Abstract
The cardiogenic homeodomain factor Nkx-2.5 and serum response factor (SRF) provide strong transcriptional coactivation of the cardiac α-actin (αCA) promoter in fibroblasts (C. Y. Chen and R. J. Schwartz, Mol. Cell. Biol. 16:6372–6384, 1996). We demonstrate here that Nkx-2.5 also cooperates with GATA-4, a dual C-4 zinc finger transcription factor expressed in early cardiac progenitor cells, to activate the αCA promoter and a minimal promoter, containing only multimerized Nkx-2.5 DNA binding sites (NKEs), in heterologous CV-1 fibroblasts. Transcriptional activity requires the N-terminal activation domain of Nkx-2.5 and Nkx-2.5 binding activity through its homeodomain but does not require GATA-4’s activation domain. The minimal interactive regions were mapped to the homeodomain of Nkx-2.5 and the second zinc finger of GATA-4. Removal of Nkx-2.5’s C-terminal inhibitory domain stimulated robust transcriptional activity, comparable to the effects of GATA-4 on wild-type Nkx-2.5, which in part facilitated Nkx-2.5 DNA binding activity. We postulate the following simple model: GATA-4 induces a conformational change in Nkx-2.5 that displaces the C-terminal inhibitory domain, thus eliciting transcriptional activation of promoters containing Nkx-2.5 DNA binding targets. Therefore, αCa promoter activity appears to be regulated through the combinatorial interactions of at least three cardiac tissue-enriched transcription factors, Nkx-2.5, GATA-4, and SRF.
Defining the molecular basis of the establishment and maintenance of cardiac muscle differentiation presents a problem in determining how a small number of tissue-restricted transcription factors can establish a very complex pattern of developmental and tissue-specific gene expression. In particular, the Drosophila tinman gene, an obligatory mesoderm determination factor, is required for the formation of the embryonic dorsal vessel, an insect equivalent of the vertebrate heart (1, 3). Mutations in the tinman gene result in the loss of heart formation in the Drosophila embryo. In addition, tinman is known to regulate NK-3 and/or bagpipe expression in the visceral mesoderm (1) and the expression of the Drosophila MEF-2 factor gene, which is required for muscle and heart cell differentiation (22). The vertebrate tinman relative, Nkx-2.5, appears to be instrumental in the patterning of the vertebrate embryonic heart as well. Nkx-2.5, a cardiac tissue-specific homeodomain factor, is expressed in early cardiac progenitor cells prior to cardiogenic differentiation and through adulthood (36, 44). Superimposed on the appearance of Nkx-2.5 in cardiac progenitor cells is the sequential expression of the cell type-restricted cardiac α-actin (αCA) and cardiac α-myosin heavy-chain (αC-MHC) genes (44). Nkx-2.5 genes identified in other vertebrates, such as the zebra fish (12, 42), Xenopus laevis (57), and chickens (54), are highly conserved and exhibit expression patterns which demarcate the heart field (12). The recent identification of avian Nkx-2.8 suggests a potential genetic redundancy during early heart development, and the role of being a vertebrate tinman homolog may be shared by at least two Nkx-2 genes (52).
There are at least seven different mouse Nkx-2 genes (27, 52), all of which contain tyrosine at position 54 in the homeodomain, which influences DNA binding site specificity (27). DNA binding assays have revealed that both Nkx-2.1, also called thyroid transcription factor (13), and Nkx-2.5 prefer DNA sequences containing 5′-TNAAGTG-3′ (10, 16, 25), also known as NKEs. Nkx-2 factors also bind with weaker avidity to a second subset of sequences that contain the 5′-TTAATT-3′ motif, which is recognized by Hox proteins (16). NKEs are present at multiple sites in the lung-specific surfactant gene promoter (4), and Ray et al. (51) showed that Nkx-2.1 and Nkx-2.5 bound the same NKEs required for mCC10 gene promoter activity. Serum response elements (SREs) of the αCA promoter served as both high-affinity (SRE2 and SRE3) and intermediate-strength (SRE1 and SRE4) binding targets of Nkx-2.5 (10). Durocher et al. (20) showed that two NKE sites in the proximal region of the cardiac atrial natriuretic factor (ANF) promoter direct high-level cardiac tissue-specific promoter activity. Gajewski et al. (22) demonstrated that two NKE promoter sites direct Drosophila mef2 expression in response to Tinman.
Nkx-2.5 serves as a modest transcription activator in transfection assays when analyzed with reporter genes carrying multimerized NKE binding sites. However, deletion of the C-terminal inhibitory domain of Nkx-2.5 stimulated transcriptional activity over 50-fold (10). Both the inhibitory domain and the highly charged activation domain of Nkx-2.5 are enriched in alanine and proline. These findings suggest that potential hydrophobic interactions between the inhibitory and activation domains might block access of transcription initiation factors to the highly charged moiety. It is likely that interactions with other co-accessory factors result in conformational changes in the Nkx-2.5 protein that cause it to become a more effective transcriptional activator. In fact, Nkx-2.5 does not appear to work in isolation but forms combinatorial binding complexes with other transcription factors, such as serum response factor (SRF). We showed that SRF is also highly enriched in embryonic cardiac progenitors (14). Transfection of the genes encoding Nkx-2.5 and SRF resulted in activation of the αCA gene in fibroblasts, providing evidence that Nkx-2.5 and SRF might be instrumental in cardiac tissue differentiation (8–11).
The zinc finger-containing GATA factors (38, 47, 49, 50) are also high in the hierarchical order of regulatory factors that might specify the cardiac cell lineage. The GATA family has been subdivided, with GATA-1, -2, and -3 being linked to hematopoiesis while GATA-4, -5, and -6 are thought to be involved in heart, gut, and blood vessel formation. Each of the six GATA proteins contains a highly conserved DNA binding domain consisting of two C-4 zinc fingers with the motif Cys-X2-Cys-X17-Cys-X2-Cys (24, 41). These two zinc fingers have been shown to direct binding to the DNA sequence element (A/T)GATA(A/G) (34, 45), although the carboxy-terminal zinc finger is sufficient for site-specific binding (63). Examination of the DNA binding site specificities of all six GATA factors indicated that they are capable of binding to the same target sequence, thus suggesting that they have the ability to substitute for one another in cells in which they are coexpressed. GATA-4 has been found to be expressed in a developmentally and lineage-specific pattern within the cardiac mesoderm and is coexpressed with Nkx-2.5 and SRF in nascent myocardial cells (41, 50). Experiments have shown that GATA-4 regulates expression of cardiac tissue-specific genes, such as cardiac troponin C (29) and αC-MHC (48). For example, injection of GATA-4 DNA into Xenopus oocytes resulted in premature expression of cardiac tissue-specific proteins (30). Forced expression of antisense GATA-4 DNA blocked expression of cardiac tissue-specific genes in P19 cells (24). GATA-4 null mice display a severe defect in the formation of the cardiac tube, which is required for the migration and folding morphogenesis of the precardiogenic splanchnic mesoderm (37, 47). Rather normal expression of cardiac tissue-specific genes was observed in these homozygous GATA-4 knockout embryos, probably reflecting redundancy of some functions in the GATA-4–GATA-5–GATA-6 subfamily.
An attractive hypothesis derived from the analysis of Tinman and Nkx-2.5 homologs is that these homeodomain factors function in phylogenetically conserved pathways in muscle cell types that do not utilize members of the MyoD family. For example, ectopic expression of Nkx-2.5 in 10T1/2 fibroblasts demonstrated that downstream targets, such as the αCA gene, were not directly activated by Nkx-2.5 alone but required the collaboration of additional factors, such as SRF (8). We asked whether GATA-4 could substitute for SRF in interactions with Nkx-2.5 and also serve as a regulator of other downstream cardiac tissue-specific genes and synthetic genes containing Nkx-2.5 DNA binding targets. We have recently shown that Nkx-2.5 and GATA-4 are capable of coactivating the ANF promoter through binding to their adjacent DNA binding sites (19). Here we demonstrate that the physical and functional interaction of Nkx-2.5 and GATA-4 results in dramatic transcriptional activation of target promoters that contain only NKEs. Since these two transcription factors are coexpressed in pre-cardiac mesoderm, it is likely that this interaction plays a significant role in the regulation of transcriptional activity during early cardiogenesis in the embryo.
MATERIALS AND METHODS
Recombinant plasmids.
The avian αCA promoter, containing the SmaI-BstEII genomic fragment from position −315 to position +15 relative to the transcription start site, was cloned adjacent to a luciferase reporter gene plasmid (αCA-Luc) as previously described (11). αCA promoter deletion mutants linked to Luc plasmids (αCA–Del-100 and αCA–Del-58) were previously described (11). The A20(3)-Luc plasmid contains three copies of a high-affinity Nkx-2.5 binding site inserted upstream of αCA–Del-58 (11). Cytomegalovirus (CMV) promoter-driven eukaryotic expression plasmids (pCGN) coding for hemagglutinin (HA) epitope-tagged murine Nkx-2.5 derivatives were constructed as previously described (10). The Nkx-2.5pm mutant, in which the invariant asparagine at position 10 in the third helix of the homeodomain was converted to a glutamine, substantially blocks DNA binding (10). Two constructs (pCGN-2.5-HDNK and pCGN-2.5-NK) corresponding to polypeptides between amino acids (aa) 122 and 230 and between aa 195 and 230 of Nkx-2.5 were generated by PCR amplification of the corresponding cDNA region followed by ligation into the XbaI to BamHI sites of pCGN. Rat GATA-4 full-length cDNA and GATA-4 deletion mutants were subcloned into the pCG eukaryotic expression vector. GATA-4 fragments excised with XbaI and BamHI and subcloned into pBluescript SK were used for in vitro transcription and translation assays. Bacterial expression vectors coding for maltose binding protein (MBP)–Nkx-2.5 fusion proteins were described by Chen et al. (8). These constructs included several deletion mutants of Nkx-2.5 subcloned into the pMAL-c2 vector (New England Biolabs [NEB]). The MBP–GATA-4 plasmid contains the full-length GATA-4 coding region in pMAL-c2 (NEB). The plasmid pCMVPA-II (59), expressing a tetramer of the Staphylococcus aureus protein A immunoglobulin G (IgG) binding domain, was a gift from Peter Uetz (European Molecular Biology Laboratory, Heidelberg, Germany). GATA-4 was excised from pCG–GATA-4 with XbaI and ligated into the same site of pCMVPA-II.
