Abstract
Cells respond to viral infection or double-stranded RNA with the transcriptional induction of a subset of alpha/beta interferon-stimulated genes by a pathway distinct from the interferon signal pathway. The transcriptional induction is mediated through a DNA sequence containing the alpha/beta interferon-stimulated response element (ISRE). We previously identified a novel transcription factor, designated double-stranded RNA-activated factor 1 (DRAF1), that recognizes this response element. The DNA-binding specificity of DRAF1 correlates with transcriptional induction, thereby distinguishing it as a positive regulator of alpha/beta interferon-stimulated genes. Two of the components of DRAF1 have now been identified as interferon regulatory factor 3 (IRF-3) and the transcriptional coactivator CREB-binding protein (CBP)/p300. We demonstrate that IRF-3 preexists in the cytoplasm of uninfected cells and translocates to the nucleus following viral infection. Translocation of IRF-3 is accompanied by an increase in serine and threonine phosphorylation. Coimmunoprecipitation analyses of endogenous proteins demonstrate an association of IRF-3 with the transcriptional coactivators CBP and p300 only subsequent to infection. In addition, antibodies to the IRF-3, CBP, and p300 molecules react with DRAF1 bound to the ISRE target site of induced genes. The cellular response that leads to DRAF1 activation and specific gene expression may serve to increase host survival during viral infection.
The interferon (IFN) system has evolved as a fundamental defense mechanism against viral infection. Cells respond to infection with the transcriptional activation of a set of antiviral genes, including the alpha/beta IFN (IFN-α/β) genes. Newly synthesized IFNs are secreted and bind to cell surface receptors to confer an antiviral biological effect (reviewed in reference 13). Cells producing IFN have an established viral infection and therefore may not be protected effectively by autocrine IFN. It is likely that the critical role of IFN in the immune system is to curb viral dissemination by functioning as a paracrine cytokine. Complementary defense mechanisms may function in a primary infected cell prior to the synthesis and action of IFN. In this report, we analyze a cellular response that may have evolved to increase host survival during the course of infection.
The effects of IFN-α/β are mediated by the induction of a specific class of genes called IFN-α/β-stimulated genes (ISGs) (reviewed in references 10, 12, 22, 28, and 44). Transcriptional activation of these genes is dependent on a DNA promoter sequence designated the IFN-α/β-stimulated response element (ISRE). The IFN-α/β signal transduction pathway that leads to the induction of these genes initiates with the activation of Janus tyrosine kinases and phosphorylation of a latent cytoplasmic transcription factor, the IFN-stimulated gene factor 3 (ISGF3). ISGF3 subsequently translocates to the nucleus and binds to the ISRE of inducible genes. ISGF3 is a multimeric factor composed of members of the signal transducer and activator of transcription factor (STAT) family, STAT1 and STAT2 (20, 43). These STATs are activated by tyrosine phosphorylation and associate with a member of the IFN regulatory factor family, p48, to form the ISGF3 complex (50). We have identified a distinct pathway that is activated in virus-infected cells, that is independent of IFN, and that leads to the formation of a novel ISRE-binding factor and the induction of ISGs (9, 11).
Clear evidence for the direct induction of ISGs in response to viral infection was provided by studies with cells that are deficient in autocrine IFN signaling, such as HEC-1B cells, which cannot respond to IFNs (21, 51). Viral infection of these cells results in the induction of a set of ISGs (2, 9, 11, 36, 52–55). To begin to elucidate the mechanism by which cells respond to virus, we identified novel cellular DNA-binding factors that are activated in response to viral infection and can recognize the ISRE (9, 11). One of the factors is activated in response to infection by a DNA tumor virus (adenovirus), by an RNA virus (Newcastle disease virus [NDV]), or by double-stranded RNA (dsRNA). It appears that viral dsRNA produced during viral transcription or replication leads to the activation of this factor. Therefore, it has been designated dsRNA-activated factor 1 (DRAF1) (9). The DNA-binding characteristics of DRAF1 correlate with specific gene induction and thereby identify DRAF1 as a positive regulator of ISG transcription independent of the action of IFN (11).
By characterizing the composition of DRAF1, we have made a major step toward understanding this response pathway. Evidence provided in this report identifies two of the subunits of DRAF1 as IFN regulatory factor 3 (IRF-3) and the transcriptional coactivator CREB-binding protein (CBP)/p300 (1, 7, 17; reviewed in references 18, 23, and 46). Both factors are present in the DRAF1 DNA-binding complex, and the association of IRF-3 with CBP or p300 is dependent on viral infection.
MATERIALS AND METHODS
Cell cultures and reagents.
HEC-1B cells and HeLa S3 cells (American Type Culture Collection [ATCC]) (hereafter referred to as HeLa cells) were grown in Dulbecco’s modified Eagle medium (DMEM) containing 10% bovine calf serum. HT1080 cells (ATCC) were grown in DMEM with 8% fetal bovine serum. THP-1 cells (ATCC) were grown in RPMI 1640 containing 8% fetal bovine serum and 50 μM 2-mercaptoethanol. Pkr0/0 murine embryo fibroblasts (MEFs) and Pkr+/+ MEFs were gifts from Bryan R. G. Williams (The Cleveland Clinic Foundation, Cleveland, Ohio) and Charles Weissmann (University of Zurich, Zurich, Switzerland) (33, 57) and were grown in DMEM containing 10% fetal bovine serum and supplemented with nonessential amino acids. Recombinant human alpha IFN (IFN-α) was provided by Hoffmann-La Roche Inc. (Nutley, N.J.) and was used at 1,000 U/ml. N-Ethylmaleimide (NEM) was obtained from Sigma. Protein phosphatase type 2A (PP2A) and okadaic acid were obtained from Upstate Biotechnology Inc. Recombinant protein tyrosine phosphatase 1B (PTP1B) was provided by Nicholas K. Tonks (Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.). Phosphoamino acid standards (phosphoserine, phosphothreonine, and phosphotyrosine) were obtained from Sigma. Thin-layer cellulose chromatography (TLC) plates were obtained from VWR Scientific. Prestained and unstained protein molecular weight standards were obtained from Gibco BRL. Unstained molecular weight standards were used for mass estimation of proteins. Anti–glutathione S-transferase (GST) antibodies, anti-CBP antibodies (A-22), anti-p300 antibodies (N-15), and specific blocking peptides were purchased from Santa Cruz Biotechnology Inc. dsRNA treatment was performed with poly(rI) · poly(rC) (Pharmacia) and with serum-free media.
Plasmid constructs.
Reverse transcription-PCR of THP-1 cell cytoplasmic RNA was used to amplify a 303-bp fragment corresponding to amino acids 107 to 208 of the human IRF-3 protein (1). This cDNA fragment was subcloned into bacterial expression plasmid pGEX2T (Pharmacia), and GST fusion proteins were used for immunization of rabbits and mice. A full-length human IRF-3 cDNA was generated by reverse transcription-PCR with primers corresponding to the 5′ and 3′ untranslated regions of the gene. The coding sequence of the IRF-3 cDNA was subcloned into pGEX2TK (Pharmacia), and the resulting GST–IRF-3 recombinant gene was cloned into pCDNA3. This plasmid was stably expressed in HT1080 cells (HT1080/GST–IRF-3 cells). The expressed GST–IRF-3 molecule and associated proteins were isolated by binding to glutathione beads (Pharmacia) for 20 min at 4°C, followed by washing in detergent-containing buffer prior to protein elution.
Viral infections.
NDV, a gift from Paula M. Pitha-Rowe (The Johns Hopkins University, Baltimore, Md.), was propagated in the allantoic cavities of 10-day-old embryonated hen eggs. Viral titers were determined by hemagglutination of chicken erythrocytes. Viral infections were performed as described previously (11). Briefly, cells were washed with serum-free media and then overlaid with 5 ml of serum-free media containing NDV at 100 hemagglutination units per ml. After 1 h, the cells were fed media containing serum. Mock infections were performed with allantoic fluid from uninfected hen eggs.
