Abstract
Transformation by simian virus 40 large T antigen (TAg) is dependent on the inactivation of cellular tumor suppressors. Transformation minimally requires the following three domains: (i) a C-terminal domain that mediates binding to p53; (ii) the LXCXE domain (residues 103 to 107), necessary for binding to the retinoblastoma tumor suppressor protein, pRB, and the related p107 and p130; and (iii) an N-terminal domain that is homologous to the J domain of DnaJ molecular chaperone proteins. We have previously demonstrated that the N-terminal J domain of TAg affects the RB-related proteins by perturbing the phosphorylation status of p107 and p130 and promoting the degradation of p130 and that this domain is required for transformation of cells that express either p107 or p130. In this work, we demonstrate that the J domain of TAg is required to inactivate the ability of each member of the pRB family to induce a G1 arrest in Saos-2 cells. Furthermore, the J domain is required to override the repression of E2F activity mediated by p130 and pRB and to disrupt p130-E2F DNA binding complexes. These results imply that while the LXCXE domain serves as a binding site for the RB-related proteins, the J domain plays an important role in inactivating their function.
Simian virus 40 (SV40) large T antigen (TAg) can transform a variety of cell types. Manifestations of the transformed phenotype include cell immortalization, growth to a high density, reduced requirement for serum, anchorage independence, and the ability to form tumors in various animal models. TAg achieves this transformation by targeting negative regulators of cell growth, including p53 and the RB family (pRB, p107, and p130). p53 and pRB are well-established tumor suppressor proteins, and their corresponding genes are commonly lost or mutated in human cancer. While p107 and p130 inactivation likely contributes to TAg-mediated transformation (see below), there is little evidence that these proteins are tumor suppressors. However, loss of p130 was recently reported in a human lung cancer cell line (12).
TAg contains at least three transforming domains. A C-terminal domain extending from approximately residue 350 to residue 550 binds to and inactivates p53 (22, 31, 42, 52). The LXCXE domain (residues 103 to 107) mediates binding to the retinoblastoma family proteins pRB, p107, and p130 (5, 6, 9, 19, 51). Mutations within TAg that disrupt binding to p53 or pRB render it unable to fully transform cells (46). An intact LXCXE domain is required for TAg to transform fibroblasts derived from Rb-1 knockout mice (3, 51). This result implies that p130 and p107, in addition to pRB, are likely to be relevant targets of TAg during the transforming process. In addition to the LXCXE and p53 binding domains, the N-terminal 82 residues of TAg, encoded by the first exon and shared with small t antigen, are required for transformation (28). Until recently, the mechanism by which the N terminus contributes to transformation was unknown.
The N terminus of TAg shares sequence homology with the J domain of the DnaJ (heat shock protein 40 [Hsp40]) family of molecular chaperones (20). The J domain consists of approximately 70 residues that bind to and stimulate the ATPase activity of specific Hsp70/DnaK family members (47). The residues histidine-proline-aspartate (HPD) are absolutely conserved within the J domain of all known DnaJ homologs (reviewed in reference 39). Substitution mutations in any of these residues render the J domain defective in activating Hsp70. Notably, all known polyomavirus large TAg homologs contain the residues HPD within the first exon (32). Several lines of evidence suggest that the N terminus of TAg behaves as a J domain. First, like cellular DnaJ proteins, SV40 TAg can bind specifically to a member of the Hsp70 family of heat shock proteins (36). Second, point mutations in the highly conserved HPD residues within the N-terminal J-domain homology region of TAg disrupt binding to Hsc70 (2). Furthermore, in an in vitro assay, the N termini of SV40 TAg and small t antigens were able to stimulate the ATPase activity of a variety of Hsp70 homologs (41). Finally, the J domains of SV40, JC virus, and BK virus could each functionally substitute for the J domain of Escherichia coli DnaJ and restore the ability of the host cell to form bacteriophage lambda plaques (21). Collectively, these results strongly suggest that the N termini of the polyomavirus large TAgs function as J domains.
We have previously demonstrated that the J domain of TAg mediates a perturbation of the phosphorylation status of p130 and p107 and induces rapid turnover of p130 (44, 45). p130 and p107, like pRB, are normally phosphorylated in a cell cycle-dependent manner in the mid- to late G1 phase (1, 26, 50). In cells expressing TAg, the normal cell cycle-dependent phosphorylation of p130 and p107 is disrupted, and only the fastest-migrating species of p130 and p107 can be detected. These effects of TAg are absolutely dependent upon an intact J domain; they are abolished by point mutations in the conserved HPD motif and are restored when the N terminus of TAg is replaced with a cellular J domain (44).
Our previous studies also indicated that the J domain of TAg contributed to TAg-mediated transformation. Mouse embryo fibroblasts (MEFs) expressing TAg with single-amino-acid substitutions in the conserved HPD motif of the J domain were unable to grow to a high cell density or in media containing reduced (1%) serum (44). However, replacement of the N terminus of TAg with intact J domains from human DnaJ homologs restored the ability of TAg to fully transform normal MEFs (44). In a separate study, TAg constructs with deletions within the N terminus or substitutions in the HPD motif were also unable to transform C3H10T1/2 and REF52 cells (41).
The studies done to date indicate that the J domain of TAg contributes to transformation and affects the phosphorylation status and stability of RB family proteins. Based on this, we considered that the J domain may contribute to other effects of TAg on the RB family. Overexpression of pRB, p107, and p130 can induce an arrest in the G1 phase of the cell cycle in a number of cell lines (4, 14, 53, 54). This G1 arrest can be overcome by expression of E2F. It can also be overcome by expression of cyclins and cdks that phosphorylate and inactivate pRB family proteins and by the viral oncoprotein adenovirus E1A (1, 4, 14, 35, 50).
At least some of the growth-suppressing properties of the pRB family are dependent on their interaction with the E2F family of transcription factors. E2F-DP heterodimers bind to specific DNA sequences found in the promoters of many genes required for cell cycle progression. RB family proteins bind to these E2F-DP complexes on DNA and repress E2F transcriptional activity during the G0/G1 phase of the cell cycle. These complexes, which can be detected by gel retardation assays, are disrupted by wild-type TAg, but not by TAg mutants that fail to bind to the RB family proteins (51).
E2F-dependent transactivation occurs during the G1/S-phase transition of the cell cycle, when the repressive RB family proteins have been inactivated by phosphorylation. Experimentally, this can be studied by assaying the ability of RB family proteins to repress transcription from an E2F promoter driving a reporter gene. In such assays, the RB-mediated repression can be relieved by expression of E2F, cyclins or cdks, or adenovirus E1A (14, 34, 35, 54).
