Abstract
Signal transducer and activator of transcription 6 (Stat6) and NF-κB are widely distributed transcription factors which are induced by different stimuli and bind to distinct DNA sequence motifs. Interleukin-4 (IL-4), which activates Stat6, synergizes with activators of NF-κB to induce IL-4-responsive genes, but the molecular mechanism of this synergy is poorly understood. Using glutathione S-transferase pulldown assays and coimmunoprecipitation techniques, we find that NF-κB and tyrosine-phosphorylated Stat6 can directly bind each other in vitro and in vivo. An IL-4-inducible reporter gene containing both cognate binding sites in the promoter is synergistically activated in the presence of IL-4 when Stat6 and NF-κB proteins are coexpressed in human embryonic kidney 293 (HEK 293) cells. The same IL-4-inducible reporter gene is also synergistically activated by the endogenous Stat6 and NF-κB proteins in IL-4-stimulated I.29μ B lymphoma cells. Furthermore, Stat6 and NF-κB bind cooperatively to a DNA probe containing both sites, and the presence of a complex formed by their cooperative binding correlates with the synergistic activation of the promoter by Stat6 and NF-κB. We conclude that the direct interaction between Stat6 and NF-κB may provide a basis for synergistic activation of transcription by IL-4 and activators of NF-κB.
Signal transducer and activator of transcription 6 (Stat6) and NF-κB belong to two distinct families of transcription factors, the STAT family and the NF-κB/Rel family, respectively. Members of both families play important roles in immune responses and in cell differentiation induced by cytokines, growth factors, and other cell activators. Stat6, like other STATs, is in the cytoplasm as a latent monomer. Upon cytokine stimulation, mainly by interleukin-4 (IL-4), Stat6 is rapidly phosphorylated by Janus kinases (Jaks) to form homodimers which enter the nucleus where Stat6 binds various target genes containing Stat6 binding sites (3, 7, 10). Unlike Stat6, NF-κB proteins are kept in the cytoplasm by association with IκB proteins. When cells are stimulated by CD40L, lipopolysaccharide (LPS), or a variety of other stimuli, NF-κB proteins are released from the associated IκB proteins and enter the nucleus, where they bind consensus κB sites in many genes (1).
Some STAT proteins bind other transcription factors or coactivators to activate transcription (2, 23, 26, 40), but Stat6 has not been shown to bind any known transcription factors. NF-κB/Rel proteins have also been shown to interact with other transcription factors or proteins of the basal transcription machinery which regulate NF-κB/Rel proteins positively or negatively in transcriptional activation (11, 13, 35, 36, 39). Recent studies suggest that Stat6 and NF-κB, activated by discrete signaling pathways, interact at some unknown point to synergistically activate transcription of certain genes that are induced in response to both signals. Examples of such genes are the germline (GL) immunoglobulin (Ig) Cɛ and Cγ1 genes, the transcription of which is necessary for class switching to IgE and IgG1 (33). Functional studies of the GL Cɛ and Cγ1 promoters indicate that both Stat6 and κB sites are required for optimal induction of transcription by IL-4 and/or by CD40L (4, 9, 15, 16, 38), but the molecular mechanism for synergistic activation of promoters by Stat6 and NF-κB proteins is not understood. A Stat6 and two or three κB sites are closely positioned in the promoters of three relatively well-characterized IL-4-responsive genes, GL Cɛ, GL Cγ1, and FcɛRII (CD23) (25), so together with the functional data, this information raises the possibility that Stat6 and NF-κB interact directly with each other. In this report, we provide physical evidence that Stat6 and NF-κB directly bind each other in vitro and in vivo as well as functional evidence that these two transcription factors cooperatively bind their cognate DNA binding sites and synergistically activate transcription. The significance of their interaction in IL-4 signaling is discussed.
MATERIALS AND METHODS
Cells and cell culture.
sIgM+ mouse B lymphoma cell line I.29μ (34) was grown at 37°C in an atmosphere of 8% CO2 in RPMI 1640 medium-20% fetal calf serum (FCS) (32). Human embryonic kidney 293 (HEK 293) cells (ATCC CRL-1573) were grown at 37°C in an atmosphere of 5% CO2 in Dulbecco’s modified Eagle’s medium-10% FCS. Sf9 insect cells were cultured at 27°C in Grace’s medium-10% FCS.
Preparation of recombinant proteins. (i) GST fusion proteins.
Mouse NF-κB p50 or p65 coding sequence was inserted 3′ and in frame to glutathione S-transferase (GST) coding sequence in the bacterial expression vector pGEX-2T (Pharmacia Biotech). The plasmid pGEX-2T-LSF was a gift from U. Hansen and E. Drouin (Dana-Farber Cancer Institute, Boston, Mass.). Escherichia coli BL21(DE3) cells (50 ml at an optical density at 600 nm of 0.8) harboring the recombinant expression vectors were induced with 1 mM isopropyl β-d-thiogalactopyranoside (IPTG; American Bioanalytical) at 30°C for 3 h. Cells were washed by resuspension and recentrifugation twice in phosphate-buffered saline (PBS) and were finally suspended in 4 ml of ice-cold suspension buffer (PBS supplemented with 1% Triton X-100, 1 mM EDTA, 1 mM dithiothreitol [DTT], and 1 mM phenylmethylsulfonyl fluoride [PMSF]). The cell suspension was sonicated, and insoluble debris was pelleted by centrifugation (12,000 × g for 15 min at 4°C). The supernatants were mixed with glutathione-agarose beads (300 μl of a 1:1 slurry in PBS) (Sigma) at 4°C for 30 min. The beads were then pelleted, washed five times with ice-cold suspension buffer and, finally, suspended in 1 ml of suspension buffer. The purity and quantity of bound full-length GST fusion proteins were examined by sodium dodecyl sulfate (SDS)–10% polyacrylamide gel electrophoresis (PAGE) followed by Coomassie blue staining.
(ii) His6-tagged rStat6.
To generate the Stat6 baculovirus expression vector, the sequence between the XhoI site and the first ATG of mouse Stat6 cDNA in mStat6-pBKS plasmid (22) was replaced by a single G and the XhoI/NotI fragment from the subsequently derived plasmid was cloned into the XhoI/NotI sites of the baculovirus transfer vector pAcHLT-C (PharMingen). The baculovirus transfer vector encoding Stat6 was cotransfected with baculovirus DNA into Sf9 cells by using the BaculoGold kit (PharMingen) to generate recombinant baculoviruses. Recombinant baculoviruses expressing human Jak2 were also generated by cotransfecting the Jak2 baculovirus transfer vector TPU 276 (27) with baculovirus DNA into Sf9 cells. Tyrosine-phosphorylated Stat6 was expressed in Sf9 cells by coinfection with both recombinant baculoviruses (27) with a multiplicity of infection of 10. Three days postinfection, nuclear extracts were prepared as described previously (29), except that EDTA, EGTA, and DTT were not included in buffer A or buffer C. Recombinant Stat6 (rStat6) was then purified from the nuclear extracts on Ni2+-nitrilotriacetic acid agarose (Qiagen) and eluted with elution buffer (12.5 mM HEPES [pH 7.9], 100 mM NaCl, 0.05% Nonidet P-40 [NP-40], 10% [vol/vol] glycerol, 2 mM β-mercaptoethanol, 1 mM PMSF) containing increasing concentrations of imidazole (50 to 500 mM). The purity of rStat6 in each eluted fraction was examined by electrophoresis on an SDS–10% PAGE gel followed by silver staining. The fractions containing rStat6 as the major band (>85% by densitometry) were pooled for the experiments.
Preparation of nuclear extracts. (i) I.29μ B cells.
Cells were left untreated or treated with recombinant mouse IL-4 (2,000 U/ml) produced by insect cells (from W. E. Paul, National Institutes of Health) for 2 h before harvesting. Cells to be treated with or without anti-IL-4 antibodies were cultured in the presence or absence of 5% hybridoma 11B11 culture supernatant (20) for 20 h before being harvested. Nuclear extracts were then prepared as described previously (29), except that 1 mM Na3VO4 was used as a supplement in buffer A and buffer C. Nuclear extracts to be treated with T-cell protein tyrosine phosphatase (TCPTP; New England Biolabs) in GST pulldown assays (see below) were prepared without Na3VO4 in buffer A and buffer C.
(ii) Transfected HEK 293 cells.
Transfection of cells was scaled up 10-fold relative to the transfection described in transient transfection assays (see below), except no reporter plasmid or internal control plasmid pfosCAT was included. Forty-eight hours posttransfection, cells were stimulated with recombinant human IL-4 (10 ng/ml) (Genzyme) for 15 min or left untreated before harvesting. Nuclear extracts were then prepared as described above.
