Abstract
Reaper (RPR), HID, and GRIM activate apoptosis in cells programmed to die during Drosophila development. We have previously shown that transient overexpression of RPR in the lepidopteran SF-21 cell line induces apoptosis and that members of the inhibitor of apoptosis (IAP) family of antiapoptotic proteins can inhibit RPR-induced apoptosis and physically interact with RPR through their BIR motifs (D. Vucic, W. J. Kaiser, A. J. Harvey, and L. K. Miller, Proc. Natl. Acad. Sci. USA 94:10183–10188, 1997). In this study, we found that transient overexpression of HID and GRIM also induced apoptosis in the SF-21 cell line. Baculovirus and Drosophila IAPs blocked HID- and GRIM-induced apoptosis and also physically interacted with them through the BIR motifs of the IAPs. The region of sequence similarity shared by RPR, HID, and GRIM, the N-terminal 14 amino acids of each protein, was required for the induction of apoptosis by HID and its binding to IAPs. When stably overexpressed by fusion to an unrelated, nonapoptotic polypeptide, the N-terminal 37 amino acids of HID and GRIM were sufficient to induce apoptosis and confer IAP binding activity. However, GRIM was more complex than HID since the C-terminal 124 amino acids of GRIM retained apoptosis-inducing and IAP binding activity, suggesting the presence of two independent apoptotic motifs within GRIM. Coexpression of IAPs with HID stabilized HID levels and resulted in the accumulation of HID in punctate perinuclear locations which coincided with IAP localization. The physical interaction of IAPs with RPR, HID, and GRIM provides a common molecular mechanism for IAP inhibition of these Drosophila proapoptotic proteins.
Programmed cell death, or apoptosis, is a genetically regulated mechanism that plays an important role in development and homeostasis in vertebrates and invertebrates (25). Three Drosophila melanogaster genes, reaper (rpr), head involution defective (hid), and grim, were identified within the chromosomal 75C1,2 region as inducers of apoptosis in cells destined to die during embryogenesis (5, 12, 30). In Drosophila, ectopic expression of rpr, hid, and grim, which encode a 65-amino-acid polypeptide (RPR), 410-amino-acid polypeptide (HID), and 138-amino-acid polypeptide (GRIM), respectively, induces cell death through activation of caspase(s) (5, 12, 15, 19, 29). rpr and grim transcripts accumulate in a defined subset of Type II neurons prior to the onset of apoptosis (20), and rpr and hid seem to act synergistically to induce programmed cell death during development of the Drosophila central nervous system midline (32). RPR, HID, and GRIM share no extensive homology, although there is resemblance between the sequences of the N-terminal 14 amino acids of these three proteins (5). The functional significance of this short homologous region remains ambiguous as mutations of the most conserved amino acid residues (28), or elimination of the whole region (4), only partially diminish the proapoptotic activity of RPR upon overexpression in insect cell cultures.
Two types of antiapoptotic proteins, caspase inhibitors and members of the inhibitors of apoptosis (IAPs) family, are known to block apoptosis in insect cells. Baculovirus P35, which normally blocks apoptosis during baculovirus replication in lepidopteran SF-21 cells (7), is a broad-spectrum caspase inhibitor (3, 31) that blocks apoptosis induced by ectopic expression of RPR, HID, or GRIM in Drosophila (5, 12, 15). The founder members of the IAP family, baculovirus Op-IAP and Cp-IAP, were identified by their ability to substitute functionally for the baculovirus P35 (2, 8, 9). In addition, baculovirus IAPs also block apoptosis induced by RPR (28), Doom (13), FADD (28), actinomycin D (8), and UV irradiation (17) in SF-21 cells. Both Op-IAP and Cp-IAP are able to block the activation of Sf-caspase-1 during baculovirus infection (23) and Cp-IAP can partially block RPR-dependent apoptosis in Drosophila developing eyes. Drosophila-encoded IAPs, D-IAP1 and D-IAP2, block HID- and RPR-induced apoptosis in Drosophila developing eyes (15). Members of the IAP family are characterized by the presence of at least one and usually two or three tandem baculovirus IAP repeat (BIR) motifs located at the amino-terminal and central portions of the protein, and most of them have a carboxy-terminal RING finger motif. Both baculovirus and Drosophila IAPs inhibit RPR-induced apoptosis in SF-21 cells (27, 28), physically interact with RPR through their BIR region (27), and alter its subcellular localization to punctate perinuclear locations which coincide with IAP localization (27). When expressed alone, RPR levels decline rapidly in the cells undergoing RPR-induced apoptosis, but coexpression with IAPs appears to stabilize RPR (27).
IAP homologs have also been identified in mammals. NAIP, one of the human IAPs, is linked to the progression of spinal muscular atrophy, which involves neuronal cell death (22). Two other mammalian IAPs, c-IAP1 and c-IAP2, bind tumor necrosis factor receptor 2 (TNFR-2)-associated factor 2 (TRAF-2) through their BIR domains (21). c-IAP1 also appears to be a component of the TNFR-1 signaling complex and may exert antiapoptotic activity by modifying signaling through TRAF-related pathways (24). Human X-chromosome-linked IAP (X-IAP) binds and directly inhibits two members of the caspase family, caspase-3 and caspase-7, in vitro (10). Expression of the most recently identified IAP homolog, human Survivin, correlates with oncogenic transformation (1). Mammalian IAPs block apoptosis in several mammalian cell lines induced by a variety of stimuli (1, 6, 11, 16, 26).
In this study we examine the induction of apoptosis by HID and GRIM in SF-21 cells. We demonstrate that baculovirus and Drosophila IAPs inhibit HID- and GRIM-induced apoptosis and physically interact with HID and GRIM through their BIR motifs. We also determine the regions of HID and GRIM which can promote apoptosis and interact with IAPs. We define a short region at the N terminus of both proteins with proapoptotic and IAP binding activities.
MATERIALS AND METHODS
Cell line and plasmid construction.