In many assays of RB family protein function, wild-type TAg can inactivate pRB, as well as p107 and p130, when these proteins have been included. The ability of TAg to inactivate pRB function requires the LXCXE domain, which mediates binding to the RB family proteins. In this study, we address the role of the J domain of TAg in the inactivation of the RB family proteins.
MATERIALS AND METHODS
Cells.
Cells were cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal clone serum (Hyclone), 100 U of penicillin per ml, and 100 μg of streptomycin per ml. MEFs expressing wild-type TAg or various TAg mutants have been described previously (44). Saos-2 cells, an Rb-1 (−/−) osteosarcoma line, were obtained from the American Type Culture Collection. Saos-2 cells were transfected by the calcium phosphate precipitation method. The cells were incubated with the precipitate for 6 h, washed twice with complete medium, and cultured for an additional 36 h. Cell cycle analysis and luciferase assays of transfected cells were performed as described previously (15).
Plasmids.
The plasmids cytomegalovirus (CMV)-RB (14), pcDNA1-HA-p130 (48), and CMVp107-HA (54) have been described previously. The TAg expression vectors pSG5-T, pSG5-K1, pSG5-H42Q, pSG5-D44N, pSG5-HSJ1-T, and pSG5-HSJ1-HQ have been described previously (44, 51). The plasmids CMVT7DP1 (23) and CMVE2F-4-HA (11) have been described previously. The luciferase reporter plasmids 3xWT-E2F (24) and dihydrofolate reductase (DHFR)-luc (pWTluc); the DHFR promoter with a mutant E2F site, pNWluc (27, 40); and the E2F-1 promoter luciferase construct containing two wild-type (pGL2-AN) or mutant E2F sites (ΔE2FA+B) have been described previously (30). The CD19 expression vector was kindly provided by T. Tedder (Duke University).
Antibodies.
Immunoprecipitations and Western blot analysis were performed as described previously (45). The following antibodies were used in this study: antihemagglutinin (HA), 12CA5 (Babco); anti-pRB, XZ77 (Santa Cruz) and 21C9 (a gift of David Cobrinik); anti-adenovirus E1A, M73; rabbit anti-mouse immunoglobulin G (Sigma); anti-p107, SD15 (Santa Cruz); anti-p130, C-20 (Santa Cruz); and anti-CD19 (provided by J. Gibbon, Dana-Farber Cancer Institute).
Gel retardation analysis.
Extracts from transfected Saos-2 cells (see Fig. 5) were prepared by a minilysis procedure (24). Extracts from MEFs expressing TAg (see Fig. 6) were prepared as previously described (29, 51). The gel retardation assays utilized an oligonucleotide probe that contained the E2F binding site of the DHFR promoter (38, 51).
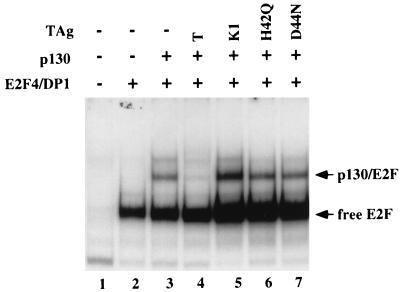
FIG. 5.
Intact LXCXE motif and J domain of TAg are required to disrupt p130-E2F DNA binding complexes. Saos-2 cells were transfected with CMVE2F-4-HA (2 μg) and CMVT7DP1 (0.5 μg) (lanes 2 to 7), pcDNA1-HA-p130 (8 μg when alone [lane 3] and 2 μg when cotransfected with a TAg construct [lanes 3 to 7]), and T, as shown by the constructs pSG5-T (6 μg [lane 4]), pSG5-K1 (6 μg [lane 5]), pSG5-H42Q (18 μg [lane 6]), and pSG5-D44N (18 μg [lane 7]). Thirty-six hours after transfection, extracts were prepared by the minilysis procedure (24), incubated with an oligonucleotide containing the DHFR promoter, and separated in a nondenaturing polyacrylamide gel.
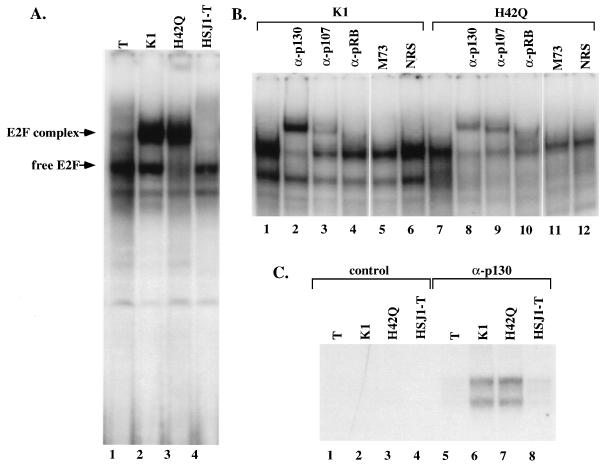
FIG. 6.
Intact LXCXE motif and J domain are required to disrupt endogenous E2F DNA binding complexes. Gel retardation assays were performed with extracts prepared from confluent MEFs that stably expressed either TAg, the LXCXE mutant K1, the J domain mutant H42Q, or the chimeric protein HSJ1-T. (A) Extracts were incubated with an oligonucleotide from the DHFR promoter as in Fig. 5. (B) The complexes observed with extracts from cells expressing K1 or H42Q (lanes 2 and 3 from panel A) were incubated with a panel of antibodies. (C) Immunoprecipitation-DOC release of E2F DNA binding activity. Extracts prepared from confluent MEFs were immunoprecipitated with the control antibody M73 (anti-adenovirus E1A [lanes 1 to 4]) or with anti-p130 antibody (lanes 5 to 8). Immune complexes were denatured and then renatured as described in Materials and Methods and incubated with the DHFR promoter as in Fig. 4.
Immunoprecipitation-DOC release experiment.
