Abstract
The model plants Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) have provided a wealth of information about genes and genetic pathways controlling the flowering process, but little is known about the corresponding pathways in legumes. The garden pea (Pisum sativum) has been used for several decades as a model system for physiological genetics of flowering, but the lack of molecular information about pea flowering genes has prevented direct comparison with other systems. To address this problem, we have searched expressed sequence tag and genome sequence databases to identify flowering-gene-related sequences from Medicago truncatula, soybean (Glycine max), and Lotus japonicus, and isolated corresponding sequences from pea by degenerate-primer polymerase chain reaction and library screening. We found that the majority of Arabidopsis flowering genes are represented in pea and in legume sequence databases, although several gene families, including the MADS-box, CONSTANS, and FLOWERING LOCUS T/TERMINAL FLOWER1 families, appear to have undergone differential expansion, and several important Arabidopsis genes, including FRIGIDA and members of the FLOWERING LOCUS C clade, are conspicuously absent. In several cases, pea and Medicago orthologs are shown to map to conserved map positions, emphasizing the closely syntenic relationship between these two species. These results demonstrate the potential benefit of parallel model systems for an understanding of flowering phenology in crop and model legume species.
The change from vegetative to reproductive growth is a critical developmental transition in the life of a plant, and the induction, expression, and maintenance of the flowering state are regulated by many external and endogenous factors. A vast number of applied and fundamental studies have demonstrated the importance of light (through daylength and light-quality effects) and temperature (through vernalization and ambient temperature effects) as the main environmental regulators of flowering. However, other factors, including nutrient status, endogenous hormones, stress, and the developmental state of the plant, can also be important. Even with respect to light and temperature, great diversity in responsiveness exists within and between different plant species. These differences are important in the adaptation of species to particular latitudinal and climatic regions, and have also been extremely important for determining the environments and agronomic regimes under which crop species can be most effectively grown.
The flowering process has been subject to detailed genetic analysis in Arabidopsis (Arabidopsis thaliana). As a small, weedy annual, Arabidopsis is responsive to a wide range of factors and has been invaluable in outlining the major genetic pathways that are likely to function in the control of flowering responses to photoperiod, vernalization, and hormone responses (Amasino, 2004; Boss et al., 2004; Putterill et al., 2004). It is likely that many of the genetic mechanisms discovered in Arabidopsis identify general themes that have been elaborated in different ways across the plant kingdom. However, plants show incredible diversity in growth habit and phenology, and it is clear that we have only scratched the surface in understanding how this diversity might be generated.
Several reports provide an illustration of the ways in which similar basic mechanisms might be adapted to produce quite different patterns of environmental response. Recent comparative studies have shown that the function of several genes involved in photoperiod responsiveness is conserved between Arabidopsis and rice (Oryza sativa), and suggest that the difference between long-day (LD)- and short-day (SD)-responsive plants results from a different regulatory interaction between two genes, CONSTANS (CO) and FLOWERING LOCUS T (FT; Hayama et al., 2003). In other cases, different genes can achieve similar patterns of environmental response. For example, in both Arabidopsis and wheat (Triticum aestivum), vernalization acts to promote flowering by repressing the expression of an important floral regulator. In Arabidopsis, this repressor is the MADS-domain protein FLOWERING LOCUS C (FLC), whereas in cereals, which do not appear to possess FLC-like genes, the corresponding role is played by an unrelated zinc-finger transcription factor (Yan et al., 2004).
With the advent of genomic approaches in a range of model plant systems, the information gained from Arabidopsis is rapidly being extended into other species. The complete sequence of the rice genome has allowed a global comparison of flowering pathways between rice and Arabidopsis (Izawa et al., 2003), and the same kind of analysis is also now possible in poplar. A number of studies have already provided detailed phylogenetic descriptions of particular flowering-related gene families and/or functional analysis of individual genes in species such as rice, barley (Hordeum vulgare), tomato (Lycopersicon esculentum), petunia (Petunia hybrida), and Antirrhinum (e.g. Carmel-Goren et al., 2003; Griffiths et al., 2003; Hayama et al., 2003; Vandenbussche et al., 2003). However, there has been only limited molecular analysis of flowering in legumes, despite the importance of flowering time in legume production systems and the availability of extensive expressed sequence tag (EST) collections for model legumes such as soybean (Glycine max) and Medicago truncatula. In soybean, precise genetic control of flowering time has been achieved using classical breeding and is essential for efficient cropping in different latitudinal and climatic regions (e.g. Curtis et al., 2000), but molecular information about the genes involved has not yet emerged into the public domain. The timing of flowering is also an agronomically important trait in many other legume species, including pea (Pisum sativum), bean (Phaseolus spp.), lentil (Lens culinaris), chickpea (Cicer arietinum), and lupin (Lupinus spp.; Huyghe, 1998), and genetic variation for flowering is being utilized in all of these (e.g. Wallace et al., 1993; Sarker et al., 1999; Kumar and van Rheenen, 2000). A better understanding of flowering control in legumes will also benefit general understanding of the flowering process. Crop and model legumes exhibit great diversity in phenology with respect to photoperiod and temperature responses, lifespan, mono/polycarpy, and the determinacy and architecture of inflorescences. For example, soybean is a vernalization-unresponsive SD species (Summerfield and Roberts, 1985), whereas both M. truncatula and Lotus japonicus are vernalization-responsive LD species (Clarkson and Russell, 1975). Both soybean and M. truncatula are annual, but another closely related Medicago species, alfalfa (Medicago sativa), and L. japonicus are perennial (Handberg and Stougaard, 1992).
It is only in the garden pea, an annual, vernalization-responsive LD species, that genetic, physiological, and molecular approaches to flowering have converged to any appreciable extent. Pioneering physiological-genetic studies through the 1970s identified a number of major flowering genes in pea and provided a model for the flowering process that incorporated vernalization, photoperiod, and mobile flowering signals (Murfet, 1985; Weller et al., 1997). The lack of subsequent progress in identifying these genes at the molecular level has meant that, until recently, it has not been possible to relate this model to those in other systems. However, several recent reports have presented mutant-based functional analyses of flowering-related genes in pea, including the photoreceptor genes PHYTOCHROME A (PHYA) and PHYB, and homologs of Arabidopsis inflorescence identity genes LEAFY (LFY), UNUSUAL FLOWER ORGAN (UFO), and APETALA1 (AP1; Hofer et al., 1997; Berbel et al., 2001; Taylor et al., 2001, 2002; Weller et al., 2001, 2004). Interestingly, the LFY ortholog in pea is not only involved in floral initiation, as it is in Arabidopsis, but also in leaf development, a function not described in Arabidopsis (Hofer et al., 1997). In other studies, the pea homologs of AP1 and PISTILLATA (PI) have been shown to fully complement the corresponding Arabidopsis mutants, despite lacking C-terminal motifs suggested to be essential for the function of the Arabidopsis genes (Yalovsky et al., 2000; Berbel et al., 2001; A. Berbel, C. Navarro, C. Ferrándiz, L. Cañas, J.-P. Beltrán, and F. Madueño, unpublished data). Other pea flowering loci, DETERMINATE (DET) and LATE FLOWERING (LF), have recently been shown to be homologs of the Arabidopsis gene TERMINAL FLOWER1 (TFL1; Foucher et al., 2003). The DET gene maintains indeterminacy of primary shoot apex, which in det mutants is converted into a determinate, secondary inflorescence. The LF gene delays flowering in a photoperiod-independent manner, and loss-of-function mutants are early flowering but retain photoperiod responsiveness. It thus appears that multiple roles of Arabidopsis TFL1 may have been differentially apportioned in different pea homologs. Overall, these reports suggest that basic flowering pathways are likely to be relatively well conserved in pea and other legumes and support the use of a candidate gene approach as a first step in identifying the molecular nature of other pea flowering genes.
We set out to define on a broad scale the extent to which genes important for the flowering process in Arabidopsis are conserved in model legumes. We found that a large proportion of Arabidopsis flowering genes are represented in legumes, and have isolated partial sequences for many of these genes from pea. Preliminary mapping analyses emphasize the close synteny between pea and Medicago and suggest some potential candidate genes for known pea mutants.
RESULTS
Identification of Flowering-Related Genes in Legumes and Isolation of Corresponding Sequences from Pea
We first compiled a list of Arabidopsis genes thought to play an important role in some aspect of the flowering process. These genes included those involved in photoperception, circadian clock function, photoperiod response, vernalization response, autonomous flowering, integration of flowering pathway signals, and the development of inflorescences and flowers, as well as a range of other flowering-related genes whose function has not been clearly categorized. We performed BLAST searches (tBLASTn) for each Arabidopsis gene in turn against gene indices for Medicago, soybean, and Lotus. BLAST hits were visually assessed for degree of amino acid conservation, and high-ranked or otherwise selected sequences were then used in tBLASTx queries of the Arabidopsis genome. EST contigs not retrieving the original Arabidopsis sequence were excluded from further analysis.
Each of the Arabidopsis genes and each of the Medicago EST contigs were also used to query the bacterial artificial chromosome (BAC) sequence database from the in-progress Medicago genome sequencing database (MtGenome at University of California, Davis). For searches with Arabidopsis genes, high-ranking hits or lower-ranking hits corresponding to short, highly conserved regions were selected by inspection. Where relevant, the full coding sequence of the gene was identified in the corresponding BAC sequence using the Arabidopsis gene structure as a guide. As in the case of the EST contigs, any sequences not returning the original Arabidopsis sequence by tBLASTn were excluded.
In some cases, this process of reciprocal BLAST searches indicated an unambiguous relationship between the Arabidopsis gene and a particular legume sequence. For example, a search with the single-copy Arabidopsis GIGANTEA (GI) gene returned a single Medicago EST contig that showed 68% identity across a 767-amino acid region and is highly likely to be the Medicago GI ortholog. In other cases where the original query was part of a gene family (e.g. MADS-box, CO-like, and FT/TFL1-like families), further analysis was necessary to assess the closest relationships and probable identity of the legume hits. This included a closer examination of sequence motifs, additional searches using less-conserved protein domains, and phylogenetic analyses.
Medicago sequences identified in this way were then used to isolate corresponding pea cDNA sequences. Degenerate primers were designed for conserved sequence blocks using CODEHOP software (Rose et al., 1998; Supplemental Table I). In some cases, the partial pea sequence obtained using these primers was used to isolate longer clones from a pea shoot cDNA library, or by RACE-PCR.
