Abstract
The human CAN gene was first identified as a target of t(6;9)(p23;q34), associated with acute myeloid leukemia and myelodysplastic syndrome, which results in the expression of a DEK-CAN fusion gene. CAN, also called NUP214, is a nuclear pore complex (NPC) protein that contains multiple FG-peptide sequence motifs. It interacts at the NPC with at least two other proteins, the nucleoporin NUP88 and hCRM1 (exportin 1), which was recently shown to function as a nuclear export receptor. Depletion of CAN in knockout mouse embryonic cells results in cell cycle arrest in G2, followed by inhibition of nuclear protein import and a block of mRNA export. We overexpressed CAN and DEK-CAN in U937 myeloid precursor cells. DEK-CAN expression did not interfere with terminal myeloid differentiation of U937 cells, whereas CAN-overexpressing cells arrested in G0, accumulated mRNA in their nuclei, and died in an apoptotic manner. Interestingly, we found that hCRM1 and import factor p97/importin β colocalized with the ectopically expressed CAN protein, resulting in depletion of both factors from the NPC. Overexpression of the C-terminal FG-repeat region of CAN, which contains the binding site for hCRM1, caused sequestering of hCRM1 in the nucleoplasm and was sufficient to inhibit cell growth and to induce apoptosis. These results confirm that CAN plays a crucial role in nucleocytoplasmic transport and imply an essential role for hCRM1 in cell growth and survival.
The recurrent chromosome translocation (6;9)(p23;q34), found in acute myeloid leukemia and myelodysplastic syndrome, fuses the coding regions of two genes, DEK and CAN (52). The resulting DEK-CAN mRNA encodes an in-frame chimeric protein that contains almost the entire DEK protein linked to the C-terminal two-thirds of CAN. DEK is a nuclear DNA-binding protein (16). CAN, also called NUP214, is a nuclear pore complex (NPC) component, or nucleoporin, and contains NPC protein-specific FG-repeat sequences (12, 30). Its deletion in mouse embryos results in cell cycle arrest in G2 followed by a block in mRNA export and inhibition of nuclear protein import (49).
The central region of CAN contains two predicted coiled-coil domains and anchors the protein to the NPC. Approximately the same domain binds NUP88, a novel NPC component of 88 kDa (13, 14). The translocation breakpoint in CAN lies in the middle of this region, and in the DEK-CAN fusion protein both nuclear envelope localization and NUP88 binding are disrupted (13). DEK-CAN is nuclear, and relocation of the C terminus of CAN from the nuclear envelope to the nucleus may contribute to the leukemogenic potential of the fusion protein (12). The C terminus of CAN, consisting of nucleoporin-specific repeats, can direct the protein to the nucleus in the absence of the NPC-binding domain (12). This relocation is mediated by binding of this part of the repeat, which is present in CAN as well as in DEK-CAN, to hCRM1, a protein of 112 kDa that was found to shuttle between the NPC and the nucleus (13, 14). hCRM1 is a member of a newly identified family of NPC-interacting proteins, which suggested that hCRM1 might function as a nucleocytoplasmic transport factor (14). Indeed, CRM1 was recently identified as an export receptor for leucine-rich nuclear export signals (NESs) (14a, 46).
The NPC is a supramolecular structure that contains multiple copies of about 100 different proteins. It mediates bidirectional transport of macromolecules between the cytoplasm and the nucleus (reviewed in references 8, 11, 38 and 42). Karyophilic proteins contain a nuclear localization signal (NLS) that is recognized by the NLS receptor, also called importin α, Srp1p, or karyopherin α (1, 2, 22, 35, 54). This complex docks to the cytoplasmic side of the NPC via p97, synonymous with importin β and karyopherin β (7, 23, 39). p97 binds to repeat-containing nucleoporins in vitro and in vivo (27, 31, 39–41). After this initial docking step, the complex is translocated through the pore via an energy-dependent process that is mediated by the small nuclear GTPase Ran and its cofactors (33, 34). At the nuclear side of the NPC, the NLS receptor and the substrate are released into the nucleoplasm while p97 remains bound to the NPC (23, 36).
Protein and ribonucleoprotein (RNP) export from the nucleus also occurs via a receptor-mediated, energy-dependent mechanism. NESs, identified in a number of proteins, are thought to play a role in this process (for reviews, see references 18, 21, and 28). Leucine-rich NESs can be transported by hCRM1, and overexpression of hCRM1 was shown to increase the export of human immunodeficiency virus type 1 (HIV-1) genomic mRNA and U small nuclear RNA (UsnRNA). Importin α was recently found to be involved in the export of capped RNA polymerase II transcripts from the nucleus, and dissociation of the importin α-RNA complex into the cytoplasm is mediated by importin β (20).
Depletion of nuclear pore components often leads to nucleocytoplasmic transport defects, growth inhibition, and cell death. Such effects have been demonstrated for several yeast mutants (10, 42), and recently for CAN/NUP214 in the mouse (49). The yeast studies have also shown that it is important to maintain the correct relative stoichiometry of NPC components, since overexpression of some components severely restricts cell growth (9, 55, 56). Overexpression of CAN and DEK-CAN in cell lines has proven to be toxic (12). To address why overexpression of these proteins is cytotoxic and to study the effects of overexpression on myeloid differentiation, we introduced inducible CAN and DEK-CAN genes into the human myeloid precursor U937 cells. Expression of the acute myelogenous leukemia-specific DEK-CAN protein did not affect the differentiation of U937 cells, whereas overexpression of CAN in U937 cells arrested them in G0, caused a defect in mRNA export and mislocalization of hCRM1 and p97, and ultimately led to apoptosis. Overexpression of the hCRM1-binding domain of CAN resulted in nuclear sequestering of hCRM1 and was sufficient to inhibit cell growth and to induce cell death.
MATERIALS AND METHODS
Expression constructs.
All the expression plasmids used in this study carry sequences encoding two copies of the influenza virus hemagglutinin (HA1) tag at the 5′ ends of their open reading frames (12) and are driven by the tetVP16-responsive promoter (24). Expression of CAN, DEK-CAN, CAN 1–1058, CAN 586–1058, CAN 816–2090, CAN 1864–2090, and CAN 1558–1840 was directed by plasmids described previously (12, 13). CAN 1140–2090 expression was directed by plasmid pHA1-Can Δ1–1139 (12). The pHA1-CAN construct (1140–1340, 1864–1912, 1984–2090) was derived from pHA1-CAN (1140–1340, 1864–2090) by an in-frame deletion of the fragment between the BamHI sites at positions 5825 and 6044 of the can cDNA. Plasmid pHA1-CAN 1864–2017 was derived by subcloning the region that encodes amino acids 1864 to 2090 from pHA1-CAN (Δ1–1139, Δ1341–1863) and inserting an oligonucleotide containing translational stops in the PstI site at position 6145 of the can cDNA. A plasmid containing the murine Bcl-xL cDNA, driven by the spleen focus-forming virus long terminal repeat, was kindly provided by G. Nuñez.
Inducible gene expression in U937T cells.
The human monoblast cell line U937 (47) and its derivatives were routinely cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum in a 37°C incubator with 5% CO2. Transfections were performed by electroporation at 0.17 kV and 960 μF on a Bio-Rad gene pulser. To generate cell lines inducibly expressing the proteins of interest, we used a two-step procedure. First, U937 cells were transfected with pUHD/TetVP16Puro (51), encoding tetVP16 under the control of the tetVP16-responsive promoter (24), thereby making tetVP16 expression tetracycline repressible and autoregulatory (45). Transfected cells were selected in 0.5 μg of puromycin per ml in the presence of 1 μg of tetracycline per ml. Single clones were examined for tetVP16 expression by RNA dot blotting of cells grown in the presence or absence of tetracycline. Of the 18 clones examined, 7 showed tetracycline-controlled expression of tetVP16. Second, clone U937T was selected for subsequent stable transfection with expression constructs containing HA1-epitope tagged cDNAs under the control of the tetVP16-responsive promoter. pHA1-CAN was cotransfected with the neomycin-selectable pMC1NeoPolyA plasmid (Stratagene), whereas all the other expression plasmids were cotransfected with the hygromycin-selectable pGEMHyg plasmid (50). Clones were selected and maintained in the presence of 1 μg of tetracycline per ml. Independent single clones were examined for tetracycline-dependent expression by Western blot analysis (43) with the anti-HA1 epitope monoclonal antibody 12CA5 (Boehringer) at 2 μg/ml. Bound antibody was visualized with a peroxidase-conjugated goat anti-mouse antibody (Jackson) and chemiluminescence reagent (New England Nuclear) used as specified by the manufacturer.