Extracts were prepared from confluent MEFs with TNN extraction buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 0.5% Nonidet P-40, 10 μg of aprotinin per ml, 10 μg of leupeptin per ml, 0.1 mM phenylmethylsulfonyl fluoride, 4 mM NaF, 0.1 mM Na orthovanadate) (24) and incubated with either an irrelevant antibody (rabbit anti-mouse immunoglobulin G and M73) or anti-p130 (C-20) and protein A-Sepharose (Pharmacia). The immune complexes were washed four times with NET-N (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40) and once with buffer A (20 mM HEPES [pH 7.5], 50 mM KCl, 1 mM MgCl2, 10% glycerol, 0.1% Nonidet P-40, 0.5 mM dithiothreitol), resuspended in 15 μl of buffer A containing 0.8% deoxycholate (DOC), and eluted on ice for 15 min. The eluate (10 μl) was treated with 15 μl of neutralization buffer (buffer A containing 1 mg of bovine serum albumin per ml, 0.1 μg of single-stranded DNA per ml, 0.8% Nonidet P-40, and 6 mM MgCl2) before incubation with the DHFR probe.
RESULTS
The J domain of TAg is required to relieve RB family-mediated G1 arrest.
It has been reported that transfection of plasmids expressing wild-type pRB, p107, or p130 in the osteosarcoma cell line Saos-2 [Rb-1(−/−)], leads to an accumulation of cells arrested in the G1 phase of the cell cycle (4, 14, 33, 37, 48, 54). This G1 arrest requires an intact RB pocket or TAg binding domain (13, 18). The arrest can be overcome by expression of E1A or E2F (34, 35, 54).
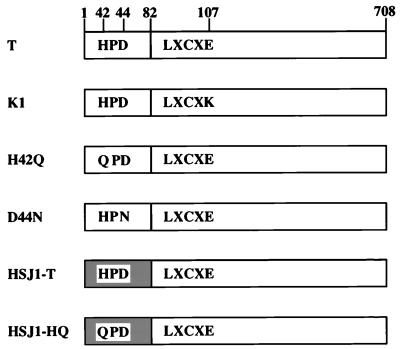
We wished to determine whether the J domain of TAg was required to overcome the G1 arrest induced by the RB family proteins. To address this, we assayed the ability of wild-type TAg and various TAg mutants to overcome the G1 arrest. The constructs are shown schematically in Fig. 1. They are wild-type TAg (T), an LXCXE mutant (K1) unable to bind to the RB family proteins, two single-residue substitutions (H42Q and D44N) within the conserved J domain HPD motif, a chimeric TAg (HSJ1-T) in which the N-terminal J domain has been deleted and replaced with the J domain from the human DnaJ homolog HSJ1, and, finally, the single-residue HPD mutant (HSJ1-HQ) of HSJ1-T.
FIG. 1.
Schematic representation of the TAg constructs used in this study. The J domain is contained entirely within the first exon of TAg (residues 1 to 82). The absolutely conserved HPD motif is indicated (residues 42 to 44). Two mutants of the HPD motif were assayed: a histidine-to glutamine substitution at residue 42 (H42Q) and a glutamate-to-asparagine substitution at residue 44 (D44N). A mutant containing a point mutation of the RB family-binding LXCXE motif, glutamine 107-to-lysine (K1), was also assayed. In the last two constructs, the first exon of TAg was deleted and replaced with the J domain of a cellular protein. The shaded box indicates the heterologous J domain from the human DnaJ homolog HSJ1 fused to residue 83 of TAg, resulting in the chimeric HSJ1-T protein. In the final construct, the histidine-to-glutamine mutation in the HPD domain was introduced into the chimeric HSJ1-T protein, resulting in the mutant chimeric protein HSJ1-HQ (44).
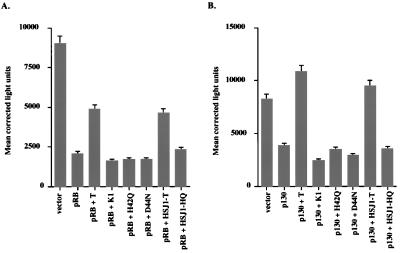
In the experiment shown in Fig. 2A, transient expression of pRB in SaoS-2 cells resulted in a 21% increase in the number of cells in the G1 phase relative to that in the control vector (defined as zero). There was a corresponding decrease in the proportion of cells in S phase (not shown). Coexpression of wild-type TAg could overcome the pRB-induced G1 arrest to a significant extent. In contrast, the RB binding domain mutant K1 did not override the G1 arrest. This indicates that binding by TAg was necessary for relief of the pRB growth-suppressive activity.
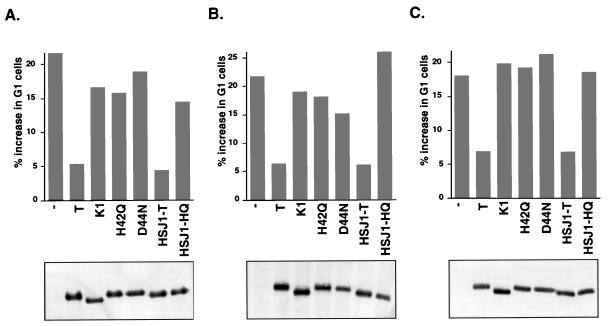
FIG. 2.
An intact LXCXE motif and J domain of TAg are required to override a G1 arrest mediated by pRB, p107, and p130. Saos-2 cells were transfected with a CD19 expression vector (1 μg), pCMV-RB (A [2 μg when alone and 0.5 μg when cotransfected with T]), pCMV-p107HA (B [6 μg when alone and 2 μg when cotransfected with T]), or pcDNA1-HA-p130 (C [8 μg when alone and 2 μg when cotransfected with T]) and the TAg constructs pSG5-T (6 μg), pSG5-K1 (6 μg), pSG5-H42Q (18 μg), pSG5-D44N (18 μg), pSG5-HSJ1-T (6 μg), and pSG5-HSJ1-HQ (18 μg). Different amounts of input DNA were used to obtain equal levels of expression of the proteins. Cells were harvested 36 h posttransfection, stained for CD19 and DNA content, and analyzed by flow cytometry (15). The percent increase in G1 cells was calculated by subtracting the value obtained with the CD19 vector alone. The experiment was repeated three times, and the results from one representative experiment are shown. The Western blot shown below each graph was prepared with extracts from the experiment illustrated. The blot was probed with an anti-TAg antibody, pAB101, and developed with alkaline phosphatase-conjugated anti-mouse antibody and nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate toluidinium salt.
To determine whether the J domain was also required to override pRB-dependent G1 arrest, several N-terminal mutant constructs were tested in this assay. The J domain mutants H42Q and D44N were unable to override the pRB-mediated growth repression (Fig. 2A), even though they retain the ability to bind to pRB (reference 44 and data not shown). Replacement of the N-terminal 82 residues of TAg with the J domain from a human DnaJ homolog, HSJ1, restored the ability of TAg to override the pRB-mediated growth arrest. The corresponding J domain mutant, HSJ1-HQ, was defective. These results demonstrate that an intact J domain of TAg is required to overcome a pRB-induced G1 arrest.
