Abstract
Using a two-step procedure involving insertional gene targeting and recombinase-mediated cassette exchange in ES cells, we have produced two lines of transgenic mice expressing a dominant-negative latency-associated myostatin propeptide under control of the myosin light chain 1F promoter and 1/3 enhancer from the TSPY locus on the Y chromosome. Males of the corresponding lines are characterized by a 5-20% increase in skeletal muscle mass. This experiment demonstrates the feasibility of a more efficient cattle production system combining superior beef production abilities for bulls and dairy abilities for cows.
Keywords: gene targeting, myostatin, Y chromosome
Intensive breeding programs implemented over the last 50 years have created cattle breeds that are highly specialized in either milk production (e.g., Holstein-Friesian and Jersey) or meat production (e.g., Angus, Hereford, Charolais, Piedmontese, and Belgian Blue). Physiological antagonisms have indeed precluded combining superior abilities for both milk and meat production in the same animal. Despite its effectiveness, the resulting production system can be considered suboptimal because of poor carcass and milk yield of dairy and beef cattle, respectively.
A more efficient alternative can be envisaged based on a specialization by sex within the same population: a breed in which cows would be of dairy type and bulls would be of beef type. To achieve this goal we propose to use genetic engineering to target trans-inactivators of myostatin (MSTN) on the Y chromosome. By doing so, males are predicted to exhibit a muscular hypertrophy akin to “double-muscling,” which, in cattle, results from naturally occurring MSTN loss-of-function mutations (1-4), whereas females will be nontransgenic and fully express their dairy potential.
To prove the feasibility of this concept, we herein describe the generation of two transgenic lines of mice in which only the males express a MSTN trans-inactivator in skeletal muscle and consequently show an increase in the weight of individuals muscles ranging from 5% to 20%.
Experimental Design
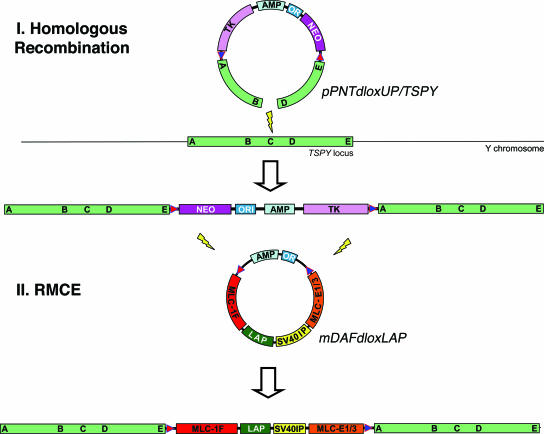
Defining an experimental strategy to generate this murine model required (i) the choice of a MSTN trans-inactivator, (ii) the choice of a targeting site on the Y chromosome, and (iii) the choice of a suitable targeting strategy. We selected the expression of the murine MSTN “latency-associated peptide” (LAP) or propeptide as a dominant-negative means to repress endogenous MSTN activity (5-9). The testis-specific protein Y-encoded (TSPY) pseudogene was chosen as a targeting site on the murine Y chromosome. Contrary to other mammalian species where TSPY genes are multicopy, the mouse TSPY is single-copy and nonfunctional despite being transcribed (10, 11). As a consequence, the murine TSPY locus is predicted to be nonessential but transcriptionally competent. After Rohozinski et al. (12), we chose insertional targeting rather than the usual replacement strategy (which has never been successfully applied to the murine Y chromosome) to insert a cassette containing a positive (neo) and a negative (HSV-tk) selectable marker flanked by heterologous lox sites into the murine Y chromosome. In a second stage, the marker cassette would then be exchanged through cremediated recombination with a cassette coding for a MSTN trans-repressor under the dependence of a strong skeletal muscle-specific promoter. Fig. 1 summarizes the main features of the proposed strategy.
Fig. 1.
Schematic representation of the targeting strategy. In a first step, an insertional targeting vector comprising a gapped homology arm (A-B/D-E) corresponding to segments of the TSPY locus, heterologous loxP sites (arrows), a positive (NEO) and negative (TK) selectable marker, an ampicilin resistance gene (AMP), and bacterial origin of replication (ORI) is targeted on the Y chromosome by homologous recombination. In a second step, the inserted vector sequences are exchanged by RMCE for a cassette coding for the murine MSTN propeptide (LAP) under the dependence of the rat myosin light chain 1F promoter (MLC-1F) and enhancer (MLC-1/3E), appended to the SV40 small tumor antigen intron and polyadenylation signal (SV40IP).
