Abstract
The RNA polymerase III factor TFIIIB forms a stable complex with DNA and can promote multiple rounds of initiation by polymerase. TFIIIB is composed of three subunits, the TATA binding protein (TBP), TFIIB-related factor (BRF), and B". Chemical footprinting, as well as mutagenesis of TBP, BRF, and promoter DNA, was used to probe the architecture of TFIIIB subunits bound to DNA. BRF bound to TBP-DNA through the nonconserved C-terminal region and required 15 bp downstream of the TATA box and as little as 1 bp upstream of the TATA box for stable complex formation. In contrast, formation of complete TFIIIB complexes required 15 bp both upstream and downstream of the TATA box. Hydroxyl radical footprinting of TFIIIB complexes and modeling the results to the TBP-DNA structure suggest that BRF and B" surround TBP on both faces of the TBP-DNA complex and provide an explanation for the exceptional stability of this complex. Competition for binding to TBP by BRF and either TFIIB or TFIIA suggests that BRF binds on the opposite face of the TBP-DNA complex from TFIIB and that the binding sites for TFIIA and BRF overlap. The positions of TBP mutations which are defective in binding BRF suggest that BRF binds to the top and N-terminal leg of TBP. One mutation on the N-terminal leg of TBP specifically affects the binding of the B" subunit.
Yeast RNA polymerase III (Pol III) is recruited to promoters by the transcription factor TFIIIB (13, 14, 46). TFIIIB is composed of three subunits, the TATA binding protein (TBP), TFIIB-related factor (BRF; also termed TFIIIB70), and a third subunit termed B". Together, these three factors form a highly stable protein-DNA complex upstream of the transcription start site at Pol III promoters. This TFIIIB-DNA complex can promote multiple rounds of initiation by polymerase (20). In vivo, TFIIIB is recruited to promoters by the factor TFIIIC (14). In purified systems in vitro, TFIIIB can be positioned at a promoter independent of TFIIIC, provided the promoter contains a functional TATA element (17, 31); an example of such a promoter is the yeast U6 promoter.
The preinitiation complexes for RNA Pol II and III and Archaea polymerase are related in several respects. All three complexes contain TBP as an essential component (41, 44). In addition, both complexes contain subunits related to the TFIIB family of proteins. The Pol II factor TFIIB contains a Zn binding site at its N terminus and a C-terminal core domain (TFIIBc) containing the TBP and DNA binding activities (1, 32). The Zn binding site of TFIIB is essential for recruitment of Pol II enzyme (3, 8, 16). The Archaea factor TFB is 32% identical and 56% similar to human TFIIB (34, 36). TFB contains a Zn binding site at its N terminus (48), and the C-terminal core domain of TFB binds TBP-DNA similarly to TFIIB (25). The Pol III factor BRF is homologous to TFIIB and TFB over the Zn binding site and core domain and is about 25% identical and 35% similar to TFIIB and TFB (9, 11, 30). However, BRF also contains a 30-kDa domain at its C terminus (the C-BRF) which is conserved only among other BRFs and appears to play a major role in interaction with TBP. In two-hybrid and affinity chromatography assays, the C-terminal domain but not the N-terminal domain interacts strongly with TBP (10, 22). In addition, it has been recently shown that the C-BRF alone can form a complex with TBP, B", and DNA (19). The role of the BRF N-terminal domain is not yet clear, although it does interact with τ131, a subunit of TFIIIC, in two-hybrid assays (10) and with C34, a subunit of Pol III assayed by affinity chromatography (22). The N-terminal region of BRF is also necessary for stimulation of transcription by TFIIIC in vitro (19).
Understanding the arrangement of the subunits and DNA in the TFIIIB complex will allow a more direct assessment of the similarity in preinitiation complexes and mechanisms of polymerase recruitment for Pol II, Pol III, and Archaea polymerase. In addition, understanding the protein-protein and protein-DNA interactions in TFIIIB will explain the great stability of the complex and how it remains stably bound during multiple rounds of initiation by polymerase. Previous work using photo-cross-linking probes incorporated into DNA has suggested the arrangement of subunits in TFIIIB (4, 5, 19, 35). BRF has been observed to cross-link over a wide distance upstream of the transcription start site between positions −39 and −12. In similar experiments, B" appears to cross-link mainly far upstream from the transcription start site between positions −30 and −38. Information from these photo-cross-linking studies is limited in that the photoprobes are at least 10 Å in length and protrude only from the major groove of the DNA. From photo-cross-linking studies, DNA mutagenesis, and DNase footprinting, TBP appears to bind 25 to 30 bp upstream from the transcription start site (18, 21). DNA in the TFIIIB complex appears bent (28); however, it is not possible to tell from the methods used if this bend is significantly greater than that observed in the TBP-DNA complex (23, 24). In this study, we used mutagenesis of TBP, BRF, and DNA as well as chemical footprinting to investigate the architecture of BRF and B" binding in the BRF-TBP-DNA and TFIIIB promoter complexes.
MATERIALS AND METHODS
Preparation of proteins.
C-BRF352 was generated by insertion of eight histidine codons and a stop codon at position 315 of the BRF coding sequence. The entire BRF coding sequence from this construct was subcloned in the BRF T7 polymerase expression vector pSH360 (11). This construct primarily produced an internal translation product beginning in the C-terminal half of BRF which migrated on sodium dodecyl sulfate-polyacrylamide gels with an apparent molecular mass of 32.5 kDa. This polypeptide was blocked to N-terminal sequencing and was identified by tryptic digestion and mass spectrometry analysis of the resulting peptides. Although we could not identify the N-terminal residue of this polypeptide, we presume that it begins at or near residue 352, analogous to the product identified by Kassavetis et al. (19). C-BRF309 was generated by subcloning the BRF coding sequence from residues 309 to 596 into plasmid pET24A (Novagen). This polypeptide was identified by N-terminal sequencing. BRF1-288 was generated by subcloning the coding sequence for BRF residues 1 to 288 with a six-His tag at the C terminus of the protein into pET24D. These plasmids were transformed to Escherichia coli BL21 and induced with 0.4 mM isopropylthiogalactopyranoside (IPTG) for 2 h (11). BRF derivatives were all purified similarly to full-length BRF (38). Full-length BRF contains a six-His tag at the C terminus; this tag does not affect in vivo function of BRF (10a). The bacteria were lysed in 6 M guanidine-HCl and bound to an Ni-nitrilotriacetic acid agarose column as specified by the manufacturer (Qiagen). The BRF derivatives were eluted from the Ni column in 6 M guanadine-HCl at pH 4.5 and neutralized to pH 7 with 1 M Tris (pH 8.8). After mixing 1:1 with 2 M urea–2 M guanidine, the eluant was applied to a Poros R2-10 reverse-phase column in 5% acetonitrile–0.1% trifluoroacetic acid. BRF derivatives eluted in an acetonitrile gradient between 10 and 90% acetonitrile. The purified proteins were dried by vacuum lyophilization at room temperature and resuspended in 6 M guanidine-HCl plus 0.1% Brij 58. Renaturation of BRF was performed as described previously (11), except that the initial dilution into renaturation buffer was increased from 1:1 to 1:9.
BRF L462S was identified in a screen for temperature-sensitive mutations in BRF (10a). This mutant was selected for further study, as it was suppressed by overexpression of TBP in vivo. The mutant was expressed in E. coli, purified as described above, and used in gel mobility shift assays as described.
