Abstract
The TATA binding protein (TBP) is a central component of the eukaryotic transcriptional machinery and is the target of positive and negative transcriptional regulators. Here we describe the cloning and biochemical characterization of an abundant human TBP-associated factor (TAF-172) which is homologous to the yeast Mot1 protein and a member of the larger Snf2/Swi2 family of DNA-targeted ATPases. Like Mot1, TAF-172 binds to the conserved core of TBP and uses the energy of ATP hydrolysis to dissociate TBP from DNA (ADI activity). Interestingly, ATP also causes TAF-172 to dissociate from TBP, which has not been previously observed with Mot1. Unlike Mot1, TAF-172 requires both TBP and DNA for maximal (∼100-fold) ATPase activation. TAF-172 inhibits TBP-driven RNA polymerase II and III transcription but does not appear to affect transcription driven by TBP-TAF complexes. As it does with Mot1, TFIIA reverses TAF-172-mediated repression of TBP. Together, these findings suggest that human TAF-172 is the functional homolog of yeast Mot1 and uses the energy of ATP hydrolysis to remove TBP (but apparently not TBP-TAF complexes) from DNA.
The TATA binding protein (TBP) is recruited to eukaryotic promoters, where it plays a central role in transcription complex assembly. TBP and a variety of TBP-associated factors (TAFs) comprise functionally distinct multisubunit complexes (17, 27, 29, 33, 46). SL1, TFIID, and TFIIIB are TBP-TAF complexes that specify assembly of RNA polymerase (pol) I, II, and III transcription complexes, respectively. SNAPc appears to be a TBP-containing complex which targets both pol II- and pol III-transcribed small nuclear RNA promoters (16, 34). B-TFIID contains TBP and a 170-kDa TAF and supports pol II transcription (39, 40).
In yeast, Mot1 has been identified as a TAF that appears to regulate transcription both negatively and positively (3, 7, 8, 18, 19, 23, 26). The MOT1 gene was identified in genetic screens for mutants that increased levels of basal transcription (8, 18, 19, 23, 26). Mot1 belongs to the Snf2/Swi2 family of conserved DNA-targeted ATPases (12), although the Mot1 ATPase can function in the absence of DNA (4). Members of this family are involved in a wide array of protein-nucleic acid transactions, including transcription, chromosome segregation, and DNA repair (5).
Mot1 was independently identified as an ATP-dependent inhibitor (ADI) of TBP-TATA interactions (2, 3). ADI activity is suppressed by TFIIA, presumably through mutual competition for TBP binding (2). Mot1 ADI activity inhibits pol II transcription in vitro, but its effectiveness might depend on the level of TFIIA present in the system. Paradoxically, Mot1-TBP complexes are distinct from TFIID complexes (28). Whether Mot1 ADI activity targets TBP alone, TFIID, or any TBP-containing complex in vivo is unclear. A genetic interaction between Mot1 and Spt3, which also interacts with TBP, has been demonstrated, suggesting that Mot1 is directly involved in TBP function (23). In addition to repression, Mot1 function has also been implicated in gene activation in vivo (23). Therefore, Mot1 might function as a negative regulator at some promoters and a positive regulator at others. Alternatively, Mot1 might function indirectly, perhaps by removing TBP from nonpromoter DNA (2). Mot1 mutants that are unable to perform this activity might affect genes differentially.
We have previously described a human TAF fraction (TAF-172) which contained components of pol III transcription factor TFIIIB (37). This fraction also possessed some properties reminiscent of Mot1 and B-TFIID. To further characterize TAF-172, we set out to clone the gene encoding it. The TAF-172 gene bears a striking resemblance to the yeast MOT1 gene. To characterize the protein, recombinant TAF-172 was produced by using a baculovirus expression system and purified to apparent homogeneity. Antibodies against TAF-172 were used to probe the abundance of TAF-172 in HeLa cells. We examined TBP–TAF-172 interactions via protein affinity chromatography and TAF-172–TBP–DNA complex formation, as well as its dissociation by ATP via the electrophoretic mobility shift assay (EMSA). The ability of TAF-172 to hydrolyze ATP and its cofactor requirements were also investigated and found to be activated by the combined action of TBP and DNA. Finally, we have used in vitro transcription assays to characterize TAF-172’s activity toward the regulation of pol II and pol III transcription. TAF-172 appears to inhibit TBP-driven but not TBP-TAF-driven pol II and pol III transcription in vitro. These findings suggest that TAF-172 targets primarily DNA-bound TBP for ATP-dependent removal from DNA.
MATERIALS AND METHODS
Solutions, DNAs, and proteins.
H buffer contained 20 mM HEPES (pH 7.5), 10% glycerol, 2 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol, and the molar KCl concentration indicated in the buffer name. TSB contained 20 mM Tris acetate (pH 8.0), 20% glycerol, 2 mM MgCl2, 200 mM potassium glutamate, 0.1 mM EDTA, 1 mM dithiothreitol, and 0.1 mM phenylmethylsulfonyl fluoride. Stop mixture contained 3 M ammonium acetate and 125-μg/ml tRNA. PSB contained 125 mM Tris-HCl (pH 6.8), 10% glycerol, 3.1% sodium dodecyl sulfate (SDS), 0.71 M β-mercaptoethanol, and 0.5-mg/ml bromphenol blue.
Plasmids pS3TI and pG6TI were previously described (6, 32). Promoter fragments for in vitro transcription were generated from pG6TI (32), pBRVA1 (45), and pGEM-U6 (22) by PCR. The TI promoter fragment was generated by cleavage of pSP72(TATA/Inr) with restriction endonuclease EcoO109 (36).
HeLa nuclear extracts and phosphocellulose fractions were prepared as previously described (30, 37). The following recombinant proteins were produce in Escherichia coli and purified as previously described: GST-TBP-N and GST-TBP-C (15), TBP (31), yeast Mot1 (4), and TFIIA (44). The B-TFIID fraction was obtained from and purified by R. Meyers and P. Sharp (Massachusetts Institute of Technology).
Purification of TAF-172 for internal peptide sequencing.
TAF-172 was immunopurified from 725 mg of the HeLa cell-derived phosphocellulose 0.1 to 0.3 step fraction (gift of D. Reinberg [University of Medicine and Dentistry of New Jersey]) with 2 mg of affinity-purified TBP antibodies as previously described (37), with the following modifications. TBP immunoprecipitates were washed with H buffer containing 0.1 M guanidine hydrochloride (GuHCl), and TAF-172 was eluted with H buffer containing 1 M GuHCl. The TAF-172 pool (∼2 μg) was dialyzed against Tris-EDTA buffer, electrophoresed on an SDS–6% polyacrylamide gel, transferred to a polyvinylidene difluoride membrane, stained with amido black, and subjected to tryptic digestion in accordance with standard protocols. Internal peptide sequencing of reverse-phase-purified peptides was performed by the Wistar Protein Microsequencing Facility (Philadelphia, Pa.).
Cloning of TAF-172.
Converging degenerate primers (GARTAYATHGCNGGNGC and GCNGGRTCYTCCATDAT), which encode the terminal regions of the sequenced peptide EVLQEYIAGADTIMEDPATR, were used in a PCR with oligo(dT)-primed human cDNA. PCR products were separated by polyacrylamide gel electrophoresis, and the expected fragment was excised and sequenced. The sequence was used to synthesize the nondegenerate probe GAGTATATTGCGGGTGCCGACACCATCATGGAAGACCCAGC. The probe was 32P end labeled and immediately used to screen a phage λgt10 human cDNA library. An initial partial clone was isolated, and its insert was subcloned into EcoRI-cut pGEM-7z (Promega) and sequenced. The clone was used in subsequent screens to isolate additional clones, one of which contained an open reading frame coding for amino acids 1 to 876 in TAF-172. Sequences in the clone and sequences coding for peptide sequences predicted to be located near the C-terminal end of TAF-172, based on alignments with MOT1, were used in a nested PCR (external primers, AGCCACATCATCTTTCG and CCARTTNACNCCRTCYTG; internal primers, TCGAGTAAACAACAATG and ACNCCRTCYTGYTGRTA) on randomly primed cDNA to obtain a 1.4-kb probe for the 3′ half of the gene. This probe contained an open reading frame coding for amino acids 816 to 1272 in TAF-172. Screens of phage libraries with this probe yielded a partial clone which contained an open reading frame coding for amino acids 976 to 1848 in TAF-172.
Affinity-purified antibodies.
