Abstract
Many small nucleolar RNAs (snoRNAs) are encoded within introns of protein-encoding genes and are released by processing of their host pre-mRNA. We have investigated the mechanism of processing of the yeast U18 snoRNA, which is found in the intron of the gene coding for translational elongation factor EF-1β. We have focused our analysis on the relationship between splicing of the EF-1β pre-mRNA and production of the mature snoRNA. Mutations inhibiting splicing of the EF-1β pre-mRNA have been shown to produce normal U18 snoRNA levels together with the accumulation of intermediates deriving from the pre-mRNA, thus indicating that the precursor is an efficient processing substrate. Inhibition of 5′→3′ exonucleases obtained by insertion of G cassettes or by the use of a rat1-1 xrn1Δ mutant strain does not impair U18 release. In the Exo− strain, 3′ cutoff products, diagnostic of an endonuclease-mediated processing pathway, were detected. Our data indicate that biosynthesis of the yeast U18 snoRNA relies on two different pathways, depending on both exonucleolytic and endonucleolytic activities: a major processing pathway based on conversion of the debranched intron and a minor one acting by endonucleolytic cleavage of the pre-mRNA.
The small nucleolar RNAs (snoRNAs) represent a wide class of short RNA molecules localized in the nucleoli of eukaryotic cells and linked to the complex process of ribosome biogenesis (reviewed in reference 28). They can be subdivided into two major classes based on their conserved structural and sequence elements (2, 40, 45). The first class consists of box C/D snoRNAs, which are characterized by a 5′-3′ terminal stem and two conserved sequence motifs, called boxes C and D, both required for their processing and stability (3, 6, 19, 53). All of them form small nucleolar ribonucleoprotein (snoRNP) complexes containing the evolutionarily conserved protein fibrillarin (3, 20, 33, 38, 46). Most of these snoRNAs contain long stretches (>10 nt) of perfect complementarity to the universal core regions of mature rRNAs (1, 41, 48), and it has been demonstrated that these antisense snoRNAs function as guide RNAs for directing site-specific 2′-O methylation of pre-rRNA (23, 32). This class also includes the phylogenetically conserved U3 and U14 and the vertebrate U8 and U22 snoRNAs, which have also been shown to directly participate in the processing reactions of the pre-rRNA (28, 49). The second class of snoRNAs is the H/ACA family (2, 12), whose members share a conserved hairpin-hinge-hairpin tail structure and short consensus sequences (H and ACA boxes); they also display short elements of complementarity to the rRNA (<10 nt) and associate with the GAR1p protein (2, 13). Recent reports demonstrate that most H/ACA snoRNPs are involved in directing site-specific isomerization of uridine into pseudouridine on pre-rRNA (11, 31). Among these, only the yeast snR30 and snR10 box H/ACA snoRNAs have been shown to be necessary for rRNA processing. The only exception to the above classification is 7-2/MRP, the RNA component of RNase MRP required for cleavage of precursor rRNA (45).
The snoRNA genes have a dichotomous genomic organization. The most abundant vertebrate snoRNAs and most of the yeast snoRNAs are transcribed from independent genes by RNA polymerase II or, in the case of 7-2/MRP, by RNA polymerase III (16). On the other hand, a large number of snoRNAs are encoded within introns of protein genes and require processing from host pre-mRNAs to be released (28). Intriguingly, most of the host genes encode proteins related to ribosome function, suggesting a possible regulatory link among all of these components (28). The presence of snoRNAs within introns also raises the new issue of relating intronic snoRNA biosynthesis to processing of the host pre-mRNA. The biosynthetic mechanism of several intron-encoded snoRNAs has been investigated in vivo and in vitro, leading to the proposal of two alternative processing models. One model predicts that the snoRNA processing reaction is accomplished through the splicing pathway by exonucleolytic trimming of the spliced host intron after it has been debranched. Such a processing mechanism was originally suggested for human U17 and U19 (22). In fact, both of these snoRNAs are released from host and nonhost spliceable precursors in vivo, suggesting that the snoRNA coding regions contain all of the signals required for faithful excision. Exonucleolytic trimming has also been proposed for some other snoRNAs, such as the U15 (47) and U20 (8) snoRNAs. A different maturation pathway, based on endonucleolytic activity, has been proposed for the Xenopus laevis U16 and U18 snoRNAs, both of which are encoded within introns of the L1 ribosomal protein gene (10, 36). It has been demonstrated that they can be produced through a pathway, alternative to splicing, which involves endonucleolytic cleavage of the pre-mRNA (5, 6). This event is driven by two independent endonucleolytic cleavages within intron sequences upstream and downstream of the snoRNA coding region, producing 5′ and 3′ cutoff products; when both cleavages occur on the same precursor, pre-snoRNA molecules are produced that still contain 5′ and 3′ trailer sequences. Finally, exonucleolytic trimming converts the processing intermediates into mature snoRNA molecules.
In this study, we have investigated the mechanism of processing of the yeast U18 snoRNA, which belongs to the family of box C/D snoRNAs. In Saccharomyces cerevisiae, this snoRNA is encoded within the intron of the gene coding for translational elongation factor EF-1β and likely functions in the methylation of 25S rRNA (23, 32). Here, we report that processing of the U18 snoRNA occurs efficiently even in the absence of host pre-mRNA splicing, showing that the entire precursor, and not only the lariat, is a substrate for this reaction. U18 snoRNA biosynthesis is not impaired when poly(G) cassettes are located upstream of the U18 coding sequences or when the major yeast 5′→3′ exonucleases (XRN1p and RAT1p) are inactivated. Under these conditions, specific cutoff products can be detected. Our results point to the involvement of an endonucleolytic activity in the processing pathway leading to release of the U18 snoRNA from the intron of the EF-1β pre-mRNA.
MATERIALS AND METHODS
Strains and media.
Growth and handling of S. cerevisiae were done by standard techniques. The strains used were CH1462 (MATα ade2 ade3 leu2 ura3 his3 can1 [24]) and D174 (MATa ade2-1 xrn1::URA3 rat1-1 [15]). For induction experiments, galactose at a final concentration of 2% was directly added to cultures grown in nonrepressive medium containing 2% raffinose and 0.05% glucose. Exonuclease-deficient strains were grown at 23°C until they reached mid-log phase and then shifted to 37°C for 1.5 h prior to galactose induction. Transcriptional pulse-chase experiments were performed as previously described (9), with minor modifications. Briefly, cultures were grown in 200 ml of selective medium containing 2% raffinose and 0.05% glucose until they reached mid-log phase, washed twice, and resuspended in the same volume of prewarmed selective medium containing 2% raffinose. After 1 h of incubation, a 10-ml aliquot was removed for an uninduced control, and galactose at a final concentration of 2% was added to start transcription, which was then repressed after 20 min by the addition of glucose to a final concentration of 4%. Aliquots (10 ml) were harvested at different times during both the induction and repression periods and quickly frozen on dry ice.
