowledgments
We thank Laura Walker and Chris Menne for technical assistance, Dr. Joe Jez for help on CHI in vitro assays and critical reading of the manuscript, and Drs. Jan Jaworski and Gene Guo for valuable discussions. We also thank Dr. Philip Urban for providing the WAT11 yeast strain, Dr. Stefan Martens for FNSII constructs, Dr. Gary Stacey for B. japonicum strains, Deana Pape for cDNAs, and Kristen Opper for in vitro expression of F3H.
This work was supported in part by the U.S. Department of Agriculture (postdoctoral fellowship no. 2002–35318–12593 to L.R.), by the Illinois-Missouri Biotechnology Alliance (grant no. 34346–13070 to O.Y.), and by the Missouri Soybean Merchandising Council (grant no. 03–242 to O.Y.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.054502.
References
- Achnine L, Blancaflor EB, Rasmussen S, Dixon RA (2004) Colocalization of L-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoid biosynthesis. Plant Cell 16: 3098–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi T, Aoki T, Ayabe S (1999) Cloning and functional expression of a cytochrome P450 cDNA encoding 2-hydroxyisoflavanone synthase involved in biosynthesis of the isoflavonoid skeleton in licorice. Plant Physiol 121: 821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S (2002) Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 18: 298–305 [DOI] [PubMed] [Google Scholar]
- Bednar RA, Hadcock JR (1988) Purification and characterization of chalcone isomerase from soybeans. J Biol Chem 263: 9582–9588 [PubMed] [Google Scholar]
- Beecher GR (2003) Overview of dietary flavonoids: nomenclature, occurrence and intake. J Nutr 133: 3248S–3254S [DOI] [PubMed] [Google Scholar]
- Bovy A, de Vos R, Kemper M, Schijlen E, Pertejo MA, Muir S, Collins G, Robinson S, Verhoeyen M, Hughes S (2002) High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1. Plant Cell 14: 2509–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier B, Coronado C, Kondorosi A, Ratet P (1995) Molecular characterization and expression of alfalfa (Medicago sativa L.) flavanone-3-hydroxylase and dihydroflavonol-4-reductase encoding genes. Plant Mol Biol 29: 773–786 [DOI] [PubMed] [Google Scholar]
- Deboo GB, Albertsen MC, Taylor LP (1995) Flavanone 3-hydroxylase transcripts and flavonol accumulation are temporally coordinate in maize anthers. Plant J 7: 703–713 [DOI] [PubMed] [Google Scholar]
- Dewick PM (1993) Isoflavonoids. In JB Harborne, ed, The Flavonoids: Advances in Research Since 1986. Chapman & Hall, London, pp 117–238
- Dixon RA, Steele CL (1999) Flavonoids and isoflavonoids: a gold mine for metabolic engineering. Trends Plant Sci 4: 394–400 [DOI] [PubMed] [Google Scholar]
- Dong Q, Schlueter SD, Brendel V (2004) PlantGDB: plant genome database and analysis tools. Nucleic Acids Res 32: D354–D359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Braun EL, Grotewold E (2001) Functional conservation of plant secondary metabolic enzymes revealed by complementation of Arabidopsis flavonoid mutants with maize genes. Plant Physiol 127: 46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gensheimer M, Mushegian A (2004) Chalcone isomerase family and fold: no longer unique to plants. Protein Sci 13: 540–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TL (1991) Flavonoid and isoflavonoid distribution in developing soybean seedling tissues and in seed and root exudates. Plant Physiol 95: 594–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E, Drummond BJ, Bowen B, Peterson T (1994) The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76: 543–553 [DOI] [PubMed] [Google Scholar]
- Hakamatsuka T, Mori K, Ishida S, Ebizuka Y, Sankawa U (1998) Purification of 2-hydroxyisoflavanone dehydratase from the cell cultures of Pueraria lobata. Phytochemistry 49: 497–505 [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, et al (2003) Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425: 191–196 [DOI] [PubMed] [Google Scholar]
- Hur S, Newby ZE, Bruice TC (2004) Transition state stabilization by general acid catalysis, water expulsion, and enzyme reorganization in Medicago savita chalcone isomerase. Proc Natl Acad Sci USA 101: 2730–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang EI, Kaneko M, Ohnishi Y, Horinouchi S (2003) Production of plant-specific flavanones by Escherichia coli containing an artificial gene cluster. Appl Environ Microbiol 69: 2699–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irani NG, Grotewold E (2003) Chalcone isomerase: more than just an enzyme? In Phytochemical Society of North America 2003 Annual Meeting, August 9–13, 2003, Peoria, IL
- Jez JM, Bowman ME, Dixon RA, Noel JP (2000) Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase. Nat Struct Biol 7: 786–791 [DOI] [PubMed] [Google Scholar]
- Jez JM, Bowman ME, Noel JP (2002) Role of hydrogen bonds in the reaction mechanism of chalcone isomerase. Biochemistry 41: 5168–5176 [DOI] [PubMed] [Google Scholar]
