Abstract
The nuclear import of proteins bearing a basic nuclear localization signal (NLS) is dependent on karyopherin α/importin α, which acts as the NLS receptor, and karyopherin β1/importin β, which binds karyopherin α and mediates the nuclear import of the resultant ternary complex. Recently, a second nuclear import pathway that allows the rapid reentry into the nucleus of proteins that participate in the nuclear export of mature mRNAs has been identified. In mammalian cells, a single NLS specific for this alternate pathway, the M9 NLS of heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1), has been described. The M9 NLS binds a transport factor related to karyopherin β1, termed karyopherin β2 or transportin, and does not require a karyopherin α-like adapter protein. A yeast homolog of karyopherin β2, termed Kap104p, has also been described and proposed to play a role in the nuclear import of a yeast hnRNP-like protein termed Nab2p. Here, we define a Nab2p sequence that binds to Kap104p and that functions as an NLS in both human and yeast cells despite lacking any evident similarity to basic or M9 NLSs. Using an in vitro nuclear import assay, we demonstrate that Kap104p can direct the import into isolated human cell nuclei of a substrate containing a wild-type, but not a defective mutant, Nab2p NLS. In contrast, other NLSs, including the M9 NLS, could not function as substrates for Kap104p. Surprisingly, this in vitro assay also revealed that human karyopherin β1, but not the Kap104p homolog karyopherin β2, could direct the efficient nuclear import of a Nab2p NLS substrate in vitro in the absence of karyopherin α. These data therefore identify a novel NLS sequence, active in both yeast and mammalian cells, that is functionally distinct from both basic and M9 NLS sequences.
The regulated movement of macromolecules into and out of the nucleus is essential for the viability of eukaryotic cells, and several distinct nuclear import and export pathways are believed to exist (reviewed in references 17 and 40). Of these, the best understood is the pathway that mediates the nuclear import of proteins bearing basic nuclear localization signals (NLSs) of the type first described for simian virus 40 (SV40) T antigen and nucleoplasmin (12, 26). Import of such proteins is initiated by the direct interaction of the basic NLS with karyopherin α (also termed importin α) (2, 19, 24, 35, 37, 50), which in turn is bound by a second component of the nuclear import machinery termed karyopherin β1 (also termed importin β) (9, 20, 35, 37). The resultant heterotrimer is then recruited to the cytoplasmic face of a nuclear pore by the direct interaction of karyopherin β1 with nucleoporins (35, 37, 43). The subsequent transition of this heterotrimeric receptor complex into the nucleus is mediated by the cellular Ran GTPase (27, 32, 34) and by a second cofactor termed p10 or NTF2 (10, 39, 41). Directionality of movement is thought to result from the ordered, sequential interaction of the karyopherin β1 subunit with specific nucleoporins (44). Protein import into the nucleus requires the expenditure of energy and this may be provided through the Ran-mediated hydrolysis of GTP (46, 49). Once the heterotrimeric import complex reaches the nucleus, dissociation is induced by the direct interaction of Ran-GTP with karyopherin β1, and this interaction also releases the three imported proteins from the nuclear pore (18, 25, 29, 36, 44). The protein cargo then remains in the nucleus while karyopherin α and β1 are recycled back to the cytoplasm.
The protein import pathway described above involves three participants with distinct roles. These are the NLS-containing protein, which simply acts as a cargo, the karyopherin β1 protein, which actually delivers the cargo to the nucleus, and karyopherin α, which serves as an adapter between the cargo and karyopherin β1. That karyopherin α has no other major role in this transport process is demonstrated by the fact that an amino-terminal ∼41-amino-acid (aa) segment of karyopherin α that directly binds to karyopherin β1 can itself serve as an NLS when attached to a carrier protein (16, 51). Therefore, karyopherin β1 can, in principle, mediate two forms of nuclear import. In the more common version, karyopherin β1 transports karyopherin α together with a cargo protein containing a basic NLS. Alternatively, in a simpler version of this pathway, karyopherin β1 can mediate the nuclear import of proteins bearing the β1 binding domain of karyopherin α, which from this perspective could be viewed as simply the NLS of karyopherin α (αNLS).
Recently a second protein import pathway that is distinct from, but similar to, the karyopherin β1 import pathway has been identified in both mammalian and yeast cells (4, 8, 15, 42, 45). The existence of this second pathway was suggested by the finding that heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1), an RNA binding protein thought to play a role in the nuclear export of mature mRNAs, contains an active NLS, termed M9, that is not recognized by karyopherin α and that has no homology to basic NLS sequences (33, 45, 48). Subsequently, several groups showed that a protein with significant (∼22%) homology to karyopherin β1, termed karyopherin β2 or transportin, could not only directly interact with the M9 NLS but also mediate the nuclear import of M9 containing protein substrates in vitro (8, 15, 42). Importantly, the karyopherin β2-mediated nuclear import of hnRNPA1 is distinct from the karyopherin β1-mediated import of basic NLS proteins in that no karyopherin α subunit is required. Instead, β2-mediated import of M9 NLS proteins appears mechanistically similar to the pathway used by karyopherin β1 to import artificial substrates containing the αNLS into the nucleus (8, 15, 42).
At present, the mammalian karyopherin β2 nuclear import pathway remains relatively poorly understood, and only two distinct protein substrates for karyopherin β2, i.e., hnRNPA1 and hnRNPF, have been identified (45). Further, only a single NLS specific for β2, i.e., the M9 NLS present in hnRNPA1, has been defined, and little information as to functionally relevant residues in the rather large (∼38-aa) M9 sequence exists (33, 48). Karyopherin β2-mediated protein import is, however, known to require the Ran cofactor (8, 38), and karyopherin β2 is also similar to karyopherin β1 in that both proteins can be shown to bind to specific nucleoporins (8). The mechanism of action of the yeast homolog of mammalian karyopherin β2, termed Kap104p, is even less well understood, although two substrate proteins for Kap104p-mediated nuclear import have been proposed (4). Interestingly, these two yeast proteins, termed Nab2p and Nab4p or Hrp1p, are both poly(A)+ RNA binding proteins that are believed to play an important role in mediating the nuclear export of mature mRNAs (5, 23). However, while Nab2p and Nab4p/Hrp1p therefore appear to be functional homologs of mammalian hnRNPs, neither protein contains any sequence with evident homology to the M9 NLS of hnRNPA1, and no NLS has been identified in either Nab2p or Nab4p/Hrp1p (4). In this paper, we report the definition of a sequence within Nab2p that binds Kap104p effectively in vivo. We further demonstrate that this Nab2p-derived sequence, while very different from the M9 NLS, can nevertheless mediate the efficient nuclear import of substrate proteins in both yeast and mammalian cells. Using an in vitro nuclear import assay, we demonstrate that yeast Kap104p can mediate the specific import of substrate proteins bearing the Nab2p NLS, but not the M9 NLS, into isolated HeLa cell nuclei. Surprisingly, this assay also revealed that karyopherin β1, but not the mammalian Kap104p homolog karyopherin β2, could also mediate the efficient nuclear import of a Nab2p NLS substrate. This report therefore identifies a novel NLS sequence specific for the yeast Kap104p nuclear import factor and also describes the first protein sequence, other than the αNLS, that is able to function as a karyopherin α-independent, karyopherin β1-dependent NLS in mammalian cells.
MATERIALS AND METHODS
Molecular clones.
The yeast KAP95 and KAP104 genes (4, 13, 28) were amplified by PCR from the genomic DNA of Saccharomyces cerevisiae Y190 (22) with primers that introduced a BamHI restriction site immediately upstream of the translation initiation codons and an XhoI site downstream of the open reading frames. The KAP104 cDNA was then inserted into the BamHI and SalI restriction sites of the yeast two-hybrid expression plasmid pGBT9 (Clontech) and the bacterial expression plasmid pQE32 (Qiagen). The resulting plasmids encode Kap104p linked to the carboxy terminus of the GAL4 DNA binding domain or to a six-histidine tag. The KAP95 cDNA was fused 3′ to a cDNA encoding glutathione S-transferase (GST) by insertion into the BamHI and SalI restriction sites of the Escherichia coli expression plasmid pGEX4T-1 (Pharmacia).