Plasmid construction.
Plasmid pBS-EFB1 containing the genomic EFB1 clone was used as the starting material for generation of all of the constructs used in this work. Part of the gene (from position −24 to position +524, with respect to the ATG codon) was amplified by PCR using oligonucleotides yU18F and E3′ (see below) and then cloned into the SacI and HindIII sites of the Bluescript plasmid to obtain pBSU18wt. Point mutations were then introduced by inverse PCR at the 5′ splice site using oligonucleotides C5F and C5B and at the branch site using oligonucleotides CbsF and I3′ to obtain pBSU18C5 and pBSU18Cbs, respectively. The introduction of a tag in the 3′ sequence of U18 was obtained by replacement of 20 nucleotides by inverse PCR using oligonucleotides U18tagF and U18tagB, and then the SacI-HindIII-tagged U18 fragments were recovered and the SacI site was blunt ended with T4 polymerase. Subsequently, the fragments were inserted into the SmaI and HindIII sites of plasmid p416GAL1 (30) to obtain the pGALU18wt, pGALU18C5, and pGALU18Cbs constructs. Mutation in the C box was introduced by inverse PCR on the tagged pBSU18 plasmids using oligonucleotides bCF and bCB; the HpaI-EcoRI fragments were then recovered and inserted into the corresponding sites of the pGALU18 plasmids. The G cassette (pG), consisting of 18 G residues, was introduced by PCR into the 5′ untranslated region (UTR) of the tagged pBSU18 plasmids by using oligonucleotides U18pG and E3′; the SpeI-EcoRI fragments were then recovered and inserted into the corresponding sites of the pGALU18 plasmids. Similarly, introduction of the G cassette into the intron was performed by PCR using oligonucleotides U18pGint and E3′ and insertion of the resulting fragment between the AflIII and EcoRI sites of the pGALU18 plasmids. To allow the expression of GALU18 constructs in the rat1-1 xrn1::URA3 strain, we changed the marker gene of plasmid p416GAL1 by inserting the ADE2 gene, recovered from plasmid Yep(ADE) (35), into the BstBI site of the URA3 gene.
Oligonucleotides.
The oligonucleotides used in the course of this work for cloning steps and/or for RNA analyses by Northern hybridization or primer extensions were named anti-tag (5′-TGCGGACTGCCTGGATGCCG-3′), bCB (5′-TTCACATAAGCGAAAAAAAGTAAT-3′), bCF (5′-CATTGAATTTAATTCTTTGGTC-3′), C5B (5′-CTTCAATGTATGACTTGTC-3′), C5F (5′-GGTATCTTCCGATTTAGT-3′), CbsF (5′-TACTACCAAGCAAAATG-3′), E3′ (5′-GCAAGCTTGTTGAACCATCTGAA-3′), E5′ (5′-TTGACGTTAAAGTATAGAGG-3′), I3′ (5′-ATGAGAACTTTTTTCTTG-3′), I5′ (5′-ACTTCCCTTTACATGTAACC-3′), ITS1 (5′-CCAGTTACGAAAATTCTTG-3′), L32 (5′-TCCTTTGAACGGACTCTCTC-3′), U1 (5′-TTCATCATAAACTTAAGTTCATTCGAATCTCCGTC-3′), U18pG (5′-AGACTAGTGGGGGGGGGGGGGGGGGGATCCCCCGGTCCAACCGA-3′), U18pGint (5′-GCACATGTGGGGGGGGGGGGGGGGGGAAGTTAACTAATAATGAT-3′), U18tagB (5′-CTGGATGCCGTGTCACTCATATCGGGGGTC-3′), U18tagF (5′-GCAGTCCGCACACAGTATCTGACGATAGCA-3′), and yU18F (5′-GCGAGCTCGTCCAACCGAATATA-3′).
RNA analysis.
RNA was extracted from exponentially growing cultures of S. cerevisiae by the hot-phenol method as previously described (39). RNA concentrations were routinely calibrated by A260 and ethidium bromide staining on formaldehyde gels and normalized by hybridization with a U1 snRNA oligonucleotide. Primer extension analyses were performed as previously described (4). For Northern blot analyses, 5 μg of total RNA was typically electrophoresed on 6% polyacrylamide–7 M urea gels and electrotransferred at 4°C to Amersham Hybond-N filters for 3 h at 380 mA in 25 mM NaPO4 (pH 6.5) buffer. All hybridizations were carried out under standard conditions. Ten picomoles of each oligonucleotide was routinely 5′ end labelled with 30 μCi of [γ-32P]ATP.
RESULTS
U18 snoRNA biosynthesis is independent of ongoing splicing.
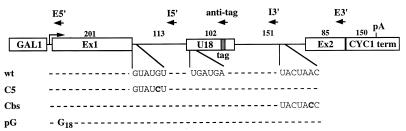
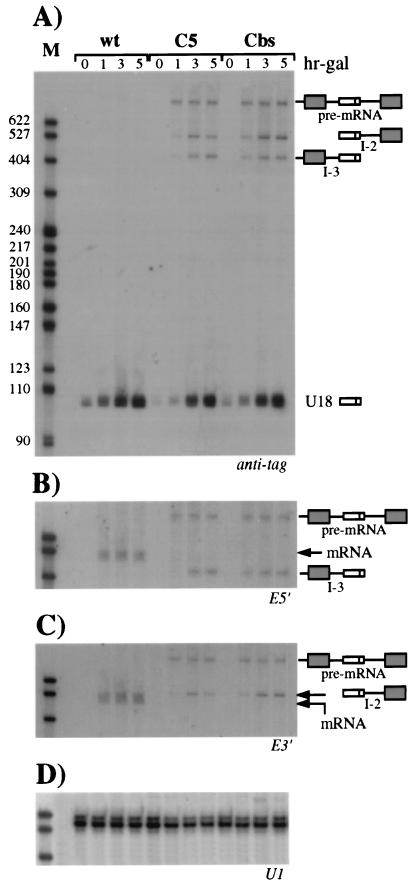
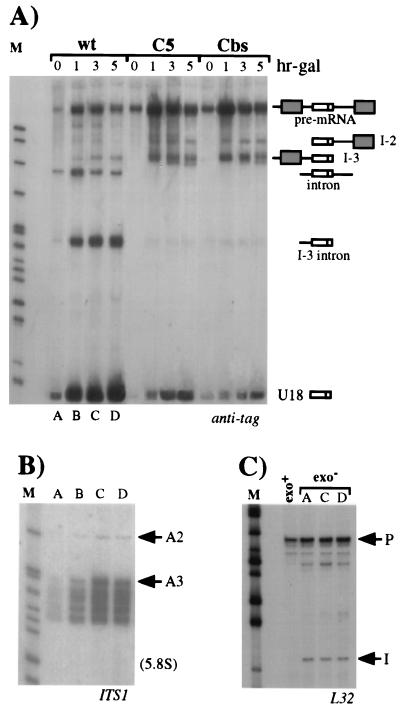
The yeast U18 snoRNA is located in the intron of the EFB1 gene (1, 28), which encodes translational elongation factor EF-1β (17). Since the EF-1β pre-mRNA has canonical splice site sequences and is likely to splice quite efficiently, it represents an interesting system for use in studying the relationship between splicing of the host pre-mRNA and biosynthesis of the intron-encoded RNA. We investigated the effect of splice site mutations on production of the U18 snoRNA: two independent single-base substitutions were introduced at the 5′ splice site and at the branch nucleotide of an episomal copy of the EFB1 gene (plasmid pGALU18wt [Fig. 1]), cloned between the GAL1 promoter and CYC1 terminator sequences of plasmid p416GAL1 (30). The invariant fifth nucleotide (underlined) of the GUAUGU consensus sequence of the 5′ splice site was substituted for a C, generating plasmid pGALU18C5 (Fig. 1). This mutation is known to strongly reduce the splicing efficiency of a precursor RNA and to trigger accumulation of pre-mRNA and splicing intermediates in vivo (27, 50). The second point mutation is carried by plasmid pGALU18Cbs and consists of the replacement of the absolutely conserved A residue (underlined) of the TACTAAC branch site with a C residue. This mutation causes a severe splicing-deficient phenotype, which results in almost undetectable levels of the spliced products and the lariat intermediate (37, 50). To monitor the processing of the U18 snoRNA from the wild-type and mutant episomal constructs, 20 nucleotides of the 3′ sequence of U18, not conserved between yeast and higher eukaryotes (36), were replaced with a tag of the same length. Each plasmid was transformed into wild-type recipient strain CH1462 (24), and expression of the episomal EFB1 gene was induced by addition