Abstract
Messenger RNA (mRNA) vaccines were highly effective against the ancestral SARS-CoV-2 strain, but the efficacy of bivalent mRNA boosters against XBB variants was substantially lower. Here, we show limited durability of neutralizing antibody (NAb) responses against XBB variants and isotype switching to immunoglobulin G4 (IgG4) responses following bivalent mRNA boosting. Bivalent mRNA boosting elicited modest XBB.1-, XBB.1.5-, and XBB.1.16-specific NAbs that waned rapidly within 3 months. In contrast, bivalent mRNA boosting induced more robust and sustained NAbs against the ancestral WA1/2020 strain, suggesting immune imprinting. Following bivalent mRNA boosting, serum antibody responses were primarily IgG2 and IgG4 responses with poor Fc functional activity. In contrast, a third monovalent mRNA immunization boosted all isotypes including IgG1 and IgG3 with robust Fc functional activity. These data show substantial immune imprinting for the ancestral spike and isotype switching to IgG4 responses following bivalent mRNA boosting, with important implications for future booster designs and boosting strategies.
Bivalent COVID-19 mRNA boosting induced limited durability of NAbs and primarily IgG4 responses with poor functionality.
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causing the coronavirus disease 2019 (COVID-19) pandemic has continued to evolve rapidly. The messenger RNA (mRNA) vaccines have shown clinical efficacy against ancestral SARS-CoV-2 (1–4), and the bivalent mRNA vaccines with the ancestral WA1/2020 and BA.5 spike immunogens have shown robust neutralizing antibody (NAb) responses against WA1/2020 but lower NAb responses against BA.5 (5–8). SARS-CoV-2 has continued to evolve with recombinant lineages, such as XBB (Fig. 1) (5, 6). Two subvariants of XBB, XBB.1.5 and XBB.1.16 (fig. S1), and further mutated variants EG.5.1 and FL.1.5.1 are now global dominant variants. The ability of XBB.1.5 to evade NAb responses has been reported recently (6, 9–11). However, the durability and functional properties of XBB antibody responses induced by bivalent mRNA boosting remain to be determined.
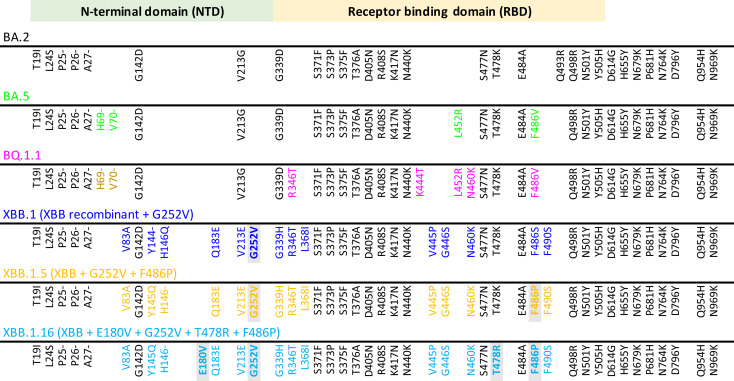
Fig. 1. Key spike mutations found in SARS-CoV-2 Omicron and XBB subvariants.
Spike amino acid substitutions in baseline sequences for BA.2, BA.5, BQ.1.1, XBB.1, XBB.1.5, and XBB.1.16 are depicted. XBB variants are recombinants between parental sequences BJ.1 (BA.2.10.1) and a sublineage of BA.2.75 (BM.1.1.1). BA.2 mutations compared with the ancestral WA1/2020 Spike are shown in black, and additional mutations relative to BA.2 are highlighted in colors corresponding to individual variants. Light gray shading indicates differences in prominent XBB lineages relative to the original recombinant founder. NTD, N-terminal domain; RBD, receptor binding domain.
Although NAbs are considered an important correlate of protection against SARS-CoV-2, the role of spike-specific T cells and other fragment crystallizable (Fc) functional antibodies in vaccine protection has received less attention. Immunoglobulin G1 (IgG1) and IgG3 responses have been widely considered to activate the immune system in the context of vaccination or infection via the classical C1q complement pathway and engagement of Fcγ receptors, and have been reported to correlate with survival following SARS-CoV-2 infection (12–14). In contrast, IgG2 does not interact substantially with the FcγRs or complement pathways (13, 15), and IgG4 interacts with the inhibitory FcγRIIB receptor (16). Both IgG2 and IgG4 have been shown to potentiate noninflammatory functions due to low Fc effector activity (17–19), with IgG4 described as a “blocking” antibody due to its ability to block excess inflammation (18, 19). Studies have shown that COVID-19 mRNA vaccines and adenoviral-vectored vaccines induce preferentially IgG1 and IgG3, with limited IgG2 and IgG4 in both humans and nonhuman primates (20–22). However, recent longitudinal follow-up studies after a second or third monovalent mRNA vaccination showed evidence of some class switching toward IgG4 (23, 24). Further evolution of IgG subclasses after a fourth mRNA vaccination remains unknown.
In this study, we characterized the magnitude, cross-reactivity, and durability of immune responses following bivalent mRNA boosting against both ancestral and XBB strains. We also evaluated the antibody isotypes and Fc functional responses after a fourth bivalent mRNA boost and a third monovalent mRNA boost.
RESULTS
Durability of humoral immune responses following bivalent mRNA boosting
We and others have recently shown that the bivalent mRNA boosters increase NAb titers to multiple SARS-CoV-2 variants (5–8, 25), but the durability of these NAb responses to the XBB subvariants remains to be determined. XBB.1.5 is a derivative of XBB.1 with the F486P mutation, and XBB.1.16 has the E180V, T478R, and F486P mutations (Fig. 1). To assess the durability of the humoral immune responses following bivalent mRNA boosting against both the ancestral and XBB variants, we determined NAb responses using a luciferase-based pseudovirus NAb assay (6, 8). Our clinical cohort included 30 participants who received the bivalent mRNA boosters, and we performed NAb assays at baseline before boosting, at week 3 after boost, and at month 3 after boost (tables S1 and S2). Between baseline and month 3, two of the study participants had SARS-CoV-2 infection, whereas 43% of study participants had a known COVID-19 infection or were nucleocapsid seropositive since the beginning of the pandemic (tables S1 and S2). We speculate that this represents an underestimate of the true rate of infection due to clinically undiagnosed and asymptomatic infections.
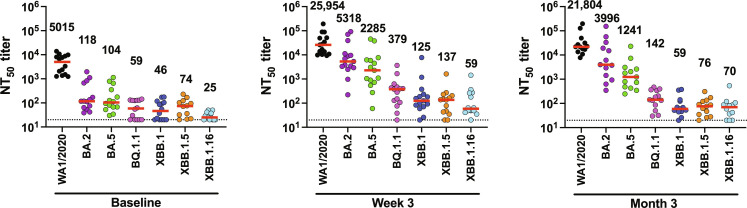
At baseline, median NAb titers to WA1/2020, BA.2, BA.5, BQ.1.1, XBB.1, XBB.1.5, and XBB.1.16 were 5015, 118, 104, 59, 46, 74, and 25, respectively, in nucleocapsid seronegative participants (Fig. 2 and fig. S2). At week 3, median NAb titers to WA1/2020, BA.2, BA.5, BQ.1.1, XBB.1, XBB.1.5, and XBB.1.16 increased to 25,954, 5318, 2285, 379, 125, 137, and 59, respectively (Fig. 2 and fig. S2), which represents a 5.2-, 45.0-, 21.9-, 6.4-, 2.7-, 1.8-, and 2.3-fold increment in median NAb titers in the respective groups. At month 3, median NAb titers to WA1/2020, BA.2, BA.5, BQ.1.1, XBB.1, XBB.1.5, and XBB.1.16 were 21,804, 3996, 1241, 142, 59, 76, and 70, respectively (Fig. 2 and fig. S2). These data suggest that bivalent mRNA boosting elicited high NAb titers against the ancestral WA1/2020 strain, moderate NAb titers against BA.5, and low NAb titers against the XBB subvariants. Moreover, whereas durability appeared relatively robust for WA1/2020 at month 3, NAb responses fell to preboost levels against XBB.1 and XBB.1.5 by month 3, suggesting more limited durability against the XBB subvariants.