Materials and Methods
Construction of the Insertional Targeting Vectors pPNTdloxUP and pPNTdloxTSPY. Two adaptors containing (i) a loxP and a SalI site and (ii) a lox2272, a PacI, and a BamHI site were ligated through their shared AflII sticky ends into a 99-bp fragment with XbaI and EcoRI overhangs, which was directionally cloned in the corresponding restriction sites of the pPNT vector to yield the pPNTdlox vector. Homology arms corresponding to nt 31165-39425 [upstream (UP)] and nt 50690-57331 (TSPY) of sequence AC069015 (encompassing the murine TSPY gene) were amplified by using the Expand Long Template PCR system (Roche, Basel, Switzerland) from R1 genomic DNA with primers containing SalI and BamHI sites, respectively, at their extremities. This approach allowed convenient cloning of the PCR products in the pPNTdlox vector to yield the pPNTdloxUP and pPNTdloxTSPY plasmids. Approximately 300-bp gaps were introduced by digestion with SacI (pPNTdloxUP) and BbvcI (pPNTdloxTSPY) followed by religation. An adaptor containing unique PmeI and AscI sites was introduced in the SacI site of pPNTdloxUP. The gapped pPNTdloxUP and pPNTdloxTSPY vectors were completely sequenced before use.
Gene Targeting in R1 ES Cells and Identification of Homologous Recombinants. Gene targeting was performed in R1 cells by using standard procedures described in refs. 13 and 14. Briefly, the targeting vectors were linearized with either AscI (pPNTdloxUP) or BbvcI (pPNTdloxTSPY), and 20 μg of the resulting products was used to electroporate 107 R1 ES cells with the addition of 25 μg/ml spermidine in the electroporation medium. Positive selection was performed by using G418 (Invitrogen) at 300 μg/ml. After picking and replica plating, colonies having undergone the expected targeting event were identified by performing PCRs with primers located in the gap and selectable markers (neo and HSV-tk). At least two PCRs were performed for each construct, exploring the right and left boundaries of the integration site, respectively. The PCRs were carried out by using the Expand Long Template PCR system. Colonies that appeared positive after PCR screening were further analyzed by Southern blotting. DNA (7.5 μg) was digested with NdeI (pPNTdloxUP) or KpnI (pPNTdloxTSPY) and electrophoresed in a 1% agorose gel before blotting on a nylon membrane by using a standard alkali transfer procedure. The filter was hybridized to a 1,154-bp tk probe excised by BamHI-XbaI digestion from the pcDNA3hsvTK vector (courtesy of F. Princen, University of Liège) according to the manufacturer's instructions (Amersham Pharmacia). Finally, colonies positive by Southern blotting were analyzed by FISH. ES cell metaphase spreads were obtained by following standard procedures described in ref. 15. The slides were treated with ribonuclease A and pepsin and fixed with 4% paraformaldehyde. Hybridization was performed at 37°C in 2× SSC buffer (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7) containing 50% formamide and 12.5% dextran sulfate. The probes were the fluorescein-labeled pPNT plasmid and a Cy3-labeled murine Y chromosome painting probe (Cambio, Cambridge, U.K.). The fluorescein signal was amplified by using the Tyramide Signal Amplification System (NEN/PerkinElmer), and the slides were counterstained with DAPI before microscopic examination.
Construction of the mDAFdloxLAP Vector. Adaptors containing loxP and lox2272 sites were cloned in the HindIII and EagI restriction sites located, respectively, upstream of the MLC1F promoter (myosin light chain, MLC) and downstream of MLC1/3E enhancer in the mDAF vector (16). Proper orientation of the lox sites for compatibility with the pPNTdlox vector was verified by sequencing. The MSTN LAP-encoding sequence was obtained by RT-PCR from total RNA extracted from skeletal muscle of 2-month-old mice by using TRIzol (Invitrogen). First-strand cDNA synthesis was carried out in a reaction volume of 20 μl starting from 2 μg of total RNA by using an oligo(dT)16 as a primer and PowerScript reverse transcriptase (BD Biosciences/Clontech). RT-PCR was performed by using MSTN LAP-specific primers, including either an EcoRI tail or a SmaI tail. The RT-PCR product was digested with EcoRI and SmaI and cloned in the corresponding sites of the mDAFdlox vector. The completed mDAFdloxLAP vector was entirely sequenced before use.
Recombinase-Mediated Cassette Exchange (RMCE). Three million cells of the RI-UP-neotk and RI-TSPY-neotk ES cell clones were coelectroporated with 25 μg of mDAFdloxLAP and 50 μg of pMCcre plasmid in a buffer containing 25 μg/ml spermidine. Gancyclovir-resistant clones (2 μM) were picked and replica-plated. Screening for the expected RMCE event was achieved by PCR with primers located in the UP and TSPY homology arms, the MLC1F promoter (PCR “A”), and MLC1/3E enhancer (PCR “B”). Clones that were positive by PCR were further analyzed by Southern blotting with HindIII restriction enzyme and the MSTN LAP as a probe. The MSTN LAP probe was obtained by PCR amplification of a 850-bp fragment from mDAFdloxLAP.