In this experiment, the amount of input DNA was adjusted for each construct to achieve equivalent levels of protein expression as assayed by Western blotting. As shown in the bottom panel of Fig. 2A, the TAg constructs were expressed at similar levels, and hence the differences in their ability to overcome a G1 arrest do not result from variation in the level of expression. Furthermore, we have previously demonstrated by pulse-chase experiments that the stabilities of the K1, H42Q, and HSJ1-T proteins are similar to that of wild-type TAg (44).
To determine whether the LXCXE and J domains of TAg were also required to relieve a G1 arrest induced by the two RB-related proteins p130 and p107 in Saos-2 cells, the same TAg constructs were cotransfected with expression plasmids for p130 or p107. As was the case for pRB, expression of p107 led to an increase in the percentage of cells in the G1 phase of the cell cycle (Fig. 2B). Expression of wild-type TAg or HSJ1-T was able to significantly override the p130-dependent G1 arrest. In contrast, neither the LXCXE mutant K1 nor the J domain mutants H42Q, D44N, and HSJ1-HQ could override the p107-mediated G1 arrest.
Finally, the ability of TAg to override a p130-dependent growth arrest in Saos-2 cells was also dependent on intact LXCXE and J domains of TAg (Fig. 2C). These results suggest that both the LXCXE and J domains are required to overcome a G1 arrest of Saos-2 cells induced by members of the RB family. The expression of the various TAgs is shown in the bottom panels of Fig. 2B and C.
The J domain of TAg is required to relieve pRB- and p130-dependent repression of E2F activity.
The ability of RB family members to induce a G1 arrest in Saos-2 cells has been correlated with their ability to repress E2F transcriptional activity (33, 37). Based on the ability of TAg to override RB family-mediated G1 arrest, it was not unreasonable to ask whether TAg could also override RB family-dependent E2F repression and, if so, whether the LXCXE and J domains were both required for this effect.
Initially, we tested the effect of expression of pRB in Saos-2 cells on the activity of a luciferase reporter construct (3xWT-E2F) that contained three E2F binding sites and a TATA box (24). As shown in Fig. 3A, expression of pRB resulted in a threefold decrease in the activity of 3xWT-E2F. Coexpression of wild-type TAg and HSJ1-T could partially override the RB-mediated transcriptional repression of this reporter. However, neither the LXCXE mutant K1 nor any of the J domain mutations could override the pRB-dependent repression of E2F activity. This demonstrates that the partial override observed with wild-type TAg requires an intact J domain as well as an intact LXCXE domain.
FIG. 3.
Intact LXCXE and J domains of TAg are required to override pRB- and p130-mediated repression of E2F activity. Thirty-five-millimeter-diameter plates of Saos-2 cells were transfected in three independent experiments with the reporter plasmids 3xWT-E2F-luciferase (A and B [0.5 μg]) and CMV–β-galactosidase and pCMVRB (A [40 ng]) or pcDNA1-HA-p130 (B [0.5 μg]) with the TAg constructs pSG5-T (1.5 μg), pSG5-K1 (1.5 μg), pSG5-H42Q (4.5 μg), pSG5-D44N (4.5 μg), pSG5-HSJ1-T (1.5 μg), and pSG5-HSJ1-T HQ (4.5 μg). Thirty-six hours after transfection, cells were extracted in situ and assayed for luciferase activity and β-galactosidase activity to correct for transfection efficiency. Shown is the average of three independently performed experiments ± standard deviation.
We next tested the ability of the pRB-related protein p130 to repress E2F transcription from this promoter. As in the case of pRB, expression of p130 repressed the activity of 3xWT-E2F in Saos-2 cells (Fig. 3B). Furthermore, wild-type TAg and HSJ1-T could efficiently override the p130-mediated transcriptional repression. The override of p130-mediated repression appeared to be more complete than the override of pRB-mediated repression. The reasons for this effect are not clear. Similar to the effects seen in Fig. 3A, the LXCXE mutant and each of the J domain mutants were unable to override the p130-mediated repression of the E2F reporter. This again indicates a requirement for both the LXCXE domain and the J domain in overcoming the transcriptional repression.
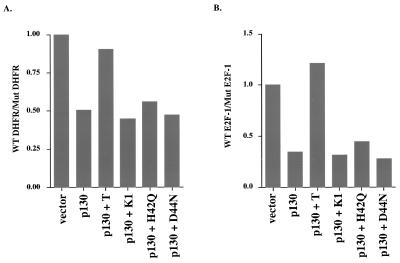
We wanted to assess the ability of TAg to override repression of E2F transcriptional activity of a more physiologically relevant promoter, since the 3xWT-E2F construct used in the experiments described above is an artificial promoter. The promoters for DHFR and E2F-1 contain specific DNA binding sites for E2F as well as several other transcription factors. The reporter activity for each of these promoters increases several fold during the G1/S-phase transition, and this increase is dependent on the E2F sites (27, 30, 40). For example, mutations in the single E2F binding site in the DHFR promoter or the two sites in the E2F-1 promoter abrogated their cell cycle-dependent increase in activity.
To test the effect of TAg on the DHFR and E2F-1 promoters, we repeated the experiment described above with p130 to repress the E2F activity (Table 1). To determine the specific effect of TAg on E2F-dependent transcription, the activity of the wild-type promoters was compared to that obtained with the corresponding mutant promoters lacking functional E2F binding sites. When p130 was expressed in Saos-2 cells, the activity of the wild-type DHFR promoter was repressed twofold relative to that of the mutant form (Fig. 4A and Table 1). Coexpression of TAg could efficiently override the repression, whereas coexpression of the LXCXE mutant K1 or the J domain mutants H42Q and D44N could not.
TABLE 1.
