Abstract
Coordinated interactions between cyclin-dependent kinases (Cdks), their target “pocket proteins” (the retinoblastoma protein [pRB], p107, and p130), the pocket protein binding E2F-DP complexes, and the Cdk inhibitors regulate orderly cell cycle progression. The cyclin D1 gene encodes a regulatory subunit of the Cdk holoenzymes, which phosphorylate the tumor suppressor pRB, leading to the release of free E2F-1. Overexpression of E2F-1 can induce apoptosis and may either promote or inhibit cellular proliferation, depending upon the cell type. In these studies overexpression of E2F-1 inhibited cyclin D1-dependent kinase activity, cyclin D1 protein levels, and promoter activity. The DNA binding domain, the pRB pocket binding region, and the amino-terminal Sp1 binding domain of E2F-1 were required for full repression of cyclin D1. Overexpression of pRB activated the cyclin D1 promoter, and a dominant interfering pRB mutant was defective in cyclin D1 promoter activation. Two regions of the cyclin D1 promoter were required for full E2F-1-dependent repression. The region proximal to the transcription initiation site at −127 bound Sp1, Sp3, and Sp4, and the distal region at −143 bound E2F-4–DP-1–p107. In contrast with E2F-1, E2F-4 induced cyclin D1 promoter activity. Differential regulation of the cyclin D1 promoter by E2F-1 and E2F-4 suggests that E2Fs may serve distinguishable functions during cell cycle progression. Inhibition of cyclin D1 abundance by E2F-1 may contribute to an autoregulatory feedback loop to reduce pRB phosphorylation and E2F-1 levels in the cell.
The cyclin D1 gene encodes a regulatory subunit of a multiprotein cyclin D1-dependent kinase (CD1K) holoenzyme complex, which phosphorylates and inactivates the tumor suppressor protein pRB (retinoblastoma protein) (15, 72). pRB phosphorylation is first detected during G1 and continues throughout the cell cycle, with the last stages occurring in G2 (8, 45). Immunoneutralization and antisense experiments have established that the abundance of cyclin D1 may be rate limiting for G1 progression in many cell types (36, 58, 59, 72). Cyclin D1 was of relatively greater importance in promoting the early G0-to-G1 transition from quiescence rather than the late G1/S phase transition, which involved primarily cyclin E (58, 59) and cyclin A (50). Phosphorylation of pRB by the CD1K complex releases a heterodimeric pRB-pocket binding complex of E2F-DP proteins, which regulate gene transcription through DNA sequences capable of binding E2F. In most cell types, high levels of E2F-1, whether induced by overexpression in cultured cells or the result of pRB gene deletion, are poorly tolerated, resulting in cellular apoptosis (30, 40, 75).
E2F-1 is a member of a family of proteins (E2F-1 to -5) which have specific domains involved in transactivation, in binding to the pocket proteins (pRB, p107, and p130), and in binding to DNA. Several differences have been observed among members of the E2F-DP family of pRB pocket binding proteins (34). Increasing evidence suggests that the E2F proteins may fall into two categories. The first group, consisting of E2F-1 to -3, shares a conserved amino-terminal cyclin A-cdk2 binding domain which is absent in E2F-4 and E2F-5. E2F-1 to -3 preferentially bind pRB (26, 63), whereas E2F-4 and E2F-5 associate with p130 in quiescent cells and with p107 in cycling cells (60, 68), and E2F-5 binds preferentially to p130 in vivo (60, 68). E2F-1 to -3 are capable of binding Sp1 (26, 63), whereas neither E2F-4 nor E2F-5 binds Sp1 (26). Dominant negative mutants of cdk3 inhibit the activity of E2F-1 to -3 but not of E2F-4 (21), and overexpression of E2F-1 to -3 in some cell types promotes S-phase entry independently of cyclin D1, whereas E2F-4 and E2F-5 cannot promote entry into S phase unless coexpressed with DP-1 (38). Together these findings suggest that distinct functions may be served by E2F-1 to -3 compared with E2F-4 and E2F-5.
Overexpression of free E2F-1 may either promote or inhibit cellular proliferation and can induce cellular apoptosis, depending on the cell type. High levels of E2F-1 inhibited growth of primary and established fibroblasts (24, 44), and ectopic Drosophila E2F expression during S phase blocked reentry of the cells into S phase in the following cycle (2), suggesting that the timing of E2F expression may be critical in determining its effects on the cell cycle. Although overexpression of E2F-1 can transform rat embryo fibroblasts (64), homozygous deletion of the E2F-1 gene in transgenic mice resulted in enhanced spontaneous tumor formation, particularly tumors of the reproductive tract, lung adenocarcinoma, and lymphomas (12, 77). Hyperplasias of testicular leydig cells, lymphoid cells, and thymocytes were a common feature of the E2F-1 knockout (KO) animals (12, 77). These findings suggest that, under certain circumstances, E2F-1 may also convey an antiproliferative and tumor suppressor function.
The induction of S-phase entry through overexpression of E2F-1 involves a mechanism that is independent of cyclin D1 and was not blocked by the cyclin-dependent kinase inhibitors (10). Overexpression of E2F-1 in REF52 cells inhibited CD1K activity via induction of a CD1K inhibitor related to p16INK4a (27). In contrast with cyclin D1, cyclin E and cyclin A are induced by E2F-1 overexpression, although the effect of E2F-1 overexpression on S-phase entry occurs independently of cdk2 activity (10). It has been proposed that the inhibition of CD1K activity by E2F-1 may function as an autoregulatory feedback loop, attenuating the proliferative and apoptotic effects of excess E2F-1 (27).
Complex transcriptional regulatory mechanisms must exist to coordinate the specific temporal profiles of cyclin and E2F mRNA induction during cell cycle progression. For example, in contrast with the induction of cyclin D1 expression, which begins early in G1 (43, 48) and decreases as cells progress into S phase, expression of E2F-1 increases at the G1/S boundary and peaks in S phase (65). Recent studies have demonstrated that autoregulatory loops occur between the cyclin-dependent kinases and their substrates, as, for example, cyclin D1 stimulates E2F-1 promoter activity (24). Thus, the E2F-1 gene is transcriptionally induced by the G1 cyclins, implying that induction of the G1 cyclins is functionally upstream of E2F-1 (24). In this study we examined further the mechanisms by which E2F-1 regulates CD1K activity and identified contrasting effects of E2F-1 and E2F-4 in regulating the cyclin D1 promoter.
MATERIALS AND METHODS
Construction of plasmid vectors.
The human cyclin D1 promoter reporter constructs (1, 70), the wild-type B-Myb promoter reporter (MybLUC) (33) and the E2F site mutant (Mybmut LUC), and the reporter gene PALUC, which contains 7 kb of the human cyclin A promoter sequence (19), have been described previously. The −163CD1LUC construct was made by PCR-directed amplification using oligonucleotides synthesized to the published sequence of the human cyclin D1 promoter (47). The cyclin D1 promoter Sp1 site between −127 and −99 was deleted by PCR-directed mutagenesis in the context of the −163-bp promoter fragment to create −163ΔSp1LUC. The cyclin D1 E2F sequences from −163 to −133, the cyclin D1 activating transcription factor (ATF) sequences from −66 to −40, the cyclin D1 Sp1-like sequences from −130 to −99, and the wild-type and mutant E2F sites from the adenovirus E2a (AdE2) promoter were synthesized as complementary strands and cloned into TK81pA3LUC to create the vectors CD1E2FLUC, CD1ATFLUC, CD1Sp1LUC, AdE2FLUC, and AdE2FmLUC (53).
Expression vectors.
The wild-type and mutant E2F–DP-1 expression vectors CMV–HA–E2F-1, CMV–DP-1 (80), CMV–E2F-1–Y411C (17, 25), pcDNA–HA–E2F-1 E132 (76), CMV–E2F-1 Δ1-88, CMV–E2F-1 Δ113-120, CMV–E2F-1 Δ206-220, and CMV–E2F-1 411/421 were generous gifts from J. R. Nevins (7). pCMV–HA–E2F-4 (14) and the wild-type and mutant pRB expression vectors CMV-pRB, RB-SE, and RB-ME (74, 80) have been described previously. The vector encoding wild-type pRB protein (phRbc-SVE) (20) was a generous gift from R. A. Weinberg. Glutathione S-transferase (GST)–DP-1(159-410) (3), GST–E2F-1(89-238) (18), and GST–E2F-4 (68) were used for the production of fusion proteins in vitro. The cDNAs encoding E2F-1 and E2F-1 E132 were isolated from CMV–HA–E2F-1 and pcDNA–HA–E2F-1 E132 (76) and cloned into the tetracycline-regulatable expression vector pBPSTR1 (51). The plasmid encoding green fluorescent protein (GFP), pEGFP-N1, was from Clontech (Palo Alto, Calif.).
Cell culture, DNA transfection, and luciferase assays were performed as previously described (52, 54). The human trophoblast cell line JEG-3, the fibroblast cell line NIH 3T3, the SAOS2 osteosarcoma cell line, and the mouse embryo fibroblasts (MEFs) derived from wild-type or E2F-1−/− mice (77) (a generous gift from Dr. L. Yamasaki) were maintained in Dulbecco modified Eagle medium (DMEM) with 10% calf serum and 1% penicillin-streptomycin. Chinese hamster ovary (CHO) cells were maintained in α-MEM with 10% fetal calf serum and 1% penicillin-streptomycin. Cells were transfected by calcium-phosphate precipitation, the media were changed after 6 h, and luciferase activity was determined after a further 24 h. The fold effect was determined for a given construct by comparison with the effect of equal molar amounts of the mutant expression plasmid or empty expression vector cassette as described in the text. Statistical analyses were performed by using the Mann-Whitney U test.
Oligodeoxyribonucleotides.