Radical missense mutations in full-length TBP were generated by site-directed mutagenesis and subcloned to pET24A (Novogen). E. coli BL21 containing these plasmids was grown to an A600 of ∼1.0 and induced with 0.4 mM IPTG for 2 h. The cells were harvested by centrifugation and lysed by sonication in 30 mM Tris (pH 7.5)–80 mM KCl–1 mM dithiothreitol–1 mM phenylmethylsulfonyl fluoride–2 mM EDTA–20% glycerol–0.05% Tween 20. The broken cells were cleared of debris by centrifugation and loaded onto Q-Sepharose Fast Flow (Pharmacia) and S-Sepharose Fast Flow columns run in tandem. After loading and washing of the columns in sonication buffer lacking Tween 20, the S-Sepharose column was eluted with a gradient of 75 to 600 mM KCl. TBP derivatives eluted at variable positions in the gradient, depending on the particular radical mutation. Fractions containing TBP were concentrated in a Centriprep 10 (Amicon).
In vitro transcription.
Pol III transcription was performed as described elsewhere (39) with whole-cell extracts from either wild-type Saccharomyces cerevisiae or strain SHY70 (40) containing the TBP I143N mutation. The transcription templates used were from plasmid pGE2 wt containing the tRNA3Leu gene (2) or pCH6 containing the U6 gene (6).
Gel mobility shift assays.
U6-major late promoter (MLP) TATA probes were created by synthesizing oligonucleotides with sequences as indicated in Fig. 1 to 3. The top strand of each oligonucleotide was phosphorylated and annealed to its complementary strand. The probe −70/+18 was synthesized by PCR using 32P-labeled oligonucleotides with indicated 5′ and 3′ ends and as a template plasmid pSL47, containing the U6 promoter from −81 to +25, except that the TATA box found at −30 in the U6 promoter was mutagenized by oligonucleotide-directed mutagenesis to TATAAAAG. Protein-DNA binding reactions were performed for 45 min in 4 mM Tris (pH 8)–60 mM KCl–5 mM MgCl2–4% glycerol–0.1% Brij 58–100 ng of poly(dG-dC)–100 μg of bovine serum albumin per ml (reaction mixtures containing TFIIBc had no Brij) in a total volume of 20 μl. Different native gel systems were used for the experiments described below to optimize the stability of the different protein-DNA complexes. For all binding reactions (except those for the TBP radical mutants), indicated probes were incubated with 1 ng of yeast TBPc (residues 61 to 180; a gift from J. Geiger) in conjunction with 15 ng of BRF, 2 ng of C-BRF352, or 4 ng of C-BRF309 and, where indicated, 12 ng of B" or with 6 ng of yeast TFIIBc (lacking residues 2 to 119). Reaction mixtures with BRF and mini-TFIIA (mTFIIA)-TBP-DNA complexes contained 1 ng of yeast TBPc and ∼0.5 ng of mTFIIA 15 (a gift from J. Geiger) in conjunction with 15 and 30 ng of BRF, 2 and 4 ng of C-BRF352, or 4 and 8 ng of C-BRF309. Gel and running buffers used for Fig. 1, 2, 3, and 5B are as described previously (37) and contain 1.5 mM (final concentration) magnesium acetate. Gel and running buffers used for Fig. 5A contained TG (25 mM Tris, 190 mM glycine [pH 8.3]) with no magnesium acetate. Gel mobility shift assays represented in Fig. 6 used gels and buffer similar to those described previously (37) but lacking EDTA and containing 0.5 mM (final concentration) magnesium acetate. These gels were run at room temperature for 40 min at 200 V.
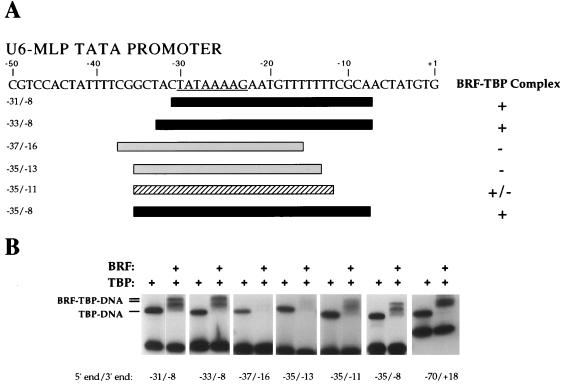
FIG. 1.
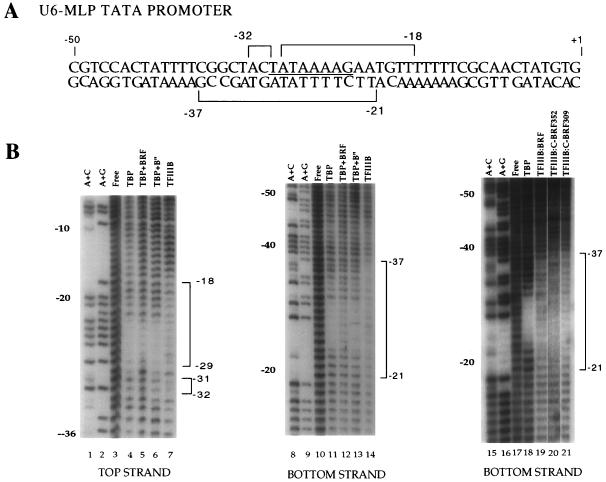
BRF requires DNA downstream of the TATA box to form a stable BRF-TBP-DNA complex. (A) The sequence of the U6-MLP TATA promoter is shown, and the asymmetric MLP TATA box is underlined. Below the sequence, the DNA probes assayed and their abilities to bind BRF-TBP are summarized. Probes able to form a stable BRF-TBP-DNA complex are black, and probes unable to form stable complexes are gray; the hatched probe forms an unstable BRF-TBP complex. (B) Gel mobility shift assays with wild-type BRF. The probe, −70/+18, contains 40 bp of DNA both upstream and downstream of the TATA box. Reaction mixtures contained 1 ng of TBPc and 15 ng of recombinant BRF.
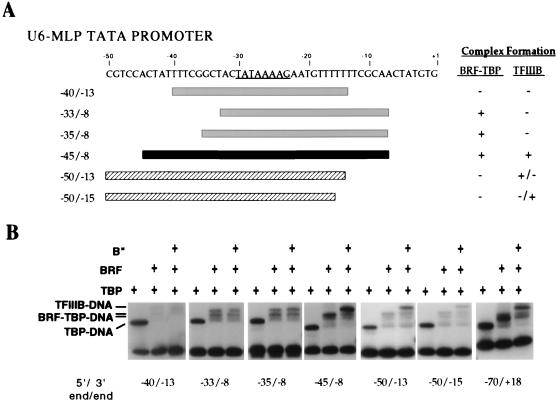
FIG. 3.
DNA upstream and downstream of the TATA box is required to form a stable TFIIIB-DNA complex. (A) Summary of DNA probes assayed and their abilities to bind BRF-TBP and recruit B" to form TFIIIB. Probes able to form a stable TFIIIB-DNA complex are black, and probes unable to form stable complexes are gray; the hatched probes form unstable TFIIIB complexes. (B) Gel mobility shift assays with wild-type BRF and B". Reaction mixtures contained 1 ng of TBPc, 15 ng of BRF, and 12 ng of B" as indicated.