GST pulldown assays.
GST fusion proteins (0.5 μg) bound to glutathione-agarose beads were incubated with nuclear extracts (10 μg) from IL-4-stimulated or unstimulated I.29μ B cells or with purified rStat6 (1 or 5 ng) after the nuclear extracts and rStat6 were subjected to different treatments. For nuclear extracts treated with TCPTP, nuclear extracts from IL-4-stimulated I.29μ B cells were treated with TCPTP in the presence or absence of 1 mM Na3VO4 at 30°C for 30 min in accordance with the manufacturer’s instructions prior to incubation with GST fusion proteins. For rStat6 treated with TCPTP, purified proteins were either pretreated with TCPTP or left untreated at 30°C for 5 h prior to incubation with GST fusion proteins. One nanogram of rStat6 was then used for binding to glutathione-agarose beads. For samples treated with ethidium bromide (EtBr), both purified rStat6 and GST fusion proteins bound to glutathione-agarose beads were preincubated with EtBr (200 μg/ml) for 30 min on ice. rStat6 (5 ng), untreated or treated with EtBr, was then used for binding to untreated or EtBr-treated GST fusion proteins (0.5 μg) bound to glutathione-agarose beads. The pulldown binding reaction was performed for 30 min at room temperature in 100 μl of binding buffer (12.5 mM HEPES [pH 7.9], 5 mM KCl, 100 mM NaCl, 0.05% NP-40, 100 μg of bovine serum albumin [BSA] per ml, 0.1 mM EDTA, 1 mM DTT, 1 mM PMSF, 1 mM Na3VO4). For those samples pretreated with EtBr, EtBr (200 μg/ml) was included in the binding buffer. The beads were then washed by resuspension and recentrifugation five times with ice-cold binding buffer without BSA and, finally, suspended and boiled for 3 min in SDS-sample buffer (125 mM Tris-HCl [pH 6.8], 20% [vol/vol] glycerol, 4% SDS, 3% DTT, 0.001% bromophenol blue). The bound Stat6 was resolved by SDS–10% PAGE, followed by Western blotting with anti-Stat6 antibody (Santa Cruz Biotechnology). For some experiments, the blots were stripped of bound antibodies and reprobed with anti-PU.1 or anti-GST antibodies (Santa Cruz Biotechnology).
Immunoprecipitation.
Nuclear extracts (100 μg) from untreated or IL-4-treated I.29μ B cells and purified rStat6 (50 ng) from Sf9 insect cells were incubated with anti-Stat6 antibodies (2 μg) in 0.5 ml of binding buffer for 1 h at 4°C. The reaction mixtures were then incubated with protein A-Sepharose beads (10 μl of a 1:1 slurry in PBS) for 1 h at 4°C, after which the beads were washed three times with RIPA buffer (10 mM Tris-HCl [pH 6.8], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS). The beads were resuspended and boiled for 3 min in SDS-sample buffer, and the samples were subjected to SDS–7.5% PAGE followed by Western blotting with antiphosphotyrosine antibody (RC20:HRPO; Transduction Laboratories) or anti-Stat6 antibody.
Coimmunoprecipitation.
I.29μ B cells (2 × 107) were cultured in methionine- and cysteine-free medium supplemented with 0.5 mCi of [35S]protein labeling mix (DuPont NEN) per ml for 3 h. Cells were then treated with LPS (50 μg/ml) (E. coli serotype O55:B5; Sigma) for 2 h, followed by stimulation with or without IL-4 (2,000 U/ml) for 30 min. Nuclear extracts were prepared as described above. Labeled nuclear extracts were precleared by incubation with protein A-Sepharose for 1 h at 4°C. The precleared nuclear extracts (180 μg each) were mixed with 2 μl of anti-Stat6 antibody and 20 μl of a 1:1 slurry (in PBS) of protein A-Sepharose, which had been preincubated with unlabeled nuclear extracts from untreated I.29μ B cells for 4 h. After incubation for 2 h at 4°C, the protein A-Sepharose beads were washed five times with the binding buffer (12.5 mM HEPES [pH 7.9], 100 mM NaCl, 0.05% NP-40, 100 μg of BSA per ml, 0.1 mM EDTA, 1 mM DTT, 1 mM PMSF, 1 mM Na3VO4). The beads were then transferred to fresh tubes, mixed with 0.5 ml of elution buffer (20 mM Tris-HCl [pH 6.8], 50 mM NaCl, 1% SDS, 5 mM DTT) and boiled for 5 min. After centrifugation, the supernatant was collected and divided into two aliquots; each one was mixed with an equal amount of dilution buffer (20 mM Tris-HCl [pH 6.8], 50 mM NaCl, 1% NP-40, 1% sodium deoxycholate), 20 μl of a 1:1 slurry of protein A-Sepharose beads which had been preincubated with unlabeled nuclear extracts as described above, and 1 μl of anti-NF-κB p50 or anti-NF-κB p65 antiserum (24). After overnight incubation at 4°C, the beads were washed five times with RIPA buffer and suspended in SDS-sample buffer. The precipitated proteins were analyzed by SDS–7.5% PAGE and fluorography.
Transient transfection assays. (i) HEK 293 cells.
The luciferase reporter plasmid (IL-4 RR)2-pFL was generated as follows. The IL-4 responsive region (RR) −124/−79 segment from the mouse GL Cɛ promoter (top strand DNA sequence, 5′-TGCCTTAGTCAACTTCCCAAGAACAGAATCAAAAGGGAACTTCCAA-3′), which contains a C/EBP, a Stat6, and a κB site (underlined), with an additional 5′ KpnI site (5′-GGTACC-3′) and an additional 3′ sequence containing a HindIII site (5′-TCGACAAGCTT-3′), was first ligated to the same fragment at the KpnI site to form a dimer, and the dimer fragment was then inserted into the HindIII site of pFL (4). The luciferase reporter plasmid containing mutated Stat6 sites was constructed by substituting DNA sequence −106/−98 of the −124/−79 segment with an XhoI sequence (5′-CCTCGAGG-3′). The reporter plasmid containing mutated κB sites was generated similarly by substituting DNA sequence −87/−80 of the −124/−79 segment with the same XhoI sequence. The mammalian expression vectors for mouse Stat6, NF-κB p50, or NF-κB p65 were derived by cloning their cDNA sequences into pcDNA3 (Invitrogen). All of the plasmids used for transfection were prepared by CsCl gradient purification. HEK 293 cells (2.5 × 105) were seeded in 5-ml cultures, and transfection was performed 24 h later by the calcium phosphate method. For each transfection, one or more expression plasmid(s) (0.5 μg each) for Stat6 or NF-κB (adjusted to 2 μg of total DNA with the empty vector pcDNA3) was cotransfected with 0.5 μg of luciferase reporter plasmid and 1 μg of internal control plasmid pfosCAT (5) for normalizing transfection efficiency. Forty hours posttransfection, cells were stimulated with or without IL-4 (10 ng/ml) for 8 h before being harvested. Preparations of cell lysates were made, and luciferase and chloramphenicol acetyltransferase (CAT) assays were performed, as described previously (4). The luciferase activity was normalized for transfection efficiency by the CAT activity.
(ii) I.29μ B cells.
Cells (5 × 107) mixed with 20 μg of reporter plasmid and 5 μg of internal control plasmid pSV2CAT (6) in 1 ml of RPMI 1640 medium were electroporated at 1,250 μF/300 V and aliquoted into two fractions, and then each fraction was cultured for 15 h in 10 ml of complete medium supplemented with IL-4 (2,000 U/ml) or not supplemented. Luciferase and CAT activities were assayed and calculated as described above.
Electrophoretic mobility shift assays (EMSAs).
A double-stranded oligonucleotide containing the GL Cɛ IL-4 RR −124/−79 segment was used as the DNA probe. DNA binding reactions were performed at room temperature for 20 min in 15-μl reaction volumes containing 5 μg of nuclear extracts, 25 fmol of 32P-end-labeled DNA probe, 2 μg of poly(dI-dC), 12.5 mM HEPES (pH 7.9), 10% (vol/vol) glycerol, 5 mM KCl, 0.1 mM EDTA, 0.05% NP-40, and 1 mM DTT. Analysis of binding complexes was performed by electrophoresis in 4 or 5% native polyacrylamide gels with 0.5× Tris-borate-EDTA buffer, followed by autoradiography. For DNA competition experiments, unlabeled competitor oligonucleotides were added into the reaction mixture at 100-fold molar excess over the probe. Antibody supershift experiments were performed with 5 μg of nuclear extracts in the binding reaction mixtures supplemented with 1 μl of the indicated antibodies and 10 μl of protein A-Sepharose (1:1 slurry). After 20 min, Sepharose beads were pelleted, and the supernatant was loaded onto the gel.