For construction of the vectors AdE2FLUC and AdE2FmTKLUC, the oligodeoxyribonucleotides containing the wild-type and mutant E2F sites from the AdE2 promoter were synthesized as complementary strands and cloned into BamHI-restricted TK81pA3LUC. The coding strand for the wild-type site (E2Fwt1) (shown with the E2F site in boldface) was 5′-AGC TTG TTT CGC GCT TAA ATT TGA GAA AGG GCG CGA AAC TAG TCA-3′, and the mutant E2F-1 sequence [E2F(1)m; shown with the mutant nucleotides lowercased], previously shown to abolish E1A-dependent transcriptional activation (73), was 5′-AGC TTG TTT Ctg aCT TAA ATT TGA GAA AGG Gtc aag AAC TAG TCA-3′. The sequences of the oligodeoxyribonucleotides used in electrophoretic mobility shift assays (EMSA) and for the construction of reporter plasmids were 5′-TCC CGG CGT CGT TTG GCG CCC GCG CCC-3′ for the cyclin D1 E2F site and 5′-TCC CCC TGC GCC CGC CCC CGC CCC CCT CCC GC-3′ for the cyclin D1 Sp1 site (−130 to −99) (47). The sequence of the consensus wild-type Sp1 site was 5′-ATT CGA TCG GGG CGG GGC GAG C-3′.
EMSA.
EMSA were performed with nuclear extracts from JEG-3 cells or cloned proteins prepared by bacterial expression or in vitro translation as previously described (69, 70). The binding buffer used in EMSA with nuclear extracts (5 to 10 μg) contained 20 mM HEPES (pH 7.4), 80 mM KCl, 1 mM MgCl2, 0.1 mM EDTA, 8.5% glycerol, and 0.2 mM dithiothreitol to which 5 to 10 fmol (20,000 cpm) of γ-32P-labelled probe and 500 ng of sonicated salmon sperm DNA were added. EMSA with the bacterially expressed GST fusion protein were performed by using 30 to 300 ng of protein in binding buffer. The bacterial expression vectors for E2F-1 [RBP3(89-238) GST–RBP(89-238)] (18) and vectors GST-E2F-2(87-244) (23) and GST–DP-1(95-410) (3) were expressed in Escherichia coli as previously described (70). Protein concentration was determined by the method of Bradford (4a) (Protein Assay Dye Reagent concentrate; Bio-Rad Laboratories, Melville, N.Y.). The purities and sizes of the eluted proteins were evaluated by Coomassie blue staining of the sodium dodecyl sulfate-polyacrylamide gels.
The antibodies used in supershift experiments included antibodies to E2F-1 (KH95 and KH20), E2F-2 (LLF2) (46), DP-1 (WTH1), pRB (XZ55), p107 (SD6 and SD15), and simian virus 40 T antigen (PAb419) (generous gifts from E. Harlow, N. Dyson, and J. Lees) and antibodies to E2F-1 (C20X), E2F-2 (C20X), E2F-4 (C108X), p130 (C20X), Sp1 (1C6X), Sp2 (K-20), Sp3 (D-20X), and Sp4 (V-20X) from Santa Cruz Biotechnology (Santa Cruz, Calif.). The protein-DNA complexes were analyzed by electrophoresis through a 5% polyacrylamide gel, with 0.25× Tris-borate-EDTA buffer (TBE) (0.045 M Tris-borate–0.001 M EDTA) with 2.5% glycerol. The gels were dried and exposed to XAR5 radiographic film.
Western blotting, cyclin D1 immune-complex assays, and flow cytometric analyses.
Western blot analysis was performed as previously described (1, 70) by using a monoclonal antibody to cyclin D1 (HD-11), an α-tubulin antibody (5H1) (6), or a cyclin-A antibody (BF683; Santa Cruz Biotechnology) and a horseradish peroxidase-conjugated anti-mouse second antibody. Reactive proteins were visualized by the enhanced chemiluminescence system (Amersham, Arlington Heights, Ill.) and quantified by densitometry.
Immunoprecipitation kinase assays were performed essentially as previously described (42, 70). Cells were transfected with expression plasmids for wild-type or mutant E2F-1 proteins and the kinase assays were performed with GST-pRB product derived from the vector pGEX-Rb (11) as the substrate (70).
Cell sorting for transfected cells using the GFP plasmid pEGFP-N1 was performed exactly as previously described (39). Flow cytometric analyses were carried out in a fluorescence-activated cell sorter (FACS) (FACStar plus; Becton Dickinson).
RESULTS
E2F-1 inhibits CD1K activity in trophoblast cells.
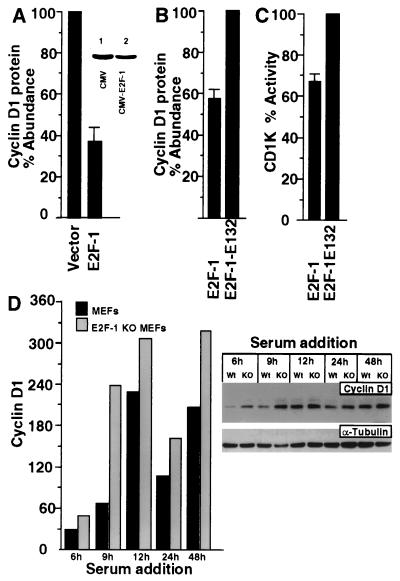
In order to examine the effect of E2F-1 on cyclin D1 protein levels and activity, transient expression studies were performed with cultured cells. Transient expression studies were conducted with FACS selection of transfected cells by using GFP as a marker (39). Cells transfected with E2F-1 and the pEGFP-N1 expression plasmid were compared with cells transfected with the empty expression vector cassette. Cells were also transfected with a tetracycline-regulated E2F-1 expression plasmid (pBPSTR1–E2F-1). FACS enrichment was performed for transfected cells, and Western blotting was performed on cells after 24 h. Cyclin D1 protein levels were inhibited 60% by the overexpression of E2F-1 compared with the effect of the empty expression vector cassette (Fig. 1A). Similar experiments were also conducted with the expression plasmid CMV–E2F-1; they demonstrated that E2F-1 inhibited cyclin D1 protein levels (Fig. 1A, inset). In contrast, E2F-1 protein levels were increased fourfold in cells transfected with the E2F-1 expression plasmid (data not shown).
FIG. 1.
E2F-1 inhibits CD1K activity in choriocarcinoma cells. (A) The E2F-1 expression vector pBPSTR1–E2F-1 was used to transfect JEG-3 cells in conjunction with the GFP plasmid pEGFP, and FACS sorting of transfected cells was performed (39). Cells transfected with E2F-1 were compared with cells transfected with the empty expression vector cassette. Western blotting was performed for cyclin D1, and the reduction in cyclin D1 protein abundance in the E2F-1-transfected cells is shown as the mean ± SEM for three separate transfections. (Inset) Similar findings were observed with the expression plasmid CMV–E2F-1. Cyclin D1 protein abundance was determined by Western blot analysis with cells transfected with empty vector (lane 1) and cells transfected with CMV–E2F-1 (lane 2). (B) Western blot analysis for cyclin D1 protein was performed with cells transfected either with the CMV-driven E2F-1 expression vector or with the DNA binding-defective mutant CMV–E2F-1 E132. (C) CD1K activity was determined in JEG-3 cells overexpressing CMV–E2F-1 or the DNA binding-defective mutant (CMV–E2F-1 E132) 48 h after transfection. Data are shown as means ± SEMs for three separate transfections. (D) Cyclin D1 Western blot analysis of protein derived from either wild-type (Wt) or E2F-1 KO MEFs. The relative abundance of cyclin D1 is shown graphically on the left, and the blots are shown on the right. The Western blot reprobed for α-tubulin is shown.
As an initial analysis of the mechanisms by which E2F-1 inhibited cyclin D1 protein, the effects of E2F-1 and the DNA binding-defective E2F-1 mutant (E2F-1 E132) were compared. Inhibition of cyclin D1 protein levels by E2F-1 was reduced 40% by the DNA binding-defective mutant (Fig. 1B) (n = 3), suggesting that the DNA binding domain of E2F-1 was required for full inhibition of cyclin D1 protein levels.
A similar analysis was performed to determine the effect of E2F-1 on CD1K activity by using an immunoprecipitation assay with pRB as the substrate. Cells were transfected with expression vectors encoding either wild-type E2F-1 or the E2F-1 mutant CMV–E2F-1–E132 (25). Cyclin D1-immune precipitation kinase assays were performed on whole-cell extracts of transfected cells. CD1K activity was reduced approximately 33% by E2F-1 (Fig. 1C) (n = 3). Similar trends were observed in CD1K activity from cells transfected with the E2F-1 expression plasmid in the absence of GFP enrichment. In these experiments, E2F-1 overexpression reduced CD1K activity by 26% at 24 h compared with the empty expression vector (data not shown). These results are consistent with and extend recent studies by Khleif et al. in which overexpression of CMV–E2F-1 inhibited CD1K activity in REF52 cells (27) by showing that the inhibition of CD1K activity by E2F-1 requires the DNA binding domain of E2F-1.
Because these studies suggested that cyclin D1 could be negatively regulated by E2F-1, we examined cyclin D1 levels in MEFs derived from mice with the E2F-1 gene homozygously deleted (77) (E2F-1 KO mice). Comparisons were made with MEFs derived from mice of the identical strain at the same passage number. Cells were grown to 15% confluence, arrested by serum deprivation for 30 h, and then treated with 20% serum. Cells were harvested at serial time points from 6 to 48 h (Fig. 1D, right panel). Western blotting comparing equal amounts of protein at each time point was performed for cyclin D1 protein levels. Cyclin D1 protein levels were increased in the E2F-1 KO MEFs compared with the wild-type MEFs. The relative abundance of cyclin D1 at each time point is shown schematically in Fig. 1D. Western blotting for α-tubulin was also performed with the same Western blot (Fig. 1D, right panel), confirming that the increase in the cyclin D1 levels was not due to differences in the amount of protein loaded. Western blotting performed for cyclin A at each time point demonstrated no difference in abundance between the parental and the E2F-1 KO MEFs (data not shown). Together these studies suggest that E2F-1 may function to inhibit cyclin D1 protein abundance and activity.
Repression of cyclin D1 promoter activity by E2F-1.