Cell culture, transfections, and luciferase assays.
CV-1 fibroblasts were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% newborn calf serum. Cells were grown to 50 to 60% confluence in 6-cm-diameter dishes and transfected with a total of 2 μg of plasmid DNA, which included 1 μg of reporter luciferase plasmid and 1 μg of a pCG-derived vector. Plasmid DNA was mixed with 8 μl of Lipofectamine (Gibco BRL) and 100 μl of DMEM for 20 min and then applied to CV-1 cells in a total volume of 1 ml of DMEM. After 18 to 24 h of incubation at 37°C, 2 ml of DMEM containing 3% horse serum and 15 μg of insulin per ml was added to the transfected cells, which were then incubated for an additional 48 h. The cells were washed with phosphate-buffered saline and lysed with 200 ml of Reporter Lysis Buffer (Promega). The lysates were scraped from dishes and centrifuged at 15,000 × g for 5 min. Supernatants were tested for luciferase activity by using a Luminometer Monolight 2010 (Analytic Luminescence Laboratory). Thirty microliters of cell extract was automatically mixed with 100 μl of luciferase substrate solution (20 mM Tris-Cl [pH 8.0], 4 mM MgSO4, 0.1 mM EDTA, 30 mM dithiothreitol [DTT], 0.5 mM ATP, 0.5 mM d-luciferin, and 0.25 mM coenzyme A), and the emitted luminescence was measured for 5 s. Protein concentrations were measured by the Bradford method (Bio-Rad) and used to normalize luciferase activities when the supernatant protein concentration varied by more than 10% between samples.
Affinity purification of MBP–Nkx-2.5 derivative proteins.
MBP fusion proteins were expressed in Escherichia coli BL21 (DE3) bacteria (Novagen). After transformation, bacteria were grown overnight in 5 ml of Luria-Bertani broth containing 100 μg of ampicillin per ml, inoculated into 100 ml of Luria-Bertani broth containing 100 μg of ampicillin per ml, and grown to an optical density at 600 nm of 0.5 to 0.6. Isopropyl-β-d-thiogalactoside was then added to a final concentration of 2 mM, and the cells were incubated for an additional 1 h at 37°C. The cells were then harvested, resuspended in 2 ml of MBP column buffer (NEB), and sonicated four times (15 s each) with a microtip. Bacterial debris was spun down at 10,000 × g for 10 min at 4°C. MBP fusion proteins were batch purified with amylose-Sepharose 4B (NEB) in accordance with the manufacturer’s recommendations. Purified amylose-bound proteins were used for in vitro binding assays. The purity and quantity of purified proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining.
DNA EMSAs.
Double-stranded oligonucleotides, including the avian cardiac actin SRE1 (nucleotides −96 to −74), a consensus Nkx-2.1 binding sequence (5′-CCACTCAAGT-3′), and a GATA binding site which was described previously (2, 56), were synthesized and used for electrophoretic mobility shift assays (EMSAs). Each reaction mixture (20 μl) contained 10 mM Tris (pH 7.5), 50 mM NaCl, 1 mM DTT, 5% glycerol, 1 mM sodium phosphate, 0.5 μg of poly(dI-dC), and purified bacterial proteins. Nuclear extract preparation was performed as described previously (11). End-labeled probes (0.02 pmol; 10,000 to 20,000 cpm) and proteins were incubated for 15 min at room temperature. For competition assays, cold competitors were incubated with the proteins for 5 min prior to the addition of probe. The DNA-protein complexes were fractionated on 5% polyacrylamide gels (acrylamide/bisacrylamide ratio, 29:1) in 0.5× Tris-borate-EDTA buffer. The gels were dried and processed for autoradiography.
Mapping of Nkx-2.5- and GATA-4-interacting domains.
CV-1 cells were transfected with pCGN–Nkx-2.5 plasmids as described above, cultured for 36 h, lysed in EBC buffer (50 mM Tris [pH 8.0]–120 mM NaCl–0.5% Nonidet P-40 containing 2 μg of aprotinin per ml, 2 μg of leupeptin per ml, 2 μg of pepstatin per ml, and 1 mM phenylmethylsulfonyl fluoride), rocked for 15 min at 4°C, and centrifuged at 12,000 × g for 5 min. An aliquot of the supernatant, containing 100 μg of total protein, was incubated with 20 μl of MBP–GATA-4 beads for 1 h at room temperature in NETN buffer (20 mM Tris [pH 8.0], 100 mM NaCl, 1 mM EDTA, 5 mM MgCl2, 1 mM DTT, 0.05% Nonidet P-40) containing 8 U of DNase I per sample. The beads were washed four times with 500 μl of NETN buffer and resuspended in Laemmli sample buffer. The supernatants were analyzed by SDS-PAGE. HA epitope-tagged Nkx proteins were detected by chemiluminescence (ECL; Amersham) after immunoblotting with a monoclonal anti-HA antibody (Boehringer Mannheim) in accordance with the manufacturer’s protocol. Mapping of GATA-4 interaction domains was performed with wild-type GATA-4 and a series of GATA-4 deletion mutants cloned into pBluescript-SKII(+), linearized with BamHI, and transcribed with T3 RNA polymerase. One microgram of in vitro-synthesized RNA was used to program a rabbit reticulocyte lysate (Promega) in accordance with the manufacturer’s recommendations. GATA-4 proteins were mixed with MBP–Nkx-2.5 beads and processed as described previously (11). Briefly, MBP or MBP–Nkx-2.5 (10 μg) was immobilized on amylose beads and incubated with 10 μl of in vitro-translated 35S-labeled wild-type or mutant GATA-4 protein in 1× binding buffer for 1 h. After being extensively washed in the binding buffer, bound proteins were eluted by boiling the beads in SDS sample buffer and resolved on SDS–10% polyacrylamide gels. The retained 35S-labeled proteins were detected by fluorography after separation by SDS-PAGE.
Affinity purification of protein complexes associated with protein A fusion proteins.
Whole-cell extracts of CV1 cells transfected with pCGN–Nkx-2.5 and plasmids expressing either protein A or protein A–GATA-4 were obtained in EBC buffer as described above. Aliquots were incubated with IgG-Sepharose beads (Pharmacia) for 15 min at 4°C and then washed four times with EBC buffer. Proteins were visualized by immunoblotting with anti-HA antibody.
RESULTS
Nkx-2.5 and GATA-4 coactivate cardiac-specific promoters.
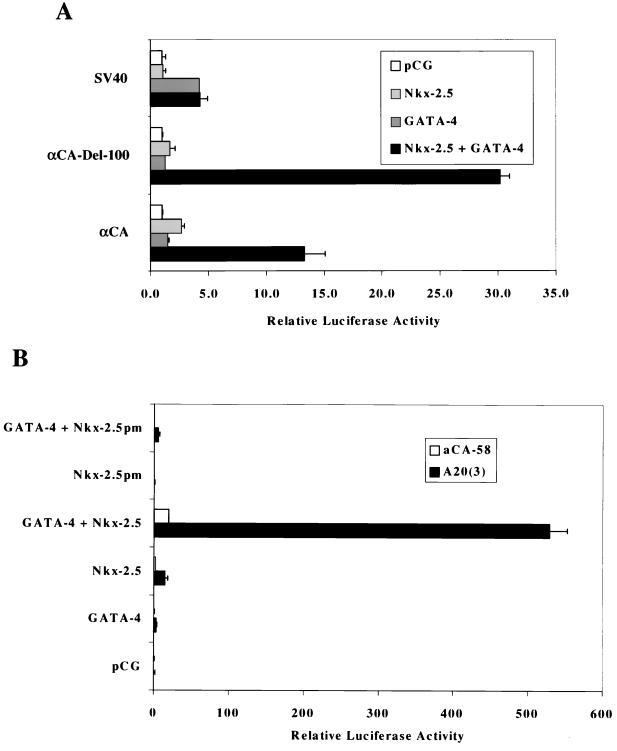
We showed that Nkx-2.5 bound to NKEs, including SREs found in the αCA promoter (11) and the A20(3)-Luc minimal promoter, which contains three copies of a high-affinity Nkx-2.5 binding element inserted upstream of the αCA–Del-58 plasmid (10). Nkx-2.5 weakly activates these promoters. We proposed that additional factors might be required to associate with Nkx-2.5 to optimally drive transcription, as recently demonstrated by the association of Nkx-2.5 with SRF, which could specify cell type-specific expression (11). We wanted to determine if GATA-4 could also collaborate with Nkx-2.5 to activate cardiac tissue-restricted promoters, since these cardiac tissue-specific transcription factors appear together during early heart development. Figure 1A shows that overexpression of either Nkx-2.5 or GATA-4 alone failed to significantly stimulate the αCA promoters in transient transfection assays in CV-1 fibroblasts. However, cotransfection of these promoters with both GATA-4 and Nkx-2.5 resulted in a 13- to 30-fold activation. The chicken αCA promoter contains four known Nkx-2.5 binding sites that are embedded within SRE1, SRE2, SRE3, and SRE4 (10). The αCA promoter has two potential GATA binding sites (at positions −304 and −161). However, EMSAs with purified GATA-4 protein did not demonstrate binding of GATA-4 to these sites (unpublished observations). Deletion of αCA promoter sequences distal to position −100, which removes potential GATA sites, still resulted in strong activation by Nkx-2.5 and GATA-4. The fact that the −100 deletion mutant showed higher activity than the 330-bp αCA promoter might also be due to the removal of inhibitory YY1 binding sites located over SRE3 and SRE2 (9, 43). In contrast, the simian virus 40 early promoter was not affected by Nkx-2.5 and was stimulated about fourfold by GATA-4. These results suggest that collaboration between Nkx-2.5 and GATA-4 is limited to the αCA promoters. Since αCA–Del-100 contains a high-affinity SRF binding site, SRE1, which also binds Nkx-2.5 (10), it may not be possible to separate GATA-4’s role with Nkx-2.5 from potential interactions with SRF.