Spodoptera frugiperda (Lepidoptera: Noctuidae) IPLB-SF-21 (SF-21) cells were maintained in TC-100 medium (Gibco BRL, Gaithersburg, Md.) supplemented with 10% fetal bovine serum (Intergen, Purchase, N.Y.) and 0.26% tryptose broth as previously described (18). All the plasmids except for the pKV-based plasmids used in in vitro binding studies are derived from pHSP70PLVI+CAT, a plasmid expressing the chloramphenicol acetyltransferase (cat) gene under the Drosophila hsp70 promoter (8). Plasmids expressing nontagged Op-iap, Cp-iap, D-iap1, D-iap2, p35, or cat, Flag-tagged rpr, Op-iap, or cat, and HA(Epi)-tagged Op-iap, Op-BIR, Op-RING, D-iap2, D-iap1-BIR, p35, or cat were previously described (8, 27, 28). In Flag-D-iap1-BIR, the cat from pHSP70PLVI+CAT is substituted with Flag-epitope tag N-terminally fused to D-iap1-BIR; in Epi-Cp-iap, Epi-D-iap1, or Epi-D-iap1-RING, cat is replaced with HA.11-epitope tag N-terminally fused to Cp-iap, D-iap1, or D-iap1-RING. The plasmid expressing hid cDNA (pHSP70PLVI+HID) was made by replacing cat from pHSP70PLVI+CAT with hid cDNA. The sequence encoding the 410-amino-acid hid open reading frame (ORF) was amplified from pHSP70PLVI+HID by PCR with Pfu polymerase (Stratagene) and the primers NHID (GAAGATCTACAATGGCCGTGCCCTTTTAT) plus CHID (ATATCCCGGGTTAACGTCTCCTGCGCTTTCAT). The resulting product was digested with BglII and XmaI and subcloned into the BglII and PstI sites of pHSP70PLVI+CAT to generate pHSP70PLVI+HID-ORF. In hid-Flag or hid-Epi, cat from pHSP70PLVI+CAT was replaced with hid-ORF C-terminally fused to either a FlagHis6-tag or an HA.11His6-tag. pHSHID-EpiHisVI+ was digested with XhoI and religated to generate pHSHIDΔ(38-335)-EpiHisVI+, which encodes the first 37 and last 74 amino acids of the 410-amino-acid hid ORF. Sequences encoding the first 37 and last 74 amino acids of the hid ORF were amplified by PCR with Pfu polymerase and the primers NHID plus MCTAGHID (GGACTAGTTGCGCTCGAGGGAAGTGG) and MNHID (GAAGATCTACAATGGCCTCGAGCAGCAGCAATAAT) plus CTAGHID (GGACTAGTTCGCGCCGCAAAGAAGC), respectively. The resulting products were digested with BglII and SpeI and subcloned into the BglII and SpeI sites of pHSHID-EpiHisVI+ to generate pHSHID(1-37)-EpiHisVI+ and pHSHID(336-410)-EpiHisVI+. Sequence encoding amino acids 2 to 53 of the 250-amino-acid cat ORF was amplified by PCR with Pfu polymerase and the primers NECOCAT (CGGAATTCGAGAAAAAAATCACTGG) plus CECOCAT (CGGAATTCCGGATGAGCATTC). The resulting product was digested with EcoRI and inserted into the EcoRI site of pHSHID(1-37)-EpiHisVI+ between the end of hid-encoding sequence and the start of the C-terminal HA.11His6-tag to generate pHSHID(1-37)C-EpiHisVI+. pHSCAT(1-53)-EpiHisVI+ was generated by subcloning the sequence encoding the first 53 amino acids of cat ORF fused to C-terminal HA.11His6-tag from pHSP70PLVI+HID(1-37)C-Epi. pHSCAT(1-53)HID(37-410)-EpiHisVI+ was generated by subcloning the sequence encoding the amino acids 37 to 410 of hid ORF into the EcoRI site of pHSCAT(1-53)-EpiHisVI+. Sequence encoding the amino acids 15 to 410 of hid ORF was amplified by PCR with Pfu polymerase (Stratagene) and the primers NMUTHID (GAAGATCTACAATGGTAGCGTCGAGTTCATC) plus CTAGHID. The resulting product was digested with BglII and SpeI and subcloned into the BglII and SpeI sites of pHSHID-EpiHisVI+ to generate pHSHIDΔ(2-14)-EpiHisVI+. The plasmid expressing grim cDNA (pHSP70PLVI+GRIM) was made by replacing cat from pHSP70PLVI+CAT with grim cDNA. The sequence encoding the 138-amino-acid grim ORF was amplified from pHSP70PLVI+GRIM by PCR with Pfu polymerase (Stratagene) and the primers NGRIM (CGGAGATCTACAATGGCCATCGCCTATTTC) plus CGRIM (CGGACTAGTTTAGTTCTCCTTGGAGGT). The resulting product was digested with BglII and SpeI and subcloned into the BglII and SpeI sites of pHSP70PLVI+CAT to generate pHSP70PLVI+GRIM-ORF. In grim-Flag or grim-Epi, cat from pHSP70PLVI+CAT was replaced with grim-ORF C-terminally fused to either a FlagHis6-tag or an HA.11His6-tag. Sequences encoding the first 16 or the first 37 amino acids of the 138-amino-acid grim ORF were amplified by PCR with Pfu polymerase and the primers NGRIM plus CDGRIM (CGACTAGTTCTGGCCAACAATTGGGC) or NGRIM plus MCTAGRIM (CGACTAGTTGCAGCAGCTGTTGCAGTC), respectively. The resulting products were digested with BglII and SpeI and subcloned into the BglII and SpeI sites of pHSHID(1-37)C-EpiHisVI+ to generate pHSGRIM(1-16)C-EpiHisVI+ and pHSGRIM(1-37)C-EpiHisVI+, respectively. Sequence encoding the amino acids 15 to 138 of the grim ORF was amplified by PCR with Pfu polymerase (Stratagene) and the primers NMUTGRIM (CGGGATCCACAATGGCCAGAAGCTATCAGC) plus CTAGRIM (CGGACTAGTGTTCTCCTTGGAGGTGGC). The resulting product was digested with BamHI and SpeI and subcloned into the BglII and SpeI sites of pHSHID-EpiHisVI+ to generate pHSGRIMΔ(2-14)-EpiHisVI+. In pKV-HID, pKV-GRIM, and pKV-Flag-D-IAP1-BIR, sequences encoding HID-Epi, GRIM with four additional methionines at the C terminus for the efficient labeling, and Flag-D-IAP1-BIR were subcloned into pKV vector (3) under control of the T7 promoter.
DNA fragmentation and viability assays.
SF-21 cells (105 per 35-mm-diameter tissue culture dish) were transfected with 2.5 μg of each of the indicated plasmids by Lipofectin-mediated transfection (Gibco BRL). The cells were heat shocked at 20 h posttransfection for 30 min at 42°C. DNA fragmentation and viability assays were described previously (8, 28).
In vivo binding assay, immunofluorescence microscopy, and immunoblot analysis.
All were performed as described previously (27).
In vitro binding assay.
The Flag-D-IAP1-BIR, 35S-labelled HID and 35S-labelled GRIM were obtained by in vitro transcription and translation by using a TNT T7-coupled reticulocyte lysate system (Promega). After translation, equivalent amounts of 35S-labelled HID or 35S-labelled GRIM were incubated with Flag-D-IAP1-BIR or empty vector (pKV) in the presence of anti-Flag affinity resin (Eastman) in Nonidet P-40 lysis buffer (27) for 4 h at 4°C with agitation. The resin was washed five times in Nonidet P-40 lysis buffer and boiled in sodium dodecyl sulfate (SDS)-sample buffer (18), and proteins were resolved by SDS-polyacrylamide gel electrophoresis and visualized by autoradiography.
RESULTS
HID and GRIM induce apoptosis in SF-21 cells.
To determine whether the Drosophila gene hid can induce apoptosis in SF-21 cells, we transiently expressed hid by transfecting the cells with a plasmid containing the entire hid cDNA under the control of the Drosophila hsp70 promoter and inducing expression by heat shocking at 20 h posttransfection. As early as 1 to 2 h after the induction of hid expression, cells began to exhibit membrane blebbing which became pronounced and was accompanied by apoptotic body formation by 8 h after induction (data not shown). Expression of hid cDNA also induced oligonucleosomal ladder formation 8 h after induction (Fig. 1B, lane 2) which was not observed in cells expressing the control cat gene (Fig. 1B, lane 1). We PCR amplified the sequence encoding only the 410-amino-acid hid ORF and expressed it under hsp70 promoter control. Transient expression of hid ORF induced cell death to the same extent as expression of the entire hid cDNA (Fig. 1B, lane 3). Next we generated constructs which express hid ORF with C-terminal HA.11-His6 (hid-Epi) or Flag-His6 (hid-Flag) tags (Fig. 1A). We tested these constructs for induction of apoptosis and found that the presence of the C-terminal tag did not affect the proapoptotic function of HID (Fig. 1B, lanes 4 and 5).