Fig. 2. NAb responses to SARS-CoV-2 Omicron and XBB subvariants.
NAb titers against the WA1/2020, BA.2, BA.5, BQ.1.1, XBB.1, XBB.1.5, and XBB.1.16 variants by luciferase-based pseudovirus neutralization assays at baseline before boosting, at week 3 following boosting, and at month 3 following boosting in nucleocapsid seronegative participants are shown. Median NAb titers (red bars) are depicted and shown numerically. LOD indicated by dotted lines represents the starting dilution used for the NAb assay, 1:20.
Minimal cellular immune responses following bivalent mRNA boosting
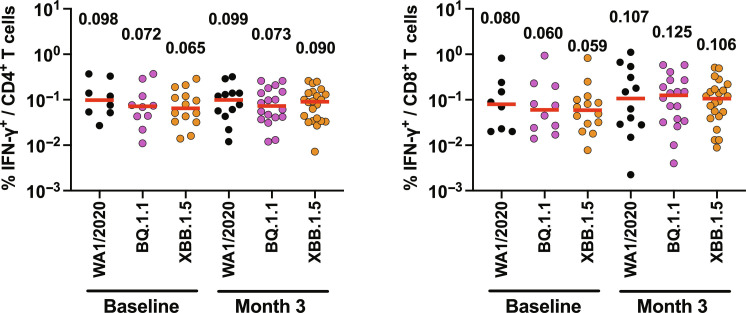
We next assessed spike-specific interferon-γ–positive (IFN-γ+) CD4+ and CD8+ T cell responses in peripheral blood mononuclear cells (PBMCs) by intracellular cytokine staining assays. Median spike-specific IFN-γ+ CD4+ T cell responses to WA1/2020, BQ.1.1, and XBB.1.5 were 0.098, 0.072, and 0.065% at baseline before boosting, and 0.099, 0.073, and 0.090% at month 3 after boost, respectively (Fig. 3). Median spike-specific IFN-γ+ CD8+ T cell responses to WA1/2020, BQ.1.1, and XBB.1.5 were 0.080, 0.060, and 0.059% at baseline before boost, and 0.107, 0.125, and 0.106% at month 3 after boost, respectively (Fig. 3). Although T cell cross-reactivity was robust, including against XBB.1.5, bivalent mRNA boosting only minimally augmented CD4+ and CD8+ T cell responses.
Fig. 3. Cellular immune responses to SARS-CoV-2 Omicron and XBB variants.
IFN-γ+ CD4+ and IFN-γ+ CD8+ T cell responses in PBMCs from vaccinees to WA1/2020, BQ.1.1, and XBB.1.5 spike-specific peptide pools by intracellular cytokine staining assays at baseline before boosting, and at month 3 following boosting are shown. Medians (red bars) are depicted and shown numerically.
IgG subclass and functional responses following bivalent mRNA boosting
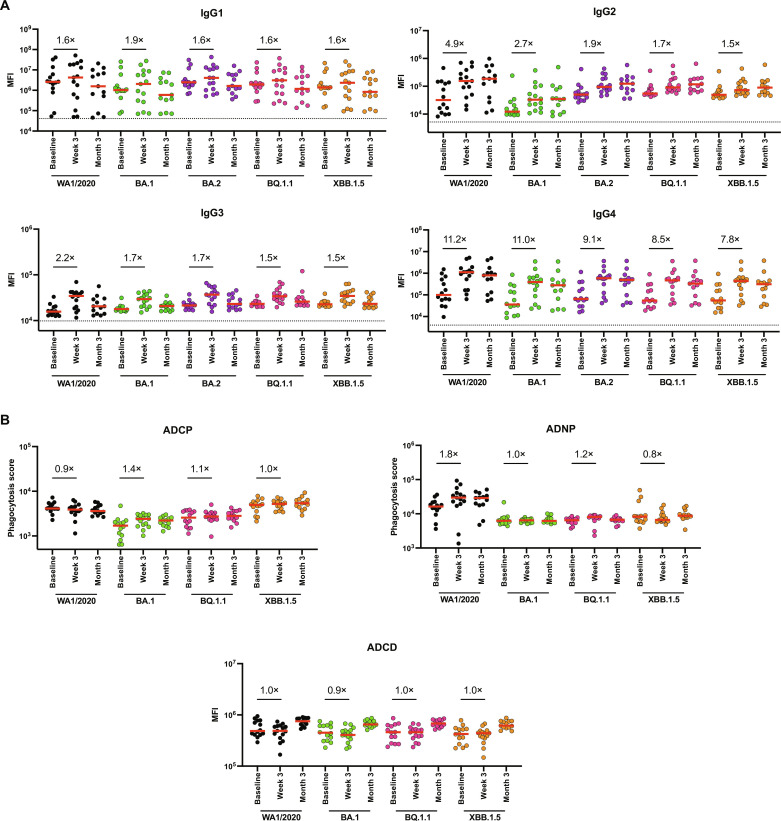
To explore in greater detail spike-specific antibody responses following bivalent mRNA boosting, we assessed IgG subclass responses (IgG1, IgG2, IgG3, and IgG4) against WA1/2020, BA.1, BA.2, BQ.1.1, and XBB.1.5 following bivalent mRNA boosting. Less than twofold increases in IgG1 responses were observed to WA1/2020, BA.1, BA.2, BQ.1.1, and XBB.1.5 at week 3 and returned to preboost levels by month 3 (Fig. 4A). A similar trend was observed with IgG3 responses. Slightly higher 4.9-, 2.7-, 1.9-, 1.7-, and 1.5-fold increases in IgG2 responses were seen to WA1/2020, BA.1, BA.2, BQ.1.1, and XBB.1.5 at week 3 (Fig. 4A). In contrast, markedly higher 11.2-, 11.0-, 9.1-, 8.5-, and 7.8-fold increases in IgG4 responses were observed to WA1/2020, BA.1, BA.2, BQ.1.1, and XBB.1.5 at week 3, and these responses were durable at month 3 (Fig. 4A). These results suggested that bivalent mRNA boosting did not substantially increase proinflammatory IgG1 and IgG3 responses but rather skewed responses primarily to isotype-switched IgG4 responses.
Fig. 4. Antibody and Fc effector function profiling by systems serology following bivalent mRNA immunization.