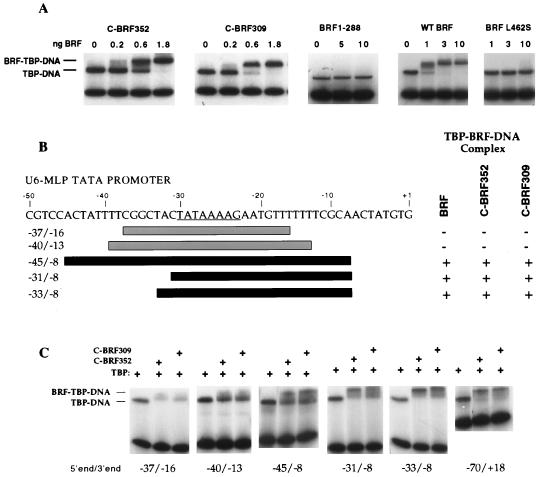
FIG. 2.
C-BRF derivatives require DNA downstream of the TATA box to form a stable complex with TBP-DNA. (A) Gel mobility shift assays using the yeast U6 promoter containing 75 bp upstream and downstream of the TATA box. The indicated amounts of C-BRF derivatives were added to reaction mixtures containing 2 ng of TBPc and the U6 promoter probe. (B) Summary of DNA probes assayed and their abilities to bind C-BRF derivatives. Probes able to form a stable BRF-TBP-DNA complex are black; probes unable to form stable complexes are gray. (C) Gel mobility shift assays with C-BRF derivatives. Reactions contain 1 ng of TBPc and either 2 ng of C-BRF352 or 4 ng of C-BRF309.
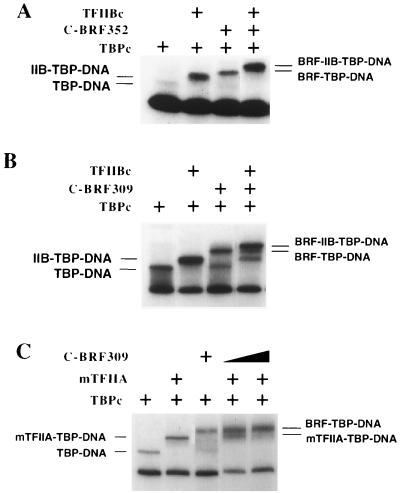
FIG. 5.
Competition assays between BRF and TFIIB and TFIIA. (A and B) Gel mobility shift assays using the U6-MLP promoter as a probe and 1 ng of TBPc with 6 ng of yeast TFIIBc (lacking residues 2 to 119) where indicated and either 2 ng of C-BRF352 or 4 ng of C-BRF309. (C) Reaction mixtures contained 1 ng of TBPc and 0.5 ng of mTFIIA where indicated and either 4 or 8 ng of C-BRF309.
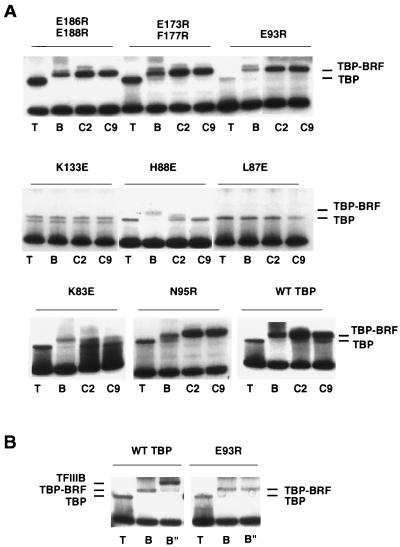
FIG. 6.
Radical point mutations in TBP affect binding of BRF and B". (A) Gel mobility shift assay of each TBP mutant, either alone (T) or with wild-type BRF (B), C-BRF352 (C2), or C-BRF309 (C9). (B) TFIIIB complex formation of TBP mutant E93R and wild-type (WT) TBP. TBP was added alone (T), with BRF (B), or with BRF and 24 ng of B" (B").
Hydroxyl radical footprints.
Hydroxyl radical cleavage reactions were carried out by adding 6 μl of cleavage solution containing 8.3 mM Fe(NH4)2(SO4)2, 16.7 mM EDTA, 18.7 mM ascorbic acid, and 2.7% H2O2 to 20-μl protein binding reaction mixtures. Reaction mixtures contained either no protein or 6 ng of yeast TBPc alone or in conjunction with 15 ng of BRF and 12 ng of B", 5 ng of C-BRF352 with 12 ng of B", or 8 ng of C-BRF309 with 12 ng of B". Binding reactions were performed as described above for mobility shift assays but with no glycerol. Cleavage was allowed to proceed for 2 min before addition of 2 μl of 50% glycerol. Reactions were loaded onto 1.5 mM (final concentration) magnesium acetate native polyacrylamide gels as described previously (37). Protein-DNA complexes were excised from gels, and the DNA was eluted into 300 μl of 90 mM Tris–92 mM boric acid–2.5 mM EDTA at pH 8.3. DNA was ethanol precipitated, resuspended in 4 μl of formamide, and after heat denaturation loaded on an 8% denaturing polyacrylamide gel. For reactions with full-length BRF where the BRF-TBP-DNA complex appeared as a doublet, the two bands of the doublet were excised together since they were too close together to separate. Probes were prepared by PCR as described above for probe −70/+18. All binding reaction mixtures contained 100,000 cpm (∼0.1 ng) of probe, of which 10,000 cpm was recovered in protein DNA complexes and loaded on the denaturing polyacrylamide gel. Quantitation was performed by PhosphorImager analysis. The ratios of intensities of bands observed for each reaction were normalized by using positions outside the protected region.
RESULTS
Formation of BRF-TBP-DNA complexes requires DNA downstream of the TATA box.
To characterize the architecture of the TFIIIB-DNA complex, we first determined the minimal segment of DNA required for formation of TBP-BRF complexes. This was measured by a method similar to that used for mapping the binding region of DNA in the TBP-TFIIB-DNA complex (27). Oligonucleotides containing different lengths of DNA upstream and/or downstream of the TATA box were synthesized, and the ability to form stable BRF-TBP-DNA complexes was assessed by gel mobility shift assays.
The TATA box found in the yeast U6 promoter (TATAAATA) is nearly symmetrical and can promote transcription in either orientation when given an appropriate transcription start site (19, 45). Because TBP (as well as TFIIIB) likely binds to this symmetrical U6 TATA box in either of two orientations, results for the wild-type U6 promoter would incorporate both orientations of assembled TBP-DNA (data not shown). We addressed this problem by mutagenesis of the U6 TATA box to the asymmetrical TATA sequence found in the adenovirus MLP (Fig. 1A). As expected, this promoter construct, U6-MLP TATA, is capable of directing Pol III transcription in vivo and in vitro, using whole-cell extracts at levels comparable to the unmodified promoter (not shown). All further analysis of TFIIIB factors was performed with this altered U6 promoter. Additionally, all assays used the conserved domain of yeast TBP (TBPc) lacking the nonconserved N-terminal region unless otherwise noted.
TBP formed stable complexes with all probes assayed since each contained an intact TATA box (Fig. 1B). Surprisingly, DNA containing only 1 bp upstream of the TATA box can support formation of a stable BRF-TBP-DNA complex (Fig. 1, probe −31/−8). The BRF-TBP-DNA complex often appears as a doublet in our gel mobility shift assays (Fig. 1). The cause of this doublet BRF-TBP-DNA complex is unknown; however, it seems dependent on the N-terminal domain of BRF, as the C-BRF does not give a doublet complex when bound to TBP-DNA (see below).
