Abstract
The telomeres of most organisms consist of short repeated sequences that can be elongated by telomerase, a reverse transcriptase complex that contains its own RNA template for the synthesis of telomere repeats. In Saccharomyces cerevisiae, the RAP1 gene encodes the major telomere binding protein Rap1p. Here we use a quantitative telomere formation assay to demonstrate that Rap1p C termini can enhance telomere formation more than 30-fold when they are located at internal sites. This stimulation is distinct from protection from degradation. Enhancement of formation required the gene for telomerase RNA but not Sir1p, Sir2p, Sir3p, Sir4p, Tel1p, or the Rif1p binding site in the Rap1p C terminus. Our data suggest that Rap1p C termini enhance telomere formation by attracting or increasing the activity of telomerase near telomeres. Earlier work suggests that Rap1p molecules at the chromosome terminus inhibit the elongation of long telomeres by blocking the access of telomerase. Our results suggest a model where a balance between internal Rap1p increasing telomerase activity and Rap1p at the termini of long telomeres controlling telomerase access maintains telomeres at a constant length.
Telomeres are the nucleoprotein complexes that protect chromosome ends from degradation and allow their complete replication. In most organisms, telomere DNA consists of an array of short TG-rich repeats. Saccharomyces cerevisiae telomeres consist of a 250- to 400-bp array of the heterogeneous repeat TG1-3 (58). Work from several laboratories has shown that these repeats are all that is required for yeast telomere function in mitosis and meiosis (6, 39, 45, 55). The number of these TG1-3 repeats, or telomere length, can be reduced by removal of the RNA primer for DNA synthesis or by degradation of the C1-3A strand at the end of S phase. The TG1-3 repeats can be elongated by the action of telomerase, a reverse transcriptase that contains an internal RNA template from which it synthesizes new TG1-3 repeats (59). Each yeast telomere contains numerous binding sites within the TG1-3 repeats for the RAP1 protein (Rap1p) (5, 53). Rap1p is required for yeast viability and functions both as a transcriptional activator of many yeast ribosomal protein and glycolytic genes and as an important component of the transcriptional silencing machinery at the yeast silent mating type cassettes and at telomeres (46).
Rap1p can be divided into two domains, a large DNA binding domain (amino acids 361 to 596 [14]) and a large C-terminal domain (amino acids 600 to 827), which contains multiple subdomains involved in transcriptional activation, transcriptional silencing, and telomere length control (4, 11, 18, 32, 50) (see Fig. 1). The Rap1p C terminus binds directly to at least three other proteins: Sir3p, Rif1p, and Rif2p (12, 36, 57). The interaction with Sir3p is important for the formation of a heterochromatin complex, consisting of Sir2p, Sir3p, Sir4p, and Rap1p, that spreads into internal regions of the chromosome to silence the transcription of nearby genes, a phenomenon known as telomere position effect, or TPE (1, 13, 33, 43, 49). Elimination of SIR2, SIR3, or SIR4 function eliminates TPE (1) and the association of Sir proteins with telomeric heterochromatin (13, 49).
FIG. 1.
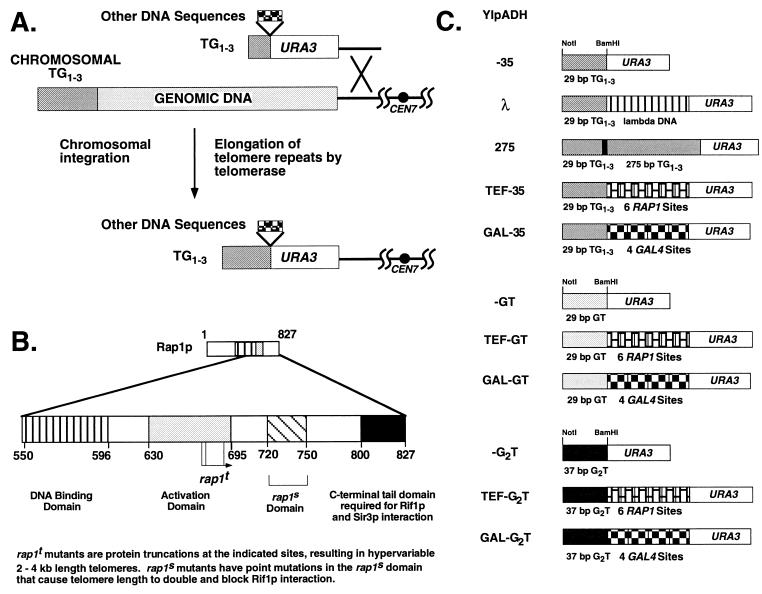
Transformation assays used to monitor telomere formation efficiency. (A) Replacement of the left telomere of chromosome VII with synthetic URA3 telomeres. Homologous recombination at the ADH4 locus replaces the terminal 17 kb of VIIL with a URA3 gene followed by different DNA inserts and 29 bp of TG1-3. Telomeres are formed when the short TG1-3 tract is elongated by de novo TG1-3 repeat addition mediated by telomerase (47) or by a recombination mechanism (see Fig. 4). The solid circle represents the centromere of chromosome VII. (B) C-terminal domain structure of the yeast telomere binding protein Rap1p based on data from several laboratories presented as in reference 27. The activation domain refers to activation of transcription; the rap1s domain refers to mutations that affect silencing at HMR and interaction with RIF1 (12); and the C-terminal domain refers to truncations that alter telomere length and telomeric silencing of established telomeres (27). (C) Synthetic telomeres used in this work. The ADH4 portion is not shown. Each telomere contains a BamHI site at the junction of the TG1-3, GT, or G2T repeats and the internal sequences.
The Rap1p C terminus also plays an important role in telomere length regulation. To date, the majority of studies have examined the effects of mutations in the Rap1p C terminus on the steady-state length of existing telomeres. Overproduction of the Rap1p C-terminal domain in vivo causes chromosomal telomeres to lengthen (4, 11, 44, 56), presumably because Rap1p interacting factors are titrated away from telomeres. Point mutations and deletion mutations in the Rap1p C terminus also cause lengthening of existing chromosomal telomeres (18, 19, 27, 50). Recent work with synthetic telomeres indicates that yeast cells measure telomere length by counting Rap1p C termini, because tethering Rap1p C termini adjacent to the terminal TG1-3 tract causes cells to maintain the terminal tract at a shorter equilibrium length (35). Experiments with the Rap1p homolog of Kluyveromyces lactis indicate that the C termini of Rap1p molecules bound to the very ends of the chromosome interact with proteins that inhibit telomere elongation, presumably by inhibiting the access of telomerase to the chromosome terminus (17). These data have led to the hypothesis that the Rap1p C terminus inhibits lengthening of established telomeres (9).
The role of Rap1p in telomere formation, the de novo addition of TG1-3 repeats onto DNA ends, has received less attention. Telomeres can be efficiently formed on linear plasmids and chromosomes in vivo when the DNA ends are capped by telomeric repeats (6, 31, 32, 41, 47, 53, 58). Previous work using TG1-3 sequences that do or do not contain Rap1p sites has suggested that Rap1p binding can increase the frequency of telomere formation on linear plasmids (32). However, since these Rap1p sites were at the ends of the TG1-3 repeats, these data could not distinguish between bound Rap1p protecting the TG1-3 sequences from exonucleolytic degradation and Rap1p actively stimulating telomere formation. Experiments with temperature-sensitive rap1 mutants grown at the highest temperature where growth is possible, where Rap1p function should be limiting, found that telomeres gradually shorten as cells grow. As with the linear-plasmid experiments, these results could not distinguish between Rap1p protecting the ends from degradation and Rap1p enhancing telomere lengthening (4, 32). Therefore, while the role of the Rap1p C terminus in limiting the elongation of existing telomeres is established, the exact role of Rap1p in telomere formation and elongation is not clear.
Here we demonstrate that the Rap1p C terminus can promote telomere formation by a mechanism that is distinct from protection from degradation. We constructed synthetic telomeres containing three different very short substrates for telomere formation (telomere seeds). Using a quantitative transformation assay, we found that internally tethered Rap1p molecules or a subset of Gal4p-Rap1p C-terminal fusions could greatly stimulate telomere formation independently of the telomere seed sequence. This enhancement of telomere formation requires most of the Rap1p C terminus, amino acids 630 to 827. This enhancement is dependent upon telomerase and is unaffected by mutations that alter the chromatin structure of steady-state telomeres. These data suggest a model in which internally located Rap1p C termini stimulate telomere formation and the elongation of short telomeres by increasing the activity of or attracting telomerase. At long telomeres, Rap1p and other molecules at the chromosome end block telomerase access, so that these two Rap1p functions form a system for maintaining telomeres at constant length.
MATERIALS AND METHODS
Strains.
Recombinant DNA manipulations were carried out in bacterial strains MC1066 (r− m+ pyrF::Tn5 trpC leuB) and JF1754 (r− m+ leuB hisB Met−). Yeast strains used were KR36-6L [MATa ade2-(1 or 101) ade8-18 ura3-52 trp1Δ1 leu2-ΔRC his3Δ] (44), the gal4Δ strain YM708 (MATα ade2-101 ura3-52 trp1Δ-901 his3-200 lys2-801 LEU2 canR gal4-542) (from Mark Johnston), and VPS106est1Δ::hisG (MATa ade2 ade3 ura3Δ trp1Δ leu2-3,112 lys2-801 can1 est1Δ::hisG) (from V. Schulz and K. Runge). The tel1Δ (YDM911) and TEL1 (YDM884) congenic strains were from Dwight Morrow and have been described elsewhere (37).