FIG. 1.
Synergistic activation of the αCA promoter and the NKE multimerized A20(3) promoter by Nkx-2.5 and GATA-4. (A) CV-1 cells were transfected with luciferase reporter genes (1 μg of DNA) containing the αCA promoter, the −100-bp deletion mutant of αCA containing a serum response element (αCA–Del-100), and the simian virus 40 (SV40) early promoter. Cotransfectants contained CMV promoter-directed expression vectors driving Nkx-2.5 or GATA-4 (400 μg of DNA) or the empty vector pCG (1 μg of DNA). Luciferase activity was assayed 3 days after transfection. The bars represent the averages of data from two independent transfections, and the error bars represent the standard deviations of corrected luciferase activity relative to that of the pCG vector in a typical experiment. (B) Reporter vectors contained either 58 bp of the αCA promoter upstream of the transcriptional start site (aCA-58) or the A20 Nkx-2.5 binding site cloned in triplicate upstream of aCA-58 [A20(3)]. Nkx-2.5pm is a mutant of Nkx-2.5 with greatly decreased DNA binding affinity.
To reduce the possibility of side effects of endogenous SRF, we chose to investigate the functional interactions between GATA-4 and Nkx-2.5 on the A20(3)-Luc minimal promoter. Figure 1B shows that GATA-4 alone did not activate A20(3)-Luc transcription while Nkx-2.5 elicited about an eightfold increase in reporter luciferase activity. However, the combination of Nkx-2.5 and GATA-4 resulted in robust transactivation, reaching upward of a 500-fold increase in activity. Moreover, this induction is dependent on the binding of Nkx-2.5 to the A20 site, because the αCA–Del-58 plasmid lacking an Nkx-2.5 binding site was not significantly activated by Nkx-2.5 or GATA-4. Coactivation requires Nkx-2.5 DNA binding activity, as shown by cotransfection experiments in which the Nkx-2.5pm mutant, which does not bind NKEs (10), failed to activate A20(3)-Luc (Fig. 1B) or αCA–Del-58 (data not shown) in combination with GATA-4.
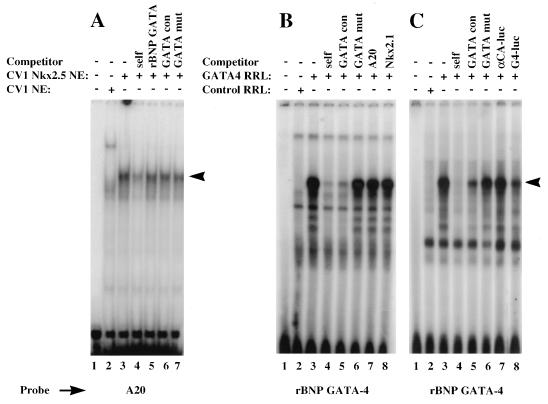
We examined the binding specificities of Nkx-2.5 and GATA-4 in the presence of the A20 site by EMSA, as shown in Fig. 2. Nuclear extracts prepared from Nkx-2.5-transfected CV-1 cells showed binding activity that was specifically competed against by a 50-fold excess of unlabeled A20 oligonucleotides. Under the same conditions, the rat BNP GATA DNA binding site was not an efficient competitor. Thus, as determined by EMSA, the A20 site effectively binds Nkx-2.5 but not GATA-4, as shown in Fig. 2A. We previously showed that SRF does not bind to multimerized A20 sites (10). Similarly, in vitro-translated GATA-4 efficiently binds to the BNP GATA element and is effectively competed against by the GATA oligonucleotide but not by a mutated GATA site or by either A20 or a high-affinity Nkx-2.1 binding site (Fig. 2B). In addition, Fig. 2C shows that consensus GATA sites were effective competitors, either as double-stranded oligonucleotides (lanes 4 and 5) or in the context of the luciferase reporter plasmid pGL2 (lane 8), whereas the luciferase plasmids containing either the αCA promoter (lane 7) or the A20(3) site (data not shown) were not efficient competitors. Therefore, Nkx-2.5 and GATA-4 factors do not display cross-binding specificities, and GATA-4 does not bind efficiently to either NKEs or cryptic sites in the reporter plasmids.
FIG. 2.
GATA-4 and Nkx-2.5 do not display cross-binding specificities. (A) GATA-4 does not bind NKEs. Nuclear extracts were prepared from CV-1 cells transfected with the Nkx-2.5 expression vector (pCGN–Nkx-2.5) or the empty vector (pCGN). Five micrograms of either the control extract (lane 2) or the Nkx-2.5 extract (lanes 3 to 7) was preincubated with 1 μg of poly(dG-dC) at room temperature for 15 min in 20 μl of 1× binding buffer; 50-fold molar excesses of unlabeled double-stranded specific (lane 4) and nonspecific (lanes 5 to 7) competitors were included in the reaction mixtures. Following the preincubation, 0.02 pmol of end-labeled A20 oligonucleotides was added, and the reaction mixtures were incubated for a further 15 min. DNA-protein complexes were resolved on a 5% polyacrylamide gel cast and run in 0.5× Tris-borate-EDTA. (B and C) Reaction conditions were similar to those described for panel A. One microliter of unprogrammed (lane 2) or GATA-4 RNA-programmed (lanes 3 to 8) rabbit reticulocyte lysate was used instead of nuclear extract. The GATA-4 binding site from the rat BNP promoter was the probe. The specific and nonspecific competitors used are indicated above the lanes. GATA con contains two GATA-4 consensus DNA binding sites (in boldface) (CACTTGATAACAGAAAGTGATAACTCT), and GATA mut has mutated GATA sites (underlined) (CACTTCTTAACAGAAAGTCTTAACTCT); αCA-luc is the αCA promoter-containing, pGL2-derived reporter vector described previously (11), and GATA-4-luc (G4-luc) is pGL2 containing three copies of the αC-MHC GATA binding site. Specific DNA-protein complexes are indicated by arrowheads.
Coactivation with GATA-4 is a property of some, but not all, NK-2 family members.
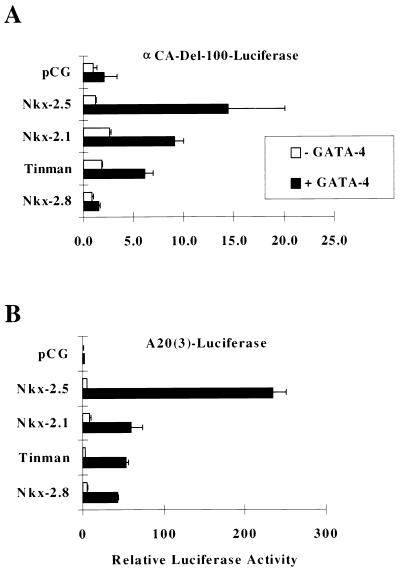
Since there are numerous NK-2-related family members, all of which are capable of efficiently binding to NKEs, we wanted to determine whether they exhibit functional properties of Nkx-2.5 and whether they are capable of coactivating gene activity with GATA-4. Figure 3 shows that maximal synergistic coactivation by Nkx-2.5 and GATA-4 occurred when expression plasmids were cotransfected with αCA-Luc and A20(3)-Luc reporter genes. The thyroid transcription factor Nkx-2.1 and the distantly related Drosophila homolog Tinman were also found to coactivate transcription of the αCa promoter with GATA-4, but at levels 60 and 44%, respectively, of the activity exhibited by Nkx-2.5 and only 26 and 22%, respectively, of the activity shown with the A20(3)-promoter. Nkx-2.8, a recently identified NK-2-related factor, barely stimulated αCA promoter activity in the presence of GATA-4 and only weakly activated the A20(3) promoter. These results indicate that Nkx-2.5 has a principal interactive role, along with GATA-4: to coactivate target promoters; this role might not be shared by other NK-2-related factors.
FIG. 3.