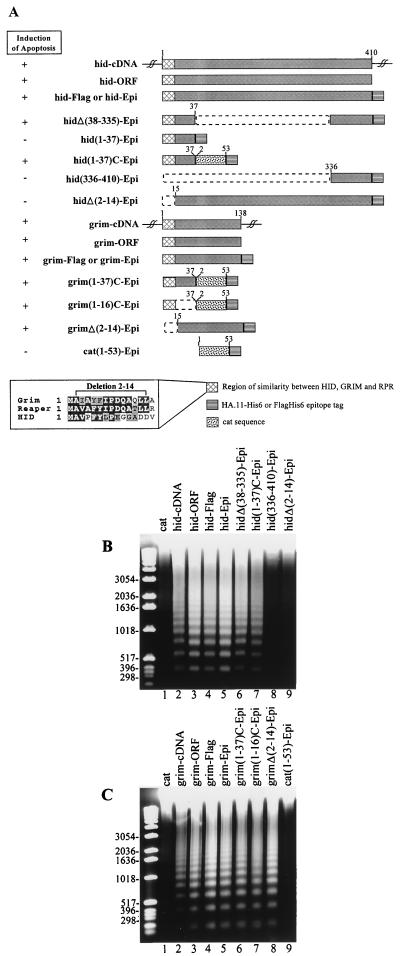
FIG. 1.
The abilities of hid and grim constructs to induce apoptosis in SF-21 cells. (A) Schematic representation of hid and grim constructs. Boxes represent coding regions, dashed lines indicate deleted regions, and single lines are untranslated sequences. Numbers on the top of the solid areas indicate amino acid position in the full-length protein. The ability of the construct to induce apoptosis is indicated by the plus or minus to the left. The abilities of hid (B) and grim (C) constructs to induce nucleolytic degradation of chromatin into oligonucleosomal ladder-sized fragments of DNA are shown. SF-21 cells were transfected with the control cat constructs (lanes 1 in panels B and C, and lane 9 in panel C) or with the hid and grim constructs indicated above each lane (lanes 2 to 9 in panel B and lanes 2 to 8 in panel C). Cellular DNA was harvested 8 h after induction and analyzed by agarose gel electrophoresis. Size markers (in base pairs) are indicated on the left.
To define the region within HID that is responsible for the induction of apoptosis, we constructed an in-frame deletion that fuses the first 37 and the last 74 amino acids of HID (Fig. 1A). This deletion, hidΔ(38-335)-Epi, induced apoptosis to the same level as hid-Epi (Fig. 1B, lane 6). In order to determine whether the first 37 or the last 74 amino acids of HID possess proapoptotic activity, we expressed the two regions independently [hid(1-37)-Epi and hid(336-410)-Epi, respectively] (Fig. 1A). However, neither of these regions induced apoptosis (Fig. 1B, lane 8; and data not shown). Immunoblot analysis revealed that hid(336-410)-Epi was expressed at detectable levels (see below), indicating that this region of HID was not sufficient to induce apoptosis. On the other hand, hid(1-37)-Epi could not be detected by immunoblot analysis (data not shown), suggesting that the expressed protein product was unstable. To increase the stability of hid(1-37)-Epi we generated a construct, hid(1-37)C-Epi (Fig. 1A), which expresses the first 37 amino acids of HID fused to the first 53 amino acids of CAT, an apoptotically neutral protein, and an HA.11-His6 tag. This construct expressed detectable levels of protein product (see below) and induced apoptosis (Fig. 1B, lane 7). To rule out any toxicity of the fusion protein, we made a construct expressing the first 53 amino acids of CAT fused to amino acids 37 to 410 of HID and an HA.11-His6 tag. This protein, CAT(1-53)HID(37-410)-Epi, did not induce apoptosis (data not shown). The N-terminal 37 amino acids of HID contain a region (amino acids 2 to 14) with similarity to the N terminus of RPR and GRIM (Fig. 1A). We, therefore, made a construct in which this region is deleted from hid-Epi, hidΔ(2-14)-Epi (Fig. 1A), and tested its ability to induce apoptosis. This deletion mutant did not induce apoptosis in SF-21 cells (Fig. 1B, lane 9) although it expressed detectable levels of protein product (see below). Thus, the first 14 amino acids of HID were required for the induction of apoptosis, and the first 37 amino acids were sufficient to induce apoptosis as long as they were stably expressed.
We investigated the ability of grim to induce apoptosis in SF-21 cells in the same way as we did for hid. grim cDNA, grim ORF, and C-terminally tagged versions of grim (grim-Epi and grim-Flag) (Fig. 1A) all induced apoptosis in SF-21 cells as documented by oligonucleosomal ladder formation (Fig. 1C, lanes 2 to 5) and membrane blebbing (data not shown). Constructs containing only the N-terminal 37 or 16 amino acids of GRIM fused to the first 53 amino acids of CAT and HA.11-His6 tag, grim(1-37)C-Epi and grim(1-16)C-Epi (Fig. 1A), also induced apoptosis (Fig. 1C, lanes 6 and 7) although the construct expressing the first 53 amino acids of CAT fused to HA.11-His6 tag (Fig. 1A), cat(1-53)-Epi, did not (Fig. 1C, lane 9). This suggested that the region of similarity among RPR, GRIM, and HID is sufficient to induce apoptosis as long as it is stably expressed. However, a construct in which the N-terminal 14 amino acids of GRIM were deleted, grimΔ(2-14)-Epi (Fig. 1A), still retained proapoptotic activity (Fig. 1C, lane 8) unlike the equivalent hid construct, hidΔ(2-14)-Epi. These results suggest that GRIM harbors another region, in addition to the 16 N-terminal amino acids, that possesses proapoptotic activity.
Inhibition of hid- and grim-induced apoptosis by p35 and iaps.
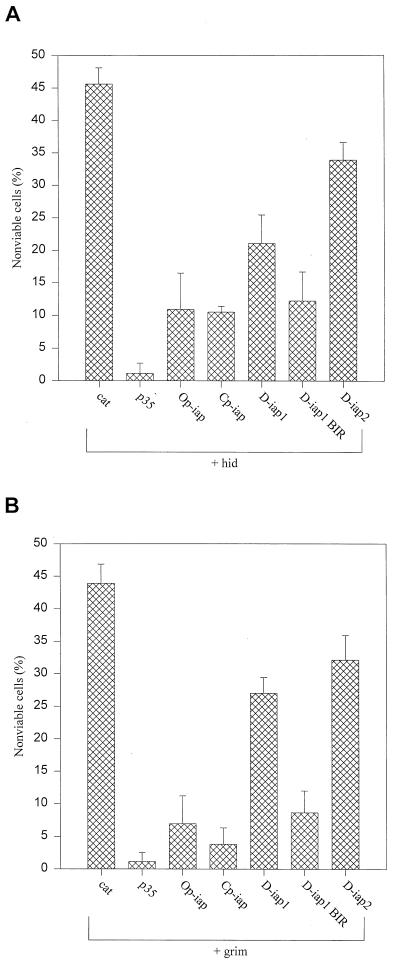
To assess the nature of the apoptotic pathway induced by hid and grim in SF-21 cells, we cotransfected cells with plasmids expressing hid-ORF or grim-ORF and several known antiapoptotic genes. Compared to cells transfected with cat gene alone, cotransfection of hid and cat and of grim and cat induced apoptosis in 46% (Fig. 2A) and 43% (Fig. 2B) of the cells, respectively. Cotransfection of hid or grim with a plasmid expressing the baculovirus gene p35, a general caspase inhibitor, almost completely inhibited hid- and grim-induced apoptosis (Fig. 2A and B). We also tested members of the iap class of antiapoptotic genes for their ability to block hid- and grim-induced apoptosis. Baculovirus Op-iap and Cp-iap and Drosophila D-iap1 and D-iap2 blocked apoptosis induced by hid and grim, although Drosophila iaps were not as effective as baculovirus iaps (Fig. 2A and B). The BIR region of D-iap1 was sufficient for the inhibition of hid- and grim-induced apoptosis and even more efficient than the full-length D-iap1 (Fig. 2A and B), which is in concordance with previously published data (15). Baculovirus and Drosophila iaps also blocked apoptosis induced by the above-described (Fig. 1A) deletion constructs of hid and grim as well as they blocked the full-length constructs (data not shown). In addition, we tested various members of the bcl-2 family (human bcl-2, human bcl-xL, Caenorhabditis elegans ced-9 and adenovirus E1B19K), but they exhibited little or no ability to block hid- and grim-induced apoptosis in SF-21 cells (data not shown).