(A) Spike-specific IgG1, IgG2, IgG3, and IgG4 responses against WA1/2020, BA.1, BA.2, BQ.1.1, and XBB.1.5 spike antigens by Luminex-based assay at baseline before boosting, week 3 following boosting, and at month 3 following boosting in nucleocapsid seronegative participants. (B) Spike-specific ADCP, ADNP, and ADCD activity against WA1/2020, BA.1, BQ.1.1, and XBB.1.5 at baseline before boosting, week 3 after boost, and at month 3 after boost is shown. Medians of the mean fluorescence intensities (MFI) in red bars are depicted, and fold changes are shown numerically. Phagocytosis score for ADCP and ADNP activity is calculated as follows: phagocytosis score = % THP-1 bead-positive cells × median fluorescent intensity of bead-positive cells/10,000. LOD indicated by dotted lines represent the median MFI of an Ebolavirus glycoprotein negative control.
Fc effector functions of antibodies have been shown to contribute to protection against SARS-CoV-2 (26–28). We therefore assessed antibody-dependent cellular phagocytosis (ADCP), antibody-dependent neutrophil phagocytosis (ADNP), antibody-dependent complement deposition (ADCD), and antibody-dependent natural killer cell activation (ADNKA) responses. Consistent with the largely negative IgG1 and IgG3 responses, bivalent mRNA boosting led to minimal or no enhanced ADCP, ADNP, and ADCD responses to WA1/2020, BA.1, BQ.1.1, and XBB.1.5 and only modest increases in ADNKA responses (Fig. 4B and fig. S3). These data show that the antibody responses following bivalent mRNA boosting were primarily IgG4 responses with limited Fc functional activity.
IgG subclass and functional responses following third monovalent mRNA boosting
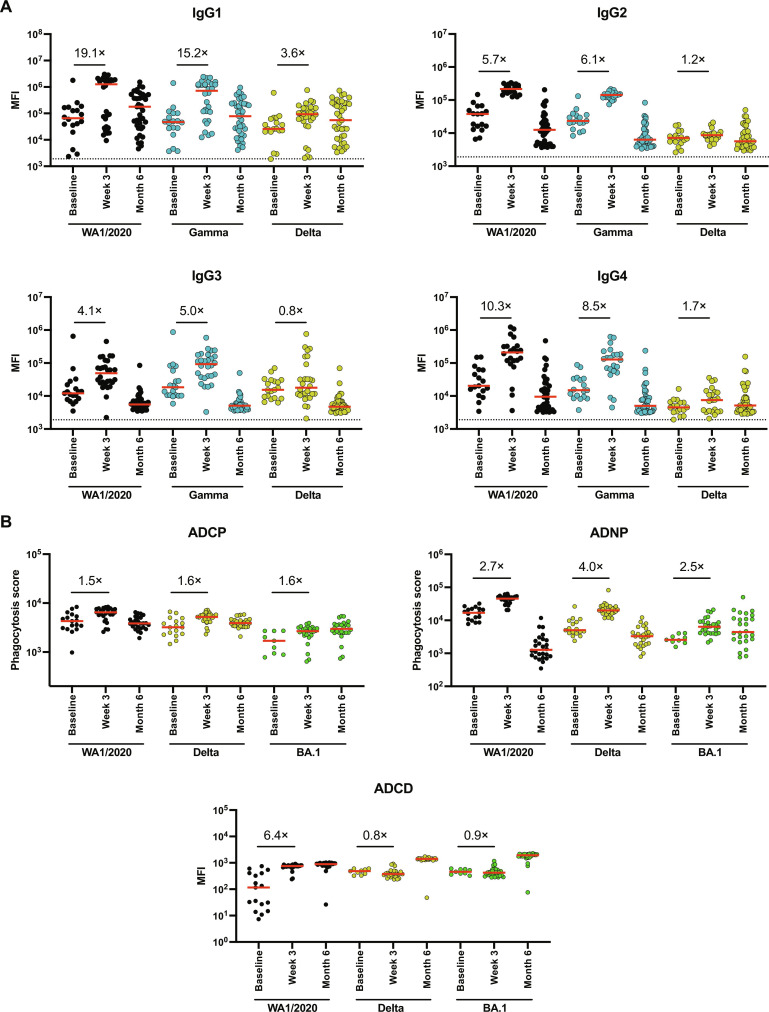
Previous reports from multiple laboratories have shown that a third monovalent mRNA immunization augmented NAbs to both WA1/2020 and Omicron variants (29–32). We assessed antibody isotypes in a separate cohort of individuals who received a third monovalent mRNA immunization (table S2). Vaccinees who received a third mRNA vaccine had robust IgG1 responses against WA1/2020 (19.1-fold), Gamma (15.2-fold), and Delta (3.6-fold) at 3 weeks after vaccination (Fig. 5A). IgG2 and IgG3 similarly increased. As previously reported (23, 24), a third monovalent mRNA immunization also led to IgG4 responses against WA1/2020 (10.3-fold), Gamma (8.5-fold), and Delta (1.7-fold) (Fig. 5A). These responses largely declined to preboost levels at month 6 following the third mRNA boost. Moreover, in contrast to the limited Fc functional activity following the fourth mRNA boost, ADCP, ADNP, and ADCD responses increased following the third mRNA boost (Fig. 5B). These data show that a third mRNA boost resulted in mixed IgG1/IgG2/IgG3/IgG4 responses with Fc functional activity.
Fig. 5. Antibody and Fc effector function profiling by systems serology following third monovalent mRNA immunization.
(A) Spike-specific IgG1, IgG2, IgG3, and IgG4 responses against spikes of WA1/2020, Gamma, and Delta variants by Luminex-based assay at baseline before boosting, at week 3 following boosting, and at month 6 following boosting in vaccinees who received third monovalent mRNA vaccines. (B) Spike-specific ADCP, ADNP, and ADCD activity against WA1/2020, Delta, and BA.1 at baseline before boosting, week 3 after boost, and month 6 after boost is shown. Medians of the MFI in red bars are depicted, and fold changes are shown numerically. Phagocytosis score for ADCP and ADNP activity is calculated as follows: phagocytosis score = % THP-1 bead-positive cells × median fluorescent intensity of bead-positive cells/10,000. LOD indicated by dotted lines represent the median MFI of an Ebolavirus glycoprotein negative control.
DISCUSSION
Studies from multiple groups have shown that bivalent mRNA boosting resulted in robust NAb responses against the ancestral WA1/2020 strain but lower NAb responses against the BA.5 strain (5–8, 11, 25). In this study, we show that the durability of NAb responses to WA1/2020 is relatively robust but that the magnitude and durability of NAb responses to XBB subvariants are more limited following bivalent mRNA boosting. This finding is consistent with recent clinical efficacy studies that have reported that the bivalent mRNA vaccines provided minimal protection against acquisition of infections that waned from 29% to 0% after 20 weeks and moderate protection against severe disease that waned from 67% to 38% after 20 weeks (33), during a time when XBB variants were predominant.
Immune imprinting likely contributed to NAb responses being directed primarily against the ancestral WA1/2020 strain following bivalent mRNA boosting (34–36). This phenomenon has also been recently described in studies where immunization with monovalent or bivalent mRNA vaccines induced limited NAbs against BA.5 and XBB variants (37–39). Moreover, boosting with monovalent BA.5 or XBB vaccines as well as breakthrough infections resulted in superior NAb responses against these variants (37, 38). Together, these data suggest that future COVID-19 boosters should exclude the ancestral WA1/2020 spike immunogen, which is the current plan for the 2023 booster.