Coactivation with GATA-4 is a property of some, but not all, NK-2 family members. CV-1 cells were transfected, as described in the legend to Fig. 1, with expression vectors coding for Nkx-2.5, Tinman, Nkx-2.1, and Nkx-2.8 in the presence of GATA-4 (black bars) or pCG (white bars). Reporter constructs were αCA–Del-100 (A) and A20(3)-Luc (B). Relative luciferase activities were assayed and reported as described in the legend to Fig. 1.
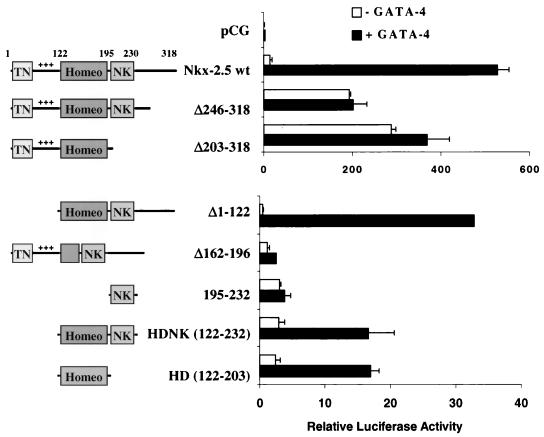
Mapping of Nkx-2.5 functional domains required for transcriptional coactivation with GATA-4.
Cotransfection of the A20(3)-Luc reporter gene with each of seven different Nkx-2.5 deletion mutants and GATA-4 was used to map the functional Nkx-2.5-interactive regions required for coactivation. The schematic diagrams illustrated in Fig. 4 show the locations of the conserved N-terminal TN domain, the charged activation domain, the homeodomain, and the NK-2-specific domain, which is found only in NK-2 relatives and Nkx-2.5 (27). In contrast to the observation that intact Nkx-2.5 requires GATA-4 for strong coactivation of the A20 minimal promoter, we found that an Nkx-2.5 C-terminal deletion mutant (Nkx-2.5Δ246–318) and a C-terminal mutant in which the NK-2-specific domain was also deleted (Nkx-2.5Δ203–318) activated transcription in CV-1 cells, without GATA-4. These results support the presence of a potent negative domain residing at the C terminus of Nkx-2.5, as was previously shown (10), and suggest that GATA-4’s primary role in interaction with Nkx-2.5 may be to counteract the repressive influences of the inhibitory domain. The use of a mutant with a deletion of the N-terminal activation domain of Nkx-2.5 (NkxΔ1–122) in cotransfections with GATA-4 resulted in a precipitous drop in transcription activity in comparison to wild-type values. However, Nkx-2.5 mutants lacking the activation domain are still able to show limited cooperation with GATA-4, as demonstrated by a 33-fold increase in reporter gene activity. These results suggest that the N-terminal domain of Nkx-2.5 is responsible for most of the transcriptional potency observed at the A20(3) site and that the activation domains of GATA-4 make a relatively modest contribution to the overall activity. Deletion of the second and third helices of the homeodomain (Δ162–196) abolished both basal and GATA-4-dependent activity by causing an inability to bind to A20 (10). In addition, since both Nkx-HD and Nkx-HDNK show about 17-fold activation with GATA-4, these results suggest that the homeodomain of Nkx-2.5 (aa 122 to 195) is sufficient for both A20 binding and modest activation with GATA-4.
FIG. 4.
Mapping of Nkx-2.5 domains required for transcriptional synergism with GATA-4. CV-1 cells were transfected with A20(3)-Luc, various mutants of Nkx-2.5, and either GATA-4 (black bars) or pCG (white bars). The numbers represent the amino acid range conserved or deleted (▵) in each mutant. The diagrams on the left represent the various conserved domains of Nkx-2.5 retained on each mutant. TN, tinman domain; +++, the positively charged N-terminal activation domain; homeo, homeobox; NK, conserved hydrophobic NK-2 motif. Luciferase activity was assayed as described in the legend to Fig. 1. Note the different scales of corrected luciferase activity for the two panels.
Deletion of the C-terminal NK-2 domain improves Nkx-2.5 homeodomain DNA binding.
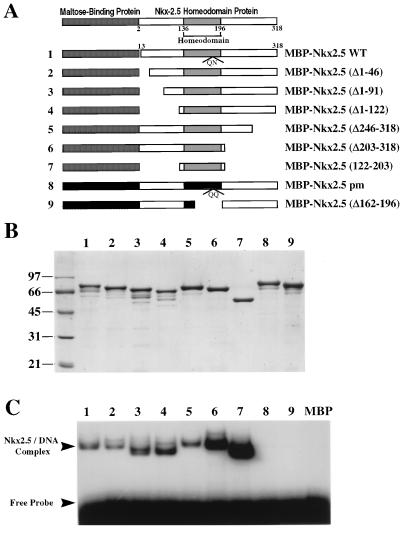
EMSAs using bacterially expressed MBP–Nkx-2.5 derivatives have shown that the homeodomain is responsible for Nkx-2.5 DNA binding activity (10). Figure 5A shows schematic diagrams of MBP–Nkx-2.5 mutant fusion proteins that were purified by amylose affinity chromatography. Approximately equal amounts of purified proteins, as shown in Fig. 5B, were analyzed by PAGE on an SDS–10% polyacrylamide gel and Coomassie blue staining. MBP–Nkx-2.5 proteins were then used to perform EMSAs with radioactive NKE oligonucleotides as shown in Fig. 5C. Each mutant that included the homeodomain region bound to the double-stranded NKE DNA probe. Removal of N-terminal sequences, as demonstrated for mutant MBP–Nkx-2.5(Δ1–91), doubled the DNA binding activity, while a deletion of a C-terminal region and the contiguous NK-2 domain (aa 203 to 246) tripled the Nkx-2.5 DNA binding activity. DNA binding activity was stimulated about 5- to 10-fold when both N-terminal and C-terminal NK-2 regions were removed (Fig. 5C). This observation is intriguing since similar binding profiles were obtained with different NKE probes, such as the A20 and Nkx-2.1/TTF-1 sites. These results suggest that both N-terminal and C-terminal regions might mask or interfere with the Nkx-2.5 homeodomain’s access to DNA.
FIG. 5.
The N-terminal and C-terminal domains inhibit Nkx-2.5 homeodomain DNA binding activity. (A) Schematic diagrams of MBP–Nkx-2.5 deletion mutants; (B) MBP–Nkx-2.5 proteins purified on amylose beads, separated by 10% PAGE, and detected by Coomassie blue staining; (C) results of an EMSA using equivalent amounts of purified MBP–Nkx-2.5 proteins and a labeled A20 oligonucleotide. EMSA conditions were as described in the legend to Fig. 2. Lane numbers in panels B and C correspond to the fusion proteins represented in panel A.
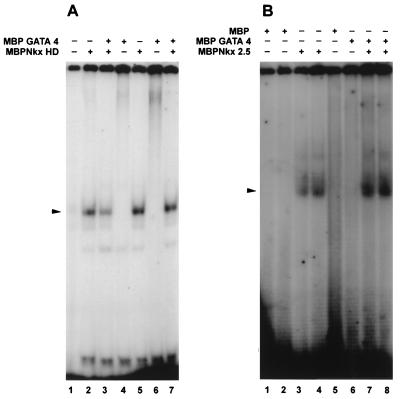
We asked if the coactivation displayed by Nkx-2.5 and GATA-4 on downstream target genes was mediated at the level of protein-protein interactions. DNA binding experiments were performed with bacterially expressed MBP–Nkx-2.5 and MBP–GATA-4 proteins to determine if protein-protein interactions would allow the formation of ternary complexes. As shown in Fig. 6A, both low (0.5-ng/μl) and high (1.25-ng/μl) concentrations of MBP–Nkx-2.5 homeodomain fusion proteins bound efficiently to NKE oligonucleotides. The addition of MBP–GATA-4 did not potentiate the binding of MBP–Nkx-2.5 homeodomain fusion proteins, and with a low homeodomain input, GATA-4 actually appeared to reduce NKE binding activity. Under these same conditions, MBP–GATA-4 was observed to facilitate the binding of a full-length MBP–Nkx-2.5 protein, as shown in Fig. 6B. Stable ternary-complex formation was not observed in the gels shown in Fig. 6, suggesting that if a ternary complex exists, it is either transient or unstable under the EMSA conditions used. However, it is possible that the interaction of GATA-4 with Nkx-2.5 induces conformational changes in the Nkx-2.5 protein leading to the functional displacement of the C-terminal inhibitory domain, thereby causing it to become a more efficient DNA binding protein.
FIG. 6.
GATA-4 facilitates DNA binding of full-length Nkx-2.5 but not the homeodomain. (A) End-labeled double-stranded A20 oligonucleotide probe was incubated with 10 or 25 ng of MBP–Nkx-2.5 homeodomain (HD), either with 250 ng of MBP–GATA-4 (lanes 3 and 7, respectively) or without MBP–GATA-4 (lanes 2 and 5, respectively), in 1× binding buffer. Lanes 4 and 6 contained only MBP–GATA-4 and A20 probe. The faint bands in lane 1 are due to spillover from lane 2. (B) MBP–Nkx-2.5 (10 or 25 ng of purified protein) was incubated with (lanes 7 and 8, respectively) or without (lanes 3 and 4, respectively) 250 ng of MBP–GATA-4 for 15 min at 30°C in 1× binding buffer. End-labeled double-stranded NKE (0.02 pmol) was added to the protein mixtures, which were subsequently incubated for an additional 15 min. Under identical conditions, 10 ng (lane 1), 25 ng (lane 2), or 250 ng (lane 5) of MBP and 250 ng of MBP–GATA-4 (lane 6) did not bind the labeled NKE probe. DNA-protein complexes are indicated by arrowheads. The gel in panel B was exposed overnight, and the one in panel A was exposed for 4 days at −70°C.