Generation and Identification of Transgenic Mice. C57BL/6J blastocysts (3.5 days old) were harvested and microinjected with targeted ES cells as described in ref. 15. Uterine transfer was performed the same day in CD1 pseudopregnant mothers by using standard procedures described in ref. 15. Individuals carrying the transgene were identified by using a multiplex PCR assay, allowing for the simultaneous amplification of an endogenous MSTN exon 1 fragment (230 bp) and a transgene-specific fragment (450 bp) spanning the junction between the MLC1F promoter and MSTN LAP sequence. The ΔMCHR1 allele in the BC-CONT line (control, CONT) was detected by using a multiplex PCR generating a 450- and 700-bp fragment for the knockout and wild-type alleles, respectively.
Weight Measurements. Live weight was recorded at 4, 5, 6, 7, 8, 9, and 10 weeks of age. Animals were killed at 10 weeks and dissected. We determined the weight of the carcass (skinned body minus head, tail, all internal organs and associated fat and connective tissue), “leg weight” (skinned leg cut at knee and tarsus level), and weights of the dissected pectoralis, triceps brachialis, and quadriceps femoris muscles.
Analysis of Transgene Expression. Total RNA was extracted from muscle and nonmuscle tissues by using TRIzol (Invitrogen). Twenty micrograms of total RNA was denatured in formaldehyde load dye (Ambion, Austin, TX), electrophoresed on a Reliant Gel System (Cambrex, Rockland, ME) in NorthernMax Mops gel running buffer (Ambion), and blotted on a positively charged nylon membrane (Amersham Pharmacia) by capillary transfer with 10 mM NaOH in 5× SSC buffer. The membrane was then hybridized overnight with 100 ng of a simian virus 40 (SV40) probe in ULTRAhyb hybridization buffer (Ambion) and washed in 0.1× SSC and 0.1% SDS-containing buffer. The SV40 probe was PCR amplified from the mDAFdloxLAP construct and labeled with 32P dCTP (Amersham Pharmacia) by random primed labeling (Invitrogen). Membranes were exposed on Hyperfilm (Amersham Pharmacia).
Morphometric Analyses. Ten-week-old mice were killed, and their quadriceps femoris were dissected and fixed in 4% buffered formaldehyde. Muscles were cut transversally at the midpoint and embedded in paraffin. Four-micrometer-wide transverse sections were made from the widest part of the muscle and stained with antibodies against collagen IV to facilitate visualization of individual fibers. Briefly, antigen was demasked by pepsin treatment for 60 min, and slides were incubated two times (1:5,000 and 1:500) with anticollagen IV rabbit polyclonal antibody AB748 (Chemicon, Temecula, CA). For each muscle section, 10 photographs were taken at ×40 magnification, these photographs being evenly dispersed throughout the section and consistently positioned across individuals. All of the entire myofibers within the microscopic field were measured by using analysis 3.2 image analysis software (Soft Imaging System, Münster, Germany), and fiber diameter was considered to be the diameter of the largest circle that could be placed within each myofiber.
Statistical Analyses. Growth curves were analyzed with the proc mixed procedure of the sas package (SAS Institute, Cary, NC). We used a mixed model including sex, genotype, sex by genotype interaction as fixed effects, and a random individual effect accounting for the covariances between repeated measurements (17). Relative muscle weights as well as myofiber diameter were analyzed separately for each sex by using the proc glm procedure of the sas package and a model including genotype as a fixed effect.
Approval by Ethics Committees. The experimental design was approved by the ethics committee of the Faculty of Veterinary Medicine, University of Liège, and conducted under license no. LA1610475 delivered by the Belgian Ministère des Classes Moyennes et de l'Agriculture.
Results
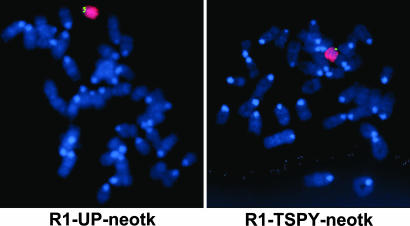
Successful Homologous Recombination on the Murine Y Chromosome By Using Insertional Targeting Vectors. We generated two distinct insertional targeting vectors by cloning (i) an 8.26-kb homology arm located 13.55 kb upstream of the TSPY pseudogene (pPNTdloxUP) and (ii) a 6.64-kb homology arm spanning the TSPY pseudogene (pPNTdloxTSPY), flanked by heterologous lox sites (loxP and lox 2272; refs. 18 and 19), in the pPNT vector providing the neo and HSV-tk cassettes (20). The homology arms were obtained by long-template PCR from genomic DNA extracted from R1 ES cells. To enhance targeting efficiency and facilitate screening, 376- and 314-bp gaps (leaving unique AscI and BbvcI restriction sites for linearization before electroporation) were generated in pPNTdloxUP and pPNTdloxTSPY, respectively. Gene targeting was performed in R1 ES cells by using standard procedures described in refs. 13 and 14. G418-resistant colonies (677 for pPNTdloxUP and 592 for pPNTdloxTSPY) were screened for successful insertion by using (i) PCR assays based on the use of vector-specific primers combined with gap-specific primers, followed by (ii) Southern blotting with a HSV-tk-specific probe and restriction enzymes cutting in the gap (pPNTdloxUP and pPNTdloxTSPY) and vector (pPNTdloxUP) and (iii) FISH by using a pPNT probe and a Y chromosome painting probe. For each construct, we were able to identify one properly targeted clone with euploid caryotype: RI-UP-neotk and RI-TSPY-neotk (Fig. 2; see also Fig. 5, which is published as supporting information on the PNAS web site).