Although serum NAb titers induced by mRNA vaccines have limited durability and cross-reactivity against variants, effector and memory CD4 and CD8 T cell responses have more robust durability and cross-reactivity (29, 30, 40, 41). Moreover, T cell responses likely contribute substantially to vaccine protection against severe disease (41–43). We observed that spike-specific CD4 and CD8 T cell responses were comparable against WA1/2020, BQ.1.1, and XBB.1.5, consistent with prior reports, but that these responses were only minimally increased by bivalent mRNA boosting. Future studies could expand T cell responses by including nonspike antigens such as the nucleocapsid or membrane (44, 45) in next-generation COVID-19 vaccines.
Binding antibody responses are generally more cross-reactive against variants than NAb responses (8, 46). Following bivalent mRNA boosting, we observed minimal increases in spike-specific IgG1 and IgG3 responses but substantial increases in IgG4 responses with poor Fc functional activity. In contrast, following third monovalent mRNA boosting, we observed major increases in all IgG isotypes with enhanced Fc functional activity. Recent reports have also showed some increase in IgG4 responses following a third monovalent mRNA immunization (23, 24), but not with an adenovirus-based vaccine (24). Our data confirm and extend these observations by showing an even more marked skewing toward IgG4 responses following a fourth mRNA immunization.
A class switch toward IgG4 happens usually when an individual is frequently exposed to an antigen (19, 47), which was demonstrated in a study with beekeepers, where IgG1 antibodies specific to phospholipase A2, a bee venom antigen, class-switched to IgG4 after 6 months of continuous bee stings (48). Similarly, in the HIV vaccine trial RV144, where participants received canarypox vector–based HIV Env vaccines and purified HIV Env gp120 protein vaccines, elevated IgG1 and IgG3 antibodies correlated with enhanced effector functions, whereas in the clinical trial VAX003, where participants received seven doses of HIV Env gp120 protein vaccines, elevated IgG2 and IgG4 levels were observed with increasing boosts and correlated with inhibition of Fc effector functions (49–51). We speculate that a class switch from IgG1 to IgG4 might occur after multiple mRNA immunizations (23). This is also in line with an observation that class switch to IgG2 or IgG4 occurs more readily from IgG1 B cells (52), potentially to balance excessive inflammation, as IgG4 is known to have inhibitory effector functions (18, 19, 53).
In summary, our data show that bivalent mRNA boosting induced only modest and transient NAb responses against XBB subvariants, suggesting immune imprinting to the ancestral WA1/2020 strain. Moreover, antibody responses were primarily IgG4 responses with limited Fc functional activity, with minimal boosting of IgG1 and IgG3 responses. Further studies are needed to elucidate the mechanisms behind the IgG4 class switching following multiple mRNA immunizations and to evaluate the possibility of tolerance induction as well. Additional studies are also needed to establish generalizability, since our cohorts were small and predominantly female. Our data demonstrate that bivalent mRNA boosting may be limited by immune imprinting and that multiple mRNA immunizations may lead to IgG4 responses with poor functional activity.
MATERIALS AND METHODS
Study population
A specimen biorepository at Beth Israel Deaconess Medical Center (BIDMC) obtained samples from individuals who received monovalent SARS-CoV-2 vaccines as well as bivalent mRNA boosters. The BIDMC institutional review board approved this study (2020P000361). All participants provided informed consent. This study included individuals from two cohorts who received three doses of mRNA vaccines (n = 33; Pfizer-BioNTech, Moderna) and the bivalent mRNA boosters (n = 30; Pfizer-BioNTech, Moderna). Participants were excluded from the immunologic assays if they had a history of SARS-CoV-2 infection or a positive nucleocapsid (N) serology by electrochemiluminescence assays or if they received immunosuppressive medications.
Pseudovirus NAb assay
NAb titers against SARS-CoV-2 variants used pseudoviruses expressing a luciferase reporter gene. In brief, the packaging construct psPAX2 (AIDS Resource and Reagent Program), luciferase reporter plasmid pLenti-CMV Puro-Luc (Addgene), and Spike protein expressing pcDNA3.1-SARS-CoV-2 SΔCT were cotransfected into human embryonic kidney (HEK) 293T cells (American Type Culture Collection, CRL_3216) with Lipofectamine 2000 (Thermo Fisher Scientific). Pseudoviruses of SARS-CoV-2 variants were generated using the Spike protein from WA1/2020 (Wuhan/WIV04/2019, GISAID accession ID: EPI_ISL_402124), Omicron BA.2 (GISAID ID: EPI_ISL_6795834.2), BA.5 (GISAID ID: EPI_ISL_12268495.2), BQ.1.1 (GISAID ID: EPI_ISL_14752457), XBB.1 (GISAID ID: EPI_ISL_15232105), XBB.1.5 (GISAID ID: EPI_ISL_16418320), and XBB.1.16 (GISAID ID: EPI_ISL_17646715). The supernatants containing the pseudotyped viruses were collected 48 hours after transfection, and pseudotyped viruses were purified by filtration with a 0.45-μm filter. To determine NAb titers in human serum, HEK293T-hACE2 cells were seeded in 96-well tissue culture plates at a density of 2 × 104 cells per well overnight. Threefold serial dilutions of heat-inactivated serum samples were prepared and mixed with 60 μl of pseudovirus. The mixture was incubated at 37°C for 1 hour before adding to HEK293T-hACE2 cells. After 48 hours, cells were lysed in Steady-Glo Luciferase Assay (Promega) according to the manufacturer’s instructions. SARS-CoV-2 neutralization titers were defined as the sample dilution at which a 50% reduction (NT50) in relative light units was observed relative to the average of the virus control wells.
Intracellular cytokine staining assay
CD4+ and CD8+ T cell responses were quantitated by pooled peptide-stimulated intracellular cytokine staining assays. Peptide pools contained 15–amino acid peptides overlapping by 11 amino acids spanning the SARS-CoV-2 WA1/2020, BQ.1.1, or XBB.1.5 Spike proteins (21st Century Biochemicals). PBMCs (106) were resuspended in 100 μl of R10 medium supplemented with CD49d monoclonal antibody (mAb) (1 μg/ml) and CD28 mAb (1 μg/ml). Each sample was assessed with mock (100 μl of R10 plus 0.5% dimethyl sulfoxide, background control), peptides (2 μg/ml), and/or phorbol myristate acetate (10 pg/ml) and ionomycin (1 μg/ml; Sigma-Aldrich) (100 μl; positive control) and incubated at 37°C for 1 hour. After incubation, 0.25 μl of GolgiStop and 0.25 μl of GolgiPlug in 50 μl of R10 were added to each well and incubated at 37°C for 8 hours and then held at 4°C overnight. The next day, the cells were washed twice with Dulbecco's phosphate-buffered saline (DPBS), stained with aqua live/dead dye for 10 min, and then stained with predetermined titers of mAbs against CD279 (clone EH12.1, BB700), CD4 (clone L200, BV711), CD27 (clone M-T271, BUV563), CD8 (clone SK1, BUV805), and CD45RA [clone 5H9, allophycocyanin (APC) H7] for 30 min. Cells were then washed twice with 2% fetal bovine serum (FBS)/DPBS buffer and incubated for 15 min with 200 μl of BD CytoFix/CytoPerm Fixation/Permeabilization solution. Cells were washed twice with 1× Perm Wash buffer (BD Perm/Wash buffer 10× in the CytoFix/CytoPerm Fixation/Permeabilization kit diluted with MilliQ water and passed through 0.22-μm filter) and stained intracellularly with mAbs against Ki67 (clone B56, BB515), interleukin-21 (IL-21) [clone 3A3-N2.1, phycoerythrin (PE)], CD69 [clone TP1.55.3, extracellular domain (ECD)], IL-10 (clone JES3-9D7, PE CY7), IL-13 (clone JES10-5A2, BV421), IL-4 (clone MP4-25D2, BV605), tumor necrosis factor–α (TNF-α) (clone Mab11, BV650), IL-17 (clone N49-653, BV750), IFN-γ (clone B27; BUV395), IL-2 (clone MQ1-17H12, BUV737), IL-6 (clone MQ2-13A5, APC), and CD3 (clone SP34.2, Alexa Fluor 700) for 30 min. Cells were washed twice with 1× Perm Wash buffer and fixed with 250 μl of freshly prepared 1.5% formaldehyde. Fixed cells were transferred to 96-well round bottom plate and analyzed by BD FACSymphony system. Data were analyzed using FlowJo v9.9.