A cDNA encoding full-length Nab2p (5) was likewise isolated from yeast genomic DNA by PCR amplification with primers introducing EcoRI (5′) and XhoI (3′) restriction sites. This cDNA was fused in frame with, and 3′ to, sequences encoding the VP16 transcription activation domain by insertion into the EcoRI and XhoI restriction sites of the yeast two-hybrid expression plasmid pVP16 (6). The pVP16-Nab2p deletion series (Fig. 1) was similarly constructed by PCR amplification of DNA fragments encoding the indicated Nab2p amino acids and inserting these fragments into the EcoRI and XhoI restriction sites of pVP16. The pVP16/M1, -M2 and -M3-Nab2p mutant expression plasmids were derived from pVP16/Nab2p(161/271) by using the Quick-Change site-directed mutagenesis protocol (Stratagene). Plasmids encoding either wild-type or mutant Nab2p aa 161 to 271 fused to the carboxy terminus of maltose binding protein (MBP) were made by transferring the relevant NAB2 cDNA fragment (EcoRI to XhoI) from the pVP16 background into the EcoRI and SalI restriction sites of pMAL-c2 (New England Biolabs).
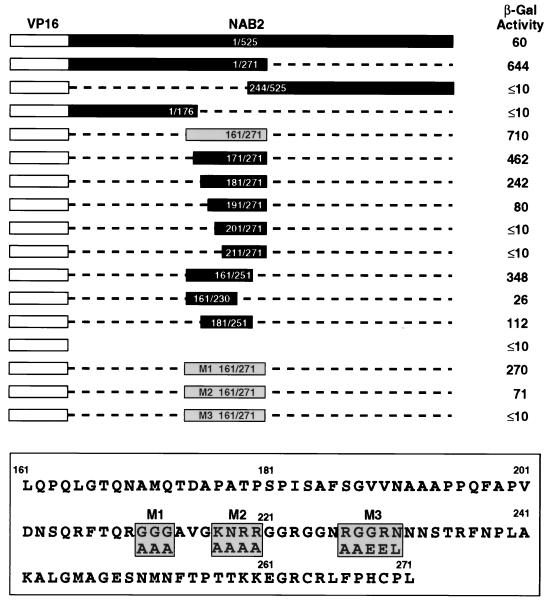
FIG. 1.
Mapping of the Kap104p binding domain of Nab2p by using the yeast two-hybrid assay. Full-length Kap104p was expressed as a fusion protein linked to the GAL4 DNA binding domain, while wild-type and mutant forms of Nab2p were expressed as fusions with the VP16 transcription activation domain. The ability of Kap104p to interact with the indicated wild-type and mutant forms of Nab2p was then assayed in the yeast two-hybrid indicator strain Y190, which contains GAL4 DNA binding sites linked to the lacZ gene, and is given at right in milli-optical density units of β-Gal activity per milliliter of yeast extract. The indicated Nab2p missense mutants M1, M2, and M3 were constructed in the context of the boxed 161-to-271 Nab2p sequence which showed high-affinity binding to Kap104p.
A plasmid encoding GST fused to full-length human karyopherin β1 (20) was kindly provided by S. Kornbluth. Plasmid pGST/αNLS, encoding the first 71 aa of human karyopherin α1 (also termed NPI-1 or hSRP-1) (47, 50) fused to the GST carboxy terminus, was made by inserting the appropriate PCR-amplified, karyopherin α1-derived DNA fragment into the EcoRI and XhoI sites of pGEX4T-1 (Pharmacia). Plasmids pMBP-MIP, encoding full-length human karyopherin β2 fused to the carboxy terminus of MBP, pGST-M9, encoding the M9 NLS fused to the GST carboxy terminus, and pGST-TNLS, encoding the SV40 large-T-antigen NLS (TNLS) fused to the GST carboxy terminus, have been described elsewhere (15). DNA fragments encoding human Ran and p10 (32, 34, 39, 41) were amplified by PCR from a HeLa cDNA library (Marathon Ready cDNA; Promega) by using primers that introduced a BamHI restriction site 5′ to the initiation codon and an XhoI restriction site 3′ of the Ran or p10 open reading frame. The amplified cDNAs were then cloned into the BamHI and XhoI restriction sites of the GST fusion protein expression plasmid pGEX 5X-1 (Pharmacia). The resulting plasmids encode GST-Ran or GST-p10 fusion proteins that are cleavable at the fusion junction by factor Xa protease. The GST-Ran/T24N point mutant expression plasmid was provided by S. Kornbluth.
To express green fluorescent protein (GFP)-Nab2p fusion proteins in yeast, a cDNA encoding GFP was isolated by PCR from pGFP-S65T (Clontech) and ligated as a BglII-EcoRI fragment to DNA fragments (EcoRI-XhoI) encoding wild-type or mutant (M3) Nab2p residues 161 to 271. These DNA sequences were then inserted 3′ to the phosphoglycerate kinase promoter in a previously described yeast expression vector that also contains phosphoglycerate kinase terminator sequences, a 2μm origin of replication, and a LEU2 selectable marker (6, 7). Equivalent mammalian expression plasmids, encoding β-galactosidase (β-Gal) fused to wild-type or mutant (M3) Nab2p aa 161 to 271, were made by ligating a cDNA encoding β-Gal (NcoI-BamHI) and the relevant Nab2p sequences (BamHI-XhoI) into the NcoI and XhoI restriction sites of the mammalian expression plasmid pBC12/CMV (11).
Yeast two-hybrid interaction analysis.
The interaction between Nab2p and Kap104p was assayed by using the yeast two-hybrid in vivo protein interaction system as previously described (6, 7, 14, 15) in the yeast indicator strain Y190 (22).
In vitro nuclear import assays.
GST (Pharmacia), MBP (New England Biolabs), and histidine (His-Tag; Qiagen)-tagged fusion proteins were purified by standard protocols following growth and induction of E. coli expression strains at 30°C. GST moieties were removed from Ran and p10 by incubation of the purified GST fusion proteins bound to glutathione-Sepharose 4B beads (Pharmacia) with factor Xa protease (Pharmacia) for 6 h at 25°C in cleavage buffer (Pharmacia). GST moieties were removed from the karyopherin β1, Kap95p, and Ran/T24N fusion proteins by incubation of proteins bound to glutathione-Sepharose 4B beads with thrombin protease (Pharmacia) in phosphate-buffered saline (PBS). Cleaved proteins were dialyzed into PBS with 5% glycerol and 0.1 mM dithiothreitol, analyzed for integrity by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and quantitated visually by comparison to molecular weight standards (Gibco-BRL) after staining with Coomassie blue G250 (Gibco-BRL). The import substrate proteins GST-M9 NLS, MBP-Nab2p NLS and mutants M1, M2, M3, GST-αNLS, and GST-TNLS were all labeled with fluorescein (FLOUS; Boehringer Mannheim) at a 20:1 molar ratio of FLOUS to protein. All recombinant proteins were concentrated (Centricon; Amicon) to 0.25 to 1.0 mg/ml and frozen in aliquots for single use at −80°C.
Import assay conditions were essentially as described elsewhere (1, 3, 15). HeLa cells were grown on 175-cm2 flasks to subconfluency in Dulbecco modified Eagle medium with 5% fetal bovine serum. The HeLa cells were then trypsinized, suspended in PBS, and harvested at 500 × g. Cells were then incubated for 10 min in cell dissociation solution (Sigma) at 37°C and subsequently washed in PBS. Cells were permeabilized with digitonin (35 μg/ml; Calbiochem) for 5 min at 4°C and then washed three times with transport buffer (1). Nuclei were then purified from cellular debris on a 2-ml 50% sucrose-PBS cushion (1,000 × g, 5 min), washed with transport buffer, and stored on ice just prior to use. Approximately 104 nuclei were used per 60 μl of assay mixture.