Plasmids.
The PvuII fragment of pADHUCAIV containing the ADH4-URA3-TG1-3 sequences (6) was ligated to the 3.3-kb EcoRV-SmaI fragment of YIp5 to form YIpADH. The TG1-3 repeats were removed from YIpADH by digestion with BamHI and NotI and were replaced with the oligonucleotide pair 35 (Table 1) to generate YIpADH-35. At high Rap1p concentrations in vitro, both Rap1p sites in this fragment can be bound simultaneously (15a). The single BamHI site between URA3 and the TG1-3 repeats was the site of insertion of six 35-bp TEF oligonucleotides (Table 1) in head-to-tail orientation, where the resulting BamHI site was closest to the NotI site, to form YIpADHTEF-35. A plasmid with the six TEF oligonucleotides in the opposite orientation was also constructed. Construction of the head-to-tail repeats will be detailed elsewhere (42a). Four GAL4 binding sites were also inserted by using four head-to-tail concatemers of the 22-bp GAL oligonucleotides (Table 1) to form YIpADHGAL-35. A 210-bp fragment of λ DNA was generated by using the PCR primers GGCCAGATCTAAAACAGGCTGAGCACGG and GGCCGGATCCGTTTCTGCGGGAAAGTGT to amplify bp 10101 to 10310 of lambda phage DNA, cleaving the product with BglII and BamHI, and cloning it into the BamHI site of YIpADH-35 to form YIpADHλ. The 275 bp of TG1-3 was isolated from pCT300 (275 bp of TG1-3 in pVZ-1, from K. Runge, R. Wellinger, J. Wright, and V. Zakian; the same sequence as Tel 270 [5]) by cleavage with HincII and SmaI and was cloned into the filled-in BamHI site of YIpADH-35 to yield YIpADH275. The 5-bp difference in length came about because we counted the 5 bp of TG sequences from the polylinker that are continuous with the telomeric sequence, while Gilson et al. (5) did not. The orientation of the 275 bp of TG1-3 repeats was the same as that of the telomere. Digestion of all of these YIpADH plasmids with SalI and NotI releases a SalI-ADH4-URA3-insert-TG1-3-NotI fragment which can replace the left telomere of chromosome VII (VIIL). To form the YIpADH-GT and YIpADH-G2T versions of YIpADH-35, YIpADHTEF-35, and YIpADHGAL-35, each of the latter three plasmids was cleaved with BamHI and NotI, and the GT and G2T oligonucleotides (Table 1) were ligated to these vectors. The correct transformants were identified by incorporation of the XhoI site. The sequences of all of these synthetic telomeres were verified by DNA sequencing. The leu2::URA3-insert-TG1-3 constructions were made by cutting YIpADH-35, YIpADHTEF-35, and YIpADHGAL-35 with HindIII and NotI, filling in the ends with the Klenow fragment of Escherichia coli DNA polymerase I, and cloning the URA3 fragment into the EcoRV site of LEU2 in pLEU2-0.9 (which contains the 0.9-kb EcoRI-SalI fragment of LEU2 in the same sites of pBR322). The yeast artificial chromosomes (YACs) were constructed by using YAC-ATA (from K. Runge and R. Wellinger; to be described elsewhere [43a]) and replacing one of two 275-bp telomeres with the SalI-NotI fragment from YIpADH-35 and YIpADHTEF-35. The 275-bp TG1-3 telomere and the test telomere are separated by 1.8 kb of HIS3 DNA for propagation in bacteria. The Gal4p-Rap1p fusions are expressed from the ADC1 promoter on a multicopy (2μm) HIS3 plasmid. The 653-800 and 653-817 fusions were constructed by introducing a stop codon followed by an XbaI site by PCR, cloning the fragments into the original vector, and verifying the products by sequencing. The remaining fusions were provided by D. Shore (11). The tlc1Δ::HIS3 allele replaces all of the TLC1 transcript (nucleotides 1 to 1306) with the HIS3 gene from pRS313 by the method of Morrow et al. (37), and the est1Δ::hisG allele replaces the internal EcoRI-to-NsiI fragment of the EST1 coding region with a 0.9-kb fragment from Salmonella typhimurium hisG. A plasmid containing the TLC1 gene was provided by M. Singer and D. Gottschling, and the EST1 gene was provided by V. Lundblad.
TABLE 1.
Oligonucleotides used for synthetic telomere constructions
| Name | Sequencesa |
|---|---|
| 35 | GATCCGGGTGTGTGGGTGTGTGGGTGTGGGTGTGC and GCCCACACACCCACACACCCACACCCACACGCCGG |
| TEF | GATCCCATTCATGTTGCACCCACACATTTAGACCA and GGTAAGTACAACGTGGGTGTGTAAATCTGGTCTAG |
| GAL | GATCCGGAGGACTGTCCTCCGA and GCCTCCTGACAGGAGGCTCTAG |
| GT | GATCCGTGTGTGTGTGTGTGTGTGTGTGTGTGTGCGGCCGCTCGAG and GCACACACACACACACACACACACACACACGCCGGCGAGCTCCCGG |
| G2T | GATCCGGTGGTGGTGGTGGTGGTGGTGGTGGTGGTGGTGGTGCGGCCGCTCGAG and GCCACCACCACCACCACCACCACCACCACCACCACCACGCCGGCGAGCTCCCGG |
Rap1p sites are boldfaced or underlined.
Quantitative telomere formation and integration.
Quantitative transformations were performed as described elsewhere (44) by transforming 5 μg of completely digested plasmid or 1 μg of the replicating plasmid YEp24 into yeast strain KR36-6L, KR36-6L bearing sir mutations (sir1::HIS3 and sir2::HIS3 insertion mutations and complete open reading frame deletions of sir3 and sir4, to be described elsewhere [43a]), or YM708 expressing different Gal4p-Rap1p fusions. To quantitate the rate of telomere formation, the number of transformants was determined, and in most cases 100 to 200 transformants of each type were transferred to a master plate lacking uracil, grown overnight at 30°C, replica plated to a plate containing uracil and histidine, grown overnight, and assayed for TPE by testing for growth on YC-Ura and 5-fluoroorotic acid plates (6). Subsequently, 5 to 10 transformants of each type predicted to have telomere formation events were checked by Southern analysis using the StuI-NsiI 3′ fragment of URA3 as a probe (see Fig. 2). For YIpADH275 and YIpADHTEF-35, 100% of all TPE-positive strains formed telomeres. For YIpADH-35, 91% of TPE-positive cells formed telomeres. For YIpADHλ, 50% of TPE-positive cells formed telomeres. Because the values in Tables 2 through 4 are based on TPE-positive cells, the fold increase in telomere formation is slightly underestimated. Integration of the YIpADH275 construction at internal loci (URA3) did not give detectable levels of TPE, i.e., did not score as a telomere formation event, in this assay (data not shown). In the cases of sir2, sir3, and sir4 mutants which do not exhibit TPE, 6 to 12 transformant colonies from each strain were analyzed by Southern hybridization to determine the fraction of telomeres formed. The fold enhancement of telomere formation is expressed as follows: number of telomeres formed by using the test telomere with an insert between URA3 and the TG tracts/number of telomeres formed by using the vector telomere bearing only URA3 and the TG tracts (e.g., number of YIpADHTEF-35 telomeres/number of YIpADH-35 telomeres). The fold differences from two to four single experiments were then averaged to obtain the final value. The range of these values is noted in each table and is ±20% or less in most cases. The number of YIpADH-35 transformants was usually greater than 40/5 μg of plasmid, and the number of YEp24 transformants was always greater than 103/μg. The numbers of YIpADH-GT and -G2T transformants were lower, usually 10 to 30/5 μg for the experiments reported in Table 4. For Table 2, the fraction of transformants that showed TPE in wild-type cells was 0.71 for YIpADH-35, 0.47 for YIpADHλ, 1.0 for YIpADHTEF-35, and 1.0 for YIpADH275. For Table 3, the fraction of transformants that showed TPE was 0.69 to 0.88 for YIpADH-35, 0.96 to 1.0 for YIpADHTEF-35, 0.64 to 0.76 for YIpADHGAL-35 in strains that do not show enhancement, and 0.90 to 1.0 for YIpADHGAL-35 in strains that do show enhancement, except for 0.75 for 653-800. Thus, the increased number of transformants parallels the increased number of telomere formation events. Cells bearing the Gal4p-Rap1p(618-827) and Gal4p-Rap1p(630-827) fusions grew poorly and gave fewer transformants per microgram for all plasmids tested. For Table 4 in strain YM708, the fraction of YIpADH-GT transformants that showed TPE was 0.54; for YIpADHTEF-GT it was 0.94, for YIpADH-G2T it was 0.38, and for YIpADHTEF-G2T it was 0.88. For Table 4 in strain YM708/Gal4-Rap1p(653-827), the fraction of YIpADH-GT transformants that showed TPE was 0.70; for YIpADHGAL-GT it was 0.99, for YIpADH-G2T it was 0.39, and for YIpADHGAL-G2T it was 0.41.
FIG. 2.