FIG. 2.
Protection against hid- and grim-induced cell death in SF-21 cells by antiapoptotic genes. The cells were cotransfected with plasmids expressing hid (A) or grim (B) and either cat, p35, Op-iap, Cp-iap, D-iap1, D-iap1-BIR, or D-iap2. Cell viability was determined by trypan blue exclusion 8 h after heat shock. The results shown are relative to viability of cells transfected with the cat gene alone (set at 100% viability) and represent at least three independent experiments for each cotransfected combination. Standard deviations are indicated by the error bars.
IAPs physically interact with HID and GRIM.
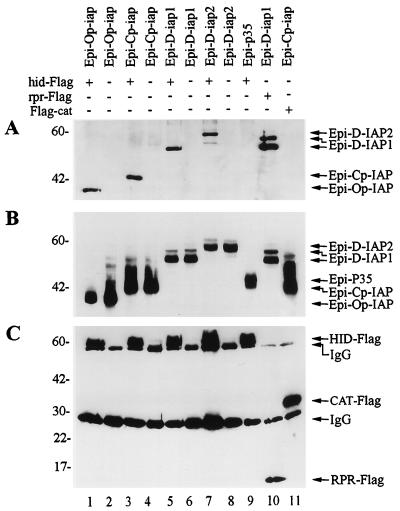
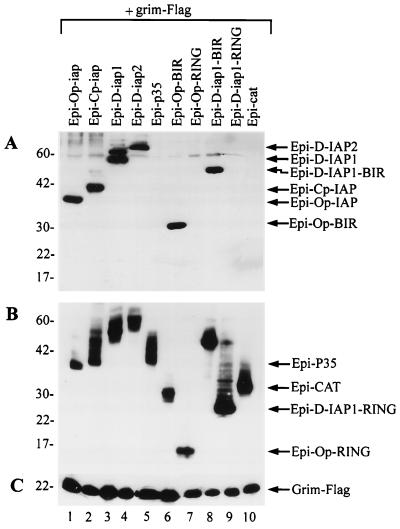
Because baculovirus and Drosophila IAPs physically interact with RPR and inhibit RPR-induced apoptosis (27), we investigated the possibility that baculovirus and Drosophila IAPs also interact with HID and GRIM. To do so, we first transiently coexpressed HID-Flag with HA-epitope-tagged versions of baculovirus IAPs, Epi-Op-IAP and Epi-Cp-IAP, or Drosophila IAPs, Epi-D-IAP1 and Epi-D-IAP2, and determined if anti-Flag antibodies could coimmunoprecipitate the IAP fusion proteins. As a negative control, we tested an HA-tagged version of the baculovirus P35 protein, Epi-P35, which blocked HID- and GRIM-induced apoptosis as well as the untagged version. We found that all IAPs coprecipitated with HID (Fig. 3A, lanes 1, 3, 5, and 7), but P35 did not (Fig. 3A, lane 9), although it was efficiently expressed (Fig. 3B, lane 9). Expression of the HA-tagged and Flag-tagged constructs in the cell lysates was confirmed for all transfections (Fig. 3B and C). Coprecipitation of Epi-D-IAP1 with RPR-Flag served as a positive control (Fig. 3A, lane 10), and coprecipitation of Epi-Cp-IAP with Flag-CAT served as an additional negative control (Fig. 3A, lane 11).
FIG. 3.
IAPs physically interact with HID. (A) SF-21 cells were transiently transfected with plasmids expressing HID-Flag, RPR-Flag, or Flag-CAT and indicated HA-epitope-tagged (Epi) genes. At 3 h after heat shock, aliquots of cell lysates were immunoprecipitated with the M2 anti-Flag monoclonal antibody resin. Coprecipitated HA-epitope-tagged constructs were detected by immunoblot analysis with the rabbit anti-HA.11 polyclonal antiserum. (B) Expression of the HA-epitope-tagged proteins was confirmed by immunoblot analysis with the mouse HA.11 monoclonal antibody of aliquots of the total cell lysates from panel A. (C) Expression of the HID-Flag, RPR-Flag, and Flag-CAT was confirmed by reprobing the membrane from panel A with the M2 anti-Flag antibody. The positions of the Flag- or HA-epitope-tagged proteins and monoclonal immunoglobulin G antibody are indicated by arrows on the right. Molecular mass markers (in kilodaltons) are shown on the left.
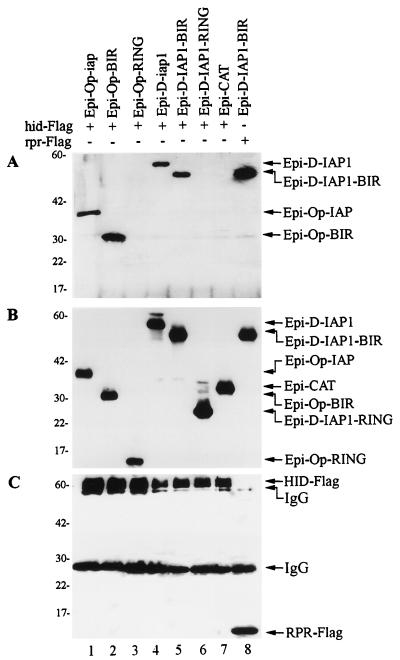
In order to determine which portion of IAPs possessed HID binding activity, we tested the ability of HA-tagged BIR and RING finger domains to individually bind HID-Flag. Epi-Op-BIR and Epi-D-IAP1-BIR coimmunoprecipitated with HID-Flag (Fig. 4A, lanes 2 and 5), but Epi-Op-RING and Epi-D-IAP1-RING did not (Fig. 4A, lanes 3 and 6). In control lanes, the HA-tagged version of bacterial CAT (Epi-CAT) did not coimmunoprecipitate with HID-Flag (Fig. 4A, lane 7), and Epi-D-IAP1-BIR coimmunoprecipitated with RPR-Flag as expected (Fig. 4A, lane 8). Expression of the HA-tagged and Flag-tagged constructs in the cell lysates was confirmed for all transfections (Fig. 4B and C).
FIG. 4.
HID interacts with the BIR region of IAPs. (A) SF-21 cells were transiently transfected with plasmids expressing HID-Flag or RPR-Flag and indicated HA-epitope-tagged (Epi) genes. At 3 h after heat shock, aliquots of cell lysates were immunoprecipitated with the M2 anti-Flag monoclonal antibody resin. Coprecipitated HA-epitope-tagged constructs were detected by immunoblot analysis with the rabbit anti-HA.11 polyclonal antiserum. (B) Expression of the HA-epitope-tagged proteins was confirmed by immunoblot analysis with the mouse HA.11 monoclonal antibody of aliquots of the total cell lysates from panel A. (C) Expression of the HID-Flag and RPR-Flag was confirmed by reprobing the membrane from panel A with the M2 anti-Flag antibody. The positions of the Flag- or HA-epitope-tagged proteins and monoclonal immunoglobulin G antibody are indicated by arrows on the right. Molecular mass markers (in kilodaltons) are shown on the left.