Effect of SV40 T antigen on p130-mediated repression of E2F-dependent promoters
| Transfection | DHFR promoter
|
E2F-1 promoter
|
||||
|---|---|---|---|---|---|---|
| Luciferase activity (RLU)a
|
RRb | Luciferase activity (RLU)a
|
RRc | |||
| Wild type | Mutant | Wild type | Mutant | |||
| Vector | 50,308 ± 5,902 | 35,769 ± 1,509 | 1.0 | 14,951 ± 808 | 20,473 ± 168 | 1.0 |
| p130 | 18,571 ± 1,460 | 26,077 ± 1,480 | 0.51 | 4,576 ± 104 | 18,241 ± 1,238 | 0.34 |
| +T | 38,712 ± 4,193 | 30,417 ± 3,462 | 0.90 | 10,786 ± 601 | 12,190 ± 609 | 1.21 |
| +K1 | 10,805 ± 695 | 17,131 ± 1,200 | 0.45 | 2,122 ± 184 | 9,186 ± 509 | 0.32 |
| +HQ | 34,411 ± 3,385 | 43,646 ± 734 | 0.56 | 7,608 ± 732 | 23,475 ± 169 | 0.44 |
| +DN | 31,311 ± 2,014 | 45,961 ± 1,823 | 0.48 | 6,170 ± 606 | 30,243 ± 2,143 | 0.28 |
Mean luciferase activity (relative light units [RLU]) from three independently performed experiments ± standard deviation.
RR, repression ratios of DHFR wild-type to mutant values normalized to that of the vector by dividing by 1.41.
RR, repression ratios of E2F-1 wild-type to mutant values normalized to that of the vector by dividing by 0.73.
FIG. 4.
Intact LXCXE and J domains of TAg are required to override p130-mediated repression of E2F activity on physiological promoters. Wild-type (WT) DHFR-luc or mutant DHFR-luc reporter (A [0.5 μg]), wild-type E2F-1 (pGL2-AN) or mutant E2F-1-luciferase (ΔE2FA+B) (B [0.5 μg]), and CMV–β-galactosidase and pcDNA1-HA-p130 (0.5 μg), with the TAg constructs pSG5-T (1.5 μg), pSG5-K1 (1.5 μg), pSG5-H42Q (4.5 μg), pSG5-D44N (4.5 μg), pSG5-HSJ1-T (1.5 μg), and pSG5-HSJ1-T HQ (4.5 μg), were transfected as described in the legend to Fig. 3. In panel A, the activity of the wild-type DHFR promoter reporter was normalized to that of a reporter containing a mutation in the E2F DNA binding sites. Similarly, in panel B, the wild-type E2F-1 promoter reporter activity was normalized to that obtained with the mutant reporter in the absence of p130. In each panel, the average of three independent experiments is shown.
Similar results were obtained with the E2F-1 promoter (Fig. 4B and Table 1). Expression of p130 led to a threefold reduction of the wild-type E2F-1 promoter (pGL2-AN) activity relative to that of the mutant promoter (ΔE2FA+B). Coexpression of TAg could efficiently override p130-mediated repression. However, coexpression of the LXCXE or J domain mutants had no effect. This is again consistent with a requirement for an intact J domain to override p130-mediated E2F repression.
The J domain of TAg is required to disrupt RB family-E2F DNA binding complexes.
RB family proteins form DNA binding complexes with the E2F family of transcription factors. We have previously reported that expression of wild-type TAg, but not an LXCXE mutant of TAg, could disrupt p130-E2F and p107-E2F DNA binding complexes (51). Since the J domain was required to relieve pRB- and p130-mediated E2F repression, we wanted to determine whether the J domain of TAg was required for the disruption of p130-E2F DNA binding complexes. To this end, we reproduced the p130-E2F DNA binding complex by transfecting its components into Saos-2 cells. Protein complexes were extracted by a protocol that preserves E2F DNA binding activity from transfected proteins while virtually eliminating E2F DNA binding activity from endogenous E2F complexes (24).
As shown in Fig. 5 (lane 2), expression of E2F-4 with DP1 led to the formation of a specific DNA binding complex. If p130 was coexpressed with E2F-4-DP1, then an additional DNA binding complex was observed (lane 3). In contrast, when TAg was coexpressed, the p130-E2F-4-DP1 complex was significantly reduced (lane 4). Coexpression of the K1 mutant had no effect (lane 5). J domain mutants of TAg retain the ability to efficiently associate with p130 via the LXCXE motif (45). Hence, we considered the possibility that these mutants may either disrupt p130-E2F DNA binding complexes or alternatively become part of the complex. However, coexpression of the J domain mutants H42Q (lane 6) and D44N (lane 7) did not disrupt the p130-E2F DNA binding complexes or change their mobility. When the extracts were incubated with a variety of anti-TAg antibodies, no change in mobility or disruption was observed in the p130-E2F complexes of these cells (data not shown). Therefore, despite an intact LXCXE domain, the J domain mutations of TAg were unable to inhibit the formation of p130-E2F DNA binding complexes. Furthermore, we have been unable to detect TAg in these complexes. The experiment in Fig. 5 indicates that an intact J domain is required to disrupt p130-E2F DNA binding complexes. This may provide a mechanistic explanation for the inability of J domain mutants to override p130-mediated repression of E2F activity (Fig. 3 and 4).
We have previously shown that stable expression of wild-type TAg, but not the LXCXE mutant K1, in MEFs greatly reduces the level of endogenous p130-E2F and p107-E2F DNA binding complexes (51). To determine whether an intact J domain was also required to disrupt endogenous E2F complexes, we prepared lysates from MEFs that had been established with various constructs of TAg (44). Extracts were prepared from growth-arrested cultures that had been confluent for at least 2 days by using an extraction procedure that preserves endogenous E2F DNA binding complexes (38). As shown in Fig. 6A, a slower-migrating complex was observed in extracts prepared from confluent cultures expressing either K1 (Fig. 6A, lane 2) or H42Q (lane 3). However, in extracts from cells expressing wild-type TAg or the HSJ1-T chimera (lanes 1 and 4), only the faster-migrating free E2F was observed. This experiment demonstrates that an intact J domain is required to disrupt endogenous E2F DNA binding complexes. The specific DNA binding activity of the indicated complexes was determined by competition with unlabeled competitor DNA (data not shown). For reasons presently unknown, there reproducibly appeared to be less free E2F DNA binding activity present in extracts prepared from cells that expressed the H42Q mutant (compare levels of free E2F in lanes 3 and 2).
To confirm that the slower-migrating complex observed in the K1- and H42Q-expressing cell lines contained pRB family proteins, antibody supershift experiments were performed with the same extracts used in Fig. 6A. An anti-p130 antibody was able to supershift the majority of the slower-migrating complex in extracts prepared from K1-expressing cells (Fig. 6B, lane 2) and H42Q-expressing cells (lane 8). An anti-p107 antibody supershifted a fraction of the complex, indicating that the complex also contained some p107 (lanes 3 and 9). An anti-pRB antibody could also supershift some of the slower-migrating complex (lanes 4 and 10). Two control antibodies, normal rabbit serum (NRS), and M73, an antibody against the adenovirus E1A protein, had no effect on the E2F DNA binding complexes (lanes 5, 6, 11, and 12). Hence, an intact J domain was required to disrupt endogenous E2F DNA binding complexes of all three pRB-related proteins in confluent MEFs.