MADS-Box Gene Family
MADS-box proteins are transcription factors that control a diverse range of developmental processes in plants (Becker and Theissen, 2003). They are characterized by a highly conserved N-terminal DNA-binding domain termed the MADS box. The MADS-box gene family contains more than 100 members in Arabidopsis and comprises five major clades, of which only one, the so-called MIKC class, has been subject to significant functional analysis (Becker and Theissen, 2003). There are 39 MIKC class genes in Arabidopsis (Parenicova et al., 2003), and these include genes acting in the control of flowering time (FLC, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 [SOC1], SHORT VEGETATIVE PHASE [SVP]), floral meristem identity (AP1, FRUITFULL [FUL], CAULIFLOWER [CAL]), floral organ identity (AP3, PI, SEPALLATA [SEP]), fruit formation and ovule identity (AGAMOUS [AG], SHATTERPROOF [SHP]), seed development (TRANSPARENT TESTA16 [TT16]), and root development (ANR1). A number of independent phylogenetic analyses have described distinct clades within the MIKC class (Becker and Theissen, 2003; Kofuji et al., 2003; Parenicova et al., 2003), although the relationship among these clades has not been convincingly resolved.
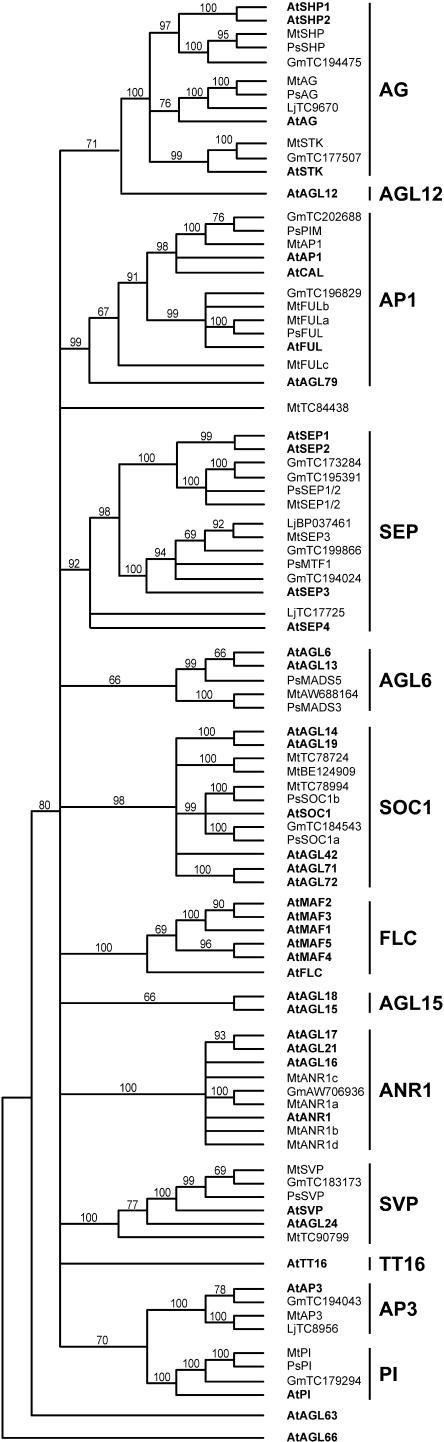
We identified 22 distinct MIKC-class MADS-box sequences from Medicago EST and genomic sequences (Supplemental Table II). The cladogram in Figure 1 shows that these sequences are distributed across 9 of the 14 groups defined by Becker and Theissen (2003). No sequences belonging to the AGAMOUS-LIKE12 [AGL12], AGL15, TM8, GGM13/TT16 (“B-sister”), or FLC groups were identified in Medicago, soybean, or Lotus databases. The first four of these clades are not well studied, but the FLC clade in Arabidopsis contains several genes with a role in regulating flowering time (Boss et al., 2004). FLC is the best characterized of these genes and is a central repressor of flowering and an important mediator of the vernalization response (Boss et al., 2004). We also tried unsuccessfully to isolate FLC homologs in pea using degenerate primers.
Figure 1.
Phylogenetic analysis of MIKC MADS-box genes in legumes and Arabidopsis. Neighbor-joining tree for amino acid sequences spanning the M, I, and K domains (185 characters) aligned with ClustalX and rooted on AtAGL66 is shown. The bootstrap values are indicated as a percentage above each branch. Ps, garden pea; Mt, M. truncatula; Lj, L. japonicus; Gm, soybean. Sequence accession numbers are: MtAG (AC137837); MtANR1a (AC123898); MtANR1b (AC126010, TC82622); MtANR1c (TC81292); MtANR1d (AC144564); MtAP1 (AC144726); MtAP3 (AC136451, TC90653); MtFULa (TC84496); MtFULb (TC82227); MtFULc (AC146650b, AL387855); MtPI (TC92737); MtSEP1/2 (AC146650a, BQ123807); MtSEP3 (AC144644); MtSHP (TC86876); MtSTK (TC93057); MtSVP (AC135848, TC87621).
The Arabidopsis AP1 gene confers A-function in the floral meristem and has additional roles in specifying inflorescence identity (Jack, 2004). The AP1 clade has four members in Arabidopsis, including the AP1 and CAL genes; FUL, which functions redundantly with AP1/CAL; and AGL79 (Parenicova et al., 2003). Unlike AP1, CAL, and FUL, AGL79 is expressed predominantly in roots (Parenicova et al., 2003), suggesting that it may differ in function from the other three genes. In Medicago, AP1 is represented by a single genomic sequence and FUL by two very similar EST sequences, MtFULa and MtFULb (Fig. 1). These ESTs were derived from flower and pod libraries. A third, more divergent Medicago EST sequence was represented by a genomic clone and a single isolate from a root cDNA library. The relationship of this gene to other members of the AP1/FUL clade was not clear, but for convenience we have referred to it as MtFULc. In pea, an AP1/CAL ortholog has been shown to correspond to the inflorescence identity gene PROLIFERATING INFLORESCENCE MERISTEM (PIM; Taylor et al., 2002). We also isolated a full-length sequence for a single pea FUL ortholog that corresponds to a partial sequence previously reported by Litt and Irish (2003).
Together with the AGL6/AGL13 group, the four Arabidopsis SEP genes form a distinct sister clade to the AP1 group in most phylogenetic analyses (e.g. Becker and Theissen, 2003; Kofuji et al., 2003). The Arabidopsis SEP proteins interact with A-, B-, and C-class MADS-box proteins to confer organ identity in all four floral whorls (Pelaz et al., 2000; Honma and Goto, 2001; Ditta et al., 2004). As in the case of AP1 and CAL, Arabidopsis SEP1 and SEP2 are very similar and are likely to reflect a recent duplication. Consistent with this interpretation, we found only two clearly SEP-like sequences in Medicago: one ortholog of SEP1/2 and another gene more closely related to SEP3. We isolated a single pea cDNA corresponding to SEP1/2, and identified the previously described pea gene MTF1 (Buchner and Boutin, 1998) as an ortholog of SEP3. In Lotus databases, we found nothing corresponding to SEP1/2 but a clear SEP3-like sequence and a more distantly related SEP sequence that may be more similar to SEP4. In soybean, both SEP1/2 and SEP3 are represented by two closely related but distinct EST contigs. The function of the Arabidopsis AGL6 and AGL13 genes has not yet been demonstrated, but these genes and closely related genes in other species are mainly expressed in floral organs and ovules (Rounsley et al., 1995; Immink et al., 2003) and can influence flowering time when ectopically expressed (e.g. Carlsbecker et al., 2004). We isolated two distinct pea genes belonging to this clade (PsMADS3 and PsMADS5) but could only identify single EST contigs in Medicago and soybean (Fig. 1; Supplemental Table II).
The SOC1/AGL20 gene was first identified as a flowering-related gene in Sinapis alba (Menzel et al., 1996). It was subsequently shown to be a direct target for regulation by CO (Samach et al., 2000) and also to have an important role in integration of signals from photoperiod, GA, and vernalization pathways (Moon et al., 2003). In Arabidopsis, the clade containing SOC1 has five other members. Nothing is yet known about the function of these other genes, but one of them (AGL14) is expressed predominantly in roots (Rounsley et al., 1995), suggesting that not all members of this clade have a role in flowering. We identified three distinct Medicago EST contigs belonging to this clade. Of these, one (TC78994) is clearly a close homolog of SOC1, while the other two are less closely related and do not correspond clearly to any other gene in the SOC1 clade. Interestingly, these two contigs are comprised of ESTs obtained predominantly from root tissue. A clear SOC1 sequence was also present in soybean. Using the Medicago sequence, we isolated two different SOC1-related sequences from pea (PsSOC1a and PsSOC1b) that are both more closely related to SOC1 than to any other member of the Arabidopsis SOC1 clade.
The SVP gene acts as a dosage-dependent repressor of flowering (Hartmann et al., 2000), whereas AGL24 is a negative regulator of floral meristem identity that is repressed by LFY and AP1 during floral meristem development (Yu et al., 2004). Two SVP-like genes were present in Medicago databases. One of these was very similar to SVP and was also represented in soybean and Lotus. We isolated a partial pea sequence of this putative SVP ortholog. The other Medicago sequence (TC90799) was somewhat less similar to SVP but could not be clearly identified as either SVP or AGL24 (Fig. 1).
We also identified legume sequences belonging to three other groups of MADS-box genes less directly relevant to an analysis of the floral transition. The AP3 and PI genes confer B-function in floral meristem identity (Jack, 2004). AP3 and PI sequences are both present in Medicago and soybean (Fig. 1), and we isolated the apparent pea ortholog of PI. The four Arabidopsis members of the AGAMOUS clade (AG/SHP1/SHP2/SEEDSTICK [STK]) are all expressed mainly in carpels/ovules and contribute to aspects of carpel and ovule identity or development (Pinyopich et al., 2003). We identified Medicago ESTs corresponding to AG, SHP1/2, and STK, and isolated pea cDNAs corresponding to AG and SHP1/2. We also obtained an additional hit on the Medicago genomic sequence (AC137828) that appeared to belong within this clade, but we could not retrieve a full MIKC coding sequence for this gene. AG is represented in Lotus, and SHP and STK genes in soybean. The Arabidopsis ANR1 clade comprises four members that are predominantly expressed in roots (Burgeff et al., 2002), and the ANR1 gene itself controls lateral root development in response to 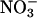 supply (Zhang and Forde, 1998). Four distinct Medicago sequences and a single soybean EST belonging to this clade were identified, but the relationship of these sequences to the Arabidopsis ANR1-like genes was not analyzed.
supply (Zhang and Forde, 1998). Four distinct Medicago sequences and a single soybean EST belonging to this clade were identified, but the relationship of these sequences to the Arabidopsis ANR1-like genes was not analyzed.