To investigate the possibility that IAPs interact with GRIM, we transiently coexpressed GRIM-Flag with HA-epitope-tagged versions of baculovirus IAPs, Epi-Op-IAP and Epi-Cp-IAP, or Drosophila IAPs, Epi-D-IAP1 and Epi-D-IAP2, and determined if anti-Flag antibodies could coimmunoprecipitate the IAP fusion proteins. We found that all expressed IAPs and the BIR region of IAPs coprecipitated with GRIM (Fig. 5A, lanes 1 to 4, 6 and 8), but the RING motifs of IAPs, P35, or CAT did not (Fig. 5A, lanes 5, 7, 9, and 10), although they were efficiently expressed (Fig. 5B, lanes 5 and 10). Expression of the HA-tagged and Flag-tagged constructs in the cell lysates was confirmed for all transfections (Fig. 5B and C).
FIG. 5.
IAPs physically interact with GRIM. (A) SF-21 cells were transiently transfected with plasmids expressing grim-Flag and indicated HA-epitope-tagged (Epi) constructs. At 3 h after heat shock, aliquots of cell lysates were immunoprecipitated with the M2 anti-Flag monoclonal antibody resin. Coprecipitated HA-epitope-tagged constructs were detected by immunoblot analysis with the rabbit anti-HA.11 polyclonal antiserum. (B) Expression of the HA-epitope-tagged proteins was confirmed by immunoblot analysis with the mouse HA.11 monoclonal antibody of aliquots of the total cell lysates from panel A. (C) Expression of GRIM-Flag was confirmed by immunoblot analysis with the M2 anti-Flag antibody of aliquots of the total cell lysates from panel A. The positions of the HA-epitope-tagged proteins and GRIM-Flag are indicated by arrows on the right. Molecular mass markers (in kilodaltons) are shown on the left.
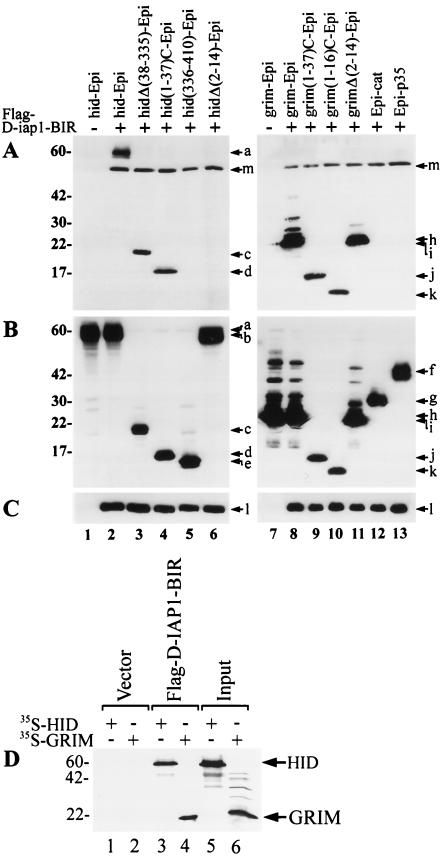
To determine which portion of HID and GRIM had IAP binding activity, we tested the ability of HA-tagged HID-ORF, HIDΔ(38-335), HID(1-37)C, HID(336-410) or HIDΔ(2-14) and GRIM-ORF, GRIM(1-37)C, GRIM(1-16)C or GRIMΔ(2-14) to individually bind Flag-tagged D-IAP1-BIR. The HID-Epi, HIDΔ(38-335)-Epi and HID(1-37)C-Epi coimmunoprecipitated with Flag-D-IAP1-BIR (Fig. 6A, lanes 2 to 4), but HID(336-410)-Epi and HIDΔ(2-14)-Epi did not (Fig. 6A, lane 5 and 6). All of the tested GRIM constructs coimmunoprecipitated with Flag-D-IAP1-BIR (Fig. 6A, lanes 8 to 11). This included grim(1-16)C-Epi containing only the first 16 amino acids of GRIM and grimΔ(2-14)-Epi lacking the N-terminal amino acids of GRIM, suggesting that there are two independent IAP-BIR binding sites in GRIM. In control lanes, Epi-P35 and Epi-CAT did not coimmunoprecipitate with Flag-D-IAP1-BIR (Fig. 6A, lanes 12 and 13). Expression of the HA-tagged and Flag-tagged constructs in the cell lysates was confirmed for all transfections (Fig. 6B and C).
FIG. 6.
IAP interaction with deletions of HID and GRIM. (A) SF-21 cells were transiently transfected with plasmids expressing Flag-D-IAP1-BIR and indicated HA-epitope-tagged (Epi) constructs. At 3 h after heat shock, aliquots of cell lysates were immunoprecipitated with the M2 anti-Flag monoclonal antibody resin. Coprecipitated HA-epitope-tagged constructs were detected by immunoblot analysis with the rabbit anti-HA.11 polyclonal antiserum. (B) Expression of the HA-epitope-tagged proteins was confirmed by immunoblot analysis with the mouse HA.11 monoclonal antibody of aliquots of the total cell lysates from panel A. (C) Expression of the Flag-D-IAP1-BIR was confirmed by immunoblot analysis with the M2 anti-Flag antibody of aliquots of the total cell lysates from panel A. The positions of the HA-epitope-tagged proteins and Flag-D-IAP1-BIR are indicated by arrows and letters as follows: a, HID-Epi; b, HIDΔ(2-14)-Epi; c, HIDΔ(38-335)-Epi; d, HID(1-37)C-Epi; e, HID(336-410)-Epi; f, Epi-P35; g, Epi-CAT; h, GRIM-Epi; i, GRIMΔ(2-14)-Epi; j, GRIM(1-37)C-Epi; k, GRIM(1-16)-Epi; l, Flag-D-IAP1-BIR; and m, D-IAP1-BIR cross-reacting with polyclonal HA antibody. (D) HID and GRIM bind D-IAP1-BIR in vitro. 35S-labelled in vitro translated HID and GRIM were added to in vitro translated unlabelled Flag-D-IAP1-BIR or empty vector (pKV) and immunoprecipitated with the M2 anti-Flag affinity resin. Coprecipitated 35S-labelled HID and GRIM were analyzed by SDS-polyacrylamide gel electrophoresis and autoradiography (lanes 1 to 4). Equivalence of input radiolabelled proteins is shown in lanes 5 and 6. Molecular mass markers (in kilodaltons) are shown on the left.
To demonstrate the association of HID and GRIM with D-IAP1-BIR in vitro, we tested the ability of 35S-labelled in vitro translated HID and GRIM to bind Flag-D-IAP1-BIR. HID and GRIM coimmunoprecipitated with in vitro-translated Flag-D-IAP1-BIR (Fig. 6D, lanes 3 and 4) but not with products from the in vitro transcription and translation of the vector itself (Fig. 6D, lanes 1 and 2). Therefore, baculovirus and Drosophila IAPs physically interact with HID and GRIM in vivo and in vitro, and the region encompassing the BIR motifs of IAPs is necessary for this protein-protein interaction.
HID and GRIM colocalize with IAPs.