As an independent assay of the ability of p130 and p107 to associate with E2F in cells expressing TAg, we performed an immunoprecipitation-DOC release experiment (Fig. 6C) (24). Lysates prepared from confluent cultures of MEFs were immunoprecipitated with an anti-p130 antibody or with a control antibody. Immune complexes were denatured by DOC, followed by renaturation in the presence of Nonidet P-40. The treated lysates were then subjected to gel shift analysis with the DHFR probe. When immunoprecipitations of p130 were treated in this manner, E2F activity was only coprecipitated with p130 in MEFs that expressed K1 (Fig. 6C; lane 6) or H42Q (lane 7). In contrast, no E2F DNA binding activity was immunoprecipitated by the p130 antibody from MEFs expressing wild-type TAg (lane 5) or HSJ1-T (lane 8). A control antibody did not precipitate E2F DNA binding activity from any of the cell lines. This confirms that an intact J domain is required to dissociate p130-E2F complexes.
DISCUSSION
Several domains participate in TAg-mediated cellular transformation, including the p53 binding domain, the LXCXE or pRB-binding motif, and the N-terminal J domain. In the present study, we determined that the N-terminal J domain cooperates with the LXCXE motif to inactivate the growth-suppressive properties of pRB, p107, and p130, as well as to inhibit the ability of pRB and p130 to repress E2F-dependent transactivation. Furthermore, the J domain as well as the LXCXE motif is required to disrupt RB family-E2F DNA binding complexes. While an intact LXCXE domain was required for each of these activities, it was not sufficient. There was an absolute requirement for an intact J domain as well. These results indicate that while LXCXE serves as a binding motif, necessary for TAg interaction with each member of the RB family, inactivation of RB family function requires the J domain as well.
The growth-inhibitory activity of pRB and p130 has been shown to correlate with their ability to bind to members of the E2F family of transcription factors and repress E2F-dependent promoters (25, 33). Results with p107 have not been as clear-cut, but E2F binding is still likely to be important for growth inhibition in most cell types (43, 53). Our results strengthen this hypothesis. We have not been able to separate genetically the ability of TAg to override RB family-mediated growth arrest from its ability to derepress E2F-dependent transcription. This suggests that the basis for the requirement for the J domain in both assays is likely to be the derepression of E2F-dependent genes. The J domain is also necessary for inactivation of p130 and p107 during TAg-mediated transformation (44). Given that p130 and p107 regulate a subset of E2F-dependent genes (16), it will be interesting to know whether these genes are derepressed by TAg in a J domain-dependent manner.
While the biochemical mechanism leading to the derepression of E2F-dependent genes is not yet fully understood, two possibilities, which are not mutually exclusive, deserve consideration. One possibility, discussed further below, is that the J domain is necessary in order for TAg to compete efficiently with E2F for binding to RB family proteins. Another possibility is that the role of the J domain is to inactivate RB family members by perturbing the stability or phosphorylation status of RB family proteins. We have previously shown these activities to be absolutely J domain dependent. Furthermore, the steady-state levels of endogenous p130 are lower in MEFs expressing wild-type TAg than in MEFs expressing J domain mutants of TAg (44).
Binding of the LXCXE motif of TAg to RB family proteins is not sufficient for disruption of RB family-E2F complexes and inactivation of their growth-suppressive properties. This is reminiscent of the adenovirus E1A oncoprotein, which also targets the RB family of proteins. Like TAg, adenovirus E1A contains an LXCXE motif essential for binding to RB family proteins (49). A second domain, contained within conserved region 1 (CR1), is also required to disrupt RB family-E2F complexes (7, 10). E1A containing a mutation in CR1 could associate with pRB-E2F and p107-E2F DNA binding complexes, but was unable to disrupt them. In this case, the CR1 mutant of E1A became part of the E2F DNA binding complexes (17). There are two significant differences between E1A and TAg. First, there is no homology between the J domain of TAg and CR1 of E1A (8). In fact, E1A does not contain any region of homology to the J domain of DnaJ proteins. Second, we have not been able to detect the J domain mutants of TAg as components of RB family-E2F complexes.
This study, along with the work on E1A, suggests that there is more to the oncoprotein-RB family protein interaction than simple binding of the LXCXE motif of the viral oncoprotein to the RB family proteins. It appears that in spite of the common LXCXE motif, the adenovirus E1A and SV40 TAg viral oncoproteins have adopted different strategies to mediate complete inactivation of the RB family proteins. The detailed study of these mechanisms should provide a new insight in the biology of the oncogenic proteins of DNA tumor viruses, as well as a new understanding of the regulation of RB family members and E2F-dependent transcription.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant CA-63113 from the National Cancer Institute and the Women’s Cancers Program of the Dana-Farber Cancer Institute.
J.Z. was supported in part by the Ministerio de Educación y Ciencia of Spain. J.A.D. is a Scholar of the Leukemia Society of America.