In contrast to TFIIB, which requires DNA 7 bp upstream and downstream of the TATA for stable binding, BRF requires DNA 12 to 15 bp downstream of the TATA box for stable binding to TBP-DNA. Although a BRF-TBP-DNA complex is visible when the DNA contains 12 bp downstream of the TATA box (Fig. 1, probe −35/−11), the BRF-TBP-DNA complex is more stable with 15 bp of DNA downstream of the TATA box (probe −35/−8). BRF is unable to form a stable complex with TBP-DNA if the DNA contains fewer than 12 bp downstream of the TATA box (probe −35/−13). Surprisingly, BRF disrupts the TBP-DNA complex with probes containing less than 12 bp downstream of the TATA box (Fig. 1B, probes −37/−16 and −35/−13). When TBP is first bound to DNA containing fewer than 12 bp of downstream DNA, addition of BRF destabilizes the TBP-DNA complex in the gel mobility shift assay. These unexpected results suggest that the TBP-BRF-DNA complex formed with the shortened DNA is unstable to electrophoresis.
It has been demonstrated that the C-terminal domain of BRF can bind TBP-DNA and that this complex can recruit B", although this complex is sensitive to challenge by high salt and heparin (19). To determine if the C-BRF bound to TBP-DNA similarly to full-length BRF, two variants of the C-BRF were generated. The first BRF variant contains BRF residues 309 to 596 (C-BRF309), and the second is an internal translation product beginning at about residue 352 and ending at residue 596 (C-BRF352) (Materials and Methods) (19). In equilibrium binding assays, both of these derivatives bound TBP-DNA with less than 1 nM affinity (Fig. 2A), which is at least threefold higher than the affinity seen for full-length BRF (Fig. 2A) (29, 37). These C-BRF derivatives also recruited B" to the complex with the same affinity as full-length BRF (not shown). In contrast, the N-terminal domain of BRF (residues 1 to 288) produced in E. coli did not form a stable complex with TBP (Fig. 2A). Finally, a single missense mutation in the C terminus of BRF (BRF L462S) isolated as a temperature-sensitive mutation (Materials and Methods) showed at least 10-fold-reduced affinity for TBP-DNA (Fig. 2A). Together, these experiments confirm that BRF interacts with TBP-DNA primarily through the nonconserved C-terminal domain.
A subset of probes were assessed for the ability to form stable complexes with TBP and either C-BRF309 or C-BRF352 (Fig. 2B). Both C-BRF309 and C-BRF352 were found to exhibit the same requirement for downstream DNA as full-length BRF. Both C-BRF derivatives could not form complexes with probes that contained either 7 or 10 bp of DNA both upstream and downstream of the TATA box (probes −37/−16 and −40/−13) but did form a complex if 15 bp of downstream DNA was present (probes −31/−8 and −33/−8).
Formation of TFIIIB-DNA complexes requires DNA upstream and downstream of the TATA box.
Probes containing different lengths of DNA upstream and/or downstream of the TATA box were also assessed for the ability to form stable TFIIIB-DNA complexes by mobility shift assays (Fig. 3). As expected from the requirement for DNA downstream of the TATA box for BRF binding, TFIIIB also requires 15 bp of DNA downstream of the TATA box for stable complex formation. In addition, formation of stable TFIIIB-DNA complexes requires 15 bp of DNA upstream of the TATA box. Extension of the upstream DNA to 20 bp allows formation of low-affinity TFIIIB complexes with just 10 bp of DNA downstream of the TATA box (Fig. 3, probe −50/−13). This result suggests that B" interacts with DNA between 15 and 20 bp upstream of the TATA box and stabilizes the interaction of BRF with the complex such that the requirement for BRF interactions with DNA is somewhat reduced.
We have previously shown that B" can form a complex with TBP-DNA at very high concentrations (20-fold higher than was used in the above-described reactions) (38). Gel mobility shift analysis with different-length probes suggests that this complex is probably not related to the usual mechanism of B" incorporation into the IIIB complex, as only very long probes, containing 40 bp of DNA upstream and downstream of the TATA box, were able to form stable B"-TBP-DNA complexes (data not shown).
Hydroxyl radical footprinting of TFIIIB factors.
To further define the residues of DNA in contact with subunits of TFIIIB, hydroxyl radical footprinting was performed on factors binding to the U6-MLP TATA promoter (Fig. 4). Hydroxyl radical ions attack sugars in the DNA backbone. The conserved core of yeast TBP protected most nucleotides in the TATA box of the U6-MLP TATA promoter from hydroxyl radical attack (Fig. 4B, lanes 4, 11, and 18). Addition of BRF to TBP-DNA did not protect any residue outside of the TATA box from hydroxyl radical cleavage (Fig. 4B, lanes 5 and 12). The same footprinting results are seen when either C-BRF309 or C-BRF352 is added to TBP-DNA complexes (data not shown). Similarly, DNase I protection experiments showed no difference between the TBP-DNA and TBP-BRF-DNA complexes (29, 36a). Ethylation interference assays also showed no difference in phosphates required for formation of TBP and TBP-BRF complexes with DNA (36a). Addition of B" to TBP-DNA also did not protect any residues outside of the TATA box from hydroxyl radical cleavage (Fig. 4B, lanes 6 and 13). In contrast, addition of both BRF and B" to the TBP-DNA complex results in a dramatic extension of the hydroxyl radical footprint 5 bp downstream of the TATA box and 2 bp upstream of the TATA box on the top strand of the U6-MLP TATA promoter (lane 7) while extending the hydroxyl radical footprint 7 bp upstream and 2 bp downstream of the TATA box on the bottom strand of the U6-MLP TATA promoter (Fig. 4, lanes 14 and 19).
FIG. 4.
Hydroxyl radical protection of TFIIIB-TBP-DNA complexes. (A) Summary of hydroxyl radical protection at U6-MLP TATA by TFIIIB. (B) Hydroxyl radical footprinting of the U6-MLP. The panel on the far right shows hydroxyl radical footprinting of the bottom strand of the U6-MLP, using either of the two C-BRF derivatives. TFIIIB indicates that TBPc, BRF, and B" were all added. The intensity of each base in each reaction was quantitated and normalized to the corresponding intensity in unbound DNA. Those bases that are reproducibly protected with the addition of TBPc, BRF, and B" are bracketed. Protection by TFIIIB was defined as a greater than 25% decrease in intensity compared with TBPc bound alone. Hydroxyl radical protection of the U6-MLP TATA nontranscribed strand with TFIIIB containing either C-BRF derivative shows similar protection to that of TFIIIB containing wild-type BRF (10a).
Hydroxyl radical footprinting was also performed on TFIIIB complexes containing the C-terminal BRF derivatives in place of full-length BRF. TFIIIB containing either wild-type BRF, C-BRF309, or C-BRF352 showed very similar protection of residues from hydroxyl radical attack on both strands of the U6-MLP TATA promoter (Fig. 4B and data not shown).
Competition with TFIIA and TFIIB maps the location of BRF within the TBP-DNA complex.
To probe the location of BRF within the BRF-TBP-DNA complex, two Pol II factors whose structures with TBP-DNA have been solved were used as competitors for BRF binding to TBP-DNA. For these experiments, we used polypeptide derivatives identical or analogous to those used in crystallography: TBPc, TFIIBc, and yeast TFIIA containing an internal deletion in the large TFIIA subunit (mTFIIA) (see Materials and Methods). Previously, it was shown that both TFIIA and TFIIB competed for binding with the complete TFIIIB complex, but the ability of BRF to compete with TFIIA and TFIIB for binding was not assessed (38).