Cell growth, DNA content analysis, and apoptosis.
For induction of tetracycline-controlled gene expression, cells were washed four times with 10 ml of phosphate-buffered saline and seeded at 105 cells/ml in complete medium containing the desired tetracycline concentration. Cell proliferation was measured by a nonradioactive cell proliferation assay (Promega) as specified by the manufacturer. The DNA content and cell cycle distribution were evaluated by flow cytometric analysis of propidium iodide-stained nuclei as described previously (37). Fragmented DNA was isolated as previously described (32) and separated on 1% agarose gels containing ethidium bromide.
Differentiation of U937 cells.
Induced DEK-CAN58 cells were cultured for 5 days in the presence of 1 ng of transforming growth factor β1 (TGFβ1) (Promega) per ml and 250 ng of 1,25-dihydroxy vitamin D3 (Biomol) per ml. The percentages of monocyte surface antigen-positive cells were evaluated by fluorescence-activated cell sorter (FACS) analysis 5 days after induction of differentiation. The following mouse monoclonal antibodies were used: anti-CD11a, anti-CD11b (MO1), anti-CD14 (MY4), anti-CD15, and anti-CD18.
Indirect immunofluorescence.
Cytospins of U937 cells were fixed, permeabilized, and immunostained as described previously (12). Primary antibodies were diluted 1:400 for anti-C-terminal CAN (CNC [12]), to 2 μg/ml for 12CA5, to 2 μg/ml for MAb3E9 (7), and 1:90 for affinity-purified anti-hCRM1 (14). Bound primary antibodies were visualized with goat secondary antibodies conjugated to fluorescein isothiocyanate (FITC; Sigma) or Texas red (U.S. Biochemicals). Images were obtained by confocal laser-scanning microscopy on a Bio-Rad MRC1000 microscope with a 60× oil objective.
Transport assays.
In vitro nuclear protein import assays were performed essentially as described previously (3), with MDBK lysate instead of reticulocyte lysate as a source of essential cytosolic factors. Import was allowed to proceed for 20 min at 27°C, with parallel reactions on ice as controls. Immediately after import, the cells were washed and fixed in 3% formaldehyde for 15 min on ice. The mean fluorescence of the accumulated APC-NLS substrate in these cells was determined by FACS analysis. To determine the intracellular distribution of polyadenylated RNA, we hybridized cells on sterile cytospins with an oligo(dT)50 probe directly coupled to FITC as described previously (49). Images were obtained by confocal laser-scanning microscopy on a Bio-Rad MRC1000 microscope with a 60× oil objective.
Electron microscopy.
CAN7 cells were induced for 60 h and then fixed for 2 h in 0.1 M phosphate buffer (pH 7.4) containing 2% glutaraldehyde. The cells were then postfixed in 1% osmium tetroxide for 1 h, dehydrated, and embedded in Spurr low-viscosity resin (Electron Microscopy Sciences). Thin sections were cut, stained with uranyl acetate and lead, and examined with a JEOL JEM-1200EX II electron microscope.
RESULTS
CAN overexpression induces G0 arrest and apoptosis.
To study the effects of CAN and DEK-CAN on the growth, survival, and differentiation of myeloid cells, we introduced inducible CAN and DEK-CAN genes into the human myeloid precursor U937 cells. First, we generated the cell line U937T, which expressed tetVP16 in a tetracycline-dependent manner (24, 45, 51) but maintained normal growth characteristics. Subsequently, this clone was stably transfected with expression constructs containing HA1-epitope tagged CAN (ttCAN) or ttDEK-CAN cDNAs under the control of the tetVP16-responsive promoter. Each cell line used for this study was selected from a number of clonal lines based on the relative expression level of the transfected gene after tetracycline withdrawal.
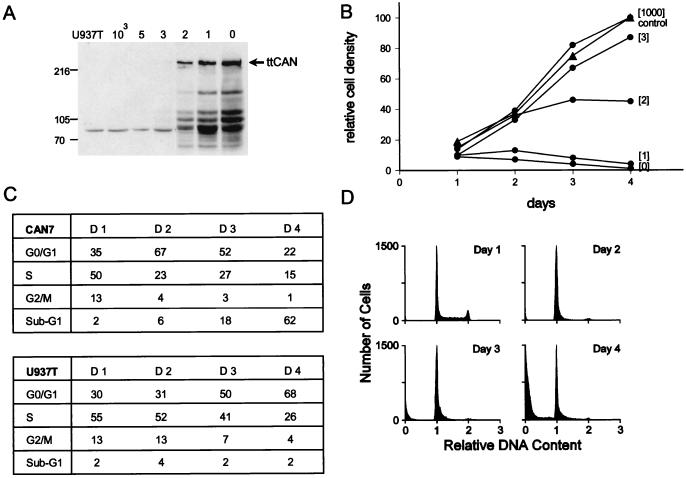
A clone that expressed relatively high levels of ttCAN upon withdrawal of tetracycline, CAN7, was selected to study the effects on cell growth of overexpression of different amounts of CAN by varying the concentrations of tetracycline in the culture medium. ttCAN was detected by immunofluorescence staining, with the anti-HA1 monoclonal antibody, as early as 10 h after induction and reached maximal levels after ∼24 h (data not shown). Western blot analysis showed that low concentrations of tetracycline in the culture medium (1 and 2 ng/ml) allowed the expression of intermediate levels of ttCAN whereas complete absence of tetracycline caused greater amounts of ttCAN to accumulate (Fig. 1A). In the presence of 3 ng of tetracycline per ml, ttCAN was barely detectable on Western blots, whereas no protein was detected in lysates from cells cultured at higher tetracycline concentrations. The growth curves of induced and uninduced CAN7 cells showed that growth inhibition was directly proportional to the amount of expressed ttCAN and inversely proportional to the tetracycline concentration in the medium (Fig. 1B). Like the parental U937T cells, the uninduced CAN7 cells continued to grow with a doubling time of about 24 h, whereas cells expressing the highest levels of ttCAN (0 and 1 ng of tetracycline/ml) stopped growing and their numbers were reduced after 3 days. Under conditions that gave expression of intermediate levels of ttCAN protein (2 ng of tetracycline per ml), we observed some increase in cell number, albeit at a significantly lower rate than that without induction.
FIG. 1.
CAN-overexpressing cells are growth inhibited. (A) Inducible expression of HA1-tagged CAN in CAN7 cells grown for 24 h in the presence of 1,000, 5, 3, 2, 1, and 0 ng of tetracycline per ml, as indicated above the lanes, was assayed by Western blot analysis of the sodium dodecyl sulfate (SDS)–6% polyacrylamide gel. Induced parental U937T cells serve as a negative control. Each lane contains lysate from 5 × 105 cells. The blot was probed with the anti-HA1 monoclonal antibody 12CA5. The arrow indicates ttCAN protein. Lysates of cells expressing large amounts of ttCAN protein show specific truncated products, which do not seem to affect the results. The sizes of molecular mass standards, run in an adjacent lane, are indicated on the left in kilodaltons. (B) Growth curves of induced CAN7 cells (•) and U937T control cells (▴). Cultures were maintained in medium containing the indicated tetracycline concentrations, and viability was measured daily by a nonradioactive proliferation assay. The relative density was calculated as a percentage of the density of uninduced cells on day 4. Mean values of triplicate determinations are plotted; the standard deviations were below 10%. This experiment is one of three that all gave similar results. (C) The cell cycle phase distribution of induced CAN7 cells (upper panel) and U937T cells (lower panel) at 1, 2, 3, and 4 days after tetracycline withdrawal was calculated from flow cytometric measurements of the DNA content. (D) Flow cytometry profiles showing DNA fluorescence of propidium iodide-stained CAN7 cell nuclei at 1, 2, 3, and 4 days after withdrawal of tetracycline. This is a representative experiment of three, all of which gave similar results.