of galactose to exponentially growing cells pregrown on raffinose. Aliquots were harvested at different times after transcriptional induction, and the accumulation of tagged U18 was assessed by Northern analysis. As shown in Fig. 2A, hybridization with the anti-tag oligonucleotide detected high levels of accumulation of U18 snoRNA expressed from the wild-type episomal EFB1 construct (wt lanes). Almost comparable levels of tagged U18 were also observed when the snoRNA was encoded within the mutant splicing substrates (C5 and Cbs lanes). Serial hybridizations of the same filter with exon probes (Fig. 2B and C) allowed the characterization of the different products visualized in panel A. As expected, in strains carrying the U18C5 and U18Cbs constructs, EF-1β pre-mRNA accumulated and no spliced mRNA was detected; moreover, no signals due to the utilization of cryptic splice sites were detected by this and other analyses (data not shown); on the contrary, mature mRNA is normally produced from the wild-type precursor (wt lanes). The anti-tag probe detected two additional products in strains carrying the splicing mutants: bands I-2 and I-3. Band I-2 was also visualized with oligonucleotide E3′, which is specific for the downstream exon (panel C), while band I-3 was revealed by oligonucleotide E5′, which recognizes the 5′ exon (panel B). From the sizes of the bands, it can be predicted that I-2 extends from the 5′ end of U18 to the 3′ end of the pre-mRNA, while I-3 extends from the 5′ end of the pre-mRNA to the 3′ end of U18 (shown in the diagrammatic representations on the right). This was confirmed by RNase H and primer extension analyses with appropriate oligonucleotides (data not shown). The stability of the I-2 and I-3 molecules, which do not possess the 5′ cap and the poly(A) tail, respectively, is very likely due to the interaction of specific factors with the conserved C/D boxes, which would prevent degradative trimming, as already described in other cases (6, 44, 54). In fact, mutants with mutations in the conserved C box accumulated neither I-2 and I-3 nor U18 (data not shown). In panel D, the hybridization on the same filter with the U1 snRNA probe is shown as a control.
FIG. 1.
Structures of EFB1 episomal constructs expressing the tagged U18 snoRNA. The diagrammatic representation shows plasmid pGALU18, containing the transcriptional unit of the EFB1 gene between the GAL1 promoter and the CYC1 terminator, and its mutant derivatives. The dark box within the U18 snoRNA represents the tag sequence. All of the mutations introduced are shown in boldface letters and are aligned with the corresponding wild-type (wt) sequences. G18 indicates the site of insertion of the poly(G) cassette. The different oligonucleotides utilized in the work are indicated by arrows in the regions where they hybridize. The lengths of the different portions of the construct are indicated in nucleotides. pA indicates the polyadenylation site within the CYC1 terminator (term).
FIG. 2.
Production of the U18 snoRNA from wild-type (wt) EFB1 and its splicing mutant derivatives. (A) Total RNA was extracted from strain CH1462 transformed with the pGALU18wt (wt lanes), pGALU18C5 (C5 lanes), and pGALU18Cbs (Cbs lanes) plasmids at different times of galactose induction (hr-gal). Samples of 5 μg were run on a 6% acrylamide–7 M urea gel, electroblotted onto a nylon membrane, and hybridized with the anti-tag oligonucleotide. The different products are indicated on the right, together with their schematic representations (dark boxes are exons, open boxes are U18, and lines are intronic sequences plus the 5′ UTR). The numbers above the lanes indicate hours of galactose induction. The same filter was rehybridized with oligonucleotides E5′ (B) and E3′ (C), which are complementary to the 5′ and 3′ exons, respectively. (D) Control hybridization to U1 RNA. Oligonucleotides used in filter hybridizations are indicated below the panels. Lanes M contained MspI-digested pBR322 plasmid DNA. Molecular sizes are indicated on the left in nucleotides.
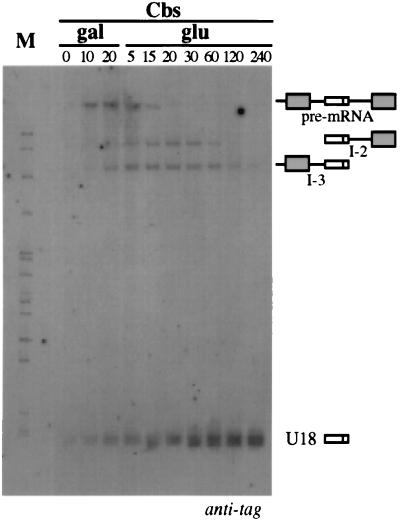
The I-2 and I-3 molecules are likely to represent U18 snoRNA biosynthetic intermediates deriving from unspliced pre-mRNA, similar to what was previously shown for the U14, U16, and U18 snoRNAs in X. laevis (6, 53, 54). To test this hypothesis, we examined the precursor-product relationship by the transcriptional pulse-chase procedure (9). Transcription from plasmid pGALU18Cbs was induced by galactose for a short time (20 min) and then repressed by glucose addition. Aliquots were collected at different time points, and the presence of the different processing products was analyzed by Northern analysis. The amount of RNA loaded in each lane was normalized with a U1 snRNA control hybridization (not shown). Figure 3 shows that the pre-mRNA was converted to the mature U18 snoRNA by a two-step reaction. The pre-mRNA reached a maximum at 20 min after induction, and at 20 min after glucose repression, it had almost disappeared. Concomitantly with the pre-mRNA decrease, the levels of the I-2 and I-3 molecules increased. The U18 snoRNA reached its maximum accumulation between 30 and 60 min, and its level remained stable until the end of the analysis (4 h), as expected for a stable RNA. Since U18 accumulation is delayed with respect to pre-mRNA disappearance but rather follows the kinetics of I-2 and I-3 accumulation, we concluded that I-2 and I-3 molecules represent U18 processing intermediates. Note that the U18 signal was visualized even before galactose addition to the medium (lane gal 0), suggesting that some transcription also occurs before induction.