Immunoassays.
Protein extracts were prepared by fractionation of cells with a Dounce homogenizer under hypotonic conditions, followed by preparation of nuclear and cytoplasmic extracts as described previously (14, 42). Alternatively, whole-cell extracts were prepared by lysing cells on ice in buffer containing 50 mM Tris (pH 7.6), 400 mM NaCl, 0.5% Nonidet P-40, 5 mM potassium EDTA, 1 mM EGTA, 10 mM sodium phosphate (pH 7.2), 50 mM sodium fluoride, and 2 mM sodium vanadate. The insoluble material was removed by centrifugation at 12,000 × g and 4°C. All extracts were prepared in the presence of 1 mM phenylmethylsulfonyl fluoride (Sigma) and 1 μg each of leupeptin, pepstatin A, and aprotinin (Boehringer Mannheim Biochemicals) per ml.
Immunoprecipitations were performed by incubating extracts with an excess of antibody to IRF-3 or control antibody for 4 h at 4°C. Immunocomplexes were bound to protein G-agarose (Gibco BRL), washed, separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), and electroblotted to an Immobilon-P transfer membrane (Millipore). Proteins were detected by immunoblotting with primary antibody, followed by anti-mouse or anti-rabbit secondary antibody (Amersham) and enhanced chemiluminescence reagents (DuPont NEN).
DNA-binding assays.
Protein extracts were analyzed for specific ISRE-binding proteins by two methods. The first was an electrophoretic mobility shift assay (EMSA) done as described previously (9, 11). Nuclear extracts were incubated with a 32P-labeled oligonucleotide probe containing the human ISG15 ISRE (41) for 20 min at room temperature. The effects of specific antibodies were tested by incubation with nuclear extracts at 4°C for 60 min prior to the addition of the probe.
Alternatively, specific ISRE-binding proteins were detected by incubation with ISRE-Sepharose beads, followed by SDS-PAGE and immunoblotting. Briefly, an ISG15 double-stranded oligonucleotide containing the ISRE (5′-GGGAAAGGGAAACCGAAACTGAA-3′) (ISRE in italic type) or a mutated double-stranded oligonucleotide (5′-GGGAAAGGGAAACCCAAACTGAA-3′) (mutation in boldface type) (9) was multimerized and coupled to CNBr-activated Sepharose 4B (Pharmacia). Approximately 5.0 mg of total cellular protein, isolated by Dounce homogenization and extraction, was incubated with ISRE-Sepharose in the presence of 100 μg of nonspecific plasmid DNA and 50 μg of salmon sperm DNA (Pharmacia) at 4°C. The affinity beads were collected by centrifugation and washed, and bound proteins were separated by SDS-PAGE, followed by immunoblotting with antibody to IRF-3.
Protein phosphatase and alkylation treatments.
Nuclear extracts were treated in vitro with recombinant PTP1B at 37°C for 1 h in buffer containing 25 mM HEPES (pH 7.0) and 0.2% 2-mercaptoethanol (Sigma). PTP1B was inhibited by the inclusion of 5 mM sodium vanadate prior to incubation. Nuclear extracts were treated in vitro with PP2A by incubation at 30°C for 1 h in buffer containing 50 mM Tris (pH 7.0), 0.1 mM CaCl2, and 1 mM MnCl2. PP2A was inhibited by the inclusion of 10 nM okadaic acid prior to incubation. For protein alkylation, nuclear extracts were incubated in the absence or presence of 10 mM NEM for 30 min at room temperature, and then dithiothreitol was added to 10 mM to inactivate any of the remaining NEM.
Metabolic labeling and phosphoamino acid analysis.
For 32P metabolic labeling, cells were incubated with 250 μCi of [32P]orthophosphate (DuPont NEN) per ml in phosphate-free media and infected with NDV or mock infected. The time of NDV infection was varied, while the total time of metabolic labeling for each sample was kept constant. Whole-cell extracts were prepared by detergent lysis, normalized for equivalent trichloroacetic acid-precipitable counts, and subjected to immunoprecipitation with either control antibody or antibody to IRF-3. SDS-PAGE gels were either fixed and dried or electroblotted to Immobilon-P membranes and exposed to film. Labeling with [35S]methionine-[35S]cysteine (DuPont NEN) at 160 μCi/ml was performed with methionine- and cysteine-free media. Detergent cell lysates were normalized for protein prior to analysis. Gels were treated with En3Hance (DuPont NEN) prior to exposure to film.
For phosphoamino acid analysis, the 32P-labeled IRF-3 and adjacent areas from control gels were excised from membranes and subjected to hydrolysis in 5.7 N HCl at 110°C as described previously (6, 8). Phosphoamino acids were identified by two-dimensional electrophoresis at pH 3.5 and pH 1.9 on a TLC plate as described previously (6, 8). Unlabeled phosphoamino acid internal standards were detected with ninhydrin staining. Radioactive bands were quantified with a PhosphorImager (445 SI; Molecular Dynamics).
RESULTS
Activation of DRAF1 by NDV infection.
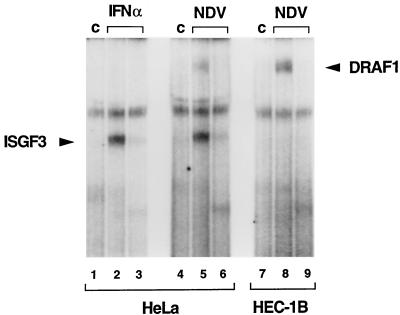
Cells respond to viral infection with the activation of ISGs by two distinct mechanisms. One mechanism involves the action of IFN-α/β cytokines produced by virus-infected cells. As previously described, stimulation of HeLa cells with IFN-α induces the appearance of the ISGF3 transcription factor. This induction is shown by an EMSA with a radiolabeled ISRE probe (Fig. 1, lane 2). Infection of HeLa cells with NDV, a paramyxovirus that produces dsRNA during the course of infection, activates two ISRE-binding transcription factors: IFN-α/β-induced ISGF3 and a novel factor, DRAF1 (Fig. 1, lane 5) (9, 11). ISGF3 activation in HeLa cells in response to NDV infection is due to autocrine IFN production. This becomes evident by NDV infection of cells unable to respond to IFN (HEC-1B cells) (21, 52). In IFN-unresponsive HEC-1B cells, DRAF1 is induced following viral infection, but ISGF3 is not (Fig. 1, lane 8). We showed previously that the appearance of DRAF1 precedes the appearance of ISGF3 in cells that respond to autocrine IFN and thereby DRAF1 functions as a direct response to virus (9, 11).
FIG. 1.
Activation of ISRE-binding factors by IFN-α and NDV. HeLa cells were either untreated (control [c]; lanes 1 and 4), treated with IFN-α for 1 h (lanes 2 and 3), or infected with NDV for 3 h (lanes 5 and 6). HEC-1B cells were either untreated (lane 7) or infected with NDV for 3 h (lanes 8 and 9). Nuclear extracts were prepared and analyzed by an EMSA with a radiolabeled ISRE probe. The specificity of the protein-DNA complexes was shown by competition with a 100-fold excess of unlabeled ISRE oligonucleotide (lanes 3, 6, and 9).
DRAF1 is sensitive to phosphatase and alkylation.