To examine if fully induced CAN7 cells arrested at a particular phase of the cell cycle, we studied the DNA content of the overexpressing cells by FACS analysis of propidium iodide-stained nuclei (Fig. 1C and D). After 2 days of CAN induction, the percentage of cells in S phase was reduced from 50 to 23% and the percentage of cells in G2/M was reduced from 13 to 4%. The percentage of cells with a diploid DNA content was increased from 35 to 67%, suggesting that the cells had arrested at G1/G0. At this time point, very few cells were scored with a sub-G1 DNA content (see below). By day 3, however, 18% of the cells showed a sub-G1 DNA content, and this fraction increased to 62% by day 4 after tetracycline withdrawal (Fig. 1C, upper panel). The increase in the percentage of hypodiploid nuclei in ttCAN-overexpressing cells indicated that the cells became apoptotic (37). Moreover, the cells displayed morphologic features of apoptosis, including DNA fragmentation (Fig. 2A), nuclear segmentation, and chromatin condensation (Fig. 2B). The cells also stained positively by the terminal deoxytransferase-mediated dUTP-biotin nick end labeling method (17), which detects double-strand DNA breaks that are indicative of apoptosis (data not shown). U937T cells grown in the absence of tetracycline continued to cycle normally. They reached confluency by day 4, resulting in more cells in G1/G0 and fewer cells in S and G2/M. In contrast to ttCAN-overexpressing cells, the percentage of cells with a sub-G1 DNA content remained low, between 2 and 4% (Fig. 1C, lower panel).
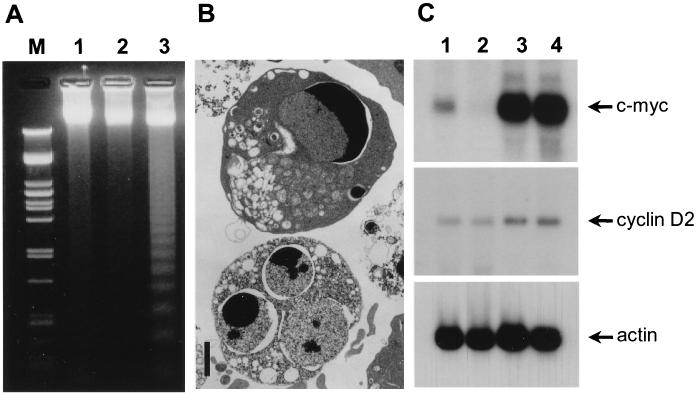
FIG. 2.
Overexpression of CAN induces G0 apoptosis in U937T cells. (A) Agarose gel electrophoresis of DNA from CAN7 cells cultured for 3 days in the absence of tetracycline (lane 3) shows internucleosomal DNA cleavage, whereas DNA from the parent U937T cells grown in the presence (lane 1) or absence (lane 2) of 1 μg of tetracycline per ml remains unfragmented. PstI-digested λ DNA served as a molecular weight marker (lane M). (B) Electron micrographs showing apoptotic CAN-overexpressing CAN7 cells after 3 days of induction. Bar, 2 μm. (C) Northern blot analysis of 15 μg of RNA isolated from the total culture (lane 1) and the FACS-sorted diploid-cell fraction (lane 2) of CAN7 cells induced for 40 h, compared to the total cultures of uninduced CAN7 cells (lane 3) and induced parental U937T cells (lane 4). The amounts of c-myc (2.4 kb; top panel), and cyclin D2 (6.0 kb; middle panel) mRNA were compared to the levels of actin mRNA (2.0 kb; bottom panel).
To determine if CAN-arrested CAN7 cells were blocked in G1 or G0, we studied their c-myc mRNA expression levels. Cycling cells express high levels of c-myc mRNA, whereas cells that exit the cell cycle and arrest in G0 do not transcribe c-myc (29). CAN7 cells were induced in the absence of tetracycline for 40 h and RNA was isolated from the total culture and from cells sorted for a diploid DNA content. RNA from total cultures of uninduced CAN7 cells and parental U937T cells was isolated to serve as a control. Northern blot analysis showed a dramatic decrease in the amount of c-myc mRNA in the ttCAN-expressing cells compared to that expressed in uninduced cells. The low level of c-myc mRNA in the total culture was derived from the fraction of cells that remained cycling, since the sorted diploid-cell fraction was negative for c-myc mRNA (Fig. 2C, top panel). A similar, albeit smaller, reduction was observed in the amounts of cyclin D2 mRNA (Fig. 2C, middle panel), which is normally present throughout the cell cycle (44). The more rapid downregulation of c-myc could be caused by the very short half-life of c-myc mRNA (53). These data strongly suggest that, prior to apoptosis, the ttCAN-overexpressing cells exited the cell cycle and arrested in G0.
Bcl-xL coexpression does not prevent apoptosis.
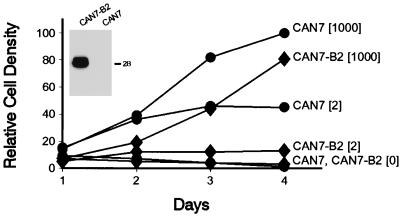
Bcl-xL, a member of the Bcl-2 family, can protect cells against a variety of apoptosis inducers (5, 19, 25). We examined whether constitutive coexpression of the Bcl-xL gene with ttCAN in CAN7 cells could inhibit CAN-induced apoptosis. A representative stably transfected CAN7 clone, CAN7-B2, expressed high levels of Bcl-xL protein and grew slower than the parental CAN7 clone (Fig. 3). This could be a direct consequence of Bcl-xL expression, since expression of this protein has been shown to prolong the G1 phase in U937 and other cell lines (6). Bcl-xL expression had no influence on growth inhibition or cell death after tetracycline was removed from several independent CAN7-Bcl-xL clones (Fig. 3), indicating that Bcl-xL could not inhibit the apoptotic cell death of induced CAN7 cells.
FIG. 3.
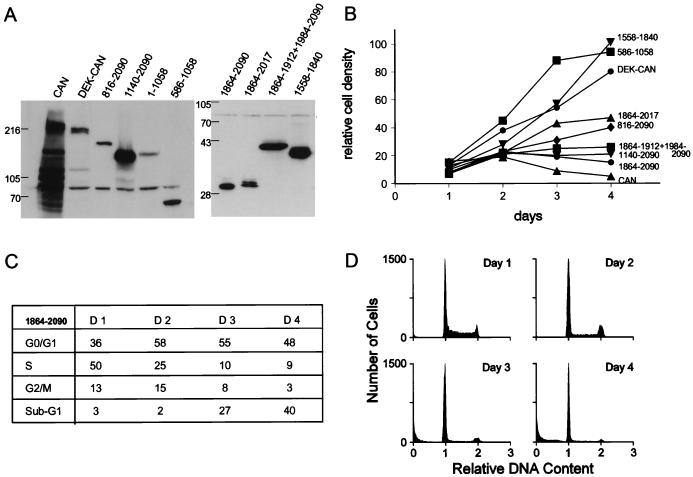
Bcl-xL coexpression does not rescue CAN-overexpressing cells. Growth curves of a representative Bcl-xL-overexpressing CAN7 clone, CAN7-B2 (⧫), compared to CAN7 (•), both grown in 1,000, 2, and 0 ng of tetracycline per ml for 4 days. Mean relative density values of triplicate cultures are plotted against time; the standard deviations were below 10%. This experiment is one of three, all of which gave similar results. Inset: Western blot of an SDS–9% polyacrylamide gel containing lysate from 5 × 105 cells per lane, probed with a mouse monoclonal antibody to Bcl-xL. The 29-kDa doublet represents the Bcl-xL protein. The position of the 28-kDa molecular mass standard is indicated on the right.
Transport defects in CAN-overexpressing cells.
We studied the subcellular localization of CAN in induced and uninduced cells by indirect immunofluorescence with the polyclonal CAN antiserum CNC (12). In the parental U937T cells, endogenous CAN levels were low and the protein localized to the nuclear envelope (Fig. 4A). During the first day after tetracycline withdrawal, ttCAN in CAN7 cells was localized mainly to the nuclear envelope and cytoplasmic speckles. The latter structures could be annulate lamellae or simply aggregates of insoluble protein (Fig. 4B). However, during the second day after induction, an increasing percentage of cells showed nuclear localization of ttCAN, and by the third day, 90% of the cells had ttCAN in the nucleus (Fig. 4C). The nuclear staining was diffuse, with a few strong dots. These results demonstrate that the loss of cell viability upon ttCAN overexpression coincides with the accumulation of ttCAN in the nucleoplasm.