CONSTANS-LIKE Gene Family
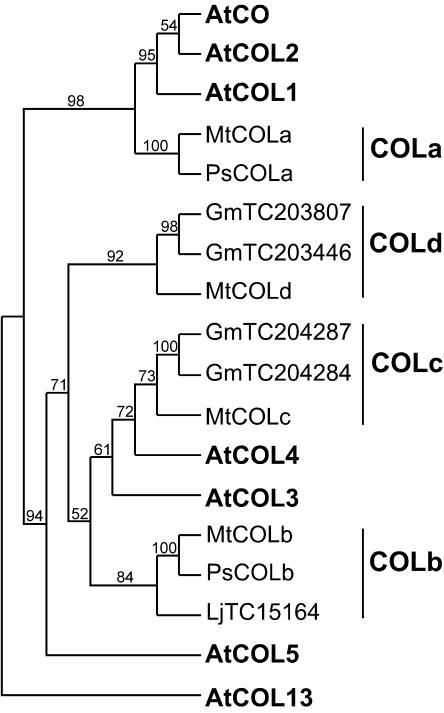
The CO gene was originally defined by an allelic series of mutants that flower late in LD and do not respond to photoperiod (Koornneef et al., 1991). CO has subsequently been shown to encode a transcription factor that plays a central role in the photoperiod detection mechanism (Putterill et al., 1995). Distinctive features of the CO protein include a zinc-finger region near the N terminus that resembles two B-box domains and a region near the C terminus designated the CCT domain (Putterill et al., 1995). CO is part of a 17-member gene family in Arabidopsis that consists of 3 broad clades, which are referred to as groups I, II, and III by Griffiths et al. (2003). Group I genes (CO and CONSTANS-LIKE1 [COL1]–COL5) have essentially same domain structure as CO, whereas group II genes (COL6–COL8 and COL16) have only a single B-box, and in group III genes (COL9–COL15) the second B-box is replaced by a more divergent zinc-finger domain (Griffiths et al., 2003). However, apart from CO itself, little is known about the function of these other CO-like genes, and we therefore restricted our focus to group I genes only. In Arabidopsis, these genes fall into two distinct groups: CO/COL1/COL2 (group Ia) and COL3/4/5 (group Ic). Three other subgroups of group I COL genes are described only from barley and rice and may be monocot specific (Griffiths et al., 2003).
We identified four group I COL sequences in Medicago databases, which included one group Ia sequence (designated MtCOLa) and three distinct group Ic sequences (designated MtCOLb–MtCOLd). A full-length cDNA corresponding to MtCOLa was isolated from pea and designated PsCOLa. The cladogram in Figure 2 shows that MtCOLa and PsCOLa genes form a sister group to Arabidopsis CO/COL1/COL2, while the three other Medicago genes fall within the COL3 to COL5 clade. One of these genes (designated MtCOLc) clusters with COL3 and COL4, whereas the other two genes (designated MtCOLb and MtCOLd) are more divergent, falling between COL3/COL4 and COL5. A single EST from Lotus shows greatest similarity to MtCOLb, whereas MtCOLc and MtCOLd each appear to be represented by a pair of closely related EST contigs in soybean (Fig. 2; Supplemental Table III). A full-length cDNA for a pea COLb sequence (PsCOLb) was also isolated independently by library screening and RACE-PCR.
Figure 2.
Phylogenetic analysis of CO-like genes in legumes and Arabidopsis. Neighbor-joining tree for a concatenation of the two B-box and CCT amino acid domains (135 characters) aligned with ClustalX and rooted on AtCOL13 is shown. Bootstrap values are indicated as a percentage above each branch. Ps, garden pea; Mt, M. truncatula; Lj, L. japonicus; Gm, soybean. Sequence accession numbers are: MtCOLa (TC86982); MtCOLb (TC87753); MtCOLc (TC78669); MtCOLd (AC127169, AW693899, TC88293).
FT/TFL1 Gene Family
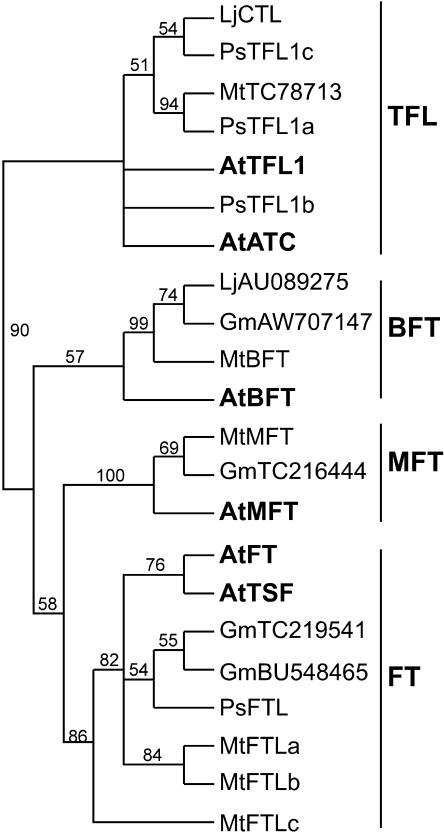
Like CO, the FT gene was originally defined by an allelic series of mutants that flower late in LD and do not respond to photoperiod (Koornneef et al., 1991). The FT gene encodes a small protein with weak similarity to the mammalian Raf kinase inhibitor protein and the phosphatidylethanolamine-binding protein (Kardailsky et al., 1999; Kobayashi et al., 1999) and is a direct regulatory target of CO (Samach et al., 2000). Recent reports have suggested that the FT protein may participate in the mobile flowering stimulus (An et al., 2004). Arabidopsis FT is a member of a six-gene family that includes another important flowering-related gene, TFL1. Unlike FT, TFL1 acts to delay the transition to flowering and also promotes the indeterminacy of the primary inflorescence (Bradley et al., 1997; Ratcliffe et al., 1998). Less is known about the function of other members of the family, but preliminary evidence suggests that they may also contribute to regulation of flowering time (Mimida et al., 2001; Yoo et al., 2004). Comparative studies of the FT/TFL1 family in tomato (Carmel-Goren et al., 2003) and rice (Izawa et al., 2002) suggest the presence of four ancient clades corresponding to Arabidopsis FT/TSF, TFL1/ATC, BROTHER OF FT AND TFL1 (BFT), and MOTHER OF FT AND TFL1 (MFT).
Figure 3 shows a cladogram derived from alignment of partial amino acid sequences from a number of FT/TFL-like genes identified in legumes. The isolation of three pea genes belonging to the TFL1 clade has been described previously (Foucher et al., 2003). Only one of these TFL-like genes (DET/TFL1a) was represented by a Medicago EST sequence. Lotus databases contained only one TFL1-like sequence that was most similar to LF/TFL1c, and no TFL-like sequences were present in soybean. MFT and BFT clades were clearly represented in both Medicago and soybean. Genes belonging to the FT/TSF clade were represented in soybean by two distinct ESTs, and in Medicago genomic sequence by three genes located on the same BAC, which we have designated FTLa, FTLb, and FTLc (Fig. 3; Supplemental Table IV). None of these three genes matched an EST sequence, and it is not yet known whether any are expressed. However, we succeeded in isolating FT-like partial cDNA sequences from pea (PsFTL) and other closely related legumes (Vicia, Lens, Trifolium) using the degenerate primer approach (Fig. 3; V. Hecht, unpublished data).
Figure 3.
Phylogenetic analysis of FT/TFL1-like genes in legumes and Arabidopsis. Neighbor-joining tree for the amino acid domain corresponding to exons 2, 3, and 4 (118 characters) aligned with ClustalX and rooted at the midpoint is shown. The bootstrap values are indicated as a percentage above each branch. Ps, garden pea; Mt, M. truncatula; Lj, L. japonicus; Gm, soybean. Sequence accession numbers are: MtBFT (AC146807); MtMFT (TC81061); MtFTLa (AC123593a); MtFTLb (AC123593b); MtFTLc (AC123593c).
Photoreceptors and Light Signaling
Light is an important regulator of the floral transition and affects flowering in several different ways (Boss et al., 2004). Two groups of photoreceptors have a well-documented role in control of flowering; the red and far-red light-absorbing phytochrome family and the blue light-absorbing cryptochrome family. Recent evidence suggests that these photoreceptors interact to control FT expression both by influencing the stability of the CO protein (Valverde et al., 2004) and by other CO-independent mechanism(s) (Cerdán and Chory, 2003). Two phytochromes, phyA and phyB, have been characterized previously in pea and shown to have substantial effects on flowering consistent with the roles of their homologs in Arabidopsis (Weller et al., 2001, 2004). PHYA and PHYB each appear to be represented by a single gene in both Medicago and Lotus databases (Table I). The situation in soybean is more complex, where four EST contigs for PHYA are present, including two that correspond to previously reported full-length cDNAs and two others that are clearly distinct. A sequence corresponding to Arabidopsis PHYE is also present in soybean. The cryptochrome family in pea has also been characterized in some detail and consists of a single CRYPTOCHROME1 (CRY1) gene and two distinct CRY2 genes that differ in expression pattern (J.D. Platten, E. Foo, F. Foucher, V. Hecht, J.B. Reid, and J.L. Weller, unpublished data). Each of these three pea CRY genes is represented in Medicago. Only one CRY2 is represented in soybean and Lotus sequence databases, but two distinct CRY1-like EST contigs are present in soybean.
Table I.