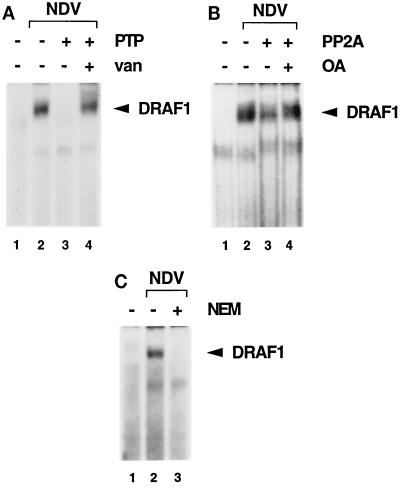
The activation of DRAF1 requires the function of a protein kinase, since the appearance of DRAF1 is blocked by the kinase inhibitors genistein and staurosporine (9). To determine whether DRAF1 is phosphorylated on tyrosine, the effect of PTP1B on DRAF1 was examined by an EMSA (Fig. 2A). Nuclear extracts were prepared from uninfected or NDV-infected HEC-1B cells and incubated in vitro with PTP1B prior to DNA binding. DRAF1 was apparent in extracts from NDV-infected cells (Fig. 2A, lane 2) but was completely abrogated by treatment with PTP1B (lane 3). The effect was inhibited by the addition of sodium vanadate, a specific inhibitor of tyrosine phosphatases (Fig. 2A, lane 4). This result suggests that DRAF1 is tyrosine phosphorylated and that this modification is required for its ability to bind to DNA. Since the STAT family also requires tyrosine phosphorylation for DNA binding, we tested the effects of specific antibodies to STAT1, STAT2, STAT3, STAT5, and STAT6 on the appearance of DRAF1 in an EMSA, but they had no effect (data not shown).
FIG. 2.
(A) Effect of a protein tyrosine phosphatase (PTP) on DRAF1. HEC-1B nuclear extracts from untreated cells (lane 1) or cells infected with NDV (lanes 2 to 4) were used in an EMSA with a radiolabeled ISRE probe. Prior to the DNA-binding reaction, the extracts were incubated in the absence (lanes 1 and 2) or in the presence (lanes 3 and 4) of recombinant PTP1B. Tyrosine phosphatase activity was inhibited by the inclusion of 5 mM sodium vanadate (van) (lane 4). (B) Effect of a serine/threonine protein phosphatase on DRAF1. HEC-1B nuclear extracts from untreated cells (lane 1) or cells infected with NDV (lanes 2 to 4) were used in an EMSA. Prior to the DNA-binding reaction, the extracts were incubated in the absence (lanes 1 and 2) or in the presence (lanes 3 and 4) of PP2A. Protein phosphatase activity was inhibited by the inclusion of 10 nM okadaic acid (OA) (lane 4). (C) Effect of protein alkylation on the appearance of DRAF1. HEC-1B nuclear extracts from untreated cells (lane 1) or cells infected with NDV (lanes 2 and 3) were used in an EMSA. Prior to the DNA-binding reaction, the extracts were either untreated (lanes 1 and 2) or treated with 10 mM NEM (lane 3).
A similar analysis was performed to examine the role of serine or threonine phosphorylation on DRAF1 activity (Fig. 2B). Nuclear extracts containing DRAF1 were incubated in the presence of PP2A, a serine- and threonine-specific protein phosphatase, before the EMSA. DRAF1 activity was partially reduced after dephosphorylation with PP2A (Fig. 2B, compare lanes 2 and 3). This effect was inhibited by the addition of okadaic acid, a specific inhibitor of PP2A (Fig. 2B, lane 4).
Although DRAF1 is similar to ISGF3 in its ability to bind to an ISRE-like target, DRAF1 does not contain the DNA-binding subunit of ISGF3, p48 (or ISGF3γ) (9). The p48 protein belongs to the IRF family of DNA-binding proteins (38, 40, 47, 50). A characteristic of IRF proteins is their sensitivity to alkylation with NEM (34). To test whether DRAF1 is also sensitive to alkylation, nuclear extracts containing DRAF1 were treated with NEM and then analyzed by an EMSA (Fig. 2C). The NEM treatment completely abrogated the DNA-binding activity of DRAF1, indicating that DRAF1 DNA binding is sensitive to alkylation with NEM (Fig. 2C, lane 3).
Antibodies to IRF-3 react with DRAF1.
Recently, several new members of the IRF family of proteins were identified. This family now includes p48, IRF-1, IRF-2, IRF-3, ICSBP, ICSAT/Pip/LSIRF/IRF-4, IRF-5 (EMBL accession number U51127), IRF-6 (EMBL accession number U73029), and IRF-7 (1, 16, 19, 23a, 25, 35, 38, 40, 50, 56, 59). Several characteristics of IRF-3 made it a candidate subunit of DRAF1. Overexpression of IRF-3 was reported to activate the expression of a reporter gene driven by an ISG promoter (1). In addition, IRF-3 is constitutively expressed in all cell types tested, whereas the expression of some of the IRF proteins is cell type restricted or is seen only following IFN stimulation (1). This ubiquitous pattern of IRF-3 expression is in accord with DRAF1 activation in a variety of cell types.
To determine whether DRAF1 contains IRF-3, the effect of a specific antibody generated against IRF-3 was tested (Fig. 3A). The immunogen used to produce the antibody corresponded to a unique region of IRF-3 that shares only three contiguous amino acids with other IRF proteins. The specific antibody does not cross-react with heterologous IRFs, such as p48, IRF-1, or IRF-2 (data not shown). Nuclear extracts were prepared from HEC-1B cells that were either untreated or infected with NDV. The nuclear extracts were incubated in the absence of added antibody (Fig. 3A, lanes 1 and 4), in the presence of control antibody (lanes 2 and 5), or in the presence of specific anti–IRF-3 antibody (lanes 3 and 6), and DRAF1 was subsequently analyzed by an EMSA. The addition of control antibody had no effect on DRAF1 (Fig. 3A, lane 5), while the specific anti–IRF-3 antibody completely abrogated its appearance (lane 6). The inclusion of a GST–IRF-3 fusion protein in the DNA-binding reaction could reverse the effect of the anti–IRF-3 antibody on the DRAF1 complex (Fig. 3B, lanes 5 and 6), but a GST-p48 fusion protein had no effect (lane 7). In addition, the depletion of IRF-3 from nuclear extracts by immunoprecipitation also depleted DRAF1 activity (data not shown). These results indicate that IRF-3 is a subunit of DRAF1.
FIG. 3.
(A) Effect of antibody to IRF-3 on DRAF1. Nuclear extracts from untreated HEC-1B cells (lanes 1 to 3) or cells infected with NDV (lanes 4 to 6) were used in an EMSA with a radiolabeled ISRE probe. Prior to the addition of the probe, either no antisera (Ab) (lanes 1 and 4), control murine antisera (c) (1:50 dilution) (lanes 2 and 5), or specific murine antisera to IRF-3 (1:50 dilution) (lanes 3 and 6) were added to the DNA-binding reaction mixtures. (B) Inhibition by GST–IRF-3 fusion protein of anti–IRF-3 antibody effects. Nuclear extracts from untreated HEC-1B cells (lane 1) or cells infected with NDV (lanes 2 to 7) were used in an EMSA. Prior to the addition of the probe, either 1 μg of control rabbit antibody (Ab) (lane 3) or 1 μg of rabbit antibody to IRF-3 (lanes 4 to 7) was added. Antibody incubation was done in the absence of added GST fusion protein (lanes 3 and 4), in the presence of 200 ng (lane 5) and 600 ng (lane 6) of GST–IRF-3 (amino acids 107 to 208), or in the presence of 200 ng of GST-p48 (amino acids 103 to 225) (lane 7). (C) Effect of NDV infection on the ability of IRF-3 to bind to the ISRE. Whole-cell extracts were prepared from HEC-1B cells that were either untreated (lanes 1 and 3) or infected with NDV for 6 h (lanes 2 and 4). The extracts were incubated with ISRE-Sepharose beads (lanes 1 and 2) or with rabbit antibody to IRF-3 (lanes 3 and 4). The bound proteins were separated by SDS-PAGE and detected by immunoblotting (IB) with a murine antibody (α) to IRF-3. IP, immunoprecipitation. Positions of prestained molecular mass standards are shown in kilodaltons on the left.