Import factors were used at ∼2 μM (final concentration); substrate proteins were used at ∼500 nM (final concentration). Ran and p10 were used at ∼4 μM each for standard import assays. For the Ran/T24N mutant assay, we used import factors at ∼1 μM, substrate at ∼200 nM, and Ran, Ran/T24N, and p10 at ∼8 μM. For the karyopherin α-Nab2p competition, MBP or MBP-Nab2p was used at ∼10 μM. All reactions included 0.1 mM GTP and an ATP regeneration system (1, 3) (Boehringer Mannheim). A control of trimethyl rhodamine isothiocyanate-labeled bovine serum albumin (Calbiochem) was added to each reaction mixture to monitor for possible nonspecific nuclear permeabilization.
Import reactions were performed for 15 min at 30°C, and the products then were immediately fixed on ice with 8% paraformaldehyde–0.25% glutaraldehyde for 10 min. Nuclei were then harvested at 250 × g for 1.5 min, resuspended in 50% PBS-Flouromount G (Southern Biotechnology Associates), mounted on slides under coverslips, and allowed to set for 15 min, and the edges were sealed with nail polish. Images were digitally captured on a Leica DMRB fluorescence microscope and converted to 8-bit gray scale with Adobe Photoshop 4.0 software.
Protein binding by recombinant Nab2p.
Recombinant, purified import factors (His-tagged Kap104p, karyopherin β1, MPB-karyopherin β2, and Kap95p) were covalently coupled at 3 mg/ml to active ester-agarose beads (Affi-Gel 10; Bio-Rad) and then used to prepare 15-μl microaffinity columns. The columns were then equilibrated by passage of 100 μl of binding buffer (10 mM HEPES [pH 7.4], 250 mM NaCl, 0.1 mM dithiothreitol, 5 mM magnesium acetate) before loading of either recombinant MBP-Nab2p(161-271) or MBP in 50 μl of binding buffer. The columns were then washed with 200 μl of binding buffer before elution with 50 μl of 1 M MgCl2. The entire flowthrough and eluate fractions for each column were analyzed on an SDS–12.5% polyacrylamide gel, and proteins were visualized by staining.
Immunofluorescence analysis.
Human 293T cells were transfected by the calcium phosphate method. Approximately 72 h after transfection, localization of the transiently expressed wild-type and mutant β-Gal–Nab2p(161-271) fusion proteins was determined by indirect immunofluorescence analysis as described previously (15), by using a 1:2,000 dilution of a monoclonal anti-β-Gal antibody (Promega) and a 1:200 dilution of a rhodamine-conjugated goat anti-mouse secondary antibody (Cappel). Cells were visualized with a Leica DMRB fluorescence microscope.
Localization of the GFP-Nab2p and GFP/M3-Nab2p fusion proteins in yeast was performed essentially as described previously (30). The yeast strain used here, termed JK9-3d a/a/α/α, was a gift of J. Heitman and is a large, tetraploid yeast derived by the mating of the previously described JK9-3d a/a and JK9-3d α/α diploid yeast strains (21). After transformation with the relevant expression plasmids and selection for transformants on leucine-deficient plates, log-phase yeast cultures were prepared and the yeast cells were fixed by treatment with 3.4% formaldehyde in PBS for 20 min, then washed with PBS, and stained with 4′,6′-diamidino-2-phenylindole (1 μg/ml; DAPI) prior to being mounted on a glass coverslip. GFP was visualized on the fluorescein channel of a Leica DMRB fluorescence microscope.
RESULTS
It has previously been demonstrated that the full-length Nab2p and Nab4p/Hrp1p proteins are able to directly interact with Kap104p in vitro and that inactivation of the Kap104p protein expression results in mislocalization of the normally nuclear Nab2p and Nab4p/Hrp1p proteins to the yeast cell cytoplasm (4). Based on these data, it was proposed that Nab2p and Nab4p/Hrp1p are substrates for a Kap104p-dependent nuclear import pathway. If this proposal is correct, then Nab2p and Nab4p/Hrp1p should contain a binding site for Kap104p that, when transferred to a heterologous protein, would function as a Kap104p-dependent NLS. To identify the proposed Kap104p binding site on Nab2p, we used the yeast two-hybrid assay for detection of protein-protein interactions (14). In this analysis, the full-length Kap104p protein was expressed as a fusion protein linked to the GAL4 DNA binding domain, while Nab2p, and various mutants of Nab2p, were expressed as fusions to the VP16 transcription activation domain (6, 7, 15).
As shown in Fig. 1, the full-length VP16-Nab2p fusion protein was able to specifically interact with Kap104p, as determined by the detection of a modest activation of β-Gal indicator gene expression in Y190 yeast indicator cells (22) expressing both the VP16-Nab2p and the GAL4-Kap104p fusion proteins but not in cells expressing either GAL4-Kap104p or VP16-Nab2p alone. Two extensive deletion mutants of the 525-aa Nab2p, which retained either residues 1 to 271 or residues 244 to 525, mapped the Kap104p binding site of Nab2p to the amino-terminal half of Nab2p (Fig. 1). Of interest, the fusion protein containing Nab2p residues 1 to 271 gave an ∼11-fold-higher level of β-Gal activity than did full-length Nab2p. While the reason for this increase is unclear, we note that expression of the full-length VP16-Nab2p fusion protein in yeast resulted in a slow-growth phenotype, suggesting that this fusion protein is somewhat toxic when overexpressed. In contrast, none of the VP16-Nab2p deletion mutants exerted any detectable effect on yeast growth (data not shown). The enhanced β-Gal activity seen with VP16-Nab2p(1-271) and certain other deletion mutants may therefore result from relief of the toxicity observed with full-length VP16-Nab2p.
Further deletion of the VP16-Nab2p fusion protein demonstrated that a 110-aa Nab2p fragment extending from residues 161 to 271 retained full Kap104p binding activity. Further amino-terminal deletion produced a gradual loss in β-Gal activity, with residues 191 to 271 displaying only ∼10% of the activity of the 161-to-271 fragment, while the 201-to-271 Nab2p fragment was inactive. Similarly, deletion from the carboxy terminus to Nab2p residue 251 reduced activity ∼2-fold, while further deletion, to residue 230, reduced β-Gal activity to ∼4% of the level seen with the 161-to-271 Nab2p fragment (Fig. 1). Based on these data, we conclude that the Kap104p binding domain on Nab2p is fully contained between residues 161 to 271, with the large majority of the binding activity localized between Nab2p residues 181 and 251. Of interest, this Kap104p binding domain of Nab2p contains three repeats of the sequence RGG, between residues 210 and 229, that have been suggested to form an RGG motif, a known RNA binding sequence (Fig. 1) (5).
To assess the importance of these three RGG repeats for Nab2p binding to Kap104p, we next constructed three missense mutations of Nab2p, in the context of the VP16-Nab2p(161-271) fusion protein, that targeted each of these three repeat elements individually (Fig. 1). Analysis of the Kap104p binding capacity of these mutants in the two-hybrid assay demonstrated that the M1 fusion protein, mutated at residues 211 to 213, retained ∼40% of the binding activity of the wild-type sequence whereas the M2 fusion protein, mutated between residues 217 and 220, retained ∼10% of Kap104p binding activity. A third Nab2p mutant, bearing a missense mutation of residues 227 to 231, failed to detectably interact with Kap104p in this in vivo protein interaction assay (Fig. 1).
The Kap104p binding domain of Nab2p functions as an NLS.