Photoreceptor, circadian, and photoperiod pathway genes
References: 1, This study; 2, J.D. Platten, E. Foo, F. Foucher, V. Hecht, J.B. Reid, and J.L. Weller (unpublished data); 3, Sato (1988); 4, Weller et al. (2001). –, Sequence not identified.
| Name
|
Arabidopsis Gene
|
Pea
|
Medicago
|
Soybean EST
|
Lotus EST
|
|||
|---|---|---|---|---|---|---|---|---|
| Gene | %a | Reference | Genomic | EST | ||||
| PHYA | AT1G09570 | PsPHYAb (M37217) | 75% (FL) | 3 | AC148406 | BE248606 | GmPHYA (L38844) | TC8897 |
| GmcPHYA (L34842) | ||||||||
| BF425620 | ||||||||
| BG406259 | ||||||||
| PHYB | AT2G18790 | PsPHYBb (AF069305) | 74% (FL) | 4 | – | AW691215 | TC227575 | TC13239c |
| PHYE | AT4G18130 | – | – | – | AW186474 | – | ||
| CRY1 | AT4G08920 | PsCRY1b (AY508969) | 75% (FL) | 2 | – | TC89497c | TC193237d | TC15430c |
| TC183546d | ||||||||
| CRY2 | AT1G04400 | PsCRY2Abd (AY508972) | 63% (FL) | 2 | – | TC78737c | TC190841 | AV417531c |
| PsCRY2Bbd (AY508974) | 59% (FL) | 2 | AC122171be | TC86685be | – | – | ||
| FKF1 | AT1G68050 | – | AC140104be | AW684990be | AI960991c | TC19023 | ||
| ZTL | AT5G57360 | – | AC148970e | TC79173e | TC198766c | – | ||
| PFT1 | AT1G25540 | PsPFT1 (AY830924) | 56% (P: 28%) | 1 | – | TC80931 | TC182513c | TC17352 |
| PIF3 | AT1G09530 | – | – | CA919209c | TC196287 | – | ||
| CCA1 | AT2G46830 | PsMYB1f (AY826730) | 44% (P: 28%) | 1 | – | – | TC196677 | TC8391 |
| LHY | AT1G01060 | PsMYB2bf (AY826731) | 72% (P: 11%) | 1 | AC150443ef | TC89350cef | TC189020c | – |
| TOC1 | AT5G61380 | PsTOC1 (AY830927) | 83% (P: 9%) | 1 | – | TC90874 | TC197211 | – |
| ELF3 | AT2G25920 | PsELF3 (AY830925) | 56% (FL) | 1 | AC122168be | TC83932be | – | – |
| ELF4 | AT2G40080 | PsELF4 (AY830926) | 82% (P: 44%) | 1 | AC145219e | TC80100e | TC181568 | – |
| ELF6 | AT5G04240 | – | AC133709be | BF644901be | GM218975 | BP064330 | ||
| GI | AT1G22770 | PsGIb (AY826733) | 70% (P: 22%) | 1 | AC148397be | TC85289be | TC17500c | TC7440c |
| CO | AT5G15840 | Details of CO gene family given in Figure 2 and Supplemental Table II. | ||||||
| FT | AT1G65480 | Details of FT/TFL1 gene family given in Figure 3 and Supplemental Table III. | ||||||
| SOC1 | AT2G45660 | Details of MADS-box gene family given in Figure 1 and Supplemental Table I. | ||||||
Identity percentage at the amino acid level; (FL), full-length cDNA; (P), partial cDNA: percentage of Arabidopsis cDNA.
Map position available.
Several EST sequences available corresponding to the same gene; see Supplemental Table VI for other accession numbers.
Multiple distinct sequences corresponding to a single Arabidopsis gene.
Medicago genomic and EST sequences correspond to the same gene.
Exact relationship to Arabidopsis gene unclear.
Recent evidence suggests that another group of flowering-related proteins may also function as photoreceptors. The Arabidopsis ZEITLUPE (ZTL), FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 (FKF1), and LOV-KELCH PROTEIN2 proteins can all influence flowering and are characterized by the presence of a flavin-binding LOV domain, suggesting a role in light perception. This possibility is supported by recent reports demonstrating a light-regulated function for both ZTL and FKF1 (Imaizumi et al., 2003; Mas et al., 2003). This family is represented in both Medicago and soybean by two distinct EST contigs, one more similar to ZTL and the other to FKF1 (Table I).
Medicago and soybean EST sequences were also identified for a number of Arabidopsis genes that affect flowering through downstream effects on photoreceptor signaling. These sequences included PHYTOCHROME AND FLOWERING TIME1 (PFT1; Cerdán and Chory, 2003), for which we also isolated a partial pea cDNA, and PHYTOCHROME INTERACTING FACTOR3 (PIF3; Zhu et al., 2000; Table I).
Photoperiod Pathway and Circadian Clock
The flowering response to photoperiod is determined by a complex interaction between circadian regulation of CO mRNA expression and light regulation of CO protein stability, which in Arabidopsis results in substantial induction of FT expression only in LD (Yanovsky and Kay, 2003; Valverde et al., 2004). Normal circadian rhythms are thus central to normal photoperiod responsiveness. In Arabidopsis, the circadian rhythm is thought to be generated by the interaction of three key proteins: two closely related myb transcription factors, LATE ELONGATED HYPOCOTYL (LHY) and CIRCADIAN CLOCK ASSOCIATED1 (CCA1), and the pseudo response-regulator TIMING OF CAB1 (TOC1; Alabadí et al., 2002). A number of other genes, including EARLY FLOWERING3 (ELF3; Hicks et al., 2001), ELF4 (Doyle et al., 2002), ELF6 (Noh et al., 2004), and GI (Fowler et al., 1999; Park et al., 1999), are reported to affect clock function in various ways, although the exact nature of their contribution is not clear (Boss et al., 2004). These genes are all represented in Medicago and soybean databases, with the exception that CCA1 and LHY are represented by a single sequence and no ELF3-related sequence was present in soybean (Table I). In pea, we identified the probable orthologs of ELF3, ELF4, and GI, and a partial sequence highly homologous to TOC1. Two nonoverlapping fragments of LHY/CCA1-like MYB sequence from pea show a similar amino acid identity to both LHY and CCA1, and are most probably derived from a single gene.
Photoperiod-Independent Pathways: Activators and Repressors of FLC
Many of the Arabidopsis genes that regulate flowering in a photoperiod-independent manner act through effects on expression of the key MADS-box gene FLC. Genes in the autonomous pathway (FCA, FY, LUMINIDEPENDENS [LD], FLOWERING LOCUS D [FLD], FVE, FPA, FLOWERING LOCUS K [FLK]) mostly appear to be involved in either epigenetic or posttranscriptional repression of FLC expression (Simpson, 2004). The Arabidopsis autonomous pathway genes are all represented in Medicago (Table II), and we were able to isolate the corresponding pea sequence for all of them except FY and FLK. All of these genes are also represented by clear homologs in soybean and Lotus, with the exception of FPA in soybean and LD in Lotus. Arabidopsis genes that mediate vernalization responsiveness, such as VERNALIZATION1 (VRN1), VRN2, and VERNALIZATION INSENSITIVE3 (VIN3), also ultimately act to repress FLC expression (Amasino, 2004; Boss et al., 2004). We isolated partial sequence for two pea VRN1 homologs, based on Medicago and soybean ESTs.
Table II.
Autonomous and vernalization pathway genes (activators and repressors of FLC)
–, Sequence not identified.
| Name
|
Arabidopsis Gene
|
Pea
|
Medicago
|
Soybean EST
|
Lotus EST
|
||
|---|---|---|---|---|---|---|---|
| Gene | %a | Genomic | EST | ||||
| FCA | AT4G16280 | PsFCAb (AY805329) | 41% (FL) | AC135100bc | BG449291bc | CB063363 | BP034279 |
| FY | AT5G13480 | – | – | TC84839 | TC17965 | TC19740 | |
| FLD | AT3G10390 | PsFLD (AY830930) | 86% (P: 31%) | – | AW586703f | TC187859 | AV777759 |
| FVE | AT2G19520 | PsFVEb (AY830931) | 76% (FL) | AC121243bc | TC87417bcd | TC227037 | BP058734d |
| FPA | AT2G43410 | PsFPAb (AY830932) | 72% (P: 98%) | – | TC84230d | – | TC19296d |
| LD | AT4G02560 | PsLDb (AY826732) | 38% (P: 75%) | – | TC91556 | AW569075d | – |
| FLK | AT3G04610 | – | – | TC89387d | TC176909 | TC13879d | |
| VIN3 | AT5G57380 | – | – | – | – | TC9019 | |
| VRN1 | AT3G18990 | PsVRN1ae (AY830928) | 67% (P: 15%) | AC137825bef | TC78901bef | TC229117ce | TC15957 |
| PsVRN1be (AY830929) | 75% (P: 15%) | TC76370c | TC220172e | ||||
| FRL1 | AT5G16320 | – | AC121232b | – | – | – | |
| FRL2 | AT1G31814 | – | AC137079b | – | – | – | |
| VIP3 | AT4G29830 | – | – | TC82282 | TC178641 | TC10222 | |
| VIP4 | AT5G61150 | – | – | TC87729 | TC176048 | – | |
| ESD4 | AT4G15880 | – | AC147012bc | TC79449bc | TC199514 | TC13506 | |
| PIE1 | AT3G12810 | – | – | – | TC194085 | AV776810 | |
Identity percentage at the amino acid level; (FL), full-length cDNA; (P), partial cDNA: percentage of Arabidopsis cDNA.
Map position available.
Medicago genomic and EST sequences correspond to the same gene.
Several EST sequences available corresponding to the same gene; see Supplemental Table VI for other accession numbers.
Multiple distinct sequences corresponding to a single Arabidopsis gene.
Exact relationship to Arabidopsis gene unclear.
The repressive effects of the autonomous and vernalization pathways on FLC expression are balanced by the action of FRIGIDA (FRI) and FRIGIDA-LIKE (FRL) genes, which combine to enhance FLC expression (Michaels et al., 2004). We failed to find any sequences closely related to FRI in any of the legume databases. However, we identified two Medicago genomic sequences encoding full-length FRL proteins (MtFRLa and MtFRLb), which respectively share 33% and 30% amino identity with FRL1 and 28 and 29% identity with FRL2.
We also identified legume sequences corresponding to a number of other Arabidopsis genes that have been reported to affect flowering at least in part through effects on FLC expression. These included VERNALIZATION INDEPENDENCE3 (VIP3), VIP4, EARLY IN SHORT DAYS4 (ESD4), and PHOTOPERIOD INDEPENDENT EARLY FLOWERING1 (PIE1; Table II).