Fig. 2.
Demonstration of the integration of the transgene on the Y chromosome for both the R1-UP-neotk and R1-TSPY-neotk clones. ES cell metaphase spreads were hybridized with a fluorescein-labeled transgene-specific pPNT probe (green) and a Cy3-labeled murine Y-specific painting probe (red), and counterstained with DAPI.
Integration of a MSTN Trans-Inactivator on the Murine Y Chromosome by Means of RMCE. The cDNA sequence coding for the murine MSTN LAP was obtained by RT-PCR from total skeletal muscle RNA and cloned into the mDAFdlox plasmid, properly placed under the dependence of the rat MLC1F promoter and 1/3 enhancer for expression in skeletal muscle. The mDAFdlox plasmid corresponds to the mDAF plasmid (16) in which we inserted a loxP and lox2272 sequence upstream of the MLC1F promoter and downstream of the MLC1/3 enhancer, respectively. Clones RI-UP-neotk and RI-TSPY-neotk were coelectroporated with the mDAFdloxLAP and pMCcre plasmids, the latter encoding the cre recombinase under the dependence of a tk promoter (21). Gancyclovir-resistant clones were screened for correct RMCE by (i) PCR assays with UP/TSPY- and MLC-specific primers followed by (ii) Southern blotting with a MSTN LAP probe. We identified 10 (RI-UP-LAP 1-10) and 4 (RI-TSPY-LAP 1-4) clones, respectively, having undergone proper RMCE (Fig. 6, which is published as supporting information on the PNAS web site).
Generation of Transgenic F1 Mice Expressing a Male-Specific MSTN Trans-Inactivator. Four of the R1-UP-LAP clones and all R1-TSPY-LAP clones were used for microinjection into recipient C57BL/6J blastocysts, followed by reimplantation in CD1 foster mothers (15). Thirteen chimeric males (of which six were 100% agouti) and one chimeric female were obtained from RI-UP-LAP clones, whereas a single, 100% agouti chimeric male was obtained from a RI-TSPY-LAP clone. The seven 100% agouti chimeric males were mated to C57BL/6J females to yield an F1 generation. We obtained 110 males and 90 females (P = 0.16) from three of the six RI-UP-LAP-derived males and 69 males and 47 females (P = 0.04) from the unique RI-TSPY-LAP-derived chimera. As expected, all F1 males were shown by a PCR assay to carry the transgene, whereas none of the F1 females did (data not shown), thereby confirming the Y-specific integration and germ-line transmission of both UP-LAP and TSPY-LAP transgenes.
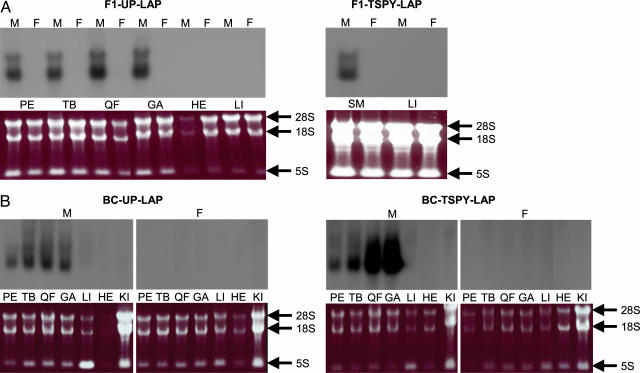
Transgene expression was assayed by Northern blotting with a SV40 probe and total RNA extracted from skeletal muscle, heart, and liver of a 13-week-old F1 male and female from each line. In both lines, transgene-specific transcripts were detected exclusively in male skeletal muscle (Fig. 3A). As expected, we found no evidence for the expression of the transgene either in the liver or in the heart.
Fig. 3.