Partial purification of DRAF1 and UV cross-linking to DNA previously had revealed a DNA-binding component of approximately 60 to 70 kDa (11). To determine if IRF-3 could be this DNA-binding subunit, we tested the binding of IRF-3 to the ISRE either before or after NDV infection by use of an immobilized ISRE affinity resin (Fig. 3C). Cytoplasmic and nuclear protein extracts were prepared and combined to represent total cellular protein. Cells were either untreated or infected with NDV, and extracts were incubated with ISRE-Sepharose beads. Proteins bound to ISRE-Sepharose were analyzed by SDS-PAGE and immunoblotting with antibody to IRF-3. The results indicated that IRF-3 was capable of binding to ISRE-Sepharose only after infection with virus (Fig. 3C, lane 2). This difference in binding capability was not due to different levels of IRF-3 in the two extracts, since a control immunoprecipitation showed equivalent levels of IRF-3 protein (Fig. 3C, lanes 3 and 4). An increase in the apparent molecular mass of IRF-3 was also noted following NDV infection. IRF-3 isolated from untreated cells migrated in SDS-PAGE with a molecular mass of approximately 62 kDa and shifted to approximately 64 kDa after infection.
IRF-3 exists in the cytoplasm and translocates to the nucleus following viral infection.
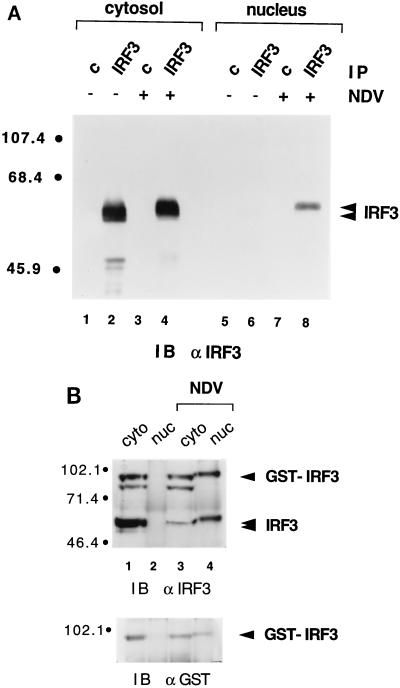
We previously demonstrated that DRAF1 activation occurs in the absence of new protein synthesis (9). Proteins comprising DRAF1 must therefore preexist within the cell. To determine IRF-3 cellular localization, cytoplasmic and nuclear extracts were prepared from HEC-1B cells that were either untreated or infected with NDV. The protein extracts were normalized for cell equivalents and assayed for IRF-3 by immunoprecipitation and immunoblotting (Fig. 4A). In uninfected cells, IRF-3 was found entirely in the cytosolic fraction (Fig. 4A, compare lanes 2 and 6). However, after viral infection, IRF-3 could be detected in the nuclear fraction, indicating that a nuclear translocation event had taken place (Fig. 4A, lane 8). The same cytoplasm-to-nucleus translocation could be seen with a GST–IRF-3 fusion protein stably expressed in cells (Fig. 4B). Following infection, the endogenous IRF-3 protein or the transfected GST–IRF-3 protein displayed slower migration in SDS-PAGE. The modification responsible for the alteration in migration appears to be a prerequisite for IRF-3 translocation to the nucleus and subsequent DNA binding.
FIG. 4.
(A) Cellular localization of IRF-3 in control and virus-infected cells. Nuclear and cytoplasmic extracts were prepared from untreated HEC-1B cells (lanes 1, 2, 5, and 6) or cells infected with NDV for 6 h (lanes 3, 4, 7, and 8). Extracts were normalized for cell equivalents and subjected to immunoprecipitation (IP) with control rabbit antibody (c) (lanes 1, 3, 5, and 7) or rabbit antibody to IRF-3 (lanes 2, 4, 6, and 8). The immunoprecipitated proteins were separated by SDS-PAGE and detected by immunoblotting (IB) with a murine antibody (α) to IRF-3. Numbers at left are in kilodaltons. (B) Cellular localization of a transfected GST–IRF-3 chimera. Nuclear (nuc) and cytoplasmic (cyto) extracts were prepared from untreated HT1080/GST–IRF-3 cells (lanes 1 and 2) or cells infected with NDV for 6 h (lanes 3 and 4). Extracts were normalized for cell equivalents and subjected to immunoprecipitation with a rabbit antibody to IRF-3. The immunoprecipitated proteins were separated by SDS-PAGE and detected by immunoblotting (IB) with a murine antibody (α) to IRF-3 (upper panel). The blot was subsequently stripped and reprobed with a rabbit antibody (α) to GST (lower panel).
Serine phosphorylation of IRF-3 increases following NDV infection.
A possible modification that could cause IRF-3 to exhibit slower migration in SDS-PAGE after viral infection is phosphorylation. To analyze the phosphorylation state of IRF-3, HEC-1B cells were labeled with [32P]orthophosphate during either mock infection or infection with NDV (Fig. 5A). Whole-cell lysates were prepared and subjected to immunoprecipitation with either a control antibody (Fig. 5A, lanes 1, 3, and 5) or antibody to IRF-3 (lanes 2, 4, and 6). In the absence of viral infection, IRF-3 was modified by phosphorylation at a low level (Fig. 5A, lane 2). At 3 h postinfection, the level of IRF-3 phosphorylation increased 1.5-fold (Fig. 5A, lane 4), and after 6 h, the phosphorylation level increased approximately 3-fold over the basal level (lane 6). Since the relative levels of IRF-3 were the same (Fig. 5A, lower panel), the results indicated that IRF-3 protein phosphorylation increased. This finding correlates with the characteristic slower migration.
FIG. 5.
(A) Increase in IRF-3 phosphorylation during viral infection. Whole-cell extracts were prepared by detergent lysis of HEC-1B cells that were metabolically labeled with [32P]orthophosphate for a total of 7 h during either mock infection (0 h) (lanes 1 and 2) or infection with NDV for 3 h (lanes 3 and 4) or 6 h (lanes 5 and 6). Extracts were subjected to immunoprecipitation (IP) with a control murine antibody (c) (lanes 1, 3, and 5) or a murine anti–IRF-3 antibody (lanes 2, 4, and 6). Immunoprecipitated proteins were denatured in SDS sample buffer, and 75% of the sample was separated by SDS-PAGE. The gel was fixed, dried, and exposed to film (upper panel). The remaining 25% was analyzed by SDS-PAGE and immunoblotting (IB) with a rabbit polyclonal antibody (α) to IRF-3 (lower panel). The positions of the prestained molecular mass standards are shown in kilodaltons on the left. (B) Effect of protein phosphatase treatment on the migration of IRF-3 in SDS-PAGE. Cytoplasmic (C) and nuclear (N) extracts were prepared from HEC-1B cells that were either untreated (lanes 1 and 2) or infected with NDV (lanes 3 to 7). A total of 100 μg of cytoplasmic extract or 35 μg of nuclear extract was incubated in the absence of phosphatase (lanes 1 to 3), in the presence of PP2A (lanes 4 and 5), or in the presence of PTP1B (PTP) (lanes 6 and 7). The activities of PP2A and PTP1B were inhibited by the inclusion of 10 nM okadaic acid (OA) (lane 5) and 5 mM sodium vanadate (van) (lane 7), respectively. Proteins were separated by SDS-PAGE and detected by immunoblotting (IB) with a murine antibody (α) to IRF-3. (C) Phosphoamino acid analysis of 32P-labeled IRF-3. Cells were metabolically labeled with [32P]orthophosphate, and lysates from mock-infected cells (upper panels) or NDV-infected cells (lower panels) were immunoprecipitated (IP) with control (c) antibody (boxes 1 and 3) or antibody to IRF-3 (boxes 2 and 4). Following SDS-PAGE, proteins were electroblotted to Immobilon-P membranes, and the area containing IRF-3 or the adjacent area from control immunoprecipitations was subjected to partial acid hydrolysis. The hydrolysates were resolved by two-dimensional electrophoresis on a TLC plate. Positions of the serine, threonine, and tyrosine phosphoamino acid internal standards are indicated by dotted circles (pS, pT, and pY, respectively), and origins are denoted by a plus sign.