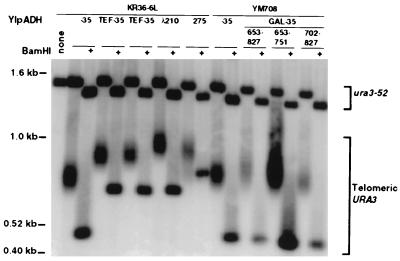
Transformants exhibiting TPE have formed telomeres from the terminal 29-bp TG1-3 tract. Representative genomic DNAs from cells transformed with the indicated telomere (Fig. 1C) that exhibited TPE were digested with either StuI or StuI plus BamHI and were analyzed by Southern hybridization using the 3′ StuI-NsiI fragment of URA3 as a probe. Telomeric URA3 is the URA3 telomere at VIIL, and ura3-52 is the chromosome V URA3 locus. The StuI-plus-BamHI digestion showed that the BamHI site was not deleted during telomere formation, indicating that de novo telomere addition was to the terminal 29-bp TG1-3 tract.
TABLE 2.
Internal Rap1p binding sites stimulate telomere formation
| Construct | Telomere | Fold increase in telomere formation efficiencya
|
||||||
|---|---|---|---|---|---|---|---|---|
| Wild type | sir1 | sir2 | sir3 | sir4 | TEL1 | tel1Δ | ||
| YIpADH-35 | URA3-TG1-3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| YIpADHTEF-35 | URA3-(Rap1p site)6-TG1-3 | 68 | 55 | 106 | 79 | 114 | 23 | 45 |
| YIpADHλ | URA3-(210 bp of λ DNA)-TG1-3 | 0.45 | 0.49 | 0.67 | 1.2 | 1.2 | 1.3 | 1.4 |
| YIpADH275 | URA3-(275 bp of TG1-3)-TG1-3 | 47 | ND | ND | ND | ND | ND | ND |
Quantitative transformation assays were performed with each construct containing the insert indicated, and the number of telomere formation events for each construct was normalized to that of the construct with no insert (YIpADH-35). Each value represents the average from three to four experiments. In each experiment, an aliquot of the same culture was individually transformed with each of the synthetic telomere-containing constructs shown. The number of telomere formation events was determined by assaying TPE (for the wild-type, sir1, TEL1, and tel1Δ strains) or by Southern analysis (for the sir2, sir3, and sir4 strains) as described in Materials and Methods and was normalized to the YIpADH-35 frequency. The sir mutations are in KR36-6L. TEL1 (YDM884) and tel1Δ::HIS3 (YDM911) are in the YPH274 background (37). The fold enhancement values shown vary by ±20% between experiments. ND, not done.
TABLE 4.
The level of telomere formation enhancement is independent of the terminal TG tracts used as substrates for telomere formation
| Construct | Telomere | Fold increase in telomere formationa with strain:
|
|
|---|---|---|---|
| YM708 | YM708/ Gal4-Rap1p (653-827) | ||
| YIpADH-35 | URA3-TG1-3 | 1 | 1 |
| YIpADHTEF-35 | URA3-(Rap1p site)6-TG1-3 | 29 | 36 |
| YIpADHGAL-35 | URA3-(Gal4p site)4-TG1-3 | 2 | 24 |
| YIpADHGAL | URA3-(Gal4p site)4 | ND | 0.09 |
| YIpADH-GTb | URA3-GT | 1 | 1 |
| YIpADHTEF-GT | URA3-(Rap1p site)6-GT | 27 | ND |
| YIpADHGAL-GT | URA3-(Gal4p site)4-GT | ND | 30 |
| YIpADH-G2Tc | URA3-G2T | 1 | 1 |
| YIpADHTEF-G2T | URA3-(Rap1p site)6-G2T | 20 | ND |
| YIpADHGAL-G2T | URA3-(Gal4p site)4-G2T | ND | 29 |
The fold increase is calculated with respect to the YIpADH plasmid that contains no insert between URA3 and the same terminal telomere seed (e.g., YIpADHTEF-GT is compared to YIpADH-GT) because different sequences seed telomere formation with different efficiencies (see the text and footnotes b and c). Values are averages from at least three experiments. YIpADHGAL was formed by releasing the telomere with a SalI-plus-BamHI digestion instead of SalI plus NotI to yield an adh4-URA3 fragment with no TG1-3 repeats (Fig. 1). Experiments were performed as for Tables 2 and 3. The TEF-35 and GAL-35 values are from Table 3. ND, not done.
The fold increase of telomere formation by YIpADH-GT compared to YIpADH-35 was 0.79 in YM708 and 0.56 in YM708/Gal4-Rap1p(653-827).
The fold enhancement of telomere formation by YIpADH-G2T compared to YIpADH-35 was 0.15 in YM708 and 0.19 in YM708/Gal4-Rap1p(653-827).
TABLE 3.
Internally bound Rap1p C termini enhance telomere formation
| Telomere | Fold increase in telomere formation over YIpADH-35a with the following Gal4-Rap1p fusion:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| None | 618-827 | 630-827 | 653-827 | 667-827 | 679-827 | 702-827 | 653-751 | 653-800 | 653-817 | rap1-12 653-827 | |
| URA3-(Rap1p site)6-TG1-3 | 29 | 34 | 36 | 36 | 32 | 43 | 42 | 37 | 44 | 40 | 35 |
| URA3-(Gal4p site)4-TG1-3 | 2 | 34 | 35 | 24 | 20 | 6.3 | 0.51 | 0.67 | 10 | 11 | 29 |
Yeast cells (YM708) expressing different Gal4p-Rap1p fusion proteins were transformed with YIpADH-35 (defined as 1), YIpADHTEF-35 [URA3-(Rap1p site)6-TG1-3], or YIpADHGAL-35 [URA3-(Gal4p site)4-TG1-3], and the enhancement of telomere formation was determined. The numbers refer to the amino acids of the 827-amino-acid Rap1p fused to Gal4p (see Materials and Methods). Results shown are averages from two to four separate experiments. The range of these values is ±20% or less. The rap1-12 C terminus contains two point mutations that abolish binding to the Rap1p interacting factor Rif1p (12).
Telomere formation on YACs was determined by transforming cells with 2 μg of DNA cut with NotI and SacI to expose the 275-bp and test telomeres (see Fig. 5). YAC transformations were performed in strain YM708 (wild type), YM708tlc1Δ::HIS3, or VPS106est1Δ::hisG. The tlc1Δ and est1Δ strains were generated by loss of a plasmid bearing the wild-type gene. YM708tlc1Δ was transformed after ∼75 divisions of growth after TLC1 plasmid loss, and VPS106 was transformed ∼35 divisions of growth after EST1 plasmid loss to allow for complete expression of the null phenotypes (23, 30). Several hundred transformants from multiple transformations (with the YAC containing the YIpADH-35 telomere, ∼2 × 104 transformants/μg for wild-type cells, ∼30/μg for est1Δ cells, and ∼80/μg for tlc1Δ cells) were obtained. Transformants were either all large colonies in wild-type cells or large colonies and small senescent colonies in tlc1Δ and est1Δ cells. The fraction of colonies in each size class was the same for YACs bearing the TEF-35 or YIpADH-35 telomeres. To determine the fraction of transformants in both size classes that formed telomeres, several (>10) transformants of each size class from each strain were mated to a TLC1 EST1 ura3 trp1 strain in order to form diploids. Diploids were then analyzed for linkage of URA3 and TRP1 and for TPE and by Southern blotting. The average fraction of transformants showing TPE (Trp+ FoaR colonies) for both size classes for the YIpADH-35 telomere was 1.0 for wild-type cells, 0.94 for tlc1Δ cells, and 0.78 for est1Δ cells, and that for the TEF-35 telomere was 1.0 for wild-type cells, 0.88 for tlc1Δ cells, and 0.88 for est1Δ cells. The fold differences in YAC telomere formation were determined by calculating the number of YAC telomere formation events for each size class and then calculating the ratio of all YAC–TEF-35 formation events to all YAC-35 formation events for each experiment. The results from two to four experiments were averaged to give the values in Table 5. Since telomere formation by the strand-copying mechanism is the same for TEF-35 and YIpADH-35 telomeres (Table 5, tlc1Δ cells), comparing the relative formation frequencies of the YIpADH-35 and TEF-35 at the left arm of chromosome VII (VIIL) and the YAC (Tables 3 and 5) indicates that the level of formation of the YIpADH-35 telomere by the strand-copying mechanism on the YAC is 2.8 times as high as that by telomerase at VIIL.
FIG. 5.
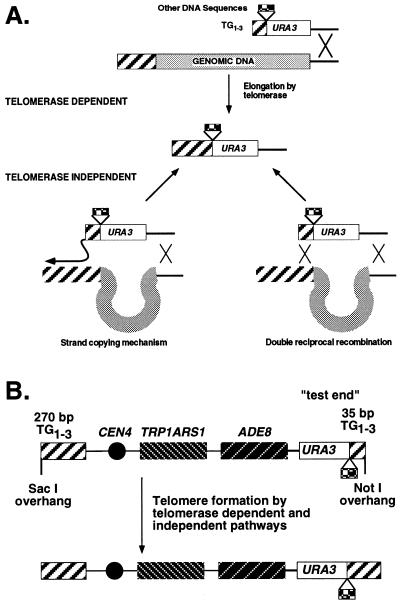
Telomere formation pathways on chromosomes and short YACs. (A) Telomeres can be formed on chromosomes by integration and subsequent elongation by telomerase or by two telomerase-independent pathways: a double reciprocal recombination event with the ADH4 locus and TG1-3 repeats (the TG1-3 repeats need not be cis to ADH4) or a nonreciprocal strand-copying event that has been shown to take place at the ends of linear plasmids during transformation (see the text). Both the telomerase-dependent and strand-copying pathways predict a single-stranded TG1-3 intermediate that is later made double stranded by DNA repair replication (60). (B) The 12-kb YAC vector for testing telomere formation in strains lacking telomerase RNA. One telomere contains 275 bp of TG1-3 sequences, while the other contains the YIpADH-35 and YIpADHTEF-35 telomeres (Fig. 1C). The close proximity of the two YAC telomeres allows the 275-bp TG1-3 repeats to serve as substrates for the test telomere in the strand-copying pathway of telomere formation (41).