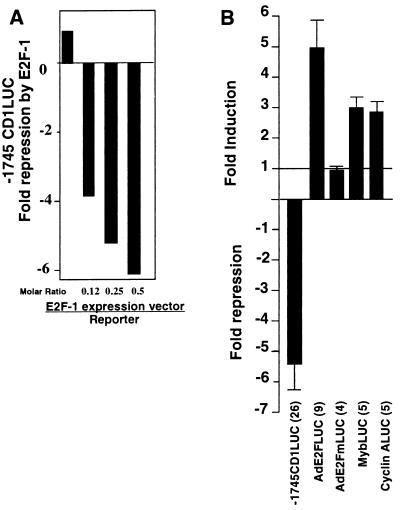
In order to investigate further the mechanisms by which E2F-1 inhibited cyclin D1 abundance, the effect of E2F-1 on cyclin D1 promoter activity was determined. Cotransfection of E2F-1 in JEG-3 cells inhibited activity of the −1745CD1LUC reporter in a dose-dependent manner (Fig. 2A). Compared with the empty expression vector cassette, overexpression of E2F-1 inhibited the full-length −1745CD1LUC reporter 5.4-fold (Fig. 2B). Repression of the −1745CD1LUC reporter was observed whether E2F-1 was overexpressed from a cytomegalovirus expression vector (Fig. 2B) or the tetracycline-regulated expression plasmid pBPSTR1 (see below). In parallel experiments E2F-1 induced a synthetic E2F-responsive reporter plasmid 4.5-fold but did not induce a similar plasmid with a mutation in the E2F site that abolished E2F binding (Fig. 2B). The Myb promoter linked to the luciferase reporter gene was induced threefold by E2F-1, and the cyclin A LUC reporter was induced threefold (Fig. 2B), consistent with the results of previous studies (9).
FIG. 2.
E2F-1 represses the cyclin D1 promoter activity. (A) The E2F-1 expression vector (CMV–E2F-1) was transfected into JEG-3 cells with the −1745CD1LUC reporter. The ratio of transfected expression vector to reporter plasmid is shown on the abscissa. A representative example from three separate experiments is shown. (B) The expression vector CMV–E2F-1 was transfected with the −1745CD1LUC reporter. Comparisons were made with the effects on the reporter plasmids AdE2FLUC, AdE2FmLUC, MybLUC, and cyclin ALUC (the cyclin A luciferase reporter gene) for the numbers of separate transfections indicated. Data are shown as means ± SEMs.
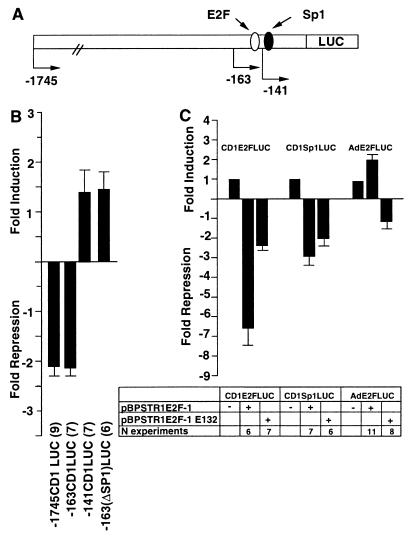
Two proximal regions of the transfected human cyclin D1 gene promoter are required for full repression by E2F-1.
The region of the cyclin D1 promoter required for regulation by E2F-1 was determined in JEG-3 cells by using a series of 5′ promoter deletions (Fig. 3A). Overexpression of E2F-1 by the vector pBPSTR1, which had inhibited cyclin D1 protein levels and CD1K activity, repressed the −1745CD1LUC reporter twofold (Fig. 3B). Deletion from −1745 to −163 did not affect repression of the cyclin D1 promoter. Deletion of the region between −163 and −141, which deletes the consensus E2F site, abolished E2F-1-mediated repression, resulting in a promoter fragment that was modestly induced by E2F-1 (Fig. 3B). In the initial description of the human cyclin D1 promoter, sequences homologous to an Sp1 binding site had been identified at −124 (47). Recent studies have demonstrated that E2F-1 is capable of regulating promoter activity through Sp1 binding sites (26, 35, 63). In order to examine the possible role of the Sp1-like sequences in E2F-1-mediated repression of the promoter, these sequences were deleted within the context of the −163-bp fragment (−163ΔSp1LUC) (Fig. 3B). Deletion of the Sp1 site abolished the repression of the cyclin D1 promoter (Fig. 3B). Together these studies suggest that the E2F binding site and the Sp1 site are required for full repression of the cyclin D1 promoter by E2F-1.
FIG. 3.
Repression of the cyclin D1 promoter by E2F-1. (A) Schematic representation of the cyclin D1 promoter, indicating the presence of the DNA sequences resembling E2F and Sp1 binding sites. (B) The wild-type or mutant pBPSTR1–E2F-1 expression vector was cotransfected with the −1745CD1LUC reporter. The expression vector (300 to 600 ng) was transfected with −1745CD1LUC (4.8 μg) or an equal amount of each of the other 5′ promoter constructs. Data are means ± SEMs for the numbers of separate transfections indicated. (C) The heterologous reporter constructs consisting of the cyclin D1 E2F site (CD1E2FLUC), the cyclin D1 Sp1 binding site (CD1Sp1LUC), and the adenovirus E2F site (AdE2FLUC) were transfected with expression plasmids encoding wild-type or mutant E2F-1 expression plasmids. Each result is shown as the mean fold repression or induction ± SEM for the number of separate experiments indicated, with comparison normalized for the effect of the empty expression vector cassette.
To determine whether the E2F or Sp1-like sequences were regulated by E2F-1, cotransfection experiments were conducted. Reporter plasmids encoding the cyclin D1 E2F site (CD1E2FLUC) or the cyclin D1 Sp1 site (CD1Sp1LUC) were examined with expression vectors encoding either wild-type E2F-1 or the E2F-1 DNA binding-defective mutant E132. Overexpression of E2F-1 by the vector pBPSTR1 repressed the cyclin D1 E2F reporter plasmid 6.5-fold (Fig. 3C). pBPSTR1–E132 repressed the cyclin D1 E2F reporter only twofold, again suggesting the E2F-1 binding domain was required for full repression. The cyclin D1 Sp1 site was repressed threefold by overexpression of E2F-1 (Fig. 3C). Overexpression of the DNA binding-defective mutant CMV–E2F-1 E132, however, did not abolish the repression (Fig. 3C). In contrast, the luciferase reporter construct containing the adenovirus E2F site (AdE2FLUC) was induced by pBPSTR1–E2F-1, and the induction was abolished by mutation of the DNA binding domain (Fig. 3C).
CMV–E2F-1 also repressed cyclin D1 E2F reporter activity sevenfold (mean ± standard error of the mean [SEM], 7.4 ± 1.8; n = 10) (data not shown). CMV–E2F-1 E132 did not significantly repress the cyclin D1 E2F reporter. The cyclin D1 Sp1 site reporter was repressed fourfold (mean ± SEM, 4.05 ± 0.4; n = 6) by CMV–E2F-1 (data not shown). Together these studies indicate that both the cyclin D1 E2F and the Sp1-like sequences are able to convey negative regulation in the presence of E2F-1.
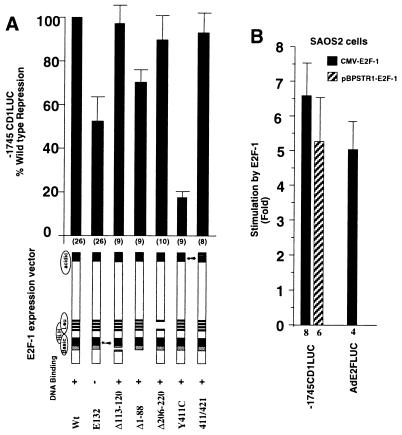
The pRB binding domain, the DNA binding domain, and the amino terminus of E2F-1 are involved in full repression of the cyclin D1 promoter.
In separate experiments we examined in further detail the domains of E2F-1 required for repression of the cyclin D1 promoter. A series of E2F-1 mutant expression plasmids was assessed in conjunction with the −1745CD1LUC reporter (Fig. 4A). The data are expressed relative to the repression of the wild-type E2F-1 expression plasmid (100%). Deletion of the DNA binding domain (E132) reduced repression by 50%. Deletion of the basic region (Δ113-120) did not significantly reduce repression of the −1745CD1LUC reporter. The amino terminus of E2F-1 has recently been shown to regulate activity through an Sp1 site (63). Deletion of the amino terminus (E2F-1 Δ1-88) reduced repression of −1745CD1LUC activity by 27% (Fig. 4A). The deletion of the leucine zipper region (Δ206-220) maintained at least 90% repression of the promoter, suggesting that this region was dispensable for the repression function. The E2F-1 mutant Y411C binds DNA but is defective in pRB- and p107-dependent function (7); in this experiment, it was severely defective in repression function, which was less than 20% that of the wild type (Fig. 4A). The E2F-1 double point mutant 411/421, which has previously been shown to be selectively defective in overcoming p107, while maintaining wild-type pRB repressor function (7), exhibited wild-type repression of cyclin D1, suggesting that the p107 repressor function was not required for repression of the cyclin D1 promoter. Together these studies indicate that the pRB binding and DNA binding domains of E2F-1 are involved in full repression of the cyclin D1 promoter. In addition, the amino-terminal region, which was involved in repression through an Sp1 site, was also required for full repression (63).
FIG. 4.
Repression of the cyclin D1 promoter by E2F-1 requires the pRB interactive domain and the amino terminus. (A) The wild-type (Wt) or mutant CMV–E2F-1 expression vectors were cotransfected with the −1745CD1LUC reporter. Expression vector (300 to 600 ng) was transfected with −1745CD1LUC (4.8 μg). Data are means ± SEMs for the numbers of separate transfections indicated. (B) The reporter constructs −1745CD1LUC and AdE2FLUC were transfected into the pRB-defective cell line SAOS2. Cotransfections were conducted with expression vectors encoding E2F-1 (CMV–E2F-1 or pBPSTR1–E2F-1). The induction of the reporters by E2F-1 is shown as the mean ± SEM for the number of separate transfections indicated.