To determine the type of amino acid phosphorylation of IRF-3 after viral infection, the effects of protein phosphatases were evaluated (Fig. 5B). Extracts were prepared from untreated or infected HEC-1B cells and incubated in the absence or presence of protein phosphatases to analyze IRF-3 migration. Cytoplasmic IRF-3 from control cells migrated at its normal position (Fig. 5B, lane 1). Nuclear extracts from control cells contained no detectable IRF-3 (Fig. 5B, lane 2). Following viral infection, IRF-3 with the characteristic slower migration appeared in nuclear extracts (Fig. 5B, lane 3). Treatment of nuclear IRF-3 with a serine- and threonine-specific phosphatase, PP2A, resulted in a return of IRF-3 to its original migration position prior to infection (Fig. 5B, lane 4). This result was specific for PP2A activity, since inclusion of okadaic acid inhibited the effect (Fig. 5B, lane 5). In contrast, treatment of nuclear IRF-3 with a tyrosine phosphatase, PTP1B, had no effect on the migration of IRF-3 (Fig. 5B, lane 6). This result indicates that IRF-3 undergoes an increase in serine and/or threonine phosphorylation after viral infection that is responsible for the reduction in the relative migration observed in SDS-PAGE.
Since treatment of DRAF1 with PTP1B inhibits its ability to bind to DNA (Fig. 2A), it remained possible that IRF-3 was modified by tyrosine phosphorylation. To further investigate the type of phosphorylation modifying IRF-3, we performed a phosphoamino acid analysis (Fig. 5C). HEC-1B cells were metabolically labeled with [32P]orthophosphate during either mock infection or infection with NDV. Whole-cell lysates were subjected to immunoprecipitation, SDS-PAGE, and electroblotting to an Immobilon-P membrane. The 32P-labeled IRF-3 protein was excised from the membrane, as was the adjacent area from the control immunoprecipitation. Proteins from the four membrane pieces were subjected to partial acid hydrolysis for various times, and the products were separated by two-dimensional TLC. A representative TLC autoradiogram is shown in Fig. 5C. IRF-3 immunoprecipitated from mock-infected cells clearly showed the presence of phosphoserine (Fig. 5C, box 2). Following NDV infection, the amount of phosphoserine in IRF-3 increased and the presence of phosphothreonine was also detected (Fig. 5C, box 4). The phosphoserine content of IRF-3 appeared to increase after infection by approximately threefold, consistent with the increase in phosphate incorporation in IRF-3 revealed by immunoprecipitation (Fig. 5A). No phosphotyrosine was detected in IRF-3 either before or after viral infection.
Activation of DRAF1 in PKR-deficient cells.
Since serine phosphorylation is involved in the modification of IRF-3 and DRAF1 in response to virus or dsRNA, the function of a dsRNA-activated kinase is indicated. For this reason, a potential role of the dsRNA-dependent protein kinase (PKR) in this signal pathway was evaluated. PKR is a serine and threonine protein kinase that becomes activated by binding to dsRNA (reviewed in references 27 and 29). It has been identified as a signal-transducing kinase in the activation of transcription factor NF-κB and has been implicated in IFN induction, the antiviral response, and the regulation of cell growth (30, 32, 33, 37, 57).
To determine whether PKR functions in the activation of IRF-3 and DRAF1, cells lacking PKR were tested for DRAF1 induction. Mice deficient in functional PKR (Pkr0/0) were generated by targeted gene disruption (57). MEFs derived from these mice (Pkr0/0) and from parental wild-type mice (Pkr+/+) were infected with NDV. Nuclear extracts were prepared and assayed for the appearance of DRAF1 activity by an EMSA (Fig. 6). Both DRAF1 and ISGF3 were activated in the Pkr+/+ and Pkr0/0 MEFs following NDV infection and, more significantly, the levels were similar in both cell types. The ISGF3 activity was likely due to autocrine IFN production during viral infection. These results suggest that PKR is not required for DRAF1 formation and binding to the ISRE.
FIG. 6.
Activation of DRAF1 in Pkr0/0 MEFs. Wild-type (Pkr+/+) MEFs or PKR-deficient (Pkr0/0) MEFs were either untreated (lanes 1 and 4) or infected with NDV for 3 h (lanes 2, 3, 5, and 6). Nuclear extracts were prepared and analyzed by an EMSA with a radiolabeled ISRE probe. The specificity of the protein-DNA complexes was shown by competition with a 100-fold excess of unlabeled ISRE oligonucleotide (lanes 3 and 6).
Association of IRF-3 with CBP and p300 following viral infection.
A previous study indicated that IRF-3 did not function as an independent transcriptional activator (1). The DNA-binding domain of a heterologous protein (GAL4) was fused to IRF-3, and the chimera was tested for GAL4-targeted gene expression after viral infection. The GAL4–IRF-3 fusion protein did not activate transcription, suggesting that IRF-3 may interact with other factors to elicit gene regulation. For this reason, we analyzed the ability of IRF-3 to physically interact with other cellular proteins following viral infection.
Untreated or NDV-infected HEC-1B cells were metabolically labeled with [35S]methionine-[35S]cysteine, and extracts were analyzed for proteins that coimmunoprecipitated with IRF-3 (Fig. 7A). Following viral infection, a prominent radiolabeled band with an apparent molecular mass of approximately 300 kDa coimmunoprecipitated with IRF-3 (Fig. 7A, lane 4). The coimmunoprecipitated protein was not present in control antibody immunocomplexes (Fig. 7A, lanes 1 and 3) or in uninfected cells (lane 2). To establish that the coimmunoprecipitated protein was of cellular origin and was also present in other cells, an analysis was performed with HeLa cells (Fig. 7B). HeLa cells were either treated with dsRNA or infected with NDV and metabolically labeled with [35S]methionine-[35S]cysteine. Immunoprecipitations with anti–IRF-3 antibody again demonstrated a coimmunoprecipitated protein of approximately 300 kDa only following dsRNA treatment or viral infection (Fig. 7B, lanes 4 and 8). The same result was obtained following infection with dl312 adenovirus (data not shown).
FIG. 7.
(A) Association of IRF-3 with a 300-kDa cellular protein following HEC-1B cell infection with NDV. Detergent cell lysates were prepared from cells following metabolic labeling with [35S]methionine-[35S]cysteine during either mock infection (lanes 1 and 2) or infection with NDV for 6 h (lanes 3 and 4). Extracts were subjected to immunoprecipitation (IP) with a control rabbit antibody (c) (lanes 1 and 3) or a rabbit anti–IRF-3 antibody (lanes 2 and 4). The immunoprecipitated proteins were separated by SDS-PAGE and detected by fluorography. The positions of the prestained molecular mass standards are shown in kilodaltons on the left. (B) Presence in HeLa cells treated with dsRNA or infected with NDV of a 300-kDa cellular protein associated with IRF-3. Detergent cell lysates were prepared from cells that were metabolically labeled with [35S]methionine-[35S]cysteine. Cells were untreated (lanes 1 and 2), treated with 150 μg of poly(rI) · poly(rC) per ml for 2 h (lanes 3 and 4), mock infected (lanes 5 and 6), or infected with NDV for 6 h (lanes 7 and 8). The immunoprecipitation analysis was performed as described in panel A. (C) Association of the IRF-3-associated 300-kDa protein with the ISRE following virus infection. Whole-cell extracts were prepared from HEC-1B cells that were metabolically labeled with [35S]methionine-[35S]cysteine and either mock infected (lanes 1 and 2) or infected with NDV for 6 h (lanes 3 and 4). The extracts were incubated with DNA-Sepharose beads prepared with native ISRE (wt) (lanes 2 and 4) or with a mutated ISRE (mt) (lanes 1 and 3). The DNA-bound proteins were detected as described in panel A. (D) Copurification of the 300-kDa protein with stably expressed GST–IRF-3 fusion protein. Detergent cell lysates were prepared from HT1080/GST–IRF-3 cells that were metabolically labeled with [35S]methionine-[35S]cysteine and either mock infected (lane 1) or infected with NDV for 6 h (lane 2). Extracts were incubated with glutathione-agarose beads and washed, and bound proteins were separated and detected as described in panel A. Asterisks indicate migration of 300-kDa proteins.