TABLE 5.
Telomere formation enhancement by Rap1p requires telomerase RNA and Est1p
| YAC | Telomere | Fold increase in telomere formationa (YAC transformation efficiency)b by strain
|
||
|---|---|---|---|---|
| Wild type | tlc1Δ | est1Δ | ||
| YIpADH-35 | URA3-TG1-3 | 1 (0.050) | 1 (0.10) | 1 (0.033) |
| TEF-35 | URA3-(Rap1p site)6-TG1-3 | 10 (0.60) | 1.2 (0.13) | 2.8 (0.075) |
Telomere formation on a 12-kb YAC (Fig. 5) was assayed in wild-type (YM708) cells, cells lacking telomerase RNA (tlc1Δ), and cells lacking a protein that coimmunoprecipitates with telomerase activity (est1Δ) (see the text). To determine the frequency of telomere formation, several transformants from each strain were mated to a TLC1 EST1 ura3 trp1 strain to form diploids. Diploids were then analyzed for telomere formation by assaying linkage of URA3 and TRP1, by assaying TPE, and by Southern blotting. The reduction of enhancement in telomere formation efficiency by the TEF-35 telomeres indicates that telomerase RNA is required for this process and that Est1p enhances it. The standard deviation of these triplicate assays was ±20% of the mean, except for the tlc1Δ TEF-35 experiment, where it was ±40%.
The average number of YAC transformants per microgram of YAC DNA normalized to the number of YEp24 transformants per microgram of YEp24 DNA for each YAC strain combination (numbers were not corrected for telomere formation events). The standard deviations for these values were ±30%.
For quantitative integration at LEU2, identical amounts of cut DNA (5 μg) were used for the leu2::URA3-insert-TG1-3 transformations (see Fig. 4). Fifty to 100 Ura+ transformants were analyzed for the Leu− phenotype, indicating correct integration at LEU2 disrupting the gene. All transformations were carried out in strain YM708 with no plasmid or with the 2μm HIS3 plasmid expressing the Gal4-Rap1p fusion bearing amino acids 653 to 827.
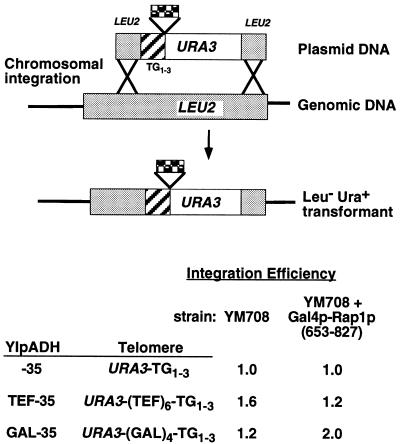
FIG. 4.
Rap1p sites do not increase the frequency of homologous integration at LEU2. To test if extra Rap1p sites increase the frequency of homologous integration during transformation, the HindIII-NotI fragments from YIpADH-35, YIpADHTEF-35, and YIpADHGAL-35 (see Table 2 and Materials and Methods) were used to disrupt LEU2 coding sequences in YM708 cells. The leu2::URA3-insert-TG1-3 constructs were used to transform LEU2 cells or LEU2 cells expressing a Gal4p-Rap1p fusion (containing Rap1p amino acids 653 to 827 [Table 3]). The number of Ura+ Leu− yeast transformants for each construct was then normalized to the YIpADH-35 amount, and the results from duplicate experiments are shown. The range of values is ±20%.
Other methods.
Yeast cell extracts were prepared by the method of Steiner et al. (48). Western analysis used a mouse monoclonal antibody (Ab) to the DNA binding domain of Gal4p (Santa Cruz), horseradish peroxidase-conjugated goat antimouse secondary Ab, and a NEN Renaissance Western Chemiluminescence kit. Protein concentrations were determined by Bradford assay (Bio-Rad). Southern blot analysis of yeast telomere length will be described in detail elsewhere (42).
RESULTS
Internal Rap1p sites enhance the frequency of telomere formation.
To assay telomere formation quantitatively, the left telomere of chromosome VII was truncated at the ADH4 locus by using homologous recombination to replace the distal sequences with the URA3 gene followed by TG1-3 sequences. Subsequent elongation of the TG1-3 repeats converts this construct to a functional yeast telomere (Fig. 1A) (6). We modified the previous construction of Gottschling et al. (6) by replacing the 125-bp fragment that contains 81 bp of TG1-3 and five Rap1p binding sites (5) with a synthetic 35-bp oligonucleotide containing two overlapping Rap1p sites (Table 1); this construct will be referred to as YIpADH-35 (Fig. 1C).
As described below, YIpADH-35 did not support efficient telomere formation upon transformation compared to other constructions. Telomere formation was defined as integration of the URA3 construction and de novo addition of TG1-3 sequences to the short 29-bp TG1-3 tract to form a chromosome end that the cell maintained over many generations. Telomere formation was monitored by transforming the same preparation of competent yeast cells with 5 μg of YIpADH-35 and other constructs and determining the number of Ura+ transformants that also exhibit TPE. The fold increase in the number of TPE-positive transformants relative to YIpADH-35 was calculated to measure the relative frequency of telomere formation. TPE is the repression of constitutive, but not induced, transcription of genes at telomeres but not at internal loci (6). For these URA3 telomeres, TPE allows these colonies to grow on medium lacking uracil and on medium containing uracil and 5-fluoroorotic acid, a toxin that kills yeast containing the URA3 gene product (6). Except where noted, 100 to 200 Ura+ transformants were picked and screened for TPE for each quantitative assay in this work. The number of transformants and telomere formation events increased as the amount of transforming DNA increased from 1 to 2 and 5 μg (Materials and Methods and data not shown). A subset of transformants that exhibited TPE was analyzed by Southern hybridization for elongation of the terminal TG1-3 repeats (representative transformants are shown in Fig. 2). More than 86% of all TPE-positive transformants examined had elongated the 29-bp TG1-3 repeats distal to URA3 (Materials and Methods). Thus, this system allows a replica plate assay for comparing the efficiency of telomere formation for different constructions.
Surprisingly, a construction containing 275 bp of TG1-3 (and at least 14 Rap1p sites [5]) in place of the 29 bp of TG1-3 (data not shown) formed telomeres 47-fold more efficiently than YIpADH-35. To determine whether the increase in telomere formation was due to either the longer TG1-3 repeats or the presence of Rap1p molecules, different inserts were placed into the BamHI site between the URA3 gene and the 29-bp TG1-3 tract. These inserts included a 210-bp fragment of lambda DNA (λ), a 275-bp fragment of TG1-3 followed by 50 bp of polylinker, and a 210-bp fragment containing six repeats of a 35-bp sequence from the TEF2 UAS, each of which contains one Rap1p binding site (TEF) (Table 1 and Fig. 1C). The number of telomere formation events was normalized to the YIpADH-35 control for each transformation, and the results of two to four experiments for each cell type were averaged. A subset of Ura+ transformants that showed TPE from each cell type in this work was always examined by Southern analysis, and in most cases these transformants contained new telomeres at the ADH4 locus that were formed by elongating the 29-bp TG1-3 sequences. Some exceptional YIpADH-275 telomeres in which the terminal 29-bp TG1-3 repeats and polylinker were deleted and telomere formation occurred by elongation of the 275-bp TG1-3 repeats were observed, but in all cases these transformants did contain a telomere distal to URA3. A detailed analysis of these exceptional transformants will be given elsewhere (42).
Constructions containing either 275 bp of internal TG1-3 sequences or six nontelomeric Rap1p binding sites (the TEF sites; see Table 1) showed, respectively, 50- and 70-fold increases in telomere formation compared to YIpADH-35 or YIpADHλ (Table 2). This enhancement of telomere formation was independent of the orientation of the nontelomeric Rap1p binding sites (data not shown). The array of six TEF sites stimulated telomere formation as well as the 275 bp of TG1-3 sequences, given the ±20% variation in fold enhancement (Table 2). Therefore, the enhancement of telomere formation was caused by Rap1p molecules and not by TG1-3 sequences.
Enhancement of telomere formation is independent of SIR genes and TEL1.
TPE is a property of telomere chromatin structure, similar to silencing at the yeast silent mating type cassettes HMRa and HMLα (1). Silencing at HMRa and HMLα requires the SIR1, SIR2, SIR3, and SIR4 genes, while TPE requires only SIR2, SIR3, and SIR4 (1), although tethering Sir1p to the telomere can increase TPE (2). Sir2p, Sir3p, and Sir4p are associated with telomere-proximal DNA (49). A folded-back telomeric chromatin structure requiring Sir2p, Sir3p, Sir4p, and Rap1p has recently been proposed to explain TPE (49). To determine if these gene products were important for this Rap1p-mediated enhancement of telomere formation, these experiments were repeated in strains bearing different sir mutations (Table 2). Telomere formation was monitored in sir2, sir3, and sir4 cells by Southern blot analysis of 6 to 12 transformants for each construction. All the sir mutants showed as much stimulation as the wild-type strain or more, indicating that these gene products are not required for this enhancement. These data indicate that telomere formation enhancement does not require SIR2, SIR3, SIR4, or the proposed folded-back telomeric chromatin structure but uses some other TPE-independent mechanism.