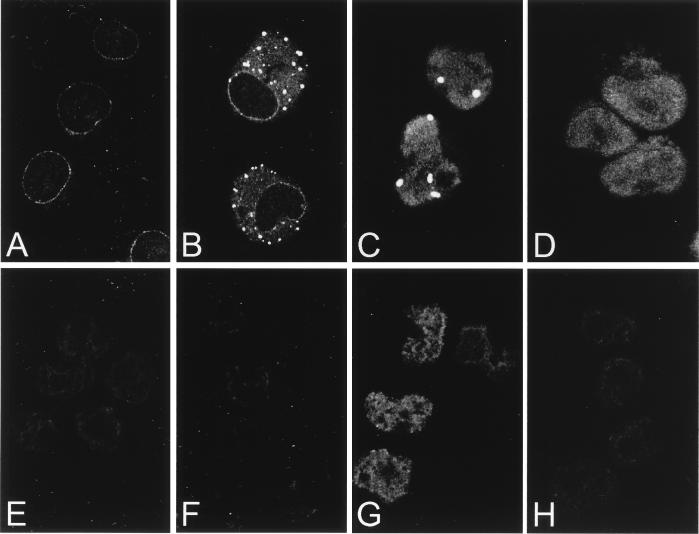
FIG. 4.
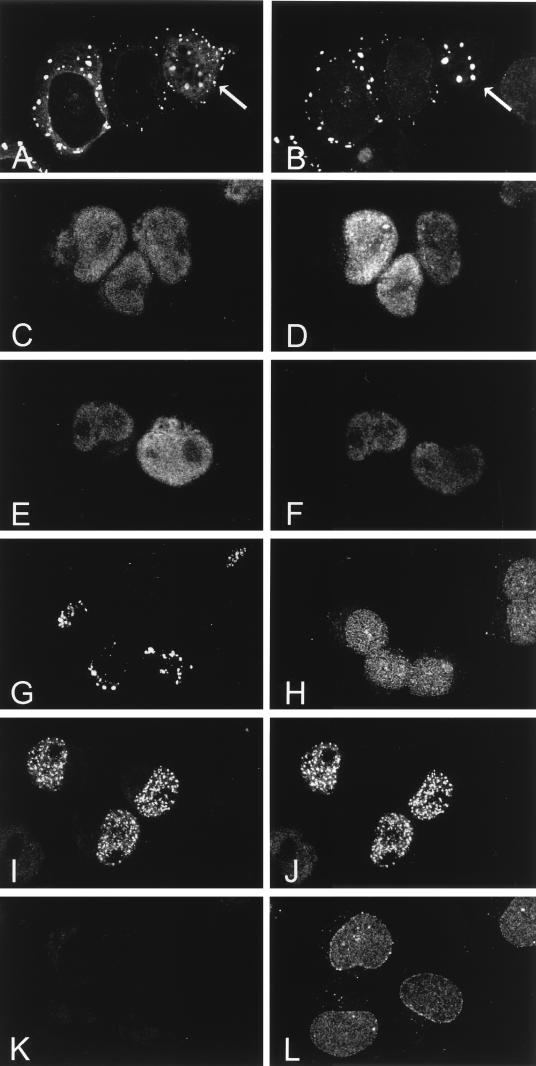
Polyadenylated RNA export defect in CAN-overexpressing cells. (A to C) Confocal images of endogenous CAN expression in U937T cells (A) and overexpressed CAN in CAN7 cells 20 h (B) and 48 h (C) after induction; the cells were stained with the anti-CAN antiserum αCNC. (D) Overexpression of HA-CAN1864-2090 in U937T cells stained with the anti-HA antibody 12CA5. (E to H) Subcellular localization of polyadenylated RNA in control U937T cells (E) and CAN7 cells after 48 h (F) and 60 h (G) of induction and in HA-CAN 1864-2090 cells after 60 h of induction (H) analyzed by in situ hybridization with an FITC-conjugated oligo(dT)50 probe.
Because CAN is a nucleoporin, we asked whether the cytotoxity of its overexpression coincided with perturbations in its NPC transport function. We measured the nuclear export capacity of the ttCAN-overexpressing cells by monitoring the localization of polyadenylated RNA by in situ hybridization with an oligo(dT)50 probe directly coupled to FITC (4). At 48 h after tetracycline withdrawal, poly(A)+ RNA in both U937T cells (Fig. 4D) and CAN7 cells (Fig. 4E) appeared to be diffusely distributed throughout the nucleoplasm and cytoplasm. In contrast, at 56 to 60 h after ttCAN induction, only CAN7 cells showed strong nuclear staining, indicating nuclear accumulation of poly(A)+ RNA (Fig. 4F). At this time point, most cells were still alive and arrested in G0. As a control, we induced apoptosis by culturing U937T cells for 48 h in the presence of 50 μg of the protein synthesis inhibitor cycloheximide per ml. In this case, apoptosis was not preceded by mRNA accumulation in the nucleus (data not shown), suggesting that the block in mRNA export observed in CAN7 cells was specific for CAN overexpression.
The nuclear protein import capacity of induced CAN7 cells was assessed in vitro by examining the accumulation of an NLS-linked fluorescent substrate, APC-NLS, in the nuclei of digitonin-permeabilized cells (3). After 24 h of tetracycline withdrawal, ttCAN-overexpressing CAN7 cells showed an import capacity similar to that of U937T cells whereas CAN7 cells that had been induced for 48 h did not appreciably import APC-NLS (data not shown). These results indicated that the inhibition of nuclear protein import in CAN-overexpressing cells coincided with the cell cycle arrest. Therefore, we compared the import capacity of CAN7 cells 2 days after tetracycline withdrawal with that of U937T cells that had reached their maximal density 4 days after seeding. In both cultures, almost 70% of the cells were arrested with a diploid DNA content (Fig. 1C). Unexpectedly, we could not detect NLS protein import in the density-arrested U937T cells either, which makes it impossible to distinguish between cell cycle arrest and CAN overexpression as the primary cause of the import defect.
Inhibition of nucleocytoplasmic transport may be caused by gross structural alterations in the NPC or nuclear envelope or by functional perturbation. Thin-section electron microscopy did not reveal structural perturbations of the NPC or nuclear envelope in CAN-overexpressing CAN7 cells after 3 days of induction (data not shown). Therefore, we assessed whether the toxicity of excess CAN resulted from functional inactivation of CAN-interacting proteins. hCRM1, a protein that coimmunoprecipitates with CAN, shuttles between the nucleus and the cytoplasm and functions as an export receptor for leucine-rich nuclear export sequences (14, 46). We used immunopurified polyclonal antiserum to detect hCRM1 (14) in combination with a monoclonal antibody, 12CA5, to the HA1 epitope to detect ttCAN in double-immunostaining experiments. Strikingly, we found that endogenous hCRM1 colocalized with overexpressed CAN in the nuclear envelopes, cytoplasm, and nuclei of CAN7 cells (Fig. 5A and B). In cells that expressed CAN in both cytoplasmic and nuclear speckles, hCRM1 preferentially colocalized with the nuclear structures (Fig. 5A and B). The accumulation of ttCAN in the nuclei of CAN7 cells after 3 days resulted in colocalization of hCRM1 in the nucleus and depletion of this factor from the nuclear envelope. We then studied the localization of the transport factor p97/importin β, which binds to CAN and other FG-repeat containing nucleoporins in vitro and localizes to the NPC and cytoplasm (7, 23, 39). We immunostained induced CAN7 cells with a monoclonal antibody to p97 (MAb3E9 7) and anti-CNC and found that p97 colocalized with ectopically expressed CAN protein in the nuclear envelope and the cytoplasmic speckles. Two days after ttCAN induction, colocalization also occurred in the nucleus, which resulted in depletion of p97 from the nuclear envelope (Fig. 6A and B).
FIG. 5.