Association of the 300-kDa cellular protein with IRF-3 could also be demonstrated with techniques that did not depend on a specific immunoprecipitating antibody. The presence of the 300-kDa protein in ISRE-DNA complexes was seen with the use of an immobilized ISRE affinity resin (as performed with IRF-3 in Fig. 3) (Fig. 7C). Cell lysates were prepared from control or NDV-infected cells metabolically labeled with [35S]methionine-[35S]cysteine, and proteins were incubated with either a native ISRE oligonucleotide linked to Sepharose beads or a mutated ISRE oligonucleotide linked to the beads. The IRF-3-associated 300-kDa protein was found in DNA complexes only with the native ISRE sequence and only following viral infection (Fig. 7C, lane 4). In addition, a GST–IRF-3 fusion protein stably expressed in HT1080 cells was tested for its ability to associate with the 300-kDa cellular protein (Fig. 7D). Cells expressing GST–IRF-3 were metabolically labeled with [35S]methionine-[35S]cysteine before or after infection. Cell lysates were prepared, and radiolabeled GST–IRF-3 was collected on glutathione beads. Analysis by SDS-PAGE demonstrated an association of the 300-kDa protein with GST–IRF-3 only following viral infection (Fig. 7C, lane 2).
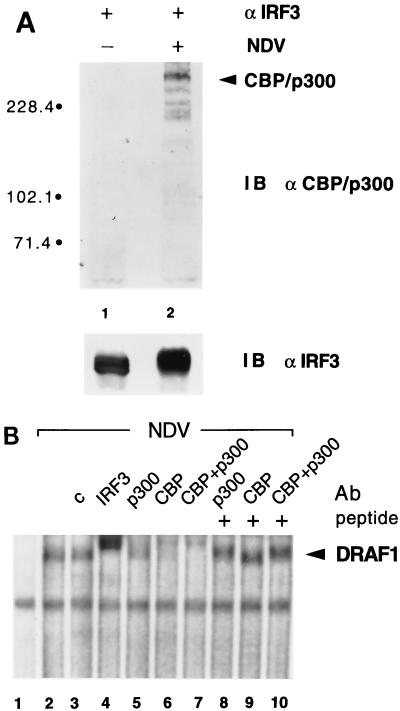
The molecular mass of the protein associated with IRF-3 prompted us to investigate the possibility of its identity with the transcriptional coactivators CBP and p300 (Fig. 8) (7, 17). An immunoprecipitation-immunoblotting assay was performed with control HEC-1B cells or cells infected with NDV (Fig. 8A). Proteins in the immunocomplexes with anti–IRF-3 antibody were separated by SDS-PAGE, transferred to an Immobilon-P membrane, and immunoblotted with antibodies to CBP and p300. Antibodies to CBP and p300 reacted with the coimmunoprecipitated protein following viral infection (Fig. 8A, lane 2). In addition to a prominent CBP/p300 protein band, there were several smaller reactive species. These may correspond to cleavage products that transfer more efficiently to the Immobilon-P membrane than the larger native molecules. The results demonstrated a physical interaction of IRF-3 with CBP and/or p300 following viral infection that is of sufficient affinity to resist detergent lysis of cells and standard immunoprecipitation techniques.
FIG. 8.
(A) Identification of the IRF-3-associated protein as CBP/p300. Detergent cell lysates were prepared from HEC-1B cells that were either mock infected (lane 1) or infected with NDV for 6 h (lane 2). Extracts were subjected to immunoprecipitation with rabbit anti–IRF-3 antibody (α). Immunoprecipitated proteins were separated by SDS-PAGE and detected by immunoblotting (IB) with a mixture of specific rabbit antibodies to p300 and CBP (upper panel) or with a murine antibody to IRF-3 (lower panel). Numbers at left are in kilodaltons. (B) Presence of CBP and p300 in the DRAF1 complex. Nuclear extracts from untreated HEC-1B cells (lane 1) or cells infected with NDV (lanes 2 to 10) were used in an EMSA with a radiolabeled ISRE probe. Prior to the addition of the probe, no antibody (Ab) (lanes 1 and 2), 2 μg of control rabbit antibody (c) (lane 3), 2 μg of specific rabbit antibody to IRF-3 (lane 4), 2 μg of specific rabbit antibody to p300 (lanes 5 and 8), 2 μg of specific rabbit antibody to CBP (lanes 6 and 9), or 1 μg of antibodies to both p300 and CBP (lanes 7 and 10) was added to the DNA-binding reaction mixtures. The effect of the anti-p300 and anti-CBP antibodies was blocked by the inclusion of 0.2 μg of control peptide(s) used as an immunogen to generate the antibodies (lanes 8 to 10).
To determine if the CBP and p300 molecules were associated with IRF-3 in the DRAF1 DNA-binding complex, the effects of specific antibodies to either CBP or p300 were evaluated (Fig. 8B). Nuclear extracts were prepared from uninfected or infected cells and used in an EMSA with the radiolabeled ISRE probe to display the DRAF1 complex. Antibodies were added to the DNA-binding reaction mixture to evaluate their ability to supershift or inhibit the appearance of DRAF1. Specific antibodies to either p300 or CBP partially inhibited the appearance of DRAF1 (Fig. 8B, lanes 5 and 6), and inclusion of both anti-CBP and anti-p300 antibodies to the binding reaction mixture completely eliminated the appearance of DRAF1 (lane 7). Similar amounts of control antibodies had no effect on DRAF1 (Fig. 8B, lane 3), and as previously demonstrated, anti–IRF-3 antibodies completely eliminated the appearance of DRAF1 (lane 4). These results confirm the presence of CBP, p300, and IRF-3 in the DRAF1 ISRE-binding complex and appear to indicate that either of the related molecules CBP and p300 can associate with IRF-3 following infection.
DISCUSSION
The cellular response to viral infection appears to result in the transcriptional induction of the IFN-α/β genes and in the direct induction of a subset of the ISGs (2, 9, 11, 36, 52–55). Viral induction of the ISGs is not dependent on autocrine IFN, since it can be seen in cells deficient in the response to IFN, such as HEC-1B cells (21, 51). We provided evidence previously that viral activation of a specific ISRE-binding factor, DRAF1, leads to the transcriptional induction of specific ISGs (9, 11). The activation of DRAF1 may be a general response to invading virus, since both a DNA virus (adenovirus) and an RNA virus (NDV) as well as dsRNA can activate DRAF1 (9, 11). Viral dsRNA appears to be generated during viral transcription or replication and to act as a mediator of DRAF1 activation. DRAF1 may function to activate antiviral genes, antiproliferative genes, and/or apoptosis. Since the appearance of DRAF1 precedes the autocrine IFN-induced appearance of ISGF3 in response to IFN, DRAF1 may afford the host some measure of defense to infection (9). Other investigators have described the appearance of another ISRE-binding factor, activated in response to vesicular stomatitis virus, VIBP (5). The relationship of this factor to DRAF1 or to another factor that we have detected in some cells, DRAF2, remains to be determined (9, 11).
To begin to elucidate the mechanism by which DRAF1 is activated, we investigated the component nature of DRAF1. IRF molecules are capable of recognizing an inner core sequence of the ISRE (25, 38, 40, 42, 50), so we turned our attention to IRFs and specifically to IRF-3. The presence of IRF-3 in DRAF1 was directly tested with specific antibody to IRF-3 in a DNA-binding reaction. Antibody to IRF-3 eliminated the appearance of DRAF1, demonstrating the presence of IRF-3 in the complex. IRF-3 appears to reside exclusively in the cytoplasm of cells and to translocate to the nucleus following viral infection. Scanning the primary amino acid sequence of IRF-3 does not reveal a consensus nuclear localization sequence (NLS) (reviewed in references 15 and 45). In this respect, IRF-3 is distinct from some IRF family members, such as IRF-1, which contain a putative NLS and localize to the nuclear compartment (31). However, it is possible that IRF-3 does contain a functional NLS that is masked prior to infection. IRF-3 may also contain a nuclear export signal (NES) that is functional only prior to infection (reviewed in reference 49). The mechanisms that control IRF-3 cellular localization remain to be determined and could depend on serine phosphorylation regulating an NLS or an NES or interactions with as-yet-unidentified factors. Since tyrosine phosphorylation appears to be required for DRAF1 DNA binding and IRF-3 does not contain phosphotyrosine, another DRAF1 subunit may be tyrosine phosphorylated.