Other Regulators of Flowering
Many additional Arabidopsis genes with substantial effects on the flowering transition have been described. The relationship of these genes to established flowering pathways is not clear in many cases. Many of them may have roles in more general cellular processes and may thus affect flowering only indirectly. A list of these genes is presented in Table III, which shows that many of the genes are also represented by legume EST sequences. We did not pursue the isolation of homologous pea sequences for these genes, except in the case of EMBRYONIC FLOWER1 (EMF1; Aubert et al., 2001), where two distinct partial pea cDNA sequences were identified. We also identified a single partial pea cDNA equivalently similar to the related Arabidopsis VRN2 and EMF2 genes (PsVEL, Table III).
Table III.
Floral promoter and repressor genes
–, Sequence not identified.
| Name
|
Arabidopsis Gene
|
Pea
|
Medicago
|
Soybean EST
|
Lotus EST
|
||
|---|---|---|---|---|---|---|---|
| Gene | %a | Genomic | EST | ||||
| FLOWERING PROMOTING FACTOR1 | AT5G24860 | – | – | TC87988b | TC182917 | TC6345 | |
| TOE1 | AT2G28550 | – | AC145164cd | TC88593cd | TC191364 | AV425482 | |
| TFL2 | AT5G17690 | – | – | TC88793 | AW471580b | – | |
| EMF1 | AT5G11530 | PsEMF1ace (AY826734) | 37% (P: 46%) | – | – | – | – |
| PsEMF1bce | 34% (P: 46%) | ||||||
| EMF2 | AT5G51230 | PsVELf (AY830933) | 58% (P: 17%) | – | TC88797 | TC184854b | TC11442 |
| ELF5 | AT5G62640 | – | – | TC89554 | TC192128b | BI417263b | |
| RELATIVE OF ELF6 | AT3G48430 | – | AC144928d | TC92858d | TC196271b | – | |
| SVP | AT2G22540 | Details of MADS-box gene family given in Figure 1 and Supplemental Table I. | |||||
| AGL24 | AT4G24540 | ||||||
Identity percentage at the amino acid level; (FL), full-length cDNA; (P), partial cDNA: percentage of Arabidopsis cDNA.
Several EST sequences available corresponding to the same gene; see Supplemental Table VI for other accession numbers.
Map position available.
Medicago genomic and EST sequences correspond to the same gene.
Multiple distinct sequences corresponding to a single Arabidopsis gene.
Exact relationship to Arabidopsis gene unclear.
Many Arabidopsis genes that act later in the floral transition to specify inflorescence or floral organ identity are members of the MADS-box gene family. Other genes important at these later stages include the LFY, UFO, and AP2 genes (Jack, 2004). Table IV shows that all of these genes are represented in pea and in legume databases.
Table IV.
Inflorescence and floral identity genes
References: 1, Hofer et al. (1997); 2, Taylor et al. (2001). –, Sequence not identified.
| Name
|
Arabidopsis Gene
|
Pea
|
Medicago
|
Soybean EST
|
Lotus EST
|
|||
|---|---|---|---|---|---|---|---|---|
| Gene | %a | Reference | Genomic | EST | ||||
| LFY | AT5G61850 | UNIb (AF010190) | 64% (FL) | 1 | AC139708c | – | BU761377 | – |
| UFO | AT1G30950 | STPb (AF004843) | 62% (FL) | 2 | AC147538 | – | – | NP645867 |
| AP2 | AT4G36920 | AP2-like (AF325506) | 52% (FL) | – | – | TC82951 | TC219011c | AV425482 |
| AG | AT4G18960 | Details of MADS-box gene family given in Figure 1 and Supplemental Table I. | ||||||
| AP1 | AT1G69120 | |||||||
| AP3 | AT3G54340 | |||||||
| PI | AT5G20240 | |||||||
| TFL1 | AT5G03840 | Details of FT/TFL1 gene family given in Figure 3 and Supplemental Table III. | ||||||
Identity percentage at the amino-acid level; (FL), full-length cDNA.
Map position available.
Several EST sequences available corresponding to the same gene; see Supplemental Table VI for other accession numbers.
Mapping
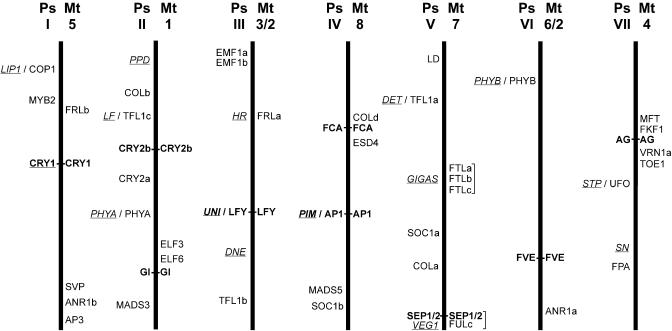
Map positions have already been determined for several flowering-related pea genes (e.g. Hall et al., 1997; Timmerman-Vaughan et al., 2002; J.D. Platten, E. Foo, F. Foucher, V. Hecht, J.B. Reid, and J.L. Weller, unpublished data). In a preliminary attempt to map other genes, we used the partial pea cDNA sequences to sequence introns from genomic DNA of parent lines for two different recombinant inbred line populations. Appropriate polymorphisms were converted to CAPS or dCAPS markers for mapping (Supplemental Table V). We mapped 13 additional genes in this way. None of the mapped pea genes were located close to previously described flowering loci, except for PsSEP1/2, which mapped in the vicinity of VEG1 (Hall et al., 1997; Fig. 4). Map positions are also available for a subset of the sequenced Medicago BACs (http://medicago.plantpath.ucdavis.edu), and this information allowed us to identify map positions for 26 of the Medicago genes. In addition, the MtCRY1 gene has recently been mapped to chromosome 5 (B. Jullier and T. Huguet, personal communication). The close synteny between pea and Medicago has recently been described in detail (Choi et al., 2004; Kaló et al., 2004) and allows the approximate chromosomal position of a gene in one species to be inferred from the position of its ortholog in the other with a reasonable degree of confidence. Consistent with this relationship, we found that nine pairs of putative orthologs (AP1/PIM, LFY/UNI, FCA, CRY1, CRY2b, FVE, GI, SEP1/2, and AG) had corresponding map positions (Fig. 4). The map positions of several additional Medicago genes (SVP, ELF3, VRN1, FTLa/b/c) indicate the probable map positions for pea genes that we isolated but were not able to map in our initial attempts through lack of polymorphism (Fig. 4).
Figure 4.
Approximate map locations of flowering genes in pea and Medicago. This figure represents diagrammatically the syntenic relationships described for pea and Medicago by Kaló et al. (2004) and Choi et al. (2004). The seven pea linkage groups are shown as thick vertical lines, and the corresponding Medicago chromosomes are indicated at the top right of each (Ps, garden pea; Mt, M. truncatula). The approximate map positions of pea genes are indicated on the left and Medicago genes on the right. Pea loci defined by mutants are underlined, and orthologous genes in the two species are shown in bold and joined by a horizontal line. Note that homologous chromosomes are normalized in length, and map positions are therefore relative for each linkage group/chromosome and each species. Actual map positions of Medicago genes are as follows: AG (AC137837; LG4-58.7), ANR1a (AC123898; LG2-1.5), ANR1b (AC126010 and TC82622; LG5-15), AP1 (AC144726; LG8-49.5), AP3 (AC136451 and TC90653; LG5-5.2), COLd (AC127169 and AW693899 and TC88293; LG8-68-70.9), CRY2b (AC122171; LG1-18.1), ELF3 (AC122168 and TC83932; LG1-51.8), ELF6 (AC133709 and BF644901; LG1-54.8), ESD4 (AC147012 and TC79449; LG8-64.2-68), FCA (AC135100 and BG449291; LG8-68), FKF1 (AC140104 and AW684990; LG4-61.6), FRLa (AC121232; LG3-37.9), FRLb (AC137079; LG5-78.1-80.3), FTLa (AC123593a; LG7-50.6), FTLb (AC123593b; LG7-50.6), FTLc (AC123593c; LG7-50.6), FULc (AC146650b and AL387855; LG7-5.1), FVE (AC121243 and TC87417 and TC91602; LG2-19.3), GI (AC148397 and TC85289; LG1-58.5), LFY (AC139708; LG3-85), MFT (AC139526 and TC81061; LG4-62.4), SEP1/2 (AC146650a; LG7-5.1), SVP (AC135848 and TC87621; LG5-16.4), TOE1 (AC145164 and TC88593; LG4-46.9), VRN1a (AC137825 and TC78901; LG4-51.4).
The map positions of Medicago genes also identified three potential candidate gene relationships that are supported by functional comparisons. The map position of the FTL gene cluster on Medicago chromosome 7 corresponds to the approximate position of the pea flowering gene GIGAS in linkage group V. Similarly, the position of the Medicago FRLa gene on chromosome 3 corresponds to the approximate position of the HR gene in pea linkage group III. Finally, two Medicago genes, a SEP1/2 ortholog and a divergent FUL-like gene (MtFULc), both map to a position at the top of Medicago chromosome 7 that corresponds to the position of the pea gene VEG1 at the bottom of linkage group V.
DISCUSSION
The recent progress in EST and genome sequencing projects from Medicago, Lotus, and soybean has allowed us to identify legume homologs for a wide range of genes potentially related to the flowering process. It is clear that this picture will continue to expand as full genome sequences become available. Even so, we found that very few Arabidopsis genes were completely unrepresented in the four legume species. Genes in this category included FLC and related MADS-AFFECTING FLOWERING genes, FRI, FWA, the recently identified SCHLAFMUTZE and SCHNARCHZAPFEN genes, and TARGET OF EAT2 (TOE2; see Boss et al., 2004). It is possible that these genes might be present but expressed at a low level in legumes and therefore are not represented in EST collections. Alternatively, it is possible that close homologs of these genes may not be present in legumes. The care necessary in drawing conclusions from EST data alone is illustrated in the example of the FT/TFL1-like gene family in tomato, for which none of the six genes were represented in extensive EST collections (Carmel-Goren et al., 2003). However, our failure to identify close FLC homologs in pea is interesting in light of the importance of this gene in Arabidopsis and the observation that FLC-like genes appear to be absent from the rice genome (Izawa et al., 2003) and have not been described outside the Brassicaceae apart from a single recent example in Petunia (Vandenbussche et al., 2003). Genes clearly corresponding to FRI were also reported to be absent from the rice genome (Izawa et al., 2003), but several FRI-related genes have recently been identified in rice, including one more closely related to FRI than to other Arabidopsis FRI-like genes (Michaels et al., 2004). Although we were unable to identify a putative FRI ortholog in Medicago, other FRI-like genes are clearly present and, as in Arabidopsis (Michaels et al., 2004), may function in a manner similar to FRI itself.