The effect of a known regulator of steady-state telomere length, Tel1p, on this enhancement of telomere formation was also assayed. Inactivating mutations of TEL1 cause cells to maintain telomeres at a new steady-state length, 50 to 100 bp of TG1-3, while still maintaining TPE (6, 8, 34, 44). Telomere formation efficiencies of YIpADH-35, YIpADHλ, and YIpADHTEF-35 (Table 2) (Materials and Methods) were determined in a congenic pair of TEL1 (YDM884) and tel1Δ (YDM911) strains (37). Telomere formation in TEL1 and tel1Δ cells, respectively, for YIpADHλ was 1.3- and 1.4-fold above that for YIpADH-35, and for YIpADHTEF-35 it was 23- and 45-fold increased. Therefore, Tel1p was not required for enhancement of telomere formation mediated by the six internal Rap1p sites. These data indicate that two properties of fully formed telomeres, TEL1-dependent length regulation (34, 44) and TPE, are not required for the Rap1p-mediated enhancement of telomere formation.
Internal Rap1p C termini are sufficient to enhance telomere formation.
Rap1p binding can induce changes in the DNA helix (5), which might play a role in the enhancement of telomere formation that we observed. To determine whether the entire Rap1p molecule bound to internal DNA sites was required for telomere formation enhancement, different portions of the Rap1p C-terminal domain fused to the DNA binding domain of Gal4p were tested for their ability to stimulate the formation of telomeres containing internal Gal4p sites. Synthetic telomeres containing four Gal4p sites, which can bind eight Gal4p molecules (YIpADHGAL-35), were transformed into cells that contained no endogenous GAL4 gene (gal4Δ cells) and were expressing different Gal4p-Rap1p C-terminal fusions, and the efficiency of telomere formation was monitored (Table 3). Fusions containing amino acids 630 to 827 of the Rap1p C terminus, including the transcription activation domain, the Rif1p-Sir3p interaction domain, and the C-terminal domain (Fig. 1B), enhanced telomere formation as well as the six TEF sites. Thus, the Rap1p C terminus tethered by a different DNA binding domain can mediate telomere formation enhancement.
The level of enhancement was not significantly altered by the rap1-12 double point mutation that abrogates Rap1p-Rif1p interaction (12) (Table 3). The high level of GAL-35 enhancement over the YIpADH-35 telomere indicates that Rif1p binding to the one or two wild-type Rap1p molecules on the 29-bp TG1-3 tract does not significantly affect telomere formation. These data indicate that Rif1p is not required for enhancing telomere formation. Rif1p is known to affect the steady-state length of established telomeres (12, 35). Thus, like the Sir proteins and Tel1p, a protein known to function in the chromatin structure and length regulation of fully formed telomeres was not required for the enhancement of telomere formation mediated by internal Rap1p C termini.
Small deletions which removed amino acids 630 to 702 of the transcriptional activation domain (11) or amino acids 800 to 827, which play a role in the length regulation of established telomeres (27), significantly reduced telomere formation enhancement (Table 3). Even though some truncations still showed enhancement compared to YIpADH-35 (e.g., 653-800 and 679-827 [Table 3]), the number of formation events was less than that obtained in cells bearing the 630-827 Gal4p-Rap1p fusion. Thus, the integrity of the entire C-terminal domain was required for maximal enhancement of telomere formation.
To rule out the possibility that the different levels of enhancement were due to differences in the stability of the Gal4p-Rap1p fusion proteins, equal amounts of cell extracts from several of the different strains tested were examined by Western blotting using a monoclonal Ab against the Gal4p DNA binding domain (Fig. 3). All the fusion proteins tested were detected. While the expression levels of different fusions varied, the 618-827 and 630-827 fusions, which showed the highest stimulation of telomere formation, were expressed at lower levels than 653-827 and 679-827 (Fig. 3A), which showed reduced levels of stimulation. The 702-827 fusion, which showed a level of telomere formation more than 35-fold lower than the 618-827 fusion (Table 3), was clearly expressed, but at a lower level than the 618-827 fusion (Fig. 3B). Thus, the large differences in enhancement were not due to large differences in expression of the fusion proteins. Conceivably, these differences may result from the ability of the Rap1p C terminus in the fusion protein to fold quickly into the structure recognized by the telomere formation machinery, and the larger fusions may fold more efficiently than the smaller fusions.
FIG. 3.
Expression of Gal4p-Rap1p fusions. Yeast cell extracts from YM708 cells expressing the indicated Gal4p-Rap1p fusion (100 μg/lane) were analyzed by Western blotting using a mouse monoclonal Ab to Gal4p amino acids 1 to 147 (see Materials and Methods). Amino acid numbers are shown as in Table 3. No plasmid, no Gal4p-Rap1p expression plasmid. The two panels show lanes from the same gel. The unusual migration of the 630-827 fusion and the multiple forms of some of the proteins have been observed in the analysis of these proteins in many different experiments (data not shown). These results indicate that differences in telomere formation enhancement (Table 3) cannot be accounted for by underexpression of various fusion proteins.
Telomere formation enhancement by internal Rap1p C termini is independent of the terminal TG tract.
Telomere formation enhancement appeared to be due to stimulation by internal Rap1p C termini, as opposed to protection from degradation, because the nontelomeric Rap1p and Gal4p-Rap1p binding sites that stimulated formation were internal to the terminal TG1-3 sequences in these telomeres. In addition, some Gal4p-Rap1p fusions failed to stimulate telomere formation, indicating that any Gal4p-Rap1p fusion binding to these sites was not sufficient to enhance telomere formation (Table 3). However, these data could not rule out a model in which the terminal TG1-3 sequences can be degraded but the internal Rap1p C termini protect the chromosome terminus from degradation until telomerase can add telomeric sequences. Previous experiments have indicated that only a small number of T or G residues are required to seed telomere formation (16, 38). Alternatively, the internal Rap1p C termini form some structure that encompasses the two Rap1p sites in the 29-bp TG1-3 tract and somehow protects the terminal TG1-3 tract from degradation. Such a structure must be distinct from the recently proposed folded-back telomere chromatin because internal Rap1p sites can enhance telomere formation in strains lacking Sir2p, Sir3p, or Sir4p and TPE (Table 2), which should lack this structure (13, 49).
To test whether internal Rap1p C termini stimulate telomere formation through a mechanism that is distinct from protection from degradation, telomeres were constructed with terminal sequences that could not be bound by Rap1p but could still allow telomere formation. These sequences, or “telomere seeds,” were 29 bp of poly(GT) or 37 bp of poly(G2T) (Table 1). Both of these sequences can be used as substrates, or seeds, for telomere formation (31). The TG1-3 seed in YIpADH-35, YIpADHTEF-35, and YIpADHGAL-35 was replaced with either the GT or G2T seed to form two new families of synthetic telomeres in order to test these different models (Fig. 1C).
If the enhancement of telomere formation was due to stimulation by internal Rap1p C termini, then the relative levels of telomere formation should be approximately equal regardless of the terminal telomere seed. However, if telomere formation enhancement is due to protection until telomerase can add TG1-3 sequences to any G or T residue, then the YIpADHGAL telomere lacking any telomere seed should also show enhanced telomere formation with respect to YIpADH-35. If telomere formation enhancement is due to Rap1p C termini forming a structure that encompasses and protects the terminal TG1-3 sequences, then the GAL and TEF telomeres with GT and G2T telomere seeds should show very little enhancement, because these terminal tracts lack the Rap1p binding sites present in the TG1-3 telomere seed.
Telomere formation using the above constructs was analyzed with the strains shown in Table 3. In the case of YIpADHGAL lacking a telomere seed, the rate of telomere formation in cells expressing the Gal4p-Rap1p(653-827) fusion was extremely low (Table 4). This result is inconsistent with the simple model in which the Rap1p C termini enhance telomere formation by protecting URA3 and the Gal4p sites from degradation until telomerase can add TG1-3 sequences to any G or T residue. This result shows that a telomere seed is required to form telomeres efficiently in this system.
The poly(GT) and poly(G2T) sequences functioned as telomere seeds in this assay but were not as efficient as TG1-3 (Table 4). In cells bearing the Gal4p-Rap1p(653-827) fusion, the relative levels of telomere formation with respect to YIpADH-35 were ∼0.6 for YIpADH-GT and ∼0.2 for YIpADH-G2T. The relative efficiencies of telomere formation for these telomere seeds alone, highest for TG1-3, intermediate for GT, and lowest for G2T, are qualitatively similar to previous results obtained with linear plasmids (31). These data show that the Rap1p binding sites in the 29-bp TG1-3 telomere seed were not required for telomere formation.
The relative levels of telomere formation enhancement were approximately equal for all constructs containing internal binding sites for Rap1p C termini regardless of the terminal telomere seed (Table 4). Compared to that in the YIpADH construct with the same telomere seed and no internal Rap1p or Gal4p sites, telomere formation enhancement was 24-fold for YIpADHGAL-35, 30-fold for YIpADHGAL-GT, and 29-fold for YIpADHGAL-G2T. These levels of enhancement are indistinguishable given the 20% variation in this assay. The fold increases in telomere formation in the YM708 strain for all three YIpADHTEF constructs were also extremely similar. These results indicate that telomere formation enhancement by internal binding sites for Rap1p C termini was independent of the presence of Rap1p sites in the terminal telomere seed. They also show that the relative level of enhancement is independent of how well the terminal tract functions in telomere formation, since the sequences seed telomere formation in the order TG1-3→GT→G2T but all show similar levels of enhancement in the TEF and GAL constructs. These results are inconsistent with the model that Rap1p C termini form a structure that protects the terminal telomere seed from degradation, because the poly(GT) and poly(G2T) sequences do not contain Rap1p sites. The simplest explanation for these data is that the internal Rap1p C termini perform some function that enhances telomere formation independent of the telomere seed, and the terminal tracts determine the absolute level of telomere formation (see Discussion).