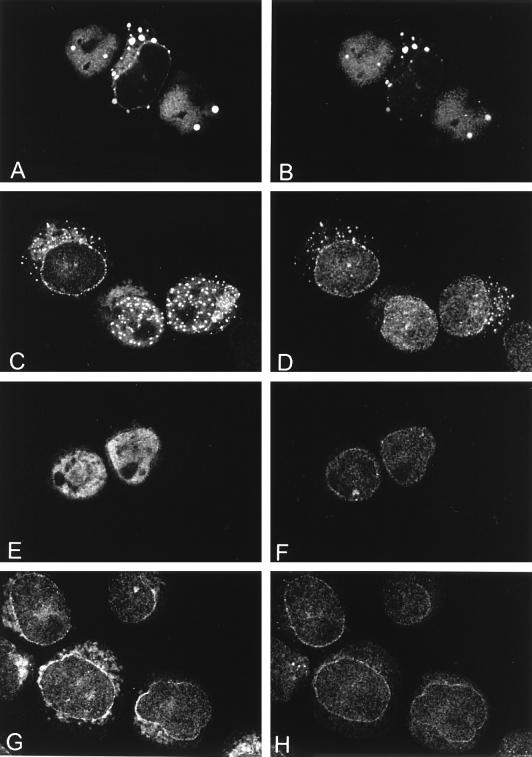
Colocalization of full-length CAN and CAN mutants with hCRM1. The subcellular distribution of hCRM1 and CAN proteins after 2 days of induction is shown. (A, C, E, G, I, and K) Confocal microscopy showing immunodetection of induced CAN and mutant proteins by an anti-HA1 monoclonal antibody followed by a goat anti-mouse Texas red-conjugated secondary antibody. (B, D, F, H, J, and L) Distribution of endogenous hCRM1 in the same cells stained with the anti-hCRM1 antiserum and a goat anti-rabbit FITC-linked antibody. hCRM1 colocalized with full-length CAN (A and B [cells indicated by arrows]), with the C-terminal FG repeat regions of CAN, CAN 1864–2090 (C and D) and CAN (1140–1340, 1864–1912, 1984–2090) (E and F), and with DEK-CAN (I and J). In contrast, hCRM1 did not colocalize with the more N-terminally located FG repeat region of CAN, represented by CAN 1558–1840 (G and H); it showed normal distribution in the nuclear membrane and the nucleoplasm, similar to that of induced U937T cells (K and L).
FIG. 6.
Double immunostaining of CAN and mutants with p97/importin β. The subcellular distribution of p97 and CAN proteins after 2 days of induction is shown. (A, C, E, and G) Confocal images of indirect immunofluorescence with the anti-CAN polyclonal antiserum anti-CNC, detected with a Texas red-conjugated goat anti-rabbit antibody. (B, D, F, and H) Confocal images of the same cells immunostained with an antibody to p97 (MAb3E9), detected with an FITC-conjugated goat anti-mouse antibody. Endogenous p97 colocalized with overexpressed CAN in the nuclear membrane and in the cytoplasmic and nucleoplasmic speckles (A and B). In cells overexpressing CAN 816–2090, only the nuclear membrane and the cytoplasmic speckles showed colocalization of the CAN mutant with p97 (C and D). Cells overexpressing CAN 1140–2090 showed normal p97 localization in the nuclear membrane (E and F), comparable to cells expressing endogenous levels of CAN (G and H).
Expression of DEK-CAN and CAN mutants in U937 cells.
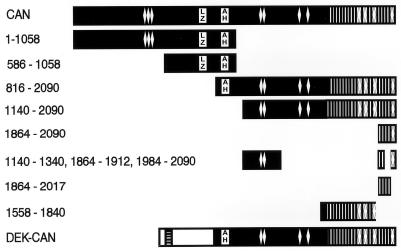
To map the region of CAN that mediated cycle arrest and apoptosis, we studied the response of U937T cells to induced expression of CAN mutants and the leukemia-specific DEK-CAN fusion protein (Fig. 7). Clones were selected for high levels of expression after induction; however, some mutants, including DEK-CAN, did not reach the expression levels of ttCAN (Fig. 8A). Most of the mutants described in this study were analyzed previously for subcellular localization and coimmunoprecipitating proteins in transient-transfection studies (13). The N terminus of CAN (CAN 1–1058) and a shorter central region (CAN 589–1058) both associate with the NPC and bind NUP88. Expression of these mutants did not affect cell growth or viability (Fig. 8B). However, the expression of C-terminal regions of CAN, such as CAN 816–2090, which localizes to the NPC, cytoplasm, and nucleus, and the nuclear mutant CAN 1140–2090, inhibited cell growth (Fig. 8B). This effect could be attributed to overexpression of the most C-terminal FG repeat-containing region (CAN 1864–2090), which was sufficient for the lethal phenotype (Fig. 8B). Importantly, this part of CAN harbors the hCRM1-binding domain. DNA histogram analysis (Fig. 8C and D) of induced CAN 1864–2090 cells showed that 2 days after tetracycline withdrawal, the percentage of cells in S phase was reduced from 50 to 25% and the percentage of cells in G0/G1 had increased from 36 to 58%. The percentage of cells with a sub-G1 DNA content was 27% after 3 days of induction and 40% after 4 days (Fig. 8C). This demonstrates that overexpression of the hCRM1-binding region is sufficient to cause cycle arrest and cell death. Although the entire cell population underwent apoptosis, the onset was slower than when the full-length CAN was overexpressed, possibly because of subtle differences in the expression levels between the two clones. CAN deletion mutants that lack part of the region necessary for hCRM1 coimmunoprecipitation [CAN (1140–1340, 1864–1912, 1984–2090) and CAN 1864–2017] still localized to the nucleus and exhibited the lethal phenotype, albeit at considerably higher expression levels (Fig. 8A and B). The FG repeat-containing region just N-terminal of the hCRM1-binding region (CAN 1558–1840) localized to the cytoplasm and did not affect cell growth (Fig. 8B). The most highly inducible DEK-CAN clone, DEK-CAN58, expressed only about 25% of the amount of CAN protein produced in CAN7, as estimated by Western blot analysis (Fig. 8A). This clone was slightly growth inhibited but did not die of apoptosis (Fig. 8B).
FIG. 7.
Overview of CAN deletion mutants. Black bars represent CAN and CAN mutant proteins; numbers on the left represent amino acid boundaries. Predicted structural motifs are represented as follows: vertical lines, FG repeats; diamonds, FXF repeats; LZ, coiled-coil 1 and adjacent leucine zipper; AH, coiled-coil 2. Horizontal stripes indicate an acidic region in the DEK sequence (white bar).
FIG. 8.
The C terminus of CAN is sufficient to inhibit cell growth. (A) Western blot analysis of SDS–6% polyacrylamide (left panel) and SDS–10% polyacrylamide (right panel) gels with lysates from 5 × 105 induced cells expressing the indicated CAN mutants and DEK-CAN. CAN (1140–1340, 1864–1912, 1984–2090) is abbreviated to CAN 1864−1912+1984−290. Blots were probed as described for Fig. 1A. (B) Growth curves of cells overexpressing selected CAN mutants and DEK-CAN grown in the absence of tetracycline for 4 days. Data shown are the mean values of triplicate cultures from one experiment of at least three independent experiments that gave similar results. (C) Cell cycle phase distribution of induced CAN 1864–2090 cells. (D) Quantitation of the DNA content by flow cytometric analysis in CAN 1864–2090 cells, overexpressing the hCRM1-binding domain, on days 1, 2, 3, and 4 after withdrawal of tetracycline.
Double-immunofluorescence staining studies of CAN mutants (Fig. 5, left panel) with hCRM1 (Fig. 5, right panel) showed that endogenous hCRM1 colocalized with CAN 816–2090 in the cytoplasm and in the nucleus (data not shown). The endogenous hCRM1 also colocalized with the hCRM1-binding domain (CAN 1864–2090) in the nucleus (Fig. 5C and D). The nuclear mutants CAN (1140–1340, 1864–1912, 1984–2090) (Fig. 5E and F) and CAN 1864–2017 (data not shown), which both lack part of the hCRM1-binding domain that is required to coimmunoprecipitate hCRM1 (13), still caused redistribution of hCRM1 to the nucleus. In contrast, CAN 1558–1840, which also does not coprecipitate hCRM1 (13), did not colocalize with hCRM1 (Fig. 5G and H) or influence cell growth. Although DEK-CAN levels were not high enough to induce cell death, the staining patterns of DEK-CAN and hCRM1 were overlapping in highly expressing cells (Fig. 5I and J). Taken together, all (partly) nuclear CAN mutants colocalize with hCRM1, suggesting that the region of CAN needed for in vivo interaction with hCRM1 is smaller than the hCRM1-binding domain identified by coimmunoprecipitation. High expression levels of these mutants resulted in sequestration of hCRM1 in the nucleus and depletion of hCRM1 from the nuclear envelope, which coincided with growth inhibition and cell death.