TFIIB interacts both with the C-terminal stirrup of TBP and with DNA on one face of the TBP-DNA complex (32). Addition of C-BRF309, C-BRF352, or full-length BRF to TFIIB-TBP-DNA produced complexes which migrate more slowly than either TBP-TFIIBc-DNA or TBP-BRF-DNA (Fig. 5A and B and data not shown). This result suggests that BRF does not interact on the same face of the TBP-DNA complex as TFIIB.
TFIIA interacts with TBP near the N-terminal stirrup and interacts with DNA upstream of the TATA box (15, 42). All BRF derivatives tested (C-BRF309, C-BRF352, and wild-type BRF) competed with TFIIA for binding to TBP-DNA (Fig. 5C and data not shown). Addition of increasing concentrations of BRF blocked the binding of TFIIA to TBP (Fig. 5C). Similarly, increasing the TFIIA concentration blocked the formation of the TBP-BRF-DNA complex, and only the faster-migrating TFIIA-TBP-DNA complex was observed (not shown). Wild-type TFIIA and mTFIIA gave similar results in competition assays (not shown). These results show that BRF likely overlaps in its position with TFIIA for binding to TBP, DNA, or both.
Mutations in the TBP N-terminal leg and a mutation on the top surface of TBP disrupt BRF binding.
Surface-exposed residues of human TBP have been extensively mutagenized to determine which residues of TBP are involved in binding the Pol II transcription factors (7, 43). In general, these results support the crystal structure models of TFIIA and TFIIB bound to TBP-DNA. TFIIB fails to bind to mutations within the C-terminal stirrup of TBP, and TFIIA fails to bind mutations in the N-terminal stirrup. Based on the work of Berk and coworkers (7), we made analogous radical mutations on the surface of full-length yeast TBP within the TFIIB and TFIIA binding regions (Fig. 6). The yeast TBP residues were chosen for mutation because (i) the analogous residues in human TBP did not affect TBP binding to DNA (7), (ii) they are near the sites of hydroxyl radical protection by TFIIIB when the results are modeled to the TBP-DNA structure (see Fig. 7), and (iii) some of the mutants are located near the surfaces of TBPs which interact with TFIIB or TFIIA. We tested an additional TBP mutation, K133E, which was shown previously to affect binding of BRF to TBP in vivo (references 9 and 12; see also Fig. 7) and in vitro (5a). The TBP mutants were expressed in E. coli and purified. Each mutant was tested in a gel mobility shift assay for binding to wild-type BRF or the two C-terminal BRF derivatives. Since all of the mutations are radical changes in TBP surface residues rather than truncations of the side chains by alanine substitution, we cannot distinguish whether BRF directly contacts these amino acid side chains or if these mutations are merely close to the location where BRF interacts with TBP.
FIG. 7.
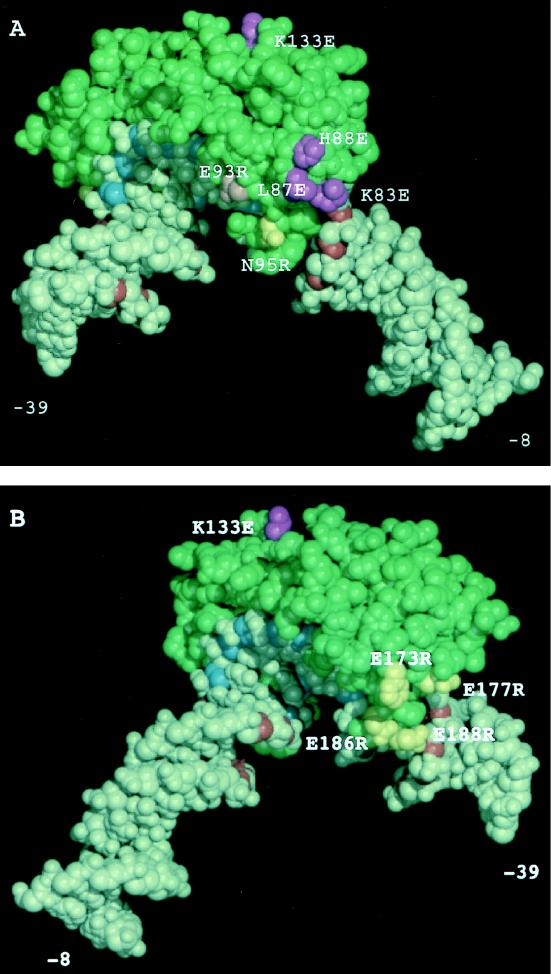
Modeling of hydroxyl radical TFIIIB footprinting, DNA deletion analysis, and TBP mutagenesis to the TBP-DNA structure. (A) View of one face of the TBP-DNA complex where TFIIA binds near the N-terminal stirrup. (B) View of the opposite face of the TBP-DNA complex where TFIIB binds to the C-terminal stirrup. The conserved domain of yeast TBP is represented in green, the sugar residues of the TATA box that are protected from hydroxyl radical cleavage by TBP alone are shown in blue, and sugar residues outside of the TATA box protected from hydroxyl radical attack by TFIIIB are shown in red. TBP side chains which when mutated show decreased binding to BRF are shown in purple; TBP side chains which when mutated show no phenotype for BRF or B" binding are yellow. The pink mutation reduces binding of B" but not BRF.
Mutations K83E, L87E, and H88E, all in the N-terminal leg of TBP, showed a defect in binding to BRF compared to wild-type TBP (Fig. 6A and 7A). This defect was similar for both full-length BRF and the C-BRF derivatives. Two mutations on this face of TBP, E93R and N95R, showed no defect in BRF binding. We also tested mutation K97E, located at the bottom of the N-terminal stirrup for BRF binding. In contrast to results with human TBP (7), this TBP mutant had a severe DNA binding defect, and we could not quantitate its effect on binding of BRF. One mutation, K133E, located on top of TBP also exhibited a defect in binding of all BRF derivatives. Finally, two double mutations on the opposite face of TBP (E186R E188R and E173R F177R) did not affect the binding of any BRF derivative (Fig. 6A and 7B). Together, the results for the radical TBP mutations support the model that the C-terminal domain of BRF binds to the top and N-terminal leg of TBP.
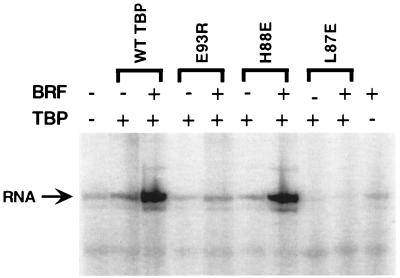
We also tested the TBP mutants defective for interaction with BRF for a defect in Pol III transcription by complementation of a whole-cell extract made from the temperature-sensitive TBP mutant I143N, which is defective for Pol I, II, and III transcription in vitro (40). Surprisingly, most of these TBP radical mutants had no striking defect for in vitro transcription by Pol III (Fig. 8 and data not shown). Rescue of transcription from this extract required the addition of both recombinant TBP and BRF. Only one of these TBP mutants, L87E, could not restore transcription in the TBP-depleted extract (Fig. 8). The result that many of the TBP mutants can still function in transcription is similar to that seen for some TBP mutants on the DNA binding surface which have a strong defect in TBP-DNA binding but little if any defect in Pol II transcription (47). A second related example is the finding that several TBP mutants are defective for interaction with TFIIB in vitro but show no transcription defect in vivo (26, 43). We speculate that strong cooperative interactions between subunits of TFIIIC, TFIIIB, Pol III, and DNA can compensate for the defects in protein-protein interactions between TBP and BRF.