Because these studies suggested that pRB binding was required for E2F-1 inhibition of cyclin D1 promoter activity, we assessed the effect of E2F-1 on cyclin D1 promoter activity in the SAOS2 osteosarcoma cell line, which is functionally pRB deficient. E2F-1, overexpressed from either the cytomegalovirus expression vector or the tetracycline-regulated expression vector pBPSTR1, induced the cyclin D1 promoter five- to sixfold (Fig. 4B). The reporter construct containing the E2F sequences from the AdE2 gene was also induced fivefold by overexpression of E2F-1 (Fig. 4B). These studies suggest that the ability of E2F-1 to repress the cyclin D1 promoter is cell type dependent and may depend upon the presence of the pRB protein.
Induction of the cyclin D1 gene promoter by pRB.
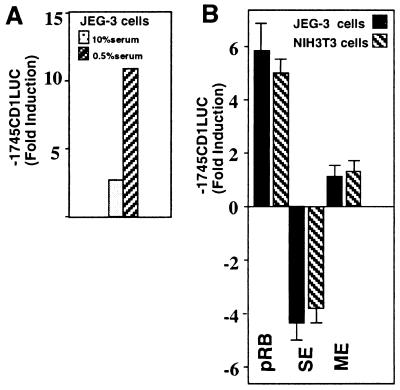
The transfection experiments using the E2F-1 mutants implicated the pRB binding domain of E2F-1 in negative regulation of cyclin D1 promoter activity, as the plasmid E2F-1 Y411C is defective in pRB interactions and failed to repress the cyclin D1 promoter. In order to examine the mechanism by which pRB regulates cyclin D1 levels, transient expression studies were carried out with wild-type and mutant pRB expression plasmids in conjunction with the −1745CD1LUC reporter construct. A carboxy-terminal fragment of pRB referred to as RB-SE has previously been shown to act as a dominant negative inhibitor of pRB function (74). The carboxy-terminal region of pRB is not conserved with p107; therefore, the dominant negative activity of the RB-SE vector is thought to be preferential or specific for inhibition of pRB function. pRB activated the cyclin D1 promoter five- to sevenfold (mean; Fig. 5). The activation of the cyclin D1 promoter by pRB was greater in 0.5% serum than in 10% serum (Fig. 5A). In randomly cycling JEG-3 or NIH 3T3 cells, overexpression of RB-SE inhibited basal cyclin D1 promoter activity, whereas overexpression of the extreme pRB carboxy terminus (amino acids 835 to 928) (RB-ME) did not affect cyclin D1 promoter activity (Fig. 5B). These results are consistent with those of previous studies demonstrating that cyclin D1 protein levels are reduced in cell lines deficient in pRB (4, 49) and that inhibition of gene expression by E2F-1 correlates with activation by pRB.
FIG. 5.
pRB activation of the cyclin D1 promoter requires the pocket binding domain. (A) The −1745CD1LUC reporter was cotransfected with expression vectors encoding pRB into JEG-3 cells. The data shown are from a representative experiment and have been adjusted for the effect of the empty vector cassette. (B) Data are the means ± SEMs from four separate transfections in which the ratio of pRB expression plasmid to reporter was 0.25:1.
In order to determine whether the induction of cyclin D1 promoter activity by pRB involved the E2F and Sp1 sequences, cotransfection experiments were conducted with the pRB expression vector (phRbc-SVE) (20). The −163CD1LUC reporter was induced 2-fold by pRB (1.9 ± 0.2; n = 11); however, the −141CD1LUC reporter was also induced 1.5-fold (1.5 ± 0.3; n = 11), and the −163ΔSp1LUC reporter was induced 1.4-fold (1.4 ± 0.2; n = 12) (data not shown). These studies suggest that the minimal pRB-responsive sequences are located within the proximal promoter. pRB is capable of regulating gene expression through interacting with a variety of different transcription factors, including c-Myc and Ets-related proteins (67), both of which have been shown to regulate cyclin D1 through proximal promoter sequences.
E2F-4 activates the cyclin D1 promoter.
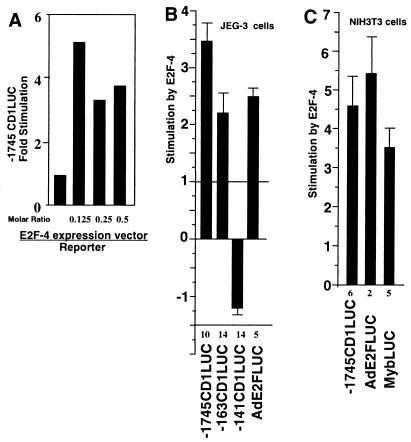
In order to determine how the different E2F proteins regulate cyclin D1 promoter activity, transient expression studies were performed with JEG-3 cells. The −1745-bp cyclin D1 LUC reporter was activated 3.5-fold by E2F-4 in JEG-3 cells (Fig. 6A and B). The induction of the cyclin D1 promoter by E2F-4 was sustained to −163 bp; however, deletion from −163 to −141 abolished induction by E2F-4 (Fig. 6B). E2F-4 also activated the AdE2 viral E2F site 2.5-fold in JEG-3 cells (Fig. 6B). In order to determine whether regulation of the cyclin D1 promoter by E2F-4 was a common feature in other cell types, studies were performed with NIH 3T3 and CHO cells. The −1745 cyclin D1 promoter was activated 4.5-fold by E2F-4 in NIH 3T3 cells (Fig. 6C) and 5-fold by E2F-4 in CHO cells (data not shown). These results were compared directly with the effect of E2F-4 on either the viral AdE2FLUC or the MybLUC reporter. E2F-4 induced the AdE2 reporter 5.5-fold and induced the Myb reporter 3.6-fold in NIH 3T3 cells (Fig. 6C). Together these studies demonstrate that in normally cycling cells, E2F-4 is capable of activating the cyclin D1 promoter, and that full induction requires sequences between −163 and −141 bp.
FIG. 6.
E2F-4 activates cyclin D1 promoter activity. JEG-3 (A and B) and NIH 3T3 (C) cells were transfected with the reporter plasmid −1745CD1LUC, −163CD1LUC, −141CD1LUC, AdE2FLUC, AdE2FmLUC, or MybLUC. Cotransfections were conducted with the E2F-4 wild-type expression plasmid. Fold effect is shown normalized for the effect of the expression vector cassette. Data in panels B and C are means ± SEMs for the numbers of experiments indicated.
The proximal E2F-1 repressor site of the cyclin D1 promoter binds Sp1–Sp3 and Sp4 proteins.
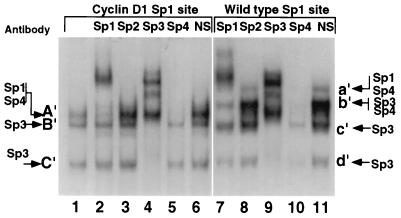
Because the Sp1 site governed a component of E2F-regulated cyclin D1 expression, the nature of the protein complexes binding to this region was assessed. Comparison was made between the Sp1-like sequences from the cyclin D1 promoter and a wild-type Sp1 site. Three complexes (A′ through C′) were formed (Fig. 7, lane 1) with the cyclin D1 Sp1 site. Band A′ was supershifted with the Sp1 antibody (Fig. 7, lane 2), bands B′ and C′ were shifted with the Sp3 antibody (Fig. 7, lane 4) and band A′ was abolished by the Sp4 antibody (Fig. 7, lane 5). None of the bands were shifted by either the Sp2 antibody (Fig. 7, lane 3), the E2F antibodies, or antibodies to pRB, p107, or p130 (data not shown). The wild-type Sp1 site formed four bands (a′ through d′) with mobilities related to, but different from, those of the complexes binding the cyclin D1 Sp1 site. Band a′ was shifted with Sp1 antibody (Fig. 7, lane 7), bands c′ and d′ were shifted with the Sp3 antibody (Fig. 7, lane 9), and bands a′ and b′ were reduced with the Sp4 antibody (Fig. 7, lane 10).
FIG. 7.
The proximal E2F-1 repressor element of the cyclin D1 promoter binds Sp proteins. The γ-32P-labelled cyclin D1 Sp1-like sequence (lanes 1 to 6) or the canonical Sp1 site (lanes 7 to 11) was incubated with JEG-3 cell nuclear extracts and specific antibodies to the Sp proteins or equal amounts of unrelated antibody (NS) as indicated above each lane. Arrows indicate the predicted proteins constituting the bands identified through supershifting or inhibition of DNA binding.
The distal E2F site of the cyclin D1 promoter binds E2F–DP-1.
In order to examine the proteins binding to this region of the cyclin D1 promoter distal E2F-1 repressor site in JEG-3 cells, nuclear extracts were incubated with the cyclin D1 E2F site or with the wild-type AdE2 E2F site (Fig. 8A), and results were compared.
FIG. 8.
The cyclin D1 promoter E2F-like sequences compete for nuclear binding with the adenovirus E2F site. (A) The γ-32P-labelled viral AdE2 E2F site probe (lanes 1 to 4) or cyclin D1 E2F site (lanes 5 to 8) was incubated with JEG-3 cell nuclear extracts and a 100-fold molar excess of cold double-stranded specific competitor for the AdE2 (Ad) or cyclin D1 (CD) E2F site or an equimolar amount of mutant AdE2 E2F site competitor (NS). Arrows indicate specific bands competed by double-stranded cognate competitor. (B) The viral AdE2 probe was incubated with nuclear extracts and antibodies (Ab) as indicated or control serum (NS). The antibodies used were KH95 (lane 2), C-20 (lane 3), KH20 (lane 4), L-20 (lane 5), C-20 (lane 6), C-108 (lane 7), PAb419 (lane 8), XZ55 (the pRB antibody) (lane 9), SD15 (lane 10), SD6 (lane 11), C-20 (lane 12), and WTH1 (lane 13).