Luminex antibody profiling
Serum samples were analyzed by customized Luminex assay to quantify the relative concentration of antigen-specific antibody subclasses, as previously described (54, 55). Briefly, SARS-CoV-2 antigens were used to profile specific humoral immune responses. Antigens were coupled to magnetic Luminex beads (Luminex Corp.) by carbodiimide–N-hydroxysuccinimide ester coupling (Thermo Fisher Scientific). Antigen-coupled beads were washed and incubated with heat-inactivated plasma samples. Heat inactivation was done at 56°C for 30 min, followed by centrifugation at 13,000 rpm for 5 min to pellet aggregates and nonsoluble material. The linear ranges of serum dilution factors were initially determined for all antibody isotypes, where samples from each group were randomly chosen and assayed against the variants to be tested. On the basis of the dilution curves generated, dilutions for specific antibody isotypes, subclasses, and Fc-binding antibodies were done as follows in 1× PBS: IgG1 = 1:2000, IgG2 = 1:250, IgG3 = 1:500, IgG4 = 1:250. The heat-inactivated plasma samples were incubated with the antigen-coupled beads for 2 hours at room temperature in 384-well plates (Greiner Bio-One) on a plate shaker. Unbound antibodies were washed away using 1× assay buffer [1× PBS (pH 7.4), 0.1% (w/v) bovine serum albumin, 0.05% (w/v) Tween 20] on the HydroSpeed 384-well plate washer for a total of three washes. Immune complexes were then detected through the addition of PE-coupled detection antibody for each subclass and/or isotype (IgG1, IgG2, IgG3, and IgG4; Southern Biotech). Plates were sealed with aluminum foil adhesive to protect from light. All detection antibody binding incubations were done at room temperature on a plate shaker. After 1 hour of incubation, plates were washed three times using 1× assay buffer. After the final wash, beads were resuspended in 40 μl of QSOL buffer (Sartorius) and flow cytometry was performed with an iQue (IntelliCyt). Analysis was performed on IntelliCyt ForeCyt (v8.1). Mean fluorescence intensity (MFI) was calculated from technical replicates for each sample for each antigen-specific antibody binding event.
Antibody-dependent complement deposition
ADCD was conducted as previously described (56). Briefly, SARS-CoV-2 antigens were coupled to magnetic Luminex beads (Luminex Corp.) similar to above. Coupled beads were incubated for 2 hours at 37°C with heat-inactivated serum samples (1:10 dilution in 1× PBS) to form immune complexes. Beads were then washed with 1× assay buffer to remove unbound immunoglobulins and contaminants. To measure ADCD of complement component 3 (C3), lyophilized guinea pig complement (Cedarlane) was diluted in gelatin veronal buffer with calcium and magnesium (GBV++) (Boston BioProducts) and added to beads mixture. Subsequently, C3 was detected with goat anti–C3–fluorescein isothiocyanate (FITC) detection antibody (Mpbio). Complement deposition is reported as MFI. Flow cytometry was performed with iQue (IntelliCyt).
ADCP and ADNP
ADCP and ADNP were conducted according to the previously described protocols (57–59). In detail, SARS-CoV-2 antigens were biotinylated and coupled to yellow and green (505/515) fluorescent NeutrAvidin-conjugated microspheres (Thermo Fisher Scientific). To form immune complexes, antigen-coupled beads were incubated for 2 hours at 37°C with heat-inactivated serum at a 1:100 dilution in 1× PBS. Beads were then washed using 1× PBS. For ADNP, granulocytes were isolated from whole blood by lysing red blood cells in Ammonium-Chloride-Potassium (ACK) lysis buffer (1:10 blood in ACK lysis buffer) for 7 min before precipitation by centrifugation. Granulocytes were washed three times with cold 1× PBS to remove serum and other contaminants, and were then resuspended at 2.5 × 105 cells/ml in R10 medium. Fifty thousand cells per well were added to each well and incubated with immune complexes for 1 hour at 37°C, 5% CO2, and cells were fixed with 4% paraformaldehyde (Alfa Aesar). Anti-CD66 Pacific blue was added to the granulocyte mixture to gate for neutrophils. For ADCP, the immune complexes were incubated for 16 to 18 hours with THP-1 cells (American Type Culture Collection): 25,000 live THP-1 cells per well at a concentration of 1.25 × 105 cells/ml in R10 medium [RPMI 1640 (Sigma-Aldrich) supplemented with 10% FBS (Sigma-Aldrich), 5% penicillin-streptomycin (50 μg/ml; Corning), 5% l-glutamine (4 mM; Corning), and 5% Hepes buffer (pH 7.2) (50 mM; Corning)] at 37°C, 5% CO2. Cellular uptake of antigen-coated microspheres in the presence of heat-inactivated serum was done as described above. Flow cytometry was performed to identify the percentage of cells that had phagocytosed beads as well as the number of beads that had been phagocytosed (phagocytosis score = % positive cells × median fluorescent intensity of positive cells/10,000). Flow cytometry was performed with 5-Laser LSR Fortessa Flow Cytometer, and analysis was performed using FlowJo V10.7.1 and RStudio V3.3.0.
Acknowledgments
Funding: We acknowledge NIH grants CA260476 (D.H.B.) and AI69309 (A.Y.C.), Gates Foundation grant INV-042469 (D.H.B. and B.K.), Massachusetts Consortium for Pathogen Readiness (D.H.B.), and the Ragon Institute (D.H.B.).
Author contributions: N.L., A.Y.C., and D.H.B. conceptualized the study. N.L., J.M., N.P.H., T.A., J.L.F., C.R.M., R.C.P., S.L.R., and N.S. constructed plasmids and performed pseudovirus neutralization assays. J.L., D.M.T., and C.W. performed ICS assays. A.Y.C., E.A.B., and M.R. enrolled study participants and assisted with sample and clinical data collection. T.M.C., X.T., and R.P.M. performed systems serology assays. B.K. performed bioinformatic analysis. N.L., and D.H.B. analyzed all data and wrote the manuscript. All authors reviewed and edited the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Correspondence and requests for materials should be addressed to D.H.B. (dbarouch@bidmc.harvard.edu).