The ability of IAPs to physically interact with HID suggested that IAPs and HID should localize to the same subcellular location when coexpressed, as do IAPs and RPR. To verify this, we expressed hid-Flag in SF-21 cells in the presence or absence of HA-tagged IAPs. In the absence of IAPs, HID localized predominantly to the cytoplasm of the cells 1 h after induction (Fig. 7A). By 4 h after induction we were not able to detect HID within the cells (Fig. 7B), although the expression of HID caused nuclear condensation and fragmentation in transfected cells (Fig. 7C). Coexpression with P35 inhibited HID-induced nuclear condensation and fragmentation, and HID was observed in the cytoplasm of transfected cells at both 1 and 4 h after induction (data not shown).
FIG. 7.
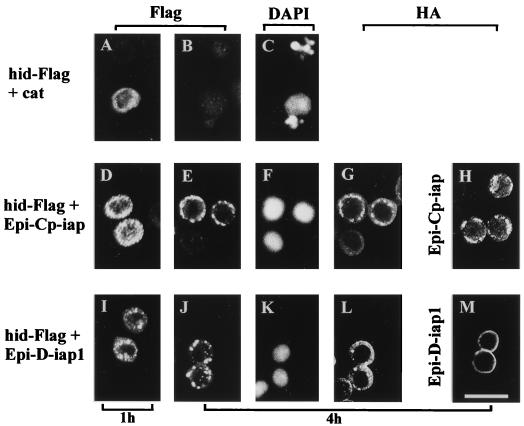
Colocalization of IAPs and HID. SF-21 cells were transiently transfected with plasmids expressing hid-Flag and cat (panels A, B, and C; B and C are the same field), hid-Flag and Epi-Cp-iap (panels D to G; E to G are the same field), Epi-Cp-iap (panel H), hid-Flag and Epi-D-iap1 (panels I to L; J to L are the same field), or Epi-D-iap1 (panel M). At 1 h (panels A, D, and I) or 4 h (panels B, C, E to H, and J to M) after heat shock, cells were fixed with formaldehyde, permeabilized, and analyzed by indirect immunofluorescence. HID-Flag was visualized with mouse M2 anti-Flag monoclonal antibody and lissamine rhodamine-conjugated goat anti-mouse immunoglobulin G plus immunoglobulin M antibody (panels A, B, D, E, I, and J). HA-epitope-tagged proteins were visualized with rabbit anti-HA.11 polyclonal antiserum and fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G antibody (panels G, H, L, and M). Nuclei were visualized by costaining with DAPI (4′,6-diamidino-2-phenylindole) (panels C, F, and K). Scale bar equals 25 μm.
Cp-IAP also blocked HID-induced nuclear condensation and fragmentation (Fig. 7F) as well as the rapid disappearance of HID (Fig. 7E). However, in this case, HID displayed punctate perinuclear localization 1 h after induction (Fig. 7D) and more pronounced localization 4 h after induction (Fig. 7E). This location of HID overlapped with the subcellular localization of Cp-IAP (Fig. 7G). The same punctate perinuclear localization was observed for Cp-IAP in the absence of HID (Fig. 7H). Similar to Cp-IAP, D-IAP1 blocked HID-induced nuclear condensation and fragmentation (Fig. 7K) and rapid disappearance of HID, and showed overlapping subcellular localization (Fig. 7J and L). Colocalization of HID and D-IAP1 was not as evident as for HID and Cp-IAP, probably because D-IAP1 displayed less punctate staining when compared with Cp-IAP (Fig. 7, compare H and M).
In addition, coexpression of HID and Op-IAP resulted in an almost identical subcellular localization pattern as that observed upon coexpression of HID and Cp-IAP, and reversing the epitope tags on HID and Cp-IAP or Op-IAP did not alter the pattern of subcellular localization when coexpressed or expressed individually (data not shown).
We also examined subcellular localization of GRIM and found that, in the absence of IAPs, GRIM induced nuclear condensation and fragmentation that were concurrent with the disappearance of GRIM from the cytoplasm of SF-21 cells, while the coexpression of IAPs blocked GRIM-induced nuclear condensation and fragmentation and resulted in accumulation of GRIM in punctate perinuclear locations which coincided with IAP localizations (data not shown).
The effect of Cp-IAP on the stability of HID.
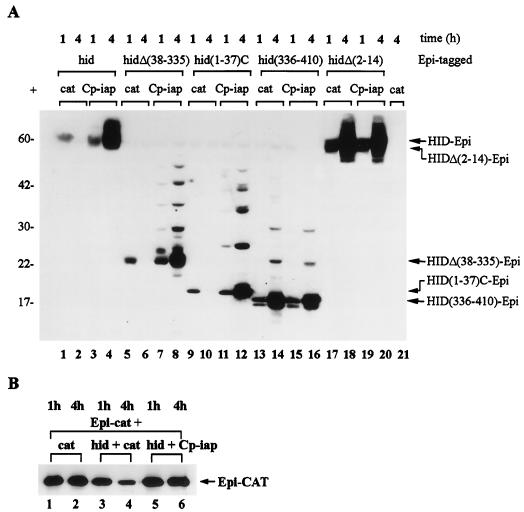
While testing the expression levels of the HID constructs, we found that we could detect HID 1 h after induction (Fig. 8A, lane 1) but not by 4 h after induction (Fig. 8A, lane 2). This finding prompted us to investigate the stability of HID constructs under conditions when HID induced apoptosis compared to conditions when HID-induced apoptosis was blocked by Cp-IAP. Expression of hid-Epi, hidΔ(38-335)-Epi, or hid(1-37)C-Epi in SF-21 cells in the absence of Cp-IAP induced apoptosis (Fig. 1B), and under these conditions proteins that were present 1 h after induction (Fig. 8A, lanes 1, 5, and 9) could not be detected by 4 h after induction (Fig. 8A, lanes 2, 6, and 10). Under conditions when Cp-IAP blocked apoptosis induced by various HID constructs, HID-Epi proteins accumulated during the 4-h period after induction, and this accumulation coincided with the appearance of the additional bands of higher molecular mass (Fig. 8A, lanes 3, 4, 7, 8, 11, and 12).
FIG. 8.
The effect of Cp-IAP on the stability of HID. (A and B) SF-21 cells were transiently transfected with plasmids expressing combinations of genes indicated above the lanes of each panel and harvested 1 h (lanes 1, 3, 5, 7, 9, 11, 13, 15, 17, and 19) or 4 h (lanes 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, and 21) after induction. Immunoblot analysis was done with the mouse anti-HA.11 monoclonal antibody, and the arrows at the right point to the major expected product for each of the expressed genes. Molecular mass markers (in kilodaltons) are shown at the left.
On the other hand, expression of hid(336-410)-Epi or hidΔ(2-14)-Epi did not induce apoptosis (Fig. 1B), and protein products were stable; HID(336-410)-Epi and HIDΔ(2-14)-Epi accumulated during the 4-h period following induction when expressed with or without Cp-IAP, and were accompanied by the appearance of higher-molecular-mass bands (Fig. 8A, lanes 13 to 20). Additional bands of higher molecular mass represent increments of 7 to 8 kDa and are probably results of posttranslational modification. As a control, we investigated the stability of apoptotically neutral protein, Epi-CAT, under the same conditions as in Fig. 8A. The amount of Epi-CAT decreased approximately two- to fourfold under conditions when HID induced apoptosis (Fig. 8B, lanes 3 and 4), and Epi-CAT was completely stabilized when HID-induced apoptosis was blocked by Cp-IAP (Fig. 8B, lanes 5 and 6). This was in sharp contrast to proapoptotic HID constructs which were completely degraded by 4 h after induction (Fig. 8A, lanes 2, 6, and 10). In additional experiments P35 was equally efficient in stabilizing HID proteins as Cp-IAP (data not shown). As deletion of the 14 N-terminal amino acids abrogated both proapoptotic activity and instability of HID, these results suggest that instability of HID is conferred by the N-terminal 37 amino acids and may be linked to the ability of these residues to induce apoptosis.