FIG. 8.
Two TBP radical mutations fail to rescue Pol III transcription in vitro. A whole-cell extract from TBP mutant I143N was supplemented with 20 ng of BRF and 20 ng of either wild-type TBP or mutant TBP as indicated. Transcription is from the tRNA2Leu promoter.
TBP mutants affecting B" interaction.
Each of the TBP mutants was also tested for the ability to incorporate B" into a TFIIIB complex. For the most part, the results were as expected: those TBP mutants which failed to form BRF-TBP-DNA complexes also failed to form TFIIIB complexes, and those TBP mutants capable of forming BRF-TBP-DNA complexes could support the addition of B". However, one TBP mutation, E93R, affected only the binding of B" (Fig. 6B and 7A). This result suggests that BRF and B" both contact TBP on the N-terminal leg. This TBP mutant was defective in transcription and could not rescue the TBP-depleted in vitro transcription extract (Fig. 8).
DISCUSSION
We have probed the arrangement of the subunits in the TFIIIB-DNA complex by using three primary methods: (i) genetic dissection using mutations in BRF, TBP, and DNA combined with gel mobility shift assays to assess binding activity, (ii) chemical probing of the protein occupancy along the promoter DNA using hydroxyl radical ion, and (iii) biochemical competition assays to probe the locations of factors binding to the TBP-DNA complex. These results lead to a model for the interaction of TFIIIB subunits and DNA.
BRF binding to TBP-DNA requires DNA downstream of the TATA box.
BRF requires an unusually long stretch of DNA downstream of the TATA box (12 to 15 bp) and as little as 1 bp upstream for formation of a stable complex with TBP-DNA. This requirement for downstream DNA is distinct from other characterized factors which interact with TBP-DNA. TFIIA requires 3 to 5 bp of DNA upstream of the TATA (15), and TFIIB (the Pol II homolog of BRF) requires 7 bp upstream and downstream for binding (27, 32). The requirement for downstream DNA is consistent with photo-cross-linking results which demonstrate that BRF in the TFIIIB complex can cross-link between positions −26 and −12 at the SUP4 promoter (4, 5, 35). Additionally, DNase footprinting of a partial complex of TFIIIC, TBP, and BRF showed BRF-dependent DNase protection downstream from the presumed position of TBP (21). The surprising finding from our results is that BRF did not require DNA upstream from the TATA box for stable complex formation. Previous photo-cross-linking studies have suggested that BRF cross-links as far upstream as −38/−39 (5, 19). There are several possible explanations for this discrepancy. First, the most likely explanation is that the photo-cross-linking probes used are quite long (at least 10 Å) and may cross-link to protein which is not in direct contact with DNA. Second, TFIIIB complexes formed at the wild-type U6 promoter in the absence of TFIIIC are heterogeneous, as they promote transcription in both orientations (19, 45). Thus, upstream cross-linking of BRF may result from “backward” TFIIIB complexes. Finally, when B" is added, BRF may undergo a conformational change which results in contacts with DNA upstream of the TATA.
Despite the clear requirement for downstream DNA in formation of stable TBP-BRF-DNA complexes, we were unable to map the interaction of BRF with any specific backbone positions in the DNA. Hydroxyl radical footprinting of TBP-BRF complexes did not reveal any close contacts between BRF and any specific sugar residues in DNA downstream of the TATA. Similarly, ethylation interference assays did not reveal any specific phosphate residues required for BRF binding outside of the TATA box. One possibility is that the interaction of BRF with downstream DNA is flexible such that if a phosphate which normally interacts with BRF is modified, BRF will interact with a different backbone position. Another alternative is that BRF interacts with the DNA bases in either the major or minor groove of downstream DNA rather than the DNA backbone.
The homologs TFIIB and BRF have been shown to bind TBP-DNA with about the same affinity (37). However, it is clear from previous work (10, 19, 22) and the results shown here that BRF binds TBP-DNA by a mechanism very different from that used by TFIIB. The TBP-DNA binding activity of BRF is confined to the C-terminal nonconserved half of BRF, a domain with greater affinity for TBP-DNA than full-length BRF. In contrast, the N-terminal domain of BRF (residues 1 to 288) does not form complexes with TBP-DNA stable to electrophoresis. The reason for the failure of the conserved N-terminal domain of BRF to interact with TBP is unknown; however, it may be partly because numerous residues in TFIIB and Archaea TFB which contact TBP and DNA (25, 32) are not conserved in BRF (only 12 of 27 of these residues are conserved between yeast BRF and TFIIB).
The TBP binding activities of full-length BRF and the C-BRF appear identical except that the C-BRF binds with higher affinity than full-length BRF. First, the C-BRF derivatives and full-length BRF have the same requirement for DNA downstream of the TATA box. Second, binding of full-length BRF and that of the C-BRF derivatives are affected identically by radical mutations in TBP.
Position of BRF in the TBP-BRF-DNA complex.
Two different types of experiments place the position of BRF on the opposite side of TBP from where the homologous factor TFIIB binds. TFIIB and BRF can simultaneously bind to TBP-DNA complexes. This finding indicates that both the C-terminal stirrup of TBP and the DNA on this face of TBP are not contacted by BRF. In addition, this result suggests that the conformation of DNA and TBP in the TBP-BRF-DNA complex is very similar to that seen in the TBP-TFIIB-DNA complex. If BRF caused a significant conformational change in either TBP or DNA, then TFIIB would not be expected to bind to this complex. In contrast, TFIIA competes for binding with all BRF derivatives. Since TFIIA binding also does not cause a significant conformational change in either TBP or DNA upon binding (15, 42), these findings suggest that the binding sites for TFIIA and BRF on TBP and/or DNA overlap.
The second line of evidence which positions BRF in the TBP-DNA complex is the location of TBP mutations which reduce the binding of BRF. No mutations tested on the C-terminal leg or stirrup of TBP affect BRF binding. In contrast, three positions on the N-terminal leg affect binding of both full-length BRF and the C-BRF derivatives. In addition, one mutant on the top surface of TBP, K133E, has a strong effect on BRF binding. Similar analysis with human TBP has demonstrated at least seven radical mutations on the top surface of TBP affect binding of yeast BRF (one of which is equivalent to K133E) (5a). These results are also consistent with the position of Pol III-specific TBP mutations which are suppressed by overexpression of BRF (9, 12, 40). Thus, it appears that the C terminus of BRF interacts with TBP on the top surface and N-terminal leg as well as with DNA at least 15 bp downstream of the TATA box.
Position of B" and BRF within the TFIIIB-DNA complex.
It was not possible to directly determine the position of B" in the TFIIIB complex because (i) B" does not bind TBP-DNA with high affinity as does BRF and (ii) BRF may undergo a conformational change upon B" binding to TBP-BRF-DNA. However, binding experiments using various lengths of DNA suggest that B" interacts upstream of the TATA box. While BRF requires DNA only downstream of the TATA box, binding of B" to this complex requires an additional 15 residues upstream. One possibility is that this upstream DNA is bound solely by B". A second possibility is that BRF undergoes a conformational change upon B" addition such that it now contacts upstream DNA. This seems less likely, as formation of the TFIIIB complex still requires downstream DNA and TBP mutations which reduce BRF binding also reduce the formation of TFIIIB complexes. A third possibility is that B" can contact DNA both upstream and downstream of the TATA. Finally, TBP could undergo a conformational change which results in interaction with upstream and/or downstream DNA. This last possibility seems least likely, as TBP has not been observed to undergo any significant conformational change in complex with DNA, not bound to DNA, or in complex with TFIIA or with TFIIB and DNA (15, 23, 24, 32, 33, 42). The suggestion that B" interacts with DNA upstream from the TATA box is also consistent with photo-cross-linking results showing B" primarily cross-linking with far upstream DNA (5). One TBP mutation also suggests a site of B" interaction with TBP in the TFIIIB complex. Mutation at TBP position 93 specifically reduces the affinity of B" for BRF-TBP-DNA. This position is on the N-terminal leg of TBP and near the mutations which affect BRF binding. It seems that both B" and BRF converge on the N-terminal leg of TBP.