We found that some genes present as single-copy genes in Arabidopsis, including GI, ELF3, and FCA, are also apparently single copy in the four model legumes. However, for many other genes, differences in duplication history are apparent. For example, a number of paralogous gene pairs in Arabidopsis are only represented by a single gene in Medicago and/or pea. This is true for several MADS-box genes (e.g. SEP1/SEP2, AP1/CAL, SHP1/SHP2) and other genes, including LHY/CCA1, group Ia COL genes, and PHYB. The converse is true for several other genes, where Medicago or pea contain multiple copies of genes that are single copy in Arabidopsis (e.g. TFL1a/c, CRY2a/b, SOC1a/b, EMF1a/b, VRN1a/b). In still other cases, independent duplications may have occurred in both legume and Arabidopsis lineages, resulting in orthologous groups of genes. Comparisons across the legume species show that individual Medicago genes were frequently represented by two very similar but distinct soybean sequences. (e.g. SEP1/2, SEP3, COLc, COLd, PHYA, CRY1). This is consistent with several reports suggesting that soybean may have undergone whole-genome or extensive segmental duplication after divergence from its last common ancestor with Medicago (Shoemaker and Specht, 1995; Blanc and Wolfe, 2004). However, these authors also suggest a more complex history of duplications in both soybean and Medicago lineages.
Integrative Mapping and Candidate Gene Identification
Of the four legume species included in this study, pea and Medicago are the two most closely related taxonomically, belonging to two sister tribes within the galegoid group. Recent reports have demonstrated extensive macrosynteny among legume genomes and a particularly close relationship between pea and Medicago (Choi et al., 2004; Kaló et al., 2004). Despite a 10-fold difference in genome size and a difference in chromosome number, the organization of pea and Medicago genomes reflects only a small number of gross rearrangements (Kaló et al., 2004). The location of nine additional ortholog pairs to corresponding map positions further supports this close relationship, while our combined mapping data from pea and Medicago suggest how it may be useful for a map-based approach to pea gene identification.
For example, map positions in pea and Medicago appear to rule out several flowering-inhibitory genes including ELF3, ELF6, SVP, ESD4 and the putative single LHY/CCA1 ortholog as candidate genes for the early flowering pea mutants sn, dne, ppd, and hr (Fig. 4). The pea GIGAS gene has been considered similar to genes in the Arabidopsis autonomous pathway because gigas mutants are late flowering but retain responsiveness to both photoperiod and vernalization (Beveridge and Murfet, 1996). However, we can now exclude many of these genes, including LD, FPA, FVE, and FCA, as candidate genes for GIGAS, and can also rule out flower-promoting genes in the Arabidopsis photoperiod pathway, such as CO, GI, CRY2, and SOC1 (Fig. 4).
On the positive side, mapping data has also suggested several potential candidate gene relationships that are supported by phenotypic similarity. Dominant HR alleles behave similarly to FLC/FRI, conferring a strong delay in flowering particularly under SD and a near-obligate requirement for LD or vernalization (Murfet, 1985; Weller et al., 1997) Although neither FRI nor FLC orthologs appear to be present in pea or Medicago, genes similar to the recently described FRI-like genes are present in Medicago, and one of these (MtFRLa) is located in a region that corresponds to the location of HR in pea. Like FRI, Arabidopsis FRL1 delays flowering by contributing to the maintenance of high FLC expression, and frl1 mutations relax the obligate LD requirement of fully wild-type Arabidopsis (Michaels et al., 2004) in a manner similar to the hr allele in pea.
The possibility that the pea GIGAS gene may correspond to one of the three colocating Medicago FT-like genes is also supported by phenotypic comparisons. The nonflowering yet clearly photoperiod-responsive phenotype of gigas mutants under certain LD conditions (Beveridge and Murfet, 1996) is better interpreted as a conditional failure to specify inflorescence identity rather than as a failure in LD perception or response, especially when compared to other mutants such as phyA that are clearly impaired in their photoperiodic response (Weller et al., 2001). This suggests that GIGAS may act relatively late in the floral transition, at the level of pathway integration and/or inflorescence identity. Also, the late or nonflowering phenotype of gigas mutants can be partially rescued by grafting to the wild type, indicating that GIGAS is somehow required for the mobile flowering stimulus (Beveridge and Murfet, 1996). Such a role has also recently been proposed for FT (An et al., 2004).
Single mutants at the pea VEG1 locus do not flower under any conditions but, like gigas mutants, still show vegetative responses to photoperiod, suggesting a complete failure to specify inflorescence identity. In Arabidopsis, a similar phenotype is seen in plants carrying multiple mutations in SEP genes (Ditta et al., 2004), while triple mutants for AP1 clade genes AP1, CAL, and FUL also completely fail to produce flowers (Ferrándiz et al., 2000). The colocation of pea VEG1 with a SEP1/2 ortholog and a FUL-like gene is therefore intriguing and warrants further examination.
Implications for the Control of Flowering in Pea
The presence of a full complement of photoperiod pathway genes in legumes and the functional conservation of some of these genes in rice suggests that the photoperiod response in pea is likely to be a fairly close variation on the Arabidopsis theme. The pea genes SN, DNE, and PPD all clearly affect photoperiod responsiveness and have graft-transmissible inhibitory effects on flowering (Weller et al., 1997). Expression analyses of newly isolated pea CO- and FT-like genes in sn, dne, and ppd mutants should help us to interpret these graft-transmissible effects in terms of the Arabidopsis photoperiod pathway. Several flower-promoting Arabidopsis photoperiod pathway genes (e.g. TOC1, ELF4) also remain as potential candidates for these pea genes.
In comparison, there is much less evidence for the wider conservation of the Arabidopsis autonomous and vernalization pathways. Although homologs of genes in these pathways are present in pea and in other species, none have been shown to affect flowering. Correspondence between the vernalization response in pea and the Arabidopsis vernalization and autonomous pathway genes is therefore less certain. In addition, a number of observations suggest that differences may exist. Firstly, vernalization in pea has graft-transmissible effects and has been suggested to act at least partly through the photoperiod (“inhibitor”) pathway (Murfet, 1985). We can now begin to test this possibility by examining whether vernalization may regulate any of the pea photoperiod pathway homologs. Secondly, as discussed above, the main regulatory target of these pathways in Arabidopsis, FLC, has yet to be identified in pea. If autonomous/vernalization pathways do regulate flowering in pea, it is possible they may converge on a distinct regulatory target. FLC-independent effects of vernalization have been reported in Arabidopsis and include the up-regulation of the floral promoting MADS-box gene AGL24 (Michaels et al., 2003). It may thus also be informative to test the effects of vernalization on the expression of a range of potentially flowering-related MADS-box genes in pea. The LF gene in pea also functions as a regulated repressor of flowering (Foucher et al., 2003), and it also remains possible that this gene may play a role analogous to FLC in the integration of flowering signals.
Concluding Remarks
This preliminary survey of legume flowering-related genes should provide a springboard for a range of further studies relating to flowering and photoperiodic responses in this important plant group. It has already been shown that pea homologs of genes such as LFY and TFL1 exhibit significant differences in function to their Arabidopsis counterparts. It is likely that further studies of the genes identified here will also give a new perspective on other characteristic aspects of flowering physiology and inflorescence architecture in pea, and may help to uncover the molecular basis for natural genetic variation controlling flowering in a range of species. Our results also highlight the potential usefulness of a comparative mapping approach to flowering gene identification in legumes, and offer the prospect of rapid transfer of information from pea and Medicago to other closely related, agronomically important species.
MATERIALS AND METHODS
Database Searches, Alignment, and Phylogenetic Analysis
Legume homologs of Arabidopsis (Arabidopsis thaliana) flowering genes were identified in tBLASTn searches against legume gene index databases at the Institute for Genomic Research (http://tigrblast.tigr.org/tgi/), the Medicago genome sequencing database (MtGenome at the University of California, Davis; http://medicago.plantpath.ucdavis.edu/BLAST/), and the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/BLAST/). Hits were validated against the Arabidopsis genome (The Arabidopsis Information Resource; http://www.arabidopsis.org/Blast/) in tBLASTx/tBLASTn searches. Validated sequences were translated and protein alignments were performed with ClustalX (Thompson et al., 1997). In some cases, alignments were manually adjusted using GENEDOC (Nicholas et al., 1997; http://www.psc.edu/biomed/genedoc). Distance and parsimony-based methods were used for phylogenetic analyses in PAUP* 4.0b10 (http://paup.csit.fsu.edu/).
Cloning and Isolation of Pea Genes
Degenerate primers for isolation of pea (Pisum sativum) genes were designed using the CODEHOP strategy (Rose et al., 1998). Conserved domains were identified from protein sequence alignments using the BLOCKMAKER application (http://blocks.fhcrc.org/blocks/make_blocks.html), and degenerate primers targeting these domains were selected from the CODEHOP software output (http://blocks.fhcrc.org/codehop.html). Selected primers were manually optimized by reference to the input legume EST sequences (see Supplemental Table I). Pea genes were cloned using first-strand cDNA as template. RNA was isolated from apical buds using the Plant RNeasy extraction kit (Qiagen, Hielden, Germany) and reverse transcribed using the Omniscript RT kit (Qiagen) with oligo(dT)12 primer. PCR fragments obtained were cloned in pGEM-T Easy (Promega, Madison, WI) and sequenced.