Identification of IRF-3 as a subunit of DRAF1 allowed us to use coimmunoprecipitation assays to search for IRF-3-associated proteins. Metabolic radiolabeling of cells revealed a coimmunoprecipitated protein with an apparent molecular mass of approximately 300 kDa. Use of characterized antibodies revealed the presence of the transcriptional coactivators CBP and p300 in immunocomplexes with IRF-3 only following viral infection and in the DRAF1 DNA-binding complex. The CBP and p300 coactivators have been shown to associate with a growing number of transcription factors (reviewed in references 18, 23, and 46). In the IFN signal pathway, CBP and p300 molecules interact with the STAT1 and STAT2 subunits of ISGF3 (4, 26, 48, 58). CBP and p300 molecules do not bind to DNA directly but have several properties that may contribute to their role in transcriptional activation: physical association with basal transcription factors, intrinsic acetyltransferase activity that appears to play a role in histone acetylation and chromatin conformation, and acetylation of specific transcription factors (3, 24, 39, 46). IRF-3 nuclear translocation and association with CBP and p300 in response to viral infection lead to the generation of DRAF1 and the activation of a subset of ISGs (11). In addition, it is possible that DRAF1 is involved in the induction of one or more of the type I IFN genes. DRAF1 may function to activate genes that play a role in host survival via either known antiviral and antiproliferative mechanisms or as-yet-unidentified mechanisms.
ACKNOWLEDGMENTS
We thank Christopher Daly for providing unpublished observations critical to the completion of the manuscript. We also thank all the members of our laboratory for help and suggestions, Carrie Mahlum and Emily Huang for technical assistance, and Dianna Berry for assisting in the production of antibodies to IRF-3. The gifts of PTP1B from Nicholas K. Tonks and PKR-deficient MEFs from Bryan R. G. Williams and Charles Weissmann are greatly appreciated. We also thank Joav Prives and Dafna Bar-Sagi for assistance with the phosphoamino acid analyses and Michael Katze for helpful discussions concerning PKR. Recombinant human IFN-α was a gift from Hoffmann-La Roche Inc.
This work was supported by grants from the National Institutes of Health (RO1CA50773 and PO1CA28146) and The Council for Tobacco Research (3717) to N.C.R.
REFERENCES
- 1.Au W-C, Moore P A, Lowther W, Juang Y-T, Pitha P M. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc Natl Acad Sci USA. 1995;92:11657–11661. doi: 10.1073/pnas.92.25.11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bandyopadhyay S K, Leonard G T, Jr, Bandyopadhyay T, Stark G R, Sen G C. Transcriptional induction by double-stranded RNA is mediated by interferon-stimulated response elements without activation of interferon-stimulated gene factor 3. J Biol Chem. 1995;270:19624–19629. doi: 10.1074/jbc.270.33.19624. [DOI] [PubMed] [Google Scholar]
- 3.Bannister A J, Kourarides T. The CBP coactivator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D’Andrea A, Livingston D M. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-α. Nature. 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 5.Bovolenta C, Lou J, Kanno Y, Park B-K, Thornton A M, Coligan J E, Schubert M, Ozato K. Vesicular stomatitis virus infection induces a nuclear DNA-binding factor specific for the interferon-stimulated response element. J Virol. 1995;69:4173–4181. doi: 10.1128/jvi.69.7.4173-4181.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle W J, van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- 7.Chrivia J C, Kwok R P S, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 8.Cooper J A, Sefton B M, Hunter T. Detection and quantification of phosphotyrosine in proteins. Methods Enzymol. 1983;99:387–402. doi: 10.1016/0076-6879(83)99075-4. [DOI] [PubMed] [Google Scholar]
- 9.Daly C, Reich N C. Double-stranded RNA activates novel factors that bind to the interferon-stimulated response element. Mol Cell Biol. 1993;13:3756–3764. doi: 10.1128/mcb.13.6.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daly C, Reich N C. Receptor to nucleus signaling via tyrosine phosphorylation of the p91 transcription factor. Trends Endocrinol Metab. 1994;5:159–164. doi: 10.1016/1043-2760(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 11.Daly C, Reich N C. Characterization of specific DNA-binding factors activated by double-stranded RNA as positive regulators of interferon-α/β-stimulated genes. J Biol Chem. 1995;270:23739–23746. doi: 10.1074/jbc.270.40.23739. [DOI] [PubMed] [Google Scholar]
- 12.Darnell J E, Jr, Kerr I M, Stark G R. JAK-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 13.DeMaeyer E, DeMaeyer-Guignard J. Interferons and other regulatory cytokines. New York, N.Y: Wiley-Interscience; 1988. [Google Scholar]
- 14.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biol Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 16.Driggers P H, Ennist D L, Gleason S L, Mak W, Marks M S, Levi B, Flanagan J R, Appella E, Ozato K. An interferon gamma-regulated protein that binds the interferon-inducible enhancer element of major histocompatibility complex class I genes. Proc Natl Acad Sci USA. 1990;87:3743–3747. doi: 10.1073/pnas.87.10.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;10:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 18.Eckner R. p300 and CBP as transcriptional regulators and targets of oncogenic events. Biol Chem. 1996;377:685–688. [PubMed] [Google Scholar]
- 19.Eisenbeis C F, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- 20.Fu X-Y, Schindler C, Improta T, Aebersold R, Darnell J E., Jr The proteins of ISGF-3, the interferon α-induced transcriptional activator, define a gene family involved in signal transduction. Proc Natl Acad Sci USA. 1992;89:7840–7843. doi: 10.1073/pnas.89.16.7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuse A, Ashino-Fuse H, Kuwata T. Binding of 125I-labeled human interferon to cell lines with low sensitivity to interferon. Gann. 1984;75:379–384. [PubMed] [Google Scholar]
- 22.Gilmour K C, Reich N C. Signal transduction and activation of gene transcription by interferons. Gene Expression. 1995;5:1–18. [PMC free article] [PubMed] [Google Scholar]
- 23.Goldman P S, Tran V K, Goodman R H. The multifunctional role of the co-activator CBP in transcriptional regulation. Recent Prog Horm Res. 1997;52:103–119. [PubMed] [Google Scholar]
- 23a.Grossman, A., H. W. Mittrücker, L. Lantonio, and T. W. Mak. Unpublished data.