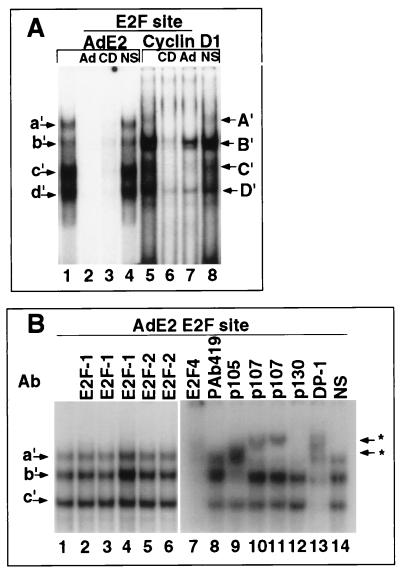
The four nuclear complexes (a′ through d′) binding the AdE2 E2F probe were competed by either the wild-type AdE2 E2F probe or the cyclin D1 E2F probe (Fig. 8A, lanes 1 to 4). The cyclin D1 E2F probe also bound four nuclear complexes (A′ through D′) with mobilities similar to those of the complexes bound by the AdE2 E2F site (Fig. 8A, lane 5). The cyclin D1 E2F site binding complexes were competed by either an excess (100-fold) of cyclin D1 E2F probe or the wild-type AdE2 E2F probe but not by mutant AdE2 E2F sequences. The AdE2 E2F site competed most efficiently for its cognate binding site (Fig. 8A; compare lanes 2 and 3), and the cyclin D1 E2F site competed most efficiently for binding to its cognate binding site (Fig. 8A; compare lanes 6 and 7).
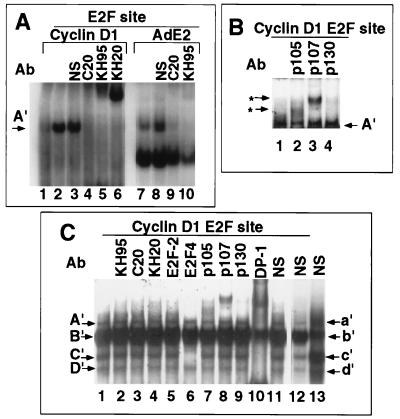
In order to establish the binding characteristics of the E2F proteins in JEG-3 cell extracts, EMSA using the E2F site from the viral AdE2a promoter and specific supershifting antibodies were performed. The E2F-1 and E2F-2 antibodies did not affect the complex (Fig. 8B, lanes 2 to 6). The E2F-4 antibody affected bands a′, b′, and c′ (Fig. 8B, lane 7). Complex b′ was shifted with the pRB antibody (Fig. 8B, lane 9), and band a′ was shifted by the addition of the p107 antibodies (Fig. 8B, lanes 10 and 11). The p130 antibody inhibited binding to band a′ (Fig. 8, lane 12), and the DP-1 antibody reduced bands b′ and c′ and a component of band a′ (Fig. 8B, lane 13). The control sera did not affect any of the complexes (Fig. 8B, lane 14). These data suggest that in these cells, band a′ of the AdE2 E2F site consists of p130, p107, E2F-4 and DP-1; band b′ consists of pRB–DP-1–E2F-4; and band c′ consists of free E2F-4–DP-1. The finding that E2F-4 forms part of a pRB complex is consistent with the findings of recent studies (14, 46, 68) showing pRB–E2F-4 complexes binding to the AdE2 E2F site.
In order to determine whether E2F-1 was capable of binding the cyclin D1 E2F site, in vitro-translated E2F-1 was incubated with the γ-32P-labelled cyclin D1 E2F site (Fig. 9A, lane 2) and the binding pattern was compared with that found with equal amounts of unprogrammed lysate (Fig. 9A, lane 1). A specific band, designated A′, was formed in the presence of E2F-1 (Fig. 9A). Supershifts were conducted with antibodies to E2F-1 (C20, KH95, and KH20). The addition of the E2F-1-specific antibodies supershifted the complex binding to the cyclin D1 E2F site (Fig. 9A, lanes 4 to 6), indicating that band A′ contains E2F-1 protein. A comparison was made with the adenovirus E2F site. In vitro-translated E2F-1 bound the adenovirus E2F site, and the E2F-1-specific antibodies supershifted the complex (Fig. 9A, lanes 9 and 10). Supershifts were then conducted with the cyclin D1 E2F site by using JEG-3 cell nuclear extracts and antibodies to pRB, p107, and p130 (Fig. 9B, lanes 2 to 4, and Fig. 9C). The E2F-1 and E2F-2 antibodies, however, did not affect the complexes binding the cyclin D1 site (Fig. 9C, lanes 2 to 5). The E2F-4 antibody shifted the complex binding band C′ (Fig. 9C, lane 6). The pRB antibody induced a partial shift (Fig. 9C, lane 7), as did the p107 and p130 antibodies (Fig. 9C, lanes 8 and 9). On shorter exposures, pRB, p107, and p130 appeared to be derived from band B′ (data not shown). The DP-1 antibody shifted components of bands A′, B′, C′, and D′ (Fig. 9C, lane 10). These studies suggest that the cyclin D1 E2F site is capable of binding E2F-1 and that in JEG-3 cell nuclear extracts, band A′ contains DP-1 and band B′ contains DP-1 with contributions from pRB, p107, and p130. Band C′ contains E2F-4–DP-1, and band D′ contains DP-1. The additional constituents contributing to binding of the cyclin D1 E2F site remain to be determined.
FIG. 9.
Binding of E2F–DP-1 proteins to the cyclin D1 promoter E2F-like sequences. (A) The γ-32P-labelled cyclin D1 E2F site (lanes 1 to 6) or the viral AdE2 E2F site (lanes 7 to 10) was incubated with either unprogrammed in vitro translate (lane 1) or in vitro translate programmed with the E2F-1 cDNA (lanes 2 to 10). Antibodies specific for E2F-1 were added as indicated above the lanes and include KH95, C-20, and KH20. The A′ band (arrow) indicates specific E2F-1 binding. NS, pAB419. (B and C) JEG-3 cell nuclear extracts were incubated with the γ-32P-labelled cyclin D1 E2F site either alone (lanes 1) or with the addition of specific antibodies. (B) Antibodies to pRB (also called p105 [XZ55]) (lane 2), p107 (SD6) (lane 3), and p130 (C-30) (lane 4). Asterisks indicate supershifted complexes. (C) Antibodies added for supershift were KH95 (lane 2), C-20 (lane 3), KH20 (lane 4), LLF2 (lane 5), C-108 (lane 6), XZ55 (lane 7), SD6 (lane 8), C-20 (lane 9), and WTH1 (lane 10). Lane 11, control serum. NS, pAB419.
DISCUSSION
In these studies overexpression of E2F-1 inhibited CD1K activity, cyclin D1 protein levels, and cyclin D1 promoter activity. The inhibition of cyclin D1 promoter activity by E2F-1 required the amino terminus, the DNA binding domain, and the pRB binding domain, while overexpression of pRB induced cyclin D1 promoter activity. Two regions of the cyclin D1 promoter were required for full repression by E2F-1. The proximal site of the cyclin D1 promoter repressed by E2F-1 bound Sp1. The E2F-1 amino terminus and DNA binding domain were both required for regulation through Sp1 binding sites, and these domains of E2F-1 were required for full repression of the cyclin D1 promoter (63). The cyclin D1 Sp1 binding site resembles a component of a cell cycle-regulated repressor element found in the cdc2 and cdc25C gene promoters (82). Although the distal cyclin D1 promoter element required for repression by E2F-1 bound in vitro-translated E2F-1, E2F-4–DP-1 and pRB–p107–p130 were identified as the binding proteins in cell nuclear extracts, suggesting that the effect of E2F-1 may be mediated indirectly. In conjunction with the known role of cyclin D1 in promoting the phosphorylation of pRB and, subsequently, the release of E2F-1, these findings suggest that cyclin D1 is a downstream target of E2F-1 repression. E2F-1 contributes to the inhibition of CD1K activity and expression through inhibition of cyclin D1 promoter activity. E2F-1 inhibition of CD1K activity may limit the overexpression of E2F-1, which can induce cellular apoptosis (22, 56, 57).
Our finding that the cyclin D1 promoter is transcriptionally induced by pRB is consistent with previous observations that the adenovirus E1A and simian virus 40 large T antigen, which antagonize the action of pRB, reduce cyclin D1 mRNA abundance (5, 37, 66). Cyclin D1 protein levels were reduced in pRB-deficient cell lines (4, 41, 49), and cyclin D1 mRNA levels were increased in cells transfected with a pRB expression vector (20). Although the pRB binding domain of E2F-1 was required for repression of cyclin D1, deletion of the cyclin D1 E2F site did not abolish pRB activation (71), and pRB is not required for all E2F-1-dependent functions (13, 62). The Sp1 transactivation function can be induced by pRB (29), and the pRB binding domain of E2F-1 was required for regulation of Sp1-dependent activity (63). In addition to Sp1, pRB is also capable of interacting with other transcription factors, including Ets and Myc proteins, which regulate activity of the proximal cyclin D1 promoter (1, 55). The mechanisms by which pRB regulates the cyclin D1 promoter remain to be fully determined.
E2F-4 induced the cyclin D1 promoter and bound the cyclin D1 E2F site. Our data are consistent with a model in which the release of E2F-4 in G0-G1 induces cyclin D1 expression early in G1, leading to the induction of CD1K activity. The phosphorylation of pRB, induction of E2F-1, and release of E2F-1 are associated with the induction of cyclin E and cyclin A (78) and with induction of G1-phase progression (25). The induction of cyclin D1 by E2F-4 is consistent with the temporal profile of induction of E2F-4 during the cell cycle. Free E2F-4 is the major complex induced during early G1-phase transition stimulated by serum addition. In previous studies, the induction of free E2F-4 preceded the induction of E2F-1 gene expression, and the G1-phase regulatory genes induced by E2F-4 previously remained to be determined (46). The differential regulation of cyclin D1 promoter activity by E2F-4 and E2F-1 is the first description of differential regulation of a target promoter by these two proteins, although E2F-4 and E2F-1 were previously shown to exhibit several functional differences. E2F-4 mRNA is expressed throughout the cell cycle and was induced by serum in human keratinocytes early in G1, preceding the induction of E2F-1 by several hours (14, 60). E2F-4 forms a major component of the E2F binding complex in quiescent cells and a component of the major free E2F activity found in cycling cells (68). E2F-4, unlike E2F-1, displays high affinity for p130 in quiescent cells and for p107 in cycling cells (68). In vitro, E2F-4 selectively binds to the pocket domains of p130 and p107 but binds the pRB pocket poorly (60). E2F-4 overcomes a p130-mediated G1 arrest more efficiently than a pRB-induced G1 blockade (68). As the major form of E2F released in response to mitogens early in G1, the release of E2F-4 would be predicted to occur coincident with the induction of cyclin D1. The distinguishable temporal profiles of activity of E2F-4 and E2F-1 suggest that they may have distinct regulatory functions during the cell cycle, conveyed through differential regulation of target genes, which in these studies, include the cyclin D1 gene.