TAF-172 sequences encoding amino acids 1 to 623 and 952 to 1858 were subcloned separately into the NdeI site of the pET16b expression vector (Novagen) by PCR. The polyhistidine-tagged proteins were expressed in E. coli BL21 (Novagen) and purified from GuHCl-solubilized inclusion bodies by nickel affinity chromatography (Pharmacia) in accordance with the manufacturer’s directions. Both proteins were injected into the same rabbit to produce polyclonal antibodies. Both proteins were coupled to Affi-Gel 10 (Bio-Rad) and used to affinity purify TAF-172 antibodies. Antibodies were eluted with a solution of 50 mM glycine (pH 2.0) and 150 mM NaCl. The antibodies were immediately neutralized with Tris-Cl (pH 8) and dialyzed against TSB (lacking dithiothreitol). Affinity-purified human TBP and TAFII250 antibodies were purified in the same manner against their cognate antigens.
Baculovirus expression and purification of TAF-172.
To generate full-length TAF-172, the two cDNA clones and the overlapping PCR product were combined by using convenient restriction sites and inserted into the vector pFastBac1 (Gibco-BRL) along with the 6× polyhistidine tag acquired from pET16b. Recombinant baculovirus was generated by using the Bac-to-Bac Baculovirus Expression kit (Gibco-BRL). Sf9 cells (5 × 108) grown in Grace’s insect medium (Gibco-BRL) supplemented with 10% fetal bovine serum were infected with recombinant TAF-172 baculovirus for 72 h and harvested by centrifugation. Cells were washed in 25 ml of PBSM (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4 · 7H2O, 1.4 mM KH2PO4, 12.5 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride) and lysed with 10 ml of AS0.6 buffer (60 mM Tris acetate [pH 8.0], 15 mM MgCl2, 20% glycerol, 0.625 mM ammonium sulfate, 1 mM phenylmethylsulfonyl fluoride, 2-μg/ml leupeptin, 2-μg/μl pepstatin A). The cell lysate was sonicated to reduce viscosity and centrifuged to pellet cell debris. Polyhistidine-tagged TAF-172 was affinity purified on a 2-ml nickel Sepharose column (Pharmacia) in accordance with the manufacturer’s directions and eluted with NE buffer (20 mM Tris acetate [pH 8.0], 10% glycerol, 2 mM MgCl2, 200 mM potassium glutamate, 400 mM imidazole). The eluate (4 ml) was chromatographed on a 150-ml Sephacryl S300 gel filtration column (Pharmacia) equilibrated with TSB. TAF-172 fractions eluting between 180 and 230 kDa (9 ml) were pooled and applied to a 0.5-ml S-Sepharose (Pharmacia) column equilibrated with TSB. TAF-172 was present in the flowthrough fraction and was stored at −80°C.
Immunoprecipitations.
Rabbit serum (1 ml) containing either TAF-172, TFIIA, or control nonspecific antibodies was incubated with protein A Sepharose (200 μl) for 2 h at 4°C with constant mixing. The resin was washed with 200 mM sodium borate solution, and antibodies were cross-linked to the resin with 10 mM dimethyl pimelimidate at 23°C for 1 h. Cross-linking was quenched with 200 mM ethanolamine (pH 7.5), and the resin was washed with H.1 buffer.
HeLa nuclear extracts (22 μg, 1 ml) were mixed with protein A Sepharose containing cross-linked TAF-172 (0.2 ml) and/or TFIIA (0.5 ml) or equivalent amounts of control antibodies at 4°C for 3.5 h. The resin was removed by centrifugation, and the extracts were stored at −80°C. The resin was washed with H1 buffer, followed by H.1 buffer, and then drained. Depletion of TBP from nuclear extracts was done similarly, except that 0.2 mg of affinity-purified TBP antibodies was used per mg of nuclear extract.
TBP–TAF-172 binding.
TAF-172 was incubated with either GST-TBP-N, GST-TBP-C, or TSB buffer alone for 2 h at 37°C. Proteins were then incubated with glutathione agarose at 4°C for an additional 1 h and washed with H1 buffer. Bound proteins were eluted with H.15 buffer containing 7.5 mM reduced glutathione. Eluted fractions were precipitated with trichloroacetic acid.
EMSA.
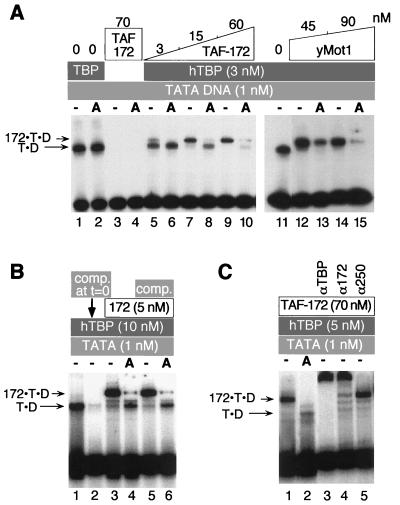
The 50-bp probe contains the adenovirus major late TATA box (TATAAAAG) and 28 bp of DNA upstream of the TATA box (2). The DNA was 32P end labeled with polynucleotide kinase and gel purified in accordance with standard protocols. In addition to TBP, TAF-172, and ATP in the amounts indicated in the corresponding figure legend (see Fig. 5), reaction mixtures contained 4 mM Tris-Cl (pH 8.0), 4% glycerol, 5 mM MgCl2, 60 mM KCl, 0.1% Brij 58, 5-μg/ml poly(dG-dC), and 100-μg/ml bovine serum albumin (2). Reaction mixtures containing TAF-172 or Mot1 and/or human TBP were incubated for 20 min at 23°C prior to loading on the gel. Samples (20 μl) were loaded onto native 6% (59:1 acrylamide-bisacrylamide ratio) polyacrylamide gels containing 1× TG buffer (25 mM Tris-Cl [pH 8.3], 190 mM glycine, 1 mM EDTA, 5 mM magnesium acetate), 2.5% (vol/vol) glycerol, and 0.5 mM dithiothreitol in running buffer containing 1× TG. Electrophoresis was continued at 35 mA for 60 to 90 min at 4°C.
FIG. 5.
ATP-mediated dissociation of TAF-172–TBP–DNA complexes as detected by EMSA. (A) Reaction mixtures contained human TBP (hTBP) (lanes 1, 2, and 5 to 15), radiolabeled TATA DNA (50 bp), and increasing concentrations of TAF-172 (lanes 5 to 10) or yeast Mot1 (lanes 12 to 15), as indicated. The letter A indicates that 0.1 mM ATP was included. Migration of TBP-DNA complexes is indicated by T·D and TAF-172/Mot1–human TBP–DNA complexes are indicated by 172·T·D. (B) TBP-DNA or TAF-172–TBP–DNA complexes were preassembled at the indicated concentrations. ATP (0.1 mM) and/or competitor (comp.) TATA DNA (250 nM) were then simultaneously added to the reaction mixtures in lanes 4 to 6, as indicated. Reactions were allowed to continue for 5 min before loading of the gel. In lane 2, competitor DNA was added at the same time as the labeled probe. (C) Reactions similar to those in panel A, except that a 106-bp TATA DNA probe was used. Where indicated, 0.6 μg of antigen affinity-purified TBP, TAF-172, or control TAFII250 antibody was added to the reaction mixture.
ATPase assay.
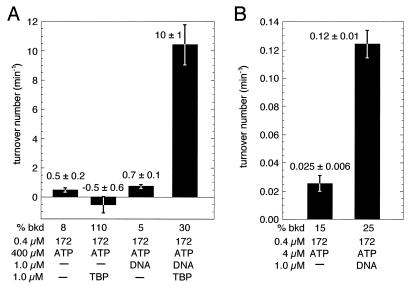
Reaction mixtures contained TBP, TAF-172, G6TI DNA (361 bp), and ATP (including 0.5-μCi/μl [α-32P]ATP) at the concentrations indicated in the corresponding figure (see Fig. 6). Reactions were performed in TSB at 30°C in a volume of 10 μl. At various times, 1-μl samples were spotted on polyethylenimine thin-layer chromatography plates, dried, and developed with a solution of 0.8 M glacial acetic acid and 0.8 M LiCl2. The plates were dried, and the radioactivity present as [α-32P]ATP and [α-32P]ADP was quantitated by a PhosphorImager and NIH Image software. The percent ATP hydrolyzed was plotted as a function of time by using Kaleidagraph software, and a global linear fit of the data was made. Standard errors are reported.
FIG. 6.
TAF-172 is a TBP-stimulated DNA-dependent ATPase. (A and B) TAF-172 was assayed for ATPase activity in the presence or absence of TBP and a 361-bp G6TI promoter DNA fragment, as indicated. Background rates of ATP hydrolysis determined in parallel reactions lacking TAF-172 were subtracted from the data; standard errors were summed. Background rates as a percentage of the rate determined in the presence of TAF-172 are indicated (% bkd).
In vitro pol II and pol III transcription assays.