In order to isolate full-length clones for PsCOLa, PsCOLb, PsELF3, PsFPA, PsFVE, and PsSOC1a, PCR fragments obtained previously were used as probe to screen a shoot apex cDNA library (see Taylor et al., 2001) using standard procedures. A full-length clone corresponding to PsCOLb was also obtained by TAIL-PCR (Liu et al., 1995) and 3′ RACE (Frohman et al., 1988) using primers PsCO-F1 (5′-GATAGAGAAGCTAGGGTTATG-3′) and PsCO-R1 (5′-CAGCATCTGGATCGGATTCG-3′). The partial cDNA sequence initially obtained for PsMYB1 was extended by 3′ RACE using primers LHY-R1 (5′-AGGATAATGAGGAAAAAGCACACC-3′) and LHY-R2 (5′-GAAATACCAGAAGTGCTCAACC-3′). The PsFCA cDNA fragment was used as a probe to screen a pea genomic library. Ten positive clones were isolated and restriction mapped revealing two classes of clones. A clone from both classes (FCA λ10 and FCA λ17) was subcloned into pUC18 and sequenced. The two lambda clones shared 97% nucleotide identity, suggesting they could represent different alleles of the same gene; therefore, only FCA λ10 was sequenced completely. To confirm the exon structure of the genomic clone, reverse transcription-PCR using primers to exon 1 (PsEx1-F; 5′- GGTTTCACTCTGTTAGCCAA-3′) and exon 9 (FCA-R9; Supplemental Table IV) was performed. Since neither of the PsFCA genomic clones contained the 3′ region of the FCA gene, 3′ RACE was performed with primers PsEx8 (5′-GATGTGGTTTTGTCAAATATTC-3′) and RACE primer (5′-GACTCGAGTCGACATCGA-3′).
In order to isolate some of the MADS-box family genes, a pea floral cDNA library was screened under low-stringency conditions with the cDNA of the Antirrhinum majus AP3 ortholog DEF (Sommer et al., 1990) as a probe. Twenty clones were isolated that represented six different MADS-box genes (PsFUL, PsMADS3, PsMAD5, PsPI, PsPIM, PsSEP1). PsAG and PsSHP sequences were isolated by screening the same library with a partial cDNA corresponding to the C-terminal region of the Antirrhinum AG ortholog PLE (Bradley et al., 1993). Two clones were isolated corresponding to a full-length cDNA of PsSHP and a partial cDNA clone of PsAG. RACE (Marathon cDNA amplification kit; CLONTECH, Palo Alto, CA) was performed to amplify the 5′ region of PsAG, following the manufacturer's instructions, with primers PM7RACE (5′-GAGACATCTGGTCCTGCCTGGCATAC-3′) and PM7RACE2 (5′-GCTGCTGAGGTTGGATGCGCTCGAAG-3′). Products were cloned and sequenced, and specific primers were designed to amplify full-length PsAG cDNA by reverse transcription-PCR, PM7BamFOR (5′-GGATCCGCTTGCAACTATGAGTTTTCC-3′) and PM/BamREV (5′-GGATCCAACTCTTTCCCTTCTCAACCGC-3′).
Mapping and Marker Development
Genomic DNA corresponding to each of the isolated pea cDNAs was sequenced from parents of two different recombinant inbred line populations (JI281 × JI399, Hall et al., 1997; cv Térese × Torsdag, Laucou et al., 1998), and CAPS markers were developed to target appropriate polymorphisms (Supplemental Table V). The PsAG gene was mapped in a similar manner using a third population (JI15 × JI399; Hall et al., 1997). The map positions of PsCOLb (PeaCO), PsMADS3 (pm3), PsPIM (peasqua), PsMADS5 (pm5), and PsSEP1/2 (pm6) have already been published (Hall et al., 1997; Timmerman-Vaughan et al., 2002).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers (in parentheses) PsAG (AY884291), PsCOLa (AY830921, AY826727), PsCOLb (AY830922, AY805328), PsELF3 (AY830925), PsELF4 (AY830926), PsEMF1a (AY826734), PsFCA (AY805329), PsFLD (AY830930), PsFTL (AY830923), PsFPA (AY830932), PsFUL (AY884287), PsFVE (AY830931), PsGI (AY826733), PsLD (AY826732), PsMADS3 (AY884288), PsMADS5 (AY884289), PsMYB1 (AY826730), PsMYB2 (AY826731), PsPFT1 (AY830924), PsPI (AY842491), PsSEP1/2 (AY884290), PsSHP (AY884292), PsSOC1a (AY830920, AY826728), PsSOC1b (AY826729), PsSVP (AY830919), PsTOC1 (AY830927), PsVEL (AY830930), PsVRN1a (AY830928), and PsVRN1b (AY830929).
Supplementary Material
Acknowledgments
We thank Natalie Conod, Reika Tanabe, and Augustine Cheong for assistance with gene isolation; Julie Hofer and Dot Steane for help with database searches and phylogenetic analysis; Bernadette Julier for information about map positions of Medicago sequences; and Carlos Alonso-Blanco and Takashi Araki for making Arabidopsis sequences available prior to publication.
This work was supported by the Australian Research Council Discovery Project (grant no. DP0210947 to J.L.W.), Génoplante (project PEA-A; C.R.), the Secretaría General del Plan Nacional de Investigación Científica y Desarrollo Tecnológico (grant no. BIO2000–0940 to J.P.B.), the Ministerio de Educación y Ciencia (fellowships to C.F. and C.N.), the European Union Grain Legumes Integrated Project (grant no. FP6–2002–FOOD–1–506223 to N.E. and C.R.), and a New Zealand FRS&T Fellowship and Marsden Fund grant (R.M.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.057018.
References
- Alabadí D, Yanovsky MJ, Más P, Harmer SL, Kay SA (2002) Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr Biol 12: 757–761 [DOI] [PubMed] [Google Scholar]
- Amasino R (2004) Vernalization, competence and the epigenetic memory of winter. Plant Cell 16: 2553–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H, Roussot C, Suárez-López P, Corbesier L, Vincent C, Piñeiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, et al (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Aubert D, Chen L, Moon YH, Martin D, Castle LA, Yang CH, Sung ZR (2001) EMF1, a novel protein involved in the control of shoot architecture and flowering in Arabidopsis. Plant Cell 13: 1865–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A, Theissen G (2003) The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol Phylogenet Evol 29: 464–489 [DOI] [PubMed] [Google Scholar]
- Berbel A, Navarro C, Ferrándiz C, Cañas L, Madueño F, Beltrán J-P (2001) Analysis of PEAM4, the pea AP1 functional homologue, supports a model for AP1-like genes controlling meristem and floral organ identity in different plant species. Plant J 25: 441–451 [DOI] [PubMed] [Google Scholar]
- Beveridge CA, Murfet IC (1996) The gigas mutant in pea is deficient in the floral stimulus. Physiol Plant 96: 637–645 [Google Scholar]
- Blanc G, Wolfe KH (2004) Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16: 1667–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss PK, Bastow RM, Mylne JS, Dean C (2004) Multiple pathways in the decision to flower: enabling, promoting, and resetting. Plant Cell 16: S18–S31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D, Carpenter R, Sommer H, Hartley N, Coen E (1993) Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell 72: 85–95 [DOI] [PubMed] [Google Scholar]
- Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E (1997) Inflorescence commitment and architecture in Arabidopsis. Science 275: 80–83 [DOI] [PubMed] [Google Scholar]
- Buchner P, Boutin JP (1998) A MADS box transcription factor of the AP1/AGL9 subfamily is expressed in the seed coat of pea (Pisum sativum) during development. Plant Mol Biol 38: 1253–1255 [DOI] [PubMed] [Google Scholar]
- Burgeff C, Liljegren SJ, Tapia-Lopez R, Yanofsky MF, Alvarez-Buylla ER (2002) MADS-box gene expression in lateral primordia, meristems and differentiated tissues of Arabidopsis thaliana roots. Planta 214: 365–372 [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Tandre K, Johanson U, Englund M, Engstrom P (2004) The MADS box gene DAL1 is a potential mediator of the juvenile-to-adult transition in Norway spruce (Picea abies). Plant J 40: 546–557 [DOI] [PubMed] [Google Scholar]
- Carmel-Goren L, Liu YS, Lifschitz E, Zamir D (2003) The SELF-PRUNING gene family in tomato. Plant Mol Biol 52: 1215–1222 [DOI] [PubMed] [Google Scholar]
- Cerdán PD, Chory J (2003) Regulation of flowering time by light quality. Nature 423: 881–885 [DOI] [PubMed] [Google Scholar]
- Choi HK, Mun JH, Kim DJ, Zhu H, Baek JM, Mudge J, Roe B, Ellis N, Doyle J, Kiss GB, et al (2004) Estimating genome conservation between crop and model legume species. Proc Natl Acad Sci USA 101: 15289–15294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson NM, Russell JS (1975) Flowering responses to vernalization and photoperiod in annual medics (Medicago spp.). Aust J Agric Res 26: 831–838 [Google Scholar]
- Curtis DF, Tanner JW, Luzzi BM, Hume DJ (2000) Agronomic and phenological differences of soybean isolines differing in maturity and growth habit. Crop Sci 40: 1624–1629 [Google Scholar]
- Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF (2004) The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol 14: 1935–1940 [DOI] [PubMed] [Google Scholar]
- Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognár L, Nagy F, Millar AJ, Amasino RM (2002) The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419: 74–77 [DOI] [PubMed] [Google Scholar]
- Ferrándiz C, Gu Q, Martienssen R, Yanofsky MF (2000) Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127: 725–734 [DOI] [PubMed] [Google Scholar]
- Foucher F, Morin J, Courtiade J, Cadioux S, Ellis N, Banfield M, Rameau C (2003) DETERMINATE and LATE FLOWERING are two TERMINAL FLOWER1/CENTRORADIALIS homologs that control distinct phases of flowering initiation and development in pea. Plant Cell 15: 2742–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J (1999) GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J 18: 4679–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman NA, Dush MK, Martin GR (1988) Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA 85: 8998–9002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths S, Dunford RP, Coupland G, Laurie DA (2003) The evolution of CONSTANS-like gene families in barley, rice and Arabidopsis. Plant Physiol 131: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall KJ, Parker JS, Ellis THN, Turner L, Knox MR, Hofer JMI, Lu J, Ferrándiz C, Hunter PJ, Taylor JD, et al (1997) The relationship between genetic and cytogenetic maps of pea. II. Physical maps of linkage mapping populations. Genome 40: 755–769 [DOI] [PubMed] [Google Scholar]
- Handberg K, Stougaard J (1992) Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. Plant J 2: 487–496 [Google Scholar]
- Hartmann U, Hoehmann S, Nettesheim K, Wisman E, Saedler H, Huijser P (2000) Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J 21: 351–360 [DOI] [PubMed] [Google Scholar]
- Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K (2003) Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422: 719–722 [DOI] [PubMed] [Google Scholar]
- Hicks KA, Albertson TM, Wagner DR (2001) EARLY FLOWERING 3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell 13: 1281–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer J, Turner L, Hellens R, Ambrose M, Matthews P, Michael A, Ellis N (1997) UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr Biol 7: 581–587 [DOI] [PubMed] [Google Scholar]
- Honma T, Goto K (2001) Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 469–471 [DOI] [PubMed] [Google Scholar]
- Huyghe C (1998) Genetics and genetic modifications of plant architecture in grain legumes: a review. Agronomie 18: 383–411 [Google Scholar]
- Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA (2003) FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426: 302–306 [DOI] [PubMed] [Google Scholar]
- Immink RGH, Ferrario S, Busscher-Lange J, Koiker M, Busscher M, Angenent GC (2003) Analysis of the petunia MADS-box transcription factor family. Mol Genet Genomics 268: 598–606 [DOI] [PubMed] [Google Scholar]
- Izawa T, Oikawa T, Sugiyama N, Tanisaka T, Yano M, Shimamoto K (2002) Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering in rice. Genes Dev 16: 2006–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T, Takahashi Y, Yano M (2003) Comparative biology comes into bloom: genomic and genetic comparison of flowering pathways in rice and Arabidopsis. Curr Opin Plant Biol 6: 113–120 [DOI] [PubMed] [Google Scholar]
- Jack T (2004) Molecular and genetic mechanisms of floral control. Plant Cell (Suppl) 16: S1–S17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaló P, Seres A, Taylor SA, Jakab J, Kevei Z, Kereszt A, Endre G, Ellis THN, Kiss GB (2004) Comparative mapping between Medicago sativa and Pisum sativum. Mol Genet Genomics 272: 235–246 [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Kofuji R, Sumikawa N, Yamasaki M, Kondo K, Ueda K, Ito M, Hasebe M (2003) Evolution and divergence of the MADS-box gene family based on genome-wide expression analysis. Mol Biol Evol 20: 1963–1977 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 229: 57–66 [DOI] [PubMed] [Google Scholar]
- Kumar J, van Rheenen HA (2000) A major gene for time of flowering in chickpea. J Hered 91: 67–68 [DOI] [PubMed] [Google Scholar]
- Laucou V, Haurogné K, Ellis N, Rameau C (1998) Genetic mapping in pea. 1. RAPD-based linkage map of Pisum sativum. Theor Appl Genet 97: 905–915 [Google Scholar]
- Litt A, Irish VF (2003) Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: implications for the evolution of floral development. Genetics 165: 821–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Mas P, Kim WY, Somers DE, Kay SA (2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570 [DOI] [PubMed] [Google Scholar]
- Menzel G, Apel K, Melzer S (1996) Identification of two MADS box genes that are expressed in the apical meristem of the long-day plant Sinapis alba in transition to flowering. Plant J 9: 399–408 [DOI] [PubMed] [Google Scholar]
- Michaels SD, Bezerra IC, Amasino RM (2004) FRIGIDA-related genes are required for the winter-annual habit in Arabidopsis. Proc Natl Acad Sci USA 101: 3281–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Ditta G, Gustafson-Brown C, Pelaz S, Yanofsky M, Amasino RM (2003) AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J 33: 867–874 [DOI] [PubMed] [Google Scholar]
- Mimida N, Goto K, Kobayashi Y, Araki T, Ahn JH, Weigel D, Murata M, Motoyoshi F, Sakamoto W (2001) Functional divergence of the TFL1-like gene family in Arabidopsis revealed by characterization of a novel homologue. Genes Cells 6: 327–336 [DOI] [PubMed] [Google Scholar]
- Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG, Lee I (2003) The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J 35: 613–623 [DOI] [PubMed] [Google Scholar]
- Murfet IC (1985) Pisum sativum. In A Halevy, ed, CRC Handbook of Flowering, Vol IV. CRC Press, Boca Raton, FL, pp 97–126
- Nicholas KB, Nicholas HB Jr, Deerfield DWI (1997) GeneDoc: analysis and visualization of genetic variation. EMBNew News 4: 14 [Google Scholar]
- Noh B, Lee SH, Kim HJ, Yi G, Shin EA, Lee M, Jung KJ, Doyle MR, Amasino RM, Noh YS (2004) Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 16: 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenicova L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, et al (2003) Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new origins to the MADS world. Plant Cell 15: 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG (1999) Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285: 1579–1582 [DOI] [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203 [DOI] [PubMed] [Google Scholar]
- Pinyopich A, Ditta GS, Savidge B, Liljegren SJ, Baumann E, Wisman E, Yanofsky MF (2003) Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424: 85–88 [DOI] [PubMed] [Google Scholar]
- Putterill J, Laurie R, Macknight R (2004) It's time to flower: the genetic control of flowering time. Bioessays 26: 363–373 [DOI] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857 [DOI] [PubMed] [Google Scholar]
- Ratcliffe OJ, Amaya I, Vincent CA, Rothstein S, Carpenter R, Coen ES, Bradley DJ (1998) A common mechanism controls the life cycle and architecture of plants. Development 125: 1609–1615 [DOI] [PubMed] [Google Scholar]
- Rose TM, Schultz ER, Henikoff JG, Pietrokovski S, McCallum CM, Henikoff S (1998) Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly-related sequences. Nucleic Acids Res 26: 1628–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounsley SD, Ditta GS, Yanofsky MF (1995) Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 7: 1259–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Sarker A, Erskine W, Sharma B, Tyagi MC (1999) Inheritance and linkage relationships of days to flower and morphological loci in lentil (Lens culinaris Medikus ssp. culinaris). J Hered 90: 270–275 [Google Scholar]
- Sato N (1988) Nucleotide sequence and expression of the phytochrome gene in Pisum sativum: differential regulation by light of multiple transcripts. Plant Mol Biol 11: 697–710 [DOI] [PubMed] [Google Scholar]
- Shoemaker RC, Specht JE (1995) Integration of the soybean molecular and classical genetic linkage groups. Crop Sci 35: 436–446 [Google Scholar]
- Simpson GG (2004) The autonomous pathway: epigenetics and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Curr Opin Plant Biol 7: 1–5 [DOI] [PubMed] [Google Scholar]
- Sommer H, Beltrán JP, Huijser P, Pape H, Lönnig WE, Saedler H, Schwartz-Sommer Z (1990) Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: the protein shows homology to transcription factors. EMBO J 9: 605–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerfield RJ, Roberts EH (1985) Glycine max. In CRC Handbook of Flowering, Vol I. CRC Press, Boca Raton, FL, pp 100–117
- Taylor S, Hofer J, Murfet I, Sollinger J, Singer S, Knox M, Ellis N (2002) PROLIFERATING INFLORESCENCE MERISTEM, a MADS-box gene that regulates floral meristem identity in pea. Plant Physiol 129: 1150–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SA, Hofer J, Murfet IC (2001) Stamina pistilloida, the pea ortholog of Fim and UFO, is required for normal development of flowers, inflorescences, and leaves. Plant Cell 13: 31–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman-Vaughan GM, Frew TJ, Russell AC, Khan T, Butler R, Gilpin M, Murray S, Falloon K (2002) QTL mapping of partial resistance to field epidemics of Ascochyta Blight of pea. Crop Sci 42: 2100–2111 [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Vandenbussche M, Zethof J, Souer E, Koes R, Tornielli GB, Pezzotti M, Ferrario S, Angenent GC, Gerats T (2003) Toward the analysis of the petunia MADS box gene family by reverse and forward transposon insertion mutagenesis approaches: B, C, and D floral organ identity functions require SEPALLATA-like MADS box genes in petunia. Plant Cell 15: 2680–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DH, Zobel RW, Yourstone KS (1993) A whole-system reconsideration of paradigms about photoperiod and temperature control of crop yield. Theor Appl Genet 86: 17–26 [DOI] [PubMed] [Google Scholar]
- Weller JL, Batge SL, Smith JJ, Kerckhoffs LHJ, Sineshchekov VA, Murfet IC, Reid JB (2004) A dominant mutation in the pea PHYA gene confers enhanced responses to light and impairs the light-dependent degradation of phytochrome A. Plant Physiol 135: 2186–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Beauchamp N, Kerckhoffs LHJ, Platten JD, Reid JB (2001) Interaction of phytochromes A and B in the control of de-etiolation and flowering in pea. Plant J 26: 283–294 [DOI] [PubMed] [Google Scholar]
- Weller JL, Reid JB, Taylor SA, Murfet IC (1997) The genetic control of flowering in pea. Trends Plant Sci 2: 412–418 [Google Scholar]
- Yalovsky S, Rodríguez-Concepción M, Bracha K, Toledo-Ortiz G, Gruissem W (2000) Prenylation of the floral transcription factor APETALA1 modulates its function. Plant Cell 12: 1257–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J (2004) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303: 1640–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky M, Kay S (2003) Living by the calendar: how plants know when to flower. Nat Rev Mol Cell Biol 4: 265–275 [DOI] [PubMed] [Google Scholar]
- Yoo SY, Kardailsky I, Lee JS, Weigel D, Ahn JH (2004) Acceleration of flowering by overexpression of MFT (MOTHER OF FT AND TFL1). Mol Cells 17: 95–101 [PubMed] [Google Scholar]
- Yu H, Ito T, Wellmer F, Meyerowitz EM (2004) Repression of AGAMOUS-LIKE 24 is a crucial step in promoting flower development. Nat Genet 36: 157–161 [DOI] [PubMed] [Google Scholar]
- Zhang H, Forde BG (1998) An Arabidopsis MADS-box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409 [DOI] [PubMed] [Google Scholar]
- Zhu YX, Tepperman JM, Fairchild CD, Quail PH (2000) Phytochrome B binds with greater apparent affinity than phytochrome A to the basic helix-loop-helix factor PIF3 in a reaction requiring the PAS domain of PIF3. Proc Natl Acad Sci USA 97: 13419–13424 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.