The present studies provide some insight into the mechanisms by which E2F-1 inhibits the cyclin D1 promoter. E2F-1-mediated inhibition of the cyclin D1 promoter involved two nuclear protein binding regions. The Sp1 binding site of the cyclin D1 promoter was required for full repression by E2F-1. In recent studies the Sp1 binding site of the cdc25C gene was shown to be an important component of a cell-cycle-dependent negative regulatory sequence (82). The repressor element in the cdc25C gene was referred to as part of a cell cycle-dependent element and was conserved with the cdc2 gene promoter (82). An Sp1 site has recently been shown to convey regulation by E2F-1 (26, 35, 63), and E2F-1 protein is capable of binding Sp1 in vitro (26, 35). In the present study, the DNA binding and pRB binding domains of E2F-1 were both required for regulation of the cyclin D1 promoter, and these domains of E2F-1 were also required for regulation of Sp1-dependent promoter activity (63). Because Sp1 is capable of binding E2F-1 but not E2F-4, the interaction with Sp1 in trans may be important in the differential regulation of the cyclin D1 promoter by E2F-1 and E2F-4. The involvement of both the E2F and Sp1 binding sites in negative regulation of the cyclin D1 promoter by E2F-1 suggests the possibility that E2F-1–Sp1 complexes together have a specific role in repression of the cyclin D1 promoter. Although E2F-1 was capable of binding to the cyclin D1 promoter E2F site, E2F-4 and not E2F-1 bound the E2F site in EMSA with cell nuclear extracts. These findings suggest that either (i) E2F-1 binding occurs to the cyclin D1 E2F site and was not detected in these assays, or (ii) the mechanism of repression through the E2F site is indirect and independent of DNA binding. In this regard, it is possible that under alternate conditions, E2F-1 binding to the cyclin D1 promoter may be detected by using cell extracts, either in specific phases of the cell cycle or during differentiation. Alternatively, in vivo footprinting (81) or deoxycholate release may identify E2F-1 interactions that are not detected by EMSA (62). Repression of cyclin D1 by E2F-1 through an indirect mechanism could involve the induction of an additional factor by E2F-1 that represses the cyclin D1 promoter through the E2F site, or the repression may be mediated through competition for a positive regulator of transcription, such as E2F-4, by heterodimerizing with a critical partner required for activation, such as DP-1.
The distal cyclin D1 promoter sequence required for transcriptional repression by E2F-1 bound E2F-4–DP-1–p107. The E2F proteins, in conjunction with their heterodimeric partners, the DPs, regulate gene transcription, at least in part, by binding the E2F site. Like the cyclin D1 gene, several other genes induced during the G1-S phase of the cell cycle are also targets of repression by E2F-1. The E2F site in the promoter of the E2F-1 and the B-Myb genes convey negative regulation by E2F-1 during S-phase transition (24, 33). The E2F binding site of the cyclin D1 promoter is homologous with but distinguishable from the E2F sites in the promoters of the B-Myb and cyclin A genes (19). In our studies, the E2F-1 DNA binding domain was required for full repression of the cyclin D1 promoter. The E2F-1 mutant Y411C was defective in repression of the cyclin D1 promoter but maintained wild-type transactivation of the AdE2 E2F site (7), indicating that the repression function and transactivation properties of E2F-1 are dissociable. Within the carboxy terminus of E2F-1, distinct mutations interfered with cyclin D1 repressor function of E2F-1, as the mutant E2F-1 411/421 repressed the cyclin D1 promoter but the E2F-1 Y411C mutant was defective in repression. In this regard, it is of interest that dissociable domains of E2F-1 are also involved in the induction of apoptosis and the ability to transactivate the AdE2 enhancer E2F site (22, 56). The E2F-1 DNA binding domain was required for E2F-1-mediated apoptosis, but the transactivation function of E2F-1 was dispensable (22, 56). Together these studies suggest that the transactivation function of E2F-1 can be separated from other properties of E2F-1, including the ability to repress cyclin D1 promoter activity and the ability to induce apoptosis. It has been proposed that cyclin D1 may contribute to either the induction (16, 31, 79) or the inhibition of apoptosis (32), depending upon the cell type. DT40 lymphoma cells, in which cyclin D1 was selectively deleted, were more prone to radiation-induced apoptosis, and the reintroduction of cyclin D1 inhibited the increase in apoptotic cells (32). In MEFs, rodent neuronal cells, and mouse mammary epithelial cells, cyclin D1 overexpression sensitizes cells to apoptosis-inducing agents (16, 31, 79). Our studies support the notion that different domains of the E2F-1 protein regulate distinct functions, and further studies will be required to determine whether the inhibition of cyclin D1 and the induction of apoptosis correlate in these cells.
The nuclear protein complexes binding the cyclin D1 E2F site were similar to yet distinguishable from the E2F site of the AdE2 gene. E2F-4 and DP-1 were the main components of the AdE2 E2F site, and the cyclin D1 promoter E2F site bands A′, B′, and C′ contained DP-1, and band C′ contained E2F-4. In conjunction with E2F-4–DP-1, both the cyclin D1 E2F site and the E2F site of the AdE2 gene promoter bound pRB, p130, and p107. The heterodimeric partner bound to DP-1 at the cyclin D1 E2F site, which contributes to the main band, B′, remains to be determined. The main cyclin D1 E2F binding complex, band B′, did not contain E2F-4, yet this band was the primary complex induced during serum- or growth factor-induced G1 phase progression (71). Recent studies have identified the presence of an E2F-DP complex which contained a novel species of E2F, which is induced during S phase (46). The role of this currently unidentified DP-binding protein, likely in the E2F family, remains to be determined and may be important in the differential regulation of the cyclin D1 gene by E2F-1 and E2F-4.
Distinguishable combinatorial interactions between the E2F proteins and their pocket proteins, the binding of these complexes to the cyclin promoters, and the induction of the activity of these complexes by the cyclins together likely contribute to the distinct temporal profiles of cyclin gene regulation during the cell cycle. In previous studies, E2F-4 binding activity to the AdE2 E2F site shifted during normal G1-phase progression from a p130 complex to p107 and pRB (46). p130 becomes hyperphosphorylated and decreases in abundance as cells pass through G1 phase and p107 abundance increases (68, 78). Expression of cyclin D1 is induced during G1-phase progression, while expression of cyclin A increases later in S phase as E2F-1 levels increase and as cyclin D1 mRNA levels decrease (37, 65). Like the cyclin D1 promoter E2F site, the cyclin A promoter variant E2F site, which is involved in cyclin E-induced expression of cyclin A, also binds E2F-4–p107 proteins (78). Both cyclin D1 (61) and cyclin E (78) can induce the cyclin A promoter. Induction of the cyclin A promoter by cyclin E is associated with unaltered p107–E2F binding at the cyclin A promoter E2F site, whereas cyclin D1 overexpression leads to dissociation of the p107–E2F complex (78). The cyclin A promoter was induced 10-fold by E2F-1 in NIH 3T3 cells (18a), but cyclin D1 promoter activity was repressed by E2F-1. The differential regulation of the cyclin D1 and cyclin A promoters by E2F-1 and the differential effects of cyclin D1 and cyclin E abundance on the composition of the proteins binding to the cyclin D1 and cyclin E promoters may contribute to the specificity in regulation of the G1 cyclin promoters during cell cycle progression. With the delineation of the functional E2F and Sp1 binding sites of the cyclin D1 promoter herein, the contribution of the subtle differences in E2F binding sites between the G1-phase cyclin promoters to differences in cell cycle-regulated transcription can now be examined in detail. Distinct E2F-pocket protein complexes may convey distinct transcriptional effects at a given E2F site, and the nature of these complexes is likely important in the differential regulation of the G1-phase cyclins and the E2F genes.
ACKNOWLEDGMENTS
We are grateful to E. Harlow, D. Heimbrook, R. Weinberg, M. Pagano, G. Draetta, D. Livingston, W. Krek, D. Ginsberg, R. Watson, J. Wang, W. Kaelin, J. Nevins, L. Bandara, and N. La Thangue for plasmids and antibodies, and to D. Gebhard for assistance with flow cytometry analysis. We thank L. Yamasaki for helpful discussions and the MEF derived from the E2F-1 KO mice.
This work was supported in part by grant 94-27 from the American Cancer Society (Illinois Division, Inc.) and 1R29CA70897-01 and R01CA75503 from the National Cancer Institute (to R.G.P.). G.W. was supported in part by a Travel Fellowship from the Aichi Health Promotion Foundation, the Owari Kenyu-kai, and the Takasu Foundation. A.R. was supported by a P.F. Sobotka postgraduate scholarship from the University of Western Australia. G.V. was a recipient of a C. J. Martin postdoctoral fellowship from the Australian National Health and Medical Research Council and an AMRAD Corporation postdoctoral award. Work at the Albert Einstein College of Medicine was also supported by Cancer Center Core National Institutes of Health grant 5-P30-CA13330-26.