Analysis of transgene expression. Assessment of transgene expression in the F1-UP-LAP and F1-TSPY-LAP (A) and BC-UP-LAP and BC-TSPY-LAP (B) transgenic lines by Northern blotting with an SV40 probe and total RNA extracted from pectoralis (PE), triceps briachialis (TB), quadriceps femoris (QF), gastrocnemius (GA), heart (HE), liver (LI) and kidney (KI) is shown. SM (skeletal muscle) corresponds to a mixture of RNA from PE, TB, QF, and GA. “M” and “F” corresponds to samples from males and females, respectively. Ethidium bromide-stained RNA gels before transfer allow for comparison of RNA quantities between lanes. 28S, 18S, and 5S correspond to the ribosomial RNAs.
Generation of Transgenic “BC” Mice to Analyze the Phenotypic Effect of the Male-Specific MSTN Trans-Inactivator. To measure the phenotypic effect of the Y-specific transgene, we mated four F1-UP-LAP males and two F1-TSPY-LAP males with 129/SV females to produce backcross (BC) animals. To generate a control population (BC-CONT), we crossed three nontransgenic males, derived from [(ΔMCHR1)R1 > C57BL/6J] chimeric males (22) mated to C57BL/6J females, with 129/SV females. We produced 184 BC-UP-LAPs (87 males and 97 females, P = 0.46), 218 BC-TSPY-LAPs (114 males and 104 females, P = 0.50), and 154 BC-CONTs (60 males and 94 females, P = 0.006).
Transgene expression in the BC animals was assayed in skeletal muscle (pectoralis, triceps brachialis, quadriceps femoris, and gastrocnemius), heart, liver, and kidney of a 13-week-old male and female for both BC-TSPY-LAP and BC-UP-LAP lines. As expected, transgene-specific transcripts were detected exclusively in skeletal muscle of male BC-TSPY-LAP and BC-UP-LAP animals. Transgene expression seemed stronger and characterized by an increasing rostro-caudal gradient in the BC-TSPY-LAP animal (Fig. 3B). Such an axial gradient has been reported for a chloramphenicol acetyltransferase transgene driven by the same regulatory elements (23).
Lack of Evidence for a Transgene-Specific Effect on Growth. BC animals were reared for 10 weeks during which they were weighed weekly. Analyzing the growth curves by using a mixed model including sex, age (weeks 4-10), genotype (UP-LAP, TSPY-LAP,or CONT), sex by genotype interaction, and random individual effects indicated that transgene genotype (both UP-LAP and TSPY-LAP) had a significant (P = 0.0004) positive effect on weight; however, the effect did not differ significantly (P = 0.96) between males and females (Fig. 7, which is published as supporting information on the PNAS web site). This observation indicates that at least part of the effect on growth is independent of transgene expression, thus probably due to polygenic background effects. Indeed, and because R1 ES cells are of 129/Sv × 129cX/Sv genotype (24), the different BC lines could exhibit phenotypic differences due to variable proportions of 129/Sv and 129cX/Sv genes.
The Transgene Causes a Male-Specific Muscular Hypertrophy. To test for an effect of transgene expression on muscle mass, we killed all BC animals at 10 weeks of age and weighed the carcass, the leg (skinned leg cut at knee and tarsus level), and a series of individual muscles. To correct for the differences in live weight observed between lines and individuals (see above), carcass, leg, and muscle weights were divided by live weight at slaughter. When analyzing males, both transgenic lines (BC-UP-LAP and BC-TSPY-LAP) exhibited highly significant increases in relative carcass, leg, and individual muscle weights when compared with the control line (BC-CONT). Carcass and leg weights were increased by ≈5%, triceps briachialis weight was increased by ≈10%, and quadriceps femoris weight was increased between ≈15% (BC-UP-LAP) and 20% (BC-TSPY-LAP) (Table 1). When comparing females, on the contrary, there was no evidence at all for an effect of genotype (UP-LAP, TSPY-LAP, or CONT) on normalized carcass, leg, or individual muscle weights (Table 1). These results strongly suggest that the effects observed in the males are caused by the transgenes. The stronger effect in quadriceps femoris (hind legs) when compared with triceps brachialis (front legs) and pectoralis in both lines supports the occurrence of a rostro-caudal gradient and corroborates the Northern blot results in the BC-TSPY-LAP line. Weights of the triceps brachialis and quadriceps femoris were slightly higher in the BC-TSPY-LAP than in the BC-UP-LAP males (P = 0.06 and 0.03, respectively), suggesting that the transgene effect is larger in the former, again corroborating the findings of the Northern blots.