RESULTS
GST-NF-κB p50 and GST-NF-κB p65 pull down Stat6 in vitro.
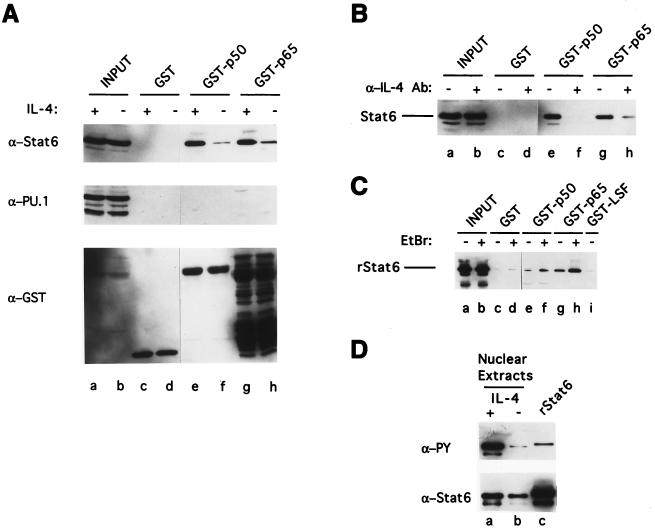
To determine whether Stat6 and NF-κB bind each other in the absence of their DNA binding sites, we tested the ability of GST-NF-κB fusion proteins to pull down Stat6. cDNAs encoding either NF-κB p50 or NF-κB p65 were fused with GST coding sequence and expressed in E. coli. These GST fusion proteins were bound to glutathione-agarose beads and incubated with nuclear extracts from I.29μ B lymphoma cells untreated or treated with IL-4. Figure 1A shows that GST-NF-κB p50 and GST-NF-κB p65 fusion proteins pull down Stat6 and that much more Stat6 is pulled down from nuclear extracts of IL-4-stimulated than of unstimulated I.29μ B cells (top panel, lanes e through h). Beads containing the GST tag alone do not pull down Stat6 (top panel, lanes c and d). To provide evidence for the specificity of this binding, we determined whether another transcription factor, PU.1, is also pulled down, by reprobing the same blot with antibody against PU.1. Figure 1A (middle panel) demonstrates that PU.1 is not pulled down by any of the GST-NF-κB fusion proteins.
FIG. 1.
Interaction of Stat6 and NF-κB p50 and NF-κB p65 in GST pulldown assays. (A) GST fusion proteins, as indicated, were incubated with nuclear extracts from I.29μ sIgM+ B lymphoma cells stimulated with IL-4 for 2 h (lanes c, e, and g) or cells left unstimulated (lanes d, f, and h). Lanes a and b contain the input proteins in the nuclear extracts (10 μg each). The bound proteins were resolved by SDS–10% PAGE, followed by Western blotting with anti-Stat6 (top panel), anti-PU.1 (middle panel), and anti-GST (bottom panel) antibodies sequentially on the same blot. In the bottom panel, the bands below the full-length protein in GST-p65 lanes are shorter forms of GST-p65 fusion proteins. Although the photograph has been cut and spliced, all the lanes are from the same blot, autoradiographed for the same time. This is also true for panels B and C. (B) Nuclear extracts from I.29μ B cells left untreated (lanes c, e, and g) or cultured in the presence of anti-IL-4 antibody (lanes d, f, and h) were incubated with GST fusion proteins as indicated. The bound Stat6 was resolved by SDS–10% PAGE, followed by Western blotting with anti-Stat6 antibody. The result from three-times-longer exposure of the film than that used for panel A is shown. Input Stat6 in the nuclear extracts (10 μg) is shown in lanes a and b. (C) Purified rStat6, untreated or treated with EtBr, was used for binding to GST fusion proteins as indicated in the absence (lanes c, e, g, and i) or presence (lanes d, f, and h) of EtBr. Lanes a and b show the inputs of pretreated rStat6 (5 ng). The bound rStat6 was subjected to SDS–10% PAGE, followed by Western blotting with anti-Stat6 antibody. (D) Nuclear extracts (100 μg) from IL-4-treated (lane a) or untreated (lane b) I.29μ B cells and purified rStat6 (50 ng) (lane c) were immunoprecipitated with anti-Stat6 antibody, and the immunoprecipitates were then subjected to SDS–7.5% PAGE, followed by Western blotting with antiphosphotyrosine antibody (top panel) and anti-Stat6 antibody (bottom panel) sequentially on the same blot.
The difference in the amounts of Stat6 pulled down from untreated and IL-4-stimulated B cells is not due to differential sample loading, because equal amounts of GST fusion proteins were detected within each set when the same blot was reprobed with anti-GST antibodies (bottom panel). These data indicate that Stat6 can bind to NF-κB proteins specifically and that this binding is increased by IL-4 treatment, although the amount of Stat6 present in the extracts was not greater after IL-4 treatment (lanes a and b).
Since I.29μ B cells constitutively express a low level of IL-4 (32), we asked whether the small amount of binding of Stat6 observed with extracts from unstimulated cells could be inhibited by treating cells with anti-IL-4 antibodies. Cells were cultured in the absence or presence of anti-IL-4 antibodies for 20 h in order to block endogenous IL-4 activity and to allow any preexisting IL-4-activated Stat6 to decay. As shown in Fig. 1B, preincubation with anti-IL-4 antibody greatly reduced binding of Stat6 to GST-NF-κB p50 or to GST-NF-κB p65, although the amounts of Stat6 in nuclear extracts from both untreated and anti-IL-4 antibody-treated cells were similar (lanes a and b). Therefore, IL-4 stimulation greatly increases the ability of Stat6 to bind to GST-NF-κB fusion proteins.
To address the possibility that the observed binding between Stat6 and GST-NF-κB fusion proteins is mediated by other proteins in the nuclear extracts and not due to direct interaction, we tested the ability of rStat6, which was purified from insect cells that were coinfected with mouse Stat6- and human Jak2-expressing recombinant baculoviruses, to bind GST-NF-κB fusion proteins. We found that purified rStat6 was pulled down by GST-NF-κB p50 or by GST-NF-κB p65 (Fig. 1C, lanes e and g). The reduced binding efficiency with rStat6 relative to nuclear Stat6 from I.29μ B cells may be due to inefficient phosphorylation of rStat6 by human Jak2 in insect cells. Evidence for this possibility was provided by the finding that rStat6 binds much less antiphosphotyrosine antibody than does nuclear Stat6 from IL-4-treated I.29μ B cells (ratio of 1:28) on a Western blot (Fig. 1D).
To determine if any contaminating E. coli DNA in the GST fusion protein preparations might serve as a bridge to pull down Stat6, the DNA intercalating agent EtBr was included in the binding reaction mixture (12). Adding EtBr (200 μg/ml) to the binding reaction mixture does not decrease the amount of pulled-down rStat6 (Fig. 1C, lanes f and h). To further control for nonspecific binding between Stat6 and other proteins, we tested whether a GST fusion of the transcription factor LSF (31) binds rStat6 and found that it does not (lane i). We conclude that rStat6 can specifically and directly interact with NF-κB proteins without DNA or other bridging molecules.
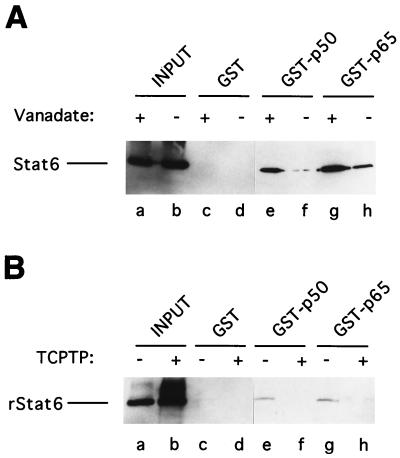
Phosphorylated tyrosine(s) in Stat6 is required for interaction between Stat6 and NF-κB p50 or p65.