REFERENCES
- 1.Beijersbergen R L, Carlée L, Kerhoven R M, Bernards R. Regulation of the retinoblastoma protein-related p107 by G1 cyclin complexes. Genes Dev. 1995;9:1340–1353. doi: 10.1101/gad.9.11.1340. [DOI] [PubMed] [Google Scholar]
- 2.Campbell K S, Mullane K P, Ibraham I A, Stubdal H, Zalvide J, Pipas J M, Silver P A, Roberts T M, Schauffhausen B S, DeCaprio J A. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes Dev. 1997;11:1098–1110. doi: 10.1101/gad.11.9.1098. [DOI] [PubMed] [Google Scholar]
- 3.Christensen J B, Imperiale M J. Inactivation of the retinoblastoma susceptibility protein is not sufficient for the transforming function of the conserved region 2-like domain of simian virus 40 large T antigen. J Virol. 1995;69:3945–3948. doi: 10.1128/jvi.69.6.3945-3948.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claudio P P, De Luca A, Howard C M, Baldi A, Firpo E J, Koff A, Paggi M G, Giordano A. Functional analysis of pRB2/p130 interaction with cyclins. Cancer Res. 1996;56:2003–2008. [PubMed] [Google Scholar]
- 5.DeCaprio J A, Ludlow J W, Figge J, Shew J-Y, Huang C-M, Lee W-H, Marsilio E, Paucha E, Livingston D M. SV40 large T antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 6.Dyson N, Bernards R, Friend S H, Gooding L R, Hassell J A, Major E O, Pipas J M, Vandyke T, Harlow E. Large T antigens of many polyomaviruses are able to form complexes with the retinoblastoma protein. J Virol. 1990;64:1353–1356. doi: 10.1128/jvi.64.3.1353-1356.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dyson N, Guida P, McCall C, Harlow E. Adenovirus E1A makes two distinct contacts with the retinoblastoma protein. J Virol. 1992;66:4606–4611. doi: 10.1128/jvi.66.7.4606-4611.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dyson N, Howley P M, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 9.Ewen M E, Xing Y, Lawrence J B, Livingston D M. Molecular cloning, chromosomal mapping, and expression of the cDNA for p107, a retinoblastoma gene product-related protein. Cell. 1991;66:1155–1164. doi: 10.1016/0092-8674(91)90038-z. [DOI] [PubMed] [Google Scholar]
- 10.Fattaey A R, Harlow E, Helin K. Independent regions of adenovirus E1A are required for binding to and dissociation of E2F-protein complexes. Mol Cell Biol. 1993;13:7267–7277. doi: 10.1128/mcb.13.12.7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ginsberg D, Vairo G, Chittenden T, Xiao Z-X, Xu G, Wydner K, DeCaprio J A, Lawrence J B, Livingston D M. E2F-4, a new member of the E2F transcription factor family, interacts with p107. Genes Dev. 1994;8:2665–2679. doi: 10.1101/gad.8.22.2665. [DOI] [PubMed] [Google Scholar]
- 12.Helin K, Holm K, Niebuhr A, Eiberg H, Tommerup N, Hougaard S, Poulsen H S, Spang-Thomsen M, Nørgaard P. Loss of the retinoblastoma protein-related p130 protein in small cell lung carcinoma. Proc Natl Acad Sci USA. 1997;94:6933–6938. doi: 10.1073/pnas.94.13.6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiebert S W. Regions of the retinoblastoma gene product required for its interaction with the E2F transcription factor are necessary for E2 promoter repression and pRB-mediated growth suppression. Mol Cell Biol. 1993;13:3384–3391. doi: 10.1128/mcb.13.6.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinds P W, Mittnacht S, Dulic V, Arnold A, Reed S I, Weinberg R A. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- 15.Hofmann F, Livingston D M. Differential effects of cdk2 and cdk3 on the control of pRB and E2F function during G1 exit. Genes Dev. 1996;10:851–861. doi: 10.1101/gad.10.7.851. [DOI] [PubMed] [Google Scholar]
- 16.Hurford R K J, Cobrinik D, Lee M-H, Dyson N. pRB and p107/p130 are required for regulated expression of different sets of E2F responsive genes. Genes Dev. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- 17.Ikeda M-A, Nevins J R. Identification of distinct roles for separate E1A domains in disruption of E2F complexes. Mol Cell Biol. 1993;13:7029–7035. doi: 10.1128/mcb.13.11.7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaelin W G, Jr, Ewen M E, Livingston D M. Definition of the minimal simian virus 40 large T antigen- and adenovirus E1A-binding domain in the retinoblastoma gene product. Mol Cell Biol. 1990;10:3761–3769. doi: 10.1128/mcb.10.7.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalderon D, Smith A E. In vitro mutagenesis of a putative DNA binding domain of SV40 large-T. Virology. 1984;139:109–137. doi: 10.1016/0042-6822(84)90334-9. [DOI] [PubMed] [Google Scholar]
- 20.Kelley W, Landry S J. Chaperone power in a virus? Trends Biochem Sci. 1994;19:277–278. doi: 10.1016/0968-0004(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 21.Kelley W L, Georgopoulos C. The T/t common exon of simian virus 40, JC, and BK polyomavirus T antigens can functionally replace the J-domain of the Escherichia coli DnaJ molecular chaperone. Proc Natl Acad Sci USA. 1997;94:3674–3684. doi: 10.1073/pnas.94.8.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kierstead T D, Tevethia M J. Association of p53 binding and immortalization of primary C57BL/6 mouse embryo fibroblasts by using simian virus 40 T-antigen mutants bearing internal overlapping deletion mutations. J Virol. 1993;67:1817–1829. doi: 10.1128/jvi.67.4.1817-1829.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krek W, Ewen M E, Shirodkar S, Arany Z, Kaelin W G, Jr, Livingston D M. Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin A-dependent protein kinase. Cell. 1994;78:161–172. doi: 10.1016/0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 24.Krek W, Livingston D M, Shirodkar S. Binding to DNA and the retinoblastoma gene product promoted by complex formation of different E2F family members. Science. 1993;262:1557–1560. doi: 10.1126/science.8248803. [DOI] [PubMed] [Google Scholar]
- 25.Lacy S, Whyte P. Identification of a p130 domain mediating interactions with cyclin A/cdk2 and cyclin E/cdk2 complexes. Oncogene. 1997;14:2395–2406. doi: 10.1038/sj.onc.1201085. [DOI] [PubMed] [Google Scholar]
- 26.Mayol X, Garriga J, Graña X. Cell cycle-dependent phosphorylation of the retinoblastoma-related protein p130. Oncogene. 1995;11:801–808. [PubMed] [Google Scholar]