- Jez JM, Noel JP (2002) Reaction mechanism of chalcone isomerase: pH dependence, diffusion control, and product binding differences. J Biol Chem 277: 1361–1369 [DOI] [PubMed] [Google Scholar]
- Jung W, Yu O, Lau SM, O'Keefe DP, Odell J, Fader G, McGonigle B (2000) Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat Biotechnol 18: 208–212 [DOI] [PubMed] [Google Scholar]
- Kimura Y, Aoki T, Ayabe S (2001) Chalcone isomerase isozymes with different substrate specificities towards 6′-hydroxy- and 6′-deoxychalcones in cultured cells of Glycyrrhiza echinata, a leguminous plant producing 5-deoxyflavonoids. Plant Cell Physiol 42: 1169–1173 [DOI] [PubMed] [Google Scholar]
- Liu CJ, Blount JW, Steele CL, Dixon RA (2002) Bottlenecks for metabolic engineering of isoflavone glycoconjugates in Arabidopsis. Proc Natl Acad Sci USA 99: 14578–14583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens S, Forkmann G (1999) Cloning and expression of flavone synthase II from Gerbera hybrids. Plant J 20: 611–618 [DOI] [PubMed] [Google Scholar]
- McKhann HI, Hirsch AM (1994) Isolation of chalcone synthase and chalcone isomerase cDNAs from alfalfa (Medicago sativa L.): highest transcript levels occur in young roots and root tips. Plant Mol Biol 24: 767–777 [DOI] [PubMed] [Google Scholar]
- Ni W, Fahrendorf T, Ballance GM, Lamb CJ, Dixon RA (1996) Stress responses in alfalfa (Medicago sativa L.): transcriptional activation of phenlpropanoid pathway genes in elicitor-induced cell suspension cultures. Plant Mol Biol 30: 427–438 [DOI] [PubMed] [Google Scholar]
- Ovadi J, Srere PA (2000) Macromolecular compartmentation and channeling. Int Rev Cytol 192: 255–280 [DOI] [PubMed] [Google Scholar]
- Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P (1996) Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol 272: 51–64 [DOI] [PubMed] [Google Scholar]
- Ro DK, Douglas CJ (2004) Reconstitution of the entry point of plant phenylpropanoid metabolism in yeast (Saccharomyces cerevisiae): implications for control of metabolic flux into the phenylpropanoid pathway. J Biol Chem 279: 2600–2607 [DOI] [PubMed] [Google Scholar]
- Sawada Y, Kinoshita K, Akashi T, Aoki T, Ayabe S (2002) Key amino acid residues required for aryl migration catalyzed by the cytochrome P450 2-hydroxyisoflavone synthase. Plant J 31: 555–564 [DOI] [PubMed] [Google Scholar]
- Shimada N, Aoki T, Sato S, Nakamura Y, Tabata S, Ayabe S (2003) A cluster of genes encodes the two types of chalcone isomerase involved in the biosynthesis of general flavonoids and legume-specific 5-deoxy(iso)flavonoids in Lotus japonicus. Plant Physiol 131: 941–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokoriyama M (1957) Interconversion of chalcones and flavanones of a phloroglucinol-type structure. J Am Chem Soc 79: 4199–4202 [Google Scholar]
- Shirley BW, Hanley S, Goodman HM (1992) Effects of ionizing radiation on a plant genome: analysis of two Arabidopsis transparent testa mutations. Plant Cell 4: 333–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit G, Puvanesarajah V, Carlson RW, Barbour WM, Stacey G (1992) Bradyrhizobium japonicum nodD1 can be specifically induced by soybean flavonoids that do not induce the nodYABCSUIJ operon. J Biol Chem 267: 310–318 [PubMed] [Google Scholar]
- Steele CL, Gijzen M, Qutob D, Dixon RA (1999) Molecular characterization of the enzyme catalyzing the aryl migration reaction of isoflavonoid biosynthesis in soybean. Arch Biochem Biophys 367: 146–150 [DOI] [PubMed] [Google Scholar]
- Subramanian S, Xu L, Lu G, Odell J, Yu O (2004) The promoters of the isoflavone synthase genes respond differentially to nodulation and defense signals in transgenic soybean roots. Plant Mol Biol 54: 623–639 [DOI] [PubMed] [Google Scholar]
- Urban P, Mignotte C, Kazmaier M, Delorme F, Pompon D (1997) Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J Biol Chem 272: 19176–19186 [DOI] [PubMed] [Google Scholar]
- Weisshaar B, Jenkins GI (1998) Phenylpropanoid biosynthesis and its regulation. Curr Opin Plant Biol 1: 251–257 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B (1999) Evidence for enzyme complexes in the phenylpropanoid and flavonoid pathways. Physiol Plant 107: 142–149 [Google Scholar]
- Yu O, Jung W, Shi J, Croes RA, Fader GM, McGonigle B, Odell JT (2000) Production of the isoflavones genistein and daidzein in non-legume dicot and monocot tissues. Plant Physiol 124: 781–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu O, McGonigle B (2005) Metabolic engineering of isoflavone biosynthesis. Adv Agron 86: 147–190 [Google Scholar]
- Yu O, Shi J, Hession AO, Maxwell CA, McGonigle B, Odell JT (2003) Metabolic engineering to increase isoflavone biosynthesis in soybean seed. Phytochemistry 63: 753–763 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.