Upon IL-4 signaling, latent Stat6 is tyrosine phosphorylated, allowing Stat6 homodimers to form. Stat6 treated with protein tyrosine phosphatase loses its ability to bind DNA (37), presumably due to the transposition of Stat6 from active dimers to latent monomers. To directly address the question of whether Stat6 also requires phosphorylated tyrosine(s) to bind NF-κB proteins, nuclear extracts from IL-4-stimulated I.29μ B cells were preincubated with TCPTP. The incubation was performed in the presence or absence of the tyrosine phosphatase inhibitor vanadate so that endogenous protein tyrosine phosphatase activity present in the nuclear extracts would not compromise the result. As shown in Fig. 2A, much more Stat6 was pulled down by GST-NF-κB p50 and GST-NF-κB p65 from vanadate-treated than from vanadate-free nuclear extracts that had been treated with TCPTP. Furthermore, TCPTP-treated rStat6 cannot bind the GST-NF-κB fusion proteins, while untreated rStat6 can (Fig. 2B). These results indicate that only the tyrosine-phosphorylated Stat6, presumably the dimer form but not the monomer form, is capable of binding to NF-κB proteins. We do not know whether phosphorylated tyrosine in Stat6 directly participates in the binding interface or sustains a binding domain structure required for protein-protein interaction. Further investigation is also required to understand whether only the phosphotyrosine residue which is responsible for Stat6 dimer formation is required or whether other tyrosines, which may also be phosphorylated upon IL-4 stimulation, are required for protein binding activity.
FIG. 2.
Requirement for the phosphotyrosines of Stat6 in GST pulldown assays. (A) Nuclear extracts from IL-4-stimulated I.29μ B cells were treated with TCPTP in the presence (lanes c, e, and g) or absence (lanes d, f, and h) of vanadate prior to incubation with the indicated GST fusion proteins. Lanes a and b show the input Stat6 in the pretreated nuclear extracts (10 μg). (B) Purified rStat6 was left untreated (lanes c, e, and g) or pretreated with TCPTP (lanes d, f, and h) prior to incubation with GST fusion proteins. rStat6 was then used for binding to the indicated GST fusion proteins. Lanes a and b show the inputs of pretreated rStat6 (1 ng). The bound Stat6 and rStat6 were resolved by SDS–10% PAGE, followed by Western blotting with anti-Stat6 antibody. The lanes are from the same blot, autoradiographed for the same time.
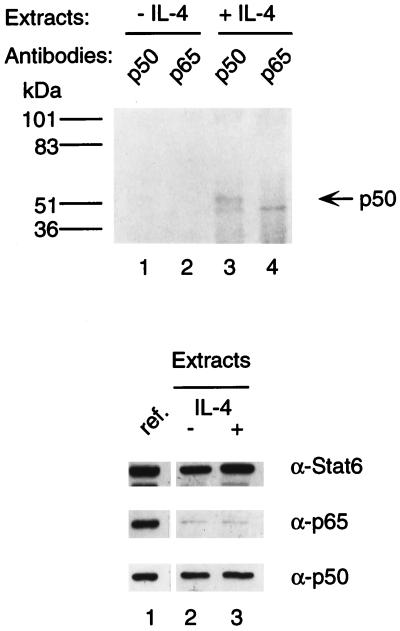
NF-κB p50 coimmunoprecipitates with Stat6 in vivo.
To determine whether Stat6-NF-κB protein complexes also exist in vivo, we tested the ability of anti-Stat6 antibody to coimmunoprecipitate NF-κB proteins from I.29μ B cells. Cells labeled with [35S]methionine and [35S]cysteine were treated with LPS to activate NF-κB, and nuclear extracts were prepared after labeled cells were further treated with IL-4 or left untreated. Immunoprecipitates obtained from nuclear extracts with anti-Stat6 antibody were dissolved and reprecipitated with anti-NF-κB p50 or anti-NF-κB p65 antiserum. As shown in Fig. 3, NF-κB p50 was coprecipitated with Stat6 from IL-4-stimulated but not from unstimulated I.29μ B cells (lanes 1 and 3). The detected NF-κB p50 in the immunoprecipitates was not due to incomplete washing, because NF-κB p50 was not detected in the immunoprecipitates when nuclear extracts from untreated cells were used (lane 1), although equal amounts of NF-κB p50 are present in nuclear extracts of IL-4-treated and untreated cells (bottom panel). Furthermore, although a substantial amount of inactive Stat6 is present in nuclear extracts from untreated cells (bottom panel), the ability of antibody to Stat6 to coprecipitate NF-κB p50 from nuclear extracts appears to require the presence of the activated Stat6 in IL-4-stimulated cells.
FIG. 3.
Interaction of Stat6 and NF-κB p50 in vivo. I.29μ B cells were labeled with [35S]methionine and [35S]cysteine for 3 h. Cells were then treated with LPS for 2 h, followed by stimulation with (lanes 3 and 4) or without (lanes 1 and 2) IL-4 for 30 min. Nuclear extracts were first immunoprecipitated with anti-Stat6 antibodies. Immunoprecipitates were dissolved, and aliquots were reprecipitated with rabbit antiserum against NF-κB p50 or p65. The precipitated proteins were analyzed by SDS–7.5% PAGE and fluorography. The positions of size markers were as indicated (in kilodaltons). Bottom panels: The contents of Stat6 and NF-κB proteins in the nuclear extracts (5 μg each) were analyzed by SDS–10% PAGE, followed by Western blotting with anti-Stat6, anti-p50, or anti-p65 antibody. The samples were analyzed on the same blot and autoradiographed for the same time as the reference samples (lane 1 of each panel) of nuclear extracts (5 μg each) from HEK 293 cells cotransfected with equal amounts of Stat6, NF-κB p50, and NF-κB p65 expression plasmids.
We could not detect, however, NF-κB p65 coimmunoprecipitated with Stat6 (Fig. 3, top panel, lanes 2 and 4). Western blotting used to determine the contents of NF-κB proteins in the 35S-labeled I.29μ nuclear extracts indicated that much less p65 than p50 was present (bottom panel). The weaker p65 signal must be due to the presence of less p65 than p50 in the I.29μ nuclear extracts rather than to a difference in the efficiency of precipitation by the two antibodies, because equal amounts of NF-κB p50 and p65 from another cell line, which had been transfected with expression constructs for these two proteins, blotted onto the same membrane showed similar image densities with anti-NF-κB p50 and anti-NF-κB p65 antibodies (bottom panel). Furthermore, both antibodies can recognize native and denatured forms of target proteins comparably well (data not shown). Consistent with this observation, Miyamoto et al. (18) found that p65 is present at lower concentrations than p50 in mature B-cell lines. Therefore, due to insufficient p65 present in the nuclear extracts, we could not determine if p65 complexes with Stat6 in vivo.
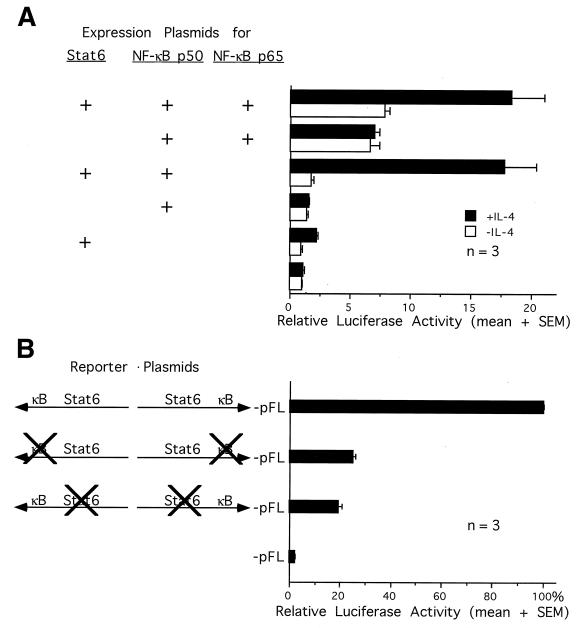
Stat6 and NF-κB proteins synergistically activate an IL-4-inducible reporter gene containing both cognate binding sites.
To determine if Stat6 and NF-κB synergistically induce transcription from a DNA segment containing adjacent Stat6 and κB sites, we performed a reporter gene assay in HEK 293 cells cotransfected with expression plasmids for Stat6 and NF-κB. HEK 293 cells were chosen because they have no detectable Stat6 and little nuclear NF-κB but do have the signaling machinery to activate Stat6 (8, 17). The reporter plasmid contains a luciferase gene driven by the minimal c-fos promoter and two copies of IL-4 RR −124/−79, each of which contains a C/EBP, a Stat6, and a κB site from the GL Cɛ promoter (4). Although the upstream copy is inverted, its orientation appears to have no effect on enhancer activity (4). The C/EBP site is required for optimal activation of this DNA segment by IL-4, although the protein which binds this site in I.29μ B cells has not been identified.