We studied the distribution of endogenous p97 (Fig. 6, right panel) in cells expressing CAN mutants (Fig. 6, left panel) and found colocalization of p97 with CAN 816–2090 in the nuclear envelope and the cytoplasmic speckles. Cells expressing this CAN mutant in nuclear speckles showed a normal p97 distribution (Fig. 6C and D). Expression of DEK-CAN (data not shown) or CAN 1040–2090 (Fig. 6E and F) did not affect p97 localization, indicating that sequences in the central region of CAN mediate this effect.
Poly(A)+ RNA in clones expressing DEK-CAN (data not shown) or the C terminus of CAN (Fig. 4H) was diffusely distributed in the nucleoplasm and cytoplasm, similar to U937T control cells (see Fig. 4D), indicating that there was no defect in mRNA export in these cells. Thus, it is unlikely that hCRM1 is involved in the export of mRNA. Digitonin-permeabilized cells overexpressing the hCRM1-binding domain were severely inhibited in APC-NLS import, comparable to CAN-overexpressing cells and density-arrested U937T cells (data not shown). Therefore, a possible additional effect on import of hCRM1 sequestering in the nucleus could not be determined.
DEK-CAN does not inhibit differentiation of U937 cells.
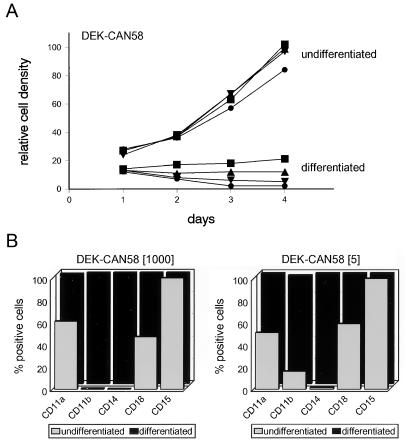
To study the effect of DEK-CAN on myeloid maturation, we induced clone DEK-CAN58 to differentiate by using a combination of transforming growth factor β1 (TGFβ1) and 1,25-dihydroxy vitamin D3 (D3) (48) after 3 days of withdrawal from tetracycline. Surprisingly, the cells died rapidly (Fig. 9A). Immunofluorescence staining showed elevated levels of DEK-CAN in these cells compared to those in undifferentiated cells (data not shown). We consistently found this effect, even when differentiation was induced by other chemicals, such as dimethyl sulfoxide and phorbol-12-myristate-13-acetate. We therefore tried partially releasing the DEK-CAN58 cells for 3 days in the presence of 5 and 10 ng of tetracycline per ml prior to inducing differentiation. Both partially induced and uninduced cells ceased to proliferate upon exposure to TGFβ1 and D3 (Fig. 9A). Compared to uninduced cells (in 1,000 ng of tetracycline per ml), viability was not affected in cells cultured with differentiation agents in 10 ng of tetracycline per ml, whereas about 50% of the cells grown in 5 ng of tetracycline per ml died. The remaining cells in these cultures expressed high levels of DEK-CAN.
FIG. 9.
Differentiation antigen expression in U937T cells expressing DEK-CAN. (A) Growth curves of DEK-CAN58 cells cultured in medium containing 1,000 (▪), 10 (▵), 5 (▾), or 0 (•) ng of tetracycline per ml, seeded at 105 cells/ml in the presence (differentiated) or absence (undifferentiated) of 1 ng of TGFβ1 per ml and 250 ng of D3 per ml. The cells were cultured in normal medium containing the indicated tetracycline concentrations for 3 days prior to the induction of differentiation. (B) DEK-CAN58 cells cultured in 1,000 ng of tetracycline per ml (no DEK-CAN expression) and 5 ng of tetracycline per ml (partial DEK-CAN induction) were differentiated for 5 days in medium containing TGFβ1 and D3 (differentiated; black bars). Undifferentiated cells cultured in 1,000 and 5 ng of tetracycline per ml are also shown (undifferentiated; gray bars). Expression of the indicated differentation antigens (CD11a, CD11b, CD14, CD15, and CD18) was evaluated by cytofluorimetry with specific monoclonal antibodies. Results are expressed as percentages of antigen-positive cells.
DEK-CAN58 cells grown in 5 and 1,000 ng of tetracycline per ml were induced to differentiate by a 5-day exposure to TGFβ1 and D3. As a measure of induction of differentiation, we monitored the increased expression of the cell surface antigens CD11a, CD11b, CD14, and CD18. The enhanced expression of these markers indicated that DEK-CAN-expressing DEK-CAN58 cells (Fig. 9B, right panel) differentiated normally to mature monocytes in a manner indistinguishable from that of tetracycline-repressed DEK-CAN58 cells (Fig. 9B, left panel). Moreover, Giemsa staining of the two differentiated cell populations showed the same morphology (data not shown), confirming that DEK-CAN expression did not inhibit the myeloid differentiation of U937 cells.
DISCUSSION
We examined the biologic properties of the CAN protein and the leukemia-specific DEK-CAN fusion protein by overexpressing these proteins in U937 myeloid precursor cells. We found that ectopic expression of CAN caused a cell cycle arrest in G0 followed by a block in mRNA export and apoptotic cell death. Overexpression of the most C-terminal FG repeat-containing region of CAN, which binds hCRM1, was sufficient to reproduce most of these effects: it induced growth arrest and apoptosis. Thus, overexpression of CAN and the hCRM1-binding domain of CAN interfered with some of the normal functions of CAN in a dominant-negative way. Considering the large number of importin β family members, the effects of the C-terminal fragment of CAN on apoptosis could conceivably be related to interfering with another transport factor besides hCRM1. However, in immunoprecipitation experiments, only hCRM1 coprecipitates with the C-terminal FG repeat (13) and is therefore most likely to be responsible. Moderate DEK-CAN expression slightly inhibited cell growth and did not interfere with the differentiation of U937 cells to mature monocytes. Because U937 cells represent an intermediate stage of monocytic development (26), it is possible that their terminal differentiation is not affected because DEK-CAN may inhibit the differentiation only of earlier myeloid precursors. Myeloid cells from patients with t(6;9) acute myeloid leukemia are partially inhibited in their differentiation pathways but are not totally blocked. Therefore, it is also possible that DEK-CAN has no effect on differentiation but affects the proliferation of early precursor cells in these patients.
CAN overexpression induces cell cycle arrest and apoptosis.
CAN-overexpressing cells and, to a lesser extent, cells overexpressing the hCRM1-interaction domain of CAN accumulated with a diploid DNA content. Arrested CAN-overexpressing cells no longer express c-myc, a proto-oncogene that is continuously expressed in proliferating cells but is downregulated when cells exit the cell cycle (15, 53). U937T cells that were mostly arrested in G0/G1 after reaching their maximal density did not import detectable levels of import substrate. Therefore, additional effects of the overexpression of CAN and the hCRM1-binding domain on nuclear protein import could not be measured. It is possible that CAN overexpression interferes with the nuclear transport of factors critical to cell growth or survival. Alternatively, proper stoichiometry of the components that make up the NPC could be necessary for the formation of new NPCs, a process that is presumably essential for growth. CAN depletion leads to cell cycle arrest in G2 (49), whereas overexpression of CAN arrested cells in G0, suggesting that CAN is essential to proper cell cycle progression. Furthermore, CAN-depleted cells still have hCRM1 in their nuclear envelopes (14), whereas the nuclear envelopes of CAN-overexpressing cells are depleted of hCRM1 (see below). It will be interesting to see how the transport function of hCRM1 could be linked to cell cycle progression.
Cells overexpressing full-length CAN or CAN mutants containing the hCRM1-binding domain die after 72 to 96 h of induction and show morphologic features of apoptosis. Coexpression of the potent survival factor Bcl-xL did not protect CAN-overexpressing cells from apoptosis, suggesting that a non-Bcl-xL-controlled pathway is activated.
hCRM1 function is essential.
hCRM1 normally localizes to the nucleus and the NPC and is regularly released from the NPC into the nucleoplasm (14). CAN mutants containing the hCRM1-binding region were, at least in part, nuclear and caused nuclear accumulation of hCRM1. Overexpression of these mutants at levels that were lethal resulted in a complete depletion of hCRM1 from the nuclear envelope. This result suggests that the excess of the nuclear hCRM1-binding domain of CAN competes with NPC-associated CAN for binding to hCRM1 and inhibits a function of hCRM1 that is essential for cell growth or survival. In the light of recent findings, this essential function of hCRM1 most probably is the export of NES-containing (ribonuclear) proteins, among which are UsnRNPs (14a, 46). Inhibition of UsnRNP export prevents their cytoplasmic modification and may eventually affect pre-mRNA splicing, which could be an effective inducer of apoptosis. It would be interesting to see if progressive inhibition of other essential nuclear transport factors also causes apoptosis or if this is specific to hCRM1. Why do cells arrest in G0 if hCRM1 is a general nuclear export factor? Besides possible defects in the transport of specific NES-containing proteins, it is conceivable that inhibition of splicing could preferentially affect the production of short-lived mRNAs, encoding proteins with a high turnover rate, such as c-MYC, that directly affect the ability of the cell to enter the G1 phase.