Table 1. Effect of the transgene on body composition and muscle weight.
| Least square means of body part and muscle weights relative to live weight ± SE, % (n)
|
Statistical significance of genotype effect and respective contrasts (effect, %)
|
||||||
|---|---|---|---|---|---|---|---|
| Body part or muscle | BC-UP-LAP | BC-TSPY-LAP | BC-CONT | Genotype effect | UP-CONT | TSPY-CONT | UP-TSPY |
| Males | |||||||
| Carcass | 41.19 ± 0.27 (24) | 40.57 ± 0.20 (44) | 39.28 ± 0.27 (25) | <0.0001 | <0.0001 (4.9) | 0.0002 (3.3) | 0.0703 (1.5) |
| Leg | 1.54 ± 0.02 (24) | 1.51 ± 0.01 (45) | 1.45 ± 0.02 (25) | <0.0004 | <0.0001 (6.2) | 0.0023 (4.1) | 0.1482 (2.0) |
| Quadriceps f. | 0.74 ± 0.01 (57) | 0.77 ± 0.01 (67) | 0.64 ± 0.01 (40) | <0.0001 | <0.0001 (15.6) | <0.0001 (20.3) | 0.0266 (−3.9) |
| Triceps b. | 0.44 ± 0.01 (57) | 0.45 ± 0.01 (68) | 0.40 ± 0.01 (35) | <0.0001 | <0.0001 (10.0) | <0.0001 (12.5) | 0.0575 (−2.2) |
| Pectoralis | 0.97 ± 0.02 (24) | 0.97 ± 0.01 (45) | 0.94 ± 0.02 (26) | 0.4149 | 0.2822 (3.2) | 0.2159 (3.2) | 0.9976 (0.0) |
| Females | |||||||
| Carcass | 39.07 ± 0.30 (27) | 39.24 ± 0.29 (29) | 38.78 ± 0.31 (25) | 0.5667 | 0.5148 (0.7) | 0.2905 (1.2) | 0.6852 (−0.4) |
| Leg | 1.47 ± 0.02 (27) | 1.46 ± 0.02 (29) | 1.44 ± 0.02 (27) | 0.3305 | 0.1882 (2.1) | 0.2064 (1.4) | 0.939 (0.7) |
| Quadriceps f. | 0.63 ± 0.01 (66) | 0.64 ± 0.01 (55) | 0.63 ± 0.01 (55) | 0.6721 | 0.4791 (0.0) | 0.4064 (1.6) | 0.8731 (−1.6) |
| Triceps b. | 0.40 ± 0.01 (65) | 0.39 ± 0.01 (55) | 0.40 ± 0.01 (52) | 0.4572 | 0.4761 (0.0) | 0.2120 (−2.5) | 0.5510 (2.6) |
| Pectoralis | 0.81 ± 0.01 (27) | 0.80 ± 0.01 (29) | 0.78 ± 0.01 (25) | 0.2193 | 0.0835 (3.8) | 0.2981 (2.6) | 0.4552 (1.2) |
The increase in weight observed for UP-LAP and TSPY-LAP females (Fig. 7) thus likely reflects a proportionate increase in weight of all organs, whereas that of UP-LAP and TSPY-LAP males involves an additional transgene-specific effect on muscle mass.
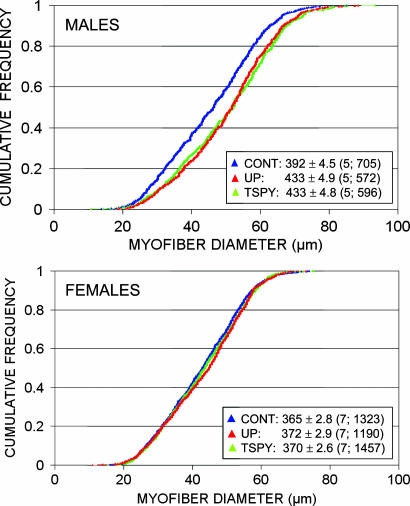
The Male-Specific Muscular Hypertrophy Is Due to an Increase in Myofiber Diameter. Transgenic expression of the MSTN propeptide has been shown to cause an increase in myofiber diameter (5, 6). To test whether a similar myofiber hypertrophy would have been induced by the transgene in BC-UP-LAP and BC-TSPY-LAP males, we performed histological examinations of transverse sections of the quadriceps femoris. We analyzed five 10-week-old males and seven females for each of the three lines (BC-UP-LAP, BC-TSPY-LAP, and BC-CONT). To ensure representativeness, we selected animals with a weight at slaughter within 0.5 g of the mean of their sex-genotype class. We determined the diameter of all myofibers within 10 consistently positioned ×40 microscopic fields for an average of 158 myofibers per individual. Fig. 4 shows the cumulative frequency distribution of myofiber diameter in males and females sorted by genotypes. A highly significant increase in myofiber diameter is seen in both BC-UP-LAP and BC-TSPY-LAP males but not in their female counterparts. Compared with BC-CONT, average myofiber diameter was increased by 10.39% (P < 0.0001) and 10.46% (P < 0.0001) in BC-UP-LAP and BC-TSPY-LAP males, respectively. Comparable figures were 1.99% (NS) and 1.35% (NS) in females.