G. Watanabe and C. Albanese contributed equally to this work.
REFERENCES
- 1.Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell R G. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- 2.Asano M, Nevins J R, Wharton R P. Ectopic E2F expression induces S phase and apoptosis in Drosophila imaginal discs. Genes Dev. 1996;10:1422–1432. doi: 10.1101/gad.10.11.1422. [DOI] [PubMed] [Google Scholar]
- 3.Bandara L R, Buck V M, Zamanian M, Johnston L H, La Thangue N B. Functional synergy between DP-1 and E2F-1 in the cell cycle-regulatory transcription factor DRTF1/E2F. EMBO J. 1993;12:4317–4324. doi: 10.1002/j.1460-2075.1993.tb06116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartkova J, Lukas J, Muller H, Lutzhoft D, Strauss M, Bartek J. Cyclin D1 protein expression and function in human breast cancer. Int J Cancer. 1994;57:351–361. doi: 10.1002/ijc.2910570311. [DOI] [PubMed] [Google Scholar]
- 4a.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Buchou T, Kranenburg O, van Dam H, Roelen D, Zantema A, Hall F L, van der Eb A J. Increased cyclin A and decreased cyclin D levels in adenovirus 5 E1A-transformed rodent cell lines. Oncogene. 1993;8:1765–1773. [PubMed] [Google Scholar]
- 6.Caceres A, Binder L I, Payne M R, Bender P, Rebhun L, Steward O. Differential subcellular localization of tubulin and the microtubule-associated protein MAP2 in brain tissue as revealed by immunocytochemistry with monoclonal hybridoma antibodies. J Neurosci. 1983;4:394–410. doi: 10.1523/JNEUROSCI.04-02-00394.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cress W D, Johnson D G, Nevins J R. A genetic analysis of the E2F1 gene distinguishes regulation by Rb, p107, and adenovirus E4. Mol Cell Biol. 1993;13:6314–6325. doi: 10.1128/mcb.13.10.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeCaprio J A, Ludlow J W, Figge J, Shew J Y, Huang C M, Lee W H, Marsilio E, Paucha E, Livingston D M. The retinoblastoma-susceptibility gene product becomes phosphorylated in multiple stages during cell cycle entry and progression. Proc Natl Acad Sci USA. 1992;89:1795–1798. doi: 10.1073/pnas.89.5.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeGregori J, Kowalik T, Nevins J R. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeGregori J, Leone G, Ohtani K, Mirone A, Nevins J R. E2F-1 accumulation bypasses a G1 arrest resulting from the inhibition of G1 cyclin-dependent kinase activity. Genes Dev. 1995;9:2873–2887. doi: 10.1101/gad.9.23.2873. [DOI] [PubMed] [Google Scholar]
- 11.Ewen M E, Sluss H K, Sherr C J, Livingston D M, Matsushime H. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell. 1993;73:487–497. doi: 10.1016/0092-8674(93)90136-e. [DOI] [PubMed] [Google Scholar]
- 12.Field S J, Tsai F-Y, Kuo F, Zubiaga A M, Kaelin W G J, Livingston D M, Orkin S H, Greenberg M E. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- 13.Flemington E K, Speck S H, Kaelin W G J. E2F-1-mediated transactivation is inhibited by complex formation with the retinoblastoma susceptibility gene product. Proc Natl Acad Sci USA. 1993;90:6914–6918. doi: 10.1073/pnas.90.15.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ginsberg D, Vairo G, Chittenden T, Xiao Z-X, Xu G, Wydner K L, DeCaprio J A, Lawrence J B, Livingston D M. E2F4, a new member of the E2F transcription family, interacts with p107. Genes Dev. 1994;8:2665–2679. doi: 10.1101/gad.8.22.2665. [DOI] [PubMed] [Google Scholar]
- 15.Goodrich D W, Wang N P, Quian Y-W, Lee E Y-H P, Lee W H. The retinoblastoma gene product regulates progression through the G1 phase of the cell cycle. Cell. 1991;67:293–302. doi: 10.1016/0092-8674(91)90181-w. [DOI] [PubMed] [Google Scholar]
- 16.Han E K-H, Begeman M, Sgambato A, Soh J-W, Doki Y, Xing W-Q, Liu W, Weinstein I B. Increased expression of cyclin D1 in a murine mammary epithelial cell line induces p27kip1, inhibits growth and enhances apoptosis. Cell Growth Differ. 1996;7:699–710. [PubMed] [Google Scholar]
- 17.Helin K, Harlow E, Fattaey A. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol Cell Biol. 1993;13:6501–6508. doi: 10.1128/mcb.13.10.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helin K, Lees J A, Vidal M, Dyson N, Harlow E, Fattaey A. A cDNA encoding a pRB-binding protein with properties of the transcription factor E2F. Cell. 1992;70:337–350. doi: 10.1016/0092-8674(92)90107-n. [DOI] [PubMed] [Google Scholar]
- 18a.Henglein, B. Unpublished data.
- 19.Henglein B, Chenivesse X, Wang J, Eick D, Brechot C. Structure and cell cycle-regulated transcription of the human cyclin A gene. Proc Natl Acad Sci USA. 1994;91:5490–5494. doi: 10.1073/pnas.91.12.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinds P W, Mittnacht S, Dulic V, Arnold A, Reed S I, Weinberg R A. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- 21.Hofmann F, Livingston D M. Differential effects of cdk2 and cdk3 on the control of pRB and E2F function during G1 exit. Genes Dev. 1996;10:851–861. doi: 10.1101/gad.10.7.851. [DOI] [PubMed] [Google Scholar]
- 22.Hsieh J-K, Frersdorf S, Kouzarides T, Martin K, Lu X. E2F1-induced apoptosis requires DNA binding but not transactivation and is inhibited by the retinoblastoma protein through direct interaction. Genes Dev. 1997;11:1840–1852. doi: 10.1101/gad.11.14.1840. [DOI] [PubMed] [Google Scholar]
- 23.Ivey-Hoyle M, Conroy R, Huber H E, Goodhart P J, Oliff A, Heimbrook D C. Cloning and characterization of E2F-2, a novel protein with the biochemical properties of transcription factor E2F. Mol Cell Biol. 1993;13:7802–7812. doi: 10.1128/mcb.13.12.7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson D G, Ohtani K, Nevins J. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 25.Johnson D G, Schwarz J K, Cress W D, Nevins J R. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 26.Karlseder J, Rotheneder H, Wintersberger E. Interaction of Sp1 with growth- and cell cycle-regulated transcription factor E2F. Mol Cell Biol. 1996;16:1659–1667. doi: 10.1128/mcb.16.4.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khleif S N, DeGregori J, Yee C L, Otterson G A, Kaye F J, Nevins J R, Howley P M. Inhibition of cyclin D-CDK4/CDK6 activity is associated with an E2F-mediated induction of cyclin kinase inhibitor activity. Proc Natl Acad Sci USA. 1996;93:4350–4354. doi: 10.1073/pnas.93.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khochbin S, Chabanas A, Albert P, Albert J, Lawrence J J. Application of bromodeoxyuridine incorporation measurements to the determination of cell distribution within the S phase of the cell cycle. Cytometry. 1988;9:499–503. doi: 10.1002/cyto.990090516. [DOI] [PubMed] [Google Scholar]
- 29.Kim S-J, Onwuta U S, Lee Y I, Li R, Botchan M R, Robbins P D. The retinoblastoma gene product regulates Sp1-mediated transcription. Mol Cell Biol. 1992;12:2455–2463. doi: 10.1128/mcb.12.6.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kowalik T F, DeGregori J, Schwarz J K, Nevins J R. E2F1 overexpression in quiescent fibroblasts leads to induction of cellular DNA synthesis and apoptosis. J Virol. 1995;69:2491–2500. doi: 10.1128/jvi.69.4.2491-2500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kranenburg O, van der Eb A J, Zantema A. Cyclin D1 is an essential mediator of apoptotic neuronal cell death. EMBO J. 1996;15:46–54. [PMC free article] [PubMed] [Google Scholar]
- 32.Lahti J M, Li H, Kidd V J. Elimination of cyclin D1 in vertebrate cells leads to an altered cell cycle phenotype which is rescued by overexpression of murine cyclins D1, D2, or D3 but not by a mutant cyclin D1. J Biol Chem. 1997;272:10859–10869. doi: 10.1074/jbc.272.16.10859. [DOI] [PubMed] [Google Scholar]
- 33.Lam E W-F, Watson R J. An E2F-binding site mediates cell-cycle regulated repression of mouse B-myb transcription. EMBO J. 1993;12:2705–2713. doi: 10.1002/j.1460-2075.1993.tb05932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.La Thangue N B. E2F and the molecular mechanisms of early cell-cycle control. Biochem Soc Trans. 1996;24:54–59. doi: 10.1042/bst0240054. [DOI] [PubMed] [Google Scholar]
- 35.Lin S-Y, Black A R, Kostic D, Pajovic S, Hoover C N, Azizkhan J C. Cell cycle-regulated association of E2F1 and Sp1 is related to their functional interaction. Mol Cell Biol. 1996;16:1668–1675. doi: 10.1128/mcb.16.4.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lukas J, Bartkova J, Rohde M, Strauss M, Bartek J. Cyclin D1 is dispensable for G1 control in retinoblastoma gene-deficient cells independently of cdk4 activity. Mol Cell Biol. 1995;15:2600–2611. doi: 10.1128/mcb.15.5.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lukas J, Pagano M, Staskova Z, Draetta G, Bartek J. Cyclin D1 protein oscillates and is essential for cell cycle progression in human tumor cell lines. Oncogene. 1994;9:707–718. [PubMed] [Google Scholar]
- 38.Lukas J, Petersen B O, Holm K, Bartek J, Helin K. Deregulated expression of E2F family members induces S-phase entry and overcomes p16INK4A-mediated growth suppression. Mol Cell Biol. 1996;16:1047–1057. doi: 10.1128/mcb.16.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lybarger L, Dempsey D, Franek K J, Chervenak R. Rapid generation and flow cytometric analysis of stable GFP-expressing cells. Cytometry. 1996;25:211–220. doi: 10.1002/(SICI)1097-0320(19961101)25:3<211::AID-CYTO2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 40.Macleod K, Hu Y, Jacks T. Loss of Rb activates both p53-dependent and independent cell death pathways in the developing mouse nervous system. EMBO J. 1996;15:6178–6188. [PMC free article] [PubMed] [Google Scholar]