The localization of overexpressed full-length CAN changed from mainly nuclear membrane and cytoplasm after the first day of induction to mainly nuclear during the second and third days of expression. HeLa cells, transiently transfected to highly overexpress CAN, showed a similar nuclear localization in 5 to 10% of the cells (12). It may be that overexpressed CAN spills over into the cytoplasm and nucleus when all of the NPC-binding sites are saturated. Transport of CAN into the nucleus could be mediated by its association with hCRM1 (13). The colocalization of CAN and hCRM1 in cytoplasmic structures suggests that complex formation has already occurred in the cytoplasm. Since hCRM1 has a half-life of approximately 24 h (14), it is unlikely that the hCRM1 observed in the cytoplasm of CAN-overexpressing cells is only newly synthesized hCRM1. Instead, these data suggest that hCRM1 travels from the nucleus to the cytoplasm, in addition to its release from the NPC into the nucleoplasm. This would be in agreement with the shuttling function of CRM1 in yeast (46). In addition, the movement of hCRM1 between the nucleus and the cytoplasm was confirmed by microinjection studies in Xenopus oocytes (14).
The region of CAN required for its colocalization with hCRM1 in the nucleus was smaller than the hCRM1-binding domain identified by coimmunoprecipitation (13), suggesting that the in vivo interaction of mutants that lack part of the binding domain is too weak to be detected by immunoprecipitation. This finding is consistent with the idea that nuclear localization of the C-terminal repeat region of CAN, which does not contain a known NLS, is mediated by hCRM1 (13).
CAN overexpression induces defects in p97 localization and mRNA export.
Overexpressed CAN colocalized with the nuclear import factor p97/importin β, initially in the nuclear membrane and cytoplasmic structures and subsequently in nuclear structures. p97 binds to CAN in vitro (39), but this interaction is not strong enough to mediate coimmunoprecipitation with CAN from cell lysates of CAN-overexpressing cells (14). Based on binding studies of NUP98 mutants and other nucleoporins, the FG repeat regions are thought to harbor the p97-binding domain (36, 40). Our results with CAN 1558–1840 and CAN 1864–2090 show that p97 does not bind to the overexpressed C-terminal CAN repeat regions alone. Instead, p97 colocalizes with full-length CAN and partly with CAN 817–2090, both of which bind to the NPC and form structures of unknown composition upon overexpression. It is possible that p97 also associates in vivo with the central region of CAN, either directly or via another protein, and that binding of p97 to CAN requires both additive interactions.
Only cells overexpressing full-length CAN demonstrated a defect in polyadenylated RNA export after 55 to 60 h of induction. These were also the only cells to sequester p97 in their nuclei. These observations may be directly linked because depletion of p97 from the cytoplasm may lead to a block in importin α import into the nucleus, thereby inhibiting its function in the export of capped RNAs (20). Studies with yeast cells overexpressing the nucleoporin gene NUP116 also suggest that p97 plays a role in mRNA export. Overexpression of the GLFG repeat region of Nup116p severely inhibits cell growth and blocks polyadenylated-RNA export (27). This region interacts with Kap95p, an essential yeast homolog of the vertebrate import factor p97, suggesting that sequestering of this factor is at least partly responsible for the phenotype (27).
In summary, our results indicate that the cytotoxicity of CAN overexpression may be caused by depleting hCRM1 from the nuclear envelope and confining it to the nucleus, thereby inhibiting the export of NES-containing substrates. Furthermore, only full-length CAN, which contains the central protein-protein interaction domain in addition to the C-terminal FG repeats, colocalizes with p97 in the nucleus and causes nuclear accumulation of polyadenylated RNA.
The mechanism by which DEK-CAN contributes to leukemogenesis remains unknown. DEK is a sequence-specific DNA-binding protein, and DEK-binding sites were recently identified in the regulatory regions of several early myeloid genes (16). DEK-CAN could exhibit altered transcriptional regulation compared with DEK, due to the presence of CAN sequences or proteins that associate with this portion of CAN, such as hCRM1. Alternatively, the redistribution of hCRM1 towards the nucleoplasm by DEK-CAN could interfere with the transport function of CAN and hCRM1. DEK-CAN is expressed at such a low level in leukemic cells that a total depletion of hCRM1 from the NPC is not to be expected but a shift in the balance of nuclear hCRM1 may have an effect on hematopoietic cell growth or differentiation.
ACKNOWLEDGMENTS
We are grateful to Dario Vignali for pUHD/TetVP16Puro, Gabriel Nuñez for the spleen focus-forming virus Bcl-xL plasmid, John Cleveland for a c-myc probe, Charles Sherr for a cyclin D2 probe, Stephen Adam for monoclonal antibody MAb318, and Maarten Fornerod for affinity-purified hCRM1 antibodies. We thank Sharon Frase and Andrea Elberger for use of the Confocal Laser Scanning Facility, UT Memphis (funded by PHS grant CLSM 1S10RR08385), Donna Davis and Gopal Murti for electron microscopic studies, Richard Ashmun for FACS analyses, Sjozef van Baal for help with the figures, Charlette Hill for secretarial assistance, and Sue Vallance for scientific editing.
These studies were supported in part by Cancer Center CORE grant CA-21765 and by the Associated Lebanese Syrian American Charities (ALSAC) of St. Jude Children’s Research Hospital.