The Nkx-2.5 homeodomain is required for DNA-independent binding to GATA-4.
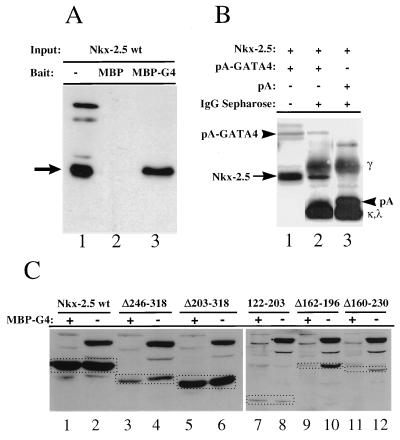
MBP–GATA-4 pull-down assays were used to investigate the interaction of Nkx-2.5 and GATA-4 in solution. Bacterially expressed MBP and MBP–GATA-4 fusion proteins were purified on amylose beads and incubated with whole-cell extracts prepared from CV-1 cells transfected with HA epitope-tagged Nkx-2.5. After extensive washing, the bound material was eluted and analyzed by SDS-PAGE and immunoblotting with an anti-HA monoclonal antibody. As shown in Fig. 7A, retention of Nkx-2.5 specifically by MBP–GATA-4 but not by MBP alone suggests that Nkx-2.5 and GATA-4 proteins associate in solution. It is important to note that endogenous CV-1 cell proteins stained with anti-HA in the input lanes, marked as a minus sign in Fig. 7, were poorly retained by MBP–GATA-4. To demonstrate that a physical association between the two proteins occurs in the cellular environment, a construct consisting of the S. aureus protein A IgG binding domain fused to the N terminus of GATA-4 was used to immobilize protein complexes associated with GATA-4 in transfected CV-1 cells. Figure 7B shows that in contrast to the isolated protein A IgG binding domain, the protein A–GATA-4 fusion protein was able to bind cotransfected HA–Nkx-2.5.
FIG. 7.
Pull-down protein association assays with MBP–GATA-4 and HA-tagged Nkx-2.5 protein extracts from transfected CV-1 cells. (A) MBP protein (lane 2) or MBP–GATA-4 fusion protein (MBP-G4) (lane 3) was purified from bacterial extracts, immobilized on amylose beads, and mixed with whole-cell extracts of CV-1 cells transfected with HA epitope-tagged wild-type Nkx-2.5 (Nkx-2.5 wt). After being washed, bound proteins were identified by immunoblotting. Lane 1 represents 25% of the input cellular extract. The arrow points to the Nkx-2.5-specific band. Other bands represent nonspecific cross-reactivity of the antibody. (B) Whole-cell extracts of CV-1 cells cotransfected with pCGN–Nkx-2.5 and expression vectors for either protein A (pA) (lane 3) or protein A–GATA-4 (pA-GATA4) (lane 2) were incubated with IgG-Sepharose beads and washed extensively. Immobilized protein complexes were eluted by boiling in SDS sample buffer and visualized by immunoblotting with anti-HA antibody. Note that proteins containing the protein A domains (arrowheads), as well as IgG heavy (γ) and light (κ,λ) chains, were visualized due to affinity for the secondary antibody. Lane 1 represents 25% of the input extract from pCGN–Nkx-2.5- and protein–GATA-4 transfected cells. (C) Immobilized MBP–GATA-4 protein (MBP-G4) purified from bacterial extract was mixed with whole-cell extract of CV-1 cells transfected with either HA epitope-tagged wild-type Nkx-2.5 (Nkx-2.5 wt) or various Nkx-2.5 mutants (see Fig. 3 for diagrams). After being washed, bound proteins were separated by SDS-PAGE (lanes 1, 3, 5, 7, 9, and 11). Lanes 2, 4, 6, 8, 10, and 12 represent 20% of the input cellular extract. Immunoblotting was performed with anti-HA antibody. Specific bands representing Nkx-2.5 mutants are identified by a broken-line box.
Mapping of the minimal interactive regions of Nkx-2.5 was done by assaying nuclear extracts prepared from CV-1 cells, transfected with CMV-driven plasmid vectors expressing Nkx-2.5 mutants (Fig. 7C), with bacterially expressed MBP–GATA-4 bound to amylose resins. We observed that mutants with an intact homeodomain (NkxΔ246–318, NkxΔ203–318, and Nkx122–203) were specifically bound by GATA-4, whereas deletion of the second and third helices of the homeodomain (Δ162–196 and Δ160–230) resulted in no binding to GATA-4. These results demonstrate that the homeodomain of Nkx-2.5 is minimally required for physical interaction with GATA-4.
GATA-4’s second zinc finger is required for synergistic activation and physical interaction with Nkx-2.5.
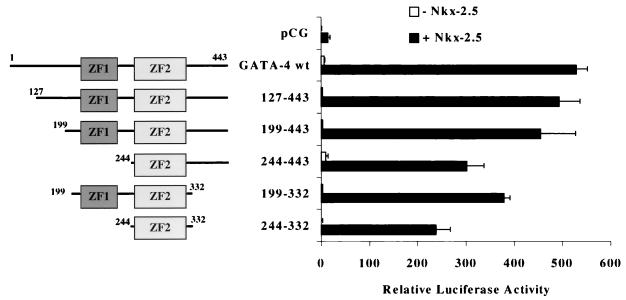
We then determined which domains of GATA-4 were sufficient for coactivation of the A20(3)-Luc promoter with Nkx-2.5 in CV-1 cells. As shown in Fig. 8, transfections with a variety of GATA-4 deletion mutants resulted in only small reductions in reporter gene activity. Those exhibiting transcriptional activity at least 200-fold over the baseline included GATA-4 mutants which retained the region from aa 244 to 331. These results indicate that this region of GATA-4, which contains the second zinc finger and the adjacent basic C-terminal extension, is sufficient for Nkx-2.5 coactivation.
FIG. 8.
Mapping GATA-4 domains required for transcriptional synergism with Nkx-2.5. CV-1 fibroblasts were transfected with A20(3)-Luc, various mutants of GATA-4, and either Nkx-2.5 (black bars) or pCG (white bars). The diagrams on the left represent the domains of GATA-4 retained in each deletion mutant. ZF1 and ZF2 refer to the amino- and carboxy-terminal zinc fingers of GATA-4, respectively. Luciferase activity was assayed as described in the legend to Fig. 1. wt, wild type.
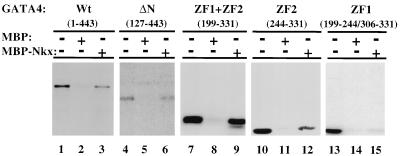
To confirm the results of transactivation assays, the same deletion mutants of GATA-4 were synthesized and labeled with 35S by in vitro translation and mixed with MBP or MBP–Nkx-2.5 protein immobilized on amylose beads as described above. The results, shown in Fig. 9, confirm earlier data indicating that the region of GATA-4 containing the second zinc finger and its C-terminal basic domain is sufficient for specific binding to Nkx-2.5 in the absence of DNA. A deletion mutant of GATA-4 containing zinc finger 1 (aa 199 to 244) fused to the C-terminal extension of zinc finger 2 (aa 306 to 332) was unable to bind Nkx-2.5 (Fig. 9, lanes 13 to 15), suggesting that zinc finger 2 (aa 244 to 306) is both necessary and sufficient for Nkx-2.5 binding and coactivation (Fig. 8).
FIG. 9.
Physical interaction between Nkx-2.5 and GATA-4 is mediated by the C-terminal zinc finger (ZF2) of GATA-4. Ten microliters of reticulocyte lysate containing in vitro-translated, 35S-labeled wild-type (Wt) GATA-4 (lanes 1 to 3), an N-terminally truncated GATA-4 mutant (ΔN) (lanes 4 to 6), GATA-4 with both zinc fingers (lanes 7 to 9), GATA-4 with only zinc finger 2 (lanes 10 to 12), or GATA-4 with only zinc finger 1 (ZF1) (lanes 13 to 15) was incubated with approximately 10 μg of MBP (lanes 2, 5, 8, 11, and 14) or MBP–Nkx-2.5 (lanes 3, 6, 9, 12, and 15). The beads were washed extensively, and the bound proteins were resolved on SDS–10% polyacrylamide gels and visualized by autoradiography. Lanes 1, 4, 7, 10, and 13 contained 10% of the in vitro translation volume used for binding reactions. After being washed, bound proteins were visualized by fluorography.
DISCUSSION
This study examined the functional and physical interactions between two cardiac tissue-restricted transcription factors, Nkx-2.5 and GATA-4, which are coexpressed during the earliest stages of cardiogenesis. Cotransfection experiments in CV-1 fibroblasts showed that a dramatic activation of cardiac tissue-specific promoters occurred only when Nkx-2.5 and GATA-4 were cotransfected, in contrast to when the transcription factors were transfected individually. This synergistic activation attained powerful levels, 500-fold over baseline, when a promoter with the trimeric Nkx binding site A20(3) was used in cotransfection assays, compared to the 15- to 20-fold activation observed with the αCA promoter (Fig. 1) and also with the αC-MHC promoter (54a). The synergistic activation levels may be amplified by the absence of inhibitory sequences in the A20(3) minimal promoter, in contrast to the more complex αCA and αC-MHC promoters. Alternatively, the high level of activation may simply reflect the increased affinity of Nkx-2.5 for the A20(3) site in comparison with the lower-affinity NKE sites present in the αCA and αC-MHC promoters. In either case, this study and earlier ones (11, 12) have demonstrated that αCa promoter activity appears to be regulated through the combinatorial interactions of at least three cardiac tissue-enriched transcription factors, Nkx-2.5, GATA-4, and SRF. In fact, we have observed that Nkx-2.5, GATA-4, and SRF are capable of physically interacting with each other and that much higher αCA promoter activity is elicited when all three factors are coexpressed (54b).