Figure 7 shows our combined results modeled on the TBP-DNA complex with 15 bp of DNA downstream of the TATA box and 9 bp upstream. One limitation of this model is that we do not know for certain if TBP and/or the DNA undergoes any conformational change in the TFIIIB complex. The simplest interpretation of our results, based on the hydroxyl radical footprinting data, is that in TFIIIB, BRF and B" together surround TBP contacting DNA both upstream and downstream of the TATA and are close to the N- and C-terminal legs of TBP. BRF also interacts along the top surface of TBP. Binding of TFIIIB subunits on both faces of the TBP-DNA complex as well as extensive protein-protein interactions between the three TFIIIB subunits probably contributes to the great stability of the TFIIIB-DNA complex. We cannot say for certain whether B" or BRF contributes to any particular sugar residues protected in the final complex. Since BRF requires DNA downstream of the TATA and can bind in the presence of TFIIB, a conservative interpretation is that BRF is responsible for the hydroxyl radical protections seen near the N-terminal stirrup of TBP and does not make DNA contacts on the opposite face of TBP. Perhaps B" is responsible for the backbone protections seen in Fig. 7B as well as those upstream of the TATA box in Fig. 7A. A clear answer to this question awaits structural or other biochemical analysis of the complex.
ACKNOWLEDGMENTS
We thank A. Berk and Y. Shen for communication of unpublished results, J. Geiger for gifts of purified proteins, M. Dolejsi (FHCRC) and B. Lane (Harvard Microchemistry) for protein sequence analysis, and J. Geiger, R. Reeder, S. Roberts, and J. Ranish for comments on the manuscript.
This work was supported by a graduate student training grant to T.C. and funds from the NIH (GM53451) and a Leukemia Society Scholar Award to S.H. S.H. is an associate investigator of the Howard Hughes Medical Institute.
The first two authors made equal contributions to this work.
REFERENCES
- 1.Bagby S, Kim S, Maldonado E, Tong K I, Reinberg D, Ikura M. Solution structure of the carboxy-terminal core domain of human TFIIB: similarity to cyclin A and interaction with TATA binding protein. Cell. 1995;82:857–867. doi: 10.1016/0092-8674(95)90483-2. [DOI] [PubMed] [Google Scholar]
- 2.Baker R E, Camier S, Sentenac A, Hall B D. Gene size differentially affects the binding of yeast transcription factor tau to two intragenic regions. Proc Natl Acad Sci USA. 1987;84:8768–8772. doi: 10.1073/pnas.84.24.8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barberis A, Muller C W, Harrison S C, Ptashne M. Delineation of two functional regions of transcription factor TFIIB. Proc Natl Acad Sci USA. 1993;90:5628–5632. doi: 10.1073/pnas.90.12.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartholomew B, Durkovich D, Kassavetis G A, Geiduschek E P. Orientation and topography of RNA polymerase III in transcription complexes. Mol Cell Biol. 1993;13:942–952. doi: 10.1128/mcb.13.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartholomew B, Kassavetis G A, Geiduschek E P. Two components of Saccharomyces cerevisiae transcription factor IIIB (TFIIIB) are stereospecifically located upstream of a tRNA gene and interact with the second-largest subunit of TFIIIC. Mol Cell Biol. 1991;11:5181–5189. doi: 10.1128/mcb.11.10.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Berk, A., and Y. Shen. Personal communication.
- 6.Brow D A, Guthrie C. Splicing a spliceosomal RNA. Nature. 1989;337:14–15. doi: 10.1038/337014a0. [DOI] [PubMed] [Google Scholar]
- 7.Bryant G O, Martel L S, Burley S K, Berk A J. Radical mutations reveal TATA-box binding protein surfaces required for activated transcription in vivo. Genes Dev. 1996;10:2491–2504. doi: 10.1101/gad.10.19.2491. [DOI] [PubMed] [Google Scholar]
- 8.Buratowski S, Zhou H. Functional domains of transcription factor TFIIB. Proc Natl Acad Sci USA. 1993;90:5633–5637. doi: 10.1073/pnas.90.12.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buratowski S, Zhou H. A suppressor of TBP mutations encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell. 1992;71:221–230. doi: 10.1016/0092-8674(92)90351-c. [DOI] [PubMed] [Google Scholar]
- 10.Chaussivert N, Conesa C, Shaaban S, Sentenac A. Complex interactions between yeast TFIIIB and TFIIIC. J Biol Chem. 1995;270:15353–15358. doi: 10.1074/jbc.270.25.15353. [DOI] [PubMed] [Google Scholar]
- 10a.Colbert, T. Unpublished data.
- 11.Colbert T, Hahn S. A yeast TFIIB-related factor involved in RNA polymerase III transcription. Genes Dev. 1992;6:1940–1949. doi: 10.1101/gad.6.10.1940. [DOI] [PubMed] [Google Scholar]
- 12.Cormack B P, Struhl K. Regional codon randomization: defining a TATA-binding protein surface required for RNA polymerase III transcription. Science. 1993;262:244–248. doi: 10.1126/science.8211143. [DOI] [PubMed] [Google Scholar]
- 13.Gabrielson O S, Sentenac A. RNA polymerase III(C) and its transcription factors. Trends Biochem Sci. 1991;16:412–416. doi: 10.1016/0968-0004(91)90166-s. [DOI] [PubMed] [Google Scholar]
- 14.Geiduschek E P, Kassavetis K A. RNA polymerase III transcription complexes. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 15.Geiger J H, Hahn S, Lee S, Sigler P B. Crystal structure of the yeast TFIIA/TBP/DNA complex. Science. 1996;272:830–836. doi: 10.1126/science.272.5263.830. [DOI] [PubMed] [Google Scholar]
- 16.Ha I, Roberts S, Maldonado E, Sun X, Kim L, Green M, Reinberg D. Multiple functional domains of human transcription factor IIB: distinct interactions with two general transcription factors and RNA polymerase II. Genes Dev. 1993;7:1021–1032. doi: 10.1101/gad.7.6.1021. [DOI] [PubMed] [Google Scholar]
- 17.Joazeiro C A, Kassavetis G A, Geiduschek E P. Identical components of yeast transcription factor IIIB are required and sufficient for transcription of TATA box-containing and TATA-less genes. Mol Cell Biol. 1994;14:2798–2808. doi: 10.1128/mcb.14.4.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joazerio C A P, Kassavetis G A, Geiduschek E P. Alternative outcomes in assembly of promoter complexes: the roles of TBP and a flexible linker in placing TFIIIB on tRNA genes. Genes Dev. 1996;10:725–739. doi: 10.1101/gad.10.6.725. [DOI] [PubMed] [Google Scholar]