- 41.Marhin W W, Hei Y-J, Chen S, Jiang Z, Gallie B L, Phillips R A, Penn L Z. Loss of pRb and Myc activation co-operate to suppress cyclin D1 and contribute to transformation. Oncogene. 1996;12:43–52. [PubMed] [Google Scholar]
- 42.Matsushime H, Quelle D E, Shurtleff S A, Shibuya M, Sherr C J, Kato J-Y. D-type cyclin-dependent kinase activity in mammalian cells. Mol Cell Biol. 1994;14:2066–2076. doi: 10.1128/mcb.14.3.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsushime H, Roussel M F, Ashmun R A, Sherr C J. Colony-stimulating factor 1 regulates novel cyclins during the G1 phase of the cell cycle. Cell. 1991;65:701–713. doi: 10.1016/0092-8674(91)90101-4. [DOI] [PubMed] [Google Scholar]
- 44.Melillo R M, Helin K, Lowy D R, Schiller J T. Positive and negative regulation of cell proliferation by E2F-1: influence of protein level and human papillomavirus oncoproteins. Mol Cell Biol. 1994;14:8241–8249. doi: 10.1128/mcb.14.12.8241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mittnacht S, Lees J A, Desai D, Morgan D O, Weinberg R A. Distinct subpopulations of the retinoblastoma protein show a distinct pattern of phosphorylation. EMBO J. 1994;13:118–127. doi: 10.1002/j.1460-2075.1994.tb06241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moberg K, Starz M A, Lees J A. E2F-4 switches from p130 to p107 and pRB in response to cell cycle reentry. Mol Cell Biol. 1996;16:1436–1449. doi: 10.1128/mcb.16.4.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Motokura T, Arnold A. The PRAD1/cyclin D1 proto-oncogene: genomic organization, 5′ DNA sequence, and sequence of a tumor-specific rearrangement breakpoint. Genes Chromosomes Cancer. 1993;7:89–95. doi: 10.1002/gcc.2870070205. [DOI] [PubMed] [Google Scholar]
- 48.Motokura T, Bloom T, Kim H G, Juppner H, Ruderman J V, Kronenberg H M, Arnold A. A novel cyclin encoded by a bcl1-linked candidate oncogene. Nature. 1991;350:512–515. doi: 10.1038/350512a0. [DOI] [PubMed] [Google Scholar]
- 49.Muller H, Lukas J, Schneider A, Warthoe P, Bartek J, Eilers M, Strauss M. Cyclin D1 expression is regulated by the retinoblastoma protein. Proc Natl Acad Sci USA. 1994;91:2945–2949. doi: 10.1073/pnas.91.8.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell-cycle. EMBO J. 1992;11:961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paulus W, Baur I, Boyce F M, Breakefield X O, Reeves S A. Self-contained, tetracycline-regulated retroviral vector system for gene delivery to mammalian cells. J Virol. 1996;70:62–67. doi: 10.1128/jvi.70.1.62-67.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pestell R G, Albanese C, Hollenberg A, Jameson J L. Transcription of the human chorionic gonadotropin α and β genes is negatively regulated by c-jun. J Biol Chem. 1994;269:31090–31096. [PubMed] [Google Scholar]
- 53.Pestell R G, Albanese C, Lee R J, Watanabe G, Moran E, Johnson J, Jameson J L. A potential role for cell-cycle control proteins in regulation of the cAMP-responsive glycoprotein hormone α-subunit gene. Cell Growth Differ. 1996;7:1337–1344. [PubMed] [Google Scholar]
- 54.Pestell R G, Albanese C, Watanabe G, Johnson J, Eklund N, Lastowiecki P, Jameson J L. Epidermal growth factor and c-Jun act via a common DNA regulatory element to stimulate transcription of the ovine P-450 cholesterol side chain cleavage (CYP11A1) promoter. J Biol Chem. 1995;270:18301–18308. doi: 10.1074/jbc.270.31.18301. [DOI] [PubMed] [Google Scholar]
- 55.Philipp A, Schneider A, Väsrik I, Finke K, Xiong Y, Beach D, Alitalo K, Eilers M. Repression of cyclin D1: a novel function of MYC. Mol Cell Biol. 1994;14:4032–4043. doi: 10.1128/mcb.14.6.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Phillips A C, Bates S, Ryan K M, Helin K, Vousden K H. Induction of DNA synthesis and apoptosis are separable functions of E2F-1. Genes Dev. 1997;11:1853–1863. doi: 10.1101/gad.11.14.1853. [DOI] [PubMed] [Google Scholar]
- 57.Qin X, Livingston D M, Kaelin W G, Jr, Adams P D. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc Natl Acad Sci USA. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Resnitzky D, Gossen M, Bujard H, Reed S I. Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol Cell Biol. 1994;14:1669–1679. doi: 10.1128/mcb.14.3.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Resnitzky D, Reed S I. Different roles for cyclins D1 and E in regulation of the G1-to-S transition. Mol Cell Biol. 1995;15:3463–3469. doi: 10.1128/mcb.15.7.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sardet C, Vidal M, Cobrinik D, Geng Y, Onufryk C, Chen A, Weinberg R A. E2F-4 and E2F-5, two members of the E2F family, are expressed in the early phases of the cell cycle. Proc Natl Acad Sci USA. 1995;92:2403–2407. doi: 10.1073/pnas.92.6.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schulze A, Zerfass K, Spitkovsky D, Berges J, Middendorp S, Jansen-Durr P, Henglein B. Cell cycle regulation of cyclin A gene transcription is mediated by a variant E2F binding site. Proc Natl Acad Sci USA. 1995;92:11264–11268. doi: 10.1073/pnas.92.24.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sellers W, Novitch B G, Miyake S, Heith A, Otterson G A, Kaye F J, Lassar A B, Kaelin W G. Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation and suppress tumor cell growth. Genes Dev. 1997;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shin E K, Tevosian S G, Yee A S. The N-terminal region of E2F-1 is required for transcriptional activation of a new class of target promoter. J Biol Chem. 1996;271:12261–12268. doi: 10.1074/jbc.271.21.12261. [DOI] [PubMed] [Google Scholar]
- 64.Singh P, Wong S H, Hong W. Overexpression of E2F-1 in rat embryo fibroblasts leads to neoplastic transformation. EMBO J. 1994;13:3329–3338. doi: 10.1002/j.1460-2075.1994.tb06635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Slansky J, Li Y, Kaelin W G, Farnham P J. A protein synthesis-dependent increase in E2F1 mRNA correlates with growth regulation of the dihydrofolate reductase promoter. Mol Cell Biol. 1993;13:1610–1618. doi: 10.1128/mcb.13.3.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spitkovsky D, Steiner P, Lukas J, Lees E, Pagano M, Schulze A, Joswig S, Picard D, Tommasino M, Eilers M, Jansen-Durr P. Modulation of cyclin gene expression by adenovirus E1A in a cell line with E1A-dependent conditional proliferation. J Virol. 1994;68:2206–2214. doi: 10.1128/jvi.68.4.2206-2214.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taya Y. RB kinases and RB-binding proteins: new points of view. Trends Biochem Sci. 1997;22:14–17. doi: 10.1016/s0968-0004(96)10070-0. [DOI] [PubMed] [Google Scholar]
- 68.Vairo G, Livingston D M, Ginsberg D. Functional interaction between E2F-4 and p130: evidence for distinct mechanisms underlying growth suppression by different RB family members. Genes Dev. 1995;9:869–881. doi: 10.1101/gad.9.7.869. [DOI] [PubMed] [Google Scholar]
- 69.Watanabe G, Howe A, Lee R J, Albanese C, Shu I-W, Karnezis A, Zon L, Kyriakis J, Rundell K, Pestell R G. Induction of cyclin D1 by simian virus 40 small tumor antigen. Proc Natl Acad Sci USA. 1996;93:12861–12866. doi: 10.1073/pnas.93.23.12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Watanabe G, Lee R J, Albanese C, Rainey W E, Batlle D, Pestell R G. Angiotensin II (AII) activation of cyclin D1-dependent kinase activity. J Biol Chem. 1996;271:22570–22577. doi: 10.1074/jbc.271.37.22570. [DOI] [PubMed] [Google Scholar]
- 71.Watanabe, G., and R. G. Pestell. Unpublished data.
- 72.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 73.Weintraub S J, Prater C A, Dean D C. Retinoblastoma protein switches the E2F site from positive to negative element. Nature. 1992;358:259–261. doi: 10.1038/358259a0. [DOI] [PubMed] [Google Scholar]
- 74.Welch P J, Wang J Y J. Disruption of retinoblastoma protein function by coexpression of its C pocket fragment. Genes Dev. 1995;9:31–46. doi: 10.1101/gad.9.1.31. [DOI] [PubMed] [Google Scholar]
- 75.Wu X, Levine A J. p53 and E2F-1 cooperate to mediate apoptosis. Proc Natl Acad Sci USA. 1994;91:3802–3806. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu G, Livingston D M, Krek W. Multiple members of the E2F transcription factor family are the products of oncogenes. Proc Natl Acad Sci USA. 1995;92:1357–1361. doi: 10.1073/pnas.92.5.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yamasaki L, Jacks T, Bronson R, Goillot E, Harlow E, Dyson N J. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell. 1996;85:537–548. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
- 78.Zerfass-Thome K, Schulze A, Zwerschke W, Vogt B, Helin K, Bartek J, Henglein B, Jansen-Durr P. p27KIP1 blocks cyclin E-dependent transactivation of cyclin A gene expression. Mol Cell Biol. 1997;17:407–415. doi: 10.1128/mcb.17.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou P, Jiang W, Weghorst C M, Weinstein I B. Overexpression of cyclin D1 enhances gene amplification. Cancer Res. 1996;56:36–39. [PubMed] [Google Scholar]
- 80.Zhu L, van der Heuvel S, Helin K, Fattaey A, Ewen M, Livingston D, Dyson N, Harlow E. Inhibition of cell proliferation by p107, a relative of the retinoblastoma protein. Genes Dev. 1993;7:1111–1125. doi: 10.1101/gad.7.7a.1111. [DOI] [PubMed] [Google Scholar]
- 81.Zwicker J, Liu N, Engeland K, Lucibello F C, Muller R. Cell cycle regulation of E2F site occupation in vivo. Science. 1996;271:1595–1597. doi: 10.1126/science.271.5255.1595. [DOI] [PubMed] [Google Scholar]
- 82.Zwicker J, Lucibello F C, Wolfraim L A, Gross C, Truss M, Engeland K, Muller R. Cell-cycle regulation of the cyclin A, cdc25C and cdc2 genes is based on a common mechanism of transcriptional repression. EMBO J. 1995;14:4514–4522. doi: 10.1002/j.1460-2075.1995.tb00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]