Figure 4A shows that in the presence of IL-4, coexpression of Stat6 and NF-κB p65/p50 synergistically activated the promoter by 18-fold, whereas expression of Stat6 or NF-κB p65/p50 alone induced the promoter activity by two- and sevenfold, respectively, relative to that of the empty expression vector. A similar promoter activity was also observed by overexpression of Stat6 and NF-κB p50 (17-fold), whereas expression of NF-κB p50 alone did not induce the promoter at all (1-fold). These data suggest that IL-4-activated Stat6 must interact with NF-κB proteins, either p65/p50 heterodimers or p50/p50 homodimers, in order to induce the IL-4 RR. This synergy does not appear to require the transactivation activity of NF-κB, since p50/p50 homodimers have been found to be incapable of transcriptional activation (28). In the absence of IL-4, Stat6 cannot induce the promoter, and expression of NF-κB alone transactivates as well as that of Stat6 and NF-κB together. Therefore, consistent with the physical evidence presented above, Stat6 must be activated by IL-4 in order to functionally synergize with NF-κB.
FIG. 4.
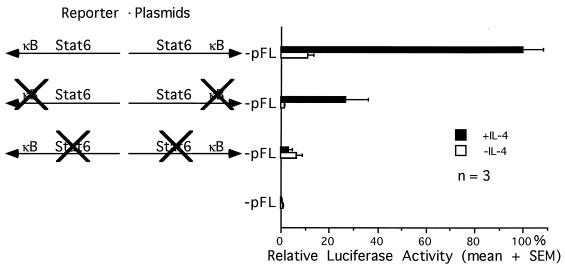
Synergy between Stat6 and NF-κB in activating an IL-4-inducible reporter gene in HEK 293 cells. (A) HEK 293 cells were transfected with one or more expression plasmids for Stat6 or NF-κB as indicated, together with the luciferase reporter plasmid (IL-4 RR)2-pFL and the control plasmid pfosCAT. Forty hours posttransfection, cells were stimulated with (▪) or without (□) IL-4 for 8 h before being harvested. The luciferase activity was normalized for transfection efficiency by the CAT activity and calculated as a relative value by assigning the activity from unstimulated cells transfected with empty vector alone a value of 1. The data are presented as means plus standard errors of the means (SEMs) from three transfection experiments. (B) HEK 293 cells were transfected with wild-type or other mutated (IL-4 RR)2-pFL reporter plasmids as pictured, together with expression plasmids for Stat6, NF-κB p50, and NF-κB p65 and the control plasmid pfosCAT. Forty hours posttransfection, cells were stimulated with IL-4 for 8 h before being harvested. The luciferase activity was normalized by the CAT activity and calculated as a percentage relative to the activity from cells transfected with the wild-type reporter plasmid. The data are presented as means plus SEMs from three transfection experiments.
To determine if Stat6 and NF-κB require their cognate binding sites in the IL-4 RR to synergistically activate the reporter gene, we performed reporter gene assays in HEK 293 cells overexpressing both Stat6 and NF-κB, by using (IL-4 RR)2-pFL reporter plasmids containing wild-type or mutated κB or Stat6 enhancer sequences. Mutations at either the κB sites or the Stat6 sites in the reporter plasmids reduced transcriptional activities by 75 and 80%, respectively (Fig. 4B). Therefore, both cognate binding sites are required for synergistic activation of the reporter gene by Stat6 and NF-κB.
Cooperative DNA binding activity correlates with functional synergy between Stat6 and NF-κB proteins.
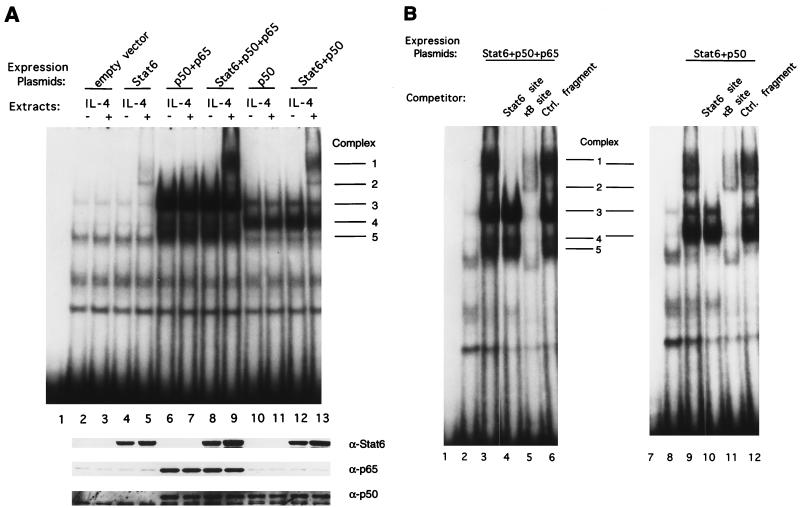
Since the synergistic activation of the promoter by Stat6 and NF-κB proteins requires both cognate DNA binding sites, we wished to determine if Stat6 and NF-κB proteins bind cooperatively to the IL-4 RR of the GL Cɛ promoter and if the presence of such protein-DNA complexes correlates with transcriptional activity. Nuclear extracts prepared from the transfected HEK 293 cells were incubated with a labeled DNA probe that contains the IL-4 RR −124/−79 sequence, and EMSAs were performed. Nuclear extracts from cells transfected with the Stat6-expressing vectors form an IL-4-inducible complex (complex 2) (Fig. 5A, top panel, lanes 4 and 5) that contains Stat6 (see below). Nuclear extracts from cells transfected with NF-κB p50 and p65 form three other non-IL-4-inducible complexes (complexes 3, 4, and 5) (lanes 6 and 7) that contain different dimers of NF-κB proteins (see below). Importantly, when plasmids encoding Stat6 and NF-κB p50 and p65 are cotransfected, a strong slow-migrating complex (complex 1) can be induced by IL-4 treatment (lanes 8 and 9). Complex 1 contains Stat6 and NF-κB proteins (presumably p50/p65 heterodimers; see below). A similar IL-4-inducible complex is also observed when Stat6 and NF-κB p50 are coexpressed in cells (lanes 12 and 13), whereas expression of NF-κB p50 alone results only in formation of the non-IL-4-inducible complex 4 (lanes 10 and 11). In lane 13, complex 1 contains Stat6 and NF-κB p50 (presumably p50/p50 homodimers; see below). In conclusion, synergistic activation of the promoter by Stat6 and NF-κB p50 or by Stat6 and NF-κB p65 and p50 is correlated with the presence of complex 1.
FIG. 5.
EMSAs demonstrating that Stat6 and NF-κB in nuclear extracts from transfected HEK 293 cells together form an IL-4-inducible complex with the IL-4 RR −124/−79 Cɛ DNA fragment. (A) DNA binding reactions were performed with nuclear extracts (5 μg) from HEK 293 cells transfected with different combinations of expression plasmids as indicated and left untreated (lanes 2, 4, 6, 8, 10, and 12) or stimulated with IL-4 for 15 min (lanes 3, 5, 7, 9, 11, and 13). Lane 1 contains probe alone. Bottom panels: Western blotting analyses of nuclear extracts (5 μg each) corresponding to the samples used in EMSA. The blot was probed with anti-Stat6, anti-NF-κB p65, and anti-NF-κB p50 antibodies sequentially. (B) DNA competition experiments were performed with nuclear extracts (5 μg) from IL-4-treated HEK 293 cells overexpressing Stat6 and NF-κB p50 and p65 (lanes 3 to 6) or Stat6 and NF-κB p50 (lanes 9 to 12). A 100-fold molar excess (2.5 pmol) of unlabeled double-stranded oligonucleotides containing a Stat6 site (lanes 4 and 10), a κB site (lanes 5 and 11), or a size-matched control DNA fragment (lanes 6 and 12) was added to the DNA binding reaction mixture. Binding reactions with nuclear extracts from empty vector-transfected cells are shown in lanes 2 and 8. Lanes 1 and 7 contain probe alone.
The amount of complex 1 exceeds that of complex 2, which contains Stat6 but no NF-κB proteins (see below), indicating that Stat6 and NF-κB proteins bind cooperatively to the IL-4 RR. The enhanced formation of complex 1 by coexpression of Stat6 and NF-κB proteins was not due to enhanced expression of either Stat6 or NF-κB proteins in the cotransfected cells, because equivalent levels of expression of Stat6, NF-κB p50, and p65 in the nuclear extracts were detected in Western blotting analyses from singly or multiply transfected cells (Fig. 5A, bottom panels).