In vitro pol II transcription reaction mixtures contained 3 mM HEPES, 9 mM Tris acetate (pH ∼7.9), 5 mM MgCl2, 10% glycerol, 15 mM KCl, 90 mM potassium glutamate, 50 μM EDTA, 0.5 mM dithiothreitol, 4 mM spermidine, 1% polyvinyl alcohol, 10-μg/ml poly(dG-dC), 1-μg/ml G6TI 361-bp promoter DNA or 10-μg/ml TI 2,487-bp DNA, 0.5 mM GTP, 0.5 mM UTP, 0.5 mM CTP, 10 μM ATP, and 4 μCi of [α-32P]ATP in a volume of 20 μl. Nuclear extracts (60 μg) and other reaction components (except nucleoside triphosphates) were added, and incubations at 30°C were continued for 20 min. Transcription was initiated by addition of nucleoside triphosphates and allowed to proceed at 30°C for 20 min. Reactions were terminated with 80 μl of stop mixture. The RNA was extracted with a phenol-chloroform mixture, precipitated with ethanol, resuspended in 90% formamide, and electrophoresed on 7 M urea–6% polyacrylamide gels. Gels were dried, and the radioactivity was visualized by using a PhosphorImager. In vitro VA1 pol III transcription reaction mixtures contained 6 mM HEPES, 4 mM Tris acetate (pH ∼7.7), 4 mM MgCl2, 7% glycerol, 50 mM KCl, 40 mM potassium glutamate, 25 μM EDTA, 0.5 mM dithiothreitol, 0.5 mM spermidine, 10-μg/ml poly(dG-dC), 1-μg/ml VA1 315-bp promoter DNA, 0.5 mM GTP, 0.5 mM UTP, 0.5 mM CTP, 10 μM ATP, and 4 μCi of [α-32P]ATP. In vitro U6 transcription reaction mixtures contained 6 mM HEPES, 4 mM Tris acetate (pH ∼7.7), 4 mM MgCl2, 7% glycerol, 45 mM KCl, 60 mM potassium glutamate, 25 μM EDTA, 0.5 mM dithiothreitol, 1 mM spermidine, 1% polyvinyl alcohol, 10-μg/ml poly(dG/dC), 1-μg/ml U6 306-bp promoter DNA, 0.5 mM GTP, 0.5 mM UTP, 0.5 mM CTP, 10 μM ATP, and 4 μCi of [α-32P]ATP.
RESULTS
Cloning of the TAF-172 gene.
HeLa nuclear extracts were fractionated over phosphocellulose, and TBP–TAF-172 complexes eluting in the P.3 fraction were immunopurified with TBP antibodies (37). TAF-172 was further fractionated by SDS-polyacrylamide gel electrophoresis, and electroblotted to a polyvinylidene difluoride membrane. Proteins were stained with amido black, and the TAF-172 band was excised and subjected to trypsin digestion. Six reverse-phase-purified peptides were sequenced. Based upon the peptide sequence, degenerate primers were synthesized and used in a PCR with human cDNA to generate a probe spanning the coding region of the peptide. λgt10 phage plaques (2 × 106) were probed to obtain a single partial cDNA clone. Additional cDNA library screens and PCRs provided the entire TAF-172 gene (see Materials and Methods). An in-frame stop codon precedes the initial methionine codon, indicating that the entire 5′ end of the open reading frame is present. The entire open reading frame encodes a 1,849-amino-acid protein with a predicted molecular mass of 206 kDa (Fig. 1A). All six sequenced peptides are present in the coding sequence.
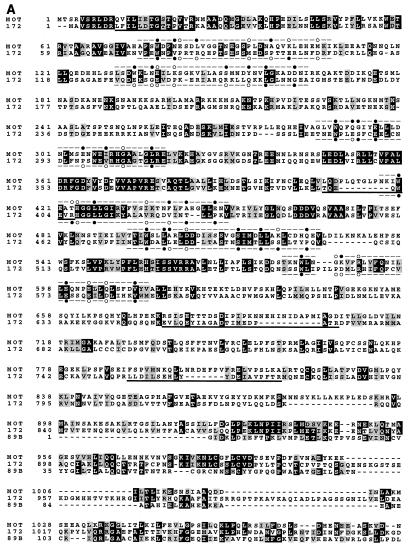
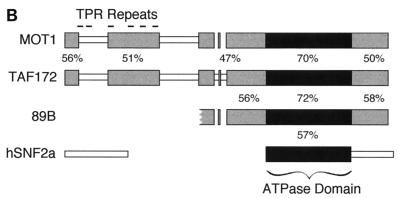
FIG. 1.
(A) Alignment of yeast Mot1, human TAF-172, and Drosophila 89B helicase. Protein sequences were aligned by using the CLUSTAL W sequence alignment program. Identical amino acids have a black background, and conserved (I-L-M-V, T-S, D-E, Q-N, K-R-H, Y-W-F, and G-A) amino acids have a gray background. Peptide sequences obtained by internal microsequencing are underlined. Previously identified TPR motifs are indicated above the Mot1 sequence, and corresponding TAF-172 regions are indicated below the TAF-172 sequence. A black dot indicates a conserved fit to the TPR consensus, an open circle indicates no match, and a dash indicates no consensus. The sequence of TAF-172 is identical to that of TAFII170 (41), except at position 945, which is a C in TAFII170 and an S in TAF-172 (accession no. AF038362) and two GenBank expressed sequence tags of TAF-172 (accession no. R07413 and T78264). (B) Schematic alignment of Drosophila 89B helicase, human TAF-172, yeast Mot1, and human Snf2a. Gray blocks depict regions of high sequence similarity. The black box represents the putative ATPase-helicase domain found in all Snf2 family members. Percent similarities were calculated by using the identical and conserved amino acid changes described above. Blocks were generated by using the MACAW sequence alignment program.
BLAST searches of protein databases (1) revealed extensive sequence similarity among human TAF-172, the yeast Mot1 protein (37% identity and 50% conservation), and the Drosophila 89B helicase gene product of a partial cDNA clone (56% identity and 66% conservation) over their entire sequence (Fig. 1). Essentially all of the similarity between TAF-172 and Mot1 is contained within five sequence blocks, suggesting that they encode functions common to both (Fig. 1B). The most highly conserved block is predicted to contain a domain common to members of the Snf2 family of DNA-targeted ATPases. TAF-172 shows no similarity to other members of the Snf2 family outside of the ATPase domain. Residues that align with Mot1’s proposed tetratricopeptide repeats (TPR) are indicated in Fig. 1A. While this report was under review, the sequence of an identical gene, that for TAFII170, was reported (41). TAFII170 binds TBP and appears to be a component of B-TFIID.
Purification of recombinant TAF-172.
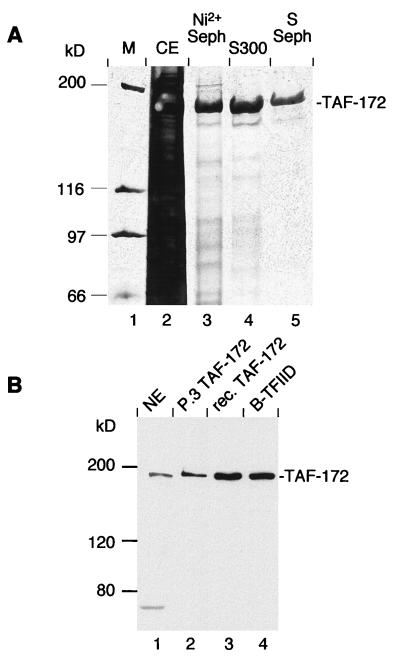
E. coli expression of TAF-172 was largely ineffective due to its insolubility and incomplete synthesis. The entire TAF-172 open reading frame was subcloned into a baculovirus vector along with a polyhistidine tag to aid in purification. Recombinant TAF-172 was highly overexpressed and soluble in infected Sf9 insect cells. TAF-172 was purified to apparent homogeneity by using nickel Sepharose, gel filtration, and S-Sepharose (Fig. 2A). All of the experiments described here used the high-purity S-Sepharose fraction.
FIG. 2.
Purification of recombinant TAF-172. (A) Sf9 insect cells were infected with recombinant baculovirus containing the TAF-172 gene and six in-frame histidine codons at the amino terminus. Equal proportions of pooled fractions generated at each stage of the purification process were analyzed on an SDS–6% polyacrylamide gel in which the proteins were stained with silver. Molecular weight markers (M) are shown in lane 1. The crude cell lysate (CE; lane 2), the Ni2+ column elution (lane 3), the Sephacryl S300 gel filtration pool (lane 4), and the S Sepharose pool (lane 5) are shown. (B) Western blot of TAF-172. HeLa nuclear extract (NE; 40 μg, lane 1), immunopurified, P.3-derived TAF-172 (20 ng, lane 2), recombinant (rec.) TAF-172 (25 ng, lane 3), and a B-TFIID containing Superdex 200 PG fraction (39) (1.2 μg, lane 4) were electrophoresed on an SDS–6% polyacrylamide gel. Proteins were electroblotted to nitrocellulose and probed with affinity-purified TAF-172 antibodies.