REFERENCES
- 1.Adam E J H, Adam S A. Identification of cytosolic factors required for nuclear location sequence-mediated binding to the nuclear envelope. J Cell Biol. 1994;125:547–555. doi: 10.1083/jcb.125.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adam S A, Gerace L. Cytosolic proteins that specifically bind nuclear localization signals are receptors for nuclear import. Cell. 1991;66:837–847. doi: 10.1016/0092-8674(91)90431-w. [DOI] [PubMed] [Google Scholar]
- 3.Adam S A, Marr R S, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amberg D C, Goldstein A L, Cole C N. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- 5.Boise L H, González-Garcia M, Postema C E, Ding L, Lindsten T, Turka L A, Mao X, Nuñez G, Thompson C B. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 6.Borner C. Diminished cell proliferation associated with the death-protective activity of Bcl-2. J Biol Chem. 1996;271:12695–12698. doi: 10.1074/jbc.271.22.12695. [DOI] [PubMed] [Google Scholar]
- 7.Chi N C, Adam E J H, Adam S A. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis L I. The nuclear pore complex. Annu Rev Biochem. 1995;64:865–896. doi: 10.1146/annurev.bi.64.070195.004245. [DOI] [PubMed] [Google Scholar]
- 9.Davis L I, Fink G R. The NUP1 gene encodes an essential component of the yeast nuclear pore complex. Cell. 1990;61:965–978. doi: 10.1016/0092-8674(90)90062-j. [DOI] [PubMed] [Google Scholar]
- 10.Doye V, Hurt E C. Genetic approaches to nuclear pore structure and function. Trends Genet. 1995;11:235–241. doi: 10.1016/s0168-9525(00)89057-5. [DOI] [PubMed] [Google Scholar]
- 11.Fabre E, Hurt E C. Nuclear transport. Curr Opin Cell Biol. 1994;6:335–342. doi: 10.1016/0955-0674(94)90023-x. [DOI] [PubMed] [Google Scholar]
- 12.Fornerod M, Boer J, van Baal S, Jaeglé M, Von Lindern M, Murti K G, Davis D, Bonten J, Buijs A, Grosveld G. Relocation of the carboxyterminal part of CAN from the nuclear envelope to the nucleus as a result of leukemia-specific chromosome rearrangements. Oncogene. 1995;10:1739–1748. [PubMed] [Google Scholar]
- 13.Fornerod M, Boer J, van Baal S, Morreau H, Grosveld G. Interaction of cellular proteins with the leukemia specific fusion proteins DEK-CAN and SET-CAN and their normal counterpart, the nucleoporin CAN. Oncogene. 1996;13:1801–1808. [PubMed] [Google Scholar]
- 14.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti K G, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRMl is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 15.Freytag S O. Enforced expression of the c-myc oncogene inhibits cell differentiation by precluding entry into a distinct predifferentiation state in G0/G1. Mol Cell Biol. 1988;8:1614–1624. doi: 10.1128/mcb.8.4.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu G, Grosveld G, Markovitz D. DEK, an autoantigen involved in a chromosomal translocation in acute myelogenous leukemia, binds to the human immunodeficiency virus type 2 enhancer. Proc Natl Acad Sci USA. 1997;94:1811–1815. doi: 10.1073/pnas.94.5.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gavrieli Y, Sherman Y, Ben-Sasson S A. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerace L. Nuclear export signals and the fast track to the cytoplasm. Cell. 1995;82:341–344. doi: 10.1016/0092-8674(95)90420-4. [DOI] [PubMed] [Google Scholar]
- 19.González-Garcia M, Garcia I, Ding L, O’Shea S, Boise L H, Thompson C B, Nuñez G. bcl-x is expressed in embryonic and postnatal neural tissues and functions to prevent neuronal cell death. Proc Natl Acad Sci USA. 1995;92:4304–4308. doi: 10.1073/pnas.92.10.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Görlich D, Draft R, Kostka S, Vogel F, Hartmann E, Laskey R A, Mattaj I W, Izaurraide E. Importin provides a link between nuclear protein import and U snRNA export. Cell. 1996;87:21–32. doi: 10.1016/s0092-8674(00)81319-7. [DOI] [PubMed] [Google Scholar]
- 21.Görlich D, Mattaj I. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 22.Görlich D, Prehn S, Laskey R A, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 23.Görlich D, Vogel F, Mills A D, Hartmann E, Laskey R A. Distinct functions for the two importin subunits in nuclear protein import. Nature. 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- 24.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracyclin-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gottschalk A R, Boise L H, Oltvai Z N, Accavitti M A, Korsmeyer S J, Quintáns J, Thompson C B. The ability of bcl-xL and bcl-2 to prevent apoptosis can be differentially regulated. Cell Death Differ. 1996;3:113–118. [PubMed] [Google Scholar]
- 26.Hilfinger J M, Clark N, Smith M, Robinson K, Markovitz D M. Differential regulation of the human immunodeficiency virus type 2 enhancer in monocytes at various stages of differentiation. J Virol. 1993;67:4448–4453. doi: 10.1128/jvi.67.7.4448-4453.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iovine M K, Watkins J L, Wente S R. The GLFG repetitive region of the nucleoporin Nup116p interacts with Kap95p, an essential yeast nuclear import factor. J Cell Biol. 1995;131:1699–1713. doi: 10.1083/jcb.131.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Izaurralde E, Mattaj I W. RNA export. Cell. 1995;81:153–159. doi: 10.1016/0092-8674(95)90323-2. [DOI] [PubMed] [Google Scholar]
- 29.Kelly K, Siebenlist U. The regulation and expression of c-myc in normal and malignant cells. Annu Rev Immunol. 1986;4:317–338. doi: 10.1146/annurev.iy.04.040186.001533. [DOI] [PubMed] [Google Scholar]
- 30.Kraemer D, Wozniak R W, Blobel G, Radu A. The human can protein, a putative oncogene product associated with myeloid leukemogenesis, is a nuclear pore complex protein that faces the cytoplasm. Proc Natl Acad Sci USA. 1994;91:1519–1523. doi: 10.1073/pnas.91.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kraemer D M, Strambio-de-Castillia C, Blobel G, Rout M P. The essential yeast nucleoporin NUP159 is located on the cytoplasmic side of the nuclear pore complex and serves in karyopherin-mediated binding of transport substrate. J Biol Chem. 1995;270:19017–19021. doi: 10.1074/jbc.270.32.19017. [DOI] [PubMed] [Google Scholar]
- 32.Martin S J, Lennon S V, Bonham A M, Cotter T G. Induction of apoptosis (programmed cell death) in human leukemic HL-60 cells by inhibition of RNA or protein synthesis. J Immunol. 1990;145:1859–1867. [PubMed] [Google Scholar]
- 33.Melchior F, Paschal B, Evans J, Gerace L. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. . (Erratum, 124:217, 1994.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore M S, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- 35.Moroianu J, Blobel G, Radu A. Previously identified protein of uncertain function is karyopherin α and together with karyopherin β docks import substrate at nuclear pore complexes. Proc Natl Acad Sci USA. 1995;92:2008–2011. doi: 10.1073/pnas.92.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moroianu J, Hijikata M, Blobel G, Radu A. Mammalian karyopherin α1β and α2β heterodimers: α1 or α2 subunit binds nuclear localization signal and β subunit interacts with peptide repeat-containing nucleoporins. Proc Natl Acad Sci USA. 1995;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicoletti I, Migliorati G, Pagliacci M C, Grignani F, Riccardi C. A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J Immunol Methods. 1991;139:271–279. doi: 10.1016/0022-1759(91)90198-o. [DOI] [PubMed] [Google Scholar]
- 38.Panté N, Aebi U. Toward the molecular details of the nuclear pore complex. J Struct Biol. 1994;113:179–189. doi: 10.1006/jsbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- 39.Radu A, Blobel G, Moore M S. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci USA. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Radu A, Moore M S, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 41.Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 42.Rout M P, Wente S R. Pores for thought: nuclear pore complex proteins. Trends Biochem Sci. 1994;4:357–363. doi: 10.1016/0962-8924(94)90085-x. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 44.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 45.Shockett P, Difilipantonio M, Hellman N, Schatz D. A modified tetracyclin regulated system provides autoregulatory inducible gene expression in cultured cells and transgenic mice. Proc Natl Acad Sci USA. 1995;92:6522–6526. doi: 10.1073/pnas.92.14.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 47.Sundström C, Nilsson K. Establishment and characterization of a human histiocytic lymphoma cell line (U-937) Int J Cancer. 1976;171:565–577. doi: 10.1002/ijc.2910170504. [DOI] [PubMed] [Google Scholar]
- 48.Testa U, Masciulli R, Tritarelli E, Pustorino R, Mariani G, Martucci R, Barberi T, Camagna A, Valtieri M, Peschle C. Transforming growth factor-beta potentiates vitamin D3-induced terminal monocytic differentiation of human leukemic cell lines. J Immunol. 1993;150:2418–2430. [PubMed] [Google Scholar]
- 49.van Deursen J, Boer J, Kasper L, Grosveld G. G2 arrest and impaired nucleocytoplasmic transport in mouse embryos lacking the proto-oncogene CAN/Nup214. EMBO J. 1996;15:5574–5583. [PMC free article] [PubMed] [Google Scholar]
- 50.van Deursen J, Lovell-Badge R, Oerlemans F, Schepens J, Wieringa B. Modulation of gene activity by consecutive gene targeting of one creatine kinase M allele in mouse embryonic stem cells. Nucleic Acids Res. 1991;19:2637–2643. doi: 10.1093/nar/19.10.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vignali D A, Carson R T, Chang B, Mittler R S, Strominger J L. The two membrane proximal domains of CD4 interact with the T cell receptor. J Exp Med. 1996;183:2097–2107. doi: 10.1084/jem.183.5.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Von Lindern M, Fornerod M, van Baal S, Jaeglé M, de Wit T, Buijs A, Grosveld G. The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, and the expression of a chimeric, leukemia-specific dek-can mRNA. Mol Cell Biol. 1992;12:1687–1697. doi: 10.1128/mcb.12.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waters C, Littlewood T, Hancock D, Moore J, Evan G. c-myc protein expression in untransformed fibroblasts. Oncogene. 1991;6:101–109. [PubMed] [Google Scholar]
- 54.Weis K, Mattaj I W, Lamond A I. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science. 1995;268:1049–1053. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]
- 55.Wente S R, Blobel G. A temperature-sensative NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J Cell Biol. 1993;123:275–284. doi: 10.1083/jcb.123.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wozniak R W, Blobel G, Rout M P. POM152 is an integral protein of the pore membrane domain of the yeast nuclear envelope. J Cell Biol. 1994;125:31–42. doi: 10.1083/jcb.125.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]