The Nab2p protein is predominantly localized to the nuclei of expressing yeast cells (4, 5) and therefore presumably contains an active NLS. To determine whether the mapped Kap104p binding domain of Nab2p can serve as an NLS in vivo, we fused DNA sequences encoding the wild-type or M3 mutant form of residues 161 to 271 of Nab2p to the 3′ end of a cDNA encoding GFP. We then expressed the resultant encoded fusion proteins, as well as wild-type GFP, in yeast cells and determined their subcellular localization. The GFP proteins were localized based on their intrinsic fluorescence, while the yeast nucleus was localized by treatment with a dye (DAPI) that binds to DNA.
As shown in Fig. 2D, the parental GFP protein shows a highly diffuse distribution in expressing yeast cells. In contrast, the GFP-Nab2p fusion protein is localized to a domain within the yeast cell (Fig. 2B) that appears to be the nucleus, based on coincident straining with DAPI (Fig. 2F). Finally, the GFP/M3-Nab2p mutant fusion protein also displays a nonrandom localization in expressing yeast cells (Fig. 2C) but is not concentrated in the nucleus, as determined by the noncoincident straining observed with DAPI (Fig. 2G).
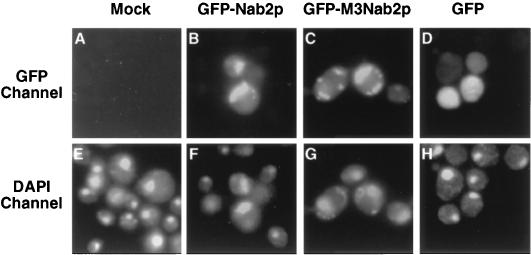
FIG. 2.
Subcellular localization in yeast cells of substrate proteins bearing wild-type and M3 mutant forms of the Kap104p binding domain of Nab2p. Yeast cells expressing wild-type GFP (D and H), a GFP fusion to Nab2p residues 161 to 271 (B and F), or a GFP fusion to the M3 mutant form of this same sequence (C and G) were fixed and stained with DAPI, a dye that binds DNA specifically. The subcellular localization of the GFP proteins was then identified in a fluorescence microscope (A through D) and compared to the localization of the DAPI-stained nucleus (E through H).
The data presented in Fig. 2 demonstrate that the wild-type Nab2p(161-271) sequence, but not the M3 mutant of this sequence, is sufficient to localize a protein to the yeast cell nucleus. This finding is consistent with the hypothesis that it is the interaction of Nab2p with Kap104p (Fig. 1) that mediates Nab2p nuclear localization. To determine whether this Nab2p NLS would also mediate nuclear uptake in mammalian cells, we expressed fusion proteins consisting of β-Gal linked to the Nab2p(161-271) sequence in mammalian cells by transient transfection. As shown in Fig. 3, the Nab2p NLS was indeed able to confer a predominantly nuclear localization on the normally cytoplasmic β-Gal protein (Fig. 3A and C). While introduction of the M3 point mutant into the Nab2p NLS blocked the nuclear localization of this β-Gal fusion protein (Fig. 3B), neither the M1 nor the M2 mutation was found to prevent Nab2p NLS function in mammalian cells (data not shown). We therefore conclude that the Nab2p NLS is active in both yeast and mammalian cells and that the M3 mutation inactivates the Nab2p NLS in both cellular contexts.
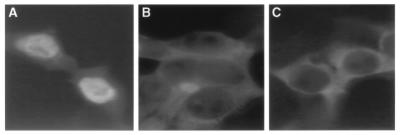
FIG. 3.
Subcellular localization in human 293T cells of substrate proteins bearing wild-type and M3 mutant forms of the Kap104p binding domain of Nab2p. Human 293T cells were transfected with expression plasmids encoding wild-type β-Gal (C) or encoding β-Gal fused to the wild-type (A) or M3 mutant (B) form of the 161-to-271 Kap104p binding domain of Nab2p. Immunofluorescence analysis, using a β-Gal-specific monoclonal antibody, revealed that linkage to the wild-type Nab2p sequence resulted in the relocalization of β-Gal to the nucleus (A), whereas the β-Gal–M3 fusion showed the same cytoplasmic localization as that seen with wild-type β-Gal (B and C). No detectable fluorescence signal was observed with mock-transfected 293T cells (data not shown).
Nuclear import of the Nab2p NLS can be mediated by Kap104p.
We next wished to demonstrate that the nuclear import of proteins bearing the Nab2p NLS in yeast cells was indeed mediated by Kap104p. Because the only established in vitro nuclear import assay relies on the use of isolated mammalian nuclei (1, 3), we first examined whether Kap104p could mediate the import of a substrate bearing the Nab2p NLS into such isolated mammalian nuclei in vitro. For this purpose, we expressed full-length Kap104p and Kap95p, the yeast karyopherin β1 homolog, in bacteria and then purified them by using standard techniques. We also generated substrate proteins bearing the M9 NLS (derived from human hnRNPA1), the αNLS (derived from human karyopherin α), the Nab2p NLS (residues 161 to 271), or the SV40 TNLS. These proteins were then incubated with isolated HeLa cell nuclei in the presence of purified human Ran, p10, and an ATP/GTP energy source.
As shown in Fig. 4, yeast Kap104p proved able to mediate the nuclear import of the Nab2p NLS substrate into isolated mammalian nuclei but failed to support the nuclear import of substrates bearing the M9 NLS, the αNLS, or the SV40 TNLS. This experiment therefore demonstrates that yeast Kap104p is able to effectively interact with the exclusively human proteins present in this in vitro system to mediate the sequence-specific nuclear import of a Nab2p NLS substrate. Although the inability of Kap104p to import the αNLS and SV40 TNLS substrates was expected, the lack of activity with the M9 NLS was perhaps surprising, in that the M9 NLS is the substrate for karyopherin β2, the human homolog of Kap104p (15, 42).
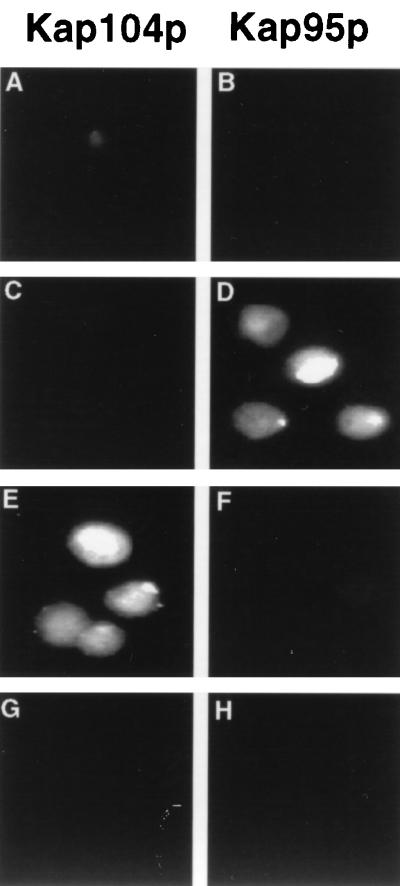
FIG. 4.
Protein import into isolated mammalian nuclei mediated by yeast karyopherin β homologs. Isolated HeLa cell nuclei were incubated with fluorescein-labeled substrate proteins consisting of GST fused to the M9 NLS (A and B), the αNLS (C and D), or the SV40 TNLS (G and H) or of MBP fused to residues 161 to 271 of Nab2p (E and F). In addition, the nuclei were incubated with either purified recombinant yeast Kap104p or Kap95p, as well as with added human Ran, p10, and a source of energy. After incubation at 30°C for 15 min, the nuclei were fixed and analyzed for nuclear import of the labeled substrate proteins.