TAF-172 polyclonal antibodies generated against E. coli-expressed TAF-172 fragments reacted with purified baculovirus-produced TAF-172 and a protein of the same size in HeLa nuclear extracts and in a P.3-derived TAF-172 fraction (Fig. 2B, lanes 1 to 3). The correspondence of the apparent molecular masses provided further evidence that the full-length TAF-172 gene had been cloned. TAF-172 antibodies also cross-reacted with a protein with a similar size in a highly purified B-TFIID fraction (lane 4) which was derived from a phosphocellulose fraction similar to that of TAF-172 and contains TAFII170 (39–41). This further confirms that these two TAFs are equivalent.
Endogenous expression and limited coassociation of TBP and TAF-172 in HeLa cells.
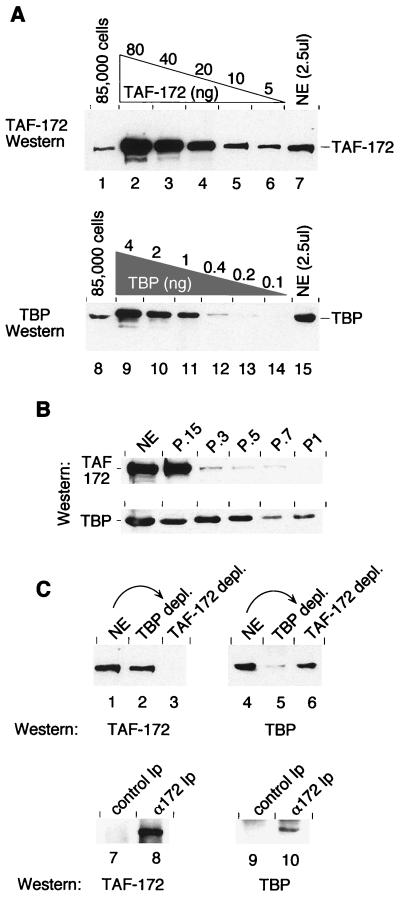
To quantitate the steady-state expression level of TAF-172 and TBP in vivo, HeLa cells were solubilized in SDS protein sample buffer, and their TAF-172 and TBP content was analyzed by Western blotting. Signals were compared against known concentrations of recombinant TAF-172 or TBP (Fig. 3A). TAF-172 appeared to be relatively abundant in HeLa cells, at an estimated 170,000 molecules per cell. TBP was estimated to be present at 200,000 molecules per cell.
FIG. 3.
Endogenous expression and coassociation of TAF-172 and TBP. (A) HeLa cells (85,000) were solubilized in protein sample buffer and electrophoresed on an SDS–6% polyacrylamide gel (lane 1) along with decreasing amounts of recombinant TAF-172 (80, 40, 20, 10, and 5 ng; lanes 2 to 6) and HeLa nuclear extract (NE; 50 μg; lane 7). Proteins were electroblotted to nitrocellulose and probed with affinity-purified TAF-172 antibodies. Equivalent amounts of HeLa cells and nuclear extracts were also electrophoresed on an SDS–7.8% polyacrylamide gel (lanes 8 and 15) along with recombinant TBP (4, 2, 1, 0.4, 0.2, and 0.1 ng; lanes 9 to 14), electroblotted, and probed with affinity-purified TBP antibodies. (B) HeLa nuclear extracts were chromatographed over phosphocellulose in H.15 buffer and serially step eluted with buffer containing 0.3, 0.5, 0.7, and 1.0 M KCl, as indicated. Equal portions of these fractions were separated on SDS–6% or–7.8% polyacrylamide gels and probed by Western blotting for TAF-172 or TBP, respectively, with affinity-purified antibodies. (C) HeLa nuclear extracts (50 μg; lanes 1 and 4), or nuclear extracts immunodepleted (depl.) of TBP (lanes 2 and 5) or TAF-172 (lanes 3 and 6) were separated on SDS–6% or–7.8% polyacrylamide gels and probed by Western blotting as indicated. Immunoprecipitates (Ip) from these depletions were eluted and treated similarly (lanes 7 to 10). The amount of immunoprecipitate used in the TBP Western blot (lanes 9 and 10) was 10 times that used for the TAF-172 Western blot (lanes 7 and 8).
We next examined the potential coassociation of TBP and TAF-172. HeLa nuclear extracts were fractionated over phosphocellulose. Five fractions were generated (P.15 or flowthrough, P.3, P.5, P.7, and P1), corresponding to the KCl concentration used in the serial step elutions. Most of the TAF-172 flowed through the phosphocellulose column (Fig. 3B), while most of the TBP remained bound, suggesting that most of the TAF-172 is not stably associated with TBP, including TFIID. The TBP that flowed through the phosphocellulose may or may not be bound to TAF-172.
The potential coassociation of TBP and TAF-172 was also examined by immunoprecipitation. Nuclear extracts were immunodepleted with affinity-purified antibodies against either TBP or TAF-172. As shown in Fig. 3C, TAF-172 antibodies depleted TAF-172 (lane 3) but had little effect on the level of TBP (lane 6). Conversely, TBP antibodies depleted TBP (lane 5) but had little effect on the level of TAF-172 (lane 2). Our previous immunoprecipitation studies with HeLa TAF-172 and TBP derived from a P.3 fraction indicate that these TBP antibodies are capable of coimmunoprecipitating stoichiometric amounts of TAF-172, which indicates that these antibodies do not disrupt stable TBP–TAF-172 interactions (37; also see Fig. 5C). Thus, it appears that the bulk of TAF-172 and TBP is not stably associated in HeLa nuclear extracts.
When the TAF-172 immunoprecipitate was examined, it contained both TAF-172 (lane 8) and small amounts of TBP (lane 9). Corroborating results were obtained with TBP immunoprecipitates (data not shown). TAFII250, which represents a marker for TFIID, was not detected in the immunoprecipitates (data not shown), indicating that TAF-172 is not stably associated with TFIID. Similar conclusions were drawn from Mot1 and yeast TFIID studies (28). Thus, although HeLa cells contain nearly as much TAF-172 as TBP, very little of the two appears to be coassociated. This low association is not due to low intrinsic stability of the complex because TBP–TAF-172 complexes immunopurified from HeLa cell extracts are stable when repeatedly washed with 1 M KCl and are dissociated only in the presence of GuHCl (37).
Association of recombinant TAF-172 with recombinant TBP.
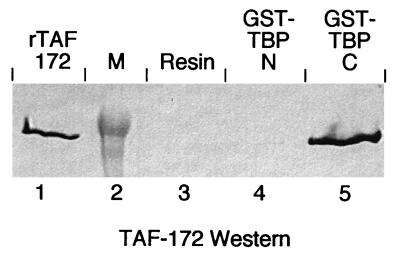
To assess whether recombinant TAF-172 binds TBP directly and to determine whether the conserved carboxyl-terminal domain of TBP is sufficient for binding, glutathione S-transferase (GST) fusion proteins containing either the conserved carboxyl-terminal domain (GST-TBP-C) or the nonconserved amino-terminal domain (GST-TBP-N) of human TBP were incubated with TAF-172. The GST fusions were then immobilized on glutathione agarose and assayed for TAF-172 retention by SDS-polyacrylamide gel electrophoresis, followed by Western blotting (Fig. 4). TAF-172 was retained in the presence of GST-TBP-C but not on resin alone or on resin containing GST-TBP-N. TAF-172 was also bound by full-length TBP (data not shown). These results demonstrate that TAF-172 binds directly to the conserved core domain of TBP and corroborates the coimmunoprecipitation data obtained with isolated HeLa TAF-172 (37). While it appears that TAF-172 does associate directly with TBP, binding was detected only after prolonged (∼3 h) incubation of the two proteins. At the concentration of proteins employed, diffusion-limited interactions, in general, proceed within seconds. This suggests that, at least in the absence of DNA, stable association of TBP and TAF-172 is, in some way, kinetically limited.
FIG. 4.
Recombinant TAF-172 binds to TBP. TAF-172 (10 μg) was incubated alone (lane 3) or with 2 μg of GST fusions containing either amino-terminal residues 1 to 163 (lane 4, GST-TBP-N) or carboxyl-terminal residues 168 to 339 (lane 5, GST-TBP-C) and glutathione agarose as described in Materials and Methods. The resin was washed, and eluted proteins were subjected to SDS–6% polyacrylamide gel electrophoresis and analyzed for TAF-172 by Western blotting. Recombinant TAF-172 (50 ng) is present in lane 1 and represents 0.5% of the input. Molecular mass markers (M) are in lane 2.
Dissociation of TBP-DNA complexes by TAF-172 and ATP.