Transfection experiments with deletion mutants showed that at least the homeobox of Nkx-2.5 and the second zinc finger of GATA-4 are required for potent transcriptional coactivation through an Nkx-2.5 binding site. Moreover, a mutant protein consisting of only the second zinc finger of GATA-4 and its C-terminal extension was sufficient for a 200-fold coactivation with Nkx-2.5. On the other hand, the N-terminal activation domain of Nkx-2.5 is responsible for most of the transcriptional activity. However, the isolated homeodomain was still able to cooperate with GATA-4 to produce a modest activation at the A20(3) site. In contrast, SRF was unable to synergistically activate the same promoter with a truncated Nkx-2.5 mutant containing only the homeobox (11). This discrepancy may relate to differences in function of the activation domains of SRF and GATA-4 when complexed with the Nkx-2.5 homeodomain. Physical mapping by pull-down assays with purified proteins confirmed that the minimal interacting domain consists of the second zinc finger of GATA-4 and the homeodomain of Nkx-2.5. It is tempting to precisely define the interactive regions of the homeodomain by testing smaller deletions. For example, the Δ162–196 mutant, with a deletion of helices II and III of the homeodomain, is unable to bind GATA-4, suggesting that these regions may be involved in binding GATA-4. However, it is also possible that improper folding of helix I of the homeodomain in the absence of helices II and III prevents it from binding GATA-4.
Since GATA-4 does not bind either the αCA or the A20(3)-Luc promoter, the synergy with Nkx-2.5 could be mediated by recruitment of GATA-4 by DNA-bound Nkx-2.5 or by an effect of GATA-4 on Nkx-2.5 that is independent of GATA-4 binding to DNA. We were unable to demonstrate a physical association between GATA-4 and Nkx-2.5 as DNA-bound heterocomplexes. We were also unable to demonstrate recruitment of GATA-4 by DNA-bound Nkx-2.5 or recruitment of Nkx-2.5 by DNA-bound GATA-4 by pull-down assays using purified MBP–Nkx-2.5, MBP–GATA-4, and biotinylated oligonucleotides containing either the A20 Nkx-2.5 binding sequence or the αC-MHC GATA binding site (54a). It is possible that the interaction between GATA-4 and Nkx-2.5 is weakened when either protein is bound to DNA.
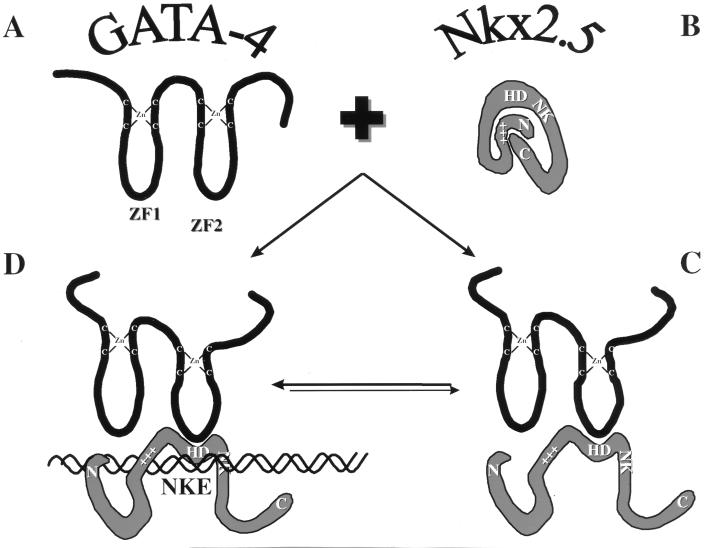
Transfection data revealed that the effect of GATA-4 on full-length Nkx-2.5 is very similar to the changes observed with C-terminally deleted Nkx-2.5 mutants. Since the C-terminal region has been previously shown to act as a transcriptional inhibitor (10), these data suggest that GATA-4 may cause a conformational change in Nkx-2.5 that relieves the negative constraints exerted by the inhibitory Nkx-2.5 C-terminal domain. What is the nature of this putative conformational change? We favor a model, shown in Fig. 10, in which Nkx-2.5 is activated through its homeodomain’s interaction with GATA-4, which then causes a change in the physical structure of Nkx-2.5. Perhaps changes in the shape of the Nkx-2.5 protein elicited by the touching of the homeodomain by GATA-4’s second zinc finger result in displacement of the inhibitory C-terminal domain, which then further improves DNA binding activity and reveals the N-terminal activation domain. In support of this model, we have observed that GATA-4 will facilitate Nkx-2.5 DNA binding activity but is unable to improve the binding efficiency of a mutant that contains only the Nkx-2.5 homeodomain (Fig. 6). Thus, interaction of GATA-4 with Nkx-2.5 might evoke strong cotransactivation by removing physical impediments that occlude the Nkx-2.5 N-terminal activation domain, by increasing DNA binding affinity, and/or by enhancing interactions with ancillary proteins. The induction of a conformational change that increases the transcriptional activity of Nkx-2.5 by the interaction of GATA-4 with Nkx-2.5 might be similar to the effects of Extradenticle and Pbx1 on the DNA binding affinity and transcriptional activity of Engrailed and Hox proteins (32, 61).
FIG. 10.
Working model for activation of Nkx-2.5 by interaction with GATA-4. GATA-4 interacts with Nkx-2.5 and induces a conformational change resulting in increased affinity for the Nkx-2.5 binding site (NKE) and increased accessibility of the activation domain (+++). (A) Isolated GATA-4 protein; (B) isolated Nkx-2.5 protein. The model postulates that in the inactivated state the Nkx-2.5 homeodomain (HD) has a low affinity for NKE due to steric hindrance by the C-terminal domain, possibly via hydrophobic interactions with the N terminus of Nkx-2.5. In addition, it is possible that the C-terminal–N-terminal interactions keep the activation domain inaccessible to the transcriptional machinery. (C) Interaction with GATA-4 would then remove this negative effect of the C terminus on Nkx-2.5 transcriptional activity. (D) NKE binding by activated Nkx-2.5–GATA-4 complexes. It is also possible that GATA-4 leaves the complex before or after NKE binding (data not shown). ZF1 and ZF2, zinc fingers 1 and 2, respectively.
Interactions between zinc finger and homeodomain proteins are not without precedent. One example is the interaction of the zinc finger protein Swi5p and the homeodomain Pho2p, which has a role in the coactivation of the yeast HO promoter (5–7, 18). Another example is the Drosophila zinc finger orphan receptor Ftz-F1, required for the developmental function of the homeobox protein Ftz; Ftz-F1 interacts with Ftz to bind cooperatively to DNA. However, these in vitro and in vivo interactions between Ftz and Ftz-F1 do not require the homeodomain of Ftz (26). In addition, mammalian Nkx-2.1 cooperates with Sp1 and Sp3 to synergistically activate the Clara cell secretory protein promoter (58). Another zinc finger protein, the glucocorticoid receptor, is able to interfere in a hormone-dependent manner with the interaction between the POU homeodomain protein Oct-1 and VP16 (39). We showed that a physical interaction between the second zinc finger of GATA-4 and the Nkx-2.5 homeodomain was sufficient to elicit robust transcriptional activity. It is not yet known how GATA-4 alters the Nkx-2.5 protein’s confirmation to unmask the N-terminal activation domain and improve Nkx-2.5 DNA binding activity, but protein-protein interactions between zinc fingers and homeodomain structural regions appear to be essential components of the process. We have also shown that other closely related NK-2 members are not equivalent in cotransactivation assays with GATA-4. The role of the NK-2-specific domain does not appear to be an important determinant for coactivation with GATA-4, since Tinman, which lacks the NK-2-specific domain, still provided rather strong coactivity.
Recent studies by Durocher et al. (19) have shown that the ANF promoter is also activated in synergy with Nkx-2.5 and GATA-4, through their binding to separate DNA elements. They have shown that GATA-6 cannot substitute for GATA-4 with regard to interaction with Nkx-2.5. In the future, it will be important to determine whether other members of the GATA family are able to interact with Nkx proteins, since combination of factors might reveal novel coactivating and corepressive pairs, especially in light of the increasing number of proteins which contain both zinc finger and homeobox motifs (17, 28). For example, Drosophila ZFH1, a factor encoded by a gene downstream of tinman, is also required for insect dorsal vessel formation, and its vertebrate homolog, ZEB, is expressed during the early stages of mammalian cardiogenesis (40). The ability to establish highly productive cooperative interactions with other cardiac tissue-restricted proteins may explain why transcription factors such as GATA-4, which show high-level expression not only in the heart but also in other tissues, such as the gut, can regulate promoters in a cardiac tissue-specific manner. This pattern of tissue- and developmentally restricted regulation by cooperation with heterologous partners is seen with many homeobox (8, 11, 33, 60, 62, 64) and zinc finger (15, 21, 31, 35, 46) proteins and is exemplified by the interaction of Sp1 and the erythroid Krüuppel-like factor with members of the GATA family (21, 23, 46) and by the cooperative interaction of Nkx-2.1 with Sp1 and Sp3 to activate a target promoter (58).