In addition, we also found that levels of GRIM declined when expressed individually and that coexpression with IAPs stabilized GRIM (data not shown).
DISCUSSION
In this study, we have analyzed the nature of the apoptotic response induced by the Drosophila genes hid and grim in a heterologous insect cell line, SF-21. A role for hid and grim in regulating apoptosis was previously established in Drosophila embryos (5, 12), developing eyes (5, 12, 15), and the central nervous system (20, 32). We have demonstrated that HID and GRIM can induce apoptosis in a lepidopteran cell line, SF-21, and investigated which portions of HID and GRIM possess proapoptotic activity by generating a series of deletions. Our deletion analysis showed that the N-terminal 37 amino acids of either HID or GRIM were sufficient to induce apoptosis if stably expressed as fusion proteins. Furthermore, the only region of sequence similarity among RPR, HID, and GRIM (14 N-terminal amino acids) is required for the induction of apoptosis by HID. In the case of GRIM, the N-terminal 16 amino acids were sufficient to induce apoptosis. The C-terminal truncations of HID and GRIM appeared to induce apoptosis efficiently, but we cannot assess the relative ability of mutant and wild-type proteins to induce apoptosis in these assays because they rely on overexpression of the genes; in vivo, the mutants may exhibit only partial activity.
Overexpression of the GRIM deletion lacking the proapoptotic N-terminal 14 amino acids is still able to induce apoptosis, suggesting that GRIM might have two independent regions with death-promoting abilities. Sequence analysis of amino acids present in this GRIM deletion and comparison with other known proapoptotic protein domains (e.g., death domain, death effector domain, BH3, caspase or pro-caspase domains) did not reveal any significant similarity, and therefore GRIM might contain a novel death-inducing region. According to the mutational analysis of RPR, deletion of RPR lacking this region of similarity (4, 28) or site-specific mutants of the most conserved residues of this region (4, 28) still retained proapoptotic activity. Thus, the existence of two independent death-promoting regions may not be unique to GRIM, and future mutational analysis of GRIM will be required to define this novel death-inducing motif.
We have demonstrated that baculovirus and Drosophila iaps physically associate with and block apoptosis induced by Drosophila proteins HID and GRIM. The region encompassing the BIR motifs of IAPs is both necessary and sufficient for this interaction. In the case of baculovirus IAPs, the RING finger motif is also important for antiapoptotic activity (8, 14) but its role is not understood at this point. The N-terminal 37 amino acids of HID and GRIM are sufficient to bind IAPs. We have not attempted to further define this region of HID. In the case of GRIM, we have found that the N-terminal 16 amino acids are sufficient to induce apoptosis if stably expressed, suggesting that this limited region of similarity between Drosophila inducers of apoptosis is important in the regulation as well as signaling of these proteins. However, a GRIM deletion lacking the N-terminal 14 amino acids still induces apoptosis and associates with IAPs indicating the presence of two distinct apoptosis-inducing motifs within GRIM. Physical association of IAPs with HID, GRIM, and RPR provides a possible mechanism by which IAPs can inhibit apoptosis induced by these Drosophila inducers of apoptosis. Through this interaction, IAPs may physically block access of HID, GRIM, and RPR to downstream effector proteins, or alternatively, this interaction may displace IAPs from binding as inhibitors to effector proteins such as caspases.
Physical interaction of HID and GRIM with Cp-IAP or D-IAP1 results in localization of HID and GRIM to the same cellular location as IAPs. As HID and GRIM show predominantly diffused cytoplasmic staining in the absence of IAPs, this localization is IAP directed. In SF-21 cells, overexpression of IAPs leads to their localization to the punctate perinuclear subcellular locations. However, that subcellular localization is more pronounced for baculovirus IAPs than for Drosophila IAPs, whose localization appears to be less punctate. We also do not exclude the possibility that HID might affect colocalization of HID and D-IAP1 as colocalization of HID and D-IAP1 is more punctate in its appearance than localization of D-IAP1 alone. IAPs have the same effect on subcellular localization of RPR (27). Therefore, the ability of IAPs to redirect RPR, HID, and GRIM to a different subcellular location and potentially block the access of these inducers of apoptosis to downstream effectors may be a component of the mechanism by which IAPs inhibit induction of apoptosis by these proapoptotic proteins.
The rapid decline in the levels of HID following induction in SF-21 cells and its accumulation and relocalization in the presence of IAPs are similar if not identical to the behavior of RPR (27) and suggest an additional level of complexity in their regulation. The N-terminal 37 amino acids of HID were sufficient to confer instability to a fusion protein, while the C-terminal portion of HID lacking the terminal 37 amino acids was stable. Since the N-terminal 37 amino acids were sufficient to initiate apoptosis, the rapid decline in HID levels may either be associated with activation of apoptosis or due to proteolytic signaling by the N-terminal residues themselves. The former is likely to be the case since expression of P35, a caspase inhibitor which does not bind to HID, was able to block the degradation of HID. In the case of RPR, we demonstrated that aspartate-specific caspases are not directly responsible for RPR degradation (27), suggesting that another protease may be activated in the process of induction of apoptosis. The rapid degradation of apoptotic inducers such as HID and RPR would allow precision control of the concentration of free inducers and finer tuning of the cellular environment. The binding of IAPs to inducers and their cellular relocalization would then serve to buffer the effect of a rapid rise in inducer concentrations which is known to precede apoptosis.
In addition to physically interacting with RPR, HID, and GRIM, IAPs also associate with another Drosophila proapoptotic protein, Doom (13), with mammalian TRAF-2 (21, 24), a protein implicated in TNF-induced signaling pathways (21, 24), and with some mammalian members of the caspase family of apoptotic effectors (10). None of these proteins appear to contain sequences related to the N-terminal 16 amino acids of RPR, HID, and GRIM. The ability of IAPs to physically interact with a variety of inducers of apoptosis and to block apoptosis induced by diverse stimuli places IAPs in a central position as sensors and inhibitors of death signals that proceed through a number of different pathways. Being at this position, IAPs would serve as cellular checkpoints allowing apoptosis to ensue only after the levels of inducers rise above the threshold determined by IAP levels.
ACKNOWLEDGMENTS
We thank Hermann Steller (Massachusetts Institute of Technology) for hid cDNA, John M. Abrams (University of Texas) for grim cDNA and Somasekar Seshagiri for helpful discussions.
This work was supported in part by Public Health Service grant AI38262 from the National Institute of Allergy and Infectious Disease to L.K.M.