The yeast Mot1 protein uses the energy of ATP hydrolysis to dissociate TBP from DNA (ADI activity) (2, 3). To determine whether TAF-172 possesses ADI activity, radiolabeled TBP-TATA DNA complexes were incubated with TAF-172 in the absence or presence of ATP and binding was examined by EMSA. TBP alone shifted the TATA probe, and ATP did not affect this interaction (Fig. 5A, lanes 1, 2, and 11). TAF-172 did not stably bind to the probe in the presence or absence of ATP (lanes 3 and 4). When TAF-172 was incubated with human TBP-TATA complexes, a slower-migrating species was observed (172-T-D in lanes 5, 7, and 9). In the presence of ATP, this complex disappeared, and the T-D complex reappeared (lanes 6, 8, and 10). Increasing TAF-172 concentrations resulted in less reappearance of the T-D complex, and at the highest concentration, very little was detected, which verifies that TAF-172 possesses ADI activity. The re-emergence of the T-D complex at the lower TAF-172 concentrations contrasts with yeast Mot1 (lanes 12 to 15) and indicates that either some of the TAF-172 dissociated from the T-D complex before it induced TBP to dissociate from DNA or that free TBP rebound to the probe.
To distinguish between these alternatives, competitor DNA was added to the reaction mixture at the same time as ATP. As shown in Fig. 5B, similar levels of the T-D complex were observed in the presence of ATP and TAF-172, irrespective of the presence of competitor DNA (compare lanes 3 and 4 with 5 and 6). When the competitor DNA was added to the reaction mixture at the same time as the DNA probe, it effectively competed for TBP binding, which indicates that sufficient competitor was present to sequester any free TBP. These data suggest that free TBP did not rebind the probe and are consistent with the interpretation that ATP hydrolysis also induces the dissociation of TAF-172 from the T-D complex.
Figure 5C verifies the composition of the 172-T-D complex. Lanes 1 and 2 recapitulate the ADI activity on a longer probe. Affinity-purified antibodies directed against either TBP or TAF-172 supershifted the 172-T-D complex (lanes 3 and 4), but control antibodies against an unrelated protein (TAFII250) did not (lane 5). Similar 172-T-D complexes were obtained with HeLa-derived TAF-172 (data not shown). When compared side by side in a variety of biochemical assays, HeLa-derived and recombinant TAF-172 behaved indistinguishably (data not shown).
TAF-172 is a TBP- and DNA-stimulated ATPase.
The isolated Mot1 ATPase domain hydrolyzes ATP in the absence of TBP and DNA cofactors at a rate of ∼25 min−1 (3). In the presence of TBP, the ATPase activity of the full-length protein is stimulated approximately fivefold (4). In the presence of saturating ATP concentrations, recombinant TAF-172 possessed little or no detectable ATPase activity in the absence of cofactors or in the presence of either TBP or a 361-bp TATA-containing DNA fragment (Fig. 6A). Strikingly, in the presence of both TBP and DNA, TAF-172 possessed potent ATPase activity, having a turnover value of ∼10 min−1. Thus, in apparent contrast to Mot1, the TAF-172 ATPase is activated synergistically by the combined action of TBP and DNA.
TBP has a fairly high affinity for nonspecific DNA (Kd, ∼300 nM) (6). At the concentration of TBP (1 μM) used in the ATPase experiments, TBP readily bound nonspecific DNA. In accordance with this, we found that the TAF-172 ATPase was activated by TBP and a DNA cofactor that lacks a TATA box (data not shown).
TAF-172 binds DNA very weakly in the absence of TBP (unpublished data). It is plausible that DNA alone can activate the TAF-172 ATPase but at a level below the sensitivity of the assay. While the use of a saturating ATP concentration (0.4 mM) ensured that ATP was not limiting the activity of the ATPase, it precluded accurate determinations of very low levels of ATP hydrolysis. To assess whether DNA alone might activate the TAF-172 ATPase, the ATP concentration was reduced 100-fold, while the level of [α-32P]ATP remained constant. This, in effect, increased the sensitivity of the assay 100-fold. As shown in Fig. 6B, TAF-172 alone possessed very weak ATPase activity (apparent turnover, ∼0.025 min−1), which might reflect its intrinsic activity or a minor amount of an ATPase contaminant in the preparation. However, addition of DNA to TAF-172 activated the ATPase approximately fivefold, indicating that DNA alone can activate the TAF-172 ATPase. This ATPase activity is nevertheless only 1% of that observed in the presence of both TBP and DNA.
TAF-172 inhibits TBP-driven, but not TBP-TAF-driven, in vitro transcription.
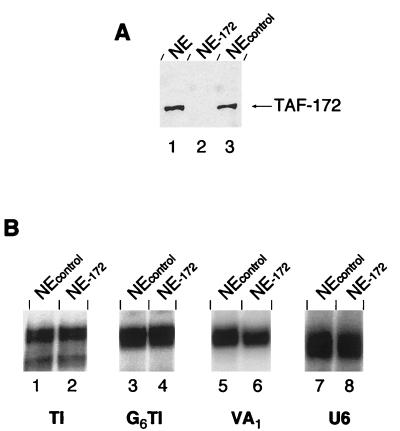
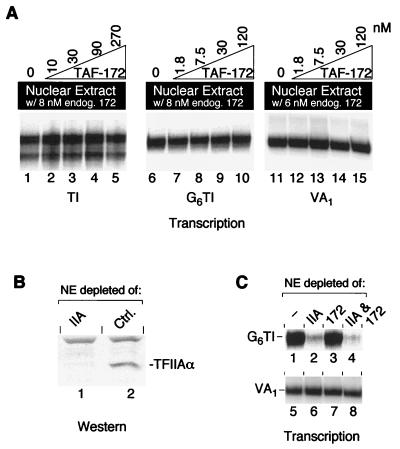
Mot1 is an inhibitor of yeast basal pol II transcription (2, 3). Initially, TAF-172 was identified as the major polypeptide present in a TAF fraction that was immunopurified with TBP antibodies and eluted with GuHCl (37). The TAF-172 fraction, in conjunction with TBP and another TAF fraction, reconstituted TFIIIB activity. The TAF-172 fraction also inhibited pol II transcription. To determine whether TAF-172 is a regulator of human pol II transcription and/or a pol III transcription factor, TAF-172 antibodies were used to immunodeplete TAF-172 from HeLa nuclear extracts. Western blotting confirmed the quantitative depletion of TAF-172 relative to control depletions (Fig. 7A).
FIG. 7.
Depletion of endogenous TAF-172 from pol II and pol III transcription reaction mixtures. (A) HeLa nuclear extract (NE; 120 μg, lane 1) and extracts immunodepleted with TAF-172 serum (lane 2) or an unrelated control serum (lane 3) were electrophoresed on an SDS–6% polyacrylamide gel, electroblotted to nitrocellulose, and probed for TAF-172. (B) TAF-172-depleted or control depleted nuclear extracts were used to transcribe 5 nM TI (lanes 1 and 2), G6TI (lanes 3 and 4), VA1 (lanes 5 and 6), or U6 (lanes 7 and 8) DNA as described in Materials and Methods. The time of exposure to the phosphor screen was varied to obtain similar signal intensities among the different promoters.
For pol II transcription, we used the synthetic basal TI promoter, which contains the adenovirus major late TATA box and the TdT initiator (32, 36). TI lacks activator binding sites. Since endogenous TAF-172 is present in nuclear extracts at concentrations similar to those of TBP, we expect depletion of TAF-172 to give rise to a greater level of transcription if, indeed, endogenous TAF-172 acts as a general transcriptional repressor. However, as shown in Fig. 7B (lanes 1 and 2), depletion of TAF-172 had no detectable effect on pol II basal transcription. Similarly, depletion of TAF-172 did not affect activated transcription of the Sp1-responsive pol II-transcribed G6TI promoter (lanes 3 and 4), the TATA-less, pol III-transcribed VA1 gene (lanes 5 and 6), or the TATA-containing pol III-transcribed U6 gene. Each is recognized by a distinct TBP-TAF complex: TFIID, TFIIIB, or SNAPc, respectively. Thus, TAF-172 does not appear to be a functional in vitro component of any of these TBP-TAF complexes.
Yeast TFIIA is inhibitory to Mot1 ADI activity, through competitive interactions with yeast TBP (2). Similar results were obtained with TAF-172, human TBP, and human TFIIA (unpublished data). Perhaps the TFIIA that is present in HeLa nuclear extracts counteracts the repressing activity of TAF-172. We explored this possibility in two ways. First, in an effort to outcompete any potential TAF-172 inhibitor, recombinant TAF-172 was titrated into a standard nuclear extract, which was then assayed for pol II and pol III transcription. As shown in Fig. 8A, addition of increasing amounts of TAF-172 had little effect on transcription from the TI (lanes 1 to 5), G6TI (lanes 6 to 10), or VA1 (lanes 11 to 15) promoter. The highest amount of recombinant TAF-172 added corresponds to approximately a 15- to 40-fold molar excess over the amount of resident TBP or TAF-172. If such a putative TAF-172 inhibitor was present and acting stoichiometrically, it would have to be present at extremely high levels. Western blotting data indicate that there is less TFIIA in nuclear extracts than TBP or TAF-172 (data not shown).