Involvement of Stat6 and NF-κB proteins in complex 1 was confirmed by DNA competition experiments with nuclear extracts from IL-4-stimulated HEK 293 cells overexpressing Stat6 and NF-κB p50 and p65 or Stat6 and NF-κB p50 (Fig. 5B). Addition of a 100-fold molar excess of unlabeled double-stranded oligonucleotides containing the Stat6 site abolished complexes 1 and 2 (lanes 4 and 10), indicating that these two complexes contain Stat6, whereas addition of a 100-fold molar excess of double-stranded κB site oligonucleotides ablated complexes 1, 3, 4, and 5 (lanes 5 and 11), indicating that these four complexes contain NF-κB proteins. Competition of each complex by Stat6 and κB sites is specific because a size-matched control DNA fragment did not compete for any of these five complexes (lanes 6 and 12).
Functional synergy between endogenous Stat6 and NF-κB proteins in I.29μ B cells correlates with cooperative DNA binding activity of a complex containing both Stat6 and NF-κB proteins.
We next asked if the endogenous Stat6 and NF-κB proteins in I.29μ B cells also synergistically activate transcription. I.29μ B cells were transiently transfected with the (IL-4 RR)2-pFL reporter plasmids, containing wild-type or mutated Stat6 or κB enhancer sequences, and cells were subsequently left untreated or cultured with mouse IL-4 for 15 h before transcriptional activities were measured. Figure 6 shows that the transcriptional activity of the wild-type reporter plasmid is induced 10-fold by IL-4 stimulation. Mutation of the Stat6 sites in the reporter plasmid not only completely abolished IL-4 inducibility but also reduced transcriptional activity by 97%. Mutations at the κB sites resulted in a 74% reduction in transcriptional activity, although the IL-4 inducibility of the promoter was not affected. These results indicate that constitutive NF-κB activity in these cells in the absence of Stat6 binding sites cannot transactivate the reporter gene (3% of wild-type activity) and that IL-4-activated Stat6 in the absence of κB sites has little transactivating ability (26% of wild-type activity). Therefore, Stat6 and NF-κB in this B-cell line synergistically activate transcription from the IL-4 RR.
FIG. 6.
Functional synergy between Stat6 and NF-κB in I.29μ B cells. I.29μ B cells were transfected with wild-type or mutated (IL-4 RR)2-pFL reporter plasmids, as pictured, together with the control plasmid pSV2CAT. Aliquots of the transfected cells were cultured with (▪) or without (□) IL-4 for 15 h before harvesting. The luciferase activity was normalized by the CAT activity and calculated as a percentage relative to the activity from IL-4-treated cells transfected with the wild-type reporter plasmid. The data are presented as means plus standard errors of the means (SEMs) from three transfection experiments.
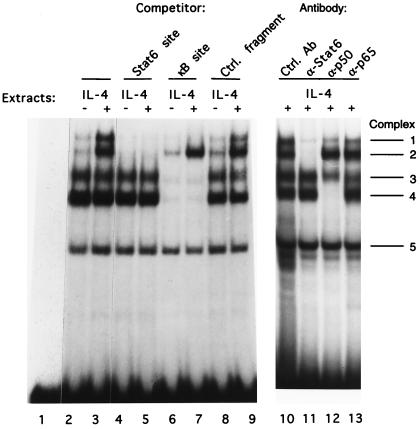
To determine if the functional synergy correlates with DNA binding activity of complexes containing Stat6 and NF-κB proteins in I.29μ nuclear extracts, we performed EMSAs, using the IL-4 RR −124/−79 DNA fragment as the probe and nuclear extracts from cells untreated or treated with IL-4. Nuclear extracts from IL-4-treated cells form two IL-4-inducible complexes (complexes 1 and 2) and three complexes not induced by IL-4 (complexes 3, 4, and 5) (Fig. 7, lanes 2 and 3). Competition experiments indicate that complexes 1 and 2 contain Stat6 and that complexes 1, 3, and 4 contain NF-κB proteins, because excess Stat6 or κB site oligonucleotides, but not an irrelevant oligonucleotide, compete for these complexes (lanes 4 to 9). Therefore, complex 1 contains both Stat6 and NF-κB proteins. Complex 5 does not contain Stat6 or NF-κB proteins, and we have not been able to identify the proteins involved in this complex. Since the amount of complex 1 is about equal to that of complex 2, which contains Stat6 but no NF-κB proteins, and since these EMSA experiments were performed in the presence of a large excess of the DNA probe, this result suggests that Stat6 and NF-κB proteins cooperatively bind the DNA fragment containing both cognate binding sites. Furthermore, their cooperative DNA binding activity appears to correlate with their functional synergy.
FIG. 7.
EMSAs demonstrating that Stat6 and NF-κB in nuclear extracts from I.29μ B cells together form an IL-4-inducible complex with the IL-4 RR −124/−79 Cɛ DNA fragment. Lanes 2 to 3: EMSAs of nuclear extracts from I.29μ B cells untreated (lane 2) or treated with IL-4 (lane 3). Lane 1 contains probe alone. Lanes 4 to 9: DNA competition experiments were performed as in Fig. 5B, except nuclear extracts from untreated (lanes 4, 6, and 8) or IL-4-stimulated (lanes 5, 7, and 9) I.29μ B cells were used in the binding reactions. The unlabeled double-stranded oligonucleotide competitors were added into the binding reaction mixtures as indicated. Lanes 10 to 13: antibody supershift experiments were performed with nuclear extracts from IL-4-treated I.29μ B cells. One microliter of the indicated antibodies was added to the DNA binding reaction.
The components of these complexes were identified by antibody supershift assays (Fig. 7, lanes 10 through 13). As predicted from the competition experiments, antibody to Stat6 abolishes complex 2 and greatly reduces complex 1, confirming the presence of Stat6 in these two complexes (lane 11). Antibody to NF-κB p50 completely eliminates complex 4 (lane 12). Antibodies to other members of the NF-κB/Rel family have not been found to supershift complex 4 (data not shown), suggesting that complex 4 contains NF-κB p50/p50 homodimers. Complex 1 is partially inhibited by anti-NF-κB p50 antibodies (lane 12) and also by antibodies to p65 (lane 13), indicating that NF-κB p50 and p65 are involved in complex 1 and suggesting that additional NF-κB dimers may be present in this complex. Complex 3 is partially inhibited by anti-NF-κB p50 and anti-NF-κB p65 and by anti-c-Rel antibodies (lanes 12 and 13 and data not shown), suggesting that different NF-κB heterodimers contribute to this complex.
In agreement with these findings, nuclear extracts from splenic B cells stimulated with IL-4 and CD40 ligand form two nuclear complexes with the IL-4/CD40 RR of the GL Cɛ promoter, and these two complexes contain both Stat6 and NF-κB/Rel proteins (9). (The IL-4/CD40 RR, −124/−56, contains the −124/−79 segment used in this study and an additional κB site at −65/−56.) Thus, the Stat6-NF-κB complexes in B cells may be responsible for synergistic activation of genes by IL-4 and NF-κB inducers.
DISCUSSION
In this study, we provide evidence that Stat6 physically interacts with NF-κB proteins and that this interaction results in synergistic activation of transcription of a gene containing both cognate sites. Although for most STATs, binding to their palindromic sites in a promoter appears to correlate with induction of transcription (30), Stat6 seems to differ in that a Stat6 binding site, even when multimerized, is not sufficient for IL-4 induction (4, 17, 30). The poor induction of transcription by IL-4-activated Stat6 alone in transient transfection experiments in HEK 293 cells (Fig. 4A) and I.29μ B cells (Fig. 6) is consistent with those previous findings. Thus, it appears that Stat6 cannot activate transcription from the IL-4 RR enhancer region of the GL Cɛ promoter without cooperating with other transcriptional activators, such as NF-κB proteins.
Interactions between STATs and other proteins have been previously reported. Examples are interaction between the Stat1-Stat2 heterodimer and p48 (23), Stat1 or Stat2 and p300/CBP (2, 40), and Stat3 and c-Jun (26). NF-κB/Rel proteins have also been shown to interact with other transcription factors or proteins involved in basal transcription machinery (11, 13, 35, 36, 39). However, direct interaction between STATs and NF-κB/Rel proteins has not been previously reported, although Stat1 and NF-κB were found to synergistically activate the IRF-1 promoter (21).