Fig. 4.
Cumulative frequency distribution of quadriceps femoris myofiber diameter in males and females of the BC-CONT (blue), BC-UP-LAP (red) and BC-TSPY-LAP (green) lines. Means and standard errors are given for each sex-genotype combination. Numbers in parentheses correspond to the number of analyzed individuals and total number of myofibers.
Discussion
This work demonstrates that it is feasible to engineer strains of mammals in which only males express a muscular hypertrophy as a result of the expression of trans-inactivators of the MSTN gene from a transgene integrated on the Y chromosome. Recent improvements in gene targeting and nuclear transfer (e.g., ref. 25) now also make this target realistic in livestock species including cattle. Progress in sequencing the bovine genome, including the Y chromosome, will greatly facilitate the selection of suitable Y-specific targeting sites in this species as well.
The effect on muscle mass observed in this work (between 5% and 20%), if achieved in livestock, would have a major economic value. The use of alternative more potent promoters and/or MSTN trans-inactivators, including catalytical small interfering RNAs, follistatin, or dominant-negative activin type II receptors, has the potential to cause stronger effects on muscle mass, thereby increasing the value of the proposed approach even more (e.g., refs. 5, 9, and 26).
In some cattle breeds [particularly the Belgian Blue breed (BBB)], “double muscling” is associated with a high incidence of dystocia, leading to the nearly systematic reliance on cesarean section in some countries. Quite obviously, this major drawback has limited the dissemination of the BBB to most countries, especially those relying on less intensive management systems. It is noteworthy that the high incidence of dystocia in BBB is due to (i) the extreme muscular hypertrophy characterizing BBB that results from the combined effect of loss-of-function mutation in the MSTN gene and additional “polygenic” effects and (ii) the extreme muscularity of the calf, but also of the cow, resulting in a narrowed pelvic channel. In the proposed scheme, calving ease is unlikely to be problematic because (i) the muscular hypertrophy will be less extreme than, for instance, in BBB, and (ii) the cows will be nontransgenic and, hence, of dairy type. In addition, one could envisage delaying expression of the MSTN trans-inactivators in order to obtain a postnatal expression of the muscular hypertrophy. Such delayed expression could be achieved by using promoters that are becoming active only in later stages of development or that are inducible through exogenous means. We have recently demonstrated the effectiveness of delayed MSTN invalidation in obtaining late-onset muscular hypertrophy by using cre-loxP-mediated conditional MSTN invalidation (27).
The availability of the RI-UP-neotk and RI-TSPY-neotk ES cell lines will allow the exploration of other applications requiring the integration of specific transgenes on the Y chromosome. Sperm sexing would be one such obvious application.
Supplementary Material
Acknowledgments
We thank Dominique Poncelet and Anne Cornet for their contribution in early phases of this project, Benoît Brouwers and Vincent Augenbron for expert technical assistance, and Carole Charlier for fruitful discussions. This work has been funded by grants from the Belgian Ministère de l'Agriculture et des Classes Moyennes and the Walloon Ministry of Agriculture.
Author contributions: D.P., L.G., and M.G. designed research; D.P. performed research; L.G., A.A., C.H., and H.D.S. contributed new reagents/analytic tools; D.P., F.F., and M.G. analyzed data; and D.P. and M.G. wrote the paper.
Abbreviations: BBB, Belgian Blue breed; BC, backcross; CONT, control; LAP, latency-associated peptide; MLC, myosin light chain; MSTN, myostatin; RMCE, recombinase-mediated cassette exchange; SV40, simian virus 40; TSPY, testis-specific protein Y-encoded; UP, upstream.