FIG. 3.
Transcriptional pulse-chase analysis of U18 processing. Northern blot analysis was performed on 5 μg of total RNA extracted from strain CH1462 containing the pGALU18Cbs construct (Fig. 1) at different times after galactose induction (gal lanes) or glucose transcriptional repression (glu lanes). The filter was hybridized with the anti-tag oligonucleotide. The different molecules are diagrammed on the right, as in Fig. 2. The numbers above the lanes indicate minutes of incubation in medium containing the indicated carbon source. Lane M contained MspI-digested pBR322 plasmid DNA.
Role of 5′→3′ exonucleases in U18 snoRNA processing.
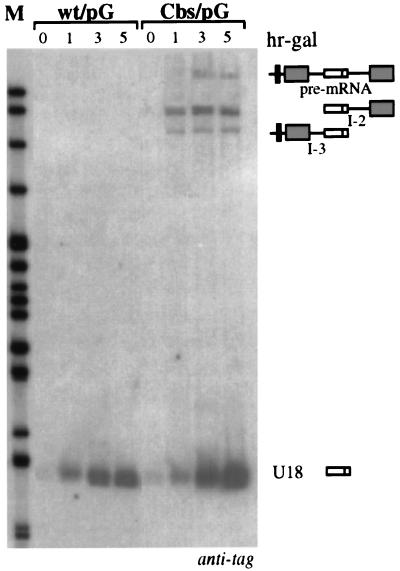
Two main processing pathways have been proposed for intron-encoded snoRNAs, both involving 5′→3′ and 3′→5′ exonucleolytic activities for the removal of 5′ and 3′ trailer sequences on either the debranched lariat or host pre-mRNA cutoff products. Since the above results show that the EF-1β pre-mRNA can be a substrate for U18 maturation, we investigated whether exonucleolytic trimming from the 5′ end of the pre-mRNA could be responsible for U18 production. If this is the case, trimming should occur after decapping of the pre-mRNA. Since poly(G) tracts form extremely stable RNA secondary structures, which represent efficient blocks to yeast 5′→3′ exonucleases (9, 35, 52), we inserted such sequences into the 5′ UTRs of the pGALU18wt and pGALU18Cbs constructs (wt/pG and Cbs/pG derivatives; Fig. 1) and looked at the accumulation of tagged U18 (Fig. 4). In strains transformed with the construct containing the branch site mutation (Cbs/pG lanes), U18 and I-2 accumulated efficiently. These results suggest the presence of processing sites between the poly(G) cassette and U18. As a control, the poly(G) cassette inserted into the wild-type construct, which produces U18 mainly through the splicing pathway, also showed an unaffected processing pattern (wt/pG lanes). The insertion of a poly(G) cassette into the 5′ portion of the intron (34 nucleotides upstream of the snoRNA coding region), also did not affect U18 accumulation (data not shown), suggesting the occurrence of processing inside the intron.
FIG. 4.
Insertion of a G cassette in the 5′ UTR does not abolish U18 processing. Northern blot analysis of 5 μg of total RNA extracted at different times of galactose induction (hr-gal) from yeast strains transformed with plasmids pGALU18wt/pG and pGALU18Cbs/pG containing the pG insertion in the 5′ UTR of the wild-type (wt) construct and of the Cbs mutant, respectively (Fig. 1). Hybridization was performed with the anti-tag oligonucleotide. The different molecules are diagrammed on the right, as in Fig. 2. The black box in the 5′ UTR represents the G cassette. The numbers above the lanes indicate hours of galactose induction. Lane M contained MspI-digested pBR322 plasmid DNA.
To better understand the role of 5′→3′ exonucleases in U18 snoRNA processing, we also made use of a yeast strain carrying mutations in the RAT1 and XRN1 genes, whose protein products have been reported to be the major 5′→3′ exonuclease activities present in yeast extracts (21, 25). Both proteins RAT1p and XRN1p have been implicated in a number of processes related to RNA metabolism in vivo, including pre-rRNA processing (15, 43) and mRNA degradation (7, 18). Plasmids pGALU18wt, pGALU18C5, and pGALU18Cbs were transformed into the temperature-sensitive rat1-1 xrn1Δ double mutant strain (15), and the effect of these mutations on tagged U18 processing was determined. Figure 5A shows a Northern analysis performed with the anti-tag oligonucleotide on total RNA extracted from cells induced for different times with galactose after 1.5 h of growth at 37°C, which is the nonpermissive temperature for rat1-1. Even in the absence of the RAT1p and XRN1p exonucleases, processing of the mature U18 snoRNA from the wild-type pre-mRNA was not impaired and proceeded with time (lanes wt). In contrast to what has been shown for the same precursor in the CH1462 strain, lack of the RAT1p and XRN1p exonucleases led to visualization of the full-length pre-mRNA and of the linearized lariat, very likely as a result of their overall decreased decay rate. In addition, the I-3 intermediate and a shortened form of the lariat, trimmed to the 3′ end of U18 (I-3 intron), as assessed by hybridizations and primer extensions with appropriate oligonucleotides, were also visualized. The presence of the I-3 intermediate (wt lanes) indicates that U18 processing from the wild-type EFB1 transcript occurs not only from the spliced intron but also with lower efficiency, from the unspliced pre-mRNA. Figure 5A shows that the U18C5 and U18Cbs precursors were also able to produce mature U18 snoRNA, even though at a lower level than the wild-type construct (C5 and Cbs lanes). Their overall processing pattern was not affected by the lack of XRN1p and RAT1p activities, the only difference being the I-2 product, which was represented less than in the Exo+ strain. The reason for this is not clear.
FIG. 5.