Stat6 synergizes with either NF-κB p50 alone or with NF-κB p65 plus NF-κB p50 to induce transcription to equivalent levels in our experiments; yet, NF-κB p65, but not NF-κB p50, contains a transactivating domain (28). Thus, the synergistic transcriptional activation by Stat6 and NF-κB may not be due to the transactivating domain of NF-κB p65. These data suggest that the binding between Stat6 and NF-κB proteins may allow transactivation by Stat6.
The binding between NF-κB and Stat6 appears to change the characteristics of Stat6 in two ways. First, it appears that the DNA binding affinity of Stat6 is substantially enhanced by interacting with NF-κB proteins. In the EMSA experiments, the amount of the IL-4-inducible complex containing both Stat6 and NF-κB (complex 1) is much greater than (in cotransfected HEK 293 cells) or approximately equal to (in I.29μ B cells) the amount of the second IL-4-inducible complex, which contains Stat6 but no NF-κB. As the binding reactions are performed in the presence of a large excess of DNA probe, these data suggest that NF-κB proteins increase the affinity of Stat6 for binding to the probe.
The second change in Stat6 that appears to occur upon binding to NF-κB is an induction of the transactivating ability of Stat6. The existence of complex 2, which contains Stat6 but no NF-κB, suggests that Stat6 can bind its cognate site well without NF-κB proteins (Fig. 7). In addition, complex 2 forms even when the adjacent C/EBP site is mutated or when a single Stat6 site is used as the DNA probe (data not shown), and thus this complex appears to contain only Stat6. However, as mentioned earlier, even when Stat6 sites are multimerized, Stat6 alone cannot transactivate, implying that binding of Stat6 to its DNA cognate sites is not sufficient to activate transcription. These data plus the fact that NF-κB p50 in the absence of p65 can synergize with Stat6 suggest that functional changes in Stat6, e.g., formation of a new protein interaction surface which results in transactivating ability, might occur upon binding to NF-κB proteins. Thus, synergistic transcriptional activation by Stat6 and NF-κB proteins appears to be achieved by both quantitative and functional changes in Stat6 bound to its cognate site.
Only the Stat6 from IL-4-treated cells appears to be capable of binding DNA or of binding NF-κB proteins in GST pulldown experiments, although similar levels of Stat6 proteins were observed by Western blotting in nuclear extracts from both untreated and IL-4-treated I.29μ B cells. The levels of Stat6 may be similar because the nuclear extraction protocol used did not completely exclude cytoplasmic Stat6 and/or because the extracts may contain decayed nuclear Stat6 which has lost the activating phosphorylations. Consistent with the latter explanation, Stat6 in nuclear extracts from IL-4-treated I.29μ B cells had a 25-fold greater level of tyrosine phosphorylation than did Stat6 in nuclear extracts from untreated cells after normalization for protein input (Fig. 1D).
The requirement for tyrosine phosphorylation of Stat6 was directly demonstrated when treatment of both IL-4-activated Stat6 and recombinant Stat6 with tyrosine phosphatases prevented binding to NF-κB (Fig. 2). Therefore, in agreement with current models of JAK/STAT signaling pathways, both the DNA binding and protein binding activities of Stat6 require IL-4 activation and the ensuing tyrosine phosphorylation of Stat6. Our analyses did not permit us to determine, however, whether the same phosphorylated tyrosine that is required for DNA binding (Y641) (17) is required for Stat6 to bind NF-κB.
The coimmunoprecipitation experiment demonstrates that Stat6 binds NF-κB p50 in the sIgM+ B lymphoma cell line I.29μ, although the binding was difficult to detect and thus might be weak. It is possible, however, that their interaction is of relatively high affinity but still difficult to detect if each of these proteins is also involved in several other interactions. For instance, the protein factors working through the C/EBP site might be potential candidates for interaction with Stat6 and NF-κB, as mutation of the C/EBP site abolishes IL-4 inducibility of the Cɛ promoter (4, 13, 36).
Although Stat6 directly binds NF-κB p65 and NF-κB p50, we could not detect binding of complexes containing both Stat6 and NF-κB when only a Stat6 site or a κB site was used as a probe in EMSAs (data not shown). Furthermore, in DNA competition experiments, we found that adding a large excess of Stat6 site or κB site competitors eliminated only complexes containing their corresponding DNA binding proteins. These data suggest that when Stat6-NF-κB protein complexes are bound to their cognate sites, their physical interaction might be weak. It is possible that the protein binding domains overlap the DNA binding domains of Stat6 and/or NF-κB proteins. A transition from a strong to a weak interaction between Stat6 and NF-κB upon binding to their cognate binding sites may allow each transcription factor to interact with the basal transcription machinery, thereby synergistically activating transcription. Cooperative DNA binding activity of complex 1 containing both Stat6 and NF-κB proteins in EMSA may simply reflect the avidity of Stat6-NF-κB complexes for DNA containing both cognate binding sites.
The presence of preformed Stat6-NF-κB complexes in the absence of DNA suggests that an additional level of regulation of IL-4 signaling may exist. Li et al. (14) showed that the expression level of p48 affects the DNA binding specificity of Stat1-Stat2 heterodimers, thereby regulating interferon-responsive gene expression. An analogous mechanism might be adopted by NF-κB proteins for regulating Stat6. In addition, the preformed complexes of activated Stat6 and NF-κB proteins may increase both the DNA binding affinity and specificity of Stat6 for gene promoters with neighboring κB sites, such as the GL Cɛ, Cγ1, and CD23 promoters.
In the sIgM+ B lymphoma cell line I.29μ, we could not detect NF-κB p65 coprecipitated with Stat6, very likely due to the low level of NF-κB p65 present in the nuclear extracts. Since we find that Stat6 can bind NF-κB p50 and NF-κB p65 equally well in vitro and that Stat6 can form complexes with NF-κB p50/p50 homodimers as well as with p65/p50 heterodimers by EMSA, it is possible that NF-κB p65 can form complexes with Stat6 in vivo when sufficient NF-κB p65 is present in the cell. However, it is also possible that Stat6 binds differentially to the different subunits of the NF-κB/Rel family in vivo. Since the subunit composition of the NF-κB/Rel family differs among different cell types and during differentiation, it is possible that the function of activated Stat6 is regulated through the binding of different NF-κB/Rel proteins, which bind to κB sites in many genes differentially and function differently on different genes (15, 19). Furthermore, cross-talk between NF-κB/Rel proteins and other transcription factor families (11, 13, 35, 36) might also modulate the function of Stat6-NF-κB complexes in transcriptional activation, especially on genes containing composite sites. Thus, a complex regulation could be generated by IL-4 signaling in a cell type-, differentiation stage-, and stimulus-dependent manner. These additional levels of regulation remain to be elucidated.
ACKNOWLEDGMENTS
We thank J. N. Ihle for the mStat6-pBKS plasmid, U. Schindler for the TPU 276 plasmid, S.-C. Lin for the NF-κB p50 and NF-κB p65 expression vectors, U. M. Hansen and E. E. Drouin for the pGEX-2T-LSF plasmid, N. R. Rice for the anti-p50 and anti-p65 antibodies, C. Peterson for valuable advice, and P. Dobner, F. He, T. Kowalik, C. Schrader, G. Qiu, and A. Yesilaltay for helpful comments on the manuscript.
This work was supported by the Council for Tobacco Research U.S.A. (no. 4019) and by National Institutes of Health grant AI23283.