To further confirm the specificity of the observed nuclear import activity of Kap104p, we also examined whether any of these NLS substrates would be imported into human nuclei by Kap95p, the yeast homolog of karyopherin β1. As shown in Fig. 4, Kap95p was able to induce the nuclear import of the αNLS substrate but was inactive with the M9, Nab2p and SV40 TNLS sequences. The inability of Kap95p to import the SV40 TNLS substrate was expected, as this import is known to also require the addition of karyopherin α to this in vitro assay (21, 37). Based on these data, we therefore conclude that nuclear import of the Nab2p NLS substrate is entirely dependent, in this in vitro assay system, on the presence of the yeast Kap104p nuclear import factor and that the Nab2p NLS is not a substrate for Kap95p.
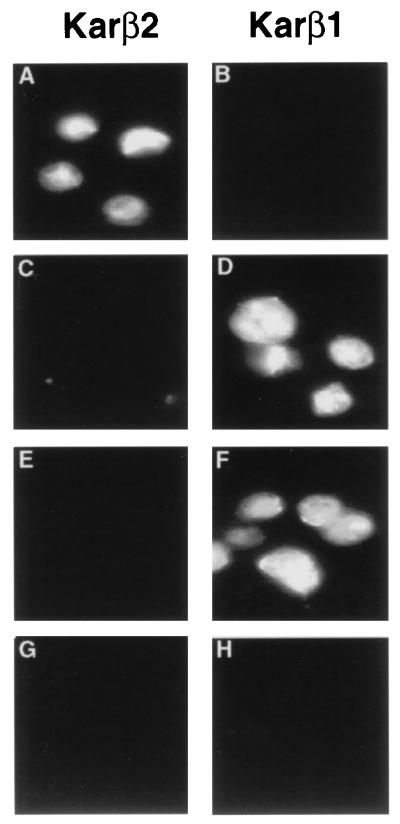
Human karyopherin β1 can mediate Nab2p NLS-dependent nuclear import.
The observation that the M9 NLS cannot serve as a substrate for nuclear import by Kap104p (Fig. 4) raised the question of whether the nuclear import of Nab2p NLS substrates in mammalian cells could be mediated by the mammalian karyopherin β2 homolog of yeast Kap104p. To examine this question, we analyzed the in vitro nuclear import of a Nab2p NLS substrate into isolated mammalian cell nuclei, using recombinant human karyopherins β2 and β1 in place of the recombinant yeast Kap104p and Kap95p tested in Fig. 4.
As shown in Fig. 5, karyopherin β2 was able to mediate the effective nuclear import of its expected NLS substrate, the M9 sequence derived from hnRNPA1, but failed to detectably mediate nuclear import of the Nab2p NLS substrate. Remarkably, however, human karyopherin β1 not only induced the nuclear import of its predicted substrate, i.e., the αNLS, but also permitted the effective import of the Nab2p NLS substrate into the nucleus. It is important to note that these reactions do not involve significant levels of karyopherin α, as demonstrated here by the lack of import of the SV40 TNLS substrate, which is dependent on not only karyopherin β1 but also karyopherin α (20, 35). In fact, addition of a cytoplasmic extract containing karyopherin α to this assay results in efficient SV40 TNLS-mediated nuclear import (data not shown). Therefore, it appears that the Nab2p NLS substrate, like the αNLS substrate, is instead imported by the action of karyopherin β1 alone.
FIG. 5.
Protein import into isolated mammalian nuclei mediated by human karyopherins β1 and β2. The in vitro nuclear import assays were performed as described for Fig. 4 except that recombinant human karyopherins β2 and β1 were substituted for their respective yeast homologs Kap104p and Kap94p. Panels are as described in the legend to Fig. 4.
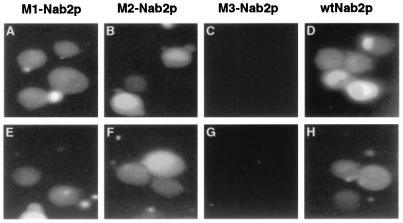
To further confirm the specificity of the in vitro nuclear import assays shown in Fig. 4 and 5, we next examined whether mutations of Nab2p would perturb import. In particular, we wished to determine whether the M3 mutation of Nab2p, which blocks Kap104p binding (Fig. 1) and also blocks the nuclear localization of Nab2p NLS substrates in both yeast and mammalian cells (Fig. 2 and 3), would also block the nuclear import of a recombinant Nab2p NLS substrate in vitro. In fact, as shown in Fig. 6, the M3 mutation effectively blocked nuclear import mediated by both yeast Kap104p and human karyopherin β1. In contrast, neither the M1 mutation nor the M2 mutation had any clear effect on the efficiency of nuclear import mediated by either factor (Fig. 6). It is therefore apparent that nuclear import of Nab2p NLS substrates by both Kap104p and karyopherin β1 is specific and, apparently, mediated by at least somewhat similar protein sequence recognition events.
FIG. 6.
Mutations of Nab2p that block Kap104p binding also block in vitro nuclear import. Fluorescein-labeled substrate proteins, consisting of MBP fused to the wild-type or M1, M2, or M3 mutant form of Nab2p aa 161 to 271 (Fig. 1), were analyzed for in vitro import into isolated mammalian nuclei, as described for Fig. 4, by using either yeast Kap104p (A to D) or human karyopherin β1 (E to H) as an import factor.
The Nab2p NLS binds karyopherin β1 directly and competes for karyopherin α binding.
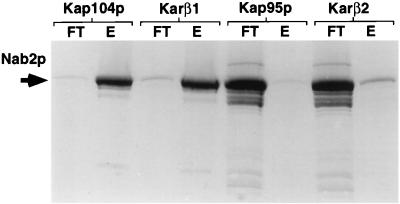
The data presented in Fig. 4 and 5 suggest that Kap104p and karyopherin β1, but not Kap95p and karyopherin β2, directly interact with the Nab2p NLS to mediate nuclear localization. To examine whether this direct interaction indeed occurs, we prepared microaffinity columns consisting of recombinant Kap104p, Kap95p, karyopherin β1, or karyopherin β2 linked to an Affi-Gel 10 matrix. Recombinant MBP-Nab2p(161-271) was then loaded onto each column, and the flowthrough was collected. After being washed with multiple volumes of binding buffer, bound proteins were eluted with 1 M MgCl2 and the eluate was collected. Flowthrough and eluate fractions from each column were analyzed by gel electrophoresis. As shown in Fig. 7, both the Kap104p and karyopherin β1 affinity columns specifically bound the MBP-Nab2p(161-271) protein, while no (Kap95p) or very little (karyopherin β2) binding was noted with the other two affinity columns. These data obtained for entirely recombinant proteins therefore confirm that the Nab2p NLS can indeed specifically interact with both yeast Kap104p and mammalian karyopherin β1.
FIG. 7.
The Nab2p NLS binds both Kap104p and karyopherin β1 (Kar β1) in vitro. Microaffinity columns (15 μl) consisting of the indicated purified recombinant import factors linked to an Affi-Gel 10 matrix were loaded with purified recombinant MBP-Nab2p(161-271) in 50 μl of binding buffer. After being washed with 200 μl of binding buffer, bound proteins were eluted in 50 μl of 1 M MgCl2. The flowthrough (FT) and eluate (E) samples (but not the wash) were then analyzed by SDS-PAGE and visualized by staining. A parallel experiment using purified MBP as a column load did not reveal binding to any of the four affinity columns (data not shown).
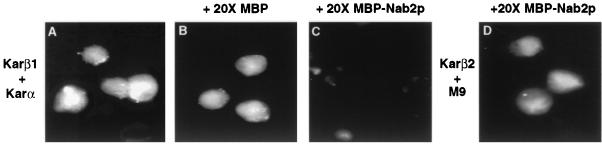
If the nuclear import of Nab2p NLS substrates by human karyopherin β1 is mediated by a direct interaction similar to the interaction of karyopherin β1 with the αNLS, then one would predict that these interactions would be competitive. To test whether this is indeed the case, we examined whether a 20-fold molar excess of the Nab2p NLS (residues 161 to 271) linked to MBP would inhibit nuclear import of an αNLS substrate by karyopherin β1. As shown in Fig. 8C, this was indeed found to be the case. This inhibition was specific, in that a 20-fold molar excess of MBP had no effect (Fig. 8B) whereas MBP-Nab2p failed to inhibit import of an M9 NLS substrate by karyopherin β2 (Fig. 8D). These data are therefore consistent with the hypothesis that the nuclear import of Nab2p NLS substrates by human karyopherin β1 occurs via a mechanism similar to the one used by karyopherin β1 to import karyopherin α.
FIG. 8.
The Nab2p NLS can specifically compete with the αNLS for karyopherin β1-mediated nuclear import. In vitro nuclear import assays, using human karyopherin β1 (Kar β1; A to C) or β2 (D) and fluorescein-labeled GST-αNLS (A to C) or GST-M9 (D), were performed as described for Fig. 5 except that unlabeled recombinant MBP (B) or recombinant MBP-Nab2p(161-271) (C and D) was added to the isolated nuclei at a 20-fold molar excess over the labeled substrate proteins prior to initiation of the import assay.
Kap104p-mediated nuclear import is Ran dependent.
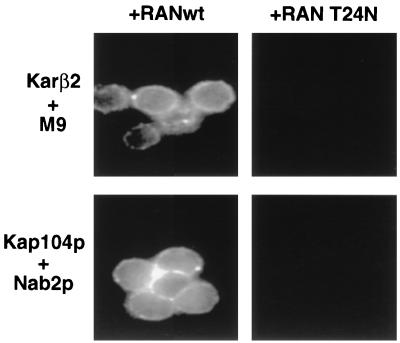
It has recently been demonstrated that nuclear import of an M9 NLS substrate by karyopherin β2 is, like all forms of nuclear import by karyopherin β1, dependent on the biological activity of the Ran GTPase (8, 38). This result is reproduced in Fig. 9, which shows that not only the karyopherin β2-mediated in vitro nuclear import but also the nuclear pore docking of an M9 NLS substrate could be blocked by addition of a dominant negative mutant of Ran (T24N) that fails to bind GTP effectively (31). In contrast, efficient nuclear import of the GST-M9 fusion protein is observed in the presence of equivalent levels of wild-type human Ran. Similarly, we observed that the Kap104p-mediated nuclear import of a Nab2p NLS substrate was also effectively blocked when the T24N Ran mutant was used in place of wild-type Ran in this in vitro assay (Fig. 9).
FIG. 9.
Ran is required for both karyopherin β2 (Kar β2)- and Kap104p-mediated nuclear import. Nuclear import assays were performed as described for Fig. 4 and 5 except that the Ran mutant T24N was substituted for wild-type Ran (RAN wt) in the two incubations visualized at the right.
DISCUSSION
Considerable progress has been made in understanding the prototypic nuclear import pathway utilized by proteins bearing basic NLS sequences (17, 39). This has included the identification of karyopherin α as the NLS receptor (19, 35, 50), the characterization of karyopherin β1 as the key transport factor in this pathway (9, 20, 35, 43), and the realization of the major role played by nucleoporins (35, 37, 43) and, particularly, by the Ran protein (25, 27, 32, 34) in mediating this protein import pathway. An interesting insight to emerge from this previous work is the realization that karyopherin β1 can actually mediate the nuclear import of at least two distinct classes of NLS sequences. In the more common pathway, karyopherin β1 binds to karyopherin α, which in turn binds to the short, basic NLS sequences commonly found on proteins requiring import into the nucleus (20, 37). In addition to this indirect import pathway, karyopherin β1 can also import protein substrates that are able to bind β1 directly, such as artificial protein substrates bearing the ∼41-aa karyopherin β1 binding domain found in karyopherin α, here termed the αNLS (16, 51). However, to this point, no natural substrates able to utilize this alternate, direct karyopherin β1 import pathway have been identified other than karyopherin α itself.
While the basic NLS nuclear import pathway is by far the best characterized, it is clear that other sequence-specific protein nuclear import pathways exist. The demonstration that hnRNPA1 contains an NLS sequence, termed M9, that is unlike basic NLS sequences and that cannot functionally interact with either karyopherin α or karyopherin β1 (33, 48) suggested that hnRNPA1 was a likely substrate for such an alternative import pathway. Subsequent work by several groups demonstrated that M9 NLS-mediated protein nuclear import was in fact dependent on a factor displaying significant homology to karyopherin β1, termed karyopherin β2 or transportin (8, 15, 42). Of note, the nuclear import of M9 NLS substrates by karyopherin β2 was found to involve the direct interaction of the M9 NLS with β2 and thus was independent of any karyopherin α-like adapter protein. Therefore, M9 NLS import by karyopherin β2 could be viewed as mechanistically similar to the karyopherin β1-mediated import of substrates bearing the αNLS.
Although evidence suggesting that karyopherin β2-mediated protein import, like karyopherin β1-mediated import, is dependent on the Ran cofactor and on specific interactions with cellular nucleoporins has been presented (8, 38), this alternate protein import pathway remains poorly understood. In particular, the M9 NLS remains the only known NLS specific for karyopherin β2, and the determinants of specificity in this rather large (∼38-aa) sequence remain essentially uncharacterized (33, 48). We were therefore intrigued by the recent identification of the yeast homolog of karyopherin β2, termed Kap104p, and by the suggestion that the yeast nuclear poly(A)+ RNA binding proteins Nab2p and Nab4p/Hrp1p were substrates for a Kap104p-mediated nuclear import pathway (4). This report was particularly interesting in that neither Nab2p nor Nab4p/Hrp1p contains any sequence with evident homology to the M9 NLS.
As a first step toward demonstrating that Nab2p indeed contained a Kap104p-dependent NLS, we sought to define a Kap104p binding domain in the 525-aa Nab2p protein by using the yeast two-hybrid assay. As shown in Fig. 1, this sequence turned out to be quite large, with maximal Kap104p binding being contained within a 110-aa segment extending from residues 161 to 271, while the core Kap104p binding domain of Nab2p appeared to map between residues 191 and 251 (60 aa). We note that the other two protein domains shown to directly bind to karyopherin β homologs are also quite large, with the β1 binding domain of karyopherin α mapped to a 41-aa segment (16, 51), while the karyopherin β2 binding domain of hnRNPA1, i.e., the M9 NLS, has been mapped to a 38-aa segment (33, 48). It therefore appears possible that NLSs able to function directly via karyopherin β homologs, as opposed to via a karyopherin α-type adapter protein, may in general be relatively large.
Previous analysis has led to the proposal that Nab2p contains three functional domains (5). These are a glutamine-rich repeat extending from residues 101 to 172, an RGG motif located between residues 210 and 229, and an extended sequence of repeated CCCH-type putative zinc finger motifs extending from residues 264 to 453. Based on the mutational analysis presented in Fig. 1, it appears that neither the glutamine-rich element nor the zinc finger repeats contribute significantly to Kap104p binding. In contrast, the putative RGG motif is centrally located in the Kap104p binding domain of Nab2p. To test whether the three RGG repeats in Nab2p actually contribute to Kap104p binding, we mutated each one in turn to generate the Nab2p point mutants M1, M2, and M3. As shown in Fig. 1, the M1 mutation only modestly inhibited Kap104p binding, while the M2 mutation reduced binding ∼10-fold. In contrast, the M3 mutation entirely eliminated Kap104p binding in vivo. While these data are therefore consistent with the hypothesis that these RGG repeats contribute to specific Kap104p binding, they do not eliminate the possibility that these sequences also participate in RNA binding by Nab2p (5).
An interesting aspect of the data presented in Fig. 1 is that we were able to readily detect an interaction between full-length Kap104p and various Nab2p derivatives in the yeast cell nucleus. In the case of karyopherin β1, karyopherin α binding is blocked in the nucleus by a high-affinity interaction of karyopherin β1 with the GTP-bound form of Ran (Ran-GTP), which is highly localized to the nucleus (18, 27, 36). Indeed, the Ran-GTP-mediated release of karyopherin α, and also of the bound NLS cargo, into the cell nucleoplasm is thought to represent the final step in the karyopherin β1-mediated protein import pathway. Therefore, the fact that we were able to readily detect the interaction of full-length Kap104p and Nab2p in the yeast cell nucleus, in the presence of endogenous Ran-GTP, suggests either that the Kap104p interaction with the Nab2p NLS is not efficiently blocked by Ran-GTP or, alternately, that the fusion of the GAL4 DNA binding domain to the amino terminus of Kap104p interferes with Ran-GTP, but not Nab2p NLS, binding to Kap104p. While Bonifaci et al. (8) have reported that Ran-GTP is unable to dissociate an M9 NLS substrate from mammalian karyopherin β2 under conditions where it readily induces dissociation of karyopherin β1 from karyopherin α, more recent data (25, 45) suggest that the karyopherin β2-M9 interaction is indeed disrupted by Ran-GTP.
A critical question was whether the Kap104p binding domain of Nab2p mapped in Fig. 1 would, in fact, function as an NLS. As shown in Fig. 2 and 3, the wild-type Nab2p(161-271) sequence was able to induce the nuclear localization of a linked substrate protein in not only yeast but also mammalian cells. In contrast, the M3 missense mutant of Nab2p, which has lost the ability to interact with Kap104p (Fig. 1), failed to promote nuclear localization in either cell system. Based on these data, one would predict that the nuclear localization of the Nab2p(161-271) substrate in yeast and human cells would be mediated, respectively, by Kap104p and by the mammalian Kap104p homolog karyopherin β2. To test whether this was indeed the case, we first examined whether yeast Kap104p and Kap95p would be active in an in vitro nuclear import assay using isolated mammalian nuclei and human forms of the Ran and p10 cofactors. In fact, as shown in Fig. 4, Kap104p was able to induce the readily detectable import of a substrate protein bearing the Kap104p binding domain of Nab2p but failed to import other candidate import substrates, including the M9 NLS substrate. Similarly, yeast Kap95p induced the specific nuclear import of a substrate bearing the human αNLS but was inactive with the other substrates tested, including the Nab2p NLS. These data are interesting for two reasons. First, they clearly demonstrate that Nab2p does indeed contain an NLS that is a specific target for the Kap104p import factor. Second, they demonstrate that both recombinant yeast Kap104p and yeast Kap95p can mediate the sequence-specific nuclear import of NLS substrates into isolated mammalian nuclei in the presence of only human cofactors. Although Kap95p has previously been shown to promote the docking of a basic NLS substrate at the nuclear pores of isolated mammalian nuclei in the presence of the yeast karyopherin α homolog Srplp (13), this is, to our knowledge, the first demonstration for Kap95p, and certainly for Kap104p, that these proteins are active as import factors in an otherwise entirely mammalian nuclear context. This result implies that most or perhaps all cofactor interactions involved in both Kap95p/karyopherin β1- and Kap104p/karyopherin β2-mediated nuclear protein import are conserved between yeast and mammalian cells. However, the finding that the M9 NLS was unable to function as a substrate for Kap104p in this in vitro assay suggests that the NLS target specificities of Kap104p and karyopherin β2 have diverged significantly over this same evolutionary time period. In fact, the Nab2p(161-271) sequence used in this analysis bears no evident similarity to the M9 NLS found in hnRNPA1 (33, 48), although both are somewhat glycine rich.
To examine whether the Nab2p NLS could be recognized by karyopherin β2, the human homolog of Kap104p, we next asked whether Nab2p would function as an in vitro NLS in the presence of karyopherin β1 or β2 (Fig. 5). In fact, although karyopherin β2, as expected, induced the efficient nuclear import of the M9 NLS, it failed to functionally interact with the Nab2p NLS. Therefore, these data further support the hypothesis that the NLS target specificities of Kap104p and karyopherin β2 have diverged to the point of incompatibility.
If the Nab2p NLS is not a substrate for karyopherin β2, then how is it localized to the nuclei of mammalian cells (Fig. 3)? Surprisingly, the data presented in Fig. 5 reveal that the Nab2p NLS forms a substrate for human karyopherin β1 acting in the absence of karyopherin α. Therefore, this Nab2p-derived sequence appears able to function as a direct karyopherin β1-dependent NLS in the manner described previously only for the αNLS (16, 51). This import reaction is specific in that it is blocked by the M3 missense mutation (Fig. 6) that also prevents the nuclear localization of a Nab2p NLS substrate in mammalian cells (Fig. 3). Like the αNLS (16, 51), the Nab2p NLS is able to directly interact with karyopherin β1 in vitro (Fig. 7), while a 20-fold molar excess of an unlabeled MBP-Nab2p NLS substrate was found to specifically block the karyopherin β1-dependent import of an αNLS substrate but did not affect the nuclear import of an M9 NLS substrate by karyopherin β2 (Fig. 8). These data are therefore consistent with the hypothesis that karyopherin β1 mediates the nuclear import of proteins bearing the αNLS and the Nab2p NLS via similar or identical mechanisms. However, comparison of the sequence of the 41-aa αNLS reported previously with the Nab2p NLS defined in this study (Fig. 1) does not reveal any clear homology, although both contain a significant number of basic residues. It is therefore possible that the αNLS and Nab2p NLS binding sites on karyopherin β1 are at least partly distinct. Nevertheless, it is interesting to speculate that the apparently similar interaction of the Nab2p NLS and the αNLS with karyopherin β1 may reflect the fact that the protein target specificities of the related karyopherin β1/Kap95 and karyopherin β2/Kap104 proteins were similar at some point in the past and may subsequently have evolved differently in the yeast and mammalian contexts. Clearly, although both Kap95p and karyopherin β1 can functionally interact with the αNLS (Fig. 4 and 5), only the mammalian form has retained the ability to interact with the Nab2p NLS. In any case, the identification of a second sequence that can function as a karyopherin β1-dependent, karyopherin α-independent NLS in mammalian cells raises the possibility that there may be other, functionally analogous NLSs that play an important role in the nuclear import of as yet undefined proteins and RNAs.
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
We thank S. Kornbluth and J. Moore for the gift of the bacterial GST-β1 and GST-Ran/T24N expression plasmids, P. Palese for the cDNA encoding karyopherin α1 (NPI-1), J. Heitman for the JK9-3d a/a/α/α yeast strain, and L. Thorne for the complete yeast Kap104p-encoding gene.
This research was supported by funds from the Howard Hughes Medical Institute.
REFERENCES
- 1.Adam E J H, Adam S A. Identification of cytosolic factors required for nuclear localization sequence-mediated binding to the nuclear envelope. J Cell Biol. 1994;125:547–555. doi: 10.1083/jcb.125.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adam S A, Gerace L. Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell. 1991;66:837–847. doi: 10.1016/0092-8674(91)90431-w. [DOI] [PubMed] [Google Scholar]
- 3.Adam S A, Sterne-Marr R E, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aitchison J D, Blobel G, Rout M P. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- 5.Anderson J T, Wilson S M, Datar K V, Swanson M S. NAB2: a yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol Cell Biol. 1993;13:2730–2741. doi: 10.1128/mcb.13.5.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogerd H P, Fridell R A, Blair W S, Cullen B R. Genetic evidence that the Tat proteins of human immunodeficiency virus types 1 and 2 can multimerize in the eukaryotic cell nucleus. J Virol. 1993;67:5030–5034. doi: 10.1128/jvi.67.8.5030-5034.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogerd H P, Fridell R A, Madore S, Cullen B R. Identification of a novel cellular co-factor for the Rev/Rex class of retroviral regulatory proteins. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- 8.Bonifaci N, Moroianu J, Radu A, Blobel G. Karyopherin β2 mediates nuclear import of a mRNA binding protein. Proc Natl Acad Sci USA. 1997;94:5055–5060. doi: 10.1073/pnas.94.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chi N C, Adam E J H, Adam S A. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corbett A H, Silver P A. The NTF2 gene encodes an essential, highly conserved protein that functions in nuclear transport in vivo. J Biol Chem. 1996;271:18477–18484. doi: 10.1074/jbc.271.31.18477. [DOI] [PubMed] [Google Scholar]
- 11.Cullen B R. Trans-activation of human immunodeficiency virus occurs via a bimodal mechanism. Cell. 1986;46:973–982. doi: 10.1016/0092-8674(86)90696-3. [DOI] [PubMed] [Google Scholar]
- 12.Dingwall C, Sharnick S V, Laskey R A. A polypeptide domain that specifies migration of nucleoplasmin into the nucleus. Cell. 1982;30:449–458. doi: 10.1016/0092-8674(82)90242-2. [DOI] [PubMed] [Google Scholar]
- 13.Enenkel C, Blobel G, Rexach M. Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J Biol Chem. 1995;270:16499–16502. doi: 10.1074/jbc.270.28.16499. [DOI] [PubMed] [Google Scholar]
- 14.Fields S, Song O-K. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 15.Fridell R A, Truant R, Thorne L, Benson R E, Cullen B R. Nuclear import of hnRNP A1 is mediated by a novel cellular cofactor related to karyopherin-β. J Cell Sci. 1997;110:1325–1331. doi: 10.1242/jcs.110.11.1325. [DOI] [PubMed] [Google Scholar]
- 16.Görlich D, Henklein P, Laskey R A, Hartmann E. A 41 amino acid motif in importin-α confers binding to importin-β and hence transit into the nucleus. EMBO J. 1996;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- 17.Görlich D, Mattaj I W. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 18.Görlich D, Pante N, Kutay U, Aebi U, Bischoff F R. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- 19.Görlich D, Prehn S, Laskey R A, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 20.Görlich D, Vogel F, Mills A D, Hartmann E, Laskey R A. Distinct functions for the two importin subunits in nuclear protein import. Nature. 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- 21.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 22.Heitman J, Movva N R, Hiestand P C, Hall M N. FK 506-binding protein proline rotamase is a target for the immunosuppressive agent FK 506 in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:1948–1952. doi: 10.1073/pnas.88.5.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henry M, Borland C Z, Bossie M, Silver P A. Potential RNA binding proteins in Saccharomyces cerevisiae identified as suppressors of temperature-sensitive mutations in NPL3. Genetics. 1996;142:103–115. doi: 10.1093/genetics/142.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imamoto N, Tachibana T, Matsubae M, Yoneda Y. A karyophilic protein forms a stable complex with cytoplasmic components prior to nuclear pore binding. J Biol Chem. 1995;270:8559–8565. doi: 10.1074/jbc.270.15.8559. [DOI] [PubMed] [Google Scholar]
- 25.Izaurralde E, Kutay U, von Kobbe C, Mattaj I W, Görlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalderon D, Roberts B L, Richardson W D, Smith A E. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 27.Koepp D M, Silver P A. A GTPase controlling nuclear trafficking: running the right way or walking RANdomly? Cell. 1996;87:1–4. doi: 10.1016/s0092-8674(00)81315-x. [DOI] [PubMed] [Google Scholar]
- 28.Koepp D M, Wong D H, Corbett A H, Silver P A. Dynamic localization of the nuclear import receptor and its interactions with transport factors. J Cell Biol. 1996;133:1163–1176. doi: 10.1083/jcb.133.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kutay U, Izaurralde E, Bischoff F R, Mattaj I W, Görlich D. Dominant-negative mutants of importin-β block multiple pathways of import and export through the nuclear pore complex. EMBO J. 1997;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim C R, Kimata Y, Oka M, Nomaguchi K, Kohno K. Thermosensitivity of green fluorescent protein fluorescence utilized to reveal novel nuclear-like compartments in a mutant nucleoporin NSP1. J Biochem. 1995;118:13–17. doi: 10.1093/oxfordjournals.jbchem.a124868. [DOI] [PubMed] [Google Scholar]
- 31.Lounsbury K M, Richards S A, Carey K L, Macara I G. Mutations within the Ran/TC4 GTPase. J Biol Chem. 1996;271:32834–32841. doi: 10.1074/jbc.271.51.32834. [DOI] [PubMed] [Google Scholar]
- 32.Melchior F, Paschal B, Evans J, Gerace L. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michael W M, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 34.Moore M S, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- 35.Moroianu J, Blobel G, Radu A. Previously identified protein of uncertain function is karyopherin α and together with karyopherin β docks import substrate at nuclear pore complexes. Proc Natl Acad Sci USA. 1995;92:2008–2011. doi: 10.1073/pnas.92.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moroianu J, Blobel G, Radu A. Nuclear protein import: Ran-GTP dissociates the karyopherin αβ heterodimer by displacing α from an overlapping binding site on β. Proc Natl Acad Sci USA. 1996;93:7059–7062. doi: 10.1073/pnas.93.14.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moroianu J, Hijikata M, Blobel G, Radu A. Mammalian karyopherin α1 β and α2 β heterodimers: α1 and α2 subunit binds nuclear localization signal and β subunit interacts with peptide repeat-containing nucleoporins. Proc Natl Acad Sci USA. 1995;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakielny S, Siomi M C, Siomi H, Michael W M, Pollard V, Dreyfuss G. Transportin: nuclear transport receptor of a novel nuclear protein import pathway. Exp Cell Res. 1996;229:261–266. doi: 10.1006/excr.1996.0369. [DOI] [PubMed] [Google Scholar]
- 39.Nehrbass U, Blobel G. Role of the nuclear transport factor p10 in nuclear import. Science. 1996;272:120–122. doi: 10.1126/science.272.5258.120. [DOI] [PubMed] [Google Scholar]
- 40.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 41.Paschal B M, Gerace L. Identification of NTF2, a cytosolic factor for nuclear import that interacts with nuclear pore complex protein p62. J Cell Biol. 1995;129:925–937. doi: 10.1083/jcb.129.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pollard V W, Michael W M, Naklelny S, Siomi M C, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 43.Radu A, Blobel G, Moore M S. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci USA. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 45.Siomi M C, Eder P S, Kataoka N, Wan L, Liu Q, Dreyfuss G. Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J Cell Biol. 1997;138:1181–1192. doi: 10.1083/jcb.138.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sweet D J, Gerace L. A GTPase distinct from Ran is involved in nuclear protein import. J Cell Biol. 1996;133:971–983. doi: 10.1083/jcb.133.5.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang P, Palese P, O’Neill R E. The NPI-1/NPI-3 (karyopherin α) binding site on the influenza A virus nucleoprotein NP is a nonconventional nuclear localization signal. J Virol. 1997;71:1850–1856. doi: 10.1128/jvi.71.3.1850-1856.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weighardt F, Biamonti G, Riva S. Nucleo-cytoplasmic distribution of human hnRNP proteins: a search for the targeting domains in hnRNP A1. J Cell Sci. 1995;108:545–555. doi: 10.1242/jcs.108.2.545. [DOI] [PubMed] [Google Scholar]
- 49.Weis K, Dingwall C, Lamond A I. Characterization of the nuclear protein import mechanism using Ran mutants with altered nucleotide binding specificities. EMBO J. 1996;15:7120–7128. [PMC free article] [PubMed] [Google Scholar]
- 50.Weis K, Mattaj I W, Lamond A I. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science. 1995;268:1049–1053. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]
- 51.Weis K, Ryder U, Lamond A I. The conserved amino-terminal domain of hSRP1α is essential for nuclear protein import. EMBO J. 1996;15:1818–1825. [PMC free article] [PubMed] [Google Scholar]