FIG. 8.
TAF-172 does not inhibit TBP-TAF-promoted in vitro transcription in the presence or absence of TFIIA. (A) HeLa nuclear extracts (100 μg in lanes 1 to 10 and 80 μg in lanes 11 to 15) containing endogenous TAF-172 (endog. 172) as indicated, were used to transcribe 5 nM TI (lanes 1 to 5), G6TI (lanes 6 to 10), or VA1 (lanes 11 to 15) DNA in the presence of increasing concentrations of recombinant TAF-172, as indicated. Time of exposure to the phosphor screen was varied to obtain similar signal intensities among the different promoters. (B) HeLa nuclear extracts (120 μg) immunodepleted with TFIIA serum (lane 1) or an unrelated control (ctrl.) serum (lane 2) were electrophoresed on an SDS–6% polyacrylamide gel, electroblotted to nitrocellulose, and probed for the α subunit of TFIIA. (C) HeLa nuclear extracts (100 μg) depleted of TFIIA, TAF-172, or TFIIA and TAF-172 were used to transcribe 5 nM G6TI (lanes 1 to 4) or VA1 (lanes 5 to 8) DNA.
The second strategy used to address whether TFIIA might counteract TAF-172 involved immunodepleting TFIIA from nuclear extracts (Fig. 8B). As expected of an important pol II transcription factor, G6TI transcription was diminished upon TFIIA depletion relative to mock-depleted or TAF-172-depleted reactions (Fig. 8C, lanes 1 to 3). In control experiments, pol III VA1 transcription was unaffected by depletion of either TAF-172 or TFIIA (lanes 5 to 8). If the function of TFIIA is to counteract transcriptional repression by TAF-172, then we would expect that depletion of both TFIIA and TAF-172 would lead to restoration of transcription. However, as shown in lane 4, this is not the case. Transcription of G6TI was no stronger than in the TFIIA-depleted reaction (lane 2), which indicated that, at least in this TFIID-based system, TFIIA does not function solely to counteract any potential TAF-172 repression.
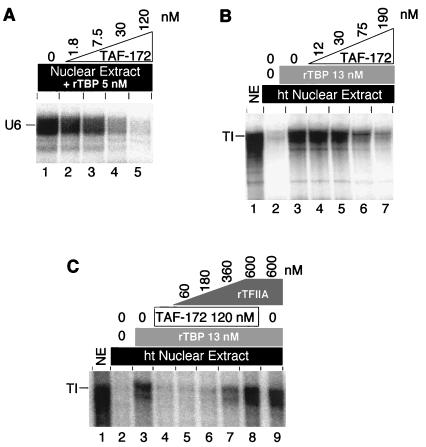
The description of yeast Mot1 as an ADI of pol II transcription was initially based upon an assay that utilized recombinant yeast TBP instead of the TFIID TBP-TAF complex (2). Perhaps TAF-172/Mot1 targets primarily TBP and not TBP-TAF complexes. The pol III-transcribed U6 gene is transcribed poorly in HeLa nuclear extracts in the absence of added TBP (34, 35). To examine whether TAF-172 might target TBP in the U6 transcription system, TAF-172 was titrated into HeLa nuclear extracts reconstituted with recombinant TBP and assayed for transcription of the U6 gene. As shown in Fig. 9A, addition of TAF-172 had a dramatic inhibitory effect on U6 transcription, which suggests that TAF-172 might target primarily TAF-free TBP.
FIG. 9.
Inhibition of TBP-promoted transcription by TAF-172. (A) HeLa nuclear extracts (100 μg) were used to transcribe 5 nM U6 DNA (lane 1) in the presence of increasing concentrations of TAF-172, as indicated. All of the reaction mixtures contained 5 nM pure recombinant human TBP (rTBP) in addition to endogenous TBP-TAF complexes. (B) HeLa nuclear extracts (NE; 60 μg) were mock treated (lane 1) or heat treated (ht; lanes 2 to 7) at 47°C for 15 min to selectively inactivate endogenous TBP (25) and used to transcribe 5 nM TI promoter DNA in the absence (lane 2) or presence (lanes 3 to 7) of pure recombinant human TBP. TBP, TI DNA, ATP (10 μM), and increasing concentrations of TAF-172, as indicated, were preincubated at 30°C for 30 min prior to the addition of nuclear extract. (C) Reactions identical to those depicted in panel B, except that increasing concentrations of pure recombinant human TFIIA (rTFIIA) were included in the reaction mixtures shown in lanes 5 to 9, as indicated.
To further test this possibility, the endogenous TBP which may be largely complexed with TAFs in HeLa nuclear extracts was specifically inactivated by heat and replaced with recombinant TBP (25). As shown in Fig. 9B, TI transcription was not reconstituted in such heat-treated extracts (lane 2) unless recombinant TBP (lane 3) was added back. When TAF-172 was titrated into the reaction, it too inhibited transcription (lanes 4 to 7). Taken together, the data suggest that TAF-172 inhibits both pol II and pol III transcription primarily through interactions with TBP and not TBP-TAF complexes. However, we cannot rule out the possibility that TAF-172 is targeted to TBP-TAF complexes through promoter-specific factors.
In yeast transcription reactions reconstituted with yeast TBP, yeast TFIIA reverses Mot1-mediated transcriptional inhibition (2). In Fig. 9C, we address whether the same is true in the human system. In transcription reactions reconstituted with human TBP and repressed by TAF-172 (lanes 1 to 4), increasing concentrations of recombinant human TFIIA reversed TAF-172-mediated repression (lanes 5 to 8). This observation is consistent with the notion that TFIIA functions, in part, by assembling TBP into an active promoter complex that resists incorporation of the TAF-172/Mot1 inhibitor (2).
DISCUSSION
TAF-172 is a human homolog of yeast Mot1.
TAF-172 is a human TBP-associated factor and a member of the Snf2/Swi2 family of conserved DNA-dependent ATPases. TAF-172 bears strong sequence similarity to the yeast Mot1 protein, and this similarity is punctuated over its entire coding sequence. Both Mot1 and TAF-172 bear strong similarity to the Drosophila 89B helicase, although only what appears to be the carboxyl-terminal half of the 89B helicase has been cloned (14). It is important to note that the 89B helicase, Mot1, and TAF-172 have not been demonstrated to be helicases. Together, these three proteins make up a distinct Mot1 subgroup within the Snf2-Swi2 family.
TAF-172 and Mot1 share several distinct blocks of homology. Based upon studies with Mot1, the most N-terminal block might interact with TBP (4). TAF-172 does not possess any motifs that are detectable by on-line algorithms. However, Mot1 has been reported to contain TPR (8) which span the first two N-terminal homology blocks, as well as the nonhomology region that separates them (Fig. 1). A number of TPR-like sequences are found at corresponding positions in TAF-172, but the loose TPR consensus and the proposed presence of TPR in nonhomology regions preclude a firm conclusion on whether such a TPR structural domain exists in this region of either protein. The C-terminal half of TAF-172 and Mot1 contains the ATPase and helicase-like domains. These domains reside in a region that has the highest degree of conservation (70%) and is the only region possessing significant homology to other Snf2 family members (∼60% conservation). TAF-172, the 89B helicase, and Mot1 share extensive sequence homology on either side of the ATPase helicase domain that is specific to the Mot1 subdomain. The function of these regions is unknown. Consistent with the idea that these homology regions are important, deletions from either end of Mot1, as well as site-specific mutations in the Mot1 ATPase domain, impair its function (3, 4).
Relationship to B-TFIID.
B-TFIID is a human TBP-TAF complex that contains a 170-kDa TAF (39, 40). While this report was under review, a clone (TAFII170) that encodes this protein was reported (41), and it is identical to TAF-172. Unlike TAF-172–TBP complexes, B-TFIID appears to support basal pol II transcription in vitro. The basis of this difference might be the relative level of TFIIA in each system. TFIIA counteracts TAF-172/Mot1 transcriptional repression in vitro through mutual competition for TBP binding (this study and reference 2).
TAF-172 is relatively abundant and appears to target primarily TAF-free TBP.