Telomere formation is not due to increased homologous recombination.
An alternative model to explain these data is that the Rap1p C termini tethered to the TEF or GAL telomere constructions increase the frequency of homologous recombination that occurs during our telomere formation assay (Fig. 1A). To test this possibility, the YIpADH-35, TEF-35, and GAL-35 telomere constructs were inserted into the LEU2 gene in vitro (Fig. 4). These constructions were used to monitor the frequency of homologous integration at the LEU2 locus in gal4Δ cells and in gal4Δ cells expressing the Gal4p-Rap1p(653-827) fusion. The differences in the frequency of homologous integration were an order of magnitude less than the 20- to 36-fold stimulation of telomere formation in the TEF and GAL telomeres (Tables 3 and 4). Thus, telomere formation enhancement was not due to increased levels of homologous integration. An experiment with telomere formation on YACs described below also rules out increased telomere-telomere recombination (54) as a mechanism for enhancement of telomere formation (see Table 5 below). The sum of these data suggests that internal Rap1p C termini stimulate telomere formation by improving the cell’s ability to lengthen or heal the chromosome terminus (see Discussion).
Enhancement of telomere formation requires telomerase and is not due to recombination-mediated telomere formation.
Telomere formation enhancement by Rap1p C termini could occur by stimulating telomerase activity or by an alternative pathway of acquiring TG1-3 repeats such as a recombination-mediated mechanism (29, 41, 47, 60) (summarized in Fig. 5A). Telomerase contains an internal RNA which templates the addition of TG1-3 repeats. The yeast telomerase RNA is encoded by the yeast TLC1 gene, deletion of which causes telomeres to shorten over successive divisions until cells die (47). Deletion of the yeast EST1 gene causes a similar phenotype (30, 52), and Est1p appears to play an important role in telomerase activity (23, 48), but not as a catalytic subunit of telomerase (3, 20, 26, 52). Two recombination pathways have been demonstrated, a nonreciprocal strand-copying mechanism (41) and a reciprocal transfer of subtelomeric repeats (29). The telomerase pathway requires TLC1 and EST1, while the recombination pathways should not, so only the recombination pathway can promote telomere formation in tlc1Δ cells. If the Rap1p C terminus-mediated enhancement of telomere formation occurs through the telomerase pathway, e.g., by increasing telomerase activity, loss of tlc1 or est1 function should reduce this enhancement and the YIpADH-35 and YIpADHTEF-35 telomeres should be formed at similar frequencies. However, if this enhancement depends on either recombination pathway, the relative telomere formation frequencies for YIpADH-35, YIpADHTEF-35, and YIpADHGAL-35 would still be the same in tlc1 or est1 mutants. To distinguish between these hypotheses, we determined whether tlc1Δ and est1Δ mutations altered Rap1p C-terminal telomere formation enhancement by forming telomeres in these mutant cells.
Telomere formation by replacement of the VIIL telomere was monitored in tlc1Δ and est1Δ mutant cells. These strains were generated by loss of a plasmid bearing the wild-type gene; the deletion strains were grown for transformation, and the transformants obtained were rescued from eventual cell death by mating to wild-type strains in order to reintroduce TLC1 or EST1 and then were scored for telomere formation (Materials and Methods). Replacement of the VIIL telomere occurred at a very low frequency in both cell types. Multiple transformations with 5 μg of DNA per transformation into tlc1Δ cells produced only 5 to 20 transformants with YIpADH-35 and 5 to 30 transformants with YIpADHTEF-35 (TEF-35), compared to ∼150 and ∼3,700 transformants, respectively, from similar experiments in wild-type cells with these constructs. Most of the YIpADH-35 transformants did not form telomeres, so the fold enhancement in telomere formation could not be determined with this approach. Consequently, a system that produced higher levels of telomere formation in tlc1Δ and est1Δ cells was required to determine if TLC1 and EST1 were required for Rap1p C terminus-mediated telomere formation enhancement. A YAC-based system was developed for these experiments.
Two 12-kb YACs bearing one telomere with 275 bp of TG1-3 sequences and one YIpADH-35 or TEF-35 telomere (Fig. 5B) were constructed. Each YAC can replicate extrachromosomally, so telomere formation is the only requirement for obtaining a transformant. In addition, the strand-copying mechanism of telomere formation was first detected on extrachromosomal linear plasmids (41). Thus, the YAC system was expected to provide a telomerase-independent means of forming telomeres so that a sufficient number of telomere formation events could be assayed to determine the contributions of TLC1 and EST1 to Rap1p-mediated telomere formation enhancement. If this enhancement occurs by stimulation of a telomerase-independent mechanism, then the stimulation of telomere formation by the TEF-35 telomere should also be detected in tlc1Δ and est1Δ cells. However, if telomerase is required for enhancement, the numbers of YIpADH-35 and TEF-35 telomere formation events will be similar.
All of the YACs tested transformed cells at much higher frequencies than the VIIL telomere replacement constructs. YACs bearing the TEF telomere showed only a 10-fold enhancement of telomere formation in wild-type (YM708) cells (Table 5). The reduced level of enhancement of telomere formation most likely reflects an increase in telomerase-independent telomere formation for the YIpADH-35 telomere by the strand-copying mechanism, which is highly efficient (41), and dilutes the differences in telomere formation frequency between the TEF-35 and YIpADH-35 telomeres (Materials and Methods). These data show that the telomere formation enhancement observed by integration at ADH4 on VIIL also occurs on YACs.
The level of telomere formation on YACs bearing the YIpADH-35 or TEF-35 telomere was indistinguishable in tlc1Δ strains. Thus, TLC1, the gene for yeast telomerase RNA, was required for Rap1p-mediated telomere formation enhancement. These data indicate that active telomerase is a required component for internal Rap1p C termini to stimulate telomere formation.
The YAC bearing the TEF-35 telomere showed 2.8-fold enhancement of telomere formation over the YAC bearing the YIpADH-35 telomere in est1Δ cells (Table 5). This enhancement represented more than a threefold reduction compared to wild-type cells. Thus, EST1 was also required for stimulating telomere formation. These data show that telomere formation enhancement by internal Rap1p C termini requires telomerase RNA and Est1p.
DISCUSSION
In this study, a quantitative transformation assay for telomere formation at VIIL with a short telomere formation substrate, or seed, has revealed a novel function for the major yeast telomere binding protein Rap1p. Previous work from many laboratories has shown that the Rap1p C terminus inhibits the lengthening of established telomeres by acting at the chromosome terminus. The data presented here show that internally located Rap1p C termini can stimulate telomere formation in a telomerase-dependent manner. Because the telomere substrates used here require elongation of the TG1-3 repeats to form a telomere, this system is a model for the stimulation of telomere elongation. Thus, internal Rap1p C termini on short telomeres most probably stimulate telomere lengthening, in contrast to the action of Rap1p at the ends of long, established telomeres (17). These results suggest a model in which a balance between the activities of internal Rap1p C termini, which stimulate telomere elongation, and those of terminal Rap1p molecules, which limit elongation, regulates telomere length (see below).
Telomere formation enhancement by internal Rap1p C termini occurred by a mechanism that was distinct from protection from degradation as judged by several criteria. First, the Rap1p C termini that enhance formation, tethered by either the Rap1p or Gal4p DNA binding domain, were located internal to the terminal TG tracts that seed telomere formation. Second, the folded-back telomere chromatin structure recently proposed to explain TPE was unlikely to be involved in the enhancement of telomere formation because cells lacking both SIR genes and TPE had as high or higher levels of telomere formation enhancement (Table 2). If the folded-back structure were involved in telomere formation enhancement by protecting the end from degradation, the level of enhancement should have been greatly reduced in sir mutants. Third, telomere formation enhancement occurred with telomere seeds that contained no Rap1p binding sites, so Rap1p binding to these terminal tracts was dispensable for enhancement. In the telomeres containing poly(GT) and poly(G2T) seeds, the only binding sites for Rap1p and Gal4p were internal to these tracts (Table 1 and Fig. 1C). Fourth, the relative level of telomere formation enhancement was independent of the sequence of the terminal tract (Table 4). Since the individual tracts formed telomeres with different frequencies (in descending order, TG1-3, GT, and G2T) but the TEF and GAL constructs showed the same fold enhancement with all three telomere seeds, the internal Rap1p C termini function independently of the terminal tract. These results are inconsistent with a model in which the Rap1p C termini form a structure which protects the terminal tract; such a model predicts that the TG1-3 tract with two Rap1p sites should show a much greater level of enhancement in the TEF-35 and GAL-35 constructs because the TG1-3 repeats would be part of such a structure. The simplest explanation for all these data is that the internal Rap1p C termini enhance telomere formation by a mechanism that is independent of the terminal tract and so is distinct from the internal Rap1p C termini protecting the terminal tract from exonucleolytic degradation.