- 27.Means A L, Slansky J E, McMahon S L, Knuth M W, Farnham P J. The HIP1 binding site is required for growth regulation of the dihydrofolate reductase gene promoter. Mol Cell Biol. 1992;12:1054–1063. doi: 10.1128/mcb.12.3.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montano X, Millikan R, Milhaven J M, Newsome D A, Ludlow J W, Arthur A K, Fanning E, Bikel I, Livingston D M. Simian virus 40 small tumor antigen and an amino-terminal domain of large tumor antigen share a common transforming function. Proc Natl Acad Sci USA. 1990;87:7448–7452. doi: 10.1073/pnas.87.19.7448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mudryj M, Hiebert S W, Nevins J R. A role for the adenovirus inducible E2F transcription factor in a proliferation dependent signal transduction pathway. EMBO J. 1990;9:2179–2184. doi: 10.1002/j.1460-2075.1990.tb07387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neuman E, Flemington E K, Sellers W R, Kaelin W G., Jr Transcription of the E2F-1 gene is rendered cell cycle dependent by E2F DNA-binding sites within its promoter. Mol Cell Biol. 1994;14:6607–6615. doi: 10.1128/mcb.14.10.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peden K W, Srinivasan A, Farber J M, Pipas J M. Mutants with changes within or near a hydrophobic region of simian virus 40 large tumor antigen are defective for binding cellular protein p53. Virology. 1989;168:13–21. doi: 10.1016/0042-6822(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 32.Pipas J M. Common and unique features of T antigens encoded by the polyomavirus group. J Virol. 1992;66:3979–3985. doi: 10.1128/jvi.66.7.3979-3985.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin X-Q, Chittenden T, Livingston D M, Kaelin W G., Jr Identification of a growth suppression domain within the retinoblastoma gene product. Genes Dev. 1992;6:953–964. doi: 10.1101/gad.6.6.953. [DOI] [PubMed] [Google Scholar]
- 34.Qin X-Q, Livingston D M, Ewen E, Sellers W R, Adams P D. Deregulated E2F-1 transcription factor expression leads to S-phase entry and p53-mediated apoptosis. Proc Natl Acad Sci USA. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin X-Q, Livingston D M, Ewen M, Sellers W R, Arany Z, Kaelin W G., Jr The transcription factor E2F-1 is a downstream target of RB action. Mol Cell Biol. 1995;15:742–755. doi: 10.1128/mcb.15.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawai E T, Butel J S. Association of a cellular heat shock protein with simian virus 40 large T antigen in transformed cells. J Virol. 1989;63:3961–3973. doi: 10.1128/jvi.63.9.3961-3973.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sellers W R, Rodgers J W, Kaelin W G., Jr A potent transrepression domain in the retinoblastoma protein induces a cell cycle arrest when bound to E2F sites. Proc Natl Acad Sci USA. 1995;92:11544–11548. doi: 10.1073/pnas.92.25.11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shirodkar S, Ewen M, DeCaprio J A, Morgan J, Livingston D M, Chittenden T. The transcription factor E2F interacts with the retinoblastoma product and a p107-cyclin A complex in a cell cycle-regulated manner. Cell. 1992;68:1–20. doi: 10.1016/0092-8674(92)90214-w. [DOI] [PubMed] [Google Scholar]
- 39.Silver P A, Way J C. Eukaryotic DnaJ homologs and the specificity of Hsp70 activity. Cell. 1993;74:5–6. doi: 10.1016/0092-8674(93)90287-z. [DOI] [PubMed] [Google Scholar]
- 40.Slansky J E, Li Y, Kaelin W G, Farnham P J. A protein synthesis-dependent increase in E2F1 mRNA correlates with growth regulation of the dihydrofolate reductase promoter. Mol Cell Biol. 1993;13:1610–1618. doi: 10.1128/mcb.13.3.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Srinivasan A, McClellan A J, Vartikar J, Marks I, Cantalupo P, Li Y, Whyte P, Rundell K, Brodsky J L, Pipas J M. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol Cell Biol. 1997;17:4761–4773. doi: 10.1128/mcb.17.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Srinivasan A, Peden K W C, Pipas J M. The large tumor antigen of simian virus 40 encodes at least two distinct transforming functions. J Virol. 1989;63:5459–5463. doi: 10.1128/jvi.63.12.5459-5463.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Starostik P, Chow K N B, Dean D C. Transcriptional repression and growth suppression by the p107 pocket protein. Mol Cell Biol. 1996;16:3606–3614. doi: 10.1128/mcb.16.7.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stubdal H, Zalvide J, Campbell K S, Schweitzer C, Roberts T M, DeCaprio J A. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol Cell Biol. 1997;17:4979–4990. doi: 10.1128/mcb.17.9.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stubdal H, Zalvide J, DeCaprio J A. Simian virus 40 large T antigen alters the phosphorylation state of the RB-related proteins p130 and p107. J Virol. 1996;70:2781–2788. doi: 10.1128/jvi.70.5.2781-2788.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson D L, Kalderon D, Smith A E, Tevethia M J. Dissociation of RB binding and anchorage-independent growth from immortalization and tumorigenicity using SV40 mutants producing N-terminally truncated large T antigens. Virology. 1990;178:15–34. doi: 10.1016/0042-6822(90)90375-2. [DOI] [PubMed] [Google Scholar]
- 47.Tsai J, Douglas M G. A conserved HPD sequence of the J-domain is necessary for YDJ1 stimulation of Hsp70 ATPase activity at a site distinct from substrate binding. J Biol Chem. 1996;271:9347–9354. doi: 10.1074/jbc.271.16.9347. [DOI] [PubMed] [Google Scholar]
- 48.Vairo G, Livingston D M, Ginsberg D. Functional interaction between E2F-4 and p130: evidence for distinct mechanisms underlying growth suppression by different retinoblastoma protein family members. Genes Dev. 1995;9:869–881. doi: 10.1101/gad.9.7.869. [DOI] [PubMed] [Google Scholar]
- 49.Whyte P, Ruley H E, Harlow E. Two regions of the adenovirus early region 1A proteins are required for transformation. J Virol. 1988;62:257–265. doi: 10.1128/jvi.62.1.257-265.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao Z-X, Ginsberg D, Ewen M, Livingston D M. Regulation of the retinoblastoma protein-related protein p107 by G1 cyclin-associated kinases. Proc Natl Acad Sci USA. 1996;93:4633–4637. doi: 10.1073/pnas.93.10.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zalvide J, DeCaprio J A. Role of pRB-related proteins in simian virus 40 large-T-antigen-mediated transformation. Mol Cell Biol. 1995;15:5800–5810. doi: 10.1128/mcb.15.10.5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu J, Rice P W, Gorsch L, Abate M, Cole C N. Transformation of a continuous rat embryo fibroblast cell line requires three separate domains of simian virus 40 large T antigen. J Virol. 1992;66:2780–2791. doi: 10.1128/jvi.66.5.2780-2791.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu L, Enders G, Lees J A, Beijersbergen R L, Bernards R, Harlow E. The pRB-related protein p107 contains two growth suppression domains: independent interactions with E2F and cyclin/cdk complexes. EMBO J. 1995;14:1904–1913. doi: 10.1002/j.1460-2075.1995.tb07182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu L, van den Heuval S, Helin K, Fattaey A, Ewen M, Livingston D M, Dyson N, Harlow E. Inhibition of cell proliferation by p107, a relative of the retinoblastoma protein. Genes Dev. 1993;7:1111–1125. doi: 10.1101/gad.7.7a.1111. [DOI] [PubMed] [Google Scholar]