Supplementary Materials
This PDF file includes:
Supplementary Methods
Figs. S1 to S3
Tables S1 to S3
REFERENCES AND NOTES
- 1.Polack F. P., Thomas S. J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J. L., Pérez Marc G., Moreira E. D., Zerbini C., Bailey R., Swanson K. A., Roychoudhury S., Koury K., Li P., Kalina W. V., Cooper D., Frenck R. W. Jr., Hammitt L. L., Türeci Ö., Nell H., Schaefer A., Ünal S., Tresnan D. B., Mather S., Dormitzer P. R., Şahin U., Jansen K. U., Gruber W. C., C4591001 Clinical Trial Group , Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 383, 2603–2615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baden L. R., el Sahly H. M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S. A., Rouphael N., Creech C. B., McGettigan J., Khetan S., Segall N., Solis J., Brosz A., Fierro C., Schwartz H., Neuzil K., Corey L., Gilbert P., Janes H., Follmann D., Marovich M., Mascola J., Polakowski L., Ledgerwood J., Graham B. S., Bennett H., Pajon R., Knightly C., Leav B., Deng W., Zhou H., Han S., Ivarsson M., Miller J., Zaks T., COVE Study Group , Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 384, 403–416 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sadoff J., Gray G., Vandebosch A., Cárdenas V., Shukarev G., Grinsztejn B., Goepfert P. A., Truyers C., Fennema H., Spiessens B., Offergeld K., Scheper G., Taylor K. L., Robb M. L., Treanor J., Barouch D. H., Stoddard J., Ryser M. F., Marovich M. A., Neuzil K. M., Corey L., Cauwenberghs N., Tanner T., Hardt K., Ruiz-Guiñazú J., le Gars M., Schuitemaker H., van Hoof J., Struyf F., Douoguih M., ENSEMBLE Study Group , Safety and efficacy of single-dose Ad26.COV2.S vaccine against Covid-19. N. Engl. J. Med. 384, 2187–2201 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naranbhai V., Garcia-Beltran W. F., Chang C. C., Berrios Mairena C., Thierauf J. C., Kirkpatrick G., Onozato M. L., Cheng J., St Denis K. J., Lam E. C., Kaseke C., Tano-Menka R., Yang D., Pavlovic M., Yang W., Kui A., Miller T. E., Astudillo M. G., Cahill J. E., Dighe A. S., Gregory D. J., Poznansky M. C., Gaiha G. D., Balazs A. B., Iafrate A. J., Comparative immunogenicity and effectiveness of mRNA-1273, BNT162b2, and Ad26.COV2.S COVID-19 vaccines. J. Infect. Dis. 225, 1141–1150 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis-Gardner M. E., Lai L., Wali B., Samaha H., Solis D., Lee M., Porter-Morrison A., Hentenaar I. T., Yamamoto F., Godbole S., Liu Y., Douek D. C., Lee F. E.-H., Rouphael N., Moreno A., Pinsky B. A., Suthar M. S., Neutralization against BA.2.75.2, BQ.1.1, and XBB from mRNA bivalent booster. N. Engl. J. Med. 388, 183–185 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller J., Hachmann N. P., Collier A. R. Y., Lasrado N., Mazurek C. R., Patio R. C., Powers O., Surve N., Theiler J., Korber B., Barouch D. H., Substantial neutralization escape by SARS-CoV-2 omicron variants BQ.1.1 and XBB.1. N. Engl. J. Med. 388, 662–664 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Q., Bowen A., Valdez R., Gherasim C., Gordon A., Liu L., Ho D. D., Antibody response to omicron BA.4–BA.5 bivalent booster. N. Engl. J. Med. 388, 567–569 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collier A.-R. Y., Miller J., Hachmann N. P., Mahan K. M., Liu J., Bondzie E. A., Gallup L., Rowe M., Schonberg E., Thai S., Barrett J., Borducchi E. N., Bouffard E., Jacob-Dolan C., Mazurek C. R., Mutoni A., Powers O., Sciacca M., Surve N., Van Wyk H., Wu C., Barouch D. H., Immunogenicity of BA.5 bivalent mRNA vaccine boosters. N. Engl. J. Med. 388, 565–567 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yue C., Song W., Wang L., Jian F., Chen X., Gao F., Shen Z., Wang Y., Wang X., Cao Y., ACE2 binding and antibody evasion in enhanced transmissibility of XBB.1.5. Lancet Infect. Dis. 23, 278–280 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uriu K., Ito J., Zahradnik J., Fujita S., Kosugi Y., Schreiber G., Genotype to Phenotype Japan (G2P-Japan) Consortium, Sato K., Enhanced transmissibility, infectivity, and immune resistance of the SARS-CoV-2 omicron XBB.1.5 variant. Lancet Infect. Dis. 23, 280–281 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q., Bowen A., Tam A. R., Valdez R., Stoneman E., Mellis I. A., Gordon A., Liu L., Ho D. D., SARS-CoV-2 neutralising antibodies after bivalent versus monovalent booster. Lancet Infect. Dis. 23, 527–528 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nimmerjahn F., Ravetch J. V., Fcγ receptors as regulators of immune responses. Nat. Rev. Immunol. 8, 34–47 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Bruhns P., Iannascoli B., England P., Mancardi D. A., Fernandez N., Jorieux S., Daëron M., Specificity and affinity of human Fcγ receptors and their polymorphic variants for human IgG subclasses. Blood 113, 3716–3725 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Zohar T., Loos C., Fischinger S., Atyeo C., Wang C., Slein M. D., Burke J., Yu J., Feldman J., Hauser B. M., Caradonna T., Schmidt A. G., Cai Y., Streeck H., Ryan E. T., Barouch D. H., Charles R. C., Lauffenburger D. A., Alter G., Compromised humoral functional evolution tracks with SARS-CoV-2 mortality. Cell 183, 1508–1519.e12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y., Krémer V., Iannascoli B., Goff O. R. L., Mancardi D. A., Ramke L., de Chaisemartin L., Bruhns P., Jönsson F., Specificity of mouse and human Fcgamma receptors and their polymorphic variants for IgG subclasses of different species. Eur. J. Immunol. 52, 753–759 (2022). [DOI] [PubMed] [Google Scholar]
- 16.Lilienthal G. M., Rahmöller J., Petry J., Bartsch Y. C., Leliavski A., Ehlers M., Potential of murine IgG1 and human IgG4 to inhibit the classical complement and Fcγ receptor activation pathways. Front. Immunol. 9, 958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vidarsson G., Dekkers G., Rispens T., IgG subclasses and allotypes: From structure to effector functions. Front. Immunol. 5, 520 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Napodano C., Marino M. P., Stefanile A., Pocino K., Scatena R., Gulli F., Rapaccini G. L., Delli Noci S., Capozio G., Rigante D., Basile U., Immunological role of IgG subclasses. Immunol. Invest. 50, 427–444 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Rispens T., Huijbers M. G., The unique properties of IgG4 and its roles in health and disease. Nat. Rev. Immunol. 