REFERENCES
- 1.Ambrosini G, Adida C, Altieri D C. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 2.Birnbaum M J, Clem R J, Miller L K. An apoptosis-inhibiting gene from a nuclear polyhedrosis virus encoding a polypeptide with Cys/His sequence motifs. J Virol. 1994;68:2521–2528. doi: 10.1128/jvi.68.4.2521-2528.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bump N J, Hackett M, Hugunin M, Seshagiri S, Brady K, Chen P, Ferenz C, Franklin S, Ghayur T, Li P, et al. Inhibition of ICE family proteases by baculovirus antiapoptotic protein p35. Science. 1995;269:1885–1888. doi: 10.1126/science.7569933. [DOI] [PubMed] [Google Scholar]
- 4.Chen P, Lee P, Otto L, Abrams J. Apoptotic activity of REAPER is distinct from signaling by the tumor necrosis factor receptor 1 death domain. J Biol Chem. 1996;271:25735–25737. doi: 10.1074/jbc.271.42.25735. [DOI] [PubMed] [Google Scholar]
- 5.Chen P, Nordstrom W, Gish B, Abrams J M. grim, a novel cell death gene in Drosophila. Genes Dev. 1996;10:1773–1782. doi: 10.1101/gad.10.14.1773. [DOI] [PubMed] [Google Scholar]
- 6.Chu Z L, McKinsey T A, Liu L, Gentry J J, Malim M H, Ballard D W. Suppression of tumor necrosis factor-induced cell death by inhibitor of apoptosis c-IAP2 is under NF-kappaB control. Proc Natl Acad Sci USA. 1997;94:10057–10062. doi: 10.1073/pnas.94.19.10057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clem R J, Fechheimer M, Miller L K. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science. 1991;254:1388–1390. doi: 10.1126/science.1962198. [DOI] [PubMed] [Google Scholar]
- 8.Clem R J, Miller L K. Control of programmed cell death by the baculovirus genes p35 and iap. Mol Cell Biol. 1994;14:5212–5222. doi: 10.1128/mcb.14.8.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crook N E, Clem R J, Miller L K. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J Virol. 1993;67:2168–2174. doi: 10.1128/jvi.67.4.2168-2174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devereaux Q L, Takahashi R, Salvesen G S, Reed J C. X-linked IAP is a direct inhibitor of cell-death proteases. Nature. 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- 11.Duckett C S, Nava V E, Gedrich R W, Clem R J, Van Dongen J L, Gilfillan M C, Shiels H, Hardwick J M, Thompson C B. A conserved family of cellular genes related to the baculovirus iap gene and encoding apoptosis inhibitors. EMBO J. 1996;15:2685–2694. [PMC free article] [PubMed] [Google Scholar]
- 12.Grether M E, Abrams J M, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- 13.Harvey A J, Bidwai A P, Miller L K. Doom, a novel product of the Drosophila mod(mdg4) gene, induces apoptosis and binds to baculovirus inhibitor-of-apoptosis proteins. Mol Cell Biol. 1997;17:2835–2843. doi: 10.1128/mcb.17.5.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harvey A J, Soliman H, Kaiser W J, Miller L K. Anti- and pro-apoptotic activities of baculovirus and Drosophila IAPs in an insect cell line. Cell Death Differ. 1997;4:733–744. doi: 10.1038/sj.cdd.4400294. [DOI] [PubMed] [Google Scholar]
- 15.Hay B A, Wassarman D A, Rubin G M. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- 16.Liston P, Roy N, Tamai K, Lefebvre C, Baird S, Cherton-Horvat G, Farahani R, McLean M, Ikeda J E, MacKenzie A, Korneluk R G. Suppression of apoptosis in mammalian cells by NAIP and a related family of IAP genes. Nature. 1996;379:349–353. doi: 10.1038/379349a0. [DOI] [PubMed] [Google Scholar]
- 17.Manji G A, Hozak R R, LaCount D J, Friesen P D. Baculovirus inhibitor of apoptosis functions at or upstream of the apoptotic suppressor P35 to prevent programmed cell death. J Virol. 1997;71:4509–4516. doi: 10.1128/jvi.71.6.4509-4516.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Reilly D R, Miller L K, Luckow V A. Baculovirus expression vectors: a laboratory manual. W. H. New York, N.Y: Freeman & Co.; 1992. [Google Scholar]
- 19.Pronk G J, Ramer K, Amiri P, Williams L T. Requirement of an ICE-like protease for induction of apoptosis and ceramide generation by REAPER. Science. 1996;271:808–810. doi: 10.1126/science.271.5250.808. [DOI] [PubMed] [Google Scholar]
- 20.Robinow S, Draizen T A, Truman J W. Genes that induce apoptosis: transcriptional regulation in identified, doomed neurons of the drosophila CNS. Dev Biol. 1997;190:206–213. doi: 10.1006/dbio.1997.8696. [DOI] [PubMed] [Google Scholar]
- 21.Rothe M, Pan M G, Henzel W J, Ayres T M, Goeddel D V. The TNFR2-TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell. 1995;83:1243–1252. doi: 10.1016/0092-8674(95)90149-3. [DOI] [PubMed] [Google Scholar]
- 22.Roy N, Mahadevan M S, McLean M, Shutler G, Yaraghi Z, Farahani R, Baird S, Besner-Johnston A, Lefebvre C, Kang X, Salih M, Aubry H, Tamai K, Guan X, Ioannou P, Crawford T O, de Jong P J, Surh L, Ikeda J, Korneluk R G, MacKenzie A. The gene for neuronal apoptosis inhibitory protein is partially deleted in individuals with spinal muscular atrophy. Cell. 1995;80:167–178. doi: 10.1016/0092-8674(95)90461-1. [DOI] [PubMed] [Google Scholar]
- 23.Seshagiri S, Miller L K. Baculovirus inhibitors of apoptosis (IAPs) block activation of Sf-caspase-1. Proc Natl Acad Sci USA. 1997;94:13606–13611. doi: 10.1073/pnas.94.25.13606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shu H, Takeuchi M, Goeddel D V. The tumor necrosis factor receptor 2 signal transducers TRAF2 and c-IAP1 are components of the tumor necrosis factor receptor 1 signaling complex. Immunology. 1996;93:13973–13978. doi: 10.1073/pnas.93.24.13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steller H. Mechanisms and genes of cellular suicide. Science. 1995;267:1445–1449. doi: 10.1126/science.7878463. [DOI] [PubMed] [Google Scholar]
- 26.Uren A G, Pakusch M, Hawkins C J, Puls K L, Vaux D L. Cloning and expression of apoptosis inhibitory protein homologs that function to inhibit apoptosis and/or bind tumor necrosis factor receptor-associated factors. Proc Natl Acad Sci USA. 1996;93:4974–4978. doi: 10.1073/pnas.93.10.4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vucic D, Kaiser W J, Harvey A J, Miller L K. Inhibition of Reaper-induced apoptosis by interaction with inhibitor of apoptosis proteins (IAPs) Proc Natl Acad Sci USA. 1997;94:10183–10188. doi: 10.1073/pnas.94.19.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vucic D, Seshagiri S, Miller L K. Characterization of reaper- and FADD-induced apoptosis in a lepidopteran cell line. Mol Cell Biol. 1997;17:667–676. doi: 10.1128/mcb.17.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White K, Tahaoglu E, Steller H. Cell killing by the Drosophila gene reaper. Science. 1996;271:805–807. doi: 10.1126/science.271.5250.805. [DOI] [PubMed] [Google Scholar]
- 30.White K, Grether M E, Abrams J M, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- 31.Xue D, Horvitz H R. Inhibition of the Caenorhabditis elegans cell-death protease CED-3 by a CED-3 cleavage site in baculovirus p35 protein. Nature. 1995;377:248–251. doi: 10.1038/377248a0. [DOI] [PubMed] [Google Scholar]
- 32.Zhou L, Schnitzler A, Agapite J, Schwartz L M, Steller H, Nambu J R. Cooperative functions of the reaper and head involution defective genes in the programmed cell death of drosophila central nervous system midline cells. Proc Natl Acad Sci USA. 1997;94:5131–5136. doi: 10.1073/pnas.94.10.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]