Possible mechanisms by which complexes of transcription factors activate cardiac tissue-specific genes include induction of conformational changes in the DNA and recruitment of additional proteins that modulate transcriptional activity. DNA bending induced by complexes of transcription factors bound to different sites in the promoter may make the promoter more accessible to the transcriptional machinery (55). This mechanism may operate in promoters, such as that of ANF, which bind both Nkx-2.5 and GATA-4 (20, 24). However, the results with the A20(3) minimal promoter, which does not directly bind GATA-4, make it difficult to attribute the dramatic synergistic activation by GATA-4 and Nkx-2.5 solely to DNA bending. On the other hand, DNA-bound transcription factors may interact to recruit additional components of the transcriptional machinery or enzymes, such as histone acetyltransferases (HATs), that modify the conformation of the transcription factors themselves or the structure of chromatin. An interesting example is the recruitment of the coactivator and HAT protein p300 by MyoD and MEF2C (53). In the context of chromatin, it is possible that cardiac tissue-specific multiprotein complexes of transcription factors are able to recruit HATs and other ancillary proteins, which can modify the chromatin structure to achieve appropriate developmental and anatomically specific regulation.
ACKNOWLEDGMENTS
This study was supported by National Institutes of Health grants P01 HL49953 and R01 HL50422.
Vishal Nigam was a Howard Hughes Medical Institute Medical Student Research Training Fellow.
REFERENCES
- 1.Azpiazu N, Frasch M. tinman and bagpipe: two homeobox genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 1993;7:1325–1340. doi: 10.1101/gad.7.7b.1325. [DOI] [PubMed] [Google Scholar]
- 2.Bellaguli N, Schildmeyer L, Schwartz R J. Organization and myogenic restricted expression of the murine serum response factor gene: a role for autoregulation. J Biol Chem. 1997;272:18222–18231. doi: 10.1074/jbc.272.29.18222. [DOI] [PubMed] [Google Scholar]
- 3.Bodmer R. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development. 1993;118:719–729. doi: 10.1242/dev.118.3.719. [DOI] [PubMed] [Google Scholar]
- 4.Bohinski R J, Di Lauro R, Whitsett J A. The lung-specific surfactant protein B gene promoter is a target for thyroid transcription factor 1 and hepatocyte nuclear factor 3, indicating common factors for organ-specific gene expression along the foregut axis. Mol Cell Biol. 1994;14:5671–5681. doi: 10.1128/mcb.14.9.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brazas R M, Bhoite L T, Murphy M D, Yu Y, Chen Y, Neklason D W, Stillman D J. Determining the requirements for cooperative DNA binding by Swi5p and Pho2p (Grf10p/Bas2p) at the HO promoter. J Biol Chem. 1995;270:29151–29161. doi: 10.1074/jbc.270.49.29151. [DOI] [PubMed] [Google Scholar]
- 6.Brazas R M, Stillman D J. Identification and purification of a protein that binds DNA cooperatively with the yeast SWI5 protein. Mol Cell Biol. 1993;13:5524–5537. doi: 10.1128/mcb.13.9.5524. . (Erratum, 13:7200.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brazas R M, Stillman D J. The Swi5 zinc-finger and Grf10 homeodomain proteins bind DNA cooperatively at the yeast HO promoter. Proc Natl Acad Sci USA. 1993;90:11237–11241. doi: 10.1073/pnas.90.23.11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen C Y, Croissant J, Majesky M, Topouzis S, McQuinn T, Frankovsky M J, Schwartz R J. Activation of the cardiac alpha-actin promoter depends upon serum response factor, Tinman homologue, Nkx-2.5, and intact serum response elements. Dev Genet. 1996;19:119–130. doi: 10.1002/(SICI)1520-6408(1996)19:2<119::AID-DVG3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 9.Chen C Y, Schwartz R J. Competition between negative acting YY1 versus positive acting serum response factor and tinman homologue Nkx-2.5 regulates cardiac α-actin promoter activity. Mol Endocrinol. 1997;11:812–822. doi: 10.1210/mend.11.6.0015. [DOI] [PubMed] [Google Scholar]
- 10.Chen C Y, Schwartz R J. Identification of novel DNA binding targets and regulatory domains of a murine tinman homeodomain factor, nkx-2.5. J Biol Chem. 1995;270:15628–15633. doi: 10.1074/jbc.270.26.15628. [DOI] [PubMed] [Google Scholar]
- 11.Chen C Y, Schwartz R J. Recruitment of the tinman homologue Nkx-2.5 by serum response factor activates cardiac α-actin gene transcription. Mol Cell Biol. 1996;16:6372–6384. doi: 10.1128/mcb.16.11.6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J N, Fishman M C. Zebrafish tinman homologue demarcates the heart field and initiates myocardial differentiation. Development. 1996;122:3809–3816. doi: 10.1242/dev.122.12.3809. [DOI] [PubMed] [Google Scholar]
- 13.Civitareale D, Lonigro R, Sinclair A J, Di Lauro R. A thyroid-specific nuclear protein essential for tissue-specific expression of the thyroglobulin promoter. EMBO J. 1989;8:2537–2542. doi: 10.1002/j.1460-2075.1989.tb08391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Croissant J D, Kim J H, Eichele G, Goering L, Lough J, Prywes R, Schwartz R J. Avian serum response factor expression restricted primarily to muscle cell lineages is required for alpha-actin gene transcription. Dev Biol. 1996;177:250–264. doi: 10.1006/dbio.1996.0160. [DOI] [PubMed] [Google Scholar]
- 15.Crossley M, Merika M, Orkin S H. Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol Cell Biol. 1995;15:2448–2456. doi: 10.1128/mcb.15.5.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Damante G, Fabbro D, Pellizzari L, Civitareale D, Guazzi S, Polycarpou-Schwartz M, Cauci S, Quadrifoglio F, Formisano S, Di Lauro R. Sequence-specific DNA recognition by the thyroid transcription factor-1 homeodomain. Nucleic Acids Res. 1994;22:3075–3083. doi: 10.1093/nar/22.15.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dawid I B, Toyama R, Taira M. LIM domain proteins. C R Acad Sci Ser III. 1995;318:295–306. [PubMed] [Google Scholar]
- 18.Dohrmann P R, Voth W P, Stillman D J. Role of negative regulation in promoter specificity of the homologous transcriptional activators Ace2p and Swi5p. Mol Cell Biol. 1996;16:1746–1758. doi: 10.1128/mcb.16.4.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durocher D, Charron F, Warren R, Schwartz R J, Nemer M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 1997;16:5687–5696. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durocher D, Chen C-Y, Ardati A, Schwartz R J, Nemer M. The atrial natriuretic factor promoter is a downstream target for Nkx-2.5 in the myocardium. Mol Cell Biol. 1996;16:4648–4655. doi: 10.1128/mcb.16.9.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer K D, Haese A, Nowock J. Cooperation of GATA-1 and Sp1 can result in synergistic transcriptional activation or interference. J Biol Chem. 1993;268:23915–23923. [PubMed] [Google Scholar]
- 22.Gajewski K, Kim Y, Lee Y M, Olson E N, Schultz R A. D-mef2 is a target for tinman activation during Drosophila heart development. EMBO J. 1997;16:515–522. doi: 10.1093/emboj/16.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gregory R C, Taxman D J, Seshasayee D, Kensinger M H, Bieker J J, Wojchowski D M. Functional interaction of GATA1 with erythroid Kruppel-like factor and Sp1 at defined erythroid promoters. Blood. 1996;87:1793–1801. [PubMed] [Google Scholar]
- 24.Grépin C, Dagnino L, Robitaille L, Haberstroh T, Antakly T, Nemer M. A hormone-encoding gene identifies a pathway for cardiac but not skeletal muscle gene transcription. Mol Cell Biol. 1994;14:3115–3129. doi: 10.1128/mcb.14.5.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guazzi S, Price M, De Felice M, Damante G, Mattei M G, Di Lauro R. Thyroid nuclear factor 1 (TTF-1) contains a homeodomain and displays a novel DNA binding specificity. EMBO J. 1990;9:3631–3639. doi: 10.1002/j.1460-2075.1990.tb07574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guichet C, Copeland J W, Erdely M, Hlousek D, Zavorsky P, Ho J, Brown S, Percival-Smith A, Krause H M, Ephrussi A. The nuclear receptor homologue Ftz-F1 and the homeodomain protein Ftz are mutually dependent cofactors. Nature. 1997;385:548–552. doi: 10.1038/385548a0. [DOI] [PubMed] [Google Scholar]
- 27.Harvey R P. NK-2 homeobox genes and heart development. Dev Biol. 1996;178:203–216. doi: 10.1006/dbio.1996.0212. [DOI] [PubMed] [Google Scholar]
- 28.Ido A, Miura Y, Tamaoki T. Activation of ATBF1, a multiple-homeodomain zinc-finger gene, during neuronal differentiation of murine embryonal carcinoma cells. Dev Biol. 1994;163:184–187. doi: 10.1006/dbio.1994.1134. [DOI] [PubMed] [Google Scholar]