- 24.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:607–614. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 25.Harada H, Fujita T, Miyamoto M, Kimura Y, Maruyama M, Furia A, Miyata T, Taniguchi T. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell. 1989;58:729–739. doi: 10.1016/0092-8674(89)90107-4. [DOI] [PubMed] [Google Scholar]
- 26.Horvai A E, Xu L, Korzus E, Brard G, Kalafus D, Mullen T-M, Rose D W, Rosenfeld M G, Glass C K. Nuclear integration of JAK/STAT and Ras/AP1 signaling by CBP and p300. Proc Natl Acad Sci USA. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hovanessian A G. The double stranded RNA activated protein kinase induced by interferon: dsRNA-PK. J Interferon Res. 1989;9:641–647. doi: 10.1089/jir.1989.9.641. [DOI] [PubMed] [Google Scholar]
- 28.Ihle J N, Witthuhn B A, Quelle F W, Yamamoto K, Silvennoinen O. Signaling through the hematopoietic cytokine receptors. Annu Rev Immunol. 1995;13:369–398. doi: 10.1146/annurev.iy.13.040195.002101. [DOI] [PubMed] [Google Scholar]
- 29.Katze M. Regulation of the interferon-induced PKR: can viruses cope? Trends Microbiol. 1995;3:75–78. doi: 10.1016/s0966-842x(00)88880-0. [DOI] [PubMed] [Google Scholar]
- 30.Koromilas A E, Roy S, Barber G N, Katze M G, Sonenberg N. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science. 1992;257:1685–1689. doi: 10.1126/science.1382315. [DOI] [PubMed] [Google Scholar]
- 31.Köster M, Schaper F, Kirchhoff S, Hauser H. Intracellular localization of STAT proteins: analysis of nuclear translocation in living cells. Eur Cytokine Netw. 1996;7:520. [Google Scholar]
- 32.Kumar A, Haque J, Lacoste J, Hiscott J, Williams B R G. Double-stranded RNA-dependent protein kinase activates transcription factor NF-κB by phosphorylating IκB. Proc Natl Acad Sci USA. 1994;91:6288–6292. doi: 10.1073/pnas.91.14.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar A, Yang Y-L, Flati V, Der S, Kadereit S, Deb A, Haque J, Reis L, Weissmann C, Williams B R G. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-κB. EMBO J. 1997;16:406–416. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levy D E, Kessler D S, Pine R, Darnell J E., Jr Cytoplasmic activation of ISGF3, the positive regulator of interferon-α-stimulated transcription, reconstituted in vitro. Genes Dev. 1989;3:1362–1371. doi: 10.1101/gad.3.9.1362. [DOI] [PubMed] [Google Scholar]
- 35.Matsuyama T, Grossman A, Mittrücker H-W, Siderovski D P, Kiefer F, Kawakami T, Richardson C D, Taniguchi T, Yoshinaga S K, Mak T W. Molecular cloning of LSIRF, a lymphoid-specific member of the interferon regulatory factor family that binds the interferon-stimulated response element (ISRE) Nucleic Acids Res. 1995;23:2127–2136. doi: 10.1093/nar/23.12.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Memet S, Besancon F, Bourgeade M-F, Thang M N. Direct induction of interferon-γ- and interferon-α/β-inducible genes by double-stranded RNA. J Interferon Res. 1991;11:131–141. doi: 10.1089/jir.1991.11.131. [DOI] [PubMed] [Google Scholar]
- 37.Meurs E, Chong K, Galabru J, Shaun N, Thomas B, Kerr I M, Williams B R G, Hovanessian A G. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- 38.Miyamoto M, Fugita T, Kimura Y, Maruyama M, Harada H, Sudo Y, Miyata T, Taniguchi T. Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to the IFN-β gene regulatory elements. Cell. 1988;54:903–913. doi: 10.1016/s0092-8674(88)91307-4. [DOI] [PubMed] [Google Scholar]
- 39.Ogryzko V V, Schlitz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivator p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–960. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 40.Pine R, Decker T, Kessler D S, Levy D E, Darnell J E., Jr Purification and cloning of interferon-stimulated gene factor 2 (ISGF2): ISGF2 (IRF-1) can bind to the promoters of both beta interferon and interferon-stimulated genes but is not a primary transcriptional activator of either. Mol Cell Biol. 1990;10:2448–2457. doi: 10.1128/mcb.10.6.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reich N, Evans B, Levy D, Fahey D, Knight E, Jr, Darnell J E., Jr Interferon-induced transcription of a gene encoding a 15-kDa protein depends on an upstream enhancer element. Proc Natl Acad Sci USA. 1987;84:6394–6398. doi: 10.1073/pnas.84.18.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reich N C, Darnell J E., Jr Differential binding of interferon-induced factors to an oligonucleotide that mediates transcriptional activation. Nucleic Acids Res. 1989;17:3415–3424. doi: 10.1093/nar/17.9.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schindler C, Fu X-Y, Improta T, Aebersold R, Darnell J E., Jr Proteins of transcription factor ISGF-3: one gene encodes the 91- and 84-kDa ISGF-3 proteins that are activated by interferon α. Proc Natl Acad Sci USA. 1992;89:7836–7839. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schindler C, Darnell J E., Jr Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 45.Schlenstedt G. Protein import into the nucleus. FEBS Lett. 1996;389:75–79. doi: 10.1016/0014-5793(96)00583-2. [DOI] [PubMed] [Google Scholar]
- 46.Shikama N, Lyon J, La Thangue N B. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol. 1997;7:230–236. doi: 10.1016/S0962-8924(97)01048-9. [DOI] [PubMed] [Google Scholar]
- 47.Taniguchi T, Harada H, Lampier M. Regulation of the interferon system and cell growth by the IRF transcription factors. J Cancer Res Clin Oncol. 1995;121:516–520. doi: 10.1007/BF01197763. [DOI] [PubMed] [Google Scholar]
- 48.Torchia J, Rose D W, Inostroza J, Kamel Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 49.Ullman K S, Powers M A, Forbes D J. Nuclear export receptors: from importin to exportin. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. [DOI] [PubMed] [Google Scholar]
- 50.Veals S A, Schindler C, Leonard D, Fu X-Y, Aebersold R, Darnell J E, Jr, Levy D E. Subunit of an alpha-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding proteins. Mol Cell Biol. 1992;12:3315–3324. doi: 10.1128/mcb.12.8.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verhaegen M, Divizia M, Vandenbussche P, Kuwata T, Content J. Abnormal behavior of interferon-induced enzymatic activities in an interferon-resistant cell line. Proc Natl Acad Sci USA. 1980;77:4479–4483. doi: 10.1073/pnas.77.8.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wathelet M G, Clauss I M, Nols C B, Content J, Huez G A. New inducers revealed by the promoter sequence analysis of two interferon-activated human genes. Eur J Biochem. 1987;169:313–321. doi: 10.1111/j.1432-1033.1987.tb13614.x. [DOI] [PubMed] [Google Scholar]
- 53.Wathelet M G, Clauss I M, Content J, Huez G A. Regulation of two interferon-inducible genes by interferon, poly (rI) · poly (rC) and viruses. Eur J Biochem. 1988;174:323–329. doi: 10.1111/j.1432-1033.1988.tb14101.x. [DOI] [PubMed] [Google Scholar]
- 54.Wathelet M G, Berr P M, Huez G A. Regulation of gene expression by cytokines and virus in human cells lacking the type-I interferon locus. Eur J Biochem. 1992;206:901–910. doi: 10.1111/j.1432-1033.1992.tb16999.x. [DOI] [PubMed] [Google Scholar]
- 55.Wu C, Ohimori Y, Bandyopadhyay S, Sen G, Hamilton T. Interferon-stimulated response element and NFκB sites cooperate to regulate double-stranded RNA-induced transcription of the IP-10 gene. J Interferon Res. 1994;14:357–363. doi: 10.1089/jir.1994.14.357. [DOI] [PubMed] [Google Scholar]
- 56.Yamagata T, Nishida J, Tanaka T, Sakai R, Mitani K, Yoshida M, Taniguchi T, Yazaki Y, Hirai H. A novel interferon regulatory factor family transcription factor, ICSAT/Pip/LSIRF, that negatively regulates the activity of interferon-regulated genes. Mol Cell Biol. 1996;16:1283–1294. doi: 10.1128/mcb.16.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Y-L, Reis L F L, Pavlovic J, Aguzzi A, Schäfer R, Kumar A, Williams B R G, Aguet M, Weissmann C. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 1995;14:6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang J J, Vinkemeier U, Guo W, Chakaravarti D, Horvath C, Darnell J E., Jr Two regions between Stat1 and CBP/p300 in interferon γ signaling. Proc Natl Acad Sci USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang L, Pagano J S. IRF-7, a new interferon regulatory factor associated with Epstein-Barr virus latency. Mol Cell Biol. 1997;17:5748–5757. doi: 10.1128/mcb.17.10.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]