Abstract
T-cell protein tyrosine phosphatase (TCPTP) exists as two forms generated by alternative splicing: a 48-kDa endoplasmic reticulum (ER)-associated form (TC48) and a 45-kDa nuclear form (TC45). To identify TCPTP substrates, we have generated substrate-trapping mutants, in which the invariant catalytic acid of TCPTP (D182) is mutated to alanine. The TCPTP D182A substrate-trapping mutants were transiently overexpressed in COS cells, and their ability to form complexes with tyrosine-phosphorylated (pTyr) proteins was assessed. No pTyr proteins formed complexes with wild-type TCPTP. In contrast, TC48-D182A formed a complex in the ER with pTyr epidermal growth factor receptor (EGFR). In response to EGF, TC45-D182A exited the nucleus and accumulated in the cytoplasm, where it bound pTyr proteins of ∼50, 57, 64, and 180 kDa. Complex formation was disrupted by vanadate, highlighting the importance of the PTP active site in the interaction and supporting the characterization of these proteins as substrates. Of these TC45 substrates, the ∼57- and 180-kDa proteins were identified as p52Shc and EGFR, respectively. We examined the effects of TC45 on EGFR signaling and observed that it did not modulate EGF-induced activation of p42Erk2. However, TC45 inhibited the EGF-induced association of p52Shc with Grb2, which was attributed to the ability of the PTP to recognize specifically p52Shc phosphorylated on Y239. These results indicate that TC45 recognizes not only selected substrates in a cellular context but also specific sites within substrates and thus may regulate discrete signaling events.
Protein tyrosine phosphatases (PTPs) are a diverse family of enzymes characterized by the consensus sequence (I/V)HCXAGXXR(S/T)G, which contains the catalytically essential Cys and Arg residues. PTPs can be subdivided into transmembrane, receptor-like, and intracellular enzymes. Intracellular PTPs are often modular molecules containing structural motifs such as Src homology 2 (SH2) domains, PEST sequences, and band 4.1 domains on either the N- or C-terminal side of their catalytic domains (55, 58). In most cases, the biological function of individual PTPs remains elusive. To understand the biological roles of this diverse family of enzymes, the identification of their physiological substrates is of paramount importance.
T-cell PTP (TCPTP) is a ubiquitous PTP originally isolated from a human peripheral T-cell cDNA library (12, 13). Although TCPTP was one of the first PTPs to be cloned, its biological function remains unknown. The TCPTP cDNA encodes a 48-kDa protein (TC48) that displays 65% sequence identity overall and 74% sequence identity within the conserved catalytic domain with PTP1B, the prototypic PTP (4, 9, 56, 57). Like PTP1B (17), TC48 is targeted to the endoplasmic reticulum (ER) by a stretch of hydrophobic residues at the extreme C terminus (12, 35). Alternative splicing of the TCPTP transcript gives rise to a 45-kDa form of the enzyme (TC45) which lacks the hydrophobic C-terminal tail (residues 382 to 418) (7, 40, 54) and is targeted to the nucleus by a bipartite nuclear localization sequence (35, 53, 54). Therefore, TC48 and TC45 have the same catalytic domain but are targeted to two distinct sites, the ER and the nucleus, respectively (35).
As a first step toward understanding the function of TCPTP, we have used a substrate-trapping approach in which the invariant PTP catalytic acid (Asp 181 in PTP1B and Asp 182 in TCPTP), which serves as a general acid catalyst to protonate the tyrosyl leaving group in the substrate, has been mutated to Ala (16, 19). We have shown previously that mutation of the PTP catalytic acid yields an enzyme in which catalytic activity is substantially impaired but affinity for substrate is largely unaffected (16, 19). These substrate-trapping mutants can thus form stable complexes with tyrosine-phosphorylated (pTyr) substrates in vivo (16). We have now expressed D182A mutant forms of TC48 and TC45 in COS cells and isolated complexes containing the mutant TCPTPs and their trapped substrates. Despite the fact that the TCPTP variants have identical catalytic domains, the TC48-D182A and TC45-D182A substrate-trapping mutants precipitated distinct pTyr proteins as well as substrates in common. We show that the TC48-D182A mutant remained associated with the ER, whereas the TC45-D182A substrate-trapping mutant underwent a change in localization, exiting the nucleus and accumulating in the cytoplasm in response to epidermal growth factor (EGF). We have demonstrated that TC48-D182A formed a complex with the EGF receptor (EGFR) in the ER, whereas in contrast, the ER-localized PTP1B recognized EGFR and an additional pTyr protein of ∼60 kDa, consistent with a difference in the intrinsic substrate specificity of these closely related enzymes. We have shown that following EGF stimulation and exit of TC45 from the nucleus, EGFR, p52Shc, and two unidentified pTyr proteins are specific substrates of TC45. Furthermore, TC45 recognized specifically p52Shc phosphorylated on tyrosine 239 but not tyrosine 317. Our data suggest that both localization and inherent substrate specificity contribute to the substrate preference displayed by TCPTP in vivo.
MATERIALS AND METHODS
Materials.
Recombinant human EGF was purchased from Genzyme Diagnostics (Cambridge, Mass.), human alpha and beta interferons (IFN-α and -β), recombinant human IFN-γ, and cycloheximide were purchased from Sigma (St. Louis, Mo.), and 4,5-dianilinophthalimide (DAPH) was purchased from Research Biochemicals International (Natick, Mass.). The following constructs were generously provided by various colleagues: glutathione S-transferase (GST)–Grb2 pMT3 and hemagglutinin (HA)-p42Erk2 pJ3H by L. Feig (Tufts University, Boston, Mass.) and J. Chernoff (Temple University, Philadelphia, Pa.), respectively, c-Src Y527F pMIKneo by R. Birge (Rockefeller University, New York, N.Y.), and RasV12 pDCR and Raf-CAAX pcDNA3 by L. Van Aelst (Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.). Antigen for the generation of the monoclonal anti-pTyr antibody G98 (subtype immunoglobulin M [IgM]) was produced as described previously (30) and provided by A. J. Rossomando (Bayer Pharmaceuticals, West Haven, Conn.). The following antibodies were kindly provided: polyclonal affinity-purified anti-TCPTP antibody 6228 (35) by J. A. Lorenzen and E. H. Fischer (University of Washington, Seattle), monoclonal anti-TCPTP antibody CF4 by D. Hill (Calbiochem Oncogene Research Products, Cambridge, Mass.), rabbit polyclonal anti-EGFR serum 1964 by G. Gill (University of California, San Diego), and anti-Src ascites D710 by J. Bolen (DNAX, Palo Alto, Calif.).
Cell culture and transfections.
COS1, HeLa, and NIH 3T3 cells were cultured at 37°C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS), penicillin (100 U/ml), and streptomycin (100 μg/ml). Where indicated, COS1 cells were serum starved for 24 h in DMEM containing 0.1% FBS, penicillin, and streptomycin. COS1 cells were seeded at 6 × 105 per 10-cm-diameter dish, or 2.0 × 105 cells per 6-cm-diameter dish, ∼24 h prior to transfection. Cells were transfected by the calcium phosphate precipitation method using TCPTP- or PTP1B-pMT2 plasmid DNA at 20 μg per 10-cm-diameter dish or 5 μg per 6-cm-diameter dish. A fine DNA-calcium phosphate precipitate was obtained, and at 5 to 6 h, cells were washed three times with phosphate-buffered saline (PBS) and supplemented with fresh DMEM containing 10% FBS. Cells were collected at 36 to 48 h posttransfection, or at 24 h posttransfection cells were serum starved for a further 24 h, and processed for either immunoprecipitation or immunofluorescence assays.
Plasmid constructs.
pBluescript II KS (Stratagene, La Jolla, Calif.) constructs encoding wild-type 48- and 45-kDa TCPTP cDNAs and the wild-type and mutant PTP1B construct in pMT2 have been described elsewhere (16, 53). Site-directed mutagenesis of the 45- and the 48-kDa TCPTP cDNAs to generate D182A mutants in pMT2 was accomplished by using the U-labeled template method of Kunkel (31) and a Bio-Rad (Hercules, Calif.) Mutagene kit. TCPTP 45-kDa pCG constructs were generated by PCR using the cloned Pfu DNA polymerase (Stratagene). The TCPTP/1B chimera construct, encoding the TCPTP catalytic domain (residues 1 to 321) and the C-terminal 35 residues of PTP1B, was generated by PCR and subcloned into the XbaI/EcoRI digested wild-type and mutant 45-kDa TCPTP pMT2 constructs. The c-Src Y527F-pJ3Ω construct was generated by subcloning the c-Src Y527F pMIKneo fragment into pBluescript and then further subcloning the HindIII/KpnI fragment from this into pJ3Ω (39). DNA encoding GST-tagged Shc (amino acids 17 to 473) and the various Shc mutants were generated by subcloning Shc into the pEBG vector (52). Y239F, Y240F, and Y317F mutants were generated by using primers that carry the desired mutations in a PCR-based mutagenesis as described previously (32). The structures of the recombinant plasmids generated were confirmed by restriction endonuclease analysis and sequencing.
Immunoprecipitation and immunoblotting.
Two 10-cm-diameter dishes of cells transfected with the wild-type and mutant TCPTP- or PTP1B-pMT2 constructs were washed three times with ice-cold PBS and lysed in 0.9 ml of IP (immunoprecipitation) lysis buffer (50 mM Tris-HCl [pH 7.5], 1% [wt/vol] Triton X-100, 200 mM NaCl, 5 mM iodoacetic acid, 5 mM NaF, leupeptin [5 μg/ml], aprotinin [5 μg/ml], pepstatin A [1 μg/ml], 1 mM benzamidine, 2 mM phenylmethylsulfonyl fluoride) on ice. Cell lysates were precleared with 0.1 ml of IgG Sorb (The Enzyme Center, Malden, Mass.) for 30 min at 4°C. Precleared lysates were subsequently centrifuged (12,000 × g for 5 min at 4°C), and TCPTP or PTP1B was immunoprecipitated with 15 μg of monoclonal anti-TCPTP antibody CF4 or 10 μl of anti-PTP1B FG6 ascites prebound to protein A-Sepharose CL-4B (Pharmacia, Uppsala, Sweden). Where indicated, TCPTP and the TCPTP/1B chimera were immunoprecipitated for 1 to 2 h at 4°C with 15 μl of rabbit polyclonal antiserum 6228 to TCPTP. The precipitates were collected by centrifugation (12,000 × g, 1 min) and washed once with ice-cold IP lysis buffer, four times with ice-cold IP lysis buffer without iodoacetic acid, and once with ice-cold 50 mM Tris-HCl (pH 7.5). Immune complexes were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to Immobilon (Millipore, Bedford, Mass.), probed with monoclonal anti-pTyr antibody G98 (subtype IgM) at a 1:1,000 dilution of ascites, and detected with peroxidase-conjugated goat anti-IgM (Cappel, Durham, N.C.) at a dilution of 1:4,000. Anti-TCPTP CF4, anti-PTP1B FG6, and anti-pTyr G98 antibodies were diluted in 20 mM Tris-HCl (pH 7.5)–500 mM NaCl–0.05% Tween 20–5% (wt/vol) nonfat dry milk. All other antibodies were diluted in 20 mM Tris-HCl (pH 7.5)–150 mM NaCl–0.1% Tween 20–5% nonfat dry milk. Monoclonal anti-TCPTP antibody CF4 was used at 1 μg/ml, polyclonal anti-Shc antibody (Transduction Laboratories, Lexington, Ky.) was used at 1 μg/ml, monoclonal anti-Grb2 antibody (Transduction Laboratories) was used at 0.25 μg/ml, rabbit anti-EGFR polyclonal antiserum 1964 was used at a dilution of 1:2,000, and monoclonal anti-EGFR antibody EGFR1 (BIOMOL Research Laboratories Inc., Plymouth Meeting, Pa.) was used at 1 μg/ml. These antibodies were detected with peroxidase-conjugated donkey anti-rabbit or sheep anti-mouse antibodies (Amersham Life Science, Cleveland, Ohio) at a dilution of 1:4,000. All blots were developed by using enhanced chemiluminescence (Amersham).
p42Erk2 kinase assays.