Lack of the major 5′→3′ exonucleases does not affect U18 production. (A) Northern analysis of 5 μg of total RNA extracted from the rat1-1 xrn1Δ double mutant strain transformed with the pGALU18wt (wt lanes), pGALU18C5 (C5 lanes), or pGALU18Cbs (Cbs lanes) construct (Fig. 1). Cultures were shifted to the nonpermissive temperature, and after 1.5 h, galactose was added to the medium (hr-gal). Hybridization was performed with the anti-tag oligonucleotide. The different products are indicated on the right as in Fig. 2. The numbers above the lanes indicate hours of galactose induction. (B) Samples (5 μg) of the same RNAs extracted from the rat1-1 xrn1Δ strain transformed with the pGALU18wt construct (lanes A through D of panel A) were electroblotted onto a separate filter and hybridized with an oligonucleotide complementary to the ITS1 region of rRNA (see Materials and Methods). The positions of the two pre-5.8S rRNA species are indicated on the right. The migration of the mature 5.8S rRNA is also indicated. (C) Primer extension analysis using oligonucleotide L32 (see Materials and Methods), which is specific for the intronic sequence of the RPL32 transcript, performed on the same RNA preparations from the rat1-1 xrn1Δ strain (exo− lanes) transformed with the pGALU18wt construct (lanes A, C, and D of panel A) and on control RNA extracted from strain CH1462 (exo+ lane). The extended products corresponding to the 5′ ends of the intron (I) and of the pre-mRNA (P) are indicated. Although not shown, DNA sequencing reactions using oligonucleotide L32 were run alongside the primer extension reactions. Lanes M contained MspI-digested pBR322 plasmid DNA.
To verify whether the XRN1p and RAT1p activities are inhibited under our experimental conditions, we analyzed processing reactions known to respond to the inactivation of the same 5′→3′ exonucleases. Since it has been previously shown (15) that 5′-extended forms of 5.8S rRNA accumulate highly in strains carrying mutations in the RAT1 and XRN1 genes, the RNAs shown in Fig. 5A (lanes A through D) were probed with an oligonucleotide specific for the ITS1 region of the rRNA. Accordingly, Fig. 5B shows that at the nonpermissive temperature, processing intermediates extending to cleavage site A3, upstream from the 5.8S rRNA, accumulated. Under the same conditions, molecules intermediate in size, extending from the 5′ end of the U18 snoRNA to the 5′ end of the intron were not detected, either by Northern analysis (Fig. 5A) or by primer extension (see below). The XRN1p and RAT1p exonucleases are also involved in intron trimming or degradation; in fact, both the EFB1 and the RPL32 introns accumulated stably as linear molecules when the rat1-1 xrn1Δ strain was shifted to the nonpermissive temperature (Fig. 5A and C, respectively). Even if it is not possible to rule out the existence of a third exonuclease, 5′→3′ exonucleases other than XRN1p and RAT1p have not been identified in yeast extracts (21, 25, 42). If present, such an exonuclease should not be very active, as shown by the accumulation of large amounts of linearized lariats; it is more probable, then, that formation of the 5′ end of the U18 snoRNA is due to endonucleolytic activity.
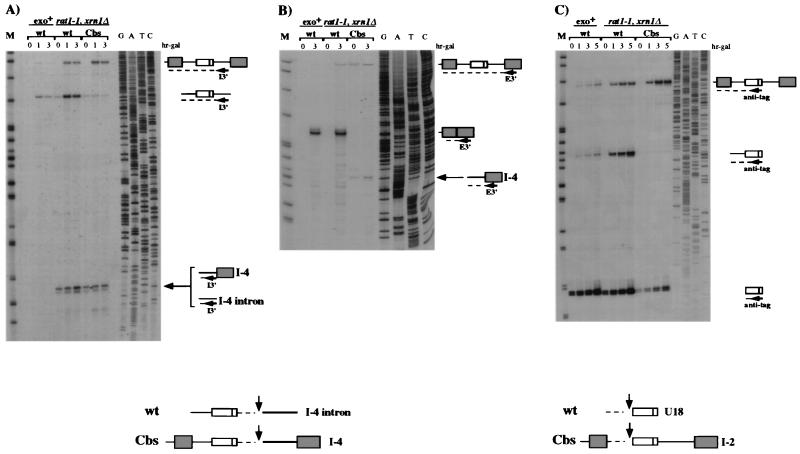
Endonucleolytic cleavage occurs 3′ of the U18 coding region.
The direct evidence for the occurrence of endonucleolytic cleavage is the identification of cutoff products. In a wild-type context, these intermediates are rapidly degraded, whereas in a rat1-1 xrn1Δ mutant strain, they should have a longer half-life since the residual degradative pathway would be the less efficient 3′→5′ exonucleolytic trimming (29). To search for the presence of 3′ cutoff molecules, we carried out primer extension analysis with an oligonucleotide hybridizing to the 3′ region of the intron (oligonucleotide I3′; Fig. 1). Figure 6A illustrates that in the rat1-1 xrn1Δ mutant strain, both the wild-type (wt lanes) and mutant (Cbs lanes) constructs revealed a strong stop of reverse transcriptase 25 nucleotides downstream from U18 (arrow) mapping at a UUAU sequence. This signal, which was absent in the control Exo+ strain (exo+ lanes), identifies the 5′ ends of 3′ cutoff molecules, which are stabilized in the rat1-1 xrn1Δ background. In the wild-type construct, this product should originate from the excised lariat (I-4 intron molecules), while in the Cbs splicing mutant, it is produced from the pre-mRNA (I-4 molecules). This was confirmed by primer extension with an oligonucleotide complementary to the 3′ exon (oligonucleotide E3′; Fig. 1), which revealed the presence of I-4 molecules with the Cbs construct (Fig. 6B). These data indicate that the I-4 intermediates can be produced either from processing of the spliced intron or from processing of the unspliced pre-mRNA. Primer extension with the anti-tag oligonucleotide revealed that in the rat1-1 xrn1Δ background, the wild-type and Cbs constructs produce a mature 5′ end of U18 without products with lengths intermediate between the 5′ end of the intron and U18, indicating that cleavage must occur very close to the 5′ end of U18 (Fig. 6C).
FIG. 6.
3′ cutoff molecules accumulate in the rat1-1 xrn1Δ mutant strain during processing of the U18 snoRNA. Primer extension analysis, using the I3′ (A), E3′ (B), or anti-tag (C) oligonucleotide, of total RNA extracted from the rat1-1 xrn1Δ double mutant strain (rat1-1 xrn1Δ lanes) transformed with the wild-type (wt) and Cbs constructs (Fig. 1) and of control RNA extracted from strain CH1462 (exo+ lanes) transformed with the wild-type construct. The positions of the oligonucleotides are shown in Fig. 1. rat1-1 xrn1Δ cells were shifted to the nonpermissive temperature, and after 1.5 h, galactose was added to the medium (hr-gal). The numbers above the lanes indicate hours of galactose induction. The different primer extension products are schematically represented on the right. Note that the signal corresponding to the 5′ end of the intron in the Cbs lanes is due to hybridization of the I3′ primer to the endogenous spliced EF-1β intron. DNA sequencing reactions using the I3′, E3′, and anti-tag oligonucleotides are also shown. Lanes M contained MspI-digested pBR322 plasmid DNA. At the bottom are schematic representations of the cutoff products that originated from 3′ and 5′ cleavage of the intron and of the pre-mRNA.