The Rap1p C terminus-mediated enhancement of telomere formation requires telomerase RNA (the product of the TLC1 locus) and is enhanced by Est1p (Table 5). When the tlc1Δ and est1Δ strains were tested for telomere formation by replacing VIIL (Fig. 1A) or on short YACs (Fig. 5), stimulation was greatly reduced in both cases. However, VIIL telomeres formed at such low rates in the tlc1Δ and est1Δ strains that, while the ∼35-fold higher level of formation of the TEF-35 telomere compared to the YIpADH-35 telomere was reduced, the final level was highly variable. This low rate of formation in tlc1Δ and est1Δ strains made it impossible to use the telomere formation assay at VIIL to determine an accurate fold enhancement. Telomere formation on short linear plasmids can use an alternative, telomerase-independent mechanism to transfer sequences between linear plasmid ends (41, 54), which can allow the YIpADH-35 or TEF-35 telomere to acquire TG1-3 sequences from the other end of the YAC (Fig. 5). We verified that telomere formation enhancement mediated by internal Rap1p molecules occurred in this system. The fold enhancement was smaller, most probably because the telomerase-independent mechanism increases the absolute number of YIpADH-35 telomere formation events. By use of short YACs, larger numbers of telomere formation events were obtained in tlc1Δ and est1Δ cells, which allowed an accurate comparison between the rates of YIpADH-35 and TEF-35 telomere formation. These data show that telomerase RNA and therefore active telomerase, as well as Est1p, must be present in the cell in order for the internal Rap1p molecules to stimulate telomere formation. Est1p is a single-stranded TG1-3 binding protein that coimmunoprecipitates with telomerase RNA and activity (23, 48, 52) but is probably not a part of the core catalytic enzyme (3, 20, 25).
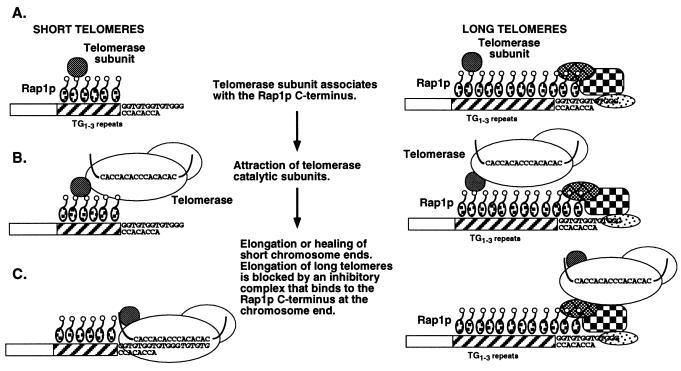
Our results suggest a model in which internal Rap1p C termini play a role in telomere length control. We propose that the Rap1p C terminus stimulates the elongation of short telomeres by increasing telomerase activity in cis. We suggest that one of the many components that can interact with the Rap1p C terminus is a protein that can either attract the telomerase enzyme or increase its activity, so we refer to this protein as the telomerase accessory subunit (Fig. 6). If the telomere is short because of chromosome breakage or degradation, the activated telomerase can add telomere repeats to the nearest available substrate (Fig. 6), which in most cases will be the chromosome terminus that need not be capped by telomere sequences (16, 38). Data from other laboratories indicate that if the telomere is long or of normal length, Rap1p molecules bound to the chromosome terminus interact with factors there to block telomerase access and prevent lengthening (4, 17, 18). In our model, these inhibitory factors would be absent from short telomeres. The simplest form of this model is that telomerase is always attracted by internal Rap1p C termini but its action at chromosome ends is determined by whether the Rap1p molecules at the chromosome terminus can block telomerase access. This model parallels the results in Table 4, where formation enhancement by internal Rap1p C termini was independent of the sequence at the chromosome terminus. A direct consequence of this model is that attracting telomerase to normal-length telomeres where lengthening is blocked is futile, and only those telomeres that are short (i.e., without the inhibiting Rap1p at the chromosome terminus) will be elongated. These “futile” cycles in wild-type cells provide a way to constantly monitor telomere length and keep it within a set range. When telomeres become too short, Rap1p at the terminus no longer inhibits telomere lengthening (see below) and elongation occurs. If telomerase always adds a constant number of TG1-3 repeats equivalent to the amount of sequence lost over several cell cycles, only a few active telomerase molecules are required to maintain all telomeres within a constant range of lengths. Those telomeres that become too long would be shortened by telomere deletion mechanisms (21).
FIG. 6.
Speculative model for Rap1p C terminus-mediated enhancement of telomere formation and length regulation. (A) A telomerase subunit (see the text) interacts with internal Rap1p C termini. (B) The telomerase subunit provides a docking site for the catalytic subunits of telomerase or serves to increase telomerase activity. (C) If a short telomere or broken chromosome end is near the bound Rap1p C termini, telomerase can bind to the free 3′ end and add telomeric repeats. The free 3′ end need only have limited homology to telomerase RNA (16, 38). If the TG1-3 repeats are long, then the Rap1p and chromosome terminus-specific factors bound at the chromosome end (e.g., Cdc13p, Stn1p, Est1p) block telomerase access to the 3′ end and no lengthening occurs (7, 17, 24, 40, 52) (see the text). Over the course of many cell divisions, the combined actions of telomerase recruitment and inhibition of elongation at only those telomeres with normal lengths maintains all chromosomal telomeres within the same length range. A model similar to the activity at short telomeres was previously proposed by Kramer and Haber (16) to describe TTGGGG-stimulated telomere formation in yeast, but the proteins involved in that stimulation were not identified (see the text).
These considerations suggest that Rap1p C termini at different locations within the telomere repeats are seen differently by the cell. These differences may reflect interactions with different sets of proteins at these sites, such as Cdc13p, Est1p, Stn1p (7, 24, 40, 52), and other single-stranded TG1-3 binding factors (15, 22). In addition, recent work has shown that yeast cells monitor the number of Rap1p C termini at each telomere (35, 42). The cell may determine if the most distal Rap1p C terminus is inhibitory by “counting” the number of Rap1p C termini from the telomere-nontelomere junction.
Our model claims that telomerase mediates the Rap1p C terminus-dependent enhancement of telomere lengthening because a telomerase accessory subunit is part of telomeric chromatin structure (Fig. 6). This model is consistent with previous results showing that overexpression of a truncated TLC1 RNA (47) and the Rap1p C terminus (56) can have similar effects on TPE, which is dependent on telomere chromatin structure (1). Presumably, both telomerase RNA and the Rap1p C terminus compete for telomere-associated proteins critical for maintaining normal telomere structure and TPE. The shorter telomeres in cells overexpressing TLC1 RNA (5a, 47) are also consistent with the model, as excess telomerase RNA is expected to compete for telomerase protein subunits. Removal of the proposed telomerase accessory subunit from internal Rap1p C termini would reduce the frequency with which telomerase is brought to or activated at the telomere, reducing the frequency of telomere elongation and causing a net shortening of telomeres.
The concept that proteins that bind at or near the telomere can directly or indirectly attract telomerase can explain several previous observations in yeast and human cells. In HeLa cells, telomere formation can be seeded by telomere repeats, but a large number of repeats is necessary for efficient formation (10). These requirements could reflect a similar property of internally bound human telomere repeat factor, TRF1, to attract or stimulate telomerase activity at short telomeres and, similarly to Rap1p, inhibit telomere elongation on existing telomeres (51). Internal TTGGGG sequences have been shown to stimulate telomere formation in yeast (16, 38). Tbf1p is a protein that binds to TTAGGG repeats in the telomere-associated X repeats that can also bind to TTGGGG (28). If Tbf1p can also attract telomerase, X repeats could form an efficient back-up mechanism for stimulating telomere formation in the event of loss of the terminal TG1-3 sequences. Telomerase would be attracted to sequences within X elements and add telomere sequences to the nearby broken end. Thus, this model can explain telomere behavior in human cells and provides a mechanism for telomere formation or chromosome “healing” in the event of catastrophic loss of telomere repeats.
ACKNOWLEDGMENTS
We thank the investigators mentioned in the text for strains and plasmids, N. Roy for the KR36-6L strains bearing sir mutations, R. Kota for unpublished data, and K. Hotmire for technical support.
K.W.R. is supported by a Junior Faculty Research Award from the American Cancer Society. This work was supported by NIH grant GM50752 to K.W.R.
REFERENCES
- 1.Aparicio O M, Billington B L, Gottschling D E. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- 2.Chien C T, Buck S, Sternglanz R, Shore D. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell. 1993;75:531–541. doi: 10.1016/0092-8674(93)90387-6. [DOI] [PubMed] [Google Scholar]
- 3.Cohn M, Blackburn E H. Telomerase in yeast. Science. 1995;269:396–400. doi: 10.1126/science.7618104. [DOI] [PubMed] [Google Scholar]
- 4.Conrad M N, Wright J H, Wolf A J, Zakian V A. RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell. 1990;63:739–750. doi: 10.1016/0092-8674(90)90140-a. [DOI] [PubMed] [Google Scholar]
- 5.Gilson E, Roberge M, Giraldo R, Rhodes D, Gasser S M. Distortion of the DNA double helix by RAP1 at silencers and multiple telomeric binding sites. J Mol Biol. 1993;231:293–310. doi: 10.1006/jmbi.1993.1283. [DOI] [PubMed] [Google Scholar]
- 5a.Gottschling, D. Personal communication.