TAF-172 is about as abundant in HeLa cells as is TBP (∼200,000 molecules). However, the majorities of TAF-172 and TBP do not appear to be stably associated with each other. This lack of association is not due to a low mutual affinity because the small amount that is associated is very resistant to repeated high-salt washes, which allowed us to immunopurify the complex >10,000-fold (37). Resin pull-down experiments between purified recombinant TBP and recombinant TAF-172 demonstrate a direct interaction between TAF-172 and the evolutionarily conserved C-terminal domain of TBP. Immunoprecipitation and column chromatography purification indicate that TAF-172 is not stably associated with TFIID, TFIIIB, or SNAPc. Consistent with this, Mot1 does not appear to be tightly associated with yeast TFIID (28).
TAF-172 does not appear to inhibit TBP-TAF-driven transcription but potently inhibits TBP-driven pol II and pol III transcription. Likewise, Mot1 inhibits yeast transcription in vitro driven by yeast TBP. Mot1 has not been tested for activity on yeast TFIID. The findings obtained with TAF-172 suggest that it targets primarily TAF-free TBP, and not TFIID, in this system. We cannot exclude the possibility that promoter-specific factors or factors missing from crude nuclear extracts target TAF-172 to TBP-TAF complexes.
The large abundance of TBP-free TAF-172 is puzzling. It is possible that TAF-172 possesses other cellular functions in addition to TBP regulation. Alternativley, the abundance of TAF-172 might be necessary to ensure a very low basal level of TBP bound randomly to chromosomal DNA, particularly since high concentrations of TAF-172 are required for maximal ATP-dependent dissociation of TBP from DNA. TBP has a relatively high affinity for nonspecific DNA (6). Once bound to nonspecific DNA, TBP can coalesce the assembly of active pol II transcription complexes in vitro (6). This is clearly not desirable in the cell, and the high levels of TAF-172 might serve to minimize this nonspecific binding. TBP-TAF complexes such as TFIID associate very poorly with DNA in the absence of TFIIA and promoter-specific activators (20, 43). Therefore, unlike TBP, TFIID is not likely to be promiscuously bound to nonpromoter DNA and thus is not a necessary target of TAF-172/Mot1.
Mechanism of action of TAF-172/Mot1 on TBP-TATA complexes.
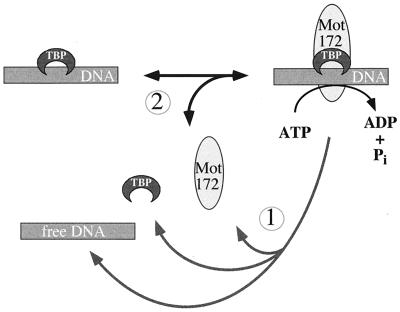
To recycle TBP-DNA complexes, TAF-172/Mot1 would be expected to possess two activities: (i) dissociation of TBP from DNA and (ii) dissociation of itself from TBP. For Mot1, this first step is ATP dependent (ADI activity) and has been well documented (2–4). At relatively low TAF-172 concentrations, ATP-dependent removal of TAF-172 from TBP appears to predominate. At higher concentrations, ADI activity predominates.
Figure 10 illustrates a simplified mechanism that might account for the action of the TAF-172/Mot1 ATPase on TBP-DNA complexes. TAF-172/Mot1 binds to TBP-DNA complexes. ATP hydrolysis leads to the dissociation of TAF-172/Mot1 and TBP from each other and from DNA (reaction 1). However, at least for TAF-172, ATP hydrolysis is not entirely coupled to reaction 1. An alternative nonproductive reaction occurs in which ATP hydrolysis leads to the dissociation of TAF-172, leaving an intact TBP-DNA complex (reaction 2). Repetitive rounds of association and dissociation of TAF-172 with the TBP-DNA complex are consistent with the high rate of ATP hydrolysis by TAF-172 only in the presence of this complex. Higher TAF-172 concentrations drive more frequent encounters with the TBP-DNA complex and provide more opportunity for TAF-172 to dissociate TBP (i.e., reaction 1). In the context of the cell, additional factors might exist which favor reaction 1 over reaction 2.
FIG. 10.
Possible ATPase cycle for TAF-172 and Mot1. ATP hydrolysis leads to either dissociation of TBP and TAF-172 from DNA (reaction 1) or just dissociation of TAF-172 (reaction 2). Note that the ATP-dependent dissociation of TAF-172 is not the reverse of the association reaction, in that ATP is not generated. Additional details are provided in the text.
Possible auxiliary factors.
A number of studies on Mot1 implicate the involvement of cofactors for Mot1 function. Mot1 does copurify with and appears to interact directly with both TBP and yeast TAFII90 (referred to as TAFII85 in reference 42). The human and Drosophila homologs of yeast TAFII90 are human TAFII100 and drosophila TAFII80, both of which are components of TFIID (10, 11, 21, 27, 33, 38). The Arabidopsis COP1 gene also encodes a homolog of these TAFs (11). Interestingly, the COP1 gene product appears to function as a developmental transcriptional repressor (9).
Yeast TAFII90 and its homologs contain β-transducin (WD40) repeats (11). Proteins with WD40 repeats tend to interact with proteins containing TPR motifs, which Mot1 reportedly contains. Such protein pairs appear to be generally involved in repression mechanisms. While drosophila TAFII80 and human TAFII100 interact with TBP and other TAF components of TFIID, these interactions do not require the WD40 repeats, suggesting that the WD40 repeats are available for interaction with other proteins (11, 21). It is plausible that TAF-172/Mot1 might interact with human TAFII100 or yeast TAFII90, thereby allowing it to associate with TFIID. The prevailing evidence from both the human and yeast systems, however, indicates that TFIID is not stably associated with TAF-172/Mot1.
The promoter-specific repressor Leu3p functions through a TBP-Mot1 complex (42), which implicates a direct cofactor involvement in Mot1-mediated repression. Since Mot1 does not repress all genes (8, 23), Mot1 might be targeted to specific promoters through direct interactions with sequence-specific factors. Genetic interactions between the yeast NOT proteins, Spt3, TFIIA, and Mot1 have been defined (7, 23). Spt3 also interacts with TBP and has homology to human TAFII18 (13, 24). Taken together, the interactions suggest that Mot1, and presumably TAF-172, normally functions in the context of numerous other proteins to regulate transcription through TBP.
ACKNOWLEDGMENTS
We thank W. Herr and R. Tjian for supplying the cDNA libraries used in the cloning, D. Reinberg for the P.3 used in the isolation of TAF-172, and R. Meyers and P. Sharp for supplying the B-TFIID fraction. We thank members of the Pugh and Auble laboratories for many fruitful discussions.
This work was supported by grants from the National Institutes of Health (GM47855 to B.F.P. and GM55763 to D.T.A.), the Searle Scholars Program/The Chicago Community Trust (B.F.P.), and the Leukemia Society of America (B.F.P.).