23, 763–778 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farkash I., Feferman T., Cohen-Saban N., Avraham Y., Morgenstern D., Mayuni G., Barth N., Lustig Y., Miller L., Shouval D. S., Biber A., Kirgner I., Levin Y., Dahan R., Anti-SARS-CoV-2 antibodies elicited by COVID-19 mRNA vaccine exhibit a unique glycosylation pattern. Cell Rep. 37, 110114 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stephenson K. E., le Gars M., Sadoff J., de Groot A. M., Heerwegh D., Truyers C., Atyeo C., Loos C., Chandrashekar A., McMahan K., Tostanoski L. H., Yu J., Gebre M. S., Jacob-Dolan C., Li Z., Patel S., Peter L., Liu J., Borducchi E. N., Nkolola J. P., Souza M., Tan C. S., Zash R., Julg B., Nathavitharana R. R., Shapiro R. L., Azim A. A., Alonso C. D., Jaegle K., Ansel J. L., Kanjilal D. G., Guiney C. J., Bradshaw C., Tyler A., Makoni T., Yanosick K. E., Seaman M. S., Lauffenburger D. A., Alter G., Struyf F., Douoguih M., van Hoof J., Schuitemaker H., Barouch D. H., Immunogenicity of the Ad26.COV2.S vaccine for COVID-19. JAMA 325, 1535–1544 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu D. Y., Gorman M. J., Yuan D., Yu J., Mercado N. B., McMahan K., Borducchi E. N., Lifton M., Liu J., Nampanya F., Patel S., Peter L., Tostanoski L. H., Pessaint L., van Ry A., Finneyfrock B., Velasco J., Teow E., Brown R., Cook A., Andersen H., Lewis M. G., Lauffenburger D. A., Barouch D. H., Alter G., Defining the determinants of protection against SARS-CoV-2 infection and viral control in a dose-down Ad26.CoV2.S vaccine study in nonhuman primates. PLOS Biol. 20, e3001609 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irrgang P., Gerling J., Kocher K., Lapuente D., Steininger P., Habenicht K., Wytopil M., Beileke S., Schäfer S., Zhong J., Ssebyatika G., Krey T., Falcone V., Schülein C., Peter A. S., Nganou-Makamdop K., Hengel H., Held J., Bogdan C., Überla K., Schober K., Winkler T. H., Tenbusch M., Class switch toward noninflammatory, spike-specific IgG4 antibodies after repeated SARS-CoV-2 mRNA vaccination. Sci. Immunol. 8, eade2798 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buhre J. S., Pongracz T., Künsting I., Lixenfeld A. S., Wang W., Nouta J., Lehrian S., Schmelter F., Lunding H. B., Dühring L., Kern C., Petry J., Martin E. L., Föh B., Steinhaus M., von Kopylow V., Sina C., Graf T., Rahmöller J., Wuhrer M., Ehlers M., mRNA vaccines against SARS-CoV-2 induce comparably low long-term IgG Fc galactosylation and sialylation levels but increasing long-term IgG4 responses compared to an adenovirus-based vaccine. Front. Immunol. 13, 1020844 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Q., Iketani S., Li Z., Liu L., Guo Y., Huang Y., Bowen A. D., Liu M., Wang M., Yu J., Valdez R., Lauring A. S., Sheng Z., Wang H. H., Gordon A., Liu L., Ho D. D., Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell 186, 279–286.e8 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartsch Y. C., Tong X., Kang J., Avendaño M. J., Serrano E. F., García-Salum T., Pardo-Roa C., Riquelme A., Cai Y., Renzi I., Stewart-Jones G., Chen B., Medina R. A., Alter G., Omicron variant spike-specific antibody binding and Fc activity are preserved in recipients of mRNA or inactivated COVID-19 vaccines. Sci. Transl. Med. 14, eabn9243 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaplonek P., Fischinger S., Cizmeci D., Bartsch Y. C., Kang J., Burke J. S., Shin S. A., Dayal D., Martin P., Mann C., Amanat F., Julg B., Nilles E. J., Musk E. R., Menon A. S., Krammer F., Saphire E. O., A. Carfi, Alter G., mRNA-1273 vaccine-induced antibodies maintain Fc effector functions across SARS-CoV-2 variants of concern. Immunity 55, 355–365.e4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorman M. J., Patel N., Guebre-Xabier M., Zhu A. L., Atyeo C., Pullen K. M., Loos C., Goez-Gazi Y., Carrion R. Jr., Tian J. H., Yuan D., Bowman K. A., Zhou B., Maciejewski S., McGrath M. E., Logue J., Frieman M. B., Montefiori D., Mann C., Schendel S., Amanat F., Krammer F., Saphire E. O., Lauffenburger D. A., Greene A. M., Portnoff A. D., Massare M. J., Ellingsworth L., Glenn G., Smith G., Alter G., Fab and Fc contribute to maximal protection against SARS-CoV-2 following NVX-CoV2373 subunit vaccine with Matrix-M vaccination. Cell Rep. Med. 2, 100405 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reinscheid M., Luxenburger H., Karl V., Graeser A., Giese S., Ciminski K., Reeg D. B., Oberhardt V., Roehlen N., Lang-Meli J., Heim K., Gross N., Baum C., Rieg S., Speer C., Emmerich F., Breisinger S., Steinmann D., Bengsch B., Boettler T., Kochs G., Schwemmle M., Thimme R., Neumann-Haefelin C., Hofmann M., COVID-19 mRNA booster vaccine induces transient CD8+ T effector cell responses while conserving the memory pool for subsequent reactivation. Nat. Commun. 13, 4631 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J., Chandrashekar A., Sellers D., Barrett J., Jacob-Dolan C., Lifton M., McMahan K., Sciacca M., VanWyk H., Wu C., Yu J., Collier A. R. Y., Barouch D. H., Vaccines elicit highly conserved cellular immunity to SARS-CoV-2 Omicron. Nature 603, 493–496 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hachmann N. P., Miller J., A.-r. Y. Collier, Barouch D. H., Neutralization escape by SARS-CoV-2 omicron subvariant BA.4.6. N. Engl. J. Med. 387, 1904–1906 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hachmann N. P., Miller J., Collier A. R. Y., Ventura J. D., Yu J., Rowe M., Bondzie E. A., Powers O., Surve N., Hall K., Barouch D. H., Neutralization escape by SARS-CoV-2 omicron subvariants BA.2.12.1, BA.4, and BA.5. N. Engl. J. Med. 387, 86–88 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin D.-Y., Xu Y., Gu Y., Zeng D., Sunny S. K., Moore Z., Durability of bivalent boosters against omicron subvariants. N. Engl. J. Med. 388, 1818–1820 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Francis T., On the doctrine of original antigenic sin. Proc. Am. Philos. Soc. 104, 572–578 (1960). [Google Scholar]
- 35.Zhang A., Stacey H. D., Mullarkey C. E., Miller M. S., Original antigenic sin: How first exposure shapes lifelong anti–influenza virus immune responses. J. Immunol. 