COS1 cells, in 10-cm-diameter dishes, were transfected with 2 μg of HA-tagged p42Erk2 pJ3H plasmid and either 15 μg of the pMT2 plasmid or 15 μg of the 45-kDa TCPTP or 45-kDa TCPTP D182A pMT2 plasmid. Cells were serum starved overnight, either left unstimulated or stimulated with 100 ng of EGF per ml for 20 min, and processed for p42Erk2 kinase assays. HA-tagged p42Erk2 was immunoprecipitated with anti-HA antibody 12CA5 (5 μl of ascites per 10-cm-diameter dish of cells) for 90 min at 4°C, and p42Erk2 kinase activity was measured by using myelin basic protein as substrate as previously described (3). Anti-HA immune complexes from these reactions were resolved by SDS-PAGE, immunoblotted with anti-HA antibodies, and quantitated by densitometry in order to normalize for HA-Erk2 activity.
Immunofluorescence.
COS1 cells were seeded onto glass coverslips, transfected, and processed at 36 to 48 h posttransfection. Cells were then washed three times with PBS, fixed for 15 min at room temperature with 3.2% paraformaldehyde in PBS, washed twice with PBS and once with 150 mM glycine in PBS, and incubated with 150 mM glycine in PBS for 10 min. Following two PBS washes, cells were permeabilized with 0.2% (wt/vol) Triton X-100 in PBS (PBST) for 2 min and blocked with 5% normal goat serum in PBST for 30 min. Monoclonal anti-TCPTP antibody CF4 or polyclonal affinity-purified anti-TCPTP antibody 6228 was applied at 0.7 or 0.8 μg/ml, respectively. Polyclonal anti-EGFR serum 1964 (directed to the intracellular EGFR domain) was applied at a dilution of 1:500. Monoclonal anti-protein disulfide isomerase (PDI; StressGen Biotechnologies Corp., Victoria, British Columbia, Canada) was applied at 10 μg/ml. Anti-HA ascites (12CA5), anti-Myc ascites (9E10), and anti-Src ascites (D710) were applied at a dilution of 1:100. Anti-pTyr-specific ascites G98 (subtype IgM) was applied at a dilution of 1:1,000. Antibodies were diluted in PBST containing 0.5% normal goat serum and incubated for 1 h at room temperature. Cells were washed three times with PBST, and fluorescein isothiocyanate (FITC)-conjugated (goat anti-rabbit, goat anti-mouse, or goat anti-mouse IgM) and Texas red-conjugated (sheep anti-mouse or goat anti-rabbit) antibodies (Cappel) were applied for 30 min at a dilution of 1:200 in PBST. Cells were washed twice with PBST and twice with PBS and incubated for 1 min with Hoechst 33258 (Molecular Probes, Eugene, Oreg.) diluted in PBS to a concentration of 2.5 μg/ml. Cells were then washed three times with PBS, and coverslips were mounted in mounting medium (6.6 mM sodium bicarbonate, 0.57 mM anhydrous sodium carbonate, 0.95 mg of p-phenylenediamine [Aldrich, Milwaukee, Wis.] per ml, 80.9% [vol/vol] glycerol [Polysciences Inc., Warrington, Pa.] [final pH of 8.0]). Immunofluorescence was visualized on a Nikon Microphot-FXA microscope.
GST precipitations.
For GST-Grb2 precipitations, COS1 cells grown in 10-cm-diameter dishes were cotransfected with 15 μg of 45-kDa TCPTP pMT2 plasmid, 45-kDa TCPTP D182A pMT2 plasmid, or pMT2 alone and 5 μg of GST-Grb2 pMT3 plasmid. Cells were serum starved and either left unstimulated or stimulated with EGF (100 ng/ml) for 20 min. Cells were then lysed in 0.9 ml of IP lysis buffer, lysates were precleared with 0.1 ml of IgG Sorb and centrifuged (12,000 × g for 5 min at 4°C), and dithiothreitol (DTT) was added to supernatants to 2 mM. GST-Grb2 was precipitated with glutathione-Sepharose 4B (Pharmacia Biotech, Piscataway, N.J.) for 60 min at 4°C, precipitates were washed with IP lysis buffer containing 2 mM DTT and then split into two, and proteins were resolved by SDS-PAGE and immunoblotted with either polyclonal anti-Shc antibody or a monoclonal anti-Grb2 antibody. The TCPTP expression levels in lysates were analyzed by immunoblotting with the anti-TCPTP CF4 antibody.
For GST-Shc precipitations, COS1 cells grown in 10-cm-diameter dishes were transfected with 5 μg of either vector, wild-type Shc, or tyrosine-to-phenylalanine Shc mutants (Y317F, Y239F, Y239F/Y240F, and Y317F/Y239F/Y240F) in pEBG. Cells were serum starved, either left unstimulated or stimulated with EGF (100 ng/ml) for 20 min, then lysed in 0.9 ml of IP lysis buffer containing 2 mM vanadate (without iodoacetic acid), and centrifuged at 12,000 × g for 20 min at 4°C. DTT was added to the supernatants to 1 mM, and GST-Shc was precipitated with glutathione-Sepharose 4B for 90 min at 4°C. Precipitates were washed with the same lysis buffer as specified above containing 1 mM DTT and split into two, and proteins were resolved by SDS-PAGE and immunoblotted with anti-pTyr G98 antibody, anti-Shc antibody, or anti-Grb2 antibody.
45-kDa TCPTP D182A/GST-Shc association assays.
COS1 cells grown in 10-cm-diameter dishes were cotransfected with 5 μg of the 45-kDa TCPTP D182A pMT2 plasmid and either 5 μg of pEBG vector control, wild-type Shc, or tyrosine-to-phenylalanine Shc mutants (Y317F, Y239F, Y239F/Y240F, and Y317F/Y239F/Y240F) in pEBG. At 24 h posttransfection, cells were serum starved for 24 h and either left unstimulated or stimulated with EGF (100 ng/ml) for 20 min. Cells were then lysed in 2.0 ml of IP lysis buffer, and lysates were precleared with IgG Sorb. Precleared lysates were centrifuged (5,000 × g for 5 min at 4°C), and TCPTP was immunoprecipitated with antibody CF4. Precipitates were resolved by SDS-PAGE and immunoblotted with anti-Shc antibody or anti-TCPTP antibody CF4. The expression levels of the GST-Shc proteins were analyzed by immunoblotting with Shc antibodies.
RESULTS
Identification of substrates of TCPTP and PTP1B.
In this study, we have used a substrate-trapping approach (16, 19) to compare the substrate recognition of the alternatively spliced and differentially localized TC48 and TC45 proteins. In addition, we have compared the substrate specificity of TC48 and the highly related enzyme PTP1B (74% sequence identity within the catalytic domain), both of which are targeted to the cytoplasmic face of membranes of the ER (17, 35).
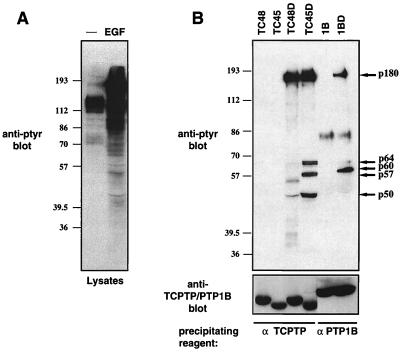
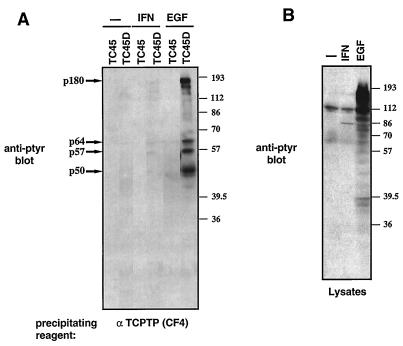
COS1 cells transiently overexpressing wild-type or substrate-trapping mutant forms of either TCPTP or PTP1B were serum starved and stimulated with EGF. Stimulation of COS1 cells with EGF resulted in a dramatic increase in the number of pTyr proteins (Fig. 1A). The expressed PTPs were precipitated with specific monoclonal antibodies to TCPTP and PTP1B, and the associated proteins were visualized by anti-pTyr immunoblotting (Fig. 1B, upper panel). We found that whereas no pTyr proteins were present in wild-type TCPTP or PTP1B immunoprecipitates, a number of pTyr proteins were precipitated by using the substrate-trapping mutants (Fig. 1B, upper panel). Wild-type and substrate-trapping mutants of TCPTP (D182A) and PTP1B (D181A) were expressed and precipitated at similar levels (Fig. 1B, lower panel). Despite the extensive similarity between TCPTP and PTP1B catalytic domains, their respective substrate-trapping mutants precipitated distinct patterns of pTyr proteins (Fig. 1B). Although the TC48-D182A, TC45-D182A, and PTP1B-D181A mutants precipitated a 180-kDa pTyr protein (p180) in common, differences were observed in the lower-molecular-mass pTyr proteins present in the complexes. Only the PTP1B-D181A mutant precipitated a 60-kDa pTyr protein (p60). In addition, the TC45-D182A mutant precipitated pTyr proteins of 50, 57, and 64 kDa (p50, p57, and p64) that were not precipitated by the TC48-D182A or PTP1B-D181A mutant (Fig. 1B). The ∼86-kDa band present in wild-type PTP1B and PTP1B-D181A mutant immunoprecipitates is IgM derived from the anti-PTP1B FG6 ascites used for precipitation and recognized by the anti-IgM peroxidase-conjugated secondary antibody.
FIG. 1.