REFERENCES
- 1.Baeuerle P A, Henkel T. Function and activation of NF-κB in the immune system. Annu Rev Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D’Andrea A, Livingston D M. Cooperation of Stat2 and p300/CBP in signaling induced by interferon-α. Nature. 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 3.Darnell J E, Jr, Kerr I M, Stark G R. Jak-Stat pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 4.Delphin S A, Stavnezer J. Characterization of an IL-4 responsive region in the immunoglobulin heavy chain ɛ promoter: regulation by NF-IL4, a C/EBP family member and NF-κB/p50. J Exp Med. 1995;181:181–192. doi: 10.1084/jem.181.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilman M Z, Wilson R N, Weinberg R A. Multiple protein-binding sites in the 5′-flanking region regulate c-fos expression. Mol Cell Biol. 1986;6:4305–4316. doi: 10.1128/mcb.6.12.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gorman G M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyl transferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou J, Schindler U, Henzel W J, Ho T C, Brasseur M, McKnight S L. An interleukin-4-induced transcription factor: IL-4 Stat. Science. 1994;265:1701–1706. doi: 10.1126/science.8085155. [DOI] [PubMed] [Google Scholar]
- 8.Hsu H, Xiong J, Goeddel D V. The TNF receptor 1-associated protein TRADD signals cell death and NF-κB activation. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 9.Iciek L A, Delphin S A, Stavnezer J. CD40 cross-linking induces IgE germline transcripts in B cells via activation of NF-κB: synergy with IL-4 induction. J Immunol. 1997;158:4769–4779. [PubMed] [Google Scholar]
- 10.Ihle J N. STATs: signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- 11.Kaszubska W, van Huijsduijnen R H, Ghersa P, DeRaemy-Schenk A-M, Chen B P C, Hai T, DeLamarter J F, Whelan J. Cyclic AMP-independent ATF family members interact with NF-κB and function in the activation of the E-selection promoter in response to cytokines. Mol Cell Biol. 1993;13:7180–7190. doi: 10.1128/mcb.13.11.7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai J-S, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LeClair K P, Blanar M A, Sharp P A. The p50 subunit of NF-κB associates with NF-IL6 transcription factor. Proc Natl Acad Sci USA. 1992;89:8145–8149. doi: 10.1073/pnas.89.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Leung S, Qureshi S, Darnell J E, Jr, Stark G R. Formation of STAT1-STAT2 heterodimers and their role in the activation of IRF-1 gene transcription by interferon-α. J Biol Chem. 1996;271:5790–5794. doi: 10.1074/jbc.271.10.5790. [DOI] [PubMed] [Google Scholar]
- 15.Lin S C, Stavnezer J. Activation of NF-κB/Rel by CD40 engagement induces the mouse germline immunoglobulin Cγ1 promoter. Mol Cell Biol. 1996;16:4591–4603. doi: 10.1128/mcb.16.9.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Messner B, Stutz A M, Albrecht B, Peiritsch S, Woisetschlager M. Cooperation of binding sites for Stat6 and NF-κB/rel in the IL-4-induced up-regulation of the human IgE germline promoter. J Immunol. 1997;159:3330–3337. [PubMed] [Google Scholar]
- 17.Mikita T, Campbell D, Wu P, Williamson K, Schindler U. Requirements for interleukin-4-induced gene expression and functional characterization of Stat6. Mol Cell Biol. 1996;16:5811–5820. doi: 10.1128/mcb.16.10.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyamoto S, Schmitt M J, Verma I M. Qualitative changes in the subunit composition of κB-binding complexes during murine B-cell differentiation. Proc Natl Acad Sci USA. 1994;91:5056–5060. doi: 10.1073/pnas.91.11.5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyamoto S, Verma I M. Rel/NF-κB/IκB story. Adv Cancer Res. 1995;66:255–292. [PubMed] [Google Scholar]
- 20.Ohara J, Paul W E. Production of a monoclonal antibody to and molecular characterization of B-cell stimulatory factor-1. Nature. 1985;315:333–336. doi: 10.1038/315333a0. [DOI] [PubMed] [Google Scholar]
- 21.Ohmori Y, Schreiber R D, Hamilton T A. Synergy between interferon-γ and tumor necrosis factor-α in transcriptional activation is mediated by cooperation between signal transducer and activator of transcription 1 and nuclear factor κB. J Biol Chem. 1997;272:14899–14907. doi: 10.1074/jbc.272.23.14899. [DOI] [PubMed] [Google Scholar]
- 22.Quelle F W, Shimoda K, Thierfelder W, Fischer C, Kim A, Ruben S M, Cleveland J L, Pierce J H, Keegan A D, Nelms K, Paul W E, Ihle J N. Cloning of murine Stat6 and human Stat6, Stat proteins that are tyrosine phosphorylated in responses to IL-4 and IL-3 but are not required for mitogenesis. Mol Cell Biol. 1995;15:3336–3343. doi: 10.1128/mcb.15.6.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qureshi S A, Salditt-Georgieff M, Darnell J E., Jr Tyrosine-phosphorylated Stat1 and Stat2 plus 48-kDa proteins all contact DNA in forming interferon-stimulated-gene factor 3. Proc Natl Acad Sci USA. 1995;92:3829–3833. doi: 10.1073/pnas.92.9.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rice N R, Ernst M K. In vivo control of NF-κB activation by IκBα. EMBO J. 1993;12:4685–4695. doi: 10.1002/j.1460-2075.1993.tb06157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richards M L, Katz D H. Analysis of the promoter elements necessary for IL-4 and anti-CD40 antibody induction of murine FcɛRII (CD23). Comparison with the germline ɛ promoter. J Immunol. 1997;158:263–272. [PubMed] [Google Scholar]
- 26.Schaefer T S, Sanders L K, Nathans D. Cooperative transcriptional activity of Jun and Stat3β, a short form of Stat3. Proc Natl Acad Sci USA. 1995;92:9097–9101. doi: 10.1073/pnas.92.20.9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schindler U, Wu P, Roth M, Brasseur M, McKnight S L. Components of a Stat recognition code: evidence for two layers of molecular selectivity. Immunity. 1995;2:687–697. doi: 10.1016/1074-7613(95)90013-6. [DOI] [PubMed] [Google Scholar]
- 28.Schmitz M L, Baeuerle P A. The p65 subunit is responsible for the strong transcription activating potential of NF-κB. EMBO J. 1991;10:3805–3817. doi: 10.1002/j.1460-2075.1991.tb04950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schreiber E, Matthias P, Muller M M, Schaffner W. Rapid detection of octamer proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seidel H M, Milocco L H, Lamb P, Darnell J E, Jr, Stein R B, Rosen J. Spacing of palindromic half sites as a determinant of selective STAT (signal transducers and activators of transcription) DNA binding and transcriptional activity. Proc Natl Acad Sci USA. 1995;92:3041–3045. doi: 10.1073/pnas.92.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shirra M K, Zhu Q, Huang H-C, Pallas D, Hansen U. One exon of the human LSF gene includes conserved regions involved in novel DNA-binding and dimerization motifs. Mol Cell Biol. 1994;14:5076–5087. doi: 10.1128/mcb.14.8.5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shockett P, Stavnezer J. Effect of cytokines on switching to IgA and α germline transcripts in the B lymphoma I.29μ: transforming growth factor-β activates transcription of the unrearranged Cα gene. J Immunol. 1991;147:4374–4383. [PubMed] [Google Scholar]
- 33.Stavnezer J. Antibody class switching. Adv Immunol. 1996;61:79–146. doi: 10.1016/s0065-2776(08)60866-4. [DOI] [PubMed] [Google Scholar]
- 34.Stavnezer J, Sirlin S, Abbott J. Induction of immunoglobulin isotype switching in cultured I.29μ B lymphoma cells: characterization of the accompanying rearrangements of heavy chain genes. J Exp Med. 1985;161:577–601. doi: 10.1084/jem.161.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stein B, Baldwin A S, Jr, Ballard D W, Greene W C, Angel P, Herrlich P. Cross-coupling of the NF-κB p65 and Fos-Jun transcription factors produces potentiated biological function. EMBO J. 1993;12:3879–3891. doi: 10.1002/j.1460-2075.1993.tb06066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stein B, Cogswell P C, Baldwin A S., Jr Functional and physical associations between NF-κB and C/EBP family members: a Rel domain-bZIP interaction. Mol Cell Biol. 1993;13:3964–3974. doi: 10.1128/mcb.13.7.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang D Z, Cherrington A, Famakin-Mosuro B, Boothby M. Independent pathways for de-repression of the mouse Ig heavy chain germ-line ɛ promoter: an IL-4 NAF/NF-IL-4 site as a context-dependent negative element. Int Immunol. 1996;8:977–989. doi: 10.1093/intimm/8.7.977. [DOI] [PubMed] [Google Scholar]
- 38.Warren W D, Berton M T. Induction of germline γ1 and ɛ Ig gene expression in murine B cells: interleukin 4 and the CD40 ligand-CD40 interaction provide distinct but synergistic signals. J Immunol. 1995;155:5637–5646. [PubMed] [Google Scholar]
- 39.Xu X, Prorock C, Ishikawa H, Maldonado E, Ito Y, Gelinas C. Functional interaction of the v-Rel and c-Rel oncoproteins with the TATA-binding protein and association with transcription factor IIB. Mol Cell Biol. 1993;13:6733–6741. doi: 10.1128/mcb.13.11.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J J, Vinkemeier U, Gu W, Chakravarti D, Horvath C M, Darnell J E., Jr Two contact regions between Stat1 and CBP/p300 in interferon γ signaling. Proc Natl Acad Sci USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]