202, 335–340 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Brown E. L., Essigmann H. T., Original antigenic sin: The downside of immunological memory and implications for COVID-19. mSphere 6, e00056-00021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yisimayi A., Song W., Wang J., Jian F., Yu Y., Chen X., Xu Y., Yang S., Niu X., Xiao T., Wang J., Zhao L., Sun H., An R., Zhang N., Wang Y., Wang P., Yu L., Lv Z., Gu Q., Shao F., Jin R., Shen Z., Xie X. S., Wang Y., Cao Y., Repeated Omicron infection alleviates SARS-CoV-2 immune imprinting. Nature 625, 148–156 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Q., Guo Y., Tam A. R., Valdez R., Gordon A., Liu L., Ho D. D., Deep immunological imprinting due to the ancestral spike in the current bivalent COVID-19 vaccine. Cell Rep. Med. 4, 101258 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schiepers A., van ’t Wout M. F. L., Greaney A. J., Zang T., Muramatsu H., Lin P. J. C., Tam Y. K., Mesin L., Starr T. N., Bieniasz P. D., Pardi N., Bloom J. D., Victora G. D., Molecular fate-mapping of serum antibody responses to repeat immunization. Nature 615, 482–489 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goel R. R., Painter M. M., Apostolidis S. A., Mathew D., Meng W., Rosenfeld A. M., Lundgreen K. A., Reynaldi A., Khoury D. S., Pattekar A., Gouma S., Kuri-Cervantes L., Hicks P., Dysinger S., Hicks A., Sharma H., Herring S., Korte S., Baxter A. E., Oldridge D. A., Giles J. R., Weirick M. E., McAllister C. M., Awofolaju M., Tanenbaum N., Drapeau E. M., Dougherty J., Long S., D’Andrea K., Hamilton J. T., McLaughlin M., Williams J. C., Adamski S., Kuthuru O., The UPenn COVID Processing Unit‡, Frank I., Betts M. R., Vella L. A., Grifoni A., Weiskopf D., Sette A., Hensley S. E., Davenport M. P., Bates P., Luning Prak E. T., Greenplate A. R., Wherry E. J., mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science 374, abm0829 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wherry E. J., Barouch D. H., T cell immunity to COVID-19 vaccines. Science 377, 821–822 (2022). [DOI] [PubMed] [Google Scholar]
- 42.Liu J., Yu J., McMahan K., Jacob-Dolan C., He X., Giffin V., Wu C., Sciacca M., Powers O., Nampanya F., Miller J., Lifton M., Hope D., Hall K., Hachmann N. P., Chung B., Anioke T., Li W., Muench J., Gamblin A., Boursiquot M., Cook A., Lewis M. G., Andersen H., Barouch D. H., CD8 T cells contribute to vaccine protection against SARS-CoV-2 in macaques. Sci. Immunol. 7, eabq7647 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barouch D. H., Covid-19 vaccines—Immunity, variants, boosters. N. Engl. J. Med. 387, 1011–1020 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vidal S. J., Collier A. R. Y., Yu J., McMahan K., Tostanoski L. H., Ventura J. D., Aid M., Peter L., Jacob-Dolan C., Anioke T., Chang A., Wan H., Aguayo R., Ngo D., Gerszten R. E., Seaman M. S., Barouch D. H., Correlates of neutralization against SARS-CoV-2 variants of concern by early pandemic Sera. J. Virol. 95, e0040421 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grifoni A., Weiskopf D., Ramirez S. I., Mateus J., Dan J. M., Moderbacher C. R., Rawlings S. A., Sutherland A., Premkumar L., Jadi R. S., Marrama D., de Silva A. M., Frazier A., Carlin A. F., Greenbaum J. A., Peters B., Krammer F., Smith D. M., Crotty S., Sette A., Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 181, 1489–1501.e15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Narowski T. M., Raphel K., Adams L. E., Huang J., Vielot N. A., Jadi R., de Silva A. M., Baric R. S., Lafleur J. E., Premkumar L., SARS-CoV-2 mRNA vaccine induces robust specific and cross-reactive IgG and unequal neutralizing antibodies in naive and previously infected people. Cell Rep. 38, 110336 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aalberse R. C., Stapel S. O., Schuurman J., Rispens T., Immunoglobulin G4: An odd antibody. Clin. Exp. Allergy 39, 469–477 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Aalberse R. C., van der Gaag R., van Leeuwen J., Serologic aspects of IgG4 antibodies. I. Prolonged immunization results in an IgG4-restricted response. J. Immunol. 130, 722–726 (1983). [PubMed] [Google Scholar]
- 49.Chung A. W., Ghebremichael M., Robinson H., Brown E., Choi I., Lane S., Dugast A.-S., Schoen M. K., Rolland M., Suscovich T. J., Mahan A. E., Liao L., Streeck H., Andrews C., Rerks-Ngarm S., Nitayaphan S., de Souza M. S., Kaewkungwal J., Pitisuttithum P., Francis D., Michael N. L., Kim J. H., Bailey-Kellogg C., Ackerman M. E., Alter G., Polyfunctional Fc-effector profiles mediated by IgG subclass selection distinguish RV144 and VAX003 vaccines. Sci. Transl. Med. 6, 228ra238 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Chung A. W., Kumar M. P., Arnold K. B., Yu W. H., Schoen M. K., Dunphy L. J., Suscovich T. J., Frahm N., Linde C., Mahan A. E., Hoffner M., Streeck H., Ackerman M. E., Elrath M. J. M., Schuitemaker H., Pau M. G., Baden L. R., Kim J. H., Michael N. L., Barouch D. H., Lauffenburger D. A., Alter G., Dissecting polyclonal vaccine-induced humoral immunity against HIV using systems serology. Cell 163, 988–998 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fischinger S., Dolatshahi S., Jennewein M. F., Rerks-Ngarm S., Pitisuttithum P., Nitayaphan S., Michael N., Vasan S., Ackerman M. E., Streeck H., Alter G., IgG3 collaborates with IgG1 and IgA to recruit effector function in RV144 vaccinees. JCI Insight 5, e140925 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Horns F., Vollmers C., Croote D., Mackey S. F., Swan G. E., Dekker C. L., Davis M. M., Quake S. R., Lineage tracing of human B cells reveals the in vivo landscape of human antibody class switching. eLife 5, e16578 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van der Zee J. S., van Swieten P., Aalberse R. C., Inhibition of complement activation by IgG4 antibodies. Clin. Exp. Immunol. 64, 415–422 (1986). [PMC free article] [PubMed] [Google Scholar]
- 54.Brown E. P., Dowell K. G., Boesch A. W., Normandin E., Mahan A. E., Chu T., Barouch D. H., Bailey-Kellogg C., Alter G., Ackerman M. E., Multiplexed Fc array for evaluation of antigen-specific antibody effector profiles. J. Immunol. Methods 443, 33–44 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown E. P., Licht A. F., Dugast A. S., Choi I., Bailey-Kellogg C., Alter G., Ackerman M. E., High-throughput, multiplexed IgG subclassing of antigen-specific antibodies from clinical samples. J. Immunol. Methods 386, 117–123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fischinger S., Fallon J. K., Michell A. R., Broge T., Suscovich T. J., Streeck H., Alter G., A high-throughput, bead-based, antigen-specific assay to assess the ability of antibodies to induce complement activation. J. Immunol. Methods 473, 112630 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ackerman M. E., Moldt B., Wyatt R. T., Dugast A. S., McAndrew E., Tsoukas S., Jost S., Berger C. T., Sciaranghella G., Liu Q., Irvine D. J., Burton D. R., Alter G., A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. J. Immunol. Methods 366, 8–19 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Butler A. L., Fallon J. K., Alter G., A sample-sparing multiplexed ADCP assay. Front. Immunol. 10, 1851 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karsten C. B., Mehta N., Shin S. A., Diefenbach T. J., Slein M. D., Karpinski W., Irvine E. B., Broge T., Suscovich T. J., Alter G., A versatile high-throughput assay to characterize antibody-mediated neutrophil phagocytosis. J. Immunol. Methods 471, 46–56 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods
Figs. S1 to S3
Tables S1 to S3