- 19.Kassavetis G A, Bardeleben C, Kumar A, Ramirez E, Geiduschek E P. Domains of the Brf component of RNA polymerase III transcription factor IIIB (TFIIIB): functions in assembly of TFIIIB-DNA complexes and recruitment of RNA polymerase to the promoter. Mol Cell Biol. 1997;17:5299–5306. doi: 10.1128/mcb.17.9.5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kassavetis G A, Braun B R, Nguyen L H, Geiduschek E P. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990;60:235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- 21.Kassavetis G A, Joazeiro C A P, Pisano M, Geiduschek E P, Colbert T, Hahn S, Blanco J A. The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell. 1992;71:1055–1064. doi: 10.1016/0092-8674(92)90399-w. [DOI] [PubMed] [Google Scholar]
- 22.Khoo B, Brophy B, Jackson S P. Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes Dev. 1994;8:2879–2890. doi: 10.1101/gad.8.23.2879. [DOI] [PubMed] [Google Scholar]
- 23.Kim J L, Nikolov D B, Burley S K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 24.Kim Y, Geiger J H, Hahn S, Sigler P B. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 25.Kosa P F, Ghosh G, DeDecker B S, Sigler P B. The 2.1-A crystal structure of an archeal preinitiation complex: TATA-box-binding protein/transcription factor (II)B core/TATA-box. Proc Natl Acad Sci USA. 1997;94:6042–6047. doi: 10.1073/pnas.94.12.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee M, Struhl K. A severely defective TATA-binding protein–TFIIB interaction does not preclude transcriptional activation in vivo. Mol Cell Biol. 1997;17:1336–1345. doi: 10.1128/mcb.17.3.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S, Hahn S. Model for binding of transcription factor TFIIB to the TBP-DNA complex. Nature. 1995;376:609–612. doi: 10.1038/376609a0. [DOI] [PubMed] [Google Scholar]
- 28.Leveillard T, Kassavetis G A, Geiduschek E P. S. cerevisiae transcription factors IIIB and IIIC bend the DNA of a tRNAGLN gene. J Biol Chem. 1991;266:5162–5168. [PubMed] [Google Scholar]
- 29.Librizzi M D, Moir R D, Brenowitz M, Willis I M. Expression and purification of the RNA polymerase III transcription specificity factor IIIB70 from Saccharomyces cerevisiae and its cooperative binding with TATA-binding protein. J Biol Chem. 1996;271:32695–32701. doi: 10.1074/jbc.271.51.32695. [DOI] [PubMed] [Google Scholar]
- 30.Lopez-De-Leon A, Librizzi M, Puglia K, Willis I M. PCF4 encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell. 1992;71:211–220. doi: 10.1016/0092-8674(92)90350-l. [DOI] [PubMed] [Google Scholar]
- 31.Margottin F, Dujardin G, Gerard M, Egly J-M, Huet J, Sentenac A. Participation of the TATA factor in transcription of the yeast U6 gene by RNA polymerase C. Science. 1991;251:424–426. doi: 10.1126/science.1989075. [DOI] [PubMed] [Google Scholar]
- 32.Nikolov D B, Chen H, Halay E D, Usheva A A, Hisatake K, Lee D K, Roeder R G, Burley S K. Crystal structure of a TFIIB/TBP/TATA element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 33.Nikolov D B, Hu S, Lin J, Gasch A, Hoffmann A, Horikoshi M, Chua N, Roeder R G, Burley S K. Crystal structure of TFIID TATA-box binding protein. Nature. 1992;360:40–46. doi: 10.1038/360040a0. [DOI] [PubMed] [Google Scholar]
- 34.Ouzounis C, Sander C. TFIIB, an evolutionary link between the transcription machineries of archaebacteria and eukaryotes. Cell. 1992;71:189–190. doi: 10.1016/0092-8674(92)90347-f. [DOI] [PubMed] [Google Scholar]
- 35.Persinger J, Bartholomew B. Mapping the contacts of yeast TFIIIB and RNA polymerase III at various distances from the major groove of DNA by DNA photoaffinity labeling. J Biol Chem. 1996;271:33039–33046. doi: 10.1074/jbc.271.51.33039. [DOI] [PubMed] [Google Scholar]
- 36.Qureshi S A, Khoo B, Baumann P, Jackson S P. Molecular cloning of the transcription factor TFIIB homolog from Sulfolobus shibatae. Proc Natl Acad Sci USA. 1995;92:6077–6081. doi: 10.1073/pnas.92.13.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36a.Roberts, S. Unpublished data.
- 37.Roberts S, Colbert T, Hahn S. TFIIIC determines RNA polymerase III specificity at the TATA-containing yeast U6 promoter. Genes Dev. 1995;9:832–833. doi: 10.1101/gad.9.7.832. [DOI] [PubMed] [Google Scholar]
- 38.Roberts S, Miller S J, Lane W S, Lee S, Hahn S. Cloning and functional characterization of the gene encoding the TFIIIB90 subunit of RNA polymerase III transcription factor TFIIIB. J Biol Chem. 1996;271:14903–14909. doi: 10.1074/jbc.271.25.14903. [DOI] [PubMed] [Google Scholar]
- 39.Schultz M C, Choe S Y, Reeder R H. Specific initiation by RNA polymerase I in a whole-cell extract from yeast. Proc Natl Acad Sci USA. 1991;88:1004–1008. doi: 10.1073/pnas.88.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schultz M C, Reeder R H, Hahn S. Variants of the TATA-binding protein can distinguish subsets of RNA polymerase I, II, and III. Cell. 1992;69:697–702. doi: 10.1016/0092-8674(92)90233-3. [DOI] [PubMed] [Google Scholar]
- 41.Struhl K. Duality of TBP, the universal transcription factor. Science. 1994;263:1103–1104. doi: 10.1126/science.8108728. [DOI] [PubMed] [Google Scholar]
- 42.Tan S, Hunziker Y, Sargent D F, Richmond T J. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature. 1996;381:127–134. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- 43.Tang H, Sun X, Reinberg D, Ebright R H. Protein-protein interactions in eukaryotic transcription initiation: structure of the pre-initiation complex. Proc Natl Acad Sci USA. 1996;93:1119–1124. doi: 10.1073/pnas.93.3.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomm M. Archaeal transcription factors and their role in transcription initiation. FEMS Microbiol Rev. 1996;18:159–171. doi: 10.1111/j.1574-6976.1996.tb00234.x. [DOI] [PubMed] [Google Scholar]
- 45.Whitehall S K, Kassavetis G A, Geiduschek E P. The symmetry of the yeast U6 RNA gene’s TATA box and the orientation of the TATA-binding protein in yeast TFIIIB. Genes Dev. 1995;9:2974–2985. doi: 10.1101/gad.9.23.2974. [DOI] [PubMed] [Google Scholar]
- 46.Willis I M. RNA polymerase III. Genes, factors and transcriptional specificity. Eur J Biochem. 1993;212:1–11. doi: 10.1111/j.1432-1033.1993.tb17626.x. [DOI] [PubMed] [Google Scholar]
- 47.Yamamoto T, Horikoshi M, Wang J, Hasegawa S, Weil P A, Roeder R G. A bipartite DNA binding domain composed of direct repeats in the TATA box binding factor TFIID. Proc Natl Acad Sci USA. 1992;89:2844–2848. doi: 10.1073/pnas.89.7.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu W, Zeng Q, Coangelo C M, Lewis M, Summers M F, Scott R A. The N-terminal domain of TFIIB from Pyrococcus furiosus forms a zinc ribbon. Nat Struct Biol. 1996;3:122–124. doi: 10.1038/nsb0296-122. [DOI] [PubMed] [Google Scholar]