- 6.Gottschling D E, Aparicio O M, Billington B L, Zakian V A. Position effect at yeast telomeres: reversible repression of polII transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 7.Grandin N, Reed S I, Charbonneau M. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 1997;11:512–527. doi: 10.1101/gad.11.4.512. [DOI] [PubMed] [Google Scholar]
- 8.Greenwell P A, Kronmal S L, Porter S E, Gassenhuber J, Obermaier B, Petes T D. TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell. 1995;82:823–829. doi: 10.1016/0092-8674(95)90479-4. [DOI] [PubMed] [Google Scholar]
- 9.Greider C W. Telomere length regulation. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 10.Hanish J P, Yanowitz J L, de Lange T. Stringent sequence requirements for the formation of human telomeres. Proc Natl Acad Sci USA. 1994;91:8861–8865. doi: 10.1073/pnas.91.19.8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardy C F J, Balderes D, Shore D. Dissection of a carboxy-terminal region of the yeast regulatory protein RAP1 with effects on both transcriptional activation and silencing. Mol Cell Biol. 1992;12:1209–1217. doi: 10.1128/mcb.12.3.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardy C F J, Sussel L, Shore D. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992;6:801–814. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- 13.Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–95. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 14.Henry Y A L, Chambers A, Tsang J S H, Kingsman A J, Kingsman S M. Characterisation of the DNA binding domain of the yeast RAP1 protein. Nucleic Acids Res. 1990;18:2617–2623. doi: 10.1093/nar/18.9.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konkel L M, Enomoto S, Chamberlain E M, McCune Z P, Iyadurai S J, Berman J. A class of single-stranded telomeric DNA-binding proteins required for Rap1p localization in yeast nuclei. Proc Natl Acad Sci USA. 1995;92:5558–5562. doi: 10.1073/pnas.92.12.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Kota, R. Unpublished data.
- 16.Kramer K M, Haber J E. New telomeres in yeast are initiated with a highly selected subset of TG1-3 repeats. Genes Dev. 1993;7:2345–2356. doi: 10.1101/gad.7.12a.2345. [DOI] [PubMed] [Google Scholar]
- 17.Krauskopf A, Blackburn E H. Control of telomere growth by interactions of RAP1 with the most distal telomere repeats. Nature. 1996;383:354–357. doi: 10.1038/383354a0. [DOI] [PubMed] [Google Scholar]
- 18.Kyrion G, Boakye K A, Lustig A J. C-terminal truncation of RAP1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5159–5173. doi: 10.1128/mcb.12.11.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kyrion G, Liu K, Liu C, Lustig A J. RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev. 1993;7:1146–1159. doi: 10.1101/gad.7.7a.1146. [DOI] [PubMed] [Google Scholar]
- 20.Lendvay T S, Morris D K, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B, Lustig A J. A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev. 1996;10:1310–1326. doi: 10.1101/gad.10.11.1310. [DOI] [PubMed] [Google Scholar]
- 22.Lin J-J, Zakian V A. Isolation and characterization of two Saccharomyces cerevisiae genes that encode proteins that bind to (TG1-3)n single strand telomeric DNA in vitro. Nucleic Acids Res. 1994;22:4906–4913. doi: 10.1093/nar/22.23.4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin J-J, Zakian V A. An in vitro assay for Saccharomyces telomerase requires EST1. Cell. 1995;81:1127–1135. doi: 10.1016/s0092-8674(05)80017-0. [DOI] [PubMed] [Google Scholar]
- 24.Lin J-J, Zakian V A. The Saccharomyces CDC13 protein is a single-stranded TG1-3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc Natl Acad Sci USA. 1996;93:13760–13765. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lingner J, Cech T R, Hughes T R, Lundblad V. Three ever shorter telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc Natl Acad Sci USA. 1997;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lingner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 27.Liu C, Mao X, Lustig A J. Mutational analysis defines a C-terminal tail domain of RAP1 essential for telomeric silencing in Saccharomyces cerevisiae. Genetics. 1994;138:1025–1040. doi: 10.1093/genetics/138.4.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Z P, Tye B K. A yeast protein that binds to vertebrate telomeres and conserved yeast telomeric junctions. Genes Dev. 1991;5:49–59. doi: 10.1101/gad.5.1.49. [DOI] [PubMed] [Google Scholar]
- 29.Lundblad V, Blackburn E H. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 30.Lundblad V, Szostak J W. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 31.Lustig A J. Hoogsteen G-G base pairing is dispensable for telomere healing in yeast. Nucleic Acids Res. 1992;20:3021–3028. doi: 10.1093/nar/20.12.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lustig A J, Kurtz S, Shore D. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science. 1990;250:549–553. doi: 10.1126/science.2237406. [DOI] [PubMed] [Google Scholar]
- 33.Lustig A J, Liu C, Zhang C, Hanish J P. Tethered Sir3p nucleates silencing at telomeres and internal loci in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2483–2495. doi: 10.1128/mcb.16.5.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lustig A J, Petes T D. Identification of yeast mutants with altered telomere structure. Proc Natl Acad Sci USA. 1986;83:1398–1402. doi: 10.1073/pnas.83.5.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marcand S, Gilson E, Shore D. A protein-counting mechanism for telomere length regulation in yeast. Science. 1997;275:986–990. doi: 10.1126/science.275.5302.986. [DOI] [PubMed] [Google Scholar]
- 36.Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 37.Morrow D M, Tagle D A, Shiloh Y, Collins F S, Heiter P. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell. 1995;82:831–840. doi: 10.1016/0092-8674(95)90480-8. [DOI] [PubMed] [Google Scholar]
- 38.Murray A W, Claus T E, Szostak J W. Characterization of two telomeric DNA processing reactions in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4642–4650. doi: 10.1128/mcb.8.11.4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murray A W, Schultes N P, Szostak J W. Chromosome length controls mitotic chromosome segregations in yeast. Cell. 1986;45:529–536. doi: 10.1016/0092-8674(86)90284-9. [DOI] [PubMed] [Google Scholar]
- 40.Nugent C I, Hughes T R, Lue N F, Lundblad V. Cdc13p: a single-stranded telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 41.Pluta A F, Zakian V A. Recombination occurs during telomere formation in yeast. Nature. 1989;337:429–433. doi: 10.1038/337429a0. [DOI] [PubMed] [Google Scholar]
- 42.Ray, A., and K. Runge. The yeast telomere length counting machinery is critically sensitive to sequences at the telomere/non-telomere junction. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 42a.Ray, A., and K. W. Runge. Unpublished data.
- 43.Renauld H, Aparicio O M, Zierath P D, Billington B L, Chhablani S K, Gottschling D E. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 1993;7:1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- 43a.Roy, N., and K. W. Runge. Submitted for publication.
- 44.Runge K W, Zakian V A. TEL2, an essential gene required for telomere length regulation and telomere position effect in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3094–3105. doi: 10.1128/mcb.16.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sandell L L, Zakian V A. Loss of a yeast telomere: arrest, recovery and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 46.Shore D. RAP1: a protean regulator in yeast. Trends Genet. 1994;10:408–412. doi: 10.1016/0168-9525(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 47.Singer M S, Gottschling D E. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 48.Steiner B R, Hidaka K, Futcher B. Association of the Est1 protein with telomerase activity in yeast. Proc Natl Acad Sci USA. 1996;93:2817–2821. doi: 10.1073/pnas.93.7.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 50.Sussel L, Shore D. Separation of transcriptional activation and silencing functions of the RAP1-encoded repressor/activator protein 1: isolation of viable mutants affecting both silencing and telomere length. Proc Natl Acad Sci USA. 1991;88:7749–7753. doi: 10.1073/pnas.88.17.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 52.Virta-Pearlman V, Morris D K, Lundblad V. Est1 has the properties of a single-stranded telomere end-binding protein. Genes Dev. 1996;10:3094–3104. doi: 10.1101/gad.10.24.3094. [DOI] [PubMed] [Google Scholar]
- 53.Wang S-S, Zakian V A. Sequencing of Saccharomyces telomeres cloned using T4 DNA polymerase reveals two domains. Mol Cell Biol. 1990;10:4415–4419. doi: 10.1128/mcb.10.8.4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang S S, Zakian V A. Telomere-telomere recombination provides an express pathway for telomere acquisition. Nature. 1990;345:456–458. doi: 10.1038/345456a0. [DOI] [PubMed] [Google Scholar]
- 55.Wellinger R J, Zakian V A. Lack of positional requirements for autonomously replicating sequence elements on artificial yeast chromosomes. Proc Natl Acad Sci USA. 1989;86:973–977. doi: 10.1073/pnas.86.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiley E A, Zakian V A. Extra telomeres, but not internal tracts of telomeric DNA, reduce transcriptional repression at Saccharomyces telomeres. Genetics. 1995;139:67–79. doi: 10.1093/genetics/139.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wotton D, Shore D. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 1997;11:748–760. doi: 10.1101/gad.11.6.748. [DOI] [PubMed] [Google Scholar]
- 58.Zakian V A. The structure and function of telomeres. Annu Rev Genet. 1989;23:579–604. doi: 10.1146/annurev.ge.23.120189.003051. [DOI] [PubMed] [Google Scholar]
- 59.Zakian V A. Structure, function and replication of Saccharomyces cerevisiae telomeres. Annu Rev Genet. 1996;30:141–172. doi: 10.1146/annurev.genet.30.1.141. [DOI] [PubMed] [Google Scholar]
- 60.Zakian V A, Runge K, Wang S S. How does the end begin? Formation and maintenance of telomeres in ciliates and yeast. Trends Genet. 1990;6:12–16. doi: 10.1016/0168-9525(90)90043-6. [DOI] [PubMed] [Google Scholar]