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Auble D T, Hahn S. An ATP-dependent inhibitor of TBP binding to DNA. Genes Dev. 1993;7:844–856. doi: 10.1101/gad.7.5.844. [DOI] [PubMed] [Google Scholar]
- 3.Auble D T, Hansen K E, Mueller C G, Lane W S, Thorner J, Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 4.Auble D T, Wang D, Post K W, Hahn S. Molecular analysis of the SNF2/SWI2 protein family member MOT1, an ATP-driven enzyme that dissociates TATA-binding protein from DNA. Mol Cell Biol. 1997;17:4842–4851. doi: 10.1128/mcb.17.8.4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson M, Laurent B C. The SNF/SWI family of global transcriptional activators. Curr Opin Cell Biol. 1994;6:396–402. doi: 10.1016/0955-0674(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 6.Coleman R A, Pugh B F. Evidence for functional binding and stable sliding of the TATA binding protein on nonspecific DNA. J Biol Chem. 1995;270:13850–13859. doi: 10.1074/jbc.270.23.13850. [DOI] [PubMed] [Google Scholar]
- 7.Collart M A. The NOT1, SPT3, and MOT1 genes functionally interact to regulate transcription at core promoters. Mol Cell Biol. 1996;16:6668–6676. doi: 10.1128/mcb.16.12.6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis J L, Kunisawa R, Thorner J. A presumptive helicase (MOT1 gene product) affects gene expression and is required for viability in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1879–1892. doi: 10.1128/mcb.12.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng X W, Matsui M, Wei N, Wagner D, Chu A M, Feldemann K A, Quail P H. COP1, an arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a Gβ homologous domain. Cell. 1992;71:791–801. doi: 10.1016/0092-8674(92)90555-q. [DOI] [PubMed] [Google Scholar]
- 10.Dubrovskaya V, Lavigne A C, Davidson I, Acker J, Staub A, Tora L. Distinct domains of hTAFII100 are required for functional interaction with transcription factor TFIIF beta (RAP30) and incorporation into the TFIID complex. EMBO J. 1996;15:3702–3712. [PMC free article] [PubMed] [Google Scholar]
- 11.Dynlacht B D, Weinzierl R O, Admon A, Tjian R. The dTAFII80 subunit of Drosophila TFIID contains β-transducin repeats. Nature. 1993;363:176–179. doi: 10.1038/363176a0. [DOI] [PubMed] [Google Scholar]
- 12.Eisen J A, Sweder K S, Hanawalt P C. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisenmann D M, Arndt K M, Ricupero S L, Rooney J W, Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 1992;6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- 14.Goldman-Levi R, Miller C, Bogoch J, Zak N B. Expanding the Mot1 subfamily: 89B helicase encodes a new Drosophila melanogaster SNF2-related protein which binds to multiple sites on polytene chromosomes. Nucleic Acids Res. 1996;24:3121–3128. doi: 10.1093/nar/24.16.3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagemeier C, Walker S, Caswell R, Kouzarides T, Sinclair J. The human cytomegalovirus 80-kilodalton but not the 72-kilodalton immediate-early protein transactivates heterologous promoters in a TATA box-dependent mechanism and interacts directly with TFIID. J Virol. 1992;66:4452–4456. doi: 10.1128/jvi.66.7.4452-4456.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henry R W, Sadowski C L, Kobayashi R, Hernandez N. A TBP-TAF complex required for transcription of human snRNA genes by RNA polymerase II and III. Nature. 1995;374:653–656. doi: 10.1038/374653a0. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 18.Jiang Y W, Stillman D J. Epigenetic effects on yeast transcription caused by mutations in an actin-related protein present in the nucleus. Genes Dev. 1996;10:604–619. doi: 10.1101/gad.10.5.604. [DOI] [PubMed] [Google Scholar]
- 19.Karnitz L, Morrison M, Young E T. Identification and characterization of three genes that affect expression of ADH2 in Saccharomyces cerevisiae. Genetics. 1992;132:351–359. doi: 10.1093/genetics/132.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi N, Boyer T G, Berk A J. A class of activation domains interacts directly with TFIIA and stimulates TFIIA-TFIID-promoter complex assembly. Mol Cell Biol. 1995;15:6465–6473. doi: 10.1128/mcb.15.11.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kokubo T, Gong D W, Yamashita S, Takada R, Roeder R G, Horikoshi M, Nakatani Y. Molecular cloning, expression, and characterization of the Drosophila 85-kilodalton TFIID subunit. Mol Cell Biol. 1993;13:7859–7863. doi: 10.1128/mcb.13.12.7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunkel G R, Maser R L, Calvet J P, Pederson T. U6 small nuclear RNA is transcribed by RNA polymerase III. Proc Natl Acad Sci USA. 1986;83:8575–8579. doi: 10.1073/pnas.83.22.8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madison J M, Winston F. Evidence that Spt3 functionally interacts with Mot1, TFIIA, and TATA-binding protein to confer promoter-specific transcriptional control in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:287–295. doi: 10.1128/mcb.17.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mengus G, May M, Jacq X, Staub A, Tora L, Chambon P, Davidson I. Cloning and characterization of hTAFII18, hTAFII20 and hTAFII28: three subunits of the human transcription factor TFIID. EMBO J. 1995;14:1520–1531. doi: 10.1002/j.1460-2075.1995.tb07138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakajima N, Horikoshi M, Roeder R G. Factors involved in specific transcription by mammalian RNA polymerase II: purification, genetic specificity, and TATA box-promoter interactions of TFIID. Mol Cell Biol. 1988;8:4028–4040. doi: 10.1128/mcb.8.10.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piatti S, Tazzi A, Pizzagalli P, Plevani P, Lucchini G. Control of DNA synthesis genes in budding yeast: involvement of the transcriptional modulator MOT1 in the expression of the DNA polymerase alpha gene. Chromosoma. 1992;102:S107–S113. doi: 10.1007/BF02451793. [DOI] [PubMed] [Google Scholar]
- 27.Poon D, Bai Y, Campbell A M, Bjorklund S, Kim Y J, Zhou S, Kornberg R D, Weil P A. Identification and characterization of a TFIID-like multiprotein complex from Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1995;92:8224–8228. doi: 10.1073/pnas.92.18.8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poon D, Campbell A M, Bai Y, Weil P A. Yeast Taf170 is encoded by MOT1 and exists in a TATA box-binding protein (TBP)–TBP-associated factor complex distinct from transcription factor IID. J Biol Chem. 1994;269:23135–23140. [PubMed] [Google Scholar]
- 29.Pugh B F. Mechanisms of transcription complex assembly. Curr Opin Cell Biol. 1996;8:303–311. doi: 10.1016/s0955-0674(96)80002-0. [DOI] [PubMed] [Google Scholar]
- 30.Pugh B F. Preparation of HeLa nuclear extracts. In: Tymms M J, editor. In vitro transcription and translation protocols. Vol. 37. Totowa, N.J: Humana Press, Inc.; 1995. pp. 349–358. [Google Scholar]
- 31.Pugh B F. Purification of the human TATA-binding protein, TBP. In: Tymms M J, editor. In vitro transcription and translation protocols. Vol. 37. Totowa, N.J: Humana Press, Inc.; 1995. pp. 359–367. [DOI] [PubMed] [Google Scholar]
- 32.Pugh B F, Tjian R. Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell. 1990;61:1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- 33.Reese J C, Apone L, Walker S S, Griffin L A, Green M R. Yeast TAFIIs in a multisubunit complex required for activated transcription. Nature. 1994;371:523–527. doi: 10.1038/371523a0. [DOI] [PubMed] [Google Scholar]
- 34.Sadowski C L, Henry R W, Lobo S M, Hernandez N. Targeting TBP to a non-TATA box cis-regulatory element: a TBP-containing complex activates transcription from snRNA promoters through the PSE. Genes Dev. 1993;7:1535–1548. doi: 10.1101/gad.7.8.1535. [DOI] [PubMed] [Google Scholar]
- 35.Simmen K A, Bernues J, Parry H D, Stunnenberg H G, Berkenstam A, Cavallini B, Egly J M, Mattaj I W. TFIID is required for in vitro transcription of the human U6 gene by RNA polymerase III. EMBO J. 1991;10:1853–1862. doi: 10.1002/j.1460-2075.1991.tb07711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smale S T, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 37.Taggart A K, Fisher T S, Pugh B F. The TATA-binding protein and associated factors are components of pol III transcription factor TFIIIB. Cell. 1992;71:1015–1028. doi: 10.1016/0092-8674(92)90396-t. [DOI] [PubMed] [Google Scholar]
- 38.Tanese N, Saluja D, Sun L, Vassallo M, Chen J L, Admon A. Molecular cloning and analysis of two subunits of the human TFIID complex: hTAFII130 and hTAFII100. Proc Natl Acad Sci USA. 1997;93:13611–13616. doi: 10.1073/pnas.93.24.13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Timmers H T, Meyers R E, Sharp P A. Composition of transcription factor B-TFIID. Proc Natl Acad Sci USA. 1992;89:8140–8144. doi: 10.1073/pnas.89.17.8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Timmers H T, Sharp P A. The mammalian TFIID protein is present in two functionally distinct complexes. Genes Dev. 1991;5:1946–1956. doi: 10.1101/gad.5.11.1946. [DOI] [PubMed] [Google Scholar]
- 41.van der Knaap J A, Borst J W, van der Vliet P C, Gentz R, Timmers H T M. Cloning of the cDNA for the TATA-binding protein-associated factorII170 subunit of transcription factor B-TFIID reveals homology to global transcription regulators in yeast and Drosophila. Proc Natl Acad Sci USA. 1997;94:11827–11832. doi: 10.1073/pnas.94.22.11827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wade P A, Jaehning J A. Transcriptional corepression in vitro: a Mot1p-associated form of TATA-binding protein is required for repression by Leu3p. Mol Cell Biol. 1996;16:1641–1648. doi: 10.1128/mcb.16.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang W, Gralla J D, Carey M. The acidic activator GAL4-AH can stimulate polymerase II transcription by promoting assembly of a closed complex requiring TFIID and TFIIA. Genes Dev. 1992;6:1716–1727. doi: 10.1101/gad.6.9.1716. [DOI] [PubMed] [Google Scholar]
- 44.Weideman C A, Netter R C, Benjamin L R, McAllister J J, Schmiedekamp L A, Coleman R A, Pugh B F. Dynamic interplay of TFIIA, TBP, and TATA DNA. J Mol Biol. 1997;271:61–75. doi: 10.1006/jmbi.1997.1152. [DOI] [PubMed] [Google Scholar]
- 45.White R J, Stott D, Rigby P W. Regulation of RNA polymerase III transcription in response to F9 embryonal carcinoma stem cell differentiation. Cell. 1989;59:1081–1092. doi: 10.1016/0092-8674(89)90764-2. [DOI] [PubMed] [Google Scholar]
- 46.Zawel L, Reinberg D. Common themes in assembly and function of eukaryotic transcription complexes. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]