- 29.Ip H S, Wilson D B, Heikinheimo M, Tang Z, Ting C-N, Simon M C, Leiden J M, Parmacek M S. The GATA-4 transcription factor transactivates the cardiac muscle-specific troponin C promoter-enhancer in nonmuscle cells. Mol Cell Biol. 1994;14:7517–7526. doi: 10.1128/mcb.14.11.7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang Y, Evans T. The Xenopus GATA-4/5/6 genes are associated with cardiac specification and can regulate cardiac-specific transcription during embryogenesis. Dev Biol. 1996;174:258–270. doi: 10.1006/dbio.1996.0071. [DOI] [PubMed] [Google Scholar]
- 31.Kawana M, Lee M-E, Quertermous E E, Quertermous T. Cooperative interaction of GATA-2 and AP1 regulates transcription of the endothelin-1 gene. Mol Cell Biol. 1995;15:4225–4231. doi: 10.1128/mcb.15.8.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knoepfler P S, Kamps M P. The pentapeptide motif of Hox proteins is required for cooperative DNA binding with Pbx1, physically contacts Pbx1, and enhances DNA binding by Pbx1. Mol Cell Biol. 1995;15:5811–5819. doi: 10.1128/mcb.15.10.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knoepfler P S, Lu Q, Kamps M P. Pbx-1 Hox heterodimers bind DNA on inseparable half-sites that permit intrinsic DNA binding specificity of the Hox partner at nucleotides 3′ to a TAAT motif. Nucleic Acids Res. 1996;24:2288–2294. doi: 10.1093/nar/24.12.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ko L J, Engel J D. DNA-binding specificities of the GATA transcription factor family. Mol Cell Biol. 1993;13:4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobayashi A, Sogawa K, Fujii-Kuriyama Y. Cooperative interaction between AhR.Arnt and Sp1 for the drug-inducible expression of CYP1A1 gene. J Biol Chem. 1996;271:12310–12316. doi: 10.1074/jbc.271.21.12310. [DOI] [PubMed] [Google Scholar]
- 36.Komuro I, Izumo S. Csx: a murine homeobox-containing gene specifically expressed in the developing heart. Proc Natl Acad Sci USA. 1993;90:8145–8149. doi: 10.1073/pnas.90.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuo C T, Morrisey E E, Anandappa R, Sigrist K, Lu M M, Parmacek M S, Soudais C, Leiden J M. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11:1048–1056. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- 38.Kuo M-H, Nadeau E T, Grayhack E J. Multiple phosphorylated forms of the Saccharomyces cerevisiae Mcm1 protein include an isoform induced in response to high salt concentrations. Mol Cell Biol. 1997;17:819–832. doi: 10.1128/mcb.17.2.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kutoh E, Strömstedt P-E, Poellinger L. Functional interference between the ubiquitous and constitutive octamer transcription factor 1 (OTF-1) and the glucocorticoid receptor by direct protein-protein interaction involving the homeo subdomain of OTF-1. Mol Cell Biol. 1992;12:4960–4969. doi: 10.1128/mcb.12.11.4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai Z-C, Rushton E, Bate M, Rubin G. Loss of function of the Drosophila zfh-1 gene results in abnormal development of mesodermally derived tissues. Proc Natl Acad Sci USA. 1993;90:4122–4126. doi: 10.1073/pnas.90.9.4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laverriere A C, MacNeill C, Mueller C, Poelmann R E, Burch J B, Evans T G. GATA-4/5/6, a subfamily of three transcription factors transcribed in developing heart and gut. J Biol Chem. 1994;269:23177–23184. [PubMed] [Google Scholar]
- 42.Lee K H, Xu Q, Breitbart R E. A new tinman-related gene, nkx2.7, anticipates the expression of nkx2.5 and nkx2.3 in zebrafish heart and pharyngeal endoderm. Dev Biol. 1996;180:722–731. doi: 10.1006/dbio.1996.0341. [DOI] [PubMed] [Google Scholar]
- 43.Lee T C, Shi Y, Schwartz R J. Displacement of BrdUrd-induced YY1 by serum response factor activates skeletal α-actin transcription in embryonic myoblasts. Proc Natl Acad Sci USA. 1992;89:9814–9818. doi: 10.1073/pnas.89.20.9814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lints T J, Parsons L M, Hartley L, Lyons I, Harvey R P. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendents. Development. 1993;119:419–431. doi: 10.1242/dev.119.2.419. [DOI] [PubMed] [Google Scholar]
- 45.Merika M, Orkin S H. DNA-binding specificity of GATA family transcription factors. Mol Cell Biol. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merika M, Orkin S H. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Krüppel family proteins Sp1 and EKLF. Mol Cell Biol. 1995;15:2437–2447. doi: 10.1128/mcb.15.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Molkentin J, Lin Q, Duncan S A, Olson E N. Requirement of the transcription factor GATA-4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 48.Molkentin J D, Kalvakolanu D V, Markham B E. Transcription factor GATA-4 regulates cardiac muscle-specific expression of the α-myosin heavy-chain gene. Mol Cell Biol. 1994;14:4947–4957. doi: 10.1128/mcb.14.7.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morrisey E E, Ip H S, Tang Z, Lu M M, Parmacek M S. GATA-5: a transcriptional activator expressed in a novel temporally and spatially-restricted pattern during embryonic development. Dev Biol. 1997;183:21–36. doi: 10.1006/dbio.1996.8485. [DOI] [PubMed] [Google Scholar]
- 50.Morrisey E E, Ip H S, Lu M M, Parmacek M S. GATA-6: a zinc finger transcription factor that is expressed in multiple cell lineages derived from lateral mesoderm. Dev Biol. 1996;177:309–322. doi: 10.1006/dbio.1996.0165. [DOI] [PubMed] [Google Scholar]
- 51.Ray M K, Chen C-Y, Schwartz R J, DeMayo F J. Transcriptional regulation of a mouse Clara cell-specific protein (mCC10) gene by the NKx transcription factor family members thyroid transcription factor 1 and cardiac muscle-specific homeobox protein (CSX) Mol Cell Biol. 1996;16:2056–2064. doi: 10.1128/mcb.16.5.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reecy J M, Yamada M, Cummings K, Sosic D, Chen C Y, Eichele G, Olson E N, Schwartz R J. Chicken Nkx-2.8: a novel homeobox gene expressed in early heart progenitor cells and pharyngeal pouch-2 and -3 endoderm. Dev Biol. 1997;188:295–311. doi: 10.1006/dbio.1997.8641. [DOI] [PubMed] [Google Scholar]
- 53.Sartorelli V, Huang J, Hamamori Y, Kedes L. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol Cell Biol. 1997;17:1010–1026. doi: 10.1128/mcb.17.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schultheiss T M, Xydas S, Lassar A B. Induction of avian cardiac myogenesis by anterior endoderm. Development. 1995;121:4203–4214. doi: 10.1242/dev.121.12.4203. [DOI] [PubMed] [Google Scholar]
- 54a.Sepulveda, J. Unpublished data.
- 54b.Sepulveda, J., V. Nigam, and R. Schwartz. Unpublished data.
- 55.Sharrocks A D, Shore P. DNA bending in the ternary nucleoprotein complex at the c-fos promoter. Nucleic Acids Res. 1995;23:2442–2449. doi: 10.1093/nar/23.13.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thuerauf D J, Hanford D S, Glembotski C C. Regulation of rat brain natriuretic peptide transcription. A potential role for GATA-related transcription factors in myocardial cell gene expression. J Biol Chem. 1994;269:17772–17775. [PubMed] [Google Scholar]
- 57.Tonissen K F, Drysdale T A, Lints T J, Harvey R P, Krieg P A. XNkx-2.5, a Xenopus gene related to Nkx-2.5 and tinman: evidence for a conserved role in cardiac development. Dev Biol. 1994;162:325–328. doi: 10.1006/dbio.1994.1089. [DOI] [PubMed] [Google Scholar]
- 58.Toonen R F, Gowan S, Bingle C D. The lung enriched transcription factor TTF-1 and the ubiquitously expressed proteins Sp1 and Sp3 interact with elements located in the minimal promoter of the rat Clara cell secretory protein gene. Biochem J. 1996;316:467–473. doi: 10.1042/bj3160467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uetz P, Zeller R. Vectors for expression of protein-A-tagged proteins in vertebrate cells. Anal Biochem. 1996;237:161–163. doi: 10.1006/abio.1996.0220. [DOI] [PubMed] [Google Scholar]
- 60.Vershon A K. Protein interactions of homeodomain proteins. Curr Opin Biotechnol. 1996;7:392–396. doi: 10.1016/s0958-1669(96)80113-3. [DOI] [PubMed] [Google Scholar]
- 61.Wilson D S, Desplan C. Homeodomain proteins. Cooperating to be different. Curr Biol. 1995;5:32–34. doi: 10.1016/s0960-9822(95)00010-8. [DOI] [PubMed] [Google Scholar]
- 62.Wolberger C. Homeodomain interactions. Curr Opin Struct Biol. 1996;6:62–68. doi: 10.1016/s0959-440x(96)80096-0. [DOI] [PubMed] [Google Scholar]
- 63.Yang H-Y, Evans T. Distinct roles for the two cGATA-1 finger domains. Mol Cell Biol. 1992;12:4562–4570. doi: 10.1128/mcb.12.10.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zappavigna V, Sartori D, Mavilio F. Specificity of HOX protein function depends on DNA-protein and protein-protein interactions, both mediated by the homeo domain. Genes Dev. 1994;8:732–744. doi: 10.1101/gad.8.6.732. [DOI] [PubMed] [Google Scholar]