References
- 1.Grobet, L., Royo Martin, L. J., Poncelet, D., Pirottin, D., Brouwers, B., Riquet, J., Schoeberlein, A., Dunner, S., Ménissier, F., Massabanda, et al. (1997) Nat. Genet. 17, 71-74. [DOI] [PubMed] [Google Scholar]
- 2.Kambadur, R., Sharma, M., Smith, T. P. L. & Bass, J. J. (1997) Genome Res. 7, 910-916. [DOI] [PubMed] [Google Scholar]
- 3.McPherron, A. C. & Lee, S.-J. (1997) Proc. Natl. Acad. Sci. USA 94, 12457-12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grobet, L., Poncelet, D., Royo Martin, L. J., Brouwers, B., Pirottin, D., Michaux, C., Menissier, F., Zanotti, M., Dunner, S. & Georges, M. (1998) Mamm. Genome 9, 210-213. [DOI] [PubMed] [Google Scholar]
- 5.Lee, S.-J. & McPherron, A. C. (2001) Proc. Natl. Acad. Sci. USA 98, 9306-9311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang, J., Ratovitski, T., Brady, J. P., Solomon, M. B., Wells, K. D. & Wall, R. J. (2001) Mol. Reprod. Dev. 60, 351-361. [DOI] [PubMed] [Google Scholar]
- 7.Thies, S. R., Chen, T., Davies, M. V., Tomkinson, K. N., Pearson, A. A., Shakey, Q. A. & Wolfman, N. M. (2001) Growth Factors 18, 251-259. [DOI] [PubMed] [Google Scholar]
- 8.Hills, J. J., Davies, M. V., Pearson, A. A., Wang, J. H., Hewick, R. H., Wolfman, N. M. & Qiu, Y. (2002) J. Biol. Chem. 277, 40735-40741. [DOI] [PubMed] [Google Scholar]
- 9.Wolfman, N. M., McPherron, A. C., Pappano, W. N., Davies, M. V., Song, K., Tomkinson, K. N., Wright, J. F., Zhao, L., Sebald, S. M., Greenspan, D. S. & Lee, S.-J. (2003) Proc. Natl. Acad. Sci. USA 100, 15842-15846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mazeyrat, S. & Mitchell, M. J. (1998) Hum. Mol. Genet. 7, 557-562. [DOI] [PubMed] [Google Scholar]
- 11.Vogel, T., Boettger-Tong, H., Nanda, I., Dechend, F., Agulnik, A. I., Bishop, C. E., Schmid, M. & Schmidtke, J. (1998) Chromosome Res. 6, 35-40. [DOI] [PubMed] [Google Scholar]
- 12.Rohozinski, J., Agoulnik, A. I., Boettger-Tong, H. L. & Bishop, C. E. (2002) Genesis 32, 1-7. [DOI] [PubMed] [Google Scholar]
- 13.Nagy, A., Rossant, J., Nagy, R., Abramow-Newerly, W. & Roder, J. C. (1993) Proc. Natl. Acad. Sci. USA 90, 8424-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torres, R. M. & Kuhn, R. (1997) Laboratory Protocols for Conditional Gene Targeting (Oxford Univ. Press, New York).
- 15.Nagy, A., Gertsenstein, M., Vintersten, K. & Behringer, R. (2003) Manipulating the Mouse Embryo: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 3rd Ed.
- 16.Rosenthal, N., Kornhauser, J. M., Donoghue, M., Rosen, K. M. & Merlie, J. P. (1989) Proc. Natl. Acad. Sci. USA 86, 7780-7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Littell, R. C., Henry, P. R. & Ammerman, C. B. (1998) J. Anim. Sci. 76, 1216-1231. [DOI] [PubMed] [Google Scholar]
- 18.Lee, G. & Saito, I. (1998) Gene 216, 55-65. [DOI] [PubMed] [Google Scholar]
- 19.Kolb, A. F. (2001) Anal. Biochem. 290, 260-271. [DOI] [PubMed] [Google Scholar]
- 20.Tybulewicz, V. L., Crawford, C. E., Jackson, P. K., Bronson, R. T. & Mulligan, R. C. (1991) Cell 65, 1153-1163. [DOI] [PubMed] [Google Scholar]
- 21.Gu, H., Zou, Y. R. & Rajewsky, K. (1993) Cell 73, 1155-1164. [DOI] [PubMed] [Google Scholar]
- 22.Adamantidis, A., Thomas, E., Foidart, A., Tyhon, A., Coumans, B., Minet, A., Tirelli, E., Seutin, V., Grisar, T. & Lakaye, B. (2005) Eur. J. Neurosci., in press. [DOI] [PubMed]
- 23.Donoghue, M. J., Patton, B. L., Sanes, J. R., Merlie, J. P. (1992) Development (Cambridge, U.K.) 116, 1101-1112. [DOI] [PubMed] [Google Scholar]
- 24.Threadgill, D. W., Yee, D., Matin, A., Nadeau, J. H. & Magnuson, T. (1997) Mamm. Genome 8, 390-393. [DOI] [PubMed] [Google Scholar]
- 25.Kuroiwa, Y., Kasinathan, P., Matsushita, H., Sathiyaselan, J., Sullivan, E. J., Kakitani, M., Tomizuka, K., Ishida, I. & Robl, J. M. (2004) Nat. Genet. 36, 775-780. [DOI] [PubMed] [Google Scholar]
- 26.Dykxhoorn, D. M., Novina, C. D. & Sharp, P. A. (2003) Nat. Rev. Mol. Cell Biol. 4, 457-467. [DOI] [PubMed] [Google Scholar]
- 27.Grobet, L., Pirottin, D., Farnir, F., Poncelet, D., Royo, L. J., Brouwers, B., Christians, E., Desmecht, D., Coignoul, F., Kahn, R., et al. (2003) Genesis 35, 227-238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.