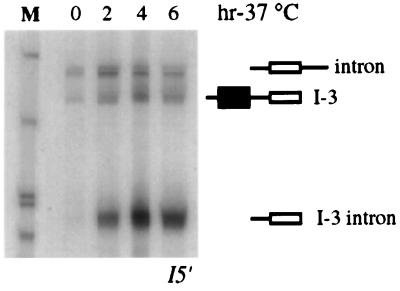
Northern analysis with an oligonucleotide hybridizing to the intronic sequence upstream of U18 was performed on RNA extracted from the untransformed rat1-1 xrn1Δ mutant strain grown at 24°C and then shifted to the nonpermissive temperature for different times. Figure 7 shows the presence of the I-3 and I-3 intron molecules (the different migration of I-3 in comparison to that in previous gels is due to the smaller size of the first exon of the endogenous EFB1 gene with respect to the ectopic constructs; see also the legend to Fig. 7). The coexistence of these products indicates that endogenous U18 snoRNA biosynthesis can also follow two different pathways based on processing from the debranched lariat and from the pre-mRNA. The existence of a splicing-independent pathway is in agreement with the recent finding that in strains deficient in debranching activity, the U18 snoRNA still accumulates, although at lower levels (34).
FIG. 7.
Endogenous U18 snoRNA processing in the rat1-1 xrn1Δ mutant strain. Northern analysis of total RNA extracted from the untransformed rat1-1 xrn1Δ double mutant strain at different times after a shift to the nonpermissive temperature (hr-37°C) and hybridized with oligonucleotide I5′ (Fig. 1). The different products are indicated on the right, as in Fig. 2. Note that the migration of the I-3 molecule is faster than that of the debranched intron with respect to previous gels. This is due to the smaller size of the first exon of the endogenous EFB1 gene with respect to the ectopic constructs (101 and 201 nt, respectively). Lane M contained MspI-digested pBR322 plasmid DNA.
DISCUSSION
A peculiar feature of most of the snoRNAs identified so far is that they are present in introns of protein-encoding genes and originate not from independent transcription but from processing of the pre-mRNAs which contain them (28). In the yeast S. cerevisiae, a subclass of snoRNAs are independently transcribed as part of polycistronic transcripts containing several snoRNAs interspersed with noncoding sequences (14, 45, 55). In both cases, the mechanism by which snoRNAs get released from their precursor molecules is still poorly understood.
Previous experiments with higher eukaryotes failed to demonstrate a unique processing mechanism leading to intronic snoRNA synthesis. Human U17 and U19 snoRNA processing studies suggested that splicing and snoRNA processing are linked and that the release of mature snoRNAs is the result of lariat debranching and subsequent exonucleolytic trimming from the ends of the linearized intron (22). In contrast, experiments with X. laevis oocytes and embryos with U14, U16, and U18 snoRNAs demonstrated that processing can take place independently of splicing (6, 10, 53). In the case of the U16 snoRNA, a detailed mutational analysis showed that intact boxes C and D are necessary for the formation of a specific complex containing fibrillarin and for committing the pre-mRNA to the processing pathway. Competition studies also indicated that if snoRNA processing is inhibited, splicing is enhanced, suggesting that the two processes compete with each other. Nevertheless, the coexistence of these two pathways does not exclude the possibility that part of the U16 snoRNA also originates through the splicing pathway. In the case of the U16 snoRNA, this peculiar splicing-processing phenotype correlates with the snoRNA being inside a poorly spliceable intron (6).
In this investigation, we extended the study of the processing of snoRNA-containing introns to the yeast S. cerevisiae U18 snoRNA, which is encoded in the intron of the EFB1 gene. To dissect the U18 biosynthetic pathway and study the relationship between splicing and processing, we used two different approaches: the first involved the analysis of snoRNA production from constructs mutated at specific splice sites, and the second made use of cis- and trans-acting mutations affecting the activity of 5′→3′ exonucleases. In both cases, the aim was to analyze the overall effect of these mutations on U18 accumulation.
We have shown that inhibition of splicing, obtained through site-specific mutagenesis of either the 5′ splice site or the branch sequence, did not affect U18 biosynthesis, thus indicating that the snoRNA can be efficiently produced by a pathway alternative to splicing. In these experiments, it was also possible to identify processing intermediates revealing the utilization of the unspliced pre-mRNA as a substrate for this alternative pathway. They correspond to pre-mRNA molecules truncated at either the 5′ or 3′ end of U18 and are analogous to the intermediates previously identified for U18 (and also U16) snoRNA processing in X. laevis (36). These molecules were also visualized in the processing of the endogenous EFB1 pre-mRNA when 5′→3′ exonucleolytic activity was blocked, indicating that processing from the unspliced pre-mRNA can also occur when splicing is not inhibited. This is consistent with independent data demonstrating that in strains deficient in debranching activity, the U18 snoRNA still accumulates (approximately 30% of the wild-type level; 34, 51).
Two different mechanisms could be responsible for U18 processing from the pre-mRNA: (i) exonucleolytic trimming from the ends of the transcript or (ii) endonucleolytic cleavage followed by trimming. The first possibility was ruled out by the use of constructs containing, in the 5′ UTR, a poly(G) cassette, which is known to represent an efficient structural block to yeast 5′→3′ exonucleases (9, 35, 52) and which should block exonucleolytic progression from the 5′ end of the pre-mRNA. This construct exhibited normal accumulation of the U18 snoRNA, indicating that some endonucleolytic activity must be involved in the production of the 5′ end of the U18 snoRNA. Utilizing the rat1-1 xrn1Δ mutant strain, we have shown that the U18 snoRNA normally accumulates from the wild-type EFB1 construct and is still present, although at lower levels, in the case of the splicing mutant derivatives. In this strain, we were also able to identify RNA products proving the occurrence of endonucleolytic cleavage 3′ of U18: a 3′ cutoff product whose 5′ end maps 25 nucleotides downstream of U18 in correspondence to a UUAU sequence was found. These molecules were not detected in Exo+ strains because they are rapidly degraded; instead, they were visualized only under conditions of 5′→3′ exonuclease inhibition. The 5′ cutoff products originating from cleavage downstream of U18 were not detected because in the rat1-1 xrn1Δ strain the 3′→5′ exonucleases are fully functional and can trim the cleaved products from the 3′ end. Since we did not detect 5′-extended forms of U18 in the rat1-1 xrn1Δ strain, we suggest the occurrence of a processing site very close to the 5′ end of U18. Interestingly, a UUAU sequence immediately precedes the 5′ end of U18. This situation is very similar to that previously described for the U16 snoRNA in X. laevis, where one cleavage site was found 15 nucleotides 3′ to the snoRNA coding region and four independent sites were identified in the upstream region, one being only three nucleotides from the 5′ end of U16.
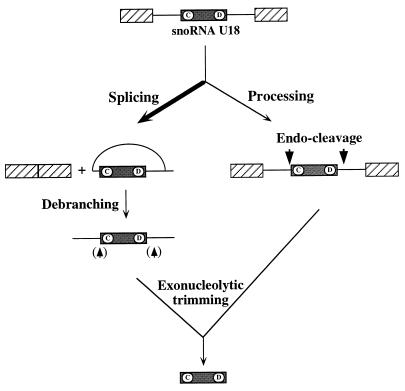
It is important to point out that while endonucleolytic cleavage is of crucial importance for the release of U18 from the splicing-independent pathway, exoribonuclease trimming would be sufficient to convert the debranched lariat to the mature snoRNA in the splicing pathway. Nevertheless, the two pathways can coexist, as demonstrated by the occurrence of endonucleolytic processing of the released intron. These data indicate that the U18 snoRNA is preferentially released from its host intron through the splicing pathway; nevertheless, a second minor pathway exists which produces U18 through processing of the entire primary transcript. When the splicing efficiency is reduced, this second pathway becomes the dominant biosynthetic mechanism (Fig. 8). Endonucleolytic processing of snoRNAs is not surprising, since many cases of poly-snoRNA transcripts have been reported in yeast and plants (14, 26, 55). Nevertheless, the physiological relationships among endonucleolytic processing, splicing, and RNA turnover of snoRNA-containing precursors inside the nucleus are essential aspects that still needs clarification. Identification of the processing factors and the elements conferring specificity on the reaction will explain how these processes are related to each other.
FIG. 8.
Schematic representation of the two alternative pathways leading to accumulation of the U18 snoRNA in yeast. The arrowheads not in parentheses indicate the positions of endonucleolytic cleavages; those in indicate cleavages identified inside the intron in the rat1-1 xrn1Δ strain.
ACKNOWLEDGMENTS
We thank Josette Banroques for hospitality in her laboratory to T.V. in the initial part of this work and Stephen Kearsey and David Tollervey for providing the Exo− strain. We also thank Alessandro Michienzi for helpful discussions, Massimo Arceci for his skillful technical help, and Fabio Riccobono and M-Medical for oligonucleotide facilities.
This work was partially supported by grants from Biotecnologie MURST 5%, C.N.R. Target Project on Biotechnology, and PRIN 40%-MURST.
REFERENCES
- 1.Bachellerie J-P, Michot B, Nicoloso M, Balakin A, Ni J, Fournier M J. Antisense snoRNAs: a family of nucleolar RNAs with long complementarities to rRNA. Trends Biochem Sci. 1995;20:261–264. doi: 10.1016/s0968-0004(00)89039-8. [DOI] [PubMed] [Google Scholar]
- 2.Balakin A G, Smith L, Fournier M J. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 3.Baserga S J, Yang X W, Steitz J A. An intact box C sequence in the U3 snRNA is required for binding of fibrillarin, the protein common to the major family of nucleolar snRNPs. EMBO J. 1991;10:2645–2651. doi: 10.1002/j.1460-2075.1991.tb07807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boorstein W R, Craig E A. Primer extension analysis of RNA. Methods Enzymol. 1989;180:347–369. doi: 10.1016/0076-6879(89)80111-9. [DOI] [PubMed] [Google Scholar]
- 5.Caffarelli E, Arese M, Santoro B, Fragapane P, Bozzoni I. In vitro study of processing of the intron-encoded U16 small nucleolar RNA in Xenopus laevis. Mol Cell Biol. 1994;14:2966–2974. doi: 10.1128/mcb.14.5.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caffarelli E, Fatica A, Prislei S, De Gregorio E, Fragapane P, Bozzoni I. Processing of the intron-encoded U16 and U18 snoRNAs: the conserved C and D boxes control both the processing reaction and the stability of the mature snoRNA. EMBO J. 1996;15:1121–1131. [PMC free article] [PubMed] [Google Scholar]
- 7.Caponigro G, Parker R. Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol Rev. 1996;60:233–249. doi: 10.1128/mr.60.1.233-249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavaillé J, Bachellerie J-P. Processing of fibrillarin-associated snoRNAs from pre-mRNA introns: an exonucleolytic process exclusively directed by the common stem-box terminal structure. Biochimie. 1996;78:443–456. doi: 10.1016/0300-9084(96)84751-1. [DOI] [PubMed] [Google Scholar]
- 9.Decker C J, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 10.Fragapane P, Prislei S, Michienzi A, Caffarelli E, Bozzoni I. A novel small nucleolar RNA (U16) is encoded inside a ribosomal protein intron and originates by processing of the pre-mRNA. EMBO J. 1993;12:2921–2928. doi: 10.1002/j.1460-2075.1993.tb05954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganot P, Bortolin M-L, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 12.Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- 13.Girard J-P, Lehtonen H, Caizergues-Ferrer M, Amalric F, Tollervey D, Lapeyre B. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J. 1992;11:673–682. doi: 10.1002/j.1460-2075.1992.tb05099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henry, Y. Personal communication.
- 15.Henry Y, Wood H, Morrissey J P, Petfalski E, Kearsey S, Tollervey D. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 1994;13:2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernandez N. Transcription of vertebrate snRNA genes and related genes. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 281–313. [Google Scholar]
- 17.Hiraga K, Suzuki K, Tsuchiya E, Miyakawa T. Cloning and characterization of the elongation factor EF-1β homologue of Saccharomyces cerevisiae. FEBS Lett. 1993;316:165–169. doi: 10.1016/0014-5793(93)81208-h. [DOI] [PubMed] [Google Scholar]
- 18.Hsu C L, Stevens A. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang G M, Jarmolowski A, Struck J C R, Fournier M J. Accumulation of U14 small nuclear RNA in Saccharomyces cerevisiae requires box C, box D, and a 5′, 3′ terminal stem. Mol Cell Biol. 1992;12:4456–4463. doi: 10.1128/mcb.12.10.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jansen R P, Hurt E C, Kern H, Lehtonen H, Carmo-Fonseca M, Lapeyre B, Tollervey D. Evolutionary conservation of the human nucleolar protein fibrillarin and its functional expression in yeast. J Cell Biol. 1991;113:715–729. doi: 10.1083/jcb.113.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kenna M, Stevens A, McCammon M, Douglas M G. An essential yeast gene with homology to the exonuclease-encoding XRN1/KEM1 gene also encodes a protein with exoribonuclease activity. Mol Cell Biol. 1993;13:341–350. doi: 10.1128/mcb.13.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiss T, Filipowicz W. Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev. 1995;9:1411–1424. doi: 10.1101/gad.9.11.1411. [DOI] [PubMed] [Google Scholar]
- 23.Kiss-László Z, Henry Y, Bachellerie J-P, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 24.Kranz J E, Holm C. Cloning by function: an alternative approach for identifying yeast homologs of genes from other organisms. Proc Natl Acad Sci USA. 1990;87:6629–6633. doi: 10.1073/pnas.87.17.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larimer F W, Hsu C L, Maupin M K, Stevens A. Characterization of the XRN1 gene encoding a 5′→3′ exoribonuclease: sequence data and analysis of disparate protein and mRNA levels of gene-disrupted yeast cells. Gene. 1992;120:51–57. doi: 10.1016/0378-1119(92)90008-d. [DOI] [PubMed] [Google Scholar]
- 26.Leader D J, Clark G P, Watters J, Beven A F, Shaw P J, Brown J W S. Clusters of multiple different small nucleolar RNA genes in plants are expressed as and processed from polycistronic pre-snoRNAs. EMBO J. 1997;16:5742–5751. doi: 10.1093/emboj/16.18.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lesser C F, Guthrie C. Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science. 1993;262:1982–1988. doi: 10.1126/science.8266093. [DOI] [PubMed] [Google Scholar]
- 28.Maxwell E S, Fournier M J. The small nucleolar RNAs. Annu Rev Biochem. 1995;35:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 29.Muhlrad D, Decker C J, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 30.Mumberg D, Müller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ni J, Tien A L, Fournier M J. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 32.Nicoloso M, Qu L-H, Michot B, Bachellerie J-P. Intron-encoded antisense small nucleolar RNAs: the characterization of nine novel species points to their role as guides for the 2′-O-ribose methylation of rRNAs. J Mol Biol. 1996;260:178–195. doi: 10.1006/jmbi.1996.0391. [DOI] [PubMed] [Google Scholar]
- 33.Peculis B A, Steitz J A. Sequence and structural elements critical for U8 snRNP function in Xenopus oocytes are evolutionarily conserved. Genes Dev. 1994;8:2241–2255. doi: 10.1101/gad.8.18.2241. [DOI] [PubMed] [Google Scholar]
- 34.Petfalski E, Dandekar T, Henry Y, Tollervey D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol Cell Biol. 1998;18:1181–1189. doi: 10.1128/mcb.18.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Presutti C, Villa T, Hall D, Pertica C, Bozzoni I. Identification of the cis-elements mediating the autogenous control of ribosomal protein L2 mRNA stability in yeast. EMBO J. 1995;14:4022–4030. doi: 10.1002/j.1460-2075.1995.tb00073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prislei S, Michienzi A, Presutti C, Fragapane P, Bozzoni I. Two different snoRNAs are encoded in introns of amphibian and human L1 ribosomal protein genes. Nucleic Acids Res. 1993;21:5824–5830. doi: 10.1093/nar/21.25.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rain J-C, Legrain P. In vivo commitment to splicing in yeast involves the nucleotide upstream from the branch site conserved sequence and the Mud2 protein. EMBO J. 1997;16:1759–1771. doi: 10.1093/emboj/16.7.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schimmang T, Tollervey D, Kern H, Frank R, Hurt E C. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 1989;8:4015–4024. doi: 10.1002/j.1460-2075.1989.tb08584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmitt M E, Brown T A, Trumpower L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith C M, Steitz J A. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- 41.Steitz J A, Tycowski K T. Small RNA chaperones for ribosome biogenesis. Science. 1995;270:1626–1627. doi: 10.1126/science.270.5242.1626. [DOI] [PubMed] [Google Scholar]
- 42.Stevens A. Purification and characterization of a Saccharomyces cerevisiae exoribonuclease which yields 5′-mononucleotides by a 5′→3′ mode of hydrolysis. J Biol Chem. 1980;255:3080–3085. [PubMed] [Google Scholar]
- 43.Stevens A, Hsu C L, Isham K R, Larimer F W. Fragments of the internal transcribed spacer 1 of pre-rRNA accumulate in Saccharomyces cerevisiae lacking 5′→3′ exoribonuclease 1. J Bacteriol. 1991;173:7024–7028. doi: 10.1128/jb.173.21.7024-7028.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terns M P, Grimm C, Lund E, Dahlberg J E. A common maturation pathway for small nucleolar RNAs. EMBO J. 1995;14:4860–4871. doi: 10.1002/j.1460-2075.1995.tb00167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 46.Tyc K, Steitz J A. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 1989;8:3113–3119. doi: 10.1002/j.1460-2075.1989.tb08463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tycowski K T, Shu M D, Steitz J A. A small nucleolar RNA is processed from an intron of the human gene encoding ribosomal protein S3. Genes Dev. 1993;6:1120–1130. doi: 10.1101/gad.7.7a.1176. [DOI] [PubMed] [Google Scholar]
- 48.Tycowski K T, Shu M D, Steitz J A. A mammalian gene with introns instead of exons generating stable RNA products. Nature. 1996;379:464–466. doi: 10.1038/379464a0. [DOI] [PubMed] [Google Scholar]
- 49.Venema J, Tollervey D. Processing of pre-ribosomal RNA in Saccharomyces cerevisiae. Yeast. 1995;11:1629–1650. doi: 10.1002/yea.320111607. [DOI] [PubMed] [Google Scholar]
- 50.Vijayraghavan U, Parker R, Tamm J, Iimura Y, Rossi J, Abelson J, Guthrie C. Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathway, particularly assembly of the spliceosome. EMBO J. 1986;5:1683–1695. doi: 10.1002/j.1460-2075.1986.tb04412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Villa T. Ph.D. thesis. Rome, Italy: University of Rome “La Sapienza”; 1998. [Google Scholar]
- 52.Vreken P, Raué H A. The rate-limiting step in yeast PGK1 mRNA degradation is an endonucleolytic cleavage in the 3′-terminal part of the coding region. Mol Cell Biol. 1992;12:2986–2996. doi: 10.1128/mcb.12.7.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watkins N J, Leverette R D, Xia L, Andrews M T, Maxwell E S. Elements essential for processing intronic U14 snoRNA are located at the termini of the mature snoRNA sequence and include conserved nucleotide boxes C and D. RNA. 1996;2:118–133. [PMC free article] [PubMed] [Google Scholar]
- 54.Xia L, Watkins N J, Maxwell E S. Identification of specific nucleotide sequences and structural elements required for intronic U14 snoRNA processing. RNA. 1997;3:17–26. [PMC free article] [PubMed] [Google Scholar]
- 55.Zagorski J, Tollervey D, Fournier M J. Characterization of an SNR gene locus in Saccharomyces cerevisiae that specifies both dispensible and essential small nuclear RNAs. Mol Cell Biol. 1988;8:3282–3290. doi: 10.1128/mcb.8.8.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]