Precipitation of TCPTP and PTP1B substrates by using the Asp→Ala trapping mutants. (A) COS1 cells were serum starved and stimulated with 100 ng of EGF per ml for 15 min at 37°C. Cells were lysed in 3× Laemmli sample buffer, and proteins were resolved by SDS-PAGE (10% gel) and immunoblotted with the anti-pTyr antibody G98. Molecular size markers (prestained from Sigma; lot 125H9408) are indicated in kilodaltons on the left. (B) COS1 cells transiently transfected with either the 48-kDa (TC48) or 45-kDa (TC45) TCPTP wild-type construct or D182A mutants (TC48D and TC45D) or with the PTP1B (1B) wild-type construct or D181A mutant (1BD) were serum starved and stimulated with EGF (100 ng/ml) for 15 min at 37°C. Cells were then lysed, and TCPTP or PTP1B immunoprecipitates were resolved by SDS-PAGE (10% gel) and immunoblotted with the anti-pTyr antibody G98 (upper panel) or with a TCPTP-specific (CF4) or PTP1B-specific (FG6) antibody (lower panel). The major pTyr proteins coprecipitating with TC48D (p180), TC45D (p180, p64, p57, and p50), or 1BD (p180 and p60) are indicated by arrows on the right, and positions of molecular size standards are shown on the left.
Although the treatment of COS1 cells with EGF stimulated the tyrosine phosphorylation of a large number of proteins (Fig. 1A), the TCPTP and PTP1B substrate-trapping mutants precipitated only a small subset of these potential substrates. Furthermore, the substrate-trapping mutant PTPs did not recognize only the most highly phosphorylated substrates but appeared to bind selectively to pTyr proteins which were minor components of the pTyr pattern in cells lysates (p60 for PTP1B-D181A; p50, p57, and p64 for the TC45-D182A mutant) (Fig. 1A and B, upper panel). These results indicate that TCPTP and PTP1B have a restricted substrate specificity in vivo and, importantly, that they exhibit differences in substrate specificity. The fact that TC48-D182A and TC45-D182A mutants, which have the same catalytic domain, recognized distinct substrates indicates that subcellular localization may play a critical role in dictating TCPTP substrate recognition in vivo.
Differential recognition of p60 by PTP1B and TC48 is due to differences in intrinsic substrate specificity.
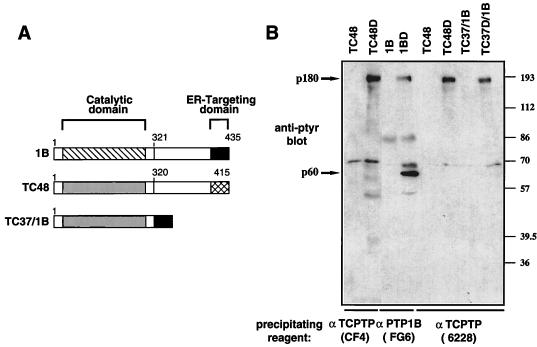
Considering that PTP1B and TC48 both localize to the ER and show 74% identity within their catalytic domains (65% identity overall) (4, 9, 13), one may have predicted that the 60-kDa PTP1B substrate would also be recognized by the TC48-D182A substrate-trapping mutant. To examine whether the apparent specificity of PTP1B for the 60-kDa substrate was attributable to differences in inherent substrate specificity, or to possible differences in the ER microenvironments in which PTP1B and TCPTP reside, we sought to relocalize the TC48-D182A mutant to the precise location of PTP1B in the ER. We generated a TCPTP/PTP1B chimera comprising the TCPTP catalytic domain (residues 1 to 320; TC37) and the extreme C-terminal 35 residues of PTP1B which are necessary and sufficient for ER localization (17) (Fig. 2A). The wild-type (TC37/1B) and D182A (TC37D/1B) chimeras were expressed transiently in COS1 cells, and the ability of the TC37D/1B chimera to associate with pTyr proteins was investigated. Although the TC37D/1B chimera was efficiently targeted to the ER by the C-terminal 35 residues of PTP1B, as assessed by its colocalization with the ER marker PDI (data not shown), it did not precipitate the PTP1B 60-kDa substrate (Fig. 2). Similar levels of TCPTP, PTP1B, and the chimeric proteins were expressed and precipitated (data not shown). These results indicate that the selectivity of PTP1B for the 60-kDa substrate is most likely attributable to inherent substrate specificity and not to differences in the ER microenvironments of TCPTP and PTP1B. A nonspecific band of ∼70 kDa is present in all immunoprecipitates, including the vector-alone control (data not shown), and therefore is not a substrate of TCPTP or PTP1B. The reverse chimera, containing the PTP1B catalytic domain (residues 1 to 321) and the TCPTP C-terminal domain (residues 320 to 415), was unstable and proteolyzed (data not shown). The results indicate that the PTP1B and TC48 exhibit differences in specificity despite their similar subcellular localization and high degree of sequence similarity. Whether the specificity of PTP1B for the 60-kDa substrate is attributable solely to its catalytic domain or also involves sequences from the noncatalytic C-terminal segment remains to be established.
FIG. 2.
PTP1B, but not TC48, exhibits inherent specificity for pTyr protein p60. (A) Schematic representation of PTP1B (1B), the 48-kDa TCPTP (TC48), and the TC37/1B chimera. (B) COS1 cells transiently transfected with the constructs indicated at the top were serum starved and stimulated with EGF for 15 min. Cells were then lysed, and immunoprecipitates were generated with the indicated antibodies. The epitope for anti-TCPTP CF4 is within residues 349 to 415 of TC48, whereas that of the anti-TCPTP 6228 epitope is within TCPTP residues 1 to 320. Immunoprecipitates were resolved by SDS-PAGE (10% gel) and immunoblotted with the anti-pTyr antibody G98. p180 and p60 substrates are indicated by arrows on the left, and molecular size standards (in kilodaltons) are on the right. The heavy ∼86-kDa band present in both wild-type and mutant 1B immunoprecipitates is IgM derived from the precipitating antibody (anti-PTP1B FG6 ascites).
Criteria for classification of pTyr proteins as PTP substrates.
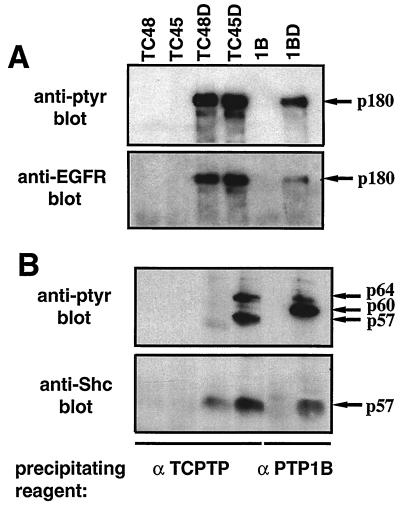
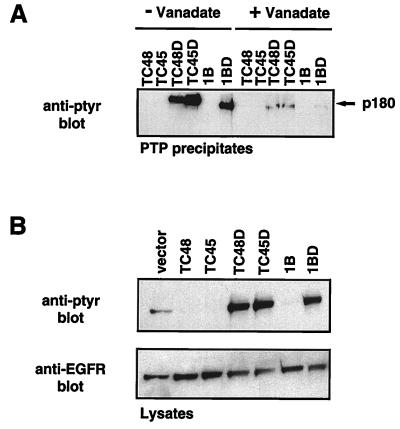
The 180-kDa substrate recognized by the TC45, TC48, and PTP1B substrate-trapping mutants was identified by immunoblot analysis as the EGFR (Fig. 3A). The following observations support the classification of EGFR as a substrate for these PTPs. First, the inclusion of vanadate, a PTP inhibitor that covalently modifies the invariant, essential nucleophilic cysteine residue at the active site (14, 28), disrupted formation of complexes between the substrate-trapping mutant PTPs and the pTyr EGFR (Fig. 4A). Similarly, addition of vanadate disrupted all of the mutant PTP-pTyr protein complexes observed in Fig. 1 (data not shown). The findings that wild-type PTPs did not precipitate the pTyr proteins or the dephosphorylated form of the EGFR and that vanadate disrupted the association of pTyr proteins with the substrate-trapping mutant PTPs indicate that the pTyr proteins that coprecipitated with the mutant enzymes are likely to be PTP substrates that interact exclusively through the active site. Second, comparison of the state of tyrosine phosphorylation of the EGFR in cell lysates following expression of either wild-type or substrate-trapping mutant PTPs (Fig. 4B) revealed that in comparison to cells expressing the vector control, the EGFR is dephosphorylated by the wild-type PTPs but protected from dephosphorylation by the substrate-trapping mutants, consistent with a direct recognition of the EGFR as substrate by these PTPs.
FIG. 3.
Interaction of substrate-trapping mutant TCPTP and pTyr EGFR and -p52Shc. (A) TCPTP (TC48 and TC45) or PTP1B (1B) or their respective Asp→Ala mutants (TC48D, TC45D, and 1BD) were immunoprecipitated, resolved by SDS-PAGE, and immunoblotted with either the anti-pTyr antibody G98 (upper panel) or anti-EGFR rabbit polyclonal antiserum 1964 (lower panel). (B) TCPTP and PTP1B wild-type or D182A immunoprecipitates were resolved by SDS-PAGE and immunoblotted with either anti-pTyr antibody G98 (upper panel) or anti-Shc antibody (lower panel). Panels A and B show different regions of different gels.
FIG. 4.
The EGFR is a substrate of TCPTP. (A) COS1 cells transiently transfected with either the wild-type 48-kDa (TC48) or 45-kDa (TC45) TCPTP or D182A mutants (TC48D and TC45D) or with the wild-type PTP1B (1B) or D181A mutant (1BD) were serum starved and stimulated with EGF (100 ng/ml) for 15 min at 37°C. Cells were lysed in the presence or absence of 2 mM vanadate, and TCPTP or PTP1B immunoprecipitates were resolved by SDS-PAGE (10% gel) and immunoblotted with anti-pTyr antibody G98. (B) COS1 cells transiently transfected with indicated plasmid constructs were serum starved and stimulated with EGF (100 ng/ml) for 15 min at 37°C. Cells were lysed in IP lysis buffer, lysates were precleared with IgG Sorb, and supernatants were resolved by SDS-PAGE (10% gel) and immunoblotted with either the anti-pTyr antibody G98 (upper panel) or anti-EGFR rabbit polyclonal antiserum 1964 (lower panel).
The TC48-D182A mutant traps the EGFR at the ER.
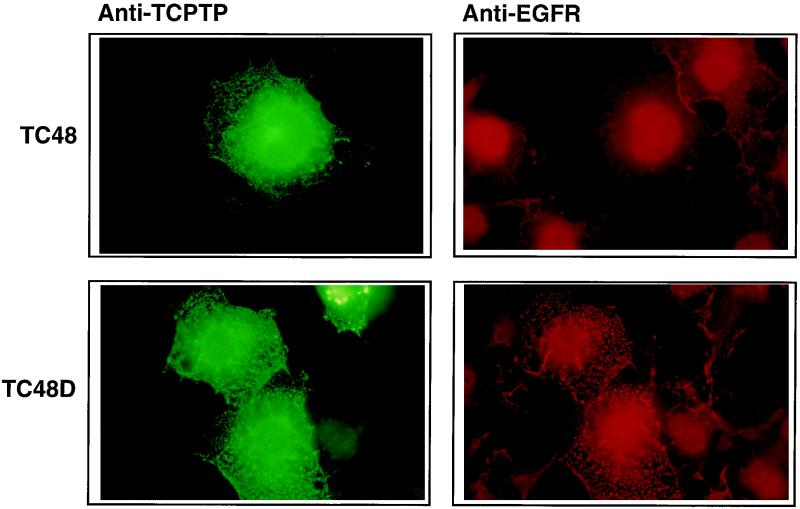
Previously, we have reported that the PTP1B-D181A mutant retains and accumulates the endogenous EGFR in an intracellular complex (16). To investigate whether the TC48-D182A mutant also results in the accumulation of EGFR, we examined the location of endogenous EGFR in randomly growing COS1 cells transfected with either TC48 or the TC48-D182A mutant. TC48 and TC48-D182A exhibited a reticular staining pattern in COS1 cells (Fig. 5) which colocalized with the ER marker PDI (data not shown). In contrast, the endogenous EGFR exhibited a uniform and diffuse staining in cells overexpressing TC48 (Fig. 5, upper right). However, in cells expressing the TC48-D182A substrate-trapping mutant, the endogenous EGFR displayed intense punctuate staining (Fig. 5, lower right) which colocalized with foci of anti-TCPTP staining (Fig. 5, lower left). Similar results were obtained for TC48-D182A in cells following serum starvation with or without EGF stimulation (data not shown). These results demonstrate that expression of the TC48-D182A substrate-trapping mutant modifies localization of EGFR, likely through a physical association which retains and accumulates the EGFR in the ER. These data illustrate the important point that the complex was formed in the intact cell and did not arise from an event that occurred postlysis.
FIG. 5.
Colocalization of the TC48-D182A substrate-trapping mutant and endogenous EGFRs. COS1 cells seeded on glass coverslips were transfected with wild-type TC48 (upper half) or D182A mutant (lower half). At 36 h posttransfection, cells were fixed with paraformaldehyde and processed for immunofluorescence as described in Materials and Methods. Cells were incubated with mouse monoclonal TCPTP CF4 antibody and anti-EGFR rabbit polyclonal antiserum 1964. Overexpressed TC48 (left) and endogenous EGFR (right) were visualized with FITC-conjugated goat anti-mouse and Texas red-conjugated goat anti-rabbit antibodies, respectively. Original magnification, ×600.
EGFR and p52Shc are specific substrates of TC45.
The 180-kDa pTyr protein precipitated by the TC45-D182A mutant was identified by immunoblot analysis as the EGFR (Fig. 3A), whereas the 57-kDa pTyr protein was identified as the adaptor protein p52Shc (Fig. 3B). Neither p52Shc nor EGFR was present in immunoprecipitates of wild-type TCPTP or PTP1B. We used a specific monoclonal antibody that recognizes all isoforms of Shc in COS1 cells (p46Shc, p52Shc, and p66Shc) but did not detect cross-reactivity with the 50- or 64-kDa pTyr protein precipitated by the TC45-D182A mutant (data not shown). p52Shc was also detected in immunoprecipitates of the TC48-D182A and PTP1B-D181A mutants (Fig. 3B); however, it was not tyrosine phosphorylated, indicating that p52Shc is not a substrate for either TC48 or PTP1B (Fig. 1B, 2, and 3B). Shc is known to associate directly with pTyr EGFR via its PTB and SH2 domains (1, 2); however, it is not tyrosine phosphorylated when cell lysates are prepared in the absence of the PTP inhibitor vanadate. In these experiments, the cells were lysed in the absence of vanadate; therefore, it is likely that the presence of unphosphorylated p52Shc in TC48-D182A and PTP1B-D181A immunoprecipitates is due to its direct association with the pTyr EGFR. In addition, although equal amounts of pTyr EGFR were precipitated by the TC45-D182A and TC48-D182A mutants (Fig. 3A), more p52Shc protein associated with the TC45-D182A mutant than the TC48-D182A mutant (Fig. 3B), presumably reflecting an EGFR-independent interaction between pTyr p52Shc and TC45-D182A.
The TC45-D182A mutant undergoes a change in localization from the nucleus to the cytoplasm following EGF stimulation.
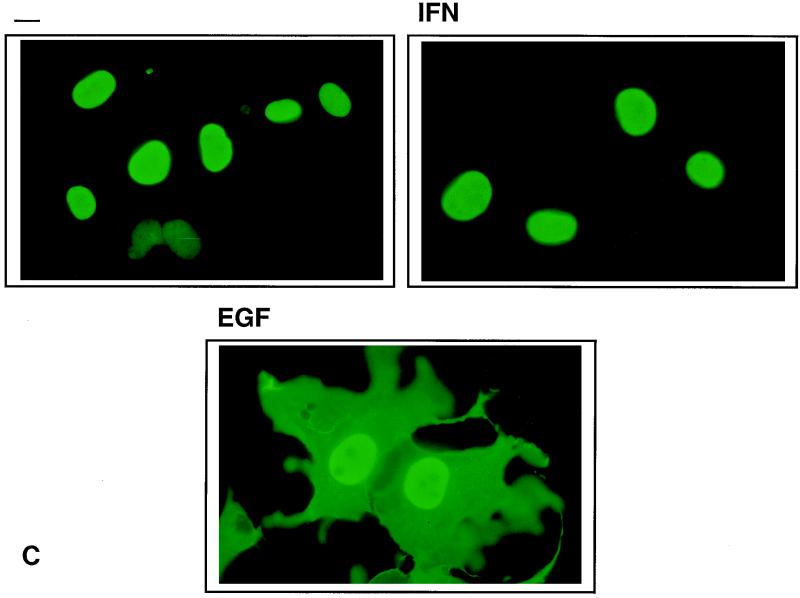
At first, the ability of the TC45-D182A mutant to associate with EGFR and p52Shc appeared to be enigmatic. Numerous studies including our own (35, 53, 54) have reported exclusive nuclear localization of TC45, whereas EGFR and Shc are known to reside outside the nucleus. We postulated that the TC45-D182A mutant may relocalize to the cytoplasm, thereby allowing its association with pTyr EGFR and Shc. To address this possibility, we correlated the precipitation of pTyr proteins with the localization of the TC45-D182A mutant in cells stimulated with IFN-α and -γ or EGF. We used IFN as a control because it stimulates the JAK/STAT pathway and results in translocation of tyrosyl-phosphorylated STATs to the nucleus (reviewed in reference 29). We found that in serum-starved cells or in cells stimulated with IFN, there was no detectable association of the TC45-D182A mutant with pTyr proteins (Fig. 6A). Notably, the TC45-D182A mutant did not precipitate the IFN-inducible, ∼86-kDa pTyr protein (Fig. 6A) which was present in the lysates (Fig. 6B) and was identified by immunoblot analysis as STAT1 (data not shown). In serum-starved cells and in IFN-stimulated cells, the TC45-D182A mutant was predominantly nuclear (Fig. 6C). In marked contrast, stimulation of COS1 cells overexpressing the TC45-D182A mutant with EGF resulted in the precipitation of the pTyr proteins described above (Fig. 6A), which correlated with a dramatic increase of TCPTP in the cytoplasm (Fig. 6C). This change in localization was transient; it occurred rapidly, within 5 min of EGF stimulation, reached a maximum within 15 to 30 min in ∼90% of cells overexpressing the TC45-D182A protein, and returned to the nucleus within 6 h (data not shown). Considering that the change in localization occurred in such a short time period, it is unlikely to represent de novo synthesis of the TC45-D182A protein.
FIG. 6.
EGF-dependent change in localization of TC45-D182A and association with pTyr substrates. COS1 cells transfected with the 45-kDa TCPTP wild type (TC45) or D182A mutant (TC45D) were serum starved and either left unstimulated (−) or stimulated with EGF (100 ng/ml) or IFN (10 ng of IFN-γ and 1,000 U of IFN-α per ml) for 15 min. (A) Cells were lysed, and overexpressed TCPTP was immunoprecipitated, resolved by SDS-PAGE (10% gel), and immunoblotted with the anti-pTyr antibody G98. The major pTyr-containing proteins coprecipitating with TC45D (p180, p64, p57, and p50) are indicated by arrows on the left. Molecular size standards (in kilodaltons) are shown on the right. (B) COS1 cells were serum starved and stimulated with either EGF or IFN as described above. Cells were lysed in 3× Laemmli sample buffer, and proteins were resolved by SDS-PAGE (10% gel) and immunoblotted with the anti-pTyr antibody G98. (C) Cells were processed for immunofluorescence by using the anti-TCPTP CF4 antibody. Original magnification, ×600.
We also observed that the TC45-D182A mutant accumulated in the cytoplasm of 30 to 50% of randomly growing cells (data not shown), indicating that a signal to trigger this event exists in randomly growing cells. Although an EGF-dependent change in localization of the TC45-D182A mutant was readily observed, a change in localization of the wild-type TC45 enzyme could not be detected by immunofluorescence. Therefore, heterokaryon assays, which are used to characterize proteins that are retained in the nucleus or that undergo nucleocytoplasmic transport (41, 44), were performed as an independent confirmation of the ability of TC45 to exit the nucleus. Randomly growing HeLa cells overexpressing TC45 and randomly growing, untransfected NIH 3T3 cells were fused to form heterokaryons in which human and mouse nuclei share a common cytoplasm. The overexpressed TCPTP was detected in both HeLa and 3T3 nuclei but not in the cytoplasm of the heterokaryons (data not shown). Since heterokaryons were generated in the presence of cycloheximide, the recovery of TC45 in 3T3 nuclei reflects shuttling of the PTP between the nucleus and cytoplasm rather than de novo synthesis. These results demonstrate that TC45 is not restricted to the nucleus, but that it can exit and therefore attain access to cytoplasmically localized substrates.
The change in TC45-D182A localization requires EGFR tyrosine kinase activity.
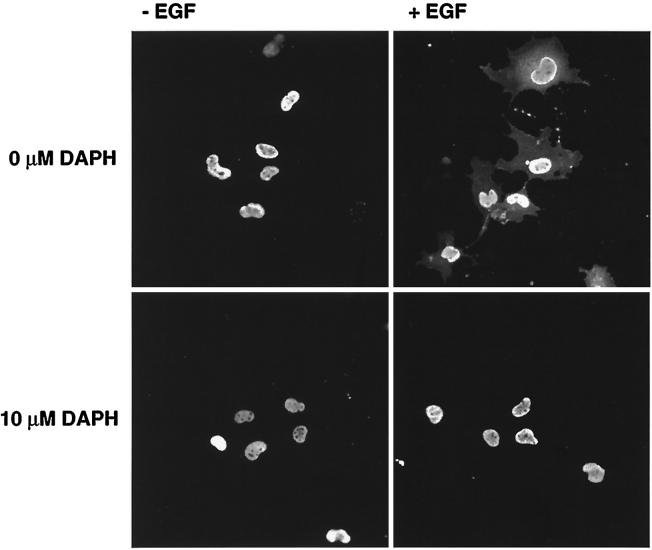
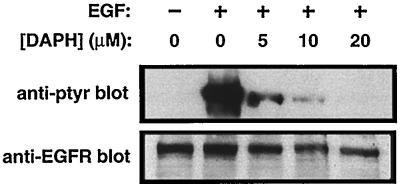
To characterize further the cytoplasmic translocation of TC45-D182A in response to EGF, we determined whether EGFR activation was required for this event to occur. COS1 cells overexpressing TC45-D182A were pretreated for 90 min with the protein tyrosine kinase (PTK) inhibitor DAPH, which at low concentrations (≤10 μM) is highly selective for the EGFR (5), and the effect of EGF on TC45-D182A localization was assessed (Fig. 7). In serum-starved COS1 cells, 10 μM DAPH significantly inhibited the EGF-induced EGFR autophosphorylation and the cytoplasmic translocation of the TC45-D182A mutant (Fig. 7). These results suggest that EGFR PTK activity is essential for the EGF-induced change in localization of TC45-D182A.
FIG. 7.
The EGFR-specific PTK inhibitor DAPH inhibits the EGF-dependent change in localization of the TC45-D182A substrate-trapping mutant. COS1 cells grown on coverslips and transfected with mutant TC45-D182A were serum starved and preincubated with 0, 5, 10, or 50 μM DAPH, the specific inhibitor of the EGFR PTK, for 90 min at 37°C and 5% CO2. Cells were either left unstimulated (−) or stimulated with EGF (100 ng/ml) for 15 min. Cells were subsequently either fixed with paraformaldehyde and processed for immunofluorescence by using the anti-TCPTP CF4 antibody or lysed in 3× Laemmli sample buffer, and proteins were resolved by SDS-PAGE (10% gel). Resolved proteins were then immunoblotted with the anti-pTyr antibody G98 or the monoclonal anti-EGFR antibody EGFR1. The top panel shows representative fields from immunofluorescence studies at a magnification of ×200.
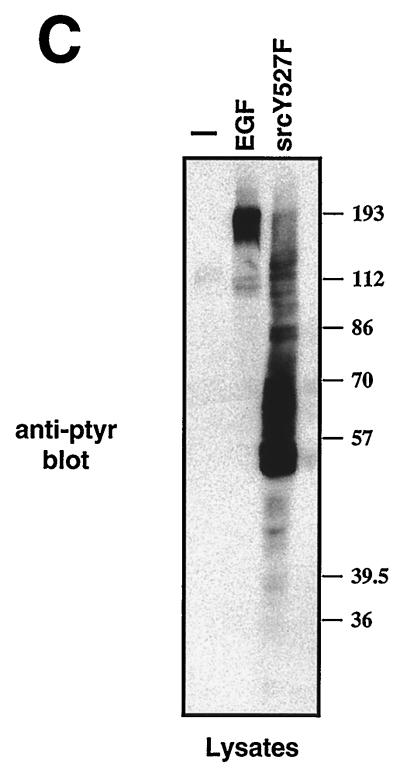
In addition, we investigated the influence of downstream targets of the EGFR on TC45-D182A localization. Pretreatment of TC45-D182A-expressing cells with rapamycin and wortmannin, which inhibit the activation of the 70-kDa S6 kinase (10) and phosphatidylinositol-3-kinase activities, respectively (23, 60), did not prevent the EGF-induced change in localization (data not shown). Coexpression of TC45-D182A with constitutively active forms of Ras (RasV12) and Raf (Raf-CAAX), which induced mitogen-activated protein kinase (MAPK) activation, did not stimulate cytoplasmic translocation in randomly growing cells in which the TC45-D182A protein was nuclear (data not shown). Consistent with previous reports (37, 59), overexpression of a constitutively active form of c-Src, Src Y527F, dramatically increased the total cellular pTyr content (Fig. 8B and C) and resulted in the phosphorylation of Shc on Y239/240 and Y317, a TCPTP substrate (data not shown). However, we found that coexpression of Src Y527F did not significantly alter localization of the TC45-D182 in serum-starved cells (Fig. 8A). These results suggest that the change in TCPTP localization may result from a specific signaling event emanating from the EGFR PTK and is not due simply to hyperactivation of signaling pathways or hyperphosphorylation of proteins on tyrosine in general.
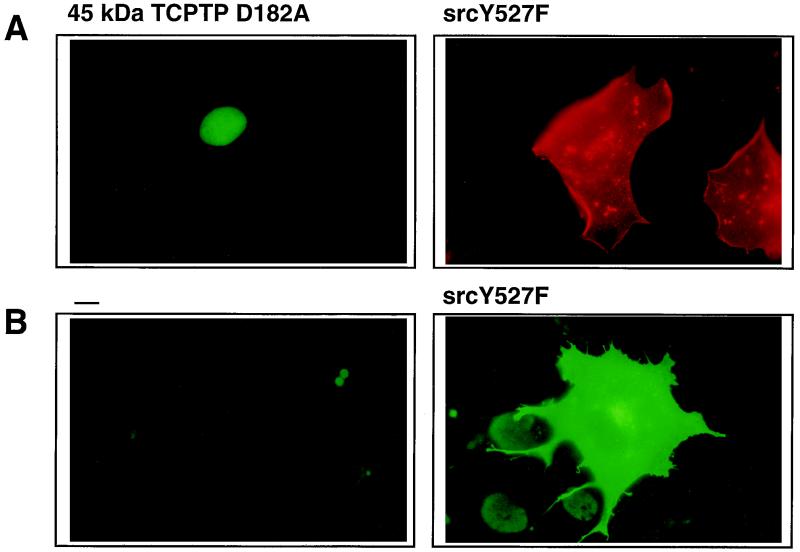
FIG. 8.
Expression of Src Y527F does not induce the change in TC45-D182A localization. COS1 cells seeded on glass coverslips in 60-mm-diameter dishes were cotransfected with either 5 μg of TC45-D182A plasmid and 1 μg of Src Y527F plasmid or transfected with 1 μg of Src Y527F plasmid alone. Transfected cells were serum starved and processed for immunofluorescence by using the TCPTP antibody CF4. (A) Cells transfected with TC45-D182A and Src Y527F were incubated with rabbit polyclonal TCPTP antibody 6228 and monoclonal anti-Src antibody D710. Overexpressed Src Y527F was visualized with Texas red-conjugated sheep anti-mouse antibodies, and TCPTP was visualized with FITC-conjugated goat anti-rabbit antibodies. (B) Cells transfected with empty vector (left) or with Src Y527F plasmid (right) were incubated with the pTyr antibody G98. pTyr was visualized with FITC-conjugated goat anti-mouse IgM. (C) COS1 cells transfected with empty vector or with Src Y527F plasmid were serum starved and either left unstimulated (−) or stimulated with EGF for 15 min. Cells were then collected in 3× Laemmli sample buffer, and proteins were resolved by SDS-PAGE (10% gel) and immunoblotted with anti-pTyr antibody G98. Molecular size standards (in kilodaltons) are shown on the right.
TC45 modulates the association of p52Shc with Grb2 but not the activation of p42Erk2.
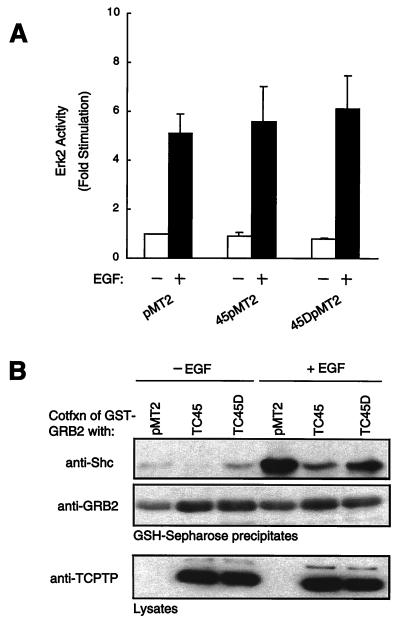
The identification of EGFR and p52Shc as potential substrates of TC45 suggests that the PTP may modulate the function of these molecules in EGFR signaling. First, we examined the effect of TCPTP on the activation of the MAPK p42Erk2 in response to EGF (Fig. 9A). COS1 cells were cotransfected with HA-tagged p42Erk2 and either the wild-type TC45 or the TC45-D182A mutant. The effects of TCPTP on the EGF-induced p42Erk2 activity were ascertained at EGF concentrations of 1 ng/ml, 10 ng/ml (data not shown), and 100 ng/ml (Fig. 9A) and at 2 min (data not shown) and 15 min (Fig. 9A) after the addition of growth factor. Although the TC45-D182A mutant precipitated pTyr EGFR and p52Shc under these conditions, it failed to modulate basal or EGF-inducible p42Erk2 activity (Fig. 9A). These results indicate that the association of the TC45-D182A mutant with the EGFR, p52Shc, p50, and p64 does not modulate p42Erk2 activation in response to EGF.
FIG. 9.
TC45 does not modulate MAPK activity but inhibits the association of p52Shc with Grb2. (A) COS1 cells were cotransfected with HA-tagged p42Erk2 plasmid and either TC45 or TC45-D182A mutant plasmid or vector alone. Transfected cells were serum starved and either left unstimulated or stimulated with EGF (100 ng/ml) for 20 min. Cells were then lysed, and HA-tagged p42Erk2 was precipitated and assayed by using myelin basic protein as substrate. Activity was normalized for HA-Erk2 protein. Values shown are arbitrary units and are means ± standard errors of three independent experiments. (B) COS1 cells were cotransfected (Cotfxn) with GST-Grb2 plasmid and either TC45 or TC45-D182A plasmid or vector alone. Transfected cells were serum starved and either left unstimulated or stimulated with EGF (100 ng/ml) for 15 min. Cells were then lysed, and GST-Grb2 was precipitated with glutathione (GSH)-Sepharose 4B. Precipitates were resolved by SDS-PAGE and immunoblotted with anti-Shc antibody or anti-Grb2 antibody. The expression level of TCPTP in lysates was analyzed by using antibody CF4.
Since the results of this study indicate that p52Shc is a substrate of TC45, we formed the hypothesis that the phosphatase may affect the association of Shc with Grb2. COS1 cells were cotransfected with GST-Grb2 and either the wild-type TC45 or the TC45-D182A mutant. GST-Grb2 was precipitated from both serum-starved and EGF-stimulated cells, and the effect of TCPTP on the association with p52Shc was assessed by immunoblotting. We found that overexpression of the wild-type TC45, and to a lesser extent the TC45-D182A mutant, impaired EGF-induced association of p52Shc with Grb2 (Fig. 9B). That the TC45-D182A inhibition of GST-Grb2/Shc association was less efficient than that of the wild-type enzyme can most likely be attributed to substrate trapping by the D182A mutant being less efficient than dephosphorylation in vivo. These results suggest that TCPTP may serve to modulate a p52Shc/Grb2-induced signaling pathway that is independent of EGF-induced p42Erk2 activation.
The TC45-D182A mutant recognizes specific sites of tyrosine phosphorylation on Shc.
Recent studies have demonstrated that Shc is phosphorylated on tyrosines 239 and 240 as well as tyrosine 317 following EGFR activation (21, 22, 24, 59). Both the Y317 and Y239 sites are capable of binding Grb2 (22, 24, 59). The presence of multiple sites of tyrosine phosphorylation on Shc suggests that it may participate in more than one signaling pathway. Indeed, the Shc phosphorylation site mutant Y317F can inhibit the EGF-induced stimulation of MAPK, whereas the Y239F/Y240F mutant does not (22), implicating Y317 and not Y239/Y240 in activation of the MAPK pathway. We tested whether TC45 recognizes Shc Y239/Y240 specifically and thus can exert an effect on Shc/Grb2 association without affecting p42Erk2 activation.
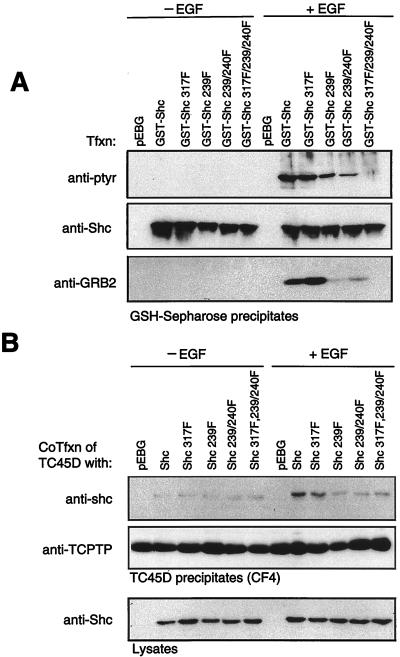
First, we examined both the phosphorylation state of Shc and the association of endogenous Grb2 following EGF stimulation (Fig. 10A). COS1 cells were transfected with wild-type or various tyrosine-to-phenylalanine mutants of a GST-Shc fusion protein (Y317F, Y239F, Y239/240F, and Y317F/Y239F/Y240F), and the state of Shc phosphorylation in GST-Shc precipitates was determined by anti-pTyr immunoblotting. Consistent with previous studies (22, 59), we observed phosphorylation of Shc in response to EGF not only on tyrosine 317 but also on tyrosines 239 and 240 (Fig. 10A). The Y→F mutations in Shc did not alter the ability of the GST-Shc mutants to associate with the EGFR (data not shown). Therefore, differential phosphorylation of the GST-Shc mutants was not due to inappropriate localization. When the same GST-Shc precipitates were immunoblotted with antibodies to Grb2, we observed that although the wild-type GST-Shc and the GST-Shc Y317F mutant associated with endogenous Grb2 to similar levels, Grb2 binding was significantly reduced in GST-Shc Y239F and Y239F/240F precipitates (Fig. 10A). These results indicate that in COS1 cells, tyrosines 239 and 240 of p52Shc are phosphorylated and that tyrosine 239 is a major Grb2 binding site (Fig. 10A). Taken together with the ability of TCPTP to inhibit the association of Grb2 with p52Shc without affecting p42Erk2 activity, these results strongly suggest that TC45 specifically recognizes Shc phosphorylated on Y239 but not Y317.
FIG. 10.
Tyrosine 239 of p52Shc is the major Grb2 binding site and is recognized by TC45-D182A. (A) COS1 cells were transfected (Tfxn) with either the wild type or the indicated tyrosine-to-phenylalanine phosphorylation site mutants of GST-Shc. Transfected cells were serum starved, either left unstimulated or stimulated with EGF (100 ng/ml) for 15 min, and lysed, and overexpressed GST fusion protein was collected on glutathione (GSH)-Sepharose 4B. Precipitates were resolved by SDS-PAGE and immunoblotted with either anti-pTyr antibody G98 or an anti-Shc or anti-Grb2 antibody. The apparent difference in the association of Grb2 with the Y239F and Y239F/Y240F GST-Shc mutants was not reproducible. (B) COS1 cells were cotransfected (CoTfxn) with the TC45-D182A mutant and the wild type or the indicated tyrosine-to-phenylalanine phosphorylation site mutants of GST-Shc. Cells were serum starved, either left unstimulated or stimulated with EGF (100 ng/ml, 15 min), and then lysed, and TC45-D182A was precipitated with specific antibodies. Precipitates were resolved by SDS-PAGE and immunoblotted with anti-Shc or anti-TCPTP (CF4) antibody. The expression level of the GST-Shc proteins in the lysates was analyzed by using an anti-Shc antibody.
To test this hypothesis, we assessed the ability of TC45-D182A to associate, in an EGF-dependent manner, with Shc mutated in the various phosphorylation sites (Fig. 10B). COS1 cells were cotransfected with the TC45-D182A mutant and the wild-type or mutant Y→F GST-Shc constructs. The amount of GST-Shc in TC45-D182A precipitates was determined following EGF stimulation by immunoblotting with anti-Shc antibodies. GST-Shc associated with TC45 in a nonspecific manner which was independent of tyrosine phosphorylation and was not observed for the endogenous p52Shc. To minimize such nonspecific interactions, we expressed TC45-D182A in lower amounts. Similar amounts of TC45-D182A were precipitated, and the Shc proteins were also expressed to similar levels in each condition (Fig. 10B). We observed that association of the TC45-D182A substrate-trapping mutant with GST-Shc and GST-Shc Y317F, but not GST-Shc Y239F, Y239F/Y240F or Y239F/Y240F/Y317F, was induced in response to EGF. The results clearly demonstrate that Y239 of Shc is required for association with the TC45-D182A substrate-trapping mutant (Fig. 10B). These results therefore reinforce the hypothesis that TC45 modulates specifically the phosphorylation of p52Shc at tyrosine 239.
DISCUSSION
Previously we reported the development of a substrate-trapping mutant for the identification of physiological PTP substrates. Mutagenesis to alanine of the essential catalytic acid residue which is invariant in the PTP family of enzymes, Asp 181 in PTP1B, resulted in a mutant enzyme which displayed a Km comparable to that of the wild-type phosphatase but a kcat which was reduced by a factor of ∼105 (16). As such, this mutant retains the ability to bind substrates but is catalytically impaired and thus forms stable enzyme-substrate complexes. In this study, we have used this approach to identify substrates for the closely related phosphatase, TCPTP.
We have demonstrated that TCPTP and PTP1B, which are promiscuous in vitro (25, 48, 56, 61), are highly specific in their substrate preference when examined in a cellular context. Although TC48 and PTP1B both localize to the ER and have 65% sequence identity overall (74% within their catalytic domains), they recognized common and distinct substrates. Both TC48-D182A and PTP1B-D181A substrate-trapping mutants precipitated the EGFR and both PTP mutants accumulated the EGFR at the ER, illustrating that complex formation occurred within the intact cell and not postlysis. Importantly, these complexes were not formed by the wild-type enzymes, nor did the wild-type enzymes accumulate the EGFR at the ER. In addition, it is known that vanadate functions as a PTP inhibitor by modifying the essential nucleophilic cysteinyl residue, characteristic of the PTP signature motif, which is located at the active site of the enzyme (14, 28). We observed that complex formation between the substrate-trapping mutant PTPs and the pTyr proteins was disrupted when cells were lysed in the presence of 2 mM vanadate, illustrating that the interaction involved the PTP active site. Therefore the association between the substrate-trapping mutants and pTyr proteins most likely reflects an enzyme-substrate interaction.
In addition to the EGFR, the PTP1B substrate-trapping mutant precipitated a pTyr protein with an apparent molecular mass of ∼60 kDa which was not precipitated by either the TC48-D182A or the TC45-D182A substrate-trapping mutant. The identity of the PTP1B 60-kDa substrate (previously referred to as p70 due to differences in Mr standards used for SDS-PAGE [16]) remains unknown. We have tested but failed to detect cross-reactivity with pTyr proteins within this molecular weight range in assays using antibodies to c-Src, c-Yes, paxillin, Sam68, p62Dok, protein kinase Cδ, Raf-1, SHP-2, and Shc. Using a TCPTP/1B chimera which relocalizes TC48 to the precise location of PTP1B in the ER, we demonstrated that the recognition of the 60-kDa substrate by PTP1B is due to inherent specificity rather than differences in the ER microenvironments of PTP1B and TCPTP. These data illustrate that intrinsic properties of individual PTPs contribute to their apparent substrate specificity.
The function of PTP1B and TC48 remains to be defined. Neither PTP1B nor TC48 affected the EGF-inducible p42Erk2 activity (data not shown), indicating that events triggered at the plasma membrane are not modulated by these ER-localized enzymes. Nevertheless, one of the functions of these highly related PTPs may be to prevent inappropriate signalling from the EGFR which may occur in a ligand-independent fashion as the nascent receptor PTK undergoes posttranslational modifications in the ER (51). Furthermore, a number of transforming, mutant receptor PTKs that display ligand-independent activity, including mutant forms of the EGFR (15) and HER2 (27), have been identified. Interestingly, there are data to suggest that some of these mutant PTKs signal while localized in the ER (15, 27). Therefore, PTPs such as PTP1B and TC48 have the potential to attenuate such inappropriate signaling events.
We have demonstrated that the differentially localized variants of TCPTP recognize distinct subsets of substrates, highlighting the additional importance of subcellular location in determining substrate specificity. In contrast to the TC48-D182A mutant, which precipitated only the EGFR, the TC45-D182A substrate-trapping mutant precipitated four pTyr proteins with apparent molecular masses of 50, 57, 64, and 180 kDa. We have identified two of the TC45 substrates as the EGFR (p180) and p52Shc (p57). The 50- and 64-kDa pTyr proteins that associated with TC45-D182A did not cross-react with antibodies that recognize all three isoforms of Shc (p46Shc, p52Shc, and p66Shc) and thus are neither p46Shc nor p66Shc (43). The sites of phosphorylation in p46Shc and p66Shc have yet to be defined. As discussed below, the phosphorylation of Shc on Y239 is essential for association with the TC45-D182A mutant. Therefore one possibility is that the p46Shc and p66Shc isoforms are not precipitated by TC45-D182A because they are not efficiently phosphorylated on the residue equivalent to Y239 in p52Shc. In addition to p52Shc and the EGFR, we also tested antibodies to c-Src, c-Yes, paxillin, Sam68, p62Dok, protein kinase Cδ, Raf-1, SHP-2, and Nck but failed to detect cross-reactivity with the 64- and 50-kDa pTyr proteins precipitated by TC45-D182A.
Although the EGFR and Shc are localized cytoplasmically, TC45 is predominantly nuclear in randomly growing cells. We observed that the pTyr proteins associated with TC45-D182A only when cells were stimulated with EGF. This correlated with the EGF-induced exit from the nucleus and accumulation of the TC45-D182A substrate-trapping mutant in the cytoplasm. We were unable to detect an accumulation of wild-type TC45 in the cytoplasm but could detect readily its ability to exit the nucleus by heterokaryon analysis (41, 44) (data not shown). Our ability to detect accumulation in the cytoplasm of the TC45-D182A substrate-trapping mutant, but not the wild-type TC45, is most likely attributable to the ability of the former to generate stable complexes with pTyr substrates following EGF stimulation which would prevent reentry into the nucleus.
Although the accumulation of the TC45-D182A substrate-trapping mutant in the cytoplasm following EGFR activation was reflected in the formation of stable complexes with pTyr substrates, it is important to note that hyperphosphorylation of proteins by constitutively activated Src (Y527F) was not sufficient to induce the cytoplasmic accumulation of the phosphatase. Indeed, the phosphorylation of the TCPTP substrate Shc on tyrosines 317 and 239/240 by the activated Src Y527F (data not shown) did not induce the accumulation of TC45-D182A in the cytoplasm. These results suggest that gratuitous tyrosine phosphorylation of proteins, including TCPTP substrates, is not sufficient to trigger the change in localization of the phosphatase, but rather that specific signaling events that are dependent on the activated EGFR may be required.
In light of the fact that the TC45-D182A substrate-trapping mutant recognized both the EGFR and p52Shc, we were surprised at first that TC45 did not modulate the MAPK pathway. EGFR activation results in transautophosphorylation of the C-terminal tail of the receptor, creating specific binding sites for the recruitment of various SH2-containing proteins, including Shc and Grb2 (2, 11, 42, 43). Grb2 associates, via its SH2 domains, directly with the receptor or to pTyr Shc, providing a link to SOS and MAPK activation (6, 33, 45–47, 49). EGFR mutants lacking the autophosphorylation sites no longer bind Grb2 but maintain their ability to phosphorylate Shc and to activate the Ras/MAPK pathway (20, 26, 34). Although numerous studies have demonstrated the importance of Grb2 in stimulating the Ras/MAPK pathway (6, 18, 36, 46), the relative importance of direct binding of Grb2 to Y1068 of the EGFR (2) compared to binding and signaling via Shc (50), remains unclear. The importance of Shc in activation of the Ras/MAPK pathway by EGF is illustrated by the fact that overexpression of the Shc Y317F mutant inhibits the EGF-induced activation of MAPK in NIH 3T3 cells expressing an EGFR in which the autophosphorylation sites have been mutated (20, 22). In addition, overexpression of p52Shc in HeLa cells stimulates EGF-induced MAPK activity (38). Considering that TC45 inhibited directly the association of p52Shc with Grb2 without affecting p42Erk2 activation, our studies suggest that Shc may participate in signaling events involving Grb2 that are independent of the MAPK pathway.
Consistent with recently published reports (21, 22, 24, 59), we found that Shc is phosphorylated not only on tyrosine 317 but also on tyrosines 239 and 240 in COS1 cells and that the Y239 site is a Grb2 binding site. We have demonstrated that TC45 modulates the association of p52Shc with Grb2 and that the TC45-D182A trapping mutant specifically recognizes Shc phosphorylated on tyrosine 239. Therefore, TC45 may modulate specifically the association of Grb2 with Y239 on p52Shc and the signaling pathways emanating from this site. However, we observed that TC45 did not affect EGF-induced activation of p42Erk2, indicating that despite the fact that Grb2 binds to pY239, the signals emanating from this site do not contribute to MAPK activation. These results provide evidence for bifurcation of signaling pathways at the level of Shc and are consistent with recent studies which have demonstrated that the Y239/Y240 site does not contribute to EGF-induced MAPK activity but nonetheless is important in EGF-induced mitogenesis (22). The Shc Y239/Y240 site also contributes to interleukin-3-dependent myc expression in the BaF3 pro-B-cell line (21). However, in NIH 3T3 cells stably overexpressing a truncated mutant EGFR which selectively phosphorylates Shc, the Y239F/Y240F Shc mutant does not modulate EGF-induced myc expression under conditions where it does inhibit mitogenesis (22). Therefore, the role of the Y239/Y240 site in EGF-induced mitogenesis remains to be elucidated. To further our understanding of TC45 function, additional studies will be needed to elucidate the role of the p52Shc Y239/Y240 site in EGF-induced signaling in combination with the identification of the 50- and 64-kDa pTyr proteins precipitated by TC45-D182A. The identification of these substrates will allow a better understanding of how the Y239/Y240 site can bind Grb2 without stimulating the MAPK cascade. In addition to Grb2, we have observed the association of pTyr proteins of ∼60 and 62 kDa with GST-Shc phosphorylated on Y239, but not Y317, following EGF stimulation (data not shown). Others have reported that phosphopeptides modeled on the Y239/Y240 site bind pTyr proteins from v-Src-transformed cells (59). Until these additional proteins are characterized, it will be difficult to evaluate the contribution of Grb2 binding at Y239 to the specificity of signaling.
In this study, we have demonstrated that the ubiquitous and closely related PTPs TCPTP and PTP1B are highly specific enzymes that recognize distinct, as well as overlapping, substrates and that localization of the alternatively spliced TCPTP variants is critical for their substrate recognition. We show that in contrast to signaling molecules such as MAPKs and the STATs, which translocate to the nucleus following growth factor or cytokine stimulation (8, 29), the 45-kDa TCPTP exits the nucleus in response to EGF and gains access to cytoplasmic substrates. One of the specific cellular substrates of the 45-kDa TCPTP is the adaptor protein p52Shc, phosphorylated at the Y239 site. Therefore, the 45-kDa TCPTP recognizes distinct sites within defined substrates and thus may regulate specific signaling events emanating from such molecules.
ACKNOWLEDGMENTS
We thank James Lorenzen and Edmond Fischer for the TCPTP 6228 antibody, David Hill for the TCPTP CF4 antibody, and Gordon Gill for the EGFR 1964 antibody. We also thank Carmelita Bautista for providing monoclonal antibodies, Robert Del Vecchio and Martha Daddario for anti-pTyr antibody screening, Salim Mamajiwalla for subcloning the c-Src Y527F mutant, Scott Walk for technical assistance, and Michael Gutch and Mike Myers for critical reading of the manuscript.
This work was supported by grant CA53840 from the National Institutes of Health. K.S.R. is supported by the Bierne Carter Foundation, and T.T. is a National Health and Medical Research Council of Australia C. J. Martin Fellow.
REFERENCES
- 1.Batzer A G, Blaikie P, Nelson K, Schlessinger J, Margolis B. The phosphotyrosine interaction domain of Shc binds an LXNPXY motif on the epidermal growth factor receptor. Mol Cell Biol. 1995;15:4403–4409. doi: 10.1128/mcb.15.8.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batzer A G, Rotin D, Skolnik E Y, Schlessinger J. Hierarchy of binding sites for Grb2 and Shc on the epidermal growth factor receptor. Mol Cell Biol. 1994;14:5192–5201. doi: 10.1128/mcb.14.8.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett A M, Hausdorff S F, Oreilly A M, Freeman R M, Neel B G. Multiple requirements for SHPTP2 in epidermal growth factor-mediated cell cycle progression. Mol Cell Biol. 1996;16:1189–1202. doi: 10.1128/mcb.16.3.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown-Shimer S, Johnson K A, Lawrence J B, Johnson C, Bruskin A, Green N R, Hill D E. Molecular cloning and chromosome mapping of the human gene encoding protein phosphotyrosyl phosphatase 1B. Proc Natl Acad Sci USA. 1990;87:5148–5152. doi: 10.1073/pnas.87.13.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchdunger E, Trinks U, Mett H, Regenass U, Muller M, Meyer T, McGlynn E, Pinna L A, Traxler P, Lydon N B. 4,5-Dianilinophthalimide: a protein-tyrosine kinase inhibitor with selectivity for the epidermal growth factor receptor signal transduction pathway and potent in vivo antitumor activity. Proc Natl Acad Sci USA. 1994;91:2334–2338. doi: 10.1073/pnas.91.6.2334. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Buday L, Downward J. Epidermal growth factor regulates p21ras through the formation of a complex of receptor, Grb2 adapter protein, and Sos nucleotide exchange factor. Cell. 1993;73:611–620. doi: 10.1016/0092-8674(93)90146-h. [DOI] [PubMed] [Google Scholar]
- 7.Champion-Arnaud P, Gesnel M C, Foulkes N, Ronsin C, Sassone-Corsi P, Breathnach R. Activation of transcription via AP-1 or CREB regulatory sites is blocked by protein tyrosine phosphatases. Oncogene. 1991;6:1203–1209. [PubMed] [Google Scholar]
- 8.Chen R H, Sarnecki C, Blenis J. Nuclear localization and regulation of erk- and rsk-encoded protein kinases. Mol Cell Biol. 1992;12:915–927. doi: 10.1128/mcb.12.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chernoff J, Schievella A R, Jost C A, Erikson R L, Neel B G. Cloning of a cDNA for a major human protein-tyrosine-phosphatase. Proc Natl Acad Sci USA. 1990;87:2735–2739. doi: 10.1073/pnas.87.7.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung J, Kuo C J, Crabtree G R, Blenis J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell. 1992;69:1227–1236. doi: 10.1016/0092-8674(92)90643-q. [DOI] [PubMed] [Google Scholar]
- 11.Cohen G B, Ren R, Baltimore D. Modular binding domains in signal transduction proteins. Cell. 1995;80:237–248. doi: 10.1016/0092-8674(95)90406-9. [DOI] [PubMed] [Google Scholar]
- 12.Cool D E, Tonks N K, Charbonneau H, Fischer E H, Krebs E G. Expression of a human T-cell protein-tyrosine-phosphatase in baby hamster kidney cells. Proc Natl Acad Sci USA. 1990;87:7280–7284. doi: 10.1073/pnas.87.18.7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cool D E, Tonks N K, Charbonneau H, Walsh K A, Fischer E H, Krebs E G. cDNA isolated from a human T-cell library encodes a member of the protein-tyrosine-phosphatase family. Proc Natl Acad Sci USA. 1989;86:5257–5261. doi: 10.1073/pnas.86.14.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denu J M, Lohse D L, Vijayalakshmi J, Saper M A, Dixon J E. Visualization of intermediate and transition-state structures in protein-tyrosine phosphatase catalysis. Proc Natl Acad Sci USA. 1996;93:2493–2498. doi: 10.1073/pnas.93.6.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ekstrand A J, Liu L, He J, Hamid M L, Longo N, Collins V P, James C D. Altered subcellular location of an activated and tumour-associated epidermal growth factor receptor. Oncogene. 1995;10:1455–1460. [PubMed] [Google Scholar]
- 16.Flint A J, Tiganis T, Barford D, Tonks N K. Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proc Natl Acad Sci USA. 1997;94:1680–1685. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frangioni J V, Beahm P H, Shifrin V, Jost C A, Neel B G. The nontransmembrane tyrosine phosphatase PTP-1B localizes to the endoplasmic reticulum via its 35 amino acid C-terminal sequence. Cell. 1992;68:545–560. doi: 10.1016/0092-8674(92)90190-n. [DOI] [PubMed] [Google Scholar]
- 18.Gale N W, Kaplan S, Lowenstein E J, Schlessinger J, Bar-Sagi D. Grb2 mediates the EGF-dependent activation of guanine nucleotide exchange on Ras. Nature. 1993;363:88–92. doi: 10.1038/363088a0. [DOI] [PubMed] [Google Scholar]
- 19.Garton A J, Flint A J, Tonks N K. Identification of p130cas as a substrate for the cytosolic protein tyrosine phosphatase PTP-PEST. Mol Cell Biol. 1996;16:6408–6418. doi: 10.1128/mcb.16.11.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gotoh N, Tojo A, Muroya K, Hashimoto Y, Hattori S, Nakamura S, Takenawa T, Yazaki Y, Shibuya M. Epidermal growth factor-receptor mutant lacking the autophosphorylation sites induces phosphorylation of Shc protein and Shc-Grb2/ASH association and retains mitogenic activity. Proc Natl Acad Sci USA. 1994;91:161–171. doi: 10.1073/pnas.91.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gotoh N, Tojo A, Shibuya M. A novel pathway from phosphorylation of tyrosine residues 239/240 of Shc, contributing to suppress apoptosis by IL-3. EMBO J. 1996;15:6197–6204. [PMC free article] [PubMed] [Google Scholar]
- 22.Gotoh N, Toyoda M, Shibuya M. Tyrosine phosphorylation sites at amino acids 239 and 240 of Shc are involved in epidermal growth factor-induced mitogenic signaling that is distinct from Ras/mitogen-activated protein kinase activation. Mol Cell Biol. 1997;17:1824–1831. doi: 10.1128/mcb.17.4.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han J W, Pearson R B, Dennis P B, Thomas G. Rapamycin, wortmannin, and the methylxanthine SQ20006 inactivate p70s6k by inducing dephosphorylation of the same subset of sites. J Biol Chem. 1995;270:21396–21403. doi: 10.1074/jbc.270.36.21396. [DOI] [PubMed] [Google Scholar]
- 24.Harmer S L, DeFranco A L. Shc contains two Grb2 binding sites needed for efficient formation of complexes with SOS in B lymphocytes. Mol Cell Biol. 1997;17:4087–4095. doi: 10.1128/mcb.17.7.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hippen K L, Jakes S, Richards J, Jena B P, Beck B L, Tabatabai L B, Ingebritsen T S. Acidic residues are involved in substrate recognition by two soluble protein tyrosine phosphatases, PTP-5 and rrbPTP-1. Biochemistry. 1993;32:12405–12412. doi: 10.1021/bi00097a019. [DOI] [PubMed] [Google Scholar]
- 26.Honegger A, Dull T J, Bellot F, Van Obberghen E, Szapary D, Schmidt A, Ullrich A, Schlessinger J. Biological activities of EGF-receptor mutants with individually altered autophosphorylation sites. EMBO J. 1988;7:3045–3052. doi: 10.1002/j.1460-2075.1988.tb03169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hudziak R M, Ullrich A. Cell transformation potential of a HER2 transmembrane domain deletion mutant retained in the endoplasmic reticulum. J Biol Chem. 1991;266:24109–24115. [PubMed] [Google Scholar]
- 28.Huyer G, Liu S, Kelly J, Moffat J, Payette P, Kennedy B, Tsaprailis G, Gresser M J, Ramachandran C. Mechanism of inhibition of protein-tyrosine phosphatases by vanadate and pervanadate. J Biol Chem. 1997;272:843–851. doi: 10.1074/jbc.272.2.843. [DOI] [PubMed] [Google Scholar]
- 29.Ihle J N. STATs: signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- 30.Kamps M P, Sefton B M. Identification of multiple novel polypeptide substrates of the v-src, v-yes, v-fps, v-ros, and v-erb-B oncogenic tyrosine protein kinases utilizing antisera against phosphotyrosine. Oncogene. 1988;2:305–315. [PubMed] [Google Scholar]
- 31.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamkin T D, Walk S F, Liu L, Damen J E, Krystal G, Ravichandran K S. Shc interaction with Src homology 2 domain containing inositol phosphatase (SHIP) in vivo requires the Shc-phosphotyrosine binding domain and two specific phosphotyrosines on SHIP. J Biol Chem. 1997;272:10396–10401. doi: 10.1074/jbc.272.16.10396. [DOI] [PubMed] [Google Scholar]
- 33.Li N, Batzer A, Daly R, Yajnik V, Skolnik E, Chardin P, Bar-Sagi D, Margolis B, Schlessinger J. Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signalling. Nature. 1993;363:85–87. doi: 10.1038/363085a0. [DOI] [PubMed] [Google Scholar]
- 34.Li N, Schlessinger J, Margolis B. Autophosphorylation mutants of the EGF-receptor signal through auxiliary mechanisms involving SH2 domain proteins. Oncogene. 1994;9:3457–3465. [PubMed] [Google Scholar]
- 35.Lorenzen J A, Dadabay C Y, Fischer E H. COOH-terminal sequence motifs target the T-cell protein tyrosine phosphatase to the ER and nucleus. J Cell Biol. 1995;131:631–643. doi: 10.1083/jcb.131.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowenstein E J, Daly R J, Batzer A G, Li W, Margolis B, Lammers R, Ullrich A, Skolnik E Y, Bar-Sagi A, Schlessinger J. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell. 1992;70:431–442. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- 37.McGlade J, Cheng A, Pelicci G, Pelicci P G, Pawson T. Shc proteins are phosphorylated and regulated by the v-src and v-fps protein-tyrosine kinases. Proc Natl Acad Sci USA. 1992;89:8869–8873. doi: 10.1073/pnas.89.19.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Migliaccio E, Mele S, Salcini A E, Pelicci G, Lai K-M V, Superti-Furga G, Pawson T, Di Fiore P P, Lanfrancone L, Pelicci P G. Opposite effects of the p52shc/p46shc and p66shc splicing isoforms on the EGF receptor-MAP kinase-fos signaling pathway. EMBO J. 1997;16:706–716. doi: 10.1093/emboj/16.4.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moorgenstern J P, Land J H. A series of mammalian expression vectors and characterization of their expression of a reporter gene in stably and transiently transfected cells. Nucleic Acids Res. 1990;18:1068. doi: 10.1093/nar/18.4.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mosinger B, Jr, Tillmann U, Westphal H, Tremblay M L. Cloning and characterization of a mouse cDNA encoding a cytoplasmic protein-tyrosine-phosphatase. Proc Natl Acad Sci USA. 1992;89:499–503. doi: 10.1073/pnas.89.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakielny S, Dreyfuss G. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J Cell Biol. 1996;134:1365–1373. doi: 10.1083/jcb.134.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pawson T. Protein modules and signalling networks. Nature. 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 43.Pelicci G, Lanfracone L, Grignani F, McGlade J, Cavallo F, Forni G, Nicoletti I, Grignani F, Pawson T, Pelicci P G. A novel transforming protein (SHC) with an SH2 domain is implicated in mitogenic signal transduction. Cell. 1992;70:93–104. doi: 10.1016/0092-8674(92)90536-l. [DOI] [PubMed] [Google Scholar]
- 44.Pinol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 45.Ravichandran K S, Lorenz U, Shoelson S E, Burakoff S J. Interaction of Shc with Grb2 regulates association of Grb2 with mSOS. Mol Cell Biol. 1995;15:593–600. doi: 10.1128/mcb.15.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rozakis-Adcock M, Fernley R, Wade J, Pawson T. The SH2 and SH3 domains of mammalian Grb2 couple the EGF receptor to the Ras activator mSos1. Nature. 1993;363:83–85. doi: 10.1038/363083a0. [DOI] [PubMed] [Google Scholar]
- 47.Rozakis-Adcock M, McGlade J, Mbamalu G, Pelicci G, Daly R, Li W, Batzer A, Thomas S, Brugge J, Pelicci P G, Schlessinger J, Pawson T. Association of the Shc and GRB2/Sem5 SH2-containing proteins is implicated in activation of the ras pathway by tyrosine kinases. Nature. 1992;360:689–692. doi: 10.1038/360689a0. [DOI] [PubMed] [Google Scholar]
- 48.Ruzzene M, Donella-Deana A, Marin O, Perich J W, Ruzza P, Borin G, Calderan A, Pinna L A. Specificity of T-cell protein tyrosine phosphatase toward phosphorylated synthetic peptides. Eur J Biochem. 1993;211:289–295. doi: 10.1111/j.1432-1033.1993.tb19897.x. [DOI] [PubMed] [Google Scholar]
- 49.Salcini A E, McGlade J, Pelicci G, Nicoletti I, Pawson T, Pelicci P G. Formation of Shc-GRB2 complexes is necessary to induce neoplastic transformation by overexpression of Shc proteins. Oncogene. 1994;9:2827–2836. [PubMed] [Google Scholar]
- 50.Sasaoka T, Langlois W J, Leitner J W, Draznin B, Olefsky J M. The signaling pathway coupling epidermal growth factor receptors to activation of p21ras. J Biol Chem. 1994;269:32621–32625. [PubMed] [Google Scholar]
- 51.Slieker L J, Martensen T M, Lane M D. Synthesis of epidermal growth factor receptor in human A431 cells. J Biol Chem. 1986;261:15233–15241. [PubMed] [Google Scholar]
- 52.Tanaka M, Gupta R, Mayer B J. Differential inhibition of signaling pathways by dominant-negative SH2/SH3 adapter proteins. Mol Cell Biol. 1995;15:6829–6837. doi: 10.1128/mcb.15.12.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tiganis T, Flint A J, Adam S A, Tonks N K. Association of the protein tyrosine phosphatase TCPTP with nuclear import factor p97. J Biol Chem. 1997;272:21548–21557. doi: 10.1074/jbc.272.34.21548. [DOI] [PubMed] [Google Scholar]
- 54.Tillmann U, Wagner J, Boerboom D, Westphal H, Tremblay M L. Nuclear localization and cell cycle regulation of a murine protein tyrosine phosphatase. Mol Cell Biol. 1994;14:3030–3040. doi: 10.1128/mcb.14.5.3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tonks N K. Protein tyrosine phosphatases and the control of cellular signaling responses. Adv Pharmacol. 1996;36:91–119. doi: 10.1016/s1054-3589(08)60578-5. [DOI] [PubMed] [Google Scholar]
- 56.Tonks N K, Diltz C D, Fischer E H. Characterization of the major protein-tyrosine-phosphatases of human placenta. J Biol Chem. 1988;263:6731–6737. [PubMed] [Google Scholar]
- 57.Tonks N K, Diltz C D, Fischer E H. Purification of the major protein-tyrosine-phosphatases of human placenta. J Biol Chem. 1988;263:6722–6730. [PubMed] [Google Scholar]
- 58.Tonks N K, Neel B G. From form to function: signaling by protein tyrosine phosphatases. Cell. 1996;87:365–368. doi: 10.1016/s0092-8674(00)81357-4. [DOI] [PubMed] [Google Scholar]
- 59.van der Geer P, Wiley S, Gish G D, Pawson T. The Shc adaptor protein is highly phosphorylated at conserved, twin tyrosine residues (Y239/240) that mediate protein-protein interactions. Curr Biol. 1996;6:1435–1444. doi: 10.1016/s0960-9822(96)00748-8. [DOI] [PubMed] [Google Scholar]
- 60.Wymann M P, Bulgarelli-Leva G, Zvelebil M J, Pirola L, Vanhaesebroeck B, Waterfield M D, Panayotou G. Wortmannin inactivates phosphoinositide 3-kinase by covalent modification of Lys-802, a residue involved in the phosphate transfer reaction. Mol Cell Biol. 1996;16:1722–1733. doi: 10.1128/mcb.16.4.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Z Y, Thieme-Sefler A M, Maclean D, McNamara D J, Dobrusin E M, Sawyer T K, Dixon J E. Substrate specificity of the protein tyrosine phosphatases. Proc Natl Acad Sci USA. 1993;90:4446–4450. doi